Abstract
The diverse eukaryotic proteins that contain zinc fingers participate in many aspects of nucleic acid metabolism, from DNA transcription to RNA degradation, post-transcriptional gene silencing, and small RNA biogenesis. These proteins can be classified into at least 30 types based on structure. In this review, we focus on the CCHC-type zinc fingers (ZCCHC), which contain an 18-residue domain with the CX2CX4HX4C sequence, where C is cysteine, H is histidine, and X is any amino acid. This motif, also named the “zinc knuckle”, is characteristic of the retroviral Group Antigen protein and occurs alone or with other motifs. Many proteins containing zinc knuckles have been identified in eukaryotes, but only a few have been studied. Here, we review the available information on ZCCHC-containing factors from three evolutionarily distant eukaryotes—Saccharomyces cerevisiae, Arabidopsis thaliana, and Homo sapiens—representing fungi, plants, and metazoans, respectively. We performed systematic searches for proteins containing the CX2CX4HX4C sequence in organism-specific and generalist databases. Next, we analyzed the structural and functional information for all such proteins stored in UniProtKB. Excluding retrotransposon-encoded proteins and proteins harboring uncertain ZCCHC motifs, we found seven ZCCHC-containing proteins in yeast, 69 in Arabidopsis, and 34 in humans. ZCCHC-containing proteins mainly localize to the nucleus, but some are nuclear and cytoplasmic, or exclusively cytoplasmic, and one localizes to the chloroplast. Most of these factors participate in RNA metabolism, including transcriptional elongation, polyadenylation, translation, pre-messenger RNA splicing, RNA export, RNA degradation, microRNA and ribosomal RNA biogenesis, and post-transcriptional gene silencing. Several human ZCCHC-containing factors are derived from neofunctionalized retrotransposons and act as proto-oncogenes in diverse neoplastic processes. The conservation of ZCCHCs in orthologs of these three phylogenetically distant eukaryotes suggests that these domains have biologically relevant functions that are not well known at present.
Electronic supplementary material
The online version of this article (10.1007/s00018-020-03518-7) contains supplementary material, which is available to authorized users.
Keywords: Zinc-knuckle, ZCCHC, RNA metabolism, Yeast, Human, Arabidopsis
Introduction
Zinc fingers are small zinc-ligating domains that make up the largest and most diverse superfamily of nucleic acid-binding proteins in eukaryotes. Zinc fingers stably fold via the coordination of one or more zinc ions (Zn2+) through cysteine (C), histidine (H), and, less frequently, aspartate (D) residues. Zinc finger-containing proteins are classified depending on the sequence and structure of their zinc finger motifs, which contain diverse combinations of C and H [1]. The classical zinc finger is the C2H2 type, which consists of two C and two H residues, and is often described as CX2–4CX12HX2–6H (where X is any amino acid) [2]. The structures of zinc fingers resemble multiple finger-shaped protrusions, through which they bind to their DNA, RNA, or protein targets [3]. The first protein with zinc fingers described was the general transcription factor TFIIIA of Xenopus laevis, a DNA- and RNA-binding protein with nine C2H2 motifs [4]. About 2% of human proteins contain zinc fingers, which have been classified into 30 different types, as described in The Human Genome Organization Gene Nomenclature Committee database (HGNC; https://www.genenames.org/).
As classified in the InterPro database (https://www.ebi.ac.uk/interpro; [5]), zinc knuckles are 18-residue, CCHC-type zinc fingers (ZCCHC) containing the CX2CX4HX4C consensus sequence (IPR025836). ZCCHC-containing factors have two short β-strands connected by a turn (the zinc knuckle) followed by a short helix or loop. The first ZCCHC was found in the murine leukemia virus (MLV) and Rous avian sarcoma virus (RSV) [6], and later in many other Group Antigen (Gag) proteins of retroviral nucleocapsids and eukaryotic retrotransposons [7]. Except for the spumaviruses, all retroviral nucleocapsid proteins have one or two ZCCHCs [8]. The two ZCCHCs of the Human immunodeficiency virus (HIV)-1 Gag protein are considered the ZCCHC prototype and are essential for viral genome packaging and infectivity [9]. Figure 1 shows a representation of the three-dimensional structure of a viral ZCCHC [10], obtained using the Visual Molecular Dynamics program [11].
Fig. 1.
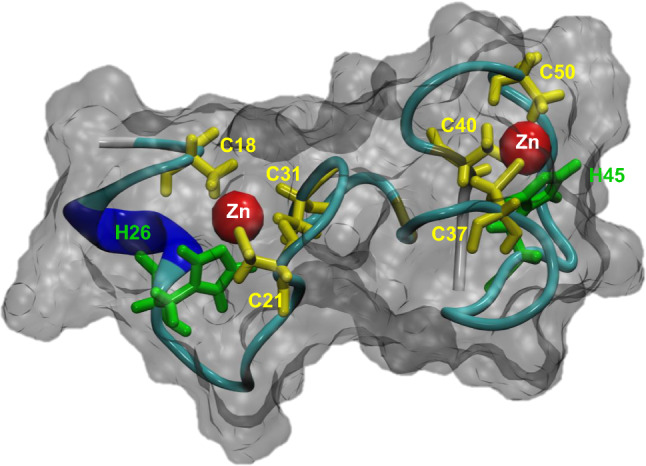
Partial three-dimensional structure of the lentiviral EIAV nucleocapsid protein NCp11. The three-dimensional structure of a peptide of 37 amino acids (QTCYNCGKPGHLSSQCRAPKVCFKCKQPGHFSKQCRS) corresponding to residues 22–58 from the lentiviral EIAV nucleocapsid protein NCp11 (Protein Data Bank identification code for the partial structure: 2BL6) is shown. This peptide contains two ZCCHCs of the CX2CX3GHX4C type, which are underlined in the sequence above, and is complexed with zinc (red spheres). Only the relevant C and H residues of the ZCCHCs are highlighted. The structure shown in this figure was determined by two-dimensional hydrogen isotope nuclear magnetic resonance spectroscopy, as previously described [10]. We obtained this image using the Visual Molecular Dynamics molecular visualization program (https://www.ks.uiuc.edu/Research/vmd/) [11]
ZCCHCs have not been found in bacteria, but a similar motif with residues of three conserved C and one H (CX4CXHX5C) is present in the C-terminal region of the essential Escherichia coli protein YjeQ [12]. YjeQ is a conserved bacterial GTPase that acts as a chaperone, facilitating the assembly of the 30S ribosomal subunit. YjeQ contributes to the folding of the decoding center for mRNA translation and binds the mature 30S subunit through its ZCCHC-like domain [13]. YjeQ depletion causes an accumulation of free 30S and 50S ribosomal subunits, and a reduction in 70S mature ribosomes [12]. YjeQ and its bacterial orthologs are considered promising targets for new antibiotics, since these proteins are required for Staphylococcus aureus to grow in experimental assays with mice (Mus sp.) [14, 15]. ZCCHCs have also been found in the DNA primase of the Archaea Pyrococcus furiosus [16].
Here, we describe our systematic search and analysis of ZCCHC-containing factors from three evolutionarily distant eukaryotes, representing fungi, plants, and metazoa, respectively: Saccharomyces cerevisiae (hereafter, yeast), Arabidopsis thaliana (Arabidopsis), and Homo sapiens (humans). To our knowledge, no exhaustive compilation or comparative analysis of ZCCHC-containing factors in highly distant organisms has been published.
Genome-wide searches for ZCCHC-containing factors
To systematically and comprehensively identify ZCCHC-containing proteins, we carried out two different types of searches for each of the three species under study: the first used the search terms “knuckle” or “CCHC”, and the second used the CX2CX4HX4C consensus sequence. We searched the UniProt Knowledgebase (UniProtKB; https://www.uniprot.org) using “knuckle” or “CCHC” and the name of the organism. Then, we repeated the search in organism-specific databases: the Saccharomyces Genome Database (https://www.yeastgenome.org), and the Arabidopsis Information Resource (TAIR; https://www.arabidopsis.org), and HGNC databases. The main problem that we found was that the results obtained from different searches were incoherent due to incomplete annotations and the presence of many duplicate, fragmented, or obsolete entries. This is likely because the UniProtKB database contains both reviewed (manually annotated) and unreviewed (computationally analyzed) records. For example, using “CCHC” and “Saccharomyces cerevisiae” as search terms on UniProtKB yielded 58 entries, many of which corresponded to the same protein from different yeast strains, and only eight of which were unique and harbored the consensus CX2CX4HX4C sequence. We then searched for the CX2CX4HX4C consensus sequence using the PatMatch pattern-matching program (https://www.yeastgenome.org/nph-patmatch), which can search for short sequences (< 30 amino acids), including ambiguous and degenerate patterns [17].
ZCCHC-containing factors encoded by the yeast genome
Using PatMatch, we obtained 19 unique sequence entries and 32 hits from 5,916 analyzed yeast sequences, several of which harbored more than one CX2CX4HX4C sequence. Eight of the 19 factors harboring a CX2CX4HX4C sequence were Gag proteins from retrotransposons. We next searched for structural information on the remaining 11 proteins (Supporting Dataset 1a) in the UniProtKB database, which compiles the information provided by three programs that predict protein domains: PROSITE (https://prosite.expasy.org), Pfam (https://pfam.xfam.org), and SMART (https://smart.embl.de). No domains or motifs were annotated for YOL029C or YER137C, two proteins of unknown function, and the CX2CX4HX4C sequences of other two proteins (DPOD/POL3 and HEL1) form part of two Cys-rich domains that are larger than that of the ZCCHC (Supporting Dataset 1a). The In Between RING finger (IBR) domains in HEL1 are ~ 50-residue C6HC zinc finger domains (CX4CX14–30CX1–4CX4CX2CX4HX4C; the sequence found with PatMatch is underlined). IBRs have been described in members of some families of E3 ubiquitin-protein ligases, and form a Gag knuckle-like zinc-binding structure [18].
We concluded that the yeast genome encodes at least seven ZCCHC-containing factors, all with a known function; six of these are involved in RNA metabolism, as discussed later (Supporting Dataset 1b; for all yeast amino acid sequences, see Supplementary Fig. 1).
ZCCHC-containing factors encoded by the Arabidopsis genome
Equivalent results to those of yeast were obtained when we used the version of the PatMatch program found in the TAIR10 database (https://www.arabidopsis.org/cgi-bin/patmatch/nph-patmatch.pl), which includes 27,416 Arabidopsis proteins. We found 135 factors with a CX2CX4HX4C sequence; for 53 of these proteins, the CX2CX4HX4C sequence was annotated in UniProtKB to form part of domains or motifs that are not, and are larger than, a ZCCHC (Supporting Dataset 1c), mainly IBRs. Nine of the 82 remaining factors were Gag proteins of retrotransposons, and the other eight harbored a DUF4283 domain of unknown function (Supporting Dataset 1d), which is frequently associated with ZCCHC and with the reverse transcriptase domain, as described in the Pfam database. We classified these factors as putative retrotransposon-derived proteins.
Four other proteins (encoded by the AT3G16350, AT3G31950, AT3G50870, and AT4G08867 genes; Arabidopsis Genome Initiative gene identifiers [19] are used throughout this review) were not annotated as containing a ZCCHC in UniProtKB or Arabidopsis-specific databases, nor were predicted by PROSITE or SMART. In addition, proteins encoded by AT3G50870 and AT4G08867 belong to families without members harboring ZCCHCs, and we considered it very unlikely that their CX2CX4HX4C sequence was a true ZCCHC (Supporting Dataset 1c). However, AT3G16350 and AT3G31950 may encode genuine ZCCHC-containing factors. AT3G16350 encodes a MYB-type transcription factor with high similarity to KUODA1 (KUA1), sharing a very similar CX2CX4HX4C sequence that is annotated in UniProtKB as a ZCCHC. In the Aramemnon database (https://aramemnon.botanik.uni-koeln.de/), AT3G31950 is clustered with four proteins of unknown function (AT4G06479, AT4G06526, AT5G29070, and AT5G34870), the last three of which are annotated as zinc knuckle (CCHC-type) proteins. Therefore, we classified these four proteins as harboring uncertain ZCCHCs and did not include them in our subsequent analyses (Supporting Dataset 1c).
Most of the 61 remaining Arabidopsis factors with a ZCCHC are nuclear proteins involved in RNA metabolism (Supporting Dataset 1d; for all Arabidopsis amino acid sequences, see Supplementary Fig. 2). Many of these are predicted by homology, and only a few have been functionally studied.
ZCCHC-containing factors encoded by the human genome
There are PatMatch versions for yeast and Arabidopsis [17] searches, but none are integrated with human protein databases. Therefore, to search for human zinc knuckle proteins, we used the ScanProsite tool (https://prosite.expasy.org/scanprosite) with the CX2CX4HX4C sequence and “Homo sapiens” as a filter for the taxonomy option. As a result, we found 245 hits in 153 sequences, including isoforms corresponding to 69 different genes. When we analyzed these proteins in UniProtKB, we found a predicted ZCCHC in 34 factors (Supporting Dataset 1e). The CX2CX4HX4C sequence was placed in a Cys-rich region in four proteins, and in the remaining 31, it was a part of a larger domain, in most cases an IBR (Supporting Dataset 1e).
The 34 human ZCCHC-containing factors that we found in UniProtKB included the 24 proteins that are annotated in the HGNC database as ZCCHC1–ZCCHC25 (https://www.genenames.org/data/genegroup/#!/group/74), with the exception of ZCCHC23, because it harbors a degenerate ZCCHC in which the H residue is substituted by asparagine (N). Most human ZCCHC-containing factors of known function are involved in messenger RNA (mRNA), microRNA (miRNA), and ribosomal RNA (rRNA) metabolism (Supporting Dataset 1f; for all human amino acid sequences, see Supplementary Fig. 3).
Most ZCCHC-containing proteins are involved in RNA metabolism
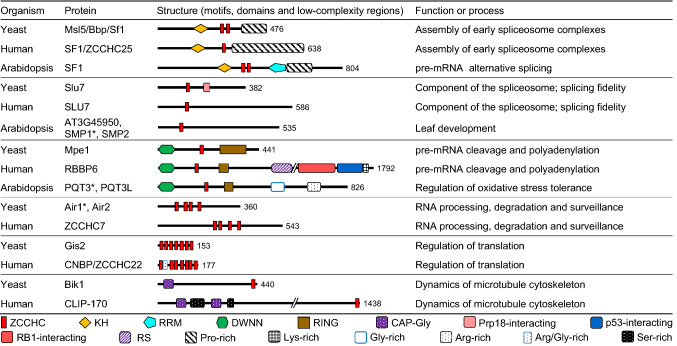
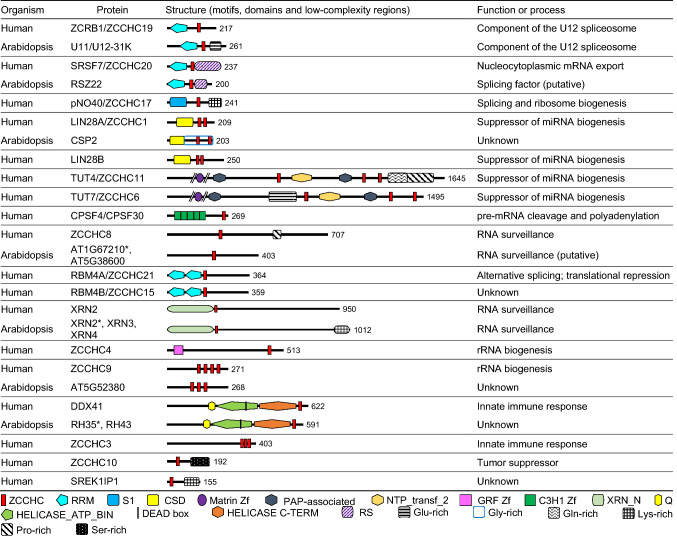
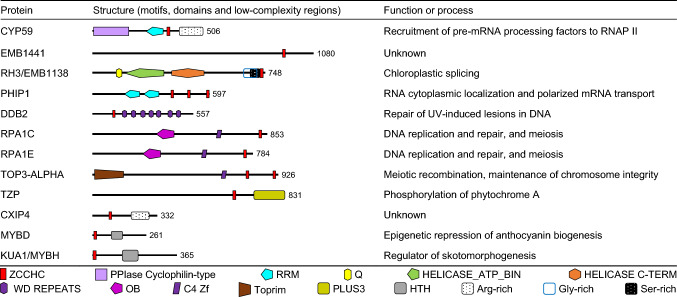
We then analyzed each of the proteins that we considered to be genuine ZCCHC-containing factors (Supporting Datasets 1b, 1d, and 1f), looking for their orthologs in the other two species under study, and using databases and the Basic Local Alignment Search Tool for proteins (BLASTP) [20]. Then, we gathered information on each protein in general and specific databases, as well as in publications. We present the proteins that have been studied to some extent in three figures: yeast ZCCHC-containing factors, with their human and Arabidopsis orthologs, in Fig. 2; the remaining human factors and their Arabidopsis orthologs in Fig. 3; and the remaining Arabidopsis factors in Fig. 4. We did not include in these figures putative orthologs not harboring ZCCHCs. We added the structural information that we found in UniProtKB: motifs, domains, and low-complexity regions (Figs. 2, 3, 4).
