Abstract
The generation of new myelin-forming oligodendrocytes in the adult CNS is critical for cognitive function and regeneration following injury. Oligodendrogenesis varies between gray and white matter regions suggesting that local cues drive regional differences in myelination and the capacity for regeneration. However, the layer- and region-specific regulation of oligodendrocyte populations is unclear due to the inability to monitor deep brain structures in vivo. Here, we harnessed the superior imaging depth of three-photon microscopy to permit long-term, longitudinal in vivo three-photon imaging of the entire cortical column and subcortical white matter. We find that cortical oligodendrocyte populations expand at a higher rate in the adult brain than those of the white matter. Following demyelination, oligodendrocyte replacement is enhanced in the white matter, while the deep cortical layers show deficits in regenerative oligodendrogenesis and the restoration of transcriptional heterogeneity. Together, our findings demonstrate that regional microenvironments regulate oligodendrocyte population dynamics and heterogeneity in the healthy and diseased brain.
Introduction
In the central nervous system (CNS), oligodendrocytes produce myelin, which enwraps neuronal axons to increase axonal conduction, fine-tune circuit timing, and provide metabolic support. In 1901, Paul Flechsig proposed that regional myelination is regulated by the functional maturation of neural circuits (fundamental law of myelogenesis1), implying that the cellular microenvironment may shape the generation of oligodendrocytes.
Throughout life, oligodendrocytes are generated via the differentiation of oligodendrocyte precursor cells (OPCs, also called oligodendrocyte progenitor cells). Recent work shows that environmental factors such as brain region and age influence OPCs to become functionally heterogenous via differential ion channel expression2. In line with these findings, the rates of OPC proliferation and differentiation differ between gray and white matter areas3,4 and white matter-derived precursors generate oligodendrocytes with a higher efficiency than gray matter-derived OPCs after transplantation5. Additionally, mature oligodendrocytes make up a heterogeneous cell population with distinct transcriptional subtypes6 that exhibit spatial preference7,8 and differential responses to injury and disease9–11. However, a complete understanding of region-specific regulation of oligodendrogenesis and transcriptional heterogeneity is limited due to the inability to monitor the population dynamics of oligodendrocytes in white matter in the living brain.
In demyelinating diseases, such as multiple sclerosis (MS), death of oligodendrocytes elicits regenerative oligodendrogenesis12 and remyelination that can restore neuronal function13,14. Historically, MS has been regarded as a disease of the white matter; however, recent work shows that MS patients have extensive gray matter lesions that correlate with late-stage disabilities15,16. Interestingly, leukocortical lesions that span white and gray matter regions display distinct pathological hallmarks suggesting that cellular microenvironments of lesions in different brain regions distinctly regulate myelin loss and repair17. Yet, the cellular dynamics of oligodendrocyte population growth, loss and regeneration remain uncharacterized across brain regions.
Advancements in multiphoton microscopy have revolutionized our understanding of cellular dynamics in intact tissues by extending imaging depths four-fold compared to conventional confocal microscopy18. For example, previous studies using longitudinal in vivo two-photon imaging in mouse primary motor cortex14 and somatosensory cortex12,19,20 revealed fundamental aspects of oligodendrogenesis in the healthy and demyelinated brain. However, due to light scattering in brain tissue these studies have been limited to the superficial ~400 microns of the cortical gray matter when extended over multiple months. The recent development and application of three-photon microscopy, which uses a longer excitation wavelength and experiences reduced scattering compared to two-photon, has permitted in vivo imaging depths greater than 1 mm at high resolution21–23. However, the high peak intensity of excitation pulses required for three-photon excitation can result in tissue damage particularly when applied repetitively24. Whether three-photon microscopy can be reliably utilized for long-term volumetric measurements of cellular behavior in the intact brain remains unclear.
Here, we asked whether the dynamics of oligodendrocyte populations differ across cortical and subcortical regions in the context of health and disease. We developed an imaging protocol that permits long-term volumetric in vivo three-photon imaging without tissue damage, cellular reactivity, or perturbation of healthy oligodendrogenesis in the adult mouse brain. This protocol enabled us to track the cellular behavior of over 13,000 individual oligodendrocytes across 2–3 months in health and disease. Longitudinal tracking of individual oligodendrocytes showed that white matter generated more oligodendrocytes per volume compared to gray matter, however, the population growth of oligodendrocytes was accelerated in the gray matter. Following cuprizone-mediated demyelination, we found that the oligodendrocytes and their subpopulations were equivalently reduced across gray and white matter, however, the white matter showed enhanced regenerative oligodendrogenesis and population replacement compared to gray matter regions. Layer-specific analyses revealed that the deep cortical layers had a reduced capacity for regenerative oligodendrogenesis and restoration of the MOL5/6 oligodendrocyte subpopulation following demyelinating injury. Together, these results indicate that regional microenvironments differentially modulate oligodendrogenesis and heterogeneous oligodendrocyte subpopulations to influence myelination and regeneration in the healthy and diseased brain.
Results
In vivo three-photon imaging of cortical gray and white matter
To determine region-specific dynamics of oligodendrocyte generation in a cortical column encompassing the six layers of the cerebral cortex and the subcortical white matter, we built a custom three-photon microscope that enabled imaging to depths of ~1000 μm across multiple months (Methods, Extended Data Fig. 1). We implanted chronic cranial windows over the posterior parietal cortex (PPC) of MOBP-EGFP transgenic mice25 where enhanced green fluorescent protein (EGFP) expression is driven by the MOBP promoter, thereby labeling all mature oligodendrocytes and their associated myelin sheaths in the gray and white matter (Extended Data Fig. 2). Two-photon imaging of the highest clarity cranial windows in MOBP-EGFP transgenic mice three weeks after implantation allowed the visualization of oligodendrocyte cell bodies to ~600 μm from the brain surface (Fig. 1a). Three-photon imaging allowed for the collection of ~400 × 400 × 1050 μm 3D volumes of EGFP-labeled oligodendrocytes encompassing the subcortical white matter (Fig. 1b, Supplementary Video 1). Because multiphoton imaging quality is dependent on a combination of signal generation and out-of-focus background, we used the signal-to-background ratio (SBR) to compare two-photon and three-photon image quality in MOBP-EGFP mice. We found that two-photon imaging had higher SBR of oligodendrocyte cell bodies compared to three-photon imaging in the superficial 100 μm of cortex due to the increased probability of two-photon excitation in regions of minimal scattering24. In contrast, three-photon imaging had increased SBR at depths greater than 400 μm beneath the brain surface compared to two-photon imaging (21.8±3.9 vs. 7.3±3.1 for 401 – 500 μm depth, Fig. 1c). This increase in image quality resulted in the detection of a higher number of oligodendrocytes in layer 5 when compared with two-photon data (78.8±10.2 vs. 41.4±10.7, Fig. 1d), highlighting that two-photon imaging only partially samples the labeled oligodendrocyte population at depths below cortical layers 1–4.
Figure 1 |. Three-photon microscopy enhances the detection of oligodendrocytes and myelin deep in the adult mouse brain.
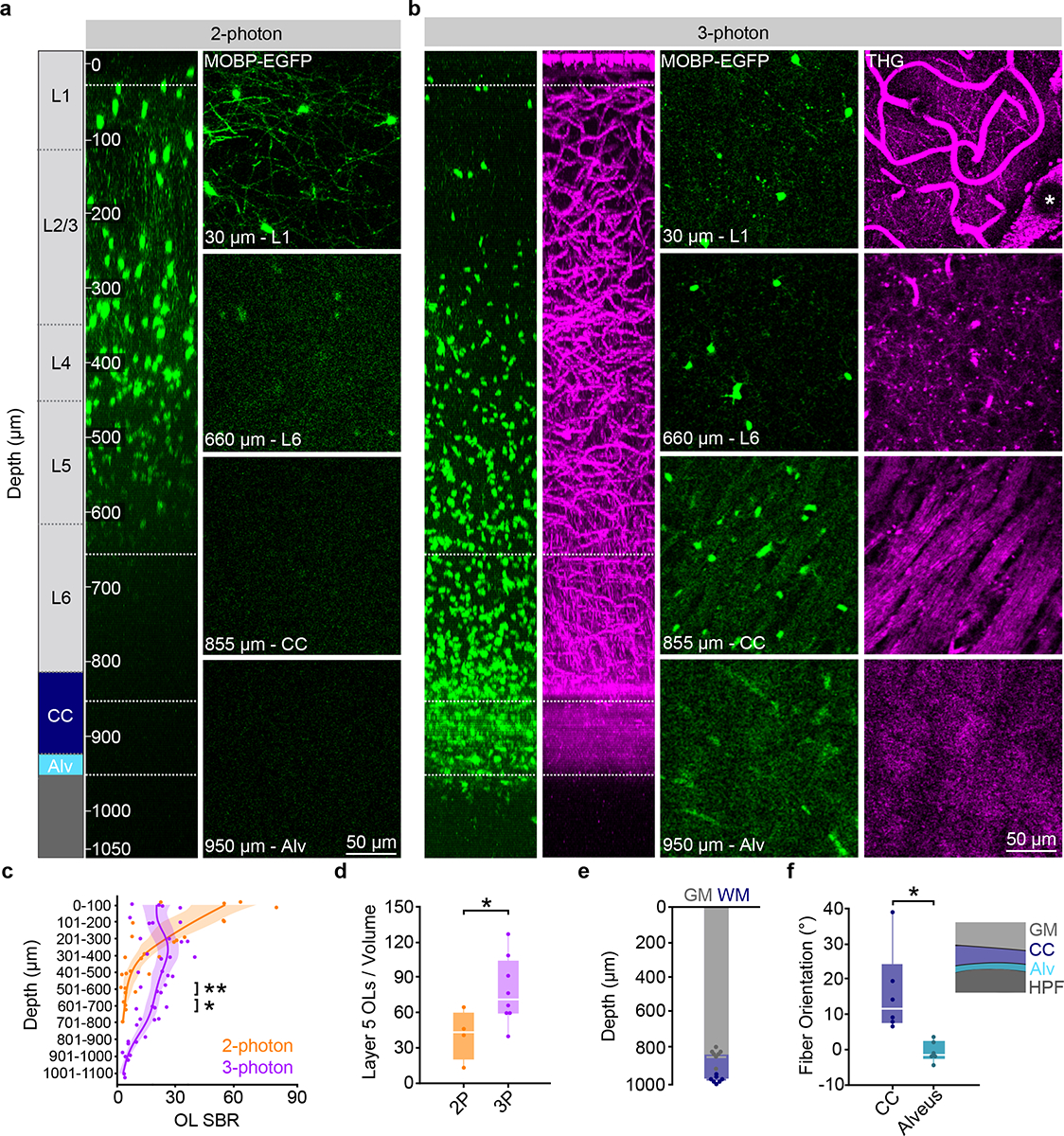
a) xz projection of a 2-photon z stack of oligodendrocytes and myelin in a high-quality chronic cranial window acquired with the Olympus 25X objective. Power was modulated from 8–160 mW. b) xz projection of a 3-photon z stack of MOBP-EGFP and third harmonic generation (THG) signal shows high SBR through the PPC, subcortical corpus callosum (CC), and the alveus of the hippocampus. The average power at the sample was modulated from 0 – 45 mW. c) Two-photon (2P) SBR of oligodendrocyte cell bodies is significantly increased in the superficial 100 μm of cortex compared to three-photon (3P, unpaired, two-tailed Student’s t-test, t(6) = −2.45, p = 0.043), while 3P imaging at depths greater than 400 μm has enhanced SBR (unpaired, two-tailed Student’s t-test, t(6) = 2.92, p = 0.027 (401 – 500 μm), t(6) = 4.72, p = 0.003 (501 – 600 μm), t(3.07) = 4.41, p = 0.021 (601 – 700 μm). d) 2P and 3P allow for the detection of similar numbers of oligodendrocytes in layers 1–4, yet significantly fewer cells are detected with 2P in layer 5 (unpaired, two-tailed t-test, t(10)=2.29, p=0.045). e) Stacked bar chart showing the cortical (gray) and total (blue) imaging depth across the PPC for n=7 mice at 10 weeks of age. f) The corpus callosum (CC) can be differentiated from the alveus by the orientation of THG-positive fibers (t(5.57)=−3.24, p=0.026) for n=6 mice. *p < 0.05, **p < 0.01, n.s., not significant; For growth curves, cubic splines approximate the measure of center and the error bars are 95% confidence intervals, bar plots represent mean±SEM, box plots represent the median, interquartile ranges and minimum/maximum values. For detailed statistics, see Supplementary Table 3.
Three-photon microscopy allowed for the collection of third harmonic generation (THG) signal, which is generated at optical interfaces of refractive index and third-order nonlinear susceptibility26,27 (Supplementary Video 1). Similar to previous studies21,28, we found that THG strongly labeled blood vessels from venules to capillaries, individual myelin sheaths, and myelinated fiber bundles in the subcortical white matter (Fig. 1b). We confirmed that THG-positive sheaths in the superficial PPC colocalized with EGFP+ sheaths in MOBP-EGFP mice and revealed a similar in vivo myelin labeling pattern to Spectral Confocal Reflectance (SCoRe) microscopy29 (Extended Data Fig. 2). Adaptive optics (AO) via image-based Zernike-mode aberration correction with a deformable mirror revealed individual THG-positive myelin sheath cross sections at depths of up to 1000 μm and increased the SBR and axial resolution of oligodendrocytes in the corpus callosum (CC, Extended Data Fig. 3). At depths of 750 – 900 μm, AO correction resulted in a 68% increase in the 3P signal of oligodendrocyte cell bodies and a 202% increase in oligodendrocyte processes. Furthermore, AO optimization allowed for a ~56% reduction in the average power applied to the sample at depth (Extended Data Fig. 3). Additionally, we used THG microscopy to determine the depth from cortical surface to white matter and to further define anatomical landmarks within the white matter. Across eight, ten-week-old mice, we found that the average distance from brain surface to white matter in the PPC was 832±15 μm, while the thickness of the white matter was 135±10 μm, for a total imaging depth of 967±6 μm (Fig. 1e). The subcortical white matter below the posterior parietal cortex consists of myelinated fiber tracts of output axons from the cortex (corpus callosum) and the hippocampus (alveus)30 (Fig. 1f). We found that the orientation of the THG+ fibers allowed for the delineation of the corpus callosum and the alveus of the hippocampal formation (15.3±4.9 vs. −0.4±1.04 degrees, respectively, Fig. 1b,f). Together, these data show that three-photon imaging provides greater imaging depth, higher imaging quality, and an additional label-free signal to study oligodendrocytes and myelination in cortical gray and white matter regions.
Optimization of excitation parameters for long-term three-photon microscopy
Although initial experiments revealed markedly enhanced depth and resolution of three-photon imaging, (Fig. 1), we found that repeated, long-term volumetric imaging using these parameters caused cumulative tissue damage that was visualized using THG imaging (Fig. 2a). This laser-induced damage resulted in oligodendrocyte cell death and a subsequent burst of oligodendrogenesis that could be predicted by a two-fold increase in the THG intensity at the previous time point (Fig. 2b–c). The onset of diffuse 1300 nm excitation-induced oligodendrocyte cell death was more rapid than reported for targeted single-oligodendrocyte ablation with 775 nm excitation31, and was sustained for up to fifty days. The injury-induced oligodendrogenesis had variable dynamics with a peak rate at ~20–40 days post-lesion (Fig. 2b,c). To develop an imaging protocol appropriate for multi-month longitudinal in vivo imaging of cell behavior, we implemented several optical and scanning modifications with the goal of reducing the average power at the sample while maintaining adequate SBR (Extended Data Fig. 4, Methods). Due to mouse-to-mouse variability in the scattering properties of the intact brain, nearly identical exponential power curves resulted in either healthy long-term imaging or lesions with oligodendrocyte cell death (Fig. 2d). Therefore, we measured mouse-specific effective attenuation lengths (EALs) in the adult posterior parietal cortex to estimate the pulse energy delivered at each focal plane as described previously21,28,32 (Extended Data Figure 5; Methods). The pulse energy at the focal plane was calculated using the equation below:
Figure 2 |. Optimization of excitation parameters permits long-term three-photon microscopy without tissue damage.
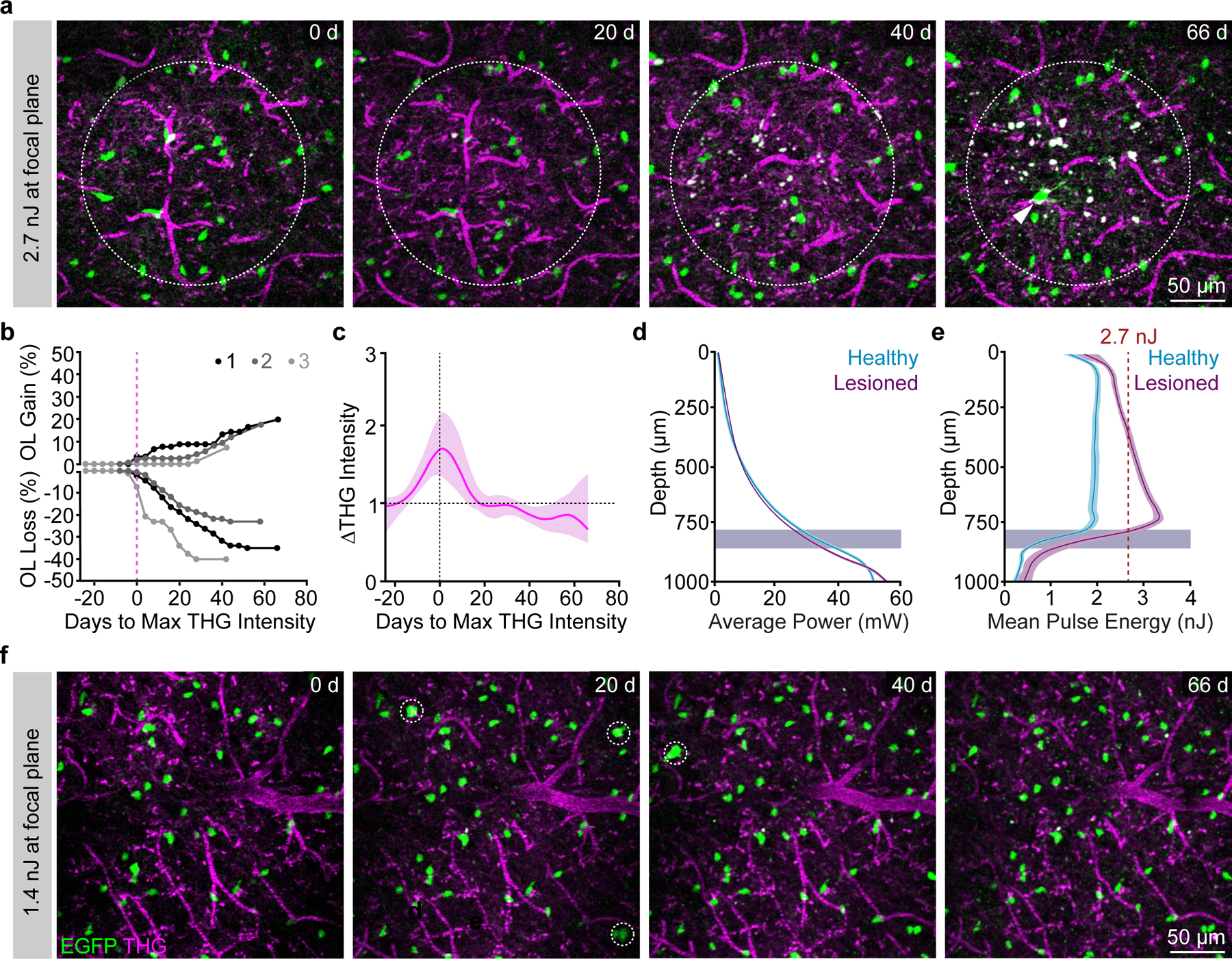
a) Example imaging time points from a mouse exposed to 2.7 nJ pulse energy at the focus. Dotted circle represents the area of the lesion as defined by an increase in THG intensity; white arrowhead shows a newly-generated OL in response to cell ablation. Depth of lesion=594 – 786 μm. b) Cumulative OL population cell loss (%) and cell gain (%) in lesioned regions shows rapid OL cell death that proceeded for up to 40 days following the tissue damage. OL regeneration was rapid and biphasic for lesion 1 (black) but delayed by ~20 days and linear for lesions 2 and 3. Mean OL population loss was 32.9±5.2 %; Mean replacement of the lost OLs was 50.0±16.9%; Mean z-width of the lesions was 149±26.1 μm (mean±SEM). c) Mean intensity of the THG signal over time in the lesion area aligned to the time point of max intensity change for n=3 lesioned mice. d) Similar exponential power vs. depth curves were applied at all time points in lesioned vs. healthy mice that underwent long-term 3P imaging. The excitation laser was blanked on the galvanometer overscan. e) The pulse energy at the focus vs. z-depth was determined for each mouse by calculating the EAL at the first time point. Mouse-specific differences in cortical EAL, and not average power (d), drove the increase in excitation that caused cellular damage. The minimum damage threshold was 2.7 nJ at the focus (red dotted line). Healthy imaged mice had a mean pulse energy at the focus of less than 2 nJ across the cortical depth (cyan). f) Example imaging time points from a healthy mouse with optimized settings and ~1.4 nJ excitation at the focus across the cortex. Note the detection of new oligodendrocytes (white dotted circles) without cell death or changes in THG intensity. For THG, power, and pulse energy curves, cubic splines approximate the measure of center and the error bars are95% confidence intervals. Data in (d-e) are from n=3 mice (Lesioned) and n=10 mice (Healthy long-term imaging).
Where is pulse energy at the focal plane (nJ), is pulse energy at the surface (nJ), Z is depth from the brain surface (μm), EAL is the measured effective attenuation lengths (gray and white matter) for each mouse. Maintaining a relatively constant pulse energy of ~2 nJ across the cortical depth (1.98±0.20 nJ; n = 11 longitudinally-imaged mice) with an absolute maximum threshold of 2.7 nJ (Fig. 2e) was sufficient to maintain high oligodendrocyte SBR and track healthy oligodendrogenesis without oligodendrocyte death (Fig. 2f). The average power modulation curves varied slightly for each mouse due to differences in the thickness and opacity of the chronic cranial window preparation (Extended Data Fig. 5). The average power modulation by depth curves that were applied over multiple months of 3P imaging without damage can be described by the exponential equation:
Where is is Average Power at the Surface (mW) and z is Z-depth from the brain surface (μm) (Extended Data Fig. 5). The entire depth of the cortex, excluding the white matter, could be imaged with less than 30 mW of average power at the surface.
