Abstract
Ovarian endometriosis is a common gynecological condition that can cause infertility in women of childbearing age. However, the pathogenesis is still unknown. We demonstrate that the carboxyl terminus of Hsc70-interacting protein (CHIP) is a negative regulator in the development of endometriosis and reduces HMGB1 expression in endometriotic cells. Meanwhile, CHIP interacts with HMGB1 and promotes its ubiquitinated degradation, thereby inhibiting aerobic glycolysis and the progression of endometriosis. Furthermore, the CHIP agonist YL-109 effectively suppresses the growth of ectopic endometrium in endometriosis mouse model, which could be a potential therapeutic approach for endometriosis. In conclusion, our data suggest that CHIP may inhibit the development of endometriosis by suppressing the HMGB1-related glycolysis.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-022-04637-z.
Keywords: CHIP, Endometriosis, Glycolysis, HMGB1, Protein modification, Ubiquitination
Introduction
Endometriosis is a disease characterized by endometrial-like tissues that grow outside of the uterine cavity. It is an estrogen-dependent gynecological disorder that affects 6–10% of reproductive-aged women worldwide [1]. The most common implantation site of endometriosis is ovary and posterior cul-de-sac [2]. Ovarian endometriosis (OEM, also called ovarian chocolate cysts) is a cyst formed by regular bleeding of the ectopic endometrium in response to ovarian hormones. The main symptoms include chronic pelvic pain, dyspareunia, dysmenorrhea, and infertility [3]. Women with endometriosis have previously reported to be more susceptible to ovarian cancer [4]. However, the causes of endometriosis are still unknown. Although endometriosis is not generally considered a malignant disorder, it does share similar characteristics with tumors, such as glycolytic metabolism [5, 6].
Glycolysis, also known as the Warburg effect, refers to the fact that cells prefer glycolysis and use this metabolic pathway to produce ATP as their primary source of energy supply, rather than oxidative phosphorylation, even in the presence of oxygen and properly functioning mitochondria. This phenomenon was prevalent in tumors and was also present in endometriosis [5]. The glycolytic pathway could produce lactic acid, and excess lactic acid promoted angiogenesis, cell invasion, cell metastasis, and immunosuppression [7, 8]. Many glycolytic vital enzymes, such as 6 Phosphofructo-2-kinase/Fructose-2,6-Biphosphatase 3 (PFKFB3), were highly expressed in endometriosis cells and contributed to the progression of endometriosis [9]. Hypoxia-inducible factor-1α (HIF-1α), a transcription factor, was a crucial regulator of glycolysis and promoted the development of endometriosis [10, 11]. However, studies on the regulation of glycolysis in endometriosis are still rare.
High mobility group protein 1 (HMGB1) is a nonhistone chromosome-binding protein. HMGB1 was previously reported to be involved in glycolysis. HMGB1 promoted pulmonary fibrosis by inducing aerobic glycolysis through upregulation of HIF-1α [12]. Moreover, HMGB1, as a new hippo pathway regulator, mediated the expression of Yes-associated protein (YAP), which contributed to aerobic glycolysis in hepatocellular carcinoma cells [13]. Besides, HMGB1 also played an important role in endometriosis. HMGB1 was previously reported to promote the formation of an immunosuppressive environment and aggravate chronic inflammation of endometrial ovarian cyst [14]. It also participated in the formation of endometriosis by regulating autophagy [15]. In addition, the HMGB1-TLR-MyD88 signaling pathway induced pain in endometriosis [16]. Numerous studies have elucidated that HMGB1 promoted endometriosis, but it was unclear whether HMGB1 affected endometriosis through the glycolytic pathway. HMGB1 can be regulated by posttranslational modifications (PTMs) including methylation [17], phosphorylation [18], acetylation [19], ADP-ribosylation [20], and glycosylation [21]. Furthermore, ubiquitination plays an important role in many diseases. However, the ubiquitin ligases that regulate HMGB1 remain largely unknown.
As an important posttranslational modification, ubiquitination regulates different cellular signaling pathways [22]. The carboxyl terminus of Hsc70-interacting protein (CHIP) encoded by the STUB1 gene is composed of 303 amino acid residues and functions of E3 ubiquitin ligase and collaborates with molecular chaperones. The N-terminus of the CHIP protein contains a TPR structural region, which regulates binding with molecular chaperone proteins. The C-terminal includes the activity of E3 ubiquitin ligase, which promotes the ubiquitin degradation of the substrate [23]. CHIP promoted ubiquitin degradation of phosphorylated protein kinase B (AKT) and maintained the stability of the intracellular pathway [24]. In gastric cancer, CHIP inhibited cell invasion, metastasis, and angiogenesis by degrading NF-κB through ubiquitination [25]. However, the biological function of CHIP in endometriosis has not been investigated and the specific regulatory mechanisms of CHIP ubiquitination need to be further explored.
Here, we identify a novel molecular mechanism between CHIP and HMGB1 involved in the development of endometriosis. We found that CHIP is a new HMGB1-binding protein that promotes ubiquitination and accelerates degradation of HMGB1. CHIP also attenuates cellular glycolysis via HMGB1, thus inhibiting the development of endometriosis. These data provide a theoretical basis for further using the CHIP as a potential therapeutic intervention in ovarian endometriosis.
Materials and methods
Tissue collection and immunohistochemical staining
The Human Investigation Committee of Weifang Medical University has approved this study. All 50 cases ovarian endometriosis eutopic and ectopic endometrium were obtained from the Department of Obstetrics and Gynecology of the Affiliated Hospital of Weifang Medical University. In addition, we obtained 50 cases of normal endometrium from patients with nonendometriosis. These patients had not received any hormone therapy for at least three months before the procedure. The informed consent of all participants was obtained. Samples were collected and paraffin-embedded for subsequent immunohistochemical experiments. The specific method used was as described previously [26]. The staining of immunohistochemistry is scored by the percentage of positive cells as well as the intensity of staining. The score of staining intensity was defined as 0 for negative, 1 for weak staining, 2 for moderate staining, and 3 for strong positive staining. The frequency of positive cells was defined as: 0 scores for less than 5%, 1 scores for 5–25%, 2 scores for 26–50%, 3 scores for 51–75%, and 4 scores for > 75%. The semi-quantitative score scale ranges from 0 to 12, with < 4 defined as negative.
The isolation of stromal cells derived from the ectopic endometrium of patients with ovarian endometriosis
Ectopic endometrial tissues were collected from patients with ovarian endometriosis to isolate and culture primary ectopic endometrial stromal cells (EESC). The specific method used was as described previously [27]. Briefly, the endometrial tissues were cut into 1 mm3 pieces and then digested with collagenase IV (Sigma, St Louis, MO, USA) at 37 °C for 1.5 h. The pieces were then separated with 76 mm and 37 mm (pore size) nylon mesh.
Cell culture and transfection
Anna Strazinski-Powitz established the endometrial epithelial cell line (11Z), kindly provided by Prof. Sun-wei Guo, Fudan University, Shanghai. HEK293T cells were cultured in DMEM (HyClone), and 11Z and EESC were maintained in DMEM/F12 medium (HyClone). The medium was supplemented with 10% FBS (HyClone) with 100 μg/mL penicillin and 100 μg/mL streptomycin, and cells were cultured at 37 °C and 5% CO2. Cells were seeded in six-well plates and transfected using Lipofectamine 2000 (Invitrogen, Shanghai, China) according to the manufacturer’s instructions.
Plasmids, antibodies
The specific constructs used for the recombinant plasmids (Flag-CHIP, HA-CHIP, Flag-HMGB1, HA-HMGB1, Flag-CHIP-K30A, HA-CHIP-K30A, Flag-CHIP-H260Q, HA-CHIP-H260Q) were as described previously [28]. The CHIP-H260Q mutant is deficient in E3 ligase activity, while the CHIP-K30A mutant showed a defect in binding to molecular chaperones [29]. The CHIP shRNA was generated with oligonucleotide 5′-CCAGCTGGAGATGGAGAGTTA-3′, and the HMGB1 shRNA was generated with oligonucleotide 5ʹ-GGACAAGGCCCGTTATGAA-3ʹ. The antibodies used are shown in Supplementary Table S1.
Cell proliferation analysis
Transfected cells (20,000–25,000 cells/plate) were replated in 24-well plates, and the cells were counted for 4 consecutive days by cell counter. The specific method used was as described previously [28]. Three independent replicate experiments were done for each experiment.
Colony formation assay
Transfected cells were reseeded in six-well plates (200–800 cells/plate) and continued to be cultured for 10–14 days. Cells were fixed with 4% paraformaldehyde for 15 min, stained with crystal violet for 15 min, and photographed with a gel imaging system (G: BOX F3 Gel Document System). The specific method used was as described previously [30]. Three independent replicate experiments were done for each experiment.
