Abstract
At the turn of the twenty-first century, fundamental changes took place in the understanding of the structure and function of proteins and then in the appreciation of the intracellular space organization. A rather mechanistic model of the organization of living matter, where the function of proteins is determined by their rigid globular structure, and the intracellular processes occur in rigidly determined compartments, was replaced by an idea that highly dynamic and multifunctional "soft matter" lies at the heart of all living things. According this “new view”, the most important role in the spatio-temporal organization of the intracellular space is played by liquid–liquid phase transitions of biopolymers. These self-organizing cellular compartments are open dynamic systems existing at the edge of chaos. They are characterized by the exceptional structural and compositional dynamics, and their multicomponent nature and polyfunctionality provide means for the finely tuned regulation of various intracellular processes. Changes in the external conditions can cause a disruption of the biogenesis of these cellular bodies leading to the irreversible aggregation of their constituent proteins, followed by the transition to a gel-like state and the emergence of amyloid fibrils. This work represents a historical overview of changes in our understanding of the intracellular space compartmentalization. It also reflects methodological breakthroughs that led to a change in paradigms in this area of science and discusses modern ideas about the organization of the intracellular space. It is emphasized here that the membrane-less organelles have to combine a certain resistance to the changes in their environment and, at the same time, show high sensitivity to the external signals, which ensures the normal functioning of the cell.
Keywords: Intrinsically disordered proteins, Membrane-less organelles, Liquid-liquid phase separation
Introduction
A living cell is an open, dynamic, multicomponent system filled with a large number of continuously interacting molecules of water, salts, osmolytes, various small molecules of organic and inorganic origin, proteins, nucleic acids, lipids, polysaccharides, etc. For the fulfillment of its vital functions, cell needs to ensure the coordinated simultaneous implementation of many biochemical reactions, and also it has to be sufficiently resistant to the external influences. The localization of various biochemical processes within the individual (and specialized) cell compartments ensures a well-coordinated regulation of the cellular metabolism. For example, the presence of a nuclear membrane not only create a well-protected “vault” for keeping precious genetic material, but also allows eukaryotic cells to separate the transcription process that takes place in the nucleus from the translation process that takes place in the cytoplasm. Furthermore, this division (or compartmentalization) of the cellular space ensures the increased rate of chemical reactions by concentrating the reaction components inside specialized compartments. It also helps excluding the components that might inhibit these reactions or simply do not participate in them. There are also compartments responsible for the segregation of components destructive to the cell.
For a long time, it was believed that the differentiation of specialized processes in a cell is provided by the presence of biological membranes, lipid bilayers with protein molecules embedded in them, as organelles bounded by such a membrane are highly stable. Such lipid bilayers are formed due to the amphiphilicity of the molecules that form them, as corresponding phospholipids contain the hydrophilic (polar) heads, which are exposed to the aqueous medium, and the hydrophobic (non-polar) tails, which are immersed into the interior of the membrane. The membranes formed by phospholipids are permeable to low molecular weight neutral and fat-soluble substances (the phenomenon described by the solubility diffusion model, also known as the Meyer–Overton rule [1–3], correlating the solubility of a substance in an organic phase with its membrane permeability) [4], but practically are impenetrable for ions and large polar molecules. The transport across biological membranes of the substances incompatible with the spontaneous solubility-based diffusion is conducted by the carrier proteins or membrane channels, the presence of which in the membrane determines the functionality of a particular cellular compartment [5]. More generally, functions of the membranes are determined by their permeability. The compartments bounded by the lipid membrane include the nucleus, the Golgi apparatus, the endoplasmic reticulum, lysosomes, and mitochondria (and chloroplasts in plant cells). Such membrane-embedded organelles are highly stable, and this provides important means for preserving constituents isolated due to the presence of the surrounding membrane for a long time in a constantly changing intracellular environment. Furthermore, these organelles are even able to maintain their structure after being isolated from the cells. Until recently, such membrane-bounded compartmentalization was considered as the only method of the intracellular space organization.
However, along with the compartments bounded by membranes, researchers have long found “cellular bodies” that do not have a membrane. In particular, the nucleolus, the best known and most studied of the membrane-less organelles (MLOs), was discovered about 200 years ago (reviewed in [6]). Over time, more and more such MLOs were discovered, first in the nucleus, and then in the cytoplasm. However, for a long time, these compartments were of interest mainly to the cell biologists, whose main efforts were directed towards the description of these organelles and attempts to somehow characterize their purposes and functional roles in the cell. In a number of cases, the same organelles were discovered and described independently by different groups of researchers and therefore received different names. Currently, there are about a hundred organelles that do not have a membrane. Until recently, no one even thought that organelles participating in completely different processes occurring in the cell could have common properties and a common mechanism of formation. As a rule, these compartments act as microreactor-fermenters, storages of target molecules, and network hubs that regulate various cellular signaling pathways.
Only quite recently, in the mid-2010s, the understanding was achieved that the formation of all these membrane-less structures is based on the reversible liquid–liquid phase separation (LLPS), leading to the significantly increased concentrations of specific biopolymers (proteins and nucleic acids) inside the MLOs as compared to the concentrations of these constituents in the environment.
Such a radical change in the understanding of the molecular mechanisms of MLO formation became possible only after the equally significant changes took place in the concepts of the protein structure and function. In fact, at the turn of the century it became obvious that globular proteins do not exhaust the variety of functional protein structures. It turned out that a number of proteins do not have an ordered structure and nevertheless perform essential functions in the cell; i.e., they are functionally active in the absence of unique structures. Such proteins are known now as intrinsically disordered. The name emphasizes that the disorderedness of their structure is an inherent property of these proteins, which is encoded in their amino acid sequences and determines their diverse functionality. It would seem that the existence of such functional intrinsically disordered proteins (IDPs) that do not have a stable conformation requires rejection of the dogma that has been prevailing for more than 120 years that the structure of a protein determines its function. In our opinion, this is not correct, as the discovery of IDPs does not negate the importance of the structure–function paradigm, but rather extends it to a more general protein structure–function continuum model [7–9]. In fact, it is simply sufficient to interpret the terms "structure" and "function" more broadly. Disorder is just one of the structural varieties. There are many flavors of protein disorder, and proteins can be disordered to different degrees.
Unlike ordered proteins, which have unique structures, structural representation of disordered proteins must include consideration of conformational ensembles, which reflect dynamic equilibrium between different conformers separated by low energy barriers. As a result, any, even rather insignificant changes in the external conditions, such as temperature, pH or ionic strength of a solution, interaction with a partner, or post-translational modifications, can lead to a significant change in the dynamic structure of an ensemble of IDP molecules. This defines the polyfunctionality of IDPs and explains why these proteins play a dominant role in the formation of MLOs. Therefore, understanding of the MLO biogenesis is tightly linked now to the process of liquid–liquid phase separation, which, in its turn, heavily relies on intrinsically disordered proteins with their structural plasticity, binding promiscuity, and ability to be engaged in polyvalent weak interactions.
It should be noted that the concept of the "organelles without a membrane" is broader than the “membrane-less organelle” concept. A mandatory attribute of the most MLOs, which researchers have long observed in the nucleus and cytoplasm of a cell, is their formation via liquid–liquid phase separation driven by polyvalent weak interactions. On the contrary, although "organelles without a membrane" (e.g., ribosome) do not have membranes, they are formed, exist, and function not due to the LLPS, but as a result of strong intermolecular interactions. Curiously, although ribosomes have a rigid well-determined 3D structure [10–13], they are formed via the folding-at-binding mechanism, where mostly disordered ribosomal proteins [14] fold at the ribosome assembly due to the specific protein–protein and protein–rRNA interactions.
The goal of this article was to represent an overview of the key discoveries and methodological breakthroughs associated with the formation of modern ideas about the organization of the intracellular space.
Early research into the organization of the intracellular space
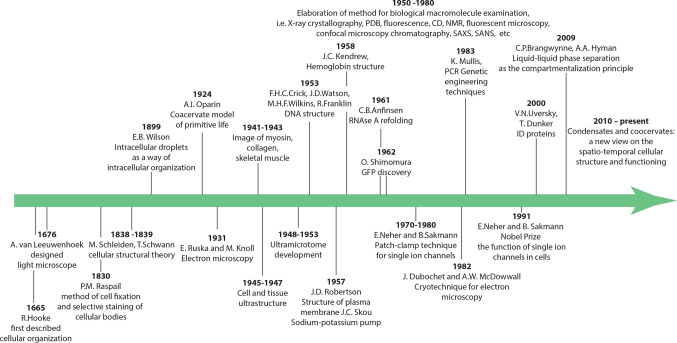
Everything has started in the middle of seventeenth century, when for the first time in the history of mankind, Robert Hooke (1635–1703) used a light microscope to describe the cellular organization of several natural objects: elderberry, dill, carrots, fly and mosquito eyes, mold, and moss [15]. Looking at the thin sections of cork under his compound microscope he noticed honey-comb like compartments. Based on these observations, he introduced the concept of "cell" derived from a Latin word “cellula”, which means a hollow space [15]. This pioneering work was paralleled by the active research of Antonie van Leeuwenhoek (1632–1723), who used lenses and a microscope of his own design to study the surrounding natural objects. It was he who first managed to attract the attention of biologists to microscopes. He was one of the first people who observe microorganisms (animalcules) and living cells, such as bacteria from ponds and tartar of the teeth, erythrocytes of a fish, spermatozoa, and protozoans [16]. However, although the resolution of the instruments designed by A. Leeuwenhoek could potentially allow him to observe some details of the intracellular structure, in his works this topic was not analyzed [17]. Therefore, the researchers of the seventeenth century confined themselves to the idea of cells as voids in a continuous mass of living tissues. The general cell theory was developed almost 175 years later, in 1838, by German botanist Matthias Jacob Schleiden (1804–1881) for plants [18], which was extended to animals in 1839, by German physiologist Theodor Schwann (1810–1882), who is considered now as a founder of modern histology by defining the cell as the basic unit of animal structure [19]. Figure 1 shows some of the most significant, in our opinion, stages in the development of biological science, which determined the modern concept of the organization of the intracellular space.
Fig. 1.
The most important stages in the development of biological science. The most significant methodological breakthroughs and scientific discoveries, which made it possible to reveal the role of the liquid–liquid phase transitions (above the time axis) and of biological lipid membranes (below the time axis) in the different compartmentalization of the intracellular space are shown
Eventually, the perception of a cell as a hollow space has changed, as some specific structures were found in these voids in a living tissue. In 1774, Felice Fontana (1730–1805) discovered nucleolus as a cellular compartment that becomes visible under the microscope during the interphase of the eukaryotic cell division (reviewed in [6]). However, the first reliable descriptions of the nucleolus were reported more than 60 years later. In fact, in 1835, Rudolf Wagner (1805–1864) described an important formation in the ovum of several mammalian species, which he called the macula germinativa, and which is known now as the nucleolus [20]. Independently, in 1837, Gabriel Gustav Valentin (1810–1883) described nucleolus as “a kind of second nucleus within the nucleus” [21]. Curiously, the nucleolus was observed well before the discovery of nucleus, which was described by Robert Brown (1773–1858) more than 50 years later, in the first half of the nineteenth century, based on the analysis of a plant cell using a microscope with achromatic lenses [22].
Some other intracellular bodies were observed as well raising a question on the internal structure of the cell, and a term “protoplasm” (the substance found inside cells) was proposed by Jan Evangelista Purkyně (1787–1869) in 1839 for the description of the "living substance" of the cell and to designate the colloid substance (cytosol) within the cell outer membrane [23]. Scientists believed that the physical foundations of life should be found in the structure of this biological substance [24]. It should be noted that already at the beginning of the twentieth century the term "protoplasm" began to gradually go out of use, since it began to go beyond the cellular structure and became associated with vitalism and such contradictory topics as primitive life, spontaneous generation of life, etc., while the attention of biologists was focused on particular elements of the cellular composition [24] (Fig. 1).
However, despite the widespread criticism of the theories of life associated with protoplasm, the study of this issue played a very important role in the understanding of the internal structure of cells and the formation of molecular biology. In what follows, the term protoplasm will be used to indicate the cytoplasm and the cell nucleus. However, for a long time, after the discovery of the nucleolus in 1774, detailed analysis of the intracellular organization was hindered by the fact that all the elements of the cytoplasm have similar optical properties and, therefore, are inaccessible to observation by conventional light microscopy. In 1879, Walther Flemming (1843–1905) expressed this observation in his study of salamander epithelial cells: “Having reached plasma, a human does not see anything at all” [25].
This issue was resolved when a method of cell fixation and selective staining of certain cellular bodies was elaborated [26]. This approach led to the emergence of a new direction of biological science, cytochemistry, and allowed discovery and description of nuclear chromatin, nucleoli, mitochondria, Golgi apparatus, endoplasmic reticulum, centrioles, and some other cytoplasmic inclusions. End of the nineteenth century was associated with the emergence of a number of theories on the structure of protoplasm: reticular, fibrillar, foam, alveolar, microsomal, and granular [24]. Furthermore, in 1899, Edmund Beecher Wilson (1856–1939) made an important observation that the researchers of the protoplasm structure often observe a fundamentally different picture of the internal structure of a cell, even when working with the same objects. In fact, observed internal structure of the cell can be very different and depends on the differences in methods used for fixation and staining of the studied cells [27]. As a result of the analysis of the literature, and having studied the internal structure of living and fixed eggs of several species of sea stars and sea urchins, Edmund Wilson came to the conclusion that the protoplasm of the eggs of all these creatures, although they differ visually, represents an emulsion consisting of a solid substance and droplets of various sizes and, apparently, of various chemical nature [27].
He was also the first to point that these droplets are liquid, being able to coalesce [27]. In fact, a fortunate combination of circumstances allowed Edmund Wilson to prove the liquid nature of the droplets forming an intracellular emulsion. This was because the microspheres in the protoplasm of the oocytes of one of the sea star species were natively pigmented. Due to this fact, during the destruction of the egg, he was able to observe how granules having the same color merge when colliding with each other, forming a larger droplet (coalescing) [27]. Interestingly, more than 100 years later, a similar approach was used by Clifford Brangwynn and Anthony Hyman to confirm the liquid nature of C. elegans P-granules in their pioneering work, which revived the interest of researchers around the world to study the role of phase transformations of biopolymers in cell life [28] (Fig. 2). Edmund Wilson was also the first to propose that the seemingly continuous matter of protoplasm consists of smaller objects that were inaccessible to the eyes of his contemporaries due to the limitations of the resolution of light microscopes of his time. Furthermore, he suggested that it is from these small and invisible elements the alveoli and fibrillar structures visible to the eye are formed, similar to the processes of coagulation of gelatin and albumin in vitro. Therefore, it seems that Edmund Beecher Wilson was the first to propose considering the phase separation of biopolymers as a way of the internal organization of the cell [27].
Fig. 2.
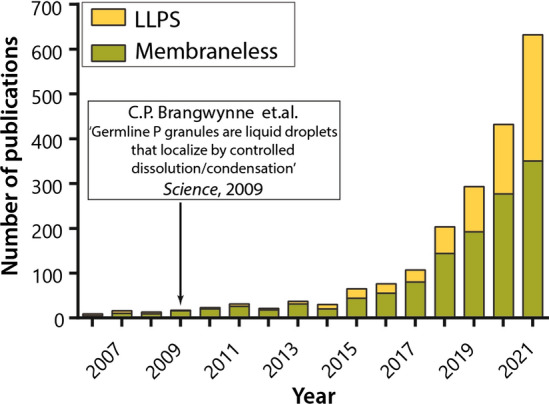
Growth in the number of publications on membrane-less organelles after the publication of the article from Brangwynne et al. [28]
These ideas where so significant that in 1924, Alexander Ivanovich Oparin (1894–1980) proposed a model, according to which the formation of liquid coacervates due to phase separations was a necessary element in the formation of primitive life [29]. In 1929, British biologist John Burdon Sanderson Haldane (1892–1964) proposed similar hypothesis of the progressive emergence of life on the primitive earth [30, 31]. However, the successes of the biochemical reductionist approach in the analyses of the cell and the advances in the study of biological membranes of the twentieth century led to the fact that the colloidal theory of the structure of protoplasm faded into the background for more than 100 years (Fig. 1). During that time, the study of phase transformations of biopolymers (proteins, polysaccharides, and nucleic acids) did not stop completely, but continued mainly by the efforts of specialists in colloidal chemistry, mainly in the interests of the food industry and pharmacology [32–37].
Studies on the principles of the intracellular organization in the twentieth century
Most advances in the analysis of the protoplasm structure in XX century are linked to the discovery and development of the electron microscopy. This technique is behind of detailed structural characterization of the plasma membrane of the cell [38] that was predicted in earlier studies based on the results of indirect methods (e.g., based on the analysis of the blood cells of man and of the rabbit, dog, guinea pig, sheep, and goat, it was stated: “It is clear that all our results fit in well with the supposition that the chromocytes are covered by a layer of fatty substances that is two molecules thick”) [39]. This technique was also used for the description of major cellular organelles surrounded by lipid membranes, such as mitochondria [40], endoplasmic reticulum [41], and Golgi apparatus [42]. In 1974, Belgian-American cell biologist and medical doctor Albert Claude (1899–1983), Belgian cytologist and biochemist Christian René Marie Joseph, Viscount de Duve (1917–2013), and Romanian-American cell biologist George Emil Pallade (1912–2008) were awarded the Nobel Prize in Physiology or Medicine for “their discoveries concerning the structural and functional organization of the cell”. In the 1970s, the actual structure of the membranes including lipid bilayer and integral and peripheral proteins was recreated using the freezing—chipping (etching) method of electron microscopy [43–45]. Already at the end of the nineteenth century, a British physiologist and biologist Charles Ernest Overton (1865–1933), who is regarded now as a pioneer of the theory of the cell membrane, discovered that the cell membrane selectively passes molecules with different properties into the cell, i.e., has a barrier function [46] and that the anesthetic potency correlates inversely with lipophilicity [2].
By the end of the twentieth century, largely due to the development of specialized approaches, such as the patch-clamp technique elaborated by Erwin Neher and Bert Sakmann in the late 1970s and early 1980s (who received The Nobel Prize in Physiology or Medicine 1991 "for their discoveries concerning the function of single ion channels in cells") [47–49] and the measurement of the conductivity of model lipid membranes (1962) [50], several mechanisms of the substance transport across biological membranes have been described in detail: passive transport, active transport, and secondary active transport [51]. In 1997, Danish chemist Jens Christian Skow (1918–2018) was awarded the Nobel Prize in Chemistry for his discovery of the sodium potassium pump [52, 53]. During this period, transporter proteins and ion channels were discovered and characterized. Based on all these developments and observations, the assumption began to prevail that biological membranes and membrane transport are the key elements for maintaining the vital activity of the cell. It is interesting that Alexander Oparin, who proposed a coacervate model of primitive organisms [29], regretted that his studies bypassed the liposomal model of the origin of life [54].
The middle of the twentieth century marked the beginning of the epoch of the biological crystallography (Fig. 1). In 1953, British molecular biologist, biophysicist, and neuroscientist Francis Harry Compton Crick (1916–2004), American molecular biologist, geneticist and zoologist James Dewey Watson, British biophysicist Maurice Hugh Frederick Wilkins (1916–2004), and English chemist and X-ray crystallographer Rosalind Franklin (1920–1958) deciphered the structure of DNA [55–57]. In 1962, The Nobel Prize in Physiology or Medicine was awarded to James Watson, Francis Crick and Maurice Wilkins for their 1953 discovery of the molecular structure of DNA, but Rosalind Franklin, who died from cancer at the age of 37, was not so honored, despite the fact that her work was central to the understanding of the molecular structures of DNA.
In 1958 and 1960, English biochemist, crystallographer, and science administrator Sir John Cowdery Kendrew (1917–1997) and British molecular biologist Max Ferdinand Perutz (1914–2002) were the first to determine the crystal structures of globular proteins, hemoglobin and myoglobin [58, 59]. In 1962, they received the Nobel Prize in Chemistry, for their work on the analysis of the structure of heme-containing proteins. Solving the first crystal structures of globular proteins was paralleled by the classic experiments of Christian Boehmer Anfinsen Jr. (1916–1995) on ribonuclease refolding [60]. This work unequivocally showed that the spatial structure of a globular protein is determined by its primary sequence, and for this research, Christian Anfinsen received the Nobel Prize in Chemistry in 1972. In the subsequent years, advances in the X-ray diffraction analysis have significantly strengthened the idea that the unique structure of a protein is strictly determined by its amino acid sequence and, in its turn, defines unique protein function.