Fig. 2.
Schematic representation of the structures of yeast ZCCHC-containing factors, with indication of their putative Arabidopsis and human orthologs. Schemes have been drawn to scale. Motifs, domains, and low-complexity regions are represented by symbols, and were obtained from the UniProtKB database and publications. The Arabidopsis Genome Initiative [19] gene code is shown for Arabidopsis proteins. In cases of several paralogs, the asterisk indicates which protein is represented
Fig. 3.
Schematic representation of the structure of human ZCCHC-containing factors with indication of their putative Arabidopsis orthologs. Other details are as described in the legend of Fig. 2
Fig. 4.
Schematic representation of the structure of Arabidopsis ZCCHC-containing factors. Other details are as described in the legend of Fig. 2
We found that the structures and functions of ZCCHC-containing factors were very diverse, harboring a wide variety of domains and motifs in addition to the ZCCHC, and participating in many different processes. As described in UniProtKB and publications, most of the ZCCHC-containing factors participate in RNA metabolism, including transcriptional elongation, polyadenylation, translation, pre-mRNA splicing, RNA export, RNA degradation, miRNA and rRNA biogenesis, and post-transcriptional gene silencing. However, when we tried to group them into functional categories, the results were barely informative, because only a few proteins had been assigned a Gene Ontology (GO) or KEGG term (results of the GO analysis are included in Supporting Datasets 1b, 1d, and 1f). In this section, we summarize the most relevant information on the proteins that play a role in RNA metabolism, mostly from studies on yeast and humans.
Yeast Msl5 is involved in the assembly of early spliceosome complexes
Pre-mRNA splicing occurs in two consecutive transesterification reactions, executed by the spliceosome to remove introns and join exons, which co-transcriptionally contribute to mRNA maturation. More than 150 trans-acting factors and five small nuclear uridine-rich ribonucleoproteins (snRNPs; U1, U2, U4, U5, and U6) participate in splicing together with other splicing factors, such as serine (S)/arginine (R)-rich (SR) proteins—which contain an Arg-Ser-rich region (the RS domain)—and heterogeneous nuclear ribonucleoproteins (hnRNPs). In addition, several intronic sequences play essential roles in splicing, including the 5′ and 3′ splice sites and the branch point (reviewed in [21–24]).
Yeast Mud synthetic-lethal 5 (Msl5), also termed Branchpoint binding protein (Bbp) and Splicing factor 1 (Sf1), specifically interacts with the seven-nucleotide branch point sequences (UACUAAC) that are found close to the 3′ splice sites of pre-mRNA introns [25], and participates in the assembly of early spliceosome complexes [26]. The human ortholog of Msl5, termed SF1 or ZCCHC25 (Fig. 2), is encoded by an essential gene and binds to the degenerate sequence CURAY (where R and Y are purine and pyrimidine, respectively) [25]. Yeast Msl5 and human SF1 interact with Mutant U1 Die 2 (Mud2) and its human ortholog U2 Auxiliary Factor 65 kDa (U2AF65) splicing factor, respectively, facilitating branch point recognition [27]. SF1 harbors an extended K homology (KH) domain, a ZCCHC, and a Pro-rich C-terminal half. Deletion assays have shown that the KH domain, but not the ZCCHC or the Pro-rich region, is required for RNA binding and pre-spliceosome assembly [28]. Although similar in structure to human SF1, Msl5 includes a second ZCCHC (Fig. 2), whose specific function has not been determined, although it does not seem to be involved in RNA binding [29]. MSL5 is also an essential gene in yeast. Conditional msl5 mutants show aberrant splicing, mainly of introns with weak consensus splice sequences, and high levels of pre-mRNAs in the cytoplasm, suggesting that Msl5 is required for the nuclear retention of unspliced and misspliced pre-mRNAs [30].
The protein encoded by AT5G51300 is considered to be the Arabidopsis homolog of yeast Msl5 and human SF1 [31], and is annotated at TAIR and UniProtKB as ARABIDOPSIS SF1 HOMOLOG (AtSF1). The protein participates in alternative splicing of pre-mRNAs and its structure is very similar to those of its yeast and human orthologs, containing a KH domain, two ZCCHCs, an RNA recognition motif (RRM), and a Pro-rich C-terminal region (Fig. 2). Unlike its SF1 and MLS5 orthologs, AT5G51300 seems to be a single-copy gene (according to our BLASTP search), which is not essential, and its loss of function causes dwarfism, early flowering, and hypersensitivity to abscisic acid [31].
Yeast Slu7 and its human ortholog are required for splicing fidelity
The yeast Pre-mRNA processing factor 18 (Prp18) stabilizes the interaction between adjacent exons and the U5 snRNA during the second step of pre-mRNA splicing [32]. Prp18 and Synergistic lethal with U5 snRNA protein 7 (Slu7) cooperate to promote the second step of splicing, and Slu7 is required to recruit Prp18 to the spliceosome [33, 34]. Yeast SLU7 is essential and encodes a protein with a ZCCHC in its N-terminal half, followed by a Prp18-interacting region (Fig. 2). Slu7 is required for splicing fidelity, since it participates in the selection of the 3′ splice site [35].
Human SLU7 also seems to be involved in splicing fidelity, and its depletion alters the use of the splicing acceptor sequence (AG), so that the spliceosome indiscriminately binds to other AG dinucleotides [36]. The ZCCHC of human SLU7 is also located in its N-terminus, within a nuclear localization signal. This signal, together with the ZCCHC, is required to retain human SLU7 in the nucleus, but not for its import from the cytoplasm [37].
Arabidopsis has three putative SLU7 orthologs encoded by AT3G45950, AT1G65660, and AT4G37120 (Fig. 2). Alignment of these three proteins with other eukaryotic SLU7 factors revealed the presence of a conserved ZCCHC [37, 38]. AT3G45950 is annotated in TAIR and UniProtKB as a “Pre-mRNA splicing Prp18-interacting factor”; AT1G65660 and AT4G37120 are annotated in UniProtKB as “Pre-mRNA-splicing factor SLU7-A” (SLU7-A) and SLU7-B, respectively, and in TAIR as SWELLMAP1 (SMP1) and SMP2, respectively, because both were identified in a screen for leaf developmental abnormalities. Loss of function of SMP genes causes a decrease in plant size, narrow and pointed leaves, and defects in leaf venation pattern. SMP1 and SMP2 are functionally redundant during development [38]. The relationship among these three Arabidopsis proteins and pre-mRNA splicing has been inferred only from their similarity to SLU7 factors, and the role of their ZCCHC has not been determined.
Human ZCRB1/ZCCH19 is a component of the minor spliceosome
The U2-type spliceosome is present in all eukaryotes and processes most introns. Many eukaryotes also contain a minor spliceosome that excises the rare U12-type introns, which represent only 0.5% of the introns in a given organism and have different donor- and branch-site consensus sequences. The U12-type spliceosome has been lost independently in distantly related species, including yeast [39]. Most of the genes containing U12-type introns contain only one of these introns, and also have several U2-type introns. Splicing is slower in U12-type than in U2-type introns, possibly allowing control of the cellular levels of mature mRNAs because inefficient splicing can cause retention of the U12 intron and mRNA decay [40].
Human zinc finger CCHC-type and RNA-binding domain-containing 1 (ZCRB1), also named ZCCHC19 and U11/U12-31K, is one of the components of the U11/U12 snRNPs of the U12 spliceosome, connecting both ends of the intron being processed [41]. The essential Arabidopsis gene AT3G10400 is the ortholog of human ZCRB1, encoding a U11/U12-31K protein, which acts as an RNA chaperone for proper splicing of U12-type introns [42]. The human and Arabidopsis U11/U12-31K proteins have 62% sequence similarity [43] and a similar structure, with an RRM domain followed by a ZCCHC (Fig. 3). Moreover, they both exclusively localize to the nucleus, as would be expected from their role in pre-mRNA splicing [41, 42]. In addition, the C-terminal half of the Arabidopsis AT3G10400 protein harbors a low-complexity region that is rich in glutamic acid (E); this region is absent from human ZCRB1. There is no yeast U11/U12-31K ortholog, since Saccharomyces cerevisiae lacks the entire U12 spliceosome.
Human SRSF7/ZCCHC20 is an SR factor involved in RNA export and decay
Members of the large and conserved SR family of RNA-binding proteins function in eukaryotic pre-mRNA metabolism and mRNA decay, export, and translation. SR proteins share a C-terminal domain, the RS domain (a low-complexity region rich in alternating S and R that in SR proteins is longer than 50 amino acids), and an RRM in their N-terminal region [44]. The RS domain confers flexibility to the SR proteins; the conformation of these proteins varies depending on the proteins or RNA molecules with which they interact [45, 46].
The best-studied role of SR proteins is in the regulation of constitutive and alternative splicing of pre-mRNAs. In constitutive splicing, SR proteins bind pre-mRNA exonic splicing enhancers through their RRM domain and recruit the spliceosome to nearby splice sites through their RS domain [47, 48]. SR factors antagonize hnRNPs, which repress alternative splicing and bind exonic and intronic splicing silencers [49].
Most SR proteins are nucleoplasmic, although some move from the nucleus to the cytoplasm, suggesting a role in mRNA export, translation control, and/or other cytoplasmic processes [46]. In the nucleus, SR proteins are usually found in multiple aggregates, termed speckles, and are frequently used as markers of these nuclear compartments. Human serine/arginine-rich splicing factor 7 (SRSF7), initially termed 9G8 and then ZCCHC20, contains an RRM domain followed by a single ZCCHC and an RS domain at its C-terminal half (Fig. 3). ZCCHC20 and SRp20 (also termed serine/arginine-rich splicing factor 3; SRSF3) are two SR proteins that show a highly dynamic behavior in their intracellular localization, shuttling between the nucleus and cytoplasm [50]. Both ZCCHC20 and SRp20 (which does not harbor a ZCCHC) bind RNA, suggesting, together with their dynamic localization, that they have a role in RNA transport. ZCCHC20 and SRp20 specifically bind to the intron-less mRNA transport element of the histone H2A, to promote its export to the cytoplasm [51]. The process of mRNA export requires the binding of any mRNA to adapter proteins that interact with receptors. For example, human mRNAs bound to ZCCHC20 and SRp20 selectively interact with nuclear RNA export factor 1 (NXF1), which is considered the predominant receptor for mRNA export from the nucleus to the cytoplasm [51].
The Arabidopsis genome encodes two subfamilies of SR factors with ZCCHCs
As with many other gene families, the number of SR family members is larger in plants (up to 22 in rice [Oryza sativa]) than in yeast (2) or humans (9). Only one of the human SR proteins, ZCCHC20, contains a ZCCHC [50]. The Arabidopsis genome encodes 19 SR proteins, belonging to six subfamilies, termed SR, RS, SC, SCL, RSZ, and RS2Z [52]. SR45 may represent an additional independent subfamily [53]. Only the members of the RSZ (RSZ21 [encoded by AT1G23860], RSZ22 [AT4G31580], and RSZ22A [AT2G24590]) and RS2Z (RS2Z32 [AT3G53500] and RSZ33 [AT2G37340]) subfamilies have one and two ZCCHCs, respectively [52, 53]. The ZCCHCs of human ZCCHC20 and its putative Arabidopsis ortholog, RSZ22, are involved in RNA recognition specificity [54, 55]. The structure of RSZ22 is very similar to that of human ZCCHC20 (Fig. 3); however, the function of Arabidopsis RSZ and RS2Z factors is not well known. Arabidopsis RSZ and RS2Z factors co-localize and accumulate in speckles that are distinct from those of other members of the SR family and splicing factors [53]. Some ZCCHC-containing proteins interact with SR factors of the SCL subfamily in yeast two-hybrid (Y2H) and co-immunoprecipitation assays [53, 55].
The localization of Arabidopsis RSZ22 is dynamic, as it moves between the nucleus and cytoplasm, and its ZCCHC is not required for its nuclear localization, but is required for it to interact with other splicing factors [56]. RSZ22 has been found in nuclear speckles and within the nucleolus, and mutations affecting its RRM or ZCCHC domains prevent it from translocating to the nucleolus.
A role for Arabidopsis RSZ33 in splicing has also been proposed based on its interactions with other putative splicing factors (RSZ22 and RSZ21) and the alterations in splicing some mRNAs, including its own, which cause its overexpression [57]. RSZ22 and RSZ21 interact with U1-70K, a component of U1 snRNP [58]. In a directed Y2H assay, RSZ33 interacted with CYCLIN-DEPENDENT PROTEIN KINASE G1 (CDKG1), which regulates the splicing of CALLOSE SYNTHASE 5 (CALS5) pre-mRNA, encoding the main synthase involved in the synthesis of the pollen callose wall [59]. As a consequence, loss of CALS5 and CDKG1 causes defects in pollen wall formation and male fertility. Based on these results, it has been proposed that splicing of CALS5 pre-mRNA is facilitated by its indirect binding to CDKG1 through RSZ33 [59].
pNO40, a human ZCCHC-containing factor, plays a double role in splicing and ribosome biogenesis
The human nucleolar protein of 40 kDa (pNO40), also named ZCCHC17, is a multifunctional protein that participates in ribosome biogenesis and splicing. pNO40 has been found in the nucleolus and cytoplasm as part of the 60S ribosomal subunit [60, 61]. pNO40 has recently been identified as repressing transcription of the 47S rDNA, which encodes the 28S, 18S, and 5.8S rRNAs, by inhibiting the binding of the basal transcription factor upstream binding factor (UBF), to the 35S rDNA promotor [62]. pNO40 interacts with SR proteins to recruit them from nuclear speckles into the nucleolus, where they are retained. Sequestration of SR factors into the nucleolus perturbs RNA metabolism, including mRNA export, as shown in cells overexpressing pNO40, which display nuclear retention of poly(A)+ RNAs and alterations of pre-mRNA splicing [63]. In addition to its ZCCHC located in the middle of the protein, human pNO40 harbors an S1 RNA-binding domain in its N-terminal half, and an extended basic domain (Lys-rich) in its C-terminal half (Fig. 3). No pNO40 ortholog has been found in yeast [61], and we did not identify any similar Arabidopsis proteins in databases or in our BLASTP analysis.
Metazoan LIN28 suppresses the biogenesis of the let-7 microRNA
Genetic screens have identified key genes controlling developmental timing in the nematode Caenorhabditis elegans, including lin-4 and let-7, the first two genes identified in eukaryotes as encoding miRNAs (reviewed in [64]). miRNAs are small (about 22 nt), non-coding RNAs that post-transcriptionally repress gene expression. miRNAs are encoded by endogenous genes that have been found in plant, animal, and protist genomes, and inhibit the translation of their complementary mRNA targets. Mature, functional miRNAs are generated from longer precursors that form stem-loop structures; these stem-loops are processed by Dicer, a member of the RNase III family of endoribonucleases. In metazoans, transcription of the genes encoding miRNAs, the MIR genes, yields a primary precursor (pri-miRNA) that is processed in the nucleus by Drosha, another type III RNase, generating a shorter precursor (pre-miRNA) that is exported to the cytoplasm, where it is then processed by Dicer to form a duplex, one strand of which is the mature miRNA (reviewed in [65]).
The lin-28 gene of C. elegans encodes an RNA-binding factor; the lin-28 mRNA is targeted by the lin-4 miRNA [31]. The LIN28 protein is highly conserved at the structural and functional levels in a variety of animals, from worms to humans. In invertebrates, lin28 is a single-copy gene, whereas all vertebrates have two paralogs, LIN28A and LIN28B [66]. Human LIN28A (also termed ZCCHC1) and LIN28B block let-7 maturation at different steps: LIN28A moves from the nucleus to the cytoplasm, where it blocks pre-let-7 processing by Dicer; and LIN28B sequesters pri‐let‐7 in the nucleolus, away from Drosha [67]. Metazoan LIN28 proteins are RNA-binding factors with two ZCCHCs and a nucleic acid-interacting domain, called the cold-shock domain (CSD; Fig. 3 and Supporting Dataset 1f) [68]. The CSD is about 70 amino acids long and includes two consensus RNA-binding domains. CSDs are found in many different proteins from a wide range of eubacteria and eukaryotes; these proteins bind RNA and single-stranded DNA, acting as RNA-chaperones [69]. CSD-containing proteins are structurally and functionally diverse, participating in multiple steps of RNA metabolism [70]. Human LIN28A and LIN28B are also termed COLD-SHOCK DOMAIN DNA-BINDING PROTEIN 1 (CSDD1) and CSDD2, respectively.
