Abstract
The hallmark of Nanos proteins is their typical (CCHC)2 zinc finger motif (zf-nanos). Animals have one to four nanos genes. For example, the fruit fly and demosponge have only one nanos gene, zebrafish and humans have three, and Fugu rubripes has four. Nanos genes are mainly known for their evolutionarily preserved role in germ cell survival and pluripotency. Nanos proteins have been reported to bind the C-terminal RNA-binding domain of Pumilio to form a post-transcriptional repressor complex. Several observations point to a link between the miRNA-mediated repression complex and the Nanos/Pumilio complex. Repression of the E2F3 oncogene product is, indeed, mediated by cooperation between the Nanos/Pumilio complex and miRNAs. Another important interaction partner of Nanos is the CCR4–NOT deadenylase complex. Besides the tissue-specific contribution of Nanos proteins to normal development, their ectopic expression has been observed in several cancer cell lines and various human cancers. An inverse correlation between the expression levels of human Nanos1 and Nanos3 and E-cadherin was observed in several cancer cell lines. Loss of E-cadherin, an important cell–cell adhesion protein, contributes to tumor invasion and metastasis. Overexpression of Nanos3 induces epithelial–mesenchymal transition in lung cancer cell lines partly by repressing E-cadherin. Other than some most interesting data from Nanos knockout mice, little is known about mammalian Nanos proteins, and further research is needed. In this review, we summarize the main roles of Nanos proteins and discuss the emerging concept of Nanos proteins as oncofetal antigens.
Keywords: Nanos, Pumilio, Germ cell specification, Cancer, Phylogeny, RNA-binding protein, RNA regulation, Multiprotein complexes, Cancer testis antigen, pRb deficiency
Introduction
Nanos was originally discovered and studied in Drosophila melanogaster (fruit fly) [1]. Nanos proteins belong to a highly conserved protein family found in both vertebrates and invertebrates. In D. melanogaster, the nanos gene was primarily found to be essential for anterior–posterior axis polarity, abdomen formation, and germ cell development [1–3]. The Nanos protein establishes a multisubunit translation-inhibitory complex with Pumilio, its RNA-binding partner. The genomes of mouse and other mammals contain three Nanos-encoding genes, Nanos1, Nanos2, and Nanos3. Nanos homologs exist in several other species, such as Caenorhabditis elegans, Xenopus laevis, and Danio rerio (summarized in Table 1). The germ stem cell function of Nanos orthologs is conserved from invertebrates to mammals such as Mus musculus (Nanos2 and Nanos3) [4] and Homo sapiens (Nanos3) [5]. Two essential characteristics of germline cells are that they can give rise to all the cell types present in the adult (totipotency) and that they are immortal, passing on their genetic information to an endless series of generations. Nanos protein expression has also been linked to increased cell migration and invasion [6, 7]. Ectopic expression of Nanos1 mRNA and Nanos3 protein has been observed in human lung carcinomas [7, 8], suggesting a functional link between Nanos proteins and lung cancer.
Table 1.
Overview of reported nanos homologs in vertebrates and invertebrates
| Scientific name | Common name | Name of nanos homolog | Reference |
|---|---|---|---|
| Invertebrates | |||
| Drosophila melanogaster | Fruit fly | nanos (nos) | [1] |
| Helobdella robusta | Leech | Hro-nos | [128] |
| Caenorhabditis elegans | Roundworm | nos1, nos2 and nos3 | [101] |
| Hydra magnipapillata | Fresh-water polyp | Cnnos1 and Cnnos2 | [129] |
| Schistocerca americana | Grasshopper | nanos | [130] |
| Gryllus domesticus | Cricket | nanos | [130] |
| Podocoryne carnea | Jellyfish | nanos | [131] |
| Nematostella vectensis | Sea anemone | NvNanos1 and NvNanos2 | [132] |
| Anopheles gambia, Anopheles stephensi and Aedes aegypti | Mosquito | Anga nos, Anst nos and Aeae nos | [133] |
| Apis mellifera | Honeybee | nanos | [134] |
| Bombyx mori | Silkmoth | nanosM, nanosN, nanosO and nanosP | [135] |
| Sycon ciliatum | Sponge | SciNanos | [136] |
| Vertebrates | |||
| Xenopus laevis | Frog | Xcat-2 and nanos3 | [137] GenBank accession number XM_018251758.1 |
| Danio rerio | Zebrafish | nos1, nos2 and nos3 | [102] GenBank accession number NM_131878.1 |
| Mus musculus | Mouse | Nanos1, Nanos2 and Nanos3 | [4, 107] |
| Homo sapiens | Human | Nanos1, Nanos2 and Nanos3 | [11] |
| Xenopus tropicalis | Frog | Xtcat-2 and nanos3 | [138] GenBank accession numbers XM_004919168.3, XM_004919167.3 |
| Rattus norvegicus | Rat | Nanos1, Nanos2 and Nanos3 | [6] |
We present a general overview of the Nanos proteins in different organisms, their structures, and their roles in development and cancer in Drosophila and mammals. Since Nanos proteins are linked to essential molecular processes and characteristics such as the cell cycle, pluripotency, epithelial–mesenchymal transition (EMT), and cell survival versus apoptosis, further research on Nanos genes and proteins could shed more light on various biological phenomena, especially cancer.
Structures of Nanos genes and proteins
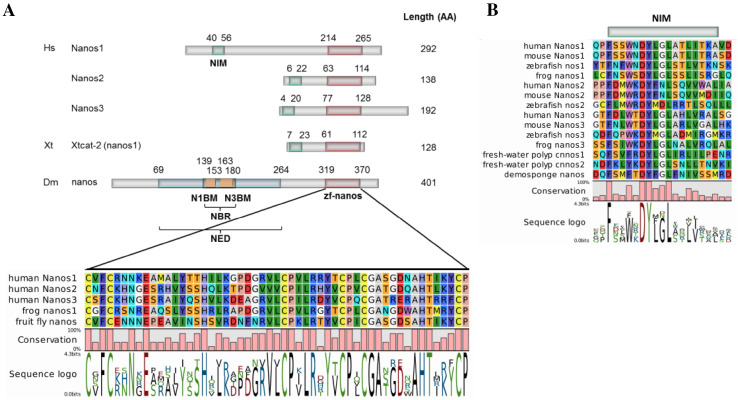
Nanos genes encode proteins with a typical carboxy-terminal zinc finger motif (CCHC)2 (Fig. 1a), which is the only domain that is evolutionarily conserved between mammalian Nanos family members and those in lower organisms such as the fruit fly and the roundworm [9]. Likewise, it is the most conserved sequence among the three mammalian Nanos paralogs (Nanos family members of the same species). This domain is crucial for Nanos function, because it mediates binding with RNA as well as with interaction partners such as Pumilio [10, 11]. Nanos proteins from vertebrates and some invertebrates (such as sponge, fresh-water polyp and jellyfish) share an additional N-terminal region of 17 amino acids (AA) called NIM (NOT1 interacting motif) [9, 12] (Fig. 1b). In contrast to the C-terminal domain (zf-nanos), the sequences of the N-terminal domains of the various Nanos proteins are not conserved.
Fig. 1.
Nanos protein domains. a All Nanos proteins contain a C-terminal (CCHC)2 zinc finger domain (zf-nanos). Nanos proteins of all vertebrates and a few invertebrates have an additional N-terminal NOT1-interacting motif (NIM). Drosophila melanogaster (Dm) has a nanos effector domain (NED) with a central region (NOT module-binding region, NBR) that can bind NOT1 and NOT3, components of the CCR4–NOT complex. N1BM, NOT1 binding motif; N3BM, NOT3 binding motif. Amino acid (AA) positions of the domains are given on top of the sequences. Hs, Homo sapiens, Xt, Xenopus tropicalis. The figure was adapted from [66]. b Alignment of the NIM domain in several vertebrate and invertebrate organisms
Zf-nanos is the only conserved domain that can be used to create a reliable phylogenetic tree. By browsing the gene and genome databases of UCSC, Ensembl, and NCBI, we observed that there is at least one nanos gene in all animals, even in the comb jellies, which are among the most ancestral animals (Table 2). Depending on the species, the genome encodes one (D. melanogaster), two (Hydra vulgaris), three (C. elegans, M. musculus, H. sapiens), or four (Fugu rubripes) nanos genes (Tables 1, 2). Most vertebrates have three nanos genes, whereas some reptiles have lost a nanos gene and birds seem to have lost two. Xenopus tropicalis has only two annotated nanos genes, although a third gene has been reported [9]. Similarly, a nanos2 gene that has not been annotated is found in stickleback.
Table 2.
