Abstract
Astronauts on board the International Space Station (ISS) are exposed to the damaging effects of microgravity and cosmic radiation. One of the most critical and sensitive districts of an organism is the eye, particularly the retina, and > 50% of astronauts develop a complex of alterations designated as spaceflight-associated neuro-ocular syndrome. However, the pathogenesis of this condition is not clearly understood. In the current study, we aimed to explore the cellular and molecular effects induced in the human retinal pigment ARPE-19 cell line by their transfer to and 3-day stay on board the ISS in the context of an experiment funded by the Agenzia Spaziale Italiana. Treatment of cells on board the ISS with the well-known bioenergetic, antioxidant, and antiapoptotic coenzyme Q10 was also evaluated. In the ground control experiment, the cells were exposed to the same conditions as on the ISS, with the exception of microgravity and radiation. The transfer of ARPE-19 retinal cells to the ISS and their living on board for 3 days did not affect cell viability or apoptosis but induced cytoskeleton remodeling consisting of vimentin redistribution from the cellular boundaries to the perinuclear area, underlining the collapse of the network of intermediate vimentin filaments under unloading conditions. The morphological changes endured by ARPE-19 cells grown on board the ISS were associated with changes in the transcriptomic profile related to the cellular response to the space environment and were consistent with cell dysfunction adaptations. In addition, the results obtained from ARPE-19 cells treated with coenzyme Q10 indicated its potential to increase cell resistance to damage.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-021-03989-2.
Keywords: Retina, Microgravity, Radiation, Retinopathy, Space flight
Introduction
Space radiation and microgravity present in spacecraft and space stations in orbit around the Earth cause several time-dependent health alterations in astronauts both during their missions and after their return to Earth. The most well-known are fluid shifts, loss of bone and muscle mass, cardiovascular imbalances, alterations in immunity, sleep interruption, and circadian misalignment, which have been described for more than two decades in a multitude of scientific reports and reviews [1–6]. At the cellular level, the main pathological effects of microgravity and cosmic radiation are structural alterations caused by free radical-mediated molecular damage [7–10].
Space exploration has now entered a new phase in which NASA, ESA, and other national space agencies are working together to plan long-term space missions in order, for example, to create lunar bases and reach other space destinations such as Mars. At this stage, assessing the risk inherent in space missions and challenging the numerous obstacles to astronaut safety are increasingly important in mission planning. In particular, since the microgravity and ionizing radiation inevitably present in the spacecraft environment represent two serious stress factors for astronaut health, the discovery of pharmacological countermeasures is clearly a mandatory prerequisite for mission planning [11, 12]. A basic principle of pharmacological research is that the most valid rational basis for identifying tools capable of blocking or inhibiting a pathological process is knowledge of its pathogenic mechanisms. This principle applies to the range of damage and health impairments known to be endured by astronauts during their lengthy space missions.
The eye, particularly the retina, is severely sensitive to radiation and microgravity damage, which eventually cause apoptotic cell death [9, 10, 13–18]. Approximately 10 years ago, NASA reported that nearly 60% of 300 astronauts returning to Earth after long-term stay aboard the International Space Station (ISS) were affected by several neuro-ophthalmic alterations, including optic disc edema, globe flattening, choroidal and retinal folds, hyperopic refractive error shifts, nerve fiber layer infarcts, and visual acuity reduction [19], comprehensively termed spaceflight associated neuro-ocular syndrome (SANS) [20–23]. The pathogenesis of SANS is not fully understood. The relevance of astronauts’ visual impairment led NASA to consider vision as one of the top health risks for long-term space flight [24].
The ISS, where astronauts are exposed to the space environment for several months [25], offers a great opportunity to directly study astronaut health impairments, but also to execute experiments aimed at unraveling the pathogenesis of damage even at the molecular level and to test the efficacy of potential countermeasures to minimize damage and therefore maximize the feasibility of space missions.
In this paper, we aimed to explore the impact exerted on both the cell structure and on the transcriptomic profile by the transfer of human retinal pigment epithelial ARPE-19 cells from the Earth to the ISS and their subsequent culture on board for 3 days. The effects of coenzyme Q10 (CoQ10) treatment, by virtue of its well-known antioxidant and anti-apoptotic abilities [26–29], are also reported. The results presented here were obtained in the context of the project “The Coenzyme Q10 (CoQ10) as Countermeasure for Retinal Damage on board the International Space Station: the CORM Project”, funded by the Agenzia Spaziale Italiana (ASI) and belonging to a group of investigations of the ESA VITA (Vitality, Innovation, Technology, Ability) mission, which have been included in the ISS expedition 52/53. To optimize the conditions for the experiment aboard the ISS, we previously carried out on-ground experiments revealing that a 3-day stay in simulated microgravity (using a rotating wall vessel bioreactor) led ARPE-19 cells to undergo apoptosis, while X-rays induced a dose-dependent DNA damage accompanied by a DNA damage response at telomeres, referred to as telomere dysfunction-induced foci accumulation [26].
Material and methods
Cell culture
Human retinal pigment epithelial ARPE-19 cells (ATCC, Manassas, VA, USA) were cultured in 50% Dulbecco’s modified Eagle’s medium (DMEM, Lonza, Basel, CH) and 50% Ham’s F12 Medium (Lonza) supplemented with 10% fetal bovine serum (FBS; # 35–015-CV; Corning, NY, USA), 100 U/mL penicillin–streptomycin (Lonza), and 2 mM L-glutamine (Lonza) in a humidified incubator at 37 °C and 5% CO2. ARPE-19 cells were frozen and shipped to the Kennedy Space Center (KSC) Space Station Processing Facility (SSPF) laboratories (Cape Canaveral, Florida, USA) months before the scheduled launch date. Two weeks before launch, the cells were thawed and cultured in the KSC laboratories. For the experiment on board the ISS, ARPE-19 cells were seeded at a density of 20,000 cells/cm2 on cell culture supports, i.e., cyclic olefin copolymer (COC) ibiTreat plastic coverslips (IBIDI, Martinsried, Germany), which allow cell adhesion that is comparable to standard cell culture flasks, flexibility, and good optical performance. Dedicated experimental units (EUs, KEU-ST, Kayser Italia, Livorno, Italy), which are electromechanical devices capable of executing cell culture protocols and are qualified for flights to the ISS, were used for the experiment on board the ISS (Fig. 1A–B). EUs contain five reservoirs: three were loaded with cell culture media (50% DMEM, 50% Ham’s F12, 10% FBS, 100 U/mL Penicillin–Streptomycin, 2 mM L-Glutamine, 25 mM Hepes [Lonza]) in the presence or absence of 10 µM CoQ10 (Sigma-Aldrich, St. Louis, USA) dissolved in a commercial vehicle to ensure cellular uptake (0.04% Kolliphor P407 micro, Sigma-Aldrich); one with Dulbecco’s phosphate-buffered saline (DPBS, added with Ca2 + /Mg2 + , Lonza) for cell rinse prior to fixation; one with RNAlater (Ambion AM7020, MA, USA) as a fixative solution. At day 1 after seeding, cell cultures on COC plastic coverslips were transferred into the cell culture chambers of the EUs, which were then introduced into Experiment Containers KIC-SL to compose the experimental hardware (EH). EHs were inserted into a passive temperature-controlled transportation container, named Biokit (Kayser Italia), containing phase-change materials preheated at 27 °C. An iButton data logger was placed on an EH to record the temperature every minute until the end of the experiment (the recorded temperature profile is shown in Fig. 1C). Three EHs were transferred to flight authorities (two containing cells not treated with CoQ10, and one containing cells treated with CoQ10) on day 2 after cell seeding. On August 14, 2017, the hardware was launched on a Dragon/Falcon 9 vector in the framework of the Space X CRS-12 mission on day 3 after seeding; it reached the ISS on day 5. On board the ISS, astronaut Paolo Nespoli manually inserted the EHs into the ESA KUBIK incubator set at 37 °C to power the EHs. Fluidic activations were automatically performed, allowing media replacement on days 6, 7, and 8 after seeding, and rinsing and fixation on day 9. In particular, fixation with RNAlater occurred 5 min after rinsing with DPBS saline solution. In total, the EHs spent 72 h inside the KUBIK facility. The samples were then manually transferred into the Minus Eighty Laboratory Freezer for ISS (MELFI) refrigerator by astronaut Paolo Nespoli and stored at -80 °C until their return to Earth on September 16, 2017. The temperature profile was downloaded from the data logger, allowing the reproduction of the same time/temperature profile as that of the ground control experiments. Once on the ground, the samples were thawed and retrieved from the EUs. COC plastic coverslips containing cells were sectioned with a scalpel while they were continuously submerged in RNAlater. Five sections were obtained from each sample: one was subjected to apoptosis evaluation through TUNEL assay, one to vimentin immunofluorescence analysis, and three to transcriptomic analysis. Images of cells were acquired by optical microscopy using an EVOS XL Core Imaging System (Thermo Fisher Scientific, USA).
Fig. 1.
A The hardware of CORM experiment: the experimental units (EUs) were integrated into experimental containers to compose the experiment hardware (EH), which were placed inside the Biokit transportation container that was launched to the International Space Station (ISS). Once onboard ISS, the EH were inserted into the ESA KUBIK incubator in the ISS Columbus module. B Schematic fluidic of the EU. C iButton data logger-derived temperature profile of the CORM experiment
Tunel
The DeadEnd Fluorometric TUNEL System kit (Promega, USA) was used to detect DNA breaks according to the manufacturer's protocol. Briefly, samples were fixed in 4% formaldehyde solution in PBS for 25 min at + 4 °C and washed twice with PBS. The cells were then permeabilized with 0.2% Triton X‐100 in PBS for 5 min. After washing with PBS, the samples were incubated in equilibration buffer from the staining kit for 10 min and then treated with staining reaction mix for 1 h at 37 °C in a humidified chamber protected from light. The reaction was arrested by incubation for 15 min in 2 × SSC. After washing with PBS, cells were stained with Hoechst 33,242 dye (40,6-diamidino-2-phenylindole; Thermo Fisher Scientific) for 10 min and mounted with ProLong Diamond Antifade Mountant (Thermo Fisher Scientific). For a positive staining control, the slides were treated after permeabilization with DNase I (10 units/mL) for 10 min. The samples were analyzed using a confocal microscope (Nikon TE2000 using EZ-C1 Software; Nikon Corp.) equipped with a 60XA/1.40 oil-immersion objective and digitally captured.
Immunofluorescence
Cells grown on plastic coverslips and treated as previously described were fixed for 5 min with ice-cold acetone. Unspecific binding sites were blocked with PBS containing 3% bovine serum albumin (BSA; Sigma-Aldrich) for 1 h at 22 ± 2 °C. The cells were then incubated overnight at 4 °C with anti-vimentin antibody (#MAB1681, Chemicon, Merck KGaA, Darmstadt, Germany) diluted 1:100 in PBS with 0.5% BSA. After washing three times with PBS-0.5% BSA, samples were incubated for 1 h at 22 ± 2 °C in the dark with fluorescein isothiocyanate-conjugated secondary antibody (anti-mouse IgM [#AP128F, Chemicon] diluted 1:200). The samples were washed three times, mounted on glass slides using Fluoromount™ aqueous mounting medium (Sigma-Aldrich), evaluated using an epifluorescence microscope (Nikon, Firenze, Italy) at 100 × magnification, and imaged using a HiRes IV digital CCD camera (DTA, Pisa, Italy).
