Abstract
Objectives
To investigate the utility of deep learning (DL) automated segmentation-based MRI radiomic features and clinical-radiological characteristics in predicting early recurrence after curative resection of single hepatocellular carcinoma (HCC).
Methods
This single-center, retrospective study included consecutive patients with surgically proven HCC who underwent contrast-enhanced MRI before curative hepatectomy from December 2009 to December 2021. Using 3D U-net-based DL algorithms, automated segmentation of the liver and HCC was performed on six MRI sequences. Radiomic features were extracted from the tumor, tumor border extensions (5 mm, 10 mm, and 20 mm), and the liver. A hybrid model incorporating the optimal radiomic signature and preoperative clinical-radiological characteristics was constructed via Cox regression analyses for early recurrence. Model discrimination was characterized with C-index and time-dependent area under the receiver operating curve (tdAUC) and compared with the widely-adopted BCLC and CNLC staging systems.
Results
Four hundred and thirty-four patients (median age, 52.0 years; 376 men) were included. Among all radiomic signatures, HCC with 5 mm tumor border extension and liver showed the optimal predictive performance (training set C-index, 0.696). By incorporating this radiomic signature, rim arterial phase hyperenhancement (APHE), and incomplete tumor “capsule,” a hybrid model demonstrated a validation set C-index of 0.706 and superior 2-year tdAUC (0.743) than both the BCLC (0.550; p < 0.001) and CNLC (0.635; p = 0.032) systems. This model stratified patients into two prognostically distinct risk strata (both datasets p < 0.001).
Conclusion
A preoperative imaging model incorporating the DL automated segmentation-based radiomic signature with rim APHE and incomplete tumor “capsule” accurately predicted early postsurgical recurrence of a single HCC.
Critical relevance statement
The DL automated segmentation-based MRI radiomic model with rim APHE and incomplete tumor “capsule” hold the potential to facilitate individualized risk estimation of postsurgical early recurrence in a single HCC.
Key Points
A hybrid model integrating MRI radiomic signature was constructed for early recurrence prediction of HCC.
The hybrid model demonstrated superior 2-year AUC than the BCLC and CNLC systems.
The model categorized the low-risk HCC group carried longer RFS.
Graphical Abstract
Keywords: Artificial intelligence, Carcinoma (hepatocellular), Recurrence, Magnetic resonance imaging, Machine learning
Introduction
Hepatocellular carcinoma (HCC) is the sixth leading type of cancer and the third most fatal malignancy worldwide [1]. Surgical resection is recommended as the first-line treatment for early-stage HCC [2, 3]. However, even after curative-intent resection, tumor recurrence occurs in ~70% of patients [1, 2], whilst early recurrence within two years accounts for > 70% of recurrence [4, 5]. Tumor burden and aggressive characteristics, such as worse tumor differentiation, microvascular invasion (MVI), and satellite nodules, have been reported to be associated with early recurrence in HCC [4, 6–8]. Nonetheless, histopathological biomarkers had limited implications for clinical decision-making in the pretreatment context. Therefore, noninvasive estimation of early recurrence risk in HCC is crucial for individualized treatment.
Magnetic resonance imaging (MRI) is instrumental in the noninvasive diagnosis and management of HCC. Several semantic MR imaging features, such as rim arterial phase hyperenhancement (APHE), arterial phase peritumoral enhancement, and hepatobiliary phase (HBP) peritumoral hypointensity, have been associated with early recurrence of HCC [8, 9]. However, these semantic features are inadequate for prognostication due to limited predictive performances and suboptimal interobserver reproducibility.
Radiomics has emerged as a new radiological technique that enables the extraction of high-throughput quantitative image features beyond inspections of naked human eyes from standard-of-care medical images, providing important insights into cancer phenotypes and tumor microenvironments that are distinct and complementary to other clinical information [10]. Previous studies have shown good predictive accuracy of MRI radiomic analyses for HCC recurrence after surgery [6, 11–13]. However, these studies generally included limited sample sizes (e.g., 48–361 patients) and utilized manual or semiautomated segmentation, which are time-consuming, labor-intensive, operator-dependent, and subject to inter-rater variability. Fortunately, with recent advances in artificial intelligence (AI) deep learning (DL) algorithms, liver and HCC lesions can be segmented in an automated manner, which may improve both efficiency and reproducibility [14–16]. DL image segmentation models enable the fully automated detection of tumor margins for fast and reproducible HCC segmentation. Accurate segmentation of liver and tumors is a critical prerequisite for subsequent quantitative analysis and holds the huge potential to standardize and improve clinical management. Nevertheless, to our knowledge, there is currently limited evidence on the utility of automated segmentation-based MRI radiomic analyses for predicting postoperative early recurrence in HCC.
Therefore, using the DL-assisted automated segmentation technique, this study aimed to develop and validate a predictive model for early recurrence based on MRI radiomic features and clinical-radiological characteristics in patients with single early-stage HCC following curative resection.
Materials and methods
This single-institution, retrospective study was approved by the institutional review board of West China Hospital of Sichuan University, with a waiver of the informed consent.
Patients
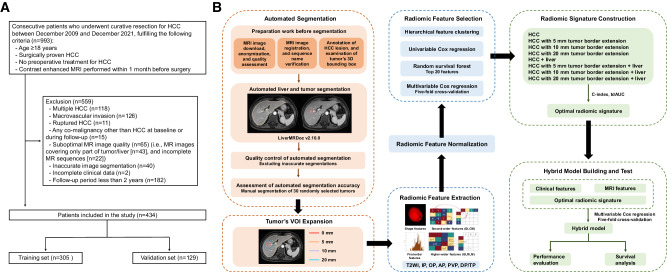
Consecutive patients who received curative resection for HCC between December 2009 and December 2021 were retrospectively recruited. The inclusion criteria were: (a) age ≥ 18 years, (b) surgically proven HCC, (c) no preoperative treatment for HCC, and (d) contrast-enhanced MRI performed within 1 month before surgery. The exclusion criteria were: (a) multiple HCC, (b) macrovascular invasion, (c) ruptured HCC, (d) any co-malignancy other than HCC at baseline or during follow-up, (e) suboptimal MR image quality (i.e., MR images covering only part of tumor/liver, and incomplete MR sequences), (f) inaccurate image segmentation (detailed below), (g) incomplete clinical data (detailed below), and (h) follow-up period less than 2 years. Eligible patients were randomly assigned to training and validation sets at a ratio of 7:3 (Fig. 1A).
Fig. 1.
Flowcharts depicting (A) the recruitment of patients and (B) the workflow of radiomics. AP, arterial phase; C-index, concordance index; DP, delayed phase; OP, opposed phase; PVP, portal venous phase; tdAUC, time-dependent area under the receiver operating characteristic curve; TP, transitional phase; T2WI, T2-weighted imaging; VOI, volume of interest; 3D, three dimensional
Baseline clinical, laboratory, and histopathological data were collected from the electronic medical records. Cirrhosis was diagnosed according to the Clinical Practice Guidelines [17]. Intraoperative ultrasound was routinely performed for each patient to detect small occult HCCs and guide the resection strategy.
Of note, 16.8% (73/434) of these patients have been reported in our prior work [18], where the primary focus was on the MRI features associated with HCC recurrence without radiomic model construction.
MRI technique
MRI was performed with various 3.0-T or 1.5-T scanners. The choice of MRI contrast agents, either extracellular or hepatobiliary, was determined by the surgeons or multidisciplinary team. MRI systems and acquisition protocols are detailed in Supplementary Material 1 and Table S1.
MRI evaluation
Two abdominal radiologists (H.Y.J. and H.W., with 8 and 5 years of experience in liver MRI, respectively) independently reviewed all deidentified MR images. They were informed of the HCC diagnosis but were blinded to other clinical, histopathological, and follow-up information. Discrepancies between the two readers were resolved by a senior abdominal radiologist with over 20 years of experience in liver MRI.
On a per-lesion basis, the following features were evaluated: (a) tumor size (cm), (b) enhancement pattern (typical vs atypical, with typical enhancement pattern referring to the presence of non-rim APHE coupled with nonperipheral “washout” [19]), (c) rim APHE, (d) corona enhancement, (e) nonsmooth tumor margin, (f) incomplete tumor “capsule,” (g) delayed central enhancement, (h) enhancing “capsule,” (i) intratumoral necrosis, (j) fat in mass, more than adjacent liver, (k) radiological cirrhosis, (l) diffuse fatty change, (m) diffuse iron overload, (n) splenomegaly, (o) ascites, (p) collateral circulation, (q) gastroesophageal varices, and (r) main portal vein diameter (cm). Definitions of the imaging features haven been described in our prior study [20].
