Summary
Background
The prognosis of lung carcinoma has changed since the discovery of molecular targets and their specific drugs. Somatic Epidermal Growth Factor Receptor (EGFR) mutations have been reported in lung carcinoma, and these mutant proteins act as substrates for targeted therapies. However, in a resource-constrained country like India, panel-based next-generation sequencing cannot be made available to the population at large. Additional challenges such as adequacy of tissue in case of lung core biopsies and locating suitable tumour tissues as a result of innate intratumoral heterogeneity indicate the necessity of an AI-based end-to-end pipeline capable of automatically detecting and learning more effective lung nodule features from CT images and predicting the probability of the EGFR-mutant. This will help the oncologists and patients in resource-limited settings to achieve near-optimal care and appropriate therapy.
Methods
The EGFR gene sequencing and CT imaging data of 2277 patients with lung carcinoma were included from three cohorts in India and a White population cohort collected from TCIA. Another cohort LIDC-IDRI was used to train the AIPS-Nodule (AIPS-N) model for automatic detection and characterisation of lung nodules. We explored the value of combining the results of the AIPS-N with the clinical factors in the AIPS-Mutation (AIPS-M) model for predicting EGFR genotype, and it was evaluated by area under the curve (AUC).
Findings
AIPS-N achieved an average AP50 of 70.19% in detecting the location of nodules within the lung region of interest during validation and predicted the score of five lung nodule properties. The AIPS-M machine learning (ML) and deep learning (DL) models achieved AUCs ranging from 0.587 to 0.910.
Interpretation
The AIPS suggests that CT imaging combined with a fully automated lung-nodule analysis AI system can predict EGFR genotype and identify patients with an EGFR mutation in a cost-effective and non-invasive manner.
Funding
This work was supported by a grant provided by Conquer Cancer Foundation of ASCO [2021IIG-5555960128] and Pfizer Products India Pvt. Ltd.
Keywords: Artificial intelligence, Radiology, Oncology, Medical imaging, Lung carcinoma, Lung cancer
Research in context.
Evidence before this study
Prior to undertaking this study, we conducted a comprehensive review of existing evidence to understand the landscape of lung carcinoma treatment, EGFR mutations, AI applications in lung nodule analysis, and the specific challenges faced in resource-constrained settings.
Sources:
-
1.Databases:
-
a.Google Scholar
-
b.PubMed
-
c.The Cancer Imaging Archive
-
d.ImageNet
-
a.
-
2.Journals:
-
a.LANCET: Digital Health
-
b.CA: A Cancer Journal for Clinicians
-
c.Nature: Modern Pathology
-
d.European Respiratory Journal
-
e.Frontiers in Immunology
-
f.Institute of Electrical and Electronics Engineers (IEEE)
-
g.Journal of Clinical Biology
-
h.Nature Reviews: Clinical Oncology
-
i.American Association for Cancer Research (AACR): Cancer Research
-
j.American Association of Physicists in Medicine: Medical Physics
-
k.Journal of Medical Imaging
-
a.
Criteria used to include or exclude studies:
-
1.
We included studies that directly addressed the research questions/objectives of our study.
-
2.
We only included peer-reviewed journal articles, conference papers, theses, dissertations, and other scholarly publications.
-
3.
The exact start and end dates of the search were 1st January 2002 and 31st July 2023.
-
4.
We included studies published in all languages.
Search terms used:
We searched for journals and books on Google Scholar and PubMed using the keywords: “EGFR mutation”, “CT images”, and “artificial intelligence”.
The quality of the evidence was assessed for risk of bias, considering study design, sample size, and methodology. The evidence highlighted the significant impact of targeted therapies based on EGFR mutations, the potential of AI in nodule analysis, and the limitations faced in real-world clinical settings, particularly in countries like India.
Added value of this study
This study contributes to the existing evidence by addressing specific gaps identified in the literature:
-
1.
Population-specific model: unlike previous research, which predominantly focused on populations of White and Chinese origins, our study centres on the Indian population. This addition is particularly important given the genetic diversity of populations and the need for population-specific models.
-
2.
Detection and characterisation of lung nodules: while most of the previous studies often concentrated solely on nodule detection, our research extends the scope to comprehensive nodule characterization and its correlation with EGFR mutational status.
-
3.
Avoids resource-intensive steps: the novel AI-based Predictive System (AIPS) introduced here offers a streamlined approach that avoids resource-intensive steps like manual image annotation and complex feature engineering, making it more practical for implementation in resource-limited settings.
Implications of all the available evidence
Collectively, the available evidence and the findings of this study have significant implications for practice, policy, and future research:
-
1.
Triaging patients for targeted therapies: the identification and prediction of EGFR mutational status through AI-based nodule characterisation can guide oncologists in effectively triaging patients for targeted therapies. This not only optimizes patient care but also has implications for resource allocation in healthcare systems.
-
2.
Population-specific model: the model’s focus on the Indian population underscores the importance of tailoring AI approaches to specific populations for improved accuracy and relevance. This study’s insights could potentially influence the development of similar AI systems for other populations, thereby advancing global healthcare practices. Future research could explore the integration of AI-based strategies into routine clinical workflows and investigate the generalizability of these findings to other populations and settings.
Introduction
The treatment of lung carcinoma has undergone a paradigm shift with the emergence of newer molecular therapies. The prognosis of patients with biomarker-driven cancer treated with targeted therapy is substantially better than patients unable to receive targeted therapies. Somatic Epidermal Growth Factor Receptor (EGFR) mutations have been reported in lung carcinoma, and these mutant proteins act as substrates for targeted therapies. The administration of EGFR-targeted therapy has revolutionised lung cancer management.1 Somatic mutation in the EGFR gene is assessed by gene sequencing of biopsied tumour tissues, which faces the challenge of making available panel-based next-generation sequencing (NGS) to the population at large in a resource-constrained country like India. Additional challenges associated with NGS include the adequacy of tissue in the case of lung core biopsies, locating suitable tumour tissues due to innate intratumoral heterogeneity,2,3 the shift in EGFR mutation status after subsequent chemotherapy,4,5 and reduced DNA quality.6
Studies have shown promising results in automatically categorising and characterising lung nodules due to combining AI with CT imaging7,8 (Appendix p 4). This provides an alternative to analysing the lung nodules with no additional cost. Despite these advancements, numerous methods have only focused on detecting nodules in CT imaging.9,10 Additionally, studies have utilised AI to extract comprehensive information from the entire lung for predicting EGFR genotype and assessing the response to targeted therapy in lung cancer but these studies have predominantly concentrated on the White and Chinese populations.11,12 For example, Wang et al. introduced an AI system that predicts lung cancer patients’ EGFR genotype and treatment outcomes by analysing CT images of the entire lung.11 As a step forward, we have done a comprehensive characterization of the nodule that can reflect EGFR genotype information and might affect therapeutic efficacy, with a primary focus on the Indian population.
