Abstract
Axon degeneration is a pathophysiological process of axonal dying and breakdown, which is characterized by several morphological features including the accumulation of axoplasmic organelles, disassembly of microtubules, and fragmentation of the axonal cytoskeleton. Autophagy, a highly conserved lysosomal-degradation machinery responsible for the control of cellular protein quality, is widely believed to be essential for the maintenance of axonal homeostasis in neurons. In recent years, more and more evidence suggests that dysfunctional autophagy is associated with axonal degeneration in many neurodegenerative diseases. Here, we review the core machinery of autophagy in neuronal cells, and provide several major steps that interfere with autophagy flux in neurodegenerative conditions. Furthermore, this review highlights the potential role of neuronal autophagy in axon degeneration, and presents some possible molecular mechanisms by which dysfunctional autophagy leads to axon degeneration in pathological conditions.
Keywords: Autophagy, Axon degeneration, Mitophagy, Wallerian degeneration
Introduction
Neurons are highly polarized cells that have morphologically distinct regions optimized for their functional roles. A typical neuron consists of a cell body and two types of processes: the dendrites and the axon. Among them, the axon is a long, slender extension that transmits electrical or chemical signals away from the neuron’s cell body. In many pathophysiological conditions, a program of axonal destruction can be activated and leads to the rapid degeneration of the whole or one segment of the axon [1, 2]. Axon degeneration is a process of axonal dying and breakdown, which is characterized by several morphological features including the accumulation of axoplasmic organelles, disassembly of microtubules, and fragmentation of the axonal cytoskeleton. Axon degeneration can be initiated by a broad range of triggers. It is not only necessary for the development, plasticity, and regeneration of neurons [3, 4], but also a prominent early event in different pathological conditions including traumatic axonal injury, toxic neuropathy, and chronic neurodegenerative diseases [5–7]. In many neurodegenerative diseases such as Parkinson’s disease (PD), amyotrophic lateral sclerosis (ALS), and Alzheimer’s disease (AD), the axon is the first damaged neuronal compartment that often occurs many years before the death of cell body [8, 9].
Over the past 30 years, great progress has been made in the understanding of axonal degeneration. Today, it is generally accepted that axon degeneration is an active process of axonal self-destruction. That was not always the case. Previously, axonal degradation was considered as a result of losing trophic factors from the soma. The classic example of axonal degeneration, Wallerian degeneration, was first described by Waller in 1850, and had been regarded as a passive axonal death for a long time. However, the discovery of a mouse strain carrying the spontaneous gain-of-function mutation Wallerian degeneration slow (Wlds) has radically challenged this dogma in 1989 [10]. The observation on Wlds mice, in which injured axons apart from cell bodies show a significant prolonged survival, revealed that axonal degeneration might be an active program under the control of axons [5, 10], although the exact mechanism still remains unclear.
At the beginning of this century, autophagy was first linked to the neurodegeneration [11]. Meanwhile, Ravikumar et al. found that aggregate-prone proteins that are typically associated with Huntington’s disease (HD) accumulated in cells treated with autophagy inhibitors [12]. Subsequently, two studies from Japanese scientists provided further evidence for this. Komatsu et al. and Hara et al. found that mice lacking the autophagy-related genes (Atg7 and Atg5) in the nervous system showed behavioral defects, axonal degeneration, and neuronal loss [13, 14]. Thus, these studies highlighted that constitutive autophagy plays a vital role in maintaining axonal homeostasis.
Autophagy, or ‘self-eating’, is a machinery of lysosome-dependent degradation of intracellular materials. Now, it is well known that autophagy is involved in a wide variety of physiological processes, such as adaptive metabolic response, embryonic development and the establishment of immunologic self-tolerance. Moreover, emerging evidence suggests that the dysregulation of autophagy may contribute to a broad spectrum of human diseases, including cancer, muscular disease and neurodegenerative disease [15, 16]. In neurons, constitutive autophagy is maintained at a relatively low level under physiological condition, which is important for the homeostasis and protein quality control in axon. In contrast, dysfunctional autophagy can lead to neurodegenerative disease and axon breakdown. Here, we review our understanding of the machinery of autophagy, consider the major steps that are interfering with autophagy process in axon degeneration, and highlight some aspects influenced by abnormal autophagy contributing to axonal destruction.
The machinery of autophagy
Autophagy is a highly conserved cellular degradative pathway that involves the delivery of cytoplasmic cargo to the lysosome. At present, three major types of autophagy have been identified, including chaperone-mediated autophagy (CMA), microautophagy, and macroautopahgy. Chaperone-mediated autophagy makes use of a chaperone protein selectively deliver the soluble cytosolic proteins into the lysosome for degradation. Microautophagy involves the direct engulfment of cytoplasmic materials at the lysosome surface through the invagination of lysosomal membrane. Macroautophagy is characterized by the formation of double-membrane autophagosome sequestering and delivering cytosolic cargoes into lysosomes for degradation [17]. Among them, macroautophagy is supposed to be the major catabolic mechanism and is most extensively investigated. This Review will focus on the pathways and processes of macroautophagy (hereafter autophagy).
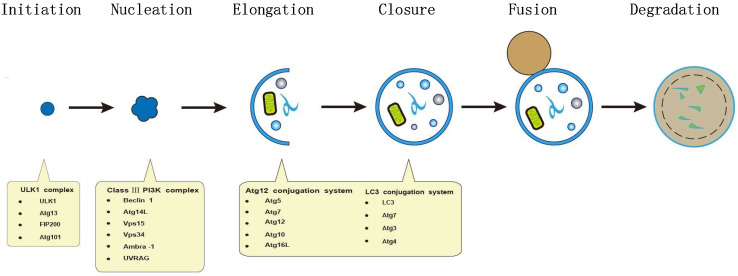
Over the past two decades, the molecular events that drive autophagy have been characterized detailedly in yeasts and mammalian cells. Nowadays, more than 30 autophagy-related (Atg) genes have been identified, which function as molecular machinery for autophagy. Briefly, autophagy initiates as an isolation membrane (IM, also called phagophore or pre-autophagosome), then gradually grows into a double-membrane autophagosome, and subsequently matures into an autolysosome after fusion with lysosomes. Finally, the autophagosome-containing cytoplasmic materials are degraded by lysosomal enzymes [18].
The formation of autophagosome
Mechanistically, the process of autophagosome formation can be divided into a series of interrelated steps including initiation, nucleation, elongation and maturation. At first, a cup-like single-membrane structure appeared. Then, it elongated into a complete double-membrane vesicle, termed autophagosome in mammals [19].
Of all the Atg proteins in autophagosome formation, the Atg1/Unc-51-like kinase 1(ULK1) complex is at the most upstream step of autophagy pathway. The process of autophagy starts from the association of ULK1, Atg13, FIP200, and Atg101. The ULK1 complex facilitates the recruitment of other related proteins to the site of autophagosome nucleation [17, 20, 21]. Furthermore, class III phosphatidylinositol 3-kinase (PI3K), Vps34, is required for the formation of autophagosomes. In mammals, class III PI3K complex consists of Beclin 1 (Atg6 homolog), Atg14L, hVps15, and hVps34 [20, 22, 23]. Herein, phosphatidylinositol 3-phosphate (PI3P), the product of Vps34, plays a vital role in the early stages of autophagy. In the meantime, the activity of Vps34 is modulated through its interaction with Beclin 1 [24]. Therefore, the Beclin 1 binding partners such as ambra-1 and UVRAG can induce autophagy. By contrast, the binding of anti-apoptotic proteins Bcl-2 or Bcl-XL to Beclin 1 can inhibit autophagy [25].
The elongation of autophagic membrane depends on two ubiquitin-like conjugating systems: the Atg12–Atg5–Atg16L system and the microtubule-associated protein 1 light chain 3 (LC3)/Atg8 system. Among them, the Atg12–Atg5–Atg16L complex is necessary for the elongation of the phagophore membrane, but it is dissociated with mature autophagosome [20, 26, 27]. In the LC3/Atg8 ubiquitin-like conjugating system, LC3 is biosynthesized as a precursor and is cleaved at its COOH terminus by Atg4B to form LC3-I. Then, cytosolic isoform LC3-I is conjugated to phosphatidylethanolamine (PE) to produce a lipidated form of LC3 (LC3-II) by Atg7 (E1-like enzyme) and Atg3 (E2-like enzyme) [28, 29]. LC3-II is specifically targeted to the elongating autophagosome membrane and remains on completed autophagosomes [30] (Fig. 1). Newly generated double-membrane autophagosomes matures after turning to autolysosomes by fusion with lysosomes. This process depends on the association of Syntaxin17 (STX17), SNAP29 and the lysosomal SNARE VAMP8 [31].
Fig. 1.
Cellular mechanism of autophagy. The autophagic pathway can be divided into separate stages, including initiation, nucleation, elongation, maturation, fusion, and degradation. The ULK1–Atg13–FIP200–Atg101 complex initiates autophagic process and facilitates the recruitment of other autophagy-related proteins to the site of autophagosome nucleation. Furthermore, the formation of autophagosomes requires the activity of the class III phosphatidylinositol 3-kinase (PI3K) consisting of Beclin 1, Atg14L, hVps15, and hVps34. Membranes originated from the ER, Golgi complex, or mitochondria compose the cup-like isolation membrane (also called phagophore). During the stage of elongation, LC3-II and the Atg12–Atg5–Atg16L complex locate on the membrane and promote phagophore expansion. In the meantime, phagophore engulfs cytoplasmic components, resulting in an autophagosome. Then, the double-membrane autophagosome merge with lysosome to form a single-membrane autolysosome. Finally, the components engulfed in autophagosome are degraded and released as small molecules
Autophagy in neurons
In neurons, the vast majority of autophagosomes formed in the distal axons. Occasionally, formation of LC3-positive vesicles could be detected in mid axons and cell bodies [32–34]. The autophagosomes in the distal axons need to be delivered to the soma where lysosomes accumulate for degradation. This centripetal movement of autophagosomes is dependent on the motor protein dynein and is a microtubule-dependent retrograde transport along axon [35].
Under both normal and pathological conditions, autophagosomes are located on microtubules by the interaction of LC3 and microtubule-associated protein 1B (MAP1B) that function as a regulator for microtubule stability [36]. Along the axon, dynein–dynactin complex, the major minus-end motor for transporting various cargo on microtubules, drive autophagosomes to undergo robust retrograde motility [32]. Specifically, newly generated autophagosomes at the distal axons usually exhibit inefficient back-and-forth bidirectional motility, because dynein and kinesin motors (including kinesin-1 and kinesin-2), both bound to autophagosomes trafficking along axons, possess competing activities [37]. To determine and maintain the direction of autophagosomes in axonal transport, motor scaffolding protein JIP1 recruitment to nascent autophagosomes plays an essential role as a switch to regulate the initiation of unidirectional movement toward the soma [37]. JIP1 binds directly to both dynein at p150Glued and kinesin 1 at kinesin heavy chain (KHC) simultaneously. Once JIP1 is recruited to autophagosomes via binding to LC3, the interaction that blocks the binding of JIP1 to KHC prevents kinesin 1 activation. Autophagosomes begin to exit out of the distal axon and continue into the mid- and proximal axon depending on JIP1 recruitment. At that time, most autophagosomes turns to move in a unidirectional retrograde manner [37].
During autophagic trafficking along axon, autophagosome can fuse with late endosome (LE) to form an amphisome in the mid-axon and then with lysosomes, thus generating autolysosomes. In addition, some autophagosomes can also fuse directly with lysosomes to form autolysosomes. It is exceptionally common that the majority of axonal autophagosomes are positive for LAMP1, an endosomal/lysosomal marker [33, 38]. However, there exists some LAMP1-negative autophagosomes in the soma that are likely synthesized locally within this region [38]. Furthermore, the formation of amphisomes is a prerequisite for retrograde movement. LE-loaded SNAPIN acts as an adaptor attaching dynein to facilitate the retrograde transport of amphisomes [39]. Amphisomes finally fuses with lysosome for digesting cargos. As autophagosomes move along the axons retrogradely, they mature and become increasingly acidified [38]. Autophagosome maturation into autolysosomes are believed to occur mostly when they reach the proximal regions of axon or the cell body [33, 38]. The primary site of autophagosome maturation is supported by the appearance of autophagosome accumulation in the soma only, not in the neurites, when lysosome function is blocked with bafilomycin A1 [38].
