Abstract
Human papillomavirus type 6 (HPV-6) is a low-risk HPV whose replication cycle, like that of all HPVs, is differentiation dependent. We have previously shown that CCAAT displacement protein (CDP) binds the differentiation-induced HPV-6 E1 promoter and negatively regulates its activity in undifferentiated cells (W. Ai, E. Toussaint, and A. Roman, J. Virol. 73:4220–4229, 1999). Using electrophoretic mobility shift assays (EMSAs), we now report that Yin Yang 1 (YY1), a multifunctional protein that can act as a transcriptional activator or repressor and that can also inhibit HPV replication in vitro, binds the HPV-6 E1 promoter. EMSAs, using subfragments of the promoter as competitors, showed that the YY1 binding site is located at the 5′ end of the E1 promoter. When a putative YY1 site was mutated, the ability of YY1 to bind was greatly decreased. The activity of the mutated E1 promoter, monitored with the reporter gene luciferase, was threefold greater than that of the wild-type promoter, suggesting that YY1 negatively regulates HPV-6 E1 promoter activity. Nuclear extracts from differentiated keratinocytes showed decreased binding of YY1 to the wild-type promoter. Consistent with this, in differentiated keratinocytes, the activity of the transfected luciferase gene transcribed from the mutated promoter was comparable to that of the wild-type promoter; both promoters were up-regulated in differentiated keratinocytes compared to undifferentiated cells. These data suggest that YY1 functions in undifferentiated keratinocytes but not in differentiated keratinocytes. Both the wild-type and mutated promoters could be negatively regulated by overexpression of a plasmid encoding CDP. Thus, both YY1 and CDP appear to be negative regulators of the differentiation-induced HPV-6 E1 promoter and thereby the HPV life cycle. In contrast, only binding of CDP was detected using the E1 promoter of the high-risk HPV-31.
Human papillomaviruses (HPVs) are a large family of DNA viruses that cause epithelial hyperproliferative lesions. Within the genital tract, HPVs are classified as high-risk or low-risk types. While infection with the high-risk viruses results in a greater-than-200-fold-increased risk for the development of high-grade squamous epithelial neoplasia, the increase in risk following infection with the low-risk viruses is approximately 10-fold (58). All HPVs have a covalently closed double-stranded genome consisting of approximately 8 kbp. They have essentially the same genomic organization, including the early region, which encodes the nonstructural viral proteins E1 through E7, and the late region, which encodes the two structural proteins L1 and L2. The genes are arranged in the following order: E6, E7, E1, E2(E4), E5, L2, and L1. Between the translation termination codon of L1 and the translational start site of E6 is the long control region, of approximately 800 bp, which contains cis elements that regulate viral DNA replication and transcription. For the high-risk viruses, the most readily detectable promoters are located upstream of the E6 and E1 open reading frames (ORFs); for the low-risk viruses, there is an additional promoter upstream of E7.
The HPV life cycle is differentiation dependent and occurs in two stages. Initial infection is thought to take place in the undifferentiated basal cell layer. The viral genome is maintained at less than 100 copies per cell, viral gene expression is limited, and no structural proteins or virus is produced. When the infected keratinocyte differentiates, there is a change in the transcription program, the genome is amplified to thousands of copies, structural proteins are synthesized, and virus is produced. Analyses of the virus life cycle have been conducted using tissue sections from human biopsies (11, 32, 65), a variety of models including infected cells or tissue grown in nude mice (19, 63, 64), and in vitro induction of differentiation by growth as organotypic (raft) cultures (8, 16, 29, 45), in medium containing high calcium (4, 21, 33) or in semisolid (methylcellulose) medium (21, 57). The increased transcription detected during this productive stage requires that the DNA be maintained episomally (23). For the high-risk viruses, DNA replication switches from a theta mode to a rolling-circle mode upon differentiation (21). However, for low-risk viruses, throughout the life cycle, only theta replicative intermediates have been detected (5).
The viral genes required for transient DNA replication are E1 and E2, which bind to the origin of replication located upstream of the E6 ORF (9, 12, 13, 22). It has recently been reported that for stable DNA replication in the nonproductive stage, the E6 and E7 proteins are required for episomal maintenance (66). Their role, however, is currently unclear. In the nonproductive stage, the E6, E7, E1, and E2 proteins are translated from unspliced or spliced messages initiated from the E6 promoter (and possibly from the E7 promoter, described only for the low-risk viruses) (11, 29, 49, 50, 54, 62, 65).
In addition, both the E1 and E2 genes are transcribed from a promoter immediately upstream of the E1 ORF (11, 35, 49). Utilization of the E1 promoter allows the E1 gene to be transcribed as the first ORF of the transcript rather than as a downstream ORF. Transcripts from this promoter also encode the late structural proteins (11, 30, 56). Expression from the E1 promoter is highly differentiation specific, in contrast to expression from the E6 and E7 promoters, which is reported either to be constitutive or to increase to a lesser extent upon differentiation (15, 26, 29, 32, 35, 50, 65). DNA amplification seen in the productive stage correlates with up-regulation of the differentiation-specific E1 promoter and an increase in the E1/E2 mRNA ratio (35, 49).
While up-regulation of the E1 promoter during differentiation is well documented, little is known about factors regulating this promoter. Repression of this promoter in undifferentiated cells would keep the level of E1 and E2 proteins low and ensure that transcripts able to encode structural proteins were also low. We recently reported that CCAAT displacement protein (CDP), the 180-kDa human homologue of the Drosophila Cut protein (28, 46), negatively regulates the E6, E7, and E1 promoters of the low-risk HPV type 6 (HPV-6) genome (Fig. 1) (1). When keratinocytes are induced to differentiate, binding of the E6, E7, and E1 promoters by CDP is no longer detectable and there is an increase in expression from all three promoters (1). This is consistent with other reports indicating that CDP functions as a repressor in undifferentiated cells but not in differentiated cells (3, 40, 42, 61).
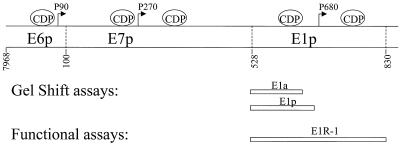
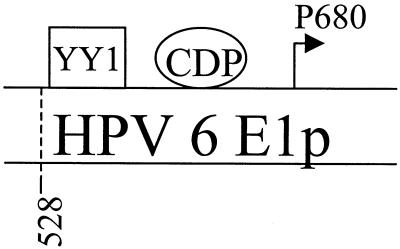
FIG. 1.
Schematic illustration of HPV-6 E6, E7, and E1 promoters. The locations of CDP binding sites for HPV-6 are shown, as are the E6, E7, and E1 transcription initiation sites (arrows) and the oligonucleotides used in the EMSAs (gel shift assays) and the functional assays. The 5′ end of E1a, E1p, and E1R-1 is at the translation start site of E7 (nt 528). The 3′ end of E1a is at the AccI site (nt 616); the 3′ end of E1p is just upstream of the E1 transcriptional start site at the DraI site (nt 670). The E1R-1 sequence ends just upstream of the E1 translational start site (nt 829).
