Abstract
Orphan nuclear receptor fetoprotein transcription factor (FTF) was previously identified as a specific regulator of the α1-fetoprotein gene during early liver development and in response to hormonal signals (L. Galarneau, J.-F. Paré, D. Allard, D. Hamel, L. Lévesque, J. D. Tugwood, S. Green, and L. Bélanger, Mol. Cell. Biol. 16:3853–3865, 1996). Here we report a functional analysis of FTF interactions with the hepatitis B virus (HBV) nucleocapsid promoter. DNA-protein-binding assays show that the HBV core promoter contains two high-affinity FTF-binding sites and a third, lower-affinity site shared with other receptors. Transfections in HepG2, Hep3B, and PLC/PRF/5 hepatoma cells using chloramphenicol acetyltransferase reporter genes with the nucleocapsid promoter linked or not linked to enhancer I indicate that FTF is a potent activator of the HBV core promoter, more efficient than HNF4α, HNF3α, HNF3β, or C/EBPα. Steroidogenic factor 1, a close FTF homolog which binds to the same DNA motif and is expressed ectopically in HepG2 cells, seems to be an even stronger inducer than FTF. Point mutations of the FTF-binding sites indicate direct FTF activatory effects on the core promoter and the use of both high-affinity sites for productive interaction between the core promoter and enhancer I. Coexpression assays further indicate that FTF and HNF4α are the most efficient partners for coactivation of the pregenomic core promoter, which may largely account for the hepatic tropism and the early amplification of HBV infection. Carboxy terminus-truncated FTF behaves as a dominant negative mutant to compete all three FTF sites and strongly deactivate core promoter interactions with enhancer I; this suggests possible new ways to interfere with HBV infection.
Viral hepatitis B is a leading cause of liver disease and primary hepatocellular carcinoma (HCC) and a leading cause of cancer deaths in populations in which hepatitis B virus (HBV) carriage is endemic (3, 7, 24). Vaccination has proven remarkably effective in preventing HBV infection (and hence HCC) in some high-risk communities (9), but efforts are also directed toward pharmacological and other means of controlling the virus. Molecular biological studies have considerably advanced our understanding of how the HBV genome operates, providing important new clues to the natural history of HBV-related diseases and, potentially, new therapeutical avenues. The HBV genome (Fig. 1) consists of ≈3.2 kb of circular DNA encoding four overlapping reading frames driven by promoter and enhancer elements which operate in a highly liver-restricted manner. The HBV nucleocapsid promoter has been especially targeted for detailed molecular analysis, for its pivotal role in the hepatotropism and early life cycle of HBV. The nucleocapsid promoter contains a basic core segment which carries two genetically distinguishable promoters, the preC and core pregenomic promoters, coordinately activated by an upstream regulatory domain (devoid of intrinsic promoter activity) extending from nucleotide (nt) 1636 to nt 1744 (43, 46). The cumulative data on the nucleocapsid promoter (and other HBV promoter and enhancer elements as well) clearly indicate that HBV hepatotropism basically reflects the combined need for several liver-enriched transcription factors in order for the HBV genome to replicate efficiently. HBV transgenes are transcribed mostly in the liver (1, 2, 23), transfected HBV transcribes and replicates better in well-differentiated hepatocellular lines (8, 19, 36, 38), and some especially virulent strains of HBV contain mutations that convert nucleocapsid promoter sequences into novel high-affinity sites for liver-type transcription factors (15, 20, 26, 32); chronic HBV hepatitis culminating in HCC (11) also implies that HBV makes sustained efficient use of liver transcription factors.
FIG. 1.
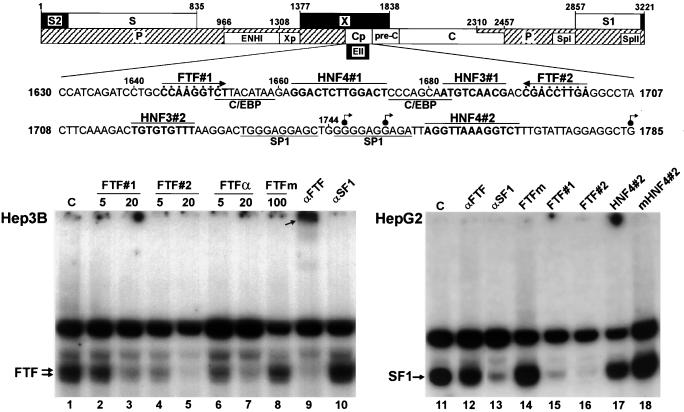
The HBV core promoter contains two high-affinity binding sites for nuclear receptor FTF (or its close relative SF1). The upper diagram displays the main structural and regulatory components of the HBV genome and a partial nucleotide sequence of the core promoter (Cp); dots mark nucleotides present in the consensus FTF-binding site. S, S1, S2, Spl, and Spll, envelope genes and promoters; X/Xp, X gene and promoter; P, DNA polymerase; C/preC, nucleocapsid gene and upstream region; ENHI and EII, enhancers I and II; angled arrows, transcription initiation sites reported for the C (nts 1745, 1751, and 1818 to 1821) and preC (nts 1785 to 1793) mRNA transcripts (29, 41, 43). Autoradiograms show EMSAs conducted with 3 μg of total nuclear proteins from Hep3B or HepG2 cells using a 32P-labeled HBV-FTF#2 oligonucleotide probe. The value above each lane is the fold molar excess of the competing unlabeled oligonucleotide. HepG2 reactions used a 50-fold molar excess of competitors. FTFα and FTFm, native and mutant FTF sites from the AFP gene promoter; HNF4#2 and mHNF4#2, wild-type and mutant HBV sequence from nt 1757 to nt 1769; C, no competitor. Lanes αFTF and αSF1 show supershift reactions using antibodies against FTF or SF1. Note that in the Hep3B reactions, αFTF completely displaces the specifically retarded complexes toward an upper band (arrow, lane 9) whereas displacement is negligible with the HepG2 extract (lane 12) (a faint supershifted band was visible in other assays). Conversely, specific bandshifts are strongly decreased by αSF1 in the HepG2 extract (lane 13) and not in the Hep3B extract (lane 10). These assays indicate highly specific occupancy of the FTF#2 nt 1689 to 1707 segment by FTF or SF1 and that FTF#1 has slightly less affinity for FTF/SF1 than does FTF#2 and HNF4#2 has less affinity than FTF#1.
