Abstract
Replication of viral RNA genomes requires the specific interaction between the replicase and the RNA template. Members of the Bromovirus and Cucumovirus genera have a tRNA-like structure at the 3′ end of their genomic RNAs that interacts with the replicase and is required for minus-strand synthesis. In Brome mosaic virus (BMV), a stem-loop structure named C (SLC) is present within the tRNA-like region and is required for replicase binding and initiation of RNA synthesis in vitro. We have prepared an enriched replicase fraction from tobacco plants infected with the Fny isolate of Cucumber mosaic virus (Fny-CMV) that will direct synthesis from exogenously added templates. Using this replicase, we demonstrate that the SLC-like structure in Fny-CMV plays a role similar to that of BMV SLC in interacting with the CMV replicase. While the majority of CMV isolates have SLC-like elements similar to that of Fny-CMV, a second group displays sequence or structural features that are distinct but nonetheless recognized by Fny-CMV replicase for RNA synthesis. Both motifs have a 5′CA3′ dinucleotide that is invariant in the CMV isolates examined, and mutational analysis indicates that these are critical for interaction with the replicase. In the context of the entire tRNA-like element, both CMV SLC-like motifs are recognized by the BMV replicase. However, neither motif can direct synthesis by the BMV replicase in the absence of other tRNA-like elements, indicating that other features of the CMV tRNA can induce promoter recognition by a heterologous replicase.
An essential feature of viral infection is the replication of the viral genome. Many of the plant and animal viruses have plus-strand RNA genomes that, following entry into cells, can readily be translated to provide the proteins required for their replication. In addition, the genomic RNA of these viruses directs the synthesis of a complementary minus strand, which serves as the template for synthesis of multiple copies of genomic and possible subgenomic plus-strand RNA (8). Brome mosaic virus (BMV) and Cucumber mosaic virus (CMV) are the type members of the Bromovirus and Cucumovirus genera, respectively, in the family Bromoviridae, which belongs to the alpha-like superfamily of viruses (19, 28). CMV and BMV have a similar genome organization, but CMV has a broader host range and a more severe economic impact (3, 29, 35). Both viruses have tripartite genomes, designated RNA1, RNA2, and RNA3. RNA1 encodes the putative protein with helicase and RNA-capping activities (5, 27), while RNA2 encodes the RNA-dependent RNA polymerase (RdRp). RNA3 is dicistronic and encodes the movement protein and the coat protein that is expressed via a subgenomic RNA. In Cucumoviruses but not Bromoviruses, an additional subgenomic RNA, 4a, that corresponds to the 3′-proximal portion of RNA2 has also been identified (13, 35). RNA4a encodes the 2b protein and may be involved in systemic infection, pathogenicity, and suppressing posttranscriptional gene silencing (7, 14, 15, 30).
Replication of the BMV and CMV genomes requires the replicase that is composed of a complex of virally encoded 1a and 2a proteins and unidentified cellular proteins. The term replicase is used to distinguish the protein complex from the subunit within the replicase that catalyzes the formation of phosphodiester bonds, the RdRp. This distinction is necessary due to the increasing number of publications that focus on recombinant RdRps (for example, see reference 53). Enriched BMV replicase preparations can accurately initiate minus-strand, plus-strand, and subgenomic RNA synthesis (2, 16, 21, 25, 31, 32, 45, 46). In vitro synthesis of CMV genomic and satellite RNAs has also been reported (23, 38, 51), but the viral sequences involved in replication have not been biochemically characterized.
The 3′ ends of BMV plus-strand RNAs have well-conserved sequences and structural features wherein the terminal 135 nucleotides (nt) can form tRNA-like structures (4, 18, 36). In vitro, the tRNA-like structure is necessary and sufficient to direct the initiation of minus-strand RNA synthesis (10, 31). A stem-loop named SLC within the BMV tRNA-like structure appears to be an addition to the canonical tRNA structure (18). BMV SLC is necessary for RNA synthesis and for interacting with the replicase (9, 17). When an 8-nt sequence containing the 3′ initiation site (CCA 3′) was attached to BMV SLC, the resultant RNA, SLC+8, was able to direct RNA synthesis (9). RNA synthesis requires the 3-nt loop (5′AUA3′) that was found to be essential for BMV RNA replication in barley protoplasts (9, 17, 26, 40). Recently a high-resolution structure for BMV SLC was determined by nuclear magnetic resonance spectroscopy (26). The structure shows a flexible stem with a large internal loop followed by a rigid stem leading to a terminal triloop with the sequence 5′AUA3′. The flexible internal loop allows efficient RNA synthesis, while the closing base pair in the stem causes the 5′-most adenylate in the triloop to be displaced from the loop by interacting with the 3′-most adenylate (26). The protruding 5′ adenylate has been hypothesized to provide a structure for replicase recognition (26). Mutating the protruding 5′ adenylate in the triloop to a guanylate proved fatal for BMV infection in plants (40) and for RNA synthesis in vitro (26). The middle nucleotide in the triloop was less important in vitro and in vivo (26, 40).
The 3′ ends of CMV plus-strand RNAs also form well-conserved tRNA-like structures (Fig. 1A) (11, 33, 41, 42, 43). An SLC-like structure (Fig. 1A) is present in the Fny isolate of CMV RNAs at approximately the same position relative to the SLC within the BMV 3′ end (nt 2137 to 2176) (11). BMV replicase can recognize and direct synthesis from a chimeric RNA containing the tRNA-like region of Fny-CMV in transfected protoplasts (39). A deletion of the CMV tRNA-like region abolishes RNA synthesis by the CMV replicase (6). Here we show that the SLC-like structure of Fny-CMV plays a role in minus-strand RNA synthesis in vitro. In addition, a second motif predicted to fold into a stem-pentaloop structure was found in several CMV isolates and can also direct RNA synthesis. Both RNA motifs require two invariant nucleotides, a cytidylate and an adenylate, for directing efficient minus-strand synthesis. The requirements for CMV core promoter recognition are compared with those for the BMV core promoter.
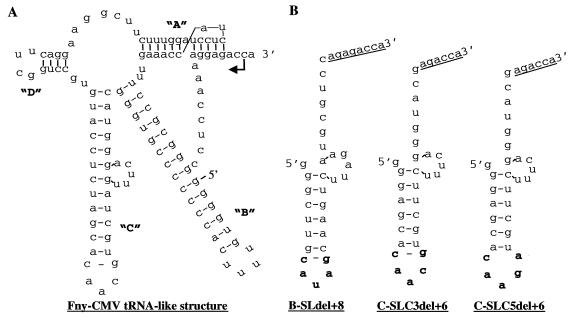
FIG. 1.
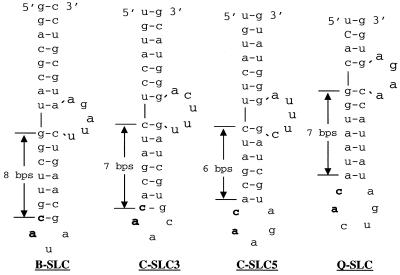
Predicted secondary structure of the Fny-CMV tRNA-like region and minimal SLC constructs. (A) Secondary structure predicted for the Fny-CMV tRNA-like region as depicted in Rizzo and Paulakaitis (42). Stem-loops are named A to D and indicated in bold letters. (B) Schematic of the predicted structure for B-SLdel+8, C-SLC3del+6, and C-SLC5del+6 using the mfold program (24). The nucleotides in the triloop and the C-G closing base pair of B-SLdel+8 and C-SLC3del+6 as well as the nucleotides contributing to the pentaloop of C-SLC5del+6 are indicated in bold.
