Abstract
The plus-sense RNA genome of Japanese encephalitis virus (JEV) contains noncoding regions (NCRs) of 95 and 585 bases at its 5′ and 3′ ends, respectively. The last 83 nucleotides of the 3′-NCR are predicted to form stable stem-loop (SL) structures. The shape of this 3′-SL structure is highly conserved among divergent flaviviruses even though only small stretches of nucleotide sequence contained within these structures are conserved. These SL structures have been predicted to function as cis-acting signals for RNA replication and as such may bind to viral and cellular proteins that may be involved in viral replication. We have studied the interaction of the JEV 3′-NCR RNA with host proteins using gel retardation assays. We show that the JEV 3′-SL structure RNA forms three complexes with proteins from the S100 cytoplasmic extract prepared from the neonatal mouse brain. These complexes could be obtained in the presence of 200 mM KCl, indicating that the RNA-protein interaction may be physiologically relevant. UV-induced cross-linking and Northwestern blotting analyses detected three proteins with apparent molecular masses of 32, 35, and 50 kDa that bound to the JEV 3′-SL structure RNA. Screening of the neonatal mouse brain cDNA library with the JEV 3′-SL structure RNA identified a 36-kDa Mov34 protein interacting with it. Competition experiments using the RNA extracted from JEV virions established that the 36-kDa Mov34 protein indeed bound to the JEV genome. Murine Mov34 belongs to a family of proteins whose members have been shown to be involved in RNA transcription and translation. It is, therefore, likely that the murine Mov34 interaction with JEV 3′-NCR has a role in RNA replication.
Japanese encephalitis virus (JEV) is a flavivirus with a plus-sense, single-stranded RNA genome of ∼11 kb. The genomic RNA contains a single open reading frame capable of encoding a polyprotein of ∼3,400 amino acids which is subsequently cleaved into three structural (capsid, pre-M, and envelope) and seven nonstructural (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) proteins. The coding region in the genome is flanked by 5′ and 3′ noncoding regions (NCRs) of 95 and 585 bases (63). The size and sequence of the 3′-NCR vary among different flaviviruses although its secondary structure comprising stable stem-loop (SL) formations is predicted to be highly conserved. The last 80 to 90 bases at the extreme 3′ end of the genome have been predicted to form stable SL structures in different flaviviruses (10, 11, 63, 65). There are only small stretches of nucleotide sequences, mostly located in the loop regions of these SL structures, that are conserved among flaviviruses (10). Conservation of the location and the shape of these SL structures in the 3′-NCR of flaviviruses indicates that they may have some functional significance.
During the course of flavivirus replication, a minus-sense RNA is synthesized from the plus-sense genomic RNA. This intermediate form of RNA serves as template for the synthesis of the plus-sense genomic RNA which is synthesized to 10- to 100-fold-higher levels than the minus-sense RNA (12, 62). Both the nucleotide sequence and the structural elements of the 3′-NCR such as the 3′-SL structure are predicted to contain cis-acting signals for the initiation of the viral RNA transcription, and thus, they may specifically interact with viral or cellular proteins during viral RNA replication (12, 71).
A number of host proteins have been shown to be associated with the RNA-dependent RNA polymerase of phage Qβ (9), brome mosaic virus (53), cucumber mosaic virus (34), tobacco mosaic virus (48), vesicular stomatitis virus (19), measles virus (44), influenza virus (46), and poliovirus (33, 43). Moreover, a variety of host proteins has been shown to interact with putative cis-acting elements of Sindbis virus (50), mouse hepatitis virus (40), hepatitis C virus (1, 16, 28, 36, 41, 66), rubella virus (51, 61), human immunodeficiency virus (6, 14, 64), and other RNA viruses (reviewed in reference 39). As for the flaviviruses, Blackwell and Brinton (8) have described binding of elongation factor 1α (EF-1α) to the 3′-SL region of the West Nile virus (WNV), which belongs to the same antigenic subgroup of flaviviruses as JEV (12). Recently, Chen et al. (15) have shown that the JEV nonstructural proteins NS3 and NS5 bind to the 3′-NCR. The NS5 protein, considered to be the RNA-dependent RNA polymerase, binds to the 83-nucleotide 3′-SL structure. However, it is not known if any of the cellular proteins interact with JEV 3′-NCR.
In this study, we have used a UV-induced protein-RNA cross-linking assay to investigate the host protein binding to the RNA representing the JEV 3′-SL structure. At least three proteins with apparent molecular masses of 32, 35, and 50 kDa from the mouse brain bound to the JEV 3′-NCR. Screening of a mouse brain cDNA expression library identified a 36-kDa Mov34 protein that bound to the JEV 3′-SL RNA. This protein belongs to a family of proteins whose members are involved in control of RNA transcription and translation.
MATERIALS AND METHODS
Virus and cells.
The GP78 strain of JEV (69, 70) was used in these studies. The virus was grown in porcine stable kidney (PS) cells obtained from the National Centre for Cell Sciences, Pune, India. PS cells were grown in Eagle's minimal essential medium (Gibco) supplemented with 10% fetal calf serum.
Construction of the cDNA clones.
Culture supernatant of JEV GP78-infected PS cells was used to isolate the viral RNA using the QIAmp viral RNA extraction kit (Qiagen) as described previously (69). Synthesis of cDNA to the 3′-NCR was carried out by a standard procedure (38) using avian myeloblastosis virus reverse transcriptase and a synthetic oligonucleotide, SV35 (TCTAGAGATCCTGTGTTCTTCCTCAC). The oligonucleotide contained an XbaI site (shown in italics) and a 21-nucleotide sequence (underlined) that was complementary to bases 10955 to 10976 located at the extreme 3′ end of the JEV RNA (69) (GenBank accession no. AF075723). A 582-nucleotide cDNA coding for the 3′-NCR RNA (spanning nucleotides 10395 to 10976) was PCR amplified using SV35 and an upstream synthetic oligonucleotide, SV52 (GGGCCCTGTGATTTAAAGTAGAAA), which contained an ApaI site followed by an 18-base JEV GP78 sequence (underlined). The PCR product was cloned in pGemT vector (Promega), and a plasmid clone in which the ApaI site of the vector and the insert were close together was identified. This recombinant plasmid was digested with ApaI to remove vector sequences and ligated to itself to generate pJE3NCR. The insert sequence and its orientation were confirmed by nucleotide sequencing. The ApaI site in this plasmid was close to the T7 promoter. Thus, RNA transcript made from this plasmid would contain a 15-nucleotide vector sequence at its 5′ end.
For the construction of pJE3SL, the cDNA made as described above was PCR amplified with SV35 and SV62 (GGGCCCGGAGATCTTCTGCTCTAT), which contained an ApaI site at its 5′ end followed by the JEV GP78 sequence between bases 10891 and 10908 (underlined). The PCR product was cloned at the ApaI site of pGemT vector as described above. The insert sequence and its orientation were confirmed by nucleotide sequencing. As the ApaI site in this plasmid was 15 bases away from the T7 transcription initiation site, the RNA transcript made from this plasmid would contain, at its 5′ end, a 15-nucleotide sequence derived from the vector.
Preparation of RNA transcripts.
For in vitro synthesis of RNA, purified plasmid DNA was linearized with XbaI. This would provide, in the synthesized RNA, a 3′ end precisely like that in the JEV genome. To generate 32P-labeled RNA, 1 μg of linearized plasmid DNA was transcribed in vitro with T7 RNA polymerase in a 20-μl reaction volume containing 40 mM Tris-Cl (pH 7.5), 6 mM MgCl2, 10 mM NaCl, 2 mM spermidine, 10 mM dithiothreitol (DTT), 0.5 mM (each) ribonucleotides (A, C, and G), 12 μM UTP, 50 μCi of [α-32P]UTP (3,000 Ci/mmol; NEN), and 20 U of RNasin (Promega). The reaction mixture was incubated for 2 h at 30°C. RNase-free DNase I (1 U/μg of DNA) was then added, and the reaction mixture was incubated at 37°C for 30 min. This was followed by one phenol-chloroform-isoamyl alcohol and one chloroform-isoamyl alcohol extraction. The aqueous phase containing the RNA was stored at −70°C. For synthesis of the cold RNA, the reaction was carried out essentially as described above except that 0.5 mM cold UTP replaced the radiolabeled UTP. RNA yield was determined by trichloroacetic acid precipitation. A specific activity of ∼108 cpm/μg was routinely obtained.
