Abstract
Devil facial tumour disease (DFTD) comprises two genetically distinct transmissible cancers (DFT1 and DFT2) endangering the survival of the Tasmanian devil (Sarcophilus harrisii) in the wild. DFT1 first arose from a cell of the Schwann cell lineage; however, the tissue-of-origin of the recently discovered DFT2 cancer is unknown. In this study, we compared the transcriptome and proteome of DFT2 tumours to DFT1 and normal Tasmanian devil tissues to determine the tissue-of-origin of the DFT2 cancer. Our findings demonstrate that DFT2 expresses a range of Schwann cell markers and exhibits expression patterns consistent with a similar origin to the DFT1 cancer. Furthermore, DFT2 cells express genes associated with the repair response to peripheral nerve damage. These findings suggest that devils may be predisposed to transmissible cancers of Schwann cell origin. The combined effect of factors such as frequent nerve damage from biting, Schwann cell plasticity and low genetic diversity may allow these cancers to develop on rare occasions. The emergence of two independent transmissible cancers from the same tissue in the Tasmanian devil presents an unprecedented opportunity to gain insight into cancer development, evolution and immune evasion in mammalian species.
Electronic supplementary material
The online version of this article (10.1007/s00018-019-03259-2) contains supplementary material, which is available to authorized users.
Keywords: Tasmanian devil, Devil facial tumour disease, Schwann cell, Transmissible cancer, Contagious cancer, DFT2
Introduction
The mammalian immune system has been shaped over millions of years to protect against infectious disease and external threats to physiological homeostasis. In mammals, naturally occurring transmissible cancers have only been identified in two species: canine transmissible venereal tumour in dogs (Canis lupus familiaris) [1] and devil facial tumour disease (DFTD) in Tasmanian devils (Sarcophilus harrisii) [2]. The recent emergence of two genetically independent transmissible cancers (DFT1 and DFT2) in the Tasmanian devil has highlighted a potential predisposition of this marsupial species to developing contagious cancers [3, 4]. Known collectively as DFTD, both DFT1 and DFT2 cancers manifest as aggressive facial tumours that are transferred between hosts as allogeneic grafts during typical biting behaviours of the Tasmanian devil [2, 3]. Although DFT1 and DFT2 are almost indistinguishable by gross examination and share similar patterns of mutation [4], karyotypic and genetic studies have revealed the independent nature of these two cancers. Specifically, complex chromosomal rearrangements and microsatellite genotypes that are identical within each clonal lineage differ between DFT1 and DFT2 [3, 5]. Karyotypic and genetic studies have also revealed two homologous copies of the X chromosome in DFT1 cells and an X and Y in DFT2, confirming that the cancers arose independently [3, 6, 7].
DFT1 was first observed in 1996 and has triggered severe population declines across most of the island state of Tasmania [8, 9]. Extensive studies into the nature and biology of this tumour have identified DFT1 as a cancer of the Schwann cell lineage based on expression of transcription factors and proteins related to the myelination pathway [10]. Periaxin (PRX), a myelin protein, has proven to be a reliable diagnostic marker for DFT1 in tissue sections [10, 11]. Indeed, it was the lack of expression of PRX in collected DFTD samples that led to the discovery of DFT2 in southern Tasmania in 2014 [3]. As DFT2 was discovered only recently, its tissue-of-origin remains unknown, although it is known to express several markers consistent with a neuroectodermal origin [4]. An understanding of the founder cells of DFT2 tumours in devils will be paramount to further investigations into the susceptibility of this endangered species to transmissible cancers.
In this study, we compared the transcriptome and proteome of DFT2 tumours to DFT1 and normal Tasmanian devil tissues to determine the tissue-of-origin of the DFT2 cancer. Our findings demonstrate that DFT2 expresses a range of Schwann cell markers and exhibits expression patterns consistent with a similar origin to the DFT1 cancer. We discuss these findings in terms of current knowledge of DFTD and propose a mechanism by which injury from biting behaviours may combine with other factors such as low genetic diversity to increase the risk of transmissible Schwann cell tumours emerging in the Tasmanian devil.
Materials and methods
Cell lines and biological samples
The DFT1 cell line C5065 was provided by A.-M. Pearse and K. Swift of the Tasmanian Department of Primary Industries, Parks, Water and Environment (DPIPWE). This cell line was previously established from a DFT1 biopsy obtained under the approval of the Animal Ethics Committee of the Tasmanian Parks and Wildlife Service (Permit No. 33/2004-5, 32/2005-6). The DFT2 cell line SN and fibroblast cell line TD344 were established from single cell suspensions obtained from tissue biopsies. All cell cultures were maintained in RPMI 1640 medium (Thermo Fisher Scientific, Waltham, USA) supplemented with 10% foetal calf serum, at 35 °C and 5% CO2 in a humidified incubator. DFTD biopsies were collected from diseased devils using a 4 mm disposable biopsy punch (Kai Medical, Singen, Germany). Other primary tissue samples were obtained from Tasmanian devils ethically euthanised for welfare reasons. All samples were collected under the approval of the University of Tasmania Animal Ethics Committee (Permit Nos. A009215, A0012513 and A0014976).
RNA sequencing and analysis
Tissue samples stored in RNAlater (Thermo Fisher Scientific, Waltham, USA) were homogenised for RNA extraction by bead-beating with 2.3 mm zirconia beads using a Mini-Beadbeater-24 (BioSpec Products, Bartlesville, USA). RNA was extracted using the RNeasy® Mini Kit (QIAGEN Bioinformatics, Hilden, Germany), according to the instructions of the manufacturer. Duplicate biological replicates were prepared for each tissue type. Due to the minimal amount of peripheral nerve tissue that was available for this project, RNA samples were combined for this tissue to generate a RNA yield sufficient for sequencing. RNA integrity was assessed using a Eukaryotic RNA 6000 Nano Kit and 2100 Bioanalyzer (Agilent, Santa Clara, USA). All samples had RIN scores greater than 9.0. 100 base-pair single-read mRNA sequencing of duplicate samples for each tissue was performed on the Illumina Hiseq-2500 platform (Illumina, San Diego, USA), as previously described [12]. Sequence quality was assessed using fastqc (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Sequencing reads were aligned to the Tasmanian devil reference genome (7.0.82) using subread [13], and counts were summarized into genes using featureCounts [14]. Technical replicates were combined and a minimum threshold of 50 counts across all samples was applied to exclude genes with extremely low expression. Expression levels were normalized by upper quartile normalisation using EDAseq [15]. Differential gene expression was calculated using limma/voom (Online resource 1) [16]. Boxplots of pairwise Euclidean distances of gene expression between DFT1 and DFT2, and between human tumours originating from the same and different tissue types were calculated using RPKM-normalised DFTD read counts and human cancer data obtained from Weinstein et al. [17]. The difference in the distribution of pairwise distances between the ‘within’ and ‘between’ tissues-of-origin groups in human cancers was calculated using a Wilcoxon rank sum test. The difference in the distribution of pairwise distances between DFTD and the human ‘within’ and ‘between’ tissue-of-origin groups was calculated using a Kolmogorov–Smirnov test. Hierarchical clustering of sample data (columns) was performed on the union of differentially expressed genes (logFC > 2.0, p < 0.05) using the R package pvclust, according to the average method and a correlation-based dissimilarity matrix. Approximately unbiased p values (AU) and bootstrap probabilities (BP) were estimated from 1000 bootstrapping iterations. Genes (rows) were clustered based on Pearson’s correlation coefficient. Heat map colour represents the number of standard deviations each data point lies from the population mean across all tissue samples (z score). For functional enrichment analysis, enriched GO terms, KEGG pathways and REACTOME pathways were clustered into functionally-associated groups based on shared genes (kappa score > 0.4) using ClueGO version 2.5 [24]. Single sample gene set enrichment analysis (ssGSEA) was performed using the GSVA R package, and gene set enrichment analysis (GSEA) was performed using GSEA 3.0 [18, 19]. Gene sets analysed using ssGSEA and GSEA were obtained from a previous study exploring the Schwann cell transcriptome during response to injury in a mouse model [20]. Enrichment of the top 50 up-regulated genes during injury was analysed in devil tissues to represent a repair Schwann cell phenotype, and the top 50 down-regulated genes were analysed to represent a myelinating Schwann cell phenotype.
