Abstract
Mini-adenoviruses (mAd) deleted of all viral coding regions represent an emerging approach for transgene expression. We have exploited the unique features of the adeno-associated virus (AAV) terminal repeats within the context of an adenovirus–adeno-associated hybrid virus (Ad/AAV) as a strategy for rapid and efficient generation of mAd. Excision and generation of mAd from the parental Ad/AAV hybrid vector was achieved in 293 cells through recombination but without selection for mAd production. Analysis of mAd isolated from 293 cells indicated that mAd DNA exists as monomer and dimer forms within the recombinant viral capsid. Formation of recombinant mAd was significantly increased using an AAV Rep78- or Rep68-expressing cell line through Rep-mediated excision utilizing the AAV terminal repeat sequences present in the Ad/AAV hybrid virus genome. The mAd viruses were infectious and able to transfer functional gene to A549 and HeLa cells. This approach is rapid and efficient, thereby providing a simplified methodology for generating mAd with functional transducing capabilities.
Considerable progress has been made in recent years toward the development of viral vectors for short- and long-term gene therapy treatment of disease. The human adenovirus (Ad) is an exciting prospect as a vector for gene delivery (1, 4, 16). Ad is a common DNA virus that naturally infects the airway epithelia as well as other tissues in the body. The life cycle of Ad has been well characterized, its genome may be easily manipulated in the laboratory, and recombinant viruses are readily grown to high titers. Ad has a wide host cell range that includes nondividing cells in vitro and in vivo. It is possible to achieve efficient gene expression in quiescent and differentiated cells. Finally, Ad is a relatively benign human virus that is associated with mild disease and, importantly, is not associated with the development of any human malignancy.
Several disadvantages exist for the use of Ad as a vector for long-term gene therapy. First, it is evident from animal studies that Ad elicits an inflammatory response shortly after infection and a subsequent cytotoxic-T-cell response directed against virus-infected cells (reviewed in reference 34). The result is immune clearance of virus-infected cells and extinction of expression of any foreign gene introduced by the recombinant viral vector. Since one would anticipate that repeated application of any gene therapy approach may be required for continued treatment of certain diseases, the rapid immune response to Ad infection severely compromises the use of this system for long-term gene therapy. It appears likely that the expression of Ad-encoded proteins, as well as a foreign transgene, leads to immune recognition (32, 37). A second disadvantage for long-term gene therapy is that the Ad has no direct means to persist in infected cells (1, 4, 16). These problems with Ad vectors apply predominantly to long-term gene therapy approaches, whereas short-term treatments of cancer (8), for example, would not necessarily be compromised and in fact may be enhanced.
An emerging strategy for viral vector development incorporates critical features from individual viruses as a means of generating unique vectors with highly specialized properties. Such “hybrid” viruses can in theory combine essential genetic elements from one virus with those of another as a mechanism of bypassing current limitations inherent in the parental vector. Viral vector chimeras were generated between Ad and retrovirus (5), Ad and Epstein-Barr virus (31), Ad and simian virus 40 (SV40) (11), and Ad and adeno-associated virus (AAV) (10, 18, 19). The unique property of the AAV terminal repeats (TRs) has allowed a number of laboratories to generate Ad/AAV hybrid vectors with unique integrating capabilities (24). Two AAV elements are required for site-specific viral DNA integration: the TRs and either of the two larger Rep polypeptides, Rep68 and Rep78. Rep proteins are also required for efficient AAV replication and excision from the host genome (2, 21, 25).
We hypothesized that the introduction of the AAV TR sequences into the Ad genome flanking a heterologous DNA insert, coupled with transient expression of the AAV Rep protein, would allow for the excision of the AAV TR/insert from the recombinant Ad genome and packaging into recombinant virions. Recently, it was demonstrated that mini-Ad (mAd) vectors are generated as by products of first-generation Ad/AAV hybrid virus amplification, but without selection for mAd production. The mAd genomes contained only the transgene flanked by AAV TRs, Ad packaging signals, and Ad inverted TRs (ITRs) (18). Here we demonstrate high-yield production of mAd using a Rep-expressing cell line (6). We have exploited the unique features of the AAV TRs within the context of an Ad/AAV hybrid virus as a means of generating a mAd vector devoid of all Ad genes. Efficient excision and generation of mAd from the parental Ad/AAV hybrid vectors was achieved using minimal AAV elements. This approach is rapid and efficient, thereby providing a simplified methodology for generating mAd with functional transducing capabilities.
MATERIALS AND METHODS
Plasmid and virus constructions.
The Ad/AAV hybrid plasmid pAd/AAV-EGFP-Neo was generated through multiple cloning manipulations beginning with plasmid pBS-TR-3D (a gift of Sergei Zolotukhin and Nick Muzyczka, University of Florida Gene Therapy Center [unpublished]). pBS-TR-3D is based in plasmid pBluescript (pBS) and contains within the pBS polylinker region the left AAV TR sequence (145 bp, A/B/B′/C/C′/A′/D) (21), a 1,300-bp fragment of stuffer DNA, and the right AAV TR with a double-D (DD) sequence (160 bp, D′/A/B/B′/C/C′/A′/D) (26, 35). The Ad5 left-end 422 bp containing the ITR and packaging domain (14) was inserted next to the left AAV TR. Ad5 DNA sequences from nucleotides (nt) 3330 to 3940 were inserted next to the AAV right TR. Finally, the 1,300-bp stuffer DNA was replaced with the EGFP-Neo expression cassettes from plasmid pTR-UF2 (38). Sequence analysis of this plasmid showed that the intact left AAV TR was lost, and only the AAV TR D sequence remained. The plasmid was linearized using a restriction site outside the Ad5 ITR and used for recombination into Ad5 dl309 according to the method of Stow (30). Virus plaques were isolated on 293 cells (12) (used to complement the deletion of the Ad5 E1 region). Virus stocks were amplified in 293 cells and confirmed by restriction endonuclease digestion and nucleotide sequence analysis of viral DNAs.
Cells and viruses.
