Abstract
A novel insulin-like growth factor (igf3), which is exclusively expressed in the gonads, has been widely identified in fish species. Recent studies have indicated that Igf3 regulates spermatogonia proliferation and differentiation in zebrafish; however, detailed information on the role of this Igf needs further in vivo investigation. Here, using Nile tilapia (Oreochromis niloticus) as an animal model, we report that igf3 is required for spermatogenesis and reproduction. Knockout of igf3 by CRISPR/Cas9 severely inhibited spermatogonial proliferation and differentiation at 90 days after hatching, the time critical for meiosis initiation, and resulted in less spermatocytes in the mutants. Although spermatogenesis continued to occur later, more spermatocytes and less spermatids were observed in the igf3−/− testes when compared with wild type of testes at adults, indicating that Igf3 regulates spermatocyte to spermatid transition. Importantly, a significantly increased occurrence of apoptosis in spermatids was observed after loss of Igf3. Therefore, igf3−/− males were subfertile with drastically reduced semen volume and sperm count. Conversely, the overexpression of Igf3 in XY tilapia enhanced spermatogenesis leading to more spermatids and sperm count. Transcriptomic analysis revealed that the absence of Igf3 resulted in dysregulation of many genes involved in cell cycle, meiosis and pluripotency regulators that are critical for spermatogenesis. In addition, in vitro gonadal culture with 17α-methyltetosterone (MT) and 11-ketotestosterone (11-KT) administration and in vivo knockout of cyp11c1 demonstrated that igf3 expression is regulated by androgens, suggesting that Igf3 acts downstream of androgens in fish spermatogenesis. Notably, the igf3 knockout did not affect body growth, indicating that this Igf specifically functions in reproduction. Taken together, our data provide genetic evidence for fish igf3 in the regulation of reproductive capacity by controlling spermatogenesis.
Electronic supplementary material
The online version of this article (10.1007/s00018-019-03439-0) contains supplementary material, which is available to authorized users.
Keywords: Growth factor, Meiosis, Androgen, Cell apoptosis, Fish
Introduction
Spermatogenesis is a complex developmental process that begins with the mitotic proliferation of spermatogonia, then proceeds through two meiotic divisions followed by spermiogenesis, during which the haploid spermatids develop into spermatozoa. These spermatozoa then undergo maturation and obtain the ability to fertilize [1, 2]. The molecular mechanisms controlling spermatogenesis in mammals are well studied [3]; however, this process in fish required further elucidation. A number of studies have demonstrated that fish spermatogenesis is well controlled by stage- and cell-specific interactions of various hormones [4–7]. In fish, as in other vertebrates, steroid hormones are the main endocrine regulators of gonadal differentiation [5]. Previous studies have demonstrated that the pituitary gonadotropins Fsh and Lh are potent steroidogenic hormones acting through their receptors (Fshr and Lhcgr) in Leydig cells, resulting in the production of estrogens, androgens, and progestins that control the different stages of spermatogenesis [8–14]. Recent studies with genome editing have revealed that mutation of the fshb gene in zebrafish delayed puberty onset but the mutants remained fertile [15]. However, infertility has been observed in two double fsh/lh and fshr/lhcgr mutants [16]. In addition, mutation of the lhcgr or lhb gene in zebrafish had no effects on male fertility [15], suggesting that Lh is not required for spermatogenesis in zebrafish. One of the main androgens, 11-ketotestosterone (11-KT), is involved in the initiation of spermatogonial proliferation toward meiosis in fish species [10, 17–20]. Mutation of the androgen receptor (ar) gene consistently disrupts spermatogenesis causing reduced sperm production [21, 22]. Progestin, 17α, 20β-dihydroxy-4-pregnen-3-one (DHP), is an essential factor for the initiation of meiosis in spermatogenetic cells of eel and tilapia [12, 14, 23, 24]. Furthermore, disruption of pgr in tilapia also results in dysregulation of spermatogenesis and leads to a significant decline in sperm count, sperm motility and fertility [25, 26]. Estrogens have been shown to stimulate the renewal of spermatogonial stem cells in male eels [11]. Additionally, exogenous estrogen treatment impairs spermatogenesis while mutation of cyp19a1a, the gene encoding aromatase, promotes spermatogenesis in fish [27–29]. Although these data strongly demonstrate that steroid hormones are important for fish spermatogenesis and fertility, the downstream underlying mechanisms have not yet been clarified. Other studies have shown that sex steroids are most likely not the only factors involved because they seem to interact with other hormones during spermatogenesis [30]. It is well known that spermatogenesis occurs in conjunction with cell proliferation, differentiation and tissue growth [31], implying that growth factors, particularly insulin-like growth factors (Igfs), might take part in these physiological processes [32–34].
The igf1 gene was found to be expressed in spermatogonia, Sertoli cells and Leydig cells in the testis of rainbow trout (Oncorhynchus mykiss), tilapia (Oreochromis niloticus) and Japanese seabass (Lateolabrax japonycus) [35–38]. Expression of igf2 was also detected in the testis of many fish species, but its cellular localization is unknown [39, 40]. The roles of Igf1 have been preliminarily elucidated through a series of in vitro studies, whereas Igf2 has rarely been investigated in fish. Using an in vitro gonadal culture system, Igf1 recombinant protein was shown to stimulate spermatogenesis in Japanese eel and tilapia testes [41, 42]. In addition, Igf1 stimulates the incorporation of thymidine into spermatogonia and primary spermatocytes from cultured spermatogenic rainbow trout testis [43, 44]. Furthermore, Igf1 stimulates DNA synthesis in spermatogonia [35]. A previous study from our group has shown that a novel insulin-like growth factor (igf3), which is distinct from the conventional igf1 and igf2, is cloned in fish species [45]. This igf is exclusively expressed in the gonads, thus implying its special role in fish reproduction [45]. Recent reports indicate that Igf3 regulates oocyte maturation in zebrafish (Danio rerio) [46–48]. An in vitro gonad culture with Igf3 recombinant protein increased the number of undifferentiated and differentiating spermatogonia and up-regulated the expression of genes related to spermatogonial differentiation and entry into meiosis [49]. Further research revealed that Igf3 activates β-catenin to promote the differentiation of Aund to Adiff spermatogonia [50]. In addition, igf3 expression was regulated by Fsh. It was concluded that Fsh promoted spermatogonial proliferation and differentiation and their entry into meiosis through up-regulation of Sertoli cell production of Igf3 [51–53]. Expression of igf3 is also weakly regulated by androgen [49]. Overall, this information results suggested that igf3 could be a major growth factor involved in regulating spermatogenesis and reproduction in fish. However, these results were mainly obtained through in vitro studies with recombinant protein or Igf receptor inhibitor treatment. Currently, there is no information available on the role of Igf3 in spermatogenesis supported by genetic studies. This is partly due to the lack of powerful genetic approaches for functional studies in fish; however, this has changed with the emergence of TALEN and CRISPR/Cas9, which have been well established in several fish species, including tilapia [54–56].
To provide genetic evidence for the functional and physiological significance of this novel igf in teleosts, we have undertaken the present study using CRISPR/Cas9 and overexpression approaches to disrupt and overexpress Igf3. Subsequently, we performed a detailed analysis of the reproductive phenotypes in tilapia. Our analysis focused on spermatogenesis, fertility and gene expression. The results from the present study provide comprehensive genetic evidence for Igf3 functions in fish spermatogenesis.
Materials and methods
Fish
The founder strain of the Nile tilapia, which was first introduced from Egypt in Africa, was obtained from Prof. Nagahama (Laboratory of Reproductive Biology, National Institute for Basic Biology, Okazaki, Japan). The fish were reared in large tanks with a circulating aerated freshwater system. Fertilized eggs were obtained by crossing XY males with normal females (XX). Animal experiments were conducted in accordance with the regulations of Guide for Care and Use of Laboratory Animals and were approved by the Committee of Laboratory Animal Experimentation at Southwest University, China.
Establishment of igf3 mutant lines by CRISPR/Cas9
The tilapia igf3 mutant lines were generated by CRISPR/Cas9 as described by our previous study [54]. Briefly, a gRNA target site containing a restriction enzyme site was selected by identifying sequences corresponding to GGN18NGG on the sense or antisense strand of igf3 using the online tool, ZIFIT Targeter (https://zifit.partners.org/zifit/Introduction.aspx). Sequences that perfectly matched the final 12 nt of the target and the NGG PAM sequence were discarded so as to avoid the off-targets [57]. Artificially synthesized gRNA and Cas9 mRNA were co-injected into one-cell stage embryos at concentration of 250 ng/μl and 500 ng/μl. Twenty embryos were randomly collected 72 h after injection. The genomic DNA used for the mutation assays was extracted from pooled control and injected embryos. DNA fragments spanning the target site were amplified using the primers listed in Supplementary Table S1. The mutations were analyzed by restriction enzyme digestion and Sanger sequencing.
F0 fish were screened by restriction enzyme digestion. Heterozygous F1 offsprings were obtained by mating F0 XY male founders with wild type (WT) XX females. The F1 fish were genotyped by the fin-clip assay and individuals with frame-shift mutations were selected. XY male and XX female siblings of the F1 generation that carried the same mutation were mated to generate homozygous F2 mutants. The genetic sex of the mutants was identified using tilapia sex specific marker-5 [58, 59].
A knockout model of cyp11c1, which is the gene encoding 11β-hydroxylase that catalyzes androgen synthesis in teleosts, was also generated by our laboratory using the above method (data not shown).
Overexpression of Igf3 in XY tilapia
As described above, igf3 is exclusively expressed in gonad tissue [45]. An in vivo overexpression experiment was performed to examine the effects of Igf3 on spermatogenesis in XY individuals according to the methods outlined in our previous study [60]. The igf3 ORF was amplified by PCR with a primer set that introduced the BamHI and EcoRI sites. The amplified fragment was digested by BamHI and EcoRI and then ligated into a pIRES-hrGFP-1a vector. The plasmid DNA of igf3-pIRES-hrGFP was diluted in filtered PBS solution to a final concentration of 100 ng/μl, and then injected into one-cell stage fertilized eggs using an SYS-PV830 injector under microscope (WPI, USA). Genomic DNA was extracted from the injected fish and then used to screen Igf3 overexpression fish at 90 and 150 days after hatching (dah). Gene specific primers based on the vector sequence were designed to screen positive fish (Supplemental Tab. S1). The testes of the WT and injected fish were examined by monitoring the GFP signal. The testes were subjected to both histological and gene expression analyses.
Sampling and histological examination
Before processing for histological examination, the body weight and age were recorded for all the sampled fish. Both testes of each fish were excised, weighed and the gonadosomatic index (GSI; the ratio between the testis weight and body weight) was calculated. For one fish, one testis was used to extract RNA for gene expression analyses. The other testis was used for histological analysis or gene expression. Samples were fixed in Bouin's solution or 4% paraformaldehyde (PFA) for 24 h at room temperature. They were then dehydrated and embedded in paraffin. Tissue blocks were sectioned at 5 μm and stained with hematoxylin and eosin.
