Abstract
Cardiovascular diseases represent one of the most important health problems of developed countries. One of the main actors involved in the onset and development of cardiovascular diseases is the increased production of reactive oxygen species that, through lipid peroxidation, protein oxidation and DNA damage, induce oxidative stress and cell death. Basic and clinical research are ongoing to better understand the endogenous antioxidant mechanisms that counteract oxidative stress, which may allow to identify a possible therapeutic targeting/application in the field of stress-dependent cardiovascular pathologies. In this context, increasing attention is paid to the glutathione/glutathione-peroxidase and to the thioredoxin/thioredoxin-reductase systems, among the most potent endogenous antioxidative systems. These key enzymes, belonging to the selenoprotein family, have a well-established function in the regulation of the oxidative cell balance. The aim of the present review was to highlight the role of selenoproteins in cardiovascular diseases, introducing the emerging cardioprotective role of endoplasmic reticulum-resident members and in particular one of them, namely selenoprotein T or SELENOT. Accumulating evidence indicates that the dysfunction of different selenoproteins is involved in the susceptibility to oxidative stress and its associated cardiovascular alterations, such as congestive heart failure, coronary diseases, impaired cardiac structure and function. Some of them are under investigation as useful pathological biomarkers. In addition, SELENOT exhibited intriguing cardioprotective effects by reducing the cardiac ischemic damage, in terms of infarct size and performance. In conclusion, selenoproteins could represent valuable targets to treat and diagnose cardiovascular diseases secondary to oxidative stress, opening a new avenue in the field of related therapeutic strategies.
Keywords: Selenoproteins, Cardiovascular diseases, Endoplasmic reticulum, Biomarkers, Selenoprotein T
Introduction
Cardiovascular diseases (CVD) and their consequences remain the most common worldwide cause of death, accounting for more than 4 million deceases each year across Europe, and representing the most important health problem of the Western world [1, 2]. The major cardiac syndromes, myocardial infarction (MI) and heart failure (HF), are responsible for a large portion of CVD-dependent deaths; in these common diseases, cell death, occurring primarily by apoptosis or necrosis, is an important component of their pathogenesis [3]. Along with other factors, reactive oxygen species (ROS) are considered the initiators of the harm in CVD, being responsible for lipid peroxidation, protein oxidation and DNA damage. An excess of ROS results in oxidative stress and may cause cell death [4]. Accordingly, oxidative stress has been associated with diverse pathophysiological events, including CVD [5, 6].
MI is a leading cause of death in its acute phase, but the long-term morbidity and mortality are also alarmingly high [7]. Early and successful restoration of myocardial reperfusion after an ischemic event represents the most effective strategy to reduce infarct size, preserving the left ventricular systolic function and improving the clinical outcome. Paradoxically, the reperfusion strategy itself may induce further myocardial damage in a phenomenon named “lethal reperfusion-induced injury” [8, 9]. The subsequent condition results in inflammation and oxidative damage through the induction of oxidative stress rather than immediate restoration of normal function [7–9]. In addition, even if performing a successful reperfusion, mortality and morbidity are significant after an acute ST-segment elevation MI (STEMI) by electrocardiography analysis in a 1-year follow-up [10]. For these reasons, additional mechanical and pharmacological cardioprotective strategies, able to limit the reperfusion injury and further enhance the benefit of reperfusion, are urgently needed.
To preserve their integrity against oxidative stress-dependent damages and to repair deleterious structural oxidative modifications, cells employ potent endogenous antioxidative systems for ROS detoxification [11]. In mammalian cells, glutathione/glutathione peroxidase and thioredoxin/thioredoxin reductase systems constitute the principal endogenous antioxidant defense line involved in multiple redox-regulated signaling pathways [12–15]. The crucial enzymes belong to the selenoprotein family and display several fundamental functions mostly related to the oxidative cell balance control [12–15].
Several selenoproteins catalyse redox reactions by involving the oxidation of sulfhydryl groups and/or reduction of disulfides [16, 17]. As ROS-detoxifying enzymes, selenoproteins represent an array of antioxidants with different specific subcellular localization and chemical reactivities [18]. In particular, different experimental evidences show that selective selenoproteins contribute to redox regulation in CVD, displaying cardioprotection and representing promising therapeutic tools in the treatment of these pathological disorders [19, 20].
This review aims to focus on the specific function played by selenoproteins in CVD and to introduce the emerging role of one of them, namely Selenoprotein T (SELENOT), in cardioprotection, as the first results indicate that this member is crucial for the cardiovascular function.
The selenoprotein family
Distinctive structural and functional features of selenocysteine (Sec)
Selenium (Se) is an essential trace element co-translationally incorporated into the polypeptide chain as component of the amino acid selenocysteine (Sec), the 21st amino acid in the genetic code, which is encoded by TGA [21, 22]. It has been observed that selenoproteins are found in all kingdoms of life including certain types of fungi and viruses [16, 23, 24]. Proteins including Sec in their polypeptide chain are defined as selenoproteins [14].
Sulfur (S) and Se are members of the chalcogen group of elements having very similar chemical and physical properties, that can be involved in similar chemical reactions, such as thiol/disulfide and thiol-disulfide-like (that is selenol/disulfide or thiol/selenosulfide when Se replaces S) exchange reactions [25]. Among the trace elements used as cofactors by enzymes, Se has unique properties; in fact, unlike other metal cofactors such as zinc and copper, Se is not only a part of the polypeptide chain in the form of an amino acid, but Sec can be also localized in the enzymatic catalytic site [16]. In selenoproteins, Sec function is partially preserved only when cysteine (Cys) replaces Sec; however, in most cases, the substitution of Sec with Cys leads to a detrimental reduction in the catalytic efficiency [17], as demonstrated by Zhong et al. [26] in a study by mutagenesis carried out on the rat thioredoxin reductase (TXNRD). The higher importance of Sec compared to Cys in protein function, is a debated issue, but a common view suggests the ability of Se to provide some superior chemical or physical properties able to improve the functions of the macromolecules [17]. Accordingly, unlike other amino acids, Sec is normally used only when required for protein function, supporting the idea that this amino acid is a key functional (catalytic) group in proteins. [16, 27, 28].
Additionally, Sec could have a protective effect in proteins. In fact, the one-electron oxidized product of Sec (selanyl radical), is less oxidizing compared to the cysteine-thiyl radical [29]. Consequently, the selanyl radical is not sufficient to alter proteins, while cysteine-thiyl radical can do this with a higher efficiency [29]. Accordingly, Bianco et al. [30] reported that, similarly to thiols, selenols are also target for nitroxyl (azanone, HNO); however, while HNO-induced modifications of thiols can be either reversible and irreversible, HNO-induced modifications of selenols appear to be only irreversible [30]. Other studies provided evidences about the biochemical significance of Sec-containing enzymes under oxidative conditions. For example, Snider et al. [31] compared the effect induced by several oxidant species [ROS, reactive nitrogen species (RNS) and reactive halogen oxidants] exposition on the Sec-containing TXNRD with respect to the Cys-ortholog TXNRD. These authors clearly demonstrated that the Sec-containing TXNRD results in a significantly higher capacity to resist to inactivation by oxidation, while the Cys-containing TXNRD was inactivated under the same conditions of oxidative stress.
Very recently, Ingold et al. [32] further contributed to the knowledge of the function exerted by Sec, providing additional evidence for the advantage to use Se in the form of Sec in proteins. To show this, they compared the selenolate- versus thiolate-based catalysis in mice with a targeted mutation of the catalytically active Sec to Cys in glutathione peroxidase 4 (GPX4), demonstrating that Sec-containing GPX4 prevents ferroptosis due to its resistance to irreversible inactivation mediated by overoxidation.
These findings indicate that, unlike thiol containing proteins, selenoproteins are resistant to irreversible oxidative modification, reflecting the advantage of selenolate- versus thiolate-based catalysis.
The higher reactivity of Sec compared to Cys is also indicated by the higher catalytic activity exerted by the Sec-containing enzymes, which are typically 100- to 1000-fold more active than their Cys mutants [33]. This could account for the presence of Sec in biological systems and might be the rationale of Sec selection over Cys in specific enzymes [17].
Biologically, the unique property of Sec is that, unlike the other amino acids, its biosynthesis always occurs on its own selenocysteine-specific tRNA, designated tRNA[Ser]Sec, that controls the expression of all the Selenoprotein family, a process that is not reported for any other tRNA species [16, 27]. The gene for tRNA[Ser]Sec is designated as Trsp [34, 35] and Sec incorporation during protein synthesis is dependent on both a UGA codon in the open reading frame and a selenocysteine insertion sequence (SECIS) in the 3′-untranslated region [34]. Since two different homozygous mutations found in the human Sep (O-phosphoserine) tRNA:Sec tRNA synthase (SEPSECS) gene, encoding for an enzyme crucially involved in the selenocysteyl-tRNA[Ser]Sec synthesis [36], are associated with progressive cerebello-cerebral atrophy, an essential functional role of Sec biosynthesis can be postulated. Other mutations in SEPSECS gene have been identified in several ethnicities in the world [37, 38]. These mutations are able to generate a phenotype which is clinically similar among all patients and is now classified as ponto-cerebellar hypoplasia type 2D (PCH2D), leading to decreased selenoprotein levels and to an augmented brain susceptibility to oxidative stress [36, 39, 40].
Mutations of two other genes involved in the Sec insertion pathway are responsible for pathological conditions. In particular, mutations in SECISBP2 (SECIS binding protein 2) gene and TRU-TCA1-1 (transfer RNA-Sec [TCA] 1-1) gene, encoding tRNA[Ser]Sec, cause a multisystem disorder with myopathic features, induced by the loss of selenoprotein N (SELENON), increased oxidative stress due to the alteration of crucial antioxidant selenoproteins (i.e. GPXs and TXNRDs) and thyroid dysfunction induced by iodothyronine deiodinases (DIOs) deficiency [39–44]. Additionally, Trsp gene knockout in mice results in embryonic lethality, indicating that selenoprotein synthesis is strictly required for mammalian life [45].
Selenoproteins and the selenoproteome
The first identified selenoprotein was the mammalian glutathione peroxidase 1 (GPX1) [46]. Subsequently, thanks to a combination of bioinformatic and experimental approaches, a large number of selenoproteins were identified in all the life kingdoms, and the full human selenoproteome, encoded by 25 selenoprotein genes was characterized [27]. Among these genes are included the glutathione peroxidases (GPXs), the thioredoxin reductases (TXNRDs) and the iodothyronine deiodinases (DIOs) [16].
GPXs are major components of the antioxidant defense and redox homeostasis, strongly involved in a wide range of physiological functions [14]. They use glutathione as a co-factor to catalyze the reduction of hydrogen peroxide (H2O2) and/or phospholipid hydroperoxide [14, 47]. In humans, there are eight genes encoding GPX enzymes and five of these are selenoproteins (GPX1–4, GPX6) containing Sec in the active site [27, 47].
TXNRDs, three of which have been identified in mammals (TXNRD 1–3) are members of the pyridine nucleotide-disulfide oxidoreductases that, together with thioredoxin (Trx) and NADPH, represent the major disulphide reduction system of the cell [15, 16]. The two most important features of TXNRDs are the N-terminal redox-active dithiol/disulfide in one subunit [48] and a 16-residue C-terminal elongation with the conserved selenothiol active-site in the adjacent subunit (sequence: -Gly-Cys-Sec-Gly-OH) [49]. This structure forms the redox-active center of the enzyme [48]. Several other proteins having these characteristic structural features are considered members of the Trx family.
The three mammalian DIOs (DIO1-3) are vitally involved in the regulation (activation/deactivation) of thyroid hormone activity by reductive deiodination [16]. DIO1 or DIO2 catalyze the activation of thyroid hormone by converting the prohormone thyroxine (3,5,3′,5′ tetraiodothyronine, T4) to the biologically active form, 3,5,3′-triiodothyronine (T3), while DIO3 catalyzes the irreversible inactivation of T3 and T4 to yield the inactive hormones 3,5-diiodo-l-thyronine (T2) and reverse T3 (rT3), respectively [50].
Overall, TXNRDs, GPXs and DIOs are the three-best characterized selenoprotein subfamilies. They have different enzymatic activities, but all require reductants to provide the electrons for their catalytic redox cycle [14].
The human selenoproteome also includes the methionine sulfoxide reductase B1 (MSRB1) that, together with TXNRD and GPX isoforms, synergically act to provide antioxidant defense [16]. While two other selenoproteins are crucially involved in selenoprotein synthesis (selenophosphate synthetase 2, SEPHS2) and transport (selenoprotein P, SELENOP) [16, 28], the role of many selenoproteins and their mechanisms of action (referring to selenoprotein W, SELENOW, selenoprotein T, SELENOT, selenoprotein H, SELENOH, selenoprotein V, SELENOV, selenoprotein I, SELENOI, selenoprotein F, SELENOF, selenoprotein M, SELENOM, selenoprotein K, SELENOK, selenoprotein S, SELENOS, selenoprotein O, SELENOO, and selenoprotein N, SELENON [16, 27], are still elusive.
However, the role of specific selenoproteins is emerging. After the complete identification of the selenoprotein family members in 2003 [27], which occurred after the Human Genome Project, their nomenclature was revised in 2016 [51]. Selenoproteins with characterized functions are designated according to their functions (GPXs, TXNRDs, DIOs, MSRB1 and SEPHS2). Selenoproteins without a clearly demonstrated enzymatic function, structure, tissue and cellular localization, are indicated by the common symbol SELENO followed by a specific letter [51]. Some of them, containing structures encompassing Trx folds (where Cys residues, normally present in the active site, are replaced by Sec residues), namely Trx-like proteins, include SELENOF, SELENOH, SELENOM, SELENOO, SELENOP, SELENOT, SELENOV, and SELENOW, and act in a Trx similar manner also contributing to the Trx system activity [47]. Considering the presence of the Trx fold, a thiol-based oxidoreductase activity has been postulated for these SELENO members [52]. Among them, SELENOP possesses unique features, i.e. it is the only selenoprotein with multiple Sec residues that facilitates Se transport between organs [53]. In addition, since SELENOP is transported across the blood–brain barrier [47, 54] it is the most commonly used selenoprotein as a marker of Se status [55, 56].
