Abstract
Protein acetylation has emerged as a means of controlling levels of mRNA synthesis in eukaryotic cells. Here we report that acetyl coenzyme A (acetyl-CoA) stimulates RNA polymerase II transcription in vitro in the absence of histones. The effect of acetyl-CoA on basal and activated transcription was studied in a human RNA polymerase II transcription system reconstituted from recombinant and highly purified transcription factors. Both basal and activated transcription were stimulated by the addition of acetyl-CoA to transcription reaction mixtures. By varying the concentrations of general transcription factors in the reaction mixtures, we found that acetyl-CoA decreased the concentration of TFIID required to observe transcription. Electrophoretic mobility shift assays and DNase I footprinting revealed that acetyl-CoA increased the affinity of the general transcription factor TFIID for promoter DNA in a TBP-associated factor (TAF)-dependent manner. Interestingly, acetyl-CoA also caused a conformational change in the TFIID-TFIIA-promoter complex as assessed by DNase I footprinting. These results show that acetyl-CoA alters the DNA binding activity of TFIID and indicate that this biologically important cofactor functions at multiple levels to control gene expression.
Transcription from natural RNA polymerase II promoters is tightly controlled by the combined actions of positive and negative regulatory factors, including site-specific activators, repressors, cofactors, and chromatin-associated proteins. For transcription of any given gene, a complex array of signals must ultimately by integrated at the promoter to set the proper level of RNA production. Protein acetylation is one such signal implicated in controlling levels of mRNA synthesis in eukaryotes. It has been shown that specific lysines in the N termini of core histones are acetylated in transcriptionally active regions of chromatin (reviewed in references 6 and 39). The prevailing model suggests that acetylation of histones relieves the repressive effects of chromatin and allows the transcription machinery access to promoters, resulting in a localized derepression of transcription (reviewed in reference 40). Several nuclear histone acetyltransferases have been identified, including Tetrahymena p55 and the human coactivators CREB binding protein (CBP), p300, P/CAF, GCN5, and TAFII250 (3, 7, 27, 29, 42). Transcriptional activators can recruit acetyltransferases to promoters, where they acetylate histones in nucleosomes (38). Conversely, deacetylases are thought to be recruited to chromatin by transcriptional repressors (reviewed in reference 31).
Histones are not the only substrates of nuclear acetyltransferases. The tumor suppressor p53 is a substrate of acetylation by p300 (22). Acetylation at specific lysines stimulates DNA binding by p53 in response to DNA damage (22, 34). TFIIE and TFIIF can be acetylated by P/CAF, p300, and TAFII250, although the functions of these posttranslational modifications have not been identified (15). HMG I(Y) is acetylated by CBP resulting in disruption of the interferon-β enhanceosome and a decrease in transcription (28). Acetylation of HMG-17 by P/CAF alters its interaction with nucleosomes (19). Acetylation of GATA-1 by p300 stimulates DNA binding and may alter the conformation of the GATA-1–DNA complex (5). The transcriptional activator EKLF is acetylated by p300 and CBP in its activation domain (44). These emerging data support a model in which protein acetylation is a general mechanism for regulating RNA polymerase II transcription in higher eukaryotes.
Structural studies combined with biochemical experiments have revealed that acetyl coenzyme A (acetyl-CoA) can alter the conformation of the proteins that it binds. It is well documented that acetyl-CoA alters the conformation and activity of metabolic enzymes when it binds as an allosteric effector (2, 43). The histone acetyltransferase activities of human GCN5 and P/CAF were found to be stabilized by acetyl-CoA (18). X-ray crystallography and biochemical experiments on the histone acetyltransferase Hat1 support the proposal that this enzyme undergoes a conformational change upon binding acetyl-CoA (9). Recently, X-ray crystallography revealed that the enzyme serotonin N-acetyltransferase undergoes a striking conformational change upon binding the substrate acetyl-CoA (20). In this case, acetyl-CoA induces the formation of a binding site for the second substrate, serotonin. Thus, acetyl-CoA is emerging as an important cofactor in transcriptional regulation, not only as a substrate for enzymatic reactions but also as a cofactor for inducing conformational changes in some of the proteins to which it binds.
In this study, we have investigated whether acetyl-CoA can act as a cofactor for human RNA polymerase II transcription in the absence of histones. To carry out these experiments, we developed an in vitro transcription system responsive to transcriptional activators that is reconstituted from recombinant and highly purified human general transcription factors. Our results show that acetyl-CoA increases both basal and activated transcription in the absence of histones and reveal that acetyl-CoA is a cofactor that stimulates the binding of TFIID to promoter DNA.
MATERIALS AND METHODS
Transcription factors.
The following recombinant proteins were expressed and purified as described previously: human TATA binding protein (TBP) (33), human TFIIA (30, 36), human TFIIB (16), human TFIIE-34 and TFIIE-56 (32), c-Jun (4), and GAL4-p53 (37). Human RAP30 and human RAP74 were expressed in Escherichia coli (1, 10, 35) and purified (24) as described previously. Human RNA polymerase II was purified from HeLa nuclear pellets through the DEAE-5PW step as described previously (26). Human TFIIH was purified from HeLa cytoplasmic extracts through the phenyl column as described previously (11). Nuclear extracts were prepared from Jurkat cells by the method of Dignam et al. (8).
A method for immunopurifying human TFIID was developed and will be described in detail elsewhere (J. Goodrich and R. Tjian, unpublished data). Briefly, HeLa cell nuclear extracts were fractionated by phosphocellulose chromatography (8). Proteins eluting between 0.5 M and 1.0 M KCl were pooled, and TFIID was further immunopurified using an anti-hTAFII130 monoclonal antibody. Immunoprecipitated material was washed extensively, and TFIID was specifically eluted from antibody beads with 2 column volumes of buffer containing an epitope peptide.
In vitro transcription.