Real-time PCR
Total RNA was extracted from the gonads of WT XY fish at 10, 30, 70, 90, 120 and 180 dah (days after hatching) for igf3 ontogeny analysis. Three parallel samples (each composed of testes from three individuals) were prepared from igf3−/− and WT XY fish at 90 and 180 dah. Total RNA was extracted and treated with DNase I to eliminate genomic DNA contamination. About 1.0 μg RNA was used to synthesize the first strand cDNA using the PrimeScript RT Master Mix Perfect Real Time Kit. Real-time PCR was performed on an ABI-7500 real-time PCR machine (Applied Biosystems, Germany) according to the SYBR Premix Ex TaqTM II protocol (Takara). Primer sequences used for real-time PCR are listed in Supplemental Tab. S1. The efficiency and specificity of primers used for qRT-PCR have been previously evaluated by our group [23, 26, 61–63]. The relative abundances of mRNA transcripts were evaluated using the following formula: RQ = 2−△△Ct [64]. The arithmetic mean of the reference gene (β-actin) copy number was used to normalize the expression values. Data are presented as the mean ± SD of the triplicates.
Immunofluorescence (IF), immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH)
The gonads of WT XY, igf3 knockout and overexpression fish were sampled at 90, 150 and 180 dah. The sections were fixed with 4% PFA for 20 min at room temperature, permeabilized with 1% Triton X-100 in PBS for 10 min and then blocked in 5% bovine serum albumin (BSA)/PBS for 30 min at room temperature. The sections were then incubated with polyclonal antibodies in 5% BSA/PBS overnight at 4 °C. The following polyclonal antibodies were prepared by our laboratory: Igf3, Amh, Gsdf, Sox30 (a gene mainly expressed in spermatid) [65], eEf1A1b (a gene specifically expressed in spermatogonia) [66], Vasa and Cyp11c1 (the gene encoding 11β-hydroxylase, the key enzyme for androgen 11-KT synthesis). The specificity of these antibodies has been analyzed previously [66, 67]. Histone H3 Phospho (S10) (pHH3) and PCNA antibodies were purchased from Cell Signaling Technology (Beverly, MA, USA). For IF, Alexa Fluor 488- and 594-conjugated secondary antibodies (Thermo Fisher scientific) were diluted to 1:500 in blocking solution and incubated with tissue overnight at 4 °C to detect the primary antibodies. The nuclei were stained by DAPI. For IHC, the second antibody (HRP-conjugated goat anti-rabbit IgG, 1:1000 dilution) was used to detect the primary antibody. Diaminobenzidine tetrachloride (DAB) was applied for the color reaction. Slides were first counterstained with hematoxylin, and then dehydrated and mounted. Images were captured under Olympus BX51 light microscope. The specificity of the Igf3 staining was confirmed by incubating sections of testis with primary antibody pre-absorbed with the antigen used to generate the antibody.
Fluorescence in situ hybridization (FISH) was performed to examine the gene expression of tilapia gonads from WT and igf3 knockout fish. Previous studies demonstrated that plzf is a marker gene of undifferentiated spermatogonia and c-kit is a marker gene of differentiated spermatogonia in dogfish, zebrafish and trout [68–70], while sycp3 is specifically localized in the spermatocytes of tilapia [23]. Probes for sycp3, plzf and c-kit antisense digoxigenin-labeled RNA strands were transcribed in vitro from a linearized pGEM-Teasy-target gene cDNA clone using the RNA labeling kit (Roche, Germany). For more sensitive fluorescence in situ hybridization detection, the tyramide signal amplification (TSA™) Plus Fluorescence Systems (NEL756, PerkinElmer Life Science) was used according to the manufacturer’s instructions. All IF and FISH positive signals were quantified using ImageJ software [26].
The fluorescence intensity was analyzed with the Image Pro Plus software. The sum integrated optical density (IOD) value of fluorescence was calculated using the Count and Measure tools of Image Pro Plus software. The fluorescence area of images was calculated with ImageJ [26].
Western blot
Gonad samples were prepared from igf3−/− and WT XY fish at 90 dah to evaluate gene expression. In addition, Gonad samples from WT XY fish at 70, 90 and 120 were also prepared. Total proteins were extracted from the gonads and diluted to a final concentration of 20 mg/ml. Western blots for Igf3 expression were performed as previously reported [67]. Gene expression levels were measured by densitometry with Fusion-CAPT software (Vilber Lourmat, France) and normalized using α-tubulin as the reference protein.
Transcriptome sequencing and analysis
RNA extracted from igf3−/− and WT XY fish testes at 90 dah were 2 × 100-bp paired-end sequenced using the HiSeq 2000 platform (Illumina). Clean reads from each library were aligned to the reference genome (Orenil1.0, https://www.ensembl.org/Oreochromis_niloticus/Info/Index) using Tophat with the default parameters. The reads per kb per million reads (RPKM) method was used to calculate the gene expression level. The assembled transcripts were merged using the reference annotation (Oreochromis niloticus: Orenil1.0.78.gtf, downloaded from Ensembl) with Cuffmerge, while differential expression analysis was performed with Cuffdiff. The corrected p value (qvalue) of 0.05 and the log2 (fold change) of 1 were set as the threshold for significant differential expression. The DEGs were classified according to the following criteria: genes meeting both “q value < 0.05” and “log2 (igf3 KO_FPKM/WT_FPKM) > 1” statistical criteria were classified as up-regulated genes; however, “q value < 0.05” and “log2 (igf3 KO _FPKM/WT_FPKM) < − 1” were classified as down-regulated genes. The Gene Ontology (GO) of the differentially expressed genes (DEGs) was analyzed by the WEGO online tool. To study the biological pathways of the identified up/down-regulated genes, we mapped these differentially expressed genes to pathways in the Kyoto Encyclopedia of Genes and Genomes (KEGG) using the KOBAS web server (https://kobas.cbi.pku.edu.cn/).
Sperm characteristics and fertility assessment
Semen of the igf3−/− (n = 3) and WT (n = 3) XY fish were collected by gonadal autopsy at 180 dah and were diluted at 1:100 with Hank’s solution before being smeared on the polylysine treated glass slides. After air-drying, the specimens were stained with Papanicolaou EA50 dye (Nobleryder, China). Morphological examination of the sperm from WT and igf3−/− XY fish was performed under an Olympus BX51 light microscope. The sperm count and sperm motility (SM) were analyzed using the Sperm Quality Analyzer (ZKPACS-E) according to previously described methods [66].
The fertility of adult igf3−/− (n = 3) and WT (n = 3) XY fish was assessed via artificial insemination. Eggs from WT XX female fish (n = 3) were divided into 6 groups (approximately 300 eggs/group). Artificial insemination was performed using sperm obtained from the igf3−/− and WT XY fish. The number of gastrula-stage embryos was counted under a light microscope to calculate the fertilization rate after 15 h of fertilization.
Cell proliferation and apoptosis assays
EdU was diluted in PBS to 1 mg/ml and injected intraperitoneally (IP; 50 μl) into igf3 knockout and overexpression fish for three days as previously described [71]. Immunolocaliztion of EdU was performed using the Click-iT EdU AlexaFluor 488 Imaging Kit per the manufacturer’s instructions (Invitrogen), followed by DAPI staining.
Terminal deoxynucleotidyl transferase (TdT) dUTP nick-end labeling (TUNEL) assays were carried out to detect apoptotic cells in the WT and igf3−/− XY fish at 90 and 180 dah using the in situ cell death detection kit (Roche, Germany) according to the manufacturer’s instructions. The cross sections were then used for Sox30 and DAPI staining. The number of TUNEL-positive cells was calculated using four cross sections. Images were acquired using a Zeiss Axio Imager Z2 microscope.
In vitro gonad culture and androgen treatment
Ex vivo organ culture was performed as previously described for zebrafish [72]. The gonads of XY tilapia at 90 dah were dissected, washed with PBS (pH 7.4), treated for 30 min with the antibiotics penicillin (1000 IU/ml) and streptomycin (800 μg/ml) for. The gonads were randomly cut into two segments (mean length = 1.0 cm) with each gonad evenly divided into two groups. One was set as the control group and the other as the 17α-methyltestosterone (MT), 11-ketotestosterone (11-KT) and trilostane (an inhibitor of 3β-HSD) treatment groups. Each group contained at least 3 testes. The primary medium was DMEM: F12 (1:1) medium (pH 7.2–7.4) amended with 10 mM HEPES, 5% fetal bovine serum, 100 IU/ml penicillin, and 100 μg/ml streptomycin. Cultures were incubated at 28 °C. After 12 h of incubation, hormone was added at different concentrations (MT 50, 100 and 200 nM; 11-KT: 100, 200 and 450 nM; trilostane: 30 μg/ml) for 4 days. The medium was changed every two days. MT 11-KT and trilostane were purchased from Sigma-Aldrich (USA). Finally, the gonads from each well were collected, washed with PBS (pH 7.4) and then used to examine igf3 expression via real-time PCR, as described above.
Serum steroid hormone assay
Blood samples were collected from the caudal veins of the igf3 knockout fish (n = 3) as well as the control fish (n = 3). The serum 11-KT level was measured using EIA Assay kits (Cayman Chemical Co, USA) following the manufacturer’s instructions.
Results
Expression profile of igf3 during spermatogenesis in XY tilapia
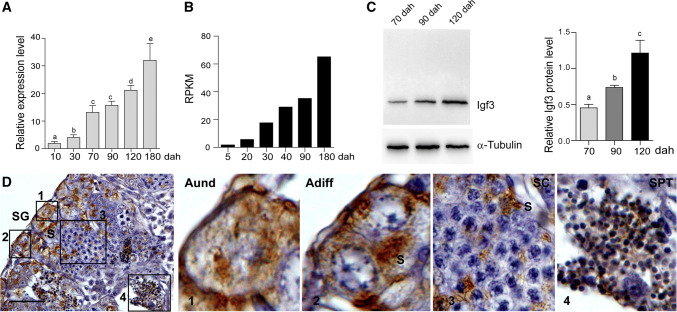
igf3 mRNA was detected in the XY gonads using real-time PCR from 10 to 180 dah. The expression level in the testes gradually increased during the evaluated period, with a remarkably increased level from 90 to 120 dah, which is the critical period for germ cell meiosis (Fig. 1a). This expression pattern was further confirmed by the gonadal transcriptome data (Fig. 1b). In addition, Western blot using protein extracted from the gonads at 70, 90 and 120 dah further demonstrated the expression pattern of Igf3 during testicular development (Fig. 1c).
Fig. 1.
Expression pattern and cellular localization of Igf3 in tilapia testis. a Expression of igf3 mRNA in the tilapia XY gonads at different developmental stages, as determined by real-time PCR. Data are expressed as the mean ± SD of three different gonadal pools at each developmental stage. Different letters indicate statistical differences at p < 0.05 as determined by one-way ANOVA followed by Tukey’s/Kramer post hoc test. b Transcriptomic analysis shows that igf3 expression was up-regulated during gonadal development. XY gonads from tilapia at 5, 20, 30, 40, 90 and 180 dah were sequenced using Illumina 2000 HiSeq technology in our previous study. A normalized measure of RPKM (reads per kb per million reads) was used to normalize the expression profiles of igf3. c Expression of the Igf3 protein in testes at 70, 90 and 120 dah was determined by Western blot. α-Tubulin was used as a loading control. d Cellular localization of the Igf3 protein in tilapia testis at 180 dah, as determined by IHC. The positive signal is denoted by the brown color. 1, 2, 3 and 4 are magnifications of the boxed areas of d. Aund type A undifferentiated spermatogonia, Adiff type A differentiated spermatogonia, SG spermatogonia, SC spermatocyte, SPT spermatid, S Sertoli cells, dah days after hatching. Scale bar 25 μm
Immunohistochemistry (IHC) was performed using testes at 180 dah to ascertain what cell populations express Igf3. Specific signals were mainly observed in the Sertoli cells surrounding the spermatogonia of the testes. Igf3 was also expressed in the type A differentiated spermatogonia. Some signals were detected in the Sertoli cells surrounding primary spermatocytes. Interestingly, specific signals were also observed in the spermatids (Fig. 1d). In contrast, no specific signal was observed in the testicular section stained with the Igf3 primary antibody pre-absorbed with the antigen used to produce antibody (Supplemental Fig. 1).
