Summary
RNA sequencing (RNA-seq) has recently been used in translational research settings to facilitate diagnoses of Mendelian disorders. A significant obstacle for clinical laboratories in adopting RNA-seq is the low or absent expression of a significant number of disease-associated genes/transcripts in clinically accessible samples. As this is especially problematic in neurological diseases, we developed a clinical diagnostic approach that enhanced the detection and evaluation of tissue-specific genes/transcripts through fibroblast-to-neuron cell transdifferentiation. The approach is designed specifically to suit clinical implementation, emphasizing simplicity, cost effectiveness, turnaround time, and reproducibility. For clinical validation, we generated induced neurons (iNeurons) from 71 individuals with primary neurological phenotypes recruited to the Undiagnosed Diseases Network. The overall diagnostic yield was 25.4%. Over a quarter of the diagnostic findings benefited from transdifferentiation and could not be achieved by fibroblast RNA-seq alone. This iNeuron transcriptomic approach can be effectively integrated into diagnostic whole-transcriptome evaluation of individuals with genetic disorders.
Keywords: RNA sequencing, RNA-seq, transcriptome, transdifferentiation, induced neuron, genetic diagnosis, neurological disorder, clinically accessible tissue, fibroblast, isoform
Our RNA-seq analysis workflow uses transdifferentiated fibroblasts to enhance the genetic diagnosis of neurological disorders. It identifies neuron-specific aberrant transcriptional events, resulting in diagnoses in 25% of cases. This demonstrates that transdifferentiation of clinically accessible tissues is a feasible approach to improve the clinical utilization of diagnostic whole transcriptome analysis.
Introduction
A prompt and accurate molecular diagnosis is critical for appropriately managing individuals with presumed Mendelian disorders. Despite advancements in molecular diagnostics through clinical exome sequencing (ES) and whole genome sequencing (WGS), more than half of these sequencing evaluations fail to yield definitive diagnoses.1,2,3,4,5,6,7,8,9,10 Various in silico prediction tools have been developed to assess the impact of a variant on gene expression and splicing to address this issue.11,12,13,14 However, without functional validation, many noncoding and splice region variants remain of uncertain clinical significance.15,16,17
Complementary to DNA sequencing, RNA sequencing (RNA-seq) has recently been utilized to detect abnormalities in the transcriptome, such as aberrant expression, splicing, and mono-allelic expression (MAE), resulting in an increase of molecular diagnostic yield by about 7.5%–36%.18,19,20,21,22,23,24,25,26,27 A significant obstacle in implementing RNA-seq for clinical diagnosis is the tissue specificity for gene expression. Adequate expression of gene(s) of interest in the study specimen is essential for any diagnostic pipeline. However, obtaining sufficient expression of tissue-specific genes in clinically accessible tissues (CATs), namely blood, fibroblasts, or muscle, can be difficult.20,28 For instance, in many Mendelian disorders characterized with intellectual and developmental disability, the underlying genetic defect may disrupt a disease-associated gene that is specifically expressed in the brain, and much less so if at all in non-neuronal cells.28 Furthermore, even when a gene has an abundant overall expression, its isoforms may be differentially detected depending on the tissue type due to tissue-specific alternative splicing, alternative cleavage, and polyadenylation events.29,30 A comprehensive analysis of RNA splicing events across different tissues revealed that approximately 40% of genes undergoing splicing were underrepresented in at least one CAT.28 Therefore, it is essential to address the limitation of tissue-specific transcript expression to improve the clinical implementation for diagnostic RNA-seq.
Transdifferentiation (direct reprogramming) of CATs into disease-relevant cell types holds promise for overcoming these obstacles. Compared to induced pluripotent stem cell (iPSC) reprogramming followed by differentiation, transdifferentiation provides a faster, more cost-effective, and potentially more genomically stable process to obtain the desired tissue types without differentiating through an intermediary pluripotency state.31,32 These attributes make transdifferentiation an attractive strategy for clinical diagnostic workflows that routinely process large volumes of samples. Human skin-derived fibroblasts can be transdifferentiated into neurons33 (iNeurons). This direct reprogramming protocol, if adapted to a diagnostic setting, can potentially enhance the detection of neuron-specific disease-associated gene expression, thereby improving the molecular diagnosis of Mendelian disorders with neurological phenotypes, which represent the most prevalent phenotypic manifestation among all genetic disorders for which clinical genetic testing is performed. Additionally, this approach can provide valuable insights into the underlying mechanisms of neurological disorders, which aids in interpreting genetic variants or for a transcriptome-first analytical approach.
In this study, we present an RNA-seq analysis workflow utilizing transdifferentiated fibroblasts to enhance the genetic diagnoses of neurological disorders. Our proposed workflow is characterized by its simplicity, cost effectiveness, and timely execution (6–8 weeks) while demonstrating its robustness. Notably, our approach effectively identified neuron-specific aberrant transcriptional events, including but not limited to aberrant splicing, aberrant expression, and mono-allelic expression, resulting in a diagnostic yield of 25.4% (18/71) in individuals with neurological disorders who were recruited to the Undiagnosed Diseases Network (UDN). Impressively, the iNeuron RNA-seq approach proved effective in providing key evidence to finalize molecular diagnoses after an uninformative RNA-seq analysis using fibroblast samples, benefiting 27.7% (5/18) individuals. Our work demonstrates that transdifferentiation of CATs is an effective and feasible approach to improve the clinical utilization of diagnostic whole transcriptomic analysis for individuals with rare genetic disorders.
Material and methods
Compendium
A cohort of 75 probands, along with 9 family members and 3 unrelated controls, was recruited from the UDN Baylor College of Medicine (BCM) clinical site. Enrollment criteria mandated that each individual present at least one primary neurological phenotype. WGS/ES data from 75 probands and 7 family members was obtained from UDN sequencing core. Fibroblast-to-neuron transdifferentiation was performed for all individuals. Four probands whose iNeuron data did not meet the quality control (QC) standards were removed from the final analysis.
Lentivirus production
HEK-293FT cells, at 70%–80% confluence, were transfected using jetPRIME (114-15, Polyplus-transfection), following the specified protocol of the transfection reagent. The pLVX-UbC-rtTA-Ngn2:2A:Ascl1 plasmid was kindly provided by Dr. Fred Gage (Addgene plasmid # 127289). To package the target plasmid, we used second-generation lentiviral packaging plasmids (gift from Dr. Didier Trono). This included two helper plasmids: psPAX2, which contained a minimal HIV enzyme set (Addgene #12260), and pMD2.g, encoding the VSV-G envelope (Addgene #12259). Lentivirus-containing supernatant was collected at 24- and 48-h points and concentrated overnight at 4°C, following the Lenti-X Concentrator (631232, Takara) protocol. The prepared virus was either used immediately or stored in aliquots at −80°C for future applications.
Transdifferentiation of fibroblasts to neurons
Human primary dermal fibroblasts established from skin punch biopsies were maintained in high-glucose DMEM medium, supplemented with 10% fetal bovine serum (FBS), 1% non-essential amino acid (NEAA), and 1% penicillin-streptomycin (P-S). The protocol for transdifferentiation of fibroblasts into neurons was adapted from the method established by Fred Gage’s group.33 Briefly, human primary dermal fibroblasts were transduced with lentivirus particles containing a TetON cassette for the doxycycline-induced overexpression of NEUROG2 and ASCL1 (pLVX-UbC-rtTA-Ngn2:2A:Ascl1, Addgene, #127289). The standard FBS in the culture medium was replaced with tetracycline-free FBS. Two days later, 2 μg/mL puromycin was added in the culture medium for 7 days. Following at least three passages after viral infection, the fibroblasts were seeded on Cultrex-coated plates at a density of 50,000/cm2. On the following day, the culture medium was switched to neuron conversion (NeuC) medium, consisting of 1:1 DMEM/F12 medium and neurobasal medium, supplemented with 1 X N2, 1 X B27, 1 X P-S, 1 μg/mL laminin (R&D, #3400-010-02), 200 μM Dibutyryl cyclic-AMP (Tocris, #1141), 0.5 μM LDN193189 (Tocris, #6053), 5 μM A83-1 (Tocirs, #2939), 3 μM CHIR99021 (Tocris, #4423), 5 μM Forskolin (Tocris, #1099), 10 μM SB431542, #1614), 1 μM Pyrintegrin (Tocris, #4978), 7.5 μM KC7F2 (Tocris, #4324), 0.175 μM ZM336372 (Cayman, #10010367), 0.1 μM AZ960 (Cayman, #16731), and 2 μg/mL doxycycline. Medium was changed 3 times a week for 3 weeks.
Immunocytochemistry
Fibroblasts infected with a vector for the overexpression of NEUROG2 and ASCL1 were plated and cultivated on glass coverslips in the presence of doxycycline for 21 days. The iNeurons were fixed using a 4% paraformaldehyde solution and then treated with a blocking buffer containing 5% goat serum, 1% bovine serum albumin, and 0.1% Triton X-100 for 30 min. Subsequently, the cells were exposed to primary antibodies overnight at 4°C. The appropriate Alexa Fluor secondary antibodies were utilized. The primary antibodies used were Tubulin β-III (1:200, R&D systems, #MAB1195) and MAP2 (1:500, Millipore, #AB5622). Nuclei were stained using DAPI (Sigma). Imaging was conducted using a Zeiss AxioVision microscope.
RNA sequencing
Total RNA was extracted from iNeurons using the RNeasy mini kit (Qiagen) following the manufacturer’s instructions with the inclusion of an on-column gDNA removal step. The integrity and quality of the RNA were assessed using the Qubit 4 Fluorometer and the Qubit RNA HS Assay Kit (ThermoFisher). Library preparation was performed with the TruSeq Stranded mRNA Library Prep Kit (Illumina). The constructed libraries underwent 150 bp paired-end sequencing at a depth of approximately 100–150 million reads per sample. The obtained sequencing data were processed utilizing the Sentieon pipeline with alignment to the GRCh38/hg38 reference sequence employing STAR-v2.7.10a. Gene expression levels were quantified using RNA-SeQC, which generated Transcripts Per Million (TPM) values for expressed genes in each sample.34,35 The processed alignment files were subsequently utilized for outlier detection. The RNA-seq data were visualized using the Integrative Genomics Viewer (IGV) software.
Amplicon-based NGS
Amplicon-based NGS is comprised of a two-step PCR library preparation workflow that generates ready to sequence libraries. After the DNA is extracted using Qiagen’s QIAamp DNA Mini Kit, the targeted region is amplified using FastStart Taq DNA Polymerase from Roche. The primers are designed to include a linker region on each end that act as a binding site for the secondary PCR primers. The amplicon from the first PCR is purified using 0.9× AMPure XP beads and is used as a template for the secondary PCR. The second pair of primers consists of flow cell binding sequences, sequencing primer sites, and barcodes that will be included into the template via PCR. The completed library is ready to sequence after a final AMPure XP bead purification of the secondary PCR product.
Differential gene expression and functional enrichment analysis
Differential gene expression analysis between 87 iNeurons and 77 cultured fibroblasts was conducted using the DESeq2 package in the R programming language. The criterion for selecting differentially expressed genes (DEGs) was defined as a fold change (FC) > 2 and adjusted p < 0.05. To gain insights into the biological functions of the upregulated DEGs in iNeurons, functional enrichment analysis was performed using the Metascape tool.36
Quality control of iNeurons
The iNeuron score (iN_Score) was calculated as the geometric mean of the TPM values of the top 500 DEGs that were up-regulated in iNeurons. To assess the relationship between ASCL1 qPCR expression levels in intermediate fibroblasts, iNeuron ASCL1 qPCR expression, and iNeuron RNA-seq ASCL1 TPM values, Spearman correlation analysis was conducted. Receiver operating characteristic (ROC) curve analysis was performed to assess whether the expression levels of ASCL1 or NEUROG2 in the iNeuron RNA-seq data could serve as predictors of transdifferentiation quality. This analysis utilized the pROC package in R. To identify potential chromosomal aneuploidies that may arise during the transdifferentiation process, copy number variation (CNV) analysis was performed on both the RNA and the DNA sequencing data. The RNAseqCNV package in R was used for RNA-based CNV analysis,37 whereas CNVpytor was utilized for the DNA-based analysis.38
qRT-PCR
RNA was transcribed into complementary DNA (cDNA) using the iScript cDNA Synthesis Kit (Bio-Rad). qRT-PCR was performed to determine mRNA levels using the SsoAdvanced Universal SYBR Green Supermix (Bio-Rad). GAPDH was used as an internal control. The relative fold change in gene expression was determined using the comparative threshold cycle ΔΔCt method.
Outlier detection
The RNA outliers detection pipeline (DROP) was utilized with a permissive threshold (OUTRIDER: p < 0.05; FRASER: p < 0.05; Delta >0.2) to identify potential outliers in gene expression.39 In order to prioritize the expression outliers, haploinsufficient genes (Z < 0), recessive genes (Z < 0), and triplosensitive genes (Z > 0) were flagged using data from ClinGen and OMIM. Potential expression or splicing outliers were cross-referenced with OMIM genes, with phenotype-driven prioritization performed by PhenoApt.40 DNA findings from ES/WGS were supplemented with predictions from SpliceAI,11 if applicable, to correlate and substantiate relevant RNA findings.