Human LIN28A binds to pre-let-7 and circumvents maturation of this miRNA by recruiting terminal uridylyltransferase 4 (TUT4), which catalyzes pre-let-7 3′ uridylation and induces its degradation, thereby preventing pre-let-7 processing by Dicer [64, 71]. TUT4, also termed ZCCHC11, and its paralog TUT7 (ZCCHC6), which plays a minor role in the oligouridylation of pre-let-7, are members of the non-canonical Poly(A) Polymerases (PAPs), belonging to the DNA polymerase β superfamily. TUT4 and TUT7 harbor three ZCCHCs. In addition to the ZCCHC, TUT4 and TUT7 also harbor a Matrin C2H2-type zinc finger, a nucleotidyltransferase (NTP_transf) domain, two Cid1 family PAP-associated domains, and Pro- and Gln-rich regions in TUT4, but a Glu-rich region in TUT7 (Fig. 3 and Supporting Dataset 1f). A specific interaction between the two ZCCHCs of mouse LIN28A and pre-let-7 is necessary and sufficient to induce its oligouridylation by TUT4 [64]. Mouse LIN28A has been found in the periphery of the endoplasmic reticulum and suppresses endoplasmic reticulum-associated translation of mRNAs that form small hairpins containing AAGNNG, AAGNG, or UGUG sequences, which are the binding targets for LIN28A [72].
Our analyses on PROSITE and PatMatch did not identify any RNA uridylyltransferase containing a ZCCHC in Arabidopsis. However, Arabidopsis HEN1 SUPPRESSOR1 (HESO1, encoded by AT2G39740) and UTP:RNA URIDYLYLTRANSFERASE 1 (URT1, encoded by AT2G45620) have been shown to function redundantly in miRNA 3′ uridylation, a process in which HESO1 plays the predominant role [73]. URT1 could be the functional ortholog of TUT7 or TUT4 because, in addition to their similar uridylation activity, it displayed the highest similarity to both factors in our BLASTP analysis: 35% and 33%, respectively. The reciprocal BLASTP, using the AT2G45620 protein to search in human databases, identified TUT7 and TUT4 as the most likely homologs. In addition, the structures of TUT4, TUT7, and URT1 are similar, sharing the NT (PF01909) and Cid1 family PAP (PF03828) domains, although URT1 lacks a ZCCHC.
The Arabidopsis genome encodes several Gly-rich proteins similar to LIN28
The similarity between GLYCINE-RICH PROTEIN2 (GRP2; encoded by AT4G38680), also named COLD SHOCK DOMAIN PROTEIN2 (CSDP2) or COLD SHOCK PROTEIN2 (CSP2), and human LIN28 has been previously described [66, 74]. CSP2 and its paralog CSP1 belong to the plant GRP superfamily of proteins, whose five families (I–V) have repeats of glycines with the GnX structure. Family IV is composed of RNA-binding GRPs (RBGs), which have one or two RNA-binding domains in their N-terminus and a 30–70% G content in the C-terminus. Four RBG subclasses have been defined based on their structure (IVa–d). Members of the IVa, b, and d subclasses contain a single RRM, and members of the IVc subclass contain a CSD in their N-terminus [75, 76]. The Arabidopsis genome encodes 19 IVa–d RGBs, seven of which belong to the IVa subclass, three to IVb, four to IVc, and five to IVd [77]. In addition to an RNA-binding domain (either RRM or CSD), members of the IVb subclass, which are termed RNA-BINDING GLYCINE-RICH PROTEIN B1 (RBGB1) to RBGB3, and members of the IVc subclass, which are termed CSP1 to CSP4, have one (RBGB) or several (seven in CSP1 and CSP3, and two in CSP2) ZCCHCs in the Gly-rich region of their C-terminus (Fig. 3 and Supplementary Fig. 2) [76].
CSP1 and CSP2 associate with polyribosomes via mRNA, display mRNA-chaperone activity, and localize to the nucleolus and the cytoplasm [78, 79]. Nucleolar localization of CSP2 is dependent on its C-terminal GR/ZCCHC region. CSP2 and its closest paralog, CSP4/GRP2B (encoded by AT2G21060), which are partially redundant, negatively regulate seed germination and tolerance to freezing and salt stress [80]. CSP3 (encoded by AT2G17870) is required for abiotic stress tolerance, because its loss of function increases sensitivity to freezing, drought, and salt stress; its overexpression increases tolerance to these stresses. However, the molecular function of CSP3 related to stress or RNA metabolism is yet to be elucidated [81].
CSP3 seems to be a versatile protein whose interactions and subcellular localization suggest a role in ribosome biogenesis and RNA processing and stability. CSP3 has been found in the nucleolus, nucleoplasm, nuclear speckles, and cytoplasm. In the nucleolus and nucleoplasm, CSP3 interacts with NUCLEOLIN1 (NUC1), ribosome biogenesis factors, and 60S ribosomal subunit proteins. Arabidopsis NUC1 regulates the transcription of the 45S rDNA and the processing of the 45S pre-rRNA, the primary precursor of the 25S, 18S, and 5.8S rRNAs [82, 83]. CSP3 also interacts with several poly(A)-binding proteins in nuclear speckles, and with the DECAPPING5 PROTEIN5 (DCP5) protein in the cytoplasm [81, 84].
Little information is available on the Arabidopsis RBGB subfamily, which is composed of three members, RBGB1–RBGB3, and exhibits RNA-chaperone activity when expressed in Escherichia coli. Transcription of RBGB2 (also termed RZ-1A) is induced by cold treatment but not by other types of abiotic stress or exogenous abscisic acid treatment, and its overexpression confers tolerance to freezing in Arabidopsis and to cold exposure in E. coli [85]. RBGB1 (RZ-1B) and RBGB3 (RZ-1C) localize in nuclear speckles. Simultaneous loss of function of RZ-1B and RZ-1C perturbs the splicing of many genes. Several aspects of the development of rz-1b rz-1c double mutants are delayed, including germination and flowering time, and their leaves are serrated. RZ-1B and RZ-1C interact in Y2H assays with some SR proteins of different subfamilies, such as RSZ21 and RSZ22, all of which contain ZCCHCs. In addition, RZ-1B and RZ-1C interact with themselves and with each other through their C-terminal domain, which is required for their localization in nuclear speckles [86].
ZCCHC-containing factors involved in pre-mRNA polyadenylation
In eukaryotic pre-mRNA maturation, which yields mature mRNAs competent for nuclear export and translation, 3′-end cleavage and polyadenylation are key steps. In mammals, about 20 factors constitute the core poly(A) machinery, which includes PAP and the conserved cleavage and polyadenylation specificity factor (CPSF) and cleavage stimulation factor (CstF) complexes, which are required for pre-mRNA cleavage, but not for polyadenylation. PAP is recruited by CPSF and catalyzes the addition of 200–250 adenosines to the 3′ end of the cleaved pre-mRNA in a template-independent fashion [87].
Yeast Mutant PCF11 extragenic suppressor 1 (Mpe1) is a component of the CPSF complex. Mpe1 is required for pre-mRNA cleavage and for polyadenylation at the bona fide site, as shown by examination of mpe1 mutants [88]. Mpe1 is an E3 ligase with a RING finger domain at its C-terminus, a variant of a ubiquitin-like (UBL) domain in the N-terminus, termed Domain With No Name (DWNN), and a ZCCHC in its central region (Fig. 2). Mutational analysis of these domains revealed that they are required for pre-mRNA polyadenylation, because the DWNN mediates protein–protein interactions within the CPSF complex, and the RING finger and the ZCCHCs mediate RNA binding [89].
The human homolog of yeast Mpe1 is the Retinoblastoma (Rb) binding protein 6 (RBBP6), also termed p53-associated cellular protein-testes derived (PACT). RBBP6 contains the same three domains as Mpe1, i.e., DWNN, ZCCHC, and RING finger. In addition, RBBP6 is four times larger than yeast Mpe1, and harbors an RS domain, followed by p53- and Rb-binding regions and a Lys-rich region in the C-terminus (Fig. 2). RBBP6 binds the p53 and Rb tumor suppressor factors, and thus interferes with the binding of p53 to DNA [90, 91]. Knockdown of Rbbp6 in mice enhances the accumulation of p53 by reducing its polyubiquitination and causes embryo lethality. Consequently, loss of RBBP6 in mice increases p53-dependent gene transcription, revealing that RBBP6 is a negative regulator of p53 [92]. Knockdown of RBBP6 in human cells with a small interfering RNA (siRNA) dramatically reduces the efficiency of 3′ pre-mRNA cleavage, causing a decrease in the abundance of mRNAs and an increase in the use of distal poly(A) sites. The decrease in mRNA levels is particularly evident for transcripts containing AU-rich elements (AREs) in their 3′-UTRs, which are naturally unstable transcripts targeted by the exosome. The DWNN is essential for binding to CstF, and, therefore, also essential for 3′ pre-mRNA cleavage, as shown by 3′ cleavage and polyadenylation assays in cells after knockdown of RBBP6 with and without transgenes expressing different regions of the RBBP6 protein [93].
The Arabidopsis genes AT4G17410 and AT5G47430 encode putative orthologs of Mpe1 and RBBP6 (Fig. 2), both of which are annotated in the Aramemnon database as “Mpe1-like components of the CPSF complex” and in the TAIR database as “encoding a CCHC-type zinc finger protein with a DWNN domain”. We found AT4G17410 and AT5G47430 to be the only Arabidopsis genes annotated as encoding a protein with a DWNN. AT4G17410 has been named PARAQUAT TOLERANCE 3 (PQT3) and is a negative regulator of oxidative stress tolerance [94]. Its plant, fungal, and protist orthologs lack the p53- and Rb-binding and SR domains [95]. However, PQT3 and its paralog AT5G47430 (PQT3-like; PQT3L), which also contain the DWNN, ZCCHC, and RING finger domains, have a Gly- and an Arg-rich region in their C-terminal half (Fig. 2). The function of PQT3 and PQTL3L in pre-mRNA cleavage and polyadenylation is not known.
The human CPSF4 (also termed CPSF30) is also a component of the CPSF complex, acting as a co-factor of the cleavage and polyadenylation machinery. CPSF4 and its animal orthologs harbor a ZCCHC in their C-terminus (Fig. 3), which is absent from fungi and plants [96, 97]. In vitro RNA-binding assays revealed that its ZCCHC enhances poly(U)-binding ability. In addition, the ZCCHC alone produces the same enhancement of poly(U) binding as the other five zinc fingers together, which are C3H1-type (CX8CX5CX3H) zinc fingers, and are present in all CPSF30/CPSF4 orthologs, including the Yeast 30 kDa homolog 1 (YTH1) [96].
Air proteins are key factors in RNA degradation by the exosome
The yeast Arginine methyltransferase-interacting RING finger protein 1 (Air1) was identified in a screen based on the Y2H assay for structures that interact with Histone methyltransferase 1 (Hmt1), the major arginine methyltransferase in yeast. Many RNA-binding proteins undergo arginine methylation by Hmt1, including histones and hnRNPs [98]. Arginine methylation by yeast Hmt1 and its human counterpart occurs in the Arg/Gly-rich C-terminal domain, termed the RGG box, of hnRNPs [99]. RGG/RG domains are encoded by all eukaryotic genomes and are the second most abundant among human RNA-binding domains [100]. RGG domains are intrinsically disordered, and mediate degenerate specificity in RNA binding. RGGs are found as single RNA-binding domains or combined with more structured domains, such as the RRM- or KH-type domains [101].
The eukaryotic exosome is a multiprotein complex with 3′ to 5′ exonucleolytic activity that processes and/or degrades normal and aberrant RNA species, and the by-products of their maturation, in both the nucleus and the cytoplasm. To be catalytically active and to recruit its RNA substrates, the exosome is assisted by cofactors. These cofactors include protein complexes that act as activators, RNA helicases, RNA-binding proteins that act as adaptors, including ZCCHC proteins, and other proteins that do not fit in any of these categories. In yeast, the Trf4/5-Air1/2-Mtr4 polyadenylation (TRAMP) and Superkiller (Ski) complexes are the best-characterized exosome cofactors, which recruit the exosome to its RNA targets in the nucleus and cytoplasm, respectively. The TRAMP and Ski complexes are required for RNA surveillance. TRAMP is also required for maturation of snRNAs and rRNAs, while the Ski complex also participates in the turnover of functional mRNAs [102].
The yeast TRAMP complex is composed of the non-canonical Trf4/5 PAP, RNA helicase Mtr4 (mRNA transport 4), and RNA-binding proteins Air1 and Air2 [103]. The number of ZCCHCs in Air1 and Air2 is not clear. These two proteins have four CX2CX4HX4C sequences (Supporting Dataset 1b), but they have been described as harboring five ZCCHCs [104, 105]. However, their second ZCCHC is not canonical, since there is an additional residue between the second C and the H (CX2CX5HX4C). In agreement with the four CX2CX4HX4C sequences present in both proteins, four ZCCHCs are described for Air1 in UniProtKB, but only three are annotated for Air2; the CX2CX5HX4C sequence is not considered a ZCCHC in either case.
Air1 and Air2 bind Trf4 and Trf5 through the ZCCHCs to recognize RNA substrates of TRAMP. The Trf4 subunit of TRAMP adds a short poly(A) tail to cryptic unstable transcripts (CUTs). It can also polyadenylate many different improperly processed or aberrant non-coding RNAs (ncRNAs) that participate in splicing [small nuclear RNAs (snRNAs)] or in ribosome biogenesis and translation [transfer RNAs (tRNAs), small nucleolar RNAs (snoRNAs), and rRNAs]. Polyadenylation of these TRAMP targets stimulates their degradation by the nuclear exosome. Mutating AIR1 and AIR2, which encode proteins with a substitution in the second C of the third and fourth ZCCHC, revealed that the second C in these motifs is critical for Air function in vivo [104].
Co-immunoprecipitation analysis coupled with mass spectrometry allowed for the discovery of two different MTR4-containing human complexes, one of which is restricted to the nucleoplasm and the second to the nucleolus. The nuclear exosome-targeting (NEXT) complex is composed of MTR4, RNA Binding Motif Protein 7 (RBM7), and ZCCHC8, which possesses a single ZCCHC that can potentially interact with RNA. Based on its dual interaction with the exosome and the spliceosome, an additional role has been proposed for the NEXT complex in the recruitment of the exosome for decay and/or processing of intron-encoded snoRNAs [106].
ZCCHC7 is the putative human ortholog of yeast Air1 and Air2, and co-purified with the nucleolar fraction of MTR4 and the exonuclease RRP6 (also known as EXOSC10). RRP6 is a catalytic subunit of the RNA exosome that is highly conserved in eukaryotes and has been found in the nucleolus and the nucleoplasm. RRP6 is required for proper maturation of ncRNA substrates, such as rRNAs, snRNAs, snoRNAs, and CUTs (reviewed in [107]). Three RRP6-like proteins (RRP6L1 to RRP6L3) have been found in Arabidopsis: RRP6L1 in the nucleus and in the nucleolar vacuole, RRP6L2 mainly in the nucleolus, and RRP6L3 in the cytoplasm, but its functional relationship with the exosome has not been determined [108].
Loss of function of ZCCHC7 causes the accumulation of by-products excised from the 5′-ETS of 47S pre-rRNA processing [109]. ZCCHC7 is a nucleolar protein, consistent with its role in rRNA biogenesis. ZCCHC7 has four ZCCHCs (Fig. 2), which mediate the interaction with PAP-associated domain-containing 5 (PAPD5) and PAPD7, the human orthologs of yeast TRF4-2 and TRF4-1, respectively, suggesting the existence of a human nucleolar TRAMP-like complex [110]. Using BLASTP, we found several Arabidopsis ZCCHC-containing proteins of unknown function with limited sequence similarity to human ZCCHC7, no one of which was a credible ZCCHC7 homolog.
AT1G67210 and AT5G38600 are annotated in the TAIR10 database as encoding proline-rich spliceosome-associated family protein/zinc knuckle (CCHC-type) family proteins, and are proposed to be ZCCHC8 homologs [111, 112] with structures similar to that of human ZCCHC8, harboring a single ZCCHC. They have also been identified in co-immunoprecipitation assays using HUA ENHANCER 2 (HEN2) as bait [112]. Arabidopsis HEN2 and MTR4 are the two Arabidopsis co-orthologs of the yeast Mtr4 helicase; HEN2 is nucleoplasmic and MTR4 is mainly nucleolar, as shown by their hybrid proteins with GFP [111, 112]. Both of these helicases associate with core components of the nuclear exosome, but most proteins that co-precipitate with MTR4-GFP are ribosomal processing factors that do not co-purify with HEN2-GFP. Plants lacking HEN2 activity accumulate many types of polyadenylated nuclear RNA species, including unspliced pre-mRNAs or misspliced mRNAs, excised introns, and by-products of the processing of ncRNAs. Based on these results, the existence of two specialized RNA helicases, acting as cofactors of the exosome, has been proposed in Arabidopsis: MTR4 assists in the nucleolus to the exosome in the maturation of rRNAs, and HEN2 in the degradation of many different RNA nuclear substrates. The functions of AT1G67210 and AT5G38600 have not yet been determined, but both may be part of two different NEXT-like complexes, together with HEN2 and RBM7, and participate in the degradation and/or processing of nuclear exosome RNA substrates [112].
