Abstract
The nuclear pore is a key structure in eukaryotes regulating nuclear-cytoplasmic transport as well as a wide range of cellular processes. Here, we report the characterization of the first Toxoplasma gondii nuclear pore protein, named TgNup302, which appears to be the orthologue of the mammalian Nup98-96 protein. We produced a conditional knock-down mutant that expresses TgNup302 under the control of an inducible tetracycline-regulated promoter. Under ATc treatment, a substantial decrease of TgNup302 protein in inducible knock-down (iKD) parasites was observed, causing a delay in parasite proliferation. Moreover, the nuclear protein TgENO2 was trapped in the cytoplasm of ATc-treated mutants, suggesting that TgNup302 is involved in nuclear transport. Fluorescence in situ hybridization revealed that TgNup302 is essential for 18S RNA export from the nucleus to the cytoplasm, while global mRNA export remains unchanged. Using an affinity tag purification combined with mass spectrometry, we identified additional components of the nuclear pore complex, including proteins potentially interacting with chromatin. Furthermore, reverse immunoprecipitation confirmed their interaction with TgNup302, and structured illuminated microscopy confirmed the NPC localization of some of the TgNup302-interacting proteins. Intriguingly, facilitates chromatin transcription complex (FACT) components were identified, suggesting the existence of an NPC-chromatin interaction in T. gondii. Identification of TgNup302-interacting proteins also provides the first glimpse at the NPC structure in Apicomplexa, suggesting a structural conservation of the NPC components between distant eukaryotes.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-017-2459-3) contains supplementary material, which is available to authorized users.
Keywords: Toxoplasma gondii, Nuclear pore complex, Nucleoporins, Gene expression, Apicomplexa
Introduction
Toxoplasma gondii is a unicellular eukaryotic pathogen. It belongs to the apicomplexan phylum, which encompasses some of the deadliest pathogens of medical and veterinary importance, including Plasmodium (the cause of malaria), Cryptosporidium (responsible for cryptosporidiosis), and Eimeria (coccidiosis). T. gondii is an obligate intracellular parasite that leads to the development of focal central nervous system infections in patients with HIV/AIDS. In addition, Toxoplasma is also a clinically important opportunistic pathogen that can cause birth defects in the offspring of newly infected mothers. The life cycle of T. gondii is complex, with multiple differentiation steps that are critical to parasite survival in human and feline hosts [1]. Although gene expression is tightly controlled in Apicomplexa, which is particularly evident during the cell cycle [2, 3], the molecular mechanisms underlying its regulation are poorly understood. The initial studies suggest that histone modifications and chromatin remodeling have an important role in chromatin structure for gene regulation [4, 5]. Eukaryotic nuclei are enclosed by the double-membrane nuclear envelope (NE), which is perforated by large protein structures termed nuclear pore complexes (NPCs), allowing a controlled bidirectional nucleocytoplasmic transport of macromolecules [6]. In eukaryotes, the organization and composition of NPCs have remained conserved throughout evolution, and they play an important role in other biological processes, such as chromosomal segregation mechanisms, mitotic spindle formation, transcription activation, and cytokinesis [7]. Each NPC is composed of multiple copies of approximately 30 different proteins known as nucleoporins (NUPs), for which the composition and structure have been largely characterized in Saccharomyces cerevisiae [8], mammals [9, 10], and Trypanosoma brucei [11, 12]. Strikingly, the description of the putative components of the NPC in T. brucei led to the discovery of a conserved protein arrangement that spans the eukaryotic kingdom [11]. Three conserved classes of NPC proteins were previously described in yeast and humans [6]: (1) membrane-bound nucleoporins, which link the nuclear pore to the NE; (2) core-scaffold NUPs, which are restricted at the structural level; and (3) NUPs that are distributed on the cytoplasmic or nucleoplasmic face of the NE. The three distinct structural arrangements found in eukaryotic core-scaffold NUPs (β-propeller fold, α-solenoid fold, and a mixture of both) were found to be conserved in T. brucei. Moreover, the sequence of some T. brucei core-scaffold NUPs also encompasses phenylalanine-glycine (FG) repeats, which is typical of NUPs [11]. The FG NUPs are known to regulate the transport of molecule through the nuclear pore by extending their FG repeats region into the channel [13]. These results point to a possible ancient inheritance of the structural components of the NPCs [11]. However, it is still unknown whether this apparent structural conservation between Opisthokonts (humans and yeast) and Discicristates (Trypanosoma) also holds true for other distant eukaryotes. Apicomplexan parasites (Alveolates) may provide another point of comparison between eukaryotic groups. However, although the dynamics of the nuclear pores were described during the cell cycle of the erythrocytic forms of P. falciparum [14] and for an individual component of the nuclear pore (PfSec13) [15], the composition of the P. falciparum NPC remains unknown. In T. gondii, the NPC has not been studied, and its components remain to be identified.
We characterized the NPC in T. gondii by identifying the protein complex associated with a conserved component of the core-scaffold NUPs. In this study, we show that TgNup302 is an essential protein that presents a typical perinuclear staining. In a conditional TgNup302 mutant, controlled bidirectional nuclear-cytoplasmic transport was severely impaired, and gene expression was also perturbed. Immunoprecipitation of proteins associated with TgNup302 provide further characterization of the NPC in T. gondii and indicate that structural conservation among eukaryotic NUPs is also true for Alveolates.
Materials and methods
Parasite tissue culture and manipulation
Toxoplasma gondii strain RHΔKu80 TaTi (a strain with ATc inducible system and high homologous recombination; [16]); tachyzoites were propagated in vitro in human foreskin fibroblasts (HFF) using Dulbeccos’s modified Eagles medium supplemented with 10% fetal calf serum (FCS), 2-mM glutamine, and 1% penicillin-streptomycin. T. gondii tachyzoites were grown in ventilated tissue culture flasks at 37 °C and 5% CO2. Transgenes were introduced by electroporation into tachyzoites of T. gondii strains and stable transformants were selected by culture in the presence of 2 µM pyrimethamine or chloramphenicol (34 mg/ml). Clonal lines were obtained by limiting dilution. Prior to RNA and protein purification, intracellular parasites were purified by sequential syringe passage with 17-gauge and 26-gauge needles and filtration through a 3-µm polycarbonate membrane filter.
Generation of transgenic T. gondii strains
The TgNup302 iKD line was generated using RH∆ku80TaTi and a plasmid containing genomic fragments encompassing 2-kb upstream the gene and 2 kb from the predicted ATG [16]. To produce myc-tagged TgNup115, TgNup134, TgNup129, TgFACT140, TgNup503, TgNup407, and TGGT1_311625 proteins by the knock-in strategy, a DNA fragment of 2-kb upstream of the stop codon from the genomic sequence was amplified from genomic DNA of ∆ku80 RH T. gondii (type I strain) and cloned in the pLIC-Myc-CAT plasmid. The plasmid (25 µg) was linearized with NheI, ApaI, NcoI, BstBI, NcoI, NsiI, and NarI, respectively, and transfected in 5.106 TgNup302 iKD tachyzoites followed by chloramphenicol selection. The sequences of all primers used in this study are listed in Supplementary Table 1.
Plaque assays and parasite growth assays
Plaque assays were performed using 6-well plates containing human fibroblast cells infected with 200 parasites per well in media with or without 1 µg/ml ATc, fixed after 7 days post-infection and labeled with a crystal violet solution.
For parasite growth assays, 8 × 105 parasites per well in a 24-well plate were incubated 4 h in normal media or media with ATc. After coverslips were incubated for 24, 48, or 72 h, fixation and staining were carried out using formaldehyde and an antibody directed against the TgENO2. The number of parasites per vacuole was counted for 100 vacuoles per condition.
Antibodies
The anti-TgENO2 rabbit [17], anti-TgChromo1 mouse [18], and anti-TgNF3 [19] mouse antibodies were used at 1:1000 and 1:200 respectively. Anti-HA rabbit (Eurogentech) and rat (Invitrogen) antibodies were used at 1:500 in IFA and in Western blots. Anti-Myc mouse (ThermoFisher) was used at 1:200 in IFA and 1:500 in Western blot.
Immunofluorescence assay, confocal imaging, and morphology Microscopy
Intracellular and extracellular parasites tachyzoites were fixed with 4% paraformaldehyde in PBS for 15 min, followed by two PBS washes. Extracellular parasites were dried on Teflon slides. Parasites were permeabilized with 0.1% Triton X-100 in PBS containing 0.1% glycine for 10 min at room temperature. Samples were blocked with FCS in the same buffer and the primary antibodies were added on parasites in the same buffer for 1h at room temperature. Secondary antibody coupled to Alexa-488 or to Alexa-594 (Molecular probes) diluted at 1:1000 was added in addition to DAPI for nucleus staining. Confocal imaging was performed with an LSM880 microscope (Zeiss) and a Plan Apochromat objective (Plan Apochromat 63×:1.40 Oil DIC M27, Zeiss). Ultrastructural morphology was performed using the conventional microscopy [19], except that 8% paraformaldehyde containing 0.01% glutaraldehyde was used for cryo-IEM.
Structured illumination microscopy (SIM)
SIM was used to obtain high-resolution images using an ElyraPS1 microscope system (Zeiss) with a 100x oil-immersion lens (alpha Plan Apochromat 100×, NA 1.46, oil immersion) and a resolution of 120 nm along the x–y-axis and 500 nm along the z-axis (PSF measured on 100 nm beads; Sampling voxel size: 0.050 µm × 0.050 µm × 0.150 µm). Three lasers (405, 488, and 561 nm) were used for excitation. SIM images were acquired with an EMCCD camera (Andor Technology Ltd., UK) and processed with the ZEN software; exposure times varied between 100 and 120 ms. Three-dimensional images were generated using a z-step of 150 nm (total thickness ~5 µm), while reconstructions and co-distributions were determined with the IMARIS software (Pearson’s coefficient). We determined the same A (red-HA signal) and B (green-Myc signal) thresholds for all parasite strains. The acquisition was performed sequentially using Zeiss Filter Sets 43HE, 38HE, and BP 420–480. Fifteen frames were acquired to reconstruct one image (five rotations × three phases, with an SIM grating period of 51 µm for the blue channel, 42 µm for the green channel, and 34 µm for the red channel). One hundred nanometer beads were imaged to measure the chromatic misalignment of our system (fit procedure by the Zen software); these parameters enabled us to further correct the alignment on each acquired multi-channel stack.
RNA FISH
Intracellular parasites were treated with or without ATc for 48 h and purified as described earlier. The parasites were fixed with 4% paraformaldehyde for 30 min and settled on RNA-treated slides as previously described [20]. Cy3-oligo d(T)40 and Cy3-18S RNA were hybridized on the slide at 50 °C overnight and washed as previously described [20, 21]. After 10-min DAPI staining, the slides were mounted on coverslips.
Cellular fractionation and Western blot for reverse immunoprecipitation
Intracellular parasites (5 × 108 tachyzoites) of the TgNup302-HA ΔKu80RHTaTi strain were purified on a 3-mm filter and washed twice with PBS. The parasite pellet was resuspended in 1 mL of NEB1 buffer (10-mM HEPES pH7.9, 1.5-mM MgCl2, 10-mM KCl, 0.5-mM DTT, 0.1-mM EDTA, 0.65% NP40, and 0.5-mM PMSF), incubated on ice for 10 min and centrifuged at 1500g for 10 min at 4 °C. The supernatant was kept as the cytoplasmic extract. The pellet was then resuspended with 100 µl of buffer NEB2 (20-mM HEPES pH 7.9, 1.5-mM MgCl2, 420-mM NaCl, 0.2-mM EDTA, 0.5-mM DTT, 25% glycerol, and 0.2-mM PMSF) for 10 min on ice and centrifuged at 12,000g for 10 min at 4 °C.
