Abstract
Maternal genes have a pivotal role in regulating metazoan early development. As such their functions have been extensively studied since the dawn of developmental biology. The temporal and spatial dynamics of their transcripts have been thoroughly described in model organisms and their functions have been undergoing heavy investigations. Yet, less is known about the evolutionary changes shaping their presence within diverse oocytes. Due to their unique maternal inheritance pattern, a high degree is predicted to be present when it comes to their expression. Insofar only limited and conflicting results have emerged around it. Here, we set out to elucidate which evolutionary changes could be detected in the maternal gene expression patterns using phylogenetic comparative methods on RNAseq data from 43 species. Using normalized gene expression values and fold change information throughout early development we set out to find the best-fitting evolutionary model. Through modeling, we find evidence supporting both the high degree of divergence and constraint on gene expression values, together with their temporal dynamics. Furthermore, we find that maternal gene expression alone can be used to explain the reproductive modes of different species. Together, these results suggest a highly dynamic evolutionary landscape of maternal gene expression. We also propose a possible functional dichotomy of maternal genes which is influenced by the reproductive strategy undertaken by examined species.
Keywords: gene expression, maternal transcripts, evolution of development, maternal zygotic transition
Introduction
Early metazoan development is characterized by fast cell divisions, which do not allow de novo transcription. Yet, the development still commences due to a set of predetermined factors present already in the oocytes. These factors are the RNA and protein products of maternal genes (Harvey 1936; Stroband et al. 1992). Among various functions, some better-known gene products of maternal genes are responsible for the maintenance of the chromatin state (Hirasawa et al. 2008; Golding et al. 2011) and in parallel the activation of the silenced zygotic genome (Bultman et al. 2006; Gu et al. 2011; Pan and Schultz 2011). Evidence also suggests roles in cell adhesion (Larue et al. 1994; de Vries et al. 2004), inhibition of polyspermy (Burkart et al. 2012), and patterning of the early embryo (Lehmann and Nusslein-Volhard 1991). Maternal genes exhibit autoregulation through molecular complexes responsible for breaking down maternal transcripts (Lykke-Andersen et al. 2008; Tsukamoto et al. 2008). Together with activation of de novo transcribed factors, the maternal genes become degraded gradually from the early embryo through a period that was defined as the maternal-to-zygotic transition (MZT) (Vastenhouw et al. 2019). Generally, the MZT can be separated into two major regulatory phases. The first phase can be characterized by post-transcriptional (Thomsen et al. 2010; Stoeckius et al. 2014) and post-translational (Krauchunas et al. 2012) regulatory events. That is, it has been shown previously that maternal transcripts possess longer 3′ untranslated regions (3′-UTRs), an observation attributed to post-transcriptional regulation converging on cis motifs found in these regions (Shen-Orr et al. 2010). During the second regulatory phase, the zygotic genome is activated, therefore the major regulatory events will shift towards a transcriptional one (Lee et al. 2014; De Iaco et al. 2017). The transition from a maternal control to a zygotic control happens on a large scale, as maternally provided transcripts can take up to three-quarters of the zygotic transcriptome (Thomsen et al. 2010). Despite its complexity and magnitude, the MZT is a highly conserved transition present in all metazoan species (Vastenhouw et al. 2019) and even some plant species (Zhao et al. 2022). Several functions have been proposed for the MZT, such as acting as an internal clock (Tadros and Lipshitz 2009), a major reprogramming event (Lee et al. 2014), or the degradation of oogenesis-specific transcripts (Pan et al. 2005).
Maternal genes fall under the umbrella term of maternal effects, a term originally coined by Mousseau and Fox (1998, page 5). According to their model, the genes falling under the maternal effect will show higher evolutionary divergences compared to the genes which have no such effects present. Furthermore, this weakened selection is further enhanced by a generational uncoupling, meaning that the population under selection will not respond to selection in the same generation, instead, there will be a lag of one generation (Kirkpatrick and Lande 1989). Empirical evidence has emerged throughout the years which suggests that the theoretical predictions might hold true (Demuth and Wade 2007; Cruickshank and Wade 2008).
Comparative studies have had a strong resurgence in the past few decades, due to the constant development of robust statistical analyses and the implementation of these into user-friendly libraries (see for example, Revell 2012; Pennell et al. 2014). In parallel, the development of high-throughput data generation has seen an unprecedented expansion. Together, the two opened the avenue for research on the evolutionary forces shaping molecular phenomena through comparative analyses. Indeed, recent studies focused on gene expression datasets in a comparative framework to decipher evolutionary forces shaping the transcriptomes. Focus has been given to mammalian organ gene expression evolution (Brawand et al. 2011; Guschanski et al. 2017; Cardoso-Moreira et al. 2019; Fukushima and Pollock 2020). Some studies ventured into elucidating narrower evolutionary questions, such as siphonophore transcriptome evolution (Munro et al. 2022) or the ovary specific gene expression in Drosophiliids (Church et al. 2023). All these studies share in common the use of phylogenetic comparative methods, an approach proposed by Felsenstein in his seminal article (Felsenstein 1985). There, Felsentsein argued against the use of standard statistical tools within comparative biology as species share a common ancestry. Standard statistical tools are ill-equipped to deal with such covariation, therefore, he proposed an alternative approach in the form of independent contrasts (Felsenstein 1985). Since then various other approaches have been developed to deal with biological data in a comparative framework (Pagel 1997, 1999; Freckleton et al. 2002; Rohlfs and Nielsen 2015).
Here, we set out to elucidate the evolutionary patterns shaping maternal gene expression evolution using phylogenetic comparative methods. Results from theoretical approaches (Mousseau and Fox 1998) and sequence based methods (Demuth and Wade 2007; Cruickshank and Wade 2008) suggest a highly dynamic evolutionary landscape of maternal genes. On contrary, more recent studies on this topic (Atallah and Lott 2018a, 2018b) point towards the maternal transcriptome being conserved. However, these studies are limited in species included and omitted the use of phylogenetic comparative methods. To determine which scenario is more plausible, we set out to study the maternal transcriptome evolution in a phylogenetic comparative framework with an expanded set of species included in the analysis. Our results suggest the presence of signals for divergence and constraint being simultaneously present in the maternal transcriptome.
Materials and Methods
All analyses performed and intermediate datasets can be found at the following github repository: mat_gene_exp_evol.
Quantification and Differential Gene Expression Analysis
Following data retrieval, preprocessing, and assemblies (see Supplementary Methods S1.4, Supplementary Material online), the quantification step was performed with the salmon's pseudoaligner algorithm (Patro et al. 2017). To ensure the highest quality of the alignment step we followed the suggestions of the developers (Supplementary Methods, Supplementary Material online). Low-quality samples were flagged based on visual inspection of principal component analysis (PCA) plots. This was further dissected using the hierarchical clustering of cross-sample Euclidean distances. Samples which were grouped outside their expected cluster (i.e. developmental stage they have been samples from) have been flagged. The low-quality samples overlapped between the two approaches and have been discarded from further analyses.
For differential gene expression analysis custom R scripts (R Core Team 2022) were written which utilized widely used libraries for downstream analyses (Supplementary Methods, Supplementary Material online and GitHub repository). Here, we define a maternal transcript as any transcript with a transcript per million (TPM) value above 2 within the oocyte stages of the sampled species. This cutoff was chosen following previous results (Wagner et al. 2013). For DGE a standard pipeline of tximport (Soneson et al. 2015) and DESeq2 (Love et al. 2014) pipeline was used. Not all species have a reported timeframe for MZT, therefore contrasting points had to be determined. Euclidean distances were quantified across the variance-stabilized samples. Compared to the oocyte stages the first developmental stage with the highest distances was searched by pairwise comparisons of the samples. This was visually confirmed by clustering the samples based on Euclidean distances. The heatmap provided visual information for shifts in the transcriptome and in conjunction with the known developmental stages the earliest major transcriptional shift was set as an anchoring point for the MZT. To account for unknown variables during data collection a surrogate variable analysis was performed using the sva (Leek et al. 2012), and these variables were incorporated in the design formula during differential gene expression analysis. Where variables apart from the developmental stages were known the design formulas were set up with these accounted for.