293, HeLa and A549 cells were maintained as monolayer cultures in Dulbecco modified Eagle medium (DMEM) containing 10% bovine calf serum (HyClone). C12 cells that carry the AAV Rep and Cap genes (6) were propagated in DMEM containing 10% heat-inactivated fetal bovine serum (HyClone). For viral infections, cells were grown to ∼75% confluency and infected with viruses at low and high multiplicities of infection (MOIs) for 1 h at 37°C. For preparation of virions, infected cell lysates were prepared by suspension of cells in Tris-buffered saline solution following four freeze-thaw cycles. Cell lysates were cleared by centrifugation at 3,000 × g at 15°C for 15 min, followed by incubation with 500 U of DNase I and 250 mg of RNase A per ml in the presence of 2 mM MgCl2 and 2 mM CaCl2 for 30 min at 37°C. Purified virus particles were prepared by centrifugation over a CsCl2 step gradient (1.4 to 1.25 g/cc of CsCl2) and rebanded by equilibrium centrifugation (1.35 g/cc of CsCl2) (34). Virus particles were quantified by lysis of dilutions in buffer containing 0.1% sodium dodecyl sulfate (SDS), and the absorbance at 260 nm was measured; 1 optical density unit at 260 nm equals 1012 particles/ml. Helper virus contamination level was determined by plaque assay on 293 cells.
Viral replication and infectious center assays.
For viral replication assays, infected cell monolayers were washed three times with Tris-buffered saline solution at 24 h after infection, and low-molecular-weight DNA was isolated by the method of Hirt (15). For the analysis of viral DNA in purified virions, 1/10-volume (50 μl) aliquots from each virus preparation were incubated in 50 mM Tris (pH 8.0), 1 mM EDTA, 0.5% SDS, and 1 mg of proteinase K per ml for 1 h at 50°C. Samples were then separated on 0.8% agarose gel and transferred to a nylon membrane (Hybond N+; Amersham). The blots were hybridized to an Ad5 left-end DNA fragment (nt 1 to 355) or to a 3.1-kbp BglII fragment (EGFP/Neo cassette) obtained from the plasmid pTRUF2 (38). mAds were assayed for infectious units (IU) using 293 cells infected with wild-type Ad as a helper virus (10 PFU/cell) coinfected with logarithmic dilutions of DNase I-treated mAds purified by CsCl2 equilibrium centrifugation. Infectious centers were scored by in situ hybridization, as described earlier (27), using EGFP/Neo as a probe. The number of infectious centers observed multiplied by the dilution factor was used in computing the titer of the IU per milliliter. This value was compared to the number of physical virus particles per milliliter determined spectrophotometrically, and the physical particle/infectious particle ratio was calculated. For wild-type Ad5, the particle/PFU ratio is 20 to 25.
GFP expression.
A549 and HeLa cells were infected with mAd (fraction E/M) at a multiplicity of 200 particles/cell or with the parental Ad/AAV-EGFP-Neo parental virus at the same MOI. At different times after infection, green fluorescent protein (GFP) fluorescence was observed using a fluorescein filter on an Axiovert 135 (Zeiss) microscope.
Electron microscopy.
For examination of viral particles by transmission electron microscopy, CsCl2-purified viruses were adsorbed onto Formvar-carbon-coated copper grids and stained with saturated solution of uranyl acetate.
RESULTS
Production of mAd in 293 cells.
We generated a recombinant Ad/AAV hybrid virus that carries the GFP (EGFP) reporter gene and the neomycin selectable marker (Neo) gene flanked by the AAV TR D-sequence on the left side and a complete AAV terminal repeat on the right side containing an additional D-sequence (TR-DD) (26, 35) (Fig. 1A). 293 cells were infected with a cellular lysate containing the Ad/AAV hybrid virus (293 cells complement the E1 deletion in the hybrid virus to allow virus replication). After DNase I digestion of a cleared, infected cell lysate, viruses were separated on a CsCl2 step gradient (1.4 to 1.25 g of CsCl2 per ml). Based on their density, two major bands were visualized (Fig. 2A, F and E/M). Electron microscopy demonstrated that the viral particles in the E/M (i.e., empty/mini) fraction have the same morphology as mature wild-type Ad (F, full fraction). Negative staining showed that viral particles found in the F fraction are homogeneously electron dense (Fig. 2B). The lighter band contained a mixture of two populations: empty and DNA-containing particles with the same size and shape as wild-type Ad (Fig. 2B). The DNA in these particles was DNase I resistant, confirming that it is packaged within the virions.
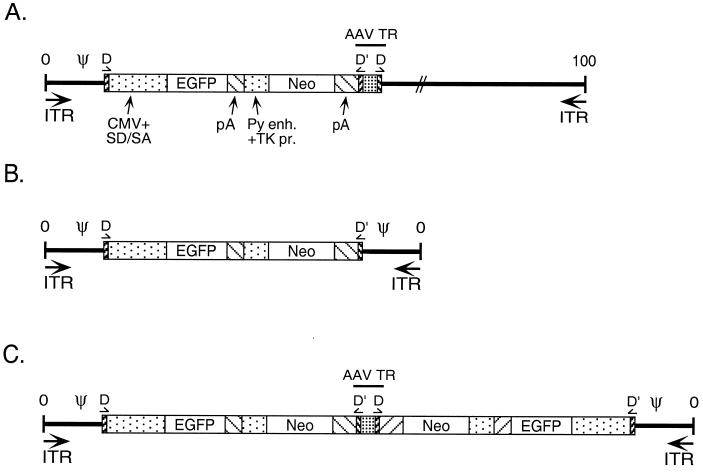
FIG. 1.
Viral genomic maps. (A) Ad/AAV hybrid virus. This virus carries, from left to right, the left end of Ad5 containing the ITR and packaging domain, the AAV TR D sequence, an EGFP/Neo expression cassette from the plasmid pTRUF2 (38) (the EGFP gene is driven from the cytomegalovirus promoter, and the Neo gene is under the control of the polyomavirus enhancer and thymidine kinase promoter; both genes ended with the SV40 polyadenylation signal), an intact AAV TR with a DD sequence (TR-DD), and the remainder of the Ad genome. Ad5 sequences located between nt 423 and 3329 are missing from this virus backbone (E1 deletion). (B) Monomeric mAd. The mAd genome contains the EGFP/Neo cassette flanked by symmetrical sequences containing the AAV TR D sequence and the Ad5 packaging domain-ITR region (nt 1 to 420). (C) Dimeric mAd. This mAd carries a duplicated monomeric genome with an intact AAV TR-DD at the internal junction of the duplication.