MATERIALS AND METHODS
Preparation of enriched replicases.
Preparation of enriched Fny-CMV replicase was adapted from the protocol in Hayes and Buck (23). Briefly, young leaves of 7-week-old tobacco (Nicotiana tabacum cv. Xanthi, nc) plants were inoculated with Fny-CMV and harvested 4 days postinoculation. One hundred grams of the infected leaves was homogenized at 4°C in 100 ml of buffer C (50 mM Tris-HCl [pH 8.2], 15 mM MgCl2, 10 mM dithiothreitol [DTT], 0.1 mM phenylmethylsulfonyl fluoride [PMSF], 40% glycerol). The homogenate was centrifuged at 300 × g for 30 min to remove large plant pieces and cell debris. The supernatant was then centrifuged at 35,000 × g for 30 min to pellet the membrane-containing material. The resulting pellet was resuspended in 20 ml of solubilization buffer (50 mM Tris-HCl [pH 8.2], 15 mM MgCl2, 10 mM DTT, 0.1 mM PMSF, 0.75% Triton X-100, 0.5 M KCl, 40% glycerol) and stirred for 1 h at 4°C. It was then centrifuged at 100,000 × g for 1 h, and the resulting supernatant was loaded onto an S400 gel filtration column (4 by 90 cm). The fractions were tested for RNA synthesis using a standard replicase assay. BMV replicase was prepared from infected barley (Hordeum vulgare) leaves as described by Sun et al. (49). Cowpea chlorotic mottle virus (CCMV) replicase was prepared from infected cowpea (Vigna unguiculata) leaves as described by Adkins and Kao (1).
Synthesis and purification of RNAs.
DNAs corresponding to the 3′ ends of plus and minus strands of CMV RNA3 were generated by PCR amplification. Pairs of oligonucleotide primers were used, one of which contained a T7 promoter to enable transcription. For cDNA copies containing the subgenomic promoter region, oligonucleotide primers used were complementary to nt 960 to 1198 of Fny-CMV RNA3. Following transcription using T7 RNA polymerase, RNAs were separated by denaturing polyacrylamide gel electrophoresis (PAGE), and the band of correct molecular mass was excised with a razor. The gel was then crushed to elute RNA in a solution containing 0.3 M sodium acetate. RNA concentrations were determined by spectrophotometry and checked for quality by staining with toluidine blue following denaturing PAGE.
RNA synthesis assay.
Replicase activity assays were carried out as described by Adkins et al. (2). Briefly, each assay consisted of a 40-μl reaction containing 20 mM sodium glutamate (pH 8.2), 12 mM DTT, 4 mM MgCl2, 0.5% (vol/vol) Triton X-100, 2 mM MnCl2, 200 μM ATP, 500 μM GTP, 200 μM UTP, 61 nM [α-32P]CTP (400 Ci/mmol, 10 mCi/ml; Amersham Inc.), the desired amount of template, and 7 μl of RdRp. Following incubation for 90 min at 30°C, the reaction products were extracted with phenol-chloroform (1:1, vol/vol) and precipitated with ethanol (6:1, vol/vol), 10 μg of glycogen, and 0.4 M (final concentration) of ammonium acetate. Loading buffer (final concentrations 45% [vol/vol] deionized formamide, 1.5% [vol/vol] glycerol, 0.04% [wt/vol] bromophenol blue, and 0.04% [wt/vol] xylene cyanol) was added, and the products were denatured by heating at 90°C for 3 min prior to electrophoresis on 5, 12, or 20% acrylamide–7 M urea denaturing gels. Gels were dried and exposed to film at −80°C, and the amount of label incorporated into newly synthesized RNAs was quantified with a PhosphorImager (Molecular Dynamics). Synthesis from each experiment was normalized to the wild-type control value within each reaction.
RESULTS
Initiation of genomic and subgenomic RNA synthesis by CMV replicase in vitro.
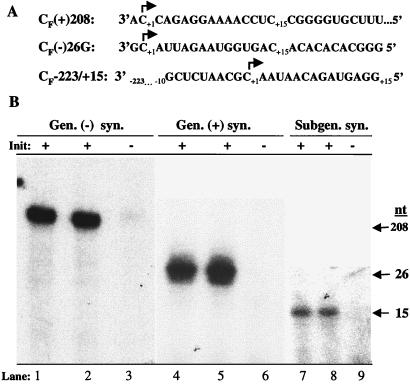
To determine the ability of the enriched Fny-CMV replicase preparation to synthesize RNA in vitro, RNAs corresponding to the genomic plus- and minus-strand 3′ ends of Fny-CMV RNA3 and subgenomic RNA were generated. For minus-strand synthesis, we made a 208-nt RNA, named CF(+)208, which contains the entire tRNA-like region of RNA3. Using the assay conditions described for BMV replicase (49), we observed that CF(+)208 was able to direct minus-strand synthesis efficiently (Fig. 2B, lanes 1 and 2). To determine whether initiation took place from the canonical CCA 3′ sequence of the CMV tRNA-like structure (normal initiation nucleotide is underlined), the cytidylates at positions +1 and +2 were both changed to guanylates. While the penultimate cytidylate is expected to be the initiation site (31), initiation of BMV minus-strand synthesis can take place from the +2 site if the penultimate cytidylate is changed (9). In CMV this mutation reduced the ability of the template to direct synthesis to near background levels (Fig. 2B, lane 3), indicating that initiation of minus-strand synthesis took place from the CCA initiation site.
FIG. 2.
Initiation of genomic plus- and minus-strand and subgenomic RNA synthesis by Fny-CMV replicase. (A) Templates used in the assay. CF(+)208 indicates a 208-nt RNA corresponding to the 3′ end of Fny-CMV RNA3. CF(−)26G indicates a 27-nt RNA with a 3′ nontemplated nucleotide that is complementary to the 5′ end of Fny-CMV RNA3. CF−223/+15 indicates part of the region complementary to the intercistronic region and the initiation cytidylate for subgenomic RNA synthesis. An arrow denotes the +1 initiation cytidylate in the three RNAs. (B) Autoradiograms of genomic and subgenomic RNA products made by the enriched Fny-CMV replicase preparation. Arrows denote the 208-nt, 26-nt, and 15-nt genomic minus-strand, genomic plus-strand, and subgenomic RNA products, respectively. RNAs with a wild-type initiation site are indicated by +, while those with the initiation site mutated to guanylate(s) are indicated by −. The genomic minus-strand, genomic plus-strand, and subgenomic RNA products were separated on 5, 12, and 20% PAGE, respectively.
To determine whether the enriched Fny-CMV replicase preparation could initiate genomic plus-strand synthesis, we generated a 27-nt RNA named CF(−)26G that corresponds to the 3′ end of the CMV RNA3 minus strand. A nontemplated nucleotide at the 3′ end is required for efficient BMV genomic plus-strand RNA synthesis (46). The CF(−)26G RNA also contained a nontemplated guanylate at the 3′ end and was able to efficiently direct genomic plus-strand synthesis (Fig. 2B, lanes 4 and 5). To determine whether initiation took place from the C at +1, it was mutated to a guanylate. The ability of the resultant RNA to direct genomic plus-strand RNA synthesis was reduced to less than 3% of the wild-type value (Fig. 2B, lane 6).