The supernatant was kept as the nuclear extract. The insoluble material was extracted using an SDS buffer (2% SDS, 10-mM Tris, and 0.2-mM PMSF) for 20 min at room temperature and centrifuged at 12,000g for 10 min, and the supernatant was kept. Then, 30 µl of Pierce anti-c-myc-tag beads were washed twice with 1X TBS (50-mM Tris-HCl, 150-mM NaCl, and 0.5-mM PMSF) with centrifugation of 4000g for 1 min between each wash. Saturation of the beads was established with 500 µL of 1X TBS and 5 µL of BSA (1 mg/mL) under stirring for 15 min at 4 °C, and then, the beads were washed twice as previously. Nuclear extract and insoluble material are added to the anti-c-Myc beads (the volume was extended to approximately 900 µl of 1X TBS to dilute the salt concentration) and incubated overnight at 4 °C under stirring. The next day, the beads were washed five times with 1X TBS-T (1X TBS, Tween 20% and 0.5 mM PMSF) and one time with 62.5-mM Tris pH 6.8 + PMSF, with centrifugation at 4000g for 1 min between each wash. Finally, the immunoprecipitated proteins from the beads were eluted with 1X DTT (Tris 0.5-M pH 6.8, SDS 20%, saccharose, DTT 1 M), warmed at 95 °C for 5 min and centrifuged at 14,000 rpm for 1 min. The supernatants were kept to perform a Western blot.
gRNA CRISPR/Cas9 screening
To generate guide RNAs (gRNAs) to disrupt specific TgNup115, TgNup302, TgNup593, TgSec13, TgNup530, TgNup37, TgNup68, TgNup216, TgNup67, TgFACT140, TGGT1_228100, TgNup129, TgNup134, TgNup407, TgNup503, and TGGT1_311625 genes, we modified the CRISPR Cas9-gRNA plasmid (pSAG1::Cas9-U6::sgUPRT plasmid) as previously described [22]. To study the impact of targeted mutations at a specific locus, RH Δhxgprt parasites (10.106) were transfected with 100 µg of CRISPR plasmids generated previously by electroporation. To estimate the frequency of CRISPR/CAS9-mediated gene disruptions, 30 µL of transfection reagent was added to HFF monolayers and analyzed by immunofluorescence staining for GFP 24 h post-transfection.
Mass-spectrometry proteomic analysis
After denaturation at 100 °C in 5% SDS, 5% β-mercaptoethanol, 1-mM EDTA, 10% glycerol, and 10-mM Tris pH 8 buffer for 3 min, protein samples were fractionated on a 10% acrylamide SDS-PAGE gel. The electrophoretic migration was stopped as soon as the protein sample entered 1 cm into the separating gel. The gel was briefly labeled with Coomassie Blue, and five bands, containing the whole sample, were cut. In gel, digestion of gel slices was performed as previously described [23].
An UltiMate 3000 RSLCnano System (Thermo Fisher Scientific) was used for separation of the protein digests. Peptides were automatically fractionated onto a commercial C18 reversed phase column (75 µm × 150 mm, 2-µm particle, PepMap100 RSLC column, Thermo Fisher Scientific, temperature 35 °C). Trapping was performed during 4 min at 5 μL/min, with solvent A (98% H2O, 2% ACN and 0.1% FA). Elution was performed using two solvents A (0,1% FA in water) and B (0.1% FA in ACN) at a flow rate of 300 nL/min. Gradient separation was 3 min at 5% B, 37 min from 5% B to 30% B, 5 min to 80% B, and maintained for 5 min. The column was equilibrated for 10 min with 5% buffer B prior to the next sample analysis.
The eluted peptides from the C18 column were analyzed by Q-Exactive instruments (Thermo Fisher Scientific). The electrospray voltage was 1.9 kV, and the capillary temperature was 275 °C. Full MS scans were acquired in the Orbitrap mass analyzer over m/z 300–1200 range with resolution 35,000 (m/z 200). The target value was 5.00E + 05. Ten most intense peaks with charge state between 2 and 4 were fragmented in the HCD collision cell with normalized collision energy of 27%, and tandem mass spectrum was acquired in the Orbitrap mass analyzer with resolution 17,500 at m/z 200. The target value was 1.00E+05. The ion selection threshold was 5.0E+04 counts, and the maximum allowed ion accumulation times were 250 ms for full MS scans and 100 ms for tandem mass spectrum. Dynamic exclusion was set to 30 s.
Proteomic data analysis
Raw data collected during nanoLC-MS/MS analyses were processed and converted into *.mgf peak list format with Proteome Discoverer 1.4 (Thermo Fisher Scientific). MS/MS data were interpreted using search engine Mascot (version 2.4.0, Matrix Science, London, UK) installed on a local server. Searches were performed with a tolerance on mass measurement of 0.2 Da for precursor and 0.2 Da for fragment ions, against a composite target decoy database (50,620 total entries) built with three strains of Toxoplasma gondii ToxoDB.org database (strains ME49, GT1, and VEG, release 12.0, September 2014, 25,264 entries) fused with the sequences of recombinant trypsin and a list of classical contaminants (46 entries). Cysteine carbamidomethylation, methionine oxidation, protein N-terminal acetylation, and cysteine propionamidation were searched as variable modifications. Up to one trypsin, missed cleavage was allowed. For each sample, peptides were filtered out according to the cut-off set for proteins hits with two or more peptides taller than seven residues, ion score >25, identity score >0, and no false positive identification.
Library preparation and RNA-Seq
RNA was extracted using Invitrogen Trizol Reagent (cat #15596018), followed by genomic DNA removal and cleaning using an RNase-free DNase. An Agilent 2100 Bioanalyzer was used to assess the integrity of the RNA samples. Only RNA samples having an RNA integrity score between 8 and 10 were used. Library preparation was performed using the TruSeq Stranded mRNA Sample Preparation kit (Illumina) according to the manufacturer’s instructions. Libraries were validated using a Fragment Analyzer and quantified by qPCR (ROCHE LightCycler 480). Clusters were generated on a flow cell within a cBot using the Cluster Generation Kit (Illumina), and libraries were sequenced as 50-bp reads on a HiSeq 2000 using a Sequence By Synthesis (SBS) technique (Illumina).
Image analysis and base calling were performed using the HiSeq Control Software and Real-Time Analysis component. Demultiplexing was performed using Illumina’s conversion software (bcl2fastq 2.17). The quality of the data was assessed using FastQC from the Babraham Institute and the Illumina software Sequence Analysis Viewer (SAV). Potential contaminants were investigated with the FastQ Screen software from the Babraham Institute.
Contamination by Escherichia coli sequences was evident in the sequenced samples. This contamination, due to a reagent from Life Technologies (SuperScript II enzyme), has been eliminated by aligning sequences (using BWA v0.7.12-r1039 [24]) to the E. coli genome and keeping the unmapped reads for downstream analysis.
RNA-seq reads were aligned to the Toxoplasma gondii genome (ToxoDB-25_TgondiiGT1_Genome.fasta from the ToxoDB Toxoplasma Genomics Resource, downloaded on 08/31/15) with a set of gene model annotations (ToxoDB-25_TgondiiGT1.gff from the ToxoDB Toxoplasma Genomics Resource, downloaded on 08/31/15) using the splice junction mapper TopHat 2.0.13 [25] (with bowtie 2.2.3 [26]). Final read alignments having more than three mismatches were discarded. Gene counting was performed using HTSeq-count 0.6.1p1 (union mode) [27]. Because the data come from a strand-specific assay, the read must be mapped to the opposite strand of the gene. Before statistical analysis, genes with less than 15 reads (combining all the analyzed samples) were filtered out. edgeR differentially expressed genes were identified using the Bioconductor [28] package edgeR [29] 3.6.7. The data were normalized using the Relative Log Expression (RLE) [30] normalization factors. Genes with adjusted p-values less than 5% (according to the FDR method from Benjamini-Hochberg) were declared differentially expressed.
Quantitative real-time PCR
All primers were designed online using Primer2 v.0.4.0 and are listed in Table S1; the cDNA samples were synthesized from total RNA samples using the RevertAid First Strand cDNA Synthesis Kit (Fermentas). qRT-PCR was carried out on an Mx3000P system (Agilent Technologies). Individual reactions were prepared with 0.5 µM of each primer, ~5 ng of cDNA and SYBR Green PCR Master Mix (Applied Biosystems, CA) to a final volume of 20 µl. All experiments were performed twice with separate biological replicates. For each experiment, reactions were performed in triplicate, and the expression of individual genes was normalized to the housekeeping tubulin gene Ct values.
Quantifications
The differences of localization for TgNup115, TgNup134, TgNup407, and TgNup503 proteins with or without ATc treatment were calculated with the Image J Software. We designed that a Macro which after manually determining the parasite edge (polygon tool and manual drawing on the phase contrast image) and the nucleus edge (threshold based on DAPI signal) is able to determine the quantity of Myc-tagged signal (in green) in the nucleus and the cytoplasm of parasites for each strain.
For each parasite, we measured the area in µm² of regions of interest (nucleus and cytoplasm) and the integrated and mean intensities in gray levels in each region. With the different values, we calculated the ratio cytoplasm signal versus the nuclear signal with or without ATc treatment for each strain. This ratio corresponds to the integrated intensity for the cytoplasm versus integrated intensity for the nucleus.
Statistics
A non-parametric Student’s T test was performed where statistical analysis was required.
Results
TgNup302 is crucial for T. gondii growth
Computational searching through the Toxoplasma genome database (toxodb.org) using the human Nup98/96 protein as a template revealed the existence of a protein (TGGT1_259640) that is conserved in most eukaryotes from yeast to mammals as an integral component of the NPCs. We constructed a phylogenetic tree, including some members of the Apicomplexa, Fungi, Plantae, and Eumetazoa, where there is a level of characterization of the NPC proteome and some direct experimental information [8, 10, 11, 31, 32]. The Nup98/96 homologs from the Apicomplexa taxon, joining representative branch species, such as Toxoplasma, Neospora, Eimeria, Theileria, and Babesia, are relatively close to Eumetazoa, followed by Plantae. Surprisingly, members of the Plasmodium species, which belong to the Apicomplexa phylum as Toxoplasma, present a distant branching taxon separated from the Apicomplexa (Fig. S1). The TgNup302 gene (TGGT1_259640) is predicted to produce a long 2894-amino-acid protein containing an autocleavage domain (Pfam domain: PF04096) that may produce two distinct N-terminal (1079 aa) and C-terminal (1620 aa) peptides after self-cleavage. The presence of GLFG repeats, a typical variant of the FG repeats present in the human Nup98 protein, at its N-terminus and a Nup96-like domain (PF12110) identified by the Pfam database at its C-terminus is typical of this protein family. This structure is conserved among eukaryotes, with Nup98/96 and NUP145 being the representative proteins in humans and yeast, respectively. In vertebrates, Nup98 binds directly to Nup96, the C-terminal half of the proteolytically processed Nup98/Nup96 polyprotein, and is a component of the Nup107-160 complex (the Nup84 complex in yeast) [9, 33]. However, the size of the unprocessed T. gondii protein (302 kDa) is much larger than in yeast (145 kDa) and in humans (194 kDa).
To better characterize the biological role of this protein, we used a promoter replacement strategy to produce a conditional knock-down (iKD) mutant strain. In this strain, the expression of the TgNup302 transcript is under the control of anhydrotetracycline (ATc) (Fig. S2) [16]. When ATc is added to the culture media, the regulatable promoter is repressed, and the TgNup302 transcript is no longer produced. We simultaneously added an HA-tag to the N-terminus of the protein to follow the protein expression (Fig. S2). Because the human and yeast homologous proteins undergo a self-proteolytic cleavage, producing two polypeptides, we also tagged the TgNup302 protein with a Myc tag at its C-terminus (Fig. S2) in the iKD strain. Therefore, we produced a strain expressing TgNup302 under the control of ATc and tagged at its N- and C-termini. The correct insertion of the construction at the TgNup302 locus was validated by genomic PCR (Fig.S2). Using anti-HA (α-HA) and anti-Myc (α-Myc) monoclonal antibodies by immunofluorescence assay (IFA), we observed that TgNup302 presented a punctate perinuclear pattern in the parasite, consistent with nuclear pore staining (Fig. 1a). Under ATc treatment, a substantial decrease in TgNup302 protein expression in iKD parasites was observed for both HA- and Myc-tagged proteins. Western blots of total protein extracts from this transgenic parasite line revealed the expression of the two polypeptides tagged with HA- or Myc-tags of the predicted size (150 and 170 kDa, respectively) (Fig. 1b) in the absence of ATc indicating the cleavage of the TgNup302 protein into two polypeptides. Notably, TbNup158, the T. brucei homologue of TgNup302, does not perform self-cleavage and remains as one polypeptide [11] and, therefore, is closer to the yeast Nup116 or Nup100 protein.
