Abstract
The transcription factor Ptf1a is a crucial helix–loop–helix (bHLH) protein selectively expressed in the pancreas, retina, spinal cord, brain, and enteric nervous system. Ptf1a is preferably assembled into a transcription trimeric complex PTF1 with an E protein and Rbpj (or Rbpjl). In pancreatic development, Ptf1a is indispensable in controlling the expansion of multipotent progenitor cells as well as the specification and maintenance of the acinar cells. In neural tissues, Ptf1a is transiently expressed in the post-mitotic cells and specifies the inhibitory neuronal cell fates, mostly mediated by downstream genes such as Tfap2a/b and Prdm13. Mutations in the coding and non-coding regulatory sequences resulting in Ptf1a gain- or loss-of-function are associated with genetic diseases such as pancreatic and cerebellar agenesis in the rodent and human. Surprisingly, Ptf1a alone is sufficient to reprogram mouse or human fibroblasts into tripotential neural stem cells. Its pleiotropic functions in many biological processes remain to be deciphered in the future.
Keywords: Pancreatic development, Retinal development, Transcriptional regulation, Cell fate specification, Acinar cells, Inhibitory neurotransmitter, GABAergic, Glycinergic, Glutamatergic, Diabetes, Somatic cell reprogramming, Inheritable
Introduction
A helix–loop–helix (HLH) protein structure motif was first described by Murre and colleagues in 1989 [1]. The typical HLH motif is characterized by a loop connecting two α-helices, among which the larger helix contains the basic amino acid residues that can bind DNA in the major groove, and the smaller helix allows dimerization by folding and packing against another helix. The HLH transcription factors can form homodimers or heterodimers, and bind to a consensus DNA sequence motif called E-box, CANNTG (N: A/C/T/G) [1, 2], or an E-box-like sequence, CAYRMK (Y: C/T; R: A/G; M: A/C; K: G/T) [3].
More than 240 HLH proteins have been identified and can be categorized into seven groups based on their tissue distributions, DNA-binding specificities, and dimerization capacities [4, 5]. Notably, the class I HLH members (E proteins) are ubiquitously expressed in almost all tissues. They assemble into heterodimers or homodimers and bind to the canonical E-box only. The class I members include E12 and E47 (alternatively spliced E2A, or TCF3), E2-2 (TCF4) and HEB (TCF12) in the vertebrates. The class II members are distributed in certain tissues specifically. Most of them cannot form homodimers, but preferably form heterodimers with class I members and bind both E-box and E-box-like sequences. The majority of HLH proteins belong to class II. The class V members, including inhibitors of DNA-binding (ID), lack the basic DNA-binding region. The ID proteins are usually low in mature cells but abundant in proliferating or differentiating cells. They are capable of forming heterodimers with E proteins or class II proteins, but unable to bind DNA and hence act as negative regulators of transcription [6, 7]. Other HLH classes will not be discussed here. The variations in E-box and E-box-like sequences and diversities in homodimers and heterodimers provide many possibilities in transcriptional control. The HLH proteins represent one of the most important transcription factor families regulating cell proliferation, specification and fate determination in development.
The pancreas-specific transcription factor 1a, Ptf1a (alias: bHLHa29, PTF1-p48, Ptf1p48, p48 DNA-binding subunit of transcription factor PTF1), is a class II HLH member. Ptf1a was named after the transcription factor PTF1 [8], which is a transcription complex composed of three distinct subunits, p75, p64 and Ptf1-p48. The p75 subunit was later identified as the HEB protein, and could be replaced by E47 in a small fraction of the PTF1 complexes [9]. HEB or E47 forms a HLH heterodimer with Ptf1a and together bind to the E-box [9]. The p64 subunit turns out to be Rbpj (CBF1/Rbpjκ/Su(H)/Lag-1, CSL) [10] that binds the TC-box (TTTCCC) consensus sequences [11]. In mature acinar cells, Rbpj is replaced by Rbpjl in the complex [12, 13]. Simultaneous binding to the bipartite cognate sites (E-box and TC-box) is required for PTF1 transcriptional activity.
Accumulating data indicate that Ptf1a plays a critical role in controlling development and physiological function of many organs, including pancreas, brain, spinal cord, retina and others. We will give a deliberate review about the past findings and the important advances.
Genomic, RNA and protein structures of Ptf1a
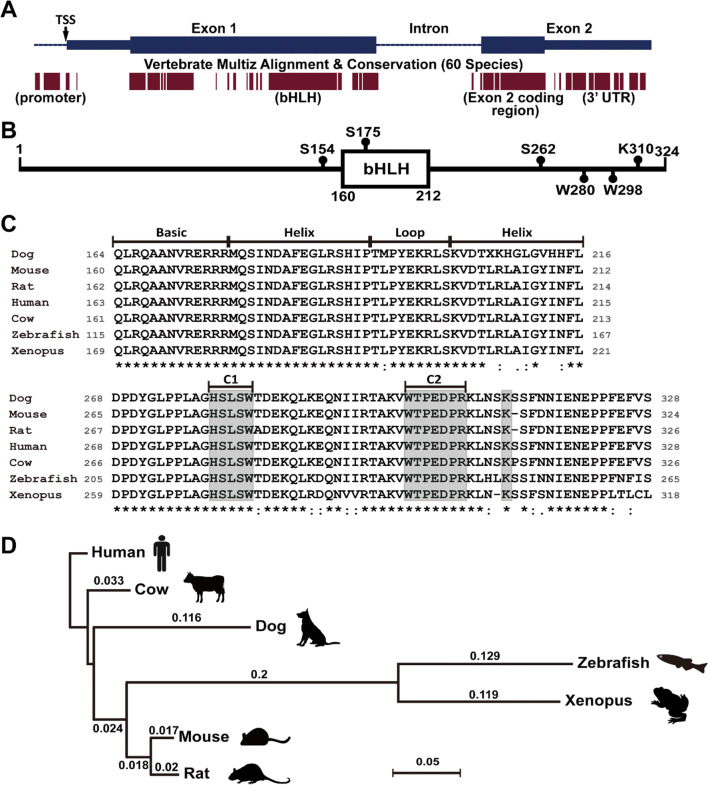
The evolution history of Ptf1a gene can at least be traced back to as early as sea urchin [14]. The sea urchin orthologue of Ptf1a is expressed in a unique population of cells in the embryonic stomach. These cells co-express exocrine cell markers and resemble vertebrate pancreatic exocrine cells in genetic regulatory network [14]. Though the function and role of Ptf1a seem to be evolutionarily preserved, its genomic sequences vary highly in many species and are not much conserved when compared with the extent of conservation of ancient transcription factor genes such as the Hox family. Typical Ptf1a genomic structure is composed of two exons and one intron as shown in the mouse genome (Fig. 1a). Alignment of 60 vertebrate Ptf1a genome sequences revealed several highly conserved regions, including the 5′ promoter region, the bHLH domain region in exon 1, the exon 2 coding region, and the 3′ untranslated region (UTR) (Fig. 1a). The conserved 5′ promoter region is very close to the transcription start site (TSS) and contains transcription factor binding sites that are critical for the regulation of transcription initiation. Hundreds of potential transcription factor binding sites within 500 bp upstream of the mouse Ptf1a TSS site are predicted by PROMO (v3) with a dissimilarity margin less or equal to 15%. It must be emphasized here that the 3′ regulatory regions are extremely important for Ptf1a transcription. One 3′ conserved enhancer region, about 400 bp located at 25 kb downstream of the coding sequence, is critical for the transcription of Ptf1a in pancreatic development. Mutations in this region are associated with pancreas agenesis and diabetes [15, 16]. Another 3′ conserved enhancer region, located at 10.8 kb downstream of the coding sequence, is necessary for tissue-specific expression of Ptf1a in the dorsal neural tube [17].
Fig. 1.
Genomic structure and protein sequences of Ptf1a. a The upper portion of the image shows the genomic structure of the mouse Ptf1a gene. The blue bars represent two exons; the heightened parts are coding regions for Ptf1a. TSS, transcription start site. The lower portion of the image shows the corresponding multiz alignment and conservation of Ptf1a DNA sequences in 60 vertebrate species by UCSC genome browser. b The mouse Ptf1a protein is composed of 324 amino acid residues. The bHLH domain and several important residues are shown. c Multiple alignment of amino acid residues of the bHLH domains (upper image) and C termini (lower image) of Ptf1a proteins from seven vertebrate species. All sequences were downloaded from NCBI. The gray boxes in the C-terminal region (C1 and C2) outline two conserved regions important for interaction with Rbpj or Rbpjl protein, and one conserved lysine residue required for ubiquitination and proteasome degradation. d The phylogenetic tree for seven vertebrate Ptf1a proteins was constructed by the neighbor-joining method. The distance was calculated with Poisson correction. Gaps were distributed proportionally. The scale bar represents a distance of 0.05
The 3′ UTR region of many genes contains various sequences including microRNA response elements (MREs), AU-rich elements (AREs), and the poly(A) tail that are crucial in gene expression regulation by influencing the localization, stability, and translation efficiency of an mRNA. The 3′ UTR in the mouse Ptf1a mRNA is 335-base long and contains a binding site for RNA-binding protein Rbms3, which indirectly affects pancreas development by regulating the translation efficiency of Ptf1a protein [18]. Scanning of Ptf1a 3′ UTR sequence with TargetScan identifies many conserved miRNA targeting sites, whose biological significance is yet to be revealed.
A typical Ptf1a protein has a bHLH domain in the middle as shown in the mouse Ptf1a which is composed of 324 amino acid residues (Fig. 1b). In the mouse Ptf1a (Fig. 1b, c), there are two conserved tryptophan residues (W280 and W298 in the C1 and C2 regions, respectively) important for interaction with Rbpj/Rbpjl [10], and three serine residues (S154, S175, S262) for possible phosphorylation (predicted by GPS3.0, http://gps.biocuckoo.org) and regulation of Ptf1a activity. The lysine residue K310 (K312 in human) has been shown to be required for E3 ubiquitin ligase TRIP12 (thyroid hormone receptor-interacting protein 12) that targets Ptf1a for ubiquitination and proteasomal degradation [19]. From zebrafish, Xenopus, mouse, rat, dog, and cow to human, alignment of the seven vertebrate Ptf1a amino acid sequences indicates that the phylogenetic tree of Ptf1a is in accordance with the evolutionary history (Fig. 1d). The alignment also reveals a couple of conserved regions, including the bHLH domain and a region in the C terminus (Fig. 1c), implying that these regions are important for Ptf1a functions. Notably, the amino acid sequences of the DNA-binding domain (the basic region plus the first helix, or the larger helix) remain unchanged in the seven species. Except for the dog, all species share exactly the same sequences in the bHLH domain. Interestingly, the dog has more evolutionary changes in Ptf1a conserved domains than any other listed mammals (Fig. 1c), mostly attributing to the accelerated process with artificial selections [20]. This raises a plausible question whether the dog Ptf1a is more adaptive than other species’ and hints at the future direction of Ptf1a evolution.
Rbpj-dependent and -independent activity of Ptf1a
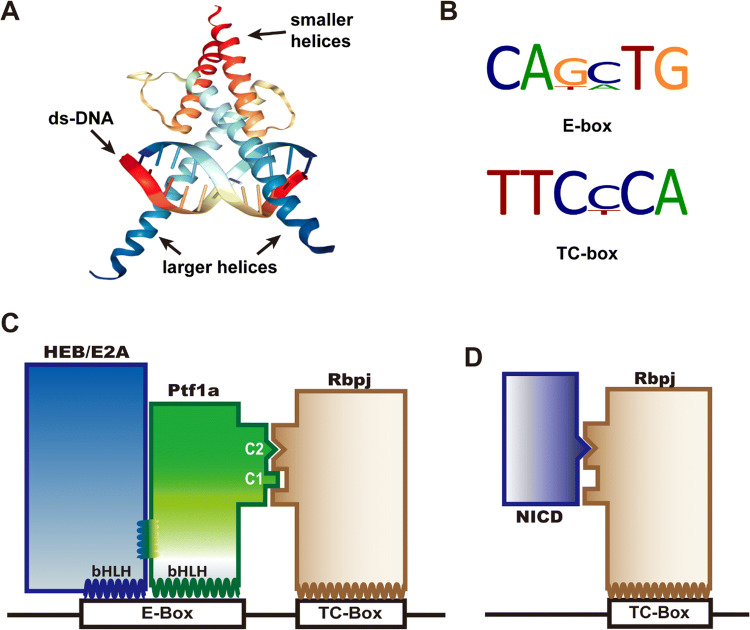
Literally, the major activity of a transcription factor is transcription regulation, which usually requires it to bind to a consensus site in the chromosome. As a bHLH domain member, Ptf1a assembles into a heterodimer with another HLH factor (preferably with an E protein) and binds to the E-box or E-box-like consensus through its larger helix (Fig. 2a, b). Depending on whether Rbpj is present in the complex, the transcription activity of Ptf1a is categorized to be Rbpj-dependent or -independent.
