Abstract
Traditional methods under sparse view for reconstruction of photoacoustic tomography (PAT) often result in significant artifacts. Here, a novel image to image transformation method based on unsupervised learning artifact disentanglement network (ADN), named PAT-ADN, was proposed to address the issue. This network is equipped with specialized encoders and decoders that are responsible for encoding and decoding the artifacts and content components of unpaired images, respectively. The performance of the proposed PAT-ADN was evaluated using circular phantom data and the animal in vivo experimental data. The results demonstrate that PAT-ADN exhibits excellent performance in effectively removing artifacts. In particular, under extremely sparse view (e.g., 16 projections), structural similarity index and peak signal-to-noise ratio are improved by ∼188 % and ∼85 % in in vivo experimental data using the proposed method compared to traditional reconstruction methods. PAT-ADN improves the imaging performance of PAT, opening up possibilities for its application in multiple domains.
Keywords: Photoacoustic tomography, Unsupervised disentanglement strategy, Mitigating artifact, Sparse view
1. Introduction
Photoacoustic tomography (PAT) is a non-invasive and non-radiological multimodal biomedical imaging technique that has gained significant attention as a promising preclinical and clinical tool [1], [2], [3], [4], [5]. By merging the superior contrast capabilities of optical imaging with the excellent resolution characteristics of acoustic imaging, this approach enables high-resolution structural and functional imaging of tissues within the body [6], [7]. PAT has been broadly employed in preclinical studies involving whole-body imaging of small animals. For instance, it has been successfully employed to map microvascular networks and explore resting-state functional connectivity in the brains of mice [8], [9], [10]. Recently, PAT has been widely adopted in clinical practice and has been utilized for human imaging purposes. Its applications encompass functional brain imaging, diagnosis of cardiovascular diseases, breast cancer and molecular imaging [11], [12], [13], [14], [15]. In PAT, the primary objective of imaging is to reconstruct the initial pressure distribution using time-resolved photoacoustic signals, which are generated by laser pulses and captured by ultrasonic transducers [16]. After absorbing the energy from a laser pulse, the tissue absorbers undergo thermoelastic expansion, generating an initial sound pressure that propagate outward. The ultrasonic signals are detected by multiple ultrasonic transducers positioned at different locations around the tissue, and the distribution of optical absorption in the tissue is reconstructed using reconstruction algorithms, such as universal back projection (UBP) [17] or time inversion (TR) [18]. These methods rely on the ideal assumption of the availability of complete measurement data from full views. However, in practical implementation, obtaining complete photoacoustic signals becomes challenging due to limitations in the bandwidth and quantity of ultrasonic transducers. The utilization of these conventional approaches in PAT sparse reconstruction may give rise to image distortion, blurriness, and subpar resolution.
To tackle these problems, a range of techniques are being investigated, including improvements in physical hardware and optimizations in reconstruction methods. Applying the acoustic deflectors [19], utilizing the spherical array system [20], or implementing the toroidal sensor arrays [21], these efforts have made improvements at the physical hardware level, thereby enhancing the imaging quality of PAT. Although these techniques can effectively enhance the imaging quality of PAT, they contribute to increased system costs and a more complex structure, consequently impeding the broad adoption. On the other hand, the imaging performance of PAT can also be enhanced by improving the reconstruction method. The utilization of the adaptive filtered back-projection algorithm [22] and the improved time reversal reconstruction algorithm based on particle swarm optimization [23] can significantly improve image quality with a reduced number of measurement positions or scanning times.
In recent years, deep learning has been extensively implemented in the realm of biomedical imaging, and its application in PAT reconstruction has demonstrated remarkable potential [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36]. At present, deep learning methods utilized for PAT reconstruction are primarily categorized into supervised learning [24], [25], [26], [27], [28] and unsupervised learning [29], [30], [31], [32], [33]. U-Net networks are the primary framework for supervised learning in PAT. Guan et al. presented the Dense Dilation U-Net for eliminating artifacts in 3D PAT [27]. Deng et al. introduced the SE-Unet network architecture for artifact removal, which demonstrated superior performance when compared to back projection algorithm [28]. A substantial amount of paired datasets is required by supervised learning-based approaches. Nevertheless, gathering a large number of ground truth images in practical scenarios is impractical.
Compared to supervised learning, unsupervised learning models are more flexible and capable of handling unlabeled samples while also identifying potential patterns and structures in the data, ultimately improving generalization performance of the mode. Vu et al. put forward Wasserstein-generated adversarial networks with gradient penalties (WGAN-GP) to remove limited-views and limited-bandwidth artifacts in PAT [30]. Shahid et al. applied ResGAN to extract and learn low-frequency features that are obscured by severe interference, aiming to recover high-quality images [31]. Over the past few years, artifact disentanglement network (ADN) has shown its effectiveness in accomplishing high-resolution image-to-image translation tasks in the field of biomedical imaging [32], [33], [34], [35]. Liao et al. introduced an ADN network designed for unsupervised disentanglement of metal artifacts from CT images [32]. Lyu et al. further utilized an improved unsupervised ADN network to successfully tackle the challenges of modality translation, artifact reduction and vertebra segmentation [33]. Motivated by these advancements, this study proposes a photoacoustic tomography artifact disentanglement network (PAT-ADN) to remove artifacts caused by sparse-view sampling. A series of simulated and in vivo experimental data were employed to assess the performance of the proposed method. The results demonstrate the efficacy of the PAT-ADN in capably removing artifacts in PAT. Particularly, when handling extremely sparse data (e.g., 16 projections), the PAT-ADN outperforms the supervised learning methods (e.g., U-Net), resulting in an improvement of approximately 15% in Structure Similarity Index (SSIM) and 14% in Peak Signal to Noise Ratio (PSNR) for circular phantom experimental data.
2. Principles and methods
2.1. Artifacts Removal based on PAT-ADN
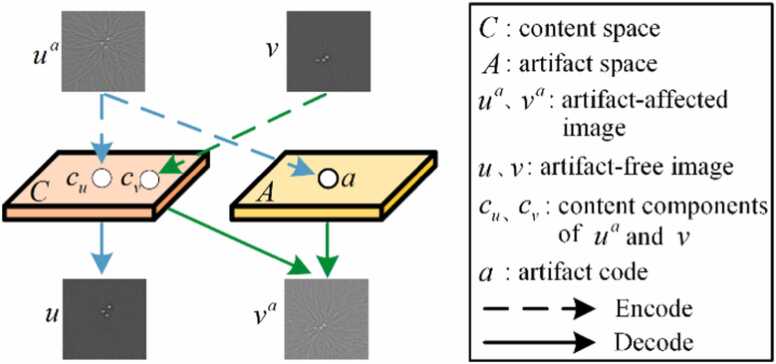
PAT image post-processing can be regarded as a challenging image-to-image translation problem, presenting the opportunity to transform artifact images into high-quality, artifact-free images. The proposed network is formulated within the framework of ADN to handle the challenge of artifact removal in PAT in an unsupervised learning approach. Let represent the domain of the PAT image with artifacts captured under sparse views, while denotes the domain of the full-view PAT. is denoted as a collection of unpaired images, and is an artifact removal model that removes artifacts from . is an artifact-free image not paired with . The process diagram for artifact disentanglement of PAT-ADN is depicted in Fig. 1. In this method, the artifact and content components are disentangled by encoding the artifact-affected image to a content space C and an artifact space A. The image , obtained from the artifact-free domain , is exclusively encoded to yield the content code . Accomplishing the decoding process of , which corresponds to the removal of artifacts from , results in obtaining an artifact-free image . Similarly, through the act of decoding the artifact code and content component , artifacts are inserted into images that are previously devoid of any artifacts, generating the image containing artifacts . It is worth noting that the proposed PAT-ADN method contributes to the learning of these encoding and decoding processes without paired datasets.