To determine if the enriched Fny-CMV replicase preparation could direct subgenomic RNA synthesis, a transcript named CF−223/+15, containing the 223-nt intercistronic region upstream of the +1C initiation site and a 15-nt template, was generated (complementary to nt 960 to 1198 of RNA3). In a separate study, it was demonstrated that the length of the sequence downstream of the +1 initiation site does not significantly affect subgenomic RNA synthesis (M.-H. Chen, M. Roossinck, and C. C. Kao, submitted for publication). CF−223/+15 was able to direct subgenomic RNA synthesis, as was revealed by the presence of the expected 15-nt product (Fig. 2B, lanes 7 and 8). A change of the +1C to a guanylate reduced synthesis of the 15-nt product to less than 3% (Fig. 2B, lane 9). The enriched preparation of Fny-CMV replicase was able to initiate RNA synthesis from all of the initiation sites used in CMV replication in vivo. Therefore, this Fny-CMV replicase preparation could be used to further elucidate the features in the RNA that are required for synthesis.
Recognition of the tRNA-like region.
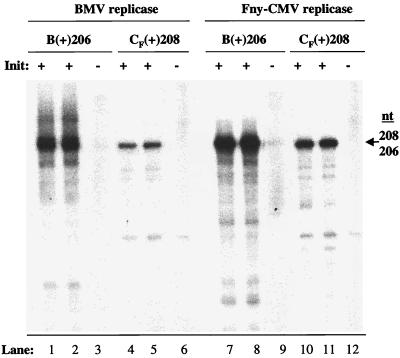
Initiation of minus-strand synthesis is the first step in replicating the RNA genome and is the subject of the remainder of this work. As there are extensive similarities between the predicted secondary structures of the BMV and CMV tRNA-like regions at the 3′ end of the genome, we sought to determine if the Fny-CMV replicase could recognize the tRNA-like region of BMV RNA3 and vice versa. Transcripts 206 or 208 nt in length and corresponding to the 3′ ends of Fny-CMV and BMV RNA3s were generated and assayed for their ability to direct minus-strand synthesis by the Fny-CMV and BMV replicases. Consistent with the in vivo results of Rao and Grantham (39), the BMV replicase was able to direct minus-strand RNA synthesis from the Fny-CMV RNA3 tRNA-like sequence [CF(+)208] as well as from its own [B(+)206] (Fig. 3, lanes 1 and 2 and 4 and 5). The Fny-CMV replicase could also use the BMV tRNA-like region to direct RNA synthesis (Fig. 3, lanes 7 and 8). All of the RNAs synthesized initiated from the canonical CCA3′ initiation site, since replacing guanylates at the +1 and +2 positions reduced synthesis to less than 3% (Fig. 3, lanes 3, 6, 9, and 12). These results show that the tRNA-like regions of genomic BMV and Fny-CMV RNAs have common motifs for initiation of minus-strand RNA synthesis.
FIG. 3.
Recognition of the tRNA-like region by BMV and Fny-CMV replicases. Autoradiogram of the 206- and 208-nt RNA products synthesized by the BMV and CMV replicases from the tRNA-like regions of BMV and CMV. Initiation-competent RNAs are indicated by +. Initiation-incompetent RNAs are indicated by −.
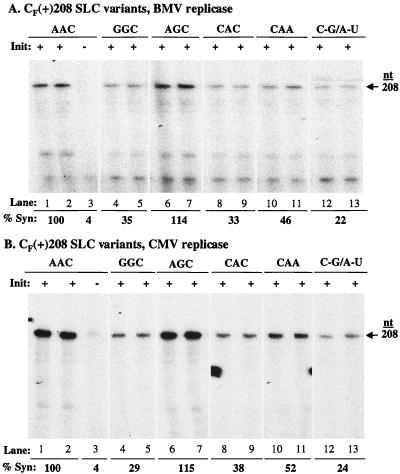
The Fny-CMV SLC-like structure, henceforth referred to as C-SLC3, is predicted to have a C-G closing base pair and a triloop of the sequence 5′AAC3′ (Fig. 1A). To determine the effect of mutations in C-SLC3 on RNA synthesis in the context of the entire tRNA-like structure, we generated 208-nt transcripts named CF(+)208 and variants thereof that contained the 3′ end of Fny-CMV RNA. Changing both the 5′ adenylates in the triloop to guanylates reduced synthesis to 35 and 29% by the BMV and Fny-CMV replicases, respectively (Fig. 4A, lanes 4 and 5, and Fig. 4B, lanes 4 and 5). Changing only the middle adenylate to a guanylate increased synthesis to 115% by both RNA replicases compared to the respective wild-type controls (Fig. 4A, lanes 6 and 7, and Fig. 4B, lanes 6 and 7). This demonstrates that the 5′-most adenylate in C-SLC3 triloop is important for directing efficient synthesis.
FIG. 4.
Effect of nucleotide substitutions within C-SLC3 in the context of the tRNA-like structure. (A) Autoradiogram of the RNA products made by BMV replicase. (B) Autoradiogram of the RNA products made by Fny-CMV replicase. The arrow identifies the 208-nt RNA product. Initiation-competent RNAs (+) initiation-incompetent RNAs (−) are indicated. The wild-type and mutated sequences of C-SLC3 are shown near the top of the autoradiogram. The effect of nucleotide substitutions on RNA synthesis is denoted as a percentage relative to the amount of synthesis directed by the wild-type sequence, indicated as AAC. All results presented are from at least three independent trials with a standard deviation of ≤12%.
To extend this observation, we replaced the 5′-most adenylate with a cytidylate and found that RNA synthesis was reduced to 33 and 38% by BMV and Fny-CMV replicases, respectively (Fig. 4A, lanes 8 and 9, and Fig. 4B, lanes 8 and 9). A triloop of 5′CAA3′ directed 46 and 52% synthesis by the BMV and Fny-CMV replicases, respectively (Fig. 4A, lanes 10 and 11, and Fig. 4B, lanes 10 and 11). This indicates that the 3′ cytidylate of the triloop may not be as critical as the 5′ adenylate in directing RNA synthesis (compare Fig. 4A and B, lanes 8 and 9 and 10 and 11). To determine the effect of the C-G closing base pair in C-SLC3 on the ability to direct RNA synthesis, the C-G was mutated to an A-U. This substitution reduced RNA synthesis to less than 25% by the BMV and Fny-CMV replicases (Fig. 4A, lanes 12 and 13, and Fig. 4B, lanes 12 and 13). These results indicate that the 5′ adenylate in the triloop and either the formation of the closing base pair or the nucleotides comprising the base pair are important for efficient minus-strand RNA synthesis in vitro in the context of the entire tRNA-like structure.
Interaction of BMV and CMV replicases with minimal SLC.