The yeast Gis2 and human CNBP and RBM4 proteins promote cap-independent translation
Expansion of CCTG repeats in the first intron of the human gene encoding the CCHC-type zinc finger nucleic acid-binding protein (CNBP), also termed Zinc finger 9 (ZNF9) and ZCCHC22, causes the autosomal dominant form of Myotonic dystrophy type 2 [113]. Human CNBP was the first cellular eukaryotic ZCCHC-containing protein described, and was found to bind to the conserved Sterol Regulatory Element present in promoters of genes encoding enzymes of the cholesterol biosynthetic pathway [114]. CNBP was later identified as an RNA-binding protein in Xenopus laevis by its ability to bind the 5′-UTR of mRNAs encoding ribosomal proteins, which suggests a role in their translational control [115, 116]. CNBP orthologs, including yeast Glucose inhibition of gluconeogenic growth suppressor 2 protein (Gis2), harbor seven ZCCHCs (Fig. 2), and interact with ribosomal proteins and the translating ribosome. Human CNBP and yeast Gis2 are located in stress granules [117], which are cytoplasmic compartments containing translationally repressed mRNAs. Stress granules form when translation initiation is impaired and function in translational repression and/or mRNA decay [118]. Human CNBP and yeast Gis2 are also considered translational activators because they promote cap-independent translation through interactions with the 5′-terminal oligopyrimidine (5′ TOP) tract of mRNAs and the translating ribosome [119, 120]. Many eukaryotic 5′ TOP mRNAs are transcribed from genes that encode factors involved in translation, including ribosomal proteins and ribosomal biogenesis factors [121]. Human CNBP enhances global transcription and translation by unfolding DNA and RNA G-quadruplex (G4) structures, located in the promoters of genes or 5′-UTRs of mRNAs, respectively [122, 123]. Although CNBP orthologs have been identified in fungi and metazoans, such as Drosophila melanogaster [124], no CNBP or Gis2 orthologs have been described in plants. Our BLASTP searches of the yeast Gis2 and human CNBP sequence against the TAIR10 protein database yielded CSP1 and CSP3 as the most similar factors in the Arabidopsis proteome.
RNA-binding domain protein 4A (RBM4A), also named ZCCHC21, and RBM4B (ZCCH15) are the human orthologs of Drosophila melanogaster LARK (low-complexity amyloid-like reversible kinked segments); LARK and the two human RBM4 factors harbor two RRM domains and a ZCCHC (Fig. 3). The RRMs and C-terminus of LARK are required for its splicing function. Deletion analysis of LARK suggests that its ZCCHC and second RRM domain act in concert in translational regulation [125]. RBM4A is a dynamic protein that shuttles between the nucleus and cytoplasm, and is involved in controlling alternative pre-mRNA splicing and translational repression. In the cytoplasm, RBM4A suppresses cap-dependent translation in unstressed conditions, but under stress, it promotes translation mediated by IRES (internal ribosome entry sites) of stress-responsive genes. In the nucleus, RBM4B is localized in nuclear speckles and the nucleolus, where it might play a role in rRNA biogenesis, since it interacts with the 40 kDa subunit of RNA polymerase I (RNAP I) in Y2H assays (reviewed in [126]). Similar to human CNBP, metazoan LARK/RBM4 proteins have been found to be DNA G4-binding proteins [127]. There is no Drosophila melanogaster LARK homolog described in Arabidopsis, and our searches for Arabidopsis homologs of the yeast LARK protein and of its human orthologs did not yield similar factors.
XRNs contribute to RNA turnover and post-transcriptional gene silencing
Eukaryotic 5′ → 3′ exoribonucleases (XRNs) contribute to RNA turnover and maintenance of RNA homeostasis, which involves degradation of a wide range of normal and defective mRNAs. XRNs also participate in the maturation of many species of ncRNAs, such as rRNAs, tRNAs, and snoRNAs. XRNs have been found in the nucleoplasm, the nucleolus, and the cytoplasm. XRNs were first identified in yeast, which has two of these enzymes: Xrn1 (also named Pacman) and Xrn2 (also named ribonucleic acid trafficking, Rat1), which act in the cytoplasm and nucleus, respectively. Yeast Xrn1 has been found predominantly in the cytoplasm, but also in the nucleus, and it has been implicated in the degradation of decapped or cleaved mRNAs, and with the processing of ncRNAs, such as snoRNAs or tRNAs [128, 129]. More recently, Xrn1 has been shown to act in the nucleus as an activator of the transcription of genes encoding unstable transcripts, many of which encode proteins involved in ribosome biogenesis and mRNA translation [130]. Xrn2/Rat1 is required for the termination of transcription by RNAP I and II [131, 132], and for the maturation of snoRNAs and rRNAs [133, 134].
The human and Arabidopsis proteomes lack an ortholog of yeast Xrn1, but they contain one and three homologs of yeast Xrn2/Rat1, respectively, all of which have a ZCCHC, unlike yeast XRNs. In addition to the ZCCHC, all XRNs, including yeast Xrn2/Rat1, harbor a 5′ → 3′ exonuclease domain in their N-terminus (Fig. 3). Human XRN2 is involved in the termination of transcription by RNA polymerase II, and it degrades nascent RNA downstream of the 3′ cleavage site [135]. Human XRN2 is also involved in the suppression of replication stress and maintenance of genomic stability [136]. The three Xrn2/Rat1 orthologs of Arabidopsis are XRN2 (encoded by AT5G42540), XRN3 (AT1G75660), and XRN4 (AT1G54490). Arabidopsis XRN2 and XRN3 are nucleolar and nucleoplasmic factors, and XRN4 is cytoplasmic [137]. XRN2 and XRN3 act in 45S pre-rRNA processing [138], and as endogenous RNA silencing suppressors, and are required for the degradation of excised miRNA loops produced during miRNA maturation. XRN4 localizes in the processing bodies with the decapping enzymes DCP1 and DCP2, all of which are involved in mRNA decay. Plants without XRN4 activity over-accumulate 3′ fragments of RNAs, including targets of miRNAs [139]. XRN4 is a suppressor of post-transcriptional gene silencing, because it degrades decapped mRNAs, preventing them from becoming templates for RNA-dependent RNA polymerases, thus producing small interfering RNAs that would enter into the post-transcriptional gene silencing pathway [140].
Human ZCCHC4 and ZCCHC9 are involved in rRNA biogenesis
Human ZCCHC4 is a nucleolar and cytoplasmic RNA m6A methyltransferase involved in the methylation of the 28S rRNA in the cytoplasm. The 28S rRNA is a component of the 60S ribosomal subunit, and the loss of ZCCHC4 reduces 60S ribosomal subunit levels and global translation, although it has no effect on mature 28S and 18S rRNA production [141]. ZCCHC4 harbors a ZCCHC and a zinc finger GRF-type, containing three conserved glycine (G), R, and phenylalanine (F) residues in the center of the domain (Fig. 3). ZCCHC4 does not have orthologs in yeast, and even though it has been claimed to be conserved in multicellular organisms, ZCCHC4 orthologs have only been analyzed in animals [141]. We did not find any Arabidopsis protein with enough similarity to be considered a credible ortholog of ZCCHC4.
ZCCHC9 is a nucleoplasmic and nucleolar protein [142]. Like Air1, Air2, and ZCCHC7 (but not ZCCHC8), ZCCHC9 harbors four ZCCHCs (Fig. 3), and its similarity to these proteins has been pointed out; however, ZCCHC9 is considered a protein without a yeast homolog [143]. ZCCHC9 is assumed to be a ribosome biogenesis factor because its knockdown causes high over-accumulation of 21S pre-rRNA, a precursor of 18S rRNA [144]. AT5G52380 encodes the Arabidopsis protein most similar to human ZCCHC9, according to the Aramemnon database and our own BLASTP analysis, but it has not been studied. The reciprocal BLASTP analysis, searching with the AT5G52380 protein in human databases, also shows that ZCCHC9 is the human protein most similar to Arabidopsis AT5G52380. This Arabidopsis protein harbors five CX2CX4HX4C sequences (Supporting Dataset 1d) recognized by the SMART program as ZCCHCs, but only three of them were predicted with PROSITE (Fig. 3).
Other Arabidopsis-specific ZCCHC-containing factors that are also involved in RNA metabolism
Arabidopsis CYCLOPHILIN59 (CYP59; encoded by AT1G53720) and its orthologs, Silencer of Germline 7 (SIG-7) of Caenorhabditis elegans and RRM-containing cyclophilin regulating transcription (Rct1) of Schizosaccharomyces pombe, participate in regulating the recruitment of pre-mRNA processing factors to RNAP II. They interact with the C-terminal domain (CTD) of the large subunit of RNAP II, which is composed of tandem repeats of the conserved heptapeptide YSPTSPS, and whose S residues are dynamically phosphorylated and dephosphorylated. The phosphorylation status of the CTD determines its ability to recruit diverse factors, including splicing factors, to RNAP II. Similar to the histone code of protein modifications that affect chromatin structure and gene expression, the existence of a CTD code that affects the recruitment of factors to RNAP II has been postulated. The CTD must be unphosphorylated to initiate transcription, but phosphorylation of S5 and S2 is required for transcriptional elongation, and phosphorylation of S2 is required for termination [145]. CYP59 and its orthologs act as peptidyl isomerases for the trans-to-cis isomerization of P residues present in the CTD heptapeptide repeats. Arabidopsis CYP59 is an RNA-binding protein that interacts with SR factors in Y2H and pull-down assays, as well as with the CTD of RNAP II, and localizes in a punctuate pattern next to splicing speckles where most nuclear SR factors are found, as well as in the nucleolus. In plants overexpressing CYP59, phosphorylation of the RNAP II CTD is reduced [146]. Arabidopsis CYP59 and its plant orthologs harbor a ZCCHC between a RRM domain and a low-complexity region in its C-terminal region (Fig. 4). ZCCHCs are absent in their yeast and animal orthologs.
Two Arabidopsis ZCCHC-containing factors, encoded by AT5G49930 and AT5G26742, were isolated because of the effects of their loss of function, which cause embryonic lethality, so they are termed EMBRYO DEFECTIVE 1441 (EMB1441) and EMB1138, respectively [147]. AT5G49930 is annotated in the Aramemnon database as “putative (yeast RQC2)-like component of ribosome-associated quality control complex”, and as the ortholog of the human Nuclear Export Mediator Factor (NEMF) in HomoloGene. NEMF is a component of the ribosome quality-control complex that facilitates the recognition and ubiquitination of stalled 60S subunits [148]. As described in HomoloGene, all NEMF orthologs in animals, plants, and yeast share two domains of unknown function, DUF814 and DUF3441, but only plant orthologs (Arabidopsis and rice) harbor a ZCCHC.
EMB1138 (AT5G26742) or RH3 and its maize ortholog are DEAD-box RNA helicases with RNA-chaperone activity that participate in chloroplastic splicing and could also be involved in the assembly of 50S ribosomal subunits in the chloroplast [149, 150]. RH3/EMB1138 is described in the HomoloGene database as the ortholog of human DExD-box helicase 50 (DDX50), which localizes in nucleoli and nuclear speckles and participates in antiviral responses [151], but only plant orthologs seem to harbor a ZCCHC (Fig. 4). RH3 is the only chloroplastic ZCCHC-containing protein that we found.
AT3G55340 encodes PHRAGMOPLASTIN-INTERACTING PROTEIN 1 (PHIP1), a plant-specific protein that seems to be required for RNA cytoplasmic localization and polarized mRNA transport, which involves the movement of ribonucleoprotein complexes along the microtubular cytoskeleton [152]. PHIP1 contains three ZCCHCs and two RRM domains (Fig. 4).
ZCCHC-containing proteins that are not associated with RNA metabolism
Yeast Bilateral karyogamy defect 1 (Bik1) and its human ortholog, CAP-Gly domain-containing linker protein 1 (CLIP1, better known as CLIP-170), play critical roles in the regulation of microtubule polymerization, stabilization, and dynamics. Both factors harbor a ZCCHC in their C-terminal regions [153] and are cytoplasmic (Fig. 2 and Supporting Dataset 1b and 1f). Our results indicate that Bik1 is the only yeast ZCCHC-containing factor with a known function that is not involved in RNA metabolism. The ZCCHC described for Bik1 is not recognized by the ScanProsite or SMART programs (Supporting Dataset 1b), but it has been found to be involved in protein–protein interactions [154]. Two ZCCHCs have been described in human CLIP-170 (Fig. 2 and Supporting Dataset 1f). The second ZCCHC of CLIP-170, which is absent from Bik1, contains an additional amino acid between the second C and the H (CX2CX5HX4C). This atypical domain is functional, as shown by site-directed mutagenesis [155]. We have not found credible Bik1 or CLIP‐170 orthologs using BLASTP searches, but several Arabidopsis factors have been proposed, based on amino acid composition instead of sequence identity [156].
We found two human ZCCHC-containing factors involved in innate immune responses: DDX41 and ZCCHC3. DDX41 is an intracellular sensor of pathogenic double-stranded DNA that activates the immune response [157]. Arabidopsis RH35 (encoded by AT5G51280) and its paralog RH43 (AT4G33370) are described in HomoloGene as orthologs of human DDX41; RH35 seems to be nuclear, and RH43 cytoplasmic, but they are not well studied. These four orthologs harbor a ZCCHC in their C-terminal regions, as described in UniProtKB and HomoloGene databases (Fig. 3 and Supporting Dataset 1d and 1f). Human ZCCHC3 is considered a co-sensor for cytosolic viral RNA and DNA [158, 159]. No other domains or motifs that three ZCCHCs located in its C-terminal region are described for ZCCHC3 in UniProtKB (Fig. 3).
Six of the human ZCCHC-containing proteins are encoded by long terminal repeat (LTR) retrotransposon-derived genes, belonging to two different families: Mammalian retrotransposon-derived (Mart) and the Paraneoplastic Ma Antigen (PNMA) (Supporting Dataset 1f). Members of these families are classified as domesticated Gag proteins, encoded by neofunctionalized retrotransposons whose genes are present in mammalian genomes and seem to have been domesticated during evolution [160]. The human genome encodes 11 Mart genes with deletions affecting the regions encoding reverse transcriptase integrase activity, but only three (PEG10 [Paternally expressed gene 10 protein], ZCCHC5, and ZCCHC16) harbor a ZCCHC [161]. The human PNMA family has 19 members, three of which harbor a ZCCHC in their C-terminal region with highly similar sequences: the paraneoplastic antigen Ma3 (PNMA3), PNMA7A (ZCCHC12), and PNMA7B (ZCCHC18). These PNMA proteins have been associated with Paraneoplastic Neuronal Disorder (PND), a non-metastatic complication of cancer; they seem to play an oncogenic role [162]. ZCCHC10 has recently also been related to cancer, with a suppressor effect instead of an oncogenic effect. ZCCHC10 binds and hence stabilizes p53 by inhibiting its interaction with murine double minute 2 (MDM2), an E3 ubiquitin-protein ligase that ubiquitinates p53 for proteasomal degradation [163].
Some other Arabidopsis factors with ZCCHCs have not been associated with RNA metabolism, such as DNA DAMAGE-BINDING PROTEIN 2 (DDB2; AT5G58760), which is involved, together with ARGONAUTE1 (AGO1), in the repair of UV-induced lesions. Arabidopsis AGO1 is the main effector of post-transcriptional gene silencing mediated by miRNAs, other endogenous siRNAs, and viral siRNAs [164]. DDB2 and AGO1 form a chromatin-bound complex, and photoproduct-derived siRNAs facilitate the recognition of UV-damaged DNA for repair [165]. REPLICATION PROTEIN A 1C (RPA1C) and RPA1E play a redundant role in DNA replication, repair, and meiosis (crossing over and repair); all plant RPA1C orthologs harbor at least one ZCCHC that is also present in all RPA1E orthologs in the Brassicaceae [166]. The ZCCHC of TOP3-alpha (TOP3α) is required for meiotic recombination and chromosome integrity, preventing extra crossovers [167]. TANDEM ZINC KNUCKLE PROTEIN (TZP; AT5G43630) is a nuclear protein that was identified as a quantitative trait locus negatively controlling morning-specific growth in Arabidopsis [168]. TZP is required for nuclear phosphorylation of phytochrome A (phyA), the primary photoreceptor for far-red light. TZP interacts with phyA through its Plus3 domain [169]. We found five genes encoding transcription factors of the MYB family (AT1G70000, AT3G16350, AT5G47390, AT5G56840, and AT5G61620), one of them with an uncertain ZCCHC, and only two of which have already been studied. MYB-LIKE DOMAIN (MYBD; AT1G70000) is involved in the epigenetic repression of anthocyanin biogenesis [170]. KUA1 (encoded by AT5G47390), also named MYB HYPOCOTYL ELONGATION-RELATED (MYBH), was first identified as a regulator of skotomorphogenesis [171], and later as a circadian clock-regulated gene that promotes leaf cell expansion, controlling the homeostasis of reactive oxygen species [172].