Overview of Nanos protein sequences predicted from genomic and transcriptomic databases
| Taxonomy | Species (common name) | Nanos gene(s) |
|---|---|---|
| Deuterostomes | ||
| Primates | Homo sapiens (human) |
Nanos1 (NP_955631) Nanos2 (NP_001025032) Nanos3 (NP_001092092) |
| Rodentia | Mus musculus (mouse) |
Nanos1 (NP_848508) Nanos2 (NP_918953) Nanos3 (NP_918948) |
| Carnivora | Canis lupus familiaris (dog) |
Nanos1 (XP_005637940) Nanos2 (XP_541547) Nanos3 (ENSCAFT00000026271.3) |
| Metatheria | Monodelphis domestica (opossum) |
Nanos1 (XP_001376960) Nanos2 (XP_007492184) Nanos3 (XP_007489148) |
| Aves | Gallus gallus (chicken) | Nanos1 (XP_015144398) |
| Reptilia | Chrysemys picta (turtle) |
Nanos1 (XP_005284212) Nanos2 (XP_005283826) Nanos3 (XP_005310751) |
| Reptilia | Alligator sinensis (alligator) |
Nanos1 (XP_014373968) Nanos3 (XP_006023461) |
| Amphibia | Xenopus tropicalis (frog) |
Nanos1 (NP_988857) Nanos3 (XP_004919224) |
| Fish | Danio rerio (zebrafish) |
Nos1 (NP_001292590) Nos2 (XP_009300191) Nos3 (NP_571953) |
| Fish | Gasterosteus aculeatus (stickleback) |
Nanos1a (ENSGACT00000022078) Nanos1b (ENSGACT00000006526) Nanos3 (ENSGACT00000025168) |
| Fish | Fugu rubripes (fugu) |
Nanos1a (XP_011609291) Nanos1b (XP_011618819) Nanos2 (XP_011606249) Nanos3 (XP_011610763) |
| Urochordata | Ciona intestinalis (sea squirt) | Nanos (XP_002130327) |
| Cephalochordata | Branchiostoma floridae (amphioxus) | Nanos (XP_002608940) |
| Hemichordata | Saccoglossus kowalevskii (acorn worm) | Nanos (NP_001161595) |
| Echinodermata | Strongylocentrotus purpuratus (sea urchin) |
Nanos1a (XP_001177221) Nanos1b (NP_001073023) |
| Protostomes | ||
| Lophotrochozoa | Aplysia californica (sea hare) |
Nanos1 (XP_005096656) Nanos2 (XP_012937610) |
| Lophotrochozoa | Helobdella robusta (leech) |
Nanos1a (XP_009018920) Nanos1b (XP_009013101) |
| Ecdysozoa | Drosophila melanogaster (fruit fly) | Nanos (Nos) (NP_476658) |
| Ecdysozoa | Caenorhabditis elegans (roundworm) |
Nos1 (NP_496358) Nos2 (NP_495452) Nos3 (NP_496101) |
| Ecdysozoa | Solenopsis invicta (red fire ant) |
Nanos1a (XP_011159578) Nanos1b (XP_011159747) Nanos1c (LOC105205302) |
| Ecdysozoa | Bombyx mori (silkmoth) |
NanosM (NP_001098700) NanosN (NP_001098702) NanosO (NP_001093314) NanosP (NP_001093313) |
| Non-bilaterian animals | ||
| Cnidaria | Nematostella vectensis (sea anemone) |
Cnnos1 (XP_001637175) Cnnos2 (XP_001641215) |
| Cnidaria | Hydra vulgaris(fresh-water polyp) |
Cnnos1 (XP_002161850) Cnnos2 (XP_002159764) |
| Cnidaria | Acropora digitifera (acroporid coral) |
Cnnos1 (XP_015755666) Cnnos2 (XP_015758550) |
| Placozoa | Trichoplax adhaerens (placozoan) | Nanos (XP_002114667) |
| Porifera | Amphimedon queenslandica (demosponge) | Nanos (XP_003384296) |
| Ctenophora | Mnemiopsis leidyi (comb jelly) |
Nanos1a (ML130210a) Nanos1b (ML22086a) |
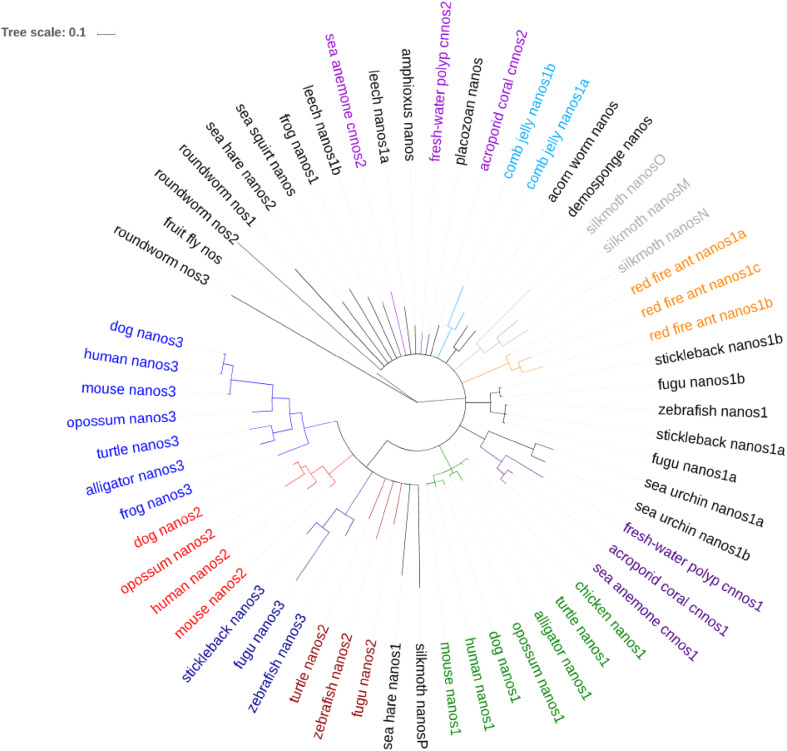
Based on the phylogenetic analysis of zf-nanos, vertebrate nanos1, -2, and -3 proteins mainly cluster together with nanos1, -2, and -3 proteins from other species (orthologs), respectively, rather than with their paralogs (Fig. 2). This indicates that the nanos gene had undergone duplications and that the resulting paralogs probably evolved new functions. Some nanos genes, such as those of H. vulgaris and other cnidarians, cannot be classified within the branches of vertebrate nanos1, -2, or -3 genes. Their nanos genes were probably duplicated independently during evolution. The nanos genes in red fire ant, fugu, and silkmoth had also undergone lineage specific duplications and this resulted in four nanos genes in the latter two animals (Table 2). The fourth nanos gene of fugu is probably a duplicated nanos1 homolog (Fig. 2).
Fig. 2.
Phylogenetic tree based on the zinc finger domain of Nanos proteins. The zf-nanos domains of metazoan nanos homologs were aligned with MUSCLE [123]. With this alignment as input, a Bayesian inference (BI) consensus tree was built using MrBayes 3 [124]. Convergence (< 0.01) was reached after 5,000,000 generations. The circular BI tree was visualized with iTOL [125]
The large sequence differences between the non-zf part of Nanos orthologs and paralogs are manifested in a major difference in protein length between Nanos sequences (Fig. 1a). The nanos gene of Drosophila encodes the largest protein sequence (401 AA), which is considerably larger than Nanos proteins from mouse and human (Nanos1: 267 and 292 AA; Nanos2: 136 and 138 AA; and Nanos3 178 and 192 AA, respectively). In Xenopus, nanos1 (Xtcat-2) comprises only 128 AA, including the 16-AA NIM region and the 52-AA zinc finger domain (Fig. 1a). These differences might be linked with different molecular interaction partners and functions.
Nanos interaction partners
Few nanos interaction partners have been identified. See Table 3 for the known interaction partners of human Nanos proteins.
Table 3.
Known interaction partners of human Nanos proteins
| Nanos protein | Interaction partner | Interaction domains | Reference |
|---|---|---|---|
| Nanos1 | Pumilio2 | zf-nanos | [11] |
| p120-catenin | N-terminal domain (including NIM) | [6] | |
| β-Catenin | nd | [6] | |
| SNAPIN | N-terminal domain and zf-nanos are needed | [28] | |
| GEMIN3 | N-terminal domain (without NIM) | [67] | |
| CNOT1 | NIM domain | [9] | |
| Nanos2 | CNOT1 | NIM domain | [9] |
| Nanos3 | CNOT1 | NIM domain | [9] |
zf-nanos zinc finger domain of nanos proteins, NIM NOT1 interacting motif; nd: not determined
Pumilio proteins
Pumilio is at the origin of the PUF family, which is named after its founders Pumilio (Pum) of D. melanogaster and FBF of C. elegans. PUF proteins are RNA-binding proteins found in eukaryotes ranging from plants to yeasts, invertebrates, and humans [13]. The number of PUF family members varies from multiple in C. elegans, Saccharomyces cerevisiae, and Arabidopsis thaliana to only one member in insects, such as D. melanogaster. Humans and mice have two Pumilio-encoding genes. They all share a highly conserved C-terminal RNA-binding domain comprising eight tandem repeats, collectively called the PUM homology domain (PUM-HD) [14]. Each repeat binds to one RNA base in the mRNA target [15].
Pumilio has been reported to bind both Pumilio-binding elements (PBEs, 5′-UGUANAUA-3′) and Nanos regulatory/response elements (NREs) in the 3′ untranslated region (UTR) of their target mRNAs and recruits, among other proteins, deadenylation and decapping proteins. The NREs are composed of two sequences, called box A (5′-GUUGU-3′) and box B (5′-AUUGUA-3′). Nanos binds to the first part of the box B sequence [16]; the last part of this NRE box B sequence shares identity with the first part of the PBE.
Tandem affinity purification (TAP) and a DNA microarray were used to identify mRNAs associated with the Pumilio protein in adult ovaries and embryos of Drosophila [17]. For this analysis, a TAP-tagged C-terminal fragment of pumilio was expressed under the control of an ovary-specific promoter. A PBE was present in 54% of the adult and 22% of the embryonic pumilio targets identified. Unlike for the human Pumilio proteins, Drosophila pumilio binds nanos mRNA in the embryo. Nonetheless, nanos mRNA lacks the UGUA(A/U/C)AUA motif [17], and another non-canonical motif in nanos mRNA was found to mediate pumilio binding [18].
Besides binding RNA, the PUM-HD domain can bind various proteins, such as nanos [10], CNOT8 [19], and DAZ [20]. Nanos binding is mediated through the loop region between the last two pumilio repeats [21]. Nanos determines the location in the embryo or the postnatal cell type where the specific translation inhibition of the nanos/pumilio complex occurs [22–24].
In Drosophila, the interaction between nanos and pumilio is stabilized by a NRE-containing RNA fragment and is, therefore, RNA-dependent [10]. However, human Nanos1 was found to interact with Pumilio2 in the absence of RNA [11]. Likewise, mouse Nanos3 was shown to interact with Pumilio in an RNA-independent manner [25]. The interaction between Xcat-2/nanos and pumilio was also confirmed in Xenopus, but RNA dependence was not investigated here [26].
Although the N-terminal sequence of PUF proteins is very variable, in some family members, it contains two conserved pumilio motifs that can be traced back from humans to Drosophila [27]. Multiple domains in the N-terminus confer repressive activity [27]. The N-terminus is also important for dimerization of Pumilio2 [11] and for specific protein interactions, such as the interaction between Pumilio2 and SNAPIN [28]. SNAPIN is a widely expressed protein that is part of BLOC-1 (biogenesis of the lysosome-related organelle complex 1) and BORC (BLOC-1-related complex) and associates with the SNARE complex [29–31]. It is involved in several functions involving intracellular vesicles, such as endosomes and lysosomes [32–34]. The relevance of the interaction between Pumilio2 and SNAPIN is unknown.
PUF proteins have the conserved role of maintaining stem cells [35–37], but other roles, such as in sperm/oocyte switch [38], long-term memory [39], and anterior–posterior patterning [22], have been acquired during evolution. PUF proteins perform these functions by post-transcriptional regulation of their targets, as reviewed in [40]. This occurs in cooperation with interaction partners such as nanos [10], CPEB [26, 41], and the CCR4–NOT complex [42]. Although PUF proteins are generally believed to repress mRNA translation by deadenylation [19] or interference with translation initiation [43], PUF proteins can also stimulate mRNA translation [44, 45]. Further research is needed to understand how these repressive and activating functions are integrated, which could vary with the target or the interaction partner, or depend on extracellular or intracellular signals.
The identification of mRNA targets of the human Pumilio proteins in HeLaS3 cancer cells led to the discovery of extensive interactions with the miRNA regulatory system [46]. Pumilio-associated mRNAs were identified using RNA immunoprecipitation followed by a microarray-based analysis. RNA sequences that specifically bind Pumilio (PBE, Pumilio-binding elements) are more likely to be located near miRNA-binding sites; similarly, mRNA targets of Pumilio are enriched in miRNA-binding sites. Links between Pumilio-mediated and miRNA-mediated repression have, indeed, been discovered [23, 47, 48]. It would be interesting to further investigate the link between the miRNA regulatory complex and the Nanos/Pumilio complex. This might reveal new interaction partners of the Nanos proteins.
In addition, many Pumilio targets are associated with pathways involved in cancer, such as angiogenesis, cell proliferation, and cell survival [46], and Pumilio-mediated regulation is, indeed, disturbed in several cancers [49–51].
