Abstract
Kaposi Sarcoma (KS) is the most common cancer in people living with HIV (PLWH) in many countries where Kaposi Sarcoma-associated herpesvirus is endemic. Treatment has changed little in twenty years, but the disease presentation has. This prospective cohort study enrolled 122 human immunodeficiency virus (HIV) positive KS patients between 2017 and 2019 in Malawi. Participants were treated with bleomycin, vincristine, and combination antiretroviral therapy, the local standard of care. One-year overall survival was 61%, and progression-free survival was 58%. The 48-week complete response rate was 35%. RNAseq (n = 78) differentiated two types of KS lesions, those with marked endothelial characteristics and those enriched in inflammatory transcripts. This suggests that different KS lesions are in different disease states consistent with the known heterogeneous clinical response to treatment. In contrast to earlier cohorts, the plasma HIV viral load of KS patients in this study was highly variable. 25% of participants had no detectable HIV; all had detectable KSHV viral load. This study affirms that many KS cases today develop in PLWH with well-controlled HIV infection and that different KS lesions have differing molecular compositions. Further studies are needed to develop predictive biomarkers for this disease.
Keywords: Kaposi Sarcoma herpesvirus, Transcriptome RNA sequencing, clinical study, Differential gene expression
Introduction
Kaposi Sarcoma (KS) is among the most common cancers in Eastern and Central Africa 1, 2. In Malawi, in 2021, KS accounted for approximately one-fifth of all cancer cases 3. Incidence rates are similar in Uganda and other KS-endemic countries 4, 5. In South Africa, KS is the most common cancer in men living with HIV (PLWH) 6. In KS-endemic countries, KS affects human immunodeficiency virus (HIV)-positive and negative individuals, males and females, and children.
First described in 1872, KS preceded the introduction of HIV into the human population. In the US and Europe, one-third of KS cases develop in PLWH on stable combination antiretroviral therapy (cART), who have suppressed HIV viral loads and near normal CD4 counts 7, 8. Classic KS was described before HIV was introduced into the human population at a time when a detailed assessment of human immune status was not possible; it clustered in specific populations, such as men of Mediterranean descent 9. Classic KS today is not necessarily associated with overt immune deficiency either 10. In KS-endemic countries, high CD4 counts provide less protection against the development of KS in PLWH compared to non-KS-endemic countries 11, 12, an indication that HIV-KS will continue to be of concern in Sub-Saharan Africa (SSA). Potential reasons for the high incidence rates of KS in the region include the high rates of Kaposi Sarcoma-associated herpesvirus (KSHV), KSHV/ HIV co-infection, suboptimal cART coverage, or co-infection of KSHV with other diseases 12.
KSHV infection necessarily precedes KS disease. KSHV is also the etiological agent for diseases that often co-exist in KS patients, such as Multicentric Castleman Disease (MCD), Primary Effusion Lymphoma (PEL), KS-Immune Reconstitution Inflammatory Syndrome (KS-IRIS), and KSHV Inflammatory Cytokine Syndrome (KICS) (reviewed in 13). In SSA, KSHV seroprevalence exceeds 80% before puberty. Here, KSHV infection often precedes HIV acquisition (except in instances of HIV mother-to-child transmission). Other endemic childhood infections, such as malaria, are common and may modulate KSHV acquisition 14.
Many HIV-positive KS patients with limited-stage disease respond to antiretroviral therapy alone initially; however, most KS patients require concurrent or subsequent cytotoxic cancer therapy 15–17, including those not infected with HIV (classic KS). Standard treatment strategies for KS are based on cytotoxic chemotherapy 18–22. Prior studies 16, 23–27 informed first-line treatment recommendations in SSA and other low- and middle-income countries (LMIC), including vincristine /bleomycin (BV) and non-liposomal doxorubicin or paclitaxel. HIV-KS is treated with pegylated liposomal doxorubicin 28–31. Overall survival (OS) and progression-free survival (PFS) in SSA are lower than in the US or Europe 27. This discrepancy is due to a combination of factors, including late presentation and insufficient access to pegylated liposomal doxorubicin (Doxil). It is unknown whether, in addition, there also exist differences in the underlying biology of KS, such as KSHV strain distribution 32, or if there are different molecular drivers of the disease. This represents a gap in our knowledge and a barrier to optimal treatment designs that this study aimed to address.
We report on a prospective cohort study of KS patients treated at the Kamuzu Central Hospital (KCH) in Lilongwe, Malawi. The motivation was that a better understanding of both clinical and molecular parameters would inform disease management and uncover novel targets for intervention. The aim was to describe KS in PLWH with access to cART and medical care typical for countries with endemic KS, i.e., childhood-acquired KSHV 33 and epidemic HIV. The primary objectives were to define the baseline characteristics, OS, PFS, and complete response (CR) at 48 weeks. All participants received bleomycin (15 IU/m2) and vincristine 2 mg (fixed dose) and initiated cART concurrently or were continued on cART. The number of chemotherapy cycles was left to the clinician’s discretion per the local standard of care. RNA-sequencing of 78 KS skin lesion biopsies was performed to identify KS subsets based on human transcript patterns. This study produced the largest database of KS transcriptome data to date.
Methods
Study design
LCCC1424 was a prospective, open-label cohort study of pathology-confirmed HIV-associated KS patients that initiated chemotherapy treatment in Malawi. Patient eligibility was defined as having histological confirmation of KS, HIV positive, on or off cART, 18 years of age or older, residing within 200 km from the study site, and having the ability to give informed consent. Pregnancy and breastfeeding were not considered exclusion criteria, given that the study was observational and with diagnostic and treatment interventions administered according to local standards of care. Prior cART experience was allowed, but previous chemotherapy was not. Participants with previously treated KS were excluded.
Eligible participants were enrolled from February 2017 to June 2019 and were followed for up to 96 weeks. Upon histological confirmation of KS diagnosis, a comprehensive baseline evaluation was performed. The baseline evaluation included the collection of two skin biopsies and baseline clinical laboratory testing. KS staging was assessed by a complete skin examination and chest radiograph, conforming to previously published criteria 34. Five marker lesions were selected and evaluated at each visit. Marker lesions were selected based on the operating procedures from concurring randomized phase III KS studies at this site 16, 27. Specifically, we identified lesions that were at least 1 cm x 1 cm, at least 1 cm away from, and distinguishable from any nearby lesions. We then choose the five largest lesions with clearly distinguishable margins. These were saved on a body map to ensure the same lesions were measured each time.
The decision to initiate chemotherapy treatment was at the physician’s discretion. All but one participant started treatment at screening. One participant was delayed in initiating treatment as the participant did not promptly report back to the clinic. Here, the enrollment date was used as the initiation date. Chemotherapy was administered according to a standardized clinic treatment algorithm and consisted of BV only, whilst cART was administered according to national guidelines. Treatment toxicity was graded using the National Cancer Institute Common Technology Criteria for Adverse Events (CTCAE) version 5.
Statistical analysis
The Kaplan-Meier method was used to estimate the OS and PFS, with the two-sided 95% CIs calculated using the log-log transformation. A Cox proportional-hazards regression model was used to estimate the hazard ratios for multiple clinical variables. Multivariate logistic regression was used to study associations between the dichotomous CR outcome and targeted clinical variables at 48 weeks. The associations between KSHV viral load and the targeted clinical variables were performed independently. The Pearson or the Spearman methods were used to calculate the correlations for continuous clinical variables. The binary clinical variables were studied using either the Wilcoxon rank sum test (minimum group size < 30) or the t-test (minimum group size ≥ 30). The normality assumption was verified by the Shapiro-Wilk test.
KSHV Viral Load
DNA was extracted using the MagNA Pure Instrument (Roche). Following DNA extraction, KSHV viral load was determined as described 35.
Immunohistochemistry
Formalin-Fixed Paraffin-Embedded blocks containing the skin biopsies were sectioned at the UNC Pathology core facility. Sections were stained for LANA following a previously described protocol 36 (Supplementary Figure 1). A dilution factor 1:100 of mouse anti-human LANA antibodies was used (Leica Biosystems Cat# NCL-L-HHV8-LNA).
Transcriptome analysis
Approximately 30 mg tissue was processed using the Qiagen, RNeasy Fibrous Tissue Mini Kit (Qiagen, cat# 74704), with 300 µl RLT, 0.05% DX Reagent, and one 3.0 mm stainless steel ball. The sample was homogenized for three minutes or until completion using the Qiagen Tissue Lyser (Qiagen, cat# 85300), and RNA was isolated according to the manufacturer’s instructions. RNA quantity and quality were verified by Qubit assay (Life Technologies) and on a 4200 Tape Station (Agilent Technologies). Library preparation and templating were automated on the Ion Chef System (Life Technologies Pub. No. MAN0013432 Rev. H.0), where libraries were diluted to 75 pM before templating on the Ion 540 chip (Thermofisher cat#. A27765). Sequencing was performed on the Ion Torrent S5 sequencer (Life Technologies Pub. No. MAN00006735 Rev: F.0.). The cleaned FASTQ files are submitted to SRA achieves. The sequencing coverage and quality statistics for each sample are summarized in Supplemental Table 1.