Another Arabidopsis factor that has been studied to some extent is CAX-INTERACTING PROTEIN 4 (CXIP4; AT2G28910), a nuclear and cytoplasmic protein that was initially identified, because its expression in yeast activates the H+/Ca2+ antiporter CAX1 [173]. However, CXIP4 was later found to be an interactor of MORPHOLOGY OF ARGONAUTE1-52 SUPPRESSED 2 (MAS2) in a screen based on the Y2H assay. MAS2 is the presumed ortholog of human NF-kappa-B-activating protein, and seems to be a key player in the regulation of rRNA synthesis that also interacts with splicing and ribosomal biogenesis factors, so CXIP4 may be involved in RNA metabolism [174].
ZCCHC sequences are conserved among orthologs
A consensus sequence has been proposed for ZCCHCs, based on the alignment of retroviral and cellular ZCCHCs: X2-C-ϕ-ψ-C-G-ψ-X-G-H-ω-(A/S)-(+)-(−)-C-P-X, where ϕ is an aromatic amino acid, ψ is a polar or charged residue able to form hydrogen bonds, ω is a hydrophobic residue, and (+) and (−) are positively and negatively charged amino acids [175]. To obtain a consensus sequence, we considered factors found in the three organisms studied here (110 factors, excluding the retrotransposon-related factors and proteins with uncertain ZCCHCs) and aligned their 198 putative ZCCHCs in two ways, either all together or grouped by species (20 from yeast, 121 from Arabidopsis, and 57 human). We then generated logos from the multiple alignments obtained, using the WebLogo program (https://weblogo.threeplusone.com/create.cgi), resulting in similar findings for the three organisms (Fig. 5) as those previously described by other authors [175] and the corresponding PROSITE profile. The PROSITE database is composed of entries describing protein domains and profiles to identify them [176–178]. PROSITE profiles provide the corresponding alignment and logo for any group of proteins that share a given domain. However, the analysis of ZCCHCs (https://prosite.expasy.org/PDOC50158) may be biased, since it includes ZCCHCs from 260 retrotransposons and 285 cellular proteins. These sequences seem to have been chosen irrespectively of their phylogenetic distances, with mammalian factors being over-represented, including orthologs of very closely related species, and those of plants being under-represented.
Fig. 5.
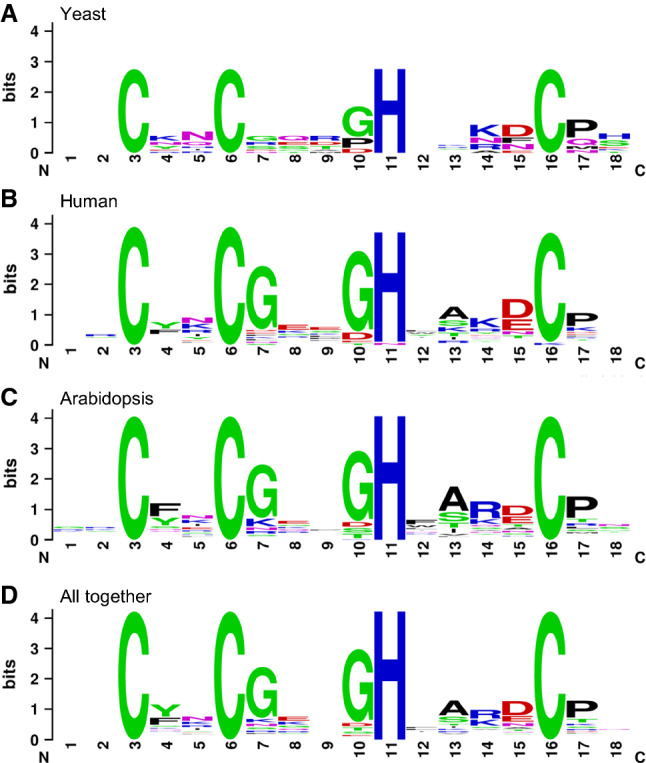
Logos representing normalized amino acid frequencies obtained from the multiple alignment of the 18 amino acids that constitute the ZCCHCs of yeast, human, and Arabidopsis proteins. Sequences were taken grouped by species (a–c) or all together (d) to generate the corresponding logos from multiple alignments, using the WebLogo version 3 software (https://weblogo.threeplusone.com/create.cgi). Uncertain and retrotransposon-related factors were not taken into account, and the resulting 198 sequences were aligned, all together or grouped by species
The results of our multiple alignments showed that in 160 (81%) sequences, G was at the 10th position, suggesting that the CX2CX3GHX4C is the ancestral sequence of ZCCHCs (Fig. 5 and Supplementary Figs. 1–4). In fact, in the InterPro database, ZCCHCs have two different accession numbers, IPR025836 for the CX2CX4HX4C sequence, and IPR025829 for CX2CX3GHX4C. We also found G in the 7th position in 140 (71%) of the sequences, and R at the 14th position in 51 of the 121 sequences of Arabidopsis (42%), an enrichment that was not found for yeast or human ZCCHCs (Supplemental Fig. 4), suggesting a lineage-specific evolution of the ZCCHCs in plants.
Tryptophan (W) displayed the most structural constraints, because it only occurred once at the 2nd, 4th, and 17th positions, but was found nineteen times at the 12th position. Hence, sequences in which any position is represented only once could be false positives or ZCCHCs with peculiar features. For example, we only found a ZCCHC-containing yeast factor with a W at its 2nd position: Bik1, the only ZCCHC-containing yeast factor that is not nuclear or involved in RNA metabolism. Its putative human ortholog, CLIP-170, harbors a tyrosine (Y), which is also an aromatic amino acid, instead of a W at the 2nd position of the ZCCHC. Therefore, the presence of these aromatic amino acids at the second position of the ZCCHCs could be necessary for them to interact with proteins instead with RNA. The AT1G75560 and AT4G00980 Arabidopsis proteins have W in the 4th and 17th positions, respectively, and both are annotated as “Zinc knuckle (CCHC-type) family protein” but have not been studied. PROSITE and SMART predicted the CX2CX4HX4C sequences present in these factors to be ZCCHCs (Supporting Dataset 1d). No human protein contained a W other than that of the 12th position of the ZCCHC.
To ascertain if there is conservation in the sequences of the ZCCHCs among functionally related factors, we constructed phylogenetic trees with the 198 ZCCHCs that we considered genuine (20 of yeast, 121 of Arabidopsis, and 57 of human ZCCHC proteins). Then, we focused our analysis on factors harboring a single ZCCHC, because the ZCCHCs of a specific protein that harbor more than one, grouped with the ZCCHCs of different proteins (Fig. 6), making it difficult to draw conclusions. We observed that most paralogs were grouped in clades, and that several clades included orthologs or co-orthologs. This was the case for human DDX41 and Arabidopsis RH34 and RH35; the yeast Mpe1, human RBBP6, and Arabidopsis PQT3 and PQT3L; the Slu7 orthologs of the three organisms; yeast Bik1 and human CLIP-170, each of which clustered with two Arabidopsis unknown proteins; and human and Arabidopsis XRN factors and U11/U12-31K spliceosomal proteins. However, we did not observe clusters among ZCCHCs from other proteins involved in the same general process (e.g., splicing, translation, and polyadenylation), except one with human SF1 and pNO40, which participate directly (SF1) or indirectly (pNO40) in pre-mRNA splicing. We also found an unexpected cluster formed by human SRSF7 and CPSF4 factors. SRSF7 is an SR factor involved in RNA export, but CPSF4 is a co-factor of the cleavage and polyadenylation machinery. However, the structures of the members of these two pairs of proteins are very different, with different domains and ZCCHC positions (Figs. 2, 3).
Fig. 6.
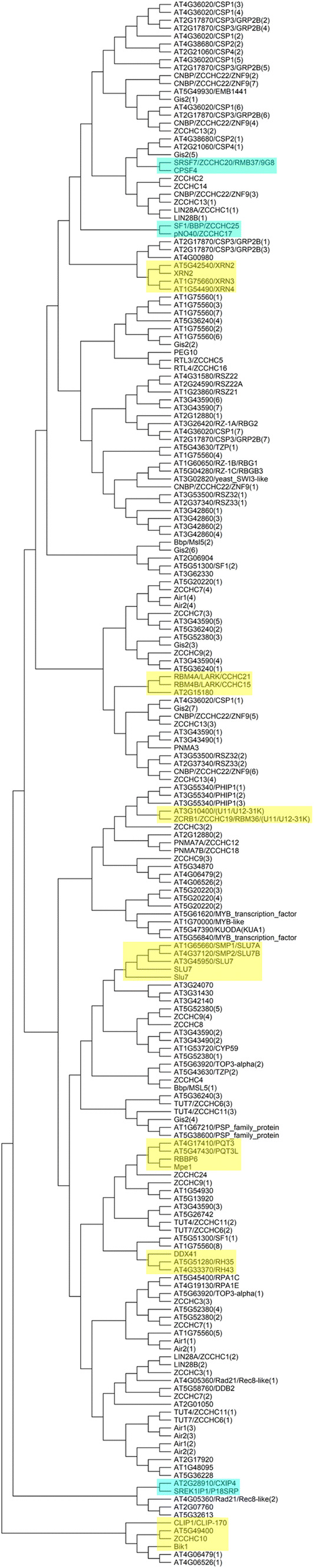
Phylogeny of the ZCCHCs of yeast, Arabidopsis, and human proteins. The unrooted tree was obtained with the MEGA X software (https://www.megasoftware.net/) from the alignment of the 18 amino acids of the 198 ZCCHCs found in yeast, human, and Arabidopsis proteins. The Neighbor-Joining algorithm was used, with bootstrap values from 1000 replicates. The position of a given ZCCHC in proteins with more than one of these motifs is indicated in brackets (1, 2, and 3 indicate first, second, and third, respectively, within the protein sequence, from its N- to its C-terminus). Clades highlighted in yellow or blue include ZCCHCs from proteins with a single ZCCHC that are known to be orthologs, or without a known functional relationship, respectively
In addition, we found the ZCCHC of AT2G15180, an unknown protein annotated as “zinc knuckle (CCHC-type) family protein”, clearly grouped with the two human RBM4 factors. In our BLASTP searches, however, we did not find any Arabidopsis protein similar to RBM4 factors, as previously mentioned. RBM4 factors harbor two RRM domains (Fig. 3), but none is annotated for AT2G15180, suggesting that is not their Arabidopsis functional ortholog. We also found a cluster formed by the plant-specific CXIP4 and human SREK1-interacting protein 1 (SREK1IP1), also known as p18 splicing regulatory protein (P18SRP), whose function is unknown, although it is known to interact with the SR splicing regulatory protein SRrp86 [179]. Only the ZCCHC is predicted in both proteins, in what seem to be similar regions, which may not be sufficient to consider them structural orthologs (Figs. 3, 4). The finding of similar CX2CX4HX4C sequences in proteins without any other structural homology may have not biological relevance.
Conclusions and perspectives
In this work, we compiled information available in public databases and the literature on ZCCHC-containing factors in yeast, Arabidopsis, and humans, which we considered representative of the Fungi, Plantae, and Animalia kingdoms, respectively. No representative of the Protista kingdom was considered because of the scarcity of functional genomics information available. We used PatMatch (for yeast and Arabidopsis) and ScanProsite (for human) to search for proteins containing the CX2CX4HX4C sequence, whose primary structures were later analyzed on UniProtKB, a database that identifies protein domains, motifs and low-complexity regions by compiling the information obtained from diverse programs, such as PROSITE, and SMART.
We found a wide array of combinations of ZCCHCs within proteins; in most cases, each protein had only a single copy, but there could be as many as eight. Moreover, they were present in any region of the protein, and could be alone or combined with other domains or motifs (Figs. 2, 3, 4). In addition, we found that many ZCCHC-containing proteins harbor low-complexity regions, which are mainly rich in P, L, G, R, or S, including RS domains that we found in proteins related to pre-mRNA splicing. Proteins with low-complexity regions are likely to have more interactors than those lacking these regions [180]; therefore, low-complexity regions could cooperate with ZCCHCs for RNA or protein binding, or have their own biological functions. Constructs expressing human CNBP recombinant proteins lacking the RG-rich region (Arg/Gly-rich) located between their first and second ZCCHC (Fig. 2) are required for high-affinity RNA binding. The proline-rich segment of ZCCHC8 (Fig. 2) is the site of interaction with the RRM domain of RBM7 in the NEXT complex of the exosome [106].
Our results obtained from PROSITE and SMART did not always coincide. For example, PROSITE predicted only three ZCCHCs in yeast Air2, but SMART predicted five, and we found four CX2CX4HX4C sequences within this protein (Supplementary Fig. 1 and Supporting Dataset 1a, 1b). Air2 is also considered by some studies to harbor five ZCCHCs, but the sequence of the second one is CX2CX5HX4C instead of CX2CX4HX4C, and its deletion does not impair its function [104]. A similar case is that of Retinitis pigmentosa 9 (RP9), which is annotated as a ZCCHC-containing factor in UniProtKB, but also harbors a CCHC sequence with a different spacing (CX2CX4HX6C). We also found in the literature that some variants of the canonical ZCCHC seem to behave as true zinc knuckles, but they are not annotated in UniProtKB, nor have they been predicted with any other program available. This is the case with the human RecQ4 helicase, which harbors an atypical ZCCHC in which the second C is substituted with an Asparagine (CNHC), while its animal (Drosophila, mouse, rat, and Xenopus) homologs harbor a canonical ZCCHC. Structural and functional analyses have shown that human and Xenopus RecQ4 bind nucleic acids in vitro, mainly forked and single-strand RNA substrates, and that the substitution of N by C, reconstituting a canonical ZCCHC, impairs binding to DNA substrates [181].
We found that the presence and number of ZCCHCs are usually conserved in orthologous proteins (Figs. 2, 3), suggesting a role that, in some cases, is involved in subcellular localization, translocation, or binding to other proteins or nucleic acids. More remarkable yet is the finding of similarly clustered ZCCHCs in evolutionarily distant orthologs (Fig. 6), which suggests selective pressure acting within highly conserved ortholog groups, probably because these specific sequences play an irreplaceable role in the process in which their proteins are involved. Phylogenetic analysis of ZCCHC sequences should allow discovery and/or discrimination of true functional paralogs and orthologs. Particularly interesting will be the genomes containing large multigene families, like those of plants.
Most ZCCHC-containing proteins are nuclear, a minority are nuclear and/or cytoplasmic, and very few are only cytoplasmic. Unexpectedly, we found an Arabidopsis ZCCHC-containing factor, the DEAD-box RNA helicase RH3, to be chloroplastic. This Arabidopsis protein and its maize co-orthologs, ZmRH3A and ZmRH3B, are involved in chloroplastic splicing and chloroplastic ribosome biogenesis [149], and database searches showed that they harbor a ZCCHC, as does their rice ortholog, encoded by Os03g61220. Most chloroplast proteins are encoded by nuclear genes, synthetized in the cytoplasm and imported into the chloroplast through their N-terminal transit peptide, which targets them to the chloroplast, where they are cleaved off. The ZCCHC at the end of the Gly-Ser-rich C-terminal region of the Arabidopsis RH3 protein (Fig. 4) likely forms part of the mature chloroplastic protein and is probably functional.
We also found six human neofunctionalized genes derived from the retrotransposon Gag protein, belonging to two different families (Supporting Dataset 1f). The proteins encoded by these genes interact with other proteins instead of RNA. The discovery of their interacting proteins allowed researchers to infer their functions [160]. Some of them seem to function in diverse tumor-related processes, promoting cell division, inhibiting apoptosis, or hindering the degradation of key proteins [182]. We found that many Arabidopsis proteins annotated as “Zinc knuckle (CCHC-type) family protein” are very similar to Ta11-like non-LTR retrotransposons, sharing the DUF4283 domain, and they could derive from retrotransposons and encode new functions, as in humans. Indeed, one of these genes, AT5G36228, is considered to be a transposable element locus whose polymorphisms in natural Arabidopsis populations have been proposed to play a role in adaptive evolution to changing environments [183].
