Abstract
G-quadruplex (G4) DNA is a type of quadruple helix structure formed by a continuous guanine-rich DNA sequence. Emerging evidence in recent years authenticated that G4 DNA structures exist both in cell-free and cellular systems, and function in different diseases, especially in various cancers, aging, neurological diseases, and have been considered novel promising targets for drug design. In this review, we summarize the detection method and the structure of G4, highlighting some non-canonical G4 DNA structures, such as G4 with a bulge, a vacancy, or a hairpin. Subsequently, the functions of G4 DNA in physiological processes are discussed, especially their regulation of DNA replication, transcription of disease-related genes (c-MYC, BCL-2, KRAS, c-KIT et al.), telomere maintenance, and epigenetic regulation. Typical G4 ligands that target promoters and telomeres for drug design are also reviewed, including ellipticine derivatives, quinoxaline analogs, telomestatin analogs, berberine derivatives, and CX-5461, which is currently in advanced phase I/II clinical trials for patients with hematologic cancer and BRCA1/2-deficient tumors. Furthermore, since the long-term stable existence of G4 DNA structures could result in genomic instability, we summarized the G4 unfolding mechanisms emerged recently by multiple G4-specific DNA helicases, such as Pif1, RecQ family helicases, FANCJ, and DHX36. This review aims to present a general overview of the field of G-quadruplex DNA that has progressed in recent years and provides potential strategies for drug design and disease treatment.
Keywords: Cancer, Aging, Telomere, G-quadruplex, G4 ligand, DNA replication, Epigenetic, Helicase
Detection of G-quadruplex DNA
Since the discovery of the right-handed B-type double helix DNA by Watson and Crick in 1953, many high-level DNA structures have been discovered, such as Z-DNA, Holliday junction DNA, and triplex DNA. In 1962, Gellert et al. first discovered that four guanosine 5′-monophosphate (GMP) molecules can form a planar helical unit, thus forming a relatively stable helical aggregate, which laid an important foundation for the subsequent discovery of G-quadruplex structures [1]. In 1988, Sen et al. found that DNA sequences similar to telomeres containing multiple guanine sequences could spontaneously fold to form a four-stranded DNA structure under certain conditions, which were called g-quadruplexes or g-tetraplexes [2]. Since then, increasing studies have discovered a series of specific DNA sequences that can form G4 structures, especially sequences located in the regulatory regions of certain oncogenes and in the telomeres.
Potential G4 sequences (PGSs) in the genome can be analyzed and identified using bioinformatics methods. Algorithms were developed to predict the putative G4 structures in the genome, with the typical G4 sequence motif being G≥3NL1G≥3NL2G≥3NL3G≥3, where NL1, NL2, and NL3 represent the loop sequences of G4, with lengths limited to 1–7 nt [3, 4]. Through bioinformatics prediction, as many as 376,000 potential G4 sequences were found in the human genome [4]. Meanwhile, PGSs in Escherichia coli [5–7], Saccharomyces cerevisiae [8, 9], and other species have also been analyzed and located, and the potential functions of these sequences have also been speculated. Currently, depending on different strategies, multiple algorithms are available to determine and analyze PGSs in the indicated DNA sequences or genomes, such as the regular expression matching tools ImGQfinder [10] and AllQuads [11], the scoring and sliding window tools G4Hunter [12–14], pqsfinder [15] and QGRS Mapper [16], and the machine learning tool Quadron [17].
Apart from bioinformatics methods, next-generation sequencing has also been adopted to map the G4 structures in the genome. Currently, two strategies based on next-generation sequencing are mainly exploited: G-quadruplex sequencing (G4-seq) and G4 chromatin immunoprecipitation sequencing (ChIP-seq). G4-seq is a combination of the genome-wide DNA polymerase-stop assay and high-throughput sequencing, and it has identified 716,310 G4 DNA sequences in the human genome [18], nearly twice that predicted using bioinformatics methods. The G4s identified via G4-seq include many non-canonical G4 structures with long loops and/or bulges, which are difficult to predict using bioinformatics methods. Subsequently, whole-genome G4 maps of 12 important model organisms and pathogens of clinical relevance, including S. cerevisiae, Arabidopsis thaliana, Mus musculus, and Homo sapiens, were generated using an improved version of G4-seq, which provided the key sequence features that determine different patterns of G4 formation and the relevance of G4 localization across genomes [19]. It should be noted that some of the G4 sites mapped via G4-seq might not form G4 structures in vivo, as the technique was performed using cell-free systems, and the effect of proteins that may alter the stability of G4 structures cannot be excluded, especially for some long-loop G4s with low stability.
G4 ChIP-seq is dependent on chromatin immunoprecipitation using G4-specific antibodies and high-throughput sequencing [20, 21]. Using the G4 structure-specific antibody BG4, it was found that G4 structures are enriched in the promoters and 5'-UTRs of highly transcribed genes, particularly in genes related to cancer, such as c-MYC [20]. In addition, some endogenous proteins, such as human XPB/XPD1 [22], ATRX [23], yeast Rif [24], and Pif1 [25], which can bind to folded G4 structures, have also been mapped to G4 structures in the human genome via ChIP-seq. G4 sites mapped through G4 ChIP-seq can represent the actual G4 structures in the cell; however, the number of G4 sites can be affected by antibody specificity and sample treatment.
Structure of G-quadruplex DNA
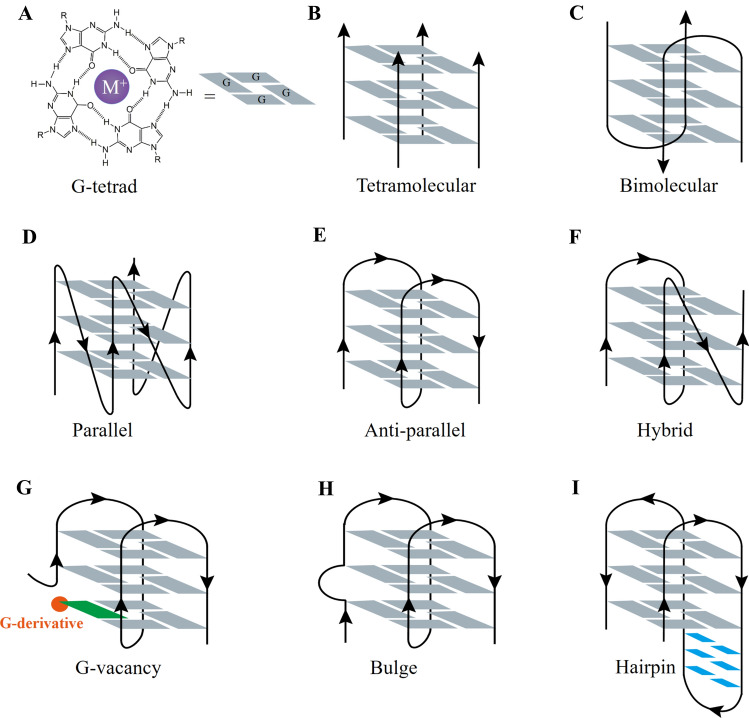
G-quadruplexes are stabilized by Hoogsteen hydrogen bonding; four guanines form a G-tetrad (Fig. 1A), then, two or more G-tetrads stack on top of each other, forming a quadruple helical structure driven by cations. According to the number of DNA strands, folded G4 can be grouped into intermolecular G4 (two or four DNA strands, Fig. 1B, C) or intramolecular G4 (one DNA strand, Fig. 1D–F) [26, 27]. Intramolecular G4 can display diverse topologies depending on the sequence difference, loop length, and ionic environment [28]. According to the orientation of the sequence in the G4 spatial structure, it can be divided into parallel, antiparallel, or hybrid structures (Fig. 1D–F). The spatial configuration of G4 has been widely studied using biophysical methods, such as NMR, X-ray, and circular dichroism spectra. As for the human telomeric G4 sequence d(AGGG(TTAGGG)3), it exhibits a canonical antiparallel state in a Na+ solution (Fig. 1E) [29]; however, in a K+ solution, it presents a mixture of parallel, antiparallel, and hybrid structures (Figs. 1D–F) [30, 31], which indicates that the same G4 sequence may have multiple complex structures in different environments. However, the effect of different topologies on the function of G4 in cells requires further study.
Fig. 1.
Structures and topologies of G-quadruplex DNA. A The chemical structure of the G-quartet. The G-quartet is formed by four guanines and is stabilized via Hoogsteen hydrogen bonding in the presence of a central cation. B, C Representative topologies of intermolecular G-quadruplex structures. D–F Schematic representation of canonical intramolecular G-quadruplex structures. According to the orientation of the G4 sequences, these can be divided into parallel, antiparallel, and hybrid G4 structures. G–I Schematic representation of non-canonical intramolecular G-quadruplex structures
In addition, an increasing number of studies regarding G4 structures with unique features, such as a G-vacancy, a bulge, or a long loop with duplex, have surfaced recently, expanding the repertoire of G-quadruplexes. The G-vacancy-bearing G-quadruplex (GVBQ) was recently excavated, which is formed by one G2 and three G3 tracts [32] (Fig. 1G). By accepting a guanine derivative, such as dGMP, GMP, or GTP, the G-vacancy can be filled up to form an intact G-tetrad, thereby forming an enhanced G4 [33, 34]. Using the bioinformatics method, approximately 220,000 potential GVBQ-forming sequences emerged in the human genome, most of which are preferentially located at the 5′-end of genes and may potentially respond to the environment in specific cellular processes [32]. The formation of GVBQ in promoters can regulate gene expression and has emerged as a novel therapeutic target. One typical example of this is the GVBQ from the promoter of the platelet-derived growth factor receptor beta (PDGFR-β) gene, which is a cell surface receptor tyrosine kinase that is closely associated with various pathologies, including atherosclerosis, fibrotic disorders, rheumatoid diseases, and cancers [35–37]. The G4 sequence in the nuclease hypersensitivity element (NHE) of the PDGFR-β promoter can form GVBQs. Intriguingly, 3'- and 5'-end G-vacancies are both present in the PDGFR-β promoter G4 sequences; also, guanine metabolites and drugs showed a conserved selectivity for the 5'-vacancy, and cGMP preferred to bind both the 3'- and 5'-end vacancies and formed two fill-in G4s with similar populations [33, 38]. Recently, a bi-functional guanine-RHAU23 peptide conjugate composed of a guanine moiety and a 23-aa G4-binding domain from the RHAU helicase has been reported to target and stabilize the GVBQs in DNA with superior specificity, providing a novel and promising alternative targeting strategy to a distinctive panel of G4s [39].
Meanwhile, bulge occurrence among G-tetrads in G4 has also been discovered in a series of G4 topologies (Fig. 1H). High-throughput sequencing of G4s in the human genome indicates that G4s with bulges are widespread, accounting for 21.6% and 30% of the total observed G4 sequences in solutions containing K+ and the G4 ligand pyridostatin, respectively [18]. The bulge varies with the base identity, size, and number per structure [40–42]. In single projection G4, the pyrimidine bulge is more common and more stable than the adenine bulge [40–42], while the guanine bulge has not yet been reported. In polynucleotide projection G4, the size of the bulge can increase to 7 nt [43]. Recently, a G4 structure containing a duplex bulge of up to 33 nt was reported, and that G4 stability slightly increased with increasing duplex bulge size, broadening the diversity of G4 topologies [44]. G4s containing bulges are widespread in gene promoters; for example, a thymine bulge G4 has been reported in the NHE of the KRAS promoter and plays a critical role in regulating KRAS signaling [45], which is a driver of many cancers by activating multiple pathways [46], especially PI3K, MAPK, PLCe, and RalGDS signaling.
The loop size of canonical G4 in bioinformatics algorithms has been defined as 1–7 nt. However, in the human genome, G4 sequences with long loops (> 7 nt) account for 21.5% of the total observed G4 in K+ and 24% in pyridostatin solutions [18]. Increasing studies have revealed the existence of hairpins or duplexes in the long loop of G4s [47, 48], forming quadruplex–duplex hybrids (Fig. 1I), which are also known as stem-loop-containing quadruplex sequences (SLQS) [49]. In the human genome, 80,307 SLQS embedded within 60,172 unique clusters were identified, most of which were strand-specifically located in promoter regions and were closely related to hundreds of brain tissue-related and cancer-associated genes [49]. Usually, the loop size has a significant impact on the stability and formation of G4 both in cell-free and cellular systems [50], but a stem loop can reinforce the stability of SLQS compared to a non-structured loop [51], thereby regulating the expression of key genes. Recently, an SLQS was reported and presented two distinct solution conformations that could co-exist in the human oncogene PIM1: form 1, containing a (3 + 1) G-tetrad core with a propeller loop, a co-axially stacked hairpin stem-loop, and a lateral loop; and form 2, containing a chair-type G-tetrad core and an adjoining G · C · G · C tetrad, with two lateral loops and a co-axially stacked hairpin stem loop [52]. The existence of a long loop can diversify the spatial configuration of G4 and provide sequence-specific (duplex binding) and scaffold-specific (G4 binding) structural elements, which can be targeted and selectively modulate gene expression.
Intracellular functions of G-quadruplex DNA
The general existence of G4s in specific regions of the genome, such as in DNA replication origins, chromosome ends, promoters, and gene transcriptional regulatory regions, determines their pivotal functions in various biological processes, including DNA replication, telomere maintenance, transcription, homologous recombination (HR), and epigenetics.
G-quadruplexes and DNA replication
G-quadruplexes affect DNA replication and genome stability
G4 DNA plays a dual role in DNA replication, as a critical component of metazoan replication origins and as an obstacle during DNA replication, leading to genome instability.
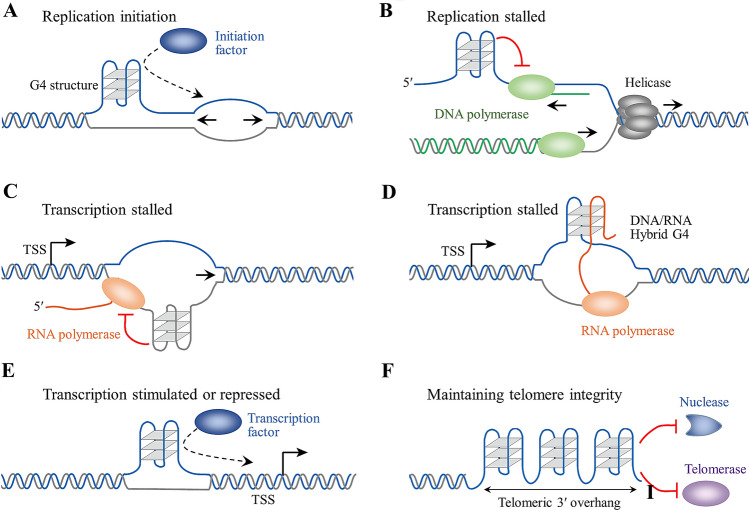
In mammalian genomes, ~ 100,000 potential DNA replication origins have been predicted, and 80%–90% of these origins contain GC-rich regions, forming origin G-rich repeated elements (OGRE), which can potentially form G-quadruplexes [53]. A previous experiment showed that G4 motifs in DNA replication origins are required for replication initiation and that affecting G4 stability also impairs the origin function (Fig. 2A) [54]. Furthermore, a recent study proclaimed that the deletion of G4 motifs in OGRE can strongly reduce the origin activity in mouse cells, and a G4 sequence from OGRE introduced into an ectopic origin-free region can establish a new functional origin [55]. The mechanism of G4-induced replication initiation may be due to the recruitment of replication initiation factors to replication origin sites, such as the origin recognition complex [56], treslin–MTBP complex [57], and the replication timing regulation protein Rif1 [58], which were reported to bind to G4 specifically both in cell-free and in cellular systems.
Fig. 2.
The biological function of G-quadruplex DNA in DNA replication, transcription, and telomere maintenance. A G4 can initiate DNA replication via the recruitment of replication initiation factors, such as the origin recognition complex, treslin-MTBP complex, and Rif1. B G4 can stall DNA polymerase during the DNA replication process, causing DNA replication disorder and genome instability. The replication of G4 can be resolved through the participation of G4-specific helicases such as Pif1, FANCJ, BLM and WRN, and TLS polymerases such as Rev1, Pol κ, Pol η, and Primpol. C G4 on the template strand can directly impede RNA polymerase during the transcription process. D G4 sequences on the non-template strand can form hybrid G4 with the newly synthesized RNA, inducing transcription termination. E The formation of G4 upstream of the TSS will usually inhibit transcription. G4 can also recruit certain transcription factors, which may either facilitate or restrain transcription. F A string of G-quadruplexes in the telomeric 3′-overhang involves telomere metabolism and telomere integrity maintenance. The G-quadruplex can protect telomeres from nuclease and interfere with telomerase activity
After initiation, DNA replication proceeds in a semi-discontinuous manner, during which the lagging strand remains single-stranded for a while and is intrinsically more prone to G4 formation. There is mounting evidence that G4 formation can obstruct the DNA replication process in both cell-free and cellular systems (Fig. 2B). Mutation of the dog-1 helicase in C. elegans was first reported for deletions in genes containing G-rich regions [59], which indicates that a deficiency in G4 unfolding may give rise to DNA synthesis disorders and genome instability. Simultaneously, mutations or deficiencies in other G4 unfolding-related proteins in the cell were authenticated to obstruct the DNA replication process, such as the helicases FANCJ [60], BLM [61], WRN [62], Pif1 [63], and DDX11[64]. The effect of G4s on DNA replication was also detected directly using cell-free systems. In a DNA polymerase-stop assay, G4s from telomeres were reported to arrest Taq DNA polymerase [65, 66] and yeast polymerase δ [67], leading to replication blockage. Furthermore, the G4 replication process by E. coli Pol I depends on the stability of G4 and the concentration of Pol I [6]; when high-stability G4s, such as four-layer G4s, occur on the template, E. coli Pol I will similarly be blocked.
Replication of G-quadruplexes
There are several pathways that can resolve the process of impeded G4 replication. A major method by which this happens is mediated by polymerase and G4-specific helicase to collaboratively replicate G4. G4-specific helicases, including FANCJ, BLM, WRN, Pif1, and RHAU, will be reviewed in the following sections. Some nucleases, such as DNA2 [68] and EXO1 [69], also facilitate the replication of G4 structures to avoid genome collapse and fork stagnation at G4 sites.
In addition, some polymerases that specifically recognize and replicate G4 structures have been observed in both cell-free and cellular systems. In recent years, increasing evidence has demonstrated that some translesion DNA synthesis (TLS) polymerases, such as Rev1, Pol κ, and Pol η, are implicated in G4 processing to promote DNA fork progression. A cell-free study showed that yeast Rev1 could replicate G-tract DNA in a template-specific manner, indicating that Rev1 is a G template-specific DNA polymerase [70] and can incorporate dCMP into the 3' end of DNA primers and function as a scaffold for proteins during TLS or base excision repair. Interacting with FANCJ helicase, REV1 can be directly recruited to G4s or through interactions mediated by PCNA to remodel oxidatively damaged G4 DNA [71]. Furthermore, human Rev1 has been shown to specifically bind G4 DNA substrates, dislodge tetrad guanines to unfold G4 DNA, and prevent G4 structures from refolding, thereby promoting fork progression [72]. It is worth noting that a loss of human Rev1 increased G4 mutation frequency > 200-fold compared to a control sequence and that hRev1 exhibited a strong affinity for parallel G4 rather than antiparallel or hybrid G4, whereas insert-2 in Rev1, a motif conserved in vertebrates but not yeast or plants, is responsible for the selective binding and accurate replication of G4 [73]. Apart from Rev1, other TLS polymerases, such as Pol κ, Pol η, and Dpo4, also participate in G4 replication [74–79]. Interestingly, the C-terminal of Rev1 may interact with Pol κ or Pol η to collaboratively replicate G4s.
Meanwhile, a novel eukaryotic DNA primase and DNA-directed polymerase, named PrimPol, has emerged in recent years and may be involved in G4 replication. PrimPol is a critical factor in DNA damage tolerance and mitochondrial DNA replication, particularly in invertebrate and human cells [80–82]. Recently, it was reported that PrimPol bound to and suppressed the formation of G4 structures, promoting the restart of DNA synthesis closely coupled to G4 replication impediments in DT40 cells [83]. PrimPol can also suppress R-loop formation in genes containing G4 and H-DNA motifs across the genome in both avian and human cells [84]. However, a recent study indicated that DNA replication by PrimPol is strongly blocked by representative stable G4 structures from human mitochondrial DNA, and that it could be overcome by the presence of Pif1 helicase in an error-prone DNA synthesis manner [85]. Therefore, cooperation between multiple proteins is indispensable for maintaining genome stability.
