Abstract
Hepatocellular carcinoma (HCC) is one of the most common malignancies worldwide, with a high mortality rate. Its dismal prognosis is attributed to late diagnosis, high risk of recurrence and drug resistance. To improve the survival of patients with HCC, new approaches are required for early diagnosis, real-time monitoring and effective treatment. Exosomes are small membranous vesicles released by most cells that contain biological molecules and play a great role in intercellular communication under physiological or pathological conditions. In cancer, exosomes from tumor cells or non-tumor cells can be taken up by neighboring or distant target cells, and the cargoes in exosomes are functional to modulate the behaviors of tumors or reshape tumor microenvironment (TME). As essential components, non-coding RNAs (ncRNAs) are selectively enriched in exosomes, and exosomal ncRNAs participate in regulating specific aspects of tumor development, including tumorigenesis, tumor metastasis, angiogenesis, immunomodulation and drug resistance. Besides, dysregulated exosomal ncRNAs have emerged as potential biomarkers, and exosomes can serve as natural vehicles to deliver tumor-suppressed ncRNAs for treatment. In this review, we briefly summarize the biology of exosomes, the functions of exosomal ncRNAs in HCC development and their potential clinical applications, including as biomarkers and therapeutic tools.
Keywords: Hepatocellular carcinoma, Exosome, Exosomal non-coding RNA, Tumor biology, Biomarker, Delivery vehicle
Introduction
Liver cancer is the sixth common malignancy and the fourth leading cause of cancer-related death worldwide in 2018 [1]. Hepatocellular carcinoma (HCC) accounts for 80% of liver cancer, and it mainly occurs in the context of chronic inflammation and fibrosis that is caused by virus hepatitis, alcohol and non-alcohol fatty disease (NAFLD) [2–4]. The 5-year survival rate of HCC is lower than 20% [5], primarily attributed to difficulty of early diagnosis via current biomarkers [6]. Consequently, most HCC patients are diagnosed at an advanced stage, accompanied with intrahepatic or distant metastasis. When diagnosed, the patients are not eligible for curative treatments (resection or transplantation) while palliative treatments are the exclusive choices [7, 8]. For example, the multi-target kinase inhibitor sorafenib can prolong the survival of patients with advanced HCC for approximately 3 months. However, the response rate of sorafenib is low (~ 10%) and most patients develop disease progression due to drug resistance [9, 10]. Moreover, even for those receiving surgical resection, recurrence is a severe problem and half of the patients with recurrence die within 1 year [11, 12]. Thus, there is an urgent need to develop new approaches of early diagnosis, real-time monitoring, and effective treatment for HCC.
Liquid biopsy is a minimally invasive technology for detecting tumor cells or nucleic acids from body fluids, including circulating tumor cell (CTC), circulating tumor DNA (ctDNA) and exosome, which help diagnose cancer, detect disease progression or therapeutic response [13]. However, the numbers of CTCs are few and half-life of ctDNA is short. Thus, increasing studies have focused on exosomes, which are abundant in biofluids and keep stable for a long time. Exosomes are small extracellular vesicle (EVs) secreted by most cells and contain bioactive cargoes (proteins, nucleic acids, lipids). In cancer, exosomes from tumor cells or non-tumor cells can transfer the cargoes to recipient cells to modify their phenotypes and reshape tumor microenvironment (TME), affecting tumor development [14]. Thus, exosomes are essential messengers in tumor progression. Moreover, exosomes are widespread in diverse biofluids, e.g., blood, urine, saliva. The concentration of exosomes in cancer patients is higher than healthy individuals, and cargoes in exosomes can reflect the origin cells and real-time diseases states. Thus, exosomes and their cargoes are under investigation as potential biomarkers contributing to diagnosis [15], prognosis [16] and treatments [17] in cancer. What’s more, since exosomes are stable and with low immunogenicity, they are exploited as vehicles for carrying drugs and anti-tumor nucleic acids to treat cancer.
Non-coding RNAs (ncRNAs) are a class of RNAs without protein-coding capability, such as microRNAs (miRNAs), long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs). By interacting with DNAs, RNAs or proteins, ncRNAs can regulate gene expression on transcription, post-transcription, epigenetic levels and modulate cellular processes [18]. Emerging evidence suggests that various ncRNAs are dysregulated in cancer and participate in complex network of biological regulation, e.g., cell proliferation, migration, apoptosis [19]. Interestingly, ncRNAs are selectively enriched in exosomes, and exosomal ncRNAs can exert biological functions in recipient cells to affect the critical processes of tumor development, e.g., tumorigenesis, tumor metastasis, angiogenesis, immunomodulation and drug resistance [20]. Besides, due to aberrantly expressed, exosomal ncRNAs represent a powerful source of biomarkers [21, 22] and novel therapeutic targets. In this review, we mainly overview the biology of exosomes, the functions of exosomal ncRNAs in HCC development and their potential clinical applications, including as biomarkers or therapeutic tools.
The characteristics and biogenesis of exosomes
Exosomes are often referred to EVs, but they should not be confused. Different from other EVs, exosomes are 30–100 nm membranous vesicles of endocytic origin that are released by live cells while apoptosis bodies are secreted during apoptosis and microvesicles (MVs) are shedding from plasma membrane directly [23]. Although exosomes were considered as cellular garbage bins to remove hazardous substance before, recent studies have identified them as important means of intercellular communication [24]. Exosomes can mediate bulk of physiological or pathological processes, e.g., antigen presentation [25], injury repair [26], and tumor metastasis [27]. The functions of exosomes depend on their contents. An array of biomacromolecules is contained in exosomes, including proteins (heat shock proteins, tetraspanin), nucleic acids (mRNAs, ncRNAs, DNAs), as well as lipids (cholesterol). In Exocarta (www.exocarta.org), 9769 proteins, 3408 mRNAs, 2838 miRNAs and 1116 lipids were identified in exosomes in multiple organisms. Besides, exRNA Atlas (exrna-atlas.org/), a data repository of the Extracellular RNA Communication Consortium (ERCC), includes 5309 small RNA sequencing and qPCR-derived exRNA profiles from human and mouse biofluids in 19 studies. And the expression of long RNA species, including 15,501 lncRNAs, 18,333 mRNAs and 58,330 circRNAs, in human blood exosomes were collected from 92 samples in exoRbase (www.exorbase.org/). The cargoes are cell-type specific, e.g., exosomes derived from B-lymphocytes expressed MHC I/II molecules and the B cell marker CD20 [28]. However, the cargoes in exosomes are not identical to those in parent cells and are often influenced by extracellular stimuli, e.g., hypoxia and low pH could induce the biogenesis of exosomes and change the contents [29, 30]. It illustrates that the biogenesis of exosomes and sorting of the cargoes is precisely regulated.
In brief, exosomes are generated as intraluminal vesicles (ILVs) that are formed by inward budding of endosomal membrane during the maturation of multivesicular bodies (MVBs), and secreted upon fusion of MVBs with plasma membrane (Fig. 1). The mechanisms of exosomal biogenesis involve multiple factors [31], and the most known regulator is endosomal sorting complex required for transport (ESCRT). There are four types of ESCRT, including ESCRT 0-III. ESCRT 0 and ESCRT I cluster-specific cargoes and target them to endosomal membrane, then ESCRT I-ESCRT II complex are involved in buddings of endosomal membrane and they recruit ESCRT III to perform scission of ILVs. Apart from ESCRT, there are also other regulators in exosomal biogenesis, e.g., tetraspanins (CD9, CD63, CD81) and some accessory proteins (ALIX, VPS4). The motility of MVBs to plasma membrane is regulated by GTPase, e.g., Rab 27a/b.
Fig. 1.
The biogenesis, secretion and uptake of exosomes. Early endosomes are formed by endocytosis. With the assistance of endosomal sorting complex required for transport (ESCRT), intraluminal vesicles (ILVs) are generated while early endosomes develop to multivesicular bodies (MVBs). Some MVBs fuse with lysosome to be degraded while other MVBs are trafficked to fuse with the plasma membrane (it is regulated by GTPase, e.g., Rab27a/b) to release ILVs to extracellular milieu, which are called exosomes. Exosomes contain bioactive cargoes derived from parent cells, including DNAs, RNAs, proteins, as well as lipids. Exosomes can be ingested by recipient cells by endocytosis or fusing with plasma membrane, or through ligand–receptor interaction to exert diverse biological functions
After secreted into extracellular space, exosomes can naturally target to neighboring or distant recipient cells. The exosomes may activate downstream signal pathways in recipient cells by ligand-receptor interaction [32]. Additionally, exosomes can be internalized through fusion with plasma membrane, phagocytosis or receptor-mediated endocytosis (clathrin dependent and independent) to deliver the cargoes into cytoplasm. The cargoes are functional in recipient cells and change the phenotypes of recipient cells, e.g., mRNAs can be translated to proteins, and miRNAs can regulate gene expression by binding to target mRNAs in recipient cells [33]. In cancer, it has been reported that tumor cells release more exosomes than normal cells, and tumor-derived exosomes (TDEs) participate in the initiation and progression of cancer while normal cell-derived exosomes can inhibit tumorigenesis, due to differential components.
Separation and characterization of exosomes and exosomal ncRNAs
To investigate the functions of exosomes in cancer biology, we need to interrogate exosomal contents. In this review, we mainly focus on ncRNAs in exosomes since they are the most abundant and crucial biomolecules derived from exosomes. First, we need to isolate exosomes from samples, e.g., conditioned culture medium, ground tissues or biofluids. Up to now, there is no standard method of exosome isolation, and five categories of exosome isolation techniques have been developed, including differential ultracentrifugation-based techniques, size-based techniques, immunocapture-based techniques, precipitation, and microfluidic-based techniques [34]. According to a worldwide survey of International Society for Extracellular Vesicles (ISEV), differential centrifugation is the most commonly used method for separation (about 56%), and density gradient centrifugation, filtration, size-exclusion chromatography (SEC), immuno-isolation, precipitation are used by 5–20% of respondents each [35]. Moreover, additional techniques that involve a wide variety of microfluidic device have been developed, such as field-flow fractionation (FFF), asymmetric flow field-flow fractionation (AFFF), and field-free viscoelastic flow. However, absolute purification or complete isolation of exosomes is an unrealistic goal yet. Current methods, including centrifugation protocols or commercial kits all cannot separate exosomes specifically and completely. Thus, the separation products are actually mixture of different EVs and other substances, e.g., MVs and lipoprotein, which may affect downstream analysis. Thus, appropriate methods should be selected depending on downstream applications and scientific question. Highly purified exosomes are needed to attribute a function or biomarker to exosomes while less pure exosomes may be acceptable when a biomarker is useful without enrichment.
To further the promise of EVs including exosomes as biomarkers or for treatment, The ISEV board members have published a minimal information for studies of extracellular vesicles (MISEV) in 2014 and update in 2018 [36]. For separation, 93% of MISEV2018 survey respondents agreed with the categorization of techniques by recovery and specificity. For example, the methods with high recovery and low specificity include precipitation and low-molecular weight filters, the methods with intermediate recovery and specificity include SEC, differential centrifugation, high-molecular weight filters and membrane-affinity columns, the methods with low recovery and high specificity include density gradient centrifugation and immuno-isolation. And there is no method with high recovery and high specificity. Combinations of these methods outperform single-method approaches. Any newly developed technique for isolation must indicate to which of the 4 recovery/specificity options. More importantly, 98% of MISEV2018 Survey respondents agreed that all the experimental details, e.g., centrifugation (g-force, rotor, ultracentrifuge, adjusted k-factor, tube type, adaptor, time, temperature, and brake), should be deposited on the EV-TRACK (http://evtrack.org/index.php) to allow reproducibility.