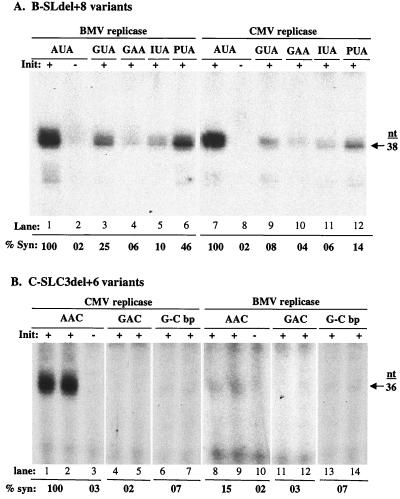
We wanted to determine if SLC is necessary and sufficient to direct RNA synthesis in the absence of the other tRNA-like elements. BMV SLdel+8, henceforth referred to as B-SLdel+8 (Fig. 1B), and derivatives had previously been characterized for the ability to direct efficient RNA synthesis by the BMV replicase (26) (Fig. 5A, lane 1). B-SLdel+8 efficiently directed synthesis by the Fny-CMV replicase (Fig. 5A, lane 7). Initiation of synthesis takes place from the canonical CCA3′ initiation site, as evidenced when a change of the +1 and +2 cytidylates to guanylates reduced synthesis to near background levels (Fig. 5A, lanes 2 and 8). Changing the 5′-most adenylate of the triloop from 5′AUA3′ to 5′GUA3′ reduced synthesis to 25 and 8% by the BMV and Fny-CMV replicases, respectively (Fig. 5A, lanes 3 and 9). Changing the triloop to 5′GAA3′ reduced synthesis to less than 6% by both replicases (Fig. 5A, lanes 4 and 10). Removal of the C6 amino moiety from the 5′ adenylate by substitution with an inosine reduced synthesis to less than 10% by both replicases (Fig. 5A, lanes 5 and 11). Consistent with the previous report of Kim et al. (26), removal of all the exocyclic groups from adenine by a substitution with a purine directed 46% synthesis by the BMV replicase, higher than that observed with guanine or inosine at this position (Fig. 5A, lanes 3 to 6). Kim et al. (26) postulated that a C6 keto group at this position could produce electrostatic repulsion of the adenine from the loop, whereas a purine without any exocyclic group would be less disruptive of these bonding interactions. A purine at the 5′-most position in the triloop did not restore RNA synthesis by the Fny-CMV replicase nearly as much (Fig. 5A, lane 12), suggesting that the two replicases have altered requirements for the exocyclic groups of adenine. Similar assays were also carried out with B-SLdel+8 and CCMV replicase, another member of the Bromovirus genus, and the results were identical to those with the BMV replicase (K. Sivakumaran and C. C. Kao, unpublished data). Taken together, these results clearly show that the 5′-most adenylate in the BMV SLC triloop (B-SLC) plays a critical role in directing efficient RNA synthesis. These results are consistent with BMV replication requirements in vivo (40) and with the idea that the BMV replicase can replicate a chimeric RNA containing the tRNA-like sequence of CMV RNA3 in transfected protoplasts (39).
FIG. 5.
Effect of nucleotide substitutions within the minimal SLC of BMV and Fny-CMV on RNA synthesis. (A) Autoradiogram of RNA products made by BMV and Fny-CMV replicases. The arrow identifies a 38-nt RNA replicase product. The different nucleotide substitutions within the triloop and the closing base pair are shown near the top of the autoradiogram. I, inosine; P, purine lacking any exocyclic groups (26). The effect of nucleotide substitutions on RNA synthesis is shown as a percentage relative to the amount of synthesis directed by B-SLdel+8. (B) Autoradiogram of RNA products made by Fny-CMV and BMV replicases. The arrow identifies a 36-nt RNA replicase product. The effect of nucleotide substitutions on RNA synthesis is shown as a percentage relative to the amount of synthesis directed by C-SLC3del+6. All results presented are from at least three independent trials with a standard deviation of <12%.
Next we wanted to determine if C-SLC3 is recognized by the BMV and Fny-CMV replicases in a similar manner. C-SLC3del+6 is a derivative of C-SLC3 that lacks 7 nt at the 5′ end and has the addition of a 6-nt sequence containing the initiation site (CCA3′) attached to the 3′ end (Fig. 1B). C-SLC3del+6 is able to direct efficient synthesis by the Fny-CMV replicase, resulting in a 36-nt product (Fig. 5B, lanes 1 and 2). Initiation of synthesis takes place from the canonical CCA3′ initiation site, since mutating the +1 and +2 cytidylates to guanylates reduced synthesis to near background levels (Fig. 5B, lane 3). Changing the triloop from 5′AAC3′ to 5′GAC3′ resulted in 2% synthesis (Fig. 5B, lanes 4 and 5). Changing the closing C-G base pair to G-C reduced synthesis to 7% (Fig. 5B, lanes 6 and 7), indicating that the 5′-most adenylate and the closing base pair are critical for synthesis. Unlike B-SLdel+8, C-SLC3del+6 and its derivatives were not efficiently recognized by BMV replicase (Fig. 5B, lanes 8 to 14), indicating that BMV replicase may require additional motifs that are lacking in C-SLC3del+6.
SLC-like sequence in CMV isolates within subgroup I.
Unlike BMV, of which only a limited number of isolates have been identified, many CMV isolates have been identified and sequenced. Based on serological evidence and nucleic acid hybridization, these isolates have been categorized into two subgroups, designated I and II (34). Phylogenetic analysis of the 5′ untranslated region as well as the coat protein open reading frame of CMV isolates suggests that subgroup I could be divided further into subgroups IA and IB (44). A compilation of the sequence encompassing the SLC-like region of 30 different subgroup I isolates revealed two general patterns (Fig. 6A and Table 1). Twenty-four of the 30 isolates examined have well-conserved sequences that could form a stem-loop structure identical to that of Fny (Fig. 6A and B), with the predominant triloop and closing base pair being identical to that of Fny. The 5′-most adenylate in the triloop and the C-G closing base pair are invariant in all CMV group I isolates (Fig. 6A). Two isolates have a cytidylate as the middle nucleotide of the triloop, while most have an adenylate. The 3′ nucleotide in the triloop showed more variability, with isolates having a cytidylate, a uridylate, or a guanylate at this position (Fig. 6A). In addition, a 4- to 5-nt pyrimidine-rich internal bulge is present within the SLC-like structure in all the isolates examined (Fig. 1A and 6A).
FIG. 6.
Alignment of the sequences containing the SLC-like region of 30 CMV strains, predicted stem-loop structures of C-SLC3 and C-SLC5, and nondenaturing and denaturing gel analyses of C-SLC3 and C-SLC5. (A) SLC-like region of 1A and 1B isolates of 30 CMV isolates. Sequence data were obtained from the GenBank accessions in Table 1. The different isolates analyzed are indicated on the left side of the figure. Isolates having a stem region followed by a triloop are shown in the C-SLC3-like group. Isolates having a stem region followed by a pentaloop are shown in the C-SLC5-like group. The invariant CA dinucleotide is shown in bold. The sequences of the putative internal bulge are shown under the bracket. (B) Schematic depicting the lower portion of the predicted structure of C-SLC3 and C-SLC5. The invariant CA dinucleotide is indicated in bold. The predicted structure was generated using the mfold program (24). (C) Nondenaturing and denaturing gel analyses of C-SLC3 and C-SLC5 RNAs. The RNAs contained a 17-nt region corresponding to the lower stem-loop region of C-SLC3 and C-SLC5. The RNAs for nondenaturing gel analysis were heated to 90°C for 2 min, cooled on ice, and electrophoresed through a nondenaturing 20% polyacrylamide gel. The RNAs are SL13, a 13-nt stem with a triloop (lane 1), C-SLC3 (lane 2), and C-SLC5 (lane 3). For denaturing gel analysis, the RNAs were analyzed on a denaturing 20% polyacrylamide gel. The RNAs are C-SLC3 (lane 4) and C-SLC5 (lane 5).
