Abstract
Automated quantification of brain tissues on MR images has greatly contributed to the diagnosis and follow-up of neurological pathologies across various life stages. However, existing solutions are specifically designed for certain age ranges, limiting their applicability in monitoring brain development from infancy to late adulthood. This retrospective study aims to develop and validate a brain segmentation model across pediatric and adult populations. First, we trained a deep learning model to segment tissues and brain structures using T1-weighted MR images from 390 patients (age range: 2–81 years) across four different datasets. Subsequently, the model was validated on a cohort of 280 patients from six distinct test datasets (age range: 4–90 years). In the initial experiment, the proposed deep learning-based pipeline, icobrain-dl, demonstrated segmentation accuracy comparable to both pediatric and adult-specific models across diverse age groups. Subsequently, we evaluated intra- and inter-scanner variability in measurements of various tissues and structures in both pediatric and adult populations computed by icobrain-dl. Results demonstrated significantly higher reproducibility compared to similar brain quantification tools, including childmetrix, FastSurfer, and the medical device icobrain v5.9 (p-value< 0.01). Finally, we explored the potential clinical applications of icobrain-dl measurements in diagnosing pediatric patients with Cerebral Visual Impairment and adult patients with Alzheimer’s Disease.
Subject terms: Neurology, Neurodegenerative diseases, Neurodevelopmental disorders, Paediatric neurological disorders, Biomedical engineering
Introduction
Neuroimaging techniques play a crucial role in advancing our understanding of the human brain, covering its structure, development, function, and pathologies1. Magnetic Resonance Imaging (MRI) stands out as a non-invasive technology to obtain high-resolution, in vivo measurements of the human brain2. Automated analysis of MR images contributes to the diagnosis of neurological pathologies across various life stages, from childhood (e.g., focal cortical dysplasia3) to late adulthood (e.g., Alzheimer’s disease4).
Quantitative assessment, exemplified by volumetric analysis, enhances the objectivity of brain interpretation compared to visual MRI scan inspection alone. Traditional techniques for brain MR image segmentation involve atlas-based methods and statistical models, such as FreeSurfer5, volBrain6, or the medical device software, icobrain v5.97,8. Nevertheless, recent progress in deep learning models, such as QuickNat9, AssemblyNet10, and FastSurfer11, has demonstrated superior performance compared to traditional methodologies, as evidenced in a recent review12.
Despite the growing role of quantitative analysis tools, additional technical and clinical validation is required4. Notably, there is a lack of validated models for robust and reliable brain quantification in multi-scanner settings, common in clinical data. Additionally, recent algorithms, including deep learning methods, are usually developed and validated using adult datasets. However, standard MRI processing methods designed for adult images may not be suitable for pediatric datasets13. Pediatric brain analysis poses unique challenges such as reduced tissue contrast, within-tissue intensity heterogeneities, and smaller regions of interest13,14. Consequently, pediatric brain analysis commonly employs specialized analysis tools like childmetrix15.
In pediatric studies, a common dilemma arises regarding the use of age-appropriate methods for different developmental stages or maintaining a consistent method across all ages16. While age-specific models are optimized for specific age ranges, their use introduces the risk of attributing age-related differences to methodological inconsistencies rather than genuine brain development or change. Particularly when monitoring patients across different transitional phases, such as from the pediatric stage through adolescence and into adulthood, there is a significant need for a general, consistent, and reliable method, eliminating reliance on multiple age-specific methods.
In this work, we develop and validate a brain segmentation pipeline across pediatric and adult populations, emphasizing the impact of heterogeneous and representative training data rather than the optimization of the deep learning architecture employed. The primary objective of this study is to explore whether a single deep learning model can be optimized to consistently quantify structural MRI across the lifespan, reflecting the distinctive neuroanatomy of each developmental stage. We hypothesize that a single deep learning model trained on datasets covering a wide age range will perform comparably to age-specific models within their respective age groups. The secondary objective is to validate the proposed pipeline’s performance in terms of reproducibility, diagnostic accuracy, and computational time. We hypothesize that the proposed deep learning-based pipeline will produce results comparable to established methods such as childmetrix, icobrain v5.9, and FastSurfer, while ensuring accurate and reproducible brain quantification across pediatric and adult populations.
Materials and methods
Datasets
Four separate datasets collectively containing 390 patients, aged between 2 and 81 years, were utilized for training. Validation was performed on a separate cohort of 280 patients from six distinct test datasets, covering an age range from 4 to 90 years. These datasets consisted of 757 T1-weighted MRI scans acquired from various manufacturers (Philips, Siemens, GE, Fujifilm) with different magnetic field strengths (1.5T/3T ∼ 32%/68%) across 21 scanners. The patients represented a diverse pathological conditions, including developmental disorders, cerebral visual impairment, depression, bipolar disorder, schizophrenia, multiple sclerosis, and Alzheimer’s disease. Table 1 presents a summary of the diverse datasets employed in this retrospective study. Further details about these datasets can be found in Appendix A.
Table 1.
The datasets utilized for model training and validation consisted of both pediatric (denoted with suffix p) and adult (denoted with suffix a) data.
| Scenario | Dataset | Subjects | Age range (years) | Diagnosis | Female (%) | Scanner type | Source |
|---|---|---|---|---|---|---|---|
| Training | 1.1.p | 157 | 5–21 | DD and HC | 40% | 3T: Siemens | CMI-HBN17 |
| 1.2.p | 66 | 2–6 | HC | 48% | 3T: GE | Calgary18 | |
| 1.3.a | 32 | Adults | VLOSLP, LOD and HC | NA | 3T: Philips | Research cohort19 | |
| 1.4.a | 135 | 16–81 | MS | 60% | 1.5–3T: Siemens, GE and Philips, 1.5T: Fujifilm | Clinical practice | |
| Accuracy | 2.p | 103 | 4–16 | BD, Sz and HC | 45% | 1.5T: GE | CANDIShare20 |
| 2.a | 30 | 18–90 | HC | 66% | 1.5T: Siemens | MICCAI201221 | |
| Reproducibility | 3.p | 70 | 6–17 | DD and HC | 44% | 3T: Siemens | NKI22 |
| 3.a | 10 | 39–57 | MS | 70% | 3T: Siemens, GE and Philips | Re3T7 | |
| Diagnosis Performance | 4.p | 21 | 4–13 | CVI and no CVI | 23% | 1.5T:Siemens and Philips, 3T: Philips | Clinical practice |
| 4.a | 46 | 58–85 | AD and HC | 54% | 1.5T: GE | MIRIAD23 |
A subset of patients were randomly selected from original training datasets. These datasets include individuals with Developmental Disorders (DD), Healthy Control (HC), Very-Late-Onset Schizophrenia-Like Psychosis (VLOSLP), Late-Onset Depression (LOD), Bipolar Disorder (BD), Schizophrenia (Sz) and Multiple Sclerosis (MS), Cerebral Visual Impairment (CVI) and Alzheimer’s Disease (AD). NA = not available.
