Abstract
Daily fluctuations in animal physiology, known as circadian rhythms, are orchestrated by a conserved molecular timekeeper, known as the circadian clock. The circadian clock forms a transcription–translation feedback loop that has emerged as a central biological regulator of many 24-h processes. Early studies of the intestine discovered that many digestive functions have a daily rhythm and that intestinal cell production was similarly time-dependent. As genetic methods in model organisms have become available, it has become apparent that the circadian clock regulates many basic cellular functions, including growth, proliferation, and differentiation, as well as cell signalling and stem cell self-renewal. Recent connections between circadian rhythms and immune system function, and between circadian rhythms and microbiome dynamics, have also been revealed in the intestine. These processes are highly relevant in understanding intestinal stem cell biology. Here we describe the circadian clock regulation of intestinal stem cells primarily in two model organisms: Drosophila melanogaster and mice. Like all cells in the body, intestinal stem cells are subject to circadian timing, and both cell-intrinsic and cell-extrinsic circadian processes contribute to their function.
Keywords: Stem cells, Digestive tract, Cellular signalling, Immunity, Cell cycle
Circadian rhythms
Circadian rhythms are 24-h recurring physiological processes such as daily sleep/wake cycles, feeding/fasting cycles, daily changes in body temperature, hormone levels, and cardiovascular function [1–3]. The word Circadian is derived from Latin “circa” (meaning “about”) and “diem” (meaning “day”). Circadian Rhythms have four main characteristics: (1) an approximately 24-h period, corresponding to the 24-h rotation of the Earth’s axis; (2) temperature-compensation, meaning they are maintained as a 24-h process under a wide range of environmental temperatures; (3) the ability to be synchronized by external cues such as light and feeding (called “zeitgebers” from German meaning “time giver”), to synchronize 24-h internal timekeeping with 24-h changes in environment; (4) the ability to persist in the absence of external cues (called “free-running”), which means the physiological changes arising from circadian clock function reflect ongoing internal cellular and molecular timing rather than a simple response to the cues [1, 2]. These characteristics differentiate circadian clock and other environmental-responsive processes, including diurnal rhythms whose daily repetition may be determined by light responses, and ultradian and infradian rhythms whose period is less or more than 24-h, respectively. Hence, circadian rhythms are 24-h oscillatory networks of molecular and cellular activity, present in animals to maximize their health and fitness under constant 24-h planetary change.
The observation that biology is synchronized to the 24-h day–night cycle and persists in the absence of external cues was first supported by experimental evidence in 1729 when French scientist Jean-Jacques d’Ortous de Mairan kept a plant in a windowless room and observed that the cyclic behaviour of leaf opening continued in complete darkness [4]. Rhythmic behaviours were documented in a wide variety of organisms, in the conidiation of fungi [5], the eclosion of fruit flies [6–9], and the locomotor activity of finches [10]. This research eventually led to the discovery of an endogenous genetic timekeeper: the study of eclosion rhythms in fruit flies, Drosophila melanogaster, unearthed three genetic mutants with disrupted rhythms, the Period (Per) mutants [9]. Mapped to the X-chromosome of Drosophila, the Per gene linked circadian rhythms to a molecular timekeeper, the circadian clock [11], and was the first demonstration that animal behaviour could be attributed to the function of a single gene.
Recent studies have implicated the working of the circadian clock system with Intestinal Stem Cell (ISC) function [12–16]. This review will explore the role of the circadian clock in regulating processes relevant to intestinal and ISC biology, in mice and Drosophila. As this field is still in its infancy, connections between the circadian clock and molecular processes important in ISCs, but not yet linked with circadian function, will be highlighted as possible directions for future work.
The circadian clock
Circadian rhythms are maintained by a cell-intrinsic molecular transcription–translation feedback cycle called the circadian clock. This molecular system influences the expression of many genes that contribute to 24-h cycles of cellular function. The circadian clock is conserved from flies to mammals and has been shown to regulate a total of ~ 40% of genes in the mouse [17], and possibly an even higher number in primates [18]. These genes vary between different tissues, suggesting circadian functions are specific depending on cell type [17, 19]. Due to the tremendous number of cellular processes it can regulate, it is perhaps not surprising that the circadian clock has been linked to many diseases [20, 21] including cancer [22–24], diabetes [25], inflammatory bowel disease [26, 27] and obesity [28, 29]. The mechanisms of how the clock fully impacts intestinal health remain to be elucidated.
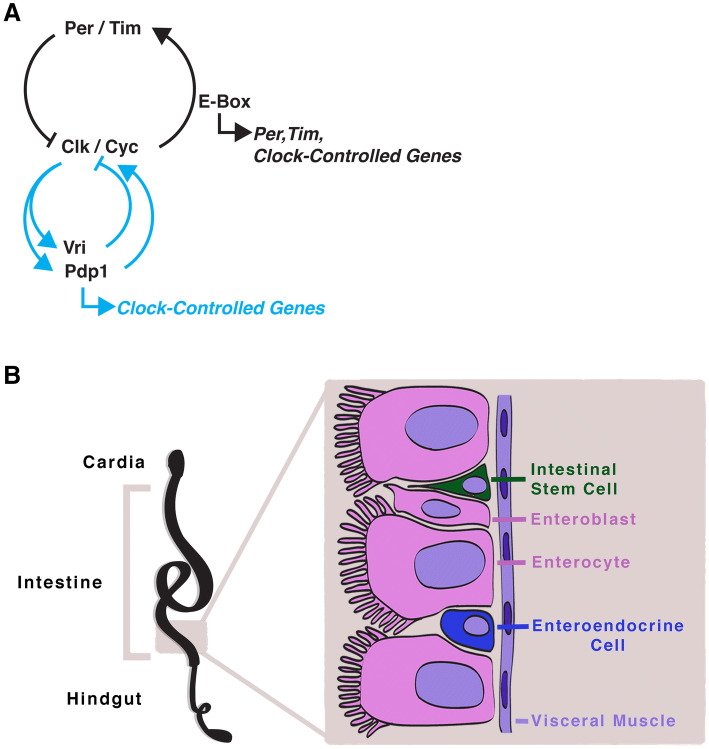
The components of the circadian clock in animals are highly conserved between Drosophila and mice. In Drosophila, the circadian clock is a transcription–translation feedback loop in which protein heterodimers, Per and Timeless (Tim), repress their own transcriptional activators, Clock (Clk) and Cycle (Cyc) (Fig. 1a). In the beginning of the day, the repressors are active, however, over the day, light-induced degradation of Tim by the photoreceptive protein Cryptochrome (Cry) releases Per-Tim inhibition. These repressors are degraded, allowing the Clk/Cyc transcriptional activators to bind to E-box (5′-CACGTG-3′) regions all over in the genome, including that of their repressors, starting the next cycle [30–34]. This process is free-running: in the absence of environmental cues, phosphorylation and degradation of Per reset the circadian clock to maintain its approximately 24-h period [35–37]. A secondary feedback loop consists of transcription factors Vrille (Vri) and PAR-domain protein 1 (Pdp1) that are transcribed by Clk/Cyc and modify Clk expression; Vri represses Clk and Pdp1 activates Clk in turn [38–41]. This second transcription/translation system is thought to confer greater robustness to the clock since expression of both the repressors and activators oscillate in opposite phases. The mechanism for temperature compensation remains unknown, although in plants the ratio of clock components may buffer rhythms in varying temperatures [42].
Fig. 1.
The Drosophila circadian clock, and intestine. a The circadian clock in Drosophila consists of the basic-helix–loop–helix (bHLH) transcription factors Clk and Cyc which bind to E-box regions in the genome driving the transcription of clock-controlled genes. The core circadian clock genes Period (Per) and Timeless (Tim) form a protein heterodimer that represses Clk/Cyc activity. A secondary stabilizing loop involving Pdp1 and Vri activates or represses Clk, respectively. Pdp1 and/or Vri could themselves regulate target genes in a rhythmic fashion as well, contributing to the overall clock-controlled gene rhythm. b The intestine in Drosophila is a pseudostratified epithelium consisting of basally located intestinal stem cells (ISC) which either self-renew to maintain the stem cell pool or divide into progenitor cells, called enteroblasts. The different epithelial cells as well as the visceral muscle are thought to serve as a niche for the ISCs in this simplified epithelium
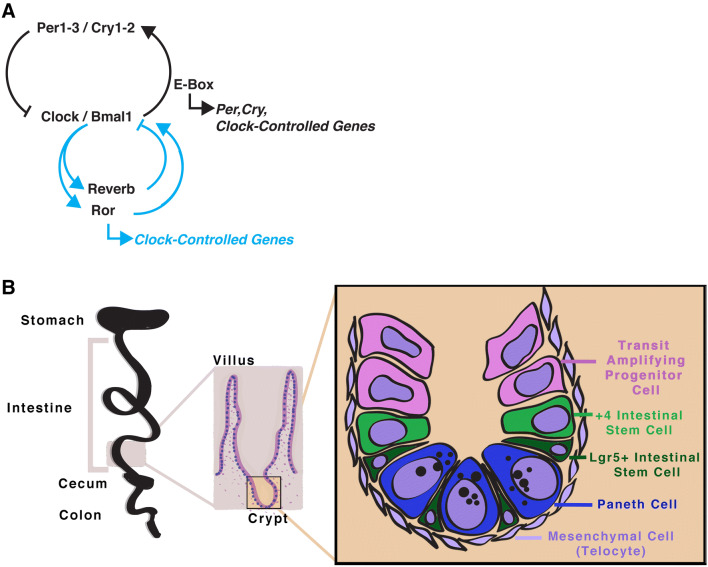
In mice, Brain and Muscle Aryl hydrocarbon receptor nuclear translocator Like protein 1 (Bmal1) and Circadian Locomotor Output Cycles Kaput (Clock) proteins similarly dimerize and transcriptionally activate Per and Cry, which in turn repress Clock/Bmal1 activity (Fig. 2a). In mammals, Tim is not part of the central clock mechanism, it is substituted by Cry itself and the transcription–translation feedback loop is otherwise similar. The mammalian clock has additional genetic redundancy, as it consists of multiple paralogs of clock components: Bmal1-2 [43], Clk1-2 or Npas2 [44–46], Per1-3 [47–51] and Cry1-2 [52–54]. As in Drosophila, the activity of Clock/Bmal1 drives its own repression, and a secondary stabilizing loop is also present, but consisting of Reverbα (also known as Nuclear Receptor Subfamily 1 Group D Member 1, Nr1d1), or Reverbβ (Nr1d2) [55], and multiple isotypes of Retinoic Acid Receptor-Related Orphan Receptor (Rorα, also known as Nr1f1, as well as Rorβ and Rorγ, with several isoforms of each of these) [56, 57], which feedback in a tissue-specific manner to drive a second opposite-phased system. Post-transcriptional and post-translational modifications of circadian clock components also play a role in regulating circadian gene expression [58]. The mechanisms of both Drosophila and mammalian circadian clocks are well-established due to decades of research [1, 2]. It is also important to mention that non-transcriptional mechanisms can establish circadian rhythms, although in this review we will primarily consider the canonical transcription–translation circadian clock system.
Fig. 2.
The mouse circadian clock, and the intestinal crypt. a The circadian clock in mice consists of basic-helix-loop-helix (bHLH) transcription factors clock Clock (or Npas2) and Bmal1 which bind to E-box regions in the genome driving the transcription of clock-controlled genes. These genes include the core circadian clock genes Per1-3 and Cry1-2 which form protein heterodimers that represses Clock/Bmal1 activity. A secondary stabilizing loop involving RORs and Reverbs (and their respective isotypes) activate or repress Bmal1, respectively. Both of these nuclear receptors could regulate their own clock-controlled target genes as well. b The intestine in mice is folded into crypts and projections into the lumen (villi). Differentiation occurs as progenitor cells move upward to the villi, and old cells are shed from the tips of the villi. The epithelium consists of rapidly dividing LGR5+ intestinal stem cells (ISC) located at the base of the crypt as well as quiescent +4 ISCs. which either self-renew to maintain the stem cell pool or divide into progenitor cells, called transit amplifying progenitor cells. Both mesenchymal cells, in particular telocytes, and Paneth cells serve as a Wnt pathway niche for the ISCs
Entrainment of the circadian clock
A major question in circadian biology is understanding how the clock is entrained by environmental cues to synchronize its activity with 24-h cycles. Light, temperature and feeding are three cues that have been shown to synchronize circadian clocks. In many animals, the circadian system is hierarchical, being composed of a central pacemaker driving the synchronization of peripheral pacemakers located inside cells throughout tissues of the body [59]. Photoperiod changes, the periods of light and darkness, are sensed by retinal cells of the eye, then sent to the central pacemaker in the brain. This central pacemaker, located in the suprachiasmatic nucleus of the hypothalamus, is thought to coordinate these peripheral clocks through neuronal signals, and the hypothalamo-pituitary-adrenal axis [60–65]. Hormones sent via this system, time the peripheral pacemakers in distant tissues, that work together to establish circadian rhythms in the body [66, 67]. Each peripheral timekeeper maintains rhythmic gene expression patterns relative to the central pacemaker [66, 68–70]. This complex and intimately connected system allows the body to overall coordinate the timing of physiological functions to anticipate the demands of the organism based on 24-h time.
Photoperiod light is sensed by opsins in mice [52, 71–75], and Cry in Drosophila [76–80]. While in mammals a hierarchical system conveys photoperiod information from the retina to peripheral tissue clocks, in Drosophila, photoperiod cues can synchronize clocks in tissues directly and independently [81, 82]. Although the mechanism for 24-h period temperature compensation is unknown, in mammalian cells [83] and in Drosophila [84] temperature changes can synchronize the circadian clock in peripheral tissues. Feeding has been shown to be an important entrainment cue in peripheral tissues in mice, such as the liver, kidney, and heart [85–88], as well as their equivalents in Drosophila [89–91]. Indeed, peripheral clocks are highly dependent on feeding time. In mice it was shown that feeding can uncouple peripheral and central clocks, as the former is directly entrained by the time of food intake while the latter is entrained by photoperiod. This likely reflects the hierarchical system of clock timing in mammals, where the central clock of the suprachiasmatic nucleus receives timing cues from light hitting the retina, and the peripheral clocks receive hormonal cues from the central clock. In vitro, serum shock [92], the glucocorticoid dexamethasone [93], forskolin [94], and insulin [95] can be used to synchronize the clock, underscoring the importance of hormones and cellular signaling in entraining clock timing. In Drosophila, hormonal inter-organ communication is not so clear, however, studies have revealed elements of circadian time-setting [90, 91], including food synchronization of peripheral tissues [16, 91, 96].
Intestinal stem cells
The intestine is a dynamic tissue that undergoes nearly continuous cellular turnover. This is most likely due to the harsh processes of nutrient digestion, that can damage intestinal cells needed for nutrient absorption, hormone release, and pathogen defense. In addition to its role in digestion, the intestine is also a barrier to the outside environment that is evolved to cope with constant bombardment of pathogens and harmful chemicals. Nearly the entire intestinal epithelium is renewed weekly in mammals, and its dynamic turnover is supported by a population of ISCs [97–99]. In animals, the intestinal epithelium consists of several cell types: (1) ISCs; (2) progenitor cells (transit amplifying cells in mice, or enteroblasts in Drosophila); (3) absorptive cells (enterocytes); (4) enteroendocrine cells; (5) secretory cells (Paneth cells, goblet cells). Secretory cells have not been found in Drosophila, whose epithelium and immune system is simplified in its cellular diversity. However, the Drosophila intestine resembles and is regulated by the same pathways of its mammalian counterpart, making this system an attractive model for basic research [100, 101].
Drosophila intestinal stem cells
The Drosophila intestine (also known as the midgut) is a pseudostratified epithelium with ISCs located near the base and differentiated cells facing the lumen (Fig. 1b). ISCs divide into daughter ISCs or enteroblasts [102–104] that differentiate into enterocytes or enteroendocrine cells [105, 106]. Drosophila ISCs are marked by the Notch pathway ligand Delta, that is involved in signaling differentiation to ISC daughter cells as they are produced [104]. The stem cell niche is well defined in the Drosophila intestine, consisting of important signals for ISC maintenance that are provided by the stem and progenitor cells themselves [107–110], as well as the visceral muscle [108, 111–117]. The Bone Morphogenic Protein (Bmp) and Wnt signaling pathways present in this system, further highlight the similarity between the Drosophila ISC system and that of mammals [101, 115, 118–122].
Mammalian intestinal stem cells
In mammals, the intestinal epithelium is arranged in a crypt-villus structure with stem cells located at the base of the crypts of Lieberkühn, and differentiating daughter cells moving upward to the tip of the villus [123] (Fig. 2b). The progenitor cells differentiate as they travel up the crypt to the villus, although differentiated Paneth cells will migrate back down to the crypt where they reside with the stem cells at the base. In mice, at least two distinct populations of stem cells are known: the +4 ISCs (sometimes also called label-retaining cells) and the crypt base columnar (CBC) cells. The CBCs are a population of quick-cycling ISCs in which Wnt signaling is important for stem cell proliferation and differentiation. The +4 ISCs, or ISCs at approximately that position in the crypt, have been separately identified by the markers Bmi1 [124], Tert [125], Hopx [126], Lrig1 [127], and most recently by Clusterin [128], however, the precise relationships between cells bearing these markers is an area of ongoing research. Some of these markers may be also expressed by CBCs [129], and the quick-cycling CBCs are specifically marked by Leucine-rich repeat-containing G-protein coupled receptor (Lgr5) [130], Olfm4 [131] and Ascl2 [132, 133]. Complex and not fully understood lineage relationships exist between these different ISC populations in this tissue, but it is thought that during baseline, uninjured conditions the Lgr5 + ISCs divide once every 1–2 days to produce the differentiated cells of the intestinal epithelium [130, 134, 135]. The +4 cells on the other hand seem to play a role during stress and regeneration. At the base of the crypt, mesenchymal cells and Paneth cells form the Lgr5+ ISC niche, by secreting Wnt pathway ligands that create a zone of high Wnt activity (described in detail below). ISCs in both Drosophila and mice have proven to be excellent, and mutually beneficial systems to inform our knowledge of basic stem cell biology.
