Abstract
Experimental evidence for a direct role of lipids in determining the structure, dynamics, and function of membrane proteins leads to the term ‘functional lipids’. In particular, the sterol molecule cholesterol modulates the activity of many membrane proteins. The precise nature of cholesterol-binding sites and the consequences of modulation of local membrane micro-viscosity by cholesterol, however, is often unknown. Here, we review the current knowledge of the interaction of cholesterol with transmembrane proteins, with a special focus on structural aspects of the interaction derived from nuclear magnetic resonance approaches. We highlight examples of the importance of cholesterol modulation of membrane protein function, discuss the specificity of cholesterol binding, and review the proposed binding motifs from a molecular perspective. We conclude with a short perspective on what could be future trends in research efforts targeted towards a better understanding of cholesterol/membrane protein interactions.
Keywords: Membrane protein, Lipid, Cholesterol, Function, NMR spectroscopy, Structure
Introduction: transmembrane proteins and their functional lipids
The biological membrane plays a vital role in a variety of biological processes, including transport, cellular recognition, adhesion, energy production, and signaling cascades. The membrane properties, which influence these processes, are a result of a complex interplay between the protein and the lipid components [1]. While it is nowadays well accepted that lipid–protein interactions are essential for the aforementioned cellular processes, only little is known about their physicochemical nature as well as their actual role in these processes [1, 2]. This is because lipids and proteins can influence one another in multiple ways [3–5]. Membrane proteins influence the structure and dynamics of lipids, such as acyl chain order, membrane thickness and elasticity, permeability, lipid-domain formation, lipid head-group orientation, and acyl chain dynamics [6]. In addition, in turn, the physicochemical nature of the membrane profoundly impacts the structure and dynamics of membrane-embedded proteins and peptides [7], and thereby modulates their function [6, 8, 9].
According to the fluid mosaic model, cellular membranes were originally thought to primarily serve as solvents for membrane proteins [10]. This view changed when the functional raft hypothesis was developed [11], and was further strengthened through the discovery of lipid molecules that act as secondary messengers [12, 13]. Nowadays, there is strong evidence that lipids play a direct role in determining the structure, dynamics, and function of membrane proteins [14], leading to the formulation of the term “functional lipids”.
The activity of a variety of ion channels, including the members of all major ion channel families, is affected by changes in membrane cholesterol levels [15]. Consistent with a functional importance of cholesterol and other lipids, crystal structures of ion channels and different membrane protein classes revealed bound lipids [16]. For example, nonan-1-ol and diacylglycerol molecules were observed in complex with the K+ channel KcsA [17]. In addition, phosphatidylglycerol and cardiolipin influence oligomerization of the voltage-dependent anion channel (VDAC) [18], which is the major ATP gate in the outer mitochondrial membrane and plays an important role in apoptosis [19]. The membrane-lipid composition also modulates the interaction of VDAC with tubulin [20], as well as the gating properties of the VDAC channel [21].
A very important lipid is cholesterol, because it influences the function of many membrane proteins. Robust and accurate experimental information on the precise nature of cholesterol-binding sites in membrane protein structures, however, is often lacking. Molecular flexibility of both protein structures and cholesterol molecules adds to the complexity of studying the interaction of cholesterol with transmembrane proteins. In addition, technical difficulties have to be overcome, such as the insolubility of cholesterol in polar environments.
Cholesterol–protein interaction motifs
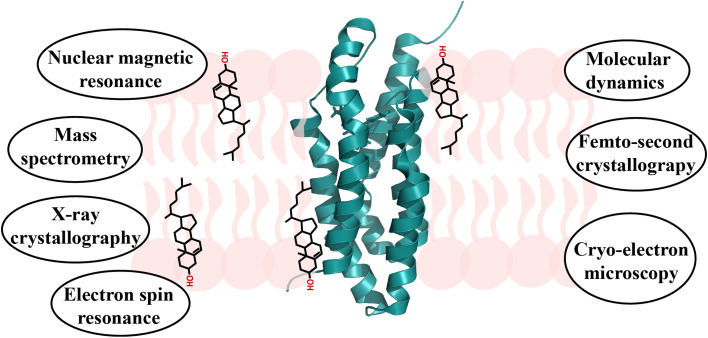
X-ray crystallography [22, 23], electron spin resonance [24–27], cryo-electron microscopy [28–30], binding assays employing site-directed mutagenesis [31–33], docking and molecular dynamics simulations [34–36], and nuclear magnetic resonance (NMR) [37] spectroscopy have been used to characterize the interaction of cholesterol with transmembrane proteins (Fig. 1). Based on these studies, different cholesterol-binding motifs in transmembrane proteins have been proposed. These include the cholesterol recognition amino acid consensus (CRAC) motif and its inverted variant, the CARC motif, the cholesterol consensus motif (CCM) motif, and so-called “tilted peptides” [38, 39]. In addition, other specific or non-universal motifs were suggested [26, 30, 34, 39–41]. The interaction of cholesterol with soluble proteins has also been studied in detail and was summarized in a recent review based on the analysis of crystal structures of proteins containing cholesterol molecules [42]. The analysis showed that the hydroxyl group of cholesterol preferentially interacts with Asn, Gln, and/or Tyr residues, whereas the hydrophobic part of cholesterol makes contacts with Leu, Ile, Val, and Phe residues. Moreover, the hydroxyl group likes to form hydrogen bonds with residues in protein α-helices, while cholesterol’s hydrophobic core often interacts with residues in β-strands or regions that do not fold into regular secondary structure [42]. As we summarize below, there appear to be commonalities with respect to protein/cholesterol interactions between soluble and transmembrane proteins, that is similar residue types and protein secondary structure elements contribute to binding to the polar and hydrophobic parts of the cholesterol molecule.
Fig. 1.
Illustration of the complex interplay of cholesterol (black) with membrane environment (pink) and proteins (green). Selected techniques used to study protein–lipid interactions are listed
An often discussed cholesterol-binding motif is CRAC [32, 38, 39], which follows the sequence composition (from N- to C-terminus): apolar leucine or valine residue, followed by one to five residues of any type, then a mandatory aromatic residue (tyrosine), followed by another segment of one to five residues, and then “capped” by a final basic lysine or arginine residue, i.e., (L/V)-X1–5-(Y)-X1–5-(K/R). Since cholesterol does not display any structural and chemical variations, the looseness in the definition of the CRAC motif is surprising and raised some skepticism about its predictive value [43, 44]. Sequence analysis identified CRAC motifs in a number of transmembrane proteins, which are known to bind cholesterol, including G-protein-coupled receptors [45–47] and the ion-channel large-conductance Ca2+-sensitive voltage-gated K+ channels (BK), nicotinic acetylcholine receptor (nAChR), and Kir2.1 [39], as well as the translocator protein TSPO [32, 48]. Consistent with the importance of the CRAC motif, single mutations within this sequence motif, such as substitution of the central tyrosine residue, attenuated cholesterol binding [44, 49, 50]. In addition, it was suggested that binding of cholesterol to the CRAC motif is energetically favorable if the motif is located within the transmembrane region, whereas binding to CRAC motifs outside of the membrane would be energetically unfavorable [38]. For example, measurements of cholesterol-dependent channel activity in combination with computational studies suggested that the membrane-adjacent CRAC motif, V444—Y450—K453, from a total of seven CRAC motifs that are present in the cytosolic domain of the BK channel, contributes to the sensitivity of the channel to cholesterol [51]. In addition, CRAC motifs of the ion channels nAChR and Kir2.1, which are located outside of the membrane bilayer, were suggested to be energetically less favorable for cholesterol binding [41, 51]. Experimental confirmation of cholesterol binding to CRAC motifs of transmembrane proteins was, however, obtained in only a few cases [31].
Very similar to the CRAC motif is the sequence motif called CARC. CARC represents an “inverted CRAC” motif with the amino acid composition (K/R)-X1–5-(Y/F)-X1–5-(L/V) [52]. The definition of CARC is even less restrictive than that of CRAC by including either tyrosine or phenylalanine in the central position. Due to the “snorkeling” effect [53], which is attributed to the burial of the side-chain of lysine (or arginine) in the hydrophobic part of the membrane and emergence of its cationic group at the membrane surface [52], the presence of the basic residue allows location of the CARC motif at the polar–apolar interface of a transmembrane domain. CARC motifs are present in the transmembrane domains of transient receptor potential vanilloid 1 channels (TRPV1) [31], nAChR [38], and Kir2.1 [41]. In TRPV1, the sequence motif R579—F582—L585 is located in transmembrane helix 5. Mutations of these three characteristic CARC residues affected the sensitivity of TRPV1 to cholesterol in measurements of capsaicin-induced currents [31]. AChR has sequence stretches, which fit to the CARC definition, in transmembrane helices 1, 3 and 4 [38]. Kir2.1 contains CARC motifs in both the cytosolic and transmembrane domain, with two of them located at the interface between the transmembrane and cytosolic region [41]. Substitution of V77 by isoleucine in the CARC motif R67—F73—V77 abolished Kir2.1’s sensitivity towards cholesterol, while mutation of R67 and F73 resulted in non-functional channels. Conversely, mutation of all three residues within a second potential CARC motif, R82—F88—L90, did not affect the cholesterol-sensitivity of Kir2.1 [41]. Experimental structural data for cholesterol binding to the proposed CARC sequence motif are currently lacking.