TABLE 1.
Strains
| Strain | GenBank accession no. or reference
|
||
|---|---|---|---|
| RNA1 | RNA2 | RNA3 | |
| Fny | D00356 | D00355 | D10538 |
| Sny | Unpublished | U66094 | |
| Y | D12537 | U66287 | AF103993 |
| Leg | D16403 | D16406 | D16405 |
| Ix | U20220 | U20218 | U20219 |
| Nt9 | D28778 | D28779 | D28780 |
| Kor | U66287 | L36251 | |
| Sd | D86330 | AB008777 | |
| O | Hayakawa et al. (22) | D10209 | D00385 |
| B | U59740 | ||
| K | S2187 | ||
| As | AF033667 | AF01329 | |
| E5 | D42080 | ||
| M8 | D49496 | ||
| E | AB042294 | ||
| Ft | D28487 | ||
| Cs | D42080 | ||
| N | Y16926 | ||
| Km | AB042294 | ||
| C | D83958 | ||
| M | D10539 | ||
| Pepo | D43800 | ||
| D8 | AB004781 | ||
An SLC motif different than the one in Fny-CMV was observed in six isolates. Based on the mfold program (24), the predicted structure for these isolates is a stem region with an internal bulge followed by a rigid stem with a 5′CAAGA3′ pentaloop (Fig. 6B). This motif is invariant in six isolates (Fig. 6A). In addition, these isolates have a pyrimidine-rich bulge (Fig. 6A). To distinguish this stem-pentaloop structure from the stem-triloop (C-SLC3), this second motif will henceforth be referred to as C-SLC5.
To determine if C-SLC5 has a distinct conformation compared to C-SLC3, 17-nt transcripts corresponding to the lower stem-loop region of Fny and Ix isolates were made for carrying out nondenaturing and denaturing gel analysis. The RNAs from Fny and Ix RNAs comigrated in a denaturing gel (Fig. 6C, lanes 4 and 5), as expected. However, in a nondenaturing gel, the Ix RNA migrates more slowly than the RNA of Fny (Fig. 6C, lanes 2 and 3), suggesting that the two RNAs exist in different RNA conformations. It should be noted that some isolates of both subgroups 1A and 1B have the C-SLC5 structure. Furthermore, some CMV strains, such as NT9, contain both motifs in different RNAs.
Recognition of the C-SLC5 structure by Fny-CMV replicase.
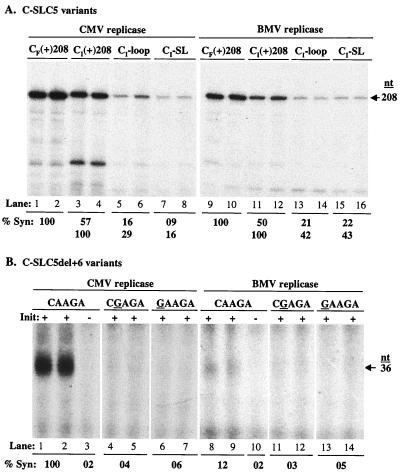
To determine if the Fny-CMV and BMV replicases recognize the C-SLC5 structure, we made a 208-nt transcript termed CI(+)208 that corresponds to the 3′ end of the Ixora isolate (Ix in Fig. 6) and carried out standard replicase assays. CI(+)208 RNA is able to direct synthesis by the Fny-CMV replicase (Fig. 7A, lanes 3 and 4), but at 57% of the level of CF(+)208. CI(+)208 RNA is also recognized by BMV replicase to direct synthesis at approximately 50% compared to CF(+)208 (Fig. 7A, lanes 9 and 10 and 11 and 12). To determine if the SLC-like loop of CI(+)208 plays a role in RNA synthesis, we mutated the sequence containing the pentaloop from 5′CAAGAUG3′ to 5′CGGGAUG3′ and to 5′AAAGAUU3′ (mutated nucleotides are underlined). These changes reduced synthesis to 29 and 16% compared to CI(+)208 (Fig. 7A, lanes 5 to 8). We observed similar results in the presence of BMV replicase, where changing the stem-loop sequence from 5′CAAGAUG3′ to 5′CGGAUG3′ or 5′AAAGAUU3′ reduced synthesis to 42 and 43%, respectively (Fig. 7A, lanes 13 and 14 and 15 and 16). Taken together, the results indicate that although the loop sequence is changed relative to that in B-SLC and C-SLC3, C-SLC5 is still recognized by BMV and Fny-CMV replicases through the nucleotides within the loop.
FIG. 7.
Effect of nucleotide substitutions within C-SLC5 on RNA synthesis. (A) Autoradiogram showing the RNA products made by Fny-CMV and BMV replicases. CF(+)208 indicates a 208-nt RNA corresponding to the 3′ end of Fny-CMV RNA3. CI(+)208 indicates a 208-nt RNA corresponding to the 3′ end of Ix-CMV RNA3. CI-loop indicates a 208-nt RNA with the 5′CAAGA3′ sequence in the pentaloop mutated to 5′CGGGA3′. CI-SL indicates a 208-nt RNA with the 5′CAAGAUG3′ stem-loop sequence mutated to 5′AAAGUAU3′. Quantification of the effect of nucleotide substitutions on RNA synthesis is shown as the percentage of three independent assays relative to the amount of synthesis directed by CF(+)208 as well as CI(+)208. (B) Autoradiogram showing the effect of nucleotide substitutions within C-SLC5del+6 on RNA synthesis. The nucleotide substitutions are indicated at the top of the autoradiogram. The effect of nucleotide substitutions on RNA synthesis is shown as a percentage relative to the amount of synthesis directed by C-SLC5del+6. All results presented are from at least three independent trials with a standard deviation of <12%.
Next we wanted to determine if the C-SLC5 elements are sufficient to initiate RNA synthesis in the absence of other tRNA-like elements. Transcripts analogous to C-SLC3del+6, termed C-SLC5del+6, which lacked a 5′ 7-nt region and had a 6-nt sequence containing the initiation site (CCA3′) attached to the 3′ end (Fig. 1B), were generated and tested. C-SLC5del+6 is efficiently recognized by Fny-CMV replicase, resulting in a 36-nt product (Fig. 7B, lanes 1 and 2). Initiation of synthesis takes place from the canonical CCA3′ initiation site, since mutating the +1 and +2 cytidylates to guanylates reduced synthesis to 3% (Fig. 7B, lane 3). To examine the role of the conserved CA dinucleotide, the conserved A was changed to a guanylate, and the resultant RNA directed synthesis at 4% relative to the wild-type control (Fig. 7B, lanes 4 and 5). Changing the conserved C to a guanylate reduced synthesis to 6% (Fig. 7B, lanes 6 and 7). The conserved CA dinucleotide is required even when the structure of the loop is altered. Like C-SLC3del+6, C-SLC5del+6 and its derivatives are not efficiently recognized by BMV replicase (Fig. 7B, lanes 8 to 14), indicating that BMV replicase may require additional motifs that are missing in both RNAs.
