Abstract
The RNA polymerase II (Pol II) pre-initiation complex (PIC) is a critical node in eukaryotic transcription regulation, and its formation is the major rate-limiting step in transcriptional activation. Diverse cellular signals borne by transcriptional activators converge on this large, multiprotein assembly and are transduced via intermediary factors termed coactivators. Cryogenic electron microscopy, multi-omics and single-molecule approaches have recently offered unprecedented insights into both the structure and cellular functions of the PIC and two key PIC-associated coactivators, Mediator and TFIID. Here, we review advances in our understanding of how Mediator and TFIID interact with activators and affect PIC formation and function. We also discuss how their functions are influenced by their chromatin environment and selected cofactors. We consider how, through its multifarious interactions and functionalities, a Mediator-containing and TFIID-containing PIC can yield an integrated signal processing system with the flexibility to determine the unique temporal and spatial expression pattern of a given gene.
Introduction
Precise spatiotemporally regulated transcription by RNA polymerase II (Pol II), the enzyme responsible for transcription of eukaryotic protein-encoding mRNAs and assorted untranslated RNA species, is the net outcome of an intricate choreography among a large number of factors1. These include a set of general transcription factors (GTFs) on which Pol II is dependent for promoter-specific transcription initiation2–4. Historically, biochemical studies aimed at reconstituting accurate, basal-level (uninduced) transcription at a paradigmatic promoter were instrumental in identifying the complete complement of GTFs, which includes TFIIA, TFIIB, TFIID, TFIIE, TFIIF and TFIIH4. At the core promoter, Pol II and these GTFs assemble into a pre-initiation complex (PIC), whose formation and function is a critical rate-limiting step in transcription4. Remarkably, in conjunction with Pol II and the other GTFs, just the TATA binding protein (TBP) subunit of the multi-subunit TFIID was originally required for basal transcription5–8. Altogether, these early studies established that the core PIC functions of recruiting and orienting Pol II at promoters and commencing phosphodiester bond formation reside within TBP and the other GTFs.
Prior biochemical studies2,4 and more recent cryogenic electron microscopy (cryo-EM) structures9–13 have collectively provided insights into how TBP and the other GTFs generate the PIC and facilitate promoter melting and initiation (Fig. 1). On a TATA-containing promoter, PIC assembly begins with TBP binding to the TATA element. TFIIA further stabilizes TBP–promoter interactions by making direct contacts with TBP and with upstream DNA backbone phosphates. Next, TFIIB slots into the TBP–promoter complex, also making specific contacts with promoter DNA upstream and downstream of the TATA box through TFIIB recognition elements (BREs). The resulting assembly serves as a platform for Pol II recruitment, which is facilitated by TFIIF. A fully functional PIC is finally generated through the incorporation of TFIIE and TFIIH. The ATP hydrolysis-dependent XPB translocase subunit of TFIIH then melts promoter DNA around the transcription start site (TSS) to allow templated formation of nascent RNA11,14–16 (Box 1). Concomitant conformational changes in the PIC (such as clamp closure, stalk movements and TFIIB linker transitions) ensue and further stabilize the resulting ‘open complex’. A separate TFIIH CDK-activating kinase (CAK) module that contains a cyclin-dependent kinase, CDK7, phosphorylates the carboxy-terminal domain (CTD) repeats of RPB1, the largest Pol II subunit. Following initiation, the nascent RNA chain overcomes several checkpoints, including abortive RNA synthesis and promoter escape, which mainly relate to dealing with otherwise stabilizing TFIIB contacts9,17.
Fig. 1 |. A paradigmatic TBP-nucleated PIC assembly pathway.
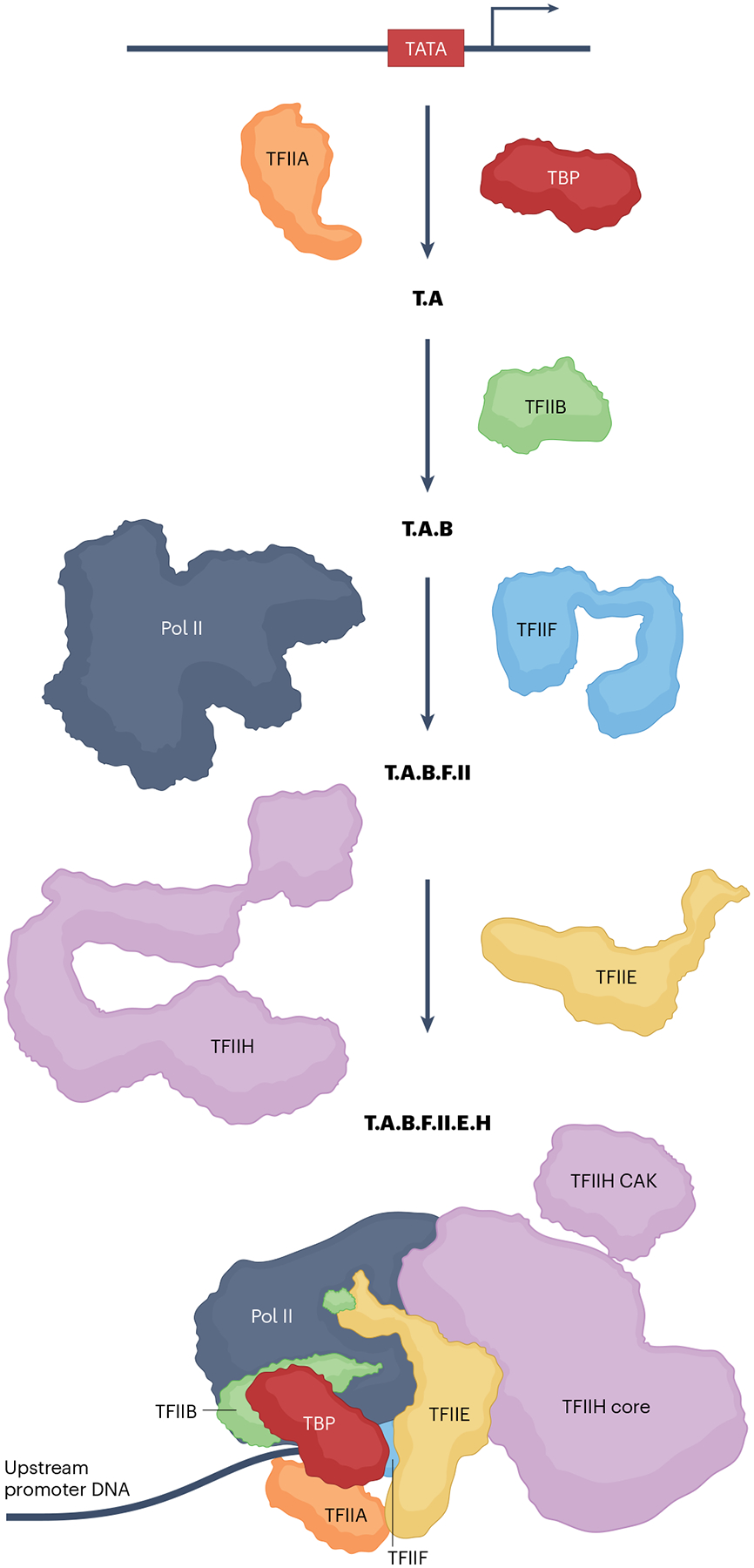
A composite TATA binding protein (TBP)-nucleated pre-initiation complex (PIC) assembly pathway on a TATA element-containing promoter, based on biochemical and structural studies of the minimal PIC (see main text and references therein). In the first step, TBP binding to the minor groove of the TATA element (red box) is stabilized by TFIIA, which makes direct contacts with TBP and with upstream DNA backbone phosphates (T.A). Additional stabilization comes from TFIIB (T.A.B), both through interactions with the T.A subcomplex as well as through interactions with TFIIB recognition element (BRE) sequences in the promoter. The amino-terminal regions of TFIIB, consisting of the zinc ribbon, B-reader and B-linker domains, recruit RNA polymerase II (Pol II) and support transcription initiation through interactions, respectively, with Pol II and the template and non-template DNA strands. T.A.B is a landing pad for Pol II in a step that is facilitated by heterodimeric TFIIF, which binds close to the Pol II cleft and causes the Pol II clamp to partially open, resulting in stabilization of the double-stranded template DNA in the cleft (T.A.B.F.II). Finally, TFIIE and TFIIH enter to yield a functional PIC (T.A.B.F.II.E.H). TFIIE makes multiple contacts with Pol II, including sites at the RPB4/7 stalk. A cluster of TFIIE winged helix domains extend out over the cleft and together with TFIIF form a protein bridge that further stabilizes the template. TFIIE is also critical in recruiting TFIIH into the PIC. Formation of the first phosphodiester bond by the Pol II active site is preceded by melting of double-stranded DNA around the transcription start site (TSS) by the TFIIH XPB translocase11,15,16. Multiple conformational changes (such as clamp closure, stalk movements and TFIIB linker transitions) in the PIC also ensue, further stabilizing the resulting ‘open complex’. The nascent RNA chain faces a steric clash with the stabilizing interactions of N-terminal regions of TFIIB, which forces Pol II to undergo reiterative abortive synthesis of short oligomeric RNAs9. Pol II promoter escape and extension of the RNA chain past a length of about 12 nucleotides entails TFIIB release from the PIC17. TFIIE and TFIIF are also released, both to relinquish the open state’s stabilizing contacts and to enable entry of elongation factors (Fig. 2). Although it fully supports basal transcription, this minimal PIC is not responsive to transcriptional activators. CAK, CDK-activating kinase.
Box 1. Dual roles of TFIIH as a general transcription factor and coactivator.
Similar to Mediator and TFIID, TFIIH is a multi-subunit complex, consisting of several functional and structural modules that contain three distinct enzymatic activities14. In addition to the kinase and XPB translocase activities discussed in the main text, the XPD subunit harbours ATPase activity that comes into play in the course of TFIIH function in transcription-coupled repair. This feature is one of several that distinguish TFIIH from other general transcription factors (GTFs). Whereas for all intents and purposes it behaves as a bona fide GTF, evidence is emerging that, similar to TFIID, TFIIH may be a coactivator doubling as a GTF. There have been reports that it is targeted by a wide range of activators, including p53 (ref. 148) and nuclear receptors149. In this regard, there is a long history of reports of GTFs as activator targets, beginning with the identification of TFIIB as the target of the viral activator VP16 (ref. 150). However, given the difficulty in segregating the essential roles of GTFs in basal transcription from any coactivator role they might harbour, it remains unclear whether these interactions are relevant in vivo. In the case of TFIIH, a recent study has shown that it serves as a bona fide coactivator for the nuclear receptor ERR in embryonic stem cells151. There is also evidence for a conditional requirement of TFIIH in vitro152–155. Also, cryogenic electron microscopy (cryo-EM) analyses revealed that yeast pre-initiation complexes (PICs) reconstituted without TFIIH have a high propensity to spontaneously form open complexes12,156, consistent with the notion that this is an intrinsic property of RNA polymerase II (Pol II), as is the case for other multi-subunit RNA polymerases, including Pol I and Pol III, as well as bacterial RNA polymerase. There is also evidence for contingent TFIIH use in vivo: TFIIH is limiting in early B cells and seems to occupy active genes only at later developmental stages157 when it may be recruited by activators to highly active genes, potentially in its capacity as a coactivator.
PIC function, in turn, is regulated by signal-bearing cell type-specific and gene-specific transcriptional activators that respond to developmental and environmental cues and bind to regulatory regions of the gene1. However, contrary to initial expectations, a complement of GTFs including TBP, TFIIA, TFIIB, TFIIE, TFIIF and TFIIH was unable to support activator-dependent transcription by Pol II in vitro18,19. Because unfractionated cell-free extracts did support activator function, such biochemical studies were the first to raise the possibility that activator signals may be mediated by ‘coactivators’ that would thus constitute a class of transcription factors that is distinct from both site-specific transcriptional activators and GTFs18,19 (Fig. 2). Through further biochemical analyses, the TBP-associated factor (TAF) subunits of TFIID20, as well as other nuclear factors, were identified as coactivators for various activators. In particular, the large, multi-subunit Mediator complex was independently identified by both genetic and biochemical approaches in yeast, and biochemically in metazoans through interactions with activators, as well as in functional screens for coactivator activity in nuclear extract-derived chromatographic fractions21–23. A large body of work has since established critical roles of TFIID and Mediator in activated transcription in eukaryotic cells. Furthermore, consistent with biochemical studies24–26, more recent genomic and genetic studies27,28 have suggested cooperative function of these factors. Somewhat unexpectedly, TFIID TAFs and Mediator were found to be closely associated with the PIC, thus blurring the originally envisaged distinction between coactivators and PIC components. Indeed, whereas TFIID is a GTF by definition, the Mediator may also arguably be regarded as one in its own right29,30.
Fig. 2 |. General principles of coactivator-dependent PIC recruitment and function.
