Abstract
Nitrogen (N) is one of the most important essential macro-elements for plant growth and development, and nitrate represents the most abundant inorganic form of N in soils. The nitrate uptake and assimilation processes are finely tuned according to the available nitrate in the surroundings as well as by the internal finely coordinated signaling pathways. The NIN-like proteins (NLPs) harbor both RWP-RK, and Phox and Bem1 (PB1) domains, and they belong to the well-characterized plant-specific RWP-RK transcription factor gene family. NLPs are known to be involved in the nitrate signaling pathway by activating downstream target genes, and thus they are implicated in the primary nitrate response in the nucleus via their RWP-RK domains. The PB1 domain is a ubiquitous protein–protein interaction domain and it comprises another regulatory layer for NLPs via the protein interactions within NLPs or with other essential components. Recently, Ca2+–Ca2+ sensor protein kinase–NLP signaling cascades have been identified and they allow NLPs to have central roles in mediating the nitrate signaling pathway. NLPs play essential roles in many aspects of plant growth and development via the finely tuned nitrate signaling pathway. Furthermore, recent studies have highlighted the emerging roles played by NLPs in the N starvation response, nodule formation in legumes, N and P interactions, and root cap release in higher plants. In this review, we consider recent advances in the identification, evolution, molecular characteristics, and functions of the NLP gene family in plant growth and development.
Electronic supplementary material
The online version of this article (10.1007/s00018-019-03164-8) contains supplementary material, which is available to authorized users.
Keywords: Interaction, NIN-like protein, Nodule formation, Nitrogen use efficiency, Phosphorus, Symbiosis
Introduction
Nitrogen (N) is a major constituent of proteins, chlorophyll, nucleotides, and hormones, and thus it has profound effects on plant growth and productivity [1–4]. Huge amounts of N fertilizer are applied to soils to maximize crop yields to meet the high requirements for food due to the growing population worldwide [5, 6]. However, less than 50% of the applied N fertilizer is absorbed by plants, depending on the soil conditions and crop species [7–10]. The input of excess N causes severe environmental pollution and produces greenhouse gases (such as N2O) that contribute to climate change [11–13]. Therefore, there is an urgent need to improve the N use efficiency (NUE) of plants to balance high crop yields with lower N fertilizer inputs [12, 14–16]. Among the various methods that can be used for improving the NUE, transgenic approaches are considered the most promising ways for meeting the current demand for a high NUE in crops, but they require a comprehensive understanding of all the processes involved with N uptake and assimilation [5, 12].
Nitrate is one of the most abundant inorganic forms of N in aerobic soils but it is also the most readily leached form of N due to its chemical nature [17]. Recent research indicates that nitrate can act as a nutritional element but also as a signaling molecule in plants [8, 18–23]. The components involved in the nitrate signaling pathway were identified in recent years [18–20]. In particular, the nitrate assimilation-specific regulator NIT2, which contains the DNA-binding RWP-RK domain, was shown to regulate nitrate signaling in Chlamydomonas [18]. Further research demonstrated that the NIT2 protein is structurally similar to the NODULE INCEPTION (NIN) proteins from legume plants [18]. The first NIN gene was identified in the legume plant Lotus japonicus and it is functionally necessary for nodule formation [24]. The NIN protein also contains a conserved RWP-RK domain but it has an additional Phox and Bem1 (PB1) domain, and it is considered the founding member of the NIN-like proteins (NLPs) [25–27]. Phylogenetic analyses determined the specific NIN proteins in legumes, and other NLPs were identified in both legumes and other non-N fixing plants, such as rice, Arabidopsis, wheat, and maize [25, 27–29]. The NIN proteins in legumes are also considered to be NLPs based on the presence of the PB1 domain [27]. In addition to the RWP-RK and PB1 domains, another conserved domain was identified in the N terminal regions of NLPs as the GAF domain [18, 27]. However, recent studies have disputed whether the GAF is a feature of NLPs because the folded GAF-related structure has not been confirmed in NLPs [30, 31]. Thus, this debate has led researchers to reconsider the conserved domain in the N terminal regions of NLPs.
Recently, several studies have contributed to our understanding of nitrate signaling-mediated N uptake and assimilation, and many key components that facilitate this process have been characterized [19, 32–35]. AtNLP6 and AtNLP7 were identified as having central roles in nitrate signaling, where the activities of AtNLP6/7 in nitrate signaling are finely tuned by the post-transcriptional regulation of the phosphorylation state [32, 36]. The phosphorylated AtNLP6/7 remain in the nucleus to activate the genes involved in the primary nitrate response (PNR) via their conserved RWP-RK domain in the presence of nitrate [32, 36]. Recently, several Ca2+-sensor protein kinases (CPKs) were identified (i.e., CPK10/30/32) that phosphorylate AtNLP6/7 as intermediates between the NRT1/PTR FAMILY 6.3/ NITRATE TRANSPORTER 1.1 (NPF6.3/NRT1.1) which mediated Ca2+ signals and the retention of AtNLP6/7 in the nucleus in the presence of nitrate, thereby facilitating the central roles of NLPs in NPF6.3/NRT1.1-Ca2+-NLP-mediated nitrate signaling [21, 32]. In addition, several essential components of this signaling pathway have been detected using genetic and molecular approaches in Arabidopsis, such as teosinte branched1/cycloidea/proliferating cell factor1-20 (TCP20), NITRATE REGULATORY GENE 2 (NRG2), and NITRATE-INDUCIBLE GARP-TYPE TRANSCRIPTIONAL REPRESSOR 1 (NIGT1) [22, 34, 37, 38]. These findings enhance our understanding of the N uptake and assimilation processes mediated by Ca2+–CPK–NLP signaling cascades in plants, as well as providing efficient candidates and strategies for improving the NUE [39]. Moreover, the roles of NLPs in the N starvation response, nodule formation, N and phosphate (P) interactions, and root cap cell release have been clarified in recent years [34, 38, 40, 41]. In this review, we focus on the NLPs in plants, including recent advances in their identification, evolution, molecular structure, and functions. This review provides timely information about the roles played by NLPs in plant growth and environmental adaptation, as well as the underlying molecular regulatory mechanisms involved.
Identification and evolutionary history of NLPs in plants
The first NIN protein was identified in the legume species L. japonicus as a crucial regulator that controls N-mediated symbiotic root nodule formation [24]. Subsequently, nine and three NLPs were found by homologous analysis in the genomes of the non-N-fixing plants Arabidopsis and rice, respectively [25]. In recent years, due to increased availability of genome information, the genome-wide identifications of NLPs have been conducted in many plants, such as Physcomitrella patens, maize, Brassica napus, wheat, and Glycine max [29, 42–46]. However, compared with other well characterized gene families in plants [47, 48], little information is available regarding the members of this gene family in sequenced plants [25, 27].
Thus, to obtain a better understanding of the NLP gene family in plants, a comprehensive analysis was conducted of the NLP gene family in 81 plant species with genome sequences available in Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html) according to the previous established method [49]. In total, 587 NLP proteins with both RWP-RK and PB1 domains were identified in 74 plant species (Tables S1 and S2). No NLP proteins were detected in seven algae species, including six green algae (Botryococcus braunii, Chlamydomonas reinhardtii, Chromochloris zofingiensis, Coccomyxa subellipsoidea C-169, Volvox carteri, and Dunaliella salina) and one red alga (Porphyra umbilicalis) (Table S1). However, one to three NLP proteins were detected in the other branch of green algae, specifically in Micromonas pusilla CCMP1545, Micromonas sp. RCC299, and Ostreococcus lucimarinus, which belong to Mamiellales (Table S1). Interestingly, unlike other green algae, Mamiellales have reduced genomes [50, 51]. Similar results have been obtained for other gene families involved in leaf development, such as YABBY and GROWTH REGULATING FACTOR (GRF) [50]. These findings suggest that the ancestral NLP proteins originated from green alga and that they formed the basal toolkit during the evolutionary history of plants [52]. NLP proteins were not identified in some of the green algae but RWP-RK proteins (such as NIT2) were detected and they have similar functions to the NLP proteins in higher plants [18, 27]. In addition, the lowest and highest numbers of NLP proteins were found in Ostreococcus lucimarinus/Marchantia polymorpha (1) and Helianthus annuus (31), respectively (Table S1). Other plants found to contain relatively high numbers of NLP proteins comprised Daucus carota (with 21), Kalanchoe laxiflora (20), Gossypium hirsutum (20), Populus trichocarpa (14), and Linum usitatissimum (14) (Table S1). The genomes of these plant species have undergone whole-genome duplication events or whole-genome triplication [53–57], which might have contributed to their higher numbers of NLP proteins.
