Abstract
Glycosyltransferases (GTases) transfer sugar moieties to proteins, lipids or existing glycan or polysaccharide molecules. GTases form an important group of enzymes in the Golgi, where the synthesis and modification of glycoproteins and glycolipids take place. Golgi GTases are almost invariably type II integral membrane proteins, with the C-terminal globular catalytic domain residing in the Golgi lumen. The enzymes themselves are divided into 103 families based on their sequence homology. There is an abundance of published crystal structures of GTase catalytic domains deposited in the Protein Data Bank (PDB). All of these represent either of the two main characteristic structural folds, GT-A or GT-B, or present a variation thereof. Since GTases can function as homomeric or heteromeric complexes in vivo, we have summarized the structural features of the dimerization interfaces in crystal structures of GTases, as well as considered the biochemical data available for these enzymes. For this review, we have considered all 898 GTase crystal structures in the Protein Data Bank and highlight the dimer formation characteristics of various GTases based on 24 selected structures.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-017-2659-x) contains supplementary material, which is available to authorized users.
Keywords: Protein, Structure, Dimerization, Biologically relevant dimer, Crystallographic dimer, Protein–protein interfaces, GTase fold
Introduction
Eukaryotic cells are coated with glycans of variable composition and structure. These glycans are covalently attached to membrane proteins and lipids as a result of glycosylation, and form the basis of various cellular recognition events needed for cell–cell contacts or in differentiating between the own and the foreign by the immune system. Glycosylation, therefore, must be a very precise process, and improper glycosylation is in many cases manifested in diseases due to impaired cellular recognition. Such diseases include congenital disorders of glycosylation, inflammation, diabetes and cancers (for recent reviews, see Hennet and Cabalzar [1], Chang and Yang [2] and Vajaria et al. [3]).
Glycan synthesis takes place in the endoplasmic reticulum and the Golgi apparatus and involves a complex interplay between a number of carbohydrate-acting enzymes, donor and acceptor substrates, nucleotide-activated sugars and their transporters. Therefore, and to ensure fidelity in glycan synthesis, there is a specific requirement for the presence of distinct sets of glycosidases and glycosyltransferases (GTases) in the cell. The latter form a huge ensemble of enzymes currently divided into 103 sequence-based families, according to the CAZy database [4] (http://www.cazy.org). They catalyse the addition of specific sugar moieties in specific sequence and chemical configuration (i.e. the linkages between sugar units and the stereochemistry of the product and the substrate—inverting or retaining) to specific acceptor molecules, which can be carbohydrates, proteins or lipids. Given the huge variety of glycan structures needed for normal cellular recognition events, it is therefore not a surprise that the total amount of different GTases in the CAZy database approaches 250.
Glycosyltransferases—the topic of this review—are almost invariably type II integral membrane proteins with a short cytoplasmic tail, a single transmembrane domain, a stem region and a globular catalytic domain located in the Golgi lumen. Due to the difficulties in both producing and crystallizing full-length type II membrane proteins, all the crystal structures of GTases thus far solved represent their soluble, globular catalytic domains.
Glycosyltransferases form homomers
GTases have been shown to form enzyme dimers, tetramers and oligomers in live cells mainly via interactions between their catalytic domains [5–7], and it has been suggested that ordered protein arrays in the trans-Golgi might contain GTases [8]. Considering that these enzymes do not use a template, a question of considerable interest is whether enzyme complex formation is part of the cellular mechanism to ensure the fidelity of glycan synthesis.
How to analyse dimerization?
For homomeric complexes, it has been shown that dimerization is the most common transition occurring during the assembly of protein complexes [9], cyclization being the next most common, while fractional transitions are the rarest. We therefore focused on dimerization interfaces, acknowledging that even if the GTases may form higher-order oligomers, dimerization would still be a biologically relevant step in the homomer formation. Even with the abundance of structural information, analysis of protein dimerization (or formation of higher-order oligomers) with the help of crystal structures is not straightforward. Protein crystals may contain more than one protein molecule in the asymmetric unit (the smallest repetitive unit of the crystal). In such cases, these two or more molecules are typically symmetrically arranged. This so-called non-crystallographic symmetry is a feature separate from the crystallographic symmetry and would not necessarily exist, if the interaction observed in the crystallized species was not due to a functional reason. Instead, the crystal unit cell and the crystal symmetry would then simply form differently. Crystal formation necessarily involves molecular contacts; therefore, the problem is to separate functionally relevant, or “physiological”, protein–protein contacts from interactions that merely bring about and maintain crystal packing. Consequently, other data including biochemical characterization of the complexes using, e.g. gel filtration, analytical ultracentrifugation or dynamic light scattering, must be taken into account.
In favourable cases, there is a well justified logical reason for the protein to form dimers, for example in the case when a ligand binding site is formed from residues located in different monomers, or when a prediction of protein–protein interactions on the basis of analysing interaction site properties can be made with high confidence. The latter approach is a very active field of research, and a great many server-based analysis tools are now freely available [10, 11]. For this review, we have reanalysed all the 898 GTase crystal structures in the Protein Data Bank (PDB, http://www.rcsb.org) [12] using the above criteria and present our view on various GTase dimers that are likely to also form functionally relevant complexes in vivo.
Selection of GTase structures to study and their structural characteristics
At the time we started this work, the contents of the CAZy data base and the PDB included a total of 898 crystal structures of GTases. After thorough analysis of all GTase families, we chose the structures of 172 unique proteins such that 44 of the 103 GTase families were represented by at least one crystal structure. 61% of all GTase crystal structures are eukaryotic, of which 40% represent human proteins. A fair number of these structures are complexes with donor nucleotide-activated sugars and/or acceptor glycans, or molecules representing only parts of them.
Based on literature, a major motivation to obtain high-quality GTase structures seems to be to get atomic resolution details of the catalytic mechanisms and ligand binding modes to use this data for drug design. GTase structures from a wide range of species are often usable for functional analysis due to the structural conservation between enzymes across species. Each coordinate entry of the PDB is filed as a separate structure, although many of the entries are redundant. This is due to structure–function studies requiring structures of proteins in several different states, including apo- and multiple holo structures with different ligands bound. An additional reason for structural redundancy is that most GTases fall into two similar fold types: GT-A and GT-B, and variants thereof, with only a limited degree of structural difference. The structural conservation is not reflected in the sequence similarity: the average sequence identity was found to be only 12 and 11% for GT-A and GT-B folds, respectively, in a set of 67 nonredundant GTase structures representing 28 families [13]. A small portion of the GTases possess neither the GT-A nor the GT-B fold, but display slightly different topological properties [14]. GTases within a given family usually share the same fold type [15].
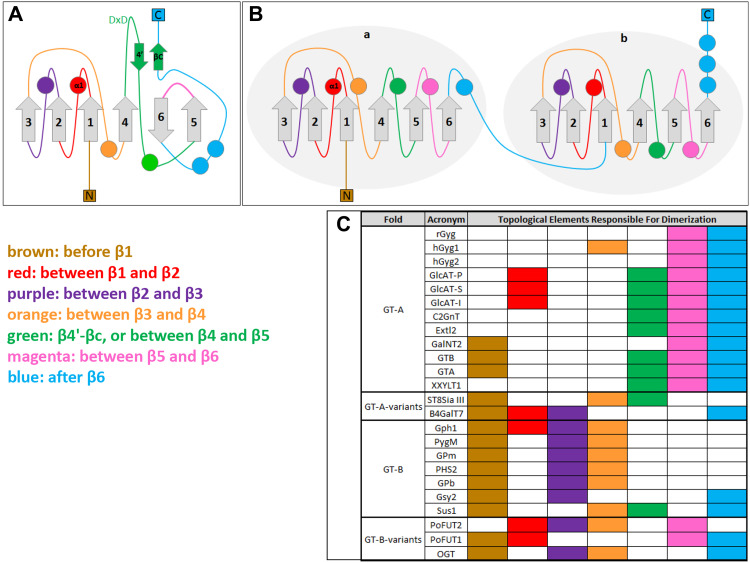
GT-A and GT-B folds have similar spatial arrangements consisting of α/β alternations, with variable N- and C-termini. Although the size of the α and β parts vary, the overall structure is always held together by a continuous central twisted β-sheet called the Rossmann fold, which is flanked by α-helices on both sides [16]. The GT-A fold contains one six-stranded β-sheet showing a 321465 topology, in which β6 is antiparallel to the other strands (Figs. 1, 2A). Insertions breaking the α/β alternation are often found between β5 and β6, and more rarely between other strands. A smaller antiparallel two-stranded β-sheet that consists of β4′ (a short strand flanking β4) and βC (a short strand in the variable C terminus) is usually present in eukaryotic GT-A folds (Fig. 2A). This two-stranded β-sheet is sometimes accompanied by parallel or antiparallel short β strands from the variable C-terminal part. Other common features of the GT-A fold are the Asp-X-Asp (also known as DxD) motif, and a divalent cation binding motif, usually flanking β4 [15, 17–19], which is needed for activity. Some GTases may occasionally lack these features and still be considered as part of the GT-A fold family.
Fig. 1.
