Abstract
Trichomonas vaginalis is a common sexually transmitted parasite that colonizes the human urogenital tract, where it remains extracellular and adheres to epithelial cells. Infections range from asymptomatic to highly inflammatory, depending on the host and the parasite strain. Despite the serious consequences associated with trichomoniasis disease, little is known about parasite or host factors involved in attachment of the parasite-to-host epithelial cells. Here, we report the identification of microvesicle-like structures (MVs) released by T. vaginalis. MVs are considered universal transport vehicles for intercellular communication as they can incorporate peptides, proteins, lipids, miRNA, and mRNA, all of which can be transferred to target cells through receptor–ligand interactions, fusion with the cell membrane, and delivery of a functional cargo to the cytoplasm of the target cell. In the present study, we demonstrated that T. vaginalis release MVs from the plasma and the flagellar membranes of the parasite. We performed proteomic profiling of these structures demonstrating that they possess physical characteristics similar to mammalian extracellular vesicles and might be selectively charged with specific protein content. In addition, we demonstrated that viable T. vaginalis parasites release large vesicles (LVs), membrane structures larger than 1 µm that are able to interact with other parasites and with the host cell. Finally, we show that both populations of vesicles present on the surface of T vaginalis are induced in the presence of host cells, consistent with a role in modulating cell interactions.
Electronic supplementary material
The online version of this article (10.1007/s00018-017-2726-3) contains supplementary material, which is available to authorized users.
Keywords: Microvesicles, Large vesicles, Cellular communication, Parasite, Trichomonas vaginalis
Introduction
The flagellated protozoan parasite Trichomonas vaginalis is the etiologic agent of trichomoniasis, the most common non-viral sexually transmitted infection worldwide with an estimated 276 million new cases annually [1]. Although asymptomatic infection is common, multiple symptoms and pathologies can arise in both men and women, including vaginitis, urethritis, prostatitis, low birth weight infants and preterm delivery, premature rupture of membranes, and infertility [2, 3]. T. vaginalis has also emerged as an important cofactor in amplifying human immunodeficiency virus (HIV) spread as individuals infected with T. vaginalis have a significantly increased incidence of human immunodeficiency virus transmission [4, 5]. In addition, T. vaginalis infection increases the risk of cervical and aggressive prostate cancer [6–9].
As an extracellular pathogen residing in the urogenital tract, T. vaginalis adherence to epithelial cells is critical for host colonization and infection establishment [10]. Despite the serious consequences associated with trichomoniasis disease, little is known about parasite or host factors involved in attachment of the parasite-to-host epithelial cells. Recently, small vesicles called exosomes have been shown to mediate both host:parasite and parasite:parasite interactions and to play a role in the attachment of T. vaginalis to host epithelial cells [11]. Exosomes are a subset of a large and diverse group of extracellular vesicles (EVs): membrane enclosed particles released by virtually any eukaryotic and prokaryotic cell [12]. EVs constitute a heterogeneous family classified by the scientific community according to the mode of biogenesis, size, and function into three major categories: (1) exosomes formed due to plasma membrane invagination into multivesicular bodies (MVB) with size ranging from 40 to 100 nm; (2) Microvesicles (MVs), also called shedding vesicles, microparticles, or ectosomes, originating from the budding and extrusion of the plasma membrane, with sizes between 50 and 1000 nm and an asymmetric structure; and (3) apoptotic bodies, with greater sizes between 50 and 5000 nm originating from cells in the process of programed cell death [13]. The study of these structures holds great interest, mainly due to their associated functions. Interestingly, both exosomes and MVs are able to mediate horizontal transfer of different molecules, including membrane and soluble proteins, nucleic acids (miRNA, DNA, and mRNA), and lipids causing modifications in the phenotype of the target cells [14–17]. Among the EVs’ subtypes, the physicochemical properties and biological function of exosomes have been well-documented during the last years [12]. However, the role and properties of MVs have been hard to establish due to experimental difficulties to obtain enriched MVs’ fraction free of exosomes. Considering that the processes that drive exosomes and MVs’ biogenesis are completely different, it is logical to hypothesize that different vesicle populations might have different functions which merit further investigations.
Here, we report that T. vaginalis form MV-like structures derived from the plasma membrane as well as from the flagella of the parasite. We set up a filtration-based approach to enrich this population and separate them from smaller exosome-like vesicles. Interestingly, although vesicles with sizes larger than 1 µm are generally considered as apoptotic bodies, recent work have described a process whereby amoeboid migration of metastatic prostate cancer cells triggered production of gigantic EVs (> 1000 to > 10,000 nm) found to emanate from large protrusions of the cellular plasma membrane [18–20]. In the present study, we found that viable T. vaginalis also release membrane vesicles larger than 1 µm (LVs) that are able to interact with other parasites and with the host cell. Finally, here, we show that both MVs and LVs present on the surface of T. vaginalis are induced in the presence of host cells, suggesting that it might be important for the parasite pathogenesis.
Materials and methods
Parasites, cell cultures, and media
Trichomonas vaginalis strains B7RC2 (PA) and Jt wild type were cultured in TYM medium supplemented with 10% horse serum, 10 U/ml penicillin, and 10 µg/ml streptomycin (Invitrogen) [21]. 100 µg/mL G418 (Invitrogen) was added to culture of TSP8-HA (TVAG_008950), TSP3-HA (TVAG_280860), and TSP1-HA (TVAG_019180)-transfected parasites [22]. Parasites were grown at 37 °C and passaged daily. The human HeLa cells were grown in DMEM complemented with 10% bovine fetal serum, 10 U/ml penicillin, and 10 µg/ml streptomycin (Invitrogen) and cultured at 37 °C/5% CO2.
Treatment with CaCl2
Trichomonas vaginalis Jt was grown in Pyrex® culture tubes (O.D. × L: 16 mm × 150 mm), containing 20 mL of TYM medium with serum (initial inoculum: 1 × 105 parasites/mL), for 30 h at 37 °C, which corresponds to the logarithmic growth phase. Next, parasites were washed three times in PBS, pH 7.2, and incubated in serum-free TYM medium with 1 mM CaCl2 at 37 °C for 30 min. For the control experiments, parasites were incubated in the absence of CaCl2. The assays were analyzed using SEM as outlined below. The percentage of parasites that contain vesicles on the cell surface was determined from counts of at least 500 parasites randomly selected per well. The results are the average of three independent experiments performed in duplicate.
SEM
Parasites either alone or after incubation with CaCl2 or HeLa cells for 30 min were washed with PBS and fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.2. The cells were then post-fixed for 15 min in 1% OsO4, dehydrated in ethanol and critical point dried with liquid CO2. The dried cells were coated with gold–palladium to a thickness of 15 nm and then observed with a Jeol JSM-5600 scanning electron microscope, operating at 15 kV.
TEM
Parasites and MVs’ sample were washed with PBS and fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.2. The cells were then post-fixed for 30 min in 1% OsO4, dehydrated in acetone, and embedded in Polybed 812. Ultra-thin sections were harvested on 300-mesh copper grids, stained with 5% uranyl acetate and 1% lead citrate, and observed with an FEI Tecnai G2 Spirit transmission electron microscope, operating at 120 kV. The images were randomly acquired with a CCD camera system (MegaView G2, Olympus, Germany).
MVs’ sample was also fixed in 2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.2, adsorbed onto charged carbon-coated grids, contrasted with 1% uranyl acetate, and examined using a transmission electron microscope.
Isolation of T. vaginalis vesicles
B7RC2 parasites (106 cells/ml) were washed and incubated for 4 h at 37 °C in TYM medium without serum, with 1 mM CaCl2 to stimulate MVs’ release. Parasites were removed by centrifugation at 3000 rpm. Then, to isolate enriched MVs, the cell free media were first filtered through 0.8 µm filter to avoid cellular debris and second through 0.2 µm filter to eliminate exosomes contamination. Importantly, when LVs were analyzed, a 3 µm filter was included instead of 0.8 µm filter obtaining a mixture of MVs and LVs. Vesicles’ fraction retrievable from the top of the filter was washed with 50 ml cold PBS and re-filtered three times. Finally, the sample was pelleted by centrifugation at 100,000 x g for 90 min, and then, the pellet was re-suspended in 200 µl cold PBS + 1X HALT protease inhibitors (Thermo Scientific).
Laser-scattering analysis
Size distribution profile of MVs’ and exosomes’ samples was determined by dynamic light scattering based on laser diffraction method using Malvern Zetasizer (Version 6.0, Malvern, USA). The EVs’ diameter was determined by measuring the back scattering intensity (1758) at 173 °C. MVs’ samples were analyzed using Zetasizer previous proteomic mass spectrometry. Three experimental runs were averaged to determine the vesicle size.
Proteomic mass spectrometry analysis
MVs’ pellet was re-suspended in a minimal volume of digestion buffer (100 mM Tris–HCl, pH 8, 8 M urea). Re-suspended proteins were reduced and alkylated by the sequential addition of 5 mM tris(2-carboxyethyl)phosphine and 10 mM iodoacetamide as described previously [23]. The samples were then digested by Lys-C (Princeton Separations) and trypsin proteases (Promega) [23]. First, Lys-C protease [~ 1:50 (w/w) ratio of enzyme:substrate] was added to each sample and incubated for 4 h at 37 °C with gentle shaking. The digests were then diluted to 2 M urea by the addition of digestion buffer lacking urea, and trypsin was added to a final enzyme:substrate ratio of 1:20 (w/w) and incubated for 8 h at 37 °C with gentle shaking. Digests were stopped by the addition of formic acid to a final concentration of 5%. Supernatants were carefully removed from the resin and analyzed further by proteomics mass spectrometry.