Wound healing assay
Cells were reseeded in a six-well plate after transfection, and the next day delineated with a medium pipette tip and photographed. The cells were continued to be cultured for 24 h. The culture medium was aspirated and discarded, washed with PBS, and photographed again. Three independent replicate experiments were done for each experiment.
Transwell invasion assay
Matrigel gel (BD Biosciences, Bedford, MA, USA) was diluted with serum-free DMEM/F12 at a concentration of 1:8, and then 40 μL of diluted BD gel was added to the upper chamber and incubated for 1 h at 37 °C to solidify the gel. The transfected cells (1 × 105) were mixed with 200 μL of serum-free DMEM/F12 medium and added to the upper chamber, and 600 μL of medium containing 10% FBS was added to the lower chamber. Twenty-four hours later, the Matrigel gel and cells were removed from the upper layer, fixed with 4% formaldehyde for 15 min, then stained with crystal violet for 10 min, photographed, and recorded the number of cells passing through the chamber. Three independent replicate experiments were done for each experiment.
Glucose consumption and lactate production
Cells were reseeded in six-well plates and replaced with serum-free DMEM/F12 medium. 12–16 h later, the supernatant was collected. The concentration of glucose and lactate was determined using a glucose (GO) assay kit (Sigma, #GAGO20-1KT) and a lactate assay kit (Biovision, #K627-100). The methods used were performed as described previously [31]. Three independent replicate experiments were done for each experiment.
Immunofluorescent analysis and proximity ligation assay (PLA)
Cells (1.2 × 105) were inoculated on cell crawlers and incubated for 24 h and then fixed with 4% paraformaldehyde. Tissue sections were hydrated and antigenically repaired. Then cells and tissue sections were treated with 0.05% Triton-100, blocked with 1% BSA for 1 h, and then incubated with primary antibody overnight at 4 °C. The samples were incubated with secondary antibody for 1 h at 37 °C and then stained with DAPI. PLA assay using Duolink® In Situ Red assay (Sigma, DUO92101) is to detect transient interactions of endogenous proteins according to the manufacturer's instructions. The specific method used was as described previously [32, 33]. The images were photographed under a fluorescence microscope (ZEISS). Three independent replicate experiments were done for each experiment.
Western blot
Cultured cells were collected and lysed on ice with lysis buffer (Beyotime, Shanghai, China, P0013), which can be left on ice for 30 min, followed by centrifugation at 12,000 rpm for 10 min at 4 °C. Keep the supernatant, add 5 × loading buffer, mix well, and heat at 100 °C for 10 min. The proteins were separated by SDS-PAGE electrophoresis, transferred to PVDF membranes, blocked in nonfat milk, and incubated overnight at 4 °C with indicated antibodies. ImageJ software was used for densitometric analysis. The specific method used was as described previously [34]. Three independent replicate experiments were done for each experiment.
Co-immunoprecipitation (Co-IP) assay
Collect the cells, add the lysate vortex, and incubate on ice for 30 min. Centrifuge at 12,000 rpm for 10 min at 4 °C, take 5% of the supernatant as input, then add beads or primary antibody as indicated, and incubate overnight at 4 °C. Add 2 × protein loading buffer and boil for 10 min. The obtained samples were subjected to subsequent western blot experiments. The specific method used was as described previously [35]. Three independent replicate experiments were done for each experiment.
GST pull-down assay
His-CHIP and GST-HMGB1 were expressed in E. coli BL (DE3), and the specific method used was as described previously [36]. Three independent replicate experiments were done for each experiment.
Ubiquitin detection
The amount of samples binding to IP was analyzed by immunoblotting with anti-HA (HA-tagged UB) or anti-ubiquitin (UB) antibodies. The specific method used was as described previously [32]. Three independent replicate experiments were done for each experiment.
Animal experiments
This experiment was approved by the Institutional Experimental Animal Review Board of Weifang Medical University. Intraperitoneal injection was used to establish a mouse model of endometriosis [37]. We used 6-week-old BALB/c female mice for this experiment. We injected estradiol benzoate (100 μg/kg) intramuscularly into the thighs of donor mice (n = 7) every two days to promote endometrial development. One week later, the uterus of the donor mice was sliced into 1 mm3 pieces, mixed well, and then performed intraperitoneal injection to the recipient mice. Two recipient mice received endometrium fragments from one donor mouse. After 1 week of establishing the endometriosis model, mice in the experimental group (n = 7) were given intraperitoneal injections of the CHIP agonist YL-109 (15 mg/kg) and mice in the control group (n = 7) were injected with normal saline twice a week for one month. Mice were executed, endometriosis-like lesions were collected and measured, and peritoneal fluid was collected for testing.
Statistical analysis
GraphPad Prism 9.0 software was used to complete the statistical analysis of all data. The obtained data were expressed as mean ± SEM. The Mann–Whitney test, Wilcoxon test, one-way ANOVA, and Student’s t test were used for data analysis. Spearman’s correlation analysis was used to assess the relationship between the two variables. Ρ-values < 0.05 were considered statistically significant. The n.s. was not significant.
Results
CHIP is downregulated in endometriotic tissues and negatively correlates with HMGB1
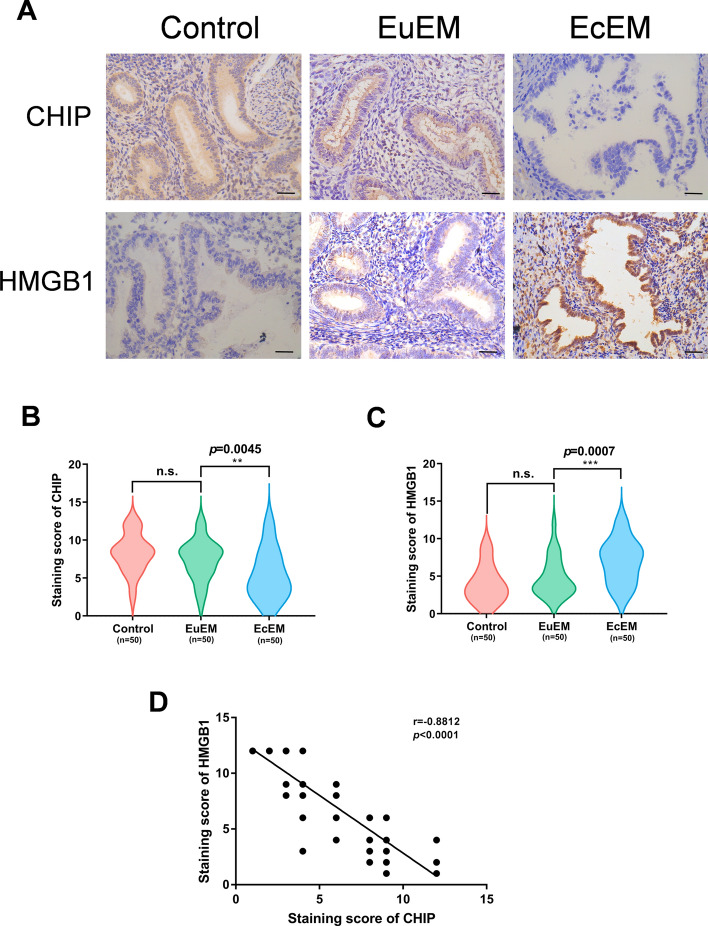
To investigate the relationship between CHIP and endometriosis, we performed immunohistochemical experiments on eutopic and ectopic endometrium from patients with endometriosis, with normal endometrium as a control. IHC staining showed that CHIP was mainly located in the cytoplasm of endometrial glandular epithelial cells (Fig. 1A). And the protein level of CHIP was significantly lower in ectopic endometrium than in normal and eutopic endometrium (Fig. 1B). In addition, we verified the expression levels of HMGB1 in the tissues. HMGB1 was more abundantly expressed in ectopic tissues (Fig. 1C) and was mainly distributed in the cytoplasm of the glandular epithelium and partially in the nucleus (Fig. 1A). The data showed that CHIP was negatively correlated with HMGB1 in endometriosis (Fig. 1D).
Fig. 1.