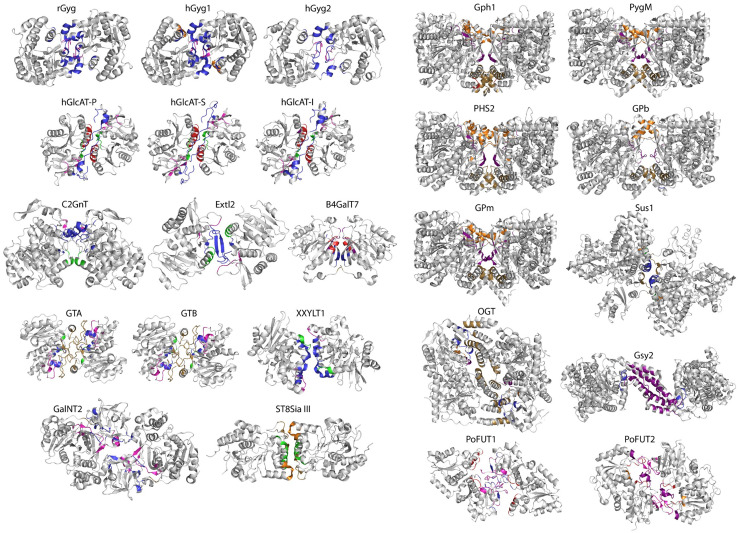
Ribbon drawings of the 24 GTase homodimeric structures comprising the research material of this study. All structures are presented in orientations which easily show the secondary structural elements in the dimer interface, with the location of interacting residues in those structural elements colour coded as follows: before β1 (brown), between β1 and β2 (red), between β2 and β3 (purple), between β3 and β4 (orange), β4′–βC/between β4 and β5 (green), between β5 and β6 (magenta), after β6 (blue). Each structure can be identified with the enzyme acronym; the same identification is used in Table 1 and in the text. GT-A fold and GT-A variant structures are on the left, while GT-B fold and GT-B variant structures are on the right
Fig. 2.
Topological elements responsible for dimerization are presented separately for GT-A and GT-B folds as topology diagrams (A, B respectively) and as a table indicating the use of each topological element by the studied GTases (C). A, B Topology of the GT-A and GT-B folds. The common structural core β-sheet is in grey with the strands numbered. The topological elements connecting the core β-strands are shown with α-helices as circles, β-strands as arrows and loops/random structure as plain lines, and colour coded as follows: before β1 (brown), between β1 and β2 (red), between β2 and β3 (purple), between β3 and β4 (orange), β4′–βC/between β4 and β5 (green), between β5 and β6 (magenta) and after β6 (blue). In C the same elements are tabulated to clarify the use of each element in dimer formation by each fold type. Colour coding is the same as in (A, B). As discussed in the text, certain topological elements are used for dimerization mainly or exclusively by GT-A enzymes, while a different set of elements is utilized by GT-Bs. Additionally, the mixed nature of the variant folds is evident
The GT-B fold consists of two separate Rossmann fold motifs, each of them consisting of a six-stranded parallel β-sheet with a 321456 topology and connected by a linker region [20] (Figs. 1, 2A). The two domains face each other, with the active site located within the resulting cleft. Some variant GTases possess a fold closely resembling the canonical GT-A or GT-B topology, but with a different order of β-strands. These variants have sometimes been regarded as new fold types, increasing the confusion in the classification. The classification we describe above is based on a common structural core shared within the dataset of the GTase structures used in this study.
The GTase structures in the CAZy database were imported, family by family, into Excel for analysis. Out of 898 crystal structures, 338 contained more than a single protein molecule in the asymmetric unit and were selected for further investigation. These 338 structures were then sorted by kingdom, species, and unique protein name. Of these, 164 were from eukaryote species, among which 82 were of human origin, representing 15 different GTases. We then set out to analyse all these human GTases in detail, including also homologues from other species when appropriate. The PDB codes of the 164 selected eukaryote GTase structures as well as the associated PDB files were gathered using a custom python script. In the case where more than one structure was available for a given protein, structural alignments were made to choose the most representative one, typically the example with the highest resolution. We did not discriminate between apo- and holoenzymes, since the local conformational changes brought about by substrate binding generally did not affect the overall fold or dimerization properties.
Our final selection contains 24 structures from 18 different GTases, representing both the main GT-A and GT-B folds and their variants (Fig. 1, Table 1). Each structure was evaluated for the likelihood of a physiological dimer being present in the asymmetric unit of the crystals using various criteria/tools (Table 1). The nature of the interface and thermodynamic properties were assessed employing the jsPISA macromolecular surface and interface calculation tool [21], Voronoi tessellation, i.e. the DiMoVo server [22], and the EPPIC [23] server. Evolutionary conservation of the interface was assessed using the InterEvol [24] server.
Table 1.
Summary of the analysis of the dimer interface of the 24 GTase structures
| Family | Organism | Acronym | PDB code | Chains | IA (Å2) | DG (kcal/mol) | BE (kcal/mol) | jsPISA | DiMoVo | EPPIC |
|---|---|---|---|---|---|---|---|---|---|---|
| Glycogenins | ocu | rGyg | 1LL0 | EF | 1205 | −23.37 | −26.92 | 64 | 0.65 | Bio |
| hsa | hGygl | 3U2U | AB | 1420 | −29.41 | −31.63 | 63 | 0.45 | Bio | |
| Glucuronyltransferases | hsa | hGyg2 | 4UEG | AB′ | 1067 | −23.43 | −24.76 | 58 | 0.51 | Bio |
| hsa | GlcAT-P | 1V84 | AB | 2049 | −21.61 | −36.12 | 72 | 0.68 | Bio | |
| hsa | GlcAT-S | 2D0J | AB | 2225 | −11.36 | −31.08 | 73 | 0.57 | Bio | |
| hsa | GlcAT-l | 3CU0 | AB | 2029 | −28.52 | −41.11 | 72 | 0.87 | Bio | |
| mmu | C2GnT | 2GAK | AB | 941 | −10.03 | −14.19 | 56 | 0.10 | Xtal | |
| N-Acetylglucosaminyltransferases | mmu | Extl2 | 1OMZ | AB | 1294 | −16.4 | −23.06 | 60 | 0.73 | Bio |
| hsa | GalNT2 | 5FV9 | AB | 1348 | −12.23 | −18.01 | 66 | 0.66 | Xtal | |
| Galactosyltransferases | hsa | GTB | 3U0X | AB | 2696 | −32.01 | −48.09 | 79 | 1.32 | Bio |
| hsa | GTA | 3U0Y | AB | 2498 | −27.12 | −41.35 | 78 | 1.27 | Bio | |
| Xylosyltransferases | mmu | XXYLT1 | 4WLM | AB | 1214 | −12.7 | −19.8 | 63 | 0.73 | Bio |
| Sialyltransferases | hsa | ST8Sia III | 5BO7 | AB | 974.7 | −8.99 | −14.62 | 59 | 0.33 | Xtal |
| Galactosyltransferases | hsa | B4GalT7 | 4IRP | AB | 757.3 | −12.18 | −14.84 | 54 | 0.67 | Bio |
| sce | Gphl | 1YGP | AB | 3355 | −28.04 | −44.12 | 71 | 0.83 | error | |
| ocu | PygM | 2IEG | AB | 2067 | −18.99 | −25.52 | 70 | 0.51 | Bio | |
| Glycogen phosphorylases | hsa | GPm | 3DDS | AB | 2507 | −26.71 | −37 | 75 | 0.69 | Bio |
| ath | PHS2 | 4BQE | AB | 2586 | −23.45 | −34.11 | 67 | 0.43 | Bio | |
| hsa | GPb | 5IKO | AC | 1471 | −7.446 | −15.29 | 57 | 0.40 | Bio | |
| Glycogen synthases | sce | Gsy2 | 3NB0 | BD | 1594 | −32.15 | −35.26 | 66 | 0.87 | Bio |
| Sucrose synthases | ath | Sus1 | 3S28 | BC | 1284 | −18.23 | −22.17 | 62 | 0.78 | Bio |
| Fucosyltransferases | hsa | PoFUT2 | 4AP5 | AB | 1717 | −15.52 | −20.71 | 62 | 0.15 | Xtal |
| cel | PoFUTl | 3ZY5 | AA′ | 1297 | −12.54 | −18.18 | 62 | 0.27 | Xtal | |
| N-Acetylglucosaminyltransferases | hsa | OGT | 4GYW | AA′ | 2058 | −12.14 | −18.07 | 60 | 0.60 | Bio |
Numerical values or assessment given by each tool is discussed in the text. Organism codes: hsa stands for Homo sapiens, ocu for Oryctolagus cuniculus, mmu for Mus musculus, sce for Saccharomyces cerevisiae, ath for Arabidopsis thaliana, cel for Caenorhabditis elegans. Chains indicate the name of the chains in the crystal structure used to analyse the dimer interface: monomers related by a crystallographic symmetry are highlighted (bold values) in contrast to monomers related by a non-crystallographic symmetry (unblod values). IA interface area; DG, solvation energy; BE total binding energy. jsPISA score is a weighted average of each of the jsPISA radar metrics, for which a value higher than 50% depicts good possibility of the interface being biologically relevant. DiMoVo score values below 0.5 predict crystal dimers, while values above 0.5 predict biological dimers. EPPIC server assessment predicts biologically relevant dimers (Bio) or crystal dimers (Xtal). For each metric, bold values indicate off-limit values suggesting a crystallographic dimer rather than a dimer of physiological relevance: IA < 800 Å2, DG > −10 kcal/mol, BE > −16 kcal/mol, jsPISA score < 50%, DiMoVo score < 0.5, EPPIC assessment for a crystal dimer
In the following paragraphs, we will first review various GTase dimers as they are described in the literature and also refer to the existing biochemical evidence of their dimerization, if such data are available. We then summarize, with the help of bioinformatic tools, their likelihood of representing physiologically relevant enzyme dimers.
GT-A folds
β-Glucuronyltransferases (PDB codes 3CU0, 1V84, 2D0J)
β-Glucuronyltransferases (EC 2.4.1.135) belong to family 43 inverting GTases, which use UDP-glucuronate as the donor substrate. They add the glucuronic acid moiety to an existing galactosyl–galactosyl–xylosyl- or galactosyl–xylosylprotein acceptor depending on the specific enzyme. Crystal structures have been solved for three of the human enzymes: glucuronyltransferase-I (GlcAT-I; PDB 3CU0) [25], glucuronyltransferase-P (GlcAT-P; PDB 1V84) [26], and glucuronyltransferase-S (GlcAT-S; PDB 2D0J) [27].