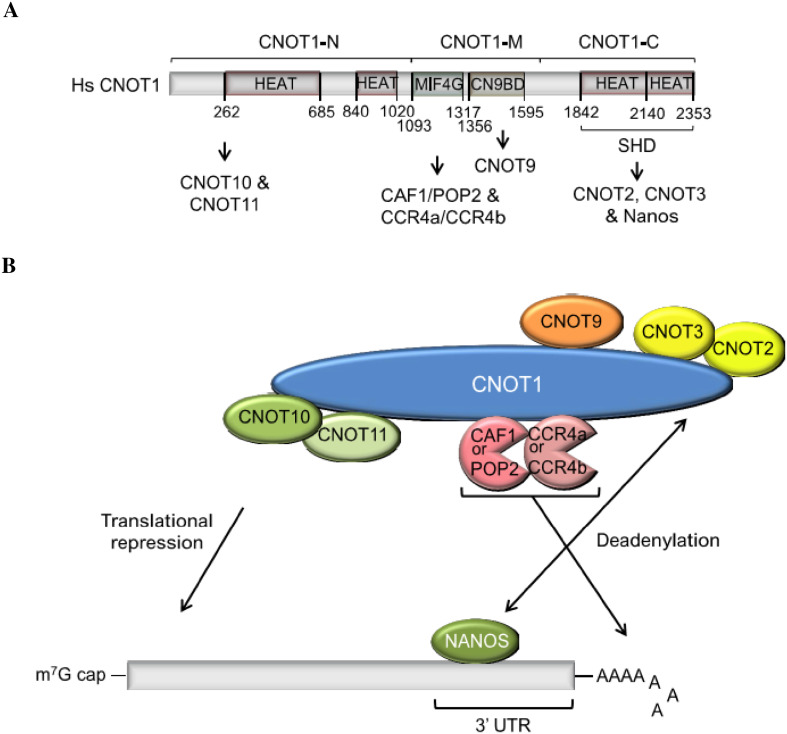
The CCR4–NOT complex
The N-terminals of all human Nanos proteins interact with the C-terminal domain of CNOT1 (Fig. 3) [9]. CNOT1 is part of the CCR4–NOT deadenylase complex, which is a common partner of Nanos proteins in some species [9, 52, 53]. The CCR4–NOT complex is a highly conserved, multisubunit complex that facilitates gene regulation in diverse ways. This complex was first studied in yeast, in which it consists of nine core proteins. Except for Caf130, homologs of these proteins exist, for instance in D. melanogaster and H. sapiens. The CNOT1 subunit is the scaffold that keeps the complex together (Fig. 3). The smaller complex, consisting of CNOT1 to -3 in humans and of Not1, Not2, and Not5 in yeast, is referred to as the NOT module [54, 55]. Proteins CCR4 and Caf1 contribute to the deadenylation activities of the CCR4–NOT protein complex (Fig. 3b). The complex can also interact with diverse proteins, such as the poly(A)-binding protein (PABP) through binding of BTG/TOB proteins [56–58], eIF4A2 [59], and proteins of the decapping complex through binding of DDX6 [60], which leads to inhibition of transcription or translation of their target genes/mRNAs, or to both. The CCR4–NOT complex is also involved in miRNA-mediated repression through binding with the GW182 protein [61, 62]. More information about the CCR4–NOT complex can be found in the following reviews [63, 64].
Fig. 3.
CNOT1 is the scaffold protein of the CCR4–NOT deadenylase complex. a Schematic representation of human CNOT1. The N-terminal region (CNOT1-N) consists of two HEAT repeats and provides binding sites for CNOT10 and CNOT11 [126]. The middle region (CNOT-M) contains the MIF4G domain, structurally related to the middle domain of eIF4G, and the CNOT9 binding domain [60], also called DUF3819 domain. The MIF4G domain binds the catalytic subunits, CAF1 or POP2 along with CCR4a or CCR4b deadenylases [127]. The C-terminal region (CNOT1-C) contains the superfamily homology domain (SHD) required for binding to CNOT2, CNOT3 [54], and Nanos proteins [9]. AA positions of the domains are given below the sequences. b Transcription and translation regulators such as Nanos and other proteins (X) bind the 3′UTR of their mRNA targets and recruit the CCR4–NOT complex. This complex stimulates deadenylation and translational repression by recruiting additional proteins. Hs, Homo sapiens
In mice, Nanos2 also binds the CCR4–NOT complex through CNOT1, but Nanos3 does this mainly by interacting with POP2 [65]. The different ways in which Nanos2 and Nanos3 interact with components of the mouse CCR4–NOT complex could be responsible for the weaker deadenylase activity of Nanos3 compared to Nanos2, and might explain why Nanos3 cannot fully compensate for loss of Nanos2, as described below. The Nanos NIM region and its interaction with the CCR4–NOT complex proved to be essential for Nanos-mediated translational repression and mRNA degradation [9]. Binding of the CCR4–NOT complex is functionally conserved: also Drosophila nanos has been shown to bind to this complex, though it has no NIM region [66]. Drosophila nanos interacts with NOT1 and NOT3 of the CCR4–NOT complex via a central region (called NBR, for NOT module-binding region) situated in the nanos effector domain (NED) [66] (Fig. 1).
Other interaction partners
Unlike the CCR4–NOT complex, other Nanos partners often differ depending on the paralog, the organism, or the mRNA target. For example, the nos-3 protein in C. elegans was found to bind the fem-3 binding factor (FBF), but neither nos-1 nor nos-2 bound FBF [38]. Another example is the Nanos1–p120-catenin interaction mediated through the NIM region, which is present in humans but not in lower organisms such as Drosophila [6]. Furthermore, human Nanos1 has been reported to interact with SNAPIN [28] and GEMIN3, an RNA DEAD box helicase [67]. GEMIN3 is a component of the SMN (Survival Motor Neuron) complex, which is essential for formation of small nuclear ribonucleoproteins (snRNPs), which are essential for correct splicing [68]. GEMIN3 was also detected in miRNP particles, and its involvement in miRNA-mediated repression has been suggested [69, 70]. The interaction between Nanos1 and GEMIN3 seems to take place in the chromatoid body of germ cells, which also contains several miRNAs and components involved in miRNA regulation, such as Dicer and Argonaute proteins [67].
Nanos functions
Nanos genes are especially known for their roles in germ cell development, which are conserved between basic model organisms and mammals. Reproductive pathways usually evolve faster than somatic pathways, which emphasize the importance of the role of nanos genes in germ cell development. Current models for discovering the target mRNAs of Nanos-containing translation-inhibitory complexes are based on identifying both NREs and PBEs in the 3′UTR of candidate transcripts.
mRNA targets of the Nanos/Pumilio complex in D. melanogaster and humans
An overview of targets of the nanos/pumilio complex in the germline of model organisms has been published by Lai and King [71]. The nanos/pumilio complex was found to repress somatic gene expression, the cell cycle, and apoptosis; this repression correlates perfectly with its function in germ cell development and survival. A short overview of the mRNA targets of Drosophila nanos discussed below and involving a PBE or NRE sequence, or both, is given in Table 4.
Table 4.
Overview of mRNA targets of Drosophila nanos
| mRNA target | Complex | Determining factor | Effect | References |
|---|---|---|---|---|
| Hunchback | Nanos/pumilio/brat | Nanos | Abdomen formation | [43, 82] |
| Cyclin B | Nanos/pumilio | Nanos | Blocking mitosis in the pole cells | [84, 85] |
| Hid | Nanos/pumilio | Nanos | Blocking apoptosis of the pole cells | [88] |
| Para | Nanos/pumilio | Pumilio | Regulates neuronal membrane excitability | [90, 91] |
| Mei-P26 | Nanos/pumilio | Nanos | Regulates self-renewal of ovarian stem cells | [53] |
| dE2F1 | Nanos/pumilio/brat | Nanos | Ensuring correct E2F regulation | [23, 49] |
Hunchback
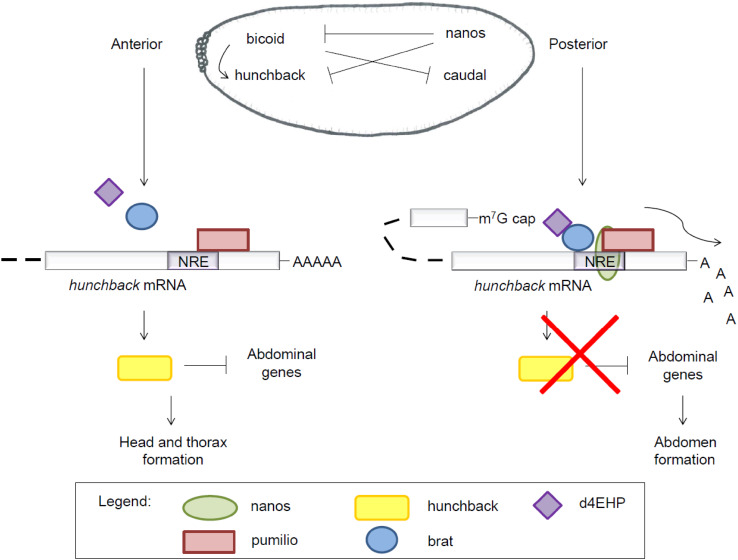
In D. melanogaster, nanos-encoding mRNA was first discovered as a maternal factor localized to the posterior pole of the unfertilized egg [1] (Fig. 4). Whereas nanos represents the abdominal determinant, the bicoid protein is the anterior determinant. Posterior localization of nanos mRNA is dependent on signals present in its 3′UTR [72, 73]. Both bicoid and nanos mRNAs are translated after fertilization. During oogenesis, several genes, such as oskar, vasa, and aub, contribute to the posterior localization of nanos RNA [74–76]. This ensures a nanos protein gradient decreasing from the posterior pole to the anterior pole. The bicoid protein activates hunchback protein expression, while nanos in association with pumilio and brain tumor (brat) represses hunchback translation (Fig. 4). This generates an anterior–posterior gradient of the hunchback protein, which blocks abdomen formation at the anterior pole, thus allowing development of the head and thorax [77]. Likewise, bicoid inhibits caudal mRNA translation, causing a posterior–anterior gradient of the caudal protein. Together, these gradients ensure correct anterior–posterior patterning of the embryo. Nanos can also repress translation of bicoid mRNA if the latter is not correctly restricted to the anterior pole [78].
Fig. 4.
Anterior–posterior patterning of the Drosophila embryo. Nanos is an important posterior determinant in Drosophila development. Nanos expression induces abdomen formation by inhibiting, in cooperation with pumilio and brat, translation of hunchback mRNA. Hunchback has two nanos response elements (NREs) in its 3′ untranslated region, but for simplicity only one is drawn. See text for further explanation
Repression of hunchback mRNA translation by nanos depends on two NREs in the 3′UTR of hunchback mRNA [78]. Pumilio, as well as nanos and brat, was found to bind these NRE sequences [16, 21, 22]. Brat was convincingly shown to bind box A of the NRE sequence [16, 79–81]. Prior RNA binding of brat or pumilio facilitates the binding of the other protein [79]. Nanos is recruited only when pumilio is bound to the NRE sequence (Fig. 4) [10]. This nanos–brat–pumilio complex blocks hunchback translation by promoting the deadenylation of hunchback mRNA [82] and inhibiting its translation [43]. Translation inhibition is mediated by brat-dependent recruitment of d4EHP, which binds the 5′ cap structure of hunchback mRNA and thereby inhibits the binding of the homologous eIF4E protein (Fig. 4).
Cyclin B
In addition to the above-mentioned role of the posterior pole plasm of the Drosophila embryo in abdomen formation, this pole is also responsible for germline formation [83]. Nanos expression is seen in the pole cells, also called primordial germ cells (PGCs), and a functional maternal nanos protein is, indeed, important for correct migration of these pole cells into the gonads and thus for germ cell formation [3, 37]. During this migration, pole cell mitosis is blocked by nanos- and pumilio-dependent repression of cyclin B RNA [84, 85]. In this case, unlike for hunchback regulation, pumilio apparently functions merely by recruiting nanos. Nanos then represses cyclin B RNA by interacting with its conserved interaction partner, the CCR4–NOT complex [86]. Though pumilio mediates nanos recruitment in vivo, the experimentally linking of nanos to the cyclin B mRNA sequence efficiently downregulates cyclin B in the absence of pumilio. Association of Nanos with cyclin B1 mRNA was found to be conserved in Xenopus [12, 26].
Hid
Nanos suppresses apoptosis and somatic gene expression in pole cells [87], by repressing translation of the pro-apoptotic head involution defective (hid) gene [88].
Para
The nanos/pumilio complex in D. melanogaster also seems to play a role in neurogenesis. Nanos and pumilio mutants and double mutants have similar effects on dendrite morphogenesis of class-III and class-IV dendritic arborization neurons [89]. Nanos was found to colocalize with RNA granules in these dendrites, which might be where the nanos/pumilio complex is located. This complex also regulates neuronal membrane excitability by repressing transcription of paralytic (para) mRNA [90–92]. Intriguingly, the PBE sequence, which is both essential and sufficient for pumilio binding, was found in the open reading frame (ORF) instead of the 3′UTR [90]. Para encodes a voltage-gated Na+ channel. Increased pumilio expression or reduced para mRNA consequently reduces voltage-gated Na+ current and membrane excitability [90, 91]. Pumilio seems to be the determining factor of the para mRNA repression by the nanos/pumilio complex [90]. Overexpression of pumilio negatively influences nanos mRNA expression, what might serve as a negative feedback mechanism, preventing excessive repression of para mRNA [17, 18, 90].