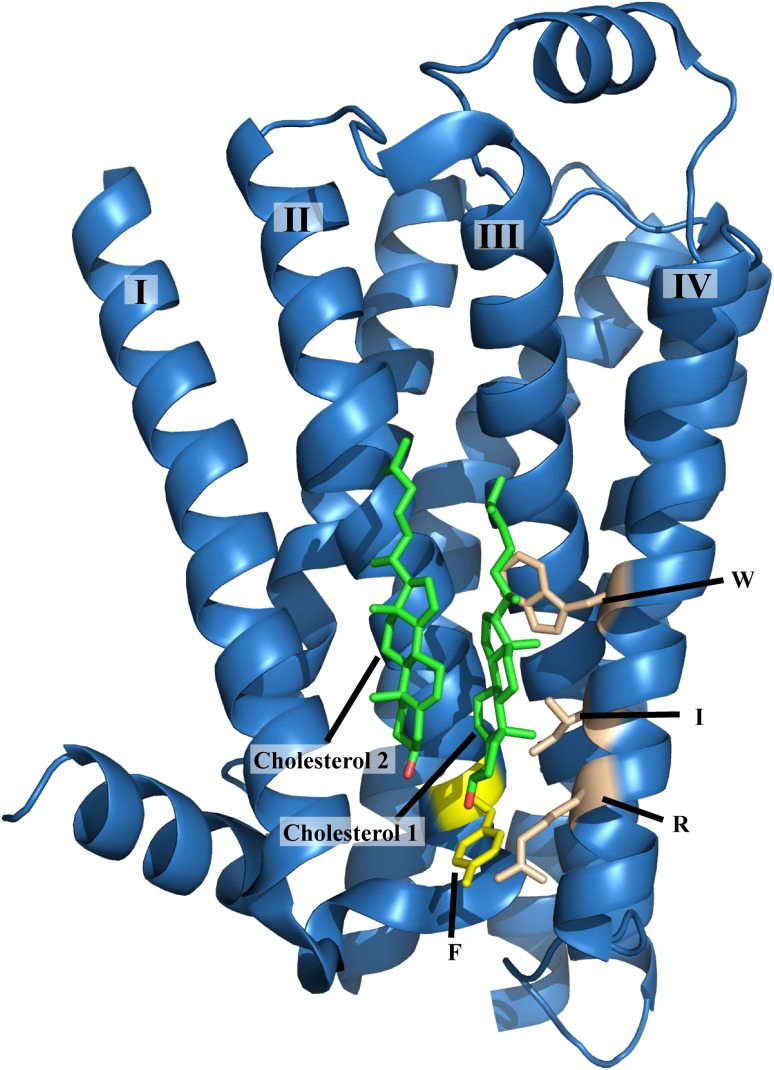
The third cholesterol-binding motif, the so-called cholesterol consensus motif (CCM), was proposed on the basis of the crystal structure of cholesterol bound to the β2-adrenergic receptor [22]. The CCM motif is predominantly found in GPCRs (Fig. 2) [22]. Unlike the proposed CRAC and CARC motifs, which include residues from one continuous segment of the protein, the CCM is a three-dimensional, experimentally validated binding motif that includes residues from adjacent helices, i.e., (W/Y)-(I/V/L)-(K/R) on one helix, and (F/Y/R) on a second helix. Notably, the residue types contributing to the CCM are similar to those in the CRAC and CARC motifs.
Fig. 2.
Cholesterol-binding motif (CCM) of the β2-adrenergic receptor (PDB code: 3D4S). Phenylalanine from one helix, and arginine, isoleucine, and tryptophan from another transmembrane helix contribute to cholesterol (green) binding
Peptides that insert in membranes or snorkel at the membrane surface have also been suggested to interact with cholesterol. Characteristic features of these peptides are their folding into helical structure upon membrane interaction and perturbation of membrane organization [38]. The distribution of hydrophobic residues within these proposed cholesterol-binding peptides is asymmetric and induces a 45° tilt with respect to the membrane plane [54]. One potential example of a tilted peptide is the Alzheimer’s β-amyloid peptide, of which residues 22EDVGSNKGAIIGLM35 bind with high affinity to cholesterol [55]. Peptide sequences with asymmetric distributions of hydrophobic residues are also present in viral fusion proteins, which require cholesterol for membrane insertion [56], suggesting that tilted peptides evolved to acquire cholesterol-binding properties to facilitate biological functions [38]. Indirect support for tilted peptide cholesterol-binding sites was furthermore provided by lipid-mixing assays and leakage of T-cell-like liposomes, and was consistent with molecular modeling predictions [57]. In addition, the protein α-synuclein, which plays an important role in Parkinson’s disease, potentially through formation of oligomeric pores in neuronal membranes [58–60], has been discussed in the context of tilted peptides. An isolated peptide corresponding to the α-synuclein sequence 67GGAVVTGVTAVA78 is toxic for cultured neurons [56] and has been suggested to bind cholesterol in a tilted orientation [61].
Besides the four cholesterol-binding motifs discussed above, transmembrane proteins might have developed other ways to interact with cholesterol. Examples thereof are the two putative non-annular-binding sites in the ion channel Kir2.1 [39, 41], as well as cholesterol interaction sites in VDAC [34, 40]. In case of Kir2.1, a combination of docking studies, all-atom molecular dynamics simulations, and functional assays on channels with site-directed mutagenesis lead to the proposition of two binding sites. The two proposed binding sites are located in the hydrophobic center of the membrane. In addition, a weaker cholesterol-binding site is potentially present at the interface between the cytosolic and the transmembrane domain of Kir2.1. Binding of cholesterol to Kir2.1 was suggested to depend on van der Waals interactions with atoms located in-between-helices [41].
The description above highlights that direct and conclusive evidence for a given cholesterol-binding site is lacking in many cases and the proposed cholesterol-binding motifs can currently at best act as a general guide to help in the design of further experiments. The strongest experimental evidence for cholesterol/transmembrane protein interactions is currently available from X-ray crystallography and cryo-electron microscopy. However, even for these very powerful techniques, it is often not possible to work with lipid environments that faithfully mimic a biological membrane. Indeed, many membrane-mimetic environments, which are used for structure determination of membrane proteins, can differ substantially in their physico-chemical properties from those of native membranes, such as hydrophobicity, monomeric concentrations of amphiphiles, dielectric properties, water concentration, and lateral pressure profile [62]. These differences can compromise the tertiary structure of transmembrane proteins, whereas the structural stability of individual transmembrane helices might be enhanced [62]. In addition, crystallization can potentially change protein and cholesterol conformations, crystal-lattice effects might affect cholesterol binding and crystallization does not capture the dynamic nature of cholesterol/transmembrane protein interactions. To mimic the natural environment of a membrane, NMR and EPR studies focusing on cholesterol/transmembrane protein interactions are best done in liposomes or small unilamellar vesicles, increasing the challenge of obtaining high-quality data. Docking and molecular dynamics simulations still lack exact force fields or suffer from other computational limitations. Functional or biophysical studies, on the other hand, are indirect and by themselves only provide circumstantial evidence.
Annular and non-annular sites for cholesterol interaction
A further classification of cholesterol-binding sites is based on their accessibility: (i) annular sites that are located directly at the transmembrane surface of the protein, and (ii) non-annular sites that are located between transmembrane helices and occluded from membrane phospholipids [39].
Annular lipids are lipids that are in direct contact with the hydrophobic surface of a membrane protein [16]. The interaction between annular lipids and transmembrane proteins occurs through hydrogen bonds, π–π and cation–π interactions, electrostatic, and van der Waals forces [63] and results in transient immobilization of the lipid. Annular-binding sites have been observed for various lipids of the cellular membrane [2]. For example, Marsh and Barrantes detected a population of immobilized lipids in the form of a two-component electron spin resonance spectrum in the vicinity of nAChR [24]. The immobilized lipids included fatty acids, steroids, and several kinds of phospholipids, suggesting that transient lipid binding was not specific [24, 64]. Subsequently, a combination of complementary surface pressure measurements suggested that nAChR interacts preferentially with sterols [65, 66]. Functional studies further revealed that increasing cholesterol levels enhance nAChR-mediated ion flux and functional activity of nAChR [67–69]. In case of the receptor serotonin1A, coarse-grained molecular dynamics simulations pointed to multiple ‘hot-spots’ for transient cholesterol binding [47]. High-occupancy sites, but with a high rate of exchange of cholesterol, were designated as annular-binding sites and were observed in both the extracellular and the intracellular side of the receptor [47, 70]. In addition, atomistic molecular dynamics simulations pointed to a large number of transient annular and non-annular-binding sites in the A2A adenosine receptor [71] and the β2-adrenergic receptor [72, 73]. Notably, cholesterol binding was not only found to be transient in these studies, but also the cholesterol molecules remained mobile in the bound state, stressing the dynamic nature of cholesterol/protein interactions.
The concept of non-annular-binding sites was developed on the basis of the observation that cholesterol does not displace annular phospholipids in nAChR [74]. In addition, electron spin resonance experiments suggested that sterols are immobilized between the transmembrane helices of nAChR [24, 74]. Non-annular cholesterol-binding regions were also proposed for the Ca2 + -ATPase of sarcoplasmic reticulum [75] and the β-adrenergic GPCR [22, 76]. Moreover, a cholesterol molecule is positioned within a groove formed by three transmembrane helices (Ia, V, and VII) of the dopamine receptor [77]. A cholesterol-binding site was furthermore identified in the 5-HT2BERG receptor [78]. In case of the human A2A adenosine receptor, three cholesterol-binding sites were observed in its crystal structure [79].
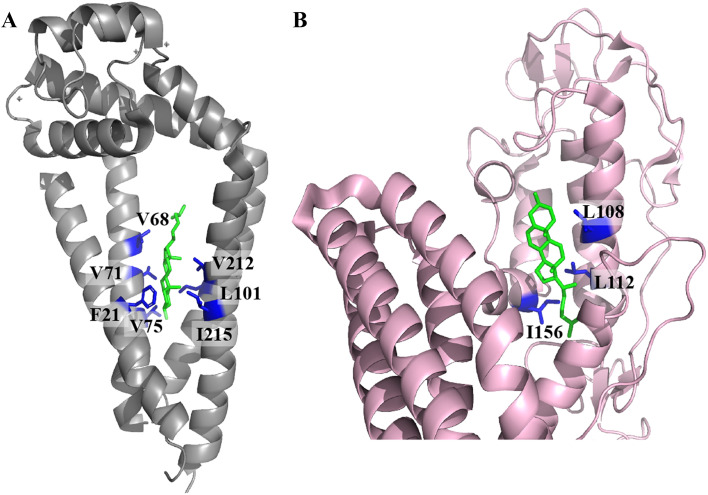
Non-annular cholesterol-binding sites were revealed by X-ray crystallography in several G-protein coupled receptors (GPCRs) (Fig. 3) [80]. For example, two cholesterol molecules were observed between the transmembrane helices of two adjacent molecules (helices I and VII in one molecule and helix I in the adjacent molecule) of the proton pumping rhodopsin ARII [81]. In addition, a cholesterol molecule was also seen in between the transmembrane helices of two μ-opiod receptors [82], and two putative non-annular cholesterol-binding regions were proposed in Kir2.1 channels [41]. Cholesterol-binding sites have also been identified in TSPO [32, 37, 48] and VDAC [34, 40]. Out of the 103 protein data bank (PDB) entries, which contain bound cholesterol molecules, 30 entries with a sequence similarity of less than 90% are listed in Table 1. Some of these proteins have cholesterol close to CRAC and CARC motifs. Many others, however, contain CRAC and/or CARC motifs, but cholesterol is bound to a different site. In addition, several proteins do not contain CRAC and/or CARC motifs although they are able to accommodate cholesterol in their structure (Table 1).