However, an impressive emerging interest has been given to the group of the endoplasmic reticulum (ER)-resident selenoproteins (SELENOF, SELENOK, SELENOM, SELENON, SELENOS, SELENOT). These selenoproteins predominantly contribute to the calcium ion (Ca2+) signaling, the protein folding and ER-associated degradation [57–61]. In particular, Hamieh et al. demonstrated that SELENOT knockdown is dramatically associated with the unfolded protein response (UPR) and ER stress and lowers ER-associated protein degradation (ERAD) and hormone production [62].
Cardiovascular role of selenoproteins
Selective selenoproteins are essential for life
It is well known that, under aerobic condition, cells continuously react with ROS, such as superoxide radical (O2·–), hydrogen peroxide (H2O2), lipid and hydroxyl radical (OH·), which derive from several metabolic reactions, and counteract their effects through a wide range of endogenous antioxidant enzymes [63]. High and persistent ROS levels lead to a dramatic imbalance between ROS and the antioxidant defence system [64]. Under these conditions, ROS can damage macromolecules due to their ability to react with specific amino acid residues present in the protein structure, thus inactivating their function, and with DNA and chromatin causing mutations or double-stranded breaks in a phenomena overall known as “oxidative damage” [65]. The current concept of “oxidative damage or oxidative stress” includes pathways also related to the “nitrosative stress” and, considering their implication in cellular and extracellular metabolic processes, to the “metabolic stress” [66]. So far, the definition of oxidative stress, firstly confined only to ROS, has been extended to RNS, such as nitric oxide (NO), peroxynitrite (ONOO−) and S-nitrosothiols (RSNO) [66]. Today, it is widely accepted that metabolic stress has been associated with diverse patho-physiological events, including CVD [6].
In this context, while the role of many selenoproteins remains to be better established and their study is under continuous investigation and development, the fundamental role exerted by selenoproteins for life is widely accepted [45, 67]. These enzymes rapidly consume and neutralize H2O2, with a consequent significant decrease of H2O2 levels and a limitation of its transmission signal space by diffusion in the cell [68].
An important contribution regarding the mechanism by which selenoproteins are involved in antioxidant defense following oxidative stress was given by Touat-Hamici et al. [69]. These authors studied, in different cell models, the regulation of the expression of several selenoproteins secondary to H2O2-induced oxidative stress, in the presence of high and low selenium concentration. Interestingly, the results demonstrated that antioxidant selenoproteins, including GPX1, GPX4, TXNRD1, SELENOS, SELENOK, SEPHS2, are up-regulated by H2O2 when selenium is limiting. Conversely, in the presence of high selenium concentration, selenoprotein expression was not sensitive or was down-regulated in response to oxidative stress [28, 70]. These observations indicate that selenium status selectively influences the response of selenoprotein expression to oxidative stress, which induces the upregulation of UGA selenocysteine recoding efficiency and relocalization of SBP2, EFsec and L30 recoding factors from cytoplasm to nucleus [69].
In vivo studies using knockout/transgenic animal models demonstrated that selenoproteins are crucially involved in protecting the cells against oxidative stress and in maintaining the redox homeostasis [71, 72].
For instance, it has been shown that, in mice, the genetic disruption of GPX4, TXNRD1, and TXNRD2 is responsible for an embryonic lethal phenotype [73–76]. In addition, depletion of the liver SELENOP gene alters the delivery of Se to peripheral target tissues, leading to growth retardation and impaired motor coordination in mice [77, 78]. Interestingly, the more recently discovered SELENOT is the only ER-resident selenoprotein so far whose genetic ablation causes embryonic lethality in mice [79, 80].
On the other hand, genetic invalidation of other selenoproteins (for example GPX1 and MSRB1 [81, 82]) revealed that they play a key role under stressful conditions.
Noteworthy, it has been observed that a reduced selenoprotein activity is counterbalanced by a cytoprotective response mediated by the transcription factor NF-E2 related factor 2 (Nrf2), which may represent an essential compensatory response for maintaining cellular redox homeostasis and viability [67, 83]. In the same study, the importance of Trsp/Nrf2 axis was further highlighted by demonstrating the key role of selenoprotein activity and of Nrf2 gene in the oxidative homeostasis of erythrocytes and in the prevention of hemolytic anemia [84].
Selective selenoproteins are required for heart development and function
As could be expected, mutations in selenoproteins and selenoenzymes have been associated with several disorders, including CVD [85]. Considering that the mouse tRNASecknockout mutant exhibits an embryonic lethal phenotype [45], it is not surprising that mutations of factors taking-part in the synthesis and co-translational incorporation of Sec, such as SEPSECS can affect the selenoprotein synthesis machinery itself [36].
Accordingly, the cell-specific alteration of selenoprotein expression, using the Cre-LoxP technology to remove the Trsp gene, revealed the essential role of selenoproteins in endothelial cell development and in proper cardiac muscle function, suggesting a direct connection between the loss of selenoprotein expression in this cell type and CVD [86]. In particular, and as reviewed by Conrad et al. [73], genetic disruption in mice of the cytosolic thioredoxin reductase (TXNRD1), the mitochondrial thioredoxin reductase (TXNRD2) and glutathione peroxidase 4 (GPX4), provokes embryonic lethality, thus revealing that selonoproteins are required not only for murine embryogenesis, but also for the regulation of cellular redox metabolism [73]. These findings are extremely relevant from a cardiovascular point of view. In fact, the knockout of TXNRD2, which exerts an essential role in hematopoiesis, heart development, and heart function, provokes congestive HF symptoms and postnatal death in a heart-specific TXNRD2-knockout mouse model [75]. It is of particular interest that these mice also showed phenotypic similarities with the histopathologic phenotype of Friedreich’s cardiomyopathy, that itself reflects the dilated congestive cardiomyopathy typical of severely Se-deficient patients [87]. Accordingly, the same authors described mutations in the TXNRD2 gene in a fraction of patients affected by dilated cardiomyopathy, thus revealing a novel mechanism for HF development linked to a crucial antioxidant enzyme involved in the regulation of cellular redox state [88]. On the other hand, Prasad et al. [89] identified homozygosity for a nonsense mutation in the TXNRD2 gene associated with familial glucocorticoid deficiency (FGD), a rare autosomal recessive disorder characterized by isolated glucocorticoid deficiency without mineralocorticoid deficiency [90]. The study was carried out in an extended consanguineous Kashmiri kindred and the results attest for the fundamental role of this selenoprotein in maintaining the adrenocortical redox state [89]. In the family involved in this study, heterozygotes individuals were clinically unaffected and, unexpectedly, neither the heterozygote nor homozygote subjects, with the absence of TXNRD2, had any evidence of cardiomyopathy, except two homozygous patients who presented cardiac anomalies, probably due to other genetic or environmental factors [38, 89].
Other studies identified a critical role for TXNRD2 in preserving the morphological and functional integrity of mitochondria in aging cardiomyocytes, pointing to a critical role of this selenoprotein in the prevention of age-related functional cardiac decline [91].
Embryonic depletion of GPX isoforms is also associated with cardiac dysfunction. Indeed, mice with homozygous null mutation for the cytosolic GPX1, the most abundant GPX, show susceptibility to oxidative stress and viral myocarditis and acceleration of cardiac hypertrophy and dysfunction [92]. Wentzel et al. [93] observed that maternal diabetes causes congenital malformations and a general altered antioxidant embryonic profile in rats, and that the decreased cardiac GPX1 levels significantly contribute to an increased risk of developing congenital cardiac malformation [93]. Forgione et al. [94] demonstrated that heterozygous deficiency of GPX1 leads to endothelial dysfunction, which in turn triggers significant structural abnormalities in vascular and cardiac tissues [94]. Furthermore, mice with GPX1 deficiency exhibit a decrease in bioavailable NO [94].
As demonstrated, the gene of human GPX1 is located on chromosome 3p21.3 and contains two exons [95, 96]. An increasing body of clinical studies showed the occurrence of different polymorphisms in GPX1 associated with negative effects on the cardiovascular system, confirming the detrimental consequences linked to GPX1 alteration in the heart. In this regard, Winter et al. [97] observed that individuals with one or two ALA6 alleles of GPX1 have a modest increased risk of coronary artery disease. Functional variants in the GPX1 gene were associated with increased intima-media thickness of carotid arteries and risk of cardiovascular and peripheral vascular diseases in type 2 diabetic patients [98]. Among other single nucleotide polymorphisms (SNPs) that have been documented for this gene, the polymorphisms of the cytosine-to-thymine (C>T) substitution (rs1050450) at codon 198 and 197 generate the Pro198Leu and Pro197Leu variations [99]. Several observations suggest a possible correlation between these polymorphisms and CVD. Accordingly, a Pro197Leu substitution of the GPX1 gene correlates with a significant genetic susceptibility to coronary-arteriosclerosis in type 2 diabetes patients [100]. In line with results obtained by Hamanishi et al. [98], Oguri et al. [101] showed that the Pro198Leu polymorphism of GPX1 significantly associated with in-stent restenosis in a Japanese population. A meta-analysis study conducted by Zhang et al. [102] revealed a significant relationship of these GPX1 variants and CVD risk in East Asian populations. and a hospital-based case–control study, in a Chinese population, indicates that the GPX1 variant 198Leu allele is significantly associated with an increased risk of CAD [103]. Although other studies to strengthen statistical significance are needed, the effect of the GPX1 Pro/Leu variants on the function of this selenoenzyme is substantial; in fact, several studies indicate a lower GPX1 activity in the Pro198Leu variant [98, 104, 105], suggesting that the correlation found between this functional variant and the increased risk of CVD, in particular CAD, might be related to a decreased GPX1 enzyme activity [104, 105].
GPX3 is the major plasma antioxidant enzyme, which controls the vascular tone and the thrombotic properties of the vascular endothelium [106]. A GPX3 deficiency inevitably leads to an impaired catalytic reduction of H2O2 and organic hydroperoxides to their corresponding alcohols [107]. Consequently, there is a decreased NO bioavailability [108, 109] that can augment platelet activation and arterial thrombosis [110]. The potential relationship between GPX3 deficiency and platelet-dependent thrombosis was addressed by Jin et al. [107], who developed a mouse model presenting a phenotype resembling that of two brothers with GPX3 deficiency, with arterial thrombosis and stroke syndromes [111]. GPX3 deficiency alters ROS metabolism, resulting in a pro-thrombotic state and vascular dysfunction, and promotes platelet-dependent arterial thrombosis [111].
These results are corroborated by the correlation found between decreased GPX3 activity and arterial thrombosis in humans [112]. Starting from idiopathic childhood stroke patients, whose clinical manifestations and thrombosis were in a familiar GPX3 and NO bioavailability reduction [111, 113], the authors hypothesized that mutation(s) or polymorphism(s) in the plasma GPX3 gene promoter may be responsible for the reduction in enzyme activity and predispose to a thrombotic disorder. Accordingly, in patients with arterial ischemic stroke, a novel GPX3 promoter haplotype reduces the gene expression with consequences typical of the lack of a crucial antithrombotic and antioxidant key enzyme [111, 113]. Of note, mice with embryonic knockdown of phospholipid hydroperoxide GPX (GPX4), the main antioxidant enzyme known to directly inhibit lipid peroxidation by reducing phospholipid hydroperoxides and lipoproteins within membranes, exhibit abnormal development of the heart, lacking the left atrium [114]. Interestingly, in support to the essential role displayed by this enzyme in the development of the cardiac system, Smith et al. [115] identified selective mutations of GPX4 as responsible of Sedaghatian type of spondylometaphyseal chondrodysplasia (SSDM), a neonatal lethal form of spondylometaphyseal dysplasia described as a new autosomal recessive disorder in 1980 [116]. Accordingly, SSDM patients die prematurely by cardio-respiratory failure [117]. In particular, patients present with severe hypotonia and cardiorespiratory problems, most die within days of birth due to respiratory failure and, as showed in an article by Aygun et al. and reference therein [117], several patients presented cardiac conduction defects, complete heart block and different structural cardiac anomalies [38, 115].
While the essential role of TXNRDs and GPXs in the heart development and function is well established, promising results have also been obtained for another selenoprotein, MSRB1 [118]. MSRB1, located in the cytosol and nucleus, reduces the enantiomer R of methionine sulfoxide, R-MetO [119–121]. In 2009, Fomenko et al. [82] found that the knock-out of MSRB1 in mice leads to potent oxidative stress in several organs, such as liver and kidney that highly express MSRB1 under physiological conditions, but to a lesser extent in the heart [82].
Glutathione peroxidases and thioredoxin reductases contribute to redox regulation and cardioprotection in cardiovascular disease
The expression of selenoproteins during cardiogenesis, and during/after stressful events, and their antioxidant function, is complex and intriguing. In cardiac adaptation to different stimuli, the evaluation of selenoprotein expression patterns and enzymatic activities under physiological and pathological conditions may provide important information about the role of this enzyme in counteracting cardiac sufferance [19]; above all, a specific role of the selenoproteomic response in protecting the heart against stress-induced damage represents an ambitious goal for basic and translational research.