Moreover, characterization of exosomes is important to show the success of isolation and demonstrate that biomarkers or functions are associated with exosomes [37]. According to MISEV 2018, the sources of EVs, e.g., number of secreting cells, volume of biofluid, should be described, and abundance of EVs should be characterized by total particle number or protein or lipid content, although none of them are exclusively associated to EVs. The purity of EVs can be measured by ratios of proteins/particles, lipids/particles or lipids/proteins. For general characterization, at least three positive protein markers of specific subtypes of EVs, including at least one transmembrane/lipid-bound protein/cytosolic protein must be evaluated, at least one negative protein marker and the presence of co-isolated components, e.g., lipoprotein or albumin, should be tested. Even more, two different but complementary techniques should be used for characterization of single vesicles, for example, images of single EVs at high resolution by electron microscopy (EM), scanning-probe microscopy (SPM), atomic-force microscopy (AFM) or super-resolution microscopy, size measured by resistive pulse sensing (RPS), or light-scattering properties, e.g., nanoparticle tracking analysis (NTA). As for exosomes, exosomes usually have cup-shaped morphologies under EM, express the markers associated with biogenesis (e.g., Tsg101, Alix, CD9/63/81, HSC70) and absence of ER markers (Calnexin), and the size of exosomes vary from 30 to 200 nm. However, many studies do not provide the adequate information of characterization yet and it needs to be improved in the future.
After separation of exosomes, we can extract exosomal ncRNAs from exosomes by RNA extracting kits or Trizol. The protocols of separation can be obtained from exRNA (http://exrna.org). Due to low concentration of exosomal RNAs, conventional methods for RNA evaluation are unavailable. For example, the results of concentration measured by Nanodrop or Qubit are inaccurate due to limitation of detection threshold. Instead, Agilent 2100 bioanalyzer can be used to evaluate the quality and quantity of exosomal RNAs. The results show that most RNA cargoes in exosomes are small RNAs which are 20–200 nucleotides in length and there is little 18S/28S rRNA. Deep sequencing reveals that the most abundant exosomal RNAs are miRNAs, and there are also fragments of mRNAs, lncRNAs or circRNAs and other types of ncRNAs, e.g., tRNAs, which are rarely explored yet [38, 39]. To identify dysregulated exosomal ncRNAs, quantitative analysis is required, and next-generation sequencing (NGS) [40], microarray [41], qPCR and digital PCR [42] can be used. A great number of exosomal ncRNAs have been reported to be dysregulated in HCC, and we will discuss their biological functions and clinical applications below.
The effects of exosomal ncRNAs on fibrosis/cirrhosis
HCC mainly occurs in a context of liver cirrhosis (LC) caused by chronic infection of hepatitis B virus (HBV) or hepatitis C virus (HCV) [2]. In the process of cancerization, ncRNA-carrying exosomes can help virus spread and disturb host immunity, leading to persistent viral infection, causing DNA damage and HCC occurrence. For example, miR-122 was enriched in serum exosomes from HCV patients, and exosomal miR-122 could help virus RNA transmission and replication between HCV-infected hepatocytes and normal hepatocytes via AGO2-miR-122-HSP90 complex [43]. Besides, serum exosomes from HCV patients could inhibit the function of natural killing (NK) cells and this effect was correlated with miR-122-5p (reduced expression of granzyme B, IGF1 receptor) and miR-222-3p (repressed expression of STAT5B) [44]. Similar situations also occur in HBV patients, caused by exosomal miR-21 and miR-29a, which suppressed the IL-12 expression in macrophages and prevented the activation of NK cells [45]. Chronic virus infection will activate hepatic stellate cells (HSCs), resulting in sustained inflammation and fibrosis. The study from Devhare PB demonstrated that HCV-infected hepatocytes could transfer exosomal miR-19a to HSCs, and miR-19a could target SOCS3 in HSC, which activated the STAT3-mediated transforming growth factor β (TGF-β) signaling pathway and promote fibrosis, which may favor the HCC tumorigenesis [46].
Additionally, NAFLD or non-alcohol steatohepatitis (NASH) has become another increasingly important disease leading to HCC. ncRNA-carrying exosomes from hepatocytes and non-hepatocytes both participate in progression of NAFLD. For example, in diet-induced steatohepatitis, the levels of exosomes increased early and reflected changes in liver histopathology. The levels of miR-122 and miR-192, two microRNAs abundant in hepatocyte increased in circulating exosomes. Thus, circulating exosomal ncRNAs may be biomarkers reflecting liver injury [47]. The elevated circulating miR-122 was associated with decreased liver miR-122, which resulted in upregulation of modulators of tissue remodel (HIF-1α, vimentin) and induced liver fibrosis [48]. Interestingly, circulating miR-122 may not be exclusively from liver. Baranova A et al. reported that adipose might actively release tumor-suppressing, anti-fibrotic miR-122 into circulation via exosomes, circulating miR-122 might infuse to liver to delay NAFLD progression and inhibit tumorigenesis [49]. Thus, detecting the levels of exosomal ncRNAs may help monitor chronic liver disease progression and evaluate the risks of HCC.
Exosomal ncRNAs and HCC progression
HCC is caused by genetic mutation and epigenetic dysregulation, involved with complex signal network. The malignant behaviors of tumor cells not only depend on themselves, but are also modulated by the interaction between tumor cells and their TME, which comprises of endothelial cells, fibroblasts, immune cells, and extracellular matrix (ECM) [50]. Apart from cell–cell contact and soluble factors, ncRNA-carrying exosomes, as signal carriers between different cells, can affect many aspects of HCC progression, including tumor metastasis, tumor angiogenesis, tumor immunity and drug resistance (Fig. 2). Understanding the roles of exosomal ncRNAs in cancer biology help develop novel anti-tumor strategy.
Fig. 2.
The interaction between HCC cells and microenvironment via exosomal ncRNAs. As functional contents, non-coding RNAs (ncRNAs) in both tumor-derived exosomes (TDEs) and non-TDEs participate in the communication between HCC cells and tumor microenvironment to promote HCC progression. Exosomal ncRNAs could promote tumor growth and invasion (miR-584, miR-93, lncRNA TUC339, decreased miR-125a/b), initiate the epithelial–mesenchymal transition (EMT) (lncRNA FAL1, decreased miR-320a), increase vascular permeability (miR-103), activate CAFs to form the pre-metastatic niche (miR-21, miR-1247-3p), affect circulating tumor cells (CTCs) seeding (miR-25-5p), promote tumor angiogenesis (miR-210, lncRNA H19), induce immunosuppressive (lncRNA TUC339), mediate drug resistance (linc-ROR, linc-VLDLR, miR-32-5p), etc.
Exosomal ncRNAs and HCC metastasis
Metastasis, the leading cause of death in cancer, is a multistep process, including tumor cells detach from primary lesion, invade the ECM, invade into circulation, survive in blood stream, extravasate from vessels, invade into secondary organs, and form metastases in target organs [51]. Exosomal ncRNAs have been demonstrated to participate in modulating sequential processes of HCC metastasis, including initiation of epithelial–mesenchymal transformation (EMT), regulation of tumor growth and invasion, increased vascular permeability, formation of pre-metastatic niches, and tumor seeding.
EMT is a conservative process that tumor cells lose their epithelial phenotypes and acquire mesenchymal phenotypes, resulting in enhanced motility and decreased cell–cell adhesion. Evidence suggests that EMT is related to initiation of metastasis and malignant transformation [52]. Both exosomal ncRNAs from tumor cells and stroma cells are potential mediators of EMT by activating relevant pathways. For example, the expression of Vimentin (mesenchymal marker) was increased while E-cadherin (epithelial marker) was decreased in HCC cells after treatment with HCC-derived exosome. lncRNA FAL1 was upregulated in HCC-derived exosomes, and lncRNA FAL1 could act as sponger of miR-1236 to increase the expression of ZEB1, which was the key transcription factors in EMT, and promote tumor metastasis [53]. Additionally, cancer-associated fibroblasts (CAFs) are known to play a great role in HCC progression, including inducing EMT [54]. The study from Zhang Z revealed that CAF-derived exosomes could induce EMT in HCC cells by activating MAPK pathways and upregulate MMP2 expression to remodel the ECM, facilitating HCC metastasis, which was associated with loss of miR-320a in CAF-derived exosomes [55].
On another hand, tumor cells can transfer oncogenic ncRNAs to recipient cells via exosome to promote tumor growth and invasion in autocrine or paracrine manners. For instance, TAK1, belonging to MAP3K subfamily, is a vital component of tumorigenesis. Selective miRNAs (e.g., miR-584) enriched in HCC-derived exosomes could be internalized by recipient HCC cells, subsequently modulated TAK1 expression to enhance HCC growth and invasion [56]. Similarly, miR-93 was upregulated in HCC patients’ serum exosomes and medium of HCC cell line, and exosomal miR-93 could stimulate the proliferation and invasion of HCC cells by targeting TIMP2/TP53INP1/CDKN1A [57]. In another study, lncRNA TUC339 was identified to be enriched in HCC-derived exosomes, and exosomal lncRNA TUC339 could promote cell proliferation and reduce cells adhesion to ECM, resulting in HCC spread [58]. Interestingly, the oncogenic effect of exosomes has been shown to not only occur by TDEs, but also non-TDEs. For example, adipose tissue could affect tumorigenesis by secreting adipokines [59]. The study from Zhang H demonstrated that plasma exosome circ-deubiquitination (circ-DB) was upregulated in HCC patients with higher body fat ratios, and exosome circ-DB derived from adipocytes could suppress miR-34a and activate the USP7/Cyclin A2 signaling pathway in HCC cells to promote HCC growth [60].
The occurrence of metastasis not only requires the enhancement of growth and invasion of tumor cells, also need tumor cells traverse the endothelial barrier. The vessels in tumor lesions have aberrant structures with increased permeability, which favor tumor metastasis. The crosstalk between cancer cells and endothelial cells, mediated by exosomal ncRNAs, takes part in impairing the vascular integrity to assist tumor cells to invade into circulation [61]. One study revealed that higher level of serum exosomal miR-103 was associated with higher metastasis potential, higher recurrence risks and shorter survival in the patients with HCC. The exosomal miR-103 secreted by HCC cells could act on endothelial cells to attenuate the endothelial junction integrity and increase vascular permeability by targeting endothelial adherens junction proteins, such as VE-Cadherin, p120-catenin, zonula occludens-1 (ZO-1), resulting in increased transendothelial invasion of HCC cells and metastasis occurrence [62].