CHIP is downregulated in endometriotic tissues and negatively correlates with HMGB1. A Expression of CHIP and HMGB1 proteins in control endometrium and in eutopic/ectopic endometrium of patients with endometriosis was detected by immunohistochemistry (scale bar, 20 µm). B, C Violin plots showing the difference in immunoreactivity of CHIP, HMGB1 in the three groups, respectively. D Spearman's correlation analysis ascertained the correlation between CHIP and HMGB1 expression in ectopic tissues. (All data represent mean ± SEM. The Mann–Whitney test, Wilcoxon test, and Spearman’s correlation analysis were used for data analysis. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001)
CHIP suppresses cell proliferation, invasion, and glycolysis in vitro and in vivo
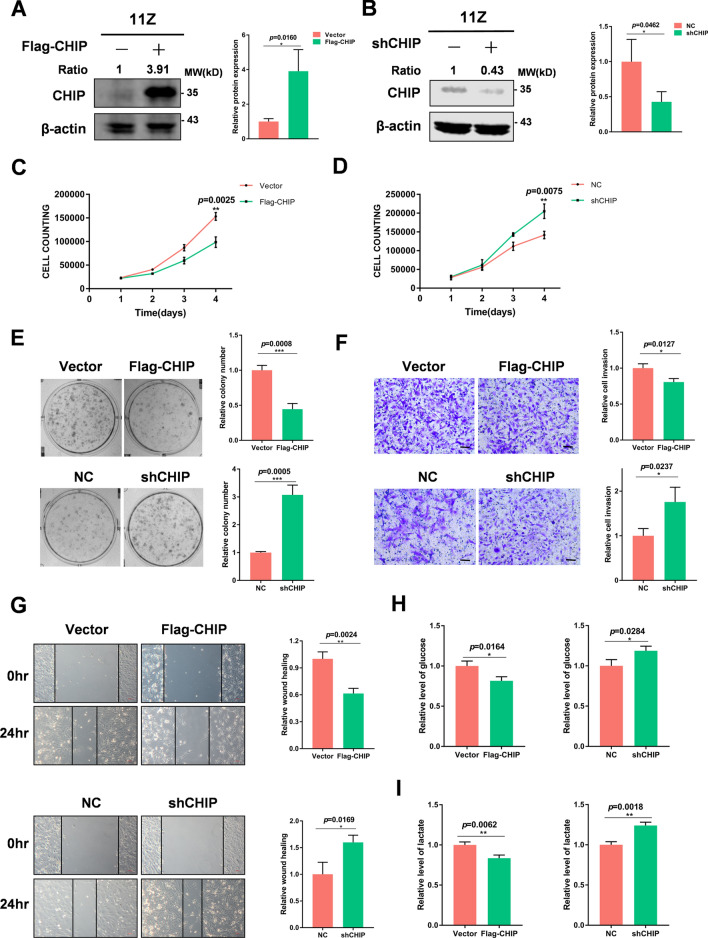
To determine the role of CHIP in endometriosis, we transfected plasmids or shRNA of CHIP into 11Z and EESC cells, respectively. We verified the transfection effect by immunoblotting (Fig. 2A, B and Supplementary Fig. 1A, B). Then we performed cell proliferation assays and colony formation assays and found that overexpression of CHIP significantly inhibited cell proliferation. In contrast, knockdown of the CHIP promoted the proliferation of endometriotic cells (Fig. 2C–E and Supplementary Fig. 1C–E). We performed transwell invasion assays and wound healing tests to determine the effect of CHIP on the cell invasion. Compared to controls, manipulation of CHIP high expression significantly inhibited the invasion and migration of endometriotic cells (Fig. 2F, G and Supplementary Fig. 1F, G). Downregulation of CHIP promoted cell migration and invasion. In addition, high expression of CHIP in cells reduced glucose consumption and lactate production. Conversely, reduced CHIP expression promoted glucose consumption and lactate production (Fig. 2H, I and Supplementary Fig. 1H, I). These results suggest that CHIP inhibits the proliferation, invasion, and glycolysis of endometriotic cells.
Fig. 2.
CHIP suppresses cell proliferation, invasion, and glycolysis in vitro. A, B Immunoblot analysis revealed overexpression and knockdown levels of CHIP in 11Z cells. Western blot was quantified by ImageJ software, and statistics were normalized to β-actin. C, D Effect of CHIP overexpression or knockdown on the proliferation of 11Z cells. E Overexpression of CHIP in 11Z cells resulted in a significant decrease in the number of colony formation. Knockdown of CHIP in 11Z cells resulted in a significant increase in the number of colony formation. F Overexpression of CHIP in 11Z cells resulted in diminished cell invasion. Knockdown of CHIP in 11Z cells resulted in enhanced cell invasion. G CHIP overexpression suppressed endometriotic cells migration. CHIP knockdown enhanced endometriotic cells migration. H, I The effects on glucose consumption and lactate production after overexpression or knockdown of CHIP are indicated, respectively. (All data represent mean ± SEM. The Student’s t test was used for data analysis. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001)
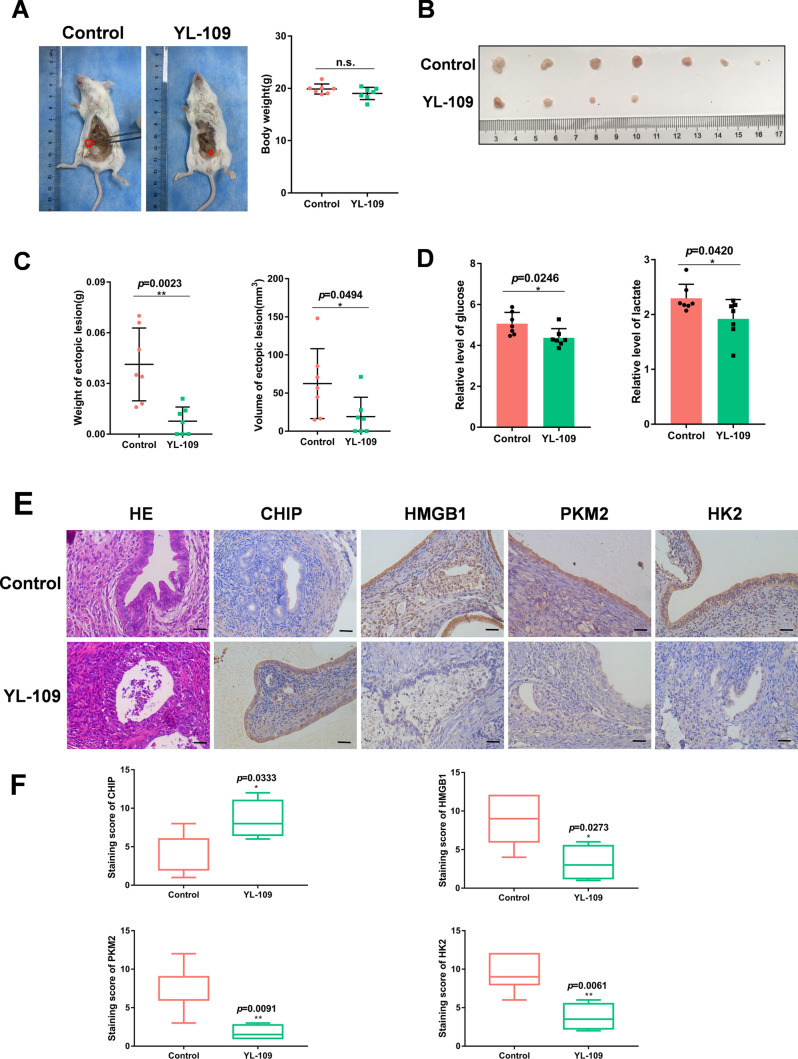
In addition, we studied the effect of YL-109 on endometriosis, a specific agonist of CHIP that induces CHIP expression through aryl hydrocarbon receptor (AhR) signaling [38]. To determine whether YL-109 inhibits the development of endometriosis in vivo, we established the endometriosis mouse model (Fig. 3A) and administered intraperitoneal injections of YL-109 to the experimental group of mice. Four weeks later, the ectopic tissues were removed for measurement and analysis (Fig. 3B). Compared to the control group, the ectopic tissues in the YL-109 treatment group were significantly suppressed in volume and weight (Fig. 3C). We also measured the glucose and lactate levels in the peritoneal fluid of mice. The results showed that the experimental group of mice consumed less glucose and produced less lactic acid (Fig. 3D). Immunohistochemical staining also showed that YL-109 caused an increase in CHIP and a decrease in HMGB1 as well as PKM2 and HK2, which were related to glycolysis (Fig. 3E, F). Taken together, these results strongly suggest that CHIP acts as a negative regulator to suppress the growth, migration, and glycolysis of endometriosis. And the CHIP agonist YL-109 has a therapeutic effect on the endometriosis mouse model.
Fig. 3.
