Abstract
Cell-surface receptors form the front line of plant immunity. The leucine-rich repeat (LRR)-receptor-like kinases SOBIR1 and BAK1 are required for the functionality of the tomato LRR-receptor-like protein Cf-4, which detects the secreted effector Avr4 of the pathogenic fungus Fulvia fulva. Here, we show that the kinase domains of SOBIR1 and BAK1 directly phosphorylate each other and that residues Thr522 and Tyr469 of the kinase domain of Nicotiana benthamiana SOBIR1 are required for its kinase activity and for interacting with signalling partners, respectively. By knocking out multiple genes belonging to different receptor-like cytoplasmic kinase (RLCK)-VII subfamilies in N. benthamiana:Cf-4, we show that members of RLCK-VII-6, −7, and −8 differentially regulate the Avr4/Cf-4-triggered biphasic burst of reactive oxygen species. In addition, members of RLCK-VII-7 play an essential role in resistance against the oomycete pathogen Phytophthora palmivora. Our study provides molecular evidence for the specific roles of RLCKs downstream of SOBIR1/BAK1-containing immune complexes.
Subject terms: Plant molecular biology, Pattern recognition receptors in plants, Plant signalling, Kinases
Cell-surface receptors form the front line of plant immunity. Here, the authors show that the RLP co-receptors SOBIR1 and BAK1 directly phosphorylate each other, leading to activation of the immune receptor complex in which RLCKs are differentially required for production of reactive oxygen species that play a role in resistance against Phytophthora palmivora.
Introduction
Plants have evolved a two-layered innate immune system to fend off invading microbes, of which the first layer is mediated by cell-surface receptors1. The two largest families of such receptors are receptor-like proteins (RLPs) and receptor-like kinases (RLKs)2. Both RLKs and RLPs detect extracellular immunogenic patterns (ExIPs), leading to extracellularly-triggered immunity (ExTI)1. ExTI is associated with various downstream immune outputs, including the rapid phosphorylation of downstream receptor-like cytoplasmic kinases (RLCKs), the swift production of reactive oxygen species (ROS), the activation of mitogen-activated protein kinase (MAPK) cascades, large-scale transcriptional reprogramming, and in some cases, the induction of programmed cell death, referred to as the hypersensitive response (HR)3.
RLKs and RLPs share the same overall structure2. However, in contrast to RLKs, RLPs lack an intracellular kinase domain for downstream signalling4. To date, the most extensively studied plant cell-surface receptors are leucine-rich repeat (LRR)-RLKs and LRR-RLPs5,6. Typically, LRR-RLKs form a complex with the regulatory LRR-RLK BRASSINOSTEROID INSENSITIVE 1-ASSOCIATED KINASE1 (BAK1) in a ligand-dependent manner7,8. A well-studied example is the Arabidopsis (Arabidopsis thaliana, At) LRR-RLK FLAGELLIN-SENSING 2 (FLS2), which specifically recognizes bacterial flagellin (or its N-terminal 22-amino acid epitope, flg22), resulting in the rapid trans-phosphorylation of the cytoplasmic kinase domains of BAK1 and FLS27–12.
LRR-RLPs constitutively associate with the LRR-RLK SUPPRESSOR OF BIR1-1 (SOBIR1), and require SOBIR1 for their function in plant immunity13–15. The tomato (Solanum lycopersicum, Sl) resistance protein Cf-4 is one of the best-studied LRR-RLPs and specifically recognizes the apoplastic effector Avr4 secreted by the pathogenic intercellular fungus Fulvia fulva, formerly known as Cladosporium fulvum16,17. Cf-4 forms a complex with SOBIR1 in a ligand-independent manner, which is essential for Cf-4-mediated resistance to F. fulva14,15. Consistent with LRR-RLKs, the LRR-RLP/SOBIR1 complex also requires the recruitment of BAK1 for the initiation of downstream signalling upon ligand perception18–20.
Protein phosphorylation, which is a swift and reversible biochemical post-translational modification, plays an important role as a versatile molecular switch in various cellular activities, including the initiation of plant immune responses5,21,22. According to whether a conserved arginine (Arg/R) is immediately preceding the highly conserved catalytic aspartate (Asp/D) in their catalytic loop, protein kinases can be subdivided into RD and non-RD kinases23. Generally, activation of RD kinases requires phosphorylation of one or more residues in their activation segment23–25. In contrast, non-RD kinases require a regulatory RD kinase, such as BAK1, to promote their phosphorylation and signalling26.
Activation of cell-surface receptor complexes subsequently triggers a suite of downstream signalling events. Increasing evidence suggests that RLCKs are the direct downstream cytoplasmic substrates of activated receptor complexes and that they fill the gap between receptors present at the cell surface and downstream signalling components21,27–30. Arabidopsis BOTRYTIS-INDUCED KINASE1 (BIK1) is one of the best-characterized RLCKs and is required for triggering ROS production upon the perception of multiple ExIPs, such as flg22, elf18 and chitin31–33. BIK1 and some other RLCKs, which are important to relay immune signalling from the cell surface to the intracellular space, all belong to Arabidopsis RLCK class VII32,34–36. RLCK-VII is composed of 46 members, and according to their sequence similarity, these members can be further divided into nine subfamilies, termed RLCK-VII-1 to RLCK-VII-937. Interestingly, flg22-, elf18- and chitin-triggered ROS production is significantly reduced in rlck-vii-5, −7 and −8 knock-out Arabidopsis plants, whereas in contrast, rlck-vii-6 knock-out plants exhibit higher flg22-induced ROS accumulation levels when compared to wild-type (WT) plants37.
The last two decades have witnessed remarkable progress in our understanding of the initiation and regulation of plant innate immunity. Nevertheless, it is largely unknown how SOBIR1 and BAK1 exactly trans-phosphorylate each other to initiate downstream signalling. Additionally, compared to the model plant Arabidopsis, little is known about the function of the various RLCK-VII subfamilies in Solanaceous plants, and it is thus far not known which RLCKs are involved in the LRR-RLP/SOBIR1-triggered immune signalling pathway.
In this work, we perform a site-directed mutagenesis screen, combined with a complementation study, in Nicotiana benthamiana:Cf-4 sobir1 plants. NbSOBIR1 threonine (Thr/T) 522, as well as its analogous residues in both tomato SOBIR1s, present in the activation segment of the kinase domain of SOBIR1, is found to play an essential role in Avr4/Cf-4-triggered immune signalling. Interestingly, in vitro phosphorylation assays demonstrate that this highly conserved Thr residue is required for SOBIR1(-like) intrinsic kinase activity. Besides, we show that SOBIR1 directly trans-phosphorylates BAK1, whereas, on the other hand, BAK1 directly trans-phosphorylates SOBIR1. These trans-phosphorylation events are proposed to eventually lead to the full activation of both SOBIR1 and BAK1, and their intrinsic kinase activity is required for these trans-phosphorylation events to take place and the initiation of downstream immune signalling. In addition to Thr522, NbSOBIR1 tyrosine (Tyr/Y) 469, as well as its analogous residues in both tomato SOBIR1s, is identified to be essential for Avr4/Cf-4-triggered MAPK activation and the initiation of the HR, but not for ROS production and its intrinsic kinase activity. By knocking out multiple candidate genes belonging to different RLCK-VII subfamilies in N. benthamiana:Cf-4, we show that members of RLCK-VII-6, −7 and −8 are differentially required for full Avr4/Cf-4-triggered biphasic ROS production. These members also play an important role in regulating flg22- and chitin-induced ROS accumulation. Additionally, members from RLCK-VII-7 are essential for the Avr4/Cf-4-induced HR and host resistance of N. benthamiana to the oomycete pathogen Phytophthora palmivora. Importantly, SOBIR1 and BAK1 directly trans-phosphorylate members of RLCK-VII-6, 7 and 8 in vitro. Our study unveils the molecular mechanism underlying the activation and signal transduction of SOBIR1/BAK1-containing immune complexes through RLCKs in N. benthamiana.
Results
Thr522, present in the activation segment of the kinase domain of NbSOBIR1, is essential for mounting Avr4/Cf-4-triggered immune signalling
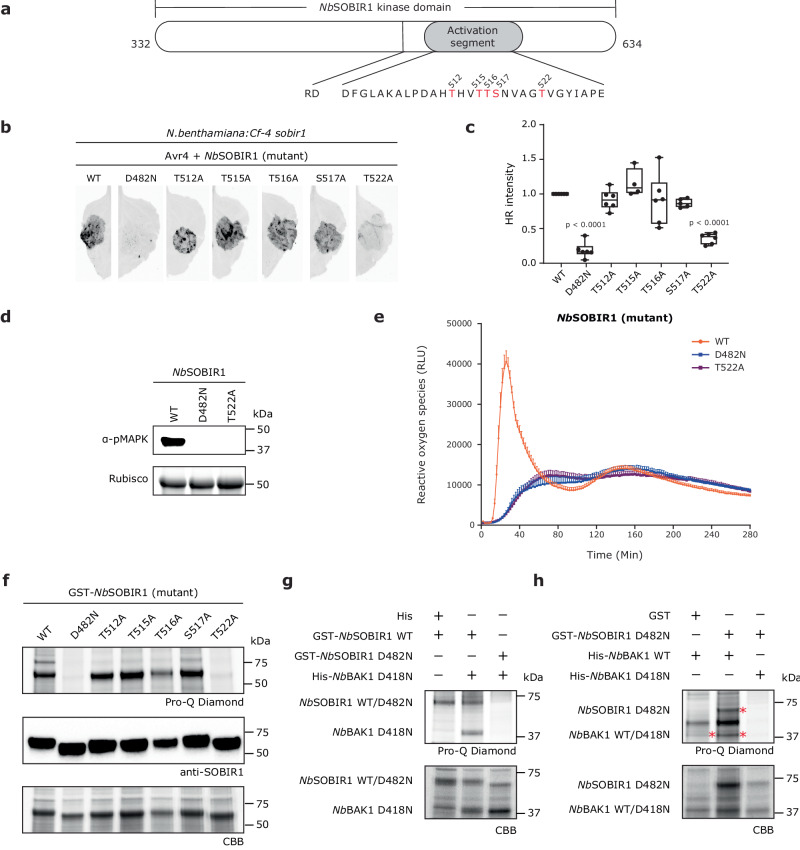
The regulatory LRR-RLK SOBIR1 is present throughout the plant kingdom, including N. benthamiana (Nb), which is a versatile experimental host plant13–15. SOBIR1 is an RD kinase, which suggests that likely SOBIR1 first requires phosphorylation of its activation segment to acquire the kinase-active conformation23,25. It has been reported that the kinase domain of AtSOBIR1 auto-phosphorylates at serine (Ser/S), Thr, and Tyr residues in vitro38, and in the activation segment of SOBIR1 of tomato and N. benthamiana such residues are present. To investigate which residue(s) in the kinase domain of SOBIR1 is(are) essential for the activation of the Cf-4/SOBIR1-triggered signalling pathway, we decided to zoom in on the activation segment of NbSOBIR114. Five potential phosphorylation sites are located in this loop of 30 amino acids, including one Ser and four Thr residues (Fig. 1a). Strikingly, these residues are highly conserved in SOBIR1 from various plant species, including tomato (Supplementary Figs. 1 and 2a).
Fig. 1. Activation of the SOBIR1/BAK1-containing immune complex by trans-phosphorylation events between SOBIR1 and BAK1.
a Schematic diagram of the kinase domain of NbSOBIR1. The amino acid sequence of the activation segment of NbSOBIR1 is shown below the diagram. Possible phosphorylation sites are denoted in red. b–e Complementation with NbSOBIR1 T522A fails to restore Avr4/Cf-4-triggered HR (b, c), MAPK activation (d), and ROS burst (e) in N. benthamiana:Cf-4 sobir1 knock-out plants. The development of HR was imaged (b) and quantified (c) at 5 dpi. Statistical significance was determined by a one-way ANOVA/Dunnett’s multiple comparison test, compared with NbSOBIR1 WT. Dots indicate individual values (centre line, median; error bar, minima and maxima; n = 6). Similar to NbSOBIR1 WT, all tested NbSOBIR1 Ser/Thr mutants fully restored the Avr4/Cf-4-triggered ROS production in this complementation study, except for NbSOBIR1 T522A. Only the results for NbSOBIR1 WT, T522A and D482N are shown. ROS production is expressed as relative light units (RLUs), and the data are represented as mean + SEM (n = 12). f Thr522 is required for the intrinsic kinase activity of NbSOBIR1. After SDS-PAGE of the E. coli lysates, the recombinant GST-NbSOBIR1 cytoplasmic kinase domain and its various mutants were stained with Coomassie brilliant blue (CBB) (bottom panel), whereas their accumulation was detected by western blotting (middle panel), and phosphorylation status was determined by performing a Pro-Q Diamond stain (top panel). g NbSOBIR1 WT directly phosphorylates kinase-dead NbBAK1 D418N. h NbBAK1 WT directly phosphorylates kinase-dead NbSOBIR1 D482N. Non-fused GST and His tags served as negative controls. Bands with the expected sizes are indicated with an asterisk. All experiments were repeated at least three times with similar results, and representative results are shown. Source data are provided as a Source Data file.
Cf-4 is functional in N. benthamiana and agro-infiltration of Avr4 in stable transgenic N. benthamiana:Cf-4 plants triggers a typical HR39. Cf-4 function is completely abolished in N. benthamiana:Cf-4 sobir1 knock-out lines, whereas complementation through transient expression of functional NbSOBIR1 restores the Avr4/Cf-4-mediated HR in such knock-out lines40. Based on these observations, we carried out site-directed mutagenesis of the activation segment of NbSOBIR1 to substitute individual Ser/Thr residues with alanine (Ala/A) residue, which lacks the phosphorylatable hydroxyl group and thereby cannot be phosphorylated. Subsequently, we performed a complementation study with the five different NbSOBIR1 mutants, taking along the NbSOBIR1 WT and the corresponding D to N (asparagine/Asn) kinase-dead mutant (D482N), as a positive and negative control, respectively41. Interestingly, in contrast to NbSOBIR1 WT but similar to NbSOBIR1 D482N, NbSOBIR1 T522A failed to restore the Avr4/Cf-4-specific HR in N. benthamiana:Cf-4 sobir1 knock-out plants (Fig. 1b). It is worth noting that this phenotype was not caused by a lack of accumulation of the NbSOBIR1 T522A protein in planta (Supplementary Fig. 3). Quantification of the intensity of the HR, which was determined by employing red light imaging42, showed that the intensity of the HR obtained upon transient co-expression of NbSOBIR1 T522A with Avr4 was significantly lower than the intensity of the HR obtained when co-expressing NbSOBIR1 WT with Avr4 (Fig. 1c). The four additional mutants of NbSOBIR1 showed a complementation capacity that was similar to NbSOBIR1 WT.