Mei-P26
Mei-P26 is a Trim-NHL (Tripartite motif and Ncl-1, HT2A, and Lin-41) protein, restricting cell growth and self-renewal of ovarian stem cells [93]. The nanos/pumilio complex targets mei-P26 mRNA in the ovarian stem cells, thereby allowing self-renewal of these stem cells. This is mediated by recruitment of the CCR4–NOT deadenylase complex [53].
E2F3
The Nanos/Pumilio complex has been shown to repress E2F3 translation in primary human fibroblasts (IMR90) [23]. E2F3 is an oncogene found to be overexpressed or dysregulated in several cancers, such as bladder [94], prostate [95], and lung cancer [96]. The E2F family includes both transcriptional activators (dE2F1 in Drosophila and E2F1 to -3 in humans) and transcriptional repressors (dE2F2 in Drosophila and E2F4 to -8 in humans).
E2F transcription factors play an important role in progression of the cell cycle and induction of apoptosis (reviewed in [97] and [98]). E2F3 mRNA contains two functional PBE sequences, and ectopic expression in IMR90 cells of any combination of a Nanos protein (Nanos1 or Nanos3) and a Pumilio protein (Pumilio1 or Pumilio2) decreased E2F3 expression levels [23]. Nanos/Pumilio-mediated regulation of E2F is conserved from Drosophila (where it regulates dE2F1 expression) to humans (where it controls the expression levels of the orthologous E2F3) [23]. Proximal to the PBEs, several miRNA seed sequences were found, and their corresponding miRNAs were shown to repress E2F3. Interestingly, this miRNA-mediated repression of E2F3 has been found to depend on the presence of these PBEs in the 3′UTR, and thus on Nanos/Pumilio-mediated regulation.
MAP3K1 and MAP2K3
MAP3K1 and MAP2K3 mRNAs are repressed by Nanos1 in combination with Pumilio1 or Pumilio2, as detailed below [49].
Functions of the nanos and pumilio proteins in Drosophila
The above-mentioned targets of the nanos/pumilio complex point out most of the known functions of the nanos protein in D. melanogaster. Furthermore, nanos RNA and protein are expressed during several stages of Drosophila oogenesis [99]. In adult ovaries, nanos is important for proliferation and survival of germline stem cells, and for cyst development [37]. Accordingly, female nanos mutants with severely reduced or no protein expression produce very few eggs [100]. Ovaries and testis from nanos-deficient embryos are devoid of germ cells. This function of nanos in germ cell development and survival is conserved in C. elegans [101] and zebrafish [102].
Also loss of pumilio causes loss of germ cells in the ovaries, and this occurs even earlier than in the ovaries of nanos mutants [37]. Other phenotypic changes caused by loss of either pumilio or nanos suggest that other nanos partners may be involved in the germline. Although, as mentioned above, the nanos/pumilio complex has been shown to regulate mei-P26, nanos, and pumilio might have other partners to regulate specific mRNA targets. For instance, interaction between cup and nanos seems to be important in the female germline [103]. Cup has been shown to be important for several functions during oogenesis [104–106].
In addition, at the pre- and post-synaptic sites of the larval neuromuscular junction, pumilio and nanos seem to have divergent functions [18]. Pumilio was found to repress GluRIIA mRNA translation and thereby stimulate the switch from GluRIIA to GluRIIB receptors, which influences the amount of current through the synapses. This regulation is even more tightly controlled, because pumilio also reduces nanos protein levels, while nanos downregulates GluRIIB.
Functions of mammalian Nanos proteins
Mouse
Unlike germ cell-specific expression of mouse Nanos2 and Nanos3 [4], mouse Nanos1 is predominantly expressed in the central nervous system [107]. Nanos1 knockout mice seem to develop normally without any obvious differences from wild-type mice [107]. Mouse Nanos3 clearly plays a role in maintaining PGCs from the migration phase onwards [4]. Nanos3-deficient mice initially have a normal number of PGCs, but these cells are gradually lost and are absent in ovaries and testes at E12.5 [4]. Ectopic expression of Nanos2 from E8.0 onwards partially counteracted the loss of both male and female germ cells in Nanos3 knockout mice, and thus partially compensated for the loss of Nanos3 [108].
Nanos2 is normally detectable only at E13.5. On the other hand, although Nanos3 is upregulated in Nanos2-null mice, male PGCs in these mice undergo apoptosis from E15.5 onwards, resulting in deficiency in male germ cells. Nanos3 transgene expression under control of the Nanos2 enhancer could not prevent this loss of spermatogonia. Nevertheless, Nanos3 transgene expression or upregulation might at least partly rescue Nanos2 deficiency. For instance, mutation of the zinc fingers in Nanos2 results in loss of Nanos3 expression and is associated with an even more severe phenotypic abnormality than complete Nanos2 deficiency [65]. Nonetheless, the inability of Nanos3 to fully compensate for Nanos2 loss indicates that these two related proteins have different functions. Nanos3 is also expressed in undifferentiated spermatogonia in the prepubertal testis [25]. By regulating the cell cycle of these spermatogonial cells, their differentiation is blocked until puberty. Given that Nanos3 interacts with Pumilio2 in spermatogonia, it is likely that also Pumilio2 is involved in this regulation [25].
Both Nanos2 and Nanos3 mouse proteins were found to be associated with ribonucleoproteins (RNPs), suggesting translational regulation. Nanos2 is also expressed in RNPs, where it recruits and represses mRNAs important for germ cell differentiation [109]. More precisely, in mouse, Nanos2 and Nanos3 are expressed in processing bodies (P-bodies) [52, 65], which are cytoplasmic mRNPs (messenger RNPs) linked with miRNA-mediated repression and containing many proteins involved in mRNA deadenylation, decapping, and decay [110, 111]. Nanos3 seems to be important for the assembly of these P-bodies in male germ cells [65], whereas Nanos2 is involved in their maintenance [52]. It would be interesting to investigate the functional association between Nanos proteins and regulatory proteins, which are generally found in the P-bodies.
Human
The first human Nanos-encoding gene was discovered in 2003 [11]. In contrast to murine nanos1 [107], human Nanos1 is not expressed in the adult brain. RT-qPCR analysis revealed Nanos1 mRNA expression in embryonic stem cells, fetal testis and ovary, and adult testis [11]. Later, others showed that Nanos1 mRNA was expressed more ubiquitously but also confirmed protein expression in fetal testis and ovary, and in adult testis [5]. However, in contrast to the original report, the latter authors also showed Nanos1 protein expression in the adult ovary. Nanos2 expression in adults was found to be restricted to the testis, in line with the findings for the mouse homologue [112]. Therefore, a possible link between NANOS2 mutations and male infertility was investigated, but the detected mutations did not seem to have a causative role in male infertility [112].
More recently, human Nanos2 was found to be expressed in the adult ovary, as well [5]. Like human Nanos1 and Nanos2, Nanos3 was found to be expressed not only in the fetal and adult testis and ovary, but also in the adult brain. Reducing Nanos3 expression levels in human embryonic stem cells significantly decreased germ cell numbers and the expression levels of genes important for germ cell development [5]. NANOS3 mutations were also studied in a cohort of sterile men, again revealing no causative role in sterility [113]. On the other hand, a plausible, pathological link has been found for NANOS3 mutations in patients with premature ovarian insufficiency [114, 115]. Unlike what has been reported for NANOS2 and NANOS3 mutations, NANOS1 mutations were convincingly linked to male infertility [116].
Nanos genes, tumor invasion, and cancer
Germ cells and cancer cells share several characteristics, such as self-renewal and rapid proliferation. Nanos genes are responsible for germline traits such as pluripotency and survival, which are also important for tumor cells. Hence, Nanos overexpression might be a logical asset for cancer tissues.
In D. melanogaster, nanos overexpression was only reported in the lethal (3) malignant brain tumor model (l(3)mbt) [117]. Nanos was only one of many genes essential in the germline that were upregulated in this model. These results point out that nanos expression is advantageous for brain tumor growth, at least in this invertebrate model.
In the mouse, an interaction between the Dmrt1 and Nanos3 genes was discovered [118]. In mice that are heterozygous for both genes, incidence of teratoma formation was significantly more elevated than in singly heterozygous mice. Like Nanos3, Dmrt1 controls male germ cell proliferation [119]. Dmrt1 additionally regulates male germ cell pluripotency by repressing Sox2.
In humans, Nanos1 is a potential effector in E-cadherin-negative cancer cells, contributing to tumor migration and invasion [6]. The mRNA expression levels of NANOS1 and CDH1 are inversely correlated in several cancer cell lines, which led to the discovery that E-cadherin represses NANOS1 [6].
Nanos3 has been found to be ectopically expressed in a variety of human cancers [120]. So far, this was further investigated only in NSCLCs, in which Nanos3 expression levels correlated with patient outcome [7]. Immunostaining of lung tumors revealed Nanos3 overexpression, particularly at the invasion front and especially in squamous cell carcinomas (SCCs). When comparing primary tumors with their metastases, Nanos3 expression levels were found to be higher in the latter. Furthermore, ectopic expression of Nanos3 has been observed in several invasive NSCLC cell lines, in which it was associated with higher invasiveness. Moreover, Nanos3 overexpression causes clear-cut EMT in human lung cancer cells, thereby reinforcing the hypothesis that ectopic Nanos expression is involved in cancer progression [7].
A likely mechanism for malignancy caused by ectopic Nanos expression involves Nanos3-mediated repression of E-cadherin, occludin, and β-catenin, combined with Nanos3-induced stimulation of expression of vimentin, slug, urokinase-type plasminogen activator (uPA), and matrix metalloproteinase-14 (MMP-14) [7]. Both transcriptional regulation (uPA, slug, and E-cadherin) and post-transcriptional regulation (MMP-14, occludin, and vimentin) have been found to be involved in these Nanos3 effects. However, Nanos3 does not bind CDH1 mRNA, suggesting that repression is at the transcriptional level. This has not yet been reported for the Nanos/Pumilio complex and should be investigated further. Nanos3 transcriptionally regulates the E-cadherin encoding CHD1 gene independently of the E-boxes in its promoter region. Other transcriptional repressors, such as Slug, Snail, and ZEB proteins, depend on these E-boxes to repress E-cadherin expression.
Remarkably, Nanos3 stabilizes vimentin mRNA by increasing its poly(A)-tail length. Furthermore, Nanos3 protects vimentin mRNA from being bound by miR-30a, which would otherwise repress translation of vimentin. This mechanism of VIM mRNA regulation is the first demonstration that binding of a Nanos protein to an mRNA sequence leads to its upregulation. Further investigation of a possible activating role for Nanos proteins is needed. Such activating role might be a specific function executed by mammalian Nanos proteins only. We must note that it has not been investigated whether Pumilio proteins are needed for the Nanos-mediated regulation of E-cadherin and vimentin. In complex organisms, the proposed Nanos role as transcriptional regulator and activator might depend as well on other interaction partners besides Pumilio. As Pumilio can act also independently of Nanos, it is conceivable that interaction of Nanos with other regulating proteins can expand its repertoire of specific mRNA targets.
