Abstract
We have characterized the mechanism by which coactivator p300 facilitates transcriptional activation mediated by the heterodimer of thyroid hormone (T3) receptor and 9-cis retinoid acid receptor (TR-RXR) in the context of chromatin. We demonstrate that, while p300 can enhance the transcriptional activation mediated by both liganded TR-RXR and GAL4-VP16, its histone acetyltransferase activity (HAT) is required for its ability to facilitate liganded TR-RXR- but not GAL4-VP16-mediated transcriptional activation. To understand how p300 is recruited by liganded TR-RXR, we have analyzed the interactions between TR-RXR and p300 as well as SRC-1 family coactivators. We show that, in contrast to a strong hormone-dependent interaction between TR-RXR and SRC-1 family coactivators, p300 displays minimal, if any, T3-dependent interaction with TR-RXR. However, p300 can be recruited by liganded TR-RXR through its interaction with SRC-1 family coactivators. Consistent with the protein-protein interaction profile described above, we demonstrate that the SRC-1 interaction domain of p300 is important for its ability to facilitate transcriptional activation mediated by TR-RXR, whereas its nuclear receptor interaction domain is dispensable. Collectively, these results reveal the functional significance of the HAT activity of p300 and define an indirect mode for the action of p300 in TR-RXR activation.
The thyroid hormone (T3) receptor (TR) is a member of the nuclear receptor superfamily and plays diverse roles in development, differentiation, homeostasis, and tumorigenesis through its ability to regulate gene expression (13, 46). Although TR can regulate transcription as a monomer or homodimer (31), TR readily forms a heterodimer with another nuclear receptor, 9-cis retinoid acid receptor (RXR) (28, 60, 61), and it is believed that the TR-RXR heterodimer is the functionally active form of TR in vivo (40).
The TR-RXR heterodimer has the capacity to repress transcription in the absence of T3 and activate transcription in the presence of the hormone (4, 16). Despite its ability to interact with general transcription factors, such as transcription factor IIB (TFIIB) and TATA-binding protein (TBP) (3, 14, 15), strong evidence indicates that additional regulatory factors are required for its repression in the absence of T3 (corepressors) as well as for its activation in the presence of T3 (coactivators). Two structurally related corepressors, SMRT (silencing mediator for retinoid and thyroid hormone receptors) and N-COR (nuclear receptor corepressor) (10, 23), have been identified; both bind to TR in the absence of T3 and facilitate repression at least in part through recruiting histone deacetylases (21, 35). On the other hand, an increasing number of coactivators have been implicated in T3-dependent activation; these include SRC-1 (39), TRIP230 (7), TIF1 (32), TIF2/GRIP1 (22, 47), RCA3/pCIP/ACTR/TRAM-1 (8, 33, 43, 44), CBP and p300 (6, 25), PCAF (5, 29), E6-AP (36), and the TRAP complex (15). Among the coactivators, CBP and p300 (38), PCAF (58), SRC-1 (41), and ACTR (8) have been shown to possess intrinsic histone acetyltransferase (HAT) activity. Acetylation of core histones in chromatin has long been proposed to facilitate transcription (1). Indeed, hyperacetylated histones are preferentially associated with transcriptionally active chromatin in mammalian cells, whereas histones in heterochromatin are hypoacetylated (20). The findings that histone acetyltransferases are associated with hormone-dependent activation and that deacetylases are involved in repression by unliganded TR-RXR or retinoid acid receptor (RAR)-RXR have led to a simple model where targeted modification of chromatin could play an important role in transcriptional control by TR-RXR (50).
Among the coactivators, SRC-1, TIF2, and RAC3 share considerable structural similarity and are collectively referred to as coactivators of the SRC-1 family. Their importance in nuclear receptor action was manifested by the diminished hormone response in cells injected with their cognate antibodies (44) and by a partial hormone insensitivity syndrome in SRC-1 null mice (57). CBP and p300 are closely related also and serve as coactivators for a variety of transcription factors, including the family of nuclear receptors. Both CBP and p300 were reported to directly interact with nuclear receptors through a conserved domain in their N termini in a ligand-dependent manner, and this direct interaction was suggested to be important for their involvement in ligand-dependent transactivation of nuclear receptors (6, 25). The importance of CBP and p300 in nuclear receptor action is highlighted by experiments showing that anti-CBP antibody selectively inhibits the transactivation of nuclear receptors in intact cells (25) and that the transactivation of RAR is defective in p300 null mice as well as in F9 cell lines, where p300 activity is inactivated by ribozyme (26, 59). Interestingly, the HAT activity of PCAF but not p300 was found to be essential for the transcriptional activation mediated by RAR (29). This result led to the notion that there is a differential requirement for the multiple HAT activities involved in transcriptional activation by different transcription factors (29).
In addition to the interaction between nuclear receptors and coactivators, multiple interactions among coactivators have been reported (25). Both CBP and p300 can interact directly with SRC-1 family coactivators (25, 44), and both CBP and p300 and SRC-1 family coactivators can interact with PCAF (41, 58). The interaction between p300 and SRC-1 family coactivators appears to be functionally important, since deletion of the SRC-1 interaction domain of p300 largely abolished its ability to mediate RAR activation (34, 48). The observed multiple interactions among coactivators have also led to the suggestion that the above coactivators may exist as preformed coactivator complexes in vivo (44). However, the PCAF complex purified recently contains neither p300 nor SRC-1 family coactivators (37).
Using Xenopus oocytes as a model system, we have previously demonstrated that chromatin organization is an essential component of the transcriptional regulation of the Xenopus thyroid receptor βA gene (TRβA) promoter by TR and its heterodimer partner RXR (52, 53, 55). In the present study, we have characterized the mechanism by which p300 modulates the transactivation of the Xenopus TRβA promoter by liganded TR-RXR. We have demonstrated here that the HAT activity of p300 is essential for its ability to enhance hormone-dependent activation by TR-RXR but not activation by GAL4-VP16 in the context of chromatin. We found, to our surprise, that the direct interaction between liganded TR-RXR and p300, if any, is hardly detectable and that the effect of p300 on TR-RXR activation is primarily mediated indirectly through its interaction with SRC-1 family coactivators.
MATERIALS AND METHODS
Plasmid constructs.
The reporter construct pTRβA5′-7-3xUAS has been described before (52). The constructs for the in vitro synthesis of GAL4-VP16, TR, and RXR mRNAs have also been described before (52). To express SRC-1, TIF2, and RAC3 in Xenopus oocytes, their full-length cDNAs were cloned into the pSP64(polyA) vector with or without the addition of a Flag tag at the N termini. pSP64(polyA)-p300 was constructed by cutting the cDNA encoding full-length human p300 from Rc/RSV-p300 with HindIII and cloning it into pSP64(polyA). All the p300 mutants were constructed based on pSP64(polyA)-p300. To construct the p300a mutant, the unique AflII site in p300 was used for the deletion of the first 243 amino acids from p300. To construct the p300b and p300c mutants, we first performed PCR using a 5′ primer encompassing a unique ApaI site (at codon 1920) and two 3′ primers which introduced a stop codon into residues 2215 and 2063, and cloned the PCR products into pBluescript II. After verification of the mutation by sequencing, the products were used to replace the 3′ DNA fragment after the unique ApaI site in pSP64(polyA)-p300. To construct p300Hm, we first cloned the unique BglII-PmlI fragment (from codons 1256 to 1943) of p300 containing the HAT domain into pBluescript II and then used site-directed mutagenesis to convert leucine 1690 and cysteine 1691 into lysine and leucine. The mutated DNA fragment was then used to replace the BglII-PmlI fragment in pSP64(polyA)-p300. Dominant negative p300 (residues 2057 to 2170) was constructed by cloning the corresponding PCR fragments into a modified pSP64(polyA) vector which contains a Flag tag. The resulting proteins have the Flag tag at their N termini. To prepare the glutathione S-transferase (GST)–TR fusion protein, the cDNA fragment encoding the entire ligand-binding domain (LBD) of Xenopus TRβA was PCR amplified and cloned into pGEX2T.
In vitro mRNA synthesis and microinjection of Xenopus oocytes.
All the constructs for in vitro transcription were linearized with a unique restriction enzyme which would cut after the poly(A) site in the pSP64(polyA) vector. After restriction digestion, DNA was deproteinated with phenol-chloroform and ethanol precipitated. The in vitro synthesis of mRNA was then carried out with an SP6 Message Machine kit from Ambion according to the manufacturer's instructions. The preparation and microinjection of Xenopus stage VI oocytes were essentially as described previously (2). To express receptor or coactivators in oocytes for protein-protein interaction analyses, we usually first dried 20 μCi of [35S]methionine with a Speed Vac and then dissolved it in 4 μl of in vitro-synthesized mRNA (400 ng/μl in diethylpyrocarbonate-treated water) or a mixture of TR-RXR (each at approximately 100 ng/μl). The resulting mixtures were then injected into the cytoplasm of stage VI oocytes (23.8 nl/oocyte), and the injected oocytes were incubated for 24 h to allow the synthesis of proteins. For transcriptional analyses, the single-stranded DNA (ssDNA) of pTRβA5′-7-3xUAS usually was injected at a concentration of 50 ng/μl in a volume of 18.4 nl/oocyte into the nuclei of oocytes 2 to 3 h after the injection of various single mRNAs or mRNA mixtures. In these cases, no [35S]methionine was coinjected, and low concentrations of TR and RXR mRNAs (10 ng/μl each) were used.