G-quadruplexes and transcription
G-quadruplexes affect gene transcription
The effect of G4 on DNA transcription depends on multiple factors, especially its orientation (on the template or non-template) and its location relative to the transcription start site (TSS). During DNA transcription, the presence of G4 on the DNA template strand blocks the movement of RNA polymerase (Fig. 2C), resulting in the arrest or even termination of transcription, directly decreasing the expression of the target genes [86, 87]. Also, G4 on the DNA non-template strand has also been reported to block the transcription process, which forms an unusually stable RNA/DNA hybrid between the G4 sequence on the non-template and the nascent RNA [88–90] (Fig. 2D). In contrast, recent studies discovered that G4 on the non-template strand can couple with and stabilize the R-loop [91], and the stabilized R-loop facilitates transcription through a mechanism involving successive rounds of R-loop formation [92]. It is worth noting that R-loop spreading caused by multiple stabilized G4 ligands can trigger the accumulation of DNA double-strand breaks (DSBs) in human cancer cells, thereby causing genome instability and activating cell apoptosis [93].
G4s have been mapped using a machine learning approach and high-throughput sequencing and were found to be enriched in cancer breakpoints and the TSSs of highly transcribed genes in human cells, particularly in genes related to cancer, such as c-MYC, BCL-2, KRAS, c-KIT, and VEGF (Table 1), further verifying the correlation among G4, transcription initiation, and the emergence of certain diseases.
Table 1.
Disease-related genes containing potential G-quadruplex structures in their promoters
| Gene | Sequence (5′–3′) | Position from TSS | Function | Disease | References |
|---|---|---|---|---|---|
| BCL-2 | P1G4: CGGGCGGGAGCGCGGCGGGCGGGCGGGCa | − 1439 to – 1412 | Apoptosis, programmed cell death | Various cancers, Alzheimer’s disease, Parkinson’s disease, stroke, and spinal cord injuries | [96, 97, 100-102] |
| Pu39: AGGGGCGGGCGCGGGAGGAAGGGGGCGGGAGCGGGGCTG | − 1489 to – 1451 | ||||
| P32: TGGGGTCCGCGACGGGGTGGGGGCTCCCGGGG | − 1906 to – 1875 | ||||
| bcl2G4-2: CCGGGCCAGGGAGCGGGGCGGAGGGGGCGGTCGGGT | − 1653 to – 1618 | ||||
| KRAS | G4near: AGGGCGGTGTGGGAAGAGGGAAGAGGGGGAGG | − 160 to – 129 | Signal transduction, cell proliferation, and cell survival | Various cancers | [45, 130] |
| G4middle: CGGGGAGAAGGAGGGGGCCGGGCCGGGCCGGCGGGGGAGGAGCGGGGGCCGGGC | − 228 to – 175 | ||||
| G4far: AGGGGTGGCTGGGGCGGTCTAGGGTGGCGAGCCGGGC | − 275 to – 239 | ||||
| c-MYC | GCGCTTATGGGGAGGGTGGGGAGGGTGGGGAAGGTGGGGAGGAGAC | − 148 to – 103 | Cell proliferation | Over 70% of cancers | [110, 111] |
| c-MYB | S1: AGGGGCGCCAGATTTGGCGGGAGGGGGAGTGT | − 80 to – 49 | Cell proliferation, cell differentiation | Colorectal tumors, leukemia, breast tumors | [253] |
| S2: CGGAGGCGGCGGGCAGGGC | − 114 to – 94 | ||||
| S3: AGGGAGGAGGGGAGGCGGCGGGACTGGGCGCGGGT | − 352 to – 318 | ||||
| S4: TGGGCCGGGCGGGGCGGGGTGGGTGGGGCCCGGGT | − 809 to – 775 | ||||
| hTERT | GGGGAGGGGCTGGGAGGGCCCGGAGGGGGCTGGGCCGGGGACCCGGGAGGGGTCGGGACGGGGCGGGG | − 90 to – 22 | Telomere maintenance | 85%–90% of cancers | [254] |
| c-KIT | c-KIT1: AGGGAGGGCGCTGGGAGGAGGGG | − 109 to – 87 | Cell growth, proliferation, migration, and survival | Gastrointestinal stromal tumors, pancreatic cancer, melanoma, and hematological neoplastic diseases | [255] |
| c-KIT2: GGGCGGGCGCGAGGGGAGGGG | − 160 to – 140 | ||||
| VEGF | GGGGCGGGCCGGGGGCGGGGTCCCGGCGGGGCGGAG | − 85 to – 50 | Angiogenesis, cell proliferation | Age-related macular degeneration, diabetes, rheumatoid arthritis, cancers | [256, 257] |
| HIF-1α | GGGCGGGGGAGAGGGGAGGGG | − 85 to – 65 | Immune response, cellular responses to hypoxia | Diabetes, cancers, vascular disease | [258] |
| PDGF | GGGGGGGGGGGGGCGGGGGCGGGGGCGGGGGAGGGG | − 82 to – 47 | Major mitogen, embryonic development, cellular differentiation, wound healing | Gliomas, sarcomas, astrocytoma, atherosclerosis, various fibrotic, neurological conditions | [35-37] |
| ILPR | (A(C/T)AGGGGT(G/C)TGGGG)40–160 | − 363 bp upstream of TSS | Insulin synthesis | Diabetes | [259] |
| C9orf72 | (GGGGCC)2–1600 | – | Autophagy | Devastating neurological diseases, including amyotrophic lateral sclerosis, frontotemporal dementia | [260, 261] |
| MET | GCGGGCGGGCGGGGCGCTGGGCT | − 48 to – 26 | Cell proliferation, anti-apoptosis, angiogenesis | Cancers of kidney, liver, stomach, breast, and brain | [262] |
| RET | AGCGGGTAGGGGCGGGGCGGGGCGGGGGCGGTCC | − 59 to – 26 | Tyrosine kinase receptor, activating MEK/ERK and PI3K/Akt | Medullary thyroid carcinomas | [135, 184] |
| DUX4 | DME1 GQ: CAGGGGATGGTGGGGCTGGGGTTGAGTGATGGGC | − 10,281 to − 10,252 | Embryonic genome activation, cell death | Facioscapulohumeral muscular dystrophy, various sarcomas, and hematological malignancies | [185] |
| D4P GQ: CGGGGTGGGGCGGGCTGTCCCAGGGGGGC | − 223 to – 195 | ||||
| PIM1 | GGGAGGGCGCGCCAGCGGGGTCGGG | – | Cooperation with c-MYC | The triple-negative breast cancer, lymphomas, hematopoietic and prostate cancers | [52] |
aThe sequences with underlines represent G-tracts in the G4s.
G4s in TSSs play diverse roles during gene transcription (Fig. 2E). One classical example is the G4 in the promoter of the BCL-2 gene, which encodes a mitochondrial membrane protein and functions by inhibiting apoptosis. BCL-2 is aberrantly overexpressed in a wide range of tumors and other diseases, including Alzheimer’s disease, Parkinson’s disease, and stroke [94]. Three promoters have been reported in BCL-2 [95]: P1, P2, and M. P1 is located 1489 to 1451 bp upstream of the TSS and is responsible for most of the expression of BCL-2; P2 and M are located between P1 and the TSS, and mainly modulate P1 activity. As a TATA-less and GC-rich promoter, P1 and its vicinity have been reported to have four disparate G4 DNA sequences (Table 1), including Pu39, bcl2G4-2, P1G4, and P32. Pu39 harbors six runs of guanine tracts and has the potential to form multiple overlapping quadruplexes with more than 15 possible combinations [96]. Pu39 functions as a transcriptional silencing element during BCL-2 expression, and the deletion or mutation of the Pu39 site in lymphoma DHL-4 cells has been shown to increase P1 activity by 2.1-fold or 2.6-fold, respectively [97]. Multiple transcription factors can bind to Pu39 and relieve its inhibitory effect on BCL-2 expression, such as specificity protein 1 (SP1) [98], Wilms’ tumor 1 (WT1) [97], and E2F [99]. Bcl2G4-2 was the second G4 sequence found near the P1 promoter, which is located 1653 to 1618 bp upstream of the TSS and can form a parallel G4 in the presence of K+; however, the stability of bcl2G4-2 appears to be much lower than that of other BCL-2 G4s, as its Tm is approximately 60 °C [100]. The potential bcl2G4-2-targeting transcription factors and their relevance in terms of BCL-2 transcription are yet to be explored. Located 1439 to 1412 bp upstream of the TSS with five runs of guanine tracts, P1G4 also functions as a BCL-2 transcription repressor, whereas in the extended BCL-2 P1 promoter region containing Pu39 and P1G4, P1G4 seems to play a more dominant role in inhibiting BCL-2 transcription in human breast cancer MCF-7 cell lines [101]. P32 is located 1906 to 1875 bp upstream of the TSS and occupies four runs of guanine tracts. Under physiological conditions with K+, P32 was identified to have an intramolecular hybrid G4 topology with high thermal stability (Tm = 82 °C) [102]. However, the inhibition of P32 via base mutations or the induction of a conformational transition with a phenanthroline derivative could cause obvious downregulation of BCL-2 expression in human lung A549 cancer cells [102], indicating that P32 may play an activating role in regulating BCL-2 expression in cellular systems. The regulation of G4s near the P1 promoter on BCL-2 expression mainly depends on the transcription factors recruited by G4, which is worthy of further study in vivo, especially for bcl2G4-2, P1G4, and P32. Also, whether there is cross talk between different G4s also deserves exploration, especially for Pu39 and P1G4, which are only 11 nt apart.
Similarly, the NHE of the oncogene KRAS also contains three putative G4 sequences, which are separated by 12 and 17 nucleotides, respectively, and are designated as G4far, G4middle, and G4near relative to the TSS (Table 1). G4near, also known as KRAS 32R, was shown to form two topologies in the 100 mM KCl solution, a hybrid G4 with a Tm ~ 72 °C, and a parallel G4 with a thymidine bulge in one strand, with a Tm ~ 55 °C [103, 104]. Pull-down assays and LC–MS/MS identified three DNA protein complexes that have an affinity for the G4near structure in human pancreatic cancer Panc-1 cells, including hnRNPA1, PARP-1, and Ku70 [103]. The hnRNPA1 was shown to bind to and destabilize the KRAS promoter G4, resulting in the upregulation of KRAS expression [105]; the same phenotype was also discovered in CCHC-type zinc-finger nucleic acid-binding protein (CNBP) [106] and myc-associated zinc finger (MAZ) [107] in later studies. In contrast, HMGB1, a ubiquitous non-histone protein involved in DNA repair, transcription, and telomere maintenance, was shown to bind to and stabilize G4near to suppress KRAS expression in Panc-1 cells [108]. In addition, the KRAS promoter G4 was associated with epigenetic mechanisms, as guanine has the lowest redox potential and can be oxidized to 8-oxoguanine (8OG) upon interaction with reactive oxygen species (ROS), impairing DNA, which will be discussed in the following section. However, based on the luciferase assay, a previous study claimed that G4middle might be a stronger repressor of promoter activity than G4near in HEK-293 cells [109], but more evidence is needed to support that claim. Furthermore, it was reported that G4far could not form an inducible and stable structure under a variety of buffer conditions in cell-free systems, while G4middle and G4near formed strong G4 structures [109], indicating that G4far may be inactive during KRAS transcription, but this needs further verification using in vivo experiments.
Another classical example is the G4 sequence in the promoter of c-MYC, which is the dominant carcinogenic driver in many cancers [110]. Located upstream of the c-MYC P1 promoter, the nuclease-hypersensitive element III1 (NHE III1) contributes to 80%–90% of c-MYC transcription and contains a 46-bp G4 sequence (named as Pu46), which occupies six G-tracts (Table 1) and forms stable G4s with multiple topologies in a cell-free system [110, 111]. Pu46 itself is critical for suppressing c-MYC expression; moreover, Pu46 can also recruit other transcription factors to regulate c-MYC transcription, including nucleoside diphosphate kinase B (NM23-H2), nucleolin, SP1, and CNBP [112]. NM23-H2 was found to maintain the single-stranded form of Pu46 to activate c-MYC transcription [113–115] and interact with CNBP, which can modulate the topological structure of G4 and unfold it [106], thereby increasing the transcription of c-MYC [116]. In contrast, nucleolin can contribute to G4 formation and stabilize Pu46 G4 to inhibit c-MYC expression [113, 117], as well as repress c-MYC transcription promoted by SP1 [118], which can recognize and bind both canonical dsDNA and G4s in the double-stranded NHE III1 [119, 120]. However, nucleolin may also act as a transcription activator in other promoters, such as VEGF [121] and ZEB1 [122]. The folding and structure of c-MYC G4 containing the G-tracts 1-2-4-5 [112], 1-2-3-4 [123], and 2-3-4-5 [124] have been reported in cell-free systems. However, the predominant G4 structure in living cells may be G4 involving G-tracts 2-3-4-5, which was verified in cells using multiple Pu22-specific small molecular probes, including a fluorescent probe named 9CI [125], a cyanine dye fluorescent probe named Cy-1 [126], and a self-assembled quinazoline–quinazolinone derivative named 4b [127]. The G-tracts 2-3-4-5 of Pu46, also known as Pu22, can fold into a parallel structure with a 1:2:1 nt loop arrangement; at present, most of the small molecule ligands targeting G4 to suppress c-MYC expression are based on this Pu22 structure.
G4 ligands targeting promoters for drug design
The structural diversity and cross talk between protein–G4 interactions originating from the G4s formed in the promoters make them attractive targets for drug design. Currently, the G4-interacting proteins database is available, providing information about proteins interacting with G4 structures [128], where more than 1000 G4 ligands have been discovered and shown in the G4 ligands database for drug discovery [129], providing a platform for the discovery and development of novel anticancer therapeutics. G4 ligands have been discovered or designed to suppress the expression of oncogenes by stabilizing G4 structures in the promoters, including porphyrin derivatives, acridine derivatives, pyridine derivatives, fluoroquinolone derivatives, among others (Table 2). Further, some natural alkaloids have been found to also target G4s and can be potential antitumor drugs, such as berberine, cryptolepine, and ellipticines.
Table 2.
G4 ligands and their therapeutic effects
| G4 ligand | Target G4 | Model | Effect | References |
|---|---|---|---|---|
| BRACO-19 | c-MYC, Telomere, KRAS | Breast cancer cells MCF-7, human epidermoid carcinoma A431 cells, human uterus carcinoma UXF1138L cells, etc | Uncapping 3′ telomere ends, inhibiting the activity of BLM and WRN, T-loop disassembly, apoptosis induction | [189–191] |
| TMPyP4 | Telomere, c-MYC, KRAS, BCL-2 | Breast cancer cells MCF7, MDA-MB-231, retinoblastoma cell, melanoma cell | Suppressing the growth and proliferation of tumor cells | [130, 131, 262–265] |
| Pyridostatin | Telomere, BRCA1, SRC | Human breast cell line MDA-MB-231, human MRC5 fibroblasts, cortical neurons, astrocytes, | Promoting growth arrest, inducing neurotoxicity, promoting DSBs | [266, 267] |
| RHPS4 | Telomere, mitochondrial G4 | SAOS-2 osteosarcoma, U251MG glioma cells, etc | Inhibition of cell proliferation, promoting DNA damage and replicative stress, promotes recombination | [194] |
| GQC-05 | c-MYC | Burkitt's lymphoma cell line CA46, acute myeloid leukemia cell lines KG-1a, CMK and TF-1 | DNA damage, apoptosis | [132–134] |
| NSC311153 | RET | Medullary thyroid carcinoma-derived TT cell line | Reduce cell viability | [136] |
| QN-1 | c-MYC |
Mouse breast carcinoma 4T1 cells, TNBC mouse model |
Cell cycle arrest and apoptosis | [137] |
| IZTZ-1 | c-MYC, | B16 melanoma cell, melanoma mouse model | Induce cell cycle arrest, apoptosis, and inhibit cell proliferation | [139] |
| DC-34 | c-MYC | Human multiple myeloma cell lines L363, KMS12PE, JIM1, etc | Cell cycle arrest | [140] |
| SYUIO-05 | c-MYC | Ramos and CA46 lymphoma cells | Dissociating the binding of NM23-H2 to the promoter | [141] |
| 7a4 | c-MYC |
RAJI, CA46, CCRF-CEM, and U266B1 cells human Burkitt’s lymphoma xenograft |
Disrupting the NM23-H2/c-MYC interaction, cell cycle arrest, and apoptosis and suppress tumor growth | [142] |
| IZCZ-3 | c-MYC | SiHa cells, human cervical squamous cancer xenograft | Provoking cell cycle arrest and apoptosis and inhibit cancer cell growth | [143] |
| CX-3543 | c-MYC | HT29 cell lines, lung carcinoma A549 cells, colorectal adenocarcinoma HCT-116 cells, breast adenocarcinoma MDA-MB-231 cells and MCF7 cells, pancreatic carcinoma MIA paca-2 cells, and acute promyelocytic leukemia HL-60, etc | Disrupting nucleolin/G4 complexes, suppressing RNA transcription, inducing apoptosis | [144, 145] |
| CX-5461 | Telomere, c-KIT1, c-MYC | Colorectal cancer DLD1 cells, ovarian cancer PEO1 cell, glioblastomas, oligodendrogliomas, oligoastrocytomas, etc | Blocking replication forks, inducing ssDNA gaps or breaks, disrupting topoisomerase II, activating the DNA damage response | [147–150, 152–155, 158] |
| Telomestatin | Telomere, c-MYB | Human lymphoma U937, glioma stem cell, etc | Dissociating telomere-binding proteins, causing telomere dysfunction in cancer cells | [172, 173, 175–177] |
| 6OTD | Telomere | Human glioblastoma U251 cells, GSC lines GBM146 and GBM157, glioblastoma U251 mouse xenografts | DNA damage, G1 cell cycle arrest, and apoptosis | [179] |
| Berberine | Telomere, RET, DUX4 | Medullary thyroid carcinomas TT cells, Rhabdomyosarcoma TE671 cells | Cell cycle arrest and apoptosis activation; inhibit muscle fibrosis, and consequently rescue muscle function | [183–185] |
| Ber8 | Telomere | Human cervical cancer cell Siha, human lung cancer cell A549, and human promyelocytic leukemia cell HL-60 | Inhibition of cell cycle arrest, cell senescence, and accelerating profound DNA damage at telomere regions | [186] |
| Ber-360A and Ber-PDS | Telomere | HeLa and HepG2 cells | Telomerase inhibition | [187] |
| EPI | Telomere | – | – | [188] |
Taking c-MYC as an example, we have summarized potential G4 ligands with physiological effects as reported in recent years. The earliest reported c-MYC G4 ligands include TMPyP4, BRACO-19, and pyridostatin, which possess the ability to suppress the growth and proliferation of multiple tumor cells including breast cancer cells, retinoblastoma cells, and melanoma cells. However, they were unable to distinguish different G4 topologies; for example, TMPyP4 has a high affinity for multiple G4s and inhibits the expression of multiple oncogenes, such as KRAS [130] and BCL-2 [131].
Ellipticine analogs, including GQC-05, NSC311153, and EPED3, have been reported to target proto-oncogenes and decrease tumor cell viability. GQC-05 was first reported as a high affinity and selective stabilizer of the c-MYC G4 [132]. In Burkitt’s lymphoma cell line CA46, GQC-05 induced cytotoxicity by altering the process of protein binding to the NHE III region of c-MYC, resulting in a corresponding decrease in c-MYC mRNA expression. Also, GQC-05 could also reduce cell viability and result in increased DNA damage and apoptosis in the acute myeloid leukemia cell lines KG-1a, CMK, and TF-1 [133]. Furthermore, GQC-05 has been shown to combine with navitoclax, a Bcl-2/Bcl-XL inhibitor, increasing its cytotoxic activity, which was more significant than either navitoclax or GQC-05 alone and more significant than navitoclax combined with cytarabine and adriamycin; however, its biological efficacy needs further verification in vivo. In addition to c-MYC G4, GQC-05 also targets the RNA G4 near the 5' splice site in Bcl-X pre-mRNA to induce apoptosis in HeLa cells [134]. NSC311153 incorporates a piperidine ring into ellipticine and achieves better G4 binding activity and stability than ellipticine [135]. In the medullary thyroid carcinoma (MTC)-derived TT cell line and a mouse MTC xenograft model, NSC311153 could target the promoter G4 and interfere with the transcription of the proto-oncogene rearranged during transfection (RET), which encodes a receptor tyrosine kinase and is related to MTC [135]. EPED3 is a highly stable and hydrophilic ellipticine analog, which initiates apoptosis at nanomolar concentrations in multiple myeloma cell lines while leaving stromal cells unharmed [136]. However, whether EPED3 targets G4s and the specific mechanism remains to be explored.