Long-term three-photon imaging does not cause tissue damage
To determine whether these modifications permitted long-term in vivo three-photon imaging without tissue damage, we assessed cellular reactivity, phototoxicity, and molecular stress via immunohistochemistry in tissue from mice that were exposed to three-photon longitudinal imagining for over 2.5 months. Mice were perfused immediately following the final day of in vivo imaging and the 385 × 385 × 1000 μm imaged regions were localized, processed for immunofluorescence, and compared to the contralateral cerebral hemisphere as described previously33,34 (Fig. 3, Extended Data Fig. 6). To assess the sensitivity and specificity of our histological damage readouts, in a separate group of mice, we induced laser lesions by saturating the THG channel at each z-plane during 3D acquisition and perfused the mice 18–24 hours later (Fig. 3a,b, Extended Data Fig. 6). These laser-induced lesions displayed an increase in the diffuse extracellular THG signal and tissue opacity, suggesting that we induced mild nonlinear- and heating-related damage (Fig. 2). First, we found no differences in the cellular reactivity of oligodendrocytes, microglia, or astrocytes between the imaged and contralateral hemispheres in healthy mice that received 2.5 months of optimized long-term 3P imaging (Extended Data Fig. 6). In contrast, positive control tissue, showed a marked increase in the density of microglia and astrocytes in the lesion sites (440.7±45.6 vs. 262.2±18.3 microglia / mm2; 290.8±76.4 vs. 142.4±49.7 astrocytes / mm2, Extended Data Fig. 6). Analyses of the RNA oxidation marker 8-hydroxyguanosine (8-OHG)35 did not reveal significant oxidative stress responses in neurons to either type of laser exposure (Extended Data Fig. 6). Next, we analyzed blood vessel coverage and pericyte density (Lectin-649, vasculature; CD13, pericytes) using automated segmentation36,37 and found no effects of long-term three-photon imaging (Extended Data Fig. 6). Interestingly, blood vessel coverage increased in acutely damaged positive control mice (22.8±3.4 vs. 16.8±2.3%) suggesting a dilation of the vasculature in response to this type of photodamage. Pericyte density was unchanged following laser injury mice, confirming that this type of photodamage does not cause cellular ablation. Together, these results suggest that while long-term, repeated three-photon excitation has the potential to induce phototoxicity, our engineering and operational controls allow for multi-month longitudinal imaging without the induction of cellular reactivity at the tissue level.
Figure 3 |. Long-term three-photon imaging does not perturb damage markers or oligodendrogenesis.
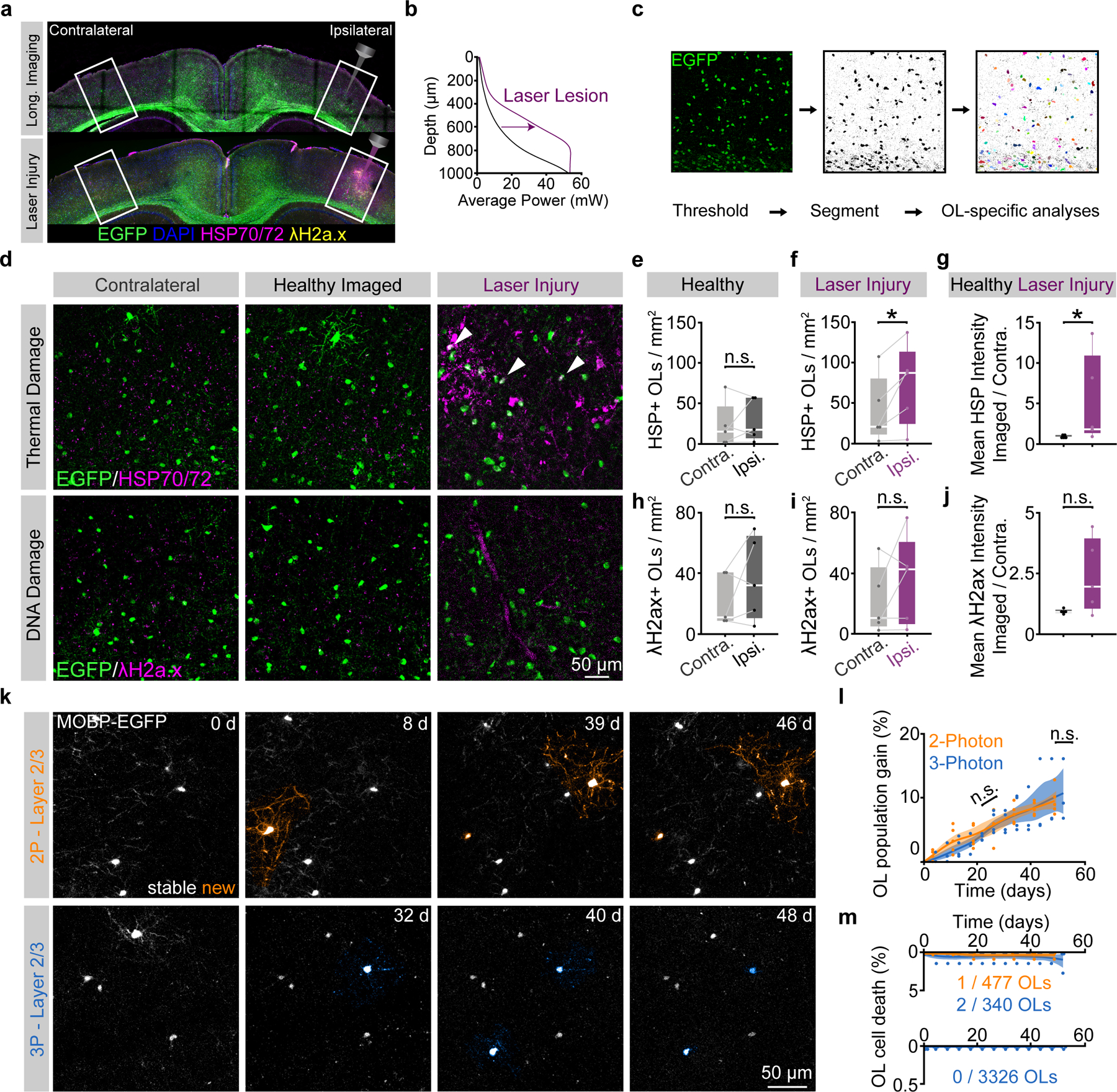
a) Coronal brain section from the imaged (right) and contralateral (left) PPC of a mouse perfused 24 hrs. following long-term 3P imaging (top) stained for MOBP-EGFP, HSP70/72 and gamma-H2ax. Brain section from the imaged and contralateral PPC of a mouse that received volumetric supra-threshold excitation to generate positive control tissue (bottom). b) Power vs. Depth curves for long-term imaging (black) vs. laser-damaged positive control tissue (magenta). c) OL segmentation and cell-specific intensity analyses. d) Images of the contralateral (left), imaged (middle) and laser-induced injury (right) regions in layer 5/6 of the PPC of MOBP-EGFP mice, stained for heat shock proteins 70/72 (top, HSP70/72, thermal damage), and phosphorylated—H2A.X (bottom, gamma-H2A.X, DNA double-strand breaks). e) No difference in HSP70/72–positive OL density between contralateral and ipsilateral hemispheres of healthy mice that underwent optimized, long-term 3P imaging. f) Increased HSP70/72–positive OL density on the ipsilateral side following laser injury (p=0.041). g) The ratio of the imaged:contralateral HSP70/72 mean intensities was increased after laser damage (p=0.037). h) No difference in λH2a.X–positive OL density between the contralateral and imaged hemispheres of healthy 3P mice. i) No difference in λH2a.X-positive OL density between the hemispheres after laser-induced injury tissue. j) No difference in the ratio of the ipsilateral:contralateral λH2a.X mean intensities in either condition. k) Longitudinal imaging of oligodendrogenesis in layer 2/3 with 2P (top, orange) and 3P (bottom, blue). l) Comparison of oligodendrogenesis in the healthy PPC with standard 2P vs. optimized 3P. No differences in OL gain between imaging modalities. m) 3P imaging does not increase mature OL death. L1–3 represents ~0–340 μm depth; L4-CC represents ~341–1000 μm depth. Data in (d-j), n=5 mice per group, (k-l), n=4 mice (3P) and n=5 mice (2P). *p < 0.05, n.s., not significant; for growth curves, cubic splines approximate the measure of center and error bars are 95% confidence intervals, box plots represent the median, interquartile ranges and minimum/maximum values. Data in (f) were compared with two-sided paired Student’s t-test, (g) two-sided Wilcoxon rank sum test; For detailed statistics, see Supplementary Table 3.
Long-term three-photon imaging does not affect healthy oligodendrogenesis or cell death
Next, we examined if long-term three-photon imaging induced molecular stress or cellular behavioral changes specifically in mature oligodendrocytes. We segmented EGFP-positive cell bodies in MOBP-EGFP transgenic tissue from long-term three-photon and acute laser injury mice and counted the density of oligodendrocytes that were positive for HSP70/72, a known marker of thermal stress, and γ-H2A.X, a marker induced by DNA damage (Fig. 3c,d; Methods). The density of HSP70/72- or γ-H2A.X-positive oligodendrocytes between the imaged and contralateral cerebral hemispheres did not differ following long-term three-photon imaging (Fig. 3e,h). Acute laser-injury tissue showed an increase in the number and fluorescence intensity of HSP70/72-positive oligodendrocytes on the exposed hemisphere (71.7±22.6 vs. 39.2±18.6 HSP+ OLs/mm2; 0.91±0.08 (healthy) vs. 5.26±2.46 (laser-injury) Fig. 3f,g), confirming that oligodendrocytes respond rapidly to thermal stress. The density and intensity of γ-H2A.X-positive oligodendrocytes, however, did not reach statistical significance (Fig. 3i–j). To determine whether three-photon longitudinal in vivo imaging affects the dynamics of oligodendrogenesis in the healthy brain, we implanted chronic cranial windows over the posterior parietal cortex in age-matched mice and compared measured rates of oligodendrogenesis in the superifical cortex following two-photon versus three-photon imaging (Fig. 3k). We found no difference in the rate or total oligodendrocyte population gain (%) between groups imaged with different modalities (two-photon = 1.7±0.2 v three-photon = 1.4±0.3% gained per week, Fig. 3l). Finally, we explored whether longitudinal three-photon imaging affected oligodendrocyte survival. Mature oligodendrocytes in the healthy brain seldomly undergo cell death, including after repeated exposure to two-photon excitation19,25,38. In the superficial cortex, we found 1/477 mature oligodendrocytes were lost in the two-photon imaging group compared to 2/340 oligodendrocytes during three-photon imaging (Fig. 3m). In regions below those accessible with longitudinal two-photon imaging (350 – 1000 μm depth), 0/3326 tracked oligodendrocytes were lost during 9.5 weeks of imaging in healthy mice (Fig. 3m). These results show that quantitative measures of health and behavior of oligodendrocytes obtained from long-term in vivo three-photon imaging in age- and region-matched mice are indistinguishable from those acquired via longitudinal two-photon imaging.
Increased population expansion of gray matter oligodendrocytes
We used long-term three-photon in vivo imaging to track oligodendrogenesis across the entire depth of the posterior parietal cortex (gray matter, GM) and the subcortical corpus callosum (white matter, WM) over multiple months in healthy mice (Fig. 4a–d). Similar to past histological studies3,4,39, we found that the white matter generated approximately 3X the number of new oligodendrocytes per volume compared to the cortical gray matter (33.8±5.5 vs. 10.9±1.3 OLs / imaging volume; 4.1±0.6 vs. 1.3±0.35 OLs / per week at 5-weeks, Fig. 4e). This difference in the density of newly-generated oligodendrocytes may be related to regional differences in oligodendrocyte precursor cell (OPC) density and proliferation40,41. To examine OPC proliferation in these regions we delivered ethynyl deoxyuridine (EdU) for one week to P70 mice (identical to onset of three-photon in vivo imaging). Tissue collection and post-hoc immunohistochemistry for Platelet-Derived Growth Factor Receptor Alpha (PDGFRα), a ubiquitous marker of oligodendrocyte precursors, and EdU were performed to assess OPC density and proliferation (Extended Data Fig. 7). We found that the density of OPCs is ~1.4X higher in the white compared to the gray matter of the PPC (248.6±23.8 vs. 181.2±7.7 OPCs/mm2). Furthermore, we found that a ~3.5X higher percentage of OPCs were EDU-positive in the white matter compared to the gray matter (51.6±6.6 vs. 14.5±2.5%). Together, these results suggest that region-specific regulation of OPC proliferation and density may support the increased number of adult-generated oligodendrocytes in the white matter.
Figure 4 |. Region-specific differences in oligodendrogenesis correlate with transcriptional heterogeneity.
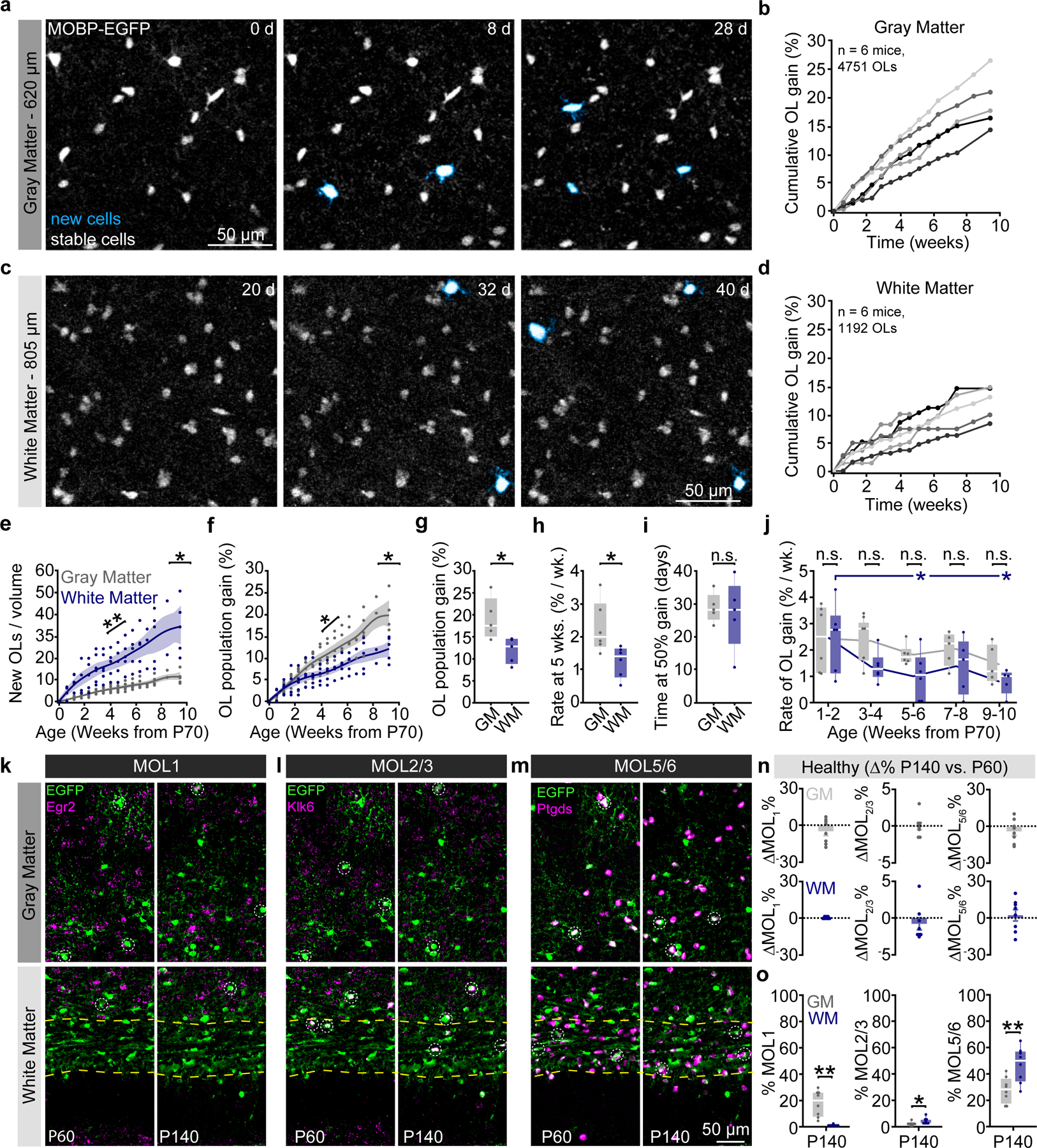
a) Oligodendrogenesis (cyan) over 4 weeks in layer 5 of the PPC. b) Cumulative OL gain curves for individual mice in the entire depth of the GM. c) Oligodendrogenesis (cyan) from weeks 3–6 in the dorsal corpus callosum. d) Cumulative OL gain curves for individual mice in the WM. e) The WM gained substantially more OLs per imaging volume at a faster rate than the GM (p=0.013 for total gain at 66d; p=0.005 for mean rate). f) The GM population experienced greater percentage growth than the WM. Significance asterisks refer to data in (g) and (h). g) Total population gain was increased in the GM vs. the WM (p=0.022). h) The rate of OL gain during the fifth week of imaging (~P100) was increased in the GM vs. WM (p=0.020). i) The time at half-maximum of the mechanistic growth curves did not differ between GM and WM. j) The rate of WM population gain decreased more rapidly with age than in the GM (p=0.037, Weeks 5–6 vs. 1–2; p=0.022, Weeks 9–10 vs. 1–2). k-m) Images of MOL1+ (EGFP/Egr1+), MOL2/3+ (EGFP/Klk6+), and MOL5/6+ (EGFP/Ptgds+) oligodendrocyte populations in the PPC and WM at P60 (left) and P140 (right). n) No age-related changes in the population proportions of OL subtypes between P60 and P140. o) Differences in transcriptional heterogeneity between the GM and WM. MOL1+ OLs represented a higher proportion of the OL population in the GM vs. WM (p=0.002), while MOL2 and MOL5/6-OLs were more abundant in the WM (p=0.018, MOL2/3; p=0.005, MOL5/6). Data in h,i were calculated from the mechanistic growth curves. Data in a–f,h,j represent n=6 mice / group, g,i, n=5 mice, n–o represent n=6 mice (P60) and n=8 mice (P140), two sections per mouse. *p<0.05, **p<0.01, n.s., not significant; for growth curves, cubic splines approximate the measure of center and error bars are 95% confidence intervals; box plots represent the median, interquartile ranges and the minimum/maximum values. Line plots connect the mean at each time point. Statistical tests were unpaired, two-sided Student’s t-tests for equal or unequal variance (e,g,h,o), Dunnett’s multiple comparisons with control (j), or two-sided Wilcoxon rank sum (o). For detailed statistics, see Supplementary Table 3.
In vivo imaging permits a unique perspective to evaluate changes in cellular behavior at the population level. We utilized this long-term approach to determine how the population of mature oligodendrocytes in gray and white matter regions changed over 2.5 months in the adult brain (Fig. 4f, Methods). In contrast to regional differences in the densities of newly-generated oligodendrocytes (Fig. 4e), the oligodendrocyte population in the adult gray matter expands by a greater percentage than the white matter (Fig. 4f). The total extent of oligodendrocyte population gain was significantly elevated in the healthy gray vs. white matter ((New OLs / Initial OLs) * 100; 19.1±2.1% vs. 12.1±1.3%, Fig. 4g). To examine the temporal dynamics of oligodendrocyte population growth in the adult gray versus white matter, we used exponential mechanistic growth modeling to describe the cell population-based decline in adult oligodendrogenesis with age42 (Extended Data Fig. 8). Oligodendrogenesis rates were similar between regions at early time points, but significantly decreased in the white matter by 15 weeks of age (2.3±0.3 vs. 1.3±0.2% per week, Fig. 4h). The time of 50% population growth did not differ between the gray and white matter (26.8±2.2 vs. 24.3±4.4 days post-P70, respectively, Fig. 4i). Binning the raw growth rate data by two-week intervals revealed that the white matter experiences a larger age-dependent decrease in population growth than the gray matter (1.0±0.4 vs. 2.5±0.5, WM, Weeks 5–6 vs. Weeks 1–2; 0.8±0.2 vs. 2.5±0.5, WM, Weeks 9–10 vs. Weeks 1–2, Fig. 4j). Together, these data reveal that gray matter oligodendrocyte population expansion remains elevated later into life than the white matter.
To examine whether regional differences in oligodendrocyte population growth were mirrored by transcriptional heterogeneity in the healthy adult mouse brain, we collected tissue from MOBP-EGFP mice at P60 and P140 for RNA in situ hybridization (ISH) of Early growth response protein 1 (Egr1), Kallikrein-6 (Klk6), and Prostaglandin D2 synthase (Ptgds). These markers represent the distinct subpopulations of mature oligodendrocytes, MOL1, MOL2/3, and MOL5/6, respectively, that differ with development, brain region, and in responses to injury6–8. We found no differences in the relative expression of these oligodendrocyte subtypes between P140 and P60 indicating that heterogeneity in these regions is largely established by P60 (Fig. 4k–n). The representation of oligodendrocyte subpopulations exhibited regional heterogeneity at P140, as the MOL1-positive oligodendrocyte population was significantly larger in the gray matter (17.2±3.4 vs. 0.6±0.2%), the MOL2/3-positive population larger in the white matter (1.8±0.5 vs. 3.8±0.8%), and the MOL5/6-positive population proportionally higher in the white matter (27.4±3.4 vs. 46.4±4.7%) (Fig. 4k–m, o). Analysis of mid-thoracic spinal cord tissue processed in parallel with the brain tissue revealed a similar pattern of spatial heterogeneity to past studies7 (Extended Data Fig. 7). These data show that there is marked regional oligodendrocyte heterogeneity in the posterior parietal cortex and subcortical white matter well into adulthood (P140).
Oligodendrocyte loss does not differ in gray or white matter after cuprizone
Histological studies of regional differences in the cuprizone-mediated demyelination model have produced varied results that differ due to dose, age, time of administration, route of delivery, sex, and/or mouse strain43–46. To examine changes to gray and white matter populations of oligodendrocytes before, during, and after demyelination, we fed ten-week-old MOBP-EGFP mice a diet of 0.2% cuprizone chow for the first 3 weeks of the ten-week imaging timeline (Fig. 5a). Our three-photon imaging parameters permitted longitudinal tracking of individual oligodendrocytes throughout the entire gray matter and the dorsal ~90% of the white matter over the entire imaging time course (Fig. 5b). The analyzed proportion of the white matter did not differ between healthy and cuprizone groups (84±8.0% vs. 86.8±3.5%, Fig. 5c). Long-term three-photon imaging allowed for the simultaneous visualization of the overlapping time courses of cuprizone demyelination and regeneration in the gray and white matter (Fig. 5d–g). Volumetric cell loss mirrored differences in the regional density of mature oligodendrocytes (Fig. 1), as the white matter lost significantly more oligodendrocytes per volume than the gray matter (157.4±37.8 vs. 36.8±6.0 OLs / imaging volume, Fig. 5h). The dynamics of population decline, however, were equivalent in the gray versus white matter (Fig. 5i), implying that the probability of survival of individual oligodendrocytes is similar across regions, as would be expected from systemic delivery of an oligodendrocyte toxin at a saturating dosage. Accordingly, we found no differences in the total oligodendrocyte population loss (75.3±6.3% vs. 75.6±7.4%), rate of oligodendrocyte population loss (9.0±0.7% vs. 9.2±1.0% loss per week), or the timing of the inflection point of loss between the gray and white matter (0.4±1.4 vs. 2.5±2.0 days post cuprizone, Fig. 5j–l). Within-groups analyses of binned rates of demyelination revealed that gray matter oligodendrocyte loss occurs slightly earlier than in the white matter, yet there were no between-groups differences in the rate of oligodendrocyte loss (9.6±1.8 vs. 2.5±0.9% per week, GM, Weeks −2–0 vs. Week −3; 18.9±1.9 vs. 2.5±0.9% per week, GM, Weeks 0–2 vs. Week −3; 22.6±3.1 vs. 2.7±1.8% per week, WM, Weeks 1–2 vs. Week −3, Fig. 5m). RNA ISH experiments showed that the prevalence of MOL1, MOL2/3, and MOL5/6-positive oligodendrocytes decreased across both the gray and white matter at four days post-cuprizone cessation (Fig. 5n–q, see Supplementary Table 3 for detailed statistics). While both surviving and newly-generated oligodendrocytes are present four days following cuprizone cessation (Fig. 5d,f), the majority of oligodendrocytes at this time point are not labeled with MOL1, MOL2/3, or MOL5/6 subpopulations markers. Despite the reduction in oligodendrocyte subpopulation markers, we found that gray and white matter heterogeneity was preserved for MOL1 and MOL2/3 but abolished for MOL5/6 (1.8±0.7 vs. 0.1±0.06%, MOL1; 0.1±0.05 vs. 1.2±0.2%, MOL2/3; 3.1±1.1 vs. 4.9±1.6%, MOL5/6, Fig. 5r). Together, these data show that cuprizone-mediated demyelination results in similar loss of oligodendrocyte populations and transcriptional heterogeneity across the posterior parietal cortex and underlying corpus callosum.
