Abstract
Fibroblast growth factor 1 (FGF1) and FGF2, the prototypic members of the FGF family of growth factors, have been implicated in a variety of physiological and pathological processes. Unlike most other FGFs, FGF1 and FGF2 are ubiquitously expressed and are not efficiently secreted. Gene knockouts in mice have previously demonstrated a role for FGF2 in brain development, blood pressure regulation, and wound healing. The relatively mild phenotypic defects associated with FGF2 deletion led to the hypothesis that the continued expression of other FGFs partially compensated for the absence of FGF2 in these mice. We now report our generation of mice lacking FGF1 and their use, in combination with our previously described FGF2 null mice, to produce mice lacking both FGF1 and FGF2. FGF1-FGF2 double-knockout mice are viable and fertile and do not display any gross phenotypic defects. In the double-knockout mice we observed defects that were similar in extent to those previously described for the FGF2 null mice. Differences in the organization of neurons of the frontal motor cortex and in the rates of wound healing were observed. We also observed in FGF2−/− mice and in FGF1-FGF2 double-knockout mice novel impairments in hematopoiesis that were similar in severity. Essentially no abnormalities were found in mice lacking only FGF1. Our results suggest that the relatively mild defects in FGF2 knockout animals are not a consequence of compensation by FGF1 and suggest highly restricted roles for both factors under normal developmental and physiological conditions.
Fibroblast growth factors (FGFs) comprise a widely expressed and multifunctional family of polypeptides. FGFs transduce signals that can regulate cell growth, migration, differentiation, or survival. The biological activity of FGFs is mediated through interactions with transmembrane tyrosine kinase receptors. Four different receptors for FGFs are known, although each is present in multiple isoforms owing to alternative splicing of the mRNA. For the most part, there is no one-to-one correspondence between FGF ligands and receptors. A given FGF may be capable of multiple receptor isoforms; conversely, any receptor variant may bind multiple FGFs (3, 8, 19).
FGF signaling has been implicated in a variety of physiological and pathological processes, ranging from angiogenesis to tumor progression. To date, however, the most clearly demonstrated role of FGF signaling is in development. Studies using knockout mice have demonstrated essential functions for FGF receptor 1 (FGFR1) and FGFR2 in early development (1, 12, 40, 41) and roles for FGFR3 in skeletal morphogenesis (9, 11). Studies of mice lacking individual FGFs reveal a variety of phenotypes which range from early embryonic lethality to very mild defects (14, 16, 17, 22, 23, 27, 30, 31, 34, 42). These findings most likely reflect the redundancy of the FGF family of ligands or their uniqueness of expression in specific tissues.
A total of 22 different FGF molecules have been described so far, although four of them (FGF-homologous factors [FHFs] FGF11 to -14) (37) may not be canonical FGFs. FGF1 and FGF2 were the first to be isolated and were originally named acidic and basic FGF, respectively, based on their isoelectric points. Despite their status as the “prototypic” FGF family members, FGF1 and FGF2 differ from most other FGFs in several important ways. FGF1 is unique among FGFs in that it binds with high affinity to all known receptor isoforms (33). Although many FGFs exhibit limited spatial or temporal expression patterns, mRNAs for FGF1 and FGF2 are detectable in a variety of tissues during both development and adulthood. FGF1 and FGF2 lack a signal peptide at their 5′ ends and are found in the cytosol; however, both factors seem to be released from cells through a nonclassical secretory pathway (6, 8). Intriguingly, both FGF1 and FGF2 have also been found in the cell nucleus. A putative nuclear localization signal has been identified at the 5′ end of the FGF1 protein (24), and alternative translation initiation sites in the 5′ region of the FGF2 gene give rise to higher-molecular-weight forms of the protein that localize to the nucleus (6, 7). The precise function(s) of these nuclear forms of FGF1 and FGF2 remains unclear.
There is evidence suggesting a role for FGF1 and FGF2 in the proper development and maintenance of neuronal tissue. Both FGFs are highly expressed in adult brain, although each factor localizes to a different neuronal subpopulation. Expression of FGF1 has been detected in sensory and motor neurons, as well as in the substantia nigra, cholinergenic neurons of the basal forebrain, and several other subcortical neuronal populations. In contrast, FGF2 expression is found primarily in astrocytes and pyramidal neurons of the hippocampus (see reference 15 and references cited therein). A potential role for FGFs in brain development is suggested by the observation that FGF2 can induce neuroectoderm formation and can establish regional neuroectodermal identities along the anteroposterior axis when provided exogenously to gastrula and early-neurula-stage frog embryos (21). In vitro, FGF2 induces astrocytes to reenter the cell cycle and induces markers of differentiation (25). FGF2 promotes the outgrowth of cultured hippocampal and cortical neurons (28) and regulates the expression of certain neurotransmitters (2). FGF2 can also stimulate division of cortical stem cells and may promote differentiation of postmitotic neurons (20, 35, 38).
FGFs have been postulated to play a major role in wound healing, with particular focus on potential roles for FGF1, FGF2, and FGF7. FGFs promote angiogenesis as well as stimulate proliferation of many cell types involved in wound healing, including endothelial cells, fibroblasts, and keratinocytes (4, 5). Topical application of FGF1 and FGF2 accelerates wound healing in a number of animal models (29, 32). Many of the cell types active in wound healing that are responsive to FGFs are themselves capable of secreting FGFs and other cytokines, raising the possibility of highly complex and regulated interactions between a variety of cell types.
Members of the FGF family have also been shown to play a role in hematopoiesis. They can synergize with hematopoietic cytokines to promote the clonal growth of hematopoietic cells in culture and antagonize the negative regulatory effects of transforming growth factor β (18). Addition of FGF to human long-term bone marrow cultures (LTBMCs) increases both the cell density of the stromal layer and the number of hematopoietic colony-forming cells in the cultures in a dose-dependent manner (36). The primary effects appear to be on the stromal cells in these cultures, but a direct effect on hematopoietic progenitors cannot be ruled out.