The GlcAT-I structure appears as a functional dimer (Fig. 1). Both monomers are required for binding to the acceptor molecule. More specifically, the oxygen and nitrogen atoms of the side chain of residue Gln318 of one monomer are at a hydrogen bonding distance from the O-6 atom of the Gal-1 moiety of the acceptor bound to the active site of the other monomer [28]. Furthermore, if the O-6 position is sulphated, the NE2 atom of Gln318 from the other monomer undergoes a conformational change and positions itself at a 3.0 Å distance from the O-4 oxygen atom of the sulphate [25]. Enzyme kinetic studies provide additional evidence in favour of a functionally relevant GlcAT-I dimer: a sulphated or a phosphorylated acceptor enhances GlcAT activity, but only if the enzyme is dimeric [25].
GlcAT-P structure [26] is highly similar to GlcAT-I. This holds true also for the dimer interface area. For example, the last β-strand, containing the Gln318 residue, extends to the active site of the other monomer, exactly as in GlcAT-I. GlcAT-P has also been shown to exist as a dimer by gel filtration under non-denaturing conditions [29], as well as by analytical ultracentrifugation, even when the N-terminal part containing the transmembrane domain is deleted [30].
GlcAT-S structure [27] was solved by using the GlcAT-P structure as the search model in molecular replacement, and the same conclusions regarding GlcAT-S dimerization could be drawn.
Glycogenins (PDB codes 1LL0, 3U2U, 4UEG)
Glycogenins (GTase family 8; EC 2.4.1.186) are autocatalytic proteins serving not only as the core of the glycogen structure, but also as enzymes catalysing the addition of the first UDP-glucose molecules in the initial phase of glycogen synthesis. In the catalysis, the stereochemistry of the added glucose is retained as α.
Several crystal structures of glycogenins have been solved: glycogenin-1 from rabbit (rGYG1; PDB 1LL0) [32] and human (PDB 3U2U) [33], as well as human glycogenin-2 (PDB 4UEG) [34, 35] serve as representative examples.
Rabbit glycogenin (rGYG1) was crystallized in two crystal forms—one containing ten molecules (five dimers) per asymmetric unit, while the other holding only one molecule per asymmetric unit. In the former crystal form (tetragonal), the monomers of the dimers are related to each other by a non-crystallographic twofold axis, creating identical dimers compared to the crystallographic dimers of the latter crystal form (orthorhombic) [32]. The decameric variant of rGYG1 is likely to be an artefact of concentrating the protein for crystallization for three reasons: (1) the purified rGYG1 was suggested to be a dimer by density gradient centrifugation [31]; (2) the active sites of glycogenin monomers in the complex would in this form be placed unfavourably with regard to the glycogen biosynthesis by the glycogen synthase; (3) the interface areas between the dimers (that form the decamer) cover only 7% of the total surface area. Thus, the decamer likely connects dimers to support crystal packing. In the orthorhombic crystal form of rGYG1, 20% of the total surface area is involved in dimer contacts, likely representing a physiologically relevant dimer as this value is typical for proteins that possess high-affinity binding with each other [36].
The ensemble of rGYG1 structures [33] with different intermediates of glycogen synthesis has revealed a “lid” domain, which guides the substrates in the narrow dimer interface. The substrates are then subjected to either intra- or intersubunit catalysis, depending on the chain length of the nascent glycan chain and steric factors in the channel. The term “intrasubunit mechanism” refers to an activity of the glycogenin monomer, while the “intersubunit mechanism” involves catalytic residues from both monomers in a glycogenin dimer. The findings by Issoglio et al. [37], who studied the mechanisms of monomeric and dimeric rabbit muscle glycogenin, fully support the above view. They found that, while a glycogenin monomer is sufficient for priming glycogen biosynthesis in vivo via the intrasubunit mechanism, the intersubunit mechanism mediated by the glycogenin dimer is needed for the full polymerization capacity of glycogenin.
Human glycogenins have been shown to form non-covalent dimers with shared enzymatic activity between monomers. All crystal forms of the human glycogenin [33] contain dimers. One of the glycogenin monomers acts as the glucose-introducing transferase, while the other serves for glucose branching in the growing glycogen chain [34, 35]. Glycogenin-1 is also co-purified with glycogenin-2, and vice versa, suggesting that the two glycogenins may also form heterodimers.
Xylosyltransferases (PDB code 4WLM)
Xyloside xylosyltransferase-1 (XXYLT1; GTase family 8; a retaining α-1,3-xylosyltransferase; EC 2.4.2.n3) catalyses the addition of an α-d-xylose to an existing xylose–glucose disaccharide to complete the synthesis of the trisaccharide O-linked to EGF-like repeats in Notch proteins [38]. XXYLT1 possesses the typical GT-A fold signature of the DxD motif to coordinate a catalytic Mn2+ ion. Human XXYLT1 has been expressed in Sf9 cells as a full-length type II membrane protein and purified [38]. It was found that XXYLT1 forms SDS-resistant homodimers linked together by a disulphide bond between the transmembrane domains. The crystal structure of the luminal catalytic domain of XXYLT1 [39] is also a dimer, with an interface area between monomers well in the range typical for functionally relevant protein–protein interactions, although the ΔG of −12.7 kcal/mol is rather low (Table 1). It was assumed that the catalytic domains provide additional dimerization contacts in XXYLT1 [39]. The active sites of the catalytic domains do not overlap with the dimer interface area, and the active sites appear to be positioned in such a way that it is consistent with the orientation of the Notch acceptor proteins.
N-Acetylglucosaminyl- and N-acetylgalactosaminyltransferases (PDB codes 2GAK, 1OMZ, 5FV9)
The crystal structures of three different N-acetylglucosaminyltransferases have been published. These are (1) core 2 β-1,6-N-acetylglucosaminyltransferase (C2GnT; GTase family 14; EC 2.4.1.102) [40], (2) α-1,4-N-acetylglucosaminyltransferase (Extl2; GTase family 64; EC 2.4.1.223) [41] from mouse, and (3) human polypeptide N-acetylgalactosaminyltransferase (GalNT2; GTase family 27; EC 2.4.1.41) [42]. Both of the glucosaminyltransferases use UDP-N-acetylglucosamine as the substrate, but they act on different acceptor glycans in different biosynthetic pathways: C2GnT adds N-acetylglucosamine to an N-acetylgalactosamine with a 1,6-linkage making the core 2 structure of mucin type O-glycans, while Extl2 produces 1,4-linked glucuronic acid and N-acetylglucosamine repeats found in heparin sulphate chains. The human galactosaminyltransferase GalNT2 uses UDP-N-acetyl-α-d-galactosamine as a substrate to add the first sugar in mucin biosynthesis.
C2GnT was found to exist both as monomers and dimers in cells [43], while the predominant form in solution (secreted in culture media) was monomeric [40, 43]. Surprisingly, in the crystal structure the two C2GnT monomers form a disulphide-bonded dimer via Cys235 residues. However, this dimer may not reflect the physiological situation, since the Cys235 is unique to the murine enzyme. The DiMoVo score for C2GnT (2GAK) is also low (Table 1), supporting the view that the observed dimer is probably a result of crystal packing. On the other hand, the jsPISA analysis suggests that the C2GnT dimer could well be a biologically relevant dimer, even without the disulphide bridge (Table 1). Of the two molecular forms, only the dimer could be crystallized. The fact that C2GnT crystal structure contains the stem domain (in addition to the catalytic domain) makes it a rare exception among the purified and crystallized GTases. Two disulphide bridges connect the stem domain to the catalytic domain, but due to high temperature factors of the stem domain and the lack of extensive contacts between the two domains, it may not represent the conformation present in the full-length protein [40].
Extl2 does not form a disulphide-bonded dimer, but the dimeric nature of the enzyme could be assigned with more confidence than for C2GnT due to the dimer interface area, the ΔG of binding and other characteristics of jsPISA interaction radar analysis (Table 1). However, no direct experimental evidence on the protein behaviour in solution exists to support this view.
GalNT2 was crystallized with three independent dimers in the asymmetric unit. Our analysis with the EPPIC server indicates that the interactions between the monomers are only crystal contacts, despite the other parameters favouring the existence of biologically relevant dimers (Table 1). Structural studies by others on the same enzyme revealed a crystallographic dimer [44] or a dimer with an interface not likely to be biologically significant [45].
The three structures described above do not superimpose well, with an r.m.s. deviation of atomic positions in pairwise comparisons ranging from 6.7 to 16.4 Å, as estimated with PyMOL (The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC.).
ABO blood group antigen glycosyltransferases (PDB codes 3U0X, 3U0Y)
ABO blood group antigens attached to membrane proteins or lipids contain a common N-acetylgalactosamine–galactose–fucose trisaccharide core, which is non-antigenic and defines the type O blood. This core structure is then modified to blood type A and B antigens upon addition of an N-acetylgalactosamine or a galactose, respectively, as a terminal sugar by a relevant glycosyltransferase (GT family 6). Several high-resolution apo- and holo structures of both blood group A specifying α-1,3-N-acetylgalactosaminyltransferase (GTA; EC 2.4.1.40) and blood group B specifying α-1,3-galactosyltransferase (GTB; EC 2.4.1.37) from humans have been solved. In addition, a chimeric enzyme (AAGlyB) capable of transferring either of the terminal sugars has been constructed and its structure solved [46]. All of these structures are highly similar, as expected given that the GTA and GTB enzymes differ only by four amino acid residues.
The GTA (PDB 3U0Y) and GTB (PDB 3U0X) structures were solved to 1.6 and 1.85 Å resolution, respectively, in complex with a GTB-specific inhibitor compound [47] and present as dimers. The respective monomers are related by twofold symmetry, which may indicate biological relevance [48]. The stem regions of the two monomers extend to form a large dimer interface dominated by random coil and mediate the physical interaction between the two type II membrane proteins. Dimer formation of the crystallizable species of GTA in solution has been experimentally verified by SDS-PAGE [49]. This type of dimer contact—formed through the stem regions—appears to be a rather unique feature of only some glycosyltransferases.