Training dataset
The training dataset comprises a wide age range, pathologies and acquisition protocols. T1-weighted images were sourced from pediatric datasets, including the Healthy Brain Network (HBN, dataset 1.1.p)17 and the Calgary Preschool MRI (dataset 1.2.p)18. Additionally, T1-weighted images of adult patients were obtained from a research cohort (dataset 1.3.a) focused on the relations between very-late-onset schizophrenia-like psychosis, hippocampal volume, early adversity, and memory function19 as well as another cohort from clinical practice (dataset 1.4.a).
Segmentation accuracy testing dataset
Two publicly available manually annotated datasets were used to validate the segmentation accuracy: the Child and Adolescent NeuroDevelopment Initiative (CANDI, dataset 2.p)20 and the MICCAI 2012 Grand Challenge and Workshop on Multi-Atlas Labeling (MICCAI2012, dataset 2.a)21. We excluded 5 images from the latter due to repeated scans of the same patient.
Reproducibility testing dataset
The reproducibility of the measurements was evaluated by analyzing two images from the same individual acquired with re-positioning within a very short time interval, ensuring no anatomical change between the two images (i.e., test and retest images). Two test-retest datasets were used to validate the reproducibility. The first dataset is a pediatric intra-scanner dataset obtained from Nathan Kline Institute (NKI, dataset 3.p)22, while the second dataset comprises 10 adult individuals who underwent two scans, using three different types of scanners (Re3T, dataset 3.a)7. Using repeated scans in multiple scanner types enables analysis for intra-scanner and inter-scanner validation.
Diagnostic performance testing dataset
The diagnostic performance is assessed using two separate datasets. The first dataset comprises pediatric patients suspected of suffering from Cerebral Visual Impairment (CVI) (dataset 4.p), approved by the local Ethical Committee of UZ Leuven, Belgium (S65276). All methods were carried out in accordance with relevant guidelines and regulations. Informed consent was obtained from all subjects or their legal guardians. Secondly, we used the Minimal Interval Resonance Imaging in Alzheimer’s Disease (MIRIAD, dataset 4.a), which includes both patients with Alzheimer’s Disease (AD) and healthy elderly individuals23.
icobrain-dl pipeline: design and development
icobrain-dl is a pipeline for brain quantification. The pipeline processes a 3D T1-weighted MR image as input and undergoes three main steps: preprocessing, brain segmentation using a deep learning model, and brain quantification. The output includes brain segmentation masks for various regions of interest (ROIs) and brain volumes.
Pre-processing
Prior to training, the images underwent several fully automated pre-processing steps. Firstly, bias-field correction was performed using the N4 inhomogeneity correction algorithm as implemented in the Advanced Normalization Tools (ANTs) toolkit24. In pediatric cases, an age-specific atlas is used to obtain the brain mask for N4 correction. Secondly, the images were affinely registered to MNI space using the algorithm in NiftyReg25. To minimize the effect of outliers, intensities were clipped at the 1st and 99th percentile. Finally, the intensities were normalized using a variation on z-scoring, this function was computed over values above the 10th percentile, with preference given to the median over the mean. The standard deviation was then computed within the 90th percentile.
Simultaneous segmentation of brain tissue and structures via a multi-head deep learning model
The proposed deep learning model is designed to perform two tasks, brain tissue segmentation and brain structural segmentation, whose labels are not mutually exclusive.
Task 1: Tissue segmentation. This task involves the segmentation of brain tissues into four distinct classes: background (i.e., not brain tissue), white matter (WM), gray matter (GM), and cerebrospinal fluid (CSF).
Task 2: Structural segmentation. This task involves the segmentation of 22 anatomical brain structures and background. A detailed list of the structures is provided in Appendix B.
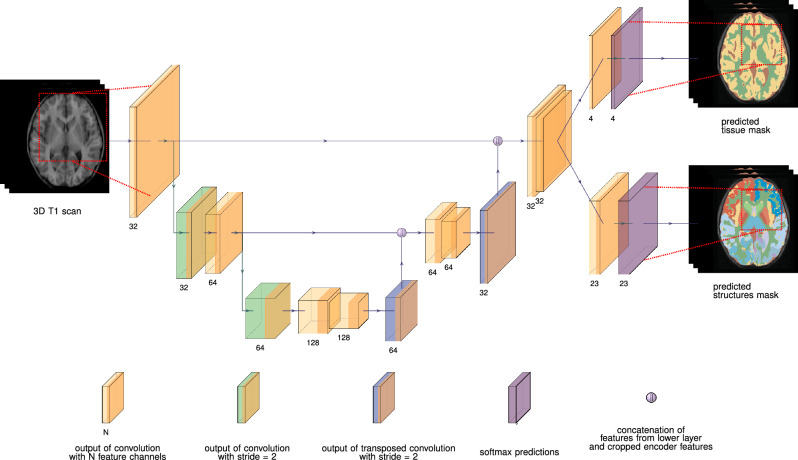
The architecture utilizes a 3D U-net backbone26, incorporating two segmentation heads. Each of both outputs is a softmax array of probability maps, where is the number of classes being predicted in task k. Moreover, certain modifications were made to the original architecture, including substituting batch normalization with weight normalization27, using leaky ReLU as the primary activation function, and using strided convolutions instead of max pooling28. Figure 1 illustrates our final architecture, while detailed information including justification for the multi-task architecture can be accessed in Appendix C.
