Abstract
The EBNA1 protein of Epstein-Barr virus (EBV) governs the replication and segregation of the viral episomes in latently infected cells and transactivates the expression of other EBV latency proteins through direct interactions with DNA sequences in the EBV latent origin of replication, oriP. To better understand how EBNA1 controls these processes, we have assessed the contribution of various EBNA1 sequences to its replication, segregation, and transactivation functions. Here we show that EBNA1 residues 325 to 376 are responsible for the transactivation activity of EBNA1. This region coincides with the DNA looping domain previously shown to mediate interactions at a distance between DNA-bound EBNA1 molecules. The same residues mediate DNA segregation but have no apparent role in DNA replication, indicating that the replication and transcription activation activities of EBNA1 are distinct. The acidic C-terminal tail of EBNA1 was not found to contribute to replication, transactivation, or segregation. We have also investigated the functional significance of two structural motifs within the DNA binding and dimerization domains of EBNA1, the proline loop and the WF motif. Although the amino acids in these motifs do not directly contact the DNA, both of these motifs were found to contribute to EBNA1 functions by increasing the DNA-binding ability of EBNA1. Mechanisms by which DNA binding is stimulated by these motifs are discussed.
Epstein-Barr virus (EBV) is a ubiquitous human gamma herpesvirus that is associated with several diseases and malignancies, including infectious mononucleosis, Burkitt's lymphoma, nasopharyngeal carcinoma, some types of Hodgkin's disease, oral hairy leukoplakia, and several types of lymphomas in immunocompromised hosts (40). EBV infection usually takes a latent form in which as many as six nuclear and three membrane proteins are expressed from the virus; the only viral protein expressed in all cases is Epstein-Barr nuclear antigen 1 (EBNA1). During latent infection, the host cell is induced to proliferate and the EBV genomes are maintained in the cell nucleus at a stable copy number as double-stranded, circular DNA episomes (reviewed in reference 50). These episomes undergo DNA replication once per cell cycle and are efficiently partitioned to the daughter cells (1, 52, 53).
Experiments designed to elucidate the EBV sequences sufficient for the maintenance of plasmids in human cells identified oriP as the viral origin of latent DNA replication and EBNA1 as the only viral protein required (53). oriP contains two functional elements, the dyad symmetry element (DS) and the family of repeats (FR) (40). The DS contains four EBNA1 binding sites and appears to be the initiation site of DNA replication (15, 33, 37, 49). The FR contains 20 EBNA1 binding sites (37) and is involved in three viral processes; it activates DNA replication from the DS, enhances transcription from several promoters, and mediates the partitioning of EBV episomes and oriP plasmids (14, 27, 38, 39). All of these activities require EBNA1 binding to the FR.
EBNA1 plays several roles in latent viral infection. First, EBNA1 activates DNA replication from oriP (19, 51). Since EBNA1 lacks enzymatic activities, origin activation is thought to involve the recruitment of host replication factors to oriP and/or the destabilization of the origin DNA (13, 32). Second, EBNA1 governs the segregation of EBV episomes and FR-containing plasmids by mediating the attachment of the FR to host cell metaphase chromosomes (11, 18, 27, 35). Third, EBNA1, when bound to the FR, transactivates the expression of viral latent gene products (14, 38, 45). Fourth, EBNA1 represses its own expression from the Qp promoter by binding to two recognition sites near this promoter (34, 41).
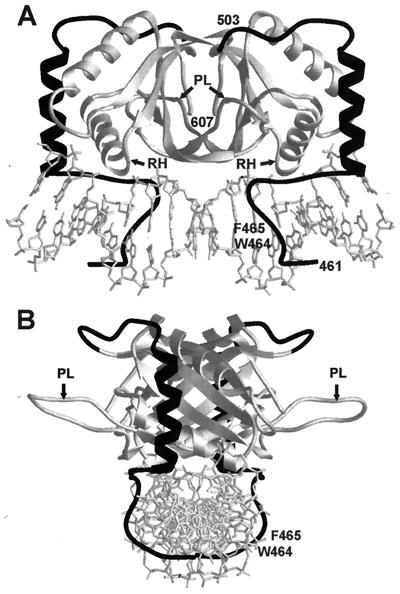
All of the EBNA1 functions require the binding of EBNA1 dimers to 18-bp palindromic recognition sites (3, 37). The EBNA1 amino acids responsible for DNA binding and dimerization colocalize to residues 459 to 607 (2, 10, 46). The crystal structure of this region of EBNA1 bound to DNA, in conjunction with biochemical data, has revealed the mechanism of the EBNA1-DNA interaction (2, 8, 10; J. Cruickshank, K. Shire, A. Davidson, A. M. Edwards, and L. Frappier, submitted for publication). The EBNA1 DNA binding and dimerization region is comprised of two domains, the core and flanking domains (Fig. 1). The core domain (amino acids 504 to 604) contains an eight-stranded antiparallel β-barrel, which forms the dimerization interface, and two α-helices per monomer. One of the helices from each monomer makes transient contacts with the major groove of the DNA, facilitating subsequent DNA interactions by the flanking domain (Cruickshank et al., submitted). The core domain is structurally homologous (root-mean-square deviation, 0.908 Å) to the DNA binding and dimerization domain of the E2 protein of bovine papillomavirus (9, 20). The structures of the two domains differ primarily by the presence of an extended proline-rich loop (termed the proline loop) in the EBNA1 core domain (Fig. 1); this loop in E2 is 9 amino acids shorter and contains no prolines. Both the exposed positioning and the proline-rich sequence of this loop suggested that the proline loop might mediate protein interactions. The flanking domain comprises an α-helix, oriented perpendicular to the DNA, and an extended chain which tunnels along the base of the minor groove of the DNA. This domain plays an important role in DNA binding; four amino acids from the flanking domain (K461, G463, R469, and K477) make a total of seven base contacts (8). The minor groove-extended chain portion of the flanking domain contains a peculiar arrangement of tryptophan and phenylalanine side chains (amino acids 464 and 465, respectively; termed the WF motif) that appears to widen the minor groove of the DNA (8).
FIG. 1.
Crystal structure of the EBNA1 DNA binding and dimerization domains bound to DNA. The structures of the EBNA core (light shading) and flanking (dark shading) domains are shown when bound to DNA, as determined by Bochkarev et al. (8). The positions of the proline loops (PL), recognition helices (RH), and WF residues (W464 and F465) are indicated. (A) View perpendicular to the DNA axis. (B) View down the axis of the DNA.
While the role of EBNA1 residues in DNA binding is reasonably well understood, the functional contribution of other regions of EBNA1 are less well defined. EBNA1 contains several unusual sequence elements (see Fig. 2A). A Gly-Ala repeat, which encompasses amino acids 101 to 325, is not required for the replication, transactivation, or segregation functions of EBNA1 but appears to enable EBNA1 to evade cytotoxic T-cell responses (7, 26). Two Gly-Arg-rich regions are present between residues 40 and 55 and residues 325 and 376. The latter region corresponds to the DNA looping or linking domain which has been shown to mediate homotypic interactions at a distance between DNA-bound EBNA1 molecules (5, 16, 24, 28), as well as heterotypic interactions with at least two cellular proteins (42, 48). This region has also been reported to bind RNA (43). The looping domain is followed by a basic nuclear localization sequence (residues 379 to 386) (2). The extreme C terminus of EBNA1 (amino acids 619 to 641) is highly acidic; this acidic tail has been reported to play roles both in transactivation (2) and in oriP plasmid maintenance (51), but assignment of its functional role has not been conclusive (23, 36, 51).
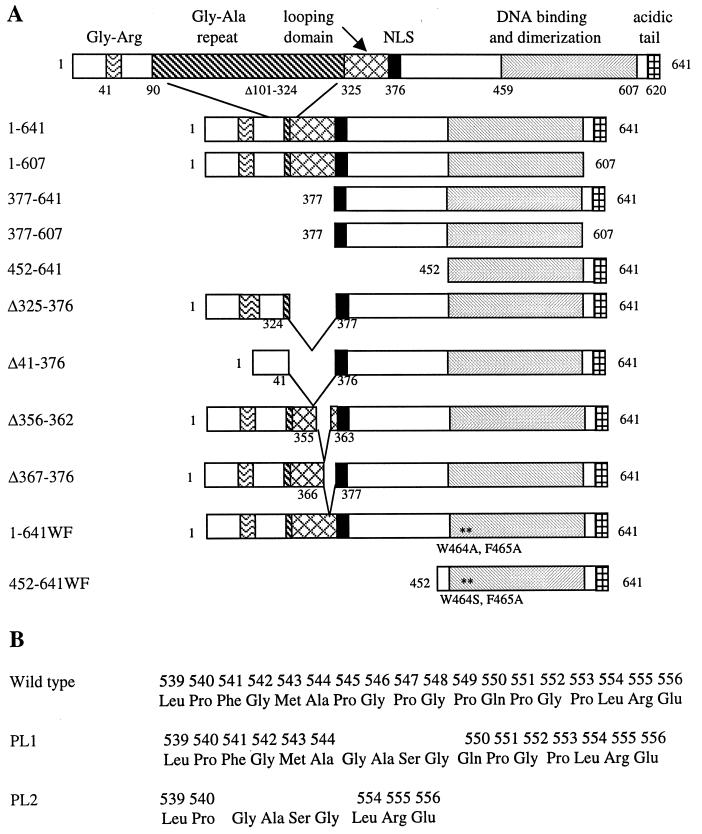
FIG. 2.