FIG. 2.
Production of mAd in 293 cells. (A) Cells were infected with Ad/AAV recombinant hybrid virus (from a third-passage virus stock) using an MOI of 10 PFU/cell. Two days after infection, cleared cellular lysates were prepared and treated with 500 U of DNase I and 250 mg of RNase A per ml. Ad/AAV and mAd viruses were separated on a CsCl2 step gradient. The lower band (F) represent full virus particles. The upper band (E/M) represents lighter particles that includes empty particles, light intermediate particles, mAd, and protein aggregates. (B) Electron microscopy analyses of Ad/AAV and mAd particles. Samples from each population were negative stained using a uranyl acetate saturated solution. The results demonstrate the presence of mature viral particles within the heavy band. The lighter band contains a mixture of full and empty particles. The bar represents 50 nm. (C) Analysis of DNA in virion samples. DNA samples were prepared from each population and analyzed by Southern blot using an EGFP/Neo probe.
During normal replication of wild-type AAV with an Ad helper virus, both monomer-length as well as dimer-length AAV genome products were observed as part of the replication pathway (2, 21, 25). DNA analysis from each virus population (Fig. 2A) demonstrated that full virus particles contained the parental Ad/AAV hybrid virus genome as a single DNA molecule of about 36 kbp (Fig. 2C). The E/M virus particles contained two small genomes of ∼4 and ∼8 kbp. Extensive characterization of these molecules by PCR, restriction enzyme digestion, and nucleotide sequence analysis demonstrated that they correspond to monomer (Fig. 1B) and dimer (Fig. 1C) forms of mAd. The approaches used to analyze the mAd genomes is depicted in Fig. 3A and B and a representative Southern blot is also shown (Fig. 3C). Restriction endonuclease digestion of monomeric and dimeric mAd genomes resulted in the release of DNA fragments of specific length whose origin was determined by hybridization with specific probes. For example, digestion with SalI generated two fragments (∼700 and ∼2.2 kbp) that were recognized by a probe corresponding to the Ad5 left end (nt 1 to 355; Fig. 3B and C). An ∼2.2-kbp fragment was observed using an EGFP-specific probe, and this fragment, as well as an ∼1.1-kbp fragment, was detected using an EGFP/Neo probe (Fig. 3A, B, and C). This model for mAd structure was further supported by comparable analyses using EcoRI, XbaI, and SmaI digestion (data not shown). EcoRI and XbaI digestion confirmed the mAd genome structure indicated by SalI digestion. The AAV TR contains two SmaI restriction sites. Digestion of the dimeric mAd genome with SmaI confirmed the integrity of the AAV TR structure. Specific nucleotide primers were used within the Ad left end and the EGFP and Neo genes to amplify DNA fragments that were predicted from the restriction mapping, and all PCR products were of the predicted size (data not shown). Finally, the precise junctions of Ad5 DNA with the EGFP/Neo expression cassette were determined by nucleotide sequence analysis of the PCR products. Collectively, these analyses confirm the structures of monomeric and dimeric mAd genomes depicted in Fig. 1 and 3.
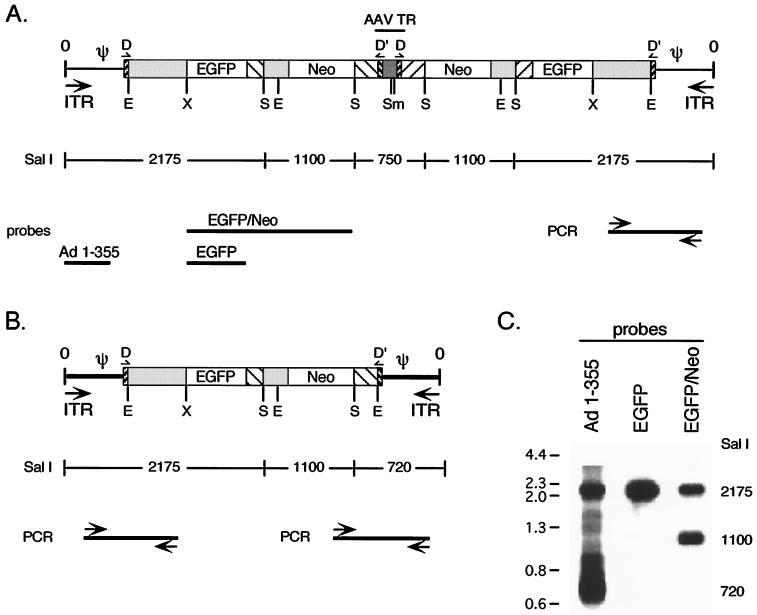
FIG. 3.
Characterization of the monomeric and dimeric mAd genomes. mAd genomes isolated from the E/M fraction (Fig. 2) were characterized by restriction endonuclease digestion and Southern blotting, by PCR using specific primer pairs, and by nucleotide sequence analysis of the PCR products. (A) Analysis of the mAd dimeric genome. (B) Analysis of the monomeric mAd genome. (C) Digestion with SalI yields distinct fragments that were identified by hybridization with probes corresponding to Ad5 nt 1 to 355, EGFP, and EGFP/Neo. The SalI restriction sites are indicated (S), and the predicted SalI cleavage pattern is shown under the schematics of the mAd genomes in panels A and B. Cleavage with other restriction enzymes was evaluated similarly using EcoRI (E), XbaI (X), and SmaI (Sm). Specific PCR products that were generated that are indicated by arrows (primers) and solid lines (products).