We aim to develop a novel, cost-effective and non-invasive AI-based strategy not only to detect but also to characterise lung nodules that may predict the EGFR mutational status (wild-type vs mutant) in lung carcinoma patients and hence effectively triage these patients requiring comprehensive molecular profiling of the EGFR-driver gene. This will help the oncologists and patients in resource-limited settings to achieve near-optimal care and appropriate therapy.
This is achieved through the fully automated AI-based Predictive System (AIPS) built using machine learning (ML) and deep learning (DL) algorithms, which is an end-to-end pipeline capable of automatically detecting and learning more effective lung nodule features from CT images and predicting the probability of the EGFR-mutant. This avoids time-consuming image annotation by radiologists, and feature engineering (complex tumour boundary segmentation or human-defined features) based on radiomics.6 Most of the studies in this field have focused primarily on the data of the White and the Chinese population,11,12 raising the need for a model trained, validated, and tested on the Indian population specifically.
Methods
Study design and participants
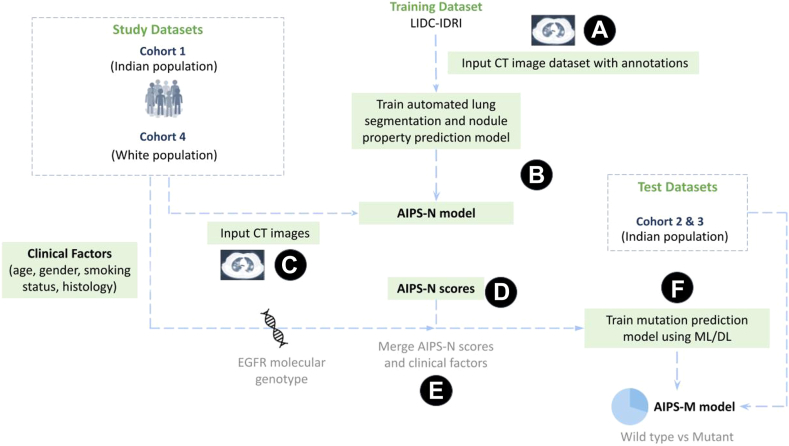
The overall workflow of the experiment is depicted in Fig. 1.
Fig. 1.
Workflow of the proposed AIPS and study design. (A) The LIDC-IDRI image and annotation dataset is downloaded from The Cancer Imaging Archive (TCIA) (B) AIPS-Nodule (AIPS-N) automated lung segmentation and nodule property prediction model is trained using the LIDC-IDRI image and annotation dataset. (C) CT images from study datasets of the Indian and White populations are fed into the trained AIPS-N model. (D) AIPS-N scores for different nodule features are calculated for each of the study datasets. (E) The AIPS-N scores, EGFR molecular genotype, and the clinical factors of the study datasets are merged. (F) Merged data is used for building the AIPS-Mutation prediction (AIPS-M) ML and DL models to ultimately predict the mutational status (wild-type or mutant) of the EGFR gene.
We included 3287 patients with lung cancer from five cohorts. Out of which, three retrospective cohorts (labelled as Cohort 1 [n = 1379], Cohort 2 [n = 591], and Cohort 3 [n = 96]) were collected from Rajiv Gandhi Cancer Institute and Research Centre, New Delhi, India (RGCI & RC) after receiving approval from the respective ethics committees. Further, Cohorts 4 and 5 were collected from two public resources - The Cancer Imaging Archive (TCIA) - comprising a White population in the USA (labelled as Cohort 4 [n = 211])13 and the Lung Image Database Consortium - Image Database Resource Initiative (LIDC-IDRI) image collection14 of 1010 patients (244,527 images) labelled as Cohort 5 [n = 1010]. EGFR gene sequencing results and lung CT images (1,582,812 images) at diagnosis time were obtained for all patients in Cohorts 1–4 (Table 1).
Table 1.
Characteristics of patients in Cohorts 1–4.
| Cohort 1 (n = 1379) | Cohort 2 (n = 591) | Cohort 3 (n = 96) | Cohort 4 (n = 211) | |
|---|---|---|---|---|
| Data source | India | India | India | USA |
| Age, years | 62.4 (23–92) | 61.7 (21–90) | 58.6 (24–86) | 67.96 (24–87) |
| Sex | ||||
| Male | 905 (65.6%) | 356 (60.2%) | 64 (66.7%) | 135 (63.9%) |
| Female | 474 (34.4%) | 235 (39.8%) | 32 (33.3%) | 76 (36.1%) |
| Smoking | ||||
| Never | 736 (54.4%) | 356 (60.2%) | 67 (69.8%) | 48 (22.7%) |
| Smoker | 643 (46.6%) | 235 (39.8%) | 29 (30.2%) | 163 (77.3%) |
| Histology | ||||
| Adenocarcinoma | 903 (65.5%) | 408 (69.0%) | 93 (96.9%) | 172 (81.5%) |
| Squamous cell carcinoma | 259 (18.8%) | 101 (17.1%) | 3 (3.1%) | 35 (16.6%) |
| Others | 217 (15.7%) | 82 (13.9%) | 0 (0.0%) | 4 (1.9%) |
| EGFR genotype | ||||
| Wild-type | 699 (50.7%) | 286 (48.4%) | 69 (71.9%) | 133 (63%) |
| Mutant | 680 (49.3%) | 305 (51.6%) | 25 (28.1%) | 38 (18%) |
| NA | 0 | 0 | 0 | 40 (19%) |
The patient characteristics are unavailable for Cohort 5.
Next, we trained, validated, and tested the AIPS-N lung segmentation and nodule feature prediction model using CT images collected from LIDC-IDRI (Cohort 5) (Fig. 1 - points A & B). Further, the CT images belonging to the Indian population (Cohorts 1–3) and the White population (Cohort 4) were fed into the trained AIPS-N model to obtain results in the form of AIPS-N scores (Fig. 1 - points C & D). The AIPS-N scores were merged with the clinical factors from the respective Cohorts (Fig. 1 - point E).
The merged dataset (AIPS-N scores merged with clinical factors) of Cohort 1 (Indian population) was split into training and internal validation subsets for model training, hyperparameter tuning, and internal validation to build AIPS-M models (Fig. 1 - point F). The merged datasets of Cohort 2 and Cohort 3 (Indian population), as well as Cohort 4 (White population), were utilised for independent testing of the AIPS-M models trained on Cohort 1. Additionally, in an entirely distinct experiment, the merged dataset of Cohort 4 (White population) was split into training, validation, and testing subsets. The inclusion criteria, data collection timeframe, data sources, and CT scanner information (manufacturer, model, and scanning parameters) for each Cohort are provided in Appendix (p 5).