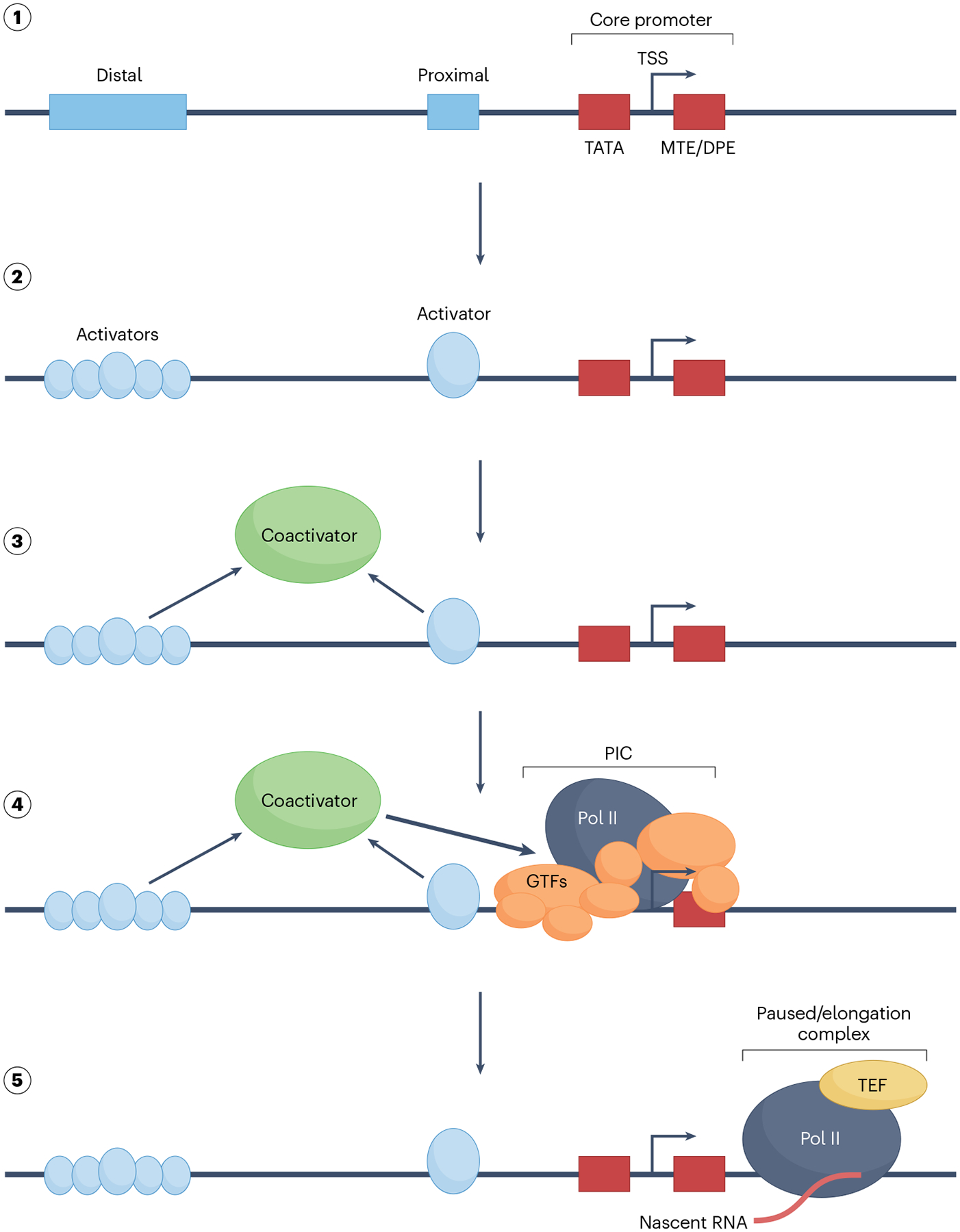
Multi-step pre-initiation complex (PIC) assembly on an idealized template (step 1) containing diverse regulatory sequence motifs that are not typically all found in a given transcription locus. Signal-bearing transcription activators can bind to cognate sites located in distal enhancers, including super-enhancers, or to sites located proximal to core promoter elements (step 2). Core promoter elements collectively specify the transcription start site (TSS). In addition to the TATA box, these may include, among others, motif ten elements (MTEs) and downstream promoter elements (DPEs). Activator binding sets off a cascade of events that include recruitment of one or more intermediary factors, depicted here as a generic ‘coactivator’ (step 3). Some coactivators play critical roles at the level of chromatin; given the focus here on the PIC, this step is not highlighted. Coactivators in turn facilitate the formation of the PIC through interactions with one or more general transcription factors (GTFs), as well as RNA polymerase II (Pol II) (step 4). Despite their historical origins as factors that promote PIC formation and function, the two coactivators discussed here, Mediator and TFIID TATA binding protein (TBP)-associated factors (TAFs), are so intimately associated with the PIC that they may justifiably be regarded as components of an expanded PIC. The PIC is tasked with ensuring precise site-specific initiation of the nascent RNA transcript and promotion of the Pol II to an elongation-competent form (step 5) following promoter escape. GTFs are also released, both to relinquish contacts that stabilize the open state and to enable entry of transcription elongation factors (TEFs), which in conjunction with other factors not discussed here have roles in promoter-proximal pausing that occurs on many genes prior to acquisition of full elongation processivity by Pol II136. PIC coactivators have the potential to regulate some of these downstream events as well.
Towards understanding how a signal transduction cascade in which Mediator and TFIID assimilate diverse activator-borne regulatory signals on the one hand and deliver processed outputs to the PIC on the other, we here review recent insights into their structure, interactions with activators and impact on the PIC. We assess coactivator roles and mechanisms in light of recent studies, especially new cryo-EM structures of these multi-subunit proteins, as well as PICs containing them. Although we consider how chromatin might affect PIC function, an important class of coactivators that includes many chromatin modulators, which also play important roles in the transcriptional history of a gene, will be largely excluded. Important studies that have mainly revealed biological roles of the coactivators in specific gene expression programmes will also not be highlighted as such. Although we emphasize studies on metazoan factors, as needed we will refer to important complementary studies in yeast systems.
Coactivator composition and architecture
Modular organization of the Mediator complex
Possessing multiple functionalities, Mediator has been implicated in numerous gene expression programmes as a central regulatory hub30,31. It is now evident that the yeast and human complexes share a large number of evolutionarily conserved orthologous subunits and display a near-identical modular structural organization31,32. However, consistent with the more demanding transcriptional programmes in metazoans, significant divergence has occurred: the larger metazoan complex possesses several metazoan-specific subunits29,30, and many otherwise conserved subunits nonetheless display non-homologous residues, mostly within intrinsically disordered regions (IDRs)29,33. As in yeast, the 26 subunits constituting the bulk complex in human are distributed among three modules designated ‘head’, ‘middle’ and ‘tail’32,34 (Fig. 3). A fourth module, known variously as the kinase or CKM module, contains a cyclin-dependent kinase (either CDK8 or CDK19), cyclin C (CCNC), MED12 (or MED12L) and MED13 (or MED13L), and associates reversibly with the Mediator complex29,30 (Fig. 3). Reconstitution of a partial, but functional, recombinant human complex from subunits of the head (MED6, MED8, MED11, MED17, MED18, MED20, MED22, MED30) and middle (MED4, MED7, MED10, MED19, MED21, MED31, MED26) modules, as well as the backbone MED14 subunit, established that these modules harbour the core effector functions of Mediator, which include interactions with Pol II and other GTFs and stimulation of basal transcription35. As discussed below, additional functionalities (especially responsiveness to select activators) are imparted, in part, by the tail subunits (MED15, MED16, MED23, MED24, MED25 and MED29), the middle subunit MED1 and, potentially, metazoan-specific head subunits MED27 and MED28, which were not included in the reconstitution. Results of genetic ablations of many Mediator subunits in mammalian cells are broadly consistent with these biochemical results36.
Fig. 3 |. Modular structure of metazoan Mediator.
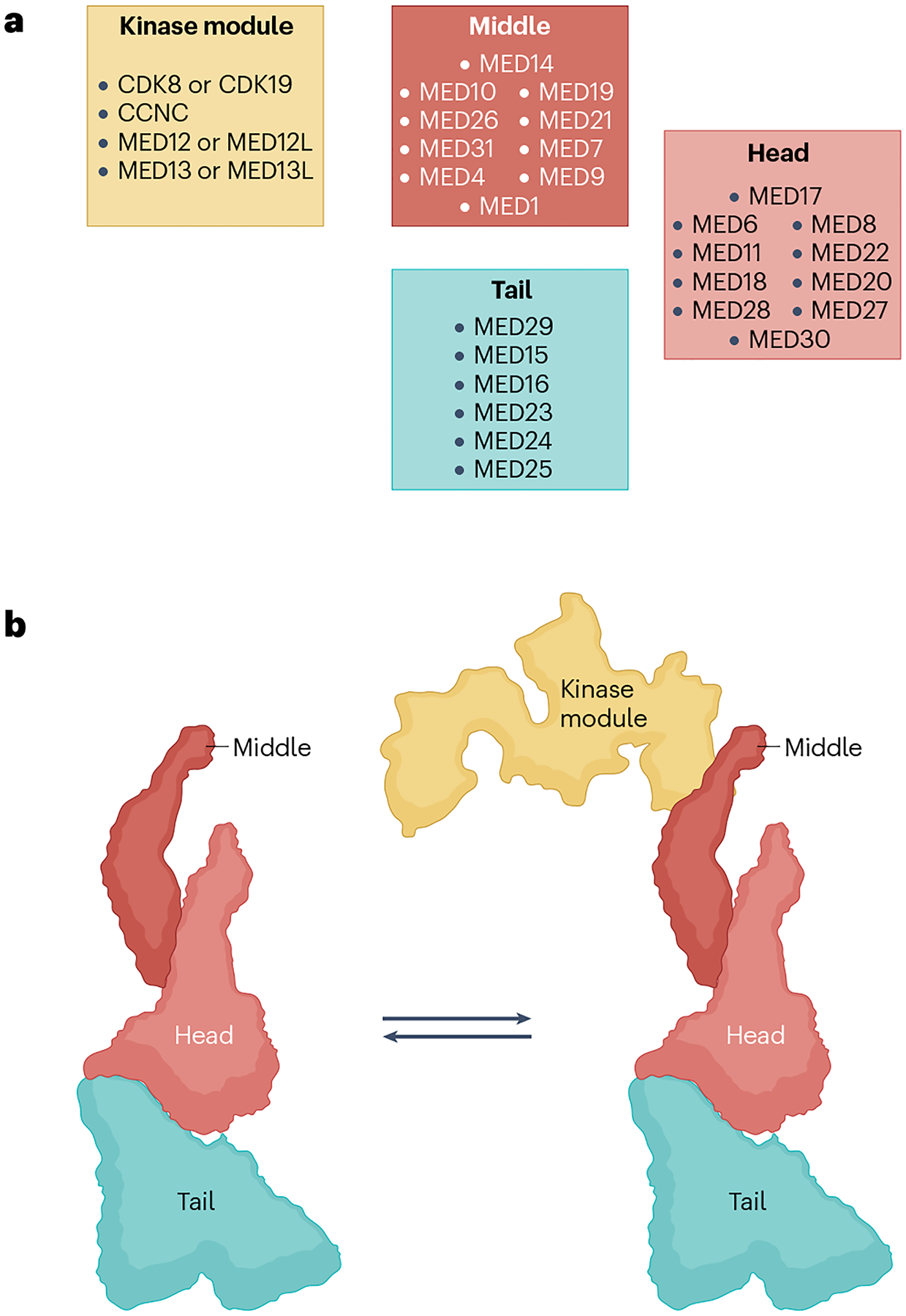
a, Human Mediator structural modules with conventional subunit assignment to the head, middle and tail modules (right). However, cryogenic electron microscopy (cryo-EM) and cross-linking–mass spectrometry studies35–46 reveal that several key subunits straddle more than one module. In particular, MED14 forms a structural backbone around which the head, middle and tail modules are organized. Similarly, whereas the bulk of MED17 resides in the head, its amino terminus is embedded in the middle. Metazoan-specific subunits MED27, MED28 and MED30 also straddle the head and tail. Also note the location of MED1 in the middle. This large subunit has not yet been visualized in its entirety, but the tail-proximal location of this activator target hot spot in Mediator structure is intriguing for models of how activation signals may be processed. The kinase module, which reversibly associates with the Mediator, is also depicted (left). As shown, three of its four constituent subunits have paralogues that can give rise to multiple permutations. b, Left: a model of the Mediator complex showing the relative location of the head, middle and tail modules. The modules can move relative to each other, resulting in several conformers of the complex (not shown). The model is based on ref. 43, which produced high-resolution structures showing detailed subunit architecture of the complex. Right: the Mediator complex in association with the kinase module. Note that this form of the Mediator can interact with activators but not with RNA polymerase II (Pol II). In metazoans, association of the kinase module with the complex is also mutually exclusive with association of the metazoan-specific MED26 middle subunit. Precisely how the two forms of the Mediator interchange is not yet known (see also Fig. 6). The kinase module is modelled after the structure in ref. 147. CDK8, cyclin-dependent kinase 8.
Recent high-resolution integrative cryo-EM and cross-linking–mass spectrometry studies of human and yeast Mediator in both the free and PIC-associated forms36–46 have suggested new models for how the complex can carry out its effector functions (see below). Among other features, the structures collectively reveal the detailed subunit and modular architecture of the complex, as well as a prominent role of MED14 as a structural and functional backbone of the Mediator that anchors each of the head, middle and tail modules. Whereas the new structures have validated that individual modules are generally autonomous units, as originally deduced from genetic studies in yeast, biochemical reconstitution and low-resolution structural studies, it is also evident that the modules are closely interlinked. Thus, for example, MED17, although regarded as a head subunit, is equally an integral middle component31,36 MED1, with important roles in nuclear receptor signalling, although localized to the middle module is actually closely associated with the tail36. Similarly, metazoan-specific MED27, MED28 and MED30 also straddle the head and tail modules36. Indeed, the high degree of interconnectedness of the tail subunits as a whole is suggestive of a mechanism for translating signals impinging on the tail into effector functions intrinsic to the head and middle modules (Mediator–Pol II and PIC interactions, see below). Finally, the range of structures observed by cryo-EM have identified multiple distinct pivot points36,41–43 around which distinct substructures can move and give rise to numerous conformational states, with implications for functional plasticity (see below).
TFIID structural modules and their rearrangements
Following its biochemical isolation from both yeast and human cells as a TBP-containing factor that supports activated transcription in vitro, detailed views of TFIID structural organization have also now been obtained by high-resolution cryo-EM and cross-linking–mass spectrometry47–49. Even at low resolution, it was evident that the circa 14 subunits comprising the human complex (TBP and TAF1–TAF13) are organized into three distinct lobes (lobes A–C)50 (Fig. 4a). Later studies in which TFIID bound to an idealized synthetic core promoter (super core promoter (SCP)), which combined numerous promoter elements (TATA, initiator (Inr), motif ten element (MTE), downstream promoter element (DPE)) that are not ordinarily all found together, revealed not only how these lobes can interact with the promoter elements but also how large-scale and coordinated lobe movements serve to deliver TBP to nucleate PIC assembly48.
Fig. 4 |. TFIID structure and dynamics.
