Abstract
DNA damage response, DNA repair and genomic instability have been under study for their role in tumor initiation and progression for many years now. More recently, next-generation sequencing on cancer tissue from various patient cohorts have revealed mutations and epigenetic silencing of various genes encoding proteins with roles in these processes. These findings, together with the unequivocal role of DNA repair in therapeutic response, have fueled efforts toward the clinical exploitation of research findings. The successful example of PARP1/2 inhibitors has also supported these efforts and led to numerous preclinical and clinical trials with a large number of small molecules targeting various components involved in DNA repair singularly or in combination with other therapies. In this review, we focus on recent considerations related to DNA damage response and new DNA repair inhibition agents. We then discuss how immunotherapy can collaborate with these new drugs and how epigenetic drugs can rewire the activity of repair pathways and sensitize cancer cells to DNA repair inhibition therapies.
Keywords: Anticancer therapy, DNA repair inhibition, Synthetic lethality
Introduction
The DNA repair machinery has evolved to cope with the endogenous and exogenous insults against the DNA of the cell. Faithful DNA repair is vital for maintaining cell and tissue homeostasis, which is evident by the fact that loss-of-function mutations in DNA repair factors cause genetic disorders and increased susceptibility to cancer. A complex network of sensors, transducers and effectors, coordinates the repair of DNA damage and ensures DNA replication fidelity.
Sensor proteins detect all types of DNA structural alterations, including nicks, gaps, double-strand breaks (DSBs), and replication lesions. Signal transducers are enzymes that control the activity of the cell cycle checkpoints and DNA repair pathways initiating signaling cascades to adjacent nucleoprotein structures. Effectors repair DNA damage and block progression through the cell cycle.
Defects in DNA repair pathways facilitate the accumulation of genomic alterations that contribute to their proliferation and survival of cancer cells. Nonetheless, tumors rely on residual DNA repair capacities to repair the damage induced by replication and genotoxic stress. Mammalian cells employ at least nine distinct pathways to repair a multitude of different genotoxic lesions: mismatch repair (MMR), base excision repair (BER), nucleotide excision repair (NER), translesion synthesis (TLS), homologous recombination (HR), non-homologous end joining (NHEJ), alternative end joining (alt-EJ), the Fanconi anemia (FA) and the O6-methylguanine DNA methyltransferase (MGMT). However, these pathways are neither completely independent of one another nor mutually exclusive processes handling different types of lesions in distinct cell cycle phases. Instead they form a precisely regulated network of multifunctional DNA repair hubs, which are involved in multiple DNA repair pathways. Multiple components of the repair network are deficient in cancer cells due to inactivating mutations or transcriptional silencing, affecting the functionality of different repair hubs and, therefore, the overall capacity for repair in conventional chemotherapeutic treatment.
Next-generation sequencing has uncovered the frequency of mutations and copy number alterations across different cancer types and demonstrated that alterations of DNA repair mechanisms are common events in carcinogenesis. However, cancer types show different preferences for inactivation of specific DNA repair processes. For instance, mutations in HR genes appear to be enriched in breast and ovarian cancer as well as in bladder cancer, cutaneous skin melanoma and chronic lymphocytic leukemia. On the other hand, certain subgroups of stomach and colorectal adenocarcinoma as well as uterine endometrial carcinoma that harbor alterations in MMR present a hypermutator phenotype with low aneuploidy. Figure 1 presents examples of genomic and transcriptomic data of four tumor types showing distinct profiles in DNA repair components involving HR and MMR [1]. Other cancer types are characterized by enrichment of DNA repair deficiencies in more than one pathway. For example, prostate cancer accumulates inactivating mutations within the HR and NER pathways. Similarly, epigenetic deregulation and promoter DNA methylation cause transcriptomic silencing in different components of repair hubs across different cancer types. With increasing understanding of the mutational landscape and epigenetic/transcriptional profile of cancer genomes and the emerging role of DNA repair in tumorigenesis, there has been a shift toward the development of drugs that target specific components of the repair machinery. Cancer cells that harbor repair deficiencies in one DNA repair pathway/component often become hyperdependent upon remaining repair pathways for survival and proliferation. Therefore, knowing the profile of deregulated components of DNA repair networks can help us target specifically the compensatory DNA repair pathways in cancer cells. Next-generation sequencing that permits whole-genome mutational analysis brings forth the dawn of precision medicine, offering the opportunity for personalized treatment strategies based on the deficiencies of patient’s DNA repair networks (Fig. 2).
Fig. 1.
Genomic and transcriptomic meta-analysis of different solid tumors. TCGA (Pancancer Atlas) mutation, expression and copy number alterations were processed using the cbioportal.org platform
Fig. 2.

Diagnostic profiling of defects associated with DNA repair in tumors. a Tools used for the molecular and histological characterization of tumor cells. b Known genetic alterations associated with compromised genome integrity in cancer cells
DNA damage response (DDR)
DDR involves recognition of double-strand breaks (DSBs) or single-strand breaks (SSBs), followed by initiation of an extended network of signaling cascades to promote DNA repair. These signal cascades can also activate cell cycle checkpoint arrest and apoptosis, and influence the aspects of DNA repair [2, 3]. These two aspects of DDR, DSB repair and checkpoint arrest cooperate to protect genomic integrity and their concomitant loss has a critical impact at the early steps of tumorigenesis. Defective DNA repair increases the mutational load and the genomic instability, whereas malfunctioning cell cycle checkpoints allow cells with DNA damage to proliferate.
DNA-PKs, ATM, ATR: initiation and activation of DDR
Like many intra-cellular signaling cascades, DNA damage signaling is driven by protein phosphorylation. Three kinases, ATM, ATR and DNA-PKs, have a principal role in activating DDR. These large proteins share similarities in domain organization and structure. Their kinase activity is located in the C-terminus and is very similar to that of the phospholipid kinase PI3K, with a specific preference to serine or threonine residue (Ser/Thr) phosphorylation [4–7]. ATM, ATR, and DNA-PKcs must be tightly regulated to prevent aberrant activation. Each kinase requires a specific protein co-factor for stable recruitment to DNA damage sites. DNA-PKc is recruited and activated at DSB ends by a Ku80-Ku70 heterodimer which plays a central role in the non-homologous end-joining (NHEJ) DSB repair pathway ([8, 9]. ATM is activated and recruited to DSBs by the MRE11-RAD50-NBS1 (MRN) complex [10]. ATR is recruited to replication protein A (RPA)-coated single strand DNA (ssDNA) by its stable binding partner ATRIP [11].
Loss of DDR components is an early event in carcinogenesis
In pre-cancerous cells, deregulation of cell cycle control and increased replication stress induce stalling and collapse of DNA replication forks, which in turn leads to DSB formation. This continuous formation of DSBs activates TP53 and DDR components such as ATM and ATR. While this activation represents a barrier to tumor progression, the loss of one or more DDR pathways and TP53 by mutations in early steps of carcinogenesis removes this barrier [12]. This is indeed supported by mutational data across major types of solid tumors. TP53, ATM and ATR are highly mutated in bladder cancer (50%, 11.2% and 4.1%), lung adenocarcinoma (51.8%, 7.9% and 5.7%) and colorectal adenocarcinoma (58.6%, 5.7% and 2.1%, respectively) [13]. Defects in ATM and ATR genes significantly contribute to cancer initiation and progression via accrual of driver mutations and generation of tumor heterogeneity on a background of genomic instability.
The concept of synthetic lethality
As mentioned above, inactivating mutations in DNA repair components are common and often lead to certain DNA repair deficiencies. Cancer cells thus become hyperdependent on remaining repair pathways for survival and proliferation. Inhibiting the rescue pathway with a specific chemical agent can provide a context for synthetic lethality [14, 15]. Synthetic lethality, a concept first described in the 1920s [16–18], refers to the event where two viable gene mutations lead to cell death when they co-occur. This concept is particularly compelling for cancer therapy, because it allows specific targeting of cancer cells that carry a gene mutation that is not found in normal cells which are therefore spared.
Targeting PARP1
Targeting BRCA1/2-deficient cancers using poly (ADP-ribose) polymerase (PARP) inhibitors (PARPi) is a model example of synthetic lethality [19]. Rucaprib (Clovis), Olaparib (AstraZeneca) and Niraparib (Tesaro Inc.) are PARP inhibitors that have been FDA approved for the maintenance treatment of recurrent gBRCAm epithelial ovarian, fallopian tube and primary peritoneal cancer (henceforth referred to as ovarian cancer) which are in a complete or partial response to platinum-based chemotherapy. Importantly, these inhibitors are the first clinically approved drugs to exploit a synthetically lethal interaction in cancer therapy. More recently, Olaparib and a new PARP inhibitor, Talazoparib, were approved for the treatment of gBRCAm recurrent breast cancer (OlympiAD; EMBRACA) and Olaparib was approved as first-line maintenance treatment in ovarian cancer patients that respond to platinum-based chemotherapy (SOLO-1).
Accumulated evidence indicates that PARP1 functions as an important DNA damage sensor protein that recognizes both SSBs and DSBs and catalyzes the formation of linear chains of ADP-ribose residues (PAR chains). The PAR chains form a platform to recruit DNA repair proteins via their PAR-binding domains at the sites of DNA damage [20, 21]. Therefore, it is considered that PARylation can prime the activation of the DNA repair cascades via recruitment of DNA damage response factors to the region near DNA lesions. Upon inhibition, the PARP enzymes which bind to DNA lesions are unable to form signaling scaffolds and dissociate from the DNA [22–24]. These trapped PARP molecules add up to the existing lesions and increase the DNA damage burden of the cell [19, 25].
Homologous recombination (HR)-deficient cells, such as cells with BRCA1/2 mutations, are overly reliant (or “addicted”) to other pathways for DNA repair, such as the NHEJ and alt-EJ [26]. The alternative modes of end joining require PARP1 to take place [27]. Therefore, when PARPs are inhibited in HR-deficient cells, there are more DNA lesions, compromised signaling activity and inability to utilize a vital DNA repair pathway. Importantly, PARPi has clinical benefit in non-BRCAm ovarian cancer patients which can be attributed to either unidentified HR deficiency, such as mutations in other HR genes, or to PARP1 actions which are independent of the alt-EJ.
Ongoing clinical trials using PARP1 inhibitors
Ongoing clinical trials examine the efficacy of licensed PARP inhibitors in various solid and hematopoietic malignancies and in combination with DNA damaging agents, and DDR, cell cycle and immune checkpoint inhibitors. The therapeutic strategies, for the most part, fall into one of the two main categories: exploitation of DNA repair defects in tumors using PARPi monotherapy or augmentation of the activity of other agents by combining them with PARPi to treat patients regardless of HR status.
The first category includes trials that use HR deficiency as a stratification biomarker for PARPi therapy through mutations in proteins directly associated with HR such as BRCA1/2 (NCT03344965 and NCT02952534), or other biomarkers suggestive of HR deficiency such as IDH mutations (NCT03561870) [28]. Among the most promising trials are POLO, in which Olaparib treatment shows benefit in BRCAm pancreatic cancer patients who responded to platinum-based chemotherapy, and TRITON2, in which preliminary results show that rucaparib monotherapy benefits BRCAm recurrent prostate cancer patients and is currently at phase III.
The second category of trials examines the efficacy of PARPi as part of a main course of combination treatment (Fig. 3). Traditionally, PARPi drug combinations have focused on DNA damaging agents and this is why the majority of combinations in current trials are with crosslinking agents, topoisomerase inhibitors and irradiation. Additionally, a number of trials aim to determine synergy of PARP inhibitors with other emerging therapeutic agents such as immune checkpoint inhibitors and angiogenesis inhibitors. The potential of immunotherapy to synergize with PARPi was demonstrated in recent literature on genomic instability and immune response [29]. Ongoing trials include ATHENA and FIRST in ovarian cancer, and KEYLYNK-010 in prostate cancer patients. On the other hand, synergism with angiogenesis inhibitors is based on their negative effect on HR, which in turn sensitizes cells to PARPi [30, 31].
Fig. 3.

Strategies for the combinatorial use of DDR inhibitors with known anticancer therapies
A large number of trials examine novel PARP inhibitors in monotherapy as well as in combination with other agents. Veliparib has currently reached phase III in multiple trials (NCT02470585, NCT02163694 and NCT02264990) in combination with paclitaxel/carboplatin for the treatment of recurrent ovarian, breast and lung cancer patients. Notably, another promising PARP inhibitor, Iniparib, which in combination with gemcitabine/carboplatin reached phase III trials in breast and lung cancer (NCT00938652, NCT01082549), was reported to have significant off-target effects which halted further testing [32].
DDR network and exploitation of synthetically lethal interactions with PARP
Due to extensive crosstalk between pathways involved in DNA damage sensing, repair and cell cycle checkpoint activation pathways, therefore, are ideal candidates for exploitation of synthetically lethal interactions. To this direction, small molecule inhibitors have been designed against ATM, ATR and DNA-PKs and tested in phase I or II clinical trials either as single agents or in combination with chemotherapy or radiotherapy [33, 34].
After DNA damage, sensor proteins such as PARP1, H2AX and sensor complexes including the Ku70/80 and MRN (MRE11/RAD50/NBS1) directly recognize the structure of DSBs and SSBs and recruit ATM, ATR and DNA-PK proteins at the break sites. Like PARP1, the Ku complex, consisting of Ku70 and Ku80 subunits, functions as a DNA sensor protein, binding DSBs after DNA damage. However, PARP1 and Ku complexes mediate DSB repair pathway choice independently and through distinct mechanisms. Binding of the Ku complex to DSBs recruits and activates the DNA-PK catalytic subunit, which facilitates NHEJ [35], whereas the binding of PARP1 promotes alt-EJ [36]. Thus, the antagonism between PARP1 and the Ku complex at DSBs may play an important role in determining repair choices. In addition to PARP1 and Ku proteins, the MRN complex acting as a DNA sensor protein has the ability to bind DNA ends, recruiting and activating ATM [37]. ATM is the main kinase responsible for phosphorylation of H2AX, which is one of the earliest steps for the recruitment of additional DDR factors [38, 39].
ATM-deficient cells exhibit enhanced sensitivity to PARPi
As mentioned above, the rationale for using PARP inhibitors to treat HR-deficient cancers in the clinic is based on the high sensitivity of BRCA1- and BRCA2-defective cells to small molecule PARP inhibitors. As such, ATM inactivation, a known cause of HR deficiency [40], has been also shown to cause sensitivity to PARP inhibition [41]. Recent preclinical studies have focused on the functional interconnection between ATM and PARP1, which also appears to contribute to enhanced PARPi sensitivity [42]. Consistent with these findings, although knockout of PARP-1 and ATM individual does not lead to embryonic lethality, double-null mice die early in embryogenesis [43]. Genome-wide studies have revealed that in patients with metastatic castration-resistant prostate cancer (mCRPC), somatic ATM alterations are detected at a high frequency (5–10%) [44]. In a 49-patient phase II clinical trial, treatment with the PARP inhibitor olaparib of patients whose prostate cancers were no longer responding to standard treatments and who had defects in DNA repair genes (BRCA1/2, ATM, Fanconi’s anemia genes, and CHEK2) led to a high response rate [45]. However, preliminary results from another clinical trial (TRITON2) indicate that mCRPC patients harboring ATM and CDK12 mutations seem to benefit less from the PARP1 inhibitor rucaparib compared to patients with BRCA1/2 loss [46].
Pairing ATR and PARP inhibitors
The partial responses to PARP inhibitors, even in BRCA1/2-mutant tumors, indicate the existence of intrinsic or acquired resistance mechanisms [47]. Such resistance mechanisms may include secondary mutations or promoter demethylation in the BRCA1/2 genes which partially restore HR activity in cancer cells. Other resistance mechanisms include point mutations in PARP1 and drug-efflux pumps affecting drug pharmacokinetics. To overcome such resistance mechanisms as well as to treat non-HR-deficient tumors, the pairing of PARP inhibitors with other DDR pathway inhibitors, such as ATR inhibitors, is ongoing.
Approximately, 50% of high-grade serous ovarian cancer (HGSOC) has defects in genes involved in HR repair [48]. Olaparib monotherapy has modest clinical benefit in recurrent BRCA1/2-mutant HGSOCs, as it results in a 40% response rate, following first-line carboplatin–taxane chemotherapy [49]. Recent studies, however, have shown that PARPi increases the dependence on ATR activity for fork stabilization, and combination of PARPi with ATR blockade is more effective than PARPi alone BRCA-mutant ovarian cancer [50, 51]. Larger prospective studies are clearly needed to confirm or refute these preliminary findings.
Synthetic lethality beyond PARP1
There is a functional crosstalk between ATM, ATR and DNA-PKs in response to DNA damage.
Whereas ATM is primarily activated in response to DSBs, ATR is mainly activated by SSBs, and for this reason older models of DNA repair placed ATM and ATR in separate and distinct repair pathways. The current model proposes that both kinases cooperate for DSB repair during ionizing radiation (IR) or genotoxic stress. Recent studies have demonstrated that ATM and the nuclease activity of Mre11 are both required for the processing of DSBs to generate the RPA-coated ssDNA that is needed for ATR recruitment and Chk1 activation at S and G2 cell cycle phase [52–54].
ATM-mediated phosphorylation of DNA-PKs at Thr2609 is critical for DNA-PKs function in DNA repair and represents another example of functional crosstalk between DNA damage response proteins [55]. It has also been proposed that ATM phosphorylation at Ser1981 can be driven by ATR following replication fork stalling or UV treatment [56]. The survival of tumor cells with impaired ATM or ATR function after DNA damage is compromised and, therefore, the crosstalk between ATM, ATR and DNA-PKs can offer a chance for synthetic lethality when one of these kinases is inactivated (by mutation, deletion or transcriptional repression) in cancer cells.
Targeting the ATM deficiency with ATR inhibitors is an emerging antitumor strategy based on the high frequency of ATM dysregulation in cancer. Somatic mutations in the ATM gene are found in many solid tumors (breast, ovarian, colorectal, and prostate) [57–59]. Regarding hematological malignancies, inactivating mutations of ATM are present in about half the patients with mantle cell lymphoma and T cell prolymphocytic leukemia [60, 61]. There is now significant preclinical evidence supporting that ATM-deficient tumors are sensitive to ATR inhibitors and, therefore, ATM has a synthetically lethal relationship with ATR in chronic lymphocytic leukemia [62], mantle cell lymphoma [63] and mammary cancer cells [64]. Importantly, ATR has also been shown to mediate replication fork stability [65]. This broad role of ATR in many aspects of genome integrity is likely one of the reasons for the success of its inhibitors in the clinic, much like PARPi. The putative clinical significance of inhibiting DNA repair proteins is highlighted by the number of ongoing clinical trials with the use of small molecule inhibitors in monotherapy settings, the majority of which tumors with known DNA repair deficiencies are selected for treatment (Table 1).