Generation of igf3 mutant lines by CRISPR/Cas9
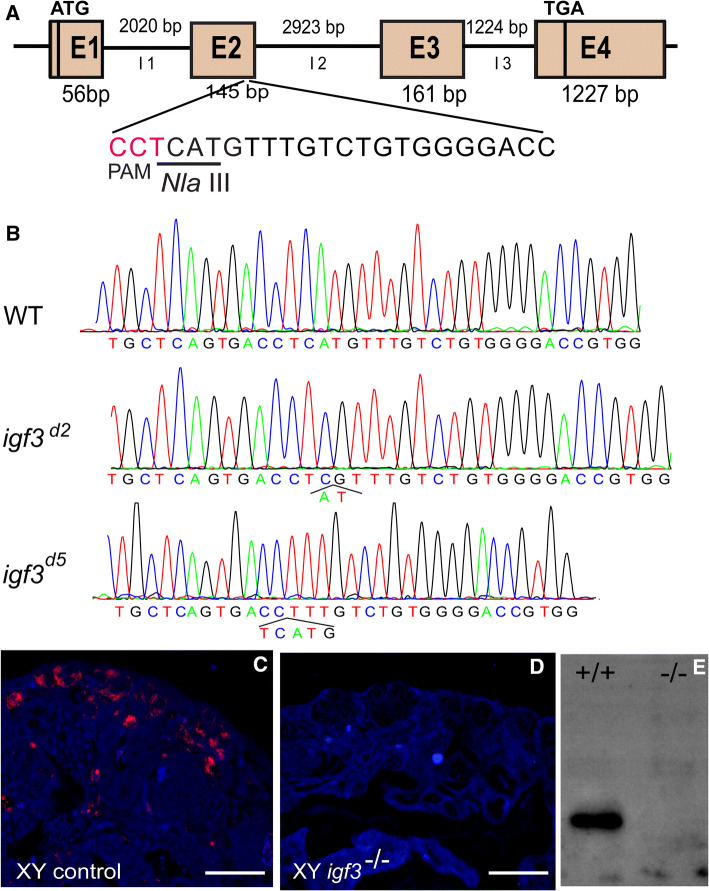
To explore the potential function of Igf3 in the testicular development of tilapia, an igf3 knockout model was generated using the CRISPR/Cas9 technology. The gene structure of the tilapia igf3 comprised of 4 exons of 56, 145, 161 and 1227 bp, and 3 introns. A target, containing an NlaIII restriction enzyme site adjacent to PAM, was selected in the second exon of igf3 (Fig. 2a). Complete digestion with NlaIII produced two fragments of 85 and 227 bp in the control group, whereas an intact DNA fragment (312 bp) was observed in embryos injected with both Cas9 mRNA and target gRNA. The mutation frequency of igf3 in the pooled embryos was approximately 31% (Supplemental Fig. S2A). Furthermore, Sanger sequencing of the uncleaved band confirmed the insertions or deletions (indels), including both in-frame and frame-shift mutations within the target site, verifying the activity of the CRISPR/gRNA in the igf3 locus (Supplemental Fig. S2B).
Fig. 2.
Generation of igf3 mutant strains of tilapia using the CRISPR/Cas9 system. a Schematic representation of gRNA targeting the igf3 locus. The gRNA was designed to target the open reading frame (exon II). The PAM (protospacer adjacent motif) site is highlighted in pink. An NlaIII restriction enzyme cutting site (underlined) in the target was used to detect the mutation. b DNA sequencing of the igf3 mutant allele of both igf3d2 and igf3d5 strains. One mutant had a 2-bp deletion, the other had a 5-bp deletion. c, d IF staining demonstrates the loss of the Igf3 protein in the testes of mutants. The Igf3 signals could be detected in the testis of the WT XY fish but not in the igf3 mutant testes at 90 dah. dah, days after hatching. Scale bar: 25 μm. e Western blot of Igf3 expression in control and mutant testes. The Igf3 protein was present in WT tilapia, whereas no band was observed in the mutants
The XY F0 igf3 deficient fish were screened by NlaIII digestion. The results indicate that 100% (32/32) of the microinjected XY fish were mutated. The mutation rate of each igf3 knockout individual was different, with mutation frequencies ranging from 27 to 95% (Supplemental Fig. S2C). The XY F0 mosaic fish were able to produce fertile sperm in the adult stages. Two independent igf3 mutant lines were established with either a 2-bp or 5-bp deletion (Fig. 2b). Both induced frame-shift mutations in the coding sequence, producing truncated proteins (Supplemental Fig. S2D). The F2 mutants were screened by NlaIII digestion (Supplemental Fig. S2E). For the two alleles, the expected Mendellian 1:2:1 ratio of the three genotypes (igf3+/+; igf3±; igf3−/−) was observed, indicating that loss of Igf3 in tilapia is not lethal (Supplemental Table S2). RT-PCR results show that almost no igf3 mRNA was amplified in the mutants (Supplemental Fig. S2F). IF results show that Igf3 was abundantly expressed in the testis of the WT XY fish, while no signal was detected in the testis of igf3−/− XY fish at 90 dah (Fig. 2c, d). Western blot further demonstrated that no Igf3 protein was detected in the testes of the igf3−/− XY fish (Fig. 2e). These data suggest that Igf3 was completely disrupted with no protein expression occurring in the knockout lines.
Loss of Igf3 impairs spermatogonia proliferation and meiosis initiation in XY tilapia
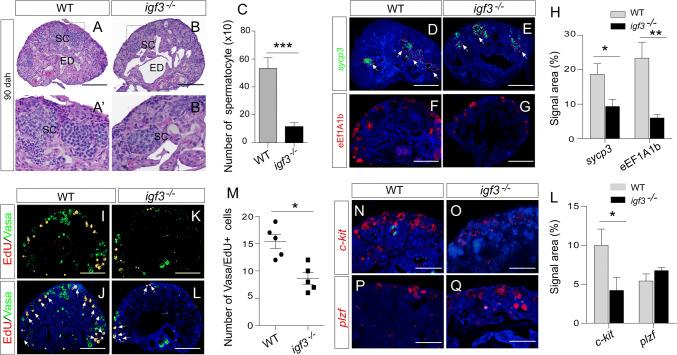
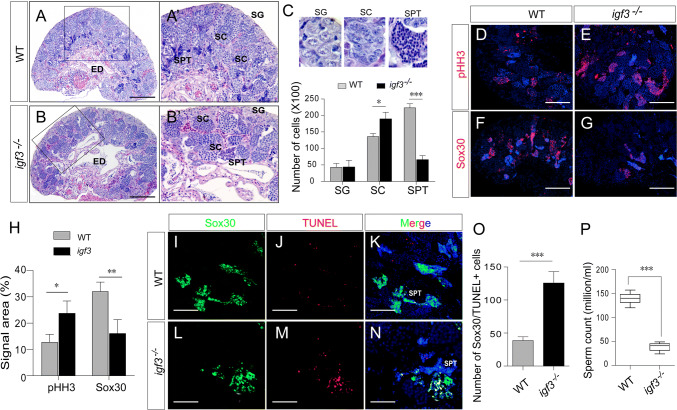
To test the role of Igf3 in testicular development, we analyzed the gonadal phenotype of the mutants at 90 dah. The growth of mutants was normal when compared with their controls, indicating that Igf3 is dispensable for tilapia growth. However, the testes weight of the igf3 mutants was significantly reduced compared with those of the WT, with an average reduction of approximately 30% (Supplemental Fig. S3A, B). Histological analysis via H&E staining showed that meiotic cells, such as spermatocytes, were observed in both the WT and mutant testes. Statistically, the number of spermatocytes was significantly decreased in the igf3−/− testes in compared with those in the WT testes (p = 0.0009; Fig. 3a–c), suggesting that meiosis is partially impaired in the mutants. We calculated the signal area of sycp3 mRNA, a marker of spermatocyte, for spermatocyte number. Consistent with the histological observations, less area of sycp3 mRNA was observed in the igf3−/− testes than that in the WT testes (Fig. 3d, e). However, the fluorescence intensity of sycp3 mRNA in the igf3−/− testis was higher than that in the WT testis (Supplemental Fig. S3C). In addition, the number of spermatogonia was significantly decreased in the igf3−/− testes when compared to that in the WT testes, as demonstrated by eEF1A1b staining, a gene specifically expressed in the spermatogonia of tilapia (Fig. 3f–h). This implies that spermatogonia development is affected after loss of Igf3. In tilapia, Vasa was highly expressed in type A undifferentiated and differentiated spermatogonia, and was decreased in spermatocytes and remained undetectable in post-meiotic cells (Supplemental Fig. S3D). The Vasa signals were significantly decreased in the igf3−/− testes when compared with that in the WT testes (Supplemental Fig. S3E), further indicating that the loss of Igf3 causes impaired germ cell development at early stage.
Fig. 3.
Knockout of igf3 leads to impairment of spermatogenesis. a–c H&E staining showed impairment of spermatogenesis in the igf3−/− testes. Quantification of cell types using histology images reflect reduced spermatocyte (SC) in the igf3−/− testes (n = 4) in comparison to the WT (n = 4). The efferent duct (ED) was enlarged in the mutants. a′, b′ Magnifications of the boxed areas of a, b, respectively. d–h IF and FISH results show that the spermatogonia marker eEF1A1b and spermatocyte marker sycp3 signals were decreased in the igf3−/− testis compared with that in the control testis. i–m EdU assay reveals that spermatogonia proliferation was affected in the igf3−/− testes. The number of EdU/Vasa positive spermatogonia (yellow) in the periphery region was significantly decreased in the igf3−/− testes when compared with controls. Vasa (green) was used to label the spermatogonia and spermatocytes of the gonads at 90 dah. n–l FISH staining shows that expression of c-kit mRNA was reduced in the igf3−/− testes while expression of plzf mRNA in the igf3−/− testes was comparable to that of the WT. Scale bars: 15 μm (a, b, d–g, i–l), 10 μm (n–q)
To analyze spermatogonia proliferation after disruption of Igf3, we monitored EdU incorporation at 90 dah, which is the stage with spermatogonia in the periphery region. Vasa/EdU positive signals were significantly decreased in the igf3−/− testes compared with the WT (Fig. 3i–m), indicating that the loss of Igf3 affects spermatogonial proliferation. To molecularly characterize spermatogenesis defects in igf3−/− testes, we performed qPCR and FISH to examine the expression of markers specific for each cell type involved in this process. At 90 dah, the expression level of plzf mRNA, which is marker of undifferentiated spermatogonia, was slightly but not significantly increased in igf3−/− testes. Expressions of the sohlh1, sohlh2, and c-kit genes present in differentiated spermatogonia were significantly decreased in igf3−/− testes (Fig. 3n–l; Supplemental Fig. S3F, G), indicating that a blockage of spermatogonia differentiation occurred in the absence of Igf3 at 90 dah. In addition, pHH3 and PCNA signals, which are two genes labeling mitotic and meiotic cells, were significantly decreased in the igf3−/− testes compared with that in the control testes, further suggesting that both mitotic and meiotic activity were decreased due to the loss of Igf3 (Supplemental Fig. S3H). IF staining and real-time PCR analyses showed that the expression level of Amh in the igf3−/− testes was comparable to that of the WT testes. However, Gsdf expression was slightly up-regulated in the igf3−/− testes compared with that of WT testes (Supplemental Fig. S4). These results indicate that spermatogonia differentiation was disrupted in the igf3−/− mutants.