Fig. 3.
Structural representations of non-annular cholesterol-binding sites in two transmembrane proteins. The cholesterol molecule is shown in green and a few residues in close proximity to cholesterol are shown in blue. PDB codes are: a 5TCX (human tetraspanin) and b selected region of 5L7D (human smoothened protein)
Table 1.
Proteins with cholesterol (PDB code: CLR) bound to their structure
| PDB ID | Protein/lipid/detergent used during purification/crystallization | Bounda | CRACb motifs | CARC motifs | Binding CRAC/CARCc |
|---|---|---|---|---|---|
| X-ray diffraction | |||||
| 1LRI | Fungal elicitor cryptogein | 1 | None | None | No |
| 5XRA | CB1/ monoolein | 1 | None | None | No |
| 2RH1 | β-2 adrenergic receptor/ monoolein | 3 | None | None | No |
| Na+/K+ ATPase/PC | |||||
| 2ZXE | α subunit | 1 | 5 | 12 | No |
| β subunit | 3 | 7 | Yes | ||
| 3WGU | α subunit | 6 | 4 | 14 | No |
| β subunit | 5 | 9 | Yes | ||
| 4XT1 | Human chemokine CX3CL1/1-oleoyl-R-glycerol | 2 | 3 | 3 | No |
| μ-opioid receptor/monoolein | |||||
| 4DKL | 1 | None | None | No | |
| 5C1M | 1 | 3 | 2 | No | |
| 4XP9 | Dopamine transporter/DDM, PE | 1 | 4 | 11 | No |
| 1N83 | Nuclear receptor ROR-alpha | 1 | 1 | 2 | Likely |
| 5LWE | Chemokine receptor type 9/DDM | 1 | 3 | 6 | Yes |
| 5I6X | Human serotonin transporter/PC, PE, PG | 1 | 6 | 5 | No |
| 5L7D | Human smoothened protein/DDM | 1 | None | None | No |
| 3N9Y | Human CYP11A1 | 2 | 7 | 6 | No |
| 5JQH | β-2 adrenoceptor/ monoacylglycerol | 2 | None | None | No |
| 4XNV | P2Y purinoceptor 1/DDM | 1 | None | None | No |
| 5IU4 | A2A adenosine receptor | 4 | None | None | No |
| 5X93 | Endothelin B receptor/ LMNG | 1 | None | None | No |
| 5WVR | OSH1 OSBP-related domain | 1 | 8 | 8 | Yes |
| 1ZHY | Oxysterol binding protein Osh4 | 1 | 3 | 11 | Yes |
| 5TCX | Human tetraspanin CD8/monoolein | 1 | 1 | 5 | No |
| 3GKI | N-terminal domain of Niemann-Pick C1 protein | 1 | None | 4 | No |
| 4BOE | Tick lipocalin japanin | 1 | 4 | 4 | Yes |
| 4OR2 | Metabotropic glutamate receptor 1/monoolein | 6 | None | None | No |
| 4IB4 | Chimera of 5-HT2B-BRIL/monoolein | 1 | None | None | No |
| 3AM6 | Proton pumping rhodopsin AR2/monoolein | 8 | 2 | 3 | No |
| 4PXZ | P2Y12 receptor/monoolein | 1 | None | None | No |
| Electron microscopy | |||||
| 5SY1 | STRA6 receptor/ lauryl maltose neopentyl glycol | 2 | 3 | 10 | No |
| 3JD8 | Niemann-Pick C1 protein | 1 | 6 | 18 | No |
| Small angle neutron scattering | |||||
| 3K2S | High density lipoprotein/POPC | 20 | 5 | 4 | No |
Included proteins have less than 90% sequence similarity (date of analysis: 29th September 2017). Cholesterol-like molecules have not been included into the search
PC 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, PE 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine, PG 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol, DDM n-dodecyl-β-d-maltopyranoside, LMNG lauryl maltose neopentyl glycol
aTotal number of cholesterol molecules bound per structure
bCRAC motifs per chain
cCholesterol motif present within 7 Å radius of one of the three defining residues of a CRAC or CARC motif
Importance of cholesterol–protein interactions
Direct binding to protein versus modulation of membrane properties
Cholesterol is known to regulate the activity of ion channels and other transmembrane proteins [39, 83]. From a mechanistic point of view, two scenarios for cholesterol—membrane protein interaction have been proposed. In the first model, cholesterol directly binds to the protein as a ligand and thus influences membrane protein function. In contrast, in the second scenario, cholesterol does not specifically bind to the protein but regulates its activity by altering the physical properties of the surrounding membrane [39, 84]. Notably, a direct interaction does not rule out the possibility that membrane properties have also been altered due to hydrophobic mismatch or change in bilayer thickness [39]. Since the physical properties of membrane bilayers can also be affected by the presence of other sterols, it is difficult to identify the specificity of cholesterol-mediated effects in such cases. The existing studies are, therefore, going into two directions, assessing whether (i) the regulatory effect of cholesterol is specific and (ii) whether cholesterol directly binds to the channel. The former typically involves the use of enantiomers of cholesterol, whereas the latter is based on direct binding essays using radiolabeled cholesterol.
Direct binding of cholesterol has been shown for the transmembrane receptor nAChR, although binding might be unspecific [85]. nAChR–cholesterol interactions stabilize the channel in the resting state, while cholesterol-induced changes in membrane thickness facilitate transitions between the uncoupled and coupled states of nAChR [86]. In addition, cholesterol was found to be required for agonist-induced opening of the GABAA receptor, up-regulating it both specifically and non-specifically [87]. Conversely, the volume-regulated anion channel (VRAC) was shown to be non-specifically down-regulated by cholesterol [88], presumably by the effects on lipid packing around the channel [89]. Cholesterol also down-regulates the activity of inwardly rectifying K+-channels (KIR), by a direct and non-stereospecific interaction with the receptor [90, 91]. Several studies furthermore revealed an inhibitory effect of cholesterol on the activity of BK channels via specific protein–sterol interactions [39, 92–94]. An inhibitory effect of cholesterol as a result of stereoselective binding was also observed for TRPV1 [31]. Notably, direct evidence for cholesterol binding through biochemical methods has so far only been obtained for a small number of channels [95–98], making it often difficult to distinguish between a direct and indirect effect of cholesterol on transmembrane protein activity.
Cholesterol and mitochondrial membrane proteins
Disorders in lipid metabolism and transport play an important role in human disease [99]. Changes in the lipid profile of a cell drastically affect the cellular metabolism and signal transduction [99, 100]. In the particular case of cancer, an up-regulation of lipid metabolism is often observed during early stages of neoplasia and is a recognized hallmark of many types of cancer [100]. Changes in the mitochondrial phospholipid membrane composition, especially in cholesterol, can be triggered by external interventions (e.g., diet) and by a range of biological events (apoptosis, disease, and aging) [101].
Cholesterol is the sole precursor of steroids, whose synthesis at the inner mitochondrial membrane (IMM) requires translocation of cholesterol from the outer mitochondrial membrane (OMM) to the IMM [102]. Improper storage and targeting of cholesterol can be toxic for cells [102]. Cholesterol translocation involves various proteins. One of the first identified cholesterol-binding proteins was the sterol carrier protein-2 (SCP-2). SCP-2 plays a role in intracellular transfer of cholesterol, including the pathway from lysosomal to mitochondrial membranes [103]. Cholesterol, successfully imported via the plasma membrane or accessed through lipid droplets, is transported to the OMM, where it remains segregated until translocation to the IMM. The latter step is rate-limiting for steroidogenesis, and is thought to involve the mitochondrial translocator protein TSPO, previously known as the peripheral-type benzodiazepine receptor [102, 104]. A study employing TSPO ligands demonstrated that ligand binding to TSPO influences cholesterol translocation from the OMM to the IMM [105]. Subsequently, the role of TSPO in cholesterol translocation was supported by experiments using a bacterial TSPO-expression system [106]. In addition, cholesterol was associated with allosteric conformational changes in TSPO [37]. Because TSPO is preferentially located at mitochondrial contact sites, it was furthermore suggested that TSPO does not function alone, but TSPO function is modulated through interactions with other proteins [102]. One of these proteins is VDAC, which binds cholesterol in vivo [107, 108] and in vitro [109], and influences cholesterol distribution in mitochondria [110]. Cholesterol is also important for VDAC gating [109], enhances the structural integrity of isolated VDAC, aids channel insertion into membranes [107, 111, 112], and promotes uniformly open channel conductance [107, 112]. In addition, cholesterol might affect VDAC’s interaction with other proteins [113]. VDAC function could thus be modulated during cancer, as well as aging and disease [101], when the content of cholesterol in the OMM increases [114, 115].
Insights into cholesterol–protein interactions through NMR spectroscopy
X-ray crystallography, NMR and electron spin resonance, and more recently cryo-electron microscopy [116], Fourier transform infrared spectroscopy [117], and femtosecond crystallography [118] have been used to study membrane protein structures and their interactions with lipids. In addition, mass spectrometry-based chemical photoaffinity labeling [119] and molecular dynamics simulations [120] were employed to characterize cholesterol binding and the impact of cholesterol on membrane protein activity.
Progress in sample preparation techniques, and methodological and technological advances have established NMR as an excellent technique to probe the interaction of two interacting species and provide atomic details of biomolecular interactions [121]. Based on these advances, detailed studies of the orientation of cholesterol and its derivatives in membrane environments have been performed by NMR [122]. NMR spectroscopy nowadays provides an important tool to study the impact of cholesterol on membrane protein structure and function.
Solution-state NMR studies of protein–cholesterol interactions
GPCRs form an important class of membrane proteins reported to bind cholesterol. Several GPCRs have been co-crystalized with cholesterol and structurally characterized by X-ray crystallography [80]. Cholesterol is known to impact the function of the GPCR β2AR, both by direct binding and indirectly by influencing the lateral pressure and order of the bilayer [123]. To obtain insight into the interaction of cholesterol with β2AR, Gater et al. performed saturation transfer difference NMR experiments on lipid cubic phase samples containing cholesterol and β2AR [124]. In addition, NMR spectroscopy supported the finding that micelle-solubilized cannabinoid receptor CB2 is structurally stabilized by cholesterol hemisuccinate, a derivative of cholesterol [125].
