Abstract
The cdc45 protein was originally identified in Saccharomyces cerevisiae and shown to be essential for initiation of eukaryotic DNA replication. Subsequent isolation and characterisation of the corresponding genes from fission yeast, Xenopus and mammals also support a replication role for the protein in these species. They further suggest that during the course of its function cdc45 interacts with a number of other replication proteins, including minichromosome maintenance proteins, the origin recognition complex and DNA polymerase α. We have cloned the gene coding for cdc45 protein from Drosophila melanogaster. We have analysed the expression pattern of the cdc45 protein throughout the cell cycle and the life cycle using a combination of indirect immunofluorescence and subcellular fractionation. Our data show that cellular localisation and developmental regulation of the protein is consistent with a role in DNA replication. DmCdc45 is predominantly expressed in proliferating cells. In addition, its subcellular location is nuclear during interphase and the protein shows association with chromatin. The chromatin-associated form of the protein shows a post-translational modification, which may be involved in control of the action of the protein. DmCdc45 shows interactions with mcm proteins, however, the interactions detected show some specificity, perhaps suggesting a preferential association with particular mcm proteins. In addition we show that a stoichiometric mcm interaction may not be obligatory for the function of cdc45 in follicle cell replication, because, unlike the mcm proteins, DmCdc45 localises to the chorion amplification foci in the follicle cells of the ovary.
INTRODUCTION
Initiation of DNA replication requires the temporally ordered interaction of a number of protein complexes at origins of replication. For higher eukaryotes the proposed pathway of protein interactions is mainly based on data obtained from Saccharomyces cerevisiae. In early G1 a pre-replicative complex (pre-RC) is found at origins of replication, components of which include: the origin recognition complex (ORC), the six member complex which specifically recognises the origin sequences (1,2); cdc6 protein (3); the minichromosome maintenance (mcm) protein family. As G1 progresses, loading of additional proteins is required to initiate DNA replication. One of these proteins is cdc45, which binds after START but before the initiation of DNA synthesis and has been proposed to be necessary for loading of DNA polymerase α.
cdc45 was originally identified in S.cerevisiae as a cold-sensitive cell cycle mutant (4). At the restrictive temperature the cdc45-1 mutant arrests in interphase with unreplicated DNA. More precise mapping of the timing of the requirement for cdc45 suggests that it is not loaded at the same time as components of the pre-RC but is loaded onto the chromatin at a later stage, prior to the initiation of DNA replication (5). cdc45 has also been shown to be needed at each replication fork individually (6). In addition to the replication defect, cdc45-1 mutants display increased recombination and chromosome loss at the semi-permissive temperature (7). The ScCdc45 gene encodes a protein with a predicted molecular weight of 74.2 kDa and a protein sequence containing no significant homology to any other protein family (5,8,9).
Genetic interactions with other replication-related proteins have been identified. Allele-specific suppression of the cdc45-1 mutant phenotype was observed with cdc46-1 (mcm5) and cdc47-1 (mcm7), but not cdc46-5 (7), and synthetic lethality was observed with orc2-1, mcm2-1 and mcm3-1 mutants (10). In addition, co-immunoprecipitation experiments have revealed interactions with ScCdc46p and ScCdc47p, although it appears that ScCdc45p is a component of only a subset of these complexes (9,11). Additional evidence for an interaction with ScCdc47p has come from two-hybrid experiments (11). ScCdc45 has also been shown to interact with other replication-related proteins. Experiments with a S.cerevisiae double mutant strain with a cold-sensitive cdc45 and a temperature-sensitive cdc7 allele show that each is dependent on the other to function (12). A cell cycle-dependent interaction between cdc45 and mcm2 has also been observed (5). In addition, loading of cdc45 onto chromatin requires functional cdc28 and is reduced in cdc6 and mcm2 mutants (5).
Proteins with homology to cdc45 have been identified from several eukaryotic species, including Tsd-2 from Ustilago maydis (13), sna41 in Schizosaccharomyces pombe (14), XCdc45 in Xenopus laevis (15), Cdc45l in mice (16) and CDC45L in humans (16,17). The behaviour of the cdc45 homologues from these species is also consistent with the protein having a role in replication. A mutant allele of the S.pombe homologue sna41 suppresses the phenotype of the MCM mutant nda4-108 (14). The human homologue, which maps to the critical region for Di George syndrome (16,17), is maximally expressed at the G1/S transition and the protein co-immunoprecipitates with Orc2p (17). Finally, in X.laevis replication extracts, loading of XCdc45 onto chromatin is controlled by cdk2 and depletion of the protein reduces DNA synthesis by 90% and prevents recruitment of DNA polymerase α onto the chromatin (14).