Preparation of cellular extracts.
Brain tissue from a suckling BALB/c mouse was harvested and washed three times with ice-cold phosphate-buffered saline. The tissue was then resuspended in cytolysis buffer (10 mM HEPES [pH 7.9], 5 mM DTT, 20% glycerol, 10 mM NaCl, 0.1 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin per ml, and 1% Triton X-100), vortexed for 30 s, and stored on ice for 15 min. The nuclei were removed by centrifugation at 2,000 × g for 5 min at 4°C. The supernatant was clarified by centrifugation at 100,000 × g for 30 min at 4°C. The resulting S100 supernatant was then transferred to a Centricon-30 microconcentrator and centrifuged as described by the manufacturer (Amicon). Concentration and buffer exchange were continued till the lysate volume was 100 to 200 μl in storage buffer containing 20 mM HEPES (pH 7.5), 100 mM NaCl, 2 mM MgCl2, 5 mM DTT, 0.1 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin per ml, and 50% glycerol. The cytoplasmic extract was stored at −70°C. The total protein concentration of the S100 extract was generally around 70 μg/μl.
Gel mobility shift assay.
About 10 μg of the S100 cytoplasmic extract was incubated in binding buffer (14 mM HEPES [pH 7.5], 6 mM Tris-Cl [pH 7.5], 1 mM EDTA, 1 mM DTT, and 60 mM KCl) with poly(I)-poly(C) (200 ng) and RNasin (1 U) in a final volume of 20 μl for 10 min at 30°C. After the addition of 3 to 6 ng of 32P-labeled RNA, the incubation was continued for 20 min at 30°C. RNA-protein complexes were electrophoresed at 4°C on a nondenaturing 5% polyacrylamide gel (acrylamide/bisacrylamide ratio, 50:1) containing 2.5% glycerol in 0.5× Tris-borate-EDTA buffer. The gel was then dried, and the complexes were visualized by autoradiography.
UV-induced cross-linking of RNA and protein.
The RNA-protein binding reaction was set up as described above. After incubation for 30 min at 30°C, the binding reaction mixture was transferred to an ice bath and irradiated with a short-wavelength (254-nm) UV lamp (4 W) held at a 3-cm distance from the reaction mixture for 30 min. After irradiation, RNase (2.5 U) was added, and the reaction mixture was incubated for 30 min at 37°C to digest unprotected RNA. The UV cross-linked products were boiled in Laemmli sample buffer for 3 min and analyzed on a discontinuous sodium dodecyl sulfate (SDS)–10% polyacrylamide gel (acrylamide/bisacrylamide ratio, 29:1). The gel was fixed in 7% acetic acid and dried. The complexes were visualized by autoradiography.
Northwestern analysis of RNA binding protein.
Protein samples were resolved on SDS–10% polyacrylamide gels and transferred to nitrocellulose membranes at 30 V for 16 h in Tris-glycine-methanol buffer at 4°C. Transferred proteins were renatured overnight at 4°C in binding buffer (14 mM HEPES [pH 7.5], 6 mM Tris-Cl [pH 7.5], 1 mM EDTA, 1 mM DTT, and 60 mM KCl) containing 1% bovine serum albumin and 16 μg of salmon testis DNA per ml. Membranes were then incubated for 30 min with 10 μg of yeast tRNA per ml at 30°C. After the addition of the 32P-labeled RNA probe, the incubation was continued for 2 h at 30°C. Incubations were carried out while shaking the membranes slowly. Nonspecifically bound radioactivity was removed by washing the membranes three times for 5 min each with binding buffer at room temperature. Protein-RNA binding was visualized by autoradiography.
cDNA library screening.
A commercially available cDNA library (Stratagene) made from BALB/c neonatal mouse brain in the Uni-ZAP XR vector was used. The library was amplified once in Escherichia coli strain XL1-Blue MRF′ and plated on NZY agar using the prescribed procedure. The plates were inverted and incubated at 42°C for 3 to 4 h until pinpoint plaques appeared. Nitrocellulose membranes were impregnated for 30 min in 20 mM isopropyl-β-d-thiogalactopyranoside (IPTG), overlaid on the plates, and incubated overnight at 37°C. The plates were chilled for 2 h at 4°C to prevent the top agarose from sticking to the nitrocellulose membrane and were air dried for 2 to 5 min on Whatman 3MM paper. The membranes were incubated at 30°C for 30 min in the binding buffer containing 14 mM HEPES (pH 7.5), 6 mM Tris-Cl (pH 7.5), 1 mM EDTA, 1 mM DTT, 60 mM KCl, and 10 μg of yeast tRNA per ml. This was followed by the addition of the probe (∼0.5 × 106 cpm/ml) and incubation at 30°C with shaking for 2 h. Nonspecifically bound radioactivity was removed by washing the membranes three times each for 5 min in the binding buffer. The membranes were air dried and exposed to X-ray film for 12 to 24 h at −70°C. Putative positive plaques were picked and purified to homogeneity in a secondary and tertiary screening round as described previously (54, 58).
Expression of histidine-tagged Mov34 in E. coli and preparation of rabbit antiserum.
Total RNA from neonatal mouse brain tissue was isolated using a commercially available RNA extraction kit (RNeasy; Qiagen). cDNA to the Mov34 gene was made using avian myeloblastosis virus reverse transcriptase and a synthetic oligonucleotide, SV173 (CAAAGCTTACTTTTTCTCCTTTTTCTC), which contained the HindIII restriction site (italics) and an 18-base complementary sequence (underlined) derived from the 3′ end of the Mov34 coding sequence (31). The Mov34 cDNA was PCR amplified using SV173 and SV172 (TGTGGATCCATGCCGGAGCTGGCGGTG), which contained Mov34-specific sequence from the 5′ end of the gene (underlined) and a BamHI restriction site (italics). The PCR was carried out using a high-fidelity enzyme mix containing the Taq and Pow polymerases (24). The PCR product was digested with restriction enzymes BamHI and HindIII and cloned in E. coli expression plasmid pQE30 (Qiagen) such that the fusion protein would contain six histidine residues at the N terminus. E. coli (M15) cells containing pQE30Mov34 plasmid were induced with IPTG, and the fusion protein was affinity purified using Ni-nitrilotriacetic acid resin (Qiagen). For preparation of the Mov34 antiserum, 100 μg of purified His-Mov34 was emulsified with complete Freund's adjuvant before injecting the rabbit intramuscularly. This was followed by a booster given a month later. The rabbit was bled 7 days later, and the serum was stored in aliquots at −70°C.
RESULTS
In vitro synthesis of RNA representing the 3′-NCR or the 3′-SL structure of JEV.
A 582-nucleotide cDNA coding for the 3′-NCR of JEV was cloned in pJE3NCR under the T7 promoter as described in Materials and Methods. The plasmid was linearized with XbaI such that the 3′ end of the in vitro-synthesized RNA would be precisely the same as in the JEV genome. Since the 3′-NCR sequence was 15 nucleotides away from the transcription initiation site, the RNA synthesized from pJE3NCR would have 15 non-JEV bases at its 5′ end derived from the vector sequence, thus giving a 597-base RNA. When transcription was carried out at 37°C, the majority of the RNA synthesized was ∼250 bases long. The full-length RNA synthesis was achieved when the transcription was carried out at 30°C. The RNA size was checked on a 5% polyacrylamide-urea gel (containing 8 M urea) using standard RNA markers (data not shown). The size of the RNA was also established by its reverse transcription and PCR amplification using oligonucleotides SV35 and SV52 (see Materials and Methods) and running the product on an agarose gel for size estimation (data not shown). A similar strategy, as described above, was used for transcription of RNA representing the 3′-SL structure of JEV. A 101-nucleotide RNA was produced, the size of which was confirmed by running it on a urea-polyacrylamide gel as described above (data not shown).