EBNA1 mutants. (A) Schematic representation of the mutants, showing pertinent regions of the protein, including the nuclear localization signal (NLS). (B) Sequences of the proline loop region in wild-type EBNA1 and the PL1 and PL2 mutants.
In this study, we have investigated the contribution of four regions of EBNA1 to the DNA replication, segregation, and transcription activation functions of the protein, namely, the looping domain, the acidic tail, and the proline loop and WF motif of the DNA-binding region.
MATERIALS AND METHODS
Escherichia coli expression constructs.
The construction of the pET15b vector expressing the EBNAWF mutant was described by Summers et al. (47). EBNA1 mutants in which amino acids 545 to 549 (PL1) and 541 to 553 (PL2) were replaced with Gly-Ala-Ser-Gly were constructed by two rounds of PCR amplification with p205 as a template (53). In the first round of PCR, two EBNA1 fragments, extending from the N terminus to the deletion site and from the deletion site to the C terminus, were amplified. Primers adjacent to the deletion site on each fragment contained NheI restriction sites at their 5′ ends, while primers that hybridized to the N and C termini of EBNA1 contained NdeI and BamHI sites, respectively. The two EBNA1 fragments were purified from agarose gels, digested with NheI, and ligated. Ligation products of the appropriate length were purified from an agarose gel and subjected to a second round of PCR with the EBNA1 N- and C-terminal primers. PCR products were gel purified, digested with NdeI and BamHI, and ligated between the NdeI and BamHI sites in pET15b (Novagen). The sequence of each clone was confirmed by DNA sequencing (MOBIX, McMaster University). To generate constructs that expressed the PL1 and PL2 substitutions in the context of amino acids 452 to 641, EBNA1 residues 452 to 641 were PCR amplified from the EBNA1 constructs described above containing the proline loop mutations. The primers used placed an NdeI site at the N terminus and a BamHI site at the C terminus of the amplified fragment. The amplified fragments were digested with NdeI and BamHI and cloned between the NdeI and BamHI sites of pET15b. The resulting constructs expressed the EBNA1 mutants as N-terminal fusions to a hexahistidine tag and thrombin protease site.
Mammalian expression constructs.
To construct the plasmids used for mammalian cell transfections, EBNA1-encoding fragments were cloned downstream of a cytomegalovirus (CMV) promoter in pcDNA3 (Invitrogen, Carlsbad, Calif.). DNA fragments encoding the wild-type protein (EBNA1–641) and EBNA1 mutants lacking amino acids 608 to 641 (EBNA1–607), 1 to 451 (EBNA452–641), 1 to 376 (EBNA377–641), or both 1 to 376 and 608 to 641 (EBNA377–607) were generated by PCR amplification of the EBNA1 gene in p205 (53) with an N-terminal primer containing either an NdeI or an NcoI site and a C-terminal primer encoding a BamHI site. The resulting EBNA1 genes lacked most of the nonessential Gly-Ala repeat region of EBNA1 in addition to the mutations listed above. The DNA fragments were digested with NdeI or NcoI, filled in with the Klenow fragment of DNA polymerase I, and then digested with BamHI. DNA fragments encoding the PL1 and PL2 mutations in the context of EBNA1 amino acids 1 to 641 (EBNA1–641PL1 and EBNA1–641PL2, respectively) were subcloned from the pET15b constructs described above by digesting with NdeI, filling in the 5′ overhang with Klenow, and then digesting with BamHI. The EBNA1–641WF mutant, in which amino acids W464 and F465 were both replaced with alanine, was constructed by two rounds of PCR amplification. In the first round, two EBNA1 fragments encoding amino acids 1 to 463 and 464 to 641 were amplified from p205 with a primer that encoded alanines at positions 464 and 465. These DNA fragments were ligated, and the ligation products were used as a template to amplify codons 1 to 641 with a C-terminal primer containing a BamHI site. The EBNAWF fragment was then digested with BamHI. All of the resulting EBNA1 fragments were cloned between the HindIII (filled in with DNA polymerase Klenow fragment) and BamHI sites of pcDNA3 (Invitrogen), downstream of the CMV promoter, and the EBNA1 genes in the resulting constructs were sequenced. The pcDNA3/EBNA1 plasmids were then modified by the addition of EBV oriP DNA sequences. A DNA fragment encoding oriP was excised from pGEMoriP (13) by digestion with BamHI and RsaI and inserted between the BglII and NruI sites of each pcDNA3/EBNA1 construct to generate pc3oriPE (where E is EBNA1 or an EBNA1 mutant). pc3oriPE plasmids expressing EBNA1 mutants Δ325–376, Δ41–376, Δ367–376, and Δ356–362 were similarly constructed as described by Shire et al. (42). pc3oriP was constructed by inserting oriP between the BglII and NruI sites of pcDNA3.
Purification of EBNA1 mutants.
EBNA452–641 and EBNA452–641WF were expressed in E. coli from pET15b constructs and purified as described by Barwell et al. (6) and Summers et al. (47), respectively. EBNA452–641PL1 and EBNA452–641PL2 were expressed from pET15b constructs in E. coli BL21(DE3) (44). Transformed cells were grown at 37°C in 2 liters of Luria-Bertani (LB) containing 100 μg of ampicillin per ml to an optical density at 600 nm of 0.60. Expression of the EBNA1 mutants was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to 1 mM, and cells were harvested 3 h postinduction. Cell pellets were rinsed in 20 mM Tris-HCl (pH 7.5)–10% sucrose and then frozen at −70°C. For protein purification, cells were thawed in 20 ml of 20 mM Tris-HCl (pH 8)–500 mM NaCl–10% glycerol–1 mM benzamidine–1 mM phenylmethylsulfonyl fluoride (PMSF)–1 mM EDTA, and the cells were lysed by sonication on ice. The lysate was clarified by centrifugation at 25,000 rpm in an SW28 rotor (Beckman) for 30 min and then passed through a 25-ml DE52 (Whatman) column equilibrated with 50 mM HEPES (pH 7.5)–350 mM NaCl–1 mM dithiothreitol (DTT)–10% glycerol. The DE52 flowthrough was diluted to a final NaCl concentration of 200 mM with buffer A (50 mM HEPES [pH 7.5], 1 mM DTT, 1 mM PMSF, 1 mM benzamadine, 10% glycerol) and applied to a 10-ml heparin-agarose column (Bio-Rad) equilibrated with buffer A plus 200 mM NaCl. The column was washed with buffer A containing 200 mM NaCl and developed with a 75-ml linear salt gradient from 200 mM to 1 M NaCl in buffer A. The fractions containing EBNA1 protein were pooled, dialyzed overnight in buffer B (50 mM HEPES [pH 7.5], 750 mM NaCl, 10% glycerol), and loaded onto a 5-ml high-pressure liquid chromatography (HPLC) metal-chelating column (PerSeptive Biosystems) that had been charged with nickel and equilibrated in buffer B plus 5 mM imidazole. The column was washed with 25 ml of buffer B containing 5 mM imidazole and then with 25 ml of buffer B containing 50 mM imidazole. EBNA1 proteins were eluted at 1 ml/min with buffer B containing 300 mM imidazole, and DTT was immediately added to the eluates to a final concentration of 10 mM. The fractions containing EBNA1 were pooled and dialyzed in buffer B plus 1 mM DTT. The histidine tag was removed from the EBNA1 protein by incubation with thrombin (1 μg/mg of protein) for 2 h at 4°C in the presence of 2.5 mM CaCl2. The removal of the histidine tag was confirmed by increased mobility on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The digested protein was diluted with buffer A to a final NaCl concentration of 200 mM and loaded onto a 1-ml HPLC Mono S column (PerSeptive Biosystems) equilibrated with buffer A plus 200 mM NaCl. The EBNA1 protein was eluted at 0.5 ml/min with buffer A containing 1 M NaCl and determined to be approximately 90% pure when examined by SDS-PAGE and Coomassie staining. EBNA1-containing fractions were aliquoted and stored at −70°C.