Under physiological conditions, only very few autophagosomes can be detected in neurons [40]. These findings initially lead to the assumption that there is a limited autophagic activity in neuronal cells. However, it is now believed that scarcity of autophagosomes in neurons is likely due to a highly efficient maturation and degradation, because blocking lysosomal degradation by inhibitory drugs results in a markedly increase of autophagosomes in primary cortical neurons [33, 41].
Alteration of autophagy in axonal degeneration
As postmitotic cells, neurons cannot dilute out aggregated proteins and damaged organelles in the cytoplasm by cell division. Thus, neurons are particularly dependent on the cellular protein-quality-control system to eliminate these toxic materials and maintain their own homeostasis. In healthy neurons, basal autophagy is constitutively active and highly efficient to maintain their viability and functionality, whereas dysfunctional autophagy might result in or contribute to axon degeneration and neuronal death [38, 42]. Nowadays, increasing evidence has demonstrated that disrupted autophagy is involved in a diverse range of neurodegenerative diseases such as AD, PD, HD, and retinal ganglion cell (RGC) death [43–47]. In fact, autophagy was initially supposed to be a possible mechanism underlying these disorders, because early studies found that there existed an increased number of autophagosomes in the brains of patients [48]. However, more recent reports support that autophagy primarily protects against the neurodegeneration. The accumulation of autophagosomes in neurodegenerative disorders only represents an adaptive response to increased protein load or is the consequence of a defect in autophagosome maturation [49, 50].
Alterations at distinct autophagy steps
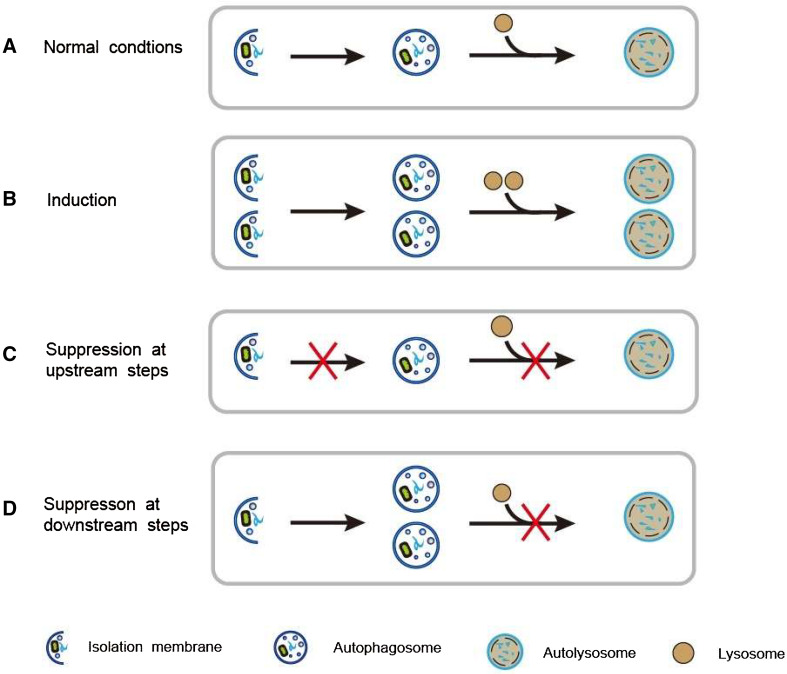
Given that autophagy is a highly dynamic process that is modulated at distinct steps, the rate of cargo degradation depends on the efficiency of autophagic flux. In detail, autophagic flux is a term describing the whole process of autophagy, including autophagosome formation, maturation, fusion with lysosomes, and degradation. The efficiency of autophagy degradation pathway mainly depends on the patency of autophagic flow rather than the number of autophagosomes. Only all the stages of autophagic flux remain unobstructed, the degradation of the substrate can be achieved. On the contrary, when the downstream of autophagic flow is blocked, the substrate could not be efficiently degraded even if autophagosome formation level is normal or even increased (Fig. 2). In neurodegenerative disorders, impairment at distinct steps of autophagic pathway from autophagosome formation to substrate degradation is likely to decrease the efficiency of autophagic degradation, thus leading to the buildup of damaged organelles or aggregated proteins.
Fig. 2.
Dynamic process of autophagic flux under different conditions. a Under normal conditions, basal autophagy occurs. b When autophagy is induced, the formation of both autophagosomes and autolysosomes increases, and autophagic flux is enhanced. c When autophagy is suppressed at upstream steps, the formation of autophagosomes and autolysosomes decreases, and autophagic flux is blocked. d When autophagy is suppressed at downstream steps, only the number of autophagosomes increases, whereas the formation of autolysosomes is blocked. Thus, autophagic flux decreases
Affects autophagosome biogenesis
Perturbed initiation of autophagy and impaired autophagy formation are widely mentioned in the chronic settings of neurodegeneration. Deregulation of key autophagy-related genes, such as Beclin 1, ATG7, and ULK1, is considered as a critical event in the above-mentioned conditions.
Beclin 1, an essential Atg protein for the initiation of autophagy, has demonstrated a markedly reduction in the brains of AD patients, which was localized into the most vulnerable brain regions of AD pathology [51]. In the meantime, increased proteolysis of Beclin 1 was also observed in AD. Caspase-mediated cleavage not only reduced the level of Beclin 1 and impaired autophagy activity, but also promoted cell apoptosis [52–54]. In addition, Ca2+-activated neutral proteases (calpains) could also cleave Beclin 1 in a similar manner [55, 56]. The perturbed initiation of autophagy was also reported in PD. It has been found that α-synuclein, an essential pathogenic protein to PD, disassociates the Beclin–Vps34 complex and perturbs autophagy. Moreover, overexpressed high-mobility group box 1 (HMGB1) is involved in α-synuclein-mediated dysfunctional autophagy by binding to Beclin1 [57].
Genetic experiments give compelling evidence that supports the ablation of critical autophagy genes resulting in axonal degeneration. Atg7 functions as E1 enzyme in the two ubiquitin-like conjugation systems to activate Atg8 and Atg12 and help them translocate to the PAS [20]. Loss of Atg7 reduces autophagy so significantly that autophagosomes are absent in the axons of Atg7-knockout Purkinje cell. Meanwhile, tissue-specific Atg7 deletion resulted in a massive loss of neurons in the cerebral and cerebellar cortices of mouse [13, 58]. Moreover, cell-specific knockout of Atg7 gene in midbrain dopamine (DA) neurons causes the formation of ubiquitinated inclusions in DA neurons and delayed neurodegeneration [59]. Similarly, neural-cell-specific Atg5−/− mice developed obvious behavioral abnormalities, which were accompanied by the accumulation of inclusion bodies in neurons [14].
ULK1 and ULK2 are necessary for the initiation of autophagy, Ulk1/2-deficient mice will die of progressive neurodegeneration perinatally. A possible mechanism for the loss of neurons is that SEC16A-dependent ER-to-Golgi trafficking of cargoes is impaired in Ulk1/2-deleted mice, and therefore, ER stress is stimulated [60]. Moreover, WIPI4 (also known as WDR45), one of mammalian orthologs for yeast Atg18 also plays a significant role in regulating autophagosome formation [61]. WIPI4 mutation is related to static encephalopathy of childhood with neurodegeneration in adulthood (SENDA) [62]. Further investigation revealed that autophagic flux was incompletely blocked at an intermediate step of autophagosome formation, which led to the accumulation of immature autophagic structures in neurons. These results were consistent with the observation in cells derived from SENDA patients [62].
Although the aetiology of neurodegenerative diseases is associated with autophagy dysfunction, enhanced induction of autophagy is commonly observed as well. For instance, ULK1 can be activated and induce autophagy in AD, because amyloid beta (Aβ), a protein central to the pathogenesis of AD, increases tribbles pseudokinase 3 (TRIB3) that inactivates mTOR [63]. Interestingly, mTOR can be sequestered in a variety of aggregate-prone protein inclusions. In HD cell model, transgenic mice, and human patient brains, the sequestration of mTOR by polyglutamine aggregates resulted in a reduced mTOR activity, and thus leading to autophagy enhancement [64, 65]. In addition, stimulated Rho-associated coiled-coilkinases (ROCK1) in AD could induce autophagosome formation via binding and phosphorylating Beclin1 and dissociating Beclin1–Bcl-2 interaction [66].
Moreover, autophagy can be induced by mutated Cu, Zn-superoxide dismutase (SOD1) and increased reactive oxygen species (ROS). About 20% of familial amyotrophic lateral sclerosis (fALS) carry the mutations in SOD1 gene. Mutant SOD1 protein misfolds and forms insoluble inclusions in motor neurons. Under the circumstances, on the one hand, autophagy can promote the degradation of misfolded proteins and damaged organelles. On the other hand, the mutant SOD1 can activate autophagy via an abnormal interaction with Beclin 1 [67–69]. ROS also plays a key role in activating class III PI3 kinase and autophagy induction in response to Aβ peptide [70]. Despite the increased autophagosome formation, substrate degradation does not always increase under these pathological conditions as expected.
In addition, autophagy initiation can be remarkably enhanced after axonal damage. In acute axonal degeneration of optic nerve, the rapid increase of Ca2+ flux released from ER is an important upstream regulator of autophagy. Nowadays, it is assumed that cytoplasmic Ca2+ might enhance autophagy activity via the activation of Ca/calmodulin-dependent protein kinase kinase-β (CaMKK-β) or Calcineurin (CaN) [71]. After application of the calcium chelator, autophagosome accumulation decreases significantly while p62-marked substrates increases remarkably [72].
Overall, although it is generally accepted that the dysfunctional autophagy is associated with neurodegeneration, the above-mentioned studies show obvious paradox phenomenon. In the case of AD, different studies suggest of different state of neuronal autophagy, activation or inhibition. This inconsistency may represent a dynamic pathophysiological process of the disease. Perhaps, the initial increase in autophagosomes and autophagic activity observed in AD models should be considered as a compensatory response to Aβ peptides, but the autophagy machinery becomes dysfunctional over time. Therefore, in the late stage of AD disease, excessive accumulation of Aβ burden will lead to autophagy dysfunction.
Defective cargo recognition
Cargo recognition failure and other defects in selective autophagy are sufficient to lead to neurodegenerative diseases. For instance, mutant huntingtin gene (mHtt) is believed to be the pathogenic cause for HD. When mHtt protein is not efficiently removed, it accumulates within neurons in the form of oligomers and aggregates [73]. Meanwhile, markedly increased autophagosomes in affected neurons are observed. However, the efficiency of autophagy-mediated proteolysis does not increase as expected, but instead the turnover of cytosolic components in HD cells is actually reduced [74]. Indeed, in cell and mouse models of HD, autophagosomes occur at normal or even increased rates, but they fail to efficiently trap cytosolic cargo in their lumen [74]. At present, the exact mechanism for the impaired ability to recognize cargo of autophagosomes remains elusive.
Recently, autophagic adaptors have attracted increasing attention. Selective autophagy targets specific cargos through a group of autophagy adaptor proteins. They recognize degradation signals on cargo proteins and bind LC3 proteins on the forming autophagosomes. Multiple lines of evidence supported that the mutation of autophagy adaptors was causally associated with neurodegenerative diseases. For example, the mutation of p62 contributes to the neurodegeneration in ALS, PD, AD, frontotemporal lobar degeneration, and childhood-onset neurodegeneration [75, 76], while mutations in OPTN have been linked to ALS, HD, and primary open-angle glaucoma [77–79]. Furthermore, mutant PINK1 and Parkin have been reported to contribute to the loss of selectivity in recognizing the cargo in PD. In familial PD cases, the mutations in PINK1 and Parkin impeded the selective degradation of damaged mitochondria via autophagic pathway, but had no obvious impact on the elimination of other organelles [80].