The disruption of spermatogenesis led us to examine cell apoptosis in the mutants. The TUNEL analysis showed that cell apoptosis in the igf3−/− testes was comparable to that in the WT testes (Supplemental Fig. S5), thereby indicating that impairment of spermatogenesis is not caused by cell apoptosis.
Loss of Igf3 alters critical mRNA expression patterns related to spermatogenesis
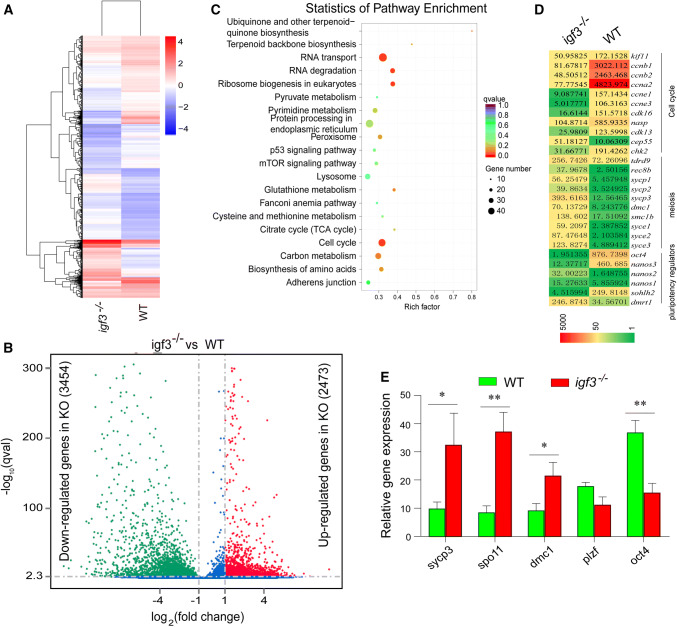
To systematically investigate gene expression changes during spermatogenesis resulting from the igf3 knockout, we compared the transcriptomes of WT and igf3−/− testes at 90 dah using RNA-seq. Transcriptomic analysis of the igf3−/− and WT testes indicated significant differences (Fig. 4a). Over 5900 DEGs were detected in the igf3−/− testes in comparison with WT testes, of which 42% (2473) were up-regulated and 58% (3454) were down-regulated, suggesting that Igf3 loss leads to a substantial change in the testicular transcriptome (Fig. 4b). KEGG analysis also showed that the DEGs between WT and igf3−/− testes were significantly clustered in the cell cycle (ccna1, ccna2, ccnb1, ccnb2, ccnb3, ccne2, cdk1, rad51), which was significantly down-regulated in the mutants (Fig. 4c). Comparative analysis of igf3−/− and WT testis transcriptomes revealed a significant up-regulation of meiosis-related genes, namely sycp1, sycp3, rec8b, dmc1, syce1, syce2 and syce3, and down-regulation of the pluripotency regulator oct4. However, expression of other pluripotency regulators dmrt1, sox2 and nanos2 in the mutant testes was significantly up-regulated compared with the WT (Fig. 4d). Consistently, real-time PCR results showed that the mRNA levels of the meiosis-related genes sycp3, dmc1, and spo11 were significantly up-regulated in the igf3−/− testes compared with that in the WT testes (Fig. 4e). This indicates that the igf3 deficiency globally influences the expression pattern of spermatogenesis-related genes that regulate multiple biological processes including spermatogonial cell maintenance, differentiation and meiosis.
Fig. 4.
Genes and molecular pathways dysregulated in Igf3-deficient testes. a Heat map of the expression of 70 Foxm1-regulated genes that were up-regulated (red) or down-regulated (blue) by two fold or more (p ≤ 0.05) in testes from Igf3-deficient tilapia relative to their expression in WT testes; presented as signal intensity normalized to the median value of signal intensities of all samples. b Volcano plots were generated using a log2 fold-change against log10 (p value) displaying the abundance of differentially expressed genes (red and green dots) of igf3−/− vs. WT fish. Red and green dots represent up-regulated (2473) and down-regulated (3454) genes in igf3−/− testes, while the blue dots represent equally expressed genes between WT and igf3−/− testes. c KEGG enrichment analysis of DEGs. Scatterplot of enriched KEGG pathways for DEG screened from igf3−/− vs. WT fish. The enrichment factor indicates the ratio of the DEGs number to the total gene number in a certain pathway. The size and color of the dots represent the gene number and the range of p values, respectively. d Comparative transcriptomic analysis between igf3−/− and wild-type testes reveal a significant up-regulation of meiosis-related genes and down-regulation of regulators of meiosis, pluripotency and the cell cycle. e Real-time PCR results show that mRNA level of the meiosis related genes sycp3, dmc1 and spo11, were significantly up-regulated in the igf3−/− testis. Data are expressed as the mean ± SD of triplicates. Differences between groups were statistically examined with unpaired two tailed Student’s t test; *p < 0.05, **p < 0.01
Loss of Igf3 greatly reduces reproductive capacity in adult XY tilapia
Adult igf3−/− XY tilapia grew normally and had a similar body weight to WT fish. However, the testes weight of igf3−/− was significantly reduced when compared with those of the WT individuals, as demonstrated by the GSI (Supplemental Fig. S6A, B). Histological analysis showed that all stages of germ cells, including spermatogonia, spermatocytes and spermatids, were present in all individuals of the mutant and WT fish. A quantitative comparison of testicular cell types revealed a significant increase in the number of spermatocytes and a drastic reduction of spermatids in the igf3−/− mutants compared to the WT, whereas the number of spermatogonia was comparable (Fig. 5a–c). This result was further confirmed by immunostaining different germ cell markers. pHH3, which was predominately expressed in the secondary spermatocytes and some proliferated germ cells of testis at 180 dah (Supplemental Fig.S6C), was more abundantly expressed in the igf3−/− testes than in the WT testes. Sox30 was predominantly detected in spermatids of the WT testes while less signal was observed in the igf3−/− testes (Fig. 5d–h). Expression of protamine, a gene specifically expressed in mature sperm, was significantly decreased in the igf3−/− testes when compared with the WT (Supplemental Fig. S6D). These results indicate that spermatogenesis activity was decreased in the igf3−/− testes when compared to the WT testes.
Fig. 5.
Knockout of igf3 resulted in reduced reproductive capacity in adult mutants. a–c H&E staining showed that all stages of germ cells including spermatogonia (SG), spermatocytes (SC) and spermatids (SPT) were present in all individuals of the mutant (n = 3) and WT (n = 3) fish. A quantitative comparison of testicular cell types revealed a significant increase in the number of spermatocytes and a drastic reduction of spermatids in the igf3−/− mutants compared to the WT. a′, b′ Magnifications of the boxed areas of a′, b, respectively. d–h IF demonstrates the increased spermatocyte and decreased spermatids in the igf3−/− mutants. pHH3 which was mainly expressed in secondary spermatocytes, was more abundantly expressed in the igf3−/− testes than that in the control testes at 180 dah. Sox30 was highly detected in spermatid of the WT testes, but less signal was observed in the igf3−/− testes. i–o The cell apoptosis indicates increased spermatid apoptosis in the igf3−/− testes. Red and green fluorescence indicate positive apoptotic signals and Sox30 positive cells, respectively. p The number of sperm was sharply reduced in igf3−/− fish when compared with the WT fish. Data are expressed as the mean ± SD of triplicates. The difference between groups was statistically examined with an unpaired two tailed Student’s t test; *p < 0.05; **p < 0.01; ***p < 0.001. Scale bars 100 μm (a, b), 50 μm (d–g), 25 μm (i–n)
The reduced spermatids in igf3−/− adult XY fish led us to examine cell apoptosis in adult mutants. The result show that spermatid apoptosis in the igf3−/− testes was increased compared with that of the WT testes, as determined by co-staining for the spermatid marker Sox30 and TUNEL signals (Fig. 5i–o). The semen volume of igf3−/− XY fish was significantly decreased compared with the WT male fish at 180 dah (Supplemental Fig. S6E). Analysis of the igf3−/− semen smear showed a drastic reduction in sperm count (Fig. 5p, Supplemental Fig. S6F, G). However, sperm morphology was normal in igf3−/− mutant and WT fish with normal flagella, and displayed vigorous flagellar activity and progressive movement (Supplemental Fig. S6H). Thus, the sperm extracted from the mutants and subsequently used for in vitro fertilization of WT eggs produced viable embryos. The fertilization rate was slightly lower than the WT XY sperm when using an equal volume of semen from the igf3−/− mutant and WT XY fish. Additionally, sperm motility was significantly decreased in the igf3−/− XY fish (Supplemental Fig. S6I, J). Therefore, spermatogenesis in igf3−/− XY fish is partially impaired and loss of Igf3 reduces the fertility of adult mutant fish.
Overexpression of Igf3 enhances spermatogenesis
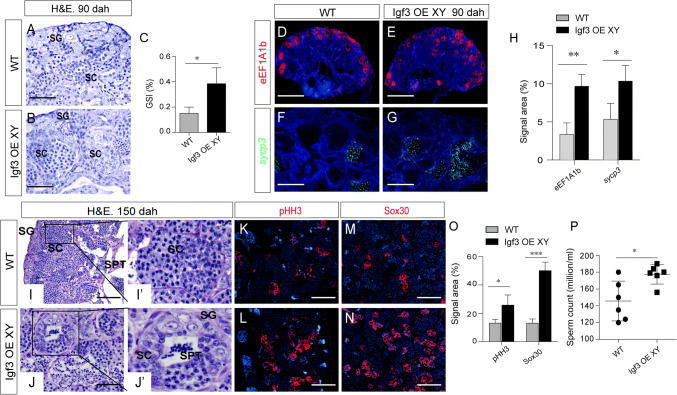
To better understand the role of Igf3 in regulating spermatogenesis, overexpression (OE) of igf3 was conducted in the XY fish using a CMV promoter-derived expression vector (Supplemental Fig. S7A). After microinjecting into one-cell stage fertilized eggs, a specific 785 bp band was amplified from the genomic DNA of the injected XY fish using genomic PCR (Supplemental Fig. S7B). When examined at 90 dah, mosaic expression of GFP was observed in the igf3 OE testes, whereas no GFP was observed in the WT testes (Supplemental Fig. S7C, D). In addition, the Igf3 protein level in the testes of overexpressed fish was significantly increased compared with the WT fish (Supplemental Fig. S7E–G). Histology at 90 dah shows that germ cells, including spermatogonia and a few spermatocytes, were observed in the WT XY fish, while more spermatogonia and spermatocytes were observed in the Igf3 overexpression fish (Fig. 6a, b). The GSI of the Igf3 overexpression fish was significantly larger than that of the WT individuals (Fig. 6c). The number of spermatogonia was significantly increased in the igf3−/− testes when compared to that in the WT testes, as demonstrated by eEF1A1b staining. In addition, the area for sycp3 signal was increased in the igf3 OE testes at 90 dah, whereas less area of sycp3 signal was observed in the WT testes (Fig. 6d–h). These results indicate that overexpression of Igf3 promotes spermatogonia differentiation and entry into meiosis.