Circadian regulation of the intestine
Nearly all cells of the body are thought to harbor circadian clock activity, and the intestine is no exception. In both Drosophila and mammals, clock gene transcriptional rhythms are present in the intestine [12, 16, 136, 137]. Experiments in vivo in Drosophila and mice, and recently in vitro using intestinal organoids, 3-dimensional cell culture models, further confirm that the circadian clock is present in the intestinal epithelium [12, 14–16]. Analysis of 24-h changes in transcript abundance has been a central test of circadian output in many different contexts, including the digestive tract. In wildtype mice, > 1000 transcripts are thought to oscillate under photoperiod [138], although free-running conditions and circadian clock mutants have not yet been tested to confirm these are a result of bona fide clock activity [138–140]. In Drosophila, over 400 genes show circadian expression rhythms, that are absent when the clock is non-functional [12]. Furthermore, one of the most highly expressed transcription factors in Drosophila ISCs is the circadian clock gene Cyc [107], the ortholog of Bmal1, which suggests that at least this one core clock gene plays an important role in these cells. The clock appears to be robust in the intestine, functioning under different diets and physiological contexts [138–140]. However, it is important to note that the transcriptional analyses thus far represent an average of all the different cell types present, and not necessarily information about cell-specific circadian rhythms in this tissue. In addition, these studies have been carried out under constant light/dark photoperiod, meaning that they have not addressed the free-running nature of the circadian clock system, and cannot completely discriminate between circadian clock target genes and light-response genes. Determining the precise functions of the clock in the intestinal epithelial cells, as well as their non-epithelial neighbors, is a problem for future research.
Despite a large number of potential transcripts, the functions of the circadian rhythms in the intestine remain poorly understood. The intestine of clock mutant Drosophila or mice has no obvious size or morphological phenotypes [12, 15, 141], yet daily rhythms in the expression and activity of enzymes and transporters for carbohydrates, peptides, and fats are very well-established [140, 142–147]. This highlights that one of the primary functions of the intestine, to absorb and digest nutrients, is probably under circadian regulation. This would be a significant role for the circadian system, likely connecting this tissue to the circadian metabolic control of the whole animal. The role of circadian rhythms in governing intestinal absorption and digestion functions have been addressed in previous reviews [148–150].
Circadian regulation of stem cells
Do stem cells themselves have a circadian clock? Early observations of circadian rhythms in mitosis and apoptosis were indeed attributed to epidermal stem cells in tongue, skin and intestinal epithelium [151]. However, studies showing circadian rhythms in stem cells, specifically, occurred later when the molecular tools needed to identify and study these cells became available. Diurnal variation in stem cell characteristics was initially shown in the haematopoetic stem cells [152, 153] and subsequently in epidermal stem cells [154, 155]. Clock genes are expressed by haematopoetic stem cells, but it is not yet clear whether circadian transcriptional cycles are present [156]. However, in the body the circadian clock regulates oscillation of the chemokine Cxcl12 and its receptor, Cxcr4 [152, 153]. These regulate extrinsic stem cell signaling, causing daily patterns of migration and homing of haematopoietic stem cells from the bone marrow to the bloodstream. Recent reports have further implicated circadian rhythms with the cell-intrinsic proliferation and self-renewal of haematopoietic stem cells, and leukemia stem cells [157, 158]. Per2 expression is increased in aged haematopoietic stem cells where it increases apoptosis and DNA-stress response [158]. In the case of leukemia stem cells, Clock and Bmal1 are required for sustained proliferation and the maintenance of the undifferentiated state [157]. In these blood cells, it is not yet clear if these are due to circadian functions, or non-circadian functions of these clock genes in different cellular pathways. In the epidermis, the role of the clock is complex since many different cell types participate in regeneration, including epidermal stem cells, and the output of the circadian system may be different depending on cell type. Hence, connecting circadian regeneration rhythms to actual stem cell activity is a challenge. As in the intestine, circadian rhythms in the mitosis of epidermal cells are present and are clock-dependent [159], and two genes, P21 and Klf9 have both been implicated as mechanisms connecting the clock to cellular proliferation [113, 160]. Migration of fibroblasts during wound closing also has a circadian regulatory role [161], suggesting several stages of skin wound healing would be time-dependent. The epidermal stem cells themselves, exhibit circadian clock function, and the loss of Bmal1 and Per1/2 increases and decreases proliferation, respectively [154]. Overall, it is clear that circadian rhythms exist in the blood and skin, and are the product of circadian clock output in many different cell types. The role of the circadian clock in stem cells in the haematopoietic and epidermal tissues has been reviewed [162–167], raising many interesting areas for future study.
In the previous examples, stem cells can be regulated by cell-extrinsic clock mechanisms, as well as cell-intrinsic ones. This raises the question of the relative contribution of extrinsic versus intrinsic mechanisms in stem cell biology, and whether stem cells need to have intrinsic circadian clock function to display time of day changes in their behaviour. Adult tissue stem cells represent a relatively undifferentiated population, that give rise to a lineage of increasingly differentiated progeny. When does the clock arise during development? The notion that the circadian system emerges as a result of differentiation is a compelling framework. In early mouse embryonic stem cells the circadian clock is completely absent [168], arising at later stages during embryonic development [169]. In the adult, however, hair follicle stem cells and muscle stem cells both have circadian clock function [154, 170]. It has been documented that adult ISCs have weak to no circadian clock activity in vitro [14], suggesting these would resemble embryonic stem cells rather than the tissue stem cells found in skin or muscle. However, in the adult Drosophila intestine, circadian clock function is present in ISCs [12, 16]. It is possible that these differences are simply species-specific, or that the physiological state of the environmental milieu, which is different in vivo than in vitro, regulates clock activity in ISCs. How cell-intrinsic circadian activity in intestinal cell lineage emerges during differentiation is an important question for future research.
Circadian regulation of intestinal stem cells
Are any ISC-related processes under similar circadian control? Tissue stem cells’ primary function is to replace the surrounding differentiated cells to maintain tissue homeostasis throughout adulthood. In the circadian field, it is generally accepted that proliferation of intestinal precursors follows 24-h cycles, based on seminal studies by Sigdestad and Potten nearly 50 years ago [148, 151, 171, 172]. These studies found that mitoses show diurnal rhythms in rodents, peaking in the early morning when the nocturnal animal begins to slow its activity and return to sleep. However, in the stem cell field, daily proliferation rhythms do not receive much attention. Indeed, early studies of intestinal tissue renewal including one by the pioneer of ISC biology, Charles Leblond, found that intestinal precursor proliferation was constant and showed no time of day dependence [97, 173, 174]. A recent study has addressed this discrepancy, showing that prior to stress, rhythms in proliferation are weak, however, under regenerative conditions where inflammation is high, cell proliferation follows clear daily rhythms [12, 15]. Together these studies hint that stem cell output is gated by the circadian clock, and modern cellular and genetic tools can now be applied to resolve the contribution of a fundamental cellular timekeeper to ISC biology.
Like many adult tissue stem cells, ISCs are located in regions of the intestine called the niche, a localized zone of cellular signaling that determines their undifferentiated status and proliferative capacity. In the mammalian intestine, homeostasis is chiefly maintained through Wnt and Bmp that regulate a balance between ISC self-renewal and differentiation [123]. In Drosophila, these same pathways contribute to ISC regenerative activity, and the overall system shares many conserved features, albeit not the exact same details [101]. For instance, Bmp can promote Drosophila ISC proliferation, while in mice it inhibits proliferation through differentiation of ISCs. Here, we will consider primarily conserved cellular processes that might be controlled by the clock in the ISC niche in both systems. In both Drosophila and mammals, during baseline, undamaged conditions, ISCs proliferate to renew the epithelium constantly, and during regenerative, post-damaged conditions ISCs are thought to further increase this regeneration. As mentioned above, certain subpopulations of ISCs in the mammalian intestinal crypt are thought to form complex lineage relationships, and these are particularly susceptible to changes from baseline to regeneration [175]. Intestinal epithelial mitoses in the Drosophila and mouse intestine shows circadian rhythms during regeneration [12, 15], which suggests that pathways which regulate ISC proliferation are regulated by the circadian clock. Since the study of circadian regulation of ISCs is a new area of research, five different possibilities that could mechanistically explain these rhythms will be discussed below, based on studies of the circadian clock in the intestine and other systems (Table 1). These five possibilities are highly relevant to both colorectal cancer and IBD pathology as well.
Table 1.
Regulatory mechanisms
| Process | Tissue/cell | Role of circadian clock | References |
|---|---|---|---|
| 1. Cell cycle/DNA repair | Brain | DNA repair by Xpa | [193] |
| Epidermal stem cells | Regulation of cell cycle (p21, Cdk4, etc.) | [154] | |
| Fibroblasts | Regulation of p16 through NONO | [183] | |
| Intestinal Stem Cells | Regulation of cell proliferation (via cell signaling) | [12, 14, 15] | |
| Intestine | Regulation of mitosis | [151, 171] | |
| Kidney | Regulation of Chk1 | [187] | |
| Liver | Regulation of Wee1, p21, DNA repair by Xpa | [177, 182, 192] | |
| Skin | Timing of DNA repair | [190, 191] | |
| 2. Self renewal (Wnt signaling) | Bone | Regulation of osteogenesis | [210] |
| Brain | Regulation of c-Myc | [216–218] | |
| Fat | Regulation of adipogenesis | [211] | |
| Intestinal stem cells | Regulation of Wnt3A in Paneth cells | [14] | |
| Intestine | Regulation of β-Catenin | [212] | |
| Liver | Regulation of c-Myc | [215] | |
| Muscle progenitor cells | Regulation of myogenesis | [208, 209] | |
| 3. Differentiation | Epidermal stem cells | Regulation of Notch, Bmp signalling | [154, 155] |
| Fat, Tendon | Regulation of Bmp signalling | [236, 237] | |
| Intestinal stem cells | Loss of Notch signalling disrupts circadian clock | [16] | |
| Keratinocytes | Regulation of Klf9 | [160] | |
| 4. Feeding and growth (insulin signaling) | Brain, liver, fibroblasts | mTOR/Bmal1 regulated protein translation | [251] |
| 5. Immune response | White blood cells | Proliferation, circulation, regulation of inflammatory response | [258, 259, 261–270, 277] |
| Intestine | Microbiome, regulation of inflammatory response (Toll-like receptors, Tnf, etc.) | [12, 15, 274–276, 279–284] | |
| Colon | Regulation of inflammatory response, and inflammasome (Nlrp3) | [26, 277, 278] |
A list of five potential regulatory mechanisms through which the circadian clock controls ISC function. References listed correspond to the papers in the text, and studies carried out on stem cell populations are emphasized
Cell cycle control of stem cell proliferation
Many cells divide approximately once per day, and a link between the circadian clock and the cell cycle has been actively researched [176]. Initial studies indicated that the cell cycle is likely to be coupled to the circadian clock in fibroblasts and in the liver [177, 178]. Live-imaging of circadian clock activity in immortalized fibroblasts revealed that the clock functions in dividing cells and helps to establish the timing of cell cycle progression, while at the same time being affected by the cell cycle during mitosis, which causes a delay in circadian clock progression [178]. The first explanation for the link between the circadian clock and the cell cycle is that, like Huygen’s pendulum, the circadian cycle of transcription/translation and the cell cycle of growth/division could exist as two oscillating processes that impact each other’s activity, in essence synchronizing them relative to one another over time in the same cell [179, 180]. This is an important concept, and the coupling of oscillators in the same cell could be a general principle linking many recurring processes with daily circadian activity. However, it is important to note that these cycles can be decoupled, and it has been shown that the cell cycle is not under any obligatory circadian clock control [181].
The regulation of the cell cycle by the clock in a top–down fashion is a second, parsimonious explanation for daily rhythms that have been observed in tissue regeneration. In the liver, Wee1 (a G2/M cell cycle checkpoint regulator) is directly controlled by the circadian clock thereby regulating the proliferation of liver cells during regeneration [177]. In hepatocytes the cell cycle inhibitor, P21, is also a circadian clock target that establishes the proliferative timing of cells [182]. This means that in the liver, multiple phases of the cell cycle appear to be downstream of the clock, thus clock mutants have abnormal regenerative output. In other tissues, other cell cycle checkpoint regulators are under circadian control, including P16, thereby influencing the timing of fibroblast division during wound healing in mice [183]. In the skin, the role of the clock is very complex. For instance, it regulates growth and the cell cycle in epidermal stem cells [154, 155, 170] that determines the propensity of epidermal stem cells to activate during regeneration or to remain dormant [154]. Interestingly, during youth the circadian clock targets genes involved in regulating epidermal cell proliferation, but during age shifts to genes involved in DNA repair and stress, indicating that the circadian program is mutable [170]. In addition to stem cells, cell cycle checkpoints in other proliferating skin cells such as hair follicle precursors are regulated by the circadian clock [113, 159], thereby influencing the growth of hair. Together, these elegant studies have shown that the clock plays cell-specific roles in diverse cells of the skin. Since the intestinal crypt of mammals houses a complex population of stem cells, as well as transit-amplifying progenitors and committed progenitors, the clock could play a highly complex role in the intestinal epithelium. In the intestine it is not yet clear if regulators such as Wee1, P21, or P16 cause circadian rhythms in proliferation, and whether these act at the levels of ISCs, or within other dividing intestinal precursors [15, 184]. Future studies will reveal the precise contributions of the circadian clock in different intestinal cells, and how or whether these interact to drive daily rhythms in overall cell cycle.
A third possibility is that the clock might not regulate the cell cycle directly, but through processes such as DNA repair that stall proliferation to correct errors. Several links between DNA damage and repair pathways and the circadian clock pathway have been established, and reviewed elsewhere [185, 186]. For instance, the mouse ortholog of the core clock gene, Tim, links DNA repair checkpoint with the circadian clock through Chk1 [187], and similarly, both Per1 and Per3 interact with Chk2 [188, 189]. This would mean that components of the circadian clock, whose levels oscillate during the day, would restrict both single and double-strand DNA repair to enable cell cycle progression at particular times of day. Although these connections have not been tested in the intestine yet, such processes could impact the timing of cell proliferation in tissues like the intestine, that undergo frequent oxidative stresses and subsequent DNA damage. Indeed, the mouse epidermis that is subject to ultraviolet light-induced DNA stress, shows time of day dependent DNA damage responses [190]. In this case, both the outcomes of DNA damage and the timing of cell proliferation that results have daily rhythms and are clock-dependent. Indeed, Xpa, a gene that mediates the excision repair of DNA nucleotides is under circadian clock control in multiple tissues, suggesting DNA repair is a fundamentally clock-regulated process [191–193]. The DNA damage response in the intestine is an interesting potential area of research that could be quite relevant in the intestine where a high metabolic rate, and frequent inflammatory events, can lead to DNA damage. However, it is important to note that the links between cell cycle control and the circadian clock have not been studied in ISCs per se, but rather other proliferating cell types, so it is not yet clear if these connections are relevant to ISCs. Indeed, in intestinal epithelial cells, a definitive link between cell cycle control and the circadian clock has not been observed to date, and it has been proposed that the correlation between the proliferation of intestinal cells and 24-h rhythms is due to other cellular processes [12, 14, 15].
Stem cell self-renewal: Wnt signaling
Wnt signaling is intimately connected to ISC biology and, through ISCs, plays a critical role in the mammalian intestine [194]. The canonical Wnt pathway proceeds from the secretion of Wnt ligand which binds to the Frizzled receptor [195], activating the coreceptors Lrp [196] and Dishevelled [197]. This inactivates a destruction complex composed of Axin, Apc, and Gsk3 to stabilize β-catenin and activate the transcription of genes including Lgr5, Axin2 and c-Myc. These target genes are thought to be present in Wnt-receiving cells, including ISCs. In the mouse intestine, Wnt signaling is boosted by R-spondins and their receptor Lgr5 to promote self-renewal of Lgr+ stem cells [198], and intestinal crypt growth and proliferation [199, 200]. In mice, a major source of Wnt ligands are telocytes, a type of mesenchymal cell [201], although Paneth cells and other cell types near the crypt also express redundant Wnt sources [194, 202–205]. In Drosophila, the ISC niche also contains redundant Wnt signals originating from the visceral muscles [206, 207] and intestinal progenitors [120]. The Wnt pathway is a conserved ISC self-renewal mechanism.