In this regard, studies by Hoffman et al. [122], provided insights in the expression and the enzymatic activity of key cardioprotective selenoproteins during oxidative stress-dependent cardiac stress. In particular, these authors reported significant increases of mRNA and protein levels of MSRB1 [123], TXNRD1, GPX3 and GPX4 in mouse models of T3- or isoproterenol-induced myocardial hypertrophy. Along with the results described above showing the cardiac abnormalities after genetic disruption of selective selenoproteins, these data suggest a significant induction of the selenoproteome response to cardiac damage, where some selenoproteins seem to be indispensable for cardioprotection in circumstances of oxidative injury.
GPXs and TXNRDs are certainly among those antioxidant enzymes most involved in cardiovascular pathologies [12–14, 124]. The direct role of GPX1 in myocardial I/R, endothelial dysfunction, athero-thrombotic vascular disease, and cardiotoxicity induced by chemotherapeutic agents has been extensively studied in in vitro and in vivo models [125]. Among the many studies related to the cardioprotective potential of GPX1, a work published in The New England Journal of Medicine by Blankenberg et al. [126] examined the protective effect of GPX1 against cardiovascular events in a large prospective cohort of patients with coronary artery disease. The authors found that low erythrocyte GPX1 activity [126] identifies patients with coronary artery disease who are at the highest risk for cardiovascular events [126]. These results indicate that the GPX1 activity assay provides a key information for identifying patients who would benefit from preventive antioxidant treatment.
In addition to GPX1, GPX3 (plasmatic isoform of GPX) also affords heart and vasculature protection against oxidative stress in a NO-dependent mechanism, contributing to preserve the platelet function in human arterial thrombosis [125]. Noteworthy a recent study carried out by Pastori et al., provides novel evidence regarding the association between reduced levels of GPX3 and increased risk of cardiovascular events in patients with atrial fibrillation. The authors also found a negative correlation between age and GPX3 levels, proposing the GPX3 decline as a possible mechanism for the enhanced cardiovascular risk in the elder population [127]. Besides, GPX4 protects lipids from oxidative damage during myocardial hypertrophy and I/R injury [125]. Finally, the roles displayed by the Trx and TXNRD system in cardiovascular events have been extensively reviewed, and the crucial participation of these redox-regulating and sensitive enzymes in the control of various cardiac functions is well established.
Cardiovascular effects of the emerging endoplasmic reticulum-resident selenoproteins
Selenoprotein S
An important role is played by the ER-associated SELENOS in cardiac function. SELENOS is also located in the plasma membrane and is involved in the retro-translocation of misfolded proteins from the ER lumen to the cytosol for degradation [128], and thus in cell protection against oxidative stress and inflammation, typical events of the misfolded protein response [129].
SELENOS seems to be particularly linked to inflammatory processes [130, 131]. Variations in the SELENOS gene influence the circulating levels of the inflammatory cytokines interleukin 1 beta (IL-1β), interleukin 6 (IL-6) and tumor necrosis factor alpha (TNF-α) [130], and the SELENOS gene expression is activated by inflammatory cytokines [131]. A case-cohort design and time-to-event analysis demonstrated that variation in the SELENOS locus may be associated with specific risk factors for CVD [132].
In 2013, in a large number of European Americans from the family-based Diabetes Heart Study, the association between SELENOS genetic variants and the risk for subclinical CVD and mortality in type 2 diabetes patients was postulated [133]. Considering the link of SELENOS gene variation and inflammatory response, the involvement of this selenoprotein in type 2 diabetes and HF should not be surprising. Accordingly, an association of SELENOS polymorphisms with coronary and carotid calcified plaque, as well as with glycemia and glycated hemoglobin (HbA1C) was observed [133].
A recent study provides evidence about selective alleles of SELENOS gene (i.e. rs34713741T) associated with an increased risk of peripheral arterial disease, while other variants (i.e. rs3877899A) seem to prevent the risk for developing abdominal aortic aneurysm [134]. Of course, all these observations deserve further genetic investigations to better clarify the role of this gene and protein in the pathogenesis of these diseases.
Selenoprotein K
Another ER transmembrane selenoprotein that appears to have multiple biological functions, including cardiac influences, is SELENOK. There is evidence that SELENOK mRNA is highly expressed in human heart and that its overexpression might reduce ROS production protecting cardiomyocytes from oxidative stress-induced by H2O2 exposure [135]. In addition, purified human SELENOK has been shown to reduce phospholipid hydroperoxides [136]. SELENOK, through interaction with other protein factors, may participate to the transport of glycosylated misfolded proteins and to the regulation of ER stress in particular cell types, contributing to ER homeostasis [137, 138]. For this reason, the effects observed in cardiomyocytes suggest a potential antioxidant action of SELENOK by regulating ER stress, a key process highly sensitive to oxidative stress.
On the other hand, Verma et al. found that SELENOK is strongly expressed in cells of the immune system, where it exerts an important role for their function and activation [139]. A knockout mice model for SELENOK revealed serious impaired activities of immune response activation, including impairment in cell migration, proliferation, and oxidative status, particularly evident for macrophages [139, 140]. It has also been observed that SELENOK deficiency inhibits the uptake of modified low density lipoprotein (LDL), which may lead to foam cell formation in primary macrophages, and inhibits the TNF-α-induced scavenger receptor CD36 surface expression [141]. These results suggest the important role of SELENOK in the expression of CD36 in macrophages during inflammation, thereby contributing to foam cell formation and atherogenesis, significant inflammatory processes also associated with cardiovascular complication [142].
However, the limited number of studies related to SELENOK in the heart, and the lack of in vivo analysis of its cardiac antioxidant potential warrant further studies to decipher its precise cardiovascular implication.
Other endoplasmic reticulum-resident selenoproteins: focus on SELENOT
The results obtained on the cardiac involvement of SELENOS and SELENOK suggest that other ER-resident selenoproteins (SELENOM, SELENON, SELENOT, and SELENOF) could be involved in ER homeostasis in the heart.
As critically reviewed by Pitts and Hoffmann [57], these selenoproteins are involved in calcium flux modulation into and from the ER lumen, in the regulation of protein folding and maturation, and in ER redox homeostasis. Through their antioxidant activity, displayed by a Sec residue in the active site, the ER-resident selenoproteins participate to fundamental biological processes such as proliferation, survival, and apoptosis [57].
Noteworthy, type 2 iodothyronine deiodinase (DIO2), a classical type-1 membrane protein residing on the ER membrane and the main T4-activating enzyme [143, 144], is very sensitive to ER stress [145]. In fact, pharmacologically induced dysregulation of the ER status triggers a rapid loss of DIO2 activity and protein via a posttranscriptional mechanism that leads to a decrease in intracellular T3 production [145].
However, from a cardiovascular point of view, limited results have been obtained so far for this subfamily of selenoproteins (particularly for SELENOM, SELENON, and SELENOF). Since ER stress (in terms of disrupted ER homoeostasis and ER altered functions) is a key process regulating death/survival mechanisms of cardiomyocytes under several types of stressful stimuli [146], the research in this field appears mandatory and may lead to an increase of the potential clinical-therapeutic impact.
Several studies indicate that ER stress is involved in the development and progression of diverse heart diseases, such as cardiac hypertrophy, ischaemic heart diseases and HF [146]. In this context, an emerging selenoprotein able to prevent ER-stress is SELENOT. After the discovery of selenoprotein R (SELENOR), which is currently known as MSRB1 [147], SELENOT was the second selonoprotein identified using a bioinformatic approach [61]. The identification of SELENOT as an up-regulated gene during neuroendocrine cell differentiation in response to the neurotrophic factor pituitary adenylate cyclase-activating polypeptide (PACAP) [148] paved the way for numerous studies. In this regard, the critical review by Anouar et al. recently updated the findings that have been reported for SELENOT during the last years [61]. SELENOT displays a thioredoxin reductase-like enzymatic activity [79] and associated with the ER membrane; in this subcellular compartment, the protein can interact with other thiol-containing proteins through its Sec active moiety [60, 62, 148]. These features strongly suggest that SELENOT exerts a key redox function by controlling protein processing in the ER, thus contributing to ER homeostasis. In particular, through its thioredoxin-like fold, SELENOT may take part to the thiol redox circuits including thiol-disulfide oxidoreductase reactions and may participate to the regulation of protein folding and maturation [61].
Accordingly, Hamieh et al. reported that SELENOT is required for adaptation to the stressful conditions of high hormone level production in endocrine cells [62]. Indeed, the knockdown of the protein in corticotrope cells resulted in an unfolded protein response (UPR), in ER stress and ER altered function, thus identifying SELENOT as an indispensable effector of hormone maturation and secretion [62]. In the cardiac ER, the protein has been identified by immunofluorescence analysis, and co-localizes with calsequestrin-2 during rat cardiogenesis [149]. Our work revealed a strong expression of SELENOT in the cardiac ER at E7 (embryonic day 7) stage of embryogenesis, which decreased at P14 (post-natal day 14) and disappeared in the adult and mature heart. However, in the adult heart exposed ex vivo to 30 min of global, no-flow ischemia, followed by 120 min of reperfusion [myocardial ischemia/reperfusion (I/R) model], SELENOT expression was significantly induced, suggesting a functional re-activation of this protein during an important oxidative burst like that generated by I/R [149]. Therefore, it is very likely that SELENOT is not required for heart function under normal conditions, but its expression can be triggered after exposure to I/R injury. An increased SELENOT expression was also observed in a neurological context after exposure to noxious toxins that are known to induce a potent oxidative stress [79], confirming the importance of this protein in pathological conditions.
Rocca et al. also highlighted an intriguing role for SELENOT in direct cardiomodulation and cardioprotection (2018) [149]. The protective role of SELENOT in the context of myocardial I/R injury was further evaluated through postconditioning I/R protocols using a SELENOT-derived peptide, PSELT (FQICVSUGYR), that encompasses the active Sec-containing redox motif (CVSU) [149]. PSELT was able to improve the cardiac performance and to reduce the infarct size after I/R insult [149]. These effects were strictly related to the catalytic motif containing the Sec residue, since an analogous peptide lacking this residue did not exert such effects [149]. The mechanism that underlies these effects consist of the activation of protective pro-survival signalling cascades and the inhibition of pro-apoptotic factors [149]. Moreover, PSELT decreased ROS and RNS formation [149], which are known to be strongly implicated in I/R tissue damage [150, 151]. However, it is still unclear if this is a direct or indirect effect of the peptide.
Notably, these findings suggest that SELENOT/PSELT could represent a therapeutic tool able to provide an antioxidant protection in individuals at high risk of coronary heart disease.
As demonstrated by Grumolato et al. [60] and reviewed by Pitts and Hoffmann [57], SELENOT upregulation is dependent on cAMP levels and intracellular calcium flux during differentiation of pheochromocytoma cells in response to PACAP. In addition, these authors found that SELENOT plays an important role, via its redox activity, in maintaining calcium homeostasis and influencing proliferation, survival and apoptosis in neuronal and endocrine cells. SELENOT can modulate calcium signaling likely through a redox mechanism involving thiol groups on calcium channels and pumps [57, 60, 152].
Future studies to improve the knowledge about the mechanism of action exerted by SELENOT in an ischemic heart may allow a better understanding of a novel adaptive pathway by which cardiomyocytes could combat the calcium overload in the ischemic heart disease.
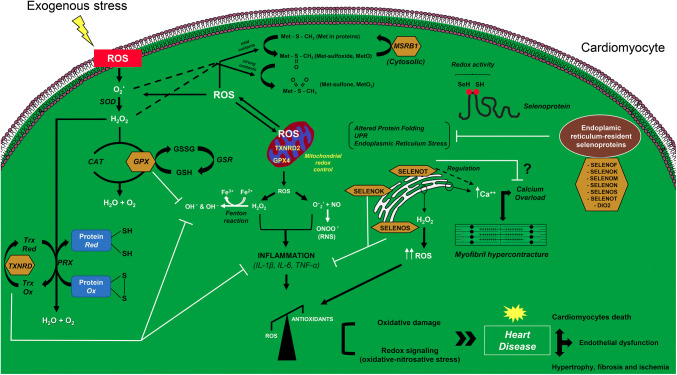
A comprehensive picture of the intracellular signaling involved in the selenoproteins action in cardiomyocytes is represented in Fig. 1.
Fig. 1.
Proposed model for the action of selenoproteins in cardiomyocytes, focusing on the redox state control in both cytosolic and mitochondrial compartments. The mechanism of action of the well-characterized selenoproteins thioredoxin reductase (TXNRD), glutathione peroxidase (GPX) and methionine sulfoxide reductase B1 (MSRB1) is illustrated. The mechanism of action of the emerging endoplasmic reticulum-resident selenoproteins in the heart, based on previous studies, is postulated. Selenoproteins are indicated with a yellow box. CAT catalase, DIO2 iodothyronine deiodinase type 2, GPX4 mitochondrial isoform of glutathione peroxidase, GSH glutathione, GSSG glutathione disulphide, GSR glutathione-disulfide reductase, H2O2 hydrogen peroxide, IL-1β interleukin-1β, IL-6 interleukin-6, Met methionine, MSRB1 methionine sulfoxide reductase B1, NO nitric oxide, O·2 superoxide radical, OH· hydroxyl radical, OH hydroxide ion, ONOO− peroxynitrite, PRX peroxiredoxin, RNS reactive nitrogen species, ROS reactive oxygen species, SELENOF selenoprotein F, SELENOK selenoprotein K, SELENOM selenoprotein M, SELENON selenoprotein N, SELENOS selenoprotein S, SELENOT selenoprotein T, SOD superoxide dismutase, TNF-α tumor necrosis factor-α, TrxOx oxidized thioredoxin, TrxRed reduced thioredoxin, TXNRD2 mitochondrial isoform of thioredoxin reductase, UPR unfolded protein response
Role of selenoprotein W
SELENOW is a 85–88 amino acid protein first identified in sheep suffering from selenium deficiency [153]. Its selenocysteine residue is located in the N-terminal portion of a relatively short functional domain [153] and different studies demonstrated a tissue-specific distribution of the protein [154]. Indeed, in sheep and primates SELENOW is highly expressed in the muscle, heart and brain [154]; on the contrary, in cardiac tissues from rodents the levels are very low [154]. SELENOW protein levels correlated with its mRNA levels but not with tissue selenium concentrations [155]. SELENOW is known to play a key antioxidative role in many cell types [156–158]. Interestingly, an alteration of SELENOW levels is able to influence the mRNA expression of other selenoproteins, maybe depending on the amount of ROS [159]. In a study performed by Yang et al. [160] on chicken embryo myocardial cells, SELENOW was sensitive to Se levels mediating, supposedly, its protective effects. Recently, it was found that a reduced Trx expression inhibits SELENOW in chicken cardiomyocytes demonstrating a close mutual relationship between these two systems [161].