Disrupts axonal transport
There is a long distance separating axonal and synaptic domains from neuronal cell bodies. The translocation and delivery of materials essential for maintaining axonal and synaptic function along axons depends highly on axonal transport. Anterograde axonal transport is responsible for the movement of membranous organelles from cell body towards axon terminals, and allows renewal of axonal proteins, whereas retrograde axonal transport returns old membrane constituents, trophic factors, exogenous material to the cell body and could represent a feedback mechanism for controlling the metabolic activity of the soma [81, 82].
Alteration of axonal transport represents an early and critical pathological change in neurodegenerative diseases such as AD, HD, PD, ALS, and spinal muscular atrophy (SMA) [83]. Under these circumstances, accumulated autophagic vacuoles observed in the dystrophic axons may be caused by impairing axonal transport [43, 84]. It has been illustrated that the accumulation of autophagosomes appears after axonal transport impairment in the Parkinsonian mimetic axonal degeneration of dopaminergic neurons by 6-hydroxydopamine (6-OHDA) treatment [85] and N-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) active derivative (MPP+) [86]. Moreover, the maturation of autophagosomes was also blocked when axonal retrograde transport is disturbed. Thus, most autophagosomes in dystrophic axon terminal are nascent [34, 36]. In this respect, damaged cellular structures involved in axonal traffic and dysfunctional motor proteins may account for ineffective axonal transport that leads to autophagosome accumulation.
Defects of motor proteins can lead to prohibition of autophagosomes moving along axons. It is found that dynein-driven endolysosomal trafficking is deficient at early asymptomatic stages in fALS-linked mice. These deficits impair the degradation of autophagic vacuoles from distal axons [43]. The mutation of critical motor proteins, such as kinesin heavy-chain gene KIF5A, dynein heavy chain, the p150Glued subunit of dynactin, the CAP-Gly domain of dynactin was observed in other neurodegenerative diseases [78, 87–89]. Moreover, the loss of dynein function resulted in the formation of protein aggregates and increased levels of the autophagosome in both cell culture and mouse HD models, compatible with impaired autophagosome-lysosome fusion [90].
Given that autophagosomes movement along axons relies on microtubule as tracks, some adverse factors that compromise the integrity of microtubule networks inevitably lead to autophagosome accumulation. In the presenilin-1 (PS1)/Aβ precursor protein (APP) transgenic mouse model, the defects in microtubule network and molecular motors resulted in impaired axonal transport, local autophagosome accumulation, and axonal dystrophy [91]. Moreover, in neurons undergoing neurofibrillary degeneration, microtubules were almost completely absent. Axonal injury result in cytoskeletal alteration, which then impacts axonal transport [92].
Axonal inclusion bodies can also impair axonal retrograde transport. For instance, extensively expressed neuronal ataxin-3 tends to aggregate into inclusion bodies that impair fiber tracts in spinocerebellar ataxia type 3 (SCA3) patients. These inclusion bodies aggregating in fiber tracts are detrimental to axonal transport and thereby contribute to the progressive neurodegeneration [93]. Recently, a new study reported that Aβ oligomers that are enriched in axons interferes with the combination of dynein motor with SNAPIN and, thus, disrupt dynein-driven retrograde transport of amphisomes [94].
Furthermore, following the acute axonal degeneration of optic nerve, increased cytoplasmic Ca2+ not only affects the formation of autophagosomes, but also disintegrate the cytoskeleton and microtubules resulting in impairment of autophagosomes clearance [72]. Therefore, the striking accumulations of immature autophagic vacuoles in dystrophic neurites of optic nerve implicates that autophagy may be enhanced significantly, or that the autophagosomes transport or maturation to lysosomes may be impaired [95]. Taken together, these findings suggest that aberrant accumulation of autophagosomes in axons observed in some pathological conditions is more likely to be caused by the disturbance of axonal transport, rather than the activation of autophagy.
Blocks the maturation and clearance of autophagosomes
As mentioned previously, increased number of autophagosomes does not always represent an enhanced autophagy activity, because the impairments in late stages of autophagic flux, in particular the blockage of autophagosome maturation and fusion also lead to the accumulation of autophagosomes. In AD patients and PS1/APP mouse model, neuronal autophagy was first induced before the extracellular deposits of amyloid beta (Aβ) peptide. Then, autophagosomes and late autophagic vacuoles significantly accumulated in dystrophic neurites, suggesting an impeded maturation of autophagic vacuoles to autolysosomes [96].
It is true that the final stage of autophagic-lysosomal degradative pathway is commonly blocked in neurodegenerative disorders. In the case of AD, the defects in lysosomal acidification contribute to the failure of autophagy-mediated protein degradation. In fact, the regulation of lysosomal pH in neurons is very sensitive to many insults [97]. For example, AD-related protein presenilin-1 mutations were reported to impair lysosome acidification and result in pathogenic protein and autophagosome accumulation in varieties of AD [98]. Furthermore, lysosomal proteolysis dysfunction can slow the axonal transport of autophagic compartments and results in the increase of immature autophagosomes. For example, in AD model of mice and primary neurons, autophagic vacuoles including autolysosomes, late endosomes, and lysosomes were observed to aggregate in focal axonal swellings by restraining lysosomal proteolysis [33, 42]. Therefore, the autophagic pathology observed in AD is likely due to impaired clearance of autophagic vesicles rather than enhanced autophagosome formation alone. [41]. In addition, aberrant autophagy accompanied by neurodegeneration is extensive in lysosomal storage diseases (LSDs), which are associated with the deficiency of lysosomal hydrolases resulting in progressive accumulation of poly-ubiquitinated protein aggregates and dysfunctional mitochondria [99]. Immature autophagosome accumulations are widely observed in Danon disease, Pompe disease, multiple sulfatase deficiency (MSD), neuronal ceroid-lipofuscinoses (NCLs), and mucopolysaccharidosis type IIIA [99–102].
In conclusion, following injury, autophagosomes can be trapped in distal axons or their maturation and degradation in the soma is blocked. Even though the autophagy initiation may be enhanced, autophagy flux in axons is compromised for deficient retrograde transport or blocked maturation and clearance.
Dual role of autophagy in axon degeneration
As mentioned above, autophagy is being considered as a pivotal mechanism for maintaining homeostasis in healthy neurons as well as a neuroprotective response when further induced in neurodegeneration conditions. Although the exact mechanism of autophagy alteration in injured neurons has not been detailed and how autophagy influences axonal degeneration is still elusive, a dual role of autophagy in axon degeneration was observed in the pathological conditions.
In neurons, basal autophagy is essential for maintenance of local homeostasis of axonal compartment and protection against axon degeneration. This protective effect appears to be related to eliminate the damaged mitochondria and protein aggregates. Indeed, suppressed autophagy may result in accumulation of abnormal proteins such as Aβ peptides and misfolded mutant SOD1 [51, 103], thus promoting protein aggregation-induced axon degeneration. Meanwhile, the beneficial role of autophagy in axon degeneration is supported by pharmacological induction of autophagy, which effectively slows the neurodegeneration process in HD and AD [65, 104]. By contrast, the inhibition of autophagy decreases the viability of dorsal root ganglia (DRG) cells and suppresses their neurite growth and branching complexity [105]. Moreover, in vitro, in vivo, and epidemiological data have demonstrated that natural agents such as resveratrol, trehalose, and quercetin protected neurons against neurotoxic chemicals or in models of neuronal injury and neurodegenerative diseases, which could be attributed to the modulation of autophagic activity [106–109]. These results agree with autophagic favorable roles in neurodegeneration.
Despite neuroprotective role of autophagy in most cases, excessive activation of autophagy has been observed in neurons undergoing cell death. In this respect, autophagy can be regarded as a molecular mechanism of programmed cell death contributing to neuron loss [110]. Though, some reviews hold that autophagy may just present an unsuccessful rescue response to cell death [111, 112]. Indeed, the exact role of activated autophagy in neuronal death remains controversial, because it is very difficult to determine whether neurons die as a direct consequence of autophagic activation or activated autophagy merely as an attempt to promote neuron survival. However, undoubtedly, alteration of autophagy predisposes cell death. Recently, there is more and more compelling evidence that supports autophagy likely plays a causative role in neuron death. Koch and Lingor reported that activation of autophagy preceded the death of RGC cells in the model of optic nerve injury [77].
Interestingly, autophagy enhancement by Atg1 overexpression can cause dendrite degeneration of class IV neurons in NMNAT heterozygotes Drosophila. By contrast, genetically blocking autophagy can suppress hypoxia-induced dendrite degeneration [113]. These results suggest a self-destructive role of autophagy. Furthermore, Koch et al. demonstrated that deficient autophagy conferred resistance to axonal degeneration, which means autophagy may play unfavorable functions in neurodegeneration. In the meantime, inhibition of autophagy by 3-methyladenine in acute conditions could slow disintegration of the axon [72, 77]. Moreover, autophagy deficiency by Atg7 ablation enhances neuronal survival in acute condition like ischemic stroke [114]. It is also reported that conditional deletion of autophagy gene Atg7 in dopamine neurons achieves a remarkable axon protection in both neurotoxic and physical injury (axotomy) models of retrograde degeneration [115]. Therefore, autophagy may play protective role in chronic disease and, in contrast, harmful role in acute disease. Indeed, it is observed that induced autophagy serves completely opposite roles in two glaucoma models: autophagy has neuroprotective effects in the chronic model and promotes RGC death in the acute one [77].
Additionally, the interaction between autophagy and other cellular mechanism make it much more complicated to define the exact role of autophagy in neurodegeneration. There are some studies reporting that autophagic activity induced by rapamycin lead to neuronal loss in ALS [84]. By contrast, when autophagy is induced by rapamycin without the drugs’ influence on immune, survival in ALS could be increased [116]. Besides the mutual influence between autophagy and other process, it also deserves consideration that activating autophagy at variant degeneration stage has different effects. For example, the inhibition of autophagy suppresses axonal degeneration within the first hours in the animal model of optic nerve injury. In contrast, the induction of autophagy at any later time promotes the survival of RGC cells [77]. Therefore, when considering the contribution of autophagy to axon destruction, the time kinetics of neurodegeneration should be taken into account.
Whether upregulation of autophagy is neuroprotective or damaging to the neuron is the subject of considerable debate. These conflicting results might be related to the variation in different disease models and different autophagic stages modulated by drugs. Indeed, obvious differences between axonal compartment and soma have been demonstrated recently. Among them, the protective effect of rapamycin was primarily observed in axons whereas the therapeutic effect of 3-MA was reported with respect to soma survival [77]. In fact, the roles of pro-survival and possible pro-cell-death place autophagy at the centre of maintaining cell and organismal homeostasis in response to various stresses. Based on current knowledge, there are several assumptions about autophagy beneficial or deleterious role: (1) it depends upon the duration of diseases which are categorized as acute and chronic; (2) it depends upon the stages of degeneration at which autophagy is induced; (3) it depends upon the intensity and duration of autophagy activation.
Potential mechanisms by which autophagy affects axonal degeneration
To date, the exact mechanisms by which autophagy is responsible for axonal degeneration have not been fully elucidated. However, mounting evidence supports that the following mechanisms may be involved in the axonal degeneration under the dysfunctional autophagy conditions.
Dysfunctional autophagy results in abnormal protein accumulation in neurons
Intracellular or extracellular aggregation of abnormal proteins is one of conspicuous pathological changes in most neurodegeneration. In eukaryotic cells, abnormal proteins or insoluble protein deposits are degraded through two major routes: the ubiquitin–proteasome system (UPS) and autophagy–lysosomal pathway. The abnormal proteins are usually recognized by molecular chaperones, which allow proteins repair or refolding. If the chaperones do not succeed in this repairing activity, they pass the abnormal protein to degradation via the UPS or chaperone-mediated autophagy. When the first two systems are compromised or cytoplasmic proteins turn into insoluble complex structures, autophagy is the only system that is able to clear these toxic products.