Digested samples were then analyzed using a LC–MS/MS platform as described previously [24, 25]. Briefly, digested samples were loaded onto a fused silica capillary column with a 5-µm electrospray tip and packed in house with 18 cm of Luna C18 3 µM particles (Phenomenex). The column was then placed in line with a Q-exactive mass spectrometer (Thermo Fisher), and peptides were fractionated using a gradient of increasing acetonitrile. Peptides were eluted directly into the mass spectrometer, where MS/MS spectra were collected. The data-dependent spectral acquisition strategy consisted of a repeating cycle of one full MS spectrum (resolution = 70,000) followed by MS/MS of the 12 most intense precursor ions from the full MS scan (resolution = 17,500) [26]. Raw data analysis was performed using the IP2 suite of software tools (Integrated Proteomics Applications, San Diego, CA). Spectra were analyzed using the ProLuCID [27] algorithm and searching against a fasta protein database consisting of all predicted open reading frames downloaded from TrichDB on January 4, 2015 [28] concatenated to a decoy database in which the amino acid sequence of each entry was reversed. The following search parameters were used: (1) precursor ion tolerance was 20 ppm; (2) fragment ion tolerance was 20 ppm; (3) cysteine carbamidomethylation was considered as a static modification; (4) peptides must be fully tryptic; and (5) no consideration was made for missed cleavages. False positive rates for peptide identifications were estimated using a decoy database approach and then filtered using the DTASelect algorithm [29–31]. XCorr and ΔCn cutoffs were identified dynamically using a linear discriminant analysis [29]. Proteins identified by at least two fully tryptic unique peptides, each with a false positive rate of less than 5%, were considered to be present in the sample. Three different sets of samples were independently analyzed. Normalized spectral abundance factor (NSAF) values including shared peptides were calculated as described and multiplied by 105 to improve readability [23]. Proteins that could not be distinguished by available peptides in any given replicate were considered as a group. The numbers in the table refer to the different groups of proteins (see supplemental Table S1).
Bioinformatics analyses
Proteins present in the MVs’ fraction were identified using BLAST tool (Basic Local Alignment Search Tool) followed by GO term enrichment according to TrichDB database [28]. Venn Diagrams were done using RStudio Version 0.99.902 (www.R-project.org). Signal peptide and transmembrane domain were annotated according to TrichDB database [28].
Immunolocalization experiments
Parasites expressing the hemagglutinin-tag (HA) version of TvTSP8, TvTSP1, and TvTSP3 were incubated at 37 °C on glass coverslips for 4 h as previously described [32]. The parasites were then fixed and permeabilized in cold methanol for 10 min. Cells were then washed and blocked with 5% fetal bovine serum (FBS) in phosphate buffered saline (PBS) for 30 min, incubated with a 1:500 dilution of anti-HA primary antibody (Covance, Emeryville, CA, USA) diluted in PBS plus 2% FBS, washed with PBS, and then incubated with a 1:5000 dilution of Alexa Fluor-conjugated secondary antibody (Molecular Probes). The coverslips were mounted onto microscope slips using ProLong Gold antifade reagent with 4′,6′-diamidino-2- phenylindole (Invitrogen). All observations were performed on a Nikon E600 epifluorescence microscope. Adobe Photoshop (Adobe Systems) and Fiji software [33] were used for image processing.
Parasite viability assays
Trichomonas vaginalis MVs display phosphatidyl-serine in their membrane detected with annexin V-FITC (data not shown). Hence, viability of parasites was analyzed using the fluorescent exclusion dye propidium iodide. For this purpose, wild-type B7RC2 (control) as well as B7RC2-transfected parasites with TvTSP8, TvTSP1, TvTSP3, and EpNEO (empty vector) were labeled with propidium iodide (PI, 20 μg/ml) at 4 °C for 10 min. PI fluorescence associated with non-viable cells was measured by flow cytometry (at λ = 544/602 nm) on FACSCalibur (Becton–Dickinson) and analyzed using Flowing software version 2.4.1 (Perttu Terho, Turku Centre for Biotechnology, Finland; www.flowingsoftware.com). Three independent experiments were performed, with three technical replicates each time.
Vesicle interaction assays
Anti-HA label
To assess interaction of LVs with parasites, TvTSP8-HA-transfected parasites were washed and incubated in TYM media without serum for 4 h at 37 °C to allow vesicle release. Later, culture media were centrifuged to remove parasites and filtered using a membrane with nominal diameter pore of 3 µm. The conditioned media (enriched in vesicles) were then collected and incubated with unstained B7RC2 wild-type parasites during 4 h at 37 °C. Coverslips were fixed and permeabilized in cold methanol for 10 min, and an immunofluorescence assay was performed as described before using anti-HA primary antibody to detect surface TvTSP8 present in donor parasites’ LVs (Covance, Emeryville, CA, USA). To assess interaction of LVs with host cells, TSP1-HA parasites were incubated at 37 °C with HeLa cells for 0.5 h, followed by 3 × PBS washes to remove unbound parasites. Cells were then fixed with paraformaldehyde for 20 min and permeabilized with Triton 0.2% for 10 min followed by immunofluorescence detection using an anti-HA tag antibody as previously described.
CMTPX label
TvTSP8-HA-transfected parasites were stained with Cell Tracker Red (CMTPX) (Thermo Fisher Scientific). After three washes to remove unbound dye, cells were incubated for 4 h at 37 °C in TYM media without serum to allow the release of vesicles. Culture media were centrifuged to remove parasites and filtered using a membrane with nominal diameter pore of 3 µm. Conditioned media were collected then and further incubated with unstained B7RC2 wild-type parasites during 4 h at 37 °C. Coverslips were then fixed and permeabilized in cold methanol for 10 min, mounted onto microscope slips using ProLong Gold antifade reagent with 4′,6′-diamidino-2-phenylindole (Invitrogen), and immediately observed on a Nikon E600 epifluorescence microscope. Adobe Photoshop (Adobe Systems) and Fiji software [33] were used for image processing.
PKH67 label: Vesicles (a mixture of MVs and LVs) released from B7RC2 parasites transfected with Empty Plasmid (EpNEO) were stained with lipophilic dye PKH-67 (Sigma Aldrich). To this end, vesicles isolated from 109 parasites were incubated with 4 µl of DilC for 4 min. The labeling was stopped adding 2 ml of PBS with 0.5% BSA. The sample was further washed thoroughly with PBS using a Vivaflow 200 100,000 MWCO PES (Sartorious Stedium) to eliminate unbound dye and pelleted by ultracentrifugation at 100,000 x g for 75 min. The pellet was then re-suspended in 100 µl of PBS and the protein concentration was estimated using Bradford reagent (Sigma Aldrich). Next, we assessed the interaction of labeled vesicles with different T. vaginalis parasites strains (B7RC2, CDC1132, G3, RU384, and SD7) by microscopy and flow cytometry. To this end, 15 µg of labeled vesicles were incubated with 105 parasites during 16 h. Importantly, the same volume of supernatant from the ultracentrifugation step was included as control to detect unspecific binding of free dye that could be present in the sample. Interaction of vesicles with parasites was observed by fluorescence microscopy and quantified using flow cytometry. To this end, part of the sample was incubated during 4 h at 37 °C to allow the parasite adhesion to microscope coverslips, fixed with 4% PFA for 15 min, and mounted onto microscope slips using ProLong Gold antifade reagent with 4′,6′-diamidino-2-phenylindole (Invitrogen) and observed on a Nikon E600 epifluorescence microscope. Adobe Photoshop (Adobe Systems) and Fiji software [33] were used for image processing. The remaining sample was fixed with PFA 4% for 15 min and analyzed by flow cytometry. Fluorescent parasites due to PKH67-labeled vesicle interaction were measured (at λ = 490/502 nm) on FACSCalibur (Becton–Dickinson) and flow cytometry data were analyzed using Flowing software version 2.4.1 (Perttu Terho, Turku Centre for Biotechnology, Finland; http://www.flowingsoftware.com). Data are expressed as percentage of fluorescent parasites among the total population of parasites analyzed (~ 5000 events per sample). Two independent experiments per triplicate were performed.
Parasite–host cell interaction
HeLa cells were seeded onto 24-well tissue culture plates in DMEM medium and allowed to form a confluent monolayer (1 × 106 cells) at 37 °C in 5% CO2. Subsequently, the HeLa cultures were washed three times with warm Hanks’ BSS and co-incubated with T. vaginalis Jt at cell ratios of 1:1 or 5:1 parasite:HeLa in DMEM without serum at 37 °C in 5% CO2 for 30 min. Prior to the co-incubation, parasites were washed three times in PBS, pH 7.2, and incubated to DMEM without serum at 37 °C for 15 min. Adhesion was confirmed by thorough washing of the cultured monolayers after their exposure to parasites. For the control experiments, parasites in the absence of host cells were analyzed. The interactions were analyzed using SEM. The percentage of parasites that contain vesicles on the cell surface was determined from counts of at least 500 parasites randomly selected per well. The results are the average of three independent experiments performed in duplicate.