Table 1.
DDR inhibitors in monotherapy clinical trials
| Agents | Target | Selected clinical trials | Disease | Selection factors | therapeutic regimen | Phase |
|---|---|---|---|---|---|---|
| FDA approved | ||||||
| Olaparib | PARP1/2 | NCT02184195 (POLO) | Metastatic pancreatic cancer | Platinum sensitivity; BRCAma | Single agent; maintenance | III |
| NCT03786796 (ORCHID) | Metastatic renal cell carcinoma | HRDb | Single agent | II | ||
| NCT03561870 (OLAGLI); PARADIGM-2 | High-grade glioma | IDHmc | Combination with low-dose chemotherapy | II | ||
| Rucaparib | PARPi | NCT02975934 (TRITON3) | Metastatic castration-resistant prostate cancer (mCRPC) | HRD | Single agent | III |
| NCT02505048 (RUBY) | Metastatic breast cancer | HRD | Single agent | III | ||
| NCT03140670 | Pancreatic adenocarcinoma | Platinum sensitivity; HRD | Single agent | II | ||
| Niraparib | PARP1/2 | NCT02655016 | Advanced ovarian cancer | Platinum sensitivity | Single agent; maintenance; 1st line | III |
| NCT03709316 | Advanced ovarian cancer | Platinum sensitivity | Single agent; maintenance; 1st line | III | ||
| NCT03601923 | Advanced pancreatic cancer | Platinum sensitivity; HRD | Single agent | II | ||
| NCT03925350 | Metastatic melanoma | HRD | Single agent | II | ||
| Talazoparib | PARP1/2 | NCT01945775 (EMBRACA) | Advanced breast cancer | Platinum sensitivity; BRCAm | Single agent | III |
| NCT02034916 (ABRAZO) | Advanced breast cancer | Platinum sensitivity; BRCAm | Single agent | II | ||
| Under evaluation | ||||||
| Veliparib | PARP1/2 | NCT02163694 | Her2-negative advanced breast cancer | BRCAm | Combination with chemotherapy; 1st-2nd line | III |
| AZD6738 | ATR | NCT03878095 | Solid tumors | IDHm | Combination with olaparib | II |
| SRA737 | CHK1 | NCT02797977 | Solid tumors | DNA repair deficiency | Combination with low-dose chemotherapy | I/II |
| LY2606368 | CHK1/2 | NCT02203513 | High-grade serous ovarian cancer (HGSOC), fallopian or primary peritoneal cancer or triple-negative breast cancer (TNBC) | BRCAm | Single agent | II |
| NCT02873975 | Solid tumors | HRD or replication stress | Single agent | II | ||
aBRCA mutated
bHomologous recombination deficient
cIDH mutated
Recent studies based on CRISPR/Cas9-mediated knockout and RNAi screens revealed that RecQ DNA helicase WRN is selectively essential in MSI models. Inactivation of WRN in microsatellite instable cells leads to lethality in vitro and in vivo, while it has no effect in microsatellite-stable cells, highlighting a new example of previously undocumented synthetically lethal interaction between MMR and the replication machinery [66, 67]. WRN is a multifunctional enzyme for genome integrity as it is involved in resolving complex DNA structures at replication forks or as a result of HR recombination. Such DNA structures are very common in MMR-deficient cells explaining the dependence of these cells to WRN activity [68].
DDR, genotoxic stress and cell cycle progression
Radiotherapy and chemotherapy with DNA-crosslinking agents that damage DNA are commonly used to treat cancer. Cisplatin, carboplatin, oxaliplatin and other similar platinum-based drugs make up a broad class of DNA-crosslinking agents that target rapidly dividing cancer cells by forming inter-strand crosslinks (ICLs) and disrupting DNA replication. IR used in radiotherapy causes multiple types of DNA damage, including DSBs and SSBs. IR also causes the formation of reactive oxygen species (ROS) which, in turn, promote the production of oxidized nucleotide adducts, such as 8-oxoguanine [69].
DDR defects in cancer can modulate the effectiveness of chemotherapy and radiotherapy
Chemotherapy and radiotherapy rely on the induction of DNA damage which is more cytotoxic for proliferating cells. Unlike normal cells, cancer cells are already burdened by genomic instability, replication stress and, by frequently malfunctioning DNA repair pathways, defects that cause genome-wide DNA damage. Because the apoptosis safety check is often inactivated, cancer cells need to repair any DNA damage to continue to proliferate. DNA damaging by chemotherapy/radiotherapy exploits this principle and this is why they have been the cornerstone of first-line therapeutic schemes for many unresectable or metastatic malignancies for decades now. Although dysregulation of the DNA damage response is associated with cancer initiation and progression, it can also result in hypersensitivity or resistance of tumors to genotoxic cancer therapy.
For instance, ovarian cancer patients with hereditary mutations in BRCA1 or BRCA2 genes that impair HR activity are more sensitive to platinum agents [70]. Notably, the appearance of compensatory mutations in BRCA1 and BRCA2 that restore HR functionality in initially cisplatin sensitive tumors is able to develop cisplatin resistance [71]. In bladder cancer, patients with impaired NER pathway due to somatic ERCC2 mutations or low ERCC1 expression are more sensitive to platinum agents [72]. Furthermore, genomic alterations in ATM and FANCC genes predict response and clinical benefit after cisplatin-based chemotherapy for muscle invasive bladder cancer [73]. NSLC patients with ERCC1-negative tumors appear to benefit from postoperative cisplatin-based chemotherapy, whereas patients with ERCC1-positive tumors do not [74]. Similarly, patients with ERCC1-negative locally advanced esophageal cancers benefit from preoperative chemoradiotherapy [75]. On the contrary, high ERCC1 expression has been positively correlated with cisplatin resistance in several human neoplasms including bladder [76], colorectal [77], gastric [78], head and neck [79–81] and ovarian cancers [82]. MMR deficiency due to mutations or downregulation of MSH2, MSH6 and MLH1 proteins has also been correlated with acquired cisplatin resistance. Functional MMR proteins recognize cisplatin adducts on DNA [83] and initiate proapoptotic signals [84] or generate gaps and strand breaks that lead to cell death [85]. Therefore, loss of MMR rescues cancer cells from apoptosis or cell death upon cisplatin treatment. Consistent with these observations, methylation-dependent silencing of MLH1 has been shown to predict poor survival in ovarian cancer patients [86]. In addition, patients with stages II and III MMR-deficient colon cancer do not benefit from fluorouracil-based adjuvant therapy [87].
Interestingly, defects in MLH1 and MSH6 are associated with increased level of translesion synthesis, which efficiently permits DNA synthesis in the presence of cisplatin adducts [88]. Replicative bypass of adducts by translesion synthesis is mediated by the coordinated activity of a specific group of DNA polymerases including POLH, POLK, and POLZ [89]. The activity of POLH and POLZ has a pivotal role on the replicative bypass of GpG adducts and is associated with cisplatin resistance [89, 90]. Defects in POLH and REV3L subunit of POLZ have been linked to increased sensitivity to cisplatin in multiple tumor cell lines [91], whereas POLH expression levels predict the survival of NSLC patients treated with platinum-based chemotherapy [92] and of metastatic gastric adenocarcinoma patients treated with oxaliplatin-based chemotherapy [93].
Despite the fact that platinum-containing chemotherapy and radiotherapy have been used in clinic for the treatment of a wide variety of solid tumors for many decades, intrinsic or acquired resistance during treatment cycles and high toxicity in patients are major obstacles that severely limit the clinical benefit.
Targeting of DDR components modulates cell cycle checkpoint activity
In the context of their role in genome integrity, ATM, ATR and DNA-PKs facilitate communication between damage recognition proteins and the cell cycle checkpoints (G1/S, intra-S and G2/M) to temporarily arrest the cell cycle and increase the opportunity for DNA repair before moving to the next stage of cell cycle. In cases of extensive DNA damage, cells permanently exit the cell cycle (senescence) or undergo programmed cell death (apoptosis) [94].
The G1/S checkpoint is believed to be controlled primarily by ATM rather than ATR. By contrast, both ATM and ATR contribute to the establishment and maintenance of the intra-S and G2/M checkpoints. The G1/S checkpoint allows the repair of DNA damage prior to the start of DNA replication, whereas the intra-S phase checkpoint can delay replication origin firing, providing time for DNA repair [95]. The G2/M checkpoint represents the last major barrier that prevents DNA damage from being transferred into mitosis which would lead to mitotic catastrophe and cell death [96].
Whereas ATM signals DSBs through the Chk2 checkpoint kinase, activated ATR in SSBs or in replication-linked DSBs signals through the Chk1 checkpoint kinase. In response to DSBs, the MRN complex recruits ATM to DSB sites. Once activated, the ATM/Chk2 pathway phosphorylates Cdc25A and p53 simultaneously within minutes promoting p53 stabilization, p21 upregulation, S-phase cell cycle arrest and activation of the p53-associated G1/S-phase checkpoint [97]. DSBs in the G2 can directly activate ATM, and indirectly, via ATM-dependent strand resection, ATR [54]. The G2/M checkpoint is initiated by the ATM/ATR-driven phosphorylation of CHK1 and CHK2 checkpoint kinases. In turn, these kinases mediate by phosphorylation (Ser216) the inhibition of Cdc25C. This prevents dephosphorylation of CDK1–cyclin B, which is required for progression into mitosis [98].
In response to SSBs at sites of DNA damage or stressed replication forks, RPA-coated ssDNA activates ATR and its binding partner ATRIP [11, 99, 100]. The kinase CHK1 is the key downstream regulator of the ATR response and is phosphorylated by ATR on Ser317 and Ser345 [65]. Activated CHK1 promotes by phosphorylation the proteasomal degradation of CDC25A and CDC25C which leads to a decrease in the CDK activity in S and G2/M cell cycle phase, thereby causing activation of intra-S and G2/M-phase checkpoints [101].
Chemical inhibition of ATM ATR, CHK1, CHK2 and Wee1 is a promising and attractive avenue for anticancer therapy because inhibition of these proteins can lead to cell cycle checkpoint abrogation (S and G2) in cancer cells. Checkpoint abrogation under genotoxic stress is associated with cell death due to excessive unrepaired DNA damage that is accumulated by rapid and unregulated cell cycle progression. In this direction, potent inhibitors of ATR, ATM, CHK1, CHK2, and WEE1 are under clinical evaluation. This family of compounds has been poorly tolerated in early monotherapy clinical trials [102, 103].
Combinatorial targeting of DDR and the G2/M checkpoint
Unrepaired DNA damage can be resolved before entering mitosis through activation of the G2 cell-cycle checkpoint. Abrogation of the G2 checkpoint allows cells with unrepaired DNA damage to enter into premature mitosis resulting in mitotic catastrophe [104–106]. A strong rationale exists for combined therapy with ATR and WEE1 inhibitors. Given that ATR inhibitors promote origin firing, replication stress and DSB generation [107–111], the parallel WEE1 inhibition can abrogate the G2 arrest in these cells allowing chromatin with unrepaired DNA damage to enter into mitosis and undergo mitotic catastrophe [112, 113].
Similarly to ATRi, PARPi also induces replication stress and DNA damage [114]. Combined WEE1 and PARP inhibition has demonstrated antitumor activity in a number of preclinical models [115, 116]. However, overlapping WEE1i and PARPi toxicity profiles hinders the development of combinations, which has been largely confirmed in early clinical trials [117]. Interestingly, the sequential administration of PARP and WEE1 inhibitors seems to maintain efficacy while ameliorating toxicity [118].
DNA repair deficiencies and immunotherapy
Advances in immunotherapy have changed the treatment landscape in many cancers. Immune checkpoint inhibitors such as anti-CTLA4 and anti-PD1/PD-L1 antibodies have demonstrated successful clinical effect in a wide range of cancers. The anti-PD-1 monoclonal antibodies, nivolumab, atezolizumab and pembrolizumab, have been shown compared to standard systemic chemotherapy, to improve overall survival in head and neck cancer, advanced melanoma, non-small cell lung cancer, and urothelial cancer [119–122]. The development of predictive biomarkers is needed to optimize patient benefit, minimize risk of toxicities and guide combination strategies. Significant therapeutic responses have recently been observed in patients presenting tumors with high mutational burden that produce substantial levels of neoantigens [123–125]. Previous studies have shown that tumor mutation burden (TMB) is associated with increased T cell cytolytic activity supporting the notion that neoantigens can drive cytotoxic T cell responses [126].
Tumor mutation burden correlates with predicted neoantigens and immune infiltration
DNA repair safeguards genomic stability and the functional loss of components involved in this process can lead to high mutational burden due to the accumulation of DNA damage. Therefore, DNA damage defects are associated with acquired somatic mutations resulting in generation of neoantigens. Neoantigens are able to trigger the activation of cytotoxic T-cells [127] and make cancer cells more immunogenic. Since the cellular response to DNA damage determines the mutational load of cancerous cells, it has become clear that DNA damage and DNA repair activity have a major impact on the interaction between the tumor and the immune system and, furthermore, that the DNA damage and repair landscape have important therapeutic implications in the context of immunotherapy. A schematic representation of the crosstalk between DNA repair and immunological response is shown in Fig. 4.
Fig. 4.
Therapeutic targeting of the DNA repair–immune response interconnection in cancer. Approved and under evaluation anticancer agents targeting distinct components of the DNA repair machinery as well as modulators of the immunological response
Previous studies have reported that specific defects in DNA repair proteins could be potential predictive biomarkers of clinical response to immune checkpoint inhibitors in a wide spectrum of tumors. The most robust current evidence supporting the association between DNA repair deficiency, the TMB and the efficiency of immune checkpoint blockade (ICB) comes from tumors with loss of MMR function. Tumors with MMR deficiency (dMMR) have high response rates to ICB, and the FDA recently approved pembroluzimab for treatment of microsatellite instability-high (MSI-H) colorectal tumors [128]. In microsatellite-unstable endometrial cancer, due to mutations in the exonuclease (‘proofreading’) domain of polymerase DNA polymerase ε (POLE), there is increased mutation burden and, as a result, a higher number of cytotoxic T tumor-infiltrating lymphocytes (TILs), compared with microsatellite-stable tumors [129]. Clinical and immunological response to immune checkpoint inhibition with pembrolizumab has been also demonstrated in hypermutated glioblastoma with POLE mutations [130].
Several reports have also linked somatic mutations (BRCA1/2, RAD51, ATM, ATR, PTEN) leading to HR repair deficiency with higher neoantigen levels and ICB response [131–134]. Similarly, in urothelial cancer, tumors harboring alterations in DNA damage response genes ATM, POLE, FANCA, ERCC2, and MSH6 were correlated with high TMB and improved clinical outcomes to ICB [135].
In human cancers, a deficiency in a DNA repair pathway can result in dependence on a compensatory DNA repair pathway [136]. HR-deficient tumors are hyperdependent on polymerase θ (POLQ)-mediated repair. POLQ appears to channel DNA repair by antagonizing HR and promoting PARP1-dependent alt-EJ repair [137]. POLQ and alternative end-joining activity have been described to be intrinsically mutagenic [138, 139], explaining the high neoantigen levels of HR-deficient tumors.
BRCA1/2-mutated ovarian tumors have been associated with higher neoantigen loads, higher T cell infiltration and improved overall survival compared to HR-proficient tumors [134]. Patients with BRCA2 mutated melanoma were found to have better response to anti-PD-1 treatment [132]. Furthermore, microsatellite-unstable tumors harboring mutations in MMR genes are characterized by TMB and enhanced response to immune therapy [124, 140].
DNA repair defects that generate intratumoral heterogeneity may confer to immune escape
An important aspect of defective DNA repair and cell cycle checkpoints is persistence of unrepaired DSBs during M phase, which lead to mis-segregation of chromosomes and chromosomal instability (CIN) [141–143]. CIN is characterized by an increased rate of gains and losses of fragments or whole chromosomes, leading to aneuploidy, and is a principal driver of tumor heterogeneity [144, 145]. Heterogeneous tumors contain multiple subclones and under selection pressure, such as chemotherapy or immunotherapy, subclones with either intrinsic or acquired resistance can survive the pressure and potentially drive disease progression.
Although ICB induces significant responses in many patients, response rates vary significantly both within and across tumor types. The use of next-generation sequencing and single-cell sequencing from different sites within the same tumor (multi-region analysis) has revealed a high degree of genetic heterogeneity within the same tumor (intratumoral heterogeneity) [146–149]. Comparative analysis of tumor subclones between the primary tumor and distant metastases from the same patient has also revealed differences, arising either from evolution of subclones that were either present at the primary tumor or through emergence of subclones that represent new branching points during the evolutionary process and the Darwinian selection. DNA repair defects and epigenetic alterations in tumor progression have a crucial role in sub-clonal evolution.
Due to intratumoral heterogeneity, PD-L1 expression levels may vary among different tumor subpopulations and, therefore, clones with low PD-L1 expression can drive immune escape. In clinic, a substantial number of patients with PD-L1 positivity (at least 40–50%) do not achieve objective response to anti-PD-1/PD-L1 therapies [150].
Oncogenic mutations may also drive immunotherapy resistance in specific subclones. Activation of the canonical WNT-β-catenin signaling pathway in melanoma cells has been shown to correlate with suppression of the chemokine CCL4, reduced T cell infiltration and resistance to anti-PD-L1 and anti-CTLA-4 mAb-based therapy [151]. Oncogenic activation of the PI3-kinase pathway due to PTEN loss in melanoma cancer cells can also promote resistance to immune therapy [152]. Similarly, activation of the epidermal growth factor receptor (EGFR) pathway has been associated with immunosuppression in lung adenocarcinoma [153].
There is growing evidence that intratumoral neoantigen heterogeneity is associated with low response to ICB therapy. Recent studies in non-small cell lung cancer (NSCLC) have demonstrated that tumors with high levels of clonal neoantigens have improved responses to ICB and that the loss of clonal neoantigens due to elimination of tumor subclones or through deletion of chromosomal regions can lead to ICB resistance [154, 155]. In addition, genomic instability can lead to evolution of immunotherapy resistant subclones that harbor somatic copy number loss of neoantigens that are critical to immune response [156]. An analysis of more than 5000 TCGA tumors across 12 tumor types demonstrated that high levels of somatic copy number alterations (SCNAs) are associated with reduced expression of cytotoxic immune cell markers [157].