When these aggregates cannot be removed by autophagy, they would interfere with normal cellular trafficking and trap still-functional proteins into themselves. In postmitotic neurons, persistence of toxic products leads to axon degeneration and cell death [50]. Impairments in autophagy are often coupled with accumulation of protein aggregates and damaged organelles, which neurons are vulnerable to different adverse insults [117]. Direct evidence that supports the role of autophagy in controlling protein quality is from transgenic mice experiments. As mentioned above, tissue-specific knockout of Atg5 or Atg7 genes in mice result in a progressive neurobehaviour abnormalities and massive neuronal loss in the cerebral and cerebellar cortices [13, 14]. Notably, Purkinje cells in mice cerebellum demonstrated an accumulation of aberrant membrane structures and polyubiquitinated proteins in the axonal swellings [58]. Therefore, it is conceivable that dysfunctional autophagy may disrupt the maintenance of axon homeostasis and thus leads to axon degeneration. In fact, in AD, PD, HD and ALS, it has been demonstrated that axonal degeneration is associated with the accumulation of misfolded proteins such as amyloid precursor protein, neurofilaments, a-synuclein-containing Lewy bodies, or huntingtin proteins [5, 118, 119].
Pharmacological evidence also indirectly supports that dysfunctional autophagy is negatively correlated to the toxic protein products clearance in neurite degeneration. For instance, autophagy-inducing chemicals could efficiently relieve the Aβ25–35-induced neurite degeneration in AD cell models [104]. Furthermore, pharmacological activation of autophagy not only reduces the level of neurodegeneration-associated proteins such as mutant huntingtin and α-synuclein, but also alleviate their neurotoxicity in Drosophila and mouse models [49].
Dysfunctional autophagy delays the clearance of impaired mitochondria
Mitochondria are the power plants inside cells that are responsible for generating ATP for cell survival through oxidative phosphorylation. Furthermore, mitochondria are implicated in critical cellular processes such as programmed cell death and the regulation of inflammatory responses [120–122]. Dysfunction of mitochondria can lead to a wide range of disorders due to the impact on cellular metabolism and production of ROS [123]. Impaired mitochondrial function and dynamics are associated with axonal degeneration in major neurodegenerative disorders such as AD and PD [124, 125].
For quality control of mitochondria in cells, mitophagy is responsible for the selective removal of superfluous and damaged mitochondria under a variety of pathophysiological conditions. At present, it is well-known that several forms of mitophagy can occur in response to mitochondria depolarization, oxidative stress, or under particular physiological scenarios such as the fertilization of oocytes and maturation of reticulocytes [126, 127]. Autophagy-dependent removal of damaged mitochondria is essential to relieve oxidative stress and prevent cytosolic and axonal damage and subsequent cell death. Nowadays, PINK1-Parkin-mediated mitophagy is the most well-characterized pathway. PINK1 is a mitochondrial serine/threonine kinase, which acts as a molecular sensor to monitor mitochondrial status and protect cells from stress-induced mitochondrial dysfunction. In healthy mitochondria, mitochondrial transmembrane potential drives PINK1 import into the inner mitochondrial membrane (IMM) by the translocase of the outer mitochondrial membrane (OMM) [128]. By contrast, mitochondrial damage causes the accumulation of PINK1 on the OMM without being imported on the IMM. Then, PINK1 recruits Parkin (PARK2) to activate mitophagy [128]. For example, in acute injured axons, damaged mitochondria are eliminated locally by neighboring lysosomes within the axon to prevent further oxidative damage [125]. By contrast, impaired mitophagy may account for the accumulation of dysfunctional mitochondrial and ROS in neuronal models of tuberous sclerosis complex (TSC), a neurodevelopmental disease caused by TSC1 or TSC2 mutations [129].
Mutations in genes that encode PINK1 and Parkin are closely related to a juvenile onset form of PD [130, 131]. At present, it is widely accepted that mutation of PINK1 and PARK2 genes plays a critical role in the pathogenesis of PD [132], because loss of PINK1 and Parkin function contributes to the accumulation of damaged mitochondria and death of dopaminergic neurons in the substantia nigra in the mesencephalon [123]. Further support for a mitochondrial etiology in Parkinson’s disease comes from the early observations that exposure to various mitochondrial toxins leads to Parkinson’s disease in humans and rodents [133]. Surprisingly, Parkinsonism is rare in primary mitochondrial diseases and PD is not characterized by the severe neurodegeneration [123]. In this regard, it is assumed that alternative mitophagy pathways may compensate for defects in PINK1 or Parkin, or that mitophagy may play a minor role in overall mitochondrial maintenance.
Mitochondrial dysfunctions and abnormal accumulation are also reported in AD. Generally, mitochondrial dysfunction can be an upstream inducer of Aβ aggregation and phosphorylated Tau, while Aβ aggregation and phosphorylated Tau can further exacerbate mitochondrial dysfunction [134]. To some extent, the accumulation of impaired mitochondria may be attributed to disturbed mitophagy. Herein, it is supposed that lysosome function is initially impaired, thereby disrupting the fusion of autophagosomes and lysosomes and inhibiting mitochondrial degradation [134]. Furthermore, haploinsufficiency of TANK-binding kinase 1 (TBK1), a novel gene that promote Parkin-PINK mitophagy by enhancing OPTN, NDP52, and p62 recruitment to impaired mitochondria, is believed to be causative for sporadic and familial ALS and frontotemporal dementia [135–137]. Mutations in valosin-containing protein (VCP), which can regulate mitophagy and autophagosome maturation, can cause multisystem degenerative disease including neuronal diseases like sporadic ALS and PD [138, 139].
It is noteworthy that impaired mitochondria and excessive production of ROS also affect autophagy activity in turn. In this respect, increased ROS can induce the autophagy by directly affecting the core autophagic machinery or indirectly regulating the autophagy-related signaling. For instance, the oxidation of a critical autophagy-related protein, Atg4, contributes to the vesicle elongation step of autophagosome formation by preventing the delipidation of LC3-II [140, 141], whereas the activation of AMPK and the induction of BNIP3 and BNIP3L can up-regulate the signals for autophagy process [142–144]. These studies manifest that autophagy activation in the setting of oxidative stress represents a negative feedback loop that keeps down the level of ROS in stressed cells.
Moreover, elevated production of ROS can disturb the nicotinamide adenine dinucleotide (NAD) pathway, which protects axon by suppressing oxidative stress [47]. This process will impair autophagic flux through decreasing SIRT1, a member of NAD+-dependent histone deacetylase family (sirtuins), and finally bring about neurodegeneration [47].
Autophagy is involved in Wallerian-like degeneration
In neurodegeneration conditions, the axon of an unhealthy neuron progressively degenerates from the distal terminal toward the cell body in a ‘dying-back’ manner, which is characterized by the granular disintegration of cytoskeleton, the appearance of myelin ovoids, and eventual breakdown of distal axons [145]. This process was defined as “Wallerian-like degeneration” because it is morphologically similar to Wallerian degeneration following axotomy.
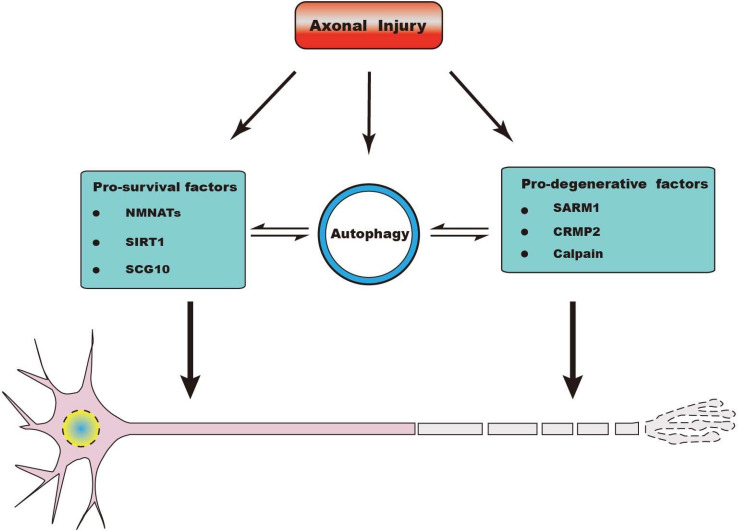
In past decades, great advances have been made in understanding the molecular mechanism of axon degeneration. An important breakthrough of Wallerian degeneration is the discovery of a spontaneous mutant mouse strain, C57BL/Wlds. The axons in Wlds mice could survive for as long as 2 weeks after transection, whereas axons in wild-type mice maintain their integrity for only 1.5 days in wild-type mice [10, 146]. Wlds protein contains the N terminus of ubiquitination factor Ufd2a and a full-length nicotinamide mononucleotide adenylyl-transferase (NMNAT) [145]. Normally, NMNAT2 is trafficked anterogradely along axons and serves as a survival factor. Disrupted axonal transport after injuries can lead to the depletion of axonal NMNAT2, which is an initiating event for injury-induced axon degeneration. Meanwhile, multiple genes necessary for Wallerian degeneration have been cloned in mouse and Drosophila. The most important finding is sterile alpha and toll-interleukin 1 receptor (TIR) motif containing 1 (SARM1), which has been identified as a key activator of Wallerian degeneration [147]. Upon injury, SARM1 is stimulated and promotes rapid loss of NAD+, which in turn leads to axonal degeneration both in neuronal cultures and in vivo. SARM1 knockout can significantly inhibit the process of axon degeneration and prevent the neuronal and axonal damage under oxidative stress [147, 148].
Recently, a crosstalk between NMNAT2 and SARM1 in axon degeneration has been observed. The TIR domain of SARM1 is required for its activity and can trigger axonal degeneration, a recent report revealed an intrinsic NADase activity in the SARM1-TIR domain [149, 150]. Meanwhile, loss of NMNAT2 may also trigger the activation of SARM1. It is supposed that the reduction of NAD+ by NMNAT2-loss stimulate SARM1 [151]. Currently, the downstream finally leading to axonal fragment of SARM1–NMNAT pathway is elusive. Gerdts [151] and Yang [152] hypothesize that mitogen-activated protein kinase (MAPK) pathway is a potential candidate. Recently, DiAntonio’s group proposed that MAPK signaling promotes axon degeneration through accelerating the turnover of NMNAT [153]. Another hypothesis demonstrates that elevated SARM1 facilitates Wallerian degeneration via activating dual leucine zipper kinase (DLK)/JNK, which subsequently stimulate calpains [154]. Calpains are usually activated in axonal degeneration by calcium influx to degrade cytoskeletal components [155]. Taken together, Wallerian degeneration is modulated by the opposing actions of pro-degenerative and pro-survival factors (Fig. 3). Among them, the pro-degenerative factors include MAPK signaling pathway and SARM1, while the main pro-survival factors is NAD+ biosynthetic enzyme NMNAT2 that can inhibit activation of the SARM1 pathway [153]. Excavating the function of NMNAT-SARM1 pathway may offer some evaluable information and help build up general understandings of neurodegeneration.
Fig. 3.
Proposed cellular pathways by which autophagy is implicated in Wallerian degeneration. Axonal injury can induce Wallerian degeneration, which is modulated by the opposing actions of pro-degenerative and pro-survival factors. It is supposed that autophagy may interact with pro-degenerative factors (SARM1, CRMP2 and calpain) and pro-survival factors (NMNATs, SIRT1 and SCG10) to modulate the process of Wallerian degeneration. However, the exact functions and underlying mechanisms of the critical molecules with respect to autophagy in Wallerian degeneration remain largely unknown
Although the causal relationships between autophagy and axonal degeneration are extensively investigated, nearly all of the researches focused on the maintenance of local homeostasis of axons. Whether autophagy is directly involved in the process of Wallerian degeneration still remains unclear. Interestingly, recent studies have demonstrated that there exist mechanistic links between neuronal autophagy and Nmnat-mediated axonal protection. In rat model of optic nerve degeneration, the axonal-protective effect of Nmnat3 was suppressed by autophagy inhibitors, whereas the autophagy activator rapamycin promoted axonal survival. These results indicated that the protection of Nmnat3 in axon degeneration was closely related to autophagy machinery [47, 156]. Similarly, the axonal-protective role of SIRT1 in the degeneration of RGCs is likely attributed to the modulation of autophagy [47, 108]. Moreover, in the Drosophila model, endogenous Nmnat can protect class IV dendritic arborization neurons from hypoxia-induced dendritic injury. However, the overexpression of Nmnat fails to prevent dendrites damage caused by Atg1 overexpression, suggesting Nmnat acts on the upstream of autophagy [113].