Like that of the HBV genome, expression of the albumin-related genes is highly restricted to hepatocytes. Among its close relatives of this four-member family (5), the α1-fetoprotein (AFP) gene is also differentially regulated in response to developmental and hormonal signals (4, 6, 13). In our analysis of AFP-specific gene regulation, we have pinpointed a critical promoter element that is absent from the other albumin genes and activated by the fetoprotein transcription factor (FTF) (The FTF designation has been approved by the Genome Database Nomenclature Committee [GDB accession no. 9837397]. In a recently proposed nomenclature, FTF corresponds to NR5A2 [Nuclear Receptors Nomenclature Committee, Letter, Cell 97:161–163, 1999].) (6, 13, 14), a nuclear receptor expressed selectively in the liver, pancreas, and intestine (13, 34). FTF is a novel member of the Drosophila fushi tarazu F1 family of orphan receptors and a close homolog of steroidogenic factor 1 (SF1), which is expressed in steroidogenic cell lineages (17). FTF is part of a transcriptional liver differentiation cascade that involves hepatocyte nuclear factor 3β (HNF3β) (34), HNF4α, and HNF1α (J.-F. Paré, S. Roy, and L. Bélanger, Abstr. 8th Biennial Int. Congr. Liver Dev. Gene Regul. Dis., abstr. 10, p. 10), and FTF emerges as the key rate-limiting factor for AFP gene activation in response to liver growth and metabolic signals.
In our initial survey of potential FTF gene targets (13), we noted that the HBV nucleocapsid promoter contains two apparent high-affinity FTF-binding sites (FTF#1 and FTF#2 in Fig. 1). This seemed of particular interest to us regarding FTF reactivity to developmental signals in a different promoter context and also because HBV functions might, perhaps, be down-regulated using FTF-directed strategies. The results presented here indicate that FTF is, indeed, a potent activator of the HBV pregenomic core promoter. Furthermore, molecular hindrance at the FTF-binding sites strongly interferes with HBV promoter-enhancer functions, suggesting possible new opportunities to antagonize HBV replication.
MATERIALS AND METHODS
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were conducted as described previously (6, 13), using total nuclear protein extracts and 32P-labeled oligonucleotide HBV-FTF#2 (1689CGACCGACCTTGAGGCCTA1707; with EcoRI overhangs) as a probe. Unlabeled oligonucleotides (with EcoRI overhangs) HBV-FTF#2 and HBV-FTF#1 (1638TCCTGCCCAAGGTCTTACAT1657), the AFP promoter FTF-binding sequence TGTTCAAGGACA (FTFα) or the nonbinding mutant sequence TGTTCAATGAAA (FTFm) (13), and the HBV-HNF4#2 sequence 1757AGGTTAAAGGTCT1769 or the mutant sequence AAATTAAAAATCT were used as competitors in EMSA reactions. Supershift assays (13) used human FTF (hFTF) (14) antiserum raised in rabbits against the hFTF extra-DNA-binding domain C-terminal domain (hFTF–glutathione S-transferase fusion protein; Pharmacia pGEX-4T3); anti-SF1 antibodies were obtained from Upstate Biotech Inc.
Gene constructs.
HBV DNA segments from luciferase vectors ABluc and ABlucΔe (29) (kindly provided by Aleem Siddiqui) were transferred into pBluescriptSK+ chloramphenicol acetyltransferase (CAT) expression vector SKCAT (Stratagene; CAT insert at HinDIII/BamHI). The 522-nt AvaI fragment (blunted) of ABlucΔe (containing the HBV core promoter) was inserted at the HinDIII site (blunted) of SKCAT, to yield HBV promoter-CAT construct HP. The 343-nt HBV enhancer I DNA segment of ABluc was amplified by PCR, fitted with 5′ KpnI and 3′ SalI sites, and cloned into vector HP digested with KpnI and SalI to generate HBV enhancer-promoter–CAT construct HEP.
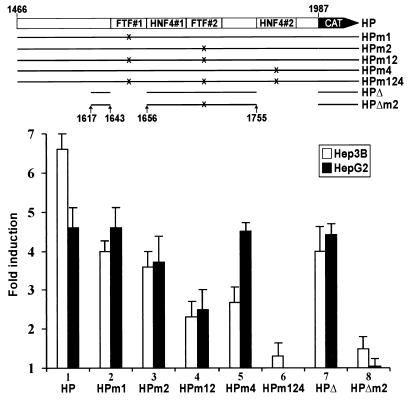
Point mutations were introduced into HBV promoter sequences FTF#1, FTF#2, and HNF4#2 using PCR-directed mutagenesis (PFU polymerase protocol of Stratagene). Nucleotide changes were inserted at G contact points needed for FTF binding to its AFP promoter site (13). Mutations m1 (1644CCAATATTT1652), m2 (1701TCAATATTG1693), and m4 (1757AAATTAAAAATCT1769) were introduced into vectors HP and HEP at either the FTF#1 (HPm1 and HEPm1), FTF#2 (HPm2 and HEPm2), or HNF4#2 (HPm4) sites, at both FTF sites (HPm12 and HEPm12), or at all three FTF and HNF4#2 sites (HPm124 and HEPm124); mutations were confirmed by sequencing. The core promoter domain was further dissected into a 127-nt DNA segment (see Fig. 3, vector HPΔ), leaving out the FTF#1 and HNF4#2 sequences and the major initiation sites for the C (nt 1818 to 1821) and preC (nt 1785 to 1793) mRNA transcripts (41, 43) but keeping two upstream C promoter initiation sites mapped by Siddiqui's group (29) (Fig. 1). Synthetic oligonucleotides overlapping within the targeted DNA region were annealed, filled with Klenow, fitted with 5′-SalI and 3′-HinDIII ends, and cloned into SalI/HinDIII-digested SKCAT. Vector HPΔm2, carrying FTF#2 mutation m2 (described above), was obtained by the same strategy.
FIG. 3.
Transient-cotransfection assays using 10 μg of FTF expression vector and 5 μg of core promoter reporter constructs. Results are averages (±1 standard deviation) of three sets of duplicate or triplicate transfections, referred to the control cotransfection with vector pCl, which was given a value of 1.
To obtain human expression vector pClhFTF, full-length human FTF cDNA (3.8 kb) was retrieved from a UniZAP-XR library (Stratagene) (14), released from the cloning vector by digestion with EcoRI and XhoI, and transferred into EcoRI/SalI-digested vector pCl from Promega. Carboxy-terminally truncated FTF vectors pClmFTFΔAF2 and pClrFTFΔLBD are mouse FTF/LRH-1 and rat FTF constructs pfΔ2 and pfΔ3 in reference 13.
Transfections.