Recently, a difluorosubstituted quinoxaline analog, named QN-1, was designed and assessed for triple-negative breast cancer (TNBC) treatment [137]. QN-1 exhibited distinctive binding to the 5′-end G-tetrad of c-MYC G4, with weaker binding to other G4s (including G4s from the telomeres, BCL-2, VEGF, HRAS, and c-KIT1), which can be distinguished from most of the reported G4 ligands. In 4T1 breast cancer cells, QN-1 selectively downregulated c-MYC transcription, resulting in cell cycle arrest and apoptosis. In a 4T1 tumor-bearing mouse model, QN-1 exhibited good in vivo anti-TNBC activity with fewer side effects [137]. Although QN-1 can target c-MYC G4 and show excellent efficacy in TNBC, it also has some inherent disadvantages, especially its high molecular weight and poor solubility in water. Therefore, depending on the properties of QN-1 and a benzothiazole-based derivative named 4I [138], another group designed a better drug-like imidazole-benzothiazole conjugate, named IZTZ-1 [139]. In a cell-free system, water-soluble IZTZ-1 showed high affinity to c-MYC Pu22 G4 (KD = 2.0 μM) by stacking on both terminal G-quartets of Pu22. IZTZ-1 exhibited the same specific stabilizing ability on c-MYC G4 as with QN-1. Intracellular assays showed that IZTZ-1 could induce cell cycle arrest, apoptosis, and inhibit cell proliferation, whereas in a melanoma mouse model, IZTZ-1 can effectively inhibit tumor growth by downregulating c-MYC expression [139].
In addition, many other c-MYC G4 target ligands have been synthesized and assessed in vitro and in vivo, including DC-34 for myeloma [140], the quindoline derivative SYUIO-05 for Ramos and CA46 lymphoma cells [141], the SYUIO-05 based quindoline derivative 7a4 for Burkitt’s lymphoma [142], and the aryl-substituted imidazole/carbazole conjugate IZCZ-3 for squamous cell carcinoma [143], which may provide many promising anticancer candidates in future clinical trials.
Currently, two G4 ligands have already entered human clinical trials: CX-3543 and CX-5461, both of which were derived from fluoroquinolones. CX-3543, also known as quarfloxin, can selectively disrupt nucleolin/G4 complexes in the nucleus, thereby suppressing the transcription of RNA polymerase I and inducing apoptosis in cancer cells [144, 145]. CX-3543 was the first G-quadruplex target drug used in human clinical trials and has progressed to phase II clinical trials for carcinoid/neuroendocrine tumors, but it was withdrawn from further trials because of bioavailability issues [146].
CX-5461, known as Pidnarulex, was reported to be highly selective for human telomeric, c-KIT1, and c-MYC G4s [147], and is a first-in-class selective rDNA transcription inhibitor [148]. CX-5461 can block replication forks and induce ssDNA gaps or breaks [149], and exerts cytotoxicity primarily via topoisomerase II poisoning [150] and the activation of the ubiquitination pathway [151]. An increasing number of studies have indicated that CX-5461 effectively targets multiple tumors. CX-5461 exhibited specific toxicity against BRCA deficiencies in cancer cells and polyclonal patient-derived xenograft models, including tumors resistant to PARP inhibition [149]. Meanwhile, CX-5461 was also reported to effectively treat aggressive acute myeloid leukemia by targeting a leukemia-initiating cell population [152] and the specific inhibition of the translation of certain metabolic regulators [153]. In combination with a single-dose X-ray, CX-5461 can enhance tumor cell killing effects in ovarian cancer cells and the CaSki cervical cancer line [154]. Besides, recent studies reported that CX-5461 was a promising therapy in the treatment of high-grade serous ovarian cancer (HGSOC), and it was shown that CX-5461 can arrest the G2/M cell cycle checkpoint in multiple HR-proficient HGSOC cell lines in combination with the clinically used TOP1 inhibitor topotecan [148], and activate the DNA damage response in combination with PARP inhibitors in HR-deficient HGSOC [155]. CX-5461 was also reported to significantly hamper the proliferation of TNBC cells and synergistically enhance the efficacy of the p53 activator APR-246 by cleaving PARP and caspase 3, while also maintaining Annexin V positivity [156]. In addition to cancers, a study corroborated that CX-5461 could prevent the development of pulmonary arterial remodeling, perivascular inflammation, pulmonary hypertension, and improved survival. CX-5461 could also partially reverse established pulmonary hypertension by inducing cell cycle arrest in human pulmonary arterial smooth muscle cells and enhancing the activity of p53 [157], highlighting its potential applications in chronic diseases.
CX-5461 is currently in advanced phase I/II clinical trials for patients with hematologic cancers (Trial ID: ACTRN12613001061729) and BRCA1/2-deficient tumors (Canadian Cancer Trials Group ID: NCT02719997) [149]. Meanwhile, the phase I dose-escalation clinical trial of CX-5461 in advanced hematologic cancers was completed in 2019, indicating that it is safe at doses associated with clinical benefit, and can attain prolonged partial response in patients with anaplastic large cell lymphoma and achieve stable disease as the best response in patients with myeloma and diffuse large B-cell lymphoma [158].
G-quadruplexes in the telomeres
G-quadruplexes affect telomere metabolism
Telomeric DNA are repetitive DNA sequences found at the ends of the chromosomes, and their lengths are closely related to cancers, aging-related diseases, type 2 diabetes, cardiovascular disease, and the human life span [159]. Telomeric DNA in humans usually contains a double-stranded repeat region 2–50 kb long that has approximately 300–8000 precise CCCTAA/TTAGGG repeats, as well as the 3′-tail single-stranded repeat region with 10–50 TTAGGG repeats, which can form G4 structures under certain conditions, thereby regulating telomere metabolism [160]. G4s in telomeres can perform multiple functions. First, G4s can act as a telomeric capping structure [161] and avoid the hydrolysis of the 3′-tail single-stranded region by nuclease (Fig. 2F), thereby guaranteeing the integrity of the telomere. However, G4 is also a potential risk factor for telomere integrity, as large-scale G4s in the double-stranded repeat region of the telomere may hinder DNA replication by DNA polymerase. When G4-specific proteins such as helicase WRN [62], Pif1 [63], and nuclease DNA2 [68], EXO1 [69] are deficient, the replication forks at telomeres may stagnate and ultimately collapse. Therefore, G4 has played a dual role in maintaining telomere integrity.
In addition, G4 affects the activity of telomerase (Fig. 2F). As a unique reverse transcriptase, telomerase consists of the telomerase reverse transcriptase and telomerase RNA template and can catalyze telomeric DNA repeats onto the 3′-ends of the linear chromosome, generating new 3′-tail single-stranded repeat regions [162]. An early study reported that the folding of telomeric DNA into G4 impedes the elongation of the ciliate Oxytricha nova telomere [163]. Multiple studies have indicated that parallel intermolecular G4 DNA can serve as a substrate for telomerases from ciliates [164] and humans [165, 166]; however, the antiparallel and hybrid G4 structures formed by telomeric overhangs impede telomerase elongation [164–166]. It is worth mentioning that the POT1-TPP1 complex can bind to and unfold all telomeric G4 topologies (parallel, antiparallel, hybrid, or two contiguous quadruplexes) through an obligatory unfolding mechanism [167, 168]. Replication protein A (RPA), located at the telomere, acts as a telomere end-binding protein and mediates the unfolding of G4s [169, 170]. After the POT1-TPP1 complex or RPA disrupts telomeric G4s, telomerase can achieve the proper elongation of the telomere.
In most normal cells, telomerase activity is almost silenced, except in some T cells or stem-like cells that need to activate reverse transcriptase activity transiently during cell proliferation. Telomerase upregulation and activation are the general characteristics of various cancers, indicating that strategies targeting telomerase activity by telomeric G4 ligands could have significant anticancer effects [171].
G4 ligands targeting telomeres for drug design
Multiple telomeric G4-targeting ligands can inhibit telomerase activity and block tumor growth and metastasis. Telomestatin is a natural macrocyclic compound isolated from Streptomyces anulatus [172] and has been reported to interact specifically with the human telomeric intramolecular G4 in the absence of monovalent cations [173, 174]. Telomestatin was demonstrated to inhibit telomerase activity and shorten telomere length, exerting pro-apoptotic and antiproliferative effects in acute leukemia [175] and multiple myeloma [176]. Telomestatin can also segregate the telomere-capping protein TRF2 from the telomere, resulting in telomeric DNA damage and effectively activating the replication stress response pathway in glioma stem cells [177]. In addition to the disruption of telomeric G4, telomestatin also targets the promoter G4 of the proto-oncogene c-Myb and reduces c-Myb expression in glioma stem cells [178]. Based on the structure of telomestatin, a new series of macrocyclic hexaoxazole-type G4 ligands (6OTD) was synthesized. Ligands with the 6OTD structure caused DNA damage, G1 cell cycle arrest, and apoptosis in human glioblastoma cells, GSC cell lines, and glioblastoma U251 mouse xenografts, and these DNA damage foci were co-localized with telomeres, indicating that 6OTD limited the growth of GSCs by targeting telomeres [179]. Compared with temozolomide, a clinical antiglioma DNA alkylating agent, 6OTD needs a lower concentration to exert its anticancer effects, preferentially affecting GSCs and telomeres, verifying that 6OTD may be a potential therapeutic against glioblastoma [179]. Meanwhile, several other 6OTD derivatives have been synthesized, including L2H2-6OTD (1a), which was reported to target RNA G4s and show cytotoxicity towards cancer cells [180], as well as the 6OTD analog 5b, which preferentially stabilizes telomeric G4s over the promoter G4s of c-KIT and KRAS [181], their effects in vivo remain to be further identified.
Berberine, a natural isoquinoline alkaloid isolated from the Chinese traditional herb Coptis chinensis and other berberis plants, has shown antiinflammatory activity in a variety of chronic diseases and anticancer potential against various human cancer cells [182]. Berberine was reported to bind to telomeric G4 and was verified as a 2:1 ligand to the G-tetrad structure model, in which two berberines were in the two binding sites and directly interacted with each tetrad [183]. However, apart from telomeric G4s, berberine can also target other non-telomeric structures. In the medullary thyroid carcinoma TT cells, berberine inhibited RET expression by more than 90%, inhibiting cell proliferation via cell cycle arrest and apoptosis activation [184]. A recent study confirmed that berberine could target the promoter G4 of the DUX4 gene, reduce DUX4 expression, inhibit muscle fibrosis, and consequently rescue muscle function in facioscapulohumeral muscular dystrophy [185]. To date, multiple berberine derivatives have been designed to have improved efficacy. Ber8, a 9-substituted berberine derivative, exhibited strong interaction with telomeric G4s and could effectively inhibit cell cycle arrest, cell senescence, and accelerate DNA damage at telomeric regions in multiple cancer cells [186]. A further study revealed that Ber8 could not only facilitate the formation of endogenous telomeric G4s in cancer cells but also disaggregate TRF1 and POT1 from the telomere and induce telomere uncapping [186]. Some berberine derivatives have been shown to exhibit telomeric G4 specificity; for example, two berberine-bisquinolinium conjugates with fluorescence response, named Ber-360A and Ber-PDS, were able to distinguish telomere double G4s from other types of G4s and dsDNA, and displayed strong telomerase inhibition and anti-tumor activity, especially in HeLa and HepG2 cells [187]. In addition, a berberine derivative named epiberberine (EPI), was first reported to specifically bind to the hybrid-2 telomeric G4, which is the major form of G4 in wild-type human telomeric DNA, and induce an unprecedented extensive four-layer binding pocket specific to the hybrid-2 G4 [188] Furthermore, EPI could convert other telomeric G4 forms, such as hybrid-1 and basket-type, to hybrid-2, which was the first such example reported [188]. However, EPI still binds weakly to dsDNA, and its antitumor activity remains to be identified in cellular systems.
In addition, several other G4 ligands which can target G4 in the promoters of oncogenes, also can stabilize telomeric G4 and exert antitumor activity, such as BRACO-19 [189–191], TMPyP4 [192], CX-5461 [193], and RHPS4 [194] (Table 2). The tight relationship between cancer, telomerase, and telomeres makes antitumor therapeutic strategies involving G4 ligands more attractive.
G-quadruplexes affect epigenetic modifications
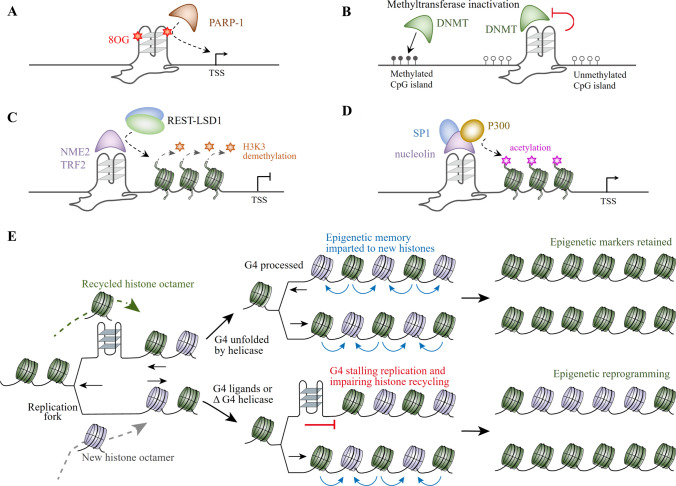
The relationship between G4 and epigenetic modifications is multifaceted. Under endogenous oxidative stress, reactive oxygen species (ROS) in cells cause DNA damage and focus on G4 sites, and different feedback mechanisms can upregulate or downregulate the expression of a large number of redox genes, resulting in oxidative DNA damage with epigenetic characteristics [195]. G4s, therefore, become epigenetic modification sites when the levels of oxidative stress become significantly increased during the inflammatory response and in cancer cells [196], as ROS can enhance the level of 7,8-dihydro-8-oxoguanine (8OG) more in G4 motifs than in non-G4 motif G-rich regions. The presence of 8OG in the G-quadruplex may lower the stability of the G4 depending on where the damage is located on its structure. The G4near of the KRAS promoter is a classical example of this. When 8OG occurs within the 11-nt loop of the G4near structure, its Tm and stability are almost unaffected; however, when 8OG is situated in the G-tetrad, both Tm and stability are greatly reduced [197]. It has been reported that the presence of 8OG on promoter G4s leads to an approximately 300% increase in gene expression [198]. Moreover, oxidized G4s that have been suggested to form unique, looped structures could also stimulate PARP-1 activity [199]. Furthermore, 8OG could enhance the binding of PARP-1 to G4near (Fig. 3A) and promote the recruitment of MAZ and hnRNPA1 to the promoter, thereby stimulating the transcription of KRAS in Panc-1 cells [200]. Certainly, 8OG on G4 can be removed through the DNA base excision repair (BER) pathway, which is initiated by 8-oxoG DNA glycosylase (OGG1) and subsequently recruits AP endonuclease 1 (APE1) to the G4 sequences and stabilize G4 folding [201]. However, a recent study indicated that OGG1 efficiently excised 8OG from oxidized G4near near the duplex but not within the G4 conformation; instead, endonuclease VIII-like 1 (Neil1) showed higher 8OG-excising activity in the G4 than the duplex DNA in Panc-1 cells [202].
Fig. 3.
G-quadruplexes can affect epigenetic modifications. A G4 can become epigenetic modification sites when the levels of oxidative stress increase. The presence of 8-oxoguanine (8OG) on the G-tetrad decreases the stability of the promoter G4, leading to an increase in gene expression. Meanwhile, 8OG could stimulate the binding of PARP-1 to the promoter G4 and promote the recruitment of MAZ and hnRNPA1 to the promoter, thereby stimulating transcription. B The formation of G4 at CpG islands can directly inhibit the activity of DNA methyltransferase after G4-preferred methyltransferase binding to the G4 structure. A typical example of this involves the DNA (cytosine-5)-methyltransferase DNMT1, DNMT3A, and DNMT3B. C G4 can affect histone epigenetics through the recruitment of proteins with histone-modifying activity, such as NME2 or TRF2, which can selectively bind to G4 and recruit the REST-LSD1 complex to remove the methylation of histone H3 Lys4 (H3K4) and inhibit gene expression. D In the promoter of ZEB1, nucleolin could bind to the G4 motif, remodel the local genomic region, facilitate the binding of SP1, and recruit P300 acetyltransferase, leading to enriched acetyl-histone H3 at the promoter, inducing gene transcription and oncogenic progression. E G4 can cause epigenetic reprogramming after the replication fork stalls at the G4 site in the presence of G4 ligands or the deficiency of G4-specific helicases. G4 structures may also impair histone recycling and lead to the localized loss of repressive chromatin, causing epigenetic reprogramming
G4 stability, position, and chromatin accessibility are closely related to CpG island methylation, where low- or non-methylated CpG islands prefer to form highly stable G4 [203]. The formation of G4 on CpG islands directly affects the activity of DNA methyltransferase (Fig. 3B) [204]. A typical example of this is DNA (cytosine-5)-methyltransferase (DNMT), which predominantly stalls and maintains cytosine methylation at CpG islands in mammals. It has been reported that human DNMT1, DNMT3A, and DNMT3B possess strong binding activity to G4 [205, 206]. It is vital to mention that the formation of the G4 structure can inhibit the methylation activity of DNMT1, thereby protecting certain CpG islands from methylation and suppressing local methylation [207].
G4 could also affect histone epigenetics by recruiting proteins with histone-modifying activity. A classic example of this is the G4-dependent recruitment of the RE1-silencing transcription factor (REST)-lysine-specific histone demethylase 1A (LSD1) repressor complex (Fig. 3C), which can remove the gene-activating monomethylation and demethylation of histone H3K4 [204]. G4 is indispensable for the occupancy of non-metastatic 2 (NME2) at the hTERT promoter as the REST-LSD1 repressor complex maintains repressive chromatin at the hTERT promoter and is dependent on NME2 [208]. In addition, telomere repeat-binding factor 2 (TRF2), which functions both inside and outside the telomere, was found to also interact with G4s in promoters [209, 210]. After recruitment in the promoter G4, TRF2 can promote the formation of the REST-coREST-LSD1-repressor complex at the p21 promoter and alter histone marks, resulting in the downregulation of p21 transcription and a reduction in the DNA damage response activation upon treatment with doxorubicin and G4-ligand 360A in cancer cells [210]. In contrast, four G4 motifs are formed in the promoter of Zinc-finger E-box binding homeobox 1 (ZEB1), which is frequently associated with cancer aggressiveness. After binding to the P1 G4, nucleolin could remodel the local genomic region, facilitate the binding of SP1, recruit P300 acetyl transferase, and enrich acetyl-histone H3 at the promoter, thereby inducing transcription and oncogenic progression (Fig. 3D) [122].
G4 causes local epigenetic reprogramming after stalling the replication fork [204, 211]. In REV1-deficient cells, DNA replication stagnation induced by G4 DNA can result in the uncoupling of DNA synthesis from histone recycling, leading to the localized loss of repressive chromatin through the preferential incorporation of newly synthesized unmodified histones during gap filling [212, 213] (Fig. 3E). G4 ligands can also hinder DNA replication and trigger local epigenetic plasticity via H3K4me3 loss and DNA cytosine methylation at the BU-1 locus of chicken DT40 cells [214]. Furthermore, a depletion of the nucleotide pool and a deficiency of G4-processing helicases, such as FANCJ, WRN, and BLM, will also induce epigenetic reprogramming and change in gene expression [215, 216]. Therefore, the replication fork stalling caused by unresolved G4 not only affects genome stability and telomere length but also impacts histone recycling and epigenetic reprogramming.