Rapid and transient activation of MAPK cascades is a critical downstream event in the resistance of plants to pathogens19,43. To determine whether Avr4/Cf-4-triggered MAPK activation in N. benthamiana also requires NbSOBIR1 Thr522, we transiently co-expressed the mutants NbSOBIR1 T522A (and NbSOBIR1 WT and D482N as a positive and negative control, respectively) with Avr4 in the leaves of an N. benthamiana:Cf-4 sobir1 knock-out line, and subsequently detected possible MAPK activation by incubating western blots of a total protein extract with p42/p44-erk antibodies. Similar to the negative kinase-dead control but in contrast to the positive WT control, complementation with NbSOBIR1 T522A failed to restore the Avr4/Cf-4-induced MAPK activation in the sobir1 knock-out line (Fig. 1d).
The swift production of ROS is another hallmark of the plant immune response and apoplastic ROS are mainly produced by nicotinamide adenine dinucleotide phosphate (NADPH) oxidases, such as the RESPIRATORY BURST OXIDASE HOMOLOGUE B (RBOHB) from N. benthamiana and tomato, which localizes at the plasma membrane43–45. We have shown that the Avr4 protein triggers a biphasic ROS burst in N. benthamiana:Cf-4, while this biphasic ROS burst is eliminated in an N. benthamiana:Cf-4 sobir1 knock-out line40. To examine whether NbSOBIR1 activation segment phosphorylation is crucial for mediating the Avr4-triggered ROS burst, we transiently expressed each NbSOBIR1 mutant in the leaves of N. benthamiana:Cf-4 sobir1 plants, after which the ROS production of discs taken from these leaves was monitored upon adding the Avr4 protein. Intriguingly, in contrast to the other four NbSOBIR1 mutants and the positive control (NbSOBIR1 WT), complementation with neither NbSOBIR1 T522A nor the negative control (NbSOBIR1 D482N) restored the Avr4-triggered ROS burst in N. benthamiana:Cf-4 sobir1 knock-out line (Fig. 1e).
Consistently, the analogous residues of NbSOBIR1 Thr522 in tomato SOBIR1, which are SlSOBIR1 Thr513 and SlSOBIR1-like Thr526, gave the same phenotype after complementation with their Thr-to-Ala mutants in N. benthamiana:Cf-4 sobir1 knock-out plants (Supplementary Fig. 2b–i). These residues are overall highly conserved in SOBIR1 from various plant species (Fig. 1a and Supplementary Fig. 2a), which suggests that this specific Thr residue might be crucial for the functionality of SOBIR1 in all plant species.
SOBIR1 and BAK1 trans-phosphorylate each other in vitro
Thr522 is located at the activation segment of the kinase domain of NbSOBIR1 and plays an important role in Cf-4/SOBIR1-initiated plant immunity. That led us to hypothesize this residue is crucial for the intrinsic kinase activity of SOBIR1, and thereby for its auto-phosphorylation. This SOBIR1 auto-phosphorylation represents step 1 in the model that was proposed by van der Burgh et al.41, in which SOBIR1 and BAK1 act together in immune signalling. To test whether this is the case, we employed in vitro phosphorylation assays. The N-terminally GST-tagged cytoplasmic kinase domain from NbSOBIR1, as well as this domain from various NbSOBIR1 mutants, were expressed in Escherichia coli, which was followed by SDS-PAGE of the E. coli lysates, Coomassie brilliant blue (CBB) staining (for determining the accumulation levels of the various mutant kinase domains) and Pro-Q staining (to determine the phosphorylation state of the different kinase domains)46. Successful production of the various NbSOBIR1 kinase domains was confirmed by western blotting, using SOBIR1 antibodies (Fig. 1f). NbSOBIR1 WT exhibited strong auto-phosphorylation activity, similar to its homologue in Arabidopsis38. Interestingly, when compared to NbSOBIR1 WT, the auto-phosphorylation activity of NbSOBIR1 T522A was completely abolished, similar to that of the kinase-dead D482N mutant, suggesting that indeed NbSOBIR1 Thr522 is essential for the intrinsic kinase activity, and thereby the auto-phosphorylation, of NbSOBIR1 (Fig. 1f). In line with this observation for NbSOBIR1 T522A, for SlSOBIR1 T513A and SlSOBIR1-like T526A also a loss of their intrinsic kinase activity was observed (Supplementary Fig. 4a, b). Altogether, these results demonstrate that this specific Thr residue is essential for SOBIR1 intrinsic kinase activity.
To elucidate whether SOBIR1 can directly phosphorylate BAK1, corresponding to the proposed SOBIR1 to BAK1 trans-phosphorylation step 2 in the model of van der Burgh et al.41, we performed an in vitro phosphorylation assay. The GST-tagged NbSOBIR1 WT cytoplasmic kinase domain, or its corresponding kinase-dead mutant, was co-expressed with the His-tagged cytoplasmic kinase domain of the NbBAK1 kinase-dead mutant. The result showed that, first of all, the kinase-dead mutant of NbBAK1, NbBAK1 D418N, of which the conserved “RD” motif in the catalytic loop is changed into “RN”, did not have intrinsic auto-phosphorylation activity, as a Pro-Q stain was negative for this mutant when combined with NbSOBIR1 D482N (Fig. 1g). However, this mutant was properly phosphorylated by kinase-active NbSOBIR1 WT, as visualized by a positive Pro-Q stain for NbBAK1 D418N (Fig. 1g). Similarly, the tomato homologue of NbBAK1, SlBAK1, was also directly phosphorylated by SlSOBIR1 WT, as well as by SlSOBIR1-like WT in vitro, as in both cases SlBAK1 D418N was properly phosphorylated. Also here, this trans-phosphorylation fully depended on the intrinsic kinase activity of SlSOBIR1 WT and SlSOBIR1-like WT, as their corresponding “RN” kinase-dead mutants did not phosphorylate SlBAK1 D418N (Supplementary Fig. 4c, d).
We next sought to determine whether BAK1 WT can directly phosphorylate SOBIR1, an event that corresponds to the proposed BAK1 to SOBIR1 trans-phosphorylation step 3 in the model of van der Burgh et al.41. Earlier, we already showed that the strong auto-phosphorylation activity of E. coli-produced NbSOBIR1 was eliminated in its “RN” kinase-dead mutant (Fig. 1f). Therefore, we performed an additional in vitro phosphorylation assay by co-expressing NbSOBIR1 D482N with either NbBAK1 WT or its kinase-dead mutant. Importantly, NbSOBIR1 D482N was phosphorylated when co-expressed with NbBAK1 WT, but not when co-expressed with NbBAK1 D418N, which demonstrates that indeed NbSOBIR1 can be trans-phosphorylated by NbBAK1 (Fig. 1h). Consistently, SlSOBIR1 D473N and SlSOBIR1-like D486N were also directly phosphorylated by SlBAK1 WT (Supplementary Fig. 4e, f). Notably, trans-phosphorylation of SOBIR1 by BAK1 also required intrinsic kinase activity of BAK1, as phosphorylation of kinase-dead SOBIR1 did not take place when co-expressed with the BAK1 kinase-dead mutant (Fig. 1h, and Supplementary Fig. 4e, f).
NbSOBIR1 Tyr469 is essential for mounting the Avr4/Cf-4-triggered HR and MAPK activation, but is not required for ROS production and its intrinsic kinase activity
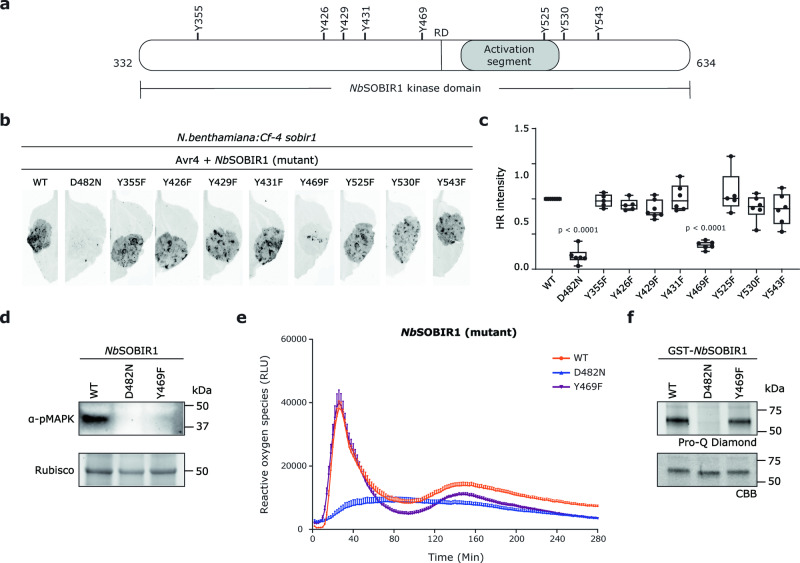
To determine whether particular Tyr residues that are present in the kinase domain of NbSOBIR1 are specifically required for the Avr4/Cf-4-triggered HR, all eight Tyr residues present in the kinase domain of NbSOBIR1 were selected to be studied (Fig. 2a). Strikingly, most of these Tyr residues are highly conserved in SOBIR1 from many different plant species (Supplementary Figs. 5 and 6). We conducted a site-directed mutagenesis involving the substitution of each of the eight Tyr residues of NbSOBIR1 by phenylalanine (Phe/F), which lacks the phosphorylatable hydroxyl group at the aromatic ring. Each NbSOBIR1 mutant was co-expressed with Avr4 in N. benthamiana:Cf-4 sobir1 knock-out plants, including WT as a positive control and D482N as a negative control. Intriguingly, in contrast to the WT, but very similar to the kinase-dead mutant, transient expression of NbSOBIR1 Y469F failed to fully complement the Avr4-triggered HR in N. benthamiana sobir1 knock-out plants (Fig. 2b). Quantification of the HR intensity that was obtained upon complementation by red light imaging42, showed that the difference in the HR intensity between NbSOBIR1 WT and NbSOBIR1 Y469F was significant (Fig. 2c). Furthermore, the remaining Tyr mutants showed a similar complementation capacity in N. benthamiana:Cf-4 sobir1 knock-out plants to the positive control (Fig. 2c).
Fig. 2. Tyr469 of the kinase domain of SOBIR1 is crucial for the Avr4/Cf-4-triggered HR and MAPK activation, but not for ROS production and intrinsic kinase activity.
a Schematic diagram of the kinase domain of NbSOBIR1, with the location of the activation segment, the RD motif, and all Tyr (Y) residues indicated. b–d Complementation with NbSOBIR1 Y469F fails to restore Avr4/Cf-4-triggered HR and MAPK activation in N. benthamiana:Cf-4 sobir1 knock-out plants. The development of an HR was imaged (b) and quantified (c) at 5 dpi. Statistical significance was determined by a one-way ANOVA/Dunnett’s multiple comparison test, compared with NbSOBIR1 WT. Dots indicate individual values (centre line, median; error bar, minima and maxima; n = 6). e Transient expression of NbSOBIR1 Y469F restores the Avr4/Cf-4-triggered ROS accumulation in N. benthamiana:Cf-4 sobir1 knock-out plants. Similar to NbSOBIR1 WT, all tested NbSOBIR1 Tyr mutants restored the Avr4/Cf-4-triggered ROS production in this complementation study. Only the results for NbSOBIR1 WT, Y469F and D482N are shown. ROS production is expressed as RLUs, and the data are represented as mean + SEM (n = 8). f NbSOBIR1 Y469F exhibits intrinsic kinase activity. All experiments were repeated at least three times with similar results, and representative results are shown. Source data are provided as a Source Data file.
To further explore the importance of NbSOBIR1 Tyr469 in Avr4/Cf-4-mediated immune signalling, we determined the occurrence of Avr4/Cf-4-induced MAPK activation in N. benthamiana:Cf-4 sobir1 knock-out plants, after co-expressing NbSOBIR1 Y469F with Avr4. NbSOBIR1 WT was taken along as a positive control, and the kinase-dead mutant was taken along as a negative control. In contrast to the positive control, but similar to the negative control, co-expression of NbSOBIR1 Y469F with Avr4 failed to restore the Avr4/Cf-4-triggered MAPK activation in N. benthamiana:Cf-4 sobir1 knock-out plants (Fig. 2d). Again, the lack of MAPK activation was not correlated with its protein accumulation in planta (Supplementary Fig. 7).
To investigate whether NbSOBIR1 Tyr469 is also required for regulating the Avr4/Cf-4-triggered ROS burst, we monitored the ROS accumulation in the leaves of N. benthamiana:Cf-4 sobir1 knock-out plants in which we transiently expressed the individual SOBIR1 mutants, upon adding Avr4 protein. Surprisingly, similar to NbSOBIR1 WT and all other Tyr-to-Phe mutants, transient expression of NbSOBIR1 Y469F also partially restored the Avr4-triggered ROS burst in N. benthamiana:Cf-4 sobir1 knock-out plants (Fig. 2e). These results suggest that NbSOBIR1 Tyr469 is not essential for the Avr4/Cf-4-triggered ROS burst.
Transient expression of NbSOBIR1 Y469F at least partially restored the Avr4-triggered ROS production in N. benthamiana:Cf-4 sobir1 knock-out plants (Fig. 2e), which led us to speculate that this Tyr-to-Phe mutant would still exhibit intrinsic kinase activity. As expected, the recombinant NbSOBIR1 Y469F still showed strong auto-phosphorylation activity in vitro (Fig. 2f). It is worth noting that we did not observe obvious differences between the staining intensities of the bands upon Pro-Q staining or between the protein mobilities of this mutant and NbSOBIR1 WT (Fig. 2f). Therefore, we conclude that NbSOBIR1 Tyr469 plays no, or only a minor role, in determining the intrinsic kinase activity of NbSOBIR1.
Consistently, the analogous residues of NbSOBIR1 Tyr469 in tomato SOBIR1s, SlSOBIR1 Tyr460 and SlSOBIR1-like Tyr473, gave a similar phenotype after complementation with their Tyr-to Phe mutants (Supplementary Figs. 6, 7 and 8). Taken together, these observations demonstrate that this particular conserved Tyr residue in the kinase domain of SOBIR1 plays an essential role in mediating specific Avr4/Cf-4-triggered plant immune responses.
