Abstract
Deep learning nowadays offers expert-level and sometimes even super-expert-level performance, but achieving such performance demands massive annotated data for training (e.g., Google’s proprietary CXR Foundation Model (CXR-FM) was trained on 821,544 labeled and mostly private chest X-rays (CXRs)). Numerous datasets are publicly available in medical imaging but individually small and heterogeneous in expert labels. We envision a powerful and robust foundation model that can be trained by aggregating numerous small public datasets. To realize this vision, we have developed Ark, a framework that accrues and reuses knowledge from heterogeneous expert annotations in various datasets. As a proof of concept, we have trained two Ark models on 335,484 and 704,363 CXRs, respectively, by merging several datasets including ChestX-ray14, CheXpert, MIMIC-II, and VinDr-CXR, evaluated them on a wide range of imaging tasks covering both classification and segmentation via fine-tuning, linear-probing, and gender-bias analysis, and demonstrated our Ark’s superior and robust performance over the state-of-the-art (SOTA) fully/self-supervised baselines and Google’s proprietary CXR-FM. This enhanced performance is attributed to our simple yet powerful observation that aggregating numerous public datasets diversifies patient populations and accrues knowledge from diverse experts, yielding unprecedented performance yet saving annotation cost. With all codes and pretrained models released at GitHub.com/JLiangLab/Ark, we hope that Ark exerts an important impact on open science, as accruing and reusing knowledge from expert annotations in public datasets can potentially surpass the performance of proprietary models trained on unusually large data, inspiring many more researchers worldwide to share codes and datasets to build open foundation models, accelerate open science, and democratize deep learning for medical imaging.
Keywords: Accruing and Reusing Knowledge, Large-scale Pretraining
1. Introduction
Deep learning nowadays offers expert-level and sometimes even super-expert-level performance, deepening and widening its applications in medical imaging and resulting in numerous public datasets for research, competitions, and challenges. These datasets are generally small as annotating medical images is challenging, but achieving superior performance by deep learning demands massive annotated data for training. For example, Google’s proprietary CXR Foundation Model (CXR-FM) was trained on 821,544 labeled and mostly private CXRs [16]. We hypothesize that powerful and robust open foundation models can be trained by aggregating numerous small public datasets. To test this hypothesis, we have chosen CXRs because they are one of the most frequently used modalities, and our research community has accumulated copious CXRs (see Table 1). However, annotations associated with these public datasets are inconsistent in disease coverage. Even when addressing the same clinical issue, datasets created at different institutions tend to be annotated differently. For example, VinDr-CXR [13] is associated with global (image-level) and local (boxed-lesions) labels, while MIMIC-CXR [4] has no expert labels per se but comes with radiology reports. ChestX-ray14 [19] and CheXpert [4] both cover 14 conditions at the image level, and their 14 conditions have overlaps but are not exactly the same. Therefore, this paper seeks to address a critical need: How to utilize a large number of publicly-available images from different sources and their readily-accessible but heterogeneous expert annotations to pretrain generic source (foundation) models that are more robust and transferable to application-specific target tasks.
Table 1.
Publicly available datasets are generally small and heterogeneously annotated. Our Ark (Fig. 1) aims to aggregate numerous datasets with heterogeneous annotations to diversify patient population, accrue knowledge from diverse experts, and meet the demand by deep learning for massive annotated training data, offering superior and robust performance (Table 2, Fig. 2 and Fig. 3) yet reducing annotation cost.
| Abbrev. | Dataset | Task | Usagea | (Pre)train/val/test |
|---|---|---|---|---|
| 1. CXPT | CheXpert [4] | classify 14 thoracic diagnoses | P|F|L|B | 223414/-/234 |
| 2. NIHC | NIH ChestX-ray14 [19] | classify 14 thoracic diseases | P|F|L|B | 75312/11212/25596 |
| 3. RSNA | RSNA Pneumonia [1] | classify lung opacity, abnormality | P|F|L | 21295/2680/2709 |
| 4. VINC | VinDr-CXR [13] | classify 6 thoracic diagnoses | P|F|L | 15000/-/3000 |
| 5. NIHS | NIH Shenzhen CXR [5] | classify tuberculosis | P|F|L | 463/65/134 |
| 6. MMIC | MIMIC-II [6] | classify 14 thoracic diagnosesb | P | 368879/2992/5159 |
| 7. NIHM | NIH Montgomery [5] | segment lungs | F | 92/15/31 |
| 8. JSRT | JSRT [17] | segment lungs, heart, clavicles | F | 173/25/49 |
| 9. VINR | VinDr-RibCXR [14] | segment 20 ribs | F | 196/-/49 |
| 10. SIIM | SIIM-ACR PTX [2] | classify pneumothoraxc | L | 10675/-/1372 |
The usage of each dataset in our experiments is denoted with P for pretraining, F for fine-tuning, L for linear probing, and B for bias study.
The labels of CXRs in MIMIC-II are derived from their corresponding radiology reports using NegBio [15] and CheXpert [4].