In addition to the few studies that addressed the identification of NLPs, evolutionary analyses of NLPs have been conducted for a limited number of plant species [25, 27]. In particular, a comprehensive phylogenetic tree was constructed using the 587 NLP proteins obtained from 74 plant species to evaluate the evolutionary history of the NLP family (Fig. 1). Similar to a previous evolutionary analysis [25], the 587 NLPs were divided into three major groups with 235, 151, and 235 proteins in Group 1, Group 2, and Group 3, respectively (Tables S1–S2 and Fig. 1). For instance, in Arabidopsis, AtNLP1–5 was assigned to Group 1, AtNLP8–9 to Group 2, and AtNLP6–7 to Group 3 (Tables S1 and S2). Similar assignments were made in earlier studies [25, 26, 43, 58]. Interestingly, the members of Group 3 appeared to have originated from green algae, whereas the proteins from Group 2 originated from mosses (Table S1). The members of Group 1 probably appeared after the division of eudicots and monocots, and they are absent from Amborella trichopoda (Table S1), which is considered the most basal lineage in angiosperms [59]. These findings suggest three separate origins for NLPs in plants, but the ancestral NLPs might originate from green algae and they include the well-known proteins AtNLP6/7.
Fig. 1.
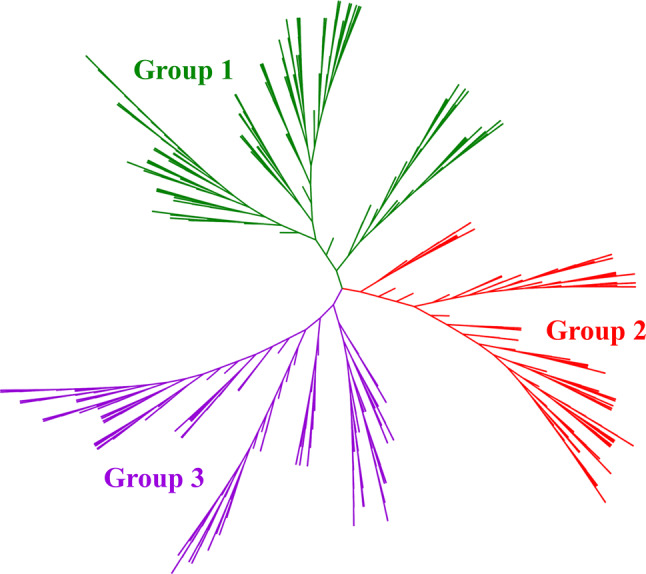
Phylogenetic tree of NLPs in plants. The tree comprises 587 NLPs from 74 plant species and it can be divided into three groups (i.e., Group 1, Group 2, and Group 3). Detailed information regarding the NLPs in these 74 plants is provided in Tables S1 and S2
Molecular structure of NLPs in plants
The typical molecular structure of NLPs contains GAF, RWP-RK, and type I/II PB1 domains [26, 27, 43]. However, according to our analysis, none of the 587 proteins analyzed harbored the GAF domain (Table S1), which differs from a previous report that many NLPs carry a GAF domain [27]. The GAF domain was first described in NIT2 together with AtNLP3 based on a sequence alignment [18], and further studies confirmed the presence of this conserved domain in the N terminal regions of NLPs [27, 44, 58]. However, does the conserved domain correspond to the well-known GAF domain? Three hidden Markov models of GAF domains, i.e., GAF (PF01590), GAF_2 (PF13185), and GAF_3 (PF13492), were used to search for the possible GAF domains in the 587 NLPs using HMMER software and they failed to detected any GAF domains in these NLPs [60]. In addition, using the protein sequence of NIT2 as a query to search against both Pfam and the NCBI’s Conserved Domain Database (CDD) [61] failed to detect any GAF domains. Previous studies have reported that the structurally characterized GAF domains can bind with low-molecular weight ligands, such as cGMP, 2-oxoglutarate, nitric oxide, and nitrate, or serve as homodimerization modules [62–65]. However, no evidence suggests that the functions of NLPs are related to the GAF domain. Therefore, the conserved domains in the N terminal regions of NLPs might not be GAF domains [30], but further studies are required to confirm this hypothesis.
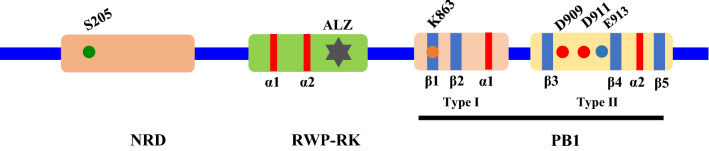
In several other recent studies, the conserved domain in the N terminal region of NLPs was designated as a nitrate-responsive domain (NRD) according to features that could allow NLPs to receive nitrate signals via this domain [30, 31]. Thus, the classical molecular structure of NLPs comprises three major domains, i.e., NRD, RWP-RK, and BP1 (Fig. 2) [30, 31]. One evolutionarily conserved site (S205) has been identified as a phosphorylation site in the NRD, where it is phosphorylated by CPK10, CPK30, and CPK32, and it is essential for the retention of AtNLP7 in the nucleus to activate nitrate-induced gene expression in the presence of nitrate [32]. The NRD is highly conserved in NLPs, but partly conserved in the specific NINs of legumes [25, 30]. However, the differences between LjNIN and LjNLP1–4 with respect to the NRD could have contribution to the loss of nitrate responsiveness by LjNINs, which may have been essential for the emergence of symbiotic N fixation in legumes [58].
Fig. 2.
Molecular structures of NLPs in plants. NLPs contain three domains: nitrate-responsive domain (NRD), RWP-RK, and Phox and Bem1 (PB1). The site S205 in NRD is an essential phosphorylation site for the nuclear retention of NLPs. RWP-RK contains two α-folds (i.e., α1 and α2) and an amphipathic leucine zipper (ALZ), and they are involved in DNA binding. The PB1 domain contains both type I and type II, and it facilitates protein–protein interactions. Type I comprises β1 (including a K residue), β2, and α1, whereas β3, β4, α2, and β5 are located in type II region. K863, D909, D911, and E913 are required for the interactions by NLPs
As a subfamily of the RWP-RK gene family, all NLPs carry the RWP-RK domain, which is the most highly conserved region in NLPs [27]. The RWP-RK domain contains two α-helices and an amphipathic leucine zipper with the conserved sequence Arg–Trp–Pro–X–Arg–Lys, which might be involved in DNA binding [24, 25, 44]. The activity of the RWP-RK domain is not required for nitrate signaling [28], but it is essential for binding to nitrate-responsive cis-elements (NREs) in the promoter regions of target genes [66].
The PB1 domain is a protein–protein interaction domain and it contains either or both the type I and type II motifs [67]. NLPs contain both type I and type II PB1 domains (type I/II PB1 domain), which can interact with type I, type II, and type I/II PB1 domains [67, 68]. There are three glutamate or aspartate residues between β3 and β4 in type I, and they are found on the rear surface of the PB1 domain [68]. The type II motif is located on the front surface of the PB1 domain and it contains an invariant lysine residue in the β1 region [31]. The interaction between two PB1 domains occurs in a front-to-back manner, with electrostatic interactions between the basic lysine residue in one PB1 domain and the acidic glutamate/aspartate residues in the other [67, 68]. The homodimerization of NLP–NLP is facilitated by the PB1 domains, and the core amino acid residues (i.e., K867, D909, D911, and E913) are essential for NLP homodimerization [31]. The homodimerization of NLPs is not required for the transactivation of nitrate-responsive genes, but it is essential for fully promoting nitrate-induced gene expression in the presence of nitrate [31]. The protein interactions between AtNLP6/7 and TCP20 in Arabidopsis, as well as between MtNIN and MtNLPs in Medicago truncatula also depend on the structure of PB1 [34, 69].