Members of N. benthamiana RLCK-VII-6, -7 and -8 are differentially required for the Avr4/Cf-4-triggered biphasic ROS burst
To search for RLCKs downstream of the Cf-4/SOBIR1/BAK1 complex in Solanaceous plants, we performed a phylogenetic analysis to identify Arabidopsis BIK1 homologues in Arabidopsis, tomato, and N. benthamiana (Supplementary Fig. 9a). All the RLCKs were further assigned to six subfamilies, which are referred to as subfamily 4, 5, 6, 7, 8 and 9, and correspond to the RLCK-VII subfamilies in Arabidopsis that were previously reported by Rao and colleagues (Supplementary Fig. 9b)37. To identify the RLCKs that play a role in cytoplasmic immune signalling downstream of the activated Cf-4/SOBIR1/BAK1 complex, and to cope with the consequences of functional gene redundancy, we attempted to implement the CRISPR/Cas9 system in N. benthamiana:Cf-4 to simultaneously target multiple RLCK-encoding genes belonging to the same subfamily (Supplementary Fig. 10 and Supplementary Table 1). N. benthamiana RLCK-VII-6 contains 14 members, which might be beyond the limit of the current multiplex CRISPR/Cas9 system (Supplementary Fig. 9b). Hence, the RLCK genes for which we had expression data and of which the expression was not down-regulated, were selected to be knocked out. Expression data were obtained from N. benthamiana:Cf-4 plants also being transgenic for the Rx resistance gene against potato virus X (PVX), in which Avr4 and the COAT PROTEIN (CP) of PVX, which matches Rx and also triggers an HR, was transiently expressed. Furthermore, the constitutively active NB-LRR PROTEIN REQUIRED FOR HR-ASSOCIATED CELL DEATH 1 (NRC1) D481V mutant, which triggers an elicitor-independent HR in N. benthamiana, was also transiently expressed (Supplementary Fig. 11 and Supplementary Table 1)47,48.
We performed ROS burst and HR assays on the transgenic plants of the T1 generation to identify the RLCK subfamilies that are required for the Avr4/Cf-4-mediated immune signalling pathway in N. benthamiana. As we observed before, the Avr4 protein triggered a biphasic ROS burst in leaf discs obtained from N. benthamiana:Cf-4 plants40. This biphasic ROS burst was generally reduced in all the rlck-vii-6 and rlck-vii-7 knock-out lines, and two of the rlck-vii-8 knock-out lines (Supplementary Fig. 12). In addition, flg22-induced ROS accumulation was dampened to different levels in all the rlck-vii-6 knock-out lines and was almost eliminated in some of the rlck-vii-7 and rlck-vii-8 knock-out lines (Supplementary Fig. 13). This observation indicates that members of RLCK-VII-6, -7, and -8 probably play an important role in regulating the Avr4/Cf-4- and flg22-triggered ROS burst in N. benthamiana. Additionally, infiltration of pure Avr4 protein in leaves of N. benthamiana:Cf-4 induces HR activation, which can be detected and quantified by red light imaging42. This HR intensity was significantly attenuated in all the rlck-vii-7 knock-out lines, but not in the other rlck-vii lines (Supplementary Fig. 14), demonstrating that in addition to their role in ROS production, members from RLCK-VII-7 might also be pivotal for the Avr4/Cf-4-triggered HR.
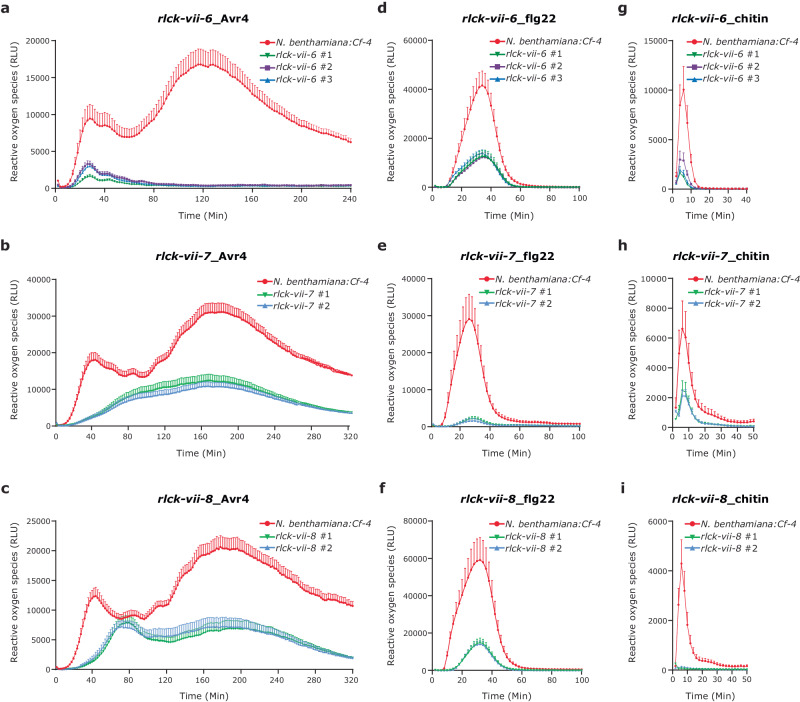
We then selected three independent homozygous rlck-vii-6 knock-out lines (rlck-vii-6 #1, rlck-vii-6 #2, and rlck-vii-6 #3), two rlck-vii-7 knock-out lines (rlck-vii-7 #1 and rlck-vii-7 #2), and two rlck-vii-8 knock-out lines (rlck-vii-8 #1 and rlck-vii-8 #2) to further verify these results (Supplementary Fig. 15). Of note, these knock-out plants did not display any obvious changes in plant overall morphology, when compared to N. benthamiana:Cf-4 (Supplementary Fig. 15). Again, we observed that the intensity of the first, early phase of the Avr4/Cf-4-triggered ROS burst was strongly reduced, whereas the second, more sustained phase of the ROS burst was completely absent in the three independent N. benthamiana:Cf-4 rlck-vii-6 knock-out lines (Fig. 3a). This observation indicates that members of RLCK-VII-6 play an important role in regulating the Avr4/Cf-4-triggered ROS burst in N. benthamiana, especially concerning the second, more sustained phase of the ROS burst. Interestingly, unlike the rlck-vii-6 knock-out plants, rlck-vii-7 knock-out plants displayed an overall reduction of the Avr4/Cf-4-triggered ROS burst, and instead of the typical biphasic ROS, only a weak and sustained ROS burst was observed (Fig. 3b). More interestingly, with rlck-vii-8 knock-out plants, even though the first and second ROS peaks triggered by Avr4/Cf-4 were strongly reduced in size, they were still distinguishable, and additionally, the first ROS peak displayed an obvious delay (Fig. 3c). These results are different from what we observed with the rlck-vii-6 and rlck-vii-7 knock-out plants, which demonstrate that there are different downstream ROS regulatory mechanisms and different RLCK-VII subfamilies are employed in different ways to regulate the Avr4/Cf-4-triggered biphasic ROS burst in N. benthamiana.
Fig. 3. Members of RLCK-VII-6, -7, and -8 differentially contribute to ROS accumulation in N. benthamiana induced by various ExIPs.
N. benthamiana:Cf-4 rlck-vii-6, rlck-vii-7, and rlck-vii-8 homozygous knock-out lines show a reduced ROS production when compared to N. benthamiana:Cf-4 (the positive control), after treatment with 0.1 μM Avr4 protein (a–c), 0.1 μM flg22 (d–f), and 10 μM chitin (g–i). ROS production is expressed as RLUs, and the data are represented as mean + SEM (n = 12 for Avr4 treatment, n = 8 for flg22 and chitin treatments). All experiments were repeated at least three times with similar results, and representative results are shown. Source data are provided as a Source Data file.
Members of RLCK-VII-6, -7, and -8 contribute to ROS accumulation in N. benthamiana induced by various ExIPs
Consistent with the aforementioned results, ROS production triggered by the flg22 peptide was strongly compromised in all these knock-out plants, especially in rlck-vii-7 knock-out plants, when compared to the positive control (Fig. 3d–f). The importance of these RLCK-VII subfamilies in positively regulating ROS accumulation led us to determine their role in regulating ROS burst induced by other ExIPs, such as chitin. Strikingly, all the independent knock-out lines also exhibited an obvious reduction in chitin-triggered ROS production, especially rlck-vii-8 knock-out lines, when compared to the positive control (Fig. 3g–i). The Arabidopsis LRR-RLPs RLP23 and RLP42 perceive a conserved 20-amino-acid peptide from necrosis and ethylene-inducing peptide 1 (NEP1)-like proteins (nlp20) and Botrytis cinerea endo-polygalacturonases (PGs and their derived peptide, pg13), respectively20,49,50. Similar to Cf-4, both RLP23 and RLP42 require SOBIR1 and BAK1 to initiate immune signalling upon recognition of their matching elicitor14,20,49,50. Interestingly, N. benthamiana plants that overexpress either RLP23 or RLP42 show sensitivity to their corresponding ExIPs. To decipher whether members of RLCK-VII-6, -7, and -8 also play a role downstream of RLP23 and RLP42, we transiently overexpressed them in the different knock-out lines, followed by adding the corresponding ligands and monitoring ROS production. Interestingly, similar to the situation with Avr4/Cf-4, both the nlp20/RLP23- and pg13/RLP42-mediated ROS burst in N. benthamiana plants was biphasic, of which the first burst was rapid with a relatively high amplitude, whereas the second burst was sustained, with relatively low amplitude (Supplementary Fig. 16). Both the nlp20/RLP23- and the pg13/RLP42-triggered ROS production was significantly impaired in all knock-out plants, when compared to the positive control (Supplementary Fig. 16). This was especially the case for the second burst in all rlck-vii-6 knock-out plants, which was completely abolished (Supplementary Fig. 16a, b). Collectively, these results indicate that the members of RLCK-VII-6, -7, and -8 are required for regulating the ROS production triggered by a broad spectrum of ExIPs in N. benthamiana.
Members of RLCK-VII-7 play an important role in the Avr4/Cf-4-induced HR and in resistance of N. benthamiana to Phytophthora palmivora
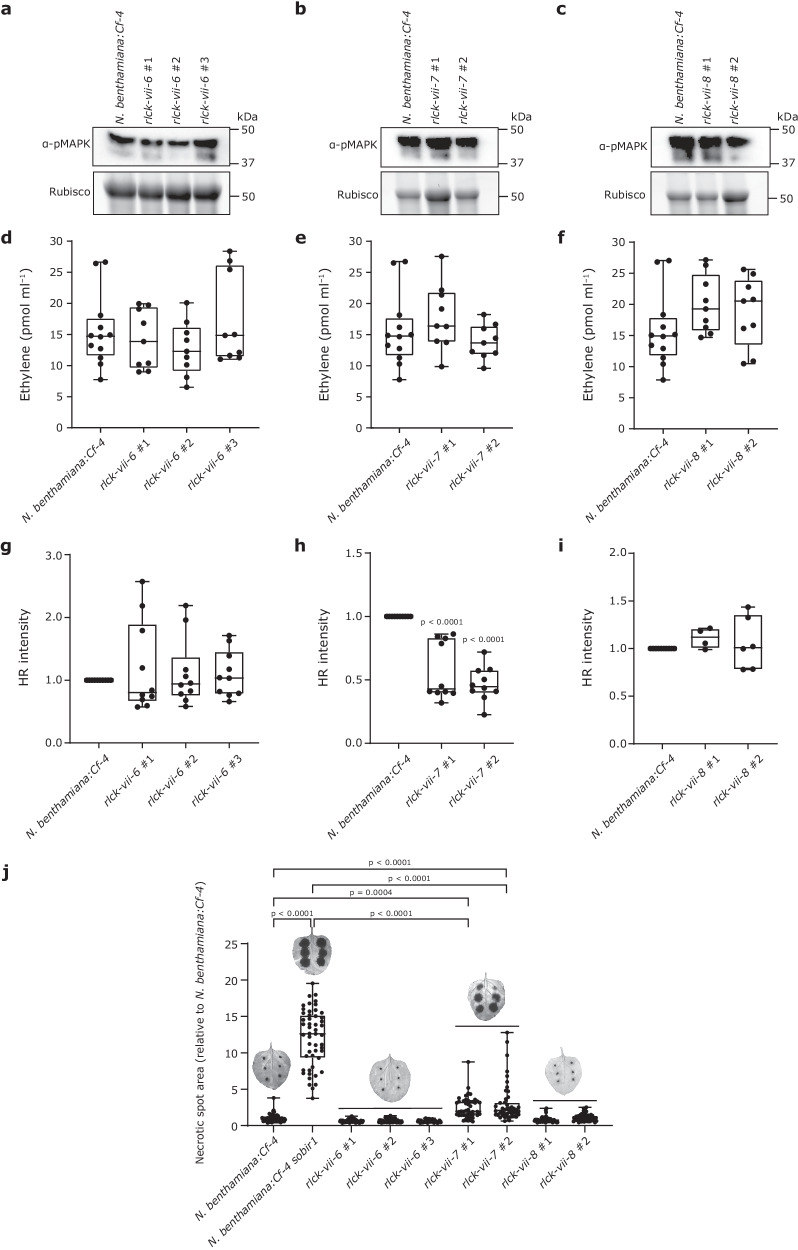
In addition to ROS production, the activation of a downstream MAPK cascade is another key signalling event of ExTI43,51. We observed that infiltration of the Avr4 protein in leaves of N. benthamiana:Cf-4 induces the activation of a MAPK cascade within five minutes (Supplementary Fig. 17). To explore whether members of RLCK-VII-6, -7, and -8 are also involved in Avr4/Cf-4-triggered MAPK activation, we infiltrated pure Avr4 protein in leaves of the different knock-out lines as well as in leaves of N. benthamiana:Cf-4 plants. The leaf samples were harvested at 15 min after Avr4 infiltration, followed by performing a western blot assay revealing MAPK activation. Intriguingly, no obvious changes in MAPK activation in all the knock-out lines were observed at the tested time point when compared to N. benthamiana:Cf-4, suggesting that these RLCK members are dispensable for the Avr4/Cf-4-triggered MAPK activation (Fig. 4a–c).
Fig. 4. Members of RLCK-VII-7 play an important role in the Avr4/Cf-4-induced HR and host resistance against Phytophthora palmivora.