Preliminary data obtained during the CDP studies (see Fig. 7 of reference 1) suggested that Yin Yang 1 (YY1) might also bind the E1 promoter. YY1 is a ubiquitously expressed 65-kDa protein which binds to elements within viral and cellular promoters and functions as both a transcriptional activator and a repressor (59, 60). It is also reported elsewhere that YY1 functions as an initiator element binding protein that directs and initiates transcription in vitro (68). The ability of YY1 to negatively regulate the E6 promoter of HPVs was first reported for HPV-18 (7) and subsequently for HPV-16 (43). More recently, Kanaya et al. (34) have reported that YY1 binding sites in the 5′ end of the long control region function as activators of HPV-31 E6 promoter-initiated transcription. YY1 DNA binding activity decreases upon differentiation of human teratocarcinoma cells and myoblasts (38, 41). Thus, we wished to determine whether the HPV-6 E1 promoter was regulated by the differentiation-dependent factor YY1.
In this study, we demonstrate that YY1 binds to the HPV-6 E1 promoter and negatively regulates the promoter activity in undifferentiated keratinocytes. When keratinocytes are induced to differentiate, YY1 is no longer functional. This evidence adds YY1 as another differentiation-dependent transcriptional repressor of the HPV-6 E1 promoter.
MATERIALS AND METHODS
Construction of recombinant plasmids.
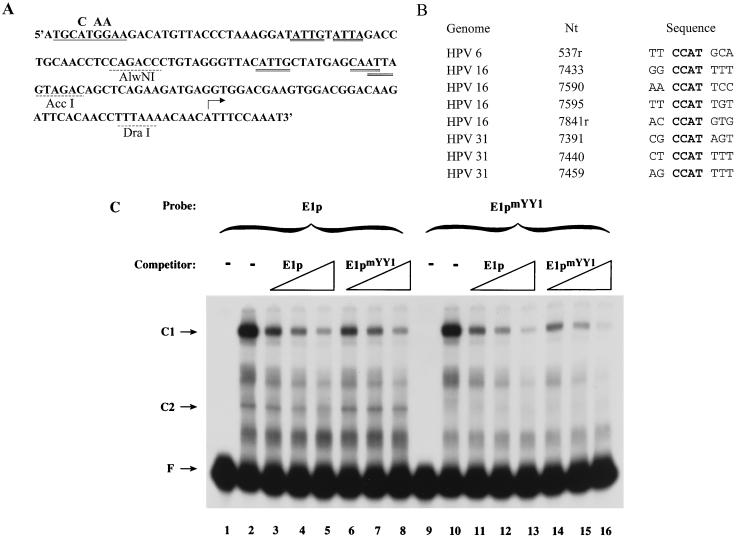
Cloning of pE1p (E1p in pUC19) and pE1R-1luc from HPV6aw50 (20) has been described previously (1) (Fig. 1). To construct E1pmYY1, two complementary oligonucleotides containing point mutations (underlined) were synthesized: YY1 mutant top, 5′ GATGCCTAAAAGACAT 3′; and YY1 mutant bottom, 5′ TAACATGTCTTTTAGGCATCTGCA 3′. These oligonucleotides were treated with T4 polynucleotide kinase and annealed to produce a PstI overhang at the 5′ end and a BbsI overhang at the 3′ end. The annealed fragment (containing nucleotides [nt] 528 to 546) was then ligated into the large fragment of pE1p, generated by cleaving pE1p with PstI and BbsI. To construct pE1R-1mYY1luc, pE1pmYY1 was cleaved with PstI and AccI, as was pE1R-1luc, and the small fragment of pE1pmYY1 was cloned into the large fragment of pE1R-1luc. HPV-31 E1R-1 (the equivalent of HPV-6 E1R-1) was amplified by PCR from pBR322.HPV31 (obtained from Laimonis A. Laimins, Northwestern University Medical School, Chicago, Ill.) using the following primers: 5′ primer, 5′ GGGCTGCAGATGCGTGGAGAAACAC 3′, and 3′ primer, 5′ GGGAAGCTTTGTAGTTACAGTCTAG 3′. The restriction sites in the primers (PstI and HindIII in the 5′ and 3′ oligonucleotides, respectively) are underlined. Prior to use, the primers were phosphorylated to facilitate ligation into a dephosphorylated vector. The amplified product was cloned into pUC19, which was previously digested with SmaI and treated with calf intestinal alkaline phosphatase. The recombinant clone contained nt 560 to 861, from the ATG of the upstream E7 ORF to the first nucleotide upstream of the ATG of the E1 ORF. 31E1p was excised from p31E1R-1 using PstI (present in the 5′ primer) and AccI (Fig. 2) digestion, recovered following gel electrophoresis, and quantitated by measuring the absorbance at 260 nm. 31E1p was used for probe preparation as published previously for HPV-6 E1p (1). All recombinant sequences were confirmed by DNA sequencing by the Biochemistry Biotechnology Facility at Indiana University School of Medicine.
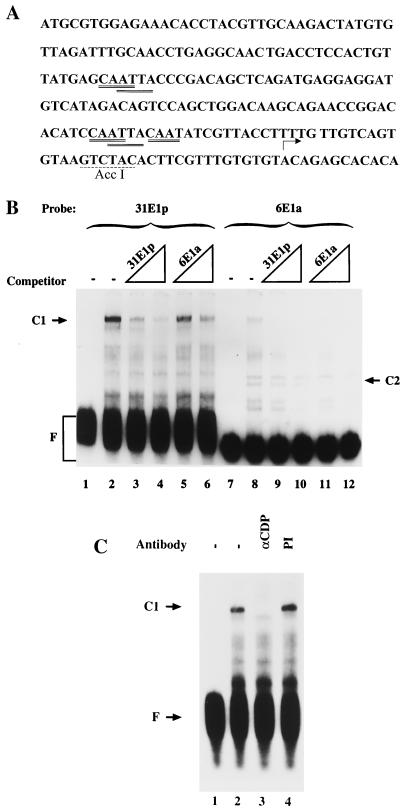
FIG. 2.
The E1 promoter of HPV-31 binds CDP. (A) The sequence of the HPV-31 E1 promoter extending from the ATG of E7 at nt 560 through the E1 transcription initiation start site at nt 742 is shown (25). The 31E1p used in EMSAs extends from the 5′ end at nt 560 to the AccI site at nt 722. Putative CDP binding sites are double underlined; the AccI site is underlined with a dashed line. (B) EMSAs were conducted using radiolabeled HPV-31 E1p (31E1p, lanes 1 to 6) or HPV-6 E1a (6E1a, lanes 7 to 12) and either no competitor (−) or a 20- or 200-fold molar excess of unlabeled 31E1p (lanes 3, 4, 9, and 10) or 6E1a (lanes 5, 6, 11, and 12). (C) EMSAs were conducted using radiolabeled 31E1p and either antiserum to CDP (αCDP) or preimmune serum (PI). C1 and C2, retarded DNA-protein complexes; F, free probe.
Cell culture and transfections.
Human keratinocytes were recovered from newborn foreskins following trypsinization and plated on a feeder layer of mitomycin-treated 3T3-J2 fibroblasts in E medium containing 10% fetal calf serum (HyClone) and 0.4 μg of hydrocortisone per ml, 0.1 nM cholera toxin, 5 μg of transferrin per ml, 2 nM 3,3′-5-triodo-l-thyronine, 5 ng of epidermal growth factor per ml, and 1× antibiotic-antimycotic solution (100 U of penicillin per ml, 0.1 mg of streptomycin per ml, and 0.25 μg of amphotericin B per ml) (all supplements from Sigma) (51, 55). After one passage in serum-free medium (SFM; Gibco/BRL) containing 100 μM gentamicin, keratinocytes were plated at 1.2 × 105 cells per well into 12-well plates in SFM with 100 μM gentamicin. Cells were transfected with a total of 2.2 μg of DNA per well, including E1R-1luc and the internal control plasmid CMVβ-galactosidase, using the Polybrene transfection procedure previously described (1, 51). Where indicated, the CDP expression plasmid (pCMVCDP) or empty vector containing only the regulatory region was also included in the transfection. Following transfection, cells were either maintained as undifferentiated cultures in SFM or switched to Dulbecco modified Eagle medium (DMEM) (with 1.8 mM Ca2+) with 10% fetal calf serum to induce differentiation, as described previously (1, 4, 21, 33).