Human transcription was determined using CLC Genomics Workbench version 21.0.5 (Qiagen) and further processed using DESeq2 37. Protein coding genes with at least ten sequence reads in at least one sample and expressed in at least half of the participants were retained for analysis. Sequence reads were randomly subsampled to a maximum of two million reads for the cell line data (SRP035883 and SRP078245). Normalized reads were further transformed using the Variance Stabilizing Transformations (VST) implemented in DESeq2. The 2,000 most variable genes (based on their coefficient of variation) were used for hierarchical clustering based on the Euclidean distance matrix and Ward’s linkage using R, version 4.1.2.
Results
Baseline characteristics
Prospective participants were screened between February 2017 and June 2019. Fifteen participants were excluded due to non-KS histology. One participant was excluded due to being HIV-negative at enrollment, and another one due to having a previous treatment of KS less than a year before enrollment. Fifteen participants were withdrawn due to incomplete enrollment. Two participants withdrew after one treatment cycle and another after four treatment cycles due to suspected side effects. The study had 122 participants with complete clinical data (Table 1).
Table 1.
Participant baseline characteristics at enrolment for a total number of participants (n = 122; interquartile range [IQR]; standard deviation [SD]).
| Total number of participants (n) | 122 |
|---|---|
| Age, years, median (IQR) | 36 (32–44) |
| Gender; men, n (%) | 98 (80) |
| Staging | |
| Tumor severity T0 (ACTG), n (%) | 18 (15) |
| Karnofsky performance status ≤ 70%, n (%) | 43 (35) |
| Illness severity S1, n (%) | 67 (55) |
| Symptoms | |
| Edema present, n (%) | 60 (51) |
| Visceral disease present, n (%) | 8 (7) |
| Oral involvement present, n (%) | 41 (35) |
| HIV Characteristics | |
| Knew HIV+ diagnosis prior to KS diagnosis, n (%) | 64 (53) |
| Months HIV+ diagnosis known, median (IQR) (n=61) | 16 (6–57) |
| On ART prior to KS diagnosis, n (%) | 61 (50) |
| Months on ART prior to diagnosis, median (IQR) (n=61) | 16 (6–57) |
| CD4 count, median (IQR) (n=115) | 197 (96–337) |
| HIV viral load log copies/ml/median (IQR) (n=116) | 3 (2–5) |
| HIV viral load suppressed < 1000 copies/ml, n (%) (n=116) | 67 (58) |
| Baseline Lab Results | |
| Hemoglobin, g/dl/mean (SD) | 11 (3) |
| Platelets, 103/µl/median (IQR) | 234 (157–313) |
| White blood cell, 103/µl/median (IQR) | 5 (4–6) |
| Absolute neutrophil count, 103/µl/ median (IQR) | 2 (1–3) |
| Creatinine, mg/dl/median (IQR) | 0.8 (0.7–0.9) |
| Bilirubin, mg/d/ median (IQR) (n=111) | 0.4 (0.3–0.5) |
The median age was 36 (IQR: 32 – 44) years; 98 (80%) participants were male. 18 (15%) participants were classified as tumor severity T0, and 104 (85%) as T1; 67 (55%) participants had an illness severity score of S1, and 55 (45%) were classified as S0. 43 (35%) participants had a Karnofsky performance status (KPS) ≤ 70. Presenting symptoms included edema in 60 (49%), visceral disease in 8 (7%), and oral involvement in 41 (34%) participants. Before diagnosis, 64 (52%) participants were aware of being HIV infected for a median of 15.7 months (IQR: 6 – 57.1). 61 (50%) participants were on cART before their KS diagnosis for a median of 15.7 months (IQR: 6 – 57.1). The median CD4 count was 197 (IQR: 96 – 337) cells/µl, and the median HIV viral load was 2.6 log10 copies/mL (IQR: 1.6 – 4.7). 67 (58%) participants had an HIV viral load of <1,000 copies/ml. For participants who were on cART before screening, the CD4 count was 212 (IQR: 98.5~345.2, n=56) cells/µl; for those not on cART, it was 176 (IQR: 75.5~332, n = 59) cells/µl. For participants who were on cART before screening, the HIV genome copy number was 1.60 (IQR: 1~3.3 n = 57) log10 copies/ml, and for those not on cART, it was 3.39 (IQR: 2.1~4.9 n= 59) log10 copies/ml.
Outcomes
The treatment outcomes are summarized in Table 2. The median number of treatment cycles was 16 (IQR: 7–17), with 66/122 (54%) participants receiving 16 or more treatment cycles. 52/122 (43%) participants discontinued treatment early, receiving less than 16 treatment cycles without achieving a complete response (CR); 23 (44%) died, and one (2%) experienced progressive disease (PD) after six treatment cycles. 24 (46%) of the 52 participants defaulted after a median of six (IQR: 2 – 9) treatment cycles, two participants were transferred to new facilities, and two dropped out due to suspected treatment toxicity. 60 (49%) participants had experienced at least one delayed or missed dose of either chemotherapy agent due to stock out. The median number of cycles for participants with one missed or delayed dose was 3 (IQR: 2 – 4). Fourteen (12%) participants had at least one cycle without bleomycin (median missed doses 2 (IQR: 1–3)), and 22 (18%) participants had at least one cycle without vincristine (median missed doses 3 (IQR 2–4)). This was an observational study with treatment determined by the treating physicians according to hospital guidelines (often dictated by local drug availability and patient needs). Most patients received chemotherapy every two weeks, which could be delayed for adverse events such as anemia, neutropenia, thrombocytopenia, or other grade 3–4 adverse events. The minimum period between cycles was never less than 14 days.
Table 2.
Clinical outcomes for the total number of participants (n), interquartile range (IQR), and Bleomycin/Vincristine (B/V)
| Variable | Sample size (n) | Value |
|---|---|---|
| Treatment Cycles | 122 | |
| Cycles received, median (IQR) | 16 (6–17 ) | |
| Received≥ 16 cycles (% ) | 66 (54 ) | |
| Reason treatment not completed (%) | 52 | |
| Died | 23 (44 ) | |
| Defaulted | 24 (46 ) | |
| Moved to another HIV treatment facility | 2 (4) | |
| Toxicity | 2 (4) | |
| Disease Progression | 1 (2) | |
| Treatment stock out (%) | 122 | |
| Had no missed doses due to stock out (B or V) | 62 (51 ) | |
| Had 1 missed dose due to stock out | 13 (11 ) | |
| Had >1 missed dose due to stock out | 47 (39 ) | |
| Drugs missed due to stock out (%) | 122 | |
| Had 1 missed dose of bleomycin | 14 (12 ) | |
| Had >1 missed dose of bleomycin | 22 (18) | |
| Had 1 missed dose of vincristine | 8 (7) | |
| Had >1 missed dose of vincristine | 28 (23) | |
| Adverse events during treatment (%) | 122 | |
| Grade 2 anemia | 14 (12 ) | |
| Grade 3/4 anemia | 4 (3) | |
| Grade 2 neutropenia | 19 (16) | |
| Grade 3/4 neutropenia | 12 (10) | |
| Grade 2 thrombocytopenia | 1 (0.8) | |
| Grade 3/4 thrombocytopenia | 0 (0) | |
| Non-hematologic grade 3/4 event | 0 (0) | |
| Had at least one delayed or reduced dose due to adverse event | 0 (0) | |
| Vital Status (% ) | 122 | |
| Alive | 67 (55 ) | |
| Died | 33 (27) | |
| Lost to Follow-Up | 22 (18) | |
| 48 weeks response (%) | 99 | |
| Complete response | 35 (35 ) | |
| Partial response | 28 (28 ) | |
| Stable disease | 4 (4) | |
| Progressive disease | 32 (32) |
The adverse effects include grade two anemia in 19 (16%) participants and grade 3/4 anemia in 4 (3%) participants. Grade two neutropenia occurred in 19 (16%) participants, and grade 3/4 neutropenia occurred in 12 (10%) participants. One participant had grade four bleomycin-induced dermatitis. There were no detected cases of grade 3 or 4 lung toxicity, neuropathy, or thrombocytopenia.