Several lines of evidence indicate that the circadian clock may regulate aspects of this important signaling pathway. A recent paper by Matsu-Ura et al. used intestinal organoids to reveal a mechanism for how Wnt signaling can be regulated by the circadian clock in the intestinal epithelium [14]. Intestinal organoids are 3-dimensional stem-cell-based cultures that recapitulate aspects of intestinal physiology, such as regeneration and differentiation of epithelial cells [200]. Using an elegant combination of the FUCCI cell cycle reporter (Fluorescence Ubiquitination-based Cell Cycle Indicator—which labels cells with green and red fluorescence depending on their stage in the cell cycle), and the TOP-FLASH Wnt reporter (which reports TCF/Lef transcriptional activity via Luciferase), Matsu-Ura et al. determined that the circadian clock regulates production of Wnt3A ligand by Paneth cells, resulting in 24-h oscillations of signaling activity that couple circadian rhythms to proliferation [14]. Although Matsu-Ura et al. did not find that ISCs themselves exhibited circadian clock activity, clock control of Wnt signaling ISC niche propagates rhythms to these cells extrinsically. Along these lines, in the epidermis, ChIP analysis has shown that Bmal1 binds rhythmically to Wnt pathway related gene promoters suggesting that the clock may modify sensitivity to Wnt signals [154]. In muscle, fat, and mesenchymal cells, the clock components Bmal1 and Reverb also have been shown to transcriptionally target various components of the Wnt pathway [208–211]. Although these studies were not carried out on stem cells, they raise interesting possibilities of how the circadian clock can regulate ISC behaviour by driving intrinsic cellular processes. The intestinal organoid system provides a means to investigate these processes and is amenable to studies of circadian clock function [13, 15]. Of note, Per2 and β-catenin have been suggested to be mutually inhibitory in the intestinal epithelium, with Per2 normally downregulating β-catenin [212], while high β-catenin levels tipping the balance to result in Per2 loss [213]. The mutual negative regulation of key components of the Wnt and circadian clock pathways in the same cell, once again underscores possible bidirectionality of cellular signaling processes and circadian clock function.
Downstream components of the Wnt signaling pathway have been shown to be regulated by the circadian clock, including target genes such as c-Myc. C-Myc is a transcription factor that regulates ISC biology [214] and shows circadian rhythms in its expression in several other tissues [215, 216]. C-myc is also interconnected with the circadian clock, like Clock/Bmal1, c-Myc binds to E-boxes in the genome where it dampens Bmal1-driven rhythms of Reverb expression [217], and/or opposes Bmal1/Clock at E-boxes [218, 219]. Of note, the circadian clock can also influence c-Myc expression through Bmal1 binding of the c-Myc promoter [218]. Overall, these studies highlight the possibility that c-Myc and the clock could oppose one another’s activity, and notably in the intestine c-Myc is highly expressed by proliferating crypt cells [220]. It is thus possible that circadian clock activity is weaker in the base of the crypt where Wnt signaling is highest, where ISCs are themselves located, in line with reports that stem cells do not have clock function [14, 168]. A bidirectional interaction between the circadian clock and c-Myc may enable a balance between circadian clock control and stem cell driven processes [221], to coordinate environmental signals with intestinal epithelial tissue renewal. Another link may exist between the Wnt pathway regulator, Gsk3b, which inhibits Wnt signal transduction but also regulates the phase of clock activity [222, 223]. However, again it is important to note that these ideas remain to be tested directly in ISCs, rather than other cell types. Circadian regulation of Wnt signaling in ISCs is a relatively unexplored area with many opportunities for future study.
Control of intestinal differentiation: a new role for the circadian clock?
The ISC niche maintains intestinal precursors in an undifferentiated and proliferative state; outside the niche, cell signaling processes cause the progeny of ISCs to differentiate. This delicate balance of ISC differentiation could be another mechanism through which the circadian clock regulates ISCs, and early studies indicated that crypt cell number exhibits daily variation [224, 225]. The Bmp pathway plays a role in intestinal morphogenesis [226–228] and is a positive regulator of differentiation that opposes Wnt signaling during ISC niche formation [229, 230]. Loss of function mutations in Bmp have been shown to increase intestinal cell proliferation in mice [231] and Drosophila [115]. In mammals, Bmp ligands are found primarily in the villus region, where differentiated intestinal cells reside, whereas the Bmp antagonist, Noggin, is found near the crypt base, where ISCs are present [228, 232–234]. In the simplified niche of Drosophila, Bmp signaling is found primarily in the visceral muscle where it regulates stem cell division following regeneration [115, 235]. Is Bmp signaling regulated by the clock? Bmp pathway components are transcriptionally targeted by Bmal1 in epidermal stem cells in mice [154, 155]. Indeed the circadian regulation of the Bmp pathway to control differentiation has been documented in both fat and tendon tissues [236, 237]. Although it has not been studied in the intestinal crypt thus far, circadian clock regulation of the Bmp pathway is a potential mechanism.
Other pathways including the Mitogen-Activated Protein Kinase (Mapk/Erk) pathway, and the Notch pathway, also influence the balance between differentiation and proliferation in the intestinal epithelium. While it is beyond the scope of this review to fully cover these, in the Drosophila intestine, Mapk/Erk signaling is well-established to regulate ISCs proliferation and rapid differentiation of progeny during the stress response [114, 238–240]. In the mouse intestine this same signaling pathway also promotes proliferation [200, 205]. The Mapk/Erk pathway interacts with the circadian clock in many systems [241], and has been recently shown to promote Tim expression, and regulate the proliferation of cancer cells [242]. Its role in integrating environmental stresses in ISCs, as well as its role in modifying the circadian clock, warrants examining whether the Mapk/Erk pathway is regulated by the circadian clock in ISCs.
The Notch pathway has a complex role in the differentiation of ISC progeny in both mammals and Drosophila, overall pushing cells toward the enterocyte cell fate in the intestine [101, 123]. Human epidermal stem cells have daily rhythms in the expression of Notch pathway components [155], and Per3 overexpression in cancer cells decreases Notch signaling components [243]. This suggests that Notch-driven ISC differentiation also could be regulated by the circadian clock. However, as is the case with Wnt signaling, these pathways can be bidirectional, as disruption of Notch signaling in Drosophila ISCs results in arrhythmic clock activity [16]. Overall, the connections between ISC differentiation, clock activity, and the regulation of cell fate by the clock are worthwhile to consider. Indeed in the epidermis, the transcription factor Klf9 establishes a precedent, as it functions downstream of the clock to drive rhythms in the differentiation of skin cells [160].
Feeding and the regulation of cell growth
The intestine receives food to digest and absorb, and it is highly sensitive to the timing of food intake. Normally the central pacemaker in the suprachiasmatic nucleus and peripheral pacemakers in the digestive tract work in concert together, with periods of activity determining the time of food intake and subsequent changes in hormone levels. However, when food is presented during periods of inactivity, peripheral digestive system clocks are synchronized independently from the central clock [85]. This is in part due to the levels of circulating insulin, which is elevated during feeding, and which is a strong entrainment cue for peripheral tissue cell clocks [93, 95]. Strikingly, it was recently shown that daily changes in proliferation of colon cells could be restored in Clock mutant mice under a restricted feeding paradigm [244]. Thus, the time of food intake can set into motion circadian-dependent signaling processes that could affect ISC biology. Insulin was recently shown to directly synchronize intestinal organoid cultures [95], making it a highly relevant candidate mechanism of how the circadian clock impact ISC activity.
In Drosophila insulin induces ISCs to grow and proliferate, because it is a signal of overall nutrient levels in the body that guides tissue size expansion [107, 245–247]. Tissue growth is particularly important during development and regeneration, and insulin signaling is critical to increase intestinal cell production at these times [245, 247]. However, in mammals the role of insulin signaling is not altogether clear. Insulin can affect mammalian ISC function, both directly and indirectly, through activation of the insulin pathway component mammalian Target of Rapamycin (mTOR). During caloric restriction, Paneth cells, that are an important part of the ISC niche, activate mTOR to augment ISC number [248]. In ISCs, mTOR can also cooperate with SIRT1 to enhance mTOR activity cell-intrinsically during calorie restriction [249]. In both of these cases, calorie restriction (and thus presumably lower insulin) paradoxically increases ISC number but not necessarily tissue size [248]. Yet another phenomenon can also occur in this complex system during another physiological context: regeneration following acute fasting. In this case, a different population of non-Lgr5+ ISCs is activated to drive proliferation through mTOR to presumably restore tissue to its normal size following a period of fasting [250]. The precise role of the insulin pathway in mammalian ISCs is not wholly consistent between Drosophila and mice, but taken together the pathway appears to regulate the ISCs of the intestinal epithelium in a context-specific fashion, and its connection to the circadian clock, feeding behaviour, and digestive tract physiology, make it an attractive candidate for future studies. Indeed, the finding that Bmal1 is phosphorylated by the mTOR target S6K, to act as a positive regulator of translation, implicates the circadian clock system with the timing of protein production from mRNA [251]. This fundamental process means that the clock not only drives rhythms in transcript abundance but, in cooperation with insulin signaling, boosts the production of proteins from these transcripts.
Inflammation and the immune system
The immune system is highly active in the digestive tract, where many types of white blood cells interact with the microbiome of the intestinal lumen. Detection of pathogens by immune system cells stimulates an inflammatory response, that subsequently affects the intestinal epithelium. In Drosophila, bacterial infection results in large changes in epithelial gene expression [252] and activation of the Janus Kinase/Signal Transducers and Activators of Transcription (Jak/Stat) pathway [253]. It is now well accepted that the Jak/Stat pathway functions in Drosophila ISCs to both promote differentiation of enterocytes [254] and rapid ISC proliferation during an inflammatory response [111, 112, 245, 253]. In this system, the cytokines Upd1-3 function to activate Stat in ISCs, coordinating the stress/infection response to ISC proliferation to replace damaged epithelial cells. In mammals, the Jak/Stat pathway is conserved, where the cytokines IL-6, IL-22, and the transcription factor Stat3 play a similar role in driving the proliferation of Lgr5+ ISCs during inflammation [255–257]. These studies reveal that the intestinal epithelium receives and translates pro-inflammatory signals into a potent ISC regenerative response.
In this context, it is important to note that many immune system functions have circadian effects including white blood cell proliferation [258] and circulation [259], and the susceptibility to infections [260, 261]. In Drosophila, the time of pathogen infection dictates survival and the immune system response downstream of the circadian clock [262, 263]. In mice, monocytes [264], macrophages [265, 266], natural killer cells [267] and T cells [268, 269] have all been shown to possess cell-intrinsic circadian clocks. This is particularly well-established in macrophages, where approximately 8% of protein-coding genes including immune response, cytokine transcription and cytokine stability have circadian rhythmicity [265]. Pro-inflammatory interleukins, such as IL-6, in macrophages mediate time-of-day-dependent inflammatory responses that determine the body’s response to infection [270]. Circadian timing of the immune system output has emerged as an important physiological mechanism, that adjusts inflammatory timing to maintain homeostasis effectively [271–273].
Several studies have recently addressed the role of inflammation in digestive tract circadian clocks. Immune system factors involved in the detection and clearance of infections, including Toll-like receptors and Cryptidins, display daily rhythms in the intestine [274, 275]. Furthermore, inflammation and cytokine expression are affected by the time of infection [276], and direct the stress response of the damaged intestinal epithelium. This results in different outcomes when mice are infected by gastrointestinal bacteria at different times of day, highlighting the circadian-dependence of the inflammatory response. Two recent studies have determined that the inflammasome, a sensor of bacterial or damage-related stresses, is regulated by the circadian clock through the inflammasome component, Nlrp3. In the liver, circadian regulation of the inflammasome in macrophages primes the sensitivity of the tissue to acute toxic insults [277], and in the colon, acute colitis is worsened due to inappropriate Nlrp3 expression [278]. In both of these cases, Nlrp3 is a direct transcriptional target of Reverb, that is itself rhythmically expressed by the core circadian clock. This means that in peripheral tissues like the intestine, the circadian clock adjusts sensitivity to pathogenic responses according to time of day. Indeed, other components of the innate immune system in the intestine have been shown to oscillate under Clock/Bmal1 according to time of day [279], and cytokines such as Tnf are also under circadian clock regulation to drive daily rhythms in intestinal epithelial proliferation [15]. This means that the intestine is likely to time the stress response as well as metabolic processes, such as digestion and feeding, although it is not clear whether to coordinate or separate these two functions. In the nearby colon, the regenerative response to acute inflammatory bowel disease has a clock-dependent phenotype as well [26]. In this case, a mouse model indicates that Per1/2 circadian clock mutants are highly susceptible to acute colitis, and have a poor regenerative response, consistent with Reverb/Nlrp3 studies [278]. Importantly, mice simply exposed to an altered photoperiod resembling shift-work show these same effects, suggesting this is in fact a bona fide circadian phenomenon rather than a non-clock role for circadian clock genes [280]. Although these studies have not yet examined the inflammatory responses of ISCs directly, it is likely that these cells respond to the timing of cytokine release from the surrounding immune system cells.
Another possible mechanism that might regulate ISCs in the intestinal epithelium, is the microbiome, a topic of growing interest in the circadian biology field. The intestinal lumen contains a tremendous population of diverse microbiota which interacts with overall animal physiology [281, 282]. Recent research has shown that intestinal microbiome composition varies over the course of the day and the microbiome can influence the host intestinal transcriptome [281]. This is in part because all animals exhibit circadian rhythms in food intake, carried out when they are active, that provide sustenance to the microbiome resident in the digestive tract. The microbiome in their turn influence host response and physiology. However, in the intestinal epithelium the clock target gene Nfil3, coordinates rhythms in the absorption of lipid nutrients, but requires microbiota to be present, revealing that microbiota is an active player in regulating circadian physiological rhythms [283]. Indeed, there are both evidence that the microbiota are required for normal circadian transcription rhythms in the intestine [284], and evidence that the clock functions independently of microbiota [139]. Many questions in this area need to be resolved, as the circadian timing of microbiotal activity clearly influences transcription and subsequent physiological responses in the intestinal epithelium [281]. A recent study has shown that microbiota is a source of important sex-specific signals, that affect male versus female daily microbial rhythms and sex-specific growth hormone production [139]. The role of the microbiome in regulating organism physiology is an exciting area of research. The precise mechanisms connecting the microbiome and animal physiology are poorly understood, and how these may affect ISC function are an open question.
Circadian rhythms and gastrointestinal disease
Human beings experience both photoperiod and food-induced circadian clock entrainment. In modern society, human circadian clocks are subject to socio-economic factors such as shiftwork, travel across time-zones, and the use of artificial light sources, that influence their synchronization. In shift workers, who regularly experience disrupted circadian rhythms, meta-analyses of clinical studies have revealed that there is an increase in various gastrointestinal disease symptoms such as pain, and inflammation [285]. In particular, two specific diseases increased in shift workers that are relevant to stem cell biology include colorectal cancer [286, 287], and inflammatory bowel disease (IBD) [288].
Colorectal cancer incidence has been examined extensively in the nursing profession, and a significant increase in cancer risk has been found in nurses that perform shift work for > 15 years [286, 289]. However, no significant trends were observed in nurses that perform sporadic shift work, or fewer years of shift-work, suggesting that only long-term circadian disruption elicits these effects. In colorectal cancer cells themselves, circadian clock genes have also been found to be dysregulated [23] with reduced Per1 levels [290–292], and mutations in Per1 [293], a core genetic component of the circadian clock. Rorα, which regulates Bmal1 expression, has also been correlated with increased risk of colorectal cancer [294]. In addition, there are reports regarding the expression of the clock genes Bmal1, Cry1 and Cry2 in colorectal cancer. Decreased Bmal1, Per1-3 and Cry2 levels have been observed in some studies [291], while others report decreased Per1 and Per3 levels and increased Clock expression [292], or increased Cry1 expression [295]. It is not clear why certain cancers show either increases or decreases in the expression of various clock gene alleles. This seemingly conflicting data could be simply due to heterogeneity within tumours, or variation between patients. It may reflect either clock gene mutation as a promoting factor in colorectal tumorigenesis during disease progression or as a passenger mutation effect of genetically unstable cancer cells. However, it is important to consider circadian dysfunction as a contributing factor to cancer growth, because the outputs of the circadian system can have multiple pro-growth and pro-metabolic effects on proliferating cells. The connection between many types of cancer and the circadian clock has been reviewed extensively, and both epidemiological and basic research suggests aberrant clock function is likely to be pro-tumorigenic [296, 297].
IBD involves the chronic inflammation of the gastrointestinal tract, that is present in two subtypes of IBD: Crohn’s disease and Ulcerative Colitis. A large (> 1000 people) occupational risk study analyzing health benefits in Germany has linked occupations with irregular shift-work to higher rates of IBD [288]. Socio-economic factors that alter circadian rhythms, such as poor sleeping habits on weekends, poor food timing, and sleep debt, have also been linked to augmented IBD symptoms in a study of 115 IBD patients [298]. This suggests that disruption of circadian rhythms is one of the many factors that may influence IBD. Transcript analysis suggests that circadian clock genes, including Per1, Npas2, Cry1, and Rorα, have altered expression in intestinal mucosa biopsies from IBD patients [299, 300], as well as lower expression of many clock genes in their peripheral blood cells [300]. Young, untreated patients recently diagnosed with IBD also show low clock gene expression in the intestinal mucosa and white blood cells [301]. Taken together, this indicates that circadian dysfunction in white blood cells is a feature of IBD, and it remains to be determined whether the same is true for the different cells of the epithelium, and surrounding lamina propria. The precise mechanisms linking circadian clock function to IBD remain to be tested, but given its role in regulating the immune system it is highly plausible that circadian disruption would increase inflammation in IBD patients [302–305]. Taken together, many studies suggest a role for circadian rhythms in gastrointestinal health and disease.
