Abstract
A microaerophilic Gram-stain-negative bacilliform bacterial strain, FB-5 T, was isolated from activated sludge in Yokohama, Japan, that exhibited filamentous growth and formed a microtube (sheath). Cells were motile using a single polar flagellum. The optimum growth temperature and pH were 30 °C and 7.5, respectively. Strain FB-5 T was catalase-negative. Peptides and amino acids were utilized as energy and carbon sources. Sugars and organic acids were not utilized. Vitamin B12 enhanced the growth of strain FB-5 T. Sulfur-dependent lithotrophic growth was possible. Major respiratory quinone was UQ-8. Major fatty acids were C16:1ω7 and C16:0. The genomic DNA G + C content was 69.16%. Phylogenetic analysis of the 16S rRNA gene suggested that strain FB-5 T belongs to the genus Sphaerotilus. The close relatives were S. natans subsup. sulfidivorans and S. natans subsup. natans with 98.0% and 97.8% similarity based on the 16S rRNA gene analysis, respectively. The genome size (6.06 Mbp) was larger than that (4.39–5.07 Mbp) of the Sphaerotilus strains. The AAI values against the related strains ranged from 71.0 to 72.5%. The range of ANI values was 81.7 − 82.5%. In addition to these distinguishable features of the genome, the core genome and dDDH analyses suggested that this strain is a novel member of the genus Sphaerotilus. Based on its physiological properties and genomic features, strain FB-5 T is considered as a novel species of the genus Sphaerotilus, for which the name S. microaerophilus sp. nov. is proposed. The type strain is FB-5 T (= JCM 35424 T = KACC 23146 T).
Supplementary Information
The online version contains supplementary material available at 10.1007/s00203-024-03991-9.
Keywords: Sphaerotilus microaerophilus sp. nov., Sheath, Microaerophile, Activated sludge
Introduction
Bacterial strains of the genera Sphaerotilus and Leptothrix are sheath-forming bacteria within the class Betaproteobacteria, which are collectively called the Sphaerotilus-Leptothrix group due to their morphological, physiological, and phylogenetic relationships (van Veen et al. 1978; Siering and Ghiorse 1996). The Sphaerotilus-Leptothrix group belongs to the family Comamonadaceae of the order Burkholderiales (Willems et al. 1991). Recently, the family Sphaerotilaceae, which is closely related to the family Comamonadaceae and consists of members of the Sphaerotilus-Leptothrix group and related genera, has been approved by the International Committee on Systematics of Prokaryotes (ICSP) (Liu et al. 2022; Oren and Göker 2023). Members of the Sphaerotilus-Leptothrix group are widely distributed in aquatic environments including streams, springs, and activated sludge (van Veen et al. 1978; Emerson and Ghiorse 1992; Gaval et al. 2003; Baskar et al. 2012; Schmidt et al. 2014). The genus Sphaerotilus comprises four species, Sphaerotilus mobilis (formerly Leptothrix mobilis), Sphaerotilus hippei, Sphaerotilus montanus, and Sphaerotilus natans (further classified as S. natans subsp. natans and S. natans subsp. sulfidivorans) for which type strains are available (Gridneva et al. 2011; Liu et al. 2022). Reclassification of S. natans subsp. sulfidivorans as Sphaerotilus sulfidivorans has been proposed in 2021 (Grabovich et al. 2021). Currently, S. sulfidivorans is a synonym of S. natans subsp. sulfidivorans according to the List of Prokaryotic names with Standing in Nomenclature (LPSN). The genus Leptothrix is characterized by its ability to oxidize manganese which is not observed in the genus Sphaerotilus (van Veen et al. 1978), excluding S. mobilis (Spring et al. 1996). L. mobilis was reclassified to the genus Sphaerotilus as S. mobilis in 2022 based on the genus boundary values of ANI (78.95–82.14%) and AAI (67.12–71.55%) proposed for the Sphaerotilus-Leptothrix group and related genera (Liu et al. 2022). The strains belonging to the non-validly described species “Leptothrix discophora” were classified as Leptothrix cholodnii and L. discophora in 1996 (Spring et al. 1996). After classification, the genus Leptothrix comprises four species: Leptothrix ochracea, L. cholodnii, L. discophora, and Leptothrix lopholea. The type strains of L. ochracea, L. lopholea, and L. cholodnii are not available from culture collections (Spring et al. 1996; Yarza et al. 2013), although L. ochracea is the type species of the genus Leptothrix (Skerman et al. 1980). Instead of the type strain, a reference strain of L. cholodnii was proposed and is available (Spring et al. 1996). No genomic data is available for the type strains of L. ochracea, L. lopholea, and L. cholodnii in the database.
In this study, we report the isolation and taxonomic characterization of a bacterial strain originating from the activated sludge of the sewage treatment plant in Yokohama exhibiting a sheathed morphology typical of the genus Sphaerotilus. Here, we describe the isolation of strain FB-5 T, a member of the genus Sphaerotilus, and propose that the isolate belongs to a novel species, S. microaerophilus sp. nov.
Materials and methods
Isolation and cultivation
Strain FB-5 T was isolated from the activated sludge of the sewage treatment plant in Yokohama, Japan. The activated sludge was suspended with water and the suspension was streaked on an agar medium (named Screening medium) followed by incubation at 25 °C for 3 days. Colony isolation was performed three times at three-day intervals using the same medium at 25 °C. Screening medium was composed of 15 g/L agar, 0.15 g/L soluble starch, 50 mg/L (NH4)2SO4, 50 mg/L K2HPO4, 50 mg/L MgSO4∙7H2O, 50 mg/L KCl, 100 mg/L CaCO3, 14 mg/L Ca(NO3)2∙4H2O, 3 mg/L FeSO4∙7H2O, 0.3 mg/L H3BO3, 0.2 mg/L CoCl2∙6H2O, 0.1 mg/L ZnSO4∙7H2O, 0.03 mg/L MnCl2∙4H2O, 0.03 mg/L Na2MoO4∙2H2O, 0.02 mg/L NiCl2, 0.2 mg/L CuCl2, 0.4 mg/L thiamin HCl, and 0.01 mg/L vitamin B12. Colonies with a rough (hairy) appearance, possibly attributable to filamentous growth, were selected using this medium. For maintains, the isolate (strain FB-5 T) was subcultured on an agar plate (named APP medium) composed of 15 g/L agar, 2 g/L Proteose-Peptone No. 3 (Difco), and 0.01 mg/L vitamin B12 at 15–25 °C in the range of one week. For taxonomic characterization, the strain was statically cultured at 25 °C for 5 days in a medium (named GPPY medium) composed of 4 g/L glucose, 2 g/L Proteose-Peptone No. 3 (Difco), 0.2 g/L yeast extract (Difco), 0.2 g/L MgSO4‧7H2O, and 0.01 mg/L vitamin B12. For short-term storage in the range of one month, subcultures were performed at 15 °C using a semi-solidified GPPY medium containing 5 g/L agar. For long-term storage, the cells statically grown in GPPY medium at 25 °C for 5 days was stored at −80 °C in the presence of 20% glycerol. Lyophilization was not performed because of a loss of viability.