Radiomic analysis
Image acquisition, preprocessing, and automated segmentation
De-identified MR images were uploaded to a commercial visualization and analysis software (LiverMRDoc; version 2.10.0; Shukun Technology Co., Ltd).
Before automated segmentation, one radiologist (H.W.) inspected all MR images in terms of the sequence names, HCC lesions, and corresponding 3D bounding boxes (i.e., the automated lesion detection annotation) on the AI software platform. To ensure accurate localization of tumors, manual adjustment was conducted for 16 patients with inaccurate 3D bounding boxes (e.g., failing to detect HCC lesions or delineate the whole tumors).
Using 3D U-net-based DL algorithms as detailed in Supplementary Material 2 and Fig. S1, automated segmentation of liver and HCC lesions was conducted on each transverse section of T2-weighted imaging (T2WI), IP, opposed phase (OP), arterial phase (AP), portal venous phase (PVP), and delayed phase (DP; for MRI with extracellular contrast agent [ECA]) or translational phase (TP; for MRI with hepatobiliary contrast agent [HCA]) images.
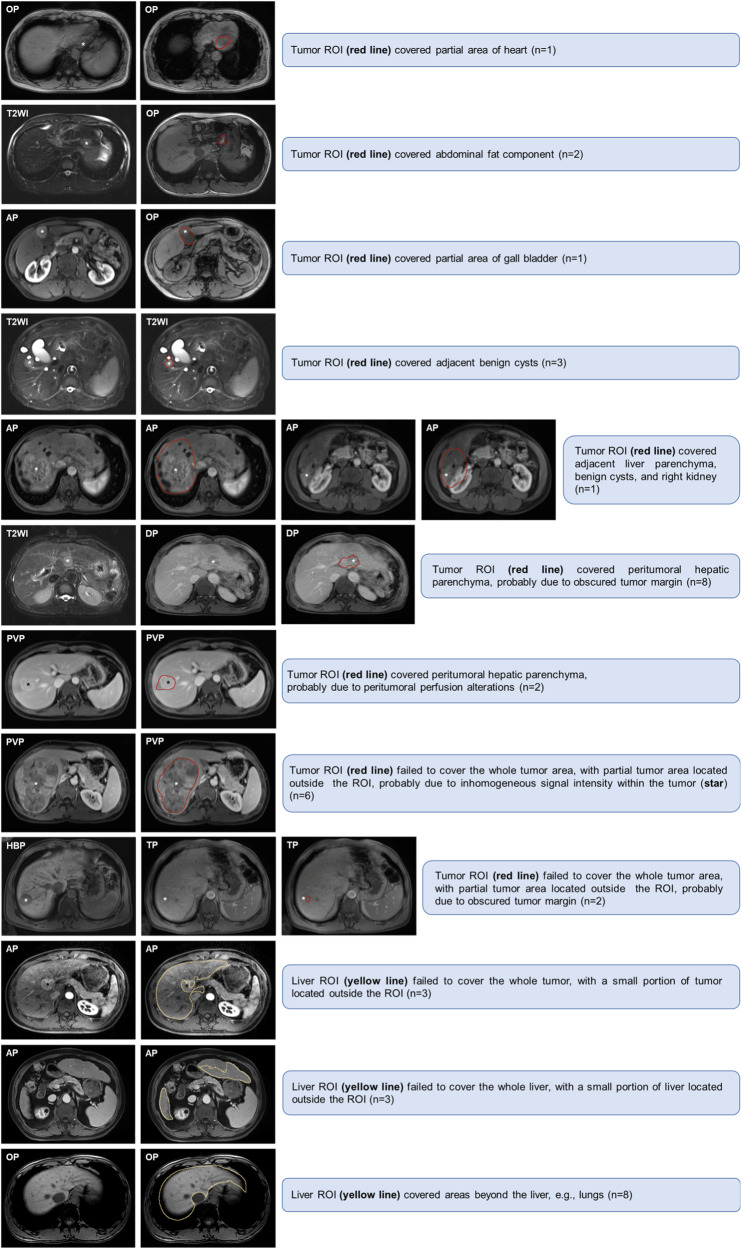
To implement the quality control, one radiologist (H.W.) visually inspected each segmented tumor and liver, and those (n = 40) with inaccurate tumor or liver segmentations on any above sequences were excluded from radiomic analyses. The exclusion criteria for inaccurate segmentation were (a) tumor region of interest (ROI) covered nontumoral areas (e.g., liver parenchyma, benign cysts, adjacent organs or tissues) (n = 18), (b) tumor ROI failed to cover the whole tumor areas (n = 8), (c) liver ROI failed to cover the whole tumor or liver areas (n = 6), and (d) liver ROI covered areas beyond the liver (n = 8). Examples of inaccurate image segmentations are presented in Fig. 2. Manual adjustment was not considered because the study aimed to examine the prognostic utility of this automated technique.
Fig. 2.
Examples of inaccurate image segmentations. AP, arterial phase; DP, delayed phase; OP, opposed phase; PVP, portal venous phase; ROI, region of interest; TP, transitional phase; T2WI, T2-weighted imaging
To assess the accuracy of automated DL segmentation, one radiologist (T.Y.Z., with 5 years of experience in liver MRI) who was unknown to the automated segmentation results manually segmented 30 randomly chosen HCC lesions using ITK-SNAP (version 3.8.0; www.itksnap.org).
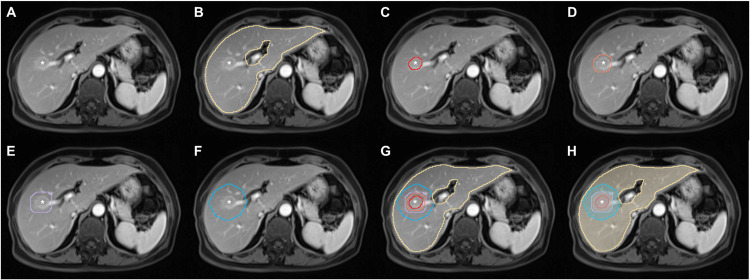
To extract radiomic features of peritumoral areas, the tumor’s 3D mask was expanded radially outwards by 5, 10, and 20 mm on each sequence using a medical research platform (UltraScholar, Version 2.0, Shukun Technology Co., Ltd, https://medresearch.shukun.net/). Accordingly, five types of VOIs were created: (a) tumor VOI, defined as the VOI covering HCC lesion; (b) three extended tumor VOIs, defined as the tumor VOI with automated extension of tumor boundaries by 5, 10, and 20 mm, respectively; and (c) liver VOI, defined as the VOI covering nontumoral liver parenchyma (Fig. 3).
Fig. 3.
An example of automated segmentation. Axial MRI scans in a 71-year-old woman demonstrate a 2.1 cm HCC (*) in segments V and VIII of the liver on (A) portal venous phase image. Automated (B) liver segmentation (yellow lines), (C) tumor segmentation (read line), tumor border extensions with (D) 5 mm (orange line), (E) 10 mm (purple line), and (F) 20 mm (blue line) to create (G, H) corresponding segmentation masks
Radiomic feature extraction
The radiomic workflow is illustrated in Fig. 1B. Detailed methods of radiomic analyses are shown in Supplementary Material 3. MR signal intensity normalization and radiomic feature extraction were performed with the PyRadiomics package (version 3.0.1; https://pyradiomics.readthedocs.io/en/v3.0.1/).
A total of 1688 features were extracted from each VOI in one sequence. Radiomic features were extracted for VOIs of tumor, tumor border extensions (5, 10, and 20 mm), and the liver, respectively.
Radiomic feature normalization and abnormal feature exclusion
Radiomic feature normalization, abnormal feature exclusion, feature selection, and radiomic signature construction were performed with R software (version 4.3.1; The R Foundation for Statistical Computing).
Values of extracted radiomic features on the training set were normalized with z scores; the means and standard deviations derived from the training set were applied to the feature normalization of the validation set. Abnormal features with a variance of 0 were excluded from further analyses.
Feature selection
After normalization and excluding abnormal features, we followed a four-step procedure to reduce dimensions and select robust radiomic features on the training set. First, intervariable collinearity was estimated by Spearman correlation analysis. For radiomic features with a Spearman’s rank correlation coefficient > 0.8, hierarchical feature clustering was performed to remove redundancy. Subsequently, univariable Cox regression analysis was performed to identify significant radiomic features associated with early recurrence. Features with a p < 0.01 were kept for further analyses. Next, random survival forest (RSF) was used to select the top 20 features. Finally, radiomic signatures were constructed by the multivariable Cox regression analysis using a backward elimination approach with five-fold cross-validation.
