Abstract
We describe here two novel mouse and human DNA polymerases: one (pol λ) has homology with DNA polymerase β while the other one (pol µ) is closer to terminal deoxynucleotidyltransferase. However both have DNA polymerase activity in vitro and share similar structural organization, including a BRCT domain, helix–loop–helix DNA-binding motifs and polymerase X domain. mRNA expression of pol λ is highest in testis and fetal liver, while expression of pol µ is more lymphoid, with highest expression both in thymus and tonsillar B cells. An unusually large number of splice variants is observed for the pol µ gene, most of which affect the polymerase domain. Expression of mRNA of both polymerases is down-regulated upon treatment by DNA damaging agents (UV light, γ-rays or H2O2). This suggests that their biological function may differ from DNA translesion synthesis, for which several DNA polymerase activities have been recently described. Possible functions are discussed.
INTRODUCTION
The process of affinity maturation of antibodies during an immune response is a unique phenomenon of adaptive mutation in which the stress-inducing agent (the immunizing antigen) triggers a mutagenic response specifically targeted to the genes involved in its recognition and clearance (the V regions of the immunoglobulin).
No molecular mechanism is known for this process, and many different models have been proposed. One of them, drawing a parallel between the immune and the bacterial SOS responses, suggests the recruitment of a DNA polymerase implicated in UV-induced mutagenesis, Rev3 (pol ζ) (1). Very recently, a number of other mammalian mutagenic DNA polymerases specifically involved in the bypass of DNA lesions have been discovered, and their possible involvement in Ig hypermutation proposed (2–5; see reviews in 6,7). For one of them however (Rad30A encoded by the XP-V gene), such an implication can be excluded, since B cells with mutated Ig genes have been isolated from patients deficient in this activity (8).
We have previously reported the particular pattern of mutation occurring on mono- or di-nucleotide repeated sequences imbedded in the Ig locus (9), either short stretches present naturally in non-coding regions, or longer runs artificially inserted in an Ig mutation substrate. The mutations generated on short repetitive sequences span the whole array of slippage events typically generated by an error-prone DNA polymerase in the absence of further correction processes, i.e. mutations, insertions, deletions and, more frequently, transient misalignments, and strongly evokes the mutation pattern reported for DNA polymerase β in vitro (10). Since knock-out experiments have excluded the role of pol β in hypermutation (11), we therefore proposed that this putative mutase could be a β-like DNA polymerase (9). DNA polymerase β belongs to the polX DNA polymerase family, a family related to a larger group of nucleotidyltransferases (12). Apart from DNA polymerase β, the only other non-viral member of this family in vertebrates is the template-independent terminal deoxynucleotidyltransferase (TdT), an enzyme with a strict lymphoid specificity and whose only documented function is the random insertion of nucleotides at the V-D and D-J junctions during the process of rearrangement of the immunoglobulin and TCR genes (13). Such untemplated nucleotide additions represent a major contribution to the overall diversity of the immune repertoire.
We therefore undertook a systematic screening of sequence databases to identify new members of the polX family. We report here the identification of two such DNA polymerases, one closely related to TdT, and the other more related to DNA polymerase β.
MATERIALS AND METHODS
cDNA isolation and sequence
PCR primers corresponding to the mouse pol λ EST (C86044) and the human pol µ EST (AA298793) were used to amplify probes from cDNA from either the mouse NSO cell line or the human Burkitt lymphoma BL2. Mouse and human pol λ probes were amplified with the same mouse-specific primers: 5′ pol λ, TTGGCAAGCGGATGGCGGAG; 3′ pol λ, GGAGGCAGCTGAAGATGCCC, using Taq polymerase (Perkin- Elmer), 64°C annealing (62°C for human cDNA) and 40 cycles. Human pol µ probe was amplified using primers: 5′ pol µ, CCCTGCCCATCCTGGAAG; 3′ pol µ, TGGAGGAAGGTACTCAAGGC (58°C annealing, 35 cycles) and used to screen both human and mouse cDNA libraries. Clontech cDNA libraries from human bone marrow and lymph nodes and from mouse spleen were used.
For pol µ, RACE amplification was performed to obtain the complete cDNA sequence (Marathon cDNA kit, Clontech). Reverse transcription was performed on 0.5 µg poly(A)+ RNA from the Ramos cell line and from CD19+CD38+ cells from human tonsils. For mouse cDNA, total RNA from 5 × 104 B220 PNAhigh Peyer’s patch B cells [purified according to Bertocci et al. (9)] was used. Reverse transcription was primed with specific primers: human-RaceA, in exon 6, CTCTCGGAGGTCATC; mouse-RaceA, in exon 2, ATCTAGCAGAGCTGG. Nested PCR amplification was performed according to manufacturer’s conditions, using two sets of specific primers: human-RaceB, GTCAGCAGTCTTCACACCGACCCG and human-RaceC, GGTTCGCAGTCCTTCCCGGTACCA; mouse-RaceB, TGGGCAGCCTGTGGGGAGAGCATC and mouse-RaceC, CAGCAGATGGCCTCCTTGGCTGAGG.
To study splicing variants, cDNAs obtained by oligo-dT-primed reverse transcription (RT–PCR kit, Stratagene) of poly(A)+ RNA from the BL2 cell line were amplified using the following primers: h-pol-µ-ATG, ATCATGCTCCCCAAACGGCGG (half EcoRV site in italics), and h-pol-µ-stop, ACGAATTCTGGAGACATTCAGTGGCCAG (EcoRI site in italics). Sequences were performed with BigDye Terminator cycle sequencing kit (Perkin-Elmer) and analyzed in an ABI 310 automated sequencer.
Genomic sequence analysis
To study gene organization, genomic phages containing the complete mouse pol µ gene were isolated from a 129 library (Stratagene). For human pol λ and pol µ genes, genomic PCR was performed using exon-specific primers and the Expand Long Template PCR System amplification kit (Boehringer) on DNA of the Ramos or BL2 cell lines.