In 1975, British biophysicist and chemist Sir Aaron Klug (1926–2018) reconstructed the three-dimensional structure of the nucleic acid–protein complex using a novel technique he elaborated, crystallographic electron microscopy [61]. He was awarded the 1982 Nobel Prize in Chemistry for this development of crystallographic electron microscopy and structural characterization of biologically important nucleic acid-protein complexes. In 1984, the structure of a large membrane protein complex, the photosynthetic reaction center from Rhodopseudomonas viridis, was determined [62], and a German biochemist Hartmut Michel received the 1988 Nobel Prize in Chemistry for this significant contribution towards the understanding of photosynthesis. In 1992, an American physician and molecular biologist Peter Agre and co-workers discovered aquaporin membrane water channels, thereby answering a long-standing biophysical question of how water crosses biological membranes specifically [63]. In 1998, an American biophysicist and neuroscientist Roderick McKinnon deciphered the structure and explained the function of the potassium transmembrane channel [64]. These were great achievements in understanding of the molecular mechanisms of membrane channel actions, and, in 2003, Peter Agre and Roderick MacKinnon shared the 2003 Nobel Prize in Chemistry for "discoveries concerning channels in cell membranes.
In parallel with structural studies, the processes of enzymatic catalysis were actively analyzed in the twentieth century as well. In 1913, a German biochemist, physical chemist, and physician Leonor Michaelis (1875–1949) and a Canadian bio-medical and medical researcher Maud Leonora Menten (1879–1960), based on their experimental analysis of the sucrose hydrolysis by the enzyme invertase, confirmed the model of enzymatic kinetics [65] proposed in 1902 by a French-Russian physical chemist and physiologist Victor Henri (1872–1940) [66], which establishes a relationship between the reaction rate and the substrate concentration, thereby reducing the activity of biological agents to the basic laws of physical chemistry. To explain the high specificity of enzymatic processes, two models were suggested as follows:
Proposed in 1894 by a German chemist Hermann Emil Louis Fischer (1852–1919) a classic "lock-and-key" hypothesis, which suggested that there is an exact fit between the substrate and the active site of the enzyme, in the same way as a key fits into a lock [67].
Proposed in 1958 by an American biochemist Daniel E. Koshland Jr. (1920–2007) an induced-fit model of substrate–protein interaction [68]. This model can be described as “hand–glove” correspondence, where the active center of the enzyme changes its conformation after binding of the substrate and thereby addresses the importance of structural flexibility of proteins for their function. In short, this model suggested that the active site of an enzyme and a substrate not necessarily have the complimentary shapes. However, a substrate can bind to an enzyme and induces a change in the structure of enzyme’s active site: the amino acids constituting the active site are tuned into a precise conformation, bringing the chemical groups of the active site into positions which enable the enzyme to perform its catalytic function most effectively [68].
More recently, Hungarian biochemists Ferenc Bruno Straub (1914–1996) and Gertrud Szabolcsi (1923–1993) proposed a fluctuation fit (or conformational selection) model suggesting selective binding of a ligand to a single conformation in the conformational ensemble [69]. According to this model, among the conformations of the dynamically fluctuating protein, the ligand selects the one, which is compatible with binding, and shifts the conformational ensemble towards this state [69]. In 1965, a French biochemist Jacques Monod (1910–1976), an American molecular biologist and biophysicist Jeffries Wyman (1901–1995), and a French neuroscientist Jean-Pierre Changeux proposed a Monod–Wyman–Changeux model postulating the existence in a protein molecule of a small number of discrete conformational states independent of ligand structure and occupancy [70]. In 1966, Daniel E. Koshland Jr. (1920–2007), a Hungarian-American biochemist George Némethy, and an American biochemist David L. Filmer proposed sequential, induced-fit binding model to explain experimental data on the interaction mechanisms of the multi-subunit proteins [71]. Here, the existence of multiple conformational states in the proteins and the ability of ligand binding to induce tertiary changes complementary to ligand structures were postulated. According to this model, ligand binding promotes “a progressive change” of conformation and the required conformation in a subunit is achieved only when the ligand is bound to it [71]. Finally, in 1981, based on the NMR analysis of the ligand binding by receptors, a British physician and pharmacologist Arnold Stanley Vincent Burgen [72] and an American biophysicist Angela M. Gronenborn [73] provided experimental evidence for the existence of the conformational selection accompanying ligand–protein interaction.
Therefore, based on all these developments, rather mechanistic ideas about principles of the organization of the living matter were formed in the twentieth century. According to these concepts, proteins are globules possessing unique crystal-like tertiary structures with strictly determined atomic positions. These unique structures of proteins determine their unique functions. The vital activities of the cell are carried out due to the strictly determined biochemical processes conducted by complex molecular machines with rigid three-dimensional structures, whereas the biological membranes of intracellular organelles serve as part of these machines and separate enzymatic processes in space.
First inconsistencies in the “harmonious picture of the universe”
Despite the general belief in the validity of protein structure–function paradigm, more and more exceptions from the rule were found, and more and more cases of “uncrystallisable” proteins or biologically active proteins without unique structures were reported [74]. Increasingly, researchers came across proteins, which upon refolding, did not form the native state, but stayed misfolded and formed aggregates. However, in most cases, these observations were taken as artifacts and explained by the errors of the experimenters.
At the same time, information was accumulating about various intracellular formations, which are known now as membrane-less organelles. Among noticeable events in this field were detailed characterization of the ultrastructure of the largest and most studied MLO—the nucleolus, which was shown to contain three main structural components: a fibrillar center, a dense fibrillar component, and a granular component [75], and which was shown to serve as a site of the ribosome biogenesis [76]. Another noticeable event of that time is a rediscovery of the Cajal (coiled) bodies (CBs), by Ariane Monneron and W. Bernhard in 1969 [77]. CBs, which are responsible for processing small nuclear and small nucleolar RNAs, were first described in 1903 by a Spanish neuroscientist, pathologist, and histologist Santiago Ramón y Cajal (1852–1934), who called them nucleolar accessory bodies because of their close association with nucleoli [78]. CBs were discovered by a “reduced silver staining method”, elaborated by Cajal as a simple and reliable procedure for demonstrating diverse cellular structures [79]. Curiously, these nuclear MLOs were (re)discovered multiple times independently by scientists from different research fields generating a realm of different names, such as "sphere organelles", "Binnenkörper (endobody)”, "nucleolar bodies", "coiled bodies", or “perichromatin fibrils” used to describe the same structure [80].
Unfortunately, the method of electron microscopy and used in the XX century methods of light microscopy did not allow observing the dynamic properties of MLOs. Because of these limitations, until the beginning of the twenty-first century, these cellular bodies were perceived as ordered macromolecular complexes or loci, in which the local concentration of any component is increased due to the work of carriers, anchoring or synthesis of these substances [81].
Paradigm shift: emerging of the protein intrinsic disorder concept
The turn of the twenty-first century was characterized by the recognition that protein functionality is not necessarily linked to unique structure. Instead, biological functions were found for many structure-less proteins, which are known now as intrinsically disordered proteins (IDPs) or hybrid proteins containing ordered domains and intrinsically disordered regions (IDRs) to emphasize that their inability to form a globular structure is an intrinsic (inherent) property of their particular polypeptide chain, due to the peculiarities of its amino acid composition and sequence [82–84]. In fact, it was shown that the ability of a polypeptide chain to form an ordered globular structure is associated with the value of the total charge (no matter what sign) and the number of hydrophobic residues [83, 85]. The lower the specific content of the amino acids with hydrophobic side chains and the greater the total charge of the polypeptide chain, the less likely this polypeptide chain will form a compact globular structure [83, 85].
It became clear that proteins can be functionally active not only in a well-folded globular state, but also in a partially or fully unfolded state [82–84, 86–97]. The ordered globular structure is mostly found in enzymes—proteins, the catalytic function of which is strictly determined and requires unique configuration of an active site. The proteins from the regulatory and signaling pathways involved in interaction with a large number of often unrelated partners require much greater structural pliability to function, and, therefore, these proteins are partially or completely unstructured in their native states [90, 98–105]. Overall, despite the lack of unique structures in IDPs/IDRs, the functional range of these proteins/regions is remarkably broad and complements functions of ordered proteins and domains [82, 84, 86, 87, 89, 90, 92, 95–97, 99, 103, 104, 106–111].
Compact globular structures are formed in proteins in aqueous media when the peculiarities of the polypeptide chain allow strong and specific intramolecular interactions. Whether a polypeptide chain can fold into a compact globular form depends on the ratio of the number of hydrophobic and charged residues that make up the amino acid composition of a protein. Many partially or completely disordered proteins can form a compact structure in complexes with their partners, with such a binding-induced (semi)ordered structure originating due to intermolecular interactions between the atoms of the polypeptide chain of the protein and the partner [14, 82–84, 90, 92–96, 102–104, 106, 112–119]. Furthermore, often, the functionality of IDPs/IDRs depends on the presence of disorder-based binding sites, called molecular recognition features (MoRFs), which are interaction-prone disordered regions that can fold at binding to specific partners [120–123]. More generally, the capability to be engaged in interactions with various binding proteins, such as other proteins, lipids, membranes, nucleic acids, polysaccharides, and small molecules of organic and inorganic origin represents an important functional feature of IDPs/IDRs. They are able to form a multitude of static complexes characterized by very unusual and complex topologies ranging from the interaction-induced local folding generating structural elements bound to the surface of a partner to folding of a whole IDP, and from wrapping around the binding partner to penetrating deep inside the binding partner, or to form semi-static or dynamic complexes [124].
Many IDPs/IDRs can form fuzzy complexes either with “the flanking fuzziness” in the bound state disordered regions flanking the interaction interface but not the interface itself, or remain disordered or characterized by “the random fuzziness” attributed to the ability to not fold at interaction and preserve highly dynamic structure in the bound state [125–131]. Some IDPs/IDPRs are able to bind to multiple partners and to gain very different structures in the bound state, and this adjustable promiscuity represents an important means for the increased complexity of the disorder-based interactomes [99, 132]. Curiously, the presence of intrinsic disorder allows formation of the most and the least stable protein complexes. Importantly, although specific disorder-based interactions often engage binding-induced folding, formation of stable complexes does not always require folding, and IDPs/IDPRs were shown to form tight complexes (with the affinity approaching picomolar levels) without gaining any ordered structure and retaining long-range flexibility and highly dynamic character [133].
Intermolecular interactions of protein molecules with each other can lead to the formation of various associates, ranging from oligomers to amorphous aggregates, and amyloid-like fibrils. For such contacts to occur, it is necessary to have hydrophobic regions of the polypeptide chain exposed to the solvent. Since IDPs/IDRs do not have unique structures, many of them would satisfy such requirements. Therefore, aggregation of partially or completely disordered proteins is a very common phenomenon, and many IDPs/IDRs are related to various human amyloidoses and other proteinopathies [134–137]. More generally, pathogenesis of many human diseases is linked to the misbehavior and deregulation of IDPs/IDRs [138–140].
At the turn of the century, it seemed that IDPs and IDRs were just an exception to a well-known and broadly accepted rule. Indeed, thanks to the advances in the X-ray diffraction analysis, by this time structural information on a large number of globular proteins had been collected in the protein data bank (PDB), and it seemed that ordered proteins constitute the bulk of protein universe. However, later it turned out that IDPs/IDPRs are not mere exceptions, but are universally present in all living organisms. It is also clear that the abundance and penetrance of disorder typically increase with an increase in organism complexity [141–144]. In fact, from 25 to 30% of eukaryotic proteins are predicted to be mostly disordered, more than half of eukaryotic proteins have long regions of disorder [141], and more than 70% of signaling proteins have long disordered regions [104]. IDPs and hybrid proteins with long IDRs could not get into PDB [145–147], since they do not easily crystallize under the traditionally used conditions.
Discovery of the protein intrinsic disorder phenomenon has opened a new page in the protein science, eventually leading to the transformation of the classical but limited protein structure–function paradigm into the more general protein–structure continuum model [7–9]. Here, the peculiarities of the amino acid sequences determine the multi-level spatiotemporal heterogeneity of IDPs and defines their mosaic structure, where different parts of a protein can be (dis)ordered to different degrees. As a result, the entire IDP can be described as a combination of foldons (independent foldable units of a protein), inducible foldons (disordered regions that can fold at least in part due to the interaction with binding partners), morphing inducible foldons (disordered regions that differently fold at interaction with different binding partners), non-foldons (non-foldable protein regions), semi-foldons (regions that are always in a semi-folded form), and unfoldons (ordered regions that have to undergo an order-to-disorder transition in order to make a protein functional) [148, 149]. Importantly, such spatiotemporal heterogeneity of IDPs defines their multifunctionality, as differently (dis)ordered regions of a protein might have well-defined and specific functions [150]. Therefore, IDPs/IDRs are structurally and functionally heterogeneous complex systems acting within the frames of the protein structure–function continuum model indicating that the actual gene–protein relationship is better described by the “one gene—many functional proteins” or “one gene—many functions” model rather than the classical (but heavily oversimplified) “one gene—one protein” paradigm [150, 151]. Furthermore, the structural flexibility of IDPs/IDRs defines the broad spectrum of means for their functional regulation and control [86, 124, 148, 152], including various post-translational modifications (PTMs) [153, 154]. Important for this review, structural pliability and the capability of IDPs/IDRs to be involved in weak multivalent interactions define the broad engagement of these proteins in the biological LLPS and MLO biogenesis [150, 155–157].
Changes in the concept of the intracellular space compartmentalization in the twenty-first century
The mechanistic theory of the protoplasm structure was shattered in 2009 by the pioneer work of Clifford P. Brangwynne, Frank Jülicher, Anthony A. Hyman and their colleagues, where the formation mechanism of the P-granules in germ cells was established [28]. This work began as a part of the Summer School for Young Scientists on New Microscopy Techniques at the Marine Biological Laboratory, Woods Hole, Massachusetts. As a learning task, the teachers of the school asked the students to observe the process of the P-granule formation in the embryos of the free-living transparent nematode Caenorhabditis elegans. Summer school students David Hole and Lindsay Moore were able to capture on video how P-granules stained with green fluorescent protein fused with each other upon collision to form a larger body (coalesced), like the oil droplets in water. The researchers realized that this behavior is not typical of solid, rigid structures. Therefore, P-granules should be considered as liquid droplets. Researchers did not immediately grasp the scope of the discovery. However, after the summer school was over, teachers Anthony A. Hyman and Clifford P. Brangwynne, who assigned the students this task, returned to this study. They conducted several more experiments that allowed them to prove that P-granules are liquid condensates and to quantitatively characterize their properties, such as viscosity and surface tension. In addition, they managed to record the processes of condensation and dissolution of these bodies. Due to these data, the authors suggested that the formation of P-granules occurs due to the spontaneous separation of a supersaturated polymer solution into two phases, one of which is enriched with a polymer substance, and the other is diluted. More importantly, the authors suggested that the mechanism of P-granule formation represents an example of the general principle of the organization of the cytoplasm via controlled liquid–liquid phase separation and may even be a mechanism of self-assembly of early life forms [28, 158]. Therefore, the results of this work brought back the forgotten hypotheses of Alexander Oparin and John Burdon Sanderson Haldane proposed about a century ago.
It is worth noting that Hyman's group was not the first group to record liquid properties of MLOs. For example, in 2005, it was proposed that Cajal bodies can be describe as “semi-liquid” spheres due to their permeability to external probe molecules, low density compared to the surrounding nucleoplasm, and surprisingly spherical shape [159]. In 2008, when studying the morphology and distribution of Zebrafish germ cell granules, it was found that these granules are capable of coalescence, although the authors of this work did not make assumptions about the state of matter of these bodies [160]. Since the liquid-like properties were earlier described for the Cajal bodies [159] and Zebrafish germ cell granules [160], the actual importance of the work of the group of Anthony A. Hyman is in the generalization of the hypothesis of the mechanism of MLO formation as the major principle of the cytoplasm organization. In addition, in their work, the authors used physical approaches available to many biological laboratories, which make it possible to quantitatively determine specific parameters characteristic of the liquid phase, such as the diffusion coefficient, surface tension, and viscosity [28]. Knowledge of these parameters makes it possible to reliably determine the state of matter of the system and compare the properties of various MLOs.
Two years later, similar studies were performed for the nucleolus [161], and later for other MLOs [162]. To date, the functional role of condensation of macromolecules in the cell has been confirmed by a large number of examples [163]. In 2012, an American group from Howard Hughes Medical Institute and University of Texas Southwestern Medical Center led by Michael K. Rosen succeeded in the in vitro reconstruction of the structure and functionality of cellular membrane-less compartments from purified protein and RNA preparations [164]. These authors showed that interactions between various multivalent macromolecules (including multi-domain proteins and RNA) in aqueous solution produce sharp liquid–liquid-demixing phase separations, generating micrometre-sized liquid droplets [164]. Using the actin-regulatory protein neural Wiskott-Aldrich syndrome protein (N-WASP) and its established biological partners NCK (also known as SH2/SH3 adaptor protein NCK-α) and phosphorylated nephrin, the authors also established that phase separation in this system coincides with a sharp increase in the activity of this system towards an actin nucleation factor, the Arp2/3 complex [164]. The authors also pointed that the LLPS is governed by the degree of nephrin phosphorylation, suggesting that PTMs can control biogenesis of the resulting biomolecular condensate [164]. Having a possibility to induce phase separation in vitro provides important means for the determination of the necessary and sufficient conditions for the formation of biomolecular condensates, as well as helps in determining the functions they perform.
Generally speaking, the separation of polymer solutions into phases is a well-known and well-studied process. In fact, the ability, under specific conditions, to undergo liquid–liquid phase transitions (LLPTs) leading to the LLPS represents a general property of various biological and non-biological polymers. For example, biocompatible aqueous two-phase systems (ATPSs, which are typically polymer-based systems) are widely used in biotechnology for multiple purposes [165, 166]. MLOs that exist as liquid droplets in cytoplasm, nucleoplasm, mitochondrial matrix, or stroma of the chloroplasts reflect the importance of phase separation in biology [155, 156, 162, 167–172]. There is a myriad of different biomolecular condensates in eukaryotic cells, the incomplete list of which includes cytoplasmic centrosomes [173], germline P-granules (germ cell granules or nuage) [28, 174], neuronal RNA granules [175], processing or P-bodies [176], and stress granules (SGs) [177], chloroplast stress granules (cpSGs) [178] and mitochondrial RNA granules [179]. In the nucleosome, there are Cajal bodies (CBs) [180], chromatin [181], cleavage bodies [182], histone locus bodies (HLBs) [183], nuclear gems or Gemini of coiled of Cajal bodies [184, 185], nuclear speckles or interchromatin granule clusters [186], nuclear pores [187], nuclear stress bodies (nSBs) [188, 189], nucleolus [190], Oct1/PTF/ transcription (OPT) domains [191], perinucleolar compartment (PNC) [192], paraspeckles [193], PML-bodies (PML oncogenic domains, PODs) [194], polycomb bodies (PcG bodies) [195], and Sam68 nuclear bodies (SNBs) [192]. The key components, characteristic size and biological role of some of listed above MLOs are given in Table 1. Therefore, for many proteins, this property to undergo LLPS was discovered long before Rosen's work. This phenomenon was especially well known to crystallographers, for whom the formation of a liquid–liquid interface is a favorable condition on the way to crystallization of a target protein. Despite this, the realization of the fact that the condensation of macromolecules with the formation of two liquid phases is the principle of the organization of the intracellular space common to all cells came only recently [196].
Table 1.