Figure 1.
The deep learning model processes a 3D T1-weighted image via a single-input, dual-output 3D convolutional neural network (CNN) to produce estimated multi-label masks for brain tissues (background, white matter, gray matter, cerebrospinal fluid) and brain structures (background + 22 brain structures). The CNN is based on the widely used 3D U-net architecture, which operates on 3D patches of the input scan. Each convolutional layer utilizes kernels, except for the two convolutional layers before the softmax layers, which use kernels. Weight normalization and leaky ReLU (slope = 0.20) are employed. The output patches have dimensions of voxels, which are smaller than the input patches’ dimensions ( voxels) due to the use of valid convolutions, mitigating off-patch-center bias.
The model was trained using a weighted sum of the per-task losses, each comprising of a soft Dice loss () and a weighted categorical cross-entropy loss (), as shown in Eq. (1).
| 1 |
We set (tissue segmentation) and (structural segmentation).
The proposed model is trained on patches of voxels from T1-weighted MR images acquired without contrast agent injection. To augment the variability in the training set, ensuring that the range of intensities and tissue contrasts is similar with those observed in multi-center, multi-scanner cohorts, we applied intensity-based data augmentation as described in Meyer et al.29. This technique uses Gaussian Mixture Modeling to change the intensity of the individual tissue components within an MR image while preserving structural information. We utilized the predefined default parameters of the public implementation of this code, available at https://github.com/icometrix/gmm-augmentation.
The model was implemented using Tensorflow 2.6 and employed He weight initialization. The training process was stopped upon detecting convergence of the validation loss. The validation set, which constituted a randomly selected 15% of the training dataset, was not utilized for optimizing the network weights. Adam optimizer was deployed with an initial learning rate of .
Efficient generation of high-quality training labels
To address the challenge of obtaining manual annotations for large datasets, we created ‘silver’ ground truth, starting from the labels predicted by icobrain v5.9 on the training datasets. Subsequently, minor manual corrections were made where necessary.
Models training scheme
We trained three deep learning models with identical architecture, each using a different set of data for training:
The icobrain-dl model was trained on both pediatric and adult data, providing the most comprehensive training dataset (i.e., datasets 1.1.p, 1.2.p, 1.3.a and 1.4.a ).
The pediatric-specific model, termed icobrain-dl-p, was exclusively trained on pediatric datasets (i.e., datasets 1.1.p and 1.2.p).
The adult-specific model, termed icobrain-dl-a, was solely trained on adult datasets (i.e., datasets 1.3.a and 1.4.a ).
Validating technical and diagnostic performance
Two sets of experiments were conducted to validate both technical and diagnostic performance, with a focus on segmentation accuracy, intra- and inter-scanner variability, and computational time.
Segmentation accuracy was evaluated through the Dice similarity coefficient (DSC) and Hausdorff distance (HD)30. DSC is a metric quantifying the overlap between two segmentation masks, with values ranging from 0 (indicating no overlap) to 100 (indicating perfect agreement). The HD measures the maximal contour distance (in millimeters) between the two masks. A smaller HD indicates greater similarity between the masks. To address the high sensitivity of the HD to outliers31, we considered the 95th percentile of the HD, denoted as HD95. In the initial experiment, DSC and HD95 calculations were performed between ground truth segmentations and both icobrain-dl and the age-specific models (icobrain-dl-p or icobrain-dl-a). Subsequently, DSC and HD95 values were computed between the icobrain-dl model and the age-specific models on datasets 2.p and 2.a.
The reproducibility of icobrain-dl was assessed by comparing it with established non-deep learning algorithms, specifically the pediatric-focused childmetrix15 and the clinically-used adult-focused medical device software icobrain v5.9, referred to as icobrain-nondl7,8. Additionally, the state-of-the-art deep learning model FastSurfer11 was included. Test-retest relative differences were computed with respect to the mean volumes across methods (dataset 3.p and 3.a), and the Wilcoxon signed-rank test was employed to identify significant differences between methods at levels of 0.01 and 0.001.
The validation of diagnostic performance serves as a proof of concept for the clinical application of the segmentation algorithm. To demonstrate the icobrain-dl’s applicability across both pediatric and adult populations, two pathologies with distinct volumetric patterns were selected. In the first experiment, the objective was to differentiate patients with CVI from those without CVI using the whole brain white matter volume (dataset 4.p), motivated by the known association between periventricular white matter damage and CVI32. The second experiment aimed to distinguish patients with AD from cognitively healthy individuals using temporal lobe cortical gray matter volume (dataset 4.a). Previous research has established the reliability of this region in discerning between AD patients and healthy controls8. Volumes from the different pipelines were normalized for head size employing the determinant of the affine transformation to the MNI atlas as a scaling factor. Head size-normalized volumes of the regions of interest (i.e., whole brain white matter and temporal lobe cortical gray matter) were used to distinguish pathology and non-pathology. Model comparisons were conducted using the area under the receiver operating characteristic curve (AUC) and the DeLong test, with a significance level of 0.0533. The assessment of accuracy, specificity, and sensitivity metrics was based on the maximum value of the Youden index.
Results
Accuracy
On the pediatric dataset 2.p, the deep learning models icobrain-dl and icobrain-dl-p exhibited comparable performance in accurately segmenting brain structures, achieving an average DSC of 82.2% and 80.8%, respectively. Their average HD95 were 3.26mm and 3.23mm. Additionally, there was a high overlap between the segmentations of icobrain-dl and the pediatric-oriented icobrain-dl-p, with an average DSC of 87.4% and HD95 1.76mm. Similar results were observed in the adult dataset 2.a, where icobrain-dl achieved an average DSC of 82.6% and HD95 of 2.27mm when compared to manual segmentations. For icobrain-dl-a, the metrics were 81.9% and 2.37mm, respectively. The average DSC between both segmentation models was 92.4% with an average HD95 of 1.02mm. Table 2 and Table 3 display the DSC and HD95 between manual ground truth segmentations and segmentations calculated by the three deep learning models.