Morphology
The morphology was examined using a JSM-7001F scanning electron microscope (SEM, JEOL, Tokyo, Japan), a JEM-2100F transmission electron microscope (TEM, JEOL, Tokyo, Japan), and a SPA-400/SPI3800N scanning probe microscope (SPM, Hitachi High-Technologies, Tokyo, Japan).
Phenotypic analyses
Unless otherwise described, the cells statically grown in GPPY medium at 25 °C for 5 days was used for phenotypic characterization. Gram-staining was performed using Favor G Nissui (Nissui Pharmaceutical Co., Ltd., Tokyo, Japan) according to the supplier’s instructions. The utilization of organic compounds as sole carbon and energy sources was determined by monitoring the increase in turbidity (absorbance at 660 nm) after triplicate subcultures in a mineral medium (named NCM medium), which was used for the characterization of the Sphaerotilus strains (Gridneva et al.2011), mainly composed of (NH4)2SO4, CaCl2, MgSO4‧7H2O, Na2S2O3, and organic compound to be tested. Sulfur-dependent lithotrophic growth was examined using batch cultures under the conditions reported by Gridneva and collaborators (Gridneva et al.2011). Sulfur-dependent lithotrophic growth was also examined using fed-batch cultures feeding with Na2S solution (1 g/L) in a 500 mL-flask (four-necked round-bottom flask containing 300 mL of medium) at 25 °C using a modified DSMZ573 medium (named m-DSMZ573 medium) under the following conditions: medium, DSMZ573 medium (without sodium acetate) supplemented with 1 mL/L Wolfe’s vitamin solution (ATCC 2094 AN 1 medium); aeration, 0.1 L/min without agitation; pH, maintained at 7.2–7.8 using a pH controller (NPH-690D, Nisshin Rika, Tokyo, Japan); Na2S feeding, H2S concentration in the exhaust was maintained at 0.2–1 ppm using a H2S sensor (FECS50-100, Figaro Engineering, Osaka, Japan) in combination with a control device composed of E5CD and G3NA-205B-UTU (OMRON, Kyoto, Japan). In the fed-batch culture, bacterial H2S consumption (removal) was calculated based on the amount of Na2S solution added to the flask to maintain the H2S concentration in the exhaust. Growth was detected as relative light units (RLU) by the ATP (including ADP and AMP) bioluminescence assay using a Lumitester Smart (Kikkoman, Tokyo, Japan). Manganese oxidation was examined in a stab culture (25 °C, one month) using MSVP agar (Emerson and Ghiorse 1992; Siering and Ghiorse 1996; Takeda et al. 2002) containing 1 mM Mn2+. L. cholodnii ATCC 51168 (= SP-6) and S. natans JCM 20382 (= ATCC 15291) were used as the positive and negative controls, respectively. Note that the subspecies affiliation of S. natans JCM 20382 is not determined (Gridneva et al. 2011). Although these strains are not the type strains, they are commonly used as references to compare their phenotypic properties with those of related isolates (Sawayama et al. 2011; Nott et al. 2020; Kashiwabara et al. 2021; Kunoh et al. 2021). Growth under aerobic conditions was examined on GPPY agar (GPPY medium solidified with 1.5% agar) and APP medium at 25 °C for 10 days. Growth under microaerophilic conditions was examined by semi-solid-state cultivation in a test tube with a diameter of 1.8 cm at 25 °C for 10 days using GPPY medium supplemented with 0.5% agar. Microaerophilic growth was also examined on GPPY agar (GPPY medium solidified with 1.5% agar) at 25 °C for 10 days using AnaeroPouch-MicroAero (Mitsubishi Gas Chemical, Tokyo, Japan). Likewise, anaerobic growth was examined using AnaeroPouch-Anaero (Mitsubishi Gas Chemical), respectively. Catalase and oxidase activities were evaluated using 3% (v/v) H2O2 and 1% (w/v) tetramethyl-p-phenylenediamine, respectively, as previously described (Smibert 1994). The following characteristics were determined using the ID test NF-18 (Nissui Pharmaceutical): nitrate reduction to nitrite; nitrite reduction to nitrogen; gelatin liquefaction; indole production; hydrolysis of esculin, urea, and arginine; decarboxylation of lysine and ornithine; and β-galactosidase activity. The effect of temperature on growth was investigated using GPPY medium at 5–50 °C (5 °C intervals). The effect of pH was investigated at 25 °C using GPPY medium at pH 3–10 (1 pH unit intervals) adjusted with HCl or NaOH. The effect of NaCl concentration was investigated at 25 °C using GPPY medium supplemented with 0, 0.05, 0.1, 0.2, 0.3, 0.4, 0.5, 1, 2, or 3% (w/v) NaCl. The growth in static cultures was checked by monitoring turbidity (absorbance at 660 nm) for 5 days.
Chemotaxonomic analysis
The cellular fatty acid composition was determined using the Sherlock Microbial Identification System (MIDI) according to the manufacturer’s instructions. The major respiratory quinones were determined using the method described by Tamaoka and collaborators (Tamaoka et al. 1983). PHB accumulation was determined using gas chromatography, as described previously (Takeda et al. 1995). For these experiments, cells grown in GPPY medium were used because strain FB-5 T did not grow in the medium used for chemotaxonomic characterization of the closely related Sphaerotilus strains (Gridneva et al. 2011; Grabovich et al. 2021).
16S rRNA gene phylogeny
The 16S rRNA gene was amplified using colony PCR with the primers 27F (Jensen et al. 2009) and 1500R (Weisburg et al. 1991) and followed by Sanger sequencing (Applied Biosystems 3730xl DNA Analyzer; Applied Biosystems, Waltham, MA, USA) in two directions. The sequence was confirmed to be the same as that extracted from the genome assembly and deposited in DDBJ/ENA/GenBank under the accession number LC775240. Phylogenetic analysis using neighbor joining (NJ), unweighted pair group method with arithmetic mean (UPGMA), maximum likelihood (ML), and maximum parsimonious (MP) methods was performed based on the 16S rRNA gene sequences. Genes for analysis were extracted from the genome sequences (Table S1) of the reference strains using Barrnap 0.9 (https://github.com/tseemann/barrnap), followed by multiple alignment (Q-INS-i algorithm) using MAFFT version 7 (Katoh et al. 2019). NJ and UPGMA trees were constructed using PHYLIP 3.698 (https://evolution.genetics.washington.edu/phylip.html) based on pairwise sequence similarities (Table S2). Similarities were calculated using blastn implemented in BLAST 2.14.0 + . MP tree was constructed using MEGA 11.0.13 (Tamura et al. 2021). For a reliable estimation, only sequences within the Sphaerotilus-Leptothrix group were used, and the root was selected using the midpoint rooting method. The topological robustness of the NJ, UPGMA, and MP trees was evaluated using a bootstrap analysis with 1000 replicates. The maximum likelihood tree was constructed using IQ-TREE 2.3.3 (Minh et al. 2020) under the TN + F + I + G4 as the best-fit nucleotide substitution model, which was selected by the ModelFinder (Kalyaanamoorthy et al. 2017) according to the Bayesian information criterion. Branching robustness was estimated using the SH-like approximate likelihood ratio test (SH-aLRT) with 1000 replicates.