Coimmunoprecipitation and GST pulldown assays.
After 24 h of incubation, the groups of oocytes coinjected with [35S]methionine and mRNAs were collected, washed with extraction buffer (20 mM HEPES [pH 7.6], 70 mM KCl, 1 mM dithiothreitol, 0.1% Nonidet P-40, 10% glycerol, 0.2 mM phenylmethylsulfonyl fluoride) twice, and homogenized in a ratio of 1 oocyte per 10 μl of extraction buffer by pipetting. The homogenates were centrifuged to remove insoluble materials and lipids. The clean extracts (20 μl) containing TR-RXR or an individual coactivator were mixed and incubated at 4°C with rotation in a final volume of 100 μl (supplemented with extraction buffer) with or without the addition of 100 nM T3. Coimmunoprecipitation was then carried out with the addition of 2 μl of anti-TR, anti-RXR, or anti-Flag (M2) antibody at 4°C for another hour with rotation, followed by incubation with 10 μl of protein A-Sepharose 4B slurry for 1 h. When anti-Flag M2 antibody was used, 5 μl of rabbit anti-mouse antibody was also added with the protein A-Sepharose slurry. After low-speed centrifugation (2,000 × g) for 1 min to remove the supernatants, the beads were washed five times for 5 min each time with extraction buffer with rotation. The beads were then resuspended in 15 μl of 2× sodium dodecyl sulfate (SDS) loading buffer and boiled at 95°C for 5 min. The samples were then analyzed by SDS–polyacrylamide gel electrophoresis (PAGE), followed by autoradiography.
In one experiment (see Fig. 9), TR-RXR and p300 were coexpressed and [35S]methionine labeled in Xenopus oocytes. Nuclei of Xenopus oocytes were isolated manually, resuspended in extraction buffer, and disrupted by pipetting. After centrifugation at 13,000 rpm in a benchtop centrifuge for 20 min at 4°C, different amounts of Xenopus oocyte nuclear extracts were mixed with 10 μl of TR-RXR–p300 oocyte extract in a final volume of 100 μl. Coimmunoprecipitation was then performed as described above, followed by autoradiography or Western analysis.
FIG. 9.
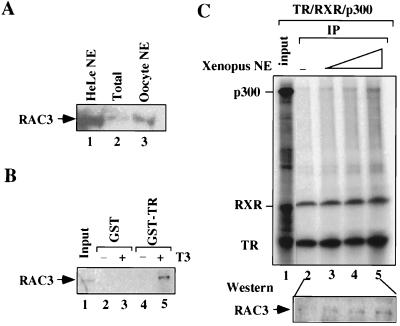
SRC-1 family coactivators in Xenopus oocytes may mediate the interaction between p300 and liganded TR-RXR. (A) Western analysis with a RAC3-specific antibody detected the presence of the putative xRAC3 in Xenopus oocytes. Lanes: 1, HeLa cell nuclear extract (NE) (control); 2, Xenopus oocyte extract; 3, Xenopus oocyte nuclear extract. (B) Putative xRAC3 bound to GST-TR in a T3-dependent manner. Input: Xenopus oocyte extract. The final concentration of T3 in the pulldown assay was 5 μM. (C) The addition of Xenopus oocyte nuclear extracts facilitated the coimmunoprecipitation of p300 with liganded TR-RXR. Oocyte extracts overexpressing p300 and TR-RXR were mixed without (lane 2) or with the addition of Xenopus oocyte nuclear extracts equivalent to 5 (lane 3), 10 (lane 4), and 20 (lane 5) oocyte nuclei in the presence of T3. The interaction between p300 and TR-RXR was then revealed by autoradiography after coimmunoprecipitation with an RXR-specific antibody as described in the legend to Fig. 4. The corresponding samples from lane 2 to lane 5 were also analyzed for the coimmunoprecipitation of xRAC3, and the result is shown in the bottom panel. IP, immunoprecipitation.
GST-TR fusion protein was purified from Escherichia coli using glutathione-Sepharose 4B affinity resin (Pharmacia) according to the manufacturer's instructions. For the GST pulldown assay, approximately 5 μg of GST-TR or GST (control) was incubated with HeLa cell nuclear extracts (500 μg) in a final volume of 200 μl or Xenopus oocyte extracts (100 μg) in a final volume of 100 μl with or without 5 μM T3 (supplemented with extraction buffer) at 4°C for 1 h with rotation. Ten microliters of glutathione beads was then added to the reaction mixtures, followed by another hour of incubation with rotation in a cold room. After low-speed centrifugation, the supernatants were set aside, and the beads were washed as described above for immunoprecipitation. The nuclear extract, the supernatant, and the bead fractions were then resolved by SDS-PAGE, followed by Western analyses with various antibodies.
Western blot analysis.
Western blot analysis was performed with a kit from Kirkegaard & Perry Laboratories, Inc., according to the manufacturer's instructions. The anti-p300 antibodies RW128 (1/500 dilution) and N-15 (1/500) were purchased from Upstate Biotechnology and Santa Cruz Biotechnology, respectively. Anti-CBP antibody A-22 (1/300) was obtained from Santa Cruz Biotechnology. Rabbit polyclonal anti-RAC3 (1/2,500) antibody was raised against RAC3 from residues 582 to 842 and is RAC3 specific. The anti–SRC-1 antibody (1/5,000) and anti-TR and anti-RXR antibodies have been described before (41, 55).
Transcriptional analysis.
Preparation of RNA and DNA from injected oocytes and transcriptional analyses by primer extension were performed as described previously (55). The internal control was the primer extension product of the endogenous histone H4 mRNA obtained with primer H4 (55). Except for one experiment (see Fig. 5) in which the CAT primer was used for primer extension and gave rise to a correctly sized product of 357 nucleotides, all primer extension analyses were carried out with primer I as described previously (55); this primer gave rise to a shorter product and usually less background. The levels of transcription were quantified by using a PhosphorImager.
FIG. 5.
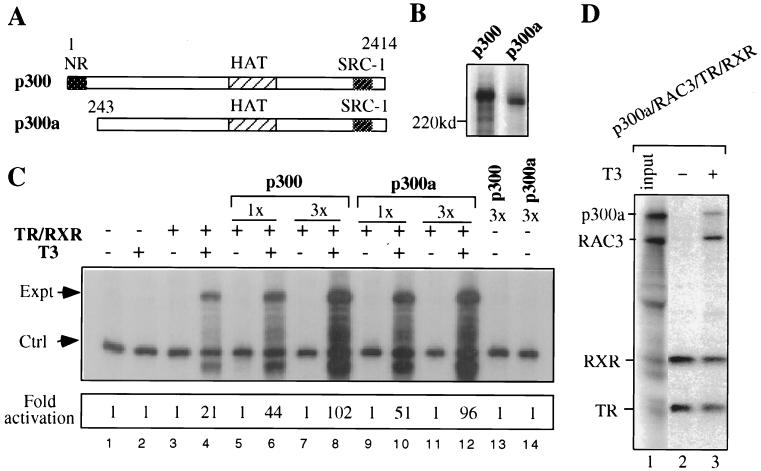
The putative nuclear receptor (NR) interaction domain of p300 is dispensable for its coactivation of TR-RXR. (A) Difference between p300 and the p300a mutant. (B) The expression of p300 and p300a in oocytes after coinjection of their mRNAs (9.6 ng/oocyte) with [35S]methionine was shown by autoradiography following SDS-PAGE. Note that the p300a protein migrated faster than the p300 protein due to the deletion of its first 243 amino acids. (C) The p300a mutant (lacking the NR interaction domain) functions as well as full-length p300 in facilitating hormone-dependent activation by TR-RXR. The injection of mRNAs encoding TR-RXR, p300, and p300a or ssDNA of the pTRβA5′-7-3xUAS reporter was done as described in the legend to Fig. 2C. The transcription analysis was also done as described in the legend to Fig. 2C, except that a different primer (CAT primer) was used here. The relative levels of transcription were quantified with a PhosphorImager. (D) The p300a mutant (lacking the NR interaction domain) can be recruited by liganded TR-RXR in the presence of RAC3. The coimmunoprecipitation experiment was performed as described in the legend to Fig. 4B, except that an oocyte extract overexpressing the p300a mutant was used.
MNase assay.