We have identified a sequence from Drosophila melanogaster with homology to cdc45. Using antisera raised against the recombinant protein we have examined expression of the protein during development and its cellular distribution at different points in the cell cycle and in developing egg chambers. These observations suggest that, as in other species, DmCdc45 is involved in normal cellular replication. In addition, they suggest a role for the protein in DNA amplification. We further present our analysis of the physical interactions that DmCdc45 makes with various other proteins involved in the initiation of DNA replication.
MATERIALS AND METHODS
Cloning and sequencing of DmCdc45
The translated GenBank EST database was screened with the S.cerevisiae cdc45p sequence using BLAST (18). Two ESTs were identified with nearly identical sequences (accession nos AA390476 and AA142241). A PCR primer was designed incorporating the presumed translational start site and used in conjunction with a reverse primer in the vector. The full coding region and 3′-untranslated region from a D.melanogaster ovarian cDNA library (a gift of R. Brent) was amplified using the Expand long template PCR system (Boehringer-Mannheim). The amplified product was cloned into pTT (Genpak). Sequencing of amplified products and cloned DNA was carried out at the Advanced Biotechnology Centre (Charing Cross and Westminster Medical School).
Screening FlyBase database with the 5′-untranslated region (5′-UTR) of the DmCdc45 cDNA identified a cosmid clone containing the 5′-end of the DmCdc45 gene. Cosmid DNA was subjected to restriction enzyme digestion and the agarose gel was blotted onto Hybond XL (Amersham) and hybridized in Church buffer (19) at 50°C with a 32P-labelled oligonucleotide containing part of the 5′-UTR sequence. The hybridising fragment was subcloned into pUC18 and sequenced as described above.
Antibodies
A 6×His–DmCdc45 fusion protein was created by cloning the cDNA into pQE30 and expressing the protein in Escherichia coli strain M15. The overexpressed 6×His-labelled protein was purified on Ni+–NTA resin under denaturing conditions using buffers recommended by the manufacturer (Qiagen). Two rabbits were inoculated and antisera were produced by Neosystem SA (Strasbourg). The resulting sera were characterised by western blotting against the purified 6×His-tagged DmCdc45 and extracts from 0–5 h D.melanogaster embryos. Attempts at affinity purification of the antibody were unsuccessful, however the specificity of the antibody was confirmed by depleting the DmCdc45 antibodies with purified bacterially expressed protein bound to nitrocellulose strips (data not shown).
Antisera for DmMcm2, 4 and 5 and for DmOrc2 and 5 were obtained by similar means against bacterially expressed 6×His-tagged partial proteins and affinity purified by standard protocols.
Polyclonal antibodies against DmMcm3 and 6 were kind gifts of M. Yamaguchi and those against D.melanogaster polymerase α were kindly provided by I. R. Lehman.
Extract preparation
Embryos were harvested and dechorionated as described (20) in 3% sodium hypochlorite. Whole cell extracts were prepared by homogenising embryos, larvae, pupae or adult flies in an equal volume of 2× Laemmli buffer, boiling and homogenising further. Embryonic extracts were further diluted in Laemmli buffer before loading on an acrylamide gel. Equivalent volumes of each fraction were separated by SDS–PAGE, blotted and probed with DmCdc45 antiserum and anti-tubulin monoclonal antibody as a control for equal loading.
Subcellular fractionation
Embryos (0–4 h) were homogenised in 3:1 (w/v) nuclear preparation buffer (15 mM Tris–HCl, pH 7.5, 15 mM NaCl, 60 mM KCl, 2 mM EDTA and 0.1% β-mercaptoethanol with protease inhibitors), passed through two layers of Miracloth and centrifuged at 11 000 g for 20 min. The supernatant (cytoplasmic fraction) was kept and the pellet (nuclear fraction) washed in nuclear preparation buffer. Nuclei were lysed in the same buffer containing 0.5% Triton X-100, centrifuged and the supernatant (nucleoplasmic fraction) kept. The pellet was sequentially extracted in nuclear preparation buffer with 0.5% Triton X-100 and 250 or 500 mM or 2 M NaCl, with extensive washing between each step. Any remaining chromatin-bound proteins were dissolved in 2× Laemmli buffer. Equivalent volumes of each fraction were separated by SDS–PAGE, blotted and probed with DmCdc45 antiserum.