Luciferase (luc) and β-galactosidase assays.
Keratinocyte extracts were prepared 40 to 48 h after transfection by using the lysis buffer and protocol from the Tropix Galacto-Light kit (Promega). The luc and β-galactosidase activities were assayed using the reagents and protocol provided by the kit. All luc activities were standardized using the β-galactosidase activities as described previously (1).
Nuclear extract preparation and electrophoretic mobility shift assays (EMSAs).
Nuclear extracts from keratinocytes were prepared using the method of Dignam et al. (14), as modified by Lee et al. (36). Extracts were made either from cells grown in SFM or from cells grown in methylcellulose, the latter treatment to induce differentiation, as described previously (1, 21, 57). EMSAs were performed as described previously (1). Briefly, 2 μg of nuclear extract, 1.0 μg of poly(dI-dC) (Pharmacia), plus or minus competitor double-stranded oligonucleotides, and monoclonal anti-YY1 (Santa Cruz) or a control isotype-matched antibody, PIN1.1 (a kind gift from Janice Blum, Indiana University School of Medicine), were incubated on ice for 15 min prior to the addition of 20,000 Cerenkov counts of 32P-labeled probe. After an additional 10-min incubation on ice, complexes and free probe were separated on 3.5% nondenaturing polyacrylamide gels. The EMSAs for Fig. 2B and C were performed with minor modifications. Nuclear extract was mixed with probe and 0.5 μg of poly(dI-dC) and incubated on ice for 15 min. The unlabeled competitors at a 20- or 200-fold molar excess, polyclonal anti-CDP antisera (a kind gift from Ellis Neufeld, Harvard University), or preimmune antisera were added and further incubated for 10 min on ice, prior to separation of complexes as described above.
RESULTS
The E1 promoter of HPV-31 binds CDP.
We previously reported that CDP negatively regulates the E1 promoter of a low-risk virus, HPV-6 (1). To determine whether this mode of regulation of the differentiation-specific promoter is conserved among low- and high-risk HPVs, the E1 promoter of the high-risk HPV-31 was radiolabeled and analyzed using EMSAs. The sequence of the HPV-31 E1 promoter is shown in Fig. 2A. Inspection of this regulatory region suggests that putative binding sites for CDP (ATTA and CAAT) are present (2, 6, 27, 34, 48). A C1 complex was formed on HPV-31 E1p (Fig. 2B, lane 2). The presence of CDP in the HPV-31 E1p C1 complex was verified by addition of CDP antisera to the EMSA (Fig. 2C, lane 3) and by competition with E36 (2), a CDP-specific oligonucleotide (data not shown). Competition experiments indicated that the affinity of the HPV-31 E1p for CDP was at least as high as that of HPV-6 E1a (Fig. 2B, lanes 2 to 6 and 8 to 12). The HPV-6 E1a oligonucleotide is smaller than the HPV-31 E1p oligonucleotide, but the majority, if not all, of the HPV-6 E1p CDP binding activity is contained within E1a (1).
A second complex, the C2 complex, was formed on HPV-6 E1a but not on HPV-31 E1p.
Inspection of the retarded bands seen with radiolabeled HPV-6 E1a and HPV-31 E1p revealed a second complex, labeled C2, formed with only HPV-6 E1a. The C2 complex was competed by an approximately 10-fold-lower molar excess of HPV-6 E1a than of HPV-31 E1p. This complex and another, the C3 complex, were previously seen in EMSAs using the radiolabeled HPV-6 E1 promoter, E1p (Fig. 1 and 3). Since these complexes appeared unique to the low-risk HPV-6 E1 promoter, we pursued the identity of the protein present in them.
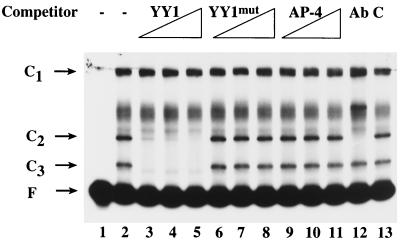
FIG. 3.
YY1 binds the HPV-6 E1 promoter. EMSAs were conducted using radiolabeled E1p (Fig. 1) and either no competitor (−) or a 20-, 50-, or 200-fold molar excess of unlabeled oligonucleotide containing a YY1 binding site (YY1), a mutated YY1 binding site (YY1mut), or an AP-4 binding site (AP-4). In lane 12, antibody to YY1 (Ab) was included in the reaction mixture, and in lane 13, a control antibody (C) was included. The YY1 and YY1mut oligonucleotides were purchased from Santa Cruz. Two complementary oligonucleotides were synthesized (Gibco/BRL) and subsequently annealed to produce the AP-4 competitor for use in EMSAs. The sequence of the top strand was 5′ GTGGTCAGCTGTAGGGCATCA 3′.
YY1 binds the HPV-6 E1 promoter.
The C2 and C3 complexes were competed by a YY1 oligonucleotide used as an unrelated competitor (1) (Fig. 3, lanes 2 through 5). When the sequence of the YY1 oligonucleotide used in competition experiments was screened by an online transcriptional factor searching program (MatInspector Version 2.2, http://transfac.gbf-braunschweig.de/TRANSFAC/index.html [52]), a potential AP-4 binding site was found in addition to a YY1 binding site. To test the hypothesis that the C2 and C3 complexes contained YY1, EMSAs were conducted using an oligonucleotide mutated in the YY1 binding site, YY1mut. To determine whether the C2 and C3 complexes contained AP-4, an AP-4-specific oligonucleotide was synthesized and used as a competitor in the EMSA. While the C2 and C3 complexes were competed with 20×, 50×, and 200× molar excess unlabeled YY1 oligonucleotide (Fig. 3, lanes 3 to 5), under the same conditions the YY1mut oligonucleotide was not able to compete the binding (Fig. 3, lanes 6 to 8), suggesting that YY1 was present in the C2 and C3 complexes. As indicated in Fig. 3 (lanes 9 to 11), 20×, 50×, and 200× molar excess unlabeled AP-4 oligonucleotide failed to eliminate binding, indicating that AP-4 is not present in the C2 or C3 complex. The presence of YY1 in the C2 and C3 complexes was further tested using YY1 monoclonal antibody in the EMSA. The C2 complex was supershifted by a monoclonal antibody directed to the N terminus of YY1 (Fig. 3, lane 12) but not by a control monoclonal antibody, PIN1.1 (Fig. 1, lane 13), indicating that YY1 was present in the C2 complex. The C3 complex, however, was not supershifted by the YY1 antibody. Given that other studies have demonstrated that, in addition to intact YY1, some nuclear extracts contain a proteolytic cleavage product of YY1 which retains DNA binding ability (44, 73), two further tests were conducted. First, when polyclonal anti-YY1 was used in the EMSA, both the C2 and C3 complexes were supershifted (data not shown), demonstrating that YY1 or its derivative is present in the C3 complex. Second, the protease inhibitors leupeptin and aprotinin were used in addition to phenylmethylsulfonyl fluoride during the preparation of nuclear extracts. In this case, only the C2 complex was observed, suggesting that the C3 complex was formed by a cleavage product of the protein present in the C2 complex (data not shown).