Development of the AIPS-nodule (AIPS-N) model
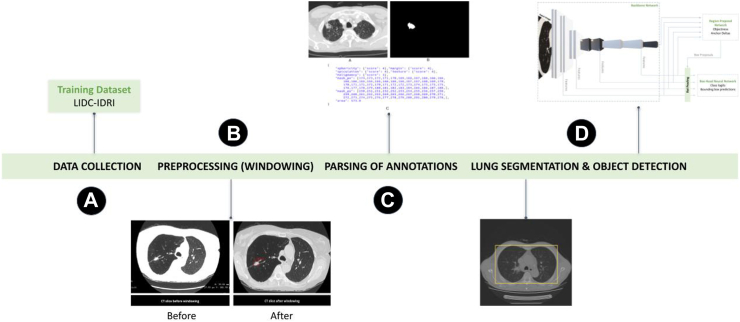
The development of the AIPS-Nodule (AIPS-N) model involved four major steps. Firstly, we downloaded the LIDC-IDRI CT image dataset (Cohort 5) containing 1010 patients (244,527 images) from TCIA.15 Next, we pre-processed these CT images by applying a technique called windowing to enhance the visibility of the lungs. Following that, we parsed the image annotations which, in our case, involved extraction of the location and features of lung nodules within the image, such as malignancy, margin, texture, sphericity, and spiculation. Next, we applied automated lung segmentation to identify and separate the lung area from the rest of the image. The parsed image annotations along with the respective pre-processed images were used to train the Faster R-CNN (region-based convolutional neural network) model. The overall workflow for developing the AIPS-N model is demonstrated in Fig. 2.
Fig. 2.
Overall workflow for developing the AIPS-N model.
Data collection
The LIDC-IDRI image dataset (Cohort 5) downloaded from TCIA15 is preprocessed to demonstrate specific anatomy and pathology in the images. The images collected from 1010 patients are in 3D-DICOM format and consist of multiple slices, which means the resolution of the images has three components - length, height, and width. We normalised the intensity values of the DICOM images to a standardised range (between 0 and 255) before applying windowing techniques. This step ensured that the object detection models received input with consistent intensity ranges across images in different cohorts and avoided any biases resulting from varying intensity scales (Appendix p 9). Additional steps were taken to ensure the representativeness of Cohort 5 for training the AIPS-N model and its subsequent generalisation to Cohorts 1–4 (Appendix p 11).
Preprocessing (windowing)
To suppress noise and irrelevant bright intensities (e.g., bones), and to demonstrate individual anatomy and pathology in lung ROI, we preprocessed the lung ROI through windowing.16 During windowing, the window width (WW set to 1500 HU), described as the range of CT numbers and the window level (WL set to −500 HU), described as the midpoint of the range of the CT numbers are adjusted to alter the slice contrast and brightness respectively for the evaluation of lung parenchyma17 (Appendix p 9).
Parsing of annotations
Every slice in a 3D image that embodies a lung cancer nodule has its corresponding annotation to locate the coordinates of the mask and features of the nodule. The coordinates and the features are a result of an image annotation process performed by four thoracic radiologists in two phases - blinded-read phase and unblinded-read phase to locate and describe all lung nodules as comprehensively as possible (Appendix p 12). The calculated inter-rater reliability (Cohen’s Kappa) is approximately 0.8448. This indicates a very strong level of agreement among the four radiologists in marking nodules greater than or equal to 3 mm. These annotations are parsed using PyLIDC Python library18 and saved into a JSON-based annotation file for each slice. The image slices, masks, and the corresponding JSON-based annotations are arranged in different folders according to the patient ID. The output directory has folders assigned with a patient ID (Appendix p 21).
We divide the image slices, masks, and the corresponding JSON-based annotations into training, validation, and testing subsets. The training subset generally contains a significant portion of the dataset. For instance, we used 70% of the total dataset for training purposes. The validation subset (15% of the total dataset) was used for testing the trained model. This is not the same as using the validation dataset for hyperparameter tuning. Instead, it is used to evaluate the model’s performance on unseen data after it has been trained. Finally, the testing subset (15% of the total dataset) allows the evaluation of the model’s generalisation and accuracy on unseen data, ensuring its reliability and effectiveness in real-world scenarios (Appendix p 33). To mitigate the influence of class imbalance on the model’s performance, we balanced the number of images in the training subset according to the class with the fewest images (Appendix p 34).
Lung segmentation and object detection
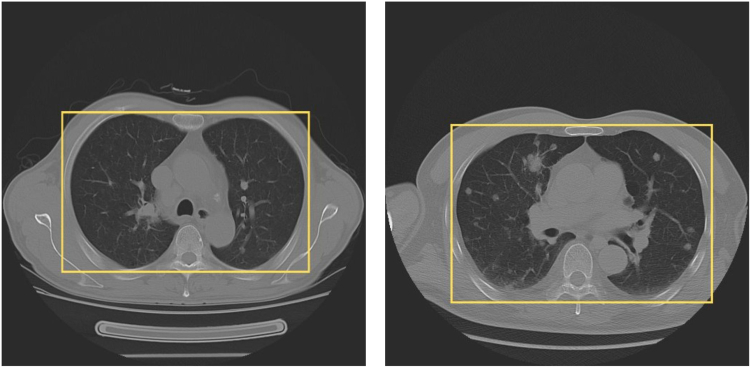
We used Facebook Research’s Detectron2 Faster R-CNN R101-FPN119 for acquiring and extracting the lung region of interest (lung ROI) and suppressing the non-lung areas in every slice of a 3D image that embodies a lung cancer nodule (Fig. 3), followed by the extraction of image features, and the training of object detection models to detect and classify lung nodules.
Fig. 3.
Lung region of interest. A yellow box is used to visually highlight and enclose the region of interest (ROI) corresponding to the lungs. This box serves as a visual reference, indicating the specific area within the image. By enclosing the lung area with a yellow box, it becomes easier to identify and focus on the relevant portion of the image for further analysis.
Detectron2 provides numerous base models19 pre-trained on a large image set such as ImageNet.20 These base models serve as the foundation for our network and are used to extract image features and train our model. One such model is the ResNet101-Feature Pyramid Network (R101-FPN) Faster R-CNN pre-trained base model. It exhibits a 42% Average Precision (AP) on the ImageNet benchmark dataset, indicating its effectiveness in detecting and classifying objects within the lung ROI, including lung nodules.