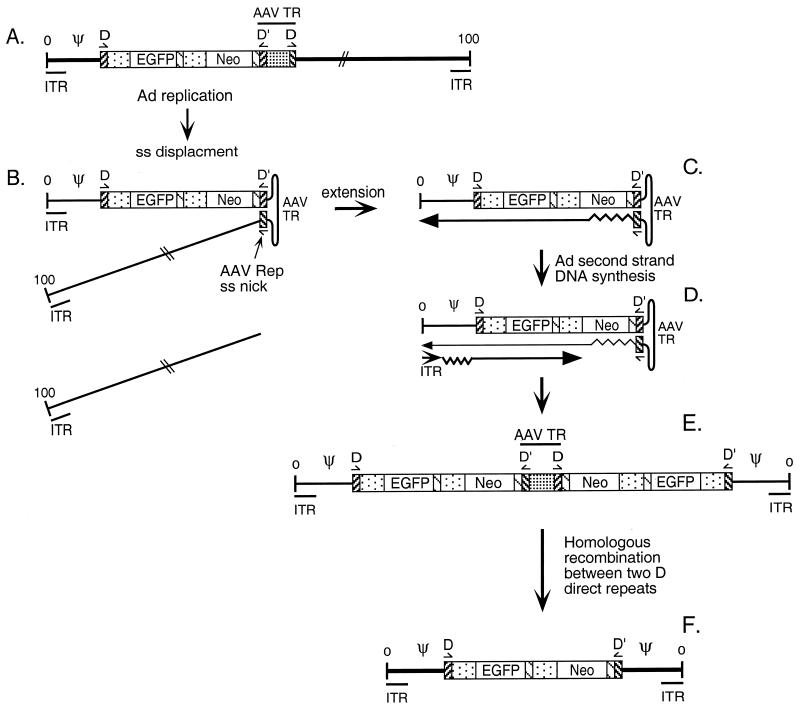
The monomer form (Fig. 1B) contained the EGFP-Neo expression cassette flanked on both sides by an identical fragment of Ad5 DNA (nt 1 to 420) containing the Ad5 ITR and packaging domain, as well as the AAV TR D sequence. The remainder of the AAV TR was missing from this mAd genome. This molecule could arise by simple homologous recombination between the AAV TR D sequences present in the parental virus genome, as proposed by Steinwaerder et al. (29) or by homologous recombination between the two AAV D direct repeats present in the dimer form (Fig. 1C). The dimeric form (Fig. 1C) contained a duplicated monomer genome where the left end of Ad5 (nt 1 to 420), the AAV TR D sequence, and the EGFP-Neo expression cassette were duplicated in an inverted manner. An intact AAV TR was present at the junction of the duplication. This molecule could have arisen from a recombination event between two internal D sequences present in the parent Ad/AAV hybrid virus or through single-strand displacement (see Fig. 6). No selection was imposed to generate the monomeric or the dimeric mAd genomes.
FIG. 6.
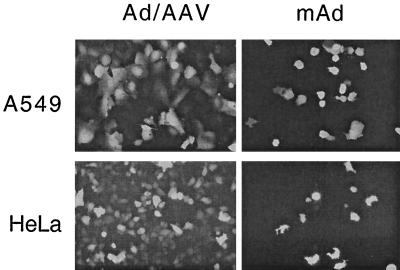
mAd infectivity and transgene expression. A549 or HeLa cells were infected with mAd at an MOI of 200 particles/cell. For a control, cells were infected with the parental Ad/AAV hybrid virus at an MOI of 200 particles/cell. At different times after infection, GFP fluorescence was observed using a fluorescein filter on an Axiovert 135 (Zeiss) microscope.
Efficient excision and replication of mAd in C12 cells.
We hypothesized that the presence of the Rep78 and Rep68 proteins during the replication cycle may improve the efficiency of mAd genome excision through the AAV TR. We first compared the replication efficiency of mAd in HeLa versus C12 cells. C12 cells are a HeLa cell derivative that inducibly express AAV Rep and Cap proteins in response to Ad infection (6). C12 cells were coinfected with the Ad/AAV hybrid virus at a low MOI (10 PFU/cell) with wild-type Ad helper (10 PFU/cell) to initiate Rep expression and replication. Replicating DNA was analyzed by Southern blot 24 h after infection using the left end of the Ad5 genome (Fig. 4A) and EGFP/Neo DNA (Fig. 4B) as probes. The Ad5 left-end probe detected both the wild-type Ad helper virus and the Ad/AAV hybrid virus and excised products, while the EGFP/Neo probe was specific for Ad/AAV hybrid virus genomes. As shown in Fig. 4, mAd genomes were produced efficiently in C12 cells in comparison to HeLa cells, suggesting minigenome formation via AAV Rep-mediated excision. Replication of the Ad/AAV hybrid virus genome and production of the mAd genome required coinfection with wild-type Ad helper virus, a finding consistent with the requirement for E1 expression for productive viral infection with E1-replacement adenoviruses. In addition, we noted the production of a small (∼2 kbp) Ad/AAV hybrid virus-specific DNA species with infections of C12 cells (arrow in Fig. 4). The size of this DNA is consistent with that expected for a single-stranded, monomeric mAd genome. A dimeric, single-stranded mAd genome would be expected to comigrate with the ∼4-kbp double-stranded, monomeric mAd genome. We did not detect the production of any EGFP-Neo-containing DNA molecules that lack the left terminus of the Ad5 genome, indicating that the presence of a single AAV D sequence at the left side of the EGFP/Neo expression cassette in conjunction with an intact AAV TR on the right side was not sufficient to give rise to AAV genomes under these experimental conditions. In addition, fractions across the entire CsCl2 equilibrium gradient were probed by Southern blot for the presence of AAV virus particles. No evidence for the production of AAV was found in C12 cells coinfected with Ad/AAV hybrid virus and wild-type Ad5.
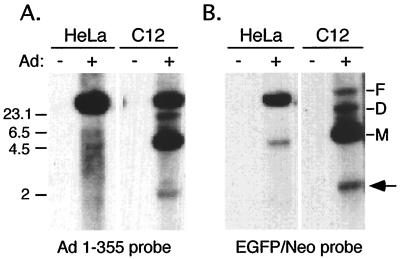
FIG. 4.
Efficient replication of mAd genome in C12 cells (6). HeLa and C12 cells were infected with CsCl2-purified Ad/AAV recombinant virus at an MOI of 10 PFU/cell with (+) or without (−) wild-type Ad5 helper virus. Replicated DNA (15) was isolated 24 h after infection and analyzed by Southern blot using Ad5 (nt 1 to 355) (A) and EGFP/Neo (B) as DNA probes. The majority of the replicated DNA in HeLa cells was full-length Ad/AAV DNA (F). In C12 cells most of the replicated DNA represents monomer (M) and dimer (D) forms of mAd genome. The arrow indicates a submonomer band that was found only in C12 cells. Markers are indicated as kilobase pairs on the left.
