Abstract
Extracellular matrix (ECM) pathologic remodeling underlies many disorders, including muscular dystrophy. Tissue decellularization removes cellular components while leaving behind ECM components. We generated “on-slide” decellularized tissue slices from genetically distinct dystrophic mouse models. The ECM of dystrophin- and sarcoglycan-deficient muscles had marked thrombospondin 4 deposition, while dysferlin-deficient muscle had excess decorin. Annexins A2 and A6 were present on all dystrophic decellularized ECMs, but annexin matrix deposition was excessive in dysferlin-deficient muscular dystrophy. Muscle-directed viral expression of annexin A6 resulted in annexin A6 in the ECM. C2C12 myoblasts seeded onto decellularized matrices displayed differential myoblast mobility and fusion. Dystrophin-deficient decellularized matrices inhibited myoblast mobility, while dysferlin-deficient decellularized matrices enhanced myoblast movement and differentiation. Myoblasts treated with recombinant annexin A6 increased mobility and fusion like that seen on dysferlin-deficient decellularized matrix and demonstrated upregulation of ECM and muscle cell differentiation genes. These findings demonstrate specific fibrotic signatures elicit effects on myoblast activity.
Keywords: Extracellular matrix, Duchenne muscular dystrophy, Limb girdle muscular dystrophy, Muscle, Annexin, Myoblast, Dysferlin
Introduction
Muscular dystrophies are defined by continual replacement of muscle fibers by fibrosis. Patterns of fibrosis appear similar across genetically distinct forms of muscular dystrophy. Mutations that disrupt the dystrophin glycoprotein complex (DGC) produce a fragile muscle plasma membrane. Duchenne muscular dystrophy (DMD) is caused by loss-of-function mutations in the dystrophin gene [1,2]. Within the DGC is the sarcoglycan complex, and recessive loss-of-function mutations in genes encoding α-, β-, δ-, or γ-sarcoglycan result in Limb Girdle muscular dystrophy (LGMD) that resembles DMD [3-5]. Excessive muscle contraction and trauma can also induce membrane rupture, requiring a quick and efficient means of repair to prevent cellular necrosis. Dysferlin is a calcium-dependent, phospholipid-binding protein that facilities muscle membrane repair [6-11], and loss-of-function mutations in DYSF, the gene encoding dysferlin, also cause LGMD [7,12]. Dysferlin mutations are associated with remarkably high serum creatine kinase levels, indicative of muscle membrane leak, as well as proximal muscle weakness and inflammatory infiltrate [13]. Patients with DYSF mutations, unlike other forms of muscular dystrophy, can often have completely normal to enhanced muscle function in early life with later onset of muscle weakness [14].
Mouse models recapitulate many of the pathological features seen in the human muscular dystrophies [15-22]. The mdx mouse harbors a single point mutation in exon 23, resulting in loss of full-length dystrophin [19]. Genetic background can shift the pathological and functional manifestations of muscular dystrophy [23,24]. Most mouse models, including mdx mice, are on C57BL substrains, and in this background these models exhibit relatively mild muscle disease. In contrast, the DBA/2 J strain intensifies membrane fragility and fibrosis in Sgcg null mice, which lack the dystrophin-associated protein γ-sarcoglycan [24]. Similarly, in the DBA/2 J strain the mdx mouse displays greater muscle damage, increased inflammation and increased intramuscular fibrosis compared to the C57BL/10 strain [20,25].
The increase in disease severity conferred by the DBA/2 J background has been attributed to genetic modifiers including latent TGF-β binding protein 4 (LTBP4); LTBP4 and the modifier osteopontin (OPN/SPP1) converge on the TGF-β pathway in a feed forward cycle [26]. Anxa6, encoding annexin A6, was also mapped as a genetic modifier of muscular dystrophy [27]. Annexin A6 (ANXA6) is a known membrane repair protein that acts as a molecular band-aid, aiding in membrane resealing, improving repair capacity upon membrane injury [27-31]. Within the muscle extracellular matrix (ECM), the role of ANXA6 is less defined but, as a protein class, annexins have been implicated in cellular migration and differentiation [32-35].
The muscle ECM influences myofiber function as well as muscle repair and regeneration. Proteins which make up the core ECM include collagens, glycoproteins, and proteoglycans. Another group of ECM proteins, termed matricellular proteins include secreted factors, regulators, and other matrix-affiliated proteins, and these proteins mediate cell-matrix interactions referred to as “dynamic reciprocity”, reflecting bi-directional interplay [36-38]. Some matricellular proteins were originally considered to be solely intracellular but were subsequently shown to have extracellular roles. For example, calrecticulin, an endoplasmic reticulum (ER)-localized protein is now recognized to have bioactive roles as a matricellular protein [39]. In healthy tissue, matricellular proteins participate in feedback loops between the matrix and cells to influence cellular behavior and maintain homeostasis. In normal muscle and after muscle injury, ECM remodeling is an essential regulator of growth, repair, and recovery. In muscular dystrophy however, there is ongoing and repetitive injury concomitant with attempted repair [40-42]. This repetitive injury results in inappropriate remodeling of the ECM and accumulation of matricellular proteins.
Analysis of the ECM composition relies on separation of the insoluble-ECM fraction from the soluble-cellular fraction of proteins. To effectively separate these two fractions, tissues need to be decellularized with complete removal of cellular proteins, allowing for enrichment of ECM proteins [43]. In situ decellularization of tissues results in the decellularization of an entire organ with retention of an intact native ECM scaffold [44,45]. One drawback to whole tissue decellularization is the lengthy exposure needed to penetrate less accessible aspects of the tissue, resulting in uneven decellularization or even tissue degeneration. An “on-slide” tissue decellularization method was optimized for skeletal muscle in which tissue slices were treated with 1 % SDS to produce decellularization and retain spatial aspects of the tissue [46].
We now applied this on-slide decellularization to evaluate the ECM composition across genetically distinct mouse models of muscular dystrophy. We identified differential ECM protein deposition between dystrophin-deficient and dysferlin-deficient skeletal muscles. Excess deposition of matricellular ECM proteins was characteristic of dystrophic muscle and was not observed in healthy muscle ECMs. We found that ECM from the more fibrotic DBA/2 J (D2) dystrophic background had the most significant increase in protein expression of decorin, periostin, thrombospondin 4 and matrix metalloproteinase 9. Interestingly, dysferlin-null muscle displayed the highest expression of matrix-associated annexin A2 and annexin A6. dECMs differentially supported C2C12 myoblast motility and differentiation. Myoblasts seeded onto dECMs from the mdx DBA/2 J background inhibited myoblast migration. Conversely, myoblast migration was enhanced on the dysferlin-null dECM compared to wildtype and mdx dystrophic scaffolds. Myoblasts seeded onto the dysferlin-null scaffolds demonstrated increased differentiation potential compared to wildtype scaffolds. Exposing myoblasts to recombinant annexin A6 was sufficient to enhance myoblast migration and movement and also increased myoblast fusion activity. Bulk RNA-sequencing of recombinant annexin A6-treated myoblasts showed upregulation of genes associated with muscle differentiation and extracellular matrix. These data illustrate that differential protein deposition into the ECM mediates cellular crosstalk in the muscular dystrophies and identify annexin A6 as an important differentiator mediating myoblast migration and differentiation.
Results
Generation of acellular myoscaffolds using detergent decellularization.
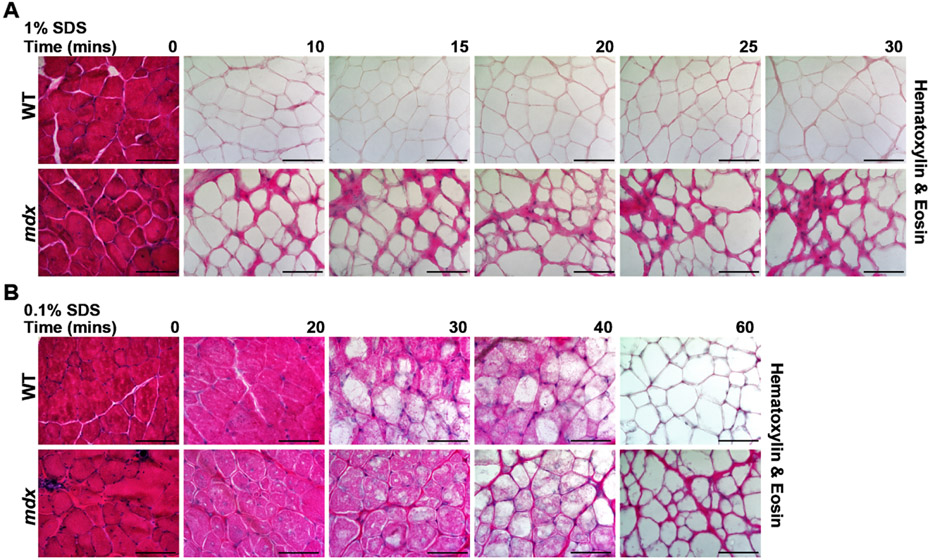
Decellularized scaffolds from heart and muscle have been studied for their ability to support cellular regeneration, but most commonly these methods have been applied to whole organs or tissues (reviewed in [47]). Recently Stearns-Reider et al. published an “on-slide” decellularization method using 30μm tissue sections exposed to 1 % SDS for 40 mins to generate dECM myoscaffolds [46]. We adapted and optimized the generation of dECM scaffolds to ensure reproducible removal of intracellular components and DNA while retaining ECM architecture and function. We evaluated a range of sodium dodecyl sulfate (SDS) exposure by incubating 25μm sections from WT and mdx quadriceps muscles in 1 % SDS for 10–30 min. Removal of the cellular components after decellularization was assessed through hematoxylin and eosin (H&E) staining, with complete decellularization observed by 10 min (Fig. 1A). Decellularization was attempted with less SDS (0.1 %) for 0–60 min (Fig. 1B; Supplemental Fig. 1). However, at the lower concentration, decellularization remained incomplete after 20–40 min. Complete decellularization was seen after a 60-minute incubation with 0.1 % SDS (Fig. 1B), but to have less potential degradation, higher SDS concentration was used at the shorter incubation time.
Fig. 1.
SDS optimization for on-slide decellularization of skeletal muscle. Representative images of Hematoxylin & Eosin (H&E) staining of wild-type (WT) and mdx quadriceps muscles that were decellularized with 1 % SDS or 0.1 % SDS for 10 to 60 min. Non-decellularized sections, treated with PBS only, served as controls (0 min). (A) Exposure of slides to 1 % SDS solution produced complete decellularization of the myoscaffolds after 10 min. (B) At 0.1 % SDS, longer incubation times were needed for decellularization, with loss of visualized cellular components achieved at 60 min. Scale bar, 100μm.
Acellular myoscaffolds retain key ECM proteins and architectural integrity.