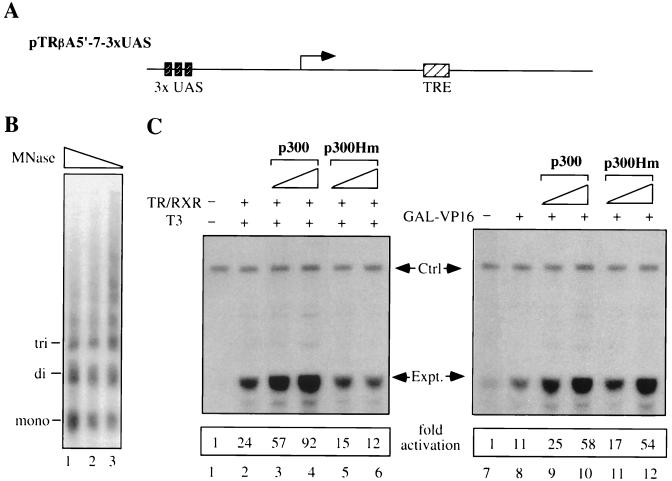
The micrococcal nuclease (MNase) assay of chromatin structure was performed as described previously (55). The resulting product (see Fig. 2B) was hybridized with end-labeled primer I.
FIG. 2.
Differential requirements for the HAT activity of p300 by liganded TR-RXR and GAL4-VP16. (A) Structure of the pTRβA5′-7-3xUAS reporter. UAS, upstream activation sequence; TRE, thyroid hormone receptor response element. (B) MNase assay showing that the injected ssDNA of pTRβA5′-7-3xUAS was converted into double-stranded DNA and assembled into regularly spaced nucleosomes. Two hours after injection, 10 oocytes were randomly picked, homogenized, and digested with 5 U of MNase for 3 (lane 1), 6 (lane 2), and 9 (lane 3) min. DNA was then purified, fractionated on a 1.5% agarose gel, blotted to a nylon filter, and probed with a 32P-labeled CAT primer. The positions of the mono-, di-, and trinucleosome lengths of DNA are indicated. (C) The HAT activity of p300 is required for its ability to enhance transcriptional activation by TR-RXR but not by GAL4-VP16. All groups of oocytes were injected with ssDNA of the pTRβA5′-7-3xUAS reporter (0.92 ng/oocyte) to establish a repressive chromatin structure. The oocytes were then injected with mRNAs encoding TR-RXR (lanes 2 to 6, 0.2 ng/oocyte) or GAL4-VP16 (lanes 8 to 12, 0.2 ng/oocyte) and p300 or p300Hm (0.6 ng/oocyte in lanes 3, 5, 9, and 11 and 1.8 ng/oocyte in lanes 4, 6, 10, and 12). The concentration of T3 in lanes 2 to 6 was 50 nM. The levels of transcription were analyzed by primer extension and quantified with a PhosphorImager. The level of transcription from the TRβA promoter (Expt.) in the control (lane 1 or 7) is arbitrarily designated as 1, and all others are expressed as fold activation in comparison to lane 1 or 7. The internal control (Ctrl) represents the level of the storage H4 mRNA in oocytes.
HAT assay.
The HAT assay was performed essentially as described previously (53), except that immunoprecipitated p300 or p300Hm (bead fraction) was used and the reaction mixtures were shaken constantly at room temperature.
RESULTS
p300 requires its HAT activity to enhance ligand-dependent activation by TR-RXR.
We established previously a T3-responsive Xenopus oocyte model system through microinjection of mRNAs encoding Xenopus TRβ and RXRα proteins and a reporter construct containing the Xenopus TRβA promoter (55). Using this model system, we have shown that TR-RXR functions as a repressor in the absence of T3 and an activator in the presence of T3 and that the assembly of the TRβA promoter into repressive chromatin, which occurs via the replication-coupled chromatin assembly pathway through injection of an ssDNA TRβA plasmid, is essential for the observation of robust activation of the TRβA promoter by TR-RXR in response to T3 (55). In addition, we have demonstrated that the addition of a histone deacetylase inhibitor, trichostatin A, can mimic the effect of T3, suggesting that acetylation of chromatin is likely to play an important role in the transactivation process mediated by liganded TR-RXR (53). Given its intrinsic HAT activity and the substantial evidence for its involvement in nuclear receptor action, we wish to characterize the mechanism by which p300 modulates transactivation by TR-RXR in the context of chromatin.
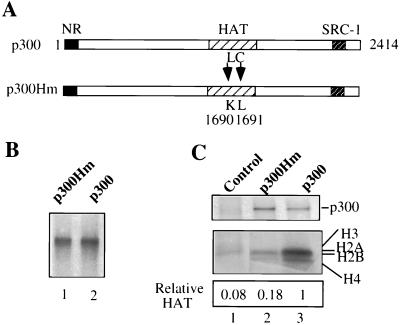
To express p300 in Xenopus oocytes, a cDNA encoding full-length human p300 was cloned into the pSP64(polyA) vector, which allows the synthesis of p300 mRNA in vitro. To test the role of the HAT activity of p300 in TR activation, we also constructed a p300 HAT mutant (p300Hm) by converting leucine 1690 to lysine and cysteine 1691 to leucine (Fig. 1A). Such mutations in CBP impair its HAT activity, most likely due to a defect in the binding of acetyl coenzyme A (29). The expression of p300 and p300Hm was first examined through coinjection of their corresponding mRNAs with [35S]methionine into groups of Xenopus oocytes. After overnight incubation, the expression of p300 and p300Hm in injected Xenopus oocytes was revealed by autoradiography following fractionation of the oocyte extracts by SDS-PAGE. As shown in Fig. 1B, both p300 and p300Hm were expressed at similar levels. The identities of both proteins were also confirmed by Western analysis with a p300-specific monoclonal antibody (Upstate Biotechnology) that recognizes an epitope in the p300 C terminus (data not shown). To ensure that p300hm has impaired HAT activity, we isolated p300 and p300Hm from the oocyte extracts by immunoprecipitation using the p300-specific antibody and performed a standard HAT assay using purified core histones as substrates. This experiment revealed that p300Hm exhibited greatly reduced HAT activity compared to p300 (Fig. 1C). These results thus allow us to test the functional importance of p300 HAT activity in transactivation mediated by TR-RXR.
FIG. 1.
Expression of p300 and its HAT mutant in Xenopus oocytes. (A) Schematic presentation of the p300 protein and p300Hm, in which leucine 1690 is converted to lysine and cysteine 1691 is converted to leucine. NR, nuclear receptor interaction domain. (B) Expression of p300 and p300Hm in oocytes. The in vitro-synthesized mRNA encoding p300 or p300Hm was coinjected with [35S]methionine into Xenopus oocytes. Oocyte extracts were prepared after overnight incubation, and the expression of p300 and p300Hm was analyzed by SDS-PAGE followed by autoradiography. (C) HAT assay showing that p300Hm has impaired HAT activity. p300 and p300Hm were isolated from oocyte extracts with a p300-specific antibody (RW128) and used for a standard HAT assay with core histones as substrates. The immunoprecipitated fraction from the extracts of oocytes injected with [35S]methionine only was used as a control (lane 1). The core histones were then resolved by SDS-PAGE (4 to 20% polyacrylamide), and the levels of acetylation were revealed by autoradiography and quantified with a densitometer.
We next analyzed whether the expression of p300 or p300Hm in oocytes could facilitate T3-dependent activation of the TRβA promoter by TR-RXR. We first established a repressive chromatin template through microinjection of pTRβA5′-7-3xUAS into Xenopus oocytes in the ssDNA form (55). The assembly of the injected reporter into chromatin in oocytes was demonstrated by a standard MNase digestion assay (Fig. 2B). As expected, the expression of TR-RXR through injection of TR-RXR mRNAs led to strong activation in the presence of T3 (Fig. 2C, compare lane 2 with lane 1). The expression of p300 further enhanced activation in a p300 mRNA dose-dependent manner (Fig. 2C, compare lanes 3 and 4 with lane 2). In contrast, no further activation was observed when p300Hm was expressed (Fig. 2C, compare lanes 5 and 6 with lane 2), even though p300Hm still maintained 10 to 20% wild-type HAT activity (Fig. 1C). Instead, a weak inhibitory effect was often observed in multiple experiments, suggesting that p300Hm may act as a dominant negative molecule to compete with endogenous p300 for TR activation. In all cases, we also examined the amount of pTRβA5′-7-3xUAS reporter DNA recovered from each group of oocytes by slot blot analysis as described previously (55). Such control experiments always revealed that a constant amount of DNA was recovered from each group of oocytes (data not shown), indicating that the difference in transcriptional activity was not due to the variation of injection. Thus, we conclude that p300 HAT activity is essential for the ability of p300 to facilitate TR activation.
We next examined whether the HAT activity of p300 is generally required for its ability to facilitate transcription in the context of chromatin. We have previously shown that GAL4-VP16 can activate the pTRβA5′-7-3xUAS reporter (which contains three upstream activation sequence [UAS] sites) assembled into repressive chromatin in Xenopus oocytes (52). We thus tested in parallel whether p300Hm could facilitate activation by GAL4-VP16. As expected, the expression of GAL4-VP16 in Xenopus oocytes by microinjection of GAL4-VP16 mRNA led to strong activation of the TRβA promoter (Fig. 2C, compare lane 8 with lane 7). The expression of p300 further enhanced activation in a dose-dependent manner (Fig. 2C, compare lanes 9 and 10 with lane 8). Interestingly, while p300Hm is defective for TR activation, it facilitated activation by GAL4-VP16 to a similar extent as wild-type p300 (Fig. 2C, compare lanes 11 and 12 with lanes 9 and 10).
