Abstract.
Human anatomy is the foundation of medical imaging and boasts one striking characteristic: its hierarchy in nature, exhibiting two intrinsic properties: (1) locality: each anatomical structure is morphologically distinct from the others; and (2) compositionality: each anatomical structure is an integrated part of a larger whole. We envision a foundation model for medical imaging that is consciously and purposefully developed upon this foundation to gain the capability of “understanding” human anatomy and to possess the fundamental properties of medical imaging. As our first step in realizing this vision towards foundation models in medical imaging, we devise a novel self-supervised learning (SSL) strategy that exploits the hierarchical nature of human anatomy. Our extensive experiments demonstrate that the SSL pretrained model, derived from our training strategy, not only outperforms state-of-the-art (SOTA) fully/self-supervised baselines but also enhances annotation efficiency, offering potential few-shot segmentation capabilities with performance improvements ranging from 9% to 30% for segmentation tasks compared to SSL baselines. This performance is attributed to the significance of anatomy comprehension via our learning strategy, which encapsulates the intrinsic attributes of anatomical structures—locality and compositionality—within the embedding space, yet overlooked in existing SSL methods. All code and pretrained models are available at GitHub.com/JLiangLab/Eden.
Keywords: Self-supervised Learning, Learning from Anatomy
1. Introduction and Related Works
Foundation models [4], such as GPT-4 [22] and DALL.E [23], pretrained via self-supervised learning (SSL), have revolutionized natural language processing (NLP) and radically transformed vision-language modeling, garnering significant public media attention [18]. But, despite the development of numerous SSL methods in medical imaging, their success in this domain lags behind their NLP counterparts. What causes these striking differences? We believe that this is because the SSL methods developed for NLP have proven to be powerful in capturing the underlying structures (foundation) of the English language; thus, a number of intrinsic properties of the language emerge naturally, as demonstrated in [19], while the existing SSL methods lack such capabilities to appreciate the foundation of medical imaging—human anatomy. Therefore, this paper is seeking to answer a fundamental question: How to learn foundation models from human anatomy in medical imaging?
Human anatomy exhibits natural hierarchies. For example, the lungs are divided into the right and left lung (see Fig. 6 in Appendix) and each lung is further divided into lobes, two on the left and three on the right lung. Each lobe is further subdivided into segments, each containing pulmonary arteries, veins, and bronchi which branch in predictable, dichotomous fashion. Consequently, anatomical structures have two important properties: locality: each anatomical structure is morphologically distinct from others; compositionality: each anatomical structure is an integrated part of a larger whole. Naturally, a subquestion is how to exploit the anatomical hierarchies for training foundation models? To this end, we devise a novel SSL training strategy, which is hierarchical, autodidactic, and coarse, resulting in a pretrained model, which is versatile, and leading to anatomical embedding, which is dense and semantics-meaningful. Our training strategy is hierarchical because it decomposes and perceives the anatomy progressively in a coarse-to-fine manner (Sect. 2.1); autodidactic because it learns from anatomy through self-supervision, thereby requiring no anatomy labeling (Sect. 2); and coarse because it generates dense anatomical embeddings without relying on pixel-level training (Sect. 3, ablation 1). The pretrained model is versatile because it is strong in generality and adaptability, resulting in performance boosts (Sect. 3.1) and annotation efficiency (Sect. 3.2) in myriad tasks. The generated anatomical embedding is dense and semantics-rich because it possesses two intrinsic properties of anatomical structures, locality (Sect. 3.3) and compositionality (Sect. 3.4), in the embedding space, both of which are essential for anatomy understanding. We call our pretrained model Adam (autodidactic dense anatomical models) because it learns autodidactically and yields dense anatomical embedding, nicknamed Eve (embedding vectors) for semantic richness (Fig. 1). We further coin our project site Eden (environment for d ense embeddings and networks), where all code, pretrained Adam and Eve are placed.
Fig. 1.
