Abstract
Pronuclear/zygotic stage is the very first stage of life. In this period, paternal pronucleus undergoes massive chromatin remodeling called “paternal reprogramming” including protamine-histone replacement and subsequent acquisition of epigenetic modifications. Although these consecutive events are required for the initiation of maternal-zygotic transition, the precise role of paternal reprogramming and its effect on subsequent embryonic development has been largely unknown to date. Recently, various new techniques, especially next-generation sequencing (NGS) and RNAi microinjection contribute to unveil the epigenetic transition from both paternal and maternal to early preimplantation embryos, suggesting not only the simple transcriptional regulation by transcription factors but also dynamic structural alteration of chromatin to initiate the wave of zygotic gene transcription. This review summarizes such recent progress for understanding the epigenetic transition in sperm and preimplantation embryos, and further argue about its transgenerational effect.
Keywords: Protamine, Histone, Epigenetic modification, Fertilization, Pronucleus, Preimplantation development, Zygotic gene activation, Transgenerational epigenetic inheritance
Introduction
The fertilized egg provides a unique environment where two nuclei, derived from either an oocyte or a sperm, exist independently in the same cytoplasm. Although these two nuclei both contain parental haploid genomes, their chromatin states are markedly different mainly because the paternal chromatin undergoes massive epigenetic reprogramming by protamine-histone exchange. Histones newly incorporated into the paternal chromatin are predominantly supplied from maternal stock, and they eventually acquire new modifications such as methylation. Most of the modifications are asymmetric between paternal and maternal pronuclei. Due to the asymmetric chromatin states, several nuclear events in each pronucleus occurs separately until the first fusion. This review first summarizes these asymmetric epigenetic events occurring in mouse zygotes, and later argues their physiological significance such as transcriptional regulation. In addition, we will also describe sperm-mediated epigenetic transgenerational inheritance, which has recently been attracting attention in the field.
The regulation of minor zygotic gene regulation
Chromatin state of zygotic pronuclei
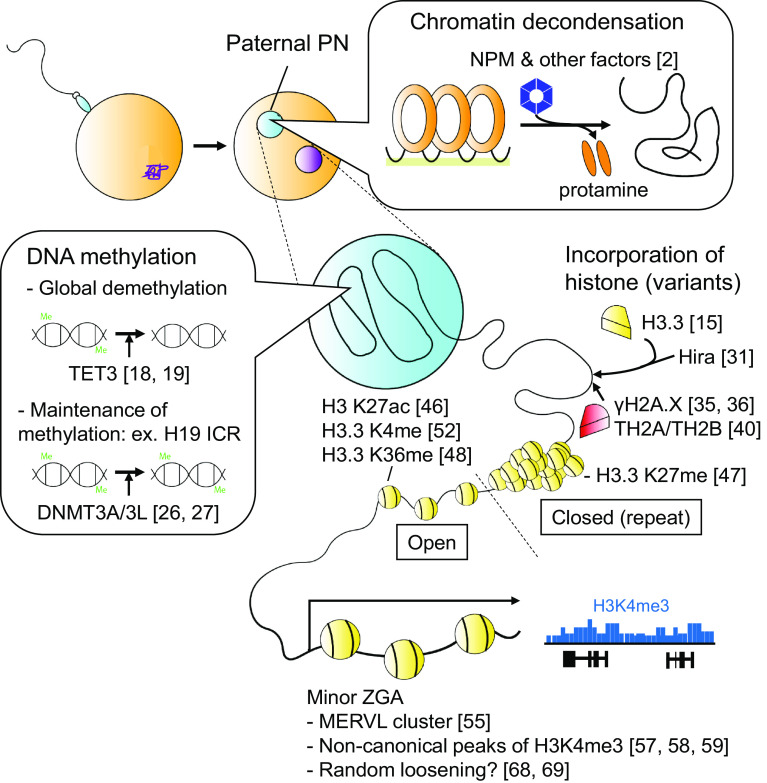
In many animal species, protamine incorporation during spermiogenesis is an essential step for proper chromatin condensation and the full fertility of sperm. In frogs, once the sperm has entered the oocyte, protamines are quickly removed from the paternal chromatin by its chaperon nucleoplasmin (NPM), which is stored in oocytes [1]. Similar observations in mice shows that Npm2-deficient oocytes fail to stimulate sperm chromatin decondensation [2] (Fig. 1). Protamine incorporation and removal are apparently a critical initiator for efficient paternal epigenetic reprogramming, as the efficiency of acquiring pups by round spermatid injection (ROSI) is significantly lower than by injecting mature spermatozoa [3]. This observation also implies that protamine-containing spermatozoa are more capable of being reprogrammed after fertilization. The details of the substantial differences between spermatids and spermatozoa in regard to efficient reprogramming have barely begun to be elucidated. So far, a study has demonstrated a failure of active DNA demethylation in ROSI-derived mouse zygotes [4]. A comparison between sperm- and spermatid-derived frog embryos recently revealed that proper epigenetic alterations such as histone methylations in sperm affect the transcription of a set of developmental genes in early embryos, implying the transgenerational effect [5].
Fig. 1.
Schematic depiction of “paternal reprogramming”. After fertilization, paternal pronucleus undergoes massive chromatin remodeling including protamine removal, acquisition of new histones and their modifications, and DNA demethylation. These events subsequently trigger minor ZGA. Reference numbers are indicated
After protamine removal, the paternal genome gradually acquires new histones, which are provided from maternal stock, and establishes a new epigenetic state, resulting in the epigenetic asymmetry between paternal and maternal pronuclei [6–15] (Fig. 1). In contrast, maternal chromatin basically maintains its modification patterns, although it is highly possible that small amounts of maternal histones are also subjected to be exchanged and acquired new modifications [16].
Among the most obvious asymmetric features between paternal and maternal pronuclei are their DNA methylation and histone 3 lysine 9 (H3K9) methylation state [7, 9, 10]. Genome-wide DNA demethylation occurs predominantly in paternal pronuclei without cell division [7], suggesting that paternal DNA demethylation is active rather than passive in this stage. In 2009, TET family proteins were identified as the first active DNA demethylases [17], and one of the family member TET3 is responsible for paternal DNA demethylation in zygotes [18, 19]. In contrast, H3K9 methylation is exclusively deposited in the maternal pronucleus [20], and is recognized by Dppa3/PGC7/Stella, which prohibit TET3 from demethylating the maternal genome by physical protection [21]. However, maternal DNA to a lesser extent is also targeted for the demethylation by TET3 [22, 23]. In maternal TET3-deficient zygotes, high level of DNA methylation and low level of DNA hydroxymethylation (an oxidation product of 5′-methylcytosine mediated by TET3) are observed in paternal pronuclei concomitant with severely impaired fecundity [18, 21]. Surprisingly, however, the transfer of a male pronucleus harvested from a maternal TET3-deficient zygote to a wild-type recipient demonstrated a comparable fecundity to the control, strongly suggesting that paternal DNA oxidation is dispensable for mouse development [24].
While the majority of the paternal genome is subjected to TET3-dependent global DNA demethylation, DNA methylation in imprinting control regions (ICRs) is faithfully maintained, and several factors including Dppa3/PGC7/Stella, TRIM28, and DNMT1 help to maintain ICR methylation in preimplantation embryos [20, 25, 26]. Interestingly, a recent study of the H19 ICR, where methylation is established by DNMT3A/3L in prospermatogonia and maintained on the paternal allele following fertilization, demonstrated that DNA methylation is re-established after fertilization by maternal DNMT3A/3L, suggesting that the continuous DNA methylation of the paternal H19 ICR is not simply due to the escape from DNA demethylation, and that gametic and zygotic DNA methylations are separable events [27] (Fig. 1).