Mouse brain cell proteins bind to the 3′-NCR of JEV.
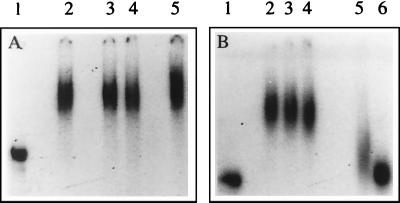
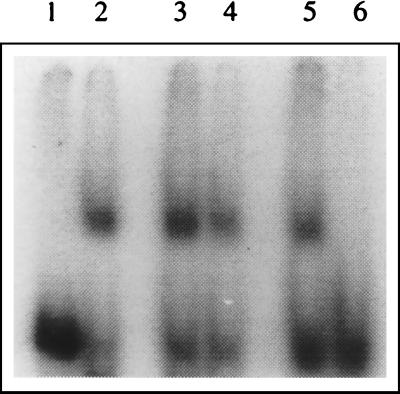
Extract from mouse brain cells was incubated with the 597-base radiolabeled RNA representing the 3′-NCR of JEV. The RNA-protein complex was resolved on a nondenaturing polyacrylamide gel (Fig. 1A). A distinct shift in RNA position was seen which indicated that the cellular protein(s) from mouse brain cells bound to the 3′-NCR of JEV. Specificity of the RNA-protein interaction was established by using nonspecific competitors such as yeast tRNA (100 ng to 10 μg) and poly(I)-poly(C) (200 ng to 1 μg) which were in great excess compared to the 3′-NCR RNA in binding reactions. These nonspecific RNAs did not interfere with the formation of the protein-RNA complex (Fig. 1A). However, an excess of unlabeled JEV 3′-NCR RNA used as a specific competitor inhibited RNA-protein complex formation (data not shown). A nonspecific transcript, made from the same vector (pGemT) as that used in the construction of pJE3NCR, did not inhibit the RNA-protein complex formation, indicating that the proteins were interacting with the 582-base JEV-specific RNA sequence in the 597-base transcript and not with the 15-base sequence that it contained from the vector backbone (data not shown). The RNA-protein interaction could be seen in the presence of as much as 200 mM KCl (Fig. 1B), indicating that the protein(s) bound to JEV 3′-NCR RNA with high affinity and the interaction may be physiologically relevant. The RNA-protein complex formation was not seen if the cell extract was treated with proteinase K before incubation with the 3′-NCR RNA (data not shown).
FIG. 1.
Binding of mouse brain cell proteins with the JEV 3′-NCR RNA. An RNA-protein binding reaction was carried out using 6 ng of radiolabeled 3′-NCR RNA (597 nucleotides long) and 10 μg of mouse brain cell S100 extract in the presence of different amounts of KCl. The RNA-protein complexes were resolved on a 5% polyacrylamide gel (acrylamide/bisacrylamide ratio, 120:1). (A) Lane 1, free probe; lanes 2, 3, and 4, binding in the presence of 200, 500, and 1,000 ng of poly(I)-poly(C), respectively; lane 5, binding in the presence of 10 μg of yeast tRNA. All of these binding reactions were carried out in the presence of 20 mM KCl. (B) Lane 1, free probe; lanes 2, 3, and 4, binding in the presence of 20, 60, and 200 mM KCl, respectively; lane 5, binding in the presence of 60 mM KCl and 1 ng of unlabeled 3′-SL RNA (101 nucleotides long); lane 6, binding in the presence of 60 mM KCl and 6 ng of unlabeled 3′-SL RNA. All of these binding reactions were carried out in the presence of 200 ng of poly(I)-poly(C).
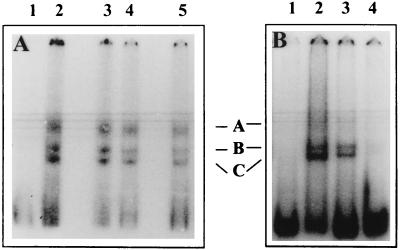
The RNA-protein complex seen above could not be resolved properly, as the RNA used in the binding reaction was 597 bp long. Therefore, it is not clear whether the complex contained one or more than one protein. In the case of WNV, Blackwell and Brinton (7) demonstrated that proteins from baby hamster kidney (BHK) cells bound to the 3′-SL structure of the viral RNA. We have, therefore, examined if the formation of the JEV 3′-NCR RNA-protein complex seen as described above was competed by the 3′-SL structure RNA of JEV. Figure 1B shows that 3′-NCR-RNA-protein complex formation was inhibited by the 101-base unlabeled RNA containing the 86-nucleotide 3′-SL structure of JEV. This indicated that the mouse brain cell proteins that bound to the 582-base 3′-NCR RNA of JEV actually bound to the 86-base 3′-SL structure. We have subsequently studied the binding of the mouse brain cell proteins to the 3′-SL structure of JEV. Figure 2 shows that the 3′-SL RNA of JEV formed at least three complexes (A, B, and C) with proteins from the mouse brain. These complexes were stable in the presence of 200 mM KCl in the binding reaction, indicating high affinity of RNA-protein interaction. The presence of a large excess of nonspecific competitors such as poly(I)-poly(C), poly(A)-poly(C)-poly(U), yeast tRNA, and plasmid DNA in the binding reaction did not inhibit the 3′-SL RNA-protein complex formation (Fig. 2A), whereas viral RNA purified from the JEV virions inhibited the complex formation (Fig. 2B), indicating that interaction of cellular proteins with the in vitro-synthesized RNA coding for the 3′-SL structure indeed represented the interaction of cellular proteins with the JEV genomic RNA.
FIG. 2.
Binding of mouse brain cell proteins with the JEV 3′-SL structure RNA. An RNA-protein binding reaction was carried out using 6 ng of radiolabeled 3′-SL structure RNA and 10 μg of mouse brain cell S100 extract in the presence of 200 mM KCl. The RNA-protein complexes were resolved on a 5% polyacrylamide gel (acrylamide/bisacrylamide ratio, 50:1). (A) Lane 1, free probe; lanes 2 to 5, binding in the presence of 500 ng of poly(I)-poly(C), poly(A)-poly(C)-poly(U), yeast tRNA, and an unrelated plasmid DNA, respectively. (B) Lane 1, free probe; lane 2, binding in the presence of 500 ng of poly(I)-poly(C); lanes 3 and 4, binding in the presence of 500 ng of poly(I)-poly(C) and the RNA extracted from 300 or 2,700 μl of culture fluid from JEV-infected cells, respectively.
UV-induced cross-linking of mouse brain cell proteins to the 3′-SL structure of JEV RNA.
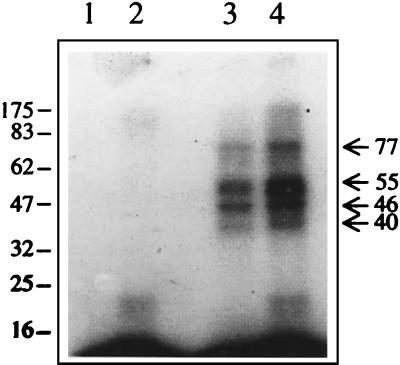
In order to characterize cellular proteins that specifically interacted with the 3′-SL structure of JEV RNA, a UV cross-linking assay was performed where proteins from the S100 extract of the mouse brain cells that interacted with radiolabeled 3′-SL structure RNA were cross-linked to it by UV irradiation. The RNA-protein complexes were subjected to RNase treatment to remove RNA sequences that may not be involved in interaction with the protein. These complexes were subsequently separated on an SDS-polyacrylamide gel. Figure 3 shows that at least four bands with apparent molecular masses of 40, 46, 55, and 77 kDa were visible when the complexes were digested with RNase T1 or RNase A. Complexes with apparent molecular masses of 46 and 55 kDa were most intense. The 55-kDa band was very intense when the complexes were digested with RNase T1 (Fig. 3). It wasn't clear if this represented more than one band. Thus, UV cross-linking identified at least four proteins that bound to the 3′-SL structure RNA. This was in contrast to only three complexes that were detected on the nondenaturing gel.