To gain insight into the interaction of cholesterol with the C-terminal domain of the amyloid precursor protein (C99), Barrett et al. performed solution-state NMR titrations in combination with alanine scanning mutagenesis for C99 solubilized in DHPC/DMPC bicelles [126]. To this end, C99 residues ranging from 690 to 710 were replaced by alanine, followed by titration with increasing concentrations of cholesterol to detect chemical shift changes in 2D 1H–15N correlation spectra. Chemical shift perturbations indicated that cholesterol binds to the GXXXG motif of C99, which is known for its ability to promote homodimerization of transmembrane helices [127].
Hiller and coworkers investigated the interaction of cholesterol with VDAC using solution-state NMR spectroscopy [109]. VDAC, which was solubilized in lauryldimethylamine oxide micelles, showed chemical shift changes in the presence of cholesterol for nine backbone amide residues belonging to β-strands 7, 8, and 11 [109]. In addition, docking and molecular dynamics simulations suggested a total of five possible binding sites for cholesterol in VDAC, with cholesterol molecules primarily located in grooves defined by ridges of hydrophobic and sometimes aromatic residues [34]. Melissa and coworkers furthermore investigated the interaction of VDAC with cholesterol using photoaffinity labeling in combination with mass spectrometry [119]. Based on these studies, it was suggested that the cholesterol-binding pocket of VDAC is localized at the functionally relevant E73 [128] and T83 residues [119].
Solid-state NMR of membrane proteins embedded into cholesterol-containing membrane environments
Solid-state NMR spectroscopy is a powerful method to study the interaction between lipids and membrane-embedded proteins. Luo and coworkers used solid-state NMR to probe the effect of membrane environment on the dynamic properties of the Influenza A M2 peptide [129]. By recording 1H–15N, 1H–13C, and 13C–15N dipolar couplings and 2H quadrupolar couplings, they showed that uniaxial diffusion of this transmembrane peptide is slowed down by two orders of magnitude (< 103 s−1) in cholesterol-rich virus envelope-mimetic membranes. In addition, the structure and dynamics of full-length influenza A M2 proteins’ were shown to be membrane-dependent [130]. Two-dimensional carbon–carbon correlation spectra of DMPC-embedded full-length AM2 showed β-strand chemical shifts for its serine, alanine, and leucine residues. In contrast, chemical shifts characteristic for α-helical secondary structure were observed in a cholesterol-rich membrane environment [130]. In agreement with the influence of cholesterol on the structural properties of transmembrane peptides and proteins [131], residues 7–11 of the human immunodeficiency virus fusion peptide convert from α-helical to β-strand conformation upon interaction with cholesterol [132].
In a recent elegant study, cholesterol binding to influenza M2 has been directly observed by high-resolution solid-state NMR using chain-fluorinated and sterol-deuterated cholesterol [133]. 13C to 19F distance measurements showed that two cholesterol molecules bind to each M2 tetramer [133]. In addition, deuterium NMR spectra of the sterol-deuterated cholesterol provided information about the orientation of cholesterol in the lipid bilayer and were used together with protein–cholesterol distances to derive a structural model of the protein/cholesterol complex [133]. This approach is particularly important, because it overcomes the problem, that chemical shift differences in the absence and presence of cholesterol can come from either direct interaction with cholesterol or cholesterol-induced changes in transmembrane protein structure.
Solid-state NMR of membrane-embedded TSPO
The translocator protein TSPO is preferentially expressed in tissues that synthesize steroids [134] and has been suggested to play an important role in cholesterol translocation from the OMM to the IMM [102]. In vitro, liposome-embedded TSPO binds cholesterol with nanomolar affinity [49]. It is, therefore, of high interest to understand how TSPO interacts with cholesterol, to which site in TSPO cholesterol binds and how cholesterol binding influences the structure and dynamics of TSPO. In addition, an open question is if and how TSPO contributes to translocation of cholesterol from the OMM to the IMM and how synthetic TSPO-specific ligands [135], which are used for diagnostics [135] or therapy [136], influence cholesterol binding.
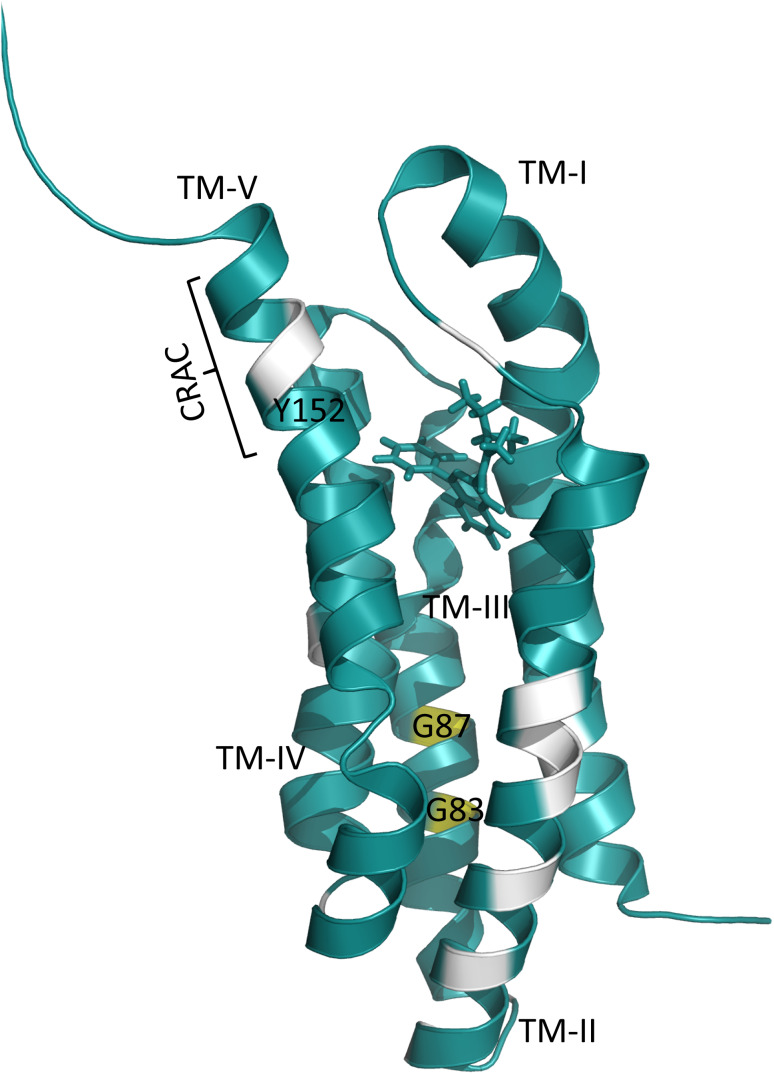
To gain insight into TSPO/ligand interactions, we determined the three-dimensional structure of TSPO from mouse using solution-state NMR [48, 137, 138]. TSPO was solubilized in the detergent fos-choline-12 and structurally stabilized through binding of the radioligand (R)-1-(2-chlorophenyl)-N-methyl-N-(1-methylpropyl)-3-isoquinoline carboxamide ((R)-PK1115). The three-dimensional structure of the TSPO/PK1115 complex revealed a tight bundle of five transmembrane helices. PK1115 is bound to a hydrophobic pocket, which is located in the interior of the structure on the cytosolic side of the lipid bilayer. The C-terminal half of transmembrane helix 5 contains a CRAC motif (residues A147–S159) and mutation of the central tyrosine in the CRAC motif abolishes cholesterol binding in vitro [49]. Side-chains of CRAC-defining residues are pointing away from the TSPO core, suggesting that these side-chains are accessible for binding to membrane-embedded cholesterol in an annular manner [48].
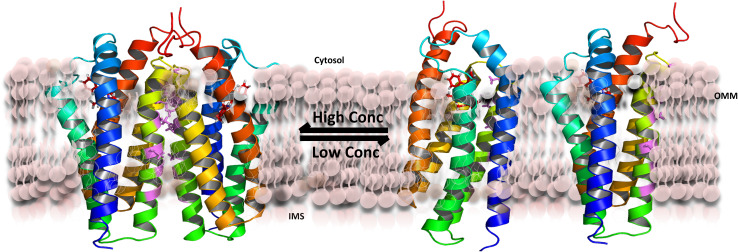
To obtain insight into the TSPO/cholesterol interplay, we reconstituted the protein into 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) liposomes [139]. Similar to mouse TSPO in fos-choline-12 micelles [48], a radioligand—in this case N-(2,5-dimethoxybenzyl)-N-(5-fluoro-2-phenoxyphenyl)acetamide (DAA1106)—increased the structural stability of the protein. A large number of high-resolution solid-state NMR experiments in combination with site-directed mutagenesis demonstrated that mouse TSPO populates a concentration-dependent monomer–dimer equilibrium in DMPC liposomes (Fig. 4). Dimer formation at high protein concentrations is mediated by 83GXXXG87 in transmembrane helix 3—a common motif for dimerization of transmembrane helices [140].
Fig. 4.
Schematic representation of the concentration-dependent monomer–dimer equilibrium of TSPO in DMPC liposomes. The presence of cholesterol favors TSPO monomers
To study the interaction of TSPO with cholesterol, additional NMR samples were prepared, in which the DMPC liposomes contained cholesterol. Because of the known ability of cholesterol to cluster [141], i.e., to make sure that there are a sufficient number of cholesterol molecules, which are able to interact with TSPO, a tenfold excess of cholesterol over protein was used. Moreover, DAA1106 was used to stabilize the TSPO fold. Comparison of solid-state NMR spectra in the absence and presence of cholesterol showed chemical shift changes in different parts of the protein. The affected residues belong to the CRAC motif in helix 5, its neighboring helix 2, as well as the dimerization interface in transmembrane helix 3 (Fig. 5). In addition, quantification of monomer/dimer species by NMR signal intensities indicated that the presence of cholesterol favors TSPO monomerization [37].
Fig. 5.
Residues that experience chemical shift perturbation upon addition of cholesterol are colored white. TM-II undergoes major chemical shift perturbation
To further support a potential cross-talk between the CRAC motif and the dimerization interface in transmembrane helix 3, residue Y152 within the CRAC motif was mutated to serine. This mutation resulted in an overall decrease of NMR spectral quality. Yet, unlike wild-type TSPO, Y152S-TSPO showed no evidence for protein dimerization. Vice versa, mutation of G87V in the dimerization interface caused chemical shift changes for residues in the CRAC motif, further strengthening the communication between the CRAC motif and the dimerization interface of TSPO and suggesting allosteric effects due to cholesterol-binding [37].