Conclusion
A healthy digestive tract is central to overall health. The high cellular turnover of the intestine in response to daily damage depends on a population of ISCs, that are highly sensitive to cell signaling pathways activated according to physiological need. Many of these pathways have shown circadian oscillations in Drosophila and mice which supports a role for the circadian clock in modulating the responsiveness of ISCs according to time of day. As well, the circadian clock has been shown to regulate a number of intrinsic cellular processes including growth, proliferation, differentiation, and DNA damage, which are likely to be, in part, downstream of cellular signals. In essence, the circadian clock regulates both external physiological processes and internal processes in cells that establishes temporal connections between a specific cell type and the body. Although a number of studies have addressed these ideas in other cell types, much work needs to be done specifically in ISCs to test these relationships. Further research will explore these possible connections, and provide critical information about the molecular details of the circadian biology of ISCs.
This work is highly relevant in understanding how the circadian clock impacts the health of the intestine, in biology, medicine, and evolution. Why does the body coordinate the timing of intestinal processes? One possibility is to increase certain physiological mechanisms to occur at optimal times of day. For instance, coordinating the time of cell growth and division with the time of activity and feeding, so that nutrients are readily available when growth is highest. Another possibility is that the circadian system coordinates the anti-phasic timing of incompatible processes. For instance, separating the timing of cell growth and division from the timing of inflammation and DNA repair. These possibilities are not mutually exclusive and speak for a role of the circadian clock in optimizing health and fitness according to a 24-h schedule that is the result of millions of years of evolution. In this sense, the study of circadian clock regulation of ISCs can be an excellent model system to address fundamental questions about the biology of circadian systems in general. These same ISC-related processes are also relevant to our understanding the connections between ISCs and diseases such as colorectal cancer and IBD.
It is clear that these studies are highly impactful in both research and medicine. Circadian biology has aptly demonstrated that, in the life sciences, the time of sample collection is a critical variable. As a gene of interest expression and activity oscillates according to time of day, interactions that occurred at one time may not be present at another. Circadian rhythms may function to synergize or separate incompatible biological processes within the same cell, or between cells of the body. As this field matures, it is important that such a fundamental system is considered in many if not most life sciences research. For medicine, these concepts provide important information for both the prevention of disease and the treatment of disease. The concept of chronotherapy refers to the application of medical treatments, such as the timing of drugs, according to an optimal physiological schedule. This has been supported experimentally [17, 306], and is an important consideration in medical practice. In addition, knowing that human activities such as shift-work, travel across time zones, and social jetlag (the weekly shifting of sleep/wake schedules from weekday to weekend) affect circadian rhythms, implies that these activities are unhealthy. This adds circadian disruption to a list of avoidable social behaviors, especially in individuals that might have a genetic propensity in developing certain diseases shown to have a circadian component. The role of the circadian clock in gastrointestinal research and health is an exciting area for future research with tremendous potential.
Acknowledgements
We thank all members of the Karpowicz lab for their help. This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (#2015-03656), the Canadian Institutes of Health Research (#388014), the Ontario Institute for Regenerative Medicine, and Crohn’s and Colitis Canada.
Abbreviations
- Apc
Adenomatous polyposis complex
- Ascl2
Achaete scute-like homolog 2
- Bmal1
Brain and muscle Aryl hydrocarbon receptor nuclear translocator like
- Bmi1
B-cell-specific moloney murine leukemia virus integration site 1
- Bmp
Bone morphogenic protein
- CBC
Crypt base columnar cell
- ChIP
Chromatin immunoprecipitation
- Chk
Checkpoint kinase
- Ck1
Casein kinase 1
- Clk
Clock
- Clock
Circadian locomotor output cycles kaput
- Cry
Cryptochrome
- Cxcl12
C-X-C chemokine ligand 12
- Cxcr4
C-X-C chemokine receptor type 4
- Cyc
Cycle
- Gsk3
Glycogen synthase kinase 3
- Hopx
Homeoboxdomain-only protein
- IBD
Inflammatory bowel disease
- IL
Interleukin
- ISC
Intestinal stem cell
- Jak/Stat
Janus kinase/signal transducers and activators of transcription
- Klf9
Kruppel-like factor 9
- Lgr5
Leucine-rich repeat-containing G-protein coupled receptor 5
- Lrig1
Leucine-rich repeats and immunoglobin-like domains-protein 1
- Lrp
Low density lipoprotein receptor
- Mapk
Mitogen-activated protein kinase
- mTOR
Mammalian target of rapamycin
- Olfm4
Olfactomedin 4
- Per
Period
- Ror
Retinoic acid receptor-related orphan receptors
- S6K
S6 kinase
- Sirt1
Sirtuin-1
- Tert1
Telomerase reverse transcriptase
- Tim
Timeless
- Tnf
Tumour necrosis factor
- Upd
Unpaired
- Vri
Vrille
- Pdp1
PAR domain protein-1
- Wee1
Wee1 G2 checkpoint kinase
- Xpa
Xeroderma pigmentosum group A
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet. 2017;18(3):164. doi: 10.1038/nrg.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hardin PE. Advances in genetics. Amsterdam: Elsevier; 2011. Molecular genetic analysis of circadian timekeeping in Drosophila; pp. 141–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turek FW. Circadian clocks: not your grandfather’s clock. Science. 2016;354(6315):992–993. doi: 10.1126/science.aal2613. [DOI] [PubMed] [Google Scholar]
- 4.de Mairan JJ. Observation botanique. Paris: Histoire de l’Académie Royale des Sciences; 1729. [Google Scholar]
- 5.Gardner GF, Feldman JF. The frq locus in Neurospora crassa: a key element in circadian clock organization. Genetics. 1980;96(4):877–886. doi: 10.1093/genetics/96.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalmus H. Periodizität und autochronie (ideochronie) als zeitregelnde eigenschaffen der organismen. Biol Gen. 1935;11:93–114. [Google Scholar]
- 7.Bunning E. Zur Kenntnis der endonomen Tagesrhythmik bei Insekten und bei Pflanzen. Ber Deut Bot Ges. 1935;53:594–623. [Google Scholar]
- 8.Pittendrigh CS. On temperature independence in the clock system controlling emergence time in Drosophila. Proc Natl Acad Sci USA. 1954;40(10):1018. doi: 10.1073/pnas.40.10.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci. 1971;68(9):2112–2116. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aschoff J. Circadian activity pattern with two peaks. Ecology. 1966;47(4):657–662. doi: 10.2307/1933949. [DOI] [Google Scholar]
- 11.Bargiello TA, Young MW. Molecular genetics of a biological clock in Drosophila. Proc Natl Acad Sci. 1984;81(7):2142–2146. doi: 10.1073/pnas.81.7.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karpowicz P, Zhang Y, Hogenesch JB, Emery P, Perrimon N. The circadian clock gates the intestinal stem cell regenerative state. Cell Rep. 2013;3(4):996–1004. doi: 10.1016/j.celrep.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore SR, Pruszka J, Vallance J, Aihara E, Matsuura T, Montrose MH, Shroyer NF, Hong CI. Robust circadian rhythms in organoid cultures from PERIOD2: LUCIFERASE mouse small intestine. Dis Models Mech. 2014;7(9):1123–1130. doi: 10.1242/dmm.014399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsu-ura T, Dovzhenok A, Aihara E, Rood J, Le H, Ren Y, Rosselot AE, Zhang T, Lee C, Obrietan K, Montrose MH, Lim S, Moore SR, Hong CI. Intercellular coupling of the cell cycle and circadian clock in adult stem cell culture. Mol Cell. 2016;64(5):900–912. doi: 10.1016/j.molcel.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stokes K, Cooke A, Chang H, Weaver DR, Breault DT, Karpowicz P. The circadian clock gene BMAL1 coordinates intestinal regeneration. Cell Mol Gastroenterol Hepatol. 2017;4(1):95–114. doi: 10.1016/j.jcmgh.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parasram K, Bernardon N, Hammoud M, Chang H, He L, Perrimon N, Karpowicz P. Intestinal Stem cells exhibit conditional circadian clock function. Stem Cell Rep. 2018;11(5):1287–1301. doi: 10.1016/j.stemcr.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci. 2014;111(45):16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mure LS, Le HD, Benegiamo G, Chang MW, Rios L, Jillani N, Ngotho M, Kariuki T, Dkhissi-Benyahya O, Cooper HM, Panda S. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science. 2018;359(6381):eaao0318. doi: 10.1126/science.aao0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109(3):307–320. doi: 10.1016/S0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 20.Bass J, Lazar MA. Circadian time signatures of fitness and disease. Science. 2016;354(6315):994–999. doi: 10.1126/science.aah4965. [DOI] [PubMed] [Google Scholar]
- 21.Panda S. Circadian physiology of metabolism. Science. 2016;354(6315):1008–1015. doi: 10.1126/science.aah4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sahar S, Sassone-Corsi P. Metabolism and cancer: the circadian clock connection. Nat Rev Cancer. 2009;9(12):886. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- 23.Momma T, Okayama H, Saitou M, Sugeno H, Yoshimoto N, Takebayashi Y, Ohki S, Takenoshita S. Expression of circadian clock genes in human colorectal adenoma and carcinoma. Oncol Lett. 2017;14(5):5319–5325. doi: 10.3892/ol.2017.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alhopuro P, Björklund M, Sammalkorpi H, Turunen M, Tuupanen S, Biström M, Niittymäki I, Lehtonen HJ, Kivioja T, Launonen V, Saharinen J, Nousiainen K, Hautaniemi S, Nuorva K, Mecklin J-P, Jarvinen H, Orntoft T, Arango D, Lehtonen R, Karhu A, Taipale J, Aaltonen LA. Mutations in the circadian gene CLOCK in colorectal cancer. Mol Cancer Res. 2010;8(7):952–960. doi: 10.1158/1541-7786.MCR-10-0086. [DOI] [PubMed] [Google Scholar]
- 25.Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, Lopez JP, Philipson LH, Bradfield CA, Crosby SD, JeBailey L, Wang X, Takahashi JS, Bass J. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466(7306):627. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pagel R, Bär F, Schröder T, Sünderhauf A, Künstner A, Ibrahim SM, Autenrieth SE, Kalies K, König P, Tsang AH, Bettenworth D, Divanovic S, Lehnert H, Fellermann K, Oster H, Derer S, Sina C. Circadian rhythm disruption impairs tissue homeostasis and exacerbates chronic inflammation in the intestine. FASEB J. 2017;31(11):4707–4719. doi: 10.1096/fj.201700141RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ananthakrishnan AN, Khalili H, Konijeti GG, Higuchi LM, de Silva P, Fuchs CS, Richter JM, Schernhammer ES, Chan AT. Sleep duration affects risk for ulcerative colitis: a prospective cohort study. Clin Gastroenterol Hepatol. 2014;12(11):1879–1886. doi: 10.1016/j.cgh.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, Eckel RH, Takahashi JS, Bass J. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308(5724):1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scott E, Carter A, Grant P. Association between polymorphisms in the Clock gene, obesity and the metabolic syndrome in man. Int J Obes. 2008;32(4):658. doi: 10.1038/sj.ijo.0803778. [DOI] [PubMed] [Google Scholar]
- 30.Allada R, White NE, So WV, Hall JC, Rosbash M. A mutant drosophila homolog of mammalian clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93(5):791–804. doi: 10.1016/S0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- 31.Hao H, Allen DL, Hardin PE. A circadian enhancer mediates PER-dependent mRNA cycling in Drosophila melanogaster. Mol Cell Biol. 1997;17(7):3687–3693. doi: 10.1128/MCB.17.7.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rutila JE, Suri V, Le M, So WV, Rosbash M, Hall JC. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell. 1998;93(5):805–814. doi: 10.1016/S0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- 33.Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TD, Weitz CJ, Takahashi JS, Kay SA. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280(5369):1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- 34.Lee C, Bae K, Edery I. PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK-CYC/dBMAL1 heterodimer without disrupting formation of the heterodimer: a basis for circadian transcription. Mol Cell Biol. 1999;19(8):5316–5325. doi: 10.1128/MCB.19.8.5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS, Young MW. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iε. Cell. 1998;94(1):97–107. doi: 10.1016/S0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- 36.Kloss B, Rothenfluh A, Young MW, Saez L. Phosphorylation of period is influenced by cycling physical associations of double-time, period, and timeless in the Drosophila clock. Neuron. 2001;30(3):699–706. doi: 10.1016/S0896-6273(01)00320-8. [DOI] [PubMed] [Google Scholar]
- 37.Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94(1):83–95. doi: 10.1016/S0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- 38.Cyran SA, Buchsbaum AM, Reddy KL, Lin M-C, Glossop NR, Hardin PE, Young MW, Storti RV, Blau J. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell. 2003;112(3):329–341. doi: 10.1016/S0092-8674(03)00074-6. [DOI] [PubMed] [Google Scholar]
- 39.Glossop NR, Houl JH, Zheng H, Ng FS, Dudek SM, Hardin PE. VRILLE feeds back to control circadian transcription of clock in the Drosophila circadian oscillator. Neuron. 2003;37(2):249–261. doi: 10.1016/S0896-6273(03)00002-3. [DOI] [PubMed] [Google Scholar]
- 40.Lim C, Chung BY, Pitman JL, McGill JJ, Pradhan S, Lee J, Keegan KP, Choe J, Allada R. Clockwork orange encodes a transcriptional repressor important for circadian-clock amplitude in Drosophila. Curr Biol. 2007;17(12):1082–1089. doi: 10.1016/j.cub.2007.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng X, Koh K, Sowcik M, Smith CJ, Chen D, Wu MN, Sehgal A. An isoform-specific mutant reveals a role of PDP1ε in the circadian oscillator. J Neurosci. 2009;29(35):10920–10927. doi: 10.1523/JNEUROSCI.2133-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gould PD, Locke JC, Larue C, Southern MM, Davis SJ, Hanano S, Moyle R, Milich R, Putterill J, Millar AJ, Hall A. The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell. 2006;18(5):1177–1187. doi: 10.1105/tpc.105.039990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103(7):1009–1017. doi: 10.1016/S0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vitaterna MH, King DP, Chang A-M, Kornhauser JM, Lowrey PL, McDonald JD, Dove WF, Pinto LH, Turek FW, Takahashi JS. Mutagenesis and mapping of a mouse gene, clock, essential for circadian behavior. Science. 1994;264(5159):719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dudley CA, Erbel-Sieler C, Estill SJ, Reick M, Franken P, Pitts S, McKnight SL. Altered patterns of sleep and behavioral adaptability in NPAS2-deficient mice. Science. 2003;301(5631):379–383. doi: 10.1126/science.1082795. [DOI] [PubMed] [Google Scholar]
- 46.DeBruyne JP, Weaver DR, Reppert SM. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat Neurosci. 2007;10(5):543. doi: 10.1038/nn1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng B, Larkin DW, Albrecht U, Sun ZS, Sage M, Eichele G, Lee CC, Bradley A. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature. 1999;400(6740):169. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]
- 48.Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, Hastings MH, Reppert SM. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288(5468):1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 49.Cermakian N, Monaco L, Pando MP, Dierich A, Sassone-Corsi P. Altered behavioral rhythms and clock gene expression in mice with a targeted mutation in the Period1 gene. EMBO J. 2001;20(15):3967–3974. doi: 10.1093/emboj/20.15.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30(2):525–536. doi: 10.1016/S0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- 51.Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, Li Q, Sun ZS, Eichele G, Bradley A, Lee CC. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105(5):683–694. doi: 10.1016/S0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