We found that ZCCHC-containing factors are involved in most steps of RNA metabolism and affect mRNAs and ncRNAs, such as rRNAs or miRNAs. On the other hand, we found very few ZCCHC-containing factors involved in processes not related to mRNA metabolism. It is possible that other ZCCHC-containing factors participate in the other processes. The vast majority of ZCCHC-containing factors discussed here are only annotated as unknown proteins; this is especially true in the case of Arabidopsis, in which 28 of the 69 proteins identified are annotated as a “zinc knuckle (CCHC-type) family protein” or “nucleic acid binding/zinc ion binding protein” (Supporting Dataset 1d). The discovery of the DNAs, RNAs, and proteins that interact with ZCCHC-containing factors, and the search for these factors in diverse eukaryotes, along with deletion analyses of ZCCHCs, will provide a better understanding of their functions and a fruitful avenue of research to provide insight into RNA metabolism across eukaryotes.
Therefore, the identification of true ZCCHC-containing factors does not seem to be straightforward, and the number of these proteins may differ from what we found. The number probably would have been higher if we had considered in this review functional ZCCHCs with atypical (retaining the ability to bind zinc, as described in UniProtKB) or degenerate (not retaining) domains. However, it would have been lower if we had taken into account not only the primary conserved sequence of the putative ZCCHCs, but also their three-dimensional structures.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We apologize to those researchers whose work could not be cited due to space constraints. We thank J. L. Micol for his critical reading, comments, and suggestions on this paper.
Funding
This work was supported by the Ministerio de Ciencia e Innovación of Spain (Grant number BIO2017-89728-R MCI/AEI/FEDER EU) and the Generalitat Valenciana (Grant number PROMETEO/2019/117), both to M.R.P.; U.A.-V. held a predoctoral fellowship (GRISOLIAP/2016/134) from the Generalitat Valenciana.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Matthews JM, Sunde M. Zinc fingers-folds for many occasions. IUBMB Life. 2002;54(6):351–355. doi: 10.1080/15216540216035. [DOI] [PubMed] [Google Scholar]
- 2.Iuchi S. Three classes of C2H2 zinc finger proteins. Cell Mol Life Sci. 2001;58(4):625–635. doi: 10.1007/PL00000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laity JH, Lee BM, Wright PE. Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol. 2001;11(1):39–46. doi: 10.1016/S0959-440X(00)00167-6. [DOI] [PubMed] [Google Scholar]
- 4.Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, Chang HY, Dosztanyi Z, El-Gebali S, Fraser M, Gough J, Haft D, Holliday GL, Huang H, Huang X, Letunic I, Lopez R, Lu S, Marchler-Bauer A, Mi H, Mistry J, Natale DA, Necci M, Nuka G, Orengo CA, Park Y, Pesseat S, Piovesan D, Potter SC, Rawlings ND, Redaschi N, Richardson L, Rivoire C, Sangrador-Vegas A, Sigrist C, Sillitoe I, Smithers B, Squizzato S, Sutton G, Thanki N, Thomas PD, Tosatto SC, Wu CH, Xenarios I, Yeh LS, Young SY, Mitchell AL. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017;45(D1):D190–D199. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henderson LE, Copeland TD, Sowder RC, Smythers GW, Oroszlan S. Primary structure of the low molecular weight nucleic acid-binding proteins of murine leukemia viruses. J Biol Chem. 1981;256(16):8400–8406. [PubMed] [Google Scholar]
- 7.Green LM, Berg JM. A retroviral Cys-Xaa2-Cys-Xaa4-His-Xaa4-Cys peptide binds metal ions: spectroscopic studies and a proposed three-dimensional structure. Proc Natl Acad Sci USA. 1989;86(11):4047–4051. doi: 10.1073/pnas.86.11.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klein DJ, Johnson PE, Zollars ES, De Guzman RN, Summers MF. The NMR structure of the nucleocapsid protein from the mouse mammary tumor virus reveals unusual folding of the C-terminal zinc knuckle. Biochemistry. 2000;39(7):1604–1612. doi: 10.1021/bi9922493. [DOI] [PubMed] [Google Scholar]
- 9.De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279(5349):384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 10.Amodeo P, Castiglione Morelli MA, Ostuni A, Battistuzzi G, Bavoso A. Structural features in EIAV NCp11: a lentivirus nucleocapsid protein with a short linker. Biochemistry. 2006;45(17):5517–5526. doi: 10.1021/bi0524924. [DOI] [PubMed] [Google Scholar]
- 11.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 12.Daigle DM, Rossi L, Berghuis AM, Aravind L, Koonin EV, Brown ED. YjeQ, an essential, conserved, uncharacterized protein from Escherichia coli, is an unusual GTPase with circularly permuted G-motifs and marked burst kinetics. Biochemistry. 2002;41(37):11109–11117. doi: 10.1021/bi020355q. [DOI] [PubMed] [Google Scholar]
- 13.Jeganathan A, Razi A, Thurlow B, Ortega J. The C-terminal helix in the YjeQ zinc-finger domain catalyzes the release of RbfA during 30S ribosome subunit assembly. RNA. 2015;21(6):1203–1216. doi: 10.1261/rna.049171.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campbell TL, Henderson J, Heinrichs DE, Brown ED. The yjeQ gene is required for virulence of Staphylococcus aureus. Infect Immun. 2006;74(8):4918–4921. doi: 10.1128/IAI.00258-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Comartin DJ, Brown ED. Non-ribosomal factors in ribosome subunit assembly are emerging targets for new antibacterial drugs. Curr Opin Pharm. 2006;6(5):453–458. doi: 10.1016/j.coph.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 16.Augustin MA, Huber R, Kaiser JT. Crystal structure of a DNA-dependent RNA polymerase (DNA primase) Nat Struct Biol. 2001;8(1):57–61. doi: 10.1038/83060. [DOI] [PubMed] [Google Scholar]
- 17.Yan T, Yoo D, Berardini TZ, Mueller LA, Weems DC, Weng S, Cherry JM, Rhee SY. PatMatch: a program for finding patterns in peptide and nucleotide sequences. Nucleic Acids Res. 2005;33(Web Server issue):262–266. doi: 10.1093/nar/gki368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beasley SA, Hristova VA, Shaw GS. Structure of the Parkin in-between-ring domain provides insights for E3-ligase dysfunction in autosomal recessive Parkinson's disease. Proc Natl Acad Sci USA. 2007;104(9):3095–3100. doi: 10.1073/pnas.0610548104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fusaro AF, Bocca SN, Ramos RL, Barroco RM, Magioli C, Jorge VC, Coutinho TC, Rangel-Lima CM, De Rycke R, Inze D, Engler G, Sachetto-Martins G. AtGRP2, a cold-induced nucleo-cytoplasmic RNA-binding protein, has a role in flower and seed development. Planta. 2007;225(6):1339–1351. doi: 10.1007/s00425-006-0444-4. [DOI] [PubMed] [Google Scholar]
- 20.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoskins AA, Moore MJ. The spliceosome: a flexible, reversible macromolecular machine. Trends Biochem Sci. 2012;37(5):179–188. doi: 10.1016/j.tibs.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer K, Koester T, Staiger D. Pre-mRNA splicing in plants: in vivo functions of RNA-binding proteins Implicated in the splicing process. Biomolecules. 2015;5(3):1717–1740. doi: 10.3390/biom5031717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi Y. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat Rev Mol Cell Biol. 2017;11:655–670. doi: 10.1038/nrm.2017.86. [DOI] [PubMed] [Google Scholar]
- 24.Shi Y. The spliceosome: a protein-directed metalloribozyme. J Mol Biol. 2017;429(17):2640–2653. doi: 10.1016/j.jmb.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 25.Berglund JA, Chua K, Abovich N, Reed R, Rosbash M. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell. 1997;89(5):781–787. doi: 10.1016/S0092-8674(00)80261-5. [DOI] [PubMed] [Google Scholar]
- 26.Kramer A. Purification of splicing factor SF1, a heat-stable protein that functions in the assembly of a presplicing complex. Mol Cell Biol. 1992;12(10):4545–4552. doi: 10.1128/mcb.12.10.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berglund JA, Abovich N, Rosbash M. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev. 1998;12(6):858–867. doi: 10.1101/gad.12.6.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rain JC, Rafi Z, Rhani Z, Legrain P, Kramer A. Conservation of functional domains involved in RNA binding and protein-protein interactions in human and Saccharomyces cerevisiae pre-mRNA splicing factor SF1. RNA. 1998;4(5):551–565. doi: 10.1017/s1355838298980335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garrey SM, Voelker R, Berglund JA. An extended RNA binding site for the yeast branch point-binding protein and the role of its zinc knuckle domains in RNA binding. J Biol Chem. 2006;281(37):27443–27453. doi: 10.1074/jbc.M603137200. [DOI] [PubMed] [Google Scholar]
- 30.Rutz B, Seraphin B. A dual role for BBP/ScSF1 in nuclear pre-mRNA retention and splicing. EMBO J. 2000;19(8):1873–1886. doi: 10.1093/emboj/19.8.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jang YH, Park HY, Lee KC, Thu MP, Kim SK, Suh MC, Kang H, Kim JK. A homolog of splicing factor SF1 is essential for development and is involved in the alternative splicing of pre-mRNA in Arabidopsis thaliana. Plant J. 2014;78(4):591–603. doi: 10.1111/tpj.12491. [DOI] [PubMed] [Google Scholar]
- 32.Crotti LB, Bacikova D, Horowitz DS. The Prp18 protein stabilizes the interaction of both exons with the U5 snRNA during the second step of pre-mRNA splicing. Genes Dev. 2007;21(10):1204–1216. doi: 10.1101/gad.1538207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones MH, Frank DN, Guthrie C. Characterization and functional ordering of Slu7p and Prp17p during the second step of pre-mRNA splicing in yeast. Proc Natl Acad Sci USA. 1995;92(21):9687–9691. doi: 10.1073/pnas.92.21.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.James SA, Turner W, Schwer B. How Slu7 and Prp18 cooperate in the second step of yeast pre-mRNA splicing. RNA. 2002;8(8):1068–1077. doi: 10.1017/s1355838202022033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frank D, Guthrie C. An essential splicing factor, SLU7, mediates 3′ splice site choice in yeast. Genes Dev. 1992;6(11):2112–2124. doi: 10.1101/gad.6.11.2112. [DOI] [PubMed] [Google Scholar]
- 36.Chua K, Reed R. The RNA splicing factor hSlu7 is required for correct 3′ splice-site choice. Nature. 1999;402(6758):207–210. doi: 10.1038/46086. [DOI] [PubMed] [Google Scholar]
- 37.Shomron N, Reznik M, Ast G. Splicing factor hSlu7 contains a unique functional domain required to retain the protein within the nucleus. Mol Biol Cell. 2004;15(8):3782–3795. doi: 10.1091/mbc.E04-02-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clay NK, Nelson T. The recessive epigenetic swellmap mutation affects the expression of two step II splicing factors required for the transcription of the cell proliferation gene STRUWWELPETER and for the timing of cell cycle arrest in the Arabidopsis leaf. Plant Cell. 2005;17(7):1994–2008. doi: 10.1105/tpc.105.032771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin CF, Mount SM, Jarmolowski A, Makalowski W. Evolutionary dynamics of U12-type spliceosomal introns. BMC Evol Biol. 2010;10:47. doi: 10.1186/1471-2148-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niemela EH, Frilander MJ. Regulation of gene expression through inefficient splicing of U12-type introns. RNA Biol. 2014;11(11):1325–1329. doi: 10.1080/15476286.2014.996454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H, Gao MX, Li L, Wang B, Hori N, Sato K. Isolation, expression, and characterization of the human ZCRB1 gene mapped to 12q12. Genomics. 2007;89(1):59–69. doi: 10.1016/j.ygeno.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 42.Kim WY, Jung HJ, Kwak KJ, Kim MK, Oh SH, Han YS, Kang H. The Arabidopsis U12-type spliceosomal protein U11/U12-31K is involved in U12 intron splicing via RNA chaperone activity and affects plant development. Plant Cell. 2010;22(12):3951–3962. doi: 10.1105/tpc.110.079103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Will CL, Schneider C, Hossbach M, Urlaub H, Rauhut R, Elbashir S, Tuschl T, Luhrmann R. The human 18S U11/U12 snRNP contains a set of novel proteins not found in the U2-dependent spliceosome. RNA. 2004;10(6):929–941. doi: 10.1261/rna.7320604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manley JL, Krainer AR. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins) Genes Dev. 2010;24(11):1073–1074. doi: 10.1101/gad.1934910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haynes C, Iakoucheva LM. Serine/arginine-rich splicing factors belong to a class of intrinsically disordered proteins. Nucleic Acids Res. 2006;34(1):305–312. doi: 10.1093/nar/gkj424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Howard JM, Sanford JR. The RNAissance family: SR proteins as multifaceted regulators of gene expression. Wiley Interdiscip Rev RNA. 2015;6(1):93–110. doi: 10.1002/wrna.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417(1):15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 48.Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6(9):1197–1211. doi: 10.1017/S1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418(6894):236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- 50.Busch A, Hertel KJ. Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip Rev RNA. 2012;3(1):1–12. doi: 10.1002/wrna.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang Y, Steitz JA. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol Cell. 2001;7(4):899–905. doi: 10.1016/S1097-2765(01)00233-7. [DOI] [PubMed] [Google Scholar]
- 52.Rauch HB, Patrick TL, Klusman KM, Battistuzzi FU, Mei W, Brendel VP, Lal SK. Discovery and expression analysis of alternative splicing events conserved among plant SR proteins. Mol Biol Evol. 2014;31(3):605–613. doi: 10.1093/molbev/mst238. [DOI] [PubMed] [Google Scholar]
- 53.Lorkovic ZJ, Hilscher J, Barta A. Co-localisation studies of Arabidopsis SR splicing factors reveal different types of speckles in plant cell nuclei. Exp Cell Res. 2008;314(17):3175–3186. doi: 10.1016/j.yexcr.2008.06.020. [DOI] [PubMed] [Google Scholar]
- 54.Cavaloc Y, Bourgeois CF, Kister L, Stevenin J. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA. 1999;5(3):468–483. doi: 10.1017/s1355838299981967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lopato S, Gattoni R, Fabini G, Stevenin J, Barta A. A novel family of plant splicing factors with a Zn knuckle motif: examination of RNA binding and splicing activities. Plant Mol Biol. 1999;39(4):761–773. doi: 10.1023/a:1006129615846. [DOI] [PubMed] [Google Scholar]
- 56.Rausin G, Tillemans V, Stankovic N, Hanikenne M, Motte P. Dynamic nucleocytoplasmic shuttling of an Arabidopsis SR splicing factor: role of the RNA-binding domains. Plant Physiol. 2010;153(1):273–284. doi: 10.1104/pp.110.154740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lopato S, Forstner C, Kalyna M, Hilscher J, Langhammer U, Indrapichate K, Lorkovic ZJ, Barta A. Network of interactions of a novel plant-specific Arg/Ser-rich protein, atRSZ33, with atSC35-like splicing factors. J Biol Chem. 2002;277(42):39989–39998. doi: 10.1074/jbc.M206455200. [DOI] [PubMed] [Google Scholar]
- 58.Golovkin M, Reddy AS. The plant U1 small nuclear ribonucleoprotein particle 70K protein interacts with two novel serine/arginine-rich proteins. Plant Cell. 1998;10(10):1637–1648. doi: 10.1105/tpc.10.10.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang XY, Niu J, Sun MX, Zhu J, Gao JF, Yang J, Zhou Q, Yang ZN. CYCLIN-DEPENDENT KINASE G1 is associated with the spliceosome to regulate CALLOSE SYNTHASE5 splicing and pollen wall formation in Arabidopsis. Plant Cell. 2013;25(2):637–648. doi: 10.1105/tpc.112.107896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chang WL, Lee DC, Leu S, Huang YM, Lu MC, Ouyang P. Molecular characterization of a novel nucleolar protein, pNO40. Biochem Biophys Res Commun. 2003;307(3):569–577. doi: 10.1016/s0006-291x(03)01208-7. [DOI] [PubMed] [Google Scholar]