Outlook
Through a combination of different experimental techniques, the interaction of cholesterol with transmembrane proteins has been characterized at increasing resolution in recent years. Cholesterol can interact with membrane proteins either at annular or non-annular sites in a specific as well as non-specific manner. In addition, cholesterol changes the physical properties of the membrane and thus is able to indirectly—without specific binding to the protein—modulate the function of membrane proteins. Important aspects of both these mechanisms are cholesterol-mediated changes in the oligomerization of membrane proteins.
The variety of reported cholesterol/protein interactions and the influence of cholesterol on membrane proteins, however, suggest that general rules with respect to protein/cholesterol interactions and their functional consequences are difficult to define. Instead, detailed studies are required that identify cholesterol-binding sites at high resolution and connect it to changes in the structure and dynamics of individual membrane proteins and functional assays with cholesterol. An important aspect of these studies could be the direct and indirect effect of cholesterol on the interactions between different membrane proteins. We believe that NMR spectroscopy, in particular solid-state NMR, can play an important role in this endeavor, because it can simultaneously probe the direct binding of cholesterol and cholesterol-induced changes in the structure and dynamics of peptides and proteins embedded in near native-like membrane environments. In addition, lipid-protein nanodiscs offer a new possibility to structural biologists to study lipid–protein interactions by NMR [142, 143]. An understanding of cholesterol/protein interactions on an atomic-scale level would not only provide more profound insight into physiological and pathophysiological signaling processes related to health and disease, but could also turn out to be valuable for the identification of new treatment strategies.
Acknowledgements
M.Z. was supported by the Deutsche Forschungsgemeinschaft Collaborative Research Center 803 (Project A11) and the European Research Council (Grant agreement number 282008).
Footnotes
Garima Jaipuria and Tina Ukmar-Godec have contributed equally.
References
- 1.Killian JA, Nyholm TKM. Peptides in lipid bilayers: the power of simple models. Curr Opin Struct Biol. 2006;16(4):473–479. doi: 10.1016/j.sbi.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 2.Huster D. Solid-state NMR spectroscopy to study protein lipid interactions. Biochim Biophys Acta Mol Cell Biol Lipids. 2014;1841(8):1146–1160. doi: 10.1016/j.bbalip.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Lee AG. How lipids affect the activities of integral membrane proteins. Biochim Biophys Acta Biomembr. 2004;1666(1–2):62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 4.McIntosh TJ, Simon SA. Roles of bilayer material properties in function and distribution of membrane proteins. Annu Rev Biophys Biomol Struct. 2006;35:177–198. doi: 10.1146/annurev.biophys.35.040405.102022. [DOI] [PubMed] [Google Scholar]
- 5.de Planque MRR, Killian JA. Protein-lipid interactions studied with designed transmembrane peptides: role of hydrophobic matching and interfacial anchoring (review) Mol Membr Biol. 2003;20(4):271–284. doi: 10.1080/09687680310001605352. [DOI] [PubMed] [Google Scholar]
- 6.Soubias O, Gawrisch K. The role of the lipid matrix for structure and function of the GPCR rhodopsin. Biochim Biophys Acta Biomembr. 2012;1818(2):234–240. doi: 10.1016/j.bbamem.2011.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khandelia H, Ipsen JH, Mouritsen OG. The impact of peptides on lipid membranes. Biochim Biophys Acta Biomembr. 2008;1778(7–8):1528–1536. doi: 10.1016/j.bbamem.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 8.Brown MF. Modulation of rhodopsin function by properties of the membrane bilayer. Chem Phys Lipid. 1994;73(1–2):159–180. doi: 10.1016/0009-3084(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 9.Mitchell DC, Niu SL, Litman BJ. Enhancement of G protein-coupled signaling by DHA phospholipids. Lipids. 2003;38(4):437–443. doi: 10.1007/s11745-003-1081-1. [DOI] [PubMed] [Google Scholar]
- 10.Singer SJ, Nicolson GL. The fluid mosaic model of the structure of cell membranes. Science. 1972;175(4023):720. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- 11.Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387(6633):569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 12.Berridge MJ, Irvine RF. Inosito phosphates and cell signaling. Nature. 1989;341(6239):197–205. doi: 10.1038/341197a0. [DOI] [PubMed] [Google Scholar]
- 13.Irvine RF. Inositol lipids in cell signalling. Curr Opin Cell Biol. 1992;4(2):212–219. doi: 10.1016/0955-0674(92)90035-b. [DOI] [PubMed] [Google Scholar]
- 14.Koshy C, Ziegler C. Structural insights into functional lipid-protein interactions in secondary transporters. Biochimica Et Biophysica Acta-General Subjects. 2015;1850(3):476–487. doi: 10.1016/j.bbagen.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 15.Levitan I, Fang Y, Rosenhouse-Dantsker A, Romanenko V (2010) Cholesterol and ion channels. In: Harris JR (ed) Cholesterol binding and cholesterol transport proteins: structure and function in health and disease, vol 51. Subcellular biochemistry, pp 509–549. 10.1007/978-90-481-8622-8_19 [DOI] [PMC free article] [PubMed]
- 16.Lee AG. Lipid-protein interactions in biological membranes: a structural perspective. Biochim Biophys Acta Biomembr. 2003;1612(1):1–40. doi: 10.1016/s0005-2736(03)00056-7. [DOI] [PubMed] [Google Scholar]
- 17.Zhou YF, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a K + channel–fab complex at 2.0 angstrom resolution. Nature. 2001;414(6859):43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 18.Betaneli V, Petrov EP, Schwille P. The role of lipids in VDAC oligomerization. Biophys J. 2012;102(3):523–531. doi: 10.1016/j.bpj.2011.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shoshan-Barmatz V, De Pinto V, Zweckstetter M, Raviv Z, Keinan N, Arbel N. VDAC, a multi-functional mitochondrial protein regulating cell life and death. Mol Aspects Med. 2010;31(3):227–285. doi: 10.1016/j.mam.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 20.Rostovtseva TK, Gurnev PA, Chen MY, Bezrukov SM. Membrane lipid composition regulates tubulin interaction with mitochondrial voltage-dependent anion channel. J Biol Chem. 2012;287(35):29589–29598. doi: 10.1074/jbc.M112.378778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rostovtseva TK, Kazemi N, Weinrich M, Bezrukov SM. Voltage gating of VDAC is regulated by nonlamellar lipids of mitochondrial membranes. J Biol Chem. 2006;281(49):37496–37506. doi: 10.1074/jbc.M602548200. [DOI] [PubMed] [Google Scholar]
- 22.Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola VP, Chien EYT, Velasquez J, Kuhn P, Stevens RC. A specific cholesterol binding site is established by the 2.8 angstrom structure of the human beta(2)-adrenergic receptor. Structure. 2008;16(6):897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwon HJ, Abi-Mosleh L, Wang ML, Deisenhofer J, Goldstein JL, Brown MS, Infante RE. Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell. 2009;137(7):1213–1224. doi: 10.1016/j.cell.2009.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marsh D, Barrantes FJ. Immobilized lipid in acetylcholine receptor-rich membranes from torpedo-marmorata. Proc Natl Acad Sci USA. 1978;75(9):4329–4333. doi: 10.1073/pnas.75.9.4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marsh D. Electron spin resonance in membrane research: protein-lipid interactions from challenging beginnings to state of the art. Eur Biophys J Biophys Lett. 2010;39(4):513–525. doi: 10.1007/s00249-009-0512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song YL, Hustedt EJ, Brandon S, Sanders CR. Competition between homodimerization and cholesterol binding to the C99 domain of the amyloid precursor protein. Biochemistry. 2013;52(30):5051–5064. doi: 10.1021/bi400735x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim SS, Upshur MA, Saotome K, Sahu ID, McCarrick RM, Feix JB, Lorigan GA, Howard KP. Cholesterol-dependent conformational exchange of the C-terminal domain of the influenza A M2 protein. Biochemistry. 2015;54(49):7157–7167. doi: 10.1021/acs.biochem.5b01065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korkhov VM, Sachse C, Short JM, Tate CG. Three-dimensional structure of TspO by electron cryomicroscopy of helical crystals. Structure. 2010;18(6):677–687. doi: 10.1016/j.str.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saibil HR. Conformational changes studied by cryo-electron microscopy. Nat Struct Biol. 2000;7(9):711–714. doi: 10.1038/78923. [DOI] [PubMed] [Google Scholar]
- 30.Taylor NMI, Manolaridis I, Jackson SM, Kowal J, Stahlberg H, Locher KP. Structure of the human multidrug transporter ABCG2. Nature. 2017;546:504. doi: 10.1038/nature22345. [DOI] [PubMed] [Google Scholar]
- 31.Picazo-Juarez G, Romero-Suarez S, Nieto-Posadas A, Llorente I, Jara-Oseguera A, Briggs M, McIntosh TJ, Simon SA, Ladron-de-Guevara E, Islas LD, Rosenbaum T. Identification of a binding motif in the S5 helix that confers cholesterol sensitivity to the TRPV1 ion channel. J Biol Chem. 2011;286(28):24966–24976. doi: 10.1074/jbc.M111.237537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Papadopoulos V. Peripheral-type benzodiazepine receptor function in cholesterol transport. Identification of a putative cholesterol recognition/interaction amino acid sequence and consensus patterns. Endocrinology. 1998;139(12):4991–4997. doi: 10.1210/en.139.12.4991. [DOI] [PubMed] [Google Scholar]