Protein extraction and digestion
Triplicate samples of C5065, SN and TD344 cell lines were resuspended in 1 ml of 100 mM triethylammonium bicarbonate (TEAB, Sigma Aldrich) containing 1% (w/v) sodium deoxycholate (SDC, Sigma Aldrich, pH7.8–8.2) and lysed at 95 °C for 5 min. Protein samples were incubated at room temperature with Benzonase nuclease (Sigma Aldrich) for 30 min to degrade RNA and DNA. Protein was recovered by removing the supernatant after centrifugation (14,000g for 5 min at 4 °C), and the protein concentration was estimated using a colorimetric bicinchoninic acid (BCA) protein assay kit (Pierce™). Protein (100 µg) from each biological sample was reduced at 65 °C for 30 min using dithiothreitol (DTT; 10 mM), then alkylated with iodoacetamide (IAA; 20 mM) at room temperature for 30 min and digested with trypsin (5 µg) overnight at 37 °C. Samples were acidified with formic acid (1% v/v) to precipitate the sodium deoxycholate. Sodium deoxycholate was removed by centrifugation (14,000×g for 5 min at 4 °C) and the peptide supernatant dried by vacuum concentration. Peptides were stored at − 20 °C and resuspended in 2% acetonitrile and 0.1% formic acid in water, before analysis by LC–MS.
High pH fractionation
For each cell line, 50 µg of protein from each of three replicates was combined to generate a pooled sample (150 µg), which was purified using a Sep-Pak C18 column (Waters) as per the manufacturer’s guidelines. Peptides were eluted in two steps; 40% acetonitrile followed by 80% acetonitrile with 0.1% formic acid. Peptides from both elution steps were dried by vacuum centrifugation then resuspended in 5 mM ammonium hydroxide and combined before separating using a ZORBAX-300SB-C18 Extend column (2.1 × 150 mm, 3.5 µm, 300 Å, Agilent) using an Agilent 1260 HPLC system equipped with a fraction collector. Peptides were eluted using a linear gradient of 5 mM ammonium hydroxide in 90% acetonitrile from 3 to 30% over 55 min, then 30–70% over 10 min at a flow rate of 0.3 ml/min. Fractions were collected every 1 min, combined into a total of 13 fractions for each cell line, then dried by vacuum centrifugation and resuspended in 2% acetonitrile with 0.1% formic acid for LC/MS analysis.
LC–MS/MS analysis and data acquisition
Nanoflow LC–MS/MS was carried out in positive mode using a Triple TOF 6600 mass spectrometer (SCIEX) equipped with an Eksigent nanoLC 400 liquid chromatography system (SCIEX) and nanoflex cHiPLC module (SCIEX). Peptides (10 µl, approx. 2 µg) were desalted with 2% acetonitrile (0.1% formic acid) at a flow rate of 5 µl/min for 5 min using a C18 trap (2 cm, 200 μm, 2.7 μm, Halo C18) for both Information Dependant Acquisition (IDA) and data independent (SWATH) experiments. For IDA, peptides were separated on a cHiPLC C18 column (15 cm × 200 μm, 3 μm, ChromXP C18CL, 120 Å, 25 °C, SCIEX) using a linear solvent gradient from 2% acetonitrile (0.1% formic acid) to 35% mobile phase B (B: 90% acetonitrile, 0.1% formic acid) at 600 nL/min over 120 min. For SWATH data was acquired using a 60 min LC gradient (5–35% mobile phase B) at 600 nl/min. Liquid chromatography eluent was subjected to positive ion nanoflow electrospray MS analysis (spray voltage 2.5 kV, curtain gas 25, GS1 20) using a nanospray III source (SCIEX) and an uncoated PicoTip Emitter (10 cm, 360 μm O.D., 20 μm I.D., New Objective). For spectral library generation in IDA mode, high pH fractions were analysed using a TOFMS survey scan (m/z 350–1500, 200 ms) followed by MS/MS analysis (100 ms each, m/z 100–1800) of the top 20 most intense precursor ions with an exclusion time of 30 s. For SWATH experiments, samples were analysed in data independent mode using m/z variable windows (100 in total) determined based on precursor m/z densities from IDA data. In SWATH mode a TOFMS survey scan was acquired (m/z 350–1500, 30 ms, high resolution setting of 30,000) followed by 100 SWATH-MS2 scans (m/z 350–1500, 30 ms, variable windows, high sensitivity setting of 15,000). Each SWATH-MS2 scan used a rolling collision energy (CE = 0.06 × m/z − 1 for +2 ions and, CE = 0.06 × m/z − 3 for +3 to 5 ions) and CE spread of 5. SWATH experiments for each digested sample were acquired in a random order with one blank injection acquired between each sample.
