Abstract
The myxomycete Didymium iridis (isolate Panama 2) contains a mobile group I intron named Dir.S956-1 after position 956 in the nuclear small subunit (SSU) rRNA gene. The intron is efficiently spread through homing by the intron-encoded homing endonuclease I-DirI. Homing endonuclease genes (HEGs) usually spread with their associated introns as a unit, but infrequently also spread independent of introns (or inteins). Clear examples of HEG mobility are however sparse. Here, we provide evidence for the transfer of a HEG into a group I intron named Dir.S956-2 that is inserted into the SSU rDNA of the Costa Rica 8 isolate of D.iridis. Similarities between intron sequences that flank the HEG and rDNA sequences that flank the intron (the homing endonuclease recognition sequence) suggest that the HEG invaded the intron during the recent evolution in a homing-like event. Dir.S956-2 is inserted into the same SSU site as Dir.S956-1. Remarkably, the two group I introns encode distantly related splicing ribozymes with phylogenetically related HEGs inserted on the opposite strands of different peripheral loop regions. The HEGs are both interrupted by small spliceosomal introns that must be removed during RNA maturation.
INTRODUCTION
Group I introns that encode homing endonuclease genes (HEGs) are parasitic genetic elements, which in genetic crosses efficiently spread into homologous positions in intron-less alleles. The process called ‘homing’ is initiated by the homing endonuclease (HE) that generates a double-stranded DNA break close to the site of intron insertion. In essence, the DNA break recruits the host cell repair and recombination machineries that in turn use intron-containing alleles as template to repair the damaged DNA. The outcome is the spread of identical copies of introns into all homologous intron-less alleles in the cell. Although unproven, an alternative RNA-based model might explain the spread of group I introns into new sites of the same or other genes. The constant spread of group I introns in nature, through recurrent gain and loss (1), has resulted in a present-day broad, but scattered intron distribution (2,3).
Most HEs contain one of four conserved protein sequence motifs. These motifs define the four families of HEGs (i.e. the LAGLIDADG, His-Cys box, HNH and GIY-YIG families). As an additional layer of complexity, the HEGs can spread independent of their associated group I splicing ribozymes (4,5). Evidence for HEG mobility comes from comparative sequence analyses and by mapping HEG insertions on the intron secondary structures. For example, in group I introns from the same or adjacent rDNA sites (e.g. introns inserted after positions L1923–L1926 in nuclear large subunit rRNA genes), closely related HEGs of the His-Cys box family are inserted into different intron secondary structure elements (i.e. terminal loops) (6). A clue to a HEG invasion was found in a group I intron in the T4 phage (4). Within the intron sequence, at the site of HEG insertion, flanking sequences very similar to that recognized by the HE were found. This observation is in agreement with a recent HEG-invasion (i.e. not an ancient HEG acquisition, see also legend for Figure 3). Given that the intron sequences that flank the HEG have no sequence-specific function, sequence similarities to the rDNA homing site are expected to rapidly degenerate by random mutations after the HEG has been inserted. It also suggests that HEG mobility is initiated in the same manner as intron homing, by a double-stranded DNA break catalyzed by the HE itself.
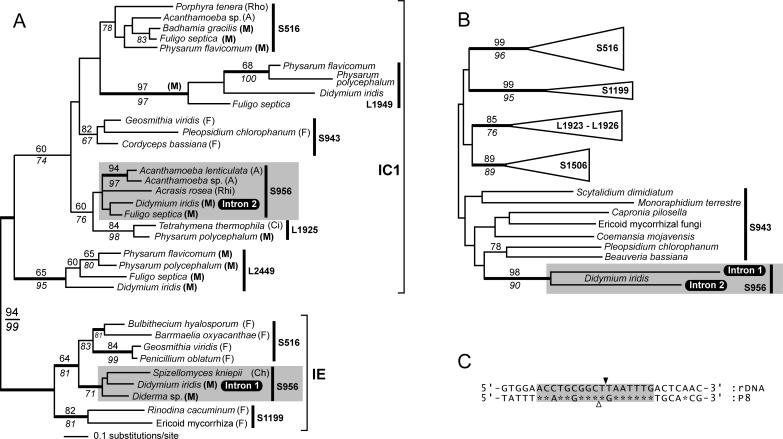
Figure 3.