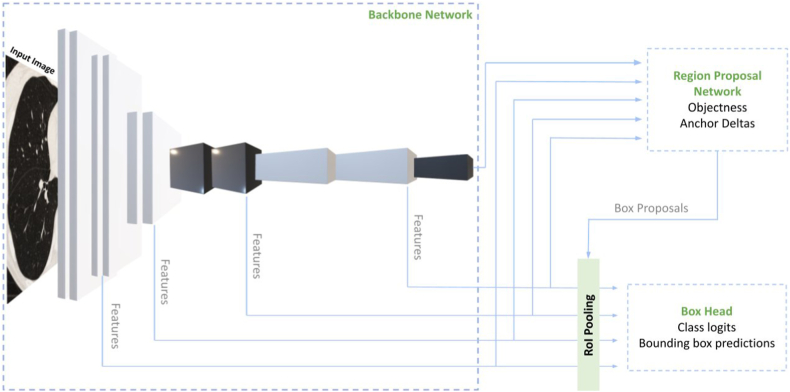
The training subset containing the annotations, masks, and image slices was fed into this pre-trained base model. The base model with the ResNet backbone extracts features from the input image and provides high-level semantic convolutional feature maps at all scales21 (Fig. 4). These feature maps contain valuable semantic information that helps identify and understand lung nodules' presence.
Fig. 4.
The three components of the AIPS-N Faster R-CNN model architecture. The model architecture consists of three primary components: the backbone network, the region proposal network (RPN), and the box head neural network. These components work together to facilitate various tasks and contribute to the overall functionality of the model. Together, these components enable the model to accurately detect and classify objects of interest.
Next, a small subnetwork called the Region Proposal Network (RPN) is used on the multi-scale feature maps. The purpose of the RPN is to predict region proposals, which are potential areas in the image that may contain objects of interest. The RPN accomplishes this by producing two outputs: objectness scores and anchor deltas. The RPN assigns an objectness score to each region proposal, indicating the probability of an object's presence in that proposed region. Higher scores suggest a higher likelihood of an object being present. Anchor deltas are predefined bounding boxes of different sizes and aspect ratios placed over the feature maps. The RPN generates anchor deltas, which represent the adjustments to the anchor box sizes and positions relative to the original image size. These deltas help refine the anchor boxes to align more accurately with the objects in the image.
Using the objectness scores and anchor deltas, the RPN generates box candidates. This process involves selecting regions with high objectness scores and applying the anchor deltas to adjust the size and position of the anchor boxes. The scales and aspect ratios are essential parameters that control the size and shape variations of the proposed boxes, allowing for the detection of objects at different scales and proportions.
The box candidates, obtained from the RPN, then undergo the RoI pooling layer. This layer reshapes the proposed regions to a standardised size, preparing them as inputs for the subsequent Box Head neural network.
The Box Head network performs two main tasks: classification and prediction of bounding box offsets. It classifies the input image by assigning object class labels to the proposed regions, determining the presence of specific objects within them. Additionally, the network predicts the precise offset values for the bounding boxes within the proposed regions, enabling accurate localization of the objects. During the training process of the Faster R-CNN model, hyperparameter configurations were kept as default from Detectron2 to optimise the model’s performance. The hyperparameters used while training the Faster R-CNN model are in the Appendix (p 13).
The workflow involved in constructing an automated lung segmentation and object detection model is performed individually for each nodule feature. Following this process, five AIPS-N feature models are obtained, one for each feature. These models can predict the location of each nodule within a lung slice, denoted by a red bounding box, within the designated lung region of interest (ROI) marked by a green rectangular box along with the predicted class. For instance, using the AIPS-N malignancy model, a prediction of 4 corresponds to “Moderately Suspicious”, while a prediction of 2 using the AIPS-N margin model corresponds to “Near Poorly Defined,” as stated in the Appendix (p 14). The AP50 value, which represents the intersection over union (IoU) threshold of 50%, is used as a measure to validate the prediction capability of the model. The Average Precision at 50% IoU (AP50) is a commonly used evaluation metric in object detection and instance segmentation tasks. It measures the accuracy of predictions by considering the overlap between predicted bounding boxes or segmentation masks and ground truth annotations.
Development of the AIPS-mutation (AIPS-M) model
EGFR mutations are reported to be identified with specific clinical factors including age, gender, smoking status, and histopathology6; therefore, diagnostic clinical factors were merged with the AIPS-N results to create the AIPS-M machine learning and deep learning (DL) based classifiers for predicting the AIPS-M score (EGFR mutation probability score) of a patient.
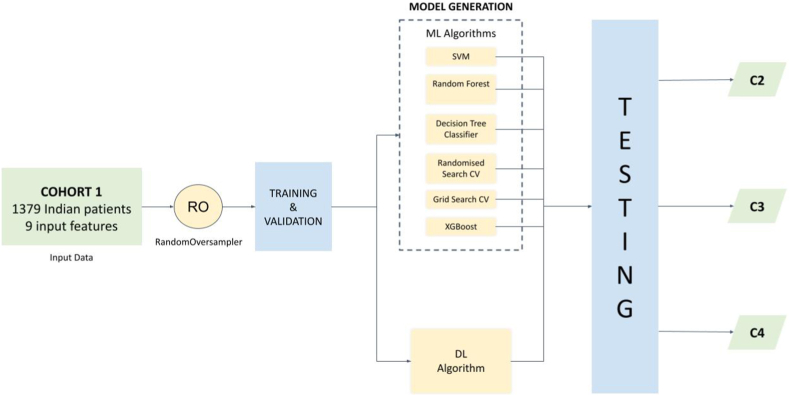
The AIPS-N scores were combined with the clinical factors of each patient, resulting in merged data with 9 input features (Appendix pp 14–15) from 1379 Indian patients in Cohort 1 (Table 1, Appendix p 35). Numerical and categorical data with missing clinical factors were imputed using mean value and value with the highest frequency respectively (Appendix p 15). Next, we used RandomOversampler to over-sample the minority class, in our case, the mutant class, by picking samples at random with replacement (Appendix p 36). The oversampled data was then split into training and validation subsets (Appendix p 37). This data was used to train and validate machine learning (ML) algorithms and the deep learning algorithm separately. A summary of the AIPS-M experiments conducted during the study is depicted in Fig. 5.
Fig. 5.
Summary of AIPS-M experiments conducted during the study.
The AIPS-N scores were combined with the clinical factors of each patient, resulting in merged data with 9 input features from 1379 Indian patients in Cohort 1. Random oversampling (RO) was applied to balance the classes, specifically oversampling the minority class (mutant class) using the RandomOversampler. The oversampled data was split into training and validation subsets. The ML models and the DL model were generated using the training and validation subsets. The trained ML models and the DL model were validated using the validation subset and tested independently using two Indian testing cohorts (Cohort 2 & Cohort 3) and a White testing cohort (Cohort 4). The performance evaluation of the models was conducted using metrics such as receiver operating characteristic curve (AUC), accuracy, recall, precision, and F1-score (Fig. 5).