Transcription reactions using the reconstituted transcription system were performed as previously described (14, 24) with the following modifications: the MgCl2 concentration was 6 mM; TFIID (∼3 ng, or as indicated in the figure legends) and TFIIA (20 ng) were added in place of recombinant TBP; and where indicated, GAL4-p53 and cJun were preincubated with promoter DNA for 5 min on ice. After the addition of general transcription factors, the reaction mixtures were transferred to 30°C for 30 min before addition of nucleoside triphosphates. RNA synthesis was allowed to proceed for 20 min at 30°C. Acetyl-CoA (Sigma or Pharmacia) at concentrations indicated in the figures and sodium butyrate (Sigma) at a final concentration of 1 mM were added to transcription reaction mixtures immediately prior to the addition of RNA polymerase II and general transcription factors. We observed that the concentration of acetyl-CoA required for maximum stimulation of transcription varied widely (100 nM to 3 μM) with different sources, lots, and ages of acetyl-CoA. We believe that this variability is associated with the fraction of the CoA in the acetylated form at the time the experiment is performed. The source of acetyl-CoA used in each experiment is indicated in the figure legends. Transcription in a Jurkat nuclear extract was carried out as described above, except that purified general transcription factors were replaced by 4 μl of nuclear extract.
G-less transcription reactions were stopped with 100 μl of a solution containing 3.1 M ammonium acetate, 10 μg of carrier yeast RNA, and 15 μg of proteinase K. After ethanol precipitation, the samples were resolved by denaturing polyacrylamide gel electrophoresis (6% polyacrylamide) (PAGE) and quantitated by PhosphorImager analysis. Reactions utilizing the (AP1)5-E1b-chloramphenicol acetyltransferase (CAT) and (GAL4)5-E1b-CAT templates were stopped with 90 μl of a solution containing 1% sodium dodecyl sulfate (SDS), 0.02 M EDTA, 0.2 M NaCl, and 10 μg of carrier yeast RNA. After extraction with phenol-chloroform and ethanol precipitation, the RNA was subjected to primer extension using a CAT-specific primer as previously described (13). Products were resolved by denaturing PAGE (8% polyacrylamide) and quantitated by PhosphorImager (Molecular Dynamics) analysis.
Mg2+-agarose mobility shift assays.
Mg2+-agarose gel shifts were performed as previously described (25). For the mobility shift assays, a 133-bp DNA fragment (containing the adenovirus major late promoter region −53 to +33) was excised from plasmid pBS-MLP (14) with EcoRI and HindIII, end labeled with T4 polynucleotide kinase in the presence of [γ-32P]ATP, and purified from an 8% nondenaturing polyacrylamide gel. TFIID and TFIIA were incubated with promoter DNA (final concentration, 0.7 nM) for 20 min at 30°C under buffer conditions that were identical to those used for transcription in a reaction volume of 10 μl. The reaction products were removed to ice and loaded onto a 1.4% agarose gel (14 cm) in 0.5× Tris-borate-EDTA (TBE) containing 5 mM magnesium acetate. Electrophoresis was performed at 4°C and 100 V for 3 h. The gels were transferred to DE81 paper (Whatman), dried, and subjected to autoradiography.
DNase I footprinting.
DNase I footprints were performed with either a 287-bp DNA fragment (−212 to +75) containing five GAL4 sites upstream of the adenovirus major late core promoter (−53 to +33) or a 230-bp DNA fragment (−152 to +78) containing plasmid DNA sequence upstream of the adenovirus major late core promoter (−53 to +33). Both DNA fragments were generated by PCR and were 32P labeled on the 5′ end of the template strand. TFIID and TFIIA (in amounts indicated in figure legends) were incubated with promoter DNA (at final concentrations indicated in the figure legends) for 20 min at 30°C under buffer conditions that were identical to those used for transcription in a reaction volume of 10 μl. Then 1 μl of a solution containing 0.15 U of DNase I (Promega) per μl and 10 mM CaCl2 was added to each reaction. After a 30-s incubation at 30°C, the reactions were stopped with 40 μl of stop solution containing 25 mM EDTA, 125 mM KCl, and 10 μg of carrier yeast RNA. SDS was added to each reaction mixture to a final concentration of 0.5%. The reaction mixtures were incubated at 65°C for 15 min and then placed on ice for 10 min. After a 10-min centrifugation at 16,000 × g in a microcentrifuge, the supernatants were transferred to new tubes. DNA was ethanol precipitated and resuspended in formamide loading buffer. Products were resolved by denaturing PAGE (8% polyacrylamide).
RESULTS
Reconstitution of a highly purified human transcription system that responds to activators.
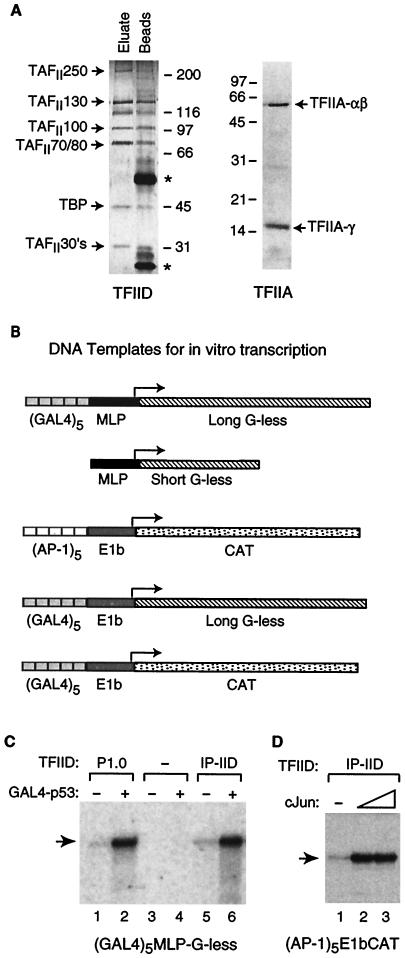
While acetylation of proteins involved in transcription has been documented, the effects of acetyl-CoA on RNA polymerase II transcription in a purified in vitro transcription system have not been investigated. To reveal mechanisms by which acetyl-CoA regulates transcription, we established a human in vitro transcription system composed of highly purified native factors (RNA polymerase II, TFIID, and TFIIH) and recombinant factors (TFIIA, TFIIB, TFIIE, and TFIIF). This system differs from one described previously (14, 24) by the addition of immunopurified human TFIID and recombinant human TFIIA in place of recombinant TBP (Fig. 1A). Human TFIID was immunopurified using an anti-hTAFII130 monoclonal antibody, washed extensively with high concentrations of salt, and eluted with an epitope peptide. The eluted TFIID shown in Fig. 1A is highly purified and is devoid of antibody heavy and light chains. The schematic in Fig. 1B depicts the five promoter templates used in transcription experiments.