Following the above preparations, the dynamics of maternal genes were determined with cut-off values for adjusted P-values of 0.05 and log2 fold change ± 2. To determine the maternal genes undergoing downregulation throughout MZT, we have used the oocyte stages as reference point and have contrasted stages after MZT against them. Where possible, stages covering the MZT have also been included into these comparisons. All genes which have an adjusted P-value < 0.05 and a log2 fold change ≤ −2 have been termed as downregulated (or degraded) maternal genes. To validate DGE results in situ hybridization-based categorizations from the Fly-FISH database (Lécuyer et al. 2007; Wilk et al. 2016) were retrieved and compared to the Drosophila melanogaster list of degraded genes.
Maternal genes were categorized according to their expression values. Four categories were defined: genes having TPM < 2 were considered not expressed as suggested previously (Wagner et al. 2013). A gene was considered as having a low expression value where 2 ≤ TPM < 100, a medium expression value where 100 ≤ TPM < 1000, and a high expression value where TPM ≥ 1000. The proportion of these categories was subjected to Fisher's exact test using custom R scripts in order to test if gene expression categories change in degraded maternal genes, maternal genes which have the degraded genes excluded and zygotic genes. We corrected for multiple testing using the approach proposed by Benjamini–Hochberg approach (Benjamini and Hochberg 1995).
Information on the gene architectural features of maternal genes is scarce to our knowledge (Heyn et al. 2014), therefore having such a dataset available would be valuable. We set out to inspect the general architectural characteristics. Furthermore, their stability is highly regulated through the binding of regulator proteins to the 3′ untranslated region (3′-UTR), therefore having an overview of such features could provide valuable information (Mishima and Tomari 2016). If there were gene models with the right information (i.e. annotated genome), we took the lengths of 3′-UTR for maternal genes that weren't degraded (i.e. persistently expressed throughout MZT). We also did this for maternal genes that were degraded and for genes that were not expressed maternally (termed as reference genes). These lengths were then directly compared between downregulated and nondownregulated maternal genes by amalgamating the information from each species. The length differences between the three categories (i.e. persistently expressed, degraded, and reference) have been subjected to statistical testing (Wilcoxon rank-sum test with Benjamini–Hochberg adjustment for false discovery rate) after logarithmic transformation (Benjamini and Hochberg 1995). This test was performed for each species separately.
Functional Enrichment
Functional enrichment analysis was done in R programming language environment (R Core Team 2022). Functional annotations were either imported from available genomic resources or assigned de novo. For the de novo annotation two strategies were used: (i) using the annotation of the de novo assembled transcripts GO terms were lifted over from the homologous gene or (ii) a web-service tool (Pannzer2) assigned high probability annotations. For the enrichment itself the enricher and enrichGO functions were utilized (Yu et al. 2012). The former was used in cases with custom gene ontological annotation databases built de novo, the latter for available annotations. If custom annotations were provided to enricher() as a background set all GO annotations retrieved for all genes per each species were used. All ontological categories were tested and considered enriched with a cut-off value of <0.05 for the adjusted P-values. Both downregulated and persistently expressed maternal genes were tested this way separately, ordering of the genes was done by the TPM values for the maternal genes.
Functional enrichments were performed on orthogroups also. In these cases, orthogroups were first annotated using the UniProt database (Bateman et al. 2021). Following the annotation, all GO terms attached to the most probable annotation for each orthogroup were retrieved using UniProtR (Soudy et al. 2020). As background set, all GO terms retrieved for all orthogroups were selected. Enrichment was performed using the hypergeometric test of enricher() mentioned above. The terms below adjusted P-values of 0.05 were selected.
Orthology Mapping
To determine orthology relationships we kept only the longest versions of genes from the translated CDSs and used OrthoFinder2 (Emms and Kelly 2015, 2019) to map them out. We performed the sequence alignments using the diamond's ultrasensitive mode (Buchfink et al. 2014), followed by clustering with the default inflation parameter, and gene trees were estimated in a multiple sequence alignment mode using MAFFT (Katoh and Standley 2013). We used a species tree retrieved from the Open Tree of Life database with the rotl (Michonneau et al. 2016; OpenTreeOfLife et al. 2019) during orthology assignments as a starting tree.
In order to use phylogenetic comparative methods, a dated species tree was required. We calibrated the species tree generated by OrthoFinder using geiger's congruify approach (Eastman et al. 2013). We accessed internal branching event timings from the TimeTree database (Kumar et al. 2017). For scaling the branch lengths, we used TreePL (Smith and O’Meara 2012). We ran the scaling process in three stages: first, an initial optimization run; second, a run testing different smoothing parameter values; and finally, a run with all parameters set to optimal values to perform the dating.
Phylogenetic Dataset Assimilation
To compare gene expressions across different species, a data matrix was created by combining orthology relationships with TPM values. The raw TPM values were first normalized within each species using edgeR's TMM method (Robinson et al. 2009), then across species using a recently proposed method (Munro et al. 2022). Following this, replicates were collapsed into a single value by calculating their means and this single value was assigned to each orthogroup for given species. If multiple genes were assigned to one orthogroup, then these paralogs were summarized into a single value represented by their mean. The values were log-transformed with a pseudocount of 0.01. To account for batch effects, available metadata was fed into limma's removeBatchEffect function (Ritchie et al. 2015).
Additionally, next to the transformed TPM values we have also utilized fold change data from our above-mentioned DGE analysis. We have extracted the log2 fold change data from our test comparing oocyte stage with the first sampled stage after MZT. The fold change data values were used directly for phylogenetic modeling as suggested previously (Dunn et al. 2013).
Evolutionary Modeling
Before fitting phylogenetic aware models, we had to ensure that the use of phylogenetic models is indeed justified. For this, we measured the phylogenetic signal present in each orthogroup utilizing Blomberg's approach (Blomberg et al. 2003). Significant values from the randomization test indicate if a phylogenetic signal is present, whilst the K metric indicates the departure from a Brownian motion assumption. A K value of 1 specifies the expected variation of the trait under a Brownian motion model. K values higher than 1 suggest that relatives resemble each other more than expected, whilst K values lower than 1 suggest more dissimilarity among relatives than expected. A driving force for the former could be selection, whilst for the latter homoplasy.
A step for classifying reproductive modes was inserted as a first step in order to use multiregime evolutionary models. The reproductive mode for each species was determined using the classification of Lodé (2012). Species with placenta and giving live birth according to this classification follow the hemotrophic viviparity mode of reproduction (such as for example Homo sapiens and Mus musculus). Oviparitic species can be characterized by internal fertilization and the embryos are supplied with high quantities of yolk (such as for example Caenorhabditis elegans and Drosophila melanogaster). Finally, ovuliparitic species utilize external fertilization with a moderate amount of yolk supplied with the oocytes. The fourth category, the histotrophic viviparity was not included in the analyzed dataset as to our knowledge there are no available datasets sufficing our criteria of inclusion. Included modes of reproduction have been mapped on the species tree using phytool`s make.simmap (Revell 2012) function. This was necessary for fitting evolutionary models that enable different rates across the mapped character states (reproductive modes in this case).
Before the model fitting step in each orthogroup the species tree was pruned for the tips which have maternal gene expression values. The model fitting for each orthogroup was performed using this pruned species tree with the normalized TPM values at each tip. All models were fit on univariate data, i.e. single orthogroup with associated expression values or fold changes. A filtering step was inserted for the minimum tree size as some phylogenetic models are sensitive to sample size (Cooper et al. 2016). Standard errors for each species were included during the model fitting. The error terms were calculated using biological replicates for each species.
Model fitting was performed using the R packages OUwie (Beaulieu et al. 2012) and geiger (Pennell et al. 2014). From the latter, white noise models were used as null models. These models do not contain phylogenetic signals within them, rather the data is best explained by a normal distribution without any covariance due to shared ancestry present. From OUwie, multiple models were included in the analysis (Table 1).
Table 1.