Collectively, our results indicate that the HAT activity of p300 is important for the ability of p300 to facilitate TR-RXR but not GAL4-VP16 activation, even though the same chromatin reporter template was used. In addition, these results indicate that the inability of p300Hm to enhance TR-RXR activation is unlikely due to a general conformation defect, since p300Hm still facilitates activation by GAL4-VP16. It should also be pointed out that the stimulatory effect of p300 on TR-RXR or GAL4-VP16 activation was observed only when p300 mRNA was injected within a certain range (less than 2 ng/oocyte). We often found that overexpression of p300 beyond that range in oocytes actually repressed both TR-RXR and GAL4-VP16 activation (data not shown), presumably resulting from an effect of “squelching” of other protein(s) involved in transcriptional activation.
Minimal direct interaction between liganded TR-RXR and p300.
The experiments described above demonstrate that p300 facilitates TR activation in the context of chromatin. We next attempted to dissect the molecular mechanisms by which p300 is involved in TR activation. Since p300 has been reported to interact with nuclear receptors as well as SRC-1 family coactivators (6, 25), it could theoretically be recruited into a TR transcriptional complex directly through its interaction with liganded TR-RXR or/and indirectly through SRC-1 family coactivators. The relative contributions of these two pathways would be governed by the relative affinities of the interaction of p300 with liganded TR-RXR as well as with SRC-1 family coactivators. We thus compared the relative affinities of the interaction of p300, SRC-1, TIF2, and RAC3 with liganded TR-RXR.
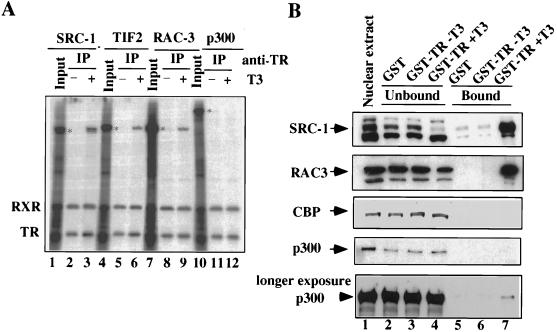
We approached this goal by expressing and labeling full-length coactivators and TR-RXR in Xenopus oocytes through coinjection of mRNAs with [35S]methionine and by examining the interaction using coimmunoprecipitation assays. We reasoned that the overexpression of coactivators in oocytes would be important for the analysis of their direct interaction with TR-RXR since, if a limiting Xenopus oocyte factor(s) is required to bridge the interaction, it would be insufficient once TR-RXR and the coactivators being tested are vastly overexpressed. To obtain high levels of expression, we not only injected oocytes with a high dose of mRNA(s) encoding each coactivator or TR-RXR (9.6 ng/oocyte) but also incubated the injected oocytes for at least 24 h before harvesting. Oocyte extracts overexpressing various coactivators were then prepared, mixed, and incubated with a TR-RXR-expressing oocyte extract in the presence or absence of 100 nM T3. The interaction between each coactivator and TR-RXR was then determined by immunoprecipitation with an RXR-specific antibody. As shown in Fig. 3A, it is clear that SRC-1, TIF2, and RAC3 all exhibited a strong hormone-dependent interaction with TR-RXR. To our surprise, no detectable T3-dependent interaction between TR-RXR and p300 was observed under the same conditions (Fig. 3A, compare lane 12 with lane 11), although p300 was well expressed. Since TR and RXR can readily form heterodimers regardless of T3, the coimmunoprecipitation of RXR with TR was, as expected, ligand independent in all cases and served as an internal control. Our results thus indicated that the interaction between p300 and liganded TR-RXR, if any, was much weaker than the interaction of liganded TR-RXR with the members of the SRC-1 family. Similar results were obtained when a TR-specific antibody was used for immunoprecipitation, indicating that the failure of coimmunoprecipitation of p300 is not likely to be due to the masking of TR epitopes by the binding of p300 (data not shown).
FIG. 3.
Minimal interaction, if any, detected between p300 and liganded TR-RXR. (A) Interaction between coactivators and TR-RXR, as assayed by coimmunoprecipitation. The expression and labeling of coactivators and receptors (TR-RXR) in Xenopus oocytes were achieved by microinjecting the individual mRNAs encoding SRC-1, TIF2, RAC3, and p300 at 9.6 ng of mRNA/oocyte or a mixture of TR and RXR mRNAs each at 1.8 ng/oocyte together with [35S]methionine. Oocyte extracts were prepared after 24 h of incubation, and the individual coactivator extracts were then mixed with TR-RXR extracts in the presence (+) or absence (−) of 100 nM T3. Coimmunoprecipitation was carried out with an RXR-specific antibody. Each input control was equivalent to approximately 50% of the immunoprecipitation (IP) samples loaded on the gel. The positions of the coactivators in the input lanes are indicated by asterisks. (B) Interaction between coactivators and TR in GST pulldown assays. HeLa cell nuclear extracts were incubated with GST-TR fusion protein or GST (control) in the absence (−) or presence (+) of 5 μM T3. The unbound (lanes 2 to 4) and bound (lanes 5 to 7) fractions, together with the nuclear extract input (lane 1), were fractionated by SDS-PAGE and analyzed by Western analysis with antibodies specific for the indicated coactivators. The input and supernatants were equivalent to about 10% of the total proteins used for GST pulldown assays. A longer exposure is shown for the p300 Western blot, revealing that a small fraction of p300 was pulled down by GST-TR in a T3-dependent manner.
The failure to detect the ligand-dependent interaction between TR-RXR and p300 is somewhat surprising, since the N-terminal domain of p300 was reported to interact with several nuclear receptors, including TR (25). To further confirm our finding, we examined the interaction of the endogenous coactivators in HeLa cell nuclear extracts with TR using a GST pulldown assay. The GST-TR fusion protein used here contains the entire LBD of Xenopus TRβA. To ensure the detection of weak interactions, HeLa cell nuclear extracts were incubated with GST-TR or GST (control) in the presence or absence of T3 under mild-stringency conditions (see Materials and Methods). The proteins bound to GST-TR or GST were then separated from the unbound proteins by affinity chromatography with glutathione-Sepharose beads, fractionated by SDS-PAGE, and analyzed by Western blotting with an antibody specific for each coactivator. As shown in Fig. 3B, SRC-1 was found to bind well to GST-TR only in the presence of a ligand (compare lane 7 with lane 6) and was almost completely depleted from the extracts (compare lane 4 to lane 1 or 3). The signals above and below the indicated SRC-1 band are most likely due to cross-reactivity of the SRC-1 antibody. Likewise, RAC3 in HeLa cell nuclear extracts was also highly enriched in the bound fraction in the presence of a ligand (Fig. 3B, compare lane 7 with lane 6). In contrast, while CBP and p300 were readily detected in the input and supernatant fractions, both CBP and p300 were hardly detected in the bound fraction with liganded GST-TR. A prolonged exposure did reveal a small fraction of p300 bound to liganded GST-TR. Since p300 could interact with SRC-1, TIF2, and RAC3 (see below), we suggest that this small fraction of p300 bound to liganded GST-TR could be due to such an indirect interaction rather than to a direct interaction with GST-TR. Nevertheless, this result is consistent with those of the coimmunoprecipitation experiments described above in that the direct interaction between p300 and liganded TR-RXR, if any, is minimal compared to the T3-dependent interaction between SRC-1 family coactivators and TR-RXR.
p300 can be recruited to liganded TR-RXR through its interaction with SRC-1 family coactivators.
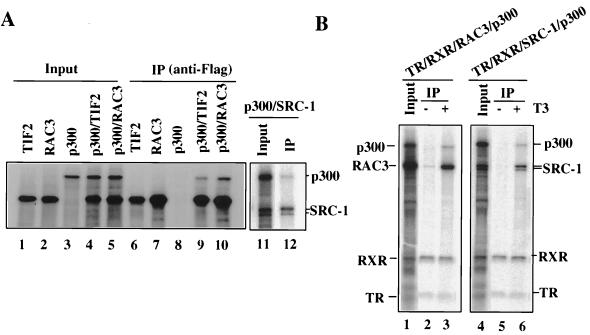
The above results suggest that the involvement of p300 in TR activation is unlikely to be a result of its direct interaction with liganded TR-RXR. Given that p300 was reported to interact with SRC-1 family coactivators (25, 44) and that liganded TR-RXR interacts with SRC-1 family coactivators (Fig. 3), we next examined whether p300 could be recruited to a liganded receptor indirectly through its interaction with SRC-1 family coactivators. We first examined the interaction between p300 and SRC-1 family coactivators. To allow a direct comparison, we introduced a Flag epitope tag into SRC-1, TIF2, and RAC3. Flag-tagged SRC-1, TIF2, and RAC3 were then expressed and radiolabeled in Xenopus oocytes by microinjection of the corresponding mRNAs, and the protein interactions with p300 were analyzed by coimmunoprecipitations with a Flag-specific antibody (M2). As shown in Fig. 4A, a considerable amount of p300 was coimmunoprecipitated with SRC-1, TIF2, and RAC3, while no p300 was pulled down in a control sample containing p300 alone. Thus, consistent with the previous reports (25, 44), our results confirm that p300 can interact directly with SRC-1, TIF2, and RAC3.