FIG. 1.
Reconstitution of a purified human RNA polymerase II transcription system that responds to activators. (A) Purified human TFIID and TFIIA. Human TFIID was purified from HeLa nuclear extracts by phosphocellulose chromatography followed by immunopurification with an anti-TAFII130 monoclonal antibody and elution with an epitope peptide in 2 column volumes of buffer. The eluted TFIID (5 μl) and protein remaining on antibody beads (5 μl) after elution were resolved by SDS-PAGE and visualized by silver staining. Asterisks depict the locations of bands containing antibody heavy and light chains. Recombinant human TFIIA was overexpressed in E. coli, purified to near homogeneity, resolved by SDS-PAGE, and visualized by staining with Coomassie brilliant blue. (B) Five DNA templates were used in the transcription experiments shown in Fig. 1 through 4. (GAL4)5-MLP-G-less consists of five GAL4 sites upstream of the adenovirus major late core promoter and a 390-bp G-less cassette. MLP-G-less(short) consists of the adenovirus major late core promoter and a 190-bp G-less cassette. (AP-1)5-E1b-CAT consists of five AP-1 sites upstream of the adenovirus E1b TATA box and the coding region for CAT. (GAL4)5-E1b-G-less consists of five GAL4 sites upstream of the adenovirus E1b TATA box and a 390-bp G-less cassette. (GAL4)5-E1b-CAT consists of five GAL4 sites upstream of the adenovirus E1b TATA box and the coding region for CAT. (C) The in vitro transcription system containing immunopurified TFIID (lanes 5 and 6) responds to the activator GAL4-p53 (30 ng). The response is similar to that observed using a crude TFIID fraction (P1.0; lanes 1 and 2), and the system is completely dependent on the addition of TFIID (lanes 3 and 4). The DNA template used in this experiment was (GAL4)5-MLP-G-less. (D) The in vitro transcription system containing immunopurified TFIID responds to the activator c-Jun (50 ng). The DNA template used in this experiment was (AP-1)5-E1b-CAT, and transcription was monitored by primer extension.
We tested this highly purified human TFIID in the reconstituted transcription system for the ability to mediate transcriptional activation. As shown in Fig. 1C, the transcription system is completely dependent on the addition of TFIID. For mediating activation by the chimeric activator GAL4-p53, the immunopurified human TFIID had activity comparable to that of a crude TFIID fraction (phosphocellulose 1 M KCl eluate [see Materials and Methods]). This transcription system also responds to other transcriptional activators, including c-Jun (Fig. 1D). Transcription with this system has been analyzed using both a G-less cassette assay (Fig. 1B and C) and primer extension (Fig. 1B and D). The in vitro transcription system is highly dependent on the addition of recombinant TFIIA, and the immunopurified TFIID shows a significant level of DNA binding in Mg2+-agarose gel shifts assays only in the presence of recombinant TFIIA (data not shown).
Acetyl-CoA stimulates basal and activated transcription in the absence of histones.
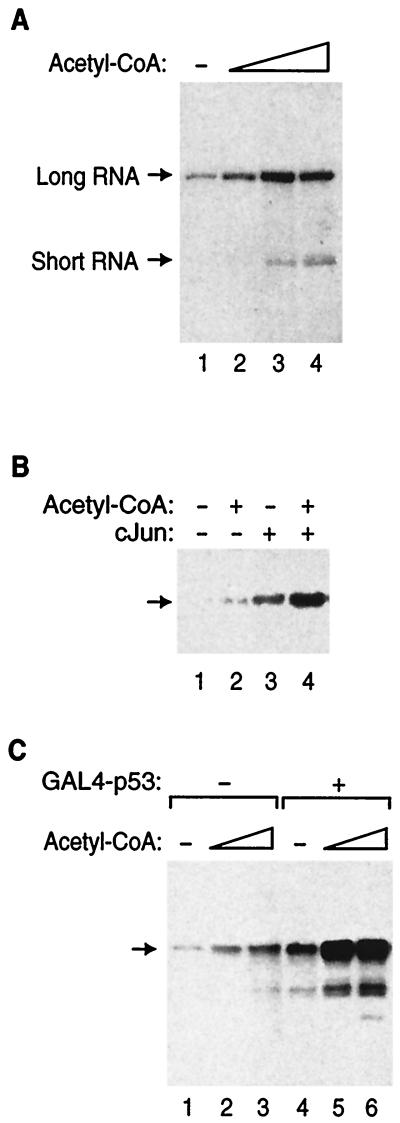
Although prevailing models predicted that chromatin templates would be needed to observe stimulation of transcription by acetyl-CoA, we hypothesized that acetyl-CoA might affect transcription in the absence of histones. We tested this hypothesis by adding increasing amounts of acetyl-CoA to transcription reaction mixtures with the adenovirus major late core promoter contained on naked DNA templates. Surprisingly, acetyl-CoA stimulated both basal transcription (short G-less transcript) and GAL4-p53-activated transcription (long G-less transcript) more than fivefold (Fig. 2A). These results demonstrate that acetyl-CoA can serve as a cofactor for RNA polymerase II transcription in the absence of histones. Stimulation of transcription by acetyl-CoA was not limited to the adenovirus major late promoter and GAL4-p53 activation but was also observed with c-Jun-activated transcription using a template containing the adenovirus E1b TATA box and upstream AP-1 sites (Fig. 2B).
FIG. 2.