Table describing the analyzed evolutionary models with the parameters included in the model
| Model | Regime | ϴ | Α | σ |
|---|---|---|---|---|
| OU1 | Single regime | ϴ | Α | σ2 |
| BM1 | Single regime | … | … | σ 2 |
| OUM | Multiple regimes | ϴ1 ϴ2 ϴ3 | α | σ2 |
| BMS | Multiple regimes | ϴ1 ϴ2 ϴ3 | … | |
| OUMV | Multiple regimes | ϴ1 ϴ2 ϴ3 | α | |
| OUMA | Multiple regimes | ϴ1 ϴ2 ϴ3 | α1 α 2 α3 | σ 2 |
| OUMVA | Multiple regimes | ϴ1 ϴ2 ϴ3 | α1 α 2 α3 |
OU1, Ornstein–Uhlenbeck model; BM, Brownian motion model; BMS, multiregime Brownian motion model; OUM, multiregime Ornstein–Uhlenbeck model; OUMV, multiregime Ornstein–Uhlenbeck model with variable σ2 parameters; OUMA, multiregime Ornstein–Uhlenbeck model with variable α parameters; OUMVA, multiregime Ornstein–Uhlenbeck model with variable α and σ2 parameters.
All selected models were fitted to each pruned orthogroup expression or fold change data. The winning models were first selected using second-order Akaike Information Criterion (AICc). After selecting the best-fitting model, a permutation test was performed in order to exclude the choice of the winning model by chance. For this, the data points were randomly shuffled across the examined species tree and the winning model was fitted to it. All AICc values from the 250 rounds of permutation were extracted and tested against the original AICc using the Kolmogorov–Smirnov test to examine if the original AICc value could be explained by chance. Models for downstream analysis were selected if their AICc values exceeded 0.5 and the permutation test returned a significant result (P-value ≤ 0.05). Parameters from models sufficing these criteria were recovered and further examined.
Parameter estimates from the best-fitting evolutionary models were extracted and subjected to further statistical testing using custom R scripts (R Core Team 2022). Significance values were calculated using a nonparametric test (Wilcoxon rank-sum test) as upon visual inspection parameter estimates did not follow a normal distribution. P-values were adjusted for false discovery rate using Benjamini–Hochberg approach (Benjamini and Hochberg 1995). Effect sizes were calculated based on the Z-statistic of the test using the formula where N stands for sample size. Furthermore, an analysis aimed to determine whether the distribution of fold change parameters exhibited a preference for positive or negative values was carried out. This was achieved by setting the null hypothesis (μ = 0) and testing the alternative hypothesis for a bias toward lesser or greater values, respectively.
Next, we sought out to explore if there is enough signal of maternal genes to set apart reproductive modes. In order to do this, we utilized phylogenetic logistic models using phylolm (Tung Ho and Ané 2014). A null hypothesis of constant dependent variables was used for the logistic models. The competing hypothesis was univariate maternal gene expression value. For each reproductive mode, a logistic model was built and tested resulting in three models for each orthogroup. Each model then could possibly classify the tested reproductive mode. Model selection was performed using both AICc values and the likelihood ratio test. For a model to be considered for downstream analysis, it had to suffice the criteria of AICc values above 0.5, a significant improvement in the fit according to the likelihood ratio test and four or more species had to be present for the tested reproductive mode. Selected models were included for downstream analysis.
Results
Maternal Gene Expressions and 3′-UTR Lengths Vary Across Metazoa
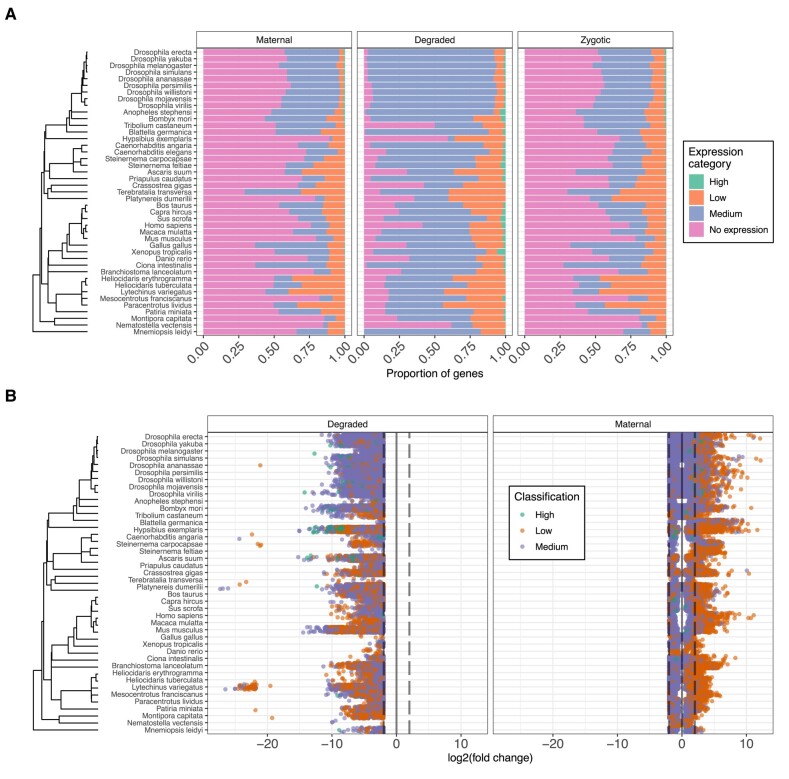
We determined the ratio of genes in the genome that had transcripts present in oocytes varied across species, on average 41% of all genes are expressed. A notable exception was T. transversa, which had 71% of all annotated genes in the oocytes. After gene expression classification, we found that a shared feature across all species is the lack of genes falling into the category of relatively high expresison. Patterns for medium or low expression genes varied across species. In Hexapoda, maternal gene expressions are skewed towards medium expression categories. In contrast, echinoderm species show enrichment in low expression categories. Maternal genes undergoing degradation during MZT are abundant in medium expression categories (Fig. 1A). Additionally, in the degraded maternal gene set the high expression category profiles are slightly more abundant (Fig. 1A). When inspecting the fold changes associated with the above-mentioned maternally expressed genes, we saw a pattern emerge (Fig. 1B). After the MZT most maternal transcripts with low expression values are downregulated. In contrast, the maternal transcripts with higher initial values show an upregulation after the MZT (Fig. 1). Enrichment analysis of maternal genes revealed terms such as RNA splicing, mRNA transport, histone acetylation, and transcription coactivator activity were significantly enriched across species (supplementary fig. S7, Supplementary Material online). We found that such features were commonly enriched across most species. For maternal genes which undergo clearance during MZT, our analysis revealed less overlapping enriched terms (supplementary figs S7 and S8, Supplementary Material online).
Fig. 1.
Maternal gene expression and fold change patterns across the metazoan tree. A) Relative proportions of maternal gene expressions across the analyzed species. Categories of gene expressions are: No expression (TPM < 2), low (2 ≥ TPM < 10), medium (10 ≥ TPM < 1000), and high (TPM ≥ 1000). B) Fold change patterns of maternal genes across the studied species. Genes with lower expression values tend to be downregulated across all included species.
The length of 3'-UTRs varies between different species, with ecdysozoan species generally having shorter 3'-UTRs compared to deuterostome species (supplementary fig. S9, Supplementary Material online). In general, persistently expressed maternal genes and downregulated maternal genes within a species have longer 3'-UTR sequences compared to other genes without a maternal expression, which may suggest that post-transcriptional regulation plays a greater role in these genes as previously suggested (Mishima and Tomari 2016). However, exceptions exist. For example, in S. carpocapsae, which have no significant differences in 3'-UTR lengths between maternally expressed and reference genes. Additionally, hexapod species show shorter 3'-UTR sequences in persistently expressed maternal genes compared to downregulated maternal genes, indicating that the stability of maternal transcripts may depend more on 3'-UTR sequences in this clade.
Phylogenetic Signal is Present in the Maternal Gene Expression Dataset and Justifies the Use of Evolutionary Models
Our analysis on the expression data where orthogroups met the species tree cutoff revealed a phylogenetic signal present in the majority of cases, justifying the use of phylogenetic comparative methods (supplementary fig. S10C, Supplementary Material online). The K-statistic estimates showed a majority of orthogroups with higher dissimilarity than expected (K < 1), while a smaller portion suggested more homogeneous expressions (K > 1) (supplementary fig. S10B, Supplementary Material online). Similar results were obtained for the fold change data, with a phylogenetic signal present in 68% of cases and a left-skewed distribution of the K-statistic.
