Abstract
Est1 is a component of yeast telomerase, and est1 mutants have senescence and telomere loss phenotypes. The exact function of Est1 is not known, and it is not homologous to components of other telomerases. We previously showed that Est1 protein coimmunoprecipitates with Tlc1 (the telomerase RNA) as well as with telomerase activity. Est1 has homology to Ebs1, an uncharacterized yeast open reading frame product, including homology to a putative RNA recognition motif (RRM) of Ebs1. Deletion of EBS1 results in short telomeres. We created point mutations in a putative RRM of Est1. One mutant was unable to complement either the senescence or the telomere loss phenotype of est1 mutants. Furthermore, the mutant protein no longer coprecipitated with the Tlc1 telomerase RNA. Mutants defective in the binding of Tlc1 RNA were nevertheless capable of binding single-stranded TG-rich DNA. Our data suggest that an important role of Est1 in the telomerase complex is to bind to the Tlc1 telomerase RNA via an RRM. Since Est1 can also bind telomeric DNA, Est1 may tether telomerase to the telomere.
Telomeres are the natural ends of linear chromosomes. Telomeres are maintained at a characteristic length by a balance between two forces, loss of telomeres during DNA replication and synthesis of telomeres by an enzyme called telomerase. Telomerase is a special reverse transcriptase which contains not only a reverse transcriptase catalytic subunit but also an RNA molecule which serves as the template for telomere elongation (11). In yeast, the catalytic subunit is called Est2 (7, 20, 25, 26), and the RNA template is called Tlc1 (39).
Telomerases from several organisms have been partially characterized (3, 7, 12, 13, 24, 26, 29, 30). In general, these complexes contain components in addition to the catalytic subunit and the RNA template (10, 12, 24, 30, 36). For the yeast Saccharomyces cerevisiae, genetic screens have identified five genes (EST1, EST2, EST3, EST4/CDC13, and TLC1) (20, 27, 39) whose mutations lead to progressive telomere shortening and eventual loss of viability (i.e., senescence). EST2 and TLC1 encode the reverse transcriptase (25, 26) and the RNA template (39), respectively. The Cdc13 or Est4 protein can bind the single-stranded G-rich telomeric sequence both in vitro and in vivo (2, 23, 32). This protein apparently caps the telomere, protecting it from nucleolytic digestion. The functions of the other two genes, EST1 and EST3, are less clear. Neither of them is required for in vitro telomerase activity (5, 25), even though mutants exhibit the same senescence phenotype as TLC1 or EST2 mutants (20). There is evidence that Est1 is associated with telomerase, since Est1 coprecipitates with Tlc1 and telomerase activity (21, 40). In addition, Est1 may be associated with the telomere since, like Cdc13, Est1 can bind single-stranded G-rich telomeric DNA in vitro (43). However, the affinity of Est1 for such DNA is low, much lower than the affinity of Cdc13. Unlike Cdc13, Est1 requires a free end for binding to DNA (43). We noticed a possible RNA-binding motif in Est1 and have studied the role of this motif with the idea that Est1 might bind Tlc1 directly.
MATERIALS AND METHODS
Yeast strains, genetic manipulations, and plasmids.
All yeast strains were derived from W303 (MATa/MATα ade2-1/ade2-1 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 can1-100/can1-100 ssd1-d/ssd1-d [psi+]) (41). Yeast media and transformation were as described previously (42).
The plasmids for generating est1Δ, ebs1Δ, est2Δ, and est3Δ strains were made as follows. The EST1 open reading frame was amplified by PCR and cloned into pT7Blue (Novagen). The NruI-EcoRV fragment of EST1 was then replaced with URA3 on a SmaI fragment. The EBS1 gene was cloned into pBSII-SK(+). The HincII-HindIII fragment of EBS1 was then replaced with URA3 on a SmaI-HindIII fragment. The est2 and est3 mutations were generated in diploid W303 by oligomer (oligo)-directed recombination (38) using the oligos shown in Table 1. Haploid est1Δ, ebs1Δ, est2Δ, and est3Δ strains were obtained by tetrad dissections of heterozygous diploids. Haploid tlc1Δ strains were made by sporulation of a tlc1Δ/TLC1 diploid (40).
TABLE 1.
Oligos used in this study
| Name | Length (nucleotides) | Sequence (5′→3′) | Purpose |
|---|---|---|---|
| EST1-5Bam | 43 | GAATTCCCGGGGATCCGTATGGATAATGAAGAAGTTAACGAAG | PCR cloning of EST1 |
| EST1-3Pst | 39 | GGGCGTCGACCTGCAGTCAAGTAGGAGTATCTGGCACTT | PCR cloning of EST1 |
| N3HAEST1 | 36 | ACCCCAGATCTTTAAATACCCATACGATGTTCCTGA | 3HA tagging of EST1 |
| BglII3HA | 26 | CCCCAGATCTGAGCAGCGTAATCTGG | 3HA tagging of EST1 |
| EST1-RNP1A | 63 | GAGAAGATATTATTTTCGGGGAATTTTCTTGCGCTAACGCTGCACTAACAGATTTTAATGACG | Making RNP1-NC |
| EST1-RNP1B | 63 | CGTCATTAAAATCTGTTAGTGCAGCGTTAGCGCAAGAAAATTCCCCGAAAATAATATCTTCTC | Making RNP1-NC |
| EST1-RNP1C | 62 | GAGAAGATATTATTTTCAAGGAATTCTCTTGCGTTAACTTTGCACTAACAGATTTTAATGAC | Making RNP1-C |
| EST1-RNP1D | 62 | GTCATTAAAATCTGTTAGTGCAAAGTTAACGCAAGAGAATTCCTTGAAAATAATATCTTCTC | Making RNP1-C |
| EST1-RNP2A | 47 | GCACTTCATTCGCCGGGTTGCTGAACGACGCGATAAATAGTCCACTG | Making RNP2-ga |
| EST1-RNP2B | 47 | CAGTGGACTATTTATCGCGTCGTTCAGCAACCCGGCGAATGAAGTGC | Making RNP2-ga |
| EST1-RNP2C | 52 | GGAAGTTTTGCACTTCATTCGCCGGATCCCTGAACGACTTGATAAATAGTCC | Making RNP2-gs |
| EST1-RNP2D | 52 | GGACTATTTATCAAGTCGTTCAGGGATCCGGCGAATGAAGTGCAAAACTTCC | Making RNP2-gs |
| EST1F511SA | 48 | CATTAACTTTGCACTAACAGATTCTAATGACGATTATGTGTATGATTC | Making F511S |
| EST1F511SB | 48 | GAATCATACACATAATCGTCATTAGAATCTGTTAGTGCAAAGTTAATG | Making F511S |
| EST1D513IA | 45 | CTTTGCACTAACAGATTTTAATATCGATTATGTGTATGATTCTCC | Making D513I |
| EST1D513IB | 45 | GGAGAATCATACACATAATCGATATTAAAATCTGTTAGTGCAAAG | Making D513I |
| EST2KOUP | 70 | ATGAAAATCTTATTCGAGTTCATTCAAGACAAGCTTGACATTGATCTACAGACCAGGGAACAAAAGCTGG | Making est2Δ |