Graphics and statistical analyses
Specific statistical considerations and the tests used are described separately for each subsection. GraphPad Prism for Windows version 7.00 and RStudio Version 0.99.902 (http://www.R-project.org) were used for graphics. Data are given as mean ± standard deviation (SD). Significance was established at P < 0.05.
Results
T. vaginalis release microvesicle-like structures
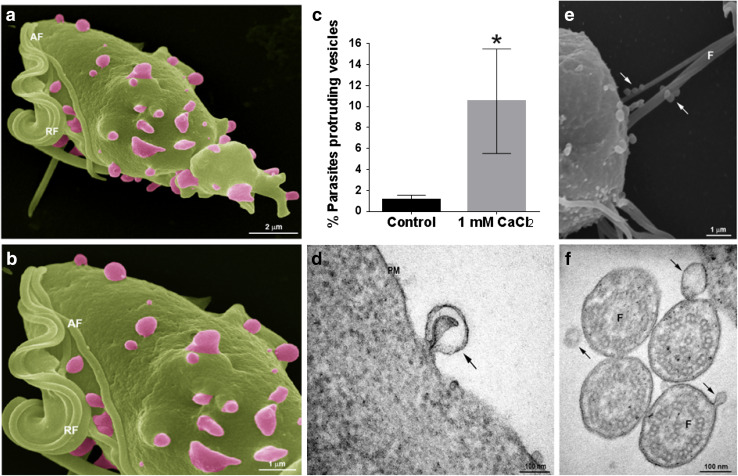
To evaluate if T. vaginalis form MVs under regular growth conditions, wild-type parasites were analyzed using SEM. Interestingly, we observed structures decorating the cell surface of 1% of examined parasites with diameters ranging from 100 to 1000 nm and different shapes (Fig. 1a, b); reminiscent to mammalian MVs [14–17]. According to the mechanism of origin, MVs are shed from the plasma membrane either at basal levels, as we observed here, or upon extracellular stimulation and concomitant elevation of intracellular calcium [34]. To evaluate if T. vaginalis share this mechanism of MVs’ formation, we incubated parasites with 1 mM CaCl2 for 30 min and examined the presence of shedding vesicles on their surface using scanning electron microscopy (SEM). Interestingly, while these shedding vesicles were not frequently observed in parasites grown in regular media, a ninefold increase in the percentage of parasites that contain MVs on the cell surface was observed upon incubation with CaCl2 (Fig. 1c). Transmission electron microscopy (TEM) analysis also revealed the protrusion of lipid bilayer surrounded vesicles from the plasma membrane of the parasites (Fig. 1d). In addition, we also observed MVs being shed from the flagellar membrane of the parasite using both SEM and TEM (Fig. 1e, f).
Fig. 1.
T. vaginalis release microvesicle-like structures. a, b Representative micrographs of MVs protruding from the plasma membrane of the parasites under growth conditions obtained by SEM. c Quantification of the percentage of parasites that contain vesicles in the cell surface upon treatment with CaCl2, and control conditions. Three independent experiments in duplicate were performed and 500 parasites were randomly counted per sample. Data are expressed as percentage of parasites with vesicles in CaCl2 treated related to untreated control parasites ± the standard deviation (SD). Unpaired Student’s t test was performed. *p < 0.05. d Representative micrograph of a MV protruding from the plasma membrane of a parasite observed by TEM. e Representative micrograph of MVs shedding from the flagella of the parasite observed using SEM. f Representative micrograph of MVs shedding from the flagella of the parasite observed using TEM. PM: plasma membrane; F: flagella. Arrows indicate MVs
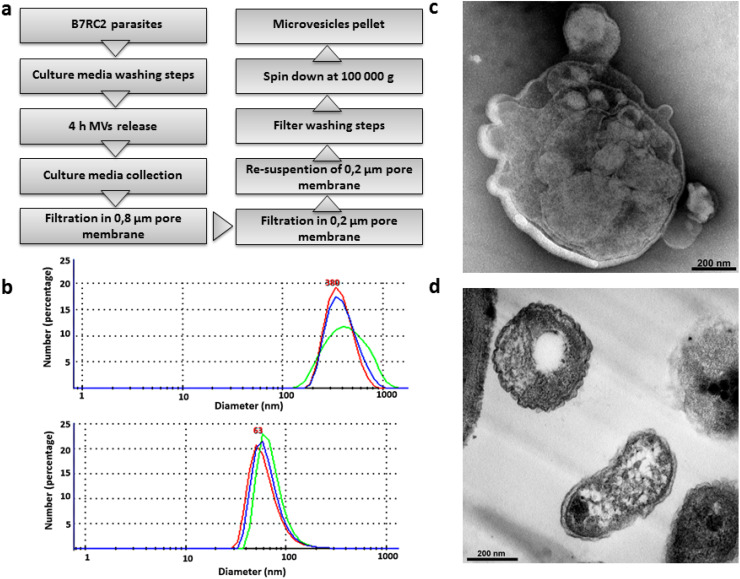
We next evaluated whether the MVs are being released by T. vaginalis. To this end, a modified version of the exosomes isolation protocol [11] was applied to isolate a population of vesicles with sizes greater than 100 nm and smaller than 1 µm (Fig. 2a). Parasites were incubated for 4 h in serum-free media to allow EVs’ release. After incubation, the samples were centrifuged to remove parasites and the supernatant was filtered through a membrane with nominal diameter pore of 800 nm to avoid cell debris contamination. Then, the sample was filtered in another 200 nm nominal diameter pore to avoid exosomes’ contamination (Fig. 2a). Particles with sizes between 800 and 200 nm, corresponding to the MVs’ fraction, were recovered from the top of the filter, washed to eliminate contamination with exosomes, and centrifuged at 100,000 g for 90 min to concentrate them (Fig. 2a). When the obtained sample was compared to a sample enriched in exosomes [11], size distribution profiling revealed a mean diameter size of 380 nm compared to the exosomes enriched sample which possess a mean diameter of 63 nm (Fig. 2b). In agreement with size profiling, examination of the preparations using TEM revealed irregular shape double membrane structures with sizes larger than exosomes (Fig. 2c, d), similar to what has been described for MVs in other cells [14–17].
Fig. 2.
Isolation of microvesicle-like structures. a Scheme of MVs’ isolation protocol. b Laser-scattering trace of isolated MVs. A mean diameter of 380 nm was measured in the MVs’ sample (top panel). In contrast, a mean diameter of 63 nm was measured in exosomes sample (bottom panel). Representative graph shows the size profile of one analyzed sample. c Negative stain TEM analysis of the enriched MVs’ sample shows vesicle within the range size of MVs. d Regular TEM analysis of the enriched MVs’ sample shows the presence of structures surrounded by membrane with different shapes and sizes ranging from 100 nm up to 1 µm
Proteomic analysis indicates that T. vaginalis MVs may be selectively charged with specific content
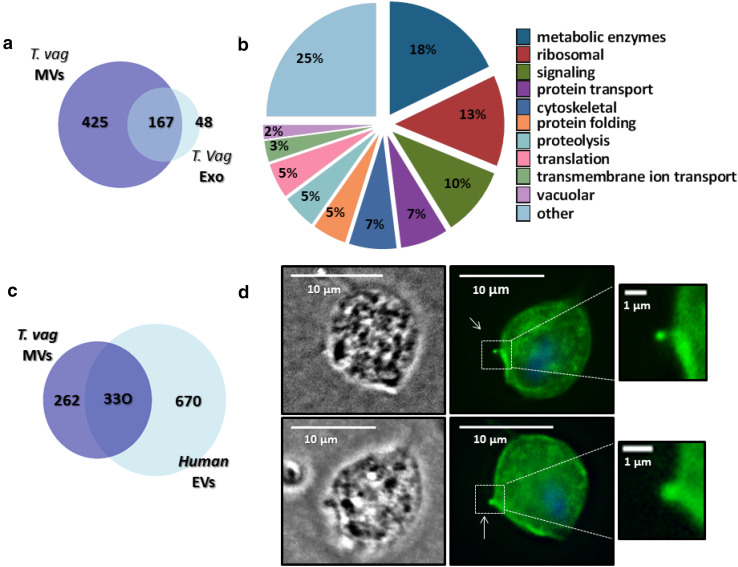
To identify the proteins present in the MV-enriched fractions, protein mass spectrometry was performed. Proteins with two or more identified peptides that were found in at least two of three mass spectrometry analyses were included in the MVs’ proteome and revealed a total of 592 proteins (Table S1). Of the 592 proteins identified, 84% are proteins with predicted domains, whereas the remaining 16% are hypothetical proteins according to BLAST analysis. As annotated in TrichDB database, 24% of proteins possess predicted transmembrane domains, and 5% have predicted signal peptide (Table S1). However, analyses of known T. vaginalis proteins indicate that many may not have the conventional N-terminal signal peptides or transmembrane domains identifiable by bioinformatics analyses [35]. We further compared the obtained proteomic data with the previously published T. vaginalis exosomes proteome [11]. Interestingly, while 39% of MVs’ proteins are shared with exosome proteome, the remaining 61% was only found in the MVs’ sample (Fig. 3a), indicating that T. vaginalis MVs may be selectively charged with specific groups of proteins, thus exerting specific functions.