G4-interacting DNA helicases
Helicases are molecular motors that hydrolyze nucleoside triphosphates to execute critical functions in various DNA metabolic processes, including DNA replication, transcription, HR, repair, and telomeric metabolism. As the formation of stable G4 during DNA replication and transcription may cause genomic instability and gene expression regulation, G4 structures must be disrupted or unfolded in a regulated manner. Therefore, G4-specific unwinding helicases have been studied in-depth through various methods in recent years. The DNA G-quadruplex unwinding helicases are listed in Table 3, among which Pif1, RecQ family helicases, FANCJ, and DHX36 are known to be closely related to human diseases and have recently been studied in depth.
Table 3.
Summary of G4 unfolding helicases
| Superfamily | Subfamily | Name | Polarity | Function | Genetic disease | References |
|---|---|---|---|---|---|---|
| SF1 | Pif1 | 5′–3′ | Telomere maintenance, DNA replication, mitochondrial genome maintenance, DSB repair, epigenetic regulation | [229, 268] | ||
| DNA2 | 5′–3′ | DNA replication, telomere maintenance | [68, 269] | |||
| UvrD | 3′–5′ | Nucleotide excision repair, mismatch repair, HR | [270, 271] | |||
| Rep | 3′–5′ | DNA replication | [272] | |||
| RecD | 5′–3′ | Double-strand-break repair | [273] | |||
| SF2 | Fe-S | FANCJ | 5′–3′ | DNA replication, DNA crosslink repair, epigenetic regulation | Fanconi anemia, breast cancer | [215, 232, 274] |
| XPD | 5′–3′ | Nucleotide excision repair, transcription regulation | Cockayne syndrome, COFS syndrome, Xeroderma Pigmentosum | [22] | ||
| DDX11 | 5′–3′ | Sister chromatid cohesion, post-replicative repair, DNA replication | Warsaw Breakage Syndrome | [64, 275, 276] | ||
| RTEL1 | 5′–3′ | Telomere maintenance, DNA replication, DNA transcription | Dyskeratosis congenita | [234, 277, 278] | ||
| RecQ | RecQ | 3′–5′ | DNA repair, HR, DNA replication | [243] | ||
| BLM | 3′–5′ | DNA repair, HR, DNA replication | Bloom syndrome | [238, 279] | ||
| WRN | 3′–5′ | DNA repair, HR, DNA replication, epigenetic regulation | Werner syndrome | [279, 280] | ||
| RecQ5 | 3′–5′ | HR, DNA replication | [244] | |||
| DEAH | DHX9 | 3′–5′ | R-loop generation, DNA replication | [281] | ||
| SF3 | SV40 T-ag | 3′–5′ | DNA replication, transcription | [282, 283] | ||
| SF4 | Twinkle | 5′–3′ | Mitochondrial DNA replication | [284] | ||
| SF5 | DHX36 | 5′–3′ | Telomere maintenance, transcription regulation | [250, 251] |
Pif1
As the founding member of superfamily 1 DNA helicase, Pif1 is evolutionarily conserved from bacteria to humans and plays a vital role in maintaining genome stability in both the nucleus and mitochondria [217, 218]. Pif1 was first found to affect the activity of telomerase by displacing telomerase from telomeric DNA in both cell-free and cellular systems [219] and promote the proliferation and suppress the apoptosis of cervical cancer cells [220]. It was then found that Pif1 can remove telomerase from the telomere ends and accelerate the cycling of telomerase for additional telomere length-dependent extensions, thereby maintaining telomere length [221]. Furthermore, some proteins were found to contribute to telomere length homeostasis in cooperation with Pif1, such as Hrq1 [222] and RPA [223].
Pif1 directly promotes DNA replication through G4 motifs. The G4 stabilizer PhenDC3 was found to cause single-strand DNA lesions and impede DNA replication at G4 sites in the cell, and this stalled DNA synthesis machinery can be resolved by the members of the Pif1 family of helicases [224]. In Pif1-deficient cells, the DNA replication fork becomes stalled, inducing DNA breakage at the G4 motif sites [25]. Furthermore, the absence of Pif1 can cause genomic instability when G4s occur at the leading strand template [225]; however, another study concluded that stalled DNA replication occurred at the lagging strand G4 sites in Pif1-deficient cells, rather than on the leading strand [226]. The differences between the two studies may stem from the differences in the G4 sequences studies, assays for characterization, and genomic location. Meanwhile, it was affirmed that the coupling between Pif1 and PCNA is indispensable for the optimal progression of the replisome through G4 motifs [226].
Pif1 preferentially binds G4 DNA relative to ssDNA, dsDNA, and a partially single-stranded duplex DNA helicase substrate, highlighting its G4 interaction specificity [227]. In a cell-free system, simulated DNA replication of an ongoing synthesis of lagging strands stalled by G4 was established and detected using single-molecule fluorescence assays, verifying that Pif1 can unfold the G4 structure sequentially in two large steps, then halt at the ss/dsDNA junction, followed by a rapid reformation of G4 and the “acrobatic” re-initiation of G4 unfolding, exhibiting its periodic patrolling activity at the stalled DNA replication sites [228, 229]. In addition, G4 unfolding mediated by Pif1 is sensitive to G4 stability, and it was found that G4 structures with short loops can be barely unfolded by Pif1 and that Pif1 preferentially unfolded antiparallel G4 rather than parallel G4 of similar stability [230, 231]. Therefore, Pif1 is a G4-specific helicase with strong G4 unwinding activity, but cooperation with other helicases or proteins may be needed when interacting with highly stable G4s.
FANCJ
FANCJ was initially ascertained as a direct interaction partner of breast cancer type 1 susceptibility protein (BRCA1); therefore, it was also termed BRCA1-associated C-terminal helicase 1 (BACH1) or BRCA1-interacting protein 1 (BRIP1) [232]. FANCJ functions in the Fanconi anemia (FA) pathway that promotes inter-strand crosslink (ICL) repair through the interplay of lesion excision, translesion synthesis, and HR; a FANCJ mutation was found to be a cause of the bone marrow failure syndrome FA. Currently, FANCJ mutations are associated with an increased risk of multiple cancers, such as breast cancer, ovarian cancer, melanoma, prostate, and hereditary colon cancers [233].
In addition to ICL repair, FANCJ was reported to dominate G4 unwinding activity in both cell-free and cellular systems, laying the foundation for its function in DNA replication stalling and epigenetic reprogramming caused by G4s. The combination of a fluorescent probe and fluorescence lifetime imaging microscopy confirmed that a decrease in FANCJ expression can increase the number of G4s in mammalian cells, implying that FANCJ plays a role in unfolding G4 structures in cells [234]. In C. elegans, a deficiency in the FANCJ ortholog dog-1 can cause exclusive deletions of G4 DNA and eventual genome instability [235], and that depletion of FANCJ caused persistent DNA replication stalling at G4 sites [60], demonstrating the vital role of FANCJ in G4 replication. FANCJ was reported to coordinate two pathways for the efficient replication of G4s in the cell, in concert with either TLS polymerase REV1 or the WRN and BLM helicases, to initiate G4 unfolding and replication [215]. FANCJ occupies an iron–sulfur (FeS) cluster, wherein mutations in this cluster have been associated with cancer predisposition. A recent study reported that FANCJ required an intact FeS cluster to unfold G4 structures on the DNA template as observed via a primer extension assay using the lagging strand DNA polymerase δ [232], and that FANCJ knockout cells expressing FeS cluster-deficient variants displayed a similar enhanced sensitivity to the G4 ligands pyridostatin and CX-5461 [232], highlighting the remarkable role of the FeS domain in G4 metabolism as executed by FANCJ. Several other G4 ligands have also been reported to affect the G4 unfolding activity by FANCJ; telomestatin, Phen-DC3, and Phen-DC6 inhibited FANCJ helicase in unimolecular G4 and in bi- or tetramolecular G4 DNA [236].
RecQ family helicases
The RecQ family helicases, including E. coli RecQ, S. cerevisiae sgs1, H. sapiens RecQ1, BLM, WRN, RecQ4, and RecQ5, widely participate in various DNA metabolic processes, including DNA repair, HE, and telomerase maintenance. Importantly, mutations in H. sapiens BLM, WRN, and RecQ4 cause Bloom, Werner, and Rothmund–Thompson syndromes, respectively, which are linked to premature aging, profound developmental abnormalities, and an increased risk of cancers [237]. RecQ family members E. coli RecQ, S. cerevisiae sgs1, and H. sapiens BLM, WRN, and RecQ5 were reported to dominate G4 unfolding activity (Table 3). As for BLM, three mechanisms have been authenticated to be involved in G4 unfolding [238]: (1) BLM unfolds G4 with a 3-ssDNA tail in three discontinuous steps via unidirectional translocation, (2) the unfolded G4 gets connected to dsDNA via ssDNA in a repetitive manner in which the same helicase remains anchored at the ss/dsDNA junction, and (3) the G4 unfolds by reeling in 5′-ssDNA. Mechanistically, the structure of a bacterial RecQ helicase bound to unfolded G4 DNA has been reported, revealing a guanine-flipping and sequestration mechanism for G4 unfolding by RecQ helicases or other G4 unwinding helicases [239].
The helicase and RNaseD C-terminal (HRDC) domain [240], which are connected to the RecQ core by a flexible linker and are unique to E. coli RecQ, S. cerevisiae sgs1, and H. sapiens BLM and WRN, have been reported to perform vital functions in G4 unfolding. Using a stopped-flow assay, the HRDC domain was reported to improve G4 unfolding in Neisseria gonorrhoeae RecQ [241]. The cooperation between the RecQ core and the HRDC domains of BLM was demonstrated during G4 binding and unfolding, where the RQC domain interaction with G4 can be stabilized via HRDC binding to ssDNA [242]. Recently, using single-molecule assays, the HRDC domain was reported to be essential for the unfolding of the G4 structure by E. coli RecQ, while another model different from BLM was proposed, in which HRDC can reinforce the association of RecQ on the DNA by interacting with the RecA core, leading to a complete and longer-lasting G4 unfolding [243]. Interestingly, H. sapiens RecQ1 and RecQ4, which do not occupy the HRDC domain intrinsically, cannot unfold G4, and RecQ5, which does not possess the HRDC domain, had G4 unfolding activity that was an order of magnitude weaker than that of BLM or WRN [244]. Therefore, the HRDC of RecQ family helicases plays a unique role in the G4 unfolding process, but its function in cells requires further exploration.
DHX36
DHX36, also known as G4 resolvase (G4R1), MLE-like protein 1 (MLEL1), or RNA helicase associated with AU-rich element (RHAU), is involved in multiple cell differentiation processes, including spermatogenesis, heart development during embryogenesis, hematopoiesis, as well as the dendritic localization of neuronal precursor microRNA [245], and functions in the regulation of RNA structures. DHX36 has extreme affinity and efficiency in binding and unfolding both RNA G4 and DNA G4 [246], regulating the expression of cancer-related genes, such as p53, PITX, YY1, VEGF, and ESR1 [247, 248], and function in breast cancer, lung cancer, and colon cancer, among others [249].
The G4 unfolding mechanism of DHX36 was revealed through its crystal structure. The G4 DNA-resolving model was proposed, stating that the negatively charged G4 DNA was tightly bound to and was partially destabilized by a positively charged structural pocket in the RecA2 and OB-like domains of Drosophila DHX36; subsequently, the G4 structure was thoroughly unfolded by the translocation activity of DHX36 [250]. Another study shed light on a parallel G4 unfolding model by DHX36 in an ATP-independent manner, in which the N-terminal specific DNA binding-induced α-helix of DHX36 together with the OB-fold-like subdomain selectively binds to parallel G4, then the G4 binding process induces the rearrangement of the helicase core, finally actuating G-quadruplex unfolding one residue at a time by pulling on the ssDNA tail [251]. Depending on the structure of the DHX36 binding to G4, a bi-functional guanine–RHAU23 peptide conjugate composed of a guanine moiety and a 23-aa G4-binding domain from the N-terminus of the RHAU helicase was designed to target and stabilize G-vacancy-bearing G4s with superior specificity, providing a novel and promising strategy to target some non-classical G4s [39].
Conclusions
The existence, structure, and function of G-quadruplex DNA structures have expanded exponentially in the last decade. With the development of detection strategies, especially through the application of high-throughput sequencing, G4 detection in cellular systems is becoming increasingly accurate. However, the topology of inherent G4 structures is highly diverse, dependent on their sequence specificity, existing ionic environment, and the accompanying molecular chaperones. Multiple G4 topologies may co-exist for the same sequence, so the investigation of the potential topologies for G4 sequences that play key roles in biological processes requires further in-depth studies. The biological effects of different G4 topologies and their transformations also merit a thorough exploration, particularly regarding their regulation of disease-related gene expression. In addition, G4-specific helicases are indispensable in organisms; apart from G4 unfolding, the biological function of helicase-mediated G4 remodeling also needs to be uncovered.
The diversity in distribution and the vital regulatory functions of G-quadruplex structures in the genome show that they are promising targets in drug design. Currently, large numbers of G4 ligands have been discovered or synthesized, but many of these do not exhibit specificity or clear structure–activity relationships in vivo, so ligands capable of targeting specific G4 structures and their therapeutic potential still require further study, especially in vivo.
Acknowledgements
None.
Author contributions
XGX and YX conceived and supervised the study and provided resources; FYT and ZZJ wrote the manuscript; FYT, MG, and XZT made the tables and created the figures; XGX, YX, and FC made manuscript revisions.
Funding
This work was supported by the Natural Science Foundation of China (Grant Number: 81970676), the key projects of the Sichuan Science and Technology Department (Grant Number: 2019YFS0537 and 2020YFS0456), the Research Startup Funding of the Affiliated Hospital of Southwest Medical University (Grant Number: 18102), Scientific Research Funding of Luzhou-Southwest Medical University (Grant Number: 2019LZXNYDJ06). The research was conducted within the context of the International Associated Laboratory ‘Helicase-mediated G-quadruplex DNA unwinding and Genome Stability’.
Data availability
Not applicable.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Fang-Yuan Teng and Zong-Zhe Jiang have contributed equally to this work.
Contributor Information
Xu-Guang Xi, Email: xxi01@ens-cachan.fr.
Yong Xu, Email: xywyll@swmu.edu.cn.