RNA isolation
RNA extraction was performed using the RNAqueous Total RNA Isolation kit (Ambion AM1912) following the manufacturer’s instructions, and 40 µL RNA was eluted. The samples were vacuum-concentrated using DNA 120 Speedvac® (ThermoSavant, USA) with low vacuum and no heating to avoid RNA fragmentation, and the samples were manually checked every 5 min until the volume reached 20 μL. Residual genomic DNA was removed using a DNA-free DNA Removal kit (Ambion AM1906) following the manufacturer’s instructions. Isolated RNA and library preparation products were quantified using Qubit 3.0 (Invitrogen, USA) and quality was assessed by capillary electrophoresis using an AATI Fragment Analyzer (Advanced Analytical Technologies, Inc., USA).
Gene expression analysis
RNA expression was quantified at Polo d’Innovazione di Genomica, Genetica e Biologia (Polo GGB, Siena, Italy) using RNAseq analysis. Total ribo-depleted cDNA libraries were prepared using SMARTer Stranded Total RNA-Seq Kit Pico Input Mammalian (Takara Bio USA, Inc.) according to the manufacturer’s instructions. Indexed DNA libraries were normalized to a concentration of 4 nM and then pooled in equal volumes to obtain a uniform read distribution. The pool was denatured and diluted to a final loading concentration of 1.4 pM according to the Illumina protocol, with the addition of 1% spike in PhiX Control v3 (Illumina, USA) as a sequencing control. To obtain a minimum of 20 million reads per sample, the pool was sequenced using a NextSeq 550 sequencer (Illumina) using a NextSeq 500/550 Mid Output v2 kit (150 cycles) (Illumina) to perform a paired-end run 2X75 bp read length.
Raw reads for each sample (three treated vs. three untreated replicates) were aligned to the human_g1k_v37 release of the GRCh37/hg19 human reference genome using the RNA-seq aligner STAR [30] (version 2.5.2b) with the default parameter settings. Count-based differential expression analysis was performed using featureCounts [31] and DeSEQ2 [32]. In detail, we counted the reads mapped to each genomic meta-feature (that is, each gene) with featureCounts (version 1.5.1), using the default parameter settings and the Ensembl Gene Transfer Format (GTF) file for the appropriate reference genome (Ensembl release 82). The count matrix from featureCounts (where each row represents an Ensembl gene, each column is a sequenced RNA library, and the values indicate the raw numbers of fragments that were uniquely assigned to the respective gene in each library) was subsequently converted into a DESeqDataSet object inside the R statistical environment. The downstream differential expression analysis in the DESeq2 R package (release 3.3) uses a generalized linear model of the form Kij ~ NB(µij,αi), where K counts (from the count matrix) for gene i and sample j are modeled using a negative binomial (NB) distribution with mean µij and gene-specific dispersion αi, which defines the relationship between the variance of the observed count and its mean value. The estimation of the dispersion values for each gene as well as the fitting of the NB distribution were performed using the DESeq function, and the log fold change (LFC) estimates, p values, and adjusted p values (Wald statistic, treated vs. untreated) were extracted using the results function using the default parameter settings. To generate even more accurate LFC estimates, we eventually calculated shrunken LFC estimates (lfcShrink function, apeglm algorithm): LFC shrinkage uses LFC estimates for all genes to shrink toward the LFC estimates of genes with little information (low counts) or high dispersion, facilitating the downstream assessment of results.
Pathway analysis
The data were analyzed using the iPathwayGuide platform in the context of pathways obtained from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Release 84.0 + /10–26, Oct 17), gene ontologies from the Gene Ontology (GO) Consortium database (2017-Nov6), miRNAs from the miRBase (Release 21), and TARGETSCAN (Targetscan version: Mouse:7.1, Human:7.1) databases. iPathwayGuide scores pathways using the impact analysis method [33], which uses two types of evidence: (i) the over-representation of differentially expressed (DE) genes in a given pathway and (ii) the perturbation of that pathway computed by propagating the measured expression changes across the pathway topology. These aspects are captured by two independent probability values, pORA and pAcc, which are then combined in a unique pathway-specific p value. pORA expresses the probability of observing the number of DE genes in a given pathway that is equal to or greater than that observed by random chance. pAcc was calculated based on the amount of total gene accumulation measured in each pathway. Once the accumulations of all gene number perturbations are computed, iPathwayGuide computes the total accumulation of the pathway as the sum of all absolute accumulations of the genes in a given pathway. The two types of evidence, pORA and pAcc, are combined into one final pathway score by calculating a p value using Fisher's method, which is then rectified for multiple comparisons using false discovery rate (FDR) or Bonferroni correction. Bonferroni correction, which is simpler and more conservative than FDR, reduces the false discovery rate by imposing a stringent threshold on each comparison adjusted for the total number of comparisons. FDR correction has more power, but only controls the family-wise false positive rate.
Results
As previously described and shown in Fig. 1, the CORM experiment was set up in the NASA KSC SSPF laboratory (Cape Canaveral, Florida, USA), where ARPE-19 cells (treated with CoQ10 or not) were seeded in a dedicated EU, which was then introduced into the EC to compose the EH [26] (Fig. 1A). The EH was included in the passive temperature-controlled transport container Biokit, which was launched to the ISS in the context of SpaceX CRS-12. Three EHs were transferred to the ISS: two containing cells not treated with CoQ10, and one containing cells treated with CoQ10. Biokit's transfer to the ISS lasted 3 days, during which the cells underwent a constant decrease in temperature up to approximately 26 °C. Once on board the ISS, the EHs were moved to the ESA KUBIK (incubator facility), set at 37 °C, allowing the automatic execution of fluidic activations according to the programmed schedule (Fig. 1B). After 3 days, the cells were fixed in RNAlater and the EHs were stored at − 80 °C in the MELFI, where they were kept until splashdown. Once in our laboratories, under freezing conditions, the EH underwent final disassembly, and the samples were inspected. Ground ARPE-19 cell cultures were also prepared as controls under normal gravity using a similar EH, following the temperature/time profile of the in-flight experiment (Fig. 1C).
Transferring ARPE-19 cells to the ISS and culturing on board for 3 days did not affect cell proliferation or apoptosis, but severely modified vimentin organization
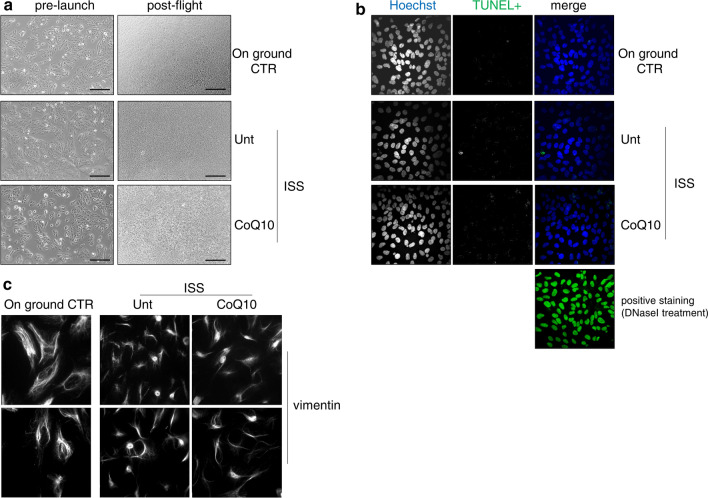
Some key cellular parameters that we first evaluated after the flight are shown in Fig. 2. The transfer of ARPE-19 cells (treated or not treated with CoQ10) to the ISS and the 3 days of incubation on board had no impact on their proliferation rate compared to ground controls. In both cases, the cells almost reached confluence and did not show any signs of damage (Fig. 2A) or apoptosis, as assessed by the TUNEL assay (Fig. 2B). However, as indicated in the representative images of immunofluorescence microscopy analysis in Fig. 2C, the organization of the vimentin network underwent profound alterations, including a marked increase in its localization in the cellular perinuclear area and a concomitant redistribution from the cellular borders, which emphasized the collapse of the vimentin intermediate filament network.
Fig. 2.
A Representative photomicrographs obtained with phase-contrast microscopy in ARPE-19 cells pre-launch and post-flight. Scale bar: 200 μm. B Evaluation of apoptosis in ARPE-19 cells by TUNEL fluorescent staining. Image size: 250 × 250 µm. C Evaluation of vimentin cellular localization in ARPE-19 cells by immunofluorescence. Image size: 100 × 100 µm
Transferring ARPE-19 cells to the ISS and culturing on board for 3 days dramatically modified the transcriptome profile
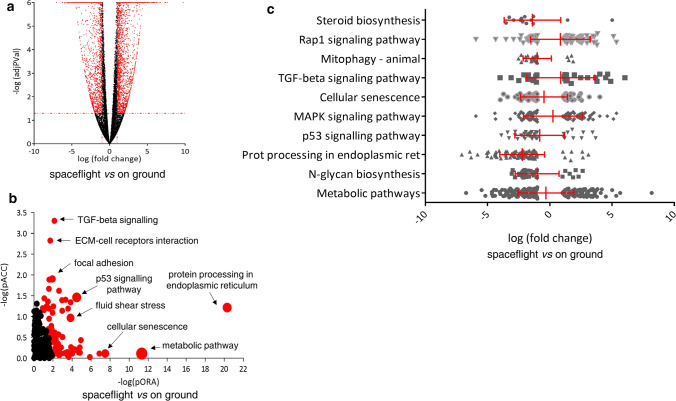
We then explored the possibility that spaceflight alters gene expression by analyzing ARPE-19 transcriptome profiles with respect to ground controls using next-generation sequencing technology, assuming a threshold of 0.05, for statistical significance (p value) and a change in the expression of a log fold with an absolute value of at least 1. Of 23,556 genes analyzed, 5555 were DE after spaceflight with respect to the ground controls (Fig. 3A and Supplementary S1). Among them, 3081 were upregulated and 2474 were downregulated. To predict the impact of the ISS environment on ARPE-19 gene expression pathways, we used the iPathwayGuide to assess the possibility that a specific pathway was perturbed and that the subsequent genes in a pathway were significantly more perturbed than the previous ones. The results predicted that 99 pathways defined in the KEGG database were significantly affected (Fig. 3B and Supplementary S2). The first ten pathways are shown in Table 1, and pathway diagrams containing coherent cascades and DE pathway genes are presented in Supplementary S3A-I. The most significantly impacted pathways were related to cellular responses to space environment adaptation/damage. The analysis of DE genes associated with perturbed pathways revealed down-modulation of metabolic pathways, N-Glycan biosynthesis, protein processing in the endoplasmic reticulum, p53 signaling, cellular senescence, mitophagy, and steroid biosynthesis, and up-modulation of MAPK, TGF-beta, and Rap1 signaling (Fig. 3C). To further explore the functional roles of the DE genes, we performed GO analysis (Fig. 4 and Supplementary S4), which demonstrated that the transfer of ARPE-19 cells to the ISS and their culturing for 3 days on board caused an alteration in response to unfolded protein, stress response of the endoplasmic reticulum, and ion binding. The first five GO terms in biological processes, molecular functions, and cellular components are listed in Table 2.
Fig. 3.