Table 2.
icobrain-dl consistently achieves high overlap in segmenting different brain structures across subject age ranges, while only minimally sacrificing accuracy and sometimes even outperforming models that are tailored for specific age ranges (icobrain-dl-p for pediatric data and icobrain-dl-a for adult data).
| Dice similarity coefficient (DSC) | ||||||
|---|---|---|---|---|---|---|
| Pediatric dataset 2.p | Adult dataset 2.a | |||||
| GT vs | icobrain-dl-p vs | GT vs | icobrain-dl-a vs | |||
| icobrain-dl | icobrain-dl-p | icobrain-dl | icobrain-dl | icobrain-dl-a | icobrain-dl | |
| WM | 84.8 (2.0) | 85.0 (2.1) | 92.3 (2.9) | 88.5 (1.1) | 85.5 (1.0) | 91.8 (1.7) |
| CGM | 83.0 (2.5) | 79.7 (3.7) | 88.2 (3.2) | 83.2 (1.6) | 82.5 (1.6) | 89.9 (2.3) |
| Lateral ventricles | 83.0 (4.6) | 83.4 (4.7) | 88.5 (6.4) | 88.7 (3.6) | 88.9 (3.7) | 93.3 (2.7) |
| Hippocampus | 78.8 (2.7) | 71.4 (11.3) | 82.1 (11.3) | 76.7 (1.8) | 77.2 (1.9) | 91.3 (1.7) |
| Caudate | 83.5 (3.5) | 82.5 (8.3) | 83.5 (9.3) | 81.8 (3.1) | 80.5 (3.2) | 91.5 (2.2) |
| Putamen | 84.8 (1.5) | 83.9 (3.1) | 89.8 (3.6) | 83.4 (2.4) | 80.7 (1.8) | 92.1 (1.6) |
| Cerebellar GM | 83.9 (2.5) | 86.9 (1.9) | 89.9 (1.9) | 78.4 (2.8) | 79.5 (2.4) | 94.1 (1.3) |
| Cerebellar WM | 76.1 (4.1) | 75.9 (4.4) | 81.1 (2.8) | 77.8 (2.8) | 77.4 (2.3) | 92.4 (1.6) |
| Thalamus | 82.6 (2.4) | 78.7 (7.0) | 90.7 (6.7) | 85.0 (1.0) | 84.5 (1.3) | 94.7 (1.0) |
We compare segmentation accuracy as measured by the Dice similarity coefficient expressed as a percentage between three deep learning models that are trained using different subsets of training data against manually created ground truth (GT) from pediatric (CANDIShare, dataset 2.p) and adult (MICCAI2012, dataset 2.a) data. The Dice similarity coefficient between the models’ predictions is also shown. The Dice similarity coefficient is reported as: mean value (standard deviation) across subjects. WM = White Matter, CGM = Cortical Gray Matter, GM = Gray Matter.
Table 3.
Summary of the Hausdorff distance 95th percentile (HD95) between ground truth (GT) and icobrain-dl or the age-specific models, and between the age-specific models and icobrain-dl.
| Hausdorff distance 95th percentile (HD95) | ||||||
|---|---|---|---|---|---|---|
| Pediatric dataset 2.p | Adult dataset 2.a | |||||
| GT vs | icobrain-dl-p vs | GT vs | icobrain-dl-a vs | |||
| icobrain-dl | icobrain-dl-p | icobrain-dl | icobrain-dl | icobrain-dl-a | icobrain-dl | |
| WM | 2.07 (0.52) | 2.08 (0.51) | 1.11 (0.21) | 1.34 (0.24) | 1.80 (0.36) | 1.01 (0.07) |
| CGM | 2.50 (0.42) | 2.81 (0.50) | 1.23 (0.35) | 1.36 (0.17) | 1.38 (0.20) | 1.00 (0.00) |
| Lateral ventricles | 6.82 (9.47) | 3.73 (6.59) | 1.62 (1.67) | 1.06 (0.14) | 1.06 (0.14) | 1.00 (0.00) |
| Hippocampus | 2.50 (0.59) | 3.43 (1.67) | 1.79 (1.38) | 1.93 (0.24) | 1.95 (0.27) | 1.00 (0.00) |
| Caudate | 1.64 (0.37) | 1.86 (1.49) | 1.77 (1.28) | 1.51 (0.21) | 1.64 (0.25) | 1.04 (0.12) |
| Putamen | 1.80 (0.27) | 2.05 (0.38) | 1.37 (0.44) | 1.52 (0.23) | 1.74 (0.19) | 1.01 (0.07) |
| Cerebellar GM | 3.29 (0.47) | 3.01 (0.57) | 2.13 (0.37) | 3.00 (0.25) | 3.03 (0.22) | 1.04 (0.12) |
| Cerebellar WM | 5.74 (1.40) | 6.72 (2.20) | 3.24 (1.09) | 6.51 (1.43) | 6.41 (0.92) | 1.16 (0.36) |
| Thalamus | 2.99 (0.37) | 3.38 (0.77) | 1.65 (1.01) | 2.24 (0.09) | 2.36 (0.20) | 1.00 (0.00) |
icobrain-dl consistently achieves high-quality segmentation of various brain structures across different age groups, as indicated by the low HD95 with comparing with GT in both datasets. The HD95 is reported as: mean value (standard deviation) in millimeters across subjects. WM = White Matter, CGM = Cortical Gray Matter, GM = Gray Matter.
These findings suggest that icobrain-dl is as effective as the age-specific models in accurately segmenting brain structures in both pediatric and adult populations.