Fig. 2.
DNA binding with Ptf1a and PTF1 complex. a The cartoon illustrates that two bHLH domains (from SCL-E47, PDB bank) bind to the major groove of DNA with their larger helices. The smaller helices fold and allow dimerization of the domains. b The common DNA binding consensus E-box and TC-box for the PTF1 complex. c Ptf1a and HEB (or E47, E12, isoforms of E2A) bind to the E-box, and Rbpj binds to the TC-box. Transcription activation requires that the PTF1 complex binds to E-box and TC-box simultaneously, which depends on Ptf1a interfacing with Rbpj via the conserved C1 and C2 regions at the C terminus. d The Notch intracellular domain (NICD) can compete with Ptf1a and E protein heterodimers for the binding sites on Rbpj protein
In most occasions, Ptf1a is associated with Rbpj (or Rbpjl) in the PTF1 complex, namely in a Rbpj-dependent manner (Fig. 2c). Fox example, in acinar development, the PTF1 complex binds a bipartite motif containing an E box and a TC box separated by preferential spacing of one, two or three helical turns of DNA [10, 21–24]. During acinar maturation and adulthood, Rbpj is replaced by Rbpjl in the PTF1 complex to maintain exocrine expression [12]. Motif analysis of the data from ChIP-seq experiments with Ptf1a and Rbpj antibodies indicates that more than 70% Ptf1a is colocalized with Rbpj/Rbpjl in the pancreas [23]. If peak calling is less stringent and the low-affinity binding events are considered, the overlap of Ptf1a and Rbpj/Rbpjl may approach 100% in the pancreas. In neural tube development, however, ChIP-seq and motif analyses reveal that the PTF1 trimer occupies barely more than 25% of peak calling events, indicating that the majority of Ptf1a protein functions independently of PTF1 complex. Further analysis shows that Rbpj-independent Ptf1a likely activates transcription in combination with Sox, Hox, Forkhead, GATA, and homeodomain family members [23], since the Ptf1a and E-protein heterodimer alone is relatively weak in activating transcription in vitro [9, 10, 25]. There are still many unanswered questions concerning Rbpj-dependent and -independent mechanisms. For example, can both mechanisms exist in the same cell? How do they coordinate with each other in cells and tissues?
Furthermore, Ptf1a represents an important component antagonizing Notch signaling [26–28]. Upon binding with its ligands (Serrate, Delta, Jagged, etc.), Notch releases its intracellular domain (NICD) through sequential proteolysis by ADAM and γ-secretase, which then translocates into the nucleus, interacts with transcription factor Rbpj (Fig. 2d), and activates the transcription of downstream genes such as Hes1 and Hes5. Beres et al. found that the C1 and C2 regions of Ptf1a, especially the tryptophan residues in the two regions, are absolutely required for cross-interaction with Rbpj [10]. NICD interacts with Rbpj through a tryptophan-containing hydrophobic motif φWφP (φ represents a hydrophobic residue) [29]. The Ptf1a C2 motif and NICD φWφP motif are structurally similar and compete to bind exclusively to the same site on Rbpj, which means that Ptf1a and NICD antagonize each other cell autonomously in a dose-dependent manner [10]. This has been demonstrated to be a very important mechanism for regulating cell fate specification during development of many tissues.
Ptf1a expression profiles
During vertebrate embryonic development, Ptf1a expression is dynamic and transient in most tissues. SAGE (serial analysis of gene expression) analysis of Ptf1a indicates that Ptf1a is expressed in the brain, retina, spinal cord, pancreas, heart, lung, liver, colon, thyroid, breast, lymph node, skin, placenta, and other major organs/tissues during development (https://cgap.nci.nih.gov/SAGE). RNA-seq data from GTEx show that Ptf1a is also expressed in the testis, ovary, stomach, small intestine, skeletal muscle, adipocyte, and other tissues (https://gtexportal.org).
In adult human tissues (data from ProteomicsDB), Ptf1a mRNA expression is much higher in pancreas, median FPKM (Fragments Per Kilobase Of Exon Per Million Fragments Mapped) value reaching 14, followed by stomach (FPKM 0.9), testis (FPKM 0.6), and cerebral cortex and prostate gland (FPKM 0.1 each). At the tissue level, Ptf1a protein abundance is highest in the ovary, median value reaching 4.52 (log10 normalized iBAQ intensity, the same below), followed by rectum (4.27), colon (4.13), and pancreas (3.35). It seems that transcription and translation of Ptf1a are not tightly coupled since the ratio of its mRNA and protein levels is not proportional among tissues. Based on the common features shared by the tissues expressing Ptf1a, an interesting question can be raised regarding whether there is a correlation between Ptf1a and cell exocrine. Notably, highest Ptf1a level is found in synovial fluid (7.18), reinforcing the idea that Ptf1a might participate in controlling synthesis and/or exocytosis of exosomes/microvesicles (including synaptic vesicles in neurons).
The expression pattern of Ptf1a, its role and associated mutant and overexpression phenotypes in mouse is listed (Table 1). Next, we will discuss these findings in detail, including the spatiotemporal expression profile of Ptf1a, its function, and its upstream and downstream signaling pathways during development of several tissues.
Table 1.
Expression profile of Ptf1a in mouse
| Organ/tissue | Developmental stages | Role of Ptf1a | Phenotypes/defects | References |
|---|---|---|---|---|
| Pancreas | E9–E13, in MPCs |
Required for acquisition of pancreatic identity from foregut endoderm cells Required for proliferation of pancreatic MPCs |
Ptf1a misexpression transforms endodermal organs into pancreas Inactivation of Ptf1a converts pancreatic MPCs into non-pancreatic cell fates. Hypomorphic Ptf1a leads to pancreatic aplasia or agenesis |
[32, 33, 35, 38] |
| E14-adult, in exocrine pancreas and acinar cells |
Promotes acinar cell fates and regulates acinar cell-specific genes Maintains acinar cell identity and its physiological functions |
High levels of Ptf1a promotes exocrine cell fates and represses endocrine cell fates Ptf1a inactivation may cause acinar cells to transform into tumor cells |
[36, 37, 119] | |
| Retina | E12–P3, in retinal precursors | Specifies horizontal and amacrine cell fates and promotes their differentiation |
Knockout of Ptf1a results in losses of horizontal and amacrine cells Ptf1a misexpression promotes horizontal and amacrine cell fates |
[59, 60, 62] |
| Spinal cord | E9.5–E13.5(?), in neuronal precursors | Specifies GABAergic and glycinergic inhibitory neuronal cell fates while repressing glutamatergic excitatory cell fates | In the absence of Ptf1a, Ptf1a-lineage cells switch fates to acquire an excitatory neuronal identity | [25, 83, 124] |
| Cerebellum | E10.5–E18.5 in neural progenitors | Specifies GABAergic cerebellar neurons including Purkinje cells, GABAergic neurons of DCN, and molecular layer interneurons while repressing glutamatergic cell fates |
Misexpression of Ptf1a converts non-Ptf1a-lineage progenitors into GABAergic instead of glutamatergic neurons Ptf1a inactivation leads to mis-specification of Ptf1a-lineage progenitors into glutamatergic neurons |
[93–96, 125] |
| Brainstem | E10.5–E13.5 in neuronal progenitors of the caudal hindbrain |
Specifies GABAergic neuronal fates in development of the nucleus of the solitary tract (nTs), the spinal trigeminal nuclei (SpV) and principal trigeminal nuclei (PrV) Specifies GABAergic and glycinergic inhibitory neurons in the cochlear nucleus. Controls the cell fate specification, development and survival of glutamatergic climbing fiber (CF) neurons and their migration to the inferior olivary nucleus (ION) |
Ptf1a inactivation results in generation of more Lmx1b-lineage viscerosensory and somatosensory neurons at the expense of Pax2-lineage GABAergic viscerosensory and somatosensory neurons, and inferior olivary neurons Development of inhibitory neurons in the cochlear nucleus is severely undermined in the absence of Ptf1a Loss of Ptf1a compromises the development and migration of CF neurons and causes some of the precursors to switch cell fates to mossy fiber neurons |
[98–100] |
| Forebrain | E10.5–E16.5, In neuronal progenitors | Ptf1a does not involve cell fate specification but confers the competence to acquire sex differentiation in the developing brain | Ptf1a-deficient male and female mice display abnormalities in reproductive organs and sexually dimorphic behaviors | [101] |
Ptf1a in pancreatic development and function
The pancreas has endocrine function to secrete insulin and glucagon which are antagonists and together regulate blood glucose homeostasis. The organ also serves as an exocrine unit to secrete amylase, elastase-1 and other important digestive enzymes. In origin, the pancreas develops from the endoderm at the foregut and midgut junction. Two separate pancreatic primordia, the dorsal and the ventral pancreas buds, arise from the junction and join together to form the pancreas.
Ptf1a RNA is detected at as early as 9 dpc (days post coitum) in the pancreas buds [25, 30, 31]. From 14 dpc and on, it seems that Ptf1a is expressed only in the exocrine part of pancreas [31]. In homozygous Ptf1a null mutant mice, acinar development was abolished, and some endocrine pancreatic cells were misallocated in the spleen [30]. Evidence from these early studies suggests that Ptf1a is a specific determinant for exocrine pancreatic cells only. However, lineage tracing studies with Ptf1aCre or Ptf1aCreERTM and R26R mice revealed that most pancreatic multipotent progenitor cells (MPCs) express Ptf1a, and are capable of differentiating into acinar, ductal or endocrine cells [32, 33]. This result is confirmed in the zebrafish using a similar approach [34]. When Ptf1a activity is totally inactivated, most of the MPCs are converted into non-pancreatic cell fates, such as gut and gall bladder [32, 34]. Conversely, Ptf1a misexpression in embryonic mouse endoderm converted the entire glandular stomach, rostral duodenum and extrahepatic biliary system into pancreas [35]. These experiments demonstrate that Ptf1a is both necessary and sufficient for acquisition of pancreatic fate from foregut endoderm. Interestingly but not surprisingly, the exocrine versus endocrine cell fate depends on the dose effect of Ptf1a. Haploinsufficiency or low levels of Ptf1a will promote endocrine cell fates while repressing exocrine cell fates, and vice versa [34, 36, 37]. These findings underscore the distinct roles of Ptf1a in the specification and expansion of pancreatic progenitor cells.
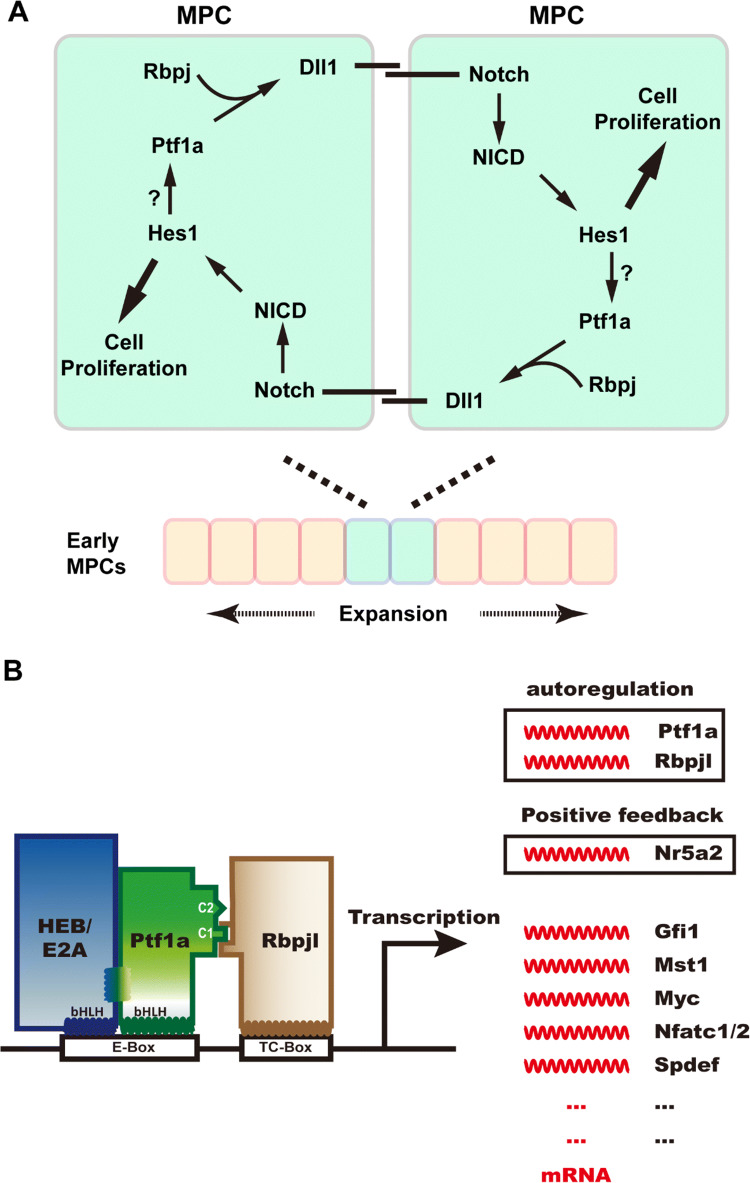
How does Ptf1a participate in the expansion of early pancreatic progenitor cells? Ptf1a is not only involved in establishing the pancreatic identity in foregut cells, Ahnfelt-Rønne et al. found that it is also indispensable in the expansion of early pancreatic MPCs by controlling Dll1 expression [38]. Using LacZ as a reporter gene in Dll1lacZ/+; Ptf1aCre/cre mice, β-gal expression (viz. Dll1 expression) was not detected in early pancreatic MPCs, indicating the requirement of Ptf1a in Dll1 expression. ChIP analysis shows that Ptf1a and/or Rbpj can bind directly to the proximal promoter region of Dll1. Dll1 binds to the Notch receptor on the cell membrane of neighbor MPCs (Fig. 3a), leading to release of NICD, which activates Notch downstream target genes such as Hes1 that mediates MPC cell proliferation. Unexpectedly, Hes1 is also required for retaining Ptf1a protein level in the MPCs [38], owing to an unknown mechanism. It is speculated that the Hes1–Ptf1a protein–protein interaction helps to stabilize the Ptf1a protein [39]. In summary, the Ptf1a and Notch pathway constitutes a positive feed-forward loop to promote early MPC proliferation and expansion in a non-cell autonomous manner (Fig. 3a). Meanwhile, another mechanism may also participate in MPC expansion. Two groups found that Ptf1a could bind to the conserved promoter of Pdx1 and activate its expression in early MPCs [40, 41]. This effect could be synergistically enhanced by Foxa1/2 [23, 42]. Ptf1a and Pdx1 together maintain the proliferation of early MPCs before cell differentiation becomes widespread.