Acetyl-CoA stimulates basal and activated transcription from naked DNA templates in vitro. (A) Acetyl-CoA stimulates basal and GAL4-p53-activated transcription from the adenovirus major late core promoter in a reconstituted transcription system. Acetyl-CoA was titrated into transcription reaction mixtures in which basal (short RNA) and GAL4-p53 activated (long RNA) transcription were monitored simultaneously. The reaction mixtures in lanes 1 to 4 contained 0, 1, 3, and 10 μM (final concentrations) acetyl-CoA (Sigma), respectively. (B) Acetyl-CoA stimulates basal and c-Jun-activated transcription from the adenovirus E1b TATA box in a reconstituted transcription system. Acetyl-CoA (Pharmacia; final concentration, 100 nM) was added to transcription reaction mixtures in the absence and presence of c-Jun (50 ng). The DNA template used in this experiment was (AP-1)5-E1b-CAT, and transcription was monitored by primer extension. (C) Acetyl-CoA stimulates basal and activated transcription in a Jurkat nuclear extract. Acetyl-CoA (Pharmacia) was added to transcription reaction mixtures to final concentrations of 37.5 nM (lanes 2 and 5) and 125 nM (lanes 3 and 6). The reaction mixtures in lanes 4 to 6 also contained GAL4-p53. The DNA template used in this experiment was (GAL4)5-E1b-CAT, and transcription was monitored by primer extension.
We asked if acetyl-CoA could stimulate basal and activated transcription in a crude nuclear extract. Since all of the general transcription factors used in the reconstituted transcription system are present in a nuclear extract, we expected that transcription in a nuclear extract would be responsive to acetyl-CoA, unless other factors specifically inhibited the acetyl-CoA stimulation of transcription or degraded the acetyl-CoA. As shown in Fig. 2C, both basal (lanes 1 to 3) and GAL4-p53-activated (lanes 4 to 6) transcription were stimulated by acetyl-CoA. The fact that these results with a crude nuclear extract are similar to those observed with the reconstituted transcription system supports the use of the reconstituted system to study the mechanism by which acetyl-CoA stimulates transcription from naked DNA templates.
Acetyl-CoA decreases the concentration of TFIID required in transcription reactions.
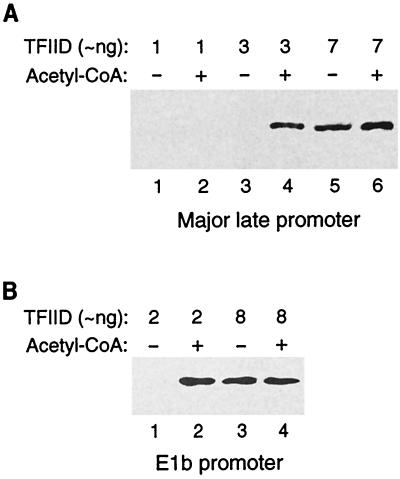
We next investigated the mechanism by which acetyl-CoA stimulated transcription from naked DNA templates. We hypothesized that acetyl-CoA might facilitate the recruitment of general transcription factors to preinitiation complexes by stimulating protein-DNA or protein-protein interactions. This hypothesis was examined by titrating general transcription factors into transcription reactions in the absence and presence of acetyl-CoA. We found that the concentration of TFIID required for transcription was decreased by the addition of acetyl-CoA with both the adenovirus major late promoter (Fig. 3A) and the adenovirus E1b promoter (Fig. 3B). Given that TFIID is responsible for promoter recognition and a number of the TFIID subunits bind promoter DNA (reviewed in references 12 and 17), the results of our in vitro transcription experiments suggest that acetyl-CoA increases the affinity of TFIID for promoter DNA.
FIG. 3.
Acetyl-CoA decreases the concentration of TFIID required for in vitro transcription from naked DNA templates. (A) TFIID was titrated into transcription reaction mixtures containing the (GAL4)5-AdMLP-G-less template and GAL4-p53 in the absence (odd lanes) and presence (even lanes) of acetyl-CoA (Sigma; final concentration, 1 μM). (B) TFIID was titrated into transcription reaction mixtures containing the (GAL4)5-E1b-G-less template and GAL4-p53 in the absence (odd lanes) and presence (even lanes) of acetyl-CoA (Sigma; final concentration, 1 μM).
Acetyl-CoA stimulates transcription and promoter DNA binding by TFIID but not TBP.
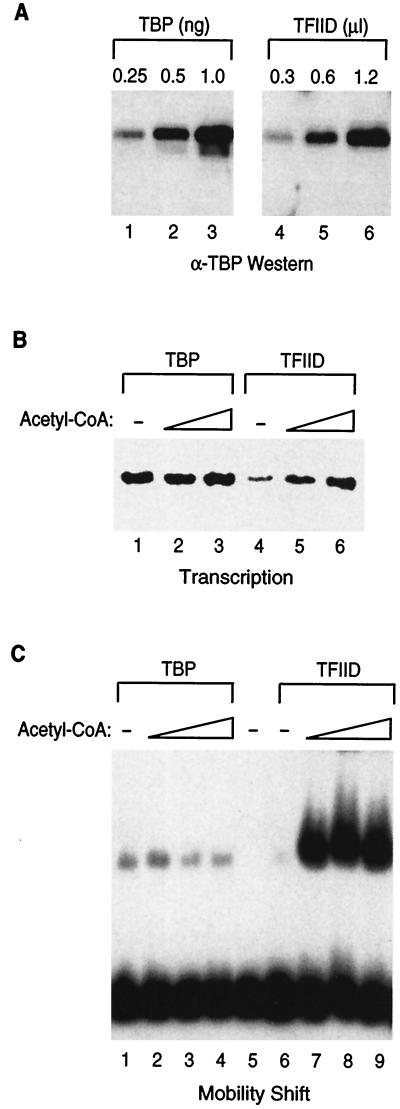
To determine whether the TBP-associated factor (TAF) subunits of TFIID were required for the response of transcription to acetyl-CoA, we investigated the effect of acetyl-CoA in a transcription system reconstituted with recombinant TBP in place of TFIID. To ensure that similar amounts of TFIID and TBP were added to the transcription reaction mixtures, we determined the relative concentrations of recombinant TBP and immunopurified TFIID by using Western blot analysis (Fig. 4A). Similar amounts of TBP and TFIID were then used in transcription assays to determine the effect of acetyl-CoA on TBP-driven transcription. As shown in Fig. 4B, the TAF subunits of TFIID were required to observe the stimulation of transcription by acetyl-CoA. It is interesting that the level of transcription observed with TFIID was lower than that observed with TBP (compare lane 4 to lane 1). Acetyl-CoA caused an increase in the level of transcription with TFIID that approached the level of transcription observed with TBP.