DNA repair targeting agents and immunotherapy
Previous reports have shown that DNA damage that arises from genotoxic stress can activate the STING pathway, an innate immune pathway activated by cytoplasmic DNA [158]. Cyclic guanosine monophosphate (GMP)-AMP synthase (cGAS) is a DNA sensor that triggers innate immune responses through production of the second messenger cyclic GMP-AMP (cGAMP), which binds and activates the adaptor protein STING. Activated STING triggers phosphorylation and nuclear translocation of IRF3 which in turn activates transcription of inflammatory genes [159, 160]. cGAS is considered to detect DNA damage derived by the formation of micronuclei during mitotic progression and leakage of DNA into the cytosol [161]. Activation of the cGAS/STING pathway leads to expression of interferons and chemokines and has been shown to attract and activate immune cells in the tumor microenvironment [162, 163].
Recently, Dunphy et al. showed that detection of genotoxic stress-induced DNA damage by ATM and PARP1 can activate the DNA sensor adaptor STING independently of the cytosolic DNA receptor cGAS and cGAMP production through a non-canonical pathway. According to this alternative pathway, the non-canonical STING signaling complex that includes the tumor suppressor p53 and the E3 ubiquitin ligase TRAF6 promotes the activation of an NF-kB-dependent transcriptional program [164].
There is pre-clinical evidence that DDR inhibitors, such as PARP inhibitors, activate the STING pathway and the type I IFN signaling in BRCA1 or ERCC1-deficient cancer cells, stimulating augmented cytotoxic-T cell infiltration [165, 166]. In addition, it has been proposed that PARP inhibition promotes adaptive overexpression of cell surface PD-L1 in these cells [167, 168]. Based on the biological evidence for the immunomodulatory function of DDR inhibitors, various ongoing clinical trials investigate the efficiency of combinatorial treatment with PARP inhibitors and immune checkpoint inhibitors targeting the PD-1 and/or PD-L1 (NCT03834519, NCT03851614, NCT03602859, NCT03308942).
In vitro and in vivo mouse experiments in pancreatic cancer showed that combination of ATM inhibition with radiation enhanced antigen presentation and type I interferon signaling. Furthermore, ATM silencing increased PD-L1 expression and increased the sensitivity of pancreatic tumors to PD-L1 blocking antibodies in association with increased tumoral CD8+ T cells and established immune memory. Interestingly, low ATM expression was inversely correlated with PD-L1 expression in clinical samples of patients with pancreatic tumors in the same study [169]. Further work is needed to uncover the mechanisms by which PARP inhibitors or other DNA repair-directed agents (such as ATM, ATR, or DNA-PKcs inhibitors) modulate the immune response and sensitivity to IC blockade.
Epigenetic factors and chromatin remodelers as components of the DNA repair machiner: the chromatin context
Recent advances in epigenetics have shed light onto the dynamic changes of chromatin modifications after DNA damage, supporting a model where specific chromatin-remodeling factors are essential components of the repair network [170]. The study of how these epigenetic and chromatin-remodeling factors modulate the chromatin dynamics and cooperate with DNA repair proteins in response to genotoxic stress is important for the identification of new synthetically lethal interactions. Therefore, pharmacological targeting of these epigenetic factors could be a new approach to enhance the efficacy of DDR inhibitors in producing DSB cytotoxicity and mitigate drug resistance.
Chromatin-remodeling and epigenetic factors mediate dynamic alterations of chromatin structure that facilitate DSB repair
Regulating the accessibility of damaged DNA to repair complexes requires a high degree of coordination between DSB repair machineries and chromatin-modifying enzymes. One of the best characterized changes in chromatin organization is the rapid formation of open chromatin structures at DSBs. The response to DSBs requires localized chromatin expansion to facilitate the assembly of repair complexes. A large number of studies, based on biochemical approaches, highlight that the chromatin expansion at the sites of damage is characterized by density loss of core and linker histones. This chromatin decompaction is thought to facilitate access of the repair machinery to damaged DNA. In this direction, recent work in human cells showed that the CHD2 remodeling activity promotes euchromatin expansion at IR-induced DSBs and the recruitment of NHEJ factors. Interestingly, recruitment of CDH2 at the sites of damage is directed by PARP1 catalytic activity [171].
Several groups have also demonstrated that the rapid formation of open chromatin is driven by an increased acetylation of histone H4 (H4K16ac) on nucleosomes, a modification that extends for many kilobases away from the break, following the spreading pattern of γH2AX [172–175]. The N-terminal tail of histone H4 interacts with the acidic patch on the surface of H2A-H2B dimers of adjacent nucleosomes. Disruption of this interaction by acetylation of histone H4 at DSBs promotes chromatin unpacking and formation of open, relaxed chromatin structures detected at DSBs [176]. Acetylation of H4K16 at DSBs is carried out by the histone acetyltransferase TIP60 and its inactivation is sufficient to block chromatin decompaction during DNA damage [175].
The chromatin state has also been shown to influence DNA repair pathway choice. After DNA damage and ATM activation, γ-H2AX recruits a number of repair factors to DNA damage sites. Among these are the E3 ubiquitin ligases RNF8 and RNF168 which generate ubiquitin marks on histones near the breaks. RNF8 promotes K63-linked polyubiquitylation of H1 linker histones [177, 178]. This polyubiquitylation is a recruitment signal for RNF168, which in turn ubiquitylates H2A-type histones at K13/K15 [177, 179, 180]. TP53BP1, a DSB pathway choice protein that actively promotes NHEJ and inhibits BRCA1-mediated HR, is strongly dependent on RNF8/RNF168-mediated chromatin ubiquitylation and binds to monoubiquitylated H2A-K15 [179].
The role of H3K36me3 histone marks in DNA mismatch repair
MMR maintains genome stability primarily by correcting single nucleotide misincorporations, and small insertion/deletion loops (IDLs) created by the DNA polymerase, improving replication fidelity [181]. In human cells, these mismatches are recognized by hMSH2–hMSH6 (hMutSα) and hMSH2–hMSH3 (hMutSβ) protein complexes. The MSH2–MSH6 complex primarily recognizes single base pair mismatches and 1–2 base IDLs while MSH2–MSH3 recognizes larger IDLs. Both MSH6 and MSH3 harbor a conserved N-terminal PIP (PCNA-interacting protein) motif and interact with the proliferating cell nuclear antigen (PCNA) in replication forks recognizing errors. Defective expression of components of these complexes leads to a mutator phenotype and MSI. Recently, it was demonstrated that H3K36me3 marks, generated by the histone methyltransferase SETD2, facilitate the binding of hMutSα complex to the chromatin [182]. Consistent with this, cells depleted of SETD2 fail to recruit hMutSα to chromatin and display a mutator phenotype characterized by MSI [182]. Lines of evidence have also highlighted a role for H3K36 dimethylation (H3K36me2) on the choice for NHEJ repair pathway [183].
Epigenetic context and nuclear architecture components impact heterochromatin DSB repair
In eukaryotic cells, euchromatin (EC) that contains the majority of the transcribed genes is loosely packed compared to heterochromatin (HC). Centromeres, pericentromeric regions, telomeres and repetitive elements comprise the constitutive heterochromatin, while developmentally silenced genes constitute the facultative heterochromatin. Constitutive heterochromatin contains histones that are hypoacetylated and hypermethylated at histone H3 lysine 9 (H3K9me2/3) and lysine 27 (H3K27me3). These histone marks are associated with anchoring to nuclear lamina [184]. The writing and maintenance of these histone modifications require a large number of epigenetic factors including the histone methyltransferases SETDB1, SUV39 (H3K9 methylation) and histone deacetylases 1 and 2 (HDAC1/2) [185]. The H3K9me2/3 histone marks are specifically recognized and bound by the heterohromatin protein 1 (HP1) which recruits other proteins such as the KAP-1 and the SMARCAD1. The KAP1 protein recruits epigenetic factors that maintain the chromatin compaction (SETDB1, HDAC1, HDAC2), while the SMARAD1 facilitates the deacetylation of newly assembled histones during replication to maintain gene silencing [186]. In response to DNA damage, the chromatin compaction in heterochromatin and the dense array of proteins that bind its domains present a barrier for DSB repair factors to access the sites of damage. Furthermore, the presence of repetitive DNA elements within heterochromatin can initiate aberrant homologous recombination events during repair, such as sister chromatid exchanges or inter-chromosomal recombination, leading to deletions, duplications, translocations, and formation of dicentric or acentric chromosomes. Therefore, heterochromatin DSB repair requires a more stringent control of HR to prevent inappropriate recombination events. Upon formation of heterochromatic DSBs, ATM phosphorylates the HP1-interacting protein Kap1 (KRAB-associated protein-1), thus reducing the strength of Kap1 interaction with damaged heterochromatin, and promoting the release of the chromatin modifier CHD3.1 (Chromodomain Helicase DNA binding protein 3). The release of CHD3.1 drives chromatin relaxation which provides access to repair complexes [187, 188].
Recent studies in Drosophila and mouse cells have shown a relocation of heterochromatin DSBs to anchoring points at the nuclear periphery. This relocation ensures safe and precise HR while preventing aberrant recombination, by isolating the DSBs and their templates away from ectopic sequences before strand invasion [189]. Notably, SUMO and the SUMO E3 ligases dPIAS and Nse2 are required to relocalize DSBs to the nuclear periphery [190, 191].
The role of chromatin remodelers in BER and NER
In eukaryotes, BER is the major pathway for the repair of alkylatively and oxidatively generated lesions such as 8-oxoguanine (8-oxoG). BER is initiated by a glycosylase, which cleaves the glycosidic bond that attaches the lesion to the sugar-phosphate backbone and generates an abasic site. Eleven glycosylases have been identified in humans and are categorized based on their structural architecture [192]. The apurinic/apyrimidinic sites are bound by the endonuclease APE1, which cleaves the DNA backbone on the 5′ side of the abasic deoxyribose phosphate, creating a nick in the DNA [193]. The synthesis step of BER employs either repair polymerase Pol β which adds a single nucleotide, or one of the processive polymerases Pol δ or Pol ε, adding up to 13 nucleotides to the 3′ hydroxyl group of the nucleotide 5′ of the nick [194]. The remaining deoxyribose phosphate is removed by the dRPase activity of Pol β, whereas the 5′ stretch of nucleotides when added by Pol δ or Pol ε is cleaved by the flap endonuclease FEN-1 [195]. The final step of BER is ligation of the nicked strand by DNA ligase IIIα in a complex with its partner protein XRCC1.
For execution of this multi-step process, it is necessary that DNA is accessible to all components of BER. Each step of BER requires access to DNA for its enzymatic activity. A number of studies in cells have demonstrated an inverse correlation between the level of chromatin compaction and BER activity [196]. Several in vitro studies also indicate that chromatin remodeler activity is sufficient to dramatically increase the BER efficiency. In vitro experiments showed that purified members of SWI/SNF subfamily and purified ISW1 and ISW2 chromatin remodelers significantly facilitate the glycosylase, APE-1 and polymerase synthesis step during BER [197–199]. It is considered that chromatin remodelers provide accessibility to BER repair proteins by either remodeling, or combined remodeling and sliding mechanisms.
NER is the major pathway for repair of bulky DNA lesions caused by UV, environmental mutagens and cancer chemotherapeutic drugs. Two distinct DNA damage recognition cascades can activate NER, depending on the location of DNA damage. Genome NER (GG-NER) is activated by helix distortions associated with DNA lesions anywhere in the genome. The main damage sensor in GG-NER is the XPC–RAD23B–CETN2 protein complex. Transcription-coupled NER (TC-NER) is activated by stalled RNA Pol II during transcript elongation by a lesion in the template strand [200].
Similarly to BER repair pathway, numerous in vitro NER assays have shown that the nucleosome structure can be a barrier to efficient NER function and purified SWI/SNF complexes increase accessibility of damaged DNA and stimulate NER repair were found to be within in vitro reconstituted mononucleosomes [201, 202]. However, it is not clear yet whether SWI/SNF complexes are involved in early NER steps facilitating XPF recruitment and lesion detection, or are recruited by XPF and promote the binding of late NER factors XPG and PCNA. The latter is supported by experiments showing that knockdown of chromatin remodelers BRG1, BRM and ARID1A/B has no effect on XPC recruitment but can impair the recruitment of late NER factors ERCC1 and XPA [203, 204].
Given the broad role of SWI/SNIF complexes in NER and BER repair pathways and the high incidence of mutations in family members across different cancer types, the exploitation of SWI/SNF deficiency-induced susceptibilities is crucial for the development efficient and precise therapies for SWI/SNF-mutated cancers.
Histone methyltransferases KMT2C/D and stalled fork stability
KMT2C and KMT2D belong to the family of mammalian mixed-lineage leukemia (MLL) genes that encode histone methyltransferases, responsible for monomethylation at H3K4 at a subset of active gene promoters and enhancers [205]. The distribution of H3K4me1 marks across cis-regulatory elements is characteristic of accessible enhancer regions and, therefore, KMT2C/D activity in enhancers is positively correlated with transcriptional activation of neighbor genes. Recent studies that focus on enhancer regulation also indicate that KMT2C/KMT2D can promote transcription independently of their methylation activity [206, 207]. Cancer genome sequencing studies have identified the histone methyltransferases KMT2C and KMT2D among the most frequently mutated genes across different types of solid tumors [13]. Besides the inactivation of these genes by loss of function mutations, their expression is significantly downregulated across different cancer types [208].
Recent studies have also uncovered an important role of KMT2C and KMT2D proteins in genome stability and in precise control of DNA repair. Chaudhuri and collogues showed that recruitment of MRE11 nuclease to stalled replication forks in HR-deficient cells is mediated by KMT2C/KMT2D [209]. Stalled replication forks are featured by exposed DNA ends in the form of ssDNA or dsDNA, which makes them susceptible to various cellular nucleases that generate HR substrates during DSB repair, including MRE11, which promote 3′–5′ short-range resection during initial steps of homologous recombination repair. The MRE11 nuclease activity at the sites of replication stress is tightly control to prevent excessive fork degradation by BRCA1/2 and RAD51. More specifically, during replication stress, BRCA1 and BRCA2 proteins relocate to stalled replication forks and promote the formation of stable RAD51 nucleoprotein filaments, thereby suppressing deleterious fork degradation mediated by the MRE11 nuclease [210, 211]. In BRCA1/2-deficient cells, stalled replication forks are unprotected from MRE11 activity and, therefore, these cells are characterized by increased genomic instability and chemosensitivity [212–214]. However, KMT2C/D inactivation in BRCA1/2-deficient tumors decreases MRE11 recruitment to stalled forks, which in turn restores fork stability, and renders BRCA1/2-deficient cells resistant to cisplatin and PARP inhibitors [209], indicating a general resistance mechanism to genotoxic stress.
Dysregulated epigenome as a driver for DNA methylation and transcriptional silencing of DNA repair genes
Comprehensive analyses of human cancer genomes over the past decade have revealed that epigenetic factors and chromatin-remodeling complexes are highly mutated in cancer. Since epigenetic factors regulate precisely gene transcription, mutations on these factors in cancer cell offer an adaptive plasticity to the transcriptome. Dysregulated epigenome in cancer is often associated with altered chromatin context in enhancers and promoters of DNA repair genes, which favors DNA methylation and transcriptional silencing. DNA methylation represents the epigenetic biomarker with the highest translational potential due to its stable nature.
Methylation of lysine 27 on histone 3 (H3K27me), a modification associated with gene repression, is a focal point of epigenetic deregulation in cancer [215]. The genetic basis of H3K27me deregulation may include either components of the H3K27 methyltransferase complex PRC2 (Polycomb repressive complex 2) and associated proteins or the H3K27 demethylase UTX [216]. Several studies have demonstrated that H3K27me3-marked genes are targets for aberrant DNA methylation in cancer cells [217, 218].
MSI has been reported in 15% of sporadic colorectal cancer cases which represent a distinct molecular subtype characterized by hypermutator phenotype and global CpG methylation [219]. Most tumors in this subtype have lost expression of the MLH1 DNA mismatch repair gene due to acquired DNA hypermethylation of the MLH1 promoter region [220]. As a result, tumors of this subtype are deficient for DNA mismatch repair (MMR) and exhibit a hypermutable phenotype.
BRCA1 and BRCA2 proteins play a basic role in the regulation and promotion of HR. Germline mutations in these genes are the most important causes of hereditary breast and ovarian cancer [221]. In addition to germline mutations, somatic mutations and epigenetic silencing of these genes occur in a variety of cancers in the general population. BRCA1 methylation is observed in approximately 11–14% of breast cancers and 5–31% of ovarian cancers [222]. BRCA1 methylation has also been detected in 18.6% and 12.1% of non-small cell lung (NSCLC) and bladder cancer (BLCA), respectively [223, 224]. BRCA2 methylation has been reported in NSCLC [225].
ATM is also epigenetically silenced in primary head and neck and breast cancers by aberrant methylation in promoter region [226, 227].
Individuals with Fanconi anemia (FA) are predisposed to develop ovarian cancer than those without FA; this is largely contributed to promoter methylation of the FANCF gene and subsequent disruption of the FA–BRCA pathway [228, 229]. Epigenetic inactivation of FANCF has also been proposed as a mechanism of sensitization to platinum chemotherapy [230].
In bladder cancer (BLCA), the excision repair cross-complementing group 1 (ERCC1) gene, encoding a key enzyme of the nucleotide excision repair (NER) pathway, and the RBBP8/CtiP gene that has an important role in HR repair exhibit a tumor-specific promoter methylation in up to 17% and 45% of BLCA patients, respectively [231]. It, therefore, is evident that epigenetic status is sufficient to cause DNA repair defects and should, therefore, be considered in patient selection for targeted therapies.
Transcriptional silencing of HR DNA repair genes due to epigenetic deregulation as predictive biomarker for response to PARP1 inhibition
Methylation of DNA repair genes seems to represent a good predictive biomarker for response to PARP inhibitors. For instance, in high-grade serous ovarian carcinoma (HGSOC), patient-derived xenografts (PDXs) with homozygous methylation of BRCA1 alleles have been found to respond to PARP1 inhibitor rucaparib. Moreover, clinical data from HGSOC patients who participated to ARIEL2 Part 1 clinical trial indicate a correlation between high levels of BRCA1 methylation in homozygous status and response to PARP inhibitors [232]. Experiments with breast cancer cell lines have also demonstrated that the status of BRCA1 methylation is correlated with sensitivity to PARP1 inhibition. In these experiments, BRCA1 silencing as well as PARPi sensitivity was abolished by the demethylating agent 5-azacytidine [233].