Gene expression analysis of ARPE-19 cells cultured on board the International Space Station compared to on ground. A The 5555 significantly differentially expressed (DE) genes (shown in red) are represented in the Volcano plot in terms of their measured expression change (x-axis) and the significance of the change (y-axis). The significance is represented in terms of the negative log (base 10) of the p value, such that genes that are more significant are plotted higher on the y-axis. The dotted lines represent the thresholds used to select the DE genes: 1 for expression change and 0.05 for significance. B Pathways perturbation vs over-representation: the pathways are plotted in terms of the two types of evidence computed by iPathwayGuide: over-representation is on the x-axis (pORA) and the total pathway accumulation is on the y-axis (pAcc). Each pathway is represented by a single dot, with significant pathways shown in red, non-significant in black, and the size of each dot is proportional to the size of the pathway it represents. Both p values are shown in terms of their negative log (base 10) values. C DE pathway genes of the most significant impacted pathways. The mean value and standard deviation are indicated in red
Table 1.
Most significantly affected pathways in ARPE-19 cells cultured on board ISS compared to on ground
| Pathway name | Pathway ID (KEGG) | p-value | p-value (FRD) | p-value (Bonferroni) |
|---|---|---|---|---|
| Metabolic pathways | 01,100 | 4.716e−12 | 1.504e−9 | 1.504e−9 |
| N-Glycan biosynthesis | 00,510 | 1.374e−7 | 2.192e−5 | 4.385e−5 |
| Protein processing in endoplasmic reticulum | 04,141 | 1.074e−6 | 1.142e−4 | 3.426e−4 |
| p53 signaling pathway | 04,115 | 1.160e−5 | 9.248e−4 | 0.004 |
| MAPK signaling pathway | 04,010 | 1.649e−5 | 9.861e−4 | 0.005 |
| Cellular senescence | 04,218 | 1.855e−5 | 9.861e−4 | 0.006 |
| TGF-beta signaling pathway | 04,350 | 4.917e−5 | 0.002 | 0.016 |
| Mitophagy—animal | 04,137 | 5.734e−5 | 0.002 | 0.018 |
| Rap1 signaling pathway | 04,015 | 8.164e−5 | 0.003 | 0.026 |
| Steroid biosynthesis | 00,100 | 9.600e−5 | 0.003 | 0.031 |
Fig. 4.
Gene Ontology analysis of differentially expressed genes in ARPE-19 cells cultured on board the International Space Station compared to on ground. The top ten Gene Ontology (GO) terms in biological processes (green), molecular functions (red), and cellular components (blue) category are listed
Table 2.
GO analysis
| Category | Identifier | Description | Count DE | Count All | p-value | p-value (FDR) | P-value (Bonferroni) |
|---|---|---|---|---|---|---|---|
| Biological Processes | GO:0,034,976 | Response to endoplasmic reticulum stress | 125 | 261 | 1,10e−11 | 1,29e−02 | 1,29e−02 |
| GO:0,034,620 | Cellular response to unfolded protein | 72 | 130 | 7,40e−11 | 4,34e−02 | 8,68e−02 | |
| GO:0,006,986 | Response to unfolded protein | 86 | 167 | 1,80e−10 | 7,04e−03 | 2,11e−06 | |
| GO:0,035,967 | Cellular response to topologically incorrect protein | 77 | 147 | 5,80e−10 | 1,70e−06 | 6,80e−06 | |
| GO:0,030,968 | Endoplasmic reticulum unfolded protein response | 67 | 123 | 8,70e−10 | 2,04e−06 | 1,02e−05 | |
| Molecular Functions | GO:0,043,167 | Ion binding | 1715 | 5615 | 2,50E−06 | 7,34E−03 | 7,34E-03 |
| GO:0,005,509 | Calcium ion binding | 217 | 610 | 4,60E-05 | 1,00E + 00 | 1,00E + 00 | |
| GO:0,004,576 | Oligosaccharyl transferase activity | 9 | 10 | 8,80E−05 | 1,00E + 00 | 1,00E + 00 | |
| GO:0,003,777 | Microtubule motor activity | 39 | 81 | 1,20E−04 | 1,00E + 00 | 1,00E + 00 | |
| GO:0,043,168 | Anion binding | 784 | 2509 | 2,60E−04 | 1,00E + 00 | 1,00E + 00 | |
| Cellular Components | GO:0,005,783 | Endoplasmic reticulum | 616 | 1615 | 2,40E−20 | 3,33e−13 | 3,33e−13 |
| GO:0,044,432 | Endoplasmic reticulum part | 463 | 1173 | 2,10E−18 | 1,46e−10 | 2,92e−11 | |
| GO:0,098,827 | Endoplasmic reticulum Subcompartment | 372 | 931 | 7,60E−16 | 3,16e−07 | 1,06e−07 | |
| GO:0,031,984 | Organelle subcompartment | 554 | 1489 | 9,10E−16 | 3,16e−07 | 1,26e−07 | |
| GO:0,005,789 | Endoplasmic reticulum membrane | 370 | 928 | 1,30E−15 | 3,61e−09 | 1,81e−08 |
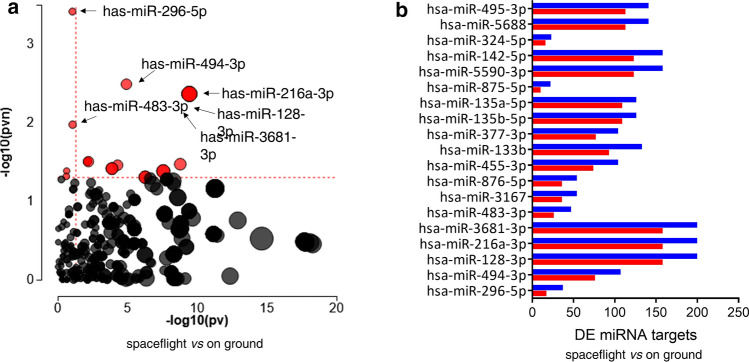
The presence of lncRNAs was screened among the DE genes, based on the lncRNAs noted on the HUGO Gene Nomenclature Committee website, which included a total of 4118 lncRNAs (http://www.genenames.org/cgi-bin/statistics) (Supplementary S5). The expression of 255 lncRNAs was deregulated in ARPE-19 cells cultured on board the ISS; 208 were upregulated and 47 were downregulated (the top ten up- and down-regulated lncRNAs are presented in Table 3). To predict active micro-RNAs (miRNAs) in ARPE-19 cells cultured on board the ISS compared to ground control, the DE genes were also analyzed to determine the presence of enriched DE targets of the miRNAs. For a given miRNA, the analysis computed the ratio between the number of DE targets and all differentially expressed targets and compared it with the ratio of all targets expressed downwards with all targets. This allowed for the calculation of the probability of observing a greater number of differentially downregulated target genes for a given miRNA only by chance. Of a total of 366 screened miRNAs, 19 achieved significant expression values, as described in Fig. 5 and Supplementary S6, with hsa-miR-296-5p, hsa-miR-494-3p, and hsa-miR-128-3p showing the best p values.
Table 3.
Top ten up- and downregulated lncRNAs in ARPE-19 cultured on board ISS
| ID symbol | lncRNA name | Chromosome location RefSeq accession |
Description | Expression (log fc) |
|---|---|---|---|---|
| HGNC:44,153 | DIO2-AS1 |
14q31.1 |
DIO2 antisense RNA 1 | 6,3 |
| HGNC:27,121 | LINC00906 |
19q12 |
Long intergenic non-protein coding RNA 906 | 5,4 |
| HGNC:45,193 | PLCE1-AS1 |
10q23.33 |
PLCE1 antisense RNA 1 | 5,2 |
| HGNC:27,437 | WDR11-AS1 |
10q26.12 |
WDR11 antisense RNA 1 | 5,1 |
| HGNC:48,727 | LINC00968 |
8q12.1 |
Long intergenic non-protein coding RNA 968 | 4,7 |
| HGNC:42,692 | LINC00370 |
13q33.3 |
Long intergenic non-protein coding RNA 370 | 4,6 |
| HGNC:27,471 | TRHDE-AS1 |
12q21.1 |
TRHDE antisense RNA 1 | 4,6 |
| HGNC:40,587 | TM4SF1-AS1 |
3q25.1 |
TM4SF1 antisense RNA 1 | 4,5 |
| HGNC:49,095 | CLSTN2-AS1 |
3q23 |
CLSTN2 antisense RNA 1 | 4,4 |
| HGNC:44,989 | LINC00842 |
10q11.22 |
Long intergenic non-protein coding RNA 842 | 4,4 |
| HGNC:44,064 | SLC7A11-AS1 |
4q28.3 |
SLC7A11 antisense RNA 1 non-coding RNA | − 3,1 |
| HGNC:40,792 | ARHGAP26-AS1 |
5q31.3 |
ARHGAP26 antisense RNA 1 | − 3,1 |
| HGNC:48,575 | LINC00888 |
3q27.1 |
Long intergenic non-protein coding RNA 888 | − 3,1 |
| HGNC:41,429 | ARHGAP26-IT1 |
5q31.3 |
ARHGAP26 intronic transcript 1 non-coding RNA | − 3,3 |
| HGNC:17,263 | PART1 |
5q12.1 AF16347 |
Prostate te androgen-regulated transcript 1 | − 3,4 |
| HGNC:48,498 | LUCAT1 |
5q14.3 |
Lung ancer associated transcript 1 | − 3,5 |
| HGNC:48,574 | LINC00887 |
3q29 |
Long intergenic non-protein coding RNA 887 | − 3,9 |
| HGNC:23,135 | RN7SL3 |
14q21.3 |
RNA component of signal recognition particle 7SL3 | − 4,6 |
| HGNC:4713 | H19 |
11p15.5 |
H19 imprinted maternally expressed transcript | − 4,6 |
| HGNC:42,812 | MRPL23-AS1 |
11p15.5 |
MRPL23 antisense RNA 1 | − 5,8 |
Fig. 5.
Bioinformatics miRNA analysis in ARPE-19 cells cultured on board the International Space Station compared to on ground. A Significant differences in the expression of specific miRNAs plotted in terms of the two types of evidence computed by iPathwayGuide: the x-axis (− log10(pv)) represents the p value based on the total number of differentially expressed (DE) target genes versus the total number of target genes and the y-axis (− log10(pvn)) is the p value based on the number of downregulated DE targets versus the total number of DE miRNA targets. Each miRNA is represented by a single dot, with significant miRNAs shown in red, non-significant in black, and the size of each dot is proportional to the size of the number of target genes for that miRNA relative to others. B List of the 19 miRNAs that reached significance, depicting the number of DE target genes that are under- (in blue) or over-expressed (in red)
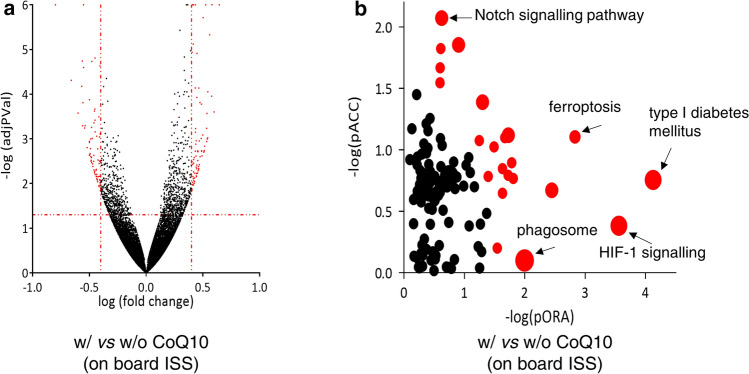
We then evaluated the role of CoQ10 treatment on gene expression in ARPE-19 cells cultured on board the ISS. Assuming a threshold of 0.05 for p and of 0.4 for fold change, of the 26,694 genes analyzed, we found 153 DE genes in cells treated with CoQ10 with respect to untreated controls (Fig. 6A and Supplementary S7). Among these DE genes, 81 were upregulated and 72 were downregulated in spaceflight ARPE-19 cells. iPathwayGuide analysis predicted that 22 pathways defined in the KEGG database were significantly affected (Fig. 6B and Supplementary S8), among which type I diabetes mellitus, HIF-1 signaling, ferroptosis, and Notch signaling pathways were the most significantly affected.