Fig. 1.
Artifact disentanglement flowchart of PAT-ADN. The artifact-affected image obtained under sparse view sampling is encoded into both artifact space A and content space C. The artifact-free image obtained under full-view sampling is encoded into content space C. Encoding and into the content space C to generate the content encoding and . Encoding into artifact space A to get artifact code . By decoding the content code , artifacts can be removed from the artifact-affected image (blue arrows ). By decoding the artifact code and content code , artifacts are introduced into artifact-free images, producing the image with artifacts (green arrows ).
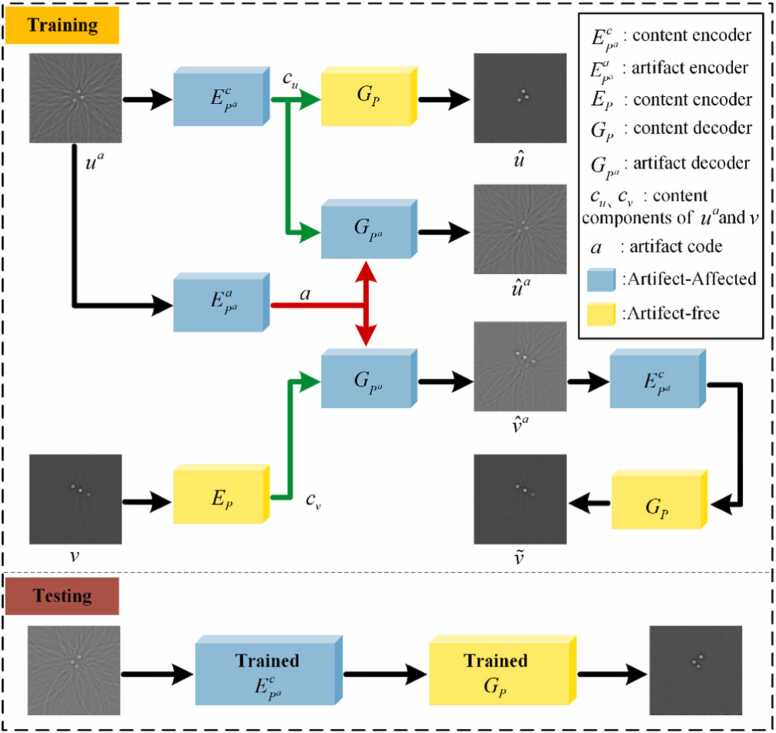
The architecture of PAT-ADN is shown in Fig. 2. Two unpaired images and are given as input images. The image denotes the expected output, recovered by eliminating artifacts from . It includes a pair of encoders for artifact-free images and for artifact-affected images. Mapping an image sample from the image domain to the latent space is the function of the encoders. A content encoder and an artifact encoder independently encode content and artifacts, respectively. The combination of these two encoders forms the encoder . The content components of and are individually mapped to the content space C by and , giving rise to the corresponding content codes and . Furthermore, the components of is transformed into the component space A by , generating the artifact code . The corresponding latent codes are represented as Eq. (1):
| (1) |
Fig. 2.
Architecture of the proposed PAT-ADN. PAT-ADN takes unpaired images and as input (). The image corresponds to the desired output, obtained after effectively removes the artifacts. The encoders consist of and , whereas and serve as the decoders. The green connection line represents the flow of content encoding to the content decoder, the red connection line represents the flow of artifact encoding to the artifact decoder, and the black connection line represents the flow of other data.
In addition, PAT-ADN also includes a pair of the decoder for artifact-free images and the decoder for artifact-affected images. The decoders are responsible for mapping a latent code from the latent space back to the image domain. receives the content code as input and generates an image that is free from artifacts. Decoding from can obtain which removes artifacts from . The images decoded by can be expressed as Eq. (2):
| (2) |
leverages the content code and artifact code as inputs to attain an image that incorporates the intended artifacts. Decoding from and results in which reconstructs from , while decoding from and may introduce artifacts to . The images decoded by is shown as Eq. (3):
| (3) |
The image can be comprehended as an integrated representation that combines both the artifacts from and the underlying content from . With the utilization of and , the artifacts can be effectively eliminated, consequently obtaining artifact-free image , as shown in Eq. (4):
| (4) |
In the training stage, an artifact image and an artifact-free image serve as inputs to the network, with the two images being non-paired. The artifact image undergoes the encoder , separating its content components , and is then reconstructed into an artifact-free image through the decoder . The artifact image , after being processed by the encoder , separates the artifact components . Then through the decoder , both and contribute to obtaining the artifact image , which is very close to . The artifact-free image , after content separation by the encoder , is combined with the artifact component . Together, they pass through the decoder , resulting in the artifact-transferred image . Subsequently, undergoes encoding with and decoding with , effectively removing the artifact and yielding the artifact-free image . In the training process, following each training iteration of the ADN network, both and are applied twice. It results in enhanced training that significantly strengthens the ability of to separate content components from artifact images and the ability of to restore images. In the testing stage, the artifact image, after being processed by the trained encoder and the trained decoder , yields the artifact-free image.