YL-109 inhibits the development of endometriosis. A–D After establishing the endometriosis mouse model, the control group was given intraperitoneal saline and the experimental group was given intraperitoneal YL-109 (15 mg/kg). After four weeks, the mice were executed to observe the growth and weight of ectopic tissues and detect the glucose and lactate levels in the peritoneal fluid. A The red circle represented the implanted ectopic tissue. B Represented the largest ectopic tissue of each mouse. E Representative photographs of H/E staining and CHIP, HMGB1, PKM2, HK2 staining of ectopic samples (scale bar, 20 µm). F Box plots showing the difference in immunoreactivity of CHIP, HMGB1, PKM2, HK2 in two groups. (All data represent mean ± SEM. The Student’s t test was used for data analysis. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001)
CHIP interacts with HMGB1
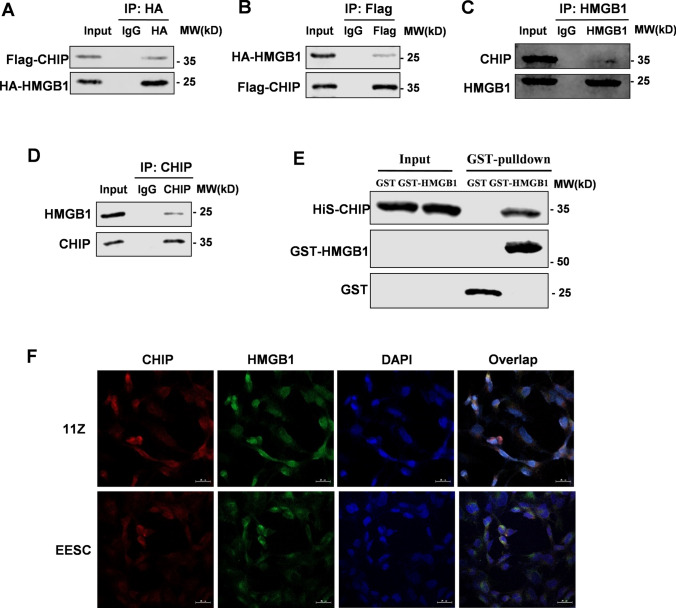
After demonstrating that CHIP and HMGB1 were negatively correlated, we further explored physical interactions between two proteins. Immunoprecipitation (Co-IP) assay detected the exogenous interaction between CHIP and HMGB1 in HEK293T cells (Fig. 4A, B). Further analysis showed that endogenous HMGB1 interacted with CHIP in endometriosis cells (Fig. 4C, D). Consistent with these results, the GST pull-down assay showed that recombinant His-CHIP could interact with GST-HMGB1 but not with GST-tagging alone (Fig. 4E). We further performed PLA experiments and verified that endogenous CHIP interacted with HMGB1 in 11Z cells (Supplementary Fig. 2A). Meanwhile, immunofluorescence experiments showed that CHIP co-localized with HMGB1 mainly in the cytoplasm of endometriotic cells (Fig. 4F). Moreover, immunofluorescence of tissue sections revealed this co-localization in ectopic endometrium of humans and mice (Supplementary Fig. 2B). Collectively, these data show that CHIP is a new binding protein for HMGB1.
Fig. 4.
CHIP interacts with HMGB1. Perform immunoprecipitation and immunoblot analysis with specified antibodies. A, B Transfected Flag-tagged CHIP and HA-tagged HMGB1 into HEK293T cells and verified their interaction by immunoprecipitation and immunoblotting. C, D Immunoprecipitation and immunoblotting to verify the interaction of endogenous CHIP and HMGB1 proteins in 11Z cells. E GST-pull down assay analysis of CHIP and HMGB1 proteins interaction using purified GST-HMGB1 and His-CHIP. F Confocal immunofluorescence microscopy was performed to analyze localization of CHIP (red) and HMGB1 (green) in endometriotic cells
CHIP downregulates HMGB1 protein stability
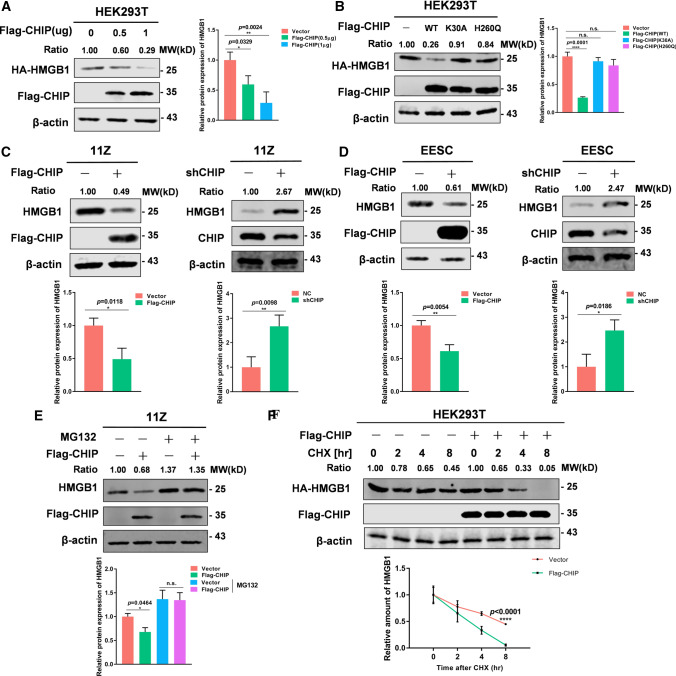
Having detected the interaction between CHIP and HMGB1, we next evaluated the effect of CHIP on HMGB1 expression levels. After transfecting Flag-CHIP and HA-HMGB1 in HEK293T cells, the results showed that CHIP dose-dependently reduced HMGB1 expression (Fig. 5A). In addition, we overexpressed WT-CHIP, H260Q CHIP mutant, or K30A CHIP mutant and detected the expression of HMGB1. We found that the two mutants of CHIP failed to reduce HMGB1 expression in HEK293T cells compared to WT-CHIP (Fig. 5B). These results suggest that CHIP destabilizes HMGB1 in the manner of E3 ligase activity and binding to molecular chaperones. We then overexpressed and knocked down CHIP in endometriotic cells to detect endogenous HMGB1 protein levels. When CHIP was transfected into 11Z and EESC cells, a decrease in endogenous HMGB1 was detected. In contrast, we performed CHIP knockdown in 11Z and EESC cells and found increased protein levels of endogenous HMGB1 (Fig. 5C, D). Meanwhile, we treated cells with the CHIP agonist YL-109 at a concentration of 10 μmol/L for 24 h. Consistent with the expected results, the protein level of HMGB1 decreased (Supplementary Fig. 3A, B).
Fig. 5.
CHIP downregulates HMGB1 protein stability. A HEK293T overexpressed Flag-CHIP (0, 0.5 or 1 μg) and HA-HMGB1 protein. Immunoblotting experiments were performed. B HEK293T overexpressed Flag-CHIP (WT or mutants) and HA-HMGB1 protein. Immunoblotting experiments were performed. C Overexpression or knockdown CHIP with Flag-CHIP or shRNA in 11Z cells, respectively. Immunoblotting experiments were performed. D Overexpression or knockdown CHIP with Flag-CHIP or shRNA in EESC cells, respectively. Immunoblotting experiments were performed. E 11Z cells were transfected with Flag-CHIP for 48 h and then treated with MG132 (20 μg/mL) for 8 h. Immunoblotting experiments were performed. F HEK293T cells with overexpression of Flag-CHIP and HA-HMGB1 were treated with CHX (100 μg/mL) for indicated time. Immunoblotting experiments were performed. (Representative western blot was quantified by ImageJ software, and statistics were normalized to β-actin. All data represent mean ± SEM. Statistical significance was analyzed with Student’s t test and one-way ANOVA. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001)
To test whether CHIP regulated the stability of HMGB1 through the proteasome pathway, we overexpressed Flag-CHIP in 11Z cells in the presence of MG132 (20 μg/mL). The results showed that MG132 eliminated the CHIP-induced downregulation of HMGB1 (Fig. 5E). Furthermore, to explore the effect of CHIP on HMGB1 half-life, we treated HEK293T cells with CHX (100 μg/mL) for indicated time. The results revealed that CHIP significantly increased the degradation of HMGB1 and shorten its half-life (Fig. 5F). We overexpressed and knocked down CHIP in 11Z cells and showed that CHIP also decreased the half-life of endogenous HMGB1 protein (Supplementary Fig. 3C, D). Altogether, these data show that CHIP accelerates HMGB1 degradation.