FIG. 4.
SRC-1 family coactivators interact with p300 and can recruit p300 to liganded TR-RXR. (A) The interaction between SRC-1 family coactivators and p300 was analyzed by coimmunoprecipitation. Oocyte extracts expressing Flag-tagged SRC-1 family coactivators and p300 were mixed as indicated, and the interaction was analyzed by coimmunoprecipitation with Flag-specific antibody (M2). Each input was equivalent to 50% of the sample loaded in the corresponding immunoprecipitation (IP) lane. Note that the p300 signal was not observed in the p300-alone control (lane 8) and was dependent on the presence of TIF2 (lane 9), RAC3 (lane 10), or SRC-1 (lane 12). (B) SRC-1 family coactivators recruit p300 to liganded TR-RXR. Oocyte extracts expressing p300 and SRC-1 or RAC3 were incubated with an oocyte extract expressing TR-RXR in the absence (lanes 2 and 5) or presence (lanes 3 and 6) of 1 μM T3. The mixtures were immunoprecipitated with an RXR-specific antibody. Each input mixture (lane 1 or 4) was equivalent to 50% of the sample loaded in the corresponding IP lane.
We next tested whether the addition of SRC-1 family coactivators could lead to the formation of ternary complexes containing p300 and liganded TR-RXR. Indeed, mixing oocyte extracts overexpressing SRC-1 or RAC3 with those containing TR-RXR and p300 led to the coimmunoprecipitation of p300 with TR-RXR in the presence but not in the absence of T3 (Fig. 4B). Similar results were observed when TIF2 was used (data not shown). These results, together with a lack of a direct interaction between p300 and liganded TR-RXR (Fig. 3), argue that the recruitment of p300 by liganded TR-RXR is primarily indirect and mediated through its interaction with SRC-1 family coactivators.
The p300 receptor interaction domain is dispensable for its role in TR activation.
Our results so far indicate an indirect model for the recruitment of p300 in T3-dependent activation by TR-RXR. Since the only reported receptor interaction domain of p300 is located at the N-terminal region (residues 1 to 100) (25), we tested the coactivator activity of a p300 mutant (p300a) lacking the putative receptor interaction domain. We first confirmed that both p300 and p300a were expressed to similar extents when similar amounts of mRNAs were injected into Xenopus oocytes (Fig. 5B). We then compared the ability of p300 and the p300a mutant to facilitate activation of the TRβA promoter by TR-RXR using a protocol like that described in the legend to Fig. 2, except that a different primer (located further downstream of the start site) was used for primer extension analysis. The expression of wild-type p300 (residues 1 to 2414) in Xenopus oocytes enhanced the transcriptional activation of liganded TR-RXR in a dose-dependent manner (Fig. 5C, compare lanes 8 and 6 with lane 4). However, the expression of p300a (residues 243 to 2414) also enhanced TR activation to a similar level as wild-type p300. Note that the stimulatory effect of p300 or p300a on TR activation is not a nonspecific effect, since the expression of p300 or p300a alone had no effect on transcription (Fig. 5C, compare lanes 13 and 14 with lane 1). In addition, we also tested whether the p300a mutant could form a complex with liganded TR-RXR in the presence of RAC3. As shown in Fig. 5D, the formation of the ternary complex containing p300a and RAC3 was observed in the presence but not in the absence of T3, while p300a alone did not bind to liganded TR-RXR (data not shown; see also Fig. 3). Thus, in agreement with the indirect model for the recruitment of p300 by liganded TR-RXR, the previously identified N-terminal receptor interaction domain of p300 is dispensable for its function in TR activation.
The p300 SRC-1 interaction domain is important for its role in TR activation.
The aforementioned results exclude a direct interaction between p300 and liganded TR-RXR as the major molecular mechanism for the involvement of p300 in TR action. Since our protein-protein interaction analyses indicated that p300 can be recruited by liganded TR-RXR indirectly through its interaction with SRC-1 family coactivators, we wished to test this indirect model experimentally. If p300 is indeed recruited into TR action through its interaction with SRC-1 family coactivators, one would predict that a p300 mutant(s) defective in interaction with SRC-1 should be compromised in its ability to facilitate TR activation and that the p300 SRC-1 interaction domain alone may function as a dominant negative molecule for TR activation.
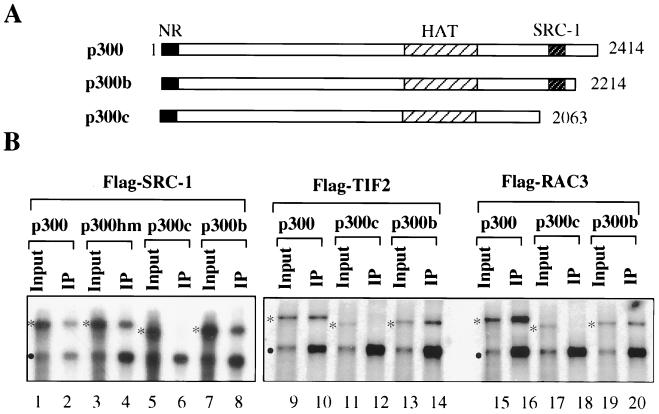
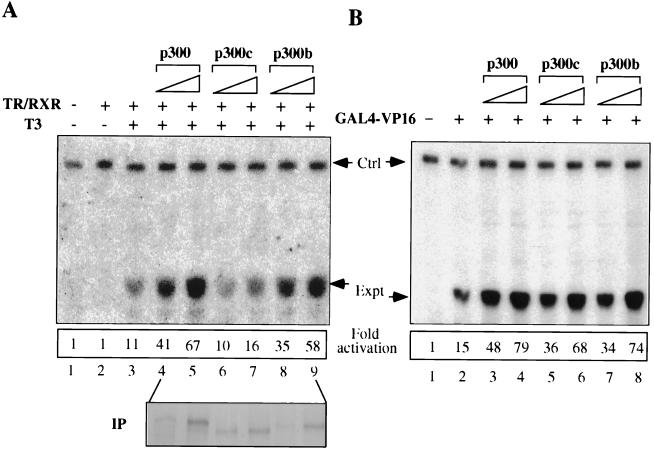
Since the SRC-1 interaction domain is located at the C terminus of p300 (6, 25), we constructed two p300 deletion mutants, p300b (residues 1 to 2214) and p300c (residues 1 to 2063), to test the first prediction. The interactions of these p300 mutants with SRC-1 family coactivators were first examined by use of the coimmunoprecipitation approach as described above. As shown in Fig. 6B, the interaction with SRC-1, TIF2, and RAC3 was maintained for p300b, while p300c did not show a detectable interaction. Note that p300Hm still interacted with SRC-1 (Fig. 6B, lane 4), although it was inactive in facilitating activation by liganded TR-RXR (Fig. 2). We next tested the ability of these p300 mutants to facilitate the T3-dependent activation of the TRβA promoter by TR-RXR. While p300b enhanced activation in a dose-dependent manner, similar to full-length p300, p300c consistently showed much reduced enhancement under the same conditions (Fig. 7A, compare lanes 6 and 7 with lanes 4 and 5 or lanes 8 and 9). Importantly, this defect was not due to the general abnormality resulting from the deletion of residues 2063 to 2414, since p300c still enhanced activation by GAL4-VP16 in a fashion similar to that of p300 and p300b (Fig. 7B). Furthermore, the impaired activation of TR-RXR by p300c was not due to the reduced level of expression, since the control immunoprecipitation experiment with an antibody specific for the N terminus of p300 (Santa Cruz Biotechnology) revealed that p300, p300b, and p300c were expressed to similar levels (Fig. 7A, lower panel, compare lane 6 with lanes 4 and 8 or lane 7 with lanes 5 and 9). These results thus support the indirect recruitment of p300 through its interaction with SRC-1 family coactivators as the primary mechanism for the effect of p300 on TR activation. Furthermore, these results underscore the differential requirement of the p300 functional domains for the activation mediated by TR-RXR and GAL4-VP16: the SRC-1 interaction domain is important for its involvement in TR activation but not for its role in activation mediated by GAL4-VP16.
FIG. 6.
Deletion of the SRC-1 interaction domain from p300 impairs its interaction with SRC-1 family coactivators. (A) Structures of the p300b and p300c mutants. NR, nuclear receptor interaction domain. (B) The p300c mutant is unable to interact with SRC-1 family coactivators (compare lanes 6 and 8, lanes 12 and 14, and lanes 18 and 20). Oocyte extracts expressing p300 and its mutants were mixed with oocyte extracts expressing individual SRC-1 family coactivators. The interactions with SRC-1 family coactivators were then analyzed by coimmunoprecipitation with the M2 antibody, specific for the Flag tag. Each input was equivalent to 20% of the sample shown in the immunoprecipitation (IP) fraction. The positions of p300 and its mutant proteins are indicated by asterisks, and the positions of SRC-1 family coactivators are indicated by dots.