On the basis of these results we decided to further investigate the formation of mAd in C12 cells. Cells were coinfected with Ad/AAV hybrid virus and wild-type Ad helper at a low MOI (10 PFU/cell) or with the Ad/AAV hybrid virus at a high MOI (250 PFU/cell) without helper virus. At a low MOI, Ad E1 mutants are defective, so a helper virus is required to ensure virus replication. At a high MOI, Ad E1 mutants are “leaky” so viral replication may occur without helper virus (22). Viral particles were separated by a step gradient (1.4 to 1.25 g of CsCl2 per ml), followed by equilibrium centrifugation (1.35 g of CsCl2 per ml). Four major bands were visualized (Fig. 5A). The densest band (F) represents intact, parental Ad/AAV hybrid virus particles and helper virus. The two light bands (E/M) were collected and analyzed together. The middle fraction (M) was novel to C12 cells coinfected with helper Ad (Fig. 5A, MOI = 10 PFU/cell with helper virus compared to MOI = 250 PFU/cell without helper virus), in comparison to the results described above with 293 cells and was analyzed separately. Electron microscopy showed a mixture of empty and DNA-containing particles in both light fractions (E/M and M) (data not shown). These particles showed the same morphology as mature wild-type virus (data not shown).
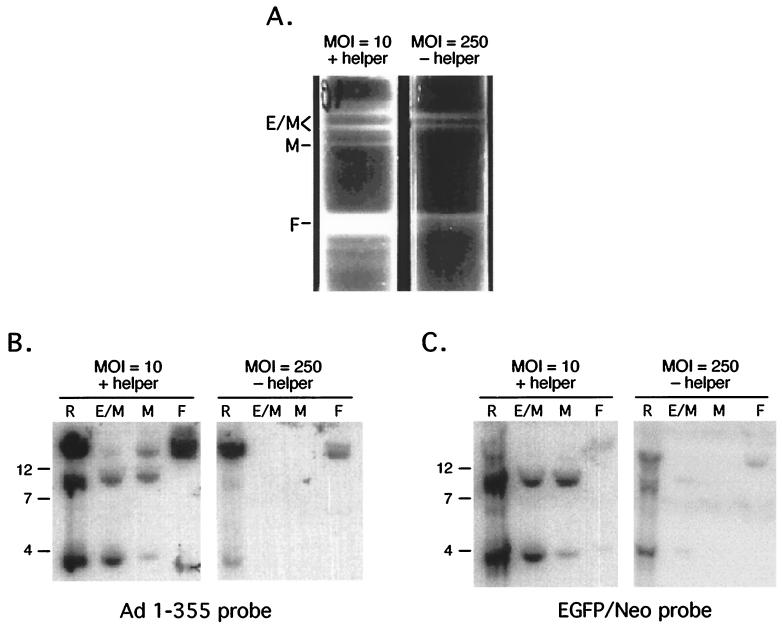
FIG. 5.
Molecular characterization of mAd. C12 cells were infected with CsCl2-purified Ad/AAV hybrid virus at an MOI of 10 PFU/cell, together with wild-type helper virus (MOI = 10 PFU/cell) or at an MOI of 250 PFU/cell without helper virus. (A) CsCl2 equilibrium ultracentrifugation of viruses produced in C12 cells. Recombinant mAds separated from the wild-type helper virus and the parental Ad/AAV hybrid viruses were separated by CsCl2 equilibrium ultracentrifugation. E/M, light virion mixture; M, middle fraction; F, full particles of Ad/AAV, heavy fraction. (B and C) Analysis of viral DNA from infected C12 cells. Replicated DNA was isolated 24 h after infection and analyzed by Southern blot (lanes R). Viral DNA was prepared from each fraction from the CsCl2 equilibrium gradient and analyzed by Southern blot (lanes E/M, M, and F). Membranes were hybridized either to Ad probe (1 to 355 bp) or to the EGFP/Neo cassette from the parent Ad/AAV. Markers are indicated in kilobase pairs on the left.
Southern blot analyses were performed to identify the DNA content of each viral population and to analyze the replicated pool of DNA in the infected cells. Blots were hybridized with a left-end Ad5 probe (nt 1 to 355) (Fig. 5B) or with the EGFP-Neo transgene cassette (Fig. 5C). When the pool of intracellular, replicated viral DNA was analyzed (lane R), the results showed that the newly formed mAd was produced far more efficiently in the presence of helper virus at low MOI than was found without helper virus at a high MOI, even though the parental hybrid virus was capable of efficient replication alone at a high MOI. Three genomic forms were generated during the replication process: ∼4 kbp corresponding to monomers, ∼8 kbp corresponding to dimers, and the high-molecular-weight form corresponding to full-length viral DNA (lane R). Quantification of the replicated and packaged products by phosphorimager analysis showed that 10% of the replicated mAd DNA molecules found in the pool of intracellular DNA were packaged into particles in comparison to 12% of the helper virus genomes that were found to be packaged into virus particles. The mAd genomes were protected from DNase I digestion and thus were completely packaged genomes.
When the DNA content of the separated virus particles was analyzed, the lighter particles (E/M) were found to contain monomers and dimers of mAd DNA (Fig. 5B, E/M fraction). Hybridization with the EGFP/Neo transgene cassette revealed that these particles were free of parental hybrid virus (Fig. 5C, E/M fraction), although some helper virus was evident in this fraction (compare the E/M fractions from Fig. 5B and C). The mAds were formed efficiently only in the presence of wild-type helper virus (Fig. 5B and 4C, E/M fraction, MOI = 10 PFU/cell versus MOI = 250 PFU/cell). At a high MOI without helper virus no mAd virus particles were detected on CsCl2 equilibrium gradients (Fig. 5A) and in the regions of the gradients corresponding to the E/M and M fractions when analyzed by Southern blot (Fig. 5B and C). Phosphorimager analysis indicated that 3% of the DNA molecules found in fraction E/M correspond to wild-type Ad (Fig. 5B, line E/M). To measure the level of infectious particles within that fraction, plaque assay on 293 cells was performed. The results demonstrated that the E/M fraction contained <0.01% contamination with infectious helper virus (data not shown).