Western blotting
Extracts were separated on 10% SDS–polyacrylamide gels and blotted onto Hybond ECL (Amersham). Membranes were blocked with 0.5% BSA in PBST (phosphate-buffered saline plus 0.1% Tween-20) for 1 h to overnight, incubated in primary antibody diluted in PBST for 1 h and washed four times in PBST. Horseradish peroxidase (HRP)-labelled goat anti-rabbit or goat anti-mouse IgG (Pierce) was diluted 1/100 000 or 1/50 000, respectively, and added to the membranes for 1 h. For immunoblots of immunoprecipitations, HRP-conjugated protein A (1/20 000) was substituted for the secondary antibody. Following further washing, bound antibody was detected using Super Signal chemiluminescence (Pierce) and X-Omat autoradiographic film (Kodak).
Immunoprecipitations
Total cell extracts were made by homogenising dechorionated 0–5 h embryos in phosphate-buffered saline (PBS) plus 0.1% Triton X-100 (1:1 w/v), passing the homogenate through two layers of Miracloth and pelleting the debris by centrifugation at 11 000 g. Antisera were incubated with protein A–Sepharose beads for 1 h, crosslinked with DMP in 0.2 M sodium borate, pH 9, washed with 0.1 M Tris–HCl, pH 8, for 1 h and equilibrated in PBS. The beads were incubated with 1.5 ml of extract for 1–2 h and washed extensively in PBS containing 0.1% Triton X-100. Bound proteins were eluted by boiling in 2× Laemmli buffer.
Immunostaining
Embryos were harvested, dechorionated in hypochlorite, rinsed in PBS containing 0.1% Triton X-100 and then fixed and stained essentially as previously (20), except that fluorescence detection was used. Prior to hybridisation, embryos were blocked in PBS, 0.1% Triton X-100 and 2% BSA for 1 h. Primary antibody (anti-DmCdc45 IgG) was added at a concentration of 1/500 in PBS, 0.1% Triton X-100 and 2% BSA and incubated at 4°C overnight. After washing in PBS plus 0.1% Triton X-100, secondary antibody (FITC-conjugated goat anti-rabbit; Vector Laboratories) was added at a concentration of 1/500 in PBST for 1 h and the embryos washed in PBST.
Embryos were counterstained with monoclonal anti-histones (1/1000; Chemicon) and monoclonal horse anti-mouse–Texas red (1/500; Vector Laboratories) following the same procedure.
Ovaries were fixed in 8% formaldehyde, extracted in PBS plus 1% Triton X-100 for 2 h and blocked as previously described (21). They were then hybridized with anti-DmCdc45 IgG or affinity-purified anti-DmOrc5 as described above and counterstained with monoclonal anti-tubulin (1/1000; Sigma)
RESULTS
A Drosophila cDNA with homology to cdc45
Searching the GenBank EST database revealed two partial Drosophila sequences with homology to human cdc45. These were used to design specific primers to obtain the 5′-end of the coding region of the Drosophila cdc45 cDNA. The full-length coding region and 3′-UTR were amplified from a Drosophila ovary cDNA library using a specific forward primer and a reverse primer located in the vector.
Sequence analysis of the PCR-amplified cDNA revealed an open reading frame encoding a peptide of 575 amino acids with a predicted molecular weight of 65.5 kDa. Comparisons of the translated sequence to other cdc45-related sequences are shown in Figure 1. The greatest similarities are with the predicted cdc45 sequences from human (39.5%), mouse (38.3%) and Xenopus (39%). The Drosophila protein is of a similar size to cdc45 in these species, in contrast to S.cerevisiae, where it is 74.5 kDa. Regions of strong conservation are apparent along the length of the protein, most notably between amino acids 18 and 58 and 212 and 248, as well as the C-terminal region (Fig. 1). Two features present in cdc45 from other species are also conserved in DmCdc45. Between amino acids 135 and 163 is an acidic domain with a poorly conserved primary sequence (unbroken line in Fig. 1). Following this region is a mostly basic domain, with similarity to a bipartite nuclear localisation signal (broken line in Fig. 1).
Figure 1.
Clustal X alignment of Drosophila (Dm), human (Hs), Xenopus (Xl) and yeast (Sc) cdc45 amino acid sequences. Black shading indicates identity between three or more species; grey shading indicates similarity. The acidic patch is indicated by an unbroken line above the sequence and the putative nuclear localisation sequence is indicated by a broken line.
