Abstract
Background:
Hydrogen sulfide (H2S) is a toxic end-product of microbial fermentation produced in the colon that may play a role in the pathogenesis of several diseases, including ulcerative colitis and colon cancer. However, the effect of diet interventions on intestinal burden of H2S gas exposure remains poorly understood.
Objective:
Determine the effect of short-term (1-week) plant- and animal-based eating patterns on ex vivo fecal H2S production in healthy human volunteers.
Methods:
The study design was an open-label, cross-over diet study and diets were self-administered. Each participant consumed two interventional diets: 1) an animal-based, low fiber (i.e. western) diet and 2) a plant-based, high fiber diet, separated by a two-week washout period. Participants collected full stool samples at the end of each week, which were processed within 2 h of collection to capture H2S production. Microfluidic qPCR (MFQPCR) was used to simultaneously quantify multiple taxonomic and functional groups involved in sulfate reduction and the fecal microbiota was characterized through high-throughput DNA sequencing.
Results:
Median H2S production was higher following the animal-based diet compared to the plant-based diet (p = 0.02; median difference 29 ppm/g, 95% CI 16e97). However, there was substantial individual variability and 2 of 11 individuals (18%) produced more H2S on the plant-based diet. Using the top and bottom quartiles of H2S percent change between animal- and plant-based diet weeks to define responders and non-responders, significant taxonomic differences were observed between the responder and non-responder cohorts.
Conclusions:
Here we report that substrate changes associated with a 1-week plant-based diet intervention resulted in lower ex vivo H2S production compared to a 1-week animal-based diet intervention in most healthy individuals. However, H2S responsiveness to diet was not uniform across the entire cohort, and potential H2S production enterotypes were characterized that may predict individualized H2S responsiveness to diet.
Keywords: Sulfate-reducing bacteria, Hydrogen sulfide, 16S rRNA gene, Dietary sulfur, Microfluidic qPCR
1. Introduction
Hydrogen sulfide (H2S) is a toxic end-product of microbial fermentation produced in the colon, which has been hypothesized to contribute to the pathogenesis of ulcerative colitis and colon cancer [1,2]. While sulfate reducing bacteria (SRB), including Desulfovibrio and Bilophila, have historically been implicated as the main drivers of colonic H2S production in the colon, diet interventions do not demonstrate a measurable effect of dietary sulfur intake on the relative abundance of SRB in feces [3]. This is likely due to the complexity of H2S production and cycling in the colon that extends beyond the contributions of SRB and dietary sulfur. Recent molecular studies have identified 43 taxa likely involved in intestinal sulfur metabolism [1], and in vitro studies have demonstrated that endogenous substrates, including mucin, taurocholic acid, and cysteine, contribute to the production of H2S [4,5]. These findings underscore the role of diet composition (e.g., plant-based, animal-based), beyond just sulfur intake, in attempts both understand and modulate colonic H2S production [6].
Controlled feeding studies demonstrate the capacity for gut microbiota composition to respond to short-term diet changes [7,8]. The effects of diet on the composition of gut microbiota, however, are difficult to measure due to strong interindividual and temporal variability [8]. This is, in large part, due to the presence of functional redundancy within the intestinal microbiota [9]. While Magee and colleagues [10] in a controlled feeding study demonstrated that changes in substrate, in the form sulfur amino acids from animal protein, increased fecal content of sulfide using the methylene blue derived measures, taxonomic composition of the gut microbiota was not determined.
To our knowledge, only two published studies have attempted direct measurement of H2S gas production capacity from stool via in vitro incubation with added substrate [4,5]. However, the effect of diet interventions on ex vivo fecal H2S gas production has not been previously published. Here we report results from a cross-over diet intervention trial using healthy human volunteers to assess the role of short-term (1-week) plant- and animal-based eating patterns on ex vivo fecal H2S production. As a secondary outcome, we also assessed whether the observed effects were driven by changes in substrate availability, gut microbiota composition, or a combination of both. Lastly, given the distinct H2S responsiveness patterns observed within the cohort we conducted a post hoc analysis to further interrogate microbial differences that may dictate diet responsiveness.
2. Methods
2.1. Dietary intervention study
This study was approved by the University of Minnesota institutional review board. Informed consent was obtained from all individual participants included in this study. Participants were recruited from among biomedical researchers and clinicians associated with the University of Minnesota Medical School Gastroenterology department. Fifteen healthy subjects consented to this study and were enrolled. Basic demographic and clinical information was obtained via questionnaires. The study design was an open-label, cross-over diet study and diets were self-administered. Participants were in regular communication with the study dietitian, and other study investigators, to ensure compliance with dietary intake and to overcome any logistical challenges of time-sensitive, complete stool sample collection.
Each participant consumed two 7 ± 2-day interventional diets: 1) an animal-based, low fiber (i.e. western) diet for which participants received low fiber diet instruction and were additionally encouraged to consume a diet higher in animal products than they might normally ingest; and 2) a plant-based, high fiber diet intervention for which participants received high fiber diet education and were provided a list of plant-based protein sources that provided similar protein intake to the animal-based, low fiber diet. Additionally, the latter group of participants were encouraged to minimize intake of animal products and to follow as close to a vegan diet as possible. Interventions were separated by a two-week washout period, during which participants were instructed to resume their usual diet. Routine assessments and sample collections were completed at 6 clinic visits over 42 days.
2.2. Dietary intake assessment
Dietary intake data was collected during each of the study periods. Three 24-h diet records were completed by participants at baseline, during the two intervention periods, and during the three washout weeks. Participants received instruction from a registered dietitian on methods for recording dietary intake. The dietitian reviewed completed diet records and obtained necessary clarification from respective participants. Analysis of 3-day diet records was done using the Nutrient Data System for Research (NDSR), version 2018. Protein sulfur was calculated using previously published equations [11].
2.3. Sample collection
Participants collected full stool samples at the end of each week (every 7 ± 2 days) in a toilet hat. Participants worked with study coordinators to ensure that stool samples were transported to the laboratory, on ice, and processed within 2 h of collection. Samples (~5 g) collected from the center of each stool sample were immediately frozen at −80 °C for subsequent DNA extraction. Additionally, participants were asked to collect 24-h urine samples during the baseline diet week and both intervention weeks. Participants collected all urine produced for a 24-h period in 3-L collection containers (VWR, Radnor, PA, USA) and samples were stored in the refrigerator. Participants were asked to begin urine collection following their first morning void and asked to note the exact time they began to complete their 24-h urine collection.
2.4. Gas measurements
The methods used for gas analysis were as previously reported [12]. Due to individual variability in stool sample size, a 1:1 ratio of stool:phosphate-buffered saline, with up to 40 g of stool was used. Mixtures were individually homogenized using a commercial blender that was pre-purged and continuously flushed with nitrogen (N2) gas. Approximately 10 g aliquots of each homogenate were transferred to individual 250 mL gas-tight septum jars (Thermo Scientific, Waltham, MA, USA), purged with N2, and incubated at 37 °C for 2 h. The 2-h incubation period was chosen based on the reported linearity of H2S in vitro gas production [4,5]. After incubation, three replicate 58 mL gas samples were removed from incubation container and injected into a separate gas-tight septum jar, to achieve a 1:5 dilution for H2S concentration measurements. H2S concentrations were measured from the second septum jar using a QRAE 3 gas monitor device (RAE Systems, San Jose, CA). H2S concentrations were corrected for dilution and reported as ppm/g dry weight of stool. Dry weight of the sample was calculated by the loss on drying method after heating at 68 °C for ≥24 h.