Figure 5 |. Cuprizone affects gray and white matter oligodendrocyte populations similarly.
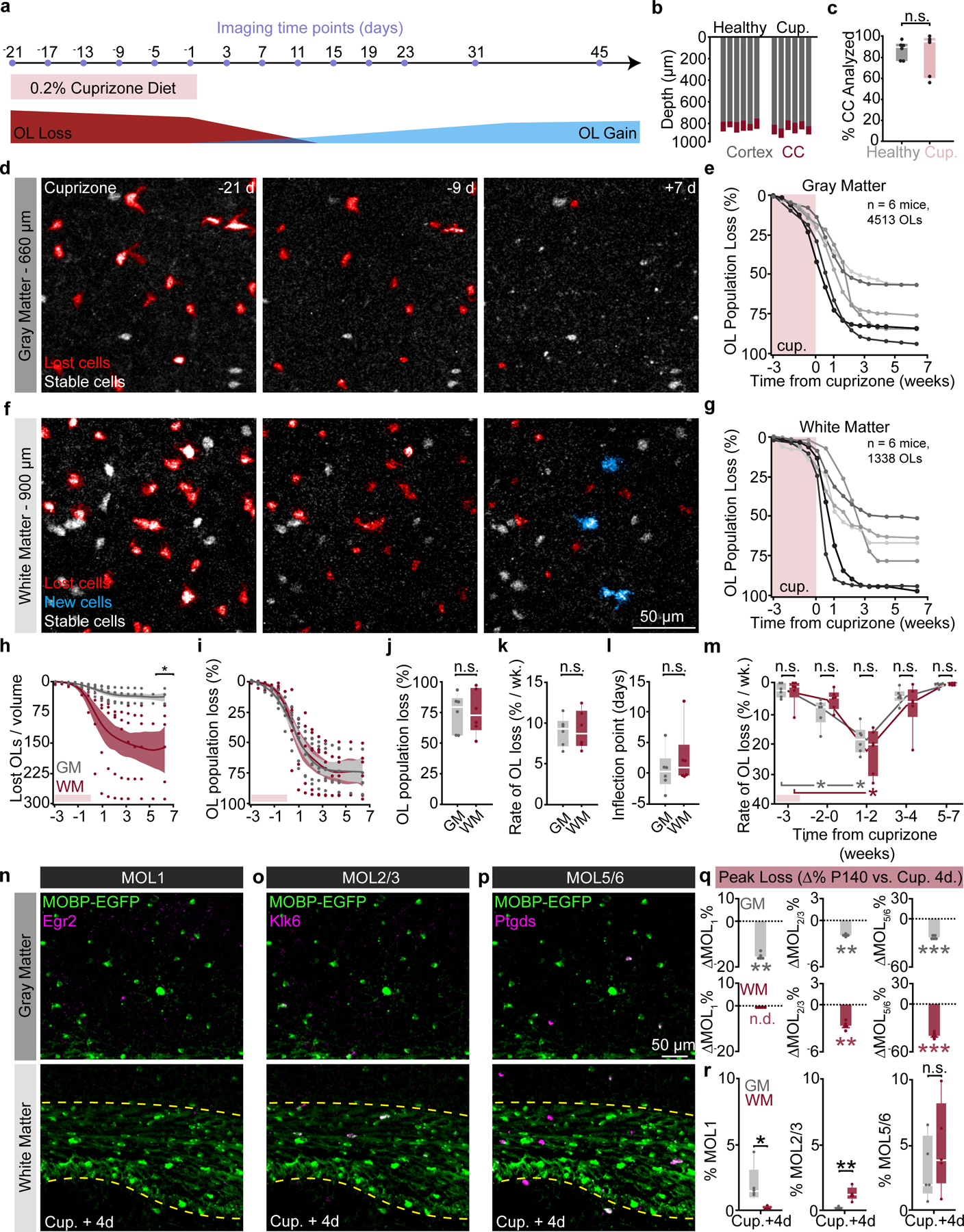
a) 3P imaging timeline to track cuprizone-induced OL loss and gain. b) Depth of PPC and subcortical WM that was imaged over 66 days. c) Percentage of the corpus callosum that was imaged/ analyzed longitudinally did not differ by group. d) Time series of cuprizone-mediated OL loss in the deep cortex. e) Cumulative OL loss (%) over time in the GM for individual mice. f) Time series of cuprizone-mediated OL loss in the WM. g) Cumulative OL loss (%) over time in the WM for individual mice. h) The number of OLs lost per 350×350×60μm imaging volume was lower in the GM vs. WM (p=0.024). i) OL population loss dynamics are similar across regions. j) No difference in the total % loss for the GM vs. WM. k) No difference in the rate of % loss for the GM vs. WM. l) No difference in the inflection point of the population loss curve. m) The rate of GM and WM population loss, binned by 2–3 week intervals. No differences between the GM and WM at each time point. OL loss increased earlier in the GM vs. WM (p=0.018, Weeks −2–0 vs. −3 GM; p=0.018, Weeks 1–2 vs. −3 GM; p=0.018, Weeks 1–2 vs. −3, WM). n-p) Images of the distribution of MOL1, MOL2/3, and MOL5/6 subpopulations in the PPC and WM at 4-days post-cuprizone. q) Cuprizone reduced expression of all three markers in both regions (Healthy P140 to Cup.+4d; p=0.007, MOL1-GM; p=0.038, MOL1-WM; p=0.004, MOL2/3-GM; p=0.004, MOL2/3-WM; p=0.0001, MOL5/6-GM; p<0.0001, MOL5/6-WM). r) GM vs. WM transcriptional heterogeneity at four days post-cuprizone (MOL1, p=0.011; MOL2/3, p=0.0097; MOL5/6, n.s.). Data in k,l were calculated from the Gompertz curve parameters. Data in a-m represent n=6 mice/group, q-r represent n=5 mice (Cup. +4d) and n=8 mice (Healthy P140), two sections per mouse. *p<0.05, **p<0.01, ***p<0.001, n.s., not significant; for growth curves, cubic splines approximate the measure of center and error bars are 95% confidence intervals, box plots represent the median, interquartile ranges and the minimum/maximum values; Line plots connect the mean at each time point. Statistical tests were unpaired, two-sided Student’s t-tests for equal/unequal variance (h,j,k,q), Steel multiple comparisons with control (m) or two-sided Wilcoxon rank sum (l,q,r). For detailed statistics, see Supplementary Table 3.
Enhanced oligodendrocyte regeneration in white compared to gray matter
Since cuprizone treatment affects oligodendrocyte populations in gray and white matter equivalently (see Fig. 5i,q), we compared the regenerative capacity of gray and white matter regions following demyelination. We analyzed the region-specific extent, rate, and timing of oligodendrogenesis following cuprizone treatment (Fig. 6a–d). Similar to previous fate-mapping studies46, we found that there were more newly-generated oligodendrocytes in the white matter compared to the gray matter following cuprizone administration for all mice analyzed (Fig. 6e, 123±47.3 vs. 13.3±2.0 OLs/imaging volume). Next, we examined the restoration of the oligodendrocyte populations in these brain regions. We found that, even when normalizing to the initial population density, the subcortical white matter showed enhanced replacement of lost oligodendrocytes over the 2.5-month imaging timeline compared to gray matter (Fig. 6f). Forty-five days following removal of cuprizone, the total oligodendrocyte replacement was 37.6±3.6% vs. 68.0±11.3% for gray versus white matter, respectively (Fig. 6g). To analyze the temporal dynamics of post-cuprizone regeneration, we used three-parameter logistic modeling as described previously14,47 (Extended Data Fig. 8). Analyzing the Gompertz growth curves of regeneration revealed that the mean rate of oligodendrocyte population replacement per week during remyelination was 5.6±0.6% versus 11.1±1.9% for gray versus white matter (Fig. 6h). Yet, the inflection point of oligodendrocyte population growth did not differ suggesting that the time course of the replacement response was similar in both regions (Fig. 6f,i). This effect was further supported by analyzing the binned oligodendrocyte growth data in which both the gray and white matter were elevated at the 3–4 week post-cuprizone time bin (8.2±1.1 vs. 3.0±0.5% per week, gray matter, Weeks 3–4 vs. Week 7; 18.1±5.1 vs. 4.5±1.3% per week, WM, Weeks 3–4 vs. Week 7) yet with a significantly higher peak in the white matter (18.1±5.1 vs. 8.2±1.1% per week, Fig. 6j). Finally, since the gray matter replaces 37.6% of lost oligodendrocytes compared to 68% replacement in the white matter, we estimated the time required to reach 100% replacement for each region. Assuming linear growth after 7 weeks post-cuprizone, ~14 weeks would be required to replace the lost oligodendrocytes in the gray matter whereas the white matter would require ~7 weeks, highlighting the reduced rate of regenerative oligodendrogenesis in the gray matter. When we examined recovery of oligodendrocyte transcriptional subpopulations via RNA ISH experiments at 7 weeks post-cuprizone cessation (age-matched to final imaging time point) we found that the MOL1 and MOL2/3 populations returned to healthy proportions in gray matter, while MOL5/6 remained decreased in both regions (−14.9±2.0%, GM; −22.6±4.2%, WM, Fig. 6n). Interestingly, the pattern of healthy gray versus white matter heterogeneity for these markers was nearly restored to healthy conditions (Fig. 6o), despite the decreased prevalence of MOL5/6 at this time point (GM vs. WM; 15.8±3.3 vs. 1.5±0.3%, MOL1; 1.2±0.4 vs. 4.1±1.2%, MOL2/3; 12.6±2.0 vs. 24.0±4.2%, MOL5/6). Together, these data show that the white matter restores the lost oligodendrocyte population more effectively than the gray matter, and bulk gray and white matter regions re-establish spatial distributions of oligodendrocyte subtypes following a demyelinating injury.
Figure 6 |. Oligodendrocyte replacement is enhanced in the white matter and partially restores regional heterogeneity.
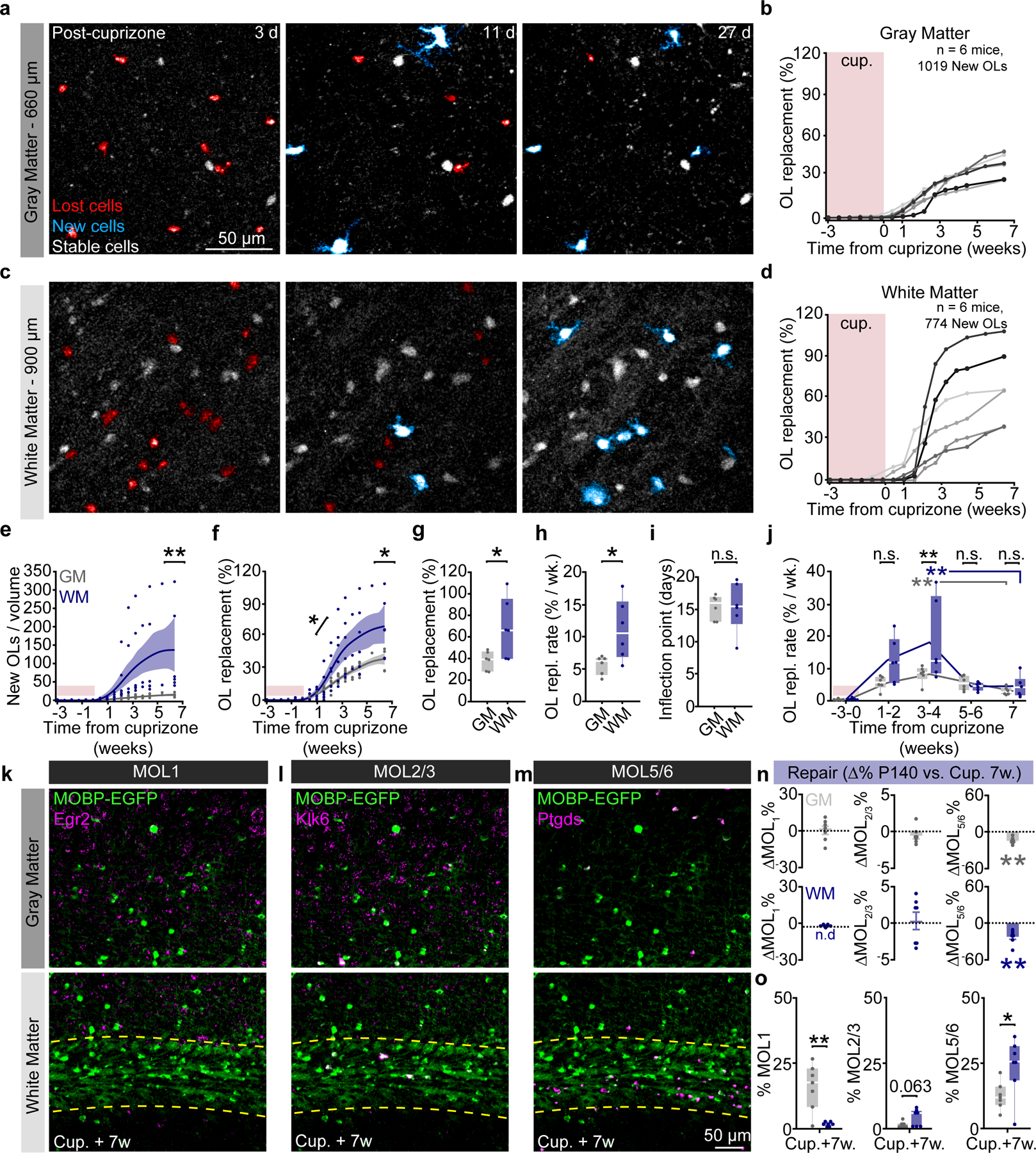
a) Timelapse of 24-day period after cuprizone cessation showing OL loss (red), followed by OL regeneration (cyan) in the deep PPC. b) Cumulative OL replacement (%-gain normalized to %-loss) across the cortex for individual mice. c) Same time points and mouse as in (a) in the corpus callosum. d) Cumulative OL replacement for individual mice in the WM. e) Density of newly-generated OLs was increased in the WM vs. GM (p=0.005). Note the high variability without normalization to OL loss. f) Cumulative OL replacement over time in WM vs. GM. Significance asterisks represent data in (g) and (h). g) OL replacement (%) is significantly increased at 45 days post-cuprizone in the WM (p=0.043). h) Rate of replacement is enhanced in the WM vs. GM (p=0.043). i) No difference in the timing of OL replacement between GM and WM. j) The rate of GM and WM population replacement binned by 1–3-week intervals with respect to cuprizone treatment. OL replacement rate is increased in the WM vs. GM at 3–4-weeks post-cuprizone (p=0.0009, black asterisks). Both the WM and the GM are significantly increased at the 3–4-week phase compared to the 7-week plateau phase (p=0.002, Weeks 3–4 vs. 7, GM; p=0.0052 Weeks 3–4 vs. 7, WM). k-m) Images of the distribution of MOL1, MOL2/3, and MOL5/6 OL populations in the PPC and WM at 7 weeks post-cuprizone. n) Change in the population proportions from healthy P140 to 7 weeks post-cuprizone (p=0.003, MOL5/6 GM; p=0.007, MOL5/6 WM). o) GM vs. WM differences in transcriptional heterogeneity 7-weeks post-cuprizone (p=0.005, MOL1; p=0.063, MOL2/3; p=0.032, MOL5/6) Data in h,i were calculated from the parameters of the Gompertz growth curves. Data in a-j represent n=6 mice per group, n-o represent n=7 mice (Cup.+7w.) and n=8 mice (Healthy P140), two sections per mouse. *p<0.05, **p<0.01, n.s., not significant; for growth curves, cubic splines approximate the measure of center and error bars are 95% confidence intervals; box plots represent the median, interquartile ranges and minimum/maximum values; line plots connect the mean at each time point. Statistical tests were unpaired, two-sided Student’s t-tests for equal or unequal variance (g, h, n, o), Dunnett’s multiple comparisons with control (within-groups, j), Two-way ANOVA with Bonferroni correction for piecewise multiple comparisons (between-groups, j) and two-sided Wilcoxon rank sum (e, n). For detailed statistics, see Supplementary Table 3.
Layer-dependent regulation of oligodendrocyte growth, loss, and regeneration
Recent studies using longitudinal in vivo two-photon imaging revealed layer-specific differences in cortical oligodendrogenesis following motor learning14 and cuprizone-mediated demyelination19, yet these studies were limited to the superficial ~300–400 μm of cortex. We utilized the enhanced imaging depth of three-photon microscopy to assess layer-specific differences in oligodendrocyte population dynamics across cortical and subcortical layers in the healthy brain and following cuprizone-mediated demyelination. Based on the myelo-, neuronal, axonal, and synaptic anatomy of the adult posterior parietal cortex (Fig. 7a), we divided gray and white matter regions into layers 1–3 (superficial cortex), layer 4 (thalamic input layer to the PPC), layer 5–6 (deep cortex), and the subcortical corpus callosum (CC). We assessed layer-dependent differences in oligodendrocyte generation and transcriptional subpopulations across four experimental time points spanning P60 to P140 in healthy and cuprizone-treated mice (Fig. 7b, Extended Data Fig. 9). In healthy mice, we found that there was increased oligodendrogenesis in L4 compared to the CC, suggesting that enhanced oligodendrogenesis in this specific cortical microenvironment drives increased population growth in the gray versus white matter (2.6±0.2 vs. 1.4±0.2% per week, Fig. 7c–d, Fig. 3). Analyses of layer-specific oligodendrocyte subpopulations showed the following patterns in the healthy posterior parietal cortex: The proportion of MOL1-positive oligodendrocytes declined with cortical depth and were largely absent from the white matter (Fig. 7e). The proportion of MOL2/3-positive oligodendrocytes increased with depth and, within the cortex, were only detected in L5–6 (Fig. 7f). The white matter had increased proportion of MOL5/6 oligodendrocytes compared to the gray matter (Fig. 7g, see Supplementary Table 3 for detailed statistics). We found that cuprizone-induced oligodendrocyte loss did not differ across cortical or subcortical layers (Fig. 7h,i). The proportion of MOL1-postive oligodendrocytes was increased in layers 1–3 compared to the corpus callosum (Fig. 7j), MOL2/3-positive oligodendrocytes were increased in the corpus callosum compared to all cortical layers (Fig. 7k), while the proportion of MOL5/6 oligodendrocytes did not differ across cortical and subcortical layers (Fig. 7l, see Supplementary Table 3 for detailed statistics). When examining oligodendrocyte regeneration seven weeks post-cuprizone removal, we found that the deep cortex (L5–6) had a decreased rate of oligodendrocyte replacement compared to the CC (5.6±0.56 vs. 11.2±1.9% per week, Fig. 7m,n). At 7 weeks post-cuprizone, the proportion of MOL1-postive oligodendrocytes was increased in layer 4 compared to the CC (Fig. 7o), MOL2/3-positive oligodendrocytes were increased in layers 5/6 and the CC compared to layers 1–4 (Fig. 7p), and the proportion of MOL5/6 oligodendrocytes in L5–6 was decreased compared to the CC (10.6±2.2 vs. 24.0±4.2%; Fig. 7q, see Supplementary Table 3 for detailed statistics).
Figure 7 |. Layer-specific differences in oligodendrogenesis correlate with changes in molecular heterogeneity.
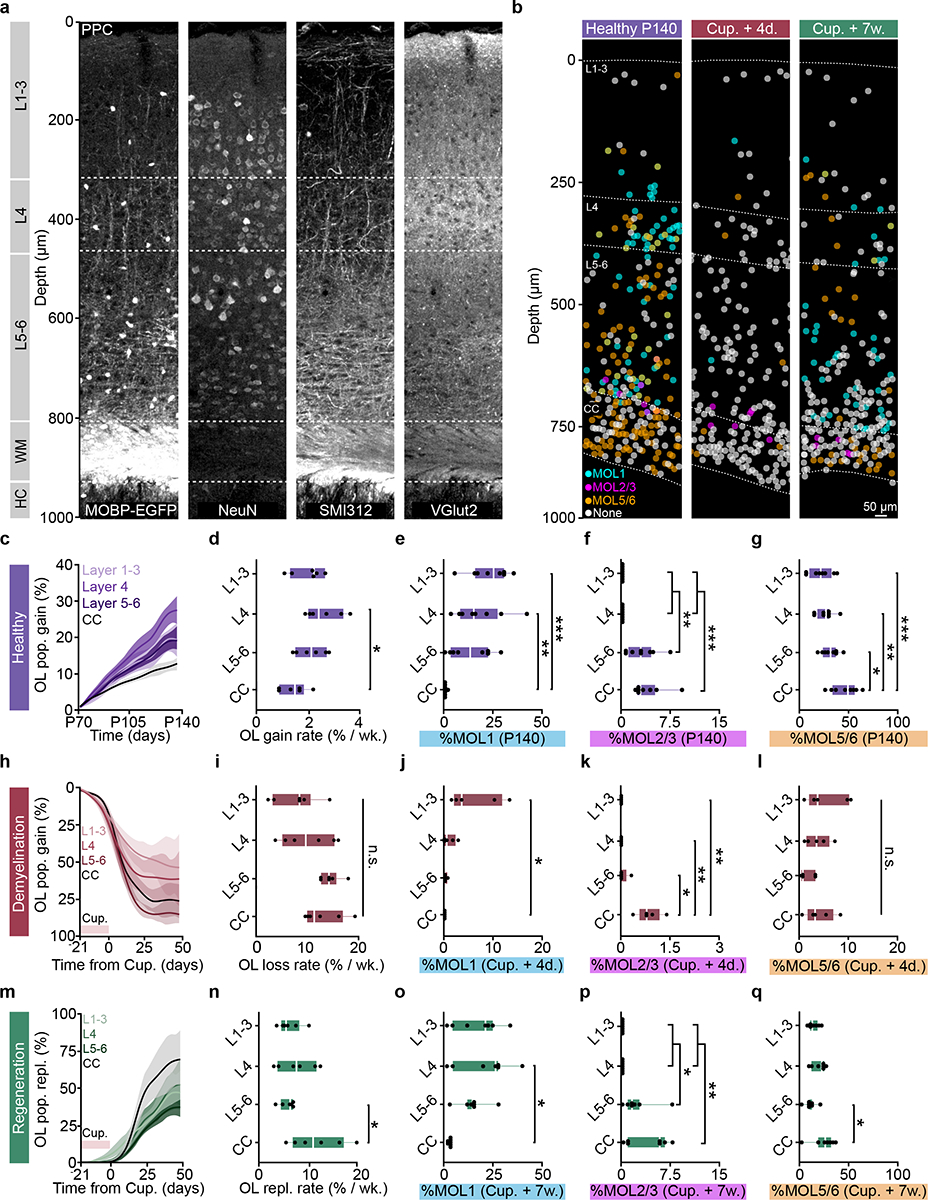
a) Myelo-, neuronal, axonal, and thalamic input architecture of the PPC and subcortical WM. Note the Vglut2-positive layer 4.b) Spatial map of MOL1 (cyan), MOL2/3(magenta), and MOL5/6 (orange) OLs across the PPC and WM at P140, four days, and seven weeks post-cuprizone removal (4 d.p.c., 7 w.p.c.) ~50% of the OLs did not express any of the markers (None, gray). c) Healthy OL growth curves plotted by sub-region across the cortical and subcortical depth. d) Layer-specific differences in healthy oligodendrogenesis (p=0.012). e) Layer-specific differences in the percentage of OLs expressing Egr1 (p=0.0004, L1–3 vs. CC; p=0.006, L4 vs. CC). f) Layer-specific differences in the percentage of OLs expressing Klk6 (p=0.0034, L4 vs. L5–6; p=0.0034, L1–3 vs. L5–6; p=0.0009, L4 vs. CC; p=0.0009, L5–6 vs. CC). g) Layer-specific differences in the percentage of OLs expressing Ptgds (p=0.042, L5–6 vs. CC; p=0.005, L4 vs. CC; p=0.0005, L1–3 vs. CC). h) OL loss curves plotted by subregion across the cortical and subcortical depth. i) No layer-specific differences in cuprizone loss rate. Note increased variability of demyelination in L1–4. j) Layer-specific differences in MOL1 at 4 d.p.c. (p=0.016). k) Layer-specific differences in MOL2/3 at 4 d.p.c. (p=0.022, L5–6 vs. CC; p=0.004, L4 vs. CC; p=0.004, L1–3 vs. CC). l) No layer-specific differences for MOL5/6 at 4 d.p.c.. m) OL replacement curves plotted by sub-region. n) OL replacement is decreased specifically in L5–6 vs. CC (p=0.035). o) Layer-specific differences in MOL1 at 7 w.p.c. (p=0.012). p) Layer-specific differences in MOL2/3 at 7 w.p.c. (p=0.025, L1–3 vs. L5–6; p=0.025, L4 vs. L5–6; p=0.003, L1–3 vs. CC; p=0.003, L4 vs. CC). q) Layer-specific differences in MOL5/6 at 7 w.p.c. (p=0.015). Data in c,d,h,I,m,n represent n=6 mice per group, data in e-g n=8 mice, 2 sections per mouse, data in j-l n=5 mice, 2 sections per mouse, o-q n=7 mice, 2 sections per mouse. *p<0.05, **p<0.01, ***p<0.001, n.s., not significant. For growth curves, cubic splines approximate the measure of center and error bars are 95% confidence intervals, box plots represent the median, interquartile ranges and minimum/maximum values. Statistical tests were One-way ANOVA with Tukey’s HSD (d, e, g, n, o, q) or Kruskal-Wallis with Dunn’s test for multiple comparisons (f, j, k, p). For detailed statistics, see Supplementary Table 3.