Phylogeny of nuclear group I introns and homing endonucleases. (A) The intron tree was generated from a 131 nt dataset using the neighbor joining (NJ) method and the Jukes–Cantor (JC) evolutionary model. Bootstrap values above the branches were calculated using the NJ method (Kimura 2 model and 2000 replicates) and values below the branches are from a maximum parsimony (MP) analysis. Values >60% are shown. Support for branches was also tested using Bayesian analysis, and thicker branches indicate ≥95% posterior probability. Introns were sampled from fungi (F), Acanthamoeba (A), Myxomycota (M), Rhodophyta (Rho), Rhizopoda (Rhi), Ciliophora (Ci) and Chytridiomycota (Ch) (classification of organisms follows that of Margulis and Schwartz (32). Introns that belong to the group IC1 or group IE subclasses are indicated, and the site of intron insertion are shown (e.g. S516, S956 and L1925). The Didymium Intron 1 and Intron 2 belong to distantly related lineages of S956 introns (shaded fields). (B) Phylogeny of His-Cys box homing endonucleases. The tree is a summary of a minimum evolution (ME)-Poisson tree. Statistical support for branches was tested by ME-Poisson (2000 replicates) and NJ-JTT (500 replicates) bootstrap analyses, and the resulting numbers are shown above and below branches, respectively. We also tested the ME-tree by calculating Bayesian posterior probabilities using the JTT model. Supported branches in the ME tree are in agreement with a previous analysis (6), and show the clustering of homing endonucleases from homologous or neighboring (i.e. L1923–L1926) sites. The close phylogenetic relationship between the Didymium homing endonucleases is strongly supported by high bootstrap values (i.e. 98 and 90) and significant posterior probabilities. (C) Comparison of sequences flanking Intron 2 and sequences flanking the Intron 2 HEG. The rDNA intron insertion site (black arrowhead) and the putative HEG insertion site (open arrowhead) are shown. Asterisks indicate sequence identity, and a 17 nt region with high identity is indicated (shaded field). The present identity in sequences supports that the HEG invaded Intron 2 in a homing-like process, and that the event took place in the recent evolution of this element [‘recent’ on an intron evolutionary time-scale can refer to events that took place thousands, or even millions of years ago (33)].
Here, we report a large group I intron (Dir.S956-2), hereafter called Intron 2, in the nuclear rDNA of the myxomycete Didymium iridis isolate ‘Costa Rica 8’. The intron encodes a group IC1 splicing ribozyme with a HEG inserted in antisense orientation into the peripheral P8 element. The homing endonuclease protein of 209 amino acids in size is active when expressed in vitro, but only after the removal of a 50-nt spliceosomal intron from the HEG. Remarkably, Intron 2 occupies the rDNA site in which we previously reported a mobile twin-ribozyme group I intron (Dir.S956-1) (7,8), hereafter called Intron 1, in the natural isolate ‘Panama 2’ of D.iridis. Our results suggest that Intron 2 is the result of a recent HEG-invasion into a group I ribozyme that resembles the Tetrahymena ribozyme in the overall structure organization.
MATERIALS AND METHODS
Didymium strains, total DNA extraction and SSU rDNA sequencing
Six Didymium isolates, including the Panama 2 (ATCC number 66541) and Costa Rica 8 (Dr Jim Clark) isolates, were used in this study (see Table 1). Didymium myxamoebas were grown on DS/2 agar plates with Escherichia coli cells as the food source (8,9), and total DNA from ∼108 cells was isolated as described by Lundblad et al. (10) and resuspended in 50 μl double-distilled water. The SSU rDNA was PCR-amplified from total DNA extracts in several overlapping fragments using multiple sets of SSU-specific oligonucleotides (contact S.J. for details). PCR products were cloned into the pGEM-T-easy cloning vector (Promega) and the DNA sequences were determined using automatic sequencing (Big Dye terminator chemistry, Applied Biosystems).
Table 1.
Complete SSU rDNA sequences of different isolates of D.iridis
| Isolate name | Group I introna | Ac. No.b |
|---|---|---|
| Honduras 1-7 | – | AJ938152 |
| Panama 2-16 | 956 | AJ938153 |
| Costa Rica 8-1 | 956 | AJ938154 |
| Costa Rica 19-1 | 1389 | AJ938151 |
| CUR 1-4 | 1389 | AJ938150 |
| HA 4-1 | – | AJ938149 |
aInsertion site for group I introns in SSU rDNA. Positions are in reference to the E.coli SSU rDNA sequence.
bGenBank accession numbers for SSU rDNA sequences.
PCR and plasmid construction
Intron 2 with some flanking rDNA exon sequences were amplified by PCR from total DNA from the Costa Rica 8 isolate using oligonuclotides OP6 (5′-GTA TGG TCG CAA GGC TG-3′) and OP92 (5′-CCA AAC GCA GGT TCA CCT A-3′), and cloned into the SmaI site of pUC18 (Sure Clone kit, Amersham Biosciences). The insert was next sub-cloned into the pGEM1 vector behind the T7 RNA polymerase promoter by cutting the insert from the pUC18 vector using EcoRI and NheI, and by ligating the gel-isolated EcoRI–NheI fragment to an EcoRI–XbaI-cut pGEM1 vector. The plasmid was named pDirS956-2.