Outcome definitions were harmonized with response category definitions (complete response, partial response (PR), stable disease, progressive disease) based on two concurring randomized phase III ACTG/AMC KS studies at this site 16, 27. The definitions below were specific to assessing the five KS marker lesions. CR was the absence of any detectable residual disease, including tumor-associated edema, persisting for at least four weeks. In patients known to have had visceral disease, an attempt at restaging with appropriate radiographic procedures should be made. PR was defined as no new oral lesions or new or progressive visceral involvement, the appearance or worsening of tumor-associated edema or effusions, or the development of five or more new cutaneous marker lesions in anatomic sites which were previously documented as having no evidence of cutaneous disease; and a 50% decrease in the sum of the products of the largest perpendicular diameters of the five cutaneous marker lesions compared to entry. PD was defined as follows: for participants with <50 cutaneous marker lesions 25% increase in the sum of the products of perpendicular diameters of the five cutaneous marker lesions compared to entry. For participants with ≥50 cutaneous marker lesions, 25% increase in the sum of the products of the perpendicular diameters of the five cutaneous marker lesions compared to entry.
The censoring date for the assessment of the vital statuses of the participants was June 30th, 2020. At censoring, 67 (55%) participants were alive, 33 (27%) participants were dead, and 22 (18%) participants were Loss-to-follow-up (LTFU). Of the 33 deaths in this set, 12 were due to PD, 12 were treatment-related, and four were due to other causes unrelated to treatment or KS. The cause of death could not be ascertained for five participants.
Among the participants with an evaluable response (n=99), 35 (35%) achieved a CR at 48 weeks based on clinical assessment, and 28 (28%) achieved a PR. Eight (8%) participants achieved stable disease (SD); 28 (28%) experienced PD. For the 69 patients who were not evaluated at exactly 48 weeks (missed appointment), the response was imputed from either the last visit that preceded the 48-week follow-up or the first visit after the 48-week follow-up, whichever was closer. Participants with baseline KPS < 70 were less likely to achieve CR at 48 weeks (p≤0.01, n = 99). No other parameters (age, gender, tumor, illness severity, edema, visceral disease, oral involvement, knew they were HIV+ before KS diagnosis, number of months HIV+ diagnosis was known, CD4 count, HIV viral load, hemoglobin, platelets, white blood cells, creatinine, or bilirubin) were significantly associated with response (Supplementary Figure 2). All participants received cART concurrent to chemotherapy according to national standards and local availability. 20 (16%) participants received dolutegravir/ lamivudine/ tenofovir (DTG/3TC/TDF). 91 (74%) participants received efavirenz/ lamivudine/ tenofovir (EFV/3TC/TDF). Eleven participants received other cART regimens. OS and PFS were estimated using the Kaplan-Meier method (Figure 1A, 1B). At one year, the OS was 61% (95%CI: 69% – 85%), and PFS was 58% (95%CI: 63% −80%), with LTFU considered a censoring event. Four variables had a statistically significant association with OS. The first variable was sex (HR = 5.19, 95%CI: 1.0 – 27); males were five times more likely to die than females; however, four times more males than females were enrolled in the study. The second variable was oral involvement (HR = 0.19, 95%CI: 0.04 – 0.96). The third variable was CD4 count (HR = 0.89, 95%CI: 0.80 – 0.99). The fourth variable was increased hemoglobin (HR = 0.79, 95%CI: 0.65 – 0.97). For each unit of hemoglobin increase (g/dL), the hazard of death decreased by 21%. No statistically significant (based on p≤0.05 after multivariate adjustments) relationships were identified between PFS hazard and any of the clinical variables. For patients with no prior cART at baseline, no statistically significant associations were made with survival outcomes due to the small sample size and, thus, lack of statistical power. Overall, almost half of the participants died within one year of diagnosis.
Figure 1:
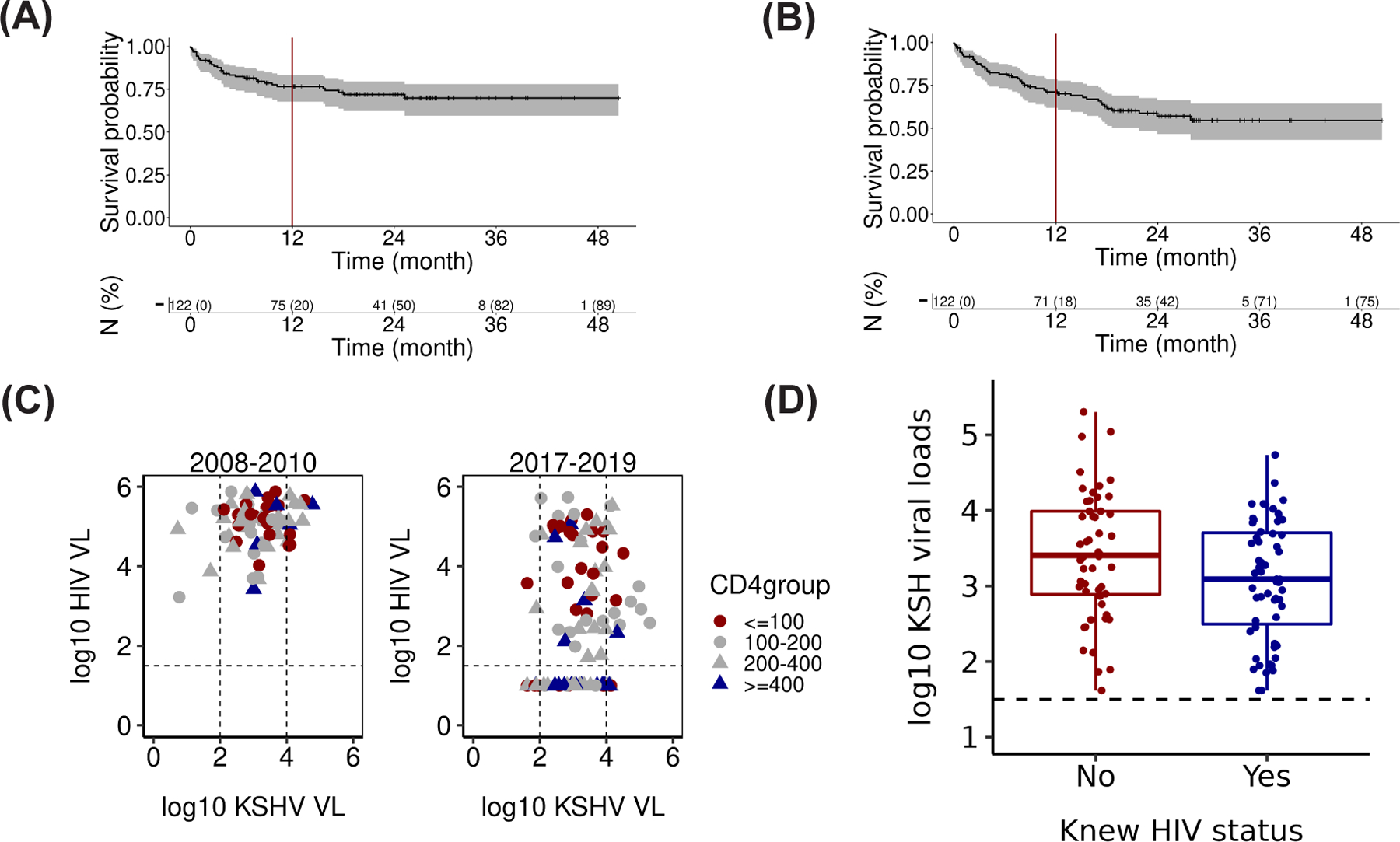
Survival curves were estimated by the Kaplan-Meier method (A) OS (n=122) and (B) PFS (n=122). There was a total of 27 events for OS and 33 events for PFS. (C) A comparison of HIV and KS viral loads and CD4 counts from a previous study (2008–2010) and this study (2017–2019). (D) Boxplot of KSHV Viral load (copies/mL, log10 scale) of subgroups defined by awareness of HIV status at least three months before KS diagnosis (variable name = “Knew HIV status,” n = 111; p ≤ 0.04).
KSHV and HIV Viral Loads
KSHV and HIV genome copy numbers (cps/mL) were determined for 111 participants at baseline (Figure 1C). We compared the HIV and KSHV genome copy numbers obtained in this enrollment period (2017–2019) to our prior study of KS patients in Malawi that were enrolled between 2008 – 2010 38. In the cART naïve 2008 – 2010 population, KSHV viral load (copies/mL, log10 scale) was 3.1 ± 0.84 (mean ± SD, n = 111). The KSHV viral load was similar in cART and chemo-naïve KS patients in 2017–2019 at 3.2 ± 0.82 (mean ± SD, n = 69). KSHV viral load was independent of CD4 count (r2 = 0.003 by the linear model, p ≤ 0.61 by F-test) in both time frames. HIV genome copy number at baseline was trimodal in the current study: 40% of KS participants had no detectable HIV in their blood (limit of detection at 50 cps/ ml), 35% had between 50 – 50,000 cps/ ml and 25% of KS participants presented with > 50,000 cps/ ml. This study thus reports a significant change to the monomodal distribution of HIV genome copies, which was 5.0 ± 0.60 (mean ± SD) log10 cps/ ml in the KS patients from 2008 – 2010. There was a trend for lower CD4 counts associated with higher HIV genome copy number (r2 = 0.18 by the linear model, p ≤ 10−5 by F-test); however, many KS patients had ≥ 400 CD4 cells/µl. In sum, a quarter of KS seen in Malawi today is no longer associated with uncontrolled HIV viral load or CD4 depletion.