- 52.Thresher RJ, Vitaterna MH, Miyamoto Y, Kazantsev A, Hsu DS, Petit C, Selby CP, Dawut L, Smithies O, Takahashi JS, Sancar A. Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science. 1998;282(5393):1490–1494. doi: 10.1126/science.282.5393.1490. [DOI] [PubMed] [Google Scholar]
- 53.Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, Hitomi K, Thresher RJ, Ishikawa T, Miyazaki J, Takahashi JS, Sancar A. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci. 1999;96(21):12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S-i, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D, Buijs RM, Bootsma D, Hoeijmakers JH, Yasui A. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398(6728):627. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 55.Preitner N, Damiola F, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110(2):251–260. doi: 10.1016/S0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 56.Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43(4):527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 57.Masana MI, Sumaya IC, Becker-Andre M, Dubocovich ML. Behavioral characterization and modulation of circadian rhythms by light and melatonin in C3H/HeN mice homozygous for the RORβ knockout. Am J Physiol Regul Integr Comp Physiol. 2007;292(6):R2357–R2367. doi: 10.1152/ajpregu.00687.2006. [DOI] [PubMed] [Google Scholar]
- 58.Koike N, Yoo S-H, Huang H-C, Kumar V, Lee C, Kim T-K, Takahashi JS. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338(6105):349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418(6901):935. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 60.Pincus G. A diurnal rhythm in the excretion of urinary ketosteroids by young men. J Clin Endocrinol. 1943;3(4):195–199. doi: 10.1210/jcem-3-4-195. [DOI] [Google Scholar]
- 61.Andrews RV, Folk GE., Jr Circadian metabolic patterns in cultured hamster adrenal glands. Comp Biochem Physiol. 1964;11(4):393–409. doi: 10.1016/0010-406X(64)90006-4. [DOI] [PubMed] [Google Scholar]
- 62.Kaneko M, Hiroshige T, Shinsako J, Dallman MF. Diurnal changes in amplification of hormone rhythms in the adrenocortical system. Am J Physiol Regul Integr Comp Physiol. 1980;239(3):R309–R316. doi: 10.1152/ajpregu.1980.239.3.R309. [DOI] [PubMed] [Google Scholar]
- 63.Kaneko M, Kaneko K, Shinsako J, Dallman MF. Adrenal sensitivity to adrenocorticotropin varies diurnally. Endocrinology. 1981;109(1):70–75. doi: 10.1210/endo-109-1-70. [DOI] [PubMed] [Google Scholar]
- 64.Kalsbeek A, Buijs RM, Van Heerikhuize JJ, Arts M, Van der Woude TP. Vasopressin-containing neurons of the suprachiasmatic nuclei inhibit corticosterone release. Brain Res. 1992;580(1–2):62–67. doi: 10.1016/0006-8993(92)90927-2. [DOI] [PubMed] [Google Scholar]
- 65.Migeon CJ, Tyler FH, Mahoney JP, Florentin AA, Castle H, Bliss EL, Samuels LT. The diurnal variation of plasma levels and urinary excretion of 17-hydroxycorticosteroids in normal subjects, night workers and blind subjects. J Clin Endocrinol Metab. 1956;16(5):622–633. doi: 10.1210/jcem-16-5-622. [DOI] [PubMed] [Google Scholar]
- 66.Yoo S-H, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong H-K, Oh WJ, Yoo OJ, Menaker M, Takahashi JS. PERIOD2: LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci. 2004;101(15):5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo H, Brewer JM, Champhekar A, Harris RB, Bittman EL. Differential control of peripheral circadian rhythms by suprachiasmatic-dependent neural signals. Proc Natl Acad Sci. 2005;102(8):3111–3116. doi: 10.1073/pnas.0409734102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zylka MJ, Shearman LP, Weaver DR, Reppert SM. Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron. 1998;20(6):1103–1110. doi: 10.1016/S0896-6273(00)80492-4. [DOI] [PubMed] [Google Scholar]
- 69.Lee C, Etchegaray J-P, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107(7):855–867. doi: 10.1016/S0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 70.Field MD, Maywood ES, O’Brien JA, Weaver DR, Reppert SM, Hastings MH. Analysis of clock proteins in mouse SCN demonstrates phylogenetic divergence of the circadian clockwork and resetting mechanisms. Neuron. 2000;25(2):437–447. doi: 10.1016/S0896-6273(00)80906-X. [DOI] [PubMed] [Google Scholar]
- 71.Hsu DS, Zhao X, Zhao S, Kazantsev A, Wang R-P, Todo T, Wei Y-F, Sancar A. Putative human blue-light photoreceptors hCRY1 and hCRY2 are flavoproteins. Biochemistry. 1996;35(44):13871–13877. doi: 10.1021/bi962209o. [DOI] [PubMed] [Google Scholar]
- 72.Todo T, Ryo H, Yamamoto K, Toh H, Inui T, Ayaki H, Nomura T, Ikenaga M. Similarity among the Drosophila (6-4) photolyase, a human photolyase homolog, and the DNA photolyase-blue-light photoreceptor family. Science. 1996;272(5258):109–112. doi: 10.1126/science.272.5258.109. [DOI] [PubMed] [Google Scholar]
- 73.Miyamoto Y, Sancar A. Vitamin B2-based blue-light photoreceptors in the retinohypothalamic tract as the photoactive pigments for setting the circadian clock in mammals. Proc Natl Acad Sci. 1998;95(11):6097–6102. doi: 10.1073/pnas.95.11.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lucas RJ, Freedman MS, Muñoz M, Garcia-Fernández J-M, Foster RG. Regulation of the mammalian pineal by non-rod, non-cone, ocular photoreceptors. Science. 1999;284(5413):505–507. doi: 10.1126/science.284.5413.505. [DOI] [PubMed] [Google Scholar]
- 75.Provencio I, Rodriguez IR, Jiang G, Hayes WP, Moreira EF, Rollag MD. A novel human opsin in the inner retina. J Neurosci. 2000;20(2):600–605. doi: 10.1523/JNEUROSCI.20-02-00600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Peschel N, Chen KF, Szabo G, Stanewsky R. Light-dependent interactions between the Drosophila circadian clock factors cryptochrome, jetlag, and timeless. Curr Biol. 2009;19(3):241–247. doi: 10.1016/j.cub.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 77.Emery P, So WV, Kaneko M, Hall JC, Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95(5):669–679. doi: 10.1016/S0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- 78.Emery P, Stanewsky R, Hall JC, Rosbash M. Drosophila cryptochromes: a unique circadian-rhythm photoreceptor. Nature. 2000;404(6777):456. doi: 10.1038/35006558. [DOI] [PubMed] [Google Scholar]
- 79.Emery P, Stanewsky R, Helfrich-Förster C, Emery-Le M, Hall JC, Rosbash M. Drosophila CRY is a deep brain circadian photoreceptor. Neuron. 2000;26(2):493–504. doi: 10.1016/S0896-6273(00)81181-2. [DOI] [PubMed] [Google Scholar]
- 80.Stanewsky R, Kaneko M, Emery P, Beretta B, Wager-Smith K, Kay SA, Rosbash M, Hall JC. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell. 1998;95(5):681–692. doi: 10.1016/S0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- 81.Plautz JD, Kaneko M, Hall JC, Kay SA. Independent photoreceptive circadian clocks throughout Drosophila. Science. 1997;278(5343):1632–1635. doi: 10.1126/science.278.5343.1632. [DOI] [PubMed] [Google Scholar]
- 82.Giebultowicz JM, Hege DM. Circadian clock in Malpighian tubules. Nature. 1997;386(6626):664. doi: 10.1038/386664a0. [DOI] [PubMed] [Google Scholar]
- 83.Buhr ED, Yoo S-H, Takahashi JS. Temperature as a universal resetting cue for mammalian circadian oscillators. Science. 2010;330(6002):379–385. doi: 10.1126/science.1195262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sehadova H, Glaser FT, Gentile C, Simoni A, Giesecke A, Albert JT, Stanewsky R. Temperature entrainment of Drosophila’s circadian clock involves the gene nocte and signaling from peripheral sensory tissues to the brain. Neuron. 2009;64(2):251–266. doi: 10.1016/j.neuron.2009.08.026. [DOI] [PubMed] [Google Scholar]
- 85.Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14(23):2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hara R, Wan K, Wakamatsu H, Aida R, Moriya T, Akiyama M, Shibata S. Restricted feeding entrains liver clock without participation of the suprachiasmatic nucleus. Genes Cells. 2001;6(3):269–278. doi: 10.1046/j.1365-2443.2001.00419.x. [DOI] [PubMed] [Google Scholar]
- 87.Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci. 2009;106(50):21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stevenson NR, Ferrigni F, Parnicky K, Day S, Fierstein JS. Effect of changes in feeding schedule on the diurnal rhythms and daily activity levels of intestinal brush border enzymes and transport systems. Biochim Biophys Acta (BBA) Biomembr. 1975;406(1):131–145. doi: 10.1016/0005-2736(75)90048-6. [DOI] [PubMed] [Google Scholar]
- 89.Oishi K, Shiota M, Sakamoto K, Kasamatsu M, Ishida N. Feeding is not a more potent Zeitgeber than the light-dark cycle in Drosophila. NeuroReport. 2004;15(4):739–743. doi: 10.1097/00001756-200403220-00034. [DOI] [PubMed] [Google Scholar]
- 90.Xu K, DiAngelo JR, Hughes ME, Hogenesch JB, Sehgal A. The circadian clock interacts with metabolic physiology to influence reproductive fitness. Cell Metab. 2011;13(6):639–654. doi: 10.1016/j.cmet.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xu K, Zheng X, Sehgal A. Regulation of feeding and metabolism by neuronal and peripheral clocks in Drosophila. Cell Metab. 2008;8(4):289–300. doi: 10.1016/j.cmet.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93(6):929–937. doi: 10.1016/S0092-8674(00)81199-X. [DOI] [PubMed] [Google Scholar]
- 93.Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schütz G, Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289(5488):2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 94.Yagita K, Okamura H. Forskolin induces circadian gene expression of rPer1, rPer2 and dbp in mammalian rat-1 fibroblasts. FEBS Lett. 2000;465(1):79–82. doi: 10.1016/S0014-5793(99)01724-X. [DOI] [PubMed] [Google Scholar]
- 95.Crosby P, Hamnett R, Putker M, Hoyle NP, Reed M, Karam CJ, Maywood ES, Stangherlin A, Chesham JE, Hayter EA, Rosenbrier-Ribeiro L, Newham P, Clevers H, Bechtold DA, O’Neill JS. Insulin/IGF-1 drives PERIOD synthesis to entrain circadian rhythms with feeding time. Cell. 2019;177(4):896.e820–909.e820. doi: 10.1016/j.cell.2019.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gill S, Le HD, Melkani GC, Panda S. Time-restricted feeding attenuates age-related cardiac decline in Drosophila. Science. 2015;347(6227):1265–1269. doi: 10.1126/science.1256682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Leblond C, Stevens C. The constant renewal of the intestinal epithelium in the albino rat. Anat Rec. 1948;100(3):357–377. doi: 10.1002/ar.1091000306. [DOI] [PubMed] [Google Scholar]
- 98.von Bertalanffy L. Principles and theory of growth. In: Nowinski WW, editor. Fundamental aspects of normal and malignant growth. Amsterdam: Elsevier; 1960. pp. 137–259. [Google Scholar]
- 99.Van Der Flier LG, Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol. 2009;71:241–260. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- 100.Casali A, Batlle E. Intestinal stem cells in mammals and Drosophila. Cell Stem Cell. 2009;4(2):124–127. doi: 10.1016/j.stem.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 101.Jiang H, Tian A, Jiang J. Intestinal stem cell response to injury: lessons from Drosophila. Cell Mol Life Sci. 2016;73(17):3337–3349. doi: 10.1007/s00018-016-2235-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.De Navascués J, Perdigoto CN, Bian Y, Schneider MH, Bardin AJ, Martínez-Arias A, Simons BD. Drosophila midgut homeostasis involves neutral competition between symmetrically dividing intestinal stem cells. EMBO J. 2012;31(11):2473–2485. doi: 10.1038/emboj.2012.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Goulas S, Conder R, Knoblich JA. The Par complex and integrins direct asymmetric cell division in adult intestinal stem cells. Cell Stem Cell. 2012;11(4):529–540. doi: 10.1016/j.stem.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ohlstein B, Spradling A. Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science. 2007;315(5814):988–992. doi: 10.1126/science.1136606. [DOI] [PubMed] [Google Scholar]
- 105.Biteau B, Jasper H. Slit/Robo signaling regulates cell fate decisions in the intestinal stem cell lineage of Drosophila. Cell Rep. 2014;7(6):1867–1875. doi: 10.1016/j.celrep.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zeng X, Hou SX. Enteroendocrine cells are generated from stem cells through a distinct progenitor in the adult Drosophila posterior midgut. Development. 2015;142(4):644–653. doi: 10.1242/dev.113357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Doupé DP, Marshall OJ, Dayton H, Brand AH, Perrimon N. Drosophila intestinal stem and progenitor cells are major sources and regulators of homeostatic niche signals. Proc Natl Acad Sci. 2018;115(48):12218–12223. doi: 10.1073/pnas.1719169115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tian H, Biehs B, Chiu C, Siebel CW, Wu Y, Costa M, de Sauvage FJ, Klein OD. Opposing activities of Notch and Wnt signaling regulate intestinal stem cells and gut homeostasis. Cell Rep. 2015;11(1):33–42. doi: 10.1016/j.celrep.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li N, Yousefi M, Nakauka-Ddamba A, Jain R, Tobias J, Epstein JA, Jensen ST, Lengner CJ. Single-cell analysis of proxy reporter allele-marked epithelial cells establishes intestinal stem cell hierarchy. Stem Cell Rep. 2014;3(5):876–891. doi: 10.1016/j.stemcr.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu X, Lu R, Wu S, Sun J. Salmonella regulation of intestinal stem cells through the Wnt/β-catenin pathway. FEBS Lett. 2010;584(5):911–916. doi: 10.1016/j.febslet.2010.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jiang H, Edgar BA. EGFR signaling regulates the proliferation of Drosophila adult midgut progenitors. Development. 2009;136(3):483–493. doi: 10.1242/dev.026955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, Edgar BA. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137(7):1343–1355. doi: 10.1016/j.cell.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lin KK, Kumar V, Geyfman M, Chudova D, Ihler AT, Smyth P, Paus R, Takahashi JS, Andersen B. Circadian clock genes contribute to the regulation of hair follicle cycling. PLoS Genet. 2009;5(7):e1000573. doi: 10.1371/journal.pgen.1000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Biteau B, Jasper H. EGF signaling regulates the proliferation of intestinal stem cells in Drosophila. Development. 2011;138(6):1045–1055. doi: 10.1242/dev.056671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Guo Z, Driver I, Ohlstein B. Injury-induced BMP signaling negatively regulates Drosophila midgut homeostasis. J Cell Biol. 2013;201(6):945–961. doi: 10.1083/jcb.201302049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dutta D, Dobson AJ, Houtz PL, Gläßer C, Revah J, Korzelius J, Patel PH, Edgar BA, Buchon N. Regional cell-specific transcriptome mapping reveals regulatory complexity in the adult Drosophila midgut. Cell Rep. 2015;12(2):346–358. doi: 10.1016/j.celrep.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 117.Zhou J, Florescu S, Boettcher A-L, Luo L, Dutta D, Kerr G, Cai Y, Edgar BA, Boutros M. Dpp/Gbb signaling is required for normal intestinal regeneration during infection. Dev Biol. 2015;399(2):189–203. doi: 10.1016/j.ydbio.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 118.Perochon J, Carroll L, Cordero J. Wnt signalling in intestinal stem cells: lessons from mice and flies. Genes. 2018;9(3):138. doi: 10.3390/genes9030138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cordero JB, Stefanatos RK, Myant K, Vidal M, Sansom OJ. Non-autonomous crosstalk between the Jak/Stat and Egfr pathways mediates Apc1-driven intestinal stem cell hyperplasia in the Drosophila adult midgut. Development. 2012;139(24):4524–4535. doi: 10.1242/dev.078261. [DOI] [PubMed] [Google Scholar]
- 120.Cordero JB, Stefanatos RK, Scopelliti A, Vidal M, Sansom OJ. Inducible progenitor-derived Wingless regulates adult midgut regeneration in Drosophila. EMBO J. 2012;31(19):3901–3917. doi: 10.1038/emboj.2012.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tian A, Jiang J. Intestinal epithelium-derived BMP controls stem cell self-renewal in Drosophila adult midgut. Elife. 2014;3:e01857. doi: 10.7554/eLife.01857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tian A, Wang B, Jiang J. Injury-stimulated and self-restrained BMP signaling dynamically regulates stem cell pool size during Drosophila midgut regeneration. Proc Natl Acad Sci. 2017;114(13):E2699–E2708. doi: 10.1073/pnas.1617790114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Beumer J, Clevers H. Regulation and plasticity of intestinal stem cells during homeostasis and regeneration. Development. 2016;143(20):3639–3649. doi: 10.1242/dev.133132. [DOI] [PubMed] [Google Scholar]