2.5. Urine sulfur measurement
Urine sulfate measurements were conducted by the Research Analytical Lab at the University of Minnesota (https://ral.cfans.umn.edu/). Urine sulfate measurements were determined by ion chromatography, using the Dionex IonPac AS18-Fast-4μm Analytical Column on an Integrion HPIC System (Thermo Scientific), with Chromeleon 7.2 software following Method 4110B, in Standard Methods for the Examination of Water and Wastewater [13].
2.6. DNA extraction, amplicon sequencing and bioinformatics
DNA was extracted from 0.25 g of thawed fecal samples using the DNeasy PowerSoil Kit (QIAGEN, Hilden, Germany), according to the manufacturer’s instructions. DNA samples were sequenced at University of Minnesota Genomics Center by using universal primers: 515f (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806r (5′-GGACTACHVGGGTWTCTAAT-3′) targeting the V4 region of the 16S rRNA gene [14]. Bar-coded sequencing was performed on the MiSeq platform (Illumina, San Diego, CA) using a 2 × 300-bp paired end protocol. All fastq files (n = 66) were deposited in the NCBI Sequence Read Archive under BioProject accession number PRJNA782460.
Amplicon sequences were analyzed using QIIME2 v.2020.8 [15]. Paired-end reads were joined, filtered and denoised using the DADA2 plugin [16]. Multiple sequence alignment was performed with MAFFT [17] and sequences were filtered to remove highly variable positions. A FastTree was used to construct and root a phylogenetic tree [18]. The naive-Bayes feature classifier in QIIME 2, trained on the SILVA database version 138 [19], was used to assign taxonomic information to each amplicon sequence variant. A sampling depth of 9498 was used for alpha and beta diversity analysis. Alpha diversity measures were calculated using observed species, Chao1, Shannon’s H and Simpson’s E indices. Bray–Curtis dissimilarity matrices were used for principal coordinates analysis. These matrices were also used to assess differences in microbial community structures using permutational multivariate analysis of variance (PERMANOVA) [20]. Linear discriminant analysis effect size (LEfSe) analysis was used to identify taxa that were overrepresented within each grouping [21].
2.7. Microfluidic qPCR
Microfluidic qPCR (MFQPCR) [22] was used to simultaneously quantify multiple taxonomic and functional groups involved in sulfate reduction. The Desulfovibrio 16S rRNA gene, the Bilophila tpa gene, and dissimilatory sulfite reductase subunit A gene (three different assays), as well as total prokaryotes (16S rRNA gene) were measured (Table 1). MFQPCR was performed using a 96.96 Dynamic Array chip (Fluidigm, San Francisco, CA, USA) and a BioMark HD System (Fluidigm) as described previously [23]. The 10× assay premix (8 μL) for each target contained 2× assay loading reagent (Fluidigm), 1× TE buffer, and 5 μM primer pair mix. The sample premix (8 μL) contained 2× SsoFast EvaGreen supermix with low ROX (Bio-Rad, Hercules, CA), 20× DNA binding dye sample Loading reagent (Fluidigm), and 3.6 μL of DNA. All assays were run in triplicate. Aliquots (5 μL) of the assay and sample pre-mixes were loaded into the MFQPCR chip and mixed using an IFC controller HX according to the manufacturer’s instructions. The chip was loaded onto BioMark HD System for real-time PCR. PCR was performed under the following conditions: 95 °C for 1 min, followed by 40 cycles of 96 °C for 10 s and 60 °C for 30 s. To ensure proper mixing of samples and assays in the chip, an additional thermal mixing protocol (70 °C for 40 min and 60 °C for 30 s) was added as the initial step, as per the manufacturer’s instructions. Each MFQPCR run included DNA standards and no-template controls. ROX was used as a passive reference dye. The threshold cycle (Ct) value was determined using the Real-Time PCR Analysis software version v.2.1.3 (Fluidigm). The standard curves were generated by linear regression analysis of the Ct values versus the known concentrations of the DNA standards (log gene copies per μl). The quantity of the target gene was calculated from the Ct values using standard curves. Only values under Ct 30, with good melt curves, were used.
Table 1.
List of microfluidic qPCR assays used to simultaneously quantify multiple taxonomic and functional groups involved in sulfate reduction, as well as total prokaryotes.
| Assay # | Taxonomic/functional group | Gene | Primer | Sequence (5′ −3′) | Reference |
|---|---|---|---|---|---|
|
| |||||
| 1 | Desulfovibrio | 16S rRNA | DSV691F | CCGTAGATATCTGGAGGAACATCAG | [24] |
| DSV826R | ACATCTAGCATCCATCGTTTACAGC | ||||
| 2 | Bilophila | tpa | tpa-FP | CGCCGGTATCGAAATCGTGA | [25] |
| tpa-RP | ATTCGCGGAAGGAGCGAGAG | ||||
| 3 | Sulfate reducers | dsrA #1a | Dsr1F+ | ACSCACTGGAAGCACGGCGG | [26] |
| Dsr1R | GTGGMRCCGTGCAKRTTGG | ||||
| 4 | Sulfate reducers | dsrA #2a | RH1-dsr-F | GCCGTTACTGTGACCAGCC | [27] |
| RH3-dsr-R | GGTGGAGCCGTGCATGTT | ||||
| 5 | Sulfate reducers | dsrA #3a | dsrA-F | CCAACATGCACGGYTCCA | [28] |
| dsrA-R | CGTCGAACTTGAACTTGAACTTGTAGG | ||||
| 6 | Total prokaryotes | 16S rRNA | 515F | GTGYCAGCMGCCGCGGTAA | [29,30] |
| 806R | GGACTACNVGGGTWTCTAAT | ||||
Different numbers indicate different primer sets targeting the same gene.
2.8. Statistical analysis
The primary endpoint of this study, effect of a short-term plant-based and animal-based eating pattern on fecal H2S production, was assessed using by Wilcoxon signed rank test given the non-normal distribution of the H2S variable. A generalized estimating equation technique was used to explore the association between diet variables and H2S production. Given the correlated nature of the data, and due to repeated measurements on subjects, the Proc GENMOD statement was used to account for subjects. This was inputted through the REPEATED statement with selection of the exchangeable working correlation structure. All statistical analyses were performed using SAS version 9.4 (SAS Institute, Cary, NC, USA). P values < 0.05 were considered significant.
3. Results
3.1. Participants and dietary intake
Two of the 15 subjects who consented and were enrolled in this study were unable to complete the study due to early research restrictions associated with the COVID-19 pandemic, and one was unable to complete due to limited availability. Moreover, while 12 subjects completed the intervention study, one participant was excluded from analyses due to an alcohol intake that exceeded the exclusion criterion (>20 g/day females and >30 g/day males) maintained throughout the study period. The final cohort of 11 subjects was predominately female (7/11; 64%) and Caucasian (9/11; 82%). Due to the open-label nature of the study, participants were intentionally evenly split between animal-based diet as the first intervention (n = 5; 45%) and a plant-based diet as the first intervention (n = 6; 55%). Median age was 32 years, range: 23e76 years; mean body mass index (BMI) was 22.6 kg/m2, range: 18.3e27.0 kg/m2.