To investigate possible cell cycle regulation of DmCdc45 transcription, the region immediately upstream of the coding region was analysed for the presence of binding sites for cell cycle-related transcription factors. The FlyBase Drosophila sequence database was screened with the 5′-end of the cDNA sequence. A single cosmid (ID no. 69D5) was identified and obtained from the HGMP Resource Centre. A 1.1 kb HindIII fragment containing the presumed 5′-UTR was identified within this cosmid by Southern blotting. A single recognition site for the transcription factor E2F was found, suggesting possible cell cycle regulation of transcription. However, no sites for the Drosophila cell cycle-related transcription factor DREF (22) were identified within 1 kb of the start of the coding region.
The Drosophila cdc45 gene localises to the X chromosome
The gene encoding DmCdc45 was localised to a single location on the X chromosome, location 1E–F, by in situ hybridisation of the cloned cDNA PCR product to polytene chromosomes (data not shown), confirming that DmCdc45 is a single copy gene.
DmCdc45 is highly expressed in the early embryo
DNA replication occurs very rapidly and from frequently spaced origins in early Drosophila embryos (23). Therefore, replication-related proteins would be expected to be present in relatively large amounts at this stage, due to maternal input of protein or RNA. In order to test this, polyclonal antisera were raised against a cdc45–hexahistidine fusion protein. Antiserum from one rabbit detected both the overexpressed protein and a single band of ∼65.5 kDa in Drosophila embryo extracts (data not shown).
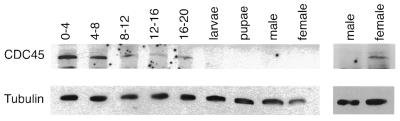
Using this antibody, expression of DmCdc45 protein was examined on immunoblots of extracts from embryos, larvae, pupae and adult flies (Fig. 2, upper), with anti-tubulin as a control for equal loading (Fig. 2, lower). Highest levels of DmCdc45 were detected in 0–4 h embryos, when DNA replication is occurring rapidly, and then decreased as the embryos aged. The protein was still detectable in embryos up to 16–20 h old, but was below detectable levels in mixed stage larvae, in pupae and in adult males. Many replication proteins also show marked increases in adult females due to the production of proteins that are maternally input into the embryo. DmCdc45 levels were increased in females, however, the relative increase was lower for this protein than for other replication proteins since it could only be detected on overloaded gels or after long exposure (Fig. 2, right).
Figure 2.
Developmental profile of DmCdc45 expression. Immunoblots of extracts from 0–4, 4–8, 8–12, 12–16 and 16–20 h embryos, mixed stage larvae, pupae and adult male and female flies were probed with anti-DmCdc45 serum (upper). A longer exposure of male and female samples is shown to the right. A monoclonal anti-tubulin antibody was used to check for equal loading of protein samples.
DmCdc45 shows physical interactions with a subset of DmMcm proteins
Genetic and physical interactions between cdc45 and mcm proteins, orc proteins and DNA polymerase have variously been reported in yeast, Xenopus and humans. To look for similar interactions in Drosophila extracts, anti-DmCdc45 and anti-DmMcm5 immunoprecipitations were performed and immunoblots hybridized with antibodies to mcm2–mcm6, orc2, orc5 and DNA polymerase α.
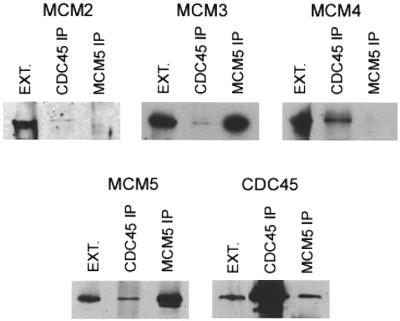
From the analysis of DmCdc45 precipitates (Fig. 3) DmMcm4 and 5 show quite strong interactions with DmCdc45; we estimate that ∼5% of the total cellular DmMcm4 and 5 is co-precipitated with DmCdc45. DmMcm2 and 3 are also detected in immunoprecipitates but at levels that are ∼10-fold less than those of DmMcm4 and 5. DmMcm6, orc2, orc5 and polymerase α were not detected in the immunoprecipitates.
Figure 3.
Immunoprecipitation with anti-DmCdc45 and anti-DmMcm5. Extracts were prepared from 0–5 h embryos and immunoprecipitations carried out using anti-DmCdc45 and anti-DmMcm5 crosslinked to protein A–Sepharose beads. Immunoblots probed with anti-DmCdc45, anti-DmMcm2, anti-DmMcm3, anti-DmMcm4 and anti-DmMcm5 sera are shown. The first lane in each panel contains an aliquot of the extract used.