Despite the broad pattern of expression and ability of FGFs to activate several FGFRs, the ability of FGF1 or FGF2 to do so in vivo may be limited due to the multiple levels of control—transcriptional, translational, and secretory—over their bioavailability. In vitro studies attributing potential biological activities to either of these FGFs must therefore be interpreted with some degree of caution. The generation of mice with homozygous deletions of specific genes has become a valuable tool in the evaluation of gene function in vivo, highlighting new roles of individual genes while revealing complex interactions among members of gene families. Several groups, including our own, have previously reported the generation and characterization of mice lacking FGF2 (14, 34, 42). Three independently derived lines of FGF2 null mice are all viable and fertile. The brains of FGF2-deficient mice contain subtle but significant reductions in the number and organization of neurons within the cortex (14, 34, 39). FGF2 null mice show various vascular defects and are hypotensive, although they retain the capacity to regulate blood pressure in response to stimuli (14, 42). A delay in the healing of full-thickness epithelial wounds in FGF2 null mice is also observed (34). The relatively mild phenotypic defects observed in FGF2 null mice have led to the hypothesis that FGF1 and FGF2 may constitute a redundant pair of FGFs with similar physiological targets and largely overlapping functions, in which case the continued expression of FGF1 would act largely to compensate for the absence of FGF2.
We now report our generation and characterization of mice lacking FGF1 and the use of these mice, in combination with our previously described FGF2−/− mice, to produce FGF1−/− FGF2−/− double knockouts. FGF1 null animals, like the FGF2 knockouts, are viable and fertile. Studies similar to those we have previously described reveal that FGF1−/− mice do not exhibit any of the phenotypic abnormalities associated with deletion of FGF2; they exhibit normal brain structure and normal rates of wound healing. In a series of new studies, we observed novel hematopoietic deficiencies in the FGF2−/− mice, while FGF1−/− mice were normal. Surprisingly, deletion of FGF1 in animals already lacking FGF2 did not seem to significantly worsen the phenotypic defects seen in the FGF2 knockouts. This observation suggests either that there is an extensive degree of redundancy and compensation within the FGF family or that FGF1 and FGF2 play rather limited roles under normal physiological conditions.
MATERIALS AND METHODS
Generation and analysis of FGF1 and FGF1-FGF2 knockout animals.
To generate FGF1-deficient mice, a clone containing FGF1's first exon was isolated from a 129SVJ mouse genomic library (Stratagene). The library was screened with the human FGF1 cDNA followed by a 120-bp NcoI-BamHI fragment containing the first exon. Two independent clones spanning approximately 12 kb were isolated. The targeting plasmid was constructed by use of the pPNT replacement vector. The XhoI and EcoRI sites of pPNT were digested, blunted, and used to insert the 2.8-kb BamHI-EcoRI 5′ arm and the 2.4-kb XbaI-NotI 3′ arm, respectively. Upon homologous recombination, a deletion of approximately 4.7 kb that includes the entire first exon results. The plasmid was linearized with NotI and purified, and 30 μg was electroporated into E14 embryonic stem (ES) cells. Transfected cells were selected in G418 (400 μg/ml) and FIAU [1-(2′-deoxy-2′-fluoro-1-β-d-arabinofuranosyl)-5-iodouracil] (0.25 μM) as previously described (34). Clones were screened by Southern analysis prior to injection into C57Bl/6 blastocysts.
For Western analysis, protein extracts from mouse tissues were prepared as previously described (34). For some immunoblots, 40 μg of total protein was directly loaded onto 15% polyacrylamide gels; alternatively, approximately 10 mg of protein was concentrated by the use of heparin-Sepharose CL-6B beads prior to electrophoresis, as previously described (34). Half of the sample (equivalent to 5 mg of total protein) was separated on a 15% polyacrylamide gel. Proteins were transferred to nitrocellulose and analyzed by using either an FGF1 (a gift of J. Schlessinger) or an FGF2 (Santa Cruz Biotechnology) polyclonal antibody and detected by using an enhanced chemiluminescence system (Amersham). Recombinant FGF1 was kindly provided by J. Schlessinger, and recombinant FGF2 was supplied by D. Rifkin.
Brain analysis.
Brain histological and immunohistochemical analyses were carried out as previously described (34), except that Nissl staining was carried out with thionine blue instead of cresyl violet.
Wound healing.
Age- and sex-matched adult mice, approximately 3 to 4 months old, were anesthetized and prepared for surgery as previously described (34). Two 6-mm-diameter full-thickness epidermal wounds were created by excision with sterile scissors. After recovery, animals were examined regularly and healing was measured semiquantitatively on a 5-point scale, with 4 indicating that the original scab was present and intact, 3 meaning that the original scab was gone but a smaller scab was present, 2 representing the lack of a scab but the presence of a wound, 1 indicating that a small wound was detectable, and 0 representing a wound completely healed. All manipulations were carried out by investigators blinded to the specimens' identities.
Blood and hematopoietic analysis.
Whole blood was obtained from anaesthetized mice via the retro-orbital venous sinus. White blood cells, red blood cells, and platelets were counted by using a model Zf Coulter Counter. The platelet counts were performed on platelet-rich plasma. Colony-forming cells (CFU-c) were assayed by culture in Iscove's modified Dulbecco medium made semisolid by the addition of methylcellulose in the presence of appropriate growth factors (StemCell Technologies, Vancouver, BC, Canada). The cultures were incubated under conditions that allow identification of burst-forming units-erythroid (BFU-e), colony-forming units-granulocyte (CFU-G), CFU-granulocyte/monocyte (CFU-GM), and CFU-granulocyte/erythroid/monocyte/megakaryocyte (CFU-GEMM) in the same dish. Three or four replicated cultures were scored after 7 to 8 days of incubation and again at 2 weeks. LTBMCs were prepared from femoral marrow as described elsewhere (13). Cultures were sampled biweekly to quantify the total number of cells and assess the number of hematopoietic progenitors present in the cultures. After 4 weeks, some cultures were irradiated (120 Gy) to suppress hematopoiesis and then recharged with stromally depleted low-density bone marrow cultures (BMCs). The stromally depleted BMCs were prepared by incubating low-density BMCs overnight in 100-mm-diameter dishes and discarding adherent cells. The nonadherent cells were added to the preformed, irradiated stromal layers. These cultures were then maintained and sampled for CFU-c as described above.