GT-A variants
Sialyltransferases (PDB code 5BO7)
ST8 α-N-acetyl-neuraminide α-2,8-sialyltransferase 3 (ST8SiaIII; EC 2.4.99) is an oligo/poly-sialylating sialyltransferase, which uses a CMP-activated sialic acid unit as a donor to add a sialic acid to a terminal position with an α-2,8 linkage on different acceptors [50]. The enzyme belongs to GTase family 29 and its crystal structure revealed a variant of the common GT-A fold [51, 52]. The ST8SiaIII structure displays a 612345 topology where all the strands are parallel (instead of 321465 with β6 antiparallel). Being active on oligo- and polysialylation, a positively charged binding pocket is needed to accommodate the negatively charged donor and acceptor molecules. The ST8SiaIII crystal structure [51] revealed that such a groove is indeed formed by patches of the surface forming the dimer interface, emphasizing that the active enzyme is by necessity a dimer. In contrast, monosialylating enzymes such as ST3GalI and ST6GalI operate on uncharged acceptor molecules and, therefore, do not need—and do not have—large positive binding areas [51, 53, 54]. ST8SiaIII’s dimer interface contains symmetrical pairs of hydrogen bonds created by residues which are not conserved in monomeric ST8SiaII and ST8SiaIV enzymes. Static light scattering experiments carried out by Volkers et al. [51] confirmed that ST8SiaIII is a dimer also in solution.
In the ST8SiaIII dimer, the two monomers are linked to each other in a manner placing the two active sites on the same side of the dimer, but about 20 Å away from the dimer interface in opposite directions. This enables both monomers to simultaneously bind a dimeric target molecule, or possibly utilize allostery in their function [51].
Galactosyltransferases (PDB code 4IRP)
β-1,4-Galactosyltransferase 7 (β4GalT7; EC 2.4.1.133) is a proteoglycan-synthesizing enzyme that adds a galactose to the second position of a growing saccharide core structure of a glycoprotein acceptor (GlcAβ1–3Galβ1–3Galβ1–4Xylβ1–O–[serine]), which already contains the initiating xylose residue. It is also a drug development target for glycosaminoglycan synthesis [55]. It belongs to GTase family 7 and its crystal structure [56] revealed a variant of the GT-A fold in which the β3 strand is replaced by a strand (β7) present in the C-terminal domain. Thus, the topology is 721465 (Fig. 2A). The monoclinic crystal had four β4GalT7 molecules in the asymmetric unit, forming two copies of a dimer. The dimeric nature of the protein is supported by the finding that the stoichiometry of UDP binding by β4GalT7 was between 0.4 and 0.6 [57]. Subsequent gel filtration analysis under native conditions provided evidence for dimer formation, suggesting that only one of the monomers in the dimer is able to bind UDP-galactose.
GT-B folds
Glycogen phosphorylases (PDB codes 1YGP, 5IKO, 4BQE, 2IEG, 3DDS)
We also included glycogen phosphorylase (GP; EC 2.4.1.1) in the list of selected enzymes, together with some others (see below), because it is classified as a member of the GT family 35. Yet, its catalytic activity differs from “classical” GTases due to the role of the enzyme in storage energy mobilization. It produces glucose-1-phosphate from linear stretches of glycogen chains by cleaving the α-1,4 glycosidic bonds. Glycogen phosphorylase is a well-known prototypic allosteric enzyme that can exist in a monomeric inactive state as well as in dimeric or tetrameric active states. It is well established that phosphorylation of a specific serine residue and binding of AMP increase the activity of the enzyme by triggering the conformational change of an unstructured loop into an α-helix and by a shift in allosteric state, respectively. The sites of both of these activation events reside near the dimer interface, as deduced from the human liver GP structure [58]. A wealth of crystallographic and biochemical evidence shows that the active unit of GPs is a dimer. The change of the oligomeric state from monomer to dimer upon activation has also recently been shown by dynamic light scattering [59].
Brain, liver and muscle isoenzyme structures of GP have been determined from human and various other organisms. The structures are highly homologous, exemplified by the 83.3% sequence identity between the isoenzymes in rabbit muscle (PDB 2IEG) [60] and human brain [59]. Despite this apparent structural identity, the dimer interface has some flexibility without affecting the activity of the enzyme. The liver isoenzyme [58] is structurally the most rigid: the dimer interface area is 3350 Å2 (PDB entry 1FA9). The corresponding values for muscle (2240 Å2) [61] and brain (1400 Å2) [59] GP dimer interfaces reflect the extent of conformational changes taking place during activation of the enzymes. The same phenomenon is also seen in the yeast GP structures [62, 63].
Inhibition of glycogen phosphorylase activity is a potential strategy for drug development, e.g. for diabetes treatment. Not surprisingly, structural studies with various ligands are gradually increasing our understanding of the dynamics and allostery of oligomeric structures of glycogen phosphorylases, e.g. rabbit muscle [64] and human liver [65, 66] variants.
Instead of glycogen phosphorylases, plants have glucan phosphorylases that belong to the same GT family 35. The Arabidopsis thaliana glucan phosphorylase PHS2 crystal structure at 1.7 Å resolution [67] revealed a dimer, in which the active site of each monomer is buried in a cavity away from the dimer interface area. The structure is also well superimposable with the glycogen phosphorylase GT-B fold enzyme structures, and can therefore be regarded with confidence as a physiologically relevant dimer.
Glycogen synthases (PDB codes 3NB0)
Glycogen synthases (EC 2.4.1.11; GT family 3) catalyse the addition of glucose units from UDP-glucose to a growing glycogen chain. Crystal structures of the yeast isoenzyme Gsy2p have been solved both in the apo state and in the glucose-6-phosphate activated state [68]. The amino acid sequence of Gsy2p is 51.7% identical (78.5% similar) to the corresponding human enzyme.
The structure of Gsy2p is an A/B/C/D tetramer, which is formed from different structurally or functionally relevant dimers: the interfaces between each monomer accommodate binding sites for either the allosteric activator glucose-6-phosphate or the donor and acceptor molecules. Each of the four monomers have a long α-helix extending from the core enzymatic domain, such that these four helices form a coiled coil arrangement in the centre of the tetramer (as seen for the B/D dimer in Fig. 1). These helices form the extensive monomer–monomer interaction surfaces seen in Table 1.
Sucrose synthase (PDB code 3S28)
Sucrose is synthesized from NDP-glucose and d-fructose by sucrose synthase (EC 2.4.1.13). Sucrose synthases are retaining GTases belonging to the GT family 4. Structural and biochemical studies of the A. thaliana enzyme AtSus1 have shown that the oligomeric state of the enzyme is linked to the regulation of its activity [69]. AtSus1 was shown to exist solely as a tetramer by analytical gel filtration. The analysis of the crystal structure using the jsPISA server revealed two types of monomer–monomer interactions responsible for the oligomerization of AtSus1: A/B (C/D) and A/D (B/C), with interface areas of 1280 and 1076 Å2, respectively. Interestingly, the GT-B domains themselves do not play any major role in forming these interactions. Instead, sucrose synthase contains separate cellular targeting and peptide binding domains, which mediate the oligomerization contacts. It appears that the transition of AtSus1 tetramers to dimers precedes the phosphorylation of Ser 167, and it has been suggested that the change in oligomerization state regulates this phosphorylation step [69]. Hardin et al. [70] have also reported that the maize enzyme exists as a dimer rather than a tetramer.
GT-B variants
Fucosyltransferases (PDB code 4AP5, 3ZY5)
Fucose is one of the sugars found either directly linked to proteins via O-linkage to a serine or threonine residue, or added as a terminal sugar on branched glycan chains. Structures of fucosyltransferases catalysing both of these types of additions have been solved.
Protein O-fucosyltransferases 1 and 2 (POFUT1 and POFUT2; EC 2.4.1.221) are inverting enzymes of GT families 65 and 68, respectively. They transfer an α-l-fucosyl residue from GDP-β-l-fucose to the hydroxyl group of serine residues in acceptor proteins.
Human POFUT2 crystal structure is known both in apo form (PDB 4AP5) and in complex with the donor substrate (PDB 4AP6) [71]. The two molecules in the asymmetric unit of the apoprotein form a non-crystallographic dimer with an extensive monomer–monomer interface of 1670 Å2. The substrate-binding cavity is formed between the two monomers such that a loop from one molecule partially covers the cavity of the other molecule. In the substrate-bound state, however, the dimer interface is reduced to 1315 Å2 due to the accommodation of the substrate. Interestingly, the structure of the enzyme–substrate complex indicated that the physiologically relevant form of POFUT2 is dimeric, since in this holoenzyme structure the dimer is formed in the same way despite holding only one molecule per asymmetric unit. Thus, a crystallographic dimer in this case seems to be identical to the biologically relevant non-crystallographic dimer simply out of necessity. POFUT2 possesses a two-domain topology, representing a variant of the GT-B fold. The first domain shows a 3217465 topology, with β5 being antiparallel to the others. The second domain shows an all-parallel 3214 topology when an α-helix replaces β5 next to β4 in an interesting deviation from the majority of the structures.
The only known crystal structure for a POFUT1 is the one of Caenorhabditis elegans enzyme (PDB 3ZY5; a complex with GDP-fucose). There is only one chain (A) in the asymmetric unit of the monoclinic unit cell, but there is a significantly large interface area (1297 Å2) with the crystallographic symmetry mate molecule (A′). Therefore, we included this putative A/A′ dimer structure in our study. The first domain in each monomer shows a 321756 topology with an antiparallel β3 strand, while the second domain shows a 32145 topology with all strands aligned in a parallel fashion. The EPPIC analysis (Table 1) indicates that the structure of POFUT1 is a crystallographic dimer, although other metrics suggest it to be a biological dimer. Interestingly, the same protein—but with a bound GDP instead of GDP-fucose—crystallizes with two molecules per asymmetric unit (PDB 3ZY3). Despite a sufficiently large interaction surface (1096 Å2), jsPISA analysis renders the structure a probable crystallographic dimer. It seems likely that POFUT1 does not form biological dimers, as also both the gel filtration chromatography and analytical ultracentrifugation data of Lira-Navarrete et al. [72] indicated that C. elegans POFUT1 is a monomeric protein.