FIG. 7.
The SRC-1 interaction domain of p300 is important for its ability to facilitate activation by liganded TR-RXR but not by GAL4-VP16. (A) The p300c mutant, but not the p300b mutant, is defective in facilitating activation by liganded TR-RXR (compare lanes 6 and 7 with lanes 8 and 9 and with lanes 4 and 5). The injection of DNA and mRNAs and primer extension analysis were as described in the legend to Fig. 2C. The levels of transcription were quantified with a PhosphorImager. To make sure all three proteins were expressed to comparable levels, the corresponding p300 and mutant mRNA samples (lower panel, lanes 4 to 9) were also injected into Xenopus oocytes together with [35S]methionine. The levels of expression were revealed by autoradiography after immunoprecipitation (IP) of p300 and its mutant proteins from the corresponding oocyte extracts with a p300-specific antibody (N-15). (B) In contrast to the results shown in panel A, the p300c mutant supported activation by GAL4-VP16 to a similar extent as p300 and the p300b mutant (compare lanes 5 and 6 with lanes 3 and 4 and with lanes 7 and 8).
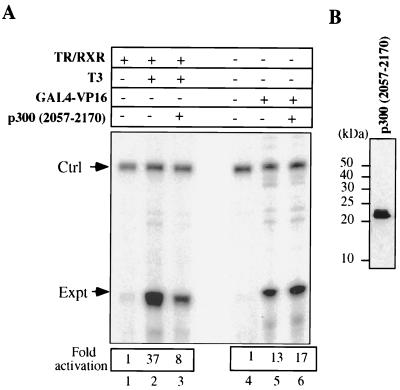
We also tested whether the SRC-1 interaction domain of p300 alone would function as a dominant negative molecule for TR activation. Indeed, overexpression of p300 (residues 2057 to 2170) containing the SRC-1 interaction domain in Xenopus oocytes reduced TR-RXR and T3-dependent activation as much as 80% without affecting activation by GAL4-VP16 (Fig. 8A). The strong inhibitory effect resulting from the overexpression of the SRC-1 interaction domain of p300 further supports the indirect model.
FIG. 8.
The expression of the SRC-1 interaction domain of p300 (residues 2057 to 2170) inhibits activation by TR-RXR but not activation by GAL4-VP16. (A) The injection of DNA and mRNAs encoding TR-RXR and GAL4-VP16 was as described in the legend to Fig. 2C. The mRNA encoding p300 (residues 2057 to 2170) was injected (2 ng/oocyte) 2 h before the other mRNAs. The levels of transcription were quantitated with a PhosphorImager. (B) Expression of p300 (residues 2057 to 2170) shown by Western analysis with M2 antibody, specific for the Flag tag. p300 (residues 2057 to 2170) has a Flag tag at the N terminus. The molecular mass markers are indicated.
As part of an effort to further substantiate the indirect model for the role of p300 in TR activation, we attempted to show a stimulatory effect on TR activation by ecotopic expression of SRC-1 family coactivators in Xenopus oocytes. So far we have yet to observe a significant stimulatory effect by expression of SRC-1 family coactivators. Injection of low doses of their mRNAs had only a minor stimulatory effect on TR activation, whereas injection of high doses of mRNAs inhibited activation (data not shown). One likely explanation is that in Xenopus oocytes the SRC-1 family coactivators are not the limiting factors relative to p300 or another factor(s) required for TR activation. Consistent with this hypothesis, the Xenopus homologue of RAC3 (xRAC3) was cloned recently, and its mRNA was found to be very abundant in early-stage Xenopus oocytes (27), suggesting that its protein level could be relatively abundant and mediate transcriptional enhancement by p300 in Xenopus oocytes. Indeed, Western blot analysis with a RAC3-specific antibody detected in a Xenopus oocyte extract a protein with a size identical to that of the mammalian RAC3 protein (Fig. 9A, compare lane 2 to lane 1). In addition, this protein was readily detected in a Xenopus nuclear extract (Fig. 9A, compare lane 3 to lane 2), indicating that this protein is a nuclear protein and is relatively abundant. To gain further evidence for the identity of this protein as xRAC3, we tested whether this Xenopus protein could bind to TR in a ligand-dependent manner using a GST-TR pulldown assay as described previously. As shown in Fig. 9B, this protein indeed bound to GST-TR but not GST (control) in a T3-dependent manner. These results thus confirm the presence as well as the relative abundance of xRAC3 in Xenopus oocytes.
The results described above, however, raise the question as to why we failed to detect the interaction between p300 and TR-RXR in coimmunoprecipitation experiments without the addition of oocyte extracts overexpressing SRC-1 family coactivators (Fig. 3). We reason that the level of SRC-1 family coactivators in Xenopus oocyte extracts could still be limiting compared to the levels of p300 and TR-RXR in the oocyte extracts used in our coimmunoprecipitation experiments. We thus tested whether the addition of Xenopus oocyte nuclear extracts could lead to the recruitment of p300 by liganded TR-RXR without further addition of oocyte extracts overexpressing SRC-1 family coactivators. As shown in Fig. 9C, the addition of an increasing amount of Xenopus oocyte nuclear extract indeed led to the increased coimmunoprecipitation of p300 with liganded TR-RXR (compare lanes 3, 4, and 5 with lane 2). As expected, the addition of the increasing amount of Xenopus oocyte nuclear extract also led to the increased coimmunoprecipitation of xRAC3 with liganded TR-RXR, as revealed by Western analysis (Fig. 9C, bottom panel). The results of this experiment are consistent with the idea that the interaction between p300 and liganded TR-RXR in natural oocytes can be mediated by SRC-1 family coactivators.
DISCUSSION
The major conclusions from these experiments are that the HAT activity of p300 is required for its ability to function as a coactivator for TR-RXR but not for GAL4-VP16 and that p300 acts as a coactivator for TR-RXR indirectly.
Differential requirement for HAT activity.
Strong genetic and biochemical evidence indicates that chromatin structure is an integral component of transcriptional regulation in eukaryotic cells (42). The assembly of DNA into chromatin generally represses transcription. How transcriptional factors overcome the repressive effect of chromatin has been a central question for transcriptional studies (17, 49). One possible mechanism is to target chromatin remodeling to regulatory regions by recruiting ATP-dependent chromatin remodeling factors (56). Consistent with this possibility, we have shown previously that liganded TR-RXR induces chromatin disruption, although the functional consequence of this targeted chromatin disruption remains to be determined (54, 55). A second mechanism involves the acetylation of chromatin. The acetylation of the lysine residues in the N-terminal tails of the histone subunits presumably weakens the constraints imposed on DNA by the core histones and thus allows the transcription machinery to gain access to regulatory DNA sequences. In agreement with this model, we have previously shown that trichostatin A, a histone deacetylase inhibitor, can activate the TRβA promoter from repressive chromatin, mimicking the activation induced by liganded TR-RXR (53). We show here that, while both liganded TR-RXR and GAL4-VP16 can stimulate transcription from repressive chromatin up to 20-fold, exogenous expression of p300 can further stimulate transcription by liganded TR-RXR and GAL4-VP16 up to 100-fold (Fig. 2C). Most importantly, we demonstrate that the HAT activity of p300 is required for its ability to function as a coactivator for TR-RXR but not for GAL4-VP16.
Recent work indicates that p300 enhances hormone-dependent activation by the estrogen receptor and the RAR from chromatin but not naked DNA template (11, 30). We show here that p300 enhances T3-dependent activation by TR-RXR from chromatin and in so doing requires its intrinsic HAT activity. Since TR-RXR binds constitutively to the TRE of the TRβA promoter in chromatin (51), we speculate that the recruitment of p300 by liganded TR-RXR could target acetylation to repressive chromatin in the promoter region and thus facilitate the assembly of RNA polymerase II preinitiation complexes. Consistent with this idea, Chen et al. demonstrated recently that hormone-induced gene expression by several nuclear receptors that they tested involves histone hyperacetylation of chromatin at hormone response elements of target genes (9). In addition, they provided evidence that hormone-induced histone hyperacetylation and transactivation are largely dependent on the HAT function of p300 (9).
It is interesting that HAT of p300 is differentially required for activation by TR-RXR and GAL4-VP16, despite the use of the same reporter. One possible explanation is that other HAT activities are utilized by GAL4-VP16 and that p300 primarily facilitates its activation by a mechanism(s) other than acetylation. This model is supported by the facts that multiple transcriptional cofactors, including GCN5, PCAF, TAF250, SRC-1, and RAC3, possess HAT activities and that p300 is a multifunctional protein (12). For instance, recent studies indicate that GAL4-VP16 can recruit both SAGA and NuA4 HAT-containing complexes to activate transcription from chromatin in vitro (24), providing an explanation for the lack of a requirement for p300 HAT activity. The second possibility is that TR-RXR itself is the substrate for p300-targeted acetylation and that its acetylation is required for its transition from a repressor in the absence of T3 to an activator in the presence of T3. A precedent for this hypothesis is p53, where modification by acetylation stimulates DNA-binding activity (19). Our previous work indicated that both TR and RXR were not acetylated by p300 under conditions where core histones and general transcription factors (TFIIE and TFIIF) were acetylated (53). Thus, we do not favor this possibility based on their lack of acetylation in vitro by p300 and the fact that TR-RXR binds constitutively to the thyroid hormone response element (TRE) in chromatin (55). The third possibility is that the HAT activity of p300 is required for activation by liganded TR-RXR but not GAL4-VP16 because of the association of unliganded TR-RXR with histone deacetylases. In this case, the HAT of p300 would presumably be required to counteract the additional repressive effect imposed on chromatin by deacetylases before transcription activation is enabled. Future work will attempt to determine whether the transcriptional activation mediated by TR-RXR and GAL4-VP16 is accompanied by an elevated level of acetylation of chromatin and whether the HAT of p300 is also required for activation by TR-RXR mutants which are defective in the recruitment of histone deacetylases.