Localization of the YY1 binding site to the 5′ end of the E1 promoter.
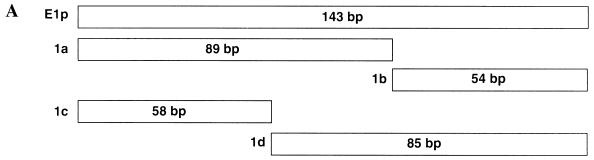
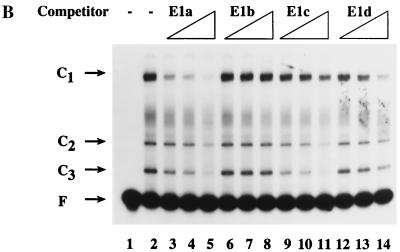
To determine the minimum fragment of the E1 promoter to which YY1 could bind, the promoter was digested as described previously with several restriction enzymes to yield four subfragments, E1a, E1b, E1c, and E1d (1) (Fig. 4A). When E1a and E1c were used as unlabeled competitors in an EMSA with radiolabeled E1 promoter, the amount of radiolabeled C2 and C3 complexes decreased (Fig. 4B, lanes 3 to 5 and 9 to 11). In contrast, addition of unlabeled E1b or E1d had little or no effect on the amount of radiolabeled C2 and C3 complex formation (Fig. 4B, lanes 6 to 8 and 12 to 14). These data suggested that the YY1 binding site resided within the E1a and E1c fragments, nt 528 to 616 and 528 to 585, respectively. In addition, a 51-bp unlabeled competitor containing nt 560 to 610 failed to decrease the amount of radiolabeled C2 and C3 complexes (data not shown). Therefore, we concluded that the YY1 binding site was located at the 5′ end of the E1 promoter between nt 528 and 560, or overlapping nt 560.
FIG. 4.
The YY1 binding site is at the 5′ end of the E1 promoter. (A) Fragments used to dissect the E1 promoter for the YY1 binding site(s) are shown. The nucleotide coordinates of E1p and E1a are provided in the legend to Fig. 1. E1c extends from nt 528 to 585; E1d extends from nt 586 to 670. (B) EMSAs were conducted using radiolabeled E1p and unlabeled E1p subfragments as competitors. The C1 complex contains CDP, most of the binding activity of which is located in E1a (1).
Mutational studies demonstrate that YY1 represses E1 promoter activity in undifferentiated keratinocytes but not in differentiated keratinocytes.
The sequence CCAT is the most frequent core sequence of YY1 binding sites (31, 71). A putative YY1 binding site with this CCAT core sequence is located at the extreme 5′ end of the E1a fragment, 5′ TTCCATGCA 3′ (antisense orientation) (Fig. 5A). A comparison of this site to the YY1 binding sites in the HPV-16 (48) and HPV-31 (34) long control region is shown in Fig. 5B. To determine whether this site was, indeed, a YY1 binding site, mutations were introduced into E1p as shown in Fig. 5A. Both E1p and the E1 promoter mutated in the YY1 binding site (E1pmYY1) were then used in EMSAs. The pattern of retarded bands visualized with the radiolabeled promoters was identical with the exception that, as predicted, no C2 complex was seen using the mutated promoter (Fig. 5B, lanes 2 and 10). Consistent with this latter observation, E1pmYY1 was not able to compete for formation of the C2 complex (Fig. 5B, lanes 6 to 8). Sequence analysis suggested that 3′ to the CCAT site might be two overlapping YY1 binding sites, one on each strand, with ACAT cores. However, since no YY1-containing complex was formed on E1pmYY1, the significance of the ACAT sites was not pursued. Both the wild-type and mutated promoters formed similar quantities of the CDP-containing complex, and both were equally effective at competing for binding to CDP, suggesting that the mutations in the YY1 binding site did not affect the ability of CDP to bind to the E1 promoter (Fig. 5B; compare lanes 3 to 5 with 6 to 8 and 11 to 13 with 14 to 16).
FIG. 5.
Mutation of the YY1 binding site results in loss of YY1 binding. (A) The sequence of the HPV-6 E1 promoter extending from the ATG of E7 at nt 528 through the E1 transcriptional start site at nt 680 is provided. The YY1 binding site identified in this study is underlined; the mutations are shown above the wild-type sequence. Putative CDP binding sites on either strand are denoted by a double underline; restriction sites are indicated by a dashed underline. (B) The alignment of the HPV-6 E1p putative YY1 binding site with CCAT-containing YY1 binding sites in HPV-16 (48) and HPV-31 (34) is shown. Nt, the nucleotide position within the genome of the first nucleotide shown under Sequence; r, antisense orientation. (C) EMSAs were conducted using radiolabeled E1p (lanes 1 to 8) or E1pmYY1 (lanes 9 to 16) and unlabeled competitor E1p (lanes 3 to 5 and 11 to 13) or E1pmYY1 (lanes 6 to 8 and 14 to 16).
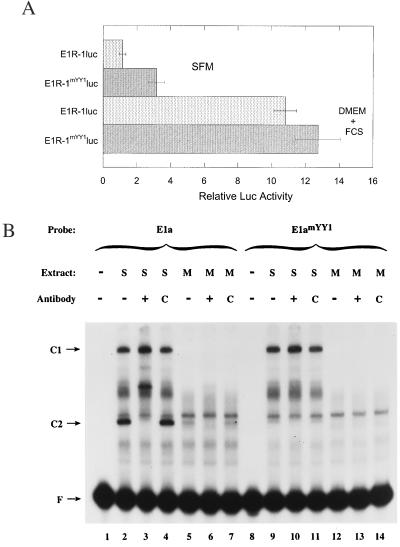
To determine the functional significance of YY1 binding to the E1 promoter, the activity of the mutated promoter was compared to that of the wild-type promoter. As described previously, the luc reporter gene was cloned at the position of the E1 translational start site (Fig. 1) (1). Keratinocytes were transfected and maintained as undifferentiated cells by incubation in SFM. After 48 h, the cells were lysed and assayed for luc activity. After correction for transfection efficiency, the relative luc activity of the mutated regulatory region (E1R-1mYY1) was compared to that of the wild-type regulatory region (E1R-1). There was threefold-greater activity with the mutated regulatory region, indicating that YY1 acts as a negative regulator in undifferentiated keratinocytes (Fig. 6A).
FIG. 6.
YY1 represses the transcription from the E1 regulatory region. (A) Keratinocytes were transfected with either the wild-type (E1R-1) or the mutated (E1R-1mYY1) regulatory region promoting expression of the reporter gene luc. Cells were grown in undifferentiated conditions (SFM) or differentiated conditions (DMEM plus fetal calf serum [FCS]) for 48 h, and cell lysates were subsequently assayed for luc activity. Activity was corrected for that of the cotransfected β-galactosidase-encoding control plasmid. Relative luc activity is plotted comparing all activities to that obtained with E1R-1luc in SFM, with the latter set to 1.0. (B) EMSAs were conducted using extracts obtained from cells grown in SFM (S, lanes 2 to 4 and 9 to 11) or in methylcellulose (M, lanes 5 to 7 and 12 to 14). +, anti-YY1; C, control antibody.