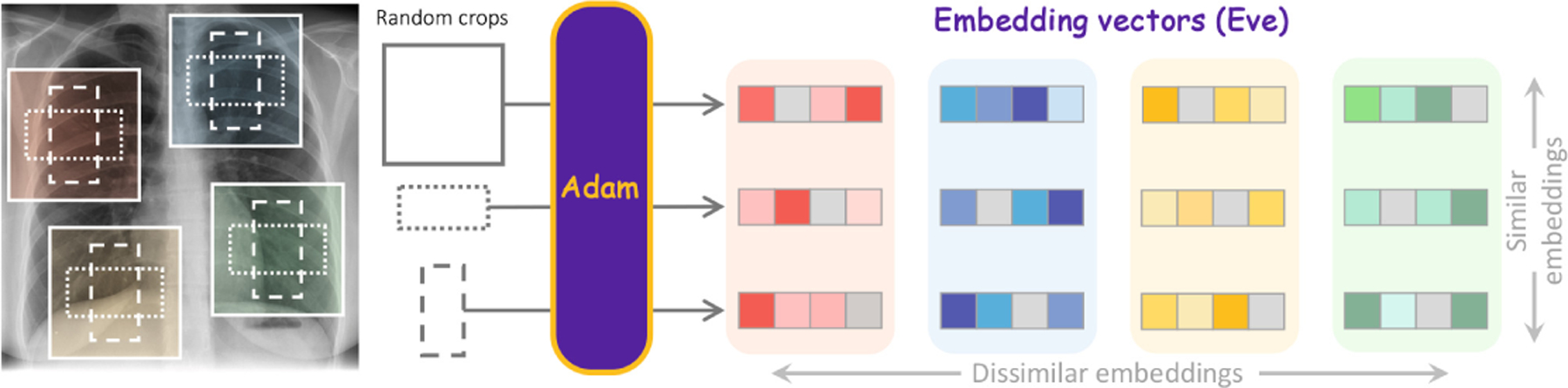
Existing SSL methods lack capabilities of “understanding” the foundation of medical imaging—human anatomy. We believe that a foundation model must be able to transform each pixel in an image (e.g., a chest X-ray) into semantics-rich numerical vectors, called embeddings, where different anatomical structures (indicated by different colored boxes) are associated with different embeddings, and the same anatomical structures have (nearly) identical embeddings at all resolutions and scales (indicated by different box shapes) across patients. Inspired by the hierarchical nature of human anatomy (Fig. 6 in Appendix), we introduce a novel SSL strategy to learn anatomy from medical images (Fig. 2), resulting in embeddings (Eve), generated by our pretrained model (Adam), with such desired properties (Fig. 4 and Fig. 8 in Appendix).
In summary, we make the following contributions: (1) A novel self-supervised learning strategy that progressively learns anatomy in a coarse-to-fine manner via hierarchical contrastive learning; (2) A new evaluation approach that facilitates analyzing the interpretability of deep models in anatomy understanding by measuring the locality and compositionality of anatomical structures in embedding space; and (3) A comprehensive and insightful set of experiments that evaluate Adam for a wide range of 9 target tasks, involving fine-tuning, few-shot learning, and investigating semantic richness of Eve in anatomy understanding.
Related Works:
(i) Self-supervised learning methods, particularly contrastive techniques [2, 16], have shown great promise in medical imaging [12, 25]. But, due to their focus on image-level features, they are sub-optimal for dense recognition tasks [28]. Recent works [10, 13] empower contrastive learning with more discriminative features via using the diversity in the local context of medical images. In contrast to them, which overlook anatomy hierarchies in their learning objectives, Adam exploits the hierarchical nature of anatomy to learn semantics-rich dense features. (ii) Anatomy learning methods integrate anatomical cues into their SSL objectives. But, GLC [6] requires spatial correspondence across images, limiting its scalability to non-aligned images. Although TransVW [11], SAM [31], and Alice [15] relax this requirement, they neglect hierarchical anatomy relations, offering no compositionality. By contrast, Adam learns consistent anatomy features without relying on spatial alignment across images (see Fig. 7 in Appendix) and captures both local and global contexts hierarchically to offer both locality and compositionality. (iii) Hierarchical SSL methods exploit transformers’ self-attention to model dependencies among image patches. But, they fail to capture anatomy relations due to inefficient SSL signals that contrast similar anatomical structures [26] or disregard relations among images [29, 30]. Adam goes beyond architecture design by introducing a learning strategy that decomposes anatomy into a hierarchy of parts for coarse-to-fine anatomy learning, and avoids semantic collision in its supervision signal.
2. Method
Our self-supervised learning strategy, depicted in Fig. 2, aims to exploit the hierarchical nature of human anatomy in order to capture not only generic but also semantically meaningful representations. The main intuition behind our learning strategy is the principle of totality in Gestalt psychology: humans commonly first recognize the prominent objects in an image (e.g., lungs) and then gradually recognize smaller details based on prior knowledge about that object (e.g., each lung is divided into lobes) [24]. Inspired by this principle, we propose a training strategy, which decomposes and perceives the anatomy progressively in a coarse-to-fine manner, aiming to learn both anatomical (local and global) contextual information and also the relative hierarchical relationships among anatomical structures. Our framework is comprised of two key components:
Fig. 2.