Insufficient oligodendrogenesis and regeneration of MOL5/6 subpopulation in deep cortex
To determine the layer-specific capacity to generate heterogeneous oligodendrocyte subpopulations across health and disease, we examined the dynamics of oligodendrogenesis and transcriptional oligodendrocyte subpopulations in healthy and cuprizone-treated mice. Compared to the healthy brain, we found that regenerative oligodendrogenesis following cuprizone treatment was increased by an order of magnitude (17% vs. 0.1–0.65%, Fig. 7d,n). Therefore, we normalized the modeled growth curves to the maximum value of oligodendrocyte gain or replacement to enable between-groups comparisons of healthy vs. regenerative oligodendrogenesis (Fig. 8a; Methods). We found that the scaled total oligodendrocyte population growth (0.3±0.03 vs. 0.5±0.1, Fig. 8a,b) and rate of growth (0.3±0.03 vs. 0.6±0.1, Fig. 7a,c) in L5–6 was decreased compared to healthy conditions, yet the growth curves had equivalent relative inflection points (Fig. 8d). Further analyses of regeneration dynamics revealed layer-dependent differences in response duration (full-width at half-maximum) and a decrease in the integrated response (area under the curve, above healthy) in L5–6 compared to the CC (19.89±2.14 vs. 11.70±2.37 days; 22.8±2.9 vs. 56.2±12.1%, Extended Data Figure 10). Next, we analyzed the loss and restoration of layer-specific oligodendrocyte heterogeneity four days and seven weeks post-cuprizone removal compared to age-matched healthy controls (Fig. 8e). To calculate the overall oligodendrocyte heterogeneity per layer, we determined the number of oligodendrocyte subpopulations that were present at a greater proportion than the minimum value in healthy mice within each layer. Four days following cuprizone removal, oligodendrocyte heterogeneity was reduced in all cortical layers whereas oligodendrocyte heterogeneity in the corpus callosum remain unchanged (Fig. 8e). Seven weeks following cuprizone removal, we found that oligodendrocyte heterogeneity across all layers was restored to healthy levels (Fig. 8e). Following regeneration, we found that layer-specific MOL1- and MOL2/3-positive subpopulations were indistinguishable from healthy controls (Fig. 8f,g). However, while MOL5/6 oligodendrocytes were regenerated in layers 1–3 and 4, we found this subpopulation remained decreased in layers 5–6 and the white matter compared to healthy conditions (10.6±2.2% vs. 31.3±3.1%, L5–6; 24.0±4.2% vs. 46.4±4.7%, CC; Fig. 8h). In the corpus callosum MOL5/6 oligodendrocytes were increased at seven weeks compared to four days following cuprizone removal (24.0±4.2% vs. 4.9±1.6%) indicating ongoing, albeit incomplete, regeneration of this subpopulation. In contrast, the proportion of MOL5/6 oligodendrocytes in layers 5/6 did not differ at seven weeks compared to four days post-cuprizone (10.6±2.2% vs. 1.9±0.8%) indicating a deficit in MOL5/6 regeneration in layers 5/6 following cuprizone-mediate demyelination. Together these results show that deep cortical layers 5–6 have pronounced deficits in regenerative oligodendrogenesis and restoration of the MOL5/6-positive oligodendrocyte subpopulation compared to other cortical and subcortical layers.
Figure 8 |. Decreased oligodendrogenesis and recovery of molecular identity in deep layers 5–6.
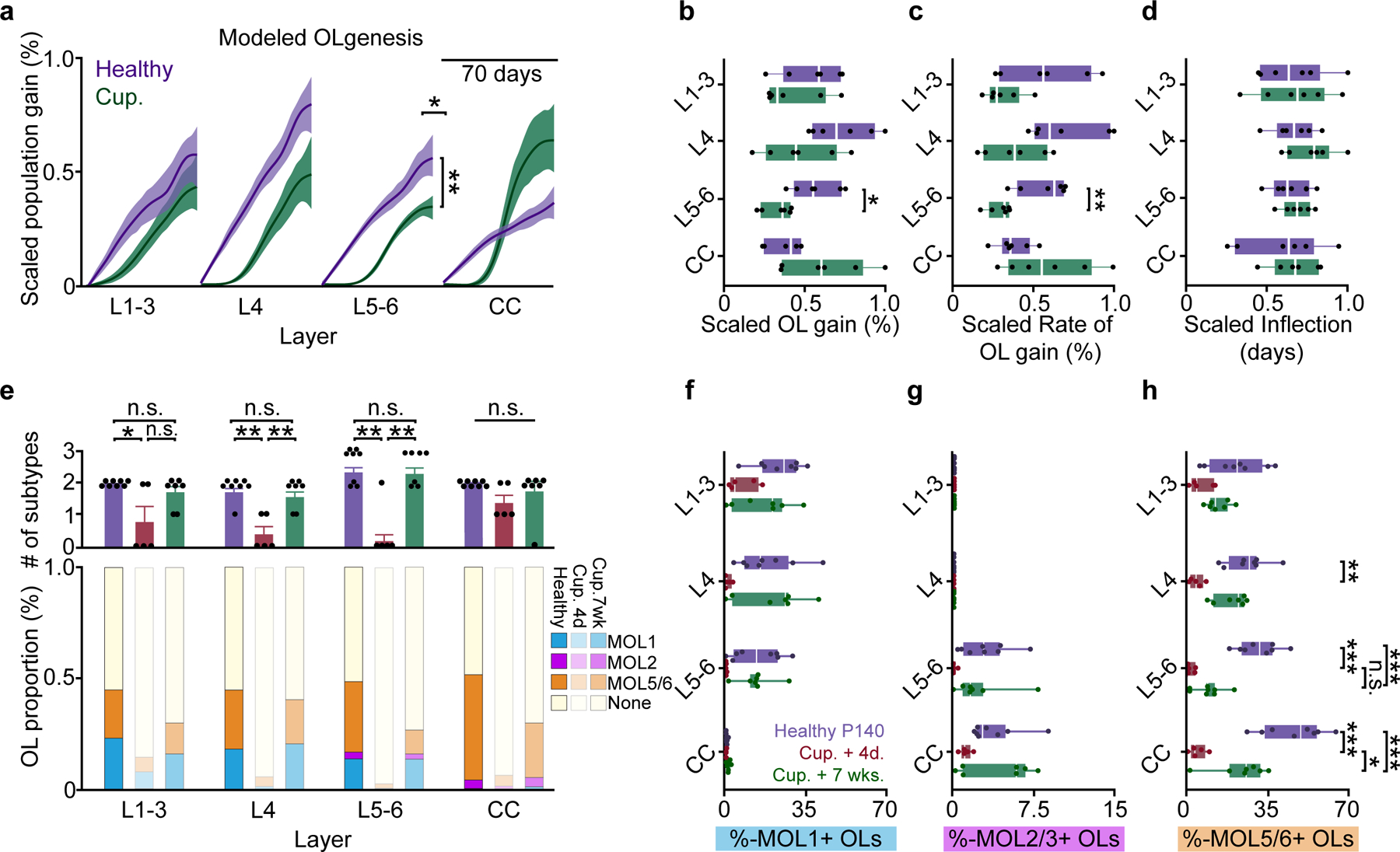
a) Modeled growth curves plotted by cortical sub-region and scaled to the maximum % gain or replacement to enable comparisons of healthy vs. regenerative oligodendrogenesis. Significance asterisks are related to data in (b–c). b) Decrease in scaled oligodendrogenesis in L5–6 post-cuprizone vs. healthy baseline, p=0.006). c) Decrease in scaled oligodendrogenesis rate in L5/6 after cuprizone (p=0.0025) d) No differences in scaled inflection points across regions. e) (Top) Differences in the number of molecular subpopulations represented at Healthy P140, Cup. +4d. and Cup. +7wk.time points (Bottom) Stacked bar chart showing changes in the relative proportion of MOL1, MOL2/3 and MOL5/6 OLs at 7-weeks post-cuprizone treatment. f) No differences in the MOL1-positive OL proportions within individual cortical layers across experimental time points. g) No differences in the MOL2/3-positive proportions within individual cortical layers across experimental time points. h) Differences in the MOL5/6 proportions within cortical layers across experimental time points (Healthy P140 (purple) vs. Cup. +4 days (red); p=0.001, L4; p=0.0001, L5–6; p=0.0001, CC). The proportion of MOL5/6 OLs returned at 7 weeks post-cuprizone only in the CC (Cup.+4d (red) vs. Cup.+7w (green), p=0.012). The MOL5/6 OL proportion returned to Healthy P140 levels in layers 1–4 but remained relatively decreased in L5–6 and the CC (Healthy P140 (purple) vs. Cup.+7wks. (green), p=0.0007, L5–6; p=0.001, CC). Data in a-d represent n=6 mice per group, data in e-h represent n=8 mice, 2 sections per mouse (P140), n=5 mice, 2 sections per mouse (Cup.+4d.) and, n=7 mice, 2 sections per mouse (Cup.+7wks.).*p<0.05, **p<0.01,***p<0.001, n.s., not significant; For growth curves, cubic splines approximate the measure of center and error bars are 95% confidence intervals, box plots represent the median, interquartile ranges and minimum/maximum values, bar plots are mean±SEM. Statistical tests were Two-way ANOVA with Bonferroni correction (between-groups comparisons, a– d), Two-way ANOVA with Tukey’s HSD (all comparisons, f-h), and Kruskal-Wallis with Dunn’s test for joint ranks (e). For detailed statistics, see Supplementary Table 3.
Discussion
The generation of myelin continues throughout life via adult oligodendrogenesis and plays essential roles in cognition and tissue regeneration following injury or disease. Exploring the regulation of oligodendrogenesis in specific brain regions is essential to understand how these processes may be harnessed to promote learning or recovery in the adult CNS. In this study, we empirically determined excitation and scanning parameters to allow for longitudinal in vivo three-photon imaging over multiple months without cellular reactivity or tissue damage. This approach enabled the analysis of oligodendrocyte generation and population dynamics in the healthy and demyelinated posterior parietal cortex and the corpus callosum at the level of individual oligodendrocytes. We confirmed previous findings via fate-mapping and EdU-labeling that the white matter generates substantially more new oligodendrocytes compared to gray matter, however, measuring the proportional change of adult gray and white matter oligodendrocyte populations over time revealed an elevated rate of population expansion in the gray matter. Following cuprizone-mediated demyelination, we found that the white matter had enhanced replacement of lost oligodendrocytes irrespective of baseline differences in oligodendrocyte density. Additional layer- and subregion-specific analyses revealed a deficiency in oligodendrocyte regeneration in the deep gray matter regions that correlated with a reduced recovery of the MOL5/6 oligodendrocyte subpopulation. As deep cortical layers are essential for transmitting cortical output signals, our results suggest that regional differences in oligodendrocyte regeneration may contribute to deficits in cognitive function observed in human MS patients. Overall, this work provides a roadmap for long-term in vivo three-photon imaging of cellular behaviors across the gray and white matter regions of the adult mouse brain.
One advantage of longitudinal measurements of cellular behaviors is that the initial population(s) of labeled cells are defined at before, during, and after interventions. This enables the analysis of population growth or decline within a defined subregion with single-cell resolution. Across development and into adulthood, the rate of oligodendrogenesis in the mouse CNS slows asynchronously across CNS regions and undergoes population decline at late stages of life4,48. Although the population density of oligodendrocytes is significantly lower in the gray matter versus the white matter, we found that the population grew at a higher rate in the cortex than in the corpus callosum in the healthy brain. These data imply that regional microenvironments regulate the carrying capacity of the oligodendrocyte population to maintain heterogenous myelination dynamics throughout life. Mature oligodendrocytes cluster into at least six specific transcriptional subpopulations in the healthy brain6 that have defined spatial preferences in the CNS7,8. Cross-sectional analyses using RNA in situ hybridization revealed gray and white matter regional differences in the proportions of MOL1, MOL2/3, and MOL5/6 oligodendrocytes that were stable across young adulthood (P60 to P140), yet it is unknown if these differences reflect specific cellular functions or simply differences in the maturation state of these populations. Furthermore, how oligodendrocyte population growth dynamics are governed to establish region-specific population patterns remains an open question. Microenvironmental cues may act at specific stages of oligodendrocyte maturation, such as OPC proliferation, differentiation, or premyelinating cell survival and integration into axonal circuits25,49. Similar to previous studies, we found that OPC proliferation is significantly higher in the subcortical white matter compared to the posterior parietal cortex3,50(Extended Data Fig. 7). As proliferation is linked to OPC differentiation or death51, these results suggest a reduced cell cycle time in the white matter that maintains homeostatic population density in response to the increased frequency of oligodendrogenesis in this region. Alternatively, regional regulation of survival of premyelinating oligodendrocytes25 in different microenvironments may regulate oligodendrocyte population dynamics. The recent discovery that specific red fluorescent proteins can be excited in the 1300 nm spectral window with high efficiencies25,52,53, opens the door to future work with dual-color, oligodendrocyte stage-specific genetic reporter approaches54 to dissect how regulation of maturational stages leads to variable population growth across cortical and subcortical regions.
Similar to lesions in MS patients, the cuprizone-mediated demyelination model results in overlapping oligodendrocyte death and regeneration55. The demyelinating injury caused by cuprizone treatment occurs in the absence of notable peripheral immune response56 making it an ideal model to evaluate the intrinsic capacity of oligodendrocyte regeneration without the confound of autoimmunity. Long-term in vivo imaging approaches are essential for analyzing the dynamics of the overlapping periods of oligodendrocyte loss and gain observed with cuprizone-mediated demyelination. Here, we showed that systemic cuprizone administration affects gray and white matter regions similarly, both in terms of oligodendrocyte cell death and changes in transcriptional heterogeneity, yet the white matter has an enhanced intrinsic capacity for regeneration of oligodendrocytes and their subpopulations compared to the gray matter. These regional differences in intrinsic capacity for regeneration highlight the need for additional insights into the regional effects of “remyelination therapies” (approaches that enhance oligodendrocyte regeneration57) to harness their full potential to increase remyelination and accelerate functional recovery following demyelinating injuries. Further work examining multicellular interactions in regional microenvironments will aid in our mechanistic understanding of differences in regional oligodendrocyte regeneration. For example, the type 1 transmembrane neuropilin-1 is specifically expressed on white matter microglia and transactivates platelet-derived growth factor receptor-ɑ to induce OPC proliferation following demyelination58. Conversely, this increased white matter regeneration could be mediated by the recruitment of Gli1-positive adult neural stem cells from the subventricular zone59. Protein–protein interaction databases and recent advances in RNA sequencing technologies will likely provide key access to ligand–receptor pairs60 that can help explore the role of multicellular interactions in oligodendrocyte population growth and regeneration.
In MS patients, in contrast to our findings with cuprizone-mediated demyelination, lesions in the white matter regions are thought to have more severe demyelination and decreased remyelination compared to gray matter lesions61,62. MS white matter lesions differ from gray matter lesions by often displaying blood-brain barrier disruption, infiltrating peripheral immune cells, and complement deposition (reviewed by Geurts and Barkhof63) suggesting that the inflammatory microenvironment of white matter may limit the regenerative capacity of oligodendrocyte populations. Indeed, mouse white matter oligodendrocyte precursor cells have delayed differentiation when exposed to a combination of interferon-γ and tumor necrosis factor-ɑ compared to gray matter-derived OPCs in vitro64. Recent findings show that OPCs can cross-present antigen and upregulate the immunoproteasome subunit PSMB8 specifically in the white matter10,65 highlighting the severe inflammatory microenvironment of the white matter suggesting that oligodendrocyte regeneration is disproportionately affected in the inflammatory context of white compared to gray matter regions in MS patients. Furthermore, regional heterogeneity of oligodendrocytes and other cell types may regulate the disease progression and regenerative capacity in a region-specific manner11. Future work using long-term in vivo three-photon imaging of oligodendrocytes in gray and white matter regions of immune-mediated demyelination models will provide important insights into the role of inflammation in regional regenerative capacity of oligodendrocyte populations. Recent longitudinal in vivo two-photon imaging following cuprizone-mediated demyelination showed that oligodendrocyte regeneration declined with depth across the superficial 300 μm of somatosensory cortex19. Our results confirm and extend these findings as we show that deep cortical layers 5–6 have reduced oligodendrocyte regeneration compared to the baseline level of healthy oligodendrogenesis. Additionally, we identified a specific oligodendrocyte subpopulation in deep layers 5–6 that fails to be reestablished following cuprizone-mediated demyelination. Since mature oligodendrocyte density increases with depth (Fig. 1d), one possibility is that increased myelin debris suppresses intrinsic mechanisms of oligodendrocyte regeneration and reestablishment of subpopulation heterogeneity in the deep cortex, but not in the white matter66. The consequences of regional deficits in oligodendrocyte regeneration and remyelination are not well understood. Cortical layers 5–6 contain the primary output neurons that project to downstream brain regions to control behavior67 and reduced oligodendrocyte regeneration in these regions could underlie the debilitating behavioral deficits experienced by MS patients such as impaired hand function68. Oligodendrocytes that closely appose layer 5 pyramidal neurons (satellite oligodendrocytes) locally shape their intrinsic excitability via buffering potassium through the glial syncytium69. Loss of these oligodendrocytes could underlie increases in neural firing rates resulting in hyperexcitability following demyelination as well as lead to a loss of local neural circuit synchronization14,70. Furthermore, regional variation in oligodendrocyte and myelin loss may lead to cell-type-specific neuron vulnerability and glial activation patterns relevant to neurodegeneration71. Remyelination is protective for damaged axons and recent work shows that cell-type specific remyelination is driven by a combination of cell type and axonal diameter20. Additional work to understand the specificity of myelin placement on neural circuits across gray and white matter in the healthy and diseased brain will greatly aid in uncovering the potential for remyelination to restore circuit and behavioral function following demyelination.
Methods
Animals
All animal experiments were conducted in accordance with protocols approved by the Animal Care and Use Committee at the University of Colorado Anschutz Medical Campus. Approximately equivalent numbers of male and female mice were used in these experiments and were kept on a 14-h light–10-h dark schedule with ad libitum access to food and water. Mice were housed at a temperature of 72±2°C with 50±20% humidity levels. All mice were randomly assigned to conditions and were precisely age-matched (±5 days) across experimental groups. C57BL/6N MOBP–EGFP (MGI:4847238) mice, which have been previously described72, were used for in vivo multiphoton imaging.
Custom three-photon microscope
The custom microscope consisted of a VIVO Multiphoton Open (3i) microscope, based on a Sutter Moveable Objective Microscope (MOM) stand, that was modified for multichannel three-photon imaging. The laser output from a regenerative amplifier with 1030 nm center wavelength, 70 W average power, <300 fs pulse duration, and adjustable repetition rate up to 2 MHz (Spirit-1030–70, Spectra Physics, 1 μJ pulse energy at 2 MHz) was wavelength converted to 1300 nm by a noncollinear optical parametric amplifier (NOPA-VIS-IR, Spectra Physics). The laser was operated at a repetition rate of 1 MHz (pulse picking = 2) and the final output power from the idler at this repetition rate was 0.8 to 1.1 W at 1300 nm. The power was modulated using a motorized rotation mount (KPRM1E/M - Ø1, Thorlabs) with a half-wave plate and Glan-Thompson prism. Beam conditioning of the NOPA output consisted of an expansion/collimating lens relay (f1 ¼ 75 mm, f2 ¼ 100 mm, Newport). The beam was expanded using a 4× reflective beam expansion telescope (BE04R, Thorlabs) to uniformly fill the deformable mirror (DM, ALPAO) and then demagnified with a telescope relay back into the microscope (f1 ¼ 500 mm, f2 ¼ 200 mm, Edmund Optics). A dual prism compressor system consisting of two 25 mm SF10 prisms cut at Brewster’s angle (10NSF10, Newport) and a gold roof mirror (HRS1015-M01, Thorlabs) was used to compensate for group delay dispersion. The beam was directed to the galvanometers (Cambridge Technologies) and through a scan lens (SL50–3P, Thorlabs), tube lens, and a 760 nm long-pass primary dichroic (Semrock). Three-photon excitation was focused through a high-NA multiphoton objective (XLPLN25XWMP2, 25x/1.05 NA, Olympus) with the correction collar set for the 0.15 mm coverslip at the surface of the sample. The fluorescent emission was separated from the excitation path by a long pass dichroic mirror, spectrally filtered (green channel 525 ∕ 50 nm, THG channel 430 ∕ 25 nm), and detected by photomultiplier tubes (H10770PA-40, Hamamatsu). The electronic signal was amplified, low-pass filtered and digitized. Laser-clocked acquisition was not implemented on the current system and 2 μs pixel dwell times were used for all experiments to allow for the collection of fluorescence from ~2 laser pulses per pixel. Data were acquired with SlideBook 2022 (Intelligent Imaging Innovations).
