SUMMARY
Corticospinal neurons (CSNs) synapse directly on spinal neurons, a diverse assortment of cells with unique structural and functional properties necessary for body movements. CSNs modulating forelimb behavior fractionate into caudal forelimb area (CFA) and rostral forelimb area (RFA) motor cortical populations. Despite their prominence, the full diversity of spinal neurons targeted by CFA and RFA CSNs is uncharted. Here, we use anatomical and RNA sequencing methods to show that CSNs synapse onto a remarkably selective group of spinal cell types, favoring inhibitory populations that regulate motoneuron activity and gate sensory feedback. CFA and RFA CSNs target similar spinal neuron types, with notable exceptions that suggest that these populations differ in how they influence behavior. Finally, axon collaterals of CFA and RFA CSNs target similar brain regions yet receive highly divergent inputs. These results detail the rules of CSN connectivity throughout the brain and spinal cord for two regions critical for forelimb behavior.
In brief
Carmona et al. comprehensively characterize the connectivity of two distinct forelimb corticospinal populations within the brain and spinal cord. Both groups preferentially target inhibitory spinal neurons but are selective in the specific neuronal subtypes they innervate. These corticospinal populations form axon collaterals in similar brain regions but receive different inputs.
Graphical Abstract
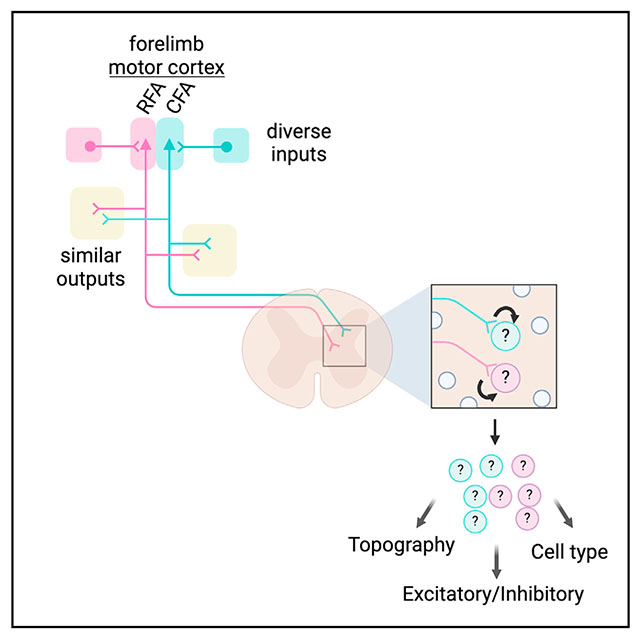
INTRODUCTION
Voluntary movement emerges from activity widely distributed throughout the nervous system.1 Signals generated by the brain recruit neuronal circuits in the spinal cord that control body muscles and regulate sensory feedback.2 These spinal circuits are derived from six major developmentally defined classes of dorsal neurons and five classes of ventral neurons, each of which comprise several transcriptionally defined subtypes with distinct structural and physiological properties.3–5 The brain shapes body movement in large part through the connections made with this diverse pool of spinal neurons, enabling countless patterns of muscle activation and regulating the structure of sensory feedback. Among the neuronal populations that project to the spinal cord, corticospinal neurons (CSNs) play an important role in this transformation.6
CSNs are the most populous descending input to the spinal cord.7 The primary axons of CSNs are widely distributed, forming terminal fields that collectively innervate nearly the entire rostro-caudal extent of the spinal cord.8–10 CSN axons also span the dorsoventral extent of the spinal gray, allowing them to synapse on diverse neuron subtypes distributed across spinal laminae. Indeed, targeted anatomical tracing efforts using transsynaptic tracing or appositional analysis have revealed several spinal neuron subtypes that receive input from CSNs, including premotor interneurons and sensory relay neurons.7,11,12 And yet, spinal neurons are incredibly diverse in identity, posing a challenge for comprehensively mapping the full matrix of corticospinal connectivity. Moreover, CSNs can be divided based on their location in the sensorimotor cortex, and the rules of spinal connectivity may differ across these populations.7 Specifically, the cell bodies of CSNs that are involved in forelimb control are distributed across two regions of the motor cortex: the caudal forelimb area (CFA) and rostral forelimb area (RFA).13 Both the CFA and RFA are involved in controlling arm and hand movements, although their exact roles are uncertain. Some suggest that RFA is more involved in grasping and CFA more involved in reaching, either through muscle-specific connectivity or activation of synergies.14 This is supported by recordings that show that RFA CSNs are more likely to be active during grasp.8 Similarly, focal cooling of RFA impairs grasp, while CFA cooling does not.15 Still, other studies have found that activity in the CFA and RFA during reach and grasp are indistinguishable16 or that transient inactivation of the CFA, rather than the RFA, produced more pronounced grasping deficits.17 While these differences may be explained by an experimental approach, a major challenge for interpreting the function of the CFA and RFA is that the spinal neuron subtypes targeted by these regions are not completely known.8,17,18 CSNs also produce robust axon collaterals that innervate many supraspinal regions, an architecture that enables coordination of behavioral signals across the entire nervous system.7,19 How CSNs in the CFA and RFA differ in their collateral connectivity with these brain regions has also been incompletely described.20 Finally, the roles of neurons are defined in part by their synaptic inputs, and we know little about the similarities or differences in brain-wide input to CFA and RFA CSNs.
Resolving these principles of connectivity is essential to understanding how descending motor control pathways enable behavior. In this study, we used a suite of transneuronal tracing, single-nucleus RNA sequencing, and synaptic electrophysiology to reveal the topographical and cell type connectivity of RFA and CFA populations. We characterized the innervation strategies of CFA and RFA CSNs in the spinal cord, revealing the cellular networks that ultimately shape descending behavioral commands. We further mapped both the brain-wide axon collateral and presynaptic input structures of CFA and RFA CSNs, discovering differences in their organization that suggest unique functional roles in behavior. Together, these results reveal the infrastructure of two unique parallel descending motor control pathways.
RESULTS
CFA and RFA CSNs target neurons in distinct spinal regions
CSNs are widely distributed across the cortical mantle, including across distinct sensorimotor cortical regions.7,21 Two major populations of motor CSNs arise from the CFA and RFA of the motor cortex.13,22 We confirmed this by injecting a retrogradely transported adeno-associated virus (AAV) encoding a fluorescent reporter (AAV-retro-FP [fluorescent protein]) into segments C3–C8 of the cervical spinal cord, home to the circuits that regulate forelimb control (Figure 1A, N = 3 mice).23 We then imaged antibody-enhanced fluorophore labeling and used the imaging analysis pipeline BrainJ to map their positions to a common brain atlas.24 Visual inspection of the dorsal surface of the brain revealed two physically separated groups of CSNs with positions corresponding to the CFA and RFA (Figure 1B).22 CFA and RFA CSNs project to overlapping, but partially distinct, regions of the spinal cord.8,9 We confirmed this using an intersectional strategy to express different fluorophores exclusively in CFA and RFA CSNs (Figure 1C, N = 6 mice, from the same mice as in Figure 3). We first injected a 1:1 cocktail of AAV-retro-Cre and AAV-retro-FlpO into the cervical spinal cord. We followed this with injections of AAV-FLEX-GFP into the CFA and AAV-FRT-tdTomato into the RFA, labeling CFA and RFA axonal projections with GFP and RFP, respectively (Figure 1C). Visual inspection of the cervical spinal cord revealed unique projection patterns of CFA and RFA CSNs. RFA CSNs appeared to target deeper regions of the spinal cord and CFA CSNs more superficial regions, consistent with previous studies (Figure 1D).8,9
Figure 1. CFA and RFA corticospinal neurons target neurons in distinct spinal regions.
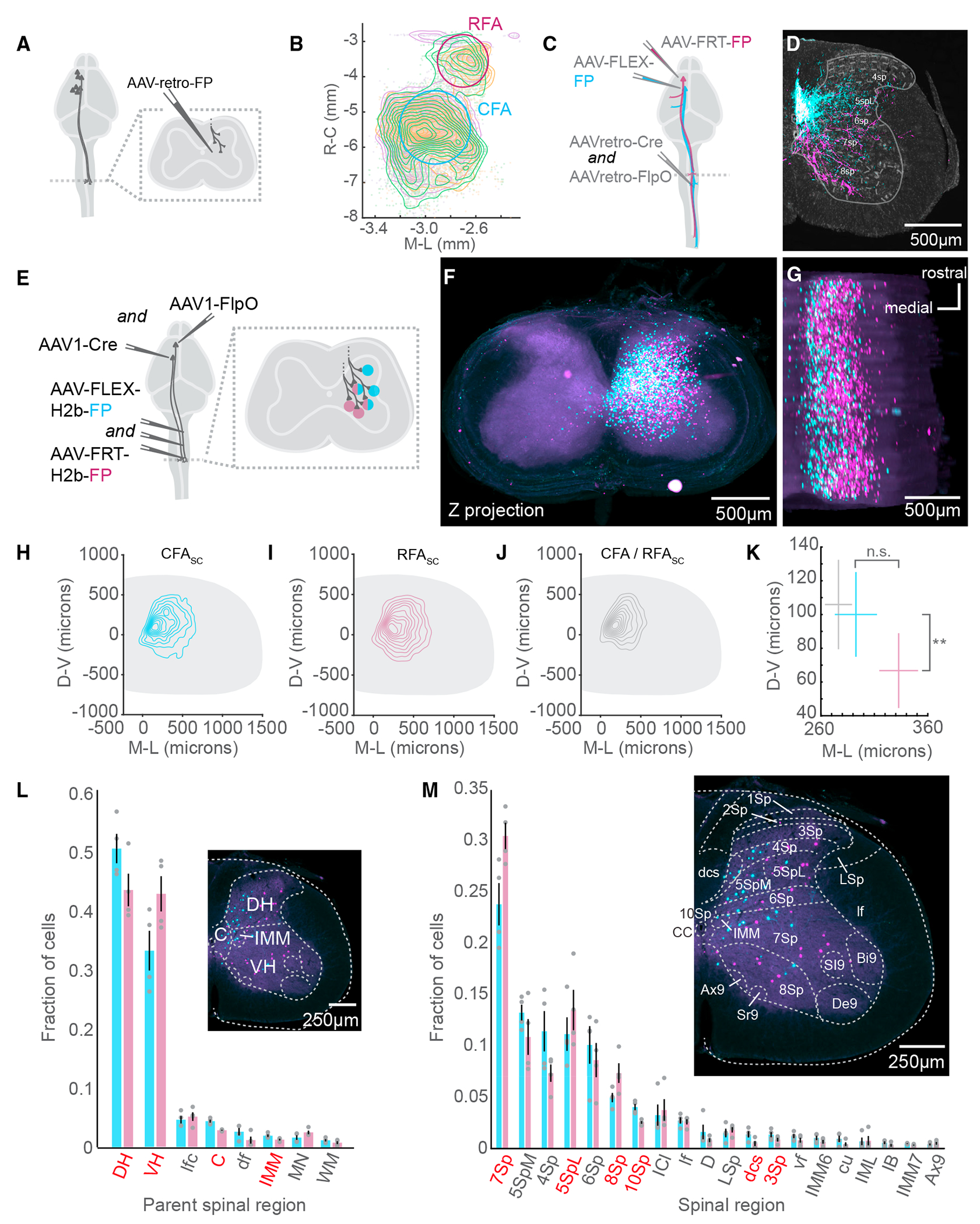
(A) Strategy to label brain-wide inputs to the cervical spinal cord.
(B) Top-down view of a 3D reconstruction of the mouse brain showing CSNs as individual points. A contour map of CSN cell bodies is superimposed. The approximate locations of CFA and RFA are indicated. The three colors indicate samples from 3 separate experiments (N = 3 mice for A and B).
(C) Strategy to simultaneously label CFA and RFA CSNs.
(D) Photomicrograph of a transverse section of the spinal cord with fluorescently labeled CFA (cyan) and RFA (magenta) axons. Neurotrace is in gray. Major spinal laminae are demarcated (representative of N = 6 mice for C and D).
(E) Strategy to simultaneously label spinal neurons that are innervated by CFA or RFA.
(F) z stack projection of aligned photomicrographs of the cervical spinal cord. CFASC is in cyan and RFASC is in magenta (representative of N = 4 mice for [F] and [G]).
(G) Rotated 3D reconstruction of a section of the cervical spinal cord.
(H) Contour plot of CFASC labeling. D-V, dorsoventral; M-L, mediolateral (N = 4 mice for [H]–[M]).
(I) Contour plot of RFASC labeling.
(J) Contour plot of double-labeled neurons (CFA/RFASC neurons).
(K) Mean centroid positions of CFASC, RFASC, and CFA/RFASC populations.
(L) The major spinal cord structures containing CFASC and RFASC neurons. Regions colored red indicate a significant difference between CFASC and RFASC fractions. The inset is a photomicrograph with major spinal structures indicated.
(M) The spinal cord laminae and nuclei containing CFASC and RFASC neurons. The inset is a photomicrograph with spinal structures indicated.
Refer to Table S1 for a complete list of the spinal cord structures and their corresponding acronyms. Data are represented as mean ± SEM. Red text in (F) and (M): p < 0.05, **p < 0.01
See also Figure S1.