NLPs play central roles in the PNR
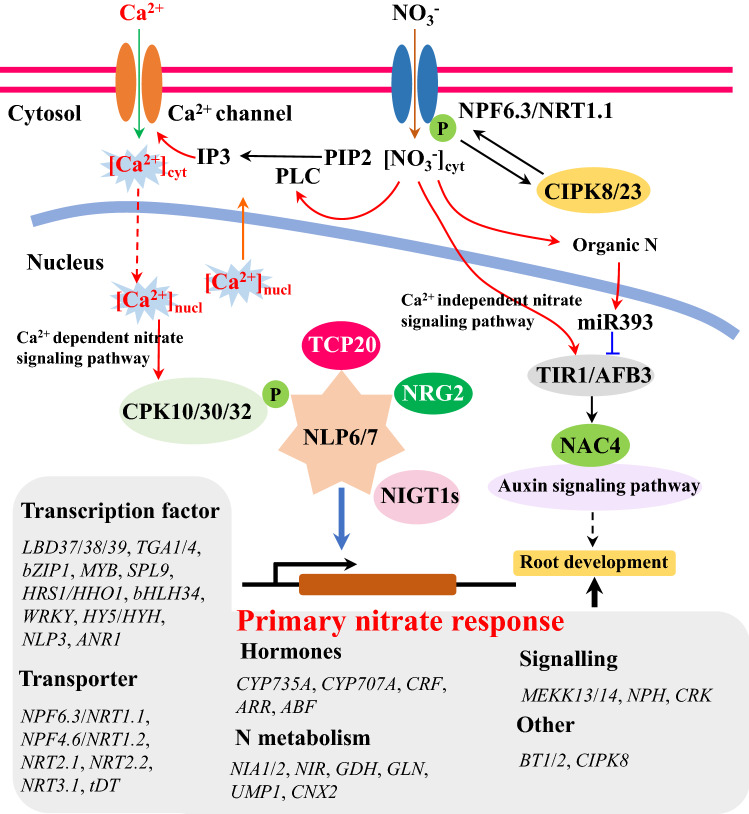
Plants can rapidly sense changes in the soil nitrate concentration without protein synthesis, but the abundances of hundreds of genes then change within minutes at the transcriptional level [70, 71]. This rapid response is defined as the PNR [71]. Plants have evolved sophisticated mechanisms based on NLPs for nitrate signaling to transform the exogenous nitrate concentration signals into endogenous gene reprogramming events related to N transport and metabolism, which shape their morphological and physiological adaptation (Fig. 3) [19, 32, 34]. AtNLP6 and AtNLP7 have been identified as key transcription factors for PNR in Arabidopsis [31]. In addition, NPF6.3/NRT1.1 is characterized as a transceptor in Arabidopsis, where it functions as both a transporter and receptor [72]. In Arabidopsis, the transport affinity of NPF6.3/NRT1.1 for nitrate is regulated by the phosphorylation status of threonine residue 101 via CALCINEURIN B-LIKE (CBL)-INTERACTING PROTEIN KINASE 8/23 (CIPK8/23) depending on the nitrate concentrations in the soil, but the nitrate-sensing ability is independent of the transport activity, and it is determined by P492L between the 10th and 11th transmembrane regions of NPF6.3/NRT1.1 [72, 73]. After sensing via NPF6.3/NRT1.1, the exogenous nitrate concentration signal can be transformed into changes in the cytosolic Ca2+ level in an NPF6.3/NRT1.1-dependent manner [21]. The nitrate-induced accumulation of cytoplasmic Ca2+ depends on the activity of phospholipase C (PLC), which is responsible for increasing the concentration of inositol 1,4,5-trisphosphate (IP3) after nitrate induction [21]. The Ca2+ channels in the plasma membrane are essential components located downstream of IP3 and they allow the Ca2+ levels to increase in the cytoplasm or nucleus [21, 32]. Thus, Ca2+ is considered to be a second messenger in the nitrate signaling pathway [20, 21].
Fig. 3.
Proposed model of NPF6.3/NRT1.1 mediated Ca2+-dependent and -independent nitrate signaling pathways in plants. NPF6.3/NRT1.1 acts as a nitrate sensor and changes in the exogenous nitrate concentrations leading to increase in the activities of PLC, thereby increasing the cytosolic IP3 levels. The increased IP3 concentration in the cytosol induces the opening of Ca2+ channels and the accumulation of cytosolic Ca2+ ([Ca2+]cyt). The signals due to the increased [Ca2+]cyt can be sensed by CPK10/30/32, which then phosphorylate NLP6/7. The phosphorylated NLP6/7 is retained in the nucleus to activate primary nitrate-responsive genes. Several other factors are key components of the Ca2+–CPK–NLP signaling cascade, including TCP20, NRG2, and NIGT1s. In addition to the Ca2+-dependent nitrate signaling pathway, another Ca2+-independent nitrate signaling pathway is mediated by NPF6.3/NRT1.1 via the TIR1/ABF3-NAC4 module to promote root development through the auxin signaling pathway
Subsequently, the nitrate-triggered Ca2+ signals are transmitted to three downstream Ca2+ sensors and effectors comprising CPK10, CPK30, and CPK32 [32]. These three CPKs can phosphorylate AtNLP7 at the conserved site Ser205 to determine the retention of AtNLP7 in the nucleus [32]. A Ser205A mutant fails to retain AtNLP7 in the nucleus and to rescue the phenotypes of the nlp7 mutant [32]. AtNLP7 must remain in the nucleus to activate PNR genes [74]. The expression level of AtNLP7 is not regulated by nitrate, but the nuclear retention of AtNLP7 is determined by nitrate [36]. In addition to AtNLP7, AtNLP6 is the closest homolog of AtNLP7 and it has the capacity for nuclear retention in a nitrate-dependent manner to trigger downstream gene reprogramming events [34]. The retention of AtNLP6/7 in the nucleus activates hundreds of genes involved with nitrate transport and metabolism [36]. Studies indicate that all NLPs have the capacity to bind with the well-known NRE sequences, which are often found in the promoter regions of nitrate-inducible genes [28, 75]. However, the efficient activation of nitrate-inducible genes by AtNLP7 does not appear to require the entire NRE because subparts of the NRE are overrepresented in the whole motif [27].
The targets of AtNLP7 were characterized by hybridizing the immunoprecipitated DNA with a whole-genome tiling array (ChIP–chip) method [36]. In total, 851 genes are targeted by AtNLP7 in Arabidopsis in response to nitrate signaling and these genes are enriched in pathways implicated in N transport and metabolism, as well as the regulation of N responses in plants [36]. Several well-known genes involved in the nitrate signaling pathway, such as ARABIDOPSIS NITRATE REGULATED1 (ANR1), LATERAL BOUNDARY DOMAIN 37/38 (LBD37/38), CIPK8, and NPF6.3/NRT1.1 [72, 76–78], are targets of AtNLP7 in the response to nitrate [36]. Recently, the yeast one-hybrid network method was employed for N-associated metabolism analysis, and AtNLP6 and AtNLP7 were identified as the first layer of transcription factors that bind directly to the genes encoding assimilation enzymes, such as COFACTOR OF NITRATE REDUCTASE AND XANTHINE DEHYDROGENASE 2 (CNX2) and NITRITE REDUCTASE 1 (NIR1) [35].
The master roles of nitrate–Ca2+–NLP as the key component of the nitrate signaling pathway have been determined [32, 74]. However, not all of the genes implicated in the PNR function in a nitrate–Ca2+–NLP-dependent manner, such as AUXIN SIGNALING F-BOX3 (AFB3) and NAC DOMAIN CONTAINING PROTEIN 4 (NAC4), thereby suggesting that another nitrate signaling pathway might be independent of Ca2+ signaling in plants [20, 21, 79]. In addition to AtNLP6/7, AtNLP3 was identified as the target of BASIC LEUCINE ZIPPER 1 (bZIP1) according to the cell-based transient assay reporting genome-wide effects of transcription factors (TARGET) method [80]. AtNLP3 belongs to the category regulated by bZIP1 without binding and it functions in a “hit-and-run” transcriptional manner to regulate the rapid response to nitrate [80].
Interestingly, a recent study proposed a model where AtNLP7 acts upstream of NPF6.3/NRT1.1 to regulate nitrate signaling by altering the expression of NPF6.3/NRT1.1 via directly binding to the promoter of NPF6.3/NRT1.1 in the presence of NH4+ [23, 33]. The expression level of NPF6.3/NRT1.1 was found to be inhibited in the Arabidopsis nlp7 mutant supplied with NH4+ and the nitrate-inducible capacities of NPF6.3/NRT1.1 were also impaired in the mutant [23]. Genetic studies suggest that NPF6.3/NRT1.1 and AtNLP7 function in the same nitrate signaling pathway, and that overexpression of the NPF6.3/NRT1.1 in the Arabidopsis nlp7 mutant completely or partially rescues the phenotypes of the Arabidopsis nlp7 mutant [23]. The roles of the NPF6.3/NRT1.1 signaling pathway in NH4+ uptake and metabolism have also been demonstrated recently [81], but it is not known whether NLPs are implicated in this signaling pathway. These findings provide a molecular basis for the interaction between nitrate and NH4+, which has been observed in many plants [82, 83].