Fig. 6.
Overexpression of igf3 enhanced spermatogenesis. a, b H&E staining shows that spermatogenesis is enhanced in the igf3 OE testes. Only spermatogonia and a few spermatocytes were observed in the WT XY fish (n = 4), while more spermatogonia and spermatocytes were observed in the igf3 OE fish (n = 4) at 90 dah. c Gonadosomatic index (GSI) of controls and igf3−/− fish (n = 3, p < 0.05). The testes of the igf3 OE fish were significantly larger than those of the control individuals. d–h IF and FISH using the spermatogonia marker eEF1A1b and the spermatocyte marker sycp3 showed more spermatogonia and spermatocytes in the igf3 OE testes. eEF1A1b and sycp3 were abundantly expressed in the igf3 OE testis at 90 dah, while less signals were observed in the WT testes. i–o Overexpression of Igf3 promotes spermatogenesis. An enlarged lumen was observed in igf3 OE testes. IF staining with pHH3 and Sox30 demonstrated that more secondary spermatocyte and spermatids accumulated in the igf3 OE fish testes compared with that of control testes. i′′, j′ Magnifications of the boxed areas of i′, j, respectively. p The number of sperm increased in igf3 OE fish compared with the WT fish. Data are expressed as the mean ± SD of triplicates. The difference between groups was statistically examined with unpaired two tailed Student’s t test; *p < 0.05; ***p < 0.001. Scale bar 10 μm (a, b, f, g), 15 μm (c, d, i, j), 50 μm (k–n)
To investigate spermatogonial DNA synthesis after overexpression of Igf3, we monitored EdU incorporation at 90 dah. Cell proliferation significantly increased in igf3 OE testes compared with the WT, indicating that Igf3 promotes spermatogonial proliferation (Supplemental Fig. S7H–J).
Histological examination revealed different stages of germ cells, including spermatogonia, spermatocytes, and spermatids, in both igf3 OE and control testes at 150 dah. Compared with the WT testis, an enlarged lumen in the OE testes was observed (Fig. 6i, j), which indicates more sperm production and intense Sertoli cell secretion into the testicular lumen. Expression of the pHH3 and Sox30 proteins was much higher in the igf3 OE testes than in the WT testes (Fig. 6k–o). The sperm count and sperm motility was significantly increased in the igf3 OE fish compared with the WT fish at 210 dah (Fig. 6p, Supplemental Fig. S7K–N). However, the body weight of igf3 OE fish was comparable to the WT (Supplemental Fig. S7O). In addition, the 11-KT level in the igf3 OE fish was comparable to WT (Supplemental Fig. S7P). These results indicate that overexpression of Igf3 enhances spermatogenesis.
Regulation of igf3 expression by androgen
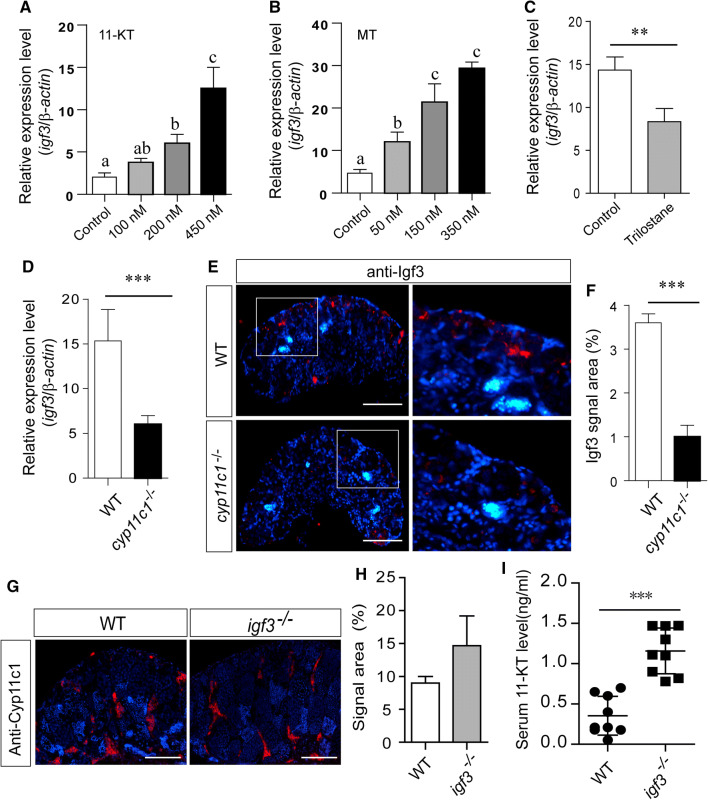
To further elucidate the relationship between testicular igf3 expression and the sex steroids that are associated with spermatogenesis regulation, we examined the effects of 11-KT and MT on the igf3 mRNA levels in the tilapia testes. Testicular fragments were cultured for 4 days with these steroids or the trilostane, an inhibitor of 3β-HSD. Under basal conditions, the expression of steroidogenic enzymes, including cyp11c1 and cyp17a1, were slightly down-regulated compared with the controls after 4 days of incubation (Supplemental Fig. 8A). Histological examination showed that the morphology of 11-KT-treated testis was similar to the controls (Supplemental Fig. 8B, C). However, the data show that igf3 mRNA expression was significantly induced by MT and 11-KT stimulation in a dose-dependent manner in the in vitro culture system (Fig. 7a, b). In contrast, igf3 mRNA was significantly down-regulated in the trilostane-treated testes when compared with the controls (Fig. 7c). Using a cyp11c1 knockout model for tilapia, expression of igf3 was examined. IF and real-time PCR data shows that the Igf3/igf3 expression level was significantly down-regulated in the cyp11c1−/− testes when compared with that of the WT testes at 180 dah (Fig. 7d–f). Real-time PCR and IF analyses show that the cyp11c1 expression level was unchanged in Igf3-deficient testes when compared with the controls (Fig. 7g, h). However, serum 11-KT was significantly increased in the Igf3-deficient XY fish when compared with the WT (Fig. 7i). These data indicate that Igf3 acts downstream of androgen.
Fig. 7.
Igf3 expression is regulated by androgen. a, b The transcript level of igf3 in testicular explants incubated with increasing concentrations of MT and 11-KT ex vivo for 4 days. Data are expressed as the mean ± SD. Different letters indicate statistical differences at p < 0.05 as determined by one-way ANOVA, followed by Tukey’s/Kramer post hoc test. c Treatment with the androgen synthesis inhibitor trilostane significantly down-regulated igf3 mRNA expression. d, e, f Real-time PCR and IF results showed that igf3 expression was significantly down-regulated in the cyp11c1−/− testes compared with that in the control testes. g, h IF analysis showed that cyp11c1 expression was not influenced in the igf3−/− testes. i Knockouts of igf3 in the XY fish resulted in elevated serum 11-KT concentrations compared with the control fish. Data are expressed as the mean ± SD of triplicates. The difference between two groups was statistically examined with unpaired two tailed Student’s t test; *p < 0.05; **p < 0.01; ***p < 0.001. Scale bar 15 μm (d), 50 μm (f)
Discussion
Igfs serve as important growth factors and are critical in a wide variety of physiological roles for vertebrate gonadal development and reproduction. Our previous study found that igf3, a novel member of the igf family, is predominantly expressed in gonadal tissue [45]. Although this novel igf was found over ten years ago, its specific role in the gonads still required further clarification by in vivo methods. In this study, we presented a number of findings that demonstrate the significance of Igf3 in male spermatogenesis and reproduction: For instance, Igf3 is specifically and abundantly expressed in the testis and its expression is dramatically increased during testes development; igf3 knockout XY fish results in male subfertility because of severe impairment of spermatogenesis; and absence of Igf3 results in dysregulation of many genes involved in the cell cycle, meiosis and pluripotency regulators. In contrast, overexpression of Igf3 in XY tilapia enhances spermatogenesis and sperm production, and Igf3 is a downstream factor of androgens involved in spermatogenesis. Thus, our report reveals the role of igf3 in fish spermatogenesis and reproduction using knockout and overexpression models (Supplemental Fig. 9).
A number of studies have previously suggested that the IGF system is involved in mammalian reproductive functions. In mice, mutation of igf1 in XY individuals results in reduced testis size and subsequent infertility due to defects in spermatogonial proliferation and differentiation. However, capacitated sperm are able to fertilize WT eggs in vitro [73, 74]. Knockouts of the igf1 receptor (igf1r) in male mice also led to a 75% reduction in testis size and daily sperm production [74]. Recent zebrafish studies showed that Igf3 increased type A undifferentiated and differentiated spermatogonia and up-regulated the expression of genes related to spermatogonial differentiation and entry into meiosis [49, 75]. In this study, in vivo ablation of igf3 in tilapia severely inhibited spermatogonial proliferation and differentiation, thereby decreasing the meiotic activity of germ cells. Although spermatogenesis resumed later in the mutants, the sperm production and semen volume were significantly reduced although the collected sperm could be used to artificially fertilize the WT eggs, which agrees with the finding in igf1 knockout mice [73]. In contrast, overexpression of Igf3 in XY fish could accelerate spermatogenesis. More spermatids were found in the adult igf3 OE testis as demonstrated by staining for Sox30, which is a spermatid marker in the mice [76]. These reproduction effects could exclude the side effects because Igf3 was predominantly expressed in the gonads, not in other tissues [45]. Although igf3 overexpression was driven by the CMV promoter, other biological process, such as body growth, was not influenced after igf3 overexpression (Supplemental Fig. S9). This also indicates that enhanced spermatogenesis is the result of Igf3 overexpression. The Igf system regulates many processes related to germ cell proliferation and differentiation. In zebrafish, Igf1 signaling is involved in primordial germ cell survival and migration [77]. Tissue cultures with tilapia Igf1 demonstrated that Igf1 is able to support spermatogenesis by stimulating both spermatogonial proliferation and the entry of spermatogonia into meiosis [42]. Moreover, in tilapia testis, igf1 could regulate spermatogenesis in conjunction with steroids [42, 78]. These results suggest that Igf3 regulates spermatogenesis activity in a similar manner as Igf1 in fish.