N. benthamiana:Cf-4 rlck-vii-6, rlck-vii-7, and rlck-vii-8 knock-out plants do not exhibit an altered Avr4/Cf-4-triggered MAPK activation (a–c) and ethylene production (d–f), when compared to N. benthamiana:Cf-4 plants. a–c, Leaf samples were harvested at 15 min after infiltration with 5 μM Avr4 protein. Total protein extracts were run on SDS gel and subjected to immunoblotting employing a p42/p44-erk antibody specifically detecting phosphorylated, and thereby activated, MAPKs (α-pMAPK). d–f Ethylene production by the various rlck knock-out plants was measured at 4 h after treatment with 2.5 μM Avr4 protein. Data points are indicated as dots from three independent experiments and plotted as box plots (centre line, median; error bar, minima and maxima; one-way ANOVA/Dunnett’s multiple comparison test; n = 9). g–i The HR intensity triggered by Avr4 (5 μM) in leaves of rlck-vii-7 knock-out plants is significantly reduced when compared to the intensity of the HR in leaves of N. benthamiana:Cf-4 plants. The HR was imaged by the Chemidoc and the HR intensity was quantified by Image Lab, at 2 dpi. Statistical significance was determined by a one-way ANOVA/Dunnett’s multiple comparison test, compared with N. benthamiana:Cf-4. Dots indicate individual values (centre line, median; error bar, minima and maxima; n = 6). j Knocking out SOBIR1 or members of RLCK-VII-7 enhances N. benthamiana susceptibility to P. palmivora. Lesion areas on leaves of 5-week-old N. benthamiana plants were quantified at 2 days post inoculation. Values are given relative to N. benthamiana:Cf-4. Statistical significance was assessed using one-way ANOVA with Tukey’s HSD (Honestly Significant Difference) post-hoc test. Dots indicate individual values (N = 45). Letters indicate statistical groupings. Experiments were repeated three times with similar results. Source data are provided as a Source Data file.
Ethylene is an important signalling molecule that orchestrates plant defence against microbial pathogens. We then examined whether this ethylene production is affected by knocking out the different RLCK-VII subfamilies. For this, independent homozygous N. benthamiana:Cf-4 rlck-vii-6, rlck-vii-7, and rlck-vii-8 knock-out lines, as well as N. benthamiana:Cf-4, were treated with 2.5 μM Avr4 protein, followed by measuring ethylene accumulation. We observed that Avr4 triggers a strong ethylene production in N. benthamiana:Cf-4. The different knock-out lines displayed no significant alterations in ethylene production (Fig. 4d–f), indicating that these RLCKs play no, or only a minor role, in regulating the Avr4/Cf-4-induced ethylene production. It is worth noting that the rlck-vii-8 knock-out lines showed a slightly increased ethylene production, implying that members of RLCK-VII-8 might be inhibitors of ethylene signalling, triggered by Avr4/Cf-4 (Fig. 4f).
To verify that members of RLCK-VII-7 are required for the Cf-4/Avr4-triggered HR in N. benthamiana, we infiltrated pure Avr4 protein in leaves of these independent homozygous knock-out lines and of N. benthamiana:Cf-4 as a control. Consistent with what we observed before, Avr4 protein infiltration revealed significant changes in the capacity to mount an Avr4/Cf-4-triggered HR of the rlck-vii-7, but not the rlck-vii-6 and rlck-vii-8 knock-out plants, when compared to N. benthamiana:Cf-4 (Fig. 4g–i). These results suggest that members of RLCK-VII-7 play an essential role in regulating the Avr4/Cf-4-triggered HR.
To investigate the actual role of these RLCKs in host defence against microbial pathogens, we inoculated leaves from all the independent rlck knock-out lines with the oomycete pathogen Phytophthora palmivora. We used N. benthamiana:Cf-4 sobir1 as a positive control, as SOBIR1 was already shown earlier to be required for resistance of tomato and N. benthamiana to several Phytophthora species52,53. In agreement with these earlier observations, N. benthamiana:Cf-4 sobir1 plants were indeed more susceptible to P. palmivora (Fig. 4j). Interestingly, we found that N. benthamiana:Cf-4 rlck-vii-7 knock-out plants were also more susceptible to P. palmivora infection than N. benthamiana:Cf-4 and the other rlck-vii knock-out plants, but to a lower extent than the N. benthamiana:Cf-4 sobir1 plants (Fig. 4j). These observations demonstrate that SOBIR1 and members of RLCK-VII-7 contribute to the immune response of N. benthamiana to P. palmivora.
SOBIR1 and BAK1 directly phosphorylate members of RLCK-VII-6, -7 and -8 in vitro
We next asked whether these important RLCK members might be trans-phosphorylated by the activated Cf-4/SOBIR1/BAK1 complex. Both SOBIR1 and BAK1 exhibit strong trans-phosphorylation activity (Fig. 1 and Supplementary Fig. 4), which provides the possibility that their kinase domains might be able to directly trans-phosphorylate these RLCKs. To verify this hypothesis, an in vitro phosphorylation assay was conducted.
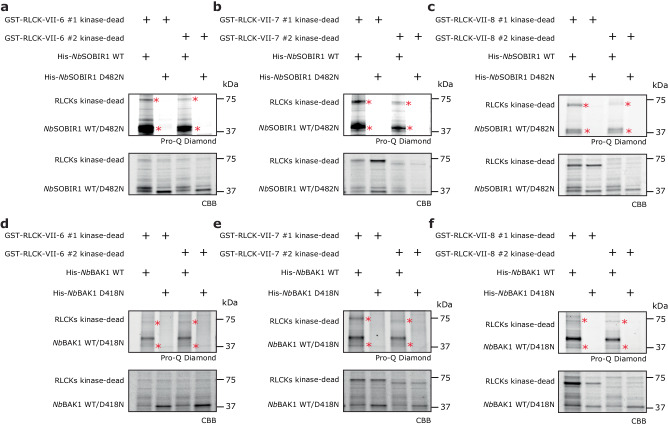
Two members were randomly selected from each of the RLCK-VII-6, -7, or -8 subfamilies, and their kinase-dead mutants were co-expressed with either the cytoplasmic kinase domain of NbSOBIR1 WT or NbBAK1 WT in E. coli, with NbSOBIR1 D482N (a kinase-dead mutant of NbSOBIR1) or NbBAK1 D418N (a kinase-dead mutant of NbBAK1), as a negative control. The proteins were run on SDS-PAGE gels and a CBB stain showed that all proteins accumulated properly in vitro (Fig. 5a–f). In line with the aforementioned observations, both NbSOBIR1 WT and NbBAK1 WT showed strong auto-phosphorylation, while their kinase-dead mutants did not. Importantly, no phosphorylated proteins were observed when the kinase-dead mutants of the selected RLCK members were co-expressed with either NbSOBIR1 D482N or NbBAK1 D418N, while these RLCKs did get phosphorylated when being co-expressed with NbSOBIR1 WT and NbBAK1 WT (Fig. 5a–f). These results demonstrate that both SOBIR1 and BAK1 indeed directly trans-phosphorylate members from the RLCK-VII subfamilies -6, -7 and -8, and that these trans-phosphorylation events rely on the kinase activity of SOBIR1 and BAK1. Likely, the trans-phosphorylation of these RLCKs by SOBIR1 and BAK1 is required for the activation of these RLCKs and subsequent initiation of the downstream signalling pathway.
Fig. 5. Members of N. benthamiana RLCK-VII-6, −7, and −8 are directly trans-phosphorylated by both SOBIR1 and BAK1 in vitro.
Two members were randomly selected from RLCK-VII-6, -7, and -8 and their kinase-dead mutants were co-expressed with either the cytoplasmic kinase domain from NbSOBIR1 WT or its D482N kinase-dead mutant (a–c), or with either the cytoplasmic kinase domain from NbBAK1 WT or its D418N kinase-dead mutant (d–f), in E. coli. After SDS-PAGE of the boiled cell lysate, the phosphorylation status of the recombinant proteins was determined by performing a Pro-Q Diamond stain (top panels), while the total proteins were stained with CBB (bottom panels). Bands with the expected sizes are indicated with a red asterisk. Experiments were repeated two times with similar results, and representative results are shown. RLCK-VII-6 #1 kinase-dead, Niben101Scf02460g01004 K110A; RLCK-VII-6 #2 kinase-dead, Niben101Scf06739g05004 K110A; RLCK-VII-7 #1 kinase-dead, Niben101Scf00712g13012 K127A; RLCK-VII-7 #2 kinase-dead, Niben101Scf01176g01025 K119A; RLCK-VII-8 #1 kinase-dead, Niben101Scf00012g00012 K109A; RLCK-VII-8 #2 kinase-dead, Niben101Scf06482g03003 K114A. Source data are provided as a Source Data file.
SOBIR1 interacts with various RLCKs in planta
To determine whether the tomato Cf-4/SOBIR1 complex actually interacts with RLCKs in planta, we tested the ability of several RLCKs from tomato to interact with SlSOBIR1 using a split-luciferase assay54. We focussed on 10 tomato RLCKs, of which the expression is at least three times upregulated at three days after inoculation of leaflets of tomato cv. Moneymaker with the necrotrophic fungal pathogen B. cinerea, as compared to mock-inoculated leaflets55. Cluc-tagged RLCKs, as well as AtBIK1-Cluc, were analysed for their ability to interact with Nluc-tagged SlSOBIR1 in planta (Supplementary Fig. 18a–l). GUS-Nluc was included as a negative control. To infer interaction, we focused on the presence of increased luminescence signals when the RLCKs were co-expressed with SlSOBIR1, as compared to GUS, in N. benthamiana. All 10 Cluc-tagged RLCKs were found to properly accumulate in leaves of N. benthamiana upon their transient expression, albeit at different levels (Supplementary Fig. 18m).
The results of the split-luciferase assay (Supplementary Fig. 18a–l) reveal that Solyc07g041940 (Sl41940) (RLCK-VII-6), Solyc11g062400 (Sl62400) (RLCK-VII-9), Solyc01g088690 (Sl88690) (RLCK-VII-4), Solyc05g007050 (Sl7050) (RLCK-VII-5), and Solyc08g077560 (Sl77560) (RLCK-VII-4) do not specifically interact with SlSOBIR1, as the luminescence signals did not really differ from the signals obtained upon their co-expression with the GUS control (Supplementary Fig. 18a, c, e, h–j). On the contrary, Solyc04g082500 (Sl82500) (RLCK-VII-7), Solyc06g062920 (Sl62920) (RLCK-VII-6), Solyc05g025820 (Sl25820) (RLCK-VII-6), Solyc01g112220 (Sl112220) (RLCK-VII-7), and Solyc06G005500 (Sl05500) (RLCK-VII-8) likely do specifically interact with SlSOBIR1, similar to AtBIK1 (Supplementary Fig. 18a, d, f, g, k, l). Sl25820 clusters with AtPBL13 and AtRIPK (Supplementary Fig. 9), which are RLCKs known to be involved in defence34,56. Sl62920 is a homologue of SlACIK1 (Supplementary Fig. 9), which has been shown before to play a role in Cf-mediated immunity to F. fulva57. Sl82500 and Sl112220 in their turn are closely related to Arabidopsis PBL30, PBL31 and PBL32, of which PBL30 (also known as CAST AWAY, CST) plays an important role in RLP1/SOBIR1-, RLP23/SOBIR1-, and RLP42/SOBIR1-mediated immunity (Supplementary Fig. 9)36. Both PBL30 and PBL31 have indeed been shown to interact with SOBIR1/EVR36,58.
Having shown, by a split-luciferase assay in N. benthamiana, that several tomato RLCKs interact in planta with SlSOBIR1, we decided to use an alternative approach to test for interaction between NbSOBIR1 and endogenous RLCKs from N. benthamiana. For this, we performed a series of TurboID (TbID)-based proximity-dependent labelling (PL)-mass spectrometry (MS) experiments59, for which we transiently expressed the fusion proteins NbSOBIR1-YFP-TbID and the control GUS-YFP-TbID in N. benthamiana:Cf-4 sobir1 knock-out plants40.
Interestingly, we identified several peptides of biotinylated RLCKs, meaning that these proteins are in close proximity to the kinase domain of NbSOBIR1 (Supplementary Fig. 19). Strikingly, the RLCK-VII-8 member Niben101Scf00012g00012, being the homologue of tomato Solyc06g005500 (Sl05500) that seems to specifically interact only with SlSOBIR1 in the split-luciferase assay (Supplementary Fig. 18a, l), was also identified. Eventually, we identified four distinct members of RLCK-VII-8, one member of RLCK-VII-6 and peptides matching RLCKs of classes IV and VIII for which we were not able to assign them to distinct subfamily members (Supplementary Fig. 19).
A complementation assay shows that transient expression of RLCK Niben101Scf00012g00012 partially restores the ROS burst
Having observed that several RLCKs interact with NbSOBIR1 in N. benthamiana, we asked whether RLCKs belonging to one particular subfamily are redundant, allowing the transient expression of one of them to restore the biphasic ROS burst, or whether each of them has a non-redundant function, requiring the expression of all subfamily members at the same time to restore this biphasic ROS burst. We chose to perform a complementation assay with the RLCK-VII-8 member Niben101Scf00012g00012 in the rlck-vii-8 #1 knock-out line and observed that transient expression of this RLCK resulted only in a partial restoration of the ROS burst, as a proper biphasic ROS was not observed (Supplementary Fig. 20). This result suggests that all members of a particular RLCK subfamily might have overlapping redundant functions in triggering ROS production, but each of them also has a specific role in mounting this part of the defence response of the plant.
Discussion
SOBIR1 and BAK1 are well-known co-receptors for LRR-RLPs, such as Cf-4, RLP23, and RLP4214,15,18,20,50. It has been reported that an RLP/SOBIR1 complex forms heterodimers with BAK1 upon its elicitation and that subsequent trans-phosphorylation events between the kinase domains of SOBIR1 and BAK1 are required for initiating downstream immune signalling41. Both SOBIR1 and BAK1 are RD kinases, suggesting the possibility of auto-phosphorylation in their activation segment. Here, our in vitro studies demonstrate that, similar to their Arabidopsis homologues, NbSOBIR1, SlSOBIR1, SlSOBIR1-like, NbBAK1 and SlBAK1 exhibit auto-phosphorylation activity (Fig. 1 and Supplementary Fig. 4)38,60,61. Furthermore, SOBIR1 indeed can directly phosphorylate BAK1, and the intrinsic kinase activity of SOBIR1 is required for this trans-phosphorylation event (Figs. 1g and 6, Supplementary Fig. 4c,d). Accordingly, BAK1 is also able to directly phosphorylate SOBIR1, which again depends on the kinase activity of BAK1 (Figs. 1h and 6, and Supplementary Fig. 4e,f). Interestingly, our observations are supported by a recent study, which shows the auto-phosphorylation of AtSOBIR1 via an intermolecular mechanism, and the trans-phosphorylation between AtSOBIR1 and AtBAK1 in vitro62. Previously, we have reported that AtSOBIR1, which constitutively activates immune responses when overexpressed in N. benthamiana, is highly phosphorylated. Kinase activity of both AtSOBIR1 and AtBAK1 is required for both the AtSOBIR1-induced constitutive cell death response and for the phosphorylation of the kinase domain of AtSOBIR141. In addition, transient co-expression of Avr4 with the dominant negative BAK1 mutants AtBAK1C408Y or AtBAK1D416N in N. benthamiana:Cf-4 results in a reduced Avr4/Cf-4-triggered HR41. Therefore, we propose that trans-phosphorylation events between the cytoplasmic kinase domains of SOBIR1 and BAK1 eventually result in the full activation of SOBIR1/BAK1-containing immune complexes, which is essential for LRR-RLP-mediated immunity. As many LRR-RLPs that are involved in plant immunity require SOBIR1 and BAK1 for their function, the model that we proposed earlier41, and is now further supported by this study and by the work of Wei and colleagues62, probably also applies to immune signalling triggered by additional RLP/SOBIR1 complexes. Moreover, increasing evidence has indicated that BAK1 promotes the activation of the receptor complex upon the perception of an ExIP by the LRRs of the matching primary receptor. However, the signalling specificity is determined by the kinase domain of the primary receptor, which is either an RLK or the constitutive RLP/SOBIR1 complex, whereas BAK1 merely acts as a general complex activator63. Therefore, after being trans-phosphorylated by BAK1, SOBIR1 is proposed to initiate the trans-phosphorylation events with downstream cytoplasmic signalling components. Such components are for example particular RLCKs, thereby triggering a specific type of immune signalling, irrespective of the RLP that is involved in the RLP/SOBIR1 complex.