Several other important factors have been implicated in NLP-mediated nitrate signaling or the response in recent years. For instance, TCP20 can interact directly with AtNLP6/7 via the histidine- and glutamine-rich domain of TCP20 and the type I/II PB1 domain of AtNLP6/7 [34]. The cellular location of AtNLP6/7-TCP20 heterodimers depends on the present of nitrate, and nitrate starvation leads to the retention of the heterodimers in the nucleus, whereas the opposite is observed when plants are supplied with nitrate [34]. Another study confirmed that AtNLP6/7-TCP20 heterodimers are essential for activation of the Cell-Cycle Progression Gene (CYCB1;1) and they promote root meristem growth in response to N starvation [34]. TCP20 functions in systemic nitrate signaling, whereas AtNLP7 acts as local nitrate signals independently of TCP20 [37]. These findings demonstrate the other roles of AtNLP in the nitrate starvation response in addition to the PNR. Moreover, the bZIP transcription factor NITRATE REGULATORY GENE 2 (NRG2) in Arabidopsis is characterized as a positive regulator located upstream of NPF6.3/NRT1.1 in nitrate signaling, where the disruption of NRG2 leads to a decrease and increase in NPF6.3/NRT1.1 and NPF7.2/NRT1.8, respectively [22]. NRG2 can interact directly with AtNLP7 in the nucleus, but this interaction does not affect the nuclear retention of AtNLP7 in response to nitrate [22]. Genetic and molecular studies indicate that NRG2 and AtNLP7 play important but nonoverlapping roles in nitrate signaling [22]. Recently, two genes from the BTB and TAZ DOMAIN PROTEIN (BT) gene family (BT1 and BT2) were identified as hub genes in the NUE regulatory network in Arabidopsis [84]. Subsequently, it was shown that these two genes act as negative regulators by repressing the expression of NRT2.1 and NRT2.4, thereby inhibiting plant growth and decreasing the NUE [84, 85]. However, the expression levels of the nitrate-inducible genes BT1 and BT2 are regulated directly by AtNLP7, thereby suggesting that BT1 and BT2 play roles downstream of NLP in the nitrate signaling pathway [85].
NLPs play crucial roles in integrating both N and P signals
More recently, NITRATE-INDUCIBLE GARP-TYPE TRANSCRIPTIONAL REPRESSOR 1 (NIGT1) was identified as a promoter of the expression of NRT2.1, but NIGT1 and AtNLP7 share different binding sites in the promoter region of NRT2.1 [38]. The AtNLP–NIGT1 cascade regulates the coordinated expression of NRT2.1 and NRT3.1/NAR2.1 in response to nitrate, and several important genes such as CYP735A2 and HY5-HOMOLOG (HYH) appear to act downstream of the AtNLP–NIGT1 cascade [38]. Thus, AtNLP–NIGT1 may function as another regulatory layer to antagonistically regulate the NLP-mediated central N signaling pathway [38]. Interestingly, PHOSPHATE STARVATION RESPONSE 1 (PHR1), which is the master regulator in P starvation [86], promotes the expression of members of the NIGT1 gene family to decrease the mRNA abundance of NRT2.1, thereby reducing the uptake of nitrate in Arabidopsis [38]. Moreover, the expression level of NIGT1/HRS1 is governed by the NPF6.3/NRT1.1-AtNLP7 regulatory module in the presence of nitrate, and the repression of primary root growth by NIGT1/HRS1 in response to P shortage depends on the nitrate signaling pathway [40, 87]. Thus, NIGT1 allows crosstalk between the N and P signaling pathways to coordinate the anabolic demands for both N and P in plants [38, 40, 87, 88].
Roles of NLPs in growth and development regulated by N nutrition
As the master regulator of nitrate signaling components, the roles played by NLP in shaping the plastic responses of plant to N availability have been investigated more extensively compared with their other functions. Studies of NLPs in Arabidopsis have demonstrated a requirement for NLPs to support normal growth and development in the response to nitrate availability.
The Arabidopsis nlp7 mutant forms a smaller rosette but the root fresh weight is unchanged, and thus the plants have a lower shoot to root fresh weight ratio compared with the wild type under full N supply conditions [89]. However, the Arabidopsis nlp7 mutant exhibits less impaired growth under limiting N conditions compared with those that receive a full N supply [89]. The Arabidopsis nlp7 mutant also exhibits delayed flowering, with longer primary roots and a higher lateral root density compared with the wild type under full nitrate condition [89]. The Arabidopsis nlp7 mutant exhibits impaired functions in the nitrate signaling pathway, thereby triggering the N starvation response in plants [89]. Indeed, the accumulation of nitrate and the reductions in the amounts of amino acids suggest that AtNLP7 has essential roles in activating genes involved with the nitrate assimilation processes [89]. Interestingly, the Arabidopsis nlp7 mutant displays greater drought resistance with reduced leaf water losses under drought treatment, which might be related to the nitrate-controlled opening of the stomata [89]. Similar phenotypes with enhanced drought resistance and a reduced stomatal aperture size were found in npf6.3/nrt1.1 mutants [90]. Considering the close relationship between AtNLP7 and NPF6.3/NRT1.1 in the nitrate signaling pathway, the similar drought resistance phenotypes of these two mutants suggest they might be involved with the same signaling pathway to regulate the plant water status in a nitrate-dependent manner. Further experiments are required to confirm the validity of this hypothesis.
The overexpression of AtNLP7 in Arabidopsis also significantly improves the growth and NUE, with enhanced photosynthesis and carbon assimilation capacities under low- and high-nitrate conditions [39]. AtNLP7-overexpressing lines exhibit changes in the root architecture with a longer primary root length and more lateral roots compared with the wild type, and the coordination of C and N assimilation results in a much improved nutrient status, which is essential for biomass accumulation and enhancing the NUE [39].
In addition, the constitutive location of AtNLP8 in the nucleus is involved with stimulating seed germination where this requires directly binding to the promoter of CYP707A2, which encodes an abscisic acid (ABA) catabolic enzyme [91], thereby reducing the ABA levels in a nitrate-dependent manner, and thus the crosstalk between nitrate signaling and ABA signaling may govern seed germination [92].
In maize, the overexpression of ZmNLP6 and ZmNLP8 in Arabidopsis nlp7-4 mutants rescues the loss of PNR phenotypes [43]. These transgenic lines have a longer primary root length and higher lateral root number compared with the wild type and/or Arabidopsis nlp7-4 mutants with a higher NUE under limited nitrate conditions, thereby confirming the roles of ZmNLP6/8 in the nitrate signaling pathway by regulating nitrate assimilation under low-nitrate conditions [33, 43]. Similar results were obtained by overexpressing ZmNLP3.1 in an nlp7-1 background in Arabidopsis [42].
Recently, it was shown that the overexpression of OsNRT1.1A/OsNPF6.3 increases the nuclear location of OsNLP3 and OsNLP4 to enhance the expression levels of genes involved with nitrate uptake and assimilation [93]. These overexpression lines have a higher NUE and yield, thereby demonstrating the important roles of OsNRT1.1A/OsNPF6.3-OsNLP3/4 in coordinating the uptake and assimilation of nitrate to improve crop yields [93].
NLPs participate in nodule formation
In M. truncatula, MtNIN has central roles in coordinating diverse symbiotic developmental processes to regulate temporal and spatial nodulation downstream of the early nod factor signaling pathway [94, 95]. MtNIN competitively inhibits ERF required for nodulation (ERN1) to repress the expression levels of Early Nodulin 11 (ENOD11) in the root epidermis and increase the mRNA levels of the cytokinin receptor Cytokinin Response 1 (CRE1) in the root cortex to integrate cytokinin signaling and nodule organogenesis processes in the roots of M. truncatula [94, 95]. In addition, MtNLP1 is retained in the nucleus in the response to nitrate, which allows it to interact with MtNIN via their homologous carboxy-terminal PB1 domains [69]. However, the interaction between MtNLP1 and MtNIN in the nucleus hinders the activation by MtNIN of genes involved in nodule formation, such as CRE1 and Nuclear Factor-Y Subunit A1 (NF-YA1), which inhibits rhizobial infection and nodule formation in a nitrate-dependent manner [69]. The roles of NF-YA1 in rhizobial infection depend on the MtNIN-centered network [96]. A remote cis-regulatory region that contains putative cytokinin response elements was identified upstream of the MtNIN gene in M. truncatula, and it is required for nodule primordium formation, where it triggers the expression of B-type response regulator PR1, thereby indicating a role for cytokinin signaling in the initiation of nodule primordium formation [97]. In addition, MtNLP4 has similar roles to MtNLP1 in the inhibition of nodule formation in response to nitrate in M. truncatula [69].
Interestingly, the NITRATE UNRESPONSIVE SYMBIOSIS 1 (NRSYM1) gene in L. japonicus encodes LjNLP4 and it also functions in the regulation of nitrate-dependent nodule formation [98]. The nrsym1 mutants fail in responding to N environment and regulating nodule number by utilizing autoregulation of nodulation (AON), which behaves as a systemic long-range signals between roots and shoots [98, 99]. NRSYM1/LjNLP4 is also retained in the nucleus in response to nitrate and it binds directly to the promoters of CLE-ROOT SIGNAL 2 (CLE-RS2) to regulate nodule formation [98]. The small secreted peptides CLE-RS2 function as root-derived signals to interact with HYPERNODULATION ABERRANT ROOT FORMATION 1 (HAR1) in shoot, and to trigger secondary shoot-derived signals that are transported back to root to negatively regulate nodule development [99, 100]. These findings suggest that NLP has essential roles in nodule formation in legumes.