The AIPS-M ML algorithms employed were support vector machine (SVM), random forest, decision tree classifier, and XGBoost. Additionally, we employed grid search cross-validation (CV) and randomised search cross-validation (CV) on the random forest model to optimise the hyperparameters of the random forest algorithm.
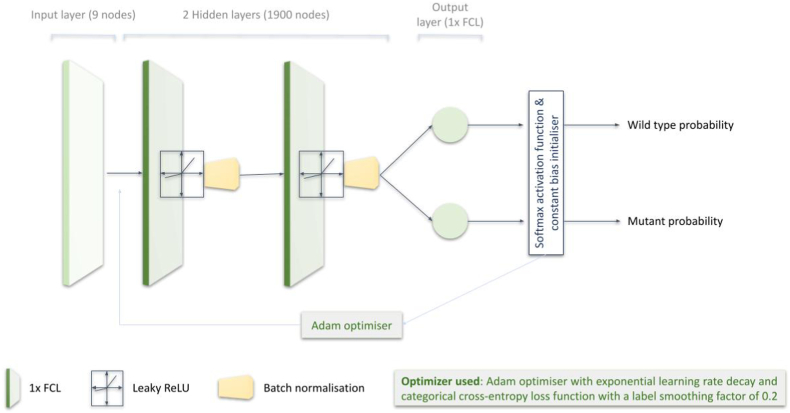
The AIPS-M DL algorithm was trained separately from the machine learning models and served as an alternative approach. Before training the DL model, we conducted hyperparameter tuning to find out the optimum value of parameters (Appendix p 16). The robustness of the DL algorithm is boosted by hyperparameter tuning, particularly when a hyperparameter affects a significant fraction of the variance.22,23 The AIPS-M DL model consists of four layers - one input layer, two hidden layers, and one output layer (Fig. 6). The total number of clinical factors and the AIPS-N scores of each nodule determines the number of nodes in the input layer (9 nodes in the input layer) (Appendix p 15). A fully connected layer (1 × FCL), an activation function (1 × leaky ReLU24), and a batch normalisation layer25 (1 × BNL) make up a single hidden layer of the DL model. The output layer is composed of a single FCL (1×) with 2 nodes. The 2 nodes are representative of the two output variables (wild-type and mutant). The probability of each output variable was calculated using softmax activation.26 During the early stages of algorithm training, the minimal loss was calculated by a constant bias initializer with a value obtained using log (total number of positive samples/total number of negative samples). Subsequently, we implemented Adam optimizer27 during training (Fig. 6).
Fig. 6.
Construction of the deep learning model.
In addition, we trained the AIPS-M models using only the clinical factors (Appendix p 15). This approach allowed us to evaluate the performance of the models in comparison to the models trained using both the clinical factors and the AIPS-N scores. By analysing the results, we gained insights into the impacts of incorporating the AIPS-N scores on the overall performance of the models.
In a separate experiment, random oversampling was applied to balance the classes in Cohort 4 (White population). Subsequently, Cohort 4 was divided into training, validation, and testing subsets. However, independent testing of Cohort 4 could not be performed due to the unavailability of data (Appendix p 17).
Role of funding source
The funder had any role in the design, conduct, analysis, or interpretation of the study, or in the decision to submit the results for publication.
Results
Results of the AIPS-N model
The preprocessing of the lung-ROI carried out through the process of windowing resulted in CT slices with adjusted contrast and brightness. The contrast between a CT slice before and after preprocessing is demonstrated in Appendix (p 22). We applied windowing to improve CT image visibility and interpretability for human observers, which is valuable for medical image analysis tasks. The use of windowing enhanced result interpretability without impacting the performance of the ML and DL models.
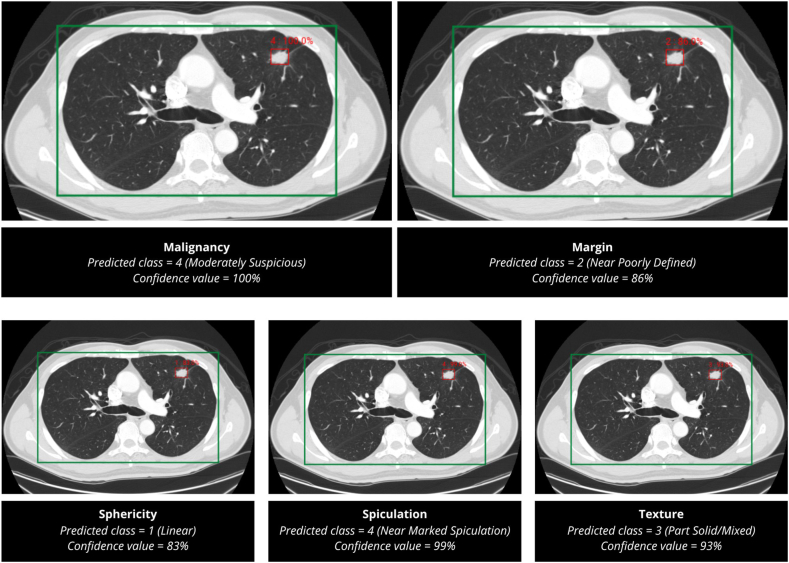
The Faster R-CNN-based AIPS-N model is trained using the participant folders containing the preprocessed CT slices, corresponding masks, and annotations (Appendix p 21). The model achieved an average AP50 value (IoU ≥ 50%) of 70.19% in predicting the location of nodules within the lung-ROI during validation. The confidence value is a machine learning probability score that indicates how confident the algorithm is that it has extracted the correct class of the nodule property. A sample prediction for all five properties is shown in Fig. 7.
Fig. 7.
Predictions made by the AIPS-N model. The AIPS-N model predicted the AIPS-N score of five properties for a CT slice embodying a lung cancer nodule. The AIPS-N scores are used to train the AIPS-M ML and DL models.
Results of the AIPS-M model
The ML algorithms trained using Cohort 1 achieved an average area under the Receiver Operating Characteristic (ROC) curve value of 0.85 in the validation subset. Among the ML algorithms, Random Forest yielded a slightly higher AUC value in the validation subset (Appendix pp 24, 38). Additionally, we tested the trained ML models using Cohort 2 and Cohort 3. Randomised Search Cross-Validation yielded a slightly higher AUC value of 0.91 (95 per cent confidence interval, 0.82–0.99) testing Cohort 2 (Appendix pp 25, 39). XGBoost yielded a slightly higher AUC value of 0.88 (95 per cent confidence interval, 0.81 to 0.95) in testing Cohort 3 (Appendix pp 26, 40). We tested the ML models trained on the Indian population (Cohort 1) on the White cohort (Cohort 4). The models achieved an average area under the receiver operating characteristic curve (AUC) value of 0.82 (Appendix pp 27, 41).