- 124.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40(7):915. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Montgomery RK, Carlone DL, Richmond CA, Farilla L, Kranendonk ME, Henderson DE, Baffour-Awuah NY, Ambruzs DM, Fogli LK, Algra S, Breault DT. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc Natl Acad Sci. 2011;108(1):179–184. doi: 10.1073/pnas.1013004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Takeda N, Jain R, LeBoeuf MR, Padmanabhan A, Wang Q, Li L, Lu MM, Millar SE, Epstein JA. Hopx expression defines a subset of multipotent hair follicle stem cells and a progenitor population primed to give rise to K6+ niche cells. Development. 2013;140(8):1655–1664. doi: 10.1242/dev.093005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Powell AE, Wang Y, Li Y, Poulin EJ, Means AL, Washington MK, Higginbotham JN, Juchheim A, Prasad N, Levy SE, Guo Y, Shyr Y, Aronow BJ, Haigis KM, Franklin JL, Coffey RJ. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 2012;149(1):146–158. doi: 10.1016/j.cell.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ayyaz A, Kumar S, Sangiorgi B, Ghoshal B, Gosio J, Ouladan S, Fink M, Barutcu S, Trcka D, Shen J, Chan K, Wrana JL, Gregorieff A. Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell. Nature. 2019;569(7754):121. doi: 10.1038/s41586-019-1154-y. [DOI] [PubMed] [Google Scholar]
- 129.Muñoz J, Stange DE, Schepers AG, Van De Wetering M, Koo BK, Itzkovitz S, Volckmann R, Kung KS, Koster J, Radulescu S, Myant K, Versteeg R, Sansom OJ, van Es J, Barker N, van Oudenaarden A, Mohammed S, Heck AJ, Clevers H. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+ 4’cell markers. EMBO J. 2012;31(14):3079–3091. doi: 10.1038/emboj.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Barker N, Van Es JH, Kuipers J, Kujala P, Van Den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449(7165):1003. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 131.Van der Flier LG, Haegebarth A, Stange DE, Van de Wetering M, Clevers H. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology. 2009;137(1):15–17. doi: 10.1053/j.gastro.2009.05.035. [DOI] [PubMed] [Google Scholar]
- 132.Schuijers J, Junker JP, Mokry M, Hatzis P, Koo B-K, Sasselli V, Van Der Flier LG, Cuppen E, van Oudenaarden A, Clevers H. Ascl2 acts as an R-spondin/Wnt-responsive switch to control stemness in intestinal crypts. Cell Stem Cell. 2015;16(2):158–170. doi: 10.1016/j.stem.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 133.van der Flier LG, van Gijn ME, Hatzis P, Kujala P, Haegebarth A, Stange DE, Begthel H, van den Born M, Guryev V, Oving I, van Es J, Barker N, Peters PJ, van de Wetering M, Clevers H. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell. 2009;136(5):903–912. doi: 10.1016/j.cell.2009.01.031. [DOI] [PubMed] [Google Scholar]
- 134.Cheng H, Leblond C. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine V. Unitarian theory of the origin of the four epithelial cell types. Am J Anat. 1974;141(4):537–561. doi: 10.1002/aja.1001410407. [DOI] [PubMed] [Google Scholar]
- 135.Bjerknes M, Cheng H. The stem-cell zone of the small intestinal epithelium. III. Evidence from columnar, enteroendocrine, and mucous cells in the adult mouse. Am J Anat. 1981;160(1):77–91. doi: 10.1002/aja.1001600107. [DOI] [PubMed] [Google Scholar]
- 136.Sládek M, Rybová M, Jindráková Z, Zemanová Z, Polidarová L, Mrnka L, O’Neill J, Pácha J, Sumová A. Insight into the circadian clock within rat colonic epithelial cells. Gastroenterology. 2007;133(4):1240–1249. doi: 10.1053/j.gastro.2007.05.053. [DOI] [PubMed] [Google Scholar]
- 137.Hoogerwerf WA, Hellmich HL, Cornélissen G, Halberg F, Shahinian VB, Bostwick J, Savidge TC, Cassone VM. Clock gene expression in the murine gastrointestinal tract: endogenous rhythmicity and effects of a feeding regimen. Gastroenterology. 2007;133(4):1250–1260. doi: 10.1053/j.gastro.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 138.Hoogerwerf WA, Sinha M, Conesa A, Luxon BA, Shahinian VB, Cornélissen G, Halberg F, Bostwick J, Timm J, Cassone VM. Transcriptional profiling of mRNA expression in the mouse distal colon. Gastroenterology. 2008;135(6):2019–2029. doi: 10.1053/j.gastro.2008.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Weger BD, Gobet C, Yeung J, Martin E, Jimenez S, Betrisey B, Foata F, Berger B, Balvay A, Foussier A, Charpagne A, Boizet-Bonhoure B, Chou CJ, Naef F, Gachon F. The mouse microbiome is required for sex-specific diurnal rhythms of gene expression and metabolism. Cell Metab. 2019;29(2):362.e368–382.e368. doi: 10.1016/j.cmet.2018.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tognini P, Murakami M, Liu Y, Eckel-Mahan KL, Newman JC, Verdin E, Baldi P, Sassone-Corsi P. Distinct circadian signatures in liver and gut clocks revealed by ketogenic diet. Cell Metab. 2017;26(3):523.e525–538.e525. doi: 10.1016/j.cmet.2017.08.015. [DOI] [PubMed] [Google Scholar]
- 141.Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 2006;20(14):1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Leibach FH, Ganapathy V. Peptide transporters in the intestine and the kidney. Annu Rev Nutr. 1996;16(1):99–119. doi: 10.1146/annurev.nu.16.070196.000531. [DOI] [PubMed] [Google Scholar]
- 143.Pan X, Hussain MM. Clock is important for food and circadian regulation of macronutrient absorption in mice. J Lipid Res. 2009;50(9):1800–1813. doi: 10.1194/jlr.M900085-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pan X, Terada T, Irie M, Saito H, Inui K-I. Diurnal rhythm of H+-peptide cotransporter in rat small intestine. Am J Physiol Gastrointest Liver Physiol. 2002;283(1):G57–G64. doi: 10.1152/ajpgi.00545.2001. [DOI] [PubMed] [Google Scholar]
- 145.Rhoads DB, Rosenbaum DH, Unsal H, Isselbacher KJ, Levitsky LL. Circadian periodicity of intestinal Na+/glucose cotransporter 1 mRNA levels is transcriptionally regulated. J Biol Chem. 1998;273(16):9510–9516. doi: 10.1074/jbc.273.16.9510. [DOI] [PubMed] [Google Scholar]
- 146.Saito M, Murakami E, Nishida T, Fujisawa Y, Suda M. Circadian rhythms in digestive enzymes in the small intestine of rats: I. Patterns of the rhythms in various regions of the small intestine. J Biochem. 1975;78(3):475–480. doi: 10.1093/oxfordjournals.jbchem.a130930. [DOI] [PubMed] [Google Scholar]
- 147.Saito M, Sato Y, Suda M. Circadian rhythm and dietary response of disaccharidase activities in isolated rat jejunum. Gastroenterology. 1978;75(5):828–831. doi: 10.1016/0016-5085(78)90465-1. [DOI] [PubMed] [Google Scholar]
- 148.Scheving LA. Biological clocks and the digestive system. Gastroenterology. 2000;119(2):536–549. doi: 10.1053/gast.2000.9305. [DOI] [PubMed] [Google Scholar]
- 149.Pacha J, Sumova A. Circadian regulation of epithelial functions in the intestine. Acta Physiol. 2013;208(1):11–24. doi: 10.1111/apha.12090. [DOI] [PubMed] [Google Scholar]
- 150.Hussain MM. Regulation of intestinal lipid absorption by clock genes. Annu Rev Nutr. 2014;34:357–375. doi: 10.1146/annurev-nutr-071813-105322. [DOI] [PubMed] [Google Scholar]
- 151.Potten CS, Al-Barwari SE, Hume WJ, Searle J. Circadian rhythms of presumptive stem cells in three different epithelia of the mouse. Cell Prolif. 1977;10(6):557–568. doi: 10.1111/j.1365-2184.1977.tb00312.x. [DOI] [PubMed] [Google Scholar]
- 152.Lucas D, Battista M, Shi PA, Isola L, Frenette PS. Mobilized hematopoietic stem cell yield depends on species-specific circadian timing. Cell Stem Cell. 2008;3(4):364–366. doi: 10.1016/j.stem.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Méndez-Ferrer S, Lucas D, Battista M, Frenette PS. Haematopoietic stem cell release is regulated by circadian oscillations. Nature. 2008;452(7186):442. doi: 10.1038/nature06685. [DOI] [PubMed] [Google Scholar]
- 154.Janich P, Pascual G, Merlos-Suárez A, Batlle E, Ripperger J, Albrecht U, Cheng H-YM, Obrietan K, Di Croce L, Benitah SA. The circadian molecular clock creates epidermal stem cell heterogeneity. Nature. 2011;480(7376):209. doi: 10.1038/nature10649. [DOI] [PubMed] [Google Scholar]
- 155.Janich P, Toufighi K, Solanas G, Luis NM, Minkwitz S, Serrano L, Lehner B, Benitah SA. Human epidermal stem cell function is regulated by circadian oscillations. Cell Stem Cell. 2013;13(6):745–753. doi: 10.1016/j.stem.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 156.Tsinkalovsky O, Rosenlund B, Laerum OD, Eiken HG. Clock gene expression in purified mouse hematopoietic stem cells. Exp Hematol. 2005;33(1):100–107. doi: 10.1016/j.exphem.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 157.Puram RV, Kowalczyk MS, de Boer CG, Schneider RK, Miller PG, McConkey M, Tothova Z, Tejero H, Heckl D, Järås M, Chen MC, Li H, Tamayo A, Cowley GS, Rozenblatt-Rosen O, Al-Shahrour F, Regev A, Ebert BL. Core circadian clock genes regulate leukemia stem cells in AML. Cell. 2016;165(2):303–316. doi: 10.1016/j.cell.2016.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wang J, Morita Y, Han B, Niemann S, Löffler B, Rudolph KL. Per2 induction limits lymphoid-biased haematopoietic stem cells and lymphopoiesis in the context of DNA damage and ageing. Nat Cell Biol. 2016;18(5):480. doi: 10.1038/ncb3342. [DOI] [PubMed] [Google Scholar]
- 159.Plikus MV, Vollmers C, de la Cruz D, Chaix A, Ramos R, Panda S, Chuong C-M. Local circadian clock gates cell cycle progression of transient amplifying cells during regenerative hair cycling. Proc Natl Acad Sci. 2013;110(23):E2106–E2115. doi: 10.1073/pnas.1215935110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Spörl F, Korge S, Jürchott K, Wunderskirchner M, Schellenberg K, Heins S, Specht A, Stoll C, Klemz R, Maier B, Wenck H, Schrader A, Kunz D, Blatt T, Kramer A. Krüppel-like factor 9 is a circadian transcription factor in human epidermis that controls proliferation of keratinocytes. Proc Natl Acad Sci. 2012;109(27):10903–10908. doi: 10.1073/pnas.1118641109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hoyle NP, Seinkmane E, Putker M, Feeney KA, Krogager TP, Chesham JE, Bray LK, Thomas JM, Dunn K, Blaikley J, O’Neill JS. Circadian actin dynamics drive rhythmic fibroblast mobilization during wound healing. Sci Transl Med. 2017;9(415):eaal2774. doi: 10.1126/scitranslmed.aal2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Paatela E, Munson D, Kikyo N. Circadian regulation in tissue regeneration. Int J Mol Sci. 2019;20(9):2263. doi: 10.3390/ijms20092263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Dierickx P, Van Laake LW, Geijsen N. Circadian clocks: from stem cells to tissue homeostasis and regeneration. EMBO Rep. 2018;19(1):18–28. doi: 10.15252/embr.201745130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Weger M, Diotel N, Dorsemans A-C, Dickmeis T, Weger BD. Stem cells and the circadian clock. Dev Biol. 2017;431(2):111–123. doi: 10.1016/j.ydbio.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 165.Plikus MV, Van Spyk EN, Pham K, Geyfman M, Kumar V, Takahashi JS, Andersen B. The circadian clock in skin: implications for adult stem cells, tissue regeneration, cancer, aging, and immunity. J Biol Rhythms. 2015;30(3):163–182. doi: 10.1177/0748730414563537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Brown SA. Circadian clock-mediated control of stem cell division and differentiation: beyond night and day. Development. 2014;141(16):3105–3111. doi: 10.1242/dev.104851. [DOI] [PubMed] [Google Scholar]
- 167.Janich P, Meng Q-J, Benitah SA. Circadian control of tissue homeostasis and adult stem cells. Curr Opin Cell Biol. 2014;31:8–15. doi: 10.1016/j.ceb.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 168.Yagita K, Horie K, Koinuma S, Nakamura W, Yamanaka I, Urasaki A, Shigeyoshi Y, Kawakami K, Shimada S, Takeda J, Uchiyama Y. Development of the circadian oscillator during differentiation of mouse embryonic stem cells in vitro. Proc Natl Acad Sci. 2010;107(8):3846–3851. doi: 10.1073/pnas.0913256107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Umemura Y, Koike N, Ohashi M, Tsuchiya Y, Meng QJ, Minami Y, Hara M, Hisatomi M, Yagita K. Involvement of posttranscriptional regulation of Clock in the emergence of circadian clock oscillation during mouse development. Proc Natl Acad Sci. 2017;114(36):E7479–E7488. doi: 10.1073/pnas.1703170114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Solanas G, Peixoto FO, Perdiguero E, Jardí M, Ruiz-Bonilla V, Datta D, Symeonidi A, Castellanos A, Welz P-S, Caballero JM, Sassone-Corsi P, Muñoz-Cánoves P, Benitah SA. Aged stem cells reprogram their daily rhythmic functions to adapt to stress. Cell. 2017;170(4):678.e620–692.e620. doi: 10.1016/j.cell.2017.07.035. [DOI] [PubMed] [Google Scholar]
- 171.Sigdestad C, Bauman J, Lesher S. Diurnal fluctuations in the number of cells in mitosis and DNA synthesis in the jejunum of the mouse. Exp Cell Res. 1969;58(1):159–162. doi: 10.1016/0014-4827(69)90126-8. [DOI] [PubMed] [Google Scholar]
- 172.Bishehsari F, Levi F, Turek FW, Keshavarzian A. Circadian rhythms in gastrointestinal health and diseases. Gastroenterology. 2016;151(3):e1–e5. doi: 10.1053/j.gastro.2016.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Bertalanffy FD. Mitotic rates and renewal times of the digestive tract epithelia in the rat. Cells Tissues Organs. 1960;40(2–3):130–148. doi: 10.1159/000141580. [DOI] [PubMed] [Google Scholar]
- 174.Pilgrim C, Erb W, Maurer W. Diurnal fluctuations in the numbers of DNA synthesizing nuclei in various mouse tissues. Nature. 1963;199(4896):863. doi: 10.1038/199863a0. [DOI] [PubMed] [Google Scholar]
- 175.e Melo FdS, de Sauvage FJ. Cellular plasticity in intestinal homeostasis and disease. Cell Stem Cell. 2018;24(1):54–64. doi: 10.1016/j.stem.2018.11.019. [DOI] [PubMed] [Google Scholar]
- 176.Borgs L, Beukelaers P, Vandenbosch R, Belachew S, Nguyen L, Malgrange B. Cell “circadian” cycle: new role for mammalian core clock genes. Cell Cycle. 2009;8(6):832–837. doi: 10.4161/cc.8.6.7869. [DOI] [PubMed] [Google Scholar]
- 177.Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302(5643):255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 178.Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119(5):693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 179.Bieler J, Cannavo R, Gustafson K, Gobet C, Gatfield D, Naef F. Robust synchronization of coupled circadian and cell cycle oscillators in single mammalian cells. Mol Syst Biol. 2014;10(7):739. doi: 10.15252/msb.20145218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Feillet C, Krusche P, Tamanini F, Janssens RC, Downey MJ, Martin P, Teboul M, Saito S, Lévi FA, Bretschneider T, van Der Horst GT, Delaunay F, Rand DA. Phase locking and multiple oscillating attractors for the coupled mammalian clock and cell cycle. Proc Natl Acad Sci. 2014;111(27):9828–9833. doi: 10.1073/pnas.1320474111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yeom M, Pendergast JS, Ohmiya Y, Yamazaki S. Circadian-independent cell mitosis in immortalized fibroblasts. Proc Natl Acad Sci. 2010;107(21):9665–9670. doi: 10.1073/pnas.0914078107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Gréchez-Cassiau A, Rayet B, Guillaumond F, Teboul M, Delaunay F. The circadian clock component BMAL1 is a critical regulator of p21WAF1/CIP1 expression and hepatocyte proliferation. J Biol Chem. 2008;283(8):4535–4542. doi: 10.1074/jbc.M705576200. [DOI] [PubMed] [Google Scholar]
- 183.Kowalska E, Ripperger JA, Hoegger DC, Bruegger P, Buch T, Birchler T, Mueller A, Albrecht U, Contaldo C, Brown SA. NONO couples the circadian clock to the cell cycle. Proc Natl Acad Sci. 2013;110(5):1592–1599. doi: 10.1073/pnas.1213317110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Polidarová L, Soták M, Sládek M, Pácha J, Sumová A, Polidarová L, Soták M, Sládek M, Pácha J, Sumová A. Temporal gradient in the clock gene and cell-cycle checkpoint kinase Wee1 expression along the gut. Chronobiol Int. 2009;26(4):607–620. doi: 10.1080/07420520902924889. [DOI] [PubMed] [Google Scholar]