Paternal histone incorporation in the early zygotic stage is independent of DNA replication. Consistent with the fact, one histone H3 variants called H3.3, whose chromatin incorporation is known to be replication-independent [28], is predominantly incorporated into paternal chromatin in this stage [15, 29]. In contrast, the canonical replication-dependent histones H3.1 and H3.2 do not appear until later zygotic stages even when they are overexpressed in zygotes [30]. Although there are only five amino acid differences between H3.3 and H3.1 (at positions 31, 87, 89, 90 and 96), these differences are critical for recognition by histone chaperones. The preferential incorporation of H3.3 into paternal chromatin is regulated by Hira, an H3.3-specific histone chaperon [31], which generally ensures the deposition to the transcriptionally active regions [28]. H3.3 is also known to localize to telomeres through the mediation of another H3.3 chaperons called ATRX. However, at least in Drosophila, ATRX-dependent H3.3 deposition is not involved in paternal chromatin assembly [32]. Surprisingly, knockdown of Hira in mouse oocytes causes not only deficiency in the incorporation of H3.3, but also a depletion of nucleosome incorporation in paternal pronuclei and subsequently, a failure in nuclear pore complex formation [33, 34]. These results indicate that the incorporation of H3.3 is a scaffold not only for nucleosome formation, but also for the entire pronuclear structure [33].
Histone H2A consists of the most diverse variants to be found among histone families. Among these variants, H2A.X is reported to be highly enriched and γ-phosphorylated (=γH2A.X) in the paternal pronucleus in the absence of aberrant DNA damage in early zygotic stages [35, 36]. Deposition of γH2A.X is more prominent in the paternal than the maternal pronucleus, which may reflect the fact that this constitutes newly assembled chromatin that needs to achieve a proper nucleosomal configuration [35]. After the first cell division, γH2A.X quickly decreases and is hardly detectable in the interphase of 2-cell embryos, whereas the H2A.X protein is continuously retained after the 2-cell stages [37]. H2A.Z, another major H2A variant, has been implicated in a functional association with H3.3, forming unstable nucleosomes that ensure transcription [38, 39]. Unlike the quick and preferential incorporation of H3.3 in the paternal pronucleus, however, H2A.Z is hardly detectable in either male and female pronuclei, and this low level persists until the 4-cell stages [16], refuting a cooperative role between H3.3 and H2A.Z in the zygote.
Other histone variants that have recently attracted great recent interest are TH2A and TH2B, which were originally identified as testis-specific H2A/H2B variants [40]. Their expression has also been demonstrated in oocytes and preimplantation embryos [40]. Th2a and Th2b genes are both located at the largest histone gene cluster, and share the same promoter [41]. Their mRNAs and proteins are abundantly expressed in spermatozoa, mature oocytes, and 1-cell embryos, and decrease after the 2-cell stage upon cell division [40]. Th2a/Th2b-double knockout (dKO) oocytes exhibits impaired preimplantation development after fertilization with wild-type sperm due to a failure of paternal genome activation during the zygotic period, and testis from the dKO males exhibit impaired spermatogenesis due to, respectively, the altered release of cohesins and incorporation of transition proteins [40, 42]. In contrast, oocytes obtained from TH2B-less females can develop without any noticeable defects after fertilization, and depletion of TH2B maintains normal spermatogenesis although functional compensation by canonical H2B, which would be expected to be replaced by TH2B in the early stage of spermatogenesis [43], suggesting that the phenotype of the Th2a/Th2b dKO mice is either a single effect of TH2A, or their compensatory effect. Importantly, exogenous expression of TH2A/TH2B together with phosphorylated NPM significantly enhances iPSC generation from mouse embryonic fibroblasts, demonstrating that TH2A and TH2B are embryonic histones that contribute to reprogramming [40].
Recent in vitro study successfully reconstituted mitotic chromatids using frog sperm as a substrate, requiring only six defined factors, including histone chaperones [44]. Although actual transition from sperm to embryonic chromatin in the zygote seems more complicated and involves many other factors, this finding provided a fundamental aspect of understanding the molecular basis for the protamine-histone transition. Related graphical illustrations are presented in Fig. 1.
Analysis of transcription pattern in minor ZGA
Mammalian preimplantation development is initiated by replacing maternal transcripts with zygotic ones, a process called the maternal-to-zygotic transition. Consistent with the asymmetric chromatin states of paternal and maternal pronuclei, transcriptional regulation in the late 1-cell stage, designated as minor zygotic gene activation (ZGA), is also known to be asymmetric. It is demonstrated that the initiation of minor ZGA in the paternal pronucleus occurs a few hours earlier than in the maternal pronucleus [45]. For decades, however, exactly how this asymmetric transcriptional regulation was established was largely unknown. It was thought that paternal chromatin is more susceptible to transcriptional activation than maternal chromatin, presumably due to the lesser extent of suppressive histone marks such as H3K9 and H3K27 methylations [13]. Indeed, active histone marks including H3.3 and H3K27 acetylation clearly exhibit a preferential deposition in the late paternal pronucleus [46]. H3K4 methylation, the most representative active histone mark, is also established in the paternal pronucleus in the middle to late zygotic stage [9].
The importance of histone methylation has been widely verified by employing loss-of-function studies with histone lysine-arginine (K-R) mutants. As in the following examples, overexpressing exogenous histones carrying K-R mutation in potentially methylated lysine residues successfully demonstrated not only the critical role of methylation of specific lysine residues, but also a distinct, incorporation-dependent role of H3.3 in early preimplantation development. For instance, exogenous expression of H3.3-K27R-GFP but not H3.1-K27R-GFP mRNA in zygotes compromised their preimplantation development concomitant with altered pericentromeric transcription and defects in chromosome segregation, suggesting an important role of H3.3 K27 methylation in pericentromeric silencing [47]. Similarly, injection of morpholino against H3.3 into zygote induced developmental arrest of preimplantation embryos, which could be rescued by the overexpression of wild-type H3.3 but not wild-type H3.1 or H3.3 K36R [48]. Although it remains unknown whether this phenotype was related to transcriptional elongation, it suggests a specific role of K36 methylated H3.3 at this stage [48]. More recently, a lysine-methionine (K-M) mutation in histone H3 was identified from pediatric glioma patients [49, 50]. In studies of cells carrying the K-M mutation, not only the exogenous mutant H3.3 but also endogenous H3 proteins, including canonical forms, were unmethylated due to the inhibition of the enzymes responsible for methylating the mutated sites [51]. By utilizing the K-M mutation instead of K-R mutation, H3.3-K4 methylation was found to be involved preferentially in the minor ZGA preferentially in paternal genome, as well as the subsequent developmental arrest [52]. This work further demonstrated that MLL3/4 is responsible for the H3.3-K4 methylation concomitant with H3K27 acetylation during the zygotic period, implying a role of enhancer activation in the paternal minor ZGA [52, 53]. However, the question of dependence for enhancer at the zygotic stage remains open as a previous study reported that transcription in the paternal pronucleus occurs independently from the enhancer [54]. More direct and concrete evidence such as Chromatin immunoprecipitation sequencing (ChIP-seq) analyses is required to argue the involvement of enhancers in zygotic transcription (ChIP-seq analysis in zygote will be mentioned in the later section).