Panels with various neurological phenotypes
The comprehensive disease-gene lists were based on NIH Genetic testing registry, NHS National Genomic Test, ClinGen, and commercially available panels.
Tissue-specific isoform activation in iNeurons
We computationally measured a subset of tissue-specific transcripts, those that are tagged by tissue-specific exons, to estimate the overall abundance of all tissue-specific isoforms. To calculate the exon-tagged isoform activation in iNeurons, we compared RNA-SeQC results from all our iNeuron samples (n = 82) and a set of fibroblast samples (n = 77). All transcripts from a gene were collapsed into an artificial transcript. The collapsed transcript was used to calculate the gene-level TPM (TPMgene), following the conventional way of computing gene-level TPM.41 Each exon from the collapsed transcript was used to calculate its individual exon-level TPM (TPM{gene, exon}). The median values for each TPM{gene, exon} and TPMgene were computed across the iNeuron and the fibroblast cohorts. Then, the relative ratio of the exon-specific fold change over the gene-level fold change was computed, denoted as λ. An equation is represented below, where TPM` denotes value from iNeurons and TPM denotes value from fibroblasts.
In this work, we considered a relative exon/gene activation ratio (λ) over two as an indication that the exon-tagged transcript is enriched in iNeurons. When the transcript is neuron enriched and has a fibroblast TPM of <1, the transcript is considered neuron specific.
Statistics
The iN_Score between iNeurons and fibroblast was assessed using a Student’s t test. p < 0.05 was deemed significant. The analyses of DEGs, exons and outlier detection are described above.
Ethics approval
The Institutional Review Boards approved the study at the National Human Genome Research Institute (15HG0130) and BCM (H-34433). Written informed consent was obtained from all study participants.
Results
Limited expression of genes associated with neurological phenotypes in CATs
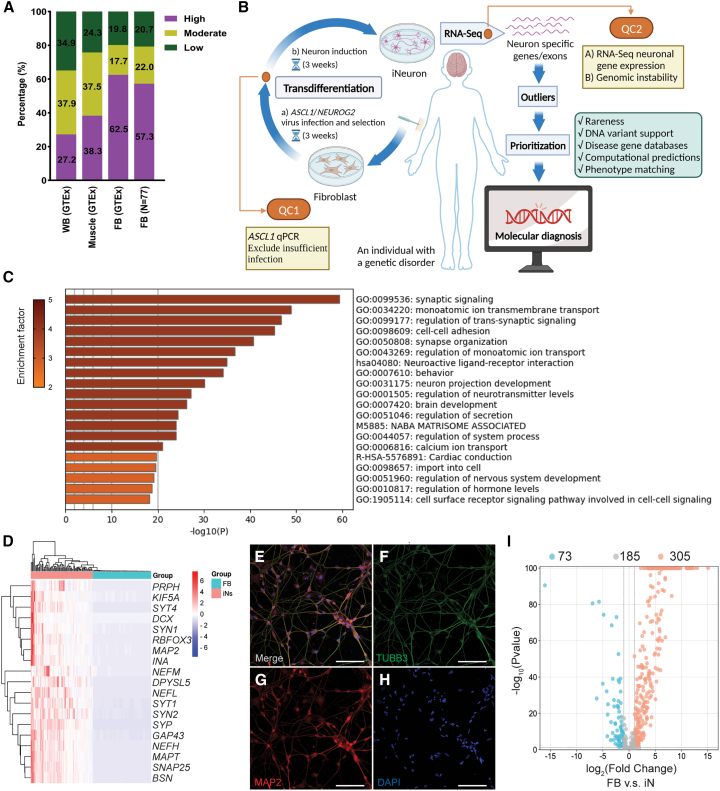
We compiled a list of 2,721 OMIM genes associated with various neurological phenotypes (OMIM-N). We first performed a computational analysis to evaluate the level of gene expression in human fibroblasts. In both the GTEx dataset and independent RNA-seq performed in fibroblasts in this study (n = 77), approximately 20% or 35% of the genes were deemed as low expression (TPM < 1) in fibroblasts or whole blood, making their detection challenging even with deeper sequencing (Figure 1A; Table S1). We next assembled eight panels of genes associated with the following neurological phenotypes: intellectual disability, brain malformations (BM), autism spectrum disorder (ASD), epilepsy, ataxia, neuropathy, neuromuscular disorder, and leukodystrophy. Approximately 10%–23% of genes in these panels are lowly expressed in fibroblasts, while the range of low-expression genes is 26%–37% in whole blood. This highlights the intrinsic limitation of using CATs for the molecular diagnosis of neurological disorders (Table S1). Interventions to activate these genes are needed to improve the representation of genes linked to Mendelian neurological disorders in RNA-seq data from CATs.
Figure 1.
Activation of low-expression OMIM-N genes in participants' fibroblasts
(A) Expression levels of OMIM-N genes in clinically assessable tissues. Gene expression levels were classified as follows: low (TPM < 1), moderate (TPM ≥ 1 and <10), and high (TPM ≥ 10). Fibroblasts (FB) RNA-seq data from GTEx and UDN participants (n = 77) were used for the assessment.
(B) Schematic of the workflow of iNeuron transdifferentiation and RNA-seq. Created with BioRender.com.
(C) Functional enrichment analysis of DEGs up-regulated in iNeurons.
(D) Robust expression of neuron-specific genes in iNeurons.
(E–H) Immunofluorescence staining of neuronal makers Tubulin β-III (TUBB3) (F) and MAP2 (G) in iNeurons. Nuclei were stained using DAPI (H). The merged image is shown in (E). Scale bar represents 100 μm.
(I) Volcano plot showing activation of low-expression OMIM-N genes.
Transdifferentiation through NEUROG2-ASCL1 manipulation
Manipulations of pro-neuronal transcription factors, microRNA, and target genes regulated by REST/NRSF have been shown to facilitate the direct conversion of human somatic cells, such as skin fibroblasts, into functional neurons.33,42,43,44 To modulate neuronal gene expression in human fibroblasts, we evaluated the effectiveness of four published neuronal induction protocols in combination with the chemical modulation of multiple cellular signaling pathways. These protocols include (1) overexpression of the pro-neuronal transcription factors NEUROG2 and ASCL133; (2) co-expression of the microRNA miR-9/9∗, miR-124, and the anti-apoptotic gene BCL2L144; (3) a combination of (1) and (2); and (4) DBD-REST-VP16,43 involving the replacement of REST/NRSF repressor domains with the activation domain of the viral activator VP16 (Figure S1).
We evaluated the four protocols in a fibroblast cell line from a healthy individual. The results revealed comparable conversion rates and optimal morphology of bipolar neurons after 21 days of induction. The protocol reported by Dr. Fred Gage’s group,33,45 combining NEUROG2-ASCL1 and a chemical cocktail, elicited the most robust activation of neuron-specific genes and genes related to various neurological phenotypes (Table S2). Extending the induction time to 28 days did not result in a significant increase in the activation of the target genes (Table S3). Therefore, this approach was selected for subsequent transdifferentiation experiments using fibroblasts from individuals with neurological disorders (Figure 1B).
Using the same fibroblast cell line, we further compared the transcriptome profiles of iNeuron with two types of human-induced pluripotent stem cell (iPSC)-derived neurons. Of the two types of iPSC-derived neurons, Neuron_1 underwent differentiation from iPSCs, while Neuron_2 was generated through the overexpression of the neuronal transcription factor Neurog2 in iPSCs. Correlation analysis revealed that the fibroblast-direct-converted iNeurons exhibited a transcriptional profile more closely aligned with hiPSC-derived neurons (r = 0.89) than with the donor’s fibroblasts (r = 0.76), indicating a cellular identity shift after transdifferentiation (Figure S2A). To evaluate the degree of similarity between the transcriptional profile of iNeurons and those of the human neural tissues, we conducted a correlation analysis comparing our iNeuron data and data from GTEx. The analysis demonstrated a high correlation between iNeurons and various neural tissues (r > 0.9), in contrast to cultured fibroblasts, which showed a weaker correlation (r < 0.7) (Figure S2A).
Transdifferentiation of individual cell lines robustly activates low-expression neurological disease-associated genes
We applied the NEUROG2-ASCL1 transdifferentiation protocol to a cohort of participants’ samples to assess its clinical utility. The study cohort includes 75 probands with at least one primary neurological phenotype who were recruited from the UDN BCM clinical site, along with 9 of their family members and 3 unrelated controls. The majority of probands (n = 63; 84.0%) were pediatric, and almost half (n = 37; 49.3%) were male (Table 1). In addition, 66.7% of probands presented with intellectual disability, while 40.0% presented with a brain malformation. A detailed summary of the prevalence of distinct neurological phenotypes is provided in Table 1. DNA sequencing data were available for 38 individuals with WGS and 37 individuals with ES. We performed skin biopsy and neuron transdifferentiation on all participants.
Table 1.
Demographic characteristics and types of neurological phenotypes included in the study
| Number | Percentage | |
|---|---|---|
| Sex | ||
| Male | 37 | 49.3 |
| Female | 38 | 50.7 |
| Age | ||
| Adult | 12 | 16.0 |
| Pediatric | 63 | 84.0 |
| Neurological phenotypes | ||
| Intellectual disability | 50 | 66.7 |
| Brain malformation | 30 | 40.0 |
| Microcephaly | 19 | 25.3 |
| Macrocephaly | 5 | 6.7 |
| Epilepsy | 31 | 41.3 |
| Hypotonia | 24 | 32.0 |
| Dystonia | 14 | 18.7 |
| Ataxia | 8 | 10.7 |
| Leukoencephalopathy | 4 | 5.3 |
| ASD | 4 | 5.3 |
| ADHD | 3 | 4.0 |
| Hearing loss | 6 | 8.0 |
| Eye disease | 14 | 18.7 |
| Neuropathy | 5 | 6.7 |
| Neuromuscular disorders | 7 | 9.3 |
More than one neurological phenotype may be present in any individual.
RNA-seq of 87 iNeurons revealed a distinct transcriptome expression profile compared to the cohort of 77 cultured fibroblasts, characterized by the up-regulation of 4,902 differentially expressed genes (DEGs) and 3,558 down-regulated DEGs. In addition, the up-regulated DEGs in iNeurons exhibited significant enrichment in functional categories related to various aspects of nervous system development, including synaptic signaling, ion transmembrane transportation, and neuron projection interaction (Figure 1C, Data S1). RNA-seq analysis also showed robust expression of a list of neuron-specific genes associated with axon formation, dendrite development, growth cone dynamics, and synapse formation (Figure 1D). Immunocytochemical staining confirmed the presence of neuronal markers such as Tubulin β-III and MAP2 (Figures 1E–1H). Consistent with expectations reported previously,33 iNeuron cells generated from transdifferentiation predominantly expressed markers of glutamatergic and GABAergic neurons, while markers for dopaminergic, serotonergic, cholinergic, and glycinergic neurons exhibited inconsistent activation (Figures S2B and S2C). In our analysis focusing on Mendelian disease-associated genes, DEG analysis between fibroblasts and iNeurons showed that more than half (54.2%, 305/563) of OMIM-N genes with low expression (TPM < 1) in fibroblasts were up-regulated in iNeurons (Figure 1I). Across eight panels of neurological disease-associated genes, the percentages of up-regulated genes varied from 53.8% to 91.7% (Table 2; Figure S3). We defined a status “activated and actionable” as a gene in iNeurons having (1) an up-regulated DEG compared to fibroblast and (2) a clinically analyzable TPM of ≥1. Of the OMIM-N genes, 23.6% (133/563) satisfied this criterion, while 15.4%–42.9% of genes from the eight neurological disease panels we assembled were deemed “activated and actionable” (Table 2). The two gene panels of neuropathy and brain malformation displayed the highest rates of “activation and actionability” (42.9% and 37.5%, respectively), while the neuromuscular disorder gene panel had the lowest rate (15.4%) (Table 2). The variability in activation rates across panels implies varying diagnostic benefits of applying this protocol for individuals with different types of neurological disorders. To provide a theoretical context starting from whole blood instead of fibroblasts, a high fraction of genes from the eight panels, ranging from 48.6% to 75.8%, reached the status of “activated and actionable” if converted to iNeurons (Table S4). Further analysis of the expression data reveals high levels of concordance between the iNeuron cohort and adult cortex samples from GTEx in the expression profiles of neurological disease-associated genes (Figure S2D).
Table 2.