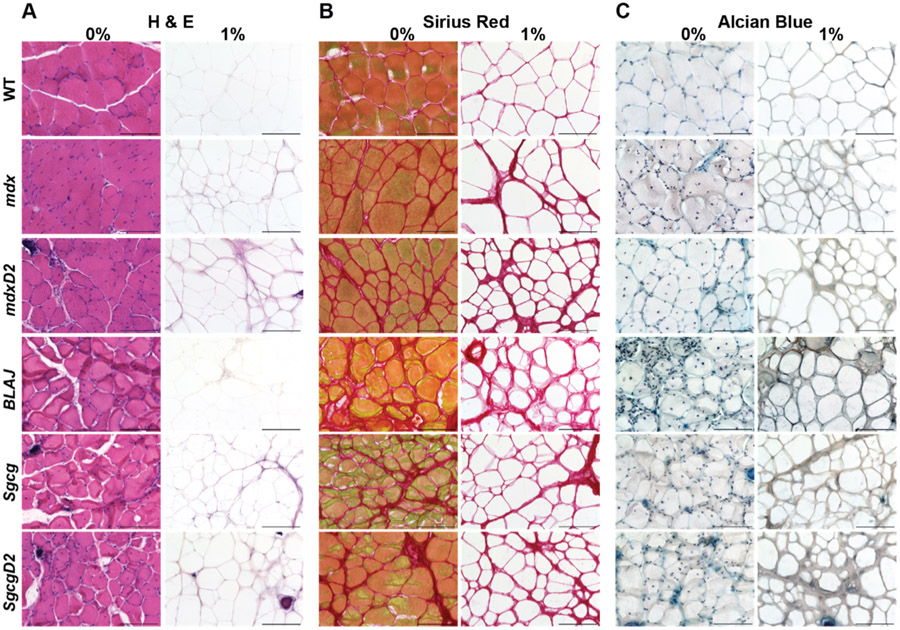
We interrogated the protein composition of dECMs from multiple muscular dystrophy mouse models including mice lacking dystrophin (mdx), sarcoglycan (Sgcg), or dysferlin (BLAJ). We also evaluated the effect of genetic background by including dystrophic models on the DBA/2 J (D2) background (mdxD2 and SgcgD2) since this background induces more intense fibrosis (Supplemental Table 1). The mutations underlying mdx and Sgcg mice disrupt sarcolemma stability, while the BLAJ model is defective in membrane repair. In each dECM model, matrix quality was assessed following decellularization to determine retention of key proteins and architectural integrity [48,49]. Complete removal of the cellular components in the dECMs across all strains was confirmed by H&E staining (Fig. 2A), and Sirius Red staining evaluated collagen deposition and integrity (Fig. 2B). Intact collagen structure was visualized within the dECMs with increased collagen deposition observed in more severe muscular dystrophy models as expected based on disease severity (mdxD2 > mdx > WT). Retention of glycosaminoglycans (GAGs) post decellularization was confirmed with Alcian Blue staining (Fig. 2C).
Fig. 2.
Acellular myoscaffolds retained ECM architectural integrity and glycosaminoglycans after on-slide decellularization. Muscle sections from WT and multiple muscular dystrophy mouse models including mdx, mdxD2, BLAJ, Sgcg, and SgcgD2 were decellularized with a 1 % SDS solution (1 %) for 10 min. Details for each mouse strain are shown in Supplemental Table 1. The non-decellularized control was incubated in PBS (0 %). Representative images of (A) H&E, (B) Sirius Red, and (C) Alcian Blue staining demonstrate removal of visualized cellular components, preservation of the dECM collagen structure and retention of glycosaminoglycans respectively. Scale bar, 100μm.
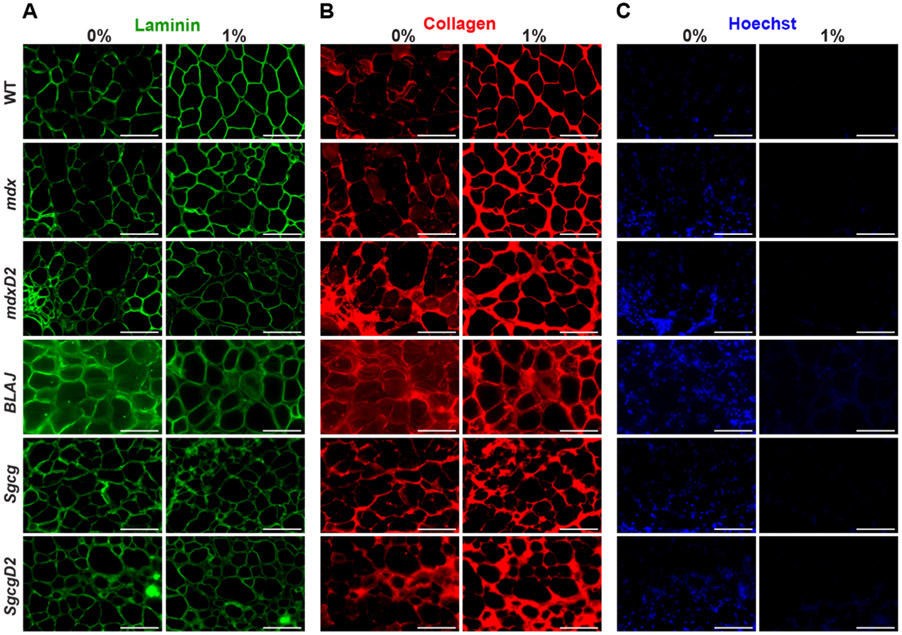
The architectural integrity of the acellular myoscaffolds was further interrogated through immunofluorescence microscopy (IFM) of laminin and collagen pre- and post-decellularization. Antibodies against laminin α2 (LAMA2) and collagen type 1 demonstrated intact laminin and collagen within the dECM scaffolds across all strains (Fig. 3A and B). Interestingly, both ECM proteins were more readily visualized in acellular scaffolds. This pattern is thought to reflect enhanced epitope exposure after cellular removal [46,50,51]. Elimination of nucleic acids with SDS was confirmed using Hoechst staining (Fig. 3C). Additionally, we confirmed the SDS method reduced DNA remnants as expected by comparing DNA content from decellularized and non-decellularized tissue sections. Exposure to 1 % SDS decellularization reduced intact DNA to less than 50 ng dsDNA per mg ECM (Supplemental Fig. 1). Not all proteins were deposited into dystrophic dECMs since there was little signal for membrane-associated proteins like dystrophin and caveolin 3 in dECMs (Supplemental Fig. 2). We selected a 10-minute incubation with 1 % SDS since it yielded intact dECMs devoid of cellular components and nucleic acids while retaining core components of the ECM and maintaining architectural integrity.
Fig. 3.
Acellular myoscaffolds from dystrophic models demonstrated excess collagen deposition in all models. Shown is immunofluorescence microscopy (IFM) of 1 % SDS decellularized myoscaffolds from WT and muscular dystrophy mouse models (mdx, mdxD2, BLAJ, Sgcg, and SgcgD2). PBS was used for a non-decellularized control (0 %). Representative images of (A) laminin-2 (α–2 chain) (B) collagen I, and (C) Hoechst staining demonstrate the intact nature of the ECM protein. All dECMs displayed brighter collagen signal, consistent with detergent-enhanced epitope unmasking. Greater collagen detection in the muscular dystrophy models was evident with broader regions of collagen deposition. Hoechst staining confirms removal of cellular DNA. Scale bar, 100μm.
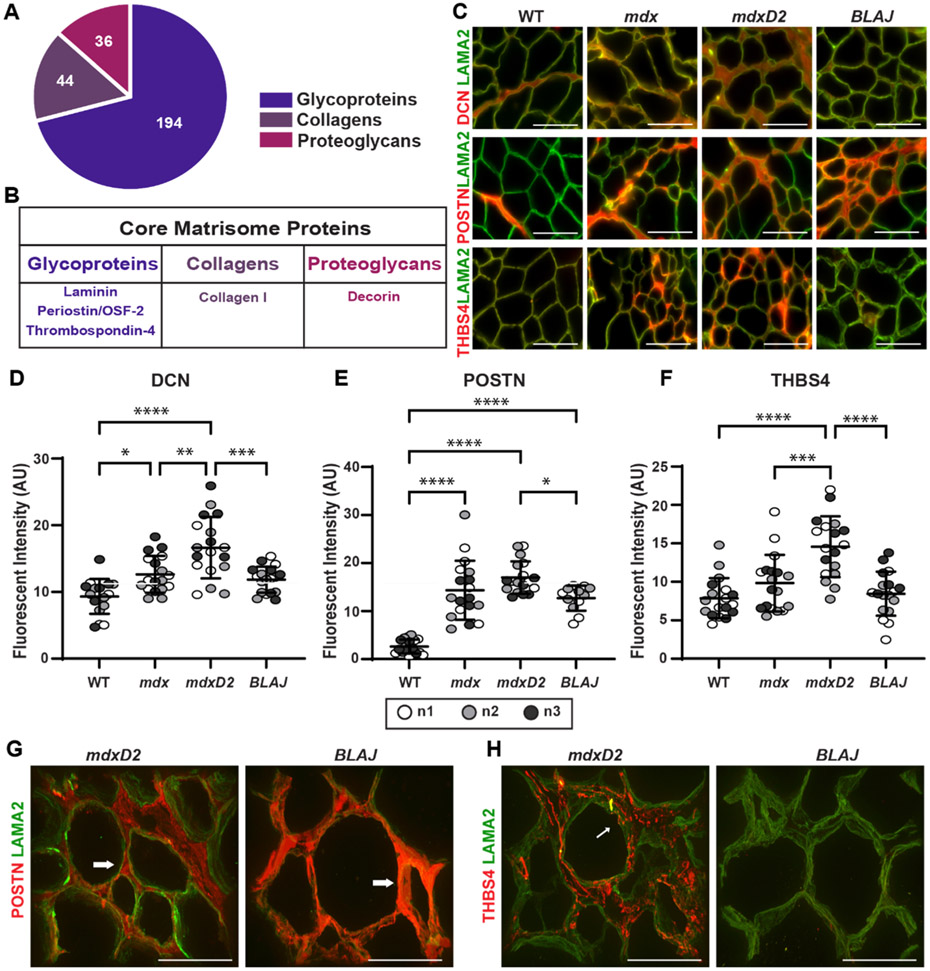
Core matrisome proteins are increased in dystrophin-deficient dECM while only periostin is increased in dysferlin-deficient dECM. ECM components can be separated into “core” and “associated” matrisome proteins [52,53]. Core matrisome proteins fall into three categories: glycoproteins (194), collagens (44), and proteoglycans (36) (Fig. 4A) [52,53]. We evaluated selected core matrisome proteins including decorin (DCN), periostin (POSTN), and thrombospondin-4 (THBS4) since each of these proteins has been implicated in muscular dystrophy pathology [54-59] (Fig. 4B). Antibodies to DCN, POSTN, and THBS4 each displayed increased signal in the dECMs from the more severely fibrotic muscular dystrophy muscles (Fig. 4C). Excess DCN, POSTN, and THBS4 were each observed in the myoscaffolds of mdxD2 compared to mdx and increased in mdx compared to WT (Fig. 4C).
Fig. 4.
Acellular myoscaffolds differentially retained core matrisome proteins decorin, periostin and thrombospondin 4 in different muscular dystrophy models. (A) Core matrisome protein distribution as defined by [60]. (B) Proteins selected for analysis in dECMs. (C) Representative IFM images from WT and mdx, mdxD2, and BLAJ dECMs showing co-staining with antibodies against decorin (DCN), periostin (POSTN), or thrombospondin-4 (THBS4) (red) with laminin-2 (α–2 chain) (LAMA2, green). DCN was observed diffusely throughout dystrophic dECMs from the DBA/2 J background (mdxD2). POSTN was increased in all muscular dystrophy subtypes. THBS4 protein expression, like DCN, was more focally upregulated in specific regions of the dystrophic myoscaffolds and was especially increased in muscular dystrophy models in the DBA/2 J background. Scale bar, 100μm. Quantitation of IFM signal demonstrated a significant increase in mean fluorescent intensity in myoscaffolds (D) DCN (WT 9.7, mdx 13.2, mdxD2 17.3, BLAJ 12.3 AU), (E) POSTN (WT 2.6, mdx 14.3, mdxD2 17.0, BLAJ 12.7 AU), and (F) THBS4 (WT 7.8, mdx 9.8, mdxD2 14.6, BLAJ 8.5 AU). n = 3 independent mice per strain (marked as n1, n2, n3), n = 6 images from 2 scaffolds per mouse. (G,H) 100x Z-stack representative images of POSTN or THBS4 (red) with LAMA2 (green) demonstrate unique protein deposition profiles on the acellular myoscaffolds. POSTN was increased in dysferlin-deficient BLAJ, nearly obliterating the LAMA2 signal (right). In contrast, POSTN signal was interspersed with LAMA2 signal in mdxD2 dECMs (left). In mdxD2 dECM, THBS4 was observed in the matrix in a vesicle-like pattern (thin white arrow). Scale bar, 50μm. Graphs show the mean with SEM bars. * p < 0.05, ** p < 0.01, *** p < 0.005, and **** p < 0.001 by one-way ANOVA.