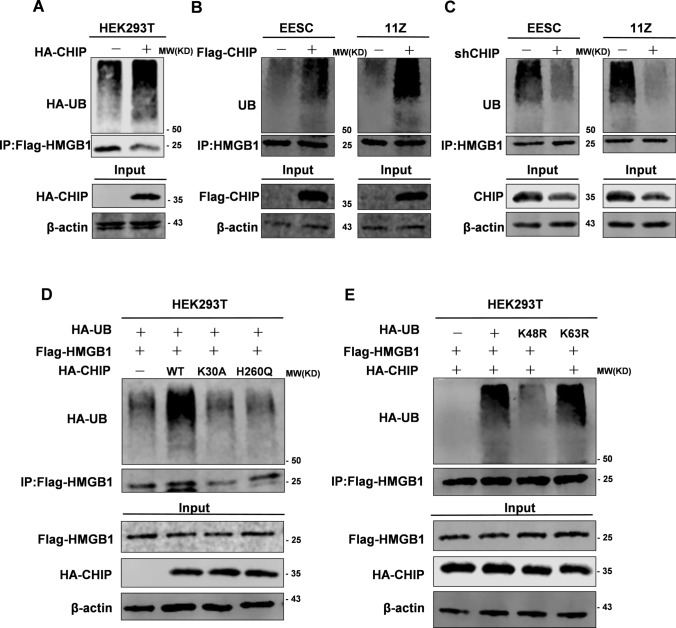
CHIP promotes HMGB1 degradation via the ubiquitin proteasome pathway
Since CHIP was an E3 ubiquitin ligase, we hypothesized that CHIP degrades HMGB1 protein levels via the ubiquitin proteasome pathway. Overexpression of Flag-CHIP increased ubiquitination level of HA-HMGB1 in HEK293T (Fig. 6A). CHIP overexpression in EESC and 11Z cells significantly promoted the ubiquitination of endogenous HMGB1 (Fig. 6B). In addition, knockdown of endogenous CHIP eliminated the ubiquitination of HMGB1 (Fig. 6C). WT-CHIP promoted ubiquitination of exogenous HMGB1 but not H260Q-CHIP or K30A-CHIP (Fig. 6D). Subsequently, we investigated the possible types of ubiquitin chains involved, and we overexpressed several ubiquitin-defective mutants (K48R and K63R). The results showed that CHIP increased Lys48-linked ubiquitination but not Lys63-linked ubiquitination (Fig. 6E). These results further support our hypothesis that CHIP degrades HMGB1 via the ubiquitin proteasome pathway.
Fig. 6.
CHIP promotes HMGB1 degradation via the ubiquitin proteasome pathway. A HEK293T cells were cotransfected with HA-CHIP, Flag-HMGB1, and HA-UB. The ubiquitylation level of HMGB1 was detected using an anti-HA antibody (HA-tagged UB). B EESC and 11Z cells were cotransfected with Flag-CHIP. The ubiquitylation level of HMGB1 was detected using an anti-ubiquitin antibody (UB). C EESC and 11Z cells were cotransfected with shCHIP. The ubiquitylation level of HMGB1 was detected using an anti-ubiquitin antibody (UB). D HEK293T cells were cotransfected with HA-CHIP (WT or mutants), Flag-HMGB1, and HA-UB. The ubiquitylation level of HMGB1 was detected using an anti-HA antibody (HA-tagged UB). E HEK293T cells were cotransfected with different ubiquitin mutants with HA-CHIP and Flag-HMGB1. The ubiquitylation level of HMGB1 was detected using an anti-HA antibody (HA-tagged UB). A–E After transfection for 48 h, the cells were treated with MG132 (20 μg/mL) for 8 h and then collected for ubiquitylation assays
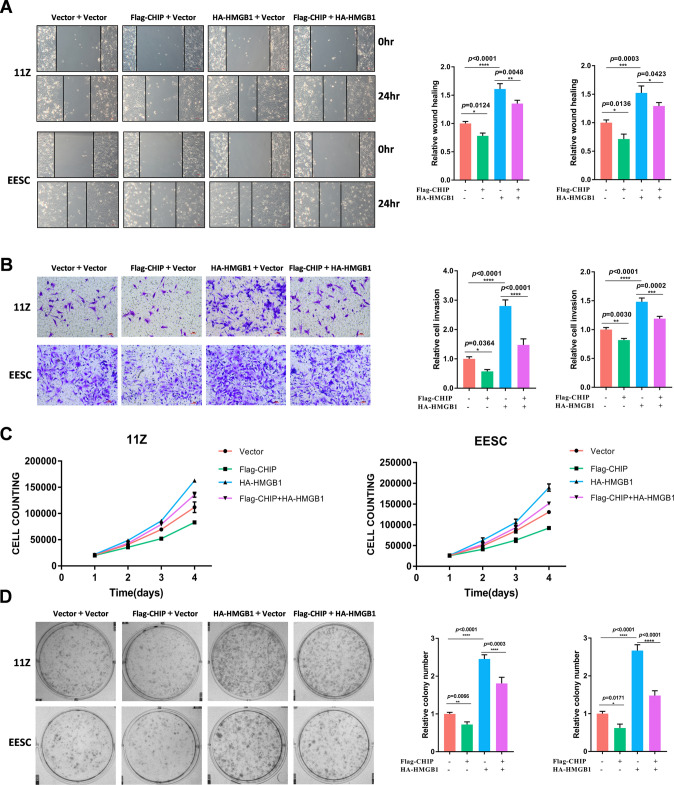
CHIP modulates cell proliferation and migration via HMGB1
To demonstrate whether CHIP affects the proliferation and migration of endometriosis via HMGB1, we manipulated cells for overexpression experiments in 11Z and EESC cells. Wound healing assay demonstrated that overexpression of CHIP significantly suppressed the migration of endometriotic cells, but this effect was counteracted by overexpression of HMGB1 (Fig. 7A). Similarly, transwell assays demonstrated that HMGB1 attenuated the effect of CHIP on cell invasion (Fig. 7B). Furthermore, overexpression of CHIP inhibited cell proliferation, while overexpression of HMGB1 reversed this function (Fig. 7C). Data from colony formation experiments consistently showed that CHIP inhibited cell proliferation by regulating HMGB1 (Fig. 7D). These results suggest that the growth and invasion suppression of CHIP is exerted through HMGB1.
Fig. 7.
CHIP modulates cell proliferation and migration via HMGB1. A–D 11Z cells were transfected with vector, Flag-CHIP, HA-HMGB1, and Flag-CHIP + HA-HMGB1, respectively. Forty-eight hours later, wounding healing assay, transwell invasion assay, cell proliferation assay, and colony formation assay were performed (scale bar, 50 µm). (All data represent mean ± SEM. One-way ANOVA was used for data analysis. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001)
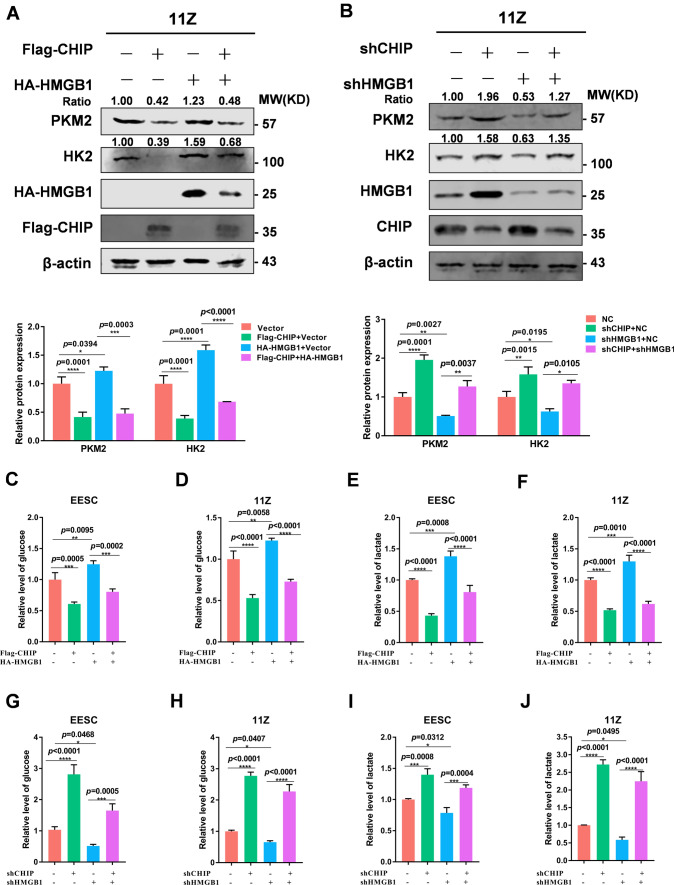
CHIP inhibits glycolysis in endometriosis via HMGB1
HMGB1 has been previously reported to promote glycolysis in disease progression. Therefore, we investigated whether CHIP regulated glycolysis in endometriosis through HMGB1. Using WB experiments, we observed the expression of PKM2 and HK2 was downregulated in CHIP overexpressing cells, while simultaneous overexpression of HMGB1 attenuated the inhibitory effect of CHIP (Fig. 8A). In contrast, knockdown of CHIP increased the expression of PKM2 and HK2, while simultaneous knockdown of HMGB1 diminished the promotion of PKM2 and HK2 (Fig. 8B). Our hypothesis was then confirmed by measuring the levels of lactate production and glucose consumption in the cell culture medium. CHIP overexpression inhibited glucose uptake and lactate production, and we found that high expression of HMGB1 rescued this effect (Fig. 8C–F). Similarly, knockdown of CHIP increased glucose consumption and lactate production, whereas this regulation can be abrogated when cells were knocked down of HMGB1 (Fig. 8G–J). The results show that CHIP affects the glycolytic process through HMGB1.