SIIM-ACR, originally for pneumothorax segmentation, is converted into a classification task for linear probing, as CXR-FM cannot be evaluated for segmentation using its only released API.
To address this need, we have developed a framework, called Ark for its ability of accruing and reusing knowledge embedded in heterogeneous expert annotations with numerous datasets, as illustrated in Fig. 1. We refer to the pretrained models with Ark as Foundation Ark or simply as Ark for short. To demonstrate Ark’s capability, we have trained two models: Ark-5 on Datasets 1–5 and Ark-6 on Datasets 1–6 (Table 1), evaluated them on a wide range of 10 tasks via fine-tuning and on 6 tasks via linear probing, and demonstrated our Ark models outperform the SOTA fully/self-supervised baselines (Table 2) and Google CXR-FM1 (Fig. 2). Ark also exhibits superior robustness over CXR-FM in mitigating underdiagnosis and reducing gender-related biases, with lower false-negative rates and greater robustness to imbalanced data (Fig. 3).
Fig. 1.
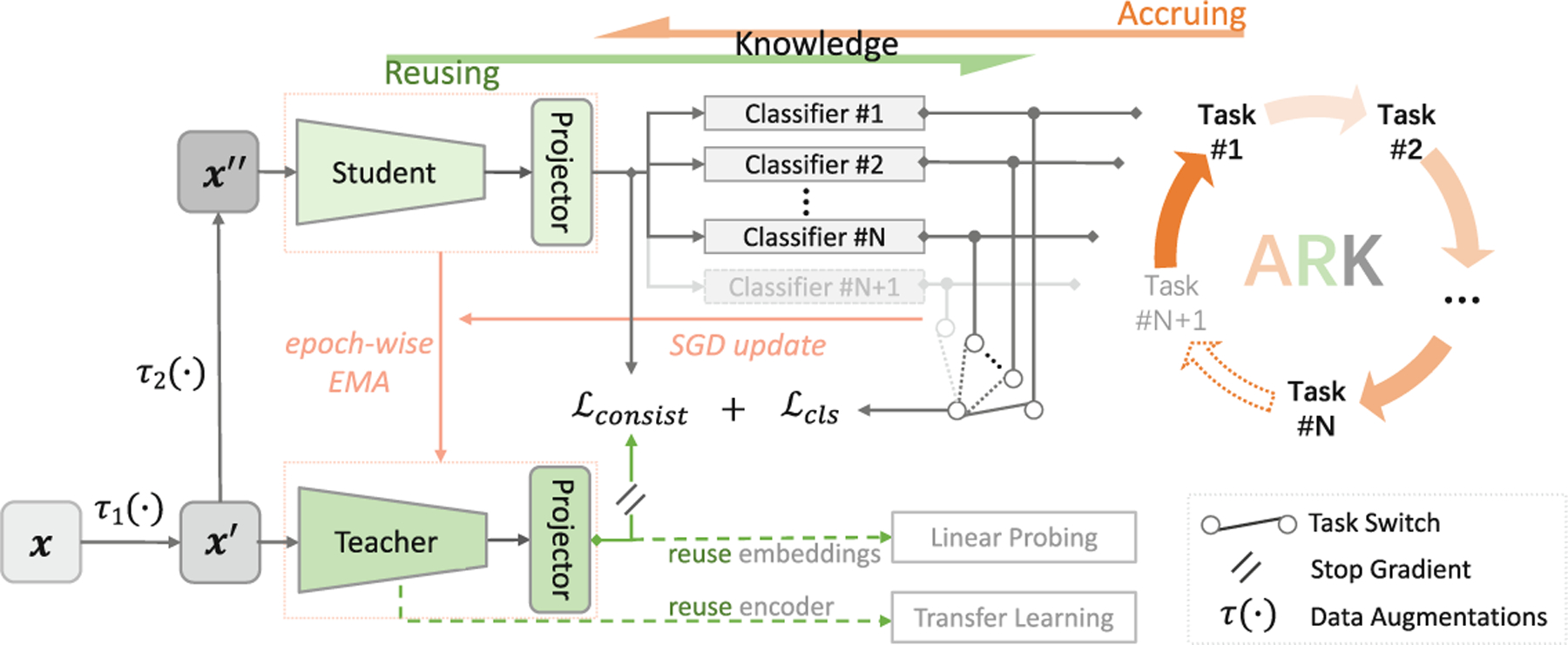
Our Ark is built on a student-teacher model with multi-task heads and trained via cyclic pretraining, aiming to accrue and reuse the expert knowledge embedded in the heterogeneous labels with numerous public datasets (see Sect.2 for details).
Table 2.