The differential requirement for HAT activity by different transcription factors has been reported before (29). In that case, the HAT activity of CBP was shown to be required for transcriptional activation by CREB but not RAR. In contrast, the HAT activity of PCAF, another coregulator for nuclear receptors, is required for ligand-dependent activation by RAR. Thus, our results differ from theirs in the requirement for p300 HAT activity in transcriptional activation by nuclear receptors. This discrepancy could result from differences in experimental conditions, such as transient transfection versus a chromatin template or differences in transcriptional systems. Indeed, Chen et al. recently reported that the HAT activities of both CBP and p300 are required for them to enhance the activation of endogenous genes in mammalian cells by the estrogen receptor (9). Thus, it is possible that the HAT activity of p300 is absolutely required for activation by nuclear receptors only when the reporter genes are assembled into a chromatin configuration. Nevertheless, the notion of the differential requirement for HAT activity by different transcription factors is enforced by our experiments in which reporter DNA was assembled into chromatin (Fig. 2A).
Is the interaction between TR and p300 direct or indirect?
p300 was shown to function as a coactivator integrator for nuclear receptors and to interact directly with nuclear receptors (6, 25). This direct interaction between p300 and nuclear receptors was believed to be functionally important. However, we provide several lines of evidence to support an indirect model for the action of p300 in TR activation. First of all, in comparison to the situation for SRC-1 family coactivators, a minimal ligand-dependent interaction, if any, is observed between p300 and TR-RXR in both the coimmunoprecipitation and GST-TR pulldown experiments (Fig. 3). Second, SRC-1 family coactivators exhibit a strong hormone-dependent interaction with TR-RXR (Fig. 3) and are required for the formation of ternary complexes containing p300 and liganded TR-RXR (Fig. 4). Furthermore, the SRC-1 interaction domain of p300 is important for p300 to serve as a coactivator for TR-RXR (Fig. 7), whereas its receptor interaction domain residing in the N terminus is dispensable.
We wish to emphasize here that our experiments do not mean that p300 cannot bind directly to liganded TR-RXR but rather that this interaction is minimal in comparison to the interaction between liganded TR-RXR and SRC-1 family coactivators (Fig. 3) and is functionally insignificant in terms of the participation of p300 in activation by liganded TR-RXR (Fig. 5). Indeed, although early work indicated that p300 can interact in a ligand-dependent manner with nuclear receptors through two LXXLL motifs in their N termini, recent results showed that the SRC-1 interaction domain but not the N-terminal receptor interaction domain is important for p300 to enhance nuclear receptor activation (34, 48). These results thus are consistent with our indirect model for the action of p300 in TR-RXR activation.
Differential pathways for the recruitment of p300.
p300 is a multifunctional protein possessing a variety of interaction domains, which allow it to interact with a wide variety of proteins (12). We show here that while deletion of the SRC-1 coactivator interaction domain impairs its activity in TR-RXR activation, it does not affect its ability to facilitate activation by GAL4-VP16. Thus, the distinct domains in the p300 protein are utilized by liganded TR-RXR and GAL4-VP16 for targeting p300 to their transcriptional activation processes. The ability of p300 to interact with multiple transcription factors simultaneously provides a mechanism for transcriptional synergy and integrational responses in cells.
While our results suggest that p300 is primarily recruited for TR activation through its interaction with SRC-1 family coactivators, other pathways may also exist for the recruitment of p300 for TR activation. Although its interaction with all three SRC-1 family coactivator members was almost completely abolished in our coimmunoprecipitation assays (Fig. 6), the p300c mutant, lacking the SRC-1 interaction domain, still retained a low level of coactivator activity for TR-RXR (Fig. 7) and did not function as a dominant negative molecule (Fig. 7). Such low activity was consistently observed over several experiments, and the expression of the p300c mutant itself had no effect on the level of transcription in the absence of TR-RXR (data not shown), suggesting that p300c could still be inefficiently recruited into T3-dependent activation. It is possible that the interaction between p300c and SRC-1 coactivators in vivo could be stabilized in the transcriptional process by other protein-protein interactions, therefore accounting for the residual coactivation observed for the p300c mutant. Alternatively, pathways independent of the SRC-1 family coactivators could also exist for the recruitment of p300 into TR activation. For instance, strong evidence exists for a functional role of the TRAP complex in TR activation (15, 18). The TRAP220 subunit in the TRAP complex was shown recently to bind p300, although the region in p300 required for this interaction has not been determined yet (45). Thus, it is possible that p300 could be involved in TR activation through its interaction with the TRAP complex. In addition, p300 could also be brought into TR activation through its interaction with PCAF (5). Nevertheless, the differential effect of the p300c mutant on TR-RXR and GAL4-VP16 activation strongly argues for the functional importance of the interaction between p300 and the SRC-1 family coactivators in TR activation.
ACKNOWLEDGMENTS
We thank Don Chen for plasmid RAC3 and P. Chambon for the TIF2 construct. We are grateful to Ming-jer Tsai, David Moore, Rainer Lanz, and Fred Pereira for critical reading of and comments on the manuscript.
This work was supported in part by NIH grant R01 DK 56324 to Jiemin Wong.
REFERENCES
- 1.Allfrey V G, Pogo B G, Littau V C, Gershey E L, Mirsky A E. Histone acetylation in insect chromosomes. Science. 1968;159:314–316. doi: 10.1126/science.159.3812.314. [DOI] [PubMed] [Google Scholar]
- 2.Almouzni G, Wolffe A P. Replication-coupled chromatin assembly is required for the repression of basal transcription in vivo. Genes Dev. 1993;7:2033–2047. doi: 10.1101/gad.7.10.2033. [DOI] [PubMed] [Google Scholar]
- 3.Baniahmad A, Ha I, Reinberg D, Tsai S, Tsai M J, O'Malley B W. Interaction of human thyroid hormone receptor beta with transcription factor TFIIB may mediate target gene derepression and activation by thyroid hormone. Proc Natl Acad Sci USA. 1993;90:8832–8836. doi: 10.1073/pnas.90.19.8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baniahmad A, Steiner C, Kohne A C, Renkawitz R. Modular structure of a chicken lysozyme silencer: involvement of an unusual thyroid hormone receptor binding site. Cell. 1990;61:505–514. doi: 10.1016/0092-8674(90)90532-j. [DOI] [PubMed] [Google Scholar]
- 5.Blanco J C, Minucci S, Lu J, Yang X J, Walker K K, Chen H, Evans R M, Nakatani Y, Ozato K. The histone acetylase PCAF is a nuclear receptor coactivator. Genes Dev. 1998;12:1638–1651. doi: 10.1101/gad.12.11.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Role of CBP/P300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 7.Chang K H, Chen Y, Chen T T, Chou W H, Chen P L, Ma Y Y, Yang-Feng T L, Leng X, Tsai M J, O'Malley B W, Lee W H. A thyroid hormone receptor coactivator negatively regulated by the retinoblastoma protein. Proc Natl Acad Sci USA. 1997;94:9040–9045. doi: 10.1073/pnas.94.17.9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 9.Chen H, Lin R J, Xie W, Wilpitz D, Evans R M. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- 10.Chen J D, Evans R M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 11.Dilworth F J, Fromental-Ramain C, Remboutsika E, Benecke A, Chambon P. Ligand-dependent activation of transcription in vitro by retinoic acid receptor alpha/retinoid X receptor alpha heterodimers that mimics transactivation by retinoids in vivo. Proc Natl Acad Sci USA. 1999;96:1995–2000. doi: 10.1073/pnas.96.5.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eckner R. p300 and CBP as transcriptional regulators and targets of oncogenic events. Biol Chem Hoppe-Seyler. 1996;377:685–688. [PubMed] [Google Scholar]
- 13.Evans R M. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fondell J D, Brunel F, Hisatake K, Roeder R G. Unliganded thyroid hormone receptor alpha can target TATA-binding protein for transcriptional repression. Mol Cell Biol. 1996;16:281–287. doi: 10.1128/mcb.16.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fondell J D, Ge H, Roeder R G. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glass C K, Lipkin S M, Devary O V, Rosenfeld M G. Positive and negative regulation of gene transcription by a retinoic acid-thyroid hormone receptor heterodimer. Cell. 1989;59:697–708. doi: 10.1016/0092-8674(89)90016-0. [DOI] [PubMed] [Google Scholar]