In addition, before the implantation of tumor cells, primary tumors can establish a favorable microenvironment in target organs to support metastases growth, which is named pre-metastatic niche. The pre-metastatic niche, with the characteristics of inflammation, immunosuppressive and angiogenesis/vascular permeability [63], can be created by TDEs that remodel stromal contents, such as activation of CAFs and recruitment of myeloid-derived suppressor cells (MDSCs) [64]. Lung is the most common site of HCC distant metastasis. The study from Fang T demonstrated the crosstalk between tumor cells and fibroblasts mediated by TDE-influenced lung metastasis of HCC. They found that the levels of serum exosomal miR-1247-3p were higher in HCC patients with lung metastasis than those without. High metastatic HCC cells could secrete exosomal miR-1247-3p to activate CAFs via targeting B4GALT3, leading to activation of β1-integrin-NF-κB signaling in fibroblasts, and activated CAFs released inflammatory cytokines (IL-6, IL-8) to form an inflammatory microenvironment, fostering lung metastasis [65].
In the final step of metastasis, CTCs will seed in the target organ and grow to form metastases. It was reported that integrin in exosomes determines organ-specific metastasis [66], and exosomal ncRNAs also affect tumor seeding. Tumor self-seeding is a process that CTCs re-infiltrate the original tumor, in which primary tumor-derived cytokines act as CTC attractants and vascular leakage favor the infiltration of CTCs [67]. The study from Liu H demonstrated that exosomes from primary HCC could be transferred to CTCs to enhance their migratory and invasive abilities, resulting in tumor self-seeding, which may breed more aggressive tumor cells and contribute to HCC progression, this effect was associated with horizon transfer of exosomal miR-25-5p by targeting leucine rich repeat containing 7 (LRRC7) [68]. It illustrated the crosstalk between primary tumor and CTCs affected HCC progression.
Exosomal ncRNAs promote angiogenesis
Active angiogenesis, supplying adequate nutrition and oxygen to tumor cells, is a critical step for tumor growth and metastasis. Due to intratumor hypoxia, tumor cells can release soluble factors, e.g., vascular endothelial growth factor (VEGF), as well as exosomes to promote pathological angiogenesis [69]. Associated exosomal ncRNAs have been investigated in HCC. For example, the levels of serum exosomal miR-210 were elevated in HCC patients and associated with microvessel density (MVD). By targeting SMAD4 and STAT6 in endothelial cells, exosomal miR-210 derived from HCC cells could promote proliferation, migration and tube formation of endothelial cells, which are essential for angiogenesis [70]. In another study, hypoxia could stimulate HCC cells to secret exosomes containing miR-155 to promote angiogenesis, and the levels of exosomal miR-155 were positively associated with the expression of VEGF and HIF-1α in HCC samples. Increased exosomal miR-155 in preoperative plasma was significantly correlated with early recurrence and poor prognosis [71]. HCC cells can also promote angiogenesis indirectly through CAFs. For example, Zhou et al. reported that HCC cells secreted exosomal miR-21 that directly targeted phosphatase and tensin homolog (PTEN) to convert HSCs to CAFs, and CAFs could promote angiogenesis by releasing angiogenic cytokine, e.g., VEGF, MMP2, MMP9, bFGF and TGF-β [72]. Additionally, the crosstalk between cancer stem cell (CSC) and endothelial cells was reported to drive tumor angiogenesis. CD90+ liver cancer stem cells released exosomes containing lncRNA H19, which could be internalized by endothelial cells, subsequently upregulated the expression of VEGF and its receptor VEGFR1 in endothelial cells, inducing angiogenesis [73]. Blocking the transfer of exosomal ncRNAs abrogates these effects, providing novel strategies for inhibiting tumor progression.
Exosomal ncRNAs modulate tumor immunity
The role of immune microenvironment cannot be ignored for tumor progression. Tumor cells can escape from immune surveillance and induce immune tolerance by multiple ways, including releasing exosomes [74]. Both ncRNAs-carrying exosomes from tumor cells and immune cells could influence immune responses [75]. On one hand, tumor cells could transfer oncogenic ncRNAs to immune cells through exosomes to induce the expansion or differentiation of immunosuppressive cells, e.g., regulator T cells (Treg), and promote apoptosis or suppress the activity of effector cells, e.g., cytotoxic T lymphocytes (CTLs) [76], leading to local or systemic immunosuppression. For example, under ER stress, HCC-derived exosomal miR-23a-3p could upregulate the expression of PD-L1 in macrophages through PTEN-PI3K/AKT pathway, which subsequently inhibited T cell function [77]. In another study, HCC-derived exosomal lncRNA TUC339 could educate the macrophages, leading to reduced pro-inflammatory cytokine production (IL-1β,TNF-α), compromised phagocytosis (CD86) and drove M2 polarization, which promoted HCC progression. By bioinformatic analysis, lncRNA TUC339 were found to be involved in regulation of CXCR chemokine receptor binding, cytokine signaling pathway, an immune system defense response [78]. Interestingly, TDEs could also activate anti-tumor immunity by presenting tumor-associated antigen (TAA) [79]. The balance between both sides of TDEs is uncertain. On another hand, ncRNA-carrying exosomes from immune cells also affect tumor progression and the effects depend on the types of immune cells. For example, exosomes from DC cells could present TAAs to CTL to activate anti-tumor immunity [80]. Exosomal miR-142 and miR-223 from primary macrophages could inhibit HCC growth by decreasing the expression of stathmin-1 (STMN1) and insulin-like growth factor-1 receptor (IGF-1R) [81]. Conversely, tumor-associated macrophages (TAMs), the most abundant immune cells in TME, released exosomes to promote HCC growth and stem cell properties by targeting CD90, associated with decreased levels of miR-125a/b [82]. In another study, exosomes derived from functional exhausted CD8+ T cells led to exhaustion of normal CD8+ T cells, and 257 dysregulated lncRNAs were identified that actively participated in the regulation of diverse process of CD8+ T cells, e.g., metabolism, biosynthetic process [83]. Thus, therapeutic intervention based on exosomal ncRNAs in immune regulation will help remodel anti-tumor immunity.
Exosomal ncRNAs mediate drug resistance
Drug resistance, an essential factor for poor prognosis, is a significant clinical problem [84]. As a research hotspot, exosomal ncRNAs have been found to be involved with drug resistance. For example, when exposed to anticancer agents, such as sorafenib, camptothecin and doxorubicin, the expression of lincRNA-VLDLR (linc-VLDLR) was increased in both HCC cells and TDEs. When incubated with TDEs, the levels of linc-VLDLR in recipient HCC cells were elevated, and linc-VLDLR could increase the expression of ATP-binding cassette, subfamily G member 2 (ABC-G2), which involved in export of chemotherapeutic agents to reduce drug-induced cell apoptosis. [85]. CSCs are also involved in drug resistance. TGF-β could enrich lincRNA-ROR (linc-ROR) within TDEs, leading to increased number of CD133+ CSCs and reduced chemotherapy-induced cell death through repression of p53, resulting in drug resistance [86]. Moreover, the drug-resistant cells could deliver exosomal ncRNAs (miR-32-5p) horizontally to drug-sensitive cells to induce EMT and drug resistance by inhibiting PTEN and activating PI3K/AKT pathway in recipient cells [87]. What’s more, exosomes from stromal cells, e.g., CAFs or TAMs also play a great role in drug resistance in other cancers although there is no study in HCC yet. These findings all support targeting exosomal ncRNAs to enhance chemosensitivity in HCC.
Exosomal ncRNAs as biomarkers for HCC
As part of liquid biopsy, exosomes are widespread in various body fluids, and ncRNAs in TDEs can reflect the information of tumors and real-time disease progression. Thus, exosomal ncRNAs are regarded as a powerful source of non-invasive biomarkers for HCC. Blood is the most frequently used source in the researches of biomarkers. The standardization of sample collection, isolation and analysis were reviewed elsewhere [88]. Compared with other forms of ncRNAs, e.g., ncRNAs in tissues or circulating free ncRNAs, circulating exosomal ncRNAs have advantages of non-invasive and stable (the lipid bilayer of exosomes can protect ncRNAs from degradation), raising great interest of researchers. Table 1 summarizes the circulating exosomal ncRNAs that serve as biomarkers in HCC.
Table 1.
Exosomal ncRNAs as biomarkers in HCC
| Function | NcRNA (expression) | Source | Method | Cohort | Year [refs.] |
|---|---|---|---|---|---|
| Diagnosis | miR-93 (↑) | Serum | qPCR | 85 HCC vs 23 healthy control | 2018 [57] |
| miR-9-3p(↓) | Serum | qPCR | 30 HCC vs 10 healthy control | 2018 [93] | |
| miR-21 (↑) | Serum | qPCR | 30 HCC vs 30 CHB, 30 healthy control | 2014 [21] | |
|
miR-122 (↑) miR-125b (↑) miR-145 (↑) miR-192 (↑) miR-194 (↑) miR-29a (↑) miR-17-5p (↑) miR-106a (↑) |
Serum | qPCR | 80 HCC vs 30 healthy control | 2019 [94] | |
|
ENSG00000258332.1(↑) LINC00635(↑) |
Serum | qPCR | 60 HCC vs 85 LC vs 96 CHB vs 60 healthy control | 2018 [95] | |
| LncRNA HEIH (↑) | Serum | qPCR | 10 HCC vs 22 LC vs 25 CHC | 2018 [22] | |
|
miR-10b (↑) miR-21 (↑) miR-122 (↓) miR-200a (↓) |
Serum (rat) | qPCR | Normal, cirrhosis, early stage and late stage of HCC | 2015 [96] | |
|
miR-18a (↑) miR-221(↑) miR-222(↑) miR-224(↑) miR-101(↓) miR-106b (↓) miR-122(↓) miR-195(↓) |
Serum | qPCR | 20 HCC vs 20 LC vs 20 CHB | 2015 [97] | |
| Predicting poor prognosis | miR-638 (↓) | Serum | qPCR | 126 HCC (retrospective) | 2018 [99] |
| miR-125b (↓) | Serum | qPCR | 128 HCC (retrospective) | 2017 [100] | |
| miR-665 (↑) | Serum | qPCR | 30 HCC (retrospective) | 2017 [101] | |
|
miR-21 (↑) lncRNA ATB (↑) |
Serum | qPCR | 79 HCC (prospective) | 2018 [102] | |
| Predicting recurrence/metastasis | miR-103 (↑) | Serum | qPCR | 85 HCC | 2018 [62] |
| miR-1247-3p (↑) | Serum | qPCR | 110 HCC | 2018 [65] | |
| miR-155 (↑) | Plasma | qPCR | 40 HCC | 2018 [71] | |
| miR-718 (↓) | Serum | Microarray | 6 HCC | 2015 [103] |
Exosomal ncRNAs as diagnostic biomarkers
Early diagnosis can improve clinical outcomes of patients with HCC since curative resection or liver transplantation is available for early HCC and 5-year survival rate can be higher than 50% [89]. Thus, screening in high-risk population, e.g., the patients with cirrhosis, chronic hepatitis B (CHB) or chronic hepatitis C (CHC), may be beneficial [90, 91]. However, due to unsatisfactory sensitivity and specificity, the efficacy of current screening tools [ultrasound and serum α-fetal protein (AFP)] was suboptimal [92]. Thus, we need new biomarkers alone or combined, with a high sensitivity and specificity, to improve the accuracy of early diagnosis.