EMSAs.
The EBNA1 binding site used in electrophoretic mobility shift assays (EMSAs) corresponds to site 1 of the DS element and was generated from the 20-bp oligomers 5′-CGGGAAGCATATGCTACCCG-3′ and 5′-CGGGTAGCATATGCTTCCCG-3′. To generate end-labeled site 1, one of the oligonucleotides was end labeled with [γ-32P]ATP and then hybridized to a twofold molar excess of the second oligonucleotide. For DNA binding reactions, purified EBNA1 proteins were titrated onto 10 fmol of radiolabeled site 1 in a 20-μl reaction containing 10 mM HEPES (pH 7.5), 5 mM MgCl2, 300 mM NaCl, and 1 μg of herring sperm DNA. Following a 20-min incubation at room temperature, 4 μl of 30% glycerol was added, and the sample run on a native 12% polyacrylamide gel in 0.5× TBE (45 mM Tris, 45 mM boric acid, 1 mM EDTA [pH 8]). Labeled DNA was visualized by autoradiography of dried gels and quantified by PhosphorImager analysis using ImageQuant software (Molecular Dynamics).
Transcription activation assays.
C33A human cervical carcinoma cells were plated in 60-mm dishes at a density of 106 cells/dish and grown for 24 h prior to transfection by the calcium phosphate coprecipitation method (17). Five micrograms of pcDNA3 plasmids encoding EBNA1 or EBNA1 mutants was combined with 2 μg of the pFRTKCAT reporter construct (a gift from Bill Sugden) (51) and 2.5 μg of herring sperm DNA in 0.25 ml of 0.25 M CaCl2. The DNA-CaCl2 solution was added dropwise to 0.25 ml of 2× HEPES-buffered saline (HBS; 50 mM HEPES, 280 mM NaCl, 1.5 mM Na2HPO4 [pH 6.95]) with vortexing. After 30 min at room temperature, the solution was added dropwise to cells in 4 ml of Dulbecco's modified Eagle's medium (DMEM), and the cells were incubated with the precipitate for 12 to 16 h at 37°C. The cells were then washed in phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4) and given fresh medium. After a 24-h incubation at 37°C, the cells were harvested. For each sample, half of the harvested cells were used in a Western blot analysis of EBNA1 expression levels, and the other half were processed for chloramphenicol acetyltransferase (CAT) assays (4). Cells for CAT assays were lysed by three rounds of freezing and thawing, and lysates were clarified by centrifugation at 14,000 rpm in a microcentrifuge. Each CAT assay mixture contained 50 μg of protein from the clarified lysates, 0.25 M Tris-HCl (pH 7.5), 0.25 mM acetyl coenzyme A, and 3 pmol of [14C]chloramphenicol (NEN) in a 150-μl reaction. The reaction mixtures were incubated at 37°C, and at various time points, 50-μl aliquots were removed and vortexed with 300 μl of ethyl acetate. After drying, the [14C]chloramphenicol was resuspended in 20 μl of ethyl acetate and spotted onto a cellulose thin-layer chromatography plate (Whatman). The plates were developed in a chloroform-methanol (95:5, vol/vol) mixture. The CAT assay products were visualized by PhosphorImager analysis of the dried plates and quantified using ImageQuant software (Molecular Dynamics). For each mutant, the amount of acetylated product produced at each time point was used to determine the acetylation rate.
Transient-replication assays.
C33A cells were plated in 100-mm dishes at 2.5 × 106 cells/dish and grown for 24 h prior to transfection. Ten micrograms of pc3oriPE constructs, expressing EBNA1 or EBNA1 mutants, was combined with 10 μg of herring sperm DNA and used to transfect the cells. Transfections were performed as described for transcription activation assays except that the volumes of the CaCl2-DNA and HBS solutions were doubled to 0.5 ml. Following removal of the DNA precipitate, the cells were washed in PBS, split into 140-mm dishes, and grown for 72 h. After harvesting, 5 × 106 cells from each plate were lysed in 700 μl of 10 mM Tris-HCl (pH 7.5)–10 mM EDTA (pH 8.0)–0.6% SDS for transient-replication analysis. High-molecular-weight DNA was precipitated in the replication assay samples by the addition of NaCl to 0.83 M and incubation overnight at 4°C (21). Low-molecular-weight DNA in the supernatant was extracted with phenol-chloroform (1:1), ethanol precipitated, and resuspended in Tris-EDTA (TE, pH 8). Half of each sample was linearized with XhoI, and 9/10 of the linearized samples were further digested with DpnI (4 U) for 2 h at 37°C. The remaining 1/10 of the linearized samples were analyzed directly by Southern blot to verify the recovery of the plasmids. DNA fragments from the restriction digests were separated on a 1% agarose gel, transferred to GeneScreen Plus (NEN), and probed with 32P-labeled pc3oriP. Radiolabeled bands were visualized by autoradiography, and linearized plasmid bands were quantified by PhosphorImager analysis using ImageQuant software (Molecular Dynamics).
Plasmid maintenance assays.
C33A cells in 100-mm dishes were transfected with 1 μg of pc3oriPE plasmids (expressing EBNA1 or EBNA1 mutants) combined with 19 μg of herring sperm DNA as described for transient-replication assays. After incubation for 12 to 16 h with the DNA precipitate, cells were washed in PBS, replated into 140-mm dishes, and grown in medium containing G418 (400 μg/ml; Gibco-BRL). Following 2 weeks of selection, 5 × 106 cells from each plate were harvested and lysed by the method of Hirt (21), while the remaining cells were frozen at −70°C for Western blot analysis. Low-molecular-weight DNA was isolated, digested with XhoI and DpnI, and Southern blotted as described for transient-replication assays. Linearized plasmid bands were visualized by autoradiography and quantified by PhosphorImager analysis using ImageQuant software (Molecular Dynamics).
Western blot analysis.
Frozen cell pellets from transcription activation, transient-replication, and plasmid maintenance assays were suspended in 100 μl of 500 mM NaCl–20 mM Tris-HCl (pH 8)–0.1% Triton–0.5 mM EDTA–1 mM PMSF–1 mM benzamadine and sonicated. The cellular debris was pelleted by centrifugation, and lysate supernatants equivalent to 30 μg of protein were separated by electrophoresis on SDS–12% polyacrylamide gels. The proteins were transferred either to a nitrocellulose (NC) filter (MilliPore) or to a polyvinylidene fluoride (PVDF) membrane (Gelman Science). After blocking, the membranes were incubated with K67 rabbit polyclonal antibody raised against EBNA452–641 (kindly provided by Jaap Middledorp), followed by secondary antibody conjugated to either peroxidase (for NC blots) or alkaline phosphatase (for PVDF blots; Kirkegaard and Perry Laboratories). The NC and PVDF blots were developed for enhanced chemiluminescence (NEN Inc.) and quantitative enhanced chemifluorescence (Amersham Inc.), respectively, by the methods of the manufacturers. Enhanced chemifluorescence-reactive bands were quantified using a Storm 860 scanner and ImageQuant software (Molecular Dynamics).
Analysis of protein folding and stability.
Circular dichroism (CD) spectroscopy was used to compare the secondary structure of EBNA452–641, EBNA452–641PL1, EBNA452–641PL2, and EBNA452–641WF. A 10 μM solution of each protein was brought to a final volume of 200 μl with PBS and scanned in a 0.1-cm cuvette using an Aviv 62A DS CD spectrometer. Samples were scanned in 1-nm steps from 300 to 200 nm at 25°C with a 1-s averaging time. The average elipticity values of five scans conducted on each protein was subtracted from that of a buffer-only scan and plotted. For protein stability studies, concentrated protein samples in 300 mM NaCl–10 mM HEPES (pH 7.5)–5 mM MgCl2 were rapidly diluted into GuHCl buffer (7.2 M guanidine hydrochloride [GuHCl], 300 mM NaCl, 10 mM HEPES [pH 7.5], 5 mM MgCl2) at 25°C to a final GuHCl concentration of 5.0 to 6.6 M. Unfolding was monitored by loss of the CD signal over time at 222 nm. The raw data were normalized and fit to the equation y = 1 − exp(−kt) using Excel (Microsoft Corp.), where t is the time in seconds, y is the fraction of the protein folded, and k is the unfolding-rate constant.