- 185.Sancar A, Lindsey-Boltz LA, Kang T-H, Reardon JT, Lee JH, Ozturk N. Circadian clock control of the cellular response to DNA damage. FEBS Lett. 2010;584(12):2618–2625. doi: 10.1016/j.febslet.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Sancar A, Lindsey-Boltz LA, Gaddameedhi S, Selby CP, Ye R, Chiou Y-Y, Kemp MG, Hu J, Lee JH, Ozturk N. Circadian clock, cancer, and chemotherapy. Biochemistry. 2014;54(2):110–123. doi: 10.1021/bi5007354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Ünsal-Kaçmaz K, Mullen TE, Kaufmann WK, Sancar A. Coupling of human circadian and cell cycles by the timeless protein. Mol Cell Biol. 2005;25(8):3109–3116. doi: 10.1128/MCB.25.8.3109-3116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Gery S, Komatsu N, Baldjyan L, Yu A, Koo D, Koeffler HP. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol Cell. 2006;22(3):375–382. doi: 10.1016/j.molcel.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 189.Im J-S, Jung B-H, Kim S-E, Lee K-H, Lee J-K. Per3, a circadian gene, is required for Chk2 activation in human cells. FEBS Lett. 2010;584(23):4731–4734. doi: 10.1016/j.febslet.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 190.Geyfman M, Kumar V, Liu Q, Ruiz R, Gordon W, Espitia F, Cam E, Millar SE, Smyth P, Ihler A, Takahashi JS, Andersen B. Brain and muscle Arnt-like protein-1 (BMAL1) controls circadian cell proliferation and susceptibility to UVB-induced DNA damage in the epidermis. Proc Natl Acad Sci. 2012;109(29):11758–11763. doi: 10.1073/pnas.1209592109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Gaddameedhi S, Selby CP, Kaufmann WK, Smart RC, Sancar A. Control of skin cancer by the circadian rhythm. Proc Natl Acad Sci. 2011;108(46):18790–18795. doi: 10.1073/pnas.1115249108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kang T-H, Lindsey-Boltz LA, Reardon JT, Sancar A. Circadian control of XPA and excision repair of cisplatin-DNA damage by cryptochrome and HERC2 ubiquitin ligase. Proc Natl Acad Sci. 2010;107(11):4890–4895. doi: 10.1073/pnas.0915085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kang T-H, Reardon JT, Kemp M, Sancar A. Circadian oscillation of nucleotide excision repair in mammalian brain. Proc Natl Acad Sci. 2009;106(8):2864–2867. doi: 10.1073/pnas.0812638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Clevers H, Loh KM, Nusse R. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science. 2014;346(6205):1248012. doi: 10.1126/science.1248012. [DOI] [PubMed] [Google Scholar]
- 195.Bhanot P, Brink M, Samos CH, Hsieh J-C, Wang Y, Macke JP, Andrew D, Nathans J, Nusse R. A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature. 1996;382(6588):225. doi: 10.1038/382225a0. [DOI] [PubMed] [Google Scholar]
- 196.Wehrli M, Dougan ST, Caldwell K, O’Keefe L, Schwartz S, Vaizel-Ohayon D, Schejter E, Tomlinson A, DiNardo S. arrow encodes an LDL-receptor-related protein essential for Wingless signalling. Nature. 2000;407(6803):527. doi: 10.1038/35035110. [DOI] [PubMed] [Google Scholar]
- 197.Kishida S, Yamamoto H, Hino S-i, Ikeda S, Kishida M, Kikuchi A. DIX domains of Dvl and Axin are necessary for protein interactions and their ability to regulate β-catenin stability. Mol Cell Biol. 1999;19(6):4414–4422. doi: 10.1128/MCB.19.6.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Yan KS, Gevaert O, Zheng GX, Anchang B, Probert CS, Larkin KA, Davies PS, Cheng Z-f, Kaddis JS, Han A, Roelf K, Calderon RI, Cynn E, Hu X, Mandleywala K, Wihelmy J, Grimes SM, Corney DC, Boutet SC, Terry JM, Belgrader P, Ziraldo SB, Mikkelsen TS, Wang F, von Furstenberg RJ, Smith NR, Chandrakesan P, May R, Chrissy MAS, Jain R, Cartwright CA, Niland JC, Hong Y-K, Carrington J, Breault DT, Epstein JA, Houchen CW, Lynch JP, Martin MG, Plevritis SK, Curtis C, Ji HP, Li L, Henning SJ, Wong MH, Kuo CJ. Intestinal enteroendocrine lineage cells possess homeostatic and injury-inducible stem cell activity. Cell Stem Cell. 2017;21(1):78.e76–90.e76. doi: 10.1016/j.stem.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Ootani A, Li X, Sangiorgi E, Ho QT, Ueno H, Toda S, Sugihara H, Fujimoto K, Weissman IL, Capecchi MR, Kuo CJ. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat Med. 2009;15(6):701. doi: 10.1038/nm.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Sato T, Vries RG, Snippert HJ, Van De Wetering M, Barker N, Stange DE, Van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt–villus structures in vitro without a mesenchymal niche. Nature. 2009;459(7244):262. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 201.Shoshkes-Carmel M, Wang YJ, Wangensteen KJ, Tóth B, Kondo A, Massassa EE, Itzkovitz S, Kaestner KH. Subepithelial telocytes are an important source of Wnts that supports intestinal crypts. Nature. 2018;557(7704):242. doi: 10.1038/s41586-018-0084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Gregorieff A, Pinto D, Begthel H, Destrée O, Kielman M, Clevers H. Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology. 2005;129(2):626–638. doi: 10.1016/j.gastro.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 203.Farin HF, Jordens I, Mosa MH, Basak O, Korving J, Tauriello DV, de Punder K, Angers S, Peters PJ, Maurice MM, Clevers H. Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature. 2016;530(7590):340. doi: 10.1038/nature16937. [DOI] [PubMed] [Google Scholar]
- 204.San Roman AK, Jayewickreme CD, Murtaugh LC, Shivdasani RA. Wnt secretion from epithelial cells and subepithelial myofibroblasts is not required in the mouse intestinal stem cell niche in vivo. Stem Cell Rep. 2014;2(2):127–134. doi: 10.1016/j.stemcr.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Sato T, Van Es JH, Snippert HJ, Stange DE, Vries RG, Van Den Born M, Barker N, Shroyer NF, Van De Wetering M, Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469(7330):415. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Lin G, Xu N, Xi R. Paracrine Wingless signalling controls self-renewal of Drosophila intestinal stem cells. Nature. 2008;455(7216):1119. doi: 10.1038/nature07329. [DOI] [PubMed] [Google Scholar]
- 207.Tian A, Benchabane H, Wang Z, Ahmed Y. Regulation of stem cell proliferation and cell fate specification by wingless/Wnt signaling gradients enriched at adult intestinal compartment boundaries. PLoS Genet. 2016;12(2):e1005822. doi: 10.1371/journal.pgen.1005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Chatterjee S, Yin H, Li W, Lee J, Yechoor VK, Ma K. The nuclear receptor and clock repressor Rev-erbα suppresses myogenesis. Sci Rep. 2019;9(1):4585. doi: 10.1038/s41598-019-41059-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Chatterjee S, Nam D, Guo B, Kim JM, Winnier GE, Lee J, Berdeaux R, Yechoor VK, Ma K. Brain and muscle Arnt-like 1 is a key regulator of myogenesis. J Cell Sci. 2013;126(10):2213–2224. doi: 10.1242/jcs.120519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.He Y, Lin F, Chen Y, Tan Z, Bai D, Zhao Q. Overexpression of the circadian clock gene rev-erbα affects murine bone mesenchymal stem cell proliferation and osteogenesis. Stem Cells Dev. 2014;24(10):1194–1204. doi: 10.1089/scd.2014.0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Guo B, Chatterjee S, Li L, Kim JM, Lee J, Yechoor VK, Minze LJ, Hsueh W, Ma K. The clock gene, brain and muscle Arnt-like 1, regulates adipogenesis via Wnt signaling pathway. FASEB J. 2012;26(8):3453–3463. doi: 10.1096/fj.12-205781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Wood PA, Yang X, Taber A, Oh E-Y, Ansell C, Ayers SE, Al-Assaad Z, Carnevale K, Berger FG, Peña MMO, Hrushesky WJ. Period 2 mutation accelerates ApcMin/+ tumorigenesis. Mol Cancer Res. 2008;6(11):1786–1793. doi: 10.1158/1541-7786.MCR-08-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Yang X, Wood PA, Ansell CM, Ohmori M, Oh E-Y, Xiong Y, Berger FG, Peña MMO, Hrushesky WJ. β-catenin induces β-TrCP-mediated PER2 degradation altering circadian clock gene expression in intestinal mucosa of ApcMin/+ mice. J Biochem. 2008;145(3):289–297. doi: 10.1093/jb/mvn167. [DOI] [PubMed] [Google Scholar]
- 214.Muncan V, Sansom OJ, Tertoolen L, Phesse TJ, Begthel H, Sancho E, Cole AM, Gregorieff A, de Alboran IM, Clevers H, Clarke AR. Rapid loss of intestinal crypts upon conditional deletion of the Wnt/Tcf-4 target gene c-Myc. Mol Cell Biol. 2006;26(22):8418–8426. doi: 10.1128/MCB.00821-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Fu L, Pelicano H, Liu J, Huang P, Lee CC. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111(1):41–50. doi: 10.1016/S0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 216.Repouskou A, Sourlingas TG, Sekeri-Pataryas KE, Prombona A. The circadian expression of c-MYC is modulated by the histone deacetylase inhibitor trichostatin A in synchronized murine neuroblastoma cells. Chronobiol Int. 2010;27(4):722–741. doi: 10.3109/07420521003786800. [DOI] [PubMed] [Google Scholar]
- 217.Altman BJ, Hsieh AL, Sengupta A, Krishnanaiah SY, Stine ZE, Walton ZE, Gouw AM, Venkataraman A, Li B, Goraksha-Hicks P, Diskin SJ, Bellovin DI, Simon MC, Rathmell JC, Lazar MA, Maris JM, Felsher DW, Hogenesch JB, Welijie AM, Dang CV. MYC disrupts the circadian clock and metabolism in cancer cells. Cell Metab. 2015;22(6):1009–1019. doi: 10.1016/j.cmet.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Repouskou A. Prombona A (2016) c-MYC targets the central oscillator gene Per1 and is regulated by the circadian clock at the post-transcriptional level. Biochim Biophys Acta (BBA) Gene Regul Mech. 1859;4:541–552. doi: 10.1016/j.bbagrm.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 219.Shostak A, Ruppert B, Ha N, Bruns P, Toprak UH, Project IM-S, Eils R, Schlesner M, Diernfellner A, Brunner M. MYC/MIZ1-dependent gene repression inversely coordinates the circadian clock with cell cycle and proliferation. Nat Commun. 2016;7:11807. doi: 10.1038/ncomms11807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Van De Wetering M, Sancho E, Verweij C, De Lau W, Oving I, Hurlstone A, Van Der Horn K, Batlle E, Coudreuse D, Haramis A-P, Tjon-Pon-Fong M, Moerer P, van den Born M, Soete G, Pals S, Eilers M, Medema R, Clevers H. The β-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111(2):241–250. doi: 10.1016/S0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 221.Matsu-ura T, Moore SR, Hong CI. WNT takes two to tango: molecular links between the circadian clock and the cell cycle in adult stem cells. J Biol Rhythms. 2018;33(1):5–14. doi: 10.1177/0748730417745913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Hirota T, Lewis WG, Liu AC, Lee JW, Schultz PG, Kay SA. A chemical biology approach reveals period shortening of the mammalian circadian clock by specific inhibition of GSK-3β. Proc Natl Acad Sci. 2008;105(52):20746–20751. doi: 10.1073/pnas.0811410106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Iitaka C, Miyazaki K, Akaike T, Ishida N. A role for glycogen synthase kinase-3β in the mammalian circadian clock. J Biol Chem. 2005;280(33):29397–29402. doi: 10.1074/jbc.M503526200. [DOI] [PubMed] [Google Scholar]
- 224.Hamilton E. Diurnal variation in proliferative compartments and their relation to cryptogenic cells in the mouse colon. Cell Prolif. 1979;12(1):91–100. doi: 10.1111/j.1365-2184.1979.tb00116.x. [DOI] [PubMed] [Google Scholar]
- 225.Al-Nafussi A, Wright N. Circadian rhythm in the rate of cellular proliferation and in the size of the functional compartment of mouse jejunal epithelium. Virchows Archiv B. 1982;40(1):71. doi: 10.1007/BF02932852. [DOI] [PubMed] [Google Scholar]
- 226.Panganiban G, Reuter R, Scott M, Hoffmann F. A Drosophila growth factor homolog, decapentaplegic, regulates homeotic gene expression within and across germ layers during midgut morphogenesis. Development. 1990;110(4):1041–1050. doi: 10.1242/dev.110.4.1041. [DOI] [PubMed] [Google Scholar]
- 227.Bitgood MJ, McMahon AP. HedgehogandBmpGenes are coexpressed at many diverse sites of cell–cell interaction in the mouse embryo. Dev Biol. 1995;172(1):126–138. doi: 10.1006/dbio.1995.0010. [DOI] [PubMed] [Google Scholar]
- 228.Batts LE, Polk DB, Dubois RN, Kulessa H. Bmp signaling is required for intestinal growth and morphogenesis. Dev Dyn Off Publ Am Assoc Anat. 2006;235(6):1563–1570. doi: 10.1002/dvdy.20741. [DOI] [PubMed] [Google Scholar]
- 229.Shyer AE, Huycke TR, Lee C, Mahadevan L, Tabin CJ. Bending gradients: how the intestinal stem cell gets its home. Cell. 2015;161(3):569–580. doi: 10.1016/j.cell.2015.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Shyer AE, Tallinen T, Nerurkar NL, Wei Z, Gil ES, Kaplan DL, Tabin CJ, Mahadevan L. Villification: how the gut gets its villi. Science. 2013;342(6155):212–218. doi: 10.1126/science.1238842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Howe JR, Bair JL, Sayed MG, Anderson ME, Mitros FA, Petersen GM, Velculescu VE, Traverso G, Vogelstein B. Germline mutations of the gene encoding bone morphogenetic protein receptor 1A in juvenile polyposis. Nat Genet. 2001;28(2):184. doi: 10.1038/88919. [DOI] [PubMed] [Google Scholar]
- 232.Haramis A-PG, Begthel H, Van Den Born M, Van Es J, Jonkheer S, Offerhaus GJA, Clevers H. De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science. 2004;303(5664):1684–1686. doi: 10.1126/science.1093587. [DOI] [PubMed] [Google Scholar]
- 233.He XC, Zhang J, Tong W-G, Tawfik O, Ross J, Scoville DH, Tian Q, Zeng X, He X, Wiedemann LM, Yuji M, Li L. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-β-catenin signaling. Nat Genet. 2004;36(10):1117. doi: 10.1038/ng1430. [DOI] [PubMed] [Google Scholar]
- 234.Kosinski C, Li VS, Chan AS, Zhang J, Ho C, Tsui WY, Chan TL, Mifflin RC, Powell DW, Yuen ST, Leung SY, Chen X. Gene expression patterns of human colon tops and basal crypts and BMP antagonists as intestinal stem cell niche factors. Proc Natl Acad Sci. 2007;104(39):15418–15423. doi: 10.1073/pnas.0707210104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Li H, Qi Y, Jasper H. Dpp signaling determines regional stem cell identity in the regenerating adult Drosophila gastrointestinal tract. Cell reports. 2013;4(1):10–18. doi: 10.1016/j.celrep.2013.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Yeung C-YC, Gossan N, Lu Y, Hughes A, Hensman JJ, Bayer ML, Kjær M, Kadler KE, Meng Q-J. Gremlin-2 is a BMP antagonist that is regulated by the circadian clock. Sci Rep. 2014;4:5183. doi: 10.1038/srep05183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Nam D, Guo B, Chatterjee S, Chen MH, Nelson D, Yechoor VK, Ma K. The adipocyte clock controls brown adipogenesis through the TGF-β and BMP signaling pathways. J Cell Sci. 2015;128(9):1835–1847. doi: 10.1242/jcs.167643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Buchon N, Broderick NA, Kuraishi T, Lemaitre B. Drosophila EGFR pathway coordinates stem cell proliferation and gut remodeling following infection. BMC Biol. 2010;8(1):152. doi: 10.1186/1741-7007-8-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Park J-S, Kim Y-S, Yoo M-A. The role of p38b MAPK in age-related modulation of intestinal stem cell proliferation and differentiation in Drosophila. Aging. 2009;1(7):637. doi: 10.18632/aging.100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Jiang H, Grenley MO, Bravo M-J, Blumhagen RZ, Edgar BA. EGFR/Ras/MAPK signaling mediates adult midgut epithelial homeostasis and regeneration in Drosophila. Cell Stem Cell. 2011;8(1):84–95. doi: 10.1016/j.stem.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Goldsmith CS, Bell-Pedersen D. Advances in genetics. Amsterdam: Elsevier; 2013. Diverse roles for MAPK signaling in circadian clocks; pp. 1–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Neilsen BK, Frodyma DE, McCall JL, Fisher KW, Lewis RE. ERK-mediated TIMELESS expression suppresses G2/M arrest in colon cancer cells. PLoS One. 2019;14(1):e0209224. doi: 10.1371/journal.pone.0209224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Zhang F, Sun H, Zhang S, Yang X, Zhang G, Su T. Overexpression of PER3 inhibits self-renewal capability and chemoresistance of colorectal cancer stem-like cells via inhibition of notch and β-catenin signaling. Oncol Res Featur Preclin Clin Cancer Ther. 2017;25(5):709–719. doi: 10.3727/096504016X14772331883976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Yoshida D, Aoki N, Tanaka M, Aoyama S, Shibata S. The circadian clock controls fluctuations of colonic cell proliferation during the light/dark cycle via feeding behavior in mice. Chronobiol Int. 2015;32(8):1145–1155. doi: 10.3109/07420528.2015.1065415. [DOI] [PubMed] [Google Scholar]