RESULTS
Generation of mice lacking FGF1.
FGF1 is a single-copy gene located on murine chromosome 18 and is organized into three relatively short coding exons separated by large introns spanning approximately 30 kb (26). Unlike the FGF2 gene, which gives rise to multiple protein isoforms through the use of alternative upstream translation initiation codons, the FGF1 gene is preceded by an in-frame stop codon and encodes a single polypeptide (3). We designed a replacement vector to delete the entire first coding exon along with several kilobases of surrounding genomic DNA. The deleted sequences were replaced with a neomycin resistance cassette (neor) driven by the mouse phosphoglycerate kinase (PGK) promoter and containing the PGK polyadenylation signal. The vector also contained a herpes simplex virus thymidine kinase (hsv-TK) gene, allowing recombinants to be screened by both positive and negative selection (Fig. 1A).
FIG. 1.
(A) The pPNT replacement vector was used to construct a targeting plasmid to delete approximately 5 kb of genomic DNA, including the entire first coding exon of FGF1 (black box). Probes used in Southern analysis are indicated. Abbreviations for restriction enzyme sites are as follows: B, BamHI; N, NcoI; Not, NotI; RI, EcoRI; RV, EcoRV; and X, XbaI. The NotI site at the 3′ end of the genomic sequence is derived from the phage DNA. The sites in parentheses were destroyed during cloning. (B) (Top) The presence of the recombined targeting plasmid in a clone (+/−) is detected by using an external probe. A normal clone (+/+) is shown for comparison. (Center and bottom) Litters from crosses of heterozygous mice contain wild-type (+/+), heterozygous (+/−), and FGF1 null (−/−) mice. Southern analysis was performed with the indicated restriction enzymes and probes. Positions of molecular size standards (in kilobases) are shown at the left. (C) Extracts from brain (lanes 2 to 4) and heart (lanes 5 to 7) tissues were analyzed for FGF1 and FGF2 expression. Tissues from wild-type (lanes 2 and 5), heterozygous (lanes 3 and 6), and FGF1 null (lanes 4 and 7) mice were assayed. (Top) Forty micrograms of whole-cell extract; immunoblotting with an anti-FGF1 polyclonal antibody. Recombinant FGF1, which contains a small N-terminal truncation, is in lane 1. The arrow indicates the migration position of FGF1. (Center) Five milligrams of total protein, concentrated by using heparin-Sepharose beads; immunoblotting with an anti-FGF1 polyclonal antibody. Recombinant FGF1 is in lane 1. The arrow indicates the migration position of FGF1. (Bottom) Five milligrams of total protein, concentrated by using heparin-Sepharose beads; immunoblotting with an anti-FGF2 polyclonal antibody. Recombinant FGF2 is in lane 1. The three unmarked arrows indicate the migration positions of the three FGF2 isoforms present in mice. The starred arrow indicates a band in lanes 5 and 6 arising from cross-reactivity of the antibody with the FGF1 present. Migration positions of molecular mass standards (in kilodaltons) are shown at the left. (D) Extracts were prepared from the brains of wild-type, FGF1−/− (FGF1 KO), FGF2−/− (FGF2 KO), and FGF1−/− FGF2−/− (Double KO) mice. (Left) Forty micrograms of whole-cell extract; immunoblotting with an anti-FGF1 polyclonal antibody. Recombinant FGF1 (rFGF1) was used as a control. (Right) Five milligrams of total protein, concentrated by using heparin-Sepharose beads; immunoblotting with an anti-FGF2 polyclonal antibody. Recombinant FGF2 was used as a control. Neither protein is present in FGF1-FGF2 double-knockout mice. The migration positions of molecular mass standards (in kilodaltons) are shown at the left.
The targeting vector was electroporated into E14 embryonic stem (ES) cells, and colonies were selected in the presence of both G418 (positive selection) and FIAU (negative selection). A total of 200 colonies were picked and screened for homologous recombination by Southern analysis with a 5′ internal probe (Fig. 1A, probe 2). Candidate clones were further analyzed by using 5′ external (probe 1) and 3′ internal (probe 3) fragments. Additional Southern analyses were carried out with neor and hsv-TK probes to confirm the presence of appropriate sequences and the absence of nonhomologous DNA (data not shown). A single clone that yielded the predicted bands in all Southern analysis was identified (Fig. 1B, top) and exhibited a normal karyotype, and this clone was injected into C57Bl/6 blastocysts. Four male chimeras were generated from this injection, and all transmitted agouti coat color to their offspring. As expected, 50% of the agouti pups were heterozygous for the FGF1 deletion, indicating that there was no lethality associated with an FGF1+/− genotype. We then intercrossed these heterozygous animals to generate FGF1 null animals (Fig. 1B, middle and lower panels).
We confirmed the absence of FGF1 protein in the FGF1 null animals through Western analysis of various tissues. Analysis of whole-cell extracts (Fig. 1C, top) from brain (lanes 2 to 4) and heart (lanes 5 to 7) tissues showed the presence of FGF1 protein in FGF1+/+ (lanes 2 and 5) and FGF1+/− (lanes 3 and 6) animals but not in FGF1−/− animals (lanes 4 and 7). We also analyzed heparin-Sepharose concentrates of whole-cell extracts (Fig. 1C, center). While FGF1 protein is easily detected in 40 μg of whole-cell extract from wild-type and heterozygous mice (top panel), it remains undetectable in 5 mg of concentrated protein from FGF1 null mice (center panel, lanes 4 and 7). As expected, FGF2 is readily detected in heparin-Sepharose concentrates from animals of all three FGF1 genotypes (bottom panel). We performed a similar analysis on a range of tissues, including skull, long bone, liver, spleen, lung, skeletal muscle, kidney, testis, and eye, and could not detect FGF1 protein in any tissues from FGF1−/− animals. We also did not observe an increase in FGF2 expression in any tissues derived from FGF1−/− animals compared to FGF1+/+ and FGF+/− mice, suggesting that FGF2 expression is not upregulated in the absence of FGF1 (data not shown).