- 61.Gueydan C, Wauquier C, De Mees C, Huez G, Kruys V. Identification of ribosomal proteins specific to higher eukaryotic organisms. J Biol Chem. 2002;277(47):45034–45040. doi: 10.1074/jbc.M208551200. [DOI] [PubMed] [Google Scholar]
- 62.Lin YM, Chu PH, Ouyang P. Ectopically expressed pNO40 suppresses ribosomal RNA synthesis by inhibiting UBF-dependent transcription activation. Biochem Biophys Res Commun. 2019 doi: 10.1016/j.bbrc.2019.06.057. [DOI] [PubMed] [Google Scholar]
- 63.Lin YM, Chu PH, Li YZ, Ouyang P. Ribosomal protein pNO40 mediates nucleolar sequestration of SR family splicing factors and its overexpression impairs mRNA metabolism. Cell Signal. 2017;32:12–23. doi: 10.1016/j.cellsig.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 64.Moss EG. Heterochronic genes and the nature of developmental time. Curr Biol. 2007;17(11):425–434. doi: 10.1016/j.cub.2007.03.043. [DOI] [PubMed] [Google Scholar]
- 65.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 66.Moss EG, Tang L. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev Biol. 2003;258(2):432–442. doi: 10.1016/s0012-1606(03)00126-x. [DOI] [PubMed] [Google Scholar]
- 67.Piskounova E, Polytarchou C, Thornton JE, LaPierre RJ, Pothoulakis C, Hagan JP, Iliopoulos D, Gregory RI. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 2011;147(5):1066–1079. doi: 10.1016/j.cell.2011.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tsialikas J, Romer-Seibert J. LIN28: roles and regulation in development and beyond. Development. 2015;142(14):2397–2404. doi: 10.1242/dev.117580. [DOI] [PubMed] [Google Scholar]
- 69.Graumann PL, Marahiel MA. A superfamily of proteins that contain the cold-shock domain. Trends Biochem Sci. 1998;23(8):286–290. doi: 10.1016/S0968-0004(98)01255-9. [DOI] [PubMed] [Google Scholar]
- 70.Mihailovich M, Militti C, Gabaldon T, Gebauer F. Eukaryotic cold shock domain proteins: highly versatile regulators of gene expression. BioEssays. 2010;32(2):109–118. doi: 10.1002/bies.200900122. [DOI] [PubMed] [Google Scholar]
- 71.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138(4):696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 72.Cho J, Chang H, Kwon SC, Kim B, Kim Y, Choe J, Ha M, Kim YK, Kim VN. LIN28A is a suppressor of ER-associated translation in embryonic stem cells. Cell. 2012;151(4):765–777. doi: 10.1016/j.cell.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 73.Wang X, Zhang S, Dou Y, Zhang C, Chen X, Yu B, Ren G. Synergistic and independent actions of multiple terminal nucleotidyl transferases in the 3′ tailing of small RNAs in Arabidopsis. PLoS Genet. 2015;11(4):e1005091. doi: 10.1371/journal.pgen.1005091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kingsley PD, Palis J. GRP2 proteins contain both CCHC zinc fingers and a cold shock domain. Plant Cell. 1994;6(11):1522–1523. doi: 10.1105/tpc.6.11.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sachetto-Martins G, Franco LO, de Oliveira DE. Plant glycine-rich proteins: a family or just proteins with a common motif? Biochim Biophys Acta. 2000;1492(1):1–14. doi: 10.1016/s0167-4781(00)00064-6. [DOI] [PubMed] [Google Scholar]
- 76.Mangeon A, Junqueira RM, Sachetto-Martins G. Functional diversity of the plant glycine-rich proteins superfamily. Plant Signal Behav. 2010;5(2):99–104. doi: 10.4161/psb.5.2.10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Krishnamurthy P, Kim JA, Jeong MJ, Kang CH, Lee SI. Defining the RNA-binding glycine-rich (RBG) gene superfamily: new insights into nomenclature, phylogeny, and evolutionary trends obtained by genome-wide comparative analysis of Arabidopsis, Chinese cabbage, rice and maize genomes. Mol Genet Genom. 2015;290(6):2279–2295. doi: 10.1007/s00438-015-1080-0. [DOI] [PubMed] [Google Scholar]
- 78.Juntawong P, Sorenson R, Bailey-Serres J. Cold shock protein 1 chaperones mRNAs during translation in Arabidopsis thaliana. Plant J. 2013;74(6):1016–1028. doi: 10.1111/tpj.12187. [DOI] [PubMed] [Google Scholar]
- 79.Sasaki K, Kim MH, Imai R. Arabidopsis COLD SHOCK DOMAIN PROTEIN2 is a RNA chaperone that is regulated by cold and developmental signals. Biochem Biophys Res Commun. 2007;364(3):633–638. doi: 10.1016/j.bbrc.2007.10.059. [DOI] [PubMed] [Google Scholar]
- 80.Sasaki K, Kim MH, Imai R. Arabidopsis COLD SHOCK DOMAIN PROTEIN 2 is a negative regulator of cold acclimation. New Phytol. 2013;198(1):95–102. doi: 10.1111/nph.12118. [DOI] [PubMed] [Google Scholar]
- 81.Kim MH, Sato S, Sasaki K, Saburi W, Matsui H, Imai R. COLD SHOCK DOMAIN PROTEIN 3 is involved in salt and drought stress tolerance in Arabidopsis. FEBS Open Biol. 2013;3:438–442. doi: 10.1016/j.fob.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pontvianne F, Matia I, Douet J, Tourmente S, Medina FJ, Echeverria M, Saez-Vasquez J. Characterization of AtNUC-L1 reveals a central role of nucleolin in nucleolus organization and silencing of AtNUC-L2 gene in Arabidopsis. Mol Biol Cell. 2007;18(2):369–379. doi: 10.1091/mbc.E06-08-0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Petricka JJ, Nelson TM. Arabidopsis nucleolin affects plant development and patterning. Plant Physiol. 2007;144(1):173–186. doi: 10.1104/pp.106.093575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim MH, Sonoda Y, Sasaki K, Kaminaka H, Imai R. Interactome analysis reveals versatile functions of Arabidopsis COLD SHOCK DOMAIN PROTEIN 3 in RNA processing within the nucleus and cytoplasm. Cell Stress Chaperones. 2013;18(4):517–525. doi: 10.1007/s12192-012-0398-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim YO, Kim JS, Kang H. Cold-inducible zinc finger-containing glycine-rich RNA-binding protein contributes to the enhancement of freezing tolerance in Arabidopsis thaliana. Plant J. 2005;42(6):890–900. doi: 10.1111/j.1365-313X.2005.02420.x. [DOI] [PubMed] [Google Scholar]
- 86.Wu Z, Zhu D, Lin X, Miao J, Gu L, Deng X, Yang Q, Sun K, Zhu D, Cao X, Tsuge T, Dean C, Aoyama T, Gu H, Qu LJ. RNA binding proteins RZ-1B and RZ-1C play critical roles in regulating pre-mRNA splicing and gene expression during development in Arabidopsis. Plant Cell. 2016;28(1):55–73. doi: 10.1105/tpc.15.00949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Neve J, Patel R, Wang Z, Louey A, Furger AM. Cleavage and polyadenylation: ending the message expands gene regulation. RNA Biol. 2017;14(7):865–890. doi: 10.1080/15476286.2017.1306171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vo LT, Minet M, Schmitter JM, Lacroute F, Wyers F. Mpe1, a zinc knuckle protein, is an essential component of yeast cleavage and polyadenylation factor required for the cleavage and polyadenylation of mRNA. Mol Cell Biol. 2001;21(24):8346–8356. doi: 10.1128/MCB.21.24.8346-8356.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee SD, Moore CL. Efficient mRNA polyadenylation requires a ubiquitin-like domain, a zinc knuckle, and a RING finger domain, all contained in the Mpe1 protein. Mol Cell Biol. 2014;34(21):3955–3967. doi: 10.1128/MCB.00077-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sakai Y, Saijo M, Coelho K, Kishino T, Niikawa N, Taya Y. cDNA sequence and chromosomal localization of a novel human protein, RBQ-1 (RBBP6), that binds to the retinoblastoma gene product. Genomics. 1995;30(1):98–101. doi: 10.1006/geno.1995.0017. [DOI] [PubMed] [Google Scholar]
- 91.Simons A, Melamed-Bessudo C, Wolkowicz R, Sperling J, Sperling R, Eisenbach L, Rotter V. PACT: cloning and characterization of a cellular p53 binding protein that interacts with Rb. Oncogene. 1997;14(2):145–155. doi: 10.1038/sj.onc.1200825. [DOI] [PubMed] [Google Scholar]
- 92.Li L, Deng B, Xing G, Teng Y, Tian C, Cheng X, Yin X, Yang J, Gao X, Zhu Y, Sun Q, Zhang L, Yang X, He F. PACT is a negative regulator of p53 and essential for cell growth and embryonic development. Proc Natl Acad Sci USA. 2007;104(19):7951–7956. doi: 10.1073/pnas.0701916104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Di Giammartino DC, Li W, Ogami K, Yashinskie JJ, Hoque M, Tian B, Manley JL. RBBP6 isoforms regulate the human polyadenylation machinery and modulate expression of mRNAs with AU-rich 3' UTRs. Genes Dev. 2014;28(20):2248–2260. doi: 10.1101/gad.245787.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Luo C, Cai XT, Du J, Zhao TL, Wang PF, Zhao PX, Liu R, Xie Q, Cao XF, Xiang CB. PARAQUAT TOLERANCE3 Is an E3 Ligase that switches off activated oxidative response by targeting histone-modifying PROTEIN METHYLTRANSFERASE4b. PLoS Genet. 2016;12(9):e1006332. doi: 10.1371/journal.pgen.1006332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pugh DJ, Ab E, Faro A, Lutya PT, Hoffmann E, Rees DJ. DWNN, a novel ubiquitin-like domain, implicates RBBP6 in mRNA processing and ubiquitin-like pathways. BMC Struct Biol. 2006;6:1. doi: 10.1186/1472-6807-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Barabino SM, Hubner W, Jenny A, Minvielle-Sebastia L, Keller W. The 30-kD subunit of mammalian cleavage and polyadenylation specificity factor and its yeast homolog are RNA-binding zinc finger proteins. Genes Dev. 1997;11(13):1703–1716. doi: 10.1101/gad.11.13.1703. [DOI] [PubMed] [Google Scholar]
- 97.Chakrabarti M, Hunt AG. CPSF30 at the interface of alternative polyadenylation and cellular signaling in plants. Biomolecules. 2015;5(2):1151–1168. doi: 10.3390/biom5021151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu Q, Dreyfuss G. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol Cell Biol. 1995;15(5):2800–2808. doi: 10.1128/mcb.15.5.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Siebel CW, Guthrie C. The essential yeast RNA binding protein Np13p is methylated. Proc Natl Acad Sci USA. 1996;93(24):13641–13646. doi: 10.1073/pnas.93.24.13641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jarvelin AI, Noerenberg M, Davis I, Castello A. The new (dis)order in RNA regulation. Cell Commun Signal. 2016;14:9. doi: 10.1186/s12964-016-0132-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ozdilek BA, Thompson VF, Ahmed NS, White CI, Batey RT, Schwartz JC. Intrinsically disordered RGG/RG domains mediate degenerate specificity in RNA binding. Nucleic Acids Res. 2017;45(13):7984–7996. doi: 10.1093/nar/gkx460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zinder JC, Lima CD. Targeting RNA for processing or destruction by the eukaryotic RNA exosome and its cofactors. Genes Dev. 2017;31(2):88–100. doi: 10.1101/gad.294769.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Houseley J, Tollervey D. The nuclear RNA surveillance machinery: the link between ncRNAs and genome structure in budding yeast? Biochim Biophys Acta. 2008;1779(4):239–246. doi: 10.1016/j.bbagrm.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 104.Fasken MB, Leung SW, Banerjee A, Kodani MO, Chavez R, Bowman EA, Purohit MK, Rubinson ME, Rubinson EH, Corbett AH. Air1 zinc knuckles 4 and 5 and a conserved IWRXY motif are critical for the function and integrity of the Trf4/5-Air1/2-Mtr4 polyadenylation (TRAMP) RNA quality control complex. J Biol Chem. 2011;286(43):37429–37445. doi: 10.1074/jbc.M111.271494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vanácová S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3(6):e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Falk S, Finogenova K, Melko M, Benda C, Lykke-Andersen S, Jensen TH, Conti E. Structure of the RBM7-ZCCHC8 core of the NEXT complex reveals connections to splicing factors. Nat Commun. 2016;7:13573. doi: 10.1038/ncomms13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fox MJ, Mosley AL. Rrp 6: Integrated roles in nuclear RNA metabolism and transcription termination. Wiley Interdiscip Rev RNA. 2016;7(1):91–104. doi: 10.1002/wrna.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lange H, Holec S, Cognat V, Pieuchot L, Le Ret M, Canaday J, Gagliardi D. Degradation of a polyadenylated rRNA maturation by-product involves one of the three RRP6-like proteins in Arabidopsis thaliana. Mol Cell Biol. 2008;28(9):3038–3044. doi: 10.1128/MCB.02064-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sudo H, Nozaki A, Uno H, Ishida Y, Nagahama M. Interaction properties of human TRAMP-like proteins and their role in pre-rRNA 5'ETS turnover. FEBS Lett. 2016;590(17):2963–2972. doi: 10.1002/1873-3468.12314. [DOI] [PubMed] [Google Scholar]
- 110.Lubas M, Christensen MS, Kristiansen MS, Domanski M, Falkenby LG, Lykke-Andersen S, Andersen JS, Dziembowski A, Jensen TH. Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell. 2011;43(4):624–637. doi: 10.1016/j.molcel.2011.06.028. [DOI] [PubMed] [Google Scholar]
- 111.Lange H, Sement FM, Gagliardi D. MTR4, a putative RNA helicase and exosome co-factor, is required for proper rRNA biogenesis and development in Arabidopsis thaliana. Plant J. 2011;68(1):51–63. doi: 10.1111/j.1365-313X.2011.04675.x. [DOI] [PubMed] [Google Scholar]
- 112.Lange H, Zuber H, Sement FM, Chicher J, Kuhn L, Hammann P, Brunaud V, Berard C, Bouteiller N, Balzergue S, Aubourg S, Martin-Magniette ML, Vaucheret H, Gagliardi D. The RNA helicases AtMTR4 and HEN2 target specific subsets of nuclear transcripts for degradation by the nuclear exosome in Arabidopsis thaliana. PLoS Genet. 2014;10(8):e1004564. doi: 10.1371/journal.pgen.1004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liquori CL, Ricker K, Moseley ML, Jacobsen JF, Kress W, Naylor SL, Day JW, Ranum LP. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293(5531):864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- 114.Rajavashisth TB, Taylor AK, Andalibi A, Svenson KL, Lusis AJ. Identification of a zinc finger protein that binds to the sterol regulatory element. Science. 1989;245(4918):640–643. doi: 10.1126/science.2562787. [DOI] [PubMed] [Google Scholar]
- 115.Pellizzoni L, Lotti F, Maras B, Pierandrei-Amaldi P. Cellular nucleic acid binding protein binds a conserved region of the 5′ UTR of Xenopus laevis ribosomal protein mRNAs. J Mol Biol. 1997;267(2):264–275. doi: 10.1006/jmbi.1996.0888. [DOI] [PubMed] [Google Scholar]
- 116.Pellizzoni L, Lotti F, Rutjes SA, Pierandrei-Amaldi P. Involvement of the Xenopus laevis Ro60 autoantigen in the alternative interaction of La and CNBP proteins with the 5′ UTR of L4 ribosomal protein mRNA. J Mol Biol. 1998;281(4):593–608. doi: 10.1006/jmbi.1998.1961. [DOI] [PubMed] [Google Scholar]
- 117.Rojas M, Farr GW, Fernandez CF, Lauden L, McCormack JC, Wolin SL. Yeast Gis2 and its human ortholog CNBP are novel components of stress-induced RNP granules. PLoS One. 2012;7(12):e52824. doi: 10.1371/journal.pone.0052824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36(6):932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Huichalaf C, Schoser B, Schneider-Gold C, Jin B, Sarkar P, Timchenko L. Reduction of the rate of protein translation in patients with myotonic dystrophy 2. J Neurosci. 2009;29(28):9042–9049. doi: 10.1523/JNEUROSCI.1983-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sammons MA, Samir P, Link AJ. Saccharomyces cerevisiae Gis2 interacts with the translation machinery and is orthogonal to myotonic dystrophy type 2 protein ZNF9. Biochem Biophys Res Commun. 2011;406(1):13–19. doi: 10.1016/j.bbrc.2011.01.086. [DOI] [PubMed] [Google Scholar]