DmMcm5 immunoprecipitations also suggested an interaction between this protein and DmCdc45, since ∼5% of total DmCdc45 present in the cell consistently co-precipitated with DmMcm5. The only other protein of those tested which was detected in the immunoprecipitates was DmMcm3, which apparently was present in near stoichiometric amounts to DmMcm5 (data not shown).
DmCdc45 is found in the nucleus
Subcellular fractionation of extracts from 0–4 h old embryos indicated that DmCdc45 is both nuclear and cytoplasmic at this stage (Fig. 4). Approximately half of all the DmCdc45 was present in the cytoplasmic fraction (Fig. 4). This could represent localisation of DmCdc45 into that subcellular compartment but may also be due to the remnants of maternal input of this protein. Half of the remaining DmCdc45 appeared to be nucleoplasmic, as it could be removed by lysing the nuclei with 0.5% Triton X-100 (Fig. 4). Nearly all of the remaining chromatin-bound DmCdc45 could be washed off by the addition of 250 mM NaCl (lane 3) to the Triton X-100-washed chromatin pellet.
Figure 4.
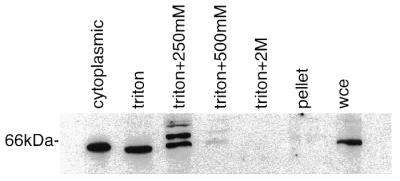
Subcellular distribution of DmCdc45 in 0–4 h embryos. Samples were separated into cytoplasmic and nuclear fractions. The nuclear fraction was subjected to successive washes in Triton X-100 plus increasing concentrations of NaCl (250 and 500 mM and 2 M) and the final pellet was solubilised in Laemmli buffer. The pattern of anti-DmCdc45 staining in unfractionated extracts is shown in the lane marked wce.
Three bands were detected in the 250 mM NaCl wash, the 65.5 kDa band, a band of equal intensity migrating ∼4 kDa higher and a much weaker band at 74 kDa. Binding to the upper bands could be competed by preincubating the antibody with overexpressed DmCdc45, suggesting that they may be related to DmCdc45. The relationship of these slower migrating bands to cdc45 is not yet clear, however, they were not removed by treatment of the fraction with calf intestinal alkaline phosphatase or λ protein phosphatase, although both of these were able to remove the phosphate groups from cdc2 (data not shown). In addition, searching of EST databases failed to reveal alternatively spliced forms of the protein similar to those seen with mammals (24).
The subcellular distribution of DmCdc45 changes during the cell cycle in Drosophila embryos
In S.cerevisiae cdc45 is nuclear throughout the cell cycle. We tested the cellular location of DmCdc45 at different stages of the Drosophila cell cycle by immunostaining fixed embryos with anti-DmCdc45 IgG and counterstaining for histones.
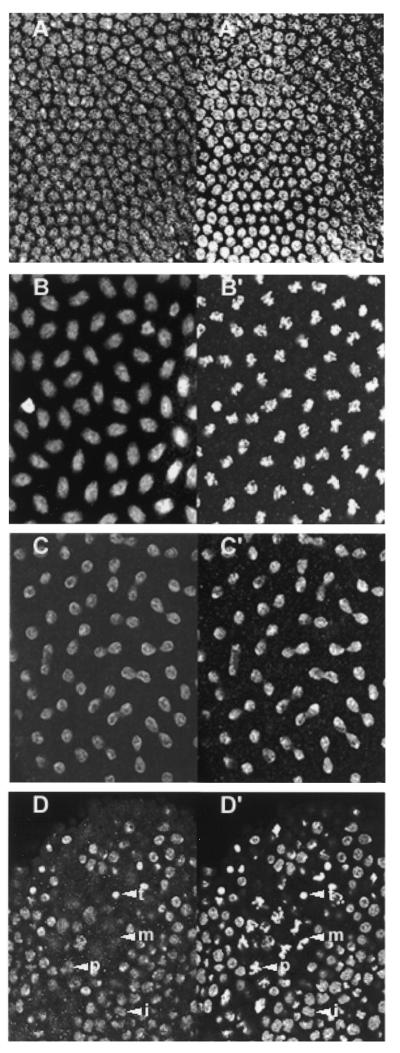
The cellular distribution of DmCdc45 changed throughout the cell cycle in the same way in both early and late embryos. In interphase cells of early embryos DmCdc45 was nuclear and staining was coincident with histone staining, suggesting that much of the protein was chromatin associated (Fig. 5A and A′). In late prophase DmCdc45 was present in the nucleus but did not appear to be associated with chromosomes (Fig. 5B and B′). DmCdc45 staining disappeared in metaphase and remained absent until late anaphase or telophase, when DmCdc45 was again seen in the nucleus, co-localising with the histones (Fig. 5C and C′). A similar pattern was observed in later embryos, in which cells were dividing asynchronously (Fig. 5D and D′).