Quantitative analysis of the fluorescence patterns of DCN, POSTN, and THBS4 protein expression in acellular myoscaffolds from WT, mdx, mdxD2, and BLAJ is shown in Fig. 4D and F. DCN, POSTN, and THBS4 intensity was significantly increased in mdxD2 acellular dECM myoscaffolds compared to mdx and WT matrices. Fluorescence intensity was also increased in mdx compared to WT for DCN and POSTN. A consistent pattern of mdxD2 > mdx > WT was observed (Fig. 4D and F). Acellular myoscaffolds generated from the BLAJ muscle had significantly less matrix protein compared to mdxD2 for all three core matrisome proteins analyzed, with protein levels trending similarly with the mdx scaffolds for POSTN and DCN (Fig. 4D and E). Interestingly, THBS4 levels in BLAJ dECMs were similar to WT THBS4 levels (Fig. 4F). WT myoscaffolds generated from the DBA/2 J strain had similar expression patterns as WTB6 with minimal DCN, POSTN, THBS4 signal, indicating the dystrophic process contributed to pathological matrix remodeling (Supplemental Fig. 3A). Quantification analysis of fluorescence area demonstrated similar results to fluorescent intensity (Supplemental Fig. 3B). Immunoblotting of cellular and urea solubilized muscle proteins isolated from WT, mdx, mdxD2, and BLAJ mice for DCN and POSTN mirrored the IFM quantitation (Supplemental Fig. 4).
Dysferlin-deficient BLAJ dECMs had a distinct pattern of dECM remodeling with only POSTN showing an increased signal compared to WT. This increased POSTN pattern in BLAJ dECMs filled the interstitial space surrounding the laminin α2 (LAMA2) staining, in contrast to the pattern seen in mdxD2 dECMs, where LAMA2 regions were interspersed with POSTN around myofibers, leaving the LAMA2 signal visible (Fig 4G). The lack of increased THBS4 was confirmed in dysferlin-deficient BLAJ dECMs, and additionally THBS4 had a vesicular pattern in dECMs from mdxD2 (Fig. 4H). These data demonstrate that core matrisomal proteins differ across different forms of muscular dystrophy.
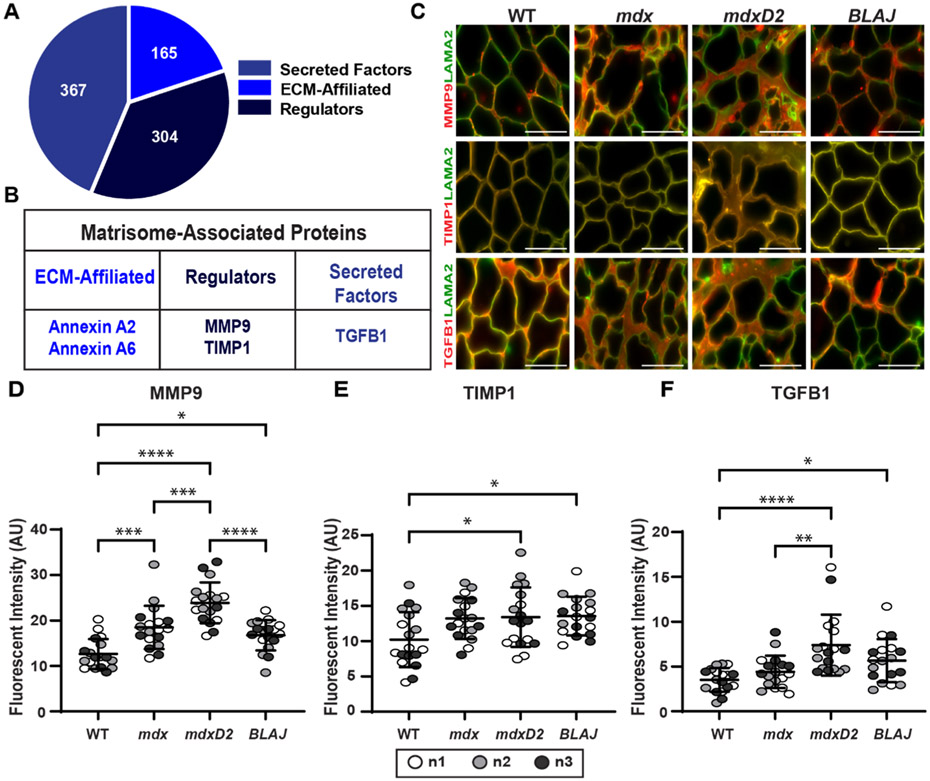
Matrisome-Associated Proteins, MMP9, TIMP-1, and TGF-β1, in acellular myoscaffolds.
The matrisome associated protein category separates into three sub-categories: Secreted Factors (367), ECM-Affiliated (165), and Regulators (304) (Fig. 5A) [52,53]. We evaluated muscular dystrophy relevant proteins from each category including annexin A2 (ANXA2), annexin A6 (ANXA6), matrix metalloproteinase-9 (MMP9), TIMP metallopeptidase inhibitor 1 (TIMP-1), and transforming growth factor beta 1 (TGF-β1) (Fig. 5B). After decellularization, acellular myoscaffolds were probed with antibodies to MMP9, TIMP-1, or TGF-β1 and with anti- LAMA2 (Fig. 5C). TIMP-1 was distributed uniformly throughout the matrix in all genetic models, while MMP9 and TGF-β1 had distinct focal patterns of distribution in dystrophic dECMs. mdxD2 had significantly greater MMP9 and TGF-β1 compared to mdx, and mdx expression levels were greater than WT (Fig. 5D and F). DBA/2 J WT myoscaffolds demonstrated similar expression patterns as WTB6 with minimal ANXA2, ANXA6, MMP2, TIMP1, and TGF-β1 signal present in the myoscaffold (Supplemental Fig. 3A). dECM myoscaffolds from dysferlin-deficient BLAJ muscle tissues had MMP9 and TGF-β1 protein levels similar to or slightly increased compared to mdx, respectively. TIMP-1 expression was increased over WT in a similar fashion across mdx, mdxD2, and BLAJ (Fig. 5D and F). Thus, acellular dECM myoscaffolds retained key core and matrisome-associated proteins and these patterns differed across different muscular dystrophy subtypes.
Fig. 5.
Acellular myoscaffolds retained matrisome-associated proteins MMP9, TIMP-1, and TGFβ–1 and demonstrate differential expression across different dECM models. (A) Matrisome-associated protein distribution as defined by [60]. (B) Selected proteins studied in dECMs. (C) Representative IFM images of dECMs co-stained with antibodies to laminin-2 (α–2 chain) (LAMA2, green) and MMP9, TIMP-1, or TGF-β1 (red). MMP9 and TGF-β1 dECM content was increased in muscular dystrophy models from the DBA/2 J background. Scale bar, 100μm. (D) MMP9 (WT 12.7, mdx 18.5, mdxD2 23.9, BLAJ 16.8 AU), (E) TIMP-1 (WT 10.2, mdx 13.2, mdxD2 13.4, BLAJ 13.6 AU), and (F) TGF-β1 (WT 3.5, mdx 4.4, mdxD2 7.4, BLAJ 5.7 AU) mean fluorescent intensity in myoscaffolds. n = 3 independent mice per strain (marked as n1, n2, n3), n = 6 images from 2 scaffolds per mouse. Graphs show the mean with SEM bars. * p < 0.05, ** p < 0.01, *** p < 0.005, and **** p < 0.001 by one-way ANOVA.
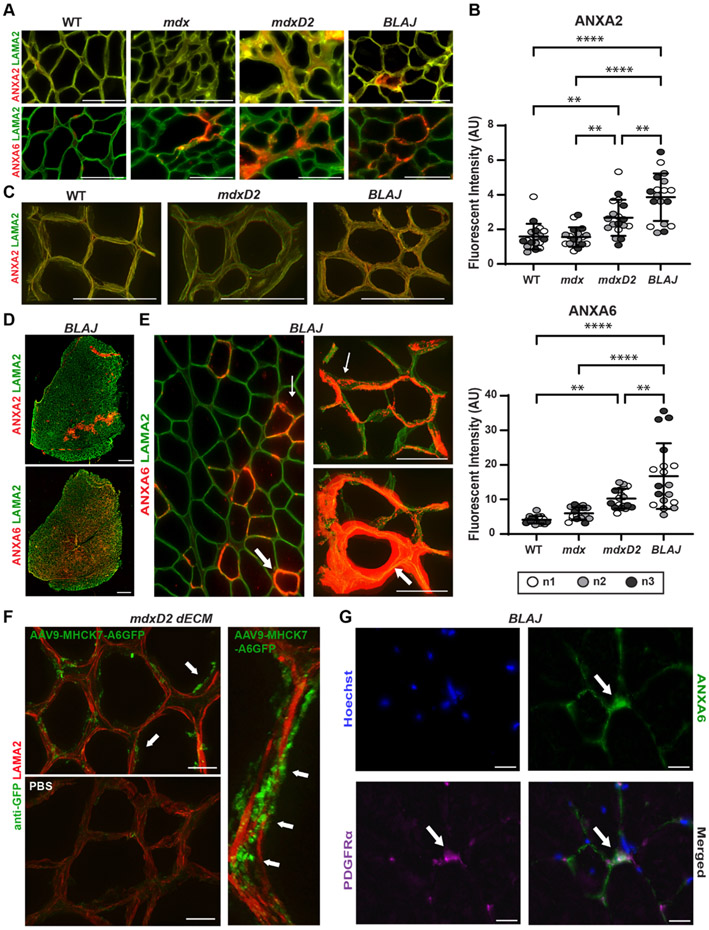
Annexins A2 and A6 were significantly increased in BLAJ dECM compared to other muscular dystrophy myoscaffolds.
The annexin protein family members, including annexin A2 and A6, belong to the matrisome-affiliated category in the subcategory of ECM-affiliated proteins [52,53,60]. Acellular dECM myoscaffolds were probed with antibodies to ANXA2 or ANXA6 and anti-laminin α2 (LAMA2) demonstrating increased ANXA2 and ANXA6 in the matrix of more severely fibrotic models of muscular dystrophy including mdxD2 and SgcgD2 (Fig. 6A; Supplemental Fig. 5). Quantitative fluorescence intensity analysis indicated that ANXA2 and ANXA6 signal in mdxD2 acellular dECM myoscaffolds was increased compared to WT, as expected (Fig. 6A and B). Interestingly, the BLAJ dECM had markedly greater ANXA2 and ANXA6 signal compared to all other strains and genotypes (Fig. 6A-C). The two annexins displayed different expression profiles with ANXA2 exhibiting lower-level, diffuse expression within the matrix compared to ANXA6, which was abundant in vesicle-like structures (Fig. 6D). Z-stack images showed the increased accumulation of ANXA6 vesicles especially in the dECM of BLAJ scaffolds (Fig. 6E; thin white arrow). However, in areas of high ANXA6 deposition, ANXA6 appeared as a solid rim over LAMA2, surrounding what remained of the acellular myofibers (Fig. 6E; thick white arrow). To investigate whether mature skeletal muscle could serve as a potential source of matrix-associated ANXA6, we generated human GFP-tagged ANXA6 utilizing an adeno-associated virus 9 (AAV9) vector driven by the striated muscle specific promotor: MHCK7 (AAV9-MHCK7-ANXA6-GFP; Supplemental Fig. 6). The increase in ANXA6 expression produced by this viral approach was only 1.1X normal, on par with upregulation seen in dystrophic muscle [61]. Mice injected with AAV9-MHCK7-ANXA6-GFP exhibited ANXA6-GFP expression in quadriceps muscle sections which was absent in PBS injected mice (Supplemental Fig. 6E). Acellular myoscaffolds generated from AAV9-injected quadriceps sections displayed ANXA6-GFP deposition in the ECM (Fig. 6F; white arrows), indicating that extracellular ANXA6 can originate from myofibers. Since ANXA6 in dECMs may derive from multiple cellular sources, we looked for co-localization with PDGFRα in native muscle since PDGFRα is enriched in fibro-adipogenic progenitor cells (FAPs). BLAJ native muscles sections were probed for ANXA6 and PDGFRα. Only a small fraction of PDGFRα co-localized with ANXA6 (Fig. 6G; white arrows), and most extracellular ANXA6 protein did not overlap with PDGFRα expression (Fig. 6G), suggesting that multiple cellular sources may be responsible for ANXA6 deposition in dystrophic muscle.