Activation of genes associated with neurological disorders in iNeurons
| Category | No. of low- expression genes in fibroblasts |
Up-regulated in iN |
Activated and actionable in iN |
||
|---|---|---|---|---|---|
| No. | % | No. | % | ||
| Neurological OMIM | 563 | 305 | 54.2 | 133 | 23.6 |
| Intellectual disability panel | 231 | 156 | 67.5 | 75 | 32.5 |
| Brain malformation panel | 24 | 22 | 91.7 | 9 | 37.5 |
| Autism spectrum disorder panel | 69 | 53 | 76.8 | 22 | 31.9 |
| Epilepsy panel | 246 | 156 | 63.4 | 72 | 29.3 |
| Ataxia panel | 292 | 176 | 60.3 | 70 | 24.0 |
| Neuropathy panel | 21 | 13 | 61.9 | 9 | 42.9 |
| Neuromuscular disorders panel | 52 | 28 | 53.8 | 8 | 15.4 |
| Leukodystrophy panel | 90 | 51 | 56.7 | 28 | 31.1 |
The infection efficiency, induction quality, and genomic integrity of iNeurons are evaluated by quality control measurements
The quality, reproducibility, and integrity of the transdifferentiation of cell lines are associated with various factors, such as lentiviral titer, genetic traits, cell viability, and technical variables. We developed two potentially interdependent metrics to evaluate the overall quality of the transdifferentiation process: (1) targeted expression levels of the two transcriptional factors ASCL1-NEUROG2 introduced by the lentiviral transfection and (2) global expression levels of genes that are expected to be activated in this protocol (Figure 2A).
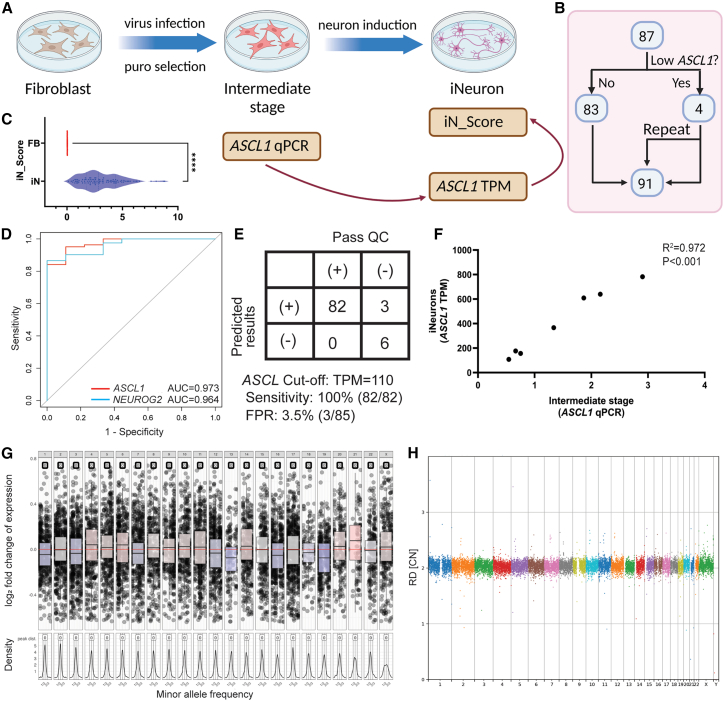
Figure 2.
Multiple quality control measurements during transdifferentiation process
(A) Schematic for determining ASCL1 qPCR as a critical step in QC to identify inadequate virus infection. Created with BioRender.com.
(B) All iNeurons samples used for QC analysis.
(C) iN_Score comparison between iNeurons and fibroblasts, represented by median with violin plot. Statistical significance denoted by ∗∗∗∗p < 0.0001.
(D) ROC curve analysis showing expression levels of ASCL1 or NEUROG2 as predictors of iNeuron quality. The area under the ROC curve (AUC) for ASCL1 is 0.973, and for NEUROG2 it is 0.964. There is no statistically significant difference between ASCL1 and NEUROG2.
(E) ASCL1 TPM cutoff of 110 in iNeurons with 100% sensitivity and 3.5% false positive rate (FPR).
(F) Correlation between ASCL1 qPCR and TPM of iNeurons. ASCL1 qPCR result is normalized to GAPDH. R2 = 0.972, p < 0.001.
(G) A representative genome-wide copy number analysis plot assessing post-induction genomic stability. RNA-based CNV calling revealed no aneuploidies or large copy number variations (CNVs) in this iNeuron sample.
(H) The post-induction genomic stability assessments are validated by further WGS analysis. WGS also detected no smaller new CNV events.
ASCL1 and NEUROG2 have median expression TPMs at 912.12 and 1250.81 in our iNeuron RNA-seq data, respectively. As expected, these two neuronal transcription factors are not expressed in our fibroblast RNA-seq data. We arbitrarily set 10% of the median iNeuron TPM for these two genes as cutoffs to indicate whether lentiviral-mediated transdifferentiation was successful. We identified four samples from the cohort that did not pass the 10% median iNeuron TPM threshold. The transdifferentiation experiment was repeated for these four samples, making it a total of 91 iNeurons samples (from 87 individuals) contributing to the entire dataset to be subject to downstream QC selection (Figure 2B).
To evaluate the global expression of genes activated in iNeurons and enhance the robustness of transdifferentiation quality, we established an iNeuron score (iN_Score), which is calculated by the geometric mean of the leading 500 up-regulated DEGs in iNeurons. The iN_Score is an effective measurement to differentiate processed iNeurons versus fibroblast cells without manipulations (median iN_Score of 2.84 in transdifferentiated cells and 0.02 in unprocessed fibroblasts, p < 0.0001, Figure 2C) based on data from our sample cohorts of 91 iNeuron lines and 77 fibroblast lines. A cutoff value for the iN_Score (0.83) was empirically determined at the tenth percentile of all scores from our iNeuron sample cohort. Nine iNeuron samples from eight individuals were identified to have poor induction quality using this criterion, and they were excluded from downstream analysis.
iN_Score offers a broader perspective than the traditional reliance on a limited set of markers, which is more prone to stochastic variation. In addition, immunostaining requires additional labor, training, and reagent costs, posing a challenge to efficient clinical implementation. To enhance the interpretability of the iN_Score, we highlighted three representative iNeuron samples, correlating their iN_Scores with the transdifferentiation efficiency deduced from staining (Figures S4A–S4D). The first sample exhibited a low transdifferentiation efficiency, falling below our QC threshold, with a low iN_Score of 0.75 and a conversion rate of ∼20%. The second sample achieved a marginally acceptable iN_Score of 1.91, corresponding to an estimated conversion rate of 40%. The third sample with a high iN_Score of 5.42 demonstrated efficiency exceeding 90% (Figures S4A–S4D). Expression levels of established mature neuron markers such as MAP2, NeuN (RBFOX3), NEFH, Tau (MAPT), SYP, and GAP43 in 3 samples also showed differences corresponding to iN_Score and transdifferentiation efficiency (Figure S4E). These findings corroborate the effectiveness of the iN_Score in accurately assessing iNeuron transdifferentiation quality and predicting conversion rates.
Of note, the original failed samples from the four specimens with low ASCL1-NEUROG2 expressions all had low iN_Scores. After repeat, three samples demonstrated improvements in lentiviral-mediated transdifferentiation (increased ASCL1 and NEUROG2 expressions) and in neuron induction (passing iN_Scores); one sample remained poorly induced despite satisfactory expressions of ASCL1 and NEUROG2. The repeated failure is potentially attributed to other confounding factors beyond lentiviral infection (Table S5).
Based on the above empirical observations, we hypothesized that the ASCL1 and NEUROG2 expressions can serve as predictive markers for the quality of iNeurons RNA-seq data. This hypothesis offers an attractive quality checkpoint at an early stage of the workflow for intervention to prevent poor-quality samples from going through the transdifferentiation procedure. The receiver operating characteristic (ROC) curve analysis indicated that both ASCL1 and NEUROG2 expression levels in the iNeuron RNA-seq data were highly predictive of a passing iN_Score (Figure 2D). By setting a TPM cutoff at 110 for ASCL1, we achieved 100% sensitivity in excluding samples with poor induction due to limited infection, with an acceptable false positive rate of 3.5% (3/85) (Figure 2E). To establish a de facto checkpoint following lentivirus infection (rather than examining expression levels later after cell culture and RNA-seq are completed), we selected seven samples to collect cell culture intermediates at the stage of 24 h post-induction. RNA was extracted for qPCR analysis of ASCL1 expression. The qPCR measurements correlated highly with the TPM of RNA-seq data from the iNeurons (R2 = 0.972, p < 0.001, Figure 2F). Therefore, a simple qPCR QC checkpoint assay targeting ASCL1 gene expression can be devised to exclude samples with low-grade infection, eliminating the need for further costly and time-consuming induction and RNA-seq analyses on a potentially failed sample.
It has been shown that genomic instabilities including aneuploidies and structural chromosome changes may arise from the preparation of iPSCs and embryonic stem cells (ESCs).46,47 To rule out such confounding factors, we performed CNV analysis using RNA-seq data from the iNeurons. This RNA-seq-based computational analysis can detect chromosomal aneuploidies (Figure S5). No significant chromosomal changes were detected in the iNeuron samples (Figure 2G), except for one sample with a possible gain of chromosome 21 (Figure S5).37 As an additional layer of validation, WGS at 50×–100× (mean 70×) coverage was performed on the intermediate fibroblasts and iNeurons derived from seven individuals. At this sequencing depth, WGS has been shown to confidently detect CNVs at the kilobase resolution, or conservatively speaking at the resolution of hundreds of kilobases.48,49 No CNVs exceeding 0.5 Mb were detected among the tested samples. The above findings confirm that no apparent structural genomic alterations arose during the transdifferentiation process (Figure 2H).
Summarizing the experiences from this study, we developed a detailed list of recommendations for QC of using iNeuron RNA-seq in genetic diagnosis. This workflow comprises two checkpoints (Figure 1B). The first checkpoint occurs during the intermediate stage, where samples from an additional well of 12-well plate can be subject to qPCR for ASCL1. This allows for the exclusion of samples with insufficient quality in initial transdifferentiation (likely reflecting efficiency of infection). The second checkpoint can be initiated during the RNA-seq data analysis before clinical interpretation. At this point the iN_Score can be calculated to ensure transdifferentiation quality. Additionally, CNV analysis is performed on the RNA-seq data to alert for unwanted culture-related genomic alterations. Overall, this rigorous QC workflow will be instrumental for clinical diagnostic laboratories considering the implementation of iNeuron RNA-seq.
iNeuron RNA-seq increased the molecular diagnostic yield
We performed a transcriptome-driven analysis based on the iNeuron RNA-seq data. Following the identification of candidate expression findings, we sought DNA variant-level validation from the WGS/ES. Independently, DNA-directed data analysis was performed as previously described1,50 to inform and complement the RNA-based analysis.
The transcriptome-driven analysis relies on intra-cohort data normalization under the assumption that samples within this cohort exhibit a high degree of genetic heterogeneity, i.e., each iNeuron sample is expected to have a unique genetic defect and thus can serve as a “control” for the rest of the samples. iNeuron is a novel sample type not represented in public databases such as GTEx. Therefore, we accumulated data from 82 specimens to power the identification of outlier events. We adopted the DROP pipeline39,51,52 to streamline the identification of expression and splicing outliers. A relatively permissive threshold in the DROP pipeline was used (OUTRIDER: p < 0.05; FRASER: p < 0.05; Delta >0.2) to allow a broader detection of potential outliers. On average, this method resulted in 526 expression outliers and 2,407 splicing outliers for each sample (Figures 3A and S6A). After limiting the analysis to known disease-associated genes in ClinGen and OMIM, we stratified and prioritized the expression outliers by matching the direction of the expression change with the expected disease mechanism, namely decreased expression (Z < 0) for findings in genes with a haploinsufficiency/autosomal-dominant mechanism or autosomal-recessive inheritance and increased expression (Z > 0) for genes with a triplosensitive disease mechanism. The resultant per sample aberrant event counts averaged at 32 for dominant trait genes and 51 for recessive trait genes (Figures 3A and S6B). Splicing outliers are similarly limited to those affecting OMIM disease-associated genes. We required that the abnormal splicing be substantiated by a DNA variant within 1 kb of the splicing junction based on the ES/WGS data (except for the TIAM1 change discussed later). On average, each sample had 821 splicing outlier-DNA variant pairs in OMIM disease-associated genes, 17 of which are supported by SpliceAI (score >0.2) (Figures 3A and S6C). Finally, by cross-referencing the candidate genes identified by OUTRIDER and FRASER and their correlation with the proband’s clinical phenotypes, we narrowed the findings down to the candidate diagnostic variants (Figure 3A). The candidate variants are manually reviewed for a final decision of their classifications.