To express the Didymium homing endonucleases encoded by Intron 1 and Intron 2, the respective HEGs were cloned into the pGEX-4T-1 expression vector (Amersham Biosciences) behind the Ptac IPTG-inducible promoter. For the Intron 1 and Intron 2 HEGs, the plasmids were named pI-DirI[SPL] and pI-DirII[SPL] (SPL for spliceosomal intron), respectively. Next, the 51 and 50 nt spliceosomal introns that interrupt the Intron 1 and Intron 2 HEGs, respectively, were removed by PCR to produce the plasmids pI-DirI and pI-DirII (contact S.J. for cloning details).
Different DNA target sequences were used to test for endonuclease activity and target site selection. Target 1 was HindIII-linearized plasmids [pGPR16 or pCPR16 (11) that contain identical inserts] that contain the intron-less rDNA allele (11). Target 2 was the intron-containing rDNA allele. For Intron 1, Target 2 was the SphI-linearized pYGAL-DirS956-1 plasmid (12) that contains the Intron 1 (i.e. Dir.S956-1) and some flanking rDNA exons. For Intron 2, Target 2 was the EcoRI-linearized pDirS956-2 plasmid (see above). All plasmid constructs were confirmed by DNA sequencing.
Protein expression and cleavage assays
The homing endonuclease-glutathione S-transferase (GST) fusion proteins were expressed in E.coli BL21 CodonPlus (DE3)-RIL cells (Stratagene) and purified from 100 ml liquid cultures with 0.1 mM ZnCl2 added, as described by Smith and Johnson (13), except that bacteria were grown at 20°C for 6 h before over-expression of proteins was induced by the addition of IPTG. Cells were harvested 6–12 h after the addition of IPTG. Successful expression of fusion proteins was monitored by western blotting using the anti-GST antibody (Amersham Biosciences). Homing endonuclease activity was tested by incubating 5 μl of affinity-purified proteins with ∼50 ng linearized target plasmids [Target 1 or Target 2 (see above)] under conditions that favor activity of the I-PpoI homing endonuclease (14) (25 mM CAPS, 25 mM CHES, pH 10.0, 1 mM DTT and 2 mM MgCl2), in a final volume of 10 μl. The cleavage reactions were incubated at 20°C for 1 h, stopped by adding 2 μl 6× T loading buffer and run on 1% 1× TBE agarose gels.
In vitro transcription and splicing
To test for self-splicing of Intron 2, the pDirS956-2 plasmid was linearized with PstI and treated with T4 DNA polymerase to create blunt ends. In vitro transcription was done by adding to the linear plasmids T7 RNA polymerase (Stratagene) and other reagents as described by the manufacturer, except that the Mg2+ concentration was lowered to 2 mM to avoid autocatalysis during transcription. For gel analysis, the transcript was labeled by the inclusion of 1 μl of [α-35S]ATP (10 μCi/μl; Amersham) in a 50 μl reaction. After 60 min of incubation at 37°C, self-splicing was initiated by adding to the RNA a buffer that contains the GTP co-factor essential for splicing (7). Samples were collected after 0–60 min of incubation and run on 5% acrylamide/8 M Urea polyacrylamide gels. RNA products were visualized by autoradiography.
Intron phylogeny
The phylogeny of S956 introns was tested by sampling introns from the two main groups of nuclear introns (i.e. the group IC1 and IE introns) and by sampling introns from different rDNA insertion sites. An alignment of the selected intron sequences was generated using BioEdit 7.01 (15) and was based on the predicted RNA secondary structure folds of the introns. The final dataset (provided as Supplementary Material) contained 131 nt positions mainly from the catalytic core. The presented neighbor joining (NJ) (Jukes–Cantor model) tree was generated using PAUP* 4.0 Beta (16). Robustness of the tree topology was tested with two different bootstrap analyses. These were a NJ-Kimura 2 (2000 replicates) analyses using PAUP and a maximum parsimony (200 replicates) analyses using MEGA 2.1 (17). In addition, a Bayesian analysis using the JC model and MrBayes 3.0B4 (18) was done to test the tree. A total of 1 000 000 generations of Metropolis-coupled Markov chain Monte Carlo was run, and trees were sampled every 100th generation. A consensus tree was generated from the 5000 last trees to find posterior probabilities.
Protein phylogeny
The I-DirII homing endonuclease sequence was added to a previously published protein dataset of nuclear homing endonucleases (6). The alignment was modified and 117 amino acid positions were chosen for phylogenetic analyses (alignment provided as Supplementary Material). The presented protein phylogeny is a minimal evolution (ME)-Poisson tree generated using MEGA. The tree topology was tested with an NJ-JTT (19) bootstrap (500 replicates) analysis using PHYLIP version 3.62 (20), and ME-Poisson bootstrap (2000 replicates) analysis using MEGA. In addition, the dataset was subjected to Bayesian analysis using MrBayes and the WAG + G model (21), as previously described for His-Cys box HEs (6).