Radiomic signature development and validation
Eight groups of radiomic signatures were built based on different combinations of radiomic features extracted from tumor, tumor border extensions, and the liver, including (a) HCC, (b) HCC with 5 mm tumor border extension, (c) HCC with 10 mm tumor border extension, (d) HCC with 20 mm tumor border extension, (e) HCC and liver, (f) HCC with 5 mm tumor border extension and liver, (g) HCC with 10 mm tumor border extension and liver, and (h) HCC with 20 mm tumor border extension and liver. The optimal radiomic signature that exhibited the highest performance was selected for building the hybrid model (detailed below).
Patient follow-up
Postoperative follow-up consisted of serum alpha-fetoprotein (AFP) level, liver biochemistry, and contrast-enhanced imaging examinations (i.e., ultrasound, computed tomography, or MRI) performed 1 month after surgery, every 3 months in the first 2 years, and every 6 months thereafter. Patients were followed up until death or the end date of this study (May 1, 2022). Early recurrence was defined as tumor recurrence within 2 years after surgery. Recurrence-free survival (RFS) was defined as the time interval from surgery to the first documented tumor recurrence or death.
Statistical analysis
Continuous variables were compared by Student’s t validation or Mann-Whitney U validation, whereas categorical variables were compared by chi-squared validation or Fisher’s exact validation, as appropriate. Interobserver agreement of MRI findings was measured with Cohen’s κ coefficient for binary features and intraclass correlation coefficient for continuous variables. The consistency between automated (A) and manual (M) segmentations was evaluated by calculating the Dice similarity coefficient (DSC), which was defined as DSC = 2 × ( | A | ∩ | M | )/( | A | ∪ | M | ) [21]. DSC is a widely used statistical metric that measures the proportion of overlapping pixels between two sets of image segmentations [22].
Model development and validation
Using the training set, a hybrid model was built by incorporating the optimal radiomic signature and clinical-radiological variables available before surgery. While controlling for age and sex, univariable Cox regression analysis was performed to identify significant predictors of early recurrence. The multicollinearity of variables was estimated by a variance inflation factor (VIF). For variables with VIF > 2, those with the largest absolute value of β coefficients were selected for further analyses. Variables with p < 0.1 in the univariable analysis following the above steps were entered into the multivariable Cox model; the final model was formulated by a backward elimination approach and the Akaike information criterion (AIC) with five-fold cross-validation.
Model discrimination and calibration were evaluated by the Harrell’s C-index [23] and calibration plot [24], respectively. The time-dependent receiver operating characteristic (tdROC) curve was used to estimate the prognostic accuracy at different time points [25]. The decision curve was plotted to measure the clinical utility of the model [26].
The hybrid model performance was compared with the widely used Barcelona Clinic Liver Cancer (BCLC) staging system [3] and the Chinese National Liver Cancer (CNLC) staging system [27].
Survival analysis
To stratify patients into high and low-risk groups for early recurrence, the optimal threshold for the proposed hybrid model was determined by X-tile software (version 3.6.1; Yale University School of Medicine). RFS was estimated by the Kaplan-Meier method and compared with the log-rank validation. Subgroup analyses were performed according to histological differentiation and MVI statuses, which were known pathological risk factors related to early recurrence of HCC [4, 7].
Statistical analyses were performed with R software (version 4.3.1; The R Foundation for Statistical Computing) or SPSS software (version 26.0; SPSS Inc.). Two-tailed p < 0.05 was considered statistically significant.
Results
Patient characteristics
A total of 434 patients (median age, 52.0 years; interquartile range [IQR], 45.0–60.0 years; 376 men) were included, with 305 (70.3%) and 129 (29.7%) patients on the training and validation sets, respectively. During a median follow-up period of 55.3 months (IQR, 39.0–79.8 months), early recurrence occurred in 32.7% (142/434) of patients.
The validation set had a higher proportion of patients with CNLC Ib stage (31.8% vs. 22.0%; p = 0.031) and more frequent diffuse iron overload (27.9% vs. 19.0%; p = 0.040) compared to the training set, whereas no differences were found in other clinical-radiological-pathological characteristics and follow-up data between the two datasets (p range, 0.223–0.963).
Patient characteristics are summarized in Table 1. MRI features and interobserver agreement are shown in Table 2.
Table 1.
Patient characteristics
| Characteristic | Whole cohort | Training set | Validation set | p value |
|---|---|---|---|---|
| (n = 434) | (n = 305) | (n = 129) | ||
| Age (y)a | 52.0 (45.0–60.0) | 51.0 (45.0–60.0) | 53.0 (45.0–60.0) | 0.736 |
| Sex | 0.587 | |||
| Women | 58 (13.4) | 39 (12.8) | 19 (14.7) | |
| Men | 376 (86.6) | 266 (87.2) | 110 (85.3) | |
| Cause of liver disease | 0.303 | |||
| HBV | 414 (95.4) | 293 (96.1) | 121 (93.8) | |
| Non-HBV | 20 (4.6) | 12 (3.9) | 8 (6.2) | |
| Cirrhosis | 0.430 | |||
| Absent | 201 (46.3) | 145 (47.5) | 56 (43.4) | |
| Present | 233 (53.7) | 160 (52.5) | 73 (56.6) | |
| Child-Pugh class | 0.586 | |||
| A | 430 (99.1) | 303 (99.3) | 127 (98.4) | |
| B | 4 (0.9) | 2 (0.7) | 2 (1.6) | |
| ALBI grade | 0.866 | |||
| 1 | 352 (81.1) | 248 (81.3) | 104 (80.6) | |
| 2 | 82 (18.9) | 57 (18.7) | 25 (19.4) | |
| BCLC stage | 0.327 | |||
| 0 | 83 (19.1) | 62 (20.3) | 21 (16.3) | |
| A | 351 (80.9) | 243 (79.7) | 108 (83.7) | |
| CNLC stage | 0.031 | |||
| Ia | 326 (75.1) | 238 (78.0) | 88 (68.2) | |
| Ib | 108 (24.9) | 67 (22.0) | 41 (31.8) | |
| Contrast agent type of MRI | 0.315 | |||
| ECA | 340 (78.3) | 235 (77.0) | 105 (81.4) | |
| HCA | 94 (21.7) | 70 (23.0) | 24 (18.6) | |
| Laboratory index | ||||
| AST (IU/L)a | 31.5 (25.0–42.0) | 31.0 (25.0–42.0) | 32.0 (26.0–42.0) | 0.352 |
| ALT (IU/L)a | 35.0 (24.0–49.8) | 34.0 (24.0–47.0) | 36.0 (24.0–54.0) | 0.310 |
| TBIL (umol/L)a | 13.6 (9.7–17.5) | 13.6 (9.6–17.4) | 13.4 (9.9–18.2) | 0.630 |
| ALB (g/L)b | 43.0 ± 4.3 | 43.0 ± 4.1 | 43.1 ± 4.6 | 0.860 |
| PLT (×10^9/L)a | 125.5 (89.0–166.0) | 124.0 (87.0–167.0) | 135.0 (90.0–166.0) | 0.496 |
| PT (S)a | 11.9 (11.3–12.6) | 11.9 (11.3–12.6) | 11.9 (11.4–12.6) | 0.829 |
| INRa | 1.0 (1.0–1.1) | 1.0 (1.0–1.1) | 1.0 (1.0–1.1) | 0.432 |
| GGT (IU/L)a | 45.0 (28.0–77.0) | 44.0 (28.0–71.0) | 46.0 (28.0–87.0) | 0.223 |
| AFP (ng/mL) | 0.137 | |||
| ≤ 400 | 333 (76.7) | 240 (78.7) | 93 (72.1) | |
| > 400 | 101 (23.3) | 65 (21.3) | 36 (27.9) | |
| Histopathological characteristics | ||||
| Tumor differentiationc | 0.805 | |||
| Well or Moderate | 291 (67.5) | 205 (67.9) | 86 (66.7) | |
| Poor | 140 (32.5) | 97 (32.1) | 43 (33.3) | |
| MVIc | 0.371 | |||
| Absent | 115 (53.0) | 77 (51.0) | 38 (57.6) | |
| Present | 102 (47.0) | 74 (49.0) | 28 (42.4) | |
| Follow-up perioda | 55.3 (39.0, 79.8) | 54.6 (39.0, 77.4) | 57.8 (39.0, 80.8) | 0.764 |
| Early recurrence rate, % | 142 (32.7) | 100 (32.8) | 42 (32.6) | 0.963 |
| RFS Rate at 24-month, %d | 67.3 (62.8, 71.8) | 67.2 (61.9, 72.5) | 67.4 (59.4, 75.4) | 0.945 |
Statistically significant p values are bold
Unless indicated otherwise, data are the number of patients, with percentages in parentheses
a Data are medians, with IRs in parentheses
b Data are means ± standard deviations
c There were 3 missing data for tumor differentiation and 217 missing data for MVI in the whole cohort
d Numbers in parentheses are 95% confidence intervals (CI)
ALB albumin, ALBI albumin-bilirubin, ALT alanine aminotransferase, AST aspartate aminotransferase, ECA extracellular contrast agent, GGT gamma-glutamyl transferase, HCA hepatobiliary contrast agent, HBV hepatitis B virus, INR international normalized ratio, PLT platelet, PT prothrombin time, TBIL total bilirubin
Table 2.