Moreover, recent reports support that autophagy is the basis for completion of Wallerian degeneration. In an in vitro model using mouse dorsal root ganglia neurons, glycogen synthase kinase 3β (GSK3β) phosphorylates and inactivates collapsing response mediator protein 2 (CRMP2), a critical mediator of promote microtubule assembly in axons, and leads to the loss of cytoskeletal integrity, thus promoting Wallerian degeneration [157]. In the meantime, GSK3β can activate Beclin 1-dependent autophagy in degenerating axons through phosphorylation of BCL2 family member MCL1, which contributes to ATP production and recruitment of phagocytes to axonal debris [158]. Furthermore, in the acute axon degeneration of rat optic nerve, the initial axonal injury results in a rapid calcium influx into the axon and activates a cascade of molecular events. On one hand, the calcium-sensitive protease calpains is activated and results in the fragmentation of microtubules and axonal disintegration [95]. On the other hand, calcium influx leads to the rapid local activation of autophagy. These findings provide a good explanation with respect to increased numbers of autophagosomes in acute axon degeneration [72, 95]. Hence, even though axonal energy supply decreases in degenerating axons for disrupted axonal transport, induced autophagy can maintain ATP level independently on mitochondrial ATP production for short time [158]. These studies supported that axonal autophagy could provide energy production required for Wallerian degeneration [158, 159], which might be important for the successful completion of Wallerian degeneration. Additionally, a recent study demonstrated that autophagy induction could stabilize microtubules by degrading microtubule disassembly protein SCG10 and promote axon regeneration after spinal cord injury [160].
In summary, the causative link between autophagy and Wallerian degeneration is beginning to be determined. However, the exact role and underlying mechanisms of autophagy in Wallerian degeneration remains largely unknown. Fundamental understanding of these processes will ultimately lead to the design of molecular therapies aimed at decreasing neurodegeneration and its sequelae.
Excessive autophagy promotes apoptosis-associated axonal degeneration
As an important adaptive catabolic process, autophagy can be activated in response to different forms of stimuli such as nutrient deficiency, hypoxia, and endoplasmic reticulum stress. However, an excessive autophagy can lead to cell death [117]. For example, autophagosome formation and lysosomal activity are increased in RGCs after intraocular pressure (IOP) [161]. In this aspect, autophagy plays a protective role in axons at first. However, when autophagy is activated in the soma with the progressive ascending in IOP, especially when IOP increases acutely, the enhancement of autophagy flux will disrupt neuron homeostasis and induce neuronal cell death rapidly [161].
Although axonal degeneration has been regarded as a mechanism that is independent from apoptosis, it can be the result of apoptotic program in neuron against adverse stimuli. This axon degeneration induced by neuronal pro-apoptotic signaling is termed as ‘axon apoptosis’ [162, 163]. It has been illustrated in global nerve growth factor (NGF) model that DLK/JNK or/and Akt pathway control neuronal apoptotic program. NGF deprivation can cause dephosphorylation of Tropomyosin receptor kinase A (TrkA), subsequently leading to activation of DLK-mediated pro-apoptotic signaling [162]. In the meantime, a major pro-survival pathway downstream of TrkA is Akt that can directly phosphorylate and inhibit DLK activity [163]. Pro-degenerative DLK and pro-survival Akt may share the same downstream via Foxo3a/c-Jun controlling cell-body-localized Puma that antagonizes Bcl-w and Bcl-xL [163]. Hence, excessive autophagy may promote axonal degeneration via stimulating somal apoptosis rapidly.
Glial autophagy and axonal degeneration
The cells of the nervous system is divided into two categories: neurons and glial cells. Undoubtedly, glia–neuron interactions play a critical role in the setting of axon injury. On the one hand, axons rely heavily on glial energy and metabolic support for their long-term survival, such as Schwann cells (SC) in the PNS and oligodendrocytes in the CNS [164]. On the other hand, glial cells such as microglia and astrocytes can help eliminate axonal fragments dependently on engulfment receptor Draper during degeneration [165].
Glial cells are sensitive to the disturbance of neurons and coordinate a sophisticated cascade of reactive events, including engagement of phagocytic programs to clear cellular debris. In a recent study, axon degeneration could induce glial responses through Draper–TRAF4–JNK pathway. This process leads to the upregulation of Draper and facilitation of glial engulfment of axonal debris [166]. Furthermore, glial InR/Akt1 activity also promotes phagocytic clearance of axonal fragmentation through STAT92E-dependent transcription of Draper [167]. In contrast, suppression of phosphoinositide-3-kinase (PI3K) signaling and reduced expression of the phagocytic receptor Draper/MEGF10 make the glial clear degenerating axons slowly [168].
Considered that PI3K and mTOR are essential effectors in the regulation of autophagy, it is assumed that autophagy is a potential digestive process in glial cells for degradating axonal fragments. After axon damage, axonal degeneration is accompanied with segmental demyelination [169]. Noteworthily, myelin sheath has been broken into some pieces observed in the cytoplasm of Schwann cells [170]. Hence, it is conceivable that activation of autophagy in Schwann cells regulates myelin destruction following axonal injury, which is further supported by the delayed myelin clearance in Schwann cells with pharmacological and genetic inhibition of autophagy [169, 170].
Furthermore, the emerging protein Sonic hedgehog (Shh) released from glial cells may be a bridge connecting glial signaling and neuronal autophagy, because Shh signaling could protect neurons against degeneration by bolstering neuronal autophagy depending on PI3K [171]. By contrast, impaired Shh signaling pathway contributes to age-related neurological disorders such as AD [172]. However, it is still unclear whether Shh bound to axonal degeneration by autophagy stimulation is exactly discharged from glia. In this respect, this intriguing relationship between glial and neuronal autophagy is worthy of further investigation.
Additionally, the role of neuroinflammation in neurodegeneration is being paid more and more attention. Especially, the activation microglia is believed to be associated with the pathogenesis of many neurodegenerative diseases. Although the inflammatory response is a self-defensive reaction against various pathogenic stimuli, chronic inflammation may become a harmful self-damaging process that can cause serious damage to host’s own tissue. Interestingly, recent studies found that activated autophagy can selectively eliminate the damaged mitochondria, thereby inhibiting the activation of inflammasomes and relieving the inflammatory response [173]. In contrast, inhibition of autophagy by genetic approaches can elevate the level of NLRP3 inflammasome, increase the production of IL-1β, IL-18, and enhance inflammatory response [120, 174]. Certainly, these findings are mainly from other disease models, however, the same response may occur in neurodegenerative diseases. Therefore, interfering with microglia autophagy may be considered as an intervention strategy for the prevention of neurodegeneration in the future.
Conclusion
Axonal degeneration is a common pathological feature in many neurodegeneration conditions, the exact molecular mechanisms underlying axon degeneration remain largely unknown. In this review, we have highlighted the autophagy defects observed in axonal degeneration and the potential mechanisms that autophagy contributes to axonal degeneration. In fact, increasing evidence supports that the roles of autophagy in degenerating axons are much more complicated than expected before. Undoubtedly, autophagy may play different roles in various stages of axon degeneration. In the meantime, there exists a complex set of interactions between autophagy and other cellular mechanisms, such as other degradation pathways including ubiquitin–proteasome system and chaperone-mediated autophagy.
Moreover, it should be taken into consideration that autophagy is a double-edged sword. Although increased autophagy is beneficial for efficient clearance of abnormal aggregations in axons, it might lead to the pathological axon loss and neuronal cell death under certain conditions. A better understanding of the causative link between dysfunctional autophagy and axon degeneration could help decipher the undergoing process in neurodegeneration and develop targeted therapeutic treatments for neurodegenerative diseases via modulating autophagy activity.
Acknowledgements
This work was supported by National Natural Science Foundation of China (No. 81673209).
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Luo L, O’Leary DD. Axon retraction and degeneration in development and disease. Annu Rev Neurosci. 2005;28:127–156. doi: 10.1146/annurev.neuro.28.061604.135632. [DOI] [PubMed] [Google Scholar]
- 2.Vargas ME, Barres BA. Why is Wallerian degeneration in the CNS so slow? Annu Rev Neurosci. 2007;30:153–179. doi: 10.1146/annurev.neuro.30.051606.094354. [DOI] [PubMed] [Google Scholar]
- 3.Scheib J, Hoke A. Advances in peripheral nerve regeneration. Nat Rev Neurol. 2013;9:668–676. doi: 10.1038/nrneurol.2013.227. [DOI] [PubMed] [Google Scholar]
- 4.Adalbert R, Morreale G, Paizs M, Conforti L, Walker SA, Roderick HL, Bootman MD, Siklos L, Coleman MP. Intra-axonal calcium changes after axotomy in wild-type and slow Wallerian degeneration axons. Neuroscience. 2012;225:44–54. doi: 10.1016/j.neuroscience.2012.08.056. [DOI] [PubMed] [Google Scholar]
- 5.Raff MC, Whitmore AV, Finn JT. Axonal self-destruction and neurodegeneration. Science. 2002;296:868–871. doi: 10.1126/science.1068613. [DOI] [PubMed] [Google Scholar]
- 6.Coleman M. Axon degeneration mechanisms: commonality amid diversity. Nat Rev Neurosci. 2005;6:889–898. doi: 10.1038/nrn1788. [DOI] [PubMed] [Google Scholar]
- 7.Neukomm LJ, Freeman MR. Diverse cellular and molecular modes of axon degeneration. Trends Cell Biol. 2014;24:515–523. doi: 10.1016/j.tcb.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischer LR, Glass JD. Axonal degeneration in motor neuron disease. Neuro-degener Dis. 2007;4:431–442. doi: 10.1159/000107704. [DOI] [PubMed] [Google Scholar]
- 9.Burke RE, O’Malley K. Axon degeneration in Parkinson’s disease. Exp Neurol. 2013;246:72–83. doi: 10.1016/j.expneurol.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lunn ER, Perry VH, Brown MC, Rosen H, Gordon S. Absence of Wallerian degeneration does not hinder regeneration in peripheral nerve. Eur J Neurosci. 1989;1:27–33. doi: 10.1111/j.1460-9568.1989.tb00771.x. [DOI] [PubMed] [Google Scholar]
- 11.Kegel KB, Kim M, Sapp E, McIntyre C, Castano JG, Aronin N, DiFiglia M. Huntingtin expression stimulates endosomal-lysosomal activity, endosome tubulation, and autophagy. J Neurosci. 2000;20:7268–7278. doi: 10.1523/JNEUROSCI.20-19-07268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 13.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 14.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 15.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 16.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parzych KR, Klionsky DJ. An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal. 2014;20:460–473. doi: 10.1089/ars.2013.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rubinsztein DC, Shpilka T, Elazar Z. Mechanisms of autophagosome biogenesis. Curr Biol. 2012;22:R29–R34. doi: 10.1016/j.cub.2011.11.034. [DOI] [PubMed] [Google Scholar]