Northern blot analysis
Clontech multi-tissue northern blots were used. An additional blot was performed using 0.5 µg poly(A)+ RNA from human thymus, total tonsils, CD19+CD38+ tonsillar cells [purified according to Pascual et al. (14)] and BL2 cells. cDNA probes containing the entire coding region were used for pol λ and pol µ and a 1 kb cDNA probe for actin. Probes were labeled by random priming (2 × 109 c.p.m./µg).
DNA damage induction in cultured cells
Ramos cells were grown in a humidified atmosphere with 5% CO2 in RPMI-glutamax (Gibco-BRL) containing antibiotics, 50 µM β-mercaptoethanol and 10% fetal calf serum (Hyclone). Prior to irradiation, cells were seeded in 6-well plates for a few hours, rinsed once with PBS and irradiated with different doses of UV light (254 nm) using a Stratalinker apparatus (Stratagene). For γ-ray treatment, cells were exposed at room temperature to a 137Cs source with an incident dose rate of 0.752 Gy/min, in a medium without serum. Following both irradiations, cells were incubated in complete medium. For oxidative damage, cells were incubated in complete medium containing 20 µM H2O2. Control and treated cells were collected at different time points, total RNA extracted using the RNeasy Mini kit (Qiagen), and oligo-dT-primed cDNA obtained with RT–PCR kit (Stratagene). Primers used for pol µ amplification are the 5′- and 3′-pol µ described above (with 30 cycles of amplification), and for pol λ: H-pol λ-5′ GTGTTCAGGGAGACAAGTGG, and H-pol λ-3′, GGCATACGTTCCAGGAAGTC (annealing 60°C, 30 cycles of amplification). Normalization was performed against actin cDNA, amplified with the following primers: 5′ actin, GCACCACACCTTCACAATG; 3′ actin, CCTGCTTTGCTGATCCACATC (55°C annealing, 25 cycles). The efficacy of the different treatments was assessed by amplification of the DNA damage induced p21WAF1 gene using primers 5′-P21, CGGTCCCGTGGACAGTGAGC and 3′-P21, AAATCTGTCAGGCTGGTCTGCC (60°C annealing, 30 cycles). Taq polymerase (Perkin-Elmer) was used for all amplifications. Semi-quantitative PCR was performed on serial dilutions of the templates. Reaction products were separated by electrophoresis on 1.5% agarose gel.
Construction of bacterial expression vectors and purification of human pol λ and pol µ from Escherichia coli
The coding regions of human pol λ and pol µ were amplified using the following primers: pol λ-ATG, GGAATTCCATATGGATCCCAGGGGTATCTTG and pol λ-His, CGGAATTCCCAGTCCCGCTAGCAGGT; pol µ-ATG-2, GGAATTCCATATGCTCCCCAAACGGCGGCGAGC and pol µ-His CGCGAATTCGGCGTTTCTCTGCTCTGGAGG, respectively (EcoRI and NdeI sites used for further cloning in italics). PCR products were double-digested with EcoRI and NdeI and cloned into P17-His Vector (kind gift of M. J. Runswick, MRC, Cambridge, UK) to place a His6-tag at the C-terminus of the recombinant protein. Recombinant pol λ and pol µ polypeptides were produced in E.coli strain BL21(DE3) harboring the plasmid P17-His-pol λ and P17-His-pol µ, respectively. Escherichia coli cultures were grown at 37°C until OD600 reached 0.8, IPTG was added to a final concentration of 1 mM and cells were incubated at 28°C for 5 h. Pelleted cells were resuspended in lysis buffer (50 mM Na2HPO4 pH 8.0, 500 mM NaCl, 10 mM β-mercaptoethanol) supplemented with protease inhibitors (Protease Inhibitor cocktail tablets, Boehringer) and 1 mg/ml lysozyme, and left 30 min on ice. Genomic DNA was sheared by sonication. Insoluble material was discarded by a 30 min centrifugation at 20 000 g. Protamine sulfate (1 mg/ml final concentration, Sigma) was added to the cleared lysate containing the soluble fraction of the induced protein, and precipitated DNA and proteins were pelleted (20 000 g, 30 min). The supernatant was applied to a Ni–NTA–agarose column (Qiagen) and washed with lysis buffer containing 20 mM imidazole. The His-tagged DNA polymerase was eluted with 300 mM imidazole, desalted on a PD10 column (Pharmacia), and loaded on a heparin–Sepharose column (Pharmacia). The protein was eluted with a 0.1–1 M NaCl gradient, and the active fractions stored at –80°C. For pol λ, the active fractions were pooled, concentrated on a 10K Microsep Concentrator (Pall Filtron), and purified further on a 15–30% glycerol gradient.
In vitro DNA polymerase assay
All reactions were conducted at 37°C for 60 min in 20 µl total volume containing 50 mM Tris–HCl (pH 7.4), 10 mM MgCl2 (or 1 mM MnCl2), 2 mM DTT, 20 mM NaCl, 0.2 mg/ml BSA, 5% glycerol and 250 µM of dNTP. For the 3′→5′ exonuclease activity assay, dNTPs were omitted. Substrate and enzyme concentrations are specified in the legend to Figure 7 and text below. TdT activity was monitored at 37°C for 60 min in the reaction conditions of the commercial calf thymus TdT (Promega), using a 17mer primer. Reactions were stopped by addition of 1/4 vol of stop solution (95% formamide, 20 mM EDTA, 0.025% xylene cyanol and 0.025% bromophenol blue), heated to 95°C for 5 min and analyzed by gel electrophoresis on a 12% denaturing polyacrylamide gel. Radiolabeled products were visualized by PhosphoImager exposure.
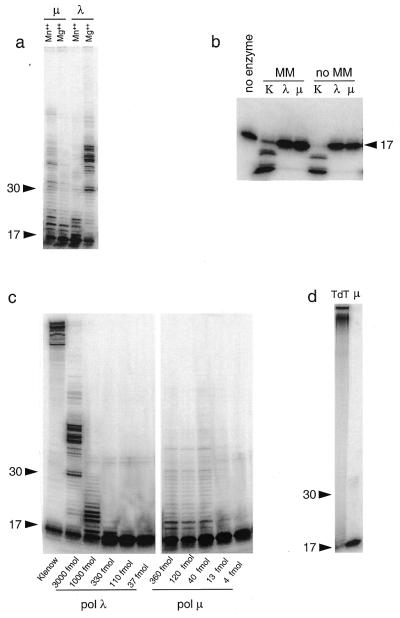
Figure 7.