It is well known that the spermatogenesis process involves four pivotal transitions that occur in physical proximity: spermatogonial differentiation, meiotic initiation, initiation of spermatid elongation and release of spermatozoa [3]. Loss of igf3 results in misregulation of genes involved in cell cycle, meiosis and pluripotency regulators. For example, transcriptomics demonstrated that undifferentiated spermatogonia-related gene expression, such as nanos2 and dmrt1, was up-regulated in igf3 mutants. However, differentiated spermatogonia-related gene expression, including c-kit, sohlh2 and dazl, were down-regulated. These data further confirm the regulatory role of spermatogonia development by igf3. At 90 dah, the meiosis activity was decreased in the mutants as shown by the reduced spermatocytes. It is well known that igf is important for regulating many genes involved in cell cycle process [79, 80]. Detailed analysis revealed that dysregulation of genes related to spermatogenesis, especially significant down-regulation of genes involved in cell cycle, could explain the decrease in number of spermatocytes even though sycp3 mRNA level was up-regulated in the mutants. In addition, expressions of meiosis related genes were also regulated by other various factors, such as androgens, retinoic acid, DHP and gonadotropins [81]. With the development of testis, more spermatocytes accumulated in the adult igf3 mutants. The up-regulation of meiosis-related genes sycp3, rec8b and dmc1 might explain why abundant spermatocytes were observed in the igf3−/− testes at 180 dah. It has also reported in mice that the defects in spermatogenesis in juvenile stage could be recovered in adult stage [82]. However, the dramatically reduced sperm count and semen volume indicates that the transition of spermatocyte to spermatid is affected after loss of Igf3. Moreover, igf3 was expressed in the spermatids, as has been seen for igf1 expression in the spermatids of mouse (Mus musculus) and medaka (Oryzias latipes) [73, 83]. Loss of Igf3 in tilapia induced spermatid apoptosis, suggesting that Igf3 is crucial for spermatid survival. These results suggest that impaired male fertility in igf3 knockout tilapia arises, at least in part, from defects during both the pre- and post-meiotic stages of spermatogenesis.
Igf3 was expressed in both the germ cells and Sertoli cells of tilapia. Further investigation is needed to study which Igf3-expressing cell type contributes to the regulation of spermatogenesis by generating conditional knockout models. The present study reports that Igf3 regulates spermatogenesis; however more research will be required to elucidate exactly how igf3 is involved. A recent study showed that Igf3 activates β-catenin signaling to stimulate spermatogonial differentiation in zebrafish [50]. Intriguingly, our previous publication using XY tilapia showed that a deficiency of Rspo1, the upstream gene of the wnt/β-catenin pathway, caused a delay in spermatogenesis and inhibition of igf3 expression [84]. In mice, Wnt/β-catenin signaling is required for proliferation of undifferentiated spermatogonia in vivo [85]. In this study, β-catenin-1 was slightly decreased in the igf3 knockout testes (data not shown). Thus, future work will examine the relationship between Igf3 and the wnt/β-catenin pathway during spermatogenesis in fish.
In teleost fish, 11-KT is considered to be the major endogenous androgenic sex hormone. Previous work showed that 11-KT induces spermatogenesis in several fish species including eel (Anguilla japonica), zebrafish, African catfish (Clarias gariepinus) and Japanese huchen (Hucho perryi). [7, 10, 86, 87]. Targeted disruption of the androgen receptor (ar) in zebrafish causes male infertility via defective spermatogenesis and also significantly decreases igf3 expression [22, 88]. In vitro studies showed that igf3 expression is also regulated by 11-KT in tilapia and zebrafish [49]. Knockouts of cyp11c1, the key gene encoding 11β-hydroxylase, in tilapia lead to significant down-regulation of igf3 expression. Most importantly, both igf3 and cyp11c1 mutants exhibit similar phenotypes in tilapia, in which both have reduced semen volume and sperm production due to the defects in spermatogenesis (unpublished data). In tilapia igf3 mutants, cyp11c1 expression was unchanged. Strangely, the 11-KT level in the igf3 mutants was significantly increased. This could also be a compensatory response due to the inhibition of spermatogenesis [21]. These studies indicate that Igf3 functions in downstream of androgen in spermatogenesis. Of course, higher androgen plasma levels could exert a negative feedback in the brain-pituitary axis which could lead to impairment of spermatogenesis and less sperm. Additional studies are required to verify that the defects in spermatogenesis caused by loss of Igf3 are not a secondary effect of increased androgen.
It is worth noting that knockouts of the igf system in mice affect body growth because reduced body size was observed in the mutants [73, 74]. However, knockouts of igf3 in tilapia did not affect body growth, indicating that Igf3 specifically functions in gonad tissue to regulate spermatogenesis in fish. Previous studies have also shown that igf3 expression is not regulated by growth hormone in zebrafish and tilapia [48, 89]. Therefore, igf3 is not involved in regulating body growth in fish.
In conclusion, the data obtained from the present study have provided strong genetic evidence that Igf3, which acts downstream of androgens, is important for spermatogenesis in tilapia. In addition, Igf3 specifically functions in reproduction but not body growth in this teleost species.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by Grants 31772830, 31602134, 31861123001 and 31630082 from the National Natural Science Foundation of China; Grant 2018YFD0900202 from the National Key Basic Research Program of China, Grant cstc2017jcyjAX0138, cstc2017shms-xdny80018 and cstc2015jcyjB80001 from the Natural Science Foundation Project of Chongqing, Chongqing Science and Technology Commission; Grant XDJK2018B026 from the Fundamental Research Funds for the Central Universities.
Abbreviations
- igf1
Insulin-like growth factor 1
- igf2
Insulin-like growth factor 2
- igf3
Insulin-like growth factor 3
- CRISPR/Cas9
Clustered regularly interspaced short palindromic repeats/clustered regularly interspaced short palindromic repeats-associated protein 9 nuclease
- WT
Wild type
- T
Testosterone
- 11-KT
11-Ketotestosterone
- DHP
17α, 20β-Dihydroxy-4-pregnen-3-one
- E2
Estradiol-17β
- PAM
Protospacer adjacent motif
- ORF
Open reading frame
- PFA
Paraformaldehyde
- IF
Immunofluorescence
- IHC
Immunohistochemistry
- FISH
Fluorescence in situ hybridization
- BSA
Bovine serum albumin
- amh
Anti-Mullerian hormone
- FDR
False discovery rate
- DEGs
Differentially expressed genes
- GO
Gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- TUNEL
Terminal deoxynucleotidyl transferase (TdT) dUTP Nick-End Labeling
- SG
Spermatogonia
- SC
Spermatocytes
- SD
Spermatids
Author contributions
MHL, CHKC and DSW conceived and designed the study, MHL, XYL, HSX, SFD, SSQ, YBL, QYZ and MMJ did the experiment, and all authors contributed to the discussion of the results and writing of the paper.
Compliance with ethical standards
Conflict of interest
The authors have nothing to disclose.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Minghui Li, Xingyong Liu, Shengfei Dai and Hesheng Xiao contributed equally to this work.
Contributor Information
Minghui Li, Email: limh@163.com.
Deshou Wang, Email: wdeshou@swu.edu.cn.
References
- 1.Griswold MD. Spermatogenesis: the commitment to meiosis. Physiol Rev. 2016;96(1):1–17. doi: 10.1152/physrev.00013.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fayomi AP, Orwig KE. Spermatogonial stem cells and spermatogenesis in mice, monkeys and men. Stem Cell Res. 2018;29:207–214. doi: 10.1016/j.scr.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Endo T, Freinkman E, de Rooij DG, Page DC. Periodic production of retinoic acid by meiotic and somatic cells coordinates four transitions in mouse spermatogenesis. Proc Natl Acad Sci USA. 2017;114(47):E10132–E10141. doi: 10.1073/pnas.1710837114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagahama Y. Endocrine regulation of gametogenesis in fish. Int J Dev Biol. 1994;38:217–229. [PubMed] [Google Scholar]
- 5.Schulz RW, de França LR, Lareyre JJ, Le Gac F, Chiarini-Garcia H, Nobrega RH, Miura T. Spermatogenesis in fish. Gen Comp Endocrinol. 2010;165(3):390–411. doi: 10.1016/j.ygcen.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Miura T, Miura C. Molecular control mechanisms of fish spermatogenesis. Fish Physiol Biochem. 2003;28:181–186. [Google Scholar]
- 7.Amer MA, Miura T, Miura C, Yamauchi K. Involvement of sex steroid hormones in the early stages of spermatogenesis in Japanese huchen (Hucho perryi) Biol Reprod. 2001;65:1057–1066. doi: 10.1095/biolreprod65.4.1057. [DOI] [PubMed] [Google Scholar]
- 8.García-López A, Bogerd J, Granneman JC, van Dijk W, Trant JM, Taranger GL, Schulz RW. Leydig cells express follicle-stimulating hormone receptors in African catfish. Endocrinology. 2009;150(1):357–365. doi: 10.1210/en.2008-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.García-López A, de Jonge H, Nóbrega RH, de Waal PP, van Dijk W, Hemrika W, Taranger GL, Bogerd J, Schulz RW. Studies in zebrafish reveal unusual cellular expression patterns of gonadotropin receptor messenger ribonucleic acids in the testis and unexpected functional differentiation of the gonadotropins. Endocrinology. 2010;151(5):2349–2360. doi: 10.1210/en.2009-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miura T, Yamauchi K, Takahashi H, Nagahama Y. Hormonal induction of all stages of spermatogenesis in vitro in the male Japanese eel (Anguilla japonica) Proc Natl Acad Sci USA. 1991;88(13):5774–5778. doi: 10.1073/pnas.88.13.5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miura T, Miura C, Ohta T, Nader MR, Todo T, Yamauchi K. Estradiol-17β stimulates the renewal of spermatogonial stem cells in males. Biochem Biophys Res Commun. 1999;264(1):230–234. doi: 10.1006/bbrc.1999.1494. [DOI] [PubMed] [Google Scholar]