RESULTS
Two self-splicing group I introns with different architecture at the same rDNA location
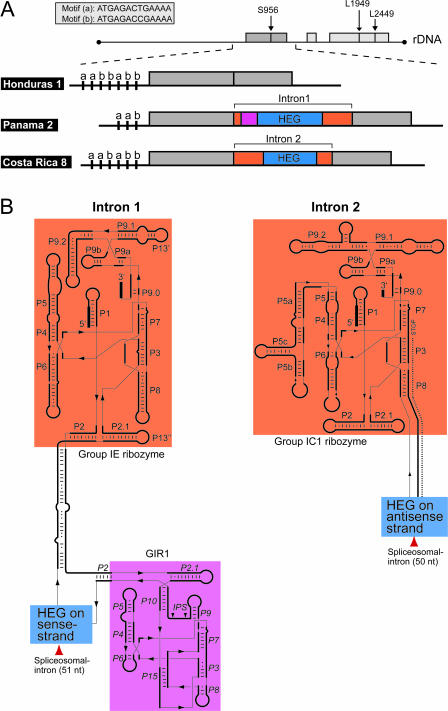
Six complete sequences of the nuclear SSU rDNA of different D.iridis isolates were determined (Table 1). Two of these sequences are interrupted by group I introns at position 956. Figure 1 shows the 20 kb Didymium extrachromosomal rDNA organization, and the predicted secondary structures of the two 956 introns. Intron 1 from the Panama 2 isolate (ATCC 66541) is organized into a twin-ribozyme fold with a small group I-like ribozyme (GIR1) and a HEG inserted into the P2 paired element of a group IE splicing ribozyme (22,23). In contrast, Intron 2 from the Costa Rica 8 isolate encodes a Tetrahymena-like group IC1 splicing ribozyme with a HEG inserted into the P8 paired element on the antisense strand. The striking differences in intron secondary structures are contrasted by the close phylogenetic relationship between the host isolates. In fact, the Panama 2 and Costa Rica 8 isolates are identical in SSU rDNA sequence [GenBank accession numbers AJ938153 (Panama 2) and AJ938154 (Costa Rica 8)], but can be distinguished by different copy numbers of a short (13 nt) motif in the 5′ upstream region of the linear extrachromosomal rDNA (Figure 1A).
Figure 1.
Properties of Didymium rDNA. (A) Schematic illustration of the small (SSU) and large (LSU) subunit ribosomal RNA genes. They are organized on 20 kb linear extrachromosomal rDNAs. In Didymium, two group I introns always interrupt the LSU rDNA at positions L1949 and L2449. The Panama 2 and Costa Rica 8 isolates are identical in SSU rDNA sequence, but can be distinguished by encoding different group I introns at position S956 (Intron 1 and Intron 2). Didymium rDNAs can also be distinguished by containing different copy numbers of 13 nt motif repeats (motifs a and b) in the 5′ upstream region. In Intron 1 and Intron 2 regions with different function are indicated by colors as described in (B). (B) Predicted secondary structures of the S956 group I introns. Splicing ribozymes (vermilion fields), HEGs (blue fields) and a group I-like ribozyme (reddish purple) are shown. Intron 1 (left) from the Panama 2 isolate contains a small group I-like ribozyme (GIR1) and a HEG that are inserted into the P2 paired element of a group IE splicing ribozyme (twin-ribozyme intron) (22,23). Intron 2 (right) from the Costa Rica 8 isolate has a HEG inserted in the antisense orientation in P8 of a Tetrahymena-like group IC1 splicing ribozyme.
Intron 1 is a well-studied autocatalytic RNA that is efficiently removed from precursor rRNAs in the absence of proteins (7,24,25). We wanted to test Intron 2 autocatalysis and cloned the intron with some flanking exon sequences behind a T7 promoter (see Materials and Methods). Transcribed precursor RNAs were incubated under conditions that favor group I ribozyme autocatalysis and samples were separated on 5% denaturing polyacrylamide gels. RNAs typical for intron self-splicing, 3′ splice-site hydrolysis and intron RNA circularization were recovered [data not shown; see Nielsen et al. (26) for the two reaction pathways typical for nuclear group I ribozymes], including unprocessed precursor RNAs, free linear and circular intron RNAs, ligated exon RNAs, 5′-exon–intron RNAs and free exon RNAs. Ligated exon RNAs and intron RNA circle junctions were amplified by RT–PCR, and directly DNA sequenced to verify the correct ligation of exons and the formation of full-length intron RNA circles, respectively (data not shown). In summary, we found that Intron 2 self-splices and generates full-length RNA circles in vitro.