Fig. 6.
Excess deposition of annexins in dysferlin deficient myoscaffolds. (A) Representative IFM images from acellular myoscaffolds generated from WT and mdx, mdxD2, and BLAJ muscles evaluated for annexin A2 (ANXA2) and annexin A6 (ANXA6) (red) with anti- laminin-2 (α–2 chain) (LAMA2, green). ANXA2 exhibits a diffuse pattern, while ANXA6 is vesicular. All scale bars are set at 100 μm. (B) ANXA2 (WT 1.6, mdx 1.6, mdxD2 2.7, BLAJ 3.9 AU) and ANXA6 (WT 4.1, mdx 6.0, mdxD2 10.2, BLAJ 16.8 AU) mean fluorescent intensity in myoscaffolds was significantly upregulated in BLAJ scaffolds, n = 3 independent mice per strain (marked as n1, n2, n3), n = 6 images from 2 scaffolds per mouse. (C) 100x Z-stack images of ANXA2 (red) with LAMA2 co-staining (green) demonstrated diffuse expression on the dECMs generated from WT, mdxD2 and BLAJ muscle. Scale bars 100 μm. (D) Tiled image of a full thickness BLAJ dECM scaffold. ANXA6 staining was present throughout the myoscaffold, while ANXA2 staining was concentrated into focal areas. Scale bars 500 μm. (E) 20x and 100x Z-stack images of ANXA6 illustrated a unique protein deposition profile on BLAJ myoscaffolds. ANXA6 (red) appeared in the matrix in a vesicle-like pattern (thin white arrow), which formed a concentrated ring of protein (thick white arrows). Scale bars 100 μm. (F) Acellular myoscaffolds were generated from AAV9-MHCK7-ANXA6-GFP and PBS treated mice. Z-stack immunofluorescent imaging revealed anti-GFP vesicular staining (green), representing the presence of muscle derived ANXA6-GFP, within the matrix of AAV9 muscle with minimal signal in PBS treated muscle. LAMA2 (red) staining marks the matrix. Scale 20 μm low magnification and 10 μm high magnification images. Scale Bars 20 μm. (G) Representative 100x IFM images of BLAJ muscle sections evaluated for co-localization of ANXA6 (Green) and PDGFRα (Purple). ANXA6 does not completely co-localize with PDGFRα. Scale Bars 20 μm. Graphs show the mean with SEM bars. * p < 0.05, ** p < 0.01, *** p < 0.005, and **** p < 0.001 by one-way ANOVA.
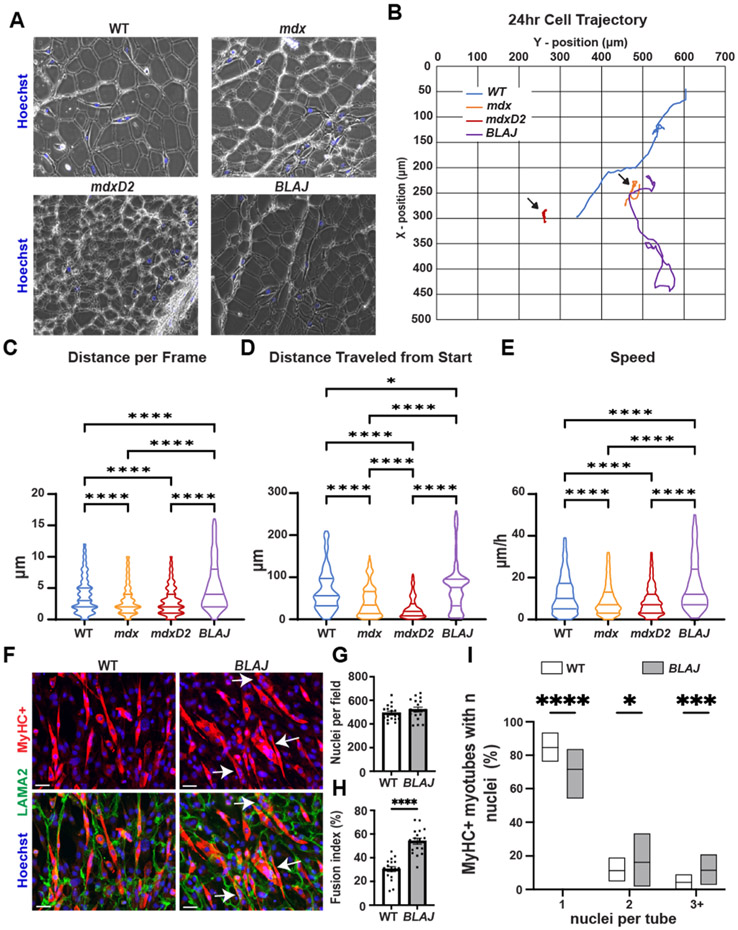
Dystrophin-deficient dECMs inhibit migration of C2C12 myoblasts seeded onto myoscaffolds while dysferlin-deficient dECMs promote C2C12 migration.
Muscle regeneration is generally impaired in severely dystrophic muscle [62], with mdxD2 mice having even more impaired muscle regeneration compared to mdx muscle [20]. Stearns-Reider showed that skeletal muscle progenitor cells derived from human pluripotent stem cells had reduced mobility and differentiation on mdx dECMs [46]. We evaluated the migration patterns of undifferentiated C2C12 myoblasts seeded onto dECMs prepared from multiple dystrophic models (Fig. 7A). C2C12 myoblasts were allowed to seed for 24 h then, cell migration was recorded for the next 24–48 h with images acquired every 20 min (experimental timeline shown in Supplemental Fig. 7). C2C12 myoblasts seeded on mdx and mdxD2 dECMs had reduced movement throughout the matrix compared to BLAJ and WT dECMs (Fig. 7B; black arrows; movement quantified in Fig. 7C). Conversely, an increase in the frame-frame distance was seen for C2C12 myoblasts seeded on BLAJ compared to WT dECMs (Fig. 7C). A similar pattern was observed in the distanced traveled from the start position by C2C12 myoblasts on the matrices with BLAJ > WT > mdx > mdxD2 (Fig. 7D). Myoblast speed was also affected by the dECM substrate. Myoblasts seeded on BLAJ dECMs demonstrated greater speed compared to WT, mdx and mdxD2 (Fig. 7E), while myoblasts seeded on mdx and mdxD2 myoscaffolds had reduced speed compared to WT scaffolds (Supplemental video 1 and 2).
Fig. 7.
mdx myoscaffolds inhibit C2C12 myoblasts movement while BLAJ myoscaffolds enhance C2C12 myoblast movement and differentiation. (A) Representative phase contrast images of C2C12 myoblasts seeded onto dECMs from WT, mdx, mdxD2, and BLAJ mice. (B) Blue (WT), orange (mdx), red (mdxD2), and purple (BLAJ) lines depict cellular movement trajectory over 24 h. Quantification of C2C12 cell movement demonstrated a significant difference in (C) distance per frame (WT 3.91, mdx 3.10, mdxD2 2.98, and BLAJ 5.41 μm), (D) distance traveled from start position (WT 69.24, mdx 43.99, mdxD2 27.12, and BLAJ 76.78 μm), and (E) speed (WT 11.89, mdx 9.31, mdxD2 8.80, and BLAJ 16.34 μm/h) among the WT and dystrophic myoscaffolds. Myoscaffolds prepared from two mice per genotype; n = 73 images per cell, with 8–11 myoblasts tracked per genotype. C2C12 cells were seeded onto WT and BLAJ dECM scaffolds and allowed to differentiate for 96 hrs. (F) Representative IFM images of MyHC+ (red) with LAMA2 (green) demonstrate increased myoblast differentiation in cells seeded on the BLAJ dECM. Scale Bars 100 μm. (G) Equal numbers of nuclei per field were present on WT or BLAJ dECMs. (H) Muscle fusion index was increased on BLAJ myoscaffolds. (I) Significantly increased percentage of MyHC + myotubes with 2 and 3+ nuclei per myotube present on BLAJ dECMs compared to WT. Graphs show the mean with SEM bars. * p < 0.5, ** p < 0.01, *** p < 0.005, and **** p < 0.001 by one-way ANOVA.
In addition to impacting cellular migration, the extracellular environment provides local cues that aid in muscle growth and regeneration influencing the fusion capacity of mononuclear myoblasts into multi-nucleated myotubes. We assessed the differentiation potential of myoblasts seeded on BLAJ acellular myoscaffolds. C2C12 myoblasts were seeded onto WT or BLAJ dECMs for 24 hrs and then serum was withdrawn to induce differentiation over a period of 96 hrs. The seeded scaffolds were subsequently fixed and stained with antibodies to myosin heavy chain 4 (MyHC), a marker of mature myotubes, and LAMA2 to visualize the matrix (Fig. 7F). Cell density was comparable between WT and BLAJ scaffolds (Fig. 7G). On BLAJ scaffolds, MyHC+ myotubes were larger and contained more nuclei per myotube, compared to MyHC+ myotubes formed on WT scaffolds (Fig. 7F-I; White arrows). MyHC+ myotubes with 2 or >3 nuclei were increased on BLAJ scaffolds compared to WT-seeded scaffolds (Fig. 7I), consistent with BLAJ scaffolds inducing greater myoblast fusion. Therefore, BLAJ scaffolds promoted enhanced myoblast migration and fusion compared to WT.
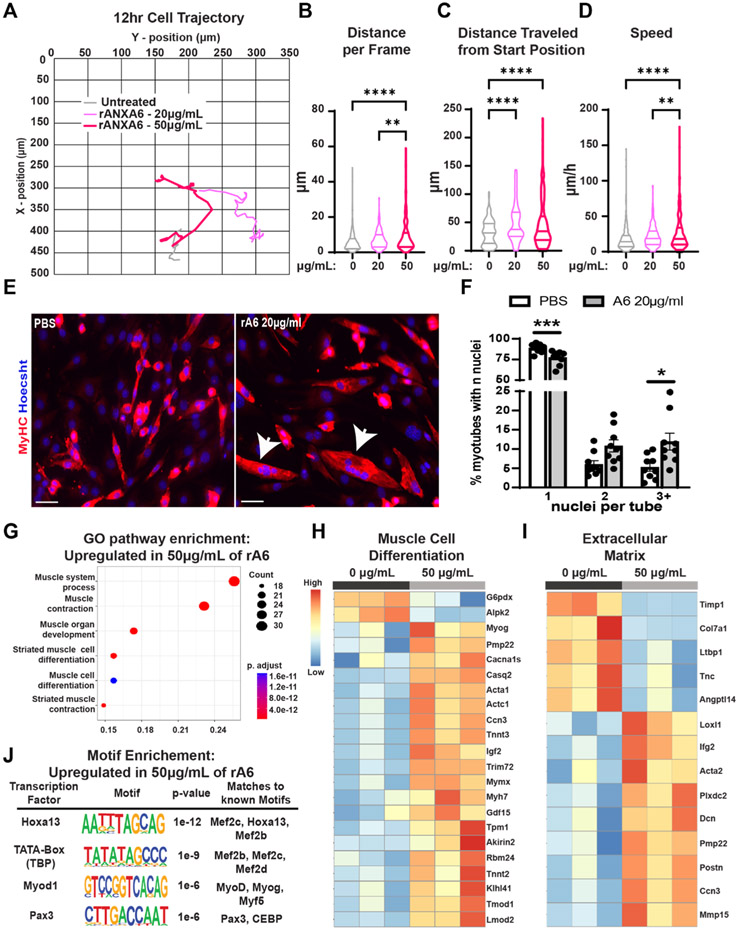
Recombinant Annexin A6 treated myoblasts demonstrate increased migration and upregulation of genes associated with muscle differentiation.