Fig. 8.
CHIP inhibits glycolysis in endometriosis via HMGB1. A 11Z cells were transfected with vector, Flag-CHIP, HA-HMGB1, and Flag-CHIP + HA-HMGB1, respectively. Cell lysate preparation for immunoblot analysis. B 11Z cells were transfected with shNC, shCHIP, shHMGB1, and shCHIP + shHMGB1, respectively. Cell lysate preparation for immunoblot analysis. C, D EESC and 11Z cells were transfected with vector, Flag-CHIP, HA-HMGB1, and Flag-CHIP + HA-HMGB1, respectively. The supernatant was collected to detect the consumption of glucose. E, F EESC and 11Z cells were transfected with vector, Flag-CHIP, HA-HMGB1, and Flag-CHIP + HA-HMGB1, respectively. The supernatant was collected to detect the production of lactate. G, H EESC and 11Z cells were treated with shNC, shCHIP, shHMGB1, and shCHIP + shHMGB1, respectively. The supernatant was collected to detect the consumption of glucose. I, J EESC and 11Z cells were treated with shNC, shCHIP, shHMGB1, and shCHIP + shHMGB1, respectively. Collection of supernatant for detection of lactate production. (Representative western blot was quantified by ImageJ software and normalized to β-actin. All data represent mean ± SEM. Statistical significance was analyzed with one-way ANOVA. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001)
Discussion
In the present study, we demonstrated that CHIP was an inhibitory factor that was low expressed in endometriosis and negatively correlated with HMGB1. CHIP downregulated glycolysis and inhibited proliferation and migration of endometriosis by promoting ubiquitination and degradation of HMGB1. To our knowledge, this is the first report to investigate the biological role of CHIP in endometriosis and to find that HMGB1 can be degraded by ubiquitination.
CHIP usually functions as a tumor suppressor, inhibiting the development of cancer. CHIP inhibited malignancy in colorectal cancer cells by targeting NF-κB signaling [39]. In lung adenocarcinoma, CHIP degraded CIB1 and inhibited subsequent EMT and tumor metastasis [40]. In our study, we demonstrated that CHIP was a negative regulator of endometriosis. Therefore, elevated CHIP in endometriosis cells inhibited cell proliferation and migration, further suppressing the development of lesions. Similarly, in the endometriosis mouse model, the CHIP agonist YL-109 significantly inhibited endometriotic lesions.
Moreover, since CHIP is an E3 ubiquitin ligase with molecular chaperone and ubiquitin ligase (E3) activity, it can perform ubiquitin degradation of substrates [41, 42]. CHIP promoted the ubiquitin degradation of ERβ [43]. And CHIP reduced the susceptibility of tumor cells to T-cell-derived IFN-γ by mediating the ubiquitinated degradation of the IFN-γ-R1/JAK1 complex [44]. Our study further confirmed that the inhibitory effect of CHIP on endometriosis was achieved by its ubiquitination and degradation of HMGB1.
HMGB1 has been previously reported to play a facilitating role in endometriosis [16, 45, 46]. Here, we discovered that CHIP interacted with HMGB1 and decreased its protein stability. Besides, CHIP shortened the half-life of endogenous and exogenous HMGB1 and promoted the degradation of HMGB1 via the ubiquitylation proteasome pathway. It has been reported that CHIP contains a TPR structural domain and a UBOX structural domain [47]. CHIP functions through them individually or in combination. The E3 ligase activity of CHIP was very necessary for ubiquitin degradation of BMAL1 to attenuate cell senescence [48]. CHIP regulated the expression of CLEC-2 protein through TPR domain and UBOX domain, which played an important role in immune response [49]. In our study, the CHIP-H260Q mutant was deficient in E3 ligase activity, while the CHIP-K30A mutant showed a defect in binding to molecular chaperones. Both mutants lost the ability to ubiquitin HMGB1. Here, we reported that TPR domain and UBOX domain were essential for CHIP to degrade HMGB1.
As reported in many studies, the development of endometriosis was closely related to glycolysis [8]. Previous studies have shown that lactic acid production was increased in the peritoneal cavity in patients with endometriosis, and the elevated lactate in the peritoneal cavity promoted cell invasion, immune escape, and angiogenesis [10]. In our experiment, we proved that CHIP regulated glycolysis through HMGB1. We detected the protein expression levels of key enzymes in glycolysis, suggesting that CHIP decreased the protein levels of PKM2 and HK2, while HMGB1 could reverse this effect. In addition, we measured glucose and lactic acid levels, and consistent with the results of western blot, CHIP relied on HMGB1 to regulate glycolysis in endometriosis.
In conclusion, our findings suggest that CHIP acts as a suppressor in ovarian endometriosis (Fig. 9). We provide new insights into the posttranslational modifications of HMGB1 and for the first time prove that HMGB1 can be degraded by ubiquitination. We also elucidate that CHIP is a new HMGB1-binding protein for targeting ubiquitination and degradation of HMGB1 and consequently inhibiting proliferation, invasion, and glycolysis in endometriosis. We may be able to exploit this novel regulatory role of CHIP with HMGB1 and glycolysis to develop new therapeutic approaches.
Fig. 9.
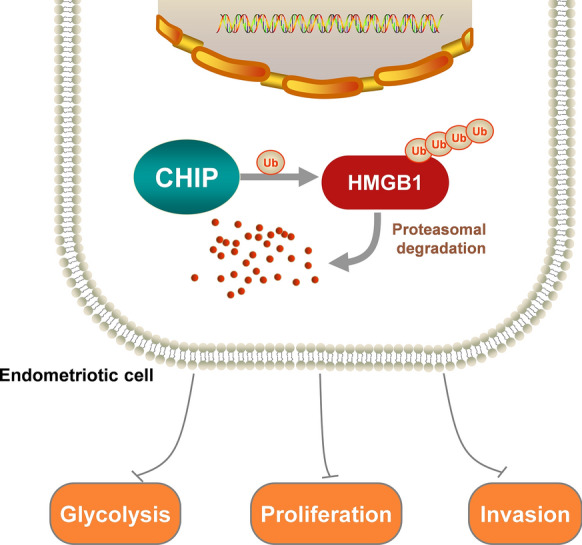
Working model. CHIP promotes the degradation of HMGB1 via the ubiquitinated proteasome pathway and inhibits glycolysis, proliferation, and invasion of endometriosis cells
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary Fig. 1. CHIP suppresses cell proliferation and invasion in vitro. A-B Immunoblot analysis revealed overexpression and knockdown levels of CHIP in EESC cell. Western blot was quantified by Image J software and statistics were normalized to β-actin. C-D Effect of CHIP overexpression or knockdown on the proliferation of EESC cells. E Overexpression of CHIP in EESC cells resulted in a significant decrease of colony formation. Knockdown of CHIP in EESC cells resulted in a significant increase of colony formation. F Overexpression of CHIP in EESC cells resulted in diminished cell invasion. Knockdown of CHIP in EESC cells resulted in enhanced cell invasion. G CHIP overexpression suppressed endometriotic cells migration. CHIP knockdown enhanced endometriotic cells migration. H-I The effects on glucose consumption and lactate production after overexpression or knockdown of CHIP are indicated, respectively. (All data represent mean ± SEM. The Student’s t-test was used for data analysis. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.) (TIF 4192 KB)
Supplementary Fig. 2. Co-localization of CHIP and HMGB1 in cells and tissues. A Representative images of in situ PLA showing the interaction between CHIP and HMGB1 in 11Z cells (scale bar, 20 µm). B Confocal immunofluorescence microscopy was performed to analyze localization of CHIP (red) and HMGB1 (green) in human and mouse ectopic endometrium (scale bar, 20 µm). (TIF 3589 KB)
Supplementary Fig. 3. CHIP decreases the protein level of HMGB1. A-B 11Z or EESC cells were treated with YL-109 (10μmo/L). Immunoblotting experiments were performed. C 11Z cells with overexpression of Flag-CHIP were treated with CHX for indicated time. Immunoblotting experiments were performed. D 11Z cells with knockdown of CHIP were treated with CHX for indicated time. Immunoblotting experiments were performed. (Representative western blot was quantified by Image J software and statistics were normalized to β-actin. All data represent mean ± SEM. Statistical significance was analyzed with Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.) (TIF 1174 KB)
Acknowledgements
We thank Prof. Sun-wei Guo (Fudan University, Shanghai) for generously providing the cell line.