Mean dietary intake for each study interval is shown in Table 2. Total energy intake was not different between any of the diet periods. As expected, percent fat, protein, animal protein, and protein sulfur were higher during the animal-based diet intervention as compared to the plant-based diet intervention. Total urine sulfate levels were higher following the animal-based diet compared to the plant-based diet (p < 0.01; 551 ± 265 mg and 314 ± 161 mg, respectively). However, due to limited compliance, complete data was only available for 6 of 11 subjects. Conversely, percent carbohydrates, total fiber, and insoluble and soluble fiber were higher during the plant-based intervention compared to the animal-based diet. In addition, and compared to baseline, while protein sulfur increased during the animal-based diet intervention, the percent carbohydrates and insoluble fiber decreased. Percent carbohydrates were also found to increase during the plant-based diet week compared to baseline.
Table 2.
Mean daily dietary intake by intervention period.
| Variable | Baseline | Animal-based | Washout (animal) | Universal | Plant-based | Washout (plant) | p-value (animal vs plant) |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Calories | 2093 ± 578 | 1859 ± 508 | 1802 ± 381 | 1988 ± 646 | 1896 ± 566 | 1989 ± 612 | 1.00 |
| Percent calories from carbohydrate (%) | 46 ± 9.3 | 36 ± 8.3a | 43 ± 6.2 | 47 ± 5.7 | 57 ± 6.0a | 44 ± 3.9 | <0.01 |
| Percent calories from fat (%) | 36 ± 6.6 | 43 ± 5.8 | 36 ± 3.9 | 34 ± 4.9 | 28.6 ± 8.5 | 36.3 ± 5.6 | <0.01 |
| Percent calories from protein (%) | 16 ± 3.9 | 20 ± 4.9 | 18 ± 4.4 | 18 ± 5.4 | 12 ± 2.5 | 18 ± 4.5 | <0.01 |
| Total carbohydrate (g) | 249 ± 86 | 171 ± 78 | 187 ± 87 | 245 ± 98 | 275 ± 76 | 223 ± 77 | 0.06 |
| Total fiber (g) | 26 ± 15 | 9 ± 4 | 22 ± 12 | 27 ± 14 | 41 ± 17 | 25 ± 17 | <0.01 |
| Insoluble (g) | 19 ± 12 | 4.9 ± 2.7a | 16 ± 9.4 | 20 ± 12 | 32 ± 14 | 17 ± 12 | <0.01 |
| Soluble (g) | 6.9 ± 3.3 | 4.4 ± 2.1 | 5.9 ± 2.5 | 6.7 ± 2.5 | 9.3 ± 4.1 | 7.5 ± 5.3 | 0.02 |
| Total fat (g) | 81 ± 35 | 89 ± 24 | 74 ± 19 | 77 ± 29 | 65 ± 33 | 84 ± 35 | 0.42 |
| Saturated fat (g) | 31 ± 13 | 33 ± 12 | 24 ± 7 | 26 ± 10 | 17 ± 13 | 28 ± 13 | 0.03 |
| Total pro (g) | 85 ± 29 | 88 ± 20 | 79 ± 12 | 84 ± 20 | 65 ± 18 | 88 ± 24 | 0.13 |
| Animal protein (g) | 54 ± 30 | 69 ± 22 | 48 ± 18 | 49 ± 24 | 2 ± 2 | 55 ± 25 | <0.01 |
| Vegetable protein (g) | 38 ± 16 | 19 ± 7 | 31 ± 13 | 37 ± 22 | 56 ± 23 | 31 ± 16 | <0.01 |
| Protein sulfur (g) | 0.7 ± 0.3 | 0.8 ± 0.3 | 0.6 ± 0.1 | 0.7 ± 0.2 | 0.4 ± 0.1a | 0.7 ± 0.2 | <0.01 |
Values presented as mean ± standard deviation.
Differs from baseline.
3.2. Hydrogen sulfide production is greater following an animal-based than a plant-based diet intervention
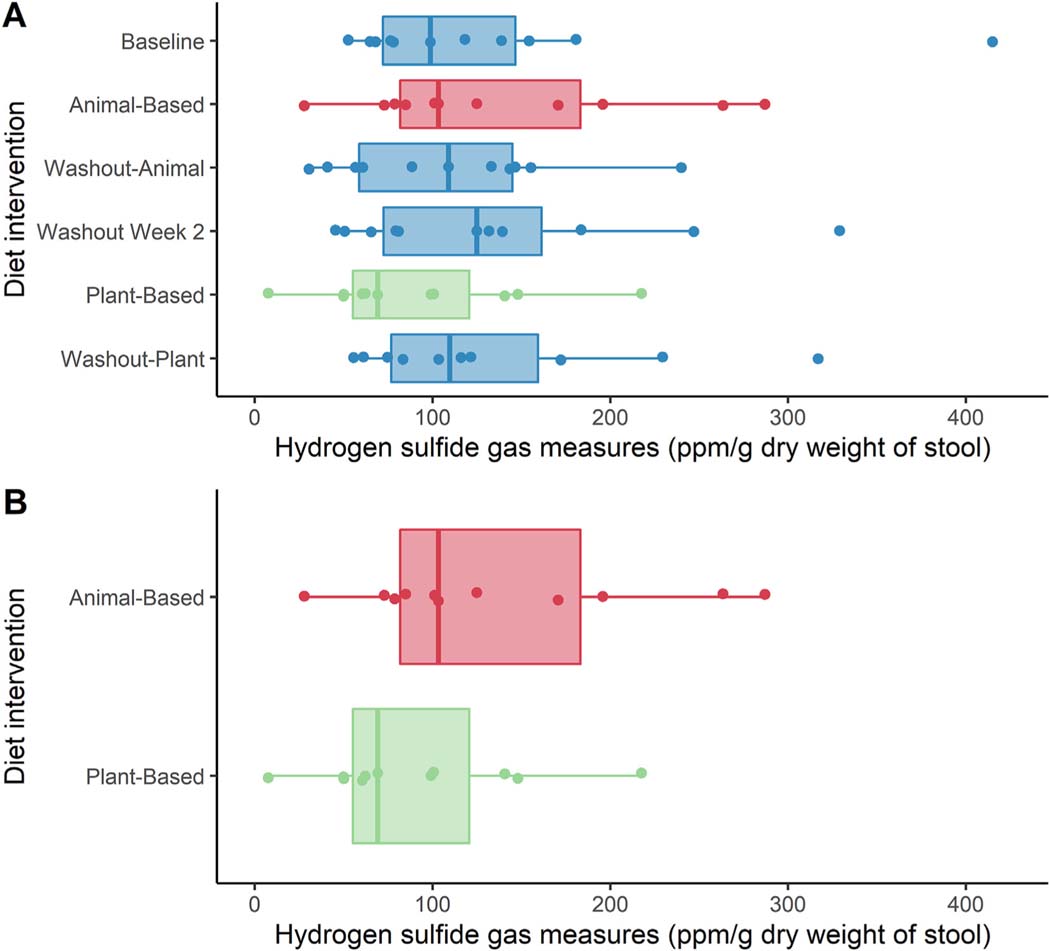
Hydrogen sulfide gas production values were obtained from 65 of the 66 full samples collected. The distribution of H2S values obtained across the entire study period is presented in Fig. 1A. Median H2S production was higher following the animal-based diet compared to the plant-based diet (p = 0.02; median difference 29 ppm/g, 95% CI 16–97) (Fig. 1B). However, there was substantial individual variability and 2 of 11 individuals (18%) produced more H2S on the plant-based diet. The percent change within an individual trended towards increased H2S on the animal-based diet, but the differences were not statistically different at α = 0.05 level (S = 20.5, p = 0.07; Median difference 32%, 95% CI 13%–65%).
Fig. 1.
A: Box plots of H2S distribution at each time point during the study interval. There was no overall difference in H2S production between diet weeks (p = 0.70), B: box plot of H2S distribution during animal- and plant-based diet weeks. Median H2S production was higher following the animal-based diet compared to the plant-based diet (p = 0.02; median difference 29 ppm/g, 95% CI 16–97).