Fig. 6.
Gene expression analysis of ARPE-19 cells cultured on board the International Space Station in the presence of absence of coenzyme Q10. A All 153 significantly differentially expressed (DE) genes (showed in red) are represented in the Volcano plot in terms of their measured expression change (x-axis) and the significance of the change (y-axis), as in Fig. 3A. The dotted lines represent the thresholds used to select the DE genes: 0.4 for expression change and 0.05 for significance. B Pathway perturbation vs over-representation. Each pathway is represented by a single dot, with significant pathways shown in red, non-significant in black, and the size of each dot is proportional to the size of the pathway it represents. Both p values are shown in terms of their negative log (base 10) values
Discussion
In the present study, we investigated the impact of the space environment on human retinal pigment epithelial cells. For this purpose, ARPE-19 cells were transferred and cultured for 3 days onboard the ISS; in addition, cells were cultured in the presence or absence of CoQ10. As a control, ARPE-19 cells were cultured on the ground under normal conditions.
Previous ground experiments on ARPE-19 cells using microgravity simulators have led to contrasting results. We previously showed that 3 days of simulated microgravity obtained by the rotating wall vessel bioreactor (RWV) increased the activity of caspase 3/7 and reduced the mitochondrial transmembrane potential, which is evidence of apoptosis induction [26]. In contrast, using a different microgravity simulator (random positioning machinery, RPM), Corydon et al. did not report apoptotic events in ARPE19 cells cultured for 5–10 days under simulated microgravity conditions [34]. In the present study, we found that the transfer of ARPE-19 cells from Cape Canaveral to the ISS and 3-day incubation on board did not affect their proliferation rate nor induce apoptosis with respect to the ground controls (Fig. 2A, B). These results could be explained by differences between microgravity simulated by the available simulator devices and between simulated microgravity and real microgravity onboard the ISS, and the use of these devices and the impact of flight hardware on cellular physiology are currently debated [35, 36]. Cells living onboard the ISS are simultaneously subjected to microgravity and space radiation, which typically do not occur in on-ground simulation experiments, where the two treatments are administered separately [37]. Indeed, the constant threat to the integrity of astronauts represented by space radiation, which causes damage to DNA directly or via the production of free radicals, and microgravity, which hampers DNA repair, have raised the opportunity to explore the potential synergies between microgravity and radiation, both in space and in Earthly analogs. The results have been conflicting, which indicates the need to perform experiments in a space environment [37]. Furthermore, in our experiment, the 3-day culturing of ARPE-19 cells on board the ISS was preceded by a 5-day permanence into the Biokit at a relatively low temperature (up to approximately 26 °C). It has been demonstrated that hypothermia elicits a strong expression of cold shock proteins in mammalian retinal cells, including retinal pigment epithelial cells [38], where they may act as protective factors against damage and prevent apoptosis [39]. Finally, it should be noted that, in our experiment, ARPE-19 cells were cultured inside the KUBIK on board the ISS for 3 days, however, due to the transfer from the ground to the ISS, the cells underwent microgravity conditions several hours before entering the KUBIK, which could have affected adaptive behavior.
Cytoskeletal disorganization and significant changes in the expression of its main components, actin, tubulin, and vimentin, both at phenotypic and genotypic levels, have been observed in various cell types subjected to altered gravity conditions [40–43] and complete destruction of the microtubule network was observed in Jurkat cells following exposure to real microgravity [44]. Similarly, in endothelial cells, a significant reduction in actin fibers was observed in response to simulated microgravity [45] and a significant increase in stress fibers following hypergravity [46]. ARPE-19 cells cultured for 5 days and 10 days on RPM revealed the appearance of two major phenotypes, i.e., adherent monolayers and 3D multicellular spheroids [34]. In addition, the reduction of F-actin filaments in favor of F-actin structures at the cell boundaries, and the downregulation of beta-actin, beta-tubulin, keratin 8, vimentin, laminin subunit beta 2, integrin beta 1, integrin beta 3, and collagen 4 gene expression have been revealed, suggesting that simulated microgravity reduces the ability of exterior structures in ARPE-19 cells to retain adhesion and stiffness [34]. Other cell types differ from ARPE-19 cells in their cytoskeleton-related responses to microgravity. For example, RPM induces the upregulation of beta actin gene expression in MCF-7 breast carcinoma [47] and in human primary chondrocytes [48] and of vimentin gene expression in FLG 29.1 leukemic cells [46] and ML-1 follicular thyroid carcinoma cells [49]. Here, we revealed a dramatic change in the cytoskeletal vimentin distribution in ARPE-19 cultured for 3 days onboard the ISS. Vimentin is a cytoskeletal protein responsible for maintaining cellular integrity and is the main constituent of intermediate filaments, which provide structural support for the cytoplasm and play an essential role in the response to mechanical stresses, such as gravity [50]. Recently, it has been demonstrated that the vimentin network maintains the resilience of the cytoplasm and enhances cytoplasmic mechanical strength and toughness under dynamic deformation via poroelastic and viscoelastic relaxation [51]. Thus, vimentin networks may reduce the risk of cell damage during major deformation events [51]. In our study, immunofluorescence analysis revealed deregulated vimentin cellular localization, mostly concentrated in the perinuclear region in cells exposed to real microgravity (Fig. 2C). The different distribution of vimentin in cells exposed to microgravity compared to the control denotes a change in the shape of the cells and suggests an alteration in the capability of cells to interact with others and with extracellular matrix components. Moreover, vimentin analysis revealed that spaceflight ARPE-19 cells contained perinuclear structures resembling aggresomes. Aggresomes are protein inclusions aggregates at the centrosome-nuclear bay, and contain proteins destined to degradation that are located within a cage comprised of intermediated filament, including vimentin [50, 52]. The formation of aggresomes is directly linked to disruption of proper protein homeostasis, in particular when elevated levels of misfolded or damaged proteins accumulate into cells [50]. This observation suggests that the real microgravity determines an alteration of protein processing in ARPE-19 cells, and is consistent with the following transcriptomic data.
The first observation that microgravity altered gene expression occurred > 20 years ago; however, since the most apparent detrimental effect of long-lasting space flights on astronauts was the strong atrophy of their musculoskeletal system, studies mainly focused on gene expression in bone and muscle cells. Subsequently, several studies have highlighted the impact of the spatial environment on gene expression in different tissues, but the real milestone for high-performance studies of the expression of entire panels of genes in response to spatial radiation and microgravity has been the recent application of genomic and proteomic approaches. Understanding the variations in gene expression in their maximum amplitude is key to unraveling the pathogenesis of damage endured by astronauts during spaceflight and, ultimately, to identify effective countermeasures. The present study found a considerable number of upregulated and downregulated DE genes (Fig. 3A). KEGG analysis, impact analysis, and GO annotation revealed that several relevant biological pathways and cellular functions were affected by space flight (Fig. 3B, C, Fig. 4). Among them, protein processing in the endoplasmic reticulum (ER) is one of the most impacted, which is also justified by the vimentin network alterations described above. The ER is a complex endomembrane system in which proteins are folded and transported to distinct cellular districts [53]. Correctly folded proteins are packaged into transport vesicles and shuttled into the Golgi system. Misfolded proteins are retained within the ER, coupled with chaperones, and if terminally misfolded, subjected to ER-associated degradation. ER accumulation of misfolded proteins determines ER stress and evokes the unfolded protein response (UPR), which aims to inhibit protein translation, degrade misfolded proteins, increase chaperone production, and eventually induce apoptosis [53]. ER stress and UPR have been correlated with microgravity in different cell types [54–56]. We found that the majority of DE genes involved in this pathway were downregulated in ARPE-19 cells cultured onboard the ISS. However, some members of the 70 kDa heat shock protein (HSP70) gene family (HSPA1A, HSPA8, and HSPA2 in particular) and E3 ligase parkin (PARK2) genes, which are involved in chaperone and E3 ubiquitin ligase activity, respectively, were significantly upregulated. Recently, interesting studies have been conducted on human endothelial cells (HUVECs). It has been reported that 4 days of HUVEC culture in RWV simulated microgravity-induced cell stress and promoted HSP70 upregulation, likely sustaining the early phases of the dynamic adaptation of HUVECs to microgravity, while HSP70 silencing impaired cell survival [57]. In addition, reduced mitochondrial function and mitochondrial content have been demonstrated [57], and the contribution of mitophagy to this adaptation, which culminates in a thrifty phenotype, has been demonstrated [41]. In addition to its role in ER metabolism, Parkin is a central regulator of mitophagy [58], another pathway found to be significantly impacted in ARPE-19 cells cultured onboard the ISS. Deleterious byproducts or oxidative stress lead to mitochondrial dysfunction, which, if the damage is unrepairable, culminates in mitochondria recognition and targeting for degradation via the autophagic process termed mitophagy [58]. Multi-omics analysis recently identified mitochondrial dysregulation as a central biological hub for the pathophysiological effects of spaceflight [59]. In this context, Parkin recognizes altered proteins on the mitochondrial outer membrane and mediates damaged mitochondrial clearance [58]. Microphthalmia-associated transcription factor (MITF) is another gene belonging to the mitophagy pathway [60] that is upregulated in ARPE-19 cells exposed to the space environment. MITF is a transcription factor responsible for retinal pigment epithelial cell development and function [61]. MITF plays a protective role against oxidative stress in ARPE-19 cells, where it upregulates antioxidant gene expression and mitochondrial biogenesis, mainly via peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC1a) and nuclear factor erythroid 2-related factor 2 (NRF2) upregulation [62].