Both Selection and Neutral Drift is Present in Maternal Gene Expression Evolution
Next, evolutionary models have been fitted to maternal gene expression datasets. Both gene expression and fold change data have been included. In total, for each orthogroup eight model fits were tested simultaneously and the best-fitting model was selected based on both AICc weights and permutation tests. The AICc weight distribution for each winning model varied. For some (BM1 and OU1), it was symmetrical at around 0.5, whilst for others it was skewed to the right (OUM, OUMA, OUMV, OUMVA, BMS) (supplementary fig. 11, Supplementary Material online). Furthermore, no apparent differences are noticeable between the fits of expression values and fold change values as they have highly similar distributions. This implies that more complicated models have better fitting models, reflecting a complicated evolutionary landscape when it comes to the expression of maternal genes.
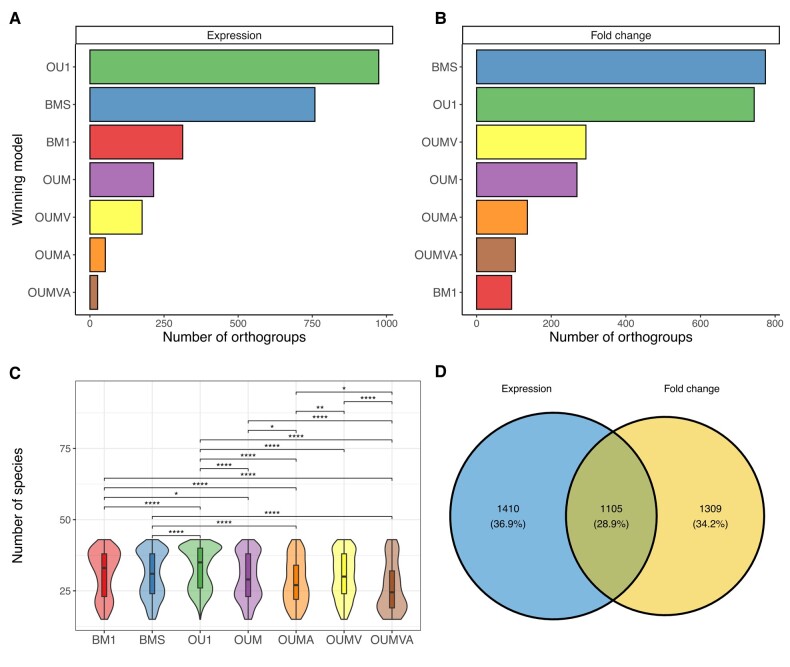
After filtering AICc weights and the significant permutation tests 3824 orthogroups remained for downstream analyses. Out of these 1105 were represented in both expression datasets and fold change datasets (Fig. 2D). The most abundant models for maternal gene expressions were the OU models with single optima and Brownian motion models with multiple σ2 values (Fig. 2A). The same was true for the fold changes between oocytes and embryos right after MZT (Fig. 2B). Models with expanded parameters, such as OUMV, OUMA, and OUMVA had been recovered more sparsely. Generally, the more parameters to be estimated the less likely the model would win. Furthermore, the parameter rich models display a tendency towards being present in orthogroups where the species numbers are sparser during analysis (Fig. 2C). Interestingly, the Brownian motion model with a single σ2 model was the least likely to win in the fold change dataset, whilst its more complicated version was the most likely to win (Fig. 2B). Most of the winning models were also associated with a significant phylogenetic signal (supplementary fig. S10A, Supplementary Material online). Models, where not all orthogroups had significant phylogenetic signals, were mostly OU and BMS models, which is not surprising as Blomberg's K test assumes a Brownian motion generating the data (Blomberg et al. 2003). Furthermore, the K-statistic, which provides an indication for the departure from the expected distribution of Brownian motion showed that BM1 models were the ones closest to the expected distribution. More complicated models showed a greater departure from the expected distribution.
Fig. 2.
The best-fitting evolutionary models. A) The number of orthogroups belonging to each tested evolutionary model in the case of the expression dataset. Only best-fitting models (AICc ≥ 0.5 and permutation test P-value ≤ 0.05) are represented. B) The same as (A) with the fold change dataset used. C) The distribution of the species present in each orthogroups after pruning for each evolutionary model tested. Their means have been compared using Wilcoxon rank-sum tests, significant results are displayed (*P-value ≤ 0.05, **P-value ≤ 0.01, ***P-value ≤ 0.001, ****P-value ≤ 0.001). D) Venn diagram of the number of orthogroups included in downstream analyses (after filtering for AICc and permutation test). In blue are the number of orthogroups for the expression dataset and in yellow are the number of orthogroups for the fold change dataset. The overlap includes orthogroups where both the expression and the fold change dataset have a best-fitting significant evolutionary model present.
Next, a gene ontological enrichment analysis was done in order to better understand the composition of gene groups following each tested evolutionary model (supplementary fig. 12, Supplementary Material online). For this, annotation was done with UniProt entries for each orthogroup using a custom script. GO terms were associated to each orthogroup based on UniProt entries and their GO term annotations. Shared functions across some models were centered around splicing events, mRNA processing, and mitochondrial processes. Orthogroups displaying sings of selection (OU1 model) were uniquely involved in processes involved in DNA replication and vesicular transport. Uniquely enriched processes in other models were that of transcription and translation regulation of orthogroups following the BMS model. As such, insight was gained about enriched functions in orthogroups following each tested evolutionary model.
Parameter Estimates of Evolutionary Models Reflect Evolutionary Patterns Across Reproductive Modes
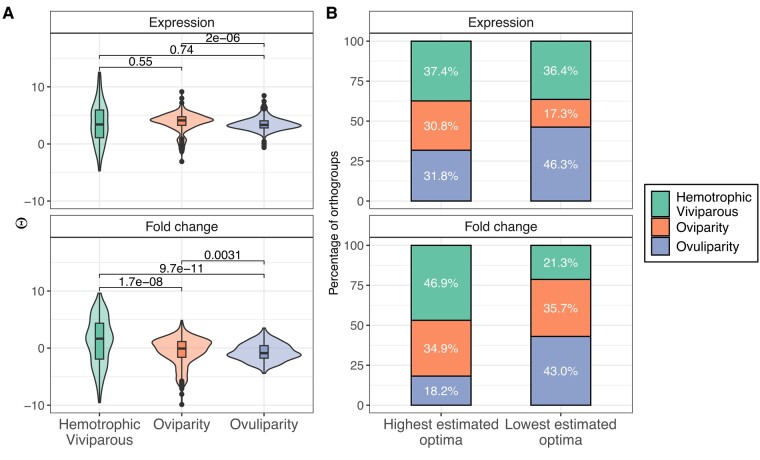
The ϴ estimates from the OUM model fittings showed wider variances for hemotrophic viviparity compared to the two other reproductive modes (Fig. 3A). Pairwise comparisons between reproductive modes for expression ϴ estimates returned overall negligible effect sizes (supplementary tables S3 and S4, Supplementary Material online). For ovuliparous and oviparous species, the fold change data showed a significant bias towards ϴ estimates below 0 (Wilcoxon rank-sum test, P-value = 6.09 × 10−12 and 8.1 × 10−3, respectively). A bias towards positive fold change values is only present in the hemotrophic viviparous species (Wilcoxon rank-sum test, P-value = 3.07 × 10−6).
Fig. 3.
ϴ estimates for orthogroups where OUM models were found to be the best-fitting model. A) Distribution of ϴ estimates for the reproductive modes in both expression dataset (above) and fold change dataset (below). Wilcoxon signed-rank test P-value results are displayed above for each pairwise comparison. B) The proportion of highest and lowest ϴ values for both datasets in all three reproductive modes. In each orthogroup, the multiple ϴ estimates have been ordered followed by selecting the highest and lowest ones.
Examining optima in various reproductive modes for each orthogroup sheds light on the regulation of maternal genes (supplementary table S5, Supplementary Material online). Notably, hemotrophic viviparity often exhibited the highest optima for fold changes (Pearson's χ2 test P-value = 6.53 × 10−7), suggesting a tendency for upregulation in maternal genes under OUM models. Conversely, ovuliparous species consistently showed the lowest number of maternal transcripts (Pearson's χ2 test P-value = 0.0164) and frequently had the lowest optima for fold changes (Pearson's χ2 test, P-value = 3.55 × 10−7). A shrinkage in relative proportions between highest and lowest optima for oviparous species is also present in the gene expression dataset (Pearson's χ2 test P-value = 0.00427).