DISCUSSION
The interactions of the BMV and CMV replicases with their homologous and heterologous core promoters for minus-strand RNA initiation were compared. A CMV replicase enriched from Fny-CMV-infected tobacco was demonstrated to initiate RNA synthesis correctly from all three classes of CMV promoters. The initiation of Fny-CMV minus-strand RNA required nucleotides within the equivalent of the BMV SLC structure. The Fny-CMV C-SLC3 resembles B-SLC in that both have a closing C-G base pair and a 5′-most adenylate in the triloop. RNA synthesis from C-SLC3 depends on the 5′-most adenylate of the CMV SLC triloop and either the C-G closing base pair or the nucleotides of this base pair. In addition to the C-SLC3 motif, alignment of the sequences of 30 CMV group I isolates revealed the existence of an alternative SLC motif with a putative 5-nt loop (C-SLC5). Both C-SLC3 and C-SLC5 were recognized by the Fny-CMV replicase in the context of the 208-nt tRNA-like structure and in minimal versions of C-SLC3 and C-SLC5. Recognition required the highly conserved cytidylate (formerly in the C-G closing base pair) and adenylate in the loop. The BMV replicase was also able to synthesize RNA from both CMV SLC motifs in the context of the whole tRNA-like structure, and it required the same CA dinucleotide. However, the BMV replicase could not synthesize RNA from the minimal CMV RNA-derived SLC motifs. These studies reveal commonalties and differences in the mechanism of minus-strand RNA synthesis by two closely related plus-strand RNA viruses.
Core promoter recognition by the BMV and CMV replicases.
The ability of the CMV tRNA-like element to direct RNA synthesis by the BMV replicase is expected. Rao and Grantham (39) demonstrated that a chimeric BMV RNA3 that contains the 3′ 300-nt RNA of Fny-CMV could replicate in barley protoplasts cotransfected with BMV RNA1 and RNA2. As an extension of the in vivo results, we observed that the Fny-CMV replicase could synthesize RNA from the BMV tRNA-like structure (Fig. 3) using the nucleotides within B-SLC that were important for recognition by the BMV replicase (Fig. 4A and B). These results demonstrate that the same stem-loop within the respective BMV and CMV tRNA-like regions comprises the minimal core promoter for minus-strand RNA synthesis.
While much similarity exists in minus-strand RNA synthesis by the BMV and CMV replicases, our results suggest that different features presented by the SLC are recognized by the two replicases. First, substitution at the 5′ position of the loop with a purine lacking exocyclic moieties restored RNA synthesis to 46% of that with B-SLdel+8 by the BMV replicase, while synthesis was only restored to 14% with the CMV replicase. This indicates that a different pattern of bond interactions takes place between the respective replicases and the conserved adenine in the loop. Second, the 5′-most adenylate recognized by both replicases likely exists in different conformations in B-SLC and C-SLC. For B-SLC, the 5′-most adenylate is displaced into the solution due to the purine-purine stacking of the 3′ adenine and the guanine of the C-G closing base pair (26). A network of H-bonds stabilizes the displaced adenylate (26). In C-SLC3, a 5′AAC3′ triloop should have weakened stacking interaction with the closing base pair. Furthermore, C-SLC5 should be in a different conformation in comparison to B-SLC since the closing C-G base pair is absent altogether and both of the conserved CA dinucleotides are in the loop (Fig. 8). It is possible that CMV replicase recognizes the conserved CA dinucleotide in a base-specific manner, while the BMV replicase normally requires the proper presentation of the 5′-most adenine.
FIG. 8.
Schematic illustrating the common features present in SLC of BMV and CMV subgroup I and II RNAs. The RNAs have a short stem region with an internal bulge, followed by a rigid stem-loop region. The conserved CA dinucleotide in CMV subgroups I and II is indicated in bold. B-SLC, C-SLC3, C-SLC5, and Q-SLC were generated using the mfold program (24).
The BMV replicase was able to use the tRNA-like structures containing C-SLC3 and C-SLC5 for RNA synthesis, but it could not use just the minimal SLC elements. This suggests that features other than SLC within the tRNA-like structure can induce the recognition of a nonhomologous SLC element. Also, for both the BMV and CMV replicases, the presence of the entire tRNA-like structure decreased the effects of the mutations observed in the minimal SLC element. There are several precedents for upstream elements contributing to the recognition of the core promoter in transcription in RNA viruses, including results from BMV. Ahlquist and colleagues demonstrated that the BMV replicase assembles in the intercistronic sequence of RNA3 (37, 47), and Chapman et al. (10) reported that the BMV RNA sequence 5′ of the tRNA-like sequence can partially suppress a mutation in the initiation site. The ability of the minimal BMV SLC to interact with the BMV replicase was reduced fourfold relative to the entire tRNA-like sequence (9). Hence, the additional sequences in the tRNA-like structure are expected to influence RNA synthesis, possibly by stabilizing replicase binding.
Recognition of C-SLC5.
Since C-SLC3 and C-SLC5 have different conformations based on nondenaturing gel electrophoresis, and since both are recognized by the CMV replicase, replicase-promoter recognition likely takes place by an induced-fit mechanism whereby the replicase and/or the RNA undergoes conformational changes after the initial interaction (20). Interaction of the BMV replicase with the CMV core elements in the CMV tRNA-like structure also likely takes place by an induced-fit mechanism. Results consistent with an induced-fit hypothesis were reported for the subgenomic promoter of BMV and CCMV (1, 47), and also for the CMV subgenomic promoter (Chen et al., submitted).
Specificity in CMV minus-strand RNA synthesis.
Despite the ability of the replicase to adjust to different features in the RNA, we emphasize that highly specific signals are likely required. Such signals are also found in the SLC-like structure of a group II CMV strain, even though the SLC-like loop of the Q-strain is predicted to have six nucleotides (Fig. 8). CMV satellite RNAs are replicated efficiently by the CMV replicase (50). An initial examination of the portion of D satellite RNA previously claimed to mimic the tRNA-like structure (50) did not reveal obvious motifs in common with the minimal SLC-like structures we found to be functional in vitro (Sivakumaran and Kao, unpublished). Experimental examination of the minimal satellite RNA signals required for RNA synthesis by the CMV replicase in vitro is under way.
It is quite possible that the CMV replicase can recognize more diverse SLC-like structures in vivo, in the context of other cis-acting elements, than what has been observed in vitro. Although in vitro studies showed a dramatic difference in the generation of RNA products from minimal plus-stranded promoters (46), preliminary results from protoplast transfections showed that CMV and BMV replicases could recognize heterologous genomic plus-strand RNA promoters (Y. Bao and M. J. Roossinck, unpublished results). It is possible that the RNA templates in vivo could associate with host factors such as RNA chaperones that affect RNA structure. The ability of CMV replicase to recognize CF(+)208 variants in vivo is being tested.
The dependence on specific signals may vary in different RNA viruses (12, 52). BMV replicase has more stringent recognition requirements than the CMV replicase. For minus-strand RNA synthesis, the CMV replicase recognized CSLC3del+6 and CSLC5del+6, while the BMV replicase did not. Initiation of the subgenomic RNAs of CMV but not BMV can take place from RNAs with different secondary structure (Chen et al., submitted), suggesting that the CMV replicase can recognize a number of RNA structures. The observation that sat RNAs are associated with infection by CMV but not BMV may be due to this reduced specificity. In turnip yellow mosaic virus, specific recognition signals may be even more relaxed in comparison to the CMV replicase, possibly due to a strong cis preference for the RNA to be replicated. An initiation site composed of 3′ACCA is apparently a key recognition element for initiation near the 3′ end by the turnip yellow mosaic virus and the Qb replicases, while other RNA structures and sequences play a more minor role in vitro (12, 13). Whether additional requirements for promoter recognition are necessary in vivo needs to be determined.