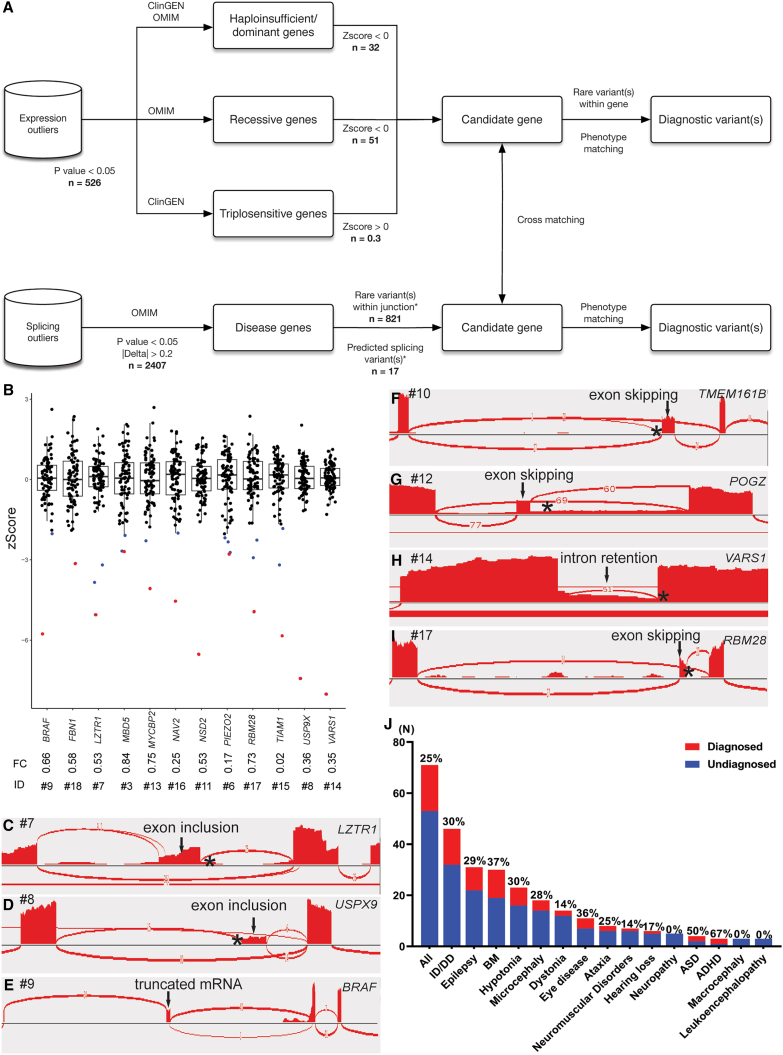
Figure 3.
The interpretation pipeline, molecular diagnostic rates, and selected diagnostic findings from the iNeuron RNA-seq workflow
(A) Analytical and interpretation pipelines used in iNeuron RNA-seq analysis. The n values indicate the average number of variants passing filtration at each step.
(B) OUTRIDER identified 12 aberrant expression outliers as diagnostic findings (BRAF, FBN1, LZTR1, MBD5, MYCBP2, NAV2, NSD2, PIEZO2, RBM28, TIAM1, USP9X, and VARS1). Each dot represents an individual participant. The blue dots represent expression outliers that fulfill the analytical filtration standards defined in (A). The red dots represent molecular diagnostic findings with integrated considerations from analytical filtrations (blue dots), phenotypic matching, and correlation with DNA variant findings. For each molecular diagnostic expression outlier (red dot), its ID and expression fold change compared to controls are listed underneath.
(C–I) FRASER detected 10 aberrant splicing events, with 7 representative events (LZTR1, USP9X, BRAF, TEME161B, POGZ, VARS1, and RBM28) illustrated here. Aberrant junctions are shown in the Sashimi plot. The asterisk represents the position of the causal variant.
(J) Molecular diagnostic yields in individuals with different neurological phenotypes.
OUTRIDER identified 12 aberrant expression levels involving the genes BRAF (MIM: 164757), FBN1 (MIM: 134797), LZTR1 (MIM: 600574), MBD5 (MIM: 611472), MYCBP2 (MIM: 610392), NAV2 (MIM: 607026)53, NSD2 (MIM: 602952), PIEZO2 (MIM: 613629), RBM28 (MIM: 612074), TIAM1 (MIM: 600687), USP9X (MIM: 300072), and VARS1 (MIM: 192150), with fold changes ranging from 0.02 to 0.84 (median 0.53), as shown in Figure 3B. All the above expression outliers can be associated with a causal DNA variant (Table 3) except for the almost diminished TIAM1 expression outlier in proband #15. Although the DNA changes in this gene cannot be confidently linked to the expression reduction, this TIAM1 expression change is considered a possible diagnostic finding because of its good disease-proband phenotype matching and the distinctive reduction in expression level (Z score = −5.83).
Table 3.
Molecular diagnostic findings from iNeurons RNA-seq
| ID | Sex | Age | Gene | Isoform | Variant DNA | Zygosity | Inheritance | Variant RNA | RNA consequence | Detection algorithma | TPM iNeuron median | TPM fibroblast median | iN required?b |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 4 years | ITPR1 | NM_001378452.1 | c.5980−17G>A | het | de novo | r.5979_5980ins[5980−15_5980−1/5979+1_5980−18,a,5980−16_5980−1] | two events: 15 nt intron inclusion at splice acceptor; entire intron inclusion | AS | 6.03 | 1.16 | yes |
| 2 | M | 22 years | DCX | NM_001195553.2 | c.946+4588G>T | hem | de novo | r.946_947ins[946+4619_947−1] | 13,549 nt intron inclusion at splice acceptor | AS | 2.31 | 0 | yes |
| 3 | F | 4 years | MBD5 | NM_001378120.1 | c.−925+35307_−557+13765delchr2:148056991_148356101del | het | maternal | r.−924_−558del | 5′ UTR exons 2–4 deletion containing 337 nt | AE (0.84), AS | 3.32 | 2.99 | yes |
| 4 | F | 7 years | CACNA1A | NM_001127222.2 | c.5015G>C | het | de novo | r.5015g>c (r.632_784del) | skewed variant allele expression at 88% fraction; skipping of exon 5 (153 nt); uncertain if the two events are related | MAE; AS | 6.48 | 2.54 | yes |
| 5 | M | 36 years | POLR3A | NM_007055.4 | c.1771−7C>G | het | paternal | r.1771_1909del/1643_1909del | two events: skipping of exon 14 (139 nt); skipping of exons 13–14 (267 nt) | AS (moderate effect) | 13.92 | 10.76 | no |
| c.3892−297_3892−221del | het | maternal | r.3891_3892ins[3892−1_3892−227/3892−74_3892−227] | two events: 227 nt intron inclusion at splice acceptor; 154 nt cryptic exon inclusion from intron 29 | AS (moderate effect) | ||||||||
| 6 | F | 5 years | PIEZO2 | NM_001378183.1 | c.5257−1G>A | het | maternal | r.5258del | 1 nt shift of splice acceptor | AE (0.17), AS (misalignment, NMD), MAE | 4.56 | 2.14 | yes |
| c.1528−1G>A | het | paternal | r.1528del | 1 nt shift of splice acceptor | AE (0.17), AS (misalignment, NMD), MAE | ||||||||
| 7 | M | 2 years | LZTR1 | NM_006767.4 | c.2178C>A | het | paternal | r.2178c>a | nonsense | not applicable | 25.41 | 36.41 | no |
| c.1943−256C>T | het | maternal | r.1942_1943ins[1942+342_1943−262/1942+360_1943−262] | two events: 117 nt cryptic exon inclusion; 99 nt cryptic exon inclusion | AE (0.53), AS | ||||||||
| 8 | F | 4 years | USP9X | NM_001039591.3 | c.1315−284G>T | mosaic | de novo | r.1314_1315ins[1315−281_1315−176/1315−281_1315−172] | two events: 106 nt or 110 nt cryptic exon inclusion | AE (0.36), AS | 43.08 | 42.06 | no |
| 9 | F | 16 years | BRAF | NM_001374258.1 | seq[GRCh38] t(5; 7)(q31.3; q34) NC_000005.10:g.140964619_qterdelins[NC_000007.14:g.140799141_qter] NC_000007.14:g.140799137_qterdelins[NC_000005.10:g.140964621qter] | het | de novo | r.-226_980:(?)/−226_980:980_981ins[980+1_980+1225]:(?) | two (or more) events: fusion gene of 5′ BRAF with sequence from 7q34; fusion gene of 5′ BRAF plus 1225 nt intron retention with sequence from 7q34; the question mark denotes uncertainty of the breakpoints | AE (0.66), AS | 6.35 | 5.83 | no |
| 10 | M | 2 years | TMEM161B | NM_153354.5 | c.800+5G>A | het | maternal | r.660_800del | skipping of exon 8 (141 nt) | AS (candidate disease-associated gene) | 5.29 | 4.06 | no |
| c.980T>C | het | paternal | r.980u>c | missense | not applicable | ||||||||
| 11 | M | 26 years | NSD2 | NM_001042424.3 | NC_000004.12:g.1869269_1873124del | het | unknown | (NSD2)x53% | expression reduction of the entire transcript | AE (0.53) | 7.70 | 13.85 | no |
| 12 | F | 4 years | POGZ | NM_015100.4 | c.2546−20T>A | het | de novo | r.2546_2570del/2545_2571ins[2545+1_2546−21,a,2546−19_2571−1] | two events: skipping of exon 18 (25 nt); retention of introns 17–18 (446 nt) | AS | 20.87 | 22.03 | no |
| 13 | M | 10 years | MYCBP2 | NM_015057.5 | c.8005C>T | het | de novo | (MYCBP2)x75% | stopgain variant likely causes NMD and skewed allele fraction | AE (0.75), MAE | 16.59 | 14.33 | no |
| 14 | F | 2 years | VARS1 | NM_006295.3 | c.3288G>T | het | paternal | r.3288delins[u,3288+1_3289−1] | entire intron 27 retention (71 nt) | AE (0.35), AS | 30.3 | 38.98 | no |
| c.2590_2592delAGCinsTGA | het | maternal | (VARS1)x35% | stopgain variant likely causes NMD and skewed allele fraction | AE (0.35), MAE | ||||||||
| 15 | F | 10 years | TIAM1 | NM_001353694.2 | c.1996−78G>A, c.1585−5707G>T, c.−188−13607G>A | het | unknown | (TIAM1)x2% | expression reduction of the entire transcript | AE (0.02) | 2.63 | 2.55 | no |
| 16 | F | 3 years | NAV2 | NM_145117.5 | c.5011_5012del | het | maternal | (NAV2)x25% | frameshift variants likely cause NMD | AE (0.25) | 3.04 | 8.67 | no |
| c.6580del | het | paternal | |||||||||||
| 17 | M | 2 years | RBM28 | NM_018077.3 | c.1745G>A | het | maternal | r.1714_1788del | variant likely destroys exon splicing enhancer and cause skipping of exon 16 (75 nt) | AE(0.73), AS | 5.26 | 5.37 | no |
| c.1489_1492dup | het | paternal | (RBM28)x73% | frameshift variants likely causes NMD | AE(0.73), MAE | ||||||||
| 18 | F | 9 years | FBN1 | NM_000138.5 | c.248−151A>G | het | paternal | r.247_248ins[248–282_248−152] | inclusion of cryptic exon (131 nt) with premature stop codon likely causing NMD | AE (0.58), AS (low abundance due to NMD), MAE | 524.4 | 517.6 | no |
Abbreviations: AE, abnormal expression; AS, abnormal splicing; MAE, mono-allelic expression; NMD, nonsense-mediated decay.
The reduction of TIAM1 expression is considered as a possible molecular diagnosis with weaker evidence compared to all other variants in this table. This is because of a lack of the matching DNA variant underlying the expression change. Three rare variants in TIAM1 are listed. However, their causal relationship with the expression reduction is uncertain.
The FBN1 variant is considered as a partial molecular diagnosis, which may partly explain the Marfanoid phenotype in this individual, but not the neurological phenotypes.
The POLR3A variants are considered as a possible molecular diagnosis because of the atypical phenotypic match.
TMEM161B54 and MYCBP255 are recently identified disease-associated genes not included in disease-associated gene databases such as OMIM at the time of the analysis. Therefore, the findings in these two individuals are considered investigational.
The phenotypes of each proband are summarized as Human Phenotype Ontology (HPO) terms (Data S2).
Content in parentheses following AE indicates the expression fold change in this individual; the content in parentheses following AS indicates that the splicing event escaped detection from the analysis pipeline but was rescued by manual analysis with special considerations.
The iNeuron process is required to activate the gene or isoform of interest to facilitate variant interpretation.
RNA-seq analyses revealed 10 aberrant splicing events involving ITPR1 (MIM: 147265), DCX (MIM: 300121), MBD5, LZTR1 (Figures 3C and S7A), USP9X (Figures 3D and S7B), BRAF (Figure 3E), TMEM161B (Figures 3F and S7C), POGZ (MIM: 614787) (Figures 3G and S8A), VARS1 (Figures 3H and S8B), and RBM28 (Figures 3I and S8C) (see further details of ITPR1, DCX, and MBD5 below). Notably, the DNA-directed analysis revealed 5 aberrant splicing events predicted by SpliceAI that were not detected by FRASER. Three of the missed variants were explained by a low abundance of the abnormal transcript, which is possibly caused by degradation from nonsense-mediated decay (NMD) and/or the leakiness of the splice variant (i.e., POLR3A [MIM: 614258] and FBN1, Figure S9). In addition, misalignment (as well as NMD) contributed to one event (PIEZO2) being missed by the DROP pipeline, in which sequencing reads supporting the abnormal splice acceptor site were misaligned into the normal splice acceptor site plus an adjacent indel (Figure S10). In total, 12 aberrant expression and 13 aberrant splicing events contributed to the final molecular diagnoses of the participants (Table 3).