Cranial window surgery
Chronic cranial windows were prepared as previously described51 with minor modifications for three-photon imaging (described below). Six- to eight-week-old mice were anesthetized with isoflurane inhalation (induction, 5%; maintenance, 1.5–2.0%, mixed with 0.5 liter per min O2) and kept at 37°C body temperature with a thermostat-controlled heating plate. After removal of the skin over the right cerebral hemisphere, the skull was cleaned and a 2 × 2 mm region of skull centered over posterior parietal cortex (1–3 mm posterior to bregma and 1–3 mm lateral) was removed using a high-speed dental drill. A piece of cover glass (VWR, No. 1) was then placed in the craniotomy and sealed with Vetbond (3M) and then dental cement (C&B Metabond). For head stabilization, a custom metal plate with a central hole of diameter = 6 mm was attached to the skull parallel to the implanted glass window, and the dental cement was drilled out around the perimeter of the window to allow objective advancement deep into the brain without headbar contact. A 5 mg / kg dose of carprofen (Zoetis) was subcutaneously administered before awakening and for two additional days for analgesia. In vivo imaging sessions began 2–3 weeks post-surgery and took place once every 4 days or weekly (see imaging timeline in Fig. 5). During imaging sessions, mice were anesthetized with isoflurane and immobilized by attaching the head plate to a custom stage (See Longitudinal three-photon imaging with adaptive optics section). The internal temperature of the mice was maintained at 37±0.5 C using a feedback temperature controller (TC-1000, CWE) and the Isoflurane was scavenged using a Minivac Gas Evacuation Unit and Charcoal Fluosorber (Harvard Apparatus).
Longitudinal two-photon imaging
Chronic cranial windows were prepared over the PPC and in vivo imaging was performed as above. 4D two-photon datasets were acquired using a Bruker Ultima Investigator upright microscope equipped with Hamamatsu GaAsP detectors and a mode-locked Ti:sapphire laser (Coherent Ultra) tuned to 920 nm. The average power at the sample was between 5 – 30 mW, corresponding to an exponential power curve of 5 – 15 mW to 80 – 120 mW with depth. At least three MOBP-EGFP-labeled mature oligodendrocytes distributed across the field of view near the surface of the brain were used to identify the same cortical area over longitudinal imaging sessions. Image stacks were acquired with Zeiss W plan-apochromat ×20/1.0 NA water immersion objective giving a volume of 444 × 444 × 350 μm (X,Y,Z; 1024 × 1024 pixels) from the cortical surface.
Long-term three-photon imaging with adaptive optics
The custom stage for three-photon imaging consisted of 3D-printed headbar clamps for anesthetized imaging affixed to a small breadboard (MB2020/M, Thorlabs) mounted on 2-axis brass goniometers (GOH-60B50, Optosigma) with the rotation center height approximately level with the cranial window. The first day of imaging (2.5–3 weeks post-surgery) consisted of the following steps: The window was flattened to be orthogonal to the excitation angle using the THG signal generated at the surface of the cranial window73–75. The tip/tilt of the stage goniometers were manually adjusted while maintaining the focal plane at the surface of the window until the field uniformity of the THG signal was maximized and did not translate significantly when moving in z. Imaging fields were chosen based on image quality at the superficial ~20 μm of the corpus callosum (~800 μm depth). Epifluorescent landmark images were taken at the end of the first time point to relocate the imaging field on subsequent time points. To correct for optical aberrations induced by the inhomogeneity of mouse brain tissue, we adopted an indirect, image-based correction technique that has been described previously22,76,77. The deformable mirror (DM, DM97–15, ALPAO) was controlled in SlideBook 2022 and custom MATLAB scripts were used to perform plotting and peak-finding for individual Zernike modes. AO correction was performed on the THG signal of the vasculature and transverse myelin sheaths in the deep cortex just above the corpus callosum (~750 μm depth) at 256 × 256 resolution, corresponding to 5.1 frames / second. To calibrate for spherical-defocus coupling, we used two-photon imaging of a small ROI surrounding a single 0.2 μm fluorescent bead in a 2-dimensional preparation. The peak intensity of the bead sample was plotted against the spherical and defocus amplitudes at each z-position and the linear relationship between the modal amplitudes was applied to the DM during correction. The raw optimization data was reviewed to confirm that the focal plane was constant at different AO configurations. Parameters for AO correction consisted of the number and order of Zernike modes for correction, DM stroke range, and the stroke increment. We corrected first for the spherical mode followed by astigmatism, coma, trefoil, and then higher-order modes with a DM range of −2 to 2 um and an increment of 0.3, which resulted in the acquisition of 196 images in ~50 seconds (including analysis time). Zernike mode settings were optimized based on the mean intensity of the THG signal, which resulted in the greatest improvements in EGFP-positive oligodendrocyte signal to background (SBR) ratio in the subcortical WM (Extended Data Fig. 3). A single AO iteration was performed for chronic structural imaging in healthy mice. The single-plane AO correction deep in the cortex resulted in a suboptimal DM pattern for signal quality in the superficial cortex of the z-column. Therefore we implemented a dynamic linear spherical aberration correction that was updated with imaging depth without the need to reoptimize at each z plane. After optimizing the DM settings the power map was set stepwise starting from the meninges. 11 power map points were set (0, 50, 100–1000 μm at 100 μm intervals) using the fluorescent counts on the THG detection channel. The power was slowly increased at 0.2 (surface) or 1% intervals while monitoring the fluorescence intensity histogram to obtain a dynamic range of signal intensity between 0–7000 counts without saturation. In cuprizone mice, the power modulation maps were reduced by ~10% during the two weeks surrounding peak demyelination (16 – 32 days post-cuprizone) to account for decreased excitation scattering after myelin loss. Large-volume z-stacks were acquired with bidirectional scanning at 512 × 512 pixels (385 × 385 μm), dwell time = 2 μs, frame averaging = 2, and z-step = 3 μm, and scan time ~.8 frames / s. The PMT gain was set to 91% for THG and 93% for GFP and a custom blackout box was built to reduce ambient light noise. Raster scanned images are only recorded during ~80% of the x-galvonometer sweep (i.e. during constant velocity), therefore the Spirit pulsed output was blanked during the overscan to reduce the average power applied to the tissue. Finally we implemented a one minute frame pause after every three minutes of z-scanning to allow for periodic heat dissipation. For subsequent imaging time points, mature oligodendrocytes in the superficial cortex were aligned to the first time point using ROI overlays around the cell body positions on day one, the spherical depth correction was applied to the DM as in day one, and the AO correction was performed daily at the same depth before acquisition.
Cuprizone-mediated demyelination
Global demyelination was induced in our C57\B6N MOBP–EGFP mice using 0.2% cuprizone (Sigma Chemical, C9012), stored in a glass desiccator at 4 °C. Cuprizone was added to powdered chow (Harlan), mixed for ~10 minutes to ensure homogeneity, and provided to mice in custom feeders (designed to minimize exposure to moisture) for 3 weeks on an ad libitum basis. Feeders were refilled every 2–3 days, and fresh cuprizone chow was prepared weekly. Healthy mice received normal powdered chow in identical feeders. Powdered chow was introduced 2–3 days before cuprizone to ensure acclimation to the powdered food. Cages were changed weekly to avoid build-up of cuprizone chow in bedding, and to minimize reuptake of cuprizone chow following cessation of diet via coprophagia. This 3-week demyelination model that has been previously described14, results in ~88% loss of MOBP-EGFP oligodendrocytes in the superficial primary motor cortex, and allows for in vivo tracking of the overlapping time courses of demyelination and oligodendrocyte regeneration. Cuprizone mice were weighed weekly during and after cuprizone feeding to confirm consumption of the drug.
Image processing and analysis
4D longitudinal image stacks were analyzed using Fiji78. Cortical layers were defined using the Allen Reference Atlas79 and in vivo THG imaging data. Layer depth was calculated for individual mice using the depth of the white matter THG signal according to the equation below to eliminate variability in mouse size, field of view location, and imaging modality:
Unprocessed 1000 μm 4D stacks were cut into substacks of layers 1–3 (0 to 356±21 μm), layers 4–6 (469±22 μm) and the subcortical WM (96±6 μm). Image registration was performed iteratively with the Poorman3Dreg plugin for lateral registration followed by the Correct3D drift plugin80 (EGFP channel, rigid body registration, 30×30×20 pixels X/Y/Z). Additional iterations of Correct3D drift were performed on high-signal ROIs as needed. The registered datasets were further processed with a median filter to remove salt and pepper noise and individual oligodendrocytes were manually tracked using custom FIJI scripts as previously reported14. Images generated for figures were brightness and contrast corrected for clarity and maximum z-projection widths are defined in the figure legends. Stable, new, and lost cells were segmented and pseudo-colored using the Lasso tool in Photoshop CS6 (Adobe).
Calculation of mouse-specific effective attenuation lengths
We measured mouse-specific effective attenuation lengths (EALs) in the adult PPC to estimate the pulse energy delivered at each focal plane as described previously21,28,32 (Extended Data Figure 5). A small percentage (<2%) of the laser power was reflected to a power meter to measure the pulse energy at the surface applied to each z-plane during volumetric imaging. Then we measured the brightest 1% of the pixels in the EGFP channel across depth, scaled these values by the cube of the intensity, and calculated the linear slope of the logarithm of the signal vs. depth plots as in Extended Data Fig. 5.
Analyses of imaging performance and white matter fiber orientation
For two-photon versus three-photon imaging comparisons, oligodendrocyte cell bodies were counted within x-y subregions greater than or equal to 200 × 200 μm2 and oligodendrocytes were only counted if SBR was greater than 2 (as measured by a line scan profile in ImageJ). For the SBR analyses, oligodendrocytes were randomly sampled throughout the imaging volume to include the full range of oligodendrocyte SBRs (i.e. dim, medium, and bright) with at least 10 oligodendrocytes / 100 μm z-depth. ROIs were drawn around the oligodendrocyte cell body and then immediately adjacent to the cell body in a background region. Single oligodendrocyte SBRs were calculated as the max intensity of the cell body / max intensity of the background. AO improvements in oligodendrocyte signal intensity were quantified in the same manner, while improvements in resolution were calculated on oligodendrocyte cell bodies or single myelin sheaths as the full width at half-maximum in the lateral or axial dimensions. Fiber orientation calculations were performed on single z-slices within the WM using the OrientationJ plugin81,82.
EdU labeling and detection
To quantify regional differences in OPC proliferation and oligodendrocyte generation, EdU (A10044, Invitrogen) was diluted in sterile saline solution (0.9% NaCl) and injected intraperitoneally from P70-P76 (matched to starting age of in vivo imaging timeline) twice daily at 5 mg per kg for a total of 7 days. The time course and 5 mg per kg doses were chosen based on previously published work83,84, with the goal of labeling a substantial number of proliferating OPCs in the gray and white matter without saturating the population within either region85 and without generating false positives due to EdU binding at sites of DNA repair86 or inducing DNA damage pathways87. EdU-labeled cells were visualized using the AlexaFluor-647 Click-iT EdU Cell Proliferation Kit for Imaging (C10340, Invitrogen). The Click-iT reaction was performed per the manufacturer specifications immediately following the blocking step in the immunofluorescence protocol.
Immunofluorescence analyses
Mice were anesthetized with an intraperitoneal injection of sodium pentobarbital (100 mg per kg body weight) and transcardially perfused with 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.0–7.4) immediately following the final imaging time point or 16–20 h post-exposure in the laser damage experiments. Brains were postfixed in 4% paraformaldehyde for 1–2 h. at 4 °C, transferred to 30% sucrose solution in PBS (pH 7.4) and stored at 4 °C for at least 24 h. Brains were extracted, frozen in TissuePlus OCT, sectioned coronally at 30 μm, and immunostained as free-floating sections. Sections were incubated in blocking solution (5% normal donkey serum, 2% bovine γ-globulin, 0.3% Triton X-100 in PBS, pH 7.4) for 1–2 h at room temperature, then overnight at 4 °C in primary antibody. When staining for Aspartoacylase (ASPA) sections were incubated with Liberate Antibody Binding Solution (L.A.B., Polysciences) for 10 minutes prior to the blocking stop. Secondary antibody incubation was performed at room temperature for 1.5 h. Sections were mounted on slides with Vectashield antifade reagent (Vector Laboratories). Images were acquired using a laser-scanning confocal microscope (Leica SP8). For laser damage experiments, mild columnar damage regions (positive control tissue) were created using identical scanning settings as in the longitudinal group, but with the power map right-shifted to saturate the THG channel. 2–3 coronal sections of the PPC spaced ~60 μm apart were chosen based on the stereotaxic coordinates of the 385 × 385 μm in vivo imaging field and aligned to the reference atlas using anatomical landmarks. Bulk intensity measurements were made as previously described34,88 on a 1000 × 1000 μm ROI drawn directly under the 385 × 385 μm imaging field, and in the identical region on the contralateral hemisphere. Irregularly-shaped lesions were cropped with a minimum area of 600 × 600 μm. For cell counts, images with a 0.56 μm pixel size were thresholded using the maximum entropy filter and then cell bodies in individual channels were segmented using FIJI’s particle analysis tool78 and then proofread manually using the ROI manager. The settings for the particle analyzer were as follows: Size filter: 40 – 2000, Circularity: 0-.7, Microglia; Size filter: 30 – 2000, Circularity: 0–1, Oligodendrocytes; Size filter: 200 – 3500, Circularity: 0–1 Neurons; Size filter: 30 – 2000, Circularity: 0–0.5; Astrocytes). For the OL-specific analyses of phototoxicity, the mean intensity of each segmented MOBP-EGFP ROI was measured together with a background ROI in the superficial cortex. Cells were considered HSP70/72- or γ–H2a.X-positive if the mean intensity within the cell body was greater than two times the background measurement. Automated segmentation of the vasculature was performed as previously described36,37. Briefly, one random section from each mouse was concatenated into a z-stack and ~15 blood vessels of varying diameters and background regions were traced for each image using the pixel classifier. The classifier was saved and batch-applied to all the images in the dataset, which were then binarized and analyzed for percentage area of vascular coverage. Pericytes were counted as above. All confocal images shown in Fig. 3 and Extended Data Fig. 6 were acquired with identical power settings, processed with a 100 pixel rolling ball background subtraction and 0.7 pixel gaussian blur, and brightness/contrast correction was applied identically across channels.
RNA in situ hybridization with immunofluorescent detection of EGFP
In situ hybridization (ISH) was performed using the commercially available RNAscope V2 kit by adapting the manufacturer’s protocol (ACD Biotechne 323100). This procedure was specifically modified to include antibody labeling of EGFP+ oligodendrocytes in the same tissue sections labeled with three mRNA probes. Probes used in this study were designed for mouse Ptgds-C1 (ACD Biotechne 492781), Klk6-C2 (ACD Biotechne 493751), and Egr2-C3 (ACD Biotechne 407871). Briefly, 20 um, PFA-fixed, tissue sections from brain and spinal cord were washed and mounted on Superfrost microscope slides. Tissue dehydration, antigen retrieval, protease treatment, and probe hybridization were carried out according to the manufacturer’s protocol. Amplification and HRP reaction steps for C2, C3, and C1 (in this order) were carried out without deviation from the manufacturer’s protocol. Briefly, slides were first washed with 1x RNA Scope wash buffer 2x for 2 minutes at room temperature before incubation with Amp 1 for 30 minutes at 40C. This process was then repeated for Amp 2 and Amp 3. Following amplification steps slides were washed 2x for 2 minutes each in RNAscope wash buffer before incubation with the respective HRP conjugate for 15 min at 40C. Next, sections were washed 2x for 2 minutes each and incubated in the corresponding fluorophore for 30 minutes at 40C (C2, TSA Vivid 570 1:1500 in TSA buffer, C3 TSA Vivid 650 1:2000 in TSA buffer, or C1 Opal TSA-DIG (Akoya) 1:750 in TSA buffer). After HRP reaction slides were washed 2x for 2 minutes, incubated in HRP blocker for 15 min at 40C, and finally washed 2x for 2 minutes. This was then repeated for the remaining 2 probes. Following the final HRP blockade, slides were incubated in blocking buffer (5%NDS, 0.3% Triton X-100 in PBS) for 60 min at room temperature. Slides were then incubated with chicken anti-GFP 1:250 (AVES, GFP-1020) for 48 hours at 4C. Slides were next washed 3x for 15 minutes in PBS. Immunohistochemistry was completed with incubation in Donkey anti-chicken 488 1:500 (Jackson, 703-545-155) for 60 minutes. ISH was completed by washing slides 3x for 15 minutes in PBS before incubation in Polaris 780 (1:125 in diluent buffer, Akoya) for 30 minutes at 40C. Finally, slides were washed 2x for 2 minutes in RNAscope wash buffer and counterstained with RNAscope DAPI for 30 seconds before cover slipping with Prolong Gold (Invitrogen, P36934) for imaging.
RNAscope image acquisition and processing
Fluorescent images used for RNAscope quantification were acquired using the Olympus DSU Spinning Disc microscope. To obtain a large field of view, adjacent images were acquired using a 20x air objective and a 10% tile overlap. A z-stack with 4 um steps was used to capture the entire thickness of each tissue section and reduce the detection of false positive labeled cells as described previously7. Images were stitched and max projected using a custom ImageJ macro based on the BaSiC plugin89.
RNAscope quantification
Following imaging, images were analyzed using Qupath Software 0.4.390. RNA expression per cell was quantified using a custom analysis pipeline. Briefly, cortical layers were measured and drawn onto each tissue section as described above (Image Processing and Analysis) to allow for quantification of mRNA puncta within cells in each individual cortical layer. Nuclei were semi-automatically segmented based on the DAPI channel. Nuclear segmentations were dilated to encompass the predicted area of each cell. Finally, mRNA puncta were segmented and assigned to DAPI+/EGFP+ cell regions for oligodendrocyte-specific quantification. Threshold values for each channel were based on the background signal in each image. Cell and puncta detection was performed on each section in Qupath and the spreadsheets containing the number, size, and area of puncta were exported and processed to calculate the percentage of each population within each cortical layer using custom R scripts (https://github.com/sbudoff/OligoRNAscopeTools). We used thresholds of 14, 12, and 7 estimate puncta per GFP positive cell of Egr2+, Klk6+, and Ptgds+, respectively, to assign oligodendrocyte populations. Single OLs could have multiple identities in these analyses. For Klk6, we observed that positive cells in the cortical sections expressed high levels of entirely clustered puncta and therefore only included cells with 1 or 2 clusters that had an estimated puncta number greater than 12. To calculate the number of oligodendrocyte classes that are present within each layer we calculated a threshold which was the lowest proportion of cells present in healthy control animals. If an animal had a proportion of oligodendrocytes higher than this threshold it was considered to have this class present within that cortical layer. We used a threshold of 5.73%, 0.394%, and 5.82%, of MOL1, MOL2/3, and MOL5/6, respectively, to determine the presence or absence of each oligodendrocyte class.
Statistics and Reproducibility
No statistical methods were used to pre-determine sample sizes, but our sample sizes were comparable to relevant publications8,10,15,80. All micrographs are representative images showing phenomena seen in multiple mice. All data were initially screened for outliers using IQR methods. All mice in a litter underwent cranial window surgery and concurrent two-photon imaging and training timelines were designated to be a “batch”. Experimental groups were replicated in multiple batches with multiple experimental groups per batch. The effects of cuprizone on MOBP-EGFP-positive oligodendrocytes did not permit blinding, however the experimenters were blinded for immunofluorescent and RNA-ISH experiments. Mice were excluded from longitudinal analysis if there was THG-positive tissue damage and OL cell death (as in Fig. 2), or if cuprizone demyelination resulted in <50% cell loss (n=1). To analyze in vivo changes in oligodendrogenesis, we used cumulative population growth and decline measures as previously described10. Briefly, oligodendrocyte gain / loss (%) was calculated as the change in oligodendrocyte number / initial cell number * 100, the rate of oligodendrocyte gain/loss (%) was calculated as the % change in oligodendrocyte number divided by a time interval (e.g. Remyelination phase), and oligodendrocyte replacement (%) as total oligodendrocyte gain / total oligodendrocyte loss * 100. The oligodendrocyte replacement (%) metric is used to control for differences in the magnitude of demyelination, as oligodendrocyte gain is strongly correlated with oligodendrocyte loss in the cuprizone model10,81,82. To analyze regional differences in the rate and timing of oligodendrogenesis, we used statistical modeling of cell growth and decline. As healthy oligodendrogenesis rates decline with age25, we found that this process was best-fit using the exponential mechanistic growth model that is commonly used to describe population growth with competition for resources91. Extended Data Fig. 8 shows the mechanistic growth curves for example mice and group means along with R2 values, which were greater than or equal to 0.91 (cortex) and 0.96 (CC) for all analyzed mice. The exponential mechanistic growth equation was calculated according to the equation below:
Where = Asymptote, = Scale, = Growth rate, and = time (days).
Cuprizone potently inhibits the survival of new oligodendrocytes when it is actively being administered in the diet and cuprizone-induced cumulative OL loss and gain curves are sigmoidal and can be effectively modeled using three-parameter logistic regression92. In this study, we used a three-parameter Gompertz model commonly used to describe tumor growth93,94 that was robust to asymmetrical differences in the timing of the inflection point. Extended Data Fig. 8 shows the Gompertz growth curves for example mice and group means along with R2 values, which were greater than or equal to 0.99 (cortex) and 0.99 (CC) for all analyzed mice. The Gompertz function was calculated according to the equation below:
Where a = asymptote, b = growth rate, c = inflection point, and t = time (days).
Because the goal of this study was to test the hypothesis that different brain regions are independent with respect to oligodendrogenesis, we used unpaired rather than paired statistics for single comparisons between gray and white matter regions for in vivo oligodendrogenesis data (Unpaired two-tailed t-test, parametric; Wilcoxon rank-sum, nonparametric). Statistical comparisons between multiple brain regions were made using one-way ANOVA followed by Tukey’s HSD post-hoc test (parametric) or Kruskal-Wallis followed by Dunn test (nonparametric). To compare oligodendrogenesis in the healthy brain to cuprizone remyelination, we scaled the datasets to the maximum value of healthy or cuprizone oligodendrogenesis. To compare healthy vs. cuprizone-induced oligodendrogenesis metrics we used full-factorial 2-way ANOVA for the interaction effect followed by multiple comparisons with Bonferroni correction. Bonferroni-adjusted significance levels are indicated in the figure legends. Normality was assessed using the Shapiro-Wilk Test and outliers were assessed by visualizing the interquartile ranges. All statistical analyses and modeling were conducted in JMP 16 (SAS).
Extended Data
Extended Data Fig. 1 |. Custom three-photon light path and modifications for longitudinal in vivo imaging.