| EST2KODW | 70 | CAGCATCATAAGCTGTCAGTATTTCATGTATTATTAGTACTAATTAACTATATGCTATAGGGCGAATTGG | Making est2Δ |
| EST2KODGUP | 19 | CCATAACTAACACGCCCTC | Diagnosis of est2Δ |
| EST2KODGDW | 21 | GGCTTATTACAAAGTTTGCGG | Diagnosis of est2Δ |
| EST3KOUP | 79 | GGGATAACAAGTAAACAATGCCGAAAGTAATTCTGGAGTCTCATTCAAAGCCAACAGACTCAGAGGGAACAAAAGCTGG | Making est3Δ |
| EST3KODW | 75 | GCCTGCAGAAGGTCATAAATATTTATATACAAATGGGAAAGTACTTAACGATCCGACTTCTATAGGGCGAATTGG | Making est3Δ |
| EST3KODGUP | 20 | GGTCAGAATGGGCGCTTGTC | Diagnosis of est3Δ |
| EST3KODGDW | 21 | CTCTAGAGGAGTACTTATCGG | Diagnosis of est3Δ |
| TELPG | 27 | GTGTGTGGGTGTGTGTGTGGGTGTGTG | Telomere probe |
Plasmid pVL242 was a gift from V. Lundblad. It contains GAL-EST1 tagged with a triple hemagglutinin (3HA) sequence and was used for the Est1 localization study shown in Fig. 1. Plasmid pJZ-3HA-EST1 was constructed as follows. pLexA-EST1 was constructed by cloning a BamHI-PstI fragment carrying EST1 into pBTM116 (2μm based, ADH1 promoter). This 2.1-kb fragment was obtained by amplifying EST1 using oligos EST1-5Bam and EST1-3Pst (Table 1). A 3HA sequence, produced by PCR using oligos N3HAEST1 and BglII3HA and template pMPY-3×HA (37), was inserted into pLexA-EST1 at the BamHI site between the LexA gene and EST1 to generate pJZ-3HA-EST1, the 3HA-tagged EST1 plasmid. Thus, EST1 is expressed at a high copy number from the ADH1 promoter. Both pLexA-EST1 and pJZ-3HA-EST1 fully complement the senescence and short-telomere phenotypes of an est1Δ strain. EST1 mutant alleles were made using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.). The oligos used for mutagenesis are listed in Table 1. The mutations were confirmed by DNA sequencing.
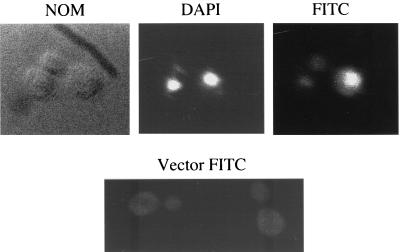
FIG. 1.
Est1 is a nuclear protein. (Top panels) Est1 tagged with 3HA was expressed from the GAL promoter (plasmid pVL242). Cells were fixed and stained with 4′,6′-diamidino-2-phenylindole (DAPI), 12CA5 (anti-HA antibody), and fluorescein isothiocyanate (FITC)-labeled secondary antibody. Cells were visualized by Nomarski optics (NOM) or by fluorescence for DAPI (DAPI) or fluorescence for FITC (FITC). About one-third of all cells examined showed strong tag- and antibody-dependent nuclear FITC staining. (Bottom panel) Control strain containing an empty vector processed for FITC staining.
Yeast cell extracts, immunoprecipitation, total RNA preparation, and Northern blot analysis.
For preparation of yeast extracts, yeast cells were grown to mid-log phase (optical density at 600 nm [OD600], about 1) with selection for plasmid-borne markers. Cells were harvested by centrifugation and washed once with ice-cold water and once with high-salt lysis buffer (HSLB) (40). All operations thereafter were carried out at 4°C. The cell pellet from 50 ml of cells at an OD600 of 1 was suspended in HSLB containing protease inhibitors and an RNase inhibitor, and about 750 μl of acid-washed glass beads was added. The cells were then either quickly frozen to −70°C and stored or broken by shaking in a Mini-Bead-Beater (Biospec). Breaking was accomplished by 30 s of vibration at the top speed twice, separated by 3 min of cooling on ice. The mixture was centrifuged at 14,000 rpm at 4°C for 5 min in an Eppendorf microcentrifuge, and the resulting supernatant was transferred into a fresh tube. The beads were washed with an additional 300 μl of the same buffer and spun to collect additional protein. The two supernatants were combined and spun again under the same conditions. The supernatant was transferred into a clean tube. The protein concentration of such cell extracts was typically 12 to 20 mg/ml, as determined by the Bradford method (Bio-Rad kit). Such extracts were used for immunoprecipitation.
Immunoprecipitation was carried out by incubating 500 to 1,000 μl of cell extract with 0.5 to 1.0 μl of 12CA5 ascitic fluid (ascitic fluid contained about 30 mg of total protein per ml) at 4°C for 60 min, followed by the addition of 10 to 20 μl of HSLB-washed protein A-agarose beads. The incubation was continued for a further 60 to 90 min. The beads were collected by a gentle, brief spin and washed four times with the same buffer but containing neither protease inhibitors nor an RNase inhibitor. The beads were suspended in 20 to 50 μl of HSLB and then split for RNA preparation and for Western blot analysis.
For RNA preparation, the immunoprecipitate was diluted fivefold with 10 mM Tris–1 mM EDTA (pH 8) and extracted with phenol-chloroform-isoamyl alcohol (25:24:1). Nucleic acids were precipitated with 0.3 M sodium acetate and 3 volumes of ethanol. This RNA was called immunoprecipitated RNA (IP-RNA).
For total RNA preparation, 3 to 5 ml of mid-log-phase cells at an OD600 of about 1 was pelleted, washed once with ice-cold water, and suspended in 250 μl of LETS buffer (0.1 M LiCl, 0.01 M EDTA, 0.01 M Tris-HCl [pH 7.4], and 0.2% sodium dodecyl sulfate in diethylpyrocarbonate-treated water). Acid-washed glass beads (300 μl) and 300 μl of phenol-chloroform were added to the cell suspension. The mixture was vortexed at the top speed for 15 s twice, with an interval of 3 min on ice. Another 200 μl of LETS buffer was added, and the mixture was vortexed briefly. The organic and aqueous phases were separated by spinning at 14,000 rpm for 5 min. The upper aqueous phase was transferred to a clean tube, and the RNA was precipitated with ethanol.
Northern analysis was carried out as follows. Total RNA (2.5 to 7.5 μg) or one-third of a sample of IP-RNA was fractionated by electrophoresis on a 6.0% formaldehyde–1.0% agarose gel and transferred to NYTRAN PLUS Nylon membranes (Schleicher & Schuell, Inc.). The blots were probed with the 32P-labeled full-length ACT1 fragment and the labeled full-length TLC1 fragment.
