Abstract
Two mRNA species are derived from the influenza C virus RNA segment six, (i) a colinear transcript containing a 374-amino-acid residue open reading frame (referred to herein as the seg 6 ORF) which is translated to yield the p42 protein, and (ii) a spliced mRNA which encodes the influenza C virus matrix (CM1) protein consisting of the first 242 amino acids of p42. The p42 protein undergoes proteolytic cleavage at a consensus signal peptidase cleavage site after residue 259, yielding the p31 and CM2 proteins. Translocation of p42 into the endoplasmic reticulum membrane occurs cotranslationally and requires the hydrophobic internal signal peptide (residues 239 to 259), as well as the predicted transmembrane domain of CM2 (residues 285 to 308). The p31 protein was found to undergo rapid degradation after cleavage from p42. Addition of the 26S proteasome inhibitor lactacystin to influenza C virus-infected or seg 6 ORF cDNA-transfected cells drastically reduced p31 degradation. Transfer of the 17-residue C-terminal region of p31 to heterologous proteins resulted in their rapid turnover. The hydrophobic nature, but not the specific amino acid sequence of the 17-amino-acid C terminus of p31 appears to act as the signal for targeting the protein to membranes and for degradation.
Influenza A, B, and C viruses encode the small (97- to 115-amino-acid) integral membrane proteins M2, NB, and CM2 respectively (27, 37). Their similarity in oligomeric form, orientation in membranes, and size of their extracellular, transmembrane, and cytoplasmic domains led to the hypothesis that these proteins perform related functions in the respective virus life cycles (36). The M2, NB, and CM2 coding regions are highly conserved among the respective A, B, and C influenza viruses that have been sequenced to date (1, 21, 45); however, little if any amino acid homology exists between the three proteins.
Influenza A virus M2 protein is packaged into virions (54) and possesses a pH-activated proton-selective ion channel activity (14, 38, 44). On entry of virions into cells via the endocytic pathway, the M2 ion channel activity acidifies the interior of virus particles, leading to disruption of protein-protein interactions between the viral matrix (M1) protein and the viral ribonucleocapsids (4, 30, 31, 55, 56). Following fusion of the viral membrane with the membrane of the endosome, the ribonucleocapsids are transported to the nucleus, where viral genome replication and transcription occurs (27). The ion channel activity of the M2 protein, during its transport to the cell surface, raises the pH of the trans-Golgi network (6, 7, 12, 13, 34, 40, 46). For influenza viruses which possess a hemagglutinin (HA) protein with a high pH threshold for undergoing protein-refolding conformational changes associated with membrane fusion, this equilibration of the lumenal pH of the trans-Golgi network with that of the cytoplasm is a prerequisite to prevent HA from fusion-activation in the wrong cellular compartment (reviewed in reference 26).
To date, no functional role or evidence of ion channel activity has been reported for the influenza C virus CM2 protein. The CM2 protein forms disulfide-linked dimers and tetramers and is oriented in membranes in an Nout-Cin orientation. CM2 contains a single site for N-linked carbohydrate addition which is frequently further modified with lactosaminoglycans. The CM2 protein is abundantly expressed at the cell surface of virus-infected and cDNA-transfected cells and is incorporated into influenza C virions (17, 36). The CM2 protein is encoded by influenza C virus RNA segment six (19). RNA segment six is initially transcribed into a colinear mRNA transcript, and a majority of this species is subsequently spliced to yield a second mRNA transcript (53). The colinear mRNA contains a 374-amino-acid open reading frame (referred to herein as the seg 6 ORF) that is translated into the precursor protein p42. Proteolytic cleavage of p42 at an internal signal peptidase cleavage site gives rise to the p31 and CM2 proteins (18, 37). The spliced mRNA encodes the matrix (CM1) protein, consisting of the N-terminal 242 residues of the seg 6 ORF followed by a stop codon introduced via mRNA splicing (53). Synthesis of the p42 protein has been detected in (i) influenza C virus-infected cells (16), (ii) seg 6 ORF cDNA-transfected cells, and (iii) in vitro translation reactions using RNA encoding the seg 6 ORF (18, 37). The N-terminal product of p42 cleavage, p31, is identical in amino acid sequence to the CM1 protein except for the presence of 17, mostly hydrophobic, amino acids at its C terminus. The p31 protein binds tightly to lipid membranes with properties more like those of the integral membrane protein CM2 than those of the peripheral membrane protein CM1 (37). In this study, we investigate further the requirements of p42 membrane insertion and we examine the fate of the p31 protein after cleavage from p42.
MATERIALS AND METHODS
Cells and viruses.
Madin-Darby canine kidney (MDCK), CV-1, and HeLa-T4 cells were grown in Dulbecco's modified Eagle's (DME) medium, containing 10% fetal bovine serum, 2 mM glutamine, and penicillin-streptomycin (P-S). Influenza C/Ann Arbor/1/50 (C/AA) virus working stocks were generated as described previously (36).
Plasmid construction and site-directed mutagenesis.
All plasmid constructs were based on the seg 6 ORF cDNA of influenza C/AA virus, whose cloning and nucleotide sequencing have been described previously (36, 37). Oligonucleotide primer sequences are available upon request. The PCR using Vent DNA polymerase (New England Biolabs, Beverly, Mass.) was used to isolate a cDNA consisting of the entire p31 ORF (amino acids 1 to 259) followed by an in-frame stop codon. This method was also used to obtain a cDNA, CM2 ΔSP, encoding the entire mature CM2 protein (amino acids 260 to 374) with a Met codon introduced immediately upstream and in-frame to initiate translation. The cDNAs for the A/M1-C17 and chloramphenicol acetyltransferase (CAT)-C17 proteins were constructed by PCR with oligonucleotide primers designed to fuse the coding sequence for the 17-amino-acid p31 C-terminal region (Trp-Leu-Val-Val-Ile-Ile-Cys-Phe-Ser-Ile-Thr-Ser-Gln-Pro-Ala-Ser-Ala) followed by an in-frame stop codon to the 3′ end of the influenza A virus M1 protein coding region (cDNA of influenza A/Puerto Rico/8/34 virus) (55) or the CAT coding region, respectively. The CAT-HA cDNA was constructed in a similar manner, fusing the coding sequence for the 12CA5 monoclonal antibody (MAb) epitope (Tyr-Pro-Tyr-Asp-Val-Pro-Asp-Tyr-Ala) followed by a stop codon to the 3′ end of the CAT coding region. All cDNAs were cloned into pGEM 3 under the control of the bacteriophage T7 promoter (36).
Site-specific mutagenesis of plasmids was performed using the unique site elimination mutagenesis kit (Pharmacia Biotech, Piscataway, N.J.). A cDNA designated pGEM p33 was constructed by changing the codon for Ala at amino acid 259 to an Arg codon and the codon for Val at amino acid 275 to a stop codon. The C terminus of p31 was truncated by 4, 8, or 13 amino acids by substituting stop codons for those encoding amino acids 256, 252, or 247, respectively. The coding sequence for amino acids 243 to 251 of the p31 protein was replaced with nine Leu codons (p31-Leu) or the codons for amino acids 94 to 102 of human synaptobrevin-2 (p31-SB) (49). The sequence of the FLAG MAb epitope was substituted for the sequence encoding amino acids 243 to 250 of p31 in p31-FLAG. A cDNA designated p31-hinge was constructed by inserting the sequence for the immunoglobulin G (IgG) hinge region (42) after the codon for amino acid 242, followed by the coding region for amino acids 239 to 374 of the p42 protein.
To facilitate the identification of altered plasmids, restriction enzyme sites were introduced into all mutagenesis primers by utilizing translationally silent nucleotide substitutions. All plasmids were sequenced using an ABI Prism 310 genetic analyzer (Applied Biosystems Inc., Foster City, Calif.). After sequencing, the cDNAs were subcloned into the eukaryotic expression vector pCAGGS (33) for transient-expression experiments.