A great number of studies suggested that circulating exosomal miRNAs could serve as potential diagnostic biomarkers, which differentiated HCC from non-HCC individuals, such as healthy control, as well as the patients with LC, CHB or CHC. For example, the levels of serum exosomal miR-93 in HCC patients were significantly higher than healthy controls (diagnostic efficacy: area under the curve, AUC = 0.825) [57]. Serum exosomes from patients with HCC contained lower levels of miR-9-3p than those from healthy controls [93]. The levels of miR-21 were increased in serum exosomes from patients with HCC than in CHB or healthy volunteers, and it was correlated with cirrhosis and tumor stages [21]. In another study, eight exosomal miRNAs (miR-122, miR-125b, miR-145, miR-192, miR-194, miR-29a, miR-17-5p, and miR-106a) had significant differences between HCC and normal serum samples. The AUC of these eight exosomal miRNAs as diagnostic biomarkers varied from 0.535 to 0.850 [94].
Apart from exosomal miRNAs, recent studies have investigated the roles of serum exosomal lncRNAs as diagnostic biomarkers for HCC. For example, one study demonstrated that the levels of serum exosomal lncRNA ENSG00000258332.1 and LINC00635 were higher in HCC patients than in patients with CHB (AUC: 0.719 and 0.750, respectively) [95]. Similarly, the increased levels of serum exosomal lncRNA HEIH were found to be able to discriminate HCV-related HCC patients from patients with HCV-induced cirrhosis or CHC [22].
Although single exosomal ncRNAs can diagnose HCC, combining panels of biomarkers will enable a more accurate prediction of HCC than single biomarker for high-risk individuals. For example, Liu H et al. detected the levels of four most dysregulated miRNAs (miR-10b, miR-21, miR-122 and miR-200a) at different stages during HCC development in rat models. The four miRNAs in serum and serum exosomes obtained more remarkable alternation than serum AFP at early HCC stages. Different combinations of AFP, exosomes and exosomal miRNAs had stronger power in predicting HCC than serum AFP alone (AUC, 0.943 vs 0.826) [96]. Moreover, ncRNAs are selectively enriched within exosomes, increasing the sensitivity of detection. The study from Sohn W demonstrated that the levels of serum exosomal miR-18a, miR-221, miR-222 and miR-224 were higher in HCC than in CHB or LC while the levels of exosomal miR-101, miR-106b, miR-122 and miR-195 were lower in HCC than in CHB. Unlike exosomal miRNAs, the levels of serum circulating miRNAs showed a smaller difference between HCC and CHB [97].
From all these studies, we can conclude that exosomal ncRNAs can be used as novel diagnostic biomarkers for HCC. However, the results among different studies may be contradictory. It might be caused by patient selection, variable etiology, sample number and detection methods. Therefore, multi-center, large-sample clinical researches are needed in the future. Even more, the development of a standard isolation method to obtain sufficient exosomes of high purity is a challenge. The present gold standard ultracentrifugation is time-consuming, equipment dependent and has low yields. The cost of commercial kits is high and the purity is variable due to different principles as said before. Recently, microfluidic systems have been developed with advantages of rapid separation and small sample requirement (even a drop of blood) [98], providing a bright future in exosome field.
Exosomal ncRNAs as prognostic and predictive biomarkers
Except for diagnosis, exosomal ncRNAs have been introduced to assess HCC prognosis. From several retrospective studies, circulating exosomal ncRNAs have been suggested as effective prognostic markers for HCC. For example, negative association of serum exosomal miR-638 with tumor size, vascular infiltration and TNM stage was observed. Decreased levels of serum exosomal miR-638 predicted poor prognosis in HCC. [99]. In another study, the levels of miR-125b were associated with tumor number, encapsulation and TNM stage. The patients with lower serum exosomal miR-125b levels showed reduced time to recurrence (TTR) and overall survival (OS). [100]. And the survival time of the exosomal miR-665 high-expression group was significantly shorter than that of the low-expression group [101]. However, the results of retrospective studies may be influenced by selection bias, and prospective studies are required. In a recent prospective study, Lee et al. reported that the OS and progression-free survival (PFS) were significantly lower in HCC patients with higher levels of exosomal miRNA-21 and lncRNA-ATB [102].
Since tumor metastasis and recurrence are crucial factors for poor prognosis, researchers may use exosomal ncRNAs to evaluate metastasis potential of tumors or predict recurrence to improve the management of HCC. For example, higher level of serum exosomal miR-103 was associated with higher metastasis potential, higher recurrence risks and shorter survival in the patients with HCC [62]. Increased level of serum exosomal miR-1247-3p was related to lung metastasis of HCC [65]. High expression of exosomal miR-155 in preoperative plasma was significantly correlated with early recurrence [71]. And the levels of preoperative serum exosomal miR-718 were lower in the patients with recurrence after liver transplantation compared with those without recurrence [103]. By detecting these circulating exosomal ncRNAs, we can learn more information about tumor biology and monitor tumor progression of high-risk patients carefully.
Another reason for poor prognosis is drug resistance. As mentioned above, linc-VLDLR, linc-ROR, and miR-32-5p in exosomes were reported to mediate the occurrence of drug resistance in vitro. We may detect circulating exosomal ncRNAs of HCC patients to predict therapeutic efficacy of anticancer drugs, although there is no such study yet. Further, researchers can dynamically monitor the levels of circulating exosomal ncRNAs at different time spots (before treatment, after treatment, disease progression) to evaluate the efficacy of treatments and real-time progression.
Therapeutic functions of exosomal ncRNAs in HCC
As TDEs can carry oncogenic ncRNAs to recipient cells to promote tumor progression, researchers may inhibit tumorigenesis by blocking the biogenesis, secretion and uptake of exosomes. However, normal cells also release exosomes to maintain physiological functions [104]. Indiscriminate suppression of exosomes biogenesis may lead to side effects. Thus, it is better to affect TDEs specifically or sort of oncogenic ncRNAs into exosomes. For example, sphingosine kinase 2 (Sphk2) is an important regulator in exosome biogenesis. Using Sphk2 siRNA-loaded nanoparticles to treat HCC cells can reduce miRNA-21 sorting into exosomes, contributing to the inhibition of tumorigenic function of exosomes [105]. The synthetic nanoparticles can be replaced by exosomes, which are regarded as natural nanoparticles, with advantages of low biotoxicity, low immunogenicity and better biocompatibility. Thus, exosomes are potential options for new therapeutic approaches in cancer.
Unmodified ncRNA-carrying exosomes from normal stem cell
It was reported that normal cells could secrete exosomal miRNAs to inhibit tumor growth [106]. Recently, researches have focused on the roles of normal stem cell and their exosomes in cancer therapy. For example, mesenchymal stem cell (MSC) was reported to migrate to TME and could affect hepatocarcinogenesis and HCC progression [107]. Evidence suggests that MSC-derived exosomes could inhibit HCC progression, which was associated with delivering tumor-suppressed ncRNAs [108, 109]. However, some studies also reported that the exosomes from MSC may promote tumor progression [110]. Thus, whether it is safe to use MSC-derived exosomes to treat cancer is uncertain. Besides, exosomal ncRNAs from another stem cells, human adult liver stem cell (HLSC) can also influence the growth of hepatoma. For example, HLSC-derived exosomes could inhibit hepatoma growth by transferring anti-tumor miRNAs to downregulate the genes involved in cell proliferation, e.g., Cyclin D1 targeted by miR-223, E2F-2 targeted by miR-31, MDR1, MIF, RAB14 targeted by miR-451, DHFR targeted by miR-24 [111]. In another study, exosomal miR-15a, miR-181b, miR-320c and miR-874 from HLSCs could decrease the expression of ITGB3, FGF1, EPHB4 and PLAU to inhibit tumor angiogenesis [112]. Thus, the anticancer therapy based on exosomes derived from normal stem cells may be one more choice.
Engineered exosomes with tumor-suppressed ncRNAs: target tumor cells or TME
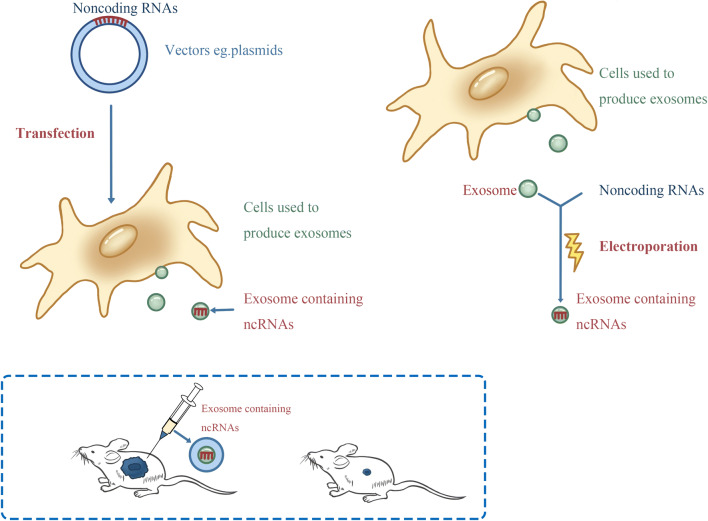
Tumor-suppressed ncRNAs are downregulated in cancer, and overexpressing tumor-suppressed ncRNAs in tumor cells inhibit tumor progression. We can package desired tumor-suppressed ncRNAs into exosomes by electroporation or transfecting the exosome-producing cells (Fig. 3), and then use the engineered exosomes to transport tumor-suppressed ncRNAs to HCC cells to reverse their malignant phenotypes. Some positive results have been obtained. For example, Lou G et al. reported that miR-122-transfected adipose mesenchymal stem cells (AMSCs) could effectively package miR-122 into secreted exosomes, and miR-122-loaded exosomes made HCC cells more sensitive to chemotherapeutic agents by repressing CCNG1, ADAM10 and IGF1R [113]. Liang G et al. used electroporation to actively load miR-26a to HEK-293T cell-derived exosomes, and miR-26a-loaded exosomes could inhibit HCC growth and invasion by inhibiting CCND2, CCNE2, CDK6 [114]. In another study, HSC-derived exosomes were loaded with miR-335-5p, and miR-335-5p-loaded exosomes could be taken up by HCC cells and inhibited HCC cell proliferation and invasion by targeting CDC42, CDK2, CSNK1G2, EIF2C2, EIF5, LIMaK1, NRG1, PLK2, TCF3, THBS1, YBX1, and ZMYND8 [115]. Moreover, it is feasible to use siRNA-loaded exosomes to treat cancer. For example, GRP78 is associated with sorafenib resistance, using the exosomes that encapsulated siRNA against GRP78 could suppress sorafenib resistance in HCC [116].
Fig. 3.
The two strategies of loading ncRNAs into exosomes. It is promising to use engineered exosomes to deliver anti-tumor ncRNAs to treat HCC. Mesenchymal stem cells (MSC) are most frequently used cells to produce exosomes in the studies. We can use vectors that contain ncRNAs to transfect MSCs, and MSCs can package the ncRNAs into secreted exosomes. Additionally, we can also use electroporation to actively load ncRNAs into MSC-derived exosomes. In assays, researches have observed that the engineered exosome-containing anti-tumor ncRNAs can inhibit tumor growth
Since TME is essential for tumor progression, targeting the stromal cells, e.g., CAFs, endothelial cells and immune cells, to remodel TME help treat cancer. For example, overexpressing miR-320a in CAFs can repress HCC metastasis by targeting PBX3 [55]. TDEs and DC-derived exosomes (DEXs) are reported to present TAAs and elicit anti-tumor immunity, forming a new class of vaccines for cancer immunotherapy [79, 80, 117]. NcRNAs are critical regulators in the immune system, influencing the differentiation, maturation, expansion and function of immune cells [118]. Thus, ncRNA-modified exosomes can be used for immunotherapy in HCC. For example, specific ncRNAs (miR-155, miR-142, and let-7i) modified TDEs could target IL-6, IL-17, IL-1b, TGFβ, SOCS1, KLRK1, IFNγ, and TLR4 to induce DC maturation and enhance their immune stimulation ability [119]. These results all suggest a novel therapeutic approach for tumor treatment.