In our experiment, transforming growth factor (TGF)-beta and p53 signaling were deregulated in ARPE-19 cells cultured onboard the ISS. While most of the DE genes involved in TGF-beta signaling were upregulated, the opposite was true for p53 signaling. Using a system biology approach to evaluate rodent transcriptomic data from GeneLab (Genelab.nasa.gov), TGF-beta and p53 were identified as the most prevalent pan-tissue signaling pathways activated in response to microgravity [63]. p53 is a major contributor to cell cycle arrest, DNA repair, and apoptosis induction in response to genotoxic or non-genotoxic damage [64]. p53 signaling is frequently activated in response to microgravity. However, heterogeneous responses have been observed depending on the cell type and culture conditions. For example, p53 pathway activation has been recently described in macrophages undergoing spaceflight or RWV simulated microgravity [65] and in lung cancer cells cultured with RPM [66], while the inhibition of p53 signaling under 3D-clinostat simulated microgravity was found in hepatoblastoma cells [67]. In ARPE-19 cells cultured onboard the ISS, we detected several p53-transactivated pro-apoptotic genes (e.g., BAX, PMAIP1, BBC3), and genes involved in cell cycle arrest (e.g., CDKN1A, GADD45A, GADD45B) and DNA repair and damage prevention (DDB2, GADD45A [67], GADD45B, SESN2) were downregulated. Similarly, we found that cellular senescence pathway genes, and in particular, most of the genes encoding senescence-associated secretory phenotype (SASP) factors [68], such as IL1A, CXCL8, SERPINE1, and IGFBP3, were significantly downregulated. In contrast, the gene coding for the SASP TGF-beta factor was upregulated. TGF-beta is a master regulator that coordinates the systemic response to microgravity at multiple biological scales [63]. TGF-beta is a well-known cytokine that belongs to a family of several protein members, including bone morphogenic proteins, growth differentiation factors, activins, and inhibins, all of which regulate a wide spectrum of cellular functions such as proliferation, apoptosis, differentiation, and immune responses [69]. TGF-beta plays a major role in retinal pigment epithelial cells, particularly in a dynamic transition along a well-differentiated, polarized epithelial to mesenchymal cells, which has been defined as retinal pigment epithelium dysfunction [70]. Functional retinal pigment epithelial cells form a single layer of polarized cells located between the photoreceptors and the choroid. Correct differentiation and polarization of the retinal pigment epithelium are essential for proper functioning. While epithelial features guarantee retinal pigment epithelium physiological homeostasis in the retina, different mechanisms have been found to be involved in retinal pigment epithelium epithelial to mesenchymal transition (EMT) and mesenchymal to epithelial transition (MET), which hinder their functions [70]. Microgravity has been previously demonstrated to trigger EMT in different cell types [65, 71]. TGF-beta signaling is one of the most potent EMT inductors and is, therefore, a major contributor to retinal pigment epithelium dysfunction [70]. High levels of vitreal TGF-beta have been detected in proliferative vitreoretinopathy patients, a pathological condition in which EMT of the retinal pigment epithelium plays an essential pathogenic role [72], which led to the evaluation of TGF-beta signaling inhibitors able to counteract EMT for the treatment of proliferative retinal diseases such as proliferative vitreoretinopathy [73]. The Hippo signaling pathway is closely interconnected with that of TGF-beta, and in our experiment, it was significantly affected in ARPE-19 cells cultured on board the ISS. This pathway regulates retinal pigment epithelial cell differentiation and is involved in EMT [74]. Connective tissue growth factor (CTGF) encodes a cysteine-rich extracellular matrix protein that acts downstream of the TGF-beta and Hippo signaling pathways [75, 76]. We found that CTGF is one of the genes overexpressed in ARPE-19 cells cultured on board the ISS. It has been hypothesized that its binding to various cell surface receptors, including integrin receptors, heparan sulfate proteoglycans, and low-density lipoprotein receptor-related proteins, enables CTGF to regulate key cellular functions, such as cell adhesion, proliferation, chemotaxis, differentiation, survival, and extracellular matrix component production [77]. A previous study indicated that CTGF increases the migratory ability of retinal pigment epithelial cells and that it is a major mediator of retinal fibrosis [78]. In addition, it has been recently demonstrated that the Hippo pathway is severely altered in a murine model of retinal degeneration, in which CTGF is markedly upregulated [76].
Analysis of DE genes revealed deregulation of the expression and activity of lncRNAs (Table 3 and Supplementary S5) and microRNAs (Fig. 5 and Supplementary S6), respectively, in ARPE-19 cells. LncRNAs are defined as transcripts longer than 200 nucleotides that generally lack protein-coding potential and can be processed like mRNAs, i.e., spliced and polyadenylated [79]. LncRNAs are involved in several cellular physiological processes, such as adaptation to stress, cell differentiation, maintenance of pluripotency, and apoptosis [79]. The correct balance of lncRNA levels is crucial for the maintenance of cellular equilibrium, and their dysregulation is associated with many disorders [79]. Variations in lncRNA expression have been documented in different cell types exposed to simulated microgravity [80–82]. Here, we demonstrated that real microgravity alters a panel of lncRNAs in ARPE-19 cells, most of which were upregulated. Considering the top ten up- and down-regulated lncRNAs, the only one involved to date in retinal cell physiology is H19, which is involved in retinal cell death and in the inflammatory response of ARPE-19 cells exposed to hyperglycemia [83]. We detected strong downregulation of H19 expression in ARPE-19 cells cultured onboard the ISS, and it has been reported that H19 downregulation correlates with TGF-beta-mediated EMT in retinal endothelial cells [84]. Similarly, another two lncRNAs downregulated in ARPE-19 cells cultured onboard the ISS, namely SLC7A11-AS1 and LUCAT1, are reported to be involved in the EMT process [85, 86]. MicroRNAs are small single-stranded non-coding RNAs, whose functions in RNA silencing and post-transcriptional regulation of gene expression are well-known [87, 88]. A recent milestone study identified a spaceflight-associated microRNA signature in response to simulated short- and long-duration spaceflight and simulated deep-space radiation conditions [89]. In addition, the panel of spaceflight-associated microRNAs has been predicted to interfere with cell signaling and metabolic pathways, partly through interaction with TGF-beta signaling [63]. Some microRNAs that were identified to be activated in ARPE-19 cells cultured on board the ISS are involved in the cell response to spaceflight. Among them, hsa-miR-296 expression was affected in HUVECs cultured in a 3D clinostat to simulate microgravity and was found to influence cell proliferation and vascular function under microgravity conditions [90]. hsa-miR-494 is upregulated in C2C12 myoblast cells subjected to clinorotation conditions and in osteoblasts isolated from tail-suspended rats, and this upregulation correlates with the inhibition of osteoclast differentiation [91]. Other miRNAs we identified are involved in the EMT process. For example, hsa-miR-145-3p, hsa-miR-324-5p, and hsa-miR142-5p act as inhibitors of EMT in different cell types [92–95]; in contrast, high expression and activation of hsa-miR 483-3p, hsa-miR-128-3p are involved in EMT induction.
CoQ10 is an essential cofactor in the electron transport chain. It is endowed with well-known protective actions with respect to different types of damage on various cell types, including those of the retina, which mainly depends on its free radical scavenging ability and the regulation of the mitochondrial permeability transition pore [29]. It is known that the choroid/retina levels of endogenous CoQ10 decrease with aging concomitantly with the progression of apoptosis-related retinal diseases [96]. We previously demonstrated the efficacy of the topical administration (via eye drops) of CoQ10 to protect retinal cell layers commissioned to apoptosis by a variety of noxious stimuli [27, 28]. On these bases, we tested the role of CoQ10 treatment in modulating the transcriptome of ARPE-19 cells cultured onboard the ISS. We found a limited number of DE genes that were subjected to KEGG and impact analyses (Fig. 6). Among the most affected biological pathways affected by CoQ10 treatment, we focused on HIF-1 signaling and ferroptosis. HIF-1 is a well-known transcription factor discovered by virtue of its role as a master regulator of oxygen homeostasis [97]. HIF-1 signaling is involved in numerous cellular responses to a variety of environmental stresses. Interestingly, it has been demonstrated that, regardless of its function as a transcription factor, HIF-1 has a protective role through its localization into mitochondria, where it reduces reactive oxygen species levels and reverses mitochondrial damage [98]. Previous studies have revealed that microgravity affects HIF-1 pathway activation. Wang et al. demonstrated that 28 days of simulated microgravity induced oxidative stress and HIF-1 activation and a panel of its downstream targets in rat hippocampus, while Vogel et al. demonstrated that HIF-1a- and HIF-1-dependent transcripts were differentially regulated under altered gravity on monocytes and macrophages during parabolic flight and suborbital ballistic rocket campaigns, whereas HIF-1-dependent gene expression adapted after 5 min of microgravity [99]. Ferroptosis is a peculiar type of iron-dependent cell death; it was first characterized in 2012 and results from the accumulation of toxic levels of lipid-reactive oxidative species [100]. Increased intracellular iron accumulation is a classical ferroptosis activator that is directly linked to the failure of glutathione (GSH)-dependent antioxidant defenses. Cellular iron metabolism includes absorption of the ferritin-Fe3 + complex by the transferrin receptor (TFRC), and it has been demonstrated that reducing iron utilization may increase the sensitivity to ferroptosis [101]. GSH production is sustained by the activity of the amino acid antiporter, SLC7A11/xCT/system xc-, which guarantees the exchange of extracellular cystine (which, in turn, generates cysteine, a limiting precursor for GSH synthesis) for intracellular glutamate [100]. It is known that inhibition of system xc- causes GSH depletion to trigger ferroptosis [102]. The direct involvement of CoQ10 in ferroptosis has been recently reported. It is known that the reduced form of CoQ10 is a potent antioxidant that counteracts lethal lipid peroxidation, thus acting as a ferroptosis suppressor. In particular, the NADH-dependent oxidoreductase, FSP1, converts non-mitochondrial oxidized to reduced CoQ10, thus furnishing a GSH-independent protective axis to counteract membrane lipid peroxidation [103]. In addition to this role, we found that CoQ10 treatment of ARPE-19 cells cultured on board the ISS reduced TFRC and induced SLC7A11 gene expression, corroborating the evidence that CoQ10 acts as a direct ferroptosis inhibitor and bona fide increases ferroptosis resistance by reducing iron cellular uptake and enhancing cystine uptake.
Conclusion
Our results indicate that the transfer of cultured ARPE-19 human retinal cells to the ISS and their 3-day living on board severely altered cytoskeleton morphology and the transcriptome profile, indicating the emergence of cellular dysfunction (Fig. 7). Although obtained in a single experiment, in which we were able to use only a limited number of ARPE-19 cells in culture to carry out the various analyses, we suggest that our results may, at least partially, reflect the response of the retinal pigment epithelial cells of astronauts' eyes subjected to space flight. The retinal pigment epithelium has peculiar functions strictly linked to its structure that are essential for normal vision [104]. Alterations in retinal pigment epithelium physiology can lead to retinopathy, which consequently affects vision. Retinal pigment epithelium dysfunction is known to be involved in the pathogenesis of several degenerative retinal diseases including age-related macular degeneration, retinitis pigmentosa, and Stargardt disease [105]. Moreover, retinal pigment epithelium dysfunction accompanies the course of non-degenerative retinal diseases, such as diabetic retinopathy [106], and is of primary importance in proliferative diseases such as proliferative vitreoretinopathy [70]. The results obtained in the present study, highlighting the potential dysfunction of ARPE-19 cells, are consistent with what has been observed in vivo in previous studies in rodents housed in spacecraft for several days [16, 107]. Interestingly, during the same SpaceX CRS-12 mission as our experiment, retinal pigment epithelium thickness reduction was observed in the retinas of mice hosted for 40 days on board the ISS [107]. Therefore, although the evidence we report here does not have an immediate impact on SANS pathogenesis, it poses an alert for the health of the astronauts' retinas, particularly in view of long-term missions, which may even extend to years. Approximately 60% of astronauts returning from long-term missions exhibit signs of SANS, the etiology of which is currently debated [19]. The causes of SANS are thought to be a mild and chronic increase in intracranial pressure, congestion of cerebral venous, and alteration of folate-dependent 1-carbon metabolism [23]. Regardless of its causes, reducing the risk of SANS is a priority for long-term spaceflight planning. Among the signs of SANS, two are particularly feared. The first is optic disc edema, which may cause severe vision loss due to the occurrence of increasing blind spots. The second is retinal and choroidal folds, which, if severe and located near the fovea, can cause distortions or visual acuity reduction [108]. The contribution of spaceflight-dependent cellular alterations of individual retinal cell types in the pathogenesis of SANS has not yet been defined. However, results from experiments conducted on cell cultures suggest that the molecular and cellular responses to microgravity and radiation could synergize with macroscopic tissue and organ alterations in the pathogenesis of SANS. A recent example of this hypothesis comes from the observation that choroidal folds in the retina of a crewmember > 5 years post-flight induced alterations in the retinal pigment epithelium [108]. Therefore, the results presented herein suggest a rational basis for future investigations aimed at identifying effective countermeasures against SANS. On this line of evidence, the treatment of ARPE-19 cells with CoQ10 on board reveals its potential to increase cell resilience towards harmful agents in the space environment. Moreover, it can be hypothesized that at least some of the gene deregulations we found in ARPE-19 cells occur in other cell types and tissues of astronauts. Based on this assumption, the evidence that important cell pathways are significantly affected by spaceflight and, if deregulated, play a central role in a variety of severe human disorders, such as cancer, cardiovascular diseases, and diabetes, confirms possible connections to astronauts’ health [109, 110].