Selenoproteins as biomarkers in cardiovascular diseases
Currently, a molecule is considered to be a biomarker if it represents a distinctive indicator of a biological process or condition [162]. A link between CVD and selenoproteins can be straightforwardly postulated since a relative Se deficiency is detected in different cardiac pathologies [163]. For example, the Se deficiency observed in patients with chronic HF [164] could be responsible of the inactivation of the selenoprotein enzymes TXNRD and GPXs [26]. Conversely, the cardiovascular protective effects of Se supplementation in populations with low serum levels have been widely discussed [165].
Owing to their properties, selenoproteins could represent good markers for cardiovascular pathologies related to oxidative stress. This is the case for preeclampsia [166], a condition characterized by high blood pressure in pregnancy that represents a leading cause of maternal and perinatal mortality and morbidity [167, 168]. Presumably, preeclampsia results from a reduced placental perfusion that increases ROS levels that, in turn, induce oxidative stress and endothelial cell dysfunction, resulting in hypertension and in the typical manifestations of preeclampsia [169]. Under normal conditions, the antioxidant selenoprotein GPX works by limiting these effects, but in preeclamptic women reduced plasma levels of Se [170, 171] and of plasma and placental GPX levels can be detected [166] and may represent useful biomarkers in this contest.
Oxidative stress has a well-known effect also in cerebrovascular events, such as stroke [172]. Reduced serum levels of SELENOP can be found in patients who underwent a stroke and are associated with a higher risk to develop this severe pathology [173]. This finding could be ascribed to the fact that SELENOP is the main Se supplier to the brain [174], and its decreased levels could reduce the protection against ROS through the Se-dependent antioxidant enzymes, leading to increased stroke risk [175]. A recent study assessed the plasma levels of SELENOP also in patients with metabolic syndrome accompanied by a history of CVD [176], and demonstrated that a decrease in circulating SELENOP levels also occurs in patients with documented CVD [176].
Thus, available data showing the potential use of selonoproteins as clinical biomarkers in cardiovascular dysfunction need to be further substantiated, but they already represent a promising starting point for translational medicine.
In addition, very recently, SELENOP plasma levels have been associated to acute MI, more precisely to its related inflammatory response and mortality in patients with cardiogenic shock [177]. In this pathological condition SELENOP levels increase in a significative manner and correlate with C-reactive protein levels, an inflammatory marker, suggesting its diagnostic potential in this context [177].
In Table 1 is reported a schematic representation of the major implications of selenoproteins in cardiovascular diseases.
Table 1.
Selenoproteins in cardiovascular diseases
| Selenoprotein | Localization | Biological function | Cardiovascular implication |
|---|---|---|---|
| TXNRD1 | Cytosol, nucleus [14]. | Oxidoreductase activity; antioxidant defense [21]. | knockout associated with embryonic lethal phenotypes; associated with hypertrophy and oxidative stress in response to pressure overload; implication in cardiovascular diseases [19, 73, 124]. |
| TXNRD2 | Mitochondria [14]. | Oxidoreductase activity; antioxidant defense [75]. | Involvement in hematopoiesis, heart development and heart function; congestive heart failure and postnatal death (knockout-model); dilated congestive cardiomyopathy (human); metabolic and contractile dysfunction [73, 75, 87, 88, 91]. |
| GPX1 | Ubiquitous, cytosol [14]. | Oxidoreductase and perodoxidase activities; antioxidant defense [14]. | Susceptibility to oxidative stress and viral myocarditis; acceleration of cardiac hypertrophy and dysfunction; increased risk of developing congenital cardiac malformation; structural abnormalities in vascular and cardiac tissues; modest increased risk of coronary artery disease; susceptibility to coronary-arteriosclerosis; protection against cardiovascular pathologies [92–94, 97, 98, 100, 122, 126]. |
| GPX3 | Plasma [14]. | Oxidoreductase and perodoxidase activities; antioxidant defense [14]. |
Decreased NO bioavailability; platelet activation and arterial thrombosis; stroke syndromes; control of the tone and the thrombotic properties of the vascular endothelium |
| GPX4 | Ubiquitous [14]. | Oxidoreductase and perodoxidase activities; antioxidant defense [14]. | knockout associated with embryonic lethal phenotypes; abnormal cardiac development; lack of the left atrium [73, 114]. |
| MSRB1 | Cytosol and nucleus [119]. | Oxidoreductase activity; antioxidant defense [14]. | Contribution to the redox control in several organs, but to a lesser extent in the heart [119]. |
| SELENOS | Endoplasmic reticulum; plasma membrane [128]. | Retro-translocation of misfolded proteins; antioxidant defense; endoplasmic reticulum stress response [128]. | Association with specific risk factors for cardiovascular diseases; risk for subclinical cardiovascular diseases and mortality in type 2 diabetes; coronary and carotid calcified plaque; risk of peripheral arterial disease [132–134]. |
| SELENOK | Endoplasmic reticulum [135]. | Antioxidant defense; regulation of endoplasmic reticulum stress [135–138]. | Association with impaired activities of immune response activation; atherogenesis; inflammation in cardiovascular diseases [139, 142]. |
| SELENOP | Plasma [14]. | Se transport; antioxidant defense [175]. | Correlation with metabolic syndrome and cardiovascular disease [176, 177]. |
| SELENOT | Endoplasmic reticulum [60]. | Antioxidant defense; intracellular Ca2+ regulation; regulation of endoplasmic reticulum stress [60, 62, 79]. | knockout associated with embryonic lethal phenotypes; increased expression in the cardiac embryogenesis; sensor of myocardial ischemia damage [79, 80, 149]. |
| SELENOW | Cytosol [52]. | Antioxidant defense [153]. | Myocardial oxidative damage [159]. |
Conclusions
Oxidative stress, due to increased ROS production or reduced ROS detoxification, is one of the leading causes of the onset and the progression of cardiovascular diseases. Potent endogenous antioxidant systems, such as selenoproteins, represent a fundamental defense mechanism able to counteract the detrimental effects of these noxious agents. Recent studies of the ER-resident selonoproteins, in particular SELENOT and its derivative peptide PSELT, pave the way for a possible therapeutic application in the ischemic heart, as post-conditioning agents. Further studies are needed to better clarify and describe the clinical potential of selenoproteins both as drugs and biomarkers, but the existing information already indicates that they represent promising factors in this field.
Acknowledgements
This work has been partially supported by “Fondazione Umberto Veronesi” Post-Doctoral Fellowship (2019) to TP and by Doctorate School in Life Science, University of Calabria.
Abbreviations
- CVDs
Cardiovascular diseases
- Cys
Cysteine
- DIO
Iodothyronine deiodinases
- ER
Endoplasmic reticulum
- ERAD
ER-associated protein degradation
- FGD
Familial glucocorticoid deficiency
- GPX
Glutathione peroxidase
- H2O2
Hydrogen peroxide
- HbA1c
Glycated hemoglobin
- HF
Heart failure
- HNO
Azanone
- I/R
Ischemia/reperfusion
- IL-1β
Interleukin-1β
- IL-6
Interleukin-6
- Met
Methionine
- MI
Myocardial infarction
- MSRB1
Methionine sulfoxide reductase B1
- NO
Nitric oxide
- O·2
Superoxide radical
- OH·
Hydroxyl radical
- ONOO-
Peroxynitrite
- PACAP
Pituitary adenylate cyclase-activating peptide
- PCH2D
Ponto-cerebellar hypoplasia type 2D
- RNS
Reactive nitrogen species
- ROS
Reactive oxygen species
- RSNO
S-Nitrosothiols
- S
Sulfur
- Se
Selenium
- Sec
Selenocysteine
- SECISBP2
SECIS binding protein 2
- SELENOH
Selenoprotein H
- SELENOI
Selenoprotein I
- SELENOK
Selenoprotein K
- SELENOM
Selenoprotein M
- SELENON
Selenoprotein N
- SELENOO
Selenoprotein O
- SELENOP
Selenoprotein P
- SELENOS
Selenoprotein S
- SELENOT
Selenoprotein T
- SELENOV
Selenoprotein V
- SELENOW
Selenoprotein W
- SELENOF
Selenoprotein F
- SEPHS2
Selenophosphate synthetase 2
- SEPSECS
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase
- SNPs
Single nucleotide polymorphisms
- STEMI
ST-segment elevation MI
- TNF-α
Tumor necrosis factor-α
- TRU-TCA1-1
Transfer RNA-Sec [TCA] 1-1
- Trx
Thioredoxin
- TXNRD
Thioredoxin reductase
- UPR
Unfolded protein response
Compliance with ethical standards
Conflict of interest
The authors declare no potential conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Carmine Rocca, Phone: +39 (0)984-492904, Email: carmine.rocca@unical.it.
Tommaso Angelone, Phone: +39 (0)984-492902/2904, Email: tommaso.angelone@unical.it.