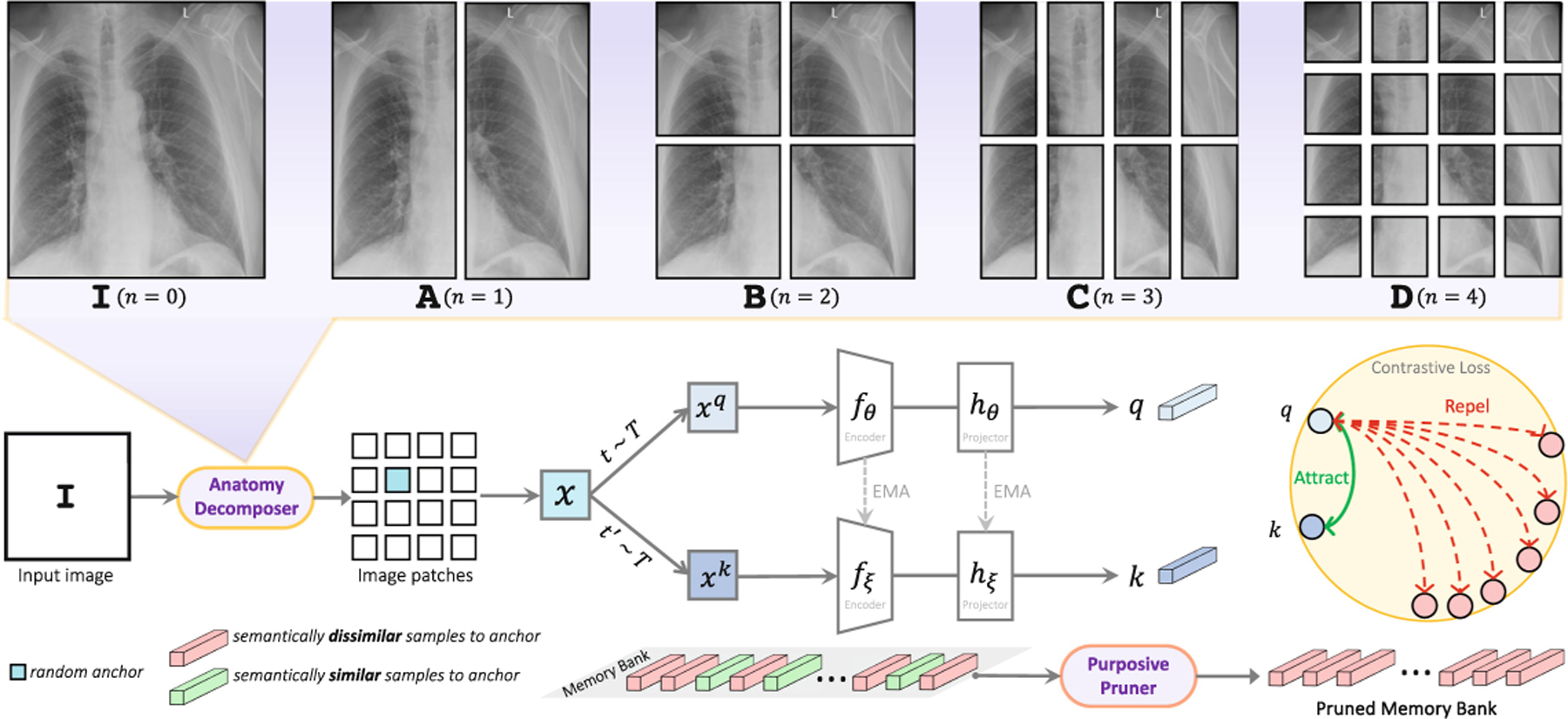
Our SSL strategy gradually decomposes and perceives the anatomy in a coarse-to-fine manner. Our Anatomy Decomposer (AD) decomposes the anatomy into a hierarchy of parts with granularity level at each training stage. Thus, anatomical structures of finer-grained granularity will be incrementally presented to the model as the input. Given image , we pass it to AD to get a random anchor . We augment to generate two views (positive samples), and pass them to two encoders to get their features. To avoid semantic collision in training objective, our Purposive Pruner removes semantically similar anatomical structures across images to anchor x from the memory bank. Contrastive loss is then calculated using positive samples’ features and the pruned memory bank. The figure shows pretraining at .
(1) Anatomy Decomposer (AD) is responsible for decomposing relevant anatomy into a hierarchy of anatomical structures to guide the model to learn hierarchical anatomical relationships in images. The AD component takes two inputs: an image and an anatomy granularity level , and generates a random anatomical structure instance . We generate anatomical structures at desired granularity level in a recursive manner. Given an image , we first split it vertically into two halves (A in Fig. 2). Then, we iteratively alternate between horizontally and vertically splitting the resulting image parts until we reach the desired granularity level (B, C, D in Fig. 2). This process results in image patches . In this set, we randomly sample an instance , which will be used as the input for training the model. As such, during the pretraining, anatomical structures at various granular levels are generated and present to the model.
(2) Purposive Pruner (PP) is responsible for compeling the model to comprehend anatomy more effectively via learning a wider range of distinct anatomical structures. Intuitively, similar anatomical structures (e.g. ribs or disks) should have similar embeddings, while also their finer-grained constituent parts (e.g. different ribs or disks) have (slightly) different embeddings. To achieve such desired embedding space, the anatomical structures need to be intelligently contrasted from each other. Our PP module, in contrast to standard contrastive learning approaches, identifies semantically similar anatomical structures in the embedding space and prevents them from being undesirably repelled. In particular, given an anchor anatomical structure randomly sampled from image , we compute the cosine similarities between features of and the ones of the points in the memory bank, and remove the samples with a similarity greater than a threshold from the memory bank. Thus, our PP prevents semantic collision, yielding a more optimal embedding space where similar anatomical structures are grouped together while distinguished from dissimilar anatomical structures.
Overall Training.