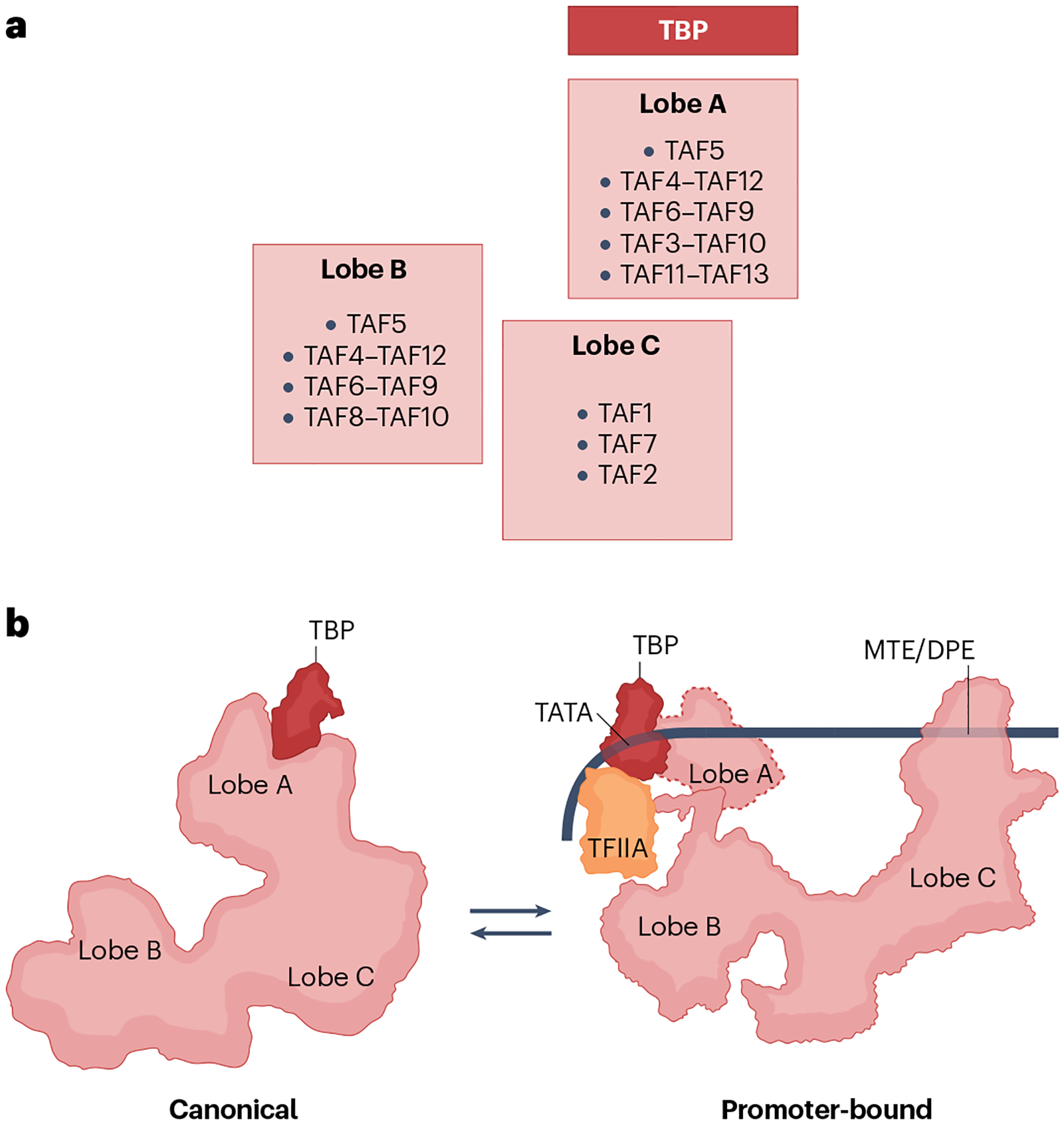
a, Subunit composition of the trilobular human TFIID structure. Several TATA binding protein (TBP)-associated factors (TAFs) exist in two copies; some TAFs also heterodimerize via histone fold-containing domains. Lobe A contains TBP as well as numerous TAFs that are also found in lobe B. Lobe C includes TAFs that have been implicated in recognition of core promoter motifs. b, As visualized by cryogenic electron microscopy (cryo-EM)47, TFIID can exist in multiple conformations. Two extreme conformations are depicted: ‘canonical’ (left), which is a relatively compact form of the TFIID complex; and promoter-bound (right), which is in one of the various ‘extended’ forms of the complex. The promoter-bound form shown is based on a cryo-EM structure that included TFIIA47, which both stabilizes the promoter complex and helps neutralize the inhibitory action of TAF1 and TAF11–TAF13. Note the dramatic relocation of TBP as a result of TFIID lobe movements. Lobe A position in the promoter-bound state is variable; it is shown here in an intermediate location (dashed outlines) based on the structure of a TFIID-containing pre-initiation complex (PIC)49. DPE, downstream promoter element; MTE, motif ten element.
In the promoter unbound state, human TFIID exhibits multiple conformations, although two — the canonical and the extended — predominate48 (Fig. 4b). Lobe A is near lobe C in the canonical state but between lobes B and C in the extended state, suggestive of a remarkable circa 100 Å displacement of the lobe (Fig. 4b). In addition to TAF5, lobe A comprises dimeric pairs of histone fold-containing TAFs (TAF3–TAF10, TAF4–TAF12, TAF6–TAF9 and TAF11–TAF13 pairs). Lobe B also contains TAF5 and the TAF4–TAF12 and TAF6–TAF9 pairs as well as a TAF8–TAF10 pair that replaces the TAF3–TAF10 pair. The bulk of lobe C comprises TAF1, TAF2 and TAF7, but the HEAT domain of the TAF6 copy in lobe A also extends to this lobe and interacts with the corresponding HEAT domain of TAF6 in lobe B. A single copy of TBP is exclusively localized to lobe A48.
In the SCP promoter-bound state (‘engaged’) (Fig. 4b), TFIID exists in a version of the extended conformation, except that TBP very prominently shifts to a lobe B-proximal location and establishes contacts with the TATA box, akin to the interactions earlier observed in TBP–promoter complexes48. Other TFIID–DNA interactions are mainly mediated through TAF1 and TAF2 contacts with the DPEs. An additional series of promoter-bound states that are also observed by cryo-EM have been interpreted as intermediates that may have roles, among others, in scanning for promoter sites48. It has been further proposed that they reflect the trajectory of TBP vis-à-vis the BC core that ultimately lands it in contact with the promoter sequences48. In the canonical and other states, the DNA binding ability of TBP is kept in check by interactions with inhibitory domains in TAF1 and the TAF11–TAF13 pair, perhaps as a mechanism for precluding promiscuous interactions. Thus, in concert with TFIIA, the pathway to delivering TBP also entails replacement of these inhibitory interactions with ultimately productive interactions with promoter DNA and other factors. The net result of these TFIID dynamics is dissociation of TBP from lobe A so that it is available to nucleate the accretion of TFIIB and other GTFs, as summarized above for the formation of a paradigmatic PIC. Therefore, one function of TFIID is as a TBP loading device, even as TAF1 and TAF2 are also critical in promoter recognition per se48.
We should note that another TAF-containing coactivator complex, SAGA, has also been proposed to fulfil a TBP loading function, at least in yeast51,52. In metazoans, where it has evolutionarily diverged considerably, including into distinct ATAC complexes, its roles in regulating the PIC are much less clear, and it may well be restricted to functioning at the level of chromatin53. Alternative TFIID complexes containing only a subset of TAFs and/or TBP-related factors (TRFs) in place of TBP have also been described54, but will not be discussed here in view of the scant mechanistic data.
Coactivator recruitment to the PIC
How TFIID and Mediator find their way into the PIC is an important question. By definition, coactivators function in concert with transcriptional activators. However, the dual nature of TFIID and Mediator as coactivators and PIC components is reflected in how they are recruited to the PIC, as each can potentially be recruited either directly or delivered via activators bound distally to the core promoter.
Activator–TFIID interactions
TFIID’s intrinsic ability to recognize distinct promoter elements allows it to make direct site-specific interactions with the DNA template as a prelude to PIC formation, as summarized above. At the same time, interactions with activators and chromatin elements (see below) can facilitate incorporation of TFIID into the PIC. Once a role for TAFs as coactivators became apparent from in vitro studies, several binary activator–TAF interactions (such as Sp1–TAF4 (ref. 55)) were identified. But following discovery of TAFs in yeast, it was found that activator-dependent transcription in living yeast cells continues unabated upon their depletion56,57. More recent studies using methods that measure nascent RNA transcripts more accurately have nonetheless concluded that in these earlier studies ‘transcriptional buffering’58, wherein reduced nascent RNA synthesis is counterbalanced by reduced RNA degradation, confounded the data, and that TAFs are in fact essential for activator function27,59. Consistent with this development, studies with selected metazoan activators, including Pygopus60 and E proteins61, have definitively revealed interactions with distinct TAFs in the context of the intact TFIID complex and, importantly, have demonstrated that they are functionally consequential. Moreover, in the case of E proteins, a mechanism involving enhanced promoter recruitment of TFIID was clearly established61. Notably, the Pygopus and E protein interactions are via the TAF4 TAFH domain that is also found in the oncogenic AML–ETO fusion protein62.
A detailed structural understanding of how activators engage TFIID has lagged. Early low-resolution cryo-EM structures of TFIID in association with diverse activators, including the tumour suppressor p53, identified distinct interactions sites across the multi-lobed complex; no gross TFIID structural changes were evident63. However, a more recent high-resolution structure of a TFIID-containing PIC on a template that also contained p53 bound to an adjacent cognate site did not reveal any interactions of p53 activation domains with TFIID or any other PIC component49. Nonetheless, this structure49, as well as other high-resolution TFIID structures47, leave open the possibility that TAFs in TFIID would be suitably positioned and available for activator targeting in the context of the PIC, as in the case of the Mediator (see below). Related, TFIID interactions with activators have the converse effect of enhancing activator binding to their cognate elements, at least for those that are located proximal to the core promoters61, indicative of cooperative binding rather than a strict recruitment of one protein by the other.
Activator–Mediator interactions
Unlike TFIID, no specific DNA binding activity has been imputed to the Mediator. Its incorporation into the PIC is therefore dependent on other DNA-binding proteins. Consistent with early in vitro experiments that revealed the ability of Mediator to support basal transcription via recruitment to the PIC22,25,64, as well as the recent structural studies that revealed its multivalent PIC interactions43,44, Mediator can in principle be recruited to the PIC in vivo independently of activators. There is also evidence from studies of ‘tailless’ Mediator in yeast cells that the coactivator can be directly recruited to PICs65,66.
Interactions with transcriptional activators that are anchored to specific loci can nonetheless be expected to greatly increase the concentration and residence time of the Mediator in the vicinity of the PIC and constitute the primary mechanism for delivering Mediator to the PIC. A large number of activators from diverse families have been shown to interact with one or the other Mediator subunit67. Many activator interactions map to Mediator tail subunits in both yeast and metazoans29,30. This led to the suggestion that the tail module is the main acceptor of regulatory signals29,30, which are subsequently transduced through the core Mediator to be translated into specific effector functions that include, among others, modulation of PIC function as discussed below. Moreover, early genetic studies in metazoans suggested that individual tail subunits might control specific transcriptional programmes68,69. Here we summarize some notable recent developments relating to activator–Mediator interactions in metazoans.
First, on a cautionary note, except for a handful of cases, not all interactions reported in the literature have been rigorously linked with an appropriate effector function; apart from a limited number of cases, it is yet to be established that identified physical activator–Mediator interactions are critical in the cell. Second, there are important exceptions to the generalization that the Mediator tail is the primary target of activators. Interactions of nuclear receptors with the middle subunit MED1 offer particularly well documented examples of interactions with defined biological consequences70. At the same time, as noted above, the tail-proximal location of MED1 might yet signify close functional coordination with the tail. Furthermore, contrary to early expectations, a given Mediator subunit may in fact serve as a regulatory node for diverse transcriptional programmes. Thus, MED1 has now been implicated in regulation by, among others, haematopoietic GATA1 (ref. 71), B cell-specific OCA-B72 (see below) and leukaemogenic E2A-PBX1 (ref. 73). Third, a distinct class of coactivators that generally are cell type-restricted coactivators and themselves do not directly bind DNA but are recruited by more ubiquitous site-specifically bound activators have also been shown to interact with Mediator. In addition to OCT2-recruited OCA-B72, these include nuclear receptor-recruited PGC-1 (ref. 74) and PRDM16 (refs. 75,76). Thus, coactivators from this class effectively furnish a supplementary, cell type-specific activation domain, which also communicates with the Mediator. Even though each of these happens to interact with MED1, there is no reason to believe that interactions of other similar coactivators will necessarily map to this subunit — especially as MED1 is not required for cell viability36,77.
Details of precisely how activators engage Mediator are just now beginning to emerge. As a prototype for nuclear receptor–Mediator interactions, NMR combined with other biophysical approaches has revealed important details underlying the interaction of the vitamin D receptor–retinoid X receptor (VDR–RXR) heterodimer with MED1 (ref. 78). Based on prior biochemical and genetic studies, the expected interaction between an LXXLL motif-containing nuclear receptor box in MED1 with the VDR AF2 helix 12 is the primary driver of the interaction. However, novel interactions, including between the VDR dimerization partner RXR and a non-nuclear receptor box region of the MED1 amino terminus, were also observed, suggestive of unexpectedly broad interaction surfaces. Similar to MED1, the tail subunit MED25 has also emerged as an activator interaction hot spot; it interacts with a subset of nuclear receptors via an LXXLL-containing domain79,80 and with the herpesvirus activator VP16 via a heterologous seven-stranded β-barrel domain (ACID)81,82. Curiously, however, one of the recent PIC cryo-EM structures, in which a Mediator preparation purified by VP16 affinity chromatography was used, failed to visualize the VP16–Mediator interaction45, perhaps because of flexible elements in both interacting partners.
Indeed, biophysical studies more intriguingly suggest a ‘free-for-all’ type of interaction between activation domains and Mediator subunits. Thus, for example, the bona fide activation domain of ETV4, an ETS family member, also interacts with the ACID domain of MED25 (ref. 83). Interestingly, the DNA-binding domain makes multiple additional contacts with other regions of MED25 (ref. 83). Even more dramatic is how the yeast activator Gcn4 makes ‘fuzzy’ interactions with MED15: the protein–protein interaction interface itself is relatively dynamic, reflecting a low intrinsic affinity, and also is oblivious to the precise orientation of the subdomains that make up the Gcn4 activation domain84. As discussed below, interaction mechanisms may also entail the numerous IDRs present in Mediator subunits, especially in metazoan subunits in which evolutionary divergence is most evident.