LC–MS/MS data analysis
Data was searched using the Paragon algorithm (SCIEX) in ProteinPilot (Version 5.0, SCIEX) in thorough ID mode with FDR calculation enabled and allowing biological modification. MS/MS spectra from all IDA experiment were searched against a reference database for the Tasmanian Devil (Sarcophilus harrisii, UniProt KB, 22, 388 entries, November 2016) to generate a spectral library. Carbamidomethyl modification of cysteine residues, and an Unused cut-off score of 1.3 (95% confidence) was used for database searches. For SWATH quantitation, ProteinPilot IDA search results for all high pH fractions were imported into PeakView2.1 with SWATH2.0 MicroApp (SCIEX) and used as a spectral library. Retention times for all Tasmanian devil facial tumour SWATH files were aligned using linear regression and by selecting five endogenous peptides across the elution profile. The top six most intense fragment ions for each peptide were extracted from the SWATH data using a maximum number of peptides of 100, 75 ppm mass tolerance, peptide confidence threshold of ≥ 0.99, and a 5 min retention time extraction window. After data processing, peptides with confidence ≥ 99% and FDR ≤ 1% (based on chromatographic feature after fragment extraction) were used for quantitation. SWATH protein area data (summed area under the curve (AUC) values for each protein) were extracted using PeakView and compared across triplicate biological samples for DFT1, DFT2 and fibroblast cell lines. Protein peak areas were log2 transformed and normalised by subtracting the total protein area for each sample. Two approaches were used to assess differentially expressed proteins including an analysis of variance (ANOVA) of the log-transformed normalised peak areas and a pairwise comparison using a two-sample Student’s t test adjusted for multiple comparisons by Benjamini–Hochberg correction (Online resource 2) [21]. Proteins with p < 0.05 (ANOVA) and an expression fold-change greater than ± 2.0 with FDR < 0.05 by t test were considered significantly altered between respective cell lines. Gene ontology analysis of significantly altered proteins was performed using DAVID version 6.8 [22, 23]. For analysis of highly expressed proteins in DFT2, enriched GO terms, KEGG pathways and REACTOME pathways were clustered into functionally-associated groups based on shared genes (kappa score > 0.4) using ClueGO version 2.5 [24]. For both analyses, protein lists were analysed using the Homo sapiens species database to overcome the paucity of biological data available for the Tasmanian devil.
Histology and immunohistochemistry
Tissue sections were fixed in 10% neutral buffered formalin and 3 µm paraffin sections were prepared and placed onto 3-aminotriethoxysilane-coated slides for staining. Standard hematoxylin and eosin (H&E) staining and immunohistochemical staining was performed as previously described [11]. The antibodies and dilutions used for immunohistochemistry are given in Online resource 3.
Availability of data and materials
The datasets supporting the conclusions of this article are available in the European Nucleotide Archive repository [http://www.ebi.ac.uk/ena/data/view/PRJEB28680 and http://www.ebi.ac.uk/ena/data/view/PRJNA416378], and the ProteomeXchange Consortium via the PRIDE partner repository [http://www.ebi.ac.uk/pride/archive/projects/PXD011209]. The remainder of the data supporting this article are included within the article and its additional files.
Results
Transcriptome analysis of DFT2 cell lines and primary tissues
DFT2 cancers are known to share a similar neuroectodermal origin to DFT1 [4], but a more precise understanding of the tissue-of-origin of DFT2 is required to understand their emergence. To determine the tissue-of-origin of DFT2 cells, we first performed deep mRNA expression analyses of DFT1 and DFT2 cell lines and primary tumour samples. Analysis of the transcriptome of DFT1 and DFT2 cell lines via RNA sequencing revealed a total of 12,632 genes (Online resource 1), with 10,118 (80.1%) expressed at similar levels (fold change < ± 4.0) across the DFT1 and DFT2 cell lines (Fig. 1a). To determine whether this variation is consistent with a similar tissue-of-origin, we computed the pairwise distance in gene expression between DFT1 and DFT2 cell lines and compared this with the pairwise distance in gene expression between publicly-available human cancer datasets of the same tissue-type and of different tissue-types (Fig. 1b) [17]. As cell lines are susceptible to biological changes in culture, we also included primary DFT1 and DFT2 biopsies in this analysis. The variation in gene expression between DFT1 and DFT2 cell lines was significantly lower than variation between human cancers of different tissue types (p = 0.006), but comparable to variation in human cancers within the same tissue type (p = 0.487) (Fig. 1b). In comparison, the variation between DFT1 and DFT2 biopsies was not significantly different to human cancers of the same (p = 0.973) or different (p = 0.263) tissue types. The increase in pairwise distance between DFT1 and DFT2 biopsies relative to cell lines could be attributable to the presence of immune infiltration in DFT2 biopsies [25]. Nonetheless, as the measured distance between DFT1 and DFT2 biopsies overlaps the distribution of pairwise distances among human cancers of the same tissue type (Online Resource 4), these data suggests that DFT1 and DFT2 could be of a similar origin.
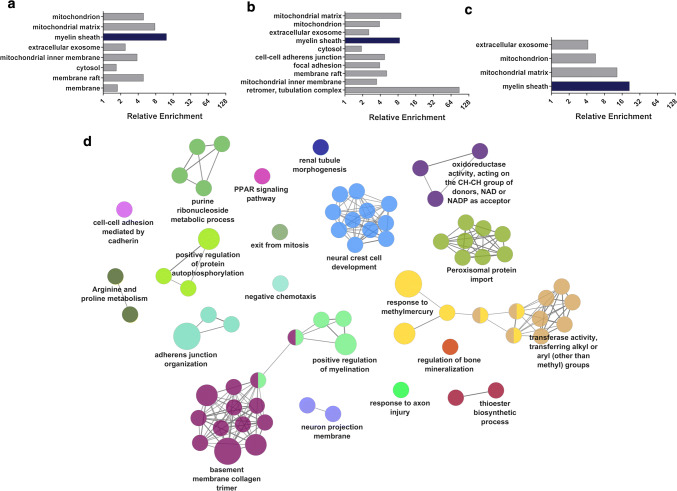
Fig. 1.
Analysis of mRNA expression variation among DFT1 and DFT2 cell lines and primary tissues. a, b Gene expression was analysed in duplicate samples of DFT1 and DFT2 cell lines by RNA sequencing. a A continuous bar plot displaying the logFC of 12,632 genes in DFT2 cell lines relative to DFT1 is shown. Genes are ranked along the x-axis from lowest logFC to highest logFC. Vertical dashed lines represent a fourfold difference in gene expression between the two tumours. b Boxplots of pairwise Euclidean distance between DFT1 and DFT2 cell lines and biopsies, and between human tumours originating from the same and different tissue types, are shown. Mean, interquartile range and outliers are indicated for each distribution. Statistical significance is defined as n.s.p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001. c, d Gene expression was analysed in primary DFT1 and DFT2 tumour samples, testis, brain, spleen, heart and peripheral nerve by RNA sequencing. Two biological replicates were individually sequenced for each tissue type except for peripheral nerve (PN), where it was necessary to pool the two replicates prior to sequencing to generate sufficient template for analysis. c Hierarchical clustering of sample data (columns) was performed on the union of differentially expressed genes (logFC > 2.0, p < 0.05) and approximately unbiased p values (AU; coloured red) and bootstrap probabilities (BP; coloured green) were estimated from 1000 bootstrapping iterations using pvclust. Genes (rows) were clustered based on Pearson’s correlation coefficient. Heat map colour represents mean gene expression standardised across tissues (z score). d Mean pairwise distance between samples was calculated from log transformed RPKM-normalised mRNA read counts, based on Pearson’s correlation coefficient. Each value represents the average of the two biological replicates
To provide definitive evidence for a similar tissue-of-origin of DFT1 and DFT2 cancers, we next compared gene expression patterns among primary DFT1 and DFT2 tumour biopsies and normal tissues from the Tasmanian devil, including testis, spleen, heart, brain and peripheral nerve (Online resource 1). Hierarchical clustering analysis of RNA sequencing data revealed that expression patterns in DFT2 tumours were most similar to DFT1 and peripheral nerve, a Schwann cell enriched tissue (Fig. 1c). Pairwise distances calculated between each tissue also demonstrated lower variation amongst DFT1, DFT2 and peripheral nerve samples than any other tissue type (Fig. 1d). These findings suggest that, like DFT1, DFT2 cancers exhibit a gene expression profile that is similar to cells of Schwann cell origin.