Figure 3. The supraspinal topography of CFA and RFA axon collaterals.
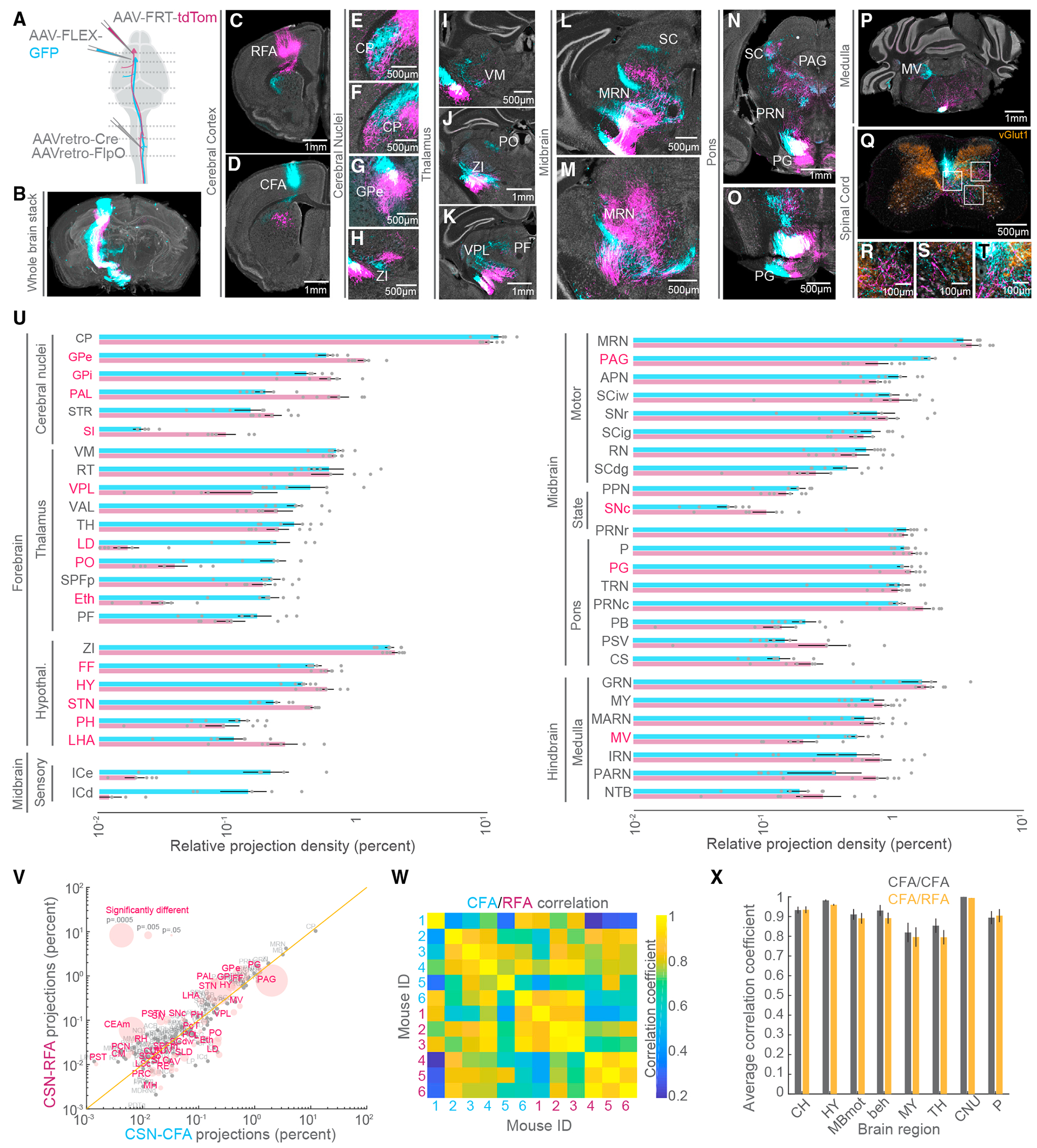
(A) Strategy to simultaneously label CFA and RFA CSNs.
(B) 3D reconstruction of a brain with CFA (cyan) and RFA (magenta) CSNs labeled (representative of N = 6 mice for B–T).
(C–T) Exemplar photomicrographs of CFA and RFA CSN labeling throughout the central nervous system. Select regions are indicated. In (Q)–(T), vGlut1 axonal boutons are immunolabeled and shown in orange.
(U) The brain regions that are targeted by CFA and RFA CSNs excluding isocortical structures (N = 6 mice for U–X). Regions are grouped by their ontology (see Table S3). Regions with significantly different fractions of CFA and RFA labeling are indicated in red. Note the log scale, given the large range of labeling.
(V) Scatterplot of regions containing CFA versus RFA projections. Regions with significantly different fractions of CFA and RFA labeling are colored red. The size of points corresponds to the p value of the comparison (paired t tests). Note the log scale, given the large range of labeling.
(W) Matrix depicting the correlation of labeling within and across all 6 mice. Quadrants correspond to CFA versus CFA (top left quadrant), RFA versus RFA (bottom right quadrant), or CFA versus RFA (bottom left and top right quadrants) labeling.
(X) The average correlation coefficients of CFA versus CFA (gray) labeling or CFA versus RFA labeling (orange) within major brain regions across all mice. CH, cerebrum; HY, hypothalamus; MBmot, motor midbrain; beh, behavioral-state-related midbrain; MY, medulla; TH, thalamus; CNU, cerebral nuclei; P, pons.
Refer to Table S3 for a complete list of the brain structures and their corresponding acronyms. Data are represented as mean ± SEM. Red text in (U) and (V): p < 0.05 See also Figure S4.
The presence of CFA and RFA CSN axons in distinct laminae of the spinal cord may reflect the innervation of topographically distinct neurons or may reflect differences in positional innervation of somatodendritic structures. We disambiguated these possibilities by labeling the spinal neurons targeted by CFA and RFA CSNs using anterograde transneuronal tracing.25,26 First, we injected a cocktail of AAV-FLEX-H2b-GFP and AAV-FLEX-H2b-RFP throughout the cervical spinal cord followed by injections of high-titer AAV1-Cre and AAV1-FlpO into CFA and RFA, respectively (Figure 1E, N = 4 mice). This led to the transneuronal spread of Cre and FlpO recombinases and the expression of GFP and RFP in spinal somata targeted by the CFA and RFA, respectively. We imaged antibody-enhanced fluorophore labeling and leveraged the image reconstruction and analysis pipeline SpinalJ to register transverse cervical spinal cord segments.27 This revealed a volume of cervical tissue with large populations of CFA-innervated spinal cord (CFASC) and RFA-innervated spinal cord (RFASC) neurons (Figures 1F and 1G, CFASC: 1,052 ± 223.96 cells, RFASC: 1,568 ± 579.12 cells). The topography of CFASC and RFASC neurons did not substantially differ across cervical spinal segments (Figure S1). Measuring the positions of CFASC and RFASC neurons revealed a great deal of overlap between these populations, including a fraction of neurons innervated by both the CFA and RFA (Figures 1H–1J, 75 ± 32.12 double-labeled cells). However, mapping the centroids of CFASC and RFASC populations across animals revealed a significant difference in the positions of innervated neurons: RFA CSNs target more ventral regions of the spinal gray, while CFA CSNs target superficial regions (Figure 1K, mean and SEM coordinates relative to the central canal; CFASC: 292.74 ± 19.70 μm medio-lateral (M/L), 100.03 ± 25.11 μm dorso-ventral (D/V); RFASC: 332.75 ± 18.15 μm M/L, 66.74 ± 22.18 μm D/V; D/V: p = 0.0081, paired t test, M/L: p = 0.1504, paired t test). Notably, double-labeled neurons settle in medial regions of the spinal cord, indicating convergent input is constrained to a select region of the spinal cord (CFA/RFASC: 276.40 ± 12.70 μm M/L, 105.91 ± 26.51 μm D/V).
The spatial coordinates of the spinal cord map onto well-defined and functionally distinct spinal laminae and nuclei.28 For example, the dorsal horn is home to several substructures involved in shaping sensory feedback, while the ventral horn is home to motor pools and the premotor neurons involved in shaping muscle activity, among other functions.23 With this knowledge, we again used SpinalJ to map the positions of GFP+ and RFP+ spinal neurons to a common spinal cord atlas. We discovered a greater fraction of RFASC neurons in the ventral horn compared to CFASC neurons (CFASC: 0.331 ± 0.033, RFASC: 0.427 ± 0.030, p = 0.0004, paired t test). Conversely, a greater fraction of CFASC neurons is found in the dorsal horn (CFASC: 0.503 ± 0.025, RFASC: 0.434 ± 0.028, p = 0.0025, paired t test) (Figure 1L). We next performed a granular analysis identifying individual laminae home to spinal neurons targeted by the CFA or RFA. Among spinal regions with labeled neurons, lamina 7 (7Sp), lateral lamina 5 (5SpL), and lamina 8 (8Sp) are home to a greater fraction of RFASC neurons compared to CFASC neurons (CFASC versus RFASC: 7Sp: 0.239 ± 0.021 versus 0.306 ± 0.013, p = 0.007; 5SpL: 0.112 ± 0.016 versus 0.135 ± 0.020, p = 0.027; 8Sp: 0.050 ± 0.005 versus 0.074 ± 0.010, p = 0.021, paired t tests), while lamina 10 (10Sp) and, to a lesser extent, lamina 3 (3Sp) contain a greater fraction of CFASC neurons (Figure 1M, 10Sp: 0.041 ± 0.003 versus 0.026 ± 0.001, p = 0.008; dorsal corticospinal tract (dcs): 0.014 ± 0.003 versus 0.005 ± 0.002, p = 0.041; 3Sp: 0.014 ± 0.003 versus 0.009 ± 0.002, p = 0.024, paired t tests). These results establish the topography of corticospinal connectivity and discover that CFA and RFA CSNs target neurons in overlapping but distinct regions of the spinal cord. Dorsal horn neurons are more likely to receive CFA input and ventral horn neurons more likely to receive RFA input, with individual spinal laminae contributing to this topographical bias.
CSNs are selective in their spinal neuron targets
The topography of spinal neurons reflects their transcriptional identity, electrophysiological properties, and synaptic connectivity. For instance, many spinal populations are defined by the expression of 2–3 transcription factors, settle in compact regions of the spinal cord, and are selective in their connectivity with sensory-motor circuits.5,29,30 The molecular and functional diversity of these spinal populations enables flexible control of motor output and precision processing of sensory feedback.31 What spinal subtypes do CSNs target? Previous efforts relied on assessing the connectivity between CSNs and individual neuronal subtypes,11,12 an approach that is difficult to scale to a more complete representation of spinal cell type diversity. Instead, we took a higher throughput approach, AnteroT-seq, by combining anterograde transneuronal cellular labeling with single-nucleus RNA sequencing. We injected high-titer AAV1-Cre into the CFA or RFA of CAG-Sun1-sfGFP (INTACT) mice,32 which express nuclear-localized GFP in the presence of Cre recombinase. This approach results in the transneuronal expression of Cre, and thus GFP, in spinal neurons targeted by the CFA or RFA (Figure 2A, N = 8 mice). We first confirmed that this approach labeled spinal neurons in a fashion consistent with the results from Figure 1. Visual inspection of antibody-enhanced INTACT labeling in spinal sections (Figure 2B, CFASC: N = 2 mice, RFASC: N = 2 mice) and volumetric reconstruction (Figure 2C, N = 1 mouse) confirmed GFP expression throughout the spinal gray. We quantified the distribution of GFP-labeled nuclei and confirmed that RFASC nuclei are more ventrally positioned compared to CFASC nuclei, like what we observed in our earlier efforts (Figure 2D, CFA: 100.67 ± 5.89 μm D/V, RFA: 60.03 ± 5.93 μm D/V, p = 1.2 × 10−6, unpaired t test of compiled data). Next, fresh-frozen cervical spinal cords were dissociated, and nuclei with GFP and DAPI fluorescence were enriched using fluorescence-activated cell sorting (FACS). Individual nuclei were profiled using SMART-Seq v.4, yielding a total of 765 and 678 nuclei post-quality control for CFA- and RFA-injected samples, respectively. We achieved a similar median gene detection for each target group with 4,769 genes for the CFA population and 4,415 genes for the RFA population. Louvain clustering of the aggregate data identified 9 clusters, which, as expected, consisted primarily of neurons (Figures S2A and S2B).
Figure 2. CSNs are selective in their spinal neuron targets.