Recently, Assay for Transposase-Accessible Chromatin with high throughput sequencing (ATAC-seq) has been applied for further qualitative and quantitative characterization of the minor ZGA, and it was found that open chromatin regions are formed as clusters in several regions, especially near the murine endogenous retrovirus-L (MERVL) gene [55]. This study also confirmed that these ATAC-seq peaks disappear with α-amanitin (an inhibitor of transcription) treatment, suggesting that they are transcription-dependent, although this study did not differentiate between paternal and maternal chromatin [55]. Similarly, genome-wide NGS study of preimplantation embryos using a low-input DNase I hypersensitivity site (DHS) mapping demonstrated that the DHS profile of the paternal genome becomes similar to that of maternal genome as early as at pronuclear stage (PN) 3, approximately 7.5 h after fertilization [56]. As PN3 is the time of the minor ZGA initiation in paternal pronuclei, this observation indicated that the paternal reprogramming for subsequent zygotic events is almost completed by this stage.
Recently, ChIP-seq was successfully adapted to allow the analysis of extremely small number of cells (~a few hundreds), and this technical development enables to directly examine the genomic localization of several histone modifications in oocytes, zygotes, and early preimplantation embryos [57–59]. Unexpectedly, these results consistently demonstrated the “non-canonical” pattern of H3K4me3, which is enriched at low levels across large genomic region (~10 kbp), and distant from transcription start sites [57–60]. This unusual epigenetic pattern is conserved from oocytes to early 2-stage embryos mainly in maternal chromatin, but paternal chromatin also exhibits similar broad distribution of H3K4me3 [59]. Interestingly, genes associated with the non-canonical H3K4me3 are related with oocyte growth and major ZGA, suggesting the role as an inherited epigenetic memory of the transcriptional state [58, 59]. However, it is also shown that the depletion of H3K4me3 in oocytes results in increased transcriptional activation, implying the role of H3K4me3 in transcriptional silencing [59]. Collectively, these genome-wide sequencing analyses clarify the transcriptional regulation of minor ZGA more unique and mysterious, thus further work is required to fully understand the precise molecular mechanisms [60]. Related graphical illustrations are presented in Fig. 1.
Putative chromatin regulators in minor ZGA
Despite such accumulation of epigenetic knowledges of the pronuclear chromatin state, the physiological importance and molecules that control the minor ZGA is not yet fully understood. In addition to MLL3/4 (mentioned previously), TIF1α has also been reported to modulate zygotic gene expression through BRG1/SNF2 in both male and female pronuclei [29]. The DHS study also identified several transcription factors, whose promoter regions are open as early as PN3 [56]. Interestingly, in these cases, knockdown of their identified factors (i.e. Mll3/4, TIF1a, and Nfya) by the siRNA injection induced developmental arrest a few stages later than their actually expressed stages, whereas complete block of transcription by α-amanitin treatment stops the development at the next embryonic stage. This implies at least two possibilities; (1) that several molecules are involved in the minor ZGA, and that inhibition of single molecule is thus insufficient to cause immediate arrest, or (2) siRNA injection is inadequate to completely suppress the target genes due to the abundance of maternally inherited mRNA/protein or limited timing of injection. Similar observation was reported in the case of somatic cell nuclear transfer (SCNT) [61]. In this study, erasure of H3K9me3 by exogenous expression of the demethylase significantly increases the efficiency of SCNT, and 49 genes were identified as their overexpression possibly improves the efficiency. One of the top-ranking genes, Zscan4d, however, failed to increase the developmental rate of SCNT embryos when it was administrated in the embryos, also suggesting that complicated gene networks rather than a sole specific factor is responsible for proper preimplantation development [61].
Transcription during the minor ZGA is subtle but substantial. However, there is little translation from these transcripts [62], suggesting the possibilities that the role of minor ZGA is not to express functional proteins, or only a few transcripts, which are critical for subsequent embryonic development, are selectively translated. Later, genome-wide transcriptome analyses in preimplantation embryos by microarray identified substantial change of transcription in 1-cell embryo, which was dramatically altered by cycloheximide (CHX: an inhibitor of translation) and α-amanitin, whereas aphidicolin (an inhibitor of DNA replication) exhibited a little effect [63]. Interestingly, there was a remarkable overlap of transcripts, which expression were commonly affected by CHX and α-amanitin, indicating that translation of maternally stored transcripts or translation of ZGA transcripts themselves is essential for the minor ZGA [64]. Further, large-scale transcriptome analysis by RNA-seq identified five transcription factors expressed in 1-cell but not in parthenogenic embryos, which possibly involved in transcriptional initiation in fertilized oocytes [65].
Interestingly, another RNA-seq analysis demonstrated that during the minor ZGA, intergenic regions are extensively expressed, and thousands of genes are transcribed at comparably low levels accompanied by inefficient mRNA splicing [66]. However, inhibition of RNA polymerase II by DRB (5,6-dichlorobenzimidazole riboside) from 1-cell to early 2-cell stages results in the developmental arrest at the 2-cell stage, suggesting that transcription at the 1-cell stages is essential for further development. These confusing observations may be interpreted as a suggestion that the transcription itself but not the products (i.e. the mRNAs or proteins) possesses a role in early embryonic development such as transcription-induced alteration of chromatin structure including the nucleosome re-positioning, histone replacement, and histone modifications required for the major ZGA at the 2-cell stage [66]. Further analysis by ATAC-seq at the early 2-cell stage corresponding to the minor ZGA also demonstrated noisy peaks [55]. However, the ATAC-seq demonstrated that open chromatin is enriched in repeat regions, especially MERVL clusters, which are highly transcribed during the minor ZGA [55], which also suggests that an open chromatin structure is one of the key factors for transactivation.
Interestingly, this idea seems to be supported by another line of evidence. There is a rare transient cell population within cultured mouse ES cells, which express high levels of transcripts found in 2-cell embryos including MERVL, designated as 2 cell-like cells (2CLCs) [67]. The 2CLCs lack the expression of pluripotent factors such as Oct4 and Sox2, and have acquired the ability to contribute to both embryonic and extraembryonic tissues, suggesting that they functionally resemble 2-cell embryos. Transcriptome analysis further identified that the knockdown of heterochromatic factors such as G9a, KAP1, and HDACs ensure the conversion from ES cells to 2CLCs, implying the link between loosened chromatin structure and 2C-like state, although it is not clear whether this is a global or target gene-specific effects [67]. Later, knockdown of CAF-1, a somatic histone H3 chaperon, in ES cells was also demonstrated to ensure the conversion to 2CLC [68], supporting the contribution of structural modulation of chromatin rather than gene-specific transcriptional regulation to the acquisition of 2C-like state. Consistently, an examination of chromatin stability in early embryos by fluorescent recovery after photobleaching (FRAP) demonstrate that highly relaxed chromatin structure is involved in totipotency of 1-cell embryos, and this looseness is lost upon differentiation during preimplantation development [69]. Further analyses and combined knowledge of early embryos and 2CLC may enable the acquisition of totipotent cells in vitro. Related graphical illustrations are presented in Fig. 1.
Sperm-retained histones: where are they?