Crosses of FGF1 heterozygotes revealed that wild-type, heterozygotes, and FGF1 null animals were born at the expected frequencies. The FGF1−/− mice appeared indistinguishable from their wild-type littermates and grew, developed, and bred normally. No significant differences were seen in the size or weight of the homozygous mutants compared to their wild-type littermates. Histopathological examination of a range of tissues from FGF1−/− animals did not reveal any striking defects, consistent with the macroscopically normal appearance and behavior of the animals. We also have not observed any disorders in elderly FGF1 null animals, suggesting that there are no long-term disorders that are degenerative in nature.
Generation of FGF1-FGF2 double-knockout animals.
One explanation for the absence of any obvious phenotypic defects in the FGF1 knockouts is the continued expression of FGF2. Similarly, the presence of only relatively minor phenotypic defects in the FGF2 knockouts could be due to the continued expression of FGF1. We therefore hypothesized that animals lacking both FGF1 and FGF2 would exhibit more severe phenotypic abnormalities than either line of single-knockout mice or would display novel defects not present in either single-knockout line. We intercrossed our FGF1 and FGF2 single-knockout mice to generate mice lacking both FGF1 and FGF2.
Crosses of FGF1−/− and FGF2−/− mice produced litters of normal size that contained exclusively FGF1+/− FGF2+/− mice, as expected. We used these double heterozygotes to generate animals lacking FGF1 and FGF2 alleles in various combinations, including animals with only a single functional allele (FGF1+/− FGF2−/− or FGF1−/− FGF2+/−) and animals null for both FGF genes (FGF1−/− FGF2−/−). All animals were born at the expected frequencies. Animals of all genotypes were grossly normal and developed and bred normally.
The absence of FGF1 and FGF2 expression in the double knockouts was confirmed by Western analysis of extracts from brains of wild-type, FGF1−/− single-knockout, FGF2−/− single-knockout, and FGF1−/− FGF2−/− double-knockout animals (Fig. 1D). For detection of FGF1, whole-cell extracts were directly loaded onto a polyacrylamide gel (left); for FGF2, proteins were first concentrated by the use of heparin-Sepharose beads (right). We readily detected FGF1 protein in wild-type and FGF2−/− animals and FGF2 expression in wild-type and FGF1−/− animals but could not detect expression of either FGF1 or FGF2 in double-knockout animals, confirming the absence of both proteins in these mice.
Brain analysis.
Both FGF1 and FGF2 are highly expressed in adult brain tissue. Although the brains of all lines of our FGF knockout mice are macroscopically indistinguishable from their wild-type counterparts, we have previously observed microscopic differences in neuronal structure in brains of FGF2−/− mice. We noted abnormalities in the cytoarchitecture of the neocortex which were most pronounced in the frontal sensorimotor area (34). Others have also reported brain abnormalities in animals lacking FGF2 (14), including decreases in numbers both neurons and glial cells (39). Because FGF1 and FGF2 have similar temporal and spatial expression patterns within the brain (15), we undertook a series of studies, similar to those previously done with the FGF2 null animals, to determine whether deletion of FGF1 resulted in comparable neuronal defects.
In our previous studies of FGF2−/− mice, a combination of Nissl staining and immunohistochemical analysis revealed a thickening of the neocortex and a coincident decrease in neuronal cell density. A similar analysis of the brains of FGF1−/− mice was therefore performed. A comparison of matched coronal sections throughout the brains of age- and sex-matched wild-type and FGF1−/− animals did not reveal any significant differences in the architecture of the cortex or in the number of neurons present. Nissl-stained sections from FGF1−/− animals were indistinguishable from those of wild-type animals (Fig. 2A and B). We next compared brains of FGF1−/− FGF2−/− mice with those of the FGF2 single knockouts to determine whether additional deletion of FGF1 in these animals resulted in more-severe defects. While we observed a thickening of the neocortex in double-knockout mice compared to wild-type animals, we did not detect more extensive changes than those seen in FGF2−/− mice (Fig. 2C and D).
FIG. 2.
Matched coronal sections of brain tissue from wild-type (A, E, and I), FGF1 null (FGF1 KO) (B, F, and J), FGF2 null (FGF2 KO) (C, G, and K), and double-knockout (Double KO) (D, H, and L) mice were microscopically analyzed. Sections were stained for Nissl substance (A to D) or analyzed by immunohistochemical techniques using antibodies against parvalbumin (E to H) or calbindin (I to L). No differences between the brains of wild-type and FGF1−/− mice were noted. Brains of FGF2−/− mice show a thickening of the motor cortex (C) and a decrease in the number of parvalbumin (G)- and calbindin (K)-positive cells. No significant further thickening of the cortex (D) or decrease in neuronal subpopulations (H and L) was apparent in the brains of FGF1−/− FGF2−/− mice.
In our previous studies of FGF2 null mice, the decrease in neurons detected by Nissl staining was mirrored by changes in neuronal subpopulations defined by the calcium-binding proteins calbindin and parvalbumin. Staining with either anticalbindin or antiparvalbumin antibodies failed to reveal any differences between wild-type and FGF1−/− animals (Fig. 2E, F, I, and J). Analysis of brain tissue from FGF2−/− and FGF1-FGF2 double-knockout mice revealed the same phenotype as that observed with Nissl staining; while clear differences between the brains of double-knockout and wild-type mice were noted, no significant differences between FGF2 single- and FGF1-FGF2 double-knockout animals could be detected (Fig. 2G, H, K, and L). We cannot, however, rule out the possibility that future studies, investigating other regions of the brain or neuronal subpopulations, will reveal differences between these groups of mice that have not been detected here.
Wound healing.
We have previously described a small but reproducible delay in the healing of full-thickness excisional wounds in FGF2−/− animals. Preliminary studies with our FGF1−/− mice indicated that deletion of FGF1 had no effect on the rate of healing of such wounds (data not shown). We were interested in determining whether FGF1-FGF2 double-knockout animals had a wound healing delay longer that that seen in FGF2 single knockouts.