RESULTS
To further understand the mechanisms by which EBNA1 functions in DNA replication, transcriptional activation, and DNA segregation, we examined the contribution of four EBNA1 elements to these processes, namely, the looping domain, the acidic tail, the WF motif, and the proline loop. For these studies, plasmids were constructed that contained oriP and a selectable neomycin resistance marker and expressed EBNA1 or an EBNA1 mutant from a CMV promoter. These constructs were used to transfect human C33A cells. The version of EBNA1 used as the wild type in these studies is one that lacks most of the Gly-Ala repeat region (Fig. 2) and has been previously shown to be functional for replication, segregation, and transcription activation activities (51, 52). To assess the replication activity of the EBNA1 mutants, transfected plasmids were recovered 3 days posttransfection and linearized and digested with DpnI. DNA replication was quantified by comparing the amount of DpnI-resistant plasmid recovered for each EBNA1 mutant with that recovered for constructs expressing wild-type EBNA1. The presence of plasmids in each sample was verified by Southern blot analysis of the linearized recovered plasmids prior to DpnI digestion. The same constructs were also used in experiments to test the long-term maintenance of the plasmids, a phenomenon that requires both DNA replication and stable segregation to the daughter cells. For these experiments, transfected cells were grown under selection for 2 weeks, and then equal numbers of cells were lysed and analyzed for the presence of unintegrated plasmid by Southern blot. The amount of plasmid DNA recovered was quantified and compared with that recovered when wild-type EBNA1 was expressed. For transcription activation assays, C33A cells were cotransfected with the EBNA1 expression constructs described above and a reporter construct which contained the CAT gene under control of the oriP FR element. Cell lysates were prepared 24 h posttransfection, and the acetylation rates were determined for each lysate using equal amounts of protein. After subtraction of background acetylation rates obtained for cell lysates lacking EBNA1, the acetylation rate for each mutant was expressed as a percentage of the acetylation rates observed for constructs expressing wild-type EBNA1.
For all experiments, Western blots were also performed to verify the expression of the EBNA1 proteins. The expression of each mutant relative to that of EBNA1–641 was not observed to vary significantly (less than twofold) from one experiment to the next. Since the transient-replication, transactivation, and plasmid maintenance assays all utilize the same EBNA1 expression constructs, the protein levels shown in Fig. 3 are representative of the expression achieved in all three assays at 24 h posttransfection. Although the transient-replication and plasmid maintenance assays involved longer transfection times, it is the initial expression levels of the EBNA1 proteins that are most relevant. Since the replication and partitioning of the EBNA1 expression construct requires functional EBNA1, at longer posttransfection times, the reduced copy number of constructs expressing nonfunctional EBNA1 mutants will result in lower EBNA1 levels per cell compared with functional EBNA1 counterparts.
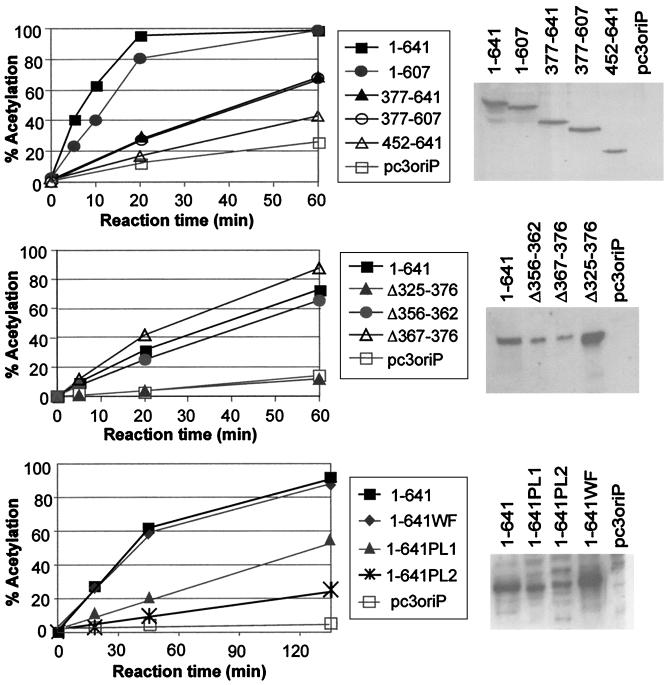
FIG. 3.
Transactivation of the CAT reporter gene by EBNA1 mutants. Constructs expressing the EBNA1 mutants indicated or no EBNA1 (pc3oriP) were cotransfected with a CAT reporter construct in which the CAT gene is under control of the oriP FR element. At 24 h posttransfection, transfected cells were lysed, and equal amounts of lysate were assayed for chloramphenicol acetylation rates. The percentage of chloramphenicol that is acetylated after various reaction times is shown on the left. To the right of each graph is a Western blot showing the expression of each EBNA1 mutant at 24 h posttransfection.
Acidic tail.
The extreme C terminus of EBNA1, amino acids 620 to 641, is highly acidic; 13 of 21 residues are aspartic or glutamic acid. The similarity of this region to acidic transactivation domains has led to the suggestion that these EBNA1 residues mediate the transcription activation activity of EBNA1. To examine this possibility, we generated an EBNA1 C-terminal truncation mutant lacking residues 608 to 641 (EBNA1–607) (Fig. 2) and compared its ability to transactivate expression of the CAT reporter with that of wild-type EBNA1 (Fig. 3A). We repeatedly found no difference in the transactivation activity of EBNA1 and EBNA1–607 (Table 1). To determine whether the acidic tail might be a transactivation domain that is redundant in the context of full-length EBNA1, we also examined the transactivation ability of an EBNA1 N-terminal truncation mutant containing only the DNA binding and dimerization domain and acidic tail (EBNA452–641) and a larger N-terminal truncation mutant spanning residues 377 to 641 (EBNA377–641). As shown in Fig. 3A and Table 1, the ability of these EBNA1 mutants to activate transcription was much lower than that observed for the wild-type protein. A small amount of activation (13% of wild-type activation) was consistently observed with EBNA377–641, but this was not mediated by the acidic tail, as its removal in EBNA377–607 did not abrogate this activation (Fig. 3A and Table 1). The low level of activity of EBNA452–641, EBNA377–641, and EBNA377–607 was not due to insufficient expression of these proteins, as all three were expressed at similar levels as wild-type EBNA1 (Fig. 3A) and at levels higher than at least one functional EBNA1 mutant (Δ367–376) (Fig. 3B). Also, EMSAs performed with extracts of the transfected cells and an EBNA1 recognition site, showed that the expression of these EBNA1 mutants resulted in the same amount of sequence-specific DNA binding activity as when wild-type EBNA1 was expressed (data not shown). Our results therefore indicate that the acidic tail is not a transactivation domain and that the major transactivation activity lies between EBNA1 residues 1 and 376.
TABLE 1.
Summary of functional activities of EBNA1 mutantsa
| EBNA1 protein | % of wild-type activity ± SD (n)
|
||
|---|---|---|---|
| Transactivation | Transient replication | Plasmid maintenance | |
| 1–641 | 100 (13) | 100 (9) | 100 (9) |
| 1–607 | 89 ± 14 (5) | 58 ± 23 (4) | 134 ± 63 (5) |
| 377–641 | 13 ± 7 (5) | 34 ± 26 (5) | 0 ± 0 (4) |
| 377–607 | 9 ± 4 (4) | NDc | ND |
| 452–641 | 4 ± 3.5 (4) | ND | ND |
| Δ325–376 | 2.5 ± 0.8 (4) | 95 ± 45b (4) | 1.6 ± 1.5b (3) |
| Δ41–376 | 6 ± 5 (4) | 47 ± 14b (4) | 0 ± 0b (3) |
| Δ356–362 | 91 ± 31 (6) | ND | 71 ± 32b (3) |
| Δ367–376 | 125 ± 13 (6) | ND | 98 ± 27b (3) |
| 1–641WF | 90 ± 22 (6) | 120 ± 57 (7) | 173 ± 109 (4) |
| 1–641PL1 | 66 ± 35 (10) | 58 ± 48 (6) | 159 ± 43 (6) |
| 1–641PL2 | 26 ± 9 (12) | 13 ± 8 (5) | 0 ± 0 (4) |
| None | N/Ad | 5 ± 4 (9) | 0 ± 0 (9) |
The transcriptional activation, transient-replication, and plasmid maintenance activities are shown as a percentage of the activity observed for EBNA1–641 in that experiment, followed by the standard deviation. The number of experiments (n) performed to generate the activities is shown in parentheses. In the transient-replication assays, wild-type EBNA1 constructs yielded 1.0 × 107 to 1.5 × 107 DpnI-resistant plasmids, as determined by comparison with a 100-pg plasmid marker. In the plasmid maintenance assays, the amount of plasmid recovered with wild-type EBNA1 varied between 1.5 and 4.5 copies/cell in different experiments and on average was 2.5 copies/cell.