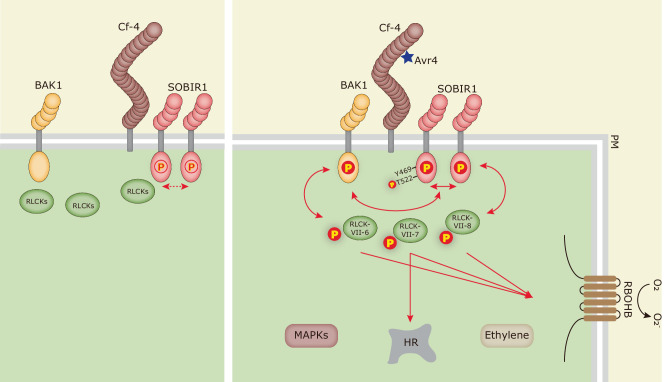
Fig. 6. Working model for the activation of the Cf-4/SOBIR1/BAK1 immune complex, and the initiation of downstream immune signalling by the activated complex.
In the absence of the Avr4 effector (left panel), the resistance protein Cf-4 constitutively interacts with SOBIR1. On the other hand, SOBIR1 constitutively forms homodimers that allow basal activation of the SOBIR1 kinase domain through cross-phosphorylation. In response to the perception of Avr4 by Cf-4 (right panel), SOBIR1 initiates strong auto-phosphorylation of its kinase domain, and the amino acid residue Thr522 is required for its intrinsic kinase activity. Meanwhile, the Cf-4/SOBIR1 complex recruits BAK1, after which trans-phosphorylation events between the cytoplasmic kinase domains of SOBIR1 and BAK1 take place. Members of the N. benthamiana RLCK-VII-6, 7, and 8 are required for the Avr4/Cf-4-triggered ROS burst, whereas members of RLCK-VII-7 are also required for the Avr4/Cf-4-induced HR. SOBIR1 Tyr469 plays an essential role in the Avr4/Cf-4-triggered HR and MAPK activation, but not in ROS accumulation and SOBIR1 intrinsic kinase activity. As this particular residue is predicted to be solvent-exposed, it is proposed that this residue regulates plant immune responses by interacting with specific downstream signalling partners. All selected RLCK-VII members can be directly trans-phosphorylated by both SOBIR1 and BAK1. Likely, these trans-phosphorylation events lead to the activation of these RLCKs, after which they can phosphorylate the RBOHB oxidase, leading to the accumulation of apoplastic ROS. Members of RLCK-VII-7 possibly phosphorylate various transcription factors or other downstream signalling components to trigger the activation of the HR. Solid arrows indicate signalling events with supporting data presented in this study, whereas dashed arrows indicate proposed events. The red open (left panel) and filled (right panel) circles with a ‘P’ inside represent low levels and high levels of phosphorylation, respectively. PM plasma membrane.
Furthermore, in this study, we show that NbSOBIR1 Thr522 and its analogous residues in tomato SOBIR1s (SlSOBIR1 Thr513 and SlSOBIR1-like Thr526), present in the activation segments of SOBIR1, are essential for their intrinsic kinase activity and thereby for the initiation of the Avr4/Cf-4-mediated immune responses (Fig. 1, and Supplementary Figs. 2 and 4). These results are further supported by a recent study, which shows that AtSOBIR1 T529A does not exhibit intrinsic kinase activity, thereby resulting in a complete loss of cell death in N. benthamiana upon its overexpression62. Interestingly, the equivalent Thr residue has been proven essential for many other RD kinases. A good example is Arabidopsis RLK CHITIN ELICITOR RECEPTOR KINASE1 (CERK1) Thr479 (Supplementary Fig. 1), which is indispensable for the activation of the AtCERK1 kinase64,65. Furthermore, the Arabidopsis LRR-RLK NUCLEAR SHUTTLE PROTEIN (NSP)-INTERACTING KINASE1 (NIK1) is a virulence target of the begomovirus NSP and is involved in plant antiviral immunity66,67. A mutation at Thr474, which is located at the activation segment of the kinase domain of NIK1 (Supplementary Fig. 1), attenuates the auto-phosphorylation of NIK1 and enhances susceptibility to the Cabbage leaf curl virus68. Therefore, this particular Thr residue might play a general role in regulating the intrinsic kinase activity of RD kinases.
In addition to Thr522, Tyr469 (as well as its analogous residues in SlSOBIR1 and SlSOBIR1-like) in the kinase domain of NbSOBIR1 has also been identified to be crucial for the Avr4/Cf-4-induced HR and MAPK activation, but not for its intrinsic kinase activity and Avr4/Cf-4-triggered ROS accumulation (Fig. 2, Supplementary Figs. 6 and 8). Strikingly, a vital role in regulating plant immunity has recently been assigned to this particular Tyr residue present in the kinase domain of several well-known RLKs69,70–73. For instance, upon the perception of elf18, the Arabidopsis LRR-RLK EFR phosphorylates at Tyr836, which is equivalent to NbSOBIR1 Tyr469, and this phosphorylation is required for the activation of EFR itself and the initiation of sequential downstream immune responses70. Arabidopsis CERK1 is the co-receptor of the fungal cell wall component chitin74,75. Phosphorylation of CERK1 Tyr428, which is also analogous to NbSOBIR1 Tyr469, is essential for chitin-triggered CERK1 activation, ROS production, MAPK activation, downstream RLCK phosphorylation, and resistance to the fungal pathogen B. cinerea69. These results collectively demonstrate the importance of this particular Tyr residue in the kinase domain of cell-surface RLKs that are involved in plant immunity. Although AtSOBIR1 has been reported to auto-phosphorylate at Tyr residues38, no phosphorylated Tyr residues have been detected by mass spectrometry (MS) in the kinase domain of AtSOBIR1, when auto-phosphorylated, or after being trans-phosphorylated by BAK1 in vitro62, or when produced in planta76. This is in contrast to the aforementioned RLKs, which can phosphorylate at this Tyr residue. Nonetheless, based on the structure of the AtSOBIR1 kinase domain, which has been determined recently62, AtSOBIR1 Tyr476 (equivalent Tyr residue of NbSOBIR1 Tyr469) is solvent-exposed, therefore, it might be easier to access by downstream components or other regulatory proteins (Supplementary Fig. 21). Therefore, this important Tyr residue in the kinase domain of SOBIR1 might not regulate plant immune responses by phosphorylation, as the Tyr469Phe mutant of SOBIR1 is still kinase-active, but by interacting with specific downstream signalling partners, such as members of RLCK-VII-7 that are required for Avr4/Cf-4-induced HR (Fig. 4h).
We observed that the kinetics of the Avr4-induced biphasic ROS burst displayed differently in N. benthamiana:Cf-4 rlck-vii-6, rlck-vii-7 and rlck-vii-8 (Fig. 3a–c). For the rlck-vii-6 knock-out lines, the second burst is specifically and completely inhibited (Fig. 3a); for the rlck-vii-7 knock-out lines, the overall ROS production is strongly attenuated, and only a weak and sustained ROS burst is triggered (Fig. 3b); whereas, for the rlck-vii-8 knock-out lines, both ROS bursts are strongly compromised, with the first burst being delayed (Fig. 3c). This raises the possibility that there are different downstream ROS regulatory mechanisms in N. benthamiana, which together determine the ROS profile. This option is supported by our observation that various RLCKs belonging to different subfamilies interact with the kinase domain of SOBIR1 in planta (Supplementary Figs. 18 and 19) and that these different RLCKs have non-redundant functions (Supplementary Fig. 20). In Arabidopsis, RESPIRATORY BURST OXIDASE HOMOLOG D (RBOHD) is engaged in extracellular ROS production, and growing evidence has suggested that RLCKs differentially regulate RBOHD activation through the differential phosphorylation of various sites in the RBOHD enzyme77. For instance, BIK1 directly phosphorylates the N-terminus of RBOHD to positively regulate ROS production in Arabidopsis, whereas PBL13, which negatively regulates RBOHD activation, directly phosphorylates the C-terminus of RBOHD. In N. benthamiana, the RBOHB homologue is responsible for the fast apoplastic ROS production during the establishment of immunity, and therefore we speculate that members from RLCK-VII-6, RLCK-VII-7 and RLCK-VII-8 phosphorylate RBOHB at different sites, thereby causing different ROS kinetics (Fig. 6)78. Furthermore, it has been reported that in N. benthamiana the first burst of the biphasic apoplastic ROS burst is mediated by swift RBOHB phosphorylation, whereas the second burst is the result of transcriptional upregulation of the RBOHB gene, a process that is mediated by activated WRKY transcription factors79. Hence, we hypothesize that members from RLCK-VII-6, upon their activation by the upstream cell-surface complex, phosphorylate RBOHB at specific sites for the swift ROS burst, and meanwhile directly or indirectly phosphorylating certain transcription factors to regulate the later phase of the ROS burst.
To determine the role of studied RLCKs in the resistance response of N. benthamiana to microbial pathogens, we challenged sobir1 knock-out plants and all the rlck knock-out plants with the oomycete pathogen P. palmivora. Our results demonstrate that SOBIR1 and members of RLCK-VII-7, the latter playing an essential role in positively regulating both the Avr4/Cf-4-triggered ROS burst and the HR, are all involved in immunity to P. palmivora, with SOBIR1 playing a more important role than all members of RLCK-VII-7 together (Fig. 4j). Interestingly, Liang and co-workers80 performed a similar experiment in Arabidopsis, inoculating higher-order rlck-vii mutants with P. capsici. They found that RLCK-VII-6 and RLCK-VII-8 members are required for resistance to this oomycete pathogen. This observation suggests that in different plant species the various RLCK-VII subfamilies play different roles in immunity. Previous studies have shown that some ExIPs (for example INF1 from P. infestans, XEG1 from P. sojae, and ParA1 from P. parasitica), are perceived by RLPs of N. benthamiana and that the HR that is triggered requires both SOBIR1 and BAK152,53,81,82. Therefore, it is very likely that also here an RLP/SOBIR1 complex, which forms the frontline of plant innate immunity, plays a prominent role in warding off invasion by P. palmivora. Indeed, it has been shown that this oomycete pathogen produces conserved elicitins that are highly homologous to for example INF183 and can potentially be recognised by the endogenous N. benthamiana LRR-RLP that is referred to as RESPONSIVE TO ELICITINS (REL)82. The various members of RLCK-VII-7 are probably, partially redundantly with RLCK-VII-6 and -8 members, involved in transducing immune signalling from the activated RLP/SOBIR1/BAK1 complex to downstream signalling partners. Understanding how plants deploy RLCKs to cope with pathogen infection will eventually contribute to breeding crops with durable disease resistance.
Methods
Plant materials
N. benthamiana:Cf-4 sobir1/sobir1-like knock-out plants were used in this study. As SOBIR1-like is not functional in N. benthamiana, we further refer to these knock-out plants as N. benthamiana:Cf-4 sobir140.
A highly efficient multiplex editing technique employed to knock-out multiple RLCK-VII subfamily members in N. benthamiana:Cf-4 has been described previously84. To screen for homozygous transformants, genomic DNA from each mutant plant was isolated by using the Phire Tissue Direct PCR Master Mix (Thermo Fisher Scientific), followed by amplifying the sgRNA-targeted regions and subsequent Sanger sequencing of the obtained PCR fragments. Primers used for genotyping can be found in Supplementary Table 2.
Plant growth conditions
All N. benthamiana plants used in this study were cultivated in a climate chamber under 15 h of light at 21 °C and 9 h of darkness at 19 °C, with a relative humidity of ~70%.