Conclusions and perspectives
In this review, we mainly summarized recent literatures on the biological functions of exosomal ncRNAs in HCC development and their clinical applications. Exosomes are small EVs from most cell types and mediate signal transduction between different cells by transporting cargoes. NcRNA-carrying exosomes from tumor cells and stroma cells can modulate the interaction between HCC cells and their microenvironments, contributing to tumor progression by regulating specific aspects, including tumor metastasis, tumor angiogenesis, tumor immunity and drug resistance. It provides us a novel horizon of tumorigenesis and potential therapeutic targets. Due to aberrant expression and ability of reflecting disease state, circulating exosomal ncRNAs can serve as diagnostic or prognostic biomarkers for HCC. Moreover, it is a promising strategy to exploit exosomes as natural nanoparticle to deliver tumor suppressor ncRNAs to target HCC cells or TME for treatment.
However, there are still some difficulties remain to be overcome. First, for liquid biopsy, we need a standard method to isolate exosomes quickly, easily and specifically, and detect the dysregulated exosomal ncRNA rapidly and cheaply. And there is no proven biomarker in large sample and multi-center researches yet. Second, for studying biological functions, since most experiments are done in vitro and culture environment cannot imitate actual conditions, maybe the cargoes and their concentration are not identical to those in our body, thus, it is uncertain whether exosomal ncRNAs really have regulator functions in vivo as in vitro. Third, for therapy, the exosome-producing cells should be selected carefully to ensure the safety of treatment. Red blood cells (RBCs) were reported as potential exosome-producing cells since they were readily available in blood banks and devoid of DNA [120]. Further, we need to learn more mechanisms about exosomal biogenesis and design an optimal system to produce more exosomes to meet therapeutic demands. Moreover, a high-efficacy and safe technology of loading ncRNAs into exosomes needs to be developed. Last but not least, targeting strategies of exosomes need to be studied further to achieve efficient delivery and avoid side effects. In all, as our understanding of exosomes improves, using exosomes in the management of HCC will become a reality in some day.
Acknowledgements
Our work is supported by National S&T Major Project (No. 2017ZX10203205), the National Natural Science Foundation of China (No. 81570589), the National Natural Science Fund for Distinguished Young Scholars of China (No. 81625003) and Changjiang Scholars Program Foundation of Chinese Ministry of Education.
Compliance with ethical standards
Conflict of interest
The authors have declared no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel R, Torre L, Jemal A. Global Cancer Statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 Countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142(6):1264–1273 e1261. doi: 10.1053/j.gastro.2011.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morgan TR, Mandayam S, Jamal MM. Alcohol and hepatocellular carcinoma. Gastroenterology. 2004;127(5 Suppl 1):S87–S96. doi: 10.1053/j.gastro.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 4.Dyson J, Jaques B, Chattopadyhay D, Lochan R, Graham J, Das D, Aslam T, Patanwala I, Gaggar S, Cole M, Sumpter K, Stewart S, Rose J, Hudson M, Manas D, Reeves HL. Hepatocellular cancer: the impact of obesity, type 2 diabetes and a multidisciplinary team. J Hepatol. 2014;60(1):110–117. doi: 10.1016/j.jhep.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 5.Allemani C, Matsuda T, Di Carlo V, Harewood R, Matz M, Niksic M, Bonaventure A, Valkov M, Johnson CJ, Esteve J, Ogunbiyi OJ, Azevedo ESG, Chen WQ, Eser S, Engholm G, Stiller CA, Monnereau A, Woods RR, Visser O, Lim GH, Aitken J, Weir HK, Coleman MP, Group CW Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 2018;391(10125):1023–1075. doi: 10.1016/s0140-6736(17)33326-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tzartzeva K, Obi J, Rich NE, Parikh ND, Marrero JA, Yopp A, Waljee AK, Singal AG. Surveillance imaging and alpha fetoprotein for early detection of hepatocellular carcinoma in patients with cirrhosis: a meta-analysis. Gastroenterology. 2018;154(6):1706–1718 e1701. doi: 10.1053/j.gastro.2018.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391(10127):1301–1314. doi: 10.1016/S0140-6736(18)30010-2. [DOI] [PubMed] [Google Scholar]
- 8.Bruix J, Reig M, Sherman M. Evidence-based diagnosis, staging, and treatment of patients with hepatocellular carcinoma. Gastroenterology. 2016;150(4):835–853. doi: 10.1053/j.gastro.2015.12.041. [DOI] [PubMed] [Google Scholar]
- 9.Bruix J, Raoul JL, Sherman M, Mazzaferro V, Bolondi L, Craxi A, Galle PR, Santoro A, Beaugrand M, Sangiovanni A, Porta C, Gerken G, Marrero JA, Nadel A, Shan M, Moscovici M, Voliotis D, Llovet JM. Efficacy and safety of sorafenib in patients with advanced hepatocellular carcinoma: subanalyses of a phase III trial. J Hepatol. 2012;57(4):821–829. doi: 10.1016/j.jhep.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 10.Cheng AL, Kang YK, Chen Z, Tsao CJ, Qin S, Kim JS, Luo R, Feng J, Ye S, Yang TS, Xu J, Sun Y, Liang H, Liu J, Wang J, Tak WY, Pan H, Burock K, Zou J, Voliotis D, Guan Z. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009;10(1):25–34. doi: 10.1016/S1470-2045(08)70285-7. [DOI] [PubMed] [Google Scholar]
- 11.Mohkam K, Dumont PN, Manichon AF, Jouvet JC, Boussel L, Merle P, Ducerf C, Lesurtel M, Rode A, Mabrut JY. No-touch multibipolar radiofrequency ablation vs. surgical resection for solitary hepatocellular carcinoma ranging from 2 to 5 cm. J Hepatol. 2018;68(6):1172–1180. doi: 10.1016/j.jhep.2018.01.014. [DOI] [PubMed] [Google Scholar]
- 12.Lim C, Bhangui P, Salloum C, Gomez-Gavara C, Lahat E, Luciani A, Compagnon P, Calderaro J, Feray C, Azoulay D. Impact of time to surgery in the outcome of patients with liver resection for BCLC 0-A stage hepatocellular carcinoma. J Hepatol. 2017 doi: 10.1016/j.jhep.2017.09.017. [DOI] [PubMed] [Google Scholar]
- 13.De Rubis G, Rajeev Krishnan S, Bebawy M. Liquid biopsies in cancer diagnosis, monitoring, and prognosis. Trends Pharmacol Sci. 2019;40(3):172–186. doi: 10.1016/j.tips.2019.01.006. [DOI] [PubMed] [Google Scholar]
- 14.Ruivo CF, Adem B, Silva M, Melo SA. The biology of cancer exosomes: insights and new perspectives. Cancer Res. 2017;77(23):6480–6488. doi: 10.1158/0008-5472.CAN-17-0994. [DOI] [PubMed] [Google Scholar]
- 15.Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N, Reissfelder C, Pilarsky C, Fraga MF, Piwnica-Worms D, Kalluri R. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523(7559):177–182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manier S, Liu CJ, Avet-Loiseau H, Park J, Shi J, Campigotto F, Salem KZ, Huynh D, Glavey SV, Rivotto B, Sacco A, Roccaro AM, Bouyssou J, Minvielle S, Moreau P, Facon T, Leleu X, Weller E, Trippa L, Ghobrial IM. Prognostic role of circulating exosomal miRNAs in multiple myeloma. Blood. 2017;129(17):2429–2436. doi: 10.1182/blood-2016-09-742296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, Yu Z, Yang J, Wang B, Sun H, Xia H, Man Q, Zhong W, Antelo LF, Wu B, Xiong X, Liu X, Guan L, Li T, Liu S, Yang R, Lu Y, Dong L, McGettigan S, Somasundaram R, Radhakrishnan R, Mills G, Lu Y, Kim J, Chen YH, Dong H, Zhao Y, Karakousis GC, Mitchell TC, Schuchter LM, Herlyn M, Wherry EJ, Xu X, Guo W. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560(7718):382–386. doi: 10.1038/s41586-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet. 2015;16(2):71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18(1):5–18. doi: 10.1038/nrc.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan Q, Yang L, Zhang X, Peng X, Wei S, Su D, Zhai Z, Hua X, Li H. The emerging role of exosome-derived non-coding RNAs in cancer biology. Cancer Lett. 2018;414:107–115. doi: 10.1016/j.canlet.2017.10.040. [DOI] [PubMed] [Google Scholar]
- 21.Wang H, Hou L, Li A, Duan Y, Gao H, Song X. Expression of serum exosomal microRNA-21 in human hepatocellular carcinoma. Biomed Res Int. 2014;2014:864894. doi: 10.1155/2014/864894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang C, Yang X, Qi Q, Gao Y, Wei Q, Han S. lncRNA-HEIH in serum and exosomes as a potential biomarker in the HCV-related hepatocellular carcinoma. Cancer Biomark. 2018;21(3):651–659. doi: 10.3233/CBM-170727. [DOI] [PubMed] [Google Scholar]
- 23.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2(8):569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 24.Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ, Geuze HJ. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183(3):1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindenbergh MFS, Stoorvogel W. Antigen presentation by extracellular vesicles from professional antigen-presenting cells. Annu Rev Immunol. 2018;36:435–459. doi: 10.1146/annurev-immunol-041015-055700. [DOI] [PubMed] [Google Scholar]
- 26.Cabral J, Ryan AE, Griffin MD, Ritter T. Extracellular vesicles as modulators of wound healing. Adv Drug Deliv Rev. 2018;129:394–406. doi: 10.1016/j.addr.2018.01.018. [DOI] [PubMed] [Google Scholar]
- 27.Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell. 2016;30(6):836–848. doi: 10.1016/j.ccell.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clayton A, Court J, Navabi H, Adams M, Mason MD, Hobot JA, Newman GR, Jasani B. Analysis of antigen presenting cell derived exosomes, based on immuno-magnetic isolation and flow cytometry. J Immunol Methods. 2001;247(1–2):163–174. doi: 10.1016/S0022-1759(00)00321-5. [DOI] [PubMed] [Google Scholar]