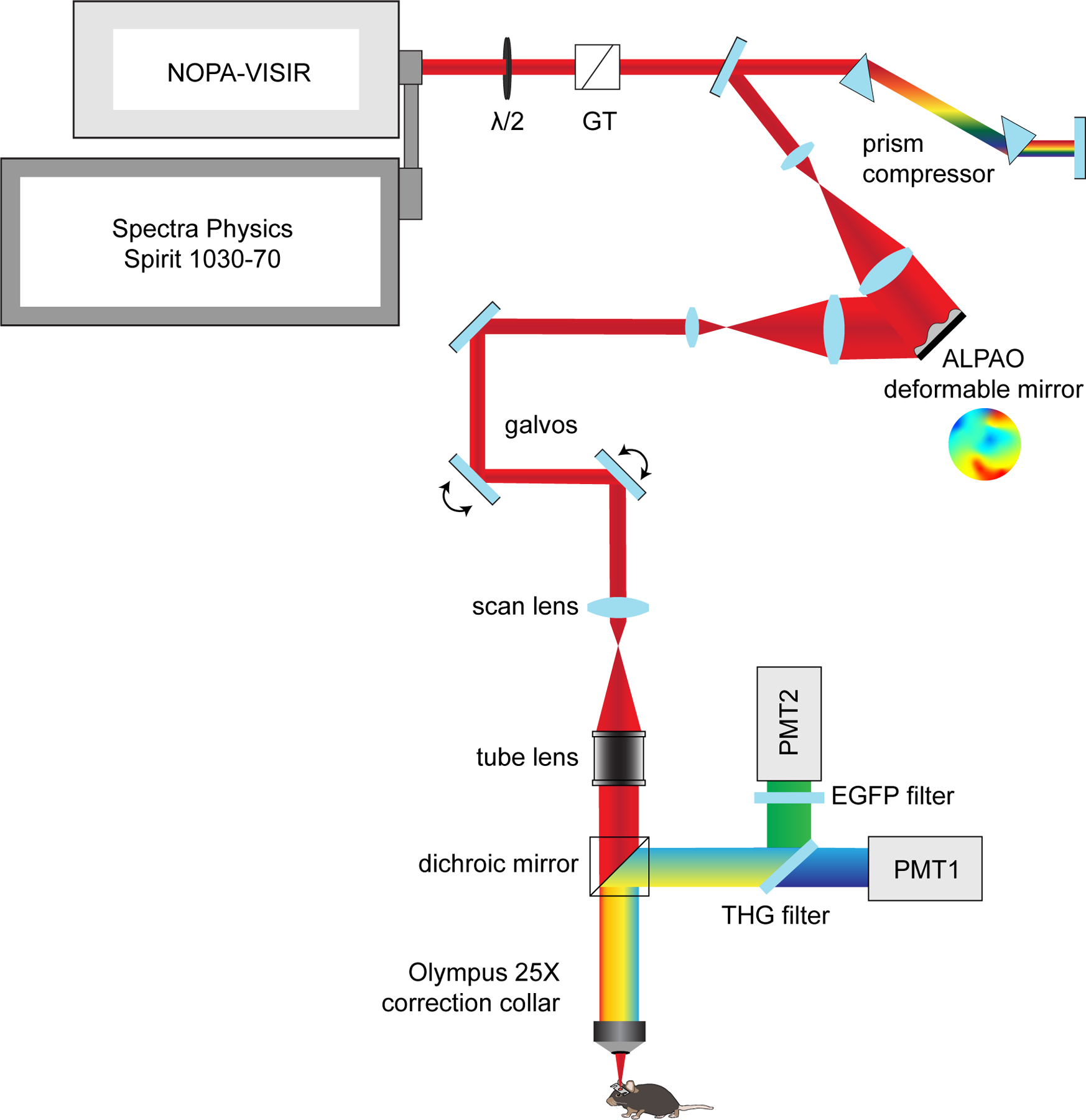
3P excitation source and light path, including motorized half-wave plate for power modulation (λ/2), Glan-Thompson polarizer (GT), dual prism compressor, beam expanding telescope, ALPAO deformable mirror, beam reduction lens relay, galvo-galvo scan mirrors, scan lens, tube lens, dichroic mirror (FF-488-di02, cut at 488 nm), and moveable objective microscope system with collection filters for third harmonic generation signal (430/25 nm, PMT1) and EGFP emission (520/70 nm, PMT2).
Extended Data Fig. 2 |. MOBP-EGFP and THG labeling of mature oligodendrocytes and myelin in the cortex and subcortical white matter.
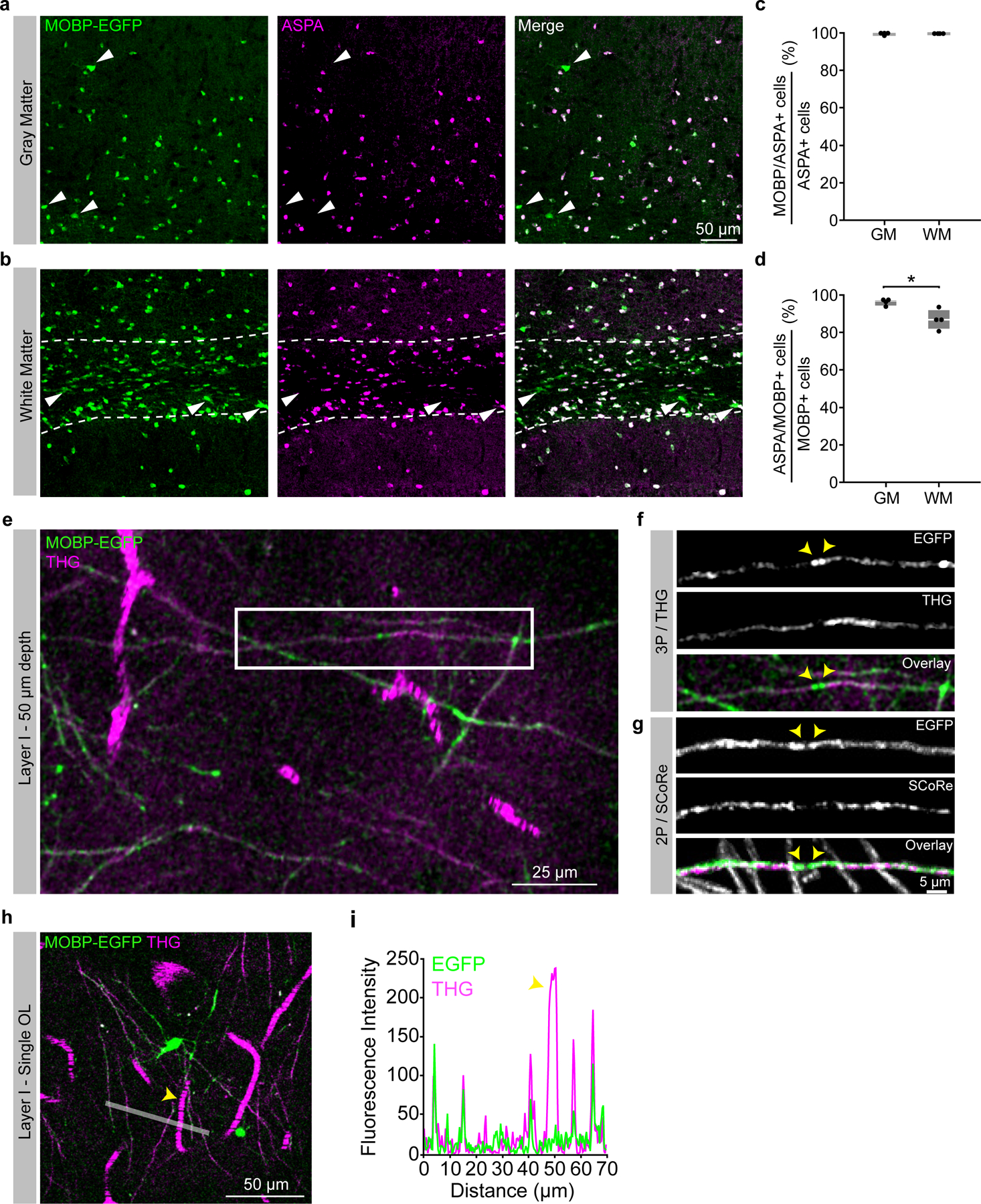
a) Confocal image of a tissue section from the deep posterior parietal cortex stained for transgenic MOBP-EGFP and ASPA. Arrowheads show large, putative newly-generated oligodendrocytes that are MOBP-EGFP-positive and ASPA-negative. b) Confocal image of ASPA/MOBP-EGFP immunofluorescence in the subcortical white matter beneath the posterior parietal cortex (dotted white lines). Arrowheads show MOBP-EGFP-positive and ASPA-negative white matter oligodendrocytes of varying sizes and brightness. c) 99.5% (cortex) and 99.6% (white matter) of ASPA-positive cells express MOBP-EGFP. n = 4 mice, 2 sections per mouse. d) 95.6% (cortex) vs. 86.5% (white matter) of MOBP-EGFP-positive oligodendrocytes were also positive for ASPA (Unpaired, two-tailed Student’s t-test for equal variance, p = 0.016). n = 4 mice, 2 sections per mouse. e) 9 μm z-projection of layer 1 in the posterior parietal cortex showing MOBP-EGFP labeling of myelin sheaths (green) and THG-labeling of myelin sheaths and blood vessels (magenta). f) Zoom image of an isolated EGFP/THG-labeled myelin sheath from the white box in (e) excited with 1300 nm three-photon excitation. g) Isolated MOBP-EGFP sheath from layer 1 of a separate mouse visualized with 920 nm two-photon excitation combined with Spectral Confocal Reflectance (SCoRe) microscopy. Note the decreased THG/SCoRe labeling at putative nodes of Ranvier in (f) and (g) (arrowheads). h) 9 μm z-projection of a single mature oligodendrocyte in layer 1 from a third mouse showing EGFP-labeled processes connected to multiple THG-labeled sheaths. i) 5-pixel line intensity plot from the white line drawn in (d) showing relative fluorescence intensity of EGFP- and THG-labeled myelin sheaths and a THG-labeled blood vessel (arrowhead). All images pixel size = 0.36 μm. *p <0.05, box plots represent the median, interquartile ranges and minimum/maximum values
Extended Data Fig. 3 |. Adaptive optics improves SBR and axial resolution in the subcortical white matter.
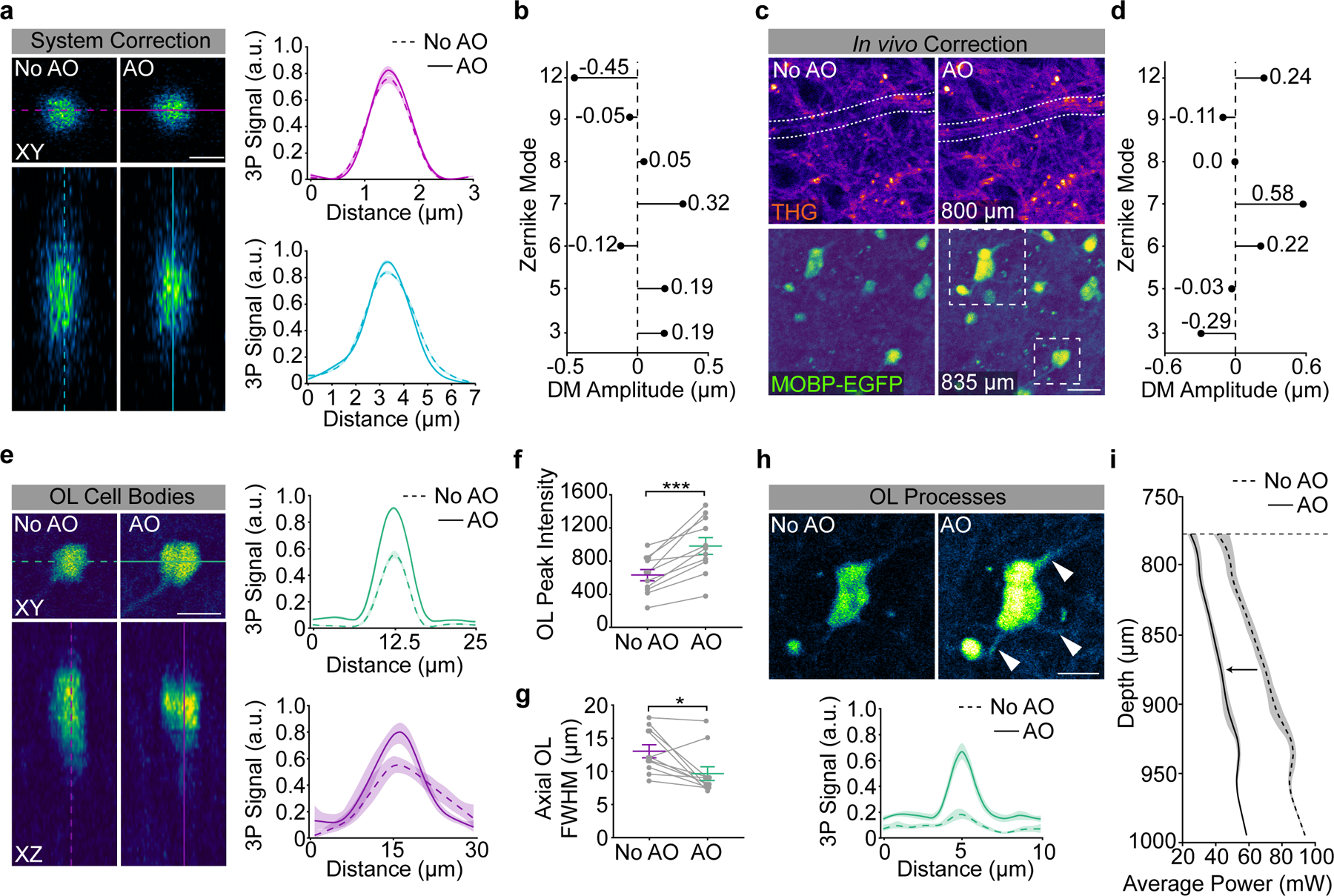
a) System adaptive optics (AO) correction with a two-dimensional 1.0 μm red polychromatic microsphere sample. Lateral (top) and axial (bottom) projections show a slight enhancement in peak signal after AO correction for system aberrations (8% increase, lateral; 9% increase, axial). The bead sample was diluted 1:10000, coverslipped with Prolong Gold, and the average power at the sample was <0.3 mW. System resolution = 0.88 μm (lateral) and 2.55 μm (axial) as calculated by the full width at half maximum of the gaussian fit after system AO correction. b) Deformable mirror (DM) amplitude plot showing the optimized stroke amplitudes (units = μm RMS) for each Zernike Mode in the system AO correction. c) In vivo, indirect, modal adaptive optics (AO) corrections were made at 775 – 825 μm depth (just above the white matter) by optimizing on the mean intensity of the third harmonic generation channel. The in vivo AO correction revealed cross sections through myelinated axons just dorsal to the corpus callosum (white dotted selection, top, 800 μm) and increased the mean intensity of EGFP-positive oligodendrocytes in the white matter ventral to the plane of the AO correction (bottom, 835 μm). Example images are single-slice images at the depth of the AO correction, 0.14 μm / pixel. d) Deformable mirror (DM) amplitude plot showing the optimized stroke amplitudes (units = μm RMS) for each Zernike Mode in the in vivo AO correction shown in (c). e) (Left) XY (top) and XZ (bottom) projections for a single EGFP-positive OL cell body from the example image shown in (c). (Right) Lateral and axial Gaussian fits of OL cell bodies from the field of view in (c). f) The peak intensity of the EGFP-oligodendrocyte signal was significantly enhanced by the AO correction (Paired, two-tailed Student’s t-test, p = 0.0002, n = 2 mice, 11 cells). g) The axial resolution of EGFP-oligodendrocyte cell bodies was significantly enhanced by the AO correction (Paired, two-tailed Student’s t-test, p = 0.014, n = 2 mice, 11 cells). h) AO correction enhances the peak intensity of OL processes by 202% (n = 2 mice, 4 processes). i) Average power vs. depth plot shows the average power at the sample through the depth of the white matter after AO correction (black, left), and the theoretical power curve (gray, right) necessary to maintain the same signal to background ratio without AO. Data are represented as individual points and mean ± SEM. Curves in a, e, and h represent the mean of the gaussian fits for each analyzed region (cell bodies in a, e, cell processes in h) with 95% confidence intervals. The intensity was integrated over a line plotted through the center of the cell structure (10 μm wide, cell bodies, 1 μm wide, processes). Zernike modes in (b), (d) = Spherical (12), Horizontal Trefoil (9), Horizontal Coma (8), Vertical Coma (7), Oblique Trefoil (6), Astigmatism (5), Oblique Astigmatism (3). For detailed statistics, see Supplementary Table 3.
Extended Data Fig. 4 |. Mechanical, scanning, and optical modifications to increase 3P signal and decrease average power.
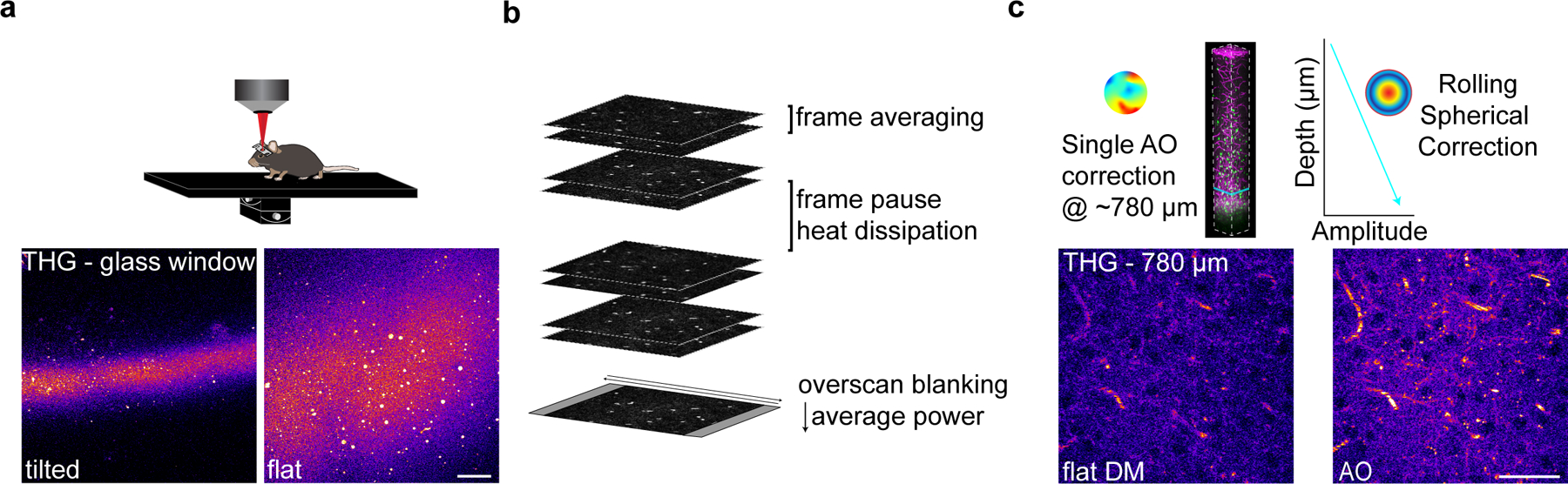
a) The third harmonic generation (THG) signal at the surface of the cranial window was used to align the preparation orthogonally to the excitation light. b) Scanning modifications to increase SBR and decrease average power to mitigate risk of tissue damage. Frame averaging was advantageous compared to increasing the pixel dwell time to reduce risk of nonlinear damage. Z-stack acquisition was paused periodically to allow for heat dissipation (1 min. pause per 3 min. scanning). Laser pulses were blanked on the galvanometer overscan to reduce the average power applied to the preparation at each z-plane. c) Imaged-based AO correction increased SBR at depth (left) and modulating the spherical aberration correction linearly with depth improved SBR throughout the imaging volume (right). For longitudinal imaging, the AO correction was made before acquiring each time point at the same z-plane just above the scattering white matter (750–850 μm depth).
Extended Data Fig. 5 |. Calculation of mouse-specific effective attenuation lengths in the posterior parietal cortex.
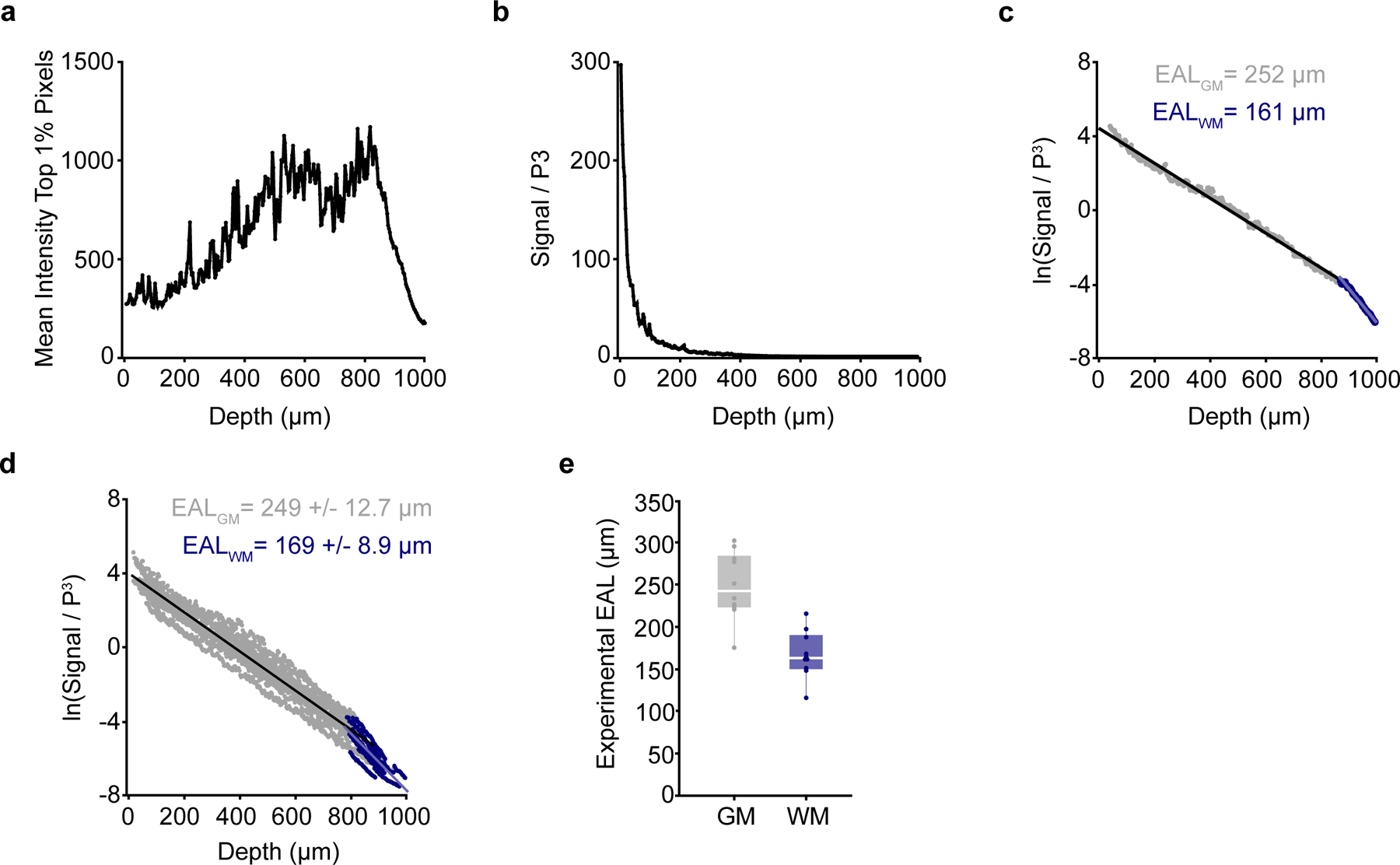
a) The mean intensity of the top 1% of the brightest pixels (experimental 3P signal) plotted with depth beneath the brain surface for an example mouse. b) Data from the mouse shown in (a) normalized to the cube of the pulse energy shows the decay of the 3P signal with depth in the mouse brain. c) Logarithm of the data in (b) shows a linear decrease with depth. The single mouse-specific effective attenuation length (EAL) can be calculated from the slope of the linear fit (gray = gray matter, blue = white matter). A steeper slope represents a shorter EAL due to increased scattering. d) Semilogarithmic plot showing gray matter and white matter EALs for n = 10 mice at the first time point of longitudinal imaging. Mean EAL (GM) = 249 +/− 12.7 μm, Mean EAL (WM) = 169 +/− 8.9 μm. e) Distributions of experimental mouse-specific EALs in the gray matter (gray) and white matter (blue). n=10 mice. Linear fits (black line plots in c,d) were calculated separately by region. Box plots represent the median, interquartile ranges and minimum/maximum values.