Examples of the various biomolecular condensates and their characteristics
| Organelle | Year of discovery | Key components | Size | Associated processes | Reference | |
|---|---|---|---|---|---|---|
| Key proteins | RNA | |||||
| Nucleolus |
1774 1835 1837 |
Fibrillarin, nucleophosmin | rRNA | 6 µm | Ribosome biogenesis | [20, 21, 190, 319, 320] |
| Cajal bodies |
1903 1969 |
Coilin, survival motor neuron protein (SMN1) |
snRNA snoRNA |
0.3–1.0 µm | Assembling spliceosomal small nuclear ribonucleoproteins | [77, 180, 321, 322] |
| PML bodies | 1960 | PML, SUMO-1, Sp100 | None | 0.1–1.0 µm | Apoptotic signaling anti-viral defense transcription regulation | [194, 258, 323] |
| Nuclear speckles | 1910 | SRSF1, SRSF2, SPOP |
Poly(A) + RNA; lncRNA MALAT1 |
0.5 µm to several µm | mRNA splicing | [186, 283, 321] |
| Cleavage bodies | 1999 | DDX1, CstF-64 |
(Pre-)U-snRNAs; 7SK snRNA C/D & H/ACA box snoRNAs TERC |
0.3–1 µm | Processing of 3′-end of pre-mRNA | [182, 324] |
| Histone locus bodies (HLBs) | 1960 | FLASH, LSM11, NPAT |
Histone mRNAs U7 snRNA U2 snRNA Y3/Y3** ncRNA |
0.3–1.0 µm | Processing of histone pre-mRNA | [183, 325] |
| Nuclear gems | 1996 | SMN, Sm proteins | None | 0.1 µm (A, C and E) or 30 μm (B, D and F) | Assembly of the spliceosomal small nuclear ribonucleoproteins (snRNPs) | [184, 185, 326] |
| Oct1/PTF/transcription (OPT) domains | 1998 | 53PB1, γH2AX, MDC1, OPT | None | 5–25 µm | Maintenance of the genome integrity | [191, 327] |
| Balbiani body* |
1845 1887 |
Bucky ball, Xvelo, Hba1, Hbg, Cup and Orb | mRNA | Up to 50 µm | Embryonic determinant that specifies germline identity by forming germ plasm (frogs and fish). For other species, Bb function(s) remains either highly hypothetical or completely unknown | [206, 328, 329] |
| Paraspeckles | 2002 |
FUS NONO SFPQ HNRNPK DAZAP1 RBM14 HNRNPH3 |
lncRNA NEAT1 | 0.5–1 µm | Storage of certain RNAs | [193, 330] |
| Polycomb bodies (PcG bodies) | 1998 | Polycomb-group proteins, PcG, Bmi1 | None | 0.2–1.5 µm | Gene silencing | [195, 331] |
| Sam68 nuclear bodies (SNBs) | 1999 |
Sam68 DBC1 SLM-1 T-STAR hnRNP L |
arcRNA | 0.3–1.0 μm | SNB may act as the regulatory factory of coupled transcription-splicing events | [192, 332–334] |
| P-bodies | 1997 |
Dcp1a Lsm proteins Rck/p54 GW182 |
Translationally repressed mRNA | 5.1–15 µm | mRNA storage and translational regulation | [175, 176, 335, 336] |
| Stress granules | 1999 |
FUS hnRNPA1 |
Poly-(A) + mRNA associated with PABP | 0.1–2.0 μm | mRNA decay and silencing | [177, 336, 337] |
The highly dynamic nature of MLOs, as well as the disordered structure of the proteins that form them, has seriously shaken the positions of the deterministic theory of the organization of the intracellular space and the linear biochemistry of intracellular processes. Currently, the attention of researchers is increasingly focused on the stochasticity of cellular processes, where the cell is considered as a system with the properties of highly dynamic "soft" matter [197].
Principles of membrane-less organelles’ formation, structure, and function
The principle components of MLOs are proteins containing intrinsically disordered domains [198], which, due to their polyvalence and high degree of conformational heterogeneity and flexibility, are capable of undergoing liquid–liquid phase separation under physiological conditions in contrast to proteins with a rigid three-dimensional structure [199]. Importantly, only weak nonspecific multiple interactions of disordered regions of scaffold proteins or RNA can be the involved in the initial stages of the MLO formation. In the work by [200], it was shown that the formation of S–S bonds and interaction of RBCC motifs of PML isoforms (which were long believed to act as driving force of PML-body formation) are possible only in the process of primary small PML-bodies maturation, when the concentration of PML-proteins (scaffold proteins of PML-bodies) is already significantly (100/1000 times) higher than their concentration in nucleus (Fig. 3). Interestingly, 25–35% of proteins in eukaryotic proteomes do not have a stable tertiary and/or secondary structure in their native state, and more than 50% of proteins contain long disordered regions in their structure.
Fig. 3.
PML-bodies formation. It is shown schematically that at the first stage, a liquid–liquid phase separation should occur, leading to a manifold increase in the concentration of scaffold proteins due to weak interactions, only after this the formation of specific interactions such as S–S bonds, RBCC motive, etc., which ultimately lead to the formation of mature MLO, in the case of PML-bodies they are large toroidal bodies are possible. Disordered regions of scaffold proteins are primarily involved in the formation of nonspecific interactions. In the case of PML proteins, these are the C-termini of PML-isoforms. Designations used in the figure are given in the lower left corner. Schematic presentation of different alternatively spliced isoforms of human PML protein are also shown: Mean disorder score determined by averaging the outputs of, IUPred2A Short IUPred2A Long, PONDR® VLXT, PONDR® VL3, PONDR® VSL2B and PONDR® FIT. The curve is superimposed with the positions of cysteine residues (black circles in N-terminal and red circles in C-terminal) and lysine residues (green circles); The generalized domain structure, indicating positions of the Ring domain (R—amino-acid residues 45–105), Box 1 (B1—residues 124–166), Box 2 (B2—residues 184–230), Coil-coil (CC—residues 229–323), Positions of the nuclear localization signal (NLS—residues 476–490), SUMO-interacting motif (SIM—residues 556–562) and nuclear export signal (NES—residues ~ 704–713, exclusive to isoform I); PML-I exons; The length of each isoform (colored line) and the length of the fragment in which the amino acid residues are the same in PML-I isoforms (black line)
Currently MLOs are defined as dynamic biomolecular condensates formed as a result LLPS. The size of MLOs extends from nanometers (signaling puncta of the adaptor protein Lat [201] to microns (e.g., nucleolus, stress granules, etc. [202–204], see Table 1). Multivalent interactions are considered to drive the MLO formation. The required multivalency may be achieved by multiple copies of folded domains (such as RRM domains) interconnected by flexible linkers or by IDRs [163] of the associated with LLPS proteins. IDRs involved in phase transitions typically have low-complexity sequences, which are repetitive and enriched in amino acids having polar (G, Q, N, S), positively charged (R, K), negatively charged (D, E) or aromatic (F, Y) side chains. The sequences often contain multiple short motifs such as YG/S-, FG-, RG-, GY-, KSPEA-, SY- and Q/N-rich regions, as well as blocks of alternating charges. Such sequence promotes weak interactions, such as charge–charge, pi–pi, and cation–pi [168]. When the polymers are separated into phases, the entropy of the system decreases only slightly compared to the decrease in the entropy of the system when the equivalent amount of their monomer units is separated into phases. Therefore, even a small decrease in the enthalpy of the system during the interaction of polymers causes a LLPS with clear boundaries between the phases. It should be borne in mind that biopolymers function in a cell under conditions of a limited free volume and a minimum of free water, i.e., under crowding conditions. These conditions significantly limit the possible conformational states of the polymer and thereby facilitate the separation of biopolymers into phases.
There is a set of physico-chemical features that define droplets as phase-separated liquid condensates in the cells: their ability to maintain spherical shape, coalescence, fluidity under shear stress, high viscosity, molecular dynamicity, assembly cooperativity, concentration-dependent size scaling, and sensitivity to 1,6-hexanediol [162, 205].
The first two signs are governed by surface tension of the dense phase, which helps the droplets to maintain a spherical shape and coalescence after touching each other. Besides, liquid condensates are distinguished by their ability to flow when subjected to shear stress. However, MLOs may exhibit also elastic properties (keep a constant deformed shape under stress). For instance, it has been shown that Balbiani bodies behave as solids [206]. Microrheology analysis revealed a high viscosity of liquid-like MLOs ranging from 1 to 103 Pa × s [162]. Another feature of MLOs is the rapid exchange of molecules within the liquid-like phase and with the surrounding milieu. The fluorescence recovery of GFP-PGL-1 in P granules occurred on a time scale of 5.9 ± 0.6 s [28]. The assembly of condensates is a cooperative process and the dense phase volume grows continuously with the macromolecule concentration increase until the system entering a single-phase regime, where the molecule’s concentration corresponds to the concentration of dense phase [204]. The sensitivity of condensates to 1,6-hexanediol (a mild perturbant of hydrophobic interactions) proposed as an indicator of LLPS is not definitive, since phase separation involves various types of interactions including pi–pi, cation–pi, and electrostatic interactions, not all of which are impaired by hexanediol [207]. The unique properties of membrane-less organelles, such as the ability to selectively concentrate or exclude certain molecules, make them implicated in a variety of cellular processes.
The concentrations of specific proteins, nucleic acids, and small molecules is locally increased within MLOs. The concentration of species inside condensates ranges from 1 to 10 µM with a partition coefficient reaching 100 [204]. This property allows MLOs to enhance the rate of cellular biochemical reactions concentrating reactants together. An illustrative example is given by the enhancement of microtubule nucleation occurring upon tubulin co-condensation with microtubule-associated protein TPX2 [208]. The ability to preferably concentrate certain molecules over others may also provide specificity in signaling or metabolic networks through accelerating some reactions over others. For instance, only one of the two substrates recruited into artificial condensates reconstituting the SUMOylation cascade was SUMOylated efficiently [209].
The preferable exclusion of some molecules from condensates (partition coefficient < 1) is expected to slow down biological reactions in the case of separation of one reaction component from another or enhance the catalytic activity of enzymes in the case of depletion of the droplets in reaction inhibitors. This is exemplified by the suppression of the auxin-dependent nuclear translocation and gene regulatory activities of auxin response factor proteins (ARF7 and ARF19) by its cytosolic sequestration [210], and the cluster stabilizing effect of the CD45 phosphatase exclusion from the condensates formed by LAT and group of adaptor proteins via reduced LAT dephosphorylation [211].
Along with the concentration regulation of the rates of biochemical reactions inside MLOs, many other physical features of condensates may affect the processes occurring in them. For example, molecular crowding inside the condensate may alter the activity of resident enzymes by decreasing the accessible to molecules volume. Additionally, the porous internal structure of the condensate can act as a filter for molecules with certain size/structure or slow down the diffusion rate of molecules inside [163]. The increased actin filament nucleation activity of N-WASP and Arp2/3 within the nephrin signaling clusters was found to be caused by the prolonged stay of the proteins on the membrane [212]. Scaffold structure within the condensate may also enhance the biochemical reaction by tethering the substrate and enzyme in close proximity. Such a mechanism has been suggested based on the analysis of the rate of SUMOylation reaction within synthetic condensates containing SUMOylation enzyme cascade [209]. The biochemical environment within the MLOs distinct from that of bulk phase may prevent protein aggregation, for example, misfolded proteins are protected by association with nucleolar proteins, such as nucleophosmin (NPM1) in the nucleolar granular component [213]. The cooperativity of the droplet formation process and the concentration-dependent scaling of the condensate size make MLOs applicable to buffering of cellular components [214]. The sensitivity of LLPS to solution properties (temperature, pH and ionic strength) define the potential for MLOs to be exploited as a means of sensing and responding to deleterious environmental conditions, such as the condensation of mRNA poly(A) binding protein Pab1 in budding yeast in response to the heat shock [215]. The viscoelastic properties of condensates allow generation of forces that can be used, for example, to deform the plasma membrane [216].
The main function of MLOs is the implementation of spatio-temporal control of biochemical reactions occurring inside the cell. For example, liquid-like RNA transport granules in neurons have been shown to serve for translocation of mRNA packaged in them from the neuron body to the ends of axons and dendrites by attaching to lysosomes through annexin A11 (Liao et al., 2019). Another example is the spatially coordinated nucleation of microtubules occurring upon the co-condensation of tubulin with microtubule-associated protein TPX2 [208]. Unlike the classical organelles surrounded by a lipid membrane, MLOs are freely exchanging their content with the environment. Unlike traditional membrane-embedded organelles, the transport of substances in and out of the MLOs is not complicated by the need of involvement of carrier proteins in this process, as well as by the requirement to have specific signals of import and export. This is because to capture the target protein inside the droplet, general physico-chemical properties are used, such as charge, polarity of the amino acids that make up the protein, etc. [168]. Another advantage of MLOs over the membrane-bound organelles is the ability of MLOs to form rapidly in response to minimal changes in the intracellular environment (temperature, pH, ionic strength, concentration of the target protein, etc.) [163]. These properties allow the cell to use phase separation to form a timely and effective response to the stress [217–219]. For example, inhibition of the Pol I-, II- and III-dependent RNA transcription by actinomycin D lead re-organization of both nuclear and cytoplasmic membrane-less organelles [220]. Importantly, the characteristics of the biochemical environment within the MLOs may differ from the conditions of the surrounding cytoplasm or nucleoplasm. This makes it possible to implement unique strategies for the regulation of biochemical reactions inside the cell. For example, inside MLOs formed by the Ddx4 protein, conditions are created that favor melting of the DNA double helix [219].
It was recently recognized that the disturbances in the maintenance of the MLO homeostasis can have detrimental consequences, e.g. leading to the formation of amyloid-like fibrils within them [217, 219, 221–226]. The aberrant liquid-to-solid phase transition and amyloid fibril formation have been shown for FUS [227], hnRNPA1 [217], TDP-43 [228], Tau [229, 230], and α-synuclein [231]. The formation of amyloid fibrils is closely associated with the development of several serious diseases, such as Alzheimer's disease, Parkinson's disease, prion diseases, amyotrophic lateral sclerosis, frontotemporal dementia (FTD), diabetes mellitus, etc. It is possible that the processes associated with the disruption of the functioning of MLOs are the cause of the development of other common diseases, including carcinogenesis [200, 232]. Therefore, it is quite obvious that studies aimed at the analysis of the principles underlying the phase transitions occurring in protein solutions will find practical applications in medicine and will make it possible to understand how to predict and correct dysfunctions of MLOs.
Stress granules, cytoplasmic MLOs formed in response to stress, are studied quite well [233]. This is partly related to the ease of managing their assembly/disassembly and the ability to simulate these processes: these MLOs appear in response to stress and dissolve after the stress removal. Transient stress-induced compartments formed via liquid–liquid phase separation most probably benefit cell survival by playing the role of temporal cellular preservation clusters for hundreds of proteins that have to be inactivated during the period of post-stress recovery, while allowing cell to save resources on their degradation and de novo synthesis. The main function of cytoplasmic stress granules is to ensure cell survival under stress conditions by preserving untranslated mRNA and a pool of specific cytoplasmic proteins, eliminating the need for their de novo synthesis, as well as to regulate the stress response [234].
The formation of stress granules is initiated by the dissociation of the polyribosomal complex, which releases into the cytoplasm untranslated mRNA associated with elements of the cell's translational machinery, in particular, with the small subunit of the ribosome [233]. mRNAs participate in the formation of stress granules both passively and actively [235]. On the one hand, a sharp increase in the concentration of mRNA in the cell cytoplasm contributes molecular crowding, thereby favoring spontaneous phase separation of disordered biopolymers [236]. On the other hand, free mRNA molecules drag scaffold proteins of stress granules (see Table 1) towards themselves through RNA recognition motifs (RRM) in these proteins, thereby increasing their local concentration to the level required for the formation of liquid condensates [237].
Stress granules are multiphase structures [238]. The scaaffold proteins of stress granules associated with mRNA molecules form a solid-like non-dynamic core of these MLOs surrounded by a layer of client proteins, which dynamically exchanges its contents with the cytoplasm [239]. The continuous exchange of the content of stress granules with the intracellular environment enables these structures to regulate the network of signaling pathways that control the cellular responses to stress [238]. Curiously, the stress granules present in yeast cells are gel-like spheroid particles practically devoid of a dynamic layer [233]. The composition and properties of stress granules are regulated by post-translational modifications of their scaffold proteins, changes in pH, salt concentration, ambient temperature, and other environmental factors [234, 238].
A change in the functional activity of stress granules is often accompanied by a change in their material state [240–242]. The course of a number of neurodegenerative diseases, including amyotrophic lateral sclerosis, Alzheimer's disease, and frontotemporal dementia, is coupled with dysregulation of the structure and properties of stress granules [243]. Incorporation into stress granules of mutant forms of proteins specific of these diseases, such as T-cell intracellular antigen-1 (TIA-1), TIA-1-related protein (TIAR), RNA-binding protein fused in sarcoma (FUS), heterogeneous nuclear ribonucleoprotein A1 (hNRNPA1), transactive response DNA binding protein 43 kDa (TDP-43), and polyadenylate-binding protein 1 (PABP1) promotes the pathological transformation of stress granules into toxic aggregates of amyloid fibrils [225, 237, 242, 244–248].
To survive stress cell activates various stress-response pathways, such as heat-shock response, while inhibiting other major signaling pathways, such as protein synthesis. Transient stress-induced compartments formed via liquid–liquid phase separation most probably benefit cell survival by playing role of temporal cellular preservation clusters for hundreds of proteins that have to be inactivated during period of post-stress recovery, while allowing cell to save resources on their degradation and resynthesis. To act as reversible stress responders, several known MLOs possess a transient nature and represent cellular adjustments to changing environmental conditions. The examples are given by stress-inducible A-bodies (amyloid bodies) and nuclear stress-bodies (nSBs), which are formed in the cell nucleus in response to severe environmental conditions. A-bodies of different composition assemble inside the nucleoli following thermal stress and hypoxia-induced extracellular acidosis. Mature A-bodies are non-dynamic solid-like condensates that consist of low-complexity RNA and proteins with fibrillation propensity. They can be distinguished from other condensates by large electron-dense fibers that can be stained with various amyloidophylic dyes [249]. However, during, their development a LLPS stage is essential. A-bodies formation is triggered by transcription of non-coding RNA derived from the ribosomal intergenic spacer DNA, called rIGSRNA. rIGSRNA contains long evolutionary conserved clusters of simple cytosine/uracil (CU) or adenine/guanine (AG) di-nucleotide repeats. The activated transcription of rIGSRNA leads to the increase in local concentration of low-complexity RNA, which induces the formation of multiple liquid-like foci most probably formed as a result of complex coacervation. Various proteins containing low complexity domains are recruited to these foci. Characteristic A-body also contain amyloid-converting motif (ACM), as well as fibrillation propensity domain. Increased concentration of such proteins driven by their interactions with low-complexity RNA initiates amyloidogenic propagation-like pathway, which leads to proteins immobilization and liquid-to-solid phase transition. The important features of A-bodies is their reversibility and the lack of toxicity. The breakdown of A-bodies is initiated by the termination of stress and is carried out in an Hsp70/90-dependent manner [250]. Typically, aggregates of amyloid fibrils formed by basic stress granule proteins are toxic to cell and are associated with development of neurodegenerative diseases [251]. However, rIGSRNA dependent amyloid bodies can be disassembled and probably represent a bright example of so called functional amyloid fibrils that can temporally store proteins in the form of amyloids.
On the other hand, nuclear stress bodies (nSBs), are formed as a result of LLPS in response to the thermal, chemical, proteotoxic, and some other types of stress and remain in a liquid-like form until disassembled during post-stress recovery [188, 252]. Like A-bodies, nSBs assembled in an RNA-dependent manner. Their formation occurs at transcription sites of satellite DNA 3, which consists of tandemly organized repeats of nucleotide sequences located on several human chromosomes, but nSBs are mainly associated with chromosome 9 (locus 9q12). It was shown that nSBs form through LLPS and their dynamic properties decrease with the duration of the stress exposure [252]. The heat shock factor 1 (HSF1) is one of the key partners of HSatIII RNA and is required for its transcription. The biological function of nSBs is most probably in resolving post-stress deformations and enhancing cellular survival. Thus, inhibition of HSF1 expression increased HeLa sensitivity to apoptosis [253]. Besides that, it has been discovered that SR proteins are translocated into nSBs, which suggests the role of nSBs in the post-stress regulation of mRNA splicing or their role as protein storage clusters [254].
Satellite DNA 3 sequences appeared relatively late in the evolution and nuclear stress bodies were not found in mammals other than primates [255]. However, in Drosophila, functionally similar formations were described and called omega speckles [256]. Omega speckles assembly depends on the long noncoding RNA Hsrω (heat shock RNA ω) transcribed from the 93D heat shock locus. In response to thermal stress, omega speckles coalesce at the Hsrω RNA transcriptional loci. It was suggested that they act as repositories for various mRNA processing factors contributing to translation inhibition and cellular post-stress recovery [257].