FIG. 4.
Acetyl-CoA stimulates transcription and TFIID binding to promoter DNA in a reaction dependent upon TAFs. (A) Western blots were carried out to determine the relative concentration of recombinant TBP (lanes 1 to 3) versus immunopurified TFIID (lanes 4 to 6), using an affinity-purified polyclonal antibody against TBP. (B) The TAF subunits of TFIID are required for acetyl-CoA stimulation of transcription from a naked DNA template in vitro. Transcription reactions were reconstituted with 0.08 ng of TBP (lanes 1 to 3) or 0.08 μl (∼2 ng) of TFIID (lanes 4 to 6). All reaction mixtures contained TFIIA, and the DNA template used was (GAL4)5-MLP-G-less. Acetyl-CoA (Pharmacia) was added to transcription reaction mixtures to a final concentration of 30 nM (lanes 2 and 5) or 100 nM (lanes 3 and 6). (C) Acetyl-CoA stimulates the binding of TFIID to promoter DNA in a TAF-dependent reaction. Mg2+-agarose mobility shift assays were performed with immunopurified TFIID, recombinant TBP, and recombinant TFIIA on a DNA fragment containing the adenovirus major late core promoter. Reaction mixtures contained 0.3 ng of TBP (lanes 1 to 4) or 0.3 μl (∼8 ng) of TFIID (lanes 6 to 9). All reaction mixtures contained 20 ng of recombinant TFIIA. Acetyl-CoA (Pharmacia) was titrated into reactions at the following final concentrations: 30 nM (lanes 2 and 7), 100 nM (lanes 3 and 8), and 300 nM (lanes 4 and 9).
We directly tested the effect of acetyl-CoA on TFIID binding by performing Mg2+-agarose mobility shift assays using the adenovirus major late core promoter. We also compared the effect of acetyl-CoA on DNA binding by TBP, again to determine if the TAF subunits of TFIID are involved in the acetyl-CoA response. Similar amounts of TBP and TFIID were used in mobility shift assays to determine the effect of acetyl-CoA on DNA binding. The binding of TFIID to the adenovirus major late promoter was dramatically stimulated by acetyl-CoA (Fig. 4C, lanes 6 through 9). In contrast, the binding of TBP to promoter DNA was not stimulated by acetyl-CoA, indicating that the TAF subunits of TFIID were required to observe the acetyl-CoA stimulation of TFIID DNA binding (Fig. 4C, lanes 1 through 4). In other experiments, we found that the binding of a partial TFIID complex containing only two subunits, TAFII250 and TBP, was not stimulated by acetyl-CoA (data not shown). Thus, TAFII250, TBP, and TFIIA were not sufficient to respond to acetyl-CoA in vitro.
Acetyl-CoA causes a conformational change in the TFIID-TFIIA-DNA complex.
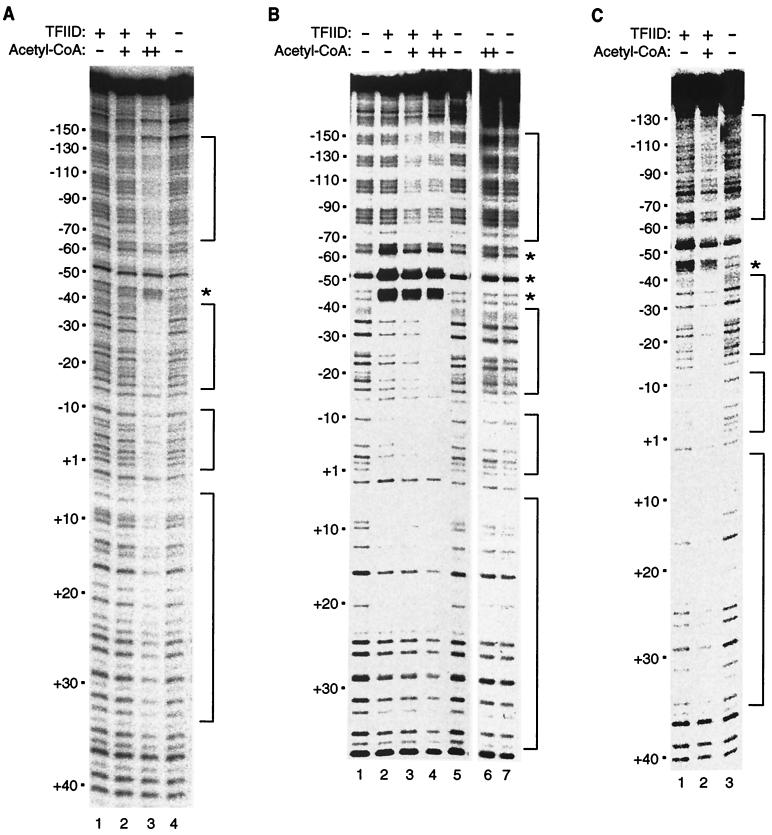
To further investigate the effect of acetyl-CoA on DNA binding by TFIID, we used DNase I footprinting. As shown in Fig. 5A, acetyl-CoA strongly stimulated TFIID binding to the adenovirus major late promoter, resulting in a DNase I footprint that extended almost continuously from positions −140 to +32 with respect to the transcription start site (+1). Bands at positions +3 and −15 were not protected from DNase I cleavage. In addition, bands in the region from positions −42 to −65 were not protected, while an enhancement of cleavage was observed at approximately −45. We conclude that acetyl-CoA stimulates promoter DNA binding by TFIID, resulting in extensive interaction of TFIID and TFIIA with promoter DNA over a region of approximately 170 bp.