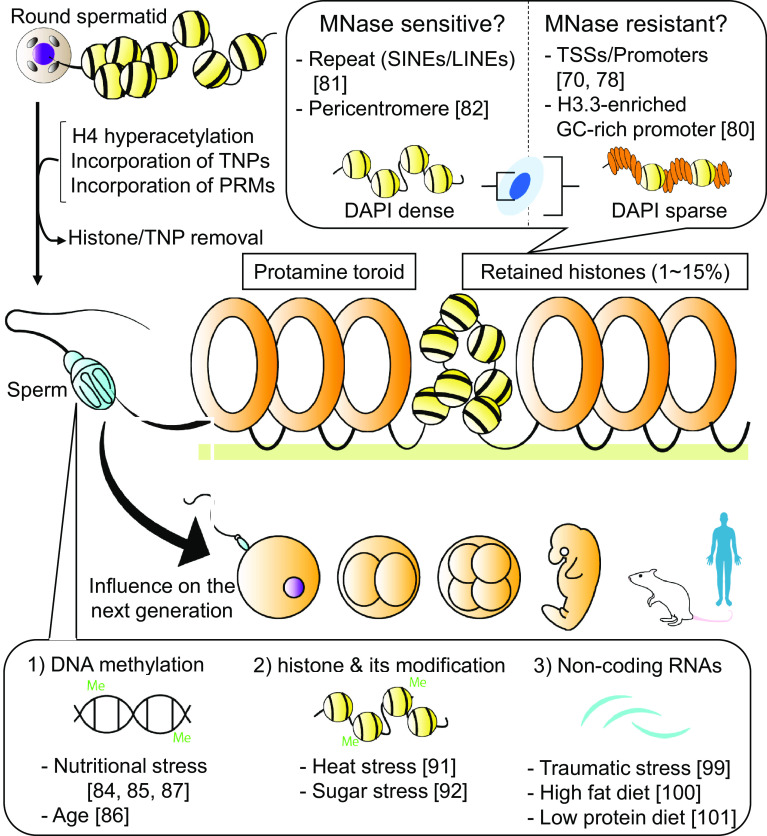
As described above, it can be easily imagined that the unique chromatin remodeling in the paternal pronucleus is tightly associated with sperm chromatin structure. In fact, this hypothesis is strongly supported by a recent study on Xenopus demonstrating the critical role of sperm-retained histones in the regulation of gene transcription in embryos [5]. On the other hand, very little histones are retained in sperm chromatin after histone-protamine replacement, although the extent varies between animal species [70]. Thus, the major question that emerges is whether histones are retained precisely within the sperm genome, and if so, where. This is particularly germane when we interpret the effects of sperm-retained histones on subsequent generations, as described in the next section.
Retention of histones in spermatozoa/mature sperm has been described in several mammalian species including human and mouse; approximately 10–15% of histones in human and 1% in mouse are retained as a heterogenous mixture of protamines and other basic proteins [70–72]. In human sperm, the discrete structures of nucleohistones and nucleoprotamines have been demonstrated, and unique toroid formation of nucleoprotamin was later suggested [71, 73]. Immunohistochemical analyses of sperm demonstrate the accumulation of histones in DAPI-dense regions, which correspond to the major satellite and pericentromeric heterochromatin demonstrated by fluorescent in situ hybridization (FISH), concomitant with hyperacetylated H4 in mouse and H3K9 trimethylation in human, suggesting that the preferential retention of sperm histones in pericentromere/repeat sequences including telomeres [74–77].
Inconsistent with these observations, however, genome-wide analyses of human sperm genome by NGS demonstrated that nucleosomes are preferentially retained in gene coding regions [78]. Interestingly, the enrichment of nucleosomes are significantly accumulated in developmental genes rather than gene-poor regions, although it is technically difficult to access pericentromere/repeat regions by NGS [70, 78, 79]. Subsequent studies of mouse sperm also revealed the enrichment of sperm nucleosomes, especially H3.3-containing nucleosomes, in promoters containing GC-rich sequences with lower DNA methylation [80]. These results also imply the participation of sperm epigenome to the transgenerational effects, especially transcriptional regulation. In contrast, similar NGS analyses performed in two other studies found that sperm nucleosomes are enriched in gene-poor regions including the centromere and repeat regions, which was further supported by the agreement of immunostaining of histones [81, 82]. This controversy may be caused by the complex sensitivity to micrococcal nuclease (MNase) treatment [82]. In addition, it has also been pointed out that inappropriate sequence data analysis may have influenced on the interpretation [83]. Further multi-dimensional investigations are required to fully understand the sperm chromatin structure, and the regularity of histone-protamine replacement during spermiogenesis. Related graphical illustrations are presented in Fig. 2.
Fig. 2.
Schematic depiction of “paternal transgenerational effect”. During spermiogenesis, sperm histones are replaced by PRMs, while 1–15% of histones are still retained in sperm chromatin. A current question is whether these histones are retained in sperm genome, and whether they have substantial roles in the next generation. Other than histones, DNA methylation and non-coding RNAs are possibly transmitted from sperm to offspring, and their involvement in the next generations are also suggested. Reference numbers are indicated
Transgenerational effects of paternal epigenome
The transgenerational inheritance of sperm epigenetic state has been receiving a large amount of attention in the field, despite the confusing situation regarding sperm chromatin structure described above. In 2010, influence of paternal diet on the gene expression and metabolism in the progeny, also designated as paternal-diet-induced intergenerational metabolic reprogramming (IGMR), was first demonstrated in rodent, strongly suggesting the non-genetic intergenerational transmission of metabolic sequelae of paternal diet through sperm [84, 85]. Regarding the molecular mechanisms, at least three possible causative factors so far have been claimed as responsible for sperm-mediated epigenetic inheritance: DNA methylation, histones (and their modifications), and RNAs. Among these, DNA methylation has been regarded as the most promising factor based on the high level of DNA methylation in sperm, as well as the more minor effect of histone-protamine replacement. Indeed, clinical cohort studies have indicated relationship between the age-associated alteration of sperm DNA methylation and the risk of certain neuropsychiatric conditions [86]. Nutritional stress also affects DNA methylation in sperm and subsequently the health condition of the offspring [87]. In one case, altered DNA methylation status in Olfr151 gene is demonstrated to be transferred to subsequent generations as “a paternal fear memory”, an odor (acetophenone)-linked fear conditioning [88]. Olfr151 is a known odorant receptor activated by acetophenone. In this study, it was demonstrated that progenies of the fear-conditioned male by acetophenone exhibit traumatic response, when they sense acetophenone without actual fear experience. This phenomenon is linked to the CpG hypomethylation in the Olfr151 gene, which is inherited from the hear-conditioned male though sperm [88]. Moreover, in zebrafish, the paternal DNA methylation pattern rather than the maternal pattern is maintained throughout early embryogenesis, and is critical for reprogramming during ZGA [89, 90].
The second factor, histone modifications, may be involved in sperm-derived transgenerational effects. In 2011, a study in Drosophila demonstrated that the stress-induced disruption of heterochromatin in gametes was transferred to subsequent generations, implying the importance of chromatin structure for epigenetic inheritance [91]. Furthermore, an IGMR study in Drosophila successfully demonstrated the link between the altered gene expression in IGMR and Polycomb/heterochromatin machinery [92]. In C. elegans, increased longevity observed in Wdr5-deficient worm can be inherited by their wild-type progeny, and multiple chromatin-modifying factors related to H3K4 and H3K9 methylations regulate transgenerational effects on fertility [93, 94]. In rodent, wild-type mouse progeny derived from sperm carrying an LSD1-overexpression allele exhibited morphological abnormalities and reduced survival rates [95]. However, there were no obvious alterations of major histone modifications in the abnormal wild-type progeny, suggesting that at least in this case, impaired histone modifications were not responsible for the sperm-mediated transgenerational abnormalities [95].
The third factor, RNA, has been in the limelight for the last few years. Initial report was demonstrated in RDE-4 deficient C. elegans, which are sensitive to flock house virus infections due to the lack of small RNA biogenesis. Surprisingly, RDE-4 deficient C. elegans can acquire an anti-viral response through viRNAs transferred from a wild-type ancestor to the progenies through their sperm [96]. Subsequently studies in C. elegans further demonstrated that the transgenerational inheritance of starvation-induced developmental arrest is also mediated by small RNA-induced gene silencing [97]. The lifespan in the F3 offspring of starved animals was also increased, suggesting that calorie restriction-induced longevity can become a transgenerational memory [97, 98]. Similarly, microRNA in mice can be a vector transmitting the effects of traumatic stress experienced by the father from the sperm to the progeny, and this can affect childrens’ behavior [99]. More recently, fragmented transfer RNAs produced and supplied by the epididymal epithelium were found to be the causative agent of the transgenerational effects of diet-induced nutritional stress [100, 101]. Importantly, these effects in mice could be reconstituted experimentally by the injection of RNA harvested from the sperm of stress-exposed fathers, although how particular transfer RNAs influence the metabolism of the offspring, and how the abnormal condition persists over the long term both remain questions to be answered. Related graphical illustrations are presented in Fig. 2.