Fig. 3.
Microvesicles’ proteomic analysis. a Venn diagram depicting proteins shared among T. vaginalis exosomes and T. vaginalis MVs. The numbers in the diagrams indicate the quantity of proteins shared or not by the different vesicles populations. b MVs’ proteins were identified using BLAST analysis and sorted into functional groups using genome annotation. Most representative groups are shown. c Venn diagram depicting proteins shared among T. vaginalis MVs’ and EVs’ list of 1000 proteins with the highest identification counts in EVpedia. The numbers in the diagrams indicate the quantity of proteins shared or not. d The localization of TvTSP8 was analyzed by immunofluorescence using parasites expressing C-terminal HA-tagged versions of TvTSP8 protein and a mouse HA-tagged antibody. Two representative images of parasites releasing plasma membrane vesicles smaller than 1 µm (magnification panels) are shown. The nucleus (blue) was stained with 4′,6′-diamidino-2-phenylindole. Arrows indicate microvesicles protruding
Among the predicted proteins with identifiable domains that allow the assignment of a predicted function according to gene ontology enrichment (GO term), 18% are enzymes related to metabolism of T. vaginalis, 13% are ribosomal related, 10% are proteins involved in signaling, 7% are cytoskeletal, and other 7% are involved in protein transport. In less abundant proportions, we found proteins related to protein folding, proteolysis, translation, transmembrane ion transport, and other cellular activities (Fig. 3b). When compared with human-compiled extracellular vesicles’ (EVs’) list of 1000 proteins with the highest identification counts in EVpedia [16, 17, 36–38], we found that T. vaginalis MVs contained homologues of approximately 56% of human EVs’ proteins (Fig. 3c). Shared proteins represent molecules commonly identified by different studies thus considered as conserved among EVs of different origins. These include cytoskeletal proteins (actin, tubulin, villin, and plastin); metabolic enzymes (GAPDH, enolase, phosphoglycerate kinase, pyruvate kinase, and lactate dehydrogenase); ribosomal proteins; heat shock proteins; 14-3-3; vesicle trafficking-related proteins (RAB proteins); and tetraspanin protein (Table S1). Importantly, ARF proteins, reported to be key players in formation, shedding [39] and cargo selection [40] in the MVs as well as endoplasmin (GP96) reported to be present exclusively in vesicles larger than exosomes [41] were also identified in our proteome.
Proteins that may have a role in T. vaginalis pathogenesis are also present in the MVs’ proteome. Two TvBspA-like proteins (TVAG_240680 and TVAG_162010) that were previously identified in the proteomic survey of surface proteins of T. vaginalis [32] are also present in the MVs’ proteome. Particularly, TVAG_240680 corresponds to the gene with the highest number of ESTs among the 11 genes found in the surface proteomic survey and has also been found in the exosome proteome [11, 32]. Considering its abundance and location, these data may be indicating that this protein may play a key role in the parasite pathogenesis. Similarly, homologues of virulence proteins characterized in Leishmania GP63-like metalloproteases (TVAG_293080 and TVAG_000880) and a tetraspanin protein (TvTSP8 and TVAG_008950) were also found in the MVs’ proteome. TvTSP8 is a transmembrane protein localized in the surface and intracellular vesicles that might have a role in parasites’ aggregation [42]. To validate the presence of this molecule in MVs, we performed an immunofluorescence using TvTSP8-HA-transfected and anti-HA antibody. Further validating our proteomic data TvTSP8 accumulates in structures smaller than 1 µm protruding from the plasma membrane of the parasite (Fig. 3d).
T. vaginalis releases vesicles larger than MVs
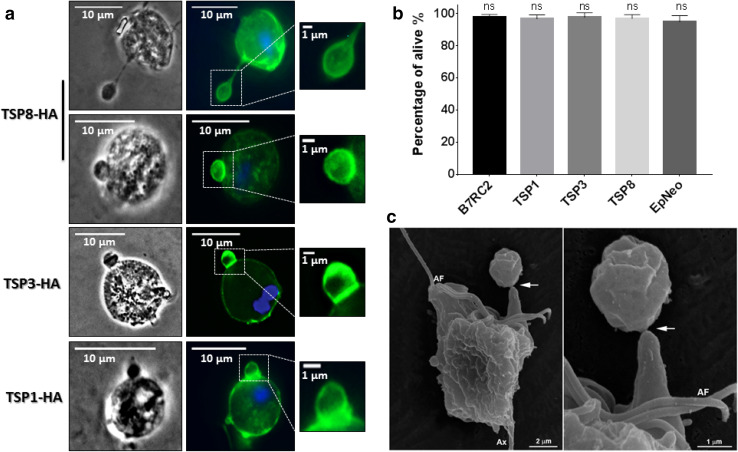
Vesicles with sizes larger than 1 µm are commonly considered as apoptotic bodies, namely, fragments of death cells [13, 43]. Interestingly, during the immunofluorescence experiments, we noted the presence of TvTSP8 in vesicles larger than 1 µm shedding from the plasma membrane of healthy TvTSP8-HA-transfected parasites (Fig. 4a). To evaluate if this is a more general observation, we decided to analyze the presence of tetraspanin proteins in larger vesicles (LVs) using parasites transfected with the surface-localized TvTSP3 and the surface and intracellular vesicle-localized TvTSP1 [42]. As can be observed in Fig. 4a, LVs shedding from the plasma membrane of TvTSP3- and TvTSP1-transfected parasites were observed. To rule out the possibility that cell death could be triggering the release of the LVs, we analyzed the viability of transfected parasites compared with non-transfected (wild-type) B7RC2 T. vaginalis strain, using propidium iodide. As observed in Fig. 4b, transfected parasites (EpNeo, TvTSP1, TvTSP3, and TvTSP8) as well as wild-type B7RC2 parasites have a viability percentage greater than 95% and no significant differences in the percentage of viability were found among them. In agreement, vesicles larger than 1 µm protruding from the plasma membrane of wild-type healthy parasites were observed using SEM (Fig. 4c), indicating that the structures observed are not an artefact of transfection and suggesting that shed LVs might normally exert an unknown function in T. vaginalis biology.
Fig. 4.
T. vaginalis release vesicles larger than regular MVs. a Cells expressing C-terminal HA-tagged versions of TvTSP8, TvTSP3, and TvTSP1 protein were stained for immunofluorescence microscopy using a mouse anti-HA antibody. The nucleus (blue) was also stained with 4´,6´-diamidino-2-phenylindole. Representative images of each transfected parasites releasing plasma membrane vesicles larger than 1 µm (LVs) (magnification panels) are shown. b Viability of EpNeo (empty vector), TvTSP1-HA, TvTSP3-HA, and TvTSP8-HA-transfected parasites as well as wild-type B7RC2 parasites was analyzed by flow cytometry using propidium iodide. Data are expressed as mean values ± standard deviation (SD) of three independent experiments with three replicates. ANOVA followed by Dunnet’s post hoc test was used to determine significant differences. ns: non-significant difference. c Representative SEM micrographs of wild-type Jt parasites also evidence the release of LVs by T. vaginalis. Arrow indicates an LV being release. AF: anterior flagella; Ax: axostyle
T. vaginalis LVs interact with parasites and host cells
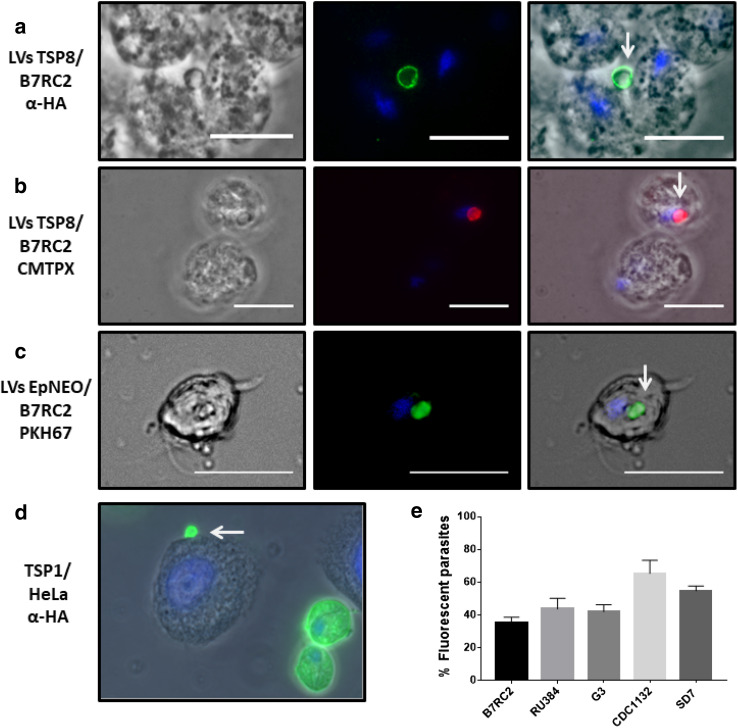
To assess a possible role of T. vaginalis LVs in cell:cell interaction, we first evaluated the capacity of LVs to interact with other parasites. To this end, LV-enriched sample was obtained from TvTSP8-HA-transfected parasites by filtering the released fraction through a membrane with nominal diameter pore of 3 µm. Then, conditioned media were incubated with wild-type B7RC2 parasites. After 4 h of incubation, an immunofluorescence assay using anti-HA antibody was performed. Interestingly, TvTSP8 positive LVs in contact with wild-type parasites were observed (Fig. 5a). As an alternative approach to validate the interaction, TvTSP8-HA-transfected parasites were labeled with Cell Tracker Red (CMTPX) and incubated to let the vesicles be released. Then, conditioned media were filtered through a membrane with nominal diameter pore of 3 µm and incubated with non-labeled B7RC2 wild-type parasites. As previously, LVs were also observed in contact with non-labeled parasites (Fig. 5b). To demonstrate that the obtained results are not dependent of the presence of the protein tag, the interaction was also evaluated with B7RC2 parasites transfected with an empty plasmid (EpNEO, which do not express any tag). Importantly, the results showed that binding was not affected by the HA tag (Fig. 5c).