FIG. 5.
Acetyl-CoA stimulates TFIID binding to the adenovirus major late promoter, resulting in an extended DNase I footprint. (A) With subsaturating amounts of TFIID, acetyl-CoA stimulates TFIID binding to promoter DNA, as observed by DNase I footprinting. Reaction mixtures (lanes 1 to 3) contained 0.3 μl (∼8 ng) of TFIID and 20 ng of recombinant TFIIA. Acetyl-CoA (Pharmacia) was added to the reaction mixtures at final concentrations of 30 nM (lane 2) or 100 nM (lane 3). The reaction mixtures contained 0.4 nM 287-bp DNA fragment (positions −212 to +75) with five GAL4 sites upstream of the adenovirus major late core promoter (positions −53 to +33) that was 32P labeled on the 5′ end of the template strand. Footprinting reactions were resolved by denaturing PAGE and analyzed with a PhosphorImager. Positions relative to the transcriptional start site (+1) are indicated on the left. Protected regions and an enhanced band are indicated on the right by brackets and an asterisk, respectively. (B) Acetyl-CoA causes an extension of the TFIID-TFIIA footprint into the region of the template DNA upstream of −70. Reaction mixtures (lanes 2 to 4) contained 0.8 μl (∼22 ng) of TFIID and 20 ng of recombinant TFIIA. Acetyl-CoA (Pharmacia) was added to the reaction mixtures at final concentrations of 30 nM (lane 3) or 100 nM (lanes 4 and 6). Reaction mixtures contained 1 nM 287-bp DNA fragment (positions −212 to +75) with five GAL4 sites upstream of the adenovirus major late core promoter (positions −53 to +33) that was 32P labeled on the 5′ end of the template strand. Footprinting reactions were resolved by denaturing PAGE and analyzed by autoradiography. Positions relative to the transcriptional start site (+1) are indicated on the left. Protected regions and enhanced bands are indicated on the right by brackets and asterisks, respectively. (C) The sequence of the upstream region is not critical for the extended DNase I footprint in response to acetyl-CoA. Reaction mixtures (lanes 1 and 2) contained 0.6 μl (∼16 ng) of TFIID and 20 ng of recombinant TFIIA. Acetyl-CoA (Pharmacia) was added at a final concentration of 30 nM (lane 2). Reaction mixtures contained 0.4 nM 230-bp DNA fragment (positions −152 to +78) with plasmid DNA sequence upstream of the adenovirus major late core promoter (positions −53 to +33) that was 32P labeled on the 5′ end of the template strand. Footprinting reactions were resolved by denaturing PAGE and analyzed with a PhosphorImager. Positions relative to the transcriptional start site (+1) are indicated on the left. Protected regions and an enhanced band are indicated on the right by brackets and an asterisk, respectively.
The protection from positions −70 to −140 was reproducibly stimulated by acetyl-CoA but was unexpected since this region contains four of the five GAL4 sites that were engineered upstream of the major late promoter. To distinguish between the effects of acetyl-CoA on the affinity of TFIID for DNA and the extended protection observed in the upstream region, we increased both the TFIID and DNA concentrations in footprinting reactions. As shown in Fig. 5B, this resulted in significant protection in the region from positions −40 to +25 in the absence of acetyl-CoA (compare lanes 1 and 2). Under these conditions, there was no evidence for protection upstream of position −70. When acetyl-CoA was included in the reaction mixtures, extension of the footprint in the region from −70 to −140 was clearly observed. To determine if the DNA sequence in the upstream region (which consists of 5 GAL4 sites) played a role in the extended DNase I footprint, we performed footprinting with a second DNA template containing the same core promoter but with plasmid DNA sequence in the upstream region. As shown in Fig. 5C, acetyl-CoA caused a DNase I footprint that extended upstream of position −70 with this DNA fragment as well. We conclude that acetyl-CoA causes a conformational change in the TFIID-TFIIA-DNA complex that results in extensive protein-DNA interactions upstream of position −70.
DISCUSSION
In this study we show that RNA polymerase II transcription is stimulated by acetyl-CoA in vitro in the absence of histones. The transcriptional stimulation results from increased affinity of TFIID for promoter DNA. Taken together with observations that transcriptional activation in vivo involves acetylation of histones and with recent discoveries that some transcriptional activators and coactivators are also targets of acetylation, our findings support a model in which acetyl-CoA acts as a cofactor in the stimulation of transcription at multiple levels.
There are several possibilities for the mechanism by which acetyl-CoA increases the binding of TFIID to promoter DNA: (i) acetylation of TFIID (or TFIIA) subunits stimulates DNA binding; (ii) acetyl-CoA stimulates the interaction between TFIID and TFIIA; and (iii) acetyl-CoA binding to TFIID induces a conformational change that results in higher-affinity DNA binding. The first possibility seemed very likely, given that TFIID contains the TAFII250 acetyltransferase and that some of the other TAFs have sequence and structural similarity to core histones (21, 23, 41). To directly determine if subunits of TFIID and TFIIA are targets of acetylation, we performed reactions using [3H]- and [14C]acetyl-CoA and looked for acetylation by filter binding and SDS-PAGE. The many experiments we performed did not provide any evidence for protein acetylation when as much as 5 pmol of highly purified TFIID and TFIIA was used. In addition, we were unable to detect acetylation of any protein in the reconstituted transcription system when acetylation reactions were performed under transcription conditions. We also performed experiments to address the second possibility, namely, that acetyl-CoA stimulated the interaction between TFIID and TFIIA. All of the experiments shown here were performed with saturating amounts of recombinant TFIIA. Addition of more TFIIA to transcription and DNA binding reaction mixtures could not substitute for acetyl-CoA (data not shown). In the absence of TFIIA, where TFIID-driven transcription and TFIID DNA binding are decreased by over 10-fold, we observed that acetyl-CoA stimulated transcription in vitro and TFIID binding in mobility shift assays (data not shown). While these results are consistent with a model in which TFIID alone responds to acetyl-CoA, we cannot be sure that the immunopurified TFIID is completely devoid of TFIIA.