Figure 5.
In situ localisation of DmCdc45 in embryonic cells. (A–D) Staining with anti-DmCdc45 IgG and FITC-labelled anti-rabbit IgG. (A′–D′) Staining with anti-histone monoclonal antibody and Texas red-conjugated anti-mouse IgG. (A–C and A′–C′) Synchronously replicating cells of early embryos at interphase (A and A′), late prophase (B and B′) and late anaphase/telophase (C and C′). (D and D′) Asynchronously dividing cells in a later stage embryo. Cells in interphase (i), prophase (p), metaphase (m) and telophase (t) are indicated.
Incubation of fixed embryos with IgG depleted of anti-cdc45 antibodies by preincubation with immobilised bacterially expressed protein produced no signal (data not shown).
The pattern of DmCdc45 staining in follicle cells is stage specific and consistent with an amplification role
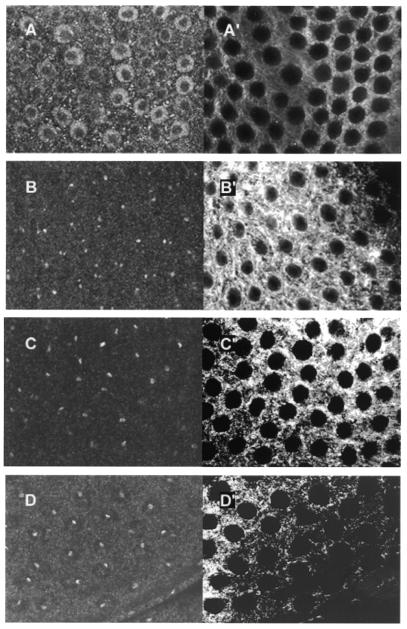
It has been previously reported that the distribution of DmOrc1 and DmOrc2 in follicle cells changes during follicle cell development and its location partially coincides with foci of amplification (21,25). The formation of orc staining foci occurs at stage 10A, prior to the detection of BrdU incorporation at sites of amplification. To determine if DmCdc45 behaves in a similar fashion, the cellular location of DmCdc45 in stage 9 and 10 follicle cells was examined. We observed a pattern of staining similar to that previously observed for DmOrc1 and DmOrc2. At stage 9 DmCdc45 staining was observed as a ring of uniform staining within the nucleus of all follicle cells (Fig. 6A and A′). In stage 10A follicle cells staining was confined to between one and three foci, with the majority of cells having two major foci (Fig. 6B and B′). By stage 10B a single, brighter focus was observed in nearly all cells, often in conjunction with a second, much weaker staining area (Fig. 6C and C′). This is similar to the pattern seen for DmOrc5 (Fig. 6D) and suggests that cdc45 is also localised to amplification foci in ovaries.
Figure 6.
In situ localisation of DmCdc45 and DmOrc5 in developing follicle cells. Left hand panels are stained with anti-DmCdc45 IgG (A–C) or anti-DmOrc5 (D) and FITC-labelled anti-rabbit IgG. Right hand panels (A′–D′) are stained with anti-tubulin monoclonal IgG and Texas red-conjugated anti-mouse IgG. DmCdc45 staining is shown at stages 9 (A), 10A (B) and 10B (C). For comparison, DmOrc5 staining is shown at stage 10B (D).
DISCUSSION
We have identified an ORF from D.melanogaster with homology to the DNA replication protein cdc45. Analysis of the primary structure of the protein showed that it contains the two domains that are conserved in all known cdc45 homologues, confirming the validity of its assignment as the Drosophila cdc45 homologue. Although the exact function of these regions is not known, mutation analysis of the corresponding region in S.cerevisiae has suggested that it contains a nuclear localisation signal (9). Of the cdc45 homologues that are known, the Drosophila protein most closely resembles vertebrate cdc45, being of a similar length and having ∼40% similarity. The presence in the 5′ upstream region of a putative binding site for the cell cycle-regulated transcription factor E2F also suggests that DmCdc45, like the vertebrate homologues, may be subject to cell cycle regulation of transcription.