2.2. Network architectures
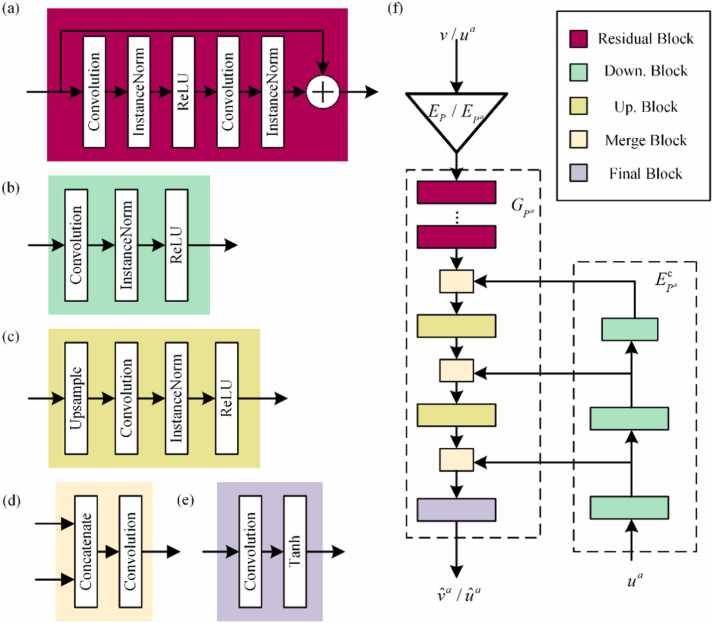
The proposed network consists of the encoders and decoders. The details of the encoder and decoder are presented in Table 1. Each building component is organized as a sequence of building blocks, comprising five distinct block types. Fig. 3 illustrates both the basic building blocks of the PAT-ADN and the detailed architecture of artifact pyramid decoding. Fig. 3(a) shows that the residual block incorporates residual connections, facilitating the consideration of low-level features in the computation of high-level features. Fig. 3(b) depicts that the down-sampling block utilizes stride convolution to decrease the dimensionality of the feature map. Fig. 3(c) demonstrates that the up-sampling block restores the feature map to its original dimension. In the merge block (Fig. 3(d)), the content feature map and the artifact feature map are connected. Fig. 3(e) exhibits the final block. Fig. 3(f) shows the intricate architecture of artifact pyramid decoding.
Table 1.
Design and structure of the building components.
| Network | Block | Channel | Count | Padding | Kernel | Stride |
|---|---|---|---|---|---|---|
| Residual | 256 | 4 | 1 | 3 | 1 | |
| Up. | 128 | 1 | 2 | 5 | 1 | |
| Up. | 64 | 1 | 2 | 5 | 1 | |
| Final | 1 | 1 | 3 | 7 | 1 | |
| Residual | 256 | 4 | 1 | 3 | 1 | |
| Merge | 256 | 1 | 0 | 1 | 1 | |
| Up. | 128 | 1 | 2 | 5 | 1 | |
| Merge | 128 | 1 | 0 | 1 | 1 | |
| Up. | 64 | 1 | 2 | 5 | 1 | |
| Merge | 64 | 1 | 0 | 1 | 1 | |
| Final | 1 | 1 | 3 | 7 | 1 | |
| Down. | 64 | 1 | 3 | 7 | 1 | |
| Down. | 128 | 1 | 1 | 4 | 2 | |
| Down. | 256 | 1 | 1 | 4 | 2 | |
| Residual | 256 | 4 | 1 | 3 | 1 | |
| Down. | 64 | 1 | 3 | 7 | 1 | |
| Down. | 128 | 1 | 1 | 4 | 2 | |
| Down. | 256 | 1 | 1 | 4 | 2 |
Fig. 3.
Building blocks of PAT-ADN and architecture details of artifact pyramid decoding. (a) Residual block. (b) Down-sampling block. (c) Up-sampling block. (d) Merging block. (e) Final block. (f) Architectural features of artifact pyramid decoding. Down. Block, down-sampling block; Up. Block, up-sampling block.
By combining artifact codes of different scales, artifact pyramid decoding achieves high-definition functionality at a relatively lower cost. comprises multiple down-sampling blocks and produces feature maps at various scales. comprises a sequence of residual blocks, merging blocks, up-sampling blocks, and final blocks.
2.3. Loss function
Learning to generate and is crucial for effectively disentangling artifacts. However, due to the absence of paired images, applying a simple regression loss is not feasible. Hence, we leverage the concept of adversarial learning [36] and introduce two discriminators and to regularize and . Then adversarial losses and can be expressed as Eq. (5):
| (5) |
Adversarial loss mitigates artifacts by promoting the resemblance of to sample from . However, obtained through this process is anatomically credible but not anatomically precise. PAT-ADN tackles the issue of anatomical accuracy by introducing the artifact consistency loss. necessitates anatomical proximity between and , without requiring complete alignment. The same principle applies to and . as shown in Eq. (6):
| (6) |
The introduction of reconstruction loss essentially encourage encoders and decoders to retain information and avoid adding any new artifacts. In particular, PAT-ADN utilizes and for encoding and decoding to achieve the results consistent with the input . To promote sharper output, L1 loss is employed. Reconstruction loss satisfies Eq. (7):
| (7) |
In this study, PAT-ADN incorporates the above four types of losses, including two adversarial losses and , an artifact consistency loss and a reconstruction loss . The weighted sum of these losses is referred to as the total loss , as shown in Eq. (8):
| (8) |
Here, represents the hyperparameter that determines the weighting of each term.
2.4. Dataset and network parameter setting
The dataset utilized in this study comprises photoacoustic tomographic images of circular phantom and in vivo mouse. The experimental data employed in this work are derived from https://doi.org/10.6084/m9.figshare.9250784[37]. The circular phantom dataset is divided into 496 images under the full view (512 projections) and an equal number of images under other sparse views (8, 16, 32, and 64 projections). The in vivo mouse abdomen dataset is divided into 274 images under the full view (512 projections) and an equal number of images under other sparse views (8, 16, 32 and 64 projections). Full-view images are used as the ground truth during the training and testing stage of the proposed method and U-Net method.
In the process of training, a small batch size of 5 and an initial learning rate of are implemented, with the number of epochs set at 50. The Adam optimization technique is employed during the training phase. For the hyperparameters in the loss function, the configuration is set as , . represents the weight of adversarial loss and . If is excessively high, it will lead the network to be more inclined to generate the content information of the image. On the contrary, if is set excessively low, the network will generate inadequate content information. Therefore, it is common to set as a baseline to balance the generated content information. indicates the weight of artifact consistency loss , influencing the composition of artifacts. signifies the weight of the reconstruction loss , which guarantees the preservation of all content information without introducing new artifacts during artifact removal. The artifact consistency loss and the reconstruction loss are considered to be equally important [32], thus setting . The GeForce RTX 2080Ti GPU with 11 GB of memory is utilized for training and testing of the PAT-AND, U-Net and CycleGAN methods.
3. Results
3.1. Results on circular phantom data
To illustrate the effectiveness of the PAT-ADN network in artifact removal, we contrast the PAT-ADN method with the highly utilized supervised deep learning technique, U-Net [38], and the unsupervised deep learning model, CycleGAN [39]. The training environment and dataset used for U-Net and CycleGAN are identical to those of PAT-ADN. SSIM and PSNR are calculated to quantitatively assess the performance of PAT-ADN.