Genomic DNA preparation and Southern blot analysis.
Genomic DNA was prepared after cells were broken with glass beads. For Southern analysis of telomere length, 1.0 μg of genomic DNA was digested with XhoI or PstI endonuclease at 37°C for 2 h. In some Southern blots (e.g., see Fig. 3 and Fig. 5C, right panel), 3 ng of DNA from an HaeII-NdeI digest of plasmid pBST3 was added to the digested genomic DNA. This digestion produced a fragment of 511 bp and a fragment of 1,436 bp that hybridize to yeast telomeric sequences and so serve as molecular size markers on Southern blots. Electrophoresis was carried out at 80 V for about 5 h. The separated DNA was transferred to NYTRAN PLUS Nylon membranes and probed with a 32P-labeled yeast telomere sequence (TELPG; Table 1).
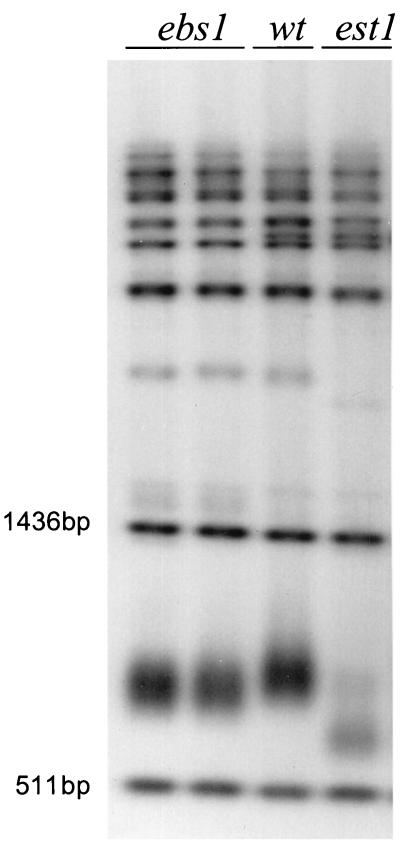
FIG. 3.
The ebs1 mutant has short telomeres. Genomic DNA was isolated from two ebs1 null mutants, a wild-type (wt) strain, and an est1 null mutant. DNA was digested with PstI, mixed with 511- and 1,436-bp size markers, and fractionated by agarose gel electrophoresis. After Southern blotting, DNA was hybridized with a 32P-labeled telomeric sequence.
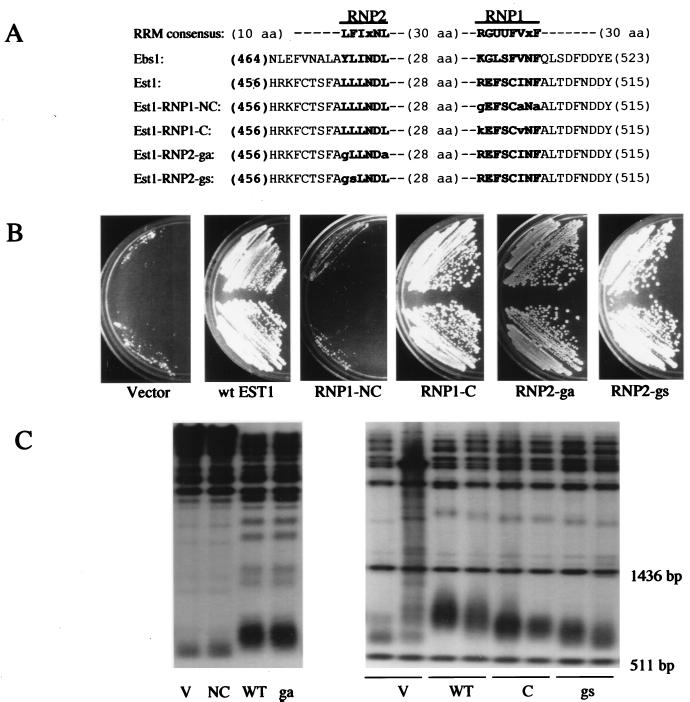
FIG. 5.
Phenotypes caused by altering the RRM region of Est1. (A) The RRM consensus sequence (1, 17) is compared to RRM-like regions in Ebs1 and Est1. The most highly conserved regions (RNP1 and RNP2) are shown in bold lettering. U, hydrophobic amino acid (aa). The amino acid changes made in the four est1 mutants are indicated by lowercase letters. To the right of RNP1 in Est1 is the sequence FNDDY; F is F511, and the first D is D513. These amino acids are altered in two previously characterized alleles of est1 (43) (see the Discussion). (B) A presenescent est1Δ strain (about 35 generations removed from the original spore clone) was transformed with plasmids based on pJZ-3HA-EST1 harboring the empty vector, wild-type (wt) EST1, or the NC, C, ga, or gs mutant allele of EST1. Two independent colonies from each transformation were streaked on YEPD plates and photographed after 60 h of growth. (C) Genomic DNA was extracted from each transformant, digested with XhoI (left panel) or PstI (right panel), and fractionated by agarose gel electrophoresis. After Southern blotting, telomeric DNA was detected with 32P-labeled oligo TELPG. V, transformants containing an empty vector; WT, wild type. In the right panel, the two sharp bands just below and above the fuzzy telomere bands are molecular size markers of 511 and 1,436 bp, respectively (see Materials and Methods).
Single-stranded DNA-binding assay.
The binding reaction was conducted with a total reaction volume of 20 μl. The binding buffer (43) contained 50 μg of poly(dI-dC) per ml. To this binding buffer was added 5.0 μl of the immunoprecipitate that had been washed once with binding buffer after the standard immunoprecipitation protocol (see above). After incubation at room temperature for 8 min, a mixture of 3.0 ng of 32P-labeled TELPG (27-mer) and 6.6 ng of 32P-labeled oligo EST1-RNP1C (62-mer) was added to the reaction, and incubation was continued for another 15 min at room temperature. The reaction was stopped by gently adding 1,000 μl of binding buffer. The beads were collected immediately, washed gently three times in binding buffer, and eventually suspended in 10 μl of binding buffer. Four microliters of loading buffer (0.25% bromophenol blue, 0.25% xylene cyanol, 30% glycerol) was added to the suspension. The mixture was heated in boiling water for 3 min before being loaded onto a 7.6% polyacrylamide–7.0 M urea–1× Tris-borate-EDTA sequencing gel. After electrophoresis, the gel was placed on Whatman paper and vacuum dried. The signal was quantified using a Molecular Dynamics PhosphorImager.
Telomerase assay.
Telomerase was purified and assayed as previously described (34). About 15 μg of each partially purified fraction was used for each telomerase assay. For the RNase A control, 50 ng of RNase A was added to the partially purified telomerase fraction; this mixture was incubated at room temperature for 5 min, and then telomerase activity was assayed.