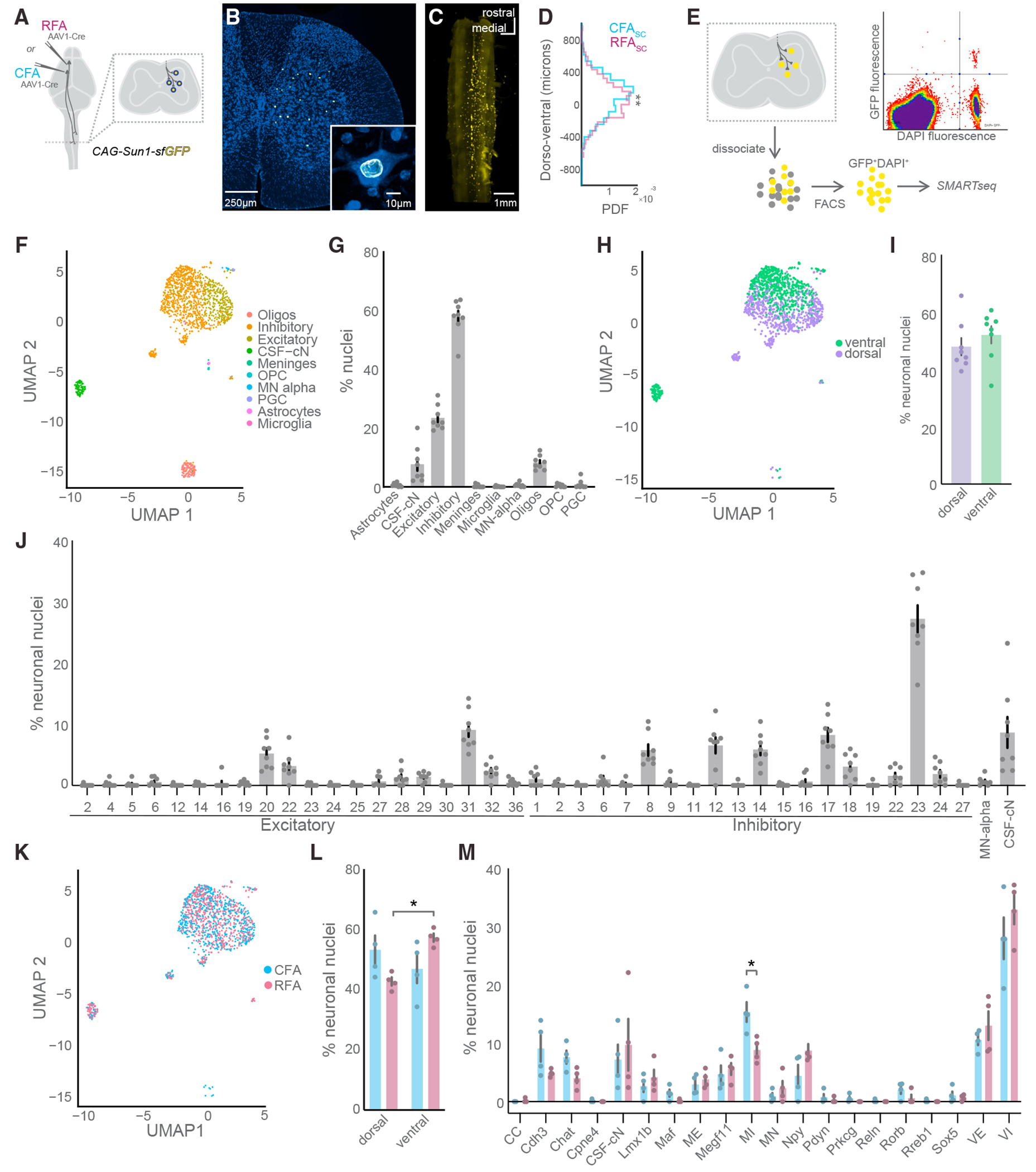
(A) Strategy to label spinal cord nuclei targeted by CFA or RFA with GFP.
(B) Exemplar photomicrograph of CFASC nuclei in the spinal cord.
(C) Light-sheet microscopy reconstruction of CFASC nuclear labeling (N = 1 mouse).
(D) Histogram of the dorsoventral distribution of CFASC and RFASC labeling (CFASC: N = 2 mice, RFASC: N = 2 mice, data compiled across mice).
(E) Strategy to isolate and perform RNA-seq on CFASC or RFASC neurons. An exemplar FACS plot is shown depicting GFP and DAPI co-labeled nuclei in the top right quadrant.
(F) Uniform manifold approximation and projection (UMAP) plot of all CSNSC sequenced nuclei passing quality control mapped to the major cell types of the harmonized spinal cord atlas from Russ et al.5 (N = 8 mice for F–M).
(G) The percentage of nuclei belonging to each major cell type across individual experiments.
(H) UMAP plot with neuronal nuclei labeled by dorsal and ventral positional identity.
(I) The positional identity of neuronal nuclei across individual experiments.
(J) The percentage of neuronal nuclei belonging to harmonized spinal neuron type clusters across individual experiments.
(K) UMAP plot of sequenced neuronal nuclei labeled by CFA or RFA experimental condition.
(L) The positional identity of CFASC and RFASC neurons across individual experiments.
(M) The percentage of neuronal nuclei belonging to the harmonized spinal neuron families across individual experiments. Oligos, oligodendrocytes; OPC, oligodendrocyte precursor cells.
Refer to Table S2 for a complete list spinal neuronal cell types, families, and their corresponding acronyms. Data are represented as mean ± SEM.*:p < 0.05, **: p < 0.01 See also Figures S2 and S3.
To determine the cell types that CSNs target at higher resolution, we mapped our complete dataset onto an atlas of spinal neuron diversity from Russ et al. comprising cross-validated RNA sequencing data compiled from several laboratories.5,33–36 This harmonized atlas catalogs spinal cell type diversity at several levels of granularity from broad neurotransmitter types to intersectional transcription factor expression that defines well-established spinal neuron subtypes (Figures 2F, S2C, and S2D). We labeled our single-nucleus transcriptomes for two classes by neurotransmitter type, glutamate for excitatory, and GABA and glycine for inhibitory (Figures 2F and S2B). First, we note a substantial preference for CSNs to contact inhibitory neurons over excitatory neurons (Figure 2G; 0.58 ± 0.02 versus 0.23 ± 0.06, p < 0.0001, Tukey’s multiple comparisons test). This result was obtained despite the relatively equal proportions of these two neuronal classes in the mouse spinal cord.37 Next, we focused solely on neuronal nuclei and identified cell types by mapping their similarity to Russ’s 69 spinal neuron clusters that include 38 excitatory clusters and 27 inhibitory clusters.5 Mapping to the broad dorsoventral position of these clusters did not show a preference in the location of the populations targeted (Figures 2H and 2I). However, we found several specific spinal cell types targeted by CSNs, including excitatory and inhibitory neurons distributed across dorsal and ventral laminae (Figure 2J). Notably, approximately 25% of all neurons belonged to the inhibitory-23 cluster (27% ± 2%), likely corresponding to the Foxp2 clade of cardinal V1 interneurons and suggesting an Ia-inhibitory interneuron identity.29 This cluster was, by a large margin, the most prominent target of CSNs. Other prominent clusters of neurons included excitatory-31 (9.07% ± 1.12%), inhibitory-12 (6.56% ± 1.32%), inhibitory-14 (5.88% ± 0.94%), and inhibitory-17 (8.26% ± 1.15%), including cell types with well-described transcriptional profiles and lineages that indicate their functional identity.38,39 By grouping these clusters into major spinal families, we identify ventral inhibitory (VI) and excitatory (VE) neurons as the most populous families of neurons targeted by CSNs (Figures S2E and S2F; Table S2). Given that ventral neurons are classified into single excitatory and inhibitory clusters each in this region because of their gradient of gene expression profiles, this was not an unexpected finding when considering the broad dorsoventral targeting of CSNs (VI: 30.48% ± 2.22%, VE: 11.81% ± 1.30%). This is followed by mid-inhibitory (MI) neurons (12.12% ± 1.53%), cerebrospinal fluid-contacting neurons (8.50% ± 2.41%), Cdh3 neurons (7.01% ± 1.33%), Chat neurons (5.84% ± 0.94%), Npy neurons (6.60% ± 1.30%), and Megf11 neurons (5.16% ± 0.85%; Figure S2F). Together, these results map the connectivity between the motor cortex and spinal neuron diversity, revealing well-defined cell types that predominate the neuronal targets of CSNs.
CFA and RFA CSNs target partially distinct cell types
CSNs are anatomically and functionally diverse, raising the question of how innervation of spinal cell types differs across populations. We next determined the spinal cell types targeted by separate populations of CFA and RFA CSNs (Figure 2K, CFASC: N = 4 mice, RFASC: N = 4 mice). We first compared these subpopulations by estimating their broad dorsoventral location using the harmonized spinal neuron atlas. RNA sequencing of CFASC and RFASC populations significantly predicted a ventral horn identity for the majority of RFASC neurons, corroborating our anatomical tracing results and indicating that RNA profiles alone may be used to predict spatial position (Figures 2L and S3A, RFA: dorsal 42.73% ± 1.38% versus ventral 57.27% ± 1.39%, p = 0.0003, unpaired t test). We next measured the fractions of CFASC and RFASC neurons that belonged to each of the major spinal neuron families (Figure S3B). While CFA and RFA CSNs targeted mostly similar spinal cell types, MI neurons make up a significantly larger fraction of CFASC neurons compared to RFASC neurons (Figures 2M and S3C, CFA: 15.37% ± 1.68% versus RFA: 8.88% ± 1.06%, p = 0.029, Sidak’s multiple comparisons test). We fractionated this comparison further, revealing that cluster inhibitory-17, which belongs to family MI, makes up a significantly larger fraction of CFASC neurons compared to RFASC neurons (Figure S3D, CFA: 10.80% ± 1.01% versus RFA: 5.73% ± 0.91%, p = 0.002, Sidak’s multiple comparisons test). This cluster, defined as deep dorsal horn neurons that co-express GABA and glycine, is implicated in sensorimotor processing and the presynaptic control of proprioceptive sensory afferents.5,39,40 In addition, cluster comparison revealed inhibitory-23 and inhibitory-12 as more prominent targets of RFA CSNs (Figure S3D, inhibitory-23, CFA: 24.64% ± 3.71% versus RFA: 29.79% ± 2.18%, p = 0.002; inhibitory-12, CFA: 4.44% ± 1.90% versus RFA: 8.69% ± 1.21%, p = 0.02, Sidak’s multiple comparisons test). Cluster inhibitory-23 is associated with type Ia inhibitory neurons of the V1 clade of ventral interneurons and is implicated in reciprocal inhibition.5,29 These results show that the CFA and RFA target mostly similar cell types in the spinal cord but also identify biases in the spinal targets of CFA and RFA CSNs, uncovering a cell type-specific connectivity matrix.
The supraspinal topography of CFA and RFA axon collaterals
CSNs control behavior through both direct projections to the spinal cord as well as collateral innervation of the brain.19,41 CSNs target regions widely distributed throughout the sensorimotor neuraxis; innervation of cell types in these regions is precisely maintained.7 We next asked whether CFA and RFA CSNs differ in their brain targets and if there are topographical features that distinguish innervation of individual brain structures. We used an intersectional strategy to specifically label CFA and RFA CSNs in the same mice. We injected AAV-retro-Cre and AAV-retro-FlpO in the cervical spinal cord, followed by AAV-FLEX-GFP into the CFA and AAV-FRT-tdTomato into the RFA (Figure 3A, N = 6 mice, the same mice as in Figure 1). We imaged antibody-enhanced GFP and tdTomato labeling and used BrainJ to register brain sections (Figure 3B). Visualizing GFP+ and tdTomato+ labeling revealed many brain regions targeted by CFA and RFA axon collaterals spanning the full rostro-caudal extent of the central nervous system, including in the spinal cord (Figures 3C–3T). We next mapped the positions of CFA and RFA CSN processes to a common brain atlas and ranked these brain regions by the fraction of total processes found in each structure (Figure 3U). We separated forebrain, midbrain, and hindbrain and ranked the brain regions belonging to subregions of these major ontologies (i.e., substantia innominata belongs to cerebral nuclei belongs to forebrain; see Table S3). Consistent with previous studies, the caudoputamen (or striatum) receives the single largest fraction of axonal collateralization (Figure 3U, CFA: 12.20% ± 1.33%, RFA: 10.38% ± 0.73%). We further compared the relative innervation of each region by CFA or RFA CSNs by leveraging the power of within-animal comparisons. While most brain regions receive indistinguishable fractions of CFA and RFA CSN input, we identified several brain regions that differed in relative innervation. These include the globus pallidus externus (Figure 3G, CFA: 0.570% ± 0.090%, RFA: 1.119% ± 0.124%, p = 0.0026, paired t test), ventral posterolateral nucleus of the thalamus (Figure 3K, CFA: 0.424% ± 0.146%, RFA: 0.151% ± 0.086%, p = 0.018, paired t test), and periaqueductal gray (Figure 3N, CFA: 2.008% ± 0.243%, RFA: 0.789% ± 0.160%, p = 7.14 × 10−4, paired t test). The magnitudes of these differences vary substantially and are not clearly a function of the total fraction of projections (Figure 3V). Further, we independently rank-ordered brain regions targeted by the CFA or RFA, revealing that the most densely innervated regions receive similar fractions of the total output of these populations (Figures S4A and S4B). Sorting RFA CSN projections by the rank order of CFA CSN projections also revealed that similar fractions of CFA and RFA CSN projections were found in the top targeted brain regions, with some notable exceptions (Figure S3C).