Fig. 3.
Ptf1a is required for early pancreatic genesis and adult acinar cell identity. a Ptf1a regulates early pancreatic MPC expansion in a positive feed-forward loop (adapted from Ahnfelt-Ronne et al. [38]). Ptf1a and Rbpj activate the expression of Notch ligand Dll1, which in turn binds to Notch receptors on the neighbor MPC, and triggers the downstream pathway of Notch. The activated Notch intracellular domain (NICD) enters the nucleus and activates transcription of target genes such as Hes1, which promotes cell proliferation. Hes1 may also maintain the Ptf1a level by stabilizing the Ptf1a structure. This positive feed-forward mechanism guarantees the proliferation and expansion of early MPCs in pancreatic primordia. b Ptf1a participates not only in the fate specification of acinar cells during development, but also in the maintenance of their physiological function and identity in the adult pancreas. The PTF1 triplex is capable of autoactivation by transactivating Ptf1a and Rbpjl expression. PTF1 activates the expression of Nr5a2 which can positively regulate the expression of Ptf1a and Rbpjl in return. PTF1 also activates the expression of many transcription factors crucial for acinar cell development and function, such as Gfi1, Mst1, Myc, Nfatc1/2, and Spdef
In later development and adulthood, Ptf1a promotes acinar differentiation and regulates acinar cell-specific gene expression (Fig. 3b) [12, 43–46], which is dependent on the PTF1 complex. PTF1 is capable of autoactivation by directly binding to the promoter regions of Ptf1a and Rbpjl and activating their transcription [12, 13, 46]. ChIP analysis with a Ptf1a antibody demonstrated that PTF1 directly regulates Nr5a2 expression by binding to its regulatory sequences [46]. In response, Nr5a2 also binds to the promoters of Ptf1a and Rbpjl and promotes their expression. The coordination of PTF1 and Nr5a2 guarantees the terminal differentiation and function of acinar cells [47]. Moreover, PTF1 controls the expression of many other transcription factor genes such as Gfi1 [48], Mst1 [49], c-Myc [50], Nfatc1/2 [51], and together regulate the growth, differentiation and physiological function of acinar cells.
There are still gaps in our understanding of how Ptf1a expression is initiated in early pancreatic organogenesis. In Hnf1b (TCF2, vHnf1)-inactivated mice, expression of Ptf1a was not detected [52]. This can be interpreted as Hnf1b as a genetically upstream regulator of Ptf1a [53]. Alternatively, it may result from the secondary effect of failure in specifying pancreatic progenitor cell identity, since Pdx1, Mnx1, Ngn3, Isl1, Hnf6, and other critical pancreatic cell markers were all dramatically downregulated or totally disappeared in the mutant. Further evidence is needed to determine if Hnf1b is able to initiate the onset of Ptf1a expression. The essential mesenchymal–epithelial interactions for the proper development of early pancreas have been established since the 1960s, and the well-studied molecules mediating the process are FGFs, especially Fgf10 from the mesenchyme cells [54–56]. Loss of function study with Fgf10 knockout mice shows that Fgf10 signaling is essential for maintaining the proliferative capacity of pancreatic progenitor cells, mostly due to its ability to maintain normal expression levels of Pdx1 [54] and Ptf1a [56]. It is likely that similar pathways from aortic endothelium or dorsal mesenchyme initiate Ptf1a expression in the pancreas bud [56, 57], but the exact mechanism needs to be deciphered.
The Ptf1a activity is also regulated post-translationally in acinar cells. The K (Lysine) acetyltransferase Kat2b (Pcaf, p/CAF) interacts with Ptf1a and enhances transcription activity of the PTF1 complex by promoting Ptf1a nuclear accumulation and its acetylation [44]. The catenin beta interacting protein 1 (Ctnnbip1, ICAT), however, interacts directly with Ptf1a and interferes with its acetylation modification by Kat2b, and thus inhibits its transcriptional activity [58]. The E3 ubiquitin–protein ligase, TRIP12 (thyroid hormone receptor interactor 12), interacts with Ptf1a and promotes its polyubiquitination and proteasomal degradation [19]. Inhibition of proteasomal degradation results in elevated levels of Ptf1a and its polyubiquitinated forms in acinar cells. Phosphorylation, SUMOylation or other modification of Ptf1a has not been explored so far.
Ptf1a in neural tissue development
During development, Ptf1a is selectively expressed in the central and peripheral nervous systems, including retina, spinal cord, brain, and enteric neural system. Unlike in pancreas development, Ptf1a is not involved in early neural progenitor proliferation in neural tissues. It is only expressed transiently in post-mitotic neural precursors and participates in cell fate specification and differentiation.
Retina
Retina is a delicate neural tissue responsible for light signal capture, transduction, modulation, and conduction. It is developed from a cluster of multipotent progenitors in the optic vesicle. The mature vertebrate retina has multiple structural layers and is composed of six major types of neurons (cone, rod, horizontal, amacrine, bipolar, and ganglion,) and one major type of glial cells (Müller cells). Among which, the cone and rod photoreceptors located in the outer nuclear layer capture photons and convert them into neuronal signals; the horizontal, bipolar and amacrine cells are interneurons positioned in the inner nuclear layer and modulate the signals from photoreceptors before passing on to ganglion cells; the ganglion cells further integrate the signals from interneurons and transmit them into the brain.
Ptf1a expression in the retina starts at around E12 (embryonic day 12) and disappears after P3 (postnatal day 3), coincident with the specification period of horizontal and amacrine cells [59, 60]. BrdU pulse labeling result shows that Ptf1a expression is restricted in the post-mitotic cells. In Ptf1a knockout retinae, the absence of Ptf1a results in a loss of all horizontal cells, GABAergic and glycinergic amacrine cells, and a temporary increase of ganglion cells and photoreceptors, indicating a cell fate switch during retinal development [59, 60]. Conversely, Ptf1a misexpression promotes the fates of horizontal cells and GABAergic and glycinergic amacrine cells at the expense of photoreceptors and ganglion cells [61, 62]. Loss- and gain-of-function results demonstrate that Ptf1a is necessary and sufficient for horizontal and amacrine cell fate commitment.
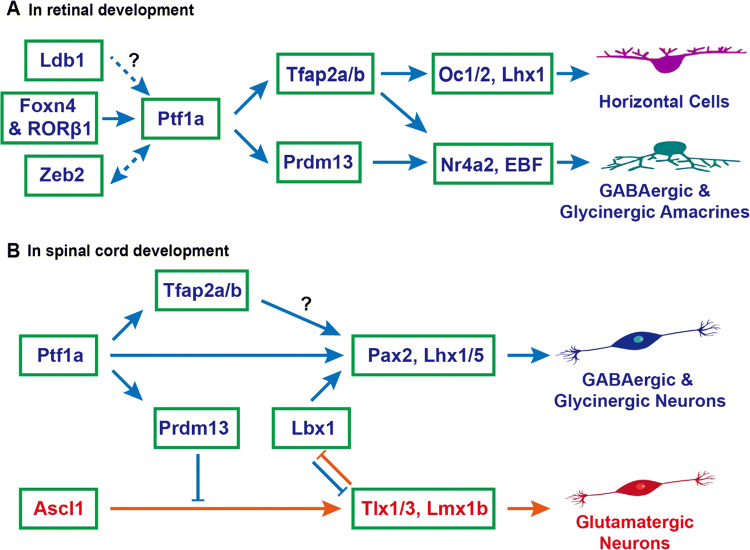
Unlike in the pancreas, the transcription factors regulating Ptf1a expression are well defined in the retina (Fig. 4a). In knockout mouse models, Foxn4-null and Ptf1a-null mice display strikingly similar phenotypes in the retina, including loss of horizontal and amacrine cells, temporary increase of ganglion cells and photoreceptors, indicating that both genes engage in the same genetic pathway [59, 63–65]. Foxn4 expression remains unchanged in Ptf1a-null retinae, but Ptf1a expression is abolished in Foxn4-null retinae, demonstrating that Ptf1a is a downstream target of Foxn4 [59]. Gain-of-function studies during chick retinogenesis further verified that misexpression of Foxn4 could greatly induce the expression of Ptf1a, while misexpression of Ptf1a inhibited the expression of Foxn4 due to negative feedback inhibition [66]. RORβ1, an isoform of the retinoid-related orphan nuclear receptor β gene (Rorb, Nr1f2), is also an upstream regulator of Ptf1a gene [67]. Knockout of RORβ1 leads to almost the same phenotypes in the mutant retinae as those in Foxn4-null and Ptf1a-null retinae. Ptf1a expression is undetectable in RORβ1-null retinae, but Foxn4 expression is not affected, indicating that Ptf1a but not Foxn4 is a downstream target of RORβ1. Data from Foxn4 knockout mice implicate that RORβ1 does not act downstream of Foxn4. Analysis of Ptf1a genomic regulatory sequences and electrophoretic mobility shift assay (EMSA) indicates that RORβ1 and Foxn4 may bind directly to the Ptf1a enhancer elements and synergistically activate Ptf1a expression [67]. However, it remains to be answered whether RORβ1 and Foxn4 form a protein complex regulating Ptf1a expression in retinal precursor cells, despite their ability to interact with each other [67]. The Zeb2 transcription factor, which is encoded by the Mowat–Wilson syndrome associated gene, has also been shown to directly bind the Ptf1a 3′ enhancer and therein controls the development of amacrine and horizontal cells by activating Ptf1a transcription [68, 69]. In the Ptf1a-null mouse retina, Zeb2 expression is downregulated, indicating that Ptf1a may regulate Zeb2 expression by positive feedback or other unknown mechanisms [69]. The LIM domain-binding protein Ldb1, along with its cofactors, may potentially be involved in regulating Ptf1a expression and thus participate in the differentiation of amacrine and horizontal cells in the retina [70].
Fig. 4.