Fig. 7.
Effects of space flight on ARPE-19 cells. The conclusions reported in the picture, except that relative to vimentin network disorganization, are based on transcriptomic data
Supplementary Information
Below is the link to the electronic supplementary material.
Funding
This project was coordinated and funded by the Italian Space Agency (ASI, grant 2016–6-U0 to M.L.) for space s tation experimentation within the national ISS utilization rights. The authors thank Fondazione Cassa di Risparmio Di Firenze (Italy) for financial support.
Data availability
All data and material used are available in the author’s labs.
Code availability (software application or custom code)
Not applicable.
Declarations
Conflict of interest
The authors declare no conflicts of interest/competing interests.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Sergio Capaccioli and Matteo Lulli contributed equally to this study.
References
- 1.Demontis GC, Germani MM, Caiani EG, Barravecchia I, Passino C, Angeloni D. Human pathophysiological adaptations to the space environment. Front Physiol. 2017;8:547. doi: 10.3389/fphys.2017.00547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strollo F, Gentile S, Strollo G, Mambro A, Vernikos J. Recent progress in space physiology and aging. Front Physiol. 2018;9:1551. doi: 10.3389/fphys.2018.01551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stavnichuk M, Mikolajewicz N, Corlett T, Morris M, Komarova SV. A systematic review and meta-analysis of bone loss in space travelers. NPJ microgravity. 2020;6:13. doi: 10.1038/s41526-020-0103-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Norsk P (2020) Adaptation of the cardiovascular system to weightlessness: Surprises, paradoxes and implications for deep space missions. Acta Physiol (Oxf) 228 (3):e13434. [DOI] [PubMed]
- 5.Crucian BE, Chouker A, Simpson RJ, Mehta S, Marshall G, Smith SM, Zwart SR, Heer M, Ponomarev S, Whitmire A, Frippiat JP, Douglas GL, Lorenzi H, Buchheim JI, Makedonas G, Ginsburg GS, Ott CM, Pierson DL, Krieger SS, Baecker N, Sams C. Immune system dysregulation during spaceflight: potential countermeasures for deep space exploration missions. Front Immunol. 2018;9:1437. doi: 10.3389/fimmu.2018.01437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grigoriev AI, Bugrov SA, Bogomolov VV, Egorov AD, Polyakov VV, Tarasov IK, Shulzhenko EB (1993) Main medical results of extended flights on space station Mir in 1986–1990. Acta astronautica 29 (8). [DOI] [PubMed]
- 7.Bradbury P, Wu H, Choi JU, Rowan AE, Zhang H, Poole K, Lauko J, Chou J. Modeling the impact of microgravity at the cellular level: implications for human disease. Front Cell Dev Biol. 2020;8:96. doi: 10.3389/fcell.2020.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beck M, Moreels M, Quintens R, Abou-El-Ardat K, El-Saghire H, Tabury K, Michaux A, Janssen A, Neefs M, Van Oostveldt P, De Vos WH, Baatout S. Chronic exposure to simulated space conditions predominantly affects cytoskeleton remodeling and oxidative stress response in mouse fetal fibroblasts. Int J Mol Med. 2014;34(2):606–615. doi: 10.3892/ijmm.2014.1785. [DOI] [PubMed] [Google Scholar]
- 9.Mao XW, Boerma M, Rodriguez D, Campbell-Beachler M, Jones T, Stanbouly S, Sridharan V, Nishiyama NC, Wroe A, Nelson GA (2018) Combined Effects of Low-Dose Proton Radiation and Simulated Microgravity on the Mouse Retina and the Hematopoietic System. Radiation research. [DOI] [PMC free article] [PubMed]
- 10.Mao XW, Pecaut MJ, Stodieck LS, Ferguson VL, Bateman TA, Bouxsein M, Jones TA, Moldovan M, Cunningham CE, Chieu J, Gridley DS. Spaceflight environment induces mitochondrial oxidative damage in ocular tissue. Radiat Res. 2013;180(4):340–350. doi: 10.1667/RR3309.1. [DOI] [PubMed] [Google Scholar]
- 11.Kast J, Yu Y, Seubert CN, Wotring VE, Derendorf H (2017) Drugs in Space: Pharmacokinetics and Pharmacodynamics in Astronauts. European journal of pharmaceutical sciences : official journal of the European Federation for Pharmaceutical Sciences 109S. [DOI] [PubMed]
- 12.Eyal S, Derendorf H (2019) Medications in Space: In Search of a Pharmacologist's Guide to the Galaxy. Pharm Res 36 (10):148. [DOI] [PubMed]
- 13.Mao XW, Boerma M, Rodriguez D, Campbell-Beachler M, Jones T, Stanbouly S, Sridharan V, Wroe A, Nelson GA. Acute effect of low-dose space radiation on mouse retina and retinal endothelial cells. Radiat Res. 2018;190(1):45–52. doi: 10.1667/RR14977.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao HW, Zhao J, Hu LN, Liang JN, Shi YY, Nie C, Qiu CY, Nan XS, Li YX, Gao FL, Liu Y, Dong Y, Luo L. Effect of long-term weightlessness on retina and optic nerve in tail-suspension rats. Int J Ophthalmol. 2016;9(6):825–830. doi: 10.18240/ijo.2016.06.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tombran-Tink J, Barnstable CJ. Space flight environment induces degeneration in the retina of rat neonates. Adv Exp Med Biol. 2006;572:417–424. doi: 10.1007/0-387-32442-9_59. [DOI] [PubMed] [Google Scholar]
- 16.Tombran-Tink J, Barnstable CJ. Space shuttle flight environment induces degeneration in the retina of rat neonates. Gravitational Space Biol Bull. 2005;18(2):97–98. [PubMed] [Google Scholar]
- 17.Roberts JE, Kukielczak BM, Chignell CF, Sik BH, Hu DN, Principato MA. Simulated microgravity induced damage in human retinal pigment epithelial cells. Mol Vis. 2006;12:633–638. [PubMed] [Google Scholar]
- 18.Mader TH, Gibson CR, Caputo M, Hunter N, Taylor G, Charles J, Meehan RT. Intraocular pressure and retinal vascular changes during transient exposure to microgravity. Am J Ophthalmol. 1993;115(3):347–350. doi: 10.1016/S0002-9394(14)73586-X. [DOI] [PubMed] [Google Scholar]
- 19.Mader TH, Gibson CR, Pass AF, Kramer LA, Lee AG, Fogarty J, Tarver WJ, Dervay JP, Hamilton DR, Sargsyan A, Phillips JL, Tran D, Lipsky W, Choi J, Stern C, Kuyumjian R, Polk JD. Optic disc edema, globe flattening, choroidal folds, and hyperopic shifts observed in astronauts after long-duration space flight. Ophthalmology. 2011;118(10):2058–2069. doi: 10.1016/j.ophtha.2011.06.021. [DOI] [PubMed] [Google Scholar]
- 20.Mader TH, Gibson CR, Otto CA, Sargsyan AE, Miller NR, Subramanian PS, Hart SF, Lipsky W, Patel NB, Lee AG. Persistent Asymmetric Optic Disc Swelling After Long-Duration Space Flight: Implications for Pathogenesis. J Neuro-Ophthalmol. 2017;37(2):133–139. doi: 10.1097/WNO.0000000000000467. [DOI] [PubMed] [Google Scholar]
- 21.Zhang LF, Hargens AR. Spaceflight-induced intracranial hypertension and visual impairment: pathophysiology and countermeasures. Physiol Rev. 2018;98(1):59–87. doi: 10.1152/physrev.00017.2016. [DOI] [PubMed] [Google Scholar]
- 22.Lee AG, Mader TH, Gibson CR, Tarver W. Space flight-associated neuro-ocular syndrome. JAMA Ophthalmol. 2017;135(9):992–994. doi: 10.1001/jamaophthalmol.2017.2396. [DOI] [PubMed] [Google Scholar]
- 23.Lee AG, Mader TH, Gibson CR, Tarver W, Rabiei P, Riascos RF, Galdamez LA, Brunstetter T. Spaceflight associated neuro-ocular syndrome (SANS) and the neuro-ophthalmologic effects of microgravity: a review and an update. NPJ Microgravity. 2020;6:7. doi: 10.1038/s41526-020-0097-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tymko MM, Boulet LM, Donnelly J. Intracranial pressure in outer space: preparing for the mission to Mars. J Physiol. 2017;595(14):4587–4588. doi: 10.1113/JP274315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alwood JS, Ronca AE, Mains RC, Shelhamer MJ, Smith JD, Goodwin TJ. From the bench to exploration medicine: NASA life sciences translational research for human exploration and habitation missions. NPJ Microgravity. 2017;3:5. doi: 10.1038/s41526-016-0002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lulli M, Cialdai F, Vignali L, Monici M, Luzzi S, Cicconi A, Cacchione S, Magi A, Di Gesualdo F, Balsamo M, Vukich M, Neri G, Donati A, Capaccioli S. The coenzyme Q10 (CoQ10) as countermeasure for retinal damage onboard the international space station: the CORM Project | SpringerLink. Microgravity Sci Technol. 2019;30(6):925–931. doi: 10.1007/s12217-018-9652-3. [DOI] [Google Scholar]
- 27.Lulli M, Witort E, Papucci L, Torre E, Schiavone N, Dal Monte M, Capaccioli S. Coenzyme Q10 protects retinal cells from apoptosis induced by radiation in vitro and in vivo. J Radiat Res. 2012;53(5):695–703. doi: 10.1093/jrr/rrs025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lulli M, Witort E, Papucci L, Torre E, Schipani C, Bergamini C, Dal Monte M, Capaccioli S. Coenzyme Q10 instilled as eye drops on the cornea reaches the retina and protects retinal layers from apoptosis in a mouse model of kainate-induced retinal damage. Invest Ophthalmol Vis Sci. 2012;53(13):8295–8302. doi: 10.1167/iovs.12-10374. [DOI] [PubMed] [Google Scholar]
- 29.Papucci L, Schiavone N, Witort E, Donnini M, Lapucci A, Tempestini A, Formigli L, Zecchi-Orlandini S, Orlandini G, Carella G, Brancato R, Capaccioli S. Coenzyme q10 prevents apoptosis by inhibiting mitochondrial depolarization independently of its free radical scavenging property. J Biol Chem. 2003;278(30):28220–28228. doi: 10.1074/jbc.M302297200. [DOI] [PubMed] [Google Scholar]
- 30.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras T (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England) 29 (1). [DOI] [PMC free article] [PubMed]
- 31.Liao Y, Smyth G, Shi K (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics (Oxford, England) 30 (7). [DOI] [PubMed]