Functional studies with mAd.
To investigate whether the mAd vector was able to infect cells and transduce gene expression, A549 and HeLa cells were infected with mAd from fraction E/M (Fig. 5A) in comparison to the parental Ad/AAV hybrid virus. In these experiments, the amounts of viruses used for infections were standardized by quantifying virus particles by measuring the optical density at 260 nm. A549 and HeLa cells were infected at an MOI of 10 PFU/cell. Expression of GFP was detected in the infected cells 24 h (data not shown) and 48 h (Fig. 6) after infection. Transgene expression was visualized in ∼15% of the mAd-infected cells. The parental hybrid virus was able to transduce GFP expression to nearly all of the cells in the culture at the same MOI. These results indicated that the mAd were less infectious than a comparable amount of the parental Ad/AAV hybrid virus. To test this directly, an infectious center assay was used. The number of infectious centers observed multiplied by the dilution factor was used in computing the titer of the IU per milliliter. This value was compared to the number of physical virus particles per milliliter as determined spectrophotometrically, and the physical particle to infectious particle ratio was calculated. This analysis demonstrated that the mAds were reduced by >100-fold in infectivity in comparison to the parental Ad/AAV hybrid virus.
In conclusion, an mAd vector was formed in C12 cells by an excision process that appears to be mediated by the AAV Rep protein. The mAd genomes that were produced exist in both monomeric and dimeric forms that are biologically active and able to transfer functional genes into target cells, although with reduced efficiency compared to the parental Ad/AAV hybrid virus.
DISCUSSION
We utilized an Ad/AAV hybrid virus containing the AAV TR sequences in a unique configuration as a technique for generating mAd vectors. This approach exploits the genetic characteristics of the AAV TRs when employed in the context of a Rep-expressing cell line. Recently, it was shown that inverted repeats inserted into the Ad E1 region could mediate predictable genomic rearrangements, resulting in new vector genomes devoid of all viral genes. These genomes contained only the transgene cassette flanked on both sides by precisely duplicated inverted repeats, Ad packaging signals, and Ad ITRs (29). As an application of this method, it was demonstrated that mAd/AAV vectors were efficiently generated as by-products of first-generation Ad/AAV vector amplification (18). In several previous attempts to generate Ads containing smaller-than-unit-length viral genomes, the packaging process was found to be inefficient when DNA molecules were <75% of the Ad genome size (3, 23). In our studies and those recently reported (18, 29) mAd genomes were found to be packaged efficiently into stable virus particles. We notice that our mAd vector and those vectors described by Leiber et al. (18) retain the Ad packaging domain duplicated at both viral termini. We would speculate that the stability of these mAd vectors, pertains to this unique configuration of packaging domains and to the viral DNA packaging process. We have tested the idea that transient expression of the AAV Rep protein will allow for efficient excision of the AAV TR/insert following subsequent replication and packaging into an mAd.
Our findings which are consistent with those of Lieber et al. (18), who demonstrated mAd production in 293 cells (Fig. 2). However, using 293 cells, we found that efficient generation of mAd was observed only after three to four serial virus amplification cycles. When CsCl2-purified parental Ad/AAV hybrid virus was used for 293 cell infections, inefficient excision of the mAd DNA from the parental Ad/AAV virus was observed, resulting in inefficient mAd production.
We improved the generation of mAd vectors using C12 cells which inducibly express the AAV Rep/Cap proteins (Fig. 3). We found that two distinct mAd forms were generated during replication of the hybrid Ad/AAV vector: a monomeric form that contains a single transgene copy (Fig. 1B) and a dimeric form that carries duplicated copies of the transgene cassette (Fig. 1C). Both forms were found in approximately equimolar ratios within the virion mixture (Fig. 3, fraction E/M). We hypothesize that these mAd genomes were formed via the mechanism depicted in Fig. 7. Ad replicates by a strand displacement mechanism, thereby releasing a single strand of viral DNA during each replication initiation event (33). The AAV TR-DD in the displaced single-strand DNA molecule would be expected to form an AAV TR secondary structure (Fig. 7B). The DD sequence may facilitate this product. The TR D sequence contains the site that the AAV Rep protein targets for endonucleolytic cleavage (2, 21, 25). Production of Rep in C12 cells following Ad infection should induce such a cleavage, resulting in release of the right end of the hybrid 2virus genome (Fig. 7B). We speculate that the apparent ∼2-kbp single-stranded, monomeric mAd genome (Fig. 3, arrow) corresponds to this cleaved product. This cleavage would yield a 3′ end within the cleaved D segment that could be extended by the Ad DNA polymerase as cellular DNA polymerase to generate a fully double-stranded molecule covalently linked at the right end (Fig. 7C). The molecule would contain an intact double-stranded left Ad ITR that could serve as a template for the Ad replication initiation complex (Fig. 7D) for second-strand DNA synthesis to generate a fully duplicated mAd genome (Fig. 7E). The proposed model is fully consistent with our analysis of the structure of the dimeric mAd genome produced in C12 cells, including the observation that the internal AAV TR sequence is intact. We did not detect the production of any EGFP-Neo-containing DNA molecules that lack the left terminus of the Ad5 genome, indicating that the presence of a single AAV D sequence at the left side of the EGFP/Neo expression cassette in conjunction with an intact AAV TR on the right side was not sufficient to give rise to AAV genomes under these experimental conditions.
FIG. 7.
Model for mAd formation. (A) Parental Ad/AAV hybrid virus. Ad replicates using a strand displacement mechanism. During that process a single strand is released. (B) The AAV TR-DD in that strand forms an AAV secondary structure. After Rep-mediated cleavage and DNA extension (B and C), the Ad second strand is synthesized (D), leading to the production of the dimer genome (E). The monomeric form of mAd (F) could be generated by homologous recombination between two AAV D direct repeats or through the mechanism proposed by Steinwaerder et al. (29).
The mAd are comparable to the new-generation “gutted” Ads, whose capability to efficiently transfer functional genes in vitro and in vivo has been demonstrated (7, 9, 17, 20, 23, 28). Like gutted vectors, mAds are devoid of all Ad genes minimizing the host immunological response. However, mAds provide distinct advantages over gutted vectors. First, mAd generation does not require stuffer fragments to maintain a certain genome size and does not require serial passage in cell lines. Second, mAd genomes retain all or part of the AAV TRs, thereby providing the potential for stable integration and long-term gene expression. Third, we demonstrate that using the mAd generation system it is possible to obtain efficient packaging of mAds with a small genome size.