The E1 promoter is up-regulated during differentiation. To determine whether YY1 acts as an activator in differentiated cells, the activities of the wild-type and mutated regulatory regions were compared in cells induced to differentiate. Keratinocytes were transfected as above and incubated under differentiating conditions (in DMEM plus fetal calf serum) (1, 4, 21, 33). After correction for transfection efficiency, the activity of the regulatory regions was compared to that of the wild-type regulatory region in undifferentiated keratinocytes (Fig. 6A). In contrast to the increased activity of the mutated regulatory region in undifferentiated conditions, the activity of both promoters was comparable in differentiated cells. As expected, both promoters were up-regulated upon differentiation.
These data suggest that YY1 plays a regulatory role in undifferentiated keratinocytes but not in differentiated keratinocytes. To determine whether the YY1 complex is formed in differentiated cells, nuclear extracts were prepared from cells transferred to methylcellulose to induce differentiation (1, 21, 57). As shown in Fig. 2 through 5, the C2 complex, which can be supershifted with antibodies to YY1, is formed in nuclear extracts from undifferentiated cells (S) (Fig. 6B, lanes 2 to 4). In contrast, this complex is greatly reduced in extracts from cells induced to differentiate (M) (Fig. 6B, lanes 5 to 7). As reported previously, the C1 complex is also formed only in nuclear extracts from undifferentiated cells (Fig. 6B, lanes 2 to 7 and 9 to 14) (1). In contrast, there are other bands that remain constant or increase upon differentiation. Thus, the absence of a functional effect of YY1 in differentiated cells is consistent with the absence of a YY1-mediated band shift detectable in EMSAs.
CDP negatively regulates the E1 promoter independently of the YY1 binding site.
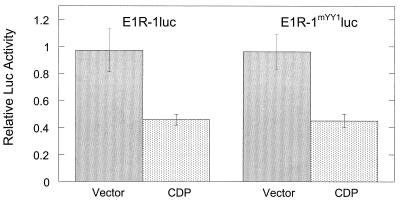
Overexpression of CDP decreases expression from the E1 regulatory region (1). The CDP binding sites have not been fully defined, but there are at least two binding sites within E1a (1). The EMSAs shown in Fig. 5B suggest that the YY1 binding site and CDP binding sites do not overlap. Functional experiments were conducted as an independent verification of this interpretation. Keratinocytes were cotransfected with the E1R-1luc or E1R-1mYY1luc plasmid and either empty vector or a CDP-expressing vector. The luc activity was normalized to an internal control and subsequently compared to the activity of the regulatory region cotransfected with empty vector. Both regulatory regions were negatively regulated to the same extent in the presence of CDP (Fig. 7). This indicates either that there is no CDP binding site overlapping the YY1 binding site since the mutations introduced into YY1 do not affect the binding of CDP to that region or that binding of CDP to this site is not critical to the activity of CDP on this regulatory region.
FIG. 7.
CDP-mediated negative regulation of the E1 regulatory region is independent of the YY1 binding site. Keratinocytes were cotransfected with either an empty vector or the same vector overexpressing CDP and a plasmid containing either the wild-type or mutated E1R-1 regulatory region upstream of the luc gene. Cells were grown in undifferentiated conditions (SFM) for 48 h, and cell lysates were subsequently assayed for luc activity. For each regulatory region, the activity was corrected for that of the internal control and relative luc activity was plotted comparing the activity in the presence of CDP to that in the presence of empty vector, the latter activity being set to 1.0.
DISCUSSION
HPV gene expression is dependent on keratinocyte differentiation. The most differentiation-dependent promoter is the E1 promoter. Little is known about the factors regulating this promoter. Presumably, upon differentiation there is a decrease in the level of transcriptional repressors and/or an increase in the level of transcriptional activators. Data presented here indicate that YY1, a differentiation-dependent transcriptional factor, binds to the HPV-6 E1 promoter and negatively regulates it in undifferentiated cells, as shown by mutational analysis. Upon keratinocyte differentiation, binding of YY1 to the E1 promoter was significantly decreased and comparable functional activity was seen with the mutated and the wild-type E1 promoters. Previously, we have shown that CDP, another differentiation-dependent transcription factor, regulated HPV-6 E1 promoter activity in a way similar to that for YY1. We have now further shown that the ability of CDP to regulate the HPV-6 E1 promoter was independent of a functional YY1 binding site. Thus, two negative regulators of transcription, which function in undifferentiated but not differentiated keratinocytes, bind the differentiation-dependent E1 promoter of low-risk HPV-6 (Fig. 8). While the E1 promoter of high-risk HPV-31 was also shown to bind CDP, no binding by YY1 was detected.
FIG. 8.
Two negative regulators, YY1 and CDP, bind the HPV-6 E1 promoter. The binding sites for CDP were described previously (1). Data supporting the presence of a YY1 binding site are provided here.
YY1, as a dual-function transcriptional factor, regulates a number of cellular and viral promoters (59, 60). There are several proposed mechanisms for YY1-mediated repression, which may not be mutually exclusive (see reviews by Shi et al. [59] and Thomas and Seto [67]. One mechanism of repression is activator displacement, in which YY1 displaces a transcriptional activator, such as AP-1 in the human gamma interferon promoter (72), and signal transducer and activator of transcription 5 in the beta-casein promoter (18, 53). Recent data suggest that, in the HPV-16 long control region, YY1 may exert part of its repressor activity by competing with Sp1 for binding (17). Another mechanism of repression is activator quenching, in which YY1 physically interacts either with a transcriptional activator or with the basal transcription machinery normally responsive to that activator. For instance, in the c-fos promoter there are two YY1 binding sites adjacent to an ATF/CREB binding site. It has been reported that YY1 can inhibit CREB-mediated transactivation either by interacting with CREB (74) or in the absence of binding to CREB (24). In HPV-16, YY1 exerts its repressor activity by inhibiting the transcriptional activity of AP-1 (48). O'Connor et al. showed that YY1 interacted with the histone acetylase CREB binding protein, a coactivator of AP-1, and postulated that, in so doing, YY1 quenched AP-1 activity (48). Finally, recent findings suggest that YY1 may function as a repressor by interacting with cofactors bearing histone deacetylase (HDAC) activity (59, 67, 70). For example, YY1 repressed transcription by tethering human RPD3, a member of the RPD3 family of HDACs, to a synthetic promoter (69). Deacetylated histones reflect more stable chromatin folding, resulting in decreased accessibility for DNA binding factors. No example of this latter mechanism has been described for YY1-mediated repression of an HPV promoter.
It is unknown how YY1 represses the HPV-6 E1 promoter. An online transcriptional factor searching program (52) did not detect any other transcription factor binding sites overlapping the YY1 binding site. However, a putative AP-1 site was detected approximately 68 nt downstream of the YY1 site, indicating that the mechanism of transactivator quenching might apply to the E1 promoter. It is also possible that YY1 recruits a cofactor that has HDAC activity, thereby inhibiting transcription. This model may apply to the other HPV-6 E1 promoter repressor, CDP. Recently, it was shown that the C terminus of CDP binds HDAC1 and that immunocomplexes of CDP possess associated HDAC activity (39). Thus, in the case of the HPV-6 E1 promoter, CDP and YY1 might independently recruit HDACs, deacetylate a component of basal transcriptional machinery, and inhibit the transcription.