Because the altered matrix composition observed in dysferlin-deficient BLAJ matrices was characterized by increased ANXA6 deposition, and this same matrix supported increased myoblast motility and fusion capacity, we assessed whether ANXA6 itself contributed to differential cellular behavior. C2C12 myoblasts cultured on standard plates were treated with 0, 20, or 50 μg/mL of recombinant ANXA6 (rANXA6) and cell migration was recorded for 12–24 h with images acquired every 20 min. C2C12 myoblast cells treated with rANXA6 had a dose-dependent increase in movement per frame (Fig. 8A; movement per frame quantified in Fig. 8B; Supplemental video 3 and 4). A similar pattern was observed in the distanced traveled from the start position by rANXA6 treated C2C12 myoblasts with 50 μg/mL > 20 μg/mL > 0 μg/mL (Fig. 8C). Myoblast speed was also affected by the rANXA6 with increasing dosage resulting in increased speed (Fig. 8D). Additionally, 20 μg/mL of rANXA6 enhanced myoblast fusion increasing the percentage of MyHC+ myotubes containing >3 nuclei relative to PBS treated myoblasts (Fig. 8E and F). While other components in the BLAJ ECM likely additionally contribute to the enhanced migration and fusion of myoblasts, these data demonstrate that ANXA6 itself mediates this enhancement.
Fig. 8.
Treatment with recombinant Annexin A6 increased myoblasts movement and differentiation and elicited a transcriptomic signature of enhanced differentiation. C2C12 cells were treated with recombinant ANXA6 (rANXA6) and tracked; (A) Grey (untreated), light pink (20 μg/mL rANXA6) and pink (50 μg/mL rANXA6) lines depict cellular movement trajectory over 12 h. Quantification of rANXA6-treated myoblasts demonstrated a significant difference in (B) distance per frame (0 μg/mL 6.01, 20 μg/mL 7.23, and 50μg/mL 12.09 μm), (C) distance traveled from start position (0 μg/mL 32.18, 20 μg/mL 49.72, and 50μg/mL 55.43 μm), and (D) speed (0 μg/mL 18.04, 20 μg/mL 21.78, and 50 μg/mL 36.20 μm). Treatments were conducted on two different passages; n = 37 images per cell, with 8 myoblasts tracked per treatment. (E) Representative IFM images of MyHC (red) with Hoechst (blue) demonstrate increased myoblast differentiation in cells treated with 20μg/mL rANXA6. Scale bars 100 μm (F) Significantly increased percentage of MyHC myotubes with >3 nuclei on BLAJ myoscaffolds compared to WT. (G) C2C12 myoblasts treated with rANXA6 increased expression of Gene Ontology terms for muscle processes and differentiation. Representative heatmaps demonstrated upregulation of genes associated with (H) muscle cell differentiation and the (I) extracellular matrix in the rANXA6 treated myoblasts. (J) HOMER motif analysis demonstrates consensus sequences for several muscle cell differentiation transcription factors. Graphs show the mean with SEM bars. * p < 0.5, ** p < 0.01, *** p < 0.005, and **** p < 0.001 by one-way ANOVA.
Myoblast fusion to myotubes is understood to reflect enhanced myoblast differentiation. To explore the effects of rANXA6 treatment on myoblasts, we conducted RNA-sequencing on C2C12 myoblasts cells treated with 0 or 50 μg/mL rANXA6 for 48 h. rANXA6 treatment separated treated myoblasts from PBS control by Principal Component Analysis (Supplemental Fig. 8A). Gene Ontology pathway analysis of rANXA6-treated myoblasts demonstrated upregulation in genes associated with muscle organ development, muscle cell differentiation and muscle contraction, consistent with the increase in migration and fusion of rANXA6 treated myoblasts (Fig. 8G). The top 20 upregulated genes in cells treated with rANXA6 include genes associated with skeletal muscle structures including Casq2, Myh3, Acta1, Tnnc2, Ckm, Mybph, Actc1, Tnnt3, and Myh1 Fig. 8H, Supplemental Fig. 8B). Other significantly upregulated genes include Myog, a muscle regulatory factor (MRF) essential for muscle development, Trim72, a muscle-enriched protein that facilitates vesicle/membrane reorganization, and Mymx, a muscle membrane fusion protein that is required for myoblast fusion. Upregulation of ECM genes was also observed in the rANXA6 treated myoblasts including Dcn and Postn (Fig. 8I). Representative heatmaps exhibiting upregulation in the treated group in several muscle processes are presented in (Supplemental Fig. 9). HOMER motif analysis of the upregulated genes revealed enrichment for binding sites known to engage transcription factors critical for muscle cell differentiation, including Hoxa13, Pax3, Mef2c, Mef2b, and Myf5 (Fig. 8J). Taken together, these data demonstrate that ANXA6 exposure increases myoblast movement, fusion, and differentiation.
Discussion
The “on-slide” dECM protocol preserves the spatial architecture and composition of the matrix. The on-slide dECM protocol removes the cellular components, increasing epitope exposure and allowing greater ECM protein accessibility and visualization [46]. Stearns-Reider et al. used this method to compare intensely fibrotic regions of mdx scaffolds with less fibrotic regions. They found that differential collagen cross-linking and laminin deposition impacted skeletal muscle progenitor cell (SMPC) motility and the ability of SMPCs to remodel severe fibrosis. We now applied the on-slide dECM method to distinct muscular dystrophy models with different molecular pathologies, and we identified key differences in dECM protein deposition across these muscular dystrophy subtypes. Our findings of impaired C2C12 mobility on severe mdxD2 scaffolds compared to less fibrotic disease models are consistent with the observations of Stearns-Reider as C2C12 myoblasts had demonstrably defective migration on mdxD2 dECMs.
We evaluated three different primary mutations that cause muscular dystrophy, and compared how these mutations shifted the myoscaffold content. Two of the primary mutations, dystrophin and γ-sarcoglycan, share a similar pathological defect, namely a fragile sarcolemma that undergoes repetitive disruption and leak. However, dystrophin itself contributes directly to the stiffness of the sarcolemma, while the sarcoglycan complex is implicated in the attachment of sarcolemma to the surrounding matrix. Thus, the finding that the dECMs similarly impact matrix remodeling supports the defect in sarcolemmal stability as the major contributor to fibrosis. Both mdx and Sgcg mutants shared myoscaffold features, including similar matrisome protein profiles and associated fibrosis, and these features were even more evident in the DBA/2 J background, which is known to produce more fibrosis [20,21,24]. In particular, these models, mdx and Sgcg, in the DBA/2 J background had significantly increased levels of core matrisomal proteins THBS4, POSTN, and DCN within the dECM scaffolds. THBS4 regulates muscle attachment at the myotendinous junction and promotes sarcolemmal stability [58,63]. THBS4 is upregulated in human muscular dystrophies and their corresponding mouse models [55,58,64,65]. Transgenic overexpression of THBS4 in muscle protects against muscular dystrophy disease progression by stabilizing the sarcolemma through enhanced trafficking of the dystrophin-glycoprotein and integrin attachment complexes [58,64]. Moreover, secretion of THBS4 was shown to be an essential component to elicit these protective effects. Secretion-deficient THBS4 resulted in reduced trafficking to the membrane and more severe muscular dystrophy. We observed THBS4 in puncta in mdxD2 myoscaffolds throughout the matrix, and these puncta likely reflect the normal secretion of THBS4 vesicles, supporting a role for THBS4 in transporting matrix-bound molecules.
We also observed upregulation of DCN in mdx and Sgcg myoscaffolds, and this upregulation was further enhanced by the DBA/2 J background. DCN is increased in the matrix of DMD muscle biopsies, likely due to the increased synthesis of DCN by DMD fibroblasts [54,56,59]. Upregulation of DCN in myoblasts was shown to improve muscle regeneration, indicating that DCN’s cell of origin impacts its effect [66]. POSTN is normally expressed at low levels in healthy tissues, but its expression is increased after acute injury and in muscular dystrophies [57,67]. Unlike THBS4, overexpression of POSTN exacerbated muscle disease progression and promoted fibrosis formation in Sgcd-null mice, a model of LGMD2F [57]. In dECMs POSTN expression was increased in the more severe mdxD2 dECM compared to the mild mdx and WT dECMs. We hypothesize that the increased TGF-β levels in this model upregulates Postn expression while Dcn is upregulated as a compensatory protective mechanism.
Dysferlin deficient muscle displays a distinct dECM content.
Dysferlin is a membrane associated protein implicated in resealing disrupted membranes, in contrast to the dystrophin complex and its role in stabilizing the sarcolemma [8,9]. Additionally, patients with DYSF mutations often have a distinct clinical course where muscle function is normal in early life unlike other forms of muscular dystrophy [14]. Similarly, mouse models lacking Dysf display a comparatively mild phenotype with certain muscles showing more pathology than others [68-70]. The pattern of increased annexin deposition and its vesicular pattern in dysferlin-deficient BLAJ muscle was only seen in dysferlin-deficiency, and this pattern is consistent with increased membrane recruitment of annexin A6 as an attempt to enhance membrane resealing. Through immunofluorescence microscopy and correlative light and electron microscopy (CLEM), annexin A6-positive vesicles have been visualized emerging from the membrane lesion seconds to minutes post membrane rupture [28,31]. The annexin vesicles identified in dECM scaffolds are plausibly derived from vesicles released from the injured myofiber repair caps, matching our finding that muscle-derived annexin deposits into the decellularized matrix. The distinct patterns of vesicular annexin A6 versus diffuse annexin A2 in the dECM scaffolds suggests the possibility that these matrix-associated proteins may have different cellular origins. Immunofluorescence imaging of 12-month-old BLAJ muscles showed annexin A2 accumulation in the interstitial space which correlated with fibroblast markers PDGFRα and perilipin-1 [71]. In contrast, we found that only a fraction of annexin A6 was expressed near PDGFRα in BLAJ muscle, further illustrating the unique contribution of the individual annexins to diseased muscle tissue.
Compared to mdxD2 dECMs, BLAJ dECMs contained less fibrosis, with laminin and collagen content similar to WT matrices. mdxD2 scaffolds inhibited myoblast migration while dysferlin-deficient myoscaffolds increased myoblast movement, migration, and fusion. Muscle differentiation is a highly coordinated process that relies on cell intrinsic and extrinsic signaling events. MyoD and MyoG are both myogenic regulatory factors (MRF) that orchestrate myoblast differentiation to myotubes with MyoD preceding MyoG in the process. The transcriptome profile of C2C12 cells treated with rANXA6 supports a more MyoG-driven process, which is consistent with upregulation of fusogenic as well as sarcomeric proteins critical for fusion and myotube growth and matching. DYSF patient-derived myoblasts were previously shown to have reduced MyoG [72]. In vivo, the upregulation of ANXA6 and its ability to enhanced myoblast differentiation and Myog expression would serve to directly counteract these deficits. The upregulation of muscle genes combined with the in vitro increase in myoblast differentiation in the presence of recombinant annexin A6, supports extracellular annexin A6 as mediating muscle repair and regeneration. Furthermore, these findings are consistent with clinical observations of DYSF patients having preserved exercise capacity in early life, and together this work highlights the importance of matrix contributions to muscle health.
Materials and methods
Animals.