Conclusion
Advancements in technologies for epigenetic studies, especially NGS and technical improvement in analyzing small-scale samples, have made study of the dynamic epigenetic transitions from gamete to embryos more accessible than ever. Genome-wide sequencing analyses in oocytes and early preimplantation embryos in particular have provided critical information for understanding reprogramming at the molecular level. However, this has also led to some confusing findings regarding sperm chromatin structure, a comprehensive understanding of which seems essential for further investigation and for understanding the transgenerational inheritance of the paternal epigenome, as well as for subsequent clinical and bioengineering applications. It is also obvious that a better understanding of paternal reprogramming will provide useful information for the technical improvement of regenerative medicine and somatic cell nuclear transfer, in the latter of which the donor nucleus is reprogrammed in the ooplasm, as similar molecules may participate in both cases.
Acknowledgements
This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) in Japan (#15H05976 to YO).
Abbreviations
- ATAC-seq
Assay for transposase-accessible chromatin with high throughput sequencing
- CHX
Cycloheximide
- ChIP-seq
Chromatin immunoprecipitation sequencing
- DHS
DNase I hypersensitivity site
- dKO
Double knock-out
- DRB
5,6-dichlorobenzimidazole riboside
- FISH
Fluorescent in situ hybridization
- FRAP
Fluorescent recovery after photobleaching
- GFP
Green fluorescent protein
- ICR
Imprinting control region
- iPSC
Induced pluripotent stem cell
- MERVL
Murine endogenous retrovirus-L
- MNase
Micrococcal nuclease
- NGS
Next-generation sequencing
- NPM
Nucleoplasmin
- PN
Pronuclear stage
- RNA-seq
RNA sequencing
- ROSI
Round spermatid injection
- SCNT
Somatic cell nuclear transfer
- ZGA
Zygotic gene activation
References
- 1.Ohsumi K, Katagiri C. Characterization of the ooplasmic factor inducing decondensation of and protamine removal from toad sperm nuclei: involvement of nucleoplasmin. Dev Biol. 1991;148(1):295–305. doi: 10.1016/0012-1606(91)90338-4. [DOI] [PubMed] [Google Scholar]
- 2.Inoue A, Ogushi S, Saitou M, Suzuki MG, Aoki F. Involvement of mouse nucleoplasmin 2 in the decondensation of sperm chromatin after fertilization. Biol Reprod. 2011;85(1):70–77. doi: 10.1095/biolreprod.110.089342. [DOI] [PubMed] [Google Scholar]
- 3.Kimura Y, Yanagimachi R. Mouse oocytes injected with testicular spermatozoa or round spermatids can develop into normal offspring. Development. 1995;121(8):2397–2405. doi: 10.1242/dev.121.8.2397. [DOI] [PubMed] [Google Scholar]
- 4.Kurotaki YK, Hatanaka Y, Kamimura S, Oikawa M, Inoue H, Ogonuki N, Inoue K, Ogura A. Impaired active DNA demethylation in zygotes generated by round spermatid injection. Hum Reprod. 2015;30(5):1178–1187. doi: 10.1093/humrep/dev039. [DOI] [PubMed] [Google Scholar]
- 5.Teperek M, Simeone A, Gaggioli V, Miyamoto K, Allen GE, Erkek S, Kwon T, Marcotte EM, Zegerman P, Bradshaw CR, Peters AH, Gurdon JB, Jullien J. Sperm is epigenetically programmed to regulate gene transcription in embryos. Genome Res. 2016;26(8):1034–1046. doi: 10.1101/gr.201541.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adenot PG, Mercier Y, Renard JP, Thompson EM. Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development. 1997;124(22):4615–4625. doi: 10.1242/dev.124.22.4615. [DOI] [PubMed] [Google Scholar]
- 7.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241(1):172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 8.Cowell IG, Aucott R, Mahadevaiah SK, Burgoyne PS, Huskisson N, Bongiorni S, Prantera G, Fanti L, Pimpinelli S, Wu R, Gilbert DM, Shi W, Fundele R, Morrison H, Jeppesen P, Singh PB. Heterochromatin, HP1 and methylation at lysine 9 of histone H3 in animals. Chromosoma. 2002;111(1):22–36. doi: 10.1007/s00412-002-0182-8. [DOI] [PubMed] [Google Scholar]
- 9.Lepikhov K, Walter J. Differential dynamics of histone H3 methylation at positions K4 and K9 in the mouse zygote. BMC Dev Biol. 2004;4:12. doi: 10.1186/1471-213X-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu H, Kim JM, Aoki F. Regulation of histone H3 lysine 9 methylation in oocytes and early pre-implantation embryos. Development. 2004;131(10):2269–2280. doi: 10.1242/dev.01116. [DOI] [PubMed] [Google Scholar]
- 11.Erhardt S, Su IH, Schneider R, Barton S, Bannister AJ, Perez-Burgos L, Jenuwein T, Kouzarides T, Tarakhovsky A, Surani MA. Consequences of the depletion of zygotic and embryonic enhancer of zeste 2 during preimplantation mouse development. Development. 2003;130(18):4235–4248. doi: 10.1242/dev.00625. [DOI] [PubMed] [Google Scholar]
- 12.Kourmouli N, Jeppesen P, Mahadevhaiah S, Burgoyne P, Wu R, Gilbert DM, Bongiorni S, Prantera G, Fanti L, Pimpinelli S, Shi W, Fundele R, Singh PB. Heterochromatin and tri-methylated lysine 20 of histone H4 in animals. J Cell Sci. 2004;117(12):2491–2501. doi: 10.1242/jcs.01238. [DOI] [PubMed] [Google Scholar]
- 13.Santos F, Peters AH, Otte AP, Reik W, Dean W. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev Biol. 2005;280(1):225–236. doi: 10.1016/j.ydbio.2005.01.025. [DOI] [PubMed] [Google Scholar]
- 14.Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14(1)):R47–58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- 15.van der Heijden GW, Dieker JW, Derijck AA, Muller S, Berden JH, Braat DD, van der Vlag J, de Boer P. Asymmetry in histone H3 variants and lysine methylation between paternal and maternal chromatin of the early mouse zygote. Mech Dev. 2005;122(9):1008–1022. doi: 10.1016/j.mod.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 16.Boskovic A, Bender A, Gall L, Ziegler-Birling C, Beaujean N, Torres-Padilla ME. Analysis of active chromatin modifications in early mammalian embryos reveals uncoupling of H2A.Z acetylation and H3K36 trimethylation from embryonic genome activation. Epigenetics. 2012;7(7):747–757. doi: 10.4161/epi.20584. [DOI] [PubMed] [Google Scholar]
- 17.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, Iqbal K, Shi YG, Deng Z, Szabo PE, Pfeifer GP, Li J, Xu GL. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477(7366):606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 19.Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334(6053):194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, Sekimoto T, Ikawa M, Yoneda Y, Okabe M, Tanaka S, Shiota K, Nakano T. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol. 2007;9(1):64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- 21.Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, Walter J. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- 22.Shen L, Inoue A, He J, Liu Y, Lu F, Zhang Y. Tet3 and DNA replication mediate demethylation of both the maternal and paternal genomes in mouse zygotes. Cell Stem Cell. 2014;15(4):459–470. doi: 10.1016/j.stem.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo F, Li X, Liang D, Li T, Zhu P, Guo H, Wu X, Wen L, Gu TP, Hu B, Walsh CP, Li J, Tang F, Xu GL. Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell. 2014;15(4):447–458. doi: 10.1016/j.stem.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 24.Inoue A, Shen L, Matoba S, Zhang Y. Haploinsufficiency, but not defective paternal 5mC oxidation, accounts for the developmental defects of maternal Tet3 knockouts. Cell Rep. 2015;10(4):463–470. doi: 10.1016/j.celrep.2014.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Messerschmidt DM, de Vries W, Ito M, Solter D, Ferguson-Smith A, Knowles BB. Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science. 2012;335(6075):1499–1502. doi: 10.1126/science.1216154. [DOI] [PubMed] [Google Scholar]