Each clinical variable was tested for association with the KSHV genome copy number at baseline in a multivariate analysis (Figure 1D). First, participants who were not aware of their HIV status at least three months before their KS diagnosis had significantly higher KSHV viral loads (3.40 ± 0.83 (mean ± SD, n = 52) compared to participants who were aware of their HIV status (3.08 ± 0.78 (mean ± SD, n = 59, p ≤ 0.05 by T-test). Second, higher hemoglobin levels (g/dL) were associated with lower KSHV viral loads, but the strength of the relationship was small (Spearman correlation coefficient r = −0.2, p ≤ 0.002). This study did not identify significant associations between CR, HIV viral loads (copies/mL, log10 scale), KSHV viral loads (copies/mL, log10 scale), and CD4 counts for participants who were HIV-positive/ cART naïve for more than three months or those who had edema at baseline.
Human transcriptome sequencing identified two subtypes of KS tumors
Targeted RNAseq yielded 95 human transcriptomes from KS biopsies with matched clinical data. Sixteen samples with low sequencing depth were removed from subsequent analyses to avoid threshold sensitivity biases, resulting in 78 samples with comparable technical quality and total cellularity. The clinical characteristics of this set were not different from the overall cohort. 11,467 genes expressed in at least half the samples with a mean expression greater than ten reads per kilobase of exon per million reads mapped (RPKM) were retained for analysis.
To determine how different the transcriptome of KS biopsies was from ‘normal’ endothelial cells, we incorporated data from DiMaio et al. 39, who described circulating endothelial colony-forming cells (ECFC) of lymphatic and blood origin, and data from Sychev et al. 40, who determined the transcriptome in Telomerase-immortalized Microvascular Endothelial Cells (TIVE) cells in both KSHV infected and uninfected cells. The t-distributed stochastic neighbor embedding (tSNE) (Figure 2 A–C) projection of samples colored by the unsupervised hierarchical clustering based on the 2,000 most differentially regulated genes easily distinguish the uninfected and KSHV-infected pure endothelial cells (colored gray and red, respectively) from the KS biopsies (colored yellow or blue). The transcripts most differentially regulated between the KS lesions and the “normal” endothelial cells include, for instance, matrix metalloproteinase MMP1, an established marker of angiogenesis. Significantly downregulated genes in the KS lesions, as compared to cell lines, were DCBLD2, RELN, FTH1, BCAR1, APLN, APOLD1, and LSS. The upregulated genes in this set of KS lesions, as compared to cell lines, included S100A12, a calcium-binding protein linked to inflammation and cancer, and adiponectin (ADIPOQ), an adipokine that regulates glucose levels and the breakdown of fatty acids. Other significantly upregulated genes included GPD1, HBB, FCGR3B, SLC4A1, ALAS2, and HBD. In the absence of single-cell data, we cannot decide whether the difference in gene transcription represents differences in the KSHV-positive tumor cells to KSHV-negative and positive endothelial cells or is the result of mixed cellularity of the patient biopsies, as compared to pure cell lines.
Figure 2:
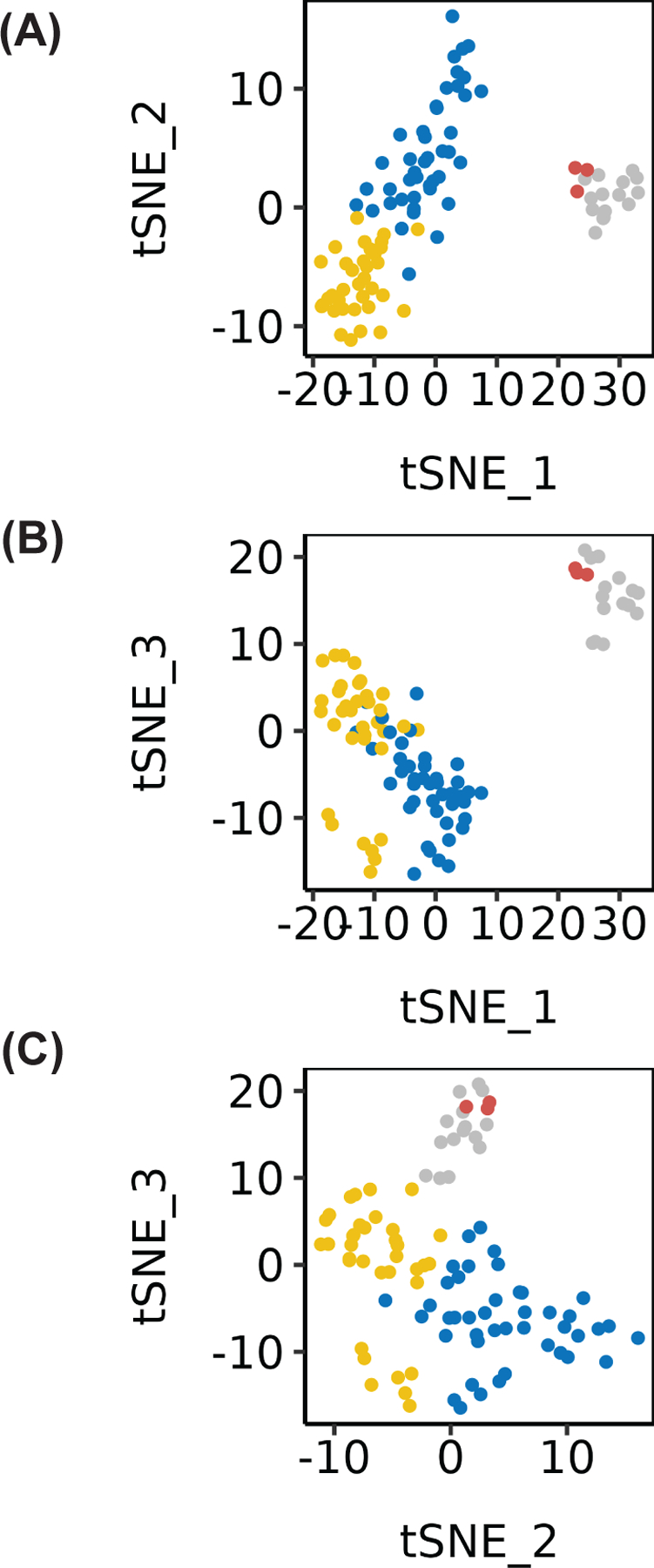
(A-C) tSNE visualizations of the 2 KS patient clusters (yellow and blue), typical uninfected endothelial cell lines (grey), and KSHV-infected TIVE cell lines (red). Both visualizations display distinct separation of normal and KSHV-infected endothelial cells vs. KS patient clusters.
Next, unsupervised hierarchical clustering was performed on the KS lesion data alone. Four potential KS tumor subtypes were identified, but due to the small sample size, we focused the analyses on the two most highly divergent subgroups of KS (Figure 3A). The first subtype included 35 samples (blue), and the second subtype 43 samples (yellow). To investigate the molecular differences between these two subtypes of KS lesions, genes were clustered using the Partitioning Around Medoids (PAM) method. Figure 3B shows a volcano plot of adjusted significance in relation to the relative mean difference in gene expression between the two clusters, blue and yellow, using the blue cluster as the reference group. The volcano plot highlights individual differentially expressed genes of greater than eight-fold mean difference between the two clusters. These were interleukin-1β (IL-1β) and CXCL5, indicative of an inflammatory microenvironment in the yellow subtype, and KIAA1199/CEMIP (cell migration inducing hyaluronidase 1). CEMIP regulates hyaluronic acid, a extracellular matrix component, and has been previously associated with KSHV infection 41. Other significantly upregulated transcripts include VAT1L, LY6H, RBP4, and PPP1R1A. ANXA8L1, Annexin A8 Like 1 protein, was the only downregulated gene in the yellow cluster and, by implication, upregulated in the blue cluster. We provisionally term the yellow cluster the “inflammatory KS subtype” and the blue cluster the “proliferative KS subtype.” Note that unsupervised clustering is driven by the sum total of all changes in gene transcription, even if anyone gene alone does not rise to individual significance in this particular experiment. No significant differences were seen between the two tumor subtypes in complete response, OS, or PFS. In sum, transcriptional profiling demonstrates substantial heterogeneity among KS lesions, with at least two major subtypes, one characterized by an inflammatory profile and the other reflecting predominantly endothelial cell components.