Our Ark-5 and Ark-6 outperform SOTA ImageNet pretrained models and the self-supervised domain-adapted model that utilizes even more training data, highlighting the importance of accruing and reusing knowledge in expert labels from diverse datasets for both classification and segmentation. With the best bolded and the second best underlined, a statistical analysis is conducted between the best vs. others, where green-highlighted boxes indicate no statistically significant difference at level p = 0.05.
| Initialization | Pretraining | Classification task | ||||
|---|---|---|---|---|---|---|
| 1. CXPT | 2. NIHC | 3. RSNA | 4. VINC | 5. NIHS | ||
| Random | - | 83.39±0.84 | 77.04±0.34 | 70.02±0.42 | 78.49±1.00 | 92.52±4.98 |
| Supervised | IN | 87.80±0.42 | 81.73±0.14 | 73.44±0.46 | 90.35±0.31 | 93.35±0.77 |
| SimMIM | IN | 88.16±0.31 | 81.95±0.15 | 73.66±0.34 | 90.24±0.35 | 94.12±0.96 |
| SimMIM | IN→CXR(926K) | 88.37±0.40 | 83.04±0.15 | 74.09±0.39 | 91.71±1.04 | 95.76±1.79 |
| Ark-5(ours) | IN→CXR(335K) | 88.73±0.20 | 82.87±0.13 | 74.73±0.59 | 94.67±0.33 | 98.92±0.21 |
| Ark-6(ours) | IN→CXR(704K) | 89.14±0.22 | 83.05±0.09 | 74.76±0.35 | 95.07±0.16 | 98.99±0.16 |
| Initialization | Pretraining | Segmentation task | ||||
| 6. NIHM | 7. JSRTLung | 8. JSRTHeart | 9. JSRTClavicle | 10. VINR | ||
| Random | - | 96.32±0.18 | 96.32±0.10 | 92.35±0.20 | 85.56±0.71 | 56.46±0.62 |
| Supervised | IN | 97.23±0.09 | 97.13±0.07 | 92.58±0.29 | 86.94±0.69 | 62.40±0.80 |
| SimMIM | IN | 97.12±0.14 | 96.90±0.08 | 93.53±0.11 | 87.18±0.63 | 61.64±0.69 |
| SimMIM | IN→CXR(926K) | 97.10±0.40 | 96.93±0.12 | 93.75±0.36 | 88.87±1.06 | 63.46±0.89 |
| Ark-5(ours) | IN→CXR(335K) | 97.65±0.17 | 97.41±0.04 | 94.16±0.66 | 90.01±0.35 | 63.96±0.30 |
| Ark-6(ours) | IN→CXR(704K) | 97.68±0.03 | 97.48±0.08 | 94.62±0.16 | 90.05±0.15 | 63.70±0.23 |
Fig. 2.
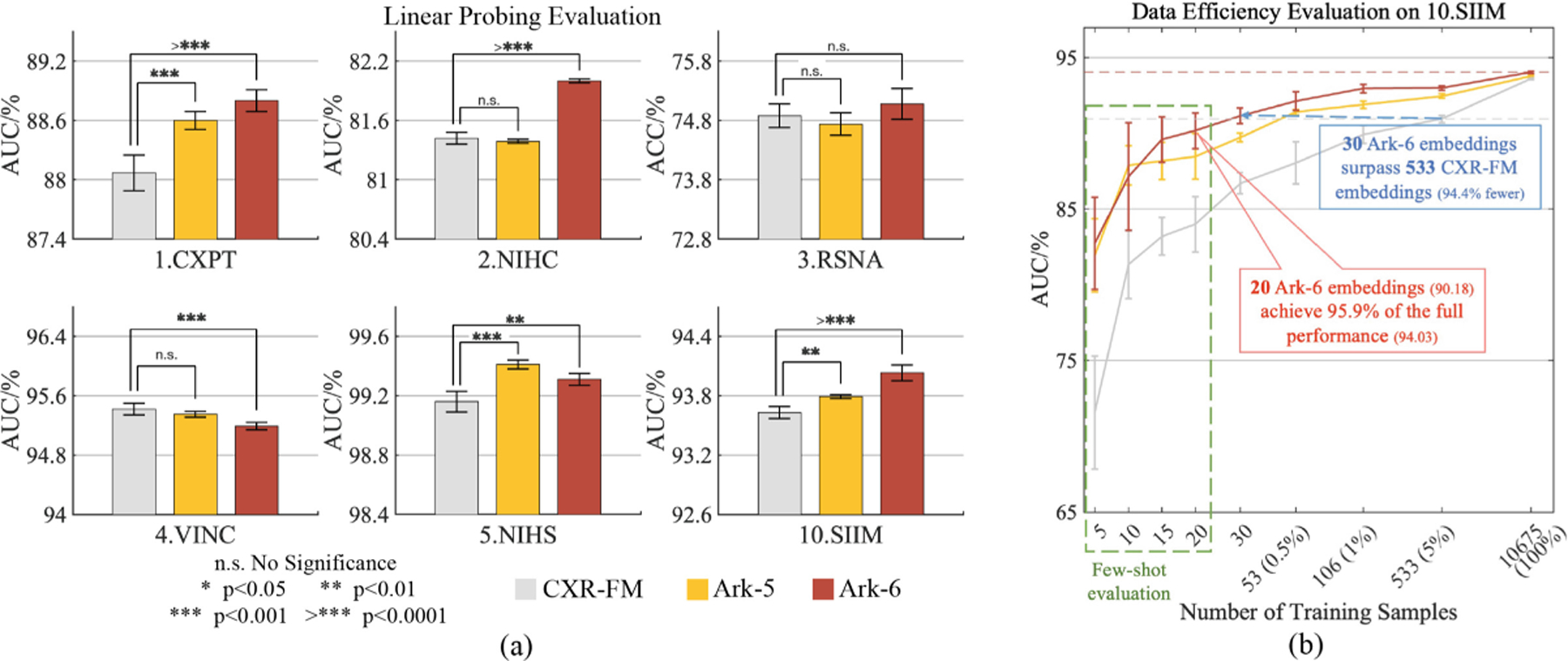
Ark-5 and Ark-6 are compared with Google CXR-FM via linear probing (a) with complete training set on six target tasks, demonstrating Ark’s superior or comparable performance and better embedding quality, and (b) with partial training sets or even few-shot samples, showcasing Ark’s outstanding performance in terms of data efficiency.