- 32.Love M, Huber W, Anders I (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 15 (12). [DOI] [PMC free article] [PubMed]
- 33.Ashan S, Drăghici S (2017) Identifying Significantly Impacted Pathways and Putative Mechanisms With iPathwayGuide. Current protocols in bioinformatics 57. [DOI] [PubMed]
- 34.Corydon TJ, Mann V, Slumstrup L, Kopp S, Sahana J, Askou AL, Magnusson NE, Echegoyen D, Bek T, Sundaresan A, Riwaldt S, Bauer J, Infanger M, Grimm D (2016) Reduced Expression of Cytoskeletal and Extracellular Matrix Genes in Human Adult Retinal Pigment Epithelium Cells Exposed to Simulated Microgravity. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology 40 (1–2). [DOI] [PubMed]
- 35.Krüger M, Pietsch J, Bauer J, Kopp S, Carvalho DTO, Baatout S, Moreels M, Melnik D, Wehland M, Egli M, Jayashree S, Kobberø SD, Corydon TJ, Nebuloni S, Gass S, Evert M, Infanger M, Grimm D (2019) Growth of Endothelial Cells in Space and in Simulated Microgravity - a Comparison on the Secretory Level. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology 52 (5). [DOI] [PubMed]
- 36.Wuest SL, Richard S, Kopp S, Grimm D, Egli M (2015) Simulated microgravity: critical review on the use of random positioning machines for mammalian cell culture BioMed research international 2015. [DOI] [PMC free article] [PubMed]
- 37.Moreno-Villanueva M, Wong M, Lu T, Zhang Y, Wu H (2017) Interplay of space radiation and microgravity in DNA damage and DNA damage response. NPJ microgravity 3. [DOI] [PMC free article] [PubMed]
- 38.Larrayoz IM, Rey-Funes M, Contartese DS, Rolón F, Sarotto A, Dorfman VB, Loidl CF, Martínez A (2016) Cold Shock Proteins Are Expressed in the Retina Following Exposure to Low Temperatures. PloS one 11 (8). [DOI] [PMC free article] [PubMed]
- 39.Rey-Funes M, Larrayoz IM, Contartese DS, Soliño M, Sarotto A, Bustelo M, Bruno M, Dorfman VB, Loidl CF, Martínez A (2017) Hypothermia Prevents Retinal Damage Generated by Optic Nerve Trauma in the Rat. Scientific reports 7 (1). [DOI] [PMC free article] [PubMed]
- 40.Lewis ML, Reynolds JL, Cubano LA, Hatton JP, Lawless BD, Piepmeier EH (1998) Spaceflight alters microtubules and increases apoptosis in human lymphocytes (Jurkat). FASEB J 12 (11). [DOI] [PubMed]
- 41.Locatelli L, Cazzaniga A, De Palma C, Castiglioni S, Maier J (2020) Mitophagy contributes to endothelial adaptation to simulated microgravity. FASEB J 34 (1). [DOI] [PubMed]
- 42.Maier JA, Cialdai F, Monici M, Morbidelli L (2015) The impact of microgravity and hypergravity on endothelial cells. BioMed Res Int 2015. [DOI] [PMC free article] [PubMed]
- 43.Prasad B, Grimm D, Strauch SM, Erzinger GS, Corydon TJ, Lebert M, Magnusson NE, Infanger M, Richter P, Krüger M (2020) Influence of microgravity on apoptosis in cells, tissues, and other systems in vivo and in vitro. Int J Mol Sci 21 (24). [DOI] [PMC free article] [PubMed]
- 44.Schatten H, Lewis ML, Chakrabarti A (2001) Spaceflight and clinorotation cause cytoskeleton and mitochondria changes and increases in apoptosis in cultured cells. Acta Astronautica 49 (3–10). [DOI] [PubMed]
- 45.Carlsson SI, Bertilaccio MT, Ballabio E, Maier J (2003) Endothelial stress by gravitational unloading: effects on cell growth and cytoskeletal organization. Biochimica et Biophysica Acta 1642 (3). [DOI] [PubMed]
- 46.Monici M, Fusi F, Paglierani M, Marziliano N, Cogoli A, Pratesi R, Bernabei PA (2006) Modeled gravitational unloading triggers differentiation and apoptosis in preosteoclastic cells. J Cell Biochem 98 (1). [DOI] [PubMed]
- 47.Kopp S, Slumstrup L, Corydon TJ, Sahana J, Aleshcheva G, Islam T, Magnusson NE, Wehland M, Bauer J, Infanger M, Grimm D (2016) Identifications of novel mechanisms in breast cancer cells involving duct-like multicellular spheroid formation after exposure to the Random Positioning Machine. Sci Rep 6. [DOI] [PMC free article] [PubMed]
- 48.Aleshcheva G, Sahana J, Ma X, Hauslage J, Hemmersbach R, Egli M, Infanger M, Bauer J, Grimm D (2013) Changes in morphology, gene expression and protein content in chondrocytes cultured on a random positioning machine. PloS One 8 (11). [DOI] [PMC free article] [PubMed]
- 49.Grimm D, Bauer J, Kossmehl P, Shakibaei M, Schöberger J, Pickenhahn H, Schulze-Tanzil G, Vetter R, Eilles C, Paul M, Cogoli A (2002) Simulated microgravity alters differentiation and increases apoptosis in human follicular thyroid carcinoma cells. FASEB J 16 (6). [DOI] [PubMed]
- 50.Morrow CS, Moore DL (2020) Vimentin's side gig: Regulating cellular proteostasis in mammalian systems. Cytoskeleton (Hoboken, NJ) 77 (11). [DOI] [PMC free article] [PubMed]
- 51.Hu J, Li Y, Hao Y, Zheng T, Gupta SK, Parada GA, Wu H, Lin S, Wang S, Zhao X, Goldman RD, Cai S, Guo M (2019) High stretchability, strength, and toughness of living cells enabled by hyperelastic vimentin intermediate filaments. Proceedings of the National Academy of Sciences of the United States of America 116 (35). [DOI] [PMC free article] [PubMed]
- 52.Kopito RR (2000) Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol 10 (12). [DOI] [PubMed]
- 53.Koksal AR, Verne GN, Zhou Q (2021) Endoplasmic reticulum stress in biological processing and disease. J Investigative Med 69 (2). [DOI] [PubMed]
- 54.Li CF, Pan YK, Gao Y, Shi F, Wang YC, Sun XQ (2019) Autophagy protects HUVECs against ER stress-mediated apoptosis under simulated microgravity. Apoptosis 24 (9–10). [DOI] [PMC free article] [PubMed]
- 55.Jiang M, Wang H, Liu Z, Lin L, Wang L, Xie M, Li D, Zhang J, Zhang R (2020) Endoplasmic reticulum stress-dependent activation of iNOS/NO-NF-κB signaling and NLRP3 inflammasome contributes to endothelial inflammation and apoptosis associated with microgravity. FASEB J 34 (8). [DOI] [PubMed]
- 56.Zhang R, Jiang M, Zhang J, Qiu Y, Li D, Li S, Liu J, Liu C, Fang Z, Cao F (2020) Regulation of the cerebrovascular smooth muscle cell phenotype by mitochondrial oxidative injury and endoplasmic reticulum stress in simulated microgravity rats via the PERK-eIF2α-ATF4-CHOP pathway. Biochimica et Biophysica Acta Molecular Basis of Disease 1866 (8). [DOI] [PubMed]
- 57.Cazzaniga A, Locatelli L, Castiglioni S, Maier JAM (2019) The dynamic adaptation of primary human endothelial cells to simulated microgravity. FASEB J 33 (5). [DOI] [PubMed]
- 58.Roca-Portoles A, Tait SWG (2021) Mitochondrial quality control: from molecule to organelle. Cel Mol Life Sci 78 (8). [DOI] [PMC free article] [PubMed]
- 59.Gertz ML, Chin CR, Tomoiaga D, MacKay M, Chang C, Butler D, Afshinnekoo E, Bezdan D, Schmidt MA, Mozsary C, Melnick A, Garrett-Bakelman F, Crucian B, Lee SMC, Zwart SR, Smith SM, Meydan C, Mason CE (2020) Multi-omic, single-cell, and biochemical profiles of astronauts guide pharmacological strategies for returning to gravity. Cell Rep 33 (10). [DOI] [PMC free article] [PubMed]
- 60.Nezich CL, Wang C, Fogel AI, Youle RJ (2015) MiT/TFE transcription factors are activated during mitophagy downstream of Parkin and Atg5. J Cell Biol 210 (3). [DOI] [PMC free article] [PubMed]
- 61.Ma X, Li H, Chen Y, Yang J, Chen H, Arnheiter H, Hou L (2019) The transcription factor MITF in RPE function and dysfunction. Progress Retinal Eye Res 73. [DOI] [PubMed]
- 62.Han S, Chen J, Hua J, Hu X, Jian S, Zheng G, Wang J, Li H, Yang J, Hejtmancik JF, Qu J, Ma X, Hou L (2020) MITF protects against oxidative damage-induced retinal degeneration by regulating the NRF2 pathway in the retinal pigment epithelium. Redox Biol 34. [DOI] [PMC free article] [PubMed]
- 63.Beheshti A, Ray S, Fogle H, Berrios D, Costes SV (2018) A microRNA signature and TGF-beta1 response were identified as the key master regulators for spaceflight response. PloS One 13 (7):e0199621. [DOI] [PMC free article] [PubMed]
- 64.Pluquet O, Hainaut P (2001) Genotoxic and non-genotoxic pathways of p53 induction. Cancer Lett 174 (1). [DOI] [PubMed]
- 65.Shi L, Tian H, Wang P, Li L, Zhang Z, Zhang J, Zhao Y (2020) Spaceflight and simulated microgravity suppresses macrophage development via altered RAS/ERK/NFκB and metabolic pathways. Cell Mol Immunol. [DOI] [PMC free article] [PubMed]
- 66.Dietz C, Infanger M, Romswinkel A, Strube F, Kraus A (2019) Apoptosis Induction and alteration of cell adherence in human lung cancer cells under simulated microgravity. Int J Mol Sci 20 (14). [DOI] [PMC free article] [PubMed]
- 67.Fukazawa T, Tanimoto K, Shrestha L, Imura T, Takahashi S, Sueda T, Hirohashi N, Hiyama E, Yuge L (2019) Simulated microgravity enhances CDDP-induced apoptosis signal via p53-independent mechanisms in cancer cells. PloS One 14 (7). [DOI] [PMC free article] [PubMed]
- 68.Kumari R, Jat P (2021) Mechanisms of cellular senescence: cell cycle arrest and senescence associated secretory phenotype. Front Cell Dev Biol 9. [DOI] [PMC free article] [PubMed]
- 69.Massagué J (2012) TGFβ signalling in context. Nat Rev Mol Cell Biol 13 (10). [DOI] [PMC free article] [PubMed]
- 70.Zhou M, Geathers JS, Grillo SL, Weber SR, Wang W, Zhao Y, Sundstrom JM (2020) Role of epithelial-mesenchymal transition in retinal pigment epithelium dysfunction. Front Cell Dev Biol 8. [DOI] [PMC free article] [PubMed]
- 71.Ranieri D, Proietti S, Dinicola S, Masiello MG, Rosato B, Ricci G, Cucina A, Catizone A, Bizzarri M, Torrisi MR (2017) Simulated microgravity triggers epithelial mesenchymal transition in human keratinocytes. Sci Rep 7 (1). [DOI] [PMC free article] [PubMed]