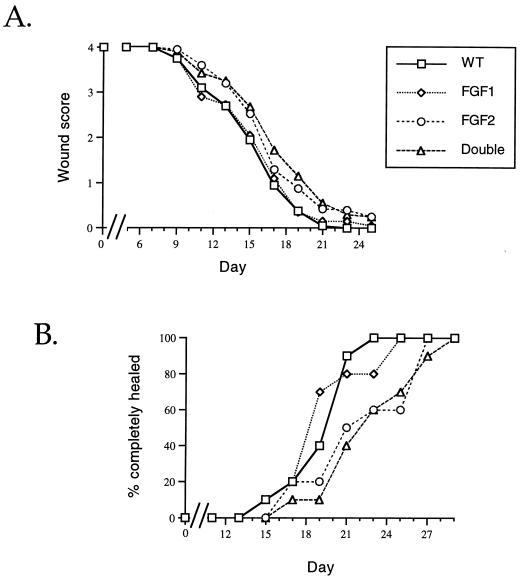
A total of 40 age- and sex-matched animals (10 each of wild type, FGF1−/−, FGF2−/−, and FGF1−/− FGF2−/−) were used to determine rates of wound healing and allow direct comparison of all groups with one another. Two 6-mm-diameter full-thickness excisional wounds were created on the back of each mouse, and the degree of wound healing was assessed visually, using a semiquantitative scale (described in Materials and Methods). At the time of wounding and throughout the experiment, all procedures and wound evaluations were performed in a genotype-blinded manner. At the end of the experiment, the data were analyzed and plotted as both percentage of animals completely healed versus time (as in the previous study) and mean degree of healing (wound score) versus time.
The wild-type and FGF1−/− animals exhibited the same degree of wound healing (Fig. 3A). The rate of wound healing in FGF2−/− mice was delayed. FGF1−/− FGF2−/− mice displayed a delay in wound healing similar to that seen in the FGF2 single knockouts. While there was a small difference in wound healing of FGF2−/− and double knockouts at days 15 and 21, it is clear that deletion of both FGF1 and FGF2 did not result in large-scale defects in wound healing. Similar results were obtained when the data were plotted as the percentages of animals in which both wounds were completely healed in the groups (Fig. 3B). Wild-type and FGF1 null animals exhibited similar rates of wound healing, and the rates of healing in FGF2−/− and FGF1-FGF2 double-knockout mice, while slower than that of the other two groups, were similar to one another. While these results do not rule out the possibility of a role for FGF1 in wound healing, any contribution it makes would seem to be smaller than that of FGF2 and beyond the limits of detection in this experiment. Additionally, any differences in rate of wound healing between FGF2−/− and double-knockout mice must also be very small.
FIG. 3.
Ten mice of each genotype were wounded on day zero, and the degree of healing was assessed visually according to a semiquantitative scale (described in Materials and Methods). Wild-type (WT) and FGF1 null mice healed at a similar rate; FGF2 null and double-knockout mice healed more slowly than wild-type mice. Data were plotted as degree of healing over time (wound score) (A) and as percentage of completely healed animals over time (B). In both cases, there was no significant further impairment in the healing of double-knockout mice compared to that of FGF2 null mice.
Hematopoiesis.
No significant differences were detected in the white blood cell, red blood cell, or platelet counts of FGF1−/−, FGF2−/−, and FGF1−/− FGF2−/− mice compared to wild-type animals. The differential counts for all of these mice were also within the normal range. Femoral bone marrow was obtained from each group of mice and cultured in semisolid medium to determine the number of steady-state hematopoietic progenitors present. Again, no differences in colony formation were observed between control BMCs and those from FGF knockout mice (data not shown).
To assess the relative ability of the bone marrow to support ongoing hematopoiesis, we prepared LTBMCs from the femurs of wild-type, FGF1−/−, and FGF2−/− mice. Stromal cells from all mice became confluent after 3 weeks in culture. At weekly intervals the nonadherent layer was sampled for myeloid progenitors and CFU-c were quantified. No statistically significant differences between cultures derived from wild-type and FGF1−/− mice were detected. On the other hand, at all time points, cultures obtained from FGF2−/− mice had fewer colonies than either wild-type or FGF1−/−-derived cultures (Fig. 4A). We also examined CFU-c in mice lacking both FGF1 and FGF2. LTBMCs were readily established, and the stromal cells grew as well as those from age-matched wild-type controls. After 4 weeks, the cultures prepared from the FGF1-FGF2 double-knockout animals showed a degree of hematopoietic impairment similar to that of the FGF2−/−-derived cultures (Fig. 4B).
FIG. 4.
(A) LTBMCs were prepared from wild-type, FGF1−/−, and FGF2−/− mice, and CFU-c were assayed at weekly intervals. No statistically significant differences between cultures derived from wild-type and FGF1−/− animals were noted, while cultures from FGF2−/− mice produced fewer myeloid progenitors. (B) LTBMCs from FGF1-FGF2 double-knockout (KO) mice exhibit diminished production of CFU-c compared to cultures from wild-type (WT) mice, but the degree of impairment is similar to that seen in cultures prepared from FGF2−/− mice. (C) LTBMCs were prepared from wild-type (WT) and FGF1-FGF2 double-knockout (DKO) mice. After the stromal layers became confluent, endogenous hematopoiesis was suppressed by irradiation. Cultures were subsequently recharged with either WT or DKO BMCs. WT stromal cells support hematopoiesis in both WT (WT on WT) and DKO (DKO on WT) BMC, while DKO stromal cells poorly support hematopoiesis in WT (WT on DKO) and DKO (DKO on DKO) BMCs.
To distinguish between effects of FGF deletion on hematopoietic stem cells and on stromal cells, LTBMCs were initiated from wild-type and FGF1−/− FGF2−/− mice. After the cultures were established, they were irradiated to suppress endogenous hematopoiesis. Cultures were subsequently recharged with fresh BMC from either wild-type or FGF1-FGF2 double-knockout mice and incubated for an additional 5 weeks. Wild-type stroma restored CFU-c production by both wild-type and double-knockout BMC, and FGF-deficient stroma was unable to restore CFU-c production by either wild-type or double-knockout BMC (Fig. 4C). This result strongly suggests that the impaired hematopoiesis in cultures derived from FGF1−/− FGF2−/− animals arises primarily from a defect in the stromal layer and is not a consequence of a defect in the hematopoietic progenitors.