References
- 1.Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc Natl Acad Sci USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334(6180):364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 3.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33(9):2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33(9):2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaplan OI, Berber B, Hekim N, Doluca O. G-quadruplex prediction in E. coli genome reveals a conserved putative G-quadruplex-Hairpin-Duplex switch. Nucleic Acids Res. 2016;44(19):9083–9095. doi: 10.1093/nar/gkw769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teng FY, Hou XM, Fan SH, Rety S, Dou SX, Xi XG. Escherichia coli DNA polymerase I can disrupt G-quadruplex structures during DNA replication. FEBS J. 2017;284(23):4051–4065. doi: 10.1111/febs.14290. [DOI] [PubMed] [Google Scholar]
- 7.Rawal P, Kummarasetti VB, Ravindran J, Kumar N, Halder K, Sharma R, Mukerji M, Das SK, Chowdhury S. Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation. Genome Res. 2006;16(5):644–655. doi: 10.1101/gr.4508806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capra JA, Paeschke K, Singh M, Zakian VA. G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae. PLoS Comput Biol. 2010;6(7):e1000861. doi: 10.1371/journal.pcbi.1000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hershman SG, Chen Q, Lee JY, Kozak ML, Yue P, Wang LS, Johnson FB. Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 2008;36(1):144–156. doi: 10.1093/nar/gkm986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varizhuk A, Ischenko D, Tsvetkov V, Novikov R, Kulemin N, Kaluzhny D, Vlasenok M, Naumov V, Smirnov I, Pozmogova G. The expanding repertoire of G4 DNA structures. Biochimie. 2017;135:54–62. doi: 10.1016/j.biochi.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Kudlicki AS. G-quadruplexes involving both strands of genomic DNA are highly abundant and colocalize with functional sites in the human genome. PLoS ONE. 2016;11(1):e0146174. doi: 10.1371/journal.pone.0146174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bedrat A, Lacroix L, Mergny JL. Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 2016;44(4):1746–1759. doi: 10.1093/nar/gkw006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brazda V, Kolomaznik J, Lysek J, Bartas M, Fojta M, Stastny J, Mergny JL. G4Hunter web application: a web server for G-quadruplex prediction. Bioinformatics. 2019;35(18):3493–3495. doi: 10.1093/bioinformatics/btz087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lacroix L. G4HunterApps. Bioinformatics. 2019;35(13):2311–2312. doi: 10.1093/bioinformatics/bty951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hon J, Martinek T, Zendulka J, Lexa M. pqsfinder: an exhaustive and imperfection-tolerant search tool for potential quadruplex-forming sequences in R. Bioinformatics. 2017;33(21):3373–3379. doi: 10.1093/bioinformatics/btx413. [DOI] [PubMed] [Google Scholar]
- 16.Kikin O, D’Antonio L, Bagga PS. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34(Web Server issue):W676–682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sahakyan AB, Chambers VS, Marsico G, Santner T, Di Antonio M, Balasubramanian S. Machine learning model for sequence-driven DNA G-quadruplex formation. Sci Rep. 2017;7(1):14535. doi: 10.1038/s41598-017-14017-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chambers VS, Marsico G, Boutell JM, Di Antonio M, Smith GP, Balasubramanian S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat Biotechnol. 2015;33(8):877–881. doi: 10.1038/nbt.3295. [DOI] [PubMed] [Google Scholar]
- 19.Marsico G, Chambers VS, Sahakyan AB, McCauley P, Boutell JM, Antonio MD, Balasubramanian S. Whole genome experimental maps of DNA G-quadruplexes in multiple species. Nucleic Acids Res. 2019;47(8):3862–3874. doi: 10.1093/nar/gkz179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansel-Hertsch R, Beraldi D, Lensing SV, Marsico G, Zyner K, Parry A, Di Antonio M, Pike J, Kimura H, Narita M, Tannahill D, Balasubramanian S. G-quadruplex structures mark human regulatory chromatin. Nat Genet. 2016;48(10):1267–1272. doi: 10.1038/ng.3662. [DOI] [PubMed] [Google Scholar]
- 21.Hansel-Hertsch R, Spiegel J, Marsico G, Tannahill D, Balasubramanian S. Genome-wide mapping of endogenous G-quadruplex DNA structures by chromatin immunoprecipitation and high-throughput sequencing. Nat Protoc. 2018;13(3):551–564. doi: 10.1038/nprot.2017.150. [DOI] [PubMed] [Google Scholar]
- 22.Gray LT, Vallur AC, Eddy J, Maizels N. G quadruplexes are genomewide targets of transcriptional helicases XPB and XPD. Nat Chem Biol. 2014;10(4):313–318. doi: 10.1038/nchembio.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Law MJ, Lower KM, Voon HP, Hughes JR, Garrick D, Viprakasit V, Mitson M, De Gobbi M, Marra M, Morris A, Abbott A, Wilder SP, Taylor S, Santos GM, Cross J, Ayyub H, Jones S, Ragoussis J, Rhodes D, Dunham I, Higgs DR, Gibbons RJ. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell. 2010;143(3):367–378. doi: 10.1016/j.cell.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 24.Kanoh Y, Matsumoto S, Fukatsu R, Kakusho N, Kono N, Renard-Guillet C, Masuda K, Iida K, Nagasawa K, Shirahige K, Masai H. Rif1 binds to G quadruplexes and suppresses replication over long distances. Nat Struct Mol Biol. 2015;22(11):889–897. doi: 10.1038/nsmb.3102. [DOI] [PubMed] [Google Scholar]
- 25.Paeschke K, Capra JA, Zakian VA. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145(5):678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haider S, Parkinson GN, Neidle S. Crystal structure of the potassium form of an Oxytricha nova G-quadruplex. J Mol Biol. 2002;320(2):189–200. doi: 10.1016/S0022-2836(02)00428-X. [DOI] [PubMed] [Google Scholar]
- 27.Schultze P, Smith FW, Feigon J. Refined solution structure of the dimeric quadruplex formed from the Oxytricha telomeric oligonucleotide d(GGGGTTTTGGGG) Structure. 1994;2(3):221–233. doi: 10.1016/s0969-2126(00)00023-x. [DOI] [PubMed] [Google Scholar]
- 28.Yuan WF, Wan LY, Peng H, Zhong YM, Cai WL, Zhang YQ, Ai WB, Wu JF. The influencing factors and functions of DNA G-quadruplexes. Cell Biochem Funct. 2020;38(5):524–532. doi: 10.1002/cbf.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Patel DJ. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1(4):263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 30.Parkinson GN, Lee MP, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417(6891):876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 31.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34(9):2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li XM, Zheng KW, Zhang JY, Liu HH, He YD, Yuan BF, Hao YH, Tan Z. Guanine-vacancy-bearing G-quadruplexes responsive to guanine derivatives. Proc Natl Acad Sci USA. 2015;112(47):14581–14586. doi: 10.1073/pnas.1516925112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang KB, Dickerhoff J, Wu G, Yang D. PDGFR-beta promoter forms a vacancy G-quadruplex that can be filled in by dGMP: solution structure and molecular recognition of guanine metabolites and drugs. J Am Chem Soc. 2020;142(11):5204–5211. doi: 10.1021/jacs.9b12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li XM, Zheng KW, Hao YH, Tan Z. Exceptionally selective and tunable sensing of guanine derivatives and analogues by structural complementation in a G-quadruplex. Angew Chem. 2016;55(44):13759–13764. doi: 10.1002/anie.201607195. [DOI] [PubMed] [Google Scholar]
- 35.Yang L, Li N, Xue Z, Liu LR, Li J, Huang X, Xie X, Zou Y, Tang H, Xie X. Synergistic therapeutic effect of combined PDGFR and SGK1 inhibition in metastasis-initiating cells of breast cancer. Cell Death Differ. 2020;27(7):2066–2080. doi: 10.1038/s41418-019-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fuchs MAA, Broeker KAE, Schrankl J, Burzlaff N, Willam C, Wagner C, Kurtz A. Inhibition of transforming growth factor beta1 signaling in resident interstitial cells attenuates profibrotic gene expression and preserves erythropoietin production during experimental kidney fibrosis in mice. Kidney Int. 2021;100(1):122–137. doi: 10.1016/j.kint.2021.02.035. [DOI] [PubMed] [Google Scholar]
- 37.Guerit E, Arts F, Dachy G, Boulouadnine B, Demoulin JB. PDGF receptor mutations in human diseases. Cell Mol Life Sci. 2021;78(8):3867–3881. doi: 10.1007/s00018-020-03753-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Onel B, Carver M, Agrawal P, Hurley LH, Yang D. The 3′-end region of the human PDGFR-beta core promoter nuclease hypersensitive element forms a mixture of two unique end-insertion G-quadruplexes. Biochim Biophys Gen Subj. 2018;1862(4):846–854. doi: 10.1016/j.bbagen.2017.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He YD, Zheng KW, Wen CJ, Li XM, Gong JY, Hao YH, Zhao Y, Tan Z. Selective targeting of guanine-vacancy-bearing G-quadruplexes by G-quartet complementation and stabilization with a guanine-peptide conjugate. J Am Chem Soc. 2020;142(26):11394–11403. doi: 10.1021/jacs.0c00774. [DOI] [PubMed] [Google Scholar]
- 40.Chan CY, Umar MI, Kwok CK. Spectroscopic analysis reveals the effect of a single nucleotide bulge on G-quadruplex structures. Chem Commun. 2019;55(18):2616–2619. doi: 10.1039/c8cc09929d. [DOI] [PubMed] [Google Scholar]
- 41.Sengar A, Vandana JJ, Chambers VS, Di Antonio M, Winnerdy FR, Balasubramanian S, Phan AT. Structure of a (3+1) hybrid G-quadruplex in the PARP1 promoter. Nucleic Acids Res. 2019;47(3):1564–1572. doi: 10.1093/nar/gky1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zivkovic ML, Rozman J, Plavec J. Structure of a DNA G-quadruplex related to osteoporosis with a G-A bulge forming a pseudo-loop. Molecules. 2020 doi: 10.3390/molecules25204867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mukundan VT, Phan AT. Bulges in G-quadruplexes: broadening the definition of G-quadruplex-forming sequences. J Am Chem Soc. 2013;135(13):5017–5028. doi: 10.1021/ja310251r. [DOI] [PubMed] [Google Scholar]
- 44.Nguyen TQN, Lim KW, Phan AT. Duplex formation in a G-quadruplex bulge. Nucleic Acids Res. 2020;48(18):10567–10575. doi: 10.1093/nar/gkaa738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ou A, Schmidberger JW, Wilson KA, Evans CW, Hargreaves JA, Grigg M, O’Mara ML, Iyer KS, Bond CS, Smith NM. High resolution crystal structure of a KRAS promoter G-quadruplex reveals a dimer with extensive poly-A pi-stacking interactions for small-molecule recognition. Nucleic Acids Res. 2020;48(10):5766–5776. doi: 10.1093/nar/gkaa262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ternet C, Kiel C. Signaling pathways in intestinal homeostasis and colorectal cancer: KRAS at centre stage. Cell Commun Signal. 2021;19(1):31. doi: 10.1186/s12964-021-00712-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karg B, Mohr S, Weisz K. Duplex-guided refolding into novel G-quadruplex (3+1) hybrid conformations. Angew Chem. 2019;58(32):11068–11071. doi: 10.1002/anie.201905372. [DOI] [PubMed] [Google Scholar]
- 48.Nguyen TQN, Lim KW, Phan AT. Folding kinetics of G-quadruplexes: duplex stem loops drive and accelerate G-quadruplex folding. J Phys Chem B. 2020;124(25):5122–5130. doi: 10.1021/acs.jpcb.0c02548. [DOI] [PubMed] [Google Scholar]
- 49.Lim KW, Jenjaroenpun P, Low ZJ, Khong ZJ, Ng YS, Kuznetsov VA, Phan AT. Duplex stem-loop-containing quadruplex motifs in the human genome: a combined genomic and structural study. Nucleic Acids Res. 2015;43(11):5630–5646. doi: 10.1093/nar/gkv355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guedin A, Gros J, Alberti P, Mergny JL. How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 2010;38(21):7858–7868. doi: 10.1093/nar/gkq639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lim KW, Khong ZJ, Phan AT. Thermal stability of DNA quadruplex-duplex hybrids. Biochemistry. 2014;53(1):247–257. doi: 10.1021/bi401161a. [DOI] [PubMed] [Google Scholar]
- 52.Tan DJY, Winnerdy FR, Lim KW, Phan AT. Coexistence of two quadruplex-duplex hybrids in the PIM1 gene. Nucleic Acids Res. 2020;48(19):11162–11171. doi: 10.1093/nar/gkaa752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prioleau MN. G-quadruplexes and DNA replication origins. Adv Exp Med Biol. 2017;1042:273–286. doi: 10.1007/978-981-10-6955-0_13. [DOI] [PubMed] [Google Scholar]
- 54.Valton AL, Hassan-Zadeh V, Lema I, Boggetto N, Alberti P, Saintome C, Riou JF, Prioleau MN. G4 motifs affect origin positioning and efficiency in two vertebrate replicators. EMBO J. 2014;33(7):732–746. doi: 10.1002/embj.201387506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prorok P, Artufel M, Aze A, Coulombe P, Peiffer I, Lacroix L, Guedin A, Mergny JL, Damaschke J, Schepers A, Cayrou C, Teulade-Fichou MP, Ballester B, Mechali M. Involvement of G-quadruplex regions in mammalian replication origin activity. Nat Commun. 2019;10(1):3274. doi: 10.1038/s41467-019-11104-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoshina S, Yura K, Teranishi H, Kiyasu N, Tominaga A, Kadoma H, Nakatsuka A, Kunichika T, Obuse C, Waga S. Human origin recognition complex binds preferentially to G-quadruplex-preferable RNA and single-stranded DNA. J Biol Chem. 2013;288(42):30161–30171. doi: 10.1074/jbc.M113.492504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumagai A, Dunphy WG. Binding of the treslin-MTBP complex to specific regions of the human genome promotes the initiation of DNA replication. Cell Rep. 2020;32(12):108178. doi: 10.1016/j.celrep.2020.108178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alavi S, Ghadiri H, Dabirmanesh B, Moriyama K, Khajeh K, Masai H. G-quadruplex binding protein Rif1, a key regulator of replication timing. J Biochem. 2021;169(1):1–14. doi: 10.1093/jb/mvaa128. [DOI] [PubMed] [Google Scholar]
- 59.Cheung I, Schertzer M, Rose A, Lansdorp PM. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat Genet. 2002;31(4):405–409. doi: 10.1038/ng928. [DOI] [PubMed] [Google Scholar]
- 60.Bosch PC, Segura-Bayona S, Koole W, van Heteren JT, Dewar JM, Tijsterman M, Knipscheer P. FANCJ promotes DNA synthesis through G-quadruplex structures. EMBO J. 2014;33(21):2521–2533. doi: 10.15252/embj.201488663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Drosopoulos WC, Kosiyatrakul ST, Schildkraut CL. BLM helicase facilitates telomere replication during leading strand synthesis of telomeres. J Cell Biol. 2015;210(2):191–208. doi: 10.1083/jcb.201410061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aggarwal M, Sommers JA, Shoemaker RH, Brosh RM., Jr Inhibition of helicase activity by a small molecule impairs Werner syndrome helicase (WRN) function in the cellular response to DNA damage or replication stress. Proc Natl Acad Sci USA. 2011;108(4):1525–1530. doi: 10.1073/pnas.1006423108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kocak E, Dykstra S, Nemeth A, Coughlin CG, Rodgers K, McVey M. The Drosophila melanogaster PIF1 helicase promotes survival during replication stress and processive DNA synthesis during double-strand gap repair. Genetics. 2019;213(3):835–847. doi: 10.1534/genetics.119.302665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Schie JJM, Faramarz A, Balk JA, Stewart GS, Cantelli E, Oostra AB, Rooimans MA, Parish JL, de Almeida EC, Dumic K, Barisic I, Diderich KEM, van Slegtenhorst MA, Mahtab M, Pisani FM, Te Riele H, Ameziane N, Wolthuis RMF, de Lange J. Warsaw breakage syndrome associated DDX11 helicase resolves G-quadruplex structures to support sister chromatid cohesion. Nat Commun. 2020;11(1):4287. doi: 10.1038/s41467-020-18066-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun D, Hurley LH. Biochemical techniques for the characterization of G-quadruplex structures: EMSA, DMS footprinting, and DNA polymerase stop assay. Methods Mol Biol. 2010;608:65–79. doi: 10.1007/978-1-59745-363-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Han H, Hurley LH, Salazar M. A DNA polymerase stop assay for G-quadruplex-interactive compounds. Nucleic Acids Res. 1999;27(2):537–542. doi: 10.1093/nar/27.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lormand JD, Buncher N, Murphy CT, Kaur P, Lee MY, Burgers P, Wang H, Kunkel TA, Opresko PL. DNA polymerase delta stalls on telomeric lagging strand templates independently from G-quadruplex formation. Nucleic Acids Res. 2013;41(22):10323–10333. doi: 10.1093/nar/gkt813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin W, Sampathi S, Dai H, Liu C, Zhou M, Hu J, Huang Q, Campbell J, Shin-Ya K, Zheng L, Chai W, Shen B. Mammalian DNA2 helicase/nuclease cleaves G-quadruplex DNA and is required for telomere integrity. EMBO J. 2013;32(10):1425–1439. doi: 10.1038/emboj.2013.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stroik S, Kurtz K, Lin K, Karachenets S, Myers CL, Bielinsky AK, Hendrickson EA. EXO1 resection at G-quadruplex structures facilitates resolution and replication. Nucleic Acids Res. 2020;48(9):4960–4975. doi: 10.1093/nar/gkaa199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haracska L, Prakash S, Prakash L. Yeast Rev1 protein is a G template-specific DNA polymerase. J Biol Chem. 2002;277(18):15546–15551. doi: 10.1074/jbc.M112146200. [DOI] [PubMed] [Google Scholar]
- 71.Lowran K, Campbell L, Popp P, Wu CG. Assembly of a G-quadruplex repair complex by the FANCJ DNA helicase and the REV1 polymerase. Genes (Basel) 2019 doi: 10.3390/genes11010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eddy S, Ketkar A, Zafar MK, Maddukuri L, Choi JY, Eoff RL. Human Rev1 polymerase disrupts G-quadruplex DNA. Nucleic Acids Res. 2014;42(5):3272–3285. doi: 10.1093/nar/gkt1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ketkar A, Smith L, Johnson C, Richey A, Berry M, Hartman JH, Maddukuri L, Reed MR, Gunderson JEC, Leung JWC, Eoff RL. Human Rev1 relies on insert-2 to promote selective binding and accurate replication of stabilized G-quadruplex motifs. Nucleic Acids Res. 2021;49(4):2065–2084. doi: 10.1093/nar/gkab041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Garcia-Exposito L, Bournique E, Bergoglio V, Bose A, Barroso-Gonzalez J, Zhang S, Roncaioli JL, Lee M, Wallace CT, Watkins SC, Opresko PL, Hoffmann JS, O’Sullivan RJ. Proteomic profiling reveals a specific role for translesion DNA polymerase eta in the alternative lengthening of telomeres. Cell Rep. 2016;17(7):1858–1871. doi: 10.1016/j.celrep.2016.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Murphy CT, Gupta A, Armitage BA, Opresko PL. Hybridization of G-quadruplex-forming peptide nucleic acids to guanine-rich DNA templates inhibits DNA polymerase eta extension. Biochemistry. 2014;53(32):5315–5322. doi: 10.1021/bi5006859. [DOI] [PubMed] [Google Scholar]