In addition to legumes, the CgNLP gene CgNIN from Casuarina glauca was characterized using both phylogenetic and transgenic approaches as an essential component implicated in root nodule symbioses with Frankia [101]. CgNIN can complement the phenotypes with failure to initiate nodule formation in nin legume mutants, and the expression of CgNIN is activated by infection with Frankia [101, 102]. The loss-of-function mutants exhibit impaired functions in terms of the nodule formation and early root hair deformation responses, and thus CgNIN may have essential roles in actinorhizal and rhizobial nodulation [101]. Other studies identified a NIN-activating factor CgNINA that activates CgNIN in the pre-infection stages of F. casuarinae infection to regulate root hair development [103].
Other functions of NLPs in plants
Interestingly, the well-known AtNLP7 has a role in root cap cell release in Arabidopsis [41, 104]. Border-like cells (BLCs) are cells located in the last layer of the root cap and they are released from the root cap in a finely tuned development-driven manner [41]. AtNLP7 mRNA is accumulated at high levels in BLCs and it is promoted by low pH [41]. Mutation of AtNLP7 allows BLCs to be released as single cells rather than the entire layer, which is related to changes in the cell wall components and the expression levels of several genes that encode cell wall-loosening enzymes [41]. Genetic analysis confirmed that the function of AtNLP7 in BLC release requires the inhibition of the expression of CELLULASE5 (CEL5) [41]. In addition, AtNLP7-mediated BLC release is not implicated in gravity sensing and root cap cell identity [104].
Conclusions and perspectives
In recent years, systems biology has been applied to identify new components and layers [105], and provides insights into global N signaling and assimilation, thereby facilitating NUE improvements in crop production [106–108]. These emerging integrated approaches are helping to understand the central roles of NLPs in the plant nitrate signaling pathways. Analyses of the evolution of NLPs have indicated the three origins of this gene family, where Group 3 has the most ancestral genes originating from green algae. The well-known AtNLP6 and AtNLP7 genes belong to Group 3. The central roles of NLPs in the uptake and assimilation of N have been confirmed by the studies of the Ca2+-dependent nitrate signaling pathway, and several key components of this regulatory network have been identified. In addition to nitrate signaling, the roles of NLPs in the N starvation response, N and P interaction, nodule formation, and root cap release have been elucidated in recent years. These results have greatly enhanced our understanding of the multiple roles of NLPs in plant growth and environmental responses, as well as the underlying molecular mechanisms involved. However, several aspects of the signaling processes facilitated by NLPs still require further investigation. For example, how do increases in the nitrate level in the cytosol enhance the activities of PLCs and what are the roles of NPF6.3/NRT1.1 in this process? Where are NLPs phosphorylated by CPKs? What are the roles of nuclear Ca2+ in the phosphorylation of NLPs? Identifying the molecular mechanisms that underlie the sensing and regulation of nitrate by answering these questions will contribute to a more comprehensive understanding of the processes involved in the uptake and assimilation of N, and the development of efficient strategies for improving the NUE in crop production. Moreover, our current knowledge of NLPs is based mainly on the studies in Arabidopsis and legumes, and no NLPs from woody plants have been functionally characterized. The growth of woody plants requires large amounts of N [109, 110] and the complex environment for annual growth may mean that the regulatory network is much more complicated than that in herbaceous plants [111]. However, the information obtained from studies of these processes in Arabidopsis could help to elucidate similar pathways in woody plants. Thus, our current knowledge of NLPs might represent only a small fraction of their diverse roles and much research is required to elucidate their more detailed features in the future.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This research was financially supported by the Research Start-up Fund for Henan Agricultural Universities (no. 30500487), Fundamental Research Funds for the Central Universities (no. 2662017QD001) and Hubei Province Natural Science Foundation of China (no. 2017CFB338). Dr. Duncan Jackson from the United Kingdom is sincerely thanked for correcting the English language in this manuscript.
Compliance with ethical standards
Conflict of interest
The authors have no conflict of interest to declare.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mu X, Chen Q, Chen F, Yuan L, Mi G. Within-leaf nitrogen allocation in adaptation to low nitrogen supply in maize during grain-filling stage. Front Plant Sci. 2016;7:699. doi: 10.3389/fpls.2016.00699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mu X, Chen Q, Chen F, Yuan L, Mi G. A RNA-seq analysis of the response of photosynthetic system to low nitrogen supply in maize leaf. Int J Mol Sci. 2017;18:2624. doi: 10.3390/ijms18091811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, Chardon F, Gaufichon L, Suzuki A. Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann Bot. 2010;105:1141–1157. doi: 10.1093/aob/mcq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tegeder M, Masclaux-Daubresse C. Source and sink mechanisms of nitrogen transport and use. New Phytol. 2018;217:35–53. doi: 10.1111/nph.14876. [DOI] [PubMed] [Google Scholar]
- 5.Schroeder JI, Delhaize E, Frommer WB, Guerinot ML, Harrison MJ, Herrera-Estrella L, Horie T, Kochian LV, Munns R, Nishizawa NK. Using membrane transporters to improve crops for sustainable food production. Nature. 2013;497:60–66. doi: 10.1038/nature11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mu X, Chen Q, Chen F, Yuan L, Mi G. Dynamic remobilization of leaf nitrogen components in relation to photosynthetic rate during grain filling in maize. Plant Physiol Biochem. 2018;129:27–34. doi: 10.1016/j.plaphy.2018.05.020. [DOI] [PubMed] [Google Scholar]
- 7.Yang L, Cao W, Thorup-Kristensen K, Bai J, Gao S, Chang D. Effect of Orychophragmus violaceus incorporation on nitrogen uptake in succeeding maize. Plant Soil Environ. 2015;61:260–265. doi: 10.17221/178/2015-PSE. [DOI] [Google Scholar]
- 8.Ibarra-Henríquez C, Fredes I, Álvarez JM, Undurraga SF, Gutiérrez RA. Nitrate signaling and early responses in Arabidopsis roots. J Exp Bot. 2017;68:2541–2551. doi: 10.1093/jxb/erx041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo J, Zhou J-J, Masclaux-Daubresse C, Wang N, Wang H, Zheng B. Morphological and physiological responses to contrasting nitrogen regimes in Populus cathayana is linked to resources allocation and carbon/nitrogen partition. Environ Exp Bot. 2019;162:247–255. doi: 10.1016/j.envexpbot.2019.03.003. [DOI] [Google Scholar]
- 10.Mu X, Chen Q, Wu X, Chen F, Yuan L, Mi G. Gibberellins synthesis is involved in the reduction of cell flux and elemental growth rate in maize leaf under low nitrogen supply. Environ Exp Bot. 2018;150:198–208. doi: 10.1016/j.envexpbot.2018.03.012. [DOI] [Google Scholar]
- 11.Mueller ND, West PC, Gerber JS, MacDonald GK, Polasky S, Foley JA. A tradeoff frontier for global nitrogen use and cereal production. Environ Res Lett. 2014;9:054002. doi: 10.1088/1748-9326/9/5/054002. [DOI] [Google Scholar]
- 12.Wan T, Xue H, Y-P Tong. Transgenic approaches for improving use efficiency of nitrogen, phosphorus and potassium in crops. J Integr Agric. 2017;16:2657–2673. doi: 10.1016/S2095-3119(17)61828-8. [DOI] [Google Scholar]
- 13.Chen Q, Soulay F, Saudemont B, Elmayan T, Marmagne A, Masclaux-Daubresse C. Overexpression of ATG8 in Arabidopsis stimulates autophagic activity and increases nitrogen remobilization efficiency and grain filling. Plant Cell Physiol. 2019;60:343–352. doi: 10.1093/pcp/pcy214. [DOI] [PubMed] [Google Scholar]