MRI characteristics and interobserver agreement
| MRI characteristic | Whole cohort | Training set | Validation set | p value | Interobserver agreementa |
|---|---|---|---|---|---|
| (n = 434) | (n = 305) | (n = 129) | |||
| Tumor size (cm)b | 3.4 (2.3–5.0) | 3.3 (2.3–4.8) | 3.6 (2.3–5.9) | 0.281 | 0.985 (0.982, 0.988) |
| Enhancement pattern | 0.571 | 0.526 (0.438, 0.613) | |||
| Typical | 308 (71.0) | 214 (70.2) | 94 (72.9) | ||
| Atypical | 126 (29.0) | 91 (29.8) | 35 (27.1) | ||
| Rim APHE | 0.574 | 0.642 (0.497, 0.787) | |||
| Absent | 408 (94.0) | 288 (94.4) | 120 (93.0) | ||
| Present | 26 (6.0) | 17 (5.6) | 9 (7.0) | ||
| Corona enhancement | 0.603 | 0.438 (0.351, 0.524) | |||
| Absent | 261 (60.1) | 181 (59.3) | 80 (62.0) | ||
| Present | 173 (39.9) | 124 (40.7) | 49 (38.0) | ||
| Nonsmooth tumor margin | 0.915 | 0.496 (0.411, 0.581) | |||
| Absent | 133 (30.6) | 93 (30.5) | 40 (31.0) | ||
| Present | 301 (69.4) | 212 (69.5) | 89 (69.0) | ||
| Incomplete tumor “capsule” | 0.703 | 0.318 (0.22, 0.415) | |||
| Absent | 129 (29.7) | 89 (29.2) | 40 (31.0) | ||
| Present | 305 (70.3) | 216 (70.8) | 89 (69.0) | ||
| Delayed central enhancement | 0.897 | 0.288 (0.055, 0.522) | |||
| Absent | 425 (97.9) | 298 (97.7) | 127 (98.4) | ||
| Present | 9 (2.1) | 7 (2.3) | 2 (1.6) | ||
| Enhancing “capsule” | 0.493 | 0.346 (0.222, 0.47) | |||
| Absent | 40 (9.2) | 30 (9.8) | 10 (7.8) | ||
| Present | 394 (90.8) | 275 (90.2) | 119 (92.2) | ||
| Intratumoral necrosis | 0.232 | 0.705 (0.635, 0.776) | |||
| Absent | 284 (65.4) | 205 (67.2) | 79 (61.2) | ||
| Present | 150 (34.6) | 100 (32.8) | 50 (38.8) | ||
| Fat in mass, more than adjacent liver | 0.284 | 0.416 (0.329, 0.502) | |||
| Absent | 269 (62.0) | 194 (63.6) | 75 (58.1) | ||
| Present | 165 (38.0) | 111 (36.4) | 54 (41.9) | ||
| Radiological cirrhosis | 0.477 | 0.628 (0.552, 0.703) | |||
| Absent | 159 (36.6) | 115 (37.7) | 44 (34.1) | ||
| Present | 275 (63.4) | 190 (62.3) | 85 (65.9) | ||
| Diffuse fatty change | 0.749 | 0.596 (0.455, 0.737) | |||
| Absent | 401 (92.4) | 281 (92.1) | 120 (93.0) | ||
| Present | 33 (7.6) | 24 (7.9) | 9 (7.0) | ||
| Diffuse iron overload | 0.040 | 0.475 (0.37, 0.581) | |||
| Absent | 340 (78.3) | 247 (81.0) | 93 (72.1) | ||
| Present | 94 (21.7) | 58 (19.0) | 36 (27.9) | ||
| Splenomegaly | 0.718 | 0.61 (0.537, 0.683) | |||
| Absent | 223 (51.4) | 155 (50.8) | 68 (52.7) | ||
| Present | 211 (48.6) | 150 (49.2) | 61 (47.3) | ||
| Ascites | 0.303 | 0.401 (0.232, 0.57) | |||
| Absent | 414 (95.4) | 293 (96.1) | 121 (93.8) | ||
| Present | 20 (4.6) | 12 (3.9) | 8 (6.2) | ||
| Collateral circulation | 0.865 | 0.406 (0.329, 0.484) | |||
| Absent | 181 (41.7) | 128 (42.0) | 53 (41.1) | ||
| Present | 253 (58.3) | 177 (58.0) | 76 (58.9) | ||
| Gastroesophageal varices | 0.839 | 0.327 (0.251, 0.403) | |||
| Absent | 195 (44.9) | 138 (45.2) | 57 (44.2) | ||
| Present | 239 (55.1) | 167 (54.8) | 72 (55.8) | ||
| Main portal vein diameter (cm)b | 1.5 (1.3–1.6) | 1.5 (1.3–1.6) | 1.5 (1.3–1.6) | 0.792 | 0.677 (0.457, 0.794) |
| Radiomic signatureb | −0.08 (−0.47–0.40) | −0.11 (−0.44–0.33) | 0.03 (−0.48–0.51) | 0.381 | … |
Statistically significant p values are bold
Unless indicated otherwise, data are the number of patients, with percentages in parentheses
a Data are ICC for continuous variables and Cohen’s κ coefficient for binary variables, with 95% confidence intervals in parentheses. Interobserver agreement was assessed by the ICC or Cohen’s κ coefficient as follows: 0.01–0.20, slight agreement; 0.21–0.40, fair agreement; 0.41–0.60, moderate agreement; 0.61–0.80, substantial agreement; and 0.81–1.00, almost perfect agreement
b Data are medians, with IRs in parentheses
ICC intraclass correlation coefficient
Evaluation of automated segmentation accuracy
DSCs for each sequence are detailed in Table S2. For 30 randomly selected HCCs (median size, 4.8 cm; IQR, 3.5–8.4 cm), the mean DSC between automated and manual tumor segmentations was 0.84 ± 0.13 (median, 0.88; IQR, 0.82–0.92) in all sequences.
Construction of radiomic signatures on the training set
The number of radiomic features in each step of feature selection on the training set is presented in Table S3. Based on the top 20 features determined by RSF, eight radiomic signatures for predicting early recurrence were constructed by multivariable Cox regression analyses (Table S4). Of these, the best performing radiomic signature for early recurrence was HCC with 5 mm tumor border extension and liver, which demonstrated a C-index of 0.696 (95%CI: 0.645, 0.746) on the training set.
There was no evidence of a difference in the C-index of the radiomic signature between MRI with extracellular contrast agent and hepatobiliary contrast agent subgroups on both training (0.673 [95%CI: 0.610, 0. 736] vs 0.743 [95%CI: 0.658, 0.828]; p = 0.210) and test (0.700 [95%CI: 0.598, 0.801] vs 0.692 [95%CI: 0.577, 0.808]; p = 0.934) sets.