Proteome analysis of DFT2 cell lines
Our transcriptomic analysis suggests that DFT2 cancers could have arisen from a similar cell of origin to DFT1. However, as protein levels can be regulated independently of mRNA, it was necessary to confirm these findings through analysis of DFT2 cells at the protein level. To examine the proteomic landscape of DFT2 tumours, we performed quantitative proteomics using the SWATH data independent acquisition (DIA) approach (Online resource 2). A primary Tasmanian devil fibroblast cell line was included in the analysis as a ‘healthy tissue’ reference line. To investigate the proteomic signatures of interest in DFT1 and DFT2 cells, we selected proteins that were expressed at least twofold higher in DFT1 or DFT2 cell lines relative to fibroblasts with FDR < 0.05. Gene ontology (GO) analysis using the proteomic database DAVID revealed significant enrichment of the GO term ‘myelin sheath’ in these DFT1 and DFT2 protein sets (Fig. 2a–b; Online resource 5). The term ‘myelin sheath’ was also highly enriched when a subset of proteins that appeared in both of these DFT1 and DFT2 protein sets was analysed (Fig. 2c; Online resource 5). Conversely, analysis of highly expressed proteins in fibroblasts relative to DFT1 or DFT2 cells failed to reveal substantial enrichment of this term (Online resource 5). To further investigate this finding, we measured for enrichment of GO terms, KEGG pathways and REACTOME pathways among the 100 most highly expressed proteins in DFT2 cells relative to fibroblasts. Clustering of these terms into functionally organised networks using ClueGO revealed that 14.1% of enriched terms were associated with ‘neural crest cell development’, while 6.1% of terms were associated with ‘positive regulation of myelin’ (Fig. 2d; Online resource 6). In comparison, other enriched terms represented basic structural and metabolic functions of cells such as collagen trimerization and transferase activity. As Schwann cells develop from the neural crest and are responsible for the myelination of peripheral nerves, these findings reflect the transcriptome data and are consistent with a Schwann cell origin of DFT2 tumours.
Fig. 2.
Proteomic analysis of DFT1 and DFT2 cell lines. Protein expression was analysed in triplicate samples of DFT1, DFT2 and fibroblast cell lines by high throughput proteomic mass spectrometry. Proteins expressed at least twofold higher in DFT1 or DFT2 cell lines relative to fibroblasts were selected for functional enrichment analysis. a–c Plots represent cellular component GO terms associated with proteins highly expressed relative to fibroblasts in a DFT1 cells, b DFT2 cells and c both DFT1 and DFT2 cells, as determined by enrichment analysis using the proteomic database DAVID. Fold enrichment of up to ten terms significantly associated with each gene set is shown (p < 0.01). The Schwann cell associated term ‘myelin sheath’ is highlighted in each graph in dark blue. d The top 100 most highly expressed genes in DFT2 cells lines relative to fibroblasts were analysed by ClueGO. Enriched GO terms, KEGG pathways and REACTOME pathways were clustered into functionally-associated groups (shown as different colours) by shared genes (kappa score > 0.4). Each group is labelled by the most significant term in the group. The size of each node represents it’s significance
Schwann cell marker expression in DFT1 and DFT2 tumours
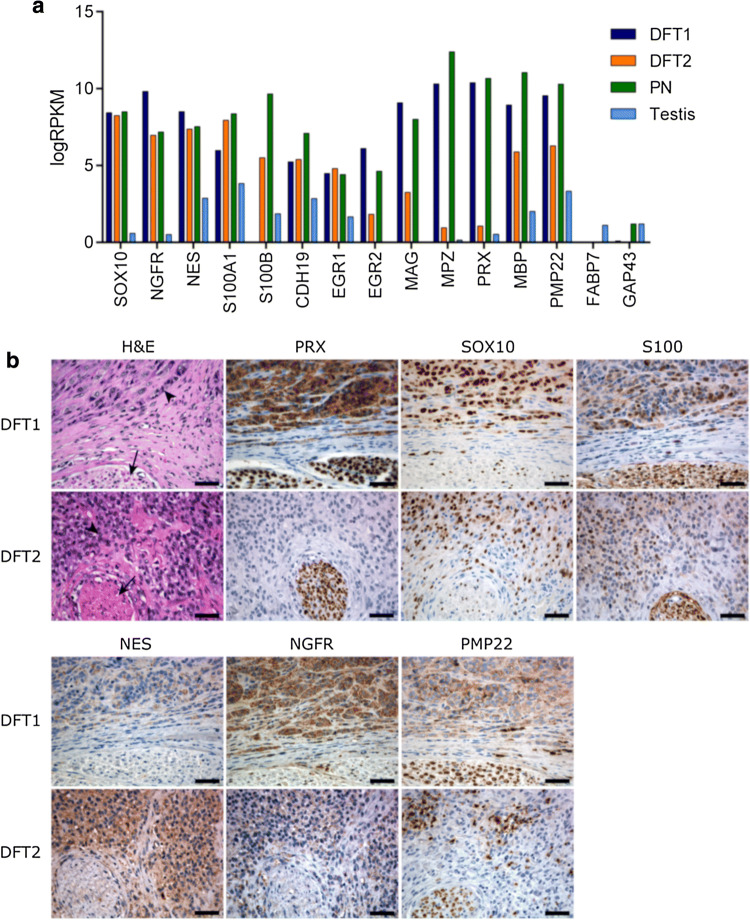
Schwann cells are a highly specialised cell type originating from the neural crest and functioning in myelination and maintenance of peripheral nerves. To reach a mature Schwann cell stage, neural crest cells undergo several differentiation steps from a Schwann cell precursor, to an immature Schwann cell, and finally to a mature myelinating or non-myelinating Schwann cell [26]. To confirm that DFT2 arose from the Schwann cell lineage, we analysed specific markers of myelination and Schwann cell differentiation in primary DFT tumours by mRNA sequencing and immunohistochemistry. At the mRNA level, important markers of Schwann cell differentiation, including SRY-box 10 (SOX10) [27, 28], nerve growth factor receptor (NGFR) [29] and the neuroepithelial marker nestin (NES) [30], were highly expressed in DFT1 and DFT2 relative to testis, a control tissue selected for its expression of a broad range of genes (Fig. 3a). Markers of precursor Schwann cells, such as growth-associated protein 43 (GAP43) and fatty acid-binding protein 7 (FABP7) [31, 32], were not highly expressed in either DFT1 or DFT2, suggesting that both tumours likely arose from a differentiated cell type. Analysis of genes required for myelination, including the key transcription factor early growth response 2 (EGR2) [33], and the structural myelin proteins PRX, myelin-associated glycoprotein (MAG) and myelin protein zero (MPZ) [34], revealed expression levels that were lower in DFT2 cells than peripheral nerve and DFT1 tissues, but higher than the testis control (Fig. 3a). This finding was supported by analysis of Schwann cell markers at the protein level by immunohistochemistry, which demonstrated lower expression of the myelin proteins PRX and peripheral myelin protein 22 (PMP22) in DFT2 sections compared to DFT1 (Fig. 3b). Other Schwann cells markers were detected at various levels at the protein level across both DFT1 and DFT2 tissue sections (Fig. 3b; Online resource 7). Importantly, expression of all markers in the surrounding normal tissue of the section was either lower or absent, demonstrating the specificity of the markers to DFT1 and DFT2 cells. Moreover, high levels of the specific neural crest lineage marker SOX10 [27, 28] were detected at the protein level in both DFT1 and DFT2 sections. Although SOX10 is expressed by other cell types arising from the neural crest including neurons, astrocytes, oligodendrocytes and melanocytes [27, 28], the absence of mRNA encoding markers of these other lineages in DFT1 and DFT2 transcriptomes supports origins of these cells from the Schwann cell lineage (Online resource 8). Due to its strong expression in both DFT1 and DFT2 tumours and restricted expression in other cell types, SOX10 could potentially be used in combination with the DFT1-only marker PRX [10, 11] to diagnose and distinguish DFT1 and DFT2.