All animals were bred and housed in a pathogen free facility. Wild-type (WT) are from the C57BL/6 strain, unless otherwise stated. C57BL/10ScSn-Dmdmdx/J (mdx), D2.B10-Dmdmdx/J (mdxD2), and Bb.A-Dysfprmd/GeneJ (BLAJ) mice were originally purchased from The Jackson Laboratory (strains #:001801, #:013141, and #:012767 respectively). Sarcoglycan γ-null (Sgcg) mice were previously generated on the C57BL/6 background and Sgcg 521ΔT (SgcgD2) were previously generated on the DBA/2 J background [16,73]. Where mice from the D2 backgrounds were used, wild-type (WTD2) mice were from the DBA/2 J strain and are indicated as such. Seven- to eight-month male and female mice were used for all experiments. Mice were bred and housed in a specific pathogen free facility on a 12-hour light/dark cycle and fed ad libitum.
Study Approval.
All procedures using mice followed Guide for the Care and Use of Laboratory Animals and were approved by Northwestern University’s Institutional Animal Care and Use Committee.
Decellularization.
Quadriceps muscle was collected, and flash frozen in liquid nitrogen. Sections were cryosectioned at 25μm (Leica CM1950) and mounted on charged Superfrost Plus Microscope Slides (cat# 1255015; Fisher Scientific). Slides were stored at −80 °C. Decellularization was performed as in Stearns-Reider et al., 2023 with the following modifications [46]. Slides were allowed to thaw to room temperature for 1 hr prior to decellularization. Slides were then placed in a 15 mL UltraPure Sodium Dodecyl Sulfate (SDS) solution (cat# 15553035; Invitrogen) diluted with PBS to 0.1 % or 1 % concentration for 60 or 10 min, respectively, under constant agitation (40 rpm). Slides were then placed in 100 mm x 100 mm square petri dishes 3 slides per dish (Fisher cat#FB0875711A) containing 13 mL of PBS (with calcium & magnesium [Cat# 21030CV; Corning]), for 15 min, then placed in a fresh 13 mL of PBS for 45 min, followed by 13 mL UltraPure Distilled H2O (cat# 10977015; Invitrogen) for 30 min, and a final 13 mL PBS wash for 45 min. All wash steps were done under constant agitation (40 rpm). Decellularized slides were either fixed for histology/immunostaining or stored in PBS.
DNA Extraction.
DNA was extracted from quadriceps muscle tissues using the DNeasy Blood & Tissue Kit (cat# 69504; Qiagen) and DNA concentration measured on a NanoDrop 2000 Spectrophotometer (Thermo Scientific). DNA electrophoresis was visualized using a Bio-Spectrum Imaging system Chemi 410 (UVP).
Histology.
Quadriceps muscle was harvested and fixed in 10 % formalin. Hematoxylin and eosin (cat# 12013B and 1070C; Newcomer Supply), Sirius Red (cat# 24901250; Polysciences, Inc), or Alcian Blue (cat#A3157; Sigma) staining was performed per manufacturer’s protocol. All images were acquired on the Keyence BZ-X810 microscope with a 40x objective and same exposure settings across all genotypes.
Protein isolation.
Tibialis anterior muscles were harvested and flash-frozen in liquid nitrogen. Tissues were ground with a mortar and pestle and lysed in Whole Tissue Lysis buffer (50 mM HEPES pH 7.5, 150 mM NaCl, 2 mM EDTA, 10 mM NaF, 10 mM Na-pyrophosphate, 10 % glycerol, 1 % Triton X-100, 1 mM phenyl-methylsulfonyl fluoride, 1x complete Mini protease inhibitor cocktail [cat # 11836170001; Roche], 1x PhosSTOP [cat# 04906837001; Roche]) and homogenized using a TissueLyser II (Qiagen) at a frequency of 30.0 1/s and time of 2.00 min/sec for 6 rounds. Protein lysates were then centrifuged at 20,000xg for 20 min to separate the soluble and insoluble fractions. The soluble supernatant layer was placed in a separate tube and the insoluble pellet layer was resuspended with an 8 M Urea buffer (8 M Urea [cat#U4883; MilliporeSigma], 2 mM EDTA, 10 mM DTT, 1x complete Mini protease inhibitor cocktail [cat # 11836170001; Roche]). The protein concentration was measured with the Quick Start Bradford 1x Dye Reagent (cat# 5000205; Bio-Rad Laboratories).
Immunoblotting.
20–25μg of protein lysate with 4x Laemmli buffer (cat# 1610747; Bio-Rad Laboratories was separated on 4–15 % Mini-PROTEAN TGX Precast Protein Gels (cat# 4561086; Bio-Rad Laboratories) and transferred to Immun-Blot PVDF Membranes for Protein Blotting (cat# 1620177; Bio-Rad Laboratories). Membranes were reversibly stained for total protein with Ponceau S Staining Solution (cat# A40000278; Thermo Scientific) to ensure transfer and equal loading prior to antibody incubation. Blocking and antibody incubations were done using StartingBlock T20 (TBS) Blocking Buffer (cat# 37543; Thermo Scientific). Primary antibodies were used at 1:1000 except for anti-thrombospondin-4 and anti-TIMP-1 which were diluted at 1:100. Secondary antibodies conjugated with horseradish peroxidase were used at 1:2500 (cat# 111035003; Jackson ImmunoResearch Laboratories). Clarity Western ECL Substrate (cat# 1705061; BioRad Laboratories) was applies to membranes and visualized using an iBright 1500 Imaging System (Invitrogen). Pierce Reversible Protein Stain Kit for PVDF Membranes (including MemCode; cat# 24585; Thermo Fisher Scientific) was used to ensure equal loading.
Immunostaining and immunofluorescence imaging.
Decellularized quadriceps cryosections (dECMs) were fixed in 4 % paraformaldehyde, rinsed, and blocked with a 1 % BSA/10 % FBS blocking buffer for 1 hour. The dECMs were incubated with the primary antibodies overnight at 4 °C at 1:100. The dECMs were then incubated with the secondary antibodies at 1:2500 for 1 hr. The dECMs were mounted with ProLong Gold Antifade Reagent (cat# P36934; Invitrogen) and images acquired on a Keyence BZ-X810 microscope with 20x, 40x, 60x and 100x objectives. Z-stack projections were acquired at 100x combined with optical sectioning. All image exposure settings were kept consistent across genotypes. Matrisome protein quantification was performed on 18 images per strain, unless otherwise noted. Specifically, protein-matrix deposition was assessed through tiled imaging of the entire cross-sectional area. From each muscle, three representative 40x images were taken from two decellularized 25 μm scaffolds per animal for a total of 6 images per animal. Images were selected to be representative containing degenerating and regenerating regions. Fluorescence intensity was quantified using ImageJ (NIH).
Antibodies.
Primary antibodies used for immunoblotting and immunostaining: anti-annexin A6 (cat# ab199422; Abcam), anti-annexin A2 (cat# 610068; BD Bioscience), anti-caveolin 3 (cat# NBP128917; Novus Bio), anti-collagen type 1 (cat# CL50151AP1; Cedarlane), anti-decorin (cat# AF1060; R&D Systems), anti-dystrophin (cat# PA137587; Invitrogen), anti-HSP90 (cat#4874; Cell Signaling Technologies), anti-laminin-2 (α–2 Chain) (cat# L0663; MilliporeSigma), anti-MMP9 (cat# ab38898; Abcam), anti-myosin 4 (cat# PA550065; Invitrogen), anti-periostin/OSF-2 (cat# NBP130042; Novus Bio), anti-Mo CD140a (PDGRFα) (cat# 17140181; Invitrogen), anti-TGF Beta 1 (cat# 218981AP; Proteintech and cat# MA515065; Invitrogen), anti-Thrombospondin-4 (cat# AF2390; R&D Systems), and anti-TIMP-1 (cat# AF980; R&D Systems). Secondary antibodies used: Alexa Fluor 488 goat anti-rat (cat# A11006; Invitrogen), Alexa Fluor 594 goat anti-rabbit (cat# A11012; Invitrogen) and Alexa Fluor 594 donkey anti-goat (cat# A11058; Invitrogen).
Adeno-associated virus production and dosing.
To achieve muscle specific expression of human annexin A6, AAV9-MHCK7-hANXA6-turboGFP was generated and packaged at Vector builder. As a viral plasmid control, liver specific expression of human annexin A6, AAV9-TBG-hANXA6-turboGFP was generated and packaged at Vector builder. At 28days of age, mdxD2 mice were injected with 5e13 vg/kg AAV9-MHCK7-hANXA6-turboGFP or PBS control in a total of 100 μl. Four weeks post injection, mice were euthanized. Tissues were harvested and stored at −80C.
Recombinant protein production.
Recombinant human annexin A6 (rANXA6) was generated in E.Coli using standard methods [74]. Protein was diluted in TBS with endotoxin levels less than 1.5 EU/mg.
Myoscaffold reseeding, live-cell imaging, and differentiation.
After decellularization as described above, MatTek chambers (cat# CCS-2; MatTek) were attached to slides mounted with skeletal muscle dECMs. The acellular myoscaffolds were incubated in DMEM proliferation media containing 10 % FBS and 1 % penicillin/streptomycin for 24 hrs prior to reseeding with C2C12 myoblasts (cat# CRL-1772; ATCC). For live-cell imaging cells were stained with Hoechst 33342 for 5-minutes and then seeded onto the acellular myoscaffolds at a density of 25,000 in 2mLs media per chamber. Seeded slides were imaged in a temperature and CO2 controlled incubation chamber (STR Stage Top Incubator; Tokai Hit) and images acquired every 20 min over 48 hrs on a Keyence BZ-X810 microscope with 20x phase contrast objectives. For recombinant annexin A6 experiments, cells were treated with 0, 20 or 50 μg/ml of recombinant annexin A6 in media. All image exposure settings were kept consistent across all genotypes. Cell tracking analysis was performed using the Dynamic Tracking software on the Keyence. For myoblast differentiation analyses C2C12s were seeded onto the decellularized myoscaffolds at a density of 150,000 in 1.5mLs media per chamber and were cultured in proliferation media for 24 hrs prior to switching to DMEM differentiation media containing 2 % Horse Serum and 1 % penicillin/streptomycin. Myoblasts were allowed to differentiate for 96 hrs and were then fixed with 4 % PFA for subsequent IFM analysis and quantification [75]. C2C12 fusion analyses were performed on 9 images per condition. Nuclei quantification was performed using Image J (NIH). Fusion index was calculated as the number of myonuclei within MyHC-positive myotubes divided by the total number of nuclei present in the field.
Bulk RNA sequencing and data analysis.
C2C12 cells were seeded at a density of 25,000 in 2mLs media per chamber. Cells were treated with 0 or 50 μg/ml of recombinant annexin A6 in proliferation media 24 hrs post seeding and were harvested 48 hrs post treatment. Cell pellets were transported to the NUSeq Core Facility at Northwestern University for RNA extraction and sequencing, where the samples were indexed and pooled for 50 bp single-end sequencing. Raw reads were mapped to mm10 using STAR aligner and gene counts were computed by HT-Seq. Read count normalization and differential expression analysis was conducted using R package DESeq2 (v1.38.3). Differentially expressed genes were selected based on adjusted p-value < 0.05. Gene Ontology analysis of differentially expressed genes was conducted using enrichGO function in R package clusterProfiler (v4.6.2). Motif analysis of differentially expressed genes was performed using HOMER [76].
Statistical Analyses.
Statistical analyses were performed with Prism (GraphPad, La Jolla, CA). Comparisons relied on 1-way ANOVA for 1 variable or 2-way ANOVA for two variables. Otherwise, unpaired two-tailed t-tests were performed. A p-value less than or equal to 0.05 was considered significant. Data were presented as single values were appropriate. Error bars represent ± standard error of the mean (SEM).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health NS047726 (EM, AD), National Institutes of Health AR052646 (EM, AD), National Institutes of Health HL061322 (EM, AD), National Institutes of Health AR048179 (RC), National Institutes of Health T32AR065972 (RC), Parent Project Muscular Dystrophy (AL), Additional funding was through Lakeside Discovery (EM, AD). We especially acknowledge the Jain Foundation for providing dysferlin-null mice from their private colony at The Jackson Laboratory.