Data from Shire et al. (42).
ND, not determined.
N/A, background acetylation seen in the absence of EBNA1 has already been subtracted from the above results (on average, 5% of 1–641 activity).
We also addressed whether the acidic tail plays a role in the DNA replication and segregation functions of EBNA1 by comparing the transient replication and long-term maintenance of the oriP plasmids expressing EBNA1–607 to that of oriP plasmids expressing wild-type EBNA1. EBNA1–607 was found to support both DNA replication (Fig. 4) and plasmid maintenance (Fig. 5). Although the transient-replication efficiency of EBNA1–607 was somewhat less than that of EBNA1, this decrease does not appear to be significant, as the plasmid maintenance activity of EBNA1–607 was as high as that of the wild-type protein (Table 1). Therefore, our results indicate that the acidic tail is not required for DNA replication or segregation.
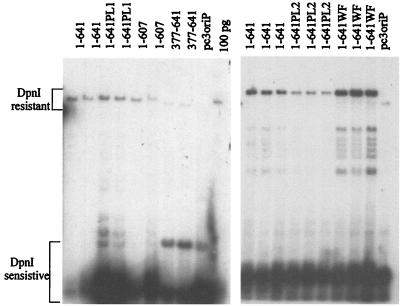
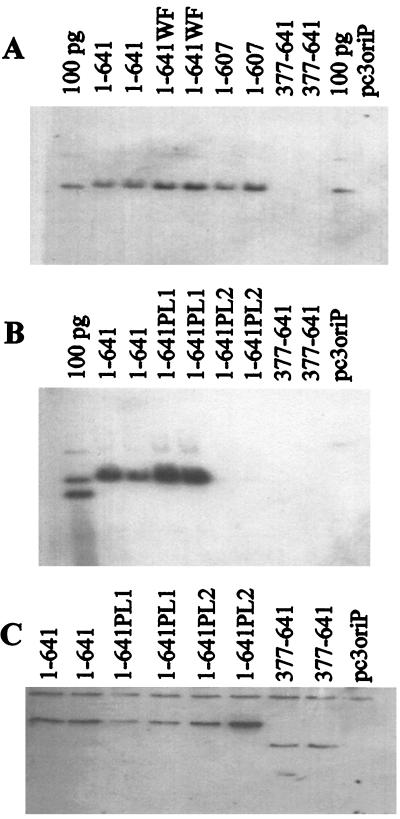
FIG. 4.
Transient DNA replication abilities of EBNA1 mutants. Plasmids containing oriP and expressing the EBNA1 protein indicated or no EBNA1 (pc3oriP) were used to transfect human cells. Three days posttransfection, the plasmids were harvested, linearized, and digested with DpnI to remove plasmid that had not undergone replication in the human cells. After agarose gel electrophoresis, the DpnI-resistant plasmid band for each EBNA1 mutant was detected by Southern blotting, and the intensity of this band was compared with that obtained with wild-type EBNA1 (1–641) to determine replication efficiency. A 100-pg marker for the linearized plasmid expressing EBNA1–641 is also shown (100 pg).
FIG. 5.
Plasmid maintenance activities of EBNA1 mutants. Human cells were transfected with plasmids containing oriP and expressing the EBNA1 protein indicated or no EBNA1 (pc3oriP) and grown under selection for 14 days. Plasmids were then isolated, linearized, digested with DpnI, and separated by agarose gel electrophoresis. (A and B) Southern blots showing the linearized plasmids recovered. Markers of 100 pg of linearized pc3oriPE1–641 (100 pg lane in panel A) or 100 pg each of pc3oriP and pc3oriPE1–641 (100 pg lane in panel B) are also shown. (C) Western blot from the experiment in panel B, showing the expression of the EBNA1 proteins after the 14-day selection. Note that for EBNA1 mutants that do not support plasmid maintenance, the EBNA1 protein is expressed from integrated copies of the plasmid.
Looping domain.
The looping domain, a Gly-Arg-rich region that spans amino acids 325 to 376, was originally defined as the region of EBNA1 that mediates interactions at a distance between DNA-bound EBNA1 dimers (16, 24). This region has subsequently been shown to mediate interactions with some cellular proteins (42, 48). To understand how protein interactions mediated by the looping domain contribute to EBNA1 functions, we constructed an EBNA1 internal deletion mutant lacking amino acids 325 to 376 (Δ325–376). In previous studies we have shown that this mutant has wild-type DNA-binding and DNA replication activity but does not support the long-term maintenance of oriP plasmids, suggesting that the deleted sequences are important for plasmid segregation (5, 42). In keeping with these findings, EBNA1 residues 377 to 641 were found to be insufficient to maintain oriP plasmids in long-term culture (Fig. 2A to C and Table 1). To determine if the looping domain also mediates transcriptional activation by EBNA1, we compared the transactivation activity of Δ325–376 with that of wild-type EBNA1. As shown in Fig. 3B, the deletion of residues 325 to 376 abrogated the transactivation activity of EBNA1, as did a larger deletion encompassing amino acids 41 to 376. These results strongly suggest that the looping domain is the transactivation domain of EBNA1, and since we have previously shown that Δ325–376 is fully active for DNA replication (42), they also indicate that the transactivation and replication functions of EBNA1 are separable.
To further examine the EBNA1 sequence requirements for transactivation, we also constructed EBNA1 mutants with smaller deletions in the looping domain (Δ356–362 and Δ367–376). Both of these mutants were found to transactivate the reporter gene at levels similar to that by wild-type EBNA1 (Fig. 3B and Table 1), indicating either that residues 325 to 355 are sufficient for transactivation or that residues 356 to 362 and 367 to 376 make redundant contributions to transactivation. With respect to the latter interpretation, we have previously observed that the looping domain contains repetitive sequences with redundant abilities to mediate protein interactions (5, 24). The Δ356–362 and Δ367–376 mutations also appear to have no significant effect on the plasmid maintenance activity of EBNA1 (Table 1) (42).
WF motif.
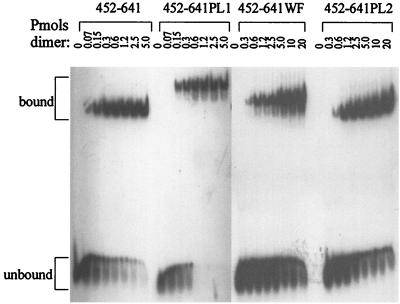
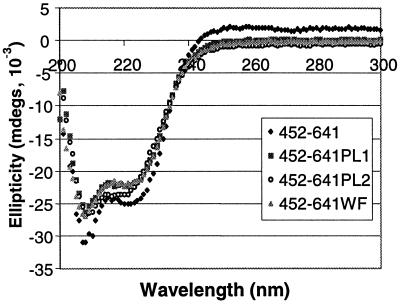
The crystal structure of the EBNA1 DNA binding and dimerization domains bound to DNA revealed a peculiar position of amino acids 464 (W) and 465 (F) (8). These residues fall within the flanking domain extended chain that inserts in the minor groove of the DNA (Fig. 1). The positioning of the aromatic side chains of WF is such that they appear to be pushing against the two DNA strands; a widening of the minor groove of 2 to 3 Å is seen at this position, suggesting that the WF side chains alter the DNA structure. We were interested in determining whether this alteration in DNA structure facilitated the initial DNA-melting step of DNA replication. To this end, we constructed EBNA1 proteins in which W464 and F465 were changed to alanines. These mutations were expressed either in the context of wild-type EBNA1 (EBNA1–641WF), for functional assays, or in the context of EBNA452–641, for assessment of DNA-binding ability. The latter protein was overproduced, purified, and used in EMSAs to determine if the mutations affected the DNA-binding activity of EBNA1. EBNA452–641WF retained sequence-specific DNA-binding activity, but the WF mutation reduced DNA-binding affinity 17-fold (Fig. 6). This reduction in DNA affinity was not due to the unfolding of the protein, as the CD spectra of EBNA452–641WF were indistinguishable from those of EBNA452–641 (Fig. 7); both proteins exhibited elliptical minima at 208 and 222 nm, in keeping with their helical content. The reduced DNA-binding affinity of EBNA452–641WF was also not due to the decreased stability of this protein, as the unfolding rates of EBNA452–641WF and EBNA452–641 in denaturant were almost identical (Table 2).
FIG. 6.