Both purified pol λ and pol µ proteins exhibit a template-dependent DNA polymerase activity in vitro. (a) In vitro assay for template-dependent DNA polymerase activity. DNA polymerase activity was assayed by extension of a 5′-radiolabeled 17mer primer annealed to single-stranded circular M13mp18, as described in Materials and Methods, in the presence of either Mg++ or Mn++ metal activators. For each reaction, 75 fmol of substrate and 75 fmol of purified protein (either pol λ or pol µ) were used. Migration positions of 17mer and 30mer oligonucleotides are marked. (b) 3′→5′ exonuclease assay. The 3′→5′ exonuclease activity was investigated in the absence of dNTP on either 3′-mismatched (MM) or 3′-matched (no MM) primer-template substrate. Klenow fragment (0.5 U) was used as positive control. (c) Processivity assay. The processivity of pol λ and pol µ was assayed with a fixed amount of substrate (25 fmol of M13 template hybridized with the 17mer primer) incubated with decreasing amounts of polymerase as specified. Klenow fragment (2 × 10–3 U) was used as control. The weak doublet observed above the 30 nt marker originates from the labeled primer (see also below). (d) Assay for terminal transferase activity. TdT activity was assessed using 0.3 pmol of 5′-labeled single-stranded 17mer primer and 75 fmol of purified pol µ, in the presence of 250 µM of the four dNTPs. Calf thymus TdT (1 U) was used as positive control.
DNA substrates
Purified 17mers, either matched GCTTGCATGCCTGCAGG or 3′-mismatched GCTTGCATGCCTGCAGC, were 5′-end-labeled using [γ-32P]ATP (Amersham Pharmacia Biotech) and T4 polynucleotide kinase (New England Biolabs). Hybridization of the primer with circular single-stranded M13mp18 (+) template was performed in 50 mM NaCl, 15 mM Tris–HCl (pH 8) and 25 mM MgCl2, at a template:primer ratio of 1:1.
RESULTS
Isolation of cDNAs encoding DNA polymerases of the polX family
Databases were searched with a degenerate motif, designed to be consensus for DNA polymerase β, TdT, and poly(A) polymerase: G(S/G)(Y/F)(N/R)(L/R)G(X)5,6D(LIVM)DXL. The 128 different peptides encoded by this motif were confronted with ESTs (expressed sequence tags) and non-redundant databases, using the advanced BLAST search program (15) and increasing the statistical significance threshold (e-value) to 1000. A modification in data submission was performed allowing their treatment in batch (L. Cocea, details available upon request). One novel sequence was sorted out from mouse and human ESTs: mouse EST C86044 from a fertilized egg library, with significant homology with DNA polymerase β (named pol λ).
A second, direct search, using the protein sequence of either human TdT or human DNA polymerase β retrieved a second sequence, human EST AA298793 from an activated T cell library, with strong homology with TdT (named pol µ). The very long 3′ untranslated region of the human sequence (see below) explains why it escaped the first database search, no sequence covering the polX motif being present in ESTs databases, which are strongly biased toward the 3′-end of expressed mRNA sequences.
Probes obtained by PCR using oligonucleotides derived from these ESTs sequences were used to screen a mouse spleen cDNA library, and human bone marrow and lymph node libraries. Complete cDNAs encoding human and mouse pol λ, and corresponding to the size of the mRNA detected (see below), were obtained from these libraries [GenBank accession numbers: AF161019 (human) and AF176099 (mouse)]. Partial cDNAs for human and mouse pol µ were obtained upon screening of the same libraries, and their sequences were completed by one round of RACE amplification of their 5′-ends [GenBank accession numbers: AF176097 (human) and AF176098 (mouse)]. The complete amino acid sequences of human pol λ and pol µ are shown in Figure 1, compared with human DNA polymerase β and human TdT, respectively. Homology between pol β and pol λ is 54% (and identity 34%), and 61% between pol µ and TdT (identity 40%). Human and mouse proteins share 84% homology (and 76% identity) for pol µ and 87% homology (82% identity) for pol λ. The strong homology observed between mouse and human sequences ceases abruptly upstream from the ATG initiator codon for both proteins, strongly suggesting that it represents their true N-terminus.
Figure 1.
Human pol µ and pol λ DNA polymerase protein sequences. (a) Comparison of human pol µ and TdT proteins performed using the advanced BLAST program. Identical residues are indicated below the alignment with (+) for conservative changes. Nuclear localization signal (NLS), BRCT domain (17), HHH motifs (16) and polX motif (Prosite database) are marked. Amino acid positions of the pol µ sequence are indicated. (b) Same comparison performed between human pol λ and DNA polymerase β. (c) Schematic domain organization of pol µ, TdT, pol λ and DNA polymerase β. Hatched HHH motifs contain residues divergent from consensus at the Gly-(hydrophobe)-Gly core positions. Amino acid (aa) length of the human proteins is indicated.
The highest region of similarity extends over the polX domain. Two HHH (helix–hairpin–helix) DNA-binding motifs align with the ones of the homologous polymerases, only one of them showing complete conservation of consensus residues (16). In contrast to DNA polymerase β, pol λ protein contains a BRCT domain at the N-terminal region [BRCA-1 C-terminal domain (17), identified by the PFAM motif search program], making the overall domain organization of TdT, pol µ and pol λ extremely similar (Fig. 1c).