Genome sequencing and analysis
The genome sequence of strain FB-5 T was determined using the following procedure: after freezing in liquid nitrogen, the cell pellet was ground, and DNA was extracted using the Wizard Genomic DNA Purification Kit (Promega, Madison, CA, USA). After purification using AMPure XP (Beckman Coulter, Brea, CA, USA) and a DNeasy Power Clean Pro Cleanup Kit (Qiagen, Venlo, Netherlands), a DNA library was constructed using the SMRTbell Express Template Prep Kit 2.0 (Pacific Biosciences, Menlo Park, CA, USA). The library was sequenced using Sequel IIe (Pacific Biosciences) in combination with the Binding Kit 2.2 (Pacific Biosciences). Based on these sequences, consensus sequences were prepared and HiFi reads were obtained by omitting sequences of low reliability (< QC 20) using SMRT Link v11 software (Pacific Biosciences). After removing short reads (< 1000 bp) using Filtlong (https://github.com/rrwick/Filtlong), HiFi reads were assembled using Flye (https://github.com/fenderglass/Flye/blob/flye/docs/USAGE.md), and accuracy was confirmed using Bandage (Wick et al. 2015) and CheckM (Parks et al. 2015). Complete genomic data, including the plasmid, were deposited in DDBJ/ENA/GenBank under accession numbers AP025730 (genome) and AP025731 (plasmid). The assembled sequences were annotated using DFAST (Tanizawa et al. 2016), and 5311 genes were identified. The basic features of the genomic data used in this study, including those of strain FB-5 T, are listed in Table S1. The contamination and completeness were estimated using CheckM2 (Chklovski et al. 2023). The dDDH value was calculated using GGDC 3.0 (Meier-Kolthoff et al. 2022) with formular 2. A heat-map style matrix was produced using TBtools 2.016 (Chen et al. 2020). Multilocus sequence analysis based on the core gene set (concatenated sequences of 92 core genes from 32 genomes) was performed using UBCG 3.0 (Na et al. 2018), and a phylogenetic tree was constructed using FastTree (Price et al. 2010) implemented in the UBCG pipeline. Branching robustness was estimated using SH-aLRT with 1000 replicates. The genome-to-genome distance was calculated by AAI using EzAAI 1.2.2 (Kim et al. 2021), and the AAI matrix was produced using TBtools 2.016. The corresponding matrix plot was generated using PHYLIP 3.698. The genome-to-genome distance was further estimated by ANI using FastANI 1.33 (Jain et al. 2018). The ANI matrix was produced using TBtools 2.016, and the corresponding matrix plot was produced using PHYLIP 3.698.
Results and discussion
Isolation and maintenance
Initially, we attempted to isolate strains of the genus Haliscomenobacter (a genus in the phylum Bacteroidota comprising sheath-forming filamentous bacteria) capable of degrading various macromolecules, including starch and hyaluronate (Mori et al. 2023), using Screening medium containing starch. After repeated streak purification, a filamentous strain was isolated as a single colony and designated as strain FB-5 T. The colonies of strain FB-5 T on Screening medium exhibited a loss of viability after 7 days of cultivation. Growth was observed on a starch-free Screening medium. No growth was observed in agar-free (liquid) Screening medium. These results suggested that the growth of the isolate was supported by organic impurities in the agar and that strain FB-5 T was not a member of the genus Haliscomenobacter. The colonies formed on APP medium were irregularly (rough) shaped (Fig. S1a). Smooth colonies were rarely observed as well (Fig. S1b). Colonies of either shape produced colonies of both shapes in subculture. A rough colony was used for taxonomic characterization and subculture.
Morphology
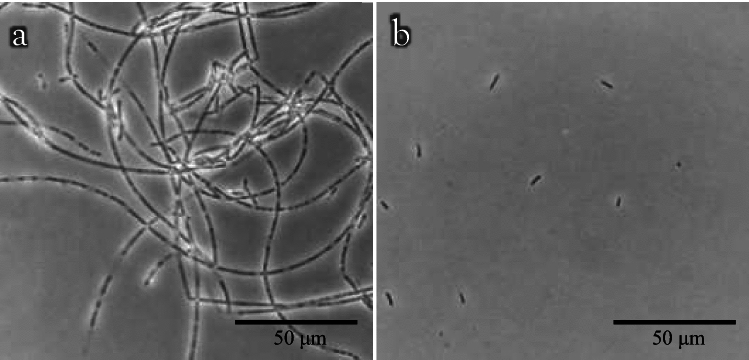
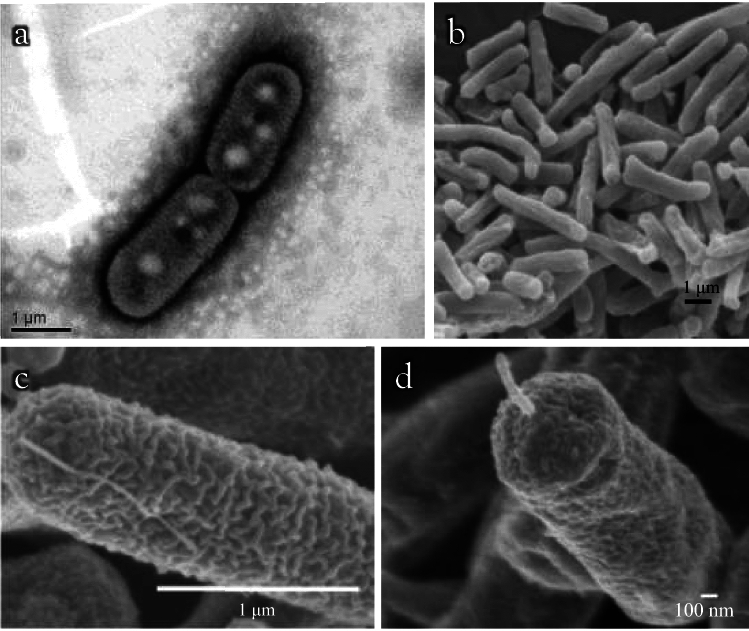
Using phase-contrast microscopy, both the filaments (lines of cells enclosed by sheaths; Fig. 1a) and single rod-shaped cells (cells without a sheath; Fig. 1b) were observed in static cultures using GPPY medium. Single rod-shaped cells occasionally exhibited motility. Cells were not connected, and filamentation was caused by sheath formation. Filamentation (sheath formation) was promoted by the addition of 0.1 g/L CaCO3 to the medium. The enhancement of sheath formation by Ca2+ and Mg2+ has been observed in L. cholodnii (Kunoh et al. 2021). However, no effect of Mg2+ for sheath formation of strain FB-5 T was suggested, as similar filamentation was observed in GPPY medium with or without MgSO4∙7H2O. Sheath formation was confirmed by SPM observation (Fig. S2). Filaments were commonly observed in the rough colonies (Fig. S1a) formed on a solid medium. In contrast, filaments were rare in the smooth colonies (Fig. S1b). Accordingly, a rough colony was selected to maintain the sheath-forming ability. Sheaths were not detected using TEM (Fig. 2a) or SEM (Fig. 2b), suggesting a weak sheath-forming capability of strain FB-5 T. The electron microscopy images (Fig. 2) revealed a cell size of 0.7–1.0 µm in width and 2.0–6.5 µm in length. The sthA (AB050638) and lthB (ACB33244) genes are essential glycosyl transferase genes for sheath formation in S. natans and L. cholodnii, respectively (Suzuki et al. 2002; Kunoh et al. 2023). A homology search using BLAST