Author contributions
TY, ZY, YS, and QW designed the study; YS, MW, and FS performed the experiments; PQ and AJ reviewed the data and advised the study; CR, ZY, and TY supervised the study; YS and QW wrote the manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (nos. 81602301 and 81972489), Natural Science Foundation of Shandong Province (no. ZR2021MH235), Shandong Province College Science and Technology Plan Project (no. J17KA254), and Clinical Research Center of Affiliated Hospital of Weifang Medical University (no. 2021wyfylcyj01).
Data availability
Enquiries about data availability should be directed to the authors.
Declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yujun Sun, Qian Wang, and Mengxue Wang have contributed equally.
Change history
3/18/2023
The missed co-corresponding author has been updated
Change history
3/17/2023
A Correction to this paper has been published: 10.1007/s00018-023-04732-9
Contributor Information
Chune Ren, Email: wyfybaby@126.com.
Zhenhai Yu, Email: tomsyu@163.com.
Tingting Yang, Email: Y402115432@163.com.
References
- 1.Burney RO, Giudice LC. Pathogenesis and pathophysiology of endometriosis. Fertil Steril. 2012;98(3):511–519. doi: 10.1016/j.fertnstert.2012.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Busacca M, Vignali M. Ovarian endometriosis: from pathogenesis to surgical treatment. Curr Opin Obstet Gynecol. 2003;15(4):321–326. doi: 10.1097/01.gco.0000084247.09900.4f. [DOI] [PubMed] [Google Scholar]
- 3.Zondervan KT, Becker CM, Missmer SA. Endometriosis. N Engl J Med. 2020;382(13):1244–1256. doi: 10.1056/NEJMra1810764. [DOI] [PubMed] [Google Scholar]
- 4.Swiersz LM. Role of endometriosis in cancer and tumor development. Ann N Y Acad Sci. 2002;955:281–292. doi: 10.1111/j.1749-6632.2002.tb02788.x. [DOI] [PubMed] [Google Scholar]
- 5.Qi XC, Zhang YX, Ji H, Wu XD, Wang FX, Xie MX, et al. Knockdown of prohibitin expression promotes glucose metabolism in eutopic endometrial stromal cells from women with endometriosis. Reprod Biomed Online. 2014;29(6):761–770. doi: 10.1016/j.rbmo.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Nicholes K, Shih IM. The origin and pathogenesis of endometriosis. Annu Rev Pathol. 2020;15:71–95. doi: 10.1146/annurev-pathmechdis-012419-032654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4(11):891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 8.Hirschhaeuser F, Sattler UG, Mueller-Klieser W. Lactate: a metabolic key player in cancer. Cancer Res. 2011;71(22):6921–6925. doi: 10.1158/0008-5472.CAN-11-1457. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Xiu J, Yang T, Ren C, Yu Z. HSF1 promotes endometriosis development and glycolysis by up-regulating PFKFB3 expression. Reprod Biol Endocrinol. 2021;19(1):86. doi: 10.1186/s12958-021-00770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiong Y, Liu Y, Xiong W, Zhang L, Liu H, Du Y, et al. Hypoxia-inducible factor 1alpha-induced epithelial-mesenchymal transition of endometrial epithelial cells may contribute to the development of endometriosis. Hum Reprod. 2016;31(6):1327–1338. doi: 10.1093/humrep/dew081. [DOI] [PubMed] [Google Scholar]
- 11.Zhang L, Xiong W, Li N, Liu H, He H, Du Y, et al. Estrogen stabilizes hypoxia-inducible factor 1alpha through G protein-coupled estrogen receptor 1 in eutopic endometrium of endometriosis. Fertil Steril. 2017;107(2):439–447. doi: 10.1016/j.fertnstert.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu J, Li J, Yu Z, Rao H, Wang S, Lan H. HMGB1 promotes HLF-1 proliferation and ECM production through activating HIF1-alpha-regulated aerobic glycolysis. Pulm Pharmacol Ther. 2017;45:136–141. doi: 10.1016/j.pupt.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 13.Chen R, Zhu S, Fan XG, Wang H, Lotze MT, Zeh HJ, 3rd, et al. High mobility group protein B1 controls liver cancer initiation through yes-associated protein-dependent aerobic glycolysis. Hepatology. 2018;67(5):1823–1841. doi: 10.1002/hep.29663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikeda M, Negishi Y, Akira S, Morita R, Takeshita T. Inflammation related to high-mobility group box-1 in endometrial ovarian cyst. J Reprod Immunol. 2021;145:103292. doi: 10.1016/j.jri.2021.103292. [DOI] [PubMed] [Google Scholar]
- 15.Huang J, Chen X, Lv Y. HMGB1 mediated inflammation and autophagy contribute to endometriosis. Front Endocrinol (Lausanne) 2021;12:616696. doi: 10.3389/fendo.2021.616696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su W, Cui H, Wu D, Yu J, Ma L, Zhang X, et al. Suppression of TLR4-MyD88 signaling pathway attenuated chronic mechanical pain in a rat model of endometriosis. J Neuroinflamm. 2021;18(1):65. doi: 10.1186/s12974-020-02066-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu S, Qian L, Ma J. Genetic alterations, RNA expression profiling and DNA methylation of HMGB1 in malignancies. J Cell Mol Med. 2022;26(15):4322–4332. doi: 10.1111/jcmm.17454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh YJ, Youn JH, Ji Y, Lee SE, Lim KJ, Choi JE, et al. HMGB1 is phosphorylated by classical protein kinase C and is secreted by a calcium-dependent mechanism. J Immunol. 2009;182(9):5800–5809. doi: 10.4049/jimmunol.0801873. [DOI] [PubMed] [Google Scholar]
- 19.Lu B, Antoine DJ, Kwan K, Lundback P, Wahamaa H, Schierbeck H, et al. JAK/STAT1 signaling promotes HMGB1 hyperacetylation and nuclear translocation. Proc Natl Acad Sci USA. 2014;111(8):3068–3073. doi: 10.1073/pnas.1316925111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Xie J, Li X, Fang J. Poly (ADP-ribosylation) of HMGB1 facilitates its acetylation and promotes HMGB1 translocation-associated chemotherapy-induced autophagy in leukaemia cells. Oncol Lett. 2020;19(1):368–378. doi: 10.3892/ol.2019.11116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim YH, Kwak MS, Park JB, Lee SA, Choi JE, Cho HS, et al. N-linked glycosylation plays a crucial role in the secretion of HMGB1. J Cell Sci. 2016;129(1):29–38. doi: 10.1242/jcs.176412. [DOI] [PubMed] [Google Scholar]
- 22.Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33(3):275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 23.Ballinger CA, Connell P, Wu Y, Hu Z, Thompson LJ, Yin LY, et al. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol. 1999;19(6):4535–4545. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su CH, Wang CY, Lan KH, Li CP, Chao Y, Lin HC, et al. Akt phosphorylation at Thr308 and Ser473 is required for CHIP-mediated ubiquitination of the kinase. Cell Signal. 2011;23(11):1824–1830. doi: 10.1016/j.cellsig.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 25.Liu F, Zhou J, Zhou P, Chen W, Guo F. The ubiquitin ligase CHIP inactivates NF-kappaB signaling and impairs the ability of migration and invasion in gastric cancer cells. Int J Oncol. 2015;46(5):2096–2106. doi: 10.3892/ijo.2015.2893. [DOI] [PubMed] [Google Scholar]
- 26.Yang T, Ren C, Lu C, Qiao P, Han X, Wang L, et al. Phosphorylation of HSF1 by PIM2 induces PD-L1 expression and promotes tumor growth in breast cancer. Cancer Res. 2019;79(20):5233–5244. doi: 10.1158/0008-5472.CAN-19-0063. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Q, Ding D, Liu X, Guo SW. Activated platelets induce estrogen receptor beta expression in endometriotic stromal cells. Gynecol Obstet Investig. 2015;80(3):187–192. doi: 10.1159/000377629. [DOI] [PubMed] [Google Scholar]
- 28.Ren C, Yang T, Qiao P, Wang L, Han X, Lv S, et al. PIM2 interacts with tristetraprolin and promotes breast cancer tumorigenesis. Mol Oncol. 2018;12(5):690–704. doi: 10.1002/1878-0261.12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang P, Li C, Li H, Yuan L, Dai H, Peng Z, et al. Ubiquitin ligase CHIP regulates OTUD3 stability and suppresses tumour metastasis in lung cancer. Cell Death Differ. 2020;27(11):3177–3195. doi: 10.1038/s41418-020-0571-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, Wang X, Wan L, Liu X, Yu H, Zhang D, et al. TIPE2 inhibits the migration and invasion of endometrial cells by targeting beta-catenin to reverse epithelial-mesenchymal transition. Hum Reprod. 2020;35(6):1377–1390. doi: 10.1093/humrep/deaa062. [DOI] [PubMed] [Google Scholar]