Phylogenetic comparison
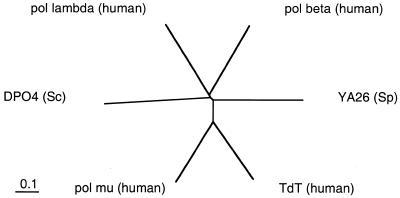
The polX signature (according to the Prosite database) is only found, among eukaryotic genomic sequences, in DNA polymerase β, TdT and two yeast proteins: a DNA polymerase β homolog in Saccharomyces cerevisiae (DPO4), and a putative TdT homolog in Schizosaccharomyces pombe (YA26). Phylogenetic comparison using the CLUSTALW program was performed between the two yeast enzymes, human DNA polymerase β, human TdT and the two new human polymerases (Fig. 2). The yeast polymerases are larger than DNA polymerase β, and their size is closer to the pol λ and pol µ proteins [506 versus 494 amino acids (aa) for YA26 and pol µ; 582 versus 575 aa for DPO4 and pol λ, whereas DNA polymerase β is 335 aa long]. Overall, the divergence of pol λ and DNA polymerase β appears quite ancient, a divergence also attested by the lack of relatedness in their exon–intron organization (see below). Furthermore, since the pol µ protein has DNA polymerase activity (see below), this suggests that the yeast YA26 protein might be a polymerase as well, and not a terminal transferase. Such untemplated activity might thus have evolved more recently from an ancient polymerase.
Figure 2.
Phylogenetic comparison of yeast and human proteins of the polX family. Alignment of sequences was performed with the CLUSTALW program using the default settings. The bar corresponds to 0.1 amino acid substitution per residue. The protein accession numbers are as follows: yeast (S.cerevisiae), DP04, P25615; yeast (S.pombe), YA26, Q09693; human DNA polymerase β, P06746; human TdT, P04053.
Tissue-specific expression
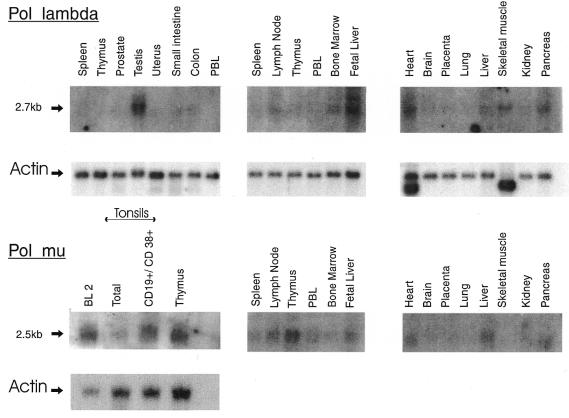
Tissue-specific expression was monitored for human pol λ and pol µ genes (Fig. 3). The size of both mRNAs detected (2.7 and 2.5 kb) matches the length of the cDNA sequences obtained, indicating that they are probably almost complete. For pol λ, the highest expression is observed in testis and fetal liver, with weaker expression, close to background level by northern blot analysis, in many different tissues. For pol µ, weak expression is also observed in a variety of tissues, with a higher representation in lymphoid tissues: among those, expression is highest in the thymus. We were struck by the fact that among 14 ESTs corresponding to human pol µ in databases (THC272006), seven belonged to the GCB1 library, representing germinal center CD20+IgD– B cells from human tonsils (these figures have evolved since: among 45 human ESTs retrieved by a BLAST homology search with the complete human pol µ mRNA sequence, 12 originate from germinal center or Burkitt lymphoma cDNA libraries). We therefore compared the expression level between thymus, total tonsils, CD19+CD38+ tonsillar cells [representing the B cell subset engaged in an immune response (14)], and a Burkitt lymphoma cell line, BL2 [corresponding to transformed CD19+CD38+ centroblasts (18)]. Expression levels were comparable between thymus and tonsillar B cells, and proportionally higher in the Burkitt lymphoma (Fig. 3) [other Burkitt cell lines, Ramos or DG75, showed similar expression levels (not shown)]. mRNA hybridization signals appear fuzzy, which is explained by the numerous splice variants with small size changes observed for this gene (see below). Tissues with the highest expression of pol µ mRNA were therefore further checked by direct sequencing to contain functional mRNA species (thymus, CD38+ tonsillar B cells and Burkitt lymphomas) (not shown).
Figure 3.
Tissue-specific expression of human pol µ and pol λ genes. Multi-tissue northern blots from Clontech were hybridized with full-length human pol µ and pol λ cDNA probes. An additional blot was hybridized, containing 0.5 µg of poly(A)+ RNA from the BL2 cell line (18), from total human tonsils and CD19+CD38+ tonsillar cells and from thymus. Control hybridization with actin is shown once for each blot. Exposure times are 3 days for pol µ and pol λ hybridizations, and 75 min for actin.
Genomic organization of human and mouse pol λ and pol µ genes
For mouse pol λ, complete genomic sequence was obtained from clone AC003694, containing 186 314 bp of mouse chromosome 19. Partial sequence for human pol λ was also available from clone AC008027, a 147 719 bp clone of chromosome 10, whose sequence is in progress. Exons 3 and 4 are missing from this sequence, and linkage has been obtained by genomic PCR from DNA of the Burkitt lymphomas BL2 and Ramos. These particular human and mouse chromosome homologs correspond to the localization of TdT. Moreover, gene mapping (GeneMap’99, NCBI) has located the human pol λ gene in the D10S540–D10S597 interval (sts-W69567), ∼10 cM from the TdT gene.
For mouse pol µ, a genomic clone was isolated from a mouse 129 phage library. Exon/intron mapping was performed either by direct sequencing (for small introns), or by PCR using exon-specific primers. For human pol µ, intron sizes and/or sequence was determined by direct genomic PCR and sequencing, using exon-specific primers (Fig. 4).
Figure 4.
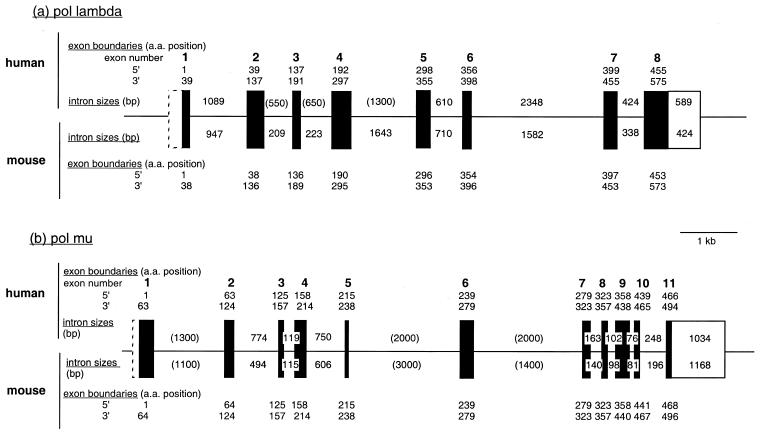
Structure of mouse and human pol λ and pol µ genes. Exon–intron organization of human pol λ and pol µ is drawn on scale with mouse genes represented below. Intron sizes are indicated (in bp). Values in parentheses are size estimates, others correspond to sequenced regions. The mouse pol λ gene has been completely sequenced (chromosome 19, accession number AC003694), and sequencing of the human pol λ gene is in progress (chromosome 10, AC008027). The amino acid positions of exon boundaries are indicated. Coding exons are represented as black boxes, and non-coding exons as open boxes (dashed boxes for 5′-untranslated regions whose exact length is not known).