showed a sthA-like gene (WP_251972251/BDI04101) with 78% identity in the genome of strain FB-5 T, whereas a lthB-like gene was not detected, suggesting that strain FB-5 T forms a Sphaerotilus-type sheath (Kondo et al. 2011; Kashiwabara et al. 2021). Sphaerotilus-type sheath is mainly composed of glucuronic acid, galactosamine and glucose (Kondo 2011; Kashiwabara et al. 2021). In contrast, Leptothrix-type sheath contains N-acetylgalactose, galactosamine, galacturonic acid, and glucosamine (Takeda et al. 2005). Purification of the sheath and subsequent sugar composition analysis are required to determine the type of sheath of strain FB-5 T, which will be our future study. Traces of a single polar flagellum (the remaining short flagellum) were rarely observed in the SEM images (Fig. 2c, d). We assume that the motility of strain FB-5 T is attributed to a single polar flagellum, as described for other members of the genera Sphaerotilus and Leptothrix (Spring et al. 1996; Grabovich et al. 2021).
Fig. 1.
Phase-contrast microscopy images of strain FB-5 T. Strain FB-5 T was statically cultivated at 25 °C for 4 days using GPPY medium. The aggregated cells were enclosed within a sheath (a) whereas non-aggregated cells remained without a sheath (b)
Fig. 2.
Transmission (a) and scanning (b-d) electron microscopy images of strain FB-5 T. The cells grown on GPPY medium were negatively stained with phosphotungstic acid and subjected to transmission electron microscopy (a). The cells for scanning electron microscopy (b-d) were fixed with glutaraldehyde and osmium tetraoxide. Coating was performed with osmium
Physiology
Strain FB-5 T was stained Gram-negative. Growth was observed in GPPY medium containing 0–0.5% NaCl but not observed in the presence of 1–3% NaCl. Growth was observed only in media containing amino acid or amino acid-based organic compounds such as peptone, Proteose-Peptone No. 3, tryptone, yeast extract, and casamino acids (Table S3). To the best of our knowledge, Proteose-Peptone No. 3 is the most suitable energy and carbon source for growth. No sugars or organic acids served as the sole growth substrate. The preference for peptides and amino acids is a distinguishing feature of strain FB-5 T in comparison to other members of the genus Sphaerotilus. Vitamin B12 enhanced the growth of the strain because its growth was poor when vitamin B12 was omitted from GPPY medium. Vitamin B12 requirements are commonly recognized in the Sphaerotilus and Leptothrix strains (Okrend and Dondero 1964; Emerson and Ghiorse 1992). Sulfur-dependent lithotrophic growth was not observed in batch culture; however, growth (increase in RLU) and consumption of H2S were detected in fed-batch culture. Continuous supply of sulfide is probably desirable for lithotrophic cultivation of strain FB-5 T. As shown in Fig. S3, manganese oxidation was not observed in strain FB-5 T. Poor growth was observed in the uppermost part of the stab culture of strain FB-5 T, suggesting the strain is facultatively microaerophilic and prefers microaerophilic conditions rather than aerobic conditions, unlike S. natans and L. cholodnii. S. natans has been reported to oxidize iron, coupled with nitrate reduction catalyzed by nitrate reductase (WP_037485935) (Park et al. 2014). The amino acid sequence of the nitrate reductase of S. natans showed 71% identity with a putative nitrate reductase (WP_251972978) of strain FB-5 T in a BLAST search, suggesting that the strain has iron-oxidizing potential. To confirm the preference of strain FB-5 T for growth under microaerophilic conditions, a semi-solid culture was performed. Poor growth was observed at the air-medium interface, whereas colonies formed 0.3–1 cm below the interface different from S. natans (Fig. S4), indicating that strain FB-5 T is microaerophilic. No growth was observed under microaerophilic and anaerobic conditions produced by AnaeroPouch-MicroAero and AnaeroPouch-Anaero, respectively. Since AnaeroPouch-MicroAero produces atmosphere of 6–12% O2, strain FB-5 T is expected to prefer O2 concentrations above 12% and less than 21%. Additionally, strain FB-5 T was cytochrome oxidase-positive same as the Sphaerotilus strains, but catalase-negative different from the Sphaerotilus strains. Nitrate was reduced to nitrite. Nitrite was not reduced to nitrogen. Gelatin was not liquefied. Indole was not produced. Esculin, urea, and arginine were not hydrolyzed. Lysine and ornithine were not decarboxylated. β-Galactosidase activity was negative. Growth was observed in a temperature range of 15–35 °C, with 30 °C being optimal. The optimum pH was 7.5, whereas growth was observed in the pH range of 7–8.
Chemotaxonomy
As shown in Table S4, the fatty acids detected from strain FB-5 T were C16:1ω7 (49.8%), C16:0 (25.4%), C12:0 (10.8%), C18:1ω7 (5.5%), and C10:0 3-hydroxy (1.4%). Although the content of C12:0 was relatively high, the overall fatty acid composition was similar to that of other Sphaerotilus strains, including S. mobilis (Spring et al. 1996; Gridneva et al. 2011). The major respiratory quinone in strain FB-5 T was UQ-8. The accumulation of PHB was confirmed. UQ-8 and PHB are commonly detected in the genus Sphaerotilus (Grabovich et al. 2021).