FIG. 3.
UV-induced cross-linking of RNA and protein. An RNA-protein binding reaction was carried out using 6 ng of radiolabeled 3′-SL structure RNA and 10 μg of mouse brain cell S100 extract in the presence of 60 mM KCl and 500 ng of poly(I)-poly(C). The RNA-protein complexes were UV cross-linked as described in Materials and Methods. After the irradiation, the free RNA was digested with RNase T1 or RNase A, and the products were separated on an SDS–10% polyacrylamide gel. Lanes 1 and 2, free RNA digested with RNase A and RNase T1, respectively; lanes 3 and 4, UV cross-linked RNA-protein complexes digested with RNase A and RNase T1, respectively. Numbers on each side of the gel indicate molecular masses in kilodaltons.
Characterization of mouse brain cell proteins binding to the 3′-SL structure RNA of JEV.
Apparent molecular weights (MWs) of the complexes seen on the SDS-polyacrylamide gel may be higher than the actual MWs of the corresponding proteins, as these complexes contain RNA covalently coupled to the protein. Northwestern blotting was carried out for an improved estimation of the MWs of proteins that bound to the 3′-SL structure RNA of JEV. Figure 4 shows specific binding of the 3′-SL structure RNA with cellular proteins with apparent molecular masses of 32, 35, and 50 kDa. Although a few other bands were also seen, these were perhaps due to the nonspecific or low-specificity binding of the cellular proteins with the 3′-SL structure RNA. The probe did not bind to any of the eight proteins used as MW markers (data not shown), indicating the specificity of the JEV 3′-SL structure RNA interaction with the three cellular proteins.
FIG. 4.
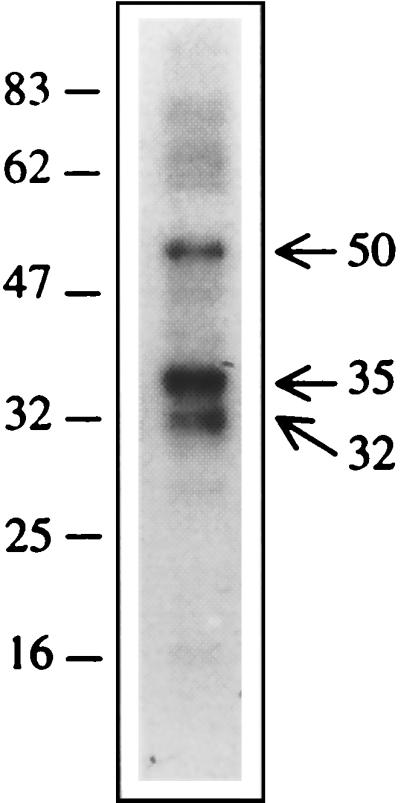
Northwestern analysis of the RNA binding proteins. Mouse brain lysate was resolved on an SDS–10% polyacrylamide gel and transferred to nitrocellulose membrane. Transferred proteins were renatured and allowed to interact with radiolabeled 3′-SL RNA (10 ng/ml) in the presence of 10 μg of yeast tRNA per ml. Three proteins with apparent molecular masses of 32, 35, and 50 kDa showed intense binding with the RNA probe. None of the proteins used as molecular weight markers bind to the RNA probe. Numbers on each side of the gel indicate molecular masses in kilodaltons.
Identification of mouse brain proteins binding to the 3′-SL structure of JEV RNA.
In order to identify the proteins that interact specifically with the 3′-SL structure RNA of JEV, we have screened a mouse brain cDNA expression library with radiolabeled 3′-SL structure RNA. About one million recombinant plaques from the library were screened to identify recombinant bacteriophage 521A2 that repeatedly bound with the 3′-SL RNA. This bacteriophage was plaque purified four times before being used for rescuing the cDNA insert in a plasmid form. Digestion of the plasmid DNA with EcoRI and subsequent nucleotide sequencing established that it had a 531-nucleotide cDNA insert. The nucleotide sequence of the cDNA insert was established (data not shown). The GenBank database homology search revealed that the insert sequence had 99.8% sequence identity with the gene coding for an already-described mouse Mov34 protein of 36 kDa (accession no. M64641). The identity at the amino acid sequence level was 100%.
Binding of Mov34 protein to 3′-SL RNA.
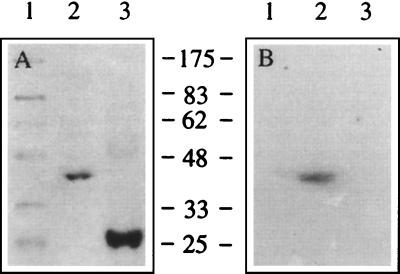
The full-length gene coding for the mouse Mov34 protein was obtained by reverse transcription-PCR using the mouse brain RNA and appropriate synthetic oligonucleotides based on the mouse Mov34 sequence (31). The protein was expressed in E. coli as a histidine-tagged fusion protein. The protein was affinity purified using the histidine tag and employed for 3′-SL RNA binding studies by Northwestern blotting (Fig. 5). The recombinant Mov34 showed clear binding with the radiolabeled RNA. None of the E. coli proteins, copurified with the histidine-tagged proteins, or the proteins used as MW markers bound to the labeled 3′-SL RNA, establishing the specificity of the Mov34 interaction with the JEV RNA. A 21-kDa buffalo growth hormone (Fig. 5) or a 51-kDa monkey zona pellucida protein (data not shown) expressed in E. coli as a histidine-tagged fusion protein did not bind with the RNA probe, indicating that the binding of the 3′-SL structure RNA with recombinant Mov34 was not simply due to the presence of the histidine tag. Allen and Miller (2) also showed that addition of the histidine tag to RB69 RegA translational repressor protein did not alter its RNA binding properties. Binding of the recombinant Mov34 to the 3′-SL RNA of JEV was also seen by gel shift (see Fig. 7). These data thus demonstrated that mouse Mov34 protein bound to the 3′-SL RNA of JEV.
FIG. 5.
Northwestern blotting of murine Mov34 expressed in E. coli. Murine Mov34 was synthesized in E. coli as a histidine-tagged protein and Northwestern blotted with radiolabeled JEV 3′-SL structure RNA synthesized in vitro. (A) Coomassie blue-stained SDS-polyacrylamide gel. Lane 1, MW markers (indicated at the right in thousands); lane 2, histidine-tagged murine Mov34; lane 3, histidine-tagged bovine growth hormone. (B) Northwestern blot of the gel in panel A.
FIG. 7.
Binding of murine Mov34 with JEV RNA. RNA-protein binding reactions were carried out using 3 ng of radiolabeled JEV 3′-SL structure RNA and 100 ng of recombinant Mov34 protein in the presence of 60 mM KCl and 500 ng of poly(I)-poly(C). Formation of the RNA-protein complex was studied in the absence or the presence of the JEV genomic RNA. The RNA-protein complexes were resolved on a 5% polyacrylamide gel (acrylamide/bisacrylamide ratio, 50:1). Lane 1, free probe; lane 2, complex formation by radiolabeled JEV 3′-SL RNA with Mov34; lanes 3 and 4, complex formation in the presence of the RNA extracted from 50 and 500 μl of culture fluid from uninfected cells, respectively; lanes 5 and 6, complex formation in the presence of the RNA extracted from 50 and 500 μl of culture fluid from JEV-infected cells, respectively.