Construction and validation of the hybrid model on the training and validation sets
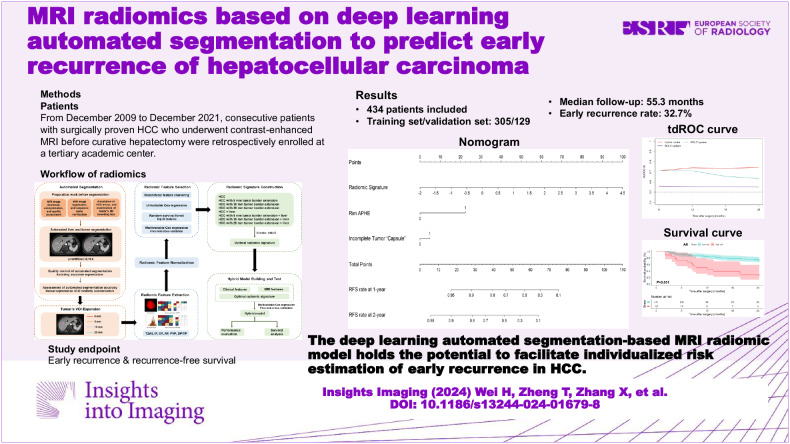
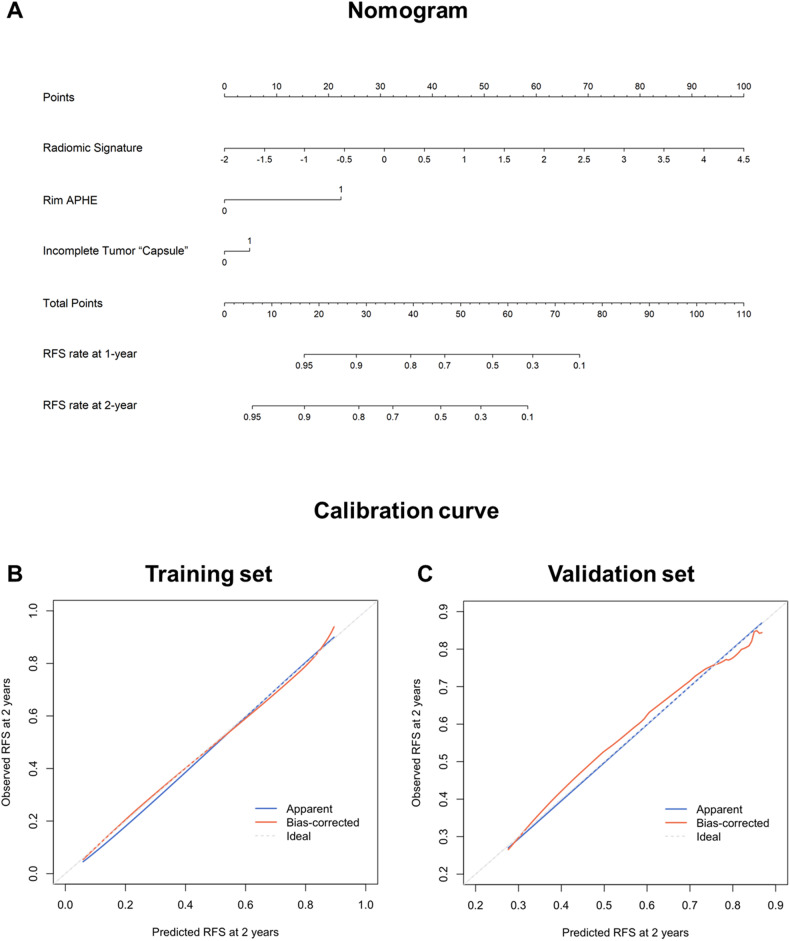
The univariable analysis identified nine variables as potential predictors for early recurrence on the training set (p range, < 0.001–0.061). On subsequent multivariable analysis, rim APHE (hazard ratio [HR] = 4.315; 95%CI: 2.384, 7.810; p < 0.001), radiomic signature (HR = 2.728; 95%CI: 2.178, 3.417; p < 0.001) and incomplete tumor “capsule” (HR = 1.370; 95%CI: 0.831, 2.258; p = 0.217) were included in the Cox model (Table 3). A hybrid model that incorporated the above predictors were constructed for predicting early recurrence and illustrated as a nomogram to provide individualized risk estimates (Fig. 4A).
Table 3.
Predictors for early recurrence based on cox regression analyses on the training set (n = 305)
| Variable | Univariable analysis | Multivariable analysis | ||
|---|---|---|---|---|
| HR | p value | HR | p value | |
| Age (> 50 y) | 0.851 (0.575, 1.262) | 0.423 | … | … |
| Sex (male) | 0.729 (0.420, 1.263) | 0.259 | … | … |
| Cause of liver disease (non-HBV) | 1.065 (0.392, 2.897) | 0.901 | … | … |
| Cirrhosis | 1.072 (0.722, 1.591) | 0.731 | … | … |
| ALBI grade (2) | 0.865 (0.513, 1.459) | 0.587 | … | … |
| AST (> 40 IU/L) | 1.140 (0.740, 1.757) | 0.552 | … | … |
| ALT (> 50 IU/L) | 0.648 (0.383, 1.094) | 0.105 | … | … |
| TBIL (> 19 umol/L) | 0.623 (0.353, 1.098) | 0.102 | … | … |
| ALB (< 40 g/L) | 0.894 (0.551, 1.451) | 0.651 | … | … |
| PLT (< 100 ×109/L) | 0.970 (0.638, 1.474) | 0.885 | … | … |
| PT (> 13 S) | 0.557 (0.270, 1.147) | 0.112 | … | … |
| INR (> 1.1) | 0.687 (0.412, 1.144) | 0.149 | … | … |
| GGT (> 60 IU/L) | 1.570 (1.043, 2.364) | 0.031 | … | … |
| AFP (> 400 ng/mL) | 1.549 (0.987, 2.432) | 0.057 | … | … |
| BCLC stage (A) | 1.550 (0.904, 2.656) | 0.111 | … | … |
| Tumor size (> 5 cm) | 2.227 (1.460, 3.398) | < 0.001 | … | … |
| Enhancement pattern (atypical) | 0.880 (0.566, 1.368) | 0.570 | … | … |
| Rim APHE | 4.670 (2.585, 8.435) | < 0.001 | 4.315 (2.384, 7.810) | < 0.001 |
| Corona enhancement | 1.599 (1.077, 2.373) | 0.020 | … | … |
| Nonsmooth tumor margin | 1.555 (0.981, 2.466) | 0.061 | … | … |
| Incomplete tumor “capsule” | 1.837 (1.123, 3.005) | 0.016 | 1.370 (0.831, 2.258) | 0.217 |
| Delayed central enhancement | 1.489 (0.469, 4,725) | 0.500 | ||
| Enhancing “capsule” | 0.716 (0.389, 1.318) | 0.284 | ||
| Intratumoral necrosis | 1.694 (1.136, 2.527) | 0.010 | ||
| Fat in mass, more than adjacent liver | 1.106 (0.738, 1.657) | 0.626 | ||
| Radiological cirrhosis | 0.914 (0.611, 1.367) | 0.661 | … | … |
| Diffuse fatty change | 0.751 (0.327, 1.723) | 0.498 | … | … |
| Diffuse iron overload | 1.011 (0.612, 1.669) | 0.966 | … | … |
| Splenomegaly | 0.932 (0.629, 1.381) | 0.725 | … | … |
| Ascites | 1.487 (0.594, 3.720) | 0.397 | … | … |
| Collateral circulation | 1.044 (0.700, 1.556) | 0.833 | … | … |
| Gastroesophageal varices | 1.183 (0.793, 1.765) | 0.411 | … | … |
| Main portal vein diameter (> 1.3 cm) | 1.491 (0.915, 2.429) | 0.109 | … | … |
| Radiomic signature | 2.773 (2.225, 3.456) | < 0.001 | 2.728 (2.178, 3.417) | < 0.001 |
Statistically significant p values are bold
Fig. 4.
A The hybrid model-based nomogram to predict early recurrence of single HCC after surgical resection. Calibration curves of the hybrid model on the (B) training and (C) validation sets, respectively
The hybrid model exhibited a C-index of 0.727 (95%CI: 0.676, 0.777) on the training set and 0.706 (95%CI: 0.630, 0.783) on the validation set. There was no evidence of a difference in the C-index of the hybrid model between MRI with extracellular contrast agent and hepatobiliary contrast agent subgroups on both training (0.699 [95%CI: 0.635, 0.764] vs 0.788 [95%CI: 0.707, 0.869]; p = 0.101) and validation (0.720 [95%CI: 0.628, 0.813] vs 0.688 [95%CI: 0.569, 0.807]; p = 0.709) sets. Calibration curves showed good agreement between the predicted survival by the hybrid model and the observed outcomes on both datasets (Fig. 4B and C).
Comparisons between the hybrid model and staging systems on the validation set
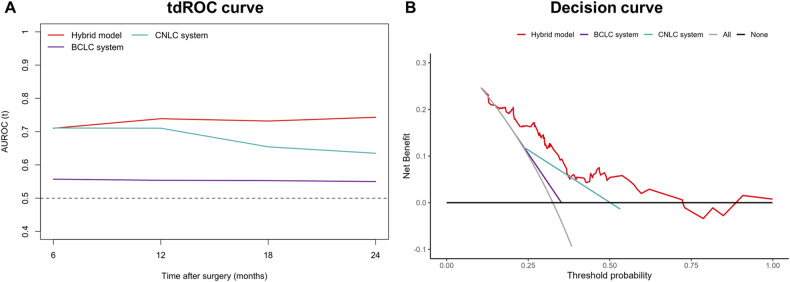
On the validation set, the C-index of the hybrid model (0.706; 95%CI: 0.630, 0.783) was higher than the BCLC system (0.543; 95%CI: 0.494, 0.592; p < 0.001) but showed no evidence of a difference from the CNLC system (0.630; 95%CI: 0.557, 0.703; p = 0.061) (Table 4). In addition, the hybrid model (0.710–0.743) demonstrated superior tdAUC to the BCLC system (0.550–0.557; p range, 0.005−< 0.001) for predicting early recurrence at 6-, 12-, 18-, and 24-month on the validation set, whereas the model (0.743) yielded a higher tdAUC at 24-month than the CNLC system (0.635; p = 0.032) but showed no evidence of a difference at other time points (p range, 0.166–0.992) (Table 4; Fig. 5A). Decision curves revealed that the hybrid model provided a larger net benefit than two staging systems on the validation set (Fig. 5B).