Transient-transfection assays were conducted with human hepatoma cell lines HepG2, Hep3B, and PLC/PRF/5 or HeLa cells (all lines were obtained from the American Type Culture Collection) using the calcium phosphate procedure previously described (6, 13). Cells (1.5 × 106 to 2.5 × 106 in 10-cm-diameter petri dishes) were maintained at 37°C with 5% CO2 in low-glucose Dulbecco's modified Eagle's medium (GIBCO) containing 10% fetal bovine serum (Wisent) and 1% penicillin-streptomycin. Cells were cotransfected with 5 μg of an HBV-CAT reporter construct, 10 μg of transcription factor expression vector, and 2.5 μg of pRSVlacZ to control for transfection efficiency (under our assay conditions, titrations with 0.1 to 30 μg of transcription factor expression vectors have shown that maximal activatory effects are generally obtained with 5 to 10 μg of vector). Cells were washed with 10 mM HEPES 16 h after transfection. CAT activities were measured by thin-layer chromatography and phosphorimaging (Storm 860 Molecular Dynamics system equipped with Imagequant software) 48 h (HepG2, Hep3B, and HeLa cells) or 72 h (PLC/PRF/5 cells) after transfection. Expression vectors for transcription factors used viral enhancer-promoter elements from murine sarcoma virus (C/EBPα), Rous sarcoma virus (HNF1α), and cytomegalovirus (FTF, FTFΔAF2, FTFΔLBD, HNF4α, HNF3α, HNF3β, SP1, and SF1).
RESULTS
The HBV core promoter contains two high-affinity FTF-binding sites.
A computer search for FTF recognition sequences in the HBV genome (using the GCG Wordsearch software) retrieved only the two candidate sequences we had noted (13) in the upstream regulatory region of the basic core promoter, one matching the FTF consensus binding site, T/CCAAGGTCA/G (HBV-FTF#2), and the other with one mismatch, CCAAGGTCt (HBV-FTF#1) (Fig. 1). To test these sequences for FTF binding in vitro, EMSAs were done with an oligonucleotide probe encompassing HBV-FTF#2 and total nuclear protein extract from Hep3B cells. Retarded protein-DNA complexes formed as expected for human FTF variants (13), and they were efficiently displaced with a 20-fold molar excess of unlabeled oligonucleotide HBV-FTF#1, HBV-FTF#2, or AFP-FTF (FTFα) (Fig. 1, lanes 3, 5, and 7) but not by a 100-fold excess of mutant AFP-FTF oligonucleotide FTFm (Fig. 1, lane 8). Furthermore, the specific bands were supershifted by anti-FTF antibodies, with no effect by antibodies against the closely related SF1 protein (Fig. 1, lanes 9 and 10). These results confirmed that the HBV core promoter contains two high-affinity FTF-binding sequences in the close vicinity of binding sites for other liver-enriched transcription factors (C/EBP, HNF3, and HNF4) (Fig. 1). The HBV-FTF#2 site displayed greater affinity for FTF than the HBV-FTF#1 site or even the strong AFP-FTF site (Kd ≈0.3 nM) (13), as shown in Fig. 1 by bandshift displacements at a low excess of competitor (lanes 2, 4, and 6). Similar EMSA results were obtained using nuclear protein extracts from PLC/PRF/5 cells (data not shown). EMSA analysis of HepG2 cells, however, revealed that HepG2 cells contain relatively little FTF and, instead, ectopically express abundant amounts of SF1. This was shown in reactions using specific anti-FTF or anti-SF1 antibodies (Fig. 1, lanes 12 and 13; see also Fig. 9A in reference 13) and confirmed by reverse transcription-PCR analyses (our unpublished results). As expected from their identical DNA-binding protein domains (13), FTF and SF1 were similar in specificity and affinity for HBV-FTF#1 or HBV-FTF#2.
FTF (or SF1) strongly activates the nucleocapsid promoter.
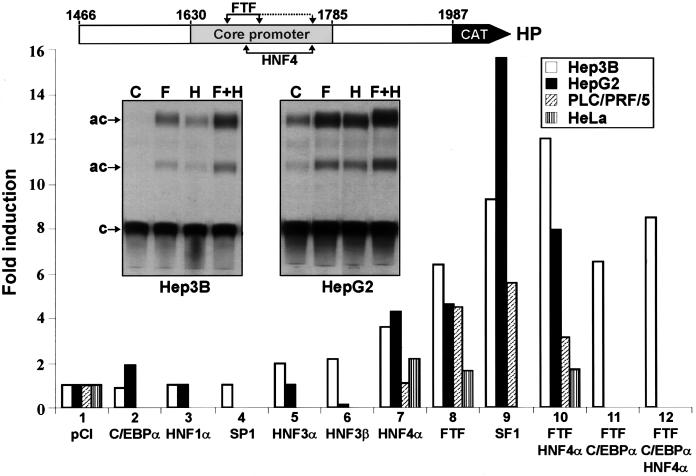
The putative FTF regulatory effect on HBV core promoter activity was assessed by transient-transfection assays with HepG2, Hep3B, and PLC/PRF/5 hepatoma cells, three human lines known to support transcription of the HBV genome (8, 19, 21, 36). We first tested the nucleocapsid promoter in a natural 0.5-kb context of contiguous DNA, without enhancer I sequences (reporter construct HP; Fig. 2). In all three hepatoma lines, cotransfection of HP with the FTF expression vector resulted in marked stimulation of HP activity (4.5- to 6.5-fold; Fig. 2, lane 8). Transfection of SF1 resulted in even stronger induction (Fig. 2, lane 9), especially in HepG2 cells (16-fold), where ectopic SF1 is already abundant (Fig. 1). Other transcription factors were then tested for coregulatory effects with FTF. Transcription factor HNF4α produced fourfold enhancement of HP reporter activity in Hep3B or HepG2 cells, which is consistent with previous studies (16, 33), and in both lines, activation by FTF and HNF4α was additive (Fig. 2, lanes 7, 8, and 10). In PLC/PRF/5 cells (less differentiated than Hep3B or HepG2 cells), the HP construct reacted poorly to HNF4α or to FTF plus HNF4α (Fig. 2, lanes 7 and 10), and in HeLa cells, only marginal stimulatory effects were observed with FTF and/or HNF4α (Fig. 2, lanes 7, 8, and 10). A well-differentiated hepatocytic environment therefore seems to be needed for efficient use of FTF and HNF4α. Factors C/EBPα, HNF1α, and HNF3α were tested in HepG2 and Hep3B cells, and factors HNF3β and SP1 were tested in Hep3B cells. C/EBPα, HNF3α, and HNF3β had some stimulatory effects (twofold; Fig. 2, lanes 2, 5, and 6), but none of the tested factors added significantly to the activation effect of FTF or HNF4α (Fig. 2, lane 11; data not shown); instead, they generally resulted in lower activation by FTF and/or HNF4α (Fig. 2, lane 12; data not shown). Thus, FTF and HNF4α were clearly the most efficient partners in coactivating the nucleocapsid promoter, which suggests productive co-occupation of their tandem binding sites (Fig. 1).