The DL algorithm trained using Cohort 1 achieved an AUC value of 0.86 in the validation subset. Additionally, we tested the trained DL model using Cohort 2 and Cohort 3 (Indian population), and Cohort 4 (White population). The AIPS-M DL model achieved an AUC value of 0.79 in both testing Cohort 2 and Cohort 3 (Appendix pp 28, 42). As previously mentioned, the developed deep learning (DL) model offers an alternative to the machine learning (ML) models. To facilitate a comprehensive comparison between the two approaches, we have included a diagram in Appendix (p 23).
In another experiment, we trained the ML and DL algorithms using only the clinical factors to evaluate their performance compared to models trained with both clinical factors and AIPS-N scores. Including AIPS-N scores led to improved performance in the machine learning models. For instance, in the testing Cohort 4, which comprised the White population, the average AUC value of the ML models trained using Cohort 1 (Indian population) increased from 0.6 to 0.9. This highlights the beneficial impact of incorporating AIPS-N scores on the predictive capabilities of the models (Appendix p 43).
The publicly available cohort 4 (White population) contained 133 wild-type class and 38 mutant-class patients. We applied RandomOversampler to the entire sample of Cohort 4 to over-sample the minority class. Next, we split Cohort 4 into training, validation, and testing subsets. AIPS-M ML algorithms trained using the training subset of Cohort 4 achieved an average AUC value of 0.81 on the validation subset (Appendix pp 29, 44) and an average AUC value of 0.85 on the testing subset (Appendix pp 30, 45).
AIPS-M DL algorithm trained using Cohort 4 achieved a validation AUC value of 0.9 and a testing AUC of 0.86 (Appendix pp 31, 46).
Case study
The following case study serves as a demonstration of the full system’s functionality, encompassing AIPS-N and AIPS-M, from a clinical perspective:
The clinical data for a patient with ID XXX637, a 71-year-old Indian male with a history of smoking was obtained from Rajiv Gandhi Cancer Institute and Research Center. The patient was included in the study based on his diagnosis of Squamous Cell Carcinoma, confirmed through histology, and the presence of the EGFR mutation, as determined by genomics data.
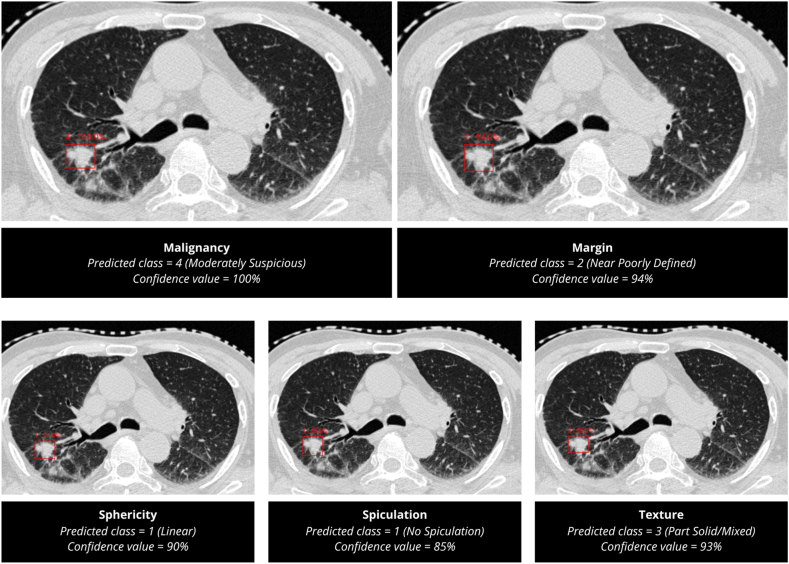
Role of AIPS-N
The AIPS-N model successfully predicted the location and characteristics of the detected nodule. It classified Sphericity and Spiculation as class 1 with confidence values of 90% and 85%, respectively. The Margin feature was predicted as class 2 with a higher confidence value of 94%. Texture analysis resulted in a prediction of class 3 with a confidence level of 93%. Lastly, the Malignancy class was predicted with a 100% confidence level (Fig. 8). A magnified version of the predictions made by the AIPS-N model is depicted in Appendix (p 32).
Fig. 8.
Predictions made by the AIPS-N model on Patient XXX637.
Role of AIPS-M
The AIPS-M models, trained using Cohort 1 (Indian population), were utilised to predict the EGFR status of Patient XXX637. According to the clinical data obtained from RGCI, the patient’s actual EGFR status is known to be ‘mutated’. To make the prediction, the models utilised both the clinical data and the AIPS-N feature scores. Remarkably, all six ML Algorithms (SVM, Random Forest, Decision Tree Algorithm, Grid Search Cross-Validation, Randomized Search Cross-Validation, and XG Boost) predicted the status as ‘mutated’, resulting in a ‘True Positive’ outcome. Similarly, the deep learning model also produced a ‘True Positive’ result (Appendix p 47). This accurate prediction of the patient's EGFR status by the models showcases the effectiveness of the applied methodologies.
Discussion
The outcome of our study suggests that regular CT imaging integrated with a fully automated lung nodule detection and characterisation AI system can predict the status of EGFR genotype and single out patients with a mutation in a cost-effective and non-invasive manner. The performance metrics of the AIPS model for both the Indian and the White population suggest that CT imaging provides information that complements clinical factors.
NGS is the benchmark diagnostic procedure for determining genotypes. However, it faces challenges due to tumour tissue heterogeneity, the changeable EGFR mutation status over time, tissue limitation in lung cores, and its cost-effectiveness in resource-constrained settings. Under such circumstances, AIPS can be applied to triage patients requiring panel-based NGS testing in a resource-constrained setting, subsequently guiding appropriate therapy. Patients confirmed to have an EGFR mutation by gene sequencing were tested using the AIPS, which showed a precise prediction of the EGFR genotype.
AIPS-N detects lung nodules and characterises five features using deep CNN. We enhanced the generalizability of our AIPS-N model by addressing systematic differences in the Cohort 5 CT images due to site, scanner, and scanning parameters. Site differences affect clinical protocols and generalizability, scanner variations impact image quality and consistency, and scanning parameter disparities influence diagnostic accuracy. Comprehensive handling of these differences ensures AI models are robust and clinically valid, increasing their effectiveness in diverse healthcare settings. We applied rescaling and windowing to Cohort 5’s images, addressing variability in imaging sources before training the AIPS-N object detection models. This helped to tackle systemic differences due to site, scanner, and scanning parameters.
While rescaling and windowing are useful techniques for addressing variability in medical imaging, they have limitations. Rescaling can lead to the loss of subtle image details, especially if extreme adjustments are made, potentially affecting diagnostic accuracy. Windowing, while enhancing certain features, may obscure others if not appropriately set, and the subjectivity in parameter selection can introduce variability. Additionally, these techniques may not fully correct for all systematic differences, such as variations in image resolution or noise levels between different scanners. Therefore, we took additional measures to prepare Cohort 5 for training the AIPS-N model and generalising to Cohorts 1–4, including consistent image format, exclusive focus on lung cancer cases, a comprehensive selection process, and data harmonisation techniques. This guarantees the model’s accuracy and applicability across varied lung cancer scenarios.