Table 4.
Comparison of performance between the hybrid model and staging systems on the validation Set (n = 129)
| Performance | Hybrid model | BCLC system | p value | CNLC system | p value |
|---|---|---|---|---|---|
| C-index | 0.706 (0.630, 0.783) | 0.543 (0.494, 0.592) | < 0.001 | 0.630 (0.557, 0.703) | 0.061 |
| 6-month tdAUC | 0.710 (0.579, 0.842) | 0.557 (0.488, 0.627) | 0.005 | 0.711 (0.590, 0.832) | 0.992 |
| 12-month tdAUC | 0.739 (0.638, 0.840) | 0.554 (0.490, 0.618) | < 0.001 | 0.710 (0.610, 0.811) | 0.635 |
| 18-month tdAUC | 0.732 (0.634, 0.830) | 0.553 (0.492, 0.614) | < 0.001 | 0.654 (0.561, 0.748) | 0.166 |
| 24-month tdAUC | 0.743 (0.653, 0.834) | 0.550 (0.489, 0.611) | < 0.001 | 0.635 (0.547, 0.723) | 0.032 |
Statistically significant p values are bold
Data in parentheses are 95% confidence intervals
Fig. 5.
Performance of the hybrid model and staging systems on the validation set. A tdROC curves at various time points of the hybrid model and staging systems on the validation set. B Decision curves of the hybrid model and staging systems on the validation set. AUROC, areas under the receiver operating characteristic
Early recurrence risk stratification on the training and validation sets
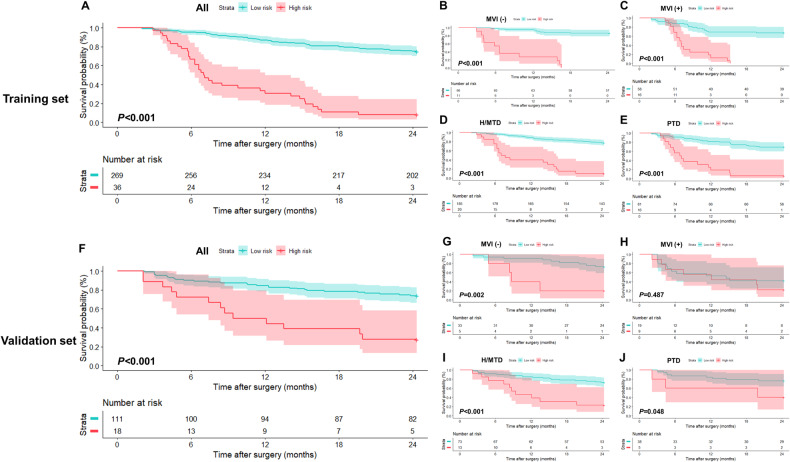
Using 1.25 as the cutoff score derived by the X-tile analysis, the hybrid model stratified all patients on the training set into two risk strata: low risk (< 1.25; n = 269; RFS rate at 24-month, 75.1%), and high risk (≥ 1.25; n = 36; RFS rate at 24-month, 8.3%) (p < 0.001) (Table 5; Fig. 6A). In each subgroup, including absence and presence of MVI, and high or moderate tumor differentiation and poor tumor differentiation, low-risk patients had significantly longer RFS than high-risk patients on the training set (p < 0.001 for all) (Table 5; Fig. 6B–E).
Table 5.
RFS rates and HRs according to each risk group defined by the hybrid model in all patients and two pathological subgroups
| Dataset and risk group | No. of patient | Event | RFS Rate at 12-month, % | RFS rate at 24-month, % | HR | p value |
|---|---|---|---|---|---|---|
| Training set | ||||||
| All | < 0.001 | |||||
| Low risk | 269 | 67 | 87.0 (83.1, 91.1) | 75.1 (70.1, 80.4) | Reference | |
| High risk | 36 | 33 | 30.6 (18.7, 50.0) | 8.3 (2.8, 24.6) | 8.588 (5.561, 13.263) | |
| MVI absent | < 0.001 | |||||
| Low risk | 66 | 9 | 95.5 (90.6, 100.0) | 86.4 (78.5, 95.1) | Reference | |
| High risk | 11 | 11 | 27.3 (10.4, 71.6) | 0.0 (NA, NA) | 19.797 (7.706, 50.860) | |
| MVI present | < 0.001 | |||||
| Low risk | 58 | 19 | 69.0 (58.0, 82.0) | 67.2 (56.2, 80.5) | Reference | |
| High risk | 16 | 16 | 12.5 (3.4, 45.7) | 0.0 (NA, NA) | 6.105 (3.028, 12.307) | |
| High or moderate tumor differentiation | < 0.001 | |||||
| Low risk | 185 | 42 | 89.2 (84.8, 93.8) | 77.3 (71.5, 83.6) | Reference | |
| High risk | 20 | 18 | 40.0 (23.4, 68.4) | 10.0 (2.7, 37.2) | 8.901 (5.029, 15.754) | |
| Poor tumor differentiation | < 0.001 | |||||
| Low risk | 81 | 25 | 81.5 (73.4, 90.4) | 69.1 (59.8, 80.0) | Reference | |
| High risk | 16 | 15 | 18.8 (6.8, 52.0) | 6.3 (0.9, 41.7) | 7.100 (3.592, 14.037) | |
| Validation set | ||||||
| All | < 0.001 | |||||
| Low risk | 111 | 29 | 84.7 (78.2, 91.7) | 73.9 (66.1, 82.5) | Reference | |
| High risk | 18 | 13 | 50.0 (31.5, 79.4) | 27.8 (13.2, 58.5) | 4.148 (2.143, 8.031) | |
| MVI absent | 0.002 | |||||
| Low risk | 33 | 9 | 90.9 (81.6, 100.0) | 72.7 (59.0, 89.6) | Reference | |
| High risk | 5 | 4 | 40.0 (13.7, 100.0) | 20.0 (3.5, 100.0) | 5.836 (1.707, 19.954) | |
| MVI present | 0.487 | |||||
| Low risk | 19 | 11 | 52.6 (34.4, 80.6) | 42.1 (24.9, 71.3) | Reference | |
| High risk | 9 | 7 | 55.6 (31.0, 99.7) | 22.2 (6.5, 75.4) | 1.395 (0.540, 3.602) | |
| High or moderate tumor differentiation | < 0.001 | |||||
| Low risk | 73 | 20 | 84.9 (77.1, 93.5) | 72.6 (63.1, 83.6) | Reference | |
| High risk | 13 | 10 | 46.2 (25.7, 83.0) | 23.1 (8.6, 62.3) | 4.517 (2.086, 9.781) | |
| Poor tumor differentiation | 0.048 | |||||
| Low risk | 38 | 9 | 84.2 (73.4, 96.6) | 76.3 (63.9, 91.1) | Reference | |
| High risk | 5 | 3 | 60.0 (29.3, 100.0) | 40.0 (13.7, 100.0) | 3.467 (0.936, 12.846) | |
Statistically significant p values are bold
Data in parentheses are 95%CIs
NA not available
Fig. 6.
Kaplan-Meier curves demonstrating differences in RFS between low (< 1.25) and high (≥ 1.25) risk strata defined by the hybrid model in all patients on the (A) training and (F) validation sets, respectively. Similar results were observed in the (B) MVI absent, (C) MVI present, (D) H/MTD, and (E) PTD subgroups on training set. Except for the (H) MVI present subgroup, two prognostically distinct risk strata were also obtained in (G, I, J) other subgroups on the validation set. H/MTD, high/moderate tumor differentiation; PTD poor tumor differentiation
Based on the above threshold, two risk strata with significantly different RFS were also obtained for all patients on the validation set (RFS rate at 24-month: 73.9% vs. 27.8%; p < 0.001) (Table 5; Fig. 6F). Additionally, low-risk patients showed significantly longer RFS compared with high-risk patients in subgroups of MVI absence (p = 0.002) (Table 5; Fig. 6G), high or moderate tumor differentiation (p < 0.001) (Table 5; Fig. 6I), and poor tumor differentiation (p = 0.048) (Table 5; Fig. 6J) on the validation set. However, there was no evidence of a difference in RFS between low- and high-risk patients in the MVI present subgroup (p = 0.487) on the validation set (Table 5; Fig. 6H).