In vitro transcription and translation.
Bacteriophage T7 RNA polymerase transcripts were synthesized from HindIII linearized plasmids, by using the T7 Message Machine kit (Ambion Inc., Austin, Tex.). RNA was translated in the presence of [35S]methionine (30 μCi per 100-μl reaction mixture) (Amersham Life Science, Arlington Heights, Ill.) using rabbit reticulocyte lysates and canine pancreatic microsomes (3 μl per reaction mixture) as described previously (36). Assays for identifying the nature of the interaction of proteins with microsomal membranes were performed as described previously (37, 55).
Infection, transfection, and metabolic labeling of cells.
MDCK cells were infected with influenza C virus at a multiplicity of infection of 1 PFU per cell for 2 h at 37°C. The inoculum was removed and the cells were then cultured at 37°C in DME medium with glutamine and P-S for 20 h.
A recombinant vaccinia virus expressing bacteriophage T7 RNA polymerase (vac-T7) was used, essentially as described previously (11). Briefly, HeLa-T4 or CV-1 cells in 35-mm-diameter tissue culture plates were infected with vac-T7 at a multiplicity of infection of 10 in OptiMEM (Gibco Life Technologies, Grand Island, N.Y.) with 0.1% bovine serum albumin for 1 h at 37°C. Plasmids (1 μg per plate) were transfected using liposomes made in our laboratory (39), and the cells were incubated for an additional 5 h posttransfection (h.p.t.) at 37°C.
Transient expression from pCAGGS plasmids was performed by transfecting 1 μg of plasmid DNA into HeLa-T4 cells in 35-mm-diameter plates using Lipofectamine Plus (Gibco Life Technologies) according to the manufacturer's protocol. Transfected cells were incubated for 18 h at 37°C prior to metabolic labeling.
For pulse-chase labeling, the cells were incubated for 30 min with DME medium deficient in methionine and cysteine (Met-Cys-DME), containing 20 mM HEPES (pH 7.1), glutamine and P-S. Cultures were labeled metabolically by replacing the medium with Met-Cys-DME containing 20 mM HEPES (pH 7.1)–glutamine–P-S and containing 35S-labeled Pro-mix (50 μCi per dish for pCAGGS or vac-T7-mediated expression or 100 μCi per dish for influenza C virus-infected cells) for 15 min at 37°C. The vac-T7 expression experiments in Fig. 1 were labeled metabolically with Met-Cys-DME containing 20 mM HEPES (pH 7.1)–glutamine–2 mM cysteine–P-S and 100 μCi of [35S]methionine per dish. The chase periods were initiated by replacing the medium with chase medium (DME medium with glutamine, 2 mM cysteine, 2 mM methionine, and P-S) and cells incubated at 37°C for various times. Cell monolayers were lysed in RIPA buffer (10 mM Tris [pH 7.4], 1% deoxycholate, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS]) (25) containing 50 mM iodoacetamide and protease inhibitors (35). The lysates were clarified by centrifugation for 15 min at 55,000 rpm in a Beckman TLA100.2 rotor and stored at −70°C. Lactacystin (Calbiochem, La Jolla, Calif.) was dissolved in ethanol at a concentration of 2 mM and stored at −20°C.
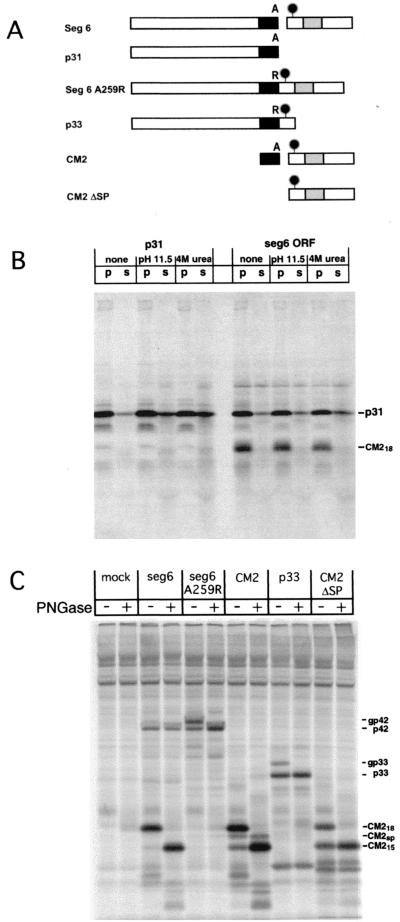
FIG. 1.
(A) Schematic diagram of influenza C virus RNA seg 6 ORF-derived cDNA constructs. Black rectangles indicate the internal signal peptide (amino acids 239 to 259), gray rectangles indicate the predicted transmembrane domain of CM2 (amino acids 285 to 308), and the black circles indicate the one utilized glycosylation site in the seg 6 ORF. The letter A (Ala) or R (Arg) indicates the amino acid at position 259, which is the −1 position relative to the predicted signal peptidase cleavage site. (B) The membrane association of p31 does not require cleavage from the p42 precursor protein. Bacteriophage T7 RNA polymerase-transcribed RNAs encoding p31 alone or seg 6 ORF were translated in the presence of canine pancreatic microsomes and subjected to the indicated treatments. The reaction mixtures were then subjected to ultracentrifugation, and the pelleted microsomes (p) were resuspended in SDS-PAGE sample buffer. The supernatants (s) were precipitated with trichloroacetic acid as described (37), before resuspension in SDS-PAGE sample buffer. The polypeptides were separated by SDS-PAGE on a 15% acrylamide gel. (C) Efficient membrane insertion requires the internal signal peptide and the CM2 transmembrane domain. HeLa-T4 cells in 35-mm-diameter tissue culture dishes were infected with vac-T7 for 1 h and then transfected with 1 μg of plasmid encoding the indicated cDNAs under control of the bacteriophage T7 RNA polymerase promoter. At 5 h.p.t., the cells were incubated in Met-Cys-DME for 30 min prior to pulse-labeling for 15 min with Met-Cys-DME containing [35S]methionine, followed by incubation for 5 min in chase medium. Cells were lysed, cellular debris was pelleted by centrifugation, and proteins were immunoprecipitated with antiserum specific for CM2 (seg 6, seg 6 A259R, CM2, and CM2 ΔSP) or CM1 (p33). The samples were divided in half and incubated without (−) or with (+) PNGase to remove N-linked carbohydrates before analysis of polypeptides by SDS-PAGE on 15% acrylamide gels. The CM2sp protein contains the signal peptide but no N-linked carbohydrates, while CM218 protein is cleaved and glycosylated (36).
Glycosylation efficiency was calculated by the formula [glycosylated protein/(unglycosylated precursor + glycosylated protein)] × 100. For calculation of the efficiency of expression of CM2 from the seg 6 ORF cDNA, the p42 radioactive signal was adjusted to reflect the amount of radioactive signal derived from the CM2 coding region of the protein (4 Met residues in mature CM2 compared to 19 Met residues in p42).
p31 antiserum.
A synthetic peptide identical to the 17-amino-acid sequence (amino acids 253 to 259) present at the C terminus of p31 but not in CM1 and containing an additional Cys residue (Cys-Trp-Leu-Val-Val-Ile-Ile-Cys-Phe-Ser-Ile-Thr-Ser-Gln-Pro-Ala-Ser-Ala) was coupled to keyhole limpet hemocyanin (Pierce, Rockford, Ill.) and used to immunize female New Zealand White rabbits (Covance Research Products, Denver, Pa.) as previously described (36).
Immunoprecipitation and glycosidase treatment.