Caenorhabditis elegans POFUT1 (424 residues in POFUT1 isoform 1) and human POFUT2 do not share considerable sequence similarity despite catalysing the same reaction: based on ExPASy homology analysis, they share 26.8% identity (49.7% similarity) over a 179 amino acid overlap. In contrast, human POFUT1 (for which no crystal structure is available yet) is identical in sequence to the human POFUT2 over the common 383 amino acid residue part.
N-acetylglucosaminyltransferases (PDB code 4GYW)
N-Acetylglucosaminyltransferase (OGT; EC 2.4.1.255) belongs to family GT41 of inverting GTases. It transfers N-acetylglucosamine from the sugar donor UDP-GlcNAc onto specific serine or threonine residues of nucleocytoplasmic proteins. It is a different GT-B variant compared to the fucosyltransferase POFUT1 described above: in addition to its GTase domain topology, it is also a considerably larger protein (1046 residues) due to its 13 tetratricopeptide repeats (TPR) containing domain. The GT-B domain topology of OGT is 3214567 for the first subdomain and 32145 for the second subdomain, with all elements parallel to each other. In the crystal structure (PDB 4GYW) [73] there is only one molecule per asymmetric unit, but molecules A and A′, which are related by crystallographic symmetry, form a dimer. In fact, the TPR domains are responsible for this dimerization. This has been shown by using the TRP domain alone in crystallization [74]. N-Acetylglucosaminyltransferase therefore seems to represent an interesting and novel variant of the GT-B fold, in addition to its unique dimerization properties.
Dimer interface analyses
The dimerization interface for each of the selected structures was analysed to review whether any similarities exist between them. We considered six different criteria: interaction surface area and energy-related metrics, amino acid composition, secondary structure composition, topology, evolutionary conservation, and active site position in the dimer structure.
Interface area and energy-related metrics
All the selected structures show an interface area larger than 800 Å2. This is commonly accepted as the minimum area for biologically relevant dimers [23, 75]. The areas vary from 941 Å2 (C2GnT) to 3355 Å2 (Gph1) (Table 1). The solvation free energy ΔG and the total binding energy vary from −7 to −32 kcal/mol and −14 to −48 kcal/mol, respectively. These three parameters are part of the jsPISA interaction radar score [21] and are as such reliable measures to assess dimerization in crystal structures. In Table 1, we also list the jsPISA score, which is a weighted average of each of the radar metrics. A value higher than 50% depicts a good probability for the interface to be biologically relevant [21].
The DiMoVo method [22] also uses the interface area as the main criterion in assessing whether the dimers are crystallographic or biologically relevant, but it also considers other criteria such as frequencies and pairwise distances of amino acids. In this way, the predictive value compared to the interface area alone is improved from 78 to even 97%. The boundary value of the DiMoVo score is 0.5; values below 0.5 quite accurately predict crystallographic dimers, while values above 0.5 predict biological dimers. Interestingly, a low DiMoVo score was obtained for hGyg1, PHS2 and GPb (Table 1) despite their good energy metrics.
The EPPIC method [23] considers evolutionary conservation as a criterion for interaction sites. In our study, all the structures with a very low DiMoVo score also scored congruently in the EPPIC assessment (Table 1).
Amino acid composition
To analyse the amino acid composition at the dimer interfaces, we calculated the ratios between the frequency of amino acids observed at the interface and the frequency of amino acids within the full-length sequence of the crystallized proteins. Alanine residues were statistically significantly absent from the interfaces, whereas arginine and proline residues were statistically overrepresented (Supp. Figure 1). This finding is in line with Hashimoto et al. [13], whose study material consisted of 73 nonredundant GTase structures representing 31 families, but were not restricted to necessarily having non-crystallographic symmetry mates in the asymmetric unit.
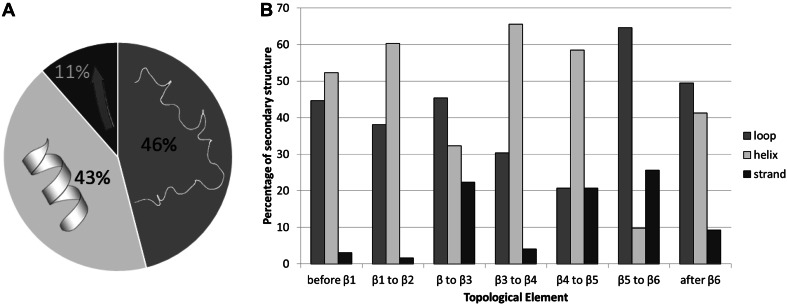
Secondary structure composition
All types of secondary structures were observed in the dimerization interfaces: α-helices, β-strands, loops and disordered regions (Fig. 3a). We analysed the secondary structure compositions of each of the topological elements responsible for dimerization contacts (Fig. 3b) and found that loops and helices are invariably the major feature. Hashimoto et al. [13] also found in their data set that β-strands were underrepresented in the dimer interfaces.
Fig. 3.
Analysis of the frequency of occurrence of secondary structure elements (α-helices, β-strands, loops and disordered regions) in the dimer interfaces of the 24 GTase homodimers of this study in the overall dataset (a) and in each topological element (b)
Topology
Topological elements responsible for dimerization were analysed by examining their position with regard to the core β-strands of GT-A and GT-B folds (Fig. 2A, B). We found features that were shared between different topological elements, as well as features that distinguish the two folds from each other (Fig. 2C).
Structures belonging to the GT-A fold were found to display a conserved dimerization interface topology, with two core dimerization elements making contacts with each other. The first element resides in the region between β5 and β6 (Fig. 2A, C, magenta); the second element is in the region after β6 (Fig. 2A, C, blue). In addition to these two core elements, some families use additional elements for dimerization (Fig. 2C). For example, glucuronyltransferases use α1 (Fig. 2A, C, red), as well as the surface created by the β4′–βC (Fig. 2A, C, green). The region between β4 and β5 is also used by N-acetylglucosaminyltransferases, galactosyltransferases and xylosyltransferases (Fig. 2A, C, green). Galactosyltransferases use amino acids located in the N-terminus of the core fold (before β1) (Fig. 2A, C, brown).
GTase structures with the GT-B fold also display similarities in the dimerization interface topology, with the nuance that the topological elements may lie on the domain “a” or domain “b” (first and second Rossmann fold domains, respectively). Glycogen phosphorylases and sucrose synthases use almost always domain “a” for dimerization, whereas glycogen synthases, fucosyltransferases and N-acetylglucosaminyltransferases use elements from both “a” and “b” domains. The first core dimerization element of the GT-B fold enzymes is the N-terminal region of the core fold, either before β1a or β1b (Fig. 2B, C, brown, blue); the second element is the region between either β2a and β3a or β2b and β3b (Fig. 2B, C, purple). The sole exception is the sucrose synthase family, which employs only the first core element and the region between β4a and β5a as an additional element (Fig. 2B, C, green). In glycogen phosphorylases and sucrose synthases, the region between β3a and β4a participates as an additional element (Fig. 2B, C, orange).
Interestingly, the structures of ST8SiaIII, B4GalT7, PoFUT1 and PoFUT2, as well as OGT, which are GT-A or GT-B fold variants, display mixed dimerization elements from both folds. PoFUT1 and PoFUT2 (GT-B variants) use the region between β5 and β6, specific to the GT-A fold dimerization interface, as well as the regions between β2 and β3, specific to the GT-B fold dimerization interface. ST8SiaIII (a GT-A variant) employs the N-terminal region before β1 and the region between β3 and β4, common to GT-B fold dimerization interface, and the region between β4 and β5 specific to the GT-A fold. In B4GalT7, the N-terminal region before β1 and the region between β2 and β3 specific to GT-B fold, as well as the C-terminal region after β6, act as core element of GT-A fold dimerization.
These data emphasize the high variability existing between the identified dimer interfaces, a phenomenon in line with the existence of multiple distinct enzyme dimers. In this regard, the lack of any consensus motifs for dimerization and the use of various topological arrangements suggest that any individual enzyme uses a specific interaction surface only for binding itself and not any nonrelevant enzyme. If the latter is the case, the end result would be a mix of all kinds of enzyme dimers and also “mixed” glycans these enzyme complexes might make. This outcome is not desirable, and seems to be prevented by highly distinct interfaces allowing only specific interactions. A similar situation must also exist between sequentially acting enzymes that are known to form heteromeric complexes with each other [7]. Whether the interfaces in the latter case are similar to those used for the formation of enzyme homodimers remains to be clarified.
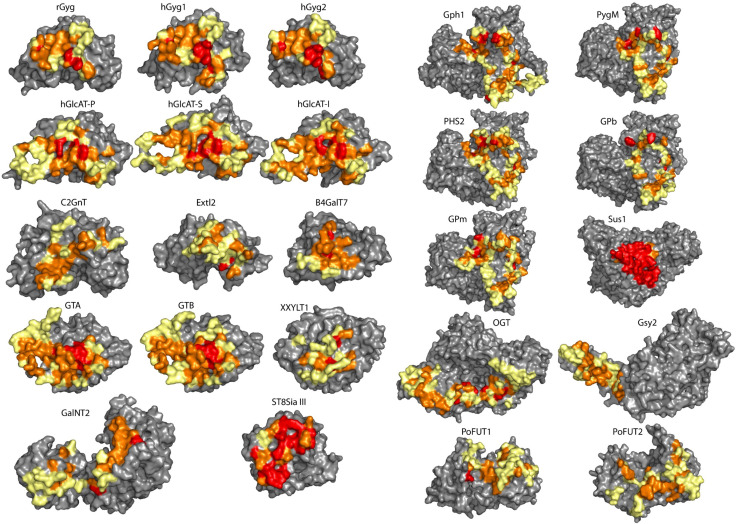
Evolutionary conservation
We also evaluated the amino acid sequence conservation in the dimerization interfaces. Briefly, multiple sequence alignments were generated by querying the sequence of each studied GTase against the OMA orthology database [76], using the InterEvolAlign server. We found various types of conservation profiles (Fig. 4), from strict conservation (red), high conservation (orange) to more diverse (yellow). The multiple sequence alignments are detailed in Suppl. Figure 2.