Generation of binary vectors for Agrobacterium tumefaciens-mediated transient transformation
The constructs pBIN-KS-35S::NbSOBIR1-eGFP (SOL2911), pBIN-KS-35S::NbSOBIR1D482N-eGFP (kinase-dead mutant) (SOL7928), pBIN-KS-35S::SlSOBIR1-eGFP (SOL2774), pBIN-KS-35S::SlSOBIR1D473N-eGFP (kinase-dead mutant) (SOL2875), pBIN-KS-35S::SlSOBIR1-like-eGFP (SOL2773), pBIN-KS-35S::SlSOBIR1-like D486N-eGFP (kinase-dead mutant) (SOL2876) and pMOG800-Avr4 have been described previously14. The codon change, resulting in a Ser/Thr-to-Ala or Tyr-to-Phe amino acid change in the kinase domain of SOBIR1s, was introduced by performing overlap extension PCR using the plasmids pENTR/D-Topo:NbSOBIR1 (SOL4064), pENTR/D-Topo:SlSOBIR1 (SOL2746), and pENTR/D-Topo:SlSOBIR1-like (SOL2745) as templates14,85. Phusion Hot Start II DNA Polymerase (Thermo Scientific) was used for the overlap extension PCR and the primers that were used are listed in Supplementary Table 2. The methylated template plasmids remaining in the PCR products were digested by DpnI (NEB), and after transformation to E. coli DH5α, the required SOBIR1 mutants carrying individual mutations were selected by Sanger sequencing, and then introduced into pBIN-KS-35S::GWY-eGFP (SOL2095; for C-terminally tagging with eGFP), by using Gateway LR Clonase II (Invitrogen). Thereby, the binary vectors pBIN-KS-35S::NbSOBIR1T512A-eGFP (SOL7909), pBIN-KS-35S::NbSOBIR1T515A-eGFP (SOL7910), pBIN-KS-35S::NbSOBIR1T516A-eGFP (SOL7911), pBIN-KS-35S::NbSOBIR1S517A-eGFP (SOL7912), pBIN-KS-35S::NbSOBIR1T522A-eGFP (SOL7913), pBIN-KS-35S::SlSOBIR1T503A-eGFP (SOL7969), pBIN-KS-35S::SlSOBIR1T506A-eGFP (SOL7970), pBIN-KS-35S::SlSOBIR1T507A-eGFP (SOL7971), pBIN-KS-35S::SlSOBIR1S508A-eGFP (SOL7972), pBIN-KS-35S::SlSOBIR1T513A-eGFP (SOL7973), pBIN-KS-35S::SlSOBIR1-likeT516A-eGFP (SOL7950), pBIN-KS-35S::SlSOBIR1-likeT519A-eGFP (SOL7951), pBIN-KS-35S::SlSOBIR1-likeT520A-eGFP (SOL7952), pBIN-KS-35S::SlSOBIR1-likeS521A-eGFP (SOL7953), pBIN-KS-35S::SlSOBIR1-likeT526A-eGFP (SOL7954), pBIN-KS-35S::NbSOBIR1Y355F-eGFP (SOL7914), pBIN-KS-35S::NbSOBIR1Y426F-eGFP (SOL7915), pBIN-KS-35S::NbSOBIR1Y429F-eGFP (SOL7916), pBIN-KS-35S::NbSOBIR1Y431F-eGFP (SOL7917), pBIN-KS-35S::NbSOBIR1Y469F-eGFP (SOL7918), pBIN-KS-35S::NbSOBIR1Y525F-eGFP (SOL7919), pBIN-KS-35S::NbSOBIR1Y530F-eGFP (SOL7920), pBIN-KS-35S::NbSOBIR1Y543F-eGFP (SOL7921), pBIN-KS-35S::SlSOBIR1Y346F-eGFP (SOL7974), pBIN-KS-35S::SlSOBIR1Y417F-eGFP (SOL7975), pBIN-KS-35S::SlSOBIR1Y420F-eGFP (SOL7976), pBIN-KS-35S::SlSOBIR1Y422F-eGFP (SOL7977), pBIN-KS-35S::SlSOBIR1Y460F-eGFP (SOL7978), pBIN-KS-35S::SlSOBIR1Y521F-eGFP (SOL7979), pBIN-KS-35S::SlSOBIR1Y522F-eGFP (SOL7980), pBIN-KS-35S::SlSOBIR1Y534F-eGFP (SOL7981), pBIN-KS-35S::SlSOBIR1Y588F-eGFP (SOL7982), pBIN-KS-35S::SlSOBIR1-likeY359F-eGFP (SOL7942), pBIN-KS-35S::SlSOBIR1-likeY430F-eGFP (SOL7943), pBIN-KS-35S::SlSOBIR1-likeY433F-eGFP (SOL7944), pBIN-KS-35S::SlSOBIR1-likeY435F-eGFP (SOL7945), pBIN-KS-35S::SlSOBIR1-likeY473F-eGFP (SOL7946), pBIN-KS-35S::SlSOBIR1-likeY529F-eGFP (SOL7947), pBIN-KS-35S::SlSOBIR1-likeY534F-eGFP (SOL7948) and pBIN-KS-35S::SlSOBIR1-likeY547F-eGFP (SOL7949), for in planta expression were obtained.
To clone the selected tomato RLCK homologues for split-luciferase assays, the open reading frames of the encoding genes were amplified from cDNA obtained from leaves of tomato cv. Moneymaker, inoculated with B. cinerea55. SlSOBIR1 was amplified from pENTR-SlSOBIR114, and GUS was amplified from pENTR-GUS (Invitrogen). PCR reactions were performed using primers with KpnI and SalI restriction sites for directional cloning into Cluc- and Nluc-vectors54 (Supplementary Table 2). The Nluc-vector includes an HA-tag for additional immunoblot analyses (Jian-Min Zhou, personal communication). After confirmation by sequencing, the vectors Solyc07G041940-Cluc (Sol7202), Solyc04G082500-Cluc (Sol7204), Solyc11G062400-Cluc (Sol7206), Solyc06G062920-Cluc (Sol7208), Solyc05G025820-Cluc (Sol7210), Solyc01G088690-Cluc (Sol7212), Solyc05G007050-Cluc (Sol7214), Solyc08G077560-Cluc (Sol7216), Solyc01G112220-Cluc (Sol7218), Solyc06G005500-Cluc (Sol7220), SlSOBIR1-Nluc (Sol6766), and GUS-Nluc (Sol6793) were transformed to A. tumefaciens strain C58C1, carrying the pCH32 helper plasmid. Generation of the AtBIK1-Cluc (Sol6625) construct was described previously86.
The bait-TbID fusion proteins were generated using a gateway-compatible 35S-YFP-TbID expression vector based on pEarleyGate101 (pEG101)87. The vectors GUS-YFP-TbID (SOL9203) and NbSOBIR1-YFP-TbID (SOL9201) were generated by LR reactions from the gateway entry vectors SOL2685 and SOL4064. The primers that were used are listed in Supplementary Table 2.
For the generation of a complementation construct for RLCK Niben101Scf00012g00012, cDNA of N. benthamiana was used to amplify the coding region of the encoding gene with Phusion Hot Start II DNA polymerase, using the primers listed in Supplementary Table 2. The purified PCR product was inserted in pENTR-TOPO and subsequently inserted in the destination vector pBIN-KS-35S::GWY-eGFP (SOL2095), with an LR reaction. The resulting complementation vector was transformed to A. tumefaciens C58C1, carrying the pCH32 helper plasmid.
A. tumefaciens-mediated transient transformation
All binary plasmids were transformed into A. tumefaciens (further referred to as Agrobacterium) strain C58C1, carrying the helper plasmid pCH32. Agrobacterium strains harbouring the transient expression constructs of RLP23 and RLP42 were received from Lisha Zhang and Thorsten Nürnberger20,50. Agrobacterium cells were harvested by centrifugation (3500 × g, 15 min) and resuspended in MMAi (1 L of MMAi: 5 g of MS salts, 1.95 g of MES, 20 g of sucrose, and 200 µM acetosyringone) to a final OD600 of 0.8. After 1 h incubation at room temperature, cultures were infiltrated into the first fully expanded N. benthamiana leaves with a 1-mL disposable syringe88.
The development of cell death was photographed and quantified by red light imaging using the ChemiDoc (Bio-Rad)42. Statistical analysis was performed using one-way ANOVA by GraphPad Prism 9.
Immunoprecipitation (IP)
The protein accumulation level of SOBIR1 mutants and various tomato RLCKs after their transient expression in leaves of N. benthamiana was determined by performing a protein immunoprecipitation assay followed by immunoblotting89. For this, leaf samples were harvested at 2 days post infiltration (dpi) and frozen in liquid nitrogen. Subsequently, the leaf samples were ground to a fine powder, after which pre-cooled extraction buffer (150 mM NaCl, 1.0% IGEPAL CA-630 [NP-40], 50 mM Tris, pH 8.0, plus one protease inhibitor tablet per 50 mL extraction buffer) was added to the leaf powder in a 1 g: 2 mL ratio and mixed thoroughly. Samples were then centrifuged at 4 °C for 15 min at 18,000 × g, and 2 mL of the supernatant was incubated with 15 µL GFP-trap_A beads at 4 °C for 1 h. Hereafter, the beads were collected by centrifugation at 1000 × g for 1 min and washed three times in 1 mL of extraction buffer. After the final wash, SDS loading buffer (200 mM Tris, pH 6.8, 8% SDS, 40% glycerol, 400 mM DTT, 2% Bromophenol blue) was added to the beads and the mixture was boiled at 95 °C for 10 min.
Subsequently, the immunoprecipitated proteins were separated on an SDS-PAGE gel and transferred to a PVDF membrane (Trans-Blot Turbo Transfer Pack, Bio-Rad), using a Trans-Blot Turbo Transfer system (Bio-Rad) (settings: 1.3 A, 25 V, 7 min). The membrane was incubated with TBS-Tween (150 mM NaCl, 20 mM Tris, pH 7.5, 0.1% tween) containing 5% milk powder at room temperature for 1 h or at 4 °C overnight. To detect GFP fusion proteins, the blots were incubated with anti-GFP-HRP (1:5000) (Miltenyi Biotec, 130-091-833). To detect Cluc-tagged proteins, blots were incubated with anti-Cluc (1 µg/mL) (Sigma, L2164) and anti-mouse-HRP (1:10,000) (GE Healthcare).
Reactive oxygen species (ROS) assay
For ROS burst assays, leaf discs were taken from 4-week-old N. benthamiana:Cf-4 and rlck knock-out plants, while for plants transiently expressing SOBIR1 mutants for the complementation studies, RLP23 or RLP42, leaf discs were collected at 24 h after ago-infiltration. Leaf discs were then floated on 80 µL of sterile water in a 96-well plate overnight and hereafter, the water in each well was replaced carefully by 50 µL of fresh sterile water. After another 1 h of incubation, 50 µL of the reaction solution, containing 100 µM of luminol (L-012, Fujifilm, Japan), 20 µg/mL horseradish peroxidase (Sigma), and the elicitor to be tested (being 0.2 µM Avr4 protein, 0.2 μM flg22, 20 μM chitohexaose, 2 μM nlp20 or 2 μM pg13), was added to each well. Subsequently, the production of luminescence was monitored with a CLARIOstar plate reader (BMG Labtech). The line charts showing the detected values of ROS were created using GraphPad Prism 9.
MAPK activation assay
Each SOBIR1 mutant was transiently co-expressed with Avr4 (OD600 = 0.8 for each binary vector) in the first fully expanded leaves of N. benthamiana:Cf-4 sobir1 plants. Leaf samples were harvested at 2 dpi, and leaf samples from the various N. benthamiana:Cf-4 rlck knock-out lines, infiltrated with a solution of 5 µM Avr4 protein, were harvested at 15 min after treatment.
Total protein was extracted and subjected to SDS-PAGE, after which an anti-p42/p44-erk antibody (NEB) was employed to detect the activated MAPKs on western blots.
Recombinant protein expression
To produce the recombinant cytoplasmic kinase domain (KD) of SOBIR1, BAK1 and various RLCKs with a GST or 6 × His tag in E. coli, the vectors pET-GST (Addgene No. 42049) and pET-15b were employed, respectively. Both vectors were linearized by PCR amplification with the primer pairs pET-GST_fw/rev and pET-15b_fw/rev (Supplementary Table 2). Meanwhile, the coding sequence of NbSOBIR1 KD, SlSOBIR1 KD, SlSOBIR1-like KD, and several selected RLCKs, as well as all their corresponding kinase-dead mutants, were PCR-amplified from the corresponding pENTR/D-Topo plasmids, using the primers containing the homologous sequence of either pET-GST or pET-15b (Supplementary Table 2). Hereafter, the linearized vector and amplified insert were recombined by using the ClonExpress II One Step Cloning kit (Vazyme, China). After transformation to E. coli DH5α, the correct expression constructs were selected by performing colony PCRs and Sanger sequencing.
For recombinant protein expression, the E. coli strain BL21 was used. Bacteria harbouring the correct expression construct were cultured at 37 °C overnight in LB liquid medium and subsequently inoculated into fresh LB medium at a ratio of 1:200 (v/v). After culturing at 37 °C for around 3 h, the bacterial population became in an exponential growth phase, with an OD600 between 0.6 and 0.8, at which IPTG was added to a final concentration of 0.5 mM, followed by incubating the culture at 22 °C overnight for protein production.
In vitro phosphorylation assay
In vitro phosphorylation assays were performed as previously described46. In brief, cells from 100 µL of the E. coli cultures that were started for protein expression were collected by centrifugation and then resuspended in 100 µL of SDS sample buffer (200 mM Tris-HCl, 8% SDS, 40% glycerol, 400 mM DTT, and 0.2% bromophenol blue), followed by boiling for 10 min. Hereafter, the samples were centrifuged for 2 min at maximum speed in an Eppendorf centrifuge, and 8 µL of the supernatant were loaded onto a precast mini-PROTEIN TGX Polyacrylamide Gel (BIORAD). After running for around 100 min at 160 V, the gel was incubated in fixation solution (50% methanol, 10% acetic acid in H2O), overnight. Next, the gel was washed in deionized water for 30 min twice, and the phosphorylated proteins were stained using a Pro-Q Diamond solution (Invitrogen). Subsequently, the staining solution was removed by washing the gel in de-staining solution (20% acetonitrile, 50 mM sodium acetate), and proteins were stained with CBB.
Phylogenetic analysis of the RLCKs from Arabidopsis, N. benthamiana, and tomato
To conduct a phylogenetic analysis of the AtBIK1 homologues in N. benthamiana, tomato, and Arabidopsis, their predicted proteomes were obtained from www.solgenomics.net and www.arabidopsis.org. Hereafter, the three predicted proteomes were independently queried for Pfam domains by using HMMER (v3.1b2; gathering cut-off)90. Sequences that contain annotated Pfam domains aside from cytoplasmic kinases (PF00069 or PF07714) were removed, and the sequences of the annotated kinase domains were extracted. Then, we took the domain PF07714 as a lead and removed the sequences that deviated in length from the kinase domain of AtBIK1. The remaining 1455 kinase domain sequences were aligned using MAFFT (v7.271)91, the alignment was subsequently trimmed using ClipKIT (v1.3.0; smart-gap)92 and a neighbour-joining phylogenetic tree was built using QuickTree (with 1000 bootstrap replicates)93 (Supplementary Fig. 9a). Next, a well-supported (>92% bootstrap support) sub-clade of putative BIK1 homologues, which comprised 123 sequences including AtBIK1, was extracted from this guide tree. Subsequently, a refined phylogenetic tree was generated with these sequences by using the maximum-likelihood (ML) phylogeny as implemented in IQ-Tree (v2.2.0)94. The 123 extracted kinase domains were re-aligned using MAFFT and trimmed as described above, and the ML phylogeny was constructed in IQ-Tree, using automatic amino acid substitution model selection (optimal model: Q.plant with five categories of rate heterogeneity)95,96. Branch support for the phylogenetic tree was obtained using ultrafast bootstrap, as well as SH-aLRT, as implemented in IQ-tree97.
Phytophthora palmivora inoculation assay
Fully expanded leaves were harvested from five-week-old N. benthamiana plants and used for inoculation with Phytophthora palmivora zoospores. P. palmivora maintenance and zoospore production were performed as described before98. Inoculations were performed by depositing 10 µL droplets containing 10,000 zoospores each on the abaxial side of the leaves. Leaves were incubated at 21 °C with a 16-h photoperiod in large Petri dishes containing wet paper to maintain moisture. The extent of colonization was monitored at 2 days post inoculation using the necrotic spot areas as a read-out for susceptibility. Necrotic areas were determined by imaging chlorophyll autofluorescence using a Chemidoc Imaging System (Bio-Rad), and the extent of colonization was quantified using ImageJ (https://imagej.nih.gov/ij).