Immunoprecipitations were performed essentially as described previously (35–37) using a rabbit serum specific for the CM2 cytoplasmic tail (final dilution, 1:100), a rabbit serum specific for p31 (final dilution 1:100), a rabbit serum specific for CM1 (final dilution, 1:50), a rabbit serum specific for CAT (final dilution, 1:100; 5 prime-3 prime, Boulder, Co.) or a MAb hybridoma (M55) supernatant specific for influenza A virus M1 protein (final dilution, 1:50) (J. Zhang and R. A. Lamb, unpublished data). To remove N-linked carbohydrate modifications from proteins, lysates were immunoprecipitated and digested for 16 h at 37°C with 0.2 U of peptide-N-glycosidase F (PNGase) (Boehringer Mannheim Corporation, Indianapolis, Ind.). Samples were then boiled in polyacrylamide gel electrophoresis (PAGE) sample buffer (35).
SDS-PAGE.
Polypeptides were analyzed on 15% acrylamide gels, as described previously (35). 14C-labeled Mr standards (Amersham Life Sciences) were loaded on gels to approximate molecular weights. The gels were fixed and dried, and radioactivity was analyzed using a Fuji BioImager 1000 and MacBas software (Fuji Medical System, Stamford, Conn.) or gels were exposed to X-Omat AR film (Eastman Kodak Co., Rochester, N.Y.) at −70°C.
Indirect immunofluorescence.
HeLa-T4 cells on glass coverslips were transfected with 1.0 μg of pCAGGS-A/M1, pCAGGS-A/M1-C17, pCAGGS-CM1, or pCAGGS-p31 plasmid DNA per 35-mm-diameter tissue culture plate and incubated for 18 h at 37°C. All subsequent steps were performed at room temperature. Cell monolayers were washed twice with phosphate-buffered saline (PBS), fixed in 1% formaldehyde in PBS for 10 min, and permeabilized with PBS containing 0.1% saponin for 10 min. A blocking solution consisting of PBS with 5% normal goat serum (Sigma Chemical Co., St. Louis, Mo.), 0.1% bovine serum albumin and 0.1% saponin was added for 30 min. The M55 MAb (anti-A/M1; final dilution, 1:10) or G32 MAb (anti-CM1; final dilution, 1:500) was added and incubated for 1 h. The cells were washed with PBS containing 0.1% saponin before addition of fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG (Jackson ImmunoResearch Laboratories, Inc., West Grove, Pa.) at a final dilution of 1:500 for 30 min. After washing with PBS containing 0.1% saponin, the coverslips were mounted onto glass slides using Vectashield (Vector Laboratories, Burlingame, Calif.) and examined on a Zeiss LSM 410 confocal microscope (Carl Zeiss Inc., Thornwood, N.Y.). All antibody dilutions were made in blocking buffer.
RESULTS
Together, the internal signal peptide and the CM2 transmembrane domain mediate efficient translocation across the endoplasmic reticulum (ER) membrane.
As compared to CM1, the p31 protein shows a much greater affinity for microsomal membranes when the proteins were translated in vitro in the presence of canine pancreatic microsomes (37). The association of p31 with membranes most likely occurs through the hydrophobic 17-amino-acid C terminus of p31, as the CM1 protein (which does not contain this amino acid sequence) dissociates from microsomes in the presence of 4 M urea, 2 M KCl, or under alkaline (pH 11.5) conditions (37). To determine whether microsome association requires the proteolytic cleavage of p31 from p42 or whether it is an intrinsic property of the p31 protein, RNAs encoding p31 or seg 6 ORF (Fig. 1A) were translated in vitro in the presence of microsomal membranes. Translation reactions were reacted under conditions (pH 11.5 or 4 M urea) which dissociate peripheral but not integral membrane proteins from microsomes, and the amount of the membrane-associated versus the membrane-dissociated p31 protein was determined (Fig. 1B). The p31 protein associated strongly with microsomes when expressed alone (90% associated in untreated lanes, 79% associated after pH 11.5 treatment, and 71% associated after 4 M urea treatment) or from the seg 6 ORF cDNA (84% associated in untreated lanes, 77% associated after pH 11.5 treatment, and 72% associated after 4 M urea treatment).
To determine whether the p31 C-terminal 17-amino-acid region completely traverses or simply binds to the ER membrane, a series of truncation-deletion mutants were constructed, utilizing the N-linked carbohydrate addition site at Asn 270 (36) as a marker for complete translocation across the ER membrane (Fig. 1A). Plasmids encoding the indicated cDNAs were transfected into vac-T7-infected HeLa-T4 cells and metabolically labeled at 5 h.p.t., and the cells were lysed after a 5-min incubation in chase medium. To identify glycosylated species, one-half of the immunoprecipitated polypeptides were treated with PNGase to remove N-linked carbohydrates before analysis by SDS-PAGE (Fig. 1C). Approximately 97% of the p42 protein was processed into glycosylated CM2 (CM218) in cells transfected with the seg 6 ORF cDNA, indicating very efficient translocation of the p42 protein into the ER membrane. Deletion of the upstream 238 amino acids of p42 (cDNA CM2) had little effect on ER translocation, as 88% of CM2 was glycosylated (CM218 compared to CM2sp and CM215). Elimination of the signal peptide cleavage site in p42 by substituting an Arg for the Ala at residue 259 (seg 6 A259R), resulted in glycosylation of 60% of the translated polypeptide (gp42 as compared to p42). This reduction in glycosylation efficiency may be due to inaccessibility of Asn 270 to the ER glycosylation machinery because of steric constraints (see below and Fig. 2A), because after in vitro translation in the presence of microsomes, the p42 protein possesses characteristics of an integral membrane protein (37).
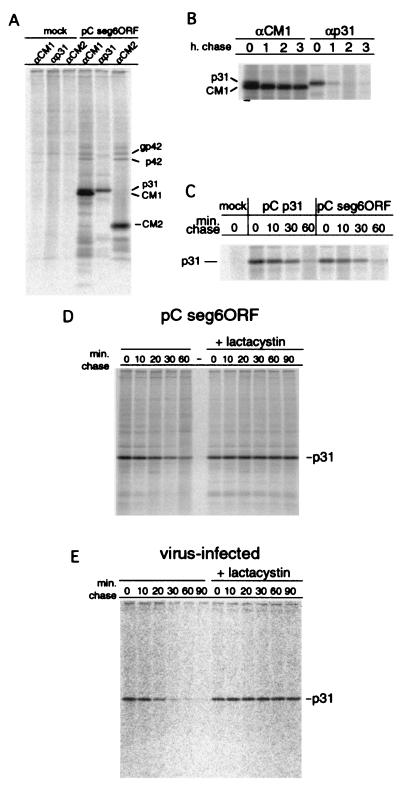
FIG. 2.
The p31 protein is degraded rapidly both after expression from cDNA and after expression in influenza C virus-infected cells. HeLa-T4 cells in 35-mm-diameter tissue culture dishes were transfected with 1 μg of plasmid DNA or mock transfected. At 18 h.p.t., the cells were incubated in Met-Cys-DME for 30 min, labeled metabolically with 35S-labeled Pro-mix for 15 min, and incubated in chase medium for the indicated times. The cells were lysed, cellular debris was pelleted by centrifugation, and the proteins were immunoprecipitated with a rabbit serum specific for CM1 (αCM1) (37), a rabbit serum specific for p31 (αp31), or a rabbit serum specific for the CM2 cytoplasmic tail (αCM2) (36). Polypeptides were analyzed by SDS-PAGE on 15% acrylamide gels. (A) Mock and pC seg 6 ORF transfected (5-min chase). (B) p 6 seg ORF transfected. (C) mock, pC p31, and pC seg 6 ORF transfected. (D) HeLa-T4 cells in 35 mm-diameter tissue culture dishes were transfected with 1 μg of pC seg 6 ORF plasmid, starved of Met and Cys at 18 h.p.t., labeled metabolically with 35S-labeled Pro-mix, and incubated in chase medium for the indicated number of minutes. Proteins were immunoprecipitated and analyzed by SDS-PAGE as described above. The 26S proteasome inhibitor lactacystin was added (10 μM final concentration) to the starve, pulse, and chase media as indicated. (E) MDCK cells were infected with influenza C/AA virus. At 20 h postinfection the cells were starved of Met and Cys, labeled metabolically with 35S-labeled Pro-mix, and incubated in chase medium for the indicated number of minutes. Proteins were immunoprecipitated and analyzed by SDS-PAGE as described above. The 26S proteasome inhibitor lactacystin was added (10 μM final concentration) to the starve, pulse, and chase media as indicated. Samples in panels C to E were immunoprecipitated with a rabbit serum specific for p31.