3.3. Taxonomic and functional gene comparison between animal- and plant-based diet interventions
The impact of diet interventions on taxonomic composition of fecal gut microbiota was examined by 16S rRNA gene sequencing. Overall a strong inter-individual variability was observed in gut microbiota (Supplementary Fig. 1). Moreover, inter-individual variability was a stronger contributor towards differences in microbial community structure than diet. Permutational multivariate analysis of variance (PERMANOVA) across the entire cohort and diet study period generated a participant R2 value of 0.76 and diet R2 value of 0.03. Limited to only the samples collected at the end of the animal- and plant-based diet weeks PERMANOVA produced similar R2 values of 0.80 and 0.03 for participant and diet, respectively. Similarly, while significant differences were observed in Shannon index values between individuals, there was no difference in alpha diversity between the animal- and plant-based diet groups. A paired Linear discriminant analysis (LDA) combined with effect size measurements (LEfSe) comparing animal-to plant-based diet weeks did not return significant findings.
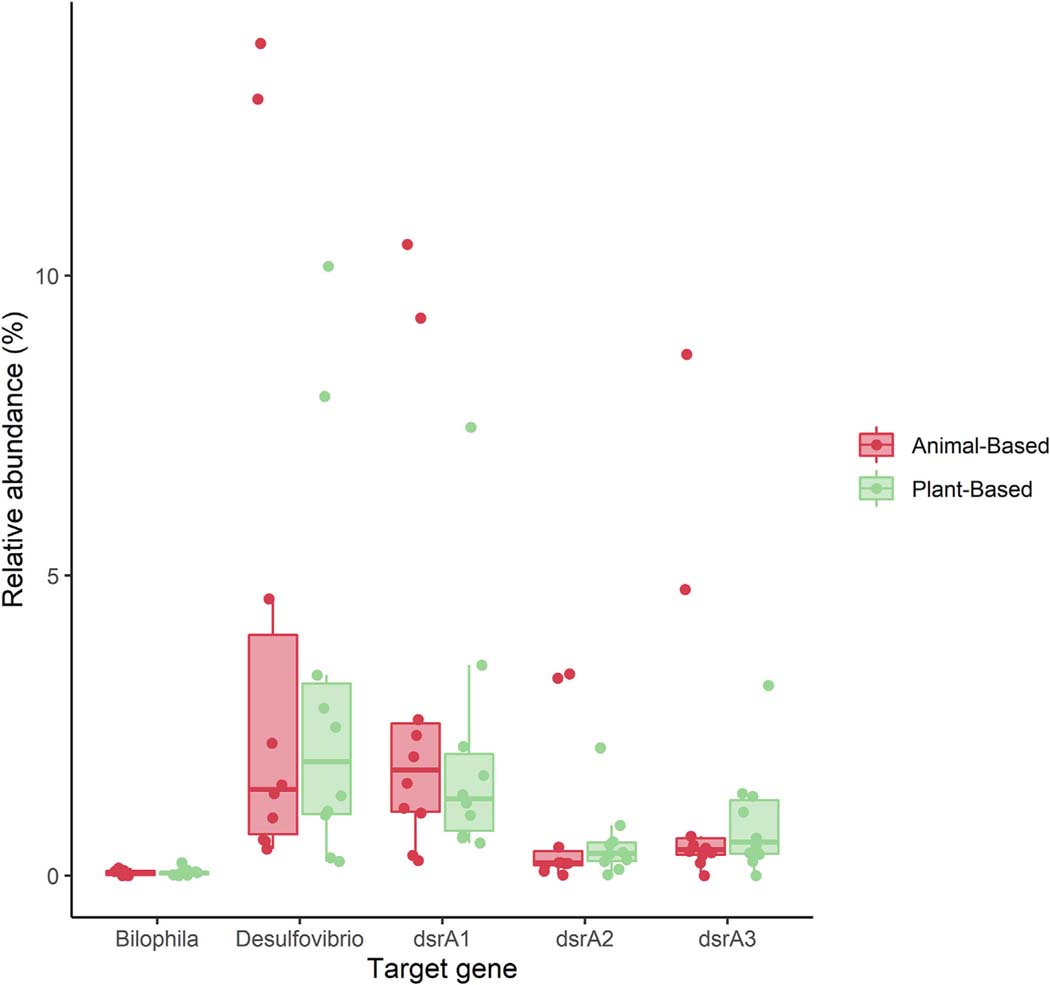
Microfluidic qPCR targeting microbial taxa (Desulfovibrio and Bilophila) and enzymes associated with sulfate reduction (dissimilatory sulfite reductase subunit A; dsrA) produced strong correlation between all qPCR targets (Supplementary Table 1). Additionally, qPCR data of all dsrA genes, as well as the genus Desulfovibrio strongly correlated with the relative abundance of Desulfovibrio, generated via 16S rRNA gene sequencing (r = 0.49–0.71). Similar results were observed when comparing qPCR and amplicon sequence data of the genus Bilophila (r = 0.548). No change in absolute- or relative abundance was observed over the course of the entire study period or when comparing animal-to plant-based diet weeks. Comparison of the relative abundance of qPCR targets between individuals consuming animal- and plant-based diets are shown in Fig. 2 and Supplementary Table 2.
Fig. 2.
Box plot of microfluidic quantitative PCR (MFQPCR) target distribution between animal- and plant-based diet weeks. There was no difference in MFQPCR targets between diet weeks.