We next sought to quantitate the similarity of labeling between CFA and RFA CSN collaterals across major brain regions (i.e., medulla, thalamus) as well as within condition reliability across mice. To this end, we created a matrix depicting the correlation coefficients of labeling across all mice (Figure 3W; see STAR Methods). Visual inspection of the correlation matrix revealed no clear structure that would suggest major differences in innervation between CFA and RFA CSNs. We next measured the correlation of CFA and RFA CSN labeling split by major brain ontologies and compared these values to a control of CFA versus CFA innervation (Figure 3X). Doing so, we found no substantial differences in the innervation of major supraspinal structures, indicating that brain-wide innervation of CFA and RFA CSNs is similar. Still, we note that the terminal fields formed by CFA and RFA CSNs often appeared topographically distinct within individual brain regions, including the caudoputamen (Figures 3E and 3F) and pontine gray (Figure 3O). These results detail the collateral output structure of CFA and RFA CSNs, revealing largely similar innervation of the brain but also some notable exceptions.
CFA and RFA CSNs are defined by unique brain-wide inputs
The information encoded in neuronal activity—and by extension neuronal function—is defined in part by synaptic inputs. Thus, revealing the inputs to CFA and RFA CSNs is essential to understanding their function. We harnessed the power of intersectional viral tracing to label inputs to CFA and RFA CSNs within the same animals. We first injected AAV-retro-Cre and AAV-retro-FlpO into the cervical spinal cord. We then injected a 1:1 mixture of AAV-FLEX-N2cG and AAV-FLEX-TVA into the RFA followed by AAV-FLEX-N2cG and AAV-FRT-TVB into the CFA. This led to the expression of rabies glycoprotein in CSNs and the expression of TVB and TVA in CFA and RFA CSNs, respectively. Two weeks later, we injected pseudotyped, G-deficient rabies viruses encoding FPs into the CFA and RFA. Specifically, we injected EnVA-N2cΔG-GFP in the RFA and EnVB-N2cΔG-tdTomato in the CFA (Figure 4A, N = 6 mice).42 This led to the expression of different fluorophores in inputs to CFA CSNs and RFA CSNs all in the same animals. We used BrainJ to align sections and map neurons onto a common atlas (Figure 4B). Notably, the lack of reciprocal connectivity between CFA and RFA CSNs was reflected in the sparseness of dual-labeled CSNs (Figures 4C and 4D). Although CFA and RFA are strongly interconnected and interact closely,43,44 these results suggest that GFP- or RFP-labeled inputs are not due to polysynaptic rabies transfer (i.e., jump across 2 synapses).44 Photomicrographs of neuronal labeling revealed widespread inputs to CFA and RFA CSNs spanning much of the brain (Figures 4E–4T). Simply with visual inspection, we observed major differences in the inputs to CFA and RFA CSNs. We quantified these differences by first independently ordering CFA or RFA inputs, revealing their distributions (Figures S5A and S5B). The substantial and consistent differences in inputs to CFA and RFA CSNs further argues against substantial polysynaptic spread. Sorting RFA labeling by the rank of inputs to CFA CSNs revealed a striking dissimilarity indicating that these two CSN populations receive substantially different inputs (Figure S5C).
Figure 4. CFA and RFA CSNs are defined by unique brain-wide inputs.
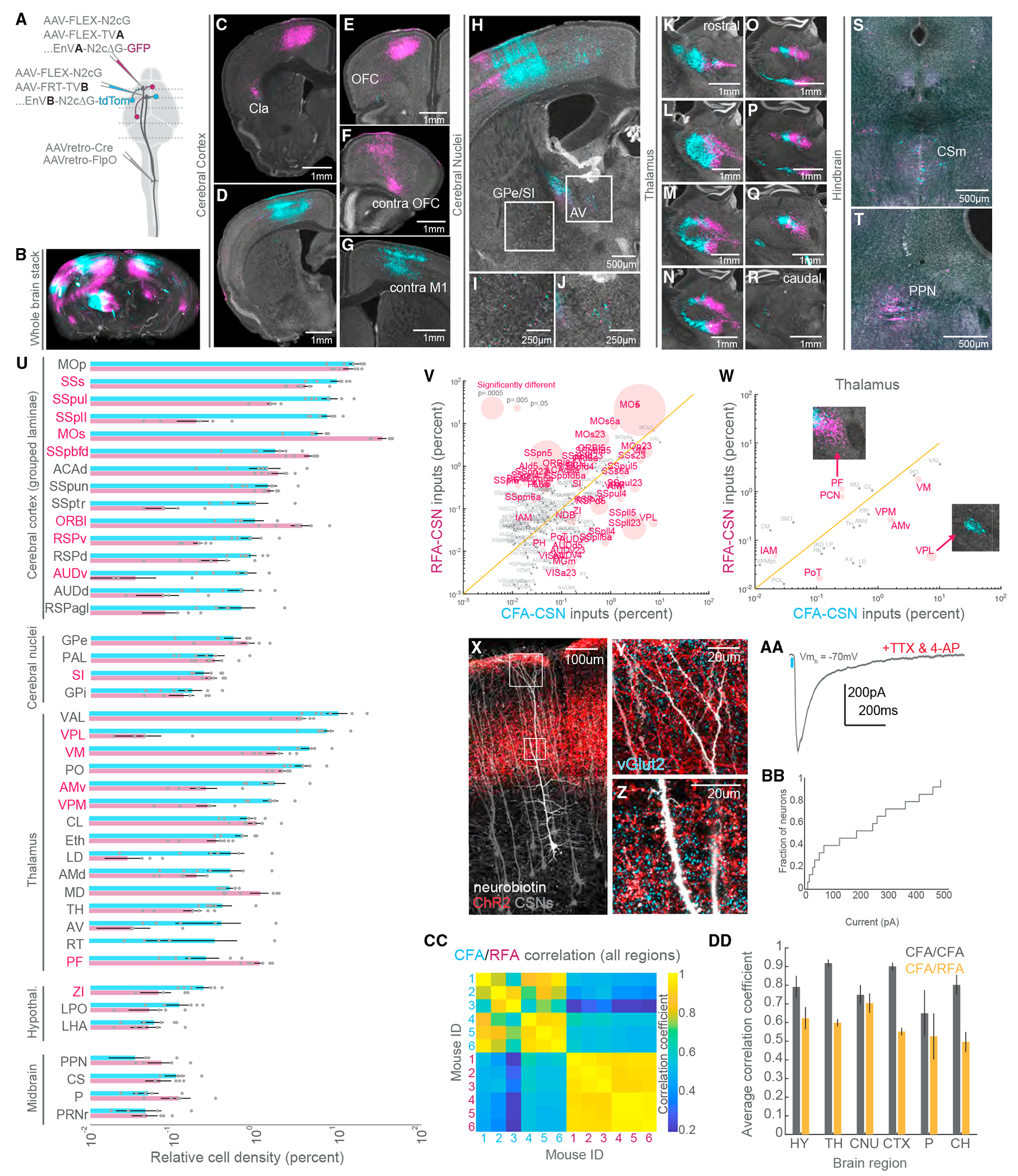
(A) Strategy to simultaneously label brain-wide inputs to CFA and RFA CSNs.
(B) 3D reconstruction of a brain with inputs to CFA (cyan) and RFA (magenta) CSNs labeled (representative of N = 6 mice).
(C–T) Exemplar photomicrographs of inputs to CFA and RFA CSNs throughout the brain. Select regions are indicated (representative of N = 6 mice for B–T).
(U) The brain regions that give rise to neurons that synapse on CFA or RFA CSNs (N = 6 mice for U–W, CC, and DD). Regions are grouped by their ontology (see Table S3). Regions with significantly different fractions of input to CFA and RFA CSNs are indicated in red. Note the log scale, given the large range of labeling.
(V) Scatterplot of regions containing inputs to CFA versus RFA CSNs. Regions with significantly different fractions of CFA and RFA inputs are colored red. The size of points corresponds to the p value of the comparison (paired t tests). Note the log scale, given the large range of labeling.
(W) Same as (V) but only showing thalamic inputs. The insets are photomicrographs illustrating the biases in thalamic input to CFA or RFA CSNs.
(X) Photomicrograph illustrating CSNs (gray), thalamocortical axons (red), and a select CSN targeted for whole-cell intracellular recording (white, representative of N = 3 mice for X–Z).
(Y) An expanded view of the superficial boxed region from (X) depicting axons surrounding the apical dendrites of CSNs. Presynaptic terminals are labeled with vGlut2 (cyan).
(Z) An expanded view of the lower boxed region from (X) depicting axons surrounding the trunk dendrites of CSNs. Presynaptic terminals are labeled with vGlut2 (cyan).
(AA) Example average EPSC evoked through optogenetic stimulation of thalamocortical axons (blue bar). Recordings are made in TTX and 4AP to isolate monosynaptic transmission.
(BB) Cumulative distribution of the current evoked through stimulation of thalamocortical axons (N = 3 mice, n = 15 neurons for AA and BB).
(CC) Matrix depicting the correlation of input labeling within and across all 6 mice. Quadrants correspond to CFA versus CFA (top left quadrant), RFA versus RFA (bottom right quadrant), or CFA versus RFA (bottom left and top right quadrants) labeling.
(DD) The average correlation coefficients of inputs to CFA versus CFA (gray) or CFA versus RFA labeling (orange) within major brain regions across all mice.
Refer to Table S3 for a complete list of the brain structures and their corresponding acronyms. Data are represented as mean ± SEM. Red text in (U)–(W): p < 0.05 See also Figures S5–S7.
What are the different brain regions that provide synaptic input to CFA versus RFA CSNs? Quantification revealed that much of the input to all CSNs arises from isocortical structures followed by substantial input from the thalamus (Figures 4U and 4V). Intriguingly, thalamic inputs to CFA or RFA CSNs are tessellated, a pattern easily appreciated in photomicrographs (Figures 4K–4R). We observed a similar patchwork structure after CFA and RFA injections of dye-conjugated cholera toxins, which are transported both in retrograde and anterograde (Figures S6A and S6B). This pattern varies along the rostro-caudal axis of the thalamus and maps onto well-defined thalamic regions, which we separately analyzed (Figure 4W). Of interest, CFA CSNs are strongly biased to receive inputs from the ventral posterolateral thalamus (VPL), among other regions, while RFA CSNs receive strong input from the parafascicular thalamus (PF), (VPL: CFA: 7.43% ± 1.55%, RFA: 0.047% ± 0.027%, p = 0.005; PF: CFA: 0.258% ± 0.114%, RFA: 1.139% ± 0.159%, p = 0.019; paired t tests).
The main subcortical input to CSNs was the thalamus. Although thalamic inputs to CSNs have been recently described,45 layer 2/3 neurons are usually considered the primary thalamo-recipient population in the agranular cortex.46,47 We sought to confirm our anatomical observations using optogenetics-assisted electrophysiology. We expressed an FP in CSNs by injecting AAV-retro-FP into the cervical spinal cord. We followed this with an injection of AAV-ChR2-FP into the caudal thalamus, resulting in the expression of ChR2 in thalamocortical axons. (Figures 4X–4Z, S7A, and S7B). We then cut acute, live brain slices through motor cortex and made whole-cell patch-clamp recordings from CSNs (N = 3 mice, n = 15 neurons). While recording at membrane holding potentials to isolate excitatory currents, we optogenetically stimulated thalamic axons and recorded excitatory postsynaptic currents (EPSCs). ChR2 stimulation evoked EPSCs in CSNs, even in the presence of tetrodotoxin (TTX) and 4-aminopyridine (4AP), to isolate monosynaptic transmission (Figures 4AA, 4BB, S7C, and S7D, 205.69 ± 44.13 pA, n = 15 cells, N = 3 mice). This confirmed thalamocorticospinal connectivity and further reveals a driving synaptic force that mediates this connectivity.
Finally, we quantified the differences in brain-wide input to CFA and RFA CSNs by measuring the correlation of labeling across mice and across brain regions, like our approach in Figures 3W and 3X. In stark contrast to axonal output labeling, inputs to CFA versus RFA CSNs were markedly uncorrelated (Figure 4CC). This result held true for several regions when we measured the average correlation coefficients separated by major brain regions (Figure 4DD, hypothalamus: 0.791 ± 0.038 versus 0.624 ± 0.062, p = 0.0315; thalamus: 0.918 ± 0.013 versus 0.599 ± 0.017, p = 0.0000; cerebral nuclei: 0.749 ± 0.033 versus 0.704 ± 0.041, p = 0.2926; cerebral cortex (CTX): 0.901 ± 0.013 versus 0.552 ± 0.020, p = 0.0000; pons: 0.650 ± 0.078 versus 0.526 ± 0.066, p = 0.1910; cerebrum: 0.802 ± 0.034 versus 0.496 ± 0.030, p = 0.0000; paired t tests). Nearly all major brain ontologies with substantial input to CSNs differ in the substructures that target CFA versus RFA CSNs. These results reveal that CFA and RFA CSNs receive substantially different brain-wide inputs, more so than their outputs, and this connectivity principle is what likely determines differential activity profiles and behavioral functions of these populations.