Extended Data Fig. 6 |. Optimized longitudinal three-photon imaging does not increase glial, neuronal, or vascular reactivity.
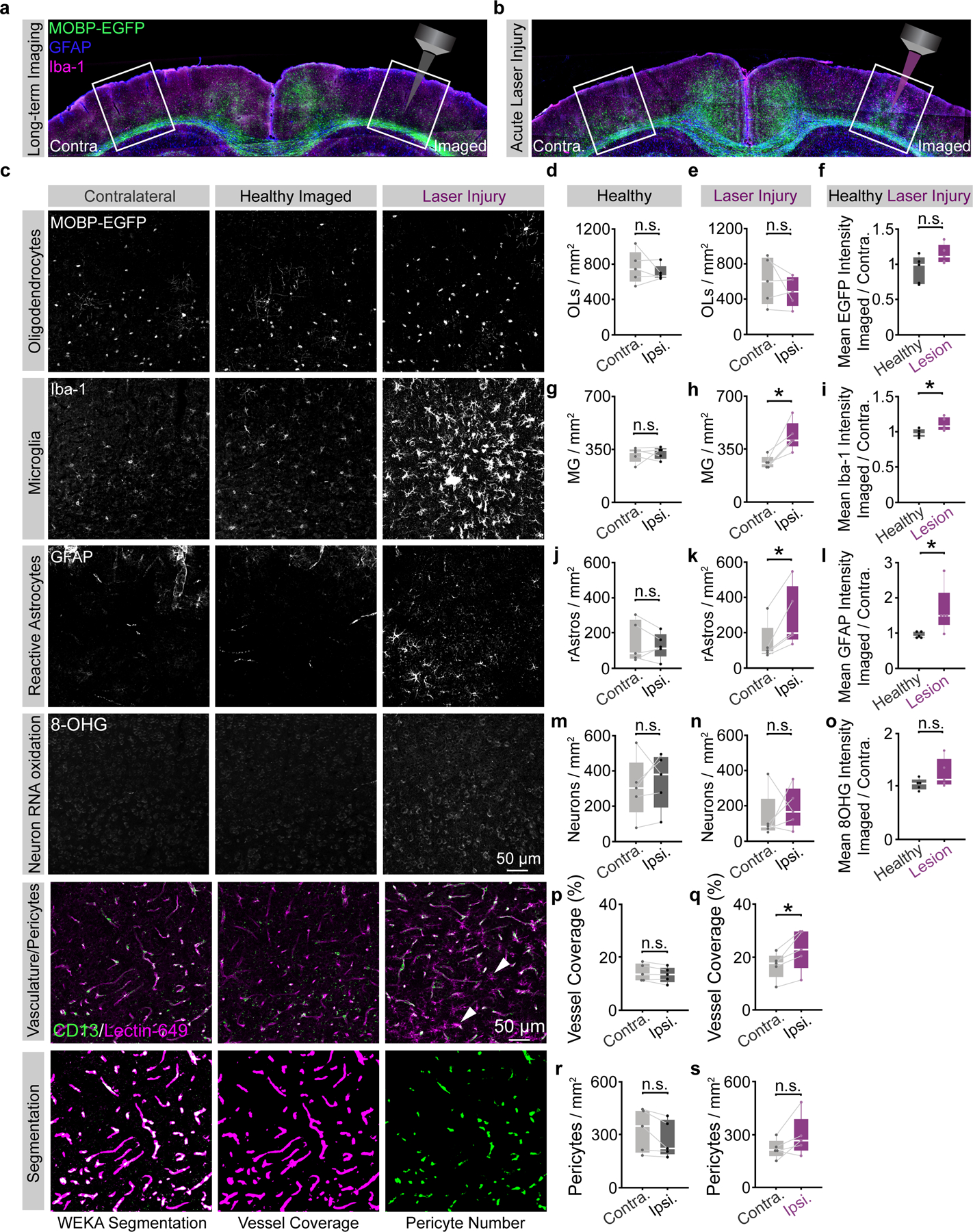
a) Related to Fig. 3. Coronal brain section from the imaged (right) and contralateral (left) posterior parietal cortex (PPC) of a mouse that was perfused 24 hrs. following 10 weeks of chronic 3P imaging. b) Coronal brain section from the imaged (right) and contralateral (left) posterior parietal cortex (PPC) of a mouse that received supra-threshold excitation across the cortex and white matter to generate laser-induced positive control tissue (laser injury, right). c) High-resolution confocal images of the contralateral (left), longitudinal imaging field (middle) and laser-induced injury (right) regions in the deep cortical layers of the PPC, stained for transgenic MOBP-EGFP expression (mature oligodendrocytes), Iba-1 (microglia), GFAP (reactive astrocytes), 8-hydroxyguanosine (8-OHG, neuronal RNA oxidation), and CD13/Lectin-49 (pericytes, vasculature, respectively). All images were taken with identical power settings, processed with a 100-pixel rolling ball background subtraction and 0.7 pixel gaussian blur, and brightness/contrast correction was applied identically across channels. The bottom row shows an example of the trainable WEKA segmentation to measure pericytes (green) and vascular coverage (magenta, see Methods). Note the Lectin-positive microglia-like cells present in the laser injury vasculature image (white arrowheads). d) No difference in the density of MOBP-EGFP OLs between the contralateral and imaged regions in healthy mice. e) No difference in the density of MOBP-EGFP OLs between the contralateral and imaged regions in laser-induced injury tissue. f) The ratio of the EGFP mean intensity for the imaged:contralateral (contra.) hemispheres was ~1 for healthy mice and did not change significantly in the positive control tissue. g) No difference in the density of Iba-1 microglia (MG) between the contralateral and imaged regions in healthy mice. h) Density of Iba-1 MG is significantly increased on the ipsilateral side of laser-induced injury tissue (p = 0.016). i) The ratio of the imaged:contralateral Iba-1 mean intensity was ~1 for healthy mice and significantly increased in positive controls (p = 0.026). j) No difference in the density of GFAP-positive reactive astrocytes (rAstros) between the contralateral and imaged regions in healthy mice. k) Density of GFAP+ rAstros is significantly increased on the damaged side in laser-induced injury tissue (p = 0.017). l) The ratio of the imaged:contralateral (contra.) GFAP mean intensity was ~1 for healthy mice and was significantly increased in positive controls (p = 0.037). m) No difference in the density of 8-OHG-positive neurons after auto-thresholding (see Methods) between the contralateral and imaged regions in healthy mice. n) No difference in the density of 8-OHG-positive neurons between contralateral and imaged regions in laser-induced injury tissue. o) No difference in the imaged:contralateral ratio of 8-OHG mean intensities between healthy vs. laser-damaged mice. p) No difference in vessel coverage (% positive pixels) between the contralateral and imaged hemispheres of healthy multi-month longitudinal 3P imaging mice. q) Increase in vessel coverage (%) on the ipsilateral hemisphere of laser-induced injury positive control mice (p = 0.022). r) No difference in the density of pericytes between the contra. and ipsi. hemispheres of healthy long-term 3P mice. s) No difference in the density of pericytes between the contra. and ipsi. hemispheres of laser-induced injury mice. *p < 0.05, **p < 0.01, n.s., not significant; n = 5 mice (healthy longitudinal), 5 mice (laser-induced injury), 2 sections, 4 hemispheres per condition. Statistical comparisons in d-e, g-h, j-k, m-n, and p-s were made with paired, two-sided Student’s t-tests (parametric) or two-sided Wilcoxon signed-rank test (nonparametric). Statistical comparisons in f,i,l,o were made with unpaired two-sided Student’s t-tests for equal/unequal variances (parametric) or two-sided Wilcoxon rank sum test (nonparametric). Box plots represent the median, interquartile ranges and minimum/maximum values. For detailed statistics, see Supplementary Table 3.
Extended Data Fig. 7 |. Immunofluorescent and in situ hybridization techniques allow for probing OPC proliferation and oligodendrocyte subpopulations in the cortex and subcortical white matter.
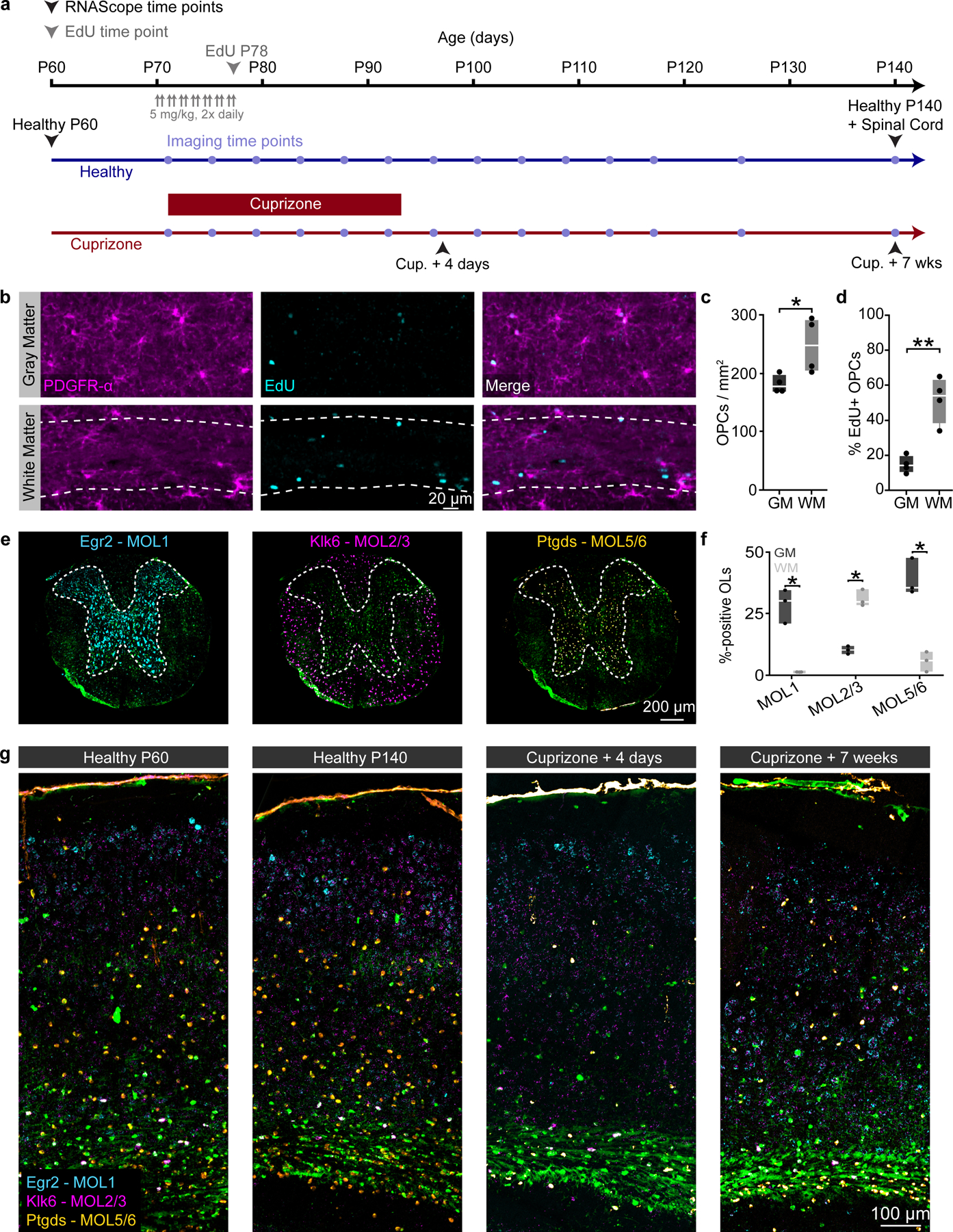
a) Experimental timeline of tissue collection for EdU and RNAScope analyses in healthy and cuprizone mice. Healthy MOBP-EGFP mice were injected with 5 mg/kg of the thymidine analog EdU twice a day (10–12 hr. interval) for seven days starting at P70 and perfused the following day. Healthy MOBP-EGFP mice were perfused at P60 and P140 to assess aging-dependent changes in adult oligodendrocyte subpopulations. MOBP-EGFP mice that were fed 0.2% cuprizone for three weeks were perfused at 4 days post-cuprizone removal (peak demyelination, matched to in vivo imaging data, Fig. 4), and 7 weeks post-cuprizone removal (regeneration, matched to the final time point of in vivo imaging, see timeline, Fig. 4). b) EdU-positive proliferated OPCs in the posterior parietal cortex (top, GM) and subcortical white matter (bottom, WM, dashed border). c) The density of PDGFR-α-positive OPCs was significantly increased in the WM compared to the GM (248.6 ± 23.8 vs. 181.2 ± 7.7, Unpaired, two-tailed Student’s t-test for equal variance, t(6) = 2.69, p = 0.036). d) The percentage PDGFR-α+/EdU+ OPCs is increased in the WM compared to the GM (Unpaired, two-tailed Student’s t-test for equal variance, 51.6 ± 6.6 vs. 14.6 ± 2.5, t(6) = 5.29, p = 0.002). e) Coronal sections from the mid-thoracic spinal cord of healthy MOBP-EGFP mice were taken at P140 and run in parallel with brain sections to confirm the labeling specificity of our oligodendrocyte subpopulation probe-set (Egr2, MOL1, cyan; Klk6, MOL2/3, magenta; Ptgds, MOL5/6, orange). f) Egr2 and Ptgds preferentially label oligodendrocytes in the spinal gray matter, while Klk6 predominantly labels oligodendrocytes in the spinal white matter (Unpaired, two-tailed students t-test for unequal variance). g) Coronal sections from the posterior parietal cortex of MOBP-EGFP mice (−1.7 to −2.3 mm posterior and 1 to 3 mm lateral to bregma) showing the pattern of OL subpopulation labeling at the experimental time points described in (a). *p < 0.05, **p<0.01, n = 4 mice, 4 sections per mouse. Boxplots represent the median, interquartile range and the minimum and maximum values. For detailed statistics, see Supplementary Table 3.
Extended Data Fig. 8 |. Modeling oligodendrocyte growth, loss, and regeneration in adult mouse cortex and white matter.
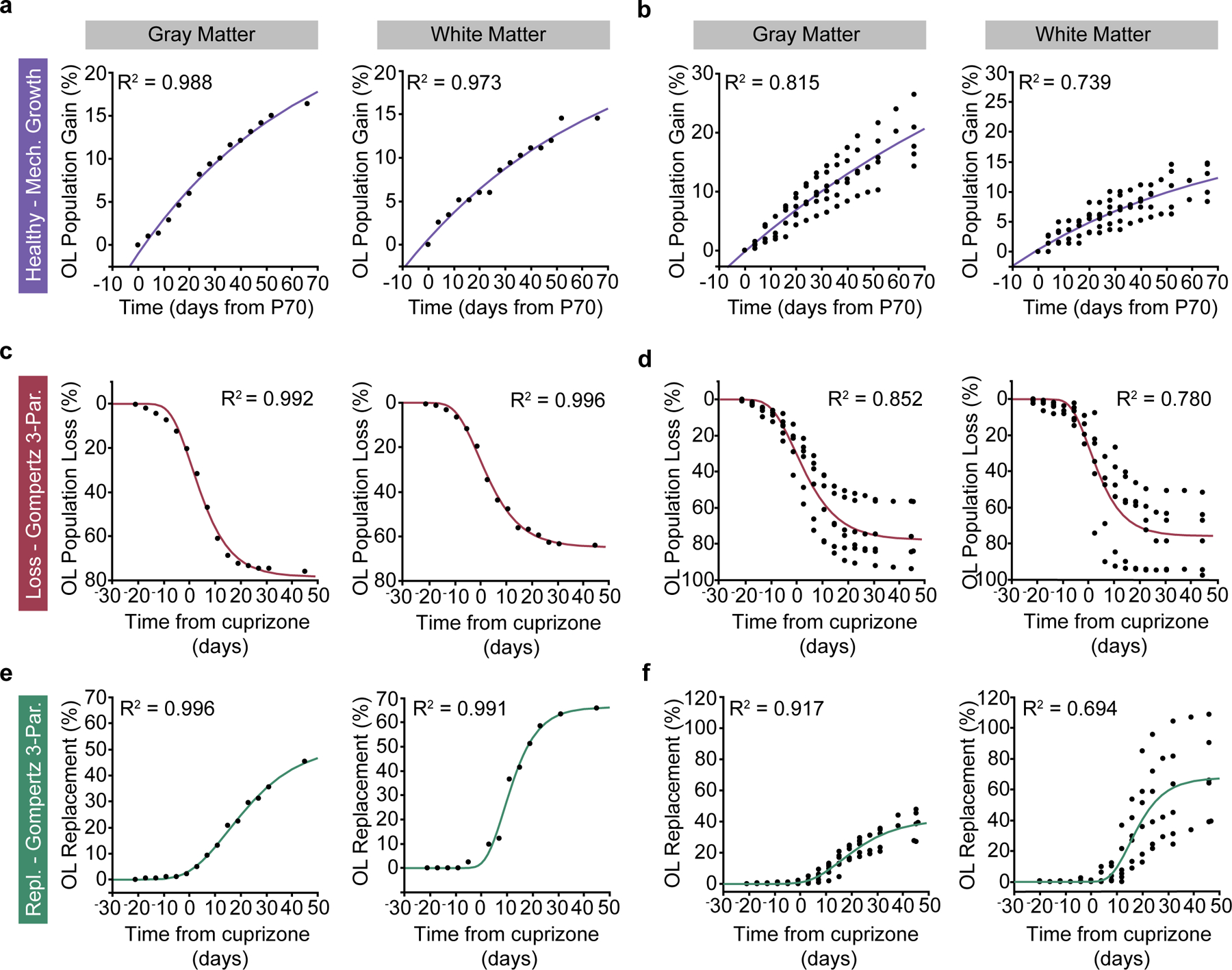
a-b) Cumulative oligodendrocyte population growth (% gain over time) in the healthy brain was modeled using asymptote-restricted exponential mechanistic growth curve-fitting. a) Cumulative OL gain (%) and mechanistic growth fit in the gray matter (left) and the white matter (right) for an example mouse. b) Cumulative OL gain (%) and mechanistic growth fits in the gray matter (left) and the white matter (right) for the healthy group (n = 6 mice). c-d) Cumulative oligodendrocyte population loss (% loss over time) due to cuprizone administration was modeled using three-parameter Gompertz Sigmoid curve-fitting. c) Cumulative OL loss (%) and three-parameter Gompertz curves in the gray matter (left) and the white matter (right) for an example mouse. d) Cumulative OL loss (%) and three-parameter Gompertz curves in the gray matter (left) and the white matter (right) for the cuprizone de/remyelination group (n = 6 mice). e-f) Cumulative oligodendrocyte population regeneration (% cell replacement over time) following cuprizone cessation was modeled using three-parameter Gompertz Sigmoid curve-fitting. e) Cumulative OL replacement (%) and three-parameter Gompertz curves in the gray matter (left) and the white matter (right) for an example mouse. f) Cumulative OL replacement (%) and three-paramter Gompertz curves in the gray matter (left) and the white matter (right) for the cuprizone de/remyelination group (n = 6 mice). Modeled rate and timing metrics in Main Figs. 4–7 were calculated by fitting curves to data from individual mice and then extracting summary data (e.g. timing of inflection point).
Extended Data Figure 9 |. Layer and region-dependent proportions of MOL1, MOL2/3, and MOL5/6 across healthy aging and cuprizone treatment.
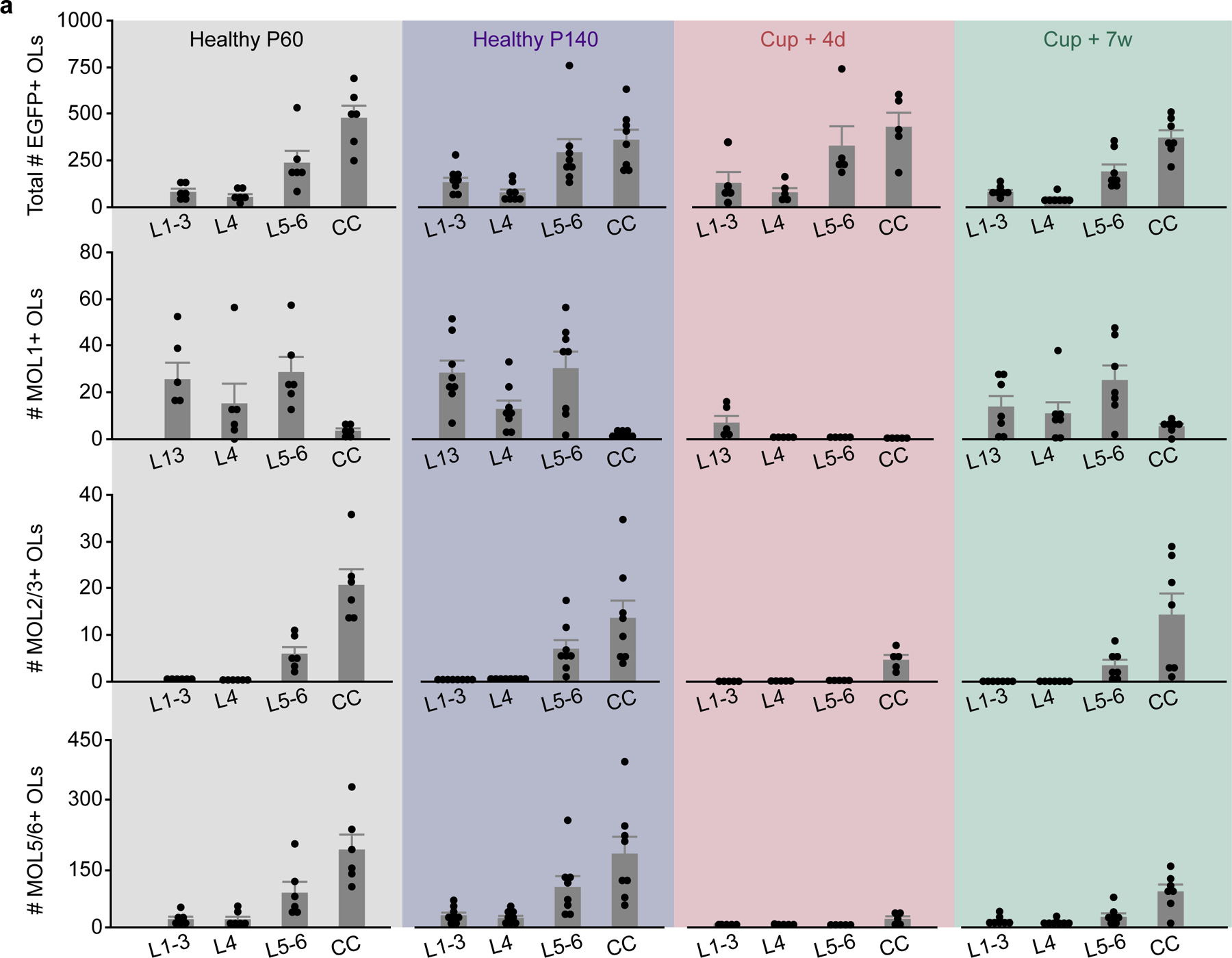
Related to Figs. 7,8. Raw counts of the total # of segmented MOBP-EGFP OLs (top), MOL1+ OLs (row 2), MOL2/3+ OLs (row 3) and MOL5/6+ OLs (bottom) across cortical and subcortical layers (x-axes) and experimental time points (shaded background colors). Data are from n=6 mice (Healthy P60), n=8 mice (Healthy P140), n=5 mice (Cup. + 4 days) and n=7 mice (Cup. + 7 weeks), 2 sections per mouse. Bar graphs represent the mean+/−SEM. Data presented in the main figures were expressed either as the percentage of total OLs for each probe (Figs. 7–8), or the change in proportion of each marker from the healthy P140 time point (Figs. 4–6).