Another feature of cdc45 homologues identified in other species is that they show physical and genetic interactions with other replication proteins. Human CDC45L can be co-immunoprecipitated with orc2L (17) and an interaction between DmCdc45 and DNA polymerase α has been observed in Xenopus replication extracts (14). We have not been able to detect an interaction of DmCdc45 with either of these proteins. Interestingly, the interaction with DNA polymerase α is also not observed in immunoprecipitations in yeast extracts (26). At this stage we are not sure whether these observations represent differences between species or simply reflect an intrinsic instability of cdc45 complexes in soluble extracts.
The interactions occurring with the mcm family of proteins do, however, seem to be conserved between species. In S.cerevisiae genetic interactions between ScCdc45 and mcm3, 5 and 7 have been observed (7,8). In addition, ScCdc45 has been identified as a component of mcm complexes along with ScMcm5 and 7 (9,11). We have observed that the Drosophila cdc45 homologue also interacts with mcm proteins. However, some variations are seen in the degree of interaction detected with individual family members. This might reflect a hierarchy of interaction with different mcms or subcomplexes thereof. Alternatively, this could again be due to the instability of soluble DmCdc45 complexes. Instability of complexes would also explain why we do not see more than 5% of each protein involved in interactions. However, similar results would also be obtained if frequent, stable interactions between cdc45 and the mcm proteins only occur when the proteins are bound to chromatin.
The interaction with the mcm proteins suggests that DmCdc45 is involved in replication of the cellular DNA. Further evidence that this is the case comes from our examination of the distribution of DmCdc45 in different stages of the cell cycle. DmCdc45 appears to enter the nucleus during anaphase/telophase and co-localise with chromatin during interphase, dissociating again prior to mitosis. Similar cell cycle-dependent distribution patterns have been observed for other Drosophila proteins known to be involved in DNA replication, such as DmMcm2 (27) and proliferating cell nuclear antigen (PCNA) (28). This pattern is also similar to that seen for human cdc45 (17) and is fully consistent with a role for cdc45 in DNA replication. In S.cerevisiae cdc45p apparently remains in the nucleus throughout the cell cycle (11), however, data obtained from chromatin fractionation experiments (5) suggest that cdc45 only associates with chromatin in interphase, which is again consistent with the changes that we observed in cellular localisation. Our immunofluorescence data are also consistent with data that we have obtained on the location of DmCdc45 using subcellular fractionation. These studies indicate that a proportion of the DmCdc45 protein is chromatin bound. Interestingly they also suggest that the chromatin-bound fraction might be modified in some way. We have not found evidence for an alternatively spliced form of the protein. In addition, the modification cannot be removed by two different types of phosphatase. Therefore, the nature of the modification remains unclear, however, the observation that it only occurs in chromatin-bound cdc45 suggests that the modification may be important in controlling the action of the protein.
Perhaps even more suggestive of a role for cdc45 in replication is its behaviour in the follicle cells of the ovary during oogenesis. During Drosophila oogenesis, ovarian follicle cells undergo cycles of endoreduplication leading to polyploidy. Following these cycles, clusters of chorion genes encoding eggshell proteins are further replicated, becoming amplified up to 80-fold (29). It has been previously shown that some proteins required for normal DNA replication are also required for chorion gene amplification. Mutations in orc2 and the transcription factor heterodimer E2F/DP affect chorion gene amplification (21,30), as does overexpression or inhibition of cyclin E (31). In addition, DmOrc1 and 2 form foci in 10A and 10B follicle cells prior to and during the period when chorion gene amplification occurs. Foci of orc staining correspond to sites of BrdU incorporation during amplification (21,25). We observed similar foci of DmCdc45 staining in follicle cells. Like the DmOrc2 foci, DmCdc45 foci initially appear at stage 10A and in stage 10B they become further defined, thus suggesting a role for DmCdc45 in the DNA replication required for amplification. This is intriguing given that similar patterns are not observed with DmMcm2 (21), with which cdc45 interacts and which is required for efficient loading of cdc45 onto chromatin in yeast (10). This suggests a role of the cdc45 protein in follicle cell replication which may be independent of a stoichiometric interaction with the mcm proteins.
Acknowledgments
ACKNOWLEDGEMENTS
Thanks are due to Stephen Kearsey, Gilles Crevel, Baz Smith and Celia Donaghue for helpful discussions and for critical reading of the manuscript. This work was supported by the Marie Curie Memorial Foundation.