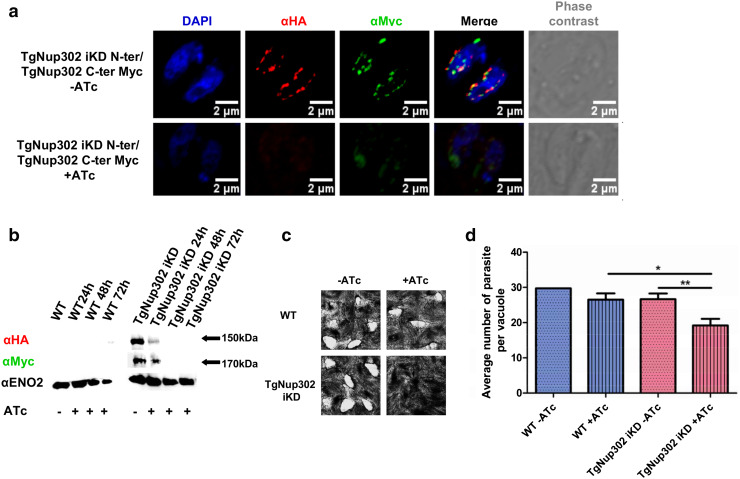
Fig. 1.
Conditional knock-down of TgNup302 and phenotypic analysis. a Localization of the TgNup302 protein in the conditional knock-down strain (tagged at its N- and C-termini) by confocal imaging using anti-HA (N-ter) and anti-Myc (C-ter) monoclonal antibodies under normal conditions (top) and after 48-hr ATc treatment (bottom). b TgNup302 double-tagged protein expression decreases under ATc treatment in the conditional knock-down strain. Immunoblots are shown for total protein extracts from the wild-type and TgNup302 iKD strain in the presence or absence of ATc for 24, 48, and 72 h. Western blots were probed with anti-HA (N-ter) and anti-Myc (C-ter) antibodies. TgENO2 was probed as a loading control. c Appropriate level of TgNup302 expression is essential for parasite growth. Plaque assays were performed with 200 parasites of wild-type and TgNup302 iKD strains with or without ATc treatment for 7 days. The wild-type strain growth was not affected by ATc treatment (top). In contrast, the growth of the conditional TgNup302 knock-down strain was dramatically impaired in the presence of ATc. d Growth assay. Parasites from the parental (blue bars) or TgNup302 iKD (red bars) strains were incubated with (crossed bars) or without ATc (dotted bars) for 48 h. The number of parasites per vacuole was scored for a minimum of 100 vacuoles. The average number of parasite is represented in this graph. The results shown are from three independent experiments. *P < 0.05; **P < 0.001
In the presence of ATc, we observed a drastic reduction in the signal from both tagged proteins, with an undetectable level after 48 h of treatment (Fig. 1b). As a first assessment of the requirement for the TgNup302 locus, we tested the ability of parasites to grow and produce plaques on host cell monolayers. After 7 days, the wild-type parasites grew normally and developed equal sized plaques in the absence or presence of ATc. Similarly, the growth of the iKD line gave rise to normal plaque numbers only in the absence of ATc. In the presence of ATc, the TgNup302-deficient parasites were unable to proliferate and form plaques (Fig. 1c), indicating a drastic impairment of growth and/or invasion of the parasites lacking the expression of TgNup302. This result was confirmed by an experiment recording the number of parasites per vacuole at a given time (Fig. 1d). While the parental strain with or without ATc showed similar growth to that of the iKD strain without ATc for 48 h, we noticed that the iKD-HA strain produced vacuoles with a smaller number of parasites. This indicates that the growth of this parasite is impaired by the absence of TgNup302 (Fig. 1d).
TgNup302 is involved in nuclear import and essential to 18S RNA export
To validate the role of TgNup302 in the nuclear import and export of macromolecular complexes, we examined the localization of a nuclear marker in the iKD strain. The previous work has established the predominant nuclear location of the glycolytic isoenzyme enolase 2 (TgENO2) [34]. We tested the localization of the TgENO2 protein by IFA using a specific antibody (α-TgENO2) in the presence and absence of ATc for 48 h. We observed predominant nuclear localization of the protein in the parental strain and in the iKD strain in the absence of ATc, as expected (Fig. 2a). However, parasites of the iKD strain grown in the presence of ATc showed a marked delocalization of TgENO2 into the cytoplasm of the parasite (Fig. 2a). This result was confirmed after quantification of the parasites which showed the nuclear or cytoplasmic localization of TgENO2 (Fig. 2b). While the iKD strain without ATc and the parental parasites exhibited predominant nuclear localization of TgENO2, most of the ATc-treated iKD parasites exhibited cytoplasmic localization, indicating a defect in the transport of this protein in the mutant parasites.
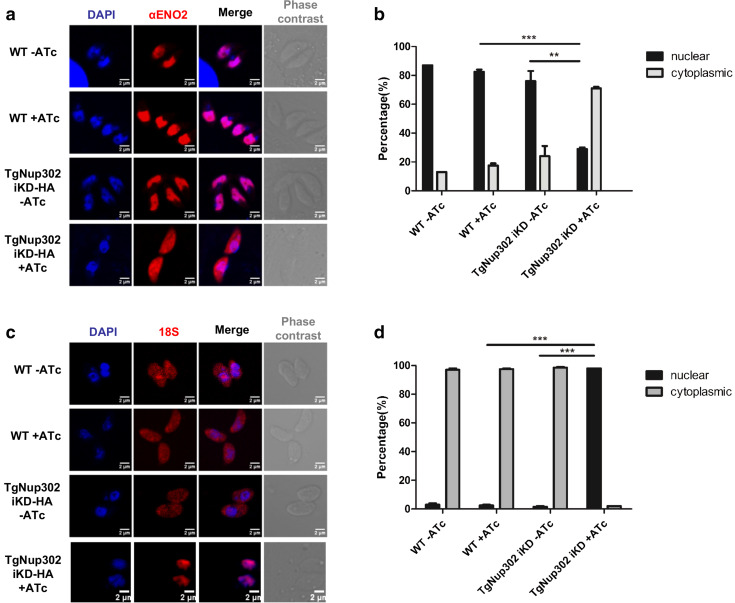
Fig. 2.
TgNup302 is involved in nuclear import and is essential for 18S RNA export. a TgNup302 is involved in the nuclear import of the TgENO2 protein. IFA was performed on paraformaldehyde-fixed intracellular parasites of the wild-type and TgNup302 iKD strains using a TgENO2 antibody (α-TgENO2) and staining of the nuclear DNA with DAPI. In the presence of ATc for 48 h, the TgENO2 protein is mislocalized in the cytoplasm of TgNup302 iKD, whereas normal nuclear localization was observed in the absence of ATc for the parental and TgNup302 iKD strains, as expected. b TgENO2 localization in intracellular parasites. The percentage of parasites displaying nuclear and cytoplasmic TgENO2 signal was determined for 100 vacuoles in the parental and TgNup302 iKD strains with or without ATc for 48 h after IFA using a α-TgENO2 staining. Predominant nuclear localization of TgENO2 was observed for the iKD strain without ATc and the parental parasites, whereas predominant cytoplasmic localization was observed for ATc-treated iKD parasites. The results shown are from three independent experiments. **P < 0.05; ***P < 0.0001. c TgNup302 is essential to 18S RNA export. RNA FISH was performed on intracellular parasites treated with or without ATc for 48 h. Parasites of the parental and TgNup302 iKD strains were hybridized with Cy3-labeled 18S oligonucleotide primers (red), and the nuclear DNA was labeled with DAPI (blue). d 18S RNA localization in extracellular parasites. The quantity of nuclear and cytoplasmic Cy3-labeled 18S signal for 100 parasites was determined for the parental and TgNup302 iKD strains with or without ATc for 48 h. In the parental strain and iKD strains without ATc treatment, most 18S RNA is cytoplasmic, while it is mainly nuclear in the iKD parasites treated with ATc. The results shown are from three independent experiments. ***P < 0.0001
To investigate whether TgNup302 is involved in mRNA export, we examined the cellular distribution of poly(A) + RNA in the parental and TgNup302 iKD strains via RNA fluorescence in situ hybridization (FISH) with labeled oligodT after 48 h with or without ATc treatment. In the parental and iKD strains, localization of the polyA + RNA was mostly cytoplasmic (Fig. S3). We also examined the localization of the 18S ribosomal RNA using a specific probe. In the wild-type and iKD strains without 48 h of ATc treatment, most 18S RNA was found to be cytoplasmic (Fig. 2c). In contrast, the 18S RNA signal was mainly nuclear in the iKD parasites treated with ATc (Fig. 2c, d). These results demonstrate that TgNup302 is essential to 18S RNA export, a typical role of the NPC. To test whether TgNup302 was involved in other proteins nuclear import, we used IFA of the parental and iKD-HA strains to evaluate the localization of a centromeric protein (TgChromo1) that was previously described to specifically bind pericentromeric heterochromatin [18]. We also tested the localization of a nucleolar marker (TgNF3). No differences in the localization of these proteins were observed between the parental and iKD-HA strains in all conditions tested (with or without ATc for 48 h) (Fig. S4).
The morphology of the parasite was assessed using electron microscopy for parental and TgNup302 iKD intracellular parasites following 48 h of growth with or without ATc treatments. No morphological differences were observed between the parental and TgNup302 iKD strains, which present a typical nucleus. ATc treatment had also no particular impact on the presence of other organelles, such as rhoptries and micronemes, for each strain, suggesting that TgNup302 is not essential for nuclear morphology (Fig. S5).
Deep sequencing suggests a role for TgNup302 in gene expression
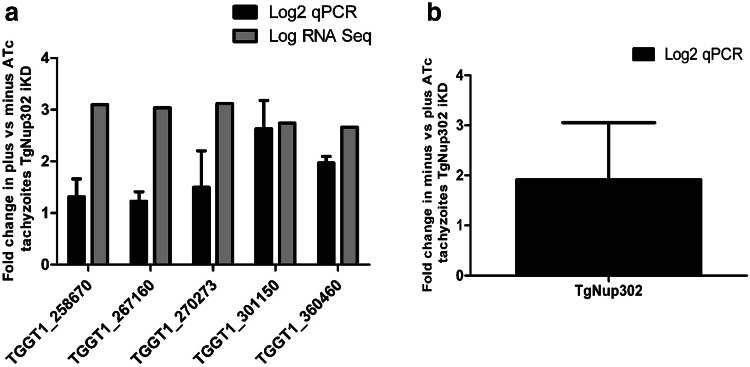
RNA-sequencing (RNA-Seq) was carried out to study gene expression dynamics in the TgNup302 iKD. Total RNA was purified from tachyzoites of the TgNup302 iKD strain grown under normal growth conditions with or without ATc treatment for 48 h (in triplicate). RNA-seq reads were aligned to the Toxoplasma gondii genome with a set of gene model annotations using the splice junction mapper TopHat 2.0.13. The data were normalized using the relative log expression (RLE) normalization factors. Genes with adjusted p-values less than 5% (according to the FDR method from Benjamini-Hochberg) were considered differentially expressed. Data analysis revealed significant changes in the transcription profile of T. gondii, with 145 genes upregulated and 65 genes downregulated (Table S2). The predominance of upregulated genes suggests a role for the NPC in the repression of gene expression. We examined the differential chromosomal distribution of differentially expressed genes and did not identify a cluster of differentially regulated genes (Fig. S6). Collectively, these data suggest that TgNup302 could play a role as a repressor and its absence modulates the transcriptional regulation of genes that are normally repressed at the tachyzoite stage of T. gondii. Validation of the RNA-Seq data was also carried out using qRT-PCR in RNA samples from the iKD TgNup302 strain with and without ATc treatment for 48 h. For that, we selected five upregulated genes with a logFC>-2 (TGGT1_258670, TGGT1_301150, TGGT1_267160, TGGT1_270273, TGGT1_360460) (Fig. 3a), and TgNup302 (TGGT1_259640) (Fig. 3b) as a control. As expected, Fig. 3a clearly shows that ATc treatment induced an increased steady-state mRNA level of all five upregulated genes, while the TgNup302 transcript was downregulated in the iKD samples treated with ATc.
Fig. 3.