Figure 3:
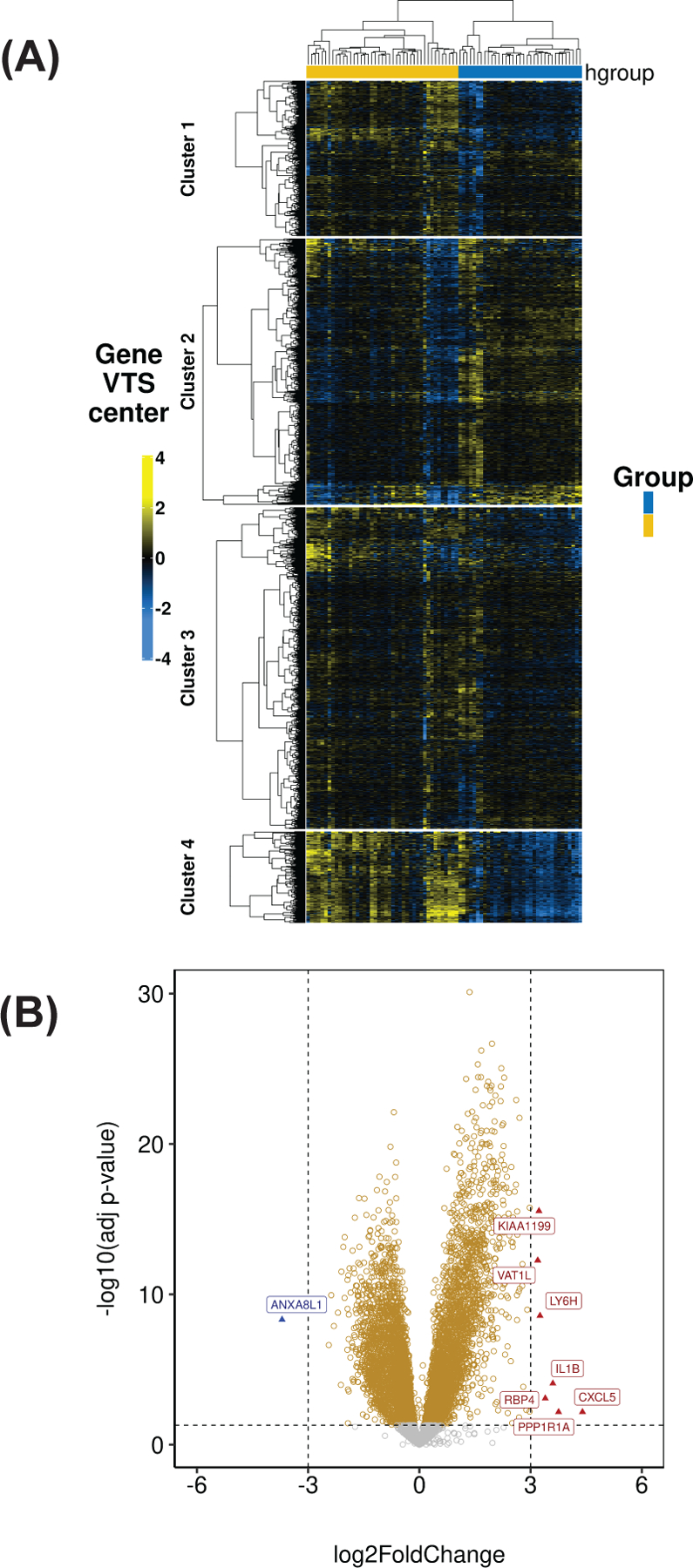
(A) A heatmap of the top 2000 most variable genes (median-centered VST transformed data from DESeq2) between the two KS subtypes. (B) Volcano plot depicting the differentially expressed genes within the two KS subtypes (blue cluster as the reference group).
Discussion
KS is a characteristic cancer for PLWH. The risk of KS is increased substantially in people living with HIV. In the US, which has low KSHV prevalence in the general population, the risk for KS in PLWH is 100-fold above that of the general population despite twenty years of cART 6, 42. The standardized incidence ratio of developing cervical cancer, considered AIDS-associated in the original CDC definition, for PLWH is in the 2-fold range, driven essentially by the large, near-universally high HPV prevalence 43. In Sub-Saharan Africa, where KSHV general population prevalence is above 50%, the estimated relative risk of developing KS in PLWH is numerically lower; however, there are many more PLWH and many more people infected with KSHV. In SSA, KS is among the five most prevalent cancers overall, irrespective of HIV status and irrespective of sex. We believe this is due to KSHV seroprevalence rates in the region being as high as 70% 44, while in the US and Europe, KSHV seroprevalence is lower and concentrated in PLWH 45.
The US standard of care, using single-agent liposomal doxorubicin (Doxil), was developed twenty years ago 15, 28, 46. Doxil is not consistently available nor affordable in many public sector hospitals in SSA. Rather BV and, more recently, paclitaxel, in combination with cART, are used 12, 24, 27. Whether pomalidomide, which was fast-tracked based on a 60% overall response rate in 28 treatment-experienced KS patients 47, will become widely affordable in SSA remains to be seen. Studies with other “imids” 48 and a second study with pomalidomide 49 support the clinical efficacy of this agent class in KS.
Here, we presented data from an observational, prospective cohort study of PLWH who present with KS aimed to define the clinical characteristics of KS patients and their responses to BV plus cART, the Malawi national standard of care. The goal was to be as inclusive as possible and to characterize a prototypical, contemporary population of PLWH who develop KS. This contrasts with prior interventional trials, which may screen out as many as half of KS patients. 122 biopsy-confirmed KS cases were enrolled between February 2017 and June 2019. All participants had to be KS treatment naïve; approximately half were on cART before enrollment (median of 15 months). This distribution sets the current study apart from earlier studies 16, 27. It represents the typical population of KS patients in recent times, i.e., after the 2016 adoption of WHO guidelines for immediate cART treatment of all HIV+ persons irrespective of CD4 count or clinical symptoms. The OS was 61% at one year, and the PFS was 58%. At 48 weeks, CR was achieved in 35% and PR in 28% (n=99). The median number of treatment cycles received was 16; however, half of the participants received sub-optimal chemotherapy due to drug shortages, which remains common in LMIC environments.
The OS and PFS were in line with prior studies. Bower et al. 15 reported a 5-year OS of 83% using cART/ liposomal Doxorubicin (Doxil) in a 140-person cohort representing a 1989 – 2013 UK population of 89% cART naïve KS patients. This is among the highest survival times reported to date in KS. None of the data from SSA approaches this result. A randomized phase III trial comparing the efficacy of BV plus cART to paclitaxel plus cART in advanced KS patients (stage T1) without prior cART obtained a one-year PFS of 44% (N = 132) and one-year OS of 80% in the BV plus cART group 27. A 2003–2009 trial by Mosam et al. 23 reported a one-year OS of 74% (n=53) using BV/ cART plus doxorubicin in T1 stage KS. Crudely approximating across studies and collection times, OS and PFS for KS in SSA have not changed in the last twenty years though admittedly, the data is minimal and often confounded by loss to follow-up.
All patients had detectable KSHV in the blood. Significantly elevated KSHV viral loads were observed in participants who did not know their HIV status at least three months before the study, reaffirming that HIV-induced immune suppression and/ or HIV directly reactivates KSHV and induces KS.
Prior studies consistently observed the existence of two HIV-KS lesion subtypes based on the KSHV gene transcription 38, 50. Limited, i.e., latent KSHV transcription was the predominant subtype; cART seemed to restrict viral transcription to the latent genes and viral miRNAs as compared to KS lesions from cART-naïve patients. This is consistent with in situ analyses, which always detect the LANA protein, but rarely other viral proteins in KS lesions 51. Here, we queried human gene transcription in KS lesions assembling the most extensive set of KS transcript data today. Differential gene expression identified two subtypes of KS lesions; one enriched in genes that induce the proliferation and migration of cells and a second enriched in inflammatory genes. Because of the targeted design for human genes only, we could not relate viral to human gene transcription.
We did not observe a difference in the survival curves or any statistically significant association with response hazard between the two KS tumor subtypes. This was expected for multiple reasons. Even though this study reports on more samples than prior studies 52–55, this is still a small data set with limited outcome data. It represents a snapshot of KS for a specific time and specific place. Second, despite multiple KS lesions on the same patient, we could biopsy only one lesion at baseline due to IRB concerns. This under-sampling likely obscured any dominant transcription pattern required to predict survival outcomes. The situation in KS contrasts with most solid tumors, where the primary tumor can be discerned and biopsied and where the primary tumor subtype predicts response. For KS, by contrast, we know that different lesions on the same patient represent different overt classes (patch, plaque, nodular), have different histological compositions 56, and respond differently to systemic treatment.
We tried to identify potential associations between clinical characteristics and gene expression within one single lesion (out of many possible) on a patient’s skin. We did not find any. This may be due to the limited power of this study. Larger studies in terms of participants and length of follow-up may uncover those. Alternatively, this may be due to the biology of KS, where single skin lesions develop independently, and it is their aggregation, as well as the lesions that develop internally in the lung and liver, as well as co-occurring MCD and PEL that drive the most robust clinical characteristics 57, 58. Unfortunately, institutional review boards frown upon extended experimental biopsies. We estimate that one would need at least as many biopsies as marker lesions to establish robust linkages between overall systemic clinical characteristics and tumor gene expression.