In the case of orthogroups where α was allowed to vary across regimes generally the optima were similar, except for ovuliparous species expression values, which were slightly lower (supplementary fig. S13C, Supplementary Material online, supplementary table S6, Supplementary Material online). No or weak differences were present in the α parameter estimates across reproductive modes (supplementary fig. S13A, Supplementary Material online, supplementary table S6, Supplementary Material online). The highest ϴ for orthogroups with OUMA models were abundant in oviparous species most commonly (supplementary fig. S13D, Supplementary Material online, supplementary table S8, Supplementary Material online). Contrary to this, the lowest ϴ characterized ovuliparous species. Supporting this pattern were the α value estimates, where the highest α values generally characterized oviparous species best (supplementary fig. S13B, Supplementary Material online, supplementary table S7, Supplementary Material online). Contrary to the lowest ϴ estimates, the lowest α values displayed a more obscure pattern. Here, no strong differences were noticeable for expression values, whilst ovuliparous species displayed the lowest α values overall for OUMA models.
Similarly to the variable alpha models, the variable σ2 models did not show in all cases strong differences between their ϴ, nor between the σ2 values (supplementary fig. S14A and C, Supplementary Material online, supplementary tables S4 and S9, Supplementary Material online). Oviparous species displayed the most frequent highest ϴ for expression values, whilst hemotrophic viviparous species most frequently had the highest optima in the fold change datasets (supplementary fig. S14D, Supplementary Material online, supplementary table 11, Supplementary Material online). The lowest optima for both expression values and fold changes were most common in hemotrophic viviparous mode. Interestingly a strong signal was noticeable for the varying σ2 estimates (supplementary fig. S14B, Supplementary Material online, supplementary table 10, Supplementary Material online). These were most frequently the lowest for oviparous species and least frequently highest.
A big proportion of the winning models followed a Brownian motion model with variable σ2 values (i.e. BMS). Here, for the parameter estimates, we noticed that the ϴ value estimates were highly variable (supplementary fig. S15C, Supplementary Material online, supplementary table 12, Supplementary Material online). Interestingly, the fitted models returned realistic estimates for the ancestral state of the ϴ values. The σ2 values displayed significant differences of moderate effect sizes (supplementary fig. S15A, Supplementary Material online, supplementary table 12, Supplementary Material online), most notably hemotrophic viviparous species had high σ2 estimates which were most frequently the highest σ2 values per orthogroup (supplementary fig. S15D, Supplementary Material online, supplementary table 13, Supplementary Material online). Oviparous species had the lowest σ2 estimates in their expression values.
Parameter estimates in the case of OU1 and BM1 models showed correlated estimates (supplementary fig. S16, Supplementary Material online). σ2 and α parameters in the case of OU1 models were strongly correlated, as were the mean expression values and the ϴ estimates for both OU1 and BM1 models. Interestingly the σ2 and α parameters negatively correlated with both the species included in the model fitting process and the mean expression values. A positive correlation was observable between mean expression values and the count of species included during model fitting. The positive correlation between mean expression and species numbers was observable across most included models (BM1, OU1, OUM), although at varying degrees (supplementary figs S16, S17, S21, Supplementary Material online). The same holds true for α and σ2 parameters (supplementary figs S16A, S17A and B, and S18A, Supplementary Material online). Only weak or no correlations are present between the parameters of BMS models (supplementary fig. S20, Supplementary Material online). A notable exception to this is the strong negative association between ϴ of the oviparous species and ovuliparous species. Correlation coefficients between the OUMA (supplementary fig. S18, Supplementary Material online) and OUMV (supplementary fig. S19, Supplementary Material online) model parameters followed the trend outlined above. In the OUMA models, the α parameters from the oviparous species do not correlate significantly with either ovuliparous or hemotrophic viviparous α parameters, nor do they significantly correlate with the σ2 parameters. A similar pattern emerged in the case of OUMV models.
Gene Expression and Fold Change Data are Sufficient to Distinguish Reproductive Modes From Each Other
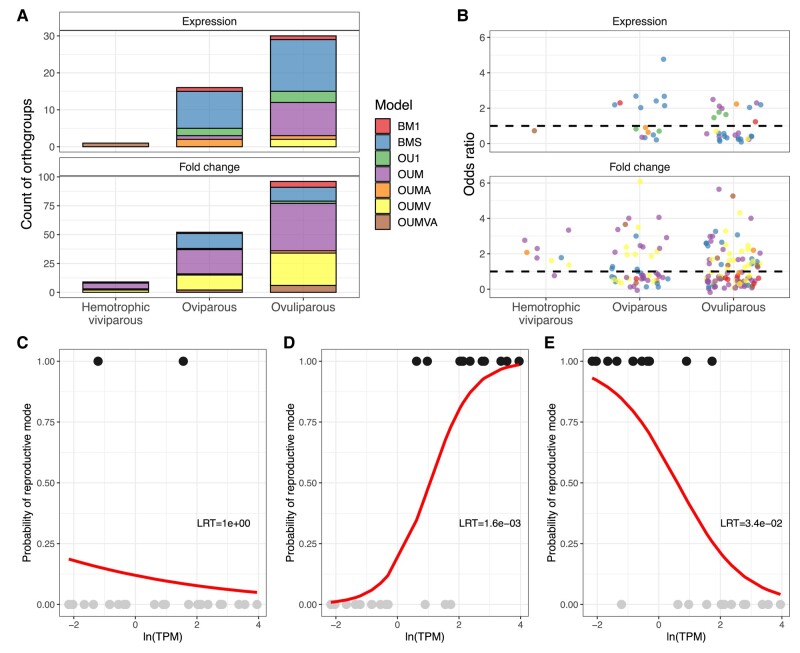
Phylogenetic logistic models were used to test if the analyzed datasets have enough signal within them to distinguish between reproductive modes. Likelihood ratio tests and AICc weights guided the selection of data points, determining that the inclusion of gene expression or fold change data enhanced model fit. Specifically, 47 models for gene expression data and 157 models for fold change data were chosen based on improved fit (see Fig. 4A). Functional enrichment was then applied to identify functions enriched in orthogroups signaling reproductive mode classification. Ovuliparous species models revealed enrichment in terms related to ribosomal and cytoskeletal functions. Additionally, hemotrophic viviparous species, identified through fold change data, showed enrichment in terms associated with nervous system development, hair follicle development, and binding activities. Apart from these enrichments, no further significant results were obtained.
Fig. 4.
Summary of phylogenetic logistic modeling. A) Number of orthogroups with sufficient phylogenetic signal present to classify reproductive mode. B) Odds ratios of phylogenetic logistic models. Dashed lines represent odds ratio of 1. Values above the line suggest that data values for that reproductive mode are higher than the other reproductive modes. Values below the dashed line suggest that data values for that reproductive mode are lower than the other reproductive modes. Phylogenetic logistic model predictions for orthogroup annotated as ribonuclease H1 (C, D, E). Separate model fits are plotted for each tested reproductive mode (hemotrophic viviparity—C, oviparity—D, and ovuliparity—E). Likelihood ratio test results against a null model are also displayed.
Overall, gene expression and fold change data can identify ovuliparity most frequently and hemotrophic viviparity the least frequently (Fig. 4A). There is also a bias in the evolutionary models of the orthogroups for each mode. For gene expression data points, the BMS models are relatively the most abundant, whilst for fold change data the OUM and OUMV models are the most prevalent (Fig. 4A). Visual inspection of logistic model odds ratios (OR) does not suggest that any phylogenetic model would contribute more likely to the classification of reproductive modes (Fig. 4B).
Since the logistic models allow for binary classification multiple logistic models were fitted to the same orthogroup, one for each reproductive more. Interestingly within the same orthogroup a logistic model could distinguish between multiple reproductive modes. Such is the case for the orthogroup annotated as ribonuclease H1, where two separate models emerged (Fig. 4D and E). In one of the models, the ovuliparous species could be distinguished by the upregulation of the ribonuclease (Fig. 4D), whilst for the oviparous species the exact opposite was noticeable (Fig. 4E). For the hemotrophic viviparity reproductive mode, the inclusion of fold change data did not significantly improve model fit (Fig. 4C).