The extent of HPV DNA replication may be regulated by affecting levels of functional replication proteins, E1 and E2, or access of these proteins to the origin of replication. We have reported that CDP negatively regulates the HPV-6 E6, E7, and E1 promoters. O'Connor et al. have recently extended these observations to show that CDP also negatively regulates the E6 promoter of high-risk HPVs, HPV-16, HPV-18, and HPV-31 (47). Here we report that, in addition, CDP binds the HPV-31 E1 promoter. Negative regulation of transcription by CDP would limit the availability of E1- and E2-containing transcripts, whether the E6, E7, or E1 promoter is used, in both high- and low-risk viruses and thereby the quantity of replication proteins. The origin of replication is located within the HPV E6 promoter (10, 12), and O'Connor et al. have presented data consistent with the interpretation that CDP also regulates interactions at the replication origin of HPV-16 and HPV-31 (47). Finally, by affecting both replication and utilization of the E1 promoter, viral DNA amplification and late gene expression would be coordinately controlled in the nonproductive environment. Thus, CDP acts as a master switch controlling and coordinating viral transcription and replication.
It appears that YY1 may be a second coordinating protein. Lee et al. (37) demonstrated that YY1 represses both HPV-18 and HPV-11 DNA replication in vitro. While the HPV-18 origin of replication contains a proximal YY1 binding site, the HPV-11 origin does not. The mechanism of repression appears to involve the interaction of YY1 with the E2 protein (37). We have demonstrated here that YY1 represses transcription from the HPV-6 E1 promoter, presumably thereby limiting the synthesis of replication proteins. Thus, at least for the low-risk viruses, YY1 may limit replication in undifferentiated cells by limiting expression of viral replication proteins and limiting access of those proteins to the replication origin. It would also ensure that templates for late gene expression, from the E1 promoter, were not made in abundance.
ACKNOWLEDGMENTS
This work was supported by NIH grant AI31494.
We thank Jean Bang and Grova Mae Lewis for excellent technical assistance and Mark Kaplan, David Skalnik, Lucinda Carr, and Michael Klemsz for helpful discussions and critical reading of the manuscript.
REFERENCES
- 1.Ai W, Toussaint E, Roman A. CCAAT displacement protein binds to and negatively regulates human papillomavirus type 6 E6, E7, and E1 promoters. J Virol. 1999;73:4220–4229. doi: 10.1128/jvi.73.5.4220-4229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andres V, Chiara M D, Mahdavi V. A new bipartite DNA-binding domain: cooperative interaction between the cut repeat and homeo domain of the cut homeo proteins. Genes Dev. 1994;8:245–257. doi: 10.1101/gad.8.2.245. [DOI] [PubMed] [Google Scholar]
- 3.Andres V, Nadal-Ginard B, Mahdavi V. Clox, a mammalian homeobox gene related to Drosophila cut, encodes DNA-binding regulatory proteins differentially expressed during development. Development. 1992;116:321–334. doi: 10.1242/dev.116.2.321. [DOI] [PubMed] [Google Scholar]
- 4.Apt D, Watts R M, Suske G, Bernard H U. High Sp1/Sp3 ratios in epithelial cells during epithelial differentiation and cellular transformation correlate with the activation of the HPV-16 promoter. Virology. 1996;224:281–291. doi: 10.1006/viro.1996.0530. [DOI] [PubMed] [Google Scholar]
- 5.Auborn K J, Little R D, Platt T H, Vaccariello M A, Schildkraut C L. Replicative intermediates of human papillomavirus type 11 in laryngeal papillomas: site of replication initiation and direction of replication. Proc Natl Acad Sci USA. 1994;91:7340–7344. doi: 10.1073/pnas.91.15.7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aufiero B, Neufeld E J, Orkin S H. Sequence-specific DNA binding of individual cut repeats of the human CCAAT displacement/cut homeodomain protein. Proc Natl Acad Sci USA. 1994;91:7757–7761. doi: 10.1073/pnas.91.16.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bauknecht T, Angel P, Royer H D, zur Hausen H H. Identification of a negative regulatory domain in the human papillomavirus type 18 promoter: interaction with the transcriptional repressor YY1. EMBO J. 1992;11:4607–4617. doi: 10.1002/j.1460-2075.1992.tb05563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bedell M A, Hudson J B, Golub T R, Turyk M E, Hosken M, Wilbanks G D, Laimins L A. Amplification of human papillomavirus genomes in vitro is dependent on epithelial differentiation. J Virol. 1991;65:2254–2260. doi: 10.1128/jvi.65.5.2254-2260.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiang C M, Ustav M, Stenlund A, Ho T F, Broker T R, Chow L T. Viral E1 and E2 proteins support replication of homologous and heterologous papillomaviral origins. Proc Natl Acad Sci USA. 1992;89:5799–5803. doi: 10.1073/pnas.89.13.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chow L T, Broker T R. Papillomavirus DNA replication. Intervirology. 1994;37:150–158. doi: 10.1159/000150373. [DOI] [PubMed] [Google Scholar]
- 11.Chow L T, Nasseri M, Wolinsky S M, Broker T R. Human papillomavirus types 6 and 11 mRNAs from genital condylomata acuminata. J Virol. 1987;61:2581–2588. doi: 10.1128/jvi.61.8.2581-2588.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Del Vecchio A M, Romanczuk H, Howley P M, Baker C C. Transient replication of human papillomavirus DNAs. J Virol. 1992;66:5949–5958. doi: 10.1128/jvi.66.10.5949-5958.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demeret C, Le Moal M, Yaniv M, Thierry F. Control of HPV 18 DNA replication by cellular and viral transcription factors. Nucleic Acids Res. 1995;23:4777–4784. doi: 10.1093/nar/23.23.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DiLorenzo T P, Steinberg B M. Differential regulation of human papillomavirus type 6 and 11 early promoters in cultured cells derived from laryngeal papillomas. J Virol. 1995;69:6865–6872. doi: 10.1128/jvi.69.11.6865-6872.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dollard S C, Wilson J L, Demeter L M, Bonnez W, Reichman R C, Broker T R, Chow L T. Production of human papillomavirus and modulation of the infectious program in epithelial raft cultures. Genes Dev. 1992;6:1131–1142. doi: 10.1101/gad.6.7.1131. [DOI] [PubMed] [Google Scholar]
- 17.Dong X P, Pfister H. Overlapping YY1- and aberrant SP1-binding sites proximal to the early promoter of human papillomavirus type 16. J Gen Virol. 1999;80:2097–2101. doi: 10.1099/0022-1317-80-8-2097. [DOI] [PubMed] [Google Scholar]
- 18.Doppler W, Welte T, Philipp S. CCAAT/enhancer-binding protein isoforms beta and delta are expressed in mammary epithelial cells and bind to multiple sites in the beta-casein gene promoter. J Biol Chem. 1995;270:17962–17969. doi: 10.1074/jbc.270.30.17962. [DOI] [PubMed] [Google Scholar]