Recently, the DNA methyltransferase inhibitors (DNMTi) and PARP inhibitors (PARPi) have been reported to act synergistically inducing cell death in acute myeloid leukemia (AML), breast and ovarian cancers [234, 235]. The most widely used DNMTis are the cytosine analogs 5-azacytidine (Aza) and 5-aza-2′-deoxycytidine (Decitabine). These analogs are incorporated into DNA during replication leading to the formation of DNMT-DNA adducts that inhibit the catalytic activity of DNMT1 leading to global DNA demethylation [236, 237]. Besides epigenetic effects, the DNMTis are also able to increase the PARP-1 trapping at the DNA damage sites, enhancing the DSBs cytotoxic effects induced by PARP inhibitors [238].
There is accumulated evidence that deregulation of epigenetic factors that promote open chromatin conformation in promoters of HR genes can confer to their transcriptional silencing in a subset of cancers. Previous studies have demonstrated the involvement of the histone methyltransferase EZH2 in inducing epigenetic silencing of RAD51 [239, 240]. Moreover, EZH2 expression was shown to be associated with the activation of RAF1–MEK signaling and expansion of breast cancer stem cells [240].
Recently our group showed that KMT2C binds the promoter region of BRCA1/2 and other HR repair genes controlling their transcription in bladder cancer cell lines. KMT2C silencing in these cell lines leads to BRCA1/2 hypo expression, HR deficiency and increased sensitivity to PARP inhibitor olaparib. Meta-analysis of TCGA RNA-seq data showed that KMT2C downregulation is associated with BRCA1/2 downregulation in bladder, head and neck, lung and colon cancer [208].
HDAC and BET inhibitors can suppress the expression of HR genes and synergize with PARPi
HDAC activity has been linked to histone deacetylation that is often associated with chromatin condensation and gene repression. They are also part of complexes involved in transcription silencing [241, 242] and it has been proposed that in cancer, HDACs activity mediates the transcriptional repression of tumor suppressor genes [243, 244]. Pharmacological inhibition of these enzymes by HDAC inhibitors causes global changes to the chromatin acetylation landscape, re-shaping the boundaries between transcriptionally active and quiescent chromatin. This results in transcriptome changes including re-expression of silent genes and repression of highly transcribed genes [245]. Interestingly, recent studies have shown that the expression of repair factors that are critical components of HR repair pathway is sensitive to HDAC inhibition. More specifically, the HDAC inhibitor vorinostat has been shown to suppress the transcription of DNA damage repair proteins, such as RAD50 and MRE11, and induce DNA DSBs in human prostate cancer cells (LNCaP), and human lung adenocarcinoma cells (A549) [225]. The ability of HDACs inhibitors to suppress HR repair leads to sensitization of cancer cells to PARP inhibition [246, 247] and to the DNA damaging agents doxorubicin and cisplatin [248, 249]. Combination of chemo/radiotherapy with HDACi in phase I and phase II clinical trials has been well tolerated and showed encouraging efficacy [250–252], while investigation of HDACi in combination with PARPi is also underway in a phase I trial (NCT03742245).
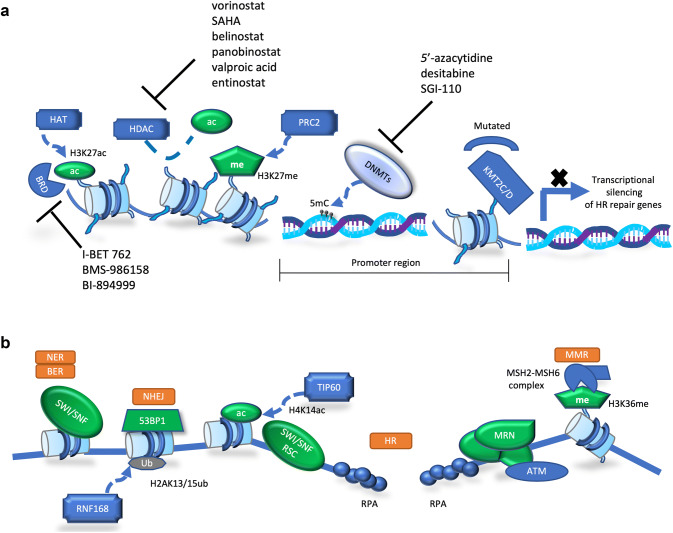
The bromodomain and extraterminal domain (BET) family comprises four members (BRD2, BRD3, BRD4, and BRDT) and is the best characterized class of acetylation readers. These proteins bind hyper-acetylated chromatin regions as active promoters or enhancers and serve as scaffolds for the recruitment of transcription factors that promote the transcription of target genes [253, 254]. BRD4 is the best characterized member of this family and has been heavily implicated in transcriptional regulation and tumorigenesis [255]. BRD4 localizes on both gene promoters and enhancers and has been shown to accumulate specifically on regulatory regions termed “super-enhancers” [256]. BET inhibitors have been shown also to impair the HR pathway by directly downregulating BRCA1 and RAD51 expression. This finding has provided a rationale to use BET inhibitors to sensitize cancer cells to PARPi, showing preclinical efficacy in breast and ovarian cancer models [257–259], and is now being further investigated in the clinic (NCT03901469). Figure 5 outlines basic aspects of epigenetic regulation of DNA repair and possible therapeutic implications, while Table 2 summarizes the clinical trials with the use of DNA repair inhibitor in combination with targeted and conventional anticancer therapies.
Fig. 5.
Therapeutic implications of the epigenetics of DNA repair. a Epigenetic regulation of HR repair components by histone modifiers, histone binding proteins and DNA modifying enzymes. b Direct involvement of epigenetic regulators in different DNA repair processes
Table 2.
Combinatorial schemes of DDR inhibitors in clinical trials
| Agents | TARGET | Selected clinical trials | Disease | Therapeutic regimen | PHASE |
|---|---|---|---|---|---|
| FDA approved | |||||
| Olaparib | PARP1/2 | NCT03150576 (PARTNER) | Triple-negative breast cancer | Combination with chemotherapy; neoadjuvant | II/III |
| NCT02446600, NCT02502266 | Platinum-sensitive high-grade ovarian cancer | Single agent and combination with angiogenesis inhibition | III | ||
| NCT03834519 (KEYLYNK-010) | Metastatic castration-resistant prostate cancer (mCRPC) | Combination with immunotherapy | III | ||
| NCT03851614 (DAPPER) | Advanced colorectal or pancreatic cancer or leiomyosarcoma | Combination with immunotherapy | II | ||
| NCT03742245 | Breast cancer | Combination with HDAC inhibition | I | ||
| Niraparib | PARP1/2 | NCT03602859 (FIRST) | High-grade non-mucinous ovarian cancer | Single agent and combination with immunotherapy; switch maintenance | III |
| NCT03308942 | Advanced non-small cell lung cancer | Single agent and combination with immunotherapy | II | ||
| NCT03574779 (OPAL) | High-grade ovarian cancer | Combination with immunotherapy and angiogenesis inhibition | II | ||
| Rucaparib | PARP1/2 | NCT03476798 | Cervical or endometrial carcinoma | Combination with angiogenesis inhibitor | II |
| Talazoparib | PARP1/2 | NCT03642132 (JAVELIN OVARIAN PARP100) | High-grade serous ovarian cancer | Maintenance after 1st line chemotherapy; single agent or combination with immunotherapy | III |
| NCT03672773 | Small cell lung cancer | Combination with low-dose chemotherapy | II | ||
| NCT03901469 | Triple negative breast cancer | Combination with BET inhibition | II | ||
| NCT02878785 | AML or R/R AML | Combination with DNA demethylating agent | I/II | ||
| Under evaluation | |||||
| Veliparib | PARP1/2 | NCT02264990 | Advanced non-small cell lung cancer | Combination with chemotherapy; 1st line | III |
| NCT02470585 | High-grade serous ovarian cancer | Combination with chemotherapy; 1st line and maintenance | III | ||
| AZD6738 | ATR | NCT03740893 (PHOENIX) | Triple-negative breast cancer | Single agent; neoadjuvant and adjuvant | II |
| NCT03682289 | Any solid tumor except ovarian cancer | Single agent or combination with olaparib | II | ||
| NCT03462342 (CAPRI) | High-grade serous ovarian cancer | Combination with olaparib | II | ||
| M6620 | ATR | NCT03896503 | Small cell lung cancer | Combination with chemotherapy | II |
| NCT02595892 | High-grade serous ovarian cancer | Combination with chemotherapy | II | ||
| NCT03517969 | Metastatic castration-resistant prostate cancer (mCRPC) | Combination with chemotherapy | II | ||
| NCT03641313 | Gastric or GEJ adenocarcinoma | Combination with chemotherapy | II | ||
| NCT03787680 (TRAP) | Metastatic castration-resistant prostate cancer (mCRPC) | Combination with olaparib | II | ||
| AZD1390 | ATM | NCT03423628 | Brain tumor—GBM or Solid tumor metastasis | Combination with radiation | I |
| AZD0156 | ATM | NCT02588105 (AToM) | Solid tumors | Single agent or combination with olaparib or chemotherapy or other | I |
| M3814 | DNA-PKcs | NCT03770689 | Rectal | Combination with chemotherapy | I/II |
| NCT03724890 | Advanced solid tumors | Combination with radiation and/or immunotherapy | I | ||
| MSC2490484A | DNA-PKcs | NCT02516813 | Advanced solid tumors | Combination with chemotherapy and/or radiation | I |
| LY3300054 | CHK1 | NCT03495323 | Advanced solid tumors | Combination with immunotherapy | I |
Concluding remarks
For many years, the study of DNA repair was fueled by the early findings indicating a tumor suppressor role for the BRCA1/2 proteins, which were commonly mutated in gynecological primarily cancers. With the identification of mutations of other DDR and DNA repair proteins in cancer, the field expanded. Nevertheless, the primary focus of these studies was the role of DDR and DNA repair proteins in early steps of carcinogenesis and particularly the resulting genomic instability. Genomic instability leads to mutagenic events which are rightfully considered to drive carcinogenesis as well as contribute to tumor heterogeneity, a feature that we know now is crucial for therapeutic resistance. We now know that the response of cells to DNA damage is a complex mechanism involving multiple networks of proteins and interconnected functions which are responsible for damage detection, cell cycle regulation and DNA repair. In this process, other chromatin modifiers and epigenetic regulators seem to play particularly important roles. Next-generation sequencing revealed that DNA repair proteins are frequently mutated in sporadic cancers from various anatomic sites. Moreover, for the first time we realized that epigenetic regulators are frequent targets of mutation events in cancer. Beyond mutations, there is accumulating evidence that gene downregulation as a result of epigenetic silencing is another mode of loss-of-function events involving loci encoding proteins that function in the context of DNA repair and also epigenetic regulation.
Physicians have been exploiting therapeutically the defects ensuing from mutations in proteins involved in DNA repair. Challenging a system with DNA repair defects with additional damage induced by genotoxic agents has been particularly successful in certain cancers with DNA repair deficiencies. Yet, the complexity of the cellular machinery in charge of DNA repair and the large number of proteins involved in this process allow cells to develop resistance to therapy. At the time, however, this complexity allows the identification of additional targets which can be targeted therapeutically in cancers carrying silenced alleles or mutated proteins with roles in DNA repair. Cell or organismal death as a result of combinatorial mutations was first described in Drosophila 100 years ago. This phenomenon, later coined as synthetic lethality, has been fueling research in the field of cancer for over a decade now. As novel inhibitors are developed and more synthetically lethal interactions are being described, promising avenues for anticancer therapy are being opened. Recent advances in our understanding of how DNA damage modulates the immune response and how repair is regulated by the epigenome have spurred interest in investigating novel drug combinations in treatment of resistant disease. Molecular characterization of each and every single tumor will provide the ground for a more personalized therapeutic exploitation of novel therapeutic options, hopefully leading to the highly praised Precision Medicine.
Acknowledgements
This work was supported by a Worldwide Cancer Research Grant (16/1217) and Horizon 2020 Grants (732309 and 801347) to AK, and a Greek General Secretariat for Research and Technology and the Hellenic Foundation for Research and Innovation (HFRI) Grant (472-EpiNotch) to TR.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Apostolos Klinakis, Email: aklinakis@bioacademy.gr.
Theodoros Rampias, Email: trampias@bioacademy.gr.
References
- 1.Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, Shen R, Taylor AM, Cherniack AD, Thorsson V, Akbani R, Bowlby R, Wong CK, Wiznerowicz M, Sanchez-Vega F, Robertson AG, Schneider BG, Lawrence MS, Noushmehr H, Malta TM, Cancer Genome Atlas, N. Stuart JM, Benz CC, Laird PW. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell. 2018;173(291–304):e6. doi: 10.1016/j.cell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kurz EU, Lees-Miller SP. DNA damage-induced activation of ATM and ATM-dependent signaling pathways. DNA Repair (Amst) 2004;3:889–900. doi: 10.1016/j.dnarep.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 3.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 4.Bannister AJ, Gottlieb TM, Kouzarides T, Jackson SP. c-Jun is phosphorylated by the DNA-dependent protein kinase in vitro; definition of the minimal kinase recognition motif. Nucleic Acids Res. 1993;21:1289–1295. doi: 10.1093/nar/21.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen YR, Lees-Miller SP, Tegtmeyer P, Anderson CW. The human DNA-activated protein kinase phosphorylates simian virus 40 T antigen at amino- and carboxy-terminal sites. J Virol. 1991;65:5131–5140. doi: 10.1128/jvi.65.10.5131-5140.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lees-Miller SP, Anderson CW. The human double-stranded DNA-activated protein kinase phosphorylates the 90-kDa heat-shock protein, hsp90 alpha at two NH2-terminal threonine residues. J Biol Chem. 1989;264:17275–17280. doi: 10.1016/S0021-9258(18)71488-9. [DOI] [PubMed] [Google Scholar]
- 7.Kim ST, Lim DS, Canman CE, Kastan MB. Substrate specificities and identification of putative substrates of ATM kinase family members. J Biol Chem. 1999;274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- 8.Gell D, Jackson SP. Mapping of protein-protein interactions within the DNA-dependent protein kinase complex. Nucleic Acids Res. 1999;27:3494–3502. doi: 10.1093/nar/27.17.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singleton BK, Torres-Arzayus MI, Rottinghaus ST, Taccioli GE, Jeggo PA. The C terminus of Ku80 activates the DNA-dependent protein kinase catalytic subunit. Mol Cell Biol. 1999;19:3267–3277. doi: 10.1128/MCB.19.5.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 11.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 12.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 13.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, Leiserson MDM, Miller CA, Welch JS, Walter MJ, Wendl MC, Ley TJ, Wilson RK, Raphael BJ, Ding L. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ashworth A. A synthetic lethal therapeutic approach: poly(ADP) ribose polymerase inhibitors for the treatment of cancers deficient in DNA double-strand break repair. J Clin Oncol. 2008;26:3785–3790. doi: 10.1200/JCO.2008.16.0812. [DOI] [PubMed] [Google Scholar]
- 15.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lucchesi JC. Synthetic lethality and semi-lethality among functionally related mutants of Drosophila melanfgaster. Genetics. 1968;59:37–44. doi: 10.1093/genetics/59.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bridges CB. The origin of variation. Am Nat. 1922;56:51–63. doi: 10.1086/279847. [DOI] [Google Scholar]
- 18.Dobzhansky T. Genetics of natural populations; recombination and variability in populations of Drosophila pseudoobscura. Genetics. 1946;31:269–290. doi: 10.1093/genetics/31.3.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lord CJ, Ashworth A. PARP inhibitors: synthetic lethality in the clinic. Science. 2017;355:1152–1158. doi: 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- 21.Stilmann M, Hinz M, Arslan SC, Zimmer A, Schreiber V, Scheidereit C. A nuclear poly(ADP-ribose)-dependent signalosome confers DNA damage-induced IkappaB kinase activation. Mol Cell. 2009;36:365–378. doi: 10.1016/j.molcel.2009.09.032. [DOI] [PubMed] [Google Scholar]
- 22.Ali AAE, Timinszky G, Arribas-Bosacoma R, Kozlowski M, Hassa PO, Hassler M, Ladurner AG, Pearl LH, Oliver AW. The zinc-finger domains of PARP1 cooperate to recognize DNA strand breaks. Nat Struct Mol Biol. 2012;19:685–692. doi: 10.1038/nsmb.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dawicki-McKenna JM, Langelier MF, DeNizio JE, Riccio AA, Cao CD, Karch KR, McCauley M, Steffen JD, Black BE, Pascal JM. PARP-1 activation requires local unfolding of an autoinhibitory domain. Mol Cell. 2015;60:755–768. doi: 10.1016/j.molcel.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eustermann S, Wu WF, Langelier MF, Yang JC, Easton LE, Riccio AA, Pascal JM, Neuhaus D. Structural basis of detection and signaling of DNA single-strand breaks by human PARP-1. Mol Cell. 2015;60:742–754. doi: 10.1016/j.molcel.2015.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pommier Y, O’Connor MJ, de Bono J. Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci Transl Med. 2016;8:362ps17. doi: 10.1126/scitranslmed.aaf9246. [DOI] [PubMed] [Google Scholar]