Fig. 5.
T. vaginalis LVs interact with other parasites and with host cell. a Conditioned media containing released vesicles from parasites transfected with TvTSP8-HA obtained using a filter with a 3 µm pore were incubated with wild-type B2RC2 parasites for 4 h. Then, an immunofluorescence assay was performed using an anti-HA antibody. The nucleus (blue) was also stained with 4′,6′-diamidino-2-phenylindole. b Parasites transfected with TvTSP8-HA were stained with Cell Tracker Red (CMTPX) and incubated for 4 h to allow vesicles’ release. After incubation, conditioned media were obtained using a filter with a 3 µm pore. Following, non-labeled wild-type B7RC2 parasites were incubated with the conditioned media and analyzed using fluorescence microscopy. The nucleus (blue) was stained with 4′,6′-diamidino-2-phenylindole. Note CMTPX labeled LVs (released by donor) interacting with non-stained wild-type parasites. c Released vesicles from empty plasmid (EpNEO)-transfected parasites were stained with PKH67 lipophilic dye (green) and incubated with B7RC2 wild-type parasites (non-labeled) for 4 h. Following, parasites were analyzed using fluorescence microscopy. The nucleus (blue) was stained with 4′,6′-diamidino-2-phenylindole. Scale bars: 10 µm. d T. vaginalis parasites transfected with TvTSP1-HA membrane tetraspanin family protein were incubated with HeLa cells for 1 h. Then, an immunofluorescence using anti-HA was performed. The nucleus (blue) was stained with 4′,6′-diamidino-2-phenylindole. Arrows indicate LVs. e Released vesicles (MVs and LVs) isolated from T. vaginalis B7RC2 stained with PKH67 lipophilic dye (green) were incubated with different parasite strains (B7RC2, RU384, G3, CDC1132, and SD7) for 16 h. Then, quantification of vesicle binding (fluorescent parasites) was performed by flow cytometry
Vesicles can potentially interact with other parasites in the population as well as host cells. Hence, to evaluate the capacity of LVs to interact with host cells, TvTSP1-HA-transfected parasites were incubated with HeLa cells and vesicle binding was evaluated using an immunofluorescence assays with an anti-HA antibody (Fig. 5d). As can be observed in Fig. 5d, we also found LVs in contact with host cell. In summary, these results are suggesting that LVs might have a role in cell:cell interaction.
T. vaginalis released vesicles are involved in cell interaction
To further evaluate if T. vaginalis released vesicles have a role in parasite:parasite interaction, PKH67-labeled vesicles (MVs and LVs) isolated from B7RC2 parasites were incubated with different wild-type T. vaginalis strains. Then, the amount of fluorescent parasites was quantified by flow cytometry to evaluate the binding capacity of isolated vesicles. As can be observed in Fig. 5e, released vesicles are able to interact with great capacity with all the T. vaginalis strains analyzed, independent of the level of adherence of the recipient strain. These results could be indicating that vesicles from T. vaginalis might have a role mediating communication between parasites. Further studies are needed to determine the nature of this interaction.
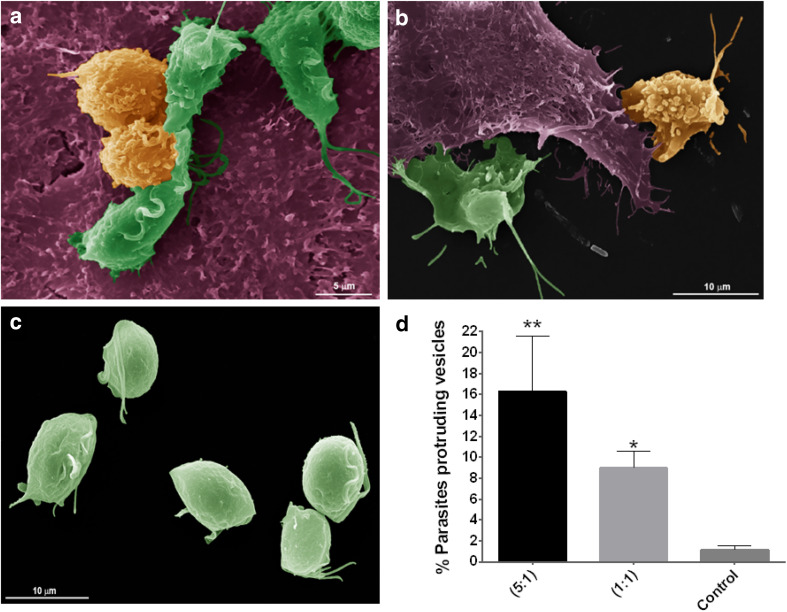
Then, to evaluate if T. vaginalis shedding vesicles have a role in host cell interaction, parasites were incubated with HeLa cells and the number of parasites that contain protruding vesicles on their cell surface was quantified using SEM (Fig. 6). Two different ratios of parasites:HeLa cells were used and parasites alone were used as control. Incubation of parasites with HeLa at 1:1 ratio resulted in a eight-fold increase in parasites protruding vesicles with different sizes (mix MVs and LVs) from their surface compared to parasites alone (Fig. 6b, c, d) showing that vesicles are being formed in response to host cell exposure. Interestingly, when we increased the ratio of parasites:HeLa cells to 5:1, an additive effect is shown as a 14-fold increase in parasites presenting protruding vesicles was observed (Fig. 6a, c, d). In concordance with our previous observation (Fig. 5), these results might be indicating that both LVs and MVs are also being formed as a sensing response to the presence of other parasites. In summary, these results are suggesting that these structures might have an important role in T. vaginalis cell communication.
Fig. 6.
T. vaginalis shed vesicles of different sizes upon exposure to host cell. Parasites were incubated with HeLa cells and the presence of shedding vesicles on their surface was analyzed using SEM. Representative micrographs show wild-type parasites containing protruding microvesicles (yellow) and parasites without microvesicles on their surface (green) upon incubation with HeLa cells (purple) at a ratio of 5:1 (a); 1:1 (b); and control parasites alone (c). d Quantification of the percentage of parasites containing shedding vesicles on the cell surface under host cell exposure. Two different ratios of parasite:host cells were used. Three independent experiments in duplicate were performed and 500 parasites were randomly counted per sample. Data are expressed as percentage of parasites releasing vesicles related to untreated control parasites ± the standard deviation (SD). ANOVA followed by Dunnet’s post hoc test was used to determine significant differences. *p < 0.05; **p < 0.005
Discussion
The present study describes structures formed by the protrusion from the plasma membrane as well as from the flagella of T. vaginalis, with different sizes, ranging from about 100 nm to a few micrometers. We observed that vesicles are being formed in response to stimulation with physiological concentration of CaCl2 [44] as well as host cell stimulation. These vesicles appear to interact with other parasites and the host cell.
The release of membrane vesicles from cell surfaces has been reported in almost all types of cells. Interestingly, we observed MVs protruding from the plasma and flagellar membrane of T. vaginalis by SEM and TEM. The latter is consistent with the recent report that MVs are released from the cilium and flagella [45]. In Chlamydomonas, Wood et al. [46] showed that cilia released EVs that contain a specific complement of proteins, different from the proteins of the isolated flagellar membranes. Similarly, Long et al. [45] isolated flagellar ectosomes from Chlamydomonas and demonstrate that flagellar ectosomes have a unique protein composition, especially being enriched in endosomal sorting complex required for transport (ESCRT) and ubiquitinated proteins. Extracellular vesicles released from the cilia of sensory neurons of Caenorhabditis elegans have the capacity to modulate mating behaviors and play a role in cell communication [47]. Whether MVs released from the flagella of T. vaginalis possess a different content than surface MVs, and whether they may play a role in communication remains still to be elucidated.