- 12.Miura T, Higuchi M, Ozaki Y, Ohta T, Miura C. Progestin is an essential factor for the initiation of the meiosis in spermatogenetic cells of the eel. Proc Natl Acad Sci USA. 2006;103(19):7333–7338. doi: 10.1073/pnas.0508419103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chauvigné F, Verdura S, Mazón MJ, Duncan N, Zanuy S, Gómez A, Cerdà J. Follicle-stimulating hormone and luteinizing hormone mediate the androgenic pathway in Leydig cells of an evolutionary advanced teleost. Biol Reprod. 2012;87(2):35. doi: 10.1095/biolreprod.112.100784. [DOI] [PubMed] [Google Scholar]
- 14.Chen SX, Bogerd J, Schoonen NE, Martijn J, de Waal PP, Schulz RW. A progestin (17α, 20β-dihydroxy-4-pregnen-3-one) stimulates early stages of spermatogenesis in zebrafish. Gen Comp Endocrinol. 2013;185:1–9. doi: 10.1016/j.ygcen.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 15.Chu L, Li J, Liu Y, Cheng CH. Gonadotropin signaling in zebrafish ovary and testis development: insights from gene knockout study. Mol Endocrinol. 2015;29(12):1743–1758. doi: 10.1210/me.2015-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Z, Lau SW, Zhang L, Ge W. Disruption of zebrafish follicle stimulating hormone receptor (fshr) but not luteinizing hormone receptor (lhcgr) gene by TALEN leads to failed follicle activation in females followed by sexual reversal to males. Endocrinology. 2015;156(10):3747–3762. doi: 10.1210/en.2015-1039. [DOI] [PubMed] [Google Scholar]
- 17.de Castro Assis LH, de Nobrega RH, Gomez-Gonzalez NE, Bogerd J, Schulz RW. Estrogen-induced inhibition of spermatogenesis in zebrafish is largely reversed by androgen. J Mol Endocrinol. 2018;60:273–284. doi: 10.1530/JME-17-0177. [DOI] [PubMed] [Google Scholar]
- 18.Cavaco JE, Bogerd J, Goos H, Schulz RW. Testosterone inhibits 11-ketotestosterone-induced spermatogenesis in African catfish (Clarias gariepinus) Biol Reprod. 2001;65:1807–1812. doi: 10.1095/biolreprod65.6.1807. [DOI] [PubMed] [Google Scholar]
- 19.Ozaki Y, Higuchi M, Miura C, Yamaguchi S, Tozawa Y, Miura T. Roles of 11beta-hydroxysteroid dehydrogenase in fish spermatogenesis. Endocrinology. 2006;147:5139–5146. doi: 10.1210/en.2006-0391. [DOI] [PubMed] [Google Scholar]
- 20.Elisio M, Chalde T, Miranda LA. Seasonal changes and endocrine regulation of pejerrey (Odontesthes bonariensis) spermatogenesis in the wild. Gen Comp Endocrinol. 2015;221:236–243. doi: 10.1016/j.ygcen.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 21.Crowder CM, Lassiter CS, Gorelick DA. Nuclear androgen receptor regulates testes organization and oocyte maturation in zebrafish. Endocrinology. 2018;159:980–993. doi: 10.1210/en.2017-00617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang H, Chen Y, Wang L, Yin Y, Li G, Guo Y, Liu Y, Lin H, Cheng CHK, Liu X. Fertility impairment with defective spermatogenesis and steroidogenesis in male zebrafish lacking androgen receptor. Biol Reprod. 2018;98(2):227–238. doi: 10.1093/biolre/iox165. [DOI] [PubMed] [Google Scholar]
- 23.Liu G, Luo F, Song Q, Wu L, Qiu Y, Shi H, Wang D, Zhou L. Blocking of progestin action disrupts spermatogenesis in Nile tilapia (Oreochromis niloticus) J Mol Endocrinol. 2014;53(1):57–70. doi: 10.1530/JME-13-0300. [DOI] [PubMed] [Google Scholar]
- 24.Wang C, Liu D, Chen W, Ge W, Hong W, Zhu Y, Chen SX. Progestin increases the expression of gonadotropins in pituitaries of male zebrafish. J Endocrinol. 2016;230(1):143–156. doi: 10.1530/JOE-16-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu Y, Liu D, Shaner ZC, Chen S, Hong W, Stellwag EJ. Nuclear progestin receptor (pgr) knockouts in zebrafish demonstrate role for pgr in ovulation but not in rapid non-genomic steroid mediated meiosis resumption. Front Endocrinol (Lausanne) 2015;6:37. doi: 10.3389/fendo.2015.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fang X, Wu L, Yang L, Song L, Cai J, Luo F, Wei J, Zhou L, Wang D. Nuclear progestin receptor (Pgr) knockouts resulted in subfertility in male tilapia (Oreochromis niloticus) J Steroid Biochem Mol Biol. 2018;182:62–71. doi: 10.1016/j.jsbmb.2018.04.011. [DOI] [PubMed] [Google Scholar]
- 27.Tang H, Chen Y, Liu Y, Yin Y, Li G, Guo Y, Liu X, Lin H. New insights into the role of estrogens in male fertility based on findings in aromatase-deficient zebrafish. Endocrinology. 2017;158(9):3042–3054. doi: 10.1210/en.2017-00156. [DOI] [PubMed] [Google Scholar]
- 28.Porseryd T, Reyhanian Caspillo N, Volkova K, Elabbas L, Källman T, Dinnétz P, Olsson PE, Porsch-Hällström I. Testis transcriptome alterations in zebrafish (Danio rerio) with reduced fertility due to developmental exposure to 17α-ethinyl estradiol. Gen Comp Endocrinol. 2018;262:44–58. doi: 10.1016/j.ygcen.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 29.Liu ZH, Chen QL, Chen Q, Li F, Li YW. Diethylstilbestrol arrested spermatogenesis and somatic growth in the juveniles of yellow catfish (Pelteobagrus fulvidraco), a fish with sexual dimorphic growth. Fish Physiol Biochem. 2018;44(3):789–803. doi: 10.1007/s10695-018-0469-1. [DOI] [PubMed] [Google Scholar]
- 30.Piferrer F. Endocrine sex control strategies for the feminization of teleost fish. Aquaculture. 2001;197:229–281. [Google Scholar]
- 31.Nakamura M, Kobayashi T, Chang XT, Nagahama Y. Gonadal sex differentiation in fish. J Exp Zool. 1998;281:362–372. [Google Scholar]
- 32.Duan C. The insulin-like growth factor system and its biological actions in fish. Am Zool. 1997;37:491–503. [Google Scholar]
- 33.Lu C, Lam HN, Menon RK. New members of the insulin family: regulators of metabolism, growth and now … reproduction. Pediatr Res. 2005;57:70R–73R. doi: 10.1203/01.PDR.0000159573.55187.CA. [DOI] [PubMed] [Google Scholar]
- 34.Reinecke M. Insulin-like growth factors and fish reproduction. Biol Reprod. 2010;82(4):656–661. doi: 10.1095/biolreprod.109.080093. [DOI] [PubMed] [Google Scholar]
- 35.Le Gac F, Loir M, Le Bail PY, Ollitrault M. Insulin-like growth factor I (IGF-I) mRNA and IGF-I receptor in trout testis and in isolated spermatogenic and Sertoli cells. Mol Reprod Dev. 1996;44:23–35. doi: 10.1002/(SICI)1098-2795(199605)44:1<23::AID-MRD3>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 36.Reinecke M, Schmid A, Ermatinger R, Loffing-Cueni D. Insulin-like growth factor I in the teleost Oreochromis mossambicus, the tilapia: gene sequence, tissue expression, and cellular localization. Endocrinology. 1997;138:3613–3619. doi: 10.1210/endo.138.9.5375. [DOI] [PubMed] [Google Scholar]
- 37.Schmid AC, Näf E, Kloas W, Reinecke M. Insulin-like growth factor-I and -II in the ovary of a bony fish, Oreochromis mossambicus, the tilapia: in situ hybridisation, immunohistochemical localisation, Northern blot and cDNA sequences. Mol Cell Endocrinol. 1999;156:141–149. doi: 10.1016/s0303-7207(99)00131-8. [DOI] [PubMed] [Google Scholar]
- 38.Berishvili G, D'Cotta H, Baroiller JF, Segner H, Reinecke M. Differential expression of IGF-I mRNA and peptide in the male and female gonad during early development of a bony fish, the tilapia Oreochromis niloticus. Gen Comp Endocrinol. 2006;146:204–210. doi: 10.1016/j.ygcen.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 39.Radaelli G, Patruno M, Maccatrozzo L, Funkenstein B. Expression and cellular localization of insulin-like growth factor-II protein and mRNA in Sparus aurata during development. J Endocrinol. 2003;178:285–299. doi: 10.1677/joe.0.1780285. [DOI] [PubMed] [Google Scholar]
- 40.Viñas J, Piferrer F. Stage-specific gene expression during fish spermatogenesis as determined by laser-capture microdissection and quantitative-PCR in sea bass (Dicentrarchus labrax) gonads. Biol Reprod. 2008;79:738–747. doi: 10.1095/biolreprod.108.069708. [DOI] [PubMed] [Google Scholar]
- 41.Nader MR, Miura T, Ando N, Miura C, Yamauchi K. Recombinant human insulin-like growth factor I stimulates all stages of 11-ketotestosteroneinduced spermatogenesis in the Japanese eel, Anguilla japonica, in vitro. Biol Reprod. 1999;61:944–947. doi: 10.1095/biolreprod61.4.944. [DOI] [PubMed] [Google Scholar]
- 42.Tokalov SV, Gutzeit HO. Spermatogenesis in testis primary cell cultures of the tilapia (Oreochromis niloticus) Dev Dyn. 2005;233:1238–1247. doi: 10.1002/dvdy.20379. [DOI] [PubMed] [Google Scholar]
- 43.Loir M, Le Gac F. Insulin-like growth factor-I and -II binding and action on DNA synthesis in rainbow trout spermatogonia and spermatocytes. Biol Reprod. 1994;51:1154–1163. doi: 10.1095/biolreprod51.6.1154. [DOI] [PubMed] [Google Scholar]
- 44.Loir M. Spermatogonia of rainbow trout, II: in vitro study of the influence of pituitary hormones, growth factors and steroids on mitotic activity. Mol Reprod Dev. 1999;53:434–442. doi: 10.1002/(SICI)1098-2795(199908)53:4<434::AID-MRD9>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 45.Wang DS, Jiao B, Hu C, Huang X, Liu Z, Cheng CH. Discovery of a gonad-specific IGF subtype in teleost. Biochem Biophys Res Commun. 2008;367:336–341. doi: 10.1016/j.bbrc.2007.12.136. [DOI] [PubMed] [Google Scholar]
- 46.Nelson SN, Van Der Kraak G. The role of the insulin-like growth factor (IGF) system in zebrafish (Danio rerio) ovarian development. Gen Comp Endocrinol. 2010;168:103–110. doi: 10.1016/j.ygcen.2010.04.021. [DOI] [PubMed] [Google Scholar]
- 47.Li J, Liu Z, Wang D, Cheng CH. Insulin-like growth factor 3 is involved in oocyte maturation of zebrafish. Biol Reprod. 2011;84:476–486. doi: 10.1095/biolreprod.110.086363. [DOI] [PubMed] [Google Scholar]
- 48.Irwin DA, Van Der Kraak G. Regulation and actions of insulin-like growth factors in the ovary of zebrafish (Danio rerio) Gen Comp Endocrinol. 2012;177(1):187–194. doi: 10.1016/j.ygcen.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 49.Nóbrega RH, Morais RD, Crespo D, de Waal PP, de França LR, Schulz RW, Bogerd J. Fsh stimulates spermatogonial proliferation and differentiation in zebrafish via Igf3. Endocrinology. 2015;156(10):3804–3817. doi: 10.1210/en.2015-1157. [DOI] [PubMed] [Google Scholar]
- 50.Safian D, Bogerd J, Schulz RW. Igf3 activates β-catenin signaling to stimulate spermatogonial differentiation in zebrafish. J Endocrinol. 2018;238(3):245–257. doi: 10.1530/JOE-18-0124. [DOI] [PubMed] [Google Scholar]
- 51.Safian D, van der Kant HJG, Crespo D, Bogerd J, Schulz RW. Follicle-stimulating hormone regulates igfbp gene expression directly or via downstream effectors to modulate Igf3 effects on zebrafish spermatogenesis. Front Endocrinol (Lausanne) 2017;8:328. doi: 10.3389/fendo.2017.00328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sambroni E, Lareyre JJ, Le Gac F. Fsh controls gene expression in fish both independently of and through steroid mediation. PLoS ONE. 2013;8(10):e76684. doi: 10.1371/journal.pone.0076684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crespo D, Assis LHC, Furmanek T, Bogerd J, Schulz RW. Expression profiling identifies Sertoli and Leydig cell genes as Fsh targets in adult zebrafish testis. Mol Cell Endocrinol. 2016;660(437):237–251. doi: 10.1016/j.mce.2016.08.033. [DOI] [PubMed] [Google Scholar]