In addition to contributing to increased local Mediator concentration near the PIC, activator–Mediator interactions have the potential to modulate Mediator function through conformational changes. Although high-resolution structural studies of activators in association with the intact Mediator complex are pending, early low-resolution images have already suggested that activators can alter Mediator conformation and, thus, its interactions with the PIC85. Given the high degree of inter-modular flexibility within the Mediator complex, it is conceivable that activators bound to the tail, or other subunits such as MED1, might indirectly impact head and middle structures implicated in interactions with Pol II and other PIC components. In this regard, given the peculiar location of MED1 by the tail–middle junction, it is particularly noteworthy that MED1 ablation yields a complex that has a higher avidity for Pol II (and also the kinase module)36, suggesting that nuclear receptor–MED1 interactions modulate Mediator–Pol II interactions.
Mediator-directed and TFIID-directed PIC formation in the context of chromatin
It has long been known that, by restricting access to the transcription machinery, nucleosomes are generally repressive for transcription86. However, the precise manner in which chromatin affects the PIC is turning out to be more nuanced. Broadly, the logic of how the PIC engages nucleosomes in the vicinity of the core promoter is dictated by the precise nucleosome configuration87. In the context of a nucleosome-depleted region, which characterizes the start site of genes that are constitutively active, such as housekeeping genes, PIC interactions with the +1 nucleosome and, potentially, also the −1 nucleosome that flank the nucleosome-depleted region may play important modulatory roles (Fig. 5). Critical roles for various TFIID TAFs in interactions with nucleosomes have come to light. A crystal structure of the tandem bromodomain repeats of metazoan TAF1, which interact with acetyl-lysine-containing histone H4 peptides, earlier suggested that TFIID can interact directly with specifically modified nucleosomes88. Subsequently, TAF3 was also found to contain PHD finger domains that interact with histone H3 trimethylated. at lysine 4 (H3K4me3), a mark that is enriched in the +1 nucleosome89. Furthermore, genes containing TATA-less core promoters seem to compensate for their diminished TBP–promoter interactions by exhibiting greater dependence on the TAF3–H3K4me3 interaction than do genes containing stronger TATA elements90. Interestingly, a lack of bromodomains in yeast TAF1 seems to be compensated by bromodomain-containing Bdf1 and Bdf2, which bind yeast TAF7 and interact with acetylated H4 (ref. 91) (but see Box 2). Similarly, lack of TAF3 PHD fingers is compensated by TAF14, which is not present in the metazoan complex and can interact with H3 acyl-lysines via its YEATS domain92. Given this intimate association of TFIID with the +1 nucleosome, together with the extended downstream TAF interactions and evidence that both TFIID and the +1 nucleosome contribute to promoter-proximal pausing (see below), it may well be that the PIC and the +1 nucleosome constitute a single extended, albeit integrated, functional unit controlling initiation and early elongation. Noteworthy in this regard is a study demonstrating that native chromatin from yeast cells actually supports higher levels of transcription in vitro than a corresponding naked DNA template93. Even though this study used TBP instead of TFIID, it is nonetheless suggestive of favourable PIC–nucleosome interactions.
Fig. 5 |. TFIID recruitment to promoters in the context of a nucleosome-depleted region.
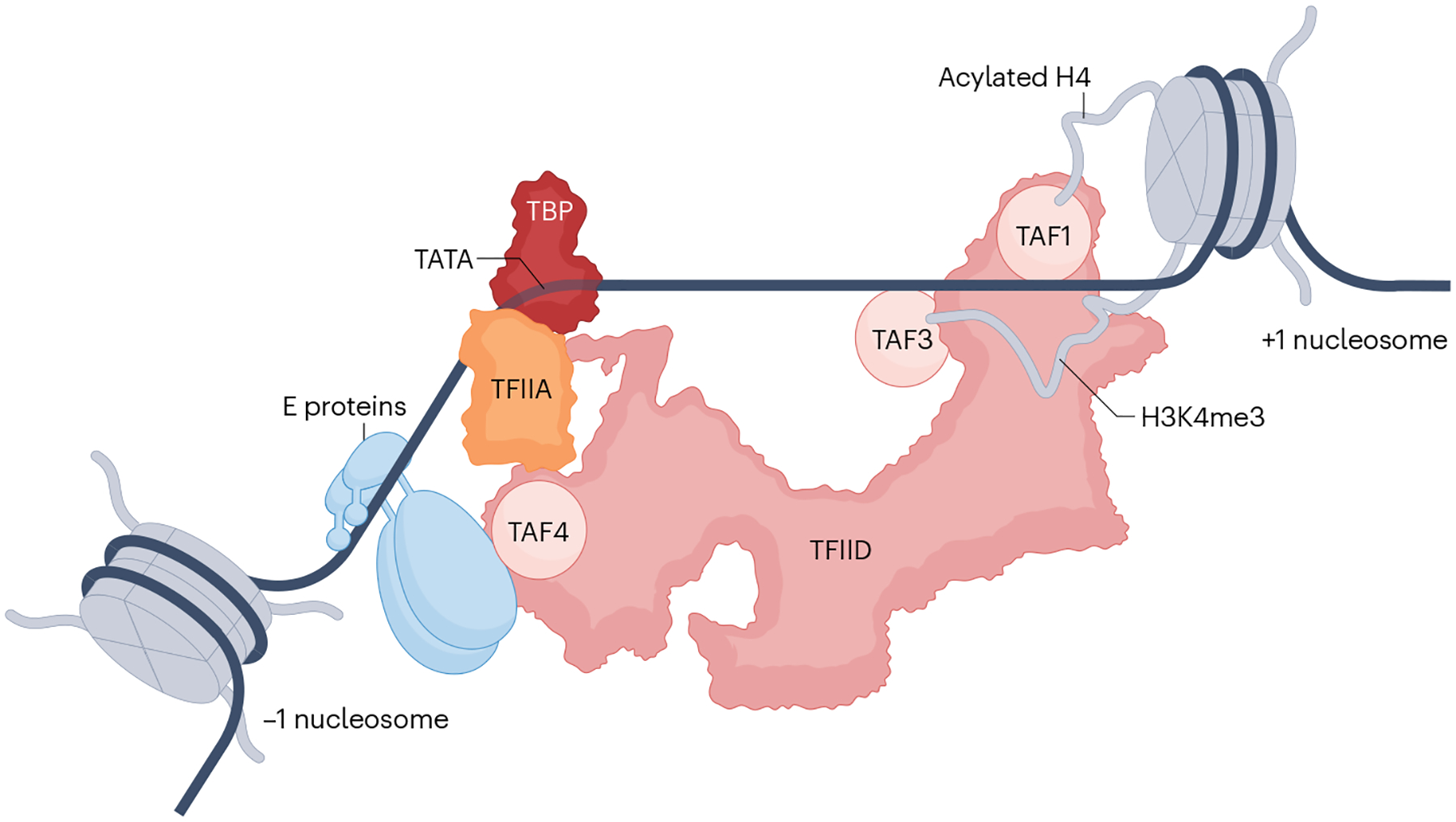
A composite model showing multiple potential interactions of TFIID just upstream of a positioned +1 nucleosome. In addition to the promoter DNA interactions (Fig. 4), TATA binding protein (TBP)-associated factors (TAFs) in TFIID can have stabilizing interactions with an activator (such as TAF4 with an E protein) bound to a proximal element or with post-translationally modified histone tails in the +1 nucleosome. Documented tail interactions include recognition of acetylated histone H4 by TAF1 and of histone H3 trimethylated at lysine 4 (H3K4me3) by TAF3. A location for the highly mobile lobe A is not specified (see Fig. 4 legend).
Box 2. Roles of Mediator-interacting and TFIID-interacting auxiliary cofactors in PIC function.
We briefly note the contribution of ubiquitous auxiliary cofactors that impinge upon the pre-initiation complex (PIC). One diverse group has been shown to have PIC coactivator activity and includes poly-ADP ribose polymerase 1, topoisomerase 1, PSIP1 (p52/75/LEDGF), PC4 and various HMG box-containing factors1,2. Because of their involvement in multiple nucleic acid transactions besides transcription, their manifold interactions with DNA, RNA and multiple PIC components, and, in the case of topoisomerase 1, dispensability of its enzymatic activity, these are best regarded as general architectural components that moonlight as PIC cofactors.
More consequential, perhaps, are the broad-acting cofactors NC2 (refs. 1,2,143,158), with specific PIC modulatory roles at the level of TFIID, and BET (bromodomain and extra-terminal) family members159, with apparent roles at the level of both TFIID and Mediator. Originally isolated biochemically as a negative cofactor, heterodimeric NC2 is currently believed to have dual functions143,158. It binds to core promoter-associated TATA binding protein (TBP) in competition with TFIIA and prevents further PIC assembly by precluding TFIIB entry. It also synergizes with Mot1, which was isolated as TBP-associated BTAF1 and is a SWI-SNF-related factor that acts in an ATP-dependent manner to dissociate TBP from promoters160. Interestingly, NC2 manifests as a positive cofactor for transcription of TATA-less genes that contain a downstream promoter element (DPE)161. It may therefore be that, through as yet uncharacterized mechanisms, Mot1 and NC2 help redirect TBP/TFIID from promoters with greater affinity for TBP to weaker promoters.
BET factors are characterized by two acetyl-lysine histone H4-interacting bromodomains similar to those in metazoan TBP-associated factor 1 (TAF1). Of the three BET family members, BRD4 is the most critical and has been implicated in both transcription initiation and early elongation across the genome159. It facilitates Mediator recruitment to both enhancers and core promoters via bromodomain interactions, potentially entailing both acetylated histones and acetylated activators162, and tends to cluster with the Mediator near active loci in condensate-like bodies. There is also evidence that BRD4 and TAF1 functionally synergize163. Importantly, it has recently become evident that yeast Bdf1/2, which were proposed to stand in for metazoan TAF1 bromodomains in targeting TFIID to acetylated H4 (see main text), may well be evolutionarily more related to metazoan BET factors, based not solely on greater amino acid sequence similarity but also on parallel functional roles with respect to both the Mediator and TFIID164. This suggests that the TAF1 bromodomains evolved to complement the bromodomains of primordial BET family members, whose effects on the PIC are more pronounced, at least in the case of the yeast Bdf1/2. Although precisely how metazoan BRD4 interacts with the Mediator and TFIID remains unknown, based on yeast studies it has been proposed that BET family proteins may facilitate nucleation of these PIC coactivators164.
Also consistent with this notion, as well as an early report of Mediator interactions with nucleosomes94, a cryo-EM structure of a Mediator-containing PIC that abuts the +1 nucleosome has very recently been published95. The structure raises the possibility that Mediator interactions via the hook structure containing MED19 and MED26 may further contribute to stabilization of the PIC, with potentially critical functional consequences, especially on weaker promoters containing non-consensus TATA-like sequences. Whereas stabilizing interactions mediated by TFIIH (p52) were also evident, predicted interactions of TFIID subunits with the nucleosome were not observed in this particular structure, likely due to the absence of the implicated modifications and because TFIID chromatin reader domains are tethered to the core via IDRs that are refractory to visualization by cryo-EM. Note that in the case of Mediator, its functional roles vis-à-vis the +1 nucleosome may extend to more indirect effects, such as in conjunction with elongation factors96 and chromatin coactivators (see below).
If, in contrast to the scenario in which the positioned +1 nucleosome is localized downstream of core promoter determinants, a nucleosome occludes these elements and thereby precludes nucleation of a PIC, then active intervention by chromatin coactivators and chaperones is necessitated. This would generally be the case for developmentally regulated and inducible genes. Numerous pathways for how ‘open chromatin’ is generated have been proposed and invoke, among other mechanisms, roles for pioneer factors that can bind their target sites even when wrapped within nucleosomes97. However, in relation to any roles for Mediator and TFIID in this process, available data remain insufficient to formulate clear-cut models. By virtue of its ability to directly bind DNA, TFIID might compete with nucleosomes for promoter occupancy98 and serve a placeholder function for the nascent PIC, just as it can bookmark loci for activation following a recent round of transcription99 or mitosis100.
By contrast, Mediator possesses certain functionalities that endow it with the potential to play a more active role in generating open chromatin. These include not only functional synergy between the histone acetyl transferase p300 and Mediator but, potentially, also Mediator-facilitated coordination between the chromatin modification and PIC formation stages of transcriptional activation. Thus, for example, Mediator facilitates formation of a ternary activator–Mediator–p300 complex that can then be resolved in several ways. In one study in which activator binding sites were juxtaposed to the core promoter, p300 autoacetylation caused p300 to dissociate from both this ternary complex and acetylated histone anchors to facilitate TFIID binding26. In another study that was more focused on oestrogen receptor function from distal enhancers, NCOA family members recruited upon hormonal induction took over at later time points from the Mediator to maintain p300 in the now-activated enhancer, perhaps thereby freeing Mediator for interaction with the PIC, as discussed below101. Nonetheless, precisely how Mediator recruited to a distal enhancer might contribute to a nucleosome-depleted region near the TSS remains unclear.
Delivery of Mediator from enhancer to core promoters
Activator binding sites are often located distally from the core promoter within enhancers that, in the case of super-enhancers, may harbour a large array of sites for diverse activators102. A notable gap in our understanding of coactivator mechanisms relates to how Mediator complexes that may initially be recruited by enhancer-bound activators are ultimately delivered to the nascent PIC at the core promoter. Some current hypotheses are discussed below.