Another example of well-researched MLOs are the PML-bodies. The study of PML-bodies has begun long before the recognition of the general principle of the formation of all MLOs via LLPS [258]. PML-bodies are polyfunctional dynamic protein organelles present in most cell types, where they are involved in the regulation of transcription, cell differentiation, stress response [259, 260]. The main protein of PML-bodies is the protein of promyelocytic leukemia (PML) [261]. As a result of alternative splicing, at least seven main isoforms of PML are generated, which differ in size and amino acid sequence of the C-terminal domain [262]. The N-terminal ordered region of the protein, which is exactly the same for all PML isoforms, contains the so-called RBCC (Ring-Box-Coil-Coil) motif consisting of several zinc-binding RING domains (Really Interesting New Gene), two B-domains (B-Boxes), as well as a leucine-rich α-helical coil-coil domain [263, 264]. The C-terminus of most PML isoforms contains a nuclear localization signal (NLS) and a SUMO-interacting motif (SIM). Together SIM and RBCC provide the possibility of specific interaction of PML with a large number of partners, and NLS is responsible for the nuclear localization of PML-bodies [265]. According to the currently accepted model of PML-body formation, oxidative dimerization of a PML isoform induces the formation of disulfide bonds between monomers of this protein and thus initiates the formation of an insoluble "aggregates", to which client proteins are recruited via SUMO/SIM interactions [259]. However, one should keep in mind that at least the following three factors are necessary for the formation of intermolecular disulfide bonds [266]:
The spatial accessibility/physical proximity of the partner cysteine residues forming a disulfide bond;
the difference between the pKa of the involved thiol groups and the pH of the local environment (with lower pH limiting reactivity and higher pH favoring increased reactivity), and
the redox environment (with lesser reactivity under more reducing conditions and greater reactivity under more oxidizing conditions)
Therefore, oxidative dimerization of PML requires high concentrations of PML molecules and enzymes that catalyze the formation of disulfide bonds “in the right place at the right time” [200]. Interactions between the regions of conserved residues of the RBCC motif also require high protein concentrations. Accordingly, the de novo formation of PML bodies requires PML precondensation [200], which can occur as a result of LLPS of PML isoforms due to the multiple weak nonspecific interactions of intrinsically disordered regions of PML isoforms.
The analysis of the amino acid sequences of PML isoforms showed that the variable C-terminal domains of almost all PML isoforms have properties characteristic of sequences potentially capable of LLPS due to electrostatic interactions [200]. The C-terminal regions of PML isoforms are highly disordered, contain a large number of charged residues and tandem repeats of the SS and PP type. The C-terminus of the PML-II isoform contains the highest proportion of disordered residues among all protein isoforms. In addition, only the C-terminal domain of PML-II is enriched with numerous RG motifs characteristic of LCD. Together with the available literature data on RBCC-free and SUMO/SIM-free incorporation of this protein into PML bodies, this allows us to conclude that the PML-II isoform can form liquid-droplet structures regardless of the interaction with other proteins [200].
It has been shown that the C-terminal domains of the PML-II and PML-V isoforms not only integrate into endogenous PML bodies, but also form dynamic liquid-droplet compartments in HeLa PML−/− cells, while partially passing into the nucleoplasm [267]. Furthermore, disruption of PML sumoylation by K490R substitution promoted mainly diffuse distribution of the C-terminal domains of the PML-II and PML-V isoforms in the HeLa cells knockout for the endogenous PML, but not in the wild-type cells. Taken together, these data confirmed the earlier hypothesis on the significant role of weak nonspecific interactions mediated by disordered C-terminal domains of PML isoforms in the formation of PML bodies.
Modern approaches to the analysis of MLOs
Despite the advances made over the past decades in the study of the principles of the formation of MLOs, many questions still remain not fully resolved. For example, how are the microscopic properties of biomolecules included into the MLO and the macroscopic properties of the resulting condensate interrelated? And also, what conditions are necessary and sufficient for the liquid–liquid phase separation to take place in the cell? A systematic study of the influence of various factors on changes in the structural properties of proteins that make up the MLOs might help answering these questions. A wide range of biophysical methods is already being used to carry out such studies.
Taking into account the fact that the first experiments to reveal the role of phase transformations of biopolymers in the cell compartmentalization were carried out within the framework of a summer school devoted to the modern methods of light microscopy [28, 158], it is not surprising that this method is the most widely used for studying MLOs. This method makes it possible to study the phase separation of biopolymers both in vivo and in vitro. Various microscopic techniques are suitable for imaging of the biomolecular condensates, including differential interference contrast (DIC) and confocal fluorescence microscopy. Since the intracellular environment is a complex colloidal system containing many contrast objects, confocal fluorescence microscopy is mainly used to study MLOs in vivo. Because of the fact that the MLOs are highly dynamic cellular bodies, an important requirement for the equipment utilized in their analysis is the ability to be used it in a time-resolved mode. In fact, the use of time-resolved microscopy makes it possible to determine/estimate various physical characteristics of condensates, such as viscosity and the value of their surface tension [268]. This is achieved by measuring the time when two small condensates coalesce into one larger droplet upon collision and by measuring the contact angle between the condensate surface and the cover slip. The surface tension is the driving force of the relaxation of the droplets shape after its deformation, while the viscosity prevents this process. Visualization of passive fusion of droplets is available to most laboratories since it does not require the usage of any specific technique. Using this method, the friction between the condensates and the glass surface, as well as the initial shape of the droplets, should be taken into account, since it can have a significant impact on the quality of the obtained data. The application of this method is complicated for samples containing a small number of droplets, since it is time-consuming to fix a statistically significant number of stochastically occurring fusing events. In addition, for droplets with low viscosity and/or high surface tension, coalescence processes are fast (usually in a millisecond scale), and detection of such events require the usage of expensive imaging devices (to increase the temporal resolution) [269]. The usage the dual-trap optical tweezers allows researchers to measure the droplets fusion time in more controllable manner and improve the data quality. The different refractive index of the condensed and dilute phases allows droplets to be optically captured using highly focused laser beam and cause them to fuse. The change in the deflection of the laser beam upon the droplets coalescence allows to detect the fusion events and to measure the relaxation time. Optical traps usually have a higher temporal resolution than video cameras used in the method of passive fusion. This allows detection of the very fast fusion events. Besides, this forced coalescence improves data collection even for samples with small number of droplets. Optical traps stimulate the droplets fusion in the bulk solution, which prevents the surface effects from interfering with data collection [269]. The dual-trap optical tweezers technique was used to study the effect of mutations on the material properties of condensates composed of FUS [227, 270] and hnRNAP1 [271], as well as the change in dynamics of RNP − RNA condensates depending on the peculiarities of the RNA and protein sequence [272]. The disadvantage of the fusion-based techniques is that they only give the ratio between the viscosity and surface tension and do not provide any explicit values.
Apparent viscosity inside the condensed phase can be evaluated by a method of passive microrheology [273]. The method consists in introducing synthetic balls of a certain size into the system in order to observe their diffusion and determine the mean square of their displacement over a certain time. The mean of apparent viscosity for Newtonian pure viscous liquids is extracted by assuming Brownian dynamics using Stokes–Einstein equation. Additionally, using this method together with droplet fusion assays permits estimation of the apparent mean of the condensate surface tension [269]. The advantages of the passive microrheology technique are the absence of the need of labeling the polymers engaged in the droplet formation, which preserve intactness of the condensates, and the availability of equipment. The method can be used even in a living cell but it requires utilization of advanced labeling techniques and super-resolution microscopy [274, 275]. The application of the passive microrheology technique is limited by the droplet size, which should be larger than the commonly used beads (20–500 nm). While interpreting the data, it is necessary to consider the possibility of nonspecific interactions between the condensate molecules and the particle surface. Frequently, particle surface passivation is required [269]. A regulatory role of salt and RNA on the viscosity of the LAF-1 droplets was tested by this method [273].
All described so far methods are limited to classifying the condensate as purely liquid or not, still they do not provide quantitative data on droplets elastic properties. However, MLOs have elastic properties along with liquid one. They are especially pronounced for those condensates serving as structural elements of the cells. Furthermore, liquid-to-solid phase transitions of some condensates are well known to be implicated in neurodegenerative diseases [217, 227, 276]. The method of active microrheology was adapted to quantify the elasticity of protein/RNA condensates [277]. This method is based on the deformation of the droplets using dual-trap optical tweezers. Two optical tweezers entrap two beads immersed inside a droplet, place them on the opposite poles of the condensate and apply controlled oscillatory stresses. The method allows determination of the complex frequency-dependent spring constant of the droplet response.
and extract complex shear modulus.
where G´ and G´´ denote the storage and loss moduli respectively. These data are then used for evaluation of the surface tension γ [277–279]. Furthermore, a viscosity and terminal relaxation time can be extracted from the complex shear modulus [277]. Storage and loss moduli allow estimation of the viscoelasticity of the material, since it is known that G” dominates for a purely viscous condensate, whereas G’ dominates for an elastic condensate [269]. The authors of this method showed that the droplets of PGL-3 (a major component of P granules, the germline granules of the nematode worm Caenorhabditis elegans) were viscoelastic rather than fully viscous, whose viscoelasticity depended on the salt concentration [277] and exhibited aging behaviors (Jawerth et al. 2020). Microrheology analysis indicated that the viscosity of liquid-like MLOs ranged from ~ 1 Pa × s for P-granules to ~ 103 Pa × s for nucleoli [162]. The approach is feasible only for droplets that is larger than 10 μm and requires the usage of beads with refractive index higher than that of the droplets [269].
Another fluorescence microscopy-based technique for assessing the liquid properties of MLOs is the method of fluorescence recovery after photobleaching (FRAP) [28]. The method is based on the irreversible photo-bleaching of the region of interest (ROI) within the droplet by a high-power laser illumination and monitoring the subsequent fluorescence recovery of the bleached region as a function of time. This approach allows researchers to determine the proportion of the mobile fraction inside the biomolecular condensate, which gives the rough estimation of the droplet viscoelasticity, the diffusion coefficient of target molecules inside the MLOs, and describes the process of substance exchange at the droplets interface. A fitting model for data analysis should be chosen with care. It strongly depends on the molecular identity of the labeled species, the size and shape of the bleached area, as well as the droplet size, the profile of the laser beam used for bleaching, the photofading, the dimensionality of the diffusion process, and the existence of the reaction processes coupled with free diffusion [269, 280]. FRAP method for studying the dynamic properties of MLOs is the easiest to use and available to most laboratories. It permits the analysis of the condensate dynamics even in the living cells. Unfortunately, FRAP faces a number of limitations. For example, it does not provide information on the material state of the condensate but only the diffusion dynamics of the labeled molecules. Namely, FRAP measurements may reveal the liquid-like behavior for one component of the condensate and the viscoelastic behavior for another. Such results are usually interpreted in the frame of a scaffold-client model [163]. Additionally, the FRAP method is difficult to apply to small droplets (< 1 μm), that are typical of living cells [272]. In such cases, fluorescence correlation spectroscopy (FCS) may be the method of choice [280]. This method allows an accurate estimation of the molecular concentrations and diffusion coefficient of individual molecules inside the biomolecular condensates. FCS is based on the measurements of the fluctuations of fluorescent intensity of labeled molecules inside femtolitre excitation volumes. Some technical limitations impede the characterization of LLPS using FCS method. Differences in the refractive indexes of the light and dense phases cause the inaccuracy in calibration of the fluorescence excitation volume using external calibration. FCS modifications, namely 2-focus FCS and Z-scan ultrafast FCS, can bypass this limitation [281, 282]. However, the availability of this method is still limited due to the high cost of ultrasensitive photodetectors required for registering the fluorescence of single molecules [207]. The FCS technique was applied to study LLSP of LAF-1 to determine the protein concentration and its diffusion coefficient in the light and dense phases [282].
The main disadvantage of standard light microscopy methods is their limited resolution, which does not allow the researcher to resolve organelles with size close to the diffraction limit (an example is RNP granules about 180 nm in size [238]) and to characterize the ultrastructure of MLOs (nuclear speckles are sub-compartmentalized into sub-speckles with a diameter of 200–500 nm, further divided into sub-compartments tens of nm in size [283]). Higher resolution of light microscopy can be achieved with the super-resolution microscopy methods [281], such as structured illumination microscopy (SIM) (XY resolution 100–130 nm) [284], stochastic optical reconstruction microscopy (STORM) (XY resolution 20 nm) [238], and lattice light sheet microscopy (LLSM) (XY resolution 150–280 nm) [285]. The application of super-resolution techniques requires the usage of specific fluorescent probes and is associated with prolonged illumination of the sample with high-intensity light [286], which may dramatically affect such sensitive systems as liquid condensates especially in living cells. Super-resolution techniques are not flexible in their application; their protocols cannot be adjusted easily in the middle of an experiment. Finally, the main disadvantage of super-resolution techniques is their cost, which is at least twice that of confocal microscopes [286].
The electron microscopy (EM) provides a resolution of 0.8 Å. The cellular ultrastructure is commonly studied by transmission electron microscopy (TEM) with negative staining. Using this method, the ultrastructure of many MLOs was studied, including nucleolus [287], nuclear speckles [288], Cajal bodies [289], etc. Despite the excellent magnification for studying the ultrastructure of MLOs, the methods of fixing biological samples applied in classical electron microscopy change the morphology of condensates beyond recognition. This is especially evident for the in vitro condensates, which barely retain their native structure upon drying. The recently developed cryoelectron microscopy (cryo-EM) method is devoid of these drawbacks. Cryo-EM is a type of transmission electron microscopy, in which a sample is examined at cryogenic temperatures. Cryo-EM is free of harsh fixation and staining of biological samples, thereby maintaining their structure native (or at least maximally close to native and minimally perturbed). This method is widely used in the analysis of soluble oligomers associated with nucleation of phase transitions as well as is applied to characterization of the gel-like structures formed upon condensates aging [281, 290, 291]. Furthermore, cryoelectron tomography gives an opportunity to study the ultrastructure of MLOs even as part of the whole organism [291, 292]. Unfortunatelly, intrinsically disordered domains being the main components of MLOs are poorly resolvable by EM [281].
The changes in the structural and dynamic properties of IDPs occuring upon phase separation in vitro have been studied by the nuclear magnetic resonance (NMR) spectroscopy. NMR may provide information on the structural features of proteins within phase separated bodies, protein interactions within the condensed phase (site of interactions, conformational changes, length scale interactions, dissociation constant), dynamics of proteins within various types of liquid-like and other mesoscale assemblies on timescales from ns to s [281]. Due to the deficiency of 13C and 15 N isotopes in nature, the saturation of proteins with these isotopes may be required to increase sensitivity of the method. To that end, recombinant proteins are expressed in bacteria grown in a minimal media supplied with these isotopes. However, the NMR experiments require a large amount of IDPs which is hardly achievable in minimal media. Furthermore, the application of the NMR method requires the use of expensive equipment, wihich is not available to every lab and can only be used by users with high technical skills. This method was applied, for instance, to detect a nearly 100-fold reduction in the rate of translational diffusion of the N-terminal LC region of Ddx4 occurring upon phase separation [284] and to identify inter-molecular interactions between the helical segment of the C-terminal LC region of TDP-43 as driving force of its phase separation [293].
The full understanding of mechanisms of MLO formation and functioning requires not only mechanical and structural characterization of phase separated systems but also the ability to predict their phase behavior and knowledge of kinetic trajectories of their formation. Each two-phase system can be described by its unique phase diagram that characterizes the conditions for the solution to undergo phase separation. Plotting phase diagrams using manual methods require performing a huge number of time- and sample-consuming experiments. Therefore, the development of high-throughput automated methods of studying phase separation is topical. The flow-based methods are promising. The recently developed method of capillary flow experiments (Capflex) allows the fast and fully automated determination of the dilute phase concentration, relative droplet size distributions, the kinetics of droplet formation and maturation into amyloid fibrils, as well as the binding affinity between the polypeptide undergoing LLPS and LLPS-modulating compounds [294]. Capflex is realized on the basis of the commercially available FIDA-1 (flow-induced dispersion analysis) instrument, which consists of a temperature-controlled tray in a sample-handling robot holding 96-well plates and a temperature-controlled chamber containing a fused silica capillary. The sample placed into the well of the plate is injected into the capillary where it undergoes LLPS due to changing of the temperature (the temperature of the capillary chamber is below the cloud point), or the environment (different composition of the solution filling the capillary); then the sample passes to detector, where the fluorescence time trace is recorded. The passing of each droplet causes a signal spike in the fluorescence time trace. The signal baseline intensity level allows determination of the protein concentration in the dilute phase and the relative peak intensity distribution closely reflects the droplet size distribution. Analyzing the fluorescence time trace also allows resolving the kinetics of droplet formation. The inbuilt function of the FIDA-1 instrument allows researchers to measure the affinity and complex size of an indicator and analyte molecule by Taylor dispersion for non-phase separated regime [294, 295]. The method is also applicable for the analysis of the preformed droplets. The dissolution of preformed droplets upon dilution reflects their liquid nature. Capflex can be easily interfaced with the automated fluid handling systems, such as the OpenTrons OT2, additionally minimizing manual interventions. This method allows 96 samples to be analyzed daily, including sample preparation time with minimal human supervision. A sample volume of 40 μl per vial is sufficient to perform two iterations per sample [294].
Even lower sample volume may be achieved by using microfluidic-based approach. For example, microfluidic device with capillary valves enables determination of saturation concentrations of five conditions with twenty replicates in a single experiment with a sample load of only 10 μl [296]. Different modes of microfluidic assay cover a number of tasks, including determining the phase diagrams of biopolymers (manipulating multiple parameters such as the protein/RNA concentration, temperature, pH, ionic strength and solution composition), observing coalescence events, studying the dynamics of protein phase transition and vizualizing the droplet aging [297]. Microfluidic tools were even applied for determining the rheological properties of condensates [298] and their composition [299]. To monitor phase transition over a long time, droplets can be entrapped in droplet storage arrays [300]. Condensates can be manipulated before, during or after their storage. Microfluidic assay is compatible with most optical detection methods [297]. The main disadvantage of microfluidic approach is the requirement of high technical skill for users. However, the recently developed capillary valving devices can be operated even by the inexperienced users [296].
Another flow-based approach has been applied to study the early stages (100 ms and 10 s) of the phase separation kinetics. The stopped-flow method is a useful and essentially automatic way for studying the kinetics of phase separation [301]. Apparent differences in kinetic traces of phase separation induced by pH jump-, urea dilution-, and maltose binding protein tag cleavage was shown using stopped-flow equipment [301]. However, the turbidity of the solution (measured by the change in absorbance at 600 nm), chosen in this study to control phase separation, is a relative characteristic and depends on the number and size of scattering particles, while not giving quantitative information about phase separation [207]. Therefore, the authors also used manual measurements of dynamic light scattering (DLS) and fluorescent microscopy to quantify the droplet number and size. It has been established that rapidly changing the pH of a solution from a carefully selected extreme to physiological value enables instantaneous induction of LLPS under near-native conditions with minimal sample dilution, whereas other commonly used approaches for inducing LLPS such as urea dilution and solubility tag cleavage may perturb kinetic schemes of the process [301].
The application of all flow-based approaches is limited to only in vitro studies. An important approach for the analysis of MLOs in living cells is based on the use of platforms that allow for controlled assembly or disassembly of cellular bodies. The design of such platforms relies on the genetic engineering techniques needed to create various combinations of proteins and their individual domains. The inclusion of fluorescent biomarkers in synthetic MLOs makes it possible to visualize and quantitatively analyze phase transitions in a cell using fluorescence confocal microscopy. In view of the highly dynamic nature of MLOs, the most acceptable way of influencing them is irradiation with light, while an activating/deactivating stimulus can be applied to a local area of the cell. To this end, several optogenetic systems have been developed, including optoDroplets [302], Corelets [303] and PixELL [304], in which the photosensitive part is represented by a photoreceptor capable of oligomerization (cryptochrome Cry2 or Cry2olig) or the formation/destruction of a complex with a partner protein (iLID/SspB and PixE/PixD) when irradiated with the light with a certain wavelength, and where the condensing part is formed by the intrinsically disordered domains or proteins containing them, which provides means for the clusterization of the system. Such platforms actually undergo concentration-dependent phase separation in the cell with the formation of liquid or gel-like bodies [302] and can be used for the analysis of the functioning of MLOs and their roles in pathological processes [305]. However, only the assembly of photo-regulated MLOs from cryptochromes is controlled by light, while disassembly is possible only due to dark relaxation [302]. It has recently been shown that MLO formed from plant phyB and PIF6 assembly and disassembly are controlled by light [306].
In same line, Corelets and PixELL platforms were used to understand the mechanisms by which the local formation of MLOs occurs and how their asymmetric distribution in the cell is maintained [303, 304]. Mechanisms of stress granule formation have been characterized using the Corelets platform [307]. A model was proposed according to which the G3BP1 protein (Ras GTPase-activating protein-binding protein 1, which is ATP- and magnesium-dependent helicase that plays an essential role in innate immunity [308]), the main component of the stress granules, through homo-dimerization forms a platform for the binding of other proteins, such as UBAP2L (Ubiquitin-associated protein 2-like), and allows the accumulation of valences (RNA-binding domains, RBDs included in G3BP and UBAP2L, as well as other components of stress granules). Phase separation is due to the interaction of a multivalent protein complex with mRNA molecules that accumulate under stress. In this work, it is indicated that intrinsically disordered domains can play not a guiding, but a regulating role in phase separation, as interaction between central disordered domains of G3BP1 inhibits the stress granule formation in the absence of RNA [236, 239]. Furthermore, intrinsically disordered domains of the G3BP1 were shown to affect the physical properties of the stress granules [236, 239].