- 20.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Bi. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki H, Kaizuka T, Mizushima N, Noda NN. Structure of the Atg101–Atg13 complex reveals essential roles of Atg101 in autophagy initiation. Nat Struct Mol Biol. 2015;22:572–580. doi: 10.1038/nsmb.3036. [DOI] [PubMed] [Google Scholar]
- 22.Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.e08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy. 2005;1:46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- 25.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 26.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 27.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 28.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2004;36:2503–2518. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M, Korolchuk VI, Lichtenberg M, Luo S, et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev. 2010;90:1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 31.Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 32.Maday S, Wallace KE, Holzbaur EL. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J Cell Biol. 2012;196:407–417. doi: 10.1083/jcb.201106120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee S, Sato Y, Nixon RA. Lysosomal proteolysis inhibition selectively disrupts axonal transport of degradative organelles and causes an Alzheimer’s-like axonal dystrophy. J Neurosci. 2011;31:7817–7830. doi: 10.1523/JNEUROSCI.6412-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yue Z. Regulation of neuronal autophagy in axon: implication of autophagy in axonal function and dysfunction/degeneration. Autophagy. 2007;3:139–141. doi: 10.4161/auto.3602. [DOI] [PubMed] [Google Scholar]
- 35.Xie R, Nguyen S, McKeehan WL, Liu L. Acetylated microtubules are required for fusion of autophagosomes with lysosomes. BMC Cell Biol. 2010;11:89. doi: 10.1186/1471-2121-11-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang QJ, Ding Y, Kohtz DS, Mizushima N, Cristea IM, Rout MP, Chait BT, Zhong Y, Heintz N, Yue Z. Induction of autophagy in axonal dystrophy and degeneration. J Neurosci. 2006;26:8057–8068. doi: 10.1523/JNEUROSCI.2261-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fu MM, Nirschl JJ, Holzbaur EL. LC3 binding to the scaffolding protein JIP1 regulates processive dynein-driven transport of autophagosomes. Dev Cell. 2014;29:577–590. doi: 10.1016/j.devcel.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maday S, Holzbaur EL. Compartment-specific regulation of autophagy in primary neurons. J Neurosci. 2016;36:5933–5945. doi: 10.1523/JNEUROSCI.4401-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng XT, Zhou B, Lin MY, Cai Q, Sheng ZH. Axonal autophagosomes recruit dynein for retrograde transport through fusion with late endosomes. J Cell Biol. 2015;209:377–386. doi: 10.1083/jcb.201412046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.e03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, Nixon RA. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer’s disease. J Neurosci. 2008;28:6926–6937. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ban BK, Jun MH, Ryu HH, Jang DJ, Ahmad ST, Lee JA. Autophagy negatively regulates early axon growth in cortical neurons. Mol Cell Biol. 2013;33:3907–3919. doi: 10.1128/MCB.00627-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie Y, Zhou B, Lin MY, Sheng ZH. Progressive endolysosomal deficits impair autophagic clearance beginning at early asymptomatic stages in fALS mice. Autophagy. 2015;11:1934–1936. doi: 10.1080/15548627.2015.1084460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, Cuervo AM. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol. 2005;64:113–122. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- 45.Li H, Li SH, Yu ZX, Shelbourne P, Li XJ. Huntingtin aggregate-associated axonal degeneration is an early pathological event in Huntington’s disease mice. J Neurosci. 2001;21:8473–8481. doi: 10.1523/JNEUROSCI.21-21-08473.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dauer W, Przedborski S. Parkinson’s disease: mechanisms and models. Neuron. 2003;39:889–909. doi: 10.1016/S0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- 47.Munemasa Y, Kitaoka Y. Autophagy in axonal degeneration in glaucomatous optic neuropathy. Prog Retin Eye Res. 2015;47:1–18. doi: 10.1016/j.preteyeres.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 48.Williams A, Jahreiss L, Sarkar S, Saiki S, Menzies FM, Ravikumar B, Rubinsztein DC. Aggregate-prone proteins are cleared from the cytosol by autophagy: therapeutic implications. Curr Top Dev Biol. 2006;76:89–101. doi: 10.1016/S0070-2153(06)76003-3. [DOI] [PubMed] [Google Scholar]
- 49.Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nat Rev Drug Discov. 2007;6:304–312. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- 50.Martinez-Vicente M, Cuervo AM. Autophagy and neurodegeneration: when the cleaning crew goes on strike. Lancet Neurol. 2007;6:352–361. doi: 10.1016/S1474-4422(07)70076-5. [DOI] [PubMed] [Google Scholar]
- 51.Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA, Small S, Spencer B, Rockenstein E, Levine B, Wyss-Coray T. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Investig. 2008;118:2190–2199. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rohn TT, Wirawan E, Brown RJ, Harris JR, Masliah E, Vandenabeele P. Depletion of Beclin-1 due to proteolytic cleavage by caspases in the Alzheimer’s disease brain. Neurobiol Dis. 2011;43:68–78. doi: 10.1016/j.nbd.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Djavaheri-Mergny M, Maiuri MC, Kroemer G. Cross talk between apoptosis and autophagy by caspase-mediated cleavage of Beclin 1. Oncogene. 2010;29:1717–1719. doi: 10.1038/onc.2009.519. [DOI] [PubMed] [Google Scholar]
- 54.Zhu Y, Zhao L, Liu L, Gao P, Tian W, Wang X, Jin H, Xu H, Chen Q. Beclin 1 cleavage by caspase-3 inactivates autophagy and promotes apoptosis. Protein Cell. 2010;1:468–477. doi: 10.1007/s13238-010-0048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Russo R, Berliocchi L, Adornetto A, Varano GP, Cavaliere F, Nucci C, Rotiroti D, Morrone LA, Bagetta G, Corasaniti MT. Calpain-mediated cleavage of Beclin-1 and autophagy deregulation following retinal ischemic injury in vivo. Cell Death Dis. 2011;2:e144. doi: 10.1038/cddis.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song F, Han X, Zeng T, Zhang C, Zou C, Xie K. Changes in beclin-1 and micro-calpain expression in tri-ortho-cresyl phosphate-induced delayed neuropathy. Toxicol Lett. 2012;210:276–284. doi: 10.1016/j.toxlet.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 57.Huang J, Yang J, Shen Y, Jiang H, Han C, Zhang G, Liu L, Xu X, Li J, Lin Z, et al. HMGB1 mediates autophagy dysfunction via perturbing Beclin1-Vps34 complex in dopaminergic cell model. Front Mol Neurosci. 2017;10:13. doi: 10.3389/fnmol.2017.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Komatsu M, Wang QJ, Holstein GR, Friedrich VL, Jr, Iwata J, Kominami E, Chait BT, Tanaka K, Yue Z. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci USA. 2007;104:14489–14494. doi: 10.1073/pnas.0701311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Friedman LG, Lachenmayer ML, Wang J, He L, Poulose SM, Komatsu M, Holstein GR, Yue Z. Disrupted autophagy leads to dopaminergic axon and dendrite degeneration and promotes presynaptic accumulation of alpha-synuclein and LRRK2 in the brain. J Neurosci. 2012;32:7585–7593. doi: 10.1523/JNEUROSCI.5809-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Joo JH, Wang B, Frankel E, Ge L, Xu L, Iyengar R, Li-Harms X, Wright C, Shaw TI, Lindsten T, et al. The noncanonical role of ULK/ATG1 in ER-to-Golgi trafficking is essential for cellular homeostasis. Mol Cell. 2016;62:491–506. doi: 10.1016/j.molcel.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lu Q, Yang P, Huang X, Hu W, Guo B, Wu F, Lin L, Kovacs AL, Yu L, Zhang H. The WD40 repeat PtdIns(3)P-binding protein EPG-6 regulates progression of omegasomes to autophagosomes. Dev Cell. 2011;21:343–357. doi: 10.1016/j.devcel.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 62.Saitsu H, Nishimura T, Muramatsu K, Kodera H, Kumada S, Sugai K, Kasai-Yoshida E, Sawaura N, Nishida H, Hoshino A et al (2013) De novo mutations in the autophagy gene WDR45 cause static encephalopathy of childhood with neurodegeneration in adulthood. Nat Genet 45:445–449, 449e441 [DOI] [PubMed]
- 63.Saleem S, Biswas SC. Tribbles pseudokinase 3 induces both apoptosis and autophagy in amyloid-beta-induced neuronal death. J Biol Chem. 2017;292:2571–2585. doi: 10.1074/jbc.M116.744730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wong ES, Tan JM, Soong WE, Hussein K, Nukina N, Dawson VL, Dawson TM, Cuervo AM, Lim KL. Autophagy-mediated clearance of aggresomes is not a universal phenomenon. Hum Mol Genet. 2008;17:2570–2582. doi: 10.1093/hmg/ddn157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O’Kane CJ, Rubinsztein DC. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 66.Hu YB, Zou Y, Huang Y, Zhang YF, Lourenco GF, Chen SD, Halliday GM, Wang G, Ren RJ. ROCK1 is associated with Alzheimer’s disease-specific plaques, as well as enhances autophagosome formation but not autophagic Abeta clearance. Front Cell Neurosci. 2016;10:253. doi: 10.3389/fncel.2016.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Crippa V, Sau D, Rusmini P, Boncoraglio A, Onesto E, Bolzoni E, Galbiati M, Fontana E, Marino M, Carra S, et al. The small heat shock protein B8 (HspB8) promotes autophagic removal of misfolded proteins involved in amyotrophic lateral sclerosis (ALS) Hum Mol Genet. 2010;19:3440–3456. doi: 10.1093/hmg/ddq257. [DOI] [PubMed] [Google Scholar]
- 68.Li L, Zhang X, Le W. Altered macroautophagy in the spinal cord of SOD1 mutant mice. Autophagy. 2008;4:290–293. doi: 10.4161/auto.5524. [DOI] [PubMed] [Google Scholar]
- 69.Nassif M, Valenzuela V, Rojas-Rivera D, Vidal R, Matus S, Castillo K, Fuentealba Y, Kroemer G, Levine B, Hetz C. Pathogenic role of BECN1/Beclin 1 in the development of amyotrophic lateral sclerosis. Autophagy. 2014;10:1256–1271. doi: 10.4161/auto.28784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lipinski MM, Zheng B, Lu T, Yan Z, Py BF, Ng A, Xavier RJ, Li C, Yankner BA, Scherzer CR, Yuan J. Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer’s disease. Proc Natl Acad Sci USA. 2010;107:14164–14169. doi: 10.1073/pnas.1009485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tong Y, Song F. Intracellular calcium signaling regulates autophagy via calcineurin-mediated TFEB dephosphorylation. Autophagy. 2015;11:1192–1195. doi: 10.1080/15548627.2015.1054594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koch JC, Knoferle J, Tonges L, Ostendorf T, Bahr M, Lingor P. Acute axonal degeneration in vivo is attenuated by inhibition of autophagy in a calcium-dependent manner. Autophagy. 2010;6:658–659. doi: 10.4161/auto.6.5.12188. [DOI] [PubMed] [Google Scholar]
- 73.Rubinsztein DC. Lessons from animal models of Huntington’s disease. Trends Genet. 2002;18:202–209. doi: 10.1016/S0168-9525(01)02625-7. [DOI] [PubMed] [Google Scholar]
- 74.Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, de Vries R, Arias E, Harris S, Sulzer D, Cuervo AM. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Haack TB, Ignatius E, Calvo-Garrido J, Iuso A, Isohanni P, Maffezzini C, Lonnqvist T, Suomalainen A, Gorza M, Kremer LS, et al. Absence of the autophagy adaptor SQSTM1/p62 causes childhood-onset neurodegeneration with ataxia, dystonia, and gaze palsy. Am J Hum Genet. 2016;99:735–743. doi: 10.1016/j.ajhg.2016.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Deng Z, Purtell K, Lachance V, Wold MS, Chen S, Yue Z. Autophagy receptors and neurodegenerative diseases. Trends Cell Biol. 2017;27:491–504. doi: 10.1016/j.tcb.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 77.Koch JC, Lingor P. The role of autophagy in axonal degeneration of the optic nerve. Exp Eye Res. 2016;144:81–89. doi: 10.1016/j.exer.2015.08.016. [DOI] [PubMed] [Google Scholar]