Fig. 3.
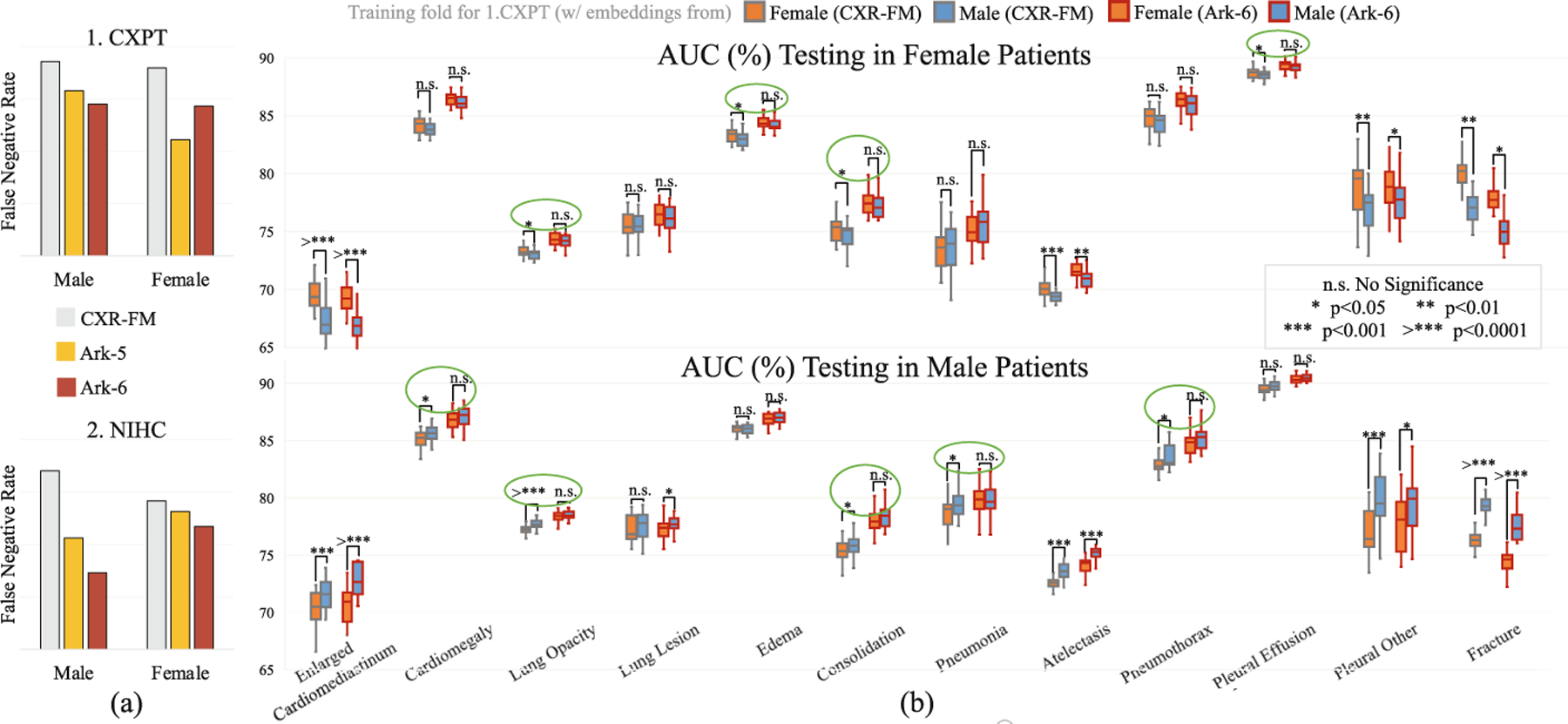
Ark models are compared with Google CXR-FM as regards false-negative rate (FNR) and gender-related bias. (a) Ark models show lower FNRs, indicating superior underdiagnosis mitigation. (b) Ark-6 demonstrates greater resilience to gender-imbalanced data. Gender bias is characterized by a significant drop in performance when training and test data are of the opposite gender, compared to when they are of the same gender (e.g., the orange whisker boxes are lower than the blue boxes in the lower-part (b)). Each green circle indicates a lung disease with gender bias by CXR-FM, as it performs differently between training on male and female data. But Ark exhibits a more robust performance, showing no significant difference on gender-segregated data.