- 26.Hirasawa R, Chiba H, Kaneda M, Tajima S, Li E, Jaenisch R, Sasaki H. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev. 2008;22(12):1607–1616. doi: 10.1101/gad.1667008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuzaki H, Okamura E, Takahashi T, Ushiki A, Nakamura T, Nakano T, Hata K, Fukamizu A, Tanimoto K. De novo DNA methylation through the 5′-segment of the H19 ICR maintains its imprint during early embryogenesis. Development. 2015;142(22):3833–3844. doi: 10.1242/dev.126003. [DOI] [PubMed] [Google Scholar]
- 28.Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9(6):1191–1200. doi: 10.1016/S1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 29.Torres-Padilla ME, Zernicka-Goetz M. Role of TIF1alpha as a modulator of embryonic transcription in the mouse zygote. J Cell Biol. 2006;174(3):329–338. doi: 10.1083/jcb.200603146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akiyama T, Suzuki O, Matsuda J, Aoki F. Dynamic replacement of histone H3 variants reprograms epigenetic marks in early mouse embryos. PLoS Genet. 2011;7(10):e1002279. doi: 10.1371/journal.pgen.1002279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loppin B, Bonnefoy E, Anselme C, Laurencon A, Karr TL, Couble P. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature. 2005;437(7063):1386–1390. doi: 10.1038/nature04059. [DOI] [PubMed] [Google Scholar]
- 32.Orsi GA, Algazeery A, Meyer RE, Capri M, Sapey-Triomphe LM, Horard B, Gruffat H, Couble P, Ait-Ahmed O, Loppin B. Drosophila Yemanuclein and HIRA cooperate for de novo assembly of H3.3-containing nucleosomes in the male pronucleus. PLoS Genet. 2013;9(2):e1003285. doi: 10.1371/journal.pgen.1003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inoue A, Zhang Y. Nucleosome assembly is required for nuclear pore complex assembly in mouse zygotes. Nat Struct Mol Biol. 2014;21(7):609–616. doi: 10.1038/nsmb.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin CJ, Koh FM, Wong P, Conti M, Ramalho-Santos M. Hira-mediated H3.3 incorporation is required for DNA replication and ribosomal RNA transcription in the mouse zygote. Dev Cell. 2014;30(3):268–279. doi: 10.1016/j.devcel.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ziegler-Birling C, Helmrich A, Tora L, Torres-Padilla ME. Distribution of p53 binding protein 1 (53BP1) and phosphorylated H2A.X during mouse preimplantation development in the absence of DNA damage. Int J Dev Biol. 2009;53(7):1003–1011. doi: 10.1387/ijdb.082707cz. [DOI] [PubMed] [Google Scholar]
- 36.Nashun B, Yukawa M, Liu H, Akiyama T, Aoki F. Changes in the nuclear deposition of histone H2A variants during pre-implantation development in mice. Development. 2010;137(22):3785–3794. doi: 10.1242/dev.051805. [DOI] [PubMed] [Google Scholar]
- 37.Wu BJ, Dong FL, Ma XS, Wang XG, Lin F, Liu HL. Localization and expression of histone H2A variants during mouse oogenesis and preimplantation embryo development. Genet Mol Res. 2014;13(3):5929–5939. doi: 10.4238/2014.August.7.8. [DOI] [PubMed] [Google Scholar]
- 38.Henikoff S, Henikoff JG, Sakai A, Loeb GB, Ahmad K. Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Res. 2009;19(3):460–469. doi: 10.1101/gr.087619.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henikoff S. Labile H3.3 + H2A.Z nucleosomes mark ‘nucleosome-free regions’. Nat Genet. 2009;41(8):865–866. doi: 10.1038/ng0809-865. [DOI] [PubMed] [Google Scholar]
- 40.Shinagawa T, Takagi T, Tsukamoto D, Tomaru C, Huynh LM, Sivaraman P, Kumarevel T, Inoue K, Nakato R, Katou Y, Sado T, Takahashi S, Ogura A, Shirahige K, Ishii S. Histone variants enriched in oocytes enhance reprogramming to induced pluripotent stem cells. Cell Stem Cell. 2014;14(2):217–227. doi: 10.1016/j.stem.2013.12.015. [DOI] [PubMed] [Google Scholar]
- 41.Huh NE, Hwang IW, Lim K, You KH, Chae CB. Presence of a bi-directional S phase-specific transcription regulatory element in the promoter shared by testis-specific TH2A and TH2B histone genes. Nucleic Acids Res. 1991;19(1):93–98. doi: 10.1093/nar/19.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shinagawa T, Huynh LM, Takagi T, Tsukamoto D, Tomaru C, Kwak HG, Dohmae N, Noguchi J, Ishii S (2015) Disruption of Th2a and Th2b genes causes defects in spermatogenesis. Development 142 (7):1287–1292. [DOI] [PubMed]
- 43.Montellier E, Boussouar F, Rousseaux S, Zhang K, Buchou T, Fenaille F, Shiota H, Debernardi A, Hery P, Curtet S, Jamshidikia M, Barral S, Holota H, Bergon A, Lopez F, Guardiola P, Pernet K, Imbert J, Petosa C, Tan M, Zhao Y, Gerard M, Khochbin S. Chromatin-to-nucleoprotamine transition is controlled by the histone H2B variant TH2B. Genes Dev. 2013;27(15):1680–1692. doi: 10.1101/gad.220095.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shintomi K, Takahashi TS, Hirano T. Reconstitution of mitotic chromatids with a minimum set of purified factors. Nat Cell Biol. 2015;17(8):1014–1023. doi: 10.1038/ncb3187. [DOI] [PubMed] [Google Scholar]
- 45.Aoki F, Worrad DM, Schultz RM. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol. 1997;181(2):296–307. doi: 10.1006/dbio.1996.8466. [DOI] [PubMed] [Google Scholar]
- 46.Hayashi-Takanaka Y, Yamagata K, Wakayama T, Stasevich TJ, Kainuma T, Tsurimoto T, Tachibana M, Shinkai Y, Kurumizaka H, Nozaki N, Kimura H. Tracking epigenetic histone modifications in single cells using Fab-based live endogenous modification labeling. Nucleic Acids Res. 2011;39(15):6475–6488. doi: 10.1093/nar/gkr343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Santenard A, Ziegler-Birling C, Koch M, Tora L, Bannister AJ, Torres-Padilla ME. Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3.3. Nat Cell Biol. 2010;12(9):853–862. doi: 10.1038/ncb2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin CJ, Conti M, Ramalho-Santos M. Histone variant H3.3 maintains a decondensed chromatin state essential for mouse preimplantation development. Development. 