3.4. Dietary intake differences between animal- and plant-based diet interventions and relationship with hydrogen sulfide production
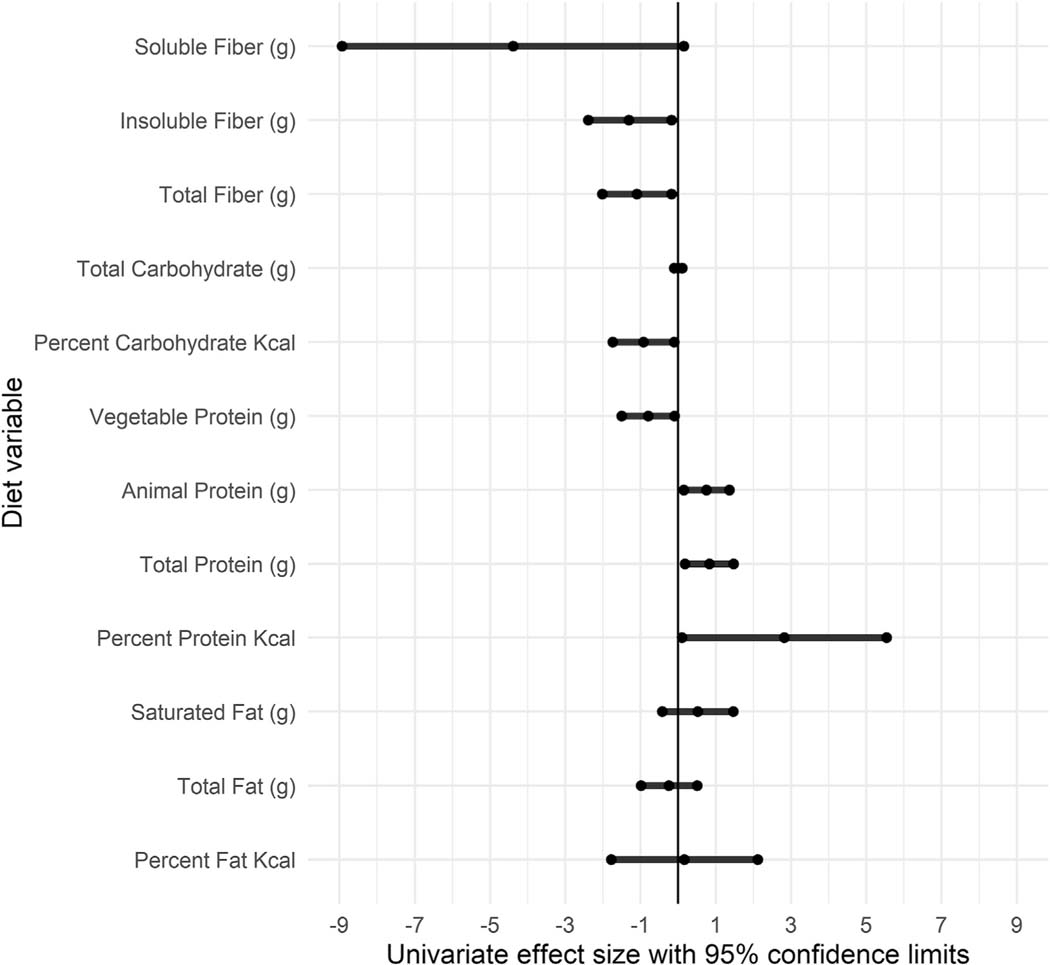
H2S production was associated with several diet variables. General estimating equation techniques accounted for individual participants to analyze the relationship between diet variables and H2S production. Results of univariate analysis (Fig. 3 and Supplementary Table 3) showed that fiber (g), insoluble fiber (g), percent carbohydrate, and vegetable protein (g) were found to negatively impact H2S measures (parameter estimates: −1.10, −1.31, −0.92, and −0.80, respectively) and percent protein, total protein (g), and animal protein (g) were found to positively impact H2S production (parameter estimates: 2.82, 0.83, 0.75, respectively). Soluble fiber (g) generated a large effect size (parameter estimate = −4.39) that was negatively associated with H2S production and trended towards significance (p = 0.06). In a multivariate model that included protein (g/day) and fiber (g/day), both variables maintained their significance with relatively unchanged effect size (Protein: 0.81, 0.16e1.47, p = 0.02 and Fiber: −1.06, −2.03 to −0.09, p = 0.03). To capture diet quality more broadly, we also included measures of the Empirical Dietary Inflammatory Potential (EDIP) [31], which positively impacted H2S production (parameter estimate: 26.42).
Fig. 3.
Univariate effect size, calculated utilizing a generalized estimating equation technique, provide quantitative measures of the size and direction of diet variables on H2S production within the entire cohort and across the entire study period.
3.5. Dietary intake and taxonomic composition differences between hydrogen sulfide responders versus non-responders
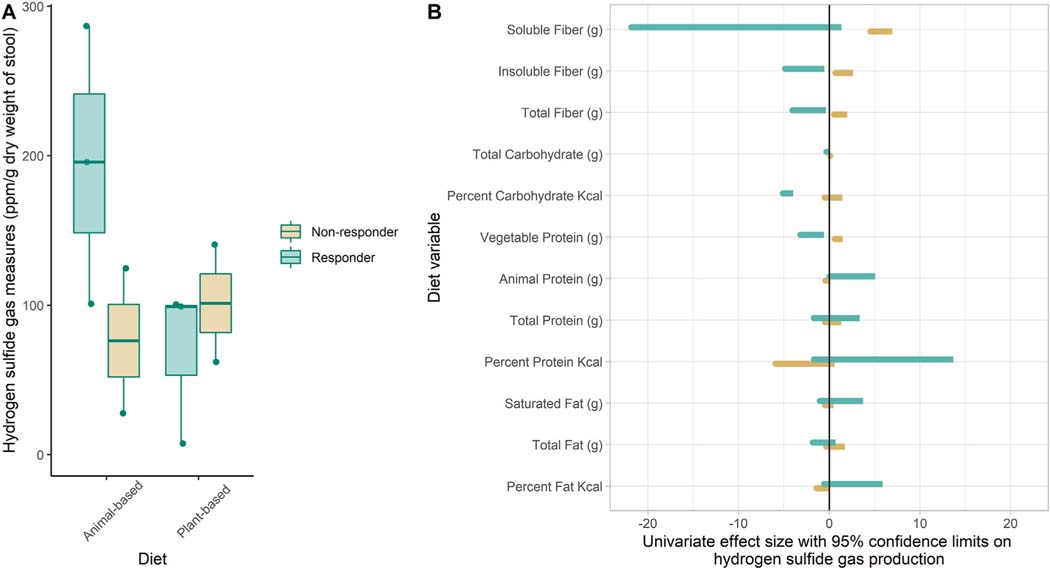
To further investigate the two participants who produced greater amounts of H2S gas at the end of the plant-based diet intervention compared to the animal-based diet, quartiles were created based on percent change in H2S production between animal- and plant-based diet weeks. Quartile 1 was composed of the two participants producing higher H2S values at the end of the plant-based diet intervention compared to the animal-based diet intervention and were characterized as “non-responders”. Quartile 4 was composed of the three participants with the largest percent change difference between animal-based and plant-based diet interventions (H2S values 50–93% lower with plant-based diet intervention) and these individuals represented “responders” (Fig. 4A).
Fig. 4.
A: Box plots of H2S distribution during animal- and plant-based diet weeks demonstrates the change in H2S production between diet weeks in the “Responder” cohort and lack of change observed in the “non-responder” cohort. B: Univariate effect size, calculated utilizing a generalized estimating equation technique, provide quantitative measures of the size and direction of diet variables on H2S production within the “responder” and “non-responder” cohorts and across the entire study period.
There was very little difference between responders and non-responders in dietary intake identified across the entire study interval or during the plant-based diet intervention (Table 3). During the animal-based diet intervention, the “non-responders” had a higher mean intake of animal protein and protein sulfur than did the “responders” (92 g/day vs 53 g/day and 0.93 g/day vs 0.56 g/day, respectively). Univariate analysis utilizing a general estimating equation technique was used to analyze the relationship between diet variables and H2S production. Results presented in Fig. 4B and Supplementary Table 4 show that fiber (g), insoluble fiber (g), soluble fiber (g), and vegetable protein (g) in the non-responder cohort had a positive impact on H2S production (parameter estimate: 1.25, 1.65, 5.75, and 1.05, respectively). In contrast, in the responder cohort, carbohydrate (g), percent carbohydrate, fiber (g), insoluble fiber (g), and vegetable protein (g) negatively impacted H2S production (parameter estimate: −0.02, −4.55, −2.21, −2.71, and −1.89, respectively) while animal protein positively impacted H2S production (parameter estimate 2.55). EDIP produced dramatically different effect sizes in non-responders and responders (parameter estimate: −9.19 and 46.23, respectively).
Table 3.