The combination of technologies for recognizing different DNA regions (CRISPR/Cas9) with the optogenetic platforms for assembling MLOs in a system called CasDrop makes it possible to understand the role of nuclear bodies in genome organization [309]. In particular, it has been shown that fusion due to the surface tension of MLOs formed in certain regions of chromosomes can lead to their convergence. It is believed that such a mechanism may underlie the formation of, for example, super-enhancers [309].
The described optogenetic tools allow reconstruction of the cellular phase separation with physiological protein concentrations and provide means to eliminate artifacts associated with protein overexpression. The caution should be exercised analyzing the data obtained using current optogenetic platforms, since the artificially introduced system multivalency may strongly affect the naturally occurring cellular processes. The carefully designed control experiments are required [207].
Conclusions
The present work represents an analysis of the literature devoted to the study of the problem of the intracellular space compartmentalization. The paper also represents the main methodological developments and describes novel experimental procedures that contributed to the formation and dissemination of the theories of cellular compartmentalization in different periods of history. It is emphasized that there are two general models describing principles of the compartmentalization within the intracellular space. The first model suggests that the vital activity of the cell is carried out mainly due to strictly determined linear biochemical processes, in the organization of which biological membranes play a key role. The second model represents the cell as a highly dynamic system with the properties of "soft matter”, in which the internal order is achieved due to the spontaneous and stochastic separation of a supersaturated intracellular polymer solution into phases. Currently, these two models of the intracellular compartmentalization are to some extent opposed to each other. However, the cell uses both mechanisms to maintain the internal organization of protoplasm [310].
Therefore, it is expected that the next period in this field will be associated with the merger of these theories and the description of the processes occurring during the interaction of MLOs with lipid membranes and their role in the cell. In fact, already in 2012, morphological changes in lipid vesicles were discovered that occurred when the vesicular walls came into the contact with the polymer condensates [311]. It was emphasized that in a system comprising lipid vesicles encapsulating aqueous two-phase polymer solutions, a variety of unusual wetting phenomena and formation of membrane nanotubes can be induced by the phase separation process and by the interactions of the lipid bilayers with the phases [311]. This phenomenon can play an important role in the organization of the intracellular transport and in determining the morphological features of the cell structure. Evidence is rapidly accumulating that the interaction of membrane-bound organelles and MLOs also plays an important role in the organization of the intracellular space [312]. For example, it is assumed that the ability to regulate liquid–liquid phase separation via the post-translational modifications of proteins that make up MLOs can play an important role in the disassembly and subsequent assembly of membrane organelles (in particular, the Golgi apparatus cisterns) during cell division. Several sets of data indicate that the physical properties of MLO (liquid, gel, amyloid-like fibrillar) can influence the formation of autophagosome membranes around them [248, 313, 314]. Many other processes within the cell are beginning to be viewed in terms of liquid–liquid phase separation. Examples of such processes include the formation of signaling complexes or adhesion complexes and tight contacts between cells, where the plasma membrane serves as a platform for the formation of supramolecular structures due to liquid–liquid phase separation [164, 211, 315]. Various functional granules are collected in certain areas of the membrane of the endoplasmic reticulum to perform certain functions, for example, folding of newly synthesized transmembrane proteins and their incorporation into the membrane (TIS granules formed by the broadly expressed RNA-binding protein TPA-induced sequence 11b (TIS11B, also known as mRNA decay activator protein ZFP36L1 [316]) or protection of functional areas of the endoplasmic reticulum under conditions of amino acid starvation (Sec granules that incorporates components of the ER exit sites and require both protein transport protein Sec23 and secretory protein Sec24AB [317] for their formation). These are far from all forms of interaction between MLOs and intracellular membranes, the mechanisms of which have yet to be investigated [318]. The development of these exciting directions can be facilitated by the improvement of the methods for three-dimensional visualization of biological objects.
Author contributions
AVF, IMG, VNU, and KKT: study conception and design, collection of literature data, data curation, formal analysis, evaluation, interpretation of data, writing and review of the manuscript, and study supervision. IAA: collection of literature, data, data curation, formal analysis, evaluation, acquisition and interpretation of data, writing and review of the manuscript, visualization; ASF, OVS, OIP, SAS: collection of literature, data, data curation, formal analysis, evaluation, acquisition and interpretation of data, writing and review of the manuscript.
Funding
This work was supported in part by a grant from Russian Science Foundation (RSCF 19-15-00107 (KKT)) and an RF President Fellowship (SP-259.2019.4 (IAA)).
Availability of data and material
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.
Declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Ethics approval and consent to participate
Not applicable.
Consent to publish
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Vladimir N. Uversky, Email: vuversky@usf.edu
Konstantin K. Turoverov, Email: kkt@incras.ru
References
- 1.Meyer H. Zur theorie der alkoholnarkose. Archiv für experimentelle Pathologie und Pharmakologie. 1899;42(2–4):109–118. doi: 10.1007/BF01834479. [DOI] [Google Scholar]
- 2.Overton CE. Studien über die Narkose: zugleich ein Beitrag zur allgemeinen Pharmakologie. Jena: G. Fischer; 1901. [Google Scholar]
- 3.Missner A, Pohl P. 110 years of the Meyer-Overton rule: predicting membrane permeability of gases and other small compounds. ChemPhysChem. 2009;10(9–10):1405–1414. doi: 10.1002/cphc.200900270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hannesschlaeger C, Horner A, Pohl P. Intrinsic membrane permeability to small molecules. Chem Rev. 2019;119(9):5922–5953. doi: 10.1021/acs.chemrev.8b00560. [DOI] [PubMed] [Google Scholar]
- 5.Peracchia C, editor. Handbook of membrane channels molecular and cellular physiology. London: Academic Press; 1994. [Google Scholar]
- 6.Pederson T. The nucleoulus. Cold Spring Harb Perspect Biol. 2011;3(3):a000638. doi: 10.1101/cshperspect.a000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uversky VN. Protein intrinsic disorder and structure-function continuum. Prog Mol Biol Transl Sci. 2019;166:1–17. doi: 10.1016/bs.pmbts.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 8.Fonin AV, Darling AL, Kuznetsova IM, Turoverov KK, Uversky VN. Multi-functionality of proteins involved in GPCR and G protein signaling: making sense of structure-function continuum with intrinsic disorder-based proteoforms. Cell Mol Life Sci. 2019;76(22):4461–4492. doi: 10.1007/s00018-019-03276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uversky VN. p53 proteoforms and intrinsic disorder: an illustration of the protein structure-function continuum concept. Int J Mol Sci. 2016 doi: 10.3390/ijms17111874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cate JH, Yusupov MM, Yusupova GZ, Earnest TN, Noller HF. X-ray crystal structures of 70S ribosome functional complexes. Science. 1999;285(5436):2095–2104. doi: 10.1126/science.285.5436.2095. [DOI] [PubMed] [Google Scholar]
- 11.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292(5518):883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 12.Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334(6062):1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 13.Yusupova G, Yusupov M. A path to the atomic-resolution structures of prokaryotic and eukaryotic ribosomes. Biochemistry (Mosc) 2021;86(8):926–941. doi: 10.1134/S0006297921080046. [DOI] [PubMed] [Google Scholar]
- 14.Peng Z, Oldfield CJ, Xue B, Mizianty MJ, Dunker AK, Kurgan L, Uversky VN. A creature with a hundred waggly tails: intrinsically disordered proteins in the ribosome. Cell Mol Life Sci. 2014;71(8):1477–1504. doi: 10.1007/s00018-013-1446-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hooke R (1665) Micrographia: or some physiological descriptions of minute bodies made by magnifying glasses with observations and inquiries thereupon. J. Martyn and J. Allestry, London
- 16.Van Leeuwenhoek A. Observations, communicated to the publisher by Mr. Antony van Leewenhoeck, in a dutch letter of the 9th Octob. 1676. here English'd: concerning little animals by him observed in rain-well-sea- and snow water; as also in water wherein pepper had lain infused. Philos Trans. 1677;12(133):821–831. doi: 10.1098/rstl.1677.0003. [DOI] [Google Scholar]
- 17.Schliwa M. The evolving complexity of cytoplasmic structure. Nat Rev Mol Cell Biol. 2002;3(4):291–296. doi: 10.1038/nrm781. [DOI] [PubMed] [Google Scholar]
- 18.Schleiden MJ (1838) Beiträge zur Phytogenesis. Archiv für Anatomie, Physiologie und wissenschaftliche Medicin. Veit, Berlin
- 19.Schwann T. Mikroscopische Untersuchungen über die Übereinstimmung in der Struktur und dem Wachsthum der Tiere und Pflanzen. Berlin: Reimer; 1839. [PubMed] [Google Scholar]
- 20.Wagner R. Einige bemerkungen und fragen über das keimbläschen (vesicular germinativa) Müller’s Archiv Anat Physiol Wissenschaft Med. 1835;268:373–377. [Google Scholar]
- 21.Valentin G. Repertorium für anatomie und physiologie: kritische darstellung fremder und ergebnisse eigener forschung. Berlin: Verlag von Veit und Comp; 1837. [Google Scholar]
- 22.Brown R. On the organs and mode of fecundation of orchidex and asclepiadea. In: Bennett JJ, editor. Miscellaneous botanical works of Robert Brown. London: Publisher for the Ray society by R. Hardwicke; 1866. pp. 511–514. [Google Scholar]
- 23.Purkinje JE (1839) ‘Uber die Analogieen in den Struktur-Elementen des thierischen und pflanzlichen Organismus. Ûbersicht der Arbeiten und Verånderungen der Schlesischen Gesellschaft fçr Vaterlåndische Cultur im Jahre, pp 81–82
- 24.Welch GR, Clegg JS. Cell versus protoplasm: revisionist history. Cell Biol Int. 2012;36(7):643–647. doi: 10.1042/CBI20120128. [DOI] [PubMed] [Google Scholar]
- 25.Flemming W. Beitrage zur Kenntniss der Zelle und ihrer Lebenserscheinungen. Arch Mikrosk Anat. 1879;16:302–436. doi: 10.1007/BF02956386. [DOI] [Google Scholar]
- 26.Raspail PM. Essai de chimie microscopique appliquée à la physiologie, ou L'art de transporter le laboratoire sur le porte-objet dans l'étude des corps organisés. Paris: L'auteur; 1830. [Google Scholar]
- 27.Wilson EB. The structure of protoplasm. Science. 1899;10(237):33–45. doi: 10.1126/science.10.237.33. [DOI] [PubMed] [Google Scholar]
- 28.Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, Hyman AA. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324(5935):1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- 29.Oparin AI. The origine of Life. Moscow: Moscow Worker; 1924. [Google Scholar]
- 30.Tirard S. J. B. S. Haldane and the origin of life. J Genet. 2017;96(5):735–739. doi: 10.1007/s12041-017-0831-6. [DOI] [PubMed] [Google Scholar]
- 31.Haldane JBS. The origin of life Rationalist Annual. 1929;148:3–10. [Google Scholar]
- 32.Overbeek JT, Voorn MJ. Phase separation in polyelectrolyte solutions; theory of complex coacervation. J Cell Physiol Suppl. 1957;49(Suppl 1):7–22. doi: 10.1002/jcp.1030490404. [DOI] [PubMed] [Google Scholar]
- 33.McMullen JN, Newton DW, Becker CH. Pectin-gelatin complex coacervates I: determinants of microglobule size, morphology, and recovery as water-dispersible powders. J Pharm Sci. 1982;71(6):628–633. doi: 10.1002/jps.2600710608. [DOI] [PubMed] [Google Scholar]
- 34.Jizomoto H. Phase separation induced in gelatin-base coacervation systems by addition of water-soluble nonionic polymers I: Microencapsulation. J Pharm Sci. 1984;73(7):879–882. doi: 10.1002/jps.2600730705. [DOI] [PubMed] [Google Scholar]
- 35.Yu S, Hu J, Pan X, Yao P, Jiang M. Stable and pH-sensitive nanogels prepared by self-assembly of chitosan and ovalbumin. Langmuir. 2006;22(6):2754–2759. doi: 10.1021/la053158b. [DOI] [PubMed] [Google Scholar]
- 36.Sarmento B, Ribeiro A, Veiga F, Sampaio P, Neufeld R, Ferreira D. Alginate/chitosan nanoparticles are effective for oral insulin delivery. Pharm Res. 2007;24(12):2198–2206. doi: 10.1007/s11095-007-9367-4. [DOI] [PubMed] [Google Scholar]
- 37.Izmailova VN, Rebinder PA. Structure formation in protein systems. Moscow: Science (Nauka); 1974. [Google Scholar]
- 38.Robertson JD. New observations on the ultrastructure of the membranes of frog peripheral nerve fibers. J Biophys Biochem Cytol. 1957;3(6):1043–1048. doi: 10.1083/jcb.3.6.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gorter E, Grendel F. On Bimolecular layers of lipoids on the chromocytes of the blood. J Exp Med. 1925;41(4):439–443. doi: 10.1084/jem.41.4.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palade GE. The fine structure of mitochondria. Anat Rec. 1952;114(3):427–451. doi: 10.1002/ar.1091140304. [DOI] [PubMed] [Google Scholar]
- 41.Palade GE, Porter KR. Studies on the endoplasmic reticulum. I. Its identification in cells in situ. J Exp Med. 1954;100(6):641–656. doi: 10.1084/jem.100.6.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dalton AJ, Felix MD. Cytologic and cytochemical characteristics of the Golgi substance of epithelial cells of the epididymis in situ, in homogenates and after isolation. Am J Anat. 1954;94(2):171–207. doi: 10.1002/aja.1000940202. [DOI] [PubMed] [Google Scholar]
- 43.Singer SJ. Biochemical organization of cell membranes. Birth Defects Orig Artic Ser. 1970;6(3):15–16. [PubMed] [Google Scholar]
- 44.Singer SJ. The molecular organization of membranes. Annu Rev Biochem. 1974;43:805–833. doi: 10.1146/annurev.bi.43.070174.004105. [DOI] [PubMed] [Google Scholar]
- 45.Singer SJ. Current concepts of molecular organization in cell membranes. Biochem Soc Trans. 1981;9(3):203–206. doi: 10.1042/bst0090203. [DOI] [PubMed] [Google Scholar]
- 46.Overton CE. Über die osmotischen Eigenschaften der lebenden Pflanzen-und Tierzelle. Zürich: Fäsi & Beer; 1895. [Google Scholar]
- 47.Sigworth FJ, Neher E. Single Na+ channel currents observed in cultured rat muscle cells. Nature. 1980;287(5781):447–449. doi: 10.1038/287447a0. [DOI] [PubMed] [Google Scholar]
- 48.Hamill OP, Marty A, Neher E, Sakmann B, Sigworth FJ. Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflug Arch. 1981;391(2):85–100. doi: 10.1007/BF00656997. [DOI] [PubMed] [Google Scholar]
- 49.Sakmann B, Neher E. Patch clamp techniques for studying ionic channels in excitable membranes. Annu Rev Physiol. 1984;46:455–472. doi: 10.1146/annurev.ph.46.030184.002323. [DOI] [PubMed] [Google Scholar]
- 50.Mueller P, Rudin DO, Tien HT, Wescott WC. Reconstitution of cell membrane structure in vitro and its transformation into an excitable system. Nature. 1962;194:979–980. doi: 10.1038/194979a0. [DOI] [PubMed] [Google Scholar]
- 51.Watson H. Biological membranes. Essays Biochem. 2015;59:43–69. doi: 10.1042/bse0590043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skou JC. The influence of some cations on an adenosine triphosphatase from peripheral nerves. Biochim Biophys Acta. 1957;23(2):394–401. doi: 10.1016/0006-3002(57)90343-8. [DOI] [PubMed] [Google Scholar]
- 53.Skou JC. Preparation from mammallian brain and kidney of the enzyme system involved in active transport of Na ions and K ions. Biochim Biophys Acta. 1962;58:314–325. doi: 10.1016/0006-3002(62)91015-6. [DOI] [PubMed] [Google Scholar]
- 54.Hyman T, Brangwynne CP. In retrospect: the origin of life. Nature. 2012;491:524–525. doi: 10.1038/491524a. [DOI] [Google Scholar]
- 55.Franklin RE, Gosling RG. Molecular configuration in sodium thymonucleate. Nature. 1953;171(4356):740–741. doi: 10.1038/171740a0. [DOI] [PubMed] [Google Scholar]
- 56.Wilkins MH, Stokes AR, Wilson HR. Molecular structure of deoxypentose nucleic acids. Nature. 1953;171(4356):738–740. doi: 10.1038/171738a0. [DOI] [PubMed] [Google Scholar]
- 57.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171(4356):737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 58.Perutz MF, Rossmann MG, Cullis AF, Muirhead H, Will G, North AC. Structure of haemoglobin: a three-dimensional Fourier synthesis at 5.5-A. Resolution, obtained by X-ray analysis. Nature. 1960;185(4711):416–422. doi: 10.1038/185416a0. [DOI] [PubMed] [Google Scholar]
- 59.Kendrew JC, Bodo G, Dintzis HM, Parrish RG, Wyckoff H, Phillips DC. A three-dimensional model of the myoglobin molecule obtained by x-ray analysis. Nature. 1958;181(4610):662–666. doi: 10.1038/181662a0. [DOI] [PubMed] [Google Scholar]
- 60.Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181(4096):223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 61.Crowther RA, Klug A. Structural analysis of macromolecular assemblies by image reconstruction from electron micrographs. Annu Rev Biochem. 1975;44:161–182. doi: 10.1146/annurev.bi.44.070175.001113. [DOI] [PubMed] [Google Scholar]
- 62.Deisenhofer J, Epp O, Miki K, Huber R, Michel H. X-ray structure analysis of a membrane protein complex. Electron density map at 3 A resolution and a model of the chromophores of the photosynthetic reaction center from Rhodopseudomonas viridis. J Mol Biol. 1984;180(2):385–398. doi: 10.1016/s0022-2836(84)80011-x. [DOI] [PubMed] [Google Scholar]
- 63.Preston GM, Carroll TP, Guggino WB, Agre P. Appearance of water channels in Xenopus oocytes expressing red cell CHIP28 protein. Science. 1992;256(5055):385–387. doi: 10.1126/science.256.5055.385. [DOI] [PubMed] [Google Scholar]
- 64.Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280(5360):69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 65.Michaelis L, Menten ML. "Die Kinetik der Invertinwirkung" [The kinetics of invertin action] Biochem Z. 1913;49(17):333–369. [Google Scholar]
- 66.Henri V. Théorie générale de l'action de quelques diastases par Victor Henri [CR Acad. Sci. Paris 135 (1902) 916-919] Comptes Rendus Biol. 2006;329(1):47–50. doi: 10.1016/j.crvi.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 67.Fischer E. Einfluss der configuration auf die wirkung der enzyme. Ber Dt Chem Ges. 1894;27:2985–2993. doi: 10.1002/cber.18940270364. [DOI] [Google Scholar]
- 68.Koshland DE., Jr Application of a theory of enzyme specificity to protein synthesis. Proc Natl Acad Sci USA. 1958;44:98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Straub FB, Szabolcsi G. O dinamicseszkih aszpektah sztukturü fermentov (On the dynamic aspects of protein structure) In: Braunstein AE, editor. Molecular biology, problems and perspectives. Nauka; 1964. pp. 182–187. [Google Scholar]
- 70.Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: a plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 71.Koshland DE, Jr, Nemethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5(1):365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- 72.Burgen AS. Conformational changes and drug action. Fed Proc. 1981;40(13):2723–2728. [PubMed] [Google Scholar]
- 73.Gronenborn AM, Clore GM, Blazy B, Baudras A. Conformational selection of syn-cAMP upon binding to the cAMP: receptor protein. FEBS Lett. 1981;136(1):160–164. doi: 10.1016/0014-5793(81)81237-9. [DOI] [PubMed] [Google Scholar]