- 26.Tutt A, Bertwistle D, Valentine J, Gabriel A, Swift S, Ross G, Griffin C, Thacker J, Ashworth A. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J. 2001;20:4704–4716. doi: 10.1093/emboj/20.17.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mansour WY, Rhein T, Dahm-Daphi J. The alternative end-joining pathway for repair of DNA double-strand breaks requires PARP1 but is not dependent upon microhomologies. Nucleic Acids Res. 2010;38:6065–6077. doi: 10.1093/nar/gkq387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sulkowski PL, Corso CD, Robinson ND, Scanlon SE, Purshouse KR, Bai H, Liu Y, Sundaram RK, Hegan DC, Fons NR, Breuer GA, Song Y, Mishra-Gorur K, De Feyter HM, de Graaf RA, Surovtseva YV, Kachman M, Halene S, Gunel M, Glazer PM, Bindra RS. 2-Hydroxyglutarate produced by neomorphic IDH mutations suppresses homologous recombination and induces PARP inhibitor sensitivity. Sci Transl Med. 2017;9:eaal2463. doi: 10.1126/scitranslmed.aal2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bever KM, Le DT. DNA repair defects and implications for immunotherapy. J Clin Investig. 2018;128:4236–4242. doi: 10.1172/JCI122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bindra RS, Schaffer PJ, Meng A, Woo J, Maseide K, Roth ME, Lizardi P, Hedley DW, Bristow RG, Glazer PM. Down-regulation of Rad51 and decreased homologous recombination in hypoxic cancer cells. Mol Cell Biol. 2004;24:8504–8518. doi: 10.1128/MCB.24.19.8504-8518.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bindra RS, Schaffer PJ, Meng A, Woo J, Maseide K, Roth ME, Lizardi P, Hedley DW, Bristow RG, Glazer PM. Alterations in DNA repair gene expression under hypoxia: elucidating the mechanisms of hypoxia-induced genetic instability. Ann N Y Acad Sci. 2005;1059:184–195. doi: 10.1196/annals.1339.049. [DOI] [PubMed] [Google Scholar]
- 32.Patel AG, Flatten KS, Schneider PA, Dai NT, McDonald JS, Poirier GG, Kaufmann SH. Enhanced killing of cancer cells by poly(ADP-ribose) polymerase inhibitors and topoisomerase I inhibitors reflects poisoning of both enzymes. J Biol Chem. 2012;287:4198–4210. doi: 10.1074/jbc.M111.296475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown JS, O’Carrigan B, Jackson SP, Yap TA. Targeting DNA repair in cancer: beyond PARP inhibitors. Cancer Discov. 2017;7:20–37. doi: 10.1158/2159-8290.CD-16-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Connor MJ. Targeting the DNA damage response in cancer. Mol Cell. 2015;60:547–560. doi: 10.1016/j.molcel.2015.10.040. [DOI] [PubMed] [Google Scholar]
- 35.Uematsu N, Weterings E, Yano K, Morotomi-Yano K, Jakob B, Taucher-Scholz G, Mari PO, van Gent DC, Chen BP, Chen DJ. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J Cell Biol. 2007;177:219–229. doi: 10.1083/jcb.200608077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol Cell Biol. 2005;25:5363–5379. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- 39.Yuan J, Chen J. MRE11-RAD50-NBS1 complex dictates DNA repair independent of H2AX. J Biol Chem. 2010;285:1097–1104. doi: 10.1074/jbc.M109.078436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bakr A, Oing C, Kocher S, Borgmann K, Dornreiter I, Petersen C, Dikomey E, Mansour WY. Involvement of ATM in homologous recombination after end resection and RAD51 nucleofilament formation. Nucleic Acids Res. 2015;43:3154–3166. doi: 10.1093/nar/gkv160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weston VJ, Oldreive CE, Skowronska A, Oscier DG, Pratt G, Dyer MJ, Smith G, Powell JE, Rudzki Z, Kearns P, Moss PA, Taylor AM, Stankovic T. The PARP inhibitor olaparib induces significant killing of ATM-deficient lymphoid tumor cells in vitro and in vivo. Blood. 2010;116:4578–4587. doi: 10.1182/blood-2010-01-265769. [DOI] [PubMed] [Google Scholar]
- 42.Aguilar-Quesada R, Munoz-Gamez JA, Martin-Oliva D, Peralta A, Valenzuela MT, Matinez-Romero R, Quiles-Perez R, Menissier-de Murcia J, de Murcia G, Ruiz de Almodovar M, Oliver FJ. Interaction between ATM and PARP-1 in response to DNA damage and sensitization of ATM deficient cells through PARP inhibition. BMC Mol Biol. 2007;8:29. doi: 10.1186/1471-2199-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Menisser-de Murcia J, Mark M, Wendling O, Wynshaw-Boris A, de Murcia G. Early embryonic lethality in PARP-1 Atm double-mutant mice suggests a functional synergy in cell proliferation during development. Mol Cell Biol. 2001;21:1828–1832. doi: 10.1128/MCB.21.5.1828-1832.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, Montgomery B, Taplin ME, Pritchard CC, Attard G, Beltran H, Abida W, Bradley RK, Vinson J, Cao X, Vats P, Kunju LP, Hussain M, Feng FY, Tomlins SA, Cooney KA, Smith DC, Brennan C, Siddiqui J, Mehra R, Chen Y, Rathkopf DE, Morris MJ, Solomon SB, Durack JC, Reuter VE, Gopalan A, Gao J, Loda M, Lis RT, Bowden M, Balk SP, Gaviola G, Sougnez C, Gupta M, Yu EY, Mostaghel EA, Cheng HH, Mulcahy H, True LD, Plymate SR, Dvinge H, Ferraldeschi R, Flohr P, Miranda S, Zafeiriou Z, Tunariu N, Mateo J, Perez-Lopez R, Demichelis F, Robinson BD, Schiffman M, Nanus DM, Tagawa ST, Sigaras A, Eng KW, Elemento O, Sboner A, Heath EI, Scher HI, Pienta KJ, Kantoff P, de Bono JS, Rubin MA, Nelson PS, Garraway LA, Sawyers CL, Chinnaiyan AM. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mateo J, Carreira S, Sandhu S, Miranda S, Mossop H, Perez-Lopez R, Nava Rodrigues D, Robinson D, Omlin A, Tunariu N, Boysen G, Porta N, Flohr P, Gillman A, Figueiredo I, Paulding C, Seed G, Jain S, Ralph C, Protheroe A, Hussain S, Jones R, Elliott T, McGovern U, Bianchini D, Goodall J, Zafeiriou Z, Williamson CT, Ferraldeschi R, Riisnaes R, Ebbs B, Fowler G, Roda D, Yuan W, Wu YM, Cao X, Brough R, Pemberton H, A’Hern R, Swain A, Kunju LP, Eeles R, Attard G, Lord CJ, Ashworth A, Rubin MA, Knudsen KE, Feng FY, Chinnaiyan AM, Hall E, de Bono JS. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med. 2015;373:1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo J, Antonarakis ES. PARP inhibition—not all gene mutations are created equal. Nat Rev Urol. 2019;16:4–6. doi: 10.1038/s41585-018-0129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pilie PG, Gay CM, Byers LA, O’Connor MJ, Yap TA. PARP inhibitors: extending benefit beyond BRCA-mutant cancers. Clin Cancer Res. 2019;25:3759–3771. doi: 10.1158/1078-0432.CCR-18-0968. [DOI] [PubMed] [Google Scholar]
- 48.Cancer Genome Atlas Research, N Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Audeh MW, Carmichael J, Penson RT, Friedlander M, Powell B, Bell-McGuinn KM, Scott C, Weitzel JN, Oaknin A, Loman N, Lu K, Schmutzler RK, Matulonis U, Wickens M, Tutt A. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376:245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- 50.Yazinski SA, Comaills V, Buisson R, Genois MM, Nguyen HD, Ho CK, Todorova Kwan T, Morris R, Lauffer S, Nussenzweig A, Ramaswamy S, Benes CH, Haber DA, Maheswaran S, Birrer MJ, Zou L. ATR inhibition disrupts rewired homologous recombination and fork protection pathways in PARP inhibitor-resistant BRCA-deficient cancer cells. Genes Dev. 2017;31:318–332. doi: 10.1101/gad.290957.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim H, George E, Ragland R, Rafail S, Zhang R, Krepler C, Morgan M, Herlyn M, Brown E, Simpkins F. Targeting the ATR/CHK1 axis with PARP inhibition results in tumor regression in BRCA-mutant ovarian cancer models. Clin Cancer Res. 2017;23:3097–3108. doi: 10.1158/1078-0432.CCR-16-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adams KE, Medhurst AL, Dart DA, Lakin ND. Recruitment of ATR to sites of ionising radiation-induced DNA damage requires ATM and components of the MRN protein complex. Oncogene. 2006;25:3894–3904. doi: 10.1038/sj.onc.1209426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cuadrado M, Martinez-Pastor B, Murga M, Toledo LI, Gutierrez-Martinez P, Lopez E, Fernandez-Capetillo O. ATM regulates ATR chromatin loading in response to DNA double-strand breaks. J Exp Med. 2006;203:297–303. doi: 10.1084/jem.20051923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 55.Chen BP, Uematsu N, Kobayashi J, Lerenthal Y, Krempler A, Yajima H, Lobrich M, Shiloh Y, Chen DJ. Ataxia telangiectasia mutated (ATM) is essential for DNA-PKcs phosphorylations at the Thr-2609 cluster upon DNA double strand break. J Biol Chem. 2007;282:6582–6587. doi: 10.1074/jbc.M611605200. [DOI] [PubMed] [Google Scholar]
- 56.Stiff T, Walker SA, Cerosaletti K, Goodarzi AA, Petermann E, Concannon P, O’Driscoll M, Jeggo PA. ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling. EMBO J. 2006;25:5775–5782. doi: 10.1038/sj.emboj.7601446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi M, Kipps T, Kurzrock R. ATM mutations in cancer: therapeutic implications. Mol Cancer Ther. 2016;15:1781–1791. doi: 10.1158/1535-7163.MCT-15-0945. [DOI] [PubMed] [Google Scholar]
- 58.Dietlein F, Thelen L, Reinhardt HC. Cancer-specific defects in DNA repair pathways as targets for personalized therapeutic approaches. Trends Genet. 2014;30:326–339. doi: 10.1016/j.tig.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 59.Schneider G, Schmidt-Supprian M, Rad R, Saur D. Tissue-specific tumorigenesis: context matters. Nat Rev Cancer. 2017;17:239–253. doi: 10.1038/nrc.2017.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boultwood J. Ataxia telangiectasia gene mutations in leukaemia and lymphoma. J Clin Pathol. 2001;54:512–516. doi: 10.1136/jcp.54.7.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Camacho E, Hernandez L, Hernandez S, Tort F, Bellosillo B, Bea S, Bosch F, Montserrat E, Cardesa A, Fernandez PL, Campo E. ATM gene inactivation in mantle cell lymphoma mainly occurs by truncating mutations and missense mutations involving the phosphatidylinositol-3 kinase domain and is associated with increasing numbers of chromosomal imbalances. Blood. 2002;99:238–244. doi: 10.1182/blood.V99.1.238. [DOI] [PubMed] [Google Scholar]
- 62.Kwok M, Davies N, Agathanggelou A, Smith E, Oldreive C, Petermann E, Stewart G, Brown J, Lau A, Pratt G, Parry H, Taylor M, Moss P, Hillmen P, Stankovic T. ATR inhibition induces synthetic lethality and overcomes chemoresistance in TP53- or ATM-defective chronic lymphocytic leukemia cells. Blood. 2016;127:582–595. doi: 10.1182/blood-2015-05-644872. [DOI] [PubMed] [Google Scholar]
- 63.Menezes DL, Holt J, Tang Y, Feng J, Barsanti P, Pan Y, Ghoddusi M, Zhang W, Thomas G, Holash J, Lees E, Taricani L. A synthetic lethal screen reveals enhanced sensitivity to ATR inhibitor treatment in mantle cell lymphoma with ATM loss-of-function. Mol Cancer Res. 2015;13:120–129. doi: 10.1158/1541-7786.MCR-14-0240. [DOI] [PubMed] [Google Scholar]
- 64.Cui Y, Palii SS, Innes CL, Paules RS. Depletion of ATR selectively sensitizes ATM-deficient human mammary epithelial cells to ionizing radiation and DNA-damaging agents. Cell Cycle. 2014;13:3541–3550. doi: 10.4161/15384101.2014.960729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chan EM, Shibue T, McFarland JM, Gaeta B, Ghandi M, Dumont N, Gonzalez A, McPartlan JS, Li T, Zhang Y, Bin Liu J, Lazaro JB, Gu P, Piett CG, Apffel A, Ali SO, Deasy R, Keskula P, Ng RWS, Roberts EA, Reznichenko E, Leung L, Alimova M, Schenone M, Islam M, Maruvka YE, Liu Y, Roper J, Raghavan S, Giannakis M, Tseng YY, Nagel ZD, D’Andrea A, Root DE, Boehm JS, Getz G, Chang S, Golub TR, Tsherniak A, Vazquez F, Bass AJ. WRN helicase is a synthetic lethal target in microsatellite unstable cancers. Nature. 2019;568:551–556. doi: 10.1038/s41586-019-1102-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Behan FM, Iorio F, Picco G, Goncalves E, Beaver CM, Migliardi G, Santos R, Rao Y, Sassi F, Pinnelli M, Ansari R, Harper S, Jackson DA, McRae R, Pooley R, Wilkinson P, van der Meer D, Dow D, Buser-Doepner C, Bertotti A, Trusolino L, Stronach EA, Saez-Rodriguez J, Yusa K, Garnett MJ. Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature. 2019;568:511–516. doi: 10.1038/s41586-019-1103-9. [DOI] [PubMed] [Google Scholar]
- 68.Kitano K. Structural mechanisms of human RecQ helicases WRN and BLM. Front Genet. 2014;5:366. doi: 10.3389/fgene.2014.00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reisz JA, Bansal N, Qian J, Zhao W, Furdui CM. Effects of ionizing radiation on biological molecules–mechanisms of damage and emerging methods of detection. Antioxid Redox Signal. 2014;21:260–292. doi: 10.1089/ars.2013.5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Byrski T, Huzarski T, Dent R, Gronwald J, Zuziak D, Cybulski C, Kladny J, Gorski B, Lubinski J, Narod SA. Response to neoadjuvant therapy with cisplatin in BRCA1-positive breast cancer patients. Breast Cancer Res Treat. 2009;115:359–363. doi: 10.1007/s10549-008-0128-9. [DOI] [PubMed] [Google Scholar]
- 71.Sakai W, Swisher EM, Karlan BY, Agarwal MK, Higgins J, Friedman C, Villegas E, Jacquemont C, Farrugia DJ, Couch FJ, Urban N, Taniguchi T. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–1120. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Van Allen EM, Mouw KW, Kim P, Iyer G, Wagle N, Al-Ahmadie H, Zhu C, Ostrovnaya I, Kryukov GV, O’Connor KW, Sfakianos J, Garcia-Grossman I, Kim J, Guancial EA, Bambury R, Bahl S, Gupta N, Farlow D, Qu A, Signoretti S, Barletta JA, Reuter V, Boehm J, Lawrence M, Getz G, Kantoff P, Bochner BH, Choueiri TK, Bajorin DF, Solit DB, Gabriel S, D’Andrea A, Garraway LA, Rosenberg JE. Somatic ERCC2 mutations correlate with cisplatin sensitivity in muscle-invasive urothelial carcinoma. Cancer Discov. 2014;4:1140–1153. doi: 10.1158/2159-8290.CD-14-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Plimack ER, Dunbrack RL, Brennan TA, Andrake MD, Zhou Y, Serebriiskii IG, Slifker M, Alpaugh K, Dulaimi E, Palma N, Hoffman-Censits J, Bilusic M, Wong YN, Kutikov A, Viterbo R, Greenberg RE, Chen DY, Lallas CD, Trabulsi EJ, Yelensky R, McConkey DJ, Miller VA, Golemis EA, Ross EA. Defects in DNA repair genes predict response to neoadjuvant cisplatin-based chemotherapy in muscle-invasive bladder cancer. Eur Urol. 2015;68:959–967. doi: 10.1016/j.eururo.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Olaussen KA, Dunant A, Fouret P, Brambilla E, Andre F, Haddad V, Taranchon E, Filipits M, Pirker R, Popper HH, Stahel R, Sabatier L, Pignon JP, Tursz T, Le Chevalier T, Soria JC, Investigators IB. DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med. 2006;355:983–991. doi: 10.1056/NEJMoa060570. [DOI] [PubMed] [Google Scholar]
- 75.Kim MK, Cho KJ, Kwon GY, Park SI, Kim YH, Kim JH, Song HY, Shin JH, Jung HY, Lee GH, Choi KD, Kim SB. Patients with ERCC1-negative locally advanced esophageal cancers may benefit from preoperative chemoradiotherapy. Clin Cancer Res. 2008;14:4225–4231. doi: 10.1158/1078-0432.CCR-07-4848. [DOI] [PubMed] [Google Scholar]
- 76.Bellmunt J, Paz-Ares L, Cuello M, Cecere FL, Albiol S, Guillem V, Gallardo E, Carles J, Mendez P, de la Cruz JJ, Taron M, Rosell R, Baselga J, Spanish Oncology Genitourinary, G Gene expression of ERCC1 as a novel prognostic marker in advanced bladder cancer patients receiving cisplatin-based chemotherapy. Ann Oncol. 2007;18:522–528. doi: 10.1093/annonc/mdl435. [DOI] [PubMed] [Google Scholar]
- 77.Shirota Y, Stoehlmacher J, Brabender J, Xiong YP, Uetake H, Danenberg KD, Groshen S, Tsao-Wei DD, Danenberg PV, Lenz HJ. ERCC1 and thymidylate synthase mRNA levels predict survival for colorectal cancer patients receiving combination oxaliplatin and fluorouracil chemotherapy. J Clin Oncol. 2001;19:4298–4304. doi: 10.1200/JCO.2001.19.23.4298. [DOI] [PubMed] [Google Scholar]
- 78.Metzger R, Leichman CG, Danenberg KD, Danenberg PV, Lenz HJ, Hayashi K, Groshen S, Salonga D, Cohen H, Laine L, Crookes P, Silberman H, Baranda J, Konda B, Leichman L. ERCC1 mRNA levels complement thymidylate synthase mRNA levels in predicting response and survival for gastric cancer patients receiving combination cisplatin and fluorouracil chemotherapy. J Clin Oncol. 1998;16:309–316. doi: 10.1200/JCO.1998.16.1.309. [DOI] [PubMed] [Google Scholar]