References
- 1.Townsend N, Wilson L, Bhatnagar P, Wickramasinghe K, Rayner M, Nichols M. Cardiovascular disease in Europe: epidemiological update. Eur Heart J. 2016;37(42):3232–3245. doi: 10.1093/eurheartj/ehw334. [DOI] [PubMed] [Google Scholar]
- 2.Lloyd-Jones D, Adams R, Carnethon M, De Simone G, Ferguson TB, Flegal K, Ford E, Furie K, Go A, Greenlund K, Haase N, Hailpern S, Ho M, Howard V, Kissela B, Kittner S, Lackland D, Lisabeth L, Marelli A, McDermott M, Meigs J, Mozaffarian D, Nichol G, O’Donnell C, Roger V, Rosamond W, Sacco R, Sorlie P, Stafford R, Steinberger J, Thom T, Wasserthiel-Smoller S, Wong N, Wylie-Rosett J, Hong Y. Heart disease and stroke statistics—2009 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2009;119(3):480–486. doi: 10.1161/CIRCULATIONAHA.108.191259. [DOI] [PubMed] [Google Scholar]
- 3.Konstantinidis K, Whelan RS, Kitsis RN. Mechanisms of cell death in heart disease. Arterioscler Thromb Vasc Biol. 2012;32(7):1552–1562. doi: 10.1161/ATVBAHA.111.224915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beckman KB, Ames BN. Endogenous oxidative damage of mtDNA. Mutat Res. 1999;424(1–2):51–58. doi: 10.1016/s0027-5107(99)00007-x. [DOI] [PubMed] [Google Scholar]
- 5.Seddon M, Looi YH, Shah AM. Oxidative stress and redox signalling in cardiac hypertrophy and heart failure. Heart. 2007;93(8):903–907. doi: 10.1136/hrt.2005.068270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griendling KK, FitzGerald GA. Oxidative stress and cardiovascular injury: part I: basic mechanisms and in vivo monitoring of ROS. Circulation. 2003;108(16):1912–1916. doi: 10.1161/01.CIR.0000093660.86242.BB. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt MR, Jespersen NR, Bøtker HE. Mechanical interventions to reduce myocardial infarct size. Heart Metab. 2016;70:8–13. [Google Scholar]
- 8.Murphy E, Steenbergen C. Mechanisms underlying acute protection from cardiac ischemia–reperfusion injury. Physiol Rev. 2008;88(2):581–609. doi: 10.1152/physrev.00024.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hausenloy DJ, Yellon DM. New directions for protecting the heart against ischaemia–reperfusion injury: targeting the reperfusion injury salvage kinase (RISK)-pathway. Cardiovasc Res. 2004;61(3):448–460. doi: 10.1016/j.cardiores.2003.09.024. [DOI] [PubMed] [Google Scholar]
- 10.Cung TT, Morel O, Cayla G, Rioufol G, Garcia-Dorado D, Angoulvant D, Bonnefoy-Cudraz E, Guérin P, Elbaz M, Delarche N, Coste P, Vanzetto G, Metge M, Aupetit JF, Jouve B, Motreff P, Tron C, Labeque JN, Steg PG, Cottin Y, Range G, Clerc J, Claeys MJ, Coussement P, Prunier F, Moulin F, Roth O, Belle L, Dubois P, Barragan P, Gilard M, Piot C, Colin P, De Poli F, Morice MC, Ider O, Dubois-Randé JL, Unterseeh T, Le Breton H, Béard T, Blanchard D, Grollier G, Malquarti V, Staat P, Sudre A, Elmer E, Hansson MJ, Bergerot C, Boussaha I, Jossan C, Derumeaux G, Mewton N, Ovize M. Cyclosporine before PCI in patients with acute myocardial infarction. N Engl J Med. 2015;373(11):1021–1031. doi: 10.1056/NEJMoa1505489. [DOI] [PubMed] [Google Scholar]
- 11.Forsberg L, de Faire U, Morgenstern R. Oxidative stress, human genetic variation, and disease. Arch Biochem Biophys. 2001;389(1):84–93. doi: 10.1006/abbi.2001.2295. [DOI] [PubMed] [Google Scholar]
- 12.Ursini F, Maiorino M, Brigelius-Flohé R, Aumann KD, Roveri A, Schomburg D, Flohé L. Diversity of glutathione peroxidases. Methods Enzymol. 1995;252:38–53. doi: 10.1016/0076-6879(95)52007-4. [DOI] [PubMed] [Google Scholar]
- 13.Raes M, Michiels C, Remacle J. Comparative study of the enzymatic defense systems against oxygen-derived free radicals: the key role of glutathione peroxidase. Free Radic Biol Med. 1987;3(1):3–7. doi: 10.1016/0891-5849(87)90032-3. [DOI] [PubMed] [Google Scholar]
- 14.Lu J, Holmgren A. Selenoproteins. J Biol Chem. 2009;284(2):723–727. doi: 10.1074/jbc.R800045200. [DOI] [PubMed] [Google Scholar]
- 15.Holmgren A. Thioredoxin. Annu Rev Biochem. 1985;54:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- 16.Labunskyy VM, Hatfield DL, Gladyshev VN. Selenoproteins: molecular pathways and physiological roles. Physiol Rev. 2014;94(3):739–777. doi: 10.1152/physrev.00039.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hondal RJ, Marino SM, Gladyshev VN. Selenocysteine in thiol/disulfide-like exchange reactions. Antioxid Redox Signal. 2013;18(13):1675–1689. doi: 10.1089/ars.2012.5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steinbrenner H, Sies H. Protection against reactive oxygen species by selenoproteins. Biochim Biophys Acta. 2009;1790(11):1478–1485. doi: 10.1016/j.bbagen.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 19.Rose AH, Hoffmann PR. Selenoproteins and cardiovascular stress. Thromb Haemost. 2015;113(3):494–504. doi: 10.1160/TH14-07-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atkins JF, Gesteland RF. The twenty-first amino acid. Nature. 2000;407(6803):463–465. doi: 10.1038/35035189. [DOI] [PubMed] [Google Scholar]
- 21.Benstoem C, Goetzenich A, Kraemer S, Borosch S, Manzanares W, Hardy G, Stoppe C. Selenium and its supplementation in cardiovascular disease what do we know? Nutrients. 2015;7(5):3094–3118. doi: 10.3390/nu7053094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Böck A, Forchhammer K, Heider J, Leinfelder W, Sawers G, Veprek B, Zinoni F. Selenocysteine: the 21st amino acid. Mol Microbiol. 1991;5(3):515–520. doi: 10.1111/j.1365-2958.1991.tb00722.x. [DOI] [PubMed] [Google Scholar]
- 23.Zoidis E, Seremelis I, Kontopoulos N, Danezis GP. Selenium-dependent antioxidant enzymes: actions and properties of selenoproteins. Antioxidants (Basel). 2018;7(5):66. doi: 10.3390/antiox7050066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mariotti M, Salinas G, Gabaldón T, Gladyshev VN. Utilization of selenocysteine in early-branching fungal phyla. Nat Microbiol. 2019;4(5):759–765. doi: 10.1038/s41564-018-0354-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wessjohann LA, Schneider A, Abbas M, Brandt W. Selenium in chemistry and biochemistry in comparison to sulfur. Biol Chem. 2007;388(10):997–1006. doi: 10.1515/BC.2007.138. [DOI] [PubMed] [Google Scholar]
- 26.Zhong L, Arnér ES, Holmgren A. Structure and mechanism of mammalian thioredoxin reductase: the active site is a redox-active selenolthiol/selenenylsulfide formed from the conserved cysteine-selenocysteine sequence. Proc Natl Acad Sci USA. 2000;97(11):5854–5859. doi: 10.1073/pnas.100114897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigó R, Gladyshev VN. Characterization of mammalian selenoproteomes. Science. 2003;300(5624):1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 28.Papp LV, Lu J, Holmgren A, Khanna KK. From selenium to selenoproteins: synthesis, identity, and their role in human health. Antioxid Redox Signal. 2007;9(7):775–806. doi: 10.1089/ars.2007.1528. [DOI] [PubMed] [Google Scholar]
- 29.Nauser T, Steinmann D, Koppenol WH. Why do proteins use selenocysteine instead of cysteine? Amino Acids. 2012;42(1):39–44. doi: 10.1007/s00726-010-0602-7. [DOI] [PubMed] [Google Scholar]
- 30.Bianco CL, Moore CD, Fukuto JM, Toscano JP. Selenols are resistant to irreversible modification by HNO. Free Radic Biol Med. 2016;99:71–78. doi: 10.1016/j.freeradbiomed.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 31.Snider GW, Ruggles E, Khan N, Hondal RJ. Selenocysteine confers resistance to inactivation by oxidation in thioredoxin reductase: comparison of selenium and sulfur enzymes. Biochemistry. 2013;52(32):5472–5481. doi: 10.1021/bi400462j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ingold I, Berndt C, Schmitt S, Doll S, Poschmann G, Buday K, Roveri A, Peng X, Porto-Freitas F, Seibt T, Mehr L, Aichler M, Walch A, Lamp D, Jastroch M, Miyamoto S, Wurst W, Ursini F, Arnér ESJ, Fradejas-Villar N, Schweizer U, Zischka H, Friedmann Angeli JP, Conrad M. Selenium utilization by GPX4 is required to prevent hydroperoxide-induced ferroptosis. Cell. 2018;172(3):409.e21–422.e21. doi: 10.1016/j.cell.2017.11.048. [DOI] [PubMed] [Google Scholar]
- 33.Kim HY, Gladyshev VN. Different catalytic mechanisms in mammalian selenocysteine- and cysteine-containing methionine-R-sulfoxide reductases. PLoS Biol. 2005;3(12):e375. doi: 10.1371/journal.pbio.0030375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berry MJ, Banu L, Chen YY, Mandel SJ, Kieffer JD, Harney JW, Larsen PR. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature. 1991;353(6341):273–276. doi: 10.1038/353273a0. [DOI] [PubMed] [Google Scholar]
- 35.Leinfelder W, Stadtman TC, Böck A. Occurrence in vivo of selenocysteyl-tRNA(SERUCA) in Escherichia coli. Effect of sel mutations. J Biol Chem. 1989;264(17):9720–9723. [PubMed] [Google Scholar]
- 36.Agamy O, Ben Zeev B, Lev D, Marcus B, Fine D, Su D, Narkis G, Ofir R, Hoffmann C, Leshinsky-Silver E, Flusser H, Sivan S, Söll D, Lerman-Sagie T, Birk OS. Mutations disrupting selenocysteine formation cause progressive cerebello-cerebral atrophy. Am J Hum Genet. 2010;87(4):538–544. doi: 10.1016/j.ajhg.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schweizer U, Fradejas-Villar N. Why 21? The significance of selenoproteins for human health revealed by inborn errors of metabolism. FASEB J. 2016;30(11):3669–3681. doi: 10.1096/fj.201600424. [DOI] [PubMed] [Google Scholar]
- 38.Fradejas-Villar N. Consequences of mutations and inborn errors of selenoprotein biosynthesis and functions. Free Radic Biol Med. 2018;127:206–214. doi: 10.1016/j.freeradbiomed.2018.04.572. [DOI] [PubMed] [Google Scholar]
- 39.Anttonen AK, Hilander T, Linnankivi T, Isohanni P, French RL, Liu Y, Simonović M, Söll D, Somer M, Muth-Pawlak D, Corthals GL, Laari A, Ylikallio E, Lähde M, Valanne L, Lönnqvist T, Pihko H, Paetau A, Lehesjoki AE, Suomalainen A, Tyynismaa H. Selenoprotein biosynthesis defect causes progressive encephalopathy with elevated lactate. Neurology. 2015;85(4):306–315. doi: 10.1212/WNL.0000000000001787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ben-Zeev B, Hoffman C, Lev D, Watemberg N, Malinger G, Brand N, Lerman-Sagie T. Progressive cerebellocerebral atrophy: a new syndrome with microcephaly, mental retardation, and spastic quadriplegia. J Med Genet. 2003;40(8):e96. doi: 10.1136/jmg.40.8.e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schoenmakers E, Agostini M, Mitchell C, Schoenmakers N, Papp L, Rajanayagam O, Padidela R, Ceron-Gutierrez L, Doffinger R, Prevosto C, Luan J, Montano S, Lu J, Castanet M, Clemons N, Groeneveld M, Castets P, Karbaschi M, Aitken S, Dixon A, Williams J, Campi I, Blount M, Burton H, Muntoni F, O’Donovan D, Dean A, Warren A, Brierley C, Baguley D, Guicheney P, Fitzgerald R, Coles A, Gaston H, Todd P, Holmgren A, Khanna KK, Cooke M, Semple R, Halsall D, Wareham N, Schwabe J, Grasso L, Beck-Peccoz P, Ogunko A, Dattani M, Gurnell M, Chatterjee K. Mutations in the selenocysteine insertion sequence-binding protein 2 gene lead to a multisystem selenoprotein deficiency disorder in humans. J Clin Investig. 2010;120(12):4220–4235. doi: 10.1172/JCI43653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dumitrescu AM, Liao XH, Abdullah MS, Lado-Abeal J, Majed FA, Moeller LC, Boran G, Schomburg L, Weiss RE, Refetoff S. Mutations in SECISBP2 result in abnormal thyroid hormone metabolism. Nat Genet. 2005;37(11):1247–1252. doi: 10.1038/ng1654. [DOI] [PubMed] [Google Scholar]
- 43.Schoenmakers E, Carlson B, Agostini M, Moran C, Rajanayagam O, Bochukova E, Tobe R, Peat R, Gevers E, Muntoni F, Guicheney P, Schoenmakers N, Farooqi S, Lyons G, Hatfield D, Chatterjee K. Mutation in human selenocysteine transfer RNA selectively disrupts selenoprotein synthesis. J Clin Investig. 2016;126(3):992–996. doi: 10.1172/JCI84747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schoenmakers E, Chatterjee K. Identification of genetic disorders causing disruption of selenoprotein biosynthesis. Methods Mol Biol. 2018;1661:325–335. doi: 10.1007/978-1-4939-7258-6_23. [DOI] [PubMed] [Google Scholar]
- 45.Bösl MR, Takaku K, Oshima M, Nishimura S, Taketo MM. Early embryonic lethality caused by targeted disruption of the mouse selenocysteine tRNA gene (Trsp) Proc Natl Acad Sci USA. 1997;94(11):5531–5534. doi: 10.1073/pnas.94.11.5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flohe L, Günzler WA, Schock HH. Glutathione peroxidase: a selenoenzyme. FEBS Lett. 1973;32(1):132–134. doi: 10.1016/0014-5793(73)80755-0. [DOI] [PubMed] [Google Scholar]