- 74.Sedzik J, Kirschner DA. Is myelin basic protein crystallizable? Neurochem Res. 1992;17(2):157–166. doi: 10.1007/BF00966794. [DOI] [PubMed] [Google Scholar]
- 75.Yasuzumi G, Sawada T, Sugihara R, Kiriyama M, Sugioka M. Electron microscope researches on the ultrastructure of nucleoli in animal tissues. Z Zellforsch Mikrosk Anat. 1958;48(1):10–23. doi: 10.1007/BF00496710. [DOI] [PubMed] [Google Scholar]
- 76.Schofer C, Weipoltshammer K. Nucleolus and chromatin. Histochem Cell Biol. 2018;150(3):209–225. doi: 10.1007/s00418-018-1696-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Monneron A, Bernhard W. Fine structural organization of the interphase nucleus in some mammalian cells. J Ultrastruct Res. 1969;27(3):266–288. doi: 10.1016/s0022-5320(69)80017-1. [DOI] [PubMed] [Google Scholar]
- 78.Cajal SR. Un sencillo metodo de coloracion selectiva del reticulo protoplasmico y sus efectos en los diversos organos nerviosos de vertebrados e invertebrados. Trab Lab Investig Biol Univ Madr. 1903;2:129–221. [Google Scholar]
- 79.Cajal SR. Un sencillo metodo de coloracion seletiva del reticulo protoplasmatico y sus efectos en los diversos organos nerviosos de vertebrados e invertebrados. Trab Lab Invest Biol(Madrid) 1903;2:129–221. [Google Scholar]
- 80.Morris GE. The Cajal body. Biochim Biophys Acta. 2008;1783(11):2108–2115. doi: 10.1016/j.bbamcr.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 81.Alberti S. Phase separation in biology. Curr Biol. 2017;27(20):R1097–R1102. doi: 10.1016/j.cub.2017.08.069. [DOI] [PubMed] [Google Scholar]
- 82.Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, Obradovic Z. Intrinsically disordered protein. J Mol Graph Model. 2001;19(1):26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 83.Uversky VN, Gillespie JR, Fink AL. Why are "natively unfolded" proteins unstructured under physiologic conditions? Proteins. 2000;41(3):415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 84.Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293(2):321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 85.Oldfield CJ, Cheng Y, Cortese MS, Brown CJ, Uversky VN, Dunker AK. Comparing and combining predictors of mostly disordered proteins. Biochemistry. 2005;44(6):1989–2000. doi: 10.1021/bi047993o. [DOI] [PubMed] [Google Scholar]
- 86.van der Lee R, Buljan M, Lang B, Weatheritt RJ, Daughdrill GW, Dunker AK, Fuxreiter M, Gough J, Gsponer J, Jones DT, Kim PM, Kriwacki RW, Oldfield CJ, Pappu RV, Tompa P, Uversky VN, Wright PE, Babu MM. Classification of intrinsically disordered regions and proteins. Chem Rev. 2014;114(13):6589–6631. doi: 10.1021/cr400525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Oldfield CJ, Dunker AK. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem. 2014;83:553–584. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- 88.Uversky VN, Dunker AK. Understanding protein non-folding. Biochim Biophys Acta. 2010;1804(6):1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008;18(6):756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 90.Uversky VN, Oldfield CJ, Dunker AK. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J Mol Recognit. 2005;18(5):343–384. doi: 10.1002/jmr.747. [DOI] [PubMed] [Google Scholar]
- 91.Tompa P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 2005;579(15):3346–3354. doi: 10.1016/j.febslet.2005.03.072. [DOI] [PubMed] [Google Scholar]
- 92.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6(3):197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 93.Uversky VN. Protein folding revisited. A polypeptide chain at the folding-misfolding-nonfolding cross-roads: which way to go? Cell Mol Life Sci. 2003;60(9):1852–1871. doi: 10.1007/s00018-003-3096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27(10):527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 95.Uversky VN. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11(4):739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Uversky VN. What does it mean to be natively unfolded? Eur J Biochem. 2002;269(1):2–12. doi: 10.1046/j.0014-2956.2001.02649.x. [DOI] [PubMed] [Google Scholar]
- 97.Dunker AK, Obradovic Z. The protein trinity–linking function and disorder. Nat Biotechnol. 2001;19(9):805–806. doi: 10.1038/nbt0901-805. [DOI] [PubMed] [Google Scholar]
- 98.Hu G, Wu Z, Uversky VN, Kurgan L. Functional analysis of human hub proteins and their interactors involved in the intrinsic disorder-enriched interactions. Int J Mol Sci. 2017 doi: 10.3390/ijms18122761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Oldfield CJ, Meng J, Yang JY, Yang MQ, Uversky VN, Dunker AK. Flexible nets: disorder and induced fit in the associations of p53 and 14–3-3 with their partners. BMC Genom. 2008;9(Suppl 1):S1. doi: 10.1186/1471-2164-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dosztanyi Z, Chen J, Dunker AK, Simon I, Tompa P. Disorder and sequence repeats in hub proteins and their implications for network evolution. J Proteome Res. 2006;5(11):2985–2995. doi: 10.1021/pr060171o. [DOI] [PubMed] [Google Scholar]
- 101.Haynes C, Oldfield CJ, Ji F, Klitgord N, Cusick ME, Radivojac P, Uversky VN, Vidal M, Iakoucheva LM. Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput Biol. 2006;2(8):e100. doi: 10.1371/journal.pcbi.0020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272(20):5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 103.Dunker AK, Brown CJ, Obradovic Z. Identification and functions of usefully disordered proteins. Adv Protein Chem. 2002;62:25–49. doi: 10.1016/s0065-3233(02)62004-2. [DOI] [PubMed] [Google Scholar]
- 104.Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323(3):573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 105.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41(21):6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 106.Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12(1):54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 107.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets: the roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272(20):5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 108.Cortese MS, Uversky VN, Dunker AK. Intrinsic disorder in scaffold proteins: getting more from less. Prog Biophys Mol Biol. 2008;98(1):85–106. doi: 10.1016/j.pbiomolbio.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Dunker AK, Uversky VN. Signal transduction via unstructured protein conduits. Nat Chem Biol. 2008;4(4):229–230. doi: 10.1038/nchembio0408-229. [DOI] [PubMed] [Google Scholar]
- 110.Dunker AK, Oldfield CJ, Meng J, Romero P, Yang JY, Chen JW, Vacic V, Obradovic Z, Uversky VN. The unfoldomics decade: an update on intrinsically disordered proteins. BMC Genom. 2008;9(Suppl 2):S1. doi: 10.1186/1471-2164-9-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Uversky VN, Dunker AK. Understanding protein non-folding. Bba-Proteins Proteom. 2010;1804(6):1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Uversky VN, Li J, Fink AL. Metal-triggered structural transformations, aggregation, and fibrillation of human alpha-synuclein. A possible molecular NK between Parkinson's disease and heavy metal exposure. J Biol Chem. 2001;276(47):44284–44296. doi: 10.1074/jbc.M105343200. [DOI] [PubMed] [Google Scholar]
- 113.Uversky VN, Permyakov SE, Zagranichny VE, Rodionov IL, Fink AL, Cherskaya AM, Wasserman LA, Permyakov EA. Effect of zinc and temperature on the conformation of the gamma subunit of retinal phosphodiesterase: a natively unfolded protein. J Proteome Res. 2002;1(2):149–159. doi: 10.1021/pr0155127. [DOI] [PubMed] [Google Scholar]
- 114.Venyaminov SY, Gudkov AT, Gogia ZV, Tumanova LG (1981) Absorption and circular dichroism spectra of individual proteins from Escherichia coli ribosomes. American Chemical Society, Pushchino, Russia
- 115.Craig TA, Veenstra TD, Naylor S, Tomlinson AJ, Johnson KL, Macura S, Juranic N, Kumar R. Zinc binding properties of the DNA binding domain of the 1,25-dihydroxyvitamin D3 receptor. Biochemistry. 1997;36(34):10482–10491. doi: 10.1021/bi970561b. [DOI] [PubMed] [Google Scholar]
- 116.Rice LM, Brennwald P, Brunger AT. Formation of a yeast SNARE complex is accompanied by significant structural changes. FEBS Lett. 1997;415(1):49–55. doi: 10.1016/S0014-5793(97)01091-0. [DOI] [PubMed] [Google Scholar]
- 117.Permyakov SE, Millett IS, Doniach S, Permyakov EA, Uversky VN. Natively unfolded C-terminal domain of caldesmon remains substantially unstructured after the effective binding to calmodulin. Proteins. 2003;53(4):855–862. doi: 10.1002/prot.10481. [DOI] [PubMed] [Google Scholar]
- 118.Fink AL. Natively unfolded proteins. Curr Opin Struct Biol. 2005;15(1):35–41. doi: 10.1016/j.sbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 119.Peng Z, Mizianty MJ, Xue B, Kurgan L, Uversky VN. More than just tails: intrinsic disorder in histone proteins. Mol Biosyst. 2012;8(7):1886–1901. doi: 10.1039/c2mb25102g. [DOI] [PubMed] [Google Scholar]
- 120.Cheng Y, Oldfield CJ, Meng J, Romero P, Uversky VN, Dunker AK. Mining alpha-helix-forming molecular recognition features with cross species sequence alignments. Biochemistry. 2007;46(47):13468–13477. doi: 10.1021/bi7012273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vacic V, Oldfield CJ, Mohan A, Radivojac P, Cortese MS, Uversky VN, Dunker AK. Characterization of molecular recognition features, MoRFs, and their binding partners. J Proteome Res. 2007;6(6):2351–2366. doi: 10.1021/pr0701411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mohan A, Oldfield CJ, Radivojac P, Vacic V, Cortese MS, Dunker AK, Uversky VN. Analysis of molecular recognition features (MoRFs) J Mol Biol. 2006;362(5):1043–1059. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- 123.Oldfield CJ, Cheng Y, Cortese MS, Romero P, Uversky VN, Dunker AK. Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry. 2005;44(37):12454–12470. doi: 10.1021/bi050736e. [DOI] [PubMed] [Google Scholar]
- 124.Uversky VN. Multitude of binding modes attainable by intrinsically disordered proteins: a portrait gallery of disorder-based complexes. Chem Soc Rev. 2011;40(3):1623–1634. doi: 10.1039/c0cs00057d. [DOI] [PubMed] [Google Scholar]
- 125.Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem Sci. 2008;33(1):2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 126.Fuxreiter M. Towards a stochastic paradigm: from fuzzy ensembles to cellular functions. Molecules. 2018;23(11):3008. doi: 10.3390/molecules23113008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Miskei M, Gregus A, Sharma R, Duro N, Zsolyomi F, Fuxreiter M. Fuzziness enables context dependence of protein interactions. FEBS Lett. 2017;591(17):2682–2695. doi: 10.1002/1873-3468.12762. [DOI] [PubMed] [Google Scholar]
- 128.Gruet A, Dosnon M, Blocquel D, Brunel J, Gerlier D, Das RK, Bonetti D, Gianni S, Fuxreiter M, Longhi S, Bignon C. Fuzzy regions in an intrinsically disordered protein impair protein-protein interactions. FEBS J. 2016;283(4):576–594. doi: 10.1111/febs.13631. [DOI] [PubMed] [Google Scholar]
- 129.Sharma R, Raduly Z, Miskei M, Fuxreiter M. Fuzzy complexes: Specific binding without complete folding. FEBS Lett. 2015;589(19 Pt A):2533–2542. doi: 10.1016/j.febslet.2015.07.022. [DOI] [PubMed] [Google Scholar]
- 130.Fuxreiter M, Tompa P. Fuzzy complexes: a more stochastic view of protein function. Adv Exp Med Biol. 2012;725:1–14. doi: 10.1007/978-1-4614-0659-4_1. [DOI] [PubMed] [Google Scholar]
- 131.Fuxreiter M. Fuzziness: linking regulation to protein dynamics. Mol Biosyst. 2012;8(1):168–177. doi: 10.1039/c1mb05234a. [DOI] [PubMed] [Google Scholar]
- 132.Alterovitz WL, Faraggi E, Oldfield CJ, Meng J, Xue B, Huang F, Romero P, Kloczkowski A, Uversky VN, Dunker AK. Many-to-one binding by intrinsically disordered protein regions. Pac Symp Biocomput. 2020;25:159–170. [PubMed] [Google Scholar]
- 133.Borgia A, Borgia MB, Bugge K, Kissling VM, Heidarsson PO, Fernandes CB, Sottini A, Soranno A, Buholzer KJ, Nettels D, Kragelund BB, Best RB, Schuler B. Extreme disorder in an ultrahigh-affinity protein complex. Nature. 2018;555(7694):61–66. doi: 10.1038/nature25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Uversky VN. Intrinsic disorder in proteins associated with neurodegenerative diseases. Front Biosci (Landmark Ed) 2009;14:5188–5238. doi: 10.2741/3594. [DOI] [PubMed] [Google Scholar]
- 135.Uversky VN. The triple power of D(3): protein intrinsic disorder in degenerative diseases. Front Biosci (Landmark Ed) 2014;19:181–258. doi: 10.2741/4204. [DOI] [PubMed] [Google Scholar]
- 136.Uversky VN. Targeting intrinsically disordered proteins in neurodegenerative and protein dysfunction diseases: another illustration of the D(2) concept. Expert Rev Proteom. 2010;7(4):543–564. doi: 10.1586/epr.10.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Uversky VN. Mysterious oligomerization of the amyloidogenic proteins. FEBS J. 2010;277(14):2940–2953. doi: 10.1111/j.1742-4658.2010.07721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Uversky VN, Oldfield CJ, Dunker AK. Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu Rev Biophys. 2008;37:215–246. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- 139.Uversky VN, Dave V, Iakoucheva LM, Malaney P, Metallo SJ, Pathak RR, Joerger AC. Pathological unfoldomics of uncontrolled chaos: intrinsically disordered proteins and human diseases. Chem Rev. 2014;114(13):6844–6879. doi: 10.1021/cr400713r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Uversky VN. Wrecked regulation of intrinsically disordered proteins in diseases: pathogenicity of deregulated regulators. Front Mol Biosci. 2014;1:6. doi: 10.3389/fmolb.2014.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ. Intrinsic protein disorder in complete genomes. Genome Inform Ser Workshop Genome Inf. 2000;11:161–171. [PubMed] [Google Scholar]
- 142.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337(3):635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 143.Xue B, Dunker AK, Uversky VN. Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J Biomol Struct Dyn. 2012;30(2):137–149. doi: 10.1080/07391102.2012.675145. [DOI] [PubMed] [Google Scholar]
- 144.Peng Z, Yan J, Fan X, Mizianty MJ, Xue B, Wang K, Hu G, Uversky VN, Kurgan L. Exceptionally abundant exceptions: comprehensive characterization of intrinsic disorder in all domains of life. Cell Mol Life Sci. 2015;72(1):137–151. doi: 10.1007/s00018-014-1661-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yan J, Cheng J, Kurgan L, Uversky VN. Structural and functional analysis of "non-smelly" proteins. Cell Mol Life Sci. 2020;77(12):2423–2440. doi: 10.1007/s00018-019-03292-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.DeForte S, Uversky VN. Resolving the ambiguity: Making sense of intrinsic disorder when PDB structures disagree. Protein Sci. 2016;25(3):676–688. doi: 10.1002/pro.2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Le Gall T, Romero PR, Cortese MS, Uversky VN, Dunker AK. Intrinsic disorder in the Protein Data Bank. J Biomol Struct Dyn. 2007;24(4):325–342. doi: 10.1080/07391102.2007.10507123. [DOI] [PubMed] [Google Scholar]
- 148.Uversky VN. Unusual biophysics of intrinsically disordered proteins. Biochim Biophys Acta. 2013;1834(5):932–951. doi: 10.1016/j.bbapap.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 149.Uversky VN. Intrinsic disorder-based protein interactions and their modulators. Curr Pharm Des. 2013;19(23):4191–4213. doi: 10.2174/1381612811319230005. [DOI] [PubMed] [Google Scholar]
- 150.Uversky VN. Functional roles of transiently and intrinsically disordered regions within proteins. FEBS J. 2015;282(7):1182–1189. doi: 10.1111/febs.13202. [DOI] [PubMed] [Google Scholar]
- 151.Uversky VN. p53 proteoforms and intrinsic disorder: an illustration of the protein structure-function continuum concept. Int J Mol Sci. 2016;17(11):1874. doi: 10.3390/ijms17111874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Habchi J, Tompa P, Longhi S, Uversky VN. Introducing protein intrinsic disorder. Chem Rev. 2014;114(13):6561–6588. doi: 10.1021/cr400514h. [DOI] [PubMed] [Google Scholar]
- 153.Iakoucheva LM, Radivojac P, Brown CJ, O'Connor TR, Sikes JG, Obradovic Z, Dunker AK. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32(3):1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pejaver V, Hsu WL, Xin F, Dunker AK, Uversky VN, Radivojac P. The structural and functional signatures of proteins that undergo multiple events of post-translational modification. Protein Sci. 2014;23(8):1077–1093. doi: 10.1002/pro.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Uversky VN. Intrinsically disordered proteins in overcrowded milieu: membrane-less organelles, phase separation, and intrinsic disorder. Curr Opin Struct Biol. 2017;44:18–30. doi: 10.1016/j.sbi.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 156.Uversky VN. Protein intrinsic disorder-based liquid-liquid phase transitions in biological systems: complex coacervates and membrane-less organelles. Adv Colloid Interface Sci. 2017;239:97–114. doi: 10.1016/j.cis.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 157.Uversky VN. Recent developments in the field of intrinsically disordered proteins: intrinsic disorder-based emergence in cellular biology in light of the physiological and pathological liquid-liquid phase transitions. Annu Rev Biophys. 2021;50(1):135–156. doi: 10.1146/annurev-biophys-062920-063704. [DOI] [PubMed] [Google Scholar]
- 158.Dolgin E. What lava lamps and vinaigrette can teach us about cell biology. Nature. 2018;555(7696):300–302. doi: 10.1038/d41586-018-03070-2. [DOI] [PubMed] [Google Scholar]
- 159.Handwerger KE, Cordero JA, Gall JG. Cajal bodies, nucleoli, and speckles in the Xenopus oocyte nucleus have a low-density, sponge-like structure. Mol Biol Cell. 2005;16(1):202–211. doi: 10.1091/mbc.e04-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Strasser MJ, Mackenzie NC, Dumstrei K, Nakkrasae LI, Stebler J, Raz E. Control over the morphology and segregation of Zebrafish germ cell granules during embryonic development. BMC Dev Biol. 2008;8:58. doi: 10.1186/1471-213X-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Brangwynne CP, Mitchison TJ, Hyman AA. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci U S A. 2011;108(11):4334–4339. doi: 10.1073/pnas.1017150108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mitrea DM, Kriwacki RW. Phase separation in biology; functional organization of a higher order. Cell Commun Signal. 2016;14:1. doi: 10.1186/s12964-015-0125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18(5):285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, Russo PS, Jiang QX, Nixon BT, Rosen MK. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483(7389):336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Albertsson PA. Partition of cell particles and macromolecules. 3. New York: Wiley; 1986. [Google Scholar]
- 166.Zaslavsky BY. Aqueous two-phase partitioning: physical chemistry and bioanalytical applications. New York: Marcel Dekker; 1994. [Google Scholar]
- 167.Brangwynne CP. Phase transitions and size scaling of membrane-less organelles. J Cell Biol. 2013;203(6):875–881. doi: 10.1083/jcb.201308087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Brangwynne CP, Tompa P, Pappu RV. Polymer physics of intracellular phase transitions. Nat Phys. 2015;11:899–904. doi: 10.1038/nphys3532. [DOI] [Google Scholar]
- 169.Uversky VN, Kuznetsova IM, Turoverov KK, Zaslavsky B. Intrinsically disordered proteins as crucial constituents of cellular aqueous two phase systems and coacervates. FEBS Lett. 2015;589(1):15–22. doi: 10.1016/j.febslet.2014.11.028. [DOI] [PubMed] [Google Scholar]