- 29.de Jong OG, Verhaar MC, Chen Y, Vader P, Gremmels H, Posthuma G, Schiffelers RM, Gucek M, van Balkom BW. Cellular stress conditions are reflected in the protein and RNA content of endothelial cell-derived exosomes. J Extracell Vesicles. 2012 doi: 10.3402/jev.v1i0.18396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King HW, Michael MZ, Gleadle JM. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer. 2012;12:421. doi: 10.1186/1471-2407-12-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19(4):213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 32.Nazarenko I, Rana S, Baumann A, McAlear J, Hellwig A, Trendelenburg M, Lochnit G, Preissner KT, Zoller M. Cell surface tetraspanin Tspan8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res. 2010;70(4):1668–1678. doi: 10.1158/0008-5472.CAN-09-2470. [DOI] [PubMed] [Google Scholar]
- 33.Mulcahy LA, Pink RC, Carter DR. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles. 2014 doi: 10.3402/jev.v3.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li P, Kaslan M, Lee SH, Yao J, Gao Z. Progress in exosome isolation techniques. Theranostics. 2017;7(3):789–804. doi: 10.7150/thno.18133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gardiner C, Di Vizio D, Sahoo S, Thery C, Witwer KW, Wauben M, Hill AF. Techniques used for the isolation and characterization of extracellular vesicles: results of a worldwide survey. J Extracell Vesicles. 2016;5:32945. doi: 10.3402/jev.v5.32945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, Ayre DC, Bach JM, Bachurski D, Baharvand H, Balaj L, Baldacchino S, Bauer NN, Baxter AA, Bebawy M, Beckham C, Bedina Zavec A, Benmoussa A, Berardi AC, Bergese P, Bielska E, Blenkiron C, Bobis-Wozowicz S, Boilard E, Boireau W, Bongiovanni A, Borras FE, Bosch S, Boulanger CM, Breakefield X, Breglio AM, Brennan MA, Brigstock DR, Brisson A, Broekman ML, Bromberg JF, Bryl-Gorecka P, Buch S, Buck AH, Burger D, Busatto S, Buschmann D, Bussolati B, Buzas EI, Byrd JB, Camussi G, Carter DR, Caruso S, Chamley LW, Chang YT, Chen C, Chen S, Cheng L, Chin AR, Clayton A, Clerici SP, Cocks A, Cocucci E, Coffey RJ, Cordeiro-da-Silva A, Couch Y, Coumans FA, Coyle B, Crescitelli R, Criado MF, D’Souza-Schorey C, Das S, Datta Chaudhuri A, de Candia P, De Santana EF, De Wever O, Del Portillo HA, Demaret T, Deville S, Devitt A, Dhondt B, Di Vizio D, Dieterich LC, Dolo V, Dominguez Rubio AP, Dominici M, Dourado MR, Driedonks TA, Duarte FV, Duncan HM, Eichenberger RM, Ekstrom K, El Andaloussi S, Elie-Caille C, Erdbrugger U, Falcon-Perez JM, Fatima F, Fish JE, Flores-Bellver M, Forsonits A, Frelet-Barrand A, Fricke F, Fuhrmann G, Gabrielsson S, Gamez-Valero A, Gardiner C, Gartner K, Gaudin R, Gho YS, Giebel B, Gilbert C, Gimona M, Giusti I, Goberdhan DC, Gorgens A, Gorski SM, Greening DW, Gross JC, Gualerzi A, Gupta GN, Gustafson D, Handberg A, Haraszti RA, Harrison P, Hegyesi H, Hendrix A, Hill AF, Hochberg FH, Hoffmann KF, Holder B, Holthofer H, Hosseinkhani B, Hu G, Huang Y, Huber V, Hunt S, Ibrahim AG, Ikezu T, Inal JM, Isin M, Ivanova A, Jackson HK, Jacobsen S, Jay SM, Jayachandran M, Jenster G, Jiang L, Johnson SM, Jones JC, Jong A, Jovanovic-Talisman T, Jung S, Kalluri R, Kano SI, Kaur S, Kawamura Y, Keller ET, Khamari D, Khomyakova E, Khvorova A, Kierulf P, Kim KP, Kislinger T, Klingeborn M, Klinke DJ, 2nd, Kornek M, Kosanovic MM, Kovacs AF, Kramer-Albers EM, Krasemann S, Krause M, Kurochkin IV, Kusuma GD, Kuypers S, Laitinen S, Langevin SM, Languino LR, Lannigan J, Lasser C, Laurent LC, Lavieu G, Lazaro-Ibanez E, Le Lay S, Lee MS, Lee YXF, Lemos DS, Lenassi M, Leszczynska A, Li IT, Liao K, Libregts SF, Ligeti E, Lim R, Lim SK, Line A, Linnemannstons K, Llorente A, Lombard CA, Lorenowicz MJ, Lorincz AM, Lotvall J, Lovett J, Lowry MC, Loyer X, Lu Q, Lukomska B, Lunavat TR, Maas SL, Malhi H, Marcilla A, Mariani J, Mariscal J, Martens-Uzunova ES, Martin-Jaular L, Martinez MC, Martins VR, Mathieu M, Mathivanan S, Maugeri M, McGinnis LK, McVey MJ, Meckes DG, Jr, Meehan KL, Mertens I, Minciacchi VR, Moller A, Moller Jorgensen M, Morales-Kastresana A, Morhayim J, Mullier F, Muraca M, Musante L, Mussack V, Muth DC, Myburgh KH, Najrana T, Nawaz M, Nazarenko I, Nejsum P, Neri C, Neri T, Nieuwland R, Nimrichter L, Nolan JP, Nolte-’t Hoen EN, Noren Hooten N, O’Driscoll L, O’Grady T, O’Loghlen A, Ochiya T, Olivier M, Ortiz A, Ortiz LA, Osteikoetxea X, Ostergaard O, Ostrowski M, Park J, Pegtel DM, Peinado H, Perut F, Pfaffl MW, Phinney DG, Pieters BC, Pink RC, Pisetsky DS, Pogge von Strandmann E, Polakovicova I, Poon IK, Powell BH, Prada I, Pulliam L, Quesenberry P, Radeghieri A, Raffai RL, Raimondo S, Rak J, Ramirez MI, Raposo G, Rayyan MS, Regev-Rudzki N, Ricklefs FL, Robbins PD, Roberts DD, Rodrigues SC, Rohde E, Rome S, Rouschop KM, Rughetti A, Russell AE, Saa P, Sahoo S, Salas-Huenuleo E, Sanchez C, Saugstad JA, Saul MJ, Schiffelers RM, Schneider R, Schoyen TH, Scott A, Shahaj E, Sharma S, Shatnyeva O, Shekari F, Shelke GV, Shetty AK, Shiba K, Siljander PR, Silva AM, Skowronek A, Snyder OL, 2nd, Soares RP, Sodar BW, Soekmadji C, Sotillo J, Stahl PD, Stoorvogel W, Stott SL, Strasser EF, Swift S, Tahara H, Tewari M, Timms K, Tiwari S, Tixeira R, Tkach M, Toh WS, Tomasini R, Torrecilhas AC, Tosar JP, Toxavidis V, Urbanelli L, Vader P, van Balkom BW, van der Grein SG, Van Deun J, van Herwijnen MJ, Van Keuren-Jensen K, van Niel G, van Royen ME, van Wijnen AJ, Vasconcelos MH, Vechetti IJ, Jr, Veit TD, Vella LJ, Velot E, Verweij FJ, Vestad B, Vinas JL, Visnovitz T, Vukman KV, Wahlgren J, Watson DC, Wauben MH, Weaver A, Webber JP, Weber V, Wehman AM, Weiss DJ, Welsh JA, Wendt S, Wheelock AM, Wiener Z, Witte L, Wolfram J, Xagorari A, Xander P, Xu J, Yan X, Yanez-Mo M, Yin H, Yuana Y, Zappulli V, Zarubova J, Zekas V, Zhang JY, Zhao Z, Zheng L, Zheutlin AR, Zickler AM, Zimmermann P, Zivkovic AM, Zocco D, Zuba-Surma EK. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1):1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boriachek K, Islam MN, Moller A, Salomon C, Nguyen NT, Hossain MSA, Yamauchi Y, Shiddiky MJA. Biological functions and current advances in isolation and detection strategies for exosome nanovesicles. Small (Weinheim an der Bergstrasse, Germany) 2018 doi: 10.1002/smll.201702153. [DOI] [PubMed] [Google Scholar]
- 38.Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, Liang M, Dittmar RL, Liu Y, Liang M, Kohli M, Thibodeau SN, Boardman L, Wang L. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319. doi: 10.1186/1471-2164-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin X, Chen Y, Chen H, Fei S, Chen D, Cai X, Liu L, Lin B, Su H, Zhao L, Su M, Pan H, Shen L, Xie D, Xie C. Evaluation of tumor-derived exosomal miRNA as potential diagnostic biomarkers for early-stage non-small cell lung cancer using next-generation sequencing. Clin Cancer Res. 2017;23(17):5311–5319. doi: 10.1158/1078-0432.CCR-17-0577. [DOI] [PubMed] [Google Scholar]
- 41.Chen J, Wang S, Jia S, Ding G, Jiang G, Cao L. Integrated analysis of long non-coding RNA and mRNA expression profile in pancreatic cancer derived exosomes treated dendritic cells by microarray analysis. J Cancer. 2018;9(1):21–31. doi: 10.7150/jca.21749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bellingham SA, Shambrook M, Hill AF. quantitative analysis of exosomal miRNA via qPCR and digital PCR. Methods Mol Biol. 2017;1545:55–70. doi: 10.1007/978-1-4939-6728-5_5. [DOI] [PubMed] [Google Scholar]
- 43.Bukong TN, Momen-Heravi F, Kodys K, Bala S, Szabo G. Exosomes from hepatitis C infected patients transmit HCV infection and contain replication competent viral RNA in complex with Ago2-miR122-HSP90. PLoS Pathog. 2014;10(10):e1004424. doi: 10.1371/journal.ppat.1004424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santangelo L, Bordoni V, Montaldo C, Cimini E, Zingoni A, Battistelli C, D’Offizi G, Capobianchi MR, Santoni A, Tripodi M, Agrati C. Hepatitis C virus direct-acting antivirals therapy impacts on extracellular vesicles microRNAs content and on their immunomodulating properties. Liver Int. 2018;38(10):1741–1750. doi: 10.1111/liv.13700. [DOI] [PubMed] [Google Scholar]
- 45.Kouwaki T, Fukushima Y, Daito T, Sanada T, Yamamoto N, Mifsud EJ, Leong CR, Tsukiyama-Kohara K, Kohara M, Matsumoto M, Seya T, Oshiumi H. Extracellular vesicles including exosomes regulate innate immune responses to hepatitis B virus infection. Front Immunol. 2016;7:335. doi: 10.3389/fimmu.2016.00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Devhare PB, Sasaki R, Shrivastava S, Di Bisceglie AM, Ray R, Ray RB. Exosome-mediated intercellular communication between hepatitis C virus-infected hepatocytes and hepatic stellate cells. J Virol. 2017 doi: 10.1128/jvi.02225-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Povero D, Eguchi A, Li H, Johnson CD, Papouchado BG, Wree A, Messer K, Feldstein AE. Circulating extracellular vesicles with specific proteome and liver microRNAs are potential biomarkers for liver injury in experimental fatty liver disease. PLoS One. 2014;9(12):e113651. doi: 10.1371/journal.pone.0113651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Csak T, Bala S, Lippai D, Satishchandran A, Catalano D, Kodys K, Szabo G. microRNA-122 regulates hypoxia-inducible factor-1 and vimentin in hepatocytes and correlates with fibrosis in diet-induced steatohepatitis. Liver Int. 2015;35(2):532–541. doi: 10.1111/liv.12633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baranova A, Maltseva D, Tonevitsky A. Adipose may actively delay progression of NAFLD by releasing tumor-suppressing, anti-fibrotic miR-122 into circulation. Obes Rev. 2019;20(1):108–118. doi: 10.1111/obr.12765. [DOI] [PubMed] [Google Scholar]