FIG. 2.
Transient-transfection assays using CAT reporter construct HP (5 μg) cotransfected with transcription factor expression vectors (each at 10 μg). Results are averages of three or four sets of duplicate or triplicate transfections, referred to control vector pCl run in parallel in each experiment and given a value of 1. Autoradiograms show CAT assays from HP cotransfection with void vector pCl (C) or expression vector FTF (F), HNF4α (H), or FTF plus HNF4α (F+H). c, chloramphenicol; ac, acetylated products. Basal HP activity in Hep3B cells, lane C, was easily detected with longer exposure.
FTF directly activates the core promoter.
To establish if FTF (or SF1) induced the core promoter via sequences FTF#1 and/or FTF#2, we used mutant reporter constructs HPm1, HPm2, and HPm12. In Hep3B cells, basal promoter activity of HPm1 or HPm2 was not significantly reduced, whereas HPm12 was inhibited by 30%. In HepG2 cells, HPm1 was unaffected while HPm2 was reduced by 50% and HPm12 was reduced by 80%. These results confirmed the use of FTF/SF1 by the core promoter under basal cell conditions, indicating a more active role for FTF#2 and the use of both FTF#1 and FTF#2 for optimal core promoter function. Mutation of FTF#1 or FTF#2 also reduced HP activation by exogenous FTF in Hep3B cells, while activation of HPm2 was slightly reduced in HepG2 cells, and double mutation of FTF#1 and FTF#2 inhibited the response to FTF by 65% in Hep3B cells and 45% in HepG2 cells (Fig. 3, lanes 2 to 4). These results indicated again that the FTF#1 and FTF#2 sites had to be simultaneously occupied for maximal activation by FTF and also that FTF induction resulted largely from direct FTF action at its two promoter-binding sites. With only one FTF site intact, the core promoter response to exogenous FTF was clearly less affected in HepG2 cells than in Hep3B cells. This might relate to abundant endogenous SF1 in HepG2 cells, allowing greater saturation of a single site and perhaps more efficient use of SF1 coactivators present in HepG2; as noted, SF1 induces the core promoter far more efficiently than FTF in HepG2 cells (compare lanes 8 and 9 in Fig. 2). While the residual basal activity of HPm12 was easily explained by its composite promoter activation domain, the residual inducibility of HPm12 by FTF (Fig. 3, lane 4) suggested that FTF might also act indirectly via other promoter regulators or that vector HP contains other functional FTF promoter sites. The most likely candidate for the latter was HNF4#2, a DR1 hormone response element (AGGTCA repeat with a 1-nt spacer); nearly canonical DR1 motifs, such as HNF4#2, form avid binding sites for HNF4 and several other nuclear receptors (33, 44), but they are also recognized by FTF (EMSA reactions indicated about 10-fold lower affinity of HNF4#2 for FTF or SF1, compared to FTF#1; Fig. 1, lane 17). We then tested HP vectors mutated in HNF4#2 (HPm4) or in FTF#1, FTF#2, and HNF4#2 (HPm124). The basal activity of HPm4 was reduced by 40% in Hep3B cells and 85% in HepG2 cells, and that of HPm124 was reduced by 60% in Hep3B cells and to an undetectable level in HepG2 cells. Induction of HPm4 by coexpressed FTF was also reduced by 60% in Hep3B cells (Fig. 3, lane 5), and FTF coexpression had only a negligible effect on HPm124 in Hep3B cells and no effect in HepG2 cells (Fig. 3, lane 6). We further tested a minimal (nt 1617 to nt 1755) upstream promoter construct lacking FTF#1 and HNF4#2 (HPΔ; Fig. 3). HPΔ responded to FTF like construct HPm1 (Fig. 3, lanes 2 and 7); when HPΔ was further mutated at the FTF#2 site (HPΔm2), all induction by FTF was again essentially eliminated (Fig. 3, lane 8). These combined results confirmed the importance of FTF#2 and were consistent with significant use of HNF4#2 by FTF/SF1 when FTF is overexpressed and especially when its higher-affinity sites are unavailable (HPm12). Under our assay conditions, it then appeared that all core promoter activation by FTF could be accounted for by its direct interaction with three FTF-binding sequences. The exact physiological role of FTF binding to HNF4#2 remains to be seen, given the abundance and higher affinity of alternate receptors competing for the DR1 motif under steady-state conditions (33, 44).
FTF effect on core promoter interactions with enhancer I.
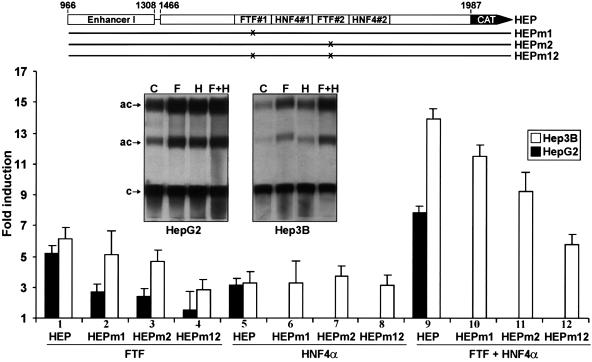
FTF effects were also examined in the context of genomic transactions between the core promoter and its cognate enhancer I domain (19, 35, 37). While the basal activity of construct HEP was about 10-fold higher than that of HP (in HepG2 or Hep3B cells) (autoradiograms in Fig. 2 and 4), FTF induction of HEP or HP was essentially the same (five- to sixfold) (Fig. 4, lane 1, versus Fig. 2, lane 8); this suggested that FTF induction of HEP was entirely due to FTF binding at the core promoter. Mutations of FTF#1 and/or FTF#2 also reduced HEP induction by FTF (Fig. 4, lanes 2 to 4), indicating that both FTF#1 and FTF#2 participated in the interaction between the core promoter and enhancer I. Contrasting with FTF, coexpression of HNF4α resulted in three- to fourfold activation of HEP or its FTF site mutants (Fig. 4, lanes 5 to 8); additive effects of FTF plus HNF4α were also proportionately maintained with all HEP constructs (Fig. 4, lanes 9 to 12). These results clearly showed that HNF4α induction of the core promoter can proceed independently from FTF/SF1. The induction by HNF4α was similar for HEP and HP constructs (Fig. 4, lanes 5 and 9; versus Fig. 2, lanes 7 and 10), also suggesting that HNF4α, like FTF, has little effect (direct or indirect) on the enhancer I segment. This is consistent with previous conclusions (33). While cotransfected FTF plus HNF4α raised HEP activity 2 logs over basal HP activity, it is noteworthy that double FTF site mutation m12 (reducing basal HP activity by as much as 80%) had little effect on the basal enhancer-promoter activity of HEP (less than 1.4-fold decrease). It is apparent that without FTF/SF1, HBV can still mount highly productive transactions with enhancer I using other core promoter factors.