We favor the third model, in which acetyl-CoA binding to TFIID induces a conformational change that results in higher-affinity DNA binding. The protection of the region from positions −70 to −140 by TFIID in the presence of acetyl-CoA is consistent with a conformational change in the TFIID-TFIIA-DNA complex. The TAFII250 acetyltransferase binds acetyl-CoA as a substrate and therefore represents a likely target for an acetyl-CoA-induced conformational change. In Mg2+-agarose mobility shift assays, however, acetyl-CoA did not stimulate DNA binding by a minimal complex containing TAFII250 and TBP, indicating that other subunits of TFIID are involved in responding to acetyl-CoA. These subunits may bind acetyl-CoA themselves or may play a direct role in DNA binding when acetyl-CoA binds TAFII250.
We observed that both basal and activated transcription were stimulated by acetyl-CoA in vitro. These studies indicate that under our minimal in vitro transcription conditions, GAL4-p53 and c-Jun do not regulate the acetyl-CoA stimulation of transcription. It remains possible, however, that activators and repressors regulate the acetyl-CoA effect on DNA binding by TFIID to chromatin templates. In addition, the concentration of acetyl-CoA in the nuclei of cells may not remain static. The observations presented here provide a scenario in which variations in the concentration of nuclear acetyl-CoA could regulate the overall level of RNA polymerase II transcription. We have been unable to find reliable published estimates of nuclear acetyl-CoA concentrations under different physiological conditions; however, it seems possible that factors such as cell cycle progression, developmental stimuli, and apoptosis will alter the concentration of nuclear acetyl-CoA, resulting in general changes in levels of mRNA transcription.
ACKNOWLEDGMENTS
We thank P. Beaurang, K. Goodrich, D. King, N. Tanese, R. Tjian, and E. Wang for contributing to the development of methods for immunopurifying functional TFIID and M. Maxon for providing TFIIE. We are grateful to N. Ahn and S. C. Galasinski for helpful discussions and for comments on the manuscript and to T. R. Cech, L. J. Kim, J. F. Kugel, I. M. Ota, and A. Pardi for comments on the manuscript.
This research was supported by Public Health Service grant GM-55235 from the National Institutes of Health, an American Cancer Society Institutional Research Grant to the University of Colorado Cancer Center, a University of Colorado Junior Faculty Development Award, and a Leukemia Society of America Special Fellowship to J.A.G.
REFERENCES
- 1.Aso T, Vasavada H A, Kawaguchi T, Germino F J, Ganguly S, Kitajima S, Weissman S M, Yasukochi Y. Characterization of cDNA for the large subunit of the transcription initiation factor TFIIF. Nature. 1992;355:461–464. doi: 10.1038/355461a0. [DOI] [PubMed] [Google Scholar]
- 2.Attwood P V, Mayer F, Wallace J C. Avidin as a probe of the conformational changes induced in pyruvate carboxylase by acetyl-CoA and pyruvate. FEBS Lett. 1986;203:191–196. doi: 10.1016/0014-5793(86)80740-2. [DOI] [PubMed] [Google Scholar]
- 3.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 4.Bohmann D, Tjian R. Biochemical analysis of transcriptional activation by Jun: differential activity of c- and v-Jun. Cell. 1989;59:709–717. doi: 10.1016/0092-8674(89)90017-2. [DOI] [PubMed] [Google Scholar]
- 5.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 6.Brownell J E, Allis C D. Special HATs for special occasions: linking histone acetylation to chromatin assembly and gene activation. Curr Opin Genet Dev. 1996;6:176–184. doi: 10.1016/s0959-437x(96)80048-7. [DOI] [PubMed] [Google Scholar]
- 7.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 8.Dignam J D, Martin P L, Shastry B S, Roeder R G. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- 9.Dutnall R N, Tafrov S T, Sternglanz R, Ramakrishnan V. Structure of the histone acetyltransferase Hat1: a paradigm for the GCN5-related N-acetyltransferase superfamily. Cell. 1998;94:427–438. doi: 10.1016/s0092-8674(00)81584-6. [DOI] [PubMed] [Google Scholar]
- 10.Finkelstein A, Kostrub C F, Li J, Chavez D P, Wang B Q, Fang S M, Greenblatt J, Burton Z F. A cDNA encoding RAP74, a general initiation factor for transcription by RNA polymerase II. Nature. 1992;355:464–467. doi: 10.1038/355464a0. [DOI] [PubMed] [Google Scholar]
- 11.Flores O, Lu H, Reinberg D. Factors involved in specific transcription initiation by RNA polymerase II: Identification and characterization of factor IIH. J Biol Chem. 1992;267:2786–2793. [PubMed] [Google Scholar]
- 12.Goodrich J A, Cutler G, Tjian R. Contacts in context: promoter specificity and macromolecular interactions in transcription. Cell. 1996;84:825–830. doi: 10.1016/s0092-8674(00)81061-2. [DOI] [PubMed] [Google Scholar]
- 13.Goodrich J A, Hoey T, Thut C J, Admon A, Tjian R. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell. 1993;75:519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]
- 14.Goodrich J A, Tjian R. Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell. 1994;77:145–156. doi: 10.1016/0092-8674(94)90242-9. [DOI] [PubMed] [Google Scholar]
- 15.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 16.Ha I, Lane W S, Reinberg D. Cloning of a human gene encoding the general transcription factor IIB. Nature. 1991;352:689–695. doi: 10.1038/352689a0. [DOI] [PubMed] [Google Scholar]