Fig. 4.
Evolutionary conservation of the amino acid sequence of the dimerization interface, visualized on each monomer (the interface facing the reader) of the 24 GTases as a colour gradient: from red (strictly conserved) through orange (high conservation) to yellow (more diverse). The residues not involved in the dimerization interface are displayed in grey. The placement of the monomers in the figure is the same as for the dimers in Fig. 1
Active site positioning
From the functional point of view, a feature of particular interest is how the active sites of the monomers relate to the dimer interface. In general, at least three possibilities exist: (1) the active sites are far away from each other, suggesting either an independent catalytic activity for both of them or that dimerization is a stabilizing factor; (2) the active sites are located close to each other to facilitate cooperative substrate binding and catalysis; or (3) the active sites overlap with the dimerization interface to provide a mechanism to regulate the enzymatic activity via dimerization.
Since not all the structures contained a substrate or any other bound ligand, we inspected donor and acceptor substrate-binding sites and the metal-binding site (for GT-A folds) as a guide to locate the active sites. In most of the GT-A folds the active site is near β4 and β4′ (Fig. 2A), while in GT-B folds it seems to be predominantly located in the linker region between the two Rossmann fold domains. In most homodimers, however, the active sites are located far away from the dimerization interface, in some cases near the opposite ends of the dimer. In contrast, even though the active sites in the glucuronyltransferase dimer reside very close to each other (20 Å away), they both are still easily accessible.
Discussion
In this review, we analysed various GTases using the available crystal structures of their globular catalytic domains to determine whether any of them represent biologically relevant dimers. Likely candidates were identified by choosing crystals with more than one molecule per asymmetric unit. Only the crystal structures of the globular catalytic domains of GTases are available, but there are good grounds to assume that these domains are responsible for, or at least contribute to, dimerization of the full-length GTases. This assumption is consistent with dimerization being a regulator of the enzymatic activity of the GTases. The fact that none of the GTases contain the dimerization signature sequence LIxxGVxxGVxxT of single-spanning transmembrane helices [77] and that their ca. 40–80 residues long stem domains appear to lack regular secondary structure provide strong support to the view that the catalytic domains have an important role in linking GTases to homodimers.
Phylogenetic analysis of GTases by Hashimoto et al. [13] indicated that certain GTase families could be classified either as “monomer families” or “dimer families”. Structures belonging to families GT44, GT7 and GT27 (GT-A fold) and GT5, GT9 and GT80 (GT-B fold) are monomers, while GT81 and GT43 (GT-A fold) and GT35 and GT23 (GT-B fold) represent homodimers. Only a few families seem to contain a mixed population of GTase oligomers. Accordingly, structures from families 35 and 43 were overrepresented in our analysis (Table 1, Fig. 1), while none of the “monomer family” structures passed the criteria used in our study. Hashimoto et al. [13] also found that, especially for the GT-B fold, homooligomer interfaces are more typically formed from helices and terminal regions or loop structures than from β-strands. A typical example for a GT-A fold enzyme is glucuronyltransferase GlcAT-I (family 43) [25], where the homodimer interface is formed from C-terminal ends including a long loop and the last α-helix: the substrate-binding sites are near the interface and acceptor substrates are in contact with both GlcAT-I monomers. Furthermore, glycogen phosphorylase (family 35) structures form homodimers via α-helices, which are missing in family 5 monomeric glycogen glucosyltransferases [13].
As discussed by Krissinel and Henrick [78], the challenge of dividing up dimers into physiological and non-physiological ones continues to exist. It is not trivial to judge a crystallized protein as a biological dimer with confidence. The main problem here is that it is still hard to define absolute values or even reliable characteristics for a biological interface; otherwise, the problem could be tackled by a bioinformatics approach. Nevertheless, the most common characteristics to assess the relevance of a dimer are the interface area (in Å2), the solvation free energy gain (kcal/mol) between the transition of isolated and interfaced structures, and the number of salt bridges or hydrogen bonds at the interface. As an example, a maximum free energy of dissociation (ΔG 0) of 15–20 kcal/mol should represent a biological dimer, and usually ten or more hydrogen bonds are found in a relevant interface. However, many dimers or higher oligomers may be transient and thus possess “weak” interactions in vivo, which may not prevail under crystallization conditions. Transient complexes with dissociation constants higher than 100 μM (ΔG 0 ≤ 5 kcal/mol) may have only a 10% probability to form crystals [79], while stable complexes can be expected to crystallize without undergoing a change in the oligomerization state. The properties of the interface itself do not completely determine the binding energy, but also depend on other factors, such as the size and shape of the complex and the entropy change. Therefore, the function of the protein should always be taken into account along with the analysis of its crystal structure. However, it is estimated that the values obtained by calculating the binding energy and the entropy of dissociation are 80% accurate for the identification of macromolecular assemblies in crystals [78].
GTases have been shown to be able not only to function as homooligomers, but also as heterooligomers [5–7]. The heterooligomers can also involve more than two GTases, forming functional multienzyme complexes [80]. To this day, however, no heteromeric complexes between two GTases have been crystallized, making analyses of their interactions impossible. Nevertheless, a few examples where a glycosyltransferase forms a complex with a non-glycosyltransferase need to be addressed here briefly. β-1,4-Galactosyltransferase 1 (β4GalT1) has been crystallized in complex with α-lactalbumin (LA) and various substrates [81]. The binding site of LA partially overlaps with the substrate-binding site, consistent with a regulatory role of the ligand in the complex: instead of an N-acetylglucosamine, a glucose is accepted for binding. A large conformational change of a critical loop region takes place upon LA binding. The other known example is the hetero-complex between EryCIII (3-alpha-mycarosylerythronolide B desosaminyl transferase), a GTase from family 1, and its partner EryCII, a cytochrome P450 family protein. The crystal structure of the EryCIII–EryCII complex has been determined [82] and it reveals a heterotetramer with an elongated quaternary organization. A homodimer of EryCIII forms the centre of the complex, while EryCII molecules reside on the periphery. It is evident in this case that the interaction surfaces for homomer and heteromer formation are located in distinct surface areas of the GTase, which is a valid observation to keep in mind for possible analogy with other heterocomplexes to be solved in the future. Conversely, as indicated earlier, glycogenins 1 and 2 (Gyg1 and Gyg2) co-purify [35], indicating that the two glycogenins may also form heterodimers. Since the crystal structures of Gyg1 and Gyg2 homodimers superimpose very well (with r.m.s. deviation of 0.865 Å), we hypothesize that the same interaction surface might be used both for homomers and for heteromers of these two GTases, which may be competing with each other.
It is also worth noting that highly specific dimerization—whether homo or heteromeric—is more likely to employ interfaces that further increase the strength of interaction. In contrast, transient interactions, with possibly a choice of interaction partners, call for interfaces that may not be clearly distinguishable from crystal contacts. This could indicate that heterooligomers, as well as some homooligomers, could be so transient that their isolation for crystallization is not favourable enough.
Lastly, it is inevitable that the data we chose—898 crystal structures of glycosyltransferases deposited in the Protein Data Bank—contain some which are physiological enzyme dimers, but happen to have crystallized with one molecule per asymmetric unit and therefore escaped our analysis. Equally well, as discussed above, it could be questioned whether some of our chosen cases are true dimers, or instead crystal artefacts—depending on the subjective weighting of criteria. However, it is neither possible nor meaningful to carefully review all the 898 available structures. We believe that the way we selected the structures, and the data we obtained, provides further support for the conclusion that glycosyltransferases can form—and do form—physiological dimers not only in crystals, but also in vivo.
Concluding remarks
The main outcomes of this review are as follows. First of all, each GTase fold type uses different topological elements for constructing their dimerization interfaces. These elements serve as fingerprints within a group of a particular fold. An interesting observation is also that variant folds can use mixed topological elements from the basic GT-A and GT-B folds. Additionally, it is typical that homodimerization does not bring the active sites of the GTase monomers close to each other. Moreover, our survey revealed that different glycosyltransferases form biologically relevant homodimeric complexes. This conclusion is supported by both biochemical and structural evidence. No heterooligomers between different glycosyltransferases have been structurally characterized, and this poses a future challenge for understanding glycosyltransferase function.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplement Fig. 1. Log-ratio of the frequency of amino acids observed at the interface and within the full-length sequence of the crystallized domains of the 24 GTase homodimers of this study. Stars indicate the statistical significance according to critical values of χ2 (PPTX 145 kb)
Supplement Fig. 2. Multiple sequence alignments of the dimerization interface for each of the 24 GTases. As the residue numbers are not sequential, they are detailed below each alignment (PPTX 2466 kb)
Acknowledgements
The financial support from the Academy of Finland (no. 285232, date 11.05.2015), University of Oulu, and Emil Aaltonen Foundation is gratefully acknowledged. We also want to acknowledge Thibaud Colas for his help with automatized scripts used for the study of the interfaces.