FIG. 4.
FTF effects on HBV core promoter-enhancer I activity. HEP reporters (5 μg) were cotransfected with 10 μg of FTF and/or 10 μg of HNF4α expression vectors (HNF4α with HEP mutants was tested in Hep3B only; lanes 6 to 8 and 10 to 12). Results are averages (±1 standard deviation) of three sets of duplicate or triplicate transfections, referred to pCl transfection, which was given a value of 1. Inset autoradiograms illustrate CAT activities recovered from HEP cotransfection with pCl (C), FTF (F), and/or HNF4α (H). c, chloramphenicol; ac, acetylated products.
AF2-truncated FTF deactivates the core promoter.
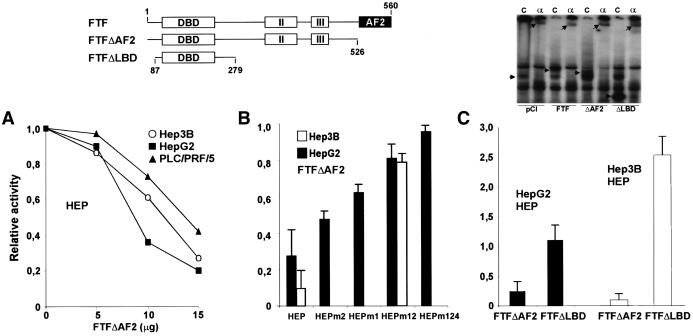
The carboxy-terminal region of the FTF protein (the activation function 2 domain) contains the hexameric amino acid motif LLIEML that is found in many other nuclear receptors and is critical for their activation. Previously, we found that AF2-truncated FTF (FTFΔAF2; diagrammed in Fig. 5) strongly suppresses AFP promoter activity in transfection assays (presumably by competing out endogenous FTF with the transcriptionally inert FTF mutant) (13). FTFΔAF2 was tested for similar dominant negative effects on basal HEP activity. In HepG2, Hep3B, or PLC/PRF/5 cells, FTFΔAF2 strongly inhibited HEP activity, down to less than 10% (Fig. 5A to C). To assess competitive effects of FTFΔAF2 at the FTF-binding sites, further assays were conducted with HEPm1, HEPm2, and HEPm12. At a concentration of FTFΔAF2 decreasing HEP activity by 70% in HepG2 cells, double FTF site mutant HEPm12 was repressed only 20% (Fig. 5B); in Hep3B cells, FTFΔAF2 (20 μg) reduced HEP activity more than 90% whereas HEPm12 activity was reduced less than 30% (Fig. 5B). These data thus support a competitive mechanism for deactivation of the nucleocapsid promoter, replacing endogenous SF1/FTF with transcriptionally nonfunctional FTFΔAF2 bound to the core promoter. Residual repression of HEPm12 by FTFΔAF2 further suggested that FTFΔAF2 might also compete out activators from the HNF4#2 site. This was tested with HepG2 cells and vector HEPm124, which had low (≈3% of that of HEP) but measurable basal activity, and no significant repression by FTFΔAF2 was found (Fig. 5B). Remarkably, deactivation of HEP by FTFΔAF2 appeared to be unsaturated under our assay conditions (Fig. 5A) and it clearly exceeded the effect expected by eliminating FTF/SF1 from enhancer-promoter transactions (as noted from the marginal change in basal levels of HEP versus HEPm12). Our interpretation is that defective FTF brought onto the core promoter disrupts alternative interactions of other promoter factors with enhancer I. Steric hindrance, more than DNA binding per se, seems to be at play, since a shorter FTF deletion mutant (FTFΔLBD) had no repressive effect and even enhanced HEP activity (Fig. 5C).
FIG. 5.
Repression of HBV enhancer-promoter activity by AF2-truncated FTF (FTFΔAF2 in the upper diagram). DBD, DNA-binding domain; II and III, receptor homology regions II and III; AF2, activation function 2. (A) Reporter construct HEP (5 μg) was cotransfected with 30 μg of the control vector pCl (reference value of 1.0) or with increasing amounts of the expression vector pClmFTFΔAF2 complemented to 30 μg with pCl. The results shown are average CAT activities recovered from three sets of duplicate transfections. (B) HepG2 cells were cotransfected with 5 μg of HEP constructs and 10 μg of pClmFTFΔAF2 or 10 μg of pCl (reference value of 1.0); Hep3B cells were cotransfected with 5 μg of HEP and 20 μg of pClmFTFΔAF2 or pCl. Results are average CAT activities (±1 standard deviation) of three triplicate transfections. (C) Cotransfections in HepG2 or Hep3B cells using 5 μg of the HEP vector and 20 μg of pClmFTFΔAF2, pClrFTFΔLBD, or pCl (reference value of 1.0). The results shown are averages (±1 standard deviation) of two sets of triplicate transfections. The autoradiogram shows bandshift assays conducted with the FTF#2 probe and nuclear protein extracts from Hep3B cells transfected with control or hemagglutinin-tagged FTF expression vectors. Lanes: C, reactions without antibodies; α, reactions with anti-FTF antibodies (upper arrows point to supershifted bands; the leftmost arrow points to endogenous FTF, and the other arrows point to the exogenous FTF products).
DISCUSSION
This study identified nuclear receptor FTF as one component operating the HBV nucleocapsid promoter, using two high-affinity FTF-binding sites for basal core promoter activity and its productive interaction with enhancer I. This is in line with early findings (46) that basal activation of the core promoter is mainly provided by the DNA segment including nts 1636 to 1703 (carrying the two FTF sites) and that removing segments FTF#1 (nt 1648 to 1668 or 1645 to 1656) or FTF#2 (nts 1687 to 1703 or 1679 to 1719) markedly reduces basic core promoter activity (10, 29, 46). Other evidence supporting the use of both the FTF#1 and FTF#2 sites includes the observation that HBV strain variants seem never to mutate FTF#2, while FTF#1 is also rarely mutated and the segment including nts 1644 to 1666 (carrying FTF#1) is frequently duplicated (15, 32): this clearly indicates a selective advantage to keeping or amplifying the two FTF sites. (It can also be noted that in HCC samples obtained from populations in which HBV is endemic, the most common gain of genetic material [≈75% of the cases] occurs by amplification of chromosome 1q [22, 40], carrying the FTF locus [14, 28]. Maintaining or enhancing FTF gene expression plausibly provides an advantage to HBV in neoplastic progression.) Recently, another group found that FTF#2 is footprinted by a liver-enriched nuclear factor, which led to the independent cloning of human FTF and to transfection assays concluding that FTF#2 is the sole element used by FTF to activate the nucleocapsid promoter (28). These experiments, however, used an HBV reporter construct from which FTF#1 was deleted and also used HeLa cells, in which transfected FTF may not make efficient use of its lower-affinity site (HNF4#2). Independent hFTF expression cloning was also achieved by yeast one-hybrid screening with the HBV segment including nts 1640 to 1663 (carrying the FTF#1 motif), and transfection assays conducted with hepatoma cells were also consistent with activation of the nt 1640 to 1663 segment by FTF (18).