The pol µ gene comprises 11 exons and shares with TdT identical exon/intron organization (19), splicing sites occurring at the same position in the aligned cDNA sequences. In contrast, the pol λ gene is split into eight exons, whose borders match neither DNA polymerase β nor TdT splicing sites. The overall size of these genes is small: ∼7.5 and 9 kb for mouse and human pol λ, respectively, and 10 kb for mouse and human pol µ.
Splice variants
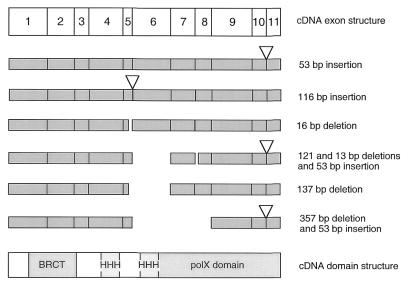
Among 11 partial human cDNAs isolated for pol µ from a lymph node library, none could encode a functional protein, due to reading frame alterations created by alternative splicing.
To study these variants, the pol µ cDNA sequence was amplified from BL2 Burkitt lymphoma cDNAs using PCR primers framing the complete sequence of the protein. A major 1500 bp product was obtained, which yielded only three sequences among 24 analyzed corresponding to the sequence shown in Figure 1. Most alterations occur after exon 5, i.e. in the second HHH domain (exon 6) and the polymerase domain (exons 6–11), and none of the variants identified contains an open reading frame covering the polymerase domain (Fig. 5). The modifications observed include: use of alternative splice junctions (e.g. at the 5/6 and 7/8 junctions, resulting in 16 and 13 bp deletions, respectively), presence of unspliced introns (e.g. intron 7–8, 118 bp) or absence of whole exons (exon 6 or exons 6–8), insertion of new exons (e.g. a 116 bp insertion encoded in intron 5–6 at the exon 5/6 junction). The splice variant the most frequently observed is a partial splicing of intron 10–11, which inserts the last 53 bp of this intron, and occurs in about half of the sequences. This insertion results in the addition of 18 aa, translation of exon 11 in the +1 reading frame (29 aa), and translation of the 3′ untranslated region up to the first stop codon encountered (96 aa), thus replacing exon 11 by a 143 aa peptide. A splice variant occurring at the homologous 10–11 exon junction has been described for mouse TdT, which results only in an in-frame 20 aa insertion without further alteration of the protein (20). However, the complete lack of sequence conservation between mouse and human pol µ 3′ untranslated regions does not argue in favor of a functional role of this major variant species.
Figure 5.
Splice variants for the human pol µ gene. Six representative examples of splice variants, isolated by RT–PCR amplification of poly(A)+ mRNA from the Ramos or the BL2 cell line with primers framing the coding sequence of pol µ, are depicted. The positions of the specific domains, BRCT, HHH and polX are indicated. Multiple events of alternate splicing are frequently observed within a cDNA (see last three examples).
For the pol λ gene, one variant species was observed among six cDNAs analyzed: it consists of a 19 bp addition at the exon 7–8 junction which prevents translation of the last exon (not shown). Among human ESTs present in databases, one (H11886) also shows an in-frame deletion of exons 5 and 6.
Protein purification and DNA polymerase activity
We overexpressed His-tagged pol λ and pol µ proteins in E.coli and purified them to near homogeneity, as judged by SDS–polyacrylamide gel electrophoresis (SDS–PAGE) and Coomassie blue staining (Fig. 6). The primer extension capacity of the purified proteins was tested using a standard DNA polymerase assay (see Materials and Methods). As shown in Figure 7a, both proteins exhibit a template-dependent polymerase activity in vitro. No elongation product could be detected when EDTA was added to the reaction (data not shown). The activity of pol λ was decreased by substituting Mg++ by Mn++, as activating metal ion (Fig. 7a), whereas pol µ activity was slightly enhanced. The processivity of these enzymes was assessed by lowering the enzyme/primer-template ratio (Fig. 7c): pol λ appears strictly distributive in this assay, whereas pol µ was more processive. When the four nucleotides were omitted from the reaction mixture, the primer, whether it displays a perfectly matched or a 3′ mismatched end with the template, remained intact in the presence of the enzymes, indicating that they lack 3′→5′ exonuclease activity (Fig. 7b). This implies that these new enzymes, like DNA polymerase β, lack proofreading activity. Moreover, given the homology between TdT and pol µ, we investigated whether this latter enzyme also exhibited non-templated nucleotide addition. Using a single-stranded 17mer DNA substrate in the presence of all four dNTPs, no terminal transferase activity was detected (Fig. 7d).
Figure 6.
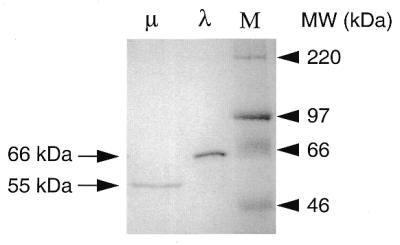
SDS–PAGE profile of purified pol λ and pol µ. The fraction purified after heparin–Sepharose chromatography was analyzed by electrophoresis on 10% polyacrylamide gel and visualized by Coomassie blue staining. Pol µ (500 ng) and pol λ (2 µg) were loaded. M indicates molecular weight markers (Rainbow marker, Amersham).