16S rRNA gene-based phylogeny
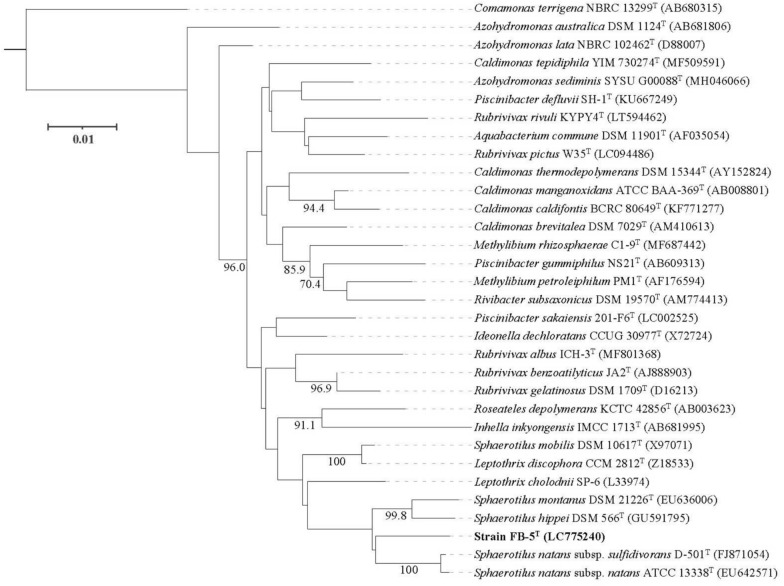
The pairwise identities of the 16S rRNA gene sequences of strains related to strain FB-5 T are listed in Table S2. As shown in the phylogenetic tree (NJ) based on the 16S rRNA gene (Fig. 3), strain FB-5 T was closely related to strains of the genus Sphaerotilus, particularly to S. natans subsup. sulfidivorans (98.0% similarity) followed by S. natans subsup. natans (97.8% similarity). Phylogenetic analysis using ML, UPGMA, and MP methods supported this result. The level of identity with S. hippei was 97.8%, with S. montanus 97.2%, and with S. mobilis 96.5%.
Fig. 3.
Phylogenetic tree (NJ) based on 16S rRNA gene sequences showing the relationship between strain FB-5 T and related strains. The percentages (> 80%) for 1000 bootstraps are shown next to the branches. The scale represents 0.01 nucleotide substitutions per site
Genomic features
The genome sequence of strain FB-5 T determined in this study exhibited a low level of contamination (0.26%), with 99.99% completeness, as shown in Table S1. The genomic DNA G + C content of strain FB-5 T was 69.16%, which was within the range (68.16 − 70.03%) of the Sphaerotilus strains. In the genome, 5,236 protein-coding and 75 non-coding genes were identified. The genome size of strain FB-5 T (6.06 Mbp) was higher than that of the Sphaerotilus strains (4.39 − 5.07 Mbp), which distinguished the isolate from related strains of the genus Sphaerotilus. Supporting the lithotrophic growth capability of strain FB-5 T in fed-batch culture, the genes required for sulfur metabolism and Calvin–Benson–Bassham cycle were observed in the genome as listed in Tables S5 and S6, respectively. Accordingly, strain FB-5 T was revealed to be lithotrophic, the same as S. natans subsp. sulfidivorans (Gridneva et al. 2011; Grabovich et al. 2021). Comparative genomic analysis was performed to further characterize the strain as a member of the Sphaerotilus-Leptothrix group.
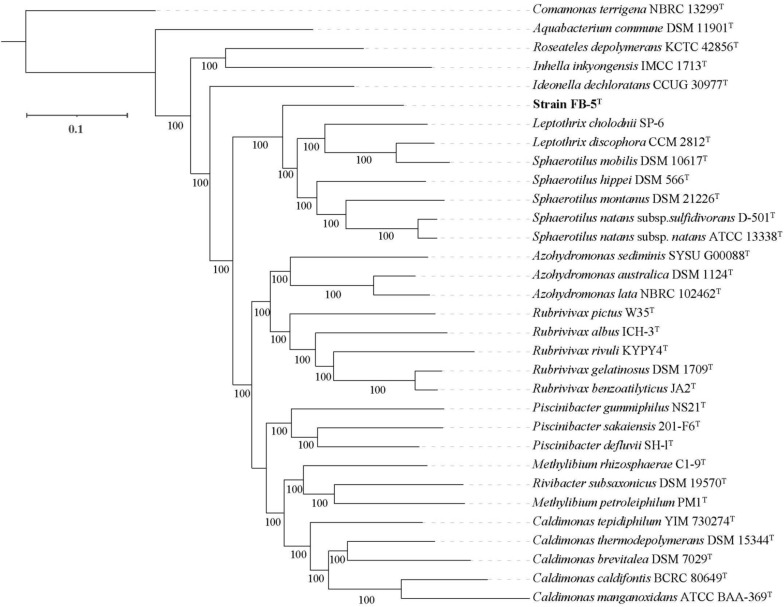
The dDDH values between strain FB-5 T and the strains of the genus Sphaerotilus ranged from 21.8 to 22.6% as shown in the heat map style matrix (Fig. S5). The values were much lower than the species cutoff value of 70% (Goris et al. 2007; Meier-Kolthoff et al. 2013), indicating that strain FB-5 T represents a new species in the genus Sphaerotilus. In the phylogenetic tree constructed based on the core genes set (Fig. 4), strain FB-5 T was located outside the clade formed by other members of the Sphaerotilus-Leptothrix group.
Fig. 4.
Phylogenetic tree (ML) based on core genome sequences showing the relationship between strain FB-5 T and related strains. The percentages (> 70%) for 1000 bootstraps are shown next to the branches. The scale represents 0.1 nucleotide substitutions per site
The AAI values against the species in the Sphaerotilus-Leptothrix group ranged from 71.0 to 72.5% (Fig. S6), with the lowest value (71.0%) against S. natans subsp. natans and the highest value (72.5%) against L. cholodnii. In the AAI matrix plot (Fig. S7), strain FB-5 T formed a clade with L. cholodnii, L. discophora, and S. mobilis, suggesting that it was closely related to these species. A neighboring clade was formed by the Sphaerotilus strains other than S. mobilis. AAI values of L. cholodnii and L. discophora against the Sphaerotilus strains, including strain FB-5 T ranged from 70.7 to 72.6% (Fig. S6). Considering the AAI relatedness and Sphaerotilus-Leptothrix group-specific genus boundary AAI value of 67.12–71.55% (Liu et al. 2022), strain FB-5 T should be classified into the genus Sphaerotilus. The ANI values between strain FB-5 T and the strains of the Sphaerotilus-Leptothrix group ranged from 81.7 to 82.5% (Fig. S8). In the ANI matrix plot (Fig. S9), strain FB-5 T was located distantly within the clades of the genera Sphaerotilus and Leptothrix, suggesting its novelty. Based on the Sphaerotilus-Leptothrix group-specific genus boundary ANI value of 78.95–82.14% (Liu et al. 2022), strain FB-5 T should be classified into the genus Sphaerotilus.