TgNup302 depletion alters gene expression. a Total RNA purified from iKD TgNup302 parasites under either control conditions or ATc treatment for 48 h were analyzed by quantitative RT-PCR (dark bars). Genes coding for the transcripts identified to be upregulated with ATc treatment in RNA-Seq were analyzed. Values are presented as log2 ratios of the signal given by the sample extracted from the ATc-treated parasites relative to those under control conditions (minus ATc). b Total RNA purified from iKD TgNup302 parasites under either control conditions or ATc treatment for 48 h were analyzed by quantitative RT-PCR. The TgNup302 gene was analyzed alongside the TgTubulin gene, which was not affected by ATc treatment. Values are presented as the Log2 ratio of ATc-treated parasites relative to those under control conditions (minus ATc)
New components of the T. gondii nuclear pore
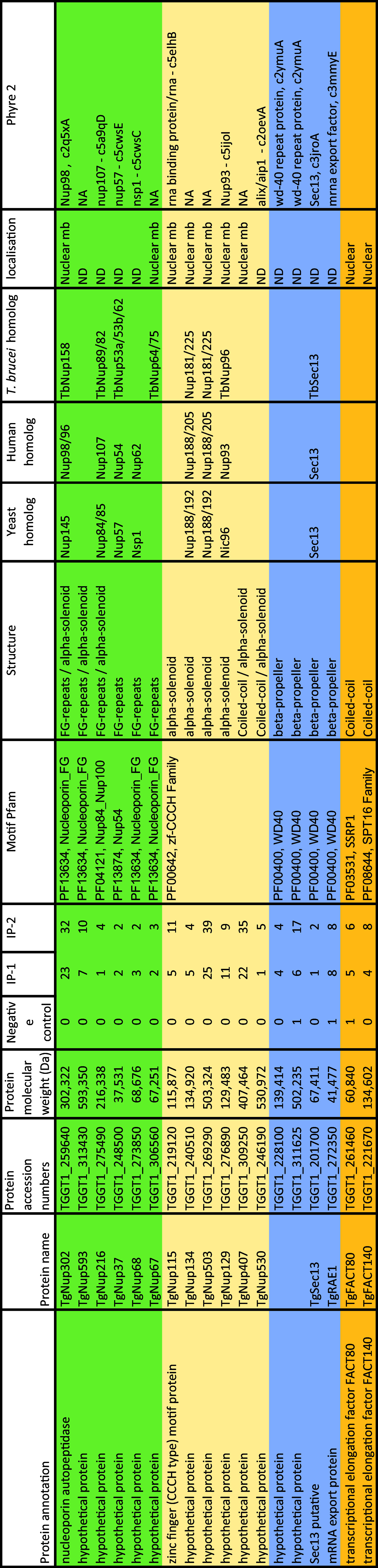
NPC components are unknown in T. gondii. To uncover the components of the NPC in T. gondii, we performed co-immunoprecipitation (co-IP) experiments on the iKD Nup302-HA parasites using α-HA antibody followed by mass-spectrometry identification of the co-IP proteins (from two experiments). As a control, we performed an α-HA immunoprecipitation on protein extracts from the parental strain, RH∆Ku80TaTi. We selected the proteins identified in both experiments but not identified in the control and those present in the control with a single peptide (Table S3 and Table 1). Among the proteins identified and listed in Table 1, we confirmed the presence of TgNup302 (TGGT1_259640) with peptides spanning both the N-terminus and the C-terminus (Fig. S7), indicating that both products of TgNup302 autocleavage are present in the IP fraction. As shown for the human Nup98/Nup96 protein [35], both polypeptides may also interact in T. gondii.
Table 1.
Identification of proteins associated with the TgNup302 protein
Identification of proteins that were copurified from nuclear extract from the parental and TgNup302 iKD N-ter-HA Tag [in duplicate (1) and (2)] strains using nano LC-MS/MS. The affinity purification was performed under high stringency conditions and protein identity was determined using a composite target decoy database (50,620 total entries) built with three strains of Toxoplasma gondii ToxoDB.org database (strains ME49, GT1 and VEG, release 12.0, September 2014, 25,264 entries). Potential TgNup302 interacting proteins were identified and sorted by their secondary structure. FG repeats containing proteins are highlighted in green, alpha-solenoid protein in orange, beta-propeller in blue, and coiled–coiled domains in yellow
ND not determined
Most of the identified proteins did not share primary sequence conservation with known nucleoporins using BLAST searches. However, a conserved structure was identified between distant eukaryotic NUPs [11], we, therefore, examined the secondary structure prediction of the selected proteins (Table 1, Fig. S8). We also searched the proteins for other motifs using the Pfam database. Among the co-immunoprecipitated proteins, five putative NUPs encompassing FG-repeat motifs (TgNup593, TgNup37, TgNup68, TgNup216, and TgNup67) were identified. We also found proteins with a β-propeller fold (TgSec13, TGGT1_311625, and TGGT1_228100), including a homolog of the yeast protein Sec13, shown to be part of the Y complex through its interaction with Nup96 [36]. Interestingly, six of the TgNup302-associated proteins (TgNup530, TgNup407, TgNup129, TgNup503, TgNup115, and TgNup134) had no recognizable Pfam motif, with the exception of TgNup115, which contains a Zinc-finger domain (CCCH). However, these proteins do contain an α-solenoid fold, a structure also found in other eukaryotic core-scaffold NUPs (Fig. S8). Moreover, two of these proteins exhibited large segments of coiled-coil structure (TgNup530, TgNup407) (Fig. S8). We further examined the sequence of these proteins using a secondary structure prediction software [37]. This allowed the identification of a domain in TgNup68 with a strong homology to the yeast Nsp1 crystal structure (Table 1). Similarly, a domain in TgNup129 was homologous to the yeast Nic96 structure (Table 1). These data indicate that the proteins composing the T. gondii NPC may have retained the same level of structural conservation as T. brucei NUPs. Notably, the size of the identified proteins substantially exceeded the size of their putative homologs. Therefore, the assigned homologs listed in Table 1 are based on the structural properties of the proteins rather than their expected size.
As expected, TgNup302 is also associated with the putative homolog of the ribonucleic acid export 1 protein (TgRae1, TGGT1_272350), which is implicated in the mRNA export pathway in other eukaryotes, which also interact with the GLFG Nup98 protein [38]. Surprisingly, we also found the two proteins (TgFACT140 and TgFACT80) corresponding to the homologs of the proteins composing the FACT complex, which is categorized as a histone chaperone critical to nucleosome reorganization during replication and transcription [25–27]. This observation indicates a potential interaction between the FACT complex and the NPC in T. gondii.
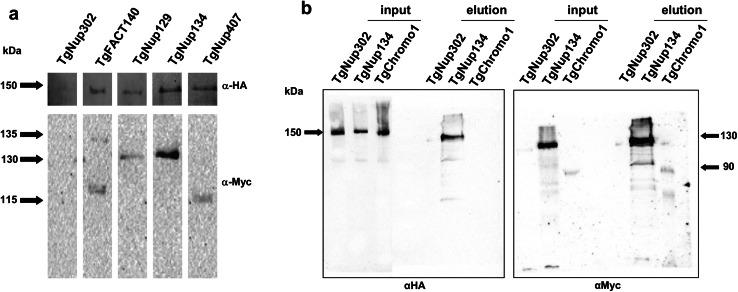
Identified proteins interact with TgNup302
To confirm the mass-spectrometry results, we performed a reverse IP of TgNup302 using the identified partner proteins. Protein extracts from parasite strains expressing each Myc-tagged protein in the TgNup302-HA iKD background were immunoprecipitated using anti-myc-tag beads. The TgNup302-HA iKD strain was used as a negative control. We then performed a Western blot using an anti-HA antibody to detect TgNup302 in the eluates of the Myc-tagged immunoprecipitates (Fig. 4). As shown in Fig. 4a, the anti-Myc antibody failed to immunoprecipitate the HA-tagged TgNup302 in the absence of Myc-tagged proteins, as expected. In contrast, we were able to confirm the interaction between TgNup302 and the TgFACT140 protein, indicating a link between the NPC and the FACT complex in T. gondii (Fig. 4a, line 2, top panel). We tested whether some of the unknown proteins with an α-solenoid fold listed in Table 1 were also able to co-immunoprecipitate TgNup302. We confirmed the presence of the TgNup302 protein in the TgNup129 co-immunoprecipitated proteins (Fig. 4a, line 3). Similarly, TgNup302 was detected after immunoprecipitation of TgNup134 and TgNup407, indicating that these proteins of unknown function may participate in the NPC (Fig. 4a, lines 4 and 5, top panel). Western blot membranes were probed with an anti-Myc antibody to detect specific Myc-tagged proteins (Fig. 4a, bottom panel). Using the anti-Myc antibody, the immunoprecipitated proteins extracted from the TgNup302 iKD parasite line showed no signal, as expected (Fig. 4a, line 1, bottom panel). We confirmed the presence of each myc-tagged protein at the expected molecular weights for TgFACT140 (135 kDa), for TgNup129 and TgNup134 (130 kDa) and for TgNup407 (115 kDa). We verified that the level of the TgNup302 protein was similar in the starting material of each strain (Figure S9a). Similarly, the presence of the myc-tagged proteins was checked for the same input samples (Figure S9b). As a control, we repeated this experiment using a strain expressing TgNup302-HA and a myc-tagged protein (TgChromo1) that was not identified by mass-spectrometry (Fig. 4b). After immunoprecipitation with anti-myc coated beads, we performed a Western blot using an anti-HA antibody to detect TgNup302 in the input and eluates (Fig. 4b left panel). We were able to confirm the interaction between TgNup302 and the TgNup134 protein (Fig. 4b, line 2, left panel). As expected, TgNup302 is not detected in the negative control as well as in the TgChromo1 co-immunoprecipitated proteins (Fig. 4b, lines 4 and 6 left panel) but is present in the input (Fig. 4b, lines 1, 2, 3, left panel). Western blot membranes were probed with an anti-Myc antibody to detect specific Myc-tagged proteins in the input and eluates (Fig. 4b, right panel). Using the anti-Myc antibody, the immunoprecipitated proteins extracted from the TgNup302 iKD parasite line showed no signal, as expected (Fig. 4b, line 1 and 4, right panel). We confirmed the presence of each myc-tagged protein in the eluates for TgChromo1 (Fig. 4b, line 6, right panel) and TgNup134 (Fig. 4b, line 5, right panel), confirming that the myc-tagged proteins were, indeed, immunoprecipitated.
Fig. 4.
Identified proteins interact with TgNup302. A Nuclear extract of ~500 × 106 parasites for the TgFACT140 (line 2), TgNup129 (line 3), TgNup134 (line 4), and TgNup407 (line 5) C-terminally myc-tagged proteins in the TgNup302-HA iKD strain was immunoprecipitated using anti-myc antibodies. Extracts from the TgNup302 HA iKD strain were immunoprecipitated using anti-myc antibodies and used as negative controls. The immunoblot was probed with anti-HA antibody (upper panel) to detect interactions between the N-terminal domain of TgNup302 and the five different proteins. The same blots were reprobed with anti-myc antibodies to verify that the myc-tagged proteins were, indeed, immunoprecipitated (bottom panel). The TgFACT140, TgNup129, TgNup134, TgNup407 C-terminally myc-tagged proteins in the TgNup302-HA iKD strain, and the TgNup302 HA iKD strain related inputs (produced from the same experiment) are shown in figure S9. b Nuclear extract of ~300 × 106 parasites for the TgNup134 (positive control) and TgChromo1 (negative control) C-terminally myc-tagged proteins in the TgNup302-HA iKD strain were immunoprecipitated using anti-Myc antibodies. Extracts from the TgNup302 HA iKD strain were immunoprecipitated using anti-Myc antibodies and used as negative controls. The immunoblot was probed with anti-HA (left panel) antibody to detect the TgNup302-HA protein in the input and eluates. The immunoblot was reprobed with an anti-myc antibody (right panel) to detect the myc-tagged proteins in the input and eluates
These results confirm the interaction between the NPC protein TgNup302 and the selected binding partners identified by mass-spectrometry.