There are limitations to this study. Ascertaining CR for KS tumors accurately requires biopsies of all indicator lesions. That was not feasible in this and other LMIC settings. To date, most IRBs will no longer approve studies requiring extensive biopsies. We were worried that insisting on this “gold” standard may introduce an enrollment bias, as many potential participants are averse to multiple biopsies. This study aimed to capture everyone with KS symptoms. Thus, the CR numbers reported here may be overly optimistic from a strictly scientific point of view. The participants and their providers reported an improvement. Given the smoldering, remitting/recurring nature of KS in the presence of cART, further studies, including quality of live measures, are indicated to determine the most meaningful response criteria in this patient population today.
The principal limitation in calculating survival outcomes was the lack of events and LTFU after one year, which reduced the statistical power in detecting late differences in survival outcomes. Out of the 78 participants with matched high-quality human transcriptome data, only eight deaths and 27 PD could be ascertained. This was expected partly due to the liberal inclusion criteria of including “all comers” and partly due to the limited resources at the site. In addition, this study had a shorter follow-up. It was a single-center experience compared to two recent multicenter, randomized phase III clinical trials that reported lower survival for HIV-associated KS in the SSA 16, 27. Considering that this study aimed to explore survival outcomes under the local standards of care, there were limitations in drug supply, treatment interruptions, and patient retention. Under these circumstances, and including the molecular heterogeneity of KS lesions, we submit that PFS and OS are more robust clinical measures than clinical response measurements developed for US AIDS patients before the availability of cART 34.
In sum, the standard of care for HIV-associated KS in the region where KS and HIV burden is the highest globally lags behind the best possible care available in the US and Europe. This study underscores the molecular diversity of individual KS lesions and KS patients. Yet, we still lack robust biomarkers to predict individual KS risk for patients on cART or individual therapy responses.
Supplementary Material
Supplementary Figure 1:
Visual gradings of LANA stains on FFPE KS skin biopsy sections. A biopsy staining negative for KSHV LANA was considered one of the study’s exclusion criteria. Representative images of (A) high, (B) medium, and (C) low LANA counts. (D-F) Control sections for each sample. (x200 magnification). Repeat staining was conducted at UNC Chapel Hill.
Supplementary Figure 2:
Logistic regression analysis to determine the effect of clinical associations on response. (A) Participants with a higher than 70% Karnofsky performance status were less likely to achieve CR at 48 weeks (p≤0.014). (B) Merging complete and partial responses revealed that a higher square root of the CD4 count is associated with achieving complete or partial response at 48 weeks.
Supplementary Table 1:
RNA sequencing. Sequencing coverage and quality statistics. a Sequencing reads were aligned to reference GRCh37. b Total RNA sequencing was performed using Ion AmpliSeq Transcriptome Human (cat# A26225) technology. Therefore virtually no reads mapped to the rRNA region (Ratio < 1:10−5 for all) c Samples with a minimum count of 10 unique gene reads were retained for further analysis.
Novelty and Impact Statement.
Kaposi Sarcoma (KS) is the most common cancer in people living with HIV (PLWH) in many countries where Kaposi Sarcoma-associated herpesvirus is endemic. No more than a handful of complete transcriptomes for this cancer have been reported, most from early AIDS KS patients in the US and Europe. We report 78 human transcriptomes from clinically annotated biopsies collected between 2017 and 2019. Two clusters of KS lesions were observed, which can be mined for novel intervention targets. Approximately half of the cohort required cytotoxic chemotherapy in addition to anti-HIV therapy. Many KS patients had undetectable HIV viral load. Sadly, overall survival was not improved compared to prior studies, as liposomal doxorubicin was unavailable.
Acknowledgments
We thank, first and foremost, the participants without whom this study would not have been possible. This work was supported by public health service grants CA254564, CA019014, and CA23958 to DPD and BD, and DE018304 to DPD. RM is a fellow of the AIDS Malignancy Consortium (CA121947). We thank Megan Perkins for immunohistochemistry.
Abbreviations:
- BV
bleomycin/vincristine
- cART
combination antiretroviral therapy
- CEMIP
cell migration inducing hyaluronidase 1
- CR
complete response
- DTG/3TC/TDF
dolutegravir/ lamivudine/ tenofovir
- EFV/3TC/TDF
efavirenz/ lamivudine/ tenofovir
- RPKM
reads per kilobase of exon per million reads mapped
- ECFC
endothelial colony-forming cells
- HIV
Human Immunodeficiency Virus
- IL-1β
Interleukin-1β
- KCH
Kamuzu Central Hospital
- KICS
KSHV Inflammatory Cytokine Syndrome
- KS
Kaposi Sarcoma
- KSHV
Kaposi Sarcoma-associated herpesvirus
- KS-IRIS
KS-Immune Reconstitution Inflammatory Syndrome
- LMIC
low and middle-income countries
- LTFU
Loss-to-follow up
- MCD
Multicentric Castleman Disease
- OS
Overall survival
- PAM
Partitioning Around Medoids
- PD
progressive disease
- PEL
Primary Effusion Lymphoma
- PFS
progression-free survival
- PLWH
people living with HIV
- PR
Partial Response
- SD
Stable Disease
- TIVE
Telomerase-immortalized Microvascular Endothelial Cells
- tSNE
t-distributed stochastic neighbor embedding
Footnotes
Author Contributions
The work reported in the paper has been performed by the authors unless clearly specified in the text.
Razia Moorad: Investigation, Data Curation, Formal Analysis, Writing – Original Draft.
Edwards Kasonkanji: Data Curation, Formal Analysis
Joe Gumulira: Investigation
Yolanda Gondwe: Project Administration
Morgan Dewey: Investigation
Yue Pan: Data curation, Formal Analysis.
Alice Peng: Data Curation, Formal Analysis.
Linda J. Pluta: Investigation
Evaristar Kudowa: Data Curation
Richard Nyasosela: Investigation
Tamiwe Tomoka: Investigation
Hannock Tweya: Investigation
Tom Heller: Investigation
Salem Gugsa: Investigation
Sam Phiri: Conceptualization, Project Administration
Dominic T Moore: Formal Analysis
Blossom Damania: Conceptualization, Funding Acquisition, Writing – Review & Editing
Matthew Painschab: Writing – Review & Editing, Supervision
Mina C. Hosseinipour: Conceptualization, Writing – Review & Editing, Project Administration, Supervision
Dirk P. Dittmer: Conceptualization, Methodology, Formal Analysis, Project Administration, Funding Acquisition, Validation, Supervision, Writing – Review & Editing
Conflict of Interest
D.P.D. has been a consultant to Thermo Fisher Scientific. The other authors declare no conflict of interest.
Ethics Statement
The study was approved by the Malawi National Health Science and Research Committee (NHSRC) and the Institutional Review Board (IRB) at the University of North Carolina at Chapel Hill. A separate informed consent for tissue collection was obtained in specific languages modeled after previous and existing cancer studies.