- 47.Gong T, Torres DJ, Berry MJ, Pitts MW. Hypothalamic redox balance and leptin signaling—emerging role of selenoproteins. Free Radic Biol Med. 2018;127:172–181. doi: 10.1016/j.freeradbiomed.2018.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sandalova T, Zhong L, Lindqvist Y, Holmgren A, Schneider G. Three-dimensional structure of a mammalian thioredoxin reductase: implications for mechanism and evolution of a selenocysteine-dependent enzyme. Proc Natl Acad Sci USA. 2001;98(17):9533–9538. doi: 10.1073/pnas.171178698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tamura K, Schimmel P. Trial for peptide bond formation using model molecules based on the interactions between the CCA sequence of tRNA and 23S rRNA. Nucleic Acids Symp Ser. 2000;44:251–252. doi: 10.1093/nass/44.1.251. [DOI] [PubMed] [Google Scholar]
- 50.Bianco AC, Salvatore D, Gereben B, Berry MJ, Larsen PR. Biochemistry, cellular and molecular biology, and physiological roles of the iodothyronine selenodeiodinases. Endocr Rev. 2012;23(1):38–89. doi: 10.1210/edrv.23.1.0455. [DOI] [PubMed] [Google Scholar]
- 51.Gladyshev VN, Arnér ES, Berry MJ, Brigelius-Flohé R, Bruford EA, Burk RF, Carlson BA, Castellano S, Chavatte L, Conrad M, Copeland PR, Diamond AM, Driscoll DM, Ferreiro A, Flohé L, Green FR, Guigó R, Handy DE, Hatfield DL, Hesketh J, Hoffmann PR, Holmgren A, Hondal RJ, Howard MT, Huang K, Kim HY, Kim IY, Köhrle J, Krol A, Kryukov GV, Lee BJ, Lee BC, Lei XG, Liu Q, Lescure A, Lobanov AV, Loscalzo J, Maiorino M, Mariotti M, Sandeep Prabhu K, Rayman MP, Rozovsky S, Salinas G, Schmidt EE, Schomburg L, Schweizer U, Simonović M, Sunde RA, Tsuji PA, Tweedie S, Ursini F, Whanger PD, Zhang Y. Selenoprotein gene nomenclature. J Biol Chem. 2016;291(46):24036–24040. doi: 10.1074/jbc.M116.756155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dikiy A, Novoselov SV, Fomenko DE, Sengupta A, Carlson BA, Cerny RL, Ginalski K, Grishin NV, Hatfield DL, Gladyshev VN. SelT, SelW, SelH, and Rdx12: genomics and molecular insights into the functions of selenoproteins of a novel thioredoxin-like family. Biochemistry. 2007;46(23):6871–6882. doi: 10.1021/bi602462q. [DOI] [PubMed] [Google Scholar]
- 53.Lescure A, Gautheret D, Carbon P, Krol A. Novel selenoproteins identified in silico and in vivo by using a conserved RNA structural motif. J Biol Chem. 1999;274(53):38147–38154. doi: 10.1074/jbc.274.53.38147. [DOI] [PubMed] [Google Scholar]
- 54.Burk RF, Hill KE, Motley AK, Winfrey VP, Kurokawa S, Mitchell SL, Zhang W. Selenoprotein P and apolipoprotein E receptor-2 interact at the blood-brain barrier and also within the brain to maintain an essential selenium pool that protects against neurodegeneration. FASEB J. 2014;28(8):3579–3588. doi: 10.1096/fj.14-252874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roman M, Jitaru P, Barbante C. Selenium biochemistry and its role for human health. Metallomics. 2014;6(1):25–54. doi: 10.1039/c3mt00185g. [DOI] [PubMed] [Google Scholar]
- 56.Hurst R, Armah CN, Dainty JR, Hart DJ, Teucher B, Goldson AJ, Broadley MR, Motley AK, Fairweather-Tait SJ. Establishing optimal selenium status: results of a randomized, double-blind, placebo-controlled trial. Am J Clin Nutr. 2010;91(4):923–931. doi: 10.3945/ajcn.2009.28169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pitts MW, Hoffmann PR. Endoplasmic reticulum-resident selenoproteins as regulators of calcium signaling and homeostasis. Cell Calcium. 2018;70:76–86. doi: 10.1016/j.ceca.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fredericks GJ, Hoffmann FW, Rose AH, Osterheld HJ, Hess FM, Mercier F, Hoffmann PR. Stable expression and function of the inositol 1,4,5-triphosphate receptor requires palmitoylation by a DHHC6/selenoprotein K complex. Proc Natl Acad Sci USA. 2014;111(46):16478–16483. doi: 10.1073/pnas.1417176111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marino M, Stoilova T, Giorgi C, Bachi A, Cattaneo A, Auricchio A, Pinton P, Zito E. SEPN1, an endoplasmic reticulum-localized selenoprotein linked to skeletal muscle pathology, counteracts hyperoxidation by means of redox-regulating SERCA2 pump activity. Hum Mol Genet. 2015;24(7):1843–1855. doi: 10.1093/hmg/ddu602. [DOI] [PubMed] [Google Scholar]
- 60.Grumolato L, Ghzili H, Montero-Hadjadje M, Gasman S, Lesage J, Tanguy Y, Galas L, Ait-Ali D, Leprince J, Guérineau NC, Elkahloun AG, Fournier A, Vieau D, Vaudry H, Anouar Y. Selenoprotein T is a PACAP-regulated gene involved in intracellular Ca2+ mobilization and neuroendocrine secretion. FASEB J. 2008;22(6):1756–1768. doi: 10.1096/fj.06-075820. [DOI] [PubMed] [Google Scholar]
- 61.Anouar Y, Lihrmann I, Falluel-Morel A, Boukhzar L. Selenoprotein T is a key player in ER proteostasis, endocrine homeostasis and neuroprotection. Free Radic Biol Med. 2018;127:145–152. doi: 10.1016/j.freeradbiomed.2018.05.076. [DOI] [PubMed] [Google Scholar]
- 62.Hamieh A, Cartier D, Abid H, Calas A, Burel C, Bucharles C, Jehan C, Grumolato L, Landry M, Lerouge P, Anouar Y, Lihrmann I. Selenoprotein T is a novel OST subunit that regulates UPR signaling and hormone secretion. EMBO Rep. 2017;18(11):1935–1946. doi: 10.15252/embr.201643504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fallab S. Reactions with molecular oxygen. New York: Angewandte Chemie International Edition in English; 1967. pp. 496–507. [Google Scholar]
- 64.Nathan C, Cunningham-Bussel A. Beyond oxidative stress: an immunologist’s guide to reactive oxygen species. Nat Rev Immunol. 2013;13(5):349–361. doi: 10.1038/nri3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hoshi T, Heinemann S. Regulation of cell function by methionine oxidation and reduction. J Physiol. 2001;531(Pt 1):1–11. doi: 10.1111/j.1469-7793.2001.0001j.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rahman T, Hosen I, Islam M, Shekhar H. Oxidative stress and human health. Adv Biosci Biotechnol. 2012;3:997–1019. doi: 10.4236/abb.2012.327123. [DOI] [Google Scholar]
- 67.Reeves MA, Hoffmann PR. The human selenoproteome: recent insights into functions and regulation. Cell Mol Life Sci. 2009;66(15):2457–2478. doi: 10.1007/s00018-009-0032-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Winterbourn CC, Hampton MB. Thiol chemistry and specificity in redox signaling. Free Radic Biol Med. 2008;45(5):549–561. doi: 10.1016/j.freeradbiomed.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 69.Touat-Hamici Z, Legrain Y, Bulteau AL, Chavatte L. Selective up-regulation of human selenoproteins in response to oxidative stress. J Biol Chem. 2014;289(21):14750–14761. doi: 10.1074/jbc.M114.551994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Papp LV, Lu J, Striebel F, Kennedy D, Holmgren A, Khanna KK. The redox state of SECIS binding protein 2 controls its localization and selenocysteine incorporation function. Mol Cell Biol. 2006;26(13):4895–4910. doi: 10.1128/MCB.02284-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kasaikina MV, Hatfield DL, Gladyshev VN. Understanding selenoprotein function and regulation through the use of rodent models. Biochim Biophys Acta. 2012;1823(9):1633–1642. doi: 10.1016/j.bbamcr.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Conrad M, Schweizer U. Unveiling the molecular mechanisms behind selenium-related diseases through knockout mouse studies. Antioxid Redox Signal. 2010;12(7):851–865. doi: 10.1089/ars.2009.2912. [DOI] [PubMed] [Google Scholar]
- 73.Conrad M. Transgenic mouse models for the vital selenoenzymes cytosolic thioredoxin reductase, mitochondrial thioredoxin reductase and glutathione peroxidase 4. Biochim Biophys Acta. 2009;1790(11):1575–1585. doi: 10.1016/j.bbagen.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 74.Jakupoglu C, Przemeck GKH, Schneider M, Moreno SG, Mayr N, Hatzopoulos AK, de Angelis MH, Wurst W, Bornkamm GW, Brielmeier M, Conrad M. Cytoplasmic thioredoxin reductase is essential for embryogenesis but dispensable for cardiac development. Mol Cell Biol. 2005;25(5):1980–1988. doi: 10.1128/MCB.25.5.1980-1988.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Conrad M, Jakupoglu C, Moreno SG, Lippl S, Banjac A, Schneider M, Beck H, Hatzopoulos AK, Just U, Sinowatz F, Schmahl W, Chien KR, Wurst W, Bornkamm GW, Brielmeier M. Essential role for mitochondrial thioredoxin reductase in hematopoiesis, heart development, and heart function. Mol Cell Biol. 2004;24(21):9414–9423. doi: 10.1128/MCB.24.21.9414-9423.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Imai H, Hirao F, Sakamoto T, Sekine K, Mizukura Y, Saito M, Kitamoto T, Hayasaka M, Hanaoka K, Nakagawa Y. Early embryonic lethality caused by targeted disruption of the mouse PHGPx gene. Biochem Biophys Res Commun. 2003;305(2):278–286. doi: 10.1016/s0006-291x(03)00734-4. [DOI] [PubMed] [Google Scholar]
- 77.Hill KE, Zhou J, McMahan WJ, Motley AK, Atkins JF, Gesteland RF, Burk RF. Deletion of selenoprotein P alters distribution of selenium in the mouse. J Biol Chem. 2003;278(16):13640–13646. doi: 10.1074/jbc.M300755200. [DOI] [PubMed] [Google Scholar]
- 78.Schomburg L, Schweizer U, Holtmann B, Flohé L, Sendtner M, Köhrle J. Gene disruption discloses role of selenoprotein P in selenium delivery to target tissues. Biochem J. 2003;370(Pt 2):397–402. doi: 10.1042/BJ20021853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boukhzar L, Hamieh A, Cartier D, Tanguy Y, Alsharif I, Castex M, Arabo A, El Hajji S, Bonnet JJ, Errami M, Falluel-Morel A, Chagraoui A, Lihrmann I, Anouar Y. Selenoprotein T exerts an essential oxidoreductase activity that protects dopaminergic neurons in mouse models of parkinson’s disease. Antioxid Redox Signal. 2016;24(11):557–574. doi: 10.1089/ars.2015.6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Castex MT, Arabo A, Bénard M, Roy V, Le Joncour V, Prévost G, Bonnet JJ, Anouar Y, Falluel-Morel A. Selenoprotein T deficiency leads to neurodevelopmental abnormalities and hyperactive behavior in mice. Mol Neurobiol. 2016;53(9):5818–5832. doi: 10.1007/s12035-015-9505-7. [DOI] [PubMed] [Google Scholar]
- 81.Cheng WH, Ho YS, Ross DA, Valentine BA, Combs GF, Lei XG. Cellular glutathione peroxidase knockout mice express normal levels of selenium-dependent plasma and phospholipid hydroperoxide glutathione peroxidases in various tissues. J Nutr. 1997;127(8):1445–1450. doi: 10.1093/jn/127.8.1445. [DOI] [PubMed] [Google Scholar]
- 82.Fomenko DE, Novoselov SV, Natarajan SK, Lee BC, Koc A, Carlson BA, Lee TH, Kim HY, Hatfield DL, Gladyshev VN. MsrB1 (methionine-R-sulfoxide reductase 1) knock-out mice: roles of MsrB1 in redox regulation and identification of a novel selenoprotein form. J Biol Chem. 2009;284(9):5986–5993. doi: 10.1074/jbc.M805770200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Suzuki T, Kelly VP, Motohashi H, Nakajima O, Takahashi S, Nishimura S, Yamamoto M. Deletion of the selenocysteine tRNA gene in macrophages and liver results in compensatory gene induction of cytoprotective enzymes by Nrf2. J Biol Chem. 2008;283(4):2021–2030. doi: 10.1074/jbc.M708352200. [DOI] [PubMed] [Google Scholar]
- 84.Kawatani Y, Suzuki T, Shimizu R, Kelly VP, Yamamoto M. Nrf2 and selenoproteins are essential for maintaining oxidative homeostasis in erythrocytes and protecting against hemolytic anemia. Blood. 2011;117(3):986–996. doi: 10.1182/blood-2010-05-285817. [DOI] [PubMed] [Google Scholar]
- 85.Rayman MP. The importance of Se to human health. Lancet. 2000;356(9225):233–241. doi: 10.1016/S0140-6736(00)02490-9. [DOI] [PubMed] [Google Scholar]
- 86.Shrimali RK, Weaver JA, Miller GF, Starost MF, Carlson BA, Novoselov SV, Kumaraswamy E, Gladyshev VN, Hatfield DL. Selenoprotein expression is essential in endothelial cell development and cardiac muscle function. Neuromuscul Disord. 2007;17(2):135–142. doi: 10.1016/j.nmd.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Puccio H, Simon D, Cossée M, Criqui-Filipe P, Tiziano F, Melki J, Hindelang C, Matyas R, Rustin P, Koenig M. Mouse models for Friedreich ataxia exhibit cardiomyopathy, sensory nerve defect and Fe-S enzyme deficiency followed by intramitochondrial iron deposits. Nat Genet. 2001;27(2):181–186. doi: 10.1038/84818. [DOI] [PubMed] [Google Scholar]
- 88.Sibbing D, Pfeufer A, Perisic T, Mannes AM, Fritz-Wolf K, Unwin S, Sinner MF, Gieger C, Gloeckner CJ, Wichmann HE, Kremmer E, Schäfer Z, Walch A, Hinterseer M, Näbauer M, Kääb S, Kastrati A, Schömig A, Meitinger T, Bornkamm GW, Conrad M, von Beckerath N. Mutations in the mitochondrial thioredoxin reductase gene TXNRD2 cause dilated cardiomyopathy. Eur Heart J. 2011;32(9):1121–1133. doi: 10.1093/eurheartj/ehq507. [DOI] [PubMed] [Google Scholar]