DISCUSSION
This study reveals both the regional and cellular spinal targets of two major populations of CSNs, as well as the input and output organization of these populations in the brain. It is first worth considering the known differences in structure and function between the CFA and RFA. Their rostro-caudal separation has led to the suggestion that the RFA may be involved in motor planning and the CFA more involved in execution. However, microstimulation, recording, and manipulation studies found mixed results suggesting that preparation and execution are encoded in both regions, while others concluded that the CFA and RFA are distinct somatotopic representations of the forelimb.8,17,22,48 Indeed, some studies have supported the notion that the RFA is specialized for grasp and the CFA is more involved in reaching.8,15,49 Other studies suggest no clear functional difference between the CFA and RFA or at least not one explained by a clear muscle-specific representation.16,17,22 One obstacle for synthesizing these results is the difference in methods used and specific circuits studied. For one, the motor cortex is incredibly diverse in cellular organization, confounding the interpretation of recordings and manipulations of large populations of cortical neurons.50–52 Even activation and inactivation of isolated cell types, including CSNs, influence activity in brain regions innervated by their collaterals.53 Moreover, because in mouse the vast majority of CSNs do not prominently synapse on motoneurons, it is not straightforward to assign muscle-specific function (i.e., digit flexion versus elbow extension) to RFA and CFA populations. Instead, their influences are defined by the interneuron populations with which they connect. Accordingly, our approach has been to first chart the logic of CFA and RFA CSN connectivity with spinal neurons so that later studies can perform recordings and manipulations informed by the circuit architecture.
We first note that CSNs formed synapses on dispersed groups of spinal neurons that span many functionally distinct laminae. Our dual anterograde transsynaptic approach revealed relatively few double-labeled neurons, and the regional specificity of this double-labeled population suggests that corticospinal convergence is restricted to medial spinal regions. However, we note that additional areas or populations may be underrepresented, as the efficacy of transneuronal labeling could be reduced in our multiviral approach. Despite the widespread innervation of spinal neurons, AnteroT-seq revealed that CSNs are remarkably selective in the spinal neuron types they target, with roughly a quarter of all CSNSC neurons belonging to a single spinal neuron cluster. Inhibitory neurons made up a large majority of CSN targets, which could reflect a prominent role for CSNs in suppressing antagonistic movements or sensory feedback.54 Additionally, this bias may prevent overexcitability through feedforward inhibition. Indeed, we find a cell type connectivity architecture that corroborates over a century of research into the inhibitory function of the pyramidal tract.41,55,56 It is intriguing to consider whether CSNs that predominantly innervate inhibitory neurons are those active at rest, like low-conduction-velocity tonically active neurons.57
CFA and RFA CSNs mostly targeted similar cell types despite the spatial separation of these populations in the sensorimotor cortex. Still, there are notable exceptions to this general trend. For example, we found a significantly larger proportion of CFASC neurons compared to RFASC neurons in the deep dorsal horn family MI and its cluster inhibitory-17, neurons thought to be involved in regulating proprioceptive afferents.39,58 This suggests that CFA CSNs, or a fraction of that population, are more involved in the filtration of sensory feedback during movement compared to RFA CSNs, perhaps constituting a reafference suppression circuit.59 Conversely, our finding that RFA CSNs targeted deeper spinal neurons indicates that these neurons may have privileged access to spinal circuits closer in location—if not in function—to motoneurons. Along those lines, more RFASC neurons than CFASC neurons belonged to the ventral inhibitory cluster inhibitory-23, a glycinergic population of putative V1 lineage. Perhaps RFA CSNs play a larger role in gating reciprocal inhibition of motor pools through synapses on Ia interneurons born of V1 lineage.60 The select recruitment of these corticospinal populations could allow for flexible alternation and co-contraction of antagonist muscles, a behavioral switch that requires co-variation of motor cortex activity.61 Future electrophysiological circuit mapping studies will be useful for probing the strength of connectivity between CSNs in CFA or RFA and these diverse spinal neurons subtypes. In addition, combining multiphoton calcium imaging with retrograde transsynaptic tracing from spinal neuron types will shed light on the differential contributions of target-specific CFA and RFA CSNs to behavior.7,61 Finally, AnteroT-seq, along with similar methods,62 will likely prove useful for mapping cell type-specific connectivity across a range of circuits in the nervous system.
In alignment with our previous study, we identify the striatum as receiving the single largest fraction of supraspinal corticospinal axon collaterals.7 Overall, CFA and RFA CSNs formed axon collaterals in the brain that were similar in their major targets so that the information encoded in these populations may be uniformly broadcast through the brain. Still, CFA and RFA CSNs may target different cell types or subregions of these brain structures. Future brain-wide transsynaptic sequencing and optogenetics-assisted circuit mapping efforts will shed light on this possibility. In stark contrast to their outputs, we identified highly divergent brain-wide inputs to CFA and RFA CSNs, a result that implies that the information encoded across populations differs. Much of the divergent input arose from the isocortex and thalamus rather than midbrain and hindbrain structures. This raises the possibility that CSNs are integrating different higher-order associative information rather than different low-level state information that would be encoded in ascending brainstem regions, where inputs were intermingled. The fact that CFA CSNs receive more inputs from somatomotor thalamic regions (i.e., VPL) compared to RFA CSNs, while RFA CSNs receive more higher-order cortical (i.e., orbital area, lateral part [ORBl]) and thalamic inputs, raises interesting possibilities. VPL serves in part as a passage for ascending spinal feedback pathways and is a major target of spinothalamic projections.63 Perhaps CFA CSNs receive proprioceptive information64 from ascending spinal and cerebellar circuits65 and rapidly integrate and transmit this information to the spinal cord to respond to changes in body position. Copies of this information could be used to suppress reafference and prevent oscillatory sensory feedback.40,58 Indeed, more CFASC than RFASC neurons belong to the inhibitory-17 cluster that probably includes GABApre neurons responsible for controlling proprioceptive reafference.
Outside of the CFA and RFA, there are several other motor and sensory cortical regions that contain CSNs, including neurons that innervate hindlimb lumbar regions of the spinal cord.66–68 These hindlimb cortical regions are important for behaviors including obstacle avoidance during locomotion.66 It will be interesting to extend our level of cellular and circuit characterization to these distinct populations and compare the spinal neurons they target. In addition, these topographically segregated populations of CSNs develop from molecularly distinct lineages with unique axon extension and connectivity strategies.69–71 Such studies show promise to shed light on how differences in cell type-specific connectivity between CFA and RFA CSNs emerge. Finally, CSN populations act to broadcast neuronal activity throughout the brain and spinal cord. This activity is complex, task dependent, and affects behavior through the neural circuits that CSNs target.7,72–74 Our study identifies the structures and cell types distributed throughout the nervous system that receive input from two essential corticospinal populations, revealing unique circuitry that dictates the influence of descending signals on motor output and sensory feedback.
Limitations of the study
Our study is limited to existing knowledge of spinal neuron diversity. As new descriptions of transcriptionally defined cell types emerge, particularly from the cervical spinal cord, it will be worthwhile to remap our data with consideration of these discoveries. Our results are also limited in technique: we used AAV1 recombinase-based transneuronal capture, a relatively new technology that works through an undetermined mechanism. While AnteroT-seq predominantly identified excitatory and inhibitory neurons as the target cells of CSNs, small populations of oligodendrocytes and, intriguingly, central canal neurons were identified with AnteroT-seq. Although we cannot rule out whether this is a byproduct of our sequencing or labeling approach, future efforts using synaptic electrophysiology and retrograde transsynaptic tracing will be valuable to confirm this unexpected connectivity. Moreover, spatial transcriptomics would be valuable for validating these results and gaining further insight into the topography of CFA and RFA cell type-specific innervation. Finally, it is possible in our dual transsynaptic rabies tracing experiments that competing expression of fluorophores underrepresents neurons that provide input to both CFA and RFA CSNs. Using non-fluorescing tags or barcoded rabies viruses would perhaps reveal more co-expression in subcortical inputs. Nevertheless, the consistent labeling of unique thalamic inputs to CFA and RFA CSNs, along with our conventional retrograde tracing experiments, supports our principal conclusion that these populations receive unique inputs.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Anders Nelson (anders.nelson@nyu.edu).
Materials availability
No new reagents have been generated.
Data and code availability
RNA sequencing data have been deposited at the Sequence Read Archive and are publicly available as of the date of publication. The accession number of the BioProject linked to this data is listed in the key resources table. All other data is available from the lead contact upon request.
All original code has been deposited at Zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken anti-EGFP | Columbia University | RRID: AB_3094577 |
| Rabbit anti-dsRed | Columbia University | RRID: AB_3086780 |
| Guinea pig anti-vGlut1 | Columbia University | RRID: AB_2665455 |
| Guinea pig anti-vGlut2 | Millipore Sigma | RRID: AB_2665454 |
| Donkey anti-chicken Alexa Fluor 488 | Jackson ImmunoResearch | RRID: AB_2340375 |
| Donkey anti-rabbit Cy3 | Jackson ImmunoResearch | RRID: AB_2307443 |
| Donkey anti-guinea pig DyLight 405 | Jackson ImmunoResearch | RRID: AB_2340470 |
| NeuroTrace 640/660 | ThermoFisher | RRID: AB_2572212 |
| Bacterial and virus strains | ||
| AAV-retro-CAG-tdTomato | Addgene | Cat# 59462-AAVrg |
| AAV1-hSyn-Cre-WPRE.hGh | Addgene | Cat# 105553-AAV1 |
| AAV1-EF1a-Flpo | Addgene | Cat# 55637-AAV1 |
| AAV-retro-hSyn-Cre-WPRE.hGh | Addgene | Cat# 105553-AAVrg |
| AAV-retro-EF1a-Flpo | Addgene | Cat# 55637-AAVrg |
| AAV2-hSyn-ChR2-EYFP-WPRE.hGH | UNC Vector Core | N/A |
| AAV-DJ-FLEX-H2B-GFP | Columbia University | N/A |
| AAV-hSyn-FlpX-H2B-dTomato | Columbia University | N/A |
| AAV6-CAG-FLEX-N2cG-mKate2.0 | Janelia Farm | N/A |
| AAV1-EF1a-FLEX-TVA-mCherry | UNC Vector Core | N/A |
| AAV2/1-CAG-FRT-TVB | Janelia Farm | N/A |
| AAV1-CAG-FLEX-GFP | UNC Vector Core | N/A |
| AAV1-EF1a-fDIO-tdTomato | Addgene | Cat# 128434-AAV1 |
| EnVA-N2c-deltaG-GFP | Thomas Jefferson | N/A |
| EnVB-N2c-deltaG-tdTomato | Janelia Farm | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Cholera Toxin Subunit Alexa Fluor 488 Conjugate | ThermoFisher | Cat# C34775 |
| Cholera Toxin Subunit Alexa Fluor 555 Conjugate | ThermoFisher | Cat# C34776 |
| Tetrodotoxin citrate | Tocris | Cat# 1069 |
| 4-Aminopyridine | Tocris | Cat# 0940 |
| TrueBlack | Biotium | Cat# 23007 |
| Deposited data | ||
| Single nucleus RNA sequencing of spinal neurons targeted by CFA or RFA | This study | NIH BioProject: PRJNA1081900 |
| Code for analysis of anatomical data | This study | Zenodo: https://doi.org/10.5281/zenodo.10712498 |
| Experimental models: Organisms/strains | ||
| C57BL/6J mice | The Jackson Laboratory | IMSR_JAX:000664 |
| B6; 129-Gt(ROSA)26Sortm5(CAG-Sun1/sfGFP)Nat/J | The Jackson Laboratory | IMSR_JAX:021039 |
| Software and algorithms | ||
| MATLAB | Mathworks | RRID:SCR_001622 |
| R | The R Project for Statistical Computing | RRID:SCR_001905 |
| Seurat | Hao et al. 202175 | RRID:SCR_016341 |
| BrainJ | Botta et al. 202024 | N/A |
| SpinalJ | Fiederling et al. 202127 | N/A |
| Prism | GraphPad | RRID:SCR_002798 |
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
All experiments and procedures were performed according to National Institutes of Health (NIH) guidelines and approved by the Institutional Animal Care and Use Committees of New York University and Columbia University. Adult mice of both sexes, aged 2–6 months, were used for all experiments. No effects of sex are reported. The strains used were C57BL6/J (Jackson Laboratories, 000664) and B6; 129-Gt(ROSA)26Sortm5(CAG-Sun1/sfGFP)Nat/J (CAG-Sun1/sfGFP, Jackson Laboratories, 021039). All mice were kept under a 12-h light-dark cycle.