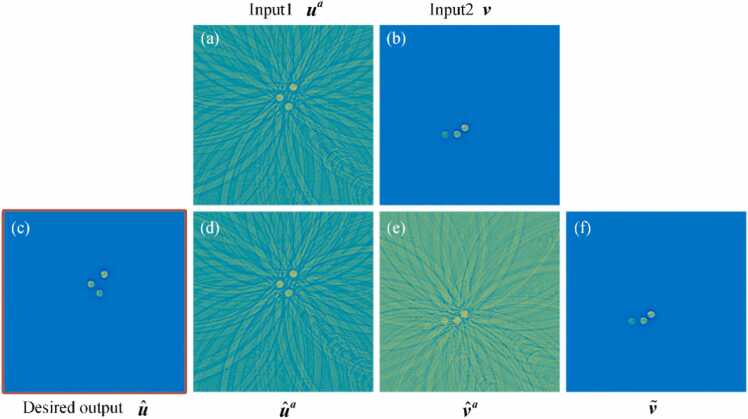
Fig. 4 illustrates the results of artifact disentanglement using the PAT-ADN method on the circular phantom. Fig. 4(a) represents the artifact image inputted into this network. Fig. 4(b) shows the artifact-free image used as input for this network. Fig. 4(c) demonstrates the artifact removal outcome achieved through the proposed method. Fig. 4(d) is the artifact image synthesized through the fusion of the artifact component and the content component. Fig. 4(e) illustrates the artifact-transferred image , obtained by transferring artifacts from to . Fig. 4(f) depicts the artifact-free image obtained by eliminating artifacts from .
Fig. 4.
Results of artifact disentanglement employing the PAT-ADN method on the circular phantom. (a) is the artifact image as the input of this network. (b) is the artifact-free image as the input of this network. (c) is the artifact removal result obtained using the proposed method. (d) is the artifact image synthesized by combining the artifact component and the content component. (e) is the artifact-transferred image obtained by transferring the artifacts from to . (f) is the artifact-free image obtained by removing artifacts from .
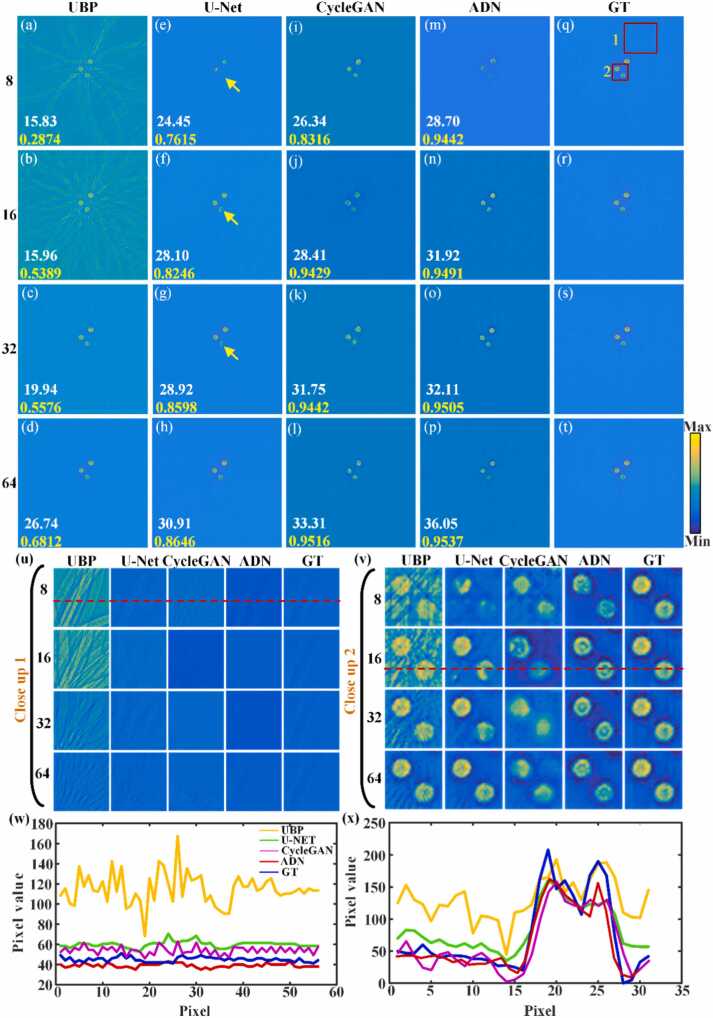
Fig. 5 compares the artifact removal capabilities of U-Net, CycleGAN and PAT-ADN in circular phantom under different projection views. In Fig. 5, the PSNR value is indicated by the white number at the bottom left, while the SSIM value is represented by the yellow number. Fig. 5(a)–(d) show reconstructed PAT images of measurement data under different projection views (e.g. 8, 16, 32 and 64 projection views) obtained using the UBP method. The PAT images reconstructed using UBP method suffer from a substantial presence of artifacts. Fig. 5(e)–(h) represent the corresponding artifact removal images obtained using U-Net. Under 64 projection views, the U-Net method eliminates a substantial quantity of artifacts. When the projection view is further sparse, the reconstruction results still exhibit some residual artifacts. And these reconstructed images suffer from details loss, as indicated by the yellow arrows in Fig. 5(e)–(g). Fig. 5(i)–(l) depict the corresponding artifact removal images obtained using CycleGAN. In comparison with the traditional U-Net method, the CycleGAN method reconstructed images with fewer artifacts. However, there still remains a minor presence of distortion in the reconstructed phantom images. Fig. 5(m)-(p) demonstrate the artifact removal images obtained using PAT-ADN, respectively. The proposed method presents excellent performance in removing artifacts under different projection views. Notably, even under highly sparse measurement conditions (8 and 16 projection views), the proposed method is also capable of substantially removing artifacts. Fig. 5(q)–(t) illustrate the same ground truth acquired using UBP under 512 projection views. Fig. 5(u) and Fig. 5(v) depict the close-up images of the red rectangles 1 and 2 in Fig. 5(q), respectively. From the close-up images, it can be observed that the results using the PAT-ADN method depict more precise details under different projection views. Fig. 5(w) is the signal distribution along the red dashed line in Fig. 5(u) under 8 projection views, while Fig. 5(x) corresponds to the signal distribution along the red dashed line in Fig. 5(v) under 16 projection views. In comparison to other traditional approaches, such as UBP, the proposed method yields a signal distribution that is more closely aligned with the ground truth, thus verifying the outstanding advantage of PAT-ADN in removing artifacts of the circular phantom under sparse projection views.
Fig. 5.
Comparison of artifact removal capabilities of U-Net, CycleGAN and PAT-ADN in circular phantom under different projections. (a)-(d) are the reconstructed results obtained using the UBP method under 8, 16, 32 and 64 projections, respectively. (e)-(h) are the artifact removal results obtained using the U-Net method under 8, 16, 32 and 64 projections, respectively. (i)-(l) are the artifact removal results obtained using the CycleGAN method under 8, 16, 32 and 64 projections, respectively. (m)-(p) are the artifact removal results obtained using the proposed method under 8, 16, 32 and 64 projections, respectively. The white numbers in the lower left corner indicate PSNR, and the yellow numbers indicate SSIM. (q)-(t) are the same ground truth images. (u) and (v) are the close-up images indicated by the red rectangles 1 and 2 in (q), respectively. (w) and (x) are the signal distribution along the red dashed lines 1 and 2 in (u) and (v) under 8 and 16 projections, respectively. GT, ground truth.