The mechanism underlying increased uPA and MMP-14 levels upon Nanos3 expression has not been elucidated. The role of the malignancy-promoting metalloprotease MMP-14 (an ECM degrading enzyme) in EMT is unmistakable. Nanos1 expression has been linked to MMP-14 induction [8]. Nanos1 is similarly overexpressed in lung carcinomas [8], where its expression is higher at the invasion front of SCCs and is linked to increased invasiveness. In addition, the expression levels of Nanos1 correlated with tumor aggressiveness (TNM stage). Evidently, identifying target mRNAs of the human Nanos1 and Nanos3 proteins could reveal more about its molecular role and how its overexpression can contribute to tumorigenesis.
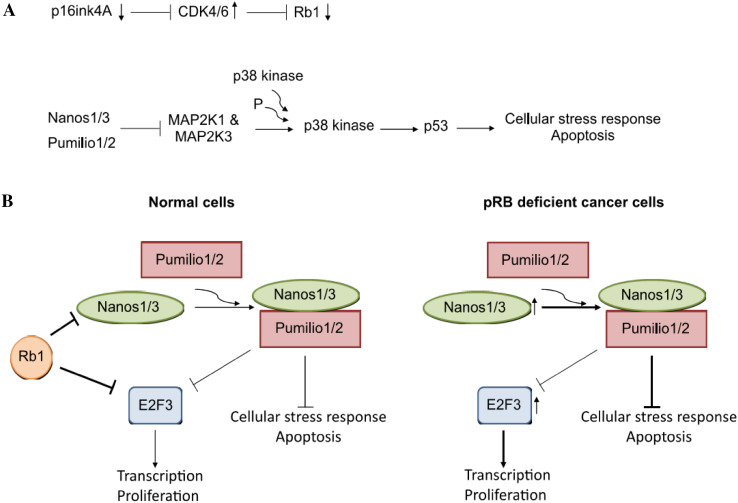
The Nanos/Pumilio complex has an interesting role in Rb1-deficient and p53 wild-type cancer cells. Functional RB1/pRb inactivation is often seen in cancers, and it can be achieved in several ways, such as E2F or CDK4/6 amplification, and inactivating mutations of p16INK4A or RB1 [121] (Fig. 5a). However, RB1/pRb inactivation can be associated with cellular stress and apoptosis, which are deleterious for cancer cell growth. Nonetheless, pRb-deficient cells often seem to evade these stress responses. pRb deletion is associated with upregulation of nanos in flies, and of NANOS1 and NANOS3 in humans [49]. Rb1 expression is needed for regulation of Nanos expression by the DREAM complex [49]. This complex, consisting of dimerization partner (DP), Rb-like, E2F, and MuvB, is evolutionarily conserved with minor variations in its components [122]. As in humans, the nanos gene is strongly bound by components of the Drosophila dREAM complex, consisting of Rb, E2F, and Myb-associated protein [49].
Fig. 5.
Rb1 deregulation in cancer cells. a Rb1 inactivation can be obtained in several ways, for example by down- or upregulation of upstream regulators. Upon loss of Rb1 the Nanos/Pumilio complex is important in cancer cells to repress p53-mediated cellular stress and apoptosis. b Schematic representation of the role of Nanos proteins in Rb1-deficient cells retaining a functional p53 protein. See text for further explanation
This inverse correlation between pRb and Nanos1 or Nanos3 expression is seen in diverse human tumor cell lines. When depleting Nanos1 in pRb-deficient cells, such as the NSCLC cell line NCI-H1666, the cell number is reduced gradually [49]. However, this was only observed in pRb-deficient cells harboring a wild-type p53 gene, suggesting that Nanos1 can repress p53-mediated inhibition of cell growth (Fig. 5b). Nanos1 was, indeed, found to downregulate MAP3K1 and MAP2K3 genes, which encode kinases upstream of p53 (Fig. 5a) [49]. In addition, Nanos1 leads to suppression of apoptosis and thus allows oncogenic growth of pRb-deficient cells. Nanos1 expression can, therefore, enable pRb-deficient cells lacking p53 mutations to evade stress responses. Though p53 is the most frequently mutated gene in human cancers, p53 mutations are rare in some cancers, such as retinoblastoma and cervical cancer. Many genes that are downregulated in retinoblastoma tumors compared to normal retinal tissue, indeed, contain PBE motifs. These genes encode proteins such as MAP3K1 and MAP2K3, which are involved in signaling and apoptotic pathways.
Conclusions and perspectives
Nanos proteins originated a long time ago and are represented in all animals. Their primary function in germ cell maintenance is generally conserved, but several other functions have been added during evolution. It would be interesting to gain a deeper understanding of these functions acquired during evolution and in which species. Nanos members form protein complexes with interaction partners such as Pumilio and the CCR4–NOT complex to mediate transcriptional and translational regulation of their target mRNAs [9, 10, 66]. Several studies reported a link between the Nanos/Pumilio complex and miRNA-mediated regulation [23, 46, 67]. The CCR4–NOT complex is also recruited by GW182 proteins and contributes to miRNA-mediated repression [61, 62]. A functional interaction between the Nanos/Pumilio complex and the miRNA regulatory complex has been reported to mediate E2F3 repression [23]. In view of the close interaction between miRNAs and the Nanos/Pumilio complex in regulating specific targets, miRNA silencing might also affect the efficiency with which the Nanos/Pumilio complex regulates these targets [23]. Further correlations between these complexes should be investigated.
Research on Nanos protein expression in cancer is limited. Given that expression of Nanos proteins is mainly restricted to the testis or to the testis and brain, and that they are overexpressed in human cancer, they are potential candidates as cancer testis antigens (CTA). In malignant tumors of epithelial origin, a key event of high diagnostic and prognostic value is inactivation or complete loss of the cell adhesion protein E-cadherin, generally during EMT. Expression levels of Nanos1 or Nanos3 proteins are inversely correlated to E-cadherin expression levels in several cancer cell lines [6], and Nanos3 was even reported to repress E-cadherin expression [7]. In addition, as the physical and functional interaction between the DREAM complex and the Nanos/Pumilio complex is conserved, this complex might play an important role in Rb-deficient cancer cells retaining a wild-type p53 (Fig. 5) [49]. In general, the Nanos/Pumilio complex modulates the expression levels of genes important in both development and disease, and most likely their influence depends on the “cellular context,” such as protein complex composition and miRNA levels.
Clearly, Nanos protein members can act as oncofetal agents in the progression of human cancers, although this should be elucidated further. Novel in vivo mouse models would be valuable for elucidating the effects of Nanos overexpression and the mechanistic pathways used by Nanos proteins to stimulate tumor progression. Identification and characterization of mRNA targets and interaction partners of mammalian Nanos proteins could also identify pathways that might be triggered in cancer cells. Furthermore, as both Nanos1 and Nanos3 play roles in lung carcinoma, the interplay between Nanos paralogs might be relevant. On the other hand, no cancer-specific expression of Nanos2 has been reported to date. In vitro and in vivo experiments could show whether Nanos2 overexpression also increases the tumorigenic potential of cancer cells.
Besides investigating the roles of Nanos proteins in cancer, the normal functions of mammalian Nanos proteins need further research. For instance, Nanos1 knockout mice seem perfectly normal. Given the function of Drosophila nanos in dendrite morphogenesis [89] and neuronal excitability [90], it could be interesting to study this in more detail in the mouse. Besides its expression in the testis, Nanos3 is also expressed in the brain, although also here no specific function has been identified. In addition, the implications of the interactions between Nanos1 and GEMIN3 and SNAPIN should be elucidated, and it would be interesting to check whether these interactions are conserved in other species.
Acknowledgements
We thank Dr. Amin Bredan for critical reading and careful editing of the manuscript, and our colleagues from Ghent University, VIB-UGent and the University of Reims (INSERM UMR-S 903) for helpful discussions. This work was supported by the Foundation against Cancer—Belgium, the Research Foundation—Flanders (FWO-Vlaanderen), and the Belgian Science Policy (Interuniversity Attraction Poles—Award IAP7/07). EDK has been a Ph.D. fellow of FWO-Vlaanderen.
References
- 1.Irish V, Lehmann R, Akam M. The Drosophila posterior-group gene nanos functions by repressing hunchback activity. Nature. 1989;338(6217):646–648. doi: 10.1038/338646a0. [DOI] [PubMed] [Google Scholar]
- 2.Wang C, Lehmann R. Nanos is the localized posterior determinant in Drosophila. Cell. 1991;66(4):637–647. doi: 10.1016/0092-8674(91)90110-K. [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi S, Yamada M, Asaoka M, Kitamura T. Essential role of the posterior morphogen nanos for germline development in Drosophila. Nature. 1996;380(6576):708–711. doi: 10.1038/380708a0. [DOI] [PubMed] [Google Scholar]
- 4.Tsuda M, Sasaoka Y, Kiso M, Abe K, Haraguchi S, Kobayashi S, Saga Y. Conserved role of nanos proteins in germ cell development. Science. 2003;301(5637):1239–1241. doi: 10.1126/science.1085222. [DOI] [PubMed] [Google Scholar]
- 5.Julaton VT, Reijo Pera RA. NANOS3 function in human germ cell development. Hum Mol Genet. 2011;20(11):2238–2250. doi: 10.1093/hmg/ddr114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strumane K, Bonnomet A, Stove C, Vandenbroucke R, Nawrocki-Raby B, Bruyneel E, Mareel M, Birembaut P, Berx G, van Roy F. E-cadherin regulates human Nanos1, which interacts with p120ctn and induces tumor cell migration and invasion. Cancer Res. 2006;66(20):10007–10015. doi: 10.1158/0008-5472.CAN-05-3096. [DOI] [PubMed] [Google Scholar]
- 7.Grelet S, Andries V, Polette M, Gilles C, Staes K, Martin AP, Kileztky C, Terryn C, Dalstein V, Cheng CW, Shen CY, Birembaut P, Van Roy F, Nawrocki-Raby B. The human NANOS3 gene contributes to lung tumour invasion by inducing epithelial–mesenchymal transition. J Pathol. 2015;237(1):25–37. doi: 10.1002/path.4549. [DOI] [PubMed] [Google Scholar]
- 8.Bonnomet A, Polette M, Strumane K, Gilles C, Dalstein V, Kileztky C, Berx G, van Roy F, Birembaut P, Nawrocki-Raby B. The E-cadherin-repressed hNanos1 gene induces tumor cell invasion by upregulating MT1-MMP expression. Oncogene. 2008;27(26):3692–3699. doi: 10.1038/sj.onc.1211035. [DOI] [PubMed] [Google Scholar]
- 9.Bhandari D, Raisch T, Weichenrieder O, Jonas S, Izaurralde E. Structural basis for the Nanos-mediated recruitment of the CCR4–NOT complex and translational repression. Genes Dev. 2014;28(8):888–901. doi: 10.1101/gad.237289.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sonoda J, Wharton RP. Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev. 1999;13(20):2704–2712. doi: 10.1101/gad.13.20.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaruzelska J, Kotecki M, Kusz K, Spik A, Firpo M, Reijo Pera RA. Conservation of a Pumilio-Nanos complex from Drosophila germ plasm to human germ cells. Dev Genes Evol. 2003;213(3):120–126. doi: 10.1007/s00427-003-0303-2. [DOI] [PubMed] [Google Scholar]