Comparison of dietary intake between hydrogen sulfide responsive and non-responsive participants.
| Variable | Overall |
Animal-based intervention |
Plant-based intervention |
|||
|---|---|---|---|---|---|---|
| Non-responder (n = 2) | Responder (n = 3) | Non-responder | Responder | Non-responder | Responder | |
|
| ||||||
| Calories | 2123 ± 513 | 2109 ± 661 | 1911 ± 105 | 1913 ± 978 | 1925 ± 851 | 2124 ± 537 |
| %CHO | 47 ± 10 | 48 ± 9.7 | 30 ± 1.4 | 40 ± 9.1 | 59 ± 3.5 | 62 ± 5.9 |
| %Fat | 36 ± 6.1 | 35 ± 7.5 | 45 ± 0 | 43 ± 5.0 | 28 ± 4.2 | 24 ± 8.0 |
| %Pro | 16 ± 5.0 | 16 ± 4.1 | 23 ± 1.4 | 17 ± 4.5 | 12 ± 3.0 | 14 ± 3.6 |
| Total CHO (g) | 257 ± 96 | 261 ± 99 | 146 ± 2 | 204 ± 141 | 283 ± 103 | 331 ± 61 |
| Total fiber | 26 ± 14 | 30 ± 20 | 7.0 ± 0.0 | 11 ± 4.0 | 39 ± 11 | 57 ± 14 |
| Insoluble | 18 ± 12 | 22 ± 16 | 2.5 ± 0.7 | 5.7 ± 3.1 | 29 ± 11 | 44 ± 12 |
| Soluble | 7.6 ± 2.4 | 7.7 ± 3.8 | 4.0 ± 0.0 | 5.0 ± 1.0 | 10 ± 0 | 13 ± 4.0 |
| Total fat (g) | 86 ± 21 | 82 ± 38 | 98 ± 5.0 | 90 ± 39 | 65 ± 38 | 64 ± 35 |
| Saturated fat (g) | 27.3 ± 9.9 | 25.8 ± 13.2 | 39.5 ± 5.0 | 35 ± 16.6 | 10.5 ± 5.0 | 11 ± 6.6 |
| Total pro (g) | 86 ± 22 | 83 ± 15 | 105 ± 1.4 | 75 ± 18 | 62 ± 15 | 82 ± 10 |
| Animal Protein | 48 ± 36 | 39 ± 22 | 92 ± 2.1a | 53 ± 13a | 1.3 ± 0.35 | 0.93 ± 0.95 |
| Vegetable protein | 38 ± 21 | 43 ± 24 | 13.3 ± 4.1 | 22.3 ± 8.9 | 60 ± 15 | 81 ± 10 |
| Protein sulfur | 0.68 ± 0.22 | 0.62 ± 0.14 | 0.93 ± 0.03a | 0.56 ± 0.15a | 0.41 ± 0.17 | 0.49 ± 0.02 |
Differs between non-responder and responder cohorts.
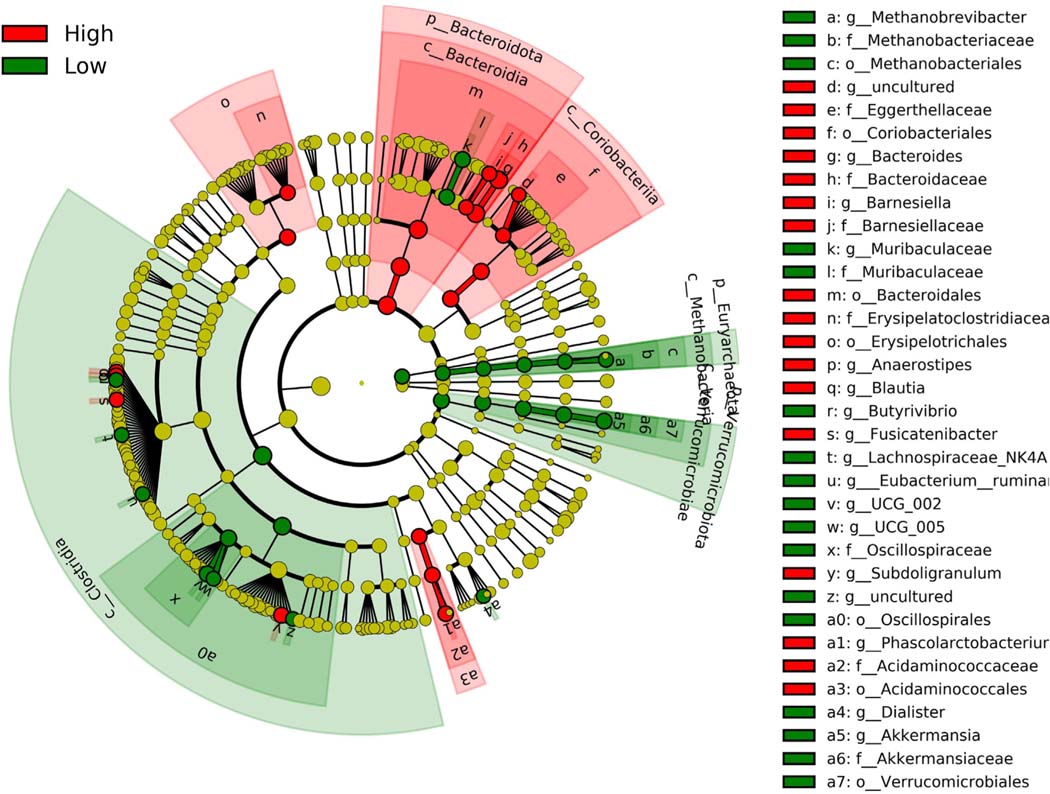
While no taxonomic changes in gut microbiota composition were observed between diet weeks in both the non-responder and responder cohorts, significant taxonomic differences were observed between responders and non-responders (Fig. 5). The non-responder cohort was enriched with members of the phyla Euryarchaeota (LDA score 3.47) and Verrucomicrobiota (LDA score 4.16) and members of the class Clostridia (LDA score 4.26). The responder cohort was enriched with the phylum Bacteroidota (previously termed Bacteroidetes; LDA score 4.42), the class Coriobacteriia (LDA score 3.93), and the orders Erysipelotrichales (LDA score 3.92) and Acidaminococcales (LDA score 3.75). The family Bacteroidaceae and genus Bacteroides had the highest LDA scores of 4.68 and were enriched in the responder cohort.
Fig. 5.
Linear discriminant analysis (LDA) combined with effect size measurements (LEfSe) comparing responders (“High”) and non-responders (“Low”). Enrichment of taxa in the responder groups are presented in red and enrichment of taxa in the non-responder group presented in green. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
A one-week plant-based diet intervention resulted in a lower ex vivo fecal H2S production compared to that produced by participants consuming an animal-based (i.e., western) diet intervention. However, this response was not uniform across all participants. The effect of short-term diet changes on ex vivo fecal H2S production appears to be related, in part, to changes in substrate availability. The competing effect of diet variables such as protein and fiber on H2S production observed in this study is largely consistent with reports from previous studies [4,5,10]. Despite this, we failed to observe a consistent effect of diet interventions on taxonomic composition of fecal gut microbiota, based on 16S rRNA gene profiling, or the abundance of several microbial taxa and genes encoding enzymes associated with sulfate reduction.
To the authors’ knowledge, this is the first study that used microfluidic quantitative PCR to simultaneously quantify multiple taxonomic and functional bacterial groups involved in sulfate reduction. Unlike previously reported by David et al., a study that was limited to a molecular analysis of a select number of studies [7], we did not demonstrate changes in sulfate reduction pathway enzymes in response to diet intervention [7]. However, we also did not modify carbohydrate intake. Although we asked participants to emphasize processed products, this still allowed for wheat products and a likely source of FOS/inulin in the diet that may not be adequately captured with the “fiber” variable. Using Gluten (grams) as a surrogate for intake of wheat products, there was no difference between diet periods in our study (p = 0.85), which may help explain the lack of observed difference in molecular targets between intervention weeks.