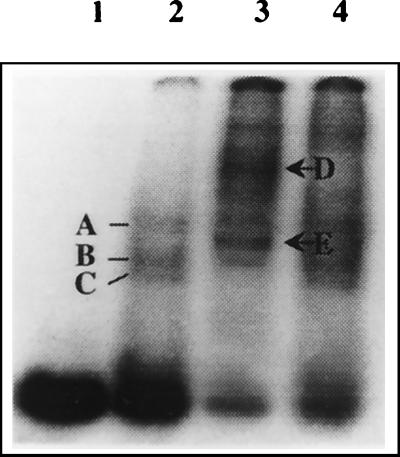
That the cellular Mov34, present in the lysate prepared from the brain cells, also bound to the 3′-SL structure RNA was demonstrated by gel mobility supershift assay that was carried out using anti-Mov34 polyclonal sera. Figure 6 shows that three RNA-protein complexes were seen when no antibody was added to the binding reaction. In the presence of the anti-Mov34 polyclonal antiserum, complexes A and B were seen; however, the lowest band (complex C) disappeared and two bands with higher MWs (indicated as D and E) appeared which were not seen when an unrelated rabbit antiserum was included in the binding reaction. The presence of a nonspecific antibody in the binding reaction did not result in the disappearance of any of the three bands characteristic of JEV 3′-SL RNA interaction with mouse brain lysate. These data indicate that Mov34 present in the mouse brain lysate indeed interacted with the JEV 3′-SL structure RNA.
FIG. 6.
Mouse brain protein binding with JEV 3′-SL structure RNA in the presence of anti-Mov34 antiserum. RNA-protein binding reactions were carried out using 6 ng of radiolabeled JEV 3′-SL structure RNA, 10 μg of mouse brain cell S100 extract, 60 mM KCl, and 500 ng of poly(I)-poly(C). These reactions were carried out in the presence or absence of the Mov34 rabbit antiserum. The RNA-protein complexes were resolved on a 5% polyacrylamide gel (acrylamide/bisacrylamide ratio, 50:1). Lane 1, free probe; lane 2, binding in the absence of the rabbit serum; lane 3, binding in the presence of the Mov34 antiserum; lane 4, binding in the presence of rabbit antiserum to an unrelated protein. In the presence of the Mov34 antiserum, besides complexes A and B, two complexes of higher mobility were seen (D and E, indicated by arrows) that were not seen in the presence of the nonspecific antiserum.
Binding of murine Mov34 to the JEV genomic RNA.
Data shown above established that the murine Mov34 protein interacted with the 3′-NCR RNA of JEV. In order to show that the murine Mov34 bound to JEV RNA, we have carried out murine Mov34 binding to the radiolabeled 3′-SL RNA in the absence or the presence of the viral genomic RNA extracted from JEV virions present in the tissue culture supernatant from the JEV-infected PS cells. Figure 7 shows that histidine-tagged Mov34 formed a complex with the 3′-SL RNA. Formation of this complex was inhibited in the presence of the virion RNA extracted from the tissue culture supernatant from the JEV-infected PS cells, whereas the RNA extracted from the tissue culture supernatant obtained from the uninfected cells did not affect the complex formation (Fig. 7). These data demonstrate that murine Mov34 bound to the JEV genome.
DISCUSSION
Flavivirus genome replication requires RNA-dependent RNA synthesis. Since eukaryotic cells do not possess the enzymatic machinery necessary for this, the virus must code for it. Considering our understanding of the DNA replication that involves a number of transcription factors besides the DNA polymerase, it is likely that RNA viruses make use of some of the cellular proteins for their genome replication. Indeed, host cell proteins have been shown to be part of the RNA replicase complex of a number of viruses (see the introduction). Moreover, for a number of plus-strand RNA viruses, host cell proteins have been shown to bind to either the 3′ or the 5′ end of the genomic RNA, although their precise role in viral RNA transcription or translation has not been established (for a review, see reference 39). In the case of flaviviruses, Blackwell and Brinton (7) showed that at least three proteins of the BHK cells with apparent molecular masses of 56, 84, and 105 kDa interacted with the 3′-NCR of WNV. Subsequently, they purified and identified the ∼56-kDa protein as the 50-kDa translation EF-1α, which also interacted with 3′-NCR of two other flaviviruses, yellow fever virus and dengue virus (8). Recently, Chen et al. (15) demonstrated the binding of JEV nonstructural proteins NS3 and NS5 to the 3′-NCR of the viral RNA and established their role in virus replication. However, these investigators did not observe any cellular protein interaction with the JEV RNA. In the present work, we have clearly demonstrated JEV RNA interaction with the cellular proteins from brains of neonatal mice. We show that at least three proteins with apparent molecular masses of 32, 35, and 50 kDa bind to the 3′-NCR of JEV genome. We have identified one of these proteins as the 36-kDa Mov34 protein.
JEV is closely related to WNV, as the two viruses belong to the same antigenic subgroup within the flavivirus family of viruses (12). However, our results for apparent MWs of cellular proteins that bind the JEV 3′-NCR appear to be different from those of Blackwell and Brinton (7) for WNV. We do, however, see a 50-kDa protein that interacts with the JEV 3′-NCR. The 50-kDa protein interacting with the WNV 3′-NCR has been identified as EF-1α, and the same protein was shown previously to interact with yellow fever and dengue viruses (8). It is, therefore, likely that the 50-kDa protein interacting with JEV 3′-NCR is EF-1α. Efforts are underway to confirm this. The difference in the apparent MWs of cellular proteins in the case of the two viruses may perhaps be related to the host cells used in the study; Blackwell and Brinton (7, 8) used BHK cells of hamster origin, whereas we have used brain cells from a neonatal mouse. In fact, there are differences in the profiles of cellular proteins, from PS cells and mouse brain cells, that bind to JEV 3′-NCR (M. Ta and S. Vrati, unpublished results).
One of the proteins that interact with the JEV 3′-NCR has been identified as the murine Mov34. The product of the Mov34 gene has been shown to be essential for embryonic viability in mice (30). Mov34 transcripts have been found in almost all cell lines and tissues examined (31). The Mov34 gene lacks TATA and CAAT boxes, and the 5′ end of the gene contains many features typical of CpG islands (31). These features and the ubiquitous expression of the Mov34 gene suggest that its product may have some sort of housekeeping role in the cell. Recently, Mahalingam et al. (42) have shown that a human homologue of murine Mov34, designated hVIP/MOV34, functions as a component of the cell cycle cascade. They showed that human immunodeficiency virus type 1 Vpr protein interacted with hVIP/MOV34 and that this interaction resulted in cell cycle arrest during the G2/M phase in human immunodeficiency virus-infected mammalian cells. In view of these observations, it is likely that murine Mov34 is required for normal progression of the cell cycle. Interestingly, Murgue et al. (45) have recently shown that dengue virus infection leads to the inhibition of human hematopoietic progenitor cell growth in vitro. Previously, Varadinova et al. (68) have shown that infection of PS cells with tick-borne encephalitis virus or with WNV leads to cell cycle arrest. We have also seen cell cycle arrest in JEV-infected PS cells (unpublished data). A parallel may thus be drawn between Mov34-mediated inhibition of the cell cycle during human immunodeficiency virus type 1 infection and that during JEV infection. It can be speculated that, subsequent to the viral infection of the cell, the JEV RNA sequesters the cellular Mov34 protein by binding it to its 3′-NCR, leading to the cell cycle arrest. This block in cell cycle may ensure that much of the cell resources are utilized by the virus for its growth.
Murine Mov34 is a 321-amino-acid protein with a calculated molecular mass of 36 kDa (31). The PROSITE search conducted on the murine Mov34 sequence found none of the known RNA binding motifs, although a number of leucine repeats can be seen between amino acids 198 and 272, and most of these leucines are spaced 6 or 13 residues apart (Fig. 8). Similar kinds of leucine repeats have been shown to be involved in binding of the hepatitis delta antigen to the hepatitis delta virus RNA (13). At the C terminus of the protein, adjacent to the leucine repeats, is a very hydrophobic domain of amino acids that is rich in KEKE motifs (55). Such sequences are also found in subunits of the multicatalytic protease or the 20S proteasome, in subunit 12 of the 26S protease, and in a variety of chaperonins including hsp90, hsp70, and calnexin (55). The KEKE motifs have been proposed to promote the association between proteins which contain them (55). Murine Mov34 shares a high degree of sequence identity with p40, a regulatory subunit of the 26S proteasome isolated from the human hepatoblastoma cell line HepG2 (67) and with the S12 subunit of the 26S proteasome of human erythrocytes (94.4 and 96% identity, respectively) (21). The murine Mov34, therefore, has been proposed to be a component of the 26S mouse proteasome (21). The 26S proteasome is a large multisubunit, multifunctional protein that is involved in degradation of ubiquitinated proteins in an ATP-dependent reaction (17, 23). Furthermore, proteasomes are involved in cell cycle control (25, 29) and in early steps of the immune response, such as major histocompatibility complex class I-restricted antigen processing (5, 26). Proteasomes are also involved in transcriptional regulation (47, 56), and recent reports suggest that they may also participate in the pathways of cellular RNA breakdown (52, 59). It is possible that the Mov34 interaction with the JEV 3′-NCR helps in binding of the proteasome with the viral RNA for its subsequent involvement in transcription or other activities listed above.