Discussion
Our findings reveal several observations that shed light on maternal gene expression during early development. Furthermore, we found evidence supporting a more intricate evolutionary pattern of maternal gene expression, compared to what was previously described (Mousseau and Fox 1998; Demuth and Wade 2007; Cruickshank and Wade 2008; Atallah and Lott 2018a, 2018b).
Potential Factors Influencing MZT Dynamics
Our assimilated datasets showed that maternal genes possess a variable expression pattern across species and clades. An apparent bias is generally present toward weakly or moderately expressed maternal genes with a big proportion of the genome. Additionally, the weakly expressed maternal genes are more likely to be degraded after the MZT, whilst moderately expressed maternal genes show a tendency towards being upregulated. This discrepancy could originate from de novo transcription from the zygotic genome. Whatever the origin of such transcripts, ultimately an increase is observable in the availability of transcripts for translation, suggesting an important role for such transcripts during and right after MZT. Our results provide further evidence that maternal genes during MZT are regulated through their 3′-UTR sequences across a wide array of clades in the Metazoan tree.
Evolutionary Model Fittings and Departure From Brownian Motion
Our evolutionary model fittings revealed a significant presence of phylogenetic signal in the expression data. This prompted further investigation into evolutionary modeling as Blomberg's K values suggested a departure from the assumption of the expected Brownian motion. We competed multiple evolutionary scenarios for each maternal gene expression data and fold change data and set out to find the best explanation for them. The assumption based on previous studies (Kirkpatrick and Lande 1989; Mousseau and Fox 1998; Demuth and Wade 2007; Cruickshank and Wade 2008) was a great deal of divergence, reflected in Brownian motion models. Some, more recent publications on drosphiliid species suggest a more dynamic landscape of maternal gene expression evolution (Atallah and Lott 2018a, 2018b). We expanded these works with a broader sampling and testing embedded in a phylogenetic framework. Our results contradict the expected divergence, rather they align more with the more recent results of a dynamic evolutionary landscape. A possible explanation for the discrepancy between nucleotide data and current results based on expression data could be attributed to the pleiotropic effects of nucleotide changes (Paaby and Rockman 2013). This effect could be alleviated by altering the expression levels of maternal genes, as this scenario would enable plasticity without affecting later developmental stages. Divergence could be achieved by varying the levels of expressions of genes, thereby introducing new possibly advantageous molecular functions in the maternal gene pool. This notion is supported by our finding that BMS and BM1 models are well represented in the expression datasets. In parallel to such divergences, a selective force maintains the expression for a set of maternal genes essential for basic cellular functions and general molecular mechanisms during early development. This is exemplified by the prevalence of OU1 models and multiregime OU models. Additionally, a potential influential point in our analyses is the omission of tips without maternal expression from the model fitting step. The potential effects are currently unknown but could be interesting to follow up.
Evolutionary Scenarios for Maternal Gene Expression and Fold Change Data
Including multiregime models in our analysis enabled us to inspect differences across different reproductive modes. OUM models provided many insights into such evolutionary differences across reproductive modes. The relatively bigger spread of ϴ estimates for hemotrophic viviparous species across all multiregime OU models suggested a scenario where due to the stable environment of the placenta variation in optima is permissible. Fold change data for such species also supports this notion, as a relatively bigger spread was present for the ϴ estimates. A slight bias towards upregulation in hemotrophic viviparous species suggests a tendency to favor the upregulation of maternal genes in such species for orthogroups following OUM models. Alternatively, spurious estimates could be explained by a sampling bias. Hemotrophic viviparous species are underrepresented in our dataset, leading to potentially spurious parameter estimates. Sampling bias could also be held accountable in OUMA and OUMV α and σ2 estimates. The overall emerging pattern for orthogroups with such models is that oviparous species have higher α and lower σ2 parameters, suggesting selective forces are more prevalent in maintaining expression values and fold changes in oviparous species. The bias could arise in the relative oversampling of drosophilid lineages with relatively less change among them compared to other lineages with fewer representatives and result in estimates reflecting this. A similar scenario could be present in orthogroups following BMS models, here the σ2 estimates are highest for hemotrophic viviparous species, which could arise from the spurious sampling mentioned above.
Evolutionary Differences Across Reproductive Modes
Oviparous species also display a slight tendency towards not being downregulated MZT, whilst ovuliparous species display a slight preference towards downregulation (Fig. 3B, supplementary fig. S11C, Supplementary Material online). This suggests that for ovuliparous embryos more maternal genes are downregulated compared to other modes of reproduction. This notion is further supported by our results showing that ovuliparous species have the lowest optima estimated most frequently. A possible explanation for this observation could be that the progeny of such species is highly susceptible to environmental stressors as their reproduction is characterized by embryos developing in the environment outside the body of the parents. In order to adapt to variable stressors selection could favor a scenario where there is less constraint on what genes are present in the maternal transcriptome. This is would be required as the early embryos are transcriptionally quiescent and in case of a new stressor transcriptional response could only be deployed later (Schulz and Harrison 2019). Furthermore, by storing the genes as transcripts would enable a quicker response due to skipping the requirement of transcription. Conversely, such a scenario would suggest that in a stable environment the expression of maternal genes is restricted to a specific set of genes required to orchestrate early developmental steps. The restriction is present as transcription is costly (Wang et al. 2015) and the energetic costs of early development are funneled to cell divisions. We found evidence supporting constraints on maternal gene expression for hemotrophic viviparous species, where embryos develop in the stable environment of the placenta. Here, we found that selection optima were the highest in some cases for expression values and upregulation of maternal genes was more likely than downregulation (Fig. 3B).
Correlation structures across parameter estimates, mean values of data, and pruned tree sizes added further details to the scenario outlined above. Across all models, the estimates suggested to varying degrees that there is a linear relationship between the pruned tree size and the mean values of data. This linear relationship was the strongest and most positive in single-regime models, meaning as more species have maternal expression associated with orthogroups, the stronger its expression values. A sensible explanation would then be if a given gene is universally required for early embryogenesis selection would favor that gene being more strongly expressed and across more species. This association is dissipated in some orthogroups following extended OU models, which further strengthens the notion outlined above. In multiregime models not all regimes have the same selective regimes present, therefore the association weakens compared to scenarios where all regimes evolve under similar circumstances. The overall positive correlation between means of expression and fold change optima estimates across all multiregime OU models further strengthens the conclusion drawn above, whereby genes with low expression show a tendency towards being downregulated during MZT. The strong overall positive correlation between α and σ2 could be potentially traced back to the sampling biases and restrictions outlined above. This is further supported by the weak negative association between pruned tree sizes, i.e. data points, and parameter estimates.
Implications of Gene Expression Differences in Reproductive Modes
Our analyses also identified genes with sufficient signal to distinguish between reproductive modes. This could have several implications. Firstly, it suggests that maternal gene expressions and developmental dynamics are influenced by the reproductive mode. Secondly, it expands further our current knowledge on the biology of oocytes and early developmental steps across species. Our result that ribonuclease H1 is upregulated in ovuliparous species and downregulated in oviparous species depicts a scenario where for species with both reproductive modes the clearance of RNAs is pivotal through ribonucleases. The origin of such ribonucleases is different, for ovuliparous species it could originate from either de novo transcription or polyadenylation of the transcripts, for oviparous species ribonuclease transcripts are a priori expressed before the MZT. Our results are limited by the scarce sampling of species. With more species sampled, we could get a finer resolution of which maternal genes are pivotal for which reproductive modes. Furthermore, these results suggest that such classifiers with more species under investigation have the potential to identify pivotal maternal genes for a priori defined clades. This approach could enable the discovery of critical maternal genes involved in primate reproduction and therefore of relevance for human reproductive health.
Proposed Hypothesis for the Evolutionary Landscape of Maternal Gene Expression
Based on our results, we propose a hypothesis for the evolutionary landscape of maternal gene expression evolution. A core set of maternal genes with functions necessary for early divisions and initiation of development are under selective constraint to be expressed across a multitude of species. This is in line with the universal requirement of cellular functions across all species during early development. This core group of maternal genes has its temporary dynamics also under constraint during MZT. Apart from this core set of maternal genes, a set of variable maternal genes could also be identified. This set varies across species and shows no constraint on its expression values. Compared to the core set of maternal genes, the variable set can be characterized by weak and variable expression, suggesting a stochastic process behind it, for example, leaky transcription. To better adapt to unpredictable environmental stressors experienced by early embryos in ovuliparous and oviparous species, it may be advantageous to produce a priori genes in a stochastic manner that can respond to those specific stressors. Therefore, selection would maintain the presence of such variably expressed maternal genes as they could still offer a selective advantage.