METHOD DETAILS
Stereotaxic viral injections
Analgesia in the form of subcutaneous injection of carprofen (5 mg per kg body weight) was administered on the day of the surgery, along with bupivacaine (2 mg per kg body weight). Mice were anesthetized with isoflurane and placed in a stereotaxic holder (Kopf). A midline incision was made to expose the skull, and a craniotomy was made over the injection site. A pulled glass pipette was filled with virus, and a Nanoject III was used to make multiple small-volume injections across into the spinal cord, with parameters that depended on the experiment and reagents used. To label CFA and RFA CSNs, 50nL each of AAV-FLEX-GFP and AAV-FRT-tdTomato were injected into CFA and RFA, respectively. Each injection was targeted to one location, approximately 700um below the pia. To label spinal neurons targeted by CFA and RFA, 400-600nL of AAV1-Cre and AAV1-FlpO were injected into CFA and RFA, respectively. We targeted CFA at 0.25mm anterior to bregma, 1.5mm lateral to bregma, and 0.65mm below the pial surface. RFA was injected at 1.8mm anterior to bregma, 1.3mm lateral to bregma, and 0.65mm below the pial surface. To label spinal nuclei for sequencing, 400-600nL of AAV1-Cre and AAV1-FlpO were injected bilaterally into CFA and RFA, respectively. For optogenetics-assisted electrophysiology, 50nL of AAV1-ChR2-GFP was injected into the thalamus, centered between PO and PF. To label inputs to CFA and RFA CSNs, the following strategy was used. Into mice that express Cre and FlpO in CSNs (see below), 50nL total of a 1:1 mixture of AAV-FLEX-N2cG and AAV-FLEX-TVA.mCherry was injected into RFA. 50nL of a 1:1 mixture of AAV-FLEX-N2cG and AAV-FRT-TVB was injected into CFA. After two weeks, 300nL of EnVA-N2cΔG-GFP was injected into RFA and 300nL of EnVB-N2cΔG-tdTomato was injected into CFA.
Spinal cord viral injections
Analgesia in the form of subcutaneous injection of carprofen (5 mg per kg body weight) was administered the day of the surgery, along with bupivacaine (2 mg per kg body weight). Mice were anesthetized with isoflurane and placed in a stereotaxic holder (Kopf). A midline incision was made to expose the spinal column. The musculature overlying the column was resected, and the T2 process was secured to minimize spinal cord movement. The tail was gently stretched with another spinal clamp to separate the vertebrae. A surgical microknife and fine forceps were used to sever the meninges, exposing the spinal cord. A pulled glass pipette was filled with virus, and a Nanoject III was used to make multiple small-volume injections across into the spinal cord, with parameters that depended on the experiment and reagents used. To label inputs to the spinal cord, 200nL of AAV-retro-mCherry was injected into each segment C3-C8 of the cervical spinal cord. Injections were split across two separate penetrations spanning the mediolateral extent of the spinal gray. Within each penetration the injection was spread across the entire superficial to ventral extent of the spinal cord. To label CFA and RFA CSNs, 200nL of a 1:1 mixture of AAV-retro-Cre and AAV-retro-FlpO was similarly injected into C3-C8 of the cervical spinal cord. To label spinal neurons targeted by CFA and RFA in the same mice, 200nL of a 1:1 mixture of AAV-FLEX-H2b-GFP and AAV-FRT-H2b-RFP was injected into C3-C8 of the cervical spinal cord. To label inputs to CFA and RFA CSNs, 200nL of a 1:1 mixture of AAV-retro-Cre and AAV-retro-FlpO was injected into C3-C8 of the cervical spinal cord. Following all injections, the skin was sutured closed, and animals were closely monitored during recovery.
Nuclear suspension and FACS
Six weeks post injection, mice were deeply anesthetized with isoflurane and transcardially perfused with a carbogenated, ice-cold artificial cerebrospinal fluid (ACSF) solution containing: 100mM NaCl, 10mM HEPES, 25mM Glucose, 75mM Sucrose, 7.5mM MgCl2, and 2.5mM KCl. A ventral laminectomy was performed, and the cervical spinal cord was rapidly isolated in carbogenated, ice-cold ACSF. The meninges were carefully removed with Vannas scissors and forceps, and the cervical spinal cord was divided into hemisections and rapidly frozen on dry ice. Samples were stored at −80C until the day of nuclear isolation. Nuclei were isolated using the 10x Chromium Nuclei Isolation Kit following the manufacturer protocol. All solutions were made fresh from each prep. DAPI was added to the samples after the final resuspension, and samples were filtered through a 30 μm filter (CellTrics, Sysmex). Fluorescence-Activated Cell Sorting was performed immediately after dissociation at the Zuckerman Institute Flow Cytometry platform using a MoFlo Astrios EQ (Beckman Coulter) sorter. Sorting was performed with 100 μm nozzle at sheath pressure of 28 PSI. Sample pressure was maintained below 28.5 PSI. The following laser lines and filters were used: For DAPI the 205 nm laser was used with a 448/59 band-pass filter. The PMT was set at 315V with an amplifier gain of 2. For GFP, the 488 nm laser was used with a 513/26 band-pass filter. The PMT was set at 370V with an amplifier gain of 2. These fluorochromes required different laser sources on different laser paths which enabled very clean separation of the emission fluorescence and no need for fluorescence compensation. The 488 laser was also used for detection of forward and side scatter. Samples were first gated of forward and side scatter to eliminate debris, electronic noise, and large particles. Doublet discrimination was performed using a combination of data plots consisting of Side Scatter height vs. Side Scatter Area, Side Scatter height vs. Side Scatter width, and DAPI Log height vs. DAPI width. DAPI and GFP double-positive nuclei were sorted using “Single Cell” mode with a drop envelope of 1. Nuclei were collected in 8 well PCR strips preloaded with lysis solution. Both the cell suspension and the collection chamber were maintained cold during sorting. PCR strips with collected nuclei were spun down immediately after sorting and frozen on dry ice.
Single-nucleus RNA-sequencing
RNA amplification, library preparation, and RNA sequencing
SMART-Seq v4 Ultra Low Input RNA Kit for Sequencing (Takara #634440) was used per manufacturer’s instructions for cDNA synthesis of single cell RNA and subsequent amplification, with the exception that all reaction volumes were reduced to 0.5x. Single cells were stored in 8-strips at −80°C in 5.25 μL of collection buffer (SMART-Seq v4 lysis buffer at 0.83x, RNase Inhibitor at 0.17 U/μl, and ERCC MIX1 at final 1x10-8 dilution as described above). Twelve to 36 8-strips were processed at a time (the equivalent of 1–3 96-well plates). At least 1 control strip was used per amplification set, containing 2 wells without cells (termed ERCC), 2 wells without cells or ERCC (termed NTC), and 2 wells of 10 pg of Mouse Whole Brain Total RNA (Zyagen, MR-201) and 2 wells of 10 pg Control RNA provided in the Takara kit.
Mouse nuclei were subjected to 22 or 23 PCR cycles after the reverse transcription step. SPRI bead (Sera-Mag Select beads GE Healthcare #29343057) purification was done using the Agilent Bravo NGS Option A instrument. A bead ratio of 1x was used (25 μL of Sera-Mag Select beads to 25 μL cDNA PCR product with 0.5 μL of 10x lysis buffer added, as per Takara instructions at 0.5x volume), and purified cDNA was eluted in 17 μL elution buffer provided by Takara. All samples were quantitated using PicoGreen on Molecular Dynamics M2 SpectraMax instrument. The samples were then run on the Agilent Fragment Analyzer (96) using the High Sensitivity NGS Fragment Analysis Kit (1bp-6000bp) to qualify cDNA size distribution. Purified cDNA was stored in 96-well plates at −20°C until library preparation.
All samples proceeded through NexteraXT DNA Library Preparation (Illumina FC131-1096) using custom 8-base Unique Design Index primers designed and manufactured by IDT (Integrated DNA Technologies). NexteraXT DNA Library prep was done at 0.2x volume on the Mantis instrument (Formulatrix). Reduction in volume was applied to input and all reagents, but otherwise the manufacturer’s instructions were followed. 50pg cDNA input was used in generating the libraries. An aliquot of all amplified cDNA samples was first normalized to 50 pg/ul with Nuclease-Free Water (Ambion), then this normalized sample aliquot was used as input material into the NexteraXT DNA Library Prep (for a total of 50pg). Before bead purification after PCR, 12.1ul of Nuclease-Free Water (Ambion) was added to the 10ul PCR reaction volume, bringing the total volume to 22.1ul post-PCR and before bead purification. SPRI Sera-Mag Select bead purification was done using the Agilent Bravo NGS Option A instrument. A bead ratio of 0.9x was used (20 μl of Sera-Mag Select beads to 22.10 μl library product, as per Illumina protocol), and all samples were eluted in 22 μL of Resuspension Buffer (Illumina). All samples were quantitated using PicoGreen using Molecular Bynamics M2 SpectraMax instrument. All samples were run on the Agilent Fragment Analyzer (96) using the High Sensitivity NGS Fragment Analysis Kit (1bp-6000bp) for sizing. Molarity was calculated for each sample using average size as reported by the Fragment Analyzer and pg/μl concentration as determined by PicoGreen. Samples (5 μL aliquot) were normalized to 2–5 nM with Nuclease-free Water (Ambion), then 2 μL from each sample within one 96-index set was pooled to a total of 192 μL at 2–5 nM concentration. Libraries were further multiplexed at 768 samples/flowcell by pooling 8 96-sample libraries using compatible Index Sets. A portion of the final library pool was sequenced on an Illumina NextSeq2000 instrument using the P2 flowcell, for a target of 500,000 reads per single cell or nucleus.
Data processing of RNA sequencing
RNA-Seq alignment and data processing was done the same way for all single cell and single nucleus RNA-Seq data, except that different versions of the genome and transcriptome were used for each species.
The raw fastq files from Illumina were trimmed using the fastqMCF program (Aronesty et al., 2011). The trimmed paired-end reads were mapped and counts generated using STAR aligner (v2.7.1a) with default settings against mouse mm10 GENCODE vM23/Ensembl 98 reference genome, downloaded from 10X cell ranger (refdata-cellranger-arc-mm10-2020-A-2.0.0). STAR uses and builds its own suffix array index which considerably accelerates the alignment step while improving on sensitivity and specificity, due to its identification of alternative splice junctions. Next, the duplicates were removed using STAR as well. Reads that did not map to the genome were then aligned to synthetic constructs (i.e., ERCC) sequences and the E.coli genome (version ASM584v2). The final results files included quantification of the uniquely mapped reads (raw exon and intron counts for the transcriptome-mapped reads). Included in the final results files are the percentages of reads mapped to the transcriptome, to ERCC spike-in controls, and to E.coli.
QC and analysis of RNA sequencing
Nuclei with <1000 genes, <100,000 total read, <75% aligned reads, >0.5 CG complexity, and >5% mitochondrial RNA were excluded from further analysis. All analysis was performed using Seurat. Mapping to data from Russ et al. was also performed with Seurat using transfer anchors.
Slice electrophysiology
Mice were deeply anesthetized with isoflurane and transcardially perfused with an ice-cold carbogenated high-magnesium (10 mM) artificial cerebrospinal fluid (ACSF). The brain was removed from the skull and glued to the stage of vibrating microtome (Leica). 300 μm coronal brain slices were cut in a bath of ice-cold, slushy, carbogenated low-calcium ACSF. Slices were immediately transferred to a 37C bath of normal ACSF containing 124 mM NaCl, 2.7 mM KCl, 2 mM CaCl2, 1.3 mM MgSO4, 26 mM NaHCO3, 1.25 mM NaH2PO4, 18 mM glucose and 0.79 mM sodium ascorbate, where they incubated for 25 min. Slices were then moved to room temperature, where they remained for the duration of the experiment. Patch electrodes (2–6 MΩ) were filled with a potassium gluconate-based internal solution containing 135 mM potassium gluconate, 2 mM MgCl2, 0.5 mM EGTA, 2 mM magnesium ATP, 0.5 mM sodium GTP, 10 mM HEPES, 10 mM phosphocreatine and 0.15% Neurobiotin. All recordings were made using a Multiclamp 700B amplifier, the output of which was digitized at 10 kHz (Digidata 1440A). Series resistance was always <30 MΩ and was compensated up to 90%. Neurons were targeted with DIC microscopy and epifluorescence when appropriate. In a subset of experiments, cell morphology was visualized through internal dialysis of 0.1 mM Alexa Fluor 594 cadaverine or 0.1 mM Alexa Fluor 488 sodium salt. ChR2-expressing axons were photostimulated using 10-ms pulses of 473-nm LED light (CoolLED) delivered through a 10× objective centered over the recording site. Brain slices were histologically processed to visualize Neurobiotin-filled cells through streptavidin-Alexa Fluor processing.
Histology and confocal imaging
Mice were anesthetized with isoflurane and transcardially perfused with ice-cold PBS followed by cold 4% paraformaldehyde. Brains and spinal cords were fixed overnight in 4% paraformaldehyde and cryopreserved in a 30% sucrose solution at 4°C for 3 days or until they sunk. Brains were embedded in Optimum Cutting Temperature Compound (OCT, Tissue-Tek) and frozen on dry ice. Spinal cords were prepared for SpinalJ processing by mounting them in dissolvable SpineRacks embedded within OCT. The spinal cord was cut into three segments comprising the cervical, thoracic, and lumbar regions. Each of these regions was further divided into 3 chunks, and these chunks were oriented rostral side downward in a 3x3 array SpineRack submerged in OCT within a mold. After SpineRacks had fully softened in OCT, they were frozen on dry ice. Sections between 50 μm and 80 μm were cut using a cryostat (Leica). Spinal cord sections were directly mounted on slides, while brain sections were deposited in 24 well plates. Tissue was rinsed several times in PBS, and permeabilized in 0.2% Triton X-100 (PBST). Immunostaining was performed with primary antibodies diluted to working concentration for 3 days at 4°C, and with secondary antibodies (Jackson ImmunoResearch) diluted 1:1000 overnight at 4°C. Brain and spinal cord slices mounted to slides were briefly incubated with TrueBlack diluted in 70% ethanol to quench lipofuscin and background autofluorescence. Confocal imaging was performed on a Zeiss 710 or Zeiss 880 using a 10x, 20x, or 40× objective.