Identified proteins localize to the T. gondii nuclear pore
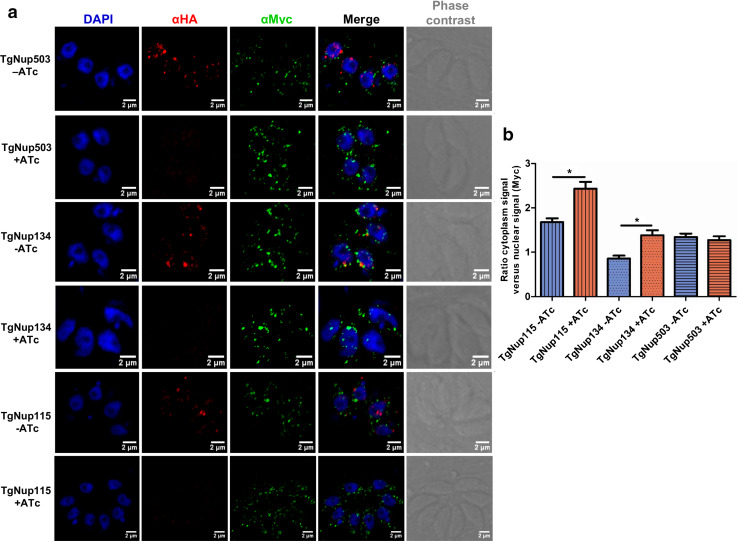
To gain more information about these unknown proteins presenting an α-solenoid fold, we tagged several of them using a Myc-tag in the iKD TgNup302-HA strain. As a control, we tagged a component of the FACT complex (TgFACT140) whose localization was anticipated to be nuclear. Using IFA, we observed that the five potential partners exhibited a TgNup302-like pattern (Figs. 5a, 6a and S9) in the absence of ATc (after 48 h of growth), indicating that they may, indeed, be components of the NPC. Therefore, we propose to rename them as TgNup proteins, as suggested in Table 1. As anticipated, the TgFACT140 protein showed strong nuclear localization that extended to the perinuclear periphery (Fig. S9). Notably, the TgNup503 protein showed a more spread pattern that extended toward the cytoplasm of the cell (Fig. 5a) and might not be connected to the nuclear envelope.
Fig. 5.
Identified proteins localize to the T. gondii nuclear pore. a Each potential partner was tagged using a myc-tag in the TgNup302-HA iKD strain. Endogenous TgNup302 iKD was labeled with rabbit monoclonal anti-HA (in red), and endogenous TgNup503, TgNup134, and TgNup115 were labeled with the mouse monoclonal anti-Myc (in green) antibody with or without ATc after 48 h of growth. b Measurement of cytoplasmic and nuclear Myc signals was determined for endogenous TgNup115 (vertical striped bars), TgNup134 (dotted bars), and TgNup503 (horizontal striped bars) potential partners without ATc treatment (blue) and after 48-h ATc treatment (red). The ratio of the cytoplasmic signal versus nuclear signal was determined. The results shown are from three independent experiments. *P < 0.05
Fig. 6.
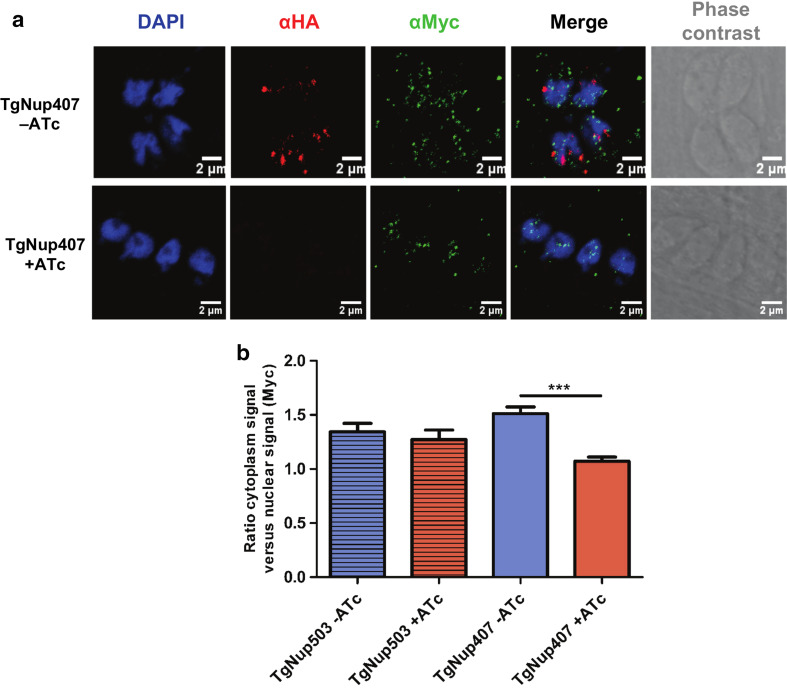
TgNup407 localization is perturbed in the TgNup302-deficient parasites. a TgNup407 protein was tagged using a Myc-tag in the TgNup302-HA iKD strain. Endogenous TgNup302 iKD was labeled with rabbit monoclonal anti-HA (in red) and endogenous TgNup407 was labeled with mouse monoclonal anti-Myc (in green) antibody with or without ATc after 48 h of growth. b Measurement of cytoplasmic and nuclear Myc signals was determined for endogenous TgNup503 (horizontal striped bars), TgNup407 (empty bars) potential partners without ATc treatment (blue), and after 48-h ATc treatment (red). The ratio of the cytoplasmic signal versus nuclear signal was determined. The results shown are from three independent experiments. ***P < 0.0001
We next examined whether the localization of these proteins was perturbed in the TgNup302-deficient parasites. We performed IFA and measured the quantity of signal in the cytoplasm and in the nucleus (as defined by DAPI staining) in the absence and presence of ATc (Fig. 5b) in multiple vacuoles. We identified two proteins (TgNup115, TgNup134) with a greater proportion of the fluorescence in the cytosol in the presence of ATc (Fig. 5b). In contrast, TgNup503 presented a distribution that remained unchanged after ATc treatment (Fig. 5b). Notably, we observed that TgNup407 localization was perinuclear in the absence of ATc and changed in the presence of ATc, with a more pronounced localization inside the nucleus (Fig. 6a). We measured the quantity of signal present at the nuclear periphery and inside the nucleus in the presence and absence of ATc (after 48 h) (Fig. 6b). We found that there was much more signal in the nucleus than at the nuclear periphery in the presence of ATc compared to in the absence of ATc (Fig. 6b). We examined the same ratio for TgNup129 protein localization, which exhibited a punctate perinuclear pattern that remained unchanged by ATc treatment (Fig. S10). Similarly, TgFACT140 exhibited a nuclear localization that was not perturbed by ATc treatment (Fig. S10).
Structured illuminated microscopy imaging of TgNup302 and its binding partners
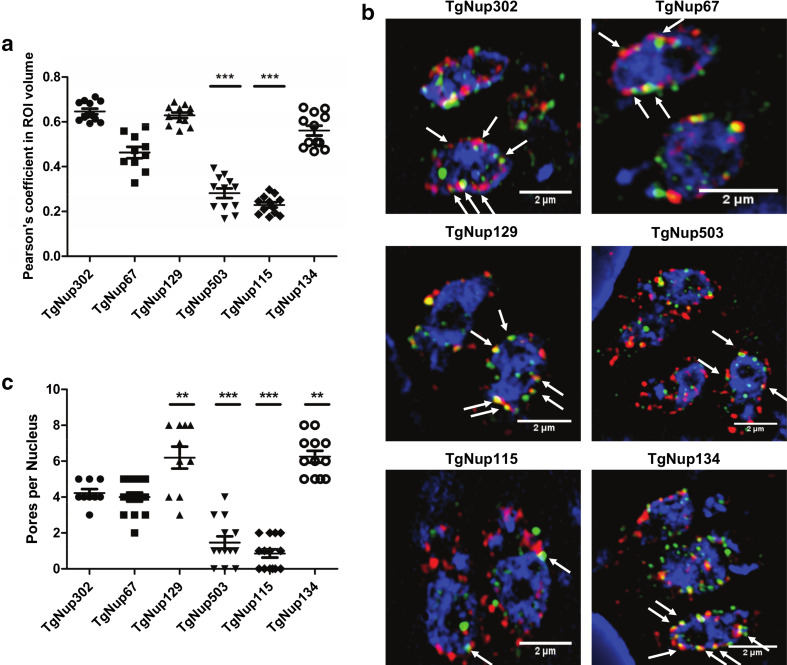
To better characterize the structure of the NPC in T. gondii, we performed structured illuminated microscopy (SIM) on selected parasite strains. For each strain, colocalization between TgNup302-HA and its myc-tagged protein partner was determined by the Imaris software (Pearson’s coefficient). Hence, we determined a Pearson’s coefficient average of 0.65 for the myc-tagged TgNup302 C-terminus, 0.46 for TgNup67, 0.63 for TgNup129, 0.28 for TgNup503, 0.23 for TgNup115, and 0.56 for TgNup134 with the HA-tagged TgNup302 protein (Fig. 7a). Using SIM microscopy, we observed for four proteins (TgNup302 C-terminus, TgNup67, TgNup129, and TgNup134) between 4 and 8 perinuclear co-distribution foci (indicated with white arrows) per parasite, representing co-distribution between HA (red) and Myc (green) staining (Fig. 7b). This may indicate that the parasite has 4 or 8 complete NPC structures per nuclei, a number that is comparable to what was observed in P. falciparum (Fig. 7c) [14, 39]. In contrast, TgNup115 and TgNup503 had lower Pearson’s coefficients, displaying a lower number of foci per parasite and indicating that these proteins may only be transiently interacting with NPCs (Fig. 7b, c).
Fig. 7.
Co-distribution of new nuclear pore proteins and TgNup302. a Co-distribution between TgNup302-N-ter HA-tag staining (in red) and myc-tag staining (in green) for TgNup302 (C-terminus), TgNup67, TgNup129, TgNup503, TgNup115, and TgNup134 knock-ins. Values are measured with the Imaris software from Structured Illumination Microscopy (SIM) (Pearson’s coefficient in ROI volume). ***P < 0.0001. b SIM images representing the co-distribution of TgNup302 (C-ter), TgNup67, TgNup129, TgNup503, TgNup115, and TgNup134 proteins tagged using a Myc-tag (in green) in the TgNup302-HA (in red) iKD strain (highlighted by white arrows). c Number of pores per nucleus in TgNup302 (C-ter), TgNup67, TgNup129, TgNup503, TgNup115, and TgNup134 knock-ins. Values are measured with the Imaris software from Structured Illumination Microscopy (SIM). ***P < 0.0001
CRISPR/Cas9 screening reveals the importance of the TgNup503 and TgNup134 proteins for nuclear-cytoplasmic shuttling
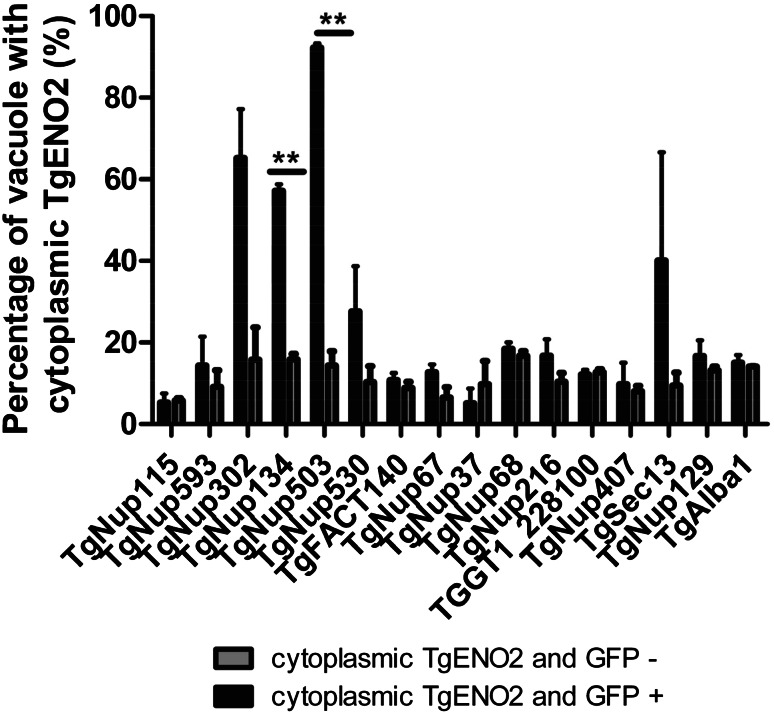
The previous studies have shown that combining CRISPR/CAS9 with sgRNAs can be used to generate site-specific double-strand breaks in the target DNA that are repaired by non-homologous-end joining, leading to short insertions and deletions that inactivate the gene [40]. Recently, the CRISPR/CAS9 system was adapted to T. gondii with a plasmid expressing a nuclear-localized CAS9 fused to green fluorescent protein (CAS9-NLS-GFP) driven by the SAG1 promoter and a single guide RNA (sgRNA) driven by the T. gondii U6 (TgU6) promoter [22]. We used CRISPR/Cas9 and a single gRNA placed downstream of the ATG to target the 15 proteins previously identified by co-immunoprecipitation with the TgNup302 protein (Table 1). As a control, we used a sgRNA targeting a non-essential gene (TgAlba1) [20]. For each construct, GFP expression was monitored to determine the transfection efficiency at 24 h after electroporation, revealing that ~30–70% of cells received the plasmid (Fig. S11). We used IFA to determine the localization of the TgENO2 protein, a nuclear marker. We determined the percentage of vacuoles that were GFP-negative and had cytoplasmic TgENO2 staining, as a control. We also scored the percentage of vacuoles that were GFP-positive (CAS9-expressing) and had cytoplasmic TgENO2 staining, illustrating the impact of the potentially mutated gene on the TgENO2 localization (Fig. 8). As expected, no differences were observed between these two scores for the parasite expressing CAS9 and the sgRNA targeting TgAlba1. Similarly, most of the targeted genes did not result in an increase in the number of vacuoles that had cytoplasmic TgENO2 and were GFP-positive. In contrast, the parasite expressing Cas9 and an sgRNA directed against the TgNup302 gene presented a drastic increase of the cytoplasmic TgENO2 signal (Fig. 8), in good concordance with the phenotype observed with the iKD strain (Fig. 2b). Interestingly, for sgRNA targeting the TgNup503 and TgNup134 genes, we observed a higher percentage (93 and 57%, respectively) of GFP-positive vacuoles with cytoplasmic TgENO2 staining than that of GFP-negative vacuoles with cytoplasmic TgENO2 staining (14 and 16%, respectively) (Fig. 8). Collectively, these results may indicate that in addition to TgNup302, the TgNup503 and TgNup134 proteins are essential for the nuclear localization of TgENO2.