We have also examined LTBMCs prepared from FGF1-FGF2 double-knockout animals ranging in age from 3 weeks to 1 year. No sign of an age-related deterioration in the capacity to support hematopoiesis in comparison to wild type mice has been seen (data not shown).
DISCUSSION
We and others have previously described a range of phenotypic defects in mice lacking FGF2, including changes in the number of neurons present in the cerebral cortex, a delay in the healing of epithelial wounds, and defects in the regulation of blood pressure (14, 34, 39, 42). While these physical and physiological consequences of FGF2 deletion are readily detectable, perhaps the most striking characteristic they share is the limited extent to which they impact development, growth, and homeostasis. The observation that FGF2 knockouts display some phenotypic defects indicates that this factor does play roles in certain developmental and physiological processes that cannot be performed by other proteins. However, because FGF2 is so widely expressed in so many tissues and has been found to induce such a wide range of responses from various cell types in vitro, the rather mild phenotypic abnormalities accompanying its deletion were somewhat surprising.
The failure of FGF2 deletion to result in widespread and extensive negative consequences might be minimized due to the continued expression of other factors, including other FGF family members. Of all the FGFs, the one that exhibits the most similarity to FGF2 in terms of structure, expression pattern, and cellular and subcellular localization is FGF1. The fact that FGF1 is capable of binding and activating all known FGFR splice variants suggests that this molecule might be able to act as a substitute for FGF2 in the knockout mice, effectively replacing FGF2 function through binding to the same receptors and eliciting similar biological responses. To address this possibility, we generated FGF1 null mice. While mice lacking FGF1 are worthy of study in their own right, we were especially interested in determining the consequences of deletion of both FGF1 and FGF2 in the same animal.
Using techniques similar to those previously employed to establish differences in brain structure and wound healing between wild-type and FGF2 knockout mice, we were unable to detect any differences between wild-type and FGF1 knockout mice. We did not observe significant differences between wild-type and FGF1 null mice in in vitro hematopoiesis, although we readily detected such differences between wild-type and FGF2 null mice. These observations suggest that any differences between FGF1 knockout mice and control animals must be smaller than those between FGF2 knockout mice and controls. However, the possibility of compensation for loss of FGF1 by FGF2 in these mice remained. We were therefore interested in determining the impact of deleting both FGF1 and FGF2. If our compensation hypothesis were correct, then we would expect deletion of FGF1 in animals already lacking FGF2 to result in a host of novel phenotypic abnormalities as well as to significantly worsen the previously observed defects.
Our analysis of FGF1-FGF2 double-knockout mice suggests that compensation does not explain the relatively mild phenotypic defects associated with deletion of either single factor, insofar as the double-knockout mice do not exhibit significantly worse defects than mice lacking only FGF2. This result demonstrates that the failure of FGF1 knockout mice to display any detectable phenotype is unlikely to be due to the continued expression of FGF2 and that the relatively mild phenotypic defects seen in the FGF2 knockout are probably not a consequence of the continued expression of FGF1.
There are several explanations for our failure to detect significant differences between wild-type and FGF1−/− mice and between FGF2−/− and FGF1−/− FGF2−/− mice. First, it is possible that we did not detect phenotypic differences arising from FGF1 deletion simply because there are none that can be seen under normal circumstances. Although we were able to rule out compensation by FGF1 or FGF2 in each of the single knockouts through generation of the double knockout, it could be that other FGFs are compensating for the absence of both FGF1 and FGF2. It need not necessarily be an FGF that is providing the compensatory signal, since other molecules may act to functionally replace the missing growth factors. If this is the case, it suggests a remarkable degree of redundancy among various FGF family members or between multiple growth factor signaling pathways.
An alternate, related explanation is that FGF1 and FGF2 play a very limited role in normal physiological processes, although they presumably perform some functions in specific situations, such as following stress or injury. In this scenario, the roles of FGF1 and FGF2 are limited during development and normal adult life. In specific situations, however, these factors play crucial roles. Support for this interpretation comes from our study of wound healing: the developmental pathways leading to the formation of skin appear normal in wild-type and knockout animals, but after wounding, repair takes longer in the FGF2−/− and FGF1-FGF2 knockout mice than in the wild type. If this explanation is valid, then only by studying the mice in the appropriate pathological state will differences between wild-type and knockout mice become apparent.
It is also possible that we failed to detect consequences of FGF1 deletion because such defects are present but not very large. Deletion of FGF1 could produce phenotypic effects below the threshold of detection by the methods employed in this report. Because we have used similar techniques to document significant differences between wild-type and FGF2−/− mice, the theoretical differences between wild-type and FGF1−/− mice, as well as between FGF2−/− and double-knockout mice, must be smaller than those between wild-type and FGF2−/− mice. The possibility of significant, but smaller, defects arising from FGF1 deletion nevertheless remains.
Finally, a more trivial explanation, but one that cannot be ignored, is that differences due to the absence of FGF1 and FGF2 are present and detectable, but not in the tissues we studied. If this were the case, then we would anticipate that analysis of other organ systems or additional physiological processes might yield differences that we have thus far been unable to detect. We believe that future studies, particularly those utilizing cultured cells derived from these animals, may reveal roles for FGF1 and FGF2 that are real, even if they are partially redundant with roles of other growth factors. For example, we have found that osteoblasts derived from FGF1-FGF2 null mice undergo premature differentiation in culture (unpublished results). This is in line with the finding that FGF signaling inhibits osteoblast differentiation (10) and suggests that FGF2 (and possibly FGF1) production by osteoblasts plays a role in regulating this process. Similarly, although FGF1-FGF2 null mice exhibit no significant hematopoietic defects, the maturation and differentiation of their hematopoietic stem cells in culture are impaired. Thus, we believe that these mice and their tissues will be useful in the assessment of the role of FGF1 and FGF2 signaling in a variety of physiological and pathological processes.
What is clear from the results obtained so far, however, is that FGF1 and FGF2 do not seem to play critical roles in development and homeostasis. Animals lacking either or both factors develop and breed normally and are largely identical to their wild-type counterparts. Important roles for FGF1 and FGF2 in a wide range of biological processes have been postulated based on a wealth of experimental evidence, including cell culture studies in vitro and transgenic approaches in vivo. While these studies have identified broad potential activities for these factors, our data suggest rather limited roles for FGF1 and FGF2 which may only be apparent under very specific conditions.