Conclusion
Table 1 summarizes the major phenotypic features of strain FB-5 T that distinguish it from known closely related species. Because strain FB-5 T did not oxidize manganese, it could be easily distinguished from S. mobilis and strains of the genus Leptothrix. The cell shape and size were almost identical to those of the known species of the genus Sphaerotilus. Sheath formation by strain FB-5 T was not particularly stable in the absence of calcium, which differs from that observed in other known species of the genus Sphaerotilus. Strain FB-5 T is microaerophilic and catalase-negative, whereas the type strains of the genus Sphaerotilus are strictly aerobic and catalase-positive. Because lithotrophic sulfur oxidation is possible, strain FB-5 T is the second lithotrophic member found after S. natans subsp. sulfidivorans in the genus Sphaerotilus. The inability of strain FB-5 T to utilize sugars and organic acids as the sole energy and carbon sources distinguished it from known species in the genus Sphaerotilus. Based on the differences in phenotypic properties and genomic features, this strain should be classified as the type strain of a new species, Sphaerotilus microaerophilus sp. nov.
Table 1.
Distinguishing characteristics of strain FB-5 T and related type strains
| Characteristics | Strain FB-5 T | S. natans subsp. natans DSM 6575 Ta | S. natans. subsp. sulfidivorans D-501 Ta | S. hippiei DSM 566 Ta | S. montanus HSTa | S. mobilis Fox-1 Ta | L. discophora LMG 8141 Ta | L. cholodnii LMG 7171 Ta | |
|---|---|---|---|---|---|---|---|---|---|
| Cell size (μm) width length |
0.7–1.0 2.0–6.5 |
1.2–2.0 2.0–6.0 |
1.0–2.0 3.9–6.0 |
0.7–1.5 2.0–6.2 |
0.8–1.4 3.3–6.0 |
0.6–0.8 1.5–12 |
0.6–0.8 2.5–12 |
0.7–1.5 2.5–15 |
|
| Sheath formation | ± b | + b | + | + | + | − b | ± | ± | |
| Microaerophilic | + | − | − | − | − | − | − | − | |
| Catalase | − | + | + | + | + | NDc | ND | ND | |
| Manganese oxidation | − | − | − | − | − | + | + | + | |
| Sulfide oxidation | + | − | + | − | − | − | − | − | |
| Growth on glucose | − | + | + | + | + | − | − | − | |
| fumarate | − | + poor growth | − | − | + | + | − | − | |
| proline | − | + | + | + | + | − | + | + | |
| pyruvate | − | + | + | + | + | − | − | − | |
| Genome size (Mbp)d | 6.06 | 4.63 | 4.39 | 4.43 | 5.07 | 4.65 | 4.65 | ND | |
| G + C content (%)e | 69.16 | 69.94 | 69.88 | 70.03 | 68.16 | 69.00 | 70.12 | 70 |
aData from Grabovich et al. (2021), Gridneva et al. (2011), and Spring et al. (1996)
b+ Positive or supported growth;− negative or did not support growth; ± sheath formation was easily lost during maintenance
cNot determined
dNo genomic data is available for L. cholodnii LMG 7171 T
eBased on genome-wide sequences other than L. cholodnii LMG 7171 T, which (mol%) was determined by the thermal denaturation method (Spring et al. 1996)
Description of Sphaerotilus microaerophilus sp. nov.
Sphaerotilus microaerophilus (mi.cro. a.e.ro'phi.lus. Gr. masc. adj. mikros, small; Gr. masc. n. aêr, air; N.L. masc. adj. philus (from Gr. masc. adj. philos), friend; N.L. masc. adj. microaerophilus, loving conditions of low air, referring to the low oxygen preference of the type strain, FB-5).
Straight rod-shaped cells with rounded ends are 0.7–1.0 × 2.0–6.5 µm in size, motile by means of a single polar flagellum. A few cells are enclosed within the sheaths. The sheath-forming ability is unstable in the absence of calcium salts. The rough colonies with fibrous edges are mostly of sheath-forming cells, whereas the smooth colonies are mostly sheathless. Facultatively microaerophilic. Semi-solidified GPPY medium is suitable for growth. Colonies are colorless. The temperature range for growth is 15–35 °C, with 30 °C being optimal. The pH range for growth is 7–8, with an optimal pH of 7.5. Aspartate, glutamate, methionine, tyrosine, peptone, yeast extract, tryptone, casamino acids, and Proteose-Peptone No. 3 are utilized in NCM medium as sole carbon sources. Ethanol, butanol, iso-butanol, propanol, glucose, glycerol, sorbitol, sorbose, arabinose, fructose, lactose, galactose, mannose, maltose, sucrose, raffinose, acetate, formate, citrate, lactate, malate, malonate, pyruvate, benzoate, oxalate, oxaloacetate, 2-oxoglutarate, succinate, fumarate, glycolate, aconitate, alanine, arginine, asparagine, cysteine, glutamine, glycine, histidine, isoleucine, leucine, lysine, phenylalanine, proline, serine, threonine, tryptophan, and valine are not utilized in NCM medium as sole carbon sources. Lithotrophic sulfur oxidation occurs in m-DSMZ573 medium with continues feeding of sulfide. Oxidase positive. Catalase negative. Nitrate is reduced to nitrite (ID test NF-18). Gelatin is not liquefied and indole is not formed (ID test NF-18). Negative for esculin hydrolysis, arginine hydrolysis, urea degradation, lysine decarboxylation, ornithine decarboxylation, and β-galactosidase (ID test NF-18). No growth in GPPY medium supplemented with 3% (w/v) NaCl. The major quinone is UQ-8. The major fatty acids are C16:1ω7, C16:0, and C12:0. The genomic DNA G + C content is 69.16%. The type strain is FB-5 T (= JCM 35424 T = KACC 23146 T) isolated from the activated sludge of a sewage treatment plant in Yokohama, Japan. The GenBank accession numbers for the 16S rRNA gene and the genome of the type strain are LC775240 and AP025730, respectively.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors express their sincere gratitude to the Yokohama City Government for providing activated sludge. The authors thank Hiromu Okazaki, Rhoji Usami, and Akane Hinago for their help with the preliminary characterization of the isolate. The authors thank Tomoaki Saito and Yui Araki for their help with the fed-batch cultivation of the isolate. The authors thank Anushka Dinesh for her help in editing this manuscript.