Fig. 8.
CRISPR-Cas9 screening reveals the importance of the TgNup503 and TgNup134 proteins for the nuclear localization of TgENO2. The TgNup503 and TgNup134 proteins are important for the nuclear localization of the TgENO2 marker. IFA was performed on paraformaldehyde-fixed intracellular parasites of TgNup115, TgNup593, TgNup302, TgNup134, TgNup503, TgNup530, TgFACT140, TgNup67, TgNup37, TgNup68, TgNup216, TGGT1_228100, TgNup407, TgSec13, TgNup129 strains, and TgAlba1 as a control using the TgENO2 antibody (α-TgENO2) and staining of the nuclear DNA with DAPI. The percentage of GFP-negative vacuoles with cytoplasmic TgENO2 and the percentage of GFP-positive vacuoles with cytoplasmic TgENO2 were determined. The results shown are from three independent experiments. **P < 0.05
Discussion
Nuclear pore complex components have widely been studied in yeast and humans. More recently, NUP proteins were uncovered in a distant eukaryote, T. brucei [11, 12]. Strikingly, structural conservation is a key feature of T. brucei core-scaffold NUPs compared to those in yeast and humans. Using T. gondii as a model Apicomplexa, we investigated the role and the composition of the nuclear pore from another distant eukaryotic branch, the Alveolates. Using a conserved nuclear pore protein (TgNup302) homolog of the NUP98/96 and NUP145 proteins in humans and yeast, respectively, we identified potential interacting proteins. Interestingly, TgNup302 has all the features of its human and yeast homologs, including the family-specific GLFG repeats. T. brucei presents only one GLFG repeat protein that does not contain an autocleavage domain, as observed for the yeast Nup100 protein. This indicates that in distant eukaryotes, the GLFG repeat motif is a key feature that was retained during evolution. Because of the stringency of our protocol and as shown by the characterization of the identified proteins, we believe that a significant number of T. gondii NUPs was identified during the course of this study. Among these proteins, we purified proteins encompassing FG (phenylalanine-glycine)-repeat motifs, which are distinctive of NPC components. In fact, it was shown that a proportion of NUPs are composed of FG repeats, which function is to mediate the passage of transport receptors and their cargos with selectively gated transport [7].
Moreover, other proteins, bearing no recognizable motifs, presented structural features that are shared with eukaryotic NUPs. We further showed that their localization corresponds to known T. gondii NUPs, indicating that they may participate in the parasite NPC. This correlates with earlier studies identifying T. brucei NUPs [11, 12]. Indeed, structural conservation led to the hypothesis of an ancient inheritance of the core-scaffold NUPs. This hypothesis is confirmed by our study identifying NUPs in Alveolates. We also noted that most of the proteins that co-immunoprecipitated with TgNup302 were very large compared with their eukaryotic counterparts. This is also true for the TgNup302 protein itself. This may indicate that fewer proteins are needed to produce a functional nuclear pore in T. gondii.
During the course of this study, we also characterized the biological function of TgNup302, demonstrating its role in the nuclear transport of proteins and rRNA. When TgNup302 was depleted, these vital functions were impaired, and as a consequence, the parasite rapidly dies. It is worth noting that the nuclear pore structure is still present in the TgNup302-depleted parasites, because some of the nuclear pore proteins exhibited normal localization (TgNup67 and TgNup129). The remaining nuclear pore structure may also still be sufficient for the active transport of the large ribonucleoprotein complexes, insuring the nuclear export of polyA + mRNAs. This indicates that TgNup302 performs specific roles in the complex, as shown for NUP98/96 and NUP145, the representative human and yeast homologs, respectively. In fact, deletion of the essential Nup145 gene results in a defect not in protein import but in polyA + RNA export [41, 42]. Moreover, NUP98 plays a specific role in RNA export from the nucleus, and it appears to be an essential component of multiple RNA export pathways [43, 44]. NUP98 functions as a docking protein for the cytosol-mediated docking of a model import substrate. The docking function was localized to the N-terminal half of NUP98, which contains all its GLFG repeats [45]. However, we cannot exclude the possibility that an undetectable amount of TgNup302 is still present at the nuclear pore after 48 h of ATc treatment, therefore, allowing the passage of mRNAs but not rRNA. We also noted that the nuclear marker TgENO2 was mainly cytoplasmic in the TgNup302-depleted parasite. However, the localization of a nucleolar marker, TgNF3, or that of a pericentromeric marker (TgChromo1), was not affected after 48 h. This may be explained by the presence of multiple nuclear import pathways in the parasite, as previously suggested by a genomic identification of potential import factors [46]. While the expected molecular weight of TgNF3 (34 kDa) would allow this protein to diffuse freely in the nucleus through the nuclear pore, TgChromo1 (98 kDa) exceeds the size cutoff (approximately 40 kDa) for passive diffusion of molecules. Alternatively, this might reflect the differences in the dynamics of nuclear import of these proteins, especially in parasites for which growth is altered by TgNup302 depletion.
Using SIM imaging, we identified two proteins (TgNup115 and TgNup503) that associate with TgNup302 to a much lesser extent. These proteins may represent transporters that transiently associate with the NPC and should be investigated further, as this class of proteins is under-represented in Apicomplexa genomes [46]. In particular, TgNup115 presents a CCCH zinc finger that may bind RNA and, therefore, play a role in RNA transport. Overall, we estimated that the number of nuclear pores in T. gondii tachyzoites to be between 4 and 8, a number that is close to what was observed in P. falciparum [14]. However, epitope-tag availability and the microscopy limitations may have hampered the identification of proteins and, therefore, may have led to an under-estimation of the pore number.
To further investigate the potential components of the T. gondii nuclear pore, we performed a CRISPR/Cas9 screen. We used the ability of the T. gondii RH strain to perform non-homologous recombination after Cas9 cleavage, leading to mutations in the target gene that may impair its expression. Using TgENO2 as a nuclear marker, we identified two other proteins (TgNup503 and TgNup134) that showed a marked defect in TgENO2 localization in addition to TgNup302. Given the TgNup503 localization and its association with TgNup302, we believe that these data reinforce the hypothesis that this protein plays a major role in the TgENO2 import and potentially other proteins. TgNup503 localization is not dependent on the presence of TgNup302, and, therefore, it may associate with other proteins of the NPC.
The TgNup134 protein strongly associates with TgNup302 and is delocalized in the absence of TgNup302. Therefore, it may associate with the NPC through its interaction with TgNup302 and may represent a peripheral NUP. TgNup134 may also have a role in nuclear import. Further studies should be performed to validate this hypothesis.
In T. gondii, the previous studies hypothesized a model of centromere sequestration, in which centromere attachment to the centrocone, a subcompartment of the nucleus, serves as an organizer of chromosome apical region location throughout the parasite cell cycle [18, 47]. We hypothesized that centromeric heterochromatin could be anchored through protein components of the nuclear membranes via nucleoporins. In the TgNup302 iKD mutant, in the presence of ATc, we showed that a marker of the pericentromeric heterochromatin is still sequestered at all times of the cell cycle, indicating that TgNup302 depletion is not sufficient to uncouple the potential link between nuclear pore and centromeric heterochromatin. However, as previously shown, TgNup302 depletion is not sufficient to fully abrogate the NPC presence at the nuclear membrane. Therefore, other proteins in the remaining NPC may still interact with centromeric heterochromatin. Moreover, TgNup302, a FG nucleoporin, may not have a direct access to chromatin as nuclear basket nucleoporin would [48]. Alternatively, proteins directly interacting with the nuclear membrane may be involved in centromere sequestration at the nuclear periphery.
Finally, RNA-seq revealed that most of the transcripts that are impacted in the iKD mutant when TgNup302 is depleted are upregulated. This indicates a potential role of the TgNup302 protein in regulating gene expression. Further experiments are needed to explore the potential role of the NPC in T. gondii gene expression. Because the parasites lacking TgNup302 are severely affected, a kinetic experiment was not performed and would be useful to identify the genes that are impacted early after the depletion of TgNup302. In higher eukaryotes, an alternative splice variant of the Nup98/96 transcript produces a shortened version of the Nup98/96 polypeptide corresponding to the Nup98 protein with a stop codon inserted after the auto-proteolytic domain. This polypeptide is found in the intranuclear pool and can interact with chromatin [49]. Such splicing variants were not detected by in the RNA-seq data available on ToxoDB. Interestingly, TgNup129 was found to be a structural homolog of the Nup93 (or Nic96 in yeast) protein. This protein was shown to interact at multiple loci on chromatin [50]. Further experiments should be performed to validate this hypothesis.
The presence of the HDAC3 protein in the IP (Table S3) may indicate that T. gondii NPCs are recruiting repression-specific complexes at the nuclear periphery. TgHDAC3 has been implicated in the control of the expression of bradyzoite-specific genes during differentiation [51]. Therefore, the NPC may have a role in maintaining the repression state for those loci. However, these genes were scattered in the genome, which may indicate that some genomic regions may contact the nuclear periphery, resulting in the repression of gene expression, a mechanism widely recognized in other eukaryotes [48] that was never investigated in T. gondii until now.
We also showed that TgNup302 is able to interact with two members of the FACT complex. Indeed, the FACT complex is involved in the reorganization of the nucleosome structure during crucial processes, such as DNA replication and transcription elongation [52]. Proteins involved in transcription elongation have already been associated with the NPC [53], but the specific role of the FACT complex at the nuclear periphery was not investigated. Notably, a genetic interaction was identified in yeast between the Pob3 gene (a homolog of TgFACT80) and the Nup100 gene, coding for a GLFG NUP (homologous to TgNup302) [54]. The FACT complex is also known to be required for proper export of mRNA either by favoring an efficient mRNA splicing [55] or through a direct interaction with mRNA export factors [56]. Therefore, we hypothesize that T. gondii, and possibly other apicomplexan parasites, may have evolved a strong link between gene transcription, elongation, splicing, and export of mRNA. The FACT complex might, therefore, facilitate the export of mRNPs through the NPC in T. gondii. This would allow, as shown for other eukaryotes, the rapid expression of genes in T. gondii.