- 19.Durst M, Bosch F X, Glitz D, Schneider A, zur Hausen H H. Inverse relationship between human papillomavirus (HPV) type 16 early gene expression and cell differentiation in nude mouse epithelial cysts and tumors induced by HPV-positive human cell lines. J Virol. 1991;65:796–804. doi: 10.1128/jvi.65.2.796-804.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farr A, Wang H, Kasher M S, Roman A. Relative enhancer activity and transforming potential of authentic human papillomavirus type 6 genomes from benign and malignant lesions. J Gen Virol. 1991;72:519–526. doi: 10.1099/0022-1317-72-3-519. [DOI] [PubMed] [Google Scholar]
- 21.Flores E R, Lambert P F. Evidence for a switch in the mode of human papillomavirus type 16 DNA replication during the viral life cycle. J Virol. 1997;71:7167–7179. doi: 10.1128/jvi.71.10.7167-7179.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frattini M G, Laimins L A. The role of the E1 and E2 proteins in the replication of human papillomavirus type 31b. Virology. 1994;204:799–804. doi: 10.1006/viro.1994.1596. [DOI] [PubMed] [Google Scholar]
- 23.Frattini M G, Lim H B, Laimins L A. In vitro synthesis of oncogenic human papillomaviruses requires episomal genomes for differentiation-dependent late expression. Proc Natl Acad Sci USA. 1996;93:3062–3067. doi: 10.1073/pnas.93.7.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galvin K M, Shi Y. Multiple mechanisms of transcriptional repression by YY1. Mol Cell Biol. 1997;17:3723–3732. doi: 10.1128/mcb.17.7.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldsborough M D, DiSilvestre D, Temple G F, Lorincz A T. Nucleotide sequence of human papillomavirus type 31: a cervical neoplasia-associated virus. Virology. 1989;171:306–311. doi: 10.1016/0042-6822(89)90545-x. [DOI] [PubMed] [Google Scholar]
- 26.Grassmann K, Rapp B, Maschek H, Petry K U, Iftner T. Identification of a differentiation-inducible promoter in the E7 open reading frame of human papillomavirus type 16 (HPV-16) in raft cultures of a new cell line containing high copy numbers of episomal HPV-16 DNA. J Virol. 1996;70:2339–2349. doi: 10.1128/jvi.70.4.2339-2349.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harada R, Berube G, Tamplin O J, Denis-Larose C, Nepveu A. DNA-binding specificity of the cut repeats from the human cut-like protein. Mol Cell Biol. 1995;15:129–140. doi: 10.1128/mcb.15.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harada R, Dufort D, Denis-Larose C, Nepveu A. Conserved cut repeats in the human cut homeodomain protein function as DNA binding domains. J Biol Chem. 1994;269:2062–2067. [PubMed] [Google Scholar]
- 29.Hummel M, Hudson J B, Laimins L A. Differentiation-induced and constitutive transcription of human papillomavirus type 31b in cell lines containing viral episomes. J Virol. 1992;66:6070–6080. doi: 10.1128/jvi.66.10.6070-6080.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hummel M, Lim H B, Laimins L A. Human papillomavirus type 31b late gene expression is regulated through protein kinase C-mediated changes in RNA processing. J Virol. 1995;69:3381–3388. doi: 10.1128/jvi.69.6.3381-3388.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hyde-DeRuyscher R P, Jennings E, Shenk T. DNA binding sites for the transcriptional activator/repressor YY1. Nucleic Acids Res. 1995;23:4457–4465. doi: 10.1093/nar/23.21.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iftner T, Oft M, Bohm S, Wilczynski S P, Pfister H. Transcription of the E6 and E7 genes of human papillomavirus type 6 in anogenital condylomata is restricted to undifferentiated cell layers of the epithelium. J Virol. 1992;66:4639–4646. doi: 10.1128/jvi.66.8.4639-4646.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones D L, Alani R M, Munger K. The human papillomavirus E7 oncoprotein can uncouple cellular differentiation and proliferation in human keratinocytes by abrogating p21Cip1-mediated inhibition of cdk2. Genes Dev. 1997;11:2101–2111. doi: 10.1101/gad.11.16.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanaya T, Kyo S, Laimins L A. The 5′ region of the human papillomavirus type 31 upstream regulatory region acts as an enhancer which augments viral early expression through the action of YY1. Virology. 1997;237:159–169. doi: 10.1006/viro.1997.8771. [DOI] [PubMed] [Google Scholar]
- 35.Klumpp D J, Laimins L A. Differentiation-induced changes in promoter usage for transcripts encoding the human papillomavirus type 31 replication protein E1. Virology. 1999;257:239–246. doi: 10.1006/viro.1999.9636. [DOI] [PubMed] [Google Scholar]
- 36.Lee K A, Bindereif A, Green M R. A small-scale procedure for preparation of nuclear extracts that support efficient transcription and pre-mRNA splicing. Gene Anal Tech. 1988;5:22–31. doi: 10.1016/0735-0651(88)90023-4. [DOI] [PubMed] [Google Scholar]
- 37.Lee K Y, Broker T R, Chow L T. Transcription factor YY1 represses cell-free replication from human papillomavirus origins. J Virol. 1998;72:4911–4917. doi: 10.1128/jvi.72.6.4911-4917.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee T C, Shi Y, Schwartz R J. Displacement of BrdUrd-induced YY1 by serum response factor activates skeletal alpha-actin transcription in embryonic myoblasts. Proc Natl Acad Sci USA. 1992;89:9814–9818. doi: 10.1073/pnas.89.20.9814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li S, Moy L, Pittman N, Shue G, Aufiero B, Neufeld E J, LeLeiko N S, Walsh M J. Transcriptional repression of the cystic fibrosis transmembrane conductance regulator gene, mediated by CCAAT displacement protein/cut homolog, is associated with histone deacetylation. J Biol Chem. 1999;274:7803–7815. doi: 10.1074/jbc.274.12.7803. [DOI] [PubMed] [Google Scholar]
- 40.Lievens P M, Donady J J, Tufarelli C, Neufeld E J. Repressor activity of CCAAT displacement protein in HL-60 myeloid leukemia cells. J Biol Chem. 1995;270:12745–12750. doi: 10.1074/jbc.270.21.12745. [DOI] [PubMed] [Google Scholar]
- 41.Liu R, Baillie J, Sissons J G, Sinclair J H. The transcription factor YY1 binds to negative regulatory elements in the human cytomegalovirus major immediate early enhancer/promoter and mediates repression in non-permissive cells. Nucleic Acids Res. 1994;22:2453–2459. doi: 10.1093/nar/22.13.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo W, Skalnik D G. CCAAT displacement protein competes with multiple transcriptional activators for binding to four sites in the proximal gp91phox promoter. J Biol Chem. 1996;271:18203–18210. doi: 10.1074/jbc.271.30.18203. [DOI] [PubMed] [Google Scholar]
- 43.May M, Dong X P, Beyer-Finkler E, Stubenrauch F, Fuchs P G, Pfister H. The E6/E7 promoter of extrachromosomal HPV16 DNA in cervical cancers escapes from cellular repression by mutation of target sequences for YY1. EMBO J. 1994;13:1460–1466. doi: 10.1002/j.1460-2075.1994.tb06400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meier V S, Groner B. The nuclear factor YY1 participates in repression of the beta-casein gene promoter in mammary epithelial cells and is counteracted by mammary gland factor during lactogenic hormone induction. Mol Cell Biol. 1994;14:128–137. doi: 10.1128/mcb.14.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meyers C, Frattini M G, Hudson J B, Laimins L A. Biosynthesis of human papillomavirus from a continuous cell line upon epithelial differentiation. Science. 1992;257:971–973. doi: 10.1126/science.1323879. [DOI] [PubMed] [Google Scholar]