- 50.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19(11):1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147(2):275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Campbell K. Contribution of epithelial-mesenchymal transitions to organogenesis and cancer metastasis. Curr Opin Cell Biol. 2018;55:30–35. doi: 10.1016/j.ceb.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li B, Mao R, Liu C, Zhang W, Tang Y, Guo Z. LncRNA FAL1 promotes cell proliferation and migration by acting as a CeRNA of miR-1236 in hepatocellular carcinoma cells. Life Sci. 2018;197:122–129. doi: 10.1016/j.lfs.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 54.Kubo N, Araki K, Kuwano H, Shirabe K. Cancer-associated fibroblasts in hepatocellular carcinoma. World J Gastroenterol. 2016;22(30):6841–6850. doi: 10.3748/wjg.v22.i30.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Z, Li X, Sun W, Yue S, Yang J, Li J, Ma B, Wang J, Yang X, Pu M, Ruan B, Zhao G, Huang Q, Wang L, Tao K, Dou K. Loss of exosomal miR-320a from cancer-associated fibroblasts contributes to HCC proliferation and metastasis. Cancer Lett. 2017;397:33–42. doi: 10.1016/j.canlet.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 56.Kogure T, Lin W, Yan I, Braconi C, Patel T. Intercellular nanovesicle-mediated microRNA transfer: a mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology. 2011;54(4):1237–1248. doi: 10.1002/hep.24504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xue X, Wang X, Zhao Y, Hu R, Qin L. Exosomal miR-93 promotes proliferation and invasion in hepatocellular carcinoma by directly inhibiting TIMP2/TP53INP1/CDKN1A. Biochem Biophys Res Commun. 2018;502(4):515–521. doi: 10.1016/j.bbrc.2018.05.208. [DOI] [PubMed] [Google Scholar]
- 58.Kogure T, Yan IK, Lin WL, Patel T. Extracellular vesicle-mediated transfer of a novel long noncoding RNA TUC339: a mechanism of intercellular signaling in human hepatocellular cancer. Genes Cancer. 2013;4(7–8):261–272. doi: 10.1177/1947601913499020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quail DF, Dannenberg AJ. The obese adipose tissue microenvironment in cancer development and progression. Nat Rev Endocrinol. 2018 doi: 10.1038/s41574-018-0126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu K, Fan Q, Li J, Ning T, Tian F, Li H, Sun W, Ying G, Ba Y. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene. 2018 doi: 10.1038/s41388-018-0619-z. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 61.Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR, Yu Y, Chow A, O’Connor ST, Chin AR, Yen Y, Wang Y, Marcusson EG, Chu P, Wu J, Wu X, Li AX, Li Z, Gao H, Ren X, Boldin MP, Lin PC, Wang SE. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25(4):501–515. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fang JH, Zhang ZJ, Shang LR, Luo YW, Lin YF, Yuan Y, Zhuang SM. Hepatoma cell-secreted exosomal microRNA-103 increases vascular permeability and promotes metastasis by targeting junction proteins. Hepatology. 2018 doi: 10.1002/hep.29920. [DOI] [PubMed] [Google Scholar]
- 63.Liu Y, Cao X. Characteristics and significance of the pre-metastatic niche. Cancer Cell. 2016;30(5):668–681. doi: 10.1016/j.ccell.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 64.Li K, Chen Y, Li A, Tan C, Liu X. Exosomes play roles in sequential processes of tumor metastasis. Int J Cancer. 2018 doi: 10.1002/ijc.31774. [DOI] [PubMed] [Google Scholar]
- 65.Fang T, Lv H, Lv G, Li T, Wang C, Han Q, Yu L, Su B, Guo L, Huang S, Cao D, Tang L, Tang S, Wu M, Yang W, Wang H. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat Commun. 2018;9(1):191. doi: 10.1038/s41467-017-02583-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, Singh S, Williams C, Soplop N, Uryu K, Pharmer L, King T, Bojmar L, Davies AE, Ararso Y, Zhang T, Zhang H, Hernandez J, Weiss JM, Dumont-Cole VD, Kramer K, Wexler LH, Narendran A, Schwartz GK, Healey JH, Sandstrom P, Labori KJ, Kure EH, Grandgenett PM, Hollingsworth MA, de Sousa M, Kaur S, Jain M, Mallya K, Batra SK, Jarnagin WR, Brady MS, Fodstad O, Muller V, Pantel K, Minn AJ, Bissell MJ, Garcia BA, Kang Y, Rajasekhar VK, Ghajar CM, Matei I, Peinado H, Bromberg J, Lyden D. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim MY, Oskarsson T, Acharyya S, Nguyen DX, Zhang XH, Norton L, Massague J. Tumor self-seeding by circulating cancer cells. Cell. 2009;139(7):1315–1326. doi: 10.1016/j.cell.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu H, Chen W, Zhi X, Chen E, Wei T, Zhang J, Shen J, Hu L, Zhao B, Feng X, Bai X, Liang T. Tumor-derived exosomes promote tumor self-seeding in hepatocellular carcinoma by transferring miRNA-25-5p to enhance cell motility. Oncogene. 2018;37(36):4964–4978. doi: 10.1038/s41388-018-0309-x. [DOI] [PubMed] [Google Scholar]
- 69.Shao C, Yang F, Miao S, Liu W, Wang C, Shu Y, Shen H. Role of hypoxia-induced exosomes in tumor biology. Mol Cancer. 2018;17(1):120. doi: 10.1186/s12943-018-0869-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lin X, Fang J, Yang X, Zhang C, Yuan Y, Zheng L, Zhuang S. Hepatocellular carcinoma cell-secreted exosomal MicroRNA-210 promotes angiogenesis in vitro and in vivo. Mol Ther Nucleic Acids. 2018;11:243–252. doi: 10.1016/j.omtn.2018.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matsuura Y, Wada H, Eguchi H, Gotoh K, Kobayashi S, Kinoshita M, Kubo M, Hayashi K, Iwagami Y, Yamada D, Asaoka T, Noda T, Kawamoto K, Takeda Y, Tanemura M, Umeshita K, Doki Y, Mori M. Exosomal miR-155 derived from hepatocellular carcinoma cells under hypoxia promotes angiogenesis in endothelial cells. Dig Dis Sci. 2018 doi: 10.1007/s10620-018-5380-1. [DOI] [PubMed] [Google Scholar]
- 72.Zhou Y, Ren H, Dai B, Li J, Shang L, Huang J, Shi X. Hepatocellular carcinoma-derived exosomal miRNA-21 contributes to tumor progression by converting hepatocyte stellate cells to cancer-associated fibroblasts. J Exp Clin Cancer Res. 2018;37(1):324. doi: 10.1186/s13046-018-0965-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Conigliaro A, Costa V, Lo Dico A, Saieva L, Buccheri S, Dieli F, Manno M, Raccosta S, Mancone C, Tripodi M, De Leo G, Alessandro R. CD90 + liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol Cancer. 2015;14:155. doi: 10.1186/s12943-015-0426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Whiteside TL. Exosomes and tumor-mediated immune suppression. J Clin Invest. 2016;126(4):1216–1223. doi: 10.1172/JCI81136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eichmuller SB, Osen W, Mandelboim O, Seliger B. Immune modulatory microRNAs involved in tumor attack and tumor immune escape. J Natl Cancer Inst. 2017 doi: 10.1093/jnci/djx034. [DOI] [PubMed] [Google Scholar]
- 76.Ye SB, Li ZL, Luo DH, Huang BJ, Chen YS, Zhang XS, Cui J, Zeng YX, Li J. Tumor-derived exosomes promote tumor progression and T-cell dysfunction through the regulation of enriched exosomal microRNAs in human nasopharyngeal carcinoma. Oncotarget. 2014;5(14):5439–5452. doi: 10.18632/oncotarget.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu J, Fan L, Yu H, Zhang J, He Y, Feng D, Wang F, Li X, Liu Q, Li Y, Guo Z, Gao B, Wei W, Wang H, Sun G. Endoplasmic reticulum stress promotes liver cancer cells to release exosomal miR-23a-3p and up-regulate PD-L1 expression in macrophages. Hepatology. 2019 doi: 10.1002/hep.30607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li X, Lei Y, Wu M, Li N. Regulation of macrophage activation and polarization by HCC-derived exosomal lncRNA TUC339. Int J Mol Sci. 2018 doi: 10.3390/ijms19102958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rao Q, Zuo B, Lu Z, Gao X, You A, Wu C, Du Z, Yin H. Tumor-derived exosomes elicit tumor suppression in murine hepatocellular carcinoma models and humans in vitro. Hepatology. 2016;64(2):456–472. doi: 10.1002/hep.28549. [DOI] [PubMed] [Google Scholar]
- 80.Lu Z, Zuo B, Jing R, Gao X, Rao Q, Liu Z, Qi H, Guo H, Yin H. Dendritic cell-derived exosomes elicit tumor regression in autochthonous hepatocellular carcinoma mouse models. J Hepatol. 2017;67(4):739–748. doi: 10.1016/j.jhep.2017.05.019. [DOI] [PubMed] [Google Scholar]
- 81.Aucher A, Rudnicka D, Davis DM. MicroRNAs transfer from human macrophages to hepato-carcinoma cells and inhibit proliferation. J Immunol. 2013;191(12):6250–6260. doi: 10.4049/jimmunol.1301728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang Y, Wang B, Xiao S, Li Y, Chen Q. miR-125a/b inhibits tumor-associated macrophages mediated in cancer stem cells of hepatocellular carcinoma by targeting CD90. J Cell Biochem. 2018 doi: 10.1002/jcb.27436. [DOI] [PubMed] [Google Scholar]
- 83.Wang X, Shen H, He Q, Tian W, Xia A, Lu XJ. Exosomes derived from exhausted CD8 + T cells impaired the anticancer function of normal CD8 + T cells. J Med Genet. 2018 doi: 10.1136/jmedgenet-2018-105439. [DOI] [PubMed] [Google Scholar]
- 84.Niu L, Liu L, Yang S, Ren J, Lai PBS. Chen GG (2017) New insights into sorafenib resistance in hepatocellular carcinoma: responsible mechanisms and promising strategies. Biochim Biophys Acta Rev Cancer. 1868;2:564–570. doi: 10.1016/j.bbcan.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 85.Takahashi K, Yan I, Wood J, Haga H, Patel T. Involvement of extracellular vesicle long noncoding RNA (linc-VLDLR) in tumor cell responses to chemotherapy. Mol Cancer Res. 2014;12(10):1377–1387. doi: 10.1158/1541-7786.MCR-13-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Takahashi K, Yan I, Kogure T, Haga H, Patel T. Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio. 2014;4:458–467. doi: 10.1016/j.fob.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fu X, Liu M, Qu S, Ma J, Zhang Y, Shi T, Wen H, Yang Y, Wang S, Wang J, Nan K, Yao Y, Tian T. Exosomal microRNA-32-5p induces multidrug resistance in hepatocellular carcinoma via the PI3 K/Akt pathway. J Exp Clin Cancer Res. 2018;37(1):52. doi: 10.1186/s13046-018-0677-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 88.Witwer KW, Buzas EI, Bemis LT, Bora A, Lasser C, Lotvall J, Nolte-’t Hoen EN, Piper MG, Sivaraman S, Skog J, Thery C, Wauben MH, Hochberg F. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013 doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Menahem B, Lubrano J, Duvoux C, Mulliri A, Alves A, Costentin C, Mallat A, Launoy G, Laurent A. Liver transplantation versus liver resection for hepatocellular carcinoma in intention to treat: an attempt to perform an ideal meta-analysis. Liver Transpl. 2017;23(6):836–844. doi: 10.1002/lt.24758. [DOI] [PubMed] [Google Scholar]