We used a modified version of the previously described exosome isolation protocol [11] to enrich vesicles larger than 100 nm and smaller than 1000 nm. The isolated vesicles were profiled by laser scattering and TEM demonstrating that the protocol used is able to separate the MVs from the exosome population. However, the yield of MVs obtained is very low which make further analysis such as proteomic survey very challenging. Recently, Benedikter et al. [48] described a method based on size exclusion chromatography that tended to have a higher yield than ultracentrifugation-based methods. Hence, this new protocol might be used for further analysis when MVs’ yield needs to be higher. Despite the difficulties, we were able to profile the protein content of MVs using mass spectrometry. Interestingly, our proteomic data revealed that 167 out of 215 proteins of exosomes were found in the MVs’ proteomic survey. This is not surprising as in many proteomic analyses, the protein content of MVs and exosomes tends to overlap. This could be explained in three different ways: (1) although several washing steps were included during the MVs’ isolation process to eliminate exosome population, it is possible that minimum level of contamination in the sample may have remained as vesicles tend to aggregate; (2) the proteins found in T. vaginalis exosomes are generally present in MVs, at least at the moment of the purification; and (3) the number of proteins identified in both proteomes is not comparable. Hence, if more proteins would have been identified in exosomes, probably, a greater overlap would have been found. Nevertheless, we found 425 proteins that were only detected in the MVs’ population. Among the proteins identified, we found proteins that participate in the MVs’ formation process (ARF and VAMP) [40] and endoplasmin, previously reported to be only present in vesicles larger than exosomes [41]. These results set the stage for further studies aimed to identify a marker to distinguish exosomes from MVs in T. vaginalis to be able to elucidate if they play different or specific functions.
Surprisingly, we observed that T. vaginalis also releases vesicles larger than regular MVs (> 1 µm). Atypically large size vesicles have been previously described in several cancer cells and denominated large oncosomes (LO) due to its large size, been specifically released by tumor cells and oncogenic material content [49]. Interestingly, tumor cells invade by transitioning to an amoeboid phenotype [50] with elliptical and blebbing morphology [51]. Importantly, these non-apoptotic membrane blebs are dynamically extruded and retracted thus enabling gliding and directional propulsion [52]. Similarly, T. vaginalis undergoes a radical change in their cell morphology during host cell attachment [53]: the free-swimming flagellate cell flattens and spreads out over the host cell tissue to become an amoeba. Whether the LVs in T. vaginalis are involved in amoebic transformation and/or movement remains to be studied. Furthermore, we also observed T. vaginalis LVs interacting with the surface of other parasites as well as with host cell. Recently, Minciacchi et al. [54] demonstrated that LO can be internalized by stromal cells through phagocytosis-like mechanism and induces a specific “reprogramming” of the fibroblasts that results in their increased ability to stimulate tube formation and tumor growth. Within this context, it is of interest to study whether LVs released by T. vaginalis are involved in modulating cellular communication.
The mechanism of MVs’ formation involves intracellular calcium mobilization with calpain activity, scramblase activation, and flippase inhibition [34]. Our results show that increased calcium levels affect the formation of MVs in T. vaginalis, similar as described in the parasite Giardia intestinalis [55]. Importantly, we have observed that vesicles protruding from parasite membranes are formed as a response to the presence of host cells, indicating that vesicles might be involved in T. vaginalis pathogenesis. Many reports have involved EVs from other protozoan parasites in cell interaction [56]. MVs from Giardia intestinalis have been shown to increase attachment of trophozoites to host cells and to be captured by human immature dendritic cells [55]. EVs originated from budding of flagellar membrane of Trypanosoma brucei mediate the transfer of virulence factor and cause host erythrocyte remodeling inducing anemia [57]. In addition, EVs released from Trypanosoma cruzi increases tissue parasitism and inflammation by stimulation of IL-4 and IL-10 synthesis, playing a central role in the acute phase of Chagas’ disease [58]. Here, we observed that both protruding surface vesicles increased upon the presence of host cells, suggesting that it might be involved in the process of communication with other parasites and/or with host cells. Whether the protrusion of vesicles plays a role in the pathogenesis process or if it is a mode of cell communication are questions that need further investigations. In this sense, proteomic analysis of MVs’ content identified proteins that might be involved in the establishment of parasite infection. These include, proteins with similarity to BspA proteins, the largest gene family encoding putative extracellular proteins in this pathogen, known to mediate adherence to host cells of mucosal bacteria [59]; metalloproteinases (GP63) implicated in Leishmania virulence [60, 61]; and the tetraspanin-like family of protein 8, TvTSP8, reported to be involved in regulating clump formation [42, 62]. Several these proteins predicted to be involved in pathogenesis have also been identified in the exosome proteome [11]. Importantly, exosomes have been demonstrated to modulate host cell immune responses and to promote parasite: parasite communication and host cell colonization [11], suggesting that transport of molecules found in MVs and exosomes might be important to establish the infection.
Over the years, different populations of vesicles released by many cell types have been identified. As information continues to growing, consensus in the classification of extracellular vesicles is still confused. To date, the most predominant classification divides extracellular vesicles into three groups on the basis of size, mode of biogenesis, and functions: exosomes (40–100 nm) formed due to membrane invagination into MVB, MVs (50 nm–1 µm) formed due to budding and extrusion of the plasma membrane, and apoptotic bodies (up to 5 µm) coming from apoptotic cells [13]. T. vaginalis EVs differ greatly in size, ranging from structures as small as exosomes, and as large as apoptotic bodies. Recently, exosomes have been reported to be as large as 250 nm and markers thought to be specific of exosomes have been found in non-exosomal vesicles [41]. Considering the results presented here as well as recent reports [63], reappraisal of EV classification may be warranted.
The present study explores a new aspect of the biology of the protozoan parasite T. vaginalis. Although there are many challenges to deal with in the study of EVs, improving our knowledge of the mechanisms involved in the release, delivery, and uptake of EVs, as well as deciphering the messages, they deliver is essential for the develop of new diagnostic tools and treatments of parasitic diseases.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1 List of microvesicles’ proteins identified by mass spectrometry. ID: official gene symbol; NSAFe: normalized spectral abundance factor; # TM domain: number of transmembrane domains (XLSX 166 kb)
Acknowledgements
We thank our colleagues in the lab for helpful discussions. This research was supported with a Grant from the Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT) Grant BID PICT 2013–1184 (NdM), a collaborative Grant from Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) and Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) (NdM and MB) and a National Institute of Health Grant (NIH) AI103182 (PJJ). NdM and VMC are researchers from the National Council of Research (CONICET) and UNSAM. YRN is a PhD fellow from CONICET. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
- MVs
Microvesicles
- miRNA
Microribonucleic acid
- mRNA
Mature ribonucleic acid
- WHO
World health organization
- HIV
Human immunodeficiency virus
- EVs
Extracellular vesicles
- MVB
Multivesicular body
- DNA
Desoxyribonucleic acid
- TYM
Trypticase, yeast extract, maltose medium
- DMEM
Dulbecco’s modified eagle medium
- PBS
Phosphate-buffered saline
- SEM
Scanning electron microscopy
- TEM
Transmission electron microscopy
- NSAF
Normalized spectral abundance factor
- GO
Gene ontology
- BLAST
Basic local alignment sequence tool
- LVs
Large vesicles
- FBS
Fetal bovine serum
- HA
Hemagglutinin
- FITC
Fluorescein isothiocyanate
- PI
Propidium iodide
- CMTPX
Cell tracker red
- SD
Standard deviation
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- BspA
Basic surface-exposed protein
- ARF
ADP-ribosylation factor
- ESTs
Expressed sequence tags