- 170.Dundr M, Misteli T. Biogenesis of nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2(12):a000711. doi: 10.1101/cshperspect.a000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhu L, Brangwynne CP. Nuclear bodies: the emerging biophysics of nucleoplasmic phases. Curr Opin Cell Biol. 2015;34:23–30. doi: 10.1016/j.ceb.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, Brangwynne CP. Coexisting liquid phases underlie nucleolar subcompartments. Cell. 2016;165(7):1686–1697. doi: 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Decker M, Jaensch S, Pozniakovsky A, Zinke A, O'Connell KF, Zachariae W, Myers E, Hyman AA. Limiting amounts of centrosome material set centrosome size in C. elegans embryos. Curr Biol. 2011;21(15):1259–1267. doi: 10.1016/j.cub.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 174.Chuma S, Hosokawa M, Tanaka T, Nakatsuji N. Ultrastructural characterization of spermatogenesis and its evolutionary conservation in the germline: germinal granules in mammals. Mol Cell Endocrinol. 2009;306(1–2):17–23. doi: 10.1016/j.mce.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 175.Kiebler MA, Bassell GJ. Neuronal RNA granules: movers and makers. Neuron. 2006;51(6):685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 176.Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol. 2007;179(3):437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wippich F, Bodenmiller B, Trajkovska MG, Wanka S, Aebersold R, Pelkmans L. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell. 2013;152(4):791–805. doi: 10.1016/j.cell.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 178.Uniacke J, Zerges W. Stress induces the assembly of RNA granules in the chloroplast of Chlamydomonas reinhardtii. J Cell Biol. 2008;182(4):641–646. doi: 10.1083/jcb.200805125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Antonicka H, Shoubridge EA. Mitochondrial RNA granules are centers for posttranscriptional RNA processing and ribosome biogenesis. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.01.030. [DOI] [PubMed] [Google Scholar]
- 180.Strzelecka M, Trowitzsch S, Weber G, Luhrmann R, Oates AC, Neugebauer KM. Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis. Nat Struct Mol Biol. 2010;17(4):403–409. doi: 10.1038/nsmb.1783. [DOI] [PubMed] [Google Scholar]
- 181.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128(4):707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 182.Li L, Roy K, Katyal S, Sun X, Bleoo S, Godbout R. Dynamic nature of cleavage bodies and their spatial relationship to DDX1 bodies, Cajal bodies, and gems. Mol Biol Cell. 2006;17(3):1126–1140. doi: 10.1091/mbc.E05-08-0768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Nizami Z, Deryusheva S, Gall JG. The Cajal body and histone locus body. Cold Spring Harb Perspect Biol. 2010;2(7):a000653. doi: 10.1101/cshperspect.a000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Matera AG, Frey MR. Coiled bodies and gems: Janus or gemini? Am J Hum Genet. 1998;63(2):317–321. doi: 10.1086/301992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Gubitz AK, Feng W, Dreyfuss G. The SMN complex. Exp Cell Res. 2004;296(1):51–56. doi: 10.1016/j.yexcr.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 186.Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4(8):605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- 187.Grossman E, Medalia O, Zwerger M. Functional architecture of the nuclear pore complex. Annu Rev Biophys. 2012;41:557–584. doi: 10.1146/annurev-biophys-050511-102328. [DOI] [PubMed] [Google Scholar]
- 188.Biamonti G, Vourc'h C. Nuclear stress bodies. Cold Spring Harb Perspect Biol. 2010;2(6):a000695. doi: 10.1101/cshperspect.a000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Biamonti G. Nuclear stress bodies: a heterochromatin affair? Nat Rev Mol Cell Biol. 2004;5(6):493–498. doi: 10.1038/nrm1405. [DOI] [PubMed] [Google Scholar]
- 190.Shav-Tal Y, Blechman J, Darzacq X, Montagna C, Dye BT, Patton JG, Singer RH, Zipori D. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol Biol Cell. 2005;16(5):2395–2413. doi: 10.1091/mbc.E04-11-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Harrigan JA, Belotserkovskaya R, Coates J, Dimitrova DS, Polo SE, Bradshaw CR, Fraser P, Jackson SP. Replication stress induces 53BP1-containing OPT domains in G1 cells. J Cell Biol. 2011;193(1):97–108. doi: 10.1083/jcb.201011083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Huang S. Review: perinucleolar structures. J Struct Biol. 2000;129(2–3):233–240. doi: 10.1006/jsbi.2000.4247. [DOI] [PubMed] [Google Scholar]
- 193.Fox AH, Lam YW, Leung AK, Lyon CE, Andersen J, Mann M, Lamond AI. Paraspeckles: a novel nuclear domain. Curr Biol. 2002;12(1):13–25. doi: 10.1016/S0960-9822(01)00632-7. [DOI] [PubMed] [Google Scholar]
- 194.Maul GG, Negorev D, Bell P, Ishov AM. Review: properties and assembly mechanisms of ND10, PML bodies, or PODs. J Struct Biol. 2000;129(2–3):278–287. doi: 10.1006/jsbi.2000.4239. [DOI] [PubMed] [Google Scholar]
- 195.Pirrotta V, Li HB. A view of nuclear Polycomb bodies. Curr Opin Genet Dev. 2012;22(2):101–109. doi: 10.1016/j.gde.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Boeynaems S, Alberti S, Fawzi NL, Mittag T, Polymenidou M, Rousseau F, Schymkowitz J, Shorter J, Wolozin B, Van Den Bosch L, Tompa P, Fuxreiter M. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 2018;28(6):420–435. doi: 10.1016/j.tcb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Turoverov KK, Kuznetsova IM, Fonin AV, Darling AL, Zaslavsky BY, Uversky VN. Stochasticity of biological soft matter: emerging concepts in intrinsically disordered proteins and biological phase separation. Trends Biochem Sci. 2019;44(8):716–728. doi: 10.1016/j.tibs.2019.03.005. [DOI] [PubMed] [Google Scholar]
- 198.Darling AL, Liu Y, Oldfield CJ, Uversky VN. Intrinsically disordered proteome of human membrane-less organelles. Proteomics. 2018;18(5–6):e1700193. doi: 10.1002/pmic.201700193. [DOI] [PubMed] [Google Scholar]
- 199.Zhou HX, Nguemaha V, Mazarakos K, Qin S. Why do disordered and structured proteins behave differently in phase separation? Trends Biochem Sci. 2018;43(7):499–516. doi: 10.1016/j.tibs.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Fonin AV, Silonov SA, Shpironok OG, Antifeeva IA, Petukhov AV, Romanovich AE, Kuznetsova IM, Uversky VN, Turoverov KK. The Role of non-specific interactions in canonical and ALT-associated PML-bodies formation and dynamics. Int J Mol Sci. 2021 doi: 10.3390/ijms22115821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Williamson DJ, Owen DM, Rossy J, Magenau A, Wehrmann M, Gooding JJ, Gaus K. Pre-existing clusters of the adaptor Lat do not participate in early T cell signaling events. Nat Immunol. 2011;12(7):655–662. doi: 10.1038/ni.2049. [DOI] [PubMed] [Google Scholar]
- 202.Maszewski J, Kwiatkowska M. Number, size, and transcriptional activity of nucleoli during different periods of interphase in antheridial filaments of Chara vulgaris L. Folia Histochem Cytobiol. 1984;22(1):9–19. [PubMed] [Google Scholar]
- 203.Anderson P, Kedersha N. Stress granules. Curr Biol. 2009;19(10):R397–R398. doi: 10.1016/J.CUB.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 204.Lyon AS, Peeples WB, Rosen MK. A framework for understanding the functions of biomolecular condensates across scales. Nat Rev Mol Cell Biol. 2021 doi: 10.1038/s41580-020-00303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Weber APSC. Evidence for and against Liquid-Liquid Phase Separation in the Nucleus. Non-coding RNA. 2019 doi: 10.3390/ncrna5040050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Boke E, Ruer M, Wühr M, Coughlin M, Lemaitre R, Gygi SP, Alberti S, Drechsel D, Hyman AA, Mitchison TJ. Amyloid-like self-assembly of a cellular compartment. Cell. 2016;166(3):637–650. doi: 10.1016/J.CELL.2016.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Alberti S, Gladfelter A, Mittag T. Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell. 2019;176(3):419–434. doi: 10.1016/j.cell.2018.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.King MR, Petry S. Phase separation of TPX2 enhances and spatially coordinates microtubule nucleation. Nat Commun. 2020;11(1):1–13. doi: 10.1038/s41467-019-14087-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Peeples W, Rosen MK. Phase separation can increase enzyme activity by concentration and molecular organization. bioRxiv. 2020 doi: 10.1101/2020.09.15.299115. [DOI] [Google Scholar]
- 210.Powers SK, Holehouse AS, Korasick DA, Schreiber KH, Clark NM, Jing H, Emenecker R, Han S, Tycksen E, Hwang I, Sozzani R, Jez JM, Pappu RV, Strader LC. Nucleo-cytoplasmic partitioning of ARF proteins controls auxin responses in Arabidopsis thaliana. Mol Cell. 2019;76(1):177–190.e175. doi: 10.1016/J.MOLCEL.2019.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Su X, Ditlev JA, Hui E, Xing W, Banjade S, Okrut J, King DS, Taunton J, Rosen MK, Vale RD. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science. 2016;352(6285):595–599. doi: 10.1126/science.aad9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Case LB, Zhang X, Ditlev JA, Rosen MK. Stoichiometry controls activity of phase-separated clusters of actin signaling proteins. Science (New York, NY) 2019;363(6431):1093–1097. doi: 10.1126/SCIENCE.AAU6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Frottin F, Schueder F, Tiwary S, Gupta R, Körner R, Schlichthaerle T, Cox J, Jungmann R, Hartl FU, Hipp MS. The nucleolus functions as a phase-separated protein quality control compartment. Science (New York, NY) 2019;365(6451):342–347. doi: 10.1126/SCIENCE.AAW9157. [DOI] [PubMed] [Google Scholar]
- 214.Deviri D, Safran SA. Physical theory of biological noise buffering by multicomponent phase separation. Proc Natl Acad Sci USA. 2021;118:25. doi: 10.1073/PNAS.2100099118/SUPPL_FILE/PNAS.2100099118.SAPP.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Riback JA, Katanski CD, Kear-Scott JL, Pilipenko EV, Rojek AE, Sosnick TR, Drummond DA. Stress-triggered phase separation is an adaptive evolutionarily tuned response. Cell. 2017;168(6):1028–1040.e1019. doi: 10.1016/J.CELL.2017.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Bergeron-Sandoval L-P, Heris HK, Chang C, Cornell CE, Keller SL, François P, Hendricks AG, Ehrlicher AJ, Pappu RV, Michnick SW. Endocytosis caused by liquid-liquid phase separation of proteins. bioRxiv. 2018 doi: 10.1101/145664. [DOI] [Google Scholar]
- 217.Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163(1):123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Jiang H, Wang S, Huang Y, He X, Cui H, Zhu X, Zheng Y. Phase transition of spindle-associated protein regulate spindle apparatus assembly. Cell. 2015;163(1):108–122. doi: 10.1016/j.cell.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, Baldwin AJ. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell. 2015;57(5):936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Andersen JS, Lam YW, Leung AKL, Ong SE, Lyon CE, Lamond AI, Mann M. Nucleolar proteome dynamics. Nature. 2005;433(7021):77–83. doi: 10.1038/NATURE03207. [DOI] [PubMed] [Google Scholar]
- 221.Kim W, Kim DY, Lee KH. RNA-binding proteins and the complex pathophysiology of ALS. Int J Mol Sci. 2021 doi: 10.3390/ijms22052598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Darling AL, Shorter J. Combating deleterious phase transitions in neurodegenerative disease. Biochim Biophys Acta Mol Cell Res. 2021;1868(5):118984. doi: 10.1016/j.bbamcr.2021.118984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Webber CJ, Lei SE, Wolozin B. The pathophysiology of neurodegenerative disease: disturbing the balance between phase separation and irreversible aggregation. Prog Mol Biol Transl Sci. 2020;174:187–223. doi: 10.1016/bs.pmbts.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Babinchak WM, Surewicz WK. Liquid-liquid phase separation and its mechanistic role in pathological protein aggregation. J Mol Biol. 2020;432(7):1910–1925. doi: 10.1016/j.jmb.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Verdile V, De Paola E, Paronetto MP. Aberrant phase transitions: side effects and novel therapeutic strategies in human disease. Front Genet. 2019;10:173. doi: 10.3389/fgene.2019.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Hofweber M, Dormann D. Friend or foe-Post-translational modifications as regulators of phase separation and RNP granule dynamics. J Biol Chem. 2019;294(18):7137–7150. doi: 10.1074/jbc.TM118.001189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162(5):1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 228.Carey JL, Guo L. Liquid-liquid phase separation of TDP-43 and FUS in physiology and pathology of neurodegenerative diseases. Front Mol Biosci. 2022 doi: 10.3389/FMOLB.2022.826719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Boyko S, Surewicz WK. Tau liquid-liquid phase separation in neurodegenerative diseases. Trends Cell Biol. 2022 doi: 10.1016/J.TCB.2022.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Darling AL, Dahrendorff J, Creodore SG, Dickey CA, Blair LJ, Uversky VN. Small heat shock protein 22 kDa can modulate the aggregation and liquid-liquid phase separation behavior of tau. Protein Sci. 2021;30(7):1350–1359. doi: 10.1002/pro.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Ray S, Singh N, Kumar R, Patel K, Pandey S, Datta D, Mahato J, Panigrahi R, Navalkar A, Mehra S, Gadhe L, Chatterjee D, Sawner AS, Maiti S, Bhatia S, Gerez JA, Chowdhury A, Kumar A, Padinhateeri R, Riek R, Krishnamoorthy G, Maji SK. α-Synuclein aggregation nucleates through liquid-liquid phase separation. Nat Chem. 2020;12(8):705–716. doi: 10.1038/s41557-020-0465-9. [DOI] [PubMed] [Google Scholar]
- 232.Boeynaems S, Tompa P, Van Den Bosch L. Phasing in on the cell cycle. Cell Div. 2018;13:1. doi: 10.1186/s13008-018-0034-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Mahboubi H. Stochaj U (2017) Cytoplasmic stress granules: Dynamic modulators of cell signaling and disease. Biochim Biophys Acta. 1863;4:884–895. doi: 10.1016/j.bbadis.2016.12.022. [DOI] [PubMed] [Google Scholar]
- 234.Kuechler ER, Budzyńska PM, Bernardini JP, Gsponer J, Mayor T. Distinct features of stress granule proteins predict localization in membraneless organelles. J Mol Biol. 2020;432(7):2349–2368. doi: 10.1016/j.jmb.2020.02.020. [DOI] [PubMed] [Google Scholar]
- 235.Khong A, Matheny T, Jain S, Mitchell SF, Wheeler JR, Parker R. The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol Cell. 2017;68(4):808–820e805. doi: 10.1016/j.molcel.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Yang P, Mathieu C, Kolaitis RM, Zhang P, Messing J, Yurtsever U, Yang Z, Wu J, Li Y, Pan Q, Yu J, Martin EW, Mittag T, Kim HJ, Taylor JP. G3BP1 Is a tunable switch that triggers phase separation to assemble stress granules. Cell. 2020;181(2):325–345e328. doi: 10.1016/j.cell.2020.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Protter DSW, Parker R. Principles and properties of stress granules. Trends Cell Biol. 2016;26(9):668–679. doi: 10.1016/j.tcb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell. 2016;164(3):487–498. doi: 10.1016/j.cell.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Guillen-Boixet J, Kopach A, Holehouse AS, Wittmann S, Jahnel M, Schlussler R, Kim K, Trussina I, Wang J, Mateju D, Poser I, Maharana S, Ruer-Gruss M, Richter D, Zhang X, Chang YT, Guck J, Honigmann A, Mahamid J, Hyman AA, Pappu RV, Alberti S, Franzmann TM. RNA-induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell. 2020;181(2):346–361 e317. doi: 10.1016/j.cell.2020.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Cao X, Jin X, Liu B. The involvement of stress granules in aging and aging-associated diseases. Aging Cell. 2020;19(4):e13136. doi: 10.1111/acel.13136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Elbaum-Garfinkle S. Matter over mind: Liquid phase separation and neurodegeneration. J Biol Chem. 2019;294(18):7160–7168. doi: 10.1074/jbc.REV118.001188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Tüű-Szabó B, Hoffka G, Duro N. Fuxreiter M (2019) Altered dynamics may drift pathological fibrillization in membraneless organelles. Biochim Biophys Acta. 1867;10:988–998. doi: 10.1016/j.bbapap.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 243.Marijan D, Tse R, Elliott K, Chandhok S, Luo M, Lacroix E, Audas TE. Stress-specific aggregation of proteins in the amyloid bodies. FEBS Lett. 2019;593(22):3162–3172. doi: 10.1002/1873-3468.13597. [DOI] [PubMed] [Google Scholar]
- 244.Marmor-Kollet H, Siany A, Kedersha N, Knafo N, Rivkin N, Danino YM, Moens TG, Olender T, Sheban D, Cohen N, Dadosh T, Addadi Y, Ravid R, Eitan C, Toth Cohen B, Hofmann S, Riggs CL, Advani VM, Higginbottom A, Cooper-Knock J, Hanna JH, Merbl Y, Van Den Bosch L, Anderson P, Ivanov P, Geiger T, Hornstein E. Spatiotemporal proteomic analysis of stress granule disassembly using APEX reveals regulation by SUMOylation and links to ALS pathogenesis. Mol Cell. 2020;80(5):876–891e876. doi: 10.1016/j.molcel.2020.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Moujaber O, Mahboubi H, Kodiha M, Bouttier M, Bednarz K, Bakshi R, White J, Larose L, Colmegna I, Stochaj U. Dissecting the molecular mechanisms that impair stress granule formation in aging cells. Biochim Biophys Acta Mol Cell Res. 2017;1864(3):475–486. doi: 10.1016/j.bbamcr.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 246.Shiina N. Liquid- and solid-like RNA granules form through specific scaffold proteins and combine into biphasic granules. J Biol Chem. 2019;294(10):3532–3548. doi: 10.1074/jbc.RA118.005423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Shin Y, Brangwynne CP. Liquid phase condensation in cell physiology and disease. Science. 2017 doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- 248.Wang Z, Zhang H. Phase separation, transition, and autophagic degradation of proteins in development and pathogenesis. Trends Cell Biol. 2019;29(5):417–427. doi: 10.1016/j.tcb.2019.01.008. [DOI] [PubMed] [Google Scholar]
- 249.Wang M, Tao X, Jacob MD, Bennett CA, Ho JJD, Gonzalgo ML, Audas TE, Lee S. Stress-induced low complexity RNA activates physiological amyloidogenesis. Cell Rep. 2018;24(7):1713–1721.e1714. doi: 10.1016/J.CELREP.2018.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Audas TE, Audas DE, Jacob MD, Ho JJD, Khacho M, Wang M, Perera JK, Gardiner C, Bennett CA, Head T, Kryvenko ON, Jorda M, Daunert S, Malhotra A, Trinkle-Mulcahy L, Gonzalgo ML, Lee S. Adaptation to stressors by systemic protein amyloidogenesis. Dev Cell. 2016;39(2):155–168. doi: 10.1016/J.DEVCEL.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Wegmann S, Eftekharzadeh B, Tepper K, Zoltowska KM, Bennett RE, Dujardin S, Laskowski PR, MacKenzie D, Kamath T, Commins C, Vanderburg C, Roe AD, Fan Z, Molliex AM, Hernandez-Vega A, Muller D, Hyman AA, Mandelkow E, Taylor JP, Hyman BT. Tau protein liquid–liquid phase separation can initiate tau aggregation. EMBO J. 2018;37(7):e98049–e98049. doi: 10.15252/EMBJ.201798049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Gaglia G, Rashid R, Yapp C, Joshi GN, Li CG, Lindquist SL, Sarosiek KA, Whitesell L, Sorger PK, Santagata S. HSF1 phase transition mediates stress adaptation and cell fate decisions. Nat Cell Biol. 2020;22(2):151–158. doi: 10.1038/s41556-019-0458-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Watanabe K, Ohtsuki T. Inhibition of HSF1 and SAFB granule formation enhances apoptosis induced by heat stress. Int J Mol Sci. 2021;22(9):4982–4982. doi: 10.3390/IJMS22094982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Ninomiya K, Adachi S, Natsume T, Iwakiri J, Terai G, Asai K, Hirose T. LncRNA-dependent nuclear stress bodies promote intron retention through SR protein phosphorylation. EMBO J. 2020;39(3):e102729–e102729. doi: 10.15252/EMBJ.2019102729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Jarmuz M, Glotzbach CD, Bailey KA, Bandyopadhyay R, Shaffer LG. The Evolution of satellite III DNA subfamilies among primates. Am J Hum Genet. 2007;80(3):495–501. doi: 10.1086/512132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Jolly C, Lakhotia SC. Human sat III and Drosophila hsrω transcripts: a common paradigm for regulation of nuclear RNA processing in stressed cells. Nucleic Acids Res. 2006;34(19):5508–5514. doi: 10.1093/NAR/GKL711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Prasanth KV, Rajendra TK, Lal AK, Lakhotia SC. Omega speckles - a novel class of nuclear speckles containing hnRNPs associated with noncoding hsr-omega RNA in Drosophila. J Cell Sci. 2000;13 Pt 19(19):3485–3497. doi: 10.1242/JCS.113.19.3485. [DOI] [PubMed] [Google Scholar]