Ptf1a signaling pathways to specify inhibitory neuronal fates in retinal and spinal cord development. a During retinal development, Foxn4 and RORβ1 jointly turn on the expression of Ptf1a, which in turn activates the expression of Tfap2a/b and Prdm13, determining the cell fates of horizontal and amacrine cells. Both GABAergic and glycinergic amacrine cells are inhibitory neurons. Zeb2 directly activates Ptf1a expression and Ptf1a may enhance Zeb2 expression during amacrine and horizontal cell development. Ldb1 complexes may also be involved in activating Ptf1a expression and vice versa. b During spinal cord development, the upstream regulator of Ptf1a is unknown. Ptf1a activates expression of downstream targets Pax2 and Lhx1/5 directly or indirectly through Tfap2a/b, specifying the progenitors towards inhibitory GABAergic and glycinergic neuronal fates. Meanwhile, Ptf1a inhibits the excitatory glutamatergic neuronal fates indirectly through the downstream gene Prdm13. The coordinate interaction between Ptf1a and Lbx1 is unclear. In cerebellar and brainstem development, the same mechanism may be also at play as in the spinal cord
Several downstream transcription factors mediating Ptf1a effects have been identified in retinal development (Fig. 4a). Using RNA-seq to compare differentially expressed genes between wild-type and Ptf1a-null retinae, a group of transcription factors, including Tfap2a, Tfap2b and Prdm13 (PR domain containing 13), were found to be significantly downregulated in the mutant retinae [62, 71]. Immunostaining with anti-Tfap2a and anti-Tfap2b antibodies showed that Tfap2a and Tfap2b proteins were undetectable in Ptf1a-null retinae [62]. Moreover, Ptf1a, Tfap2a and Tfap2b are expressed in post-mitotic cells, and have partially redundant functions in the differentiation of horizontal and amacrine cells [72]. Compound knockout of Tfap2a and Tfap2b in mouse retinae leads to similar phenotypes, i.e., loss of horizontal and amacrine cells, as seen in Ptf1a-null retinae [72]. In vivo and in vitro gain-of-function studies showed that overexpression of Ptf1a could induce the expression of Tfap2a and Tfap2b, and moreover, overexpression of Tfap2a and Tfap2b phenocopied those of Ptf1a overexpression [62]. The loss- and gain-of-function results established Tfap2a and Tfap2b as two major downstream targets of Ptf1a. In comparison, Prdm13 is present only in amacrine precursors and mature amacrine cells but not in horizontal precursors or mature horizontal cells. Correspondingly, Prdm13 deficiency only affects amacrine cells [73, 74]. By analysis of spatiotemporal expression patterns and loss-of-function phenotypes, we could place Oc1/2 [75, 76], Lhx1 [77, 78], EBF [79], and Nr4a2 [80] further downstream of Tfap2a/b and Prdm13 as shown in Fig. 4a.
The question remains as to whether Ptf1a activity is also dependent on Rbpj during retinal development. There are discrepancies in published results. In Rbpj homozygous conditional knockout retinae obtained by the Pax6 α-Cre driver, the development of amacrine and horizontal cells seems unaffected [81], whereas the two cell types are missing or drastically decreased in Ptf1a-null retinae, implying that the Ptf1a activity does not or minimally relies on Rbpj during retinal development. However, in Rbpj homozygous mutants conditionally knocked out by the Chx10-Cre driver, the development of amacrine and horizontal cells was disrupted and both populations dropped dramatically in the mutant retinae [82], suggesting that Ptf1a activity may be partially dependent on Rbpj. The discrepancies cannot be easily reconciled with the differential expression patterns of Cre recombinase. The latter result is more persuasive since the former investigated only a subset of amacrine cells, the calbindin + and calretinin + ones, with no quantitative data to support the claim. In contrast, the latter examined using a pan-amacrine cell marker, syntaxin, with solid statistical data. In agreement with this, in gain-of-function experiments in the chick retinae, it is found that interaction with Rbpj is a prerequisite for Ptf1a to specify retinal horizontal and amacrine cell fates [66]. Ectopic expression of Ptf1a was sufficient to promote the fates of Prox + horizontal cells and Tfap2a + amacrine neurons. When the interacting motifs C1 and C2 or C2 alone was mutated, Ptf1aΔC2 or Ptf1aΔC1ΔC2 lost the ability to specify these cell fates. In conclusion, Ptf1a activity is dependent on Rbpj during retinal development, but we still cannot rule out the importance of Rbpj-independent activity.
Spinal cord
The spinal cord is a part of the central nervous system that relays efferent motor neuron signals and afferent sensory neuron information. It is also the site controlling central pattern generators and simple neural reflex circuits. Developmentally, spinal cord is derived from neural ectoderm and neural tube.
By immunostaining, the expression of Ptf1a was detected at E10 in the neural tube and was limited to post-mitotic cells as well [83]. Specifically, Ptf1a is present in GABAergic neuronal precursors in spinal cord dorsal horns, and is required for fate determination of the GABAergic cell population [83]. The absence of Ptf1a results in a complete loss of inhibitory GABAergic neurons and a concomitant increase of excitatory glutamatergic neurons in the dorsal horn regions. Notably, many inhibitory neurons in the dorsal horn express glycinergic markers, which are also controlled by Ptf1a [84]. One explanation is that some dorsal horn GABAergic neurons co-release glycine [85, 86]. Thus, glycinergic neurons constitute a subset of GABAergic neurons, unlike their complete segregation in the retina. The imbalance of inhibitory and excitatory neuronal activity could interfere with primary sensory afferents and lead to sensory disorders, emphasizing the importance of Ptf1a in specifying inhibitory sensory neuronal fates.
As revealed by ChIP and other analyses, Ptf1a activity is mediated in both Rbpj-dependent and -independent manners in neural tube development [23]. Using gain-of-function studies, ChIP-seq, RNA-seq, and other approaches, a PR domain containing gene, Prdm13, was identified to be a direct downstream target of Ptf1a and Rbpj complex at the dorsal neural tube in the mouse and Xenopus [87, 88]. Prdm13 and Ptf1a display an overlapping expression pattern in the region. Overexpression of Ptf1a induces upregulation of Prdm13, and overexpression of Prdm13 phenocopies that of Ptf1a. Ptf1a protein binds to several genomic regulatory sequences of Prdm13 in a Rbpj-dependent manner. The ability of Ptf1a to inhibit the glutamatergic neuronal fate is mostly mediated by Prdm13, which suppresses the transcription of Tlx1, Tlx3 and other Ascl1 downstream genes by directly interacting with Ascl1 (Fig. 4b) [87]. The ability of Prdm13 to induce Pax2-positive GABAergic cell fates is likely an indirect effect, for example, by indirectly uplifting Lbx1 expression, which biases toward generation of GABAergic neurons.
Are Tfap2a and Tfap2b also downstream targets of Ptf1a in spinal cord development? Tfap2b is expressed in the dorsal horn with a pattern highly overlapping with that of Pax2. Its expression is completely absent in the Ptf1a knockout neural tube [89], indicating that Tfap2b is also a downstream gene of Ptf1a in neural tube development (Fig. 4b). Consistent with this, RNA-seq data showed that Tfap2b was downregulated in E11.5 Ptf1a-null neural tubes [90]. As a transcription activator, Tfap2b is speculated to activate the transcription of Lbx1 or Pax2, but this needs to be verified by further studies. Tfap2a is essential for early neural crest formation and growth. Whether it acts downstream of Ptf1a in later cell fate specification remains to be determined. Nevertheless, during cerebellar development in which Ptf1a has a similar role, it is found that both Tfap2a and Tfap2b act downstream of Ptf1a and upstream of Pax2 to determine inhibitory neuronal fates [91]. It is therefore highly possible that this is also the case during spinal cord development.
The upstream genetic regulator of Ptf1a is currently unknown in spinal cord development; however, it must be quite different from the ones in the retina. Though Foxn4 is expressed in progenitors of the neural tube, it is located only in the ventral region and does not participate in the regulation of Ptf1a as it does in the retina [92]. Similarly, the Rorb expression pattern revealed by RNA in situ hybridization assay indicates that RORβ1 is not an upstream regulator of Ptf1a either in the neural tube [89]. Genome sequence analysis indicated that a conserved enhancer region located 10.8 kb 3′ of Ptf1a coding region is required for tissue-specific expression of Pf1a in the dorsal neural tube. In this region, a DNA-binding motif for paired homeodomain (pd-hd) protein is indispensable for activating Ptf1a expression [17], implying that one of the paired homeodomain proteins is an upstream regulator of Ptf1a. Though Pax6 could bind to the motif to activate transcription in vitro, it is unlikely the upstream regulator of Ptf1a since their expression patterns are undoubtedly different from each other in the spinal cord.
Brain
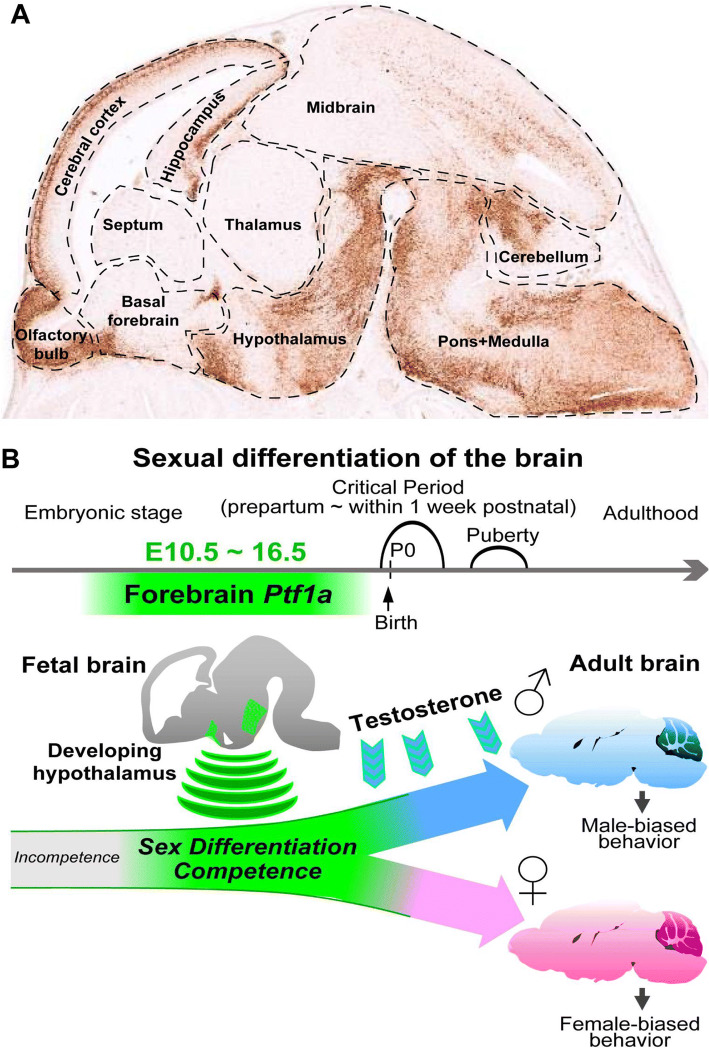
The brain is the most important organ governing cognition, intelligence and logical thinking. Anatomically, the mammalian brain can be divided into the forebrain, midbrain and hindbrain, each of them can be further subdivided both anatomically and functionally. For example, the forebrain is composed of diencephalon (thalamus and hypothalamus) and telencephalon (cerebrum). The hindbrain contains the pons and cerebellum. At developmental stage E15.5, BAC transgenic mouse and RNA in situ hybridization data from http://Gensat.org show that Ptf1a is highly expressed in the olfactory bulb, cerebral cortex, hippocampus, and hypothalamus of the forebrain, in the pons, medulla and cerebellum of the hindbrain, as well as in the midbrain (Fig. 5a). Except in the cerebellum, brainstem and forebrain, the role of Ptf1a is currently unknown in these brain regions during development.
Fig. 5.
Ptf1a participates in brain development and cell fate reprogramming. a The image shows the expression pattern of GFP/Ptf1a (brownish signal) in the E15.5 brain (sagittal section) of a BAC transgenic mouse line harboring Ptf1a regulatory sequences and GFP as a reporter gene (image adapted from http://GENSAT.org). Ptf1a is modestly or strongly expressed in many regions in the forebrain, midbrain and hindbrain. b Around perinatal stage is the critical period when male brain is exposed to testosterone signal and female exposed to non-testosterone signals to establish sexual differentiation of the brain. Forebrain Ptf1a confers the sex differentiation competence during brain development. Ptf1a-deficient mice display sexually dimorphic behavior abnormalities.
Image adapted from Fujiyama et al. [101]
The cerebellum mainly involves in motor control, for example, movement coordination, precision and timing accuracy. In cerebellar development, the Ptf1a-positive ventricular zone (VZ) progenitors give rise to GABAergic cerebellar neurons, such as Purkinje cells, GABAergic neurons of DCN (deep cerebellar nuclei) and molecular layer interneurons [93, 94]. Ectopic expression of Ptf1a in Ptf1a-negative VZ progenitors or rhombic lip progenitors converts them into GABAergic instead of glutamatergic neurons [93, 95]. On the other hand, in the absence of Ptf1a, those Ptf1a lineage VZ progenitors were either mis-specified into glutamatergic neurons and/or migrated and contributed to the ventral brainstem, or committed to apoptosis, eventually leading to postnatal cerebellar agenesis [93, 94, 96, 97]. Similar to that in the retina and spinal cord, the Ptf1a-Tfap2a/b transcriptional cascade has been shown to be the force driving inhibitory neuronal differentiation in the cerebellum [91]. It remains to be determined whether there also exists the Ptf1a–Prdm13 cascade in the cerebellum.