AIPS-M predicts the EGFR genotype using machine learning and deep learning. Machine Learning encompasses a diverse range of algorithms utilised for solving various data problems. Data scientists emphasise that no universally optimal algorithm can address every problem effectively. The selection of a suitable algorithm depends on various factors such as the nature of the problem at hand, the number of variables involved, and the most suitable model for the specific task.27 In our specific case, we found that the Grid Search Cross-Validation and Random Forest algorithms exhibited slightly superior performance in the testing cohorts of the Indian and White populations as compared to other algorithms. The trained deep learning algorithm exhibited promising performance, indicating its effectiveness in capturing complex patterns within the data. Additionally, we applied various techniques to avoid data leakage, which is a critical issue in machine learning training that can lead to overestimation of model performance and invalid results. These techniques include train-validation split, cross-validation, feature engineering, and data imputation (Appendix p 18).
Our analysis reveals two prominent distinctions between Cohort 2 [n = 591] and Cohort 3 [n = 96]. Firstly, Cohort 3 comprises significantly fewer samples than Cohort 2. Secondly, Cohort 3 exhibits a class imbalance, where positive and negative cases are unevenly represented. This imbalance can impact precision and F1 scores, contributing to the comparatively poorer precision and F1 score in Cohort 3, as compared to Cohort 2 (Appendix p 19).
We assessed the generalisability of the AIPS model because the EGFR mutation rate differs between ethnicities.28, 29, 30 Moreover, there are no mutation prediction models such as the AIPS-M trained on the Indian population, with most models primarily trained on Chinese and White populations. Hence, we used data from an Indian population to train the AIPS-M model and data from a White population and another Indian population to test the model; AIPS produced promising results in both populations. We also trained the AIPS-M ML and DL models using Cohort 4 (White population). Customising AIPS-M models for Indian and White populations acknowledges population-specific impacts on model performance. This strategy optimises accuracy by accounting for unique traits. Decision hinges on data availability and potential imaging variations (Appendix p 20).
We built the AIPS model to eliminate the need for laborious CT imaging annotation and identification of adenocarcinoma because it was trained using all types of lung cancer.6,8 This preserves its fully automated functionality and makes it more convenient for use in clinical practice. Most significantly, we found that analysis of lung nodule characteristics could play a role in lung cancer diagnosis.
Our research has several limitations to consider. Firstly, due to the absence of external annotated data, we divided Cohort 5 into training, validation, and an internal testing cohort. While this internal test set assesses the model’s performance on unseen data, it’s not fully independent. The study acknowledges this limitation, as it can’t replace the need for an entirely independent test set due to the lack of external validation data. Secondly, the Indian datasets were sourced solely from one institution, which hinders the generalizability of our findings to other settings and may not fully encompass the diversity of lung cancer cases. Moreover, we initially applied the Random Oversampler technique to the entire sample of Cohort 4, potentially introducing data leakage in the test set. We recognize that limiting oversampling exclusively to the training set is a well-established practice known to improve the assessment of model performance. Despite using multiple types of imaging equipment to generate data at RGCI, potential issues might arise when applying the platform to imaging data from different instruments or manufacturers if proper data harmonisation techniques are not employed.
To enhance targeted therapy, it’s essential to analyse genes beyond EGFR, such as ALK, KRAS, and ROS simultaneously, especially in resource-limited settings to save valuable resources. Incorporating a substantial number of datasets from the Indian population is expected to bolster the system’s performance. Additionally, we emphasise that the integration of data from various imaging devices enhances the robustness of our trained models, provided that all images undergo consistent and standardised preprocessing. Future research directions should prioritise external validation, standardised protocols, comparative analyses, longitudinal assessments, and validation across diverse populations.
In conclusion, AIPS provides a non-invasive method to predict EGFR genotype through the analysis of lung nodules detected in CT images, which reveals that genotype information can be extracted from the lung nodules.
Contributors
U.B., S.N., S.K.N, and K.R. designed research; K.R. supervised the research; U.B., S.N., S.P., M.S., A.B., and A.M. provided clinical, radiological, histopathological, and genomics datasets; S.K.N., K.R., J.T.J., T.S., N.A., and V.K. compiled information from many different sources; S.K.N. and K.R. created algorithms and framework for AI-based work; S.K.N., K.R., J.T.J., T.S., and P.P., performed research; S.K.N., K.R., T.S., P.P., N.A., and V.K. conducted testing and validation of AI models; S.K.N., and K.R. wrote the paper; K.R. provided the computational infrastructure for conducting AI-based work.
Data sharing statement
Data in the TCIA cohort is publicly available at https://wiki.cancerimagingarchive.net/display/Public/NSCLC+Radiogenomics. Other datasets used in this study belong to Rajiv Gandhi Cancer Institute and Research Centre, New Delhi, India (RGCI & RC). For researchers who want to access these datasets for academic research purposes, please direct your request to the corresponding author (krawal@amity.edu). The code used in this study including the AIPS-N lung segmentation and nodule detection model, the AIPS-M machine learning and deep learning models, and the trained weights of these models are available at https://github.com/rawallab8 and can be accessed by contacting the corresponding author (krawal@amity.edu).
Declaration of interests
Preeti P was supported by a fellowship received from the Indian Council of Medical Research (ICMR), Govt. of India [BMI/12 (66)/2021 2021-6442]. Authors declare no competing interests.
Acknowledgements
The authors would like to acknowledge Amity Institute of Biotechnology, Amity University, Uttar Pradesh. The high-end computational facility used in this work was developed and maintained from grants obtained from Kleberg Foundation, USA; Department of Biotechnology (DBT), Govt. of India [BT/PR17252/BID/7/708/2016]; SERB, Govt. of India [File Number: CVD/2020/000842]; We are thankful to Priyanka Ray Choudhury for her contribution to the conduct of this study.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.lansea.2024.100352.