Effect of proline loop and WF mutations on the DNA-binding ability of EBNA1. Purified EBNA452–641 containing the wild-type sequence or the PL1, PL2, or WF mutation was titrated onto a single EBNA1 recognition site, and bound and unbound DNA was separated by native PAGE.
FIG. 7.
Proline loop and WF mutations do not disrupt protein folding. Shown are CD spectra of purified EBNA452–641 containing the wild-type sequence or the PL1, PL2, or WF mutation. All the proteins exhibit elliptical minima at 208 and 222 nm, indicative of their helical content.
TABLE 2.
Effect of proline loop and WF mutations on EBNA1 stability
| Protein | GuHCl concn (M) | k (1/s)a | T1/2 (min)b |
|---|---|---|---|
| 452–641 | 6.6 | 4.1 × 10−4 | 28 |
| 452–641PL1 | 6.6 | 3.0 × 10−3 | 4 |
| 6.0 | 1.3 × 10−3 | 9 | |
| 5.0 | 9.6 × 10−5 | 120 | |
| 452–641PL2 | 6.6 | 9.5 × 10−3 | 1 |
| 6.0 | 3.4 × 10−3 | 4 | |
| 5.0 | 1.2 × 10−4 | 100 | |
| 452–641WF | 6.6 | 3.6 × 10−4 | 32 |
Unfolding rate constant.
Time required for half of the EBNA1 protein to unfold at the concentration of GuHCl indicated.
Since the WF mutation did not abrogate sequence-specific DNA binding, we examined the potential role of these residues in DNA replication by assaying the transient-replication and long-term maintenance activities of this mutant. The results clearly showed that EBNA1–641WF is capable of replicating and maintaining oriP plasmids at levels at or above that of EBNA1 (Fig. 4 and 5 and Table 1). This mutant was also found to transactivate transcription at levels similar to the wild-type protein (Fig. 3C and Table 1). We conclude that, despite the decreased DNA-binding ability associated with the WF mutation, the expression level of EBNA1–641WF is sufficient to allow DNA binding in this system, and that the WF motif does not otherwise contribute to DNA replication, segregation, or transactivation.
Proline loop.
The proline loop (amino acids 540 to 555) is part of the core DNA binding domain of EBNA1; it does not directly contact the DNA but extends outward from the body of the domain (Fig. 1B) (8, 9). Both the exposed positioning of this loop and its proline-rich sequence, which resembles proline-rich activation domains, suggest that it might mediate interactions with other proteins. To explore the contribution of the proline loop to EBNA1 functions, we generated two proline loop mutations; one in which five amino acids at the tip of the loop were replaced with a four-amino-acid flexible linker (PL1), and a second in which 13 amino acids of the loop were replaced with the four-amino-acid flexible linker (PL2) (Fig. 2B). To determine whether the PL1 and PL2 mutations disrupted the folding and DNA-binding ability of the EBNA1 DNA binding and dimerization domains, these mutations were first generated in the context of EBNA452–641, overproduced in E. coli, and purified. The CD spectra of the purified proteins were then compared with that of EBNA452–641 to determine whether the mutations had disrupted protein folding. As shown in Fig. 7, the spectra of all three proteins were indistinguishable, indicating that the PL1 and PL2 mutations did not disrupt the folding of EBNA1.
We then compared the DNA-binding affinity of EBNA452–641PL1 and EBNA452–641PL2 for an EBNA1 recognition site with that of EBNA452–641. The PL1 mutation had no significant effect on DNA-binding activity, while the PL2 mutation reduced DNA affinity 14-fold (Fig. 6); apparent Kd values were 8, 11, and 116 nM for the wild-type, PL1, and PL2 proteins, respectively. Although the DNA-binding affinity of EBNA452–641PL2 was reduced relative to the wild-type protein, it was very similar to that of EBNA452–641WF (apparent Kd of 140 nM). We then examined whether the reduction in DNA binding associated with EBNA452–641PL2 was due to decreased protein stability. To this end, we compared the unfolding rates in denaturant of EBNA452–641PL1 and EBNA452–641PL2 with that of EBNA452–641 (Table 2). EBNA452–641PL1 and EBNA452–641PL2 were found to have very similar stabilities, and both were less stable to the GuHCl denaturant than EBNA452–641. Although EBNA452–641PL1 and EBNA452–641PL2 unfolded faster than the wild-type protein, both of the mutants were still very stable proteins, with half of the protein remaining folded even after approximately 2 h in 5 M GuHCl. Therefore, the stabilities of EBNA452–641PL1 and EBNA452–641PL2 are not likely to be a factor affecting DNA-binding ability in vitro or functionality in vivo.
Having shown that the PL1 and PL2 mutations did not disrupt protein folding, cause the proteins to become unstable, or abrogate sequence-specific DNA binding, we then proceeded to test the functionality of full-length EBNA1 proteins containing these mutations. EBNA1–641PL1 was found to maintain plasmids at or above wild-type levels and to transactivate gene expression at levels close to the wild-type level (Fig. 3C and 5 and Table 1). Therefore this mutant appears to be active for DNA replication, segregation, and transactivation. EBNA1–641PL2, however, did not support plasmid maintenance and exhibited only low levels of transient-replication and transactivation activity (Fig. 3C, 4, and 5 and Table 1). To determine whether this apparent lack of activity might be due to lack of DNA binding caused by low expression levels of EBNA1–641PL2 coupled with its lower DNA affinity, we performed quantitative Western blots on aliquots of transfected cells collected for transactivation or transient-replication assays. The expression level of EBNA1–641PL2 was then compared with that of EBNA1–641WF. Because the PL2 and WF mutations had similar effects on DNA-binding affinity and EBNA1–641WF was fully functional, we reasoned that if the expression level of EBNA1–641PL2 was as high as or higher than that of EBNA1–641WF, then the protein was present at sufficient levels to bind to the EBNA1 recognition sites. However, quantitative Western blots performed at 24 h posttransfection showed that EBNA1–641PL2 was expressed at 34 to 45% of the level of EBNA1–641WF (data not shown), raising the distinct possibility that the lack of activity associated with the PL2 mutation was due to insufficient DNA binding by EBNA1–641PL2. Therefore, the only function that we have been able to assign to the proline loop is a contribution to DNA binding.
DISCUSSION
In efforts to further understand the mechanisms by which EBNA1 functions, we have examined the functional contributions of four unusual sequence elements of EBNA1: the looping domain, the acidic tail, the proline loop, and the WF motif. We have shown that deletion of amino acids 325 to 376 abrogates the transcriptional activation activity of EBNA1 without affecting the DNA replication function, indicating that the looping domain plays an essential role in transactivation. The importance of this region for transactivation is also supported by the studies of Yates and Camiolo (51), Wang et al. (48), and Mackey and Sugden (29). Since we have previously shown that the 325 to 376 mutation does not disrupt the DNA-binding ability of EBNA1 (5), the functional contribution of the looping domain is likely in mediating protein-protein interactions. Indeed, we and others have shown previously that the looping domain has a propensity to mediate interactions at a distance between DNA-bound EBNA1 molecules as well as interactions with at least two cellular factors (5, 16, 24, 30, 42, 48). It seems likely, therefore, that the looping domain may function by mediating DNA-looping interactions between FR-bound EBNA1 molecules and one or more components of the basal transcription machinery at the promoter, or alternatively by tethering cellular transactivation proteins to the FR. It has also been reported that part of the transactivation activity associated with EBNA1 is due to increased nuclear uptake of FR-containing plasmids (25), but whether or not the looping domain participates in this process is not yet known. This increased uptake may involve the interaction of EBNA1 with the nuclear import factor Rch1/importin α, since EBNA1 has been shown to physically interact with this protein (12, 22).
Although residues 325 to 376 play a critical role in transcription activation, deletion of these sequences has no detectable effect on the transient-replication activity of EBNA1 (42). Therefore, the transactivation and replication activities of EBNA1 are separable and likely occur by different mechanisms. The EBNA1 residues outside of the DNA binding and dimerization region that are important for DNA replication are not yet clear. Although fully active for DNA replication, the Δ325–376 mutant is unable to maintain oriP plasmids in long-term culture, indicating that this mutant is defective in DNA segregation (42). The transactivation and segregation activities of EBNA1 therefore appear to be coincident. EBNA1 is thought to govern the segregation of oriP plasmids by mediating their attachment to a component of the host mitotic chromosomes. In keeping with this model, EBNA1 looping-domain sequences have been shown to be important for the attachment of EBNA1 to mitotic chromosomes (31; H. Wu, D. F. J. Ceccarelli, and L. Frappier, submitted for publication). Recent studies suggest that the component of the host chromosome to which EBNA1 attaches is the cellular factor EBP2; EBP2 binds to the EBNA1 looping-domain sequences and appears to colocalize with EBNA1 on host mitotic chromosomes (42; Wu et al., submitted).