The ER translocation of proteins containing either the p42 internal signal peptide or the CM2 transmembrane domain alone was then assessed (Fig. 1C). The p33 cDNA, which encodes the first 274 amino acids of p42 as well as an Ala-to-Arg substitution at amino acid 259, was glycosylated at 20% efficiency (gp33 compared to p33), indicating only limited translocation of the C-terminal region. A cDNA containing the coding region for amino acids 260 to 374 (CM2 ΔSP) was glycosylated at 52% efficiency (CM218 compared to CM215), indicating that only half of the newly synthesized polypeptides were inserted into the ER membrane in the proper orientation. Taken together, these data indicate that proper and efficient membrane translocation occurs only with proteins containing both the internal signal peptide and the CM2 transmembrane domain. In addition, translocation of p42 occurred cotranslationally and required an N-ethyl maleimide-sensitive component in the canine pancreatic microsomes (data not shown), presumably the signal recognition particle (SRP).
Rapid, proteasome-dependent degradation of p31 when expressed either from cDNA or in influenza C virus-infected cells.
An antiserum specific to the p31 protein was generated by immunizing rabbits with a peptide corresponding to the 17 C-terminal amino acids unique to p31 compared to the CM1 protein. HeLa-T4 cells were transfected with a eukaryotic expression plasmid containing the seg 6 ORF or mock transfected, followed by metabolic labeling at 18 h.p.t. The cells were incubated in chase medium for 5 min before lysis and immunoprecipitation with rabbit serum specific for recombinant CM1 protein (37), a peptide corresponding to the cytoplasmic tail of CM2 (36), or the 17-amino-acid peptide corresponding to the C terminus of p31 (Fig. 2A). The CM1-specific serum immunoprecipitated two major polypeptide species from pC seg 6 ORF cDNA transfected cells: CM1 (faster migrating, predominant species) and p31 (slower migrating, less abundant species). The p31 C terminus-specific serum immunoprecipitated exclusively the slower migrating band, indicating a high specificity for p31. A small amount of p42 protein was immunoprecipitated by the CM1- and CM2-specific serum, but not by the p31-specific serum (Fig. 2A). The reason for the lack of reactivity of the p31 serum with p42 is not known, but the internal signal peptide may not be accessible to antibodies due to steric hindrance when the CM2 transmembrane domain is present in the same polypeptide chain (Fig. 1C). No major polypeptides were immunoprecipitated from mock-transfected cells by any of the antisera.
The stability of p31, when expressed from pC seg 6 ORF, was determined by transfecting HeLa-T4 cells with pC seg 6 ORF and pulse-labeling the cells at 18 h.p.t., followed by incubation in chase medium for varying periods. The cells were lysed and the proteins were immunoprecipitated with CM1- or p31-specific serum, and polypeptides were analyzed by SDS-PAGE. The CM1 protein was detected readily following incubation in chase medium for up to 3 h, whereas p31 was difficult to detect after a 1-h chase period (Fig. 2B). To determine if the reduction in the amount of p31 correlated with cleavage from p42, HeLa-T4 cells were transfected with pC p31 or pC seg 6 ORF, pulse-labeled at 18 h.p.t., and incubated in chase medium for the indicated number of minutes. The cells were lysed, the proteins were immunoprecipitated with p31-specific serum, and the polypeptides were separated by SDS-PAGE. The amount of p31 protein decreased rapidly over 60 min of incubation in chase medium when expressed alone or as a product of p42 proteolytic cleavage (Fig. 2C). In either case, one-half of the p31 present after the pulse-label was undetectable after approximately 20 min.
The most likely explanation for the loss of antibody-reactive p31 is rapid degradation. The vast majority of proteolytic degradation in eukaryotic cells occurs by the 26S proteasome pathway; therefore, the effect of the 26S proteasome inhibitor lactacystin (8, 9) on the stability of p31 was determined. HeLa-T4 cells were transfected with pC seg 6 ORF, pulse-labeled at 18 h.p.t., and incubated in chase medium for the indicated number of minutes; the cells were lysed; the proteins were immunoprecipitated with p31-specific serum; and the polypeptides were separated by SDS-PAGE. It was found that addition of lactacystin to pC seg 6 ORF-transfected HeLa-T4 cells prevented the loss of p31 (Fig. 2D), suggesting the protein was degraded via the 26S proteasome pathway. In influenza C virus-infected MDCK cells, it was found that p31 was degraded with similar kinetics (20 to 25 min for 50% of the protein to be degraded) as when expressed from cDNA and that lactacystin prevented p31 degradation (Fig. 2E).
Transfer of the 17-amino-acid C terminus of p31 to the C terminus of influenza A virus M1 protein or the CAT protein results in rapid degradation.
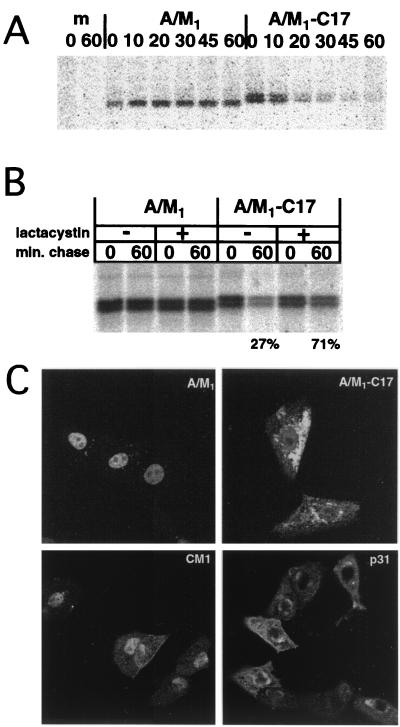
As the 17 C-terminal residues of p31 distinguish the rapidly degraded p31 protein from the relatively stable CM1 protein, it seemed plausible that these residues contained a degradation signal. To test this hypothesis, a chimeric protein, A/M1-C17 was constructed which contained the 17-amino-acid C terminus of p31 added to the C terminus of the influenza A virus M1 protein. The M1 protein was chosen as a reporter protein because it has similar biochemical properties to the CM1 protein but it has little sequence identity (5, 56). HeLa-T4 cells were transfected with pC A/M1 or pC A/M1-C17, pulse-labeled at 18 h.p.t., incubated in chase medium for the indicated number of minutes, and lysed. The proteins were immunoprecipitated with an M1-specific MAb, and the polypeptides were separated by SDS-PAGE. The M1-C17 chimeric protein was observed to undergo rapid degradation compared to the M1 protein (Fig. 3A), suggesting that the 17-residue C-terminal region of p31 can serve as a degradation signal when transferred to another protein. The M1-C17 protein migrated as a doublet for unknown reasons. The proteasome inhibitor lactacystin inhibited the degradation of A/M1-C17 (27% A/M1-C17 remaining without lactacystin, 71% remaining with lactacystin) (Fig. 3B), indicating the protein was degraded by the 26S proteasome.