- 76.Pope-Varsalona H, Liu FJ, Guzik L, Opresko PL. Polymerase eta suppresses telomere defects induced by DNA damaging agents. Nucleic Acids Res. 2014;42(21):13096–13109. doi: 10.1093/nar/gku1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Eddy S, Maddukuri L, Ketkar A, Zafar MK, Henninger EE, Pursell ZF, Eoff RL. Evidence for the kinetic partitioning of polymerase activity on G-quadruplex DNA. Biochemistry. 2015;54(20):3218–3230. doi: 10.1021/acs.biochem.5b00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Eddy S, Tillman M, Maddukuri L, Ketkar A, Zafar MK, Eoff RL. Human translesion polymerase kappa exhibits enhanced activity and reduced fidelity two nucleotides from G-quadruplex DNA. Biochemistry. 2016;55(37):5218–5229. doi: 10.1021/acs.biochem.6b00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Berroyer A, Alvarado G, Larson ED. Response of Sulfolobus solfataricus Dpo4 polymerase in vitro to a DNA G-quadruplex. Mutagenesis. 2019;34(3):289–297. doi: 10.1093/mutage/gez010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Quinet A, Tirman S, Jackson J, Svikovic S, Lemacon D, Carvajal-Maldonado D, Gonzalez-Acosta D, Vessoni AT, Cybulla E, Wood M, Tavis S, Batista LFZ, Mendez J, Sale JE, Vindigni A. PRIMPOL-mediated adaptive response suppresses replication fork reversal in BRCA-deficient cells. Mol Cell. 2020;77(3):461–474.e9. doi: 10.1016/j.molcel.2019.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bailey LJ, Bianchi J, Doherty AJ. PrimPol is required for the maintenance of efficient nuclear and mitochondrial DNA replication in human cells. Nucleic Acids Res. 2019;47(8):4026–4038. doi: 10.1093/nar/gkz056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tirman S, Cybulla E, Quinet A, Meroni A, Vindigni A. PRIMPOL ready, set, reprime! Crit Rev Biochem Mol Biol. 2021;56(1):17–30. doi: 10.1080/10409238.2020.1841089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schiavone D, Jozwiakowski SK, Romanello M, Guilbaud G, Guilliam TA, Bailey LJ, Sale JE, Doherty AJ. PrimPol is required for replicative tolerance of G quadruplexes in vertebrate cells. Mol Cell. 2016;61(1):161–169. doi: 10.1016/j.molcel.2015.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Svikovic S, Crisp A, Tan-Wong SM, Guilliam TA, Doherty AJ, Proudfoot NJ, Guilbaud G, Sale JE. R-loop formation during S phase is restricted by PrimPol-mediated repriming. EMBO J. 2019 doi: 10.15252/embj.201899793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Butler TJ, Estep KN, Sommers JA, Maul RW, Moore AZ, Bandinelli S, Cucca F, Tuke MA, Wood AR, Bharti SK, Bogenhagen DF, Yakubovskaya E, Garcia-Diaz M, Guilliam TA, Byrd AK, Raney KD, Doherty AJ, Ferrucci L, Schlessinger D, Ding J, Brosh RM. Mitochondrial genetic variation is enriched in G-quadruplex regions that stall DNA synthesis in vitro. Hum Mol Genet. 2020;29(8):1292–1309. doi: 10.1093/hmg/ddaa043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Broxson C, Beckett J, Tornaletti S. Transcription arrest by a G quadruplex forming-trinucleotide repeat sequence from the human c-myb gene. Biochemistry. 2011;50(19):4162–4172. doi: 10.1021/bi2002136. [DOI] [PubMed] [Google Scholar]
- 87.Smestad JA, Maher LJ., 3rd Relationships between putative G-quadruplex-forming sequences, RecQ helicases, and transcription. BMC Med Genet. 2015;16:91. doi: 10.1186/s12881-015-0236-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Belotserkovskii BP, Liu R, Tornaletti S, Krasilnikova MM, Mirkin SM, Hanawalt PC. Mechanisms and implications of transcription blockage by guanine-rich DNA sequences. Proc Natl Acad Sci USA. 2010;107(29):12816–12821. doi: 10.1073/pnas.1007580107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Belotserkovskii BP, Neil AJ, Saleh SS, Shin JH, Mirkin SM, Hanawalt PC. Transcription blockage by homopurine DNA sequences: role of sequence composition and single-strand breaks. Nucleic Acids Res. 2013;41(3):1817–1828. doi: 10.1093/nar/gks1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Belotserkovskii BP, Soo Shin JH, Hanawalt PC. Strong transcription blockage mediated by R-loop formation within a G-rich homopurine-homopyrimidine sequence localized in the vicinity of the promoter. Nucleic Acids Res. 2017;45(11):6589–6599. doi: 10.1093/nar/gkx403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lim G, Hohng S. Single-molecule fluorescence studies on cotranscriptional G-quadruplex formation coupled with R-loop formation. Nucleic Acids Res. 2020;48(16):9195–9203. doi: 10.1093/nar/gkaa695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee CY, McNerney C, Ma K, Zhao W, Wang A, Myong S. R-loop induced G-quadruplex in non-template promotes transcription by successive R-loop formation. Nat Commun. 2020;11(1):3392. doi: 10.1038/s41467-020-17176-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.De Magis A, Manzo SG, Russo M, Marinello J, Morigi R, Sordet O, Capranico G. DNA damage and genome instability by G-quadruplex ligands are mediated by R loops in human cancer cells. Proc Natl Acad Sci USA. 2019;116(3):816–825. doi: 10.1073/pnas.1810409116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang L, Lu Z, Zhao X. Targeting Bcl-2 for cancer therapy. Biochim Biophys Rev Cancer. 2021;1876(1):188569. doi: 10.1016/j.bbcan.2021.188569. [DOI] [PubMed] [Google Scholar]
- 95.Bredow S, Juri DE, Cardon K, Tesfaigzi Y. Identification of a novel Bcl-2 promoter region that counteracts in a p53-dependent manner the inhibitory P2 region. Gene. 2007;404(1–2):110–116. doi: 10.1016/j.gene.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cheng Y, Tang Q, Li Y, Zhang Y, Zhao C, Yan J, You H. Folding/unfolding kinetics of G-quadruplexes upstream of the P1 promoter of the human BCL-2 oncogene. J Biol Chem. 2019;294(15):5890–5895. doi: 10.1074/jbc.RA119.007516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Heckman C, Mochon E, Arcinas M, Boxer LM. The WT1 protein is a negative regulator of the normal bcl-2 allele in t(14;18) lymphomas. J Biol Chem. 1997;272(31):19609–19614. doi: 10.1074/jbc.272.31.19609. [DOI] [PubMed] [Google Scholar]
- 98.Seto M, Jaeger U, Hockett RD, Graninger W, Bennett S, Goldman P, Korsmeyer SJ. Alternative promoters and exons, somatic mutation and deregulation of the Bcl-2-Ig fusion gene in lymphoma. EMBO J. 1988;7(1):123–131. doi: 10.1002/j.1460-2075.1988.tb02791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gomez-Manzano C, Mitlianga P, Fueyo J, Lee HY, Hu M, Spurgers KB, Glass TL, Koul D, Liu TJ, McDonnell TJ, Yung WK. Transfer of E2F–1 to human glioma cells results in transcriptional up-regulation of Bcl-2. Cancer Res. 2001;61(18):6693–6697. [PubMed] [Google Scholar]
- 100.Onyshchenko MI, Gaynutdinov TI, Englund EA, Appella DH, Neumann RD, Panyutin IG. Stabilization of G-quadruplex in the BCL2 promoter region in double-stranded DNA by invading short PNAs. Nucleic Acids Res. 2009;37(22):7570–7580. doi: 10.1093/nar/gkp840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Onel B, Carver M, Wu G, Timonina D, Kalarn S, Larriva M, Yang D. A new G-quadruplex with hairpin loop immediately upstream of the human BCL2 P1 promoter modulates transcription. J Am Chem Soc. 2016;138(8):2563–2570. doi: 10.1021/jacs.5b08596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sun H, Xiang J, Shi Y, Yang Q, Guan A, Li Q, Yu L, Shang Q, Zhang H, Tang Y, Xu G. A newly identified G-quadruplex as a potential target regulating Bcl-2 expression. Biochim Biophys Acta. 2014;1840(10):3052–3057. doi: 10.1016/j.bbagen.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 103.Cogoi S, Paramasivam M, Spolaore B, Xodo LE. Structural polymorphism within a regulatory element of the human KRAS promoter: formation of G4-DNA recognized by nuclear proteins. Nucleic Acids Res. 2008;36(11):3765–3780. doi: 10.1093/nar/gkn120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cogoi S, Xodo LE. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006;34(9):2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chu PC, Yang MC, Kulp SK, Salunke SB, Himmel LE, Fang CS, Jadhav AM, Shan YS, Lee CT, Lai MD, Shirley LA, Bekaii-Saab T, Chen CS. Regulation of oncogenic KRAS signaling via a novel KRAS-integrin-linked kinase-hnRNPA1 regulatory loop in human pancreatic cancer cells. Oncogene. 2016;35(30):3897–3908. doi: 10.1038/onc.2015.458. [DOI] [PubMed] [Google Scholar]
- 106.David AP, Pipier A, Pascutti F, Binolfi A, Weiner AMJ, Challier E, Heckel S, Calsou P, Gomez D, Calcaterra NB, Armas P. CNBP controls transcription by unfolding DNA G-quadruplex structures. Nucleic Acids Res. 2019;47(15):7901–7913. doi: 10.1093/nar/gkz527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cogoi S, Zorzet S, Rapozzi V, Geci I, Pedersen EB, Xodo LE. MAZ-binding G4-decoy with locked nucleic acid and twisted intercalating nucleic acid modifications suppresses KRAS in pancreatic cancer cells and delays tumor growth in mice. Nucleic Acids Res. 2013;41(7):4049–4064. doi: 10.1093/nar/gkt127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Amato J, Madanayake TW, Iaccarino N, Novellino E, Randazzo A, Hurley LH, Pagano B. HMGB1 binds to the KRAS promoter G-quadruplex: a new player in oncogene transcriptional regulation? Chem Commun. 2018;54(68):9442–9445. doi: 10.1039/c8cc03614d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Morgan RK, Batra H, Gaerig VC, Hockings J, Brooks TA. Identification and characterization of a new G-quadruplex forming region within the kRAS promoter as a transcriptional regulator. Biochim Biophys Acta. 2016;1859(2):235–245. doi: 10.1016/j.bbagrm.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 110.Madden SK, de Araujo AD, Gerhardt M, Fairlie DP, Mason JM. Taking the Myc out of cancer: toward therapeutic strategies to directly inhibit c-Myc. Mol Cancer. 2021;20(1):3. doi: 10.1186/s12943-020-01291-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chaudhuri R, Bhattacharya S, Dash J, Bhattacharya S. Recent update on targeting c-MYC G-quadruplexes by small molecules for anticancer therapeutics. J Med Chem. 2021;64(1):42–70. doi: 10.1021/acs.jmedchem.0c01145. [DOI] [PubMed] [Google Scholar]
- 112.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci USA. 2002;99(18):11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sengupta P, Bhattacharya A, Sa G, Das T, Chatterjee S. Truncated G-quadruplex isomers cross-talk with the transcription factors to maintain homeostatic equilibria in c-MYC transcription. Biochemistry. 2019;58(15):1975–1991. doi: 10.1021/acs.biochem.9b00030. [DOI] [PubMed] [Google Scholar]
- 114.Shan C, Yan JW, Wang YQ, Che T, Huang ZL, Chen AC, Yao PF, Tan JH, Li D, Ou TM, Gu LQ, Huang ZS. Design, synthesis, and evaluation of isaindigotone derivatives to downregulate c-myc transcription via disrupting the interaction of NM23-H2 with G-quadruplex. J Med Chem. 2017;60(4):1292–1308. doi: 10.1021/acs.jmedchem.6b01218. [DOI] [PubMed] [Google Scholar]
- 115.Wang YQ, Huang ZL, Chen SB, Wang CX, Shan C, Yin QK, Ou TM, Li D, Gu LQ, Tan JH, Huang ZS. Design, synthesis, and evaluation of new selective NM23-H2 binders as c-MYC transcription inhibitors via disruption of the NM23-H2/G-quadruplex interaction. J Med Chem. 2017;60(16):6924–6941. doi: 10.1021/acs.jmedchem.7b00421. [DOI] [PubMed] [Google Scholar]
- 116.Chen S, Su L, Qiu J, Xiao N, Lin J, Tan JH, Ou TM, Gu LQ, Huang ZS, Li D. Mechanistic studies for the role of cellular nucleic-acid-binding protein (CNBP) in regulation of c-myc transcription. Biochim Biophys Acta. 2013;1830(10):4769–4777. doi: 10.1016/j.bbagen.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 117.Wu R, Li L, Bai Y, Yu B, Xie C, Wu H, Zhang Y, Huang L, Yan Y, Li X, Lin C. The long noncoding RNA LUCAT1 promotes colorectal cancer cell proliferation by antagonizing nucleolin to regulate MYC expression. Cell Death Dis. 2020;11(10):908. doi: 10.1038/s41419-020-03095-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rigo R, Palumbo M, Sissi C. G-quadruplexes in human promoters: a challenge for therapeutic applications. Biochim Biophys Acta Gen Subj. 2017;186(5 Pt B):1399–1413. doi: 10.1016/j.bbagen.2016.12.024. [DOI] [PubMed] [Google Scholar]
- 119.Raiber EA, Kranaster R, Lam E, Nikan M, Balasubramanian S. A non-canonical DNA structure is a binding motif for the transcription factor SP1 in vitro. Nucleic Acids Res. 2012;40(4):1499–1508. doi: 10.1093/nar/gkr882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tsukakoshi K, Saito S, Yoshida W, Goto S, Ikebukuro K. CpG methylation changes G-quadruplex structures derived from gene promoters and interaction with VEGF and SP1. Molecules. 2018 doi: 10.3390/molecules23040944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Uribe DJ, Guo K, Shin YJ, Sun D. Heterogeneous nuclear ribonucleoprotein K and nucleolin as transcriptional activators of the vascular endothelial growth factor promoter through interaction with secondary DNA structures. Biochemistry. 2011;50(18):3796–3806. doi: 10.1021/bi101633b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dutta A, Maji N, Sengupta P, Banerjee N, Kar S, Mukherjee G, Chatterjee S, Basu M. Promoter G-quadruplex favours epigenetic reprogramming-induced atypical expression of ZEB1 in cancer cells. Biochim Biophys Acta Gen Subj. 2021;1865(8):129899. doi: 10.1016/j.bbagen.2021.129899. [DOI] [PubMed] [Google Scholar]
- 123.Sun D, Hurley LH. The importance of negative superhelicity in inducing the formation of G-quadruplex and i-motif structures in the c-Myc promoter: implications for drug targeting and control of gene expression. J Med Chem. 2009;52(9):2863–2874. doi: 10.1021/jm900055s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dickerhoff J, Dai J, Yang D. Structural recognition of the MYC promoter G-quadruplex by a quinoline derivative: insights into molecular targeting of parallel G-quadruplexes. Nucleic Acids Res. 2021;49(10):5905–5915. doi: 10.1093/nar/gkab330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhai Q, Gao C, Ding J, Zhang Y, Islam B, Lan W, Hou H, Deng H, Li J, Hu Z, Mohamed HI, Xu S, Cao C, Haider SM, Wei D. Selective recognition of c-MYC Pu22 G-quadruplex by a fluorescent probe. Nucleic Acids Res. 2019;47(5):2190–2204. doi: 10.1093/nar/gkz059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sun R, Guo X, Yang D, Tang Y, Lu J, Sun H. c-Myc G-quadruplex is sensitively and specifically recognized by a fluorescent probe. Talanta. 2021;226:122125. doi: 10.1016/j.talanta.2021.122125. [DOI] [PubMed] [Google Scholar]
- 127.Deiana M, Chand K, Jamroskovic J, Das RN, Obi I, Chorell E, Sabouri N. A site-specific self-assembled light-up rotor probe for selective recognition and stabilization of c-MYC G-quadruplex DNA. Nanoscale. 2020;12(24):12950–12957. doi: 10.1039/d0nr03404e. [DOI] [PubMed] [Google Scholar]
- 128.Mishra SK, Tawani A, Mishra A, Kumar A. G4IPDB: a database for G-quadruplex structure forming nucleic acid interacting proteins. Sci Rep. 2016;6:38144. doi: 10.1038/srep38144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li Q, Xiang JF, Yang QF, Sun HX, Guan AJ, Tang YL. G4LDB: a database for discovering and studying G-quadruplex ligands. Nucleic Acids Res. 2013;41(Database issue):D1115–1123. doi: 10.1093/nar/gks1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.D’Aria F, Pagano B, Petraccone L, Giancola C. KRAS promoter G-quadruplexes from sequences of different length: a physicochemical study. Int J Mol Sci. 2021 doi: 10.3390/ijms22010448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Del Toro M, Bucek P, Avino A, Jaumot J, Gonzalez C, Eritja R, Gargallo R. Targeting the G-quadruplex-forming region near the P1 promoter in the human BCL-2 gene with the cationic porphyrin TMPyP4 and with the complementary C-rich strand. Biochimie. 2009;91(7):894–902. doi: 10.1016/j.biochi.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 132.Brown RV, Danford FL, Gokhale V, Hurley LH, Brooks TA. Demonstration that drug-targeted down-regulation of MYC in non-Hodgkins lymphoma is directly mediated through the promoter G-quadruplex. J Biol Chem. 2011;286(47):41018–41027. doi: 10.1074/jbc.M111.274720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Montoya JJ, Turnidge MA, Wai DH, Patel AR, Lee DW, Gokhale V, Hurley LH, Arceci RJ, Wetmore C, Azorsa DO. In vitro activity of a G-quadruplex-stabilizing small molecule that synergizes with Navitoclax to induce cytotoxicity in acute myeloid leukemia cells. BMC Cancer. 2019;19(1):1251. doi: 10.1186/s12885-019-6464-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Weldon C, Dacanay JG, Gokhale V, Boddupally PVL, Behm-Ansmant I, Burley GA, Branlant C, Hurley LH, Dominguez C, Eperon IC. Specific G-quadruplex ligands modulate the alternative splicing of Bcl-X. Nucleic Acids Res. 2018;46(2):886–896. doi: 10.1093/nar/gkx1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tian E, Landowski TH, Stephens OW, Yaccoby S, Barlogie B, Shaughnessy JD., Jr Ellipticine derivative NSC 338258 represents a potential new antineoplastic agent for the treatment of multiple myeloma. Mol Cancer Ther. 2008;7(3):500–509. doi: 10.1158/1535-7163.MCT-07-0524. [DOI] [PubMed] [Google Scholar]
- 136.Kumarasamy VM, Sun D. Demonstration of a potent RET transcriptional inhibitor for the treatment of medullary thyroid carcinoma based on an ellipticine derivative. Int J Oncol. 2017;51(1):145–157. doi: 10.3892/ijo.2017.3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hu MH, Wu TY, Huang Q, Jin G. New substituted quinoxalines inhibit triple-negative breast cancer by specifically downregulating the c-MYC transcription. Nucleic Acids Res. 2019;47(20):10529–10542. doi: 10.1093/nar/gkz835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kumar R, Chand K, Bhowmik S, Das RN, Bhattacharjee S, Hedenstrom M, Chorell E. Subtle structural alterations in G-quadruplex DNA regulate site specificity of fluorescence light-up probes. Nucleic Acids Res. 2020;48(3):1108–1119. doi: 10.1093/nar/gkz1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wu TY, Huang Q, Huang ZS, Hu MH, Tan JH. A drug-like imidazole-benzothiazole conjugate inhibits malignant melanoma by stabilizing the c-MYC G-quadruplex. Bioorg Chem. 2020;99:103866. doi: 10.1016/j.bioorg.2020.103866. [DOI] [PubMed] [Google Scholar]
- 140.Calabrese DR, Chen X, Leon EC, Gaikwad SM, Phyo Z, Hewitt WM, Alden S, Hilimire TA, He F, Michalowski AM, Simmons JK, Saunders LB, Zhang S, Connors D, Walters KJ, Mock BA, Schneekloth JS., Jr Chemical and structural studies provide a mechanistic basis for recognition of the MYC G-quadruplex. Nat Commun. 2018;9(1):4229. doi: 10.1038/s41467-018-06315-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ou TM, Lin J, Lu YJ, Hou JQ, Tan JH, Chen SH, Li Z, Li YP, Li D, Gu LQ, Huang ZS. Inhibition of cell proliferation by quindoline derivative (SYUIQ-05) through its preferential interaction with c-myc promoter G-quadruplex. J Med Chem. 2011;54(16):5671–5679. doi: 10.1021/jm200062u. [DOI] [PubMed] [Google Scholar]
- 142.Liu HY, Chen AC, Yin QK, Li Z, Huang SM, Du G, He JH, Zan LP, Wang SK, Xu YH, Tan JH, Ou TM, Li D, Gu LQ, Huang ZS. New disubstituted quindoline derivatives inhibiting Burkitt’s lymphoma cell proliferation by impeding c-MYC transcription. J Med Chem. 2017;60(13):5438–5454. doi: 10.1021/acs.jmedchem.7b00099. [DOI] [PubMed] [Google Scholar]
- 143.Hu MH, Wang YQ, Yu ZY, Hu LN, Ou TM, Chen SB, Huang ZS, Tan JH. Discovery of a new four-leaf clover-like ligand as a potent c-MYC transcription inhibitor specifically targeting the promoter G-quadruplex. J Med Chem. 2018;61(6):2447–2459. doi: 10.1021/acs.jmedchem.7b01697. [DOI] [PubMed] [Google Scholar]
- 144.Drygin D, Siddiqui-Jain A, O’Brien S, Schwaebe M, Lin A, Bliesath J, Ho CB, Proffitt C, Trent K, Whitten JP, Lim JK, Von Hoff D, Anderes K, Rice WG. Anticancer activity of CX-3543: a direct inhibitor of rRNA biogenesis. Cancer Res. 2009;69(19):7653–7661. doi: 10.1158/0008-5472.CAN-09-1304. [DOI] [PubMed] [Google Scholar]
- 145.Yao YX, Xu BH, Zhang Y. CX-3543 promotes cell apoptosis through downregulation of CCAT1 in colon cancer cells. Biomed Res Int. 2018;2018:9701957. doi: 10.1155/2018/9701957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Balasubramanian S, Hurley LH, Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat Rev Drug Discov. 2011;10(4):261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sullivan HJ, Chen B, Wu C. Molecular dynamics study on the binding of an anticancer DNA G-quadruplex stabilizer, CX-5461, to human telomeric, c-KIT1, and c-Myc G-quadruplexes and a DNA duplex. J Chem Inf Model. 2020;60(10):5203–5224. doi: 10.1021/acs.jcim.0c00632. [DOI] [PubMed] [Google Scholar]