REFERENCES
- 1.Bell S.P., Kobayashi,R. and Stillman,B. (1993) Science, 262, 1844–1849. [DOI] [PubMed] [Google Scholar]
- 2.Diffley J.F., Cocker,J.H., Dowell,S.J., Harwood,J. and Rowley,A. (1995) J. Cell Sci., 19 (suppl.), 67–72. [DOI] [PubMed] [Google Scholar]
- 3.Liang C., Weinreich,M. and Stillman,B. (1995) Cell, 81, 667–676. [DOI] [PubMed] [Google Scholar]
- 4.Moir D., Stewart,S.E., Osmond,B.C. and Botstein,D. (1982) Genetics, 100, 547–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zou L. and Stillman,B. (1998) Science, 280, 593–596. [DOI] [PubMed] [Google Scholar]
- 6.Tercero J.A., Labib,K. and Diffley,J.F.X. (2000) EMBO J., 19, 2082–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hennessy K.M., Lee,A., Chen,E. and Botstein,D. (1991) Genes Dev., 5, 958–969. [DOI] [PubMed] [Google Scholar]
- 8.Hardy C.F. (1997) Gene, 187, 239–246. [DOI] [PubMed] [Google Scholar]
- 9.Hopwood B. and Dalton,S. (1996) Proc. Natl Acad. Sci. USA, 93, 12309–12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou L., Mitchell,J. and Stillman,B. (1997) Mol. Cell. Biol., 17, 553–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalton S. and Hopwood,B. (1997) Mol. Cell. Biol., 17, 5867–5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Owens J.C., Detweiler,C.S. and Li,J.J. (1997) Proc. Natl Acad. Sci. USA, 94, 12521–12526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Onel K. and Holloman,W.K. (1997) Mol. Gen. Genet., 253, 463–468. [DOI] [PubMed] [Google Scholar]
- 14.Miyake S. and Yamashita,S. (1998) Genes Cells, 3, 157–166. [DOI] [PubMed] [Google Scholar]
- 15.Mimura S. and Takisawa,H. (1998) EMBO J., 17, 5699–5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaikh T.H., Gottlieb,S., Sellinger,B., Chen,F., Roe,B.A., Oakey,R.J., Emanuel,B.S. and Budarf,M.L. (1999) Mamm. Genome, 10, 322–326. [DOI] [PubMed] [Google Scholar]
- 17.Saha P., Thome,K.C., Yamaguchi,R., Hou,Z., Weremowicz,S. and Dutta,A. (1998) J. Biol. Chem., 273, 18205–18209. [DOI] [PubMed] [Google Scholar]
- 18.Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 20.Ashburner M. (1989) Drosophila: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Royzman I., Austin,R.J., Bosco,G., Bell,S.P. and Orr-Weaver,T.L. (1999) Genes Dev., 13, 827–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirose F., Yamaguchi,M., Handa,H., Inomata,Y. and Matsukage,A. (1993) J. Biol. Chem., 268, 2092–2099. [PubMed] [Google Scholar]
- 23.Blumenthal A.B., Kriegstein,H.J. and Hogness,D.S. (1974) Cold Spring Harbor Symp. Quant. Biol., 38, 205–223. [DOI] [PubMed] [Google Scholar]
- 24.Kukimoto I., Igaki,H. and Kanda,T. (1999) Eur. J. Biochem., 265, 936–943. [DOI] [PubMed] [Google Scholar]
- 25.Asano M. and Wharton,R.P. (1999) EMBO J., 18, 2435–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aparicio O.M., Stout,A.M. and Bell,S.P. (1999) Proc. Natl Acad. Sci. USA, 96, 9130–9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su T.T. and O’Farrell,P.H. (1997) J. Cell Biol., 139, 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamaguchi M., Hayashi,Y., Hirose,F., Matsuoka,S., Moriuchi,T., Shiroishi,T., Moriwaki,K. and Matsukage,A. (1991) Nucleic Acids Res., 19, 2403–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calvi B.R., Lilly,M.A. and Spradling,A.C. (1998) Genes Dev., 12, 734–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landis G., Kelley,R., Spradling,A.C. and Tower,J. (1997) Proc. Natl Acad. Sci. USA, 94, 3888–3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calvi B.R. and Spradling,A.C. (1999) Methods Companion Methods Enzymol., 18, 407–417. [Google Scholar]