- 33.Li H, Yao ZX, Degenhardt B, Teper G, Papadopoulos V. Cholesterol binding at the cholesterol recognition/interaction amino acid consensus (CRAC) of the peripheral-type benzodiazepine receptor and inhibition of steroidogenesis by an HIV TAT-CRAC peptide. Proc Natl Acad Sci USA. 2001;98(3):1267–1272. doi: 10.1073/pnas.031461598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiser BP, Salari R, Eckenhoff RG, Brannigan G. Computational investigation of cholesterol binding sites on mitochondrial VDAC. J Phys Chem B. 2014;118(33):9852–9860. doi: 10.1021/jp504516a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henin J, Salari R, Murlidaran S, Brannigan G. A predicted binding site for cholesterol on the GABA(A) receptor. Biophys J. 2014;106(9):1938–1949. doi: 10.1016/j.bpj.2014.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brannigan G, Henin J, Law R, Eckenhoff R, Klein ML. Embedded cholesterol in the nicotinic acetylcholine receptor. Proc Natl Acad Sci USA. 2008;105(38):14418–14423. doi: 10.1073/pnas.0803029105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaipuria G, Leonov A, Giller K, Vasa SK, Jaremko L, Jaremko M, Linser R, Becker S, Zweckstetter M. Cholesterol-mediated allosteric regulation of the mitochondrial translocator protein structure. Nat Commun. 2017 doi: 10.1038/ncomms14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fantini J, Yahi N, Garmy N. Cholesterol accelerates the binding of Alzheimer’s beta-amyloid peptide to ganglioside GM1 through a universal hydrogen-bond-dependent sterol tuning of glycolipid conformation. Front Physiol. 2013 doi: 10.3389/fphys.2013.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levitan I, Singh DK, Rosenhouse-Dantsker A. Cholesterol binding to ion channels. Front Physiol. 2014 doi: 10.3389/fphys.2014.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cross TA, Sharma M, Yi M, Zhou HX. Influence of solubilizing environments on membrane protein structures. Trends Biochem Sci. 2011;36(2):117–125. doi: 10.1016/j.tibs.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenhouse-Dantsker A, Noskov S, Durdagi S, Logothetis DE, Levitan I. Identification of novel cholesterol-binding regions in Kir2 channels. J Biol Chem. 2013;288(43):31154–31164. doi: 10.1074/jbc.M113.496117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bukiya AN, Dopico AM. Common structural features of cholesterol binding sites in crystallized soluble proteins. J Lipid Res. 2017;58(6):1044–1054. doi: 10.1194/jlr.R073452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palmer M. Cholesterol and the activity of bacterial toxins. FEMS Microbiol Lett. 2004;238(2):281–289. doi: 10.1016/j.femsle.2004.07.059. [DOI] [PubMed] [Google Scholar]
- 44.Epand RM. Cholesterol and the interaction of proteins with membrane domains. Prog Lipid Res. 2006;45(4):279–294. doi: 10.1016/j.plipres.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 45.Jafurulla M, Tiwari S, Chattopadhyay A. Identification of cholesterol recognition amino acid consensus (CRAC) motif in G-protein coupled receptors. Biochem Biophys Res Commun. 2011;404(1):569–573. doi: 10.1016/j.bbrc.2010.12.031. [DOI] [PubMed] [Google Scholar]
- 46.Oddi S, Dainese E, Fezza F, Lanuti M, Barcaroli D, De Laurenzi V, Centonze D, Maccarrone M. Functional characterization of putative cholesterol binding sequence (CRAC) in human type-1 cannabinoid receptor. J Neurochem. 2011;116(5):858–865. doi: 10.1111/j.1471-4159.2010.07041.x. [DOI] [PubMed] [Google Scholar]
- 47.Sengupta D, Chattopadhyay A. Identification of cholesterol binding sites in the serotonin(1A) receptor. J Phys Chem B. 2012;116(43):12991–12996. doi: 10.1021/jp309888u. [DOI] [PubMed] [Google Scholar]
- 48.Jaremko Ł, Jaremko M, Giller K, Becker S, Zweckstetter M. Structure of the mitochondrial translocator protein in complex with a diagnostic ligand. Science. 2014;343(6177):1363. doi: 10.1126/science.1248725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jamin N, Neumann JM, Ostuni MA, Vu TKN, Yao ZX, Murail S, Robert JC, Giatzakis C, Papadopoulos V, Lacapere JJ. Characterization of the cholesterol recognition amino acid consensus sequence of the peripheral-type benzodiazepine receptor. Mol Endocrinol. 2005;19(3):588–594. doi: 10.1210/me.2004-0308. [DOI] [PubMed] [Google Scholar]
- 50.Epand RF, Thomas A, Brasseur R, Vishwanathan SA, Hunter E, Epand RM. Juxtamembrane protein segments that contribute to recruitment of cholesterol into domains. Biochemistry. 2006;45(19):6105–6114. doi: 10.1021/bi060245+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singh AK, McMillan J, Bukiya AN, Burton B, Parrill AL, Dopico AM. Multiple cholesterol recognition/interaction amino acid consensus (CRAC) motifs in cytosolic C tail of Slo1 subunit determine cholesterol sensitivity of Ca2 + - and voltage-gated K + (BK) channels. J Biol Chem. 2012;287(24):20509–20521. doi: 10.1074/jbc.M112.356261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baier CJ, Fantini J, Barrantes FJ. Disclosure of cholesterol recognition motifs in transmembrane domains of the human nicotinic acetylcholine receptor. Sci Rep. 2011 doi: 10.1038/srep00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strandberg E, Killian JA. Snorkeling of lysine side chains in transmembrane helices: how easy can it get? FEBS Lett. 2003;544(1–3):69–73. doi: 10.1016/s0014-5793(03)00475-7. [DOI] [PubMed] [Google Scholar]
- 54.Lins L, Decaffmeyer M, Thomas A, Brasseur R. Relationships between the orientation and the structural properties of peptides and their membrane interactions. Biochim Biophys Acta Biomembr. 2008;1778(7–8):1537–1544. doi: 10.1016/j.bbamem.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 55.Di Scala C, Yahi N, Lelievre C, Garmy N, Chahinian H, Fantini J. Biochemical identification of a linear cholesterol-binding domain within Alzheimer’s beta amyloid peptide. ACS Chem Neurosci. 2013;4(3):509–517. doi: 10.1021/cn300203a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fantini J, Carlus D, Yahi N. The fusogenic tilted peptide (67–78) of alpha-synuclein is a cholesterol binding domain. Biochim Biophys Acta Biomembr. 2011;1808(10):2343–2351. doi: 10.1016/j.bbamem.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 57.Charloteaux B, Lorin A, Crowet JM, Stroobant V, Lins L, Thomas A, Brasseur R. The N-terminal 12 residue long peptide of HIV gp41 is the minimal peptide sufficient to induce significant T-cell-like membrane destabilization in vitro. J Mol Biol. 2006;359(3):597–609. doi: 10.1016/j.jmb.2006.04.018. [DOI] [PubMed] [Google Scholar]
- 58.van Rooijen BD, Claessens M, Subramaniam V. Membrane interactions of oligomeric alpha-synuclein: potential role in Parkinson’s disease. Curr Protein Pept Sci. 2010;11(5):334–342. doi: 10.2174/138920310791330659. [DOI] [PubMed] [Google Scholar]
- 59.Tosatto L, Andrighetti AO, Plotegher N, Antonini V, Tessari I, Ricci L, Bubacco L, Serra MD. Alpha-synuclein pore forming activity upon membrane association. Biochim Biophys Acta Biomembr. 2012;1818(11):2876–2883. doi: 10.1016/j.bbamem.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 60.Fantini J, Yahi N. Molecular insights into amyloid regulation by membrane cholesterol and sphingolipids: common mechanisms in neurodegenerative diseases. Expert Rev Mol Med. 2010 doi: 10.1017/s1462399410001602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Crowet JM, Lins L, Dupiereux I, Elmoualija B, Lorin A, Charloteaux B, Stroobant V, Heinen E, Brasseur R. In silico tilted properties of the 67–78 fragment of alpha-synuclein are responsible for membrane destabilization and neurotoxicity. Proteins Struct Funct Bioinform. 2007;68(4):936–947. doi: 10.1002/prot.21483. [DOI] [PubMed] [Google Scholar]
- 62.Zhou HX, Cross TA. Influences of membrane mimetic environments on membrane protein structures. Annu Rev Biophys. 2013;42:361–392. doi: 10.1146/annurev-biophys-083012-130326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Soubias O, Gawrisch K. Rhodopsin–lipid interactions studied by NMR. In: Conn PM (ed) G protein coupled receptors: modeling, activation, interactions and virtual screening. Methods Enzymol. 2013;522:209–227. doi: 10.1016/b978-0-12-407865-9.00012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marsh D, Watts A, Barrantes FJ. Phospholipid chain immobilization and steroid rotational immobilization in acetylcholine receptor-rich membranes from torpedo-marmorata. Biochem Biophys Acta. 1981;645(1):97–101. doi: 10.1016/0005-2736(81)90516-2. [DOI] [PubMed] [Google Scholar]
- 65.Popot JL, Demel RA, Sobel A, Vandeenen LLM, Changeux JP. Interaction of acetylcholine (nicotinic) receptor protein from torpedo-marmorata electric organ with monolayers of pure lipids. Eur J Biochem. 1978;85(1):27–42. doi: 10.1111/j.1432-1033.1978.tb12209.x. [DOI] [PubMed] [Google Scholar]
- 66.Ellena JF, Blazing MA, McNamee MG. Lipid–protein interactions in reconstituted membranes containing acetylcholine-receptor. Biochemistry. 1983;22(24):5523–5535. doi: 10.1021/bi00293a012. [DOI] [PubMed] [Google Scholar]
- 67.Dalziel AW, Rollins ES, McNamee MG. The effect of cholesterol on agonist-induced flux in reconstituted acetylcholine-receptor vesicles. FEBS Lett. 1980;122(2):193–196. doi: 10.1016/0014-5793(80)80435-2. [DOI] [PubMed] [Google Scholar]
- 68.Barrantes FJ. Structural basis for lipid modulation of nicotinic acetylcholine receptor function. Brain Res Rev. 2004;47(1–3):71–95. doi: 10.1016/j.brainresrev.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 69.Barrantes FJ. Cholesterol effects on nicotinic acetylcholine receptor. J Neurochem. 2007;103:72–80. doi: 10.1111/j.1471-4159.2007.04719.x. [DOI] [PubMed] [Google Scholar]