The application of the RNA-directed and the DNA-complemented analysis approach assisted in the genetic diagnosis of 18 in probands presenting with various neurological disorders that were enrolled in the UDN, accounting for 25.4% of the total cohort (Figure 3J). The diagnostic findings included four with aberrant expression, five with aberrant splicing, and eight with both aberrant expression and splicing. Furthermore, one participant (CACNA1A [MIM: 601011]) displayed an unbalanced expression of a heterozygous missense variant allele, potentially indicating the presence of another modifying variant that requires confirmation through WGS. The variant types and inheritance for the diagnostic findings include seven de novo variants, seven sets of compound heterozygous variants, and five variants inherited from a parent or of unknown inheritance (Table 3). Notably, the de novo translocation resulted in a truncated BRAF protein lacking the kinase domain, and the protocadherin-alpha gene cluster on chromosome 5 was also identified as a possible candidate for further study (Figure S11).
Neuron induction is required for the identification of the molecular diagnoses in 27.7% (5/18) of probands, with four individuals benefiting from activations of the expression of neuronal genes, while one from the activation of a neuron-specific isoform (Table 3). This methodology demonstrated a range of diagnostic rates in individuals with various neurological phenotypes. There was a notably enhanced molecular diagnostic yield in individuals with brain malformation (37%, 11/30), intellectual disability (30%, 14/46), and epilepsy (29%, 9/31); other less represented phenotypic groups on their list of phenotypes include ADHD (67%, 2/3), autism spectrum disorder (ASD; 50%, 2/4), eye diseases (36%, 4/11), and hypotonia (30%, 7/23) (Figure 3J).
Molecular diagnoses achieved through iNeuron RNA-seq by activation of neurological disease-associated genes
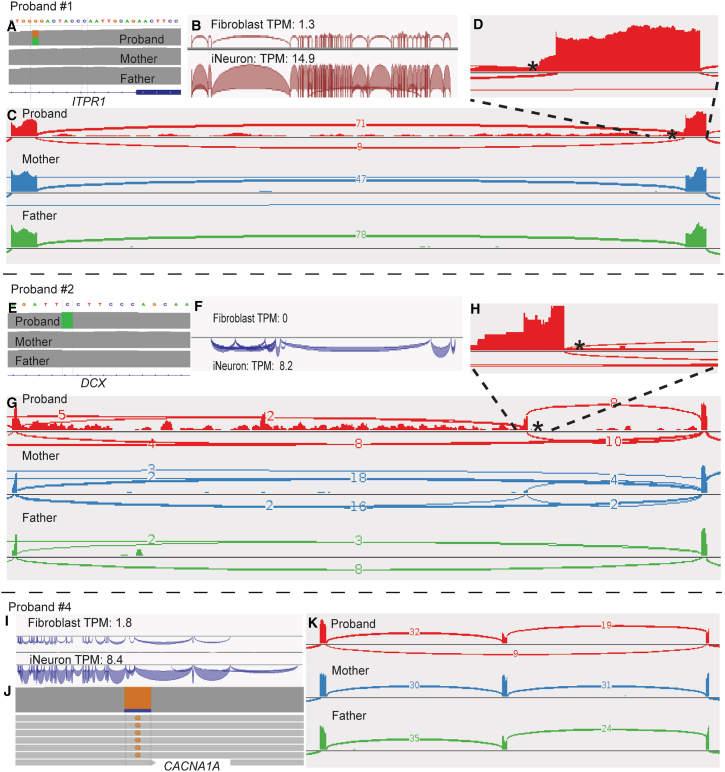
iNeuron RNA-seq conferred a significant advantage in enhancing the expression of OMIM-N genes, confirming the molecular diagnoses in five probands. In subject #1, FRASER analysis identified aberrant splicing in ITPR1 in a 4-year-old male with a history of congenital iris hypoplasia, delays in gross motor development, hypotonia, ataxia, and mild dysmorphic features. Trio GS analysis confirmed a novel de novo heterozygous intronic variant in ITPR1 (GenBank: NM_001378452.1; c.5980−17G>A), which is predicted to have a moderate impact on splicing (SpliceAI score 0.57) (Figure 4A). ITPR1 is not well expressed in fibroblasts; RNA-seq on fibroblasts has previously been attempted but yielded inconclusive results due to the low expression of ITPR1.56 Upon neuron induction, the expression of ITPR1 increased 12-fold (Figures 4B and S12A), allowing for a high-confidence detection of the 15-nucleotide retention from intron 45 in approximately 50% of the reads (Figures 4C, 4D, and S12A). The result of the 15-nucleotide retention is consistent with that obtained previously by targeted PCR amplification and Sanger sequencing from the cDNA.56 Notably, retention of the entire intron 45 was observed at a low level in the proband iNeruon RNA-seq data (Figure 4C), which is a finding that has not been previously observed from the fibroblast RNA results. Defects in ITPR1 cause Gillespie syndrome (MIM: 206700) with either autosomal-dominant or autosomal-recessive inheritance, through distinct molecular mechanisms. The c.5980−17G>A variant was presumed to have a dominant-negative effect on ITPR1 channel function, leading to the autosomal dominant form of Gillespie syndrome.56,57,58 This assumption was made because (1) the variant is de novo and (2) no additional rare variants were found using our analysis pipeline in the DNA sequencing data that can contribute to a bi-allelic model. The precise RNA-level splicing consequences provided by iNeuron RNA-seq enable further potential investigations into the molecular mechanism and disease inheritance of the ITPR1 defect in this family, which is required for downstream studies if the family were interested in pursuing molecular therapies such as antisense oligonucleotide therapy.
Figure 4.
Molecular diagnoses achieved by iNeuron RNA-seq through elevated expression of OMIM-N genes
(A) Trio WGS reveals a de novo heterozygous intronic variant in ITPR1 (GenBank: NM_001378452.1; c.5980−17G>A).
(B) Significant increase in expression of ITPR1 after transdifferentiation.
(C) iNeuron RNA-seq demonstrates a 15-nucleotide retention from intron 45 in approximately 50% of the reads, as well as complete intron 45 retention in a small fraction of reads.
(D) Zoomed in view of the 15-nucleotide retention.
(E) Trio WGS identifies a novel hemizygous deep intronic variant (GenBank: NM_001195553.2; c.946+4588G>T) in DCX.
(F) DCX is not expressed in fibroblasts but is activated in iNeurons.
(G) Sashimi plot displays inclusion of a 13,549 bp cryptic exon from intron, positioned −31 bp from the intronic variant of DCX.
(H) Zoomed-in view of the new splice junction at the intron retention.
(I) Significant increase in expression of CACNA1A after transdifferentiation.
(J) Skewed expression of the variant allele in CACNA1A.
(K) Sashimi plot reveals exon skipping located 124 kb away from the missense variant. Variants are indicated by asterisks.
iNeuron RNA-seq also enables the detection of defects in neurological genes not expressed in fibroblasts. In subject #2, FRASER detected an aberrant splicing event in DCX, a neuron-specific gene that plays essential roles in neuronal migration and establishment of the six-layer organization in the cerebral cortex.59,60,61 This 23-year-old male presented with a history of intellectual disability, epilepsy, and cortical malformation. Analysis of whole blood WGS data revealed a novel de novo deep intronic variant (GenBank: NM_001195553.2; c.946+4588G>T) (Figure 4E). Defects in DCX have been associated with neuronal migration disorders, lissencephaly, X-linked (MIM: 300067). Males with DCX-related lissencephaly typically have profound intellectual disabilities, developmental delay, epileptic seizures, and cerebral palsy.62,63 SpliceAI suggested a possible splicing effect of the c.946+4588G>T variant but with a low score of 0.11, which is considered uninformative and would result in the variant being filtered out in most DNA-based analysis pipelines. RNA-seq analysis is warranted to clarify the splicing consequence and thus the clinical significance of the deep intronic variant. However, DCX is not expressed in fibroblasts (Figures 4F and S12B). iNeuron RNA-seq was performed, which resulted in sufficient detection of DCX expression. RNA-seq uncovered retention of 13,549 nt intronic sequences, located at −31 bp from the intronic variant, which is expected to create an out-of-frame cryptic exon and lead to a frameshift consequence (Figures 4G, 4H, and S12B). The RNA-seq data serve as key supporting evidence to classify the c.946+4588G>T variant as likely pathogenic. Unexpectedly, from the mother’s iNeurons RNA-seq data, we observed a small number of abnormal junction reads. Re-inspection of the WGS data from her whole blood showed no abnormal reads at 64× coverage. The discrepancy led us to suspect potential tissue-limited mosaicism that manifests in the mother’s fibroblast. We performed amplicon-based next-generation sequencing on the mother’s fibroblast. The result revealed a variant at 29% fraction in DNA from fibroblasts and 2% in DNA from blood, implicating that the family is at risk for gonadal mosaicism for the likely pathogenic DCX variant (Figure S13).
iNeuron RNA-seq revealed an event of allele-specific expression, contributing to a more comprehensive understanding of the molecular disease mechanisms. Proband #4 is a 7-year-old female presenting with severe ataxia, progressive cerebellar hypoplasia, hypotonia, and global developmental delays. ES analysis revealed a de novo heterozygous missense variant in CACNA1A (GenBank: NM_001127222.2; c.5015G>C [p.Arg1672Pro]), which is considered to be causative because of the broad phenotypic match. However, this participant was considered to be on the most severe end of the clinical spectrum for CACNA1A-related disorders, especially when compared to a series of other individuals with similar de novo missense CACNA1A variants; furthermore, this participant did not respond to acetazolamide treatment, which is effective in therapy to other individuals with CACNA1A defects.64,65,66 Functional studies performed in a fly model gleaned insights into the molecular pathology mechanisms on a protein level, suggesting a gain-of-function mechanism.64
RNA-seq analysis from individual-derived cell lines is needed to provide a perspective of the disease presentation mechanism from a transcriptional level. Fibroblast-derived RNA-seq data were deemed uninformative due to the low expression of CACNA1A (TPM = 1.8) (Figure 4I). The TPM of CACNA1A was boosted to 8.4 in iNeurons. RNA-seq on the iNeurons showed an unexpected skewed variant fraction of 88% for the c.5015G>C variant, indicating an overexpression of the variant allele or a reduced expression of the reference allele (Figures 4J and S12C). Intriguingly, an exon skipping located 124 kb away from the missense variant was identified by FRASER (Figures 4K and S12D). The skipped exon cannot be phased with the missense variant based on the current short-read data, but the combined observations warrant additional investigations to pinpoint to potential cis or trans modifiers that modulate the disease phenotype variability and therapy response.
Activation of neuron-specific isoforms increases the molecular diagnostic yield
When evaluating the expression of a gene in a tissue or a sample, most current researchers typically calculate the TPM value on a gene level, rather than breaking it down into isoform-level values. In the GTEx portal, the gene-level TPMs are computed from RNA-SeQC, which uses an artificial collapsed transcript as the gene basis of the calculation. When a tissue-specific isoform is critical for disease pathogenesis but differs little from other ubiquitous isoforms, the presence or absence of a tissue-specific transcript can be unintentionally masked if the gene-level TPM is used in the analysis.