Fig. 3.
Analysis of Schwann cell marker expression in DFT1 and DFT2 tumours. a Gene expression was analysed in peripheral nerve (PN), DFT1 and DFT2 tumour samples and testis by RNA sequencing. Average RPKM-normalised read counts of Schwann cell lineage markers and myelination markers were plotted for each tissue. b Representative tissue sections from DFT1 and DFT2 tumours were stained with H&E or antibodies against the Schwann cell markers PRX, SOX10, S100 (multiple gene isoforms), NES, NGFR and PMP22. Serial sections were chosen to include tumour cells (indicated by an arrowhead) and a peripheral nerve bundle (indicated by an arrow) as a Schwann cell enriched tissue. Scale bars represent 50 µm
Differential analysis of DFT1 and DFT2 tumours
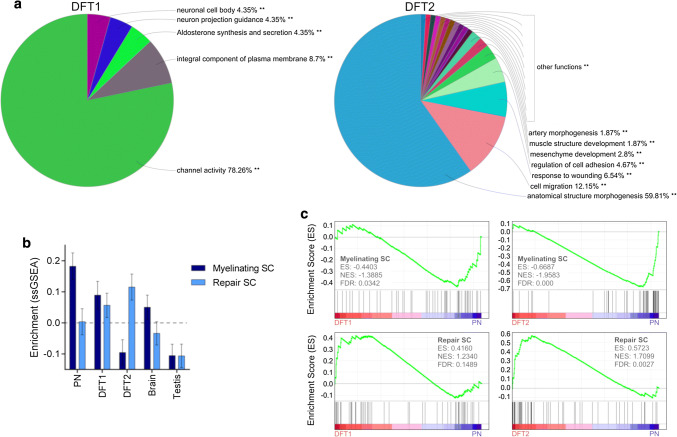
Although both DFT1 and DFT2 cells are of Schwann cell origin, the suppression of myelin-specific genes in DFT2 cells could indicate a difference in the pathways that contribute to tumorigenesis in these cancers. To understand the differences between DFT1 and DFT2 tumours, we analysed for enrichment of GO terms, KEGG pathways and REACTOME pathways among the top 500 up-regulated and down-regulated genes between DFT1 and DFT2 cell lines. Clustering of these terms into functionally-associated groups using ClueGO revealed that 78.26% of enriched terms in the DFT1 gene set were associated with ‘channel activity’ (Fig. 4a; Online resource 9), a function thought to be important for communication between Schwann cells and nerve axons [35]. In comparison, enriched terms associated with the DFT2 gene set represented functions such as ‘anatomical structure morphogenesis’, ‘cell migration’ and ‘response to wounding’. These terms are consistent with known phenotypic changes that occur in Schwann cells during peripheral nerve injury, including suppression of myelination pathways, activation of wound repair responses and differentiation into invasive mesenchymal-like cells primed for nerve remodelling [36–38]. To further test if these repair pathways are implicated in DFT2 tumorigenesis, we performed an unsupervised single sample gene set enrichment analysis (ssGSEA) using gene sets previously shown to be up- or down-regulated in a mouse model of Schwann cell injury [20]. This analysis demonstrated increased enrichment of Schwann cell repair genes and decreased enrichment of genes associated with myelinating Schwann cells in DFT2 biopsies relative to other tissues (Fig. 4b; Online resource 10). In comparison, peripheral nerve tissue exhibited enrichment of repair Schwann cell genes that was comparable to the brain control tissue, while DFT1 exhibited a phenotype that was intermediate to both DFT2 and PN. These findings were supported by GSEA analysis, which revealed significant enrichment of repair Schwann cell genes (normalised enrichment score = 1.71, FDR = 0.003), and negative enrichment of myelinating Schwann cell genes (normalised enrichment score = − 0.67, FDR < 0.001) in DFT2 biopsies relative to PN (Fig. 4c). In comparison, DFT1 biopsies exhibited only a marginal negative enrichment of myelinating Schwann cell genes relative to PN samples (normalised enrichment score = − 0.44, FDR = 0.034), and no significant enrichment of repair Schwann cell genes (normalised enrichment score = 0.42, FDR = 0.149). Given this evidence for enrichment of repair Schwann cell genes and functions in DFT2 cell lines and tumour biopsies, these pathways could be implicated in DFT2 tumorigenesis.
Fig. 4.