Split-luciferase assay
To confirm the proper accumulation of the various tomato RLCKs, fused to the C-terminal part of the luciferase enzyme (Cluc), each binary expression vector was agro-infiltrated with the silencing suppressor P19 into a whole leaf of N. benthamiana, at an OD600 of 1.0 both for the expression vector and P19. At 3 dpi the leaves were harvested and a protein immunoprecipitation assay, followed by immunoblotting using anti luciferase antibody (Sigma, L2164), was performed as described before89.
Split-luciferase assays were performed using agro-infiltration of combinations of SlSOBIR1, AtFLS2 or GUS, fused to the N-terminal part of the luciferase enzyme (Nluc), and selected tomato RLCKs, fused to the C-terminal part of the luciferase enzyme (Cluc) at an OD600 of 0.5, as described before54. At 3 dpi, leaves were imaged with the abaxial side up using a ChemiDoc (Bio-Rad) device. For this, the leaves were sprayed with luciferin (1 mM luciferin (Biovision, sodium salt 7902-100), dissolved in Milli-Q, supplemented with 1/5000 (v/v) Silwet L-77 (Lehle Seeds, VIS-30)). The leaves were kept in the dark for 5 min to reduce autofluorescence, and luminescence was subsequently detected using the following settings: no illumination, no filter, 2 × 2 binning, and an exposure time of 20 min. A colorimetric image was also made and merged with the luciferase picture.
Transient expression for TurboID (TbID)-based proximity-dependent labelling and streptavidin-pull down for mass spectrometry analysis
Samples were prepared in triplicate, expressing each TbID-tagged bait protein in 15 N. benthamiana:Cf-4 sobir1 plants, using one leaf per plant and eventually combining the agro-infiltrated leaves of five plants to generate each individual sample. Two days after agro-infiltration for transient expression of the GUS-YFP-TbID and NbSOBIR1-YFP-TbID bait constructs in N. benthamiana sobir1, the leaves were infiltrated with a solution containing biotin (200 μM, pH = 8, 10 mM MES) and harvested after one hour. For each replicate, the five treated leaves were harvested, combined, wrapped in aluminium foil, and kept in liquid nitrogen or at −80 °C until further use. The frozen samples were ground to a fine powder in liquid nitrogen using a mortar and pestle. Then, the pulverized samples were transferred to pre-cooled 50 mL V-shape tubes, weighed, and resuspended in 2 mL/g of extraction buffer (EB; pH = 8.0, 150 mM NaCl, 1.0% [v/v] IGEPAL®CA-630 (=NP-40), 50 mM Tris, Sigma protease inhibitor cocktail = 1 tablet per 50 mL). For this resuspension, the frozen samples with the EB were vortexed at room temperature and kept on ice once melted. Then, the samples were centrifuged at 18,000 × g for 30 min at 4 °C in a Sigma 4–16 K centrifuge.
To prevent saturation of the streptavidin beads with the free biotin remaining in the samples, 10 mL of the centrifuged protein extracts were desalted using PD MiniTrap PD-10 desalting columns (GE Healthcare). This was done at 4 °C, following the manufacturer’s gravity protocol for removing salt.
The desalted protein extracts were then transferred to pre-cooled 15 mL tubes and incubated for one hour at 4 °C with 10 rotations per minute in an SB3 tube rotator (STUART), with 200 μL of Dynabeads™ MyOne™ Streptavidin C1 (washed before use according to the manufacturer’s protocol). After incubation, the beads were washed three times with 1 mL of EB, without NP-40, after which they were resuspended in 45 μL of EB buffer.
Sample preparation for proteomics by mass spectrometry
While still on the beads, the disulphide bonds in the captured proteins were reduced by adding 5 μL of DTT (150 mM) and incubating the samples at 45 °C for 30 min. The sulfhydryl groups were subsequently alkylated by adding 6 μL of acrylamide (200 mM) and incubating the samples µfor 10 min at room temperature.
The peptides to be measured by mass spectrometry were subsequently released from the streptavidin-coated beads by tryptic digestion. For this, a stock solution of trypsin (0.5 μg/μL of trypsin in 1 mM HCl, pH 3) was diluted 100 times in ABC buffer (ammonium bicarbonate, 50 mM, pH = 8) and 100 μL of the diluted trypsin solution was added to each sample. The samples were then incubated overnight at room temperature, with mild agitation, after which they were acidified to pH = 3 using trifluoroacetic acid and cleaned up using μColumns according to the method published by Wendrich and co-workers99.
LC–MS/MS analysis
For the LC–MS/MS analysis, the peptides were separated by reversed phase nano liquid chromatography, using a Thermo nLC1000 system equipped with a home-made C18 nanoLC column and they were measured using an Orbitrap Exploris 480 mass spectrometer. The peptide spectra were searched in Maxquant (version 2.0.3.0) 438, using the Andromeda search engine 439 with label-free quantification (LFQ), against the version Niben1.0.1 of the N. benthamiana proteome dataset, including the protein sequence of Cf-4 (O50025), Avr4 (Q00363), LTI6b (AT3G05890), GUS (P05804) and frequently occurring contaminants.
The identified protein groups were then analysed using Perseus (version 1.6.2.3) 358. Reverse and contaminant proteins, and those only identified by matching, were filtered out. Then, protein groups identified in less than three replicate samples were also filtered out. The LFQ values were log2 transformed, and the missing values were assigned assuming a normal distribution. The relative protein quantitation of the samples relative to the controls was calculated applying both-sided Student’s t-tests, using a permutation-based adjustment (FDR = 0.05, 250 randomizations, and S0 set to 0.1).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
The authors thank Bert Essenstam from Unifarm for excellent plant care. Laurens Deurhof and Gabriel Lorencini Fiorin are acknowledged for technical assistance. We thank Xiaoqian Shi-Kunne for her help with retrieving the protein sequences of the SOBIR1 homologues from different plant species. We thank Jim Renema, Max Pluis, Agnes Omabour Hagan and Amber van Loosbroek for their help with performing some of the experiments. W.R.H. Huang is supported by the China Scholarship Council (CSC) (201706990001). S.L.V. is supported by the Peruvian Council for Science, Technology and Technological Innovation (CONCYTEC) and its executive unit FONDECYT (119-2017-FONDECYT).
Author contributions
W.R.H.H., A.M.vd B., and M.H.A.J.J. conceived and designed the experiments. W.R.H.H., C.B., S.L.V., H.L., A.M.vd B., L.Z., Y.W., and E.E. executed the experimental work, data acquisition and analysis. S.B. performed LC-MS/MS and assisted with the analysis of the acquired data. C.K. and F.F. generated rlck knock-out lines; M.F.S. performed phylogenetic analysis of BIK1 homologues; J.W. and T.N. provided input; W.R.H.H., M.S., E.E., J.S. and M.H.A.J.J. wrote the manuscript with contributions from all authors.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
The authors declare that all data supporting the findings of this study are available within the manuscript and the Supplementary Files or are available from the corresponding authors upon request. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
9/10/2024
A Correction to this paper has been published: 10.1038/s41467-024-52322-5
Contributor Information
Wen R. H. Huang, Email: Wen.Huang@tsl.ac.uk
Matthieu H. A. J. Joosten, Email: matthieu.joosten@wur.nl
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-48313-1.
References
- 1.van der Burgh, A. M. & Joosten, M. H. A. J. Plant immunity: thinking outside and inside the box. Trends Plant Sci.24, 587–601 (2019). 10.1016/j.tplants.2019.04.009 [DOI] [PubMed] [Google Scholar]
- 2.Monaghan, J. & Zipfel, C. Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol.15, 349–357 (2012). 10.1016/j.pbi.2012.05.006 [DOI] [PubMed] [Google Scholar]
- 3.Tsuda, K. & Katagiri, F. Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol.13, 459–465 (2010). 10.1016/j.pbi.2010.04.006 [DOI] [PubMed] [Google Scholar]
- 4.Chisholm, S. T., Coaker, G., Day, B. & Staskawicz, B. J. Host-microbe interactions: shaping the evolution of the plant immune response. Cell124, 803–814 (2006). 10.1016/j.cell.2006.02.008 [DOI] [PubMed] [Google Scholar]
- 5.Kong, L., Rodrigues, B., Kim, J. H., He, P. & Shan, L. More than an on-and-off switch: post-translational modifications of plant pattern recognition receptor complexes. Curr. Opin. Plant Biol.63, 102051 (2021). 10.1016/j.pbi.2021.102051 [DOI] [PubMed] [Google Scholar]
- 6.Lee, D. H., Lee, H. S. & Belkhadir, Y. Coding of plant immune signals by surface receptors. Curr. Opin. Plant Biol.62, 102044 (2021). 10.1016/j.pbi.2021.102044 [DOI] [PubMed] [Google Scholar]
- 7.Heese, A. et al. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl Acad. Sci. USA104, 12217–12222 (2007). 10.1073/pnas.0705306104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chinchilla, D., Shan, L., He, P., de Vries, S. & Kemmerling, B. One for all: the receptor-associated kinase BAK1. Trends Plant Sci.14, 535–541 (2009). 10.1016/j.tplants.2009.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gómez-Gómez, L. & Boller, T. FLS2: an LRR receptor–like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell5, 1003–1011 (2000). 10.1016/S1097-2765(00)80265-8 [DOI] [PubMed] [Google Scholar]
- 10.Gómez-Gómez, L., Bauer, Z. & Boller, T. Both the extracellular leucine-rich repeat domain and the kinase activity of FLS2 are required for flagellin binding and signaling in Arabidopsis. Plant Cell13, 1155–1163 (2001). 10.1105/tpc.13.5.1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chinchilla, D. et al. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature448, 497–500 (2007). 10.1038/nature05999 [DOI] [PubMed] [Google Scholar]
- 12.Sun, Y. et al. Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science342, 624–628 (2013). 10.1126/science.1243825 [DOI] [PubMed] [Google Scholar]
- 13.Gao, M. et al. Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe6, 34–44 (2009). 10.1016/j.chom.2009.05.019 [DOI] [PubMed] [Google Scholar]
- 14.Liebrand, T. W. H. et al. Receptor-like kinase SOBIR1/EVR interacts with receptor-like proteins in plant immunity against fungal infection. Proc. Natl Acad. Sci. USA110, 13228 (2013). 10.1073/pnas.1220015110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liebrand, T. W. H., van den Burg, H. A. & Joosten, M. H. A. J. Two for all: receptor-associated kinases SOBIR1 and BAK1. Trends Plant Sci.19, 123–132 (2014). 10.1016/j.tplants.2013.10.003 [DOI] [PubMed] [Google Scholar]
- 16.Joosten, M. H. A. J., Cozijnsen, T. J. & De Wit, P. J. G. M. Host resistance to a fungal tomato pathogen lost by a single base-pair change in an avirulence gene. Nature367, 384–386 (1994). 10.1038/367384a0 [DOI] [PubMed] [Google Scholar]
- 17.Thomas, C. M. et al. Characterization of the tomato Cf-4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf-4 and Cf-9. Plant Cell9, 2209 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Postma, J. et al. Avr4 promotes Cf-4 receptor-like protein association with the BAK1/SERK3 receptor-like kinase to initiate receptor endocytosis and plant immunity. New Phytol.210, 627–642 (2016). 10.1111/nph.13802 [DOI] [PubMed] [Google Scholar]
- 19.DeFalco, T. A. & Zipfel, C. Molecular mechanisms of early plant pattern-triggered immune signaling. Mol. Cell81, 3449–3467 (2021). 10.1016/j.molcel.2021.07.029 [DOI] [PubMed] [Google Scholar]
- 20.Albert, I. et al. An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. Nat. Plants1, 15140 (2015). 10.1038/nplants.2015.140 [DOI] [PubMed] [Google Scholar]
- 21.Macho, A. P. & Zipfel, C. Plant PRRs and the activation of innate immune signaling. Mol. Cell54, 263–272 (2014). 10.1016/j.molcel.2014.03.028 [DOI] [PubMed] [Google Scholar]
- 22.Mithoe, S. C. & Menke, F. L. Regulation of pattern recognition receptor signalling by phosphorylation and ubiquitination. Curr. Opin. Plant Biol.45, 162–170 (2018). 10.1016/j.pbi.2018.07.008 [DOI] [PubMed] [Google Scholar]
- 23.Johnson, L. N., Noble, M. E. M. & Owen, D. J. Active and inactive protein kinases: structural basis for regulation. Cell85, 149–158 (1996). 10.1016/S0092-8674(00)81092-2 [DOI] [PubMed] [Google Scholar]
- 24.Nolen, B., Taylor, S. & Ghosh, G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol. Cell15, 661–675 (2004). 10.1016/j.molcel.2004.08.024 [DOI] [PubMed] [Google Scholar]
- 25.Oliver, A. W., Knapp, S. & Pearl, L. H. Activation segment exchange: a common mechanism of kinase autophosphorylation? Trends Biochem. Sci.32, 351–356 (2007). 10.1016/j.tibs.2007.06.004 [DOI] [PubMed] [Google Scholar]
- 26.Dardick, C., Schwessinger, B. & Ronald, P. Non-arginine-aspartate (non-RD) kinases are associated with innate immune receptors that recognize conserved microbial signatures. Curr. Opin. Plant Biol.15, 358–366 (2012). 10.1016/j.pbi.2012.05.002 [DOI] [PubMed] [Google Scholar]
- 27.Lin, W., Ma, X., Shan, L. & He, P. Big roles of small kinases: the complex functions of receptor-like cytoplasmic kinases in plant immunity and development. J. Integr. Plant Biol.55, 1188–1197 (2013). 10.1111/jipb.12071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang, D., Wang, G. & Zhou, J. M. Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell29, 618–637 (2017). 10.1105/tpc.16.00891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bi, G. et al. Receptor-like cytoplasmic kinases directly link diverse pattern recognition receptors to the activation of mitogen-activated protein kinase cascades in Arabidopsis. Plant Cell30, 1543–1561 (2018). 10.1105/tpc.17.00981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou, J.-M. & Zhang, Y. Plant immunity: danger perception and signaling. Cell181, 978–989 (2020). 10.1016/j.cell.2020.04.028 [DOI] [PubMed] [Google Scholar]
- 31.Lu, D. et al. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl Acad. Sci. USA107, 496–501 (2010). 10.1073/pnas.0909705107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang, J. et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe7, 290–301 (2010). 10.1016/j.chom.2010.03.007 [DOI] [PubMed] [Google Scholar]