Slide scanning and anatomical reconstructions
Sections were imaged using an AZ100 automated slide scanning microscope equipped with a 4× 0.4-NA objective (Nikon). Image processing and analysis using BrainJ or SpinalJ was as previously described. Briefly, a seven-pixel rolling-ball filter was used on all images to reduce background signal and a machine-learning pixel classification approach using Ilastik was used to identify cell bodies and neuronal processes. To map the location of these structures to an annotated brain or spinal cord atlas, 3D image registration was performed using Elastix relative to a reference brain or spinal cord. The coordinates of detected cells and processes were then projected into the Allen Brain Atlas Common Coordinate Framework. Visualizations of the data were performed in ImageJ, and subsequent analyses were performed in MATLAB using custom software. Correlation matrices were generated in MATLAB by running a correlation between 1) CFA v. RFA within mouse, 2) CFA v. CFA across mice, and 3) CFA v RFA across mice. The input vectors were either the proportion of labeling in each brain region (Figures 3W and 4CC) or the proportion of labeling in major brain structures (Figures 3X and 4DD).
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical tests and significance values are reported in the Results. Sample sizes are indicated in the Results or figure legends. Data is mean ± standard error of mean (SEM) unless otherwise indicated. Statistical tests were performed in MATLAB and Prism. Significance is defined as p < 0.05. No data was excluded from this study.
Supplementary Material
Highlights.
Forelimb corticospinal neurons (CSNs) preferentially innervate inhibitory spinal neurons
Caudal (CFA) and rostral (RFA) CSNs target distinguishable spinal populations
CFA and RFA CSNs send axon collaterals to many similar brain regions
CFA and RFA CSNs receive input from distinct brain regions
ACKNOWLEDGMENTS
We are grateful to L. Hammond and F. Fiederling for creating exceptional brain and spinal cord image analysis pipelines as well as for helping with these tools. We thank I. Shieren and the Zuckerman Institute’s Flow Cytometry Platform for assistance with FACS. We thank S. Brenner-Morton for antibodies. Imaging was performed in part with support from the Zuckerman Cellular Imaging Platform at Columbia University and the National Institutes of Health (NIH 1S10OD023587-01). CVS-N2c rabies viruses were produced by the Center for Neuroanatomy with Neurotropic Viruses, supported by P40 OD010996. A.N. is supported by an NIH Pathway to Independence Award (R00NS118053). L.M.C. is supported by an NIH Pathway to Independence Award (K99NS127857). R.M.C. is supported by 5U19NS104649 and the Allen Institute for Brain Science.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2024.113993.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Arber S, and Costa RM (2018). Connecting neuronal circuits for movement. Science 360, 1403–1404. 10.1126/science.aat5994. [DOI] [PubMed] [Google Scholar]
- 2.Lemon RN (2008). Descending pathways in motor control. Annu. Rev. Neurosci 31, 195–218. 10.1146/annurev.neuro.31.060407.125547. [DOI] [PubMed] [Google Scholar]
- 3.Jessell TM (2000). Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat. Rev. Genet 1, 20–29. 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- 4.Lu DC, Niu T, and Alaynick WA (2015). Molecular and cellular development of spinal cord locomotor circuitry. Front. Mol. Neurosci 8, 25. 10.3389/fnmol.2015.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russ DE, Cross RBP, Li L, Koch SC, Matson KJE, Yadav A, Alkaslasi MR, Lee DI, Le Pichon CE, Menon V, and Levine AJ (2021). A harmonized atlas of mouse spinal cord cell types and their spatial organization. Nat. Commun 12, 5722. 10.1038/s41467-021-25125-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Porter R, and Lemon R (1993). Corticospinal Function and Voluntary Movement (Clarendon Press; Oxford University Press). [Google Scholar]
- 7.Nelson A, Abdelmesih B, and Costa RM (2021). Corticospinal populations broadcast complex motor signals to coordinated spinal and striatal circuits. Nat. Neurosci 24, 1721–1732. 10.1038/s41593-021-00939-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Liu Y, Li X, Zhang Z, Yang H, Zhang Y, Williams PR, Alwahab NSA, Kapur K, Yu B, et al. (2017). Deconstruction of Corticospinal Circuits for Goal-Directed Motor Skills. Cell 171, 440–455.e14. 10.1016/j.cell.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin JH (1996). Differential spinal projections from the forelimb areas of the rostral and caudal subregions of primary motor cortex in the cat. Exp. Brain Res 108, 191–205. 10.1007/BF00228094. [DOI] [PubMed] [Google Scholar]
- 10.Morecraft RJ, Ge J, Stilwell-Morecraft KS, McNeal DW, Pizzimenti MA, and Darling WG (2013). Terminal distribution of the corticospinal projection from the hand/arm region of the primary motor cortex to the cervical enlargement in rhesus monkey. J. Comp. Neurol 521, 4205–4235. 10.1002/cne.23410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ueno M, Nakamura Y, Li J, Gu Z, Niehaus J, Maezawa M, Crone SA, Goulding M, Baccei ML, and Yoshida Y (2018). Corticospinal Circuits from the Sensory and Motor Cortices Differentially Regulate Skilled Movements through Distinct Spinal Interneurons. Cell Rep. 23, 1286–1300.e7. 10.1016/j.celrep.2018.03.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levine AJ, Lewallen KA, and Pfaff SL (2012). Spatial organization of cortical and spinal neurons controlling motor behavior. Curr. Opin. Neurobiol 22, 812–821. 10.1016/j.conb.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neafsey EJ, and Sievert C (1982). A second forelimb motor area exists in rat frontal cortex. Brain Res. 232, 151–156. 10.1016/0006-8993(82)90617-5. [DOI] [PubMed] [Google Scholar]
- 14.Bizzi E, Cheung VCK, d’Avella A, Saltiel P, and Tresch M (2008). Combining modules for movement. Brain Res. Rev 57, 125–133. 10.1016/j.brainresrev.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown AR, and Teskey GC (2014). Motor Cortex Is Functionally Organized as a Set of Spatially Distinct Representations for Complex Movements. J. Neurosci 34, 13574–13585. 10.1523/Jneurosci.2500-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hyland B. (1998). Neural activity related to reaching and grasping in rostral and caudal regions of rat motor cortex. Behav. Brain Res 94, 255–269. 10.1016/s0166-4328(97)00157-5. [DOI] [PubMed] [Google Scholar]
- 17.Morandell K, and Huber D (2017). The role of forelimb motor cortex areas in goal directed action in mice. Sci. Rep 7, 15759. 10.1038/s41598-017-15835-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang F, Rouiller EM, and Wiesendanger M (1993). Modulation of sustained electromyographic activity by single intracortical microstimuli: comparison of two forelimb motor cortical areas of the rat. Somatosens. Mot. Res 10, 51–61. 10.3109/08990229309028823. [DOI] [PubMed] [Google Scholar]
- 19.Kita T, and Kita H (2012). The subthalamic nucleus is one of multiple innervation sites for long-range corticofugal axons: a single-axon tracing study in the rat. J. Neurosci 32, 5990–5999. 10.1523/JNEUROSCI.5717-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rouiller EM, Moret V, and Liang F (1993). Comparison of the connectional properties of the two forelimb areas of the rat sensorimotor cortex: support for the presence of a premotor or supplementary motor cortical area. Somatosens. Mot. Res 10, 269–289. 10.3109/08990229309028837. [DOI] [PubMed] [Google Scholar]
- 21.Wang Z, Romanski A, Mehra V, Wang Y, Brannigan M, Campbell BC, Petsko GA, Tsoulfas P, and Blackmore MG (2022). Brain-wide analysis of the supraspinal connectome reveals anatomical correlates to functional recovery after spinal injury. Elife 11, e76254. 10.7554/eLife.76254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tennant KA, Adkins DL, Donlan NA, Asay AL, Thomas N, Kleim JA, and Jones TA (2011). The organization of the forelimb representation of the C57BL/6 mouse motor cortex as defined by intracortical microstimulation and cytoarchitecture. Cereb. Cortex 21, 865–876. 10.1093/cercor/bhq159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watson C, Paxinos G, Kayalioglu G, and Heise C (2009). Atlas of the mouse spinal cord. In Spinal Cord: A Christopher and Dana Reeve Foundation Text and Atlas (Elsevier), pp. 308–379. 10.1016/B978-0-12-374247-6.50020-1. [DOI] [Google Scholar]
- 24.Botta P, Fushiki A, Vicente AM, Hammond LA, Mosberger AC, Gerfen CR, Peterka D, and Costa RM (2020). An Amygdala Circuit Mediates Experience-Dependent Momentary Arrests during Exploration. Cell 183, 605–619.e22. 10.1016/j.cell.2020.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zingg B, Chou XL, Zhang ZG, Mesik L, Liang F, Tao HW, and Zhang LI (2017). AAV-Mediated Anterograde Transsynaptic Tagging: Mapping Corticocollicular Input-Defined Neural Pathways for Defense Behaviors. Neuron 93, 33–47. 10.1016/j.neuron.2016.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zingg B, Peng B, Huang J, Tao HW, and Zhang LI (2020). Synaptic Specificity and Application of Anterograde Transsynaptic AAV for Probing Neural Circuitry. J. Neurosci 40, 3250–3267. 10.1523/JNEUROSCI.2158-19.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fiederling F, Hammond LA, Ng D, Mason C, and Dodd J (2021). SpineRacks and SpinalJ for efficient analysis of neurons in a 3D reference atlas of the mouse spinal cord. STAR Protoc. 2, 100897. 10.1016/j.xpro.2021.100897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rexed B. (1954). A cytoarchitectonic atlas of the spinal cord in the cat. J. Comp. Neurol 100, 297–379. 10.1002/cne.901000205. [DOI] [PubMed] [Google Scholar]
- 29.Bikoff JB, Gabitto MI, Rivard AF, Drobac E, Machado TA, Miri A, Brenner-Morton S, Famojure E, Diaz C, Alvarez FJ, et al. (2016). Spinal Inhibitory Interneuron Diversity Delineates Variant Motor Microcircuits. Cell 165, 207–219. 10.1016/j.cell.2016.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gabitto MI, Pakman A, Bikoff JB, Abbott LF, Jessell TM, and Paninski L (2016). Bayesian Sparse Regression Analysis Documents the Diversity of Spinal Inhibitory Interneurons. Cell 165, 220–233. 10.1016/j.cell.2016.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zholudeva LV, and Lane MA (2023). Spinal Interneurons: Plasticity after Spinal Cord Injury (Academic Press, an imprint of Elsevier; ). [Google Scholar]
- 32.Mo A, Mukamel EA, Davis FP, Luo C, Henry GL, Picard S, Urich MA, Nery JR, Sejnowski TJ, Lister R, et al. (2015). Epigenomic Signatures of Neuronal Diversity in the Mammalian Brain. Neuron 86, 1369–1384. 10.1016/j.neuron.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, Häring M, Braun E, Borm LE, La Manno G, et al. (2018). Molecular Architecture of the Mouse Nervous System. Cell 174, 999–1014.e22. 10.1016/j.cell.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayashi M, Hinckley CA, Driscoll SP, Moore NJ, Levine AJ, Hilde KL, Sharma K, and Pfaff SL (2018). Graded Arrays of Spinal and Supraspinal V2a Interneuron Subtypes Underlie Forelimb and Hindlimb Motor Control. Neuron 97, 869–884.e5. 10.1016/j.neuron.2018.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Häring M, Zeisel A, Hochgerner H, Rinwa P, Jakobsson JET, Lönnerberg P, La Manno G, Sharma N, Borgius L, Kiehn O, et al. (2018). Neuronal atlas of the dorsal horn defines its architecture and links sensory input to transcriptional cell types. Nat. Neurosci 21, 869–880. 10.1038/s41593-018-0141-1. [DOI] [PubMed] [Google Scholar]