- 121.Iadevaia V, Caldarola S, Tino E, Amaldi F, Loreni F. All translation elongation factors and the e, f, and h subunits of translation initiation factor 3 are encoded by 5′-terminal oligopyrimidine (TOP) mRNAs. RNA. 2008;14(9):1730–1736. doi: 10.1261/rna.1037108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.David AP, Pipier A, Pascutti F, Binolfi A, Weiner AMJ, Challier E, Heckel S, Calsou P, Gomez D, Calcaterra NB, Armas P. CNBP controls transcription by unfolding DNA G-quadruplex structures. Nucleic Acids Res. 2019;47(15):7901–7913. doi: 10.1093/nar/gkz527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Benhalevy D, Gupta SK, Danan CH, Ghosal S, Sun HW, Kazemier HG, Paeschke K, Hafner M, Juranek SA. The human CCHC-type zinc finger nucleic acid-binding protein binds G-Rich elements in target mRNA coding sequences and promotes translation. Cell Rep. 2017;18(12):2979–2990. doi: 10.1016/j.celrep.2017.02.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Antonucci L, D'Amico D, Di Magno L, Coni S, Di Marcotullio L, Cardinali B, Gulino A, Ciapponi L, Canettieri G. CNBP regulates wing development in Drosophila melanogaster by promoting IRES-dependent translation of dMyc. Cell Cycle. 2014;13(3):434–439. doi: 10.4161/cc.27268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.McNeil GP, Schroeder AJ, Roberts MA, Jackson FR. Genetic analysis of functional domains within the Drosophila LARK RNA-binding protein. Genetics. 2001;159(1):229–240. doi: 10.1093/genetics/159.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Markus MA, Morris BJ. RBM4: a multifunctional RNA-binding protein. Int J Biochem Cell Biol. 2009;41(4):740–743. doi: 10.1016/j.biocel.2008.05.027. [DOI] [PubMed] [Google Scholar]
- 127.Niu K, Xiang L, Jin Y, Peng Y, Wu F, Tang W, Zhang X, Deng H, Xiang H, Li S, Wang J, Song Q, Feng Q. Identification of LARK as a novel and conserved G-quadruplex binding protein in invertebrates and vertebrates. Nucleic Acids Res. 2019;47(14):7306–7320. doi: 10.1093/nar/gkz484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Parker R. RNA degradation in Saccharomyces cerevisae. Genetics. 2012;191(3):671–702. doi: 10.1534/genetics.111.137265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol. 2004;11(2):121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 130.Medina DA, Jordan-Pla A, Millan-Zambrano G, Chavez S, Choder M, Perez-Ortin JE. Cytoplasmic 5'–3' exonuclease Xrn1p is also a genome-wide transcription factor in yeast. Front Genet. 2014;5:1. doi: 10.3389/fgene.2014.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Luo W, Johnson AW, Bentley DL. The role of Rat1 in coupling mRNA 3'-end processing to transcription termination: implications for a unified allosteric-torpedo model. Genes Dev. 2006;20(8):954–965. doi: 10.1101/gad.1409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.El Hage A, Koper M, Kufel J, Tollervey D. Efficient termination of transcription by RNA polymerase I requires the 5′ exonuclease Rat1 in yeast. Genes Dev. 2008;22(8):1069–1081. doi: 10.1101/gad.463708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Petfalski E, Dandekar T, Henry Y, Tollervey D. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol Cell Biol. 1998;18(3):1181–1189. doi: 10.1128/mcb.18.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Geerlings TH, Vos JC, Raue HA. The final step in the formation of 25S rRNA in Saccharomyces cerevisiae is performed by 5′ → 3′exonucleases. RNA. 2000;6(12):1698–1703. doi: 10.1017/s1355838200001540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.West S, Gromak N, Proudfoot NJ. Human 5′ → 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432(7016):522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- 136.Morales JC, Richard P, Patidar PL, Motea EA, Dang TT, Manley JL, Boothman DA. XRN2 links transcription termination to DNA damage and replication stress. PLoS Genet. 2016;12(7):e1006107. doi: 10.1371/journal.pgen.1006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kastenmayer JP, Green PJ. Novel features of the XRN-family in Arabidopsis: evidence that AtXRN4, one of several orthologs of nuclear Xrn2p/Rat1p, functions in the cytoplasm. Proc Natl Acad Sci USA. 2000;97(25):13985–13990. doi: 10.1073/pnas.97.25.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zakrzewska-Placzek M, Souret FF, Sobczyk GJ, Green PJ, Kufel J. Arabidopsis thaliana XRN2 is required for primary cleavage in the pre-ribosomal RNA. Nucleic Acids Res. 2010;38(13):4487–4502. doi: 10.1093/nar/gkq172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Souret FF, Kastenmayer JP, Green PJ. AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol Cell. 2004;15(2):173–183. doi: 10.1016/j.molcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 140.Gazzani S, Lawrenson T, Woodward C, Headon D, Sablowski R. A link between mRNA turnover and RNA interference in Arabidopsis. Science. 2004;306(5698):1046–1048. doi: 10.1126/science.1101092. [DOI] [PubMed] [Google Scholar]
- 141.Ma H, Wang X, Cai J, Dai Q, Natchiar SK, Lv R, Chen K, Lu Z, Chen H, Shi YG, Lan F, Fan J, Klaholz BP, Pan T, Shi Y, He C. N(6-)Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat Chem Biol. 2019;15(1):88–94. doi: 10.1038/s41589-018-0184-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhou A, Zhou J, Yang L, Liu M, Li H, Xu S, Han M, Zhang J. A nuclear localized protein ZCCHC9 is expressed in cerebral cortex and suppresses the MAPK signal pathway. J Genet Genom. 2008;35(8):467–472. doi: 10.1016/s1673-8527(08)60064-8. [DOI] [PubMed] [Google Scholar]
- 143.Sanudo M, Jacko M, Rammelt C, Vanacova S, Stefl R. 1H, 13C, and 15N chemical shift assignments of ZCCHC9. Biomol NMR Assign. 2011;5(1):19–21. doi: 10.1007/s12104-010-9257-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tafforeau L, Zorbas C, Langhendries JL, Mullineux ST, Stamatopoulou V, Mullier R, Wacheul L, Lafontaine DL. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol Cell. 2013;51(4):539–551. doi: 10.1016/j.molcel.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 145.Hsin JP, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012;26(19):2119–2137. doi: 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gullerova M, Barta A, Lorkovic ZJ. AtCyp59 is a multidomain cyclophilin from Arabidopsis thaliana that interacts with SR proteins and the C-terminal domain of the RNA polymerase II. RNA. 2006;12(4):631–643. doi: 10.1261/rna.2226106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tzafrir I, Pena-Muralla R, Dickerman A, Berg M, Rogers R, Hutchens S, Sweeney TC, McElver J, Aux G, Patton D, Meinke D. Identification of genes required for embryo development in Arabidopsis. Plant Physiol. 2004;135(3):1206–1220. doi: 10.1104/pp.104.045179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Shao S, Brown A, Santhanam B, Hegde RS. Structure and assembly pathway of the ribosome quality control complex. Mol Cell. 2015;57(3):433–444. doi: 10.1016/j.molcel.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Asakura Y, Galarneau E, Watkins KP, Barkan A, van Wijk KJ. Chloroplast RH3 DEAD box RNA helicases in maize and Arabidopsis function in splicing of specific group II introns and affect chloroplast ribosome biogenesis. Plant Physiol. 2012;159(3):961–974. doi: 10.1104/pp.112.197525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gu L, Xu T, Lee K, Lee KH, Kang H. A chloroplast-localized DEAD-box RNA helicaseAtRH3 is essential for intron splicing and plays an important role in the growth and stress response in Arabidopsis thaliana. Plant Physiol Biochem. 2014;82:309–318. doi: 10.1016/j.plaphy.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 151.Han P, Ye W, Lv X, Ma H, Weng D, Dong Y, Cheng L, Chen H, Zhang L, Xu Z, Lei Y, Zhang F. DDX50 inhibits the replication of dengue virus 2 by upregulating IFN-beta production. Arch Virol. 2017;162(6):1487–1494. doi: 10.1007/s00705-017-3250-3. [DOI] [PubMed] [Google Scholar]
- 152.Ma L, Xie B, Hong Z, Verma DP, Zhang Z. A novel RNA-binding protein associated with cell plate formation. Plant Physiol. 2008;148(1):223–234. doi: 10.1104/pp.108.120527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Miller RK, D'Silva S, Moore JK, Goodson HV. The CLIP-170 orthologue Bik1p and positioning the mitotic spindle in yeast. Curr Top Dev Biol. 2006;76:49–87. doi: 10.1016/S0070-2153(06)76002-1. [DOI] [PubMed] [Google Scholar]
- 154.Blake-Hodek KA, Cassimeris L, Huffaker TC. Regulation of microtubule dynamics by Bim1 and Bik1, the budding yeast members of the EB1 and CLIP-170 families of plus-end tracking proteins. Mol Biol Cell. 2010;21(12):2013–2023. doi: 10.1091/mbc.E10-02-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Weisbrich A, Honnappa S, Jaussi R, Okhrimenko O, Frey D, Jelesarov I, Akhmanova A, Steinmetz MO. Structure-function relationship of CAP-Gly domains. Nat Struct Mol Biol. 2007;14(10):959–967. doi: 10.1038/nsmb1291. [DOI] [PubMed] [Google Scholar]
- 156.Gardiner J, Overall R, Marc J. Putative Arabidopsis homologues of metazoan coiled-coil cytoskeletal proteins. Cell Biol Int. 2011;35(8):767–774. doi: 10.1042/CBI20100719. [DOI] [PubMed] [Google Scholar]
- 157.Jiang Y, Zhu Y, Liu ZJ, Ouyang S. The emerging roles of the DDX41 protein in immunity and diseases. Protein Cell. 2017;8(2):83–89. doi: 10.1007/s13238-016-0303-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lian H, Zang R, Wei J, Ye W, Hu MM, Chen YD, Zhang XN, Guo Y, Lei CQ, Yang Q, Luo WW, Li S, Shu HB. The zinc-finger protein ZCCHC3 binds RNA and facilitates viral RNA sensing and activation of the RIG-I-like receptors. Immunity. 2018;49(3):438–448. doi: 10.1016/j.immuni.2018.08.014. [DOI] [PubMed] [Google Scholar]
- 159.Lian H, Wei J, Zang R, Ye W, Yang Q, Zhang XN, Chen YD, Fu YZ, Hu MM, Lei CQ, Luo WW, Li S, Shu HB. ZCCHC3 is a co-sensor of cGAS for dsDNA recognition in innate immune response. Nat Commun. 2018;9(1):3349. doi: 10.1038/s41467-018-05559-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Campillos M, Doerks T, Shah PK, Bork P. Computational characterization of multiple Gag-like human proteins. Trends Genet. 2006;22(11):585–589. doi: 10.1016/j.tig.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 161.Brandt J, Schrauth S, Veith AM, Froschauer A, Haneke T, Schultheis C, Gessler M, Leimeister C, Volff JN. Transposable elements as a source of genetic innovation: expression and evolution of a family of retrotransposon-derived neogenes in mammals. Gene. 2005;345(1):101–111. doi: 10.1016/j.gene.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 162.Pang SW, Lahiri C, Poh CL, Tan KO. PNMA family: Protein interaction network and cell signalling pathways implicated in cancer and apoptosis. Cell Signal. 2018;45:54–62. doi: 10.1016/j.cellsig.2018.01.022. [DOI] [PubMed] [Google Scholar]
- 163.Ning Y, Hui N, Qing B, Zhuo Y, Sun W, Du Y, Liu S, Liu K, Zhou J. ZCCHC10 suppresses lung cancer progression and cisplatin resistance by attenuating MDM2-mediated p53 ubiquitination and degradation. Cell Death Dis. 2019;10(6):414. doi: 10.1038/s41419-019-1635-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Mallory A, Vaucheret H. Form, function, and regulation of ARGONAUTE proteins. Plant Cell. 2010;22(12):3879–3889. doi: 10.1105/tpc.110.080671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Schalk C, Cognat V, Graindorge S, Vincent T, Voinnet O, Molinier J. Small RNA-mediated repair of UV-induced DNA lesions by the DNA DAMAGE-BINDING PROTEIN 2 and ARGONAUTE 1. Proc Natl Acad Sci USA. 2017;114(14):E2965–E2974. doi: 10.1073/pnas.1618834114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Aklilu BB, Culligan KM. Molecular evolution and functional diversification of replication protein A1 in plants. Front Plant Sci. 2016;7:33. doi: 10.3389/fpls.2016.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Séguéla-Arnaud M, Choinard S, Larchevêque C, Girard C, Froger N, Crismani W, Mercier R. RMI1 and TOP3alpha limit meiotic CO formation through their C-terminal domains. Nucleic Acids Res. 2017;45(4):1860–1871. doi: 10.1093/nar/gkw1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Loudet O, Michael TP, Burger BT, Le Mette C, Mockler TC, Weigel D, Chory J. A zinc knuckle protein that negatively controls morning-specific growth in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105(44):17193–17198. doi: 10.1073/pnas.0807264105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhang S, Li C, Zhou Y, Wang X, Li H, Feng Z, Chen H, Qin G, Jin D, Terzaghi W, Gu H, Qu LJ, Kang D, Deng XW, Li J. TANDEM ZINC-FINGER/PLUS3 is a key component of phytochrome A signaling. Plant Cell. 2018;30(4):835–852. doi: 10.1105/tpc.17.00677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Nguyen NH, Jeong CY, Kang GH, Yoo SD, Hong SW, Lee H. MYBD employed by HY5 increases anthocyanin accumulation via repression of MYBL2 in Arabidopsis. Plant J. 2015;84(6):1192–1205. doi: 10.1111/tpj.13077. [DOI] [PubMed] [Google Scholar]
- 171.Kwon Y, Kim JH, Nguyen HN, Jikumaru Y, Kamiya Y, Hong SW, Lee H. A novel Arabidopsis MYB-like transcription factor, MYBH, regulates hypocotyl elongation by enhancing auxin accumulation. J Exp Bot. 2013;64(12):3911–3922. doi: 10.1093/jxb/ert223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lu D, Wang T, Persson S, Mueller-Roeber B, Schippers JH. Transcriptional control of ROS homeostasis by KUODA1 regulates cell expansion during leaf development. Nat Commun. 2014;5:3767. doi: 10.1038/ncomms4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Cheng NH, Liu JZ, Nelson RS, Hirschi KD. Characterization of CXIP4, a novel Arabidopsis protein that activates the H+/Ca2+ antiporter, CAX1. FEBS Lett. 2004;559(1–3):99–106. doi: 10.1016/S0014-5793(04)00036-5. [DOI] [PubMed] [Google Scholar]
- 174.Sánchez-García AB, Aguilera V, Micol-Ponce R, Jover-Gil S, Ponce MR. Arabidopsis MAS2, an essential gene that encodes a homolog of animal NF-kappa B Activating Protein, is involved in 45S ribosomal DNA silencing. Plant Cell. 2015;27(7):1999–2015. doi: 10.1105/tpc.15.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Armas P, Calcaterra NB. Retroviral Zinc knuckles in eukaryotic cellular proteins. In: Ciofani R, Makrlik L, editors. Zinc fingers: structure, properties, and applications. Hauppauge: Nova Biomedical Publishers; 2012. pp. 51–80. [Google Scholar]
- 176.de Castro E, Sigrist CJ, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34(Web Server issue):W362–W365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sigrist CJ, De Castro E, Langendijk-Genevaux PS, Le Saux V, Bairoch A, Hulo N. ProRule: a new database containing functional and structural information on PROSITE profiles. Bioinformatics. 2005;21(21):4060–4066. doi: 10.1093/bioinformatics/bti614. [DOI] [PubMed] [Google Scholar]
- 178.Sigrist CJ, Cerutti L, Hulo N, Gattiker A, Falquet L, Pagni M, Bairoch A, Bucher P. PROSITE: a documented database using patterns and profiles as motif descriptors. Brief Bioinform. 2002;3(3):265–274. doi: 10.1093/bib/3.3.265. [DOI] [PubMed] [Google Scholar]
- 179.Heese K, Fujita M, Akatsu H, Yamamoto T, Kosaka K, Nagai Y, Sawada T. The splicing regulatory protein p18SRP is down-regulated in Alzheimer's disease brain. J Mol Neurosci. 2004;24(2):269–276. doi: 10.1385/JMN:24:2:269. [DOI] [PubMed] [Google Scholar]
- 180.Coletta A, Pinney JW, Solis DY, Marsh J, Pettifer SR, Attwood TK. Low-complexity regions within protein sequences have position-dependent roles. BMC Syst Biol. 2010;4:43. doi: 10.1186/1752-0509-4-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Marino F, Mojumdar A, Zucchelli C, Bhardwaj A, Buratti E, Vindigni A, Musco G, Onesti S. Structural and biochemical characterization of an RNA/DNA binding motif in the N-terminal domain of RecQ4 helicases. Sci Rep. 2016;6:21501. doi: 10.1038/srep21501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Xie T, Pan S, Zheng H, Luo Z, Tembo KM, Jamal M, Yu Z, Yu Y, Xia J, Yin Q, Wang M, Yuan W, Zhang Q, Xiong J. PEG10 as an oncogene: expression regulatory mechanisms and role in tumor progression. Cancer Cell Int. 2018;18:112. doi: 10.1186/s12935-018-0610-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Li ZW, Hou XH, Chen JF, Xu YC, Wu Q, Gonzalez J, Guo YL. Transposable Elements contribute to the adaptation of Arabidopsis thaliana. Genome Biol Evol. 2018;10(8):2140–2150. doi: 10.1093/gbe/evy171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.