- 12.Lai F, Zhou Y, Luo X, Fox J, King ML. Nanos1 functions as a translational repressor in the Xenopus germline. Mech Dev. 2011;128(1–2):153–163. doi: 10.1016/j.mod.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wickens M, Bernstein D, Crittenden S, Luitjens C, Kimble J. PUF proteins and 3′UTR regulation in the Caenorhabditis elegans germ line. Cold Spring Harb Symp Quant Biol. 2001;66:337–343. doi: 10.1101/sqb.2001.66.337. [DOI] [PubMed] [Google Scholar]
- 14.Zamore PD, Williamson JR, Lehmann R. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA. 1997;3(12):1421–1433. [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, McLachlan J, Zamore PD, Hall TM. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110(4):501–512. doi: 10.1016/S0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- 16.Arvola RM, Weidmann CA, Tanaka Hall TM, Goldstrohm AC. Combinatorial control of messenger RNAs by Pumilio, Nanos and brain tumor proteins. RNA Biol. 2017;14:1445–1456. doi: 10.1080/15476286.2017.1306168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerber AP, Luschnig S, Krasnow MA, Brown PO, Herschlag D. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc Natl Acad Sci USA. 2006;103(12):4487–4492. doi: 10.1073/pnas.0509260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menon KP, Andrews S, Murthy M, Gavis ER, Zinn K. The translational repressors Nanos and Pumilio have divergent effects on presynaptic terminal growth and postsynaptic glutamate receptor subunit composition. J Neurosci. 2009;29(17):5558–5572. doi: 10.1523/JNEUROSCI.0520-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldstrohm AC, Hook BA, Seay DJ, Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol. 2006;13(6):533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- 20.Moore FL, Jaruzelska J, Fox MS, Urano J, Firpo MT, Turek PJ, Dorfman DM, Pera RA. Human Pumilio-2 is expressed in embryonic stem cells and germ cells and interacts with DAZ (Deleted in AZoospermia) and DAZ-like proteins. Proc Natl Acad Sci USA. 2003;100(2):538–543. doi: 10.1073/pnas.0234478100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weidmann CA, Qiu C, Arvola RM, Lou TF, Killingsworth J, Campbell ZT, Tanaka Hall TM, Goldstrohm AC. Drosophila Nanos acts as a molecular clamp that modulates the RNA-binding and repression activities of Pumilio. Elife. 2016;5:e17096. doi: 10.7554/eLife.17096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murata Y, Wharton RP. Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell. 1995;80(5):747–756. doi: 10.1016/0092-8674(95)90353-4. [DOI] [PubMed] [Google Scholar]
- 23.Miles WO, Tschop K, Herr A, Ji JY, Dyson NJ. Pumilio facilitates miRNA regulation of the E2F3 oncogene. Genes Dev. 2012;26(4):356–368. doi: 10.1101/gad.182568.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weidmann CA, Raynard NA, Blewett NH, Van Etten J, Goldstrohm AC. The RNA binding domain of Pumilio antagonizes poly-adenosine binding protein and accelerates deadenylation. RNA. 2014;20(8):1298–1319. doi: 10.1261/rna.046029.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lolicato F, Marino R, Paronetto MP, Pellegrini M, Dolci S, Geremia R, Grimaldi P. Potential role of Nanos3 in maintaining the undifferentiated spermatogonia population. Dev Biol. 2008;313(2):725–738. doi: 10.1016/j.ydbio.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 26.Nakahata S, Katsu Y, Mita K, Inoue K, Nagahama Y, Yamashita M. Biochemical identification of Xenopus Pumilio as a sequence-specific cyclin B1 mRNA-binding protein that physically interacts with a Nanos homolog, Xcat-2, and a cytoplasmic polyadenylation element-binding protein. J Biol Chem. 2001;276(24):20945–20953. doi: 10.1074/jbc.M010528200. [DOI] [PubMed] [Google Scholar]
- 27.Weidmann CA, Goldstrohm AC. Drosophila Pumilio protein contains multiple autonomous repression domains that regulate mRNAs independently of Nanos and brain tumor. Mol Cell Biol. 2012;32(2):527–540. doi: 10.1128/MCB.06052-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ginter-Matuszewska B, Spik A, Rembiszewska A, Koyias C, Kupryjanczyk J, Jaruzelska J. The SNARE-associated component SNAPIN binds PUMILIO2 and NANOS1 proteins in human male germ cells. Mol Hum Reprod. 2009;15(3):173–179. doi: 10.1093/molehr/gap004. [DOI] [PubMed] [Google Scholar]
- 29.Ilardi JM, Mochida S, Sheng ZH. Snapin: a SNARE-associated protein implicated in synaptic transmission. Nat Neurosci. 1999;2(2):119–124. doi: 10.1038/5673. [DOI] [PubMed] [Google Scholar]
- 30.Falcon-Perez JM, Starcevic M, Gautam R, Dell’Angelica EC. BLOC-1, a novel complex containing the pallidin and muted proteins involved in the biogenesis of melanosomes and platelet-dense granules. J Biol Chem. 2002;277(31):28191–28199. doi: 10.1074/jbc.M204011200. [DOI] [PubMed] [Google Scholar]
- 31.Pu J, Schindler C, Jia R, Jarnik M, Backlund P, Bonifacino JS. BORC, a multisubunit complex that regulates lysosome positioning. Dev Cell. 2015;33(2):176–188. doi: 10.1016/j.devcel.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan PY, Tian JH, Sheng ZH. Snapin facilitates the synchronization of synaptic vesicle fusion. Neuron. 2009;61(3):412–424. doi: 10.1016/j.neuron.2008.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Somanath S, Partridge CJ, Marshall C, Rowe T, Turner MD. Snapin mediates insulin secretory granule docking, but not trans-SNARE complex formation. Biochem Biophys Res Commun. 2016;473(2):403–407. doi: 10.1016/j.bbrc.2016.02.123. [DOI] [PubMed] [Google Scholar]
- 34.Khatamzas E, Hipp MM, Gaughan D, Pichulik T, Leslie A, Fernandes RA, Muraro D, Booth S, Zausmer K, Sun MY, Kessler B, Rowland-Jones S, Cerundolo V, Simmons A. Snapin promotes HIV-1 transmission from dendritic cells by dampening TLR8 signaling. EMBO J. 2017;36(20):2998–3011. doi: 10.15252/embj.201695364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang B, Gallegos M, Puoti A, Durkin E, Fields S, Kimble J, Wickens MP. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997;390(6659):477–484. doi: 10.1038/37297. [DOI] [PubMed] [Google Scholar]
- 36.Lin H, Spradling AC. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124(12):2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- 37.Forbes A, Lehmann R. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development. 1998;125(4):679–690. doi: 10.1242/dev.125.4.679. [DOI] [PubMed] [Google Scholar]
- 38.Kraemer B, Crittenden S, Gallegos M, Moulder G, Barstead R, Kimble J, Wickens M. NANOS-3 and FBF proteins physically interact to control the sperm-oocyte switch in Caenorhabditis elegans. Curr Biol. 1999;9(18):1009–1018. doi: 10.1016/S0960-9822(99)80449-7. [DOI] [PubMed] [Google Scholar]
- 39.Dubnau J, Chiang AS, Grady L, Barditch J, Gossweiler S, McNeil J, Smith P, Buldoc F, Scott R, Certa U, Broger C, Tully T. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr Biol. 2003;13(4):286–296. doi: 10.1016/S0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- 40.Quenault T, Lithgow T, Traven A. PUF proteins: repression, activation and mRNA localization. Trends Cell Biol. 2011;21(2):104–112. doi: 10.1016/j.tcb.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 41.Nakahata S, Kotani T, Mita K, Kawasaki T, Katsu Y, Nagahama Y, Yamashita M. Involvement of Xenopus Pumilio in the translational regulation that is specific to cyclin B1 mRNA during oocyte maturation. Mech Dev. 2003;120(8):865–880. doi: 10.1016/S0925-4773(03)00160-6. [DOI] [PubMed] [Google Scholar]
- 42.Van Etten J, Schagat TL, Hrit J, Weidmann CA, Brumbaugh J, Coon JJ, Goldstrohm AC. Human Pumilio proteins recruit multiple deadenylases to efficiently repress messenger RNAs. J Biol Chem. 2012;287(43):36370–36383. doi: 10.1074/jbc.M112.373522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cho PF, Gamberi C, Cho-Park YA, Cho-Park IB, Lasko P, Sonenberg N. Cap-dependent translational inhibition establishes two opposing morphogen gradients in Drosophila embryos. Curr Biol. 2006;16(20):2035–2041. doi: 10.1016/j.cub.2006.08.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pique M, Lopez JM, Foissac S, Guigo R, Mendez R. A combinatorial code for CPE-mediated translational control. Cell. 2008;132(3):434–448. doi: 10.1016/j.cell.2007.12.038. [DOI] [PubMed] [Google Scholar]
- 45.Kaye JA, Rose NC, Goldsworthy B, Goga A, L’Etoile ND. A 3′UTR pumilio-binding element directs translational activation in olfactory sensory neurons. Neuron. 2009;61(1):57–70. doi: 10.1016/j.neuron.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Galgano A, Forrer M, Jaskiewicz L, Kanitz A, Zavolan M, Gerber AP. Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PLoS One. 2008;3(9):e3164. doi: 10.1371/journal.pone.0003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nolde MJ, Saka N, Reinert KL, Slack FJ. The Caenorhabditis elegans pumilio homolog, puf-9, is required for the 3′UTR-mediated repression of the let-7 microRNA target gene, hbl-1. Dev Biol. 2007;305(2):551–563. doi: 10.1016/j.ydbio.2007.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JA, Elkon R, Agami R. A Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol. 2010;12(10):1014–1020. doi: 10.1038/ncb2105. [DOI] [PubMed] [Google Scholar]
- 49.Miles WO, Korenjak M, Griffiths LM, Dyer MA, Provero P, Dyson NJ. Post-transcriptional gene expression control by NANOS is up-regulated and functionally important in pRb-deficient cells. EMBO J. 2014;33(19):2201–2215. doi: 10.15252/embj.201488057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miles WO, Lembo A, Volorio A, Brachtel E, Tian B, Sgroi D, Provero P, Dyson N. Alternative polyadenylation in triple-negative breast tumors allows NRAS and c-JUN to bypass PUMILIO posttranscriptional regulation. Cancer Res. 2016;76(24):7231–7241. doi: 10.1158/0008-5472.CAN-16-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fernandez S, Risolino M, Mandia N, Talotta F, Soini Y, Incoronato M, Condorelli G, Banfi S, Verde P. miR-340 inhibits tumor cell proliferation and induces apoptosis by targeting multiple negative regulators of p27 in non-small cell lung cancer. Oncogene. 2015;34(25):3240–3250. doi: 10.1038/onc.2014.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suzuki A, Igarashi K, Aisaki K, Kanno J, Saga Y. NANOS2 interacts with the CCR4–NOT deadenylation complex and leads to suppression of specific RNAs. Proc Natl Acad Sci USA. 2010;107(8):3594–3599. doi: 10.1073/pnas.0908664107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Joly W, Chartier A, Rojas-Rios P, Busseau I, Simonelig M. The CCR4 deadenylase acts with Nanos and Pumilio in the fine-tuning of Mei-P26 expression to promote germline stem cell self-renewal. Stem Cell Rep. 2013;1(5):411–424. doi: 10.1016/j.stemcr.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boland A, Chen Y, Raisch T, Jonas S, Kuzuoglu-Ozturk D, Wohlbold L, Weichenrieder O, Izaurralde E. Structure and assembly of the NOT module of the human CCR4–NOT complex. Nat Struct Mol Biol. 2013;20(11):1289–1297. doi: 10.1038/nsmb.2681. [DOI] [PubMed] [Google Scholar]