Data Availability Statement
The data supporting this study’s findings are openly available in the NCBI BioProject Database (https://www.ncbi.nlm.nih.gov/bioproject), linked to the BioProject accession number PRJNA947563. Other data that support the findings of this study are available from the corresponding author upon request
References
- 1.Motlhale M, Sitas F, Bradshaw D, Chen WC, Singini MG, de Villiers CB, Lewis CM, Muchengeti M, Waterboer T, Mathew CG, Newton R, Singh E. Epidemiology of Kaposi’s sarcoma in sub-Saharan Africa. Cancer Epidemiol 2022;78: 102167. [DOI] [PubMed] [Google Scholar]
- 2.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71: 209–49. [DOI] [PubMed] [Google Scholar]
- 3.Banda JC, Muula AS. Burden of chronic disease comorbidities among cancer patients at Queen Elizabeth and Kamuzu Central Hospitals in Malawi: an exploratory cross-sectional study. Pan Afr Med J 2021;40: 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santiago JC, Goldman JD, Zhao H, Pankow AP, Okuku F, Schmitt MW, Chen LH, Hill CA, Casper C, Phipps WT, Mullins JI. Intra-host changes in Kaposi sarcoma-associated herpesvirus genomes in Ugandan adults with Kaposi sarcoma. PLoS Pathog 2021;17: e1008594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sallah N, Palser AL, Watson SJ, Labo N, Asiki G, Marshall V, Newton R, Whitby D, Kellam P, Barroso I. Genome-Wide Sequence Analysis of Kaposi Sarcoma-Associated Herpesvirus Shows Diversification Driven by Recombination. J Infect Dis 2018;218: 1700–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruffieux Y, Muchengeti M, Olago V, Dhokotera T, Bohlius J, Egger M, Rohner E. Age and cancer incidence in 5.2 million people with HIV: the South African HIV Cancer Match study. Clin Infect Dis 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maurer T, Ponte M, Leslie K. HIV-associated Kaposi’s sarcoma with a high CD4 count and a low viral load. N Engl J Med 2007;357: 1352–3. [DOI] [PubMed] [Google Scholar]
- 8.Krown SE, Lee JY, Dittmer DP, Consortium AM. More on HIV-associated Kaposi’s sarcoma. N Engl J Med 2008;358: 535–6; author reply 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cesarman E, Damania B, Krown SE, Martin J, Bower M, Whitby D. Kaposi sarcoma. Nat Rev Dis Primers 2019;5: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krown SE, Dittmer DP, Cesarman E. Pilot study of oral valganciclovir therapy in patients with classic Kaposi sarcoma. J Infect Dis 2011;203: 1082–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.IeDEA AI-dCPWGf, EuroCoord Ci. Comparison of Kaposi Sarcoma Risk in Human Immunodeficiency Virus-Positive Adults Across 5 Continents: A Multiregional Multicohort Study. Clin Infect Dis 2017;65: 1316–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burger H, Ismail Z, Taljaard JJ. Establishing a multidisciplinary AIDS-associated Kaposi’s sarcoma clinic: Patient characteristics, management and outcomes. S Afr Med J 2018;108: 1059–65. [DOI] [PubMed] [Google Scholar]
- 13.Dittmer DP, Damania B. Kaposi sarcoma-associated herpesvirus: immunobiology, oncogenesis, and therapy. J Clin Invest 2016;126: 3165–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sabourin KR, Daud I, Ogolla S, Labo N, Miley W, Lamb M, Newton R, Whitby D, Rochford R. Malaria Is Associated With Kaposi Sarcoma-Associated Herpesvirus Seroconversion in a Cohort of Western Kenyan Children. J Infect Dis 2021;224: 303–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bower M, Dalla Pria A, Coyle C, Andrews E, Tittle V, Dhoot S, Nelson M. Prospective stage-stratified approach to AIDS-related Kaposi’s sarcoma. J Clin Oncol 2014;32: 409–14. [DOI] [PubMed] [Google Scholar]
- 16.Hosseinipour MC, Kang M, Krown SE, Bukuru A, Umbleja T, Martin JN, Orem J, Godfrey C, Hoagland B, Mwelase N, Langat D, Nyirenda M, et al. As-Needed Vs Immediate Etoposide Chemotherapy in Combination With Antiretroviral Therapy for Mild-to-Moderate AIDS-Associated Kaposi Sarcoma in Resource-Limited Settings: A5264/AMC-067 Randomized Clinical Trial. Clin Infect Dis 2018;67: 251–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krown SE, Borok MZ, Campbell TB, Casper C, Dittmer DP, Hosseinipour MC, Mitsuyasu RT, Mosam A, Orem J, Phipps WT. Stage-stratified approach to AIDS-related Kaposi’s sarcoma: implications for resource-limited environments. J Clin Oncol 2014;32: 2512–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herce ME, Kalanga N, Wroe EB, Keck JW, Chingoli F, Tengatenga L, Gopal S, Phiri A, Mailosi B, Bazile J, Beste JA, Elmore SN, et al. Excellent clinical outcomes and retention in care for adults with HIV-associated Kaposi sarcoma treated with systemic chemotherapy and integrated antiretroviral therapy in rural Malawi. J Int AIDS Soc 2015;18: 19929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mtonga W, Mujajati A, Munkombwe D, Kalungia A, Muungo LT, West J, Wood C, Ngalamika O. Therapeutic Outcomes in AIDS-Associated Kaposi’s Sarcoma Patients on Antiretroviral Therapy Treated with Chemotherapy at Two Tertiary Hospitals in Lusaka, Zambia. Curr HIV Res 2018;16: 231–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rohner E, Kasaro M, Msadabwe-Chikuni SC, Stinson K, Mohamed Z, Tweya H, Egger M, Bohlius J. Treatment and outcome of AIDS-related Kaposi sarcoma in South Africa, Malawi and Zambia: an international comparison. Pan Afr Med J 2017;28: 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bekolo CE, Soumah MM, Tiemtore OW, Diallo A, Yuma JD, Di Stefano L, Metcalf C, Cisse M. Assessing the outcomes of HIV-infected persons receiving treatment for Kaposi sarcoma in Conakry-Guinea. BMC Cancer 2017;17: 806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chalya PL, Mbunda F, Rambau PF, Jaka H, Masalu N, Mirambo M, Mushi MF, Kalluvya SE. Kaposi’s sarcoma: a 10-year experience with 248 patients at a single tertiary care hospital in Tanzania. BMC Res Notes 2015;8: 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mosam A, Shaik F, Uldrick TS, Esterhuizen T, Friedland GH, Scadden DT, Aboobaker J, Coovadia HM. A randomized controlled trial of highly active antiretroviral therapy versus highly active antiretroviral therapy and chemotherapy in therapy-naive patients with HIV-associated Kaposi sarcoma in South Africa. J Acquir Immune Defic Syndr 2012;60: 150–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freeman EE, Busakhala N, Regan S, Asirwa FC, Wenger M, Seth D, Moon KC, Semeere A, Maurer T, Wools-Kaloustian K, Bassett I, Martin J. Real-world use of chemotherapy for Kaposi’s sarcoma in a large community-based HIV primary care system in Kenya. BMC Cancer 2020;20: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwartsmann G, Sprinz E, Kromfield M, Kalakun L, Sander E, Prolla G, Di Leone L, Gerhardt L, Mans DR. Clinical and pharmacokinetic study of oral etoposide in patients with AIDS-related Kaposi’s sarcoma with no prior exposure to cytotoxic therapy. J Clin Oncol 1997;15: 2118–24. [DOI] [PubMed] [Google Scholar]
- 26.Busakhala NW, Waako PJ, Strother MR, Keter AK, Kigen GK, Asirwa FC, Loehrer PJ. Randomized Phase IIA Trial of Gemcitabine Compared With Bleomycin Plus Vincristine for Treatment of Kaposi’s Sarcoma in Patients on Combination Antiretroviral Therapy in Western Kenya. J Glob Oncol 2018;4: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krown SE, Moser CB, MacPhail P, Matining RM, Godfrey C, Caruso SR, Hosseinipour MC, Samaneka W, Nyirenda M, Busakhala NW, Okuku FM, Kosgei J, et al. Treatment of advanced AIDS-associated Kaposi sarcoma in resource-limited settings: a three-arm, open-label, randomised, non-inferiority trial. Lancet 2020;395: 1195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Northfelt DW, Dezube BJ, Thommes JA, Miller BJ, Fischl MA, Friedman-Kien A, Kaplan LD, Du Mond C, Mamelok RD, Henry DH. Pegylated-liposomal doxorubicin versus doxorubicin, bleomycin, and vincristine in the treatment of AIDS-related Kaposi’s sarcoma: Results of a randomized phase III clinical trial. Journal of Clinical Oncology 1998;16: 2445–51. [DOI] [PubMed] [Google Scholar]
- 29.Goebel FD, Goldstein D, Goos M, Jablonowski H, Stewart JS. Efficacy and safety of Stealth liposomal doxorubicin in AIDS-related Kaposi’s sarcoma. The International SL-DOX Study Group. Br J Cancer 1996;73: 989–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stewart S, Jablonowski H, Goebel FD, Arasteh K, Spittle M, Rios A, Aboulafia D, Galleshaw J, Dezube BJ. Randomized comparative trial of pegylated liposomal doxorubicin versus bleomycin and vincristine in the treatment of AIDS-related Kaposi’s sarcoma. International Pegylated Liposomal Doxorubicin Study Group. J Clin Oncol 1998;16: 683–91. [DOI] [PubMed] [Google Scholar]
- 31.Welles L, Saville MW, Lietzau J, Pluda JM, Wyvill KM, Feuerstein I, Figg WD, Lush R, Odom J, Wilson WH, Fajardo MT, Humphrey RW, et al. Phase II trial with dose titration of paclitaxel for the therapy of human immunodeficiency virus-associated Kaposi’s sarcoma. J Clin Oncol 1998;16: 1112–21. [DOI] [PubMed] [Google Scholar]