FIG. 8.
Leucine repeats and the KEKE motifs in murine Mov34. The murine Mov34 sequence (31) has been shown where leucine repeats have been boxed. Many of the leucine repeats are separated by 6 or 13 residues. The C-terminal sequence containing the KEKE motifs has been underlined.
Murine Mov34 interaction with the JEV RNA may have functional relevance for viral RNA transcription, as it belongs to a family of proteins that may participate in the regulation of transcription and translation initiation (3, 4). For example, eukaryotic translation initiation factors eIF3p47 and eIFp40, which are members of the Mov34 family of proteins, share significant amino acid identity with murine Mov34 in their N-terminal halves (3, 4, 35). Furthermore, Mov34 homologues human JAB1 and Schizosaccharomyces pombe pad1 have been shown to selectively potentiate transcription via binding to gene regulatory protein AP1 (18, 60). Thus, there is considerable evidence showing involvement of homologues of Mov34 protein in regulation of eukaryotic transcription, translation, and protein degradation (see reference 3). The reverse of this also seems to be true: a modulator of human immunodeficiency virus TAT-dependent transcriptional activation is identical to the S7 subunit of the 26S proteasome (20), and protein synthesis elongation factor EF-1α has been shown to be essential for ubiquitin-mediated degradation of certain proteins by the 26S proteasome (27). Interestingly, EF-1α has been shown to interact with the 5′ end of poliovirus genomic RNA (33) and 3′ ends of turnip yellow mosaic virus RNA (37) and WNV RNA (8). The role of EF-1α in replication of the viral RNA genome is not clear at this stage, although it has been suggested that it plays a role in targeting the viral RNA to a suitable microenvironment for efficient replication. We have not yet established the biological significance of murine Mov34 interaction with JEV RNA, but considering its homology to a number of proteins from the Mov34 family that are involved in control of transcription, it is likely to play a role in JEV RNA replication. Besides, the murine Mov34 may be responsible for protein-RNA or protein-protein interactions that may be required for subsequent assembly of viral replicase complex.
EF-1α, which binds to the 3′-NCR of WNV RNA, is a translation elongation factor. Therefore, its role in transcription and viral RNA replication is not clear. The majority of the flavivirus RNA-dependent RNA polymerase activity has been shown to be associated with the endoplasmic reticulum membrane fraction (22, 32). It has been suggested that EF-1α attaches to the endoplasmic reticulum membrane via a posttranslational modification. Blackwell and Brinton (8), therefore, suggested that EF-1α may be involved in targeting the WNV RNA to the endoplasmic reticulum membranes for subsequent replication. It appears that proteasomes with different subunit compositions are found in different locations within the cell (57), including the membranes of the endoplasmic reticulum (49). In view of these data, it is possible that the murine Mov34, which is proposed to be a proteasome component (21), directs the transport and localization of JEV RNA to a cellular microenvironment that may be suitable for virus replication.
ACKNOWLEDGMENTS
This work was supported by the core grant from the Department of Biotechnology, Government of India, to the National Institute of Immunology, New Delhi. The work was also supported by grant no. BT/PRO151/MED/09/028/96 and BT/PRO152/MED/09/029/96 from the Department of Biotechnology to S.V.
We thank L. C. Garg for the histidine-tagged bovine growth hormone and S. K. Gupta for the histidine-tagged zona pellucida protein. We thank Sher Ali and Sudip Ghosh for help with preparation of the figures and Sandip K. Basu for support and encouragement.
REFERENCES
- 1.Ali N, Siddiqui A. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc Natl Acad Sci USA. 1997;94:2249–2254. doi: 10.1073/pnas.94.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen S V, Miller E S. RNA-binding properties of in vitro expressed histidine-tagged RB69 RegA translational repressor protein. Anal Biochem. 1999;269:32–37. doi: 10.1006/abio.1999.4025. [DOI] [PubMed] [Google Scholar]
- 3.Aravind L, Ponting C P. Homologues of 26S proteasome subunits are regulators of transcription and translation. Protein Sci. 1998;7:1250–1254. doi: 10.1002/pro.5560070521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asano K, Vornlocher H P, Richter-Cook N J, Merrick W C, Hinnebusch A G, Hershey J W. Structure of cDNA encoding human eukaryotic initiation factor 3 subunits. Possible roles in RNA binding and macromolecular assembly. J Biol Chem. 1997;272:27042–27052. doi: 10.1074/jbc.272.43.27042. [DOI] [PubMed] [Google Scholar]
- 5.Belich M P, Trowsdale J. Proteasome and class I antigen processing and presentation. Mol Biol Rep. 1995;21:53–56. doi: 10.1007/BF00990971. [DOI] [PubMed] [Google Scholar]
- 6.Black A C, Luo J, Chun S, Bakker A, Faser J K, Rosenblatt J D. Specific binding of polypyrimidine tract binding protein and hnRNP A1 to HIV-1 CRS elements. Virus Genes. 1996;12:275–285. doi: 10.1007/BF00284648. [DOI] [PubMed] [Google Scholar]
- 7.Blackwell J L, Brinton M A. BHK cell proteins that bind to the 3′ stem-loop structure of the West Nile virus genome RNA. J Virol. 1995;69:5650–5658. doi: 10.1128/jvi.69.9.5650-5658.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blackwell J L, Brinton M A. Translation elongation factor-1 alpha interacts with the 3′ stem-loop region of West Nile virus genomic RNA. J Virol. 1997;71:6433–6444. doi: 10.1128/jvi.71.9.6433-6444.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blumenthal T, Carmichael G G. RNA replication: function and structure of Qβ-replicase. Annu Rev Biochem. 1979;48:525–548. doi: 10.1146/annurev.bi.48.070179.002521. [DOI] [PubMed] [Google Scholar]
- 10.Brinton M A, Fernandez A V, Dispoto J H. The 3′-nucleotides of flavivirus genomic RNA form a conserved secondary structure. Virology. 1986;153:113–121. doi: 10.1016/0042-6822(86)90012-7. [DOI] [PubMed] [Google Scholar]
- 11.Brinton M A, Wilson W D, Shi P Y, Veal J M, Zhong Y Y. Evidence for the existence of a pseudoknot structure at the 3′-terminus of the flavivirus genomic RNA. Biochemistry. 1996;35:4222–4230. doi: 10.1021/bi952398v. [DOI] [PubMed] [Google Scholar]
- 12.Chambers T J, Hahn C S, Galler R, Rice C M. Flavivirus genome organisation, expression and replication. Annu Rev Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 13.Chang M-F, Sun C-Y, Chen C-J, Chang S C. Functional motifs of delta antigen essential for RNA binding and replication of hepatitis delta virus. J Virol. 1993;67:2529–2536. doi: 10.1128/jvi.67.5.2529-2536.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang Y-N, Kenan D J, Keene J D, Gatignol A, Jeang K-T. Direct interactions between autoantigen La and human immunodeficiency virus leader RNA. J Virol. 1994;68:7008–7020. doi: 10.1128/jvi.68.11.7008-7020.1994. . (Erratum, 69:618, 1995.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen C J, Kuo M D, Chien L J, Hsu S L, Wang Y M, Lin J H. RNA-protein interactions: involvement of NS3, NS5, and 3′-noncoding region of Japanese encephalitis virus genomic RNA. J Virol. 1997;71:3466–3473. doi: 10.1128/jvi.71.5.3466-3473.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung R T, Kaplan L M. Heterogeneous nuclear ribonucleoprotein I (hnRNP-I/PTB) selectively binds the conserved 3′ terminus of hepatitis C viral RNA. Biochem Biophys Res Commun. 1999;254:351–362. doi: 10.1006/bbrc.1998.9949. [DOI] [PubMed] [Google Scholar]