- 14.Mandal VK, Sharma N, Raghuram N. Molecular targets for improvement of crop nitrogen use efficiency: current and emerging options. Engineering nitrogen utilization in crop plants. New York: Springer; 2018. pp. 77–93. [Google Scholar]
- 15.Mu X, Chen F, Wu Q, Chen Q, Wang J, Yuan L, Mi G. Genetic improvement of root growth increases maize yield via enhanced post-silking nitrogen uptake. Eur J Agron. 2015;63:55–61. doi: 10.1016/j.eja.2014.11.009. [DOI] [Google Scholar]
- 16.Luo J, Zhou J-J. Growth performance, photosynthesis, and root characteristics are associated with nitrogen use efficiency in six poplar species. Environ Exp Bot. 2019;164:40–51. doi: 10.1016/j.envexpbot.2019.04.013. [DOI] [Google Scholar]
- 17.Wang Y-Y, Cheng Y-H, Chen K-E, Tsay Y-F. Nitrate transport, signaling, and use efficiency. Annu Rev Plant Biol. 2018;69:85–122. doi: 10.1146/annurev-arplant-042817-040056. [DOI] [PubMed] [Google Scholar]
- 18.Camargo A, Llamas Á, Schnell RA, Higuera JJ, González-Ballester D, Lefebvre PA, Fernández E, Galván A. Nitrate signaling by the regulatory gene NIT2 in Chlamydomonas. Plant Cell. 2007;19:3491–3503. doi: 10.1105/tpc.106.045922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fredes I, Moreno S, Díaz FP, Gutiérrez RA. Nitrate signaling and the control of Arabidopsis growth and development. Curr Opin Plant Biol. 2019;47:112–118. doi: 10.1016/j.pbi.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 20.Armijo G, Gutiérrez RA. Emerging players in the nitrate signaling pathway. Mol Plant. 2017;10:1019–1022. doi: 10.1016/j.molp.2017.07.006. [DOI] [PubMed] [Google Scholar]
- 21.Riveras E, Alvarez JM, Vidal EA, Oses C, Vega A, Gutiérrez RA. The calcium ion is a second messenger in the nitrate signaling pathway of Arabidopsis. Plant Physiol. 2015;169:1397–1404. doi: 10.1104/pp.15.00961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu N, Wang R, Zhao L, Zhang C, Li Z, Lei Z, Liu F, Guan P, Chu Z, Crawford NM. The Arabidopsis NRG2 protein mediates nitrate signaling and interacts with and regulates key nitrate regulators. Plant Cell. 2016;28:485–504. doi: 10.1105/tpc.15.00567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao L, Zhang W, Yang Y, Li Z, Li N, Qi S, Crawford NM, Wang Y. The Arabidopsis NLP7 gene regulates nitrate signaling via NRT1.1–dependent pathway in the presence of ammonium. Sci Rep. 2018;8:1487. doi: 10.1038/s41598-018-20038-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schauser L, Roussis A, Stiller J, Stougaard J. A plant regulator controlling development of symbiotic root nodules. Nature. 1999;402:191–195. doi: 10.1038/46058. [DOI] [PubMed] [Google Scholar]
- 25.Schauser L, Wieloch W, Stougaard J. Evolution of NIN-like proteins in Arabidopsis, rice, and Lotus japonicus. J Mol Evol. 2005;60:229–237. doi: 10.1007/s00239-004-0144-2. [DOI] [PubMed] [Google Scholar]
- 26.Yokota K, Hayashi M. Function and evolution of nodulation genes in legumes. Cell Mol Life Sci. 2011;68:1341–1351. doi: 10.1007/s00018-011-0651-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chardin C, Girin T, Roudier F, Meyer C, Krapp A. The plant RWP-RK transcription factors: key regulators of nitrogen responses and of gametophyte development. J Exp Bot. 2014;65:5577–5587. doi: 10.1093/jxb/eru261. [DOI] [PubMed] [Google Scholar]
- 28.Konishi M, Yanagisawa S. Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat Commun. 2013;4:1617. doi: 10.1038/ncomms2621. [DOI] [PubMed] [Google Scholar]
- 29.Kumar A, Batra R, Gahlaut V, Gautam T, Kumar S, Sharma M, Tyagi S, Singh KP, Balyan HS, Pandey R, Gupta PK. Genome-wide identification and characterization of gene family for RWP-RK transcription factors in wheat (Triticum aestivum L.) PLoS One. 2018;13:e0208409. doi: 10.1371/journal.pone.0208409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konishi M, Yanagisawa S. Emergence of a new step towards understanding the molecular mechanisms underlying nitrate-regulated gene expression. J Exp Bot. 2014;65:5589–5600. doi: 10.1093/jxb/eru267. [DOI] [PubMed] [Google Scholar]
- 31.Konishi M, Yanagisawa S. The role of protein–protein interactions mediated by the PB1 domain of NLP transcription factors in nitrate-inducible gene expression. BMC Plant Biol. 2019;19:90. doi: 10.1186/s12870-019-1692-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.K-H Liu, Niu Y, Konishi M, Wu Y, Du H, Chung HS, Li L, Boudsocq M, McCormack M, Maekawa S. Discovery of nitrate–CPK–NLP signalling in central nutrient–growth networks. Nature. 2017;545(7654):311–316. doi: 10.1038/nature22077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao L, Liu F, Crawford N, Wang Y. Molecular regulation of nitrate responses in plants. Int J Mol Sci. 2018;19:2039. doi: 10.3390/ijms19072039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guan P, Ripoll J-J, Wang R, Vuong L, Bailey-Steinitz LJ, Ye D, Crawford NM. Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc Natl Acad Sci USA. 2017;114:2419–2424. doi: 10.1073/pnas.1615676114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaudinier A, Rodriguez-Medina J, Zhang L, Olson A, Liseron-Monfils C, Bågman A-M, Foret J, Abbitt S, Tang M, Li B. Transcriptional regulation of nitrogen-associated metabolism and growth. Nature. 2018;563:259–264. doi: 10.1038/s41586-018-0656-3. [DOI] [PubMed] [Google Scholar]
- 36.Marchive C, Roudier F, Castaings L, Bréhaut V, Blondet E, Colot V, Meyer C, Krapp A. Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat Commun. 2013;4:1713. doi: 10.1038/ncomms2650. [DOI] [PubMed] [Google Scholar]
- 37.Guan P, Wang R, Nacry P, Breton G, Kay SA, Pruneda-Paz JL, Davani A, Crawford NM. Nitrate foraging by Arabidopsis roots is mediated by the transcription factor TCP20 through the systemic signaling pathway. Proc Natl Acad Sci USA. 2014;111:15267–15272. doi: 10.1073/pnas.1411375111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maeda Y, Konishi M, Kiba T, Sakuraba Y, Sawaki N, Kurai T, Ueda Y, Sakakibara H, Yanagisawa S. A NIGT1-centred transcriptional cascade regulates nitrate signalling and incorporates phosphorus starvation signals in Arabidopsis. Nat Commun. 2018;9:1376. doi: 10.1038/s41467-018-03832-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu L-H, Wu J, Tang H, Yuan Y, Wang S-M, Wang Y-P, Zhu Q-S, Li S-G, Xiang C-B. Overexpression of Arabidopsis NLP7 improves plant growth under both nitrogen-limiting and-sufficient conditions by enhancing nitrogen and carbon assimilation. Sci Rep. 2016;6:27795. doi: 10.1038/srep27795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Medici A, Marshall-Colon A, Ronzier E, Szponarski W, Wang R, Gojon A, Crawford NM, Ruffel S, Coruzzi GM, Krouk G. AtNIGT1/HRS1 integrates nitrate and phosphate signals at the Arabidopsis root tip. Nat Commun. 2015;6:6274. doi: 10.1038/ncomms7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karve R, Suárez-Román F, Iyer-Pascuzzi AS. The transcription factor NIN-LIKE PROTEIN7 controls border-like cell release. Plant Physiol. 2016;171:2101–2111. doi: 10.1104/pp.16.00453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Z, Zhang L, Sun C, Gu R, Mi G, Yuan L. Phylogenetic, expression and functional characterizations of the maize NLP transcription factor family reveal a role in nitrate assimilation and signaling. Physiol Plant. 2018;163:269–281. doi: 10.1111/ppl.12696. [DOI] [PubMed] [Google Scholar]
- 43.Cao H, Qi S, Sun M, Li Z, Yang Y, Crawford NM, Wang Y. Overexpression of the maize ZmNLP6 and ZmNLP8 can complement the Arabidopsis nitrate regulatory mutant nlp7 by restoring nitrate signaling and assimilation. Front Plant Sci. 2017;8:1703. doi: 10.3389/fpls.2017.01703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu M, Chang W, Fan Y, et al. Genome-wide identification and characterization of NODULE-INCEPTION-Like Protein (NLP) family genes in Brassica napus. Int J Mol Sci. 2018;19:2270. doi: 10.3390/ijms19082270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ge M, Liu Y, Jiang L, Wang Y, Lv Y, Zhou L, Liang S, Bao H, Zhao H. Genome-wide analysis of maize NLP transcription factor family revealed the roles in nitrogen response. Plant Growth Regul. 2018;84:95–105. doi: 10.1007/s10725-017-0324-x. [DOI] [Google Scholar]