- 17.Gregory P D, Horz W. Chromatin and transcription—how transcription factors battle with a repressive chromatin environment. Eur J Biochem. 1998;251:9–18. doi: 10.1046/j.1432-1327.1998.2510009.x. [DOI] [PubMed] [Google Scholar]
- 18.Gu W, Malik S, Ito M, Yuan C X, Fondell J D, Zhang X, Martinez E, Qin J, Roeder R G. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol Cell. 1999;3:97–108. doi: 10.1016/s1097-2765(00)80178-1. [DOI] [PubMed] [Google Scholar]
- 19.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 20.Hebbes T R, Thorne A W, Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinzel T, Lavinsky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 22.Hong H, Kohli K, Trivedi A, Johnson D L, Stallcup M R. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci USA. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horlein A J, Naar A M, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass C K, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor corepressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 24.Ikeda K, Steger D J, Eberharter A, Workman J L. Activation domain-specific and general transcription stimulation by native histone acetyltransferase complexes. Mol Cell Biol. 1999;19:855–863. doi: 10.1128/mcb.19.1.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 26.Kawasaki H, Eckner R, Yao T P, Taira K, Chiu R, Livingston D M, Yokoyama K K. Distinct roles of the co-activators p300 and CBP in retinoic-acid-induced F9-cell differentiation. Nature. 1998;393:284–289. doi: 10.1038/30538. [DOI] [PubMed] [Google Scholar]
- 27.Kim H J, Lee S K, Na S Y, Choi H S, Lee J W. Molecular cloning of xSRC-3, a novel transcription coactivator from Xenopus, that is related to AIB1, p/CIP, and TIF2. Mol Endocrinol. 1998;12:1038–1047. doi: 10.1210/mend.12.7.0139. [DOI] [PubMed] [Google Scholar]
- 28.Kliewer S A, Umesono K, Mangelsdorf D J, Evans R M. Retinoid X receptor interacts with nuclear receptors in retinoic acid, thyroid hormone and vitamin D3 signalling. Nature. 1992;355:446–449. doi: 10.1038/355446a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T M, Glass C K, Rosenfeld M G. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 30.Kraus W L, Kadonaga J T. p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev. 1998;12:331–342. doi: 10.1101/gad.12.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lazar M A, Berrodin T J, Harding H P. Differential DNA binding by monomeric, homodimeric, and potentially heteromeric forms of the thyroid hormone receptor. Mol Cell Biol. 1991;11:5005–5015. doi: 10.1128/mcb.11.10.5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Douarin B, vom Baur E, Zechel C, Heery D, Heine M, Vivat V, Gronemeyer H, Losson R, Chambon P. Ligand-dependent interaction of nuclear receptors with potential transcriptional intermediary factors (mediators) Philos Trans R Soc London Ser B. 1996;351:569–578. doi: 10.1098/rstb.1996.0056. [DOI] [PubMed] [Google Scholar]
- 33.Li H, Gomes P J, Chen J D. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc Natl Acad Sci USA. 1997;94:8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McInerney E M, Rose D W, Flynn S E, Westin S, Mullen T M, Krones A, Inostroza J, Torchia J, Nolte R T, Assa-Munt N, Milburn M V, Glass C K, Rosenfeld M G. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 1998;12:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 36.Nawaz Z, Lonard D M, Smith C L, Lev-Lehman E, Tsai S Y, Tsai M J, O'Malley B W. The Angelman syndrome-associated protein, E6-AP, is a coactivator for the nuclear hormone receptor superfamily. Mol Cell Biol. 1999;19:1182–1189. doi: 10.1128/mcb.19.2.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogryzko V V, Kotani T, Zhang X, Schiltz R L, Howard T, Yang X J, Howard B H, Qin J, Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- 38.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 39.Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 40.Puzianowska-Kuznicka M, Damjanovski S, Shi Y B. Both thyroid hormone and 9-cis retinoic acid receptors are required to efficiently mediate the effects of thyroid hormone on embryonic development and specific gene regulation in Xenopus laevis. Mol Cell Biol. 1997;17:4738–4749. doi: 10.1128/mcb.17.8.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 42.Struhl K. Chromatin structure and RNA polymerase II connection: implications for transcription. Cell. 1996;84:179–182. doi: 10.1016/s0092-8674(00)80970-8. [DOI] [PubMed] [Google Scholar]
- 43.Takeshita A, Cardona G R, Koibuchi N, Suen C S, Chin W W. TRAM-1, a novel 160-kDa thyroid hormone receptor activator molecule, exhibits distinct properties from steroid receptor coactivator-1. J Biol Chem. 1997;272:27629–27634. doi: 10.1074/jbc.272.44.27629. [DOI] [PubMed] [Google Scholar]
- 44.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 45.Treuter E, Johansson L, Thomsen J S, Warnmark A, Leers J, Pelto-Huikko M, Sjoberg M, Wright A P, Spyrou G, Gustafsson J A. Competition between thyroid hormone receptor-associated protein (TRAP) 220 and transcriptional intermediary factor (TIF) 2 for binding to nuclear receptors. Implications for the recruitment of TRAP and p160 coactivator complexes. J Biol Chem. 1999;274:6667–6677. doi: 10.1074/jbc.274.10.6667. [DOI] [PubMed] [Google Scholar]
- 46.Tsai M J, O'Malley B W. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 47.Voegel J J, Heine M J, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 48.Westin S, Kurokawa R, Nolte R T, Wisely G B, McInerney E M, Rose D W, Milburn M V, Rosenfeld M G, Glass C K. Interactions controlling the assembly of nuclear-receptor heterodimers and co-activators. Nature. 1998;395:199–202. doi: 10.1038/26040. [DOI] [PubMed] [Google Scholar]
- 49.Wolffe A P. Transcriptional control. Sinful repression. Nature. 1997;387:16–17. doi: 10.1038/387016a0. [DOI] [PubMed] [Google Scholar]
- 50.Wolffe A P, Wong J, Pruss D. Activators and repressors: making use of chromatin to regulate transcription. Genes Cells. 1997;2:291–302. doi: 10.1046/j.1365-2443.1997.1260323.x. [DOI] [PubMed] [Google Scholar]
- 51.Wong J, Li Q, Levi B Z, Shi Y B, Wolffe A P. Structural and functional features of a specific nucleosome containing a recognition element for the thyroid hormone receptor. EMBO J. 1997;16:7130–7145. doi: 10.1093/emboj/16.23.7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong J, Liang V C, Sachs L M, Shi Y B. Transcription from the thyroid hormone-dependent promoter of the Xenopus laevis thyroid hormone receptor betaA gene requires a novel upstream element and the initiator, but not a TATA box. J Biol Chem. 1998;273:14186–14193. doi: 10.1074/jbc.273.23.14186. [DOI] [PubMed] [Google Scholar]
- 53.Wong J, Patterton D, Imhof A, Guschin D, Shi Y B, Wolffe A P. Distinct requirements for chromatin assembly in transcriptional repression by thyroid hormone receptor and histone deacetylase. EMBO J. 1998;17:520–534. doi: 10.1093/emboj/17.2.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wong J, Shi Y B, Wolffe A P. Determinants of chromatin disruption and transcriptional regulation instigated by the thyroid hormone receptor: hormone-regulated chromatin disruption is not sufficient for transcriptional activation. EMBO J. 1997;16:3158–3171. doi: 10.1093/emboj/16.11.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wong J, Shi Y B, Wolffe A P. A role for nucleosome assembly in both silencing and activation of the Xenopus TR beta A gene by the thyroid hormone receptor. Genes Dev. 1995;9:2696–2711. doi: 10.1101/gad.9.21.2696. [DOI] [PubMed] [Google Scholar]
- 56.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 57.Xu J, Qiu Y, DeMayo F J, Tsai S Y, Tsai M J, O'Malley B W. Partial hormone resistance in mice with disruption of the steroid receptor coactivator-1 (SRC-1) gene. Science. 1998;279:1922–1925. doi: 10.1126/science.279.5358.1922. [DOI] [PubMed] [Google Scholar]
- 58.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 59.Yao T P, Oh S P, Fuchs M, Zhou N D, Ch'ng L E, Newsome D, Bronson R T, Li E, Livingston D M, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 60.Yu V C, Delsert C, Andersen B, Holloway J M, Devary O V, Naar A M, Kim S Y, Boutin J M, Glass C K, Rosenfeld M G. RXR beta: a coregulator that enhances binding of retinoic acid, thyroid hormone, and vitamin D receptors to their cognate response elements. Cell. 1991;67:1251–1266. doi: 10.1016/0092-8674(91)90301-e. [DOI] [PubMed] [Google Scholar]
- 61.Zhang X K, Hoffmann B, Tran P B, Graupner G, Pfahl M. Retinoid X receptor is an auxiliary protein for thyroid hormone and retinoic acid receptors. Nature. 1992;355:441–446. doi: 10.1038/355441a0. [DOI] [PubMed] [Google Scholar]