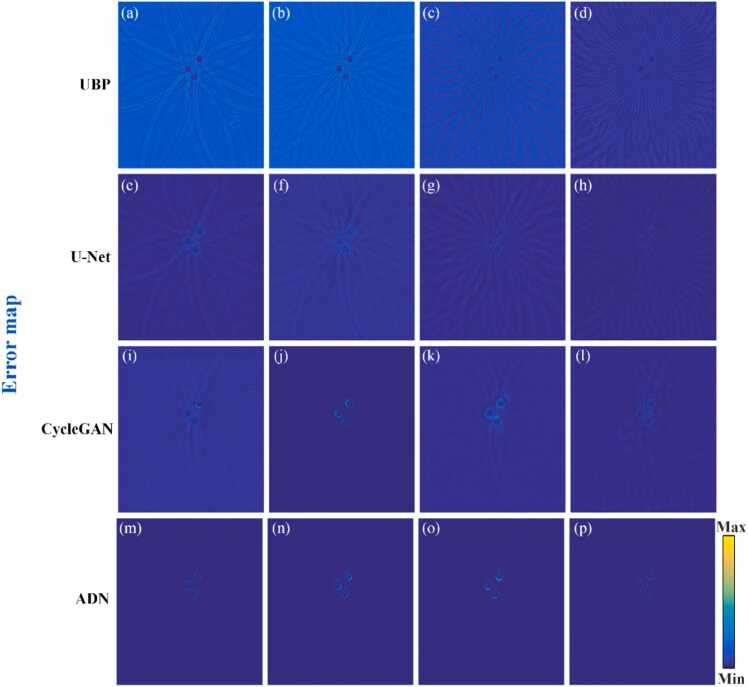
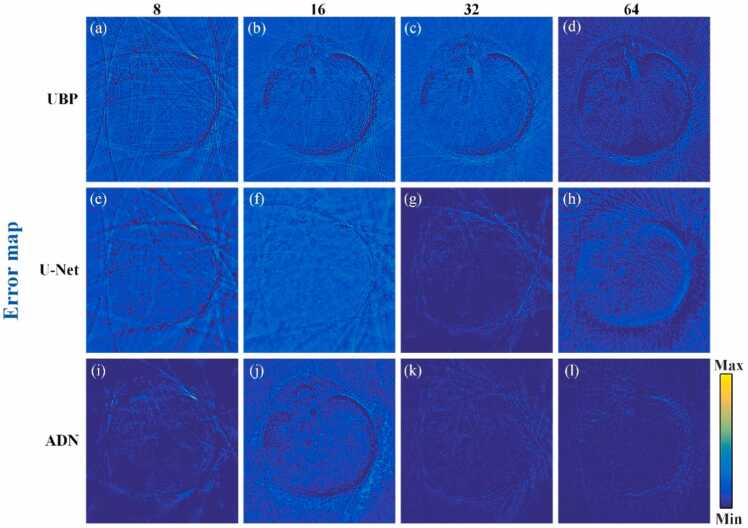
Fig. 6(a)–(p) show the error map corresponding to Fig. 5(a)–(p), indicating the difference between the ground truth and the recovered images by the mentioned methods under projection views of 8, 16, 32 and 64. The reconstructed results obtained using the PAT-ADN method closely resemble the ground truth exhibiting minimal artifacts. This observation indicates that the reconstructed images using the proposed method possess higher quality than the other three methods. From a quantitative analysis perspective, under 64 projection views, the reconstructed result using PAT-ADN method achieves a PSNR of 36.05 dB and an SSIM of 0.9537, which are 5.14 dB and 0.0891 higher, respectively, compared to the U-Net method. As the number of projection views becomes sparser (e.g., 8 projection views), the reconstructed results using PAT-ADN method reveals a ∼9 % elevation in PSNR and a ∼14 % advancement in SSIM over the reconstructed results using CycleGAN. It further demonstrates that the proposed method can efficiently removing artifacts under extremely sparse view conditions.
Fig. 6.
Error map obtained from the recovered results in the circular phantom data under different projections. (a)-(p) are the error maps corresponding to Fig. 5(a)–(p).
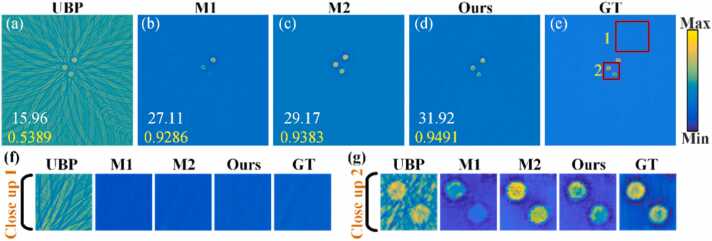
Through an ablative experiment on the Loss function, this study validates the efficacy of the PAT-ADN network design. The experimental results in Fig. 7 demonstrate a comparison of the performance between PAT-ADN and its two variants (M1, M2). M1 refers to a model trained solely with adversarial loss . M2 refers to a model trained with both adversarial loss and reconstruction loss . For both qualitative and quantitative analysis, all experiments were performed using circular phantom experimental data under 16 projection views. In Fig. 7, the reconstructed PAT images exhibit fewer artifacts using M1 and M2 methods than using UBP method. Fig. 7(f) and Fig. 7(g) depict the close-up images of the red rectangles 1 and 2 in Fig. 7(e). It’s obvious that the results reconstructed using M1 method still exhibit some persistent artifacts and even detail loss. The reconstructed structures are relatively more complete compared to M1 method, yet some artifacts still remain, as shown in Fig. 7(c). In Fig. 7(d), the results using PAT-ADN method maintain a substantial amount of details while eliminating artifacts. Quantitative analysis of the reconstruction results of models trained with different combinations of loss functions is presented in Table 2, demonstrating that the proposed method achieves a 9.43 % improvement in PSNR and a 1.15 % improvement in SSIM compared to M2. The above results indicate that the PAT-ADN model trained with adversarial loss , reconstruction loss , and artifact consistency loss exhibits significantly superior artifact removal capabilities compared to M1-M2 methods. This demonstrates the effectiveness of the proposed PAT-ADN network.
Fig. 7.
Comparison of the artifact removal capabilities of different variants of PAT-ADN on circular phantom under 16 projections. (a) is the reconstructed results obtained using the UBP method under 16 projections. (b) is the artifact removal results obtained using M1 method. (c) is the artifact removal results obtained using M2 method. (d) is the artifact removal results obtained using the PAT-ADN method under 16 projections. (e) is ground truth image. (f) and (g) are the close-up images indicated by the red rectangles 1 and 2 in (e), respectively. GT, ground truth.
Table 2.