Reproducibility
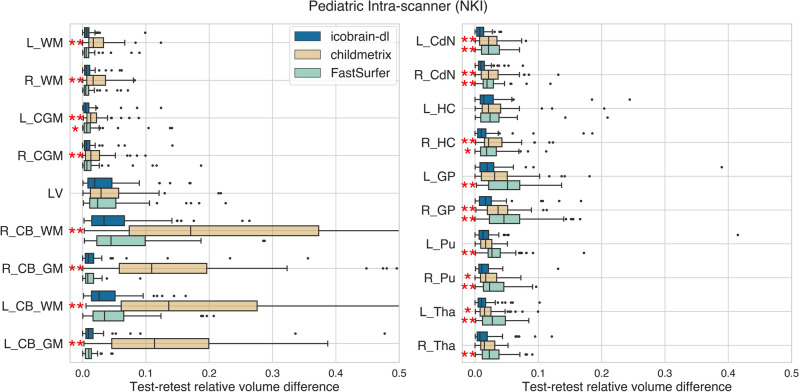
The segmentations generated by icobrain-dl systematically had lower test-retest volume differences for the pediatric intra-scanner setting (dataset 3.p) than childmetrix and FastSurfer, as illustrated in Figure 2. For most structures, these test-retest differences from icobrain-dl were significantly lower than the comparable methods ().
Figure 2.
The icobrain-dl measurements exhibited statistically significantly lower test-retest errors than both childmetrix and FastSurfer across a majority of regions for pediatric cases (dataset 3.p) in intra-scanner settings, as quantified by relative test-retest volume differences. Legend: * = , **= according to Wilcoxon signed-rank tests comparing icobrain-dl to either childmetrix or FastSurfer. To ensure overall figure readability, certain boxplots have been cropped. L = left, R = right, WM = white matter, CGM = cortical gray matter, LV = lateral ventricles, CB = cerebellum, CdN = caudate nucleus, HC = hippocampus, GP = globus pallidus, Pu = putamen, Tha = thalamus.
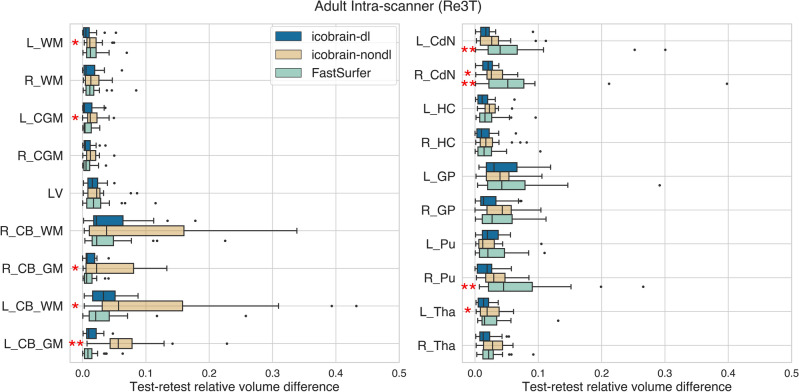
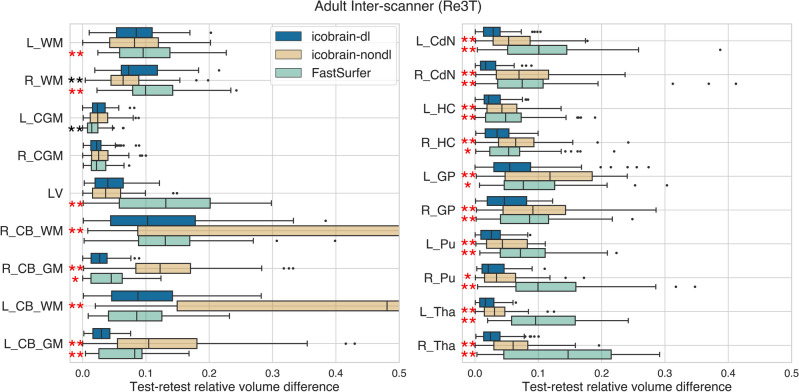
A similar pattern of lower test-retest volume differences provided by icobrain-dl was observed in adults (dataset 3.a) for intra-scanner and inter-scanner settings (see Figure 3 and 4). Specifically, in the inter-scanner setting, icobrain-dl outperformed icobrain-nondl and FastSurfer, except in the right white matter and left cortical gray matter. Notably, icobrain-dl produced significantly lower inter-scanner test-retest errors () across all substructures, including the caudate nucleus, hippocampus, globus pallidus, putamen, and thalamus.
Figure 3.
The icobrain-dl measurements exhibited equal or lower test-retest errors than icobrain-nondl and FastSurfer for adult cases in the intra-scanner settings, as quantified by intra-scanner relative test-retest volume differences. Legend: * = , **= according to Wilcoxon signed-rank tests comparing icobrain-dl to either icobrain-nondl or FastSurfer. L = left, R = right, WM = white matter, CGM = cortical gray matter, LV = lateral ventricles, CB = cerebellum, CdN = caudate nucleus, HC = hippocampus, GP = globus pallidus, Pu = putamen, Tha = thalamus.
Figure 4.
The icobrain-dl measurements exhibited statistically significantly lower test-retest errors than icobrain-nondl and FastSurfer across all the subcortical structures (right) for adult cases (dataset 3.a) in inter-scanner settings, as quantified by relative test-retest volume differences. The asterisk colour indicates the better performing method (red = icobrain-dl, black = state-of-the-art). To ensure overall figure readability, certain boxplots have been cropped. L = left, R = right, WM = white matter, CGM = cortical gray matter, LV = lateral ventricles, CB = cerebellum, CdN = caudate nucleus, HC = hippocampus, GP = globus pallidus, Pu = putamen, Tha = thalamus.
Diagnostic performance
The performance of icobrain-dl in detecting pediatric patients with CVI surpassed childmetrix (AUC of 0.48) and FastSurfer (AUC of 0.60), with an AUC of 0.69, as shown in Table 4. There was no statistically significant difference between icobrain-dl and FastSurfer in terms of AUC. Nevertheless, icobrain-dl exhibited significantly superior performance compared to childmetrix ().
Table 4.