- 89.Prasad R, Chan LF, Hughes CR, Kaski JP, Kowalczyk JC, Savage MO, Peters CJ, Nathwani N, Clark AJ, Storr HL, Metherell LA. Thioredoxin reductase 2 (TXNRD2) mutation associated with familial glucocorticoid deficiency (FGD) J Clin Endocrinol Metab. 2014;99(8):E1556–E1563. doi: 10.1210/jc.2013-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Clark AJ, Chan LF, Chung TT, Metherell LA. The genetics of familial glucocorticoid deficiency. Best Pract Res Clin Endocrinol Metab. 2009;23(2):159–165. doi: 10.1016/j.beem.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 91.Kiermayer C, Northrup E, Schrewe A, Walch A, de Angelis MH, Schoensiegel F, Zischka H, Prehn C, Adamski J, Bekeredjian R, Ivandic B, Kupatt C, Brielmeier M. Heart-specific knockout of the mitochondrial thioredoxin reductase (Txnrd2) induces metabolic and contractile dysfunction in the aging myocardium. J Am Heart Assoc. 2015;4(7):e002153. doi: 10.1161/JAHA.115.002153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lubos E, Loscalzo J, Handy DE. Glutathione peroxidase-1 in health and disease: from molecular mechanisms to therapeutic opportunities. Antioxid Redox Signal. 2011;15(7):1957–1997. doi: 10.1089/ars.2010.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wentzel P, Gäreskog M, Eriksson UJ. Decreased cardiac glutathione peroxidase levels and enhanced mandibular apoptosis in malformed embryos of diabetic rats. Diabetes. 2008;57(12):3344–3352. doi: 10.2337/db08-0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Forgione MA, Cap A, Liao R, Moldovan NI, Eberhardt RT, Lim CC, Jones J, Goldschmidt-Clermont PJ, Loscalzo J. Heterozygous cellular glutathione peroxidase deficiency in the mouse: abnormalities in vascular and cardiac function and structure. Circulation. 2002;106(9):1154–1158. doi: 10.1161/01.cir.0000026820.87824.6a. [DOI] [PubMed] [Google Scholar]
- 95.Ishida K, Morino T, Takagi K, Sukenaga Y. Nucleotide sequence of a human gene for glutathione peroxidase. Nucleic Acids Res. 1987;15(23):10051. doi: 10.1093/nar/15.23.10051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kiss C, Li J, Szeles A, Gizatullin RZ, Kashuba VI, Lushnikova T, Protopopov AI, Kelve M, Kiss H, Kholodnyuk ID, Imreh S, Klein G, Zabarovsky ER. Assignment of the ARHA and GPX1 genes to human chromosome bands 3p21.3 by in situ hybridization and with somatic cell hybrids. Cytogenet Cell Genet. 1997;79(3–4):228–230. doi: 10.1159/000134729. [DOI] [PubMed] [Google Scholar]
- 97.Winter JP, Gong Y, Grant PJ, Wild CP. Glutathione peroxidase 1 genotype is associated with an increased risk of coronary artery disease. Coron Artery Dis. 2003;14(2):149–153. doi: 10.1097/00019501-200304000-00007. [DOI] [PubMed] [Google Scholar]
- 98.Hamanishi T, Furuta H, Kato H, Doi A, Tamai M, Shimomura H, Sakagashira S, Nishi M, Sasaki H, Sanke T, Nanjo K. Functional variants in the glutathione peroxidase-1 (GPx-1) gene are associated with increased intima-media thickness of carotid arteries and risk of macrovascular diseases in japanese type 2 diabetic patients. Diabetes. 2004;53(9):2455–2460. doi: 10.2337/diabetes.53.9.2455. [DOI] [PubMed] [Google Scholar]
- 99.Moscow JA, Schmidt L, Ingram DT, Gnarra J, Johnson B, Cowan KH. Loss of heterozygosity of the human cytosolic glutathione peroxidase I gene in lung cancer. Carcinogenesis. 1994;15(12):2769–2773. doi: 10.1093/carcin/15.12.2769. [DOI] [PubMed] [Google Scholar]
- 100.Nemoto M, Nishimura R, Sasaki T, Hiki Y, Miyashita Y, Nishioka M, Fujimoto K, Sakuma T, Ohashi T, Fukuda K, Eto Y, Tajima N. Genetic association of glutathione peroxidase-1 with coronary artery calcification in type 2 diabetes: a case control study with multi-slice computed tomography. Cardiovasc Diabetol. 2007;7(6):23. doi: 10.1186/1475-2840-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Oguri M, Kato K, Hibino T, Yokoi K, Segawa T, Matsuo H, Watanabe S, Nozawa Y, Murohara T, Yamada Y. Genetic risk for restenosis after coronary stenting. Atherosclerosis. 2007;194(2):e172–e178. doi: 10.1016/j.atherosclerosis.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 102.Zhang JX, Wang ZM, Zhang JJ, Zhu LL, Gao XF, Chen SL. Association of glutathione peroxidase-1 (GPx-1) rs1050450 Pro198Leu and Pro197Leu polymorphisms with cardiovascular risk: a meta-analysis of observational studies. J Geriatr Cardiol. 2014;11(2):141–150. doi: 10.3969/j.issn.1671-5411.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tang NP, Wang LS, Yang L, Gu HJ, Sun QM, Cong RH, Zhou B, Zhu HJ, Wang B. Genetic variant in glutathione peroxidase 1 gene is associated with an increased risk of coronary artery disease in a Chinese population. Clin Chim Acta. 2008;395(1–2):89–93. doi: 10.1016/j.cca.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 104.Ravn-Haren G, Olsen A, Tjønneland A, Dragsted LO, Nexø BA, Wallin H, Overvad K, Raaschou-Nielsen O, Vogel U. Associations between GPX1 Pro198Leu polymorphism, erythrocyte GPX activity, alcohol consumption and breast cancer risk in a prospective cohort study. Carcinogenesis. 2006;27(4):820–825. doi: 10.1093/carcin/bgi267. [DOI] [PubMed] [Google Scholar]
- 105.Hu YJ, Diamond AM. Role of glutathione peroxidase 1 in breast cancer: loss of heterozygosity and allelic differences in the response to selenium. Cancer Res. 2003;63(12):3347–3351. [PubMed] [Google Scholar]
- 106.Takebe G, Yarimizu J, Saito Y, Hayashi T, Nakamura H, Yodoi J, Nagasawa S, Takahashi K. A comparative study on the hydroperoxide and thiol specificity of the glutathione peroxidase family and selenoprotein P. J Biol Chem. 2002;277(43):41254–41258. doi: 10.1074/jbc.M202773200. [DOI] [PubMed] [Google Scholar]
- 107.Jin RC, Mahoney CE, Coleman Anderson L, Ottaviano F, Croce K, Leopold JA, Zhang YY, Tang SS, Handy DE, Loscalzo J. Glutathione peroxidase-3 deficiency promotes platelet-dependent thrombosis in vivo. Circulation. 2011;123(18):1963–1973. doi: 10.1161/CIRCULATIONAHA.110.000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang GR, Zhu Y, Halushka PV, Lincoln TM, Mendelsohn ME. Mechanism of platelet inhibition by nitric oxide: in vivo phosphorylation of thromboxane receptor by cyclic GMP-dependent protein kinase. Proc Natl Acad Sci USA. 1998;95(9):4888–4893. doi: 10.1073/pnas.95.9.4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lubos E, Handy DE, Loscalzo J. Role of oxidative stress and nitric oxide in atherothrombosis. Front Biosci. 2008;13:5323–5344. doi: 10.2741/3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Loscalzo J. Nitric oxide insufficiency, platelet activation, and arterial thrombosis. Circ Res. 2001;88(8):756–762. doi: 10.1161/hh0801.089861. [DOI] [PubMed] [Google Scholar]
- 111.Freedman JE, Loscalzo J, Benoit SE, Valeri CR, Barnard MR, Michelson AD. Decreased platelet inhibition by nitric oxide in two brothers with a history of arterial thrombosis. J Clin Investig. 1996;97(4):979–987. doi: 10.1172/JCI118522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Voetsch B, Jin RC, Bierl C, Benke KS, Kenet G, Simioni P, Ottaviano F, Damasceno BP, Annichino-Bizacchi JM, Handy DE, Loscalzo J. Promoter polymorphisms in the plasma glutathione peroxidase (GPx-3) gene: a novel risk factor for arterial ischemic stroke among young adults and children. Stroke. 2007;38(1):41–49. doi: 10.1161/01.STR.0000252027.53766.2b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kenet G, Freedman J, Shenkman B, Regina E, Brok-Simoni F, Holzman F, Vavva F, Brand N, Michelson A, Trolliet M, Loscalzo J, Inbal A. Plasma glutathione peroxidase deficiency and platelet insensitivity to nitric oxide in children with familial stroke. Arterioscler Thromb Vasc Biol. 1999;19(8):2017–2023. doi: 10.1161/01.atv.19.8.2017. [DOI] [PubMed] [Google Scholar]
- 114.Borchert A, Wang CC, Ufer C, Schiebel H, Savaskan NE, Kuhn H. The role of phospholipid hydroperoxide glutathione peroxidase isoforms in murine embryogenesis. J Biol Chem. 2006;281(28):19655–19664. doi: 10.1074/jbc.M601195200. [DOI] [PubMed] [Google Scholar]
- 115.Smith AC, Mears AJ, Bunker R, Ahmed A, MacKenzie M, Schwartzentruber JA, Beaulieu CL, Ferretti E, FORGE Canada Consortium. Majewski J, Bulman DE, Celik FC, Boycott KM, Graham GE. Mutations in the enzyme glutathione peroxidase 4 cause Sedaghatian-type spondylometaphyseal dysplasia. J Med Genet. 2014;51(7):470–474. doi: 10.1136/jmedgenet-2013-102218. [DOI] [PubMed] [Google Scholar]
- 116.Sedaghatian MR. Congenital lethal metaphyseal chondrodysplasia: a newly recognized complex autosomal recessive disorder. Am J Med Genet. 1980;6(4):269–274. doi: 10.1002/ajmg.1320060403. [DOI] [PubMed] [Google Scholar]
- 117.Aygun C, Celik FC, Nural MS, Azak E, Kucukoduk S, Ogur G, Incesu L. Simplified gyral pattern with cerebellar hypoplasia in Sedaghatian type spondylometaphyseal dysplasia: a clinical report and review of the literature. Am J Med Genet A. 2012;158A(6):1400–1405. doi: 10.1002/ajmg.a.35306. [DOI] [PubMed] [Google Scholar]
- 118.Moskovitz J, Bar-Noy S, Williams WM, Requena J, Berlett BS, Stadtman ER. Methionine sulfoxide reductase (MsrA) is a regulator of antioxidant defense and lifespan in mammals. Proc Natl Acad Sci USA. 2001;98(23):12920–12925. doi: 10.1073/pnas.231472998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Moskovitz J. Methionine sulfoxide reductases: ubiquitous enzymes involved in antioxidant defense, protein regulation, and prevention of aging-associated diseases. Biochim Biophys Acta. 2005;1703(2):213–219. doi: 10.1016/j.bbapap.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 120.Kryukov GV, Kryukov VM, Gladyshev VN. New mammalian selenocysteine-containing proteins identified with an algorithm that searches for selenocysteine insertion sequence elements. J Biol Chem. 1999;274(48):33888–33897. doi: 10.1074/jbc.274.48.33888. [DOI] [PubMed] [Google Scholar]
- 121.Aachmann FL, Sal LS, Kim HY, Marino SM, Gladyshev VN, Dikiy A. Insights into function, catalytic mechanism, and fold evolution of selenoprotein methionine sulfoxide reductase B1 through structural analysis. J Biol Chem. 2010;285(43):33315–33323. doi: 10.1074/jbc.M110.132308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hoffmann FW, Hashimoto AS, Lee BC, Rose AH, Shohet RV, Hoffmann PR. Specific antioxidant selenoproteins are induced in the heart during hypertrophy. Arch Biochem Biophys. 2011;512(1):38–44. doi: 10.1016/j.abb.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kim HY, Gladyshev VN. Methionine sulfoxide reductases: selenoprotein forms and roles in antioxidant protein repair in mammals. Biochem J. 2007;407(3):321–329. doi: 10.1042/BJ20070929. [DOI] [PubMed] [Google Scholar]
- 124.Yamamoto M, Yang G, Hong C, Liu J, Holle E, Yu X, Wagner T, Vatner SF, Sadoshima J. Inhibition of endogenous thioredoxin in the heart increases oxidative stress and cardiac hypertrophy. J Clin Investig. 2003;112(9):1395–1406. doi: 10.1172/JCI17700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Münzel T, Camici GG, Maack C, Bonetti NR, Fuster V, Kovacic JC. Impact of oxidative stress on the heart and vasculature: part 2 of a 3-part series. J Am Coll Cardiol. 2017;70(2):212–229. doi: 10.1016/j.jacc.2017.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Blankenberg S, Rupprecht HJ, Bickel C, Torzewski M, Hafner G, Tiret L, Smieja M, Cambien F, Meyer J, Lackner KJ, AtheroGene Investigators Glutathione peroxidase 1 activity and cardiovascular events in patients with coronary artery disease. N Engl J Med. 2003;349(17):1605–1613. doi: 10.1056/NEJMoa030535. [DOI] [PubMed] [Google Scholar]
- 127.Pastori D, Pignatelli P, Farcomeni A, Menichelli D, Nocella C, Carnevale R, Violi F. Aging-related decline of glutathione peroxidase 3 and risk of cardiovascular events in patients with atrial fibrillation. J Am Heart Assoc. 2016;5(9):e003682. doi: 10.1161/JAHA.116.003682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429(6994):841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- 129.Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature. 2008;454(7203):455–462. doi: 10.1038/nature07203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Curran JE, Jowett JB, Elliott KS, Gao Y, Gluschenko K, Wang J, Abel Azim DM, Cai G, Mahaney MC, Comuzzie AG, Dyer TD, Walder KR, Zimmet P, MacCluer JW, Collier GR, Kissebah AH, Blangero J. Genetic variation in selenoprotein S influences inflammatory response. Nat Genet. 2005;37(11):1234–1241. doi: 10.1038/ng1655. [DOI] [PubMed] [Google Scholar]