Patients with gastrointestinal diseases, such as ulcerative colitis (UC), often receive inconsistent information concerning their diet [32]. Often the only advice patients receive is to consume a low fiber diet in the midst of a UC flare, but recent National Health and Nutrition Examination Survey (NHANES) data from 2013 to 2018 found that mean intake of dietary fiber is already low, with <10% of all adults achieving recommended daily levels of dietary fiber intake [33].
A western diet is characterized by a reduced fiber intake resulting from an increased intake of processed foods and animal protein. Defined diets that have been proposed for management of inflammatory bowel disease – which encompasses UC – tend to contrast a western diet pattern encouraging a less-processed, higher fiber diet pattern [34–38]. Two studies recently demonstrated a benefit to increased fiber intake in management of UC [39,40]. Fiber has been previously shown to be negatively associated with fecal H2S production when added as a substrate [5] and we also demonstrate a similar effect with dietary intake. Therefore, H2S modulation in UC patients may be one possible mechanistic effect of implementation of a plant-based diet, but this requires further research.
The role of diet and its impact on health and chronic disease can also be assessed through diet score or quantitative methods. Here we used the empirical dietary inflammatory index (EDIP) [31] and found a strong positive relationship between the EDIP score and H2S production. The EDIP score has been associated with circulating inflammatory markers in adults [41] and its relationship with H2S production provides a possible microbiota linked mechanism that requires further exploration. Interestingly, the relationship between the EDIP score and H2S measures was found to be particularly pronounced in the high H2S production potential quartile (parameter estimate 46.23) compared to the low H2S production potential quartile (parameter estimate −9.19), suggesting that gut microbiota may serve as a modulating factor individualizing response to diet.
It is likely that enrichment of specific taxa within an individual is needed to produce elevated amounts of H2S or to have a high H2S production capacity [1]. As observed in this study, the cohort of “non-responders” did not have a predictable H2S response to diet variables. While this may indicate a lack of amino acid degradation capacity, our findings show only limited agreement with in vitro work inoculating human feces with protein and peptones [42]. In contrast, the responders in our cohort were enriched with taxa representing the Lachnospiraceae and Ruminococcaceae, which is in agreement with some of the findings of Amaretti et el [42]. However, we failed to demonstrate a diet-induced enrichment of protein degrading taxa including members of the phylum Proteobacteria and orders including the Enterobacteriales and Betaproteobacteriales. It is important to recognize, however, that our responder and non-responder cohorts were defined according to H2S response to diet rather than strictly response to protein, which also includes SRBs.
Developing an improved understanding of the role that the gut microbiome plays in the paradoxical response to dietary interventions is a critical step in precision nutrition, which has recently been identified as a research and funding priority by the National Institutes of Health (NIH). Our findings supporting a role of inter-individual gut microbiome variability in H2S responsiveness to diet add to recent work demonstrating inter-individual gut microbiome variability as a predictor of post-prandial metabolic parameters [43]. Further research is needed to elucidate gut microbiome characteristics capable of predicting diet responsiveness that would allow for development of the individualized diet patterns, which form the foundation of precision nutrition.
4.1. Limitations
Although ex vivo H2S measurements are intended to provide a surrogate measurement of colonic H2S production, they are unable to replicate what is truly occurring in vivo. This, in large part, is likely due to disruption of food matrices and plant cell walls due to homogenization that allows access to microbial substrates that would not have occurred in vivo. That said, however, homogenization likely results in an improved measure of microbial H2S production potential given the expectant increase in substrate availability.
5. Conclusion
Here we report that a 1-week plant-based diet intervention resulted in lower ex vivo H2S production compared to a 1-week animal-based diet intervention in most healthy individuals. No significant changes in taxonomic composition of fecal gut microbiota, measured using 16S rRNA gene profiling, were identified that possibly explain the change in H2S production, suggesting it was primarily driven by dietary intake (i.e., substrate availability). Protein was found to have a positive effect and fiber a negative effect on H2S production. However, H2S responsiveness to diet was not uniform across the entire cohort and potential H2S production enterotypes were characterized that may predict individualized H2S responsiveness to diet. Future studies that characterize the effects of dietary intake on the microbial community structure and metabolic output of the gut microbiota in different disease states, e.g., inflammatory bowel disease, obesity, as well as broader cohorts of healthy controls are needed. This work is needed to establish microbiome-based diagnostics that could inform personalized nutritional strategies that could optimize disease management.
Supplementary Material
Acknowledgments
University of Minnesota Center Research Analytical Lab for conducting urine sulfate measurements. We also thank the University of Minnesota Supercomputing Institute for providing resources.
Funding
Healthy Foods Healthy Lives (A.Kh); Achieving Cures Together (A.Kh); the University of Minnesota MnDRIVE Initiative (MJS); Allen Foundation (MJS).
Abbreviations:
- H2S
hydrogen sulfide
- SRB
sulfate reducing bacteria
- N2
nitrogen
- PERMANOVA
permutational multivariate analysis of variance
- LEfSe
linear discriminant analysis effect size
- LDA
linear discriminant analysis
- EDIP
empirical dietary inflammatory potential
- UC
ulcerative colitis
- IBD
inflammatory bowel disease
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.clnu.2022.03.028.
Conflict of interest
The authors declare that they have no conflicts of interest.
References
- [1].Nguyen LH, Ma W, Wang DD, Cao Y, Mallick H, Gerbaba TK, et al. Association between sulfur-metabolizing bacterial communities in stool and risk of distal colorectal cancer in men. Gastroenterology 2020;158(5):1313–25. 10.1053/j.gastro.2019.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rowan FE, Docherty NG, Coffey JC, O’Connell PR. Sulphate-reducing bacteria and hydrogen sulphide in the aetiology of ulcerative colitis. Br J Surg 2009;96(2):151–8. 10.1002/bjs.6454. [DOI] [PubMed] [Google Scholar]
- [3].Dostal Webster A, Staley C, Hamilton MJ, Huang M, Fryxell K, Erickson R, et al. Influence of short-term changes in dietary sulfur on the relative abundances of intestinal sulfate-reducing bacteria. Gut Microbes 2019;10(4):447–57. 10.1080/19490976.2018.1559682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Levine J, Ellis CJ, Furne JK, Springfield J, Levitt MD. Fecal hydrogen sulfide production in ulcerative colitis. Am J Gastroenterol 1998;93(1):83–7. 10.1111/j.1572-0241.1998.083_c.x. [DOI] [PubMed] [Google Scholar]