ACKNOWLEDGMENTS
We thank Michael Ittmann and Magdalena Sastre for helpful discussions and Earl Nonon for help in the wound healing experiments, as well as the personnel of the NYU Medical Center Transgenic/ES Cell Chimera Facility.
This investigation was supported by Public Health Service grant CA42568 from the National Cancer Institute.
REFERENCES
- 1.Arman E, Haffner-Krausz R, Chen Y, Heath J K, Lonai P. Targeted disruption of fibroblast growth factor (FGF) receptor 2 suggests a role for FGF signaling in pregastrulation mammalian development. Proc Natl Acad Sci USA. 1998;95:5082–5087. doi: 10.1073/pnas.95.9.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnea A, Cho G. Basic fibroblast growth factor selectively amplifies the functional state of neurons producing neuropeptide Y but not somatostatin in cultures of fetal brain cells: evidence for a cooperative interaction with insulin-like growth factor-I. Endocrinology. 1993;133:1895–1898. doi: 10.1210/endo.133.4.8104779. [DOI] [PubMed] [Google Scholar]
- 3.Basilico C, Moscatelli D. The FGF family of growth factors and oncogenes. Adv Cancer Res. 1992;59:115–165. doi: 10.1016/s0065-230x(08)60305-x. [DOI] [PubMed] [Google Scholar]
- 4.Bennett N T, Schultz G S. Growth factors and wound healing: biochemical properties of growth factors and their receptors. Am J Surg. 1993;165:728–737. doi: 10.1016/s0002-9610(05)80797-4. [DOI] [PubMed] [Google Scholar]
- 5.Bennett N T, Schultz G S. Growth factors and wound healing. II. Roles in normal and chronic wound healing. Am J Surg. 1993;166:74–81. doi: 10.1016/s0002-9610(05)80589-6. [DOI] [PubMed] [Google Scholar]
- 6.Bikfalvi A, Klein S, Pintucci G, Rifkin D B. Biological roles of fibroblast growth factor-2. Endocr Rev. 1997;18:26–45. doi: 10.1210/edrv.18.1.0292. [DOI] [PubMed] [Google Scholar]
- 7.Bugler B, Amalric F, Prats H. Alternative initiation of translation determines cytoplasmic or nuclear localization of basic fibroblast growth factor. Mol Cell Biol. 1991;11:573–577. doi: 10.1128/mcb.11.1.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burgess W H, Maciag T. The heparin-binding (fibroblast) growth factor family of proteins. Annu Rev Biochem. 1989;58:575–606. doi: 10.1146/annurev.bi.58.070189.003043. [DOI] [PubMed] [Google Scholar]
- 9.Colvin J S, Bohne B A, Harding G W, McEwen D G, Ornitz D M. Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nat Genet. 1996;12:390–397. doi: 10.1038/ng0496-390. [DOI] [PubMed] [Google Scholar]
- 10.Debiais F, Hott M, Graulet A M, Marie P J. The effects of fibroblast growth factor-2 on human neonatal calvaria osteoblastic cells are differentiation stage specific. J Bone Miner Res. 1998;13:645–654. doi: 10.1359/jbmr.1998.13.4.645. [DOI] [PubMed] [Google Scholar]
- 11.Deng C, Wynshaw-Boris A, Zhou F, Kuo A, Leder P. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell. 1996;84:911–921. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- 12.Deng C X, Wynshaw-Boris A, Shen M M, Daugherty C, Ornitz D M, Leder P. Murine FGFR-1 is required for early postimplantation growth and axial organization. Genes Dev. 1994;8:3045–3057. doi: 10.1101/gad.8.24.3045. [DOI] [PubMed] [Google Scholar]
- 13.Dexter T M, Testa N G. In vitro methods in haemopoiesis and lymphopoiesis. J Immunol Methods. 1980;38:177–190. doi: 10.1016/0022-1759(80)90266-5. [DOI] [PubMed] [Google Scholar]
- 14.Dono R, Texido G, Dussel R, Ehmke H, Zeller R. Impaired cerebral cortex development and blood pressure regulation in FGF-2-deficient mice. EMBO J. 1998;17:4213–4225. doi: 10.1093/emboj/17.15.4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eckenstein F P. Fibroblast growth factors in the nervous system. J Neurobiol. 1994;25:1467–1480. doi: 10.1002/neu.480251112. [DOI] [PubMed] [Google Scholar]
- 16.Feldman B, Poueymirou W, Papaioannou V E, DeChiara T M, Goldfarb M. Requirement of FGF-4 for postimplantation mouse development. Science. 1995;267:246–249. doi: 10.1126/science.7809630. [DOI] [PubMed] [Google Scholar]
- 17.Floss T, Arnold H H, Braun T. A role for FGF-6 in skeletal muscle regeneration. Genes Dev. 1997;11:2040–2051. doi: 10.1101/gad.11.16.2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gabrilove J L, Wong G, Bollenbacher E, White K, Kojima S, Wilson E L. Basic fibroblast growth factor counteracts the suppressive effect of transforming growth factor-β1 on human myeloid progenitor cells. Blood. 1993;81:909–915. [PubMed] [Google Scholar]
- 19.Galzie Z, Kinsella A R, Smith J A. Fibroblast growth factors and their receptors. Biochem Cell Biol. 1997;75:669–685. [PubMed] [Google Scholar]
- 20.Ghosh A, Greenberg M E. Distinct roles for bFGF and NT-3 in the regulation of cortical neurogenesis. Neuron. 1995;15:89–103. doi: 10.1016/0896-6273(95)90067-5. [DOI] [PubMed] [Google Scholar]
- 21.Goldfarb M. Functions of fibroblast growth factors in vertebrate development. Cytokine Growth Factor Rev. 1996;7:311–325. doi: 10.1016/s1359-6101(96)00039-1. [DOI] [PubMed] [Google Scholar]