Appendix ASupplementary data
References
- 1.Ladanyi M., Pao W. Lung adenocarcinoma: guiding EGFR-targeted therapy and beyond. Mod Pathol. 2008;21:S16–S22. doi: 10.1038/modpathol.3801018. [DOI] [PubMed] [Google Scholar]
- 2.Bi W.L., Hosny A., Schabath M.B., et al. Artificial intelligence in cancer imaging: clinical challenges and applications. CA Cancer J Clin. 2019;69(2):127–157. doi: 10.3322/caac.21552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lambin P., Leijenaar R.T.H., Deist T.M., et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 4.Bai H., Wang Z., Chen K., et al. Influence of chemotherapy on EGFR mutation status among patients with non–small-cell lung cancer. J Clin Oncol. 2012;30:3077–3083. doi: 10.1200/JCO.2011.39.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mu W., Jiang L., Zhang J., et al. Non-invasive decision support for NSCLC treatment using PET/CT radiomics. Nat Commun. 2020;11:5228. doi: 10.1038/s41467-020-19116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rios Velazquez E., Parmar C., Liu Y., et al. Somatic mutations drive distinct imaging phenotypes in lung cancer. Cancer Res. 2017;77:3922–3930. doi: 10.1158/0008-5472.CAN-17-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen W., Zhou M., Yang F., et al. Multi-crop convolutional neural networks for lung nodule malignancy suspiciousness classification. Pattern Recogn. 2017;61:663–673. [Google Scholar]
- 8.Wang S., Zhou M., Liu Z., et al. Central focused convolutional neural networks: developing a data-driven model for lung nodule segmentation. Med Image Anal. 2017;40:172–183. doi: 10.1016/j.media.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chamberlin J., Kocher M.R., Waltz J., et al. Automated detection of lung nodules and coronary artery calcium using artificial intelligence on low-dose CT scans for lung cancer screening: accuracy and prognostic value. BMC Med. 2021;19:55. doi: 10.1186/s12916-021-01928-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dandil E., Cakiroglu M., Eksi Z., Ozkan M., Kurt O.K., Canan A. 2014 6th international conference of soft computing and pattern recognition (SoCPaR) [internet] IEEE; Tunis, Tunisia: 2014. Artificial neural network-based classification system for lung nodules on computed tomography scans.http://ieeexplore.ieee.org/document/7008037/ pp. 382–386. [cited 2023 July 24]. Available from: [Google Scholar]
- 11.Wang S., Yu H., Gan Y., et al. Mining whole-lung information by artificial intelligence for predicting EGFR genotype and targeted therapy response in lung cancer: a multicohort study. Lancet Digital Health. 2022;4:e309–e319. doi: 10.1016/S2589-7500(22)00024-3. [DOI] [PubMed] [Google Scholar]
- 12.Wang S., Shi J., Ye Z., et al. Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur Respir J. 2019;53 doi: 10.1183/13993003.00986-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bakr S., Gevaert O., Echegaray S., et al. A radiogenomic dataset of non-small cell lung cancer. Sci Data. 2018;5 doi: 10.1038/sdata.2018.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Armato S.G., McLennan G., Bidaut L., et al. The lung image database Consortium (LIDC) and image database resource initiative (IDRI): a Completed reference database of lung nodules on CT scans: the LIDC/IDRI thoracic CT database of lung nodules. Med Phys. 2011;38:915–931. doi: 10.1118/1.3528204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark K., Vendt B., Smith K., et al. The cancer imaging archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26:1045–1057. doi: 10.1007/s10278-013-9622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baba Y., Murphy A. Radiopaedia.org. Radiopaedia.org. 2017. Windowing (CT) [DOI] [Google Scholar]
- 17.Hovinga M., Sprengers R., Kauczor H.-U., Schaefer-Prokop C. In: Multidetector-row CT of the thorax. Schoepf U.J., Meinel F.G., editors. Springer International Publishing; Cham: 2016. CT imaging of interstitial lung diseases; pp. 105–130. [Google Scholar]
- 18.Hancock M.C., Magnan J.F. Lung nodule malignancy classification using only radiologist-quantified image features as inputs to statistical learning algorithms: probing the Lung Image Database Consortium dataset with two statistical learning methods. J Med Imag. 2016;3 doi: 10.1117/1.JMI.3.4.044504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.facebookresearch/detectron2. 2022. https://github.com/facebookresearch/detectron2/blob/7c2c8fb168a2093ce06a531c1208fba48d2984ec/MODEL_ZOO.md published online Oct 13.
- 20.Deng J., Dong W., Socher R., Li L.-J., Li Kai, Fei-Fei Li. 2009 IEEE conference on computer vision and pattern recognition. IEEE; Miami, FL: 2009. ImageNet: a large-scale hierarchical image database; pp. 248–255. [Google Scholar]
- 21.Lin T.-Y., Dollar P., Girshick R., He K., Hariharan B., Belongie S. 2017. Feature Pyramid networks for object detection; pp. 2117–2125. [Google Scholar]
- 22.van Rijn J.N., Hutter F. Proceedings of the 24th ACM SIGKDD international conference on knowledge discovery & data mining. ACM; London United Kingdom: 2018. Hyperparameter importance across datasets; pp. 2367–2376. [Google Scholar]
- 23.Rawal K., Sinha R., Nath S.K., et al. Vaxi-DL: a web-based deep learning server to identify potential vaccine candidates. Comput Biol Med. 2022;145 doi: 10.1016/j.compbiomed.2022.105401. [DOI] [PubMed] [Google Scholar]
- 24.Maas A.L., Hannun A.Y., Ng A.Y. Proceedings of the 30th International Conference on Machine Learning. 2013. Rectifier nonlinearities improve neural network acoustic models. (Vol. 30, No. 1, p. 3) [Google Scholar]
- 25.Ioffe S., Szegedy C. Proceedings of the 32nd international conference on machine learning. PMLR; 2015. Batch normalization: accelerating deep network training by reducing internal covariate shift; pp. 448–456. [Google Scholar]
- 26.Nwankpa C., Ijomah W., Gachagan A., Marshall S. 2018. Activation functions: comparison of trends in practice and research for deep learning. [DOI] [Google Scholar]
- 27.Kingma D.P., Ba J. Adam: a method for stochastic optimization. arXiv. 2014 preprint arXiv:1412.6980. [Google Scholar]
- 28.Recondo G., Facchinetti F., Olaussen K.A., Besse B., Friboulet L. Making the first move in EGFR-driven or ALK-driven NSCLC: first-generation or next-generation TKI? Nat Rev Clin Oncol. 2018;15:694–708. doi: 10.1038/s41571-018-0081-4. [DOI] [PubMed] [Google Scholar]
- 29.Wu Y.-L., Cheng Y., Zhou J., et al. Tepotinib plus gefitinib in patients with EGFR-mutant non-small-cell lung cancer with MET overexpression or MET amplification and acquired resistance to previous EGFR inhibitor (INSIGHT study): an open-label, phase 1b/2, multicentre, randomised trial. Lancet Respir Med. 2020;8:1132–1143. doi: 10.1016/S2213-2600(20)30154-5. [DOI] [PubMed] [Google Scholar]
- 30.Leonetti A., Sharma S., Minari R., Perego P., Giovannetti E., Tiseo M. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br J Cancer. 2019;121:725–737. doi: 10.1038/s41416-019-0573-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.