Small spliceosomal introns interrupt functional Didymium HEGs
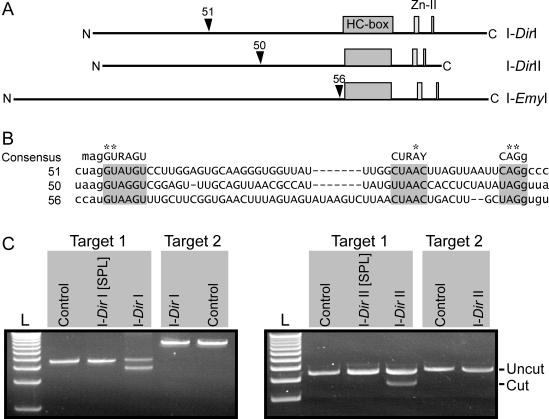
Intron 1 encodes the active HE I-DirI that promotes mobility of the intron-HEG element (8). Interestingly, the HEG is interrupted by a small (51 nt) spliceosomal intron that is removed from the mature HE mRNA (9). Our sequence analyses of Intron 2 identified a spliceosomal intron-like sequence of 50 nt that interrupts (introduces a frame-shift) the HEG. Removal of the 50 nt sequence generates an open reading frame, which hypothetically encodes a protein of 209 amino acids. Figure 2A schematically shows the organization of the HEG, and the positioning of the 50 nt HEG-intervening sequence relative to the HE protein sequence. Evidence for that the 50 nt sequence is a spliceosomal intron comes from high sequence similarity to the 51 and 56 nt spliceosomal/spliceosomal-like introns located within the Intron 1-associated HEG and a HEG from a Ericoid mycorrhizal fungi (6), respectively (Figure 2B). In addition, the 50 nt sequence shows strong sequence similarity to the consensus for spliceosomal introns in mammalian genes (Figure 2B). Functional evidence comes from expression of the Intron 2 HEG (see below), and that the 50 nt are removed from HE mRNAs in vivo (A. Vader and S.D. Johansen, unpublished data).
Figure 2.
Properties of Didymium His-Cys box HEGs. (A) The HEGs encoding the I-DirI and I-DirII homing endonucleases are interrupted by spliceosomal introns of 51 and 50 nt in size, respectively. A similar spliceosomal intron-like sequence has been identified in the I-EmyI HEG (6). The positioning of the HEG intervening sequences relative to the primary protein structure is shown (arrowheads), as are the N- and C-terminal ends. (B) Sequence alignment of spliceosomal introns (upper case letters) in nuclear HEGs (as described in A), with some flanking HEG sequences (lower case letters). The 51, 50 and 56 nt spliceosomal introns that interrupt the I-DirI, I-DirII and I-EmyI HEGs, respectively, share strong similarity to the mammalian spliceosomal intron consensus (on top) (31), including invariable positions (asterisks). Consensus nucleotides are A or C (M), A or G (R) and C or U (Y). (C) Endonucleolytic activity of homing endonucleases. Fused to the glutathione-S-transferase partner the homing endonucleases encoded by Intron 1 and Intron 2 were expressed with (I-DirI[SPL] and I-DirII[SPL], respectively) or without (I-DirI and I-DirII, respectively) interrupting spliceosomal introns. Affinity-purified fusion proteins were incubated with two different DNA targets under conditions that favor activity of the I-PpoI homing endonuclease (14). Target 1 contained the intron-lacking Didymium SSU rDNA, whereas Target 2 contained the Didymium SSU rDNA interrupted by the respective group I intron (i.e. Intron 1 or Intron 2). Endonuclease activity was recognized by cleavage of the target plasmid generating 3.83 and 0.77 kb fragments (0.77 kb band not shown). As a negative control, the DNA was incubated in the absence of proteins.
We wanted to address if the removal of HEG-intervening sequences support the production of functional HE proteins. The 51 nt spliceosomal intron in the Panama 2 isolate (i.e. the 51 nt spliceosomal intron is located within the ‘Intron 1’ group I intron) is positioned in-frame with the HE ORF and a previous study demonstrated that the extended endonuclease I-DirI[SPL] that contains the additional 17 amino acid residues (encoded by the 51 nt spliceosomal intron) is marginally active (8). The Intron 1 and Intron 2 HEGs, with or without a spliceosomal intron were cloned into the pGEX-T4-1 expression vector to generate the pI-DirI[SPL] (with the 51 nt) and pI-DirI (lacks the 51 nt) constructs for Intron 1, and the pI-DirII[SPL] (with the 50 nt) and pI-DirII (lacks the 50 nt) constructs for Intron 2. The I-DirI/I-DirI[SPL] HEs and the putative I-DirII/I-DirII[SPL] HEs were expressed in E.coli. Expression was monitored by western blots using antibodies against the GST fusion partner (data not shown). After protein induction with IPTG, the cells were lysed and the GST-HE fusion proteins were affinity-purified on GST agarose beads and incubated with linearized target plasmids. Two different DNA targets were used. Target 1 contained the intron-lacking Didymium SSU rDNA whereas Target 2 contained the Didymium SSU rDNA interrupted by the respective group I intron. Group I intron-minus alleles are natural targets for HEs, whereas intron-containing alleles are resistant to HE cleavage at the intron insertion site. Figure 2C shows an ethidium bromide-stained agarose gel of target plasmids incubated in the absence or presence of HE proteins. Under the conditions that we used, only HE proteins derived from uninterrupted HEGs (I-DirI and I-DirII) were active. The HEs generate double-strand DNA breaks in group I intron-minus rDNA targets (Target 1) at or near the site of intron insertion. Target 2, which contains the interrupted rDNA was resistant to cleavage. In conclusion, our endonuclease cleavage assay shows that Didymium Intron 1 and Intron 2 encode the functional homing endonucleases I-DirI and I-DirII, respectively. DNA cleavage was not detected for the I-DirI[SPL] and I-DirII[SPL] proteins, which supports that HEG-spliceosomal introns must be removed during RNA maturation.