In recent years, the biophysical phenomenon of liquid–liquid phase separation has been invoked in models for activator–coactivator interactions, as well as in maintenance of enhancers (especially super-enhancers) and promoters in close proximity within subnuclear compartments that might facilitate coactivator–PIC interactions103. Indeed, Pol II and Mediator clusters have been observed in mammalian cells and have been interpreted in terms of active transcriptional hubs with a propensity to form liquid–liquid phase-separated condensates104,105. Several factors potentially underlie this clustering. It could result from multiple Mediator-tethering activators in a single enhancer106. Juxtaposition of multiple transcription units with their enhancers and promoters into confined subnuclear zones could amplify the clustering effect103. Critically, IDRs in Mediator subunits, especially in those that have diverged considerably from their yeast orthologues33,67,107, as well as in Pol II108,109, activation domains67 and, potentially, TFIID TAFs, could foster a local environment in which multivalent protein–protein interactions can occur. However, whereas activation domains and selected Mediator subunits, including MED1 with its large IDR, can at elevated concentrations generate condensates that display typical liquid–liquid phase separation behaviour105, it remains uncertain how this relates to enhancer function in cells. Real-time single-cell fluorescence microscopy of synthetic activator arrays has indicated that formation of phase-separated droplets in fact has no, or even an inhibitory, effect on their transactivation potential110. Furthermore, nanoscopic measurements of Pol II molecules at single gene loci suggest that its numbers may not be high enough to reach critical thresholds for phase separation111. Nonetheless, regardless of the underlying biophysics, these numbers (circa 10 per locus) remain consistent with factor clustering and with the hypothesis that multivalent IDR interactions serve to enhance local factor concentration to facilitate downstream PIC formation.
A stable cohesin-mediated looping model in which activator-bound Mediator contacts the PIC was also proposed earlier112. Studies in yeast, in which the upstream activator binding sites (UASs) are generally in relative proximity to the core promoter, further suggested a ‘drawbridge’ model in which a kinase module-containing Mediator is first recruited to the UAS followed by transient contacts between UAS-anchored Mediator and the PIC with concomitant loss of the kinase module, whose association with the core Mediator is incompatible with Pol II interactions113–115 (Fig. 6). However, other studies showed that in mammalian cells, acute degradation of Mediator does not generally result in disruption of stable promoter–enhancer contacts36,116. Moreover, cohesin depletion had minimal effects on gene expression117. Thus, newer models are being considered in which Mediator is not per se the physical link between the enhancer and the promoter. Rather, Mediator complexes diffusing to the PIC in the constrained space of chromatin loops generated by as yet unidentified architectural mechanisms might still retain a ‘memory’ of prior contact with enhancer activators36,118 (Fig. 6). This memory could be propagated either through long-term changes, such as compositional changes resulting from kinase module eviction and Pol II association, activator-induced conformational changes (above) or post-translational modifications118. Alternatively, enhancer-released Mediator might still retain one or more of the activators, which dynamically bind to their cognate sites119 and which could continue to maintain Mediator in a PIC-interacting conformation.
Fig. 6 |. Pathways for delivery of Mediator from distal enhancers to the PIC.
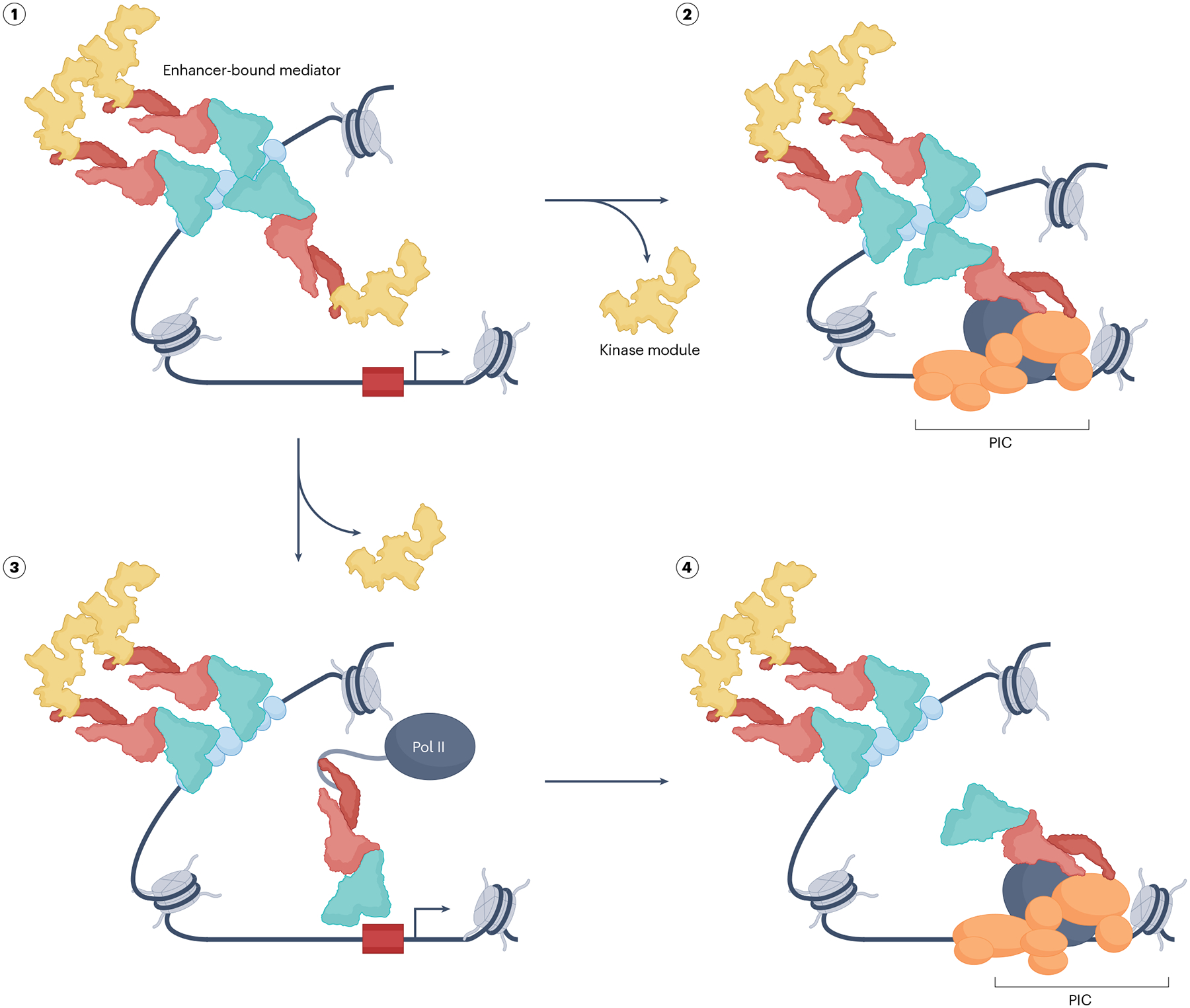
Two general models of how Mediator that has been recruited to a distal enhancer (state 1) can be delivered to the pre-initiation complex (PIC). Multiple kinase module-associated Mediator complexes, illustrative of Mediator clustering at a super-enhancer, are depicted bound to an array of activators. In one pathway (state 2), one of these complexes participates in bridging the enhancer and promoter where it facilitates PIC assembly after having ejected the kinase module. This long-range interaction may be transient and facilitated by architectural factors that generate and stabilize DNA loops. Alternatively, an enhancer-recruited Mediator complex may detach from the chromatin (state 3) after undergoing some form of ‘activation’ following activator interaction (see main text). Such an altered Mediator would be capable of interaction with RNA polymerase II (Pol II) (via its RPB1 carboxy-terminal domain (CTD)), whose Mediator association is mutually exclusive with that of the kinase module. This complex would be able to readily diffuse to a nearby nascent PIC (state 4) within the confines of a chromatin topological structure generated by as yet uncharacterized architectural factors. Among other possibilities, an interesting variant of this pathway imagines that an activator-bound Mediator dissociates from the activator’s cognate site in the enhancer and goes on to facilitate PIC assembly. Not highlighted is the likely scenario that in contrast to the enhancer-bound Mediator, which is enriched in the kinase module, the PIC-associated Mediator might be preferentially associated with MED26. In all these models, but especially in the diffusion model (state 3), it is not clear how promiscuous enhancer–promoter interactions are minimized. In addition to chromatin topology and related enhancer condensates (see main text), activity gradients emanating from the enhancer as a point source and regulated by cycles of post-translational modifications and reversals could ensure that functionally active diffusing coactivators do not stray too far away from the cognate promoter118. See main text for how Mediator might also contribute to chromatin remodelling at the promoter if it is occluded by a nucleosome.
Establishment and function of coactivator-directed PICs
In principle, each of the steps in the PIC pathway can be targeted for regulation by coactivators acting upon it. Furthermore, it is also possible that the coactivators might engender alternate PIC assembly pathways. Reminiscent of an earlier PIC reconstitution in vitro that suggested a non-canonical PIC assembly pathway120, recent single-molecule studies in yeast extracts have revealed a branched pathway in which Pol II, TFIIF and TFIIE can preassemble and cluster at the UAS in an activator-dependent manner — and, thus, presumably also in a coactivator-dependent manner — thereby creating a reservoir of PIC subcomplexes poised for delivery to the nascent PIC121. Similarly, cryogenic electron tomography analysis of a yeast PIC assembled on divergent promoters has suggested that partial PICs containing Mediator, Pol II, TFIIB and TFIIF can initially be recruited to activators prior to transfer to the core promoter and concomitant or subsequent recruitment of the other GTFs122. In the case of metazoan coactivators, important insights into how they affect PIC structure and function have come from recent cryo-EM structures containing the Mediator in the context of a TBP-nucleated PIC44,45, as well as TFIID-nucleated PICs with43 or without the Mediator49.
Impact of TAFs on the PIC
Building on the earlier promoter-bound TFIID structures47,48, one study that focused on TFIID-nucleated PICs on templates with distinct core promoter architectures49 has revealed interesting variations in assembly pathways. As predicted from earlier studies, interactions between additional TFIID TAF subunits and a range of core promoter elements other than TATA contribute substantially to the PIC nucleation event49. Furthermore, comparison of intermediate PICs that have not yet incorporated TFIIE and TFIIH shows distinct template configurations dependent on whether the promoter contains both the TATA and the DPE (as in the synthetic SCP), or either just the TATA or the DPE (as in most natural promoters). On the SCP, the intermediate PIC undergoes a much more extensive stepwise reorganization relative to the TATA-only and TATA-less promoters. Interestingly, once TFIIE and TFIIH enter the PIC, the initial promoter-specific differences are ironed out and the templates are equally well positioned for transcription initiation. Notably, this includes the previously observed TBP-induced bending of the promoter DNA, which has now been observed regardless of the presence of the TATA box, broadly consistent with the observation in yeast of equivalent TBP occupancy at both TATA and TATA-less promoters123. Thus, whereas the pathways leading to the final PICs are quite different dependent on the promoter architecture, the end products are similar, if not identical49. This raises the strong possibility that these pathways themselves may be subject to regulation, especially as TATA box presence in promoters strongly correlates with tissue-specific expression of target genes124. One important outcome of having a TFIID-nucleated PIC, as opposed to a TBP PIC, is the marked effect on the stabilization of TFIIH, especially via a network of interactions around XPB43. The new studies suggest that through an interplay between TFIID and TFIIH that entails partial loss of TAF contacts with downstream promoter DNA to allow TFIIH–DNA interactions, both the XPB translocase and CAK are ultimately well positioned for their respective catalytic activities43,49 (Fig. 7; see also below). Thus, in addition to delivering TBP to the promoter and promoting PIC nucleation via TAF–DNA interactions, TFIID also functions as a PIC scaffold that facilitates initiation.
Fig. 7 |. A TFIID-containing and Mediator-containing PIC.
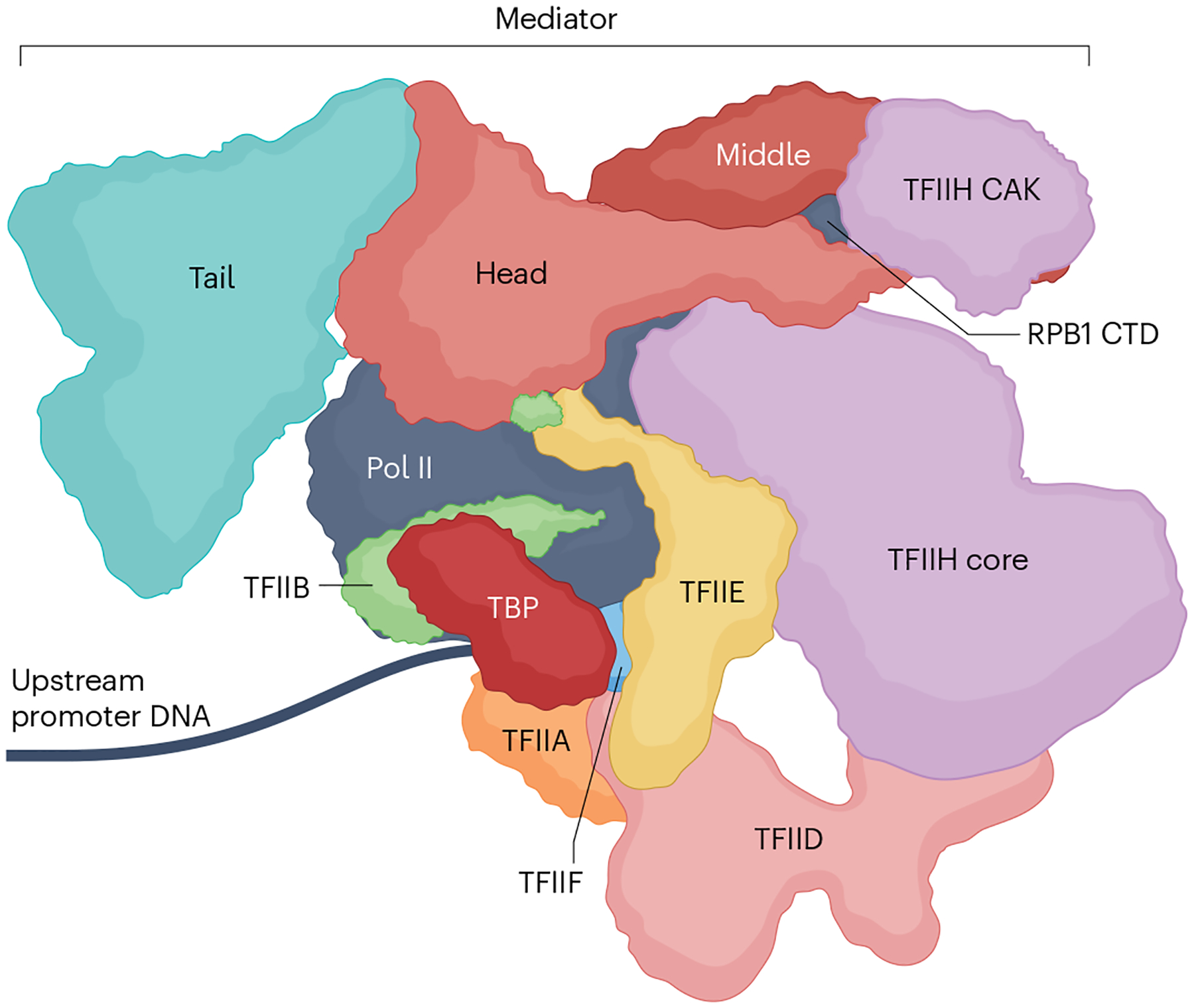
A model for a TFIID-containing and Mediator-containing pre-initiation complex (PIC) adapted from ref. 43. Note the additional stabilizing interactions relative to a TATA binding protein (TBP)-nucleated minimal PIC that does not contain Mediator (Fig. 1). Highlighted here is the stabilization of the RPB1 carboxy-terminal domain (CTD), but numerous other subtle, and perhaps dynamic, interactions between the coactivators, RNA polymerase II (Pol II) and general transcription factors (GTFs) also take place, as discussed and referenced in the main text. Additional stabilizing interactions of the PIC might come from promoter proximally bound activators and the downstream +1 nucleosome via TFIID (Fig. 5) and Mediator (not shown; see text). Other general cofactors that may be fulfilling architectural and other roles may also contribute but are not shown (Box 2). CAK, CDK-activating kinase.