- 17.Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 18.Claret F X, Hibi M, Dhut S, Toda T, Karin M. A new group of conserved coactivators that increase the specificity of AP-1 transcription factors. Nature. 1996;383:453–457. doi: 10.1038/383453a0. [DOI] [PubMed] [Google Scholar]
- 19.Das T, Mathur M, Gupta A K, Janssen G M, Banerjee A K. RNA polymerase of vesicular stomatitis virus specifically associates with translation elongation factor-1 alphabetagamma for its activity. Proc Natl Acad Sci USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dubiel W, Ferrel K, Rechsteiner M. Subunits of the regulatory complex of the 26S protease. Mol Biol Rep. 1995;21:27–34. doi: 10.1007/BF00990967. [DOI] [PubMed] [Google Scholar]
- 21.Dubiel W, Ferrell K, Dumdey R, Standera S, Prehn S, Rechsteiner M. Molecular cloning and expression of subunit 12: a non-MCP and non-ATPase subunit of the 26S protease. FEBS Lett. 1995;363:97–100. doi: 10.1016/0014-5793(95)00288-k. [DOI] [PubMed] [Google Scholar]
- 22.Edward Z, Takegami T. Localization and functions of Japanese encephalitis virus nonstructural proteins NS3 and NS5 for viral RNA synthesis in the infected cells. Microbiol Immunol. 1993;37:239–243. doi: 10.1111/j.1348-0421.1993.tb03206.x. [DOI] [PubMed] [Google Scholar]
- 23.Eytan E, Ganoth D, Armon T, Hershko A. ATP-dependent incorporation of 20S protease into the 26S complex that degrades proteins conjugated to ubiquitin. Proc Natl Acad Sci USA. 1989;86:7751–7757. doi: 10.1073/pnas.86.20.7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frey B, Suppmann B. Demonstration of the Expand PCR system's greater fidelity and higher yields with a lacI-based PCR fidelity assay. Biochemica. 1995;2:8–9. [Google Scholar]
- 25.Ghislain M, Udvardy A, Mann C. S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature. 1993;366:358–362. doi: 10.1038/366358a0. [DOI] [PubMed] [Google Scholar]
- 26.Goldberg A L, Rock K L. Proteolysis, proteasomes and antigen presentation. Nature. 1992;357:375–379. doi: 10.1038/357375a0. [DOI] [PubMed] [Google Scholar]
- 27.Gonen H, Smith C E, Siegel N R, Kahana C, Marrick W C, Chakraburty K, Schwartz A L, Ciechanover A. Protein synthesis elongation factor EF-1α is essential for ubiquitin-dependent degradation of certain Nα-acetylated proteins and may be substituted for by the bacterial elongation factor EF-Tu. Proc Natl Acad Sci USA. 1994;91:7648–7652. doi: 10.1073/pnas.91.16.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gontarek R R, Gutshall L L, Herold K M, Tsai J, Sathe G M, Mao J, Prescott C, Del Vecchio A M. hnRNP C and polypyrimidine tract-binding protein specifically interact with pyrimidine-rich region within the 3′NTR of the HCV RNA genome. Nucleic Acids Res. 1999;27:1457–1463. doi: 10.1093/nar/27.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gordon C, McGurk G, Dillon P, Rosen C, Hastie N D. Defective mitosis due to a mutation in the gene for a fission yeast 26S protease subunit. Nature. 1993;366:355–357. doi: 10.1038/366355a0. [DOI] [PubMed] [Google Scholar]
- 30.Gridley T, Gray D A, Orr-Weaver T, Soriano P, Baryon D E, Franke U, Jaenisch R. Molecular analysis of the Mov34 mutation: transcript disrupted by proviral integration in mice is conserved in Drosophila. Development. 1990;109:235–242. doi: 10.1242/dev.109.1.235. [DOI] [PubMed] [Google Scholar]
- 31.Gridley T, Jaenisch R, Gendron-Maguire M. The murine Mov34 gene: full-length cDNA and genomic organization. Genomics. 1991;11:501–507. doi: 10.1016/0888-7543(91)90056-k. [DOI] [PubMed] [Google Scholar]
- 32.Grun J B, Brinton M. Dissociation of NS5 from cell fractions containing West Nile virus-specific polymerase activity. J Virol. 1987;61:3641–3644. doi: 10.1128/jvi.61.11.3641-3644.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harris K S, Xiang W, Alexander L, Lane W S, Paul A V, Wimmer E. Interaction of poliovirus polypeptide 3CDpol with the 5′ and the 3′ termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J Biol Chem. 1994;269:27004–27014. [PubMed] [Google Scholar]
- 34.Hayes R J, Buck K W. Complete replication of a eukaryotic virus RNA in vitro by a purified RNA-dependent RNA polymerase. Cell. 1990;63:363–368. doi: 10.1016/0092-8674(90)90169-f. [DOI] [PubMed] [Google Scholar]
- 35.Hershey J W, Asano K, Naranda T, Vomlocher H P, Hanachi P, Marrick W C. Conservation and diversity in the structure of translation initiation factor eIF3 from humans and yeast. Biochimie. 1996;78:903–907. doi: 10.1016/s0300-9084(97)86711-9. [DOI] [PubMed] [Google Scholar]
- 36.Ito T, Lai M M C. Determination of the secondary structure of and cellular protein binding to the 3′-untranslated region of the hepatitis C virus RNA genome. J Virol. 1997;71:8698–8706. doi: 10.1128/jvi.71.11.8698-8706.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joshi R L, Ravel J M, Haenni A L. Interaction of turnip yellow mosaic virus val-tRNA with eukaryotic elongation factor EF-1α. Search for a function. EMBO J. 1986;5:1143–1148. doi: 10.1002/j.1460-2075.1986.tb04339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krug M S, Berger S L. First strand cDNA synthesis primed with oligo(dT) Methods Enzymol. 1987;152:316–325. doi: 10.1016/0076-6879(87)52036-5. [DOI] [PubMed] [Google Scholar]
- 39.Lai M M C. Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA-dependent RNA transcription. Virology. 1998;244:1–12. doi: 10.1006/viro.1998.9098. [DOI] [PubMed] [Google Scholar]
- 40.Li H-P, Zhang X, Duncan R, Comai L, Lai M M C. Heterogeneous nuclear riboprotein A1 binds to the transcription-regulatory region of mouse hepatitis virus RNA. Proc Natl Acad Sci USA. 1997;94:9544–9549. doi: 10.1073/pnas.94.18.9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo G. Cellular proteins binds to the poly(U) tract of the 3′ untranslated region of hepatitis C virus RNA genome. Virology. 1999;256:105–118. doi: 10.1006/viro.1999.9639. [DOI] [PubMed] [Google Scholar]
- 42.Mahalingam S, Ayyavoo V, Patel M, Kieber-Emmons T, Kao G D, Mushel R J, Weiner D B. HIV-1 Vpr interacts with a human 34-kDa mov34 homologue, a cellular factor linked to the G2/M phase transition of the mammalian cell cycle. Proc Natl Acad Sci USA. 1998;95:3419–3424. doi: 10.1073/pnas.95.7.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McBride A E, Schelegel A, Kirkegaard K. Human protein Sam68 relocalization and interaction with poliovirus RNA polymerase in infected cells. Proc Natl Acad Sci USA. 1996;93:2296–2301. doi: 10.1073/pnas.93.6.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moyer S A, Baker S C, Horikami S M. Host cell proteins required for measles virus reproduction. J Gen Virol. 1990;71:775–763. doi: 10.1099/0022-1317-71-4-775. [DOI] [PubMed] [Google Scholar]