Analysis of differentially expressed genes and functions among DFT1 and DFT2 tumours. a Gene expression was analysed in DFT1 and DFT2 cell lines by RNA sequencing. The top 500 most highly expressed genes in DFT1 cell lines relative to DFT2, and DFT2 cell lines relative to DFT1, were analysed by ClueGO. Enriched GO terms, KEGG pathways and REACTOME pathways were clustered into functionally-associated groups (shown as different colours) by shared genes (kappa score > 0.4). Each segment of the pie chart represents the percentage of significantly-enriched terms (p < 0.001) that clustered with each group. Groups are labelled by their most significant term. b Unsupervised enrichment analysis (ssGSEA) of gene sets differentially expressed during the Schwann cell (SC) repair response (as measured in a mouse model by Arthur-Farraj et al. [20]) was performed across PN, DFT1, DFT2, brain and testis tissues. An average enrichment score for the myelinating Schwann cell phenotype and the repair Schwann cell phenotype is plotted, with error bars representing the standard error of enrichment within each tissue-type across 50 specific genes. c Enrichment analysis (GSEA) of the above repair and myelinating gene sets was performed among DFT1 and DFT2 tissues relative to PN. Enrichment plots are shown for each comparison. The barcode plot indicates the position of genes rank-sorted according to differential expression, with over- and under-expression represented by red and blue colouring. An enrichment score (ES), normalised enrichment score (NES) and false discovery rate (FDR) q value is given for each comparison
Discussion
Given the rarity of transmissible cancers in mammalian species, the independent emergence of DFT1 and DFT2 highlights a susceptibility of the Tasmanian devil to cancers of this nature. To further understand this phenomenon, we sought to identify the tissue-of-origin of the newly arisen DFT2 tumour using ‘omics-based approaches including RNA sequencing and quantitative proteomics. This approach incorporated the high sensitivity of RNA sequencing with the functionally informative nature of proteomic mass spectrometry to provide a comprehensive snapshot of the phenotype of the DFT2 cancer. The integration of these datasets revealed common patterns of gene and protein expression among both DFT1 and DFT2 cell lines and biopsies, demonstrating a Schwann cell origin of the two tumours. This finding suggests that the susceptibility of devils to transmissible cancer is cell-lineage specific, providing a basis for understanding the emergence of DFT cancers.
While many factors likely contributed to the development of DFT1 and DFT2 in the Tasmanian devil, it has been hypothesised that disruption to the healing response within wounds could be a key factor in promoting Schwann cell oncogenesis in the devil [4]. Indeed, the innervated vibrissae on the facial regions of devils are vastly enriched with Schwann cells, and devils frequently obtain facial wounds from fighting and scavenging on sharp bone fragments. Schwann cells are highly plastic and act as ‘guardians’ of the peripheral nerve, undergoing a functional shift from myelin maintenance to myelin regeneration during wound repair [37]. Schwann cell plasticity during injury is controlled by Hippo signalling pathways [39], and involves regulation of a number of genes, including repression of myelin proteins [36, 37]. Strikingly, components of Hippo signalling pathways and downstream platelet-derived growth factor (PDGF) and ErbB2/ErbB3 pathways, have been implicated as key drivers of DFT1 and DFT2 tumorigenesis in other studies [4, 40, 41]. Furthermore, our study demonstrates gene and protein expression profiles in DFT2 tumours that are consistent with activation of these repair pathways. These findings support a mechanism by which the high frequency of facial injury and subsequent transition of Schwann cells to a repair phenotype in the Tasmanian devil may increase the likelihood of aberrations occurring in these cells, therefore giving rise to facial tumours on rare occasions. The potential for these Schwann cell cancers to become transmissible is perhaps increased in devils due to their unusual biting behaviors, which provide an uncontrolled route of tumour transfer [42]. Tissue injury and chronic inflammation are linked to a variety of cancer types in humans, and several inflammatory cytokines and growth factors released during wound healing can exert pro-oncogenic effects [43]. These factors include transforming growth factor β (TGFβ), a tumour-promoting cytokine expressed within DFT1 tumours [44] that acts as a key driver of Schwann cell repair in the wound environment [38]. Additionally, activation of repair pathways in DFTD cells could favour survival during cancer emergence. Indeed, the proliferative and invasive properties of mesenchymal-like repair Schwann cells [38] are likely to be indispensable for DFT2 progression.
Although evidence suggests that Schwann cell plasticity and injury could contribute to the specificity of DFT1 and DFT2 to the Schwann cell lineage, there are likely to be additional factors that play a role to the emergence of these cancers in devils. A genetic susceptibility of devils to solid tumours is evident from historical post-mortem data from the San Diego Zoo, which reveals an incidence of non-DFTD cancer in captive devils that was at least two times higher than any other measured species and ten times higher than average [45, 46]. Furthermore, recent analysis has revealed that non-DFTD neoplasia is the most common cause of mortality in captive Tasmanian devils, responsible for approximately 40% of fatalities [47]. This predisposition of devils to cancer may be enhanced by low genetic diversity across the population due to historical population collapses induced by climate fluctuation and culling by early European settlers [4, 5, 48]. Low genetic diversity could also play a role in DFTD transmission by allowing transferred tumour cells to go unnoticed by the immune system. Indeed, DFT2 tumours selectively express non-polymorphic major histocompatibility complex class I (MHCI) molecules that could be undetectable in genetically similar hosts [25]. Analysis of DFT1 cells suggests that tumour evolution and loss of this MHCI expression over time allows for the eventual transfer of the tumours into more diverse devil populations [25, 49].