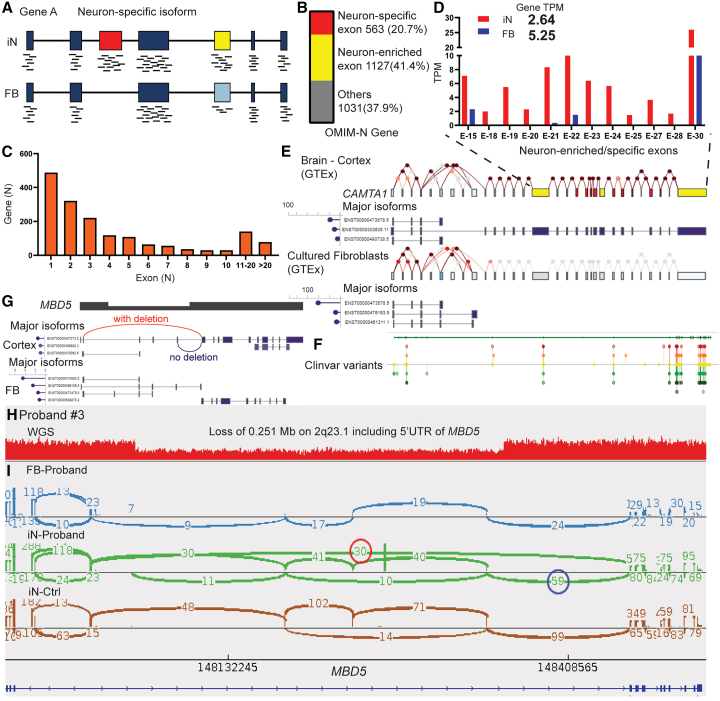
We hypothesize that the iNeuron RNA-seq platform provides an advantage over fibroblast analysis by enhancing neuron-specific isoform expressions. Given the challenges in accurately reconstructing full-length isoforms from our short-read RNA-seq data, we limited the analysis to tissue-specific isoforms that are tagged by tissue-specific exons, i.e., a tissue-specific isoform encompassing one or more exons that are differentially expressed compared to other exons in that tissue (Figure 5A). Comparative analysis of our iNeuron and fibroblast data pools revealed 41,473 neuron-enriched exons from 11,420 genes. Notably, we found that 62.1% (1,690/2,721) of the OMIM-N genes had at least one activated exon-tagged isoform, totaling 9,356 exons (Figure 5B), with a median of 3 exons per gene. About three-quarters (74.3%) of these identified genes contained 1–5 neuron-enriched exons (Figure 5C). Furthermore, we identified 936 neuron-specific exons (from 563 genes) from the neuron-enriched OMIM-N genes (Figure 5B, Data S3). These exons demonstrated low expression in fibroblasts but were activated following neuron induction, highlighting the exclusive detection of these exons and associated junctions in iNeurons. For example, although fibroblasts have higher CAMTA1 (MIM: 611501) TPM values than iNeurons on the gene level (5.25 and 2.64 respectively), exon level expression analysis reveals that multiple neuron-specific exons are successfully activated in our iNeuron data for CAMTA1 (Figure 5D). These exons are exclusively expressed in the brain, as evidenced by GTEx data (Figure 5E). Activation of these exons is critical for disease-oriented analysis, as demonstrated by the clustering of clinically relevant variants reported on ClinVar (Figure 5F). The capability to activate neuron-specific isoforms positions iNeurons as a valuable diagnostic tool for previously unresolved challenging cases.
Figure 5.
Enhanced molecular diagnosis with iNeuron RNA-seq through detection of neuron-enriched exons
(A) Schematic diagram illustrating that neuron-enriched and/or neuron-specific isoforms can be detected in iNeurons.
(B) Identification from iNeuron RNA-seq of 9,356 neuron-enriched exons (from 1,690 OMIM-N genes), including 936 neuron-specific exons (from 563 OMIM-N genes). The term “other” refers to the remaining OMIM-N genes that do not include neuron-enriched exons.
(C) Distribution of OMIM-N genes based on the number of neuron-enriched exons when comparing iNeurons with fibroblasts.
(D–F) The neuron-specific isoform in CAMTA1 is detected by iNeuron RNA-seq. The expected distribution of neuron- and fibroblast-specific isoforms are illustrated in (E) based on data from GTEx. Multiple exons toward the end of the transcript are shown to be highly enriched (yellow) or specific (red) to neurons. Interrogation of these exons in the neuronal isoform is important as illustrated by the distribution of clinically relevant variants from ClinVar in (F). Exon-level RNA-seq data from iNeurons, summarized in (D), demonstrate successful activation of the neuron-enriched and neuron-specific exons. Note that gene-level TPM (2.64 for iNeurons and 5.25 for fibroblasts) can disguise the activation of tissue-specific isoforms.
(G) Schematic showing the aberrant junction (red) caused by the 5′ UTR deletion of MBD5, which can be detected only in the neuron-specific long isoform. The alternative junction (blue) corresponds to the normal allele with no deletion.
(H) Detection of a heterozygous deletion (0.25 Mb) on 2q23.1, involving noncoding exons of MBD5 through WGS.
(I) Sashimi plot depicting the presence of abnormal junctions (red circle) exclusively in proband’s iNeurons, but not in fibroblasts. The number of junction reads in the red and the blue circles represent abundance of alleles with and without the deletion.
In our cohort, a molecular diagnosis was achieved in MBD5 leveraging the detection of the neuron-specific isoform from the iNeuron data; interpretations using the fibroblast RNA-Seq alone would have resulted in a potentially misleading conclusion. Subject #3 is a 4-year-old girl with seizures, abnormal brain MRI results, dysmorphic features, unusual metabolic profiles, and multiple members on the maternal side of the family with behavioral issues. WGS revealed a maternally inherited heterozygous deletion of exons 2–4 (NM_001378120.1) within the 5′ UTR region of MBD5 (Figures 5G and 5H). Haploinsufficiency of MBD5 has been linked to neurodevelopmental disorders characterized by intellectual disability, developmental delay, seizures, and abnormal behavioral features (Intellectual developmental disorder, autosomal dominant 1 [MIM: 156200]).67,68,69 However, the consequence of this noncoding deletion is unclear. RNA-Seq was performed on the proband fibroblasts. While MBD5 is expressed at an adequate level in fibroblasts (TPM = 8.98), we did not detect any gross expression change or abnormal junctions. The absence of gross expression change suggests that the deletion has either no effect or a mild effect on the transcription of MBD5. The apparent lack of abnormal junctions conflicted with the presence of the deletion, leading to confusions in the overall data interpretation.
Analysis of MBD5 expression data on GTEx revealed that a long isoform predominates in human brains, whereas several shorter isoforms constitute the expression of MBD5 in cultured fibroblasts (Figure S14). Only when an isoform is long enough to span over the deletion are the abnormal junctions expected to be formed. As such, we predict that (1) the different compositions of isoforms in neurons versus fibroblasts result in different shortened transcripts caused by the deletion, and (2) it is likely more meaningful to study the impact on the long neuronal isoform to evaluate the clinical significance of the deletion. We analyzed the iNeuron RNA-Seq from our cohort and identified 12 neuron-enriched exons in MBD5, which is consistent with expectations from the GTEx neuronal isoform (ENST00000407073.5) and suggests activation of the desired transcript in iNeurons (Figure S14). After neuronal induction, the abnormal junctions representing the deletion can now be detected in Subject #3 by FRASER. An abnormal expression of MBD5 is also detected by OUTRIDER, with a moderate fold change at 0.84 (Figure 3B). Leveraging the fact that iNeurons have the long neuronal isoform encompassing the deletion, we can phase the transcripts to the allele with deletion and the other allele without, which facilitates expression quantification on the specific allele with deletion (Figure 5G). Phasing the reads can help estimating the expression change specific to the deletion allele. By comparing the read counts from the two alleles with each other, we noted that the maternal allele with the deletion [30 junction reads (red circle)] has reduced expression compared to the paternal allele without the deletion [59 junction reads (blue circle)] (Figures 5G–5I). This implicates that the 5′UTR deletion possibly led to a mild expression decrease of MBD5. Taken together, the activation of a neuron-specific isoform in Subject #3 resolved misleading negative data from fibroblast RNA-Seq, contextualized disease mechanism interpretation with tissue-appropriate isoforms, and provided an allele-specific strategy to quantify the mild expression reduction.
Discussion
In this study, we established an iNeuron RNA-Seq diagnostic workflow that successfully activated neuronal genes and yielded results of potential diagnostic value in 25.4% (18/71) in a cohort of individuals presenting with various neurological disorders. The creation of iNeurons was crucial in five of these individuals, as the critical gene/isoform had low to no expression in fibroblasts. In an attempt to benchmark the degree of neuronal conversion, we compiled a list of genes linked to Mendelian disorders with a neurological phenotypic component and concluded that about half of these genes with a low expression can be effectively activated. The iNeurons used in our experiments represent an artificial “neuron-like” cell line with a snapshot at a specific time point, which cannot reconstitute the granularity of gene expressions from different neural or neuronal subtypes of cells and the temporospatial complexity of their arrangement in the human developmental brain.70,71,72,73,74 Notably, a subset of genes associated with neurological phenotypes, particularly those expressed in glial cells and neural progenitor cells, and region-specific genes, such as those found in the midbrain, hindbrain, and spinal cord, were not well activated (data not shown). In addition, a small number of genes are not bona fide “neuronal” but ended up in the neurological OMIM gene list because they cause neurological phenotypes secondarily. We envision that several strategies can be implemented to improve the scale of neurological gene coverage in a transdifferentiation RNA-seq workflow, such as optimization of the current protocol, complementation from alternative cell conversion protocols targeting different neural cell types, reprogramming into neural progenitor cells and branching the differentiation into various neural cells, and collection at different differentiation time points.
Although alternative splicing is particularly ubiquitous and highly conserved in the nervous system tissues of vertebrates, and has been associated with various neurological diseases,75,76,77,78 few causal relationships have been established between particular neuronal isoforms and Mendelian disorders. We demonstrate that neuron-specific isoforms can be effectively activated from the neuron induction. The MBD5 5′ UTR deletion illustrates that accessibility to neuron-specific isoforms directly benefits molecular diagnosis and potentially enables exploration into unrecognized disease mechanisms. The successful reconstitution of neuron-specific isoforms, although challenging to benchmark with the current knowledge, provides assurance that not only genetics but also epigenetics is reasonably established to model the disease mechanism in the personalized cellular environment. As more experience is accumulated regarding the efficacy and safety of implementing the iNeuron diagnostic workflow, we reason that attempts can be made to utilize the individual-derived iNeuron cell lines for molecular diagnosis stratification and therapy screening.
In contrast to iPSC-derived neurons, the transdifferentiation approach involves fewer induction steps, skips the stem cell-like stage, and better preserves the epigenetic signatures.33,45,79,80,81 It holds significant potential for disease modeling, personalized drug testing, and the development of autologous cell therapy.79,80 However, its use in clinical diagnostics is still in its early stages. One of our primary motivations in developing the iNeuron RNA-Seq workflow is to ensure its suitability for implementation into a high-volume clinical diagnostic operation. In addition to showing the clinical utility and technical robustness, it is crucial to demonstrate that the protocol is well designed so testing laboratories can properly manage the turnaround time, consumable cost, and technologists’ training requirements. Compared to the conventional approach to differentiate neuronal cells through iPSC reprogramming, the cell culture of the transdifferentiation method can be accomplished within eight weeks – substantial time is saved by omitting the intermediate iPSC procedures, although further shortening of the processing time would still make it more attractive to clinical users. The shortened cell culture time translates to direct savings in the culture consumables; the per use cost of cell culture reagents is also lower than that of the iPSC culture. The overall cost of cell culture is substantially lower than that from the iPSC-neuron differentiation approach. In our experience, the per sample cell culture reagent cost is approximately $400. Collectively, the reasonable processing time and the low reagent cost opens a viable channel for clinical laboratories to consider adopting the transdifferentiation workflow.
Conventional iPSC reprogramming and neuronal differentiation requires specialized training for technologists to pick colonies, maintain pluripotency, and select neural progenitor cells. The procedure of transdifferentiation can be accomplished with minimum cell culture experience.80,81 Furthermore, we established QC metrics to provide transparent measurements of the quality and reproducibility of the iNeuron generations, which alleviates the pressures for clinical laboratory technologists to make experience-based judgements. Our recommended QC checkpoints include qPCR analysis for ASCL1 expression and computation of an iNeuron score from RNA-seq data. These QC assays provide results that are easily quantifiable and replaces the more labor-intensive approaches such as immunostaining. In practice, implementation of the QC checkpoints can effectively rule out potentially failing samples and thus save time and reagent/sequencing cost.
Introducing cellular engineering into the diagnostic workflow provides a functional and tissue-informed view of rare disease-causing variants in a non-invasive manner, which represents a unique opportunity to realize personalized precision medicine in a Petri dish. Our proof-of-concept study on neurological genetic disorders serves as a framework that can be applied to target other genetic disorders, such as targeting cardiovascular disorders through cardiomyocyte transdifferentiation.82,83 In the past decade, the rapid development of medical genomics has fueled the widespread implementation of laboratory genomics sequencing, which generated a tremendous wealth of clinical big data that has propelled many discoveries in genomic science. Individual-derived cell manipulation is the reasonable next step for clinical genetics laboratories to pursue to improve patient care, and, if implemented clinically at a large scale, can generate paradigm-shifting large data to transform the study of genomic sciences.
Data and code availability
UDN sequencing data are available through dbGaP (accession: phs001232.v2.p1) and the UDN Gateway. Phenotype data with flagged genes of interest have been submitted to Phenome Central. DNA variants thought to contribute to the molecular diagnoses of the patients have been submitted to ClinVar.