- 79.Handra-Luca A, Hernandez J, Mountzios G, Taranchon E, Lacau-St-Guily J, Soria JC, Fouret P. Excision repair cross complementation group 1 immunohistochemical expression predicts objective response and cancer-specific survival in patients treated by Cisplatin-based induction chemotherapy for locally advanced head and neck squamous cell carcinoma. Clin Cancer Res. 2007;13:3855–3859. doi: 10.1158/1078-0432.CCR-07-0252. [DOI] [PubMed] [Google Scholar]
- 80.Hsu DS, Lan HY, Huang CH, Tai SK, Chang SY, Tsai TL, Chang CC, Tzeng CH, Wu KJ, Kao JY, Yang MH. Regulation of excision repair cross-complementation group 1 by Snail contributes to cisplatin resistance in head and neck cancer. Clin Cancer Res. 2010;16:4561–4571. doi: 10.1158/1078-0432.CCR-10-0593. [DOI] [PubMed] [Google Scholar]
- 81.Bauman JE, Austin MC, Schmidt R, Kurland BF, Vaezi A, Hayes DN, Mendez E, Parvathaneni U, Chai X, Sampath S, Martins RG. ERCC1 is a prognostic biomarker in locally advanced head and neck cancer: results from a randomised, phase II trial. Br J Cancer. 2013;109:2096–2105. doi: 10.1038/bjc.2013.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dabholkar M, Bostick-Bruton F, Weber C, Bohr VA, Egwuagu C, Reed E. ERCC1 and ERCC2 expression in malignant tissues from ovarian cancer patients. J Natl Cancer Inst. 1992;84:1512–1517. doi: 10.1093/jnci/84.19.1512. [DOI] [PubMed] [Google Scholar]
- 83.Mello JA, Acharya S, Fishel R, Essigmann JM. The mismatch-repair protein hMSH2 binds selectively to DNA adducts of the anticancer drug cisplatin. Chem Biol. 1996;3:579–589. doi: 10.1016/S1074-5521(96)90149-0. [DOI] [PubMed] [Google Scholar]
- 84.Hawn MT, Umar A, Carethers JM, Marra G, Kunkel TA, Boland CR, Koi M. Evidence for a connection between the mismatch repair system and the G2 cell cycle checkpoint. Cancer Res. 1995;55:3721–3725. [PubMed] [Google Scholar]
- 85.Moggs JG, Szymkowski DE, Yamada M, Karran P, Wood RD. Differential human nucleotide excision repair of paired and mispaired cisplatin-DNA adducts. Nucleic Acids Res. 1997;25:480–491. doi: 10.1093/nar/25.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gifford G, Paul J, Vasey PA, Kaye SB, Brown R. The acquisition of hMLH1 methylation in plasma DNA after chemotherapy predicts poor survival for ovarian cancer patients. Clin Cancer Res. 2004;10:4420–4426. doi: 10.1158/1078-0432.CCR-03-0732. [DOI] [PubMed] [Google Scholar]
- 87.Sargent DJ, Marsoni S, Monges G, Thibodeau SN, Labianca R, Hamilton SR, French AJ, Kabat B, Foster NR, Torri V, Ribic C, Grothey A, Moore M, Zaniboni A, Seitz JF, Sinicrope F, Gallinger S. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28:3219–3226. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bassett E, Vaisman A, Tropea KA, McCall CM, Masutani C, Hanaoka F, Chaney SG. Frameshifts and deletions during in vitro translesion synthesis past Pt-DNA adducts by DNA polymerases beta and eta. DNA Repair (Amst) 2002;1:1003–1016. doi: 10.1016/S1568-7864(02)00150-7. [DOI] [PubMed] [Google Scholar]
- 89.Shachar S, Ziv O, Avkin S, Adar S, Wittschieben J, Reissner T, Chaney S, Friedberg EC, Wang Z, Carell T, Geacintov N, Livneh Z. Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J. 2009;28:383–393. doi: 10.1038/emboj.2008.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Alt A, Lammens K, Chiocchini C, Lammens A, Pieck JC, Kuch D, Hopfner KP, Carell T. Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase eta. Science. 2007;318:967–970. doi: 10.1126/science.1148242. [DOI] [PubMed] [Google Scholar]
- 91.Wittschieben JP, Reshmi SC, Gollin SM, Wood RD. Loss of DNA polymerase zeta causes chromosomal instability in mammalian cells. Cancer Res. 2006;66:134–142. doi: 10.1158/0008-5472.CAN-05-2982. [DOI] [PubMed] [Google Scholar]
- 92.Ceppi P, Novello S, Cambieri A, Longo M, Monica V, Lo Iacono M, Giaj-Levra M, Saviozzi S, Volante M, Papotti M, Scagliotti G. Polymerase eta mRNA expression predicts survival of non-small cell lung cancer patients treated with platinum-based chemotherapy. Clin Cancer Res. 2009;15:1039–1045. doi: 10.1158/1078-0432.CCR-08-1227. [DOI] [PubMed] [Google Scholar]
- 93.Teng KY, Qiu MZ, Li ZH, Luo HY, Zeng ZL, Luo RZ, Zhang HZ, Wang ZQ, Li YH, Xu RH. DNA polymerase eta protein expression predicts treatment response and survival of metastatic gastric adenocarcinoma patients treated with oxaliplatin-based chemotherapy. J Transl Med. 2010;8:126. doi: 10.1186/1479-5876-8-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bartek J, Lukas J. DNA damage checkpoints: from initiation to recovery or adaptation. Curr Opin Cell Biol. 2007;19:238–245. doi: 10.1016/j.ceb.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 95.Koundrioukoff S, Polo S, Almouzni G. Interplay between chromatin and cell cycle checkpoints in the context of ATR/ATM-dependent checkpoints. DNA Repair (Amst) 2004;3:969–978. doi: 10.1016/j.dnarep.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 96.Castedo M, Perfettini JL, Roumier T, Andreau K, Medema R, Kroemer G. Cell death by mitotic catastrophe: a molecular definition. Oncogene. 2004;23:2825–2837. doi: 10.1038/sj.onc.1207528. [DOI] [PubMed] [Google Scholar]
- 97.Lavin MF. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene. 2007;26:7749–7758. doi: 10.1038/sj.onc.1210880. [DOI] [PubMed] [Google Scholar]
- 98.Bartek J, Lukas J. Mammalian G1- and S-phase checkpoints in response to DNA damage. Curr Opin Cell Biol. 2001;13:738–747. doi: 10.1016/S0955-0674(00)00280-5. [DOI] [PubMed] [Google Scholar]
- 99.Cortez D, Guntuku S, Qin J, Elledge SJ. ATR and ATRIP: partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- 100.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Donzelli M, Draetta GF. Regulating mammalian checkpoints through Cdc25 inactivation. EMBO Rep. 2003;4:671–677. doi: 10.1038/sj.embor.embor887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Do K, Wilsker D, Ji J, Zlott J, Freshwater T, Kinders RJ, Collins J, Chen AP, Doroshow JH, Kummar S. Phase I study of single-agent AZD1775 (MK-1775), a Wee1 kinase inhibitor, in patients with refractory solid tumors. J Clin Oncol. 2015;33:3409–3415. doi: 10.1200/JCO.2014.60.4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.McNeely S, Beckmann R, Bence Lin AK. CHEK again: revisiting the development of CHK1 inhibitors for cancer therapy. Pharmacol Ther. 2014;142:1–10. doi: 10.1016/j.pharmthera.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 104.Haynes B, Murai J, Lee JM. Restored replication fork stabilization, a mechanism of PARP inhibitor resistance, can be overcome by cell cycle checkpoint inhibition. Cancer Treat Rev. 2018;71:1–7. doi: 10.1016/j.ctrv.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kawabe T. G2 checkpoint abrogators as anticancer drugs. Mol Cancer Ther. 2004;3:513–519. [PubMed] [Google Scholar]
- 106.Starr ER, Margiotta JF. Pituitary adenylate cyclase activating polypeptide induces long-term, transcription-dependent plasticity and remodeling at autonomic synapses. Mol Cell Neurosci. 2017;85:170–182. doi: 10.1016/j.mcn.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 107.Shechter D, Costanzo V, Gautier J. ATR and ATM regulate the timing of DNA replication origin firing. Nat Cell Biol. 2004;6:648–655. doi: 10.1038/ncb1145. [DOI] [PubMed] [Google Scholar]
- 108.Moiseeva TN, Yin Y, Calderon MJ, Qian C, Schamus-Haynes S, Sugitani N, Osmanbeyoglu HU, Rothenberg E, Watkins SC, Bakkenist CJ. An ATR and CHK1 kinase signaling mechanism that limits origin firing during unperturbed DNA replication. Proc Natl Acad Sci USA. 2019;116:13374–13383. doi: 10.1073/pnas.1903418116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Moiseeva T, Hood B, Schamus S, O’Connor MJ, Conrads TP, Bakkenist CJ. ATR kinase inhibition induces unscheduled origin firing through a Cdc7-dependent association between GINS and And-1. Nat Commun. 2017;8:1392. doi: 10.1038/s41467-017-01401-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ammazzalorso F, Pirzio LM, Bignami M, Franchitto A, Pichierri P. ATR and ATM differently regulate WRN to prevent DSBs at stalled replication forks and promote replication fork recovery. EMBO J. 2010;29:3156–3169. doi: 10.1038/emboj.2010.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chanoux RA, Yin B, Urtishak KA, Asare A, Bassing CH, Brown EJ. ATR and H2AX cooperate in maintaining genome stability under replication stress. J Biol Chem. 2009;284:5994–6003. doi: 10.1074/jbc.M806739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.De Witt Hamer PC, Mir SE, Noske D, Van Noorden CJ, Wurdinger T. WEE1 kinase targeting combined with DNA-damaging cancer therapy catalyzes mitotic catastrophe. Clin Cancer Res. 2011;17:4200–4207. doi: 10.1158/1078-0432.CCR-10-2537. [DOI] [PubMed] [Google Scholar]
- 113.Geenen JJJ, Schellens JHM. molecular pathways: targeting the protein kinase Wee1 in cancer. Clin Cancer Res. 2017;23:4540–4544. doi: 10.1158/1078-0432.CCR-17-0520. [DOI] [PubMed] [Google Scholar]
- 114.Jelinic P, Levine DA. New insights into PARP inhibitors’ effect on cell cycle and homology-directed DNA damage repair. Mol Cancer Ther. 2014;13:1645–1654. doi: 10.1158/1535-7163.MCT-13-0906-T. [DOI] [PubMed] [Google Scholar]
- 115.Lallo A, Frese KK, Morrow CJ, Sloane R, Gulati S, Schenk MW, Trapani F, Simms N, Galvin M, Brown S, Hodgkinson CL, Priest L, Hughes A, Lai Z, Cadogan E, Khandelwal G, Simpson KL, Miller C, Blackhall F, O’Connor MJ, Dive C. The combination of the PARP inhibitor olaparib and the WEE1 inhibitor AZD1775 as a new therapeutic option for small cell lung cancer. Clin Cancer Res. 2018;24:5153–5164. doi: 10.1158/1078-0432.CCR-17-2805. [DOI] [PubMed] [Google Scholar]
- 116.Parsels LA, Karnak D, Parsels JD, Zhang Q, Velez-Padilla J, Reichert ZR, Wahl DR, Maybaum J, O’Connor MJ, Lawrence TS, Morgan MA. PARP1 trapping and DNA replication stress enhance radiosensitization with combined WEE1 and PARP inhibitors. Mol Cancer Res. 2018;16:222–232. doi: 10.1158/1541-7786.MCR-17-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pilie PG, Tang C, Mills GB, Yap TA. State-of-the-art strategies for targeting the DNA damage response in cancer. Nat Rev Clin Oncol. 2019;16:81–104. doi: 10.1038/s41571-018-0114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fang Y, McGrail DJ, Sun C, Labrie M, Chen X, Zhang D, Ju Z, Vellano CP, Lu Y, Li Y, Jeong KJ, Ding Z, Liang J, Wang SW, Dai H, Lee S, Sahni N, Mercado-Uribe I, Kim TB, Chen K, Lin SY, Peng G, Westin SN, Liu J, O’Connor MJ, Yap TA, Mills GB. Sequential therapy with PARP and WEE1 inhibitors minimizes toxicity while maintaining efficacy. Cancer Cell. 2019;35(851–867):e7. doi: 10.1016/j.ccell.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ferris RL, Blumenschein G, Jr, Fayette J, Guigay J, Colevas AD, Licitra L, Harrington K, Kasper S, Vokes EE, Even C, Worden F, Saba NF, Iglesias Docampo LC, Haddad R, Rordorf T, Kiyota N, Tahara M, Monga M, Lynch M, Geese WJ, Kopit J, Shaw JW, Gillison ML. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med. 2016;375:1856–1867. doi: 10.1056/NEJMoa1602252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Reck M, Rodriguez-Abreu D, Robinson AG, Hui R, Csoszi T, Fulop A, Gottfried M, Peled N, Tafreshi A, Cuffe S, O’Brien M, Rao S, Hotta K, Leiby MA, Lubiniecki GM, Shentu Y, Rangwala R, Brahmer JR, Investigators, K Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med. 2016;375:1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 121.Robert C, Schachter J, Long GV, Arance A, Grob JJ, Mortier L, Daud A, Carlino MS, McNeil C, Lotem M, Larkin J, Lorigan P, Neyns B, Blank CU, Hamid O, Mateus C, Shapira-Frommer R, Kosh M, Zhou H, Ibrahim N, Ebbinghaus S, Ribas A, investigators, K Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med. 2015;372:2521–2532. doi: 10.1056/NEJMoa1503093. [DOI] [PubMed] [Google Scholar]
- 122.Rosenberg JE, Hoffman-Censits J, Powles T, van der Heijden MS, Balar AV, Necchi A, Dawson N, O’Donnell PH, Balmanoukian A, Loriot Y, Srinivas S, Retz MM, Grivas P, Joseph RW, Galsky MD, Fleming MT, Petrylak DP, Perez-Gracia JL, Burris HA, Castellano D, Canil C, Bellmunt J, Bajorin D, Nickles D, Bourgon R, Frampton GM, Cui N, Mariathasan S, Abidoye O, Fine GD, Dreicer R. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387:1909–1920. doi: 10.1016/S0140-6736(16)00561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjord JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinski M, Jager N, Jones DT, Jones D, Knappskog S, Kool M, Lakhani SR, Lopez-Otin C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt AN, Valdes-Mas R, van Buuren MM, Veer L, Vincent-Salomon A, Waddell N, Australian Pancreatic Cancer Genome I, Consortium IBC, Consortium IM-S. Yates LR, PedBrain I, Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, Biedrzycki B, Donehower RC, Zaheer A, Fisher GA, Crocenzi TS, Lee JJ, Duffy SM, Goldberg RM, de la Chapelle A, Koshiji M, Bhaijee F, Huebner T, Hruban RH, Wood LD, Cuka N, Pardoll DM, Papadopoulos N, Kinzler KW, Zhou S, Cornish TC, Taube JM, Anders RA, Eshleman JR, Vogelstein B, Diaz LA., Jr PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 126.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017;377:2500–2501. doi: 10.1056/NEJMc1713444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kelderman S, Schumacher TN, Kvistborg P. Mismatch repair-deficient cancers are targets for anti-PD-1 therapy. Cancer Cell. 2015;28:11–13. doi: 10.1016/j.ccell.2015.06.012. [DOI] [PubMed] [Google Scholar]
- 129.Howitt BE, Shukla SA, Sholl LM, Ritterhouse LL, Watkins JC, Rodig S, Stover E, Strickland KC, D’Andrea AD, Wu CJ, Matulonis UA, Konstantinopoulos PA. Association of polymerase e-mutated and microsatellite-instable endometrial cancers with neoantigen load, number of tumor-infiltrating lymphocytes, and expression of PD-1 and PD-L1. JAMA Oncol. 2015;1:1319–1323. doi: 10.1001/jamaoncol.2015.2151. [DOI] [PubMed] [Google Scholar]
- 130.Johanns TM, Miller CA, Dorward IG, Tsien C, Chang E, Perry A, Uppaluri R, Ferguson C, Schmidt RE, Dahiya S, Ansstas G, Mardis ER, Dunn GP. Immunogenomics of hypermutated glioblastoma: a patient with germline POLE deficiency treated with checkpoint blockade immunotherapy. Cancer Discov. 2016;6:1230–1236. doi: 10.1158/2159-8290.CD-16-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Clarke B, Tinker AV, Lee CH, Subramanian S, van de Rijn M, Turbin D, Kalloger S, Han G, Ceballos K, Cadungog MG, Huntsman DG, Coukos G, Gilks CB. Intraepithelial T cells and prognosis in ovarian carcinoma: novel associations with stage, tumor type, and BRCA1 loss. Mod Pathol. 2009;22:393–402. doi: 10.1038/modpathol.2008.191. [DOI] [PubMed] [Google Scholar]
- 132.Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, Berent-Maoz B, Pang J, Chmielowski B, Cherry G, Seja E, Lomeli S, Kong X, Kelley MC, Sosman JA, Johnson DB, Ribas A, Lo RS. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.McAlpine JN, Porter H, Kobel M, Nelson BH, Prentice LM, Kalloger SE, Senz J, Milne K, Ding J, Shah SP, Huntsman DG, Gilks CB. BRCA1 and BRCA2 mutations correlate with TP53 abnormalities and presence of immune cell infiltrates in ovarian high-grade serous carcinoma. Mod Pathol. 2012;25:740–750. doi: 10.1038/modpathol.2011.211. [DOI] [PubMed] [Google Scholar]
- 134.Strickland KC, Howitt BE, Shukla SA, Rodig S, Ritterhouse LL, Liu JF, Garber JE, Chowdhury D, Wu CJ, D’Andrea AD, Matulonis UA, Konstantinopoulos PA. Association and prognostic significance of BRCA1/2-mutation status with neoantigen load, number of tumor-infiltrating lymphocytes and expression of PD-1/PD-L1 in high grade serous ovarian cancer. Oncotarget. 2016;7:13587–13598. doi: 10.18632/oncotarget.7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Teo MY, Seier K, Ostrovnaya I, Regazzi AM, Kania BE, Moran MM, Cipolla CK, Bluth MJ, Chaim J, Al-Ahmadie H, Snyder A, Carlo MI, Solit DB, Berger MF, Funt S, Wolchok JD, Iyer G, Bajorin DF, Callahan MK, Rosenberg JE. Alterations in DNA damage response and repair genes as potential marker of clinical benefit from PD-1/PD-L1 blockade in advanced urothelial cancers. J Clin Oncol. 2018;36:1685–1694. doi: 10.1200/JCO.2017.75.7740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kennedy RD, D’Andrea AD. DNA repair pathways in clinical practice: lessons from pediatric cancer susceptibility syndromes. J Clin Oncol. 2006;24:3799–3808. doi: 10.1200/JCO.2005.05.4171. [DOI] [PubMed] [Google Scholar]