Quantitative comparison of different variants trained with different combinations of the loss functions of PAT-ADN.
| Method |
Metrics |
|
|---|---|---|
| PSNR | SSIM | |
| M1 (only) | 27.11 | 0.9286 |
| M2 (M1 with) | 29.17 | 0.9383 |
| Ours (PAT-ADN) | 31.92 | 0.9491 |
3.2. Results on in vivo data
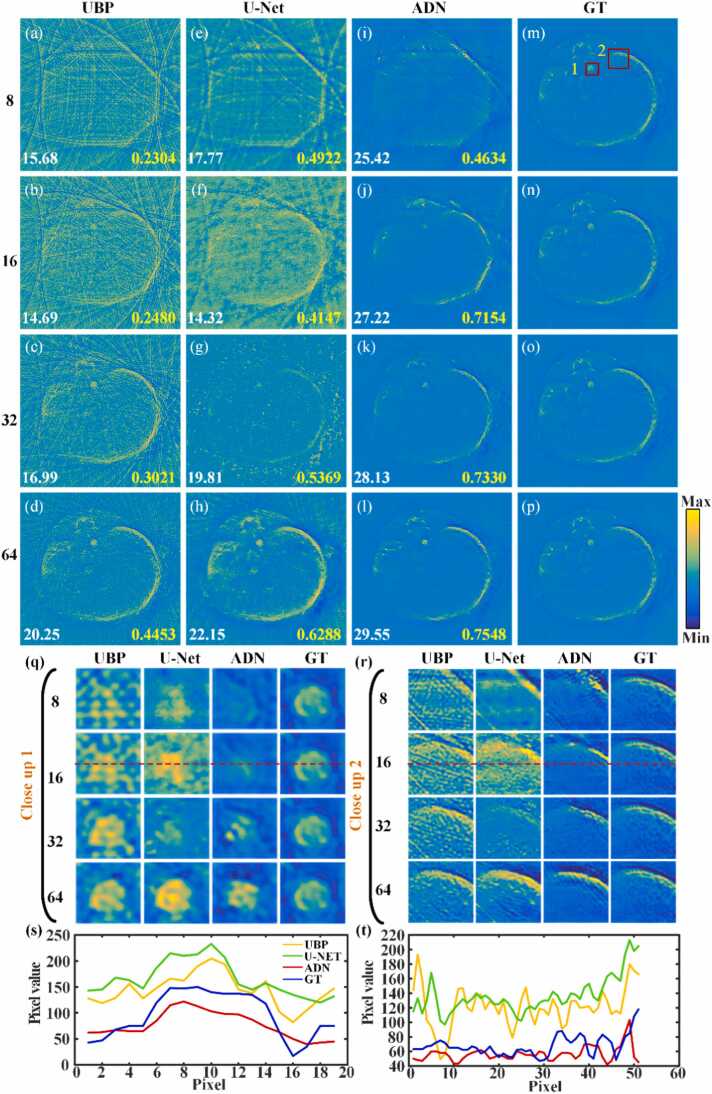
The artifact removal performance of the PAT-ADN method is further validated in in vivo data, the results are shown in Fig. 8. Fig. 8(a)–(d) depict a mouse abdominal image reconstructed using the UBP method with 8, 16, 32 and 64 projections, respectively. From these images, it is evident that the reconstructed images exhibit serious artifacts, making it challenging to discern image details. Fig. 8(e)–(h) illustrate the corresponding artifact removal images obtained using the U-Net method. Fig. 8(i)–(l) demonstrate the artifact removal images obtained using the PAT-ADN method. The PAT-ADN method demonstrates a favorable effect in removing artifacts under various projection conditions. It is obvious that even under highly sparse measurement conditions (8 and 16 projections), the method effectively suppresses artifacts. In Fig. 8, the PSNR value is indicated by the white number at the bottom left, while the SSIM value is represented by the yellow number at the bottom right. Fig. 8(m)–(p) represent the same ground truth acquired using UBP under 512 projections. Fig. 8(q) and (r) are the close-up images indicated by the red rectangles 1 and 2 in Fig. 8(m), respectively. The close-up images reveal that the PAT-ADN method outperforms both the U-Net method and the UBP method in terms of restoring edge details. Fig. 8(s) and (t) are the signal distribution along the red dashed lines 1 and 2 in Fig. 8(q) and (r) under extremely sparse views (16 projections), respectively. Obviously, the signal distribution obtained by PAT-ADN is closer to the ground truth, which verifying the feasibility of the proposed method in artifacts removal for the in vivo data.
Fig. 8.
Comparison of artifact removal capabilities of U-Net and PAT-ADN in in vivo data under different projections. (a)-(d) are the reconstructed results obtained using the UBP method under 8, 16, 32 and 64 projections, respectively. (e)-(h) are the artifact removal results obtained using the U-Net method under 8, 16, 32 and 64 projections, respectively. (i)-(l) are the artifact removal results obtained using the proposed method under 8, 16, 32 and 64 projections, respectively. The white number in the lower left corner indicates PSNR, and the yellow number in the lower right corner indicates SSIM. (m)-(p) are the same ground truth images. (q) and (r) are the close-up images indicated by the red rectangles 1 and 2 in (m), respectively. (s)-(t) show the signal distribution along the red dashed lines in (q) and (r), respectively. GT, ground truth.
Fig. 9(a)-(l) show the error map corresponding to Fig. 8(a)–(l), indicating the difference between the ground truth and the recovered images by the mentioned methods above under projections of 8, 16, 32 and 64. It is visually evident from the error map that the recovered images obtained using the PAT-ADN method have a close resemblance to the ground truth images. The experimental results further demonstrate that the unsupervised PAT-ADN method is capable of effectively removing artifacts in PAT in terms of SSIM and PSNR. Under 32 projections, the PSNR and SSIM of the proposed method can reach 28.13 dB and 0.7330, respectively, which is 11.14 dB and 0.1961 higher than that of U-Net method. When the number of projection views is further reduced to 16, the PSNR and SSIM are 27.22 dB and 0.7154, respectively, which is 12.9 dB and 0.3007 higher than that of U-Net. The above results confirm the powerful effect of the proposed method in removing artifacts in photoacoustic tomographic images.
Fig. 9.
Error map obtained from the recovered results in in vivo data under different projections. (a)-(l) are the error maps corresponding to Fig. 8(a)–(l).
4. Conclusion and discussion
A novel image domain transformation method, named PAT-ADN, was proposed to address the issue of artifacts caused by sparse-view sampling in PAT. The proposed network performs disentanglement of PAT artifact-affected images into artifact space and content space, preserving the content domain, resulting in artifact-free images. During the training phase, an artifact image and an artifact-free image, both unpaired, are provided as input. The artifact image undergoes content encoding to isolate its content component, which is then reconstructed into a new artifact-free image through content decoding. During the testing phase, the artifact image is processed by a trained set of encoders and decoders to directly generate artifact-free images. The performance of the proposed PAT-ADN was evaluated using circular phantom data and the animal in vivo experimental data. For the circular phantom data, the proposed method performed excellently under extremely sparse views (e.g. 8 projections). The SSIM and PSNR of the proposed method are 0.9442 and 28.70 dB, respectively, representing improvements of 0.1827 and 4.25 dB compared to the U-Net method. For the in vivo experimental data, this method showcases advancements over the U-Net method. Under 16 projections, the SSIM and PSNR values of this method reach 0.7154 and 27.22 dB, respectively, exhibiting enhancements of 0.3007 and 12.9 dB compared to the U-Net method. These results illustrate the exceptional artifact removal capability of the PAT-ADN method, even in the case of highly sparse views. This significant advancement enhances the quality of PAT images, providing greater flexibility for various applications of PAT.