FIG. 3.
Transfer of the p31 C-terminal region to the influenza A virus M1 protein results in rapid degradation and changes in M1 protein intracellular distribution. (A) HeLa-T4 cells in 35-mm-diameter tissue culture dishes were transfected with 1 μg of pCAGGS A/M1 (pC A/M1) or pCAGGS A/M1-C17 (pC A/M1-C17) or were mock transfected (m). At 18 h.p.t., the cells were incubated in Met-Cys-DME for 30 min, labeled metabolically with 35S-labeled Pro-mix for 15 min, and incubated in chase medium for the indicated times. The cells were lysed, cellular debris was pelleted by centrifugation, the proteins were immunoprecipitated with an M1-specific MAb (M55), and polypeptides were analyzed by SDS-PAGE on 15% acrylamide gels. (B) The experiment was performed as for panel A, except for inclusion of the proteasome inhibitor lactacystin (final concentration, 10 μM) in the starve, pulse, and chase media as indicated. (C) HeLa-T4 cells grown on glass coverslips in 35-mm-diameter tissue culture dishes were transfected with 1 μg of pC A/M1, pC A/M1-C17, pC CM1, or pC p31 and at 18 h.p.t. were fixed with 1% formaldehyde. The cells were permeabilized and incubated with an M1-specific MAb (pC A/M1 and pC A/M1-C17) or a CM1-specific MAb (pC CM1 and pC p31) followed by a goat α-mouse FITC-conjugated secondary antibody, and staining was visualized by confocal microscopy.
The intracellular localization of the proteins was determined by transfecting HeLa-T4 cells grown on glass coverslips with the pC A/M1, pC A/M1-C17, pC CM1, or pC p31 plasmids. At 18 h.p.t., the cells were fixed with 1% formaldehyde, permeabilized, and incubated with an M1- or CM1-specific MAb followed by a goat anti-mouse FITC-conjugated secondary antibody, and staining was visualized by confocal microscopy. The M1 and CM1 proteins localized predominantly to the nucleus of cells after expression from cDNA. In contrast, the A/M1-C17 and p31 proteins localized to the cytosol with some perinuclear staining evident (Fig. 3C). The intracellular localization was not altered upon incubation with lactacystin for 2 h before analysis (data not shown). Thus, the data suggest that the presence of the C-terminal 17 amino acids of p31 alone is responsible for increased degradation and altered intracellular localization of the p31 and A/M1-C17 proteins.
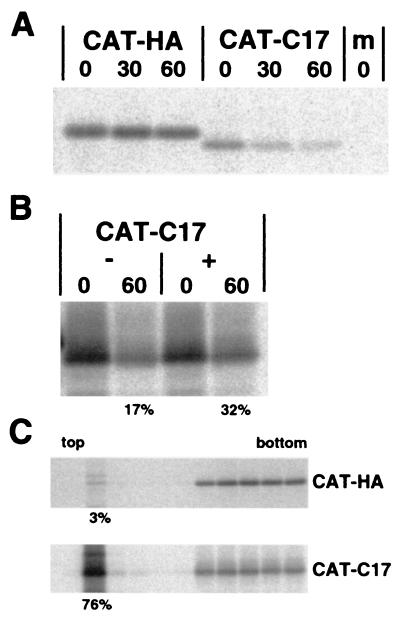
The ability of the C-terminal 17 amino acids of p31 to mediate degradation of an unrelated, nonviral protein was examined by adding this sequence to the 3′ end of the CAT ORF. As a control for C-terminal tagging, the epitope for the anti-influenza A virus HA MAb 12CA5 was added to CAT. CAT-C17 was rapidly degraded compared to the CAT-HA protein (Fig. 4A). However, this degradation was only partially inhibited by lactacystin (17% of CAT-C17 remaining without lactacystin, 32% remaining with lactacystin after a 60-min incubation in chase medium) (Fig. 4B), suggesting the CAT-C17 protein was being degraded by at least one other protease besides the 26S proteasome. As seen in Fig. 4A, the CAT-C17 protein consistently migrated faster than CAT-HA when analyzed by SDS-PAGE, despite its being eight amino acids longer. The ability of CAT-C17 and CAT-HA to associate with membranes was analyzed by sucrose gradient flotation of cellular membranes treated under alkaline (pH 11.5) conditions. The CAT-C17 protein was tightly associated with membranes (76% membrane associated) (Fig. 4C) compared to CAT-HA (3% membrane associated), providing further evidence that the C17 sequence serves as a membrane-targeting and degradation signal.
FIG. 4.
Transfer of the C-terminal 17 amino acids of p31 to the CAT protein results in increased membrane binding and rapid degradation which is partially inhibited by lactacystin. (A) CV-1 cells in 35-mm-diameter tissue culture dishes were infected with vac-T7 for 1 h and then transfected with 1 μg of plasmid encoding the indicated cDNAs under control of the bacteriophage T7 RNA polymerase promoter. At 5 h.p.t., the cells were incubated in Met-Cys-DME for 30 min prior to pulse-labeling for 15 min with Met-Cys-DME containing 50 μCi of 35S-labeled Pro-mix per dish. The cells were incubated in chase medium for the indicated number of minutes and lysed, cellular debris was pelleted by centrifugation, and proteins were immunoprecipitated with antisera specific for CAT. (B) The experiment was performed as in panel A, except for inclusion of the proteasome inhibitor lactacystin (final concentration, 10 μM) in the starve, pulse, and chase media as indicated. (C) CV-1 cells in 60-mm-diameter tissue culture dishes were infected with vac-T7 for 1 h and then transfected with 2.5 μg of plasmid encoding the indicated cDNAs under control of the bacteriophage T7 RNA polymerase promoter. At 5 h.p.t., the cells were incubated in Met-Cys-DME for 30 min prior to pulse-labeling for 30 min with Met-Cys-DME containing 50 μCi of 35S-labeled Pro-mix per dish. The cells were washed with PBS, swelled in hypotonic buffer (10 mM Tris-HCl [pH 7.4], 1 mM MgCl2), resuspended by scraping, and disrupted with 25 strokes in a Dounce homogenizer. Na2CO3 (pH 11.5) was added to a final concentration of 100 mM and incubated for 30 min at 4°C. The suspension was adjusted to 65% sucrose (4-ml volume), transferred to an SW41 centrifuge tube, and overlaid with 4 ml of 35% sucrose and 2 ml of 10% sucrose. After ultracentrifugation at 35,000 rpm for 18 h in an SW41 rotor, the samples were fractionated from the top (1 ml per fraction), adjusted to 1× RIPA buffer, and immunoprecipitated with an anti-CAT rabbit serum, and the polypeptides were separated by SDS-PAGE on 15% acrylamide gels.
Degradation of p31 requires the presence of a core group of hydrophobic amino acids within its 17-residue C terminus.
The specific nature of the degradation signal in the 17-residue C terminus of p31 was characterized further by constructing a set of mutant p31 proteins (Fig. 5; also see Materials and Methods). HeLa-T4 cells were transfected with the cDNAs, and cells were pulse-labeled at 18 h.p.t. and incubated in chase medium for the indicated number of minutes. The cells were lysed, proteins were immunoprecipitated with CM1-specific serum, and polypeptides were analyzed by SDS-PAGE. The degradation kinetics of mutant p31 proteins containing truncations of four or eight residues from the C terminus of p31 was found to be similar to that of wild-type p31 (Fig. 5). In contrast, deletion of 13 residues from the p31 C terminus, which deletes a significant number of hydrophobic residues, resulted in a protein that was relatively stable with 65% of the initial protein remaining after a 60-min incubation in chase medium. To determine the contribution of the core nine hydrophobic amino acids in the C terminus of p31 towards protein degradation, these residues were replaced with either nine leucine residues (p31-Leu), the core hydrophobic amino acids of the C-terminal membrane-anchoring sequence of synaptobrevin (p31-SB), or the hydrophilic amino acids comprising the epitope for the FLAG MAb (p31-FLAG). As shown in Fig. 5, the p31-Leu and p31-SB protein constructs were degraded rapidly while the p31-FLAG construct was quite stable over 60 min of incubation in chase medium.