- 137.Ceccaldi R, Liu JC, Amunugama R, Hajdu I, Primack B, Petalcorin MI, O’Connor KW, Konstantinopoulos PA, Elledge SJ, Boulton SJ, Yusufzai T, D’Andrea AD. Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature. 2015;518:258–262. doi: 10.1038/nature14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Boulton SJ, Jackson SP. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 1996;15:5093–5103. doi: 10.1002/j.1460-2075.1996.tb00890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Schimmel J, Kool H, van Schendel R, Tijsterman M. Mutational signatures of non-homologous and polymerase theta-mediated end-joining in embryonic stem cells. EMBO J. 2017;36:3634–3649. doi: 10.15252/embj.201796948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS, Wong F, Azad NS, Rucki AA, Laheru D, Donehower R, Zaheer A, Fisher GA, Crocenzi TS, Lee JJ, Greten TF, Duffy AG, Ciombor KK, Eyring AD, Lam BH, Joe A, Kang SP, Holdhoff M, Danilova L, Cope L, Meyer C, Zhou S, Goldberg RM, Armstrong DK, Bever KM, Fader AN, Taube J, Housseau F, Spetzler D, Xiao N, Pardoll DM, Papadopoulos N, Kinzler KW, Eshleman JR, Vogelstein B, Anders RA, Diaz LA., Jr Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Anand S, Penrhyn-Lowe S, Venkitaraman AR. AURORA-A amplification overrides the mitotic spindle assembly checkpoint, inducing resistance to Taxol. Cancer Cell. 2003;3:51–62. doi: 10.1016/S1535-6108(02)00235-0. [DOI] [PubMed] [Google Scholar]
- 142.Hernando E, Nahle Z, Juan G, Diaz-Rodriguez E, Alaminos M, Hemann M, Michel L, Mittal V, Gerald W, Benezra R, Lowe SW, Cordon-Cardo C. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 2004;430:797–802. doi: 10.1038/nature02820. [DOI] [PubMed] [Google Scholar]
- 143.Saldivar JC, Hamperl S, Bocek MJ, Chung M, Bass TE, Cisneros-Soberanis F, Samejima K, Xie L, Paulson JR, Earnshaw WC, Cortez D, Meyer T, Cimprich KA. An intrinsic S/G2 checkpoint enforced by ATR. Science. 2018;361:806–810. doi: 10.1126/science.aap9346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bakhoum SF, Landau DA. Chromosomal instability as a driver of tumor heterogeneity and evolution. Cold Spring Harb Perspect Med. 2017;7:a029611. doi: 10.1101/cshperspect.a029611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jallepalli PV, Lengauer C. Chromosome segregation and cancer: cutting through the mystery. Nat Rev Cancer. 2001;1:109–117. doi: 10.1038/35101065. [DOI] [PubMed] [Google Scholar]
- 146.Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Harbst K, Lauss M, Cirenajwis H, Isaksson K, Rosengren F, Torngren T, Kvist A, Johansson MC, Vallon-Christersson J, Baldetorp B, Borg A, Olsson H, Ingvar C, Carneiro A, Jonsson G. Multiregion whole-exome sequencing uncovers the genetic evolution and mutational heterogeneity of early-stage metastatic melanoma. Cancer Res. 2016;76:4765–4774. doi: 10.1158/0008-5472.CAN-15-3476. [DOI] [PubMed] [Google Scholar]
- 148.Cleary AS, Leonard TL, Gestl SA, Gunther EJ. Tumour cell heterogeneity maintained by cooperating subclones in Wnt-driven mammary cancers. Nature. 2014;508:113–117. doi: 10.1038/nature13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.de Bruin EC, McGranahan N, Mitter R, Salm M, Wedge DC, Yates L, Jamal-Hanjani M, Shafi S, Murugaesu N, Rowan AJ, Gronroos E, Muhammad MA, Horswell S, Gerlinger M, Varela I, Jones D, Marshall J, Voet T, Van Loo P, Rassl DM, Rintoul RC, Janes SM, Lee SM, Forster M, Ahmad T, Lawrence D, Falzon M, Capitanio A, Harkins TT, Lee CC, Tom W, Teefe E, Chen SC, Begum S, Rabinowitz A, Phillimore B, Spencer-Dene B, Stamp G, Szallasi Z, Matthews N, Stewart A, Campbell P, Swanton C. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346:251–256. doi: 10.1126/science.1253462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mandal R, Chan TA. Personalized oncology meets immunology: the path toward precision immunotherapy. Cancer Discov. 2016;6:703–713. doi: 10.1158/2159-8290.CD-16-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Spranger S, Spaapen RM, Zha Y, Williams J, Meng Y, Ha TT, Gajewski TF. Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci Transl Med. 2013;5:200ra116. doi: 10.1126/scitranslmed.3006504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Peng W, Chen JQ, Liu C, Malu S, Creasy C, Tetzlaff MT, Xu C, McKenzie JA, Zhang C, Liang X, Williams LJ, Deng W, Chen G, Mbofung R, Lazar AJ, Torres-Cabala CA, Cooper ZA, Chen PL, Tieu TN, Spranger S, Yu X, Bernatchez C, Forget MA, Haymaker C, Amaria R, McQuade JL, Glitza IC, Cascone T, Li HS, Kwong LN, Heffernan TP, Hu J, Bassett RL, Jr, Bosenberg MW, Woodman SE, Overwijk WW, Lizee G, Roszik J, Gajewski TF, Wargo JA, Gershenwald JE, Radvanyi L, Davies MA, Hwu P. Loss of PTEN promotes resistance to T cell-mediated immunotherapy. Cancer Discov. 2016;6:202–216. doi: 10.1158/2159-8290.CD-15-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Akbay EA, Koyama S, Carretero J, Altabef A, Tchaicha JH, Christensen CL, Mikse OR, Cherniack AD, Beauchamp EM, Pugh TJ, Wilkerson MD, Fecci PE, Butaney M, Reibel JB, Soucheray M, Cohoon TJ, Janne PA, Meyerson M, Hayes DN, Shapiro GI, Shimamura T, Sholl LM, Rodig SJ, Freeman GJ, Hammerman PS, Dranoff G, Wong KK. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov. 2013;3:1355–1363. doi: 10.1158/2159-8290.CD-13-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Anagnostou V, Smith KN, Forde PM, Niknafs N, Bhattacharya R, White J, Zhang T, Adleff V, Phallen J, Wali N, Hruban C, Guthrie VB, Rodgers K, Naidoo J, Kang H, Sharfman W, Georgiades C, Verde F, Illei P, Li QK, Gabrielson E, Brock MV, Zahnow CA, Baylin SB, Scharpf RB, Brahmer JR, Karchin R, Pardoll DM, Velculescu VE. Evolution of neoantigen landscape during immune checkpoint blockade in non-small cell lung cancer. Cancer Discov. 2017;7:264–276. doi: 10.1158/2159-8290.CD-16-0828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, Jamal-Hanjani M, Wilson GA, Birkbak NJ, Hiley CT, Watkins TB, Shafi S, Murugaesu N, Mitter R, Akarca AU, Linares J, Marafioti T, Henry JY, Van Allen EM, Miao D, Schilling B, Schadendorf D, Garraway LA, Makarov V, Rizvi NA, Snyder A, Hellmann MD, Merghoub T, Wolchok JD, Shukla SA, Wu CJ, Peggs KS, Chan TA, Hadrup SR, Quezada SA, Swanton C. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Takeda K, Nakayama M, Hayakawa Y, Kojima Y, Ikeda H, Imai N, Ogasawara K, Okumura K, Thomas DM, Smyth MJ. IFN-gamma is required for cytotoxic T cell-dependent cancer genome immunoediting. Nat Commun. 2017;8:14607. doi: 10.1038/ncomms14607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Davoli T, Uno H, Wooten EC, Elledge SJ. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science. 2017;355:eaaf8399. doi: 10.1126/science.aaf8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Mackenzie KJ, Carroll P, Martin CA, Murina O, Fluteau A, Simpson DJ, Olova N, Sutcliffe H, Rainger JK, Leitch A, Osborn RT, Wheeler AP, Nowotny M, Gilbert N, Chandra T, Reijns MAM, Jackson AP. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature. 2017;548:461–465. doi: 10.1038/nature23449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Klarquist J, Hennies CM, Lehn MA, Reboulet RA, Feau S, Janssen EM. STING-mediated DNA sensing promotes antitumor and autoimmune responses to dying cells. J Immunol. 2014;193:6124–6134. doi: 10.4049/jimmunol.1401869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Woo SR, Corrales L, Gajewski TF. Innate immune recognition of cancer. Annu Rev Immunol. 2015;33:445–474. doi: 10.1146/annurev-immunol-032414-112043. [DOI] [PubMed] [Google Scholar]
- 164.Dunphy G, Flannery SM, Almine JF, Connolly DJ, Paulus C, Jonsson KL, Jakobsen MR, Nevels MM, Bowie AG, Unterholzner L. Non-canonical activation of the DNA sensing adaptor STING by ATM and IFI16 mediates NF-kappaB signaling after nuclear DNA damage. Mol Cell. 2018;71(745–760):e5. doi: 10.1016/j.molcel.2018.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chabanon RM, Muirhead G, Krastev DB, Adam J, Morel D, Garrido M, Lamb A, Henon C, Dorvault N, Rouanne M, Marlow R, Bajrami I, Cardenosa ML, Konde A, Besse B, Ashworth A, Pettitt SJ, Haider S, Marabelle A, Tutt AN, Soria JC, Lord CJ, Postel-Vinay S. PARP inhibition enhances tumor cell-intrinsic immunity in ERCC1-deficient non-small cell lung cancer. J Clin Investig. 2019;129:1211–1228. doi: 10.1172/JCI123319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Pantelidou C, Sonzogni O, De Oliveria Taveira M, Mehta AK, Kothari A, Wang D, Visal T, Li MK, Pinto J, Castrillon JA, Cheney EM, Bouwman P, Jonkers J, Rottenberg S, Guerriero JL, Wulf GM, Shapiro GI. PARP inhibitor efficacy depends on CD8(+) T-cell recruitment via intratumoral STING pathway activation in BRCA-deficient models of triple-negative breast cancer. Cancer Discov. 2019;9:722–737. doi: 10.1158/2159-8290.CD-18-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Jiao S, Xia W, Yamaguchi H, Wei Y, Chen MK, Hsu JM, Hsu JL, Yu WH, Du Y, Lee HH, Li CW, Chou CK, Lim SO, Chang SS, Litton J, Arun B, Hortobagyi GN, Hung MC. PARP inhibitor upregulates PD-L1 expression and enhances cancer-associated immunosuppression. Clin Cancer Res. 2017;23:3711–3720. doi: 10.1158/1078-0432.CCR-16-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sen T, Rodriguez BL, Chen L, Corte CMD, Morikawa N, Fujimoto J, Cristea S, Nguyen T, Diao L, Li L, Fan Y, Yang Y, Wang J, Glisson BS, Wistuba II, Sage J, Heymach JV, Gibbons DL, Byers LA. Targeting DNA damage response promotes antitumor immunity through STING-mediated T-cell activation in small cell lung cancer. Cancer Discov. 2019;9:646–661. doi: 10.1158/2159-8290.CD-18-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhang Q, Green MD, Lang X, Lazarus J, Parsels JD, Wei S, Parsels LA, Shi J, Ramnath N, Wahl DR, Pasca di Magliano M, Frankel TL, Kryczek I, Lei YL, Lawrence TS, Zou W, Morgan MA. Inhibition of ATM increases interferon signaling and sensitizes pancreatic cancer to immune checkpoint blockade therapy. Cancer Res. 2019;79(15):3940–3951. doi: 10.1158/0008-5472.CAN-19-0761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Soria G, Polo SE, Almouzni G. Prime, repair, restore: the active role of chromatin in the DNA damage response. Mol Cell. 2012;46:722–734. doi: 10.1016/j.molcel.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 171.Luijsterburg MS, de Krijger I, Wiegant WW, Shah RG, Smeenk G, de Groot AJL, Pines A, Vertegaal ACO, Jacobs JJL, Shah GM, van Attikum H. PARP1 links CHD2-mediated chromatin expansion and H3.3 deposition to DNA repair by non-homologous end-joining. Mol Cell. 2016;61:547–562. doi: 10.1016/j.molcel.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Downs JA, Allard S, Jobin-Robitaille O, Javaheri A, Auger A, Bouchard N, Kron SJ, Jackson SP, Cote J. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol Cell. 2004;16:979–990. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 173.Jha S, Shibata E, Dutta A. Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol Cell Biol. 2008;28:2690–2700. doi: 10.1128/MCB.01983-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR, 3rd, Abmayr SM, Washburn MP, Workman JL. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 175.Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8:91–99. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- 176.Kruhlak MJ, Celeste A, Dellaire G, Fernandez-Capetillo O, Muller WG, McNally JG, Bazett-Jones DP, Nussenzweig A. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol. 2006;172:823–834. doi: 10.1083/jcb.200510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Mattiroli F, Vissers JH, van Dijk WJ, Ikpa P, Citterio E, Vermeulen W, Marteijn JA, Sixma TK. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell. 2012;150:1182–1195. doi: 10.1016/j.cell.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 178.Thorslund T, Ripplinger A, Hoffmann S, Wild T, Uckelmann M, Villumsen B, Narita T, Sixma TK, Choudhary C, Bekker-Jensen S, Mailand N. Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage. Nature. 2015;527:389–393. doi: 10.1038/nature15401. [DOI] [PubMed] [Google Scholar]
- 179.Fradet-Turcotte A, Canny MD, Escribano-Diaz C, Orthwein A, Leung CC, Huang H, Landry MC, Kitevski-LeBlanc J, Noordermeer SM, Sicheri F, Durocher D. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature. 2013;499:50–54. doi: 10.1038/nature12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Gatti M, Pinato S, Maspero E, Soffientini P, Polo S, Penengo L. A novel ubiquitin mark at the N-terminal tail of histone H2As targeted by RNF168 ubiquitin ligase. Cell Cycle. 2012;11:2538–2544. doi: 10.4161/cc.20919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 182.Li F, Mao G, Tong D, Huang J, Gu L, Yang W, Li GM. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. Cell. 2013;153:590–600. doi: 10.1016/j.cell.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Fnu S, Williamson EA, De Haro LP, Brenneman M, Wray J, Shaheen M, Radhakrishnan K, Lee SH, Nickoloff JA, Hromas R. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc Natl Acad Sci USA. 2011;108:540–545. doi: 10.1073/pnas.1013571108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Towbin BD, Gonzalez-Sandoval A, Gasser SM. Mechanisms of heterochromatin subnuclear localization. Trends Biochem Sci. 2013;38:356–363. doi: 10.1016/j.tibs.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 185.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Rowbotham SP, Barki L, Neves-Costa A, Santos F, Dean W, Hawkes N, Choudhary P, Will WR, Webster J, Oxley D, Green CM, Varga-Weisz P, Mermoud JE. Maintenance of silent chromatin through replication requires SWI/SNF-like chromatin remodeler SMARCAD1. Mol Cell. 2011;42:285–296. doi: 10.1016/j.molcel.2011.02.036. [DOI] [PubMed] [Google Scholar]
- 187.Goodarzi AA, Kurka T, Jeggo PA. KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat Struct Mol Biol. 2011;18:831–839. doi: 10.1038/nsmb.2077. [DOI] [PubMed] [Google Scholar]
- 188.Iyengar S, Ivanov AV, Jin VX, Rauscher FJ, 3rd, Farnham PJ. Functional analysis of KAP1 genomic recruitment. Mol Cell Biol. 2011;31:1833–1847. doi: 10.1128/MCB.01331-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Tsouroula K, Furst A, Rogier M, Heyer V, Maglott-Roth A, Ferrand A, Reina-San-Martin B, Soutoglou E. Temporal and spatial uncoupling of DNA double strand break repair pathways within mammalian heterochromatin. Mol Cell. 2016;63:293–305. doi: 10.1016/j.molcel.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 190.Chiolo I, Minoda A, Colmenares SU, Polyzos A, Costes SV, Karpen GH. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 2011;144:732–744. doi: 10.1016/j.cell.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ryu T, Spatola B, Delabaere L, Bowlin K, Hopp H, Kunitake R, Karpen GH, Chiolo I. Heterochromatic breaks move to the nuclear periphery to continue recombinational repair. Nat Cell Biol. 2015;17:1401–1411. doi: 10.1038/ncb3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Brooks SC, Adhikary S, Rubinson EH, Eichman BF. Recent advances in the structural mechanisms of DNA glycosylases. Biochim Biophys Acta. 2013;1834:247–271. doi: 10.1016/j.bbapap.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Robertson AB, Klungland A, Rognes T, Leiros I. DNA repair in mammalian cells: base excision repair: the long and short of it. Cell Mol Life Sci. 2009;66:981–993. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Lieber MR. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. BioEssays. 1997;19:233–240. doi: 10.1002/bies.950190309. [DOI] [PubMed] [Google Scholar]
- 196.Amouroux R, Campalans A, Epe B, Radicella JP. Oxidative stress triggers the preferential assembly of base excision repair complexes on open chromatin regions. Nucleic Acids Res. 2010;38:2878–2890. doi: 10.1093/nar/gkp1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Menoni H, Gasparutto D, Hamiche A, Cadet J, Dimitrov S, Bouvet P, Angelov D. ATP-dependent chromatin remodeling is required for base excision repair in conventional but not in variant H2A.Bbd nucleosomes. Mol Cell Biol. 2007;27:5949–5956. doi: 10.1128/MCB.00376-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Menoni H, Shukla MS, Gerson V, Dimitrov S, Angelov D. Base excision repair of 8-oxoG in dinucleosomes. Nucleic Acids Res. 2012;40:692–700. doi: 10.1093/nar/gkr761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Nakanishi S, Prasad R, Wilson SH, Smerdon M. Different structural states in oligonucleosomes are required for early versus late steps of base excision repair. Nucleic Acids Res. 2007;35:4313–4321. doi: 10.1093/nar/gkm436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Scharer OD. Nucleotide excision repair in eukaryotes. Cold Spring Harb Perspect Biol. 2013;5:a012609. doi: 10.1101/cshperspect.a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Gaillard H, Fitzgerald DJ, Smith CL, Peterson CL, Richmond TJ, Thoma F. Chromatin remodeling activities act on UV-damaged nucleosomes and modulate DNA damage accessibility to photolyase. J Biol Chem. 2003;278:17655–17663. doi: 10.1074/jbc.M300770200. [DOI] [PubMed] [Google Scholar]