This performance enhancement is attributed to a simple yet powerful observation that aggregating numerous public datasets costs nearly nothing but enlarges data size, diversifies patient populations, and accrues expert knowledge from a large number of sources worldwide; thereby offering unprecedented performance yet reducing annotation cost. More important, Ark is fundamentally different from self-supervised learning (SSL) and federated learning (FL) in concept. SSL can naturally handle images from different sources, but their associated expert annotations are left out of pretraining [10]. Clearly, every bit of expert annotation counts, conveying valuable knowledge. FL can utilize data with annotations from different sources, typically involving homogeneous labels, but it mainly concerns data privacy [12]. By contrast, Ark focuses on heterogeneous expert annotations with public data with no concern for data privacy and employs centralized training, which usually offers better performance with the same amount of data and annotation than distributed training as in FL.
Through this work, we have made the following contributions: (1) An idea that aggregates public datasets to enlarge and diversify training data; (2) A student-teacher model with multi-task heads via cyclic pretraining that accrues expert knowledge from existing heterogeneous annotations to achieve superior and robust performance yet reduce annotation cost; (3) Comprehensive experiments that evaluate our Ark via fine-tuning, linear-probing, and few-shot learning on a variety of target tasks, demonstrating Ark’s better generalizability and transferability in comparison with SOTA methods and Google CXR-FM; and (4) Empirical analyses for a critical yet often overlooked aspect of medical imaging models—robustness to underdiagnosis and gender imbalance, highlighting Ark significantly enhances reliability and safety in clinical decision-making.
2. Accruing and Reusing Knowledge
Our Ark aims to learn superior and robust visual representations from large-scale aggregated medical images by accruing and reusing the expert knowledge embedded in all available heterogeneous labels. The following details our Ark.
Accruing Knowledge into the Student via Cyclic Pretraining.
A significant challenge with training a single model using numerous datasets created for different tasks is label inconsistency (i.e., heterogeneity) (see Table 3 in Appendix). Manually consolidating heterogeneous labels from different datasets would be a hassle. To circumvent this issue, for each task, we introduce a specific classifier, called task head, to learn from its annotation and encode the knowledge into the model. A task head can be easily plugged into Ark, making Ark scalable to additional tasks. With multi-task heads, Ark can learn from multiple tasks concurrently or cyclically. In concurrent pretraining, a mini-batch is formed by randomly sampling an equal number of images from each dataset, and the loss for each image is computed based on its associated dataset id and labels. This idea is intuitive, but the model hardly converges; we suspect that the loss summation over all task heads simultaneously weakens gradients for back-propagation, causing confusion in weight updating. We opt for cyclic pretraining by iterating through all datasets sequentially in each round to accrue expert knowledge from all available annotations, a strategy that, we have found, stabilizes Ark’s pretraining and accelerates its convergence.
Accruing Knowledge into the Teacher via Epoch-Wise EMA.
To further summarize the accrued knowledge and accumulate the learning experiences in the historical dimension, we introduce into Ark a teacher model that shares the same architecture with the student. The teacher is updated using exponential moving average (EMA) [18] based on the student’s one epoch of learning at the end of each task. Eventually, the expert knowledge embedded in all labels and all historical learning experiences are accrued in the teacher model for further reuse in the cyclic pretraining and for future application-specific target tasks.
Reusing Accrued Knowledge from the Student to Bolster Cyclic Pretraining.
If the model learns from multiple tasks sequentially, it may “forget” the previously learned knowledge, and its performance on an old task may degrade catastrophically [7]. This problem is addressed naturally in Ark by cyclic pretraining, where the model revisits all the tasks in each round and reuses all knowledge accrued from the previous rounds and tasks to strengthen its learning from the current and future tasks. That is, by regularly reviewing the accrued knowledge through task revisitation, Ark not only prevents forgetting but also enables more efficient and effective learning from multiple tasks iteratively.
Reusing Accrued Knowledge from the Teacher to Mitigate Forgetting.
To leverage the accumulated knowledge of the teacher model as an additional self-supervisory signal, we incorporate a consistency loss between the student and the teacher, as shown in Fig. 1. To enhance this supervision, we introduce projectors in Ark that map the outputs of the student and teacher encoders to the same feature space. This further reinforces the feedback loop between the student and teacher models, facilitating the transfer of historical knowledge from the teacher to the student as a reminder to mitigate forgetting.