Down-regulation of both pol λ and pol µ mRNA expression in response to DNA damaging agents
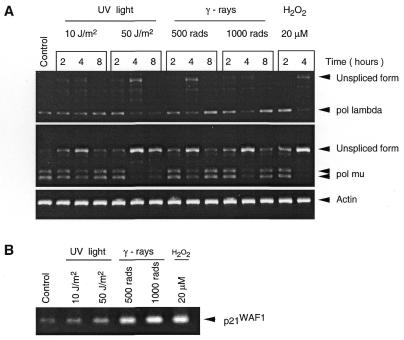
In order to investigate possible roles for the two isolated DNA polymerases, we examined whether they are involved and/or regulated during DNA repair processes. Ramos cells were treated with different DNA damaging agents, including UV light, ionizing radiation and H2O2 as an inducer of oxidative damage, to activate different DNA repair pathways. To monitor the efficacy of the different treatments, we used as control the cyclin-dependent kinase inhibitor 1 gene (p21WAF1), whose expression is induced in response to DNA damage in order to block DNA replication by preventing the G1→S transition and allow the DNA repair machinery to remove the damage (21). The results shown in Figure 8 indicate that, in parallel to the up-regulation of the p21WAF1 mRNA, pol µ, and to a lesser extent, pol λ gene expression is down-regulated upon cell exposure to all types of DNA damaging agents, in a dose and time-dependent fashion. This down-regulation, as judged by semi-quantitative RT–PCR amplification, correlates with an accumulation of the unspliced RNA species, particularly for the pol µ gene, and not with a reduced transcription. DNA contamination was excluded, since no PCR products were obtained under similar amplification conditions when reverse transcriptase was omitted (data not shown). In the case of UV light, down-regulation of mRNA expression is observed up to 8 h at 50 J/m2. For γ-rays, it peaks at 4 h and returns to control level by 8 h, probably because DNA repair is achieved. In the case of oxidative damage where cells are kept in medium containing H2O2, the down-regulation is maintained and more pronounced, mature pol λ mRNA being strongly reduced and pol µ mRNA almost undetectable after 4 h, and the effect being still observed at 20 h (not shown). Similar results were obtained with the BL2 cell line (not shown).
Figure 8.
Pol µ and pol λ are down-regulated upon exposure to DNA damaging agents. (A) Following the DNA damaging treatment, Ramos cells were collected, total RNA extracted and semi quantitative RT–PCR performed, using the actin gene as control. The results of non-saturating PCR amplification conditions are illustrated. The various treatments are indicated. The size of the expected cDNA fragments, and of the unspliced forms is marked. For pol µ, the 5′ and 3′ primers are located in exons 9 and 11, respectively. The amplified doublet (246 and 299 bp) corresponds to the spliced mRNA with or without the 53 bp addition (see splicing variants). The unspliced form (570 bp) contains introns 9–10 and 10–11. For pol λ, the 5′ and 3′ primers located in exons 4 and 6, respectively, amplify a 360 bp fragment. The unspliced form (∼1 kb) corresponds to the presence of the sole intron 5–6, the fully unspliced species with both introns present being too large (∼2.3 kb) to be efficiently amplified in this assay. (B) Expression level of p21WAF1 gene was estimated by PCR amplification of the cDNAs from the 4 h time point of the different treatments.
DISCUSSION
To uncover new members of the polX family, we confronted a degenerate motif, G(S/G)(Y/F)(N/R)(L/R)G(X)5,6D(LIVM)DXL, consensus for DNA polymerase β, TdT and poly(A) polymerase, with EST databases. This systematic approach was completed by a direct homology search, using the complete protein sequence of both human TdT and DNA polymerase β. Two new sequences were retrieved, the corresponding cDNAs isolated, and the complete human and mouse protein sequence and gene structure established.
One of these proteins, pol λ, has more homology with DNA polymerase β (54%), while the second one, pol µ, has strong homology with TdT (61%). Their structural organization is similar and includes the polX domain, two helix–loop–helix motifs corresponding to a non-sequence-specific DNA recognition motif, and one domain BRCT, which is observed in numerous DNA damage-responsive cell cycle checkpoint proteins (16). They both therefore structurally resemble TdT and not DNA polymerase β which is a smaller polymerase lacking a BRCT domain.
Highest mRNA expression of pol λ is observed in testis and fetal liver, and lower expression is detectable in a wide range of tissues. For pol µ, expression is highest in thymus and tonsillar B cells, suggesting a preferential expression in proliferating lymphoid cells. A weak expression is also detectable in a variety of different tissues.
Minor variant forms of expressed mRNA produced by alternative splicing have been described for DNA polymerase β and mouse TdT (20,22), and, similarly, splice variant forms, probably non-functional due to the major alterations they create in the polymerase domain of the protein, also exist for the pol λ gene. For the pol µ gene however, the mRNA encoding the complete protein turned out to be the minor species, representing ∼10% only of the mRNAs present in a cell, whereas the variant splice forms represent the major population. Most of these variations occur in the exons encoding the second HHH motif and the polymerase domain and therefore alter the polymerase structure, either because they introduce premature stop codons, or because they change the reading frame of the polymerase domain. The preponderance, both in normal lymphoid tissues and in transformed cell lines, of mRNA species containing premature stop codons suggests that, in addition to the inherent unusual propensity of this transcript to undergo multiple splicing events, the mRNAs produced strikingly escape the process of non-sense-mediated decay (23).
Both proteins were overexpressed in E.coli with a histidine-tag and purified near homogeneity, to assess their enzymatic properties in vitro. Surprisingly, with regard to their sequence homology, both exhibited template-dependent polymerase activity, and no terminal transferase activity was observed for pol µ. In a recent publication describing identification of the pol µ gene, Dominguez et al. (24) detected a weak terminal transferase activity, several orders of magnitude lower than its template-dependent activity. Both enzymes lack 3′→5′ exonuclease activity and are thus devoid of proofreading capacity, as DNA polymerase β is. Their fidelity in DNA synthesis thus relies on their intrinsic capacity in discriminating for correct nucleotide incorporation. In their parallel investigation, Dominguez et al. reported that pol µ has a remarkably low fidelity in vitro, when assayed in the presence of Mn++ ions (24).