RESULTS
Est1 is a nuclear protein.
If Est1 is a component of active telomerase at telomeres, then it ought to be a nuclear protein. On the other hand, if Est1 is less directly involved in telomerase function, then it might have some other location. We overexpressed a functional, 3HA-tagged version of Est1 and localized it by immunofluorescence. All the Est1-dependent immunofluorescence appeared to be in the nucleus (Fig. 1).
The association of Est1 with Tlc1 does not depend on EST2 or EST3.
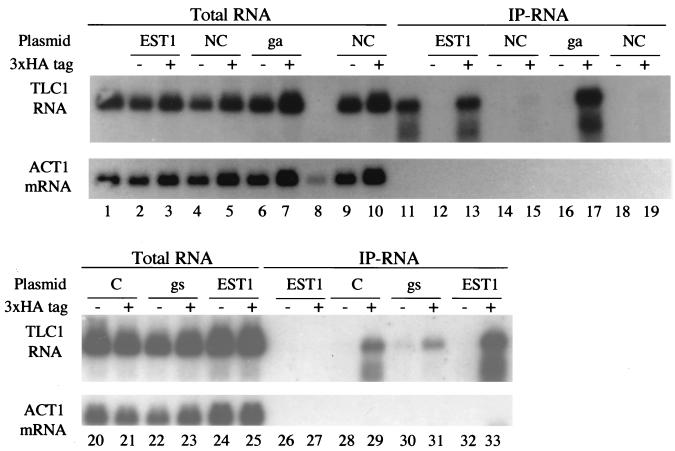
We have previously shown that the Est1 protein is associated with the Tlc1 RNA (40). To find out whether this interaction depends on other telomerase components, we examined the Est1-Tlc1 association in est2 and est3 deletion strains. A functional, 3HA-tagged version of Est1 was expressed in yeast and immunoprecipitated. Immunoprecipitates were processed for Northern blotting, and the presence of Tlc1 and control RNAs was assayed. Figure 2 shows the result from a typical experiment. The Tlc1 RNA was detected in the tagged Est1 immunoprecipitates from wild-type, est2, and est3 cells. This association was specific, since control ACT1 mRNA was not detected in the 3HA-tagged Est1 immunoprecipitate, and no RNA was detected in the untagged Est1 immunoprecipitate. The Tlc1 RNA signal revealed by Northern analysis is not due to any DNA contamination, since RNase A treatment completely eliminates the signal (see Fig. 6, lanes 26 and 27). These results indicate that the association of Est1 and Tlc1 does not require Est2 or Est3, the only other known components of the telomerase complex. Thus, the interaction of Est1 with Tlc1 may be direct.
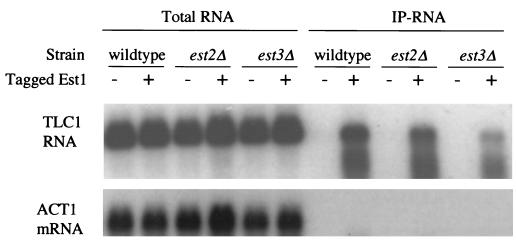
FIG. 2.
The association of Tlc1 RNA with Est1 does not depend on EST2 or EST3. The amount of Tlc1 RNA (top panel) or ACT1 mRNA (bottom panel) in various experiments was assayed by Northern blotting. “Total RNA” shows the total RNA from wild-type, est2, or est3 strains, each of which either carries (+) or does not carry (−) tagged Est1 expressed from the ADH1 promoter (pJZ-3HA-EST1). “IP-RNA” shows the RNA associated with 3HA-tagged Est1 after the latter was immunoprecipitated from tagged (+) or untagged control (−) strains with monoclonal antibody 12CA5. The amount of Tlc1 precipitated was somewhat smaller in the est2 and est3 strains than in the wild-type strain in all three of three experiments.
FIG. 6.
Association of Tlc1 RNA with Est1 RRM mutant proteins in vivo. Total RNA and immunoprecipitated RNA (IP-RNA) were made from est1 cells expressing wild-type or mutant forms of either untagged (−) or 3HA-tagged (+) Est1 from the ADH1 promoter (plasmid pJZ-3HA-EST1 and derivatives). The amounts of Tlc1 RNA and ACT1 mRNA in total RNA or coimmunoprecipitated with Est1 were assayed by Northern blotting. Two independent transformants of Est1-RNP1-NC were assayed. Controls included RNA from a tlc1 deletion mutant (lane 8), RNA from a strain carrying 3HA-tagged Est1 expressed from the genomic EST1 locus (lanes 1 and 11), and IP-RNA treated with RNase A before loading (lanes 26 and 27).
The coprecipitation of Tlc1 with Est1 was consistently less efficient in est2 or est3 mutant cells than in wild-type cells (Fig. 2). This result could be a sign that Tlc1-Est1 complex formation in vivo is aided by Est2 and Est3, although they are not required. Alternatively, the relatively poor coprecipitation of Tlc1 in the est mutants could be due to the sickness of the est cells, which in our experiments led to somewhat lower protein concentrations in cell lysates.
Ebs1 is a homolog of Est1.
Database searches show that the Est1 protein has 48% similarity (27% identity) to the protein encoded by the poorly characterized gene EBS1 (open reading frame YDR206w). This similarity is spread throughout the two proteins but is most pronounced in a 100-amino-acid region centered on a putative RNA recognition motif (RRM) in Ebs1 (see below). Interestingly, disruption of EBS1 resulted in slightly shortened telomeres (Fig. 3), although there was no senescence phenotype. Furthermore, there was an indication that est1 ebs1 double mutants had a slightly stronger senescence phenotype than est1 single mutants alone. est1 single-mutant spore clones show a variable time to senescence, but est1 ebs1 double mutants always senesce as fast as the fastest-senescing est1 single mutants. These results suggest that EBS1 is slightly redundant with EST1 for telomere maintenance.
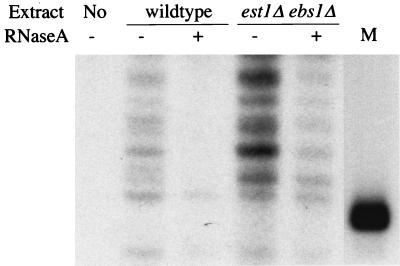
It has previously been shown that protein extracts from est1 mutants have telomerase activity in vitro (5). Since we now believe that EBS1 may be a homolog of EST1, we wondered whether this in vitro telomerase activity in est1 strains might have required EBS1. Therefore, we made extracts from an est1 ebs1 double mutant. These extracts also contained telomerase activity (Fig. 4), supporting the previous conclusion that EST1 function is not essential for in vitro telomerase activity.
FIG. 4.
The EBS1 gene is not essential for telomerase activity in vitro. Telomerase was partially purified (34) from a wild-type strain and from an est1 ebs1 double mutant. Telomerase activity was assayed (34) in the absence (−) or presence (+) of 50 ng of RNase A. Two to three times as much protein was used in the est1 ebs1 assay as in the wild-type assay, accounting for the apparently higher level of telomerase activity. Lane M, end-labeled 28-mer which serves as a marker for the input telomeric primer oligo.