Ark has the following properties:
Knowledge-centric. Annotating medical images by radiologists for deep learning is a process of transferring their in-depth knowledge and expertise in interpreting medical images and identifying abnormalities to a medium that is accessible for computers to learn. Ark’s superior and robust performance is attributed to the accumulation of expert knowledge conveyed through medical imaging annotations from diverse expert sources worldwide. At the core of Ark is acquiring and sharing knowledge: “knowledge is power” (Mac Flecknoe) and “power comes not from knowledge kept but from knowledge shared” (Bill Gates).
Label-agnostic, task-scalable and annotation-heterogeneous. Ark is label-agnostic as it does not require prior label “understanding” of public datasets, but instead uses their originally-provided labels. It is designed with pluggable multi-task heads and cyclic pretraining to offer flexibility and scalability for adding new tasks without manually consolidating heterogeneous labels or training task-specific controllers/adapters [22]. Therefore, Ark intrinsically handles the annotation heterogeneity across different datasets.
Application-versatile. Ark trains versatile foundation models by utilizing a large number of publicly-available images from diverse sources and their readily-accessible diagnostic labels. As shown in Sect.3, Ark models are more robust, generalizable, and transferable to a wide range of application-specific target tasks across diseases (e.g., pneumothorax, tuberculosis, cardiomegaly) and anatomies (e.g., lung, heart, rib), highlighting Ark’s versatility.
3. Experiments and Results
Our Ark-5 and Ark-6 take the base version of the Swin transformer (Swin-B) [9] as the backbone, feature five and six independent heads based on the pretraining tasks and their classes, and are pretrained on Datasets 1–5 and 1–6, respectively, with all validation and test data excluded to avoid test-image leaks. In the following, both models are evaluated via transfer learning (in Sects.3.1 and 3.2) on a wide range of 10 common, yet challenging, tasks on 8 publicly available datasets, encompassing various thoracic diseases and diverse anatomy. To provide a more comprehensive evaluation, we conduct linear probing (in Sect.3.3) and analyze gender biases (in Sect.3.4) on the Ark models in comparison with Google CXR-FM. Pretraining and evaluation protocols are detailed in Appendix E.
3.1. Ark Outperforms SOTA Fully/Self-supervised Methods on Various Tasks for Thoracic Disease Classification
Experimental Setup:
To demonstrate the performance improvements achieved through Ark pretraining, we compare the Ark models with SOTA fully-supervised and self-supervised models [9,21] that were pretrained on ImageNet. We also include a comparison with a SOTA domain-adapted model [10] that was first pretrained on ImageNet and then on a large-scale domain-specific dataset comprising 926,028 CXRs from 13 different sources. All downstream models share the same Swin-B backbone, where the encoder is initialized using the pretrained weights and a task-specific classification head is re-initialized based on the number of classes for the target task. We fine-tune all layers in the downstream models under the same experimental setup. We also report the results of training the downstream models from scratch (random initialization) as the performance lower bound. Note that Google CXR-FM cannot be included for comparison as it is not publicly released for fine-tuning.
Results and Analysis:
As shown in Table 2, our Ark models consistently outperform the SOTA fully/self-supervised ImageNet pretrained models on all target tasks. These results highlight the benefit of leveraging additional domain-relevant data in pretraining to reduce the domain gap and further improve the model’s performance on target tasks. Furthermore, compared with the self-supervised domain-adapted model that utilizes 926K CXRs for pretraining, Ark models yield significantly superior performance on Dataset 1, 3–5 with only 335K CXRs, and on-par performance on 2.NIHC with 704K CXRs. These results demonstrate the superiority of Ark that accrues and reuses the knowledge retained in heterogeneous expert annotations from multiple datasets, emphasizing the importance of learning from expert labels. Moreover, we observe that Ark-6 consistently outperforms Ark-5, indicating the importance of incorporating more data and annotations from diverse datasets in pretraining.
3.2. Ark Provides Generalizable Representations for Segmentation Tasks
Experimental Setup:
To evaluate the generalizability of Ark’s representations, we transfer the Ark models to five segmentation tasks involving lungs, heart, clavicles, and ribs, and compare their performance with three SOTA fully/self-supervised models. We build the segmentation network upon UperNet [20], which consists of a backbone network, a feature pyramid network, and a decoder network. We implement the backbone network with Swin-B and initialize it with the pretrained weights from the Ark and those aforementioned SOTA models. The remaining networks are randomly initialized. We then fine-tune all layers in the segmentation models under the same experimental setup.
Results and Analysis:
As seen in Table 2, Ark models achieve significantly better performance than the SOTA models, demonstrating that Ark learned generalizable representations for delineating organs and bones in CXR. This superior performance is achieved by pretraining using large-scale CXRs and various disease labels from diverse datasets. Clinically, certain thoracic abnormalities can be diagnosed by examining the edges of the lungs, heart, clavicles, or ribs in CXR. For instance, a pneumothorax can be detected by observing a visible “visceral pleural line” along part or all of the length of the lateral chest wall [11]. Cardiomegaly can be diagnosed when the heart appears enlarged, with maximum diameter of the heart exceeding a pre-defined cardiothoracic ratio [19]. Fractures can be identified when the edges of the clavicles or ribs appear abnormally displaced or the bone cortex appears offset [3]. Therefore, leveraging diagnostic information from disease labels during pretraining enables Ark models to better capture the nuanced and varied pathological patterns, strengthening the models’ ability to represent anatomically specific features that reflect abnormal conditions in various oragns or bones. By contrast, the SimMIM (IN→CXR(926K)) model is pretrained with a self-supervised masked image modeling proxy task, which may use many clues to reconstruct the masked patches that are not necessarily related to pathological conditions, leading to lower performance despite training on more images.