- 45.Murgue B, Cassar O, Guigon M, Chungue E. Dengue virus inhibits human hematopoietic progenitor growth in vitro. J Infect Dis. 1997;175:1497–1501. doi: 10.1086/516486. [DOI] [PubMed] [Google Scholar]
- 46.O'Neill R, Palese P. Cis-acting signals and trans-acting factors involved in influenza virus RNA synthesis. Infect Agents Dis. 1994;3:77–84. [PubMed] [Google Scholar]
- 47.Orian A, Whiteside S, Israel A, Stancovski I, Schwartz A L, Ciechanover A. Ubiquitin-mediated processing of NF-kappa B transcriptional activator precursor p105. Reconstitution of a cell-free system and identification of the ubiquitin-carrier protein, E2, and a novel ubiquitin-protein ligase, E3, involved in conjugation. J Biol Chem. 1995;270:21707–21714. doi: 10.1074/jbc.270.37.21707. [DOI] [PubMed] [Google Scholar]
- 48.Osman T A M, Buck K W. The tobacco mosaic virus RNA polymerase complex contains a plant protein related to the RNA binding subunit of yeast eIF3. J Virol. 1997;71:6075–6082. doi: 10.1128/jvi.71.8.6075-6082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palmer A, Rivett A J, Thomson S, Hendil K B, Butcher G W, Fuertes G, Knecht E. Subpopulations of proteasomes in rat liver nuclei, microsomes and cytosol. Biochem J. 1996;316:401–407. doi: 10.1042/bj3160401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paradigon N, Strauss J H. Mosquito homolog of the La autoantigen binds to Sindbis virus RNA. J Virol. 1996;70:1173–1187. doi: 10.1128/jvi.70.2.1173-1181.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pogue G P, Hofmann J, Duncann R, Best J M, Etherington J, Sontheimer R D, Nakhasi H L. Autoantigens interact with cis-acting elements of rubella virus RNA. J Virol. 1996;70:6269–6277. doi: 10.1128/jvi.70.9.6269-6277.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pouch M N, Petit F, Buri J, Briand Y, Schmid H P. Identification and initial characterization of a specific proteasome (prosome) associated RNase activity. J Biol Chem. 1995;270:22023–22028. doi: 10.1074/jbc.270.37.22023. [DOI] [PubMed] [Google Scholar]
- 53.Quadt R, Kao C C, Browning K S, Hershberger R P, Ahlquist P. Characterization of a host protein associated with brome mosaic virus RNA-dependent RNA polymerase. Proc Natl Acad Sci USA. 1993;90:1498–1502. doi: 10.1073/pnas.90.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quian Z, Wilusz J. Cloning of a cDNA encoding an RNA binding protein by screening expression libraries using a Northwestern strategy. Anal Biochem. 1993;212:547–554. doi: 10.1006/abio.1993.1367. [DOI] [PubMed] [Google Scholar]
- 55.Realini C, Rogers S W, Rechsteiner M. KEKE motifs: proposed roles in protein-protein association and presentation of peptides by MHC class I receptors. FEBS Lett. 1994;348:109–113. doi: 10.1016/0014-5793(94)00569-9. [DOI] [PubMed] [Google Scholar]
- 56.Richter-Ruoff B, Wolf D H, Hochstrasser H. Degradation of the yeast MAT alpha 2 transcriptional regulator is mediated by the proteasome. FEBS Lett. 1994;354:50–52. doi: 10.1016/0014-5793(94)01085-4. [DOI] [PubMed] [Google Scholar]
- 57.Rivett A J. Intracellular distribution of proteasomes. Curr Opin Immunol. 1998;10:110–114. doi: 10.1016/s0952-7915(98)80040-x. [DOI] [PubMed] [Google Scholar]
- 58.Sagesser R, Martinez E, Tsagris M, Tabler M. Detection and isolation of RNA-binding proteins by RNA-ligand screening of a cDNA expression library. Nucleic Acids Res. 1997;25:3816–3822. doi: 10.1093/nar/25.19.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmid H P, Pouch M N, Petit F, Dadet M H, Badaoui S, Boissonnet G, Buri J, Norris V, Briand Y. Relationships between proteasomes and RNA. Mol Biol Rep. 1995;21:43–47. doi: 10.1007/BF00990969. [DOI] [PubMed] [Google Scholar]
- 60.Shimanuki M, Saka Y, Yanagida M, Toda T. A novel essential fission yeast gene pad1+ positively regulates pap1(+)-dependent transcription and is implicated in the maintenance of chromosome structure. J Cell Sci. 1995;108:569–579. doi: 10.1242/jcs.108.2.569. [DOI] [PubMed] [Google Scholar]
- 61.Singh N K, Atreya C D, Nakhasi H L. Identification of calreticulin as a rubella virus RNA binding protein. Proc Natl Acad Sci USA. 1994;91:12770–12774. doi: 10.1073/pnas.91.26.12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stollar V, Schlesinger R W, Stevens T M. Studies on the nature of dengue viruses. III. RNA synthesis in cells infected with type 2 dengue virus. Virology. 1967;33:650–658. doi: 10.1016/0042-6822(67)90065-7. [DOI] [PubMed] [Google Scholar]
- 63.Sumiyoshi H, Mori C, Fuke I, Morita K, Kuhara S, Kondou J, Kikuchi Y, Nagamatu H, Igarashi A. Complete nucleotide sequence of the Japanese encephalitis virus genome RNA. Virology. 1987;161:497–510. doi: 10.1016/0042-6822(87)90144-9. [DOI] [PubMed] [Google Scholar]
- 64.Svitkin Y V, Pause A, Sonenberg N. La autoantigen alleviates translational repression by the 5′ leader sequence of the human immunodeficiency virus type 1 mRNA. J Virol. 1994;68:7001–7007. doi: 10.1128/jvi.68.11.7001-7007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takegami T, Washizu M, Yasui K. Nucleotide sequence at the 3′ end of Japanese encephalitis virus genomic RNA. Virology. 1986;152:483–486. doi: 10.1016/0042-6822(86)90152-2. [DOI] [PubMed] [Google Scholar]
- 66.Tsuchihara K, Tanaka T, Hijikata M, Kuge S, Toyoda H, Nomoto A, Yamamoto N, Shimotohno K. Specific interaction of polypyrimidine tract-binding protein with the extreme 3′-terminal structure of the hepatitis C virus genome, the 3′X. J Virol. 1997;71:6720–6726. doi: 10.1128/jvi.71.9.6720-6726.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsurumi C, DeMartino G N, Slaughter C A, Shimbara N, Tanaka K. cDNA cloning of p40, a regulatory subunit of the human 26S proteasome, and a homolog of the Mov-34 gene product. Biochem Biophys Res Commun. 1995;210:600–608. doi: 10.1006/bbrc.1995.1701. [DOI] [PubMed] [Google Scholar]
- 68.Varadinova T, Gresikova M, Batikova M. Influence of tick-borne encephalitis and West Nile viruses on the chromosomes of pig kidney cells. Acta Virol. 1983;27:238–244. [PubMed] [Google Scholar]
- 69.Vrati S, Giri R K, Razdan A, Malik P. Complete nucleotide sequence of an Indian strain of Japanese encephalitis virus: sequence comparison with other strains and phylogenetic analysis. Am J Trop Med Hyg. 1999;61:677–680. doi: 10.4269/ajtmh.1999.61.677. [DOI] [PubMed] [Google Scholar]
- 70.Vrati S, Agarwal V, Malik P, Wani S A, Saini M. Molecular characterization of an Indian isolate of Japanese encephalitis virus that shows an extended lag phase during growth. J Gen Virol. 1999;80:1665–1671. doi: 10.1099/0022-1317-80-7-1665. [DOI] [PubMed] [Google Scholar]
- 71.You S, Padmanabhan R. A novel in vitro replication system for Dengue virus. Initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J Biol Chem. 1999;274:33714–33722. doi: 10.1074/jbc.274.47.33714. [DOI] [PubMed] [Google Scholar]