Acknowledgments
The research was supported by National Institutes of Health Common Fund (U01HG007709 and U01HG007942) and the grant from National Human Genome Research Institute (R35HG011311). The project was also supported by the BCM Intellectual and Developmental Disabilities Research Center that is funded by the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health under Award Number P50HD103555 for use of the Human Stem Cell and Neuronal Differentiation Core facility and the Clinical Translational Core facility. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
Conceptualization, S. Li, J.C.S., P.L.; methodology, S. Li, S.Z., J.C.S., A.B.; investigation, S. Li, S.Z., J.C.S., J.S., M.T.T.N., A.K., P.P.H., M.P.; resources, K.C.W., L.C.B., M.W.-H., S.K., W.J.C., G.D.C., S. Lalani, C.A.B., K.M., H.-T.C., L.P., L.E., C.M.E., B.L.; data curation, S. Li, S.Z., J.C.S., M.B.N., Z.L.; visualization: S. Li, S.Z., J.C.S.; funding acquisition, P.L., C.M.E., B.L.; project administration, J.A.R., S. Li, J.C.S., C.M.E., B.L.; supervision, P.L., J.A.R., A.B., S.C.S.N., C.M.E., B.L.; writing – original draft, S. Li, P.L.; writing – review & editing, S. Li, P.L., S.Z., J.A.R., A.B., C.M.E., B.L., H.-T.C., K.C.W., K.M., G.D.C.
Declaration of interests
Baylor College of Medicine (BCM) and Miraca Holdings Inc. have formed a joint venture with shared ownership and governance of Baylor Genetics (BG), which performs genetic testing and derives revenue. P.L. and C.M.E. are employees of BCM and derive support through a professional services agreement with BG.
Published: April 8, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2024.03.007.
Web resources
OMIM, https://www.omim.org/
Supplemental information
References
- 1.Liu P., Meng L., Normand E.A., Xia F., Song X., Ghazi A., Rosenfeld J., Magoulas P.L., Braxton A., Ward P., et al. Reanalysis of Clinical Exome Sequencing Data. N. Engl. J. Med. 2019;380:2478–2480. doi: 10.1056/NEJMc1812033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Y., Muzny D.M., Reid J.G., Bainbridge M.N., Willis A., Ward P.A., Braxton A., Beuten J., Xia F., Niu Z., et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N. Engl. J. Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Splinter K., Adams D.R., Bacino C.A., Bellen H.J., Bernstein J.A., Cheatle-Jarvela A.M., Eng C.M., Esteves C., Gahl W.A., Hamid R., et al. Effect of Genetic Diagnosis on Patients with Previously Undiagnosed Disease. N. Engl. J. Med. 2018;379:2131–2139. doi: 10.1056/NEJMoa1714458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wright C.F., Campbell P., Eberhardt R.Y., Aitken S., Perrett D., Brent S., Danecek P., Gardner E.J., Chundru V.K., Lindsay S.J., et al. Genomic Diagnosis of Rare Pediatric Disease in the United Kingdom and Ireland. N. Engl. J. Med. 2023;388:1559–1571. doi: 10.1056/NEJMoa2209046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lionel A.C., Costain G., Monfared N., Walker S., Reuter M.S., Hosseini S.M., Thiruvahindrapuram B., Merico D., Jobling R., Nalpathamkalam T., et al. Improved diagnostic yield compared with targeted gene sequencing panels suggests a role for whole-genome sequencing as a first-tier genetic test. Genet. Med. 2018;20:435–443. doi: 10.1038/gim.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shashi V., Schoch K., Spillmann R., Cope H., Tan Q.K.-G., Walley N., Pena L., McConkie-Rosell A., Jiang Y.-H., Stong N., et al. A comprehensive iterative approach is highly effective in diagnosing individuals who are exome negative. Genet. Med. 2019;21:161–172. doi: 10.1038/s41436-018-0044-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee H., Deignan J.L., Dorrani N., Strom S.P., Kantarci S., Quintero-Rivera F., Das K., Toy T., Harry B., Yourshaw M., et al. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA. 2014;312:1880–1887. doi: 10.1001/jama.2014.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monies D., Abouelhoda M., Assoum M., Moghrabi N., Rafiullah R., Almontashiri N., Alowain M., Alzaidan H., Alsayed M., Subhani S., et al. Lessons Learned from Large-Scale, First-Tier Clinical Exome Sequencing in a Highly Consanguineous Population. Am. J. Hum. Genet. 2019;105:879. doi: 10.1016/j.ajhg.2019.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Sanden B.P.G.H., Schobers G., Corominas Galbany J., Koolen D.A., Sinnema M., van Reeuwijk J., Stumpel C.T.R.M., Kleefstra T., de Vries B.B.A., Ruiterkamp-Versteeg M., et al. The performance of genome sequencing as a first-tier test for neurodevelopmental disorders. Eur. J. Hum. Genet. 2023;31:81–88. doi: 10.1038/s41431-022-01185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Ligt J., Willemsen M.H., van Bon B.W.M., Kleefstra T., Yntema H.G., Kroes T., Vulto-van Silfhout A.T., Koolen D.A., de Vries P., Gilissen C., et al. Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 11.Jaganathan K., Kyriazopoulou Panagiotopoulou S., McRae J.F., Darbandi S.F., Knowles D., Li Y.I., Kosmicki J.A., Arbelaez J., Cui W., Schwartz G.B., et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell. 2019;176:535–548.e24. doi: 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 12.Sundaram L., Gao H., Padigepati S.R., McRae J.F., Li Y., Kosmicki J.A., Fritzilas N., Hakenberg J., Dutta A., Shon J., et al. Predicting the clinical impact of human mutation with deep neural networks. Nat. Genet. 2018;50:1161–1170. doi: 10.1038/s41588-018-0167-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ioannidis N.M., Rothstein J.H., Pejaver V., Middha S., McDonnell S.K., Baheti S., Musolf A., Li Q., Holzinger E., Karyadi D., et al. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am. J. Hum. Genet. 2016;99:877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stenson P.D., Mort M., Ball E.V., Evans K., Hayden M., Heywood S., Hussain M., Phillips A.D., Cooper D.N. The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum. Genet. 2017;136:665–677. doi: 10.1007/s00439-017-1779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soemedi R., Cygan K.J., Rhine C.L., Wang J., Bulacan C., Yang J., Bayrak-Toydemir P., McDonald J., Fairbrother W.G. Pathogenic variants that alter protein code often disrupt splicing. Nat. Genet. 2017;49:848–855. doi: 10.1038/ng.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cummings B.B., Marshall J.L., Tukiainen T., Lek M., Donkervoort S., Foley A.R., Bolduc V., Waddell L.B., Sandaradura S.A., O’Grady G.L., et al. Improving genetic diagnosis in Mendelian disease with transcriptome sequencing. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aal5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kremer L.S., Bader D.M., Mertes C., Kopajtich R., Pichler G., Iuso A., Haack T.B., Graf E., Schwarzmayr T., Terrile C., et al. Genetic diagnosis of Mendelian disorders via RNA sequencing. Nat. Commun. 2017;8 doi: 10.1038/ncomms15824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonorazky H.D., Naumenko S., Ramani A.K., Nelakuditi V., Mashouri P., Wang P., Kao D., Ohri K., Viththiyapaskaran S., Tarnopolsky M.A., et al. Expanding the Boundaries of RNA Sequencing as a Diagnostic Tool for Rare Mendelian Disease. Am. J. Hum. Genet. 2019;104:466–483. doi: 10.1016/j.ajhg.2019.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frésard L., Smail C., Ferraro N.M., Teran N.A., Li X., Smith K.S., Bonner D., Kernohan K.D., Marwaha S., Zappala Z., et al. Identification of rare-disease genes using blood transcriptome sequencing and large control cohorts. Nat. Med. 2019;25:911–919. doi: 10.1038/s41591-019-0457-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murdock D.R., Dai H., Burrage L.C., Rosenfeld J.A., Ketkar S., Müller M.F., Yépez V.A., Gagneur J., Liu P., Chen S., et al. Transcriptome-directed analysis for Mendelian disease diagnosis overcomes limitations of conventional genomic testing. J. Clin. Invest. 2021;131 doi: 10.1172/JCI141500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yépez V.A., Gusic M., Kopajtich R., Mertes C., Smith N.H., Alston C.L., Ban R., Beblo S., Berutti R., Blessing H., et al. Clinical implementation of RNA sequencing for Mendelian disease diagnostics. Genome Med. 2022;14:38. doi: 10.1186/s13073-022-01019-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee H.-F., Chi C.-S., Tsai C.-R. Diagnostic yield and treatment impact of whole-genome sequencing in paediatric neurological disorders. Dev. Med. Child Neurol. 2021;63:934–938. doi: 10.1111/dmcn.14722. [DOI] [PubMed] [Google Scholar]
- 25.Dekker J., Schot R., Bongaerts M., de Valk W.G., van Veghel-Plandsoen M.M., Monfils K., Douben H., Elfferich P., Kasteleijn E., van Unen L.M.A., et al. Web-accessible application for identifying pathogenic transcripts with RNA-seq: Increased sensitivity in diagnosis of neurodevelopmental disorders. Am. J. Hum. Genet. 2023;110:251–272. doi: 10.1016/j.ajhg.2022.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oquendo C.J., Wai H.A., Rich W., Bunyan D.J., Thomas N.S., Hunt D., Lord J., Douglas A.G.L., Baralle D. RNA sequencing uplifts diagnostic rate in undiagnosed rare disease patients. medRxiv. 2023 doi: 10.1101/2023.07.05.23292254. Preprint at. [DOI] [Google Scholar]
- 27.Maddirevula S., Kuwahara H., Ewida N., Shamseldin H.E., Patel N., Alzahrani F., AlSheddi T., AlObeid E., Alenazi M., Alsaif H.S., et al. Analysis of transcript-deleterious variants in Mendelian disorders: implications for RNA-based diagnostics. Genome Biol. 2020;21:145. doi: 10.1186/s13059-020-02053-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aicher J.K., Jewell P., Vaquero-Garcia J., Barash Y., Bhoj E.J. Mapping RNA splicing variations in clinically accessible and nonaccessible tissues to facilitate Mendelian disease diagnosis using RNA-seq. Genet. Med. 2020;22:1181–1190. doi: 10.1038/s41436-020-0780-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B. Alternative Isoform Regulation in Human Tissue Transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bj B. Alternative splicing: new insights from global analyses. Cell. 2006;126 doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 31.Wang H., Yang Y., Liu J., Qian L. Direct cell reprogramming: approaches, mechanisms and progress. Nat. Rev. Mol. Cell Biol. 2021;22:410–424. doi: 10.1038/s41580-021-00335-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi K., Yamanaka S. A decade of transcription factor-mediated reprogramming to pluripotency. Nat. Rev. Mol. Cell Biol. 2016;17:183–193. doi: 10.1038/nrm.2016.8. [DOI] [PubMed] [Google Scholar]
- 33.Herdy J., Schafer S., Kim Y., Ansari Z., Zangwill D., Ku M., Paquola A., Lee H., Mertens J., Gage F.H. Chemical modulation of transcriptionally enriched signaling pathways to optimize the conversion of fibroblasts into neurons. Elife. 2019;8 doi: 10.7554/eLife.41356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeLuca D.S., Levin J.Z., Sivachenko A., Fennell T., Nazaire M.-D., Williams C., Reich M., Winckler W., Getz G. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28:1530–1532. doi: 10.1093/bioinformatics/bts196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Graubert A., Aguet F., Ravi A., Ardlie K.G., Getz G. RNA-SeQC 2: efficient RNA-seq quality control and quantification for large cohorts. Bioinformatics. 2021;37:3048–3050. doi: 10.1093/bioinformatics/btab135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bařinka J., Hu Z., Wang L., Wheeler D.A., Rahbarinia D., McLeod C., Gu Z., Mullighan C.G. RNAseqCNV: analysis of large-scale copy number variations from RNA-seq data. Leukemia. 2022;36:1492–1498. doi: 10.1038/s41375-022-01547-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suvakov M., Panda A., Diesh C., Holmes I., Abyzov A. CNVpytor: a tool for copy number variation detection and analysis from read depth and allele imbalance in whole-genome sequencing. GigaScience. 2021;10:giab074. doi: 10.1093/gigascience/giab074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yépez V.A., Mertes C., Müller M.F., Klaproth-Andrade D., Wachutka L., Frésard L., Gusic M., Scheller I.F., Goldberg P.F., Prokisch H., Gagneur J. Detection of aberrant gene expression events in RNA sequencing data. Nat. Protoc. 2021;16:1276–1296. doi: 10.1038/s41596-020-00462-5. [DOI] [PubMed] [Google Scholar]