- [5].Yao CK, Rotbart A, Ou JZ, Kalantar-Zadeh K, Muir JG, Gibson PR. Modulation of colonic hydrogen sulfide production by diet and mesalazine utilizing a novel gas-profiling technology. Gut Microbes 2018;9(6):510–22. 10.1080/19490976.2018.1451280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Teigen LM, Geng Z, Sadowsky MJ, Vaughn BP, Hamilton MJ, Khoruts A. Dietary factors in sulfur metabolism and pathogenesis of ulcerative colitis. Nutrients 2019;11(4). 10.3390/nu11040931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014;505(7484):559–63. 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011;334(6052):105–8. 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tian L, Wang XW, Wu AK, Fan Y, Friedman J, Dahlin A, et al. Deciphering functional redundancy in the human microbiome. Nat Commun 2020;11(1): 6217. 10.1038/s41467-020-19940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Magee EA, Richardson CJ, Hughes R, Cummings JH. Contribution of dietary protein to sulfide production in the large intestine: an in vitro and a controlled feeding study in humans. Am J Clin Nutr 2000;72(6):1488–94. 10.1093/ajcn/72.6.1488. [DOI] [PubMed] [Google Scholar]
- [11].Magee EA, Curno R, Edmond LM, Cummings JH. Contribution of dietary protein and inorganic sulfur to urinary sulfate: toward a biomarker of inorganic sulfur intake. Am J Clin Nutr 2004;80(1):137e42. 10.1093/ajcn/80.1.137. [DOI] [PubMed] [Google Scholar]
- [12].Teigen L, Mathai PP, Matson M, Lopez S, Kozysa D, Kabage AJ, et al. Methanogen abundance thresholds capable of differentiating in vitro methane production in human stool samples. Dig Dis Sci 2020. 10.1007/s10620-020-06721-5. [DOI] [PubMed] [Google Scholar]
- [13].APHA. Method 4110B ion chromatography with chemical suppression of eluent conductivity, 2020. In: Standard methods for the examination of water and wastewater 23rd ed. 2017. 1015 Fifteenth Street, NW, Washington, DC: 20005. [Google Scholar]
- [14].Gohl DM, Vangay P, Garbe J, MacLean A, Hauge A, Becker A, et al. Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat Biotechnol 2016;34(9):942–9. 10.1038/nbt.3601. [DOI] [PubMed] [Google Scholar]
- [15].Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 2019;37(8):852–7. 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: high-resolution sample inference from illumina amplicon data. Nat Methods 2016;13(7):581–3. 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 2013;30(4): 772–80. 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 2010;5(3):e9490. 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 2013;41(Database issue):D590–6. 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Anderson MJ. Permutational multivariate analysis of variance (PERMANOVA). Wiley StatsRef: Statistics Reference Online:1–15. [Google Scholar]
- [21].Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol 2011;12(6): R60. 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Spurgeon SL, Jones RC, Ramakrishnan R. High throughput gene expression measurement with real time PCR in a microfluidic dynamic array. PLoS One 2008;3(2):e1662. 10.1371/journal.pone.0001662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Mathai PP, Dunn HM, Venkiteshwaran K, Zitomer DH, Maki JS, Ishii S, et al. A microfluidic platform for the simultaneous quantification of methanogen populations in anaerobic digestion processes. Environ Microbiol 2019;21(5): 1798–808. 10.1111/1462-2920.14589. [DOI] [PubMed] [Google Scholar]
- [24].Fite A, Macfarlane GT, Cummings JH, Hopkins MJ, Kong SC, Furrie E, et al. Identification and quantitation of mucosal and faecal desulfovibrios using real time polymerase chain reaction. Gut 2004;53(4):523–9. 10.1136/gut.2003.031245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Natividad JM, Lamas B, Pham HP, Michel ML, Rainteau D, Bridonneau C, et al. Bilophila wadsworthia aggravates high fat diet induced metabolic dysfunctions in mice. Nat Commun 2018;9(1):2802. 10.1038/s41467-018-05249-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kondo R, Nedweel DB, Purdy KJ, Silva SQ. Detection and enumeration of sulphate-reducing bacteria in estuarine sediments by competitive PCR. Geomicrobiol J 2004;21:145–57. 10.1080/01490450490275307. [DOI] [Google Scholar]
- [27].Ben-Dov E, Brenner A, Kushmaro A. Quantification of sulfate-reducing bacteria in industrial wastewater, by real-time polymerase chain reaction (PCR) using dsrA and apsA genes. Microb Ecol 2007;54(3):439–51. 10.1007/s00248-007-9233-2. [DOI] [PubMed] [Google Scholar]
- [28].Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 2012;487(7405):104–8. 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Apprill A, McNally S, Parsons R, Weber L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol 2015;75(2):129–37. 10.3354/ame01753. [DOI] [Google Scholar]
- [30].Parada AE, Needham DM, Fuhrman JA. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol 2016;18(5):1403–14. 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- [31].Tabung FK, Smith-Warner SA, Chavarro JE, Wu K, Fuchs CS, Hu FB, et al. Development and validation of an empirical dietary inflammatory index. J Nutr 2016;146(8):1560–70. 10.3945/jn.115.228718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Holt DQ, Strauss BJ, Moore GT. Patients with inflammatory bowel disease and their treating clinicians have different views regarding diet. J Hum Nutr Diet 2017;30(1):66–72. 10.1111/jhn.12400. [DOI] [PubMed] [Google Scholar]
- [33].Miketinas D, Tucker W, Patterson M, Douglas C. Usual dietary fiber intake in US adults with diabetes: NHANES 2013–2018. Curr Dev Nutr 2021;5(2):1061. 10.1093/cdn/nzab053_054. [DOI] [Google Scholar]
- [34].Chiba M, Abe T, Tsuda H, Sugawara T, Tsuda S, Tozawa H, et al. Lifestyle-related disease in Crohn’s disease: relapse prevention by a semi-vegetarian diet. World J Gastroenterol 2010;16(20):2484–95. 10.3748/wjg.v16.i20.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Konijeti GG, Kim N, Lewis JD, Groven S, Chandrasekaran A, Grandhe S, et al. Efficacy of the autoimmune protocol diet for inflammatory bowel disease. Inflamm Bowel Dis 2017;23(11):2054–60. 10.1097/MIB.0000000000001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lewis JD, Sandler RS, Brotherton C, Brensinger C, Li H, Kappelman MD, et al. A randomized trial comparing the specific carbohydrate diet to a Mediterranean diet in adults with Crohn’s disease. Gastroenterology 2021;161(3). 10.1053/j.gastro.2021.05.047. 837–852 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Olendzki BC, Silverstein TD, Persuitte GM, Ma Y, Baldwin KR, Cave D. An antiinflammatory diet as treatment for inflammatory bowel disease: a case series report. Nutr J 2014;13:5. 10.1186/1475-2891-13-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sigall Boneh R, Sarbagili Shabat C, Yanai H, Chermesh I, Ben Avraham S, Boaz M, et al. Dietary therapy with the Crohn’s disease exclusion diet is a successful strategy for induction of remission in children and adults failing biological therapy. J Crohns Colitis 2017;11(10):1205–12. 10.1093/ecco-jcc/jjx071. [DOI] [PubMed] [Google Scholar]
- [39].Fritsch J, Garces L, Quintero MA, Pignac-Kobinger J, Santander AM, Fernandez I, et al. Low-fat, high-fiber diet reduces markers of inflammation and dysbiosis and improves quality of life in patients with ulcerative colitis. Clin Gastroenterol Hepatol 2021;19(6):1189–1199 e30. 10.1016/j.cgh.2020.05.026. [DOI] [PubMed] [Google Scholar]
- [40].Valcheva R, Koleva P, Martinez I, Walter J, Ganzle MG, Dieleman LA. Inulin-type fructans improve active ulcerative colitis associated with microbiota changes and increased short-chain fatty acids levels. Gut Microbes 2019;10(3):334–57. 10.1080/19490976.2018.1526583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Tabung FK, Smith-Warner SA, Chavarro JE, Fung TT, Hu FB, Willett WC, et al. An empirical dietary inflammatory pattern score enhances prediction of circulating inflammatory biomarkers in adults. J Nutr 2017;147(8):1567–77. 10.3945/jn.117.248377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Amaretti A, Gozzoli C, Simone M, Raimondi S, Righini L, Perez-Brocal V, et al. Profiling of protein degraders in cultures of human gut microbiota. Front Microbiol 2019;10:2614. 10.3389/fmicb.2019.02614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Berry SE, Valdes AM, Drew DA, Asnicar F, Mazidi M, Wolf J, et al. Human postprandial responses to food and potential for precision nutrition. Nat Med 2020;26(6):964–73. 10.1038/s41591-020-0934-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.