Est1 has a functionally important RRM.
Computer analysis of the Ebs1 protein showed that it contained a good match to an RRM (Fig. 5). The RRM in Ebs1 led us to a putative RRM in Est1 (Fig. 5). Although this putative Est1 RRM has only a poor match to a consensus RRM, it is also true that RRMs of this class can share structural similarity without highly conserved sequence similarity, and it is the structural similarity that is important (1, 4, 17). An RRM consists of 70 to 90 amino acid residues (1, 4, 17). RNP1 and RNP2 are two important and more highly conserved segments within an RRM. The RRM crystal structures of human hnRNP C and U1 snRNP A show that the RRM forms a module of four antiparallel beta sheets on the surface and two alpha helices underneath (a βαββαβ secondary structure). RNP1 and RNP2 form the two central antiparallel beta sheets and are directly involved in single-stranded RNA interactions.
On the basis of sequence and structural information, we created two mutations in the putative Est1 RNP1 (Fig. 5A). The first was called RNP1-NC; this is a triple substitution which replaces three structurally important residues with three dissimilar amino acids. Thus, these are nonconservative (NC) changes, and the RNP1-NC mutation would be expected to reduce or eliminate RRM function. The second mutation was called RNP1-C; this is a double substitution which replaces two structurally important residues with similar amino acids found in the RNP1 motifs of some proteins. Thus, these are conservative (C) changes, and if this region of Est1 is truly an RRM, then RNP1-C might not reduce RRM function. These mutant forms of Est1 were tagged with a 3HA epitope.
The est1-rnp1-nc mutant failed to complement either the senescence or the telomere shortening of est1Δ strains (Fig. 5B and C). Furthermore, when Est1-RNP1-NC was immunoprecipitated, no Tlc1 RNA could be detected in the immunoprecipitate (Fig. 6, lanes 15 and 19; two individual transformants). This was not because of a lack of Est1-RNP1-NC protein, because Western analysis showed that equivalent amounts of wild-type Est1 and Est1-RNP1-NC were immunoprecipitated (data not shown). These results suggest that the est1-rnp1-nc mutation eliminates or severely impairs the ability of Est1 to bind to Tlc1, and this change correlates with a loss of EST1 function.
In contrast, the est1-rnp1-c mutant, in which two amino acids are substituted with conserved residues expected to function in the context of an RNP1 motif (Fig. 5), complements the est1Δ phenotype for viability and gives telomeres only slightly shorter than the wild type (Fig. 5). Est1-RNP1-C coimmunoprecipitates with Tlc1 only slightly less well than it does with wild-type Est1 (Fig. 6). Again, the ability to coimmunoprecipitate with Tlc1 is correlated with genetic function.
We also made two mutations in the RNP2 region of Est1. Few mutagenesis studies have been done with the RNP2 region of other RRMs, so it was not clear what phenotypes our RNP2 mutations should create. The RNP2-ga mutation makes two nonconservative changes; the first change is at the beginning of the beta strand, and the second is after the end of the beta strand, in the following loop. The RNP2-ga mutant is wild type for the three phenotypes assayed (cell viability, telomere length, and the Tlc1 RNA association) (Fig. 5 and 6). The RNP2-gs mutation makes two nonconservative changes; the first, again, is at the beginning of the beta strand, and the second (L to S) is in the beta strand at a well-conserved hydrophobic residue that appears to be important for the interaction with RNA, at least in some RRMs (17). The RNP2-gs mutant has significantly shortened telomeres, but it rescues the senescence of est1Δ strains (Fig. 5). This mutant protein has a reduced ability to associate with Tlc1 (Fig. 6).
In all these experiments, the ability of mutant proteins to maintain telomere length was highly correlated with their ability to coimmunoprecipitate with Tlc1 RNA (Table 2).
TABLE 2.
Summary of EST1 RRM mutants
| EST1 allele | Phenotypea
|
||
|---|---|---|---|
| Viability | Tlc1 RNA binding | Telomere length | |
| Wild type | +++ | +++ | +++ |
| RNP1-NC | −−− | −−− | −−− |
| RNP1-C | +++ | ++ | ++ |
| RNP2-ga | +++ | +++ | +++ |
| RNP2-gs | +++ | + | + |
+++, wild-type behavior; ++ or +, EST1 activity slightly or considerably less than wild-type activity, respectively; −−−, no EST1 activity (i.e., no complementation of an est1 mutant).
The EST1-RNP1-NC mutant has dominant negative phenotypes.
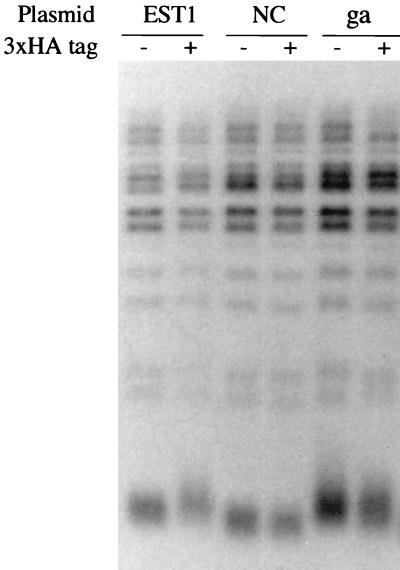
The failure of the triple substitution, est1-rnp1-nc, to complement an est1 mutation raised the question of whether the mutations might have grossly disturbed protein structure (although the fact that Est1 and Est1-RNP1-NC proteins were found equally abundant by Western analysis argues against this notion). However, we found that wild-type EST1 cells transformed with overexpressed est1-rnp1-nc had at least three phenotypes. First, they grew slowly (data not shown). Second, as assayed with a Coulter Channelyzer, average cell size was abnormally large, and microscopic examination showed that some cells were large dumbbells, typical of DNA damage arrest (data not shown). Third, these cells had relatively short telomeres (Fig. 7). These dominant phenotypes suggest that the mutant Est1-RNP1-NC protein retains some functions of wild-type Est1 and consequently disturbs normal telomere maintenance. This notion in turn suggests that the disruption of the putative RRM causes a fairly specific defect in the protein as opposed to a general defect in protein folding.
FIG. 7.
The est1 rnp1 nc mutant has a dominant short-telomere phenotype. Genomic DNA from an EST1 W303α strain also expressing plasmid-borne EST1 or est1-rnp1-nc or est1-rnp2-ga from the ADH1 promoter (pJZ-3HA-EST1 and derivatives) was digested with XhoI and analyzed by Southern blotting with 32P-labeled TELPG as a probe.
The Est1-RNP1-NC mutant can bind single-stranded telomeric DNA.