The proposed method has superior performance in detecting pediatric patients with Cerebral Visual Impairment (CVI) from those without CVI using the white matter volume normalized for head size (dataset 4.p) and comparable high performance in detecting adult patients with Alzheimer’s Disease from age-matched controls using the cortical grey matter of the temporal lobe normalized for head size (MIRIAD, dataset 4.a).
| Pediatric dataset 4.p | Adult dataset 4.a | |||||
|---|---|---|---|---|---|---|
| icobrain-dl | childmetrix | FastSurfer | icobrain-dl | icobrain-nondl | FastSurfer | |
| AUC | 0.69 | 0.48 | 0.60 | 0.99 | 0.98 | 0.98 |
| Accuracy | 0.71 | 0.57 | 0.67 | 0.96 | 0.93 | 0.96 |
| Specificity | 0.86 | 0.57 | 0.86 | 0.91 | 0.91 | 0.96 |
| Sensitivity | 0.64 | 0.57 | 0.57 | 1 | 0.96 | 0.96 |
AUC = area under the curve.The AUC obtained by icobrain-dl was significantly higher than the AUC obtained by childmetrix () while there was no statistically significant difference between icobrain-dl and FastSurfer, and icobrain-dl and icobrain v5.9 using the DeLong test. The accuracy, specificity, and sensitivity metrics are assessed at the maximum value of the Youden index.
In supporting the classification of AD patients from age-matched controls, the icobrain-dl demonstrated comparable high performance in terms of accuracy, sensitivity, and specificity. The AUC for icobrain-dl was 0.99, icobrain-nondl was 0.98, and FastSurfer was 0.98, with no statistically significant difference.
Computational time
On average, the proposed method took approximately 5 minutes to complete the entire pipeline when running on a server without a GPU (amazon web services cloud environment c6i.2xlarge, 8vCPU and 16GiB of Memory RAM) while the pipeline based on FastSurfer requires nearly 6 minutes on a GPU server (cloud environment p2.xlarge, NVIDIA Tesla K80 (12 GiB), 4vCPU and 61GiB of Memory RAM). In contrast, the non-deep learning approaches childmetrix and icobrain v5.9 running on a server without a GPU (cloud environment c6i.2xlarge, 8vCPU and 16GiB of Memory RAM) required on average 24 minutes and 27 minutes.
Qualitative results
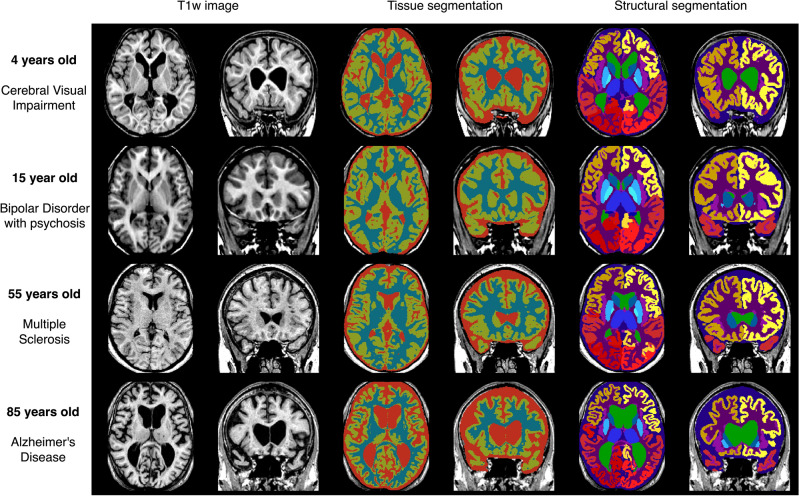
Figure 5 illustrates the segmentation results of icobrain-dl in test patients across the lifespan, with ages ranging from 4 to 85 years old. These qualitative results demonstrate the model’s robustness to diverse pathological conditions and scans with differing intensities and contrasts.
Figure 5.
Examples of segmentations of icobrain-dl on test patients with different ages and pathologies. The pipeline accurately quantifies brain tissues and structures despite variations in age, pathology, and intensity contrast, capturing anatomical variability such as the cortical atrophy patterns characteristic of patients with Alzheimer’s Disease.
Discussion
This study introduces icobrain-dl, a deep learning-based pipeline capable of performing quantitative assessment of brain tissues and structures across pediatric and adult populations.
The pipeline was developed and validated using T1-weighted images obtained from various scan vendors with different magnetic field strengths. The dataset includes patients across a broad age range with various pathological conditions. Evaluation of the proposed pipeline included segmentation accuracy and reproducibility assessments, along with an exploration of its clinical application through diagnostic performance and computational efficiency.
In contrast to methods tailored for specific age ranges, such as childmetrix for children or icobrain-nondl and FastSurfer for adults, icobrain-dl provides quantitative brain measurements across the human lifespan, from early childhood (i.e., 4 years old) to maturation and older age, within a single deep learning model. Previous experiments have shown the accuracy performance of adult-trained models in pediatric data9,10. However, in this study, we explicitly included pediatric data to train the model and observed that it does not compromise the performance on scans from adult subjects, and vice versa. Furthermore, the inclusion of a pediatric cohort allowed the deep learning model to learn and adapt to challenges associated with brain development, including reduced tissue contrast, within-tissue intensity heterogeneities, and smaller regions of interest. The proposed single deep learning model eliminates the need for multiple age-specific segmentation models, enabling consistent measurements across transitional phases, such as from the pediatric stage through adolescence to adulthood. This facilitates the creation of a reference standard for human brain development, essential for quantifying developmental changes, interpreting deviations, and identifying patterns of anatomical differences in neurological and psychiatric disorders that manifest during various stages of development and aging34.
High reproducibility is crucial for accurately measuring brain changes and atrophy35. The proposed icobrain-dl, was compared with state-of-the-art brain segmentation models, including childmetrix15, FastSurfer11 and the medical device software icobrain-nondl (i.e., icobrain v5.97,8). The results demonstrated overall superior reproducibility assessed in pediatric intra-scanner and adult intra- and inter-scanner scenarios, particularly in the adult inter-scanner setting, with significantly lower variability observed in all brain substructures (p < 0.01). This improvement can be attributed to the diverse sources of T1-weighted images used in training, along with the integration of a data augmentation algorithm. This algorithm enhanced the variability of training data in terms of intensity and contrast, which has been proven to be particularly beneficial for repetitions in different scanners (i.e., inter-scanner)29.