In conclusion, we uncovered novel members of the nuclear pore in T. gondii and also identified novel proteins that may have a role in the nuclear-cytoplasmic transport of proteins. TgNup302 is an essential protein that may be used as a tool to investigate the role of the T. gondii nuclear pore in regulating gene expression.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1 Oligonucleotides used in this study (XLS 31 KB)
Table S2 RNA-seq results. Presents all the genes up or downregulated with a FDR of 0.05 (XLS 94 KB)
Table S3 Mass-spectrometry results. The proteins highlighted in orange met the following criteria: less than 1 peptide in the control experiment and present in the two IP experiments (XLS 226 KB)
Fig. S1 T. gondii TgNup302 is evolutionarily conserved among Eukaryote. Phylogenetic tree of T. gondii TgNup302 homologues based on ClustalW alignment of sequences identified by BLASTp searches using the entire sequence of TgTgNup302 gene against 15 apicomplexan parasites (brown), 5 Fungi (green), 3 Eumetazoa (blue) and 3 Plantae (purple). The tree was reconstructed by maximum likelihood (ML) analyses with MEGA6 software. Five hundred bootstrap pseudo-replicates were used to give statistical support to the clades of the maximum likelihood topology. Scale bar reflects number of substitutions per site. Numbers in the nodes of the tree reflect the percentage of bootstrap replicates supporting each node (PDF 39 KB)
Fig. S2 Construction of the TgNup302 iKD strain. (A) Schematic of the genetic approach used to produce the conditional knock-down strain by a promoter replacement strategy. After promoter replacement, the expression of TgNup302 is under the control of anhydrotetracyline (ATc). The TgNup302 gene was HA-tagged at its 5’ (Top panel). (B) PCR was used to confirm the correct integration of the plasmid and the creation of the recombinant locus using primer pairs i and ii (sequences are in TableS1 of supplementary data). (C) Double HA-tagged (N-terminal) / Myc-tagged (C-terminal) strain for the TgNup302 gene was produced by introducing a myc tag at the 3’ end of the gene (PDF 54 KB)
Fig. S3 PolyA+ RNA were mostly cytoplasmic in the parental and iKD strains. RNA FISH was performed on intracellular parasites. Parasites of the parental and TgNup302 iKD strains were hybridized with Cy3-labeled polyA+ oligonucleotides primers (red), and the nuclear DNA was labeled with DAPI (blue) (PDF 284 KB)
Fig. S4 Conditional expression of TgNup302 has no impact on TgChromo1 and TgNF3 localization. Endogenous TgChromo1 and TgNF3 were labeled with the mouse monoclonal anti-Myc and 488-nm Alexa goat secondary antibody in parental and TgNup302 iKD strains with or without ATc treatment for 48hr (PDF 867 KB)
Fig. S5 Ultrastructure of Toxoplasma gondii. Intracellular T. gondii tachyzoite showing the nucleus (N) for the parental RHΔKu80 TaTi and iKD TgNup302 strains with or without ATc treatment. Bar=500 nm (PDF 1529 KB)
Fig. S6 Chromosomal distribution of genes differentially regulated in the TgNUP1 iKD strain. The chromosomal position and distribution of genes that were identified as differentially expressed, upregulated (top panel), and downregulated (bottom panel), in TgNup302 iKD after an ATc treatment for 48hr (PDF 777 KB)
Fig. S7 Peptides identified for TgNup302. Peptides recovered from the immunoprecipitation of TgNup302 and mapped onto the TgNup302 sequence are highlighted in yellow. The autocatalytic domain sequence is underlined (PDF 132 KB)
Fig. S8 Predicted secondary structure features, fold, and location for validated TgNups. The horizontal black line represents the polypeptide length of the proteins. The y-axis indicates the confidence score of the predicted secondary structure element. Predicted α-helices are indicated in blue, predicted β-sheets in orange, and predicted coiled-coil regions are in red arrows. The green arrows indicate FG repeats (PDF 353 KB)
Fig. S9 Identified proteins interact with TgNup302. (a) 5 µl of nuclear extract (from ~500x106 parasites) conserved before the immunoprecipitation (inputs) for the TgFACT140 (line 2), TgNup134 (line 3), TgNup129 (line 4) and TgNup407 (line 5) C-terminally myc-tagged proteins in the TgNup302-HA iKD strain were analyzed by Western blot. The immunoblot was probed with anti-HA antibody to detect the presence of the TgNup302-HA protein in each strain before immunoprecipitation. (b) Immunoblot was reprobed with an anti-myc antibody to detect the myc-tagged proteins: TgFACT140 (134 kDa), TgNup129 (129 kDa), TgNup134 (134 kDa), and TgNup407 (115 kDa) (PDF 91 KB)
Fig. S10 New components of the T. gondii nuclear pore. Each potential partner was tagged using a Myc-tag in the TgNup302-HA iKD strain. Endogenous TgNup302 iKD was labeled with the rabbit monoclonal anti-HA (in red) and endogenous TgFACT140, TgNup129 were labeled with the mouse monoclonal anti-Myc (in green) antibody with or without ATc after 48hr of growth (PDF 446 KB)
Fig. S11 Transfection efficiency for the Crisp-Cas9 screening. For each construction, the percentage of vacuole with a positive GFP expression was monitored to determine the transfection efficiency at 24 h after electroporation, revealing that ~30 to 70% of cells received the plasmid. TgAlba1 is a negative control (PDF 49 KB)
Acknowledgements
The authors would like to thank Dr. Valerie Doye for helpful discussions and Dr. R. Walker for critically reading the manuscript. We also thank Ludovic Huot for checking the integrity of RNA samples, Etienne Dewailly for electronic microscopy, Antonino Bongiovanni for his help with microscopy data analyses, and Quentin Deveuve for phylogenetic tree recommendations. The authors also thank the BioImaging Center Lille for access to instruments. This work was supported by Centre National de la Recherche Scientifique (CNRS), Institut National de la Santé et de la Recherche Médicale (INSERM), grants from the French National Research Agency (ANR) [Grant Number ANR-13-JSV3-0006-01 to MG and ANR-11-LABX-0024 to Plateforme Protéomique et Peptides Modifiés (P3M)], the Fonds Européen de Développement Economique Régionale (13003300-42405 Labex Parafrap to P3M) and the Métropole Européenne de Lille (MEL).
Abbreviations
- ATc
AnhydroTetraCycline
- Co-IP
Co-immunoprecipitation
- FACT
Facilitates chromatin transcription
- FISH
Fluorescence in situ hybridization
- IFA
Immunofluorescence assay
- iKD
Conditional knock-down
- NPC
Nuclear pore complex
- NUP
Nucleoporin
- RNA-Seq
RNA-sequencing
- Sg RNA
Single guide RNA
- SIM
Structured illumination microscopy
- Tg
Toxoplasma gondii
References
- 1.Kim K, Weiss LM. Toxoplasma gondii: the model apicomplexan. Int J Parasitol. 2004;34:423–432. doi: 10.1016/j.ijpara.2003.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behnke MS, Radke JB, Smith AT, Sullivan WJ, White MW. The transcription of bradyzoite genes in Toxoplasma gondii is controlled by autonomous promoter elements. Mol Microbiol. 2008;68:1502–1518. doi: 10.1111/j.1365-2958.2008.06249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behnke MS, Wootton JC, Lehmann MM, Radke JB, Lucas O, Nawas J, Sibley LD, White MW. Coordinated progression through two subtranscriptomes underlies the tachyzoite cycle of Toxoplasma gondii . PLoS One. 2010;5:e12354. doi: 10.1371/journal.pone.0012354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sullivan WJ, Hakimi MA. Histone mediated gene activation in Toxoplasma gondii . Mol Biochem Parasitol. 2006;148:109–116. doi: 10.1016/j.molbiopara.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 5.Gissot M, Kelly KA, Ajioka JW, Greally JM, Kim K. Epigenomic modifications predict active promoters and gene structure in Toxoplasma gondii . PLoS Pathog. 2007;3:e77. doi: 10.1371/journal.ppat.0030077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, Sali A, Rout MP. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 7.Capelson M, Hetzer MW. The role of nuclear pores in gene regulation, development and disease. EMBO Rep. 2009;10:697–705. doi: 10.1038/embor.2009.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vasu SK, Forbes DJ. Nuclear pores and nuclear assembly. Curr Opin Cell Biol. 2001;13:363–375. doi: 10.1016/S0955-0674(00)00221-0. [DOI] [PubMed] [Google Scholar]
- 10.Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeGrasse JA, DuBois KN, Devos D, Siegel TN, Sali A, Field MC, Rout MP, Chait BT. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol Cell Proteom. 2009;8:2119–2130. doi: 10.1074/mcp.M900038-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Obado SO, Brillantes M, Uryu K, Zhang W, Ketaren NE, Chait BT, Field MC, Rout MP. Interactome mapping reveals the evolutionary history of the nuclear pore complex. PLoS Biol. 2016;14:e1002365. doi: 10.1371/journal.pbio.1002365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim RYH, Fahrenkrog B, Köser J, Schwarz-Herion K, Deng J. U. Aebi, Nanomechanical Basis of Selective Gating by the Nuclear Pore Complex. Science. 2007;318:640–643. doi: 10.1126/science.1145980. [DOI] [PubMed] [Google Scholar]
- 14.Weiner A, Dahan-Pasternak N, Shimoni E, Shinder V, von Huth P, Elbaum M, Dzikowski R. 3D nuclear architecture reveals coupled cell cycle dynamics of chromatin and nuclear pores in the malaria parasite Plasmodium falciparum . Cell Microbiol. 2011;13:967–977. doi: 10.1111/j.1462-5822.2011.01592.x. [DOI] [PubMed] [Google Scholar]
- 15.Dahan-Pasternak N, Nasereddin A, Kolevzon N, Pe’er M, Wong W, Shinder V, Turnbull L, Whitchurch CB, Elbaum M, Gilberger TW, Yavin E, Baum J, Dzikowski R. PfSec13 is an unusual chromatin-associated nucleoporin of Plasmodium falciparum that is essential for parasite proliferation in human erythrocytes. J Cell Sci. 2013;126:3055–3069. doi: 10.1242/jcs.122119. [DOI] [PubMed] [Google Scholar]
- 16.Sheiner L, Demerly JL, Poulsen N, Beatty WL, Lucas O, Behnke MS, White MW, Striepen B. A systematic screen to discover and analyze apicoplast proteins identifies a conserved and essential protein import factor. PLoS Pathog. 2011;7:e1002392. doi: 10.1371/journal.ppat.1002392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dzierszinski F, Mortuaire M, Dendouga N, Popescu O, Tomavo S. Differential expression of two plant-like enolases with distinct enzymatic and antigenic properties during stage conversion of the protozoan parasite Toxoplasma gondii . J Mol Biol. 2001;309:1017–1027. doi: 10.1006/jmbi.2001.4730. [DOI] [PubMed] [Google Scholar]
- 18.Gissot M, Walker R, Delhaye S, Huot L, Hot D, Tomavo S. Toxoplasma gondii chromodomain protein 1 binds to heterochromatin and colocalises with centromeres and telomeres at the nuclear periphery. PLoS One. 2012;7:e32671. doi: 10.1371/journal.pone.0032671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olguin-Lamas A, Madec E, Hovasse A, Werkmeister E, Callebaut I, Slomianny C, Delhaye S, Mouveaux T, Schaeffer-Reiss C, Van Dorsselaer A, Tomavo S. A novel Toxoplasma gondii nuclear factor TgNF3 is a dynamic chromatin-associated component, modulator of nucleolar architecture and parasite virulence. PLoS Pathog. 2011;7:e1001328. doi: 10.1371/journal.ppat.1001328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gissot M, Walker R, Delhaye S, Alayi TD, Huot L, Hot D, Callebaut I, Schaeffer-Reiss C, Dorsselaer AV, Tomavo S. Toxoplasma gondii Alba proteins are involved in translational control of gene expression. J Mol Biol. 2013;425:1287–1301. doi: 10.1016/j.jmb.2013.01.039. [DOI] [PubMed] [Google Scholar]
- 21.Thompson J. In situ detection of RNA in blood- and mosquito-stage malaria parasites. Methods Mol Med. 2002;72:225–233. doi: 10.1385/1-59259-271-6:225. [DOI] [PubMed] [Google Scholar]