- 54.Li M, Yang H, Zhao J, Fang L, Shi H, Li M, Sun Y, Zhang X, Jiang D, Wang D. Efficient and heritable gene targeting in tilapia by CRISPR/Cas9. Genetics. 2014;197:591–599. doi: 10.1534/genetics.114.163667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li MH, Yang HH, Li MR, Sun YL, Jiang XL, Xie QP, Wang TR, Shi HJ, Sun LN, Zhou LY, Wang DS. Antagonistic roles of Dmrt1 and Foxl2 in sex differentiation via estrogen production in tilapia as demonstrated by TALENs. Endocrinology. 2013;154(12):4814–4825. doi: 10.1210/en.2013-1451. [DOI] [PubMed] [Google Scholar]
- 56.Li MH, Wang DS. Gene editing nuclease and its application in tilapia. Sci Bull. 2017;62:165–173. doi: 10.1016/j.scib.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 57.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun YL, Jiang DN, Zeng S, Hu CJ, Ye K, Yang C, Yang SJ, Li MH, Wang DS. Screening and characterization of sex-linked DNA markers and marker assisted selection in the Nile tilapia (Oreochromis niloticus) Aquaculture. 2014;433:19–27. [Google Scholar]
- 59.Li M, Sun Y, Zhao J, Shi H, Zeng S, Ye K, Jiang D, Zhou LY, Sun L, Tao W, Nagahama Y, Kocher TD, Wang D. A tandem duplicate of anti-Müllerian hormone with a missense SNP on the Y chromosome is essential for male sex determination in Nile tilapia, Oreochromis niloticus. PLoS Genet. 2015;11(11):e1005678. doi: 10.1371/journal.pgen.1005678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang DS, Kobayashi T, Zhou LY, Paul-Prasanth B, Ijiri S, Sakai F, Okubo K, Morohashi K, Nagahama Y. Foxl2 up-regulates aromatase gene transcription in a female-specific manner by binding to the promoter as well as interacting with ad4 binding protein/steroidogenic factor 1. Mol Endocrinol. 2007;21:712–725. doi: 10.1210/me.2006-0248. [DOI] [PubMed] [Google Scholar]
- 61.Li M, Wu F, Gu Y, Wang T, Wang H, Yang S, Sun Y, Zhou L, Huang X, Jiao B, Cheng CH, Wang D. Insulin-like growth factor 3 regulates expression of genes encoding steroidogenic enzymes and key transcription factors in the Nile tilapia gonad. Biol Reprod. 2012;86:1–10. doi: 10.1095/biolreprod.111.096248. [DOI] [PubMed] [Google Scholar]
- 62.Qiu Y, Sun S, Charkraborty T, Wu L, Sun L, Wei J, Nagahama Y, Wang D, Zhou L. Figla favors ovarian differentiation by antagonizing spermatogenesis in a teleosts, Nile tilapia (Oreochromis niloticus) PLoS ONE. 2015;10(4):e0123900. doi: 10.1371/journal.pone.0123900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ijiri S, Kaneko H, Kobayashi T, Wang DS, Sakai F, Paul-Prasanth B, Nakamura M, Nagahama Y. Sexual dimorphic expression of genes in gonads during early differentiation of a teleost fish, the Nile tilapia Oreochromis niloticus. Biol Reprod. 2008;78(2):333–341. doi: 10.1095/biolreprod.107.064246. [DOI] [PubMed] [Google Scholar]
- 64.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−△△Ct method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 65.Han F, Wang Z, Wu F, Liu Z, Huang B, Wang D. Characterization, phylogeny, alternative splicing and expression of Sox30 gene. BMC Mol Biol. 2010;11:98. doi: 10.1186/1471-2199-11-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen J, Jiang D, Tan D, Fan Z, Wei Y, Li M, Wang D. Heterozygous mutation of eEF1A1b resulted in spermatogenesis arrest and infertility in male tilapia, Oreochromis niloticus. Sci Rep. 2017;7:43733. doi: 10.1038/srep43733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang X, Li M, Ma H, Liu X, Shi H, Li M, Wang D. Mutation of foxl2 or cyp19a1a results in female to male Sex reversal in XX Nile tilapia. Endocrinology. 2017;158(8):2634–2647. doi: 10.1210/en.2017-00127. [DOI] [PubMed] [Google Scholar]
- 68.Bosseboeuf A, Gautier A, Auvray P, Mazan S, Sourdaine P. Characterization of spermatogonial markers in the mature testis of the dogfish (Scyliorhinus canicula L.) Reproduction. 2013;147(1):125–139. doi: 10.1530/REP-13-0316. [DOI] [PubMed] [Google Scholar]
- 69.Ozaki Y, Saito K, Shinya M, Kawasaki T, Sakai N. Evaluation of Sycp3, Plzf and cyclin B3 expression and suitability as spermatogonia and spermatocyte markers in zebrafish. Gene Expr Patterns. 2011;11(5–6):309–315. doi: 10.1016/j.gep.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 70.Bellaiche J, Lareyre JJ, Cauty C, Yano A, Allemand I, Le Gac F. Spermatogonial stem cell quest: nanos2, marker of a subpopulation of undifferentiated A spermatogonia in trout testis. Biol Reprod. 2014;90(4):79. doi: 10.1095/biolreprod.113.116392. [DOI] [PubMed] [Google Scholar]
- 71.Thomas JL, Morgan GW, Dolinski KM, Thummel R. Characterization of the pleiotropic roles of Sonic Hedgehog during retinal regeneration in adult zebrafish. Exp Eye Res. 2018;166:106–115. doi: 10.1016/j.exer.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leal MC, de Waal PP, García-López A, Chen SX, Bogerd J, Schulz RW. Zebrafish primary testis tissue culture: an approach to study testis function ex vivo. Gen Comp Endocrinol. 2009;162:134–138. doi: 10.1016/j.ygcen.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 73.Baker J, Hardy MP, Zhou J, Bondy C, Lupu F, Bellvé AR, Efstratiadis A. Effects of an Igf1 gene null mutation on mouse reproduction. Mol Endocrinol. 1996;10(7):903–918. doi: 10.1210/mend.10.7.8813730. [DOI] [PubMed] [Google Scholar]
- 74.Pitetti JL, Calvel P, Zimmermann C, Conne B, Papaioannou MD, Aubry F, Cederroth CR, Urner F, Fumel B, Crausaz M, Docquier M, Herrera PL, Pralong F, Germond M, Guillou F, Jégou B, Nef S. An essential role for insulin and IGF1 receptors in regulating Sertoli cell proliferation, testis size, and FSH action in mice. Mol Endocrinol. 2013;27(5):814–827. doi: 10.1210/me.2012-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Morais RDVS, Crespo D, Nóbrega RH, Lemos MS, van de Kant HJG, de França LR, Male R, Bogerd J, Schulz RW. Antagonistic regulation of spermatogonial differentiation in zebrafish (Danio rerio) by Igf3 and Amh. Mol Cell Endocrinol. 2017;454:112–124. doi: 10.1016/j.mce.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 76.Zhang D, Xie D, Lin X, Ma L, Chen J, Zhang D, Wang Y, Duo S, Feng Y, Zheng C, Jiang B, Ning Y, Han C. The transcription factor SOX30 is a key regulator of mouse spermiogenesis. Development. 2018;145:11. doi: 10.1242/dev.164723. [DOI] [PubMed] [Google Scholar]
- 77.Schlueter PJ, Sang X, Duan C, Wood AW. Insulin-like growth factor receptor 1b is required for zebrafish primordial germ cell migration and survival. Dev Biol. 2007;305(1):377–387. doi: 10.1016/j.ydbio.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Baroiller JF, D'Cotta H, Shved N, Berishvili G, Toguyeni A, Fostier A, Eppler E, Reinecke M. Oestrogen and insulin-like growth factors during the reproduction and growth of the tilapia Oreochromis niloticus and their interactions. Gen Comp Endocrinol. 2014;205:142–150. doi: 10.1016/j.ygcen.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 79.Mitchell GC, Fillinger JL, Sittadjody S, Avila JL, Burd R, Limesand KH. IGF1 activates cell cycle arrest following irradiation by reducing binding of ΔNp63 to the p21 promoter. Cell Death Dis. 2010;1:e50. doi: 10.1038/cddis.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rubinfeld H, Kammer A, Cohen O, Gorshtein A, Cohen ZR, Hadani M, Werner H, Shimon I. IGF1 induces cell proliferation in human pituitary tumors—functional blockade of IGF1 receptor as a novel therapeutic approach in non-functioning tumors. Mol Cell Endocrinol. 2014;390(1–2):93–101. doi: 10.1016/j.mce.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 81.Crespo D, Assis LHC, van de Kant HJG, de Waard S, Safian D, Lemos MS, Bogerd J, Schulz RW. Endocrine and local signaling interact to regulate spermatogenesis in zebrafish: follicle-stimulating hormone, retinoic acid and androgens. Development. 2019;146:21. doi: 10.1242/dev.178665. [DOI] [PubMed] [Google Scholar]
- 82.Tong MH, Yang QE, Davis JC, Griswold MD. Retinol dehydrogenase 10 is indispensible for spermatogenesis in juvenile males. Proc Natl Acad Sci USA. 2013;110(2):543–548. doi: 10.1073/pnas.1214883110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yuan C, Chen K, Zhu Y, Yuan Y, Li M. Medaka igf1 identifies somatic cells and meiotic germ cells of both sexes. Gene. 2018;642:423–429. doi: 10.1016/j.gene.2017.11.037. [DOI] [PubMed] [Google Scholar]
- 84.Wu L, Yang P, Luo F, Wang D, Zhou L. R-spondin1 signaling pathway is required for both the ovarian and testicular development in a teleosts, Nile tilapia (Oreochromis niloticus) Gen Comp Endocrinol. 2016;230–231:177–185. doi: 10.1016/j.ygcen.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 85.Takase HM, Nusse R. Paracrine Wnt/β-catenin signaling mediates proliferation of undifferentiated spermatogonia in the adult mouse testis. Proc Natl Acad Sci USA. 2016;113(11):E1489–E1497. doi: 10.1073/pnas.1601461113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.de Waal PP, Leal MC, García-López A, Liarte S, de Jonge H, Hinfray N, Brion F, Schulz RW, Bogerd J. Oestrogen-induced androgen insufficiency results in a reduction of proliferation and differentiation of spermatogonia in the zebrafish testis. J Endocrinol. 2009;202(2):287–297. doi: 10.1677/JOE-09-0050. [DOI] [PubMed] [Google Scholar]
- 87.Cavaco JE, Vilrokx C, Trudeau VL, Schulz RW, Goos HJ. Sex steroids and the initiation of puberty in male African catfish (Clarias gariepinus) Am J Physiol. 1998;275(6):R1793–R1802. doi: 10.1152/ajpregu.1998.275.6.R1793. [DOI] [PubMed] [Google Scholar]
- 88.Yu G, Zhang D, Liu W, Wang J, Liu X, Zhou C, Gui J, Xiao W. Zebrafish androgen receptor is required for spermatogenesis and maintenance of ovarian function. Oncotarget. 2018;9(36):24320–24334. doi: 10.18632/oncotarget.24407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Berishvili G, Baroiller JF, Eppler E, Reinecke M. Insulin-like growth factor-3 (IGF-3) in male and female gonads of the tilapia: development and regulation of gene expression by growth hormone (GH) and 17β-ethinylestradiol (EE2) Gen Comp Endocrinol. 2010;167:128–134. doi: 10.1016/j.ygcen.2010.01.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.