The development of transmissible cancers in mammals is thought to be a rare event, occurring only when certain factors combine to overcome robust protective defences. Here we have reported that in the Tasmanian devil this has occurred not once, but twice, in founder cells from the Schwann cell lineage. While future studies will focus on the management of DFT1 and DFT2 in wild Tasmanian devil populations, the emergence of further DFTDs is also a possibility, and may increase the complexity of the conservation management of this iconic species. Further investigations into the remarkable development of DFTDs from Schwann cells in the devil will allow for identification of strategies to mitigate this risk.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors wish to thank Narelle Phillips for immunohistochemistry and Anne-Maree Pearse and Kate Swift of the DPIPWE for provision of the DFT1 cell line used in this manuscript. We also thank the Walter and Eliza Hall Institute for Medical Research computational biology division for their bioinformatics expertise and assistance, and Maximilian Stammnitz and Young Mi Kwon for useful discussion. Aspects of this research were conducted at the Australian Proteome Analysis Facility (APAF) under the Australian Government’s National Collaborative Research Infrastructure Scheme. Research support was provided by the Australian Research Council (DP180100520), the University of Tasmania Foundation through funds raised by the Save the Tasmanian Devil Appeal and a joint research initiative between the University of Tasmania and Bioplatforms Australia.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Murchison EP, Wedge DC, Alexandrov LB, Fu B, Martincorena I, Ning Z, Tubio JMC, Werner EI, Allen J, De Nardi AB, Donelan EM, Marino G, Fassati A, Campbell PJ, Yang F, Burt A, Weiss RA, Stratton MR. Transmissible [corrected] dog cancer genome reveals the origin and history of an ancient cell lineage. Science. 2014;343(6169):437–440. doi: 10.1126/science.1247167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pearse AM, Swift K. Allograft theory: transmission of devil facial-tumour disease. Nature. 2006;439(7076):549. doi: 10.1038/439549a. [DOI] [PubMed] [Google Scholar]
- 3.Pye RJ, Pemberton D, Tovar C, Tubio JM, Dun KA, Fox S, Darby J, Hayes D, Knowles GW, Kreiss A, Siddle HV, Swift K, Lyons AB, Murchison EP, Woods GM. A second transmissible cancer in Tasmanian devils. Proc Natl Acad Sci USA. 2016;113(2):374–379. doi: 10.1073/pnas.1519691113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stammnitz MR, Coorens THH, Gori KC, Hayes D, Fu B, Wang J, Martin-Herranz DE, Alexandrov LB, Baez-Ortega A, Barthorpe S, Beck A, Giordano F, Knowles GW, Kwon YM, Hall G, Price S, Pye RJ, Tubio JMC, Siddle HVT, Sohal SS, Woods GM, McDermott U, Yang F, Garnett MJ, Ning Z, Murchison EP. the origins and vulnerabilities of two transmissible cancers in Tasmanian devils. Cancer Cell. 2018;33(4):607–619.e615. doi: 10.1016/j.ccell.2018.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siddle HV, Kreiss A, Eldridge MD, Noonan E, Clarke CJ, Pyecroft S, Woods GM, Belov K. Transmission of a fatal clonal tumor by biting occurs due to depleted MHC diversity in a threatened carnivorous marsupial. Proc Natl Acad Sci USA. 2007;104(41):16221–16226. doi: 10.1073/pnas.0704580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murchison EP, Schulz-Trieglaff OB, Ning Z, Alexandrov LB, Bauer MJ, Fu B, Hims M, Ding Z, Ivakhno S, Stewart C, Ng BL, Wong W, Aken B, White S, Alsop A, Becq J, Bignell GR, Cheetham RK, Cheng W, Connor TR, Cox AJ, Feng ZP, Gu Y, Grocock RJ, Harris SR, Khrebtukova I, Kingsbury Z, Kowarsky M, Kreiss A, Luo S, Marshall J, McBride DJ, Murray L, Pearse AM, Raine K, Rasolonjatovo I, Shaw R, Tedder P, Tregidgo C, Vilella AJ, Wedge DC, Woods GM, Gormley N, Humphray S, Schroth G, Smith G, Hall K, Searle SM, Carter NP, Papenfuss AT, Futreal PA, Campbell PJ, Yang F, Bentley DR, Evers DJ, Stratton MR. Genome sequencing and analysis of the Tasmanian devil and its transmissible cancer. Cell. 2012;148(4):780–791. doi: 10.1016/j.cell.2011.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deakin JE, Bender HS, Pearse AM, Rens W, O’Brien PC, Ferguson-Smith MA, Cheng Y, Morris K, Taylor R, Stuart A, Belov K, Amemiya CT, Murchison EP, Papenfuss AT, Graves JA. Genomic restructuring in the Tasmanian devil facial tumour: chromosome painting and gene mapping provide clues to evolution of a transmissible tumour. PLoS Genet. 2012;8(2):e1002483. doi: 10.1371/journal.pgen.1002483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lazenby BT, Tobler MW, Brown WE, Hawkins CE, Hocking GJ, Hume F, Huxtable S, Iles P, Jones ME, Lawrence C. Density trends and demographic signals uncover the long-term impact of transmissible cancer in Tasmanian devils. J Appl Ecol. 2018;55(3):1368–1379. doi: 10.1111/1365-2664.13088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hawkins CE, Baars C, Hesterman H, Hocking G, Jones ME, Lazenby B, Mann D, Mooney N, Pemberton D, Pyecroft S. Emerging disease and population decline of an island endemic, the Tasmanian devil Sarcophilus harrisii. Biol Conserv. 2006;131(2):307–324. doi: 10.1016/j.biocon.2006.04.010. [DOI] [Google Scholar]
- 10.Murchison EP, Tovar C, Hsu A, Bender HS, Kheradpour P, Rebbeck CA, Obendorf D, Conlan C, Bahlo M, Blizzard CA, Pyecroft S, Kreiss A, Kellis M, Stark A, Harkins TT, Marshall Graves JA, Woods GM, Hannon GJ, Papenfuss AT. The Tasmanian devil transcriptome reveals Schwann cell origins of a clonally transmissible cancer. Science. 2010;327(5961):84–87. doi: 10.1126/science.1180616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tovar C, Obendorf D, Murchison EP, Papenfuss AT, Kreiss A, Woods GM. Tumor-specific diagnostic marker for transmissible facial tumors of Tasmanian devils: immunohistochemistry studies. Vet Pathol. 2011;48(6):1195–1203. doi: 10.1177/0300985811400447. [DOI] [PubMed] [Google Scholar]
- 12.Patchett AL, Wilson R, Charlesworth JC, Corcoran LM, Papenfuss AT, Lyons BA, Woods GM, Tovar C. Transcriptome and proteome profiling reveals stress-induced expression signatures of imiquimod-treated Tasmanian devil facial tumor disease (DFTD) cells. Oncotarget. 2018;9(22):15895–15914. doi: 10.18632/oncotarget.24634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41(10):e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 15.Risso D, Schwartz K, Sherlock G, Dudoit S. GC-content normalization for RNA-seq data. BMC Bioinform. 2011;12:480. doi: 10.1186/1471-2105-12-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Law CW, Chen Y, Shi W, Smyth GK. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arthur-Farraj PJ, Morgan CC, Adamowicz M, Gomez-Sanchez JA, Fazal SV, Beucher A, Razzaghi B, Mirsky R, Jessen KR, Aitman TJ. Changes in the coding and non-coding transcriptome and dna methylome that define the Schwann cell repair phenotype after nerve injury. Cell Rep. 2017;20(11):2719–2734. doi: 10.1016/j.celrep.2017.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu JX, Song X, Pascovici D, Zaw T, Care N, Krisp C, Molloy MP. SWATH mass spectrometry performance using extended peptide MS/MS assay libraries. Mol Cell Proteom. 2016;15(7):2501–2514. doi: 10.1074/mcp.M115.055558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 23.da Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pages F, Trajanoski Z, Galon J. ClueGO: a cytoscape plug-into decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caldwell A, Coleby R, Tovar C, Stammnitz MR, Kwon YM, Owen RS, Tringides M, Murchison EP, Skjodt K, Thomas GJ, Kaufman J, Elliott T, Woods GM, Siddle HV. The newly-arisen devil facial tumour disease 2 (DFT2) reveals a mechanism for the emergence of a contagious cancer. Elife. 2018 doi: 10.7554/elife.35314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jessen KR, Mirsky R. The origin and development of glial cells in peripheral nerves. Nat Rev Neurosci. 2005;6(9):671–682. doi: 10.1038/nrn1746. [DOI] [PubMed] [Google Scholar]