- 70.Sengupta D, Chattopadhyay A. Molecular dynamics simulations of GPCR–cholesterol interaction: an emerging paradigm. Biochim Biophys Acta Biomembr. 2015;1848(9):1775–1782. doi: 10.1016/j.bbamem.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 71.Lee JY, Lyman E. Predictions for cholesterol interaction sites on the A(2A) adenosine receptor. J Am Chem Soc. 2012;134(40):16512–16515. doi: 10.1021/ja307532d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Prasanna X, Chattopadhyay A, Sengupta D. Cholesterol modulates the dimer interface of the beta(2)-adrenergic receptor via cholesterol occupancy sites. Biophys J. 2014;106(6):1290–1300. doi: 10.1016/j.bpj.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cang XH, Du Y, Mao YY, Wang YY, Yang HY, Jiang HL. Mapping the functional binding sites of cholesterol in beta(2)-adrenergic receptor by long-time molecular dynamics simulations. J Phys Chem B. 2013;117(4):1085–1094. doi: 10.1021/jp3118192. [DOI] [PubMed] [Google Scholar]
- 74.Jones OT, McNamee MG. Annular and nonannular binding-sites for cholesterol associated with the nicotinic acetylcholine-receptor. Biochemistry. 1988;27(7):2364–2374. doi: 10.1021/bi00407a018. [DOI] [PubMed] [Google Scholar]
- 75.Simmonds AC, East JM, Jones OT, Rooney EK, McWhirter J, Lee AG. Annular and non-annular binding-sites on the (Ca2 + + Mg2 +)-ATPase. Biochem Biophys Acta. 1982;693(2):398–406. doi: 10.1016/0005-2736(82)90447-3. [DOI] [PubMed] [Google Scholar]
- 76.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-resolution crystal structure of an engineered human beta(2)-adrenergic G protein-coupled receptor. Science. 2007;318(5854):1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Penmatsa A, Wang KH, Gouaux E. X-ray structure of dopamine transporter elucidates antidepressant mechanism. Nature. 2013;503(7474):85. doi: 10.1038/nature12533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wacker D, Wang C, Katritch V, Han GW, Huang XP, Vardy E, McCorvy JD, Jiang Y, Chu MH, Siu FY, Liu W, Xu HE, Cherezov V, Roth BL, Stevens RC. Structural features for functional selectivity at serotonin receptors. Science. 2013;340(6132):615–619. doi: 10.1126/science.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu W, Chun E, Thompson AA, Chubukov P, Xu F, Katritch V, Han GW, Roth CB, Heitman LH, Ijzerman AP, Cherezov V, Stevens RC. Structural basis for allosteric regulation of GPCRs by sodium ions. Science. 2012;337(6091):232–236. doi: 10.1126/science.1219218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yeagle PL. Non-covalent binding of membrane lipids to membrane proteins. Biochim Biophys Acta Biomembr. 2014;1838(6):1548–1559. doi: 10.1016/j.bbamem.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 81.Wada T, Shimono K, Kikukawa T, Hato M, Shinya N, Kim SY, Kimura-Someya T, Shirouzu M, Tamogami J, Miyauchi S, Jung KH, Kamo N, Yokoyama S. Crystal structure of the eukaryotic light-driven proton-pumping rhodopsin, acetabularia rhodopsin II, from marine alga. J Mol Biol. 2011;411(5):986–998. doi: 10.1016/j.jmb.2011.06.028. [DOI] [PubMed] [Google Scholar]
- 82.Manglik A, Kruse AC, Kobilka TS, Thian FS, Mathiesen JM, Sunahara RK, Pardo L, Weis WI, Kobilka BK, Granier S. Crystal structure of the mu-opioid receptor bound to a morphinan antagonist. Nature. 2012;485(7398):321–U370. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Burger K, Gimpl G, Fahrenholz F. Regulation of receptor function by cholesterol. Cell Mol Life Sci. 2000;57(11):1577–1592. doi: 10.1007/pl00000643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brown MF. Soft matter in lipid–protein interactions. Ann Rev Biophys. 2017;46:379–410. doi: 10.1146/annurev-biophys-070816-033843. [DOI] [PubMed] [Google Scholar]
- 85.Addona GH, Sandermann H, Kloczewiak MA, Miller KW. Low chemical specificity of the nicotinic acetylcholine receptor sterol activation site. Biochim Biophys Acta Biomembr. 2003;1609(2):177–182. doi: 10.1016/s0005-2736(02)00685-5. [DOI] [PubMed] [Google Scholar]
- 86.Dacosta CJB, Dey L, Therien JPD, Baenziger JE. A distinct mechanism for activating uncoupled nicotinic acetylcholine receptors. Nat Chem Biol. 2013;9(11):701. doi: 10.1038/nchembio.1338. [DOI] [PubMed] [Google Scholar]
- 87.Sooksawate T, Simmonds MA. Effects of membrane cholesterol on the sensitivity of the GABA(A) receptor to GABA in acutely dissociated rat hippocampal neurones. Neuropharmacology. 2001;40(2):178–184. doi: 10.1016/s0028-3908(00)00159-3. [DOI] [PubMed] [Google Scholar]
- 88.Klausen TK, Hougaard C, Hoffmann EK, Pedersen SF. Cholesterol modulates the volume-regulated anion current in Ehrlich–Lettre ascites cells via effects on Rho and F-actin. Am J Physiol Cell Physiol. 2006;291(4):C757–C771. doi: 10.1152/ajpcell.00029.2006. [DOI] [PubMed] [Google Scholar]
- 89.Romanenko VG, Rothblat GH, Levitan I. Sensitivity of volume-regulated anion current to cholesterol structural analogues. J Gen Physiol. 2004;123(1):77–87. doi: 10.1085/jgp.200308882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Romanenko VG, Rothblat GH, Levitan I. Modulation of endothelial inward-rectifier K + current by optical isomers of cholesterol. Biophys J. 2002;83(6):3211–3222. doi: 10.1016/S0006-3495(02)75323-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Romanenko VG, Fang Y, Byfield F, Travis AJ, Vandenberg CA, Rothblat GH, Levitan I. Cholesterol sensitivity and lipid raft targeting of Kir2.1 channels. Biophys J. 2004;87(6):3850–3861. doi: 10.1529/biophysj.104.043273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bolotina V, Omelyanenko V, Heyes B, Ryan U, Bregestovski P. Variations of membrane cholesterol alter the kinetics of Ca2+ -dependent K + channels and membrane fluidity in vascular smooth-muscle cells. Pflugers Arch Eur J Physiol. 1989;415(3):262–268. doi: 10.1007/bf00370875. [DOI] [PubMed] [Google Scholar]
- 93.Bukiya AN, Belani JD, Rychnovsky S, Dopico AM. Specificity of cholesterol and analogs to modulate BK channels points to direct sterol-channel protein interactions. J Gen Physiol. 2011;137(1):93–110. doi: 10.1085/jgp.201010519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bukiya AN, Vaithianathan T, Toro L, Dopico AM. The second transmembrane domain of the large conductance, voltage- and calcium-gated potassium channel beta(1) subunit is a lithocholate sensor. FEBS Lett. 2008;582(5):673–678. doi: 10.1016/j.febslet.2008.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gimpl G, Burger K, Fahrenholz F. Cholesterol as modulator of receptor function. Biochemistry. 1997;36(36):10959–10974. doi: 10.1021/bi963138w. [DOI] [PubMed] [Google Scholar]
- 96.Infante RE, Wang ML, Radhakrishnan A, Kwon HJ, Brown MS, Goldstein JL. NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc Natl Acad Sci USA. 2008;105(40):15287–15292. doi: 10.1073/pnas.0807328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Radhakrishnan A, Sun LP, Kwon HJ, Brown MS, Goldstein JL. Direct binding of cholesterol to the purified membrane region of SCAP: mechanism for a sterol-sensing domain. Mol Cell. 2004;15(2):259–268. doi: 10.1016/j.molcel.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 98.Singh DK, Shentu TP, Enkvetchakul D, Levitan I. Cholesterol regulates prokaryotic Kir channel by direct binding to channel protein. Biochim Biophys Acta Biomembr. 2011;1808(10):2527–2533. doi: 10.1016/j.bbamem.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Maxfield FR, Tabas I. Role of cholesterol and lipid organization in disease. Nature. 2005;438(7068):612–621. doi: 10.1038/nature04399. [DOI] [PubMed] [Google Scholar]
- 100.Schug ZT, Frezza C, Galbraith LCA, Gottlieb E. The music of lipids: how lipid composition orchestrates cellular behaviour. Acta Oncol. 2012;51(3):301–310. doi: 10.3109/0284186x.2011.643823. [DOI] [PubMed] [Google Scholar]
- 101.Monteiro JP, Oliveira PJ, Jurado AS. Mitochondrial membrane lipid remodeling in pathophysiology: a new target for diet and therapeutic interventions. Prog Lipid Res. 2013;52(4):513–528. doi: 10.1016/j.plipres.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 102.Rone MB, Fan JJ, Papadopoulos V. Cholesterol transport in steroid biosynthesis: role of protein–protein interactions and implications in disease states. Biochim Biophys Acta Mol Cell Biol Lipids. 2009;1791(7):646–658. doi: 10.1016/j.bbalip.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schroeder F, Atshaves BP, McIntosh AL, Gallegos AM, Storey SM, Parr RD, Jefferson JR, Ball JM, Kier AB. Sterol carrier protein-2: new roles in regulating lipid rafts and signaling. Biochim Biophys Acta Mol Cell Biol Lipids. 2007;1771(6):700–718. doi: 10.1016/j.bbalip.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gut P, Zweckstetter M, Banati RB. Lost in translocation: the functions of the 18-kD translocator protein. Trends Endocrinol Metab. 2015;26(7):349–356. doi: 10.1016/j.tem.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Krueger KE, Papadopoulos V. Peripheral-type benzodiazepine receptors mediate translocation of cholesterol from outer to inner mitochondrial-membranes in adrenocortical-cells. J Biol Chem. 1990;265(25):15015–15022. [PubMed] [Google Scholar]
- 106.Lacapere JJ, Delavoie F, Li H, Peranzi G, Maccario J, Papadopoulos V, Vidic B. Structural and functional study of reconstituted peripheral benzodiazepine receptor. Biochem Biophys Res Commun. 2001;284(2):536–541. doi: 10.1006/bbrc.2001.4975. [DOI] [PubMed] [Google Scholar]
- 107.Depinto V, Benz R, Palmieri F. Interaction of non-classical detergents with the mitochondrial porin—a new purification procedure and charcterization of the pore-forming unit. Eur J Biochem. 1989;183(1):179–187. doi: 10.1111/j.1432-1033.1989.tb14911.x. [DOI] [PubMed] [Google Scholar]