- 90.Mourad A, Deuffic-Burban S, Ganne-Carrie N, Renaut-Vantroys T, Rosa I, Bouvier AM, Launoy G, Cattan S, Louvet A, Dharancy S, Trinchet JC, Yazdanpanah Y, Mathurin P. Hepatocellular carcinoma screening in patients with compensated hepatitis C virus (HCV)-related cirrhosis aware of their HCV status improves survival: a modeling approach. Hepatology. 2014;59(4):1471–1481. doi: 10.1002/hep.26944. [DOI] [PubMed] [Google Scholar]
- 91.Choi Debra T., Kum Hye-Chung, Park Sulki, Ohsfeldt Robert L., Shen Yu, Parikh Neehar D., Singal Amit G. Hepatocellular Carcinoma Screening Is Associated With Increased Survival of Patients With Cirrhosis. Clinical Gastroenterology and Hepatology. 2019;17(5):976-987.e4. doi: 10.1016/j.cgh.2018.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Moon AM, Weiss NS, Beste LA, Su F, Ho SB, Jin GY, Lowy E, Berry K, Ioannou GN. No Association Between Screening for Hepatocellular Carcinoma and Reduced Cancer-Related Mortality in Patients With Cirrhosis. Gastroenterology. 2018;155(4):1128–1139 e1126. doi: 10.1053/j.gastro.2018.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tang J, Li Y, Liu K, Zhu Q, Yang WH, Xiong LK, Guo DL. Exosomal miR-9-3p suppresses HBGF-5 expression and is a functional biomarker in hepatocellular carcinoma. Minerva Med. 2018;109(1):15–23. doi: 10.23736/S0026-4806.17.05167-9. [DOI] [PubMed] [Google Scholar]
- 94.Xue X, Zhao Y, Wang X, Qin L, Hu R. Development and validation of serum exosomal microRNAs as diagnostic and prognostic biomarkers for hepatocellular carcinoma. J Cell Biochem. 2019;120(1):135–142. doi: 10.1002/jcb.27165. [DOI] [PubMed] [Google Scholar]
- 95.Xu H, Chen Y, Dong X, Wang X. Serum exosomal long noncoding RNAs ENSG00000258332.1 and LINC00635 for the diagnosis and prognosis of hepatocellular carcinoma. Cancer Epidemiol Biomarkers Prev. 2018;27(6):710–716. doi: 10.1158/1055-9965.epi-17-0770. [DOI] [PubMed] [Google Scholar]
- 96.Liu W, Ren L, Wang X, Wang T, Zhang N, Gao Y, Luo H, Navarro-Alvarez N, Tang L. Combination of exosomes and circulating microRNAs may serve as a promising tumor marker complementary to alpha-fetoprotein for early-stage hepatocellular carcinoma diagnosis in rats. J Cancer Res Clin Oncol. 2015;141(10):1767–1778. doi: 10.1007/s00432-015-1943-0. [DOI] [PubMed] [Google Scholar]
- 97.Sohn W, Kim J, Kang SH, Yang SR, Cho J-Y, Cho HC, Shim SG, Paik Y-H. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp Mol Med. 2015;47(9):e184. doi: 10.1038/emm.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ellett F, Jorgensen J, Marand AL, Liu YM, Martinez MM, Sein V, Butler KL, Lee J, Irimia D. Diagnosis of sepsis from a drop of blood by measurement of spontaneous neutrophil motility in a microfluidic assay. Nat Biomed Eng. 2018;2(4):207–214. doi: 10.1038/s41551-018-0208-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shi M, Jiang Y, Yang L, Yan S, Wang Y, Lu X. Decreased levels of serum exosomal miR-638 predict poor prognosis in hepatocellular carcinoma. J Cell Biochem. 2018;119(6):4711–4716. doi: 10.1002/jcb.26650. [DOI] [PubMed] [Google Scholar]
- 100.Liu W, Hu J, Zhou K, Chen F, Wang Z, Liao B, Dai Z, Cao Y, Fan J, Zhou J. Serum exosomal miR-125b is a novel prognostic marker for hepatocellular carcinoma. Onco Targets Ther. 2017;10:3843–3851. doi: 10.2147/OTT.S140062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Qu Z, Wu J, Wu J, Ji A, Qiang G, Jiang Y, Jiang C, Ding Y. Exosomal miR-665 as a novel minimally invasive biomarker for hepatocellular carcinoma diagnosis and prognosis. Oncotarget. 2017;8(46):80666–80678. doi: 10.18632/oncotarget.20881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee YR, Kim G, Tak WY, Jang SY, Kweon YO, Park JG, Lee HW, Han YS, Chun JM, Park SY, Hur K. Circulating exosomal non-coding RNAs as prognostic biomarkers in human hepatocellular carcinoma. Int J Cancer. 2018 doi: 10.1002/ijc.31931. [DOI] [PubMed] [Google Scholar]
- 103.Sugimachi K, Matsumura T, Hirata H, Uchi R, Ueda M, Ueo H, Shinden Y, Iguchi T, Eguchi H, Shirabe K, Ochiya T, Maehara Y, Mimori K. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br J Cancer. 2015;112(3):532–538. doi: 10.1038/bjc.2014.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Admyre C, Bohle B, Johansson SM, Focke-Tejkl M, Valenta R, Scheynius A, Gabrielsson S. B cell-derived exosomes can present allergen peptides and activate allergen-specific T cells to proliferate and produce TH2-like cytokines. J Allergy Clin Immunol. 2007;120(6):1418–1424. doi: 10.1016/j.jaci.2007.06.040. [DOI] [PubMed] [Google Scholar]
- 105.Liang J, Zhang X, He S, Miao Y, Wu N, Li J, Gan Y. Sphk2 RNAi nanoparticles suppress tumor growth via downregulating cancer cell derived exosomal microRNA. J Control Release. 2018;286:348–357. doi: 10.1016/j.jconrel.2018.07.039. [DOI] [PubMed] [Google Scholar]
- 106.Kosaka N, Iguchi H, Yoshioka Y, Hagiwara K, Takeshita F, Ochiya T. Competitive interactions of cancer cells and normal cells via secretory microRNAs. J Biol Chem. 2012;287(2):1397–1405. doi: 10.1074/jbc.M111.288662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yin Z, Jiang K, Li R, Dong C, Wang L. Multipotent mesenchymal stromal cells play critical roles in hepatocellular carcinoma initiation, progression and therapy. Mol Cancer. 2018;17(1):178. doi: 10.1186/s12943-018-0926-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bruno S, Collino F, Deregibus MC, Grange C, Tetta C, Camussi G. Microvesicles derived from human bone marrow mesenchymal stem cells inhibit tumor growth. Stem Cells Dev. 2013;22(5):758–771. doi: 10.1089/scd.2012.0304. [DOI] [PubMed] [Google Scholar]
- 109.Alzahrani FA, El-Magd MA, Abdelfattah-Hassan A, Saleh AA, Saadeldin IM, El-Shetry ES, Badawy AA, Alkarim S. Potential effect of exosomes derived from cancer stem cells and MSCs on progression of DEN-induced HCC in rats. Stem Cells Int. 2018;2018:8058979. doi: 10.1155/2018/8058979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang X, Sai B, Wang F, Wang L, Wang Y, Zheng L, Li G, Tang J, Xiang J. Hypoxic BMSC-derived exosomal miRNAs promote metastasis of lung cancer cells via STAT3-induced EMT. Mol Cancer. 2019;18(1):40. doi: 10.1186/s12943-019-0959-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fonsato V, Collino F, Herrera MB, Cavallari C, Deregibus MC, Cisterna B, Bruno S, Romagnoli R, Salizzoni M, Tetta C, Camussi G. Human liver stem cell-derived microvesicles inhibit hepatoma growth in SCID mice by delivering antitumor microRNAs. Stem Cells. 2012;30(9):1985–1998. doi: 10.1002/stem.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lopatina T, Grange C, Fonsato V, Tapparo M, Brossa A, Fallo S, Pitino A, Herrera-Sanchez MB, Kholia S, Camussi G, Bussolati B. Extracellular vesicles from human liver stem cells inhibit tumor angiogenesis. Int J Cancer. 2018 doi: 10.1002/ijc.31796. [DOI] [PubMed] [Google Scholar]
- 113.Lou G, Song X, Yang F, Wu S, Wang J, Chen Z, Liu Y. Exosomes derived from miR-122-modified adipose tissue-derived MSCs increase chemosensitivity of hepatocellular carcinoma. J Hematol Oncol. 2015;8:122. doi: 10.1186/s13045-015-0220-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liang G, Kan S, Zhu Y, Feng S, Feng W, Gao S. Engineered exosome-mediated delivery of functionally active miR-26a and its enhanced suppression effect in HepG2 cells. Int J Nanomed. 2018;13:585–599. doi: 10.2147/IJN.S154458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang F, Li L, Piontek K, Sakaguchi M, Selaru FM. Exosome miR-335 as a novel therapeutic strategy in hepatocellular carcinoma. Hepatology. 2018;67(3):940–954. doi: 10.1002/hep.29586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li H, Yang C, Shi Y, Zhao L. Exosomes derived from siRNA against GRP78 modified bone-marrow-derived mesenchymal stem cells suppress Sorafenib resistance in hepatocellular carcinoma. J Nanobiotechnol. 2018;16(1):103. doi: 10.1186/s12951-018-0429-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li J, Huang S, Zhou Z, Lin W, Chen S, Chen M, Ye Y. Exosomes derived from rAAV/AFP-transfected dendritic cells elicit specific T cell-mediated immune responses against hepatocellular carcinoma. Cancer Manag Res. 2018;10:4945–4957. doi: 10.2147/CMAR.S178326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136(1):26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 119.Taghikhani A, Hassan ZM, Ebrahimi M, Moazzeni SM. microRNA modified tumor-derived exosomes as novel tools for maturation of dendritic cells. J Cell Physiol. 2018 doi: 10.1002/jcp.27626. [DOI] [PubMed] [Google Scholar]
- 120.Usman WM, Pham TC, Kwok YY, Vu LT, Ma V, Peng B, Chan YS, Wei L, Chin SM, Azad A, He AB, Leung AYH, Yang M, Shyh-Chang N, Cho WC, Shi J, Le MTN. Efficient RNA drug delivery using red blood cell extracellular vesicles. Nat Commun. 2018;9(1):2359. doi: 10.1038/s41467-018-04791-8. [DOI] [PMC free article] [PubMed] [Google Scholar]