Intron 1 and Intron 2 encode distantly related splicing ribozymes, but closely related HEGs
To evaluate the evolutionary history of Intron 1 and Intron 2, we generated a DNA sequence alignment of 32 group I introns. Available introns at SSU site 956 as well as introns from several other rDNA sites from myxomycetes (plasmodial slime molds like Didymium), Acanthamoeba, fungi and algae were included in the final dataset. A total of 131 nt from the ribozyme core region were used to infer intron phylogeny and the remaining peripheral regions were excluded (highly divergent regions). Figure 3A shows the intron tree. The deep phylogenetic separation between group IC1 and group IE introns and the generally close phylogenetic relationship between introns at homologous rDNA sites have previously been reported (27–29), and are supported by our analyses. Our tree also provides evidence for two separate lineages of both SSU 516 (29) and SSU 956 introns. Intron 1 groups with IE-type 956 introns, whereas Intron 2 clusters with IC1-type 956 introns (see also Figure 1). Therefore, we conclude that the two Didymium introns are distantly related. The finding is contrasted by two group IC1 introns at position 1925 in LSU rDNA. The 1925 prototype intron from Tetrahymena is remarkably similar to the homologous and mobile intron in Physarum polycephalum even though they are found in distantly related organisms. Tetrahymena is a ciliate (alveolate) whereas Physarum is a plasmodial (true) slime mold. Similar introns in different hosts are best explained by intron spread across species barriers (lateral transfer).
The evolution of nuclear HEs has been addressed (6). We wanted to study the evolution of the two Didymium HEGs and added to a published protein dataset (6) the Intron 2 HE (I-DirII). Figure 3B shows the HE minimal evolution distance tree, which contains supported branches and HE-lineages found in the original analysis. As a consequence, the evolution of nuclear HEs in general will not be discussed here. Interestingly, the Intron 1 and Intron 2 HEs share a sister-group relationship that is strongly supported by bootstrap and Bayesian analyses.
DISCUSSION
We studied an intriguing case of two very differently organized group I introns at position 956 in the SSU rDNA of two closely related (i.e. identical in SSU rDNA sequence) isolates of the plasmodial slime mold D.iridis. The mobile Intron 1 (Dir.S956-1) from the Panama 2 isolate has a well-studied twin-ribozyme architecture and encodes the active HE I-DirI. Here, we report Intron 2 (Dir.S956-2) from the Costa Rica 8 isolate. It encodes a Tetrahymena-like group IC1 splicing ribozyme distantly related to the Intron 1 splicing ribozyme with a functional His-Cys box HEG (encodes the active I-DirII HE protein) inserted on the anti-sense strand of L8. Interestingly, the Intron 1 and Intron 2-derived HEs are closely related, and sequence similarity between intron-flanking and HEG-flanking sequences in Intron 2 support the recent insertion of the HEG into the P8 terminal loop.
Homologous group I introns with separate origins in D.iridis
The biochemical properties (7,24–26) and biological functions (8,9,26,30) of Intron 1 are known from the literature. Here, we report Intron 2 that interrupts the SSU rRNA gene at the same location as Intron 1, in a closely related isolate of D.iridis. A phylogeny of nuclear group I introns sampled preferentially from myxomycetes (but also contains introns from other unicellular eukaryotes) is shown in Figure 3A. The tree provides support for two different lineages of SSU 956 introns. Intron 1 forms a clade with other group IE-type 956 introns, whereas Intron 2 clusters with group IC1-type 956 introns. This is the only example that we are aware of, in which two very different group I splicing ribozymes are found at the identical rDNA site in closely related isolates of the same host species. Because both of the Didymium introns are mobile (or inferred mobile), have different evolutionary histories and belong to separate lineages of introns, Intron 1 and Intron 2 were most likely acquired from different sources.