Among the other factors tested here, HNF4α was clearly the most efficient FTF partner for costimulation of the core promoter. These two single factors, highly expressed together only in hepatocytes, may thus largely account for the hepatotropism of HBV infection. However, the core promoter also clearly needs other liver factors, since FTF and HNF4α were poorly effective in HeLa cells. The other factors tested had low activatory effects and, in fact, reduced the action of FTF and/or HNF4α. As suggested by others (29), this may reflect protein displacement from overlapping chromatin domains and suboptimal use of the stronger FTF/HNF4 activatory sites. Spacing between their four high-affinity binding sites would predictably avoid binding interference between FTF and HNF4 but not that between most of the other combinations of factors tested (Fig. 1).
FTF action could then be exerted on three functional domains intertwined in the nt 1630 to 1820 HBV segment, the C and preC promoters and enhancer II (Fig. 2) (30, 38, 42, 45). Ting's group (46) has shown that the DNA region including nts 1636 to 1703 coordinately enhances the synthesis of preC and C transcripts; it therefore seemed likely that FTF would activate both the C and preC promoters, and this was recently borne out in HBV/FTF cotransfection experiments conducted with HuH7 cells (18). One functional difference between the preC and C promoter domains is that the preC promoter is repressed by HNF4 (presumably because HNF4#2 overlaps the preC TATA-like sequence) (44). Additive effects of FTF and HNF4α might thus be taken as FTF being principally involved with pregenomic core promoter function and playing a particularly important role in the early life cycle and systemic load of HBV. This would also be consistent with the apparent lack of FTF recognition sequences in other HBV regulatory domains and would not preclude the possibility that the core promoter could use alternate factors at later stages of infection to avoid excessive squelching of FTF or because new viral products would favor other factors (10). The enhancer II issue is also intricate. Although evidence has been produced (45) for full enhancer II effects using nts 1646 to 1668 and 1704 to 1715 (i.e., bypassing FTF, HNF4, and HNF3 sites), enhancer II has more generally been defined as carrying one or both of the FTF recognition sequences (30, 36, 38, 42); furthermore, point mutations have indicated that HNF3#2 is essential both to core promoter activity and to enhancer II effects on a heterologous promoter (27). It thus seemed reasonable to think that FTF might likewise serve both core promoter and enhancer II functions, and again, recent differential analyses of HBV transcripts following HBV/FTF cotransfections indicate that such is the case (18).
The present work adds to the growing evidence that HBV regulatory domains can use alternate sets of factors to adapt efficiently to changing hepatocytic states. It also illustrates some limits of HBV transfection analyses using hepatoma cells. Here, double FTF site promoter mutants were only marginally affected in the presence of enhancer I, which may be quite misleading with regard to the actual role of FTF with intact functional sites at preneoplastic stages. Also, a high SF1 level with a low C/EBPα level (12) in HepG2 cells might mimic high pregenomic activity at early viral stages, whereas a lower FTF-to-C/EBPα ratio in Hep3B cells could rather mimic viral load conditions favoring alternate core activators or enhancer II activity. The very finding of SF1 in HepG2 cells also illustrates how aberrant gene expression in tumor cells may obscure homeostatic processes occurring in hepatocytes. In spite of these intricacies, the bulk of current data seems to make a compelling case for a preponderant role of HNF4 and FTF in tightly controlling the pregenomic core promoter and, hence, early amplification of HBV infection. In that regard, aggressive strains of HBV frequently convert HNF4#2 to a high-affinity HNF1 site (15, 20, 26), also showing the selective advantage of a genetic element that escapes potential negative regulators of HNF4#2 (44).
Resolution of whether receptor signalization pathways reaching HNF4 or FTF can now be effectively manipulated against HBV infection remains a challenging prospect, considering how HBV could switch activators and adapt to new liver conditions. Also, while specific signals conveyed by FTF to the AFP promoter might plausibly be sensed by the HBV core promoter, certainly not all FTF-dependent AFP functions are reproduced in the HBV context; glucocorticoids, in particular, inhibit AFP via FTF (13) but had no discernible FTF-dependent effects on our HBV constructs. The increasing diversity of FTF-inducible genes (25, 31, 34; Paré et al., Abstr. 8th Biennial Int. Congr. Liver Dev. Gene Regul. Dis.) also suggests that FTF may well respond to different signals to optimize its action at a given locus, none of which signals, however, may be sufficient to quench FTF activity if interrupted. The present work also predicts limited success of antisense or similar strategies simply removing FTF from action. More encouraging results were obtained here with mutant FTFΔAF2, causing dramatic inhibition of HBV enhancer-promoter functions. This effect clearly differs from simply removing FTF/SF1 from action since no comparable decrease was incurred in basal HEP activity by a mutation eliminating the two high-affinity FTF-binding sites. Notably, the shorter FTFΔLBD mutant did not repress HBV whereas it totally suppressed AFP promoter activity (13) (presumably because FTF at the AFP locus is essential to couple the AFP promoter with its distal enhancer) (6, 39). Our interpretation favors steric hindrance whereby the longer FTF mutant occupies the HBV core promoter as an inert complex that also hinders alternative protein interactions with the enhancer domain. Dominant negative factors may thus create new opportunities to tackle functional redundancies of HBV regulatable functions; liver cells might also be spared from deleterious dominant negative effects by alternating HBV target sites. Conceivably, adenovirus or adeno-associated virus vectors could serve to drive dominant negative effectors into hepatocytes in vivo; a recently described preclinical murine model of HBV infection (23) might be particularly useful to address this type of therapeutical strategy.
ACKNOWLEDGMENTS
This work was supported by a studentship (S.G.) and grant MT-6478 (L.B.) from the Medical Research Council of Canada.
We thank A. Siddiqui, R. Costa, F. Sladek, M. V. Govindan, J. Milbrandt, and A. Anderson for providing expression plasmids; Lise Lévesque and Julie Vézina for collaboration; Manuel Caruso for helpful discussions; and Marie-France Voyer and Denise Rioux for excellent secretarial assistance.