- 148.Yan S, Xuan J, Brajanovski N, Tancock MRC, Madhamshettiwar PB, Simpson KJ, Ellis S, Kang J, Cullinane C, Sheppard KE, Hannan KM, Hannan RD, Sanij E, Pearson RB, Chan KT. The RNA polymerase I transcription inhibitor CX-5461 cooperates with topoisomerase 1 inhibition by enhancing the DNA damage response in homologous recombination-proficient high-grade serous ovarian cancer. Br J Cancer. 2021;124(3):616–627. doi: 10.1038/s41416-020-01158-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Xu H, Di Antonio M, McKinney S, Mathew V, Ho B, O’Neil NJ, Santos ND, Silvester J, Wei V, Garcia J, Kabeer F, Lai D, Soriano P, Banath J, Chiu DS, Yap D, Le DD, Ye FB, Zhang A, Thu K, Soong J, Lin SC, Tsai AH, Osako T, Algara T, Saunders DN, Wong J, Xian J, Bally MB, Brenton JD, Brown GW, Shah SP, Cescon D, Mak TW, Caldas C, Stirling PC, Hieter P, Balasubramanian S, Aparicio S. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat Commun. 2017;8:14432. doi: 10.1038/ncomms14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bruno PM, Lu M, Dennis KA, Inam H, Moore CJ, Sheehe J, Elledge SJ, Hemann MT, Pritchard JR. The primary mechanism of cytotoxicity of the chemotherapeutic agent CX-5461 is topoisomerase II poisoning. Proc Natl Acad Sci USA. 2020;117(8):4053–4060. doi: 10.1073/pnas.1921649117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Masud T, Soong C, Xu H, Biele J, Bjornson S, McKinney S, Aparicio S. Ubiquitin-mediated DNA damage response is synthetic lethal with G-quadruplex stabilizer CX-5461. Sci Rep. 2021;11(1):9812. doi: 10.1038/s41598-021-88988-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hein N, Cameron DP, Hannan KM, Nguyen NN, Fong CY, Sornkom J, Wall M, Pavy M, Cullinane C, Diesch J, Devlin JR, George AJ, Sanij E, Quin J, Poortinga G, Verbrugge I, Baker A, Drygin D, Harrison SJ, Rozario JD, Powell JA, Pitson SM, Zuber J, Johnstone RW, Dawson MA, Guthridge MA, Wei A, McArthur GA, Pearson RB, Hannan RD. Inhibition of Pol I transcription treats murine and human AML by targeting the leukemia-initiating cell population. Blood. 2017;129(21):2882–2895. doi: 10.1182/blood-2016-05-718171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kusnadi EP, Trigos AS, Cullinane C, Goode DL, Larsson O, Devlin JR, Chan KT, De Souza DP, McConville MJ, McArthur GA, Thomas G, Sanij E, Poortinga G, Hannan RD, Hannan KM, Kang J, Pearson RB. Reprogrammed mRNA translation drives resistance to therapeutic targeting of ribosome biogenesis. EMBO J. 2020;39(21):e105111. doi: 10.15252/embj.2020105111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ismael M, Webb R, Ajaz M, Kirkby KJ, Coley HM. The targeting of RNA polymerase I transcription using CX-5461 in combination with radiation enhances tumour cell killing effects in human solid cancers. Cancers. 2019 doi: 10.3390/cancers11101429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sanij E, Hannan KM, Xuan J, Yan S, Ahern JE, Trigos AS, Brajanovski N, Son J, Chan KT, Kondrashova O, Lieschke E, Wakefield MJ, Frank D, Ellis S, Cullinane C, Kang J, Poortinga G, Nag P, Deans AJ, Khanna KK, Mileshkin L, McArthur GA, Soong J, Berns E, Hannan RD, Scott CL, Sheppard KE, Pearson RB. CX-5461 activates the DNA damage response and demonstrates therapeutic efficacy in high-grade serous ovarian cancer. Nat Commun. 2020;11(1):2641. doi: 10.1038/s41467-020-16393-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Makhale A, Nanayakkara D, Raninga P, Khanna KK, Kalimutho M. CX-5461 enhances the efficacy of APR-246 via induction of DNA damage and replication stress in triple-negative breast cancer. Int J Mol Sci. 2021 doi: 10.3390/ijms22115782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Xu X, Feng H, Dai C, Lu W, Zhang J, Guo X, Yin Q, Wang J, Cui X, Jiang F. Therapeutic efficacy of the novel selective RNA polymerase I inhibitor CX-5461 on pulmonary arterial hypertension and associated vascular remodelling. Br J Pharmacol. 2021;178(7):1605–1619. doi: 10.1111/bph.15385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Khot A, Brajanovski N, Cameron DP, Hein N, Maclachlan KH, Sanij E, Lim J, Soong J, Link E, Blombery P, Thompson ER, Fellowes A, Sheppard KE, McArthur GA, Pearson RB, Hannan RD, Poortinga G, Harrison SJ. First-in-human RNA polymerase I transcription inhibitor CX-5461 in patients with advanced hematologic cancers: results of a phase I dose-escalation study. Cancer Discov. 2019;9(8):1036–1049. doi: 10.1158/2159-8290.CD-18-1455. [DOI] [PubMed] [Google Scholar]
- 159.Cheng F, Carroll L, Joglekar MV, Januszewski AS, Wong KK, Hardikar AA, Jenkins AJ, Ma RCW. Diabetes, metabolic disease, and telomere length. Lancet Diabetes Endocrinol. 2021;9(2):117–126. doi: 10.1016/S2213-8587(20)30365-X. [DOI] [PubMed] [Google Scholar]
- 160.Rhodes D, Lipps HJ. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015;43(18):8627–8637. doi: 10.1093/nar/gkv862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kosiol N, Juranek S, Brossart P, Heine A, Paeschke K. G-quadruplexes: a promising target for cancer therapy. Mol Cancer. 2021;20(1):40. doi: 10.1186/s12943-021-01328-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhai LT, Rety S, Chen WF, Song ZY, Auguin D, Sun B, Dou SX, Xi XG. Crystal structures of N-terminally truncated telomerase reverse transcriptase from fungidouble dagger. Nucleic Acids Res. 2021;49(8):4768–4781. doi: 10.1093/nar/gkab261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zahler AM, Williamson JR, Cech TR, Prescott DM. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350(6320):718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 164.Oganesian L, Moon IK, Bryan TM, Jarstfer MB. Extension of G-quadruplex DNA by ciliate telomerase. EMBO J. 2006;25(5):1148–1159. doi: 10.1038/sj.emboj.7601006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Moye AL, Porter KC, Cohen SB, Phan T, Zyner KG, Sasaki N, Lovrecz GO, Beck JL, Bryan TM. Telomeric G-quadruplexes are a substrate and site of localization for human telomerase. Nat Commun. 2015;6:7643. doi: 10.1038/ncomms8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Paudel BP, Moye AL, Abou Assi H, El-Khoury R, Cohen SB, Holien JK, Birrento ML, Samosorn S, Intharapichai K, Tomlinson CG, Teulade-Fichou MP, Gonzalez C, Beck JL, Damha MJ, van Oijen AM, Bryan TM. A mechanism for the extension and unfolding of parallel telomeric G-quadruplexes by human telomerase at single-molecule resolution. Elife. 2020 doi: 10.7554/eLife.56428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chaires JB, Gray RD, Dean WL, Monsen R, DeLeeuw LW, Stribinskis V, Trent JO. Human POT1 unfolds G-quadruplexes by conformational selection. Nucleic Acids Res. 2020;48(9):4976–4991. doi: 10.1093/nar/gkaa202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Xu M, Axhemi A, Malgowska M, Chen Y, Leonard D, Srinivasan S, Jankowsky E, Taylor DJ. Active and passive destabilization of G-quadruplex DNA by the telomere POT1-TPP1 complex. J Mol Biol. 2021;433(7):166846. doi: 10.1016/j.jmb.2021.166846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ray S, Qureshi MH, Malcolm DW, Budhathoki JB, Celik U, Balci H. RPA-mediated unfolding of systematically varying G-quadruplex structures. Biophys J. 2013;104(10):2235–2245. doi: 10.1016/j.bpj.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Fernandes CAH, Morea EGO, Dos Santos GA, da Silva VL, Vieira MR, Viviescas MA, Chatain J, Vadel A, Saintome C, Fontes MRM, Cano MIN. A multi-approach analysis highlights the relevance of RPA-1 as a telomere end-binding protein (TEBP) in Leishmania amazonensis. Biochim Biophys Acta Gen Subj. 2020;1864(7):129607. doi: 10.1016/j.bbagen.2020.129607. [DOI] [PubMed] [Google Scholar]
- 171.Guterres AN, Villanueva J. Targeting telomerase for cancer therapy. Oncogene. 2020;39(36):5811–5824. doi: 10.1038/s41388-020-01405-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Shin-ya K, Wierzba K, Matsuo K, Ohtani T, Yamada Y, Furihata K, Hayakawa Y, Seto H. Telomestatin, a novel telomerase inhibitor from Streptomyces anulatus. J Am Chem Soc. 2001;123(6):1262–1263. doi: 10.1021/ja005780q. [DOI] [PubMed] [Google Scholar]
- 173.Kim MY, Vankayalapati H, Shin-Ya K, Wierzba K, Hurley LH. Telomestatin, a potent telomerase inhibitor that interacts quite specifically with the human telomeric intramolecular G-quadruplex. J Am Chem Soc. 2002;124(10):2098–2099. doi: 10.1021/ja017308q. [DOI] [PubMed] [Google Scholar]
- 174.Kim MY, Gleason-Guzman M, Izbicka E, Nishioka D, Hurley LH. The different biological effects of telomestatin and TMPyP4 can be attributed to their selectivity for interaction with intramolecular or intermolecular G-quadruplex structures. Cancer Res. 2003;63(12):3247–3256. [PubMed] [Google Scholar]
- 175.Nakajima A, Tauchi T, Sashida G, Sumi M, Abe K, Yamamoto K, Ohyashiki JH, Ohyashiki K. Telomerase inhibition enhances apoptosis in human acute leukemia cells: possibility of antitelomerase therapy. Leukemia. 2003;17(3):560–567. doi: 10.1038/sj.leu.2402825. [DOI] [PubMed] [Google Scholar]
- 176.Shammas MA, Reis RJS, Li C, Koley H, Hurley LH, Anderson KC, Munshi NC. Telomerase inhibition and cell growth arrest after telomestatin treatment in multiple myeloma. Clin Cancer Res. 2004;10(2):770–776. doi: 10.1158/1078-0432.ccr-0793-03. [DOI] [PubMed] [Google Scholar]
- 177.Hasegawa D, Okabe S, Okamoto K, Nakano I, Shin-ya K, Seimiya H. G-quadruplex ligand-induced DNA damage response coupled with telomere dysfunction and replication stress in glioma stem cells. Biochem Biophys Res Commun. 2016;471(1):75–81. doi: 10.1016/j.bbrc.2016.01.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Miyazaki T, Pan Y, Joshi K, Purohit D, Hu B, Demir H, Mazumder S, Okabe S, Yamori T, Viapiano M, Shin-ya K, Seimiya H, Nakano I. Telomestatin impairs glioma stem cell survival and growth through the disruption of telomeric G-quadruplex and inhibition of the proto-oncogene, c-Myb. Clin Cancer Res. 2012;18(5):1268–1280. doi: 10.1158/1078-0432.CCR-11-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Nakamura T, Okabe S, Yoshida H, Iida K, Ma Y, Sasaki S, Yamori T, Shin-Ya K, Nakano I, Nagasawa K, Seimiya H. Targeting glioma stem cells in vivo by a G-quadruplex-stabilizing synthetic macrocyclic hexaoxazole. Sci Rep. 2017;7(1):3605. doi: 10.1038/s41598-017-03785-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yasuda M, Ma Y, Okabe S, Wakabayashi Y, Su D, Chang YT, Seimiya H, Tera M, Nagasawa K. Target identification of a macrocyclic hexaoxazole G-quadruplex ligand using post-target-binding visualization. Chem Commun. 2020;56(85):12905–12908. doi: 10.1039/d0cc04957c. [DOI] [PubMed] [Google Scholar]
- 181.Ma Y, Iida K, Sasaki S, Hirokawa T, Heddi B, Phan AT, Nagasawa K. Synthesis and telomeric G-quadruplex-stabilizing ability of macrocyclic hexaoxazoles bearing three side chains. Molecules. 2019 doi: 10.3390/molecules24020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Yu-Ru X, Fang-Yuan T, Xiao-Zhen T, Hong-Ting Z, Yong X. Anti-inflammatory activities of berberine in the treatment of metabolic disorders by regulating the gut microbiota. Prog Biochem Biophys. 2020;47(08):835–843. doi: 10.16476/j.pibb.2020.0137. [DOI] [Google Scholar]
- 183.Bazzicalupi C, Ferraroni M, Bilia AR, Scheggi F, Gratteri P. The crystal structure of human telomeric DNA complexed with berberine: an interesting case of stacked ligand to G-tetrad ratio higher than 1:1. Nucleic Acids Res. 2013;41(1):632–638. doi: 10.1093/nar/gks1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kumarasamy VM, Shin YJ, White J, Sun D. Selective repression of RET proto-oncogene in medullary thyroid carcinoma by a natural alkaloid berberine. BMC Cancer. 2015;15:599. doi: 10.1186/s12885-015-1610-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ciszewski L, Lu-Nguyen N, Slater A, Brennan A, Williams HEL, Dickson G, Searle MS, Popplewell L. G-quadruplex ligands mediate downregulation of DUX4 expression. Nucleic Acids Res. 2020;48(8):4179–4194. doi: 10.1093/nar/gkaa146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Xiong YX, Su HF, Lv P, Ma Y, Wang SK, Miao H, Liu HY, Tan JH, Ou TM, Gu LQ, Huang ZS. A newly identified berberine derivative induces cancer cell senescence by stabilizing endogenous G-quadruplexes and sparking a DNA damage response at the telomere region. Oncotarget. 2015;6(34):35625–35635. doi: 10.18632/oncotarget.5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Liao TC, Ma TZ, Chen SB, Cilibrizzi A, Zhang MJ, Li JH, Zhou CQ. Human telomere double G-quadruplex recognition by berberine-bisquinolinium imaging conjugates in vitro and cells. Int J Biol Macromol. 2020 doi: 10.1016/j.ijbiomac.2020.04.171. [DOI] [PubMed] [Google Scholar]
- 188.Lin C, Wu G, Wang K, Onel B, Sakai S, Shao Y, Yang D. Molecular recognition of the hybrid-2 human telomeric G-quadruplex by epiberberine: insights into conversion of telomeric G-quadruplex structures. Angew Chem. 2018;57(34):10888–10893. doi: 10.1002/anie.201804667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Asamitsu S, Obata S, Yu Z, Bando T, Sugiyama H. Recent progress of targeted G-quadruplex-preferred ligands toward cancer therapy. Molecules. 2019 doi: 10.3390/molecules24030429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Burger AM, Dai F, Schultes CM, Reszka AP, Moore MJ, Double JA, Neidle S. The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res. 2005;65(4):1489–1496. doi: 10.1158/0008-5472.CAN-04-2910. [DOI] [PubMed] [Google Scholar]
- 191.Gowan SM, Harrison JR, Patterson L, Valenti M, Read MA, Neidle S, Kelland LR. A G-quadruplex-interactive potent small-molecule inhibitor of telomerase exhibiting in vitro and in vivo antitumor activity. Mol Pharmacol. 2002;61(5):1154–1162. doi: 10.1124/mol.61.5.1154. [DOI] [PubMed] [Google Scholar]
- 192.Grand CL, Han H, Munoz RM, Weitman S, Von Hoff DD, Hurley LH, Bearss DJ. The cationic porphyrin TMPyP4 down-regulates c-MYC and human telomerase reverse transcriptase expression and inhibits tumor growth in vivo. Mol Cancer Ther. 2002;1(8):565–573. [PubMed] [Google Scholar]
- 193.Li G, Shen J, Cao J, Zhou G, Lei T, Sun Y, Gao H, Ding Y, Xu W, Zhan Z, Chen Y, Huang H. Alternative splicing of human telomerase reverse transcriptase in gliomas and its modulation mediated by CX-5461. J Exp Clin Cancer Res. 2018;37(1):78. doi: 10.1186/s13046-018-0749-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Berardinelli F, Tanori M, Muoio D, Buccarelli M, di Masi A, Leone S, Ricci-Vitiani L, Pallini R, Mancuso M, Antoccia A. G-quadruplex ligand RHPS4 radiosensitizes glioblastoma xenograft in vivo through a differential targeting of bulky differentiated- and stem-cancer cells. J Exp Clin Cancer Res. 2019;38(1):311. doi: 10.1186/s13046-019-1293-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Fleming AM, Burrows CJ. On the irrelevancy of hydroxyl radical to DNA damage from oxidative stress and implications for epigenetics. Chem Soc Rev. 2020;49(18):6524–6528. doi: 10.1039/d0cs00579g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ngoi NY, Liew AQ, Chong SJF, Davids MS, Clement MV, Pervaiz S. The redox-senescence axis and its therapeutic targeting. Redox Biol. 2021;45:102032. doi: 10.1016/j.redox.2021.102032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Cogoi S, Ferino A, Miglietta G, Pedersen EB, Xodo LE. The regulatory G4 motif of the Kirsten ras (KRAS) gene is sensitive to guanine oxidation: implications on transcription. Nucleic Acids Res. 2018;46(2):661–676. doi: 10.1093/nar/gkx1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Fleming AM, Ding Y, Burrows CJ. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc Natl Acad Sci USA. 2017;114(10):2604–2609. doi: 10.1073/pnas.1619809114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Edwards AD, Marecki JC, Byrd AK, Gao J, Raney KD. G-quadruplex loops regulate PARP-1 enzymatic activation. Nucleic Acids Res. 2021;49(1):416–431. doi: 10.1093/nar/gkaa1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Cinque G, Ferino A, Pedersen EB, Xodo LE. Role of poly [ADP-ribose] polymerase 1 in activating the Kirsten ras (KRAS) gene in response to oxidative stress. Int J Mol Sci. 2020 doi: 10.3390/ijms21176237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Roychoudhury S, Pramanik S, Harris HL, Tarpley M, Sarkar A, Spagnol G, Sorgen PL, Chowdhury D, Band V, Klinkebiel D, Bhakat KK. Endogenous oxidized DNA bases and APE1 regulate the formation of G-quadruplex structures in the genome. Proc Natl Acad Sci USA. 2020;117(21):11409–11420. doi: 10.1073/pnas.1912355117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ferino A, Xodo LE. Effect of DNA glycosylases OGG1 and Neil1 on oxidized G-rich motif in the KRAS promoter. Int J Mol Sci. 2021 doi: 10.3390/ijms22031137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Jara-Espejo M, Line SR. DNA G-quadruplex stability, position and chromatin accessibility are associated with CpG island methylation. FEBS J. 2020;287(3):483–495. doi: 10.1111/febs.15065. [DOI] [PubMed] [Google Scholar]
- 204.Varshney D, Spiegel J, Zyner K, Tannahill D, Balasubramanian S. The regulation and functions of DNA and RNA G-quadruplexes. Nat Rev Mol Cell Biol. 2020;21(8):459–474. doi: 10.1038/s41580-020-0236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Li J, He G, Mu C, Wang K, Xiang Y. Assay of DNA methyltransferase 1 activity based on uracil-specific excision reagent digestion induced G-quadruplex formation. Anal Chim Acta. 2017;986:131–137. doi: 10.1016/j.aca.2017.07.021. [DOI] [PubMed] [Google Scholar]
- 206.Cree SL, Fredericks R, Miller A, Pearce FG, Filichev V, Fee C, Kennedy MA. DNA G-quadruplexes show strong interaction with DNA methyltransferases in vitro. FEBS Lett. 2016;590(17):2870–2883. doi: 10.1002/1873-3468.12331. [DOI] [PubMed] [Google Scholar]
- 207.Mao SQ, Ghanbarian AT, Spiegel J, Martinez Cuesta S, Beraldi D, Di Antonio M, Marsico G, Hansel-Hertsch R, Tannahill D, Balasubramanian S. DNA G-quadruplex structures mold the DNA methylome. Nat Struct Mol Biol. 2018;25(10):951–957. doi: 10.1038/s41594-018-0131-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Saha D, Singh A, Hussain T, Srivastava V, Sengupta S, Kar A, Dhapola P, Dhople V, Ummanni R, Chowdhury S. Epigenetic suppression of human telomerase (hTERT) is mediated by the metastasis suppressor NME2 in a G-quadruplex-dependent fashion. J Biol Chem. 2017;292(37):15205–15215. doi: 10.1074/jbc.M117.792077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Purohit G, Mukherjee AK, Sharma S, Chowdhury S. Extratelomeric binding of the telomere binding protein TRF2 at the PCGF3 promoter is G-quadruplex motif-dependent. Biochemistry. 2018;57(16):2317–2324. doi: 10.1021/acs.biochem.8b00019. [DOI] [PubMed] [Google Scholar]
- 210.Hussain T, Saha D, Purohit G, Kar A, Kishore Mukherjee A, Sharma S, Sengupta S, Dhapola P, Maji B, Vedagopuram S, Horikoshi NT, Horikoshi N, Pandita RK, Bhattacharya S, Bajaj A, Riou JF, Pandita TK, Chowdhury S. Transcription regulation of CDKN1A (p21/CIP1/WAF1) by TRF2 is epigenetically controlled through the REST repressor complex. Sci Rep. 2017;7(1):11541. doi: 10.1038/s41598-017-11177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Spiegel J, Adhikari S, Balasubramanian S. The structure and function of DNA G-quadruplexes. Trends Chem. 2020;2(2):123–136. doi: 10.1016/j.trechm.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Sarkies P, Reams C, Simpson LJ, Sale JE. Epigenetic instability due to defective replication of structured DNA. Mol Cell. 2010;40(5):703–713. doi: 10.1016/j.molcel.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Schiavone D, Guilbaud G, Murat P, Papadopoulou C, Sarkies P, Prioleau MN, Balasubramanian S, Sale JE. Determinants of G quadruplex-induced epigenetic instability in REV1-deficient cells. EMBO J. 2014;33(21):2507–2520. doi: 10.15252/embj.201488398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Guilbaud G, Murat P, Recolin B, Campbell BC, Maiter A, Sale JE, Balasubramanian S. Local epigenetic reprogramming induced by G-quadruplex ligands. Nat Chem. 2017;9(11):1110–1117. doi: 10.1038/nchem.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Sarkies P, Murat P, Phillips LG, Patel KJ, Balasubramanian S, Sale JE. FANCJ coordinates two pathways that maintain epigenetic stability at G-quadruplex DNA. Nucleic Acids Res. 2012;40(4):1485–1498. doi: 10.1093/nar/gkr868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Papadopoulou C, Guilbaud G, Schiavone D, Sale JE. Nucleotide pool depletion induces G-quadruplex-dependent perturbation of gene expression. Cell Rep. 2015;13(11):2491–2503. doi: 10.1016/j.celrep.2015.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Dai YX, Chen WF, Liu NN, Teng FY, Guo HL, Hou XM, Dou SX, Rety S, Xi XG. Structural and functional studies of SF1B Pif1 from Thermus oshimai reveal dimerization-induced helicase inhibition. Nucleic Acids Res. 2021;49(7):4129–4143. doi: 10.1093/nar/gkab188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Li S, Wang H, Jehi S, Li J, Liu S, Wang Z, Truong L, Chiba T, Wang Z, Wu X. PIF1 helicase promotes break-induced replication in mammalian cells. EMBO J. 2021;40(8):e104509. doi: 10.15252/embj.2020104509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Boule JB, Vega LR, Zakian VA. The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature. 2005;438(7064):57–61. doi: 10.1038/nature04091. [DOI] [PubMed] [Google Scholar]