- 55.Bhaskar V, Roudko V, Basquin J, Sharma K, Urlaub H, Seraphin B, Conti E. Structure and RNA-binding properties of the Not1-Not2-Not5 module of the yeast Ccr4–Not complex. Nat Struct Mol Biol. 2013;20(11):1281–1288. doi: 10.1038/nsmb.2686. [DOI] [PubMed] [Google Scholar]
- 56.Bogdan JA, Adams-Burton C, Pedicord DL, Sukovich DA, Benfield PA, Corjay MH, Stoltenborg JK, Dicker IB. Human carbon catabolite repressor protein (CCR4)-associative factor 1: cloning, expression and characterization of its interaction with the B-cell translocation protein BTG1. Biochem J. 1998;336(Pt 2):471–481. doi: 10.1042/bj3360471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morel AP, Sentis S, Bianchin C, Le Romancer M, Jonard L, Rostan MC, Rimokh R, Corbo L. BTG2 antiproliferative protein interacts with the human CCR4 complex existing in vivo in three cell-cycle-regulated forms. J Cell Sci. 2003;116(Pt 14):2929–2936. doi: 10.1242/jcs.00480. [DOI] [PubMed] [Google Scholar]
- 58.Ezzeddine N, Chang TC, Zhu W, Yamashita A, Chen CY, Zhong Z, Yamashita Y, Zheng D, Shyu AB. Human TOB, an antiproliferative transcription factor, is a poly(A)-binding protein-dependent positive regulator of cytoplasmic mRNA deadenylation. Mol Cell Biol. 2007;27(22):7791–7801. doi: 10.1128/MCB.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meijer HA, Kong YW, Lu WT, Wilczynska A, Spriggs RV, Robinson SW, Godfrey JD, Willis AE, Bushell M. Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science. 2013;340(6128):82–85. doi: 10.1126/science.1231197. [DOI] [PubMed] [Google Scholar]
- 60.Chen Y, Boland A, Kuzuoglu-Ozturk D, Bawankar P, Loh B, Chang CT, Weichenrieder O, Izaurralde E. A DDX6-CNOT1 complex and W-binding pockets in CNOT9 reveal direct links between miRNA target recognition and silencing. Mol Cell. 2014;54(5):737–750. doi: 10.1016/j.molcel.2014.03.034. [DOI] [PubMed] [Google Scholar]
- 61.Braun JE, Huntzinger E, Fauser M, Izaurralde E. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol Cell. 2011;44(1):120–133. doi: 10.1016/j.molcel.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 62.Chekulaeva M, Mathys H, Zipprich JT, Attig J, Colic M, Parker R, Filipowicz W. miRNA repression involves GW182-mediated recruitment of CCR4–NOT through conserved W-containing motifs. Nat Struct Mol Biol. 2011;18(11):1218–1226. doi: 10.1038/nsmb.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Collart MA, Panasenko OO. The Ccr4–not complex. Gene. 2012;492(1):42–53. doi: 10.1016/j.gene.2011.09.033. [DOI] [PubMed] [Google Scholar]
- 64.Shirai YT, Suzuki T, Morita M, Takahashi A, Yamamoto T. Multifunctional roles of the mammalian CCR4–NOT complex in physiological phenomena. Front Genet. 2014;5:286. doi: 10.3389/fgene.2014.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Suzuki A, Niimi Y, Saga Y. Interaction of NANOS2 and NANOS3 with different components of the CNOT complex may contribute to the functional differences in mouse male germ cells. Biol Open. 2014;3(12):1207–1216. doi: 10.1242/bio.20149308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Raisch T, Bhandari D, Sabath K, Helms S, Valkov E, Weichenrieder O, Izaurralde E. Distinct modes of recruitment of the CCR4–NOT complex by Drosophila and vertebrate Nanos. EMBO J. 2016;35(9):974–990. doi: 10.15252/embj.201593634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ginter-Matuszewska B, Kusz K, Spik A, Grzeszkowiak D, Rembiszewska A, Kupryjanczyk J, Jaruzelska J. NANOS1 and PUMILIO2 bind microRNA biogenesis factor GEMIN3, within chromatoid body in human germ cells. Histochem Cell Biol. 2011;136(3):279–287. doi: 10.1007/s00418-011-0842-y. [DOI] [PubMed] [Google Scholar]
- 68.Meister G, Buhler D, Laggerbauer B, Zobawa M, Lottspeich F, Fischer U. Characterization of a nuclear 20S complex containing the survival of motor neurons (SMN) protein and a specific subset of spliceosomal Sm proteins. Hum Mol Genet. 2000;9(13):1977–1986. doi: 10.1093/hmg/9.13.1977. [DOI] [PubMed] [Google Scholar]
- 69.Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16(6):720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nelson PT, Hatzigeorgiou AG, Mourelatos Z. miRNP:mRNA association in polyribosomes in a human neuronal cell line. RNA. 2004;10(3):387–394. doi: 10.1261/rna.5181104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lai F, King ML. Repressive translational control in germ cells. Mol Reprod Dev. 2013;80(8):665–676. doi: 10.1002/mrd.22161. [DOI] [PubMed] [Google Scholar]
- 72.Gavis ER, Lehmann R. Translational regulation of nanos by RNA localization. Nature. 1994;369(6478):315–318. doi: 10.1038/369315a0. [DOI] [PubMed] [Google Scholar]
- 73.Bergsten SE, Gavis ER. Role for mRNA localization in translational activation but not spatial restriction of nanos RNA. Development. 1999;126(4):659–669. doi: 10.1242/dev.126.4.659. [DOI] [PubMed] [Google Scholar]
- 74.Gavis ER, Curtis D, Lehmann R. Identification of cis-acting sequences that control nanos RNA localization. Dev Biol. 1996;176(1):36–50. doi: 10.1006/dbio.1996.9996. [DOI] [PubMed] [Google Scholar]
- 75.Ephrussi A, Lehmann R. Induction of germ cell formation by oskar. Nature. 1992;358(6385):387–392. doi: 10.1038/358387a0. [DOI] [PubMed] [Google Scholar]
- 76.Becalska AN, Kim YR, Belletier NG, Lerit DA, Sinsimer KS, Gavis ER. Aubergine is a component of a nanos mRNA localization complex. Dev Biol. 2011;349(1):46–52. doi: 10.1016/j.ydbio.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hulskamp M, Schroder C, Pfeifle C, Jackle H, Tautz D. Posterior segmentation of the Drosophila embryo in the absence of a maternal posterior organizer gene. Nature. 1989;338(6217):629–632. doi: 10.1038/338629a0. [DOI] [PubMed] [Google Scholar]
- 78.Wharton RP, Struhl G. RNA regulatory elements mediate control of Drosophila body pattern by the posterior morphogen nanos. Cell. 1991;67(5):955–967. doi: 10.1016/0092-8674(91)90368-9. [DOI] [PubMed] [Google Scholar]
- 79.Loedige I, Stotz M, Qamar S, Kramer K, Hennig J, Schubert T, Loffler P, Langst G, Merkl R, Urlaub H, Meister G. The NHL domain of BRAT is an RNA-binding domain that directly contacts the hunchback mRNA for regulation. Genes Dev. 2014;28(7):749–764. doi: 10.1101/gad.236513.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Loedige I, Jakob L, Treiber T, Ray D, Stotz M, Treiber N, Hennig J, Cook KB, Morris Q, Hughes TR, Engelmann JC, Krahn MP, Meister G. The crystal structure of the NHL domain in complex with RNA reveals the molecular basis of Drosophila Brain-Tumor-mediated gene regulation. Cell Rep. 2015;13(6):1206–1220. doi: 10.1016/j.celrep.2015.09.068. [DOI] [PubMed] [Google Scholar]
- 81.Laver JD, Li X, Ray D, Cook KB, Hahn NA, Nabeel-Shah S, Kekis M, Luo H, Marsolais AJ, Fung KYY, Hughes TR, Westwood JT, Sidhu SS, Morris Q, Lipshitz HD, Smibert CA. Brain tumor is a sequence-specific RNA-binding protein that directs maternal mRNA clearance during the Drosophila maternal-to-zygotic transition. Genome Biol. 2015;16:94. doi: 10.1186/s13059-015-0659-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wreden C, Verrotti AC, Schisa JA, Lieberfarb ME, Strickland S. Nanos and pumilio establish embryonic polarity in Drosophila by promoting posterior deadenylation of hunchback mRNA. Development. 1997;124(15):3015–3023. doi: 10.1242/dev.124.15.3015. [DOI] [PubMed] [Google Scholar]
- 83.Illmensee K, Mahowald AP. Transplantation of posterior polar plasm in Drosophila. Induction of germ cells at the anterior pole of the egg. Proc Natl Acad Sci USA. 1974;71(4):1016–1020. doi: 10.1073/pnas.71.4.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dalby B, Glover DM. Discrete sequence elements control posterior pole accumulation and translational repression of maternal cyclin B RNA in Drosophila. EMBO J. 1993;12(3):1219–1227. doi: 10.1002/j.1460-2075.1993.tb05763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Asaoka-Taguchi M, Yamada M, Nakamura A, Hanyu K, Kobayashi S. Maternal pumilio acts together with nanos in germline development in Drosophila embryos. Nat Cell Biol. 1999;1(7):431–437. doi: 10.1038/15666. [DOI] [PubMed] [Google Scholar]
- 86.Kadyrova LY, Habara Y, Lee TH, Wharton RP. Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development. 2007;134(8):1519–1527. doi: 10.1242/dev.002212. [DOI] [PubMed] [Google Scholar]
- 87.Hayashi Y, Hayashi M, Kobayashi S. Nanos suppresses somatic cell fate in Drosophila germ line. Proc Natl Acad Sci USA. 2004;101(28):10338–10342. doi: 10.1073/pnas.0401647101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sato K, Hayashi Y, Ninomiya Y, Shigenobu S, Arita K, Mukai M, Kobayashi S. Maternal Nanos represses hid/skl-dependent apoptosis to maintain the germ line in Drosophila embryos. Proc Natl Acad Sci USA. 2007;104(18):7455–7460. doi: 10.1073/pnas.0610052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ye B, Petritsch C, Clark IE, Gavis ER, Jan LY, Jan YN. Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr Biol. 2004;14(4):314–321. doi: 10.1016/j.cub.2004.01.052. [DOI] [PubMed] [Google Scholar]
- 90.Muraro NI, Weston AJ, Gerber AP, Luschnig S, Moffat KG, Baines RA. Pumilio binds para mRNA and requires Nanos and Brat to regulate sodium current in Drosophila motoneurons. J Neurosci. 2008;28(9):2099–2109. doi: 10.1523/JNEUROSCI.5092-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mee CJ, Pym EC, Moffat KG, Baines RA. Regulation of neuronal excitability through pumilio-dependent control of a sodium channel gene. J Neurosci. 2004;24(40):8695–8703. doi: 10.1523/JNEUROSCI.2282-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schweers BA, Walters KJ, Stern M. The Drosophila melanogaster translational repressor pumilio regulates neuronal excitability. Genetics. 2002;161(3):1177–1185. doi: 10.1093/genetics/161.3.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Neumuller RA, Betschinger J, Fischer A, Bushati N, Poernbacher I, Mechtler K, Cohen SM, Knoblich JA. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature. 2008;454(7201):241–245. doi: 10.1038/nature07014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Feber A, Clark J, Goodwin G, Dodson AR, Smith PH, Fletcher A, Edwards S, Flohr P, Falconer A, Roe T, Kovacs G, Dennis N, Fisher C, Wooster R, Huddart R, Foster CS, Cooper CS. Amplification and overexpression of E2F3 in human bladder cancer. Oncogene. 2004;23(8):1627–1630. doi: 10.1038/sj.onc.1207274. [DOI] [PubMed] [Google Scholar]