Supplementary Material
Acknowledgments
These results are part of a project that has received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No 764840. Computational analyses were carried out on the servers provided by Michael Sars Centre. We appreciate the provided infrastructure. We are also thankful for Dr. Stephan Q. Schneider's research group for providing the RNA-seq dataset for Platynereis dumerilii early development upon request. We are thankful for Aina Børve for her help throughout the project. Furthermore, we have to acknowledge the members of IGNITE-ITN for all the discussions contributing to the project. We are also grateful for the constructive feedback on the manuscript draft by Catriona Munro. Furthermore, we appreciate the comments from the reviewers as these comments improved significantly the publication.
Contributor Information
Ferenc Kagan, Department of Biological Sciences, University of Bergen, Bergen, Norway.
Andreas Hejnol, Department of Biological Sciences, University of Bergen, Bergen, Norway; Faculty of Biological Sciences, Friedrich Schiller University, Institute for Zoology and Evolutionary Research, Jena, Germany.
Supplementary Material
Supplementary material is available at Molecular Biology and Evolution online.
Data Availability
The data underlying this article can be accessed through Zenodo (https://doi.org/10.5281/zenodo.8374018). All scripts utilized throughout the publication can be accessed through the master branch on the GitHub repository (https://github.com/fka21/mat_gene_exp_evol.git).
References
- Atallah J, Lott SE. Conservation and evolution of maternally deposited and zygotic transcribed mRNAs in the early Drosophila embryo. PLoS Genet. 2018a:14(12):1–27. 10.1371/journal.pgen.1007838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atallah J, Lott SE. Evolution of maternal and zygotic mRNA complements in the early Drosophila embryo. PLoS Genet. 2018b:14(12):e1007838. 10.1371/journal.pgen.1007838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Martin MJ, Orchard S, Magrane M, Agivetova R, Ahmad S, Alpi E, Bowler-Barnett EH, Britto R, Bursteinas B, et al. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021:49(D1):D480–D489. 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu JM, Jhwueng D-C, Boettiger C, O’Meara BC. Modeling stabilizing selection: expanding the Ornstein–Uhlenbeck model of adaptive evolution. Evolution. 2012:66(8):2369–2383. 10.1111/j.1558-5646.2012.01619.x. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B (Methodol). 1995:57(1):289–300. 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- Blomberg SP, Garland JR, Ives AR. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution. 2003:57(4):717–745. 10.1111/j.0014-3820.2003.tb00285.x. [DOI] [PubMed] [Google Scholar]
- Brawand D, Soumillon M, Necsulea A, Julien P, Csárdi G, Harrigan P, Weier M, Liechti A, Aximu-Petri A, Kircher M, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011:478(7369):343–348. 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2014:12(1):59–60. 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- Bultman SJ, Gebuhr TC, Pan H, Svoboda P, Schultz RM, Magnuson T. Maternal BRG1 regulates zygotic genome activation in the mouse. Genes Dev. 2006:20(13):1744–1754. 10.1101/gad.1435106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkart AD, Xiong B, Baibakov B, Jiménez-Movilla M, Dean J. Ovastacin, a cortical granule protease, cleaves ZP2 in the zona pellucida to prevent polyspermy. J Cell Biol. 2012:197(1):37–44. 10.1083/jcb.201112094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso-Moreira M, Halbert J, Valloton D, Velten B, Chen C, Shao Y, Liechti A, Ascenção K, Rummel C, Ovchinnikova S, et al. Gene expression across mammalian organ development. Nature. 2019:571(7766):505–509. 10.1038/s41586-019-1338-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church SH, Munro C, Dunn CW, Extavour CG. The evolution of ovary-biased gene expression in Hawaiian Drosophila. PLoS Genet. 2023:19(1):e1010607. 10.1371/journal.pgen.1010607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper N, Thomas GH, Venditti C, Meade A, Freckleton RP. A cautionary note on the use of Ornstein Uhlenbeck models in macroevolutionary studies. Biol J Linn Soc. 2016:118(1):64–77. 10.1111/bij.12701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruickshank T, Wade MJ. Microevolutionary support for a developmental hourglass: gene expression patterns shape sequence variation and divergence in Drosophila. Evol Dev. 2008:10(5):583–590. 10.1111/j.1525-142X.2008.00273.x. [DOI] [PubMed] [Google Scholar]
- De Iaco A, Planet E, Coluccio A, Verp S, Duc J, Trono D. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat Genet. 2017:49(6):941–945. 10.1038/ng.3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries WN, Evsikov AV, Haac BE, Fancher KS, Holbrook AE, Kemler R, Solter D, Knowles BB. Maternal β-catenin and E-cadherin in mouse development. Development. 2004:131(18):4435–4445. 10.1242/dev.01316. [DOI] [PubMed] [Google Scholar]
- Demuth JP, Wade MJ. Maternal expression increases the rate of bicoid evolution by relaxing selective constraint. Genetica. 2007:129(1):37–43. 10.1007/s10709-006-0031-4. [DOI] [PubMed] [Google Scholar]
- Dunn CW, Luo X, Wu Z. Phylogenetic analysis of gene expression. Integr Comp Biol. 2013:53(5):847–856. 10.1093/icb/ict068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman JM, Harmon LJ, Tank DC. Congruification: support for time scaling large phylogenetic trees. Methods Ecol Evol. 2013:4(7):688–691. 10.1111/2041-210X.12051. [DOI] [Google Scholar]
- Emms DM, Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015:16(1):157. 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019:20(1):238. 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985:125(1):1–15. 10.1086/284325. [DOI] [PubMed] [Google Scholar]
- Freckleton RP, Harvey PH, Pagel M. Phylogenetic analysis and comparative data: a test and review of evidence. Am Nat. 2002:160(6):712–726. 10.1086/343873. [DOI] [PubMed] [Google Scholar]
- Fukushima K, Pollock DD. Amalgamated cross-species transcriptomes reveal organ-specific propensity in gene expression evolution. Nat Commun. 2020:11(1):4459. 10.1038/s41467-020-18090-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding MC, Williamson GL, Stroud TK, Westhusin ME, Long CR. Examination of DNA methyltransferase expression in cloned embryos reveals an essential role for Dnmt1 in bovine development. Mol Reprod Dev. 2011:78(5):306–317. 10.1002/mrd.21306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu T-P, Guo F, Yang H, Wu H-P, Xu G-F, Liu W, Xie Z-G, Shi L, He X, Jin S, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011:477(7366):606–610. 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- Guschanski K, Warnefors M, Kaessmann H. The evolution of duplicate gene expression in mammalian organs. Genome Res. 2017:27(9):1461–1474. 10.1101/gr.215566.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey EB. Parthenogenetic merogony or cleavage without nuclei in Arbacia punctulata. Biol Bull. 1936:71(1):101–121. 10.2307/1537411. [DOI] [Google Scholar]
- Heyn P, Kircher M, Dahl A, Kelso J, Tomancak P, Kalinka AT, Neugebauer KM. The earliest transcribed zygotic genes are short, newly evolved, and different across species. Cell Rep. 2014:6(2):285–292. 10.1016/j.celrep.2013.12.030. [DOI] [PubMed] [Google Scholar]
- Hirasawa R, Chiba H, Kaneda M, Tajima S, Li E, Jaenisch R, Sasaki H. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev. 2008:22(12):1607–1616. 10.1101/gad.1667008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Ls, Ané C. A linear-time algorithm for Gaussian and non-Gaussian trait evolution models. Syst Biol. 2014:63(3):397–408. 10.1093/sysbio/syu005. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013:30(4):772–780. 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M, Lande R. The evolution of maternal characters. Evolution. 1989:43(3):485–503. 10.2307/2409054. [DOI] [PubMed] [Google Scholar]
- Krauchunas AR, Horner VL, Wolfner MF. Protein phosphorylation changes reveal new candidates in the regulation of egg activation and early embryogenesis in D. melanogaster. Dev Biol. 2012:370(1):125–134. 10.1016/j.ydbio.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. 2017:34(7):1812–1819. 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