- GP63
Glycoprotein 63
- VAMP
Vesicle-associated membrane proteins
- LO
Large oncosome
- PM
Plasma membrane
- F
Flagella
- AF
Anterior flagella
- Ax
Axostyle
- TMD
Transmembrane domain
- TrichDB
Trichomonas genomic resource
References
- 1.WHO . Baseline report on global sexually transmitted infection surveillance. Geneva: World Health Organization; 2012. [Google Scholar]
- 2.Fichorova RN. Impact of T. vaginalis infection on innate immune responses and reproductive outcome. J Reprod Immunol. 2009;83(1–2):185–189. doi: 10.1016/j.jri.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swygard H, Miller WC, Kaydos-Daniels SC, Cohen MS, Leone PA, Hobbs MM, Sena AC. Targeted screening for Trichomonas vaginalis with culture using a two-step method in women presenting for STD evaluation. Sex Transm Dis. 2004;31(11):659–664. doi: 10.1097/01.OLQ.0000143091.95094.73. [DOI] [PubMed] [Google Scholar]
- 4.McClelland RS, Sangare L, Hassan WM, Lavreys L, Mandaliya K, Kiarie J, Ndinya-Achola J, Jaoko W, Baeten JM. Infection with Trichomonas vaginalis increases the risk of HIV-1 acquisition. J Infect Dis. 2007;195(5):698–702. doi: 10.1086/511278. [DOI] [PubMed] [Google Scholar]
- 5.Van Der Pol B, Kwok C, Pierre-Louis B, Rinaldi A, Salata RA, Chen PL, van de Wijgert J, Mmiro F, Mugerwa R, Chipato T, Morrison CS. Trichomonas vaginalis infection and human immunodeficiency virus acquisition in African women. J Infect Dis. 2008;197(4):548–554. doi: 10.1086/526496. [DOI] [PubMed] [Google Scholar]
- 6.Gander S, Scholten V, Osswald I, Sutton M, van Wylick R. Cervical dysplasia and associated risk factors in a juvenile detainee population. J Pediatr Adolesc Gynecol. 2009;22(6):351–355. doi: 10.1016/j.jpag.2009.01.070. [DOI] [PubMed] [Google Scholar]
- 7.Stark JR, Judson G, Alderete JF, Mundodi V, Kucknoor AS, Giovannucci EL, Platz EA, Sutcliffe S, Fall K, Kurth T, Ma J, Stampfer MJ, Mucci LA. Prospective study of Trichomonas vaginalis infection and prostate cancer incidence and mortality: physicians’ health study. J Natl Cancer Inst. 2009;101(20):1406–1411. doi: 10.1093/jnci/djp306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sutcliffe S, Alderete JF, Till C, Goodman PJ, Hsing AW, Zenilman JM, De Marzo AM, Platz EA. Trichomonosis and subsequent risk of prostate cancer in the prostate cancer prevention trial. Int J Cancer. 2009;124(9):2082–2087. doi: 10.1002/ijc.24144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Twu O, Dessi D, Vu A, Mercer F, Stevens GC, de Miguel N, Rappelli P, Cocco AR, Clubb RT, Fiori PL, Johnson PJ. Trichomonas vaginalis homolog of macrophage migration inhibitory factor induces prostate cell growth, invasiveness, and inflammatory responses. Proc Natl Acad Sci USA. 2014;111(22):8179–8184. doi: 10.1073/pnas.1321884111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwebke JR, Burgess D. Trichomoniasis. Clinical microbiology reviews. 2004;17(4):794–803. doi: 10.1128/CMR.17.4.794-803.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Twu O, de Miguel N, Lustig G, Stevens GC, Vashisht AA, Wohlschlegel JA, Johnson PJ. Trichomonas vaginalis exosomes deliver cargo to host cells and mediate hostratioparasite interactions. PLoS Pathog. 2013;9(7):e1003482. doi: 10.1371/journal.ppat.1003482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thery C. Exosomes: secreted vesicles and intercellular communications. F1000 Biol Reports. 2011;3:15. doi: 10.3410/B3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalra H, Simpson RJ, Ji H, Aikawa E, Altevogt P, Askenase P, Bond VC, Borras FE, Breakefield X, Budnik V, Buzas E, Camussi G, Clayton A, Cocucci E, Falcon-Perez JM, Gabrielsson S, Gho YS, Gupta D, Harsha HC, Hendrix A, Hill AF, Inal JM, Jenster G, Kramer-Albers EM, Lim SK, Llorente A, Lotvall J, Marcilla A, Mincheva-Nilsson L, Nazarenko I, Nieuwland R, Nolte-’t Hoen EN, Pandey A, Patel T, Piper MG, Pluchino S, Prasad TS, Rajendran L, Raposo G, Record M, Reid GE, Sanchez-Madrid F, Schiffelers RM, Siljander P, Stensballe A, Stoorvogel W, Taylor D, Thery C, Valadi H, van Balkom BW, Vazquez J, Vidal M, Wauben MH, Yanez-Mo M, Zoeller M, Mathivanan S. Vesiclepedia: a compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012;10(12):e1001450. doi: 10.1371/journal.pbio.1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon YJ, Kim OY, Gho YS. Extracellular vesicles as emerging intercellular communicasomes. BMB Reports. 2014;47(10):531–539. doi: 10.5483/BMBRep.2014.47.10.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shifrin DA, Jr, Demory Beckler M, Coffey RJ, Tyska MJ. Extracellular vesicles: communication, coercion, and conditioning. Mol Biol Cell. 2013;24(9):1253–1259. doi: 10.1091/mbc.e12-08-0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi DS, Kim DK, Kim YK, Gho YS. Proteomics of extracellular vesicles: exosomes and ectosomes. Mass Spectrom Rev. 2015;34(4):474–490. doi: 10.1002/mas.21420. [DOI] [PubMed] [Google Scholar]
- 17.Choi DS, Kim DK, Kim YK, Gho YS. Proteomics, transcriptomics and lipidomics of exosomes and ectosomes. Proteomics. 2013;13(10–11):1554–1571. doi: 10.1002/pmic.201200329. [DOI] [PubMed] [Google Scholar]
- 18.Di Vizio D, Kim J, Hager MH, Morello M, Yang W, Lafargue CJ, True LD, Rubin MA, Adam RM, Beroukhim R, Demichelis F, Freeman MR. Oncosome formation in prostate cancer: association with a region of frequent chromosomal deletion in metastatic disease. Can Res. 2009;69(13):5601–5609. doi: 10.1158/0008-5472.CAN-08-3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Vizio D, Morello M, Dudley AC, Schow PW, Adam RM, Morley S, Mulholland D, Rotinen M, Hager MH, Insabato L, Moses MA, Demichelis F, Lisanti MP, Wu H, Klagsbrun M, Bhowmick NA, Rubin MA, D’Souza-Schorey C, Freeman MR. Large oncosomes in human prostate cancer tissues and in the circulation of mice with metastatic disease. Am J Pathol. 2012;181(5):1573–1584. doi: 10.1016/j.ajpath.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morello M, Minciacchi VR, de Candia P, Yang J, Posadas E, Kim H, Griffiths D, Bhowmick N, Chung LW, Gandellini P, Freeman MR, Demichelis F, Di Vizio D. Large oncosomes mediate intercellular transfer of functional microRNA. Cell Cycle. 2013;12(22):3526–3536. doi: 10.4161/cc.26539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clark CG, Diamond LS. Methods for cultivation of luminal parasitic protists of clinical importance. Clin Microbiol Rev. 2002;15(3):329–341. doi: 10.1128/CMR.15.3.329-341.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delgadillo MG, Liston DR, Niazi K, Johnson PJ. Transient and selectable transformation of the parasitic protist Trichomonas vaginalis . Proc Natl Acad Sci USA. 1997;94(9):4716–4720. doi: 10.1073/pnas.94.9.4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Florens L, Carozza MJ, Swanson SK, Fournier M, Coleman MK, Workman JL, Washburn MP. Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods. 2006;40(4):303–311. doi: 10.1016/j.ymeth.2006.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wohlschlegel JA. Identification of SUMO-conjugated proteins and their SUMO attachment sites using proteomic mass spectrometry. Methods Mol Biol. 2009;497:33–49. doi: 10.1007/978-1-59745-566-4_3. [DOI] [PubMed] [Google Scholar]
- 25.Kaiser P, Wohlschlegel J. Identification of ubiquitination sites and determination of ubiquitin-chain architectures by mass spectrometry. Methods Enzymol. 2005;399:266–277. doi: 10.1016/S0076-6879(05)99018-6. [DOI] [PubMed] [Google Scholar]
- 26.Kelstrup CD, Young C, Lavallee R, Nielsen ML, Olsen JV. Optimized fast and sensitive acquisition methods for shotgun proteomics on a quadrupole orbitrap mass spectrometer. J Proteom Res. 2012;11(6):3487–3497. doi: 10.1021/pr3000249. [DOI] [PubMed] [Google Scholar]
- 27.Xu T, Park SK, Venable JD, Wohlschlegel JA, Diedrich JK, Cociorva D, Lu B, Liao L, Hewel J, Han X, Wong CC, Fonslow B, Delahunty C, Gao Y, Shah H, Yates JR., 3rd ProLuCID: an improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J Proteom. 2015;129:16–24. doi: 10.1016/j.jprot.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aurrecoechea C, Brestelli J, Brunk BP, Carlton JM, Dommer J, Fischer S, Gajria B, Gao X, Gingle A, Grant G, Harb OS, Heiges M, Innamorato F, Iodice J, Kissinger JC, Kraemer E, Li W, Miller JA, Morrison HG, Nayak V, Pennington C, Pinney DF, Roos DS, Ross C, Stoeckert CJ, Jr, Sullivan S, Treatman C, Wang H. GiardiaDB and TrichDB: integrated genomic resources for the eukaryotic protist pathogens Giardia lamblia and Trichomonas vaginalis . Nucleic Acids Res. 2009;37 (Database issue):526–530. doi: 10.1093/nar/gkn631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cociorva D, D LT, Yates JR (2007) Validation of tandem mass spectrometry database search results using DTASelect. In: Baxevanis AD et al (eds) Current protocols in bioinformatics/editorial board, Chap 13, pp 13–14. 10.1002/0471250953.bi1304s16 [DOI] [PubMed]