The PAT-ADN model demonstrates certain generalizability and holds great potential for clinical applications. The datasets used in the experiments all generated from the same kind of imaging objects within the same system. By expanding the datasets, the generalizability of the method can be further improved to accommodate clinical scenarios. The expanded dataset should include full-view clinical PAT images from various samples obtained by different PAT systems.
The core idea of PAT-ADN for artifact removal lies in integrating adversarial training with an encoder-decoder network to learn about the data distribution of image content and latent representation of artifacts, in order to improve disentanglement effect between content and artifact components. Through training, the encoder-decoder network develops the ability to separate artifact components from artifact images while maintaining the integrity of the content components. As a result, the proposed PAT-ADN network can effectively reduce artifacts without compromising image quality.
In conclusion, this method mainly achieves high-quality artifact removal in a relatively short time by training both encoders and decoders. As the network training does not focus on specific artifact structures, it treats irregular data distributions in the images as artifacts and separates them from other content components. The proposed PAT-ADN network is applicable to other reconstruction tasks, such as limited-view sampling reconstruction and reconstruction under low-bandwidth conditions. Actually, the PAT-ADN network can transform artifact-free images into artifacts images. This provides a new approach to dataset creation, thus reducing the difficulty of obtaining a large amount of training data. The proposed method will further expand the clinical applications of photoacoustic imaging, including eliminating motion artifacts caused by respiratory motion and early diagnosis of breast cancer.
CRediT authorship contribution statement
Tianle Li: Writing – review & editing, Writing – original draft, Validation, Software, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Shangkun Hou: Writing – review & editing, Writing – original draft, Visualization, Validation, Software, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Wenhua Zhong: Writing – review & editing, Writing – original draft, Visualization, Validation, Software, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Xianlin Song: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization. Zilong Li: Data curation, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. Hongyu Zhang: Writing – review & editing, Writing – original draft, Validation, Methodology, Formal analysis, Data curation, Conceptualization, Software, Visualization. Qiegen Liu: Writing – review & editing, Writing – original draft, Validation, Supervision, Resources, Project administration, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization. Guijun Wang: Data curation, Investigation, Software, Validation, Visualization, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors thank Jiabin Lin from School of Information Engineering, Nanchang University for helpful discussions. This work was supported by National Natural Science Foundation of China (62265011, 62122033); Jiangxi Provincial Natural Science Foundation (20224BAB212006, 20232BAB202038); National Key Research and Development Program of China (2023YFF1204302).
Disclosures
The authors declare no conflicts of interest.
Code, Data, and Materials
The code is provided in https://github.com/yqx7150/PAT-ADN, and the data involved in the main text is also provided.
Biographies

Wenhua Zhong received the bachelor degree in Electronic Information Engineering from Jiangxi Science and Technology Normal University, Nanchang, China. He is currently studying in Nanchang University for master’s degree in Electronic Information. His research interests include computer vision, deep learning, photoacoustic microscopy and photoacoustic tomography.

Tianle Li is currently studying for the bachelor degree in Computer Science in Nanchang University, Nanchang, China. His research interests include photoacoustic tomography, deep learning and digital image processing.

Shangkun Hou is currently studying for bachelor degree in Communication Engineering in Nanchang University, Nanchang, China. His research interests include image processing, artificial intelligence, photoacoustic tomography and holographic imaging.

Hongyu Zhang is currently studying for the bachelor degree in Electronic Information Engineering in Nanchang University, Nanchang, China. His research interests include photoacoustic tomography, deep learning and medical image processing.

Zilong Li received the bachelor degree in Electronic Information Engineering from Guilin University of Electronic Technology, Guilin, China. He is currently studying in Nanchang University for master’s degree in Electronic Information Engineering. His research interests include optical imaging, deep learning and photoacoustic tomography.

Guijun Wang received the bachelor degree in Electronic Information Science and Technology from Nanchang Hangkong University, Nanchang, China. He is currently studying in Nanchang University for master’s degree in Electronic Information Engineering. His research interests include image processing, deep learning and photoacoustic tomography.

Qiegen Liu received his PhD degree in Biomedical Engineering from Shanghai Jiao Tong University, Shanghai, China in 2012. Currently, he is a professor at Nanchang University. He is the winner of Excellent Young Scientists Fund. He has published more than 50 publications and serves as committee member of several international and domestic academic organizations. His research interests include artificial intelligence, computational imaging and image processing.

Xianlin Song received his Ph.D. degree in Optical Engineering from Huazhong University of Science and Technology, China in 2019. He joined School of Information Engineering, Nanchang University as an assistant professor in Nov. 2019. He has published more than 20 publications and given more than 15 invited presentations at international conferences. His research topics include optical imaging, biomedical imaging and photoacoustic imaging.
Data availability
Data will be made available on request.