FIG. 5.
A core group of hydrophobic amino acids signals the rapid degradation of p31. The altered p31 proteins and their C-terminal residues are indicated, with amino acid changes or insertions underlined. All mutations were made in the p31 cDNA except for p31-hinge, which was constructed in the seg 6 ORF cDNA. HeLa-T4 cells in 35-mm-diameter tissue culture dishes were transfected with 1 μg of pCAGGS expression vector containing the indicated cDNAs. At 18 h.p.t., the cells were incubated in Met-Cys-DME for 30 min, labeled metabolically with 35S-labeled Pro-mix for 15 min, and incubated in chase medium for the indicated times (minutes). The cells were lysed, cellular debris was pelleted by centrifugation, proteins were immunoprecipitated with a CM1-specific rabbit serum, and polypeptides were analyzed by SDS-PAGE on 15% acrylamide gels. Quantitation of the total remaining protein at 60 min of chase relative to the amount present at 0 min of chase is shown (% remaining). Immunoprecipitations were performed with a rabbit serum specific for CM1 for all mutant proteins except p31-hinge, for which a rabbit serum specific for p31 was used.
The data suggest that only the hydrophobic nature and not the specific amino acid sequence of the p31 C terminus is required to mediate rapid protein degradation. It is not known if the CM1 protein domain within p31 folds into the same conformation as native CM1 protein. It is possible that the native folding of the CM1 domain is prevented in p31 due to close proximity to the membrane and thus the entire protein is misfolded. At the present time conformation-specific antibodies to probe the folded state of CM1 are not available. Thus, in a rational attempt to allow proper folding of the CM1 protein, a proline-rich flexible linker region from IgG was inserted between the C terminus of CM1 and the 17 residues specific to the C terminus of p31. It was found that insertion of the flexible hinge domain did not prevent the rapid degradation of the protein (Fig. 5). Taken together, the data suggest that a core group of seven hydrophobic residues signals the rapid degradation of p31.
DISCUSSION
The influenza C virus RNA segment 6 colinear mRNA transcript which contains the seg 6 ORF (374 amino acid residues) is translated to yield a precursor protein, p42, which on insertion into microsomal membrane is subsequently cleaved by signal peptidase to form the membrane associated N-terminal p31 protein and the CM2 integral membrane protein (18, 37). The efficient membrane translocation of p42 (i) requires the internal signal peptide that becomes the C terminus of p31; (ii) requires the hydrophobic domain that becomes the transmembrane domain of CM2; (iii) requires an N-ethyl maleimide sensitive component, presumably the SRP; and (iv) occurs cotranslationally in vitro. Taken together, the data indicate the p42 protein undergoes insertion into the ER membrane via the classical pathway utilized by virtually all mammalian cell proteins containing signal peptides or signal anchor sequences (reviewed in reference 22): this is despite the uncommon position of the signal peptide, which is located 239 amino acids from the N terminus of the protein.
When expressed alone from cDNA, p31 associates tightly with membranes. This interaction appears to occur between the hydrophobic C-terminal region of p31 and the inner leaflet of the lipid bilayer. The p33 protein construct, which consists of a 16-residue C-terminal extension to p31 (as well as an amino acid substitution which eliminates the signal peptidase cleavage site) underwent N-linked glycosylation poorly, suggesting that complete translocation of the C-terminal region did not occur efficiently. In contrast, members of the SNARE family of cellular proteins possess C-terminal hydrophobic domains which anchor them to membranes. Insertion of these proteins into the ER membrane occurs via an SRP-independent, posttranslational pathway which requires ATP (24, 49). The C-terminal hydrophobic domains of SNAP-25 and synaptobrevin span the lipid bilayer, as judged by the efficient utilization of N-linked glycosylation sites engineered into the C terminus. Determining the structural and/or sequence differences between the p31 and SNARE protein C-terminal domains which mediate complete translocation across the lipid bilayer could provide some insights into the mechanism responsible for the anchoring of SNARE proteins to lipid bilayers.
Interestingly, when the CM2 protein was expressed without an N-terminal signal sequence, approximately half of the molecules were glycosylated, implying the protein behaves as a type III integral membrane protein. This is reminiscent of the behavior of the influenza A virus M2 and the influenza B virus NB integral membrane proteins, neither of which contains N-terminal signal sequences and which both contain only a single hydrophobic domain, yet they are oriented in the plasma membrane in an Nout-Cin type I integral membrane protein orientation (28, 52).
We have speculated previously that the requirement of both the signal peptide and transmembrane domain for efficient translocation may reflect the formation of a loop intermediate which interacts with the Sec61 translocon (37). The data presented here continue to support this hypothesis. The reduced glycosylation of p42 as well as the inability of a rabbit serum generated against a peptide corresponding to the C-terminal 17 amino acids of p31 (the internal signal peptide) to immunoprecipitate p42 could result from severe structural constraints placed on the amino acid sequence between the signal peptide and the CM2 transmembrane domain when cleavage after amino acid 259 does not occur.
The p31 protein underwent rapid degradation after cleavage from p42. Cleavage at the signal peptidase consensus sequence is not required for degradation, as p31 expressed alone or p31 expressed from the seg 6 ORF cDNA were degraded with nearly identical kinetics. The 17-amino-acid C-terminal region of p31 was able to target the influenza A virus M1 protein for rapid degradation when fused in frame to the C terminus of the M1 protein, suggesting this 17-residue sequence contains a transferable degradation signal. Since the M1 protein itself binds tightly to membranes (55), we speculate that the addition of the C-terminal 17 amino acids of p31 to M1 changes the nature of the membrane binding of the chimeric protein, rather than simply increasing the membrane affinity of the protein. This degradation signal does not have to reside at the C terminus of the protein to mediate degradation, as the p42 protein itself undergoes rapid degradation (data not shown), but this has not been explored rigorously. Extensive mutagenesis of the 17 amino acid p31 C-terminal region suggests the signal specifying degradation is a region of hydrophobicity and not a specific amino acid sequence. It is possible that the hydrophobic nature of the 17-amino-acid sequence or the tight membrane association of proteins containing this amino acid sequence may preclude proper folding of other domains of the protein. The increased membrane binding and rapid degradation of the CAT-C17 chimeric protein lends support to this hypothesis. However, a flexible hinge known in other constructs to permit independent folding of linked domains (2, 42), placed between the 17-amino-acid C terminus of p31 and the coding region of CM1 did not change significantly the degradation rate of the chimeric protein.
The degradation of p31 is inhibited by lactacystin, indicating involvement of the 26S proteasome pathway. However, we were unable to detect polyubiquitinated forms of p31, even in the presence of proteasome inhibitors. Virtually all proteins that undergo proteasome-mediated degradation acquire polyubiquitin chains on the amide groups of lysine residues, (reviewed in reference 3). However, ornithine decarboxylase (29, 32, 47) and Stat5 (48) as well as p21Cip1 (41) appear to be degraded via the 26S proteasome in an ubiquitin-independent manner. Whether p31 degradation occurs via a similar mechanism requires further investigation. The degradation of integral membrane proteins such as the major histocompatibility complex class I heavy chain proteins is mediated by the cytomegalovirus US2 and US11 genes (43, 50, 51), as well as the herpes simplex virus ICP47 gene (10, 15, 20). This process requires the export or retrograde translocation of the major histocompatibility complex class I proteins from the ER membrane into the cytoplasm, where proteasome-mediated degradation occurs (reviewed in reference 23). Whether ATP or Sec61 is required to actively dislocate p31 from cellular membranes before degradation by the 26S proteasome has not been explored.