Extended Data Figure 10 |. Layer-dependent differences in the temporal dynamics of cuprizone induced loss and regeneration.
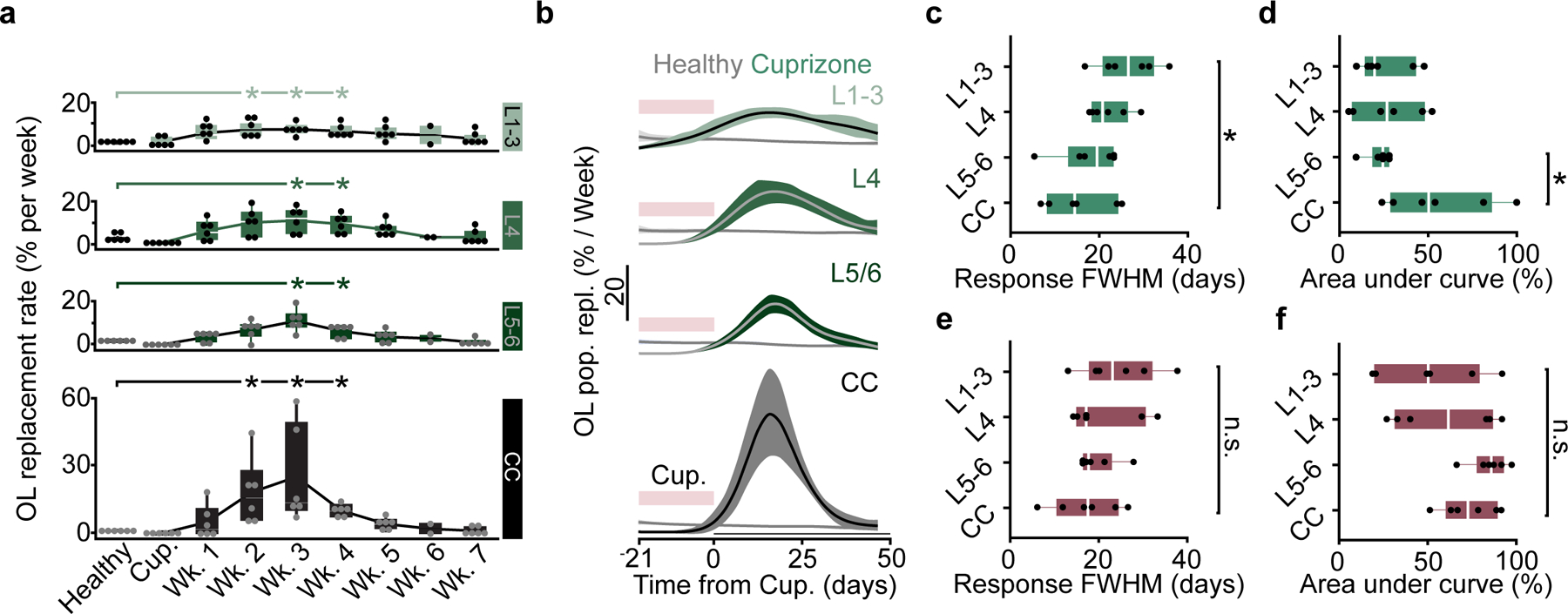
a-e) Related to Fig. 7. a) OL population replacement (%) plotted over weekly time bins following cuprizone cessation for each anatomically defined region. Replacement rate is increased from healthy baseline from 2–4 weeks post-cuprizone in L1–3 and CC, and 3–4 weeks post-cuprizone in L4 and L5/6 (Kruskal-Wallis test followed by Steel method for comparison with control, p = 0.034 for L1–3, Weeks 2–4 vs. Healthy; p = 0.034 for L4 Weeks 3–4 vs. Healthy; p = 0.034 for L5–6, Weeks 3–4 vs. Healthy; p = 0.034 for CC Weeks 2–4 vs. Healthy). n=6 mice (cuprizone), 2 mice were imaged at an extra time point during Week 6. b) First-derivative of the modeled growth curves for healthy baseline and OL replacement (Mechanistic Growth, Gompertz 3P, respectively) showing the response duration of regeneration across regions. c) The regeneration response duration, calculated as the full width at half maximum of the rate curves in (b), is significantly longer in the superficial layers 1–3 compared to the CC (One-way ANOVA followed by Tukey’s HSD, p = 0.048). d) The magnitude of the regeneration response, as calculated by the area under the curve above the healthy baseline rate, is significantly suppressed in L5–6 compared to the CC (One-way ANOVA followed by Tukey’s HSD, p = 0.037). e-f) No significant differences in the duration or magnitude of the cuprizone cell loss response across regions. Data in (a) were calculated based on the raw % change in cell population from baseline. Data in b-f were derived from the modeled and scaled growth curves. n = 6 mice (cuprizone), n=6 mice (healthy). *p < 0.05, **p < 0.01, n.s. = not significant; for growth curves, cubic splines approximate the measure of center and error bars are 95% confidence intervals; box plots represent the median, interquartile ranges and minimum/maximum values. For detailed statistics, see Supplementary Table 3.
Supplementary Material
Acknowledgements
We thank Mike Hall for machining expertise, Andrew Scallon and the CU Anschutz Optogenetics and Neural Engineering Core (P30NS048154) for 3D printing and stage design, past and current members of the Hughes Lab and the CU Anschutz Myelin Group for discussions, Drs. Julie Siegenthaler and Stephanie Bonney (Andy Shih Lab) for helpful discussions on the vascular and pericyte analyses. Michael Cammer for assistance with the histogram measurement ImageJ macros (NYU Langone Health Imaging Core). Dominik Stitch and the University of Colorado Anschutz Medical Campus Advanced Light Microscopy Core (ALMC) Facility for assistance with RNA-Scope imaging that was performed in the ALMC of the NeuroTechnology Center which is supported in part by Rocky Mountain Neurological Disorders Core Grant (P30 NS048154) and by Diabetes Research Center Grant (P30 DK116073). Funding was provided by the National Institutes of Health NINDS (F31NS120540) to MAT. Funding was provided by the University of Colorado Department of Cell and Developmental Biology Pilot Grant, the Whitehall Foundation, and the National Multiple Sclerosis Society (RG-1701-26733) and National Institutes of Health NINDS (NS115975, NS125230, NS132859) to EGH. Funding was provided by the NINDS (R01 NS118188 and UF1 NS116241) and the National Science Foundation (BCS-1926676) to EAG and DR. Funding was provided by the National Institutes of Health NEI (R01 EY030841) to APP.
Footnotes
Code availability
Code for analysis associated with the manuscript is available at https://github.com/EthanHughesLab/; RNA Scope analysis scripts are available at https://github.com/sbudoff/. The code to measure the brightest n% of pixels through a z-stack or time-series was modified from macro scripts available via the NYU Imaging Core website https://microscopynotes.com/imagej/.
Competing Interests
KK is a co-founder and part-owner of 3i. The other authors declare no competing financial interests.
Data and materials availability
All data that support the findings, tools, and reagents will be shared on an unrestricted basis; requests should be directed to the corresponding author.
References
- 1.Flechsig Of Leipsic, P. DEVELOPMENTAL (MYELOGENETIC) LOCALISATION OF THE CEREBRAL CORTEX IN THE HUMAN SUBJECT. The Lancet 158, 1027–1030 (1901). [Google Scholar]
- 2.Spitzer SO et al. Oligodendrocyte Progenitor Cells Become Regionally Diverse and Heterogeneous with Age. Neuron 101, 459–471.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dimou L, Simon C, Kirchhoff F, Takebayashi H & Götz M Progeny of Olig2-Expressing Progenitors in the Gray and White Matter of the Adult Mouse Cerebral Cortex. J. Neurosci 28, 10434–10442 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang SH, Fukaya M, Yang JK, Rothstein JD & Bergles DE NG2+ CNS Glial Progenitors Remain Committed to the Oligodendrocyte Lineage in Postnatal Life and following Neurodegeneration. Neuron 68, 668–681 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Viganò F, Möbius W, Götz M & Dimou L Transplantation reveals regional differences in oligodendrocyte differentiation in the adult brain. Nat Neurosci 16, 1370–1372 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Marques S et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science 352, 1326–1329 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Floriddia EM et al. Distinct oligodendrocyte populations have spatial preference and different responses to spinal cord injury. Nat Commun 11, 5860 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hilscher MM et al. Spatial and temporal heterogeneity in the lineage progression of fine oligodendrocyte subtypes. BMC Biol 20, 122 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pandey S et al. Disease-associated oligodendrocyte responses across neurodegenerative diseases. Cell Reports 40, 111189 (2022). [DOI] [PubMed] [Google Scholar]
- 10.Falcão AM et al. Disease-specific oligodendrocyte lineage cells arise in multiple sclerosis. Nature Medicine 24, 1837 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jäkel S et al. Altered human oligodendrocyte heterogeneity in multiple sclerosis. Nature 566, 543 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snaidero N et al. Myelin replacement triggered by single-cell demyelination in mouse cortex. Nat Commun 11, 4901 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith KJ, Blakemore WF & Mcdonald WI Central remyelination restores secure conduction. Nature 280, 395–396 (1979). [DOI] [PubMed] [Google Scholar]
- 14.Bacmeister CM et al. Motor learning promotes remyelination via new and surviving oligodendrocytes. Nature Neuroscience 23, 819–831 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bø L, Vedeler CA, Nyland HI, Trapp BD & Mørk SJ Subpial Demyelination in the Cerebral Cortex of Multiple Sclerosis Patients. Journal of Neuropathology & Experimental Neurology 62, 723–732 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Kutzelnigg A et al. Cortical demyelination and diffuse white matter injury in multiple sclerosis. Brain 128, 2705–2712 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Peterson JW, Bö L, Mörk S, Chang A & Trapp BD Transected neurites, apoptotic neurons, and reduced inflammation in cortical multiple sclerosis lesions. Ann Neurol. 50, 389–400 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Helmchen F & Denk W Deep tissue two-photon microscopy. Nat Methods 2, 932–940 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Orthmann-Murphy J et al. Remyelination alters the pattern of myelin in the cerebral cortex. eLife 9, e56621 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Call CL & Bergles DE Cortical neurons exhibit diverse myelination patterns that scale between mouse brain regions and regenerate after demyelination. Nat Commun 12, 4767 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horton NG et al. In vivo three-photon microscopy of subcortical structures within an intact mouse brain. Nat Photon 7, 205–209 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Streich L et al. High-resolution structural and functional deep brain imaging using adaptive optics three-photon microscopy. Nat Methods 18, 1253–1258 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ouzounov DG et al. In vivo three-photon imaging of activity of GCaMP6-labeled neurons deep in intact mouse brain. Nat Meth 14, 388–390 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang T, Wang T & Xu C Three-photon neuronal imaging in deep mouse brain. Optica, OPTICA 7, 947–960 (2020). [Google Scholar]
- 25.Hughes EG, Orthmann-Murphy JL, Langseth AJ & Bergles DE Myelin remodeling through experience-dependent oligodendrogenesis in the adult somatosensory cortex. Nature Neuroscience 21, 696–706 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Squier J, Muller M, Brakenhoff G & Wilson KR Third harmonic generation microscopy. Opt Express 3, 315–324 (1998). [DOI] [PubMed] [Google Scholar]
- 27.Débarre D et al. Imaging lipid bodies in cells and tissues using third-harmonic generation microscopy. Nat Methods 3, 47–53 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Yildirim M, Sugihara H, So PTC & Sur M Functional imaging of visual cortical layers and subplate in awake mice with optimized three-photon microscopy. Nature Communications 10, 177 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schain AJ, Hill RA & Grutzendler J Label-free in vivo imaging of myelinated axons in health and disease with spectral confocal reflectance microscopy. Nat Med 20, 443–449 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paxinos G & Franklin KBJ Paxinos and Franklin’s the Mouse Brain in Stereotaxic Coordinates. (Academic Press, 2019). [Google Scholar]
- 31.Chapman TW, Olveda GE, Bame X, Pereira E & Hill RA Oligodendrocyte death initiates synchronous remyelination to restore cortical myelin patterns in mice. Nat Neurosci 26, 555–569 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yildirim M et al. Quantitative third-harmonic generation imaging of mouse visual cortex areas reveals correlations between functional maps and structural substrates. Biomed. Opt. Express, BOE 11, 5650–5673 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Podgorski K & Ranganathan G Brain heating induced by near-infrared lasers during multiphoton microscopy. Journal of Neurophysiology 116, 1012–1023 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang T et al. Quantitative analysis of 1300-nm three-photon calcium imaging in the mouse brain. eLife 9, e53205 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nunomura A et al. RNA Oxidation Is a Prominent Feature of Vulnerable Neurons in Alzheimer’s Disease. J. Neurosci 19, 1959–1964 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arganda-Carreras I et al. Trainable Weka Segmentation: a machine learning tool for microscopy pixel classification. Bioinformatics 33, 2424–2426 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Schager B & Brown CE Susceptibility to capillary plugging can predict brain region specific vessel loss with aging. J Cereb Blood Flow Metab 40, 2475–2490 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tripathi RB et al. Remarkable Stability of Myelinating Oligodendrocytes in Mice. Cell Reports 21, 316–323 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu X et al. Age-dependent fate and lineage restriction of single NG2 cells. Development 138, 745–753 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dawson MRL, Polito A, Levine JM & Reynolds R NG2-expressing glial progenitor cells: an abundant and widespread population of cycling cells in the adult rat CNS. Molecular and Cellular Neuroscience 24, 476–488 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Rosenberg SS, Kelland EE, Tokar E, De la Torre AR & Chan JR The geometric and spatial constraints of the microenvironment induce oligodendrocyte differentiation. Proc Natl Acad Sci U S A 105, 14662–14667 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hill RA, Li AM & Grutzendler J Lifelong cortical myelin plasticity and age-related degeneration in the live mammalian brain. Nature Neuroscience 21, 683–695 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skripuletz T et al. Cortical demyelination is prominent in the murine cuprizone model and is strain-dependent. Am J Pathol 172, 1053–1061 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gudi V et al. Regional differences between grey and white matter in cuprizone induced demyelination. Brain Res 1283, 127–138 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Wergeland S, Torkildsen Ø, Myhr K-M, Mørk SJ & Bø L The cuprizone model: regional heterogeneity of pathology. APMIS 120, 648–657 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Baxi EG et al. Lineage tracing reveals dynamic changes in oligodendrocyte precursor cells following cuprizone-induced demyelination. Glia 65, 2087–2098 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tjørve KMC & Tjørve E The use of Gompertz models in growth analyses, and new Gompertz-model approach: An addition to the Unified-Richards family. PLoS One 12, e0178691 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang F et al. Myelin degeneration and diminished myelin renewal contribute to age-related deficits in memory. Nature Neuroscience 23, 481–486 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thornton MA & Hughes EG Neuron-oligodendroglia interactions: Activity-dependent regulation of cellular signaling. Neuroscience Letters 727, 134916 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Young KM et al. Oligodendrocyte Dynamics in the Healthy Adult CNS: Evidence for Myelin Remodeling. Neuron 77, 873–885 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hughes EG, Kang SH, Fukaya M & Bergles DE Oligodendrocyte progenitors balance growth with self-repulsion to achieve homeostasis in the adult brain. Nat Neurosci 16, 668–676 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hontani Y, Xia F & Xu C Multicolor three-photon fluorescence imaging with single-wavelength excitation deep in mouse brain. Science Advances 7, eabf3531 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thornton MA et al. Characterization of red fluorescent reporters for dual-color in vivo three-photon microscopy. NPh 9, 031912 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hughes EG, Orthmann-Murphy JL, Langseth AJ & Bergles DE Myelin remodeling through experience-dependent oligodendrogenesis in the adult somatosensory cortex. Nat Neurosci 21, 696–706 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Albert M, Antel J, Brück W & Stadelmann C Extensive Cortical Remyelination in Patients with Chronic Multiple Sclerosis. Brain Pathology 17, 129–138 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Remington LT, Babcock AA, Zehntner SP & Owens T Microglial recruitment, activation, and proliferation in response to primary demyelination. Am J Pathol 170, 1713–1724 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Franklin RJM & ffrench-Constant C Regenerating CNS myelin — from mechanisms to experimental medicines. Nature Reviews Neuroscience 18, nrn.2017.136 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Sherafat A, Pfeiffer F, Reiss AM, Wood WM & Nishiyama A Microglial neuropilin-1 promotes oligodendrocyte expansion during development and remyelination by trans-activating platelet-derived growth factor receptor. Nature Communications 12, 2265 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Samanta J et al. Inhibition of Gli1 mobilizes endogenous neural stem cells for remyelination. Nature 526, 448–452 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Armingol E, Officer A, Harismendy O & Lewis NE Deciphering cell–cell interactions and communication from gene expression. Nat Rev Genet 22, 71–88 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chang A et al. Cortical remyelination: A new target for repair therapies in multiple sclerosis. Ann Neurol. 72, 918–926 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Strijbis EMM, Kooi E-J, van der Valk P & Geurts JJG Cortical Remyelination Is Heterogeneous in Multiple Sclerosis. J Neuropathol Exp Neurol 76, 390–401 (2017). [DOI] [PubMed] [Google Scholar]
- 63.Geurts JJG & Barkhof F Grey matter pathology in multiple sclerosis. Lancet Neurol 7, 841–851 (2008). [DOI] [PubMed] [Google Scholar]
- 64.Lentferink DH, Jongsma JM, Werkman I & Baron W Grey matter OPCs are less mature and less sensitive to IFNγ than white matter OPCs: consequences for remyelination. Sci Rep 8, 2113 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kirby L et al. Oligodendrocyte precursor cells present antigen and are cytotoxic targets in inflammatory demyelination. Nat Commun 10, 3887 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kotter MR, Li W-W, Zhao C & Franklin RJM Myelin Impairs CNS Remyelination by Inhibiting Oligodendrocyte Precursor Cell Differentiation. J. Neurosci 26, 328–332 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harris KD & Shepherd GMG The neocortical circuit: themes and variations. Nat Neurosci 18, 170–181 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Madsen MAJ et al. Linking lesions in sensorimotor cortex to contralateral hand function in multiple sclerosis: a 7 T MRI study. Brain awac203 (2022) doi: 10.1093/brain/awac203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Battefeld A, Klooster J & Kole MHP Myelinating satellite oligodendrocytes are integrated in a glial syncytium constraining neuronal high-frequency activity. Nature Communications 7, 11298 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dubey M et al. Myelination synchronizes cortical oscillations by consolidating parvalbumin-mediated phasic inhibition. eLife 11, e73827 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schirmer L et al. Neuronal vulnerability and multilineage diversity in multiple sclerosis. Nature 573, 75–82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hughes EG, Orthmann-Murphy JL, Langseth AJ & Bergles DE Myelin remodeling through experience-dependent oligodendrogenesis in the adult somatosensory cortex. Nature Neuroscience 21, 696–706 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rodríguez C et al. An adaptive optics module for deep tissue multiphoton imaging in vivo. Nat Methods 18, 1259–1264 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ren C & Komiyama T Wide-field calcium imaging of cortex-wide activity in awake, head-fixed mice. STAR Protoc 2, 100973 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Squier JA & Müller M Third-harmonic generation imaging of laser-induced breakdown in glass. Appl. Opt., AO 38, 5789–5794 (1999). [DOI] [PubMed] [Google Scholar]
- 76.Sinefeld D et al. Three-Photon Adaptive Optics for Mouse Brain Imaging. Frontiers in Neuroscience 16, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Champelovier D et al. Image-based adaptive optics for in vivo imaging in the hippocampus. Sci Rep 7, 42924 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lein ES et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007). [DOI] [PubMed] [Google Scholar]
- 80.Thevenaz P, Ruttimann UE & Unser M A pyramid approach to subpixel registration based on intensity. IEEE Transactions on Image Processing 7, 27–41 (1998). [DOI] [PubMed] [Google Scholar]
- 81.Rezakhaniha R et al. Experimental investigation of collagen waviness and orientation in the arterial adventitia using confocal laser scanning microscopy. Biomech Model Mechanobiol 11, 461–473 (2012). [DOI] [PubMed] [Google Scholar]
- 82.Püspöki Z, Storath M, Sage D & Unser M Transforms and Operators for Directional Bioimage Analysis: A Survey. Adv Anat Embryol Cell Biol 219, 69–93 (2016). [DOI] [PubMed] [Google Scholar]
- 83.Chehrehasa F, Meedeniya ACB, Dwyer P, Abrahamsen G & Mackay-Sim A EdU, a new thymidine analogue for labelling proliferating cells in the nervous system. J Neurosci Methods 177, 122–130 (2009). [DOI] [PubMed] [Google Scholar]
- 84.Zeng C et al. Evaluation of 5-ethynyl-2’-deoxyuridine staining as a sensitive and reliable method for studying cell proliferation in the adult nervous system. Brain Res 1319, 21–32 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Young KM Oligodendrocyte dynamics in the healthy adult CNS: evidence for myelin remodeling. Neuron 77, 873–885 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cieślar-Pobuda A & Łos MJ Prospects and limitations of “Click-Chemistry”-based DNA labeling technique employing 5-ethynyl-2′deoxyuridine (EdU). Cytometry Part A 83, 977–978 (2013). [DOI] [PubMed] [Google Scholar]
- 87.Kohlmeier F, Maya-Mendoza A & Jackson DA EdU induces DNA damage response and cell death in mESC in culture. Chromosome Res 21, 87–100 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Podgorski K & Ranganathan G Brain heating induced by near-infrared lasers during multiphoton microscopy. Journal of Neurophysiology 116, 1012–1023 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Peng T et al. A BaSiC tool for background and shading correction of optical microscopy images. Nat Commun 8, 14836 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bankhead P et al. QuPath: Open source software for digital pathology image analysis. Sci Rep 7, 16878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Maino JL & Kearney MR Testing mechanistic models of growth in insects. Proceedings of the Royal Society B: Biological Sciences 282, 20151973 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bacmeister CM et al. Motor learning promotes remyelination via new and surviving oligodendrocytes. Nat Neurosci 23, 819–831 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Laird AK Dynamics of Tumour Growth. Br J Cancer 18, 490–502 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Benzekry S et al. Classical Mathematical Models for Description and Prediction of Experimental Tumor Growth. PLOS Computational Biology 10, e1003800 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data that support the findings, tools, and reagents will be shared on an unrestricted basis; requests should be directed to the corresponding author.