- 46.Neufeld E J, Skalnik D G, Lievens P M, Orkin S H. Human CCAAT displacement protein is homologous to the Drosophila homeoprotein, cut. Nat Genet. 1992;1:50–55. doi: 10.1038/ng0492-50. [DOI] [PubMed] [Google Scholar]
- 47.O'Connor M J, Stunkel W, Koh C H, Zimmermann H, Bernard H U. The differentiation-specific factor CDP/Cut represses transcription and replication of human papillomaviruses through a conserved silencing element. J Virol. 2000;74:401–410. [PMC free article] [PubMed] [Google Scholar]
- 48.O'Connor M J, Tan S H, Tan C H, Bernard H U. YY1 represses human papillomavirus type 16 transcription by quenching AP-1 activity. J Virol. 1996;70:6529–6539. doi: 10.1128/jvi.70.10.6529-6539.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ozbun M A, Meyers C. Human papillomavirus type 31b E1 and E2 transcript expression correlates with vegetative viral genome amplification. Virology. 1998;248:218–230. doi: 10.1006/viro.1998.9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ozbun M A, Meyers C. Temporal usage of multiple promoters during the life cycle of human papillomavirus type 31b. J Virol. 1998;72:2715–2722. doi: 10.1128/jvi.72.4.2715-2722.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pattison S, Skalnik D G, Roman A. CCAAT displacement protein, a regulator of differentiation-specific gene expression, binds a negative regulatory element within the 5′ end of the human papillomavirus type 6 long control region. J Virol. 1997;71:2013–2022. doi: 10.1128/jvi.71.3.2013-2022.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raught B, Khursheed B, Kazansky A, Rosen J. YY1 represses beta-casein gene expression by preventing the formation of a lactation-associated complex. Mol Cell Biol. 1994;14:1752–1763. doi: 10.1128/mcb.14.3.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Remm M, Remm A, Ustav M. Human papillomavirus type 18 E1 protein is translated from polycistronic mRNA by a discontinuous scanning mechanism. J Virol. 1999;73:3062–3070. doi: 10.1128/jvi.73.4.3062-3070.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rheinwald J G. Serial cultivation of normal human epidermal keratinocytes. Methods Cell Biol. 1980;21A:229–254. doi: 10.1016/s0091-679x(08)60769-4. [DOI] [PubMed] [Google Scholar]
- 56.Rotenberg M O, Chow L T, Broker T R. Characterization of rare human papillomavirus type 11 mRNAs coding for regulatory and structural proteins, using the polymerase chain reaction. Virology. 1989;172:489–497. doi: 10.1016/0042-6822(89)90191-8. [DOI] [PubMed] [Google Scholar]
- 57.Ruesch M N, Stubenrauch F, Laimins L A. Activation of papillomavirus late gene transcription and genome amplification upon differentiation in semisolid medium is coincident with expression of involucrin and transglutaminase but not keratin-10. J Virol. 1998;72:5016–5024. doi: 10.1128/jvi.72.6.5016-5024.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schiffman M H, Bauer H M, Hoover R N, Glass A G, Cadell D M, Rush B B, Scott D R, Sherman M E, Kurman R J, Wacholder S. Epidemiologic evidence showing that human papillomavirus infection causes most cervical intraepithelial neoplasia. J Natl Cancer Inst. 1993;85:958–964. doi: 10.1093/jnci/85.12.958. [DOI] [PubMed] [Google Scholar]
- 59.Shi Y, Lee J S, Galvin K M. Everything you have ever wanted to know about Yin Yang 1…. Biochim Biophys Acta. 1997;1332:F49–F66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- 60.Shrivastava A, Calame K. An analysis of genes regulated by the multi-functional transcriptional regulator Yin Yang-1. Nucleic Acids Res. 1994;22:5151–5155. doi: 10.1093/nar/22.24.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Skalnik D G, Strauss E C, Orkin S H. CCAAT displacement protein as a repressor of the myelomonocytic-specific gp91-phox gene promoter. J Biol Chem. 1991;266:16736–16744. [PubMed] [Google Scholar]
- 62.Smotkin D, Prokoph H, Wettstein F O. Oncogenic and nononcogenic human genital papillomaviruses generate the E7 mRNA by different mechanisms. J Virol. 1989;63:1441–1447. doi: 10.1128/jvi.63.3.1441-1447.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sterling J C, Skepper J N, Stanley M A. Immunoelectron microscopical localization of human papillomavirus type 16 L1 and E4 proteins in cervical keratinocytes cultured in vivo. J Investig Dermatol. 1993;100:154–158. doi: 10.1111/1523-1747.ep12462790. [DOI] [PubMed] [Google Scholar]
- 64.Stoler M H, Whitbeck A, Wolinsky S M, Broker T R, Chow L T, Howett M K, Kreider J W. Infectious cycle of human papillomavirus type 11 in human foreskin xenografts in nude mice. J Virol. 1990;64:3310–3318. doi: 10.1128/jvi.64.7.3310-3318.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stoler M H, Wolinsky S M, Whitbeck A, Broker T R, Chow L T. Differentiation-linked human papillomavirus types 6 and 11 transcription in genital condylomata revealed by in situ hybridization with message-specific RNA probes. Virology. 1989;172:331–340. doi: 10.1016/0042-6822(89)90135-9. [DOI] [PubMed] [Google Scholar]
- 66.Thomas J T, Hubert W G, Ruesch M N, Laimins L A. Human papillomavirus type 31 oncoproteins E6 and E7 are required for the maintenance of episomes during the viral life cycle in normal human keratinocytes. Proc Natl Acad Sci USA. 1999;96:8449–8454. doi: 10.1073/pnas.96.15.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thomas M J, Seto E. Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene. 1999;236:197–208. doi: 10.1016/s0378-1119(99)00261-9. [DOI] [PubMed] [Google Scholar]
- 68.Usheva A, Shenk T. YY1 transcriptional initiator: protein interactions and association with a DNA site containing unpaired strands. Proc Natl Acad Sci USA. 1996;93:13571–13576. doi: 10.1073/pnas.93.24.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang W M, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc Natl Acad Sci USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang W M, Yao Y L, Sun J M, Davie J R, Seto E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J Biol Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- 71.Yant S R, Zhu W, Millinoff D, Slightom J L, Goodman M, Gumucio D L. High affinity YY1 binding motifs: identification of two core types (ACAT and CCAT) and distribution of potential binding sites within the human beta globin cluster. Nucleic Acids Res. 1995;23:4353–4362. doi: 10.1093/nar/23.21.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ye J, Cippitelli M, Dorman L, Ortaldo J R, Young H A. The nuclear factor YY1 suppresses the human gamma interferon promoter through two mechanisms: inhibition of AP1 binding and activation of a silencer element. Mol Cell Biol. 1996;16:4744–4753. doi: 10.1128/mcb.16.9.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ye J, Young H A, Ortaldo J R, Ghosh P. Identification of a DNA binding site for the nuclear factor YY1 in the human GM-CSF core promoter. Nucleic Acids Res. 1994;22:5672–5678. doi: 10.1093/nar/22.25.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou Q, Gedrich R W, Engel D A. Transcriptional repression of the c-fos gene by YY1 is mediated by a direct interaction with ATF/CREB. J Virol. 1995;69:4323–4330. doi: 10.1128/jvi.69.7.4323-4330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]