- 72.Chen Z, Shao Y, Li X (2015) The roles of signaling pathways in epithelial-to-mesenchymal transition of PVR. Mol Vis 21. [PMC free article] [PubMed]
- 73.Chen Y, Wu B, He JF, Chen J, Kang ZW, Liu D, Luo J, Fang K, Leng X, Tian H, Xu J, Jin C, Zhang J, Wang J, Zhang J, Ou Q, Lu L, Gao F, Xu GT (2021) Effectively intervening epithelial-mesenchymal transition of retinal pigment epithelial cells with a combination of ROCK and TGF-β signaling inhibitors. Investigative Ophthalmol Vis Sc 62 (4). [DOI] [PMC free article] [PubMed]
- 74.Du Y, Chen Q, Huang L, Wang S, Yin X, Zhou L, Ye Z, Ren X, Cai Y, Ding X, Ouyang H, Li X, Ju H (2018) VEGFR2 and VEGF-C Suppresses the epithelial-mesenchymal transition Via YAP in Retinal Pigment Epithelial Cells. Curr Mol Med 18 (5). [DOI] [PubMed]
- 75.Wilson SE (2021) TGF beta -1, -2 and -3 in the modulation of fibrosis in the cornea and other organs. Experimental eye research 207. [DOI] [PubMed]
- 76.Moon S, Lee S, Caesar JA, Pruchenko S, Leask A, Knowles JA, Sinon J, Chaqour B (2020) A CTGF-YAP Regulatory Pathway Essential for Angiogenesis and Barriergenesis in the Retina. iScience 23 (6). [DOI] [PMC free article] [PubMed]
- 77.Lau LF (2016) Cell surface receptors for CCN proteins. J Cell Commun Signal 10 (2). [DOI] [PMC free article] [PubMed]
- 78.Hinton DR, He S, Jin ML, Barron E, Ryan SJ (2002) Novel growth factors involved in the pathogenesis of proliferative vitreoretinopathy. Eye (London, England) 16 (4). [DOI] [PubMed]
- 79.Di Gesualdo F, Capaccioli S, Lulli M. A pathophysiological view of the long non-coding RNA world. Oncotarget. 2014;5(22):10976–10996. doi: 10.18632/oncotarget.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang Y, Wang K, Zhang L, Tan Y, Hu Z, Dang L, Zhou H, Li G, Wang H, Zhang S, Shi F, Cao X, Zhang G (2020) Targeted overexpression of the long noncoding RNA ODSM can regulate osteoblast function in vitro and in vivo. Cell death & disease 11 (2). [DOI] [PMC free article] [PubMed]
- 81.Fu H, Su F, Zhu J, Zheng X, Ge C (2020) Effect of simulated microgravity and ionizing radiation on expression profiles of miRNA, lncRNA, and mRNA in human lymphoblastoid cells. Life sciences in space research 24. [DOI] [PubMed]
- 82.Thiel CS, Hauschild S, Huge A, Tauber S, Lauber BA, Polzer J, Paulsen K, Lier H, Engelmann F, Schmitz B, Schütte A, Layer LE, Ullrich O (2017) Dynamic gene expression response to altered gravity in human T cells. Scientific reports 7 (1). [DOI] [PMC free article] [PubMed]
- 83.Luo R, Li L, Hu YX, Xiao F (2021) LncRNA H19 inhibits high glucose-induced inflammatory responses of human retinal epithelial cells by targeting miR-19b to increase SIRT1 expression. Kaohsiung J Med Sci 37 (2). [DOI] [PMC free article] [PubMed]
- 84.Thomas AA, Biswas S, Feng B, Chen S, Gonder J, Chakrabarti S (2019) lncRNA H19 prevents endothelial-mesenchymal transition in diabetic retinopathy. Diabetologia 62 (3). [DOI] [PubMed]
- 85.Liang J, Liao J, Liu T, Wang Y, Wen J, Cai N, Huang Z, Xu W, Li G, Ding Z, Zhang B (2020) Comprehensive analysis of TGF-β-induced mRNAs and ncRNAs in hepatocellular carcinoma. Aging 12 (19). [DOI] [PMC free article] [PubMed]
- 86.Zhang LC, Wei ZB, Tang SF (2020) Knockdown of the long noncoding RNA LUCAT1 inhibits high-glucose-induced epithelial-mesenchymal transition through the miR-199a-5p-ZEB1 axis in human renal tubular epithelial cells. BioMed Res Int 2020. [DOI] [PMC free article] [PubMed]
- 87.Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116 (2). [DOI] [PubMed]
- 88.Witort E, Lulli M, Carloni V, Capaccioli S (2013) Anticancer activity of an antisense oligonucleotide targeting TRADD combined with proteasome inhibitors in chemoresistant hepatocellular carcinoma cells. J Chemotherapy (Florence, Italy) 25 (5). [DOI] [PubMed]
- 89.Malkani S, Chin CR, Cekanaviciute E, Mortreux M, Okinula H, Tarbier M, Schreurs AS, Shirazi-Fard Y, Tahimic CGT, Rodriguez DN, Sexton BS, Butler D, Verma A, Bezdan D, Durmaz C, MacKay M, Melnick A, Meydan C, Li S, Garrett-Bakelman F, Fromm B, Afshinnekoo E, Langhorst BW, Dimalanta ET, Cheng-Campbell M, Blaber E, Schisler JC, Vanderburg C, Friedländer MR, McDonald JT, Costes SV, Rutkove S, Grabham P, Mason CE, Beheshti A (2020) Circulating miRNA Spaceflight Signature Reveals Targets for Countermeasure Development. Cell Rep 33 (10). [DOI] [PMC free article] [PubMed]
- 90.Kasiviswanathan D, Chinnasamy Perumal R, Bhuvaneswari S, Kumar P, Sundaresan L, Philip M, Puthenpurackal Krishnankutty S, Chatterjee S (2020) Interactome of miRNAs and transcriptome of human umbilical cord endothelial cells exposed to short-term simulated microgravity. NPJ Microgravity 6. [DOI] [PMC free article] [PubMed]
- 91.Qin W, Liu L, Wang Y, Wang Z, Yang A, Wang T (2019) Mir-494 inhibits osteoblast differentiation by regulating BMP signaling in simulated microgravity. Endocrine 65 (2). [DOI] [PubMed]
- 92.Gong L, Wu X, Li X, Ni X, Gu W, Wang X, Ji H, Hu L, Zhu L (2020) S1PR3 deficiency alleviates radiation-induced pulmonary fibrosis through the regulation of epithelial-mesenchymal transition by targeting miR-495–3p. J Cell Physiol 235 (3). [DOI] [PubMed]
- 93.Ma Y, Duan J, Hao X (2020) Down-regulated HDAC3 elevates microRNA-495–3p to restrain epithelial-mesenchymal transition and oncogenicity of melanoma cells via reducing TRAF5. J Cell Mol Med 24 (22). [DOI] [PMC free article] [PubMed]
- 94.Jiang H, Huang G, Zhao N, Zhang T, Jiang M, He Y, Zhou X, Jiang X (2018) Long non-coding RNA TPT1-AS1 promotes cell growth and metastasis in cervical cancer via acting AS a sponge for miR-324–5p. J Exp Clin Cancer Res CR 37 (1). [DOI] [PMC free article] [PubMed] [Retracted]
- 95.Zhang Q, Liu H, Zhang J, Shan L, Yibureyimu B, Nurlan A, Aerxiding P, Luo Q (2020) MiR-142–5p suppresses lung cancer cell metastasis by targeting Yin Yang 1 to regulate epithelial-mesenchymal transition. Cell Reprogramming 22 (6). [DOI] [PubMed]
- 96.Qu J, Kaufman Y, Washington I (2009) Coenzyme Q10 in the human retina. Investigative Ophthalmol Vis Sci 50 (4). [DOI] [PubMed]
- 97.Semenza GL (2000) HIF-1: mediator of physiological and pathophysiological responses to hypoxia. J Appl Physiol (Bethesda, Md: 1985) 88 (4). [DOI] [PubMed]
- 98.Li WY, Zhou HZ, Chen Y, Cai XF, Tang H, Ren JH, Wai Wong VK, Kwan Law BY, Cheng ST, Yu HB, Cai HY, Chen WX, Tang N, Zhang WL, Tao NN, Yang QX, Ren F, He L, Jiang H, Huang AL, Chen J. NAD(P)H: quinone oxidoreductase 1 overexpression in hepatocellular carcinoma potentiates apoptosis evasion through regulating stabilization of X-linked inhibitor of apoptosis protein. Cancer Lett. 2019;451:156–167. doi: 10.1016/j.canlet.2019.02.053. [DOI] [PubMed] [Google Scholar]
- 99.Vogel J, Thiel CS, Tauber S, Stockmann C, Gassmann M, Ullrich O (2019) Expression of Hypoxia-Inducible Factor 1α (HIF-1α) and Genes of Related Pathways in Altered Gravity. Int J Mol Sci 20 (2). [DOI] [PMC free article] [PubMed]
- 100.Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS, Morrison B, Stockwell BR (2012) Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149 (5). [DOI] [PMC free article] [PubMed]
- 101.Alvarez SW, Sviderskiy VO, Terzi EM, Papagiannakopoulos T, Moreira AL, Adams S, Sabatini DM, Birsoy K, Possemato R (2017) NFS1 undergoes positive selection in lung tumours and protects cells from ferroptosis. Nature 551 (7682). [DOI] [PMC free article] [PubMed]
- 102.Dixon SJ, Patel DN, Welsch M, Skouta R, Lee ED, Hayano M, Thomas AG, Gleason CE, Tatonetti NP, Slusher BS, Stockwell BR (2014) Pharmacological inhibition of cystine-glutamate exchange induces endoplasmic reticulum stress and ferroptosis. eLife 3. [DOI] [PMC free article] [PubMed]
- 103.Santoro M (2020) The Antioxidant Role of Non-mitochondrial CoQ10: Mystery Solved! Cell metabolism 31 (1). [DOI] [PubMed]
- 104.Yang S, Zhou J, Li D (2021) Functions and diseases of the retinal pigment epithelium. Front Pharmacol 12. [DOI] [PMC free article] [PubMed]
- 105.Zarbin M (2016) Cell-Based Therapy for degenerative retinal disease. Trends Mol Med 22 (2). [DOI] [PubMed]
- 106.Xia T, Rizzolo LJ (2017) Effects of diabetic retinopathy on the barrier functions of the retinal pigment epithelium. Vis Res 139. [DOI] [PubMed]
- 107.Overbey EG, da Silveira WA, Stanbouly S, Nishiyama NC, Roque-Torres GD, Pecaut MJ, Zawieja DC, Wang C, Willey JS, Delp MD, Hardiman G, Mao XW (2019) Spaceflight influences gene expression, photoreceptor integrity, and oxidative stress-related damage in the murine retina. Scientific reports 9 (1). [DOI] [PMC free article] [PubMed]
- 108.Patel ZS, Brunstetter TJ, Tarver WJ, Whitmire AM, Zwart SR, Smith SM, Huff JL (2020) Red risks for a journey to the red planet: The highest priority human health risks for a mission to Mars. NPJ Microgravity 6 (1). [DOI] [PMC free article] [PubMed]
- 109.Iosim S, MacKay M, Westover C, Mason CE (2019) Translating current biomedical therapies for long duration, deep space missions. Precision clinical medicine 2 (4). [DOI] [PMC free article] [PubMed]
- 110.Rea G, Cristofaro F, Pani G, Pascucci B, Ghuge SA, Corsetto PA, Imbriani M, Visai L, Rizzo AM (2016) Microgravity-driven remodeling of the proteome reveals insights into molecular mechanisms and signal networks involved in response to the space flight environment. J Proteomics 137. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and material used are available in the author’s labs.
Not applicable.