ACKNOWLEDGMENTS
Helpful discussions and encouragement from IU cereal killers and editing by L. Kao are much appreciated.
The Kao lab is supported by the NSF (MCB9507344) and USDA (9702126) and a fellowship from the Samuel Robert Noble Foundation. Y.B. and M.J.R. are supported by funding from the Noble Foundation. C.C.K. acknowledges Linda and Jack Gill Fellowship.
REFERENCES
- 1.Adkins S, Kao C C. Subgenomic RNA promoters dictate the mode of recognition by bromoviral RNA-dependent RNA polymerase. Virology. 1998;252:1–8. doi: 10.1006/viro.1998.9449. [DOI] [PubMed] [Google Scholar]
- 2.Adkins S, Siegel R, Sun J H, Kao C C. Minimal templates directing accurate initiation of subgenomic RNA synthesis in vitro by the brome mosaic virus RNA-dependent RNA polymerase. RNA. 1997;3:634–647. [PMC free article] [PubMed] [Google Scholar]
- 3.Ahlquist P. Bromovirus RNA replication and transcription. Curr Opin Genet Dev. 1992;2:71–76. doi: 10.1016/s0959-437x(05)80325-9. [DOI] [PubMed] [Google Scholar]
- 4.Ahlquist P, Dasgupta R, Kaesberg P. Near identity of 3′ RNA secondary structure in bromo-viruses and cucumber mosaic virus. Cell. 1981;23:183–189. doi: 10.1016/0092-8674(81)90283-x. [DOI] [PubMed] [Google Scholar]
- 5.Ahola T, Ahlquist P. Putative RNA capping activities encoded by brome mosaic virus: methylation and covalent binding of guanylate by replicase protein 1a. J Virol. 1999;73:10061–10069. doi: 10.1128/jvi.73.12.10061-10069.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boccard F, Baulcombe D. Mutational analysis of cis-acting sequences and gene function in RNA3 of cucumber mosaic virus. Virology. 1993;193:563–578. doi: 10.1006/viro.1993.1165. [DOI] [PubMed] [Google Scholar]
- 7.Brigneti G, Voinnet O, Li L H, Ding S W, Baulcombe D C. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 1998;17:6739–6746. doi: 10.1093/emboj/17.22.6739. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Buck K W. Comparison of the replication of positive-strand RNA viruses of plants and animals. Adv Virus Res. 1996;47:159–251. doi: 10.1016/S0065-3527(08)60736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapman M, Kao C C. A minimal RNA promoter for minus-strand RNA synthesis by the brome mosaic virus polymerase complex. J Mol Biol. 1999;286:709–720. doi: 10.1006/jmbi.1998.2503. [DOI] [PubMed] [Google Scholar]
- 10.Chapman M, Rao A L N, Kao C C. Sequences 5′ of the conserved tRNA-like promoter modulate initiation of minus-strand synthesis by the brome mosaic virus RNA-dependent RNA polymerase. Virology. 1998;252:458–467. doi: 10.1006/viro.1998.9473. [DOI] [PubMed] [Google Scholar]
- 11.Davis C, Symons R H. Further implications for the evolutionary relationships between tripartite plant viruses based on cucumber mosaic virus RNA3. Virology. 1988;165:216–224. doi: 10.1016/0042-6822(88)90675-7. [DOI] [PubMed] [Google Scholar]
- 12.Deiman B A L M, Verlaan P W G, Pleij C W A. In vitro transcription by the turnip yellow mosaic virus RNA polymerase: a comparison with the alfalfa mosaic virus and brome mosaic virus replicases. J Virol. 2000;74:264–271. doi: 10.1128/jvi.74.1.264-271.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding S W, Anderson B J, Haase H R, Symons R H. New overlapping gene encoded by the cucumber mosaic virus genome. Virology. 1994;198:5762–5772. doi: 10.1006/viro.1994.1071. [DOI] [PubMed] [Google Scholar]
- 14.Ding S W, Li W X, Symons R H. A novel naturally occurring hybrid gene encoded by a plant virus facilitates long distance virus movement. EMBO J. 1995;14:5762–5772. doi: 10.1002/j.1460-2075.1995.tb00265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding S W, Shi B, Li W X, Symons R H. An interspecies hybrid is significantly more virulent than either parental virus. Proc Natl Acad Sci USA. 1996;93:7470–7474. doi: 10.1073/pnas.93.15.7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dreher T W, Hall T C. Mutational analysis of the sequence and structural requirements in brome mosaic virus RNA for minus-strand promoter activity. J Mol Biol. 1988;201:31–40. doi: 10.1016/0022-2836(88)90436-6. [DOI] [PubMed] [Google Scholar]
- 17.Dreher T W, Bujarski J J, Hall T C. Mutant viral RNAs synthesized in vitro show altered aminoacylation and replicase template activities. Nature. 1984;311:171–175. doi: 10.1038/311171a0. [DOI] [PubMed] [Google Scholar]
- 18.Felden B, Florentz C, Giege R, Westhof E. Solution structure of the 3′-end of brome mosaic virus genomic RNAs: conformational mimicry with canonical tRNAs. J Mol Biol. 1994;234:508–531. doi: 10.1006/jmbi.1994.1010. [DOI] [PubMed] [Google Scholar]
- 19.Goldbach R. Genome similarities between plant and animal RNA viruses. Microbiol Sci. 1987;4:197–202. [PubMed] [Google Scholar]
- 20.Gubser C C, Variani G. Structure of the polyadenylation regulatory element of the human U1A pre-mRNA 3′-untranslated region and interaction with the U1A protein. Biochemistry. 1996;35:2253–2267. doi: 10.1021/bi952319f. [DOI] [PubMed] [Google Scholar]
- 21.Hardy S F, German T L, Loesch-Fries L S, Hall T C. Highly active template-specific RNA-dependent RNA polymerase from barley leaves infected with brome mosaic virus. Proc Natl Acad Sci USA. 1979;76:4956–4960. doi: 10.1073/pnas.76.10.4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayakawa T, Mizukami M, Nakamura I, Suzuki M. Cloning and sequencing of RNA-1 cDNA from cucumber mosaic virus strain O. Gene. 1989;85:533–540. doi: 10.1016/0378-1119(89)90448-4. [DOI] [PubMed] [Google Scholar]
- 23.Hayes R J, Buck K W. Complete replication of a eukaryotic virus RNA in vitro by a purified RNA-dependent RNA polymerase. Cell. 1990;63:363–368. doi: 10.1016/0092-8674(90)90169-f. [DOI] [PubMed] [Google Scholar]
- 24.Jaeger J A, Turner D H, Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kao C, Sun J. The RNA-dependent RNA polymerase of a (+)-strand RNA virus uses oligoribonucleotide primers to initiate (−)-strand RNA synthesis. J Virol. 1996;70:6826–6830. doi: 10.1128/jvi.70.10.6826-6830.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim C-H, Kao C C, Tinoco I. RNA structural elements that determine specificity in the interaction between a viral RNA promoter and viral replicase. Nat Struct Biol. 2000;7:415–423. doi: 10.1038/75202. [DOI] [PubMed] [Google Scholar]