- 17.Hernandez N. TBP, a universal eukaryotic transcription factor? Genes Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- 18.Herrera J E, Bergel M, Yang X J, Nakatani Y, Bustin M. The histone acetyltransferase activity of human GCN5 and PCAF is stabilized by coenzymes. J Biol Chem. 1997;272:27253–27258. doi: 10.1074/jbc.272.43.27253. [DOI] [PubMed] [Google Scholar]
- 19.Herrera J E, Sakaguchi K, Bergel M, Trieschmann L, Nakatani Y, Bustin M. Specific acetylation of chromosomal protein HMG-17 by PCAF alters its interaction with nucleosomes. Mol Cell Biol. 1999;19:3466–3473. doi: 10.1128/mcb.19.5.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hickman A B, Namboodiri M A, Klein M A, Dyda F. The structural basis of ordered substrate binding by serotonin N-acetyltransferase: enzyme complex at 1.8 Å resolution with bisubstrate analog. Cell. 1999;97:361–369. doi: 10.1016/s0092-8674(00)80745-x. [DOI] [PubMed] [Google Scholar]
- 21.Hoffmann A, Chiang C-M, Oelgeschlager T, Xie X, Burley S K, Nakatani Y, Roeder R. A histone octamer-like structure within TFIID. Nature. 1996;380:356–359. doi: 10.1038/380356a0. [DOI] [PubMed] [Google Scholar]
- 22.Imhof A, Yang X-J, Ogryzko V V, Nakatani Y, Wolffe A P, Ge H. Acetylation of general transcription factors by histone acetyltransferases. Curr Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 23.Kokubo T, Gond D-W, Wootton J C, Horikoshi M, Roeder R G, Nakatani Y. Molecular cloning of Drosophila TFIID subunits. Nature. 1994;367:484–487. doi: 10.1038/367484a0. [DOI] [PubMed] [Google Scholar]
- 24.Kugel J F, Goodrich J A. Promoter escape limits the rate of transcription from the adenovirus major late promoter on negatively supercoiled templates. Proc Natl Acad Sci USA. 1998;95:9232–9237. doi: 10.1073/pnas.95.16.9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lieberman P M, Berk A J. A mechanism for TAFs in transcriptional activation: activation domain enhancement of TFIID-TFIIA-promoter DNA complex formation. Genes Dev. 1994;8:995–1006. doi: 10.1101/gad.8.9.995. [DOI] [PubMed] [Google Scholar]
- 26.Lu H, Flores O, Weinmann R, Reinberg D. The nonphosphorylated form of RNA polymerase II preferentially associates with the preinitiation complex. Proc Natl Acad Sci USA. 1991;88:10004–10008. doi: 10.1073/pnas.88.22.10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizzen C A, Yang X-J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 28.Munshi N, Merika M, Yie J, Senger K, Chen G, Thanos D. Acetylation of HMG I(Y) by CBP turns off IFN beta expression by disrupting the enhanceosome. Mol Cell. 1998;2:457–467. doi: 10.1016/s1097-2765(00)80145-8. [DOI] [PubMed] [Google Scholar]
- 29.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 30.Ozer J, Moore P A, Bolden A H, Lee A, Rosen C A, Lieberman P M. Molecular cloning of the small (gamma) subunit of human TFIIA reveals functions critical for activated transcription. Genes Dev. 1994;8:2324–2335. doi: 10.1101/gad.8.19.2324. [DOI] [PubMed] [Google Scholar]
- 31.Pazin M J, Kadonaga J T. What's up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 32.Peterson M G, Inostroza J, Maxon M E, Flores O, Admon A, Reinberg D, Tjian R. Structure and functional properties of human general transcription factor IIE. Nature. 1991;354:369–373. doi: 10.1038/354369a0. [DOI] [PubMed] [Google Scholar]
- 33.Peterson M G, Tanese N, Pugh B F, Tjian R. Functional domains and upstream activation properties of cloned human TATA binding protein. Science. 1990;248:1625–1630. doi: 10.1126/science.2363050. [DOI] [PubMed] [Google Scholar]
- 34.Sakaguchi K, Herrera J E, Saito S, Miki T, Bustin M, Vassilev A, Anderson C W, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sopta M, Burton Z, Greenblatt J. Structure and associated DNA-helicase activity of a general transcription initiation factor that binds to RNA polymerase II. Nature. 1989;341:410–414. doi: 10.1038/341410a0. [DOI] [PubMed] [Google Scholar]
- 36.Sun X, Ma D, Sheldon M, Yeung K, Reinberg D. Reconstitution of human TFIIA activity from recombinant polypeptides: a role in TFIID-mediated transcription. Genes Dev. 1994;8:2336–2348. doi: 10.1101/gad.8.19.2336. [DOI] [PubMed] [Google Scholar]
- 37.Thut C J, Chen J-L, Klemm R, Tjian R. p53 transcriptional activation mediated by TAFII40 and TAFII60. Science. 1995;267:100–104. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- 38.Utley R T, Ikeda K, Grant P A, Cote J, Steger D J, Eberharter A, John S, Workman J L. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 39.Wolffe A P, editor. The nucleosome. Vol. 1. Greenwich, Conn: JAI Press Ltd.; 1995. [Google Scholar]
- 40.Workman J L, Kingston R E. Alterations of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 41.Xie X, Kokubo T, Cohen S L, Mirza U A, Hoffmann A, Chait B T, Roeder R G, Nakatani Y, Burley S K. Structural similarity between TAFs and the heterotetrameric core of the histone octamer. Nature. 1996;380:316–322. doi: 10.1038/380316a0. [DOI] [PubMed] [Google Scholar]
- 42.Yang X-J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 43.Yoshinaga T. Phosphoenolpyruvate carboxylase of Escherichia coli. Studies on multiple conformational states elicited by allosteric effectors with fluorescent probe, 1-anilinonaphthalene-8-sulfonate. Biochim Biophys Acta. 1976;452:566–579. doi: 10.1016/0005-2744(76)90208-4. [DOI] [PubMed] [Google Scholar]
- 44.Zhang W, Bieker J J. Acetylation and modulation of erythroid Kruppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc Natl Acad Sci USA. 1998;95:9855–9860. doi: 10.1073/pnas.95.17.9855. [DOI] [PMC free article] [PubMed] [Google Scholar]