3.3. Ark Offers Embeddings with Superior Quality over Google CXR-FM
Experimental Setup:
To highlight the benefits of learning from more detailed diagnostic disease labels, we compare our Ark models with Google CXR-FM. CXR-FM was trained on a large dataset of 821,544 CXRs from three different sources, but with coarsened labels (normal or abnormal). By contrast, our Ark models are trained with less data, but aims to fully utilize all labels provided by experts in the original datasets. Furthermore, Ark models employ a much smaller backbone (88M parameters) compared with CXR-FM using EfficientNet-L2 (480M parameters). Since Google CXR-FM is not released and cannot be finetuned, we resorted to its released API to generate the embeddings (information-rich numerical vectors) for all images in the target tasks. For the sake of fairness, we also generated the embeddings from Ark’s projector, whose dimension is the same as Google’s. To evaluate the quality of the learned representations of these models, we conduct linear probing by training a simple linear classifier for each target task. The performance of both models is evaluated on six target tasks, including an unseen dataset, 10.SIIM, where the images have not been previously seen by the Ark models during pretraining. Additionally, we perform the same evaluation on 10.SIIM with partial training sets or even few-shot samples to further demonstrate the high quality of our Ark models’ embeddings.
Results and Analysis:
Figure 2(a) shows that Ark-6 outperforms CXR-FM significantly on Dataset 1, 2, 5 and 10, and performs comparably to CXR-FM on 3.RSNA. Similarly, Ark-5 performs better than CXR-FM on Dataset 1, 5 and 10, while performing comparably on the remaining tasks. Moreover, Fig. 2(b) shows that both Ark-5 and Ark-6 consistently outperform CXR-FM in small data regimes, highlighting the superiority of Ark’s embeddings, which carry richer information that can be utilized more efficiently. These results demonstrate that Ark models learn higher-quality representations with less pretraining data while employing a much smaller backbone than CXR-FM, highlighting that learning from more granular diagnostic labels, such as Ark, is superior to learning from coarsened normal/abnormal labels.
3.4. Ark Shows a Lower False-Negative Rate and Less Gender Bias
Experimental Setup:
Underdiagnosis can lead to delayed treatment in health-care settings and can have serious consequences. Hence, the false-negative rate (FNR) is a critical indicator of the robustness of a computer-aided diagnosis (CAD) system. Furthermore, population-imbalanced data can train biased models, adversely affecting diagnostic performance in minority populations. Therefore, a robust CAD system should provide a low false-negative rate and strong resilience to biased training data. To demonstrate the robustness of our Ark models in comparison with Google CXR-FM, we first compute the FNRs in terms of gender on 1.CXPT and 2.NIHC. We further investigate gender biases in Ark-6 and CXR-FM on 1.CXPT using gender-exclusive training sets. We follow the train/test splits in [8] to ensure a balanced number of cases per class in 40 male/female-only folds. We train linear classifiers on those folds using embeddings from Ark-6 and CXR-FM, and then evaluate these classifiers on the corresponding male/female-only test splits. The biased model will show significant differences in performance when training and test data are of the opposite gender. We detail this setup in Appendix E.
Results and Analysis:
Figure 3(a) illustrates that Ark models have lower FNRs than CXR-FM for both genders on both tasks, demonstrating that Ark models are less likely to underdiagnose disease conditions than CXR-FM. In Fig. 3(b), the biases in the pretrained models are measured by performance differences between linear classifiers trained on male-only and female-only embeddings. The upper part of Fig. 3(b) depicts the results of testing on female-only sets, where the classifiers trained on male-only embeddings generally perform poorly compared with those trained on female embeddings, revealing gender biases due to data imbalance. Among the 12 diseases, the classifiers trained with Google’s embeddings have unbiased performances for only 4 diseases, whereas those using Ark-6’s embeddings perform in an unbiased fashion with no significant differences for the 8 diseases. The same situation occurs when testing is performed on male patients as shown in the lower part of Fig. 3(b). The gender bias analysis demonstrates that Ark has greater robustness to the extremely imbalanced data that contributes to gender bias in computer-aided diagnosis.
4. Conclusions and Future Work
We have developed Foundation Ark, the first open foundation model, that realizes our vision: accruing and reusing knowledge retained in heterogeneous expert annotations with numerous datasets offers superior and robust performance. Our experimental results are strong on CXRs, and we plan to extend Ark to other modalities. We hope Ark’s performance encourages researchers worldwide to share codes and datasets big or small for creating open foundation models, accelerating open science, and democratizing deep learning for medical imaging.