- 202.Hara R, Sancar A. The SWI/SNF chromatin-remodeling factor stimulates repair by human excision nuclease in the mononucleosome core particle. Mol Cell Biol. 2002;22:6779–6787. doi: 10.1128/MCB.22.19.6779-6787.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Zhao Q, Wang QE, Ray A, Wani G, Han C, Milum K, Wani AA. Modulation of nucleotide excision repair by mammalian SWI/SNF chromatin-remodeling complex. J Biol Chem. 2009;284:30424–30432. doi: 10.1074/jbc.M109.044982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Kothandapani A, Gopalakrishnan K, Kahali B, Reisman D, Patrick SM. Downregulation of SWI/SNF chromatin remodeling factor subunits modulates cisplatin cytotoxicity. Exp Cell Res. 2012;318:1973–1986. doi: 10.1016/j.yexcr.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci USA. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Dorighi KM, Swigut T, Henriques T, Bhanu NV, Scruggs BS, Nady N, Still CD, 2nd, Garcia BA, Adelman K, Wysocka J. Mll3 and Mll4 facilitate enhancer RNA synthesis and transcription from promoters independently of H3K4 monomethylation. Mol Cell. 2017;66(568–576):e4. doi: 10.1016/j.molcel.2017.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Rickels R, Herz HM, Sze CC, Cao K, Morgan MA, Collings CK, Gause M, Takahashi YH, Wang L, Rendleman EJ, Marshall SA, Krueger A, Bartom ET, Piunti A, Smith ER, Abshiru NA, Kelleher NL, Dorsett D, Shilatifard A. Histone H3K4 monomethylation catalyzed by Trr and mammalian COMPASS-like proteins at enhancers is dispensable for development and viability. Nat Genet. 2017;49:1647–1653. doi: 10.1038/ng.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Rampias T, Karagiannis D, Avgeris M, Polyzos A, Kokkalis A, Kanaki Z, Kousidou E, Tzetis M, Kanavakis E, Stravodimos K, Manola KN, Pantelias GE, Scorilas A, Klinakis A (2019) The lysine-specific methyltransferase KMT2C/MLL3 regulates DNA repair components in cancer. EMBO Rep. 10.15252/embr.201846821 [DOI] [PMC free article] [PubMed]
- 209.Ray Chaudhuri A, Callen E, Ding X, Gogola E, Duarte AA, Lee JE, Wong N, Lafarga V, Calvo JA, Panzarino NJ, John S, Day A, Crespo AV, Shen B, Starnes LM, de Ruiter JR, Daniel JA, Konstantinopoulos PA, Cortez D, Cantor SB, Fernandez-Capetillo O, Ge K, Jonkers J, Rottenberg S, Sharan SK, Nussenzweig A. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature. 2016;535:382–387. doi: 10.1038/nature18325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hashimoto Y, Ray Chaudhuri A, Lopes M, Costanzo V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat Struct Mol Biol. 2010;17:1305–1311. doi: 10.1038/nsmb.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–979. doi: 10.1016/S1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 212.Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Taglialatela A, Alvarez S, Leuzzi G, Sannino V, Ranjha L, Huang JW, Madubata C, Anand R, Levy B, Rabadan R, Cejka P, Costanzo V, Ciccia A. Restoration of replication fork stability in BRCA1- and BRCA2-deficient cells by inactivation of SNF2-family fork remodelers. Mol Cell. 2017;68(414–430):e8. doi: 10.1016/j.molcel.2017.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Nichol JN, Dupere-Richer D, Ezponda T, Licht JD, Miller WH., Jr H3K27 methylation: a focal point of epigenetic deregulation in cancer. Adv Cancer Res. 2016;131:59–95. doi: 10.1016/bs.acr.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Ezponda T, Licht JD. Molecular pathways: deregulation of histone h3 lysine 27 methylation in cancer-different paths, same destination. Clin Cancer Res. 2014;20:5001–5008. doi: 10.1158/1078-0432.CCR-13-2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, Bergman Y, Simon I, Cedar H. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 218.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, Bollen M, Esteller M, Di Croce L, de Launoit Y, Fuks F. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 219.Simons CC, Hughes LA, Smits KM, Khalid-de Bakker CA, de Bruine AP, Carvalho B, Meijer GA, Schouten LJ, van den Brandt PA, Weijenberg MP, van Engeland M. A novel classification of colorectal tumors based on microsatellite instability, the CpG island methylator phenotype and chromosomal instability: implications for prognosis. Ann Oncol. 2013;24:2048–2056. doi: 10.1093/annonc/mdt076. [DOI] [PubMed] [Google Scholar]
- 220.Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- 221.Arai M, Utsunomiya J, Miki Y. Familial breast and ovarian cancers. Int J Clin Oncol. 2004;9:270–282. doi: 10.1007/s10147-004-0423-3. [DOI] [PubMed] [Google Scholar]
- 222.Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer. 2004;4:814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 223.Harada H, Miyamoto K, Yamashita Y, Nakano K, Taniyama K, Miyata Y, Ohdan H, Okada M. Methylation of breast cancer susceptibility gene 1 (BRCA1) predicts recurrence in patients with curatively resected stage I non-small cell lung cancer. Cancer. 2013;119:792–798. doi: 10.1002/cncr.27754. [DOI] [PubMed] [Google Scholar]
- 224.Yu J, Zhu T, Wang Z, Zhang H, Qian Z, Xu H, Gao B, Wang W, Gu L, Meng J, Wang J, Feng X, Li Y, Yao X, Zhu J. A novel set of DNA methylation markers in urine sediments for sensitive/specific detection of bladder cancer. Clin Cancer Res. 2007;13:7296–7304. doi: 10.1158/1078-0432.CCR-07-0861. [DOI] [PubMed] [Google Scholar]
- 225.Lee MN, Tseng RC, Hsu HS, Chen JY, Tzao C, Ho WL, Wang YC. Epigenetic inactivation of the chromosomal stability control genes BRCA1, BRCA2, and XRCC5 in non-small cell lung cancer. Clin Cancer Res. 2007;13:832–838. doi: 10.1158/1078-0432.CCR-05-2694. [DOI] [PubMed] [Google Scholar]
- 226.Ai L, Vo QN, Zuo C, Li L, Ling W, Suen JY, Hanna E, Brown KD, Fan CY. Ataxia-telangiectasia-mutated (ATM) gene in head and neck squamous cell carcinoma: promoter hypermethylation with clinical correlation in 100 cases. Cancer Epidemiol Biomark Prev. 2004;13:150–156. doi: 10.1158/1055-9965.EPI-082-3. [DOI] [PubMed] [Google Scholar]
- 227.Vo QN, Kim WJ, Cvitanovic L, Boudreau DA, Ginzinger DG, Brown KD. The ATM gene is a target for epigenetic silencing in locally advanced breast cancer. Oncogene. 2004;23:9432–9437. doi: 10.1038/sj.onc.1208092. [DOI] [PubMed] [Google Scholar]
- 228.Marsit CJ, Liu M, Nelson HH, Posner M, Suzuki M, Kelsey KT. Inactivation of the Fanconi anemia/BRCA pathway in lung and oral cancers: implications for treatment and survival. Oncogene. 2004;23:1000–1004. doi: 10.1038/sj.onc.1207256. [DOI] [PubMed] [Google Scholar]
- 229.Wang Z, Li M, Lu S, Zhang Y, Wang H. Promoter hypermethylation of FANCF plays an important role in the occurrence of ovarian cancer through disrupting Fanconi anemia-BRCA pathway. Cancer Biol Ther. 2006;5:256–260. doi: 10.4161/cbt.5.3.2380. [DOI] [PubMed] [Google Scholar]
- 230.Lim SL, Smith P, Syed N, Coens C, Wong H, van der Burg M, Szlosarek P, Crook T, Green JA. Promoter hypermethylation of FANCF and outcome in advanced ovarian cancer. Br J Cancer. 2008;98:1452–1456. doi: 10.1038/sj.bjc.6604325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Mijnes J, Veeck J, Gaisa NT, Burghardt E, de Ruijter TC, Gostek S, Dahl E, Pfister D, Schmid SC, Knuchel R, Rose M. Promoter methylation of DNA damage repair (DDR) genes in human tumor entities: RBBP8/CtIP is almost exclusively methylated in bladder cancer. Clin Epigenet. 2018;10:15. doi: 10.1186/s13148-018-0447-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Kondrashova O, Topp M, Nesic K, Lieschke E, Ho GY, Harrell MI, Zapparoli GV, Hadley A, Holian R, Boehm E, Heong V, Sanij E, Pearson RB, Krais JJ, Johnson N, McNally O, Ananda S, Alsop K, Hutt KJ, Kaufmann SH, Lin KK, Harding TC, Traficante N, deFazio A, McNeish IA, Bowtell DD, Swisher EM, Dobrovic A, Wakefield MJ, Scott CL. Methylation of all BRCA1 copies predicts response to the PARP inhibitor rucaparib in ovarian carcinoma. Nat Commun. 2018;9:3970. doi: 10.1038/s41467-018-05564-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Veeck J, Ropero S, Setien F, Gonzalez-Suarez E, Osorio A, Benitez J, Herman JG, Esteller M. BRCA1 CpG island hypermethylation predicts sensitivity to poly(adenosine diphosphate)-ribose polymerase inhibitors. J Clin Oncol. 2010;28:e563–e564. doi: 10.1200/JCO.2010.30.1010. [DOI] [PubMed] [Google Scholar]
- 234.Orta ML, Hoglund A, Calderon-Montano JM, Dominguez I, Burgos-Moron E, Visnes T, Pastor N, Strom C, Lopez-lazaro M, Helleday T. The PARP inhibitor Olaparib disrupts base excision repair of 5-aza-2′-deoxycytidine lesions. Nucleic Acids Res. 2014;42:9108–9120. doi: 10.1093/nar/gku638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Pulliam N, Fang F, Ozes AR, Tang J, Adewuyi A, Keer H, Lyons J, Baylin SB, Matei D, Nakshatri H, Rassool FV, Miller KD, Nephew KP. An effective epigenetic-PARP inhibitor combination therapy for breast and ovarian cancers independent of BRCA mutations. Clin Cancer Res. 2018;24:3163–3175. doi: 10.1158/1078-0432.CCR-18-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ghoshal K, Datta J, Majumder S, Bai S, Kutay H, Motiwala T, Jacob ST. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol Cell Biol. 2005;25:4727–4741. doi: 10.1128/MCB.25.11.4727-4741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 237.Stresemann C, Brueckner B, Musch T, Stopper H, Lyko F. Functional diversity of DNA methyltransferase inhibitors in human cancer cell lines. Cancer Res. 2006;66:2794–2800. doi: 10.1158/0008-5472.CAN-05-2821. [DOI] [PubMed] [Google Scholar]
- 238.Muvarak NE, Chowdhury K, Xia L, Robert C, Choi EY, Cai Y, Bellani M, Zou Y, Singh ZN, Duong VH, Rutherford T, Nagaria P, Bentzen SM, Seidman MM, Baer MR, Lapidus RG, Baylin SB, Rassool FV. Enhancing the cytotoxic effects of PARP inhibitors with DNA demethylating agents—a potential therapy for cancer. Cancer Cell. 2016;30:637–650. doi: 10.1016/j.ccell.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Chang CJ, Yang JY, Xia W, Chen CT, Xie X, Chao CH, Woodward WA, Hsu JM, Hortobagyi GN, Hung MC. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-beta-catenin signaling. Cancer Cell. 2011;19:86–100. doi: 10.1016/j.ccr.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Zeidler M, Varambally S, Cao Q, Chinnaiyan AM, Ferguson DO, Merajver SD, Kleer CG. The Polycomb group protein EZH2 impairs DNA repair in breast epithelial cells. Neoplasia. 2005;7:1011–1019. doi: 10.1593/neo.05472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Clark MD, Marcum R, Graveline R, Chan CW, Xie T, Chen Z, Ding Y, Zhang Y, Mondragon A, David G, Radhakrishnan I. Structural insights into the assembly of the histone deacetylase-associated Sin3L/Rpd3L corepressor complex. Proc Natl Acad Sci USA. 2015;112:E3669–E3678. doi: 10.1073/pnas.1421784112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Hudson GM, Watson PJ, Fairall L, Jamieson AG, Schwabe JW. Insights into the recruitment of class IIa histone deacetylases (HDACs) to the SMRT/NCoR transcriptional repression complex. J Biol Chem. 2015;290:18237–18244. doi: 10.1074/jbc.M115.661058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Gui CY, Ngo L, Xu WS, Richon VM, Marks PA. Histone deacetylase (HDAC) inhibitor activation of p21WAF1 involves changes in promoter-associated proteins, including HDAC1. Proc Natl Acad Sci USA. 2004;101:1241–1246. doi: 10.1073/pnas.0307708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Huang BH, Laban M, Leung CH, Lee L, Lee CK, Salto-Tellez M, Raju GC, Hooi SC. Inhibition of histone deacetylase 2 increases apoptosis and p21Cip1/WAF1 expression, independent of histone deacetylase 1. Cell Death Differ. 2005;12:395–404. doi: 10.1038/sj.cdd.4401567. [DOI] [PubMed] [Google Scholar]
- 245.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Chao OS, Goodman OB., Jr Synergistic loss of prostate cancer cell viability by coinhibition of HDAC and PARP. Mol Cancer Res. 2014;12:1755–1766. doi: 10.1158/1541-7786.MCR-14-0173. [DOI] [PubMed] [Google Scholar]
- 247.Min A, Im SA, Kim DK, Song SH, Kim HJ, Lee KH, Kim TY, Han SW, Oh DY, Kim TY, O’Connor MJ, Bang YJ. Histone deacetylase inhibitor, suberoylanilide hydroxamic acid (SAHA), enhances anti-tumor effects of the poly (ADP-ribose) polymerase (PARP) inhibitor olaparib in triple-negative breast cancer cells. Breast Cancer Res. 2015;17:33. doi: 10.1186/s13058-015-0534-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Lopez G, Liu J, Ren W, Wei W, Wang S, Lahat G, Zhu QS, Bornmann WG, McConkey DJ, Pollock RE, Lev DC. Combining PCI-24781, a novel histone deacetylase inhibitor, with chemotherapy for the treatment of soft tissue sarcoma. Clin Cancer Res. 2009;15:3472–3483. doi: 10.1158/1078-0432.CCR-08-2714. [DOI] [PubMed] [Google Scholar]
- 249.Maiso P, Colado E, Ocio EM, Garayoa M, Martin J, Atadja P, Pandiella A, San-Miguel JF. The synergy of panobinostat plus doxorubicin in acute myeloid leukemia suggests a role for HDAC inhibitors in the control of DNA repair. Leukemia. 2009;23:2265–2274. doi: 10.1038/leu.2009.182. [DOI] [PubMed] [Google Scholar]
- 250.Munster P, Marchion D, Bicaku E, Schmitt M, Lee JH, DeConti R, Simon G, Fishman M, Minton S, Garrett C, Chiappori A, Lush R, Sullivan D, Daud A. Phase I trial of histone deacetylase inhibition by valproic acid followed by the topoisomerase II inhibitor epirubicin in advanced solid tumors: a clinical and translational study. J Clin Oncol. 2007;25:1979–1985. doi: 10.1200/JCO.2006.08.6165. [DOI] [PubMed] [Google Scholar]
- 251.Ramalingam SS, Kummar S, Sarantopoulos J, Shibata S, LoRusso P, Yerk M, Holleran J, Lin Y, Beumer JH, Harvey RD, Ivy SP, Belani CP, Egorin MJ. Phase I study of vorinostat in patients with advanced solid tumors and hepatic dysfunction: a National Cancer Institute Organ Dysfunction Working Group study. J Clin Oncol. 2010;28:4507–4512. doi: 10.1200/JCO.2010.30.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Ramalingam SS, Parise RA, Ramanathan RK, Lagattuta TF, Musguire LA, Stoller RG, Potter DM, Argiris AE, Zwiebel JA, Egorin MJ, Belani CP. Phase I and pharmacokinetic study of vorinostat, a histone deacetylase inhibitor, in combination with carboplatin and paclitaxel for advanced solid malignancies. Clin Cancer Res. 2007;13:3605–3610. doi: 10.1158/1078-0432.CCR-07-0162. [DOI] [PubMed] [Google Scholar]
- 253.Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, Felletar I, Volkmer R, Muller S, Pawson T, Gingras AC, Arrowsmith CH, Knapp S. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Moriniere J, Rousseaux S, Steuerwald U, Soler-Lopez M, Curtet S, Vitte AL, Govin J, Gaucher J, Sadoul K, Hart DJ, Krijgsveld J, Khochbin S, Muller CW, Petosa C. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature. 2009;461:664–668. doi: 10.1038/nature08397. [DOI] [PubMed] [Google Scholar]
- 255.Donati B, Lorenzini E, Ciarrocchi A. BRD4 and Cancer: going beyond transcriptional regulation. Mol Cancer. 2018;17:164. doi: 10.1186/s12943-018-0915-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Mio C, Gerratana L, Bolis M, Caponnetto F, Zanello A, Barbina M, Di Loreto C, Garattini E, Damante G, Puglisi F. BET proteins regulate homologous recombination-mediated DNA repair: BRCAness and implications for cancer therapy. Int J Cancer. 2019;144:755–766. doi: 10.1002/ijc.31898. [DOI] [PubMed] [Google Scholar]
- 258.Sun C, Yin J, Fang Y, Chen J, Jeong KJ, Chen X, Vellano CP, Ju Z, Zhao W, Zhang D, Lu Y, Meric-Bernstam F, Yap TA, Hattersley M, O’Connor MJ, Chen H, Fawell S, Lin SY, Peng G, Mills GB. BRD4 inhibition is synthetic lethal with PARP inhibitors through the induction of homologous recombination deficiency. Cancer Cell. 2018;33(401–416):e8. doi: 10.1016/j.ccell.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Yang L, Zhang Y, Shan W, Hu Z, Yuan J, Pi J, Wang Y, Fan L, Tang Z, Li C, Hu X, Tanyi JL, Fan Y, Huang Q, Montone K, Dang CV, Zhang L. Repression of BET activity sensitizes homologous recombination-proficient cancers to PARP inhibition. Sci Transl Med. 2017;9:eaal1645. doi: 10.1126/scitranslmed.aal1645. [DOI] [PMC free article] [PubMed] [Google Scholar]