The brainstem is the most primitive portion of the brain. It connects the brain and spinal cord and relays sensory and motor signals. It also contains vital nuclei with diverse neuronal populations that regulate a wide range of life processes such as digestion, breath and heartbeat. For example, the nucleus of the solitary tract (nTs) relays baroreceptor, cardiac, pulmonary and other vagal afferents. The principal sensory trigeminal nuclei (PrV) and spinal trigeminal nuclei (SpV) process facial sensory information. The inferior olivary nuclei (ION) and cochlear nuclei handle motor coordination and sounds, respectively. It is found that Ptf1a plays a crucial role in the development of these nuclei (Table 1) [98–100]. Since these nuclei all arise from the neighboring rhombomeres, the molecular mechanism underlying Ptf1a regulation is much the same and similar to those in spinal cord and cerebellum, except in development of ION. Basically, the majority of evidence supports that the major function of Ptf1a is to promote GABAergic and glycinergic inhibitory neuronal cell fates while suppressing excitatory neuronal cell fates during development of these nuclei. However, in the development of ION, Ptf1a promotes the fates of climbing fiber (CF) neurons but suppresses mossy fiber (MF) neuronal fates [99]. This is quite unusual since CF and MF neurons are both glutamatergic excitatory neurons. It suggests that other TF(s) coexisted with Ptf1a may outweigh Ptf1a to specify excitatory neurons.
Surprisingly, unlike in other neural tissues, not all Ptf1a+ cells are post-mitotic cells; some of them are co-expressed with Ki67 in the forebrain [101]. Lineage tracing indicated that forebrain Ptf1a-lineage developed into a variety of neuronal subtypes including glutamatergic, dopaminergic, GABAergic and peptidergic neurons, in contrast to restricted inhibitory neuronal fates in the retina, spinal cord, cerebellum and cochlear nucleus [59, 83, 93, 100, 101]. It seems that forebrain Ptf1a is not involved in the excitatory versus inhibitory cell specification and fate determination, since its deletion neither alters the cell numbers nor the cell subtypes. However, Ptf1a-deficient mice exhibit complex abnormalities in sexually dimorphic behaviors and reproductive organs in both sexes, revealing a critical role of Ptf1a in regulating sexual differentiation of the brain (Fig. 5b). Ptf1a could alter cellular expression of sexually biased genes through cell autonomous or non-cell autonomous manners in the developing hypothalamus. One of the key downregulated genes in the Ptf1a-deficient hypothalamus is Kiss1 that is crucial in regulating brain sexual development through kisspeptin-GPR54 signaling pathway [102]. The expression of Ptf1a common downstream gene Prdm13 is also dramatically downregulated. However, its connection to brain sex differentiation needs further studies. Brain masculinization or feminization does not occur by exposure or non-exposure of testosterone in Ptf1a-deficient males or females, respectively, suggesting that Ptf1a confers the competence to acquire sex differentiation in the developing brain.
Enteric nervous system
The enteric nervous system (ENS) is often called the second brain, reflecting its importance, complexity and independence from the CNS. The ENS is composed of interconnecting ganglia within the myenteric and submucosal plexuses of the gut wall. Its neurons and glias arise from neural crest cells that migrate from vagal and sacral levels of the neural axis. The ENS includes sensory neurons, motor neurons and interneurons, and is capable of autonomous functions. Many neurotransmitters have been identified in the ENS neurons, such as acetylcholine, dopamine, NOS, GABA, and serotonin, most of which are identical to those found in the CNS [103]. Many researchers consider ENS as a repertoire of neurotransmitters, and diffusion, leak or uptake of the neurotransmitters into the blood stream could distribute them to the whole body. Some of the neurotransmitters can enter and/or exit the blood–brain barrier (BBB) via transporters on the brain capillary endothelial cells (BCECs), i.e., serotonin via SERT [104], GABA via GAT2/BGT-1 [105].
Some transcription factors such as Phox2b, Sox10, Ascl1, and Hand2 have been shown to be crucial in ENS development. Phox2b is essential for all autonomic ganglion formation and its deletion leads to enteric aganglionosis [106]. Sox10 is important to maintain the undifferentiated state of migrating crest cells [107]. Ascl1 promotes neurogenesis and has a role opposite to that of Sox10 [108]. Hand2 is required for late stage neurogenesis and expression of a subset of cell type-specific markers, especially vasoactive intestinal polypeptide (VIP) [109]. Genetic lineage analysis in zebrafish shows that Ptf1a is present in a subset of enteric neurons, most of which express serotonin [110], one of the major inhibitory neurotransmitters. The exact role of Ptf1a is not yet elucidated in the enteric neurons. It would be worthwhile to investigate if Ptf1a and Ascl1 are the major controllers contributing to inhibitory and excitatory ENS neurons, respectively.
Ptf1a mutations and diseases
The mutations discussed here represent naturally occurred spontaneous mutations, not genetically manipulated ones. Unlike many transcription factors with partially or fully redundant function replaceable by family members, to our knowledge, Ptf1a is very unique and indispensable. Mutations in Ptf1a coding regions and non-coding regulatory sequences often lead to genetic diseases in animals and human beings. The known disease-causing mutation sites and their associated defects are presented in Fig. 6 and Table 2.
Fig. 6.
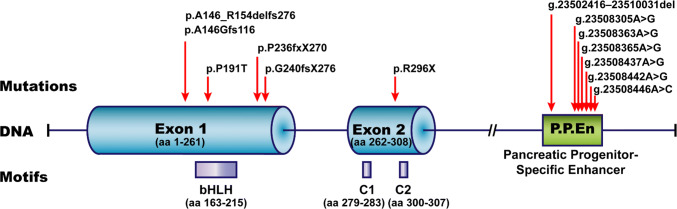
Known disease-causing mutations in human PTF1A. The human genomic (DNA) structure of PTF1A gene is illustrated (nonproportionally). Exons 1 and 2 with the corresponding numbers of coded amino acid residues, and the 3′ downstream pancreatic progenitor-specific enhancer are highlighted. The disease-causing mutations at protein level (p.xxx) or genomic level (g.xxx) are listed on top of the DNA structure. Red arrows point to the mutation sites. The critical peptide motifs, bHLH, C1 and C2, and their positions in PTF1A protein, are placed below the corresponding DNA structure
Table 2.
Ptf1a spontaneous mutations and diseases
| Species | Mutation position | Mutation type | Hetero- or homozygous | Diseases/defects | References |
|---|---|---|---|---|---|
| Mouse | A retrotransposon insertion at 12 kb upstream of Ptf1a | Insertional mutation causing Ptf1a gain-of-function | Heterozygous | Danforth’s short tail, characterized by spinal defects, kidney agenesis and anorectal malformations | [111–113, 126] |
| Human | p.R296X | Nonsense mutation causing PTF1A truncation | Homozygous | Diabetes mellitus, cerebellar agenesis, and more | [97] |
| p.P236fsX270 | Insertional mutation causing PTF1A frameshift and truncation | Homozygous | Diabetes mellitus, cerebellar agenesis, and more | [97] | |
| p.G240fsX276 | Frameshift causing PTF1A truncation | Homozygous | Diabetes mellitus, cerebellar agenesis, craniofacial defects, irregular breathing and more | [114] | |
| p.A146_R154delfsX115 | Deletional mutation causing PTF1A frameshift and truncation | Homozygous | Diabetes mellitus, cerebellar agenesis, craniofacial defects, irregular breathing, optic atrophy and more | [115] | |
| p.P191T | Missense mutation causing hypomorphic PTF1A | Homozygous | Diabetes mellitus | [116] | |
| A conserved regulatory sequence at 25 kb downstream of PTF1A | Point mutations or deletions in the distal pancreatic-specific enhancer | Homozygous, or compound heterozygous | Pancreatic agenesis | [15, 16] | |
| p.A146GfsX116 and g.23508442A > G | A frameshift and truncation mutation in PTF1A coding region in the chromosome. A point mutation at distal enhancer in the sister chromosome | Compound heterozygous | Pancreatic agenesis | [117] | |
| Unknown | Unknown mutation(s) leading to hypomorphic PTF1A in acinar cells | Unknown, possibly homozygous or compound heterozygous | Pancreatic cancer | [118–120] |
The Danforth’s short tail (Sd) is a semi-dominant mutation in the mouse, characterized by spinal defects, kidney agenesis and anorectal malformations. The Sd mutant phenotypes resemble symptoms seen in human caudal malformation syndromes, including urorectal septum malformation, caudal regression, VACTERL association, and persistent cloaca [111]. A few laboratories recently demonstrated that Sd is caused by up to tenfold overexpression of Ptf1a in the notochord, hindgut, cloaca, and mesonephros, due to an early retrotransposon insertion in the 12 kb conserved domain upstream of Ptf1a [111–113]. This raises an intriguing question of whether human caudal malformation syndromes are also caused by overexpression of Ptf1a. In addition, Sd mouse provides an excellent genetic tool for the study of Ptf1a gain-of-function effects.
The association of PTF1A with human diseases was first reported in 2004. Two families from Pakistan and Northern Europe with permanent diabetes mellitus were diagnosed as pancreatic and cerebellar agenesis [97]. Both families were found to have truncated mutations in PTF1A. In family 1, affected individuals had homozygous nonsense mutation p.R296X in PTF1A, leading to loss of the C2 motif that is required for interaction with Rbpj (Figs. 1c, 2c). In family 2, an insertion mutation c.705insG caused frameshift (p.P236fsX270) and premature truncation of PTF1A protein at codon 270, resulting in a loss of both C1 and C2 motifs. More mutations in PTF1A have since been identified [114–116]. It appears that the severity of the symptoms is associated with the severity of the mutations. Some hypomorphic PTF1A mutations may only cause isolated pancreatic aplasia but without any cerebellar phenotypes. For example, the p.P191T missense mutation in the bHLH domain affects the dimerization of PTF1A with E proteins, resulting in isolated pancreatic aplasia or neonatal diabetes mellitus, but the patients show no obvious neurodevelopmental defects [116]. In addition to symptoms caused by pancreatic and cerebellar hypoplasia, optic atrophy was recorded in a patient with PTF1A deletion and frameshift [115]. No associated symptoms in the spinal cord and urorectal system have been reported in human patients.
The contribution of non-coding variants to human diseases remains a challenge to gauge. However, mutations in the PTF1A non-coding enhancer region are found to cause human diseases. Analysis of whole genome sequencing data from patient families with isolated pancreatic agenesis uncovered different recessive mutations in a 400-bp enhancer region located at 25 kb downstream of PTF1A [15, 16]. This enhancer is required to target PTF1A in human embryonic pancreatic progenitor cells, and mutations in the enhancer leads to its inactivation and PTF1A deprivation in these cells. One rare case of neonatal diabetes mellitus and pancreatic agenesis was also reported with compound heterozygous mutation of PTF1A [117]. The patient had a deletional and frameshift mutation in the PTF1A coding region on one chromosome, and a point mutation in the 400-bp enhancer region on another chromosome. Given the fact that there are other enhancers for PTF1A specific expression in the spinal cord, it will be challenging to discover the association of these enhancer mutations with some sensorineural diseases, if there is any.
Additionally, downregulation of PTF1A is associated with pancreatic intraepithelial neoplasia (PanIN), the first stage of pancreatic ductal adenocarcinoma (PDAC) [118–120]. PTF1A is required to maintain the identity of acinar cells, and downregulation of PTF1A in these cells cause them to transdifferentiate into ductal cells. The underlying molecular mechanism of PTF1a downregulation and development of PanIN/PDAC remains poorly understood. However, it is tempting to propose that small molecules or drugs targeting PTF1A upregulation may provide an entry point for the treatment of these diseases.
Ptf1a in cell fate reprogramming and transdifferentiation
Given the importance of Ptft1a in the establishment of pancreatic identity, researchers wondered if Ptf1a could be used in the pursuit of cell fate reprogramming. Indeed, transient ectopic expression of Ptf1a in the mouse embryonic stem cells (ESCs) triggered the cells to launch pancreatic programs and express Pdx1 [121]. The differentiated cells further formed cells of all three pancreatic lineages, the acinar, endocrine and duct cells.