References
- 1.Hennet T, Cabalzar J. Congenital disorders of glycosylation: a concise chart of glycocalyx dysfunction. Trends Biochem Sci. 2015;40:377–384. doi: 10.1016/j.tibs.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Chang SC, Yang WV. Hyperglycemia, tumorigenesis, and chronic inflammation. Crit Rev Oncol Hematol. 2016;108:146–153. doi: 10.1016/j.critrevonc.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Vajaria BN, Patel PS. Glycosylation: a hallmark of cancer? Glycoconj J. 2017;34:147–156. doi: 10.1007/s10719-016-9755-2. [DOI] [PubMed] [Google Scholar]
- 4.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hassinen A, Rivinoja A, Kauppila A, Kellokumpu S. Golgi N-glycosyltransferases form both homo- and heterodimeric enzyme complexes in live cells. J Biol Chem. 2010;285:17771–17777. doi: 10.1074/jbc.M110.103184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hassinen A, et al. Functional organization of Golgi N- and O-glycosylation pathways involves pH-dependent complex formation that is impaired in cancer cells. J Biol Chem. 2011;286:38329–38340. doi: 10.1074/jbc.M111.277681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kellokumpu S, Hassinen A, Glumoff T. Glycosyltransferase complexes in eukaryotes: long-known, prevalent but still unrecognized. Cell Mol Life Sci. 2016;73:305–325. doi: 10.1007/s00018-015-2066-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engel BD, et al. In situ structural analysis of Golgi intracisternal protein arrays. Proc Natl Acad Sci USA. 2015;112:11264–11269. doi: 10.1073/pnas.1515337112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahnert SE, Marsh JA, Hernandez H, Robinson CV, Teichmann SA. Principles of assembly reveal a periodic table of protein complexes. Science. 2015;350:aaa2245. doi: 10.1126/science.aaa2245. [DOI] [PubMed] [Google Scholar]
- 10.Keskin O, Tuncbag N, Gursoy A. Predicting protein–protein interactions from the molecular to the proteome level. Chem Rev. 2016;116:4884–4909. doi: 10.1021/acs.chemrev.5b00683. [DOI] [PubMed] [Google Scholar]
- 11.Bahadur RP, Zacharias M. The interface of protein–protein complexes: analysis of contacts and prediction of interactions. Cell Mol Life Sci. 2008;65:1059–1072. doi: 10.1007/s00018-007-7451-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berman HM, et al. The protein data bank. Acta Crystallogr D Biol Crystallogr. 2002;58:899–907. doi: 10.1107/S0907444902003451. [DOI] [PubMed] [Google Scholar]
- 13.Hashimoto K, Madej T, Bryant SH, Panchenko AR. Functional states of homooligomers: insights from the evolution of glycosyltransferases. J Mol Biol. 2010;399:196–206. doi: 10.1016/j.jmb.2010.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henrissat B, Sulzenbacher G, Bourne Y. Glycosyltransferases, glycoside hydrolases: surprise, surprise! Curr Opin Struct Biol. 2008;18:527–533. doi: 10.1016/j.sbi.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 15.Breton C, Fournel-Gigleux S, Palcic MM. Recent structures, evolution and mechanisms of glycosyltransferases. Curr Opin Struct Biol. 2012;22:540–549. doi: 10.1016/j.sbi.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 16.Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 17.Breton C, Bettler E, Joziasse DH, Geremia RA, Imberty A. Sequence–function relationships of prokaryotic and eukaryotic galactosyltransferases. J Biochem. 1998;123:1000–1009. doi: 10.1093/oxfordjournals.jbchem.a022035. [DOI] [PubMed] [Google Scholar]
- 18.Breton C, Imberty A. Structure/function studies of glycosyltransferases. Curr Opin Struct Biol. 1999;9:563–571. doi: 10.1016/S0959-440X(99)00006-8. [DOI] [PubMed] [Google Scholar]
- 19.Breton C, Snajdrova L, Jeanneau C, Koca J, Imberty A. Structures and mechanisms of glycosyltransferases. Glycobiology. 2006;16:29R–37R. doi: 10.1093/glycob/cwj016. [DOI] [PubMed] [Google Scholar]
- 20.Lesk AM. Systematic representation of protein folding patterns. J Mol Graph. 1995;13:159–164. doi: 10.1016/0263-7855(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 21.Krissinel E. Stock-based detection of protein oligomeric states in jsPISA. Nucleic Acids Res. 2015;43:W314–W319. doi: 10.1093/nar/gkv314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernauer J, Bahadur RP, Rodier F, Janin J, Poupon A. DiMoVo: a Voronoi tessellation-based method for discriminating crystallographic and biological protein-protein interactions. Bioinformatics. 2008;24:652–658. doi: 10.1093/bioinformatics/btn022. [DOI] [PubMed] [Google Scholar]
- 23.Duarte JM, Srebniak A, Scharer MA, Capitani G. Protein interface classification by evolutionary analysis. BMC Bioinf. 2012;13:334-2105-13-334. doi: 10.1186/1471-2105-13-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faure G, Andreani J, Guerois R. InterEvol database: exploring the structure and evolution of protein complex interfaces. Nucleic Acids Res. 2012;40:D847–D856. doi: 10.1093/nar/gkr845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tone Y, et al. 2-O-Phosphorylation of xylose and 6-O-sulfation of galactose in the protein linkage region of glycosaminoglycans influence the glucuronyltransferase-I activity involved in the linkage region synthesis. J Biol Chem. 2008;283:16801–16807. doi: 10.1074/jbc.M709556200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kakuda S, et al. Structural basis for acceptor substrate recognition of a human glucuronyltransferase, GlcAT-P, an enzyme critical in the biosynthesis of the carbohydrate epitope HNK-1. J Biol Chem. 2004;279:22693–22703. doi: 10.1074/jbc.M400622200. [DOI] [PubMed] [Google Scholar]
- 27.Shiba T, et al. Crystal structure of GlcAT-S, a human glucuronyltransferase, involved in the biosynthesis of the HNK-1 carbohydrate epitope. Proteins. 2006;65:499–508. doi: 10.1002/prot.21118. [DOI] [PubMed] [Google Scholar]
- 28.Pedersen LC, et al. Heparan/chondroitin sulfate biosynthesis. Structure and mechanism of human glucuronyltransferase I. J Biol Chem. 2000;275:34580–34585. doi: 10.1074/jbc.M007399200. [DOI] [PubMed] [Google Scholar]
- 29.Terayama K, et al. Purification and characterization of a glucuronyltransferase involved in the biosynthesis of the HNK-1 epitope on glycoproteins from rat brain. J Biol Chem. 1998;273:30295–30300. doi: 10.1074/jbc.273.46.30295. [DOI] [PubMed] [Google Scholar]
- 30.Jessell TM, Hynes MA, Dodd J. Carbohydrates and carbohydrate-binding proteins in the nervous system. Annu Rev Neurosci. 1990;13:227–255. doi: 10.1146/annurev.ne.13.030190.001303. [DOI] [PubMed] [Google Scholar]
- 31.Pitcher J, Smythe C, Campbell DG, Cohen P. Identification of the 38-kDa subunit of rabbit skeletal muscle glycogen synthase as glycogenin. Eur J Biochem. 1987;169:497–502. doi: 10.1111/j.1432-1033.1987.tb13637.x. [DOI] [PubMed] [Google Scholar]
- 32.Gibbons BJ, Roach PJ, Hurley TD. Crystal structure of the autocatalytic initiator of glycogen biosynthesis, glycogenin. J Mol Biol. 2002;319:463–477. doi: 10.1016/S0022-2836(02)00305-4. [DOI] [PubMed] [Google Scholar]
- 33.Chaikuad A, et al. Conformational plasticity of glycogenin and its maltosaccharide substrate during glycogen biogenesis. Proc Natl Acad Sci USA. 2011;108:21028–21033. doi: 10.1073/pnas.1113921108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nilsson J, et al. LC-MS/MS characterization of combined glycogenin-1 and glycogenin-2 enzymatic activities reveals their self-glucosylation preferences. Biochim Biophys Acta. 2014;1844:398–405. doi: 10.1016/j.bbapap.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 35.Mu J, Roach PJ. Characterization of human glycogenin-2, a self-glucosylating initiator of liver glycogen metabolism. J Biol Chem. 1998;273:34850–34856. doi: 10.1074/jbc.273.52.34850. [DOI] [PubMed] [Google Scholar]
- 36.Janin J, Chothia C. The structure of protein–protein recognition sites. J Biol Chem. 1990;265:16027–16030. [PubMed] [Google Scholar]
- 37.Issoglio FM, Carrizo ME, Romero JM, Curtino JA. Mechanisms of monomeric and dimeric glycogenin autoglucosylation. J Biol Chem. 2012;287:1955–1961. doi: 10.1074/jbc.M111.287813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sethi MK, et al. Molecular cloning of a xylosyltransferase that transfers the second xylose to O-glucosylated epidermal growth factor repeats of notch. J Biol Chem. 2012;287:2739–2748. doi: 10.1074/jbc.M111.302406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu H, et al. Notch-modifying xylosyltransferase structures support an SNi-like retaining mechanism. Nat Chem Biol. 2015;11:847–854. doi: 10.1038/nchembio.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pak JE, et al. X-ray crystal structure of leukocyte type core 2 beta1,6-N-acetylglucosaminyltransferase. Evidence for a convergence of metal ion-independent glycosyltransferase mechanism. J Biol Chem. 2006;281:26693–26701. doi: 10.1074/jbc.M603534200. [DOI] [PubMed] [Google Scholar]
- 41.Pedersen LC, et al. Crystal structure of an alpha 1,4-N-acetylhexosaminyltransferase (EXTL2), a member of the exostosin gene family involved in heparan sulfate biosynthesis. J Biol Chem. 2003;278:14420–14428. doi: 10.1074/jbc.M210532200. [DOI] [PubMed] [Google Scholar]
- 42.Ghirardello M, et al. Glycomimetics targeting glycosyltransferases: synthetic, computational and structural studies of less-polar conjugates. Chemistry. 2016;22:7215–7224. doi: 10.1002/chem.201600467. [DOI] [PubMed] [Google Scholar]
- 43.El-Battari A, et al. Different glycosyltransferases are differentially processed for secretion, dimerization, and autoglycosylation. Glycobiology. 2003;13:941–953. doi: 10.1093/glycob/cwg117. [DOI] [PubMed] [Google Scholar]