Our framework consists of two twin backbones and , and projection heads and and are updated by back-propagation, while and are updated by exponential moving average (EMA) of and hθ parameters, respectively. We use a memory bank to store the embeddings of negative samples , where is the memory bank size. For learning anatomy in a coarse-to-fine manner, we progressively increase the anatomical structures granularity. Thus, at each training stage, anatomical structures with granularity level will be presented to the model. Given input image and data granularity level , we pass them to our AD to get a random anatomical structure . We apply an augmentation function on to generate two views and , which are then processed by backbones and projection heads to generate latent features and . Then, we pass and MB to our PP to remove false negative samples for anchor , resulting in pruned memory bank , which is used to compute the InfoNCE [7] loss , where is a temperature hyperparameter, is size of , and . Our AD module enables the model to first learn anatomy at a coarser-grained level, and then use this acquired knowledge as effective contextual clues for learning more fine-grained anatomical structures, reflecting anatomical structures compositionality in its embedding space. Our PP module enables the model to learn a semantically-structured embedding space that preserves anatomical structures locality by removing semantic collision from the model’s learning objective. The pretrained model derived by our training strategy (Adam) can not only be used as a basis for myriad target tasks via adaptation (fine-tuning), but also its embedding vectors (Eve) show promises to be used standalone without adaptation for other tasks like landmark detection.
3. Experiments and Results
Pretraining and Fine-Łuning Settings:
We use unlabeled training images of ChestX-ray14 [27] and EyePACS [8] for pretraining and follow [7] in pretraining settings: SGD optimizer with an initial learning rate of 0.03, weight decay 1e-4, SGD momentum 0.9, cosine decaying scheduler, and batch size 256. The input anatomical structures are resized to 224 × 224; augmentations include random crop, color jitter, Gaussian blur, and rotation. We use data granularity level () up to 4 and pruning threshold (ablation in Appendix). Following [10, 16], we adopt ResNet-50 as the backbone. For fine-tuning, we (1) use the pretrained encoder followed by a task-specific head for classification tasks, and a U-Net network for segmentation tasks where the encoder is initialized with the pretrained backbone; (2) fine-tune all downstream model’s params; (3) run each method 10 times on each task and report statistical significance analysis.
Downstream Łasks and Baselines:
We evaluate Adam on a myraid of 9 tasks on ChestX-ray14 [27], Shenzhen [14], VinDr-CXR [20], VinDR-Rib [21], SIIM-ACR [1], SCR [9], ChestX-Det [17], and DRIVE [5], covering various challenging tasks, diseases, and organs. We compare Adam with SOTA image- (MoCo-v2 [7]), patch- (TransVW [11], VICRegL [3], DenseCL [28]), and pixel-level (PCRL [32], DiRA [10], Medical-MAE [29], SimMIM [30]) SSL methods.
1). Adam provides generalizable representations for a variety of tasks.
To showcase the significance of anatomy learning via our SSL approach and its impact on representation learning, we compare transfer learning performance of Adam to 8 recent SOTA SSL methods with diverse objectives, as well as 2 fully-supervised models pretrained on ImageNet and ChestX-ray14 datasets, in 8 downstream tasks. As seen in Fig. 3, (i) our Adam consistently outperforms the SOTA dense SSL methods (VICRegL & DenseCL) as well as the SOTA medical SSL methods (PCRL & DiRA), and achieves superior or comparable performance compared to fully-supervised baselines; (ii) our