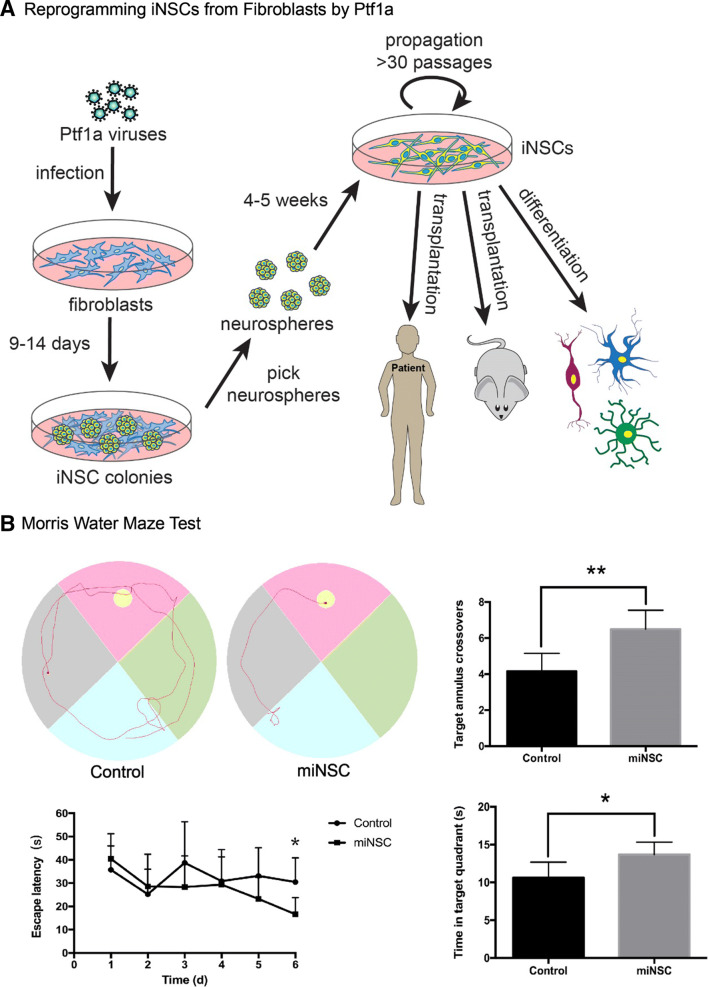
Reprogramming somatic cells and transdifferentiating them into other cell types is also a common practice in stem cell research. We had hypothesized that ectopic expression of Ptf1a in fibroblasts would convert them into pancreatic lineages or inhibitory neurons. When we overexpressed Ptf1a in human or mouse fibroblasts, surprisingly, these somatic cells were reprogrammed into neurospheres containing neural stem cells (NSCs) instead of pancreatic lineages [122]. The induced NSCs (iNSCs) were capable of differentiating into various types of neurons, astrocytes and oligodendrocytes in differentiation culture media (Fig. 7a). The underlying reprogramming mechanism is also dependent on the Ptf1a–Rbpj interaction, since mutation of the interaction site on Ptf1a (W298A), or knockdown of Rbpj, would abolish or drastically decrease the reprogramming activity.
Fig. 7.
Ptf1a alone is sufficient to reprogram human and mouse fibroblasts into induced neural stem cells (iNSCs). a After infected with lentiviruses expressing Ptf1a, human or mouse fibroblasts were transformed into neurospheres. Individual neurosphere can be further expanded and form typical single layered iNSCs in culture dishes. The iNSCs have the tripotence to differentiate into various types of neurons, astrocytes and oligodendrocytes both in vitro and in vivo. The iNSCs constitute an ideal cell source for preclinical transplantation experiments and clinical studies. b To determine the therapeutic effect of the Ptf1a-derived iNSCs, they were transplanted into the hippocampus of the Alzheimer disease mouse models. After training for 5 days, the iNSCs-transplanted mice spent much less time (bottom left) to find the target platform in the Morris water maze test. An example of travel pathways for a control mouse and an iNSC-transplanted mouse (top left) was given. Target annulus crossovers revealed that iNSC-transplanted mice displayed a preference for the target platform location (top right). Moreover, they also spent more time in the target quadrant (bottom right). These results demonstrate that iNSC transplantation improves the memory and cognitive functions of the AD mouse models
The Ptf1a-reprogrammed iNSCs also have the tripotence to differentiate into neurons, astrocytes and oligodendrocytes when transplanted in vivo. After injected into the mouse hippocampus, Ptf1a-reprogrammed mouse or human iNSCs survived and successfully integrated into the local tissue [122]. The integrated cells differentiated into many astrocytes, some oligodendrocytes, and some inhibitory and excitatory neurons that established synaptic connections with neighbor endogenous neurons. These results have demonstrated that Ptf1a-induced iNSCs have the differentiation tripotence both in intro and in vivo. Moreover, in the Aβ1-40 injected or APP/PS1 Alzheimer’s disease (AD) mouse models, implanted iNSCs significantly improved spatial learning and memory and cognitive dysfunction of the AD mouse models as evaluated by behavioral tests [122] (Fig. 7b). Despite the promising results in animal models, the question remains whether Ptf1a-induced iNSCs have any therapeutic potential in treating neurodegenerative diseases in preclinical and clinical settings.
There are several advantages for Ptf1a to reprogram iNSCs compared with other approaches [122]. (1) As a non-tumorigenic factor, Ptf1a is much safer than Sox2 or Zfp521 or other factors that are tumor-prone. (2) Ptf1a is more efficient in reprogramming iNSCs. Ptf1a could convert 0.5% fibroblasts into neurospheres. In contrast, Sox2 alone could only reach less than 0.1% [123]. The efficiency is even lower with multiple transcription factors. (3) Ptf1a-induced iNSCs possess stronger self-renewal ability and could be passaged more than 40 generations without any aging signs, while iNSCs derived from a few other approaches have only limited passaging ability. (4) Ptf1a-induced iNSCs are more efficient in directed differentiation. They are capable of generating 83.3% of neurons, 87.3% of astrocytes or 26.6% of oligodendrocytes in respective differentiation media. In comparison, Sox2-induced iNSCs could only generate 67% of neurons and 25% of astrocytes [123]. (5) Ptf1a-induced iNSCs are closer to endogenous NSCs. RNA-seq data demonstrated that Ptf1a-induced iNSCs have a higher correlation coefficient with SCR029 cells, a well-characterized mouse cortical NSCs, than those of NS5 cells (mouse ES cell-derived NSCs) and ciNSCs (NSCs chemically induced from MEFs). Therefore, Ptf1a-induced iNSCs may provide a better cell source for disease cell modeling, drug screening and cell replacement therapies.
Conclusions and perspectives
Although many advances have been made toward understanding various aspects of the important transcription factor Ptf1a in the past 30 years, beyond its role in the development of pancreas, spinal cord, retina, cerebellum, brainstem, and forebrain, many uncharted territories still remain.
Ptf1a is highly expressed in the stomach, colon and rectum along the digestive tract. In these organs, Ptf1a is possibly expressed specifically in neurons in the local ganglion plexus, controlling muscle motility and secretion of digestive enzymes, as indicated in the ENS [110]. However, it cannot be ruled out that Ptf1a may participate in the specification of other cell types beyond neuroglial cells, either cell autonomously or non-cell autonomously, as demonstrated by the anorectal developmental defects in Sd mice [111–113]. Sd mice also display kidney defects, indicating that Ptf1a affects kidney development as well. Ptf1a is also highly expressed in the auxiliary genital organs such as ovary, testis and prostate gland. Ptf1a in these organs might have a similar role as in the pancreas, or an alternative role as in the intestine. Further studies are necessary to distinguish these possibilities using the Ptf1a knockout and Sd mice.
As aforementioned, the role of Ptf1a is unexplored in the cerebral cortex, hippocampus, and other important brain regions. Does Ptf1a also determine the inhibitory versus excitatory neuronal fates in these regions? Does Ptf1a control those neurons involved in memory, learning, feelings, or space localization? Does it have any potential to promote regeneration of neurons damaged in Alzheimer’s, Parkinson’s or other neurodegenerative diseases? It would be enticing and thrilling to address these questions.
In brief, here we have discussed the past findings about and insights into the crucial functions of the transcription factor Ptf1a gained since its identification. In particular, we have enumerated its genomic and protein structures, expression profiles, upstream and downstream regulators, mutations and diseases, somatic cell reprogramming activities, and so on. We have also introduced some unsolved issues about Ptf1a that may inspire colleagues to explore uncharted territories and achieve many more exciting findings in the future.
Acknowledgements
We thank Evelyn Shiang for help with the artwork and many online databases and resources for providing useful data and tools. Some of them are listed in the context. We thank all the colleagues working on related subjects and apologize for unintentional omission of many important findings. This work is supported in part by the National Key R&D Program of China (2017YFA0104100), National Basic Research Program (973 Program) of China (2015CB964600), National Natural Science Foundation of China (81670862 and 81721003), Science and Technology Planning Projects of Guangdong Province (2017B030314025), and the Fundamental Research Funds of the State Key Laboratory of Ophthalmology, Sun Yat-sen University to MX, and the National Natural Science Foundation of China (31871497) to KJ.
Funding
The authors declare no financial interests.
Contributor Information
Kangxin Jin, Phone: (+8620) 66678731, Email: kxjin@yahoo.com, Email: jinkx@mail.sysu.edu.cn.
Mengqing Xiang, Phone: (+8620) 66677329, Email: xiangmq3@mail.sysu.edu.cn.