- 27.Mollaaghababa R, Pavan WJ. The importance of having your SOX on: role of SOX10 in the development of neural crest-derived melanocytes and glia. Oncogene. 2003;22(20):3024–3034. doi: 10.1038/sj.onc.1206442. [DOI] [PubMed] [Google Scholar]
- 28.Britsch S, Goerich DE, Riethmacher D, Peirano RI, Rossner M, Nave KA, Birchmeier C, Wegner M. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 2001;15(1):66–78. doi: 10.1101/gad.186601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bentley CA, Lee KF. p75 is important for axon growth and schwann cell migration during development. J Neurosci. 2000;20(20):7706–7715. doi: 10.1523/JNEUROSCI.20-20-07706.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lendahl U, Zimmerman LB, McKay RD. CNS stem cells express a new class of intermediate filament protein. Cell. 1990;60(4):585–595. doi: 10.1016/0092-8674(90)90662-X. [DOI] [PubMed] [Google Scholar]
- 31.Curtis R, Stewart HJ, Hall SM, Wilkin GP, Mirsky R, Jessen KR. GAP-43 is expressed by nonmyelin-forming Schwann cells of the peripheral nervous system. J Cell Biol. 1992;116(6):1455–1464. doi: 10.1083/jcb.116.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurtz A, Zimmer A, Schnutgen F, Bruning G, Spener F, Muller T. The expression pattern of a novel gene encoding brain-fatty acid binding protein correlates with neuronal and glial cell development. Development. 1994;120(9):2637–2649. doi: 10.1242/dev.120.9.2637. [DOI] [PubMed] [Google Scholar]
- 33.Topilko P, Schneider-Maunoury S, Levi G, Baron-Van Evercooren A, Chennoufi AB, Seitanidou T, Babinet C, Charnay P. Krox-20 controls myelination in the peripheral nervous system. Nature. 1994;371(6500):796–799. doi: 10.1038/371796a0. [DOI] [PubMed] [Google Scholar]
- 34.Patzig J, Jahn O, Tenzer S, Wichert SP, de Monasterio-Schrader P, Rosfa S, Kuharev J, Yan K, Bormuth I, Bremer J, Aguzzi A, Orfaniotou F, Hesse D, Schwab MH, Mobius W, Nave KA, Werner HB. Quantitative and integrative proteome analysis of peripheral nerve myelin identifies novel myelin proteins and candidate neuropathy loci. J Neurosci. 2011;31(45):16369–16386. doi: 10.1523/jneurosci.4016-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fields RD. A new mechanism of nervous system plasticity: activity-dependent myelination. Nat Rev Neurosci. 2015;16(12):756–767. doi: 10.1038/nrn4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arthur-Farraj PJ, Latouche M, Wilton DK, Quintes S, Chabrol E, Banerjee A, Woodhoo A, Jenkins B, Rahman M, Turmaine M, Wicher GK, Mitter R, Greensmith L, Behrens A, Raivich G, Mirsky R, Jessen KR. c-Jun reprograms Schwann cells of injured nerves to generate a repair cell essential for regeneration. Neuron. 2012;75(4):633–647. doi: 10.1016/j.neuron.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jessen KR, Mirsky R. The repair Schwann cell and its function in regenerating nerves. J Physiol. 2016;594(13):3521–3531. doi: 10.1113/jp270874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clements MP, Byrne E, Camarillo Guerrero LF, Cattin AL, Zakka L, Ashraf A, Burden JJ, Khadayate S, Lloyd AC, Marguerat S, Parrinello S. The wound microenvironment reprograms schwann cells to invasive mesenchymal-like cells to drive peripheral nerve regeneration. Neuron. 2017;96(1):98–114.e117. doi: 10.1016/j.neuron.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mindos T, Dun XP, North K, Doddrell RD, Schulz A, Edwards P, Russell J, Gray B, Roberts SL, Shivane A, Mortimer G, Pirie M, Zhang N, Pan D, Morrison H, Parkinson DB. Merlin controls the repair capacity of Schwann cells after injury by regulating Hippo/YAP activity. J Cell Biol. 2017;216(2):495–510. doi: 10.1083/jcb.201606052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor RL, Zhang Y, Schoning JP, Deakin JE. Identification of candidate genes for devil facial tumour disease tumourigenesis. Sci Rep. 2017;7(1):8761. doi: 10.1038/s41598-017-08908-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kosack L, Wingelhofer B, Popa A, Orlova A, Agerer B, Vilagos B, Majek P, Parapatics K, Lercher A, Ringler A, Klughammer J, Smyth M, Khamina K, Baazim H, de Araujo ED, Rosa DA, Park J, Tin G, Ahmar S, Gunning PT, Bock C, Siddle HV, Woods GM, Kubicek S, Murchison EP, Bennett KL, Moriggl R, Bergthaler A. The ERBB-STAT3 axis drives Tasmanian devil facial tumor disease. Cancer Cell. 2019;35(1):125–139.e129. doi: 10.1016/j.ccell.2018.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamede RK, McCallum H, Jones M. Biting injuries and transmission of Tasmanian devil facial tumour disease. J Anim Ecol. 2013;82(1):182–190. doi: 10.1111/j.1365-2656.2012.02025.x. [DOI] [PubMed] [Google Scholar]
- 43.Kuraishy A, Karin M, Grivennikov SI. Tumor promotion via injury- and death-induced inflammation. Immunity. 2011;35(4):467–477. doi: 10.1016/j.immuni.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morris K, Belov K. Does the devil facial tumour produce immunosuppressive cytokines as an immune evasion strategy? Vet Immunol Immunopathol. 2013;153(1–2):159–164. doi: 10.1016/j.vetimm.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 45.Griner LA. Neoplasms in Tasmanian devils (Sarcophilus harrisii) J Natl Cancer Inst. 1979;62(3):589–595. doi: 10.1093/jnci/62.3.589. [DOI] [PubMed] [Google Scholar]
- 46.Abegglen LM, Caulin AF, Chan A, Lee K, Robinson R, Campbell MS, Kiso WK, Schmitt DL, Waddell PJ, Bhaskara S, Jensen ST, Maley CC, Schiffman JD. Potential mechanisms for cancer resistance in elephants and comparative cellular response to DNA damage in humans. JAMA. 2015;314(17):1850–1860. doi: 10.1001/jama.2015.13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peck SJ, Michael SA, Knowles G, Davis A, Pemberton D. Cause of mortality and severe morbidity requiring euthanasia in captive Tasmanian devils (Sarcophilus harrisii) in Tasmania. Aust Vet J. 2019;97(4):89–92. doi: 10.1111/avj.12797. [DOI] [PubMed] [Google Scholar]
- 48.Bruniche-Olsen A, Jones ME, Austin JJ, Burridge CP, Holland BR. Extensive population decline in the Tasmanian devil predates European settlement and devil facial tumour disease. Biol Lett. 2014;10(11):20140619. doi: 10.1098/rsbl.2014.0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siddle HV, Kreiss A, Tovar C, Yuen CK, Cheng Y, Belov K, Swift K, Pearse AM, Hamede R, Jones ME, Skjodt K, Woods GM, Kaufman J. Reversible epigenetic down-regulation of MHC molecules by devil facial tumour disease illustrates immune escape by a contagious cancer. Proc Natl Acad Sci USA. 2013;110(13):5103–5108. doi: 10.1073/pnas.1219920110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the conclusions of this article are available in the European Nucleotide Archive repository [http://www.ebi.ac.uk/ena/data/view/PRJEB28680 and http://www.ebi.ac.uk/ena/data/view/PRJNA416378], and the ProteomeXchange Consortium via the PRIDE partner repository [http://www.ebi.ac.uk/pride/archive/projects/PXD011209]. The remainder of the data supporting this article are included within the article and its additional files.