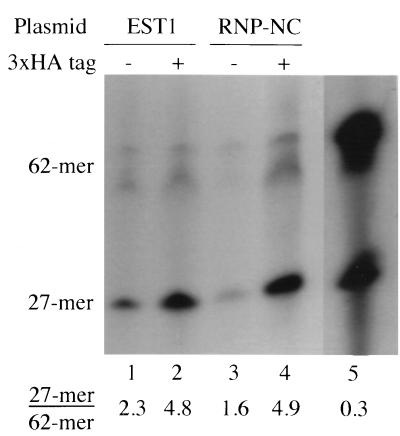
Virta-Pearlman et al. reported that Est1 protein can bind to yeast single-stranded TG-rich telomeric DNA, albeit with low affinity (43). In some proteins, RRMs can bind to single-stranded DNA (8, 14, 16, 31, 35). In fact, single-stranded TG-rich telomeric DNA may be particularly good at interacting with RRMs (8, 16, 31). Thus, we wished to know whether the RRM of Est1 was responsible for the DNA-binding activity detected by Virta-Pearlman et al. (43). We developed a single-stranded DNA-binding assay (see Materials and Methods). This assay compares the ability of immunoprecipitated 3HA-tagged Est1 protein to bind two different single-stranded DNAs, one with a telomeric sequence and one with an irrelevant sequence. The assay has a high background (i.e., there is significant binding of telomeric sequences even in the absence of Est1); nevertheless, immunoprecipitates from tagged Est1 strains reproducibly bind more telomeric sequence than do immunoprecipitates from untagged control strains. Figure 8 shows the results of one typical experiment. Immunoprecipitates from the tagged Est1 strain retained two- to threefold more telomeric sequence (oligomer TELPG; 27-mer) than immunoprecipitates from the untagged control strain (Fig. 8). Seven independent experiments (Table 3) were evaluated by analysis of variance (ANOVA), and the difference between the tagged and untagged strains was significant at the 0.05 level. The immunoprecipitated Est1 protein from tlc1Δ cells gave similar results (data not shown), indicating that the Tlc1 RNA bound to Est1 in vivo does not significantly interfere with the DNA-binding capacity of Est1.
FIG. 8.
The Est1-RNP1-NC mutant protein can bind single-stranded telomeric DNA. Monoclonal antibody 12CA5 and protein A beads were mixed with extract from tagged (+) or untagged (−) Est1 strains (lanes 1 to 4). After washing was done, immunoprecipitates were challenged with a mixture of two end-labeled oligos, a 62-mer (EST1-RNP1C) of irrelevant sequence (Table 1) and a TG-rich, telomeric 27-mer (TELPG) (Table 1). Immunoprecipitates were then washed, and the bound oligos were assayed by phosphorimaging after gel electrophoresis. Lane 5 shows 1/20 the amount of the 27-mer–62-mer mixture that was added to the immunoprecipitates in the other lanes. The ratio of 27-mer to 62-mer is shown beneath the lanes.
TABLE 3.
Summary of yeast single-stranded telomeric DNA bindinga
| Est1 protein | Ratio of 27-mer to 62-mer
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Data from expt:
| ||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | Avg | |
| Est1 | 1.7 | 1.8 | 2.3 | 2.3 | 2.4 | 2.1 | 1.4 | 2.0 |
| 3HA-tagged Est1 | 4.4 | 5.1 | 5.4 | 4.8 | 2.7 | 4.4 | 2.2 | 4.1 |
| Est1-RNP1-NC | 1.6 | 1.6 | 2.0 | 1.2 | 1.6 | |||
| 3HA-tagged Est1-RNP1-NC | 2.9 | 4.9 | 4.4 | 1.7 | 3.5 | |||
Data from 11 independent pairs of experiments are presented as the ratio of 27-mer (telomeric oligo) to 62-mer (nontelomeric oligo). Seven of these pairs of experiments compared the single-stranded DNA-binding activity of 12CA5 immunoprecipitates from untagged and tagged wild-type Est1 strains. The average ratio for untagged Est1 was 2.0; the average ratio for tagged Est1 was 4.1. By ANOVA, this difference was found statistically significant at the 0.05 level. The other four pairs of experiments compared the single-stranded DNA-binding activity of 12CA5 immunoprecipitates from untagged and tagged Est1-RNP1-NC strains. Again, ANOVA showed that the ratios (1.6 versus 3.5) were significantly different. In addition to the experiments shown here, there were one pair of experiments with wild-type Est1 and one pair of experiments with Est1-RNP1-NC (both of these pairs were done on the same day); they were excluded from the analysis because of abnormally high ratios in all parts of the experiments, including controls.
Figure 8 also shows the result from one typical experiment with the immunoprecipitated Est1-RNP1-NC mutant protein. Est1-RNP1-NC binds single-stranded TG-rich telomeric DNA just as well as does wild-type Est1. Again, the difference between tagged Est1-RNP-NC and untagged Est1-RNP-NC in multiple experiments (Table 3) was found statistically significant by an ANOVA. This result suggests that the RNA-binding motif is not needed for the single-stranded DNA-binding activity of Est1. We note, however, that this result does not directly address the issue of whether the wild-type RRM of Est1 is capable of binding single-stranded G-rich DNA. Because our experiments have been done with Est1 immunoprecipitated from yeast cells, the RRM-independent interaction between single-stranded telomeric oligos and immunoprecipitated Est1 could be due to some protein associated with Est1 and not necessarily to direct binding of single-stranded DNA to Est1. It is also possible that mutant RRM retains the ability to bind single-stranded DNA, even though it binds RNA very poorly.
Two other alleles of est1 separate the single-stranded DNA-binding function from the RNA-binding function.
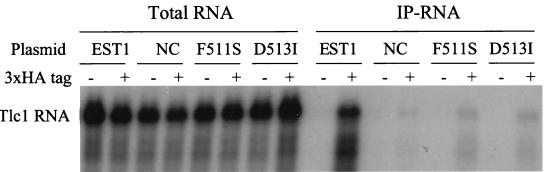
There are two previously known nonfunctional alleles of est1 that happen to fall within the RRM. These are est1F511S and est1D513I (F511 and D513 are shown in Fig. 5A) (43). These alleles are phenotypically similar to est1-rnp1-nc in that they fail to complement an est1 deletion, and they cause telomere shortening when overexpressed. However, both mutant proteins, when purified from Escherichia coli, have wild-type ability to bind single-stranded DNA in vitro (43). We have tested both of these mutant proteins for RNA binding by immunoprecipitation. Comparable amounts of wild-type and mutant proteins were immunoprecipitated (data not shown), but only the wild-type protein caused coimmunoprecipitation of substantial amounts of Tlc1 (Fig. 9). Thus, these alleles, like est1-rnp-nc, separate the RNA-binding function of Est1 from the single-stranded DNA-binding function.
FIG. 9.