References
- 1.Murre C, et al. Interactions between heterologous helix–loop–helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58(3):537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 2.Chaudhary J, Skinner MK. Basic helix–loop–helix proteins can act at the E-box within the serum response element of the c-fos promoter to influence hormone-induced promoter activation in Sertoli cells. Mol Endocrinol. 1999;13(5):774–786. doi: 10.1210/mend.13.5.0271. [DOI] [PubMed] [Google Scholar]
- 3.Grove CA, et al. A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell. 2009;138(2):314–327. doi: 10.1016/j.cell.2009.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murre C, et al. Structure and function of helix–loop–helix proteins. Biochim Biophys Acta. 1994;1218(2):129–135. doi: 10.1016/0167-4781(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 5.Massari ME, Murre C. Helix–loop–helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20(2):429–440. doi: 10.1128/MCB.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benezra R, et al. The protein Id: a negative regulator of helix–loop–helix DNA binding proteins. Cell. 1990;61(1):49–59. doi: 10.1016/0092-8674(90)90214-Y. [DOI] [PubMed] [Google Scholar]
- 7.Wang LH, Baker NE. E proteins and ID proteins: helix–loop–helix partners in development and disease. Dev Cell. 2015;35(3):269–280. doi: 10.1016/j.devcel.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cockell M, et al. Identification of a cell-specific DNA-binding activity that interacts with a transcriptional activator of genes expressed in the acinar pancreas. Mol Cell Biol. 1989;9(6):2464–2476. doi: 10.1128/MCB.9.6.2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rose SD, et al. The role of PTF1-P48 in pancreatic acinar gene expression. J Biol Chem. 2001;276(47):44018–44026. doi: 10.1074/jbc.M106264200. [DOI] [PubMed] [Google Scholar]
- 10.Beres TM, et al. PTF1 is an organ-specific and Notch-independent basic helix–loop–helix complex containing the mammalian Suppressor of Hairless (RBP-J) or its paralogue, RBP-L. Mol Cell Biol. 2006;26(1):117–130. doi: 10.1128/MCB.26.1.117-130.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rose SD, MacDonald RJ. Evolutionary silencing of the human elastase I gene (ELA1) Hum Mol Genet. 1997;6(6):897–903. doi: 10.1093/hmg/6.6.897. [DOI] [PubMed] [Google Scholar]
- 12.Masui T, et al. Replacement of Rbpj with Rbpjl in the PTF1 complex controls the final maturation of pancreatic acinar cells. Gastroenterology. 2010;139(1):270–280. doi: 10.1053/j.gastro.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masui T, et al. Early pancreatic development requires the vertebrate Suppressor of Hairless (RBPJ) in the PTF1 bHLH complex. Genes Dev. 2007;21(20):2629–2643. doi: 10.1101/gad.1575207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perillo M, et al. A pancreatic exocrine-like cell regulatory circuit operating in the upper stomach of the sea urchin Strongylocentrotus purpuratus larva. BMC Evol Biol. 2016;16(1):117. doi: 10.1186/s12862-016-0686-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weedon MN, et al. Recessive mutations in a distal PTF1A enhancer cause isolated pancreatic agenesis. Nat Genet. 2014;46(1):61–64. doi: 10.1038/ng.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonc EN, et al. Variable phenotype of diabetes mellitus in siblings with a homozygous PTF1A enhancer mutation. Horm Res Paediatr. 2015;84(3):206–211. doi: 10.1159/000435782. [DOI] [PubMed] [Google Scholar]
- 17.Mona B, et al. Regulating the dorsal neural tube expression of Ptf1a through a distal 3′ enhancer. Dev Biol. 2016;418(1):216–225. doi: 10.1016/j.ydbio.2016.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu CK, et al. Rbms3, an RNA-binding protein, mediates the expression of Ptf1a by binding to its 3′UTR during mouse pancreas development. DNA Cell Biol. 2012;31(7):1245–1251. doi: 10.1089/dna.2012.1619. [DOI] [PubMed] [Google Scholar]
- 19.Hanoun N, et al. The E3 ubiquitin ligase thyroid hormone receptor-interacting protein 12 targets pancreas transcription factor 1a for proteasomal degradation. J Biol Chem. 2014;289(51):35593–35604. doi: 10.1074/jbc.M114.620104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Driscoll CA, Macdonald DW, O’Brien SJ. From wild animals to domestic pets, an evolutionary view of domestication. Proc Natl Acad Sci USA. 2009;106(Suppl 1):9971–9978. doi: 10.1073/pnas.0901586106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masui T, et al. Transcriptional autoregulation controls pancreatic Ptf1a expression during development and adulthood. Mol Cell Biol. 2008;28(17):5458–5468. doi: 10.1128/MCB.00549-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meredith DM, et al. Multiple transcriptional mechanisms control Ptf1a levels during neural development including autoregulation by the PTF1-J complex. J Neurosci. 2009;29(36):11139–11148. doi: 10.1523/JNEUROSCI.2303-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meredith DM, et al. Program specificity for Ptf1a in pancreas versus neural tube development correlates with distinct collaborating cofactors and chromatin accessibility. Mol Cell Biol. 2013;33(16):3166–3179. doi: 10.1128/MCB.00364-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson N, et al. RNA profiling and chromatin immunoprecipitation-sequencing reveal that PTF1a stabilizes pancreas progenitor identity via the control of MNX1/HLXB9 and a network of other transcription factors. Mol Cell Biol. 2012;32(6):1189–1199. doi: 10.1128/MCB.06318-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Obata J, et al. p48 subunit of mouse PTF1 binds to RBP-Jkappa/CBF-1, the intracellular mediator of Notch signalling, and is expressed in the neural tube of early stage embryos. Genes Cells. 2001;6(4):345–360. doi: 10.1046/j.1365-2443.2001.00422.x. [DOI] [PubMed] [Google Scholar]
- 26.Petcherski AG, Kimble J. LAG-3 is a putative transcriptional activator in the C. elegans Notch pathway. Nature. 2000;405(6784):364–368. doi: 10.1038/35012645. [DOI] [PubMed] [Google Scholar]
- 27.Kovall RA. More complicated than it looks: assembly of Notch pathway transcription complexes. Oncogene. 2008;27(38):5099–5109. doi: 10.1038/onc.2008.223. [DOI] [PubMed] [Google Scholar]
- 28.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kovall RA, Hendrickson WA. Crystal structure of the nuclear effector of Notch signaling, CSL, bound to DNA. EMBO J. 2004;23(17):3441–3451. doi: 10.1038/sj.emboj.7600349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krapp A, et al. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev. 1998;12(23):3752–3763. doi: 10.1101/gad.12.23.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krapp A, et al. The p48 DNA-binding subunit of transcription factor PTF1 is a new exocrine pancreas-specific basic helix–loop–helix protein. EMBO J. 1996;15(16):4317–4329. doi: 10.1002/j.1460-2075.1996.tb00806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawaguchi Y, et al. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet. 2002;32(1):128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- 33.Pan FC, et al. Spatiotemporal patterns of multipotentiality in Ptf1a-expressing cells during pancreas organogenesis and injury-induced facultative restoration. Development. 2013;140(4):751–764. doi: 10.1242/dev.090159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim DY, et al. Functional regulation of FoxO1 in neural stem cell differentiation. Cell Death Differ. 2015;22(12):2034–2045. doi: 10.1038/cdd.2015.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willet SG, et al. Dominant and context-specific control of endodermal organ allocation by Ptf1a. Development. 2014;141(22):4385–4394. doi: 10.1242/dev.114165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dong PD, et al. Graded levels of Ptf1a differentially regulate endocrine and exocrine fates in the developing pancreas. Genes Dev. 2008;22(11):1445–1450. doi: 10.1101/gad.1663208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hesselson D, Anderson RM, Stainier DY. Suppression of Ptf1a activity induces acinar-to-endocrine conversion. Curr Biol. 2011;21(8):712–717. doi: 10.1016/j.cub.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahnfelt-Ronne J, et al. Ptf1a-mediated control of Dll1 reveals an alternative to the lateral inhibition mechanism. Development. 2012;139(1):33–45. doi: 10.1242/dev.071761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghosh B, Leach SD. Interactions between hairy/enhancer of split-related proteins and the pancreatic transcription factor Ptf1-p48 modulate function of the PTF1 transcriptional complex. Biochem J. 2006;393(Pt 3):679–685. doi: 10.1042/BJ20051063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiebe PO, et al. Ptf1a binds to and activates area III, a highly conserved region of the Pdx1 promoter that mediates early pancreas-wide Pdx1 expression. Mol Cell Biol. 2007;27(11):4093–4104. doi: 10.1128/MCB.01978-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyatsuka T, et al. Ptf1a and RBP-J cooperate in activating Pdx1 gene expression through binding to Area III. Biochem Biophys Res Commun. 2007;362(4):905–909. doi: 10.1016/j.bbrc.2007.08.076. [DOI] [PubMed] [Google Scholar]
- 42.Gao N, et al. Dynamic regulation of Pdx1 enhancers by Foxa1 and Foxa2 is essential for pancreas development. Genes Dev. 2008;22(24):3435–3448. doi: 10.1101/gad.1752608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holmstrom SR, et al. LRH-1 and PTF1-L coregulate an exocrine pancreas-specific transcriptional network for digestive function. Genes Dev. 2011;25(16):1674–1679. doi: 10.1101/gad.16860911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodolosse A, et al. p/CAF modulates the activity of the transcription factor p48/Ptf1a involved in pancreatic acinar differentiation. Biochem J. 2009;418(2):463–473. doi: 10.1042/BJ20080293. [DOI] [PubMed] [Google Scholar]
- 45.Jiang Z, et al. Exdpf is a key regulator of exocrine pancreas development controlled by retinoic acid and ptf1a in zebrafish. PLoS Biol. 2008;6(11):e293. doi: 10.1371/journal.pbio.0060293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoang CQ, et al. Transcriptional maintenance of pancreatic acinar identity, differentiation, and homeostasis by PTF1A. Mol Cell Biol. 2016;36(24):3033–3047. doi: 10.1128/MCB.00358-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hale MA, et al. The nuclear hormone receptor family member NR5A2 controls aspects of multipotent progenitor cell formation and acinar differentiation during pancreatic organogenesis. Development. 2014;141(16):3123–3133. doi: 10.1242/dev.109405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qu X, et al. Growth factor independence-1 (Gfi1) is required for pancreatic acinar unit formation and centroacinar cell differentiation. Cell Mol Gastroenterol Hepatol. 2015;1(2):233–247.e1. doi: 10.1016/j.jcmgh.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao T, et al. Hippo signaling regulates differentiation and maintenance in the exocrine pancreas. Gastroenterology. 2013;144(7):1543–1553.e9. doi: 10.1053/j.gastro.2013.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bonal C, et al. Pancreatic inactivation of c-Myc decreases acinar mass and transdifferentiates acinar cells into adipocytes in mice. Gastroenterology. 2009;136(1):309–319.e9. doi: 10.1053/j.gastro.2008.10.015. [DOI] [PubMed] [Google Scholar]
- 51.Chen NM, et al. NFATc1 links EGFR signaling to induction of Sox9 transcription and acinar-ductal transdifferentiation in the pancreas. Gastroenterology. 2015;148(5):pp. 1024–1034 e9. doi: 10.1053/j.gastro.2015.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haumaitre C, et al. Lack of TCF2/vHNF1 in mice leads to pancreas agenesis. Proc Natl Acad Sci USA. 2005;102(5):1490–1495. doi: 10.1073/pnas.0405776102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zaret KS. Genetic programming of liver and pancreas progenitors: lessons for stem-cell differentiation. Nat Rev Genet. 2008;9(5):329–340. doi: 10.1038/nrg2318. [DOI] [PubMed] [Google Scholar]
- 54.Bhushan A, et al. Fgf10 is essential for maintaining the proliferative capacity of epithelial progenitor cells during early pancreatic organogenesis. Development. 2001;128(24):5109–5117. doi: 10.1242/dev.128.24.5109. [DOI] [PubMed] [Google Scholar]
- 55.Ye F, Duvillie B, Scharfmann R. Fibroblast growth factors 7 and 10 are expressed in the human embryonic pancreatic mesenchyme and promote the proliferation of embryonic pancreatic epithelial cells. Diabetologia. 2005;48(2):277–281. doi: 10.1007/s00125-004-1638-6. [DOI] [PubMed] [Google Scholar]
- 56.Jacquemin P, et al. An endothelial-mesenchymal relay pathway regulates early phases of pancreas development. Dev Biol. 2006;290(1):189–199. doi: 10.1016/j.ydbio.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 57.Yoshitomi H, Zaret KS. Endothelial cell interactions initiate dorsal pancreas development by selectively inducing the transcription factor Ptf1a. Development. 2004;131(4):807–817. doi: 10.1242/dev.00960. [DOI] [PubMed] [Google Scholar]
- 58.Campos ML, et al. ICAT is a novel Ptf1a interactor that regulates pancreatic acinar differentiation and displays altered expression in tumours. Biochem J. 2013;451(3):395–405. doi: 10.1042/BJ20120873. [DOI] [PubMed] [Google Scholar]
- 59.Fujitani Y, et al. Ptf1a determines horizontal and amacrine cell fates during mouse retinal development. Development. 2006;133(22):4439–4450. doi: 10.1242/dev.02598. [DOI] [PubMed] [Google Scholar]
- 60.Nakhai H, et al. Ptf1a is essential for the differentiation of GABAergic and glycinergic amacrine cells and horizontal cells in the mouse retina. Development. 2007;134(6):1151–1160. doi: 10.1242/dev.02781. [DOI] [PubMed] [Google Scholar]
- 61.Dullin JP, et al. Ptf1a triggers GABAergic neuronal cell fates in the retina. BMC Dev Biol. 2007;7:110. doi: 10.1186/1471-213X-7-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jin K, et al. Tfap2a and 2b act downstream of Ptf1a to promote amacrine cell differentiation during retinogenesis. Mol Brain. 2015;8:28. doi: 10.1186/s13041-015-0118-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li S, et al. Foxn4 controls the genesis of amacrine and horizontal cells by retinal progenitors. Neuron. 2004;43(6):795–807. doi: 10.1016/j.neuron.2004.08.041. [DOI] [PubMed] [Google Scholar]