2013;140(17):3624–3634. doi: 10.1242/dev.095513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, Hovestadt V, Albrecht S, Kool M, Nantel A, Konermann C, Lindroth A, Jager N, Rausch T, Ryzhova M, Korbel JO, Hielscher T, Hauser P, Garami M, Klekner A, Bognar L, Ebinger M, Schuhmann MU, Scheurlen W, Pekrun A, Fruhwald MC, Roggendorf W, Kramm C, Durken M, Atkinson J, Lepage P, Montpetit A, Zakrzewska M, Zakrzewski K, Liberski PP, Dong Z, Siegel P, Kulozik AE, Zapatka M, Guha A, Malkin D, Felsberg J, Reifenberger G, von Deimling A, Ichimura K, Collins VP, Witt H, Milde T, Witt O, Zhang C, Castelo-Branco P, Lichter P, Faury D, Tabori U, Plass C, Majewski J, Pfister SM, Jabado N. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482(7384):226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 50.Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, Zhang J, Gajjar A, Dyer MA, Mullighan CG, Gilbertson RJ, Mardis ER, Wilson RK, Downing JR, Ellison DW, Zhang J, Baker SJ, St. Jude Children. ’. s Research Hospital-Washington University Pediatric Cancer Genome P Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44(3):251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340(6134):857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aoshima K, Inoue E, Sawa H, Okada Y. Paternal H3K4 methylation is required for minor zygotic gene activation and early mouse embryonic development. EMBO Rep. 2015;16(7):803–812. doi: 10.15252/embr.201439700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Herz HM, Mohan M, Garruss AS, Liang K, Takahashi YH, Mickey K, Voets O, Verrijzer CP, Shilatifard A. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev. 2012;26(23):2604–2620. doi: 10.1101/gad.201327.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wiekowski M, Miranda M, DePamphilis ML. Requirements for promoter activity in mouse oocytes and embryos distinguish paternal pronuclei from maternal and zygotic nuclei. Dev Biol. 1993;159(1):366–378. doi: 10.1006/dbio.1993.1248. [DOI] [PubMed] [Google Scholar]
- 55.Wu J, Huang B, Chen H, Yin Q, Liu Y, Xiang Y, Zhang B, Liu B, Wang Q, Xia W, Li W, Li Y, Ma J, Peng X, Zheng H, Ming J, Zhang W, Zhang J, Tian G, Xu F, Chang Z, Na J, Yang X, Xie W. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature. 2016;534(7609):652–657. doi: 10.1038/nature18606. [DOI] [PubMed] [Google Scholar]
- 56.Lu F, Liu Y, Inoue A, Suzuki T, Zhao K, Zhang Y. Establishing chromatin regulatory landscape during mouse preimplantation development. Cell. 2016;165(6):1375–1388. doi: 10.1016/j.cell.2016.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu X, Wang C, Liu W, Li J, Li C, Kou X, Chen J, Zhao Y, Gao H, Wang H, Zhang Y, Gao Y, Gao S. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature. 2016;537(7621):558–562. doi: 10.1038/nature19362. [DOI] [PubMed] [Google Scholar]
- 58.Dahl JA, Jung I, Aanes H, Greggains GD, Manaf A, Lerdrup M, Li G, Kuan S, Li B, Lee AY, Preissl S, Jermstad I, Haugen MH, Suganthan R, Bjoras M, Hansen K, Dalen KT, Fedorcsak P, Ren B, Klungland A. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature. 2016;537(7621):548–552. doi: 10.1038/nature19360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang B, Zheng H, Huang B, Li W, Xiang Y, Peng X, Ming J, Wu X, Zhang Y, Xu Q, Liu W, Kou X, Zhao Y, He W, Li C, Chen B, Li Y, Wang Q, Ma J, Yin Q, Kee K, Meng A, Gao S, Xu F, Na J, Xie W. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature. 2016;537(7621):553–557. doi: 10.1038/nature19361. [DOI] [PubMed] [Google Scholar]
- 60.Vaquerizas JM, Torres-Padilla ME. Developmental biology: Panoramic views of the early epigenome. Nature. 2016;537(7621):494–496. doi: 10.1038/nature19468. [DOI] [PubMed] [Google Scholar]
- 61.Matoba S, Liu Y, Lu F, Iwabuchi KA, Shen L, Inoue A, Zhang Y. Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell. 2014;159(4):884–895. doi: 10.1016/j.cell.2014.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schultz RM. Regulation of zygotic gene activation in the mouse. Bioessays. 1993;15(8):531–538. doi: 10.1002/bies.950150806. [DOI] [PubMed] [Google Scholar]
- 63.Zeng F, Baldwin DA, Schultz RM. Transcript profiling during preimplantation mouse development. Dev Biol. 2004;272(2):483–496. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 64.Hamatani T, Carter MG, Sharov AA, Ko MS. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell. 2004;6(1):117–131. doi: 10.1016/S1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- 65.Park SJ, Komata M, Inoue F, Yamada K, Nakai K, Ohsugi M, Shirahige K. Inferring the choreography of parental genomes during fertilization from ultralarge-scale whole-transcriptome analysis. Genes Dev. 2013;27(24):2736–2748. doi: 10.1101/gad.227926.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abe K, Yamamoto R, Franke V, Cao M, Suzuki Y, Suzuki MG, Vlahovicek K, Svoboda P, Schultz RM, Aoki F. The first murine zygotic transcription is promiscuous and uncoupled from splicing and 3′ processing. EMBO J. 2015;34(11):1523–1537. doi: 10.15252/embj.201490648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487(7405):57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ishiuchi T, Enriquez-Gasca R, Mizutani E, Boskovic A, Ziegler-Birling C, Rodriguez-Terrones D, Wakayama T, Vaquerizas JM, Torres-Padilla ME. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat Struct Mol Biol. 2015;22(9):662–671. doi: 10.1038/nsmb.3066. [DOI] [PubMed] [Google Scholar]
- 69.Ooga M, Fulka H, Hashimoto S, Suzuki MG, Aoki F. Analysis of chromatin structure in mouse preimplantation embryos by fluorescent recovery after photobleaching. Epigenetics. 2016;11(1):85–94. doi: 10.1080/15592294.2015.1136774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, Beisel C, Schubeler D, Stadler MB, Peters AH. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol. 2010;17(6):679–687. doi: 10.1038/nsmb.1821. [DOI] [PubMed] [Google Scholar]
- 71.Gatewood JM, Cook GR, Balhorn R, Bradbury EM, Schmid CW. Sequence-specific packaging of DNA in human sperm chromatin. Science. 1987;236(4804):962–964. doi: 10.1126/science.3576213. [DOI] [PubMed] [Google Scholar]
- 72.Pittoggi C, Renzi L, Zaccagnini G, Cimini D, Degrassi F, Giordano R, Magnano AR, Lorenzini R, Lavia P, Spadafora C. A fraction of mouse sperm chromatin is organized in nucleosomal hypersensitive domains enriched in retroposon DNA. J Cell Sci. 1999;112(20):3537–3548. doi: 10.1242/jcs.112.20.3537. [DOI] [PubMed] [Google Scholar]
- 73.Ward WS, Partin AW, Coffey DS. DNA loop domains in mammalian spermatozoa. Chromosoma. 1989;98(3):153–159. doi: 10.1007/BF00329678. [DOI] [PubMed] [Google Scholar]
- 74.Govin J, Escoffier E, Rousseaux S, Kuhn L, Ferro M, Thevenon J, Catena R, Davidson I, Garin J, Khochbin S, Caron C. Pericentric heterochromatin reprogramming by new histone variants during mouse spermiogenesis. J Cell Biol. 2007;176(3):283–294. doi: 10.1083/jcb.200604141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Meyer-Ficca ML, Lonchar JD, Ihara M, Bader JJ, Meyer RG. Alteration of poly(ADP-ribose) metabolism affects murine sperm nuclear architecture by impairing pericentric heterochromatin condensation. Chromosoma. 2013;122(4):319–335. doi: 10.1007/s00412-013-0416-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van de Werken C, van der Heijden GW, Eleveld C, Teeuwssen M, Albert M, Baarends WM, Laven JS, Peters AH, Baart EB. Paternal heterochromatin formation in human embryos is H3K9/HP1 directed and primed by sperm-derived histone modifications. Nat Commun. 2014;5:5868. doi: 10.1038/ncomms6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zalenskaya IA, Bradbury EM, Zalensky AO. Chromatin structure of telomere domain in human sperm. Biochem Biophys Res Commun. 2000;279(1):213–218. doi: 10.1006/bbrc.2000.3917. [DOI] [PubMed] [Google Scholar]