Lastly, the detection of p31 in influenza C virus-infected cells further supports proteolytic cleavage of p42 as the mechanism for CM2 production. While we have been unable to demonstrate the presence of p42 in influenza C virus-infected cells (data not shown), it has been detected by others (16). The rapid degradation of p31 may indicate the protein does not have a function in the influenza C virus life cycle. Proteolytic cleavage of CM2 from a polypeptide precursor, followed by rapid degradation of the upstream polypeptide species represents a method of gene regulation not utilized by any other member of the Orthomyxoviridae family. Therefore, it provides yet another coding strategy employed by these viruses to maximize their genome coding capacity while minimizing genome size.
ACKNOWLEDGMENTS
We thank all members of the Lamb laboratory for useful discussions and K. Nakamura, Yamagata University School of Medicine, Yamagata, Japan, for the generous gift of anti-CM1 MAbs.
This research was supported by research grant R37 AI-20201 (R.A.L.) and fellowship F32 AI-10382 (A.P.) from the National Institute of Allergy and Infectious Diseases. A.P. was an Associate and R.A.L. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Air G M, Laver W G, Luo M, Stray S J, Legrone G, Webster R G. Antigenic, sequence, and crystal variation in influenza B neuraminidase. Virology. 1990;177:578–587. doi: 10.1016/0042-6822(90)90523-t. [DOI] [PubMed] [Google Scholar]
- 2.Boerger A L, Snitkovsky S, Young J A. Retroviral vectors preloaded with a viral receptor-ligand bridge protein are targeted to specific cell types. Proc Natl Acad Sci USA. 1999;96:9867–9872. doi: 10.1073/pnas.96.17.9867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonifacino J S, Weissman A M. Ubiquitin and the control of protein fate in the secretory and endocytic pathways. Annu Rev Cell Dev Biol. 1998;14:19–57. doi: 10.1146/annurev.cellbio.14.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bui M, Whittaker G, Helenius A. Effect of M1 protein and low pH on nuclear transport of influenza virus ribonucleoproteins. J Virol. 1996;70:8391–8401. doi: 10.1128/jvi.70.12.8391-8401.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buonagurio D A, Nakada S, Fitch W M, Palese P. Epidemiology of influenza C virus in man: multiple evolutionary lineages and low rate of change. Virology. 1986;153:12–21. doi: 10.1016/0042-6822(86)90003-6. [DOI] [PubMed] [Google Scholar]
- 6.Ciampor F, Bayley P M, Nermut M V, Hirst E M, Sugrue R J, Hay A J. Evidence that the amantadine-induced, M2-mediated conversion of influenza A virus hemagglutinin to the low pH conformation occurs in an acidic trans Golgi compartment. Virology. 1992;188:14–24. doi: 10.1016/0042-6822(92)90730-d. [DOI] [PubMed] [Google Scholar]
- 7.Ciampor F, Thompson C A, Grambas S, Hay A J. Regulation of pH by the M2 protein of influenza A viruses. Virus Res. 1992;22:247–258. doi: 10.1016/0168-1702(92)90056-f. [DOI] [PubMed] [Google Scholar]
- 8.Dick L R, Cruikshank A A, Destree A T, Grenier L, McCormack T A, Melandri F D, Nunes S L, Palombella V J, Parent L A, Plamondon L, Stein R L. Mechanistic studies on the inactivation of the proteasome by lactacystin in cultured cells. J Biol Chem. 1997;272:182–188. doi: 10.1074/jbc.272.1.182. [DOI] [PubMed] [Google Scholar]
- 9.Fenteany G, Standaert R F, Lane W S, Choi S, Corey E J, Schreiber S L. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 10.Fruh K, Ahn K, Djaballah H, Sempe P, van Endert P M, Tampe R, Peterson P A, Yang Y. A viral inhibitor of peptide transporters for antigen presentation. Nature. 1995;375:415–418. doi: 10.1038/375415a0. [DOI] [PubMed] [Google Scholar]
- 11.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grambas S, Bennett M S, Hay A J. Influence of amantadine resistance mutations on the pH regulatory function of the M2 protein of influenza A viruses. Virology. 1992;191:541–549. doi: 10.1016/0042-6822(92)90229-i. [DOI] [PubMed] [Google Scholar]
- 13.Grambas S, Hay A J. Maturation of influenza A virus hemagglutinin—estimates of the pH encountered during transport and its regulation by the M2 protein. Virology. 1992;190:11–18. doi: 10.1016/0042-6822(92)91187-y. [DOI] [PubMed] [Google Scholar]
- 14.Helenius A. Unpacking the incoming influenza virus. Cell. 1992;69:577–578. doi: 10.1016/0092-8674(92)90219-3. [DOI] [PubMed] [Google Scholar]
- 15.Hill A, Jugovic P, York I, Russ G, Bennink J, Yewdell J, Ploegh H, Johnson D. Herpes simplex virus turns off the TAP to evade host immunity. Nature. 1995;375:411–415. doi: 10.1038/375411a0. [DOI] [PubMed] [Google Scholar]
- 16.Hongo S, Gao P, Sugawara K, Muraki Y, Matsuzaki Y, Tada Y, Kitame F, Nakamura K. Identification of a 374 amino acid protein encoded by RNA segment 6 of influenza C virus. J Gen Virol. 1998;79:2207–2213. doi: 10.1099/0022-1317-79-9-2207. [DOI] [PubMed] [Google Scholar]
- 17.Hongo S, Sugawara K, Muraki Y, Kitame F, Nakamura K. Characterization of a second protein (CM2) encoded by RNA segment 6 of influenza C virus. J Virol. 1997;71:2786–2792. doi: 10.1128/jvi.71.4.2786-2792.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hongo S, Sugawara K, Muraki Y, Matsuzaki Y, Takashita E, Kitame F, Nakamura K. Influenza C virus CM2 protein is produced from a 374-amino-acid protein (P42) by signal peptidase cleavage. J Virol. 1999;73:46–50. doi: 10.1128/jvi.73.1.46-50.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hongo S, Sugawara K, Nishimura H, Muraki Y, Kitame F, Nakamura K. Identification of a second protein encoded by influenza C virus RNA segment 6. J Gen Virol. 1994;75:3503–3510. doi: 10.1099/0022-1317-75-12-3503. [DOI] [PubMed] [Google Scholar]
- 20.Hughes E A, Hammond C, Cresswell P. Misfolded major histocompatibility complex class I heavy chains are translocated into the cytoplasm and degraded by the proteasome. Proc Natl Acad Sci USA. 1997;94:1896–1901. doi: 10.1073/pnas.94.5.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito T, Gorman O T, Kawaoka Y, Bean W J, Jr, Webster R G. Evolutionary analysis of the influenza A virus M gene with comparison of the M1 and M2 proteins. J Virol. 1991;65:5491–5498. doi: 10.1128/jvi.65.10.5491-5498.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson A E, van Waes M A. The translocon: a dynamic gateway at the ER membrane. Annu Rev Cell Dev Biol. 1999;15:799–842. doi: 10.1146/annurev.cellbio.15.1.799. [DOI] [PubMed] [Google Scholar]
- 23.Kopito R R. ER quality control: the cytoplasmic connection. Cell. 1997;88:427–430. doi: 10.1016/s0092-8674(00)81881-4. [DOI] [PubMed] [Google Scholar]
- 24.Kutay U, Ahnert-Hilger G, Hartmann E, Wiedenmann B, Rapoport T A. Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J. 1995;14:217–223. doi: 10.1002/j.1460-2075.1995.tb06994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamb R A, Etkind P R, Choppin P W. Evidence for a ninth influenza viral polypeptide. Virology. 1978;91:60–78. doi: 10.1016/0042-6822(78)90355-0. [DOI] [PubMed] [Google Scholar]
- 26.Lamb R A, Holsinger L J, Pinto L H. The influenza A virus M2 ion channel protein and its role in the influenza virus life cycle. In: Wimmer E, editor. Receptor-mediated virus entry into cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 303–321. [Google Scholar]