- 108.Hulce JJ, Cognetta AB, Niphakis MJ, Tully SE, Cravatt BF. Proteome-wide mapping of cholesterol-interacting proteins in mammalian cells. Nat Methods. 2013;10(3):259–264. doi: 10.1038/nmeth.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hiller S, Garces RG, Malia TJ, Orekhov VY, Colombini M, Wagner G. Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science. 2008;321(5893):1206–1210. doi: 10.1126/science.1161302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Campbell AM, Chan SHP. The voltage dependent anion channel affects mitochondrial cholesterol distribution and function. Arch Biochem Biophys. 2007;466(2):203–210. doi: 10.1016/j.abb.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 111.Pfaller R, Freitag H, Harmey MA, Benz R, Neupert W. A water-soluble form of porin from the mitochondrial outer-membrane of neurospora-crassa—properties and relationship to the biosynthetic precursor form. J Biol Chem. 1985;260(13):8188–8193. [PubMed] [Google Scholar]
- 112.Popp B, Schmid A, Benz R. Role of sterols in the functional reconstitution of water-soluble mitochondrial porins from different organisms. Biochemistry. 1995;34(10):3352–3361. doi: 10.1021/bi00010a026. [DOI] [PubMed] [Google Scholar]
- 113.Pastorino JG, Hoek JB. Regulation of hexokinase binding to VDAC. J Bioenerg Biomembr. 2008;40(3):171–182. doi: 10.1007/s10863-008-9148-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Montero J, Morales A, Llacuna L, Lluis JM, Terrones O, Basanez G, Antonsson B, Prieto J, Garcia-Ruiz C, Colell A, Fernandez-Checa JC. Mitochondrial cholesterol contributes to chemotherapy resistance in hepatocellular carcinoma. Can Res. 2008;68(13):5246–5256. doi: 10.1158/0008-5472.can-07-6161. [DOI] [PubMed] [Google Scholar]
- 115.Rouslin W, Macgee J, Gupte S, Wesselman A, Epps DE. Mitochondrial cholesterol content and membrane-properties in porcine myocardial ischemia. Am J Physiol. 1982;242(2):H254–H259. doi: 10.1152/ajpheart.1982.242.2.H254. [DOI] [PubMed] [Google Scholar]
- 116.Liao MF, Cao EH, Julius D, Cheng YF. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504(7478):107. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Guler G, Gartner RM, Ziegler C, Mantele W. Lipid–protein interactions in the regulated betaine symporter BetP probed by infrared spectroscopy. J Biol Chem. 2016;291(9):4295–4307. doi: 10.1074/jbc.M114.621979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu W, Wacker D, Gati C, Han GW, James D, Wang DJ, Nelson G, Weierstall U, Katritch V, Barty A, Zatsepin NA, Li DF, Messerschmidt M, Boutet S, Williams GJ, Koglin JE, Seibert MM, Wang C, Shah STA, Basu S, Fromme R, Kupitz C, Rendek KN, Grotjohann I, Fromme P, Kirian RA, Beyerlein KR, White TA, Chapman HN, Caffrey M, Spence JCH, Stevens RC, Cherezov V. Serial femtosecond crystallography of G protein-coupled receptors. Science. 2013;342(6165):1521–1524. doi: 10.1126/science.1244142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Budelier MM, Cheng WWL, Bergdoll L, Chen ZW, Janetka JW, Abramson J, Krishnan K, Mydock-McGrane L, Covey DF, Whitelegge JP, Evers AS. Photoaffinity labeling with cholesterol analogues precisely maps a cholesterol-binding site in voltage-dependent anion channel-1. J Biol Chem. 2017;292(22):9294–9304. doi: 10.1074/jbc.M116.773069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Grouleff J, Irudayam SJ, Skeby KK, Schiott B. The influence of cholesterol on membrane protein structure, function, and dynamics studied by molecular dynamics simulations. Biochim Biophys Acta Biomembr. 2015;1848(9):1783–1795. doi: 10.1016/j.bbamem.2015.03.029. [DOI] [PubMed] [Google Scholar]
- 121.Meyer B, Peters T. NMR spectroscopy techniques for screening and identifying ligand binding to protein receptors. Angew Chem Int Ed. 2003;42(8):864–890. doi: 10.1002/anie.200390233. [DOI] [PubMed] [Google Scholar]
- 122.Scheidt HA, Muller P, Herrmann A, Huster D. The potential of fluorescent and spin-labeled steroid analogs to mimic natural cholesterol. J Biol Chem. 2003;278(46):45563–45569. doi: 10.1074/jbc.M303567200. [DOI] [PubMed] [Google Scholar]
- 123.Prasanna X, Chattopadhyay A, Sengupta D. Role of lipid-mediated effects in beta(2)-adrenergic receptor dimerization. In: Chakrabarti A, Surolia A (eds) Biochemical roles of eukaryotic cell surface macromolecules. Adv Exp Med Biol. 2015;842:247–261. doi: 10.1007/978-3-319-11280-0_16. [DOI] [PubMed] [Google Scholar]
- 124.Gater DL, Saurel O, Iordanov I, Liu W, Cherezov V, Milon A. Two classes of cholesterol binding sites for the beta(2)AR revealed by thermostability and NMR. Biophys J. 2014;107(10):2305–2312. doi: 10.1016/j.bpj.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vukoti K, Kimura T, Macke L, Gawrisch K, Yeliseev A. Stabilization of functional recombinant cannabinoid receptor CB2 in detergent micelles and lipid bilayers. PLoS ONE. 2012 doi: 10.1371/journal.pone.0046290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Barrett PJ, Song YL, Van Horn WD, Hustedt EJ, Schafer JM, Hadziselimovic A, Beel AJ, Sanders CR. The amyloid precursor protein has a flexible transmembrane domain and binds cholesterol. Science. 2012;336(6085):1168–1171. doi: 10.1126/science.1219988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Senes A, Ubarretxena-Belandia I, Engelman DM. The C alpha-H center dot center dot center dot O hydrogen bond: a determinant of stability and specificity in transmembrane helix interactions. Proc Natl Acad Sci USA. 2001;98(16):9056–9061. doi: 10.1073/pnas.161280798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bayrhuber M, Meins T, Habeck M, Becker S, Giller K, Villinger S, Vonrhein C, Griesinger C, Zweckstetter M, Zeth K. Structure of the human voltage-dependent anion channel. Proc Natl Acad Sci USA. 2008;105(40):15370–15375. doi: 10.1073/pnas.0808115105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Luo WB, Cady SD, Hong M. Immobilization of the influenza A M2 transmembrane peptide in virus envelope-mimetic lipid membranes: a solid-state NMR investigation. Biochemistry. 2009;48(27):6361–6368. doi: 10.1021/bi900716s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liao SY, Fritzsching KJ, Hong M. Conformational analysis of the full-length M2 protein of the influenza A virus using solid-state NMR. Protein Sci. 2013;22(11):1623–1638. doi: 10.1002/pro.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tang M, Hong M. Structure and mechanism of beta-hairpin antimicrobial peptides in lipid bilayers from solid-state NMR spectroscopy. Mol BioSyst. 2009;5(4):317–322. doi: 10.1039/b820398a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zheng Z, Yang R, Bodner ML, Weliky DP. Conformational flexibility and strand arrangements of the membrane-associated HIV fusion peptide trimer probed by solid-state NMR spectroscopy. Biochemistry. 2006;45(43):12960–12975. doi: 10.1021/bi0615902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Elkins MR, Williams JK, Gelenter MD, Dai P, Kwon B, Sergeyev IV, Pentelute BL, Hong M. Cholesterol-binding site of the influenza M2 protein in lipid bilayers from solid-state NMR. Proc Natl Acad Sci. 2017 doi: 10.1073/pnas.1715127114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Papadopoulos V. Peripheral-type benzodiazepine diazepam binding inhibitor receptor—biological role in steroidogenic cell-function. Endocr Rev. 1993;14(2):222–240. doi: 10.1210/er.14.2.222. [DOI] [PubMed] [Google Scholar]
- 135.Rupprecht R, Papadopoulos V, Rammes G, Baghai TC, Fan JJ, Akula N, Groyer G, Adams D, Schumacher M. Translocator protein (18 kDa) (TSPO) as a therapeutic target for neurological and psychiatric disorders. Nat Rev Drug Discov. 2010;9(12):971–988. doi: 10.1038/nrd3295. [DOI] [PubMed] [Google Scholar]
- 136.Nothdurfter C, Baghai TC, Schule C, Rupprecht R. Translocator protein (18 kDa) (TSPO) as a therapeutic target for anxiety and neurologic disorders. Eur Arch Psychiatry Clin Neurosci. 2012;262:S107–S112. doi: 10.1007/s00406-012-0352-5. [DOI] [PubMed] [Google Scholar]
- 137.Jaremko L, Jaremko M, Giller K, Becker S, Zweckstetter M. Conformational flexibility in the transmembrane protein TSPO. Chemistry (Eur J) 2015;21(46):16555–16563. doi: 10.1002/chem.201502314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jaremko M, Jaremko L, Giller K, Becker S, Zweckstetter M. Structural integrity of the A147T polymorph of mammalian TSPO. ChemBioChem. 2015;16(10):1483–1489. doi: 10.1002/cbic.201500217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Delavoie F, Li H, Hardwick M, Robert JC, Giatzakis C, Peranzi G, Yao ZX, Maccario J, Lacapere JJ, Papadopoulos V. In vivo and in vitro peripheral-type benzodiazepine receptor polymerization: functional significance in drug ligand and cholesterol binding. Biochemistry. 2003;42(15):4506–4519. doi: 10.1021/bi0267487. [DOI] [PubMed] [Google Scholar]
- 140.Russ WP, Engelman DM. The GxxxG motif: a framework for transmembrane helix–helix association. J Mol Biol. 2000;296(3):911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 141.Hianik T, Haburcak M. Clustering of cholesterol in DMPC bilayers as indicated by membrane mechanical properties. Gen Physiol Biophys. 1993;12(3):283–291. [PubMed] [Google Scholar]
- 142.Denisov IG, Sligar SG. Nanodiscs for structural and functional studies of membrane proteins. Nat Struct Mol Biol. 2016;23(6):481–486. doi: 10.1038/nsmb.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Martinez D, Decossas M, Kowal J, Frey L, Stahlberg H, Dufourc EJ, Riek R, Habenstein B, Bibow S, Loquet A. Lipid internal dynamics probed in nanodiscs. ChemPhysChem. 2017;18(19):2651–2657. doi: 10.1002/cphc.201700450. [DOI] [PMC free article] [PubMed] [Google Scholar]