- 46.Koi S, Hisanaga T, Sato K, Shimamura M, Yamato KT, Ishizaki K, Kohchi T, Nakajima K. An evolutionarily conserved plant RKD factor controls germ cell differentiation. Curr Biol. 2016;26:1775–1781. doi: 10.1016/j.cub.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 47.Luo J, Zhou J-J, Zhang J-Z. Aux/IAA gene family in plants: molecular structure, regulation, and function. Int J Mol Sci. 2018;19:259. doi: 10.3390/ijms19010259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou J-J, Luo J. The PIN-FORMED auxin efflux carriers in plants. Int J Mol Sci. 2018;19:2759. doi: 10.3390/ijms19092759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo J, Liang Z, Wu M, Mei L. Genome-wide identification of BOR genes in poplar and their roles in response to various environmental stimuli. Environ Exp Bot. 2019;164:101–113. doi: 10.1016/j.envexpbot.2019.04.006. [DOI] [Google Scholar]
- 50.Worden AZ, Lee J-H, Mock T, Rouzé P, Simmons MP, Aerts AL, Allen AE, Cuvelier ML, Derelle E, Everett MV. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science. 2009;324:268–272. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]
- 51.Derelle E, Ferraz C, Rombauts S, Rouzé P, Worden AZ, Robbens S, Partensky F, Degroeve S, Echeynié S, Cooke R. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci USA. 2006;103:11647–11652. doi: 10.1073/pnas.0604795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Floyd SK, Bowman JL. The ancestral developmental tool kit of land plants. Int J Plant Sci. 2007;168:1–35. doi: 10.1086/509079. [DOI] [Google Scholar]
- 53.Iorizzo M, Ellison S, Senalik D, et al. A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nat Genet. 2016;48:657–666. doi: 10.1038/ng.3565. [DOI] [PubMed] [Google Scholar]
- 54.Li F, Fan G, Lu C, et al. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat Biotechnol. 2015;33:524–530. doi: 10.1038/nbt.3208. [DOI] [PubMed] [Google Scholar]
- 55.Tuskan GA, Difazio S, Jansson S, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 56.Wang Z, Hobson N, Galindo L, et al. The genome of flax (Linum usitatissimum) assembled de novo from short shotgun sequence reads. Plant J. 2012;72:461–473. doi: 10.1111/j.1365-313X.2012.05093.x. [DOI] [PubMed] [Google Scholar]
- 57.Badouin H, Gouzy J, Grassa CJ, et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature. 2017;546:148–152. doi: 10.1038/nature22380. [DOI] [PubMed] [Google Scholar]
- 58.Suzuki W, Konishi M, Yanagisawa S. The evolutionary events necessary for the emergence of symbiotic nitrogen fixation in legumes may involve a loss of nitrate responsiveness of the NIN transcription factor. Plant Signal Behav. 2013;8:e25975. doi: 10.4161/psb.25975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chase MW, Christenhusz M, Fay M, Byng J, Judd WS, Soltis D, Mabberley D, Sennikov A, Soltis PS, Stevens PF. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 2016;181:1–20. doi: 10.1111/boj.12385. [DOI] [Google Scholar]
- 60.Finn RD, Clements J, Arndt W, Miller BL, Wheeler TJ, Schreiber F, Bateman A, Eddy SR. HMMER web server: 2015 update. Nucleic Acids Res. 2015;43:W30–W38. doi: 10.1093/nar/gkv397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2016;45:D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ho YSJ, Burden LM, Hurley JH. Structure of the GAF domain, a ubiquitous signaling motif and a new class of cyclic GMP receptor. EMBO J. 2000;19:5288–5299. doi: 10.1093/emboj/19.20.5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Niemann V, Koch-Singenstreu M, Neu A, Nilkens S, Götz F, Unden G, Stehle T. The NreA protein functions as a nitrate receptor in the Staphylococcal nitrate regulation system. J Mol Biol. 2014;426:1539–1553. doi: 10.1016/j.jmb.2013.12.026. [DOI] [PubMed] [Google Scholar]
- 64.Shi R, McDonald L, Cygler M, Ekiel I. Coiled-coil helix rotation selects repressing or activating state of transcriptional regulator DhaR. Structure. 2014;22:478–487. doi: 10.1016/j.str.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 65.Möglich A, Ayers RA, Moffat K. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure. 2009;17:1282–1294. doi: 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Soyano T, Shimoda Y, Hayashi M. NODULE INCEPTION antagonistically regulates gene expression with nitrate in Lotus japonicus. Plant Cell Physiol. 2015;56:368–376. doi: 10.1093/pcp/pcu168. [DOI] [PubMed] [Google Scholar]
- 67.Korasick DA, Chatterjee S, Tonelli M, Dashti H, Lee SG, Westfall CS, Fulton DB, Andreotti AH, Amarasinghe GK, Strader LC. Defining a two-pronged structural model for PB1 (Phox/Bem1p) domain interaction in plant auxin responses. J Biol Chem. 2015;290:12868–12878. doi: 10.1074/jbc.M115.648253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sumimoto H, Kamakura S, Ito T. Structure and function of the PB1 domain, a protein interaction module conserved in animals, fungi, amoebas, and plants. Sci STKE. 2007;2007:re6. doi: 10.1126/stke.4012007re6. [DOI] [PubMed] [Google Scholar]
- 69.Lin J-S, Li X, Luo ZL, Mysore KS, Wen J, Xie F. NIN interacts with NLPs to mediate nitrate inhibition of nodulation in Medicago truncatula. Nat Plants. 2018;4:942–952. doi: 10.1038/s41477-018-0261-3. [DOI] [PubMed] [Google Scholar]
- 70.Deng M, Moureaux T, Caboche M. Tungstate, a molybdate analog inactivating nitrate reductase, deregulates the expression of the nitrate reductase structural gene. Plant Physiol. 1989;91:304–309. doi: 10.1104/pp.91.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gowri G, Kenis JD, Ingemarsson B, Redinbaugh MG, Campbell WH. Nitrate reductase transcript is expressed in the primary response of maize to environmental nitrate. Plant Mol Biol. 1992;18:55–64. doi: 10.1007/BF00018456. [DOI] [PubMed] [Google Scholar]
- 72.Ho C-H, Lin S-H, Hu H-C, Tsay Y-F. CHL1 functions as a nitrate sensor in plants. Cell. 2009;138:1184–1194. doi: 10.1016/j.cell.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 73.Hu HC, Wang YY, Tsay YF. AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J. 2009;57:264–278. doi: 10.1111/j.1365-313X.2008.03685.x. [DOI] [PubMed] [Google Scholar]
- 74.Krouk G. Nitrate signalling: calcium bridges the nitrate gap. Nat Plants. 2017;3:17095. doi: 10.1038/nplants.2017.95. [DOI] [PubMed] [Google Scholar]
- 75.Konishi M, Yanagisawa S. An NLP-binding site in the 3’flanking region of the nitrate reductase gene confers nitrate-inducible expression in Arabidopsis thaliana (L.) Heynh. Soil Sci Plant Nutr. 2013;59:612–620. doi: 10.1080/00380768.2013.809602. [DOI] [Google Scholar]
- 76.Zhang H, Forde BG. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science. 1998;279:407–409. doi: 10.1126/science.279.5349.407. [DOI] [PubMed] [Google Scholar]
- 77.Rubin G, Tohge T, Matsuda F, Saito K, Scheible W-R. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell. 2009;21:3567–3584. doi: 10.1105/tpc.109.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gojon A, Nacry P, Davidian J-C. Root uptake regulation: a central process for NPS homeostasis in plants. Curr Opin Plant Biol. 2009;12:328–338. doi: 10.1016/j.pbi.2009.04.015. [DOI] [PubMed] [Google Scholar]
- 79.Vidal EA, Álvarez JM, Moyano TC, Gutiérrez RA. Transcriptional networks in the nitrate response of Arabidopsis thaliana. Curr Opin Plant Biol. 2015;27:125–132. doi: 10.1016/j.pbi.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 80.Para A, Li Y, Marshall-Colón A, Varala K, Francoeur NJ, Moran TM, Edwards MB, Hackley C, Bargmann BO, Birnbaum KD. Hit-and-run transcriptional control by bZIP1 mediates rapid nutrient signaling in Arabidopsis. Proc Natl Acad Sci USA. 2014;111:10371–10376. doi: 10.1073/pnas.1404657111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jian S, Liao Q, Song H, Liu Q, Lepo JE, Guan C, Zhang J, Ismail AM, Zhang Z. NRT1.1-related NH4+ toxicity is associated with a disturbed balance between NH4+ uptake and assimilation. Plant Physiol. 2018;178:1473–1488. doi: 10.1104/pp.18.00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Luo J, Qin J, He F, Li H, Liu T, Polle A, Peng C, Luo Z-B. Net fluxes of ammonium and nitrate in association with H+ fluxes in fine roots of Populus popularis. Planta. 2013;237:919–931. doi: 10.1007/s00425-012-1807-7. [DOI] [PubMed] [Google Scholar]