- 30.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4(3):207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 31.Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteom Res. 2002;1(1):21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Miguel N, Lustig G, Twu O, Chattopadhyay A, Wohlschlegel JA, Johnson PJ. Proteome analysis of the surface of Trichomonas vaginalis reveals novel proteins and strain-dependent differential expression. Mol Cell Proteom. 2010;9(7):1554–1566. doi: 10.1074/mcp.M000022-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19(2):43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 35.Gardner WA, Jr, Culberson DE, Bennett BD. Trichomonas vaginalis in the prostate gland. Arch Pathol Lab Med. 1986;110(5):430–432. [PubMed] [Google Scholar]
- 36.Kim DK, Lee J, Kim SR, Choi DS, Yoon YJ, Kim JH, Go G, Nhung D, Hong K, Jang SC, Kim SH, Park KS, Kim OY, Park HT, Seo JH, Aikawa E, Baj-Krzyworzeka M, van Balkom BW, Belting M, Blanc L, Bond V, Bongiovanni A, Borras FE, Buee L, Buzas EI, Cheng L, Clayton A, Cocucci E, Dela Cruz CS, Desiderio DM, Di Vizio D, Ekstrom K, Falcon-Perez JM, Gardiner C, Giebel B, Greening DW, Gross JC, Gupta D, Hendrix A, Hill AF, Hill MM, Nolte-’t Hoen E, Hwang DW, Inal J, Jagannadham MV, Jayachandran M, Jee YK, Jorgensen M, Kim KP, Kim YK, Kislinger T, Lasser C, Lee DS, Lee H, van Leeuwen J, Lener T, Liu ML, Lotvall J, Marcilla A, Mathivanan S, Moller A, Morhayim J, Mullier F, Nazarenko I, Nieuwland R, Nunes DN, Pang K, Park J, Patel T, Pocsfalvi G, Del Portillo H, Putz U, Ramirez MI, Rodrigues ML, Roh TY, Royo F, Sahoo S, Schiffelers R, Sharma S, Siljander P, Simpson RJ, Soekmadji C, Stahl P, Stensballe A, Stepien E, Tahara H, Trummer A, Valadi H, Vella LJ, Wai SN, Witwer K, Yanez-Mo M, Youn H, Zeidler R, Gho YS. EVpedia: a community web portal for extracellular vesicles research. Bioinformatics. 2015;31(6):933–939. doi: 10.1093/bioinformatics/btu741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim DK, Kang B, Kim OY, Choi DS, Lee J, Kim SR, Go G, Yoon YJ, Kim JH, Jang SC, Park KS, Choi EJ, Kim KP, Desiderio DM, Kim YK, Lotvall J, Hwang D, Gho YS. EVpedia: an integrated database of high-throughput data for systemic analyses of extracellular vesicles. J Extracell Vesicles. 2013 doi: 10.3402/jev.v2i0.20384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim DK, Lee J, Simpson RJ, Lotvall J, Gho YS. EVpedia: a community web resource for prokaryotic and eukaryotic extracellular vesicles research. Semin Cell Dev Biol. 2015;40:4–7. doi: 10.1016/j.semcdb.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 39.Muralidharan-Chari V, Clancy J, Plou C, Romao M, Chavrier P, Raposo G, D’Souza-Schorey C. ARF6-regulated shedding of tumor cell-derived plasma membrane microvesicles. Curr Biol. 2009;19(22):1875–1885. doi: 10.1016/j.cub.2009.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.D’Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7(5):347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 41.Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M, Thery C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci USA. 2016;113(8):E968–E977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coceres VM, Alonso AM, Nievas YR, Midlej V, Frontera L, Benchimol M, Johnson PJ, de Miguel N. The C-terminal tail of tetraspanin proteins regulates their intracellular distribution in the parasite Trichomonas vaginalis . Cell Microbiol. 2015;17(8):1217–1229. doi: 10.1111/cmi.12431. [DOI] [PubMed] [Google Scholar]
- 43.Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35(4):495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wagner G, Levin RJ. Electrolytes in vaginal fluid during the menstrual cycle of coitally active and inactive women. J Reprod Fertil. 1980;60(1):17–27. doi: 10.1530/jrf.0.0600017. [DOI] [PubMed] [Google Scholar]
- 45.Long H, Zhang F, Xu N, Liu G, Diener DR, Rosenbaum JL, Huang K. Comparative analysis of ciliary membranes and ectosomes. Curr Biol. 2016;26(24):3327–3335. doi: 10.1016/j.cub.2016.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wood CR, Huang K, Diener DR, Rosenbaum JL. The cilium secretes bioactive ectosomes. Curr Biol. 2013;23(10):906–911. doi: 10.1016/j.cub.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J, Silva M, Haas LA, Morsci NS, Nguyen KC, Hall DH, Barr MM. C. elegans ciliated sensory neurons release extracellular vesicles that function in animal communication. Curr Biol. 2014;24(5):519–525. doi: 10.1016/j.cub.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benedikter BJ, Bouwman FG, Vajen T, Heinzmann ACA, Grauls G, Mariman EC, Wouters EFM, Savelkoul PH, Lopez-Iglesias C, Koenen RR, Rohde GGU, Stassen FRM. Ultrafiltration combined with size exclusion chromatography efficiently isolates extracellular vesicles from cell culture media for compositional and functional studies. Sci Reports. 2017;7(1):15297. doi: 10.1038/s41598-017-15717-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Minciacchi VR, Freeman MR, Di Vizio D. Extracellular vesicles in cancer: exosomes, microvesicles and the emerging role of large oncosomes. Semin Cell Dev Biol. 2015;40:41–51. doi: 10.1016/j.semcdb.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolf K, Mazo I, Leung H, Engelke K, von Andrian UH, Deryugina EI, Strongin AY, Brocker EB, Friedl P. Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J Cell Biol. 2003;160(2):267–277. doi: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oppel F, Muller N, Schackert G, Hendruschk S, Martin D, Geiger KD, Temme A. SOX2-RNAi attenuates S-phase entry and induces RhoA-dependent switch to protease-independent amoeboid migration in human glioma cells. Mol Cancer. 2011;10:137. doi: 10.1186/1476-4598-10-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fackler OT, Grosse R. Cell motility through plasma membrane blebbing. J Cell Biol. 2008;181(6):879–884. doi: 10.1083/jcb.200802081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lal K, Noel CJ, Field MC, Goulding D, Hirt RP. Dramatic reorganisation of Trichomonas endomembranes during amoebal transformation: a possible role for G-proteins. Mol Biochem Parasitol. 2006;148(1):99–102. doi: 10.1016/j.molbiopara.2006.02.022. [DOI] [PubMed] [Google Scholar]
- 54.Minciacchi VR, Spinelli C, Reis-Sobreiro M, Cavallini L, You S, Zandian M, Li X, Mishra R, Chiarugi P, Adam RM, Posadas EM, Viglietto G, Freeman MR, Cocucci E, Bhowmick NA, Di Vizio D. MYC mediates large oncosome-induced fibroblast reprogramming in prostate cancer. Can Res. 2017;77(9):2306–2317. doi: 10.1158/0008-5472.CAN-16-2942. [DOI] [PubMed] [Google Scholar]
- 55.Evans-Osses I, Mojoli A, Monguio-Tortajada M, Marcilla A, Aran V, Amorim M, Inal J, Borras FE, Ramirez MI. Microvesicles released from Giardia intestinalis disturb host-pathogen response in vitro. Eur J Cell Biol. 2017;96(2):131–142. doi: 10.1016/j.ejcb.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 56.Marti M, Johnson PJ. Emerging roles for extracellular vesicles in parasitic infections. Curr Opin Microbiol. 2016;32:66–70. doi: 10.1016/j.mib.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Szempruch AJ, Sykes SE, Kieft R, Dennison L, Becker AC, Gartrell A, Martin WJ, Nakayasu ES, Almeida IC, Hajduk SL, Harrington JM. Extracellular vesicles from Trypanosoma brucei mediate virulence factor transfer and cause host anemia. Cell. 2016;164(1–2):246–257. doi: 10.1016/j.cell.2015.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trocoli Torrecilhas AC, Tonelli RR, Pavanelli WR, da Silva JS, Schumacher RI, de Souza W, e Silva NC, de Almeida Abrahamsohn I, Colli W, Manso Alves MJ. Trypanosoma cruzi: parasite shed vesicles increase heart parasitism and generate an intense inflammatory response. Microbes Infect. 2009;11(1):29–39. doi: 10.1016/j.micinf.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 59.Noel CJ, Diaz N, Sicheritz-Ponten T, Safarikova L, Tachezy J, Tang P, Fiori PL, Hirt RP. Trichomonas vaginalis vast BspA-like gene family: evidence for functional diversity from structural organisation and transcriptomics. BMC Genom. 2010;11:99. doi: 10.1186/1471-2164-11-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hirt RP, Noel CJ, Sicheritz-Ponten T, Tachezy J, Fiori PL. Trichomonas vaginalis surface proteins: a view from the genome. Trends Parasitol. 2007;23(11):540–547. doi: 10.1016/j.pt.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 61.Yao C, Donelson JE, Wilson ME. The major surface protease (MSP or GP63) of Leishmania sp. biosynthesis, regulation of expression, and function. Mol Biochem Parasitol. 2003;132(1):1–16. doi: 10.1016/S0166-6851(03)00211-1. [DOI] [PubMed] [Google Scholar]
- 62.Lustig G, Ryan CM, Secor WE, Johnson PJ. Trichomonas vaginalis contact-dependent cytolysis of epithelial cells. Infect Immun. 2013;81(5):1411–1419. doi: 10.1128/IAI.01244-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eloise Pariset VA, Millet Arnaud. Extracellular vesicles: isolation methods. Advance Biosystems. 2017 doi: 10.1002/adbi.201700040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 List of microvesicles’ proteins identified by mass spectrometry. ID: official gene symbol; NSAFe: normalized spectral abundance factor; # TM domain: number of transmembrane domains (XLSX 166 kb)