In this study, we also investigated the role of the acidic C-terminal tail of EBNA1. This region has previously been reported to be a transactivation domain and to play a role in DNA segregation (2, 51). Our results clearly show that the acidic tail is not required for the replication, segregation, or transcription activation functions of EBNA1, nor does it support significant levels of transactivation in the absence of the DNA looping domain. The lack of transactivation activity of the acidic tail is also supported by the data obtained in other laboratories (23, 36, 51). Our finding that the acidic tail is not required for segregation is contrary to the findings of Yates and Camiolo (51). We believe that the reason for this discrepancy is due to differences in the position of the EBNA1 C-terminal truncation; the EBNA1 construct used by Yates and Camiolo extended to residue 619, whereas our mutant was truncated at residue 607. The 607 to 619 sequence, when exposed after removal of the acidic tail, may affect the results obtained by either decreasing EBNA1 expression levels, destabilizing the protein, or otherwise interfering with protein-protein interactions. Since our results indicate that the acidic tail is not required for the replication, transactivation, or segregation functions of EBNA1, the question remains why it is present in the protein. We speculate that during viral infection, these sequences may either regulate cellular protein expression or affect the EBNA1 levels in the cell by increasing the expression or stability of EBNA1. The latter possibility may not be a factor in our experimental system, in which high levels of EBNA1 are constantly expressed from the CMV promoter.
We have explored the contribution to EBNA1 functions of two interesting structural motifs of the EBNA1 DNA binding and dimerization domains, the proline loop and the WF motif. Our results indicate that neither motif plays a direct role in DNA replication, segregation, or transactivation but that both contribute to the DNA-binding ability of EBNA1. The EBNA1-DNA cocrystal structure, when combined with the results of biochemical DNA-binding studies using EBNA1 point mutants, indicates that the recognition helices of the EBNA1 core DNA binding domain make important sequence-specific DNA contacts and may position the protein so that the flanking-domain extended chain can be loaded into the minor groove (8; Cruickshank et al., submitted). The DNA contacts mediated by the recognition helices are transient and appear to be released once the flanking-domain extended chain is in place in the minor groove. The WF sequence falls within this minor groove extended chain but was not thought to contribute to DNA binding since these residues were not observed to make DNA contacts. Our finding that the point mutations of the WF residues reduced DNA-binding affinity 17-fold suggests that the WF motif plays a role in loading the extended chain in the minor groove of the DNA.
Our studies with the proline loop mutants have shown that DNA-binding ability is not affected when residues 545 to 549 are replaced by a flexible linker (PL1) but is affected when residues 541 to 553 are replaced by the same linker (PL2). The decreased length of the loop in the PL2 mutant was not expected to disrupt the structure of the rest of the DNA-binding domain, as the loop length in PL2 is identical to that in the structurally homologous domain of the E2 papillomavirus protein (9, 20). In keeping with this assumption, we have shown that the effect of the PL2 mutation on DNA binding was not due to lack of protein folding or decreased protein stability. Instead, it is likely that this effect was due to the deletion of residues G542 and P553, both of which were observed in the EBNA1-DNA crystal structure to participate in H-bond interactions with residues adjacent to the recognition helices (8). Our results suggest that these interactions affect the positioning of the recognition helices and that their disruption inhibits the transient binding of these helices to EBNA1 recognition sites.
ACKNOWLEDGMENTS
We gratefully acknowledge Kathy Shire for construction of EBNA1–641WF, Tina Avolio-Hunter for construction of Δ325–376 and Δ41–376, Angela Flemming for construction and purification of the EBNA452–641WF protein, Alexey Bochkarev for Fig. 1, and Alan Davidson and Jennifer Cruickshank for assistance with CD analyses. We also thank Bill Sugden for pFRTKCAT, Jaap Middeldorp for anti-EBNA1 antiserum, and John Hassell and Aled Edwards for helpful comments throughout the course of this work.
This work was supported by a grant from the National Cancer Institute of Canada. L.F. is a Medical Research Council of Canada Scientist.
REFERENCES
- 1.Adams A. Replication of latent Epstein-Barr virus genomes. J Virol. 1987;61:1743–1746. doi: 10.1128/jvi.61.5.1743-1746.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambinder R F, Mullen M, Chang Y, Hayward G S, Hayward S D. Functional domains of Epstein-Barr nuclear antigen EBNA-1. J Virol. 1991;65:1466–1478. doi: 10.1128/jvi.65.3.1466-1478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambinder R F, Shah W A, Rawlins D R, Hayward G S, Hayward S D. Definition of the sequence requirements for binding of the EBNA-1 protein to its palindromic target sites in Epstein-Barr virus DNA. J Virol. 1990;64:2369–2379. doi: 10.1128/jvi.64.5.2369-2379.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley and Sons; 1991. [Google Scholar]
- 5.Avolio-Hunter T M, Frappier L. Mechanistic studies on the DNA linking activity of the Epstein-Barr nuclear antigen 1. Nucleic Acids Res. 1998;26:4462–4470. doi: 10.1093/nar/26.19.4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barwell J, Bochkarev A, Pfuetzner R, Tong H, Yang D, Frappier L, Edwards A. Purification and crystallization of the DNA-binding and dimerization domain of the Epstein-Barr virus nuclear antigen 1. J Biol Chem. 1995;270:20556–20559. doi: 10.1074/jbc.270.35.20556. [DOI] [PubMed] [Google Scholar]
- 7.Blake N, Lee S, Redchenko I, Thomas W, Steven N, Leese A, Steigerwald-Mullen P, Kurilla M G, Frappier L, Rickinson A. Human CD8+ T cell responses to EBV EBNA1: HLA class I presentation of the (GLY-ALA) containing protein requires exogenous processing. Immunity. 1997;7:791–802. doi: 10.1016/s1074-7613(00)80397-0. [DOI] [PubMed] [Google Scholar]
- 8.Bochkarev A, Barwell J, Pfuetzner R, Bochkareva E, Frappier L, Edwards A M. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin binding protein, EBNA1, bound to DNA. Cell. 1996;84:791–800. doi: 10.1016/s0092-8674(00)81056-9. [DOI] [PubMed] [Google Scholar]
- 9.Bochkarev A, Barwell J, Pfuetzner R, Furey W, Edwards A, Frappier L. Crystal structure of the DNA binding domain of the Epstein-Barr virus origin binding protein EBNA1. Cell. 1995;83:39–46. doi: 10.1016/0092-8674(95)90232-5. [DOI] [PubMed] [Google Scholar]
- 10.Chen M-R, Middeldorp J M, Hayward S D. Separation of the complex DNA binding domain of EBNA-1 into DNA recognition and dimerization subdomains of novel structure. J Virol. 1993;67:4875–4885. doi: 10.1128/jvi.67.8.4875-4885.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delecluse H-J, Bartnizke S, Hammerschmidt W, Bullerdiek J, Bornkamm G W. Episomal and integrated copies of Epstein-Barr virus coexist in Burkitt's lymphoma cell lines. J Virol. 1993;67:1292–1299. doi: 10.1128/jvi.67.3.1292-1299.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer N, Kremmer E, Lautscham G, Mueller-Lantzsch N, Grasser F A. Epstein-Barr virus nuclear antigen 1 forms a complex with the nuclear transporter karyopherin α2. J Biol Chem. 1997;272:3999–4005. doi: 10.1074/jbc.272.7.3999. [DOI] [PubMed] [Google Scholar]
- 13.Frappier L, O'Donnell M. Overproduction, purification and characterization of EBNA1, the origin binding protein of Epstein-Barr virus. J Biol Chem. 1991;266:7819–7826. [PubMed] [Google Scholar]
- 14.Gahn T, Sugden B. An EBNA1-dependent enhancer acts from a distance of 10 kilobase pairs to increase expression of the Epstein-Barr virus LMP gene. J Virol. 1995;69:2633–2636. doi: 10.1128/jvi.69.4.2633-2636.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gahn T A, Schildkraut C L. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell. 1989;58:527–535. doi: 10.1016/0092-8674(89)90433-9. [DOI] [PubMed] [Google Scholar]