Adam demonstrates a significant performance improvement over TransVW, which is specifically designed for learning recurring anatomical structures across patients. This emphasizes the effectiveness of our coarse-to-fine approach in capturing both local and global context of anatomical structures hierarchically, in contrast to TransVW which learns them at a fixed level; and (iii) our Adam remains superior to ViT-based SSL methods such as Medical-MAE and SimMIM, which divide the input image into smaller patches and utilize self-attention to model patch dependencies. This underscores the importance of our learning strategy in effectively modeling the hierarchical relationships among anatomical structures.
Fig. 3.
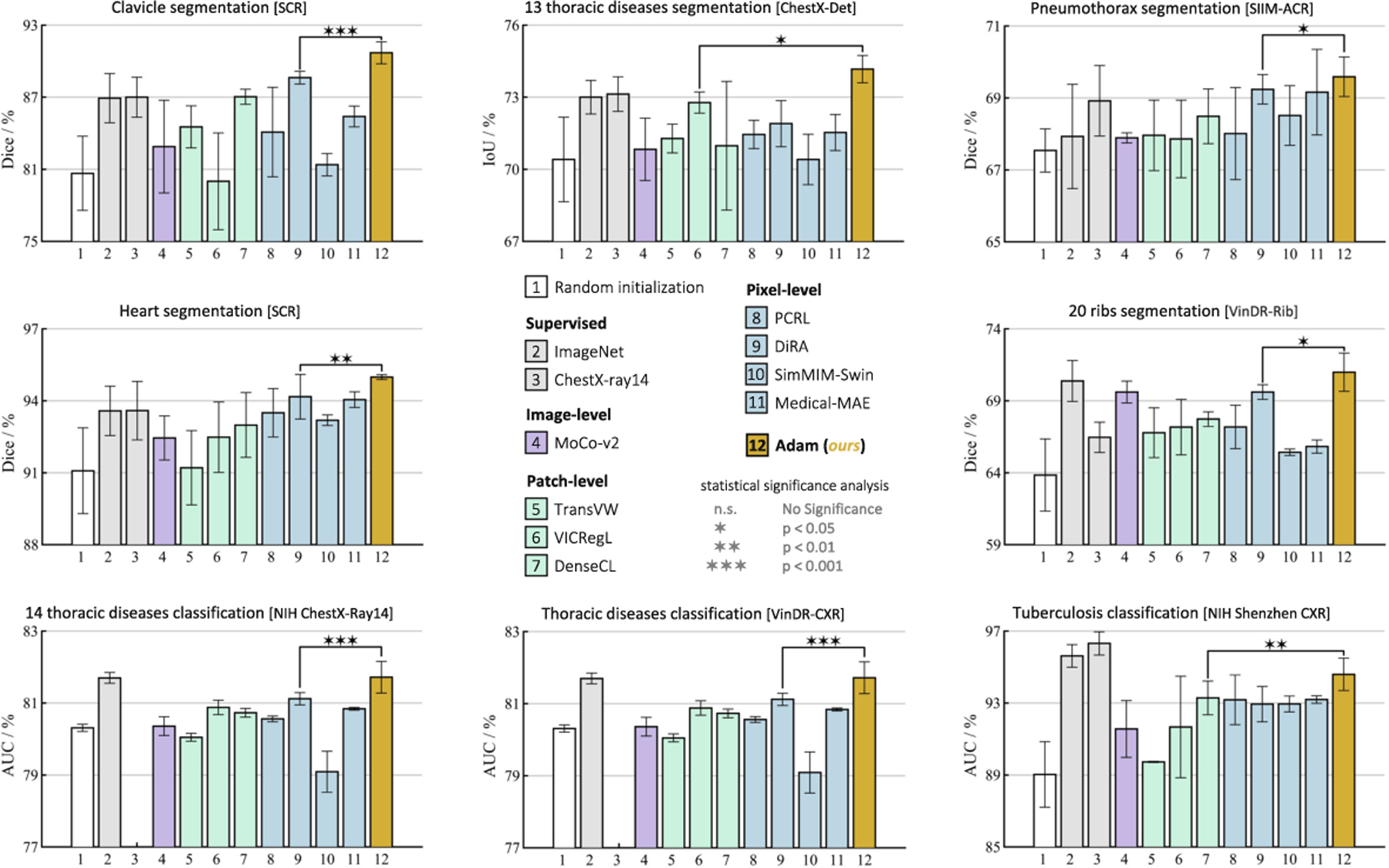
Adam provides superior performance over fully/self-supervised methods. All SSL methods are pretrained on ChestX-ray14 dataset. Statistical significance analysis (p < 0.05) was conducted between Adam and the top SSL baseline in each task.
2). Adam enhances annotation efficiency, revealing promise for few-shot learning.
To dissect robustness of our representations, we compare Adam with top-performing SSL methods from each baseline group, based on Fig. 3, in limited data regimes. We conduct experiments on Heart and Clavicle segmentation tasks, and fine-tune the pretrained models using a few shots of labeled data (3, 6, 12, and 24) randomly sampled from each dataset. As seen in Table 1, Adam not only demonstrates superior performance against baselines by a large margin (Green nums.) but also maintains consistent behavior with minimal performance drop as labeled data decreases, compared to baselines. We attribute Adam’s superior representations over baselines, as seen in Fig. 3 and Table 1, to its ability to learn the anatomy by preserving locality and compositionality of anatomical structures in its embedding space, as is exemplified in the following.
Table 1.
Few-shot transfer on two medical segmentation tasks. Adam provides outstandingly better performance compared with SSL baselines. Green numbers show Adam’s performance boosts compared with the second-best method in each task/shot.
| Method | SCR-Heart [Dice(%)] | SCR-Clavicle [Dice(%)] | ||||||
|---|---|---|---|---|---|---|---|---|
| 3-shot | 6-shot | 12-shot | 24-shot | 3-shot | 6-shot | 12-shot | 24-shot | |
| MoCo-v2 | 44.84 | 59.97 | 69.90 | 79.69 | 23.77 | 29.24 | 38.07 | 44.47 |
| DenseCL | 64.88 | 74.43 | 75.79 | 80.06 | 36.43 | 51.31 | 63.03 | 69.13 |
| DiRA | 63.76 | 64.47 | 76.10 | 81.42 | 31.42 | 38.59 | 66.81 | 73.06 |
| Adam (ours) | 84.35 (↑19) | 86.70 (↑12) | 89.79 (↑14) | 90.45 (↑9) | 66.69 (↑30) | 79.41 (↑28) | 83.96 (↑17) | 84.76 (↑12) |
3). Adam preserves anatomical structures locality.
We investigate Adam’s ability to reflect locality of anatomical structures in its embedding space against existing SSL baselines. To do so, we (1) create a dataset of 1,000 images (from ChestX-ray14 dataset) with 10 distinct anatomical landmarks manually annotated by human experts in each image, (2) extract 224 × 224 patches around each landmark across images, (3) extract latent features of each landmark instance using each pretrained model under study and then pass them through a global average pooling layer, and (4) visualize the features by using t-SNE. As seen in Fig. 4.1, existing SSL methods lack the ability in discriminating different anatomical structures, causing ambiguous embedding spaces. In contrast, Adam excels in distinguishing various anatomical landmarks, yielding well-separated clusters in its embedding space. This highlights Adam’s ability to learn a rich semantic embedding space where distinct anatomical structures have unique embeddings, and identical structures share near-identical embeddings across patients.