Comparison of these two new polymerases with DNA polymerase β, TdT and two yeast polX polymerases (the β-like polIV in S.cerevisiae and a putative TdT-like enzyme in S.pombe) indicated that the divergence between DNA polymerase β, pol λ and pol µ is quite ancient, the lack of similarity in the intron–exon structure further attesting such early divergence. The chromosomal localization of the pol λ gene (chromosome 19 in the mouse, and 10 in human at 10 cM from TdT) indicates on the other hand that this gene has conserved a genetic linkage with TdT, and not with DNA polymerase β [whose localization is on mouse and human chromosomes 8 (25)]. Divergence of pol µ and TdT is in contrast much more recent, and, considering the template-dependent activity of these various proteins, one may wonder whether the template-independence of the TdT might have evolved recently from ancient polymerase structure.
Mouse and human pol λ and pol µ proteins show a very strong conservation throughout their sequence (87 and 82% homology between human and mouse pol λ and pol µ, respectively), suggesting that these enzymes assume a specific function under strong selective pressure. Moreover, such a function is obviously not redundant with the one assumed by DNA polymerase β, since mice deficient for this latter enzyme have an embryonic lethal phenotype (26). Apart from DNA replication, DNA polymerases are essential for repair of all types of DNA damage but also for many other DNA transactions such as, e.g., non-homologous end-joining, which is exemplified in lymphoid cells by the V(D)J recombination process. Furthermore, it is well established that in bacteria many polymerases are induced or up-regulated upon exposure to UV light or to external stress (see reviews 6,7). In mammalian cells too, recent studies have indicated that expression of some DNA polymerases is specifically altered following DNA stress. DNA polymerase β is up-regulated in response to H2O2- and LPS-mediated oxidative damage, and also following treatment with the alkylating agent N-methyl-N′-nitro-N-nitrosoguanidine (27,28) Expression of DNA polymerase ζ is increased following exposure to UV radiation (29), and DNA polymerase δ is up-regulated upon induction of V(D)J recombination (30).
Expression of pol λ and pol µ genes in response to various types of DNA damage was thus studied: UV light, γ-rays and H2O2. Surprisingly, in each case mRNA expression of both polymerases was markedly reduced, the effect being still more pronounced for pol µ. In the case of the pol µ gene, the down-regulation of the mRNA appears to occur mainly at the post-transcriptional level, a striking accumulation of unspliced pre-mRNA species being observed in all cases of damage induction. It is thus unlikely that these two new enzymes may be actively involved in any of these repair pathways: repair of UV-damaged or oxidized bases, or repair of double-strand breaks. Similarly, no specific up-regulation of their expression was observed upon induction of Ig gene rearrangement in a rearrangement-inducible Abelson cell line (31) (data not shown).
The in vitro assays indicate that these two enzymes have a low processivity and are proofreading-deficient. Their possible involvement in the process of hypermutation of Ig genes was therefore tested by overexpression in the Ramos Burkitt lymphoma cell line, which shows constitutive, ongoing mutation of its Ig variable genes (32). No alteration in the Ig gene mutation pattern could be observed after stable transfection and strong overexpression of the mRNAs encoding either of these two enzymes (data not shown). Such a direct assay however may not be conclusive if other limiting factors are controlling the mutation process (access to the target, occurrence of breaks in DNA, etc.). In the case of the pol µ enzyme, still another possibility could be that a minor splicing variant, differing from the cDNA transfected, could be the active form, allowing for proper sub-cellular localization and/or protein interaction.
We have described in this paper two new DNA polymerases, identified on the basis of their belonging to the polX family. Several other mammalian polymerases have been recently identified by their relatedness with the bacterial and yeast lesion bypass enzymes UmuC/DinB/Rad30 (reviewed in 6,7). None of these various DNA polymerases so far show a tissue-specificity comparable to other lymphoid-specific enzymes, like TdT or RAG-1 and RAG-2 genes, which could attribute to them the unique function of an Ig gene mutase. However, all of them show various degrees of infidelity that make them potential candidates to achieve the specific mutation rate observed at the Ig locus in a mutating B cell (10–3–10–4/bp/cell generation) (33–35). B-cell-specific factors would thus be required to target them at the right time to the right gene. Establishing whether or not any of these new DNA polymerases assume the specific function of the Ig mutase will probably await their gene inactivation, either in mice or in Burkitt lymphoma cell lines undergoing Ig gene hypermutation in vitro (18,32).
NOTE ADDED IN PROOF
Bacterial expression of a modified form of pol µ with its catalytic site mutated has revealed recently that the normal pol µ enzyme produced in E.coli according to the conditions described here is inactive, and that the activity isolated is of bacterial origin. Preliminary experiments suggest that an active pol µ enzyme can be obtained following the specific conditions of Boule et al. (36) for bacterial expression of mouse TdTL.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by grants from the Human Frontier Science Program, the Association pour la Recherche contre le Cancer, the APEX program of the Institut National de la Santé et de la Recherche Médicale and the Fondation Princesse Grace. S.A. is supported by a Poste Vert from I.N.S.E.R.M., A.F. by a fellowship from the Fondation de France, S.S. is a fellow of Ecole Normale Supérieure and N.G. has been supported by a fellowship from the Fondation pour la Recherche Médicale.