- Larue L, Ohsugi M, Hirchenhain J, Kemler R. E-cadherin null mutant embryos fail to form a trophectoderm epithelium. Proc Natl Acad Sci USA. 1994:91(17):8263–8267. 10.1073/pnas.91.17.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Giraldez AJ. Zygotic genome activation during the maternal-to-zygotic transition. Annu Rev Cell Dev Biol. 2014:30(1):581–613. 10.1146/annurev-cellbio-100913-013027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The SVA package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012:28(6):882–883. 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann R, Nusslein-Volhard C. The maternal gene nanos has a central role in posterior pattern formation of the Drosophila embryo. Development. 1991:112(3):679–691. 10.1242/dev.112.3.679. [DOI] [PubMed] [Google Scholar]
- Lécuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM . Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007:131(1):174–187. 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Lodé T. Oviparity or viviparity? That is the question… Reprod Biol. 2012:12(3):259–264. 10.1016/j.repbio.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014:15(12):550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen K, Gilchrist MJ, Grabarek JB, Das P, Miska E, Zernicka-Goetz M. Maternal argonaute 2 is essential for early mouse development at the maternal-zygotic transition. Mol Biol Cell. 2008:19(10):4383–4392. 10.1091/mbc.e08-02-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michonneau F, Brown JW, Winter DJ. Rotl: an R package to interact with the open tree of life data. Methods Ecol Evol. 2016:7(12):1476–1481. 10.1111/2041-210X.12593. [DOI] [Google Scholar]
- Mishima Y, Tomari Y. Codon usage and 3’ UTR length determine maternal mRNA stability in zebrafish. Mol Cell. 2016:61(6):874–885. 10.1016/j.molcel.2016.02.027. [DOI] [PubMed] [Google Scholar]
- Mousseau TA, Fox CW. The adaptive significance of maternal effects. Trends Ecol Evol (Amst). 1998:13(10):403–407. 10.1016/S0169-5347(98)01472-4. [DOI] [PubMed] [Google Scholar]
- Munro C, Zapata F, Howison M, Siebert S, Dunn CW. Evolution of gene expression across species and specialized zooids in siphonophora. Mol Biol Evol. 2022:39(2):msac027. 10.1093/molbev/msac027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OpenTree et al. “Open Tree of Life Taxonomy” 10.5281/zenodo.3937750. [DOI] [Google Scholar]
- Paaby AB, Rockman MV. The many faces of pleiotropy. Trends Genet. 2013:29(2):66–73. 10.1016/j.tig.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel M. Inferring evolutionary processes from phylogenies. Zool Scr. 1997:26(4):331–348. 10.1111/j.1463-6409.1997.tb00423.x. [DOI] [Google Scholar]
- Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999:401(6756):877–884. 10.1038/44766. [DOI] [PubMed] [Google Scholar]
- Pan H, O’Brien MJ, Wigglesworth K, Eppig JJ, Schultz RM. Transcript profiling during mouse oocyte development and the effect of gonadotropin priming and development in vitro. Dev Biol. 2005:286(2):493–506. 10.1016/j.ydbio.2005.08.023. [DOI] [PubMed] [Google Scholar]
- Pan H, Schultz RM. SOX2 modulates reprogramming of gene expression in two-cell mouse embryos1. Biol Reprod. 2011:85(2):409–416. 10.1095/biolreprod.111.090886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017:14(4):417–419. 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennell MW, Eastman JM, Slater GJ, Brown JW, Uyeda JC, Fitzjohn RG, Alfaro ME, Harmon LJ. Geiger v2.0: an expanded suite of methods for fitting macroevolutionary models to phylogenetic trees. Bioinformatics. 2014:30(15):2216–2218. 10.1093/bioinformatics/btu181. [DOI] [PubMed] [Google Scholar]
- R Core Team . 2022. R: A Language and Environment for Statistical Computing. https://www.R-project.org/
- Revell LJ. Phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol. 2012:3(2):217–223. 10.1111/j.2041-210X.2011.00169.x. [DOI] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015:43(7):e47. 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. Edger: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009:26(1):139–140. 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlfs RV, Nielsen R. Phylogenetic ANOVA: the expression variance and evolution model for quantitative trait evolution. Syst Biol. 2015:64(5):695–708. 10.1093/sysbio/syv042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz KN, Harrison MM. Mechanisms regulating zygotic genome activation. Nat Rev Genet. 2019:20(4):221–234. 10.1038/s41576-018-0087-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Orr SS, Pilpel Y, Hunter CP. Composition and regulation of maternal and zygotic transcriptomes reflects species-specific reproductive mode. Genome Biol. 2010:11(6):R58. 10.1186/gb-2010-11-6-r58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, O’Meara BC. treePL: divergence time estimation using penalized likelihood for large phylogenies. Bioinformatics. 2012:28(20):2689–2690. 10.1093/bioinformatics/bts492. [DOI] [PubMed] [Google Scholar]
- Soneson C, Love MI, Robinson MD. Differential analyses for RNA-Seq: transcript-level estimates improve gene-level inferences. F1000Res. 2015:4:1521. 10.12688/f1000research.7563.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soudy M, Anwar AM, Ahmed EA, Osama A, Ezzeldin S, Mahgoub S, Magdeldin S. Uniprotr: retrieving and visualizing protein sequence and functional information from Universal Protein Resource (UniProt knowledgebase). J Proteomics. 2020:213:103613. 10.1016/j.jprot.2019.103613. [DOI] [PubMed] [Google Scholar]
- Stoeckius M, Grün D, Kirchner M, Ayoub S, Torti F, Piano F, Herzog M, Selbach M, Rajewsky N. Global characterization of the oocyte-to-embryo transition in C. aenorhabditis elegans uncovers a novel m RNA clearance mechanism. EMBO J. 2014:33(16):1751–1766. 10.15252/embj.201488769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroband HWJ, te Krounie G, van Gestel W. Differential susceptibility of early steps in carp (Cyrinus carpio) development to α-amanitin. Rouxs Arch Dev Biol. 1992:202(1):61–65. 10.1007/BF00364597. [DOI] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009:136(18):3033–3042. 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- Thomsen S, Anders S, Janga SC, Huber W, Alonso CR. Genome-wide analysis of mRNA decay patterns during early Drosophila development. Genome Biol. 2010:11(9):R93. 10.1186/gb-2010-11-9-r93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto S, Kuma A, Mizushima N. The role of autophagy during the oocyte-to-embryo transition. Autophagy. 2008:4(8):1076–1078. 10.4161/auto.7065. [DOI] [PubMed] [Google Scholar]
- Vastenhouw NL, Cao WX, Lipshitz HD. The maternal-to-zygotic transition revisited. Development. 2019:146(11):dev161471. 10.1242/dev.161471. [DOI] [PubMed] [Google Scholar]
- Wagner GP, Kin K, Lynch VJ. A model based criterion for gene expression calls using RNA-Seq data. Theory Biosci. 2013:132(3):159–164. 10.1007/s12064-013-0178-3. [DOI] [PubMed] [Google Scholar]
- Wang G-Z, Hickey SL, Shi L, Huang H-C, Nakashe P, Koike N, Tu BP, Takahashi JS, Konopka G. Cycling transcriptional networks optimize energy utilization on a genome scale. Cell Rep. 2015:13(9):1868–1880. 10.1016/j.celrep.2015.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk R, Hu J, Blotsky D, Krause HM. Diverse and pervasive subcellular distributions for both coding and long noncoding RNAs. Genes Dev. 2016:30(5):594–609. 10.1101/gad.276931.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012:16(5):284–287. 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P, Shi C, Wang L, Sun M. The parental contributions to early plant embryogenesis and the concept of maternal-to-zygotic transition in plants. Curr Opin Plant Biol. 2022:65:102144. 10.1016/j.pbi.2021.102144. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article can be accessed through Zenodo (https://doi.org/10.5281/zenodo.8374018). All scripts utilized throughout the publication can be accessed through the master branch on the GitHub repository (https://github.com/fka21/mat_gene_exp_evol.git).