- 32.Moorad R, Juarez A, Landis JT, Pluta LJ, Perkins M, Cheves A, Dittmer DP. Whole-genome sequencing of Kaposi sarcoma-associated herpesvirus (KSHV/HHV8) reveals evidence for two African lineages. Virology 2022;568: 101–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.El-Mallawany NK, Mehta PS, Kamiyango W, Villiera J, Peckham-Gregory EC, Kampani C, Krysiak R, Sanders MK, Caro-Vegas C, Eason AB, Ahmed S, Schutze GE, et al. KSHV viral load and Interleukin-6 in HIV-associated pediatric Kaposi sarcoma-Exploring the role of lytic activation in driving the unique clinical features seen in endemic regions. Int J Cancer 2019;144: 110–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krown SE, Metroka C, Wernz JC. Kaposi’s sarcoma in the acquired immune deficiency syndrome: a proposal for uniform evaluation, response, and staging criteria. AIDS Clinical Trials Group Oncology Committee. J Clin Oncol 1989;7: 1201–7. [DOI] [PubMed] [Google Scholar]
- 35.Caro-Vegas C, Sellers S, Host KM, Seltzer J, Landis J, Fischer WA 2nd, Damania B, Dittmer DP. Runaway Kaposi Sarcoma-associated herpesvirus replication correlates with systemic IL-10 levels. Virology 2020;539: 18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eason AB, Sin SH, Shah M, Yuan H, Phillips DJ, Droste M, Shamshiev A, Dittmer DP. DLX1008 (brolucizumab), a single-chain anti-VEGF-A antibody fragment with low picomolar affinity, leads to tumor involution in an in vivo model of Kaposi Sarcoma. PLoS One 2020;15: e0233116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hosseinipour MC, Sweet KM, Xiong J, Namarika D, Mwafongo A, Nyirenda M, Chiwoko L, Kamwendo D, Hoffman I, Lee J, Phiri S, Vahrson W, et al. Viral profiling identifies multiple subtypes of Kaposi’s sarcoma. MBio 2014;5: e01633–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DiMaio TA, Wentz BL, Lagunoff M. Isolation and characterization of circulating lymphatic endothelial colony forming cells. Exp Cell Res 2016;340: 159–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sychev ZE, Hu A, DiMaio TA, Gitter A, Camp ND, Noble WS, Wolf-Yadlin A, Lagunoff M. Integrated systems biology analysis of KSHV latent infection reveals viral induction and reliance on peroxisome mediated lipid metabolism. PLoS Pathog 2017;13: e1006256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dai L, Bai L, Lin Z, Qiao J, Yang L, Flemington EK, Zabaleta J, Qin Z. Transcriptomic analysis of KSHV-infected primary oral fibroblasts: The role of interferon-induced genes in the latency of oncogenic virus. Oncotarget 2016;7: 47052–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yarchoan R, Uldrick TS. HIV-Associated Cancers and Related Diseases. N Engl J Med 2018;378: 1029–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahale P, Engels EA, Coghill AE, Kahn AR, Shiels MS. Cancer Risk in Older Persons Living With Human Immunodeficiency Virus Infection in the United States. Clin Infect Dis 2018;67: 50–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dollard SC, Butler LM, Jones AM, Mermin JH, Chidzonga M, Chipato T, Shiboski CH, Brander C, Mosam A, Kiepiela P, Hladik W, Martin JN. Substantial regional differences in human herpesvirus 8 seroprevalence in sub-Saharan Africa: insights on the origin of the “Kaposi’s sarcoma belt”. International journal of cancer Journal international du cancer 2010;127: 2395–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Labo N, Miley W, Benson CA, Campbell TB, Whitby D. Epidemiology of Kaposi’s sarcoma-associated herpesvirus in HIV-1-infected US persons in the era of combination antiretroviral therapy. AIDS 2015;29: 1217–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cianfrocca M, Lee S, Von Roenn J, Tulpule A, Dezube BJ, Aboulafia DM, Ambinder RF, Lee JY, Krown SE, Sparano JA. Randomized trial of paclitaxel versus pegylated liposomal doxorubicin for advanced human immunodeficiency virus-associated Kaposi sarcoma: evidence of symptom palliation from chemotherapy. Cancer 2010;116: 3969–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Polizzotto MN, Uldrick TS, Wyvill KM, Aleman K, Peer CJ, Bevans M, Sereti I, Maldarelli F, Whitby D, Marshall V, Goncalves PH, Khetani V, et al. Pomalidomide for Symptomatic Kaposi’s Sarcoma in People With and Without HIV Infection: A Phase I/II Study. J Clin Oncol 2016;34: 4125–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reid EG, Shimabukuro KA, Moore P, Ambinder RF, Bui JD, Han S, Martinez-Maza O, Dittmer DP, Aboulafia DM, Chiao EY, Maurer T, Baiocchi RA, et al. AMC-070: Lenalidomide is Safe and Effective in HIV-associated Kaposi Sarcoma. Clin Cancer Res 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramaswami R, Polizzotto MN, Lurain K, Wyvill KM, Widell A, George J, Goncalves P, Steinberg SM, Whitby D, Uldrick TS, Yarchoan R. Safety, activity, and long-term outcomes of pomalidomide in the treatment of Kaposi sarcoma among individuals with or without HIV infection. Clin Cancer Res 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dittmer DP. Restricted Kaposi’s sarcoma (KS) herpesvirus transcription in KS lesions from patients on successful antiretroviral therapy. MBio 2011;2: e00138–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dupin N, Fisher C, Kellam P, Ariad S, Tulliez M, Franck N, van Marck E, Salmon D, Gorin I, Escande JP, Weiss RA, Alitalo K, et al. Distribution of human herpesvirus-8 latently infected cells in Kaposi’s sarcoma, multicentric Castleman’s disease, and primary effusion lymphoma. Proc Natl Acad Sci U S A 1999;96: 4546–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tso FY, Kossenkov AV, Lidenge SJ, Ngalamika O, Ngowi JR, Mwaiselage J, Wickramasinghe J, Kwon EH, West JT, Lieberman PM, Wood C. RNA-Seq of Kaposi’s sarcoma reveals alterations in glucose and lipid metabolism. PLoS Pathog 2018;14: e1006844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hong YK, Foreman K, Shin JW, Hirakawa S, Curry CL, Sage DR, Libermann T, Dezube BJ, Fingeroth JD, Detmar M. Lymphatic reprogramming of blood vascular endothelium by Kaposi sarcoma-associated herpesvirus. Nat Genet 2004;36: 683–5. [DOI] [PubMed] [Google Scholar]
- 54.Mutlu AD, Cavallin LE, Vincent L, Chiozzini C, Eroles P, Duran EM, Asgari Z, Hooper AT, La Perle KM, Hilsher C, Gao SJ, Dittmer DP, et al. In vivo-restricted and reversible malignancy induced by human herpesvirus-8 KSHV: a cell and animal model of virally induced Kaposi’s sarcoma. Cancer Cell 2007;11: 245–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang HW, Trotter MW, Lagos D, Bourboulia D, Henderson S, Makinen T, Elliman S, Flanagan AM, Alitalo K, Boshoff C. Kaposi sarcoma herpesvirus-induced cellular reprogramming contributes to the lymphatic endothelial gene expression in Kaposi sarcoma. Nat Genet 2004;36: 687–93. [DOI] [PubMed] [Google Scholar]
- 56.Radu O, Pantanowitz L. Kaposi sarcoma. Arch Pathol Lab Med 2013;137: 289–94. [DOI] [PubMed] [Google Scholar]
- 57.Tomoka T, Painschab MS, Montgomery ND, Seguin R, Mulenga M, Kaimila B, Kasonkanji E, Zuze T, Nyasosela R, Nyirenda R, Chikasema M, Tewete B, et al. A prospective description of HIV-associated multicentric Castleman disease in Malawi. Haematologica 2019;104: e215–e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dhungel BM, Montgomery ND, Painschab MS, Mulenga M, Tomoka T, Kaimila B, Zuze T, Kasonkanji E, Kampani C, Chimzimu F, Randall C, Krysiak R, et al. ‘Discovering’ primary effusion lymphoma in Malawi. AIDS 2018;32: 2264–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1:
Visual gradings of LANA stains on FFPE KS skin biopsy sections. A biopsy staining negative for KSHV LANA was considered one of the study’s exclusion criteria. Representative images of (A) high, (B) medium, and (C) low LANA counts. (D-F) Control sections for each sample. (x200 magnification). Repeat staining was conducted at UNC Chapel Hill.
Supplementary Figure 2:
Logistic regression analysis to determine the effect of clinical associations on response. (A) Participants with a higher than 70% Karnofsky performance status were less likely to achieve CR at 48 weeks (p≤0.014). (B) Merging complete and partial responses revealed that a higher square root of the CD4 count is associated with achieving complete or partial response at 48 weeks.
Supplementary Table 1:
RNA sequencing. Sequencing coverage and quality statistics. a Sequencing reads were aligned to reference GRCh37. b Total RNA sequencing was performed using Ion AmpliSeq Transcriptome Human (cat# A26225) technology. Therefore virtually no reads mapped to the rRNA region (Ratio < 1:10−5 for all) c Samples with a minimum count of 10 unique gene reads were retained for further analysis.
Data Availability Statement
The data supporting this study’s findings are openly available in the NCBI BioProject Database (https://www.ncbi.nlm.nih.gov/bioproject), linked to the BioProject accession number PRJNA947563. Other data that support the findings of this study are available from the corresponding author upon request