Abbreviations
- AAI
Average amino acid identity
- ANI
Average nucleotide identity
- dDDH
Digital DNA-DNA hybridization
- PHB
Polyhydroxybutyrate
Author contributions
MT conceived the study. TN prepared samples for isolation. MT and TM isolated the strain. TN, SM, HK, and JB determined the partial nucleotide sequence. SC, NM, SN, and MA performed genomic analysis. MS, SN, and TM performed electron microscopy. SN, RA, MA, AH, and MT were involved in phenotypic characterization. SN, SC, RA, and MT wrote the manuscript. TN and JB edited the manuscript. All the authors have reviewed and approved the final manuscript.
Funding
Open Access funding provided by Yokohama National University. This work was supported by JSPS KAKENHI (grant number 23K05052).
Data availability
The near-complete 16S rRNA gene sequence of strain FB-5 T can be obtained in GenBank/EMBL/DDBJ accession number LC775240. The whole-genome data for strain FB-5 T were deposited in DDBJ/ENA/GenBank under the accession numbers AP025730 (genome) and AP025731 (plasmid). Strain FB-5 T was deposited in the Japan Collection of Microorganisms (JCM) and Korean Agricultural Culture Collection (KACC) as JCM 35424 T and KACC 23146 T, respectively.
Declarations
Conflict of interest
The authors declare that there were no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Baskar S, Baskar R, Thorseth IH, Ovreås L, Pedersen RB. Microbially induced iron precipitation associated with a neutrophilic spring at Borra Caves, Vishakhapatnam, India. Astrobiology. 2012;12:327–346. doi: 10.1089/ast.2011.0672. [DOI] [PubMed] [Google Scholar]
- Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- Chklovski A, Parks DH, Woodcroft BJ, Tyson GW. CheckM2: a rapid, scalable and accurate tool for assessing microbial genome quality using machine learning. Nat Methods. 2023;20:1203–1212. doi: 10.1038/s41592-023-01940-w. [DOI] [PubMed] [Google Scholar]
- Emerson D, Ghiorse WC. Isolation, cultural maintenance, and taxonomy of a sheath-forming strain of Leptothrix discophora and characterization of manganese-oxidizing activity associated with the sheath. Appl Environ Microbiol. 1992;58:4001–4010. doi: 10.1128/aem.58.12.4001-4010.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaval G, Pernelle JJ. Impact of the repetition of oxygen deficiencies on the filamentous bacteria proliferation in activated sludge. Water Res. 2003;37:1991–2000. doi: 10.1016/S0043-1354(02)00421-9. [DOI] [PubMed] [Google Scholar]
- Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- Grabovich MY, Smolyakov DD, Beletsky AV, Mardanov V, Gureeva MV, Markov ND, Rudenko TS, Ravin NV. Reclassification of Sphaerotilus natans subsp. sulfidivorans Gridneva et al. 2011 as Sphaerotilus sulfidivorans sp. nov. and comparative genome analysis of the genus Sphaerotilus. Arch Microbiol. 2021;203:1595–1599. doi: 10.1007/s00203-020-02158-6. [DOI] [PubMed] [Google Scholar]
- Gridneva E, Chernousova E, Dubinina G, Akimov V, Kuever J, Detkova E, Grabovich M. Taxonomic investigation of representatives of the genus Sphaerotilus: descriptions of Sphaerotilus montanus sp. nov., Sphaerotilus hippei sp. nov., Sphaerotilus natans subsp. natans subsp. nov. and Sphaerotilus natans subsp. sulfidivorans subsp. nov., and an emended description of the genus Sphaerotilus. Int J Syst Evol Microbiol. 2011;61:916–925. doi: 10.1099/ijs.0.023887-0. [DOI] [PubMed] [Google Scholar]
- Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:1–8. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen MP, Ardö Y, Vogensen FK. Isolation of cultivable thermophilic lactic acid bacteria from cheeses made with mesophilic starter and molecular comparison with dairy-related Lactobacillus helveticus strains. Lett Appl Microbiol. 2009;49:396–402. doi: 10.1111/j.1472-765X.2009.02673.x. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwabara D, Kondo K, Usami R, Kan D, Kawamura I, Kawasaki Y, Sato M, Nittami T, Suzuki I, Katahira M, Takeda M. Structural determination of the sheath-forming polysaccharide of Sphaerotilus montanus using thiopeptidoglycan lyase which recognizes the 1,4 linkage between α-d-GalN and β-d-GlcA. Int J Biol Macromol. 2021;183:992–1001. doi: 10.1016/j.ijbiomac.2021.05.001. [DOI] [PubMed] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Park S, Chun J. Introducing EzAAI: a pipeline for high throughput calculations of prokaryotic average amino acid identity. J Microbiol. 2021;59:476–480. doi: 10.1007/s12275-021-1154-0. [DOI] [PubMed] [Google Scholar]
- Kondo K, Takeda M, Ejima W, Kawasaki Y, Umezu T, Yamada M, Koizumi J, Mashima T, Katahira M. Study of a novel glycoconjugate, thiopeptidoglycan, and a novel polysaccharide lyase, thiopeptidoglycan lyase. Int J Biol Macromol. 2011;48:256–262. doi: 10.1016/j.ijbiomac.2010.11.009. [DOI] [PubMed] [Google Scholar]
- Kunoh T, Yamamoto T, Sugimoto S, Ono E, Nomura N, Utada AS. Leptothrix cholodnii response to nutrient limitation. Front Microbiol. 2021;12:691563. doi: 10.3389/fmicb.2021.691563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunoh T, Yamamoto T, Ono E, Sugimoto S, Takabe K, Takeda M, Utada AS, Nomura N. Identification of lthB, a gene encoding a putative glycosyltransferase family 8 protein required for Leptothrix sheath formation. Appl Environ Microbiol. 2023;89:e0191922. doi: 10.1128/aem.01919-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Du J, Pei T, Du H, Feng GD, Zhu H. Genome-based taxonomic classification of the closest-to Comamonadaceae group supports a new family Sphaerotilaceae fam nov and taxonomic revisions. Syst Appl Microbiol. 2022;45:126352. doi: 10.1016/j.syapm.2022.126352. [DOI] [PubMed] [Google Scholar]
- Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff JP, Carbasse JS, Peinado-Olarte RL, Göker M. TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acid Res. 2022;50:D801–D807. doi: 10.1093/nar/gkab902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Masuzawa N, Kondo K, Nakanishi Y, Chida S, Uehara D, Katahira M, Takeda M. A heterodimeric hyaluronate lyase secreted by the activated sludge bacterium Haliscomenobacter hydrossis. Biosci Biotechnol Biochem. 2023;87:256–266. doi: 10.1093/bbb/zbac207. [DOI] [PubMed] [Google Scholar]