- 44.Lira-Navarrete E, et al. Dynamic interplay between catalytic and lectin domains of GalNAc-transferases modulates protein O-glycosylation. Nat Commun. 2015;6:6937. doi: 10.1038/ncomms7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fritz TA, Raman J, Tabak LA. Dynamic association between the catalytic and lectin domains of human UDP-GalNAc:polypeptide alpha-N-acetylgalactosaminyltransferase-2. J Biol Chem. 2006;281:8613–8619. doi: 10.1074/jbc.M513590200. [DOI] [PubMed] [Google Scholar]
- 46.Jorgensen R, et al. Structures of a human blood group glycosyltransferase in complex with a photo-activatable UDP-Gal derivative reveal two different binding conformations. Acta Crystallogr Funct Struct Biol Commun. 2014;70:1015–1021. doi: 10.1107/S2053230X1401259X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jorgensen R, Grimm LL, Sindhuwinata N, Peters T, Palcic MM. A glycosyltransferase inhibitor from a molecular fragment library simultaneously interferes with metal ion and substrate binding. Angew Chem Int Ed Engl. 2012;51:4171–4175. doi: 10.1002/anie.201108345. [DOI] [PubMed] [Google Scholar]
- 48.Schuman B, et al. Cysteine-to-serine mutants dramatically reorder the active site of human ABO(H) blood group B glycosyltransferase without affecting activity: structural insights into cooperative substrate binding. J Mol Biol. 2010;402:399–411. doi: 10.1016/j.jmb.2010.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee HJ, et al. Structural basis for the inactivity of human blood group O2 glycosyltransferase. J Biol Chem. 2005;280:525–529. doi: 10.1074/jbc.M410245200. [DOI] [PubMed] [Google Scholar]
- 50.Foley DA, Swartzentruber KG, Colley KJ. Identification of sequences in the polysialyltransferases ST8Sia II and ST8Sia IV that are required for the protein-specific polysialylation of the neural cell adhesion molecule. NCAM J Biol Chem. 2009;284:15505–15516. doi: 10.1074/jbc.M809696200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Volkers G, et al. Structure of human ST8SiaIII sialyltransferase provides insight into cell-surface polysialylation. Nat Struct Mol Biol. 2015;22:627–635. doi: 10.1038/nsmb.3060. [DOI] [PubMed] [Google Scholar]
- 52.Audry M, et al. Current trends in the structure-activity relationships of sialyltransferases. Glycobiology. 2011;21:716–726. doi: 10.1093/glycob/cwq189. [DOI] [PubMed] [Google Scholar]
- 53.Rao F, et al. Structural insight into mammalian sialyltransferases. Nat Struct Biol. 2009;16:1186–1188. doi: 10.1038/nsmb.1685. [DOI] [PubMed] [Google Scholar]
- 54.Kuhn B, et al. The structure of human alpha-2,6-sialyltransferase reveals the binding mode of complex glycans. Acta Cryst D. 2013;69:1826–1838. doi: 10.1107/S0907444913015412. [DOI] [PubMed] [Google Scholar]
- 55.Saliba M, et al. Probing the acceptor active site organization of the human recombinant beta1,4-galactosyltransferase 7 and design of xyloside-based inhibitors. J Biol Chem. 2015;290:7658–7670. doi: 10.1074/jbc.M114.628123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsutsui Y, Ramakrishnan B, Qasba PK. Crystal structures of beta-1,4-galactosyltransferase 7 enzyme reveal conformational changes and substrate binding. J Biol Chem. 2013;288:31963–31970. doi: 10.1074/jbc.M113.509984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Daligault F, et al. Thermodynamic insights into the structural basis governing the donor substrate recognition by human beta1,4-galactosyltransferase 7. Biochem J. 2009;418:605–614. doi: 10.1042/BJ20081093. [DOI] [PubMed] [Google Scholar]
- 58.Rath VL, et al. Activation of human liver glycogen phosphorylase by alteration of the secondary structure and packing of the catalytic core. Mol Cell. 2000;6:139–148. doi: 10.1016/S1097-2765(05)00006-7. [DOI] [PubMed] [Google Scholar]
- 59.Mathieu C, et al. Insights into brain glycogen metabolism: the structure of human brain glycogen phosphorylase. J Biol Chem. 2016;291:18072–18083. doi: 10.1074/jbc.M116.738898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leonidas DD, et al. Control of phosphorylase b conformation by a modified cofactor: crystallographic studies on R-state glycogen phosphorylase reconstituted with pyridoxal 5′-diphosphate. Protein Sci. 1992;1:1112–1122. doi: 10.1002/pro.5560010905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lukacs CM, et al. The crystal structure of human muscle glycogen phosphorylase a with bound glucose and AMP: an intermediate conformation with T-state and R-state features. Proteins. 2006;63:1123–1126. doi: 10.1002/prot.20939. [DOI] [PubMed] [Google Scholar]
- 62.Lin K, Rath VL, Dai SC, Fletterick RJ, Hwang PK. A protein phosphorylation switch at the conserved allosteric site in GP. Science. 1996;273:1539–1542. doi: 10.1126/science.273.5281.1539. [DOI] [PubMed] [Google Scholar]
- 63.Lin K, Hwang PK, Fletterick RJ. Distinct phosphorylation signals converge at the catalytic center in glycogen phosphorylases. Structure. 1997;5:1511–1523. doi: 10.1016/S0969-2126(97)00300-6. [DOI] [PubMed] [Google Scholar]
- 64.Birch AM, et al. Development of potent, orally active 1-substituted-3,4-dihydro-2-quinolone glycogen phosphorylase inhibitors. Bioorg Med Chem Lett. 2007;17:394–399. doi: 10.1016/j.bmcl.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 65.Thomson SA, et al. Anthranilimide based glycogen phosphorylase inhibitors for the treatment of type 2 diabetes. Part 3: X-ray crystallographic characterization, core and urea optimization and in vivo efficacy. Bioorg Med Chem Lett. 2009;19:1177–1182. doi: 10.1016/j.bmcl.2008.12.085. [DOI] [PubMed] [Google Scholar]
- 66.Rath VL, et al. Human liver glycogen phosphorylase inhibitors bind at a new allosteric site. Chem Biol. 2000;7:677–682. doi: 10.1016/S1074-5521(00)00004-1. [DOI] [PubMed] [Google Scholar]
- 67.O’Neill EC, et al. Sugar-coated sensor chip and nanoparticle surfaces for the in vitro enzymatic synthesis of starch-like materials. Chem Sci. 2014;5:341–350. doi: 10.1039/C3SC51829A. [DOI] [Google Scholar]
- 68.Baskaran S, Roach PJ, DePaoli-Roach AA, Hurley TD. Structural basis for glucose-6-phosphate activation of glycogen synthase. Proc Natl Acad Sci USA. 2010;107:17563–17568. doi: 10.1073/pnas.1006340107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng Y, Anderson S, Zhang Y, Garavito RM. The structure of sucrose synthase-1 from Arabidopsis thaliana and its functional implications. J Biol Chem. 2011;286:36108–36118. doi: 10.1074/jbc.M111.275974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hardin SC, Duncan KA, Huber SC. Determination of structural requirements and probable regulatory effectors for membrane association of maize sucrose synthase 1. Plant Physiol. 2006;141:1106–1119. doi: 10.1104/pp.106.078006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen CI, et al. Structure of human POFUT2: insights into thrombospondin type 1 repeat fold and O-fucosylation. EMBO J. 2012;31:3183–3197. doi: 10.1038/emboj.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lira-Navarrete E, et al. Structural insights into the mechanism of protein O-fucosylation. PLoS One. 2011;6:e25365. doi: 10.1371/journal.pone.0025365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lazarus MB, Nam Y, Jiang J, Sliz P, Walker S. Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature. 2011;469:564–567. doi: 10.1038/nature09638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jinek M, et al. The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin alpha. Nat Struct Mol Biol. 2004;11:1001–1007. doi: 10.1038/nsmb833. [DOI] [PubMed] [Google Scholar]
- 75.Bahadur RP, Chakrabarti P, Rodier F, Janin J. Dissecting subunit interfaces in homodimeric proteins. Proteins. 2003;53:708–719. doi: 10.1002/prot.10461. [DOI] [PubMed] [Google Scholar]
- 76.Altenhoff AM, et al. The OMA orthology database in 2015: function predictions, better plant support, synteny view and other improvements. Nucleic Acids Res. 2015;44:D27–D38. doi: 10.1093/nar/gku1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lemmon MA, Treutlein HR, Adams PD, Brunger AT, Engelman DM. A dimerization motif for transmembrane alpha-helices. Nat Struct Biol. 1994;1:157–163. doi: 10.1038/nsb0394-157. [DOI] [PubMed] [Google Scholar]
- 78.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 79.Krissinel E. Crystal contacts as nature’s docking solutions. J Comput Chem. 2010;31:133–143. doi: 10.1002/jcc.21303. [DOI] [PubMed] [Google Scholar]
- 80.Noffz C, Keppler-Ross S, Dean N. Hetero-oligomeric interactions between early glycosyltransferases of the dolichol cycle. Glycobiology. 2009;19:472–478. doi: 10.1093/glycob/cwp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ramakrishnan B, Gasba PK. Crystal structure of lactose synthase reveals a large conformational change in its catalytic component, the β1,4-galactosyltransferase-I. J Mol Biol. 2001;310:205–218. doi: 10.1006/jmbi.2001.4757. [DOI] [PubMed] [Google Scholar]
- 82.Moncrieffe MC, et al. Structure of the glycosyltransferase EryCIII in complex with its activating P450 Homologue EryCII. J Mol Biol. 2012;415:92–101. doi: 10.1016/j.jmb.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement Fig. 1. Log-ratio of the frequency of amino acids observed at the interface and within the full-length sequence of the crystallized domains of the 24 GTase homodimers of this study. Stars indicate the statistical significance according to critical values of χ2 (PPTX 145 kb)
Supplement Fig. 2. Multiple sequence alignments of the dimerization interface for each of the 24 GTases. As the residue numbers are not sequential, they are detailed below each alignment (PPTX 2466 kb)