- 22.Shen B, Brown KM, Lee TD, Sibley LD. Efficient gene disruption in diverse strains of Toxoplasma gondii using CRISPR/CAS9. MBio. 2014;5:e01114–01114. doi: 10.1128/mBio.01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miguet L, Béchade G, Fornecker L, Zink E, Felden C, Gervais C, Herbrecht R, Van Dorsselaer A, van Dorsselaer A, Mauvieux L, Sanglier-Cianferani S. Proteomic analysis of malignant B-cell derived microparticles reveals CD148 as a potentially useful antigenic biomarker for mantle cell lymphoma diagnosis. J Proteome Res. 2009;8:3346–3354. doi: 10.1021/pr801102c. [DOI] [PubMed] [Google Scholar]
- 24.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JYH, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamura K, Fukao Y, Iwamoto M, Haraguchi T, Hara-Nishimura I. Identification and characterization of nuclear pore complex components in Arabidopsis thaliana . Plant Cell. 2010;22:4084–4097. doi: 10.1105/tpc.110.079947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamura K, Hara-Nishimura I. The molecular architecture of the plant nuclear pore complex. J Exp Bot. 2013;64:823–832. doi: 10.1093/jxb/ers258. [DOI] [PubMed] [Google Scholar]
- 33.Fontoura BM, Blobel G, Matunis MJ. A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J Cell Biol. 1999;144:1097–1112. doi: 10.1083/jcb.144.6.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferguson DJP, Parmley SF, Tomavo S. Evidence for nuclear localisation of two stage-specific isoenzymes of enolase in Toxoplasma gondii correlates with active parasite replication. Int J Parasitol. 2002;32:1399–1410. doi: 10.1016/S0020-7519(02)00129-7. [DOI] [PubMed] [Google Scholar]
- 35.Griffis ER, Xu S, Powers MA. Nup98 localizes to both nuclear and cytoplasmic sides of the nuclear pore and binds to two distinct nucleoporin subcomplexes. Mol Biol Cell. 2003;14:600–610. doi: 10.1091/mbc.E02-09-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Enninga J, Levay A, Fontoura BMA. Sec13 shuttles between the nucleus and the cytoplasm and stably interacts with Nup96 at the nuclear pore complex. Mol Cell Biol. 2003;23:7271–7284. doi: 10.1128/MCB.23.20.7271-7284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pritchard CE, Fornerod M, Kasper LH, van Deursen JM. RAE1 is a shuttling mRNA export factor that binds to a GLEBS-like NUP98 motif at the nuclear pore complex through multiple domains. J Cell Biol. 1999;145:237–254. doi: 10.1083/jcb.145.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guizetti J, Martins RM, Guadagnini S, Claes A, Scherf A. Nuclear pores and perinuclear expression sites of var and ribosomal DNA genes correspond to physically distinct regions in Plasmodium falciparum . Eukaryot Cell. 2013;12:697–702. doi: 10.1128/EC.00023-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fabre E, Boelens WC, Wimmer C, Mattaj IW, Hurt EC. Nup145p is required for nuclear export of mRNA and binds homopolymeric RNA in vitro via a novel conserved motif. Cell. 1994;78:275–289. doi: 10.1016/0092-8674(94)90297-6. [DOI] [PubMed] [Google Scholar]
- 42.Dockendorff TC, Heath CV, Goldstein AL, Snay CA, Cole CN. C-terminal truncations of the yeast nucleoporin Nup145p produce a rapid temperature-conditional mRNA export defect and alterations to nuclear structure. Mol Cell Biol. 1997;17:906–920. doi: 10.1128/MCB.17.2.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Powers MA, Forbes DJ, Dahlberg JE, Lund E. The vertebrate GLFG nucleoporin, Nup98, is an essential component of multiple RNA export pathways. J Cell Biol. 1997;136:241–250. doi: 10.1083/jcb.136.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Radu A, Blobel G, Moore MS. Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc Natl Acad Sci USA. 1995;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Radu A, Moore MS, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- 46.Frankel MB, Knoll LJ. The ins and outs of nuclear trafficking: unusual aspects in apicomplexan parasites. DNA Cell Biol. 2009;28:277–284. doi: 10.1089/dna.2009.0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brooks CF, Francia ME, Gissot M, Croken MM, Kim K, Striepen B. Toxoplasma gondii sequesters centromeres to a specific nuclear region throughout the cell cycle. Proc Natl Acad Sci USA. 2011;108:3767–3772. doi: 10.1073/pnas.1006741108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lemaître C, Bickmore WA. Chromatin at the nuclear periphery and the regulation of genome functions. Histochem Cell Biol. 2015;144:111–122. doi: 10.1007/s00418-015-1346-y. [DOI] [PubMed] [Google Scholar]
- 49.Liang Y, Hetzer MW. Functional interactions between nucleoporins and chromatin. Curr Opin Cell Biol. 2011;23:65–70. doi: 10.1016/j.ceb.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown CR, Kennedy CJ, Delmar VA, Forbes DJ, Silver PA. Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes Dev. 2008;22:627–639. doi: 10.1101/gad.1632708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bougdour A, Maubon D, Baldacci P, Ortet P, Bastien O, Bouillon A, Barale J-C, Pelloux H, Ménard R, Hakimi M-A. Drug inhibition of HDAC3 and epigenetic control of differentiation in Apicomplexa parasites. J Exp Med. 2009;206:953–966. doi: 10.1084/jem.20082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reinberg D, Sims RJ. de FACTo nucleosome dynamics. J Biol Chem. 2006;281:23297–23301. doi: 10.1074/jbc.R600007200. [DOI] [PubMed] [Google Scholar]
- 53.Tous C, Rondón AG, García-Rubio M, González-Aguilera C, Luna R, Aguilera A. A novel assay identifies transcript elongation roles for the Nup84 complex and RNA processing factors. EMBO J. 2011;30:1953–1964. doi: 10.1038/emboj.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JLY, Toufighi K, Mostafavi S, Prinz J, St Onge RP, VanderSluis B, Makhnevych T, Vizeacoumar FJ, Alizadeh S, Bahr S, Brost RL, Chen Y, Cokol M, Deshpande R, Li Z, Lin Z-Y, Liang W, Marback M, Paw J, San Luis B-J, Shuteriqi E, Tong AHY, van Dyk N, Wallace IM, Whitney JA, Weirauch MT, Zhong G, Zhu H, Houry WA, Brudno M, Ragibizadeh S, Papp B, Pál C, Roth FP, Giaever G, Nislow C, Troyanskaya OG, Bussey H, Bader GD, Gingras A-C, Morris QD, Kim PM, Kaiser CA, Myers CL, Andrews BJ, Boone C. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burckin T, Nagel R, Mandel-Gutfreund Y, Shiue L, Clark TA, Chong J-L, Chang T-H, Squazzo S, Hartzog G, Ares M. Exploring functional relationships between components of the gene expression machinery. Nat Struct Mol Biol. 2005;12:175–182. doi: 10.1038/nsmb891. [DOI] [PubMed] [Google Scholar]
- 56.Hautbergue GM, Hung M-L, Walsh MJ, Snijders APL, Chang C-T, Jones R, Ponting CP, Dickman MJ, Wilson SA. UIF, a new mRNA export adaptor that works together with REF/ALY, requires FACT for recruitment to mRNA. Curr Biol. 2009;19:1918–1924. doi: 10.1016/j.cub.2009.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Oligonucleotides used in this study (XLS 31 KB)
Table S2 RNA-seq results. Presents all the genes up or downregulated with a FDR of 0.05 (XLS 94 KB)
Table S3 Mass-spectrometry results. The proteins highlighted in orange met the following criteria: less than 1 peptide in the control experiment and present in the two IP experiments (XLS 226 KB)
Fig. S1 T. gondii TgNup302 is evolutionarily conserved among Eukaryote. Phylogenetic tree of T. gondii TgNup302 homologues based on ClustalW alignment of sequences identified by BLASTp searches using the entire sequence of TgTgNup302 gene against 15 apicomplexan parasites (brown), 5 Fungi (green), 3 Eumetazoa (blue) and 3 Plantae (purple). The tree was reconstructed by maximum likelihood (ML) analyses with MEGA6 software. Five hundred bootstrap pseudo-replicates were used to give statistical support to the clades of the maximum likelihood topology. Scale bar reflects number of substitutions per site. Numbers in the nodes of the tree reflect the percentage of bootstrap replicates supporting each node (PDF 39 KB)
Fig. S2 Construction of the TgNup302 iKD strain. (A) Schematic of the genetic approach used to produce the conditional knock-down strain by a promoter replacement strategy. After promoter replacement, the expression of TgNup302 is under the control of anhydrotetracyline (ATc). The TgNup302 gene was HA-tagged at its 5’ (Top panel). (B) PCR was used to confirm the correct integration of the plasmid and the creation of the recombinant locus using primer pairs i and ii (sequences are in TableS1 of supplementary data). (C) Double HA-tagged (N-terminal) / Myc-tagged (C-terminal) strain for the TgNup302 gene was produced by introducing a myc tag at the 3’ end of the gene (PDF 54 KB)
Fig. S3 PolyA+ RNA were mostly cytoplasmic in the parental and iKD strains. RNA FISH was performed on intracellular parasites. Parasites of the parental and TgNup302 iKD strains were hybridized with Cy3-labeled polyA+ oligonucleotides primers (red), and the nuclear DNA was labeled with DAPI (blue) (PDF 284 KB)
Fig. S4 Conditional expression of TgNup302 has no impact on TgChromo1 and TgNF3 localization. Endogenous TgChromo1 and TgNF3 were labeled with the mouse monoclonal anti-Myc and 488-nm Alexa goat secondary antibody in parental and TgNup302 iKD strains with or without ATc treatment for 48hr (PDF 867 KB)
Fig. S5 Ultrastructure of Toxoplasma gondii. Intracellular T. gondii tachyzoite showing the nucleus (N) for the parental RHΔKu80 TaTi and iKD TgNup302 strains with or without ATc treatment. Bar=500 nm (PDF 1529 KB)
Fig. S6 Chromosomal distribution of genes differentially regulated in the TgNUP1 iKD strain. The chromosomal position and distribution of genes that were identified as differentially expressed, upregulated (top panel), and downregulated (bottom panel), in TgNup302 iKD after an ATc treatment for 48hr (PDF 777 KB)
Fig. S7 Peptides identified for TgNup302. Peptides recovered from the immunoprecipitation of TgNup302 and mapped onto the TgNup302 sequence are highlighted in yellow. The autocatalytic domain sequence is underlined (PDF 132 KB)
Fig. S8 Predicted secondary structure features, fold, and location for validated TgNups. The horizontal black line represents the polypeptide length of the proteins. The y-axis indicates the confidence score of the predicted secondary structure element. Predicted α-helices are indicated in blue, predicted β-sheets in orange, and predicted coiled-coil regions are in red arrows. The green arrows indicate FG repeats (PDF 353 KB)
Fig. S9 Identified proteins interact with TgNup302. (a) 5 µl of nuclear extract (from ~500x106 parasites) conserved before the immunoprecipitation (inputs) for the TgFACT140 (line 2), TgNup134 (line 3), TgNup129 (line 4) and TgNup407 (line 5) C-terminally myc-tagged proteins in the TgNup302-HA iKD strain were analyzed by Western blot. The immunoblot was probed with anti-HA antibody to detect the presence of the TgNup302-HA protein in each strain before immunoprecipitation. (b) Immunoblot was reprobed with an anti-myc antibody to detect the myc-tagged proteins: TgFACT140 (134 kDa), TgNup129 (129 kDa), TgNup134 (134 kDa), and TgNup407 (115 kDa) (PDF 91 KB)
Fig. S10 New components of the T. gondii nuclear pore. Each potential partner was tagged using a Myc-tag in the TgNup302-HA iKD strain. Endogenous TgNup302 iKD was labeled with the rabbit monoclonal anti-HA (in red) and endogenous TgFACT140, TgNup129 were labeled with the mouse monoclonal anti-Myc (in green) antibody with or without ATc after 48hr of growth (PDF 446 KB)
Fig. S11 Transfection efficiency for the Crisp-Cas9 screening. For each construction, the percentage of vacuole with a positive GFP expression was monitored to determine the transfection efficiency at 24 h after electroporation, revealing that ~30 to 70% of cells received the plasmid. TgAlba1 is a negative control (PDF 49 KB)