- 95.Foster CS, Falconer A, Dodson AR, Norman AR, Dennis N, Fletcher A, Southgate C, Dowe A, Dearnaley D, Jhavar S, Eeles R, Feber A, Cooper CS. Transcription factor E2F3 overexpressed in prostate cancer independently predicts clinical outcome. Oncogene. 2004;23(35):5871–5879. doi: 10.1038/sj.onc.1207800. [DOI] [PubMed] [Google Scholar]
- 96.Cooper CS, Nicholson AG, Foster C, Dodson A, Edwards S, Fletcher A, Roe T, Clark J, Joshi A, Norman A, Feber A, Lin D, Gao Y, Shipley J, Cheng SJ. Nuclear overexpression of the E2F3 transcription factor in human lung cancer. Lung Cancer. 2006;54(2):155–162. doi: 10.1016/j.lungcan.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 97.Bracken AP, Ciro M, Cocito A, Helin K. E2F target genes: unraveling the biology. Trends Biochem Sci. 2004;29(8):409–417. doi: 10.1016/j.tibs.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 98.Iaquinta PJ, Lees JA. Life and death decisions by the E2F transcription factors. Curr Opin Cell Biol. 2007;19(6):649–657. doi: 10.1016/j.ceb.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang C, Dickinson LK, Lehmann R. Genetics of nanos localization in Drosophila. Dev Dyn. 1994;199(2):103–115. doi: 10.1002/aja.1001990204. [DOI] [PubMed] [Google Scholar]
- 100.Lehmann R, Nusslein-Volhard C. The maternal gene nanos has a central role in posterior pattern formation of the Drosophila embryo. Development. 1991;112(3):679–691. doi: 10.1242/dev.112.3.679. [DOI] [PubMed] [Google Scholar]
- 101.Subramaniam K, Seydoux G. nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development. 1999;126(21):4861–4871. doi: 10.1242/dev.126.21.4861. [DOI] [PubMed] [Google Scholar]
- 102.Koprunner M, Thisse C, Thisse B, Raz E. A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev. 2001;15(21):2877–2885. doi: 10.1101/gad.212401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Verrotti AC, Wharton RP. Nanos interacts with cup in the female germline of Drosophila. Development. 2000;127(23):5225–5232. doi: 10.1242/dev.127.23.5225. [DOI] [PubMed] [Google Scholar]
- 104.Keyes LN, Spradling AC. The Drosophila gene fs(2)cup interacts with otu to define a cytoplasmic pathway required for the structure and function of germ-line chromosomes. Development. 1997;124(7):1419–1431. doi: 10.1242/dev.124.7.1419. [DOI] [PubMed] [Google Scholar]
- 105.Wilhelm JE, Hilton M, Amos Q, Henzel WJ. Cup is an eIF4E binding protein required for both the translational repression of oskar and the recruitment of Barentsz. J Cell Biol. 2003;163(6):1197–1204. doi: 10.1083/jcb.200309088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nakamura A, Sato K, Hanyu-Nakamura K. Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev Cell. 2004;6(1):69–78. doi: 10.1016/S1534-5807(03)00400-3. [DOI] [PubMed] [Google Scholar]
- 107.Haraguchi S, Tsuda M, Kitajima S, Sasaoka Y, Nomura-Kitabayashid A, Kurokawa K, Saga Y. nanos1: a mouse nanos gene expressed in the central nervous system is dispensable for normal development. Mech Dev. 2003;120(6):721–731. doi: 10.1016/S0925-4773(03)00043-1. [DOI] [PubMed] [Google Scholar]
- 108.Suzuki A, Tsuda M, Saga Y. Functional redundancy among Nanos proteins and a distinct role of Nanos2 during male germ cell development. Development. 2007;134(1):77–83. doi: 10.1242/dev.02697. [DOI] [PubMed] [Google Scholar]
- 109.Zhou Z, Shirakawa T, Ohbo K, Sada A, Wu Q, Hasegawa K, Saba R, Saga Y. RNA binding protein Nanos2 organizes post-transcriptional buffering system to retain primitive state of mouse spermatogonial stem cells. Dev Cell. 2015;34(1):96–107. doi: 10.1016/j.devcel.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 110.Liu J, Rivas FV, Wohlschlegel J, Yates JR, 3rd, Parker R, Hannon GJ. A role for the P-body component GW182 in microRNA function. Nat Cell Biol. 2005;7(12):1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Macfarlane LA, Murphy PR. MicroRNA: biogenesis, function and role in cancer. Curr Genom. 2010;11(7):537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kusz KM, Tomczyk L, Sajek M, Spik A, Latos-Bielenska A, Jedrzejczak P, Pawelczyk L, Jaruzelska J. The highly conserved NANOS2 protein: testis-specific expression and significance for the human male reproduction. Mol Hum Reprod. 2009;15(3):165–171. doi: 10.1093/molehr/gap003. [DOI] [PubMed] [Google Scholar]
- 113.Kusz K, Tomczyk L, Spik A, Latos-Bielenska A, Jedrzejczak P, Pawelczyk L, Jaruzelska J. NANOS3 gene mutations in men with isolated sterility phenotype. Mol Reprod Dev. 2009;76(9):804. doi: 10.1002/mrd.21070. [DOI] [PubMed] [Google Scholar]
- 114.Wu X, Wang B, Dong Z, Zhou S, Liu Z, Shi G, Cao Y, Xu Y. A NANOS3 mutation linked to protein degradation causes premature ovarian insufficiency. Cell Death Dis. 2013;4:e825. doi: 10.1038/cddis.2013.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Santos MG, Machado AZ, Martins CN, Domenice S, Costa EM, Nishi MY, Ferraz-de-Souza B, Jorge SA, Pereira CA, Soardi FC, de Mello MP, Maciel-Guerra AT, Guerra-Junior G, Mendonca BB. Homozygous inactivating mutation in NANOS3 in two sisters with primary ovarian insufficiency. Biomed Res Int. 2014;2014:787465. doi: 10.1155/2014/787465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kusz-Zamelczyk K, Sajek M, Spik A, Glazar R, Jedrzejczak P, Latos-Bielenska A, Kotecki M, Pawelczyk L, Jaruzelska J. Mutations of NANOS1, a human homologue of the Drosophila morphogen, are associated with a lack of germ cells in testes or severe oligo-astheno-teratozoospermia. J Med Genet. 2013;50(3):187–193. doi: 10.1136/jmedgenet-2012-101230. [DOI] [PubMed] [Google Scholar]
- 117.Janic A, Mendizabal L, Llamazares S, Rossell D, Gonzalez C. Ectopic expression of germline genes drives malignant brain tumor growth in Drosophila. Science. 2010;330(6012):1824–1827. doi: 10.1126/science.1195481. [DOI] [PubMed] [Google Scholar]
- 118.Krentz AD, Murphy MW, Zhang T, Sarver AL, Jain S, Griswold MD, Bardwell VJ, Zarkower D. Interaction between DMRT1 function and genetic background modulates signaling and pluripotency to control tumor susceptibility in the fetal germ line. Dev Biol. 2013;377(1):67–78. doi: 10.1016/j.ydbio.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Krentz AD, Murphy MW, Kim S, Cook MS, Capel B, Zhu R, Matin A, Sarver AL, Parker KL, Griswold MD, Looijenga LH, Bardwell VJ, Zarkower D. The DM domain protein DMRT1 is a dose-sensitive regulator of fetal germ cell proliferation and pluripotency. Proc Natl Acad Sci USA. 2009;106(52):22323–22328. doi: 10.1073/pnas.0905431106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Grelet S (2014) Implication de Nanos-3 dans l’invasion tumorale broncho-pulmonaire, Ph.D. thesis (Dissertation). University of Reims Champagne-Ardenne
- 121.Di Fiore R, D’Anneo A, Tesoriere G, Vento R. RB1 in cancer: different mechanisms of RB1 inactivation and alterations of pRb pathway in tumorigenesis. J Cell Physiol. 2013;228(8):1676–1687. doi: 10.1002/jcp.24329. [DOI] [PubMed] [Google Scholar]
- 122.Sadasivam S, DeCaprio JA. The DREAM complex: master coordinator of cell cycle-dependent gene expression. Nat Rev Cancer. 2013;13(8):585–595. doi: 10.1038/nrc3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh34032/5/1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 125.Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23(1):127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 126.Bawankar P, Loh B, Wohlbold L, Schmidt S, Izaurralde E. NOT10 and C2orf29/NOT11 form a conserved module of the CCR4–NOT complex that docks onto the NOT1 N-terminal domain. RNA Biol. 2013;10(2):228–244. doi: 10.4161/rna.23018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Petit AP, Wohlbold L, Bawankar P, Huntzinger E, Schmidt S, Izaurralde E, Weichenrieder O. The structural basis for the interaction between the CAF1 nuclease and the NOT1 scaffold of the human CCR4–NOT deadenylase complex. Nucleic Acids Res. 2012;40(21):11058–11072. doi: 10.1093/nar/gks883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pilon M, Weisblat DA. A nanos homolog in leech. Development. 1997;124(9):1771–1780. doi: 10.1242/dev.124.9.1771. [DOI] [PubMed] [Google Scholar]
- 129.Mochizuki K, Sano H, Kobayashi S, Nishimiya-Fujisawa C, Fujisawa T. Expression and evolutionary conservation of nanos-related genes in Hydra. Dev Genes Evol. 2000;210(12):591–602. doi: 10.1007/s004270000105. [DOI] [PubMed] [Google Scholar]
- 130.Lall S, Ludwig MZ, Patel NH. Nanos plays a conserved role in axial patterning outside of the Diptera. Curr Biol. 2003;13(3):224–229. doi: 10.1016/S0960-9822(03)00045-9. [DOI] [PubMed] [Google Scholar]
- 131.Torras R, Yanze N, Schmid V, Gonzalez-Crespo S. nanos expression at the embryonic posterior pole and the medusa phase in the hydrozoan Podocoryne carnea. Evol Dev. 2004;6(5):362–371. doi: 10.1111/j.1525-142X.2004.04044.x. [DOI] [PubMed] [Google Scholar]
- 132.Torras R, Gonzalez-Crespo S. Posterior expression of nanos orthologs during embryonic and larval development of the anthozoan Nematostella vectensis. Int J Dev Biol. 2005;49(7):895–899. doi: 10.1387/ijdb.051980rt. [DOI] [PubMed] [Google Scholar]
- 133.Calvo E, Walter M, Adelman ZN, Jimenez A, Onal S, Marinotti O, James AA. Nanos (nos) genes of the vector mosquitoes, Anopheles gambiae, Anopheles stephensi and Aedes aegypti. Insect Biochem Mol Biol. 2005;35(7):789–798. doi: 10.1016/j.ibmb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 134.Dearden PK. Germ cell development in the Honeybee (Apis mellifera); vasa and nanos expression. BMC Dev Biol. 2006;6:6. doi: 10.1186/1471-213X-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nakao H, Matsumoto T, Oba Y, Niimi T, Yaginuma T. Germ cell specification and early embryonic patterning in Bombyx mori as revealed by nanos orthologues. Evol Dev. 2008;10(5):546–554. doi: 10.1111/j.1525-142X.2008.00270.x. [DOI] [PubMed] [Google Scholar]
- 136.Leininger S, Adamski M, Bergum B, Guder C, Liu J, Laplante M, Brate J, Hoffmann F, Fortunato S, Jordal S, Rapp HT, Adamska M. Developmental gene expression provides clues to relationships between sponge and eumetazoan body plans. Nat Commun. 2014;5:3905. doi: 10.1038/ncomms4905. [DOI] [PubMed] [Google Scholar]
- 137.Mosquera L, Forristall C, Zhou Y, King ML. A mRNA localized to the vegetal cortex of Xenopus oocytes encodes a protein with a nanos-like zinc finger domain. Development. 1993;117(1):377–386. doi: 10.1242/dev.117.1.377. [DOI] [PubMed] [Google Scholar]
- 138.Sekizaki H, Takahashi S, Tanegashima K, Onuma Y, Haramoto Y, Asashima M. Tracing of Xenopus tropicalis germ plasm and presumptive primordial germ cells with the Xenopus tropicalis DAZ-like gene. Dev Dyn. 2004;229(2):367–372. doi: 10.1002/dvdy.10448. [DOI] [PubMed] [Google Scholar]