DDBJ/EMBL/GenBank accession nos+ To whom correspondence should be addressed. Tel: +33 1 40 61 56 83; Fax: +33 1 40 61 55 90; Email: reynaud@infobiogen.fr Present addresses: Laurentiu Cocea, Amgen Research Institute, 620 University Avenue, Suite 706, Toronto, Ontario M5G 2C1, Canada Neetu Gupta, CNRS UMR 1773, Institut Pasteur, 25 rue du Docteur Roux, 75724 Paris cedex 15, France AF161019, AF176097, AF176098 and AF176099
REFERENCES
- 1.Diaz M., Velez,J., Singh,M., Cerny,J. and Flajnik,M.F. (1999) Int. Immunol., 11, 825–833. [DOI] [PubMed] [Google Scholar]
- 2.Masutani C., Kusumoto,R., Yamada,A., Dohmae,N., Yokoi,M., Yuasa,M., Araki,M., Iwai,S., Takio,K. and Hanaoka,F. (1999) Nature, 399, 700–704. [DOI] [PubMed] [Google Scholar]
- 3.Gerlach V.L., Aravind,L., Gotway,G., Schultz,R.A., Koonin,E.V. and Friedberg,E.C. (1999) Proc. Natl Acad. Sci. USA, 96, 11922–11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McDonald J.P., Rapic-Otrin,V., Epstein,J.A., Broughton,B.C., Wang,X., Lehmann,A.R., Wolgemuth,D.J. and Woodgate,R. (1999) Genomics, 60, 20–30. [DOI] [PubMed] [Google Scholar]
- 5.Ogi T., Kato,T.,Jr, Kato,T. and Ohmori,H. (1999) Genes Cells, 4, 607–618. [DOI] [PubMed] [Google Scholar]
- 6.Radman M. (1999) Nature, 401, 866–869. [DOI] [PubMed] [Google Scholar]
- 7.Woodgate R. (1999) Genes Dev., 13, 2191–2195. [DOI] [PubMed] [Google Scholar]
- 8.Kim N., Kage,K., Matsuda,F., Lefranc,M.-P. and Storb,U. (1997) J. Exp. Med., 186, 413–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertocci B., Quint,L., Delbos,F., Garcia,C., Reynaud,C.-A. and Weill,J.-C. (1998) Immunity, 9, 257–265. [DOI] [PubMed] [Google Scholar]
- 10.Kunkel T.A. and Soni,A. (1988) J. Biol. Chem., 263, 14784–14789. [PubMed] [Google Scholar]
- 11.Esposito G., Texido,G., Betz,U.A.K., Gu,H., Müller,W., Klein,U. and Rajewsky,K. (2000) Proc. Natl Acad. Sci. USA, 97, 1166–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aravind L. and Koonin,E.V. (1999) Nucleic Acids Res., 27, 1609–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alt F.W. and Baltimore,D. (1982) Proc. Natl Acad. Sci. USA, 79, 4118–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pascual V., Liu,Y.-J., Magalski,A., de Bouteiller,O., Banchereau,J. and Capra,J.D. (1994) J. Exp. Med., 180, 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altschul S.F., Madden,T.L., Schaffen,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doherty A.J., Serpell,L.C. and Ponting,C.P. (1996) Nucleic Acids Res., 24, 2488–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bork P., Hofmann,K., Bucher,P., Neuwald,A.F., Altschul,S.F. and Koonin,E.V. (1997) FASEB J., 11, 68–76. [PubMed] [Google Scholar]
- 18.Denépoux S., Razanajaona,D., Blanchard,D., Meffre,G., Capra,J.D., Banchereau,J. and Lebecque,S. (1997) Immunity, 6, 35–46. [DOI] [PubMed] [Google Scholar]
- 19.Riley L.K., Morrow,J.K., Danton,M.J. and Coleman,M.S. (1988) Proc. Natl Acad. Sci. USA, 85, 2489–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doyen N., Fanton d’Andon,M., Bentolila,L.A., Nguyen,Q.T. and Rougeon,F. (1993) Nucleic Acids Res., 21, 1187–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harper J., Adami,G., Wei,N., Keyomarsi,K. and Elledge,S. (1993) Cell, 75, 805–816. [DOI] [PubMed] [Google Scholar]
- 22.Chyan Y.-J., Strauss,P.R., Wood,T.G. and Wilson,S.H. (1996) DNA Cell Biol., 15, 653–659. [DOI] [PubMed] [Google Scholar]
- 23.Li S. and Wilkinson,M.F. (1998) Immunity, 8, 135–141. [DOI] [PubMed] [Google Scholar]
- 24.Dominguez O., Ruiz,J.F., Lain de Vera,T., Garcia-Diaz,M., Gonzalez,M.A., Kirchhoff,T., Martinez,C., Bernad,A. and Blanco,L. (2000) EMBO J., 19, 1731–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McBride O.W., Kozak,C.A. and Wilson,S.H. (1990) Cytogenet. Cell Genet., 53, 108–111. [DOI] [PubMed] [Google Scholar]
- 26.Gu H., Marth,J.D., Orban,P.C., Mossmann,H. and Rajewsky,K. (1994) Science, 265, 103–106. [DOI] [PubMed] [Google Scholar]
- 27.Chen K.H., Yakes,F.M., Srivastava,D.K., Singhal,R.K., Sobol,R.W., Horton,J.K., Van Houten,B. and Wilson,S.H. (1998) Nucleic Acids Res., 26, 2001–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narayan S., Beard,W.A. and Wilson,S.H. (1995) Biochemistry, 34, 73–80. [DOI] [PubMed] [Google Scholar]
- 29.Gibbs P.E., McGregor,W.G., Maher,V.M., Nisson,P. and Lawrence,C.W. (1998) Proc. Natl Acad. Sci. USA, 95, 6876–6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jessberger R., Schär,P., Robins,P., Ferrari,E., Riwar,B. and Hübscher,U. (1997) Nucleic Acids Res., 25, 289–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y.Y., Wang,L.C., Huang,M.S. and Rosenberg,S. (1994) Genes Dev., 8, 688–697. [DOI] [PubMed] [Google Scholar]
- 32.Sale J.E. and Neuberger,M.S. (1998) Immunity, 9, 859–869. [DOI] [PubMed] [Google Scholar]
- 33.Johnson R.E., Washington,M.T., Prakash,S. and Prakash,L. (2000) J. Biol. Chem., 275, 7447–7450. [DOI] [PubMed] [Google Scholar]
- 34.Matsuda T., Bebenek,K., Masutani,C., Hanaoka,F. and Kunkel,T.A. (2000) Nature, 404, 1011–1013. [DOI] [PubMed] [Google Scholar]
- 35.Johnson R.E., Prakash,S. and Prakash,L. (2000) Proc. Natl Acad. Sci. USA, 97, 3838–3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boule J.B., Rougeon,F. and Papanicolaou,C. (2000), J. Biol. Chem., in press. [Google Scholar]