- 220.Wang J, Zhu X, Ying P, Zhu Y. PIF1 affects the proliferation and apoptosis of cervical cancer cells by influencing TERT. Cancer Manag Res. 2020;12:7827–7835. doi: 10.2147/CMAR.S265336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Li JR, Yu TY, Chien IC, Lu CY, Lin JJ, Li HW. Pif1 regulates telomere length by preferentially removing telomerase from long telomere ends. Nucleic Acids Res. 2014;42(13):8527–8536. doi: 10.1093/nar/gku541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Nickens DG, Rogers CM, Bochman ML. The Saccharomyces cerevisiae Hrq1 and Pif1 DNA helicases synergistically modulate telomerase activity in vitro. J Biol Chem. 2018;293(37):14481–14496. doi: 10.1074/jbc.RA118.004092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Wang YR, Guo TT, Zheng YT, Lai CW, Sun B, Xi XG, Hou XM. Replication protein A plays multifaceted roles complementary to specialized helicases in processing G-quadruplex DNA. iScience. 2021;24(5):102493. doi: 10.1016/j.isci.2021.102493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Obi I, Rentoft M, Singh V, Jamroskovic J, Chand K, Chorell E, Westerlund F, Sabouri N. Stabilization of G-quadruplex DNA structures in Schizosaccharomyces pombe causes single-strand DNA lesions and impedes DNA replication. Nucleic Acids Res. 2020;48(19):10998–11015. doi: 10.1093/nar/gkaa820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Lopes J, Piazza A, Bermejo R, Kriegsman B, Colosio A, Teulade-Fichou MP, Foiani M, Nicolas A. G-quadruplex-induced instability during leading-strand replication. EMBO J. 2011;30(19):4033–4046. doi: 10.1038/emboj.2011.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Dahan D, Tsirkas I, Dovrat D, Sparks MA, Singh SP, Galletto R, Aharoni A. Pif1 is essential for efficient replisome progression through lagging strand G-quadruplex DNA secondary structures. Nucleic Acids Res. 2018;46(22):11847–11857. doi: 10.1093/nar/gky1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Sanders CM. Human Pif1 helicase is a G-quadruplex DNA-binding protein with G-quadruplex DNA-unwinding activity. Biochem J. 2010;430(1):119–128. doi: 10.1042/BJ20100612. [DOI] [PubMed] [Google Scholar]
- 228.Zhou R, Zhang J, Bochman ML, Zakian VA, Ha T. Periodic DNA patrolling underlies diverse functions of Pif1 on R-loops and G-rich DNA. Elife. 2014;3:e02190. doi: 10.7554/eLife.02190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Hou XM, Wu WQ, Duan XL, Liu NN, Li HH, Fu J, Dou SX, Li M, Xi XG. Molecular mechanism of G-quadruplex unwinding helicase: sequential and repetitive unfolding of G-quadruplex by Pif1 helicase. Biochem J. 2015;466(1):189–199. doi: 10.1042/BJ20140997. [DOI] [PubMed] [Google Scholar]
- 230.Wang L, Wang QM, Wang YR, Xi XG, Hou XM. DNA-unwinding activity of Saccharomyces cerevisiae Pif1 is modulated by thermal stability, folding conformation, and loop lengths of G-quadruplex DNA. J Biol Chem. 2018;293(48):18504–18513. doi: 10.1074/jbc.RA118.005071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Byrd AK, Bell MR, Raney KD. Pif1 helicase unfolding of G-quadruplex DNA is highly dependent on sequence and reaction conditions. J Biol Chem. 2018;293(46):17792–17802. doi: 10.1074/jbc.RA118.004499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Odermatt DC, Lee WTC, Wild S, Jozwiakowski SK, Rothenberg E, Gari K. Cancer-associated mutations in the iron-sulfur domain of FANCJ affect G-quadruplex metabolism. PLoS Genet. 2020;16(6):e1008740. doi: 10.1371/journal.pgen.1008740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Calvo JA, Fritchman B, Hernandez D, Persky NS, Johannessen CM, Piccioni F, Kelch BA, Cantor SB. Comprehensive mutational analysis of the BRCA1-associated DNA helicase and tumor-suppressor FANCJ/BACH1/BRIP1. Mol Cancer Res. 2021;19(6):1015–1025. doi: 10.1158/1541-7786.MCR-20-0828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Summers PA, Lewis BW, Gonzalez-Garcia J, Porreca RM, Lim AHM, Cadinu P, Martin-Pintado N, Mann DJ, Edel JB, Vannier JB, Kuimova MK, Vilar R. Visualising G-quadruplex DNA dynamics in live cells by fluorescence lifetime imaging microscopy. Nat Commun. 2021;12(1):162. doi: 10.1038/s41467-020-20414-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Kruisselbrink E, Guryev V, Brouwer K, Pontier DB, Cuppen E, Tijsterman M. Mutagenic capacity of endogenous G4 DNA underlies genome instability in FANCJ-defective C. elegans. Curr Biol. 2008;18(12):900–905. doi: 10.1016/j.cub.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 236.Bharti SK, Sommers JA, George F, Kuper J, Hamon F, Shin-ya K, Teulade-Fichou MP, Kisker C, Brosh RM., Jr Specialization among iron-sulfur cluster helicases to resolve G-quadruplex DNA structures that threaten genomic stability. J Biol Chem. 2013;288(39):28217–28229. doi: 10.1074/jbc.M113.496463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Lu H, Davis AJ. Human RecQ helicases in DNA double-strand break repair. Front Cell Dev Biol. 2021;9:640755. doi: 10.3389/fcell.2021.640755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Wu WQ, Hou XM, Li M, Dou SX, Xi XG. BLM unfolds G-quadruplexes in different structural environments through different mechanisms. Nucleic Acids Res. 2015;43(9):4614–4626. doi: 10.1093/nar/gkv361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Voter AF, Qiu Y, Tippana R, Myong S, Keck JL. A guanine-flipping and sequestration mechanism for G-quadruplex unwinding by RecQ helicases. Nat Commun. 2018;9(1):4201. doi: 10.1038/s41467-018-06751-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Teng FY, Jiang ZZ, Huang LY, Guo M, Chen F, Hou XM, Xi XG, Xu Y. A toolbox for site-specific labeling of RecQ helicase with a single fluorophore used in the single-molecule assay. Front Mol Biosci. 2020;7(253):586450. doi: 10.3389/fmolb.2020.586450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Cahoon LA, Manthei KA, Rotman E, Keck JL, Seifert HS. Neisseria gonorrhoeae RecQ helicase HRDC domains are essential for efficient binding and unwinding of the pilE guanine quartet structure required for pilin antigenic variation. J Bacteriol. 2013;195(10):2255–2261. doi: 10.1128/JB.02217-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Chatterjee S, Zagelbaum J, Savitsky P, Sturzenegger A, Huttner D, Janscak P, Hickson ID, Gileadi O, Rothenberg E. Mechanistic insight into the interaction of BLM helicase with intra-strand G-quadruplex structures. Nat Commun. 2014;5(1):5556. doi: 10.1038/ncomms6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Teng FY, Wang TT, Guo HL, Xin BG, Sun B, Dou SX, Xi XG, Hou XM. The HRDC domain oppositely modulates the unwinding activity of E. coli RecQ helicase on duplex DNA and G-quadruplex. J Biol Chem. 2020;295(51):17646–17658. doi: 10.1074/jbc.RA120.015492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Budhathoki JB, Maleki P, Roy WA, Janscak P, Yodh JG, Balci H. A comparative study of G-quadruplex unfolding and DNA reeling activities of human RECQ5 helicase. Biophys J. 2016;110(12):2585–2596. doi: 10.1016/j.bpj.2016.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Heddi B, Cheong VV, Schmitt E, Mechulam Y, Phan AT. Recognition of different base tetrads by RHAU (DHX36): X-ray crystal structure of the G4 recognition motif bound to the 3′-end tetrad of a DNA G-quadruplex. J Struct Biol. 2020;209(1):107399. doi: 10.1016/j.jsb.2019.10.001. [DOI] [PubMed] [Google Scholar]
- 246.Schult P, Paeschke K. The DEAH helicase DHX36 and its role in G-quadruplex-dependent processes. Biol Chem. 2021;402(5):581–591. doi: 10.1515/hsz-2020-0292. [DOI] [PubMed] [Google Scholar]
- 247.Huang W, Smaldino PJ, Zhang Q, Miller LD, Cao P, Stadelman K, Wan M, Giri B, Lei M, Nagamine Y, Vaughn JP, Akman SA, Sui G. Yin Yang 1 contains G-quadruplex structures in its promoter and 5′-UTR and its expression is modulated by G4 resolvase 1. Nucleic Acids Res. 2012;40(3):1033–1049. doi: 10.1093/nar/gkr849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Booy EP, Howard R, Marushchak O, Ariyo EO, Meier M, Novakowski SK, Deo SR, Dzananovic E, Stetefeld J, McKenna SA. The RNA helicase RHAU (DHX36) suppresses expression of the transcription factor PITX1. Nucleic Acids Res. 2014;42(5):3346–3361. doi: 10.1093/nar/gkt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Zeng Y, Qin T, Flamini V, Tan C, Zhang X, Cong Y, Birkin E, Jiang WG, Yao H, Cui Y. Identification of DHX36 as a tumour suppressor through modulating the activities of the stress-associated proteins and cyclin-dependent kinases in breast cancer. Am J Cancer Res. 2020;10(12):4211–4233. [PMC free article] [PubMed] [Google Scholar]
- 250.Chen WF, Rety S, Guo HL, Dai YX, Wu WQ, Liu NN, Auguin D, Liu QW, Hou XM, Dou SX, Xi XG. Molecular mechanistic insights into drosophila DHX36-mediated G-quadruplex unfolding: a structure-based model. Structure. 2018;26(3):403–415.e4. doi: 10.1016/j.str.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 251.Chen MC, Tippana R, Demeshkina NA, Murat P, Balasubramanian S, Myong S, Ferre-D’Amare AR. Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36. Nature. 2018;558(7710):465–469. doi: 10.1038/s41586-018-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Liu Q, Wang Q, Lv C, Liu Z, Gao H, Chen Y, Zhao G. Brucine inhibits proliferation of glioblastoma cells by targeting the G-quadruplexes in the c-Myb promoter. J Cancer. 2021;12(7):1990–1999. doi: 10.7150/jca.53689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Monsen RC, DeLeeuw L, Dean WL, Gray RD, Sabo TM, Chakravarthy S, Chaires JB, Trent JO. The hTERT core promoter forms three parallel G-quadruplexes. Nucleic Acids Res. 2020;48(10):5720–5734. doi: 10.1093/nar/gkaa107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Ceschi S, Largy E, Gabelica V, Sissi C. A two-quartet G-quadruplex topology of human KIT2 is conformationally selected by a perylene derivative. Biochimie. 2020;179:77–84. doi: 10.1016/j.biochi.2020.09.015. [DOI] [PubMed] [Google Scholar]
- 255.Bilgen E, Cetinkol OP. Doxorubicin exhibits strong and selective association with VEGF Pu22 G-quadruplex. Biochim Biophys Acta Gen Subj. 2020;1864(12):129720. doi: 10.1016/j.bbagen.2020.129720. [DOI] [PubMed] [Google Scholar]
- 256.Moccia F, Riccardi C, Musumeci D, Leone S, Oliva R, Petraccone L, Montesarchio D. Insights into the G-rich VEGF-binding aptamer V7t1: when two G-quadruplexes are better than one! Nucleic Acids Res. 2019;47(15):8318–8331. doi: 10.1093/nar/gkz589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.De Armond R, Wood S, Sun D, Hurley LH, Ebbinghaus SW. Evidence for the presence of a guanine quadruplex forming region within a polypurine tract of the hypoxia inducible factor 1alpha promoter. Biochemistry. 2005;44(49):16341–16350. doi: 10.1021/bi051618u. [DOI] [PubMed] [Google Scholar]
- 258.Dhakal S, Yu Z, Konik R, Cui Y, Koirala D, Mao H. G-quadruplex and i-motif are mutually exclusive in ILPR double-stranded DNA. Biophys J . 2012;102(11):2575–2584. doi: 10.1016/j.bpj.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Zamiri B, Mirceta M, Bomsztyk K, Macgregor RB, Jr, Pearson CE. Quadruplex formation by both G-rich and C-rich DNA strands of the C9orf72 (GGGGCC)8*(GGCCCC)8 repeat: effect of CpG methylation. Nucleic Acids Res. 2015;43(20):10055–10064. doi: 10.1093/nar/gkv1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Fumagalli L, Young FL, Boeynaems S, De Decker M, Mehta AR, Swijsen A, Fazal R, Guo W, Moisse M, Beckers J, Dedeene L, Selvaraj BT, Vandoorne T, Madan V, van Blitterswijk M, Raitcheva D, McCampbell A, Poesen K, Gitler AD, Koch P, Berghe PV, Thal DR, Verfaillie C, Chandran S, Van Den Bosch L, Bullock SL, Van Damme P. C9orf72-derived arginine-containing dipeptide repeats associate with axonal transport machinery and impede microtubule-based motility. Sci Adv. 2021 doi: 10.1126/sciadv.abg3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Yan J, Zhao X, Liu B, Yuan Y, Guan Y. An intramolecular G-quadruplex structure formed in the human MET promoter region and its biological relevance. Mol Carcinog. 2016;55(5):897–909. doi: 10.1002/mc.22330. [DOI] [PubMed] [Google Scholar]
- 262.Ji N, Shi HQ, Fang XY, Wu ZY. Exploring the interaction of G-quadruplex and porphyrin derivative by single protein nanopore sensing interface. Anal Chim Acta. 2020;1106:126–132. doi: 10.1016/j.aca.2020.01.053. [DOI] [PubMed] [Google Scholar]
- 263.Konieczna N, Romaniuk-Drapala A, Lisiak N, Toton E, Paszel-Jaworska A, Kaczmarek M, Rubis B. Telomerase inhibitor TMPyP4 alters adhesion and migration of breast-cancer cells MCF7 and MDA-MB-231. Int J Mol Sci. 2019 doi: 10.3390/ijms20112670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Mikami-Terao Y, Akiyama M, Yuza Y, Yanagisawa T, Yamada O, Kawano T, Agawa M, Ida H, Yamada H. Antitumor activity of TMPyP4 interacting G-quadruplex in retinoblastoma cell lines. Exp Eye Res. 2009;89(2):200–208. doi: 10.1016/j.exer.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 265.Rapozzi V, Zorzet S, Zacchigna M, Della Pietra E, Cogoi S, Xodo LE. Anticancer activity of cationic porphyrins in melanoma tumour-bearing mice and mechanistic in vitro studies. Mol Cancer. 2014;13:75. doi: 10.1186/1476-4598-13-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Moruno-Manchon JF, Koellhoffer EC, Gopakumar J, Hambarde S, Kim N, McCullough LD, Tsvetkov AS. The G-quadruplex DNA stabilizing drug pyridostatin promotes DNA damage and downregulates transcription of Brca1 in neurons. Aging. 2017;9(9):1957–1970. doi: 10.18632/aging.101282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Rodriguez R, Miller KM, Forment JV, Bradshaw CR, Nikan M, Britton S, Oelschlaegel T, Xhemalce B, Balasubramanian S, Jackson SP. Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat Chem Biol. 2012;8(3):301–310. doi: 10.1038/nchembio.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Paeschke K, Bochman ML, Garcia PD, Cejka P, Friedman KL, Kowalczykowski SC, Zakian VA. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013;497(7450):458–462. doi: 10.1038/nature12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Chai W, Zheng L, Shen B. DNA2, a new player in telomere maintenance and tumor suppression. Cell Cycle. 2013;12(13):1985–1986. doi: 10.4161/cc.25306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Saha T, Shukla K, Thakur RS, Desingu A, Nagaraju G. Mycobacterium tuberculosis UvrD1 and UvrD2 helicases unwind G-quadruplex DNA. FEBS J. 2019;286(11):2062–2086. doi: 10.1111/febs.14798. [DOI] [PubMed] [Google Scholar]
- 271.Shukla K, Thakur RS, Ganguli D, Rao DN, Nagaraju G. Escherichia coli and Neisseria gonorrhoeae UvrD helicase unwinds G4 DNA structures. Biochem J. 2017;474(21):3579–3597. doi: 10.1042/BCJ20170587. [DOI] [PubMed] [Google Scholar]
- 272.Paul T, Voter AF, Cueny RR, Gavrilov M, Ha T, Keck JL, Myong S. E. coli Rep helicase and RecA recombinase unwind G4 DNA and are important for resistance to G4-stabilizing ligands. Nucleic Acids Res. 2020;48(12):6640–6653. doi: 10.1093/nar/gkaa442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Xue ZY, Wu WQ, Zhao XC, Kumar A, Ran X, Zhang XH, Zhang Y, Guo LJ. Single-molecule probing the duplex and G4 unwinding patterns of a RecD family helicase. Int J Biol Macromol. 2020;164:902–910. doi: 10.1016/j.ijbiomac.2020.07.158. [DOI] [PubMed] [Google Scholar]
- 274.Wu CG, Spies M. G-quadruplex recognition and remodeling by the FANCJ helicase. Nucleic Acids Res. 2016;44(18):8742–8753. doi: 10.1093/nar/gkw574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Cali F, Bharti SK, Di Perna R, Brosh RM, Jr, Pisani FM. Tim/Timeless, a member of the replication fork protection complex, operates with the Warsaw breakage syndrome DNA helicase DDX11 in the same fork recovery pathway. Nucleic Acids Res. 2016;44(2):705–717. doi: 10.1093/nar/gkv1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Lerner LK, Holzer S, Kilkenny ML, Svikovic S, Murat P, Schiavone D, Eldridge CB, Bittleston A, Maman JD, Branzei D, Stott K, Pellegrini L, Sale JE. Timeless couples G-quadruplex detection with processing by DDX11 helicase during DNA replication. EMBO J. 2020;39(18):e104185. doi: 10.15252/embj.2019104185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Kotsantis P, Segura-Bayona S, Margalef P, Marzec P, Ruis P, Hewitt G, Bellelli R, Patel H, Goldstone R, Poetsch AR, Boulton SJ. RTEL1 regulates G4/R-loops to avert replication-transcription collisions. Cell Rep. 2020;33(12):108546. doi: 10.1016/j.celrep.2020.108546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Wu W, Bhowmick R, Vogel I, Ozer O, Ghisays F, Thakur RS, Sanchez de Leon E, Richter PH, Ren L, Petrini JH, Hickson ID, Liu Y. RTEL1 suppresses G-quadruplex-associated R-loops at difficult-to-replicate loci in the human genome. Nat Struct Mol Biol. 2020;27(5):424–437. doi: 10.1038/s41594-020-0408-6. [DOI] [PubMed] [Google Scholar]
- 279.Zhu M, Wu W, Togashi Y, Liang W, Miyoshi Y, Ohta T. HERC2 inactivation abrogates nucleolar localization of RecQ helicases BLM and WRN. Sci Rep. 2021;11(1):360. doi: 10.1038/s41598-020-79715-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Wu WQ, Hou XM, Zhang B, Fosse P, Rene B, Mauffret O, Li M, Dou SX, Xi XG. Single-molecule studies reveal reciprocating of WRN helicase core along ssDNA during DNA unwinding. Sci Rep. 2017;7:43954. doi: 10.1038/srep43954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Chakraborty P, Grosse F. Human DHX9 helicase preferentially unwinds RNA-containing displacement loops (R-loops) and G-quadruplexes. DNA Repair. 2011;10(6):654–665. doi: 10.1016/j.dnarep.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 282.Tuesuwan B, Kern JT, Thomas PW, Rodriguez M, Li J, David WM, Kerwin SM. Simian virus 40 large T-antigen G-quadruplex DNA helicase inhibition by G-quadruplex DNA-interactive agents. Biochemistry. 2008;47(7):1896–1909. doi: 10.1021/bi701747d. [DOI] [PubMed] [Google Scholar]
- 283.Plyler J, Jasheway K, Tuesuwan B, Karr J, Brennan JS, Kerwin SM, David WM. Real-time investigation of SV40 large T-antigen helicase activity using surface plasmon resonance. Cell Biochem Biophys. 2009;53(1):43–52. doi: 10.1007/s12013-008-9038-z. [DOI] [PubMed] [Google Scholar]
- 284.Bharti SK, Sommers JA, Zhou J, Kaplan DL, Spelbrink JN, Mergny JL, Brosh RM., Jr DNA sequences proximal to human mitochondrial DNA deletion breakpoints prevalent in human disease form G-quadruplexes, a class of DNA structures inefficiently unwound by the mitochondrial replicative Twinkle helicase. J Biol Chem. 2014;289(43):29975–29993. doi: 10.1074/jbc.M114.567073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.