- 16.Goldsmith K, Bendell L, Frappier L. Identification of EBNA1 amino acid sequences required for the interaction of the functional elements of the Epstein-Barr virus latent origin of DNA replication. J Virol. 1993;67:3418–3426. doi: 10.1128/jvi.67.6.3418-3426.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;5:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 18.Harris A, Young B D, Griffin B E. Random association of Epstein-Barr virus genomes with host cell metaphase chromosomes in Burkitt's lymphoma-derived cell lines. J Virol. 1985;56:328–332. doi: 10.1128/jvi.56.1.328-332.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrison S, Fisenne K, Hearing J. Sequence requirements of the Epstein-Barr virus latent origin of DNA replication. J Virol. 1994;68:1913–1925. doi: 10.1128/jvi.68.3.1913-1925.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hegde R S, Grossman S R, Laimins L A, Sigler P B. Crystal structure at 1.7Å of the bovine papillomavirus-1 E2 DNA-binding protein bound to its DNA target. Nature. 1992;359:505–512. doi: 10.1038/359505a0. [DOI] [PubMed] [Google Scholar]
- 21.Hirt B. Selective extraction of polyoma DNA from infected mouse cell culture. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 22.Kim A L, Maher M, Hayman J B, Ozer J, Zerby D, Yates J L, Lieberman P M. An imperfect correlation between DNA replication activity of Epstein-Barr virus nuclear antigen 1 (EBNA1) and binding to the nuclear import receptor, Rch1/importin α. Virology. 1997;239:340–351. doi: 10.1006/viro.1997.8874. [DOI] [PubMed] [Google Scholar]
- 23.Kirchmaier A L, Sugden B. Dominant-negative inhibitors of EBNA1 of Epstein-Barr virus. J Virol. 1997;71:1766–1775. doi: 10.1128/jvi.71.3.1766-1775.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laine A, Frappier L. Identification of Epstein-Barr nuclear antigen 1 protein domains that direct interactions at a distance between DNA-bound proteins. J Biol Chem. 1995;270:30914–30918. doi: 10.1074/jbc.270.52.30914. [DOI] [PubMed] [Google Scholar]
- 25.Langle-Rouault F, Patzel V, Benavente A, Taillez M, Silvestre N, Bompard A, Sczakiel G, Jacobs E, Rittner K. Up to 100-fold increase of apparent gene expression in the presence of Epstein-Barr virus oriP sequences and EBNA1: implications of the nuclear import of plasmids. J Virol. 1998;72:6181–6185. doi: 10.1128/jvi.72.7.6181-6185.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levitskaya J, Coram M, Levitsky V, Imreh S, Steigerwald-Mullen P, Klein G, Kurilla M G, Masucci M G. Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature. 1995;375:685–688. doi: 10.1038/375685a0. [DOI] [PubMed] [Google Scholar]
- 27.Lupton S, Levine A J. Mapping of genetic elements of Epstein-Barr virus that facilitate extrachromosomal persistence of Epstein-Barr virus-derived plasmids in human cells. Mol Cell Biol. 1985;5:2533–2542. doi: 10.1128/mcb.5.10.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mackey D, Middleton T, Sugden B. Multiple regions within EBNA1 can link DNAs. J Virol. 1995;69:6199–6208. doi: 10.1128/jvi.69.10.6199-6208.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mackey D, Sugden B. The linking regions of EBNA1 are essential for its support of replication and transcription. Mol Cell Biol. 1999;19:3349–3359. doi: 10.1128/mcb.19.5.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mackey D, Sugden B. Studies on the mechanism of DNA linking by Epstein-Barr virus nuclear antigen 1. J Biol Chem. 1997;272:29873–29879. doi: 10.1074/jbc.272.47.29873. [DOI] [PubMed] [Google Scholar]
- 31.Marechal V, Dehee A, Chikhi-Brachet R, Piolot T, Coppey-Moisan M, Nicolas J. Mapping EBNA1 domains involved in binding to metaphase chromosomes. J Virol. 1999;73:4385–4392. doi: 10.1128/jvi.73.5.4385-4392.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Middleton T, Sugden B. EBNA1 can link the enhancer element to the initiator element of the Epstein-Barr virus plasmid origin of DNA replication. J Virol. 1992;66:489–495. doi: 10.1128/jvi.66.1.489-495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niller H H, Glaser G, Knuchel R, Wolf H. Nucleoprotein complexes and DNA 5′-ends at oriP of Epstein-Barr virus. J Biol Chem. 1995;270:12864–12868. doi: 10.1074/jbc.270.21.12864. [DOI] [PubMed] [Google Scholar]
- 34.Nonkwelo C, Skinner J, Bell A, Rickinson A, Sample J. Transcription start site downstream of the Epstein-Barr virus (EBV) Fp promoter in early-passage Burkitt lymphoma cells define a fourth promoter for expression of the EBV EBNA1 protein. J Virol. 1996;70:623–627. doi: 10.1128/jvi.70.1.623-627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petti L, Sample C, Kieff E. Subnuclear localization and phosphorylation of Epstein-Barr virus latent infection nuclear proteins. Virology. 1990;176:563–574. doi: 10.1016/0042-6822(90)90027-o. [DOI] [PubMed] [Google Scholar]
- 36.Polvino-Bodnar M, Schaffer P A. DNA binding activity is required for EBNA1-dependent transcriptional activation and DNA replication. Virology. 1992;187:591–603. doi: 10.1016/0042-6822(92)90461-w. [DOI] [PubMed] [Google Scholar]
- 37.Rawlins D R, Milman G, Hayward S D, Hayward G S. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA1) to clustered sites in the plasmid maintenance region. Cell. 1985;42:859–868. doi: 10.1016/0092-8674(85)90282-x. [DOI] [PubMed] [Google Scholar]
- 38.Reisman D, Sugden B. trans activation of an Epstein-Barr viral transcriptional enhancer by the Epstein-Barr viral nuclear antigen 1. Mol Cell Biol. 1986;6:3838–3846. doi: 10.1128/mcb.6.11.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reisman D, Yates J, Sugden B. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol Cell Biol. 1985;5:1822–1832. doi: 10.1128/mcb.5.8.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2397–2446. [Google Scholar]
- 41.Sample J, Henson E B D, Sample C. The Epstein-Barr virus nuclear protein 1 promoter active in type I latency is autoregulated. J Virol. 1992;66:4654–4661. doi: 10.1128/jvi.66.8.4654-4661.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shire K, Ceccarelli D F J, Avolio-Hunter T M, Frappier L. EBP2, a human protein that interacts with sequences of the Epstein-Barr nuclear antigen 1 important for plasmid maintenance. J Virol. 1999;73:2587–2595. doi: 10.1128/jvi.73.4.2587-2595.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Snudden D K, Hearing J, Smith P R, Grasser F A, Griffin B E. EBNA1, the major nuclear antigen of Epstein-Barr virus, resembles ‘RGG’ RNA binding proteins. EMBO J. 1994;13:4840–4847. doi: 10.1002/j.1460-2075.1994.tb06810.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 45.Sugden B, Warren N. A promoter of Epstein-Barr virus that can function during latent infection can be transactivated by EBNA-1, a viral protein required for viral DNA replication during latent infection. J Virol. 1989;63:2644–2649. doi: 10.1128/jvi.63.6.2644-2649.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Summers H, Barwell J A, Pfuetzner R A, Edwards A M, Frappier L. Cooperative assembly of EBNA1 on the Epstein-Barr virus latent origin of replication. J Virol. 1996;70:1228–1231. doi: 10.1128/jvi.70.2.1228-1231.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Summers H, Fleming A, Frappier L. Requirements for EBNA1-induced permanganate sensitivity of the Epstein-Barr virus latent origin of DNA replication. J Biol Chem. 1997;272:26434–26440. doi: 10.1074/jbc.272.42.26434. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y, Finan J E, Middeldorp J M, Hayward S D. P32/TAP, a cellular protein that interacts with EBNA-1 of Epstein-Barr virus. Virology. 1997;236:18–29. doi: 10.1006/viro.1997.8739. [DOI] [PubMed] [Google Scholar]
- 49.Wysokenski D A, Yates J L. Multiple EBNA1-binding sites are required to form an EBNA1-dependent enhancer and to activate a minimal replicative origin within oriP of Epstein-Barr virus. J Virol. 1989;63:2657–2666. doi: 10.1128/jvi.63.6.2657-2666.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yates J L. Epstein-Barr virus DNA replication. In: DePamphilis M, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 751–773. [Google Scholar]
- 51.Yates J L, Camiolo S M. Dissection of DNA replication and enhancer activation functions of Epstein-Barr virus nuclear antigen 1. Cancer Cells. 1988;6:197–205. [Google Scholar]
- 52.Yates J L, Guan N. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J Virol. 1991;65:483–488. doi: 10.1128/jvi.65.1.483-488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yates J L, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]