The recent transfer of a homing endonuclease gene
To establish the evolutionary history of the HEGs hosted by Intron 1 and Intron 2, we generated a HE phylogeny by adding the Didymium HE sequences to a previously published protein dataset. Figure 3B shows the protein tree and provides support for the close phylogenetic relationship between the two 956 HEs despite that the host splicing ribozymes (i.e. Intron 1 and Intron 2) are very different. Recently, it has become more apparent that the evolution of group I introns and their associated HEGs is more dynamic than previously anticipated [reviewed in (3)]. Comparative sequence analyses support that HEGs can insert into group I introns and spread independent of their associated group I ribozyme partners. Our data provide additional insights into HEG transfers. A closer look at the sequence that flanks the HEG-insertion in Intron 2 reveals a remarkable match (identity in 14 of 17 flanking positions) to the rDNA sequence that flank the 956 intron (Figure 3C). HE-mediated intron-spread (or ‘homing’) requires a 15–25 nt target region at the site of intron insertion. Our finding suggests that a recent homing-like event resulted in the insertion of a HEG into Intron 2. A final clue to the phylogenetic connection between the Intron 1 and Intron 2 HEGs is that they are both interrupted by a small spliceosomal intron (51 nt in Intron 1 and 50 nt in Intron 2) located in the same region (but not identical site) of the respective HEGs. Spliceosomal introns are currently found in three HEGs only (see Figure 2A and B). Interestingly, the three HEGs are encoded by introns at SSU positions 943 and 956, and also cluster on the protein phylogeny (although without support). These features may indicate an evolutionary link between the HEGs [HEGs in neighboring introns are often closely related; see Haugen et al. (3)].
Finally, to allow HEG transfer between different group I introns at the same rDNA site, different rDNA copies must be present at the same time in one single cell. In nature different lineages of rDNA (and many copies of linear extrachromosomal rDNA) co-exist in Didymium during mating between haploid amoebas [to generate a syncytium (a cell with multiple nuclei)], or after fusion of diploid cells [to generate a heterokaryon (a cell with genetically different nuclei)]. These stages in the life cycle of plasmodial slime molds provide a natural platform for group I introns and HEGs to spread between rDNA copies. Different Didymium strains of different mating types can be manipulated in experimental crosses and the system is therefore well suited to study group I intron mobility.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We wish to thank Kari Haugli for DNA sequencing and growth of myxamoebas, and Jim Clark for generously providing Didymium isolates. This work was supported by grants from The Norwegian Research Council (S.J and P.H), The Norwegian Cancer Society (S.J.) and The Aakre Foundation for Cancer Research (S.J.). Funding to pay the Open Access publication charges for this article was provided by University of Tromsø, IMB.
Conflict of interest statement. None declared.
REFERENCES
- 1.Goddard M.R., Burt A. Recurrent invasion and extinction of a selfish gene. Proc. Natl Acad. Sci. USA. 1999;96:13880–13885. doi: 10.1073/pnas.96.24.13880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cannone J.J., Subramanian S., Schnare M.N., Collett J.R., D'Souza L.M., Du Y., Feng B., Lin N., Madabusi L.V., Muller K.M., Pande N., Shang Z., Yu N., Gutell R.R. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haugen P., Simon D., Bhattacharya D. The natural history of group I introns. Trends Genet. 2005;21:111–119. doi: 10.1016/j.tig.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Loizos N., Tillier E.R., Belfort M. Evolution of mobile group I introns: recognition of intron sequences by an intron-encoded endonuclease. Proc. Natl Acad. Sci. USA. 1994;91:11983–11987. doi: 10.1073/pnas.91.25.11983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haugen P., Bhattacharya D. The spread of LAGLIDADG homing endonuclease genes in rDNA. Nucleic Acids Res. 2004;32:2049–2057. doi: 10.1093/nar/gkh520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haugen P., Reeb V., Lutzoni F., Bhattacharya D. The evolution of homing endonuclease genes and group I introns in nuclear rDNA. Mol. Biol. Evol. 2004;21:129–140. doi: 10.1093/molbev/msh005. [DOI] [PubMed] [Google Scholar]
- 7.Decatur W.A., Einvik C., Johansen S., Vogt V.M. Two group I ribozymes with different functions in a nuclear rDNA intron. EMBO J. 1995;14:4558–4568. doi: 10.1002/j.1460-2075.1995.tb00135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johansen S., Elde M., Vader A., Haugen P., Haugli K., Haugli F. In vivo mobility of a group I twintron in nuclear ribosomal DNA of the myxomycete Didymium iridis. Mol. Microbiol. 1997;24:737–745. doi: 10.1046/j.1365-2958.1997.3921743.x. [DOI] [PubMed] [Google Scholar]