- 245.Amcheslavsky A, Jiang J, Ip YT. Tissue damage-induced intestinal stem cell division in Drosophila. Cell Stem Cell. 2009;4(1):49–61. doi: 10.1016/j.stem.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Choi NH, Lucchetta E, Ohlstein B. Nonautonomous regulation of Drosophila midgut stem cell proliferation by the insulin-signaling pathway. Proc Natl Acad Sci. 2011;108(46):18702–18707. doi: 10.1073/pnas.1109348108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.O’Brien LE, Soliman SS, Li X, Bilder D. Altered modes of stem cell division drive adaptive intestinal growth. Cell. 2011;147(3):603–614. doi: 10.1016/j.cell.2011.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Yilmaz ÖH, Katajisto P, Lamming DW, Gültekin Y, Bauer-Rowe KE, Sengupta S, Birsoy K, Dursun A, Yilmaz VO, Selig M, Neielsen GP, Mino-Kenudson M, Zukerberg LR, Bhan AK, Deshpande V, Sabatini DM. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature. 2012;486(7404):490. doi: 10.1038/nature11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Igarashi M, Guarente L. mTORC1 and SIRT1 cooperate to foster expansion of gut adult stem cells during calorie restriction. Cell. 2016;166(2):436–450. doi: 10.1016/j.cell.2016.05.044. [DOI] [PubMed] [Google Scholar]
- 250.Richmond CA, Shah MS, Deary LT, Trotier DC, Thomas H, Ambruzs DM, Jiang L, Whiles BB, Rickner HD, Montgomery RK, Tovaglieri A, Carlone DL, Breault DT. Dormant intestinal stem cells are regulated by PTEN and nutritional status. Cell Rep. 2015;13(11):2403–2411. doi: 10.1016/j.celrep.2015.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Lipton JO, Yuan ED, Boyle LM, Ebrahimi-Fakhari D, Kwiatkowski E, Nathan A, Güttler T, Davis F, Asara JM, Sahin M. The circadian protein BMAL1 regulates translation in response to S6K1-mediated phosphorylation. Cell. 2015;161(5):1138–1151. doi: 10.1016/j.cell.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Buchon N, Broderick NA, Chakrabarti S, Lemaitre B. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. 2009;23(19):2333–2344. doi: 10.1101/gad.1827009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Buchon N, Broderick NA, Poidevin M, Pradervand S, Lemaitre B. Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe. 2009;5(2):200–211. doi: 10.1016/j.chom.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 254.Beebe K, Lee W-C, Micchelli CA. JAK/STAT signaling coordinates stem cell proliferation and multilineage differentiation in the Drosophila intestinal stem cell lineage. Dev Biol. 2010;338(1):28–37. doi: 10.1016/j.ydbio.2009.10.045. [DOI] [PubMed] [Google Scholar]
- 255.Jeffery V, Goldson AJ, Dainty JR, Chieppa M, Sobolewski A. IL-6 signaling regulates small intestinal crypt homeostasis. J Immunol. 2017;199(1):304–311. doi: 10.4049/jimmunol.1600960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Lindemans CA, Calafiore M, Mertelsmann AM, O’Connor MH, Dudakov JA, Jenq RR, Velardi E, Young LF, Smith OM, Lawrence G, Ivanov JA, Fu Y-Y, Takashima S, Gua G, Martin ML, O’Rourke KP, Lo Y-H, Mokry M, Romera-Hernandez M, Cupedo T, Dow LE, Nieuwenhuis EE, Shroyer NF, Liu C, Kolesnick R, van den Brink MR, Hanash AM. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature. 2015;528(7583):560. doi: 10.1038/nature16460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Oshima H, Kok S-Y, Nakayama M, Murakami K, Voon DC-C, Kimura T, Oshima M. Stat3 is indispensable for damage-induced crypt regeneration but not for Wnt-driven intestinal tumorigenesis. FASEB J. 2018;33(2):1873–1886. doi: 10.1096/fj.201801176R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Arjona A, Sarkar DK. Circadian oscillations of clock genes, cytolytic factors, and cytokines in rat NK cells. J Immunol. 2005;174(12):7618–7624. doi: 10.4049/jimmunol.174.12.7618. [DOI] [PubMed] [Google Scholar]
- 259.Scheiermann C, Kunisaki Y, Lucas D, Chow A, Jang J-E, Zhang D, Hashimoto D, Merad M, Frenette PS. Adrenergic nerves govern circadian leukocyte recruitment to tissues. Immunity. 2012;37(2):290–301. doi: 10.1016/j.immuni.2012.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Halberg F (1960) Temporal coordination of physiologic function. In: Cold Spring Harbor symposia on quantitative biology,1960, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, pp 289-310 [DOI] [PubMed]
- 261.Shackelford PG, Feigin RD. Periodicity of susceptibility to pneumococcal infection: influence of light and adrenocortical secretions. Science. 1973;182(4109):285–287. doi: 10.1126/science.182.4109.285. [DOI] [PubMed] [Google Scholar]
- 262.Stone EF, Fulton BO, Ayres JS, Pham LN, Ziauddin J, Shirasu-Hiza MM. The circadian clock protein timeless regulates phagocytosis of bacteria in Drosophila. PLoS Pathog. 2012;8(1):e1002445. doi: 10.1371/journal.ppat.1002445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Allen VW, O’Connor RM, Ulgherait M, Zhou CG, Stone EF, Hill VM, Murphy KR, Canman JC, William WJ, Shirasu-Hiza MM. Period-regulated feeding behavior and TOR signaling modulate survival of infection. Curr Biol. 2016;26(2):184–194. doi: 10.1016/j.cub.2015.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Nguyen KD, Fentress SJ, Qiu Y, Yun K, Cox JS, Chawla A. Circadian gene Bmal1 regulates diurnal oscillations of Ly6Chi inflammatory monocytes. Science. 2013;341(6153):1483–1488. doi: 10.1126/science.1240636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Keller M, Mazuch J, Abraham U, Eom GD, Herzog ED, Volk H-D, Kramer A, Maier B. A circadian clock in macrophages controls inflammatory immune responses. Proc Natl Acad Sci. 2009;106(50):21407–21412. doi: 10.1073/pnas.0906361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Hayashi M, Shimba S, Tezuka M. Characterization of the molecular clock in mouse peritoneal macrophages. Biol Pharm Bull. 2007;30(4):621–626. doi: 10.1248/bpb.30.621. [DOI] [PubMed] [Google Scholar]
- 267.Arjona A, Sarkar DK. Evidence supporting a circadian control of natural killer cell function. Brain Behav Immun. 2006;20(5):469–476. doi: 10.1016/j.bbi.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 268.Fortier EE, Rooney J, Dardente H, Hardy M-P, Labrecque N, Cermakian N. Circadian variation of the response of T cells to antigen. J Immunol. 2011;187(12):6291–6300. doi: 10.4049/jimmunol.1004030. [DOI] [PubMed] [Google Scholar]
- 269.Bollinger T, Leutz A, Leliavski A, Skrum L, Kovac J, Bonacina L, Benedict C, Lange T, Westermann J, Oster H, Solbach W. Circadian clocks in mouse and human CD4+ T cells. PLoS One. 2011;6(12):e29801. doi: 10.1371/journal.pone.0029801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Gibbs JE, Blaikley J, Beesley S, Matthews L, Simpson KD, Boyce SH, Farrow SN, Else KJ, Singh D, Ray DW, Loudon AS. The nuclear receptor REV-ERBα mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc Natl Acad Sci. 2012;109(2):582–587. doi: 10.1073/pnas.1106750109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Curtis AM, Bellet MM, Sassone-Corsi P, O’Neill LA. Circadian clock proteins and immunity. Immunity. 2014;40(2):178–186. doi: 10.1016/j.immuni.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 272.Scheiermann C, Gibbs J, Ince L, Loudon A. Clocking into immunity. Nat Rev Immunol. 2018;18(7):423. doi: 10.1038/s41577-018-0008-4. [DOI] [PubMed] [Google Scholar]
- 273.Man K, Loudon A, Chawla A. Immunity around the clock. Science. 2016;354(6315):999–1003. doi: 10.1126/science.aah4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Froy O, Chapnik N. Circadian oscillation of innate immunity components in mouse small intestine. Mol Immunol. 2007;44(8):1954–1960. doi: 10.1016/j.molimm.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 275.Froy O, Chapnik N, Miskin R. Mouse intestinal cryptdins exhibit circadian oscillation. FASEB J. 2005;19(13):1920–1922. doi: 10.1096/fj.05-4216fje. [DOI] [PubMed] [Google Scholar]
- 276.Bellet MM, Deriu E, Liu JZ, Grimaldi B, Blaschitz C, Zeller M, Edwards RA, Sahar S, Dandekar S, Baldi P, George MD, Raffatellu M, Sassone-Corsi P. Circadian clock regulates the host response to Salmonella. Proc Natl Acad Sci. 2013;110(24):9897–9902. doi: 10.1073/pnas.1120636110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Pourcet B, Zecchin M, Ferri L, Beauchamp J, Sitaula S, Billon C, Delhaye S, Vanhoutte J, Mayeuf-Louchart A, Thorel Q, Hass J, Eeckhoute J, Dombrowicz D, Duhem C, Boulinguiez A, Lancel S, Sebti Y, Burris T, Staels B, Duez H. Nuclear receptor subfamily 1 group D member 1 regulates circadian activity of NLRP3 inflammasome to reduce the severity of fulminant hepatitis in mice. Gastroenterology. 2018;154(5):1449.e1420–1464.e1420. doi: 10.1053/j.gastro.2017.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Wang S, Lin Y, Yuan X, Li F, Guo L, Wu B. REV-ERBα integrates colon clock with experimental colitis through regulation of NF-κB/NLRP3 axis. Nat Commun. 2018;9(1):4246. doi: 10.1038/s41467-018-06568-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Mukherji A, Kobiita A, Ye T, Chambon P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell. 2013;153(4):812–827. doi: 10.1016/j.cell.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 280.Preuss F, Tang Y, Laposky AD, Arble D, Keshavarzian A, Turek FW. Adverse effects of chronic circadian desynchronization in animals in a “challenging” environment. Am J Physiol Regul Integr Comp Physiol. 2008;295(6):R2034–R2040. doi: 10.1152/ajpregu.00118.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Thaiss CA, Levy M, Korem T, Dohnalová L, Shapiro H, Jaitin DA, David E, Winter DR, Gury-BenAri M, Tatirovsky E, Tuganbaev T, Federici S, Zmora N, Zeevi D, Dori-Bachash M, Pevsner-Rischer M, Kartvelishvily E, Brandis A, Harmelin A, Shibolet O, Halpern Z, Honda K, Amit I, Segal E, Elinav E. Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell. 2016;167(6):1495.e1412–1510.e1412. doi: 10.1016/j.cell.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 282.Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, Abramson L, Katz MN, Korem T, Zmora N, Kuperman Y, Biton I, Gilad S, Harmelin A, Sharpiro H, Halpern Z, Segal E, Elinav E. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 2014;159(3):514–529. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 283.Wang Y, Kuang Z, Yu X, Ruhn KA, Kubo M, Hooper LV. The intestinal microbiota regulates body composition through NFIL3 and the circadian clock. Science. 2017;357(6354):912–916. doi: 10.1126/science.aan0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Leone V, Gibbons SM, Martinez K, Hutchison AL, Huang EY, Cham CM, Pierre JF, Heneghan AF, Nadimpalli A, Hubert N, Zale E, Wang Y, Huang Y, Theriault B, Dinner AR, Musch MW, Kudsk KA, Prendergast BJ, Gilbert JA, Chang EB. Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe. 2015;17(5):681–689. doi: 10.1016/j.chom.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Caruso CC, Lusk SL, Gillespie BW. Relationship of work schedules to gastrointestinal diagnoses, symptoms, and medication use in auto factory workers. Am J Ind Med. 2004;46(6):586–598. doi: 10.1002/ajim.20099. [DOI] [PubMed] [Google Scholar]
- 286.Schernhammer ES, Laden F, Speizer FE, Willett WC, Hunter DJ, Kawachi I, Fuchs CS, Colditz GA. Night-shift work and risk of colorectal cancer in the nurses’ health study. J Natl Cancer Inst. 2003;95(11):825–828. doi: 10.1093/jnci/95.11.825. [DOI] [PubMed] [Google Scholar]
- 287.Knutsson A, Bøggild H. Gastrointestinal disorders among shift workers. Scand J Work Environ Health. 2010;36(2):85–95. doi: 10.5271/sjweh.2897. [DOI] [PubMed] [Google Scholar]
- 288.Sonnenberg A. Occupational distribution of inflammatory bowel disease among German employees. Gut. 1990;31(9):1037–1040. doi: 10.1136/gut.31.9.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Papantoniou K, Devore EE, Massa J, Strohmaier S, Vetter C, Yang L, Shi Y, Giovannucci E, Speizer F, Schernhammer ES. Rotating night shift work and colorectal cancer risk in the nurses’ health studies. Int J Cancer. 2018;143(11):2709–2717. doi: 10.1002/ijc.31655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Mostafaie N, Kállay E, Sauerzapf E, Bonner E, Kriwanek S, Cross HS, Huber KR, Krugluger W. Correlated downregulation of estrogen receptor beta and the circadian clock gene Per1 in human colorectal cancer. Mol Carcinog. 2009;48(7):642–647. doi: 10.1002/mc.20510. [DOI] [PubMed] [Google Scholar]
- 291.Mazzoccoli G, Panza A, Valvano M, Palumbo O, Carella M, Pazienza V, Biscaglia G, Tavano F, Di Sebastiano P, Andriulli A, Piepoli A. Clock gene expression levels and relationship with clinical and pathological features in colorectal cancer patients. Chronobiol Int. 2011;28(10):841–851. doi: 10.3109/07420528.2011.615182. [DOI] [PubMed] [Google Scholar]
- 292.Oshima T, Takenoshita S, Akaike M, Kunisaki C, Fujii S, Nozaki A, Numata K, Shiozawa M, Rino Y, Tanaka K, Masuda M, Imada T. Expression of circadian genes correlates with liver metastasis and outcomes in colorectal cancer. Oncol Rep. 2011;25(5):1439–1446. doi: 10.3892/or.2011.1207. [DOI] [PubMed] [Google Scholar]
- 293.Battaglin F, Xiu J, Winerip M, Goldberg RM, Philip PA, Seeber A, Puccini A, Tokunaga R, Naseem M, Soni S, McSkane M, Berger MD, Barzi A, Zhang W, Hwang JJ, Shields AF, Marshall J, Korn WM, Lenz H-J. Circadian clock gene PER1 mutations in colorectal cancer (CRC) J Clin Oncol. 2018;15:12106. doi: 10.1200/JCO.2018.36.15_suppl.12106. [DOI] [Google Scholar]
- 294.Gu D, Li S, Ben S, Du M, Chu H, Zhang Z, Wang M, Zhang Z-F, Chen J. Circadian clock pathway genes associated with colorectal cancer risk and prognosis. Arch Toxicol. 2018;92(8):2681–2689. doi: 10.1007/s00204-018-2251-7. [DOI] [PubMed] [Google Scholar]
- 295.Yu H, Meng X, Wu J, Pan C, Ying X, Zhou Y, Liu R, Huang W. Cryptochrome 1 overexpression correlates with tumor progression and poor prognosis in patients with colorectal cancer. PLoS One. 2013;8(4):e61679. doi: 10.1371/journal.pone.0061679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Masri S, Sassone-Corsi P. The emerging link between cancer, metabolism, and circadian rhythms. Nat Med. 2018;24(12):1795–1803. doi: 10.1038/s41591-018-0271-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Sulli G, Lam MTY, Panda S. Interplay between circadian clock and cancer: new frontiers for cancer treatment. Trends cancer. 2019;5(8):475–494. doi: 10.1016/j.trecan.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Chakradeo PS, Keshavarzian A, Singh S, Dera AE, Esteban JPG, Lee AA, Burgess HJ, Fogg L, Swanson GR. Chronotype, social jet lag, sleep debt and food timing in inflammatory bowel disease. Sleep Med. 2018;52:188–195. doi: 10.1016/j.sleep.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Palmieri O, Mazzoccoli G, Bossa F, Maglietta R, Palumbo O, Ancona N, Corritore G, Latiano T, Martino G, Rubino R, Biscaglia G, Scimeca D, Carella M, Annese V, Andriulli A, Latiano A. Systematic analysis of circadian genes using genome-wide cDNA microarrays in the inflammatory bowel disease transcriptome. Chronobiol Int. 2015;32(7):903–916. doi: 10.3109/07420528.2015.1050726. [DOI] [PubMed] [Google Scholar]
- 300.Liu X, Yu R, Zhu L, Hou X, Zou K. Bidirectional regulation of circadian disturbance and inflammation in inflammatory bowel disease. Inflamm Bowel Dis. 2017;23(10):1741–1751. doi: 10.1097/MIB.0000000000001265. [DOI] [PubMed] [Google Scholar]
- 301.Weintraub Y, Cohen S, Chapnik N, Ben-Tov A, Yerushalmy-Feler A, Dotan I, Tauman R, Froy O. Clock gene disruption is an initial manifestation of inflammatory bowel disease. Clin Gastroenterol Hepatol. 2019 doi: 10.1016/j.cgh.2019.04.013. [DOI] [PubMed] [Google Scholar]
- 302.Ali T, Choe J, Awab A, Wagener TL, Orr WC. Sleep, immunity and inflammation in gastrointestinal disorders. World J Gastroenterol. 2013;19(48):9231–9239. doi: 10.3748/wjg.v19.i48.9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Sobolewska-Włodarczyk A, Włodarczyk M, Szemraj J, Stec-Michalska K, Fichna J, Wiśniewska-Jarosińska M. Circadian rhythm abnormalities—association with the course of inflammatory bowel disease. Pharmacol Rep. 2016;68(4):847–851. doi: 10.1016/j.pharep.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 304.Swanson GR, Burgess HJ. Sleep and circadian hygiene and inflammatory bowel disease. Gastroenterol Clin. 2017;46(4):881–893. doi: 10.1016/j.gtc.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 305.Gombert M, Carrasco-Luna J, Pin-Arboledas G, Codoñer-Franch P. The connection of circadian rhythm to inflammatory bowel disease. Trans Res. 2018;206:107–118. doi: 10.1016/j.trsl.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 306.Antoch MP, Kondratov RV, Takahashi JS. Circadian clock genes as modulators of sensitivity to genotoxic stress. Cell Cycle. 2005;4(7):901–907. doi: 10.4161/cc.4.7.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]