References
- 1.Attia A.B.E., et al. A review of clinical photoacoustic imaging: current and future trends. Photoacoustics. 2019;16 doi: 10.1016/j.pacs.2019.100144]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tong X., et al. Non-invasive 3D photoacoustic tomography of angiographic anatomy and hemodynamics of fatty livers in rats. Adv. Sci. 2023;10(2):2005759. doi: 10.1002/advs.202205759]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steinberg I., et al. Photoacoustic clinical imaging. Photoacoustics. 2019;14:77–98. doi: 10.1016/j.pacs.2019.05.001]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin L., et al. High-speed three-dimensional photoacoustic computed tomography for preclinical research and clinical translation. Nat. Commun. 2021;12(1):882. doi: 10.1038/s41467-021-21232-1]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Upputuri P.K., Pramanik M. Recent advances toward preclinical and clinical translation of photoacoustic tomography: a review. J. Biomed. Opt. 2016;22(4) doi: 10.1117/1.JBO.22.4.041006]. [DOI] [PubMed] [Google Scholar]
- 6.Fu Q., et al. Photoacoustic imaging: contrast agents and their biomedical applications. Adv. Mater. 2019;31(6) doi: 10.1002/adma.201805875]. [DOI] [PubMed] [Google Scholar]
- 7.Yao J., Xia J., Wang L.V. Multiscale functional and molecular photoacoustic tomography. Ultrason. Imaging. 2016;38(1):44–62. doi: 10.1177/0161734615584312]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia J., Wang L.V. Small-animal whole-body photoacoustic tomography: a review. IEEE Trans. Biomed. Eng. 2013;61(5):1380–1389. doi: 10.1109/TBME.2013.2283507]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plumb A.A., et al. Rapid volumetric photoacoustic tomographic imaging with a Fabry-Perot ultrasound sensor depicts peripheral arteries and microvascular vasomotor responses to thermal stimuli. Eur. Radiol. 2018;28:1037–1045. doi: 10.1007/s00330-017-5080-9]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang P., et al. High-resolution deep functional imaging of the whole mouse brain by photoacoustic computed tomography in vivo. J. Biophotonics. 2018;11(1) doi: 10.1002/jbio.201700024]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Na S., Wang L.V. Photoacoustic computed tomography for functional human brain imaging. Biomed. Opt. Express. 2021;12(7):4056–4083. doi: 10.1364/BOE.423707]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu M., et al. Advanced ultrasound and photoacoustic imaging in cardiology. Sensors. 2021;21(23):7947. doi: 10.3390/s21237947]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mehrmohammadi M., et al. Photoacoustic imaging for cancer detection and staging. Curr. Mol. Imaging. 2013;2(1):89–105. doi: 10.2174/2211555211302010010]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin L., et al. Single-breath-hold photoacoustic computed tomography of the breast. Nat. Commun. 2018;9(1):2352. doi: 10.1038/s41467-018-04576-z]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao J., Wang L.V. Recent progress in photoacoustic molecular imaging. Curr. Opin. Chem. Biol. 2018;45:104–112. doi: 10.1016/j.cbpa.2018.03.016]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L.V., Yao J. A practical guide to photoacoustic tomography in the life sciences. Nat. Methods. 2016;13(8):627–638. doi: 10.1038/nmeth.3925]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu M., Wang L.V. Universal back-projection algorithm for photoacoustic computed tomography. Proc. SPIE. 2015;5697:251–254. doi: 10.1103/PhysRevE.71.016706]. [DOI] [PubMed] [Google Scholar]
- 18.Treeby B.E., Zhang E.Z., Cox B.T. Photoacoustic tomography in absorbing acoustic media using time reversal. Inverse Probl. 2010;26(11) doi: 10.1088/0266-5611/26/11/115003]. [DOI] [Google Scholar]
- 19.Kalva S.K., Pramanik M. Use of acoustic reflector to make a compact photoacoustic tomography system. J. Biomed. Opt. 2017;22(2) doi: 10.1117/1.JBO.22.2.026009]. 026009-026009. [DOI] [PubMed] [Google Scholar]
- 20.Estrada H., et al. Spherical array system for high-precision transcranial ultrasound stimulation and optoacoustic imaging in rodents. IEEE Trans. Ultrason. Ferroelectr. Freq. Control. 2021;68(1):107–115. doi: 10.1109/TUFFC.2020.2994877]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang H., et al. A new deep learning network for mitigating limited-view and under-sampling artifacts in ring-shaped photoacoustic tomography. Comput. Med. Imaging Graph. 2020;84 doi: 10.1016/j.compmedimag.2020.101720]. [DOI] [PubMed] [Google Scholar]
- 22.Huang H., et al. An adaptive filtered back-projection for photoacoustic image reconstruction. Med. Phys. 2015;42(5):2169–2178. doi: 10.1118/1.4915532]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun M., et al. Time reversal reconstruction algorithm based on PSO optimized SVM interpolation for photoacoustic imaging. Math. Probl. Eng. 2015;2015 doi: 10.1155/2015/795092]. [DOI] [Google Scholar]
- 24.Sahiner B., et al. Deep learning in medical imaging and radiation therapy. Med. Phys. 2019;46(1):e1–e36. doi: 10.1002/mp.13264]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim M.W., et al. Deep-learning image reconstruction for real-time photoacoustic system. IEEE Trans. Med. Imaging. 2020;39(11):3379–3390. doi: 10.1109/TMI.2020.2993835]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allman D., et al. Deep neural networks to remove photoacoustic reflection artifacts in ex vivo and in vivo tissue, in 2018. IEEE Int. Ultrason. Symp. . (IUS) 2018:1–4. doi: 10.1109/ULTSYM.2018.8579723]. [DOI] [Google Scholar]
- 27.Guan S., et al. Dense dilated UNet: deep learning for 3D photoacoustic tomography image reconstruction. arXiv Prepr. arXiv. 2021;2104(03130) doi: 10.48550/arXiv.2104.03130]. [DOI] [Google Scholar]
- 28.Deng J., et al. Unet-based for Photoacoustic Imaging Artifact Removal. Proc. IEEE Int. Ultrason. Symp. (IUS) 2018:1–4. doi: 10.1364/3D.2020.JTh2A.44]. [DOI] [Google Scholar]
- 29.Kirchner T., Gröhl J., Maier-Hein L. Context encoding enables machine learning-based quantitative photoacoustics. J. Biomed. Opt. 2018;23(5) doi: 10.1117/1.JBO.25.4.049802]. 0560008-056008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vu T., et al. A generative adversarial network for artifact removal in photoacoustic computed tomography with a linear-array transducer. Exp. Biol. Med. 2020;245(7):597–605. doi: 10.1177/1535370220914285]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shahid H., et al. Feasibility of a generative adversarial network for artifact removal in experimental photoacoustic imaging. Ultrasound Med. Biol. 2022;48(8):1628–1643. doi: 10.1016/j.ultrasmedbio.2022.04.008]. [DOI] [PubMed] [Google Scholar]
- 32.Liao H., et al. ADN: artifact disentanglement network for unsupervised metal artifact reduction. IEEE Trans. Med. Imaging. 2019;39(3):634–643. doi: 10.1109/TMI.2019.2933425]. [DOI] [PubMed] [Google Scholar]
- 33.Lyu Y., et al. Joint unsupervised learning for the vertebra segmentation, artifact reduction and modality translation of CBCT images. arXiv. 2020;(2001):00339. doi: 10.48550/arXiv.2001.00339]. [DOI] [Google Scholar]
- 34.Ruan Y., et al. QS-ADN: Quasi-supervised artifact disentanglement network for low-dose CT image denoising by local similarity among unpaired data. Phys. Med. Biol. 2023;68(20) doi: 10.1088/1361-6560/acf9da]. [DOI] [PubMed] [Google Scholar]
- 35.Ranzini M.B.M., et al. Combining multimodal information for metal artefact reduction: an unsupervised deep learning framework. arXiv. 2020;09321(2004) doi: 10.1109/ISBI45749.2020.9098633]. [DOI] [Google Scholar]
- 36.Goodfellow I., et al. Generative adversarial nets. Adv. Neural Inf. Process. Syst. 2014;27 doi: 10.48550/arXiv.1406.2661]. [DOI] [Google Scholar]
- 37.Davoudi N., Deán-Ben X., Razansky D. Deep learning optoacoustic tomography with sparse data. Nat. Mach. Intell. 2019;1(10):453–460. doi: 10.1038/s42256-019-0095-3]. [DOI] [Google Scholar]
- 38.Ronneberger O., Fischer P., Brox T. U-Net: convolutional networks for biomedical image segmentation. MICCAI. 2015;3(18):234–241. doi: 10.1007/978-3-319-24574-4_28]. [DOI] [Google Scholar]
- 39.Lu M., et al. Artifact removal in photoacoustic tomography with an unsupervised method. Biomed. Opt. Express. 2021;12(10):6284–6299. doi: 10.1364/BOE.434172]. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.