- 9.Vader A., Nielsen H., Johansen S. In vivo expression of the nucleolar group I intron-encoded I-DirI homing endonuclease involves the removal of a spliceosomal intron. EMBO J. 1999;18:1003–1013. doi: 10.1093/emboj/18.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lundblad E.W., Einvik C., Rønning S., Haugli K., Johansen S. Twelve group I introns in the same pre-rRNA transcript of the myxomycete Fuligo septica: RNA processing and evolution. Mol. Biol. Evol. 2004;21:1283–1293. doi: 10.1093/molbev/msh126. [DOI] [PubMed] [Google Scholar]
- 11.Elde M., Haugen P., Willassen N.P., Johansen S. I-NjaI, a nuclear intron-encoded homing endonuclease from Naegleria, generates a pentanucleotide 3′ cleavage-overhang within a 19 base-pair partially symmetric DNA recognition site. Eur. J. Biochem. 1999;259:281–288. doi: 10.1046/j.1432-1327.1999.00035.x. [DOI] [PubMed] [Google Scholar]
- 12.Birgisdottir A.B., Johansen S. Site-specific reverse splicing of a HEG-containing group I intron in ribosomal RNA. Nucleic Acids Res. 2005;33:2042–2051. doi: 10.1093/nar/gki341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith D.B., Johnson K.S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 14.Lowery R., Hung L., Knoche K., Bandziulis R. Properties of I-PpoI: a rare-cutting intron-encoded endonuclease. Promega Notes. 1992;38:8–12. [Google Scholar]
- 15.Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 16.Swofford D.L. PAUP*: Phylogenetic Analysis Using Parsimony (* and other methods) Sunderland, MA: Sinauer; 2002. Version 4.0b8. [Google Scholar]
- 17.Kumar S., Tamura K., Jakobsen I.B., Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 18.Huelsenbeck J.P., Ronquist F. MrBayes: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 19.Jones D.T., Taylor W.R., Thornton J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 20.Felsenstein J. PHYLIP (phylogeny inference package). Seattle: Department of Genome Sciences, University of Washington; 2002. Version 3.6a3. [Google Scholar]
- 21.Whelan S., Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 22.Einvik C., Elde M., Johansen S. Group I twintrons: genetic elements in myxomycete and schizopyrenid amoeboflagellate ribosomal DNAs. J. Biotechnol. 1998;64:63–74. doi: 10.1016/s0168-1656(98)00104-7. [DOI] [PubMed] [Google Scholar]
- 23.Johansen S., Einvik C., Nielsen H. DiGIR1 and NaGIR1: naturally occurring group I-like ribozymes with unique core organization and evolved biological role. Biochimie. 2002;84:905–912. doi: 10.1016/s0300-9084(02)01443-8. [DOI] [PubMed] [Google Scholar]
- 24.Johansen S., Vogt V.M. An intron in the nuclear ribosomal DNA of Didymium iridis codes for a group I ribozyme and a novel ribozyme that cooperate in self-splicing. Cell. 1994;76:725–734. doi: 10.1016/0092-8674(94)90511-8. [DOI] [PubMed] [Google Scholar]
- 25.Haugen P., Andreassen M., Birgisdottir A.B., Johansen S. Hydrolytic cleavage by a group I intron ribozyme is dependent on RNA structures not important for splicing. Eur. J. Biochem. 2004;271:1015–1024. doi: 10.1111/j.1432-1033.2004.04003.x. [DOI] [PubMed] [Google Scholar]
- 26.Nielsen H., Fiskaa T., Birgisdottir A.B., Haugen P., Einvik C., Johansen S. The ability to form full-length intron RNA circles is a general property of nuclear group I introns. RNA. 2003;9:1464–1475. doi: 10.1261/rna.5290903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikoh N., Fukatsu T. Evolutionary dynamics of multiple group I introns in nuclear ribosomal RNA genes of endoparasitic fungi of the genus Cordyceps. Mol. Biol. Evol. 2001;18:1631–1642. doi: 10.1093/oxfordjournals.molbev.a003952. [DOI] [PubMed] [Google Scholar]
- 28.Bhattacharya D., Friedl T., Helms G. Vertical evolution and intragenic spread of lichen-fungal group I introns. J. Mol. Evol. 2002;55:74–84. doi: 10.1007/s00239-001-2305-x. [DOI] [PubMed] [Google Scholar]
- 29.Haugen P., Coucheron D.H., Rønning S.B., Haugli K., Johansen S. The molecular evolution and structural organization of self-splicing group I introns at position 516 in nuclear SSU rDNA of myxomycetes. J. Eukaryot. Microbiol. 2003;50:283–292. doi: 10.1111/j.1550-7408.2003.tb00135.x. [DOI] [PubMed] [Google Scholar]
- 30.Vader A., Johansen S., Nielsen H. The group I-like ribozyme DiGIR1 mediates alternative processing of pre-rRNA transcripts in Didymium iridis. Eur. J. Biochem. 2002;23:5804–5812. doi: 10.1046/j.1432-1033.2002.03283.x. [DOI] [PubMed] [Google Scholar]
- 31.Kreivi J.P., Lamond A.I. RNA splicing: unexpected spliceosome diversity. Curr. Biol. 1996;6:802–805. doi: 10.1016/s0960-9822(02)00599-7. [DOI] [PubMed] [Google Scholar]
- 32.Margulis L., Schwartz K.V. Five Kingdoms. An Illustrated Guide to the Phyla of Life on Earth, 3rd edn. New York: W.H. Freeman and Company; 1998. [Google Scholar]
- 33.Logsdon J.M., Jr, Stoltzfus A., Doolittle W.F. Molecular evolution: recent cases of spliceosomal intron gain? Curr. Biol. 1998;8:R560–R563. doi: 10.1016/s0960-9822(07)00361-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.