- 22.Guo L, Degenstein L, Fuchs E. Keratinocyte growth factor is required for hair development but not for wound healing. Genes Dev. 1996;10:165–175. doi: 10.1101/gad.10.2.165. [DOI] [PubMed] [Google Scholar]
- 23.Hebert J M, Rosenquist T, Gotz J, Martin G R. FGF5 as a regulator of the hair growth cycle: evidence from targeted and spontaneous mutations. Cell. 1994;78:1017–1025. doi: 10.1016/0092-8674(94)90276-3. [DOI] [PubMed] [Google Scholar]
- 24.Imamura T, Engleka K, Zhan X, Tokita Y, Forough R, Roeder D, Jackson A, Maier J A M, Hla T, Maciag T. Recovery of mitogenic activity of a growth factor mutant with a nuclear translocation sequence. Science. 1990;249:1567–1570. doi: 10.1126/science.1699274. [DOI] [PubMed] [Google Scholar]
- 25.Kniss D A, Burry R W. Serum and fibroblast growth factor stimulate quiescent astrocytes to re-enter the cell cycle. Brain Res. 1988;439:281–288. doi: 10.1016/0006-8993(88)91485-0. [DOI] [PubMed] [Google Scholar]
- 26.Madia F, Hackshaw K V, Chiu I-M. Cloning and characterization of the mouse Fgf-1 gene. Gene. 1996;179:231–236. doi: 10.1016/s0378-1119(96)00365-4. [DOI] [PubMed] [Google Scholar]
- 27.Mansour S L, Goddard J M, Capecchi M R. Mice homozygous for a targeted disruption of the proto-oncogene int-2 have developmental defects in the tail and inner ear. Development. 1993;117:13–28. doi: 10.1242/dev.117.1.13. [DOI] [PubMed] [Google Scholar]
- 28.Matsuda S, Saito H, Nishiyama N. Effect of basic fibroblast growth factor on neurons cultured from various regions of postnatal rat brain. Brain Res. 1990;520:310–316. doi: 10.1016/0006-8993(90)91720-2. [DOI] [PubMed] [Google Scholar]
- 29.Mellin T N, Cashen D E, Ronan J J, Murphy B S, DiSalvo J, Thomas K A. Acidic fibroblast growth factor accelerates dermal wound healing in diabetic mice. Investig Dermatol. 1995;104:850–855. doi: 10.1111/1523-1747.ep12607026. [DOI] [PubMed] [Google Scholar]
- 30.Meyers E N, Lewandoski M, Martin G R. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat Genet. 1998;18:136–141. doi: 10.1038/ng0298-136. [DOI] [PubMed] [Google Scholar]
- 31.Min H, Danilenko D M, Scully S A, Bolon B, Ring B D, Tarpley J E, DeRose M, Simonet W S. Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes Dev. 1998;12:3156–3161. doi: 10.1101/gad.12.20.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okumura M, Okuda T, Nakamura T, Yajima M. Acceleration of wound healing in diabetic mice by basic fibroblast growth factor. Biol Pharm Bull. 1996;19:530–535. doi: 10.1248/bpb.19.530. [DOI] [PubMed] [Google Scholar]
- 33.Ornitz D M, Xu J, Colvin J S, McEwen D G, MacArthur C A, Coulier F, Gao G, Goldfarb M. Receptor specificity of the fibroblast growth factor family. J Biol Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- 34.Ortega S, Ittmann M, Tsang S H, Ehrlich M, Basilico C. Neuronal defects and delayed wound healing in mice lacking fibroblast growth factor 2. Proc Natl Acad Sci USA. 1998;95:5672–5677. doi: 10.1073/pnas.95.10.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qian X, Davis A A, Goderie S K, Temple S. FGF2 concentration regulates the generation of neurons and glia from multipotent cortical stem cells. Neuron. 1997;18:81–93. doi: 10.1016/s0896-6273(01)80048-9. [DOI] [PubMed] [Google Scholar]
- 36.Quito F L, Beh J, Bashayan O, Basilico C, Basch R S. Effects of fibroblast growth factor-4 (k-FGF) on long-term cultures of human bone marrow cells. Blood. 1996;87:1282–1291. [PubMed] [Google Scholar]
- 37.Smallwood P M, Munoz-Sanjuan I, Tong P, Macke J P, Hendry S H, Gilbert D J, Copeland N G, Jenkins N A, Nathans J. Fibroblast growth factor (FGF) homologous factors: new members of the FGF family implicated in nervous system development. Proc Natl Acad Sci USA. 1996;93:9850–9857. doi: 10.1073/pnas.93.18.9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Temple S, Qian bFGF, neurotrophins, and the control or cortical neurogenesis. Neuron. 1995;15:249–252. doi: 10.1016/0896-6273(95)90030-6. [DOI] [PubMed] [Google Scholar]
- 39.Vaccarino F M, Schwartz M L, Raballo R, Nilsen J, Rhee J, Zhou M, Doetschman T, Coffin J D, Wyland J J, Hung Y T. Changes in cerebral cortex size are governed by fibroblast growth factor during embryogeneisis. Nat Neurosci. 1999;2:246–253. doi: 10.1038/6350. [DOI] [PubMed] [Google Scholar]
- 40.Xu X, Weinstein M, Li C, Naski M, Cohen R I, Ornitz R M, Leder P, Deng C. Fibroblast growth factor receptor 2 (FGFR2)-mediated reciprocal regulation loop between FGF8 and FGF10 is essential for limb induction. Development. 1998;125:753–765. doi: 10.1242/dev.125.4.753. [DOI] [PubMed] [Google Scholar]
- 41.Yamaguchi T P, Harpal K, Henkemeyer M, Rossant J. fgfr-1 is required for embryonic growth and mesodermal patterning during mouse gastrulation. Genes Dev. 1994;8:3032–3044. doi: 10.1101/gad.8.24.3032. [DOI] [PubMed] [Google Scholar]
- 42.Zhou M, Sutliff R L, Paul R J, Lorenz J N, Hoying J B, Haudenschild C C, Yin M, Coffin J D, Kong L, Kranias E G, Luo W, Boivin G P, Duffy J J, Pawlowski S A, Doetschman T. Fibroblast growth factor 2 control of vascular tone. Nat Med. 1998;4:201–207. doi: 10.1038/nm0298-201. [DOI] [PMC free article] [PubMed] [Google Scholar]