One limitation of the approach described here is the use of wild-type Ad5 helper virus to complement the E1 genes needed for efficient replication of the Ad/AAV hybrid virus. While contamination of the purified mAd preparation with helper virus was low (<0.01%), for gene therapy this still represents a significant level of contamination. One way to overcome this problem would be to use an E1-complementing cell line that inducibly expresses the AAV Rep proteins (36). Alternatively, 293 cells could be used to complement the E1 deficiency and Rep provided by coinfection with wild-type AAV. With this approach, mAds may be purified from contaminating AAV particles by CsCl2 equilibrium centrifugation. A second limitation for the production of mAd vectors is the reduced transduction efficiency. During maturation of adenovirus particles, the viral protease within immature capsids cleaves precursors of viral proteins such as pTP, pVI, pVII, pVIII, and pIIIa and, in addition, it is known that viral entry into host cells requires functional protease within the mature virion (reference 13 and references therein). An explanation for the reduced infectivity of mAd may be the reduced packaging of protease into virus particles and/or the reduced activity of the protease that is packaged. The cosedimentation of mAd particles with empty and light intermediate Ad particles has confounded our efforts to examine protease activity within mAd virions, but this remains an important question that may be addressed when a helper-virus-free system for mAd production is in place.
This study demonstrates proof of the concept that a mAd vector can be efficiently generated through a genetic selection in vivo. We have improved the method by which mAd vectors may be generated by using the AAV Rep protein in a Rep-expressing cell line to select for a genetic rearrangement of the parental Ad/AAV hybrid virus. The new mAd vector is biologically active and able to stably package and transduce small genomes.
ACKNOWLEDGMENTS
Z.S. and D.V.G. contributed equally to this work.
We thank Mena Ostapchuk, Ece Erturk, Amy Ostrom, and Jared Evans for critical reviews of the manuscript and many helpful discussions. We thank Gia Feeney for excellent technical assistance. We thank Emily Huang for assistance during the viral preparations and Susanne Mirold for her role during the early stage of this work. We are very grateful to Sergei Zolotukhin and Nick Muzyczka for the gift of a plasmid containing the AAV TR and DD sequence and to Philip Johnson for C12 cells.
This research was supported by Public Health Service grants to P.H. (AI41636) and W.F.B. (HL53665). W.F.B. is an established investigator of the American Heart Association.
REFERENCES
- 1.Benihoud K, Yeh P, Perricaudet M. Adenovirus vectors for gene delivery. Curr Opin Biotechnol. 1999;10:440–447. doi: 10.1016/s0958-1669(99)00007-5. [DOI] [PubMed] [Google Scholar]
- 2.Berns K I, Giraud C. Adenovirus and adeno-associated virus as vectors for gene therapy. Ann NY Acad Sci. 1995;772:95–104. doi: 10.1111/j.1749-6632.1995.tb44735.x. [DOI] [PubMed] [Google Scholar]
- 3.Bett A J, Prevec L, Graham F L. Packaging capacity and stability of human adenovirus type 5 vectors. J Virol. 1993;67:5911–5921. doi: 10.1128/jvi.67.10.5911-5921.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenner M. Gene transfer by adenovectors. Blood. 1999;94:3965–3967. [PubMed] [Google Scholar]
- 5.Caplen N J, Higginbotham J N, Scheel J R, Vahanian N, Yoshida Y, Hamada H, Blaese R M, Ramsey W J. Adeno-retroviral chimeric viruses as in vivo transducing agents. Gene Ther. 1999;6:454–459. doi: 10.1038/sj.gt.3300835. [DOI] [PubMed] [Google Scholar]
- 6.Clark K R, Voulgaropoulou F, Johnson P R. A stable cell line carrying adenovirus-inducible rep and cap genes allows for infectivity titration of adeno-associated virus vectors. Gene Ther. 1996;3:1124–1132. [PubMed] [Google Scholar]
- 7.Clemens P R, Kochanek S, Sunada Y, Chan S, Chen H H, Campbell K P, Caskey C T. In vivo muscle gene transfer of full-length dystrophin with an adenoviral vector that lacks all viral genes. Gene Ther. 1996;3:965–972. [PubMed] [Google Scholar]
- 8.Crystal R G. In vivo and ex vivo gene therapy strategies to treat tumors using adenovirus gene transfer vectors. Cancer Chemother Pharmacol. 1999;43(Suppl.):590–599. doi: 10.1007/s002800051105. [DOI] [PubMed] [Google Scholar]
- 9.Fisher K J, Choi H, Burda J, Chen S J, Wilson J M. Recombinant adenovirus deleted of all viral genes for gene therapy of cystic fibrosis. Virology. 1996;217:11–22. doi: 10.1006/viro.1996.0088. [DOI] [PubMed] [Google Scholar]
- 10.Fisher K J, Kelley W M, Burda J F, Wilson J M. A novel adenovirus-adeno-associated virus hybrid vector that displays efficient rescue and delivery of the AAV genome. Hum Gene Ther. 1996;7:2079–2087. doi: 10.1089/hum.1996.7.17-2079. [DOI] [PubMed] [Google Scholar]
- 11.Gluzman Y, Van Doren K. Palindromic adenovirus type 5-simian virus 40 hybrid. J Virol. 1983;45:91–103. doi: 10.1128/jvi.45.1.91-103.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 13.Greber U F, Webster P, Weber J, Helenius A. The role of the adenovirus protease on virus entry into cells. EMBO J. 1996;15:1766–1777. [PMC free article] [PubMed] [Google Scholar]
- 14.Hearing P, Samulski R J, Wishart W L, Shenk T. Identification of a repeated sequence element required for efficient encapsidation of the adenovirus type 5 chromosome. J Virol. 1987;61:2555–2558. doi: 10.1128/jvi.61.8.2555-2558.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 16.Kochanek S. High-capacity adenoviral vectors for gene transfer and somatic gene therapy. Hum Gene Ther. 1999;10:2451–2459. doi: 10.1089/10430349950016807. [DOI] [PubMed] [Google Scholar]