- 78.Reid E, Kloos M, Ashley-Koch A, Hughes L, Bevan S, Svenson IK, Graham FL, Gaskell PC, Dearlove A, Pericak-Vance MA, et al. A kinesin heavy chain (KIF5A) mutation in hereditary spastic paraplegia (SPG10) Am J Hum Genet. 2002;71:1189–1194. doi: 10.1086/344210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, Kinoshita Y, Kamada M, Nodera H, Suzuki H, et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–226. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- 80.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hirokawa N, Takemura R. Molecular motors and mechanisms of directional transport in neurons. Nat Rev Neurosci. 2005;6:201–214. doi: 10.1038/nrn1624. [DOI] [PubMed] [Google Scholar]
- 82.Goldstein LS, Yang Z. Microtubule-based transport systems in neurons: the roles of kinesins and dyneins. Annu Rev Neurosci. 2000;23:39–71. doi: 10.1146/annurev.neuro.23.1.39. [DOI] [PubMed] [Google Scholar]
- 83.Morfini GA, Burns M, Binder LI, Kanaan NM, LaPointe N, Bosco DA, Brown RH, Jr, Brown H, Tiwari A, Hayward L, et al. Axonal transport defects in neurodegenerative diseases. J Neurosci. 2009;29:12776–12786. doi: 10.1523/JNEUROSCI.3463-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang X, Li L, Chen S, Yang D, Wang Y, Zhang X, Wang Z, Le W. Rapamycin treatment augments motor neuron degeneration in SOD1(G93A) mouse model of amyotrophic lateral sclerosis. Autophagy. 2011;7:412–425. doi: 10.4161/auto.7.4.14541. [DOI] [PubMed] [Google Scholar]
- 85.Lu X, Kim-Han JS, Harmon S, Sakiyama-Elbert SE, O’Malley KL. The Parkinsonian mimetic, 6-OHDA, impairs axonal transport in dopaminergic axons. Mol Neurodegener. 2014;9:17. doi: 10.1186/1750-1326-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim-Han JS, Antenor-Dorsey JA, O’Malley KL. The parkinsonian mimetic, MPP+, specifically impairs mitochondrial transport in dopamine axons. J Neurosci. 2011;31:7212–7221. doi: 10.1523/JNEUROSCI.0711-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Farrer MJ, Hulihan MM, Kachergus JM, Dachsel JC, Stoessl AJ, Grantier LL, Calne S, Calne DB, Lechevalier B, Chapon F, et al. DCTN1 mutations in Perry syndrome. Nat Genet. 2009;41:163–165. doi: 10.1038/ng.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hafezparast M, Klocke R, Ruhrberg C, Marquardt A, Ahmad-Annuar A, Bowen S, Lalli G, Witherden AS, Hummerich H, Nicholson S, et al. Mutations in dynein link motor neuron degeneration to defects in retrograde transport. Science. 2003;300:808–812. doi: 10.1126/science.1083129. [DOI] [PubMed] [Google Scholar]
- 89.Puls I, Oh SJ, Sumner CJ, Wallace KE, Floeter MK, Mann EA, Kennedy WR, Wendelschafer-Crabb G, Vortmeyer A, Powers R, et al. Distal spinal and bulbar muscular atrophy caused by dynactin mutation. Ann Neurol. 2005;57:687–694. doi: 10.1002/ana.20468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ravikumar B, Acevedo-Arozena A, Imarisio S, Berger Z, Vacher C, O’Kane CJ, Brown SD, Rubinsztein DC. Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat Genet. 2005;37:771–776. doi: 10.1038/ng1591. [DOI] [PubMed] [Google Scholar]
- 91.Sanchez-Varo R, Trujillo-Estrada L, Sanchez-Mejias E, Torres M, Baglietto-Vargas D, Moreno-Gonzalez I, De Castro V, Jimenez S, Ruano D, Vizuete M, et al. Abnormal accumulation of autophagic vesicles correlates with axonal and synaptic pathology in young Alzheimer’s mice hippocampus. Acta Neuropathol. 2012;123:53–70. doi: 10.1007/s00401-011-0896-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Takagishi Y, Katanosaka K, Mizoguchi H, Murata Y. Disrupted axon-glia interactions at the paranode in myelinated nerves cause axonal degeneration and neuronal cell death in the aged Caspr mutant mouse shambling. Neurobiol Aging. 2016;43:34–46. doi: 10.1016/j.neurobiolaging.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 93.Seidel K, den Dunnen WF, Schultz C, Paulson H, Frank S, de Vos RA, Brunt ER, Deller T, Kampinga HH, Rub U. Axonal inclusions in spinocerebellar ataxia type 3. Acta Neuropathol. 2010;120:449–460. doi: 10.1007/s00401-010-0717-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tammineni P, Cai Q. Defective retrograde transport impairs autophagic clearance in Alzheimer disease neurons. Autophagy. 2017;13:982–984. doi: 10.1080/15548627.2017.1291114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Knoferle J, Koch JC, Ostendorf T, Michel U, Planchamp V, Vutova P, Tonges L, Stadelmann C, Bruck W, Bahr M, Lingor P. Mechanisms of acute axonal degeneration in the optic nerve in vivo. Proc Natl Acad Sci USA. 2010;107:6064–6069. doi: 10.1073/pnas.0909794107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yu WH, Cuervo AM, Kumar A, Peterhoff CM, Schmidt SD, Lee JH, Mohan PS, Mercken M, Farmery MR, Tjernberg LO, et al. Macroautophagy–a novel Beta-amyloid peptide-generating pathway activated in Alzheimer’s disease. J Cell Biol. 2005;171:87–98. doi: 10.1083/jcb.200505082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wolfe DM, Lee JH, Kumar A, Lee S, Orenstein SJ, Nixon RA. Autophagy failure in Alzheimer’s disease and the role of defective lysosomal acidification. Eur J Neurosci. 2013;37:1949–1961. doi: 10.1111/ejn.12169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, Wolfe DM, Martinez-Vicente M, Massey AC, Sovak G, et al. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 2010;141:1146–1158. doi: 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Settembre C, Fraldi A, Jahreiss L, Spampanato C, Venturi C, Medina D, de Pablo R, Tacchetti C, Rubinsztein DC, Ballabio A. A block of autophagy in lysosomal storage disorders. Hum Mol Genet. 2008;17:119–129. doi: 10.1093/hmg/ddm289. [DOI] [PubMed] [Google Scholar]
- 100.Tanaka Y, Guhde G, Suter A, Eskelinen EL, Hartmann D, Lullmann-Rauch R, Janssen PM, Blanz J, von Figura K, Saftig P. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 2000;406:902–906. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- 101.Fukuda T, Roberts A, Ahearn M, Zaal K, Ralston E, Plotz PH, Raben N. Autophagy and lysosomes in Pompe disease. Autophagy. 2006;2:318–320. doi: 10.4161/auto.2984. [DOI] [PubMed] [Google Scholar]
- 102.Koike M, Shibata M, Waguri S, Yoshimura K, Tanida I, Kominami E, Gotow T, Peters C, von Figura K, Mizushima N, et al. Participation of autophagy in storage of lysosomes in neurons from mouse models of neuronal ceroid-lipofuscinoses (Batten disease) Am J Pathol. 2005;167:1713–1728. doi: 10.1016/S0002-9440(10)61253-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Onesto E, Rusmini P, Crippa V, Ferri N, Zito A, Galbiati M, Poletti A. Muscle cells and motoneurons differentially remove mutant SOD1 causing familial amyotrophic lateral sclerosis. J Neurochem. 2011;118:266–280. doi: 10.1111/j.1471-4159.2011.07298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang Y, Chen S, Zhang J, Li C, Sun Y, Zhang L, Zheng X. Stimulation of autophagy prevents amyloid-beta peptide-induced neuritic degeneration in PC12 cells. J Alzheimers Dis. 2014;40:929–939. doi: 10.3233/JAD-132102. [DOI] [PubMed] [Google Scholar]
- 105.Clarke JP, Mearow K. Autophagy inhibition in endogenous and nutrient-deprived conditions reduces dorsal root ganglia neuron survival and neurite growth in vitro. J Neurosci Res. 2016;94:653–670. doi: 10.1002/jnr.23733. [DOI] [PubMed] [Google Scholar]
- 106.Costa LG, Garrick JM, Roque PJ, Pellacani C. Mechanisms of neuroprotection by quercetin: counteracting oxidative stress and more. Oxid Med Cell Longev. 2016;2016:2986796. doi: 10.1155/2016/2986796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem. 2007;282:5641–5652. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 108.Kim D, Nguyen MD, Dobbin MM, Fischer A, Sananbenesi F, Rodgers JT, Delalle I, Baur JA, Sui G, Armour SM, et al. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer’s disease and amyotrophic lateral sclerosis. EMBO J. 2007;26:3169–3179. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wu Y, Li X, Zhu JX, Xie W, Le W, Fan Z, Jankovic J, Pan T. Resveratrol-activated AMPK/SIRT1/autophagy in cellular models of Parkinson’s disease. Neurosignals. 2011;19:163–174. doi: 10.1159/000328516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tsujimoto Y, Shimizu S. Another way to die: autophagic programmed cell death. Cell Death Differ. 2005;12(Suppl 2):1528–1534. doi: 10.1038/sj.cdd.4401777. [DOI] [PubMed] [Google Scholar]
- 111.Shen S, Kepp O, Kroemer G. The end of autophagic cell death? Autophagy. 2012;8:1–3. doi: 10.4161/auto.8.1.16618. [DOI] [PubMed] [Google Scholar]
- 112.Shen HM, Codogno P. Autophagic cell death: Loch Ness monster or endangered species? Autophagy. 2011;7:457–465. doi: 10.4161/auto.7.5.14226. [DOI] [PubMed] [Google Scholar]
- 113.Wen Y, Zhai RG, Kim MD. The role of autophagy in Nmnat-mediated protection against hypoxia-induced dendrite degeneration. Mol Cell Neurosci. 2013;52:140–151. doi: 10.1016/j.mcn.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xie C, Ginet V, Sun Y, Koike M, Zhou K, Li T, Li H, Li Q, Wang X, Uchiyama Y, et al. Neuroprotection by selective neuronal deletion of Atg7 in neonatal brain injury. Autophagy. 2016;12:410–423. doi: 10.1080/15548627.2015.1132134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cheng HC, Kim SR, Oo TF, Kareva T, Yarygina O, Rzhetskaya M, Wang C, During M, Talloczy Z, Tanaka K, et al. Akt suppresses retrograde degeneration of dopaminergic axons by inhibition of macroautophagy. J Neurosci. 2011;31:2125–2135. doi: 10.1523/JNEUROSCI.5519-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Staats KA, Hernandez S, Schonefeldt S, Bento-Abreu A, Dooley J, Van Damme P, Liston A, Robberecht W, Van Den Bosch L. Rapamycin increases survival in ALS mice lacking mature lymphocytes. Mol Neurodegener. 2013;8:31. doi: 10.1186/1750-1326-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Antonioli M, Di Rienzo M, Piacentini M, Fimia GM. Emerging mechanisms in initiating and terminating autophagy. Trends Biochem Sci. 2017;42:28–41. doi: 10.1016/j.tibs.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 118.Salminen A, Kaarniranta K, Kauppinen A, Ojala J, Haapasalo A, Soininen H, Hiltunen M. Impaired autophagy and APP processing in Alzheimer’s disease: the potential role of Beclin 1 interactome. Prog Neurobiol. 2013;106–107:33–54. doi: 10.1016/j.pneurobio.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 119.Lim J, Yue Z. Neuronal aggregates: formation, clearance, and spreading. Dev Cell. 2015;32:491–501. doi: 10.1016/j.devcel.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Weinberg SE, Sena LA, Chandel NS. Mitochondria in the regulation of innate and adaptive immunity. Immunity. 2015;42:406–417. doi: 10.1016/j.immuni.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Green DR, Galluzzi L, Kroemer G. Mitochondria and the autophagy–inflammation–cell death axis in organismal aging. Science. 2011;333:1109–1112. doi: 10.1126/science.1201940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Scheibye-Knudsen M, Fang EF, Croteau DL, Wilson DM, 3rd, Bohr VA. Protecting the mitochondrial powerhouse. Trends Cell Biol. 2015;25:158–170. doi: 10.1016/j.tcb.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Khandelwal PJ, Herman AM, Hoe HS, Rebeck GW, Moussa CE. Parkin mediates beclin-dependent autophagic clearance of defective mitochondria and ubiquitinated Abeta in AD models. Hum Mol Genet. 2011;20:2091–2102. doi: 10.1093/hmg/ddr091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ashrafi G, Schwarz TL. PINK1- and PARK2-mediated local mitophagy in distal neuronal axons. Autophagy. 2015;11:187–189. doi: 10.1080/15548627.2014.996021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sato M, Sato K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science. 2011;334:1141–1144. doi: 10.1126/science.1210333. [DOI] [PubMed] [Google Scholar]