Abbreviations:
- ECM
Extracellular matrix
- dECM
decellularized extracellular matrix
- DMD
Duchenne Muscular Dystrophy
- LGMD
Limb Girdle Muscular Dystrophy
- SDS
sodium dodecyl sulfate
Footnotes
TEASER: Fibrosis in muscular dystrophy has differential effects on myoblasts
Declaration of competing interest
Northwestern University filed provisional patents #62/783,619 and #63/309,925 on behalf of the authors (ARD and EMM). EMM is or has been a consultant to Amgen, AstraZeneca, Cytokinetics, PepGen, Pfizer, and Tenaya Therapeutics and is the CEO of Ikaika Therapeutics. ARD is the CSO of Ikaika Therapeutics
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.matbio.2024.04.001.
Data availability
Data will be made available on request.
References
- [1].Rando TA, The dystrophin-glycoprotein complex, cellular signaling, and the regulation of cell survival in the muscular dystrophies, Muscl. Nerve 24 (12) (2001) 1575–1594. [DOI] [PubMed] [Google Scholar]
- [2].Duan D, Goemans N, Takeda S, Mercuri E, Aartsma-Rus A, Duchenne muscular dystrophy, Nat. Rev. Dis. Primer 7 (1) (2021) 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Liu W, Pajusalu S, Lake NJ, Zhou G, Ioannidis N, Mittal P, Johnson NE, Weihl CC, Williams BA, Albrecht DE, Rufibach LE, Lek M, Estimating prevalence for limb-girdle muscular dystrophy based on public sequencing databases, Genet. Med 21 (11) (2019) 2512–2520. [DOI] [PubMed] [Google Scholar]
- [4].Nigro V, Savarese M, Genetic basis of limb-girdle muscular dystrophies: the 2014 update, Acta Myol. 33 (1) (2014) 1–12. [PMC free article] [PubMed] [Google Scholar]
- [5].Taghizadeh E, Rezaee M, Barreto GE, Sahebkar A, Prevalence, pathological mechanisms, and genetic basis of limb-girdle muscular dystrophies: a review, J. Cell Physiol 234 (6) (2019) 7874–7884. [DOI] [PubMed] [Google Scholar]
- [6].Davis DB, Doherty KR, Delmonte AJ, McNally EM, Calcium-sensitive phospholipid binding properties of normal and mutant ferlin C2 domains, J. Biol. Chem 277 (25) (2002) 22883–22888. [DOI] [PubMed] [Google Scholar]
- [7].Bashir R, Britton S, Strachan T, Keers S, Vafiadaki E, Lako M, Richard I, Marchand S, Bourg N, Argov Z, Sadeh M, Mahjneh I, Marconi G, Passos-Bueno MR, Moreira Ede S, Zatz M, Beckmann JS, Bushby K, A gene related to Caenorhabditis elegans spermatogenesis factor fer-1 is mutated in limb-girdle muscular dystrophy type 2B, Nat. Genet 20 (1) (1998) 37–42. [DOI] [PubMed] [Google Scholar]
- [8].Bansal D, Miyake K, Vogel SS, Groh S, Chen CC, Williamson R, McNeil PL, Campbell KP, Defective membrane repair in dysferlin-deficient muscular dystrophy, Nature 423 (6936) (2003) 168–172. [DOI] [PubMed] [Google Scholar]
- [9].Defour A, Van der Meulen JH, Bhat R, Bigot A, Bashir R, Nagaraju K, Jaiswal JK, Dysferlin regulates cell membrane repair by facilitating injury-triggered acid sphingomyelinase secretion, Cell Death. Dis 5 (2014) e1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lek A, Evesson FJ, Sutton RB, North KN, Cooper ST, Ferlins: regulators of vesicle fusion for auditory neurotransmission, receptor trafficking and membrane repair, Traffic 13 (2) (2012) 185–194. [DOI] [PubMed] [Google Scholar]
- [11].McDade JR, Archambeau A, Michele DE, Rapid actin-cytoskeleton-dependent recruitment of plasma membrane-derived dysferlin at wounds is critical for muscle membrane repair, FASEB J. 28 (8) (2014) 3660–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Liu J, Aoki M, Illa I, Wu C, Fardeau M, Angelini C, Serrano C, Urtizberea JA, Hentati F, Hamida MB, Bohlega S, Culper EJ, Amato AA, Bossie K, Oeltjen J, Bejaoui K, McKenna-Yasek D, Hosier BA, Schurr E, Arahata K, de Jong PJ, Brown RH Jr., Dysferlin, a novel skeletal muscle gene, is mutated in Miyoshi myopathy and limb girdle muscular dystrophy, Nat. Genet 20 (1) (1998) 31–36. [DOI] [PubMed] [Google Scholar]
- [13].Contreras-Cubas C, Barajas-Olmos F, Frayre-Martinez MI, Siordia-Reyes G, Guizar-Sanchez CC, Garcia-Ortiz H, Orozco L, Baca V, Dysferlinopathy misdiagnosed with juvenile polymyositis in the pre-symptomatic stage of hyperCKemia: a case report and literature review, BMC. Med. Genom 15 (1) (2022) 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Klinge L, Aboumousa A, Eagle M, Hudson J, Sarkozy A, Vita G, Charlton R, Roberts M, Straub V, Barresi R, Lochmüller H, Bushby K, New aspects on patients affected by dysferlin deficient muscular dystrophy, J. Neurol. Neurosurg. Psychiatry 81 (9) (2010) 946–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ceco E, Bogdanovich S, Gardner B, Miller T, DeJesus A, Earley JU, Hadhazy M, Smith LR, Barton ER, Molkentin JD, McNally EM, Targeting latent TGFbeta release in muscular dystrophy, Sci. Transl. Med 6 (259) (2014), 259ra144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hack AA, Ly CT, Jiang F, Clendenin CJ, Sigrist KS, Wollmann RL, McNally EM, Gamma-sarcoglycan deficiency leads to muscle membrane defects and apoptosis independent of dystrophin, J. Cell Biol 142 (5) (1998) 1279–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ho M, Post CM, Donahue LR, Lidov HG, Bronson RT, Goolsby H, Watkins SC, Cox GA, Brown RH Jr., Disruption of muscle membrane and phenotype divergence in two novel mouse models of dysferlin deficiency, Hum. Mol. Genet 13 (18) (2004) 1999–2010. [DOI] [PubMed] [Google Scholar]
- [18].Lostal W, Bartoli M, Bourg N, Roudaut C, Bentaib A, Miyake K, Guerchet N, Fougerousse F, McNeil P, Richard I, Efficient recovery of dysferlin deficiency by dual adeno-associated vector-mediated gene transfer, Hum. Mol. Genet 19 (10) (2010) 1897–1907. [DOI] [PubMed] [Google Scholar]
- [19].Sicinski P, Geng Y, Ryder-Cook AS, Barnard EA, Darlison MG, Barnard PJ, The molecular basis of muscular dystrophy in the mdx mouse: a point mutation, Science (1979) 244 (4912) (1989) 1578–1580. [DOI] [PubMed] [Google Scholar]
- [20].Coley WD, Bogdanik L, Vila MC, Yu Q, Van Der Meulen JH, Rayavarapu S, Novak JS, Nearing M, Quinn JL, Saunders A, Dolan C, Andrews W, Lammert C, Austin A, Partridge TA, Cox GA, Lutz C, Nagaraju K, Effect of genetic background on the dystrophic phenotype in mdx mice, Hum. Mol. Genet 25 (1) (2016) 130–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hammers DW, Hart CC, Matheny MK, Wright LA, Armellini M, Barton ER, Sweeney HL, The D2.mdx mouse as a preclinical model of the skeletal muscle pathology associated with Duchenne muscular dystrophy, Sci. Rep 10 (1) (2020) 14070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].van Putten M, Lloyd EM, de Greef JC, Raz V, Willmann R, Grounds MD, Mouse models for muscular dystrophies: an overview, Dis. Model. Mech 13 (2) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gordish-Dressman H, Willmann R, Dalle Pazze L, Kreibich A, van Putten M, Heydemann A, Bogdanik L, Lutz C, Davies K, Demonbreun AR, Duan D, Elsey D, Fukada SI, Girgenrath M, Patrick Gonzalez J, Grounds MD, Nichols A, Partridge T, Passini M, Sanarica F, Schnell FJ, Wells DJ, Yokota T, Young CS, Zhong Z, Spurney C, Spencer M, De Luca A, Nagaraju K, Aartsma-Rus A, Of Mice and Measures": a Project to Improve How We Advance Duchenne Muscular Dystrophy Therapies to the Clinic, J. Neuromuscul. Dis 5 (4) (2018) 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Heydemann A, Huber JM, Demonbreun A, Hadhazy M, McNally EM, Genetic background influences muscular dystrophy, Neuromuscul. Disord 15 (9–10) (2005) 601–609. [DOI] [PubMed] [Google Scholar]
- [25].Aartsma-Rus A, van Putten M, Assessing functional performance in the mdx mouse model, J. Vis. Exp 85 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Quattrocelli M, Capote J, Ohiri JC, Warner JL, Vo AH, Earley JU, Hadhazy M, Demonbreun AR, Spencer MJ, McNally EM, Genetic modifiers of muscular dystrophy act on sarcolemmal resealing and recovery from injury, PLoS. Genet 13 (10) (2017) e1007070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Swaggart KA, Demonbreun AR, Vo AH, Swanson KE, Kim EY, Fahrenbach JP, Holley-Cuthrell J, Eskin A, Chen Z, Squire K, Heydemann A, Palmer AA, Nelson SF, McNally EM, Annexin A6 modifies muscular dystrophy by mediating sarcolemmal repair, Proc. Natl. Acad. Sci. U.S.A 111 (16) (2014) 6004–6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Croissant C, Gounou C, Bouvet F, Tan S, Bouter A, Annexin-A6 in membrane repair of human skeletal muscle cell: a role in the cap subdomain, Cells 9 (7) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Boye TL, Maeda K, Pezeshkian W, Sonder SL, Haeger SC, Gerke V, Simonsen AC, Nylandsted J, Annexin A4 and A6 induce membrane curvature and constriction during cell membrane repair, Nat. Commun 8 (1) (2017) 1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Demonbreun AR, Quattrocelli M, Barefield DY, Allen MV, Swanson KE, McNally EM, An actin-dependent annexin complex mediates plasma membrane repair in muscle, J. Cell Biol 213 (6) (2016) 705–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Demonbreun AR, Fallon KS, Oosterbaan CC, Bogdanovic E, Warner JL, Sell JJ, Page PG, Quattrocelli M, Barefield DY, McNally EM, Recombinant annexin A6 promotes membrane repair and protects against muscle injury, J. Clin. Invest 129 (11) (2019) 4657–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Belvedere R, Bizzarro V, Popolo A, Dal Piaz F, Vasaturo M, Picardi P, Parente L, Petrella A, Role of intracellular and extracellular annexin A1 in migration and invasion of human pancreatic carcinoma cells, BMC Cancer 14 (2014) 961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bizzarro V, Belvedere R, Dal Piaz F, Parente L, Petrella A, Annexin A1 induces skeletal muscle cell migration acting through formyl peptide receptors, PLoS One 7 (10) (2012) e48246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Garcia-Melero A, Reverter M, Hoque M, Meneses-Salas E, Koese M, Conway JR, Johnsen CH, Alvarez-Guaita A, Morales-Paytuvi F, Elmaghrabi YA, Pol A, Tebar F, Murray RZ, Timpson P, Enrich C, Grewal T, Rentero C, Annexin A6 and Late Endosomal Cholesterol Modulate Integrin Recycling and Cell Migration, J. Biol. Chem 291 (3) (2016) 1320–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Grewal T, Hoque M, Conway JRW, Reverter M, Wahba M, Beevi SS, Timpson P, Enrich C, Rentero C, Annexin A6-A multifunctional scaffold in cell motility, Cell Adh. Migr 11 (3) (2017) 288–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bissell MJ, Hall HG, Parry G, How does the extracellular matrix direct gene expression? J. Theor. Biol 99 (1) (1982) 31–68. [DOI] [PubMed] [Google Scholar]