REFERENCES
- 1.Araki K, Miyazaki J-I, Hino O, Tomita N, Chisaka O, Matsubara K, Yamamura K-I. Expression and replication of hepatitis B virus genome in transgenic mice. Proc Natl Acad Sci USA. 1989;86:207–211. doi: 10.1073/pnas.86.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babinet C, Farza H, Morello D, Hadchouel M, Pourcel C. Specific expression of hepatitis B surface antigen (HBsAg) in transgenic mice. Science. 1985;230:1160–1163. doi: 10.1126/science.3865370. [DOI] [PubMed] [Google Scholar]
- 3.Beasley R P, Hwang L Y, Lin C C, Chien C S. Hepatocellular carcinoma and hepatitis B virus. A prospective study of 22,707 men in Taiwan. Lancet. 1981;ii:1129–1133. doi: 10.1016/s0140-6736(81)90585-7. [DOI] [PubMed] [Google Scholar]
- 4.Bélanger L, Hamel D, Lachance L, Dufour D, Tremblay M, Gagnon P M. Hormonal regulation of α1 foetoprotein. Nature. 1975;256:657–659. doi: 10.1038/256657a0. [DOI] [PubMed] [Google Scholar]
- 5.Bélanger L, Roy S, Allard D. New albumin gene 3′ adjacent to the α1-fetoprotein locus. J Biol Chem. 1994;269:5481–5484. [PubMed] [Google Scholar]
- 6.Bernier D, Thomassin H, Allard D, Guertin M, Hamel D, Blaquière M, Beauchemin M, LaRue H, Estable-Puig M, Bélanger L. Functional analysis of developmentally regulated chromatin-hypersensitive domains carrying the α1-fetoprotein gene promoter and the albumin/α1-fetoprotein intergenic enhancer. Mol Cell Biol. 1993;13:1619–1633. doi: 10.1128/mcb.13.3.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blumberg B S. The current state of the prevention of HBV infection, the carrier state and hepatocellular carcinoma. Res Virol. 1997;148:91–94. doi: 10.1016/s0923-2516(97)89890-1. [DOI] [PubMed] [Google Scholar]
- 8.Chang C, Jeng K-S, Hu C-P, Lo S J, Su T-S, Ting L-P, Chou C-K, Han S-H, Pfaff E, Salfeld J, Schaller H. Production of hepatitis B virus in vitro by transient expression of cloned HBV DNA in a hepatoma cell line. EMBO J. 1987;6:675–680. doi: 10.1002/j.1460-2075.1987.tb04807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang M H, Chen C J, Lai M S, Hsu H M, Wu T C, Kong M S, Liang D C, Shau W Y, Chen D S. Universal hepatitis B vaccination in Taiwan and the incidence of hepatocellular carcinoma in children. Taiwan childhood hepatoma study group. N Engl J Med. 1997;336:1855–1859. doi: 10.1056/NEJM199706263362602. [DOI] [PubMed] [Google Scholar]
- 10.Choi B H, Park G T, Rho H M. Interaction of hepatitis B viral X protein and CCAAT/enhancer-binding protein α synergistically activates the hepatitis B viral enhancer II/pregenomic promoter. J Biol Chem. 1999;274:2858–2865. doi: 10.1074/jbc.274.5.2858. [DOI] [PubMed] [Google Scholar]
- 11.Dunsford H A, Sell S, Chisari F V. Hepatocarcinogenesis due to chronic liver cell injury in hepatitis B virus transgenic mice. Cancer Res. 1990;50:3400–3407. [PubMed] [Google Scholar]
- 12.Friedman A D, Landschulz W H, McKnight S L. CCAAT/enhancer binding protein activates the promoter of the serum albumin gene in cultured hepatoma cells. Genes Dev. 1989;3:1314–1322. doi: 10.1101/gad.3.9.1314. [DOI] [PubMed] [Google Scholar]
- 13.Galarneau L, Paré J-F, Allard D, Hamel D, Lévesque L, Tugwood J D, Green S, Bélanger L. The α1-fetoprotein locus is activated by a nuclear receptor of the Drosophila FTZ-F1 family. Mol Cell Biol. 1996;16:3853–3865. doi: 10.1128/mcb.16.7.3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galarneau L, Drouin R, Bélanger L. Assignment of the fetoprotein transcription factor gene (FTF) to human chromosome band 1q32.11 by in situ hybridization. Cytogenet Cell Genet. 1998;82:269–270. doi: 10.1159/000015116. [DOI] [PubMed] [Google Scholar]
- 15.Günther S, Piwon N, Iwanska A, Schilling R, Meisel H, Will H. Type, prevalence, and significance of the core promoter/enhancer II mutations in hepatitis B viruses from immunosuppressed patients with severe liver disease. J Virol. 1996;70:8318–8331. doi: 10.1128/jvi.70.12.8318-8331.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo W, Chen M, Yen T S B, Ou J-H. Hepatocyte-specific expression of the hepatitis B virus core promoter depends on both positive and negative regulation. Mol Cell Biol. 1993;13:443–448. doi: 10.1128/mcb.13.1.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ikeda Y, Lala D S, Luo X, Kim E, Moisan M-P, Parker K L. Characterization of the mouse FTZ-F1 gene, which encodes a key regulator of steroid hydroxylase gene expression. Mol Endocrinol. 1993;7:852–860. doi: 10.1210/mend.7.7.8413309. [DOI] [PubMed] [Google Scholar]
- 18.Ishida H, Ueda K, Ohkawa K, Kanazawa Y, Hosui A, Nakanishi F, Mita E, Kasahara A, Sasaki Y, Hori M, Hayashi N. Identification of multiple transcription factors, HLF, FTF, and E4BP4, controlling hepatitis B virus enhancer II. J Virol. 2000;74:1241–1251. doi: 10.1128/jvi.74.3.1241-1251.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jameel S, Siddiqui A. The human hepatitis B virus enhancer requires trans-acting cellular factor(s) for activity. Mol Cell Biol. 1986;6:710–715. doi: 10.1128/mcb.6.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kidd-Ljunggren K, Öberg M, Kidd A H. Hepatitis B virus X gene 1751 to 1764 mutations: implications for HBeAg status and disease. J Gen Virol. 1997;78:1469–1478. doi: 10.1099/0022-1317-78-6-1469. [DOI] [PubMed] [Google Scholar]
- 21.Knowles B B, Howe C C, Aden D P. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science. 1980;209:497–499. doi: 10.1126/science.6248960. [DOI] [PubMed] [Google Scholar]
- 22.Kusano N, Shiraishi K, Kubo K, Oga A, Okita K, Sasaki K. Genetic aberrations detected by comparative genomic hybridization in hepatocellular carcinomas: their relationship to clinicopathological features. Hepatology. 1999;29:1858–1862. doi: 10.1002/hep.510290636. [DOI] [PubMed] [Google Scholar]
- 23.Larkin J, Clayton M, Sun B, Perchonock C E, Morgan J L, Siracusa L D, Michaels F H, Feitelson M A. Hepatitis B virus transgenic mouse model of chronic liver disease. Nat Med. 1999;5:907–912. doi: 10.1038/11347. [DOI] [PubMed] [Google Scholar]