Supplementary Material
Footnotes
Supplementary Information The online version contains supplementary material available at https://doi.org/10.1007/978-3-031-43907-0_62.
References
- 1.RSNA pneumonia detection challenge (2018). https://www.kaggle.com/c/rsna-pneumonia-detection-challenge
- 2.SIIM-ACR pneumothorax segmentation (2019). https://kaggle.com/competitions/siim-acr-pneumothorax-segmentation
- 3.Collins J: Chest wall trauma. J. Thorac. Imaging 15(2), 112–119 (2000) [DOI] [PubMed] [Google Scholar]
- 4.Irvin J, et al. : CheXpert: a large chest radiograph dataset with uncertainty labelsand expert comparison. In: Proceedings of the AAAI Conference on Artificial Intelligence, vol. 33, pp. 590–597 (2019) [Google Scholar]
- 5.Jaeger S, Candemir S, Antani S, Wáng YXJ, Lu PX, Thoma G: Twopublic chest x-ray datasets for computer-aided screening of pulmonary diseases. Quant. Imaging Med. Surg 4(6), 475 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson AE, et al. : MIMIC-CXR, a de-identified publicly available database ofchest radiographs with free-text reports. Sci. Data 6(1), 1–8 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kemker R, McClure M, Abitino A, Hayes T, Kanan C: Measuring catastrophic forgetting in neural networks. In: Proceedings of the AAAI Conference on Artificial Intelligence, vol. 32 (2018) [Google Scholar]
- 8.Larrazabal AJ, Nieto N, Peterson V, Milone DH, Ferrante E: Gender imbalance in medical imaging datasets produces biased classifiers for computer-aided diagnosis. Proc. Natl. Acad. Sci 117(23), 12592–12594 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Z, et al. : Swin transformer: hierarchical vision transformer using shifted windows. In: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 10012–10022 (2021) [Google Scholar]
- 10.Ma D, et al. : Benchmarking and boosting transformers for medical image classification. In: Kamnitsas K, et al. (eds.) Domain Adaptation and Representation Transfer, DART 2022. Lecture Notes in Computer Science, vol. 13542, pp. 12–22. Springer, Cham: (2022). 10.1007/978-3-031-16852-9_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mason RJ, et al. : Murray and Nadel’s Textbook of Respiratory Medicine E-Book: 2-Volume Set. Elsevier Health Sciences (2010) [Google Scholar]
- 12.McMahan B, Moore E, Ramage D, Hampson S, y Arcas BA: Communication-efficient learning of deep networks from decentralized data. In: Artificial Intelligence and Statistics, pp. 1273–1282. PMLR (2017) [Google Scholar]
- 13.Nguyen HQ, et al. : VinDr-CXR: an open dataset of chest x-rays with radiologist’s annotations. Sci. Data 9(1), 429 (2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen HC, Le TT, Pham HH, Nguyen HQ: VinDr-RibCXR: a benchmark dataset for automatic segmentation and labeling of individual ribs on chest x-rays. In: Medical Imaging with Deep Learning (2021) [Google Scholar]
- 15.Peng Y, Wang X, Lu L, Bagheri M, Summers R, Lu Z: NegBio: a high-performance tool for negation and uncertainty detection in radiology reports. AMIA Summits Transl. Sci. Proc 2018, 188 (2018) [PMC free article] [PubMed] [Google Scholar]
- 16.Sellergren AB, et al. : Simplified transfer learning for chest radiography models using less data. Radiology 305(2), 454–465 (2022) [DOI] [PubMed] [Google Scholar]
- 17.Shiraishi J, et al. : Development of a digital image database for chest radiographs with and without a lung nodule: receiver operating characteristic analysis of radiologists’ detection of pulmonary nodules. Am. J. Roentgenol 174(1), 71–74 (2000) [DOI] [PubMed] [Google Scholar]
- 18.Tarvainen A, Valpola H: Mean teachers are better role models: weight-averaged consistency targets improve semi-supervised deep learning results. In: Advances in Neural Information Processing Systems, vol. 30 (2017) [Google Scholar]
- 19.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM: ChestX-ray8: hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2097–2106 (2017) [Google Scholar]
- 20.Xiao T, Liu Y, Zhou B, Jiang Y, Sun J: Unified perceptual parsing forscene understanding. In: Ferrari V, Hebert M, Sminchisescu C, Weiss Y (eds.) ECCV 2018. LNCS, vol. 11209, pp. 432–448. Springer, Cham: (2018). 10.1007/978-3-030-01228-1_26 [DOI] [Google Scholar]
- 21.Xie Z, et al. : SimMIM: a simple framework for masked image modeling. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 9653–9663 (2022) [Google Scholar]
- 22.Zhu Z, Kang M, Yuille A, Zhou Z: Assembling existing labels from publicdatasets to diagnose novel diseases: Covid-19 in late 2019. In: NeurIPS Workshop on Medical Imaging meets NeurIPS (2022) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