Volumetric imaging biomarkers provided by icobrain-dl required good accuracy, specificity and sensitivity to be used as a metric for diagnosis (e.g., distinguishing patients with Alzheimer’s vs. healthy controls). The proposed pipeline exhibited comparable diagnostic performance to state-of-the-art methods, achieving the highest AUC for both clinical conditions. It is important to note that the purpose of the diagnostic performance scenario was to compare different methods using the same measurement, rather than to identify clinically relevant imaging biomarkers for specific pathologies. Future studies will explore the potential of volumetric imaging biomarkers to enhance our understanding of the underlying mechanisms of diseases and improve their diagnosis, particularly in complex and partly understood conditions like CVI. This involves increasing sample sizes and considering factors such as sexual dimorphism36 and age-dependent developmental trajectories13.
The proposed pipeline also analyses the images faster than traditional segmentation approaches, aligning with findings from previous studies employing deep learning models9,11. However, in contrast with previous deep learning models, the proposed model deployed a lightweight deep learning architecture, consisting of relatively few layers. This design choice aimed to reduce the computational complexity, facilitating model inference on CPU-only platforms and ensuring efficient segmentation without incurring the elevated economical costs associated with GPU usage. The reduced processing time avoids creating additional bottlenecks in the radiological workflow.
The annotation protocols used to establish the ground truth of brain structures may vary across datasets, potentially differing from our definition of brain structure borders. This discrepancy could explain the higher overlap observed between models than the overlap between models and ground truth. Notably, icobrain-dl and the age-specific models are trained on datasets with overlapping patients and employ the same annotation protocol.
The icobrain-dl pipeline is designed to use T1-weighted images to analyse the structural anatomy of the brain. Currently, its application is limited to conditions characterized by non-mass effects due to the absence of multimodal data, such as fluid-attenuated inversion recovery (FLAIR) images. However, future iterations of icobrain-dl aim to integrate multimodal data, thereby expanding its utility to cover a broader spectrum of pathologies.
The proposed deep learning model covers the human lifespan, starting at 4 years of age. The period preceding this age is the most dynamic phase of postnatal human brain development37. Maturation processes, including myelination, notably influence T1-weighted image contrasts, for instance, shifting from hypointense white matter in newborns to hyperintese in 2-year-old infants, making the development of a reliable segmentation model a very complex task. Hence, additional exploration is required to incorporate quantification of brain segmentation during this initial phase of brain development.
Conclusion
The proposed deep learning-based pipeline, icobrain-dl, is capable of quantifying brain tissues and structures across the human lifespan beginning at 4 years of age. Extensive validation in clinically relevant settings has demonstrated its ability to provide accurate and reproducible volume quantification of relevant brain anatomical structures from T1-weighted images.
By offering a unified solution from early childhood to maturation and older age, icobrain-dl has the potential to significantly enhance research and clinical applications in monitoring brain development and diagnosing neurological conditions.
Supplementary Information
Acknowledgements
The PARENT project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Maria Skłodowska Curie-Innovative Training Network 2020, Grant Agreement No 956394. We wish to thank Elien Bollen for her contribution in curating the data. We also express our gratitude to all investigators involved in collecting and providing free access to the datasets. This manuscript reflects the views of the authors and may not reflect the opinions or views of the database providers.
Author contributions
J.S. Investigation, Methodology, Validation, Formal analysis, Writing—original draft. M.I.M. Conceptualization, Methodology, Software. S.V.E Methodology, Software, Writing—review & editing. T.V.P. Conceptualization, Methodology, Writing—review & editing T.B. Supervision, Writing—review & editing, Funding acquisition. D.M.S. Supervision, Methodology, Conceptualization, Writing—review & editing. E.O. Supervision, Writing—review & editing, Funding acquisition.
Data and code availability
Further details regarding the publicly available datasets analyzed in the current study can be found in Appendix A. Additional datasets analyzed during the current study can be made available from the corresponding author with the permission of a third party upon reasonable request. The code employed in this study is not publicly accessible due to commercial restrictions but is available from the corresponding author upon reasonable request.
Competing interests
The following authors are employed (or have been employed at the time of performing the work relevant for this paper) by icometrix: J.S, M.I.M, S.V.E, T.V.P., T.B., D.M.S. E.O. declares no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-61798-6.
References
- 1.Smith SM, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23:S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 2.Mills KL, Tamnes CK. Methods and considerations for longitudinal structural brain imaging analysis across development. Dev. Cogn. Neurosci. 2014;9:172–190. doi: 10.1016/j.dcn.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Urbach H, et al. within a minute detection of focal cortical dysplasia. Neuroradiology. 2022;64:715–726. doi: 10.1007/s00234-021-02823-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pemberton HG, et al. Technical and clinical validation of commercial automated volumetric MRI tools for dementia diagnosis-a systematic review. Neuroradiology. 2021;63:1773–1789. doi: 10.1007/s00234-021-02746-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischl B. Freesurfer. Neuroimage. 2012;62:774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manjón JV, Coupé P. volBrain: An online MRI brain volumetry system. Front. Neuroinform. 2016;10:30. doi: 10.3389/fninf.2016.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jain S, et al. Automatic segmentation and volumetry of multiple sclerosis brain lesions from MR images. NeuroImage Clin. 2015;8:367–375. doi: 10.1016/j.nicl.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Struyfs H, et al. Automated MRI volumetry as a diagnostic tool for Alzheimer’s disease: Validation of icobrain dm. NeuroImage Clin. 2020;26:102243. doi: 10.1016/j.nicl.2020.102243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roy AG, et al. QuickNAT: A fully convolutional network for quick and accurate segmentation of neuroanatomy. Neuroimage. 2019;186:713–727. doi: 10.1016/j.neuroimage.2018.11.042. [DOI] [PubMed] [Google Scholar]