- 17.Kochanek S, Clemens P R, Mitani K, Chen H H, Chan S, Caskey C T. A new adenoviral vector: replacement of all viral coding sequences with 28 kb of DNA independently expressing both full-length dystrophin and beta-galactosidase. Proc Natl Acad Sci USA. 1996;93:5731–5736. doi: 10.1073/pnas.93.12.5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lieber A, Steinwaerder D S, Carlson C A, Kay M A. Integrating adenovirus–adeno-associated virus hybrid vectors devoid of all viral genes. J Virol. 1999;73:9314–9324. doi: 10.1128/jvi.73.11.9314-9324.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu X L, Clark K R, Johnson P R. Production of recombinant adeno-associated virus vectors using a packaging cell line and a hybrid recombinant adenovirus. Gene Ther. 1999;6:293–299. doi: 10.1038/sj.gt.3300807. [DOI] [PubMed] [Google Scholar]
- 20.Morsy M A, Gu M, Motzel S, Zhao J, Lin J, Su Q, Allen H, Franlin L, Parks R J, Graham F L, Kochanek S, Bett A J, Caskey C T. An adenoviral vector deleted for all viral coding sequences results in enhanced safety and extended expression of a leptin transgene. Proc Natl Acad Sci USA. 1998;95:7866–7871. doi: 10.1073/pnas.95.14.7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muzyczka N. Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr Top Microbiol Immunol. 1992;158:97–129. doi: 10.1007/978-3-642-75608-5_5. [DOI] [PubMed] [Google Scholar]
- 22.Nevins J R. Mechanism of activation of early viral transcription by the adenovirus E1A gene product. Cell. 1981;26:213–220. doi: 10.1016/0092-8674(81)90304-4. [DOI] [PubMed] [Google Scholar]
- 23.Parks R J, Graham F L. A helper-dependent system for adenovirus vector production helps define a lower limit for efficient DNA packaging. J Virol. 1997;71:3293–3298. doi: 10.1128/jvi.71.4.3293-3298.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Recchia A, Parks R J, Lamartina S, Toniatti C, Pieroni L, Palombo F, Ciliberto G, Graham F L, Cortese R, La Monica N, Colloca S. Site-specific integration mediated by a hybrid adenovirus/adeno-associated virus vector. Proc Natl Acad Sci USA. 1999;96:2615–2620. doi: 10.1073/pnas.96.6.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rolling F, Samulski R J. AAV as a viral vector for human gene therapy. Generation of recombinant virus. Mol Biotechnol. 1995;3:9–15. doi: 10.1007/BF02821330. [DOI] [PubMed] [Google Scholar]
- 26.Ryan J H, Zolotukhin S, Muzyczka N. Sequence requirements for binding of Rep68 to the adeno-associated virus terminal repeats. J Virol. 1996;70:1542–1553. doi: 10.1128/jvi.70.3.1542-1553.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sandalon Z, Dalyot-Herman N, Oppenheim A B, Oppenheim A. In vitro assembly of SV40 virions and pseudovirions: vector development for gene therapy. Hum Gene Ther. 1997;8:843–849. doi: 10.1089/hum.1997.8.7-843. [DOI] [PubMed] [Google Scholar]
- 28.Schiedner G, Morral N, Parks R J, Wu Y, Koopmans S C, Langston C, Graham F L, Beaudet A L, Kochanek S. Genomic DNA transfer with a high-capacity adenovirus vector results in improved in vivo gene expression and decreased toxicity. Nat Genet. 1998;18:180–183. doi: 10.1038/ng0298-180. [DOI] [PubMed] [Google Scholar]
- 29.Steinwaerder D S, Carlson C A, Lieber A. Generation of adenovirus vectors devoid of all viral genes by recombination between inverted repeats. J Virol. 1999;73:9303–9313. doi: 10.1128/jvi.73.11.9303-9313.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stow N D. Cloning a DNA fragment from the left-hand terminus of the adenovirus type 2 genome and its use in site-directed mutagenesis. J Virol. 1981;37:171–180. doi: 10.1128/jvi.37.1.171-180.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan B T, Wu L, Berk A J. An adenovirus-Epstein-Barr virus hybrid vector that stably transforms cultured cells with high efficiency. J Virol. 1999;73:7582–7589. doi: 10.1128/jvi.73.9.7582-7589.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tripathy S K, Black H B, Goldwasser E, Leiden J M. Immune responses to transgene-encoded proteins limit the stability of gene expression after injection of replication-defective adenovirus vectors. Nat Med. 1996;2:545–550. doi: 10.1038/nm0596-545. [DOI] [PubMed] [Google Scholar]
- 33.van der Vliet P C. Adenovirus DNA replication. Curr Top Microbiol Immunol. 1995;199:1–30. doi: 10.1007/978-3-642-79499-5_1. [DOI] [PubMed] [Google Scholar]
- 34.Wold W S, Doronin K, Toth K, Kuppuswamy M, Lichtenstein D L, Tollefson A E. Immune responses to adenoviruses: viral evasion mechanisms and their implications for the clinic. Curr Opin Immunol. 1999;11:380–386. doi: 10.1016/S0952-7915(99)80064-8. [DOI] [PubMed] [Google Scholar]
- 35.Xiao X, Xiao W, Li J, Samulski R J. A novel 165-base-pair terminal repeat sequence is the sole cis requirement for the adeno-associated virus life cycle. J Virol. 1997;71:941–948. doi: 10.1128/jvi.71.2.941-948.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Q, Chen F, Trempe J P. Characterization of cell lines that inducibly express the adeno-associated virus Rep proteins. J Virol. 1994;68:4847–4856. doi: 10.1128/jvi.68.8.4847-4856.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y, Su Q, Wilson J M. Role of viral antigens in destructive cellular immune responses to adenovirus vector-transduced cells in mouse lungs. J Virol. 1996;70:7209–7212. doi: 10.1128/jvi.70.10.7209-7212.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zolotukhin S, Potter M, Hauswirth W W, Guy J, Muzyczka N. A “humanized” green fluorescent protein cDNA adapted for high-level expression in mammalian cells. J Virol. 1996;70:4646–4654. doi: 10.1128/jvi.70.7.4646-4654.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]