Fig. 4.
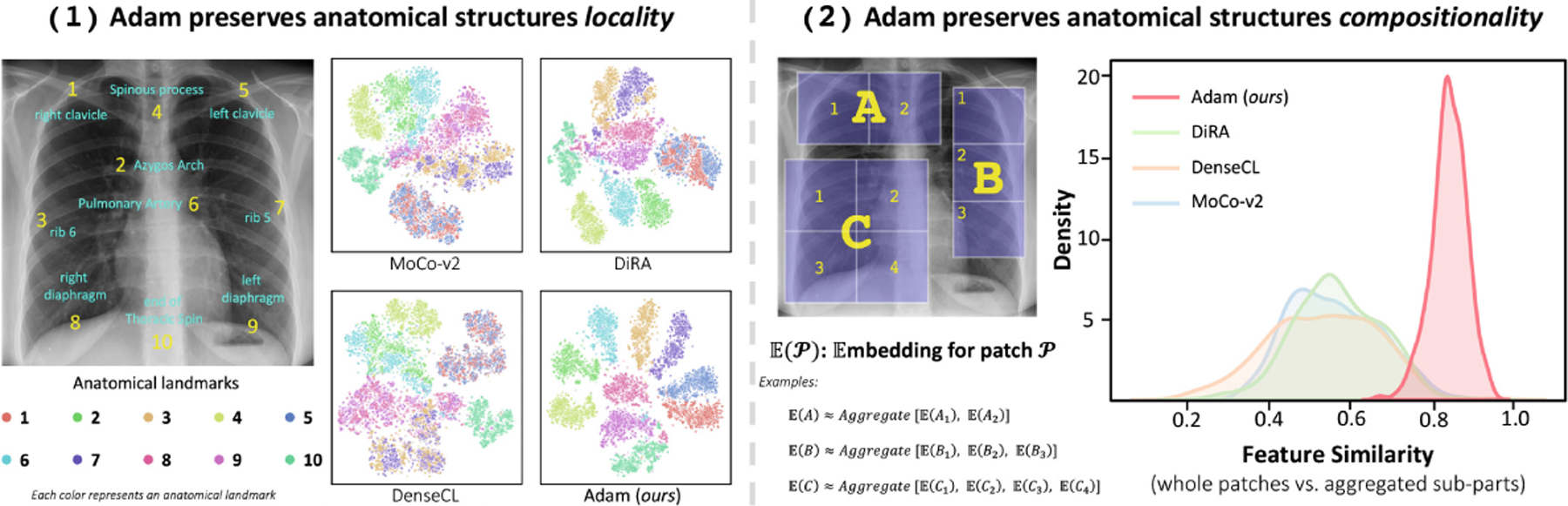
Adam preserves locality and compositionality properties, which are intrinsic to anatomical structures and critical for understanding anatomy, in its embedding space.
4). Adam preserves anatomical structures compositionality.
The embedding of a whole should be equal or close to the sum of the embedding of its each part (see examples in Fig. 4.2). To investigate Adam’s ability to reflect compositionality of anatomical structures in its embedding space against existing SSL baselines, we (1) extract random patches from test images of ChestX-ray14, and decompose each patch into 2, 3, or 4 non-overlapping sub-patches, (2) resize each extracted patch and its sub-patches to 224 × 224 and then extract their features using each pretrained model under study, (3) compute cosine similarity between the embedding of each patch and the aggregate of the embeddings of its sub-patches, and (4) visualize the similarity distributions with Gaussian kernel density estimation (KDE). As seen in Fig. 4.2, Adam’s distribution is not only narrower and taller than baselines, but also the mean of similarity value between embedding of whole patches and their aggregated sub-parts is closer to 1.
Ablation 1: Eve’s accuracy in anatomy understanding was studied by visualizing dense correspondence between (i) an image and its augmented views and (ii) different images. Given two images, we divide them into grids of patches and extract their features and using Adam’s pretrained model. For each feature vector in , we find its correspondence in based on highest cosine similarity; for clarity, we show some of the high-similarity matches (≥0.8) in Fig. 5.1. As seen, Eve has accurate dense anatomical representations, mapping semantically similar structures, regardless of their differences. Although Adam is not explicitly trained for this purpose, these results show its potential for landmark detection and image registration applications, as an emergent property.
Fig. 5.