Mediator interactions with Pol II and GTFs
Inclusion of the Mediator in the PIC has major effects on the latter’s structure, with implications for function. Structures of Mediator-containing PICs nucleated with either TBP44,45 or TFIID43 show extensive Mediator–PIC interfaces. Potentially underscoring the dynamic nature of these interactions and the existence of intermediates, some variations in Mediator–PIC structures have been observed by different groups. Overall, the interfaces include: interactions of the Pol II stalk with the head module (MED8, MED22) in a putative early intermediate that are further strengthened through association with repeat motifs in MED14 in a more mature PIC44,45; potentially transient interactions of the MED18 and MED20 subunits of the head with RPB3/RPB11 and Pol II dock in the vicinity of both the active site-proximal TFIIB B-ribbon and the TFIIE E-ribbon43,45,125; interactions of a Mediator substructure formed by MED1, MED4 and MED9 with the Pol II RPB8 (refs. 43–45); and additional interactions of the MED14-containing hook structure of the Mediator with TFIIH CAK and of head module subunits with the TFIIH core, which essentially ‘sandwich’ the hook43,44. An additional protein sandwich arises from interactions between two CTD repeats of Pol II RPB1 and subunits of the Mediator head and middle modules that optimally position these repeats for eventual phosphorylation by TFIIH CAK, the catalytic CDK7 subunit of which can also be seen interacting with a CTD unit43,45. More recently, a yeast TBP-nucleated Mediator–PIC structure captured up to 11 CTD repeats, suggestive of much more extensive contacts between Mediator and these repeats126. Thus, similar to TFIID, Mediator contributes to stabilization of TFIIH, attesting further to functional cooperativity between TFIID and Mediator in establishing the PIC. Indeed, during transitions that yield a putative mature PIC, movements of TFIID lobes and the TFIIH XPB-containing core lead to concerted movements of Mediator, TFIIE and the Pol II stalk43. The net result is stabilization of the TFIIH core and its optimal placement for its DNA translocase-dependent promoter melting activity43,127,128.
Reciprocal PIC-induced changes in the conformation of the Mediator are also reported. Free Mediator can exist in either an ‘extended’ or a ‘bent’ conformation43. However, PIC-bound Mediator is predominantly in the bent conformation, indicating that the PIC either facilitates the extended to bent transition or is selective for the bent form. Additional modular reorganization entails concerted movements of the head and middle domains arising from interactions with Pol II subunits43. Notably, however, TFIID lobe dynamics that deliver TBP to the promoter in the context of a Mediator–PIC appear similar to those seen in TFIID–promoter complexes48. Moreover, despite indications of functional cooperativity between TFIID and Mediator from biochemical and genomic analyses24–27, no direct TFIID–Mediator interactions are evident from the published structures. Although the possibility that direct interactions might occur in potential PIC intermediates that have not yet been visualized, especially given the large-scale lobe movements in TFIID, it is also likely that cooperativity results mainly from their combined roles in supporting various aspects of PIC function, as noted above for their effect on TFIIH (Fig. 7).
The Mediator–PIC interactions described above are predicted to have important consequences for PIC function in addition to the TFIIH effects discussed below. Given the apparently rapid turnover of TFIIB in single-molecule studies done in the absence of Mediator129, the potential stabilization of TFIIB within the PIC could further enhance its productive lifetime and ensure efficient RNA synthesis. Indeed, in human cell nuclear extracts in which transcription is normally absolutely dependent on the Mediator, this requirement can be bypassed if excess TFIIB is included in the reactions130. In addition to PIC instability potentially arising from the intrinsic tendency of GTFs to turn over, various factors, including Pol II-associated Gdown1, can interfere with PIC formation by occluding TFIIB and TFIIF131. Through its various stabilizing interactions, Mediator allows the negative effects of such factors to be efficiently neutralized. On the basis of a Mediator-containing but TAF-lacking yeast PIC structure, it was suggested that Mediator effects may be limited to facilitating PIC formation39. However, the latest data collectively leave open the possibility of effects at the level of PIC remodelling. Insofar as Mediator provides a sequestered environment for the PIC to undergo remodelling and mature into a fully functional form, it may be helpful to view it as a chaperone-like entity. It is therefore noteworthy that engagement of the Mediator with the PIC unexpectedly leads initially to a non-functional PIC132. Only once energy is expended by the TFIIH XPB translocase, potentially in concert with the promoter melting step, is the PIC fully activated132. At the same time, TFIIH-dependent RPB1 CTD phosphorylation also contributes to Mediator release from the PIC113,114,133,134, providing an additional basis for how stabilization of the TFIIH kinase module by Mediator and TFIID could facilitate efficient transition to the early elongation form.
Post-initiation roles of PIC coactivators
Incorporation of TFIID and Mediator into the PIC has consequences that extend beyond facilitating PIC formation. Although the fate of the TFIID subunits TAF1 and TAF2 in lobe C, which bind to DPEs43,48,49, following transcription initiation remains unclear, especially in view of the dynamic interplay with TFIIH described above43, a clash between the extended TFIID–promoter interface and Pol II molecules that are escaping from the promoter is predicted. Although studies have yet to address whether additional TFIID rearrangements occur, the downstream interactions might contribute to promoter-proximal pausing135. Whether release of Pol II paused as a result of downstream TAFs is dependent on release factors that have thus far been implicated136 is also unclear, although interactions between a leading candidate for pause release, the super elongation complex (SEC) and multiple TAFs have been described137. However, studies in the yeast system conversely suggest that the downstream TAF interactions might persist — or be restored following transient disruption — after a pioneer round of transcription to thereby facilitate transcription reinitiation99.
Evidence for Mediator roles post initiation has also emerged. The metazoan-specific middle module subunit MED26 was previously shown to interact with both the SEC138 and the related little elongation complex (LEC)139 via an N-terminal domain with homology to the transcription elongation factor (TEF) TFIIS. Although not evident in cryo-EM structures of TFIID-containing and Mediator-containing PICs43, this same region of MED26 was found to interact with TFIID138, leading to the suggestion, albeit as yet unproven, that MED26 orchestrates a transition of the PIC to an early elongation complex that is less prone to pausing. Interestingly, the Mediator kinase module has also been implicated in SEC recruitment140. Because PIC-bound Mediator does not contain the kinase module — the presence of MED26 and Pol II on the one hand and the kinase module on the other being mutually exclusive64,141 — this interaction may relate to a different SEC pathway. Although the mechanism remains unclear, a close interrelationship between Mediator and Pol II elongation is also evident from recent rapid degron-based cellular depletion studies in which elimination of the backbone MED14 subunit unexpectedly did not lead to complete shutdown of transcription owing to partial compensation by the SEC116. Moreover, in vitro studies have indicated that functional synergy between Mediator and TFIIS can facilitate Pol II transit through the +1 nucleosome that may abut the PIC96. Notably, TFIIS has been construed to be a component of an expanded PIC in yeast that may facilitate the initiation to elongation transition142,143. Altogether, even though there is no evidence that either Mediator or TFIID can travel with Pol II along gene bodies, mechanisms exist that can project their functional ranges beyond PIC assembly and initiation.
Conclusions and perspectives
As summarized in this Review, the Mediator-directed and TFIID-directed Pol II PIC is structurally and functionally quite distinct from a minimal PIC composed of TBP, TFIIA, TFIIB, TFIIE, TFIIF and TFIIH. This expanded PIC integrates multiple components for optimized sensing of diverse regulatory inputs and generation of the appropriate response. Thanks to recent developments, we now have a much better grasp of the inner workings of the machinery that relays the regulatory signal across the various components that include activators, coactivators, GTFs, Pol II and, potentially, the +1 nucleosome. In addition to specifying where transcription starts, a key effector function of the PIC is to control formation of the first few phosphodiester bonds by the active site of the Pol II enzyme and to ensure the latter’s efficient conversion into an early elongating form; thus, the complexity of the PIC is undoubtedly the result of an evolutionary need for greater control and precision of this critical step in transcription. Mediator and TFIID TAFs furnish extra regulatory layers that not only confer enhanced stability to the system, as is often highlighted, but importantly also enhance functional flexibility.
Given the wide range of signals impinging upon different loci, the question arises as to the degree to which PIC responses are customizable. Even as we now appreciate the general mechanistic principles underlying PIC function at the level of a binary on–off switch, we only partially understand how individual signals are absorbed and transduced into precisely calibrated outputs. Recent cell-based studies have emphasized that transcription occurs in bursts whose magnitude and frequency are the major determinants of the expression levels of a given locus144. Similarly, promoter-proximal pausing and pause release have been proposed as critical rate-limiting steps136. Yet, it is the rate of the steps collectively leading to formation of the initial phosphodiester bond by the Pol II active site that will ultimately determine the burst size and frequency, as well as how many Pol II molecules arrive at a pause site145. Thus, a desideratum in the field is to work out the complete mechanistic basis for precisely how an integrated transcriptional response is elicited by a PIC at an individual locus, from the arrival of the activation signal to its final modulatory effect on the Pol II active site.
An outstanding question in this regard pertains to the number of functional coactivator-mediated effector states the PIC can acquire in response to diverse inputs. It is already clear that conformational flexibilities of the Mediator and TFIID and their propensity to interact with diverse factors, whether via conventional interaction mechanisms or more unorthodox mechanisms entailing IDRs, can provide a physical basis for the functional states (see also Box 2). With its nearly 30 subunits, the Mediator is theoretically capable of accessing a large number of conformational states and may be critical in imparting at least that many distinct effector outcomes to the PIC. Although some of the variant structures revealed by cryo-EM clearly are intermediates43, others may reflect functionally relevant effector states. However, for technical reasons these studies have not yet captured the entire spectrum of possible states, which are likely to be very dynamic. For example, in addition to the absence of evidence for TFIID–Mediator contacts, there are indications from genetic studies in yeast of interactions that cannot be explained by current structures146.
The understudied phenomenon of PIC coactivator heterogeneity and its capacity to confer additional functional states may also be relevant in output customization. Variations in TFIID composition were mentioned above. Similarly, several Mediator subunits have separately encoded paralogues, and incorporation of one or another into the complex can determine the spectrum of affected genes, as exemplified by paralogues CDK8 and CDK19 (refs. 140,141). Others are translated from alternatively spliced forms, which can also lead to microheterogeneity within the bulk Mediator population with functional consequences, as is suggested from a conformational change in the tail module dependent on whether one or the other alternatively spliced version of MED16 is incorporated43. Ultimately, PIC dynamics will necessarily have to be worked out in the context of the activators that control the coactivators that, in turn, control the PIC. As highlighted here, also related is the outstanding question of precisely how distal enhancer-bound activators transmit their effects to the PIC over long distances.
In addition to delineating the complete signal transduction pathways at representative loci, the hope is that critical, perhaps unique, branchpoints will be identified that will facilitate development of customizable therapeutic modalities for diseases arising from transcriptional dysregulation. Based on the advances in just the past decade, these goals look to be achievable in the foreseeable future. Rapidly evolving cryo-EM technology, especially in conjunction with concomitant advances in cross-linking–mass spectrometry and allied integrated structural approaches, should continue to lead the way in illuminating PIC and coactivator dynamics. Ongoing development of powerful tools that allow visualization of factors at the single-molecule level both in vitro and at individual loci in living cells should nicely complement these studies. Importantly, time-tested biochemical approaches, especially now that they can be coupled with sensitive and quantitative mass spectroscopy and other biophysical approaches, should continue to provide rigorous functional tests of hypotheses arising from these analyses.
Acknowledgements
The authors apologize to colleagues whose work could not be cited directly owing to space constraints. The authors thank past and current members of their laboratory for their many contributions to the authors’ understanding of the pre-initiation complex (PIC) and its coactivators. The authors are thankful to M. Gnädig, K. Ito and T. Onikubo for critical reading of the manuscript. The authors’ work was supported by National Institutes of Health (NIH) grants CA234575, CA273709 and AI148387.