Representative images of a patient with HCC at high risk of early recurrence determined by the hybrid model are shown in Fig. S2.
Discussion
In this study, using DL-assisted automated segmentation, we developed and validated a hybrid model, which integrated MRI radiomic signature and two imaging features (rim APHE and incomplete tumor “capsule”), to predict early recurrence for patients with single HCC following curative resection. The model demonstrated superior prognostic accuracy (validation set 2-year tdAUC, 0.743) and better clinical utility compared to the BCLC (0.550; p < 0.001) and CNLC (0.635; p = 0.032) systems. Moreover, patients in the model-predicted high-risk group exhibited worse RFS than those in the low-risk group.
Accurate and reproducible segmentation is the cornerstone of radiomic analyses. However, most existing radiomics-based assessment of HCC recurrence utilized manual or semiautomated segmentation, reducing both efficiency and reproducibility. In our study, the consistency between the automated and manual tumor segmentations was satisfactory, with the mean DSC of 0.85 ± 0.12. Moreover, in a manual segmentation-based study comprising 167 single HCC, Kim et al reported that a combined clinicopathologic-radiomic model with the 3-mm border extension achieved the highest performance (C-index: 0.716) for predicting early postsurgical recurrence [6]. In our study, the automated segmentation-based hybrid model achieved similar validation set discrimination (C-index: 0.706). These initial findings offered a promising prospect for using DL-assisted automated segmentation, which might be more effective and reproducible, to standardize the development of radiomic models, thereby facilitating their translation into clinical practice.
Recently, a few studies have shown the potential utility of automated segmentation-based CT or MRI radiomics in predicting postsurgical recurrence of HCC [28, 29]. For instance, Wang et al employed a DL model to automatically segment tumors on arterial phase images in the external cohort (n = 31) and reported that an MRI-based radiomic-clinical model achieved good accuracy for predicting postsurgical recurrence [29]. To the best of our knowledge, our study is the first study in the literature to use DL-assisted automated segmentations of both the liver and tumor for MRI radiomics-based evaluation of HCC early recurrence. In comparison to previous studies, our research included a larger number of patients (n = 434), thereby enhancing the reliability and robustness of the results presented. In addition, to determine the optimal radiomic signature, we comprehensively investigated the prognostic impact of tumor, tumor border extensions, and the liver. Therefore, our final radiomic signature may convey more abundant information to achieve accurate prognostication.
The biological rationales underlying the association between the hybrid model and HCC early recurrence are not fully understood. In our study, the radiomic signature of HCC with 5 mm tumor border extension and liver exhibited the highest performance for predicting early recurrence of HCC. We speculated that this model may comprehensively capture the whole spectrums of aggressive tumor features, peritumoral microenvironments, and liver morphological and functional characteristics. Rim APHE has been shown to be correlated with tumor aggressiveness, e.g., proliferative subtype, MVI, hypoxic and fibrotic tumor microenvironments, and increased stemness of HCC [30–32]. An incomplete tumor “capsule”, indicative of infiltrative tumor growth, has been identified as an imaging marker for predicting MVI and high BRAF and RAF1 expression in HCC, which can promote tumor invasion and metastasis [33–35].
Remarkably, a small subset of our cases (n = 40) encountered challenges during automated segmentation and were excluded from radiomic analyses. These inaccurate segmentations could arise from various factors, such as the inhomogeneous signal intensity within the tumor, the obscured tumor margin, and additional signal interferences introduced by peritumoral liver parenchyma, adjacent benign cysts, or organs (e.g., heart, gall bladder, and right kidney). Hence, further refinement of the DL algorithm is warranted to improve the automated segmentation performance. If its accuracy and generalizability can be further validated and improved on large-scale multicenter populations, this advanced technique would probably become an applicable workflow in routine clinical practice.
Our study had several limitations. First, the retrospective design may have led to unavoidable selection bias. Second, the hybrid model was not validated in an independent external cohort, which is crucial to check the generalizability of the model. Thus, future multicenter studies are needed to test the applicability of our model in different populations. Third, the interobserver agreement for incomplete tumor “capsule” was only fair (Cohen’s κ coefficient, 0.316), which may reduce the reproducibility of the model. However, as an intrinsic limitation of subjective manual assessment, future studies are warrant to identify approaches for reducing interreader variability, such as utilizing more standardized image criteria or development of computer-aided feature interpretation. Fourth, the vast majority of included patients (95.4%) had HBV infection; hence, our results may not pertain to patients with other etiologies (e.g., hepatitis C virus infection and alcohol abuse). Fifth, we did not construct a postoperative model for predicting HCC early recurrence due to a large number of missing data for key pathological features (e.g., MVI and tumor differentiation). Nonetheless, the purpose of our study was to establish a noninvasive model for assisting in clinical decision-making before treatment. Finally, we did not investigate the prognostic risk stratification ability of the hybrid model across different pathological subtypes of HCC due to insufficient data. Future radiomic studies are encouraged to look at this issue.
In conclusion, using DL-assisted automated segmentation, we proposed a hybrid model by incorporating MRI-based radiomic signature, rim APHE, and incomplete tumor “capsule” for prediction of HCC early recurrence after resection. The model demonstrated superior predictive performance than two widely used staging systems, holding the potential to facilitate individualized risk estimation of postsurgical early recurrence in a single HCC.
Supplementary information
Acknowledgements
We would like to thank Professor Weixia Chen, from the Department of Radiology, West China Hospital of Sichuan University, for her guidance in MRI evaluation during this research.
Abbreviations
- AI
Artificial intelligence
- APHE
Arterial phase hyperenhancement
- BCLC
Barcelona clinic liver cancer
- C-index
Concordance index
- CNLC
Chinese National Liver Cancer
- DL
Deep learning
- DSCs
Dice similarity coefficients
- HCC
Hepatocellular carcinoma
- HR
Hazard ratio
- IQR
Interquartile range
- MRI
Magnetic resonance imaging
- MVI
Microvascular invasion
- RFS
Recurrence-free survival
- RSF
Random survival forest
- tdAUC
Time-dependent area under the receiver operating characteristic curve
- tdROC
Time-dependent receiver operating characteristic
- VIF
Variance inflation factor
- VOI
Volume of interest
Author contributions
Conceptualization: HW, TYZ, XLZ, HYJ. Methodology: HW, TYZ, XLZ, YAW, YDC, CZ, DFJ, BTW, HG, HYJ. Software: XLZ, YAW, YDC, CZ, DFJ, BTW, HG. Validation: HW, XLZ, YAW, YDC, CZ, DFJ, BTW, HG. Formal analysis: HW, XLZ, YAW, YDC, CZ, DFJ, BTW, HG. Data curation: HW, TYZ, YDC, HYJ. Funding acquisition: HYJ, BS. Project administration: HYJ, BS. Supervision: HYJ, BS. Visualization: HW, XLZ, YAW. Writing-original draft: HW, TYZ. Writing-review & editing: HW, TYZ, XLZ, YAW, YDC, CZ, DFJ, BTW, HG, HYJ, BS. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 82101997, U22A20343), the China Postdoctoral Science Foundation (Grant No. 2023T160448), the 1.3.5 project for disciplines of excellence, West China Hospital, Sichuan University (Grant No. ZYJC21012, ZYGD22004), Hainan Province Clinical Medical Center, and the Post-doctoral Station Development Project of Sanya.
Declarations
Ethics approval and consent to participate
The study was approved by the Biomedical Ethics Review Committee of West China Hospital, Sichuan University (Approval Numbers: 2022-651) with a waived requirement for informed consent (retrospective design).
Consent for publication
Not applicable.
Competing interests
Four authors of this manuscript declare a relationship with the following companies: XZ, CZ, and DJ are employees of Shukun Technology Co., Ltd. HJ is a stock owner of Kanghong Technology Co., Ltd. BS is a Deputy Editor for Insights into Imaging. They were not involved in the selection or review process for this article. The remaining authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article. The Shukun employees developed the deep learning automated segmentation software that was provided to our study work under master research agreement. The Shukun employees had no access to the clinical data and did not participate in image interpretation.
Footnotes
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hong Wei and Tianying Zheng contributed equally to this work.
Contributor Information
Hanyu Jiang, Email: hanyu_jiang@foxmail.com.
Bin Song, Email: songlab_radiology@163.com.
Supplementary information
The online version contains supplementary material available at 10.1186/s13244-024-01679-8.