- 36.Rosenberg AB, Roco CM, Muscat RA, Kuchina A, Sample P, Yao Z, Graybuck LT, Peeler DJ, Mukherjee S, Chen W, et al. (2018). Single-cell profiling of the developing mouse brain and spinal cord with splitpool barcoding. Science 360, 176–182. 10.1126/science.aam8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sathyamurthy A, Johnson KR, Matson KJE, Dobrott CI, Li L, Ryba AR, Bergman TB, Kelly MC, Kelley MW, and Levine AJ (2018). Massively Parallel Single Nucleus Transcriptional Profiling Defines Spinal Cord Neurons and Their Activity during Behavior. Cell Rep. 22, 2216–2225. 10.1016/j.celrep.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tashima R, Koga K, Yoshikawa Y, Sekine M, Watanabe M, Tozaki-Saitoh H, Furue H, Yasaka T, and Tsuda M (2021). A subset of spinal dorsal horn interneurons crucial for gating touch-evoked pain-like behavior. Proc. Natl. Acad. Sci. USA 118, e2021220118. 10.1073/pnas.2021220118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koch SC, Del Barrio MG, Dalet A, Gatto G, Gunther T, Zhang J, Seidler B, Saur D, Schule R, and Goulding M (2017). RORbeta Spinal Interneurons Gate Sensory Transmission during Locomotion to Secure a Fluid Walking Gait. Neuron 96, 1419–1431. 10.1016/j.neuron.2017.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fink AJP, Croce KR, Huang ZJ, Abbott LF, Jessell TM, and Azim E (2014). Presynaptic inhibition of spinal sensory feedback ensures smooth movement. Nature 509, 43–48. 10.1038/nature13276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramón y Cajal S. (1909). Histologie du système nerveux de l’homme & des vertébrés, française rev. & mise à jour par l’auteur, de l’espagnol par L. Azoulay (Maloine). [Google Scholar]
- 42.Reardon TR, Murray AJ, Turi GF, Wirblich C, Croce KR, Schnell MJ, Jessell TM, and Losonczy A (2016). Rabies Virus CVS-N2c(Delta G) Strain Enhances Retrograde Synaptic Transfer and Neuronal Viability. Neuron 89, 711–724. 10.1016/j.neuron.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alyahyay M, Kalweit G, Kalweit M, Karvat G, Ammer J, Schneider A, Adzemovic A, Vlachos A, Boedecker J, and Diester I (2023). Mechanisms of Premotor-Motor Cortex Interactions during Goal Directed Behavior. Preprint at bioRxiv. 10.1101/2023.01.20.524944. [DOI] [Google Scholar]
- 44.Hira R, Ohkubo F, Tanaka YR, Masamizu Y, Augustine GJ, Kasai H, and Matsuzaki M (2013). In vivo optogenetic tracing of functional corticocortical connections between motor forelimb areas. Front Neural Circuit 7, 55. 10.3389/fncir.2013.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suter BA, and Shepherd GMG (2015). Reciprocal interareal connections to corticospinal neurons in mouse M1 and S2. J. Neurosci 35, 2959–2974. 10.1523/JNEUROSCI.4287-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shepherd GMG (2009). Intracortical cartography in an agranular area. Front. Neurosci 3, 337–343. 10.3389/neuro.01.030.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strick PL, and Sterling P (1974). Synaptic termination of afferents from the ventrolateral nucleus of the thalamus in the cat motor cortex. A light and electron microscopy study. J. Comp. Neurol 153, 77–106. 10.1002/cne.901530107. [DOI] [PubMed] [Google Scholar]
- 48.Resta F, Montagni E, de Vito G, Scaglione A, Allegra Mascaro AL, and Pavone FS (2022). Large-scale all-optical dissection of motor cortex connectivity shows a segregated organization of mouse forelimb representations. Cell Rep. 41, 111627. 10.1016/j.celrep.2022.111627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kleim JA, Barbay S, and Nudo RJ (1998). Functional reorganization of the rat motor cortex following motor skill learning. J. Neurophysiol 80, 3321–3325. 10.1152/jn.1998.80.6.3321. [DOI] [PubMed] [Google Scholar]
- 50.BRAIN Initiative Cell Census Network BICCN (2021). A multimodal cell census and atlas of the mammalian primary motor cortex. Nature 598, 86–102. 10.1038/s41586-021-03950-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang M, Eichhorn SW, Zingg B, Yao Z, Cotter K, Zeng H, Dong H, and Zhuang X (2021). Spatially resolved cell atlas of the mouse primary motor cortex by MERFISH. Nature 598, 137–143. 10.1038/s41586-021-03705-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muñoz-Castañeda R, Zingg B, Matho KS, Chen X, Wang Q, Foster NN, Li A, Narasimhan A, Hirokawa KE, Huo B, et al. (2021). Cellular anatomy of the mouse primary motor cortex. Nature 598, 159–166. 10.1038/s41586-021-03970-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolff SB, and Ölveczky BP (2018). The promise and perils of causal circuit manipulations. Curr. Opin. Neurobiol 49, 84–94. 10.1016/j.conb.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andersen P, Eccles JC, and Sears TA (1962). Presynaptic inhibitory action of cerebral cortex on the spinal cord. Nature 194, 740–741. 10.1038/194740a0. [DOI] [PubMed] [Google Scholar]
- 55.Marie P. (1892). Leçons sur les maladies de la moelle (G. Masson) [Google Scholar]
- 56.Marshall C. (1936). The functions of the pyramidal tracts. Q. Rev. Biol 11, 35–56. 10.1086/394499. [DOI] [Google Scholar]
- 57.Evarts EV (1966). Pyramidal tract activity associated with a conditioned hand movement in the monkey. J. Neurophysiol 29, 1011–1027. 10.1152/jn.1966.29.6.1011. [DOI] [PubMed] [Google Scholar]
- 58.Tomatsu S, Kim G, Kubota S, and Seki K (2023). Presynaptic gating of monkey proprioceptive signals for proper motor action. Nat. Commun 14, 6537. 10.1038/s41467-023-42077-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Crapse TB, and Sommer MA (2008). Corollary discharge across the animal kingdom. Nat. Rev. Neurosci 9, 587–600. 10.1038/nrn2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lundberg A, and Voorhoeve P (1962). Effects from the pyramidal tract on spinal reflex arcs. Acta Physiol. Scand 56, 201–219. 10.1111/j.1748-1716.1962.tb02498.x. [DOI] [PubMed] [Google Scholar]
- 61.Warriner CL, Fageiry S, Saxena S, Costa RM, and Miri A (2022). Motor cortical influence relies on task-specific activity covariation. Cell Rep. 40, 111427. 10.1016/j.celrep.2022.111427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsai NY, Wang F, Toma K, Yin C, Takatoh J, Pai EL, Wu K, Matcham AC, Yin L, Dang EJ, et al. (2022). Trans-Seq maps a selective mammalian retinotectal synapse instructed by Nephronectin. Nat. Neurosci 25, 659–674. 10.1038/s41593-022-01068-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cliffer KD, Burstein R, and Giesler GJ Jr. (1991). Distributions of spinothalamic, spinohypothalamic, and spinotelencephalic fibers revealed by anterograde transport of PHA-L in rats. J. Neurosci 11, 852–868. 10.1523/JNEUROSCI.11-03-00852.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hodge CJ Jr., and Apkarian AV (1990). The spinothalamic tract. Crit. Rev. Neurobiol 5, 363–397. [PubMed] [Google Scholar]
- 65.Dacre J, Colligan M, Clarke T, Ammer JJ, Schiemann J, Chamosa-Pino V, Claudi F, Harston JA, Eleftheriou C, Pakan JMP, et al. (2021). A cerebellar-thalamocortical pathway drives behavioral context-dependent movement initiation. Neuron 109, 2326–2338.e8. 10.1016/j.neuron.2021.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Drew T, Jiang W, and Widajewicz W (2002). Contributions of the motor cortex to the control of the hindlimbs during locomotion in the cat. Brain Res. Brain Res. Rev 40, 178–191. 10.1016/s0165-0173(02)00200-x. [DOI] [PubMed] [Google Scholar]
- 67.Kamiyama T, Kameda H, Murabe N, Fukuda S, Yoshioka N, Mizukami H, Ozawa K, and Sakurai M (2015). Corticospinal tract development and spinal cord innervation differ between cervical and lumbar targets. J. Neurosci 35, 1181–1191. 10.1523/JNEUROSCI.2842-13.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moreno-Lopez Y, Bichara C, Delbecq G, Isope P, and Cordero-Erausquin M (2021). The corticospinal tract primarily modulates sensory inputs in the mouse lumbar cord. Elife 10, e65304. 10.7554/eLife.65304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sahni V, Shnider SJ, Jabaudon D, Song JHT, Itoh Y, Greig LC, and Macklis JD (2021). Corticospinal neuron subpopulation-specific developmental genes prospectively indicate mature segmentally specific axon projection targeting. Cell Rep. 37, 109843. 10.1016/j.celrep.2021.109843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gu Z, Ueno M, Klinefelter K, Mamidi M, Yagi T, and Yoshida Y (2019). Skilled Movements in Mice Require Inhibition of Corticospinal Axon Collateral Formation in the Spinal Cord by Semaphorin Signaling. J. Neurosci 39, 8885–8899. 10.1523/JNEUROSCI.2832-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Song JHT, Ruven C, Patel P, Ding F, Macklis JD, and Sahni V (2023). Cbln1 Directs Axon Targeting by Corticospinal Neurons Specifically toward Thoraco-Lumbar Spinal Cord. J. Neurosci 43, 1871–1887. 10.1523/JNEUROSCI.0710-22.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peters AJ, Lee J, Hedrick NG, O’Neil K, and Komiyama T (2017). Reorganization of corticospinal output during motor learning. Nat. Neurosci 20, 1133–1141. 10.1038/nn.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Currie SP, Ammer JJ, Premchand B, Dacre J, Wu Y, Eleftheriou C, Colligan M, Clarke T, Mitchell L, Faisal AA, et al. (2022). Movement-specific signaling is differentially distributed across motor cortex layer 5 projection neuron classes. Cell Rep. 39, 110801. 10.1016/j.celrep.2022.110801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Barrett JM, Martin ME, and Shepherd GMG (2022). Manipulation-specific cortical activity as mice handle food. Curr. Biol 32, 4842–4853.e6. 10.1016/j.cub.2022.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hao Y, Stuart T, Kowalski MH, Choudhary S, Hoffman P, Hartman A, Srivastava A, Molla G, Madad S, Fernandez-Granda C, and Satija R (2024). Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol 42, 293–304. 10.1038/s41587-023-01767-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data have been deposited at the Sequence Read Archive and are publicly available as of the date of publication. The accession number of the BioProject linked to this data is listed in the key resources table. All other data is available from the lead contact upon request.
All original code has been deposited at Zenodo and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Chicken anti-EGFP | Columbia University | RRID: AB_3094577 |
| Rabbit anti-dsRed | Columbia University | RRID: AB_3086780 |
| Guinea pig anti-vGlut1 | Columbia University | RRID: AB_2665455 |
| Guinea pig anti-vGlut2 | Millipore Sigma | RRID: AB_2665454 |
| Donkey anti-chicken Alexa Fluor 488 | Jackson ImmunoResearch | RRID: AB_2340375 |
| Donkey anti-rabbit Cy3 | Jackson ImmunoResearch | RRID: AB_2307443 |
| Donkey anti-guinea pig DyLight 405 | Jackson ImmunoResearch | RRID: AB_2340470 |
| NeuroTrace 640/660 | ThermoFisher | RRID: AB_2572212 |
| Bacterial and virus strains | ||
| AAV-retro-CAG-tdTomato | Addgene | Cat# 59462-AAVrg |
| AAV1-hSyn-Cre-WPRE.hGh | Addgene | Cat# 105553-AAV1 |
| AAV1-EF1a-Flpo | Addgene | Cat# 55637-AAV1 |
| AAV-retro-hSyn-Cre-WPRE.hGh | Addgene | Cat# 105553-AAVrg |
| AAV-retro-EF1a-Flpo | Addgene | Cat# 55637-AAVrg |
| AAV2-hSyn-ChR2-EYFP-WPRE.hGH | UNC Vector Core | N/A |
| AAV-DJ-FLEX-H2B-GFP | Columbia University | N/A |
| AAV-hSyn-FlpX-H2B-dTomato | Columbia University | N/A |
| AAV6-CAG-FLEX-N2cG-mKate2.0 | Janelia Farm | N/A |
| AAV1-EF1a-FLEX-TVA-mCherry | UNC Vector Core | N/A |
| AAV2/1-CAG-FRT-TVB | Janelia Farm | N/A |
| AAV1-CAG-FLEX-GFP | UNC Vector Core | N/A |
| AAV1-EF1a-fDIO-tdTomato | Addgene | Cat# 128434-AAV1 |
| EnVA-N2c-deltaG-GFP | Thomas Jefferson | N/A |
| EnVB-N2c-deltaG-tdTomato | Janelia Farm | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Cholera Toxin Subunit Alexa Fluor 488 Conjugate | ThermoFisher | Cat# C34775 |
| Cholera Toxin Subunit Alexa Fluor 555 Conjugate | ThermoFisher | Cat# C34776 |
| Tetrodotoxin citrate | Tocris | Cat# 1069 |
| 4-Aminopyridine | Tocris | Cat# 0940 |
| TrueBlack | Biotium | Cat# 23007 |
| Deposited data | ||
| Single nucleus RNA sequencing of spinal neurons targeted by CFA or RFA | This study | NIH BioProject: PRJNA1081900 |
| Code for analysis of anatomical data | This study | Zenodo: https://doi.org/10.5281/zenodo.10712498 |
| Experimental models: Organisms/strains | ||
| C57BL/6J mice | The Jackson Laboratory | IMSR_JAX:000664 |
| B6; 129-Gt(ROSA)26Sortm5(CAG-Sun1/sfGFP)Nat/J | The Jackson Laboratory | IMSR_JAX:021039 |
| Software and algorithms | ||
| MATLAB | Mathworks | RRID:SCR_001622 |
| R | The R Project for Statistical Computing | RRID:SCR_001905 |
| Seurat | Hao et al. 202175 | RRID:SCR_016341 |
| BrainJ | Botta et al. 202024 | N/A |
| SpinalJ | Fiederling et al. 202127 | N/A |
| Prism | GraphPad | RRID:SCR_002798 |