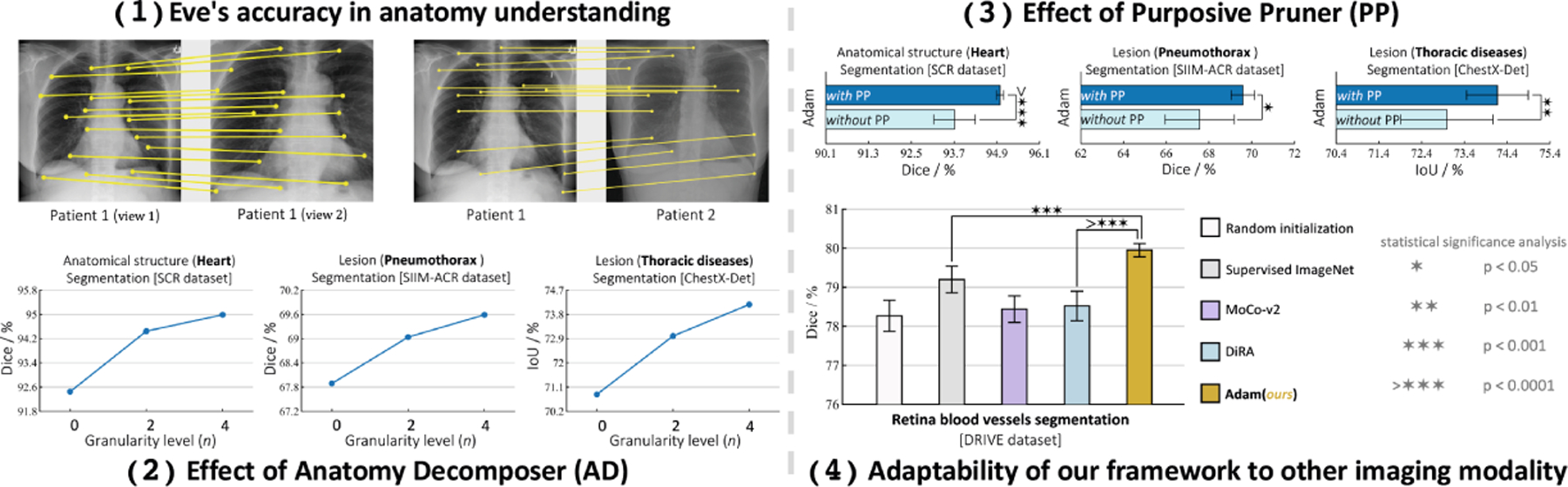
Ablation studies on (1) Eve’s accuracy in anatomy understanding, (2) effect of anatomy decomposer, (3) effect of purposive pruner, and (4) adaptability of our framework to other imaging modalities.
Ablation 2: Effect of Anatomy Decomposer was studied by gradually increasing pretraining data granularity from coarse-grained anatomy () to finer levels (up to ) and fine-tuning the models on downstream tasks. As seen in Fig. 5.2, gradual increment of data granularity consistently improves the performance across all tasks. This suggests that our coarse-to-fine learning strategy deepens the model’s anatomical knowledge.
Ablation 3: Effect of Purposive Pruner was studied by comparing a model with and without PP (i.e. contrasting an anchor with all negative pairs in the memory bank) during pretraining. Figure 5.3 shows PP leads to significant performance boosts across all tasks, highlighting its key role in enabling the model to capture more discriminative features by removing noisy contrastive pairs.
Ablation 4: Adaptability of our framework to other imaging modalities was explored by utilizing fundoscopy photography images in EyePACS as pretraining data, which possess complex structures due to the diverse variations in retinal anatomy. As depicted in Fig. 5.4, Adam provides superior performance by 1.4% (p < 0.05) in the blood vessel segmentation task compared to the top-performing SSL methods that also leverage the same pretraining images. This highlights the importance of effectively learning the anatomy and also showcases the potential applicability of our method to various imaging modalities.
4. Conclusion and Future Work
A key contribution of ours lies in crafting a novel SSL strategy that underpins the development of powerful self-supervised models foundational to medical imaging via learning anatomy. Our training strategy progressively learns anatomy in a coarse-to-fine manner via hierarchical contrastive learning. Our approach yields highly generalizable pretrained models and anatomical embeddings with essential properties of locality and compositionality, making them semantically meaningful for anatomy understanding. In future, we plan to apply our strategy to provide dense anatomical models for major imaging modalities and protocols.
Supplementary Material
Footnotes
Supplementary Information The online version contains supplementary material available at https://doi.org/10.1007/978-3-031-45857-6_10.