- [37].Bornstein P, Sage EH, Matricellular proteins: extracellular modulators of cell function, Curr. Opin. Cell Biol 14 (5) (2002) 608–616. [DOI] [PubMed] [Google Scholar]
- [38].Schultz GS, Davidson JM, Kirsner RS, Bornstein P, Herman IM, Dynamic reciprocity in the wound microenvironment, Wound Repair. Regen 19 (2) (2011) 134–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Gold LI, Eggleton P, Sweetwyne MT, Van Duyn LB, Greives MR, Naylor SM, Michalak M, Murphy-Ullrich JE, Calreticulin: non-endoplasmic reticulum functions in physiology and disease, FASEB J. 24 (3) (2010) 665–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Dowling P, Gargan S, Murphy S, Zweyer M, Sabir H, Swandulla D, Ohlendieck K, The dystrophin node as integrator of cytoskeletal organization, lateral force transmission, fiber stability and cellular signaling in skeletal muscle, Proteomes 9 (1) (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Rayagiri SS, Ranaldi D, Raven A, Mohamad Azhar NIF, Lefebvre O, Zammit PS, Borycki AG, Basal lamina remodeling at the skeletal muscle stem cell niche mediates stem cell self-renewal, Nat. Commun 9 (1) (2018) 1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Mazala DA, Novak JS, Hogarth MW, Nearing M, Adusumalli P, Tully CB, Habib NF, Gordish-Dressman H, Chen YW, Jaiswal JK, Partridge TA, TGF-beta-driven muscle degeneration and failed regeneration underlie disease onset in a DMD mouse model, JCI. Insight 5 (6) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Didangelos A, Yin X, Mandal K, Baumert M, Jahangiri M, Mayr M, Proteomics characterization of extracellular space components in the human aorta, Mol. Cell Proteom 9 (9) (2010) 2048–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Mayorca-Guiliani AE, Madsen CD, Cox TR, Horton ER, Venning FA, Erler JT, ISDoT: in situ decellularization of tissues for high-resolution imaging and proteomic analysis of native extracellular matrix, Nat. Med 23 (7) (2017) 890–898. [DOI] [PubMed] [Google Scholar]
- [45].Mayorca-Guiliani AE, Willacy O, Madsen CD, Rafaeva M, Heumuller SE, Bock F, Sengle G, Koch M, Imhof T, Zaucke F, Wagener R, Sasaki T, Erler JT, Reuten R, Decellularization and antibody staining of mouse tissues to map native extracellular matrix structures in 3D, Nat. Protoc 14 (12) (2019) 3395–3425. [DOI] [PubMed] [Google Scholar]
- [46].Stearns-Reider KM, Hicks MR, Hammond KG, Reynolds JC, Maity A, Kurmangaliyev YZ, Chin J, Stieg AZ, Geisse NA, Hohlbauch S, Kaemmer S, Schmitt LR, Pham TT, Yamauchi K, Novitch BG, Wollman R, Hansen KC, Pyle AD, Crosbie RH, Myoscaffolds reveal laminin scarring is detrimental for stem cell function while sarcospan induces compensatory fibrosis, NPJ. Regen. Med 8 (1) (2023) 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Tan YH, Helms HR, Nakayama KH, Decellularization strategies for regenerating cardiac and skeletal muscle tissues, Front. Bioeng. Biotechnol 10 (2022) 831300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Crapo PM, Gilbert TW, Badylak SF, An overview of tissue and whole organ decellularization processes, Biomaterials 32 (12) (2011) 3233–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Gilpin A, Yang Y, Decellularization strategies for regenerative medicine: from processing techniques to applications, Biomed. Res. Int 2017 (2017) 9831534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Baptista PM, Siddiqui MM, Lozier G, Rodriguez SR, Atala A, Soker S, The use of whole organ decellularization for the generation of a vascularized liver organoid, Hepatology 53 (2) (2011) 604–617. [DOI] [PubMed] [Google Scholar]
- [51].Hussein KH, Park KM, Ghim JH, Yang SR, Woo HM, Three dimensional culture of HepG2 liver cells on a rat decellularized liver matrix for pharmacological studies, J. Biomed. Mater. Res. B Appl. Biomater 104 (2) (2016) 263–273. [DOI] [PubMed] [Google Scholar]
- [52].Naba A, Clauser KR, Ding H, Whittaker CA, Carr SA, Hynes RO, The extracellular matrix: tools and insights for the “omics” era, Matrix Biol. 49 (2016) 10–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Shao X, Taha IN, Clauser KR, Gao Y, Naba A, MatrisomeDB: the ECM-protein knowledge database, Nucl. Acid. Res 48 (D1) (2019) D1136–D1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Caceres S, Cuellar C, Casar JC, Garrido J, Schaefer L, Kresse H, Brandan E, Synthesis of proteoglycans is augmented in dystrophic mdx mouse skeletal muscle, Eur. J. Cell Biol 79 (3) (2000) 173–181. [DOI] [PubMed] [Google Scholar]
- [55].Chen YW, Zhao P, Borup R, Hoffman EP, Expression profiling in the muscular dystrophies: identification of novel aspects of molecular pathophysiology, J. Cell Biol 151 (6) (2000) 1321–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Fadic R, Mezzano V, Alvarez K, Cabrera D, Holmgren J, Brandan E, Increase in decorin and biglycan in Duchenne Muscular Dystrophy: role of fibroblasts as cell source of these proteoglycans in the disease, J. Cell Mol. Med 10 (3) (2006) 758–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Lorts A, Schwanekamp JA, Baudino TA, McNally EM, Molkentin JD, Deletion of periostin reduces muscular dystrophy and fibrosis in mice by modulating the transforming growth factor-beta pathway, Proc. Natl. Acad. Sci. u S. a 109 (27) (2012) 10978–10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Vanhoutte D, Schips TG, Kwong JQ, Davis J, Tjondrokoesoemo A, Brody MJ, Sargent MA, Kanisicak O, Yi H, Gao QQ, Rabinowitz JE, Volk T, McNally EM, Molkentin JD, Thrombospondin expression in myofibers stabilizes muscle membranes, Elife 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Zanotti S, Negri T, Cappelletti C, Bernasconi P, Canioni E, Di Blasi C, Pegoraro D, Angelini C, Ciscato P, Prelie A, Mantegazza R, Morandi L, Mora M, Decorin and biglycan expression is differentially altered in several muscular dystrophies, Brain 128 (11) (2005) 2546–2555. Pt. [DOI] [PubMed] [Google Scholar]
- [60].Naba A, Clauser KR, Hoersch S, Liu H, Carr SA, Hynes RO, The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices, Mol. Cell Proteom 11 (4) (2012). M111 014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Demonbreun AR, Allen MV, Warner JL, Barefield DY, Krishnan S, Swanson KE, Earley JU, McNally EM, Enhanced muscular dystrophy from loss of dysferlin is accompanied by impaired annexin a6 translocation after sarcolemmal disruption, Am. J. Pathol 186 (6) (2016) 1610–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Wallace GQ, McNally EM, Mechanisms of muscle degeneration, regeneration, and repair in the muscular dystrophies, Annu. Rev. Physiol 71 (2009) 37–57. [DOI] [PubMed] [Google Scholar]
- [63].Subramanian A, Wayburn B, Bunch T, Volk T, Thrombospondin-mediated adhesion is essential for the formation of the myotendinous junction in Drosophila, Development 134 (7) (2007) 1269–1278. [DOI] [PubMed] [Google Scholar]
- [64].Brody MJ, Vanhoutte D, Schips TG, Boyer JG, Bakshi CV, Sargent MA, York AJ, Molkentin JD, Defective flux of thrombospondin-4 through the secretory pathway impairs cardiomyocyte membrane stability and causes cardiomyopathy, Mol. Cell Biol 38 (14) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Subramanian A, Schilling TF, Thrombospondin-4 controls matrix assembly during development and repair of myotendinous junctions, Elife 3 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Li Y, Li J, Zhu J, Sun B, Branca M, Tang Y, Foster W, Xiao X, Huard J, Decorin gene transfer promotes muscle cell differentiation and muscle regeneration, Mol. Ther 15 (9) (2007) 1616–1622. [DOI] [PubMed] [Google Scholar]
- [67].Hamilton DW, Functional role of periostin in development and wound repair: implications for connective tissue disease, J. Cell Commun. Signal 2 (1–2) (2008) 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Demonbreun AR, Rossi AE, Alvarez MG, Swanson KE, Deveaux HK, Earley JU, Hadhazy M, Vohra R, Walter GA, Pytel P, McNally EM, Dysferlin and myoferlin regulate transverse tubule formation and glycerol sensitivity, Am. J. Pathol 184 (1) (2014) 248–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Nagy N, Nonneman RJ, Llanga T, Dial CF, Riddick NV, Hampton T, Moy SS, Lehtimaki KK, Ahtoniemi T, Puolivali J, Windish H, Albrecht D, Richard I, Hirsch ML, Hip region muscular dystrophy and emergence of motor deficits in dysferlin-deficient Bla/J mice, Physiol. Rep 5 (6) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Terrill JR, Radley-Crabb HG, Iwasaki T, Lemckert FA, Arthur PG, Grounds MD, Oxidative stress and pathology in muscular dystrophies: focus on protein thiol oxidation and dysferlinopathies, FEBS. J. 280 (17) (2013) 4149–4164. [DOI] [PubMed] [Google Scholar]
- [71].Hogarth MW, Defour A, Lazarski C, Gallardo E, Diaz Manera J, Partridge TA, Nagaraju K, Jaiswal JK, Fibroadipogenic progenitors are responsible for muscle loss in limb girdle muscular dystrophy 2B, Nat. Commun 10 (1) (2019) 2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].de Luna N, Gallardo E, Soriano M, Dominguez-Perles R, de la Torre C, Rojas-Garcia R, Garcia-Verdugo JM, Illa I, Absence of dysferlin alters myogenin expression and delays human muscle differentiation "in vitro", J. Biol. Chem 281 (25) (2006) 17092–17098. [DOI] [PubMed] [Google Scholar]
- [73].Demonbreun AR, Wyatt EJ, Fallon KS, Oosterbaan CC, Page PG, Hadhazy M, Quattrocelli M, Barefield DY, McNally EM, A gene-edited mouse model of limb-girdle muscular dystrophy 2C for testing exon skipping, Dis. Model. Mech 13 (2) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Demonbreun AR, Bogdanovic E, Vaught LA, Reiser NL, Fallon KS, Long AM, Oosterbaan CC, Hadhazy M, Page PG, Joseph PRB, Cowen G, Telenson AM, Khatri A, Sadleir KR, Vassar R, McNally EM, A conserved annexin A6-mediated membrane repair mechanism in muscle, heart, and nerve, JCI. Insight 7 (14) (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].O’Brien JG, Willis AB, Long AM, Kwon J, Lee G, Li FW, Page PG, Vo AH, Hadhazy M, Spencer MJ, Crosbie RH, Demonbreun AR, McNally EM, The super-healing MRL strain promotes muscle growth in muscular dystrophy through a regenerative extracellular matrix, JCI. Insight 9 (3) (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Boeva V, Analysis of genomic sequence motifs for deciphering transcription factor binding and transcriptional regulation in eukaryotic cells, Front. Genet 7 (2016) 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.