- 258.Riviere M, Bernhard W. Examen au microscope électronique de la tumeur VX2 du lapin domestique dérivée du papillome de Shope. Bull Cancer. 1960;47:570–584. [PubMed] [Google Scholar]
- 259.Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8(12):1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- 260.Corpet A, Kleijwegt C, Roubille S, Juillard F, Jacquet K, Texier P, Lomonte P. PML nuclear bodies and chromatin dynamics: catch me if you can! Nucleic Acids Res. 2020;48(21):11890–11912. doi: 10.1093/nar/gkaa828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Weidtkamp-Peters S, Lenser T, Negorev D, Gerstner N, Hofmann TG, Schwanitz G, Hoischen C, Maul G, Dittrich P, Hemmerich P. Dynamics of component exchange at PML nuclear bodies. J Cell Sci. 2008;121(Pt 16):2731–2743. doi: 10.1242/jcs.031922. [DOI] [PubMed] [Google Scholar]
- 262.Nisole S, Maroui MA, Mascle XH, Aubry M, Chelbi-Alix MK. Differential roles of PML isoforms. Front Oncol. 2013;3:125. doi: 10.3389/fonc.2013.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Li Y, Ma X, Chen Z, Wu H, Wang P, Wu W, Cheng N, Zeng L, Zhang H, Cai X, Chen SJ, Chen Z, Meng G. B1 oligomerization regulates PML nuclear body biogenesis and leukemogenesis. Nat Commun. 2019;10(1):3789. doi: 10.1038/s41467-019-11746-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Wang P, Benhenda S, Wu H, Lallemand-Breitenbach V, Zhen T, Jollivet F, Peres L, Li Y, Chen SJ, Chen Z, de Thé H, Meng G. RING tetramerization is required for nuclear body biogenesis and PML sumoylation. Nat Commun. 2018;9(1):1277. doi: 10.1038/s41467-018-03498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Li C, Peng Q, Wan X, Sun H, Tang J. C-terminal motifs in promyelocytic leukemia protein isoforms critically regulate PML nuclear body formation. J Cell Sci. 2017;130(20):3496–3506. doi: 10.1242/jcs.202879. [DOI] [PubMed] [Google Scholar]
- 266.Rajpal G, Arvan P. Chapter 236—disulfide bond formation. In: Kastin AJ, editor. Handbook of biologically active peptides. 2. Boston: Academic Press; 2013. pp. 1721–1729. [Google Scholar]
- 267.Fonin AV, Silonov SA, Fefilova AS, Stepanenko OV, Gavrilova AA, Petukhov AV, Romanovich AE, Modina AL, Zueva TS, Nedelyaev EM, Pleskach NM, Kuranova ML, Kuznetsova IM, Uversky VN, Turoverov KK. New evidence of the importance of weak interactions in the formation of PML-bodies. Int J Mol Sci. 2022 doi: 10.3390/ijms23031613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Le Ferrand H, Duchamp M, Gabryelczyk B, Cai H, Miserez A. Time-resolved observations of liquid-liquid phase separation at the nanoscale using in situ liquid transmission electron microscopy. J Am Chem Soc. 2019;141(17):7202–7210. doi: 10.1021/jacs.9b03083. [DOI] [PubMed] [Google Scholar]
- 269.Alshareedah I, Kaur T, Banerjee PR (2021) Methods for characterizing the material properties of biomolecular condensates. In: Methods in enzymology, vol 646. 10.1016/bs.mie.2020.06.009 [DOI] [PMC free article] [PubMed]
- 270.Wang J, Choi JM, Holehouse AS, Lee HO, Zhang X, Jahnel M, Maharana S, Lemaitre R, Pozniakovsky A, Drechsel D, Poser I, Pappu RV, Alberti S, Hyman AA. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell. 2018 doi: 10.1016/j.cell.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Gui X, Luo F, Li Y, Zhou H, Qin Z, Liu Z, Gu J, Xie M, Zhao K, Dai B, Shin WS, He J, He L, Jiang L, Zhao M, Sun B, Li X, Liu C, Li D. Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly. Nat Commun. 2019 doi: 10.1038/s41467-019-09902-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Alshareedah I, Kaur T, Ngo J, Seppala H, Kounatse LAD, Wang W, Moosa MM, Banerjee PR. Interplay between short-range attraction and long-range repulsion controls reentrant liquid condensation of ribonucleoprotein-RNA complexes. J Am Chem Soc. 2019 doi: 10.1021/jacs.9b03689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CC, Eckmann CR, Myong S, Brangwynne CP. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc Natl Acad Sci U S A. 2015;112(23):7189–7194. doi: 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Hansen AS, Woringer M, Grimm JB, Lavis LD, Tjian R, Darzacq X. Robust model-based analysis of single-particle tracking experiments with spot-on. Elife. 2018 doi: 10.7554/eLife.33125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Shen H, Tauzin LJ, Baiyasi R, Wang W, Moringo N, Shuang B, Landes CF. Single particle tracking: from theory to biophysical applications. Chem Rev. 2017 doi: 10.1021/acs.chemrev.6b00815. [DOI] [PubMed] [Google Scholar]
- 276.Babinchak WM, Haider R, Dumm BK, Sarkar P, Surewicz K, Choi JK, Surewicz WK. The role of liquid-liquid phase separation in aggregation of the TDP-43 low-complexity domain. J Biol Chem. 2019 doi: 10.1074/jbc.RA118.007222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Jawerth LM, Ijavi M, Ruer M, Saha S, Jahnel M, Hyman AA, Jülicher F, Fischer-Friedrich E. Salt-dependent rheology and surface tension of protein condensates using optical traps. Phys Rev Lett. 2018 doi: 10.1103/PhysRevLett.121.258101. [DOI] [PubMed] [Google Scholar]
- 278.Zhou HX. Determination of condensate material properties from droplet deformation. J Phys Chem B. 2020 doi: 10.1021/acs.jpcb.0c06230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Zhou HX. Viscoelasticity of biomolecular condensates conforms to the Jeffreys model. J Chem Phys. 2021 doi: 10.1063/5.0038916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Taylor NO, Wei MT, Stone HA, Brangwynne CP. Quantifying dynamics in phase-separated condensates using fluorescence recovery after photobleaching. Biophys J. 2019 doi: 10.1016/j.bpj.2019.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Mitrea DM, Chandra B, Ferrolino MC, Gibbs EB, Tolbert M, White MR, Kriwacki RW. Methods for physical characterization of phase-separated bodies and membrane-less organelles. J Mol Biol. 2018;430(23):4773–4805. doi: 10.1016/j.jmb.2018.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Wei MT, Elbaum-Garfinkle S, Holehouse AS, Chen CCH, Feric M, Arnold CB, Priestley RD, Pappu RV, Brangwynne CP. Phase behaviour of disordered proteins underlying low density and high permeability of liquid organelles. Nat Chem. 2017 doi: 10.1038/NCHEM.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Spector DL, Lamond AI. Nuclear speckles. Cold Spring Harb Perspect Biol. 2011;3(2):a000646–a000646. doi: 10.1101/cshperspect.a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Brady JP, Farber PJ, Sekhar A, Lin YH, Huang R, Bah A, Nott TJ, Chan HS, Baldwin AJ, Forman-Kay JD, Kay LE. Structural and hydrodynamic properties of an intrinsically disordered region of a germ cell-specific protein on phase separation. Proc Natl Acad Sci USA. 2017 doi: 10.1073/pnas.1706197114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH. Phase separation drives heterochromatin domain formation. Nature. 2017 doi: 10.1038/nature22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Ulbrich R (2017) Beyond the limit: the world of super-resolution microscopy. Am Lab 49(8)
- 287.Raška I. Oldies but goldies: searching for Christmas trees within the nucleolar architecture. Trends Cell Biol. 2003 doi: 10.1016/j.tcb.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 288.Swift H. Studies on nuclear fine structure. Brookhaven Symp Biol. 1959 doi: 10.1016/0014-4827(65)90200-4. [DOI] [PubMed] [Google Scholar]
- 289.Gall JG, Bellini M, Za W, Murphy C. Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol Biol Cell. 1999;10:12. doi: 10.1091/mbc.10.12.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Franzmann TM, Jahnel M, Pozniakovsky A, Mahamid J, Holehouse AS, Nüske E, Richter D, Baumeister W, Grill SW, Pappu RV, Hyman AA, Alberti S. Phase separation of a yeast prion protein promotes cellular fitness. Science. 2018 doi: 10.1126/science.aao5654. [DOI] [PubMed] [Google Scholar]
- 291.Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, Pozniakovski A, Poser I, Maghelli N, Royer LA, Weigert M, Myers EW, Grill S, Drechsel D, Hyman AA, Alberti S. A Liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162(5):1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 292.Schaffer M, Pfeffer S, Mahamid J, Kleindiek S, Laugks T, Albert S, Engel BD, Rummel A, Smith AJ, Baumeister W, Plitzko JM. A cryo-FIB lift-out technique enables molecular-resolution cryo-ET within native Caenorhabditis elegans tissue. Nat Methods. 2019;16(8):757–762. doi: 10.1038/s41592-019-0497-5. [DOI] [PubMed] [Google Scholar]
- 293.Conicella AE, Zerze GH, Mittal J, Fawzi NL. ALS mutations disrupt phase separation mediated by α-helical structure in the TDP-43 low-complexity C-terminal domain. Structure. 2016 doi: 10.1016/j.str.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Stender EGP, Ray S, Norrild RK, Larsen JA, Petersen D, Farzadfard A, Galvagnion C, Jensen H, Buell AK, Norrild RK. Capillary flow experiments for thermodynamic and kinetic characterization of protein liquid-liquid phase separation. Nat Commun. 2021 doi: 10.1038/s41467-021-27433-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Otzen DE, Buell AK, Jensen H. Microfluidics and the quantification of biomolecular interactions. Curr Opin Struct Biol. 2021 doi: 10.1016/j.sbi.2021.02.006. [DOI] [PubMed] [Google Scholar]
- 296.Bremer A, Mittag T, Heymann M. Microfluidic characterization of macromolecular liquid-liquid phase separation. bioRxiv. 2020 doi: 10.1101/2020.06.16.154518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Linsenmeier M, Kopp MRG, Stavrakis S, de Mello A, Arosio P. Analysis of biomolecular condensates and protein phase separation with microfluidic technology. Biochim Biophys Acta Mol Cell Res. 2021 doi: 10.1016/j.bbamcr.2020.118823. [DOI] [PubMed] [Google Scholar]
- 298.Taylor N, Elbaum-Garfinkle S, Vaidya N, Zhang H, Stone HA, Brangwynne CP. Biophysical characterization of organelle-based RNA/protein liquid phases using microfluidics. Soft Matter. 2016 doi: 10.1039/C6SM01087C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Dormann D. FG-nucleoporins caught in the act of liquid-liquid phase separation. J Cell Biol. 2020 doi: 10.1083/jcb.201910211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Rho HS, Gardeniers H. Microfluidic Droplet-Storage Array. Micromachines. 2020;11(6):608–608. doi: 10.3390/MI11060608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Van Lindt J, Bratek-Skicki A, Nguyen PN, Pakravan D, Durán-Armenta LF, Tantos A, Pancsa R, Van Den Bosch L, Maes D, Tompa P. A generic approach to study the kinetics of liquid–liquid phase separation under near-native conditions. Commun Biol. 2021;4(1):77–77. doi: 10.1038/s42003-020-01596-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Shin Y, Berry J, Pannucci N, Haataja MP, Toettcher JE, Brangwynne CP. Spatiotemporal control of intracellular phase transitions using light-activated optodroplets. Cell. 2017;168(1–2):159–171e114. doi: 10.1016/j.cell.2016.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Bracha D, Walls MT, Wei MT, Zhu L, Kurian M, Avalos JL, Toettcher JE, Brangwynne CP. Mapping local and global liquid phase behavior in living cells using photo-oligomerizable seeds. Cell. 2018;175(6):1467–1480e1413. doi: 10.1016/j.cell.2018.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Dine E, Gil AA, Uribe G, Brangwynne CP, Toettcher JE. Protein phase separation provides long-term memory of transient spatial stimuli. Cell Syst. 2018;6(6):655–663 e655. doi: 10.1016/j.cels.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Zhu L, Richardson TM, Wacheul L, Wei MT, Feric M, Whitney G, Lafontaine DLJ, Brangwynne CP. Controlling the material properties and rRNA processing function of the nucleolus using light. Proc Natl Acad Sci U S A. 2019;116(35):17330–17335. doi: 10.1073/pnas.1903870116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Fonin AV, Antifeeva IA, Shpironok OG, Stepanenko OV, Silonov SA, Stepanenko OV, Antifeev IE, Romanovich AE, Kuznetsova IM, Kim J-I, Uversky VN, Turoverov KK. Photo-dependent membrane-less organelles formed from plant phyB and PIF6 proteins in mammalian cells. Int J Biol Macromol. 2021;176:325–331. doi: 10.1016/j.ijbiomac.2021.02.075. [DOI] [PubMed] [Google Scholar]
- 307.Sanders DW, Kedersha N, Lee DSW, Strom AR, Drake V, Riback JA, Bracha D, Eeftens JM, Iwanicki A, Wang A, Wei MT, Whitney G, Lyons SM, Anderson P, Jacobs WM, Ivanov P, Brangwynne CP. Competing protein-RNA interaction networks control multiphase intracellular organization. Cell. 2020;181(2):306–324 e328. doi: 10.1016/j.cell.2020.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Liu ZS, Cai H, Xue W, Wang M, Xia T, Li WJ, Xing JQ, Zhao M, Huang YJ, Chen S, Wu SM, Wang X, Liu X, Pang X, Zhang ZY, Li T, Dai J, Dong F, Xia Q, Li AL, Zhou T, Liu ZG, Zhang XM, Li T. G3BP1 promotes DNA binding and activation of cGAS. Nat Immunol. 2019;20(1):18–28. doi: 10.1038/s41590-018-0262-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Shin Y, Chang YC, Lee DSW, Berry J, Sanders DW, Ronceray P, Wingreen NS, Haataja M, Brangwynne CP. Liquid nuclear condensates mechanically sense and restructure the genome. Cell. 2018;175(6):1481–1491e1413. doi: 10.1016/j.cell.2018.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Aguilera-Gomez A, Rabouille C. Membrane-bound organelles versus membrane-less compartments and their control of anabolic pathways in Drosophila. Dev Biol. 2017;428(2):310–317. doi: 10.1016/j.ydbio.2017.03.029. [DOI] [PubMed] [Google Scholar]
- 311.Dimova R, Lipowsky R. Lipid membranes in contact with aqueous phases of polymer solutions. Soft Matter. 2012;8(24):6409–6415. doi: 10.1039/C2SM25261A. [DOI] [Google Scholar]
- 312.Nesterov SV, Ilyinsky NS. Uversky VN (2021) Liquid-liquid phase separation as a common organizing principle of intracellular space and biomembranes providing dynamic adaptive responses. Biochim Biophys Acta Mol Cell Res. 1868;11:119102. doi: 10.1016/j.bbamcr.2021.119102. [DOI] [PubMed] [Google Scholar]
- 313.Yamasaki A, Alam JM, Noshiro D, Hirata E, Fujioka Y, Suzuki K, Ohsumi Y, Noda NN. Liquidity is a critical determinant for selective autophagy of protein condensates. Mol Cell. 2020;77(6):1163–1175e1169. doi: 10.1016/j.molcel.2019.12.026. [DOI] [PubMed] [Google Scholar]
- 314.Zhang G, Wang Z, Du Z, Zhang H. mTOR regulates phase separation of PGL granules to modulate their autophagic degradation. Cell. 2018;174(6):1492–1506e1422. doi: 10.1016/j.cell.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 315.Beutel O, Maraspini R, Pombo-Garcia K, Martin-Lemaitre C, Honigmann A. Phase separation of zonula occludens proteins drives formation of tight junctions. Cell. 2019;179(4):923–936e911. doi: 10.1016/j.cell.2019.10.011. [DOI] [PubMed] [Google Scholar]
- 316.Ma W, Mayr C. A membraneless organelle associated with the endoplasmic reticulum enables 3'UTR-mediated protein-protein interactions. Cell. 2018;175(6):1492–1506e1419. doi: 10.1016/j.cell.2018.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Zacharogianni M, Aguilera-Gomez A, Veenendaal T, Smout J, Rabouille C. A stress assembly that confers cell viability by preserving ERES components during amino-acid starvation. Elife. 2014 doi: 10.7554/eLife.04132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Zhao YG, Zhang H. Phase Separation in Membrane Biology: The Interplay between Membrane-Bound Organelles and Membraneless Condensates. Dev Cell. 2020;55(1):30–44. doi: 10.1016/j.devcel.2020.06.033. [DOI] [PubMed] [Google Scholar]
- 319.Lafontaine DLJ, Riback JA, Bascetin R, Brangwynne CP. The nucleolus as a multiphase liquid condensate. Nat Rev Mol Cell Biol. 2021;22(3):165–182. doi: 10.1038/s41580-020-0272-6. [DOI] [PubMed] [Google Scholar]
- 320.Yoneda M, Nakagawa T, Hattori N, Ito T. The nucleolus from a liquid droplet perspective. J Biochem. 2021;170(2):153–162. doi: 10.1093/jb/mvab090. [DOI] [PubMed] [Google Scholar]
- 321.y Cajal SR (1910) El núcleo de las células piramidales del cerebro humano y de algunos mamíferos
- 322.Machyna M, Heyn P, Neugebauer KM. Cajal bodies: where form meets function. WIREs RNA. 2013;4(1):17–34. doi: 10.1002/wrna.1139. [DOI] [PubMed] [Google Scholar]
- 323.Lallemand-Breitenbach V, de Thé H. PML nuclear bodies: from architecture to function. Curr Opin Cell Biol. 2018;52:154–161. doi: 10.1016/j.ceb.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 324.Schul W, Van Der Kraan I, Matera AG, Van Driel R, De Jong L. Nuclear domains enriched in RNA 3'-processing factors associate with coiled bodies and histone genes in a cell cycle-dependent manner. Mol Biol Cell. 1999;10(11):3815–3824. doi: 10.1091/mbc.10.11.3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Callan HG, Lloyd L. Lampbrush chromosomes of crested newts Triturus cristatus (Laurenti) Philos Trans R Soc Lond Ser B Biol Sci. 1960;243(702):135–219. doi: 10.1098/RSTB.1960.0007. [DOI] [Google Scholar]
- 326.Liu Q, Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 1996 doi: 10.1002/j.1460-2075.1996.tb00725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Pombo A, Cuello P, Schul W, Yoon JB, Roeder RG, Cook PR, Murphy S. Regional and temporal specialization in the nucleus: a transcriptionally-active nuclear domain rich in PTF, Oct1 and PIKA antigens associates with specific chromosomes early in the cell cycle. EMBO J. 1998;17(6):1768–1778. doi: 10.1093/EMBOJ/17.6.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Kloc M, Bilinski S, Etkin LD. The Balbiani body and germ cell determinants: 150 years later. Curr Top Dev Biol. 2004;59:1–36. doi: 10.1016/S0070-2153(04)59001-4. [DOI] [PubMed] [Google Scholar]
- 329.Kloc M, Jedrzejowska I, Tworzydlo W, Bilinski SM. Balbiani body, nuage and sponge bodies–term plasm pathway players. Arthropod Struct Dev. 2014;43(4):341–348. doi: 10.1016/J.ASD.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 330.Fox AH, Nakagawa S, Hirose T, Bond CS. Paraspeckles: where long noncoding RNA meets phase separation. Trends Biochem Sci. 2018;43(2):124–135. doi: 10.1016/j.tibs.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 331.Saurin AJ, Shiels C, Williamson J, Satijn DPE, Otte AP, Sheer D, Freemont PS. The human polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J Cell Biol. 1998;142(4):887–898. doi: 10.1083/JCB.142.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Chen T, Boisvert FM, Bazett-Jones DP, Richard S. A role for the GSG domain in localizing Sam68 to novel nuclear structures in cancer cell lines. Mol Biol Cell. 1999;10(9):3015–3015. doi: 10.1091/MBC.10.9.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Rajan P, Dalgliesh C, Bourgeois CF, Heiner M, Emami K, Clark EL, Bindereif A, Stevenin J, Robson CN, Leung HY, Elliott DJ. Proteomic identification of heterogeneous nuclear ribonucleoprotein L as a novel component of SLM/Sam68 nuclear bodies. BMC Cell Biol. 2009;10(1):1–13. doi: 10.1186/1471-2121-10-82/FIGURES/6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Mannen T, Yamashita S, Tomita K, Goshima N, Hirose T. The Sam68 nuclear body is composed of two RNase-sensitive substructures joined by the adaptor HNRNPL. J Cell Biol. 2016;214(1):45–59. doi: 10.1083/JCB.201601024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Bashkirov VI, Scherthan H, Solinger JA, Buerstedde J-M, Heyer W-D. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J Cell Biol. 1997;136(4):761–773. doi: 10.1083/jcb.136.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Riggs CL, Kedersha N, Ivanov P, Anderson P. Mammalian stress granules and P bodies at a glance. J Cell Sci. 2020 doi: 10.1242/jcs.242487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147(7):1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.