- 33.Wan, W. L. et al. Comparing Arabidopsis receptor kinase and receptor protein‐mediated immune signaling reveals BIK1‐dependent differences. New Phytol.221, 2080–2095 (2019). 10.1111/nph.15497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin, Z. J., Liebrand, T. W., Yadeta, K. A. & Coaker, G. PBL13 is a serine/threonine protein kinase that negatively regulates Arabidopsis immune responses. Plant Physiol.169, 2950–2962 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo, X. et al. Tyrosine phosphorylation of the lectin receptor-like kinase LORE regulates plant immunity. EMBO J.39, e102856 (2020). 10.15252/embj.2019102856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pruitt, R. N. et al. The EDS1-PAD4-ADR1 node mediates Arabidopsis pattern-triggered immunity. Nature598, 495–499 (2021). 10.1038/s41586-021-03829-0 [DOI] [PubMed] [Google Scholar]
- 37.Rao, S. et al. Roles of receptor-like cytoplasmic kinase vii members in pattern-triggered immune signaling. Plant Physiol.177, 1679–1690 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leslie, M. E., Lewis, M. W., Youn, J.-Y., Daniels, M. J. & Liljegren, S. J. The EVERSHED receptor-like kinase modulates floral organ shedding in Arabidopsis. Development137, 467–476 (2010). 10.1242/dev.041335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gabriëls, S. H. E. J. et al. cDNA-AFLP combined with functional analysis reveals novel genes involved in the hypersensitive response. Mol. Plant-Microbe Interact.19, 567–576 (2006). 10.1094/MPMI-19-0567 [DOI] [PubMed] [Google Scholar]
- 40.Huang, W., Schol, C., Villanueva, S. L., Heidstra, R. & Joosten, M. Knocking out SOBIR1 in Nicotiana benthamiana abolishes functionality of transgenic receptor-like protein Cf-4. Plant Physiol.185, 290–294 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Burgh, A. M., Postma, J., Robatzek, S. & Joosten, M. H. A. J. Kinase activity of SOBIR1 and BAK1 is required for immune signalling. Mol. Plant Pathol.20, 410–422 (2019). 10.1111/mpp.12767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Landeo Villanueva, S., Malvestiti, M. C., van Ieperen, W., Joosten, M. H. A. J. & van Kan, J. A. L. Red light imaging for programmed cell death visualization and quantification in plant-pathogen interactions. Mol. Plant Pathol.22, 361–372 (2021). 10.1111/mpp.13027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu, X., Feng, B., He, P. & Shan, L. From chaos to harmony: responses and signaling upon microbial pattern recognition. Annu. Rev. Phytopathol.55, 109–137 (2017). 10.1146/annurev-phyto-080516-035649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qi, J., Wang, J., Gong, Z. & Zhou, J. M. Apoplastic ROS signaling in plant immunity. Curr. Opin. Plant Biol.38, 92–100 (2017). 10.1016/j.pbi.2017.04.022 [DOI] [PubMed] [Google Scholar]
- 45.Waszczak, C., Carmody, M. & Kangasjärvi, J. Reactive oxygen species in plant signaling. Annu. Rev. Plant Biol.69, 209–236 (2018). 10.1146/annurev-arplant-042817-040322 [DOI] [PubMed] [Google Scholar]
- 46.Taylor, I., Seitz, K., Bennewitz, S. & Walker, J. C. A simple in vitro method to measure autophosphorylation of protein kinases. Plant Methods9, 22 (2013). 10.1186/1746-4811-9-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gabriëls, S. H. E. J. et al. An NB-LRR protein required for HR signalling mediated by both extra- and intracellular resistance proteins. Plant J.50, 14–28 (2007). 10.1111/j.1365-313X.2007.03027.x [DOI] [PubMed] [Google Scholar]
- 48.Tameling, W. I. L. et al. RanGAP2 mediates nucleocytoplasmic partitioning of the NB-LRR immune receptor Rx in the Solanaceae, thereby dictating Rx function. Plant Cell22, 4176–4194 (2010). 10.1105/tpc.110.077461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, L. et al. Fungal endopolygalacturonases are recognized as microbe-associated molecular patterns by the Arabidopsis receptor-like protein RESPONSIVENESS TO BOTRYTIS POLYGALACTURONASES1. Plant Physiol.164, 352–364 (2014). 10.1104/pp.113.230698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, L. et al. Distinct immune sensor systems for fungal endopolygalacturonases in closely related Brassicaceae. Nat. Plants7, 1254–1263 (2021). 10.1038/s41477-021-00982-2 [DOI] [PubMed] [Google Scholar]
- 51.Zhang, M., Su, J., Zhang, Y., Xu, J. & Zhang, S. Conveying endogenous and exogenous signals: MAPK cascades in plant growth and defense. Curr. Opin. Plant Biol.45, 1–10 (2018). 10.1016/j.pbi.2018.04.012 [DOI] [PubMed] [Google Scholar]
- 52.Peng, K. C., Wang, C. W., Wu, C. H., Huang, C. T. & Liou, R. F. Tomato SOBIR1/EVR homologs are involved in elicitin perception and plant defense against the oomycete pathogen Phytophthora parasitica. Mol. Plant-Microbe Interact.28, 913–926 (2015). 10.1094/MPMI-12-14-0405-R [DOI] [PubMed] [Google Scholar]
- 53.Domazakis, E. et al. The ELR-SOBIR1 complex functions as a two-component RLK to mount defense against Phytophthora infestans. Mol. Plant-Microbe Interact.31, 795–802 (2018). 10.1094/MPMI-09-17-0217-R [DOI] [PubMed] [Google Scholar]
- 54.Chen, H. et al. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol.146, 323–324 (2008). 10.1104/pp.107.111740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.You, Y. Host Resistance Mechanism and Fungal Infection Strategies in the Botrytis cinerea-Tomato Interaction (Wageningen University, 2022).
- 56.Liu, J., Elmore, J. M., Lin, Z. J. D. & Coaker, G. A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe9, 137–146 (2011). 10.1016/j.chom.2011.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rowland, O. et al. Functional analysis of Avr9/Cf-9 rapidly elicited genes identifies a protein kinase, ACIK1, that is essential for full Cf-9-dependent disease resistance in tomato. Plant Cell17, 295–310 (2005). 10.1105/tpc.104.026013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burr, C. A. et al. CAST AWAY, a membrane-associated receptor-like kinase, inhibits organ abscission in Arabidopsis. Plant Physiol.156, 1837–1850 (2011). 10.1104/pp.111.175224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, Y. et al. TurboID-based proximity labeling reveals that UBR7 is a regulator of N NLR immune receptor-mediated immunity. Nat. Commun.10, 3252 (2019). 10.1038/s41467-019-11202-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li, J. et al. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell110, 213–222 (2002). 10.1016/S0092-8674(02)00812-7 [DOI] [PubMed] [Google Scholar]
- 61.Schwessinger, B. et al. Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet.7, e1002046 (2011). 10.1371/journal.pgen.1002046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei, X. et al. Structural analysis of receptor-like kinase SOBIR1 revealed mechanisms that regulate its phosphorylation-dependent activation. Plant Commun.3, 100301 (2022). 10.1016/j.xplc.2022.100301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hohmann, U. et al. Constitutive activation of leucine-rich repeat receptor kinase signaling pathways by BAK1-interacting receptor-like kinase 3 chimera. Plant Cell32, 3311–3323 (2020). 10.1105/tpc.20.00138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suzuki, M. et al. Autophosphorylation of specific threonine and tyrosine residues in Arabidopsis CERK1 is essential for the activation of chitin-induced immune signaling. Plant Cell Physiol.57, 2312–2322 (2016). 10.1093/pcp/pcw150 [DOI] [PubMed] [Google Scholar]
- 65.Suzuki, M. et al. AtCERK1 phosphorylation site S493 contributes to the transphosphorylation of downstream components for chitin-induced immune signaling. Plant Cell Physiol.60, 1804–1810 (2019). 10.1093/pcp/pcz096 [DOI] [PubMed] [Google Scholar]
- 66.Mariano, A. C. et al. Identification of a novel receptor-like protein kinase that interacts with a geminivirus nuclear shuttle protein. Virology318, 24–31 (2004). 10.1016/j.virol.2003.09.038 [DOI] [PubMed] [Google Scholar]
- 67.Santos, A. A., Lopes, K. V. G., Apfata, J. A. C. & Fontes, E. P. B. NSP-interacting kinase, NIK: a transducer of plant defence signalling. J. Exp. Bot.61, 3839–3845 (2010). 10.1093/jxb/erq219 [DOI] [PubMed] [Google Scholar]
- 68.Santos, A. A., Carvalho, C. M., Florentino, L. H., Ramos, H. J. O. & Fontes, E. P. B. Conserved threonine residues within the A-loop of the receptor NIK differentially regulate the kinase function required for antiviral signaling. PLoS ONE4, e5781 (2009). 10.1371/journal.pone.0005781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu, J. et al. A tyrosine phosphorylation cycle regulates fungal activation of a plant receptor Ser/Thr kinase. Cell Host Microbe23, 241–253 (2018). 10.1016/j.chom.2017.12.005 [DOI] [PubMed] [Google Scholar]
- 70.Macho, A. P. et al. A bacterial tyrosine phosphatase inhibits plant pattern recognition receptor activation. Science343, 1509–1512 (2014). [DOI] [PubMed]
- 71.Perraki, A. et al. Phosphocode-dependent functional dichotomy of a common co-receptor in plant signalling. Nature561, 248–252 (2018). [DOI] [PMC free article] [PubMed]
- 72.Luo, X. et al. Tyrosine phosphorylation of the lectin receptor-like kinase LORE regulates plant immunity. EMBO J. 39, e102856 (2020). [DOI] [PMC free article] [PubMed]
- 73.Mühlenbeck, H. et al. Allosteric activation of the co-receptor BAK1 by the EFR receptor kinase initiates immune signaling. eLife12, RP92110 (2024). [DOI] [PMC free article] [PubMed]
- 74.Miya, A. et al. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. PNAS104, 19613–19618 (2007). 10.1073/pnas.0705147104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wan, J. et al. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell20, 471–481 (2008). 10.1105/tpc.107.056754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van der Burgh, A. M. et al. SOBIR1-Containing Immune Complexes at the Plant Cell Surface: Partners and Signalling (Wageningen University, 2018).
- 77.Kadota, Y. et al. Quantitative phosphoproteomic analysis reveals common regulatory mechanisms between effector- and PAMP-triggered immunity in plants. New Phytol.221, 2160–2175 (2019). 10.1111/nph.15523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yoshioka, H. et al. Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell15, 706–718 (2003). 10.1105/tpc.008680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Adachi, H. et al. WRKY transcription factors phosphorylated by MAPK regulate a plant immune NADPH oxidase in Nicotiana benthamiana. Plant Cell27, 2645–2663 (2015). 10.1105/tpc.15.00213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liang, X. et al. A Phytophthora capsici RXLR effector targets and inhibits the central immune kinases to suppress plant immunity. New Phytol.232, 264–278 (2021). 10.1111/nph.17573 [DOI] [PubMed] [Google Scholar]
- 81.Wang, Y. et al. Leucine-rich repeat receptor-like gene screen reveals that Nicotiana RXEG1 regulates glycoside hydrolase 12 MAMP detection. Nat. Commun.9, 594 (2018). 10.1038/s41467-018-03010-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen, Z. et al. Convergent evolution of immune receptors underpins distinct elicitin recognition in closely related Solanaceous plants. Plant Cell35, 1186–1201 (2023). 10.1093/plcell/koad002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Evangelisti, E. et al. Time-resolved dual transcriptomics reveal early induced Nicotiana benthamiana root genes and conserved infection-promoting Phytophthora palmivora effectors. BMC Biol.15, 1–24 (2017). 10.1186/s12915-017-0379-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stuttmann, J. et al. Highly efficient multiplex editing: one-shot generation of 8x Nicotiana benthamiana and 12x Arabidopsis mutants. Plant J.106, 8–22 (2021). 10.1111/tpj.15197 [DOI] [PubMed] [Google Scholar]
- 85.Liu, H. & Naismith, J. H. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol.8, 91 (2008). 10.1186/1472-6750-8-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li, L. et al. The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe15, 329–338 (2014). 10.1016/j.chom.2014.02.009 [DOI] [PubMed] [Google Scholar]
- 87.Kim, T.-W. et al. Application of TurboID-mediated proximity labeling for mapping a GSK3 kinase signaling network in Arabidopsis. Preprint at BioRxiv, 10.1101/636324 (2019).
- 88.Van der Hoorn, R. A. L., Laurent, F., Roth, R. & De Wit, P. J. G. M. Agroinfiltration is a versatile tool that facilitates comparative analyses of Avr9/Cf-9-induced and Avr4/Cf-4-induced necrosis. Mol. Plant-Microbe Interact.13, 439–446 (2000). 10.1094/MPMI.2000.13.4.439 [DOI] [PubMed] [Google Scholar]
- 89.Liebrand, T. W. H. et al. Endoplasmic reticulum-quality control chaperones facilitate the biogenesis of Cf receptor-like proteins involved in pathogen resistance of tomato. Plant Physiol.159, 1819–1833 (2012). 10.1104/pp.112.196741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Eddy, S. R. Profile hidden Markov models. Bioinformatics14, 755–763 (1998). 10.1093/bioinformatics/14.9.755 [DOI] [PubMed] [Google Scholar]
- 91.Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol.30, 772–780 (2013). 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Steenwyk, J. L., Buida, T. J. 3rd, Li, Y., Shen, X. X. & Rokas, A. ClipKIT: a multiple sequence alignment trimming software for accurate phylogenomic inference. PLoS Biol.18, e3001007 (2020). 10.1371/journal.pbio.3001007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Howe, K., Bateman, A. & Durbin, R. QuickTree: building huge Neighbour-Joining trees of protein sequences. Bioinformatics18, 1546–1547 (2002). 10.1093/bioinformatics/18.11.1546 [DOI] [PubMed] [Google Scholar]
- 94.Nguyen, L. T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol.32, 268–274 (2015). 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A. & Jermiin, L. S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods14, 587–589 (2017). 10.1038/nmeth.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Minh, B. Q., Dang, C. C., Vinh, L. S. & Lanfear, R. QMaker: fast and accurate method to estimate empirical models of protein evolution. Syst. Biol.70, 1046–1060 (2021). 10.1093/sysbio/syab010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q. & Vinh, L. S. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol.35, 518–522 (2018). 10.1093/molbev/msx281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Evangelisti, E. et al. Hydrodynamic shape changes underpin nuclear rerouting in branched hyphae of an oomycete pathogen. mBio10, 10.1128/mBio.01516-19 (2019). [DOI] [PMC free article] [PubMed]
- 99.Wendrich, J. R., Boeren, S., Moller, B. K., Weijers, D. & De Rybel, B. In vivo identification of plant protein complexes using IP-MS/MS. Methods Mol. Biol.1497, 147–158 (2017). 10.1007/978-1-4939-6469-7_14 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the manuscript and the Supplementary Files or are available from the corresponding authors upon request. Source data are provided with this paper.