The Est1 F511S and D513I mutant proteins are defective in binding Tlc1 RNA. Total RNA (left) and IP-RNA (right) were made from est1 cells expressing wild-type or mutant forms of either untagged (−) or 3HA-tagged (+) Est1 from the ADH1 promoter (plasmid pJZ-3HA-EST1 and derivatives). The amount of Tlc1 RNA in total RNA or coimmunoprecipitated with Est1 was assayed by Northern blotting. The amount of immunoprecipitated protein was assayed by Western blotting (data not shown).
DISCUSSION
We have shown previously that Est1 is in a complex with active telomerase. Here we report that Est1 binds to Tlc1 RNA. This binding is very likely direct, as it requires neither EST2 nor EST3. When mutations are made in the RRM of Est1, Est1 partially or wholly loses its ability to associate with Tlc1. Furthermore, the severity of this association defect is correlated with the severity of the telomere shortening and senescence phenotypes of each mutant.
Previously, it has been shown that Est1 is a single-stranded DNA-binding protein in vitro with specificity for TG-rich telomeric sequences (43). In many cases, RRMs can bind both single-stranded RNA and single-stranded DNA. In particular, it seems quite common to find RRMs capable of binding TG-rich single-stranded DNA (8, 15, 16, 18, 19, 22, 28, 31, 33, 35). This fact raises the issue of whether the previously observed binding to TG-rich single-stranded DNA was due to the RRM of Est1. One argument that this might be the case is that the deletion of amino acids 435 to 565 of Est1 greatly diminished single-stranded DNA binding (43) and removed the RRM.
However, there are a number of strong arguments that single-stranded DNA binding probably is not dependent on the RRM. First, Est1 binds d(TGTGTGGG)3 but does not bind the corresponding polyribonucleotide, r(UGUGUGGG)3 (43), contrary to the expectation if an RRM were responsible for binding single-stranded DNA (8, 16). Second, a 1,000- to 10,000-fold molar excess of Tlc1 RNA fails to prevent single-stranded telomeric DNA from binding to Est1 (43). Third, two missense alleles that fall within the RRM of EST1, est1F511S and est1D513I, lack the ability to bind Tlc1 RNA (Fig. 9) but retain the ability to bind single-stranded DNA (43). These mutations thus separate the single-stranded DNA-binding function from the Tlc1 RNA-binding function. These two alleles have phenotypes strikingly similar to that of est1-rnp1-nc, as expected if their defect were specific for binding Tlc1. Fourth, Est1-RNP1-NC immunoprecipitated from yeast cells (possibly together with associated proteins) can specifically bind single-stranded telomeric DNA (Fig. 8 and Table 3), even though this protein is defective for Tlc1 binding (Fig. 6).
Thus, it appears that Est1 binds to the yeast telomerase RNA, Tlc1, via an RRM, and binds to single-stranded telomeric DNA via some other nearby motif. Therefore, as suggested by Virta-Pearlman et al. (43), Est1 could serve as a bridge between telomerase and telomere ends. That is, by simultaneously binding telomerase RNA through the RRM and binding telomeric DNA through a second motif, it could help anchor telomerase at the telomere. Recently, strong evidence for this bridge model has been provided by Evans and Lundblad (9), who showed that a Cdc13-Est2 fusion could entirely bypass the need for Est1.
The bridge model is consistent with the observation that est1 mutants have telomerase activity in vitro (where substrate telomeric oligos are provided at high concentrations) but nevertheless suffer telomere shortening in vivo (where telomere concentrations are low). That is, the need for Est1 may be apparent only at low substrate concentrations; the effect of Est1 may be to reduce the Km of the telomerase reaction. The affinity of Est1 for telomeric DNA is, however, quite low (43); perhaps this interaction is regulated by cell cycle position, telomere length, or other proteins (such as Cdc13) to help regulate steady-state telomere length.
Homologs of Est1 have not been found in other organisms. However, telomerases from other organisms do have protein components in addition to the reverse transcriptase subunit. In Tetrahymena, proteins p80 and p95 are associated with telomerase, and p80 can be cross-linked to the RNA component in vivo (6). A human protein called TP1 (also called TLP1) is associated with human telomerase and has significant homology to Tetrahymena p80 (12, 30). Furthermore TP1 can interact with the RNA component of human telomerase in the yeast-based three-hybrid assay, suggesting that TP1 binds to RNA directly. Thus, Tetrahymena, human, and perhaps other telomerases may contain RNA-binding proteins with functions analogous to the function of Est1. Since RNA-binding motifs are often short and are not all of the RNP1 or RNP2 type, functional homologs of Est1 will not necessarily have any protein sequence similarity to Est1. Moreover, it is not clear that tethering of telomerase to telomeres must occur through the RNA template, as it apparently does in S. cerevisiae. Instead, in some organisms, tethering might occur through the reverse transcriptase catalytic subunit.
ACKNOWLEDGMENTS
We thank V. Lundblad, S. Evans, and A. Krainer for helpful discussions and for communicating results prior to publication. We also thank V. Lundblad for plasmid pVL242 (GAL-EST1).
This work was supported by the Council for Tobacco Research (grant 4574) and by the U.S. Army Breast Cancer Research Program (grant DAMD17-97-1-7315).