Glossary
- +1 Nucleosome
A precisely positioned nucleosome located just downstream from the core promoter. Together with an upstream localized−1 nucleosome, it delimits the boundaries of a nucleosome-depleted region. It is not to be confused with the transcription start site (TSS), which is sometimes also referred to as the +1 site
- Basal transcription
Baseline levels of transcription that might occur in the absence of activation signals
- Bromodomain
A conserved protein domain that can recognize acetylated lysine residues. It is found in many chromatin readers, as well as BET (bromodomain and extra-terminal) family members such as BRD4
- Core promoter
A regulatory region within a transcriptional locus that consists of numerous elements that collectively or singly specify the transcription start site (TSS) by recruiting and orienting the pre-initiation complex (PIC). Elements include the TATA box, initiator (Inr), motif ten element (MTE) and downstream promoter element (DPE), among others
- Enhancers
Regulatory elements that carry binding sites for multiple transcriptional activators. Typically, enhancers are found at distal locations relative to the transcription start site (TSS), although some are located more proximally. Variants include super-enhancers, which are characterized by a large number of activator binding sites. In yeast, the upstream activator binding site (UAS) serves the same purpose
- Initiation
The formation of the first phosphodiester bond by pre-initiation complex (PIC)-associated RNA polymerase II (Pol II) following ATP hydrolysis-dependent promoter melting
- Intrinsically disordered regions
(IDRs). Regions in a protein that lack any defined three-dimensional structure, at least in the absence of an interacting protein. They have a high propensity to form higher-order structures through weak multivalent interactions
- Pausing
Transient stalling of transcribing RNA polymerase II (Pol II) soon after promoter clearance that is regulated by a combination of pause-inducing and pause-release factors
- Phase separation
A tendency of two immiscible liquids to separate into distinct phases. In the transcription field, the term largely refers to the tendency of some activators and coactivators that contain intrinsically disordered regions (IDRs) to form condensates when forced into situations where they can cluster, for example on super-enhancers
- Pol II stalk
A prominent feature of RNA polymerase II (Pol II) composed mainly of RPB4 and RPB7 subunits. It can regulate other Pol II elements (for example, the clamp) and acts as a hub for the interaction of multiple pre-initiation complex (PIC) components
- Promoter escape
A series of post-initiation events that occur as a prelude to RNA polymerase II (Pol II) entry into the elongation mode and result in relinquishing of stabilizing interactions that anchor the pre-initiation complex (PIC). This process is closely related to promoter clearance
- RPB1 CTD
The large unstructured carboxy-terminal domain (CTD) of the largest RNA polymerase II (Pol II) subunit. In human, it consists of 52 heptapeptide repeats with the consensus sequence YSPTSPS. The repeat units are subject to phosphorylation at multiple residues. In the context of the pre-initiation complex (PIC), Ser5 is the predominant target of the cyclin-dependent kinase (CDK)-activating kinase (CAK) module of TFIIH
- YEATS domain
One of several domains that can recognize acylated lysine residues. So called because of its original identification in the chromatin reading modules of Yaf9, ENL, AF9, TAF1 and Sas5
Footnotes
This article is dedicated to the memory of Michael R. Green.
Competing interests
The authors declare no competing interests.
References
- 1.Roeder RG Role of general and gene-specific cofactors in the regulation of eukaryotic transcription. Cold Spring Harb. Symp. Quant. Biol 63, 201–218 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Thomas MC & Chiang CM The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol 41, 105–178 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Schier AC & Taatjes DJ Structure and mechanism of the RNA polymerase II transcription machinery. Genes Dev. 34, 465–488 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roeder RG The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci 21, 327–335 (1996). [PubMed] [Google Scholar]
- 5.Buratowski S et al. Function of a yeast TATA element-binding protein in a mammalian transcription system. Nature 334, 37–42 (1988). [DOI] [PubMed] [Google Scholar]
- 6.Horikoshi M et al. Cloning and structure of a yeast gene encoding a general transcription initiation factor TFIID that binds to the TATA box. Nature 341, 299–303 (1989). [DOI] [PubMed] [Google Scholar]
- 7.Hoey T et al. Isolation and characterization of the Drosophila gene encoding the TATA box binding protein, TFIID. Cell 61, 1179–1186 (1990). [DOI] [PubMed] [Google Scholar]
- 8.Kao CC et al. Cloning of a transcriptionally active human TATA binding factor. Science 248, 1646–1650 (1990). [DOI] [PubMed] [Google Scholar]
- 9.Kostrewa D et al. RNA polymerase II–TFIIB structure and mechanism of transcription initiation. Nature 462, 323–330 (2009). [DOI] [PubMed] [Google Scholar]
- 10.He Y et al. Structural visualization of key steps in human transcription initiation. Nature 495, 481–486 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]; This seminal study uses cryo-EM to visualize intermediates in the assembly pathway of a TBP-nucleated PIC that has previously been biochemically described.
- 11.He Y et al. Near-atomic resolution visualization of human transcription promoter opening. Nature 533, 359–365 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plaschka C et al. Transcription initiation complex structures elucidate DNA opening. Nature 533, 353–358 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Murakami K et al. Structure of an RNA polymerase II preinitiation complex. Proc. Natl Acad. Sci. USA 112, 13543–13548 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nogales E & Greber BJ High-resolution cryo-EM structures of TFIIH and their functional implications. Curr. Opin. Struct. Biol 59, 188–194 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aibara S, Schilbach S & Cramer P Structures of mammalian RNA polymerase II pre-initiation complexes. Nature 594, 124–128 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Schilbach S et al. Structure of RNA polymerase II pre-initiation complex at 2.9 Å defines initial DNA opening. Cell 184, 4064–4072 e28 (2021). [DOI] [PubMed] [Google Scholar]; Together with Aibara et al. (2021), this work provides high-resolution structural views of PICs that have been trapped in intermediate states and sheds new light on the mechanism of promoter melting through the TFIIH translocase.
- 17.Pal M, Ponticelli AS & Luse DS The role of the transcription bubble and TFIIB in promoter clearance by RNA polymerase II. Mol. Cell 19, 101–110 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Pugh BF & Tjian R Mechanism of transcriptional activation by Sp1: evidence for coactivators. Cell 61, 1187–1197 (1990). [DOI] [PubMed] [Google Scholar]
- 19.Hoffman A et al. Highly conserved core domain and unique N terminus with presumptive regulatory motifs in a human TATA factor (TFIID). Nature 346, 387–390 (1990). [DOI] [PubMed] [Google Scholar]
- 20.Goodrich JA & Tjian R TBP–TAF complexes: selectivity factors for eukaryotic transcription. Curr. Opin. Cell Biol 6, 403–409 (1994). [DOI] [PubMed] [Google Scholar]
- 21.Malik S & Roeder RG Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem. Sci 25, 277–283 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Myers LC & Kornberg RD Mediator of transcriptional regulation. Annu. Rev. Biochem 69, 729–749 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Lee TI & Young RA Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet 34, 77–137 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Guermah M, Malik S & Roeder RG Involvement of TFIID and USA components in transcriptional activation of the human immunodeficiency virus promoter by NF-κB and Sp1. Mol. Cell Biol 18, 3234–3244 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baek HJ et al. Requirement of TRAP/mediator for both activator-independent and activator-dependent transcription in conjunction with TFIID-associated TAF(II)s. Mol. Cell Biol 22, 2842–2852 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Black JC et al. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol. Cell 23, 809–818 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Grunberg S et al. Mediator binding to UASs is broadly uncoupled from transcription and cooperative with TFIID recruitment to promoters. EMBO J. 35, 2435–2446 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun F et al. The Pol II preinitiation complex (PIC) influences Mediator binding but not promoter–enhancer looping. Genes Dev. 35, 1175–1189 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richter WF et al. The Mediator complex as a master regulator of transcription by RNA polymerase II. Nat. Rev. Mol. Cell. Biol 23, 732–749 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malik S & Roeder RG The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet 11, 761–772 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allen BL & Taatjes DJ The Mediator complex: a central integrator of transcription. Nat. Rev. Mol. Cell Biol 16, 155–166 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsai KL et al. Subunit architecture and functional modular rearrangements of the transcriptional mediator complex. Cell 157, 1430–1444 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper provides the first detailed insights into subunit and modular organization of the Mediator core complex by a combination of cryo-EM, biochemical and genetic approaches.
- 33.Toth-Petroczy A et al. Malleable machines in transcription regulation: the Mediator complex. PLoS Comput. Biol 4, e1000243 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X et al. Redefining the modular organization of the core Mediator complex. Cell Res. 24, 796–808 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cevher MA et al. Reconstitution of active human core Mediator complex reveals a critical role of the MED14 subunit. Nat. Struct. Mol. Biol 21, 1028–1034 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study, a core human Mediator complex that retains many of its effector functions is recombinantly generated, allowing assessment of roles of individual subunits, including a pivotal architectural and functional role for MED14.
- 36.El Khattabi L et al. A pliable Mediator acts as a functional rather than an architectural bridge between promoters and enhancers. Cell 178, 1145–1158.e20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plaschka C, Nozawa K & Cramer P Mediator architecture and RNA polymerase II interaction. J. Mol. Biol 428, 2569–2574 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Robinson PJ et al. Molecular architecture of the yeast Mediator complex. ELife 4, e08719 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson PJ et al. Structure of a complete Mediator–RNA polymerase II pre-initiation complex. Cell 166, 1411–1422.e16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study is one of the first to describe a Mediator-containing PIC combining cryo-EM and cross-linking–mass spectroscopy-based integrative modelling.
- 40.Nozawa K, Schneider TR & Cramer P Core mediator structure at 3.4 Å extends model of transcription initiation complex. Nature 545, 248–251 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Tsai KL et al. Mediator structure and rearrangements required for holoenzyme formation. Nature 544, 196–201 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao H et al. Structure of mammalian Mediator complex reveals tail module architecture and interaction with a conserved core. Nat. Commun 12, 1355 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen X et al. Structures of the human mediator and mediator-bound preinitiation complex. Science 372, eabg0635 (2021). [DOI] [PubMed] [Google Scholar]; This landmark study reveals, using cryo-EM, the most complete views to date of the mammalian PIC assembled with TFIID, Mediator, Pol II and all the GTFs; it allows direct assessment of how the PIC is impacted by its two associated coactivators.
- 44.Rengachari S et al. Structure of the human mediator–RNA polymerase II pre-initiation complex. Nature 594, 129–133 (2021). [DOI] [PubMed] [Google Scholar]
- 45.Abdella R et al. Structure of the human Mediator-bound transcription preinitiation complex. Science 72, 52–56 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]; Together with Rengacahari et al. (2021), this work provides structural insights into how Mediator impacts the PIC, albeit in the context of TBP.
- 46.Zhang H et al. Mediator structure and conformation change. Mol. Cell 81, 1781–1788.e4 (2021). [DOI] [PubMed] [Google Scholar]
- 47.Louder RK et al. Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Nature 531, 604–609 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patel AB et al. Structure of human TFIID and mechanism of TBP loading onto promoter DNA. Science 362, eaau8872 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; Together with Louder et al. (2016), this work first reveals how TFIID engages with the various elements that constitute a core promoter and the dramatic conformational changes it undergoes during the process.
- 49.Chen X et al. Structural insights into preinitiation complex assembly on core promoters. Science 372, eaba8490 (2021). [DOI] [PubMed] [Google Scholar]; This important cryo-EM study of TFIID promoter complexes with diverse core promoters further reveals interesting promoter sequence-dependent dynamics and variations in how TFIID interacts with the promoter.
- 50.Cianfrocco MA et al. Human TFIID binds to core promoter DNA in a reorganized structural state. Cell 152, 120–131 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Papai G et al. Structure of SAGA and mechanism of TBP deposition on gene promoters. Nature 577, 711–716 (2020). [DOI] [PubMed] [Google Scholar]
- 52.Wang H et al. Structure of the transcription coactivator SAGA. Nature 577, 717–720 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Timmers HTM SAGA and TFIID: friends of TBP drifting apart. Biochim. Biophys. Acta Gene Regul. Mech 1864, 194604 (2021). [DOI] [PubMed] [Google Scholar]
- 54.Deato MD & Tjian R An unexpected role of TAFs and TRFs in skeletal muscle differentiation: switching core promoter complexes. Cold Spring Harb. Symp. Quant. Biol 73, 217–225 (2008). [DOI] [PubMed] [Google Scholar]
- 55.Hoey T et al. Molecular cloning and functional analysis of Drosophila TAF110 reveal properties expected of coactivators. Cell 72, 247–260 (1993). [DOI] [PubMed] [Google Scholar]
- 56.Moqtaderi Z et al. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature 383, 188–191 (1996). [DOI] [PubMed] [Google Scholar]
- 57.Walker SS et al. Transcription activation in cells lacking TAFIIS. Nature 383, 185–188 (1996). [DOI] [PubMed] [Google Scholar]
- 58.Timmers HTM & Tora L Transcript buffering: a balancing act between mRNA synthesis and mRNA degradation. Mol. Cell 72, 10–17 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Warfield L et al. Transcription of nearly all yeast RNA polymerase II-transcribed genes is dependent on transcription factor TFIID. Mol. Cell 68, 118–129.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wright KJ & Tjian R Wnt signaling targets ETO coactivation domain of TAF4/TFIID in vivo. Proc. Natl Acad. Sci. USA 106, 55–60 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen WY et al. A TAF4 coactivator function for E proteins that involves enhanced TFIID binding. Genes Dev. 27, 1596–1609 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study provides evidence for TFIID coactivator functions via direct activator–TAF interactions in the context of the native TFIID complex.