- 31.Han X, Ren C, Lu C, Qiao P, Yang T, Yu Z. Deubiquitination of MYC by OTUB1 contributes to HK2 mediated glycolysis and breast tumorigenesis. Cell Death Differ. 2022 doi: 10.1038/s41418-022-00971-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ren C, Han X, Lu C, Yang T, Qiao P, Sun Y, et al. Ubiquitination of NF-kappaB p65 by FBXW2 suppresses breast cancer stemness, tumorigenesis, and paclitaxel resistance. Cell Death Differ. 2022;29(2):381–392. doi: 10.1038/s41418-021-00862-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu Z, Huang L, Qiao P, Jiang A, Wang L, Yang T, et al. PKM2 Thr454 phosphorylation increases its nuclear translocation and promotes xenograft tumor growth in A549 human lung cancer cells. Biochem Biophys Res Commun. 2016;473(4):953–958. doi: 10.1016/j.bbrc.2016.03.160. [DOI] [PubMed] [Google Scholar]
- 34.Yang T, Ren C, Qiao P, Han X, Wang L, Lv S, et al. PIM2-mediated phosphorylation of hexokinase 2 is critical for tumor growth and paclitaxel resistance in breast cancer. Oncogene. 2018;37(45):5997–6009. doi: 10.1038/s41388-018-0386-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu C, Ren C, Yang T, Sun Y, Qiao P, Wang D, et al. A noncanonical role of fructose-1, 6-bisphosphatase 1 is essential for inhibition of Notch1 in breast cancer. Mol Cancer Res. 2020;18(5):787–796. doi: 10.1158/1541-7786.MCR-19-0842. [DOI] [PubMed] [Google Scholar]
- 36.Lu C, Ren C, Yang T, Sun Y, Qiao P, Han X, et al. Fructose-1, 6-bisphosphatase 1 interacts with NF-kappaB p65 to regulate breast tumorigenesis via PIM2 induced phosphorylation. Theranostics. 2020;10(19):8606–8618. doi: 10.7150/thno.46861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang S, Xiao F, Guo SW, Zhang T. Tetramethylpyrazine retards the progression and fibrogenesis of endometriosis. Reprod Sci. 2022;29(4):1170–1187. doi: 10.1007/s43032-021-00813-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hiyoshi H, Goto N, Tsuchiya M, Iida K, Nakajima Y, Hirata N, et al. 2-(4-Hydroxy-3-methoxyphenyl)-benzothiazole suppresses tumor progression and metastatic potential of breast cancer cells by inducing ubiquitin ligase CHIP. Sci Rep. 2014;4:7095. doi: 10.1038/srep07095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Ren F, Wang Y, Feng Y, Wang D, Jia B, et al. CHIP/Stub1 functions as a tumor suppressor and represses NF-κB-mediated signaling in colorectal cancer. Carcinogenesis. 2014;35(5):983–991. doi: 10.1093/carcin/bgt393. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y, Zhou Y, Zhang P, Li X, Duan C, Zhang C. CHIP-mediated CIB1 ubiquitination regulated epithelial-mesenchymal transition and tumor metastasis in lung adenocarcinoma. Cell Death Differ. 2021;28(3):1026–1040. doi: 10.1038/s41418-020-00635-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Hohfeld J, et al. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3(1):93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 42.Wang T, Wang W, Wang Q, Xie R, Landay A, Chen D. The E3 ubiquitin ligase CHIP in normal cell function and in disease conditions. Ann N Y Acad Sci. 2020;1460(1):3–10. doi: 10.1111/nyas.14206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen LJ, Hu B, Han ZQ, Zhu JH, Fan X, Chen XX, et al. BAG2-mediated inhibition of CHIP expression and overexpression of MDM2 contribute to the initiation of endometriosis by modulating estrogen receptor status. Front Cell Dev Biol. 2020;8:554190. doi: 10.3389/fcell.2020.554190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Apriamashvili G, Vredevoogd DW, Krijgsman O, Bleijerveld OB, Ligtenberg MA, de Bruijn B, et al. Ubiquitin ligase STUB1 destabilizes IFNgamma-receptor complex to suppress tumor IFNgamma signaling. Nat Commun. 2022;13(1):1923. doi: 10.1038/s41467-022-29442-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jaeger-Lansky A, Schmidthaler K, Kuessel L, Gstottner M, Waidhofer-Sollner P, Zlabinger GJ, et al. Local and systemic levels of cytokines and danger signals in endometriosis-affected women. J Reprod Immunol. 2018;130:7–10. doi: 10.1016/j.jri.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 46.Shimizu K, Kamada Y, Sakamoto A, Matsuda M, Nakatsuka M, Hiramatsu Y. High expression of high-mobility group box 1 in menstrual blood: implications for endometriosis. Reprod Sci. 2017;24(11):1532–1537. doi: 10.1177/1933719117692042. [DOI] [PubMed] [Google Scholar]
- 47.Zhang HT, Zeng LF, He QY, Tao WA, Zha ZG, Hu CD. (2016) The E3 ubiquitin ligase CHIP mediates ubiquitination and proteasomal degradation of PRMT5. Biochim Biophys Acta. 1863;2:335–346. doi: 10.1016/j.bbamcr.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ullah K, Chen S, Lu J, Wang X, Liu Q, Zhang Y, et al. The E3 ubiquitin ligase STUB1 attenuates cell senescence by promoting the ubiquitination and degradation of the core circadian regulator BMAL1. J Biol Chem. 2020;295(14):4696–4708. doi: 10.1074/jbc.RA119.011280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shao M, Li L, Song S, Wu W, Peng P, Yang C, et al. E3 ubiquitin ligase CHIP interacts with C-type lectin-like receptor CLEC-2 and promotes its ubiquitin-proteasome degradation. Cell Signal. 2016;28(10):1530–1536. doi: 10.1016/j.cellsig.2016.07.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. CHIP suppresses cell proliferation and invasion in vitro. A-B Immunoblot analysis revealed overexpression and knockdown levels of CHIP in EESC cell. Western blot was quantified by Image J software and statistics were normalized to β-actin. C-D Effect of CHIP overexpression or knockdown on the proliferation of EESC cells. E Overexpression of CHIP in EESC cells resulted in a significant decrease of colony formation. Knockdown of CHIP in EESC cells resulted in a significant increase of colony formation. F Overexpression of CHIP in EESC cells resulted in diminished cell invasion. Knockdown of CHIP in EESC cells resulted in enhanced cell invasion. G CHIP overexpression suppressed endometriotic cells migration. CHIP knockdown enhanced endometriotic cells migration. H-I The effects on glucose consumption and lactate production after overexpression or knockdown of CHIP are indicated, respectively. (All data represent mean ± SEM. The Student’s t-test was used for data analysis. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.) (TIF 4192 KB)
Supplementary Fig. 2. Co-localization of CHIP and HMGB1 in cells and tissues. A Representative images of in situ PLA showing the interaction between CHIP and HMGB1 in 11Z cells (scale bar, 20 µm). B Confocal immunofluorescence microscopy was performed to analyze localization of CHIP (red) and HMGB1 (green) in human and mouse ectopic endometrium (scale bar, 20 µm). (TIF 3589 KB)
Supplementary Fig. 3. CHIP decreases the protein level of HMGB1. A-B 11Z or EESC cells were treated with YL-109 (10μmo/L). Immunoblotting experiments were performed. C 11Z cells with overexpression of Flag-CHIP were treated with CHX for indicated time. Immunoblotting experiments were performed. D 11Z cells with knockdown of CHIP were treated with CHX for indicated time. Immunoblotting experiments were performed. (Representative western blot was quantified by Image J software and statistics were normalized to β-actin. All data represent mean ± SEM. Statistical significance was analyzed with Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.) (TIF 1174 KB)
Data Availability Statement
Enquiries about data availability should be directed to the authors.