REFERENCES
- 1.Birney E, Kumar S, Krainer A R. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourns B D, Alexander M K, Smith A M, Zakian V A. Sir proteins, Rif proteins, and Cdc13p bind Saccharomyces telomeres in vivo. Mol Cell Biol. 1998;18:5600–5608. doi: 10.1128/mcb.18.9.5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryan T M, Sperger J M, Chapman K B, Cech T R. Telomerase reverse transcriptase genes identified in Tetrahymena thermophila and Oxytricha trifallax. Proc Natl Acad Sci USA. 1998;95:8479–8484. doi: 10.1073/pnas.95.15.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 5.Cohn M, Blackburn E H. Telomerase in yeast. Science. 1995;269:396–400. doi: 10.1126/science.7618104. [DOI] [PubMed] [Google Scholar]
- 6.Collins K, Kobayashi R, Greider C W. Purification of Tetrahymena telomerase and cloning of genes encoding the two protein components of the enzyme. Cell. 1995;81:677–686. doi: 10.1016/0092-8674(95)90529-4. [DOI] [PubMed] [Google Scholar]
- 7.Counter C M, Meyerson M, Eaton E N, Weinberg R A. The catalytic subunit of yeast telomerase. Proc Natl Acad Sci USA. 1997;94:9202–9207. doi: 10.1073/pnas.94.17.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ding J, Hayashi M K, Zhang Y, Manche L, Krainer A R, Xu R M. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev. 1999;13:1102–1115. doi: 10.1101/gad.13.9.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans S K, Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- 10.Gandhi L, Collins K. Interaction of recombinant Tetrahymena telomerase proteins p80 and p95 with telomerase RNA and telomeric DNA substrates. Genes Dev. 1998;12:721–733. doi: 10.1101/gad.12.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greider C W. Telomere length regulation. Annu Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 12.Harrington L, McPhail T, Mar V, Zhou W, Oulton R, Bass M B, Arruda I, Robinson M O. A mammalian telomerase-associated protein. Science. 1997;275:973–977. doi: 10.1126/science.275.5302.973. [DOI] [PubMed] [Google Scholar]
- 13.Harrington L, Zhou W, McPhail T, Oulton R, Yeung D S, Mar V, Bass M B, Robinson M O. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev. 1997;11:3109–3115. doi: 10.1101/gad.11.23.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikeda M, Arai K, Masai H. CTBP1/RBP1, a Saccharomyces cerevisiae protein which binds to T-rich single-stranded DNA containing the 11-bp core sequence of autonomously replicating sequence, is a poly(deoxypyrimidine)-binding protein. Eur J Biochem. 1996;238:38–47. doi: 10.1111/j.1432-1033.1996.0038q.x. [DOI] [PubMed] [Google Scholar]
- 15.Ishikawa F, Matunis M J, Dreyfuss G, Cech T R. Nuclear proteins that bind the pre-mRNA 3′ splice site sequence r(UUAG/G) and the human telomeric DNA sequence d(TTAGGG)n. Mol Cell Biol. 1993;13:4301–4310. doi: 10.1128/mcb.13.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnston S D, Lew J E, Berman J. Gbp1p, a protein with RNA recognition motifs, binds single-stranded telomeric DNA and changes its binding specificity upon dimerization. Mol Cell Biol. 1999;19:923–933. doi: 10.1128/mcb.19.1.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenan D J, Query C C, Keene J D. RNA recognition: towards identifying determinants of specificity. Trends Biochem Sci. 1991;16:214–220. doi: 10.1016/0968-0004(91)90088-d. [DOI] [PubMed] [Google Scholar]
- 18.Konkel L M, Enomoto S, Chamberlain E M, McCune-Zierath P, Iyadurai S J, Berman J. A class of single-stranded telomeric DNA-binding proteins required for Rap1p localization in yeast nuclei. Proc Natl Acad Sci USA. 1995;92:5558–5562. doi: 10.1073/pnas.92.12.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LaBranche H, Dupuis S, Ben-David Y, Bani M R, Wellinger R J, Chabot B. Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nat Genet. 1998;19:199–202. doi: 10.1038/575. [DOI] [PubMed] [Google Scholar]
- 20.Lendvay T S, Morris D K, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin J J, Zakian V A. An in vitro assay for Saccharomyces telomerase requires EST1. Cell. 1995;81:1127–1135. doi: 10.1016/s0092-8674(05)80017-0. [DOI] [PubMed] [Google Scholar]
- 22.Lin J J, Zakian V A. Isolation and characterization of two Saccharomyces cerevisiae genes that encode proteins that bind to (TG1-3)n single strand telomeric DNA in vitro. Nucleic Acids Res. 1994;22:4906–4913. doi: 10.1093/nar/22.23.4906. . (Erratum, 22:5516.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin J J, Zakian V A. The Saccharomyces CDC13 protein is a single-strand TG1-3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc Natl Acad Sci USA. 1996;93:13760–13765. doi: 10.1073/pnas.93.24.13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lingner J, Cech T R. Purification of telomerase from Euplotes aediculatus: requirement of a primer 3′ overhang. Proc Natl Acad Sci USA. 1996;93:10712–10717. doi: 10.1073/pnas.93.20.10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lingner J, Cech T R, Hughes T R, Lundblad V. Three even shorter telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc Natl Acad Sci USA. 1997;94:11190–11195. doi: 10.1073/pnas.94.21.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lingner J, Hughes T R, Shevchenko A, Mann M, Lundblad V, Cech T R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 27.Lundblad V, Szostak J W. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- 28.McKay S J, Cooke H. A protein which specifically binds to single stranded TTAGGGn repeats. Nucleic Acids Res. 1992;20:1387–1391. doi: 10.1093/nar/20.6.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamura T M, Morin G B, Chapman K B, Weinrich S L, Andrews W H, Lingner J, Harley C B, Cech T R. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 30.Nakayama J, Saito M, Nakamura H, Matsuura A, Ishikawa F. TLP1: a gene encoding a protein component of mammalian telomerase is a novel member of WD repeats family. Cell. 1997;88:875–884. doi: 10.1016/s0092-8674(00)81933-9. [DOI] [PubMed] [Google Scholar]
- 31.Newberry E P, Latifi T, Towler D A. The RRM domain of MINT, a novel Msx2 binding protein, recognizes and regulates the rat osteocalcin promoter. Biochemistry. 1999;38:10678–10690. doi: 10.1021/bi990967j. [DOI] [PubMed] [Google Scholar]
- 32.Nugent C I, Hughes T R, Lue N F, Lundblad V. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 33.Petracek M E, Konkel L M, Kable M L, Berman J. A Chlamydomonas protein that binds single-stranded G-strand telomere DNA. EMBO J. 1994;13:3648–3658. doi: 10.1002/j.1460-2075.1994.tb06672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prescott J, Blackburn E H. Telomerase RNA mutations in Saccharomyces cerevisiae alter telomerase action and reveal nonprocessivity in vivo and in vitro. Genes Dev. 1997;11:528–540. doi: 10.1101/gad.11.4.528. [DOI] [PubMed] [Google Scholar]
- 35.Reim I, Stanewsky R, Saumweber H. The puff-specific RRM protein NonA is a single-stranded nucleic acid binding protein. Chromosoma. 1999;108:162–172. doi: 10.1007/s004120050365. [DOI] [PubMed] [Google Scholar]
- 36.Saito T, Matsuda Y, Suzuki T, Hayashi A, Yuan X, Saito M, Nakayama J, Hori T, Ishikawa F. Comparative gene mapping of the human and mouse TEP1 genes, which encode one protein component of telomerases. Genomics. 1997;46:46–50. doi: 10.1006/geno.1997.5005. [DOI] [PubMed] [Google Scholar]
- 37.Schneider B L, Seufert W, Steiner B, Yang Q H, Futcher A B. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- 38.Schneider B L, Steiner B, Seufert W, Futcher A B. pMPY-ZAP: a reusable polymerase chain reaction-directed gene disruption cassette for Saccharomyces cerevisiae. Yeast. 1996;12:129–134. doi: 10.1002/(sici)1097-0061(199602)12:2<129::aid-yea891>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 39.Singer M S, Gottschling D E. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 40.Steiner B R, Hidaka K, Futcher B. Association of the Est1 protein with telomerase activity in yeast. Proc Natl Acad Sci USA. 1996;93:2817–2821. doi: 10.1073/pnas.93.7.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas B J, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 42.Tyers M, Tokiwa G, Futcher B. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 1993;12:1955–1968. doi: 10.1002/j.1460-2075.1993.tb05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Virta-Pearlman V, Morris D K, Lundblad V. Est1 has the properties of a single-stranded telomere end-binding protein. Genes Dev. 1996;10:3094–3104. doi: 10.1101/gad.10.24.3094. [DOI] [PubMed] [Google Scholar]