- 62.Zhang J et al. E protein silencing by the leukemogenic AML1–ETO fusion protein. Science 305, 1286–1289 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Liu WL et al. Structures of three distinct activator–TFIID complexes. Genes Dev. 23, 1510–1521 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Malik S et al. Structural and functional characterization of PC2 and RNA polymerase II-associated subpopulations of metazoan Mediator. Mol. Cell Biol 25, 2117–2129 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Knoll ER et al. Role of the pre-initiation complex in Mediator recruitment and dynamics. ELife 7, e39633 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Warfield L et al. Yeast Mediator facilitates transcription initiation at most promoters via a tail-independent mechanism. Mol. Cell 82, 4033–4048.e7 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boija A et al. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–1855.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang F et al. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature 442, 700–704 (2006). [DOI] [PubMed] [Google Scholar]
- 69.Stevens JL et al. Transcription control by E1A and MAP kinase pathway via Sur2 mediator subunit. Science 296, 755–758 (2002). [DOI] [PubMed] [Google Scholar]
- 70.Ge K et al. Transcription coactivator TRAP220 is required for PPARγ2-stimulated adipogenesis. Nature 417, 563–567 (2002). [DOI] [PubMed] [Google Scholar]
- 71.Stumpf M et al. The mediator complex functions as a coactivator for GATA-1 in erythropoiesis via subunit Med1/TRAP220. Proc. Natl Acad. Sci. USA 103, 18504–18509 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chu CS et al. Unique immune cell coactivators specify locus control region function and cell stage. Mol. Cell 80, 845–861.e10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee YL et al. Mediator subunit MED1 is required for E2A-PBX1-mediated oncogenic transcription and leukemic cell growth. Proc. Natl Acad. Sci. USA 118, e1922864118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen W, Yang Q & Roeder RG Dynamic interactions and cooperative functions of PGC-1α and MED1 in TRα-mediated activation of the brown-fat-specific UCP-1 gene. Mol. Cell 35, 755–768 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Iida S et al. PRDM16 enhances nuclear receptor-dependent transcription of the brown fat-specific Ucp1 gene through interactions with Mediator subunit MED1. Genes Dev. 29, 308–321 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harms MJ et al. PRDM16 binds MED1 and controls chromatin architecture to determine a brown fat transcriptional program. Genes Dev. 29, 298–307 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ito M et al. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol. Cell 5, 683–693 (2000). [DOI] [PubMed] [Google Scholar]
- 78.Belorusova AY et al. Molecular determinants of MED1 interaction with the DNA bound VDR–RXR heterodimer. Nucleic Acids Res. 48, 11199–11213 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee HK et al. MED25 is distinct from TRAP220/MED1 in cooperating with CBP for retinoid receptor activation. EMBO J. 26, 3545–3557 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Han EH, Rha GB & Chi YI MED25 is a mediator component of HNF4α-driven transcription leading to insulin secretion in pancreatic β-cells. PLoS ONE 7, e44007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Milbradt AG et al. Structure of the VP16 transactivator target in the Mediator. Nat. Struct. Mol. Biol 18, 410–415 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vojnic E et al. Structure and VP16 binding of the Mediator Med25 activator interaction domain. Nat. Struct. Mol. Biol 18, 404–409 (2011). [DOI] [PubMed] [Google Scholar]
- 83.Currie SL et al. ETV4 and AP1 transcription factors form multivalent interactions with three sites on the MED25 activator-interacting domain. J. Mol. Biol 429, 2975–2995 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tuttle LM et al. Gcn4–Mediator specificity is mediated by a large and dynamic fuzzy protein–protein complex. Cell Rep. 22, 3251–3264 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Taatjes DJ et al. Structure, function, and activator-induced conformations of the CRSP coactivator. Science 295, 1058–1062 (2002). [DOI] [PubMed] [Google Scholar]
- 86.Hu Z et al. A matter of access. nucleosome disassembly from gene promoters is the central goal transcriptional activators. Transcription 5, e29355 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cairns BR The logic of chromatin architecture and remodelling at promoters. Nature 461, 193–198 (2009). [DOI] [PubMed] [Google Scholar]
- 88.Jacobson RH et al. Structure and function of a human TAFII250 double bromodomain module. Science 288, 1422–1425 (2000). [DOI] [PubMed] [Google Scholar]
- 89.Vermeulen M et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131, 58–69 (2007). [DOI] [PubMed] [Google Scholar]
- 90.Lauberth SM et al. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell 152, 1021–1036 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Matangkasombut O et al. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev. 14, 951–962 (2000). [PMC free article] [PubMed] [Google Scholar]
- 92.Andrews FH et al. The essential role of acetyllysine binding by the YEATS domain in transcriptional regulation. Transcription 7, 14–20 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nagai S et al. Chromatin potentiates transcription. Proc. Natl Acad. Sci. USA 114, 1536–1541 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhu X et al. Histone modifications influence mediator interactions with chromatin. Nucleic Acids Res. 39, 8342–8354 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen X et al. Structures of +1 nucleosome-bound PIC–Mediator complex. Science 378, 62–68 (2022). [DOI] [PubMed] [Google Scholar]
- 96.Nock A et al. Mediator-regulated transcription through the +1 nucleosome. Mol. Cell 48, 837–848 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zaret KS Pioneer transcription factors initiating gene network changes. Annu. Rev. Genet 54, 367–385 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Workman JL & Roeder RG Binding of transcription factor TFIID to the major late promoter during in vitro nucleosome assembly potentiates subsequent initiation by RNA polymerase II. Cell 51, 613–622 (1987). [DOI] [PubMed] [Google Scholar]
- 99.Joo YJ et al. Downstream promoter interactions of TFIID TAFs facilitate transcription reinitiation. Genes Dev. 31, 2162–2174 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Teves SS et al. A stable mode of bookmarking by TBP recruits RNA polymerase II to mitotic chromosomes. eLife 7, e35621 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Murakami S, Nagari A & Kraus WL Dynamic assembly and activation of estrogen receptor α enhancers through coregulator switching. Genes Dev. 31, 1535–1548 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Blobel GA et al. Testing the super-enhancer concept. Nat. Rev. Genet 22, 749–755 (2021). [DOI] [PubMed] [Google Scholar]
- 103.Sabari BR, Dall’Agnese A & Young RA Biomolecular condensates in the nucleus. Trends Biochem. Sci 45, 961–977 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cho WK et al. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sabari BR et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shrinivas K et al. Enhancer features that drive formation of transcriptional condensates. Mol. Cell 75, 549–561.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zamudio AV et al. Mediator condensates localize signaling factors to key cell identity genes. Mol. Cell 76, 753–766.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Boehning M et al. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol 25, 833–840 (2018). [DOI] [PubMed] [Google Scholar]
- 109.Guo YE et al. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 572, 543–548 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Trojanowski J et al. Transcription activation is enhanced by multivalent interactions independent of phase separation. Mol. Cell 82, 1878–1893.e10 (2022). [DOI] [PubMed] [Google Scholar]
- 111.Li J et al. Single-molecule nanoscopy elucidates RNA polymerase II transcription at single genes in live cells. Cell 178, 491–506.e28. (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kagey MH et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Petrenko N et al. Mediator undergoes a compositional change during transcriptional activation. Mol. Cell 64, 443–454 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jeronimo C et al. Tail and kinase modules differently regulate core mediator recruitment and function in vivo. Mol. Cell 64, 455–466 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Malik S & Roeder RG Mediator: a drawbridge across the enhancer–promoter divide. Mol. Cell 64, 433–434 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jaeger MG et al. Selective Mediator dependence of cell-type-specifying transcription. Nat. Genet 52, 719–727 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; This comprehensive genetic analysis of the Mediator in human cells uses rapid degradation approaches to monitor how individual subunits impact various transcriptional parameters.
- 117.Rao SSP et al. Cohesin loss eliminates all loop domains. Cell 171, 305–320.e24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Karr JP et al. The transcription factor activity gradient (TAG) model: contemplating a contact-independent mechanism for enhancer–promoter communication. Genes Dev. 36, 7–16 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.McNally JG et al. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science 287, 1262–1265 (2000). [DOI] [PubMed] [Google Scholar]
- 120.Murakami K et al. Formation and fate of a complete 31-protein RNA polymerase II transcription preinitiation complex. J. Biol. Chem 288, 6325–6332 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Baek I et al. Single-molecule studies reveal branched pathways for activator-dependent assembly of RNA polymerase II pre-initiation complexes. Mol. Cell 81, 3576–3588.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study uses single-molecule microscopy to reveal a novel branched PIC assembly pathway in which TFIIE and TFIIF may preassemble with enhancer-bound activators prior to entry into the PIC.
- 122.Gorbea Colon JJ et al. Structural basis of a transcription pre-initiation complex on a divergent promoter. Mol. Cell 83, 574–588.e11 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rhee HS & Pugh BF Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483, 295–301 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Haberle V & Stark A Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell Biol 19, 621–637 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Plaschka C et al. Architecture of the RNA polymerase II–Mediator core initiation complex. Nature 518, 376–380 (2015). [DOI] [PubMed] [Google Scholar]
- 126.Schilbach S et al. Yeast PIC–Mediator structure with RNA polymerase II C-terminal domain. Proc. Natl Acad. Sci. USA 120, e2220542120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fishburn J et al. Double-stranded DNA translocase activity of transcription factor TFIIH and the mechanism of RNA polymerase II open complex formation. Proc. Natl Acad. Sci. USA 112, 3961–3966 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kim TK, Ebright RH & Reinberg D Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science 288, 1418–1422 (2000). [DOI] [PubMed] [Google Scholar]
- 129.Zhang Z et al. Rapid dynamics of general transcription factor TFIIB binding during preinitiation complex assembly revealed by single-molecule analysis. Genes Dev. 30, 2106–2118 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Baek HJ, Kang YK & Roeder RG Human mediator enhances basal transcription by facilitating recruitment of transcription factor IIB during preinitiation complex assembly. J. Biol. Chem 281, 15172–15181 (2006). [DOI] [PubMed] [Google Scholar]
- 131.Jishage M et al. Architecture of Pol II(G) and molecular mechanism of transcription regulation by Gdown1. Nat. Struct. Mol. Biol 25, 859–867 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Malik S, Molina H & Xue Z PIC activation through functional interplay between mediator and TFIIH. J. Mol. Biol 429, 48–63 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sogaard TM & Svejstrup JQ Hyperphosphorylation of the C-terminal repeat domain of RNA polymerase II facilitates dissociation of its complex with mediator. J. Biol. Chem 282, 14113–14120 (2007). [DOI] [PubMed] [Google Scholar]
- 134.Wong KH, Jin Y & Struhl K TFIIH phosphorylation of the Pol II CTD stimulates mediator dissociation from the preinitiation complex and promoter escape. Mol. Cell 54, 601–612 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fant CB et al. TFIID enables RNA polymerase II promoter-proximal pausing. Mol. Cell 78, 785–793.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Core L & Adelman K Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev. 33, 960–982 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yadav D et al. Multivalent role of human TFIID in recruiting elongation components at the promoter-proximal region for transcriptional control. Cell Rep. 26, 1303–1317. e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Takahashi H et al. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 146, 92–104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Takahashi H et al. MED26 regulates the transcription of snRNA genes through the recruitment of little elongation complex. Nat. Commun 6, 5941 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Galbraith MD et al. HIF1A employs CDK8-mediator to stimulate RNAPII elongation in response to hypoxia. Cell 153, 1327–1339 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sato S et al. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol. Cell 14, 685–691 (2004). [DOI] [PubMed] [Google Scholar]
- 142.Ranish JA, Yudkovsky N & Hahn S Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 13, 49–63 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sikorski TW & Buratowski S The basal initiation machinery: beyond the general transcription factors. Curr. Opin. Cell Biol 21, 344–351 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tantale K et al. A single-molecule view of transcription reveals convoys of RNA polymerases and multi-scale bursting. Nat. Commun 7, 12248 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ehrensberger AH, Kelly GP & Svejstrup JQ Mechanistic interpretation of promoter-proximal peaks and RNAPII density maps. Cell 154, 713–715 (2013). [DOI] [PubMed] [Google Scholar]; This work is a highly useful mathematical resource that allows assessment of the relative contributions of transcription initiation per se versus subsequent Pol II pausing to overall activity of a given gene.
- 146.Eychenne T et al. Functional interplay between Mediator and TFIIB in preinitiation complex assembly in relation to promoter architecture. Genes Dev. 30, 2119–2132 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Li YC et al. Structure and noncanonical Cdk8 activation mechanism within an Argonaute-containing Mediator kinase module. Sci. Adv 7, eabd4484 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Di Lello P et al. Structure of the Tfb1/p53 complex: insights into the interaction between the p62/Tfb1 subunit of TFIIH and the activation domain of p53. Mol. Cell 22, 731–740 (2006). [DOI] [PubMed] [Google Scholar]
- 149.Rochette-Egly C et al. Stimulation of RARα activation function AF-1 through binding to the general transcription factor TFIIH and phosphorylation by CDK7. Cell 90, 97–107 (1997). [DOI] [PubMed] [Google Scholar]
- 150.Lin YS et al. Binding of general transcription factor TFIIB to an acidic activating region. Nature 353, 569–571 (1991). [DOI] [PubMed] [Google Scholar]
- 151.Nakadai T et al. Two target gene activation pathways for orphan ERR nuclear receptors. Cell Res. 33, 165–183 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tyree CM et al. Identification of a minimal set of proteins that is sufficient for accurate initiation of transcription by RNA polymerase II. Genes Dev. 7, 1254–1265 (1993). [DOI] [PubMed] [Google Scholar]
- 153.Parvin JD & Sharp PA DNA topology and a minimal set of basal factors for transcription by RNA polymerase II. Cell 73, 533–540 (1993). [DOI] [PubMed] [Google Scholar]
- 154.Goodrich JA & Tjian R Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell 77, 145–156 (1994). [DOI] [PubMed] [Google Scholar]
- 155.Holstege FC et al. The requirement for the basal transcription factor IIE is determined by the helical stability of promoter DNA. EMBO J. 14, 810–819 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Dienemann C et al. Promoter distortion and opening in the RNA polymerase II cleft. Mol. Cell 73, 97–106.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 157.Kouzine F et al. Global regulation of promoter melting in naive lymphocytes. Cell 153, 988–999 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Tora L & Timmers HT The TATA box regulates TATA-binding protein (TBP) dynamics in vivo. Trends Biochem. Sci 35, 309–314 (2010). [DOI] [PubMed] [Google Scholar]
- 159.Wu SY & Chiang CM The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. J. Biol. Chem 282, 13141–13145 (2007). [DOI] [PubMed] [Google Scholar]
- 160.van Werven FJ et al. Cooperative action of NC2 and Mot1p to regulate TATA-binding protein function across the genome. Genes Dev. 22, 2359–2369 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hsu JY et al. TBP, Mot1, and NC2 establish a regulatory circuit that controls DPE-dependent versus TATA-dependent transcription. Genes Dev. 22, 2353–2358 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bhagwat AS et al. BET bromodomain inhibition releases the mediator complex from select cis-regulatory elements. Cell Rep. 15, 519–530 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sdelci S et al. Mapping the chemical chromatin reactivation landscape identifies BRD4–TAF1 cross-talk. Nat. Chem. Biol 12, 504–510 (2016). [DOI] [PubMed] [Google Scholar]
- 164.Donczew R & Hahn S BET family members Bdf1/2 modulate global transcription initiation and elongation in Saccharomyces cerevisiae. ELife 10, e69619 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]