References
- 1.SIIM-ACR pneumothorax segmentation (2019). https://www.kaggle.com/c/siim-acr-pneumothorax-segmentation/ [Google Scholar]
- 2.Azizi S, et al. : Big self-supervised models advance medical image classification. In: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 3478–3488 (2021) [Google Scholar]
- 3.Bardes A, Ponce J, LeCun Y: VICRegl: self-supervised learning of local visual features. In: Advances in Neural Information Processing Systems, vol. 35, pp. 8799–8810 (2022) [Google Scholar]
- 4.Bommasani R, et al. : On the opportunities and risks of foundation models. ArXiv (2021). https://crfm.stanford.edu/assets/report.pdf [Google Scholar]
- 5.Budai A, Bock R, Maier A, Hornegger J, Michelson G: Robust vessel segmentation in fundus images. Int. J. Biomed. Imaging 2013, 154860 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaitanya K, Erdil E, Karani N, Konukoglu E: Contrastive learning of global and local features for medical image segmentation with limited annotations. In: Advances in Neural Information Processing Systems, vol. 33, pp. 12546–12558 (2020) [Google Scholar]
- 7.Chen X, Fan H, Girshick R, He K: Improved baselines with momentum contrastive learning (2020) [Google Scholar]
- 8.Cuadros J, Bresnick G: EyePACS: an adaptable telemedicine system for diabetic retinopathy screening. Diabetes Sci. Technol. 3(3), 509–516 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Ginneken B, Stegmann M, Loog M: Segmentation of anatomical structures in chest radiographs using supervised methods: a comparative study on a public database. Med. Image Anal. 10(1), 19–40 (2006) [DOI] [PubMed] [Google Scholar]
- 10.Haghighi F, Hosseinzadeh Taher MR, Gotway MB, Liang J: DiRA: discriminative, restorative, and adversarial learning for self-supervised medical image analysis. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 20824–20834 (2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haghighi F, Taher MRH, Zhou Z, Gotway MB, Liang J: Transferable visual words: exploiting the semantics of anatomical patterns for self-supervised learning. IEEE Trans. Med. Imaging 40(10), 2857–2868 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hosseinzadeh Taher MR, Haghighi F, Feng R, Gotway MB, Liang J: A systematic benchmarking analysis of transfer learning for medical image analysis. In: Domain Adaptation and Representation Transfer, and Affordable Healthcare and AI for Resource Diverse Global Health, pp. 3–13 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hosseinzadeh Taher MR, Haghighi F, Gotway MB, Liang J: CAiD: context-aware instance discrimination for self-supervised learning in medical imaging. In: Proceedings of The 5th International Conference on Medical Imaging with Deep Learning. Proceedings of Machine Learning Research, vol. 172, pp. 535–551 (2022) [PMC free article] [PubMed] [Google Scholar]
- 14.Jaeger S, Candemir S, Antani S, Wáng YXJ, Lu PX, Thoma G: Two public chest x-ray datasets for computer-aided screening of pulmonary diseases. Quant. Imaging Med. Surg. 4(6) (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang Y, Sun M, Guo H, Yan K, Lu L, Xu M: Anatomical invariance modeling and semantic alignment for self-supervised learning in 3D medical image segmentation. arXiv (2023) [Google Scholar]
- 16.Kaku A, Upadhya S, Razavian N: Intermediate layers matter in momentum contrastive self supervised learning. In: Advances in Neural Information Processing Systems, pp. 24063–24074 (2021) [Google Scholar]
- 17.Lian J, et al. : A structure-aware relation network for thoracic diseases detection and segmentation. IEEE Trans. Med. Imaging 40(8), 2042–2052 (2021) [DOI] [PubMed] [Google Scholar]
- 18.Manjoo F: How Do You Know a Human Wrote This. The New York Times (2020) [Google Scholar]
- 19.Manning CD, Clark K, Hewitt J, Khandelwal U, Levy O: Emergent linguistic structure in artificial neural networks trained by self-supervision. Proc. Natl. Acad. Sci. 117(48), 30046–30054 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen HQ, Lam K, Le LT, et al. : VinDr-CXR: an open dataset of chest x-rays with radiologist’s annotations. Sci. Data 9, 429 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen HC, Le TT, Pham HH, Nguyen HQ: VinDr-RibCXR: a benchmark dataset for automatic segmentation and labeling of individual ribs on chest X-rays. In: Medical Imaging with Deep Learning (2021) [Google Scholar]
- 22.OpenAI: GPT-4 technical report (2023) [Google Scholar]
- 23.Ramesh A, et al. : Zero-shot text-to-image generation. In: Proceedings of the 38th International Conference on Machine Learning, vol. 139, pp. 8821–8831 (2021) [Google Scholar]
- 24.Sun Y, Hu J, Shi J, Sun Z: Progressive decomposition: a method of coarse-to-fine image parsing using stacked networks. Multimedia Tools Appl. 79(19–20), 13379–13402 (2020) [Google Scholar]
- 25.Tajbakhsh N, Roth H, Terzopoulos D, Liang J: Guest editorial annotation-efficient deep learning: the holy grail of medical imaging. IEEE Trans. Med. Imaging 40(10), 2526–2533 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang Y, et al. : Self-supervised pre-training of swin transformers for 3D medical image analysis. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 20730–20740 (2022) [Google Scholar]
- 27.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, et al. : ChestX-ray8: hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 2097–2106 (2017) [Google Scholar]
- 28.Wang X, Zhang R, Shen C, Kong T, Li L: Dense contrastive learning for self-supervised visual pre-training. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 3024–3033 (2021) [Google Scholar]
- 29.Xiao J, Bai Y, Yuille A, Zhou Z: Delving into masked autoencoders for multi-label thorax disease classification. In: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), pp. 3588–3600 (2023) [Google Scholar]
- 30.Xie Z, et al. : SimMIM: a simple framework for masked image modeling. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 9653–9663 (2022) [Google Scholar]
- 31.Yan K, et al. : SAM: self-supervised learning of pixel-wise anatomical embeddings in radiological images. IEEE Trans. Med. Imaging 41(10), 2658–2669 (2022) [DOI] [PubMed] [Google Scholar]
- 32.Zhou HY, Lu C, Yang S, Han X, Yu Y: Preservational learning improves self-supervised medical image models by reconstructing diverse contexts. In: Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), pp. 3499–3509 (2021) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