- Na SI, Kim YO, Yoon SH, Ha SM, Baek I, Chun J. UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J Microbiol. 2018;56:280–285. doi: 10.1007/s12275-018-8014-6. [DOI] [PubMed] [Google Scholar]
- Nott MA, Driscoll HE, Takeda M, Vangala M, Corsi SR, Tighe SW. Advanced biofilm analysis in streams receiving organic deicer runoff. PLoS ONE. 2020;22:e0227567. doi: 10.1371/journal.pone.0227567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okrend H, Dondero NC. Requirement of Sphaerotilus for cyanocobalamin. J Bacteriol. 1964;87:286–292. doi: 10.1128/jb.87.2.286-292.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A, Göker M. Sphaerotilaceae fam. Nov. in validation list no. 209. valid publication of new names and new combinations effectively published outside the IJSEM. Int J Syst Evol Microbiol. 2023;73:005709. doi: 10.1099/ijsem.0.005709. [DOI] [PubMed] [Google Scholar]
- Park S, Kim DH, Lee JH, Hur HG. Sphaerotilus natans encrusted with nanoball-shaped Fe(III) oxide minerals formed by nitrate-reducing mixotrophic Fe(II) oxidation. FEMS Microbiol Ecol. 2014;90:68–77. doi: 10.1111/1574-6941.12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawayama M, Suzuki T, Hashimoto H, Kasai H, Furutani M, Miyata N, Kunoh H, Takada J. Isolation of a Leptothrix strain, OUMS1, from ocherous deposits in groundwater. Curr Microbiol. 2011;63:173–180. doi: 10.1007/s00284-011-9957-6. [DOI] [PubMed] [Google Scholar]
- Schmidt B, Sánchez LA, Fretschner T, Kreps G, Ferrero MA, Siñeriz F, Szewzyk U. Isolation of Sphaerotilus-Leptothrix strains from iron bacteria communities in Tierra del Fuego wetlands. FEMS Microbiol Ecol. 2014;90:454–466. doi: 10.1111/1574-6941.12406. [DOI] [PubMed] [Google Scholar]
- Siering PL, Ghiorse WC. Phylogeny of the Sphaerotilus-Leptothrix group inferred from morphological comparisons, genomic fingerprinting, and 16S ribosomal DNA sequence analyses. Int J Syst Bacteriol. 1996;46:173–182. doi: 10.1099/00207713-46-1-173. [DOI] [PubMed] [Google Scholar]
- Skerman VBD, McGowan V, Sneath PHA. Approved lists of bacterial names. Int J Syst Bacteriol. 1980;30:225–420. doi: 10.1099/00207713-30-1-225. [DOI] [PubMed] [Google Scholar]
- Smibert RM. Phenotypic characterization. In: Gerhardt P, editor. Methods for general and molecular bacteriology. Washington: American Society for Microbiology; 1994. pp. 607–654. [Google Scholar]
- Spring S, Kampfer P, Ludwig W, Schleifer KH. Polyphasic characterization of the genus Leptothrix: new descriptions of Leptothrix mobilis sp. nov. and Leptothrix discophora sp. nov. nom. rev. and emended description of Leptothrix cholodnii emend. Syst Appl Microbiol. 1996;19:634–643. doi: 10.1016/S0723-2020(96)80036-1. [DOI] [Google Scholar]
- Suzuki T, Kanagawa T, Kamagata Y. Identification of a gene essential for sheathed structure formation in Sphaerotilus natans, a filamentous sheathed bacterium. Appl Environ Microbiol. 2002;68:365–371. doi: 10.1128/AEM.68.1.365-371.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda M, Matsuoka H, Hamana H, Hikuma M. Biosynthesis of poly-3-hydroxybutyrate by Sphaerotilus natans. Appl Microbiol Biotechnol. 1995;43:31–34. doi: 10.1007/BF00170618. [DOI] [Google Scholar]
- Takeda M, Kamagata Y, Ghiorse WC, Hanada S, Koizumi J. Caldimonas manganoxidans gen. nov., sp. nov., a poly(3-hydroxybutyrate)-degrading, manganese-oxidizing thermophile. Int J Syst Evol Microbiol. 2002;52:895–900. doi: 10.1099/00207713-52-3-895. [DOI] [PubMed] [Google Scholar]
- Takeda M, Makita H, Ohno K, Nakahara Y, Koizumi J. Structural analysis of the sheath of a sheathed bacterium, Leptothrix cholodnii. Int J Biol Macromol. 2005;37:92–98. doi: 10.1016/j.ijbiomac.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Tamaoka J, Katayama-Fujimura Y, Kuraishi H. Analysis of bacterial menaquinone mixtures by high performance liquid chromatography. J Appl Bacteriol. 1983;54:31–36. doi: 10.1111/j.1365-2672.1983.tb01297.x. [DOI] [Google Scholar]
- Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38:3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanizawa Y, Fujisawa T, Kaminuma E, Nakamura Y, Arita M. DFAST and DAGA: web-based integrated genome annotation tools and resources. Biosci Microbiota Food Health. 2016;35:173–184. doi: 10.12938/bmfh.16-003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Veen WL, Mulder EG, Deinema MH. The Sphaerotilus-Leptothrix group of bacteria. Microbiol Rev. 1978;42:329–256. doi: 10.1128/mr.42.2.329-356.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick RR, Schultz MB, Zobel J, Holt KE. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 2015;31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems A, De Ley J, Gillis M, Kersters K. Comamonadaceae, a new family encompassing the acidovorans rRNA complex, including Variovorax paradoxus gen. nov., comb. nov., for Alcaligenes paradoxus (Davis 1969) Int J Syst Bacteriol. 1991;41:445–450. doi: 10.1099/00207713-41-3-445. [DOI] [Google Scholar]
- Yarza P, Spröer C, Swiderski J, Mrotzek N, Spring S, et al. Sequencing orphan species initiative (SOS): Filling the gaps in the 16S rRNA gene sequence database for all species with validly published names. Syst Appl Microbiol. 2013;36:69–73. doi: 10.1016/j.syapm.2012.12.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The near-complete 16S rRNA gene sequence of strain FB-5 T can be obtained in GenBank/EMBL/DDBJ accession number LC775240. The whole-genome data for strain FB-5 T were deposited in DDBJ/ENA/GenBank under the accession numbers AP025730 (genome) and AP025731 (plasmid). Strain FB-5 T was deposited in the Japan Collection of Microorganisms (JCM) and Korean Agricultural Culture Collection (KACC) as JCM 35424 T and KACC 23146 T, respectively.