- 131.Gao Y, Hannan NR, Wanyonyi S, Konstantopolous N, Pagnon J, Feng HC, Jowett JB, Kim KH, Walder K, Collier GR. Activation of the selenoprotein SEPS1 gene expression by pro-inflammatory cytokines in HepG2 cells. Cytokine. 2006;33(5):246–251. doi: 10.1016/j.cyto.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 132.Alanne M, Kristiansson K, Auro K, Silander K, Kuulasmaa K, Peltonen L, Salomaa V, Perola M. Variation in the selenoprotein S gene locus is associated with coronary heart disease and ischemic stroke in two independent Finnish cohorts. Hum Genet. 2007;122(3–4):355–365. doi: 10.1007/s00439-007-0402-7. [DOI] [PubMed] [Google Scholar]
- 133.Cox AJ, Lehtinen AB, Xu J, Langefeld CD, Freedman BI, Carr JJ, Bowden DW. Polymorphisms in the selenoprotein S gene and subclinical cardiovascular disease in the Diabetes Heart Study. Acta Diabetol. 2013;50(3):391–399. doi: 10.1007/s00592-012-0440-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Strauss E, Tomczak J, Staniszewski R, Oszkinis G. Associations and interactions between variants in selenoprotein genes, selenoprotein levels and the development of abdominal aortic aneurysm, peripheral arterial disease, and heart failure. PLoS One. 2018;13(9):e0203350. doi: 10.1371/journal.pone.0203350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lu C, Qiu F, Zhou H, Peng Y, Hao W, Xu J, Yuan J, Wang S, Qiang B, Xu C, Peng X. Identification and characterization of selenoprotein K: an antioxidant in cardiomyocytes. FEBS Lett. 2006;580(22):5189–5197. doi: 10.1016/j.febslet.2006.08.065. [DOI] [PubMed] [Google Scholar]
- 136.Liu J, Zhang Z, Rozovsky S. Selenoprotein K form an intermolecular diselenide bond with unusually high redox potential. FEBS Lett. 2014;588(18):3311–3321. doi: 10.1016/j.febslet.2014.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Du S, Zhou J, Jia Y, Huang K. SelK is a novel ER stress-regulated protein and protects HepG2 cells from ER stress agent-induced apoptosis. Arch Biochem Biophys. 2010;502(2):137–143. doi: 10.1016/j.abb.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 138.Shchedrina VA, Everley RA, Zhang Y, Gygi SP, Hatfield DL, Gladyshev VN. Selenoprotein K binds multiprotein complexes and is involved in the regulation of endoplasmic reticulum homeostasis. J Biol Chem. 2011;286(50):42937–42948. doi: 10.1074/jbc.M111.310920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Verma S, Hoffmann FW, Kumar M, Huang Z, Roe K, Nguyen-Wu E, Hashimoto AS, Hoffmann PR. Selenoprotein K knockout mice exhibit deficient calcium flux in immune cells and impaired immune responses. J Immunol. 2011;186(4):2127–2137. doi: 10.4049/jimmunol.1002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Huang Z, Hoffmann FW, Fay JD, Hashimoto AC, Chapagain ML, Kaufusi PH, Hoffmann PR. Stimulation of unprimed macrophages with immune complexes triggers a low output of nitric oxide by calcium-dependent neuronal nitric-oxide synthase. J Biol Chem. 2012;287(7):4492–4502. doi: 10.1074/jbc.M111.315598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Meiler S, Baumer Y, Huang Z, Hoffmann FW, Fredericks GJ, Rose AH, Norton RL, Hoffmann PR, Boisvert WA. Selenoprotein K is required for palmitoylation of CD36 in macrophages: implications in foam cell formation and atherogenesis. J Leukoc Biol. 2013;93(5):771–780. doi: 10.1189/jlb.1212647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Weber C, Zernecke A, Libby P. The multifaceted contributions of leukocyte subsets to atherosclerosis: lessons from mouse models. Nat Rev Immunol. 2008;8(10):802–815. doi: 10.1038/nri2415. [DOI] [PubMed] [Google Scholar]
- 143.Baqui M, Botero D, Gereben B, Curcio C, Harney JW, Salvatore D, Sorimachi K, Larsen PR, Bianco AC. Human type 3 iodothyronine selenodeiodinase is located in the plasma membrane and undergoes rapid internalization to endosomes. J Biol Chem. 2003;278(2):1206–1211. doi: 10.1074/jbc.M210266200. [DOI] [PubMed] [Google Scholar]
- 144.Arrojo E, Drigo R, Bianco AC. Type 2 deiodinase at the crossroads of thyroid hormone action. Int J Biochem Cell Biol. 2011;43(10):1432–1441. doi: 10.1016/j.biocel.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Arrojo e Drigo R, Fonseca TL, Castillo M, Salathe M, Simovic G, Mohacsik P, Gereben B, Bianco AC. Endoplasmic reticulum stress decreases intracellular thyroid hormone activation via an eIF2a-mediated decrease in type 2 deiodinase synthesis. Mol Endocrinol. 2011;25:2065–2075. doi: 10.1210/me.2011-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wang S, Binder P, Fang Q, Wang Z, Xiao W, Liu W, Wang X. Endoplasmic reticulum stress in the heart: insights into mechanisms and drug targets. Br J Pharmacol. 2018;175(8):1293–1304. doi: 10.1111/bph.13888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kryukov GV, Kumar RA, Koc A, Sun Z, Gladyshev VN. Selenoprotein R is a zinc-containing stereo-specific methionine sulfoxide reductase. Proc Natl Acad Sci USA. 2002;99(7):4245–4250. doi: 10.1073/pnas.072603099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Grumolato L, Elkahloun AG, Ghzili H, Alexandre D, Coulouarn C, Yon L, Salier JP, Eiden LE, Fournier A, Vaudry H, Anouar Y. Microarray and suppression subtractive hybridization analyses of gene expression in pheochromocytoma cells reveal pleiotropic effects of pituitary adenylate cyclase-activating polypeptide on cell proliferation, survival, and adhesion. Endocrinology. 2003;144(6):2368–2379. doi: 10.1210/en.2002-0106. [DOI] [PubMed] [Google Scholar]
- 149.Rocca C, Boukhzar L, Granieri MC, Alsharif I, Mazza R, Lefranc B, Tota B, Leprince J, Cerra MC, Anouar Y, Angelone T. A selenoprotein T-derived peptide protects the heart against ischaemia/reperfusion injury through inhibition of apoptosis and oxidative stress. Acta Physiol (Oxf) 2018;223(4):e13067. doi: 10.1111/apha.13067. [DOI] [PubMed] [Google Scholar]
- 150.Hausenloy DJ, Yellon DM. Myocardial ischemia–reperfusion injury: a neglected therapeutic target. J Clin Investig. 2013;123(1):92–100. doi: 10.1172/JCI62874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Venardos KM, Perkins A, Headrick J, Kaye DM. Myocardial ischemia–reperfusion injury, antioxidant enzyme systems, and selenium: a review. Curr Med Chem. 2007;14(14):1539–1549. doi: 10.2174/092986707780831078. [DOI] [PubMed] [Google Scholar]
- 152.Li JP, Zhou JX, Wang Q, Gu GQ, Yang SJ, Li CY, Qiu CW, Deng GZ, Guo MY. Se Enhances MLCK activation by regulating selenoprotein T (SelT) in the gastric smooth muscle of rats. Biol Trace Elem Res. 2016;173(1):116–125. doi: 10.1007/s12011-016-0620-8. [DOI] [PubMed] [Google Scholar]
- 153.Whanger PD. Selenoprotein expression and function-selenoprotein W. Biochim Biophys Acta. 2009;1790(11):1448–1452. doi: 10.1016/j.bbagen.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 154.Whanger PD, Ip C, Polan CE, Uden PC, Welbaum G. Tumorigenesis, metabolism, speciation, bioavailability, and tissue deposition of selenium in selenium-enriched ramps (Allium tricoccum) J Agric Food Chem. 2000;48(11):5723–5730. doi: 10.1021/jf000739s. [DOI] [PubMed] [Google Scholar]
- 155.Gu QP, Sun Y, Ream LW, Whanger PD. Selenoprotein W accumulates primarily in primate skeletal muscle, heart, brain and tongue. Mol Cell Biochem. 2000;204(1–2):49–56. doi: 10.1023/a:1007065829068. [DOI] [PubMed] [Google Scholar]
- 156.Låg M, Refsnes M, Lilleaas EM, Holme JA, Becher R, Schwarze PE. Role of mitogen activated protein kinases and protein kinase C in cadmium induced apoptosis of primary epithelial lung cells. Toxicology. 2005;211:253–264. doi: 10.1016/j.tox.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 157.Yi JH, Park SW, Kapadia R, Vemuganti R. Role of transcription factors in mediating post-ischemic cerebral inflammation and brain damage. Neurochem Int. 2007;50:1014–1027. doi: 10.1016/j.neuint.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Haidong Y, Wei L, Wenchao Z, Ruifeng F, Xia Z, Pervez Ahmed K, Ziwei Z, Shiwen X. Different responses of selenoproteins to the altered expression of selenoprotein W in chicken myoblasts. RSC Adv. 2014;4:64032. [Google Scholar]
- 159.Liu W, Yao H, Zhao W, Shi Y, Zhang Z, Xu S. Selenoprotein W was correlated with the protective effect of selenium on chicken myocardial cells from oxidative damage. Biol Trace Elem Res. 2016;171(2):419–426. doi: 10.1007/s12011-015-0529-7. [DOI] [PubMed] [Google Scholar]
- 160.Yang J, Hamid S, Liu Q, Cai J, Xu S, Zhang Z. Gene expression of selenoproteins can be regulated by thioredoxin(Txn) silence in chicken cardiomyocytes. J Inorg Biochem. 2017;177:118–126. doi: 10.1016/j.jinorgbio.2017.08.027. [DOI] [PubMed] [Google Scholar]
- 161.Kim YJ, Chai YG, Ryu JC. Selenoprotein W as molecular target of methylmercury in human neuronal cells is down-regulated by GSH depletion. Biochem Biophys Res Commun. 2005;330:1095–1102. doi: 10.1016/j.bbrc.2005.03.080. [DOI] [PubMed] [Google Scholar]
- 162.Sunde RA. mRNA transcripts as molecular biomarkers in medicine and nutrition. J Nutr Biochem. 2010;21(8):665–670. doi: 10.1016/j.jnutbio.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Oropeza-Moe M, Wisløff H, Bernhoft A. Se deficiency associated porcine and human cardiomyopathies. J Trace Elem Med Biol. 2015;31:148–156. doi: 10.1016/j.jtemb.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 164.de Lorgeril M, Salen P, Accominotti M, Cadau M, Steghens JP, Boucher F, de Leiris J. Dietary and blood antioxidants in patients with chronic heart failure. Insights into the potential importance of Se in heart failure. Eur J Heart Fail. 2001;3(6):661–669. doi: 10.1016/s1388-9842(01)00179-9. [DOI] [PubMed] [Google Scholar]
- 165.Mistry HD, Wilson V, Ramsay MM, Symonds ME, Broughton Pipkin F. Reduced Se concentrations and glutathione peroxidase activity in preeclamptic pregnancies. Hypertension. 2008;52(5):881–888. doi: 10.1161/HYPERTENSIONAHA.108.116103. [DOI] [PubMed] [Google Scholar]
- 166.Sibai B, Dekker G, Kupferminc M. Pre-eclampsia. Lancet. 2005;365:785–799. doi: 10.1016/S0140-6736(05)17987-2. [DOI] [PubMed] [Google Scholar]
- 167.Broughton Pipkin F. Risk factors for preeclampsia. N Engl J Med. 2001;344:925–926. doi: 10.1056/NEJM200103223441209. [DOI] [PubMed] [Google Scholar]
- 168.Hubel CA. Oxidative stress in the pathogenesis of preeclampsia. Proc Soc Exp Biol Med. 1999;222:222–235. doi: 10.1177/153537029922200305. [DOI] [PubMed] [Google Scholar]
- 169.Atamer Y, Kocyigit Y, Yokus B, Atamer A, Erden AC. Lipid peroxidation, antioxidant defense, status of trace metals and leptin levels in preeclampsia. Eur J Obstet Gynecol Reprod Biol. 2005;119(60):66. doi: 10.1016/j.ejogrb.2004.06.033. [DOI] [PubMed] [Google Scholar]
- 170.Rayman MP, Bode P, Redman CW. Low Se status is associated with the occurrence of the pregnancy disease preeclampsia in women from the United Kingdom. Am J Obstet Gynecol. 2003;189:1343–1349. doi: 10.1067/s0002-9378(03)00723-3. [DOI] [PubMed] [Google Scholar]
- 171.Bulgan Kilicdag E, Ay G, Celik A, Ustundag B, Ozercan I, Simsek M. Oxidant-antioxidant system changes relative to placental-umbilical pathology in patients with preeclampsia. Hypertens Pregnancy. 2005;24:147–157. doi: 10.1081/PRG-200059863. [DOI] [PubMed] [Google Scholar]
- 172.Paravicini TM, Drummond GR, Sobey CG. Reactive oxygen species in the cerebral circulation: physiological roles and therapeutic implications for hypertension and stroke. Drugs. 2004;64:2143–2157. doi: 10.2165/00003495-200464190-00001. [DOI] [PubMed] [Google Scholar]
- 173.Arteel GE, Mostert V, Oubrahim H, Briviba K, Abel J, Sies H. Protection by selenoprotein P in human plasma against peroxynitrite mediated oxidation and nitration. Biol Chem. 1998;379:1201–1205. [PubMed] [Google Scholar]
- 174.Schomburg L, Schweizer U, Holtmann B, Flohe L, Sendtner M, Kohrle J. Gene disruption discloses role of selenoprotein P in Se delivery to target tissues. Biochem J. 2003;370:397–402. doi: 10.1042/BJ20021853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Koyama H, Abdulah R, Ohkubo T, Imai Y, Satoh H, Nagai K. Depressed serum selenoprotein P: possible new predicator of increased risk for cerebrovascular events. Nutr Res. 2009;29(2):94–99. doi: 10.1016/j.nutres.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 176.Gharipour M, Sadeghi M, Salehi M, Behmanesh M, Khosravi E, Dianatkhah M, Haghjoo Javanmard S, Razavi R, Gharipour A. Association of expression of selenoprotein P in mRNA and protein levels with metabolic syndrome in subjects with cardiovascular disease: Results of the Selenegene study. J Gene Med. 2017;19(3):e2945. doi: 10.1002/jgm.2945. [DOI] [PubMed] [Google Scholar]
- 177.Büttner P, Obradovic D, Wunderlich S, Feistritzer HJ, Holzwirth E, Lauten P, Fuernau G, de Waha-Thiele S, Desch S, Thiele H. Selenoprotein P in myocardial infarction with cardiogenic shock. Shock. 2019 doi: 10.1097/SHK.0000000000001342. [DOI] [PubMed] [Google Scholar]