- 83.Hachiya T, Sakakibara H. Interactions between nitrate and ammonium in their uptake, allocation, assimilation, and signaling in plants. J Exp Bot. 2016;68:2501–2512. doi: 10.1093/jxb/erw449. [DOI] [PubMed] [Google Scholar]
- 84.Araus V, Vidal EA, Puelma T, Alamos S, Mieulet D, Guiderdoni E, Gutiérrez RA. Members of BTB gene family of scaffold proteins suppress nitrate uptake and nitrogen use efficiency. Plant Physiol. 2016;171:1523–1532. doi: 10.1104/pp.15.01731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sato T, Maekawa S, Konishi M, Yoshioka N, Sasaki Y, Maeda H, Ishida T, Kato Y, Yamaguchi J, Yanagisawa S. Direct transcriptional activation of BT genes by NLP transcription factors is a key component of the nitrate response in Arabidopsis. Biochem Biophys Res Commun. 2017;483:380–386. doi: 10.1016/j.bbrc.2016.12.135. [DOI] [PubMed] [Google Scholar]
- 86.Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Pérez-Pérez J, Solano R, Leyva A, Paz-Ares J. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 2010;6:e1001102. doi: 10.1371/journal.pgen.1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Menz J, Li Z, Schulze WX, Ludewig U. Early nitrogen-deprivation responses in Arabidopsis roots reveal distinct differences on transcriptome and (phospho-) proteome levels between nitrate and ammonium nutrition. Plant J. 2016;88:717–734. doi: 10.1111/tpj.13272. [DOI] [PubMed] [Google Scholar]
- 88.Liu H, Yang H, Wu C, Feng J, Liu X, Qin H, Wang D. Overexpressing HRS1 confers hypersensitivity to low phosphate-elicited inhibition of primary root growth in Arabidopsis thaliana. J Integr Plant Biol. 2009;51:382–392. doi: 10.1111/j.1744-7909.2009.00819.x. [DOI] [PubMed] [Google Scholar]
- 89.Castaings L, Camargo A, Pocholle D, Gaudon V, Texier Y, Boutet-Mercey S, Taconnat L, Renou JP, Daniel-Vedele F, Fernandez E. The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J. 2009;57:426–435. doi: 10.1111/j.1365-313X.2008.03695.x. [DOI] [PubMed] [Google Scholar]
- 90.Guo F-Q, Young J, Crawford NM. The nitrate transporter AtNRT1. 1 (CHL1) functions in stomatal opening and contributes to drought susceptibility in Arabidopsis. Plant Cell. 2003;15:107–117. doi: 10.1105/tpc.006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Okamoto M, Kuwahara A, Seo M, Kushiro T, Asami T, Hirai N, Kamiya Y, Koshiba T, Nambara E. CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol. 2006;141:97–107. doi: 10.1104/pp.106.079475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yan D, Easwaran V, Chau V, Okamoto M, Ierullo M, Kimura M, Endo A, Yano R, Pasha A, Gong Y. NIN-like protein 8 is a master regulator of nitrate-promoted seed germination in Arabidopsis. Nat Commun. 2016;7:13179. doi: 10.1038/ncomms13179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang W, Hu B, Yuan D, Liu Y, Che R, Hu Y, Ou S, Liu Y, Zhang Z, Wang H. Expression of the nitrate transporter gene OsNRT1.1A/OsNPF6.3 confers high yield and early maturation in rice. Plant Cell. 2018;30:638–651. doi: 10.1105/tpc.17.00809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Marsh JF, Rakocevic A, Mitra RM, Brocard L, Sun J, Eschstruth A, Long SR, Schultze M, Ratet P, Oldroyd GE. Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 2007;144:324–335. doi: 10.1104/pp.106.093021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vernié T, Kim J, Frances L, Ding Y, Sun J, Guan D, Niebel A, Gifford ML, de Carvalho-Niebel F, Oldroyd GE. The NIN transcription factor coordinates diverse nodulation programs in different tissues of the Medicago truncatula root. Plant Cell. 2015;27:3410–3424. doi: 10.1105/tpc.15.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu C-W, Breakspear A, Guan D, Cerri MR, Abbs K, Jiang S, Robson FC, Radhakrishnan G, Roy S, Bone C. NIN acts as a network hub controlling a growth module required for rhizobial infection. Plant Physiol. 2019;179:1704–1722. doi: 10.1104/pp.18.01572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu J, Rutten L, Limpens E, van der Molen T, van Velzen R, Chen R, Chen Y, Geurts R, Kohlen W, Kulikova O. A remote cis-regulatory region is required for NIN expression in the pericycle to initiate nodule primordium formation in Medicago truncatula. Plant Cell. 2019;31:68–83. doi: 10.1105/tpc.18.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nishida H, Tanaka S, Handa Y, Ito M, Sakamoto Y, Matsunaga S, Betsuyaku S, Miura K, Soyano T, Kawaguchi M. A NIN-LIKE PROTEIN mediates nitrate-induced control of root nodule symbiosis in Lotus japonicus. Nat Commun. 2018;9:499. doi: 10.1038/s41467-018-02831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nishida H, Suzaki T. Two negative regulatory systems of root nodule symbiosis: how are symbiotic benefits and costs balanced? Plant Cell Physiol. 2018;59:1733–1738. doi: 10.1093/pcp/pcy102. [DOI] [PubMed] [Google Scholar]
- 100.Nishida H, Suzaki T. Nitrate-mediated control of root nodule symbiosis. Curr Opin Plant Biol. 2018;44:129–136. doi: 10.1016/j.pbi.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 101.Clavijo F, Diedhiou I, Vaissayre V, Brottier L, Acolatse J, Moukouanga D, Crabos A, Auguy F, Franche C, Gherbi H. The Casuarina NIN gene is transcriptionally activated throughout Frankia root infection as well as in response to bacterial diffusible signals. New Phytol. 2015;208:887–903. doi: 10.1111/nph.13506. [DOI] [PubMed] [Google Scholar]
- 102.Chabaud M, Gherbi H, Pirolles E, Vaissayre V, Fournier J, Moukouanga D, Franche C, Bogusz D, Tisa LS, Barker DG. Chitinase-resistant hydrophilic symbiotic factors secreted by Frankia activate both Ca2+ spiking and NIN gene expression in the actinorhizal plant Casuarina glauca. New Phytol. 2016;209(1):86–93. doi: 10.1111/nph.13732. [DOI] [PubMed] [Google Scholar]
- 103.Cissoko M, Hocher V, Gherbi H, et al. Actinorhizal signaling molecules: Frankia root hair deforming factor shares properties with NIN inducing factor. Front Plant Sci. 2018;9:1494. doi: 10.3389/fpls.2018.01494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Karve RA, Iyer-Pascuzzi AS. Further insights into the role of NIN-LIKE PROTEIN 7 (NLP7) in root cap cell release. Plant Signal Behav. 2018;13:e1414122. doi: 10.1080/15592324.2017.1414122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Luo J, Xia W, Cao P, Za Xiao, Zhang Y, Liu M, Zhan C, Wang N. Integrated transcriptome analysis reveals plant hormones jasmonic acid and salicylic acid coordinate growth and defense responses upon fungal infection in poplar. Biomolecules. 2019;9:12. doi: 10.3390/biom9010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gutiérrez RA. Systems biology for enhanced plant nitrogen nutrition. Science. 2012;336:1673–1675. doi: 10.1126/science.1217620. [DOI] [PubMed] [Google Scholar]
- 107.Ueda Y, Yanagisawa S. Delineation of nitrogen signaling networks: computational approaches in the big data era. Mol Plant. 2019;12:50–152. doi: 10.1016/j.molp.2019.01.008. [DOI] [PubMed] [Google Scholar]
- 108.Varala K, Marshall-Colón A, Cirrone J, Brooks MD, Pasquino AV, Léran S, Mittal S, Rock TM, Edwards MB, Kim GJ. Temporal transcriptional logic of dynamic regulatory networks underlying nitrogen signaling and use in plants. Proc Natl Acad Sci USA. 2018;115:6494–6499. doi: 10.1073/pnas.1721487115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Luo J, Li H, Liu T, Polle A, Peng C, Luo Z-B. Nitrogen metabolism of two contrasting poplar species during acclimation to limiting nitrogen availability. J Exp Bot. 2013;64:4207–4224. doi: 10.1093/jxb/ert234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Luo J, Zhou J, Li H, Shi W, Polle A, Lu M, Sun X, Luo Z-B. Global poplar root and leaf transcriptomes reveal links between growth and stress responses under nitrogen starvation and excess. Tree Physiol. 2015;35:1283–1302. doi: 10.1093/treephys/tpv091. [DOI] [PubMed] [Google Scholar]
- 111.Luo J, Shi W, Li H, Janz D, Luo Z-B. The conserved salt-responsive genes in the roots of Populus × canescens and Arabidopsis thaliana. Environ Exp Bot. 2016;129:48–56. doi: 10.1016/j.envexpbot.2015.12.008. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.