- 127.Martinez-Vicente M. Neuronal mitophagy in neurodegenerative diseases. Front Mol Neurosci. 2017;10:64. doi: 10.3389/fnmol.2017.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nguyen TN, Padman BS, Lazarou M. Deciphering the molecular signals of PINK1/Parkin mitophagy. Trends Cell Biol. 2016;26:733–744. doi: 10.1016/j.tcb.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 129.Ebrahimi-Fakhari D, Saffari A, Wahlster L, Di Nardo A, Turner D, Lewis TL, Jr, Conrad C, Rothberg JM, Lipton JO, Kolker S, et al. Impaired mitochondrial dynamics and mitophagy in neuronal models of tuberous sclerosis complex. Cell Rep. 2016;17:1053–1070. doi: 10.1016/j.celrep.2016.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 131.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 132.Hernandez DG, Reed X, Singleton AB. Genetics in Parkinson disease: Mendelian versus non-Mendelian inheritance. J Neurochem. 2016;139(Suppl 1):59–74. doi: 10.1111/jnc.13593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Martinez TN, Greenamyre JT. Toxin models of mitochondrial dysfunction in Parkinson’s disease. Antioxid Redox Signal. 2012;16:920–934. doi: 10.1089/ars.2011.4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kerr JS, Adriaanse BA, Greig NH, Mattson MP, Cader MZ, Bohr VA, Fang EF. Mitophagy and Alzheimer’s disease: cellular and molecular mechanisms. Trends Neurosci. 2017;40:151–166. doi: 10.1016/j.tins.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Matsumoto G, Shimogori T, Hattori N, Nukina N. TBK1 controls autophagosomal engulfment of polyubiquitinated mitochondria through p62/SQSTM1 phosphorylation. Hum Mol Genet. 2015;24:4429–4442. doi: 10.1093/hmg/ddv179. [DOI] [PubMed] [Google Scholar]
- 136.Heo JM, Ordureau A, Paulo JA, Rinehart J, Harper JW. The PINK1–PARKIN mitochondrial ubiquitylation pathway drives a program of OPTN/NDP52 recruitment and TBK1 activation to promote mitophagy. Mol Cell. 2015;60:7–20. doi: 10.1016/j.molcel.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Freischmidt A, Muller K, Ludolph AC, Weishaupt JH, Andersen PM. Association of mutations in TBK1 with sporadic and familial amyotrophic lateral sclerosis and frontotemporal dementia. JAMA Neurol. 2017;74:110–113. doi: 10.1001/jamaneurol.2016.3712. [DOI] [PubMed] [Google Scholar]
- 138.Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, Gibbs JR, Brunetti M, Gronka S, Wuu J, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Spina S, Van Laar AD, Murrell JR, Hamilton RL, Kofler JK, Epperson F, Farlow MR, Lopez OL, Quinlan J, DeKosky ST, Ghetti B. Phenotypic variability in three families with valosin-containing protein mutation. Eur J Neurol. 2013;20:251–258. doi: 10.1111/j.1468-1331.2012.03831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Li L, Chen Y, Gibson SB. Starvation-induced autophagy is regulated by mitochondrial reactive oxygen species leading to AMPK activation. Cell Signal. 2013;25:50–65. doi: 10.1016/j.cellsig.2012.09.020. [DOI] [PubMed] [Google Scholar]
- 141.Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zmijewski JW, Banerjee S, Bae H, Friggeri A, Lazarowski ER, Abraham E. Exposure to hydrogen peroxide induces oxidation and activation of AMP-activated protein kinase. J Biol Chem. 2010;285:33154–33164. doi: 10.1074/jbc.M110.143685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouyssegur J, Mazure NM. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol. 2009;29:2570–2581. doi: 10.1128/MCB.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Decuypere JP, Parys JB, Bultynck G. Regulation of the autophagic bcl-2/beclin 1 interaction. Cells. 2012;1:284–312. doi: 10.3390/cells1030284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yan T, Feng Y, Zhai Q. Axon degeneration: mechanisms and implications of a distinct program from cell death. Neurochem Int. 2010;56:529–534. doi: 10.1016/j.neuint.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 146.Beirowski B, Adalbert R, Wagner D, Grumme DS, Addicks K, Ribchester RR, Coleman MP. The progressive nature of Wallerian degeneration in wild-type and slow Wallerian degeneration (WldS) nerves. BMC Neurosci. 2005;6:6. doi: 10.1186/1471-2202-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Osterloh JM, Yang J, Rooney TM, Fox AN, Adalbert R, Powell EH, Sheehan AE, Avery MA, Hackett R, Logan MA, et al. dSarm/Sarm1 is required for activation of an injury-induced axon death pathway. Science. 2012;337:481–484. doi: 10.1126/science.1223899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Summers DW, DiAntonio A, Milbrandt J. Mitochondrial dysfunction induces Sarm1-dependent cell death in sensory neurons. J Neurosci. 2014;34:9338–9350. doi: 10.1523/JNEUROSCI.0877-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Essuman K, Summers DW, Sasaki Y, Mao X, DiAntonio A, Milbrandt J. The SARM1 Toll/interleukin-1 receptor domain possesses intrinsic NAD+ cleavage activity that promotes pathological axonal degeneration. Neuron. 2017;93(1334–1343):e1335. doi: 10.1016/j.neuron.2017.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gerdts J, Brace EJ, Sasaki Y, DiAntonio A, Milbrandt J. SARM1 activation triggers axon degeneration locally via NAD(+) destruction. Science. 2015;348:453–457. doi: 10.1126/science.1258366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gerdts J, Summers DW, Milbrandt J, DiAntonio A. Axon self-destruction: new links among SARM1, MAPKs, and NAD+ metabolism. Neuron. 2016;89:449–460. doi: 10.1016/j.neuron.2015.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yang J, Wu ZH, Renier N, Simon DJ, Uryu K, Park DS, Greer PA, Tournier C, Davis RJ, Tessier-Lavigne M. Pathological axonal death through a MAPK cascade that triggers a local energy deficit. Cell. 2015;160:161–176. doi: 10.1016/j.cell.2014.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Walker LJ, Summers DW, Sasaki Y, Brace EJ, Milbrandt J, DiAntonio A. MAPK signaling promotes axonal degeneration by speeding the turnover of the axonal maintenance factor NMNAT2. Life. 2017;6:e22540. doi: 10.7554/eLife.22540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gamage KK, Cheng I, Park RE, Karim MS, Edamura K, Hughes C, Spano AJ, Erisir A, Deppmann CD. Death receptor 6 promotes wallerian degeneration in peripheral axons. Curr Biol. 2017;27:890–896. doi: 10.1016/j.cub.2017.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yang J, Weimer RM, Kallop D, Olsen O, Wu Z, Renier N, Uryu K, Tessier-Lavigne M. Regulation of axon degeneration after injury and in development by the endogenous calpain inhibitor calpastatin. Neuron. 2013;80:1175–1189. doi: 10.1016/j.neuron.2013.08.034. [DOI] [PubMed] [Google Scholar]
- 156.Kitaoka Y, Munemasa Y, Kojima K, Hirano A, Ueno S, Takagi H. Axonal protection by Nmnat3 overexpression with involvement of autophagy in optic nerve degeneration. Cell Death Dis. 2013;4:e860. doi: 10.1038/cddis.2013.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wakatsuki S, Saitoh F, Araki T. ZNRF1 promotes Wallerian degeneration by degrading AKT to induce GSK3B-dependent CRMP2 phosphorylation. Nat Cell Biol. 2011;13:1415–1423. doi: 10.1038/ncb2373. [DOI] [PubMed] [Google Scholar]
- 158.Wakatsuki S, Tokunaga S, Shibata M, Araki T. GSK3B-mediated phosphorylation of MCL1 regulates axonal autophagy to promote Wallerian degeneration. J Cell Biol. 2017;216:477–493. doi: 10.1083/jcb.201606020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Conforti L, Gilley J, Coleman MP. Wallerian degeneration: an emerging axon death pathway linking injury and disease. Nat Rev Neurosci. 2014;15:394–409. doi: 10.1038/nrn3680. [DOI] [PubMed] [Google Scholar]
- 160.He M, Ding Y, Chu C, Tang J, Xiao Q, Luo ZG. Autophagy induction stabilizes microtubules and promotes axon regeneration after spinal cord injury. Proc Natl Acad Sci USA. 2016;113(40):11324–11329. doi: 10.1073/pnas.1611282113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lin WJ, Kuang HY. Oxidative stress induces autophagy in response to multiple noxious stimuli in retinal ganglion cells. Autophagy. 2014;10:1692–1701. doi: 10.4161/auto.36076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Geden MJ, Deshmukh M. Axon degeneration: context defines distinct pathways. Curr Opin Neurobiol. 2016;39:108–115. doi: 10.1016/j.conb.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Simon DJ, Pitts J, Hertz NT, Yang J, Yamagishi Y, Olsen O, Tesic Mark M, Molina H, Tessier-Lavigne M. Axon degeneration gated by retrograde activation of somatic pro-apoptotic signaling. Cell. 2016;164:1031–1045. doi: 10.1016/j.cell.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Peles E, Salzer JL. Molecular domains of myelinated axons. Curr Opin Neurobiol. 2000;10:558–565. doi: 10.1016/S0959-4388(00)00122-7. [DOI] [PubMed] [Google Scholar]
- 165.Yaron A, Schuldiner O. Common and divergent mechanisms in developmental neuronal remodeling and dying back neurodegeneration. Curr Biol. 2016;26:R628–R639. doi: 10.1016/j.cub.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lu TY, MacDonald JM, Neukomm LJ, Sheehan AE, Bradshaw R, Logan MA, Freeman MR. Axon degeneration induces glial responses through Draper-TRAF4-JNK signalling. Nat Commun. 2017;8:14355. doi: 10.1038/ncomms14355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Musashe DT, Purice MD, Speese SD, Doherty J, Logan MA. Insulin-like signaling promotes glial phagocytic clearance of degenerating axons through regulation of draper. Cell Rep. 2016;16:1838–1850. doi: 10.1016/j.celrep.2016.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Purice MD, Speese SD, Logan MA. Delayed glial clearance of degenerating axons in aged Drosophila is due to reduced PI3K/Draper activity. Nat Commun. 2016;7:12871. doi: 10.1038/ncomms12871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Jang SY, Shin YK, Park SY, Park JY, Lee HJ, Yoo YH, Kim JK, Park HT. Autophagic myelin destruction by Schwann cells during Wallerian degeneration and segmental demyelination. Glia. 2016;64:730–742. doi: 10.1002/glia.22957. [DOI] [PubMed] [Google Scholar]
- 170.Gomez-Sanchez JA, Carty L, Iruarrizaga-Lejarreta M, Palomo-Irigoyen M, Varela-Rey M, Griffith M, Hantke J, Macias-Camara N, Azkargorta M, Aurrekoetxea I, et al. Schwann cell autophagy, myelinophagy, initiates myelin clearance from injured nerves. J Cell Biol. 2015;210:153–168. doi: 10.1083/jcb.201503019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Petralia RS, Schwartz CM, Wang YX, Kawamoto EM, Mattson MP, Yao PJ. Sonic hedgehog promotes autophagy in hippocampal neurons. Biol Open. 2013;2:499–504. doi: 10.1242/bio.20134275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yao PJ, Petralia RS, Mattson MP. Sonic Hedgehog signaling and hippocampal neuroplasticity. Trends Neurosci. 2016;39:840–850. doi: 10.1016/j.tins.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhong ZY, Umemura A, Sanchez-Lopez E, Liang S, Shalapour S, Wong J, He F, Boassa D, Perkins G, Ali SR, et al. NF-kappa B restricts inflammasome activation via elimination of damaged mitochondria. Cell. 2016;164:896–910. doi: 10.1016/j.cell.2015.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lupfer C, Thomas PG, Anand PK, Vogel P, Milasta S, Martinez J, Huang G, Green M, Kundu M, Chi H, et al. Receptor interacting protein kinase 2-mediated mitophagy regulates inflammasome activation during virus infection. Nat Immunol. 2013;14:480–488. doi: 10.1038/ni.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]