References
- 1.Singal AG, Llovet JM, Yarchoan M, et al. AASLD Practice Guidance on prevention, diagnosis, and treatment of hepatocellular carcinoma. Hepatology. 2023;78:1922–1965. doi: 10.1097/hep.0000000000000466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Llovet JM, Kelley RK, Villanueva A, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7:6. doi: 10.1038/s41572-020-00240-3. [DOI] [PubMed] [Google Scholar]
- 3.Reig M, Forner A, Rimola J, et al. BCLC strategy for prognosis prediction and treatment recommendation: the 2022 update. J Hepatol. 2022;76:681–693. doi: 10.1016/j.jhep.2021.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imamura H, Matsuyama Y, Tanaka E, et al. Risk factors contributing to early and late phase intrahepatic recurrence of hepatocellular carcinoma after hepatectomy. J Hepatol. 2003;38:200–207. doi: 10.1016/S0168-8278(02)00360-4. [DOI] [PubMed] [Google Scholar]
- 5.Portolani N, Coniglio A, Ghidoni S, et al. Early and late recurrence after liver resection for hepatocellular carcinoma: prognostic and therapeutic implications. Ann Surg. 2006;243:229–235. doi: 10.1097/01.sla.0000197706.21803.a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim S, Shin J, Kim DY, Choi GH, Kim MJ, Choi JY. Radiomics on gadoxetic acid-enhanced magnetic resonance imaging for prediction of postoperative early and late recurrence of single hepatocellular carcinoma. Clin Cancer Res. 2019;25:3847–3855. doi: 10.1158/1078-0432.CCR-18-2861. [DOI] [PubMed] [Google Scholar]
- 7.Shinkawa H, Tanaka S, Kabata D, et al. The prognostic impact of tumor differentiation on recurrence and survival after resection of hepatocellular carcinoma is dependent on tumor size. Liver Cancer. 2021;10:461–472. doi: 10.1159/000517992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.An C, Kim DW, Park YN, Chung YE, Rhee H, Kim MJ. Single hepatocellular carcinoma: preoperative MR imaging to predict early recurrence after curative resection. Radiology. 2015;276:433–443. doi: 10.1148/radiol.15142394. [DOI] [PubMed] [Google Scholar]
- 9.Wei H, Jiang H, Zheng T, et al. LI-RADS category 5 hepatocellular carcinoma: preoperative gadoxetic acid-enhanced MRI for early recurrence risk stratification after curative resection. Eur Radiol. 2021;31:2289–2302. doi: 10.1007/s00330-020-07303-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 11.Zhao Y, Wu J, Zhang Q, et al. Radiomics analysis based on multiparametric MRI for predicting early recurrence in hepatocellular carcinoma after partial hepatectomy. J Magn Reson Imaging. 2021;53:1066–1079. doi: 10.1002/jmri.27424. [DOI] [PubMed] [Google Scholar]
- 12.Hectors SJ, Lewis S, Besa C, et al. MRI radiomics features predict immuno-oncological characteristics of hepatocellular carcinoma. Eur Radiol. 2020;30:3759–3769. doi: 10.1007/s00330-020-06675-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao X, Yang H, Luo X, et al. A cox nomogram for assessing recurrence free survival in hepatocellular carcinoma following surgical resection using dynamic contrast-enhanced MRI radiomics. J Magn Reson Imaging. 2023;58:1930–1941. doi: 10.1002/jmri.28725. [DOI] [PubMed] [Google Scholar]
- 14.Li X, Chen H, Qi X, Dou Q, Fu CW, Heng PA. H-DenseUNet: hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans Med Imaging. 2018;37:2663–2674. doi: 10.1109/TMI.2018.2845918. [DOI] [PubMed] [Google Scholar]
- 15.Raman AG, Jones C, Weiss CR. Machine learning for hepatocellular carcinoma segmentation at MRI: radiology in training. Radiology. 2022;304:509–515. doi: 10.1148/radiol.212386. [DOI] [PubMed] [Google Scholar]
- 16.Zheng R, Wang Q, Lv S, et al. Automatic liver tumor segmentation on dynamic contrast enhanced MRI using 4D information: deep learning model based on 3D convolution and convolutional LSTM. IEEE Trans Med Imaging. 2022;41:2965–2976. doi: 10.1109/TMI.2022.3175461. [DOI] [PubMed] [Google Scholar]
- 17.Yoshiji H, Nagoshi S, Akahane T, et al. Evidence-based clinical practice guidelines for Liver Cirrhosis 2020. J Gastroenterol. 2021;56:593–619. doi: 10.1007/s00535-021-01788-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei H, Jiang H, Qin Y, et al. Comparison of a preoperative MR-based recurrence risk score versus the postoperative score and four clinical staging systems in hepatocellular carcinoma: a retrospective cohort study. Eur Radiol. 2022;32:7578–7589. doi: 10.1007/s00330-022-08811-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei J, Jiang H, Zeng M, et al. Prediction of microvascular invasion in hepatocellular carcinoma via deep learning: a multi-center and prospective validation study. Cancers (Basel) 2021;13:2368. doi: 10.3390/cancers13102368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei H, Fu F, Jiang H, et al. Development and validation of the OSASH score to predict overall survival of hepatocellular carcinoma after surgical resection: a dual-institutional study. Eur Radiol. 2023;33:7631–7645. doi: 10.1007/s00330-023-09725-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carillo V, Cozzarini C, Perna L, et al. Contouring variability of the penile bulb on CT images: quantitative assessment using a generalized concordance index. Int J Radiat Oncol Biol Phys. 2012;84:841–846. doi: 10.1016/j.ijrobp.2011.12.057. [DOI] [PubMed] [Google Scholar]
- 22.Langkilde F, Masaba P, Edenbrandt L et al (2024) Manual prostate MRI segmentation by readers with different experience: a study of the learning progress. Eur Radiol. 10.1007/s00330-023-10515-4 [DOI] [PMC free article] [PubMed]
- 23.Uno H, Cai T, Pencina MJ, D’Agostino RB, Wei LJ. On the C-statistics for evaluating overall adequacy of risk prediction procedures with censored survival data. Stat Med. 2011;30:1105–1117. doi: 10.1002/sim.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balachandran VP, Gonen M, Smith JJ, DeMatteo RP. Nomograms in oncology: more than meets the eye. Lancet Oncol. 2015;16:e173–e180. doi: 10.1016/S1470-2045(14)71116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56:337–344. doi: 10.1111/j.0006-341X.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 26.Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making. 2006;26:565–574. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou J, Sun H, Wang Z, et al. Guidelines for the diagnosis and treatment of hepatocellular carcinoma (2019 edition) Liver Cancer. 2020;9:682–720. doi: 10.1159/000509424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L, Wu M, Zhu C, et al. Ensemble learning based on efficient features combination can predict the outcome of recurrence-free survival in patients with hepatocellular carcinoma within three years after surgery. Front Oncol. 2022;12:1019009. doi: 10.3389/fonc.2022.1019009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Song D, Wang W, et al. Data-driven assisted decision making for surgical procedure of hepatocellular carcinoma resection and prognostic prediction: development and validation of machine learning models. Cancers (Basel) 2023;15:1784. doi: 10.3390/cancers15061784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bao Y, Li JX, Zhou P, et al. Identifying proliferative hepatocellular carcinoma at pretreatment CT: implications for therapeutic outcomes after transarterial chemoembolization. Radiology. 2023;308:e230457. doi: 10.1148/radiol.230457. [DOI] [PubMed] [Google Scholar]
- 31.Hong SB, Choi SH, Kim SY, et al. MRI features for predicting microvascular invasion of hepatocellular carcinoma: a systematic review and meta-analysis. Liver Cancer. 2021;10:94–106. doi: 10.1159/000513704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rhee H, An C, Kim HY, Yoo JE, Park YN, Kim MJ. Hepatocellular carcinoma with irregular rim-like arterial phase hyperenhancement: more aggressive pathologic features. Liver Cancer. 2019;8:24–40. doi: 10.1159/000488540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu DS, Siripongsakun S, Kyong Lee J, et al. Complete tumor encapsulation on magnetic resonance imaging: a potentially useful imaging biomarker for better survival in solitary large hepatocellular carcinoma. Liver Transpl. 2013;19:283–291. doi: 10.1002/lt.23597. [DOI] [PubMed] [Google Scholar]
- 34.Lei Z, Li J, Wu D, et al. Nomogram for preoperative estimation of microvascular invasion risk in hepatitis B virus-related hepatocellular carcinoma within the Milan criteria. JAMA Surg. 2016;151:356–363. doi: 10.1001/jamasurg.2015.4257. [DOI] [PubMed] [Google Scholar]
- 35.Dong Z, Huang K, Liao B, et al. Prediction of sorafenib treatment-related gene expression for hepatocellular carcinoma: preoperative MRI and histopathological correlation. Eur Radiol. 2019;29:2272–2282. doi: 10.1007/s00330-018-5882-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.