- 27.Lamb R A, Krug R M. Orthomyxoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Press; 1996. pp. 1353–1394. [Google Scholar]
- 28.Lamb R A, Zebedee S L, Richardson C D. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell. 1985;40:627–633. doi: 10.1016/0092-8674(85)90211-9. [DOI] [PubMed] [Google Scholar]
- 29.Mahaffey D, Rechsteiner M. Discrimination between ubiquitin-dependent and ubiquitin-independent proteolytic pathways by the 26S proteasome subunit 5a. FEBS Lett. 1999;450:123–125. doi: 10.1016/s0014-5793(99)00456-1. [DOI] [PubMed] [Google Scholar]
- 30.Martin K, Helenius A. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell. 1991;67:117–130. doi: 10.1016/0092-8674(91)90576-k. [DOI] [PubMed] [Google Scholar]
- 31.Martin K, Helenius A. Transport of incoming influenza virus nucleocapsids into the nucleus. J Virol. 1991;65:232–244. doi: 10.1128/jvi.65.1.232-244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, Tanaka K, Ichihara A. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature. 1992;360:597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- 33.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants by a novel eukaryotic vector. Gene. 1991;108:193–200. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 34.Ohuchi M, Cramer A, Vey M, Ohuchi R, Garten W, Klenk H-D. Rescue of vector-expressed fowl plague virus hemagglutinin in biologically active form by acidotropic agents and coexpressed M2 protein. J Virol. 1994;68:920–926. doi: 10.1128/jvi.68.2.920-926.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paterson R G, Lamb R A. The molecular biology of influenza viruses and paramyxoviruses. In: Davidson A, Elliott R M, editors. Molecular virology: a practical approach. Oxford, United Kingdom: IRL Oxford University Press; 1993. pp. 35–73. [Google Scholar]
- 36.Pekosz A, Lamb R A. The CM2 protein of influenza C virus is an oligomeric integral membrane glycoprotein structurally analogous to influenza A virus M2 and influenza B virus NB proteins. Virology. 1997;237:439–451. doi: 10.1006/viro.1997.8788. [DOI] [PubMed] [Google Scholar]
- 37.Pekosz A, Lamb R A. Influenza C virus CM2 integral membrane glycoprotein is produced from a polypeptide precursor by cleavage of an internal signal sequence. Proc Natl Acad Sci USA. 1998;95:13233–13238. doi: 10.1073/pnas.95.22.13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinto L H, Holsinger L J, Lamb R A. Influenza virus M2 protein has ion channel activity. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- 39.Rose J K, Bonagurio B, Whitt M A. A new cationic liposome reagent mediating nearly quantitative transfection of animal cells. BioTechniques. 1991;10:520–525. [PubMed] [Google Scholar]
- 40.Sakaguchi T, Leser G P, Lamb R A. The ion channel activity of the influenza virus M2 protein affects transport through the Golgi apparatus. J Cell Biol. 1996;133:733–747. doi: 10.1083/jcb.133.4.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheaf R J, Singer J D, Swanger J, Smitherman M, Roberts J M, Clurman B E. Proteasomal turnover of p21Cip1 does not require p21Cip1 ubiquitination. Mol Cell. 2000;5:403–410. doi: 10.1016/s1097-2765(00)80435-9. [DOI] [PubMed] [Google Scholar]
- 42.Snitkovsky S, Young J A. Cell-specific viral targeting mediated by a soluble retroviral receptor-ligand fusion protein. Proc Natl Acad Sci USA. 1998;95:7063–7068. doi: 10.1073/pnas.95.12.7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Story C M, Furman M H, Ploegh H L. The cytosolic tail of class I MHC heavy chain is required for its dislocation by the human cytomegalovirus US2 and US11 gene products. Proc Natl Acad Sci USA. 1999;96:8516–85121. doi: 10.1073/pnas.96.15.8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sugrue R J, Hay A J. Structural characteristics of the M2 protein of the influenza A viruses: evidence that it forms a tetrameric channel. Virology. 1991;180:617–624. doi: 10.1016/0042-6822(91)90075-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tada Y, Hongo S, Muraki Y, Sugawara K, Kitame F, Nakamura K. Evolutionary analysis of influenza C virus M genes. Virus Genes. 1997;15:53–59. doi: 10.1023/a:1007915215958. [DOI] [PubMed] [Google Scholar]
- 46.Takeuchi K, Lamb R A. Influenza virus M2 protein ion channel activity stabilizes the native form of fowl plague virus hemagglutinin during intracellular transport. J Virol. 1994;68:911–919. doi: 10.1128/jvi.68.2.911-919.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tokunaga F, Goto T, Koide T, Murakami Y, Hayashi S, Tamura T, Tanaka K, Ichihara A. ATP- and antizyme-dependent endoproteolysis of ornithine decarboxylase to oligopeptides by the 26 S proteasome. J Biol Chem. 1994;269:17382–17385. [PubMed] [Google Scholar]
- 48.Wang D, Moriggl R, Stravopodis D, Carpino N, Marine J-C, Teglund S, Feng J, Ihle J N. A small amphipathic alpha helical region is required for transcriptional activities and proteasome-dependent turnover of the tyrosine-phosphorylated Stat5. EMBO J. 2000;19:392–399. doi: 10.1093/emboj/19.3.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whitley P, Grahn E, Kutay U, Rapoport T A, von Heijne G. A 12-residue-long polyleucine tail is sufficient to anchor synaptobrevin to the endoplasmic reticulum membrane. J Biol Chem. 1996;271:7583–7586. doi: 10.1074/jbc.271.13.7583. [DOI] [PubMed] [Google Scholar]
- 50.Wiertz E J, Jones T R, Sun L, Bogyo M, Geuze H J, Ploegh H L. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell. 1996;84:769–779. doi: 10.1016/s0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- 51.Wiertz E J, Tortorella D, Bogyo M, Yu J, Mothes W, Jones T R, Rapoport T A, Ploegh H L. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature. 1996;384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- 52.Williams M A, Lamb R A. Determination of the orientation of an integral membrane protein and sites of glycosylation by oligonucleotide-directed mutagenesis: influenza B virus NB glycoprotein lacks a cleavable signal sequence and has an extracellular NH2-terminal region. Mol Cell Biol. 1986;6:4317–4328. doi: 10.1128/mcb.6.12.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamashita M, Krystal M, Palese P. Evidence that the matrix protein of influenza C virus is coded for by a spliced mRNA. J Virol. 1988;62:3348–3355. doi: 10.1128/jvi.62.9.3348-3355.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zebedee S L, Lamb R A. Influenza A virus M2 protein: monoclonal antibody restriction of virus growth and detection of M2 in virions. J Virol. 1988;62:2762–2772. doi: 10.1128/jvi.62.8.2762-2772.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang J, Lamb R A. Characterization of the membrane association of the influenza virus matrix protein in living cells. Virology. 1996;225:255–266. doi: 10.1006/viro.1996.0599. [DOI] [PubMed] [Google Scholar]
- 56.Zhirnov O P. Isolation of matrix protein M1 from influenza viruses by acid-dependent extraction with nonionic detergent. Virology. 1992;186:324–330. doi: 10.1016/0042-6822(92)90090-c. [DOI] [PubMed] [Google Scholar]