- 40.Chen Z., Zheng Y., Yang Y., Huang Y., Zhao S., Zhao H., Yu C., Dong X., Zhang Y., Wang L., et al. PhenoApt leverages clinical expertise to prioritize candidate genes via machine learning. Am. J. Hum. Genet. 2022;109:270–281. doi: 10.1016/j.ajhg.2021.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li B., Ruotti V., Stewart R.M., Thomson J.A., Dewey C.N. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics. 2010;26:493–500. doi: 10.1093/bioinformatics/btp692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vasan L., Park E., David L.A., Fleming T., Schuurmans C. Direct Neuronal Reprogramming: Bridging the Gap Between Basic Science and Clinical Application. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.681087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Immaneni A., Lawinger P., Zhao Z., Lu W., Rastelli L., Morris J.H., Majumder S. REST-VP16 activates multiple neuronal differentiation genes in human NT2 cells. Nucleic Acids Res. 2000;28:3403–3410. doi: 10.1093/nar/28.17.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Victor M.B., Richner M., Hermanstyne T.O., Ransdell J.L., Sobieski C., Deng P.-Y., Klyachko V.A., Nerbonne J.M., Yoo A.S. Generation of human striatal neurons by microRNA-dependent direct conversion of fibroblasts. Neuron. 2014;84:311–323. doi: 10.1016/j.neuron.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herdy J.R., Traxler L., Agarwal R.K., Karbacher L., Schlachetzki J.C.M., Boehnke L., Zangwill D., Galasko D., Glass C.K., Mertens J., Gage F.H. Increased post-mitotic senescence in aged human neurons is a pathological feature of Alzheimer’s disease. Cell Stem Cell. 2022;29:1637–1652.e6. doi: 10.1016/j.stem.2022.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lund R.J., Närvä E., Lahesmaa R. Genetic and epigenetic stability of human pluripotent stem cells. Nat. Rev. Genet. 2012;13:732–744. doi: 10.1038/nrg3271. [DOI] [PubMed] [Google Scholar]
- 47.Liu P., Kaplan A., Yuan B., Hanna J.H., Lupski J.R., Reiner O. Passage number is a major contributor to genomic structural variations in mouse iPSCs. Stem Cell. 2014;32:2657–2667. doi: 10.1002/stem.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gross A.M., Ajay S.S., Rajan V., Brown C., Bluske K., Burns N.J., Chawla A., Coffey A.J., Malhotra A., Scocchia A., et al. Copy-number variants in clinical genome sequencing: deployment and interpretation for rare and undiagnosed disease. Genet. Med. 2019;21:1121–1130. doi: 10.1038/s41436-018-0295-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trost B., Walker S., Wang Z., Thiruvahindrapuram B., MacDonald J.R., Sung W.W.L., Pereira S.L., Whitney J., Chan A.J.S., Pellecchia G., et al. A Comprehensive Workflow for Read Depth-Based Identification of Copy-Number Variation from Whole-Genome Sequence Data. Am. J. Hum. Genet. 2018;102:142–155. doi: 10.1016/j.ajhg.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ee B., W Z., F, V.-B., A, T.-R., P L., Ja B. Combined Genome Sequencing and RNA Analysis Reveals and Characterizes a Deep Intronic Variant in IGHMBP2 in a Patient With Spinal Muscular Atrophy With Respiratory Distress Type 1. Pediatr. Neurol. 2021;114 doi: 10.1016/j.pediatrneurol.2020.09.011. [DOI] [PubMed] [Google Scholar]
- 51.Brechtmann F., Mertes C., Matusevičiūtė A., Yépez V.A., Avsec Ž., Herzog M., Bader D.M., Prokisch H., Gagneur J. OUTRIDER: A Statistical Method for Detecting Aberrantly Expressed Genes in RNA Sequencing Data. Am. J. Hum. Genet. 2018;103:907–917. doi: 10.1016/j.ajhg.2018.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mertes C., Scheller I.F., Yépez V.A., Çelik M.H., Liang Y., Kremer L.S., Gusic M., Prokisch H., Gagneur J. Detection of aberrant splicing events in RNA-seq data using FRASER. Nat. Commun. 2021;12:529. doi: 10.1038/s41467-020-20573-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Accogli A., Lu S., Musante I., Scudieri P., Rosenfeld J.A., Severino M., Baldassari S., Iacomino M., Riva A., Balagura G., et al. Loss of Neuron Navigator 2 Impairs Brain and Cerebellar Development. Cerebellum. 2023;22:206–222. doi: 10.1007/s12311-022-01379-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Akula S.K., Marciano J.H., Lim Y., Exposito-Alonso D., Hylton N.K., Hwang G.H., Neil J.E., Dominado N., Bunton-Stasyshyn R.K., Song J.H.T., et al. TMEM161B regulates cerebral cortical gyration, Sonic Hedgehog signaling, and ciliary structure in the developing central nervous system. Proc. Natl. Acad. Sci. USA. 2023;120 doi: 10.1073/pnas.2209964120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.AlAbdi L., Desbois M., Rusnac D.-V., Sulaiman R.A., Rosenfeld J.A., Lalani S., Murdock D.R., Burrage L.C., Undiagnosed Diseases Network. Billie Au P.Y., et al. Loss-of-function variants in MYCBP2 cause neurobehavioural phenotypes and corpus callosum defects. Brain. 2023;146:1373–1387. doi: 10.1093/brain/awac364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Keehan L., Jiang M.-M., Li X., Marom R., Dai H., Murdock D., Liu P., Hunter J.V., Heaney J.D., Robak L., et al. A Novel De Novo Intronic Variant in ITPR1 Causes Gillespie Syndrome. Am. J. Med. Genet. 2021;185:2315–2324. doi: 10.1002/ajmg.a.62232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van de Leemput J., Chandran J., Knight M.A., Holtzclaw L.A., Scholz S., Cookson M.R., Houlden H., Gwinn-Hardy K., Fung H.-C., Lin X., et al. Deletion at ITPR1 underlies ataxia in mice and spinocerebellar ataxia 15 in humans. PLoS Genet. 2007;3:e108. doi: 10.1371/journal.pgen.0030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang L., Chardon J.W., Carter M.T., Friend K.L., Dudding T.E., Schwartzentruber J., Zou R., Schofield P.W., Douglas S., Bulman D.E., Boycott K.M. Missense mutations in ITPR1 cause autosomal dominant congenital nonprogressive spinocerebellar ataxia. Orphanet J. Rare Dis. 2012;7:67. doi: 10.1186/1750-1172-7-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.des Portes V., Pinard J.M., Billuart P., Vinet M.C., Koulakoff A., Carrié A., Gelot A., Dupuis E., Motte J., Berwald-Netter Y., et al. A novel CNS gene required for neuronal migration and involved in X-linked subcortical laminar heterotopia and lissencephaly syndrome. Cell. 1998;92:51–61. doi: 10.1016/s0092-8674(00)80898-3. [DOI] [PubMed] [Google Scholar]
- 60.Francis F., Koulakoff A., Boucher D., Chafey P., Schaar B., Vinet M.C., Friocourt G., McDonnell N., Reiner O., Kahn A., et al. Doublecortin is a developmentally regulated, microtubule-associated protein expressed in migrating and differentiating neurons. Neuron. 1999;23:247–256. doi: 10.1016/s0896-6273(00)80777-1. [DOI] [PubMed] [Google Scholar]
- 61.Kim M.H., Cierpicki T., Derewenda U., Krowarsch D., Feng Y., Devedjiev Y., Dauter Z., Walsh C.A., Otlewski J., Bushweller J.H., Derewenda Z.S. The DCX-domain tandems of doublecortin and doublecortin-like kinase. Nat. Struct. Biol. 2003;10:324–333. doi: 10.1038/nsb918. [DOI] [PubMed] [Google Scholar]
- 62.Hehr U., Uyanik G., Aigner L., Couillard-Despres S., Winkler J. In: DCX-Related Disorders. GeneReviews®, Adam M.P., Everman D.B., Mirzaa G.M., Pagon R.A., Wallace S.E., Bean L.J., Gripp K.W., Amemiya A., editors. University of Washington, Seattle; 1993. [PubMed] [Google Scholar]
- 63.Buchsbaum I.Y., Cappello S. Neuronal migration in the CNS during development and disease: insights from in vivo and in vitro models. Dev. Camb. Engl. 2019;146:dev163766. doi: 10.1242/dev.163766. [DOI] [PubMed] [Google Scholar]
- 64.Luo X., Rosenfeld J.A., Yamamoto S., Harel T., Zuo Z., Hall M., Wierenga K.J., Pastore M.T., Bartholomew D., Delgado M.R., et al. Clinically severe CACNA1A alleles affect synaptic function and neurodegeneration differentially. PLoS Genet. 2017;13 doi: 10.1371/journal.pgen.1006905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pietrobon D. CaV2.1 channelopathies. Pflügers Archiv. 2010;460:375–393. doi: 10.1007/s00424-010-0802-8. [DOI] [PubMed] [Google Scholar]
- 66.Tonelli A., D’Angelo M.G., Salati R., Villa L., Germinasi C., Frattini T., Meola G., Turconi A.C., Bresolin N., Bassi M.T. Early onset, non fluctuating spinocerebellar ataxia and a novel missense mutation in CACNA1A gene. J. Neurol. Sci. 2006;241:13–17. doi: 10.1016/j.jns.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 67.Mullegama S.V., Mendoza-Londono R., Elsea S.H. In: MBD5 Haploinsufficiency. GeneReviews®, Adam M.P., Everman D.B., Mirzaa G.M., Pagon R.A., Wallace S.E., Bean L.J., Gripp K.W., Amemiya A., editors. University of Washington, Seattle; 1993. [PubMed] [Google Scholar]
- 68.Talkowski M.E., Mullegama S.V., Rosenfeld J.A., van Bon B.W.M., Shen Y., Repnikova E.A., Gastier-Foster J., Thrush D.L., Kathiresan S., Ruderfer D.M., et al. Assessment of 2q23.1 microdeletion syndrome implicates MBD5 as a single causal locus of intellectual disability, epilepsy, and autism spectrum disorder. Am. J. Hum. Genet. 2011;89:551–563. doi: 10.1016/j.ajhg.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Williams S.R., Mullegama S.V., Rosenfeld J.A., Dagli A.I., Hatchwell E., Allen W.P., Williams C.A., Elsea S.H. Haploinsufficiency of MBD5 associated with a syndrome involving microcephaly, intellectual disabilities, severe speech impairment, and seizures. Eur. J. Hum. Genet. 2010;18:436–441. doi: 10.1038/ejhg.2009.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kang H.J., Kawasawa Y.I., Cheng F., Zhu Y., Xu X., Li M., Sousa A.M.M., Pletikos M., Meyer K.A., Sedmak G., et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kirsch L., Chechik G. On Expression Patterns and Developmental Origin of Human Brain Regions. PLoS Comput. Biol. 2016;12 doi: 10.1371/journal.pcbi.1005064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nat. Rev. Neurosci. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rubenstein J.L.R. Annual Research Review: Development of the cerebral cortex: implications for neurodevelopmental disorders. JCPP (J. Child Psychol. Psychiatry) 2011;52:339–355. doi: 10.1111/j.1469-7610.2010.02307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hill R.S., Walsh C.A. Molecular insights into human brain evolution. Nature. 2005;437:64–67. doi: 10.1038/nature04103. [DOI] [PubMed] [Google Scholar]
- 75.Dredge B.K., Polydorides A.D., Darnell R.B. The splice of life: alternative splicing and neurological disease. Nat. Rev. Neurosci. 2001;2:43–50. doi: 10.1038/35049061. [DOI] [PubMed] [Google Scholar]
- 76.Raj T., Li Y.I., Wong G., Humphrey J., Wang M., Ramdhani S., Wang Y.-C., Ng B., Gupta I., Haroutunian V., et al. Integrative transcriptome analyses of the aging brain implicate altered splicing in Alzheimer’s disease susceptibility. Nat. Genet. 2018;50:1584–1592. doi: 10.1038/s41588-018-0238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Voineagu I., Wang X., Johnston P., Lowe J.K., Tian Y., Horvath S., Mill J., Cantor R.M., Blencowe B.J., Geschwind D.H. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tollervey J.R., Curk T., Rogelj B., Briese M., Cereda M., Kayikci M., König J., Hortobágyi T., Nishimura A.L., Zupunski V., et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat. Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jopling C., Boue S., Izpisua Belmonte J.C. Dedifferentiation, transdifferentiation and reprogramming: three routes to regeneration. Nat. Rev. Mol. Cell Biol. 2011;12:79–89. doi: 10.1038/nrm3043. [DOI] [PubMed] [Google Scholar]
- 80.Mollinari C., Zhao J., Lupacchini L., Garaci E., Merlo D., Pei G. Transdifferentiation: a new promise for neurodegenerative diseases. Cell Death Dis. 2018;9:830–839. doi: 10.1038/s41419-018-0891-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu Z., Su S., Zhou S., Yang W., Deng X., Sun Y., Li L., Li Y. How to reprogram human fibroblasts to neurons. Cell Biosci. 2020;10:116. doi: 10.1186/s13578-020-00476-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ieda M., Fu J.-D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B.G., Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cao N., Huang Y., Zheng J., Spencer C.I., Zhang Y., Fu J.-D., Nie B., Xie M., Zhang M., Wang H., et al. Conversion of human fibroblasts into functional cardiomyocytes by small molecules. Science. 2016;352:1216–1220. doi: 10.1126/science.aaf1502. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
UDN sequencing data are available through dbGaP (accession: phs001232.v2.p1) and the UDN Gateway. Phenotype data with flagged genes of interest have been submitted to Phenome Central. DNA variants thought to contribute to the molecular diagnoses of the patients have been submitted to ClinVar.