- 10.Coupé P, et al. AssemblyNet: A large ensemble of CNNs for 3D whole brain MRI segmentation. Neuroimage. 2020;219:117026. doi: 10.1016/j.neuroimage.2020.117026. [DOI] [PubMed] [Google Scholar]
- 11.Henschel L, et al. Fastsurfer-a fast and accurate deep learning based neuroimaging pipeline. Neuroimage. 2020;219:117012. doi: 10.1016/j.neuroimage.2020.117012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jyothi P, Singh AR. Deep learning models and traditional automated techniques for brain tumor segmentation in MRI: A review. Artif. Intell. Rev. 2023;56:2923–2969. doi: 10.1007/s10462-022-10245-x. [DOI] [Google Scholar]
- 13.Phan TV, Smeets D, Talcott JB, Vandermosten M. Processing of structural neuroimaging data in young children: Bridging the gap between current practice and state-of-the-art methods. Dev. Cogn. Neurosci. 2018;33:206–223. doi: 10.1016/j.dcn.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phan TV, et al. Evaluation of methods for volumetric analysis of pediatric brain data: the childmetrix pipeline versus adult-based approaches. NeuroImage Clin. 2018;19:734–744. doi: 10.1016/j.nicl.2018.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phan TV, et al. Structural brain dynamics across reading development: A longitudinal MRI study from kindergarten to grade 5. Hum. Brain Mapp. 2021;42:4497–4509. doi: 10.1002/hbm.25560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turesky TK, Vanderauwera J, Gaab N. Imaging the rapidly developing brain: Current challenges for MRI studies in the first five years of life. Dev. Cogn. Neurosci. 2021;47:100893. doi: 10.1016/j.dcn.2020.100893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alexander LM, et al. Data descriptor: An open resource for transdiagnostic research in pediatric mental health and learning disorders. Sci. Data. 2017 doi: 10.1038/sdata.2017.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paniukov D, Lebel RM, Giesbrecht G, Lebel C. Calgary cerebral blood flow increases across early childhood. NeuroImage. 2020 doi: 10.1016/j.neuroimage.2019.116224. [DOI] [PubMed] [Google Scholar]
- 19.Van Assche L, et al. Hippocampal volume as a vulnerability marker for late onset psychosis: Associations with memory function and childhood trauma. Schizophr. Res. 2020;224:201–202. doi: 10.1016/j.schres.2020.08.004. [DOI] [PubMed] [Google Scholar]
- 20.Kennedy DN, et al. CANDIShare: A resource for pediatric neuroimaging data. Neuroinformatics. 2012;10:319–322. doi: 10.1007/s12021-011-9133-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landman, B. & Warfield, S. Miccai 2012 workshop on multi-atlas labeling. In MICCAI Grand Challenge and Workshop on Multi-Atlas Labeling (CreateSpace Independent Publishing Platform, Nice, France, 2012).
- 22.Nooner KB, et al. The NKI-Rockland sample: A model for accelerating the pace of discovery science in psychiatry. Front. Neurosci. 2012;6:152. doi: 10.3389/fnins.2012.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malone IB, et al. Miriad-public release of a multiple time point Alzheimer’s MR imaging dataset. Neuroimage. 2013;70:33–36. doi: 10.1016/j.neuroimage.2012.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tustison NJ, et al. N4ITK: Improved N3 bias correction. IEEE Trans. Med. Imaging. 2010;29:1310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ourselin S, Roche A, Subsol G, Pennec X, Ayache N. Reconstructing a 3D structure from serial histological sections. Image Vis. Comput. 2001;19:25–31. doi: 10.1016/S0262-8856(00)00052-4. [DOI] [Google Scholar]
- 26.Çiçek, Ö., Abdulkadir, A., Lienkamp, S. S., Brox, T. & Ronneberger, O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2016, 424–432 (Springer International Publishing, Cham, 2016).
- 27.Salimans, T. & Kingma, D. P. Weight normalization: A simple reparameterization to accelerate training of deep neural networks. Adv. Neural Inf. Process. Syst., 901–909 (2016).
- 28.Isensee F, Jaeger PF, Kohl SA, Petersen J, Maier-Hein KH. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods. 2021;18:203–211. doi: 10.1038/s41592-020-01008-z. [DOI] [PubMed] [Google Scholar]
- 29.Meyer MI, et al. A contrast augmentation approach to improve multi-scanner generalization in MRI. Front. Neurosci. 2021 doi: 10.3389/FNINS.2021.708196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taha AA, Hanbury A. Metrics for evaluating 3D medical image segmentation: Analysis, selection, and tool. BMC Med. Imaging. 2015;15:1–28. doi: 10.1186/s12880-015-0068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Isensee F, et al. Automated brain extraction of multisequence MRI using artificial neural networks. Hum. Brain Mapp. 2019;40:4952–4964. doi: 10.1002/hbm.24750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ortibus, E., Fazzi, E. & Dale, N. Cerebral visual impairment and clinical assessment: The European perspective. In Seminars in Pediatric Neurology, 31, 15–24 (Elsevier, 2019). [DOI] [PubMed]
- 33.Sun X, Xu W. Fast implementation of Delong’s algorithm for comparing the areas under correlated receiver operating characteristic curves. IEEE Signal Process. Lett. 2014;21:1389–1393. doi: 10.1109/LSP.2014.2337313. [DOI] [Google Scholar]
- 34.Bethlehem RA, et al. Brain charts for the human lifespan. Nature. 2022;604:525–533. doi: 10.1038/s41586-022-04554-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo C, Ferreira D, Fink K, Westman E, Granberg T. Repeatability and reproducibility of freeSurfer, FSL-SIENAX and SPM brain volumetric measurements and the effect of lesion filling in multiple sclerosis. Eur. Radiol. 2019;29:1355–1364. doi: 10.1007/s00330-018-5710-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.López-Ojeda W, Hurley RA. Sexual dimorphism in brain development: Influence on affective disorders. J. Neuropsychiatry Clin. Neurosci. 2021;33:A4–85. doi: 10.1176/appi.neuropsych.21030067. [DOI] [PubMed] [Google Scholar]
- 37.Li G, et al. Mapping region-specific longitudinal cortical surface expansion from birth to 2 years of age. Cereb. Cortex. 2013;23:2724–2733. doi: 10.1093/cercor/bhs265. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Further details regarding the publicly available datasets analyzed in the current study can be found in Appendix A. Additional datasets analyzed during the current study can be made available from the corresponding author with the permission of a third party upon reasonable request. The code employed in this study is not publicly accessible due to commercial restrictions but is available from the corresponding author upon reasonable request.