- 78.Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460(7254):473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Arpanahi A, Brinkworth M, Iles D, Krawetz SA, Paradowska A, Platts AE, Saida M, Steger K, Tedder P, Miller D. Endonuclease-sensitive regions of human spermatozoal chromatin are highly enriched in promoter and CTCF binding sequences. Genome Res. 2009;19(8):1338–1349. doi: 10.1101/gr.094953.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Erkek S, Hisano M, Liang CY, Gill M, Murr R, Dieker J, Schubeler D, van der Vlag J, Stadler MB, Peters AH. Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa. Nat Struct Mol Biol. 2013;20(7):868–875. doi: 10.1038/nsmb.2599. [DOI] [PubMed] [Google Scholar]
- 81.Samans B, Yang Y, Krebs S, Sarode GV, Blum H, Reichenbach M, Wolf E, Steger K, Dansranjavin T, Schagdarsurengin U. Uniformity of nucleosome preservation pattern in Mammalian sperm and its connection to repetitive DNA elements. Dev Cell. 2014;30(1):23–35. doi: 10.1016/j.devcel.2014.05.023. [DOI] [PubMed] [Google Scholar]
- 82.Carone BR, Hung JH, Hainer SJ, Chou MT, Carone DM, Weng Z, Fazzio TG, Rando OJ. High-resolution mapping of chromatin packaging in mouse embryonic stem cells and sperm. Dev Cell. 2014;30(1):11–22. doi: 10.1016/j.devcel.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Royo H, Stadler MB, Peters AH. Alternative Computational Analysis Shows No Evidence for Nucleosome Enrichment at Repetitive Sequences in Mammalian Spermatozoa. Dev Cell. 2016;37(1):98–104. doi: 10.1016/j.devcel.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 84.Carone BR, Fauquier L, Habib N, Shea JM, Hart CE, Li R, Bock C, Li C, Gu H, Zamore PD, Meissner A, Weng Z, Hofmann HA, Friedman N, Rando OJ. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2010;143(7):1084–1096. doi: 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ng SF, Lin RC, Laybutt DR, Barres R, Owens JA, Morris MJ. Chronic high-fat diet in fathers programs beta-cell dysfunction in female rat offspring. Nature. 2010;467(7318):963–966. doi: 10.1038/nature09491. [DOI] [PubMed] [Google Scholar]
- 86.Jenkins TG, Aston KI, Pflueger C, Cairns BR, Carrell DT. Age-associated sperm DNA methylation alterations: possible implications in offspring disease susceptibility. PLoS Genet. 2014;10(7):e1004458. doi: 10.1371/journal.pgen.1004458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lambrot R, Xu C, Saint-Phar S, Chountalos G, Cohen T, Paquet M, Suderman M, Hallett M, Kimmins S. Low paternal dietary folate alters the mouse sperm epigenome and is associated with negative pregnancy outcomes. Nat Commun. 2013;4:2889. doi: 10.1038/ncomms3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dias BG, Ressler KJ. Parental olfactory experience influences behavior and neural structure in subsequent generations. Nat Neurosci. 2014;17(1):89–96. doi: 10.1038/nn.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Potok ME, Nix DA, Parnell TJ, Cairns BR. Reprogramming the maternal zebrafish genome after fertilization to match the paternal methylation pattern. Cell. 2013;153(4):759–772. doi: 10.1016/j.cell.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jiang L, Zhang J, Wang JJ, Wang L, Zhang L, Li G, Yang X, Ma X, Sun X, Cai J, Zhang J, Huang X, Yu M, Wang X, Liu F, Wu CI, He C, Zhang B, Ci W, Liu J. Sperm, but not oocyte, DNA methylome is inherited by zebrafish early embryos. Cell. 2013;153(4):773–784. doi: 10.1016/j.cell.2013.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Seong KH, Li D, Shimizu H, Nakamura R, Ishii S. Inheritance of stress-induced, ATF-2-dependent epigenetic change. Cell. 2011;145(7):1049–1061. doi: 10.1016/j.cell.2011.05.029. [DOI] [PubMed] [Google Scholar]
- 92.Ost A, Lempradl A, Casas E, Weigert M, Tiko T, Deniz M, Pantano L, Boenisch U, Itskov PM, Stoeckius M, Ruf M, Rajewsky N, Reuter G, Iovino N, Ribeiro C, Alenius M, Heyne S, Vavouri T, Pospisilik JA. Paternal diet defines offspring chromatin state and intergenerational obesity. Cell. 2014;159(6):1352–1364. doi: 10.1016/j.cell.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 93.Greer EL, Maures TJ, Ucar D, Hauswirth AG, Mancini E, Lim JP, Benayoun BA, Shi Y, Brunet A. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature. 2011;479(7373):365–371. doi: 10.1038/nature10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Greer EL, Beese-Sims SE, Brookes E, Spadafora R, Zhu Y, Rothbart SB, Aristizabal-Corrales D, Chen S, Badeaux AI, Jin Q, Wang W, Strahl BD, Colaiacovo MP, Shi Y. A histone methylation network regulates transgenerational epigenetic memory in C. elegans . Cell Rep. 2014;7(1):113–126. doi: 10.1016/j.celrep.2014.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Siklenka K, Erkek S, Godmann M, Lambrot R, McGraw S, Lafleur C, Cohen T, Xia J, Suderman M, Hallett M, Trasler J, Peters AH, Kimmins S. Disruption of histone methylation in developing sperm impairs offspring health transgenerationally. Science. 2015;350(6261):aab2006. doi: 10.1126/science.aab2006. [DOI] [PubMed] [Google Scholar]
- 96.Rechavi O, Minevich G, Hobert O. Transgenerational inheritance of an acquired small RNA-based antiviral response in C. elegans . Cell. 2011;147(6):1248–1256. doi: 10.1016/j.cell.2011.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rechavi O, Houri-Ze’evi L, Anava S, Goh WS, Kerk SY, Hannon GJ, Hobert O. Starvation-induced transgenerational inheritance of small RNAs in C. elegans . Cell. 2014;158(2):277–287. doi: 10.1016/j.cell.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425(6954):191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 99.Gapp K, Jawaid A, Sarkies P, Bohacek J, Pelczar P, Prados J, Farinelli L, Miska E, Mansuy IM. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat Neurosci. 2014;17(5):667–669. doi: 10.1038/nn.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, Feng GH, Peng H, Zhang X, Zhang Y, Qian J, Duan E, Zhai Q, Zhou Q. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351(6271):397–400. doi: 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- 101.Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun F, Song L, Carone BR, Ricci EP, Li XZ, Fauquier L, Moore MJ, Sullivan R, Mello CC, Garber M, Rando OJ. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351(6271):391–396. doi: 10.1126/science.aad6780. [DOI] [PMC free article] [PubMed] [Google Scholar]