- 64.Luo H, et al. Forkhead box N4 (Foxn4) activates Dll4-Notch signaling to suppress photoreceptor cell fates of early retinal progenitors. Proc Natl Acad Sci USA. 2012;109(9):E553–E562. doi: 10.1073/pnas.1115767109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xiang M, Li S. Foxn4: a multi-faceted transcriptional regulator of cell fates in vertebrate development. Sci China Life Sci. 2013;56(11):985–993. doi: 10.1007/s11427-013-4543-8. [DOI] [PubMed] [Google Scholar]
- 66.Lelievre EC, et al. Ptf1a/Rbpj complex inhibits ganglion cell fate and drives the specification of all horizontal cell subtypes in the chick retina. Dev Biol. 2011;358(2):296–308. doi: 10.1016/j.ydbio.2011.07.033. [DOI] [PubMed] [Google Scholar]
- 67.Liu H, et al. An isoform of retinoid-related orphan receptor beta directs differentiation of retinal amacrine and horizontal interneurons. Nat Commun. 2013;4:1813. doi: 10.1038/ncomms2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Menuchin-Lasowski Y, et al. Sip1 regulates the generation of the inner nuclear layer retinal cell lineages in mammals. Development. 2016;143(15):2829–2841. doi: 10.1242/dev.136101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wei W et al (2018) Requirement of the Mowat-Wilson syndrome gene Zeb2 in the differentiation and maintenance of non-photoreceptor cell types during retinal development. Mol Neurobiol. 10.1007/s12035-018-1186-6 [DOI] [PubMed]
- 70.Xiao D, Jin K, Xiang M (2018) Necessity and sufficiency of Ldb1 in the generation, differentiation and maintenance of non-photoreceptor cell types during retinal development. Front Mol Neurosci 11:271 [DOI] [PMC free article] [PubMed]
- 71.Jin K. Transitional progenitors during vertebrate retinogenesis. Mol Neurobiol. 2017;54(5):3565–3576. doi: 10.1007/s12035-016-9899-x. [DOI] [PubMed] [Google Scholar]
- 72.Bassett EA, et al. Overlapping expression patterns and redundant roles for AP-2 transcription factors in the developing mammalian retina. Dev Dyn. 2012;241(4):814–829. doi: 10.1002/dvdy.23762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Watanabe S, et al. Prdm13 regulates subtype specification of retinal amacrine interneurons and modulates visual sensitivity. J Neurosci. 2015;35(20):8004–8020. doi: 10.1523/JNEUROSCI.0089-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Goodson NB, et al. Prdm13 is required for Ebf3+ amacrine cell formation in the retina. Dev Biol. 2018;434(1):149–163. doi: 10.1016/j.ydbio.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sapkota D, et al. Onecut1 and Onecut2 redundantly regulate early retinal cell fates during development. Proc Natl Acad Sci USA. 2014;111(39):E4086–E4095. doi: 10.1073/pnas.1405354111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu F, et al. Onecut1 is essential for horizontal cell genesis and retinal integrity. J Neurosci. 2013;33(32):13053–13065, 13065a. doi: 10.1523/JNEUROSCI.0116-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Poche RA, et al. Lim1 is essential for the correct laminar positioning of retinal horizontal cells. J Neurosci. 2007;27(51):14099–14107. doi: 10.1523/JNEUROSCI.4046-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Margeta MA. Transcription factor Lim1 specifies horizontal cell laminar position in the retina. J Neurosci. 2008;28(15):3835–3836. doi: 10.1523/JNEUROSCI.0535-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jin K, et al. Early B-cell factors are required for specifying multiple retinal cell types and subtypes from postmitotic precursors. J Neurosci. 2010;30(36):11902–11916. doi: 10.1523/JNEUROSCI.2187-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jiang H, Xiang M. Subtype specification of GABAergic amacrine cells by the orphan nuclear receptor Nr4a2/Nurr1. J Neurosci. 2009;29(33):10449–10459. doi: 10.1523/JNEUROSCI.3048-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Riesenberg AN, et al. Rbpj cell autonomous regulation of retinal ganglion cell and cone photoreceptor fates in the mouse retina. J Neurosci. 2009;29(41):12865–12877. doi: 10.1523/JNEUROSCI.3382-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zheng MH, et al. The transcription factor RBP-J is essential for retinal cell differentiation and lamination. Mol Brain. 2009;2:38. doi: 10.1186/1756-6606-2-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Glasgow SM, et al. Ptf1a determines GABAergic over glutamatergic neuronal cell fate in the spinal cord dorsal horn. Development. 2005;132(24):5461–5469. doi: 10.1242/dev.02167. [DOI] [PubMed] [Google Scholar]
- 84.Huang M, et al. Ptf1a, Lbx1 and Pax2 coordinate glycinergic and peptidergic transmitter phenotypes in dorsal spinal inhibitory neurons. Dev Biol. 2008;322(2):394–405. doi: 10.1016/j.ydbio.2008.06.031. [DOI] [PubMed] [Google Scholar]
- 85.Todd AJ. GABA and glycine in synaptic glomeruli of the rat spinal dorsal horn. Eur J Neurosci. 1996;8(12):2492–2498. doi: 10.1111/j.1460-9568.1996.tb01543.x. [DOI] [PubMed] [Google Scholar]
- 86.Todd AJ, Sullivan AC. Light microscope study of the coexistence of GABA-like and glycine-like immunoreactivities in the spinal cord of the rat. J Comp Neurol. 1990;296(3):496–505. doi: 10.1002/cne.902960312. [DOI] [PubMed] [Google Scholar]
- 87.Wapinski OL, et al. Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell. 2013;155(3):621–635. doi: 10.1016/j.cell.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hanotel J, et al. The Prdm13 histone methyltransferase encoding gene is a Ptf1a–Rbpj downstream target that suppresses glutamatergic and promotes GABAergic neuronal fate in the dorsal neural tube. Dev Biol. 2014;386(2):340–357. doi: 10.1016/j.ydbio.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 89.Wildner H, et al. Genome-wide expression analysis of Ptf1a- and Ascl1-deficient mice reveals new markers for distinct dorsal horn interneuron populations contributing to nociceptive reflex plasticity. J Neurosci. 2013;33(17):7299–7307. doi: 10.1523/JNEUROSCI.0491-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Borromeo MD, et al. A transcription factor network specifying inhibitory versus excitatory neurons in the dorsal spinal cord. Development. 2014;141(14):2803–2812. doi: 10.1242/dev.105866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zainolabidin N, et al. Distinct Activities of Tfap2A and Tfap2B in the Specification of GABAergic Interneurons in the Developing Cerebellum. Front Mol Neurosci. 2017;10(281):281. doi: 10.3389/fnmol.2017.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li S, et al. Foxn4 acts synergistically with Mash1 to specify subtype identity of V2 interneurons in the spinal cord. Proc Natl Acad Sci USA. 2005;102(30):10688–10693. doi: 10.1073/pnas.0504799102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hoshino M, et al. Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron. 2005;47(2):201–213. doi: 10.1016/j.neuron.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 94.Pascual M, et al. Cerebellar GABAergic progenitors adopt an external granule cell-like phenotype in the absence of Ptf1a transcription factor expression. Proc Natl Acad Sci USA. 2007;104(12):5193–5198. doi: 10.1073/pnas.0605699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yamada M, et al. Specification of spatial identities of cerebellar neuron progenitors by ptf1a and atoh1 for proper production of GABAergic and glutamatergic neurons. J Neurosci. 2014;34(14):4786–4800. doi: 10.1523/JNEUROSCI.2722-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Millen KJ, et al. Transformation of the cerebellum into more ventral brainstem fates causes cerebellar agenesis in the absence of Ptf1a function. Proc Natl Acad Sci USA. 2014;111(17):E1777–E1786. doi: 10.1073/pnas.1315024111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sellick GS, et al. Mutations in PTF1A cause pancreatic and cerebellar agenesis. Nat Genet. 2004;36(12):1301–1305. doi: 10.1038/ng1475. [DOI] [PubMed] [Google Scholar]
- 98.Iskusnykh IY, Steshina EY, Chizhikov VV. Loss of Ptf1a leads to a widespread cell-fate misspecification in the brainstem, affecting the development of somatosensory and viscerosensory nuclei. J Neurosci. 2016;36(9):2691–2710. doi: 10.1523/JNEUROSCI.2526-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yamada M, et al. Origin of climbing fiber neurons and their developmental dependence on Ptf1a. J Neurosci. 2007;27(41):10924–10934. doi: 10.1523/JNEUROSCI.1423-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fujiyama T, et al. Inhibitory and excitatory subtypes of cochlear nucleus neurons are defined by distinct bHLH transcription factors, Ptf1a and Atoh1. Development. 2009;136(12):2049–2058. doi: 10.1242/dev.033480. [DOI] [PubMed] [Google Scholar]
- 101.Fujiyama T, et al. Forebrain Ptf1a is required for sexual differentiation of the brain. Cell Rep. 2018;24(1):79–94. doi: 10.1016/j.celrep.2018.06.010. [DOI] [PubMed] [Google Scholar]
- 102.Nakamura N et al (2016) Neonatal kisspeptin is steroid-independently required for defeminisation and peripubertal kisspeptin-induced testosterone is required for masculinisation of the brain: a behavioural study using Kiss1 knockout rats. J Neuroendocrinol 28. 10.1111/jne.12409 [DOI] [PubMed]
- 103.Sasselli V, Pachnis V, Burns AJ. The enteric nervous system. Dev Biol. 2012;366(1):64–73. doi: 10.1016/j.ydbio.2012.01.012. [DOI] [PubMed] [Google Scholar]
- 104.Wakayama K, et al. Localization of norepinephrine and serotonin transporter in mouse brain capillary endothelial cells. Neurosci Res. 2002;44(2):173–180. doi: 10.1016/S0168-0102(02)00120-7. [DOI] [PubMed] [Google Scholar]
- 105.Takanaga H, et al. GAT2/BGT-1 as a system responsible for the transport of γ-aminobutyric acid at the mouse blood–brain barrier. J Cereb Blood Flow Metab. 2001;21(10):1232–1239. doi: 10.1097/00004647-200110000-00012. [DOI] [PubMed] [Google Scholar]
- 106.Pattyn A, et al. The homeobox gene Phox2b is essential for the development of autonomic neural crest derivatives. Nature. 1999;399(6734):366–370. doi: 10.1038/20700. [DOI] [PubMed] [Google Scholar]
- 107.Watanabe Y, et al. Sox10 and Itgb1 interaction in enteric neural crest cell migration. Dev Biol. 2013;379(1):92–106. doi: 10.1016/j.ydbio.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 108.Memic F, et al. Ascl1 is required for the development of specific neuronal subtypes in the enteric nervous system. J Neurosci. 2016;36(15):4339–4350. doi: 10.1523/JNEUROSCI.0202-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lei J, Howard MJ. Targeted deletion of Hand2 in enteric neural precursor cells affects its functions in neurogenesis, neurotransmitter specification and gangliogenesis, causing functional aganglionosis. Development. 2011;138(21):4789–4800. doi: 10.1242/dev.060053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Uribe RA, Gu T, Bronner ME. A novel subset of enteric neurons revealed by ptf1a:GFP in the developing zebrafish enteric nervous system. Genesis. 2016;54(3):123–128. doi: 10.1002/dvg.22927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vlangos CN, et al. Next-generation sequencing identifies the Danforth’s short tail mouse mutation as a retrotransposon insertion affecting Ptf1a expression. PLoS Genet. 2013;9(2):e1003205. doi: 10.1371/journal.pgen.1003205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Semba K, et al. Ectopic expression of Ptf1a induces spinal defects, urogenital defects, and anorectal malformations in Danforth’s short tail mice. PLoS Genet. 2013;9(2):e1003204. doi: 10.1371/journal.pgen.1003204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lugani F, et al. A retrotransposon insertion in the 5′ regulatory domain of Ptf1a results in ectopic gene expression and multiple congenital defects in Danforth’s short tail mouse. PLoS Genet. 2013;9(2):e1003206. doi: 10.1371/journal.pgen.1003206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tutak E, et al. A Turkish newborn infant with cerebellar agenesis/neonatal diabetes mellitus and PTF1A mutation. Genet Couns. 2009;20(2):147–152. [PubMed] [Google Scholar]
- 115.Al-Shammari M, et al. A novel PTF1A mutation in a patient with severe pancreatic and cerebellar involvement. Clin Genet. 2011;80(2):196–198. doi: 10.1111/j.1399-0004.2010.01613.x. [DOI] [PubMed] [Google Scholar]
- 116.Houghton JA, et al. Isolated pancreatic aplasia due to a hypomorphic PTF1A mutation. Diabetes. 2016;65(9):2810–2815. doi: 10.2337/db15-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gabbay M, et al. Pancreatic agenesis due to compound heterozygosity for a novel enhancer and truncating mutation in the PTF1A gene. J Clin Res Pediatr Endocrinol. 2017;9(3):274–277. doi: 10.4274/jcrpe.4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Krah NM, et al. The acinar differentiation determinant PTF1A inhibits initiation of pancreatic ductal adenocarcinoma. Elife. 2015;4:e07125. doi: 10.7554/eLife.07125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Aichler M, et al. Origin of pancreatic ductal adenocarcinoma from atypical flat lesions: a comparative study in transgenic mice and human tissues. J Pathol. 2012;226(5):723–734. doi: 10.1002/path.3017. [DOI] [PubMed] [Google Scholar]
- 120.Naqvi AAT, Hasan GM, Hassan MI. Investigating the role of transcription factors of pancreas development in pancreatic cancer. Pancreatology. 2018;18(2):184–190. doi: 10.1016/j.pan.2017.12.013. [DOI] [PubMed] [Google Scholar]
- 121.Nair GG, Vincent RK, Odorico JS. Ectopic Ptf1a expression in murine ESCs potentiates endocrine differentiation and models pancreas development in vitro. Stem Cells. 2014;32(5):1195–1207. doi: 10.1002/stem.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Xiao D, et al. Direct reprogramming of fibroblasts into neural stem cells by single non-neural progenitor transcription factor Ptf1a. Nat Commun. 2018;9(1):2865. doi: 10.1038/s41467-018-05209-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ring KL, et al. Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell. 2012;11(1):100–109. doi: 10.1016/j.stem.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang J, et al. A role for dystonia-associated genes in spinal GABAergic interneuron circuitry. Cell Rep. 2017;21(3):666–678. doi: 10.1016/j.celrep.2017.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Seto Y, et al. Temporal identity transition from Purkinje cell progenitors to GABAergic interneuron progenitors in the cerebellum. Nat Commun. 2014;5:3337. doi: 10.1038/ncomms4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hamilton BA. Retrotransposon activates ectopic Ptf1 expression: a short tail. PLoS Genet. 2013;9(2):e1003331. doi: 10.1371/journal.pgen.1003331. [DOI] [PMC free article] [PubMed] [Google Scholar]