- 27.Kong F, Sivakumaran K, Kao C. The N-terminal half of the brome mosaic virus 1a protein has RNA capping-associated activities: specificity for GTP and S-adenosylmethionine. Virology. 1999;259:200–210. doi: 10.1006/viro.1999.9763. [DOI] [PubMed] [Google Scholar]
- 28.Koonin E V, Dolja V V. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit Rev Biochem Mol Biol. 1993;28:375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- 29.Lane L C. Bromoviruses. In: Kurstak E, editor. Handbook of plant virus infections and comparative diagnosis. Amsterdam, The Netherlands: Elsevier/North-Holland Press; 1981. pp. 333–376. [Google Scholar]
- 30.Mayers C N, Palukaitis P, Carr J P. Subcellular distribution analysis of the cucumber mosaic virus 2b protein. J Gen Virol. 2000;81:219–226. doi: 10.1099/0022-1317-81-1-219. [DOI] [PubMed] [Google Scholar]
- 31.Miller W A, Bujarski J J, Dreher T W, Hall T C. Minus-strand initiation by brome mosaic virus replicase within the 3′ tRNA-like structure of native and modified RNA templates. J Mol Biol. 1986;187:537–546. doi: 10.1016/0022-2836(86)90332-3. [DOI] [PubMed] [Google Scholar]
- 32.Miller W A, Dreher T W, Hall T C. Synthesis of brome mosaic virus subgenomic RNA in vitro by internal initiation on (−)-strand genomic RNA. Nature. 1985;313:68–70. doi: 10.1038/313068a0. [DOI] [PubMed] [Google Scholar]
- 33.Nitta N, Takanami Y, Kuwata T, Kubo S. Inoculation with RNAs 1 and 2 of cucumber mosaic virus induces viral RNA replicase activity in tobacco mesophyl protoplasts. J Gen Virol. 1988;69:2695–2700. [Google Scholar]
- 34.Palukaitis P, Roossinck M J, Dietzgen R G, Francki I B. Cucumber mosaic virus. Adv Virus Res. 1992;41:281–348. doi: 10.1016/s0065-3527(08)60039-1. [DOI] [PubMed] [Google Scholar]
- 35.Peden K W C, Symons R H. Cucumber mosaic virus contains a functionally divided genome. Virology. 1973;53:487–492. doi: 10.1016/0042-6822(73)90232-8. [DOI] [PubMed] [Google Scholar]
- 36.Perret V, Florentz C, Dreher T, Giege R. Structural analogies between the 3′ tRNA-like structure of brome mosaic virus RNA and yeast tRNATyr revealed by protection studies with yeast tyrosyl-tRNA synthetase. Eur J Biochem. 1989;185:331–339. doi: 10.1111/j.1432-1033.1989.tb15120.x. [DOI] [PubMed] [Google Scholar]
- 37.Quadt R, Ishikawa M, Janda M, Ahlquist P. Formation of brome mosaic virus RNA-dependent RNA polymerase in yeast requires coexpression of viral proteins and viral RNA. Proc Natl Acad Sci USA. 1995;92:4892–4896. doi: 10.1073/pnas.92.11.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quadt R, Jaspars E M J. Characterization of cucumber mosaic virus RNA-dependent RNA polymerase. FEBS Lett. 1991;279:273–276. doi: 10.1016/0014-5793(91)80166-z. [DOI] [PubMed] [Google Scholar]
- 39.Rao A L N, Grantham G L. Amplification in vivo of brome mosaic virus RNAs bearing 3′ noncoding region from cucumber mosaic virus. Virology. 1994;204:478–481. doi: 10.1006/viro.1994.1559. [DOI] [PubMed] [Google Scholar]
- 40.Rao A L N, Hall T C. Recombination and polymerase error facilitate restoration of infectivity in brome mosaic virus. J Virol. 1993;67:969–979. doi: 10.1128/jvi.67.2.969-979.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rietveld K, Pleij C W A, Bosch L. Three-dimensional models of the tRNA-like 3′-termini of some plant viral RNAs. EMBO J. 1983;2:1079–1085. doi: 10.1002/j.1460-2075.1983.tb01549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rizzo T M, Palukaitis P. Nucleotide sequence and evolutionary relationships of cucumber mosaic virus (CMV) strains: CMV RNA1. J Gen Virol. 1989;70:1–11. doi: 10.1099/0022-1317-70-1-1. [DOI] [PubMed] [Google Scholar]
- 43.Rizzo T M, Palukaitis P. Nucleotide sequence and evolutionary relationships of cucumber mosaic virus (CMV) strains: CMV RNA2. J Gen Virol. 1988;69:1777–1787. doi: 10.1099/0022-1317-69-8-1777. [DOI] [PubMed] [Google Scholar]
- 44.Roossinck M J, Zhang L, Hellwald K H. Rearrangements in the 5′ nontranslated region and phylogenetic analysis of cucumber mosaic virus RNA3 indicate radial evolution of three subgroups. J Virol. 1999;73:6752–6758. doi: 10.1128/jvi.73.8.6752-6758.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siegel R W, Bellon L, Beigelman L, Kao C C. Moieties in an RNA promoter specifically recognized by a viral RNA-dependent RNA polymerase. Proc Natl Acad Sci USA. 1998;95:11613–11618. doi: 10.1073/pnas.95.20.11613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sivakumaran K, Kao C C. Initiation of genomic positive-strand synthesis from DNA and RNA templates by a viral RNA-dependent RNA polymerase. J Virol. 1999;64:6415–6423. doi: 10.1128/jvi.73.8.6415-6423.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stawicki S S, Kao C C. Spatial perturbations within an RNA promoter specifically recognized by a viral RNA-dependent RNA polymerase (RdRp) reveal that RdRp can adjust its promoter binding sites. J Virol. 1999;73:198–204. doi: 10.1128/jvi.73.1.198-204.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sullivan M L, Ahlquist P. A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J Virol. 1999;73:2622–2632. doi: 10.1128/jvi.73.4.2622-2632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun J H, Adkins S, Faurote G, Kao C C. Initiation of (−)-strand RNA synthesis catalyzed by the BMV RNA-dependent RNA polymerase: synthesis of oligoribonucleotides. Virology. 1996;225:1–12. doi: 10.1006/viro.1996.0622. [DOI] [PubMed] [Google Scholar]
- 50.Wu G, Kaper J M. Competition of viral and satellite RNAs of cucumber mosaic virus for replication in vitro by viral RNA-dependent RNA polymerase. Res Virol. 1995;146:61–67. doi: 10.1016/0923-2516(96)80590-5. [DOI] [PubMed] [Google Scholar]
- 51.Wu G, Kaper J M, Jaspers E M J. Replication of cucumber mosaic virus satellite RNA in vitro by an RNA-dependent RNA polymerase from virus infected tobacco. FEBS Lett. 1991;292:213–221. doi: 10.1016/0014-5793(91)80870-9. [DOI] [PubMed] [Google Scholar]
- 52.Yoshinari S, Nagy P D, Simon A E, Dreher T W. CCA initiation boxes without unique promoter elements support in vitro transcription by three viral RNA-dependent RNA polymerase. RNA. 2000;6:698–707. doi: 10.1017/s1355838200992410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhong W, Uss A, Ferrari E, Lau J Y N, Hong Z. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J Virol. 2000;74:2017–2022. doi: 10.1128/jvi.74.4.2017-2022.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]