- 24.Lee W M. Hepatitis B virus infection. N Engl J Med. 1997;337:1733–1745. doi: 10.1056/NEJM199712113372406. [DOI] [PubMed] [Google Scholar]
- 25.Lee Y-K, Parker K L, Choi H-S, Moore D D. Activation of the promoter of the orphan receptor SHP by orphan receptors that bind DNA as monomers. J Biol Chem. 1999;274:20869–20873. doi: 10.1074/jbc.274.30.20869. [DOI] [PubMed] [Google Scholar]
- 26.Li J, Buckwold V E, Hon M-W, Ou J-H. Mechanism of suppression of hepatitis B virus precore RNA transcription by a frequent double mutation. J Virol. 1999;73:1239–1244. doi: 10.1128/jvi.73.2.1239-1244.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li M, Xie Y, Wu X, Kong Y, Wang Y. HNF3 binds and activates the second enhancer, ENII, of hepatitis B virus. Virology. 1995;214:371–378. doi: 10.1006/viro.1995.0046. [DOI] [PubMed] [Google Scholar]
- 28.Li M, Xie Y-H, Kong Y-Y, Wu X, Zhu L, Wang Y. Cloning and characterization of a novel human hepatocyte transcription factor, hB1F, which binds and activates enhancer II of hepatitis B virus. J Biol Chem. 1998;273:29022–29031. doi: 10.1074/jbc.273.44.29022. [DOI] [PubMed] [Google Scholar]
- 29.López-Cabrera M, Letovsky J, Hu K-Q, Siddiqui A. Multiple liver-specific factors bind to the hepatitis B virus core/pregenomic promoter: trans-activation and repression by CCAAT/enhancer binding protein. Proc Natl Acad Sci USA. 1990;87:5069–5073. doi: 10.1073/pnas.87.13.5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.López-Cabrera M, Letovsky J, Hu K-Q, Siddiqui A. Transcriptional factor C/EBP binds to and transactivates the enhancer element II of the hepatitis B virus. Virology. 1991;183:825–829. doi: 10.1016/0042-6822(91)91019-d. [DOI] [PubMed] [Google Scholar]
- 31.Nitta M, Ku S, Brown C, Okamoto A Y, Shan B. CPF: an orphan nuclear receptor that regulates liver-specific expression of the human cholesterol 7α-hydroxylase gene. Proc Natl Acad Sci USA. 1999;96:6660–6665. doi: 10.1073/pnas.96.12.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pult I, Chouard T, Wieland S, Klemenz R, Yaniv M, Blum H E. A hepatitis B virus mutant with a new hepatocyte nuclear factor 1 binding site emerging in transplant-transmitted fulminant hepatitis B. Hepatology. 1997;25:1507–1515. doi: 10.1002/hep.510250633. [DOI] [PubMed] [Google Scholar]
- 33.Raney A K, Johnson J L, Palmer C N A, McLachlan A. Members of the nuclear receptor superfamily regulate transcription from the hepatitis B virus nucleocapsid promoter. J Virol. 1997;71:1058–1071. doi: 10.1128/jvi.71.2.1058-1071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rausa F, Galarneau L, Bélanger L, Costa R. The nuclear receptor fetoprotein transcription factor is coexpressed with its target gene HNF-3β in the developing murine liver intestine and pancreas. Mech Dev. 1999;89:185–188. doi: 10.1016/s0925-4773(99)00209-9. [DOI] [PubMed] [Google Scholar]
- 35.Shaul Y, Rutter W J, Laub O. A human hepatitis B viral enhancer element. EMBO J. 1985;4:427–430. doi: 10.1002/j.1460-2075.1985.tb03646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su H, Yee J-K. Regulation of hepatitis B virus gene expression by its two enhancers. Proc Natl Acad Sci USA. 1992;89:2708–2712. doi: 10.1073/pnas.89.7.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tognoni A, Cattaneo R, Serfling E, Schaffner W. A novel expression selection approach allows precise mapping of the hepatitis B virus enhancer. Nucleic Acids Res. 1985;13:7457–7472. doi: 10.1093/nar/13.20.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Chen P, Wu X, Sun A-L, Wang H, Zhu Y-A, Li Z-P. A new enhancer element, ENII, identified in the X gene of hepatitis B virus. J Virol. 1990;64:3977–3981. doi: 10.1128/jvi.64.8.3977-3981.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wen P, Crawford N, Locker J. A promoter-linked coupling region required for stimulation of α-fetoprotein transcription by distant enhancers. Nucleic Acids Res. 1993;21:1911–1918. doi: 10.1093/nar/21.8.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wong N, Lai P, Lee S-W, Fan S, Pang E, Liew C-T, Sheng Z, Lau J W-Y, Johnson P J. Assessment of genetic changes in hepatocellular carcinoma by comparative genomic hybridization analysis. Relationship to disease stage, tumor size, and cirrhosis. Am J Pathol. 1999;154:37–43. doi: 10.1016/S0002-9440(10)65248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yaginuma K, Shirakata Y, Kobayashi M, Koike K. Hepatitis B virus (HBV) particles are produced in a cell culture system by transient expression of transfected HBV DNA. Proc Natl Acad Sci USA. 1987;84:2678–2682. doi: 10.1073/pnas.84.9.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yee J K. A liver-specific enhancer in the core promoter region of human hepatitis B virus. Science. 1989;246:658–661. doi: 10.1126/science.2554495. [DOI] [PubMed] [Google Scholar]
- 43.Yu X, Mertz J E. Promoters for synthesis of the pre-C and pregenomic mRNAs of human hepatitis B virus are genetically distinct and differentially regulated. J Virol. 1996;70:8719–8726. doi: 10.1128/jvi.70.12.8719-8726.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu X, Mertz J E. Differential regulation of the pre-C and pregenomic promoters of human hepatitis B virus by members of the nuclear receptor superfamily. J Virol. 1997;71:9366–9374. doi: 10.1128/jvi.71.12.9366-9374.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuh C-H, Ting L-P. C/EBP-like proteins binding to the functional box-α and box-β of the second enhancer of hepatitis B virus. Mol Cell Biol. 1991;11:5044–5052. doi: 10.1128/mcb.11.10.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuh C-H, Chang Y-L, Ting L-P. Transcriptional regulation of precore and pregenomic RNAs of hepatitis